Abstract
Alternative splicing is an RNA processing mechanism that affects most genes in human, contributing to disease mechanisms and phenotypic diversity. The regulation of splicing involves an intricate network of cis-regulatory elements and trans-acting factors. Due to their high sequence specificity, cis-regulation of splicing can be altered by genetic variants, significantly affecting splicing outcomes. Recently, multiple methods have been applied to understanding the regulatory effects of genetic variants on splicing. However, it is still challenging to go beyond apparent association to pinpoint functional variants. To fill in this gap, we utilized large-scale data sets of the Genotype-Tissue Expression (GTEx) project to study genetically modulated alternative splicing (GMAS) via identification of allele-specific splicing events. We demonstrate that GMAS events are shared across tissues and individuals more often than expected by chance, consistent with their genetically driven nature. Moreover, although the allelic bias of GMAS exons varies across samples, the degree of variation is similar across tissues versus individuals. Thus, genetic background drives the GMAS pattern to a similar degree as tissue-specific splicing mechanisms. Leveraging the genetically driven nature of GMAS, we developed a new method to predict functional splicing-altering variants, built upon a genotype-phenotype concordance model across samples. Complemented by experimental validations, this method predicted >1000 functional variants, many of which may alter RNA-protein interactions. Lastly, 72% of GMAS-associated SNPs were in linkage disequilibrium with GWAS-reported SNPs, and such association was enriched in tissues of relevance for specific traits/diseases. Our study enables a comprehensive view of genetically driven splicing variations in human tissues.
High-throughput sequencing technologies are enabling identification of an extraordinary number of genetic variants in the human genome (Reuter et al. 2015). These data provide a foundation to elucidate the genetic underpinnings of human diseases or phenotypic traits. Many genome-wide studies have been conducted to uncover associations between the genetic variants and complex traits (Buniello et al. 2019). However, moving from associations to revealing the underlying mechanisms remains a significant challenge. Genetic variants could affect many aspects of gene expression or function, which is a major determinant of phenotypic diversity (Manning and Cooper 2017). Until recently, research efforts have been focused on variants that may impose epigenetic or transcriptional regulation. However, it is increasingly recognized that genetic variants also play critical roles in modulating post-transcriptional mechanisms, such as alternative splicing (Wang and Cooper 2007; Sterne-Weiler and Sanford 2014).
Alternative splicing is an essential mechanism in eukaryotic gene expression, contributing to many aspects of phenotypic complexity and disease mechanisms (Hsiao et al. 2016b). Splicing is regulated by an intricate network of trans-factors and cis-regulatory elements (Hsiao et al. 2016b). Thus, it is not surprising that genetic variants may alter different aspects of splicing regulation, such as the cis-regulatory motifs, trans-factor expression or function, and the interactions between these players (Wang and Cooper 2007; Sterne-Weiler and Sanford 2014). Indeed, quantitative trait loci (QTL) mapping in lymphoblastoid cell lines suggested that splicing QTLs and expression QTLs are comparable in their effects on complex traits (Li et al. 2016; Ferraro et al. 2020).
Both computational and experimental methods have been developed to reveal splicing-disrupting genetic variants (Hsiao et al. 2016b; Rhine et al. 2019; Rowlands et al. 2019). Computationally, applications of machine learning methods have yielded promising results (Barash et al. 2010). Recently, performance improvements were achieved using deep learning to predict splice site usage directly from nucleotide sequence (Xiong et al. 2015; Bretschneider et al. 2018; Cheng et al. 2019; Jaganathan et al. 2019). However, these methods still present challenges in interpretability, and it is difficult to determine whether the features being used are biologically relevant. Experimentally, massively parallel reporter assays have enabled large-scale screens of functional variants in splicing (Ke et al. 2011; Rosenberg et al. 2015; Soemedi et al. 2017; Adamson et al. 2018; Cheung et al. 2019). However, due to the limited insert size cloned into the reporters, the splicing outcome may not always recapitulate endogenous splicing patterns. Additionally, these reporter assays can only be performed in one cell type at a time. In general, it remains a great challenge, both computationally and experimentally, to identify causal genetic variants specific to each tissue type.
In this study, we carried out global analyses of allele-specific alternative splicing using RNA-seq data from a large panel of human tissues and individuals generated by the GTEx project (The GTEx Consortium 2015). Compared to machine learning methods, allele-specific analysis is a data-driven approach that requires little prior knowledge about splicing regulatory mechanisms. The advantage of this approach includes its applicability to a single RNA-seq data set. In addition, it compares the alternative alleles of a heterozygous SNP in the same cellular environment in the same subject. Thus, the method controls for tissue conditions, trans-acting factors, global epigenetic effects, and other environmental influences.
Our lab previously developed allele-specific analysis methods to identify allele-specific splicing events, also called genetically modulated alternative splicing (GMAS) (Li et al. 2012; Hsiao et al. 2016a). Here, in addition to applying these methods to the GTEx data, we developed a new method to infer functional SNPs underlying the GMAS events. The analysis of GTEx data using these methods allowed a detailed view of GMAS across tissues and individuals and their potential functional relevance in human diseases and traits.
Results
Overview of genetic modulation of alternative splicing in GTEx data
We first applied our previously published method (Li et al. 2012) to identify GMAS events. Briefly, this method examines allelic biases in reads covering heterozygous SNPs in each gene. By comparing the allele-specific expression patterns of all heterozygous SNPs in a gene and their associations with alternative splicing, the method identifies SNPs that are associated with allele-specific splicing patterns (Methods). Although this method does not pinpoint the functional SNPs regulating splicing, it captures exons (namely GMAS exons) that are under such genetic regulation. Therefore, the SNPs with allelic bias located in the GMAS exons are named tag SNPs. Using this method, we analyzed a total of 7822 GTEx RNA-seq data sets, across 47 tissues and 515 donors, following a few quality control filters (Methods).
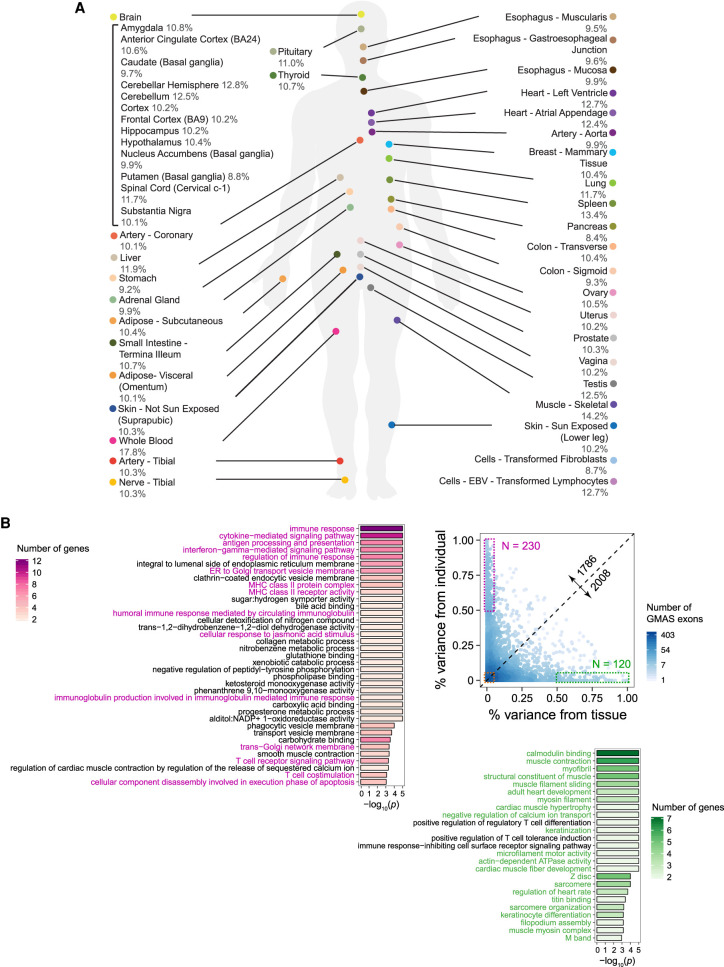
Across all tissues, a total of 12,331 exons were identified as GMAS exons, associated with 18,894 heterozygous tag SNPs (Methods), where one GMAS exon may be associated with multiple tag SNPs. We focused on GMAS events that are common to multiple samples by requiring each GMAS exon be present in ≥3 samples (across all tissues and individuals). A total of 4941 GMAS exons (7404 tag SNPs) were retained (Supplemental Table S1). For each tissue, an average of 10% of all testable exons (defined based on read coverage requirements) (see Methods) were identified as GMAS exons (Fig. 1A). This percentage is highest in whole blood (17.8%), which may reflect existence of a high level of genetic modulation of splicing, consistent with the sQTL results in the GTEx study (The GTEx Consortium 2015). Note that the prevalence of GMAS per tissue may be affected by sample size and sequencing depth (Supplemental Fig. S1A). Using subsets of samples that match the number of samples and the sequencing depth per sample in brain substantia nigra (the tissue with the least samples), we observed that blood is still among those with a high percentage of GMAS events (second to testis) (Supplemental Fig. S1B).
Figure 1.
The landscape of GMAS exons in human tissues. (A) % of GMAS exons among all testable exons in each tissue (averaged across individuals). (B) The variability of GMAS patterns across tissues and individuals (Methods). Each dot represents an exon, and the colors represent the number of overlapping dots. This analysis only included GMAS exons that exist in ≥2 individuals per tissue and ≥2 tissues per individual. The numbers along the diagonal line show the number of GMAS exons above and below the line, respectively. GO terms enriched among genes in the high variability groups (boxed) are shown. Color intensity represents the number of genes associated with each significant GO term. The P-values were estimated based on 10,000 randomizations of control genes (i.e., genes hosting alternatively spliced exons that were tested for GMAS) matching gene length and GC content of the test genes (Hsiao et al. 2016a). The significant cutoff of the P-value was set to be 1/(number of total GO terms considered).
In each tissue, the most prevalent type of alternative splicing for GMAS events is skipped exons (SEs), accounting for about 80% of all events, followed by retained introns (RIs, ∼10%) (Supplemental Fig. S2A). The difference in splicing levels of GMAS exons in individuals homozygous for the reference versus variant allele is generally lower than 50% (Supplemental Fig. S2B), although some brain regions showed larger differences relative to other tissues. Note that the distributions for alternative splice site exons may not be reliable given the small number of events.
GMAS patterns vary across tissues and individuals to a similar degree
Given the data sets from many individuals and a large panel of tissues, we first examined the global variability in GMAS patterns depending on these two variables. To segregate the impact of tissues and individuals on GMAS, we used a linear mixed model that includes these two variables and a number of confounding factors (age, ethnicity, and gender) (Methods). We observed equivalent levels of dependence of GMAS on tissues and individuals (Fig. 1B). This result is in stark contrast to previous findings that both gene expression and splicing in general predominantly vary depending on tissue types instead of individuals (Melé et al. 2015). Nevertheless, our result is not surprising because GMAS, by definition, consists of splicing events modulated by genetic variants that can be individual-specific. In turn, this result confirms the validity of the reported GMAS events. Our observation highlights that genetic background drives the splicing patterns of GMAS exons to a similar degree as tissue-specific splicing mechanisms, a previously underappreciated aspect.
Genes that contain GMAS exons with high tissue variance or high individual variance have substantially different function (Fig. 1B). The first group of genes is enriched in Gene Ontology (GO) terms associated with biophysical properties of the cells, especially related to heart or skeletal muscle function (e.g., sarcomere organization, cardiac muscle development, and cytoskeleton organization). This finding supports that alternative splicing is an important aspect contributing to the vast spectrum of biophysical properties of different cell types (Wang et al. 2008). In contrast, genes harboring GMAS exons with large variability across individuals are often involved in immune response and signaling pathways. This observation suggests that the individual variability in immune or stress response (Aguirre-Gamboa et al. 2016; Schirmer et al. 2016; Ter Horst et al. 2016) is partly accounted for by splicing variations driven by genetic backgrounds. For genes with GMAS exons with low variability across both tissues and individuals, the most significant GO terms are related to essential cellular processes (Supplemental Fig. S3), which may reflect existence of selection against splicing variability in essential genes.
GMAS patterns are shared between tissues or individuals
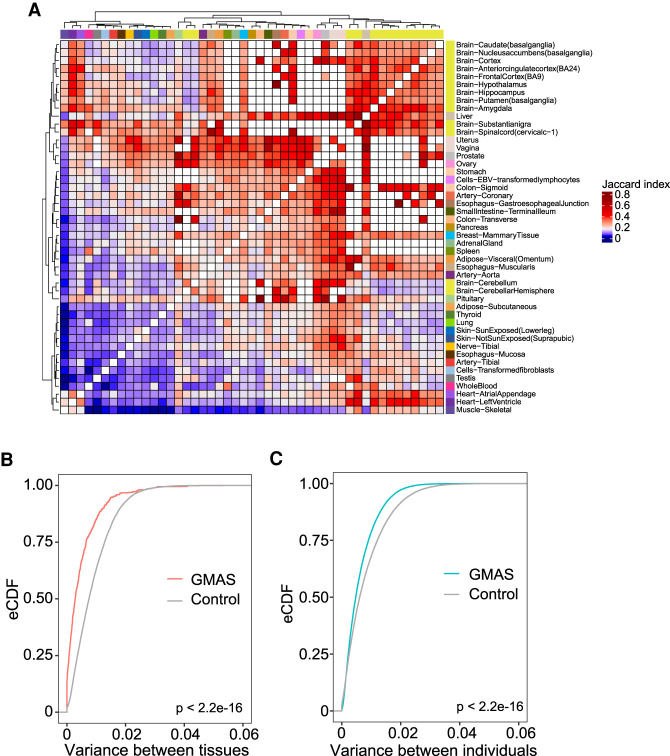
To better understand the tissue specificity of GMAS events, we next investigated the extent of overlap of GMAS exons between tissues (Methods). We observed that biologically related tissues, such as brain regions, heart and skeletal muscles, and reproductive tissues (uterus and vagina), formed clear clusters (Fig. 2A). Most brain regions shared about 25%–43% of GMAS exons with one another, with the exception of cerebellum and cerebellar hemisphere. These two regions were previously reported as outliers with distinctly higher splicing factor expression than other brain regions (Melé et al. 2015). Consistently, we observed that these two brain regions shared the most GMAS exons with each other and much less with other regions (Supplemental Fig. S4).
Figure 2.
Comparison of GMAS patterns across tissues or individuals. (A) Heat map of the Jaccard indices of GMAS exons between each pair of tissues (Methods). White boxes correspond to tissue pairs with <10 common testable exons. (B) Empirical cumulative distribution function (eCDF) of variances across tissues in the allelic biases of tag SNPs of GMAS exons for all individuals. Controls were included for comparison purposes (Methods). The P-value was calculated using the Kolmogorov–Smirnov test. (C) Similar to B but for variance across individuals per tissue.
Next, we asked whether GMAS patterns are shared between distinct tissues more than expected by chance. For this analysis, we focused on 26 representative tissues to remove redundant ones that are highly similar to each other (Methods). Each exon was required to be testable in at least 10 individuals and five representative tissues of a specific individual. We observed that the allelic bias of the GMAS tag SNPs was more similar between tissues of the same individual than expected by chance (Fig. 2B). Similarly, for the same tissue, the GMAS-associated allelic bias is also shared among individuals to a greater extent than expected by chance (Fig. 2C). These results suggest that genetic variants are important drivers for GMAS patterns, and tissue-specific effects may play a relatively less dominant role. This observation is consistent with the data in Figure 1B where the majority of GMAS exons showed relatively small variability across tissues or individuals, with those that are tissue- or individual-specific being the minority.
Inferring functional SNPs for GMAS events
Because genetic background is a main driver for GMAS events, an important task is to pinpoint the functional genetic variants underlying these events. Note that the tag SNPs identified with the GMAS events are not necessarily functional as they could be in LD with the functional SNPs. To infer the functional SNPs, we developed a new method that combines allele-specific analysis of one data set with population-level variation in GMAS patterns, namely, concordance-based analysis of GMAS (cGMAS).
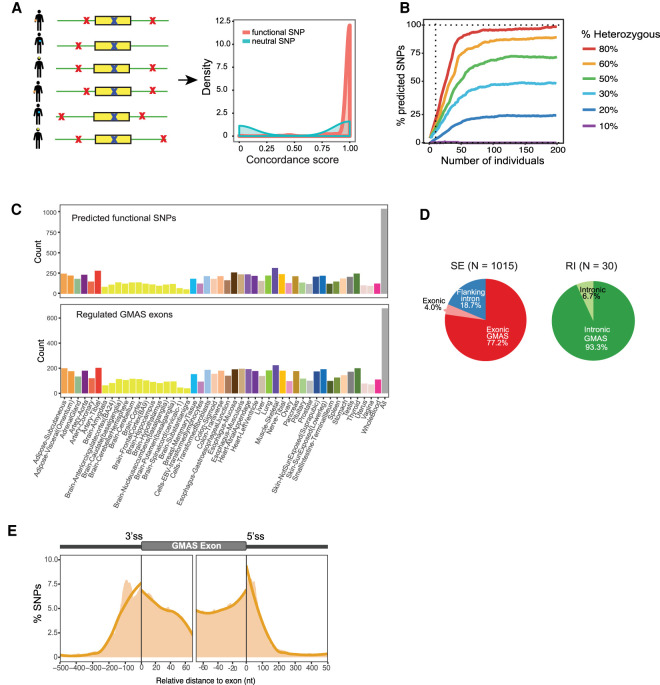
Leveraging the genetically driven nature of GMAS, cGMAS is built upon the rationale that a functional SNP, if it exists as a heterozygous SNP, should always lead to allele-specific splicing pattern (i.e., GMAS) in the corresponding data set. Thus, we expect to observe concordance between the genotype of a functional SNP and the splicing patterns of a GMAS exon across different individuals. As illustrated in Figure 3A (details in Methods), the cGMAS method considers as candidate functional SNPs all heterozygous SNPs in GTEx individuals located in the proximity of GMAS events. For each candidate SNP, a concordance score (Si) was calculated between its genotype and the GMAS pattern in each individual where the SNP genotype is available. In particular, the GMAS pattern was represented by the allelic imbalance at the tag SNP initially identified with the GMAS event (Supplemental Fig. S5). Subsequently, the distribution of Si over all individuals was analyzed using a Gaussian Mixture Model (GMM) to identify prominent peaks. The significance of each peak was evaluated via randomization of the Si values. The functionality of the candidate SNP was determined based on the number and Si values of significant peaks (FDR ≤ 0.05) detected in the above procedure (Supplemental Fig. S5).
Figure 3.
Prediction of functional SNPs for GMAS events. (A) Functional SNPs are predicted by considering candidate SNPs (red crosses) in the vicinity of a GMAS exon, including the tag SNP itself (blue crosses). Concordance among the allelic ratios of the tag SNP in all samples is calculated as described in Methods (with hypothetical distributions shown). (B) The percentage of SNPs predicted given the number of individuals in the simulated testing cohort (Methods). Different percentages of individuals with the heterozygous genotype were simulated. Vertical dotted line marks 10 individuals. (C) Top: Number of predicted functional SNPs per tissue. Bottom: Number of GMAS exons with predicted functional SNPs per tissue. The rightmost bar (All) corresponds to predictions made by pooling samples from all tissues. (D) Left pie chart: predicted functional SNPs in the exonic or intronic regions of SE (skipped exons). Exonic GMAS: the functional SNP is also the exonic GMAS tag SNP. The rest of the functional SNPs were classified into the “Exonic” or “Flanking intron” group. Right pie chart: for retained introns (RI). Intronic GMAS: the functional SNP is also the GMAS tag SNP. N refers to the number of functional SNPs for each group. No functional SNPs were predicted for alternative 5′ or 3′ ss exons. (E) Densities of predicted functional SNPs near the exon-intron boundaries of their associated GMAS exons (SEs only). The number of functional SNPs was normalized by the total number of testable SNPs at each nucleotide position. Orange curve is the fitted trend line of the shaded area that represents the SNP density at single-nucleotide resolution.
The ability to identify functional SNPs via cGMAS is expected to depend on the number of individuals that possess the GMAS pattern of a given exon. To carry out a power analysis for this method, we simulated 100 hypothetical GMAS exons with functional SNPs that occur in a varying number of individuals (Methods). In addition, we varied the fraction of the simulated individuals that harbor a heterozygous genotype at each functional SNP (Methods). As expected, greater predictive power was achieved if more individuals had the GMAS event (Fig. 3B). The proportion of individuals that had heterozygous alleles at the candidate SNP (i.e., heterozygous ratio) also affected power, where higher heterozygous ratios led to increased power.
Functional SNPs for GMAS events in GTEx individuals
We applied cGMAS to analyze the GTEx data in two ways: separately for individual tissues and collectively using data of all tissues. Because the number of data sets from each tissue is limited, the latter analysis is associated with increased predictive power. Although tissue-specific functional SNPs may not be identifiable, the pooled analysis could detect SNPs that function relatively ubiquitously across tissues. These analyses together identified 1045 putative functional SNPs corresponding to 677 GMAS exons (FDR ≤ 0.05) (Fig. 3C). These SNPs had 16%–24% of overlap with known sQTLs, depending on the method and data set used for sQTL analyses (Lappalainen et al. 2013; ’t Hoen et al. 2013; The GTEx Consortium 2015; Supplemental Fig. S6). Among the 1045 predicted functional SNPs, 1015 were associated with exon skipping events and the remaining 30 were associated with intron retention. Because alternative 5′ and 3′ splice site events generally occurred in few individuals, functional SNPs were not predicted for these types of splicing.
Among the putative functional SNPs, about 78% (812) coincided with the GMAS tag SNPs. The rest of the SNPs were located within the same exons as the GMAS exon or in the flanking introns (Fig. 3D). In addition, 23 (2.2%) putative functional SNPs resided in the 5′ splice sites (ss), and 26 (2.5%) in the 3′ ss. The alternative alleles of these SNPs caused a significant difference in the splice site strength (Supplemental Fig. S7). In general, putative functional SNPs demonstrated a positional bias towards enrichment near the splice sites of skipped exons (Fig. 3E), consistent with the expectation that regulatory elements of splicing tend to locate in close proximity to splice sites. Note that because the other types of alternative exons had relatively small numbers of events, they were not included in this analysis.
Experimental support of functional SNPs for GMAS
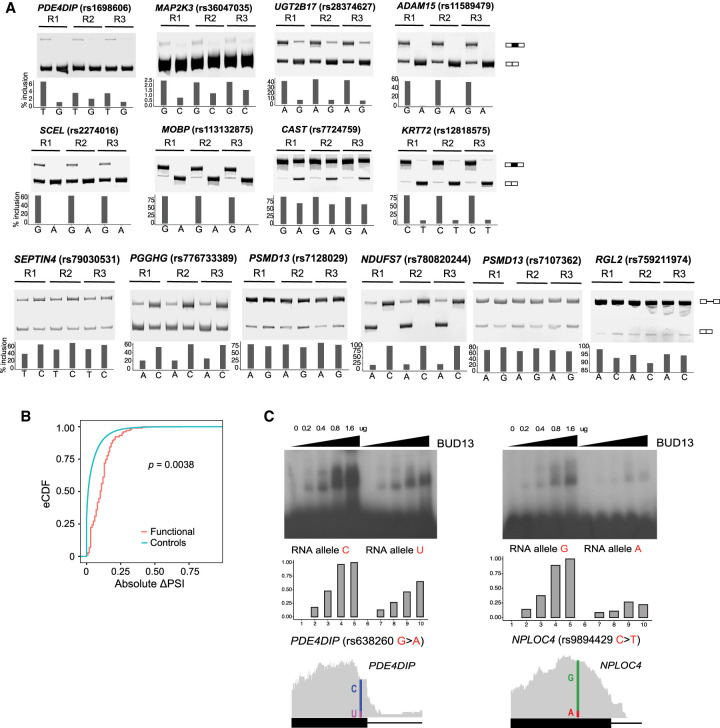
To support the predicted functional SNPs, we performed minigene reporter experiments using a splicing reporter from a previous study (Xiao et al. 2009). For each candidate SNP, we created two versions of the minigene construct, harboring the reference and variant alleles, respectively (Supplemental Table S2). Once expressed in cells, minigenes containing functional SNPs are expected to show a significant splicing difference between the two versions. Using this system in HeLa cells, we tested 14 predicted functional SNPs, eight associated with exon skipping events (PDE4DIP, MAP2K3, UGT2B17, ADAM15, SCEL, MOBP, CAST, and KRT72) and six with intron retention (SEPTIN4, PGGHG, PSMD13 [×2], NDUFS7, and RGL2) (Fig. 4A). These minigenes were further tested in U87, K562, and/or HEK293 cells (Supplemental Fig. S8A). All 14 SNPs were confirmed to lead to allele-specific splicing patterns. The observed differences in allelic effect were replicated across multiple cell lines (except for the event in MAP2K3), which highlights again that GMAS events tend to be shared across cell types. These results strongly support the predicted functionality of these SNPs.
Figure 4.
Experimental support of predicted functional SNPs. (A) Minigene experiments validating predicted functional SNPs for GMAS in triplicates (R1–3). The inclusion levels (% inclusion) of the skipped exons or retained introns were estimated from the band intensities of the PAGE gel. Note that, for illustration purposes, the y-axis scales are different for different genes. (B) eCDF of the absolute changes in the PSI values of GMAS exons upon KD of splicing factors associated with putative functional SNPs (based on motif analysis or eCLIP overlap). All splicing factors in Supplemental Figure S9A with ENCODE KD RNA-seq data are included here. The P-value was calculated using the Kolmogorov–Smirnov test. (C) EMSA validating allele-specific binding of BUD13 to putative functional SNPs. The amount of BUD13 protein used in the experiment is illustrated above the gel. BUD13 eCLIP reads (gray) are shown below the gel, where the locations of the SNPs are labeled with vertical colored lines. The fraction of reads supporting either allele at the functional SNP position is delineated.
It is expected that many functional SNPs may disrupt the interaction between splicing factors and their cis-regulatory motifs (Hsiao et al. 2016b). Among the putative functional SNPs, 492 were predicted to alter the binding motifs of known splicing factors (Cook et al. 2011; Ray et al. 2013) (using our previous motif analysis method [Hsiao et al. 2016a]) or overlap the binding sites of splicing factors in the ENCODE eCLIP data sets (Supplemental Fig. S9A; Van Nostrand et al. 2020). For these SNPs, we observed that the splicing of their associated GMAS exons showed significant changes upon splicing factor knockdown (KD) compared to random testable alternatively spliced exons as controls (ENCODE RNA-seq data) (Fig. 4B; Van Nostrand et al. 2020), supporting the functional roles of the splicing factors.
Furthermore, 31 putative functional SNPs were testable for allele-specific binding (ASB) using the ENCODE eCLIP data in our previous study (Yang et al. 2019), 18 (58%) of which had significant ASB supporting their functional roles. To experimentally confirm the ASB patterns, we carried out electrophoretic mobility shift assays (EMSA, or gel shift) for BUD13, the protein with the largest number of eCLIP peaks overlapping putative functional SNPs (Supplemental Fig. S9A,B). We focused on two candidate functional SNPs and asked whether BUD13 binds to the alternative alleles with different strength. Two versions of the RNA sequences were synthesized harboring the alternative alleles of each SNP. As shown in Figure 4C, the binding of BUD13 to target RNAs was stronger with increasing protein input. The alternative alleles of the SNPs demonstrated visible differences in their binding to the protein, supporting the functional impact of these SNPs. It should be noted that the binding motifs of BUD13 may be quite diverse (Feng et al. 2019), explaining the seemingly different nucleotide sequences of the two BUD13 targets in Figure 4C. Consistent with the EMSA data, the two SNPs were also predicted as ASB SNPs for BUD13 via eCLIP-seq analysis of K562 cells (Fig. 4C; Yang et al. 2019). In addition, minigene experiments in our recently published study confirmed the functional role of rs638250 in splicing regulation (Supplemental Fig. S9C; Yang et al. 2019). In general, BUD13 may regulate many more GMAS events, supported by the relatively high correlation between BUD13 expression and the allelic ratios of GMAS SNPs (Supplemental Fig. S9D).
GMAS events are enriched in disease-relevant genes and regions
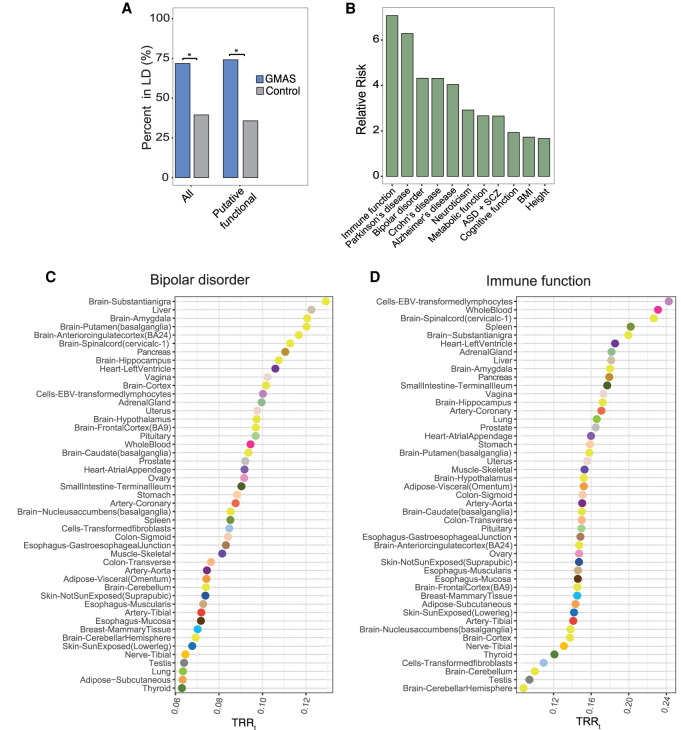
To examine the disease relevance of GMAS events, we first asked whether GMAS events are significantly associated with GWAS loci. Specifically, we examined whether the predicted functional SNPs underlying GMAS were in LD with GWAS SNPs (and within 200 kb in distance) (Methods). As controls, random variants from non-GMAS genes were sampled and analyzed relative to GWAS SNPs similarly. We observed that 74% (774) of GMAS functional SNPs were in LD with GWAS SNPs, a percentage significantly higher than that among control SNPs (P < 2.2 × 10−16) (Fig. 5A). Note that similar results were observed when including all GMAS events, not limited to those with putative functional SNPs (72%, 5317 GMAS SNPs, P < 2.2 × 10−16) (Fig. 5A). This result is expected because GMAS, by definition, is driven by genetic variants, and the functional SNPs (even if not identifiable here due to lack of power) are usually located close to the regulated exon (Fig. 3E). Overall, these observations support the likely disease relevance of GMAS events.
Figure 5.
Functional relevance of GMAS events. (A) Proportions of all GMAS SNPs and putative functional SNPs in LD with (and within 200 kb of) GWAS SNPs. Controls were random dbSNPs in genes that do not harbor GMAS SNPs. (*) P < 2.2 × 10−16 (Fisher's exact test). (B) Relative risk of GMAS SNPs in LD with selected trait/disease, defined as the ratio between the values in A of the GMAS and control groups for all GMAS SNPs. (ASD) Autism spectrum disorder, (SCZ) schizophrenia, (BMI) body mass index. All P-values < 2.2 × 10−16 (Fisher's exact test). (C,D) Trait-relevance ratio of each tissue (TRRt) defined as the proportion of GMAS SNPs identified in each tissue that were in LD with GWAS SNPs, for bipolar disorder and immune function, respectively.
We further examined the GMAS-GWAS relationship for specific traits/diseases. For each trait/disease, we repeated the above LD analysis and calculated the enrichment of all GMAS SNPs relative to control SNPs that are located in LD regions of GWAS SNPs (defined as relative risk) (Methods: Fig. 5B). A number of traits/diseases, such as immune function, Parkinson's disease, and bipolar disorder, demonstrated significantly high enrichment (Fig. 5B). Similar results were obtained when repeating this analysis using predicted functional SNPs only (Supplemental Fig. S10), although some differences do exist likely due to reduced power given the inevitably smaller number of GMAS events with predicted functional SNPs than the set of all GMAS events. GMAS SNPs associated with immune function consistently had the highest relative enrichment, in line with the known prevalence of alternative splicing in the immune system (Lynch 2004). The enriched association of GMAS SNPs with neurological function or related diseases suggests that splicing may have close relevance in these processes. Complex traits such as height and body mass index (BMI) had the lowest relative risk (although still significant), indicating that splicing likely contributes the least to their underlying biological mechanisms among those considered here.
An interesting question is whether the GMAS events were identified in tissues relevant to their associated GWAS traits/diseases. To this end, we defined a trait-relevance ratio (TRR) to evaluate the proportion of GMAS SNPs in each tissue that were in LD with GWAS SNPs for a given trait/disease (Methods). This analysis revealed some interesting insights. For example, for bipolar disorder, brain tissues had the highest TRRs among all tissues, consistent with the nature of the disease (Fig. 5C; Supplemental Fig. S11A). In contrast, TRRs were highest in lymphocytes and whole blood for immune function-associated GMAS SNPs (Fig. 5D; Supplemental Fig. S11B), both with immune relevance. In addition, GMAS SNPs associated with metabolic function had the highest TRRs in tissues (liver and adrenal gland) of close relevance to metabolism (Supplemental Fig. S11C,D). Neuroticism- and cognitive function-related GMAS SNPs were observed with high TRRs in brain tissues (Supplemental Fig. S11E–H). Note that the above TRR enrichment is unlikely solely due to tissue-specific expression of the GMAS-associated genes, as the above results still hold after removing GMAS events in known tissue-specific genes (Supplemental Fig. S12A–V; Yang et al. 2018). Thus, these observations are highly consistent with the expected tissues of relevance of the traits/diseases, supporting the potential involvement of GMAS in related functional processes. For other traits, the top tissues with high TRR values were more diverse or nonintuitive (Supplemental Fig. S11I–T). It is likely that genetically driven splicing alteration is not a primary contributor, or alternatively, these traits/diseases are complex and involve biological processes in a wide range of tissues.
Discussion
We report a comprehensive study of allele-specific alternative splicing (a.k.a. GMAS) in human tissues. Using GTEx data sets, we identified thousands of GMAS events, encompassing 4941 exons and 7404 SNPs. The multifaceted nature of the data allowed an examination of the GMAS landscape across tissues and individuals. We observed that the allele-specific pattern of GMAS events varied to similar degrees across tissues and individuals. It is well established that alternative splicing demonstrates high tissue specificity, which enables segregation of samples by tissue types rather than per individual (Barbosa-Morais et al. 2012; Merkin et al. 2012; Melé et al. 2015). In contrast, our analysis showed that, for genetically regulated splicing events, the genetic contribution to splicing variability is equivalent to that contributed by tissue specificity. As tissue specificity is often imposed by trans-acting regulators, our results suggest that cis- and trans-regulatory mechanisms have similar degrees of impact on the variability of GMAS.
In general, GMAS events can be shared across tissues or individuals or demonstrate high tissue- or individual-specificity (Figs. 1B, 2). We observed that GMAS events overall are shared more significantly than expected by chance across tissues or individuals (that share the same genotype) (Fig. 2B,C). This result is consistent with previous literature that genetically driven splicing profiles tend to be common to different cell or tissue types (Li et al. 2012; The GTEx Consortium 2015; Hsiao et al. 2016a). This is expected because genetic determinants are the most important factor for such splicing events. On the other hand, there do exist many GMAS events that are highly individual- or tissue-specific (Fig. 1B). Genes with individual-specific GMAS exons are often involved in immune-related processes. This observation not only highlights the impact of an individual's genetic makeup on the immune system but also identifies splicing as a potential mechanism through which the phenotypic effects of genetic variants are manifested. In contrast, genes containing GMAS exons with high tissue variability are involved in heart or skeletal muscle function, supporting the particular importance of alternative splicing in the biophysical properties and functions of cells (Kelemen et al. 2013).
Leveraging the GTEx genotype information and GMAS events, we developed a new method to pinpoint functional SNPs that regulate splicing. Specifically, our method appraises the concordance between the allelic bias of a candidate SNP and the splicing pattern of an alternatively spliced region, as represented by the allelic signature of the tag GMAS SNP. The key factor that determines the performance of our method is the “heterozygous ratio” of a candidate functional SNP among the testing cohort. Our method demonstrates high predictive power when many individuals have heterozygous alleles at the candidate SNP locus. Within the GTEx cohort, we were able to predict over 1000 functional SNPs for GMAS, and the quality of our predictions was confirmed by the enrichment of functional SNPs near the splice sites, a popular metric used to examine the splicing relevance of a SNP. This method can be generally applied to any data set encompassing large populations to expand the repertoire of functional SNPs that regulate splicing.
Many large-scale efforts have been devoted to understanding the functional relevance of SNPs in the human genome. To date, the GWAS catalog has documented hundreds of thousands of phenotype-associated SNPs from over 3500 publications (Buniello et al. 2019). Yet, many traits were found to associate with noncoding or intergenic SNPs that do not alter the protein sequences, which makes GWAS interpretation challenging. We observed that a high fraction of GMAS events are associated with SNPs in LD with GWAS loci, suggesting that these GWAS-reported SNP-trait associations may be related to dysregulation of splicing. This observation is further substantiated by the GMAS enrichment in tissues of expected relevance for a number of GWAS traits (e.g., bipolar disorder, metabolic and immune function). Our study indicates that allele-specific splicing analysis is an effective means to discover functionally relevant genetic variants that may contribute to disease mechanisms. Future studies can leverage long-read sequencing technologies to better characterize GMAS. This may be achieved through improved haplotyping and incorporation of isoform level ASE events that would otherwise be missed by short reads. The use of long reads may also make it easier to detect functional cis-regulatory variants for splicing (Deonovic et al. 2017).
Methods
Preprocessing of GTEx RNA-seq data and identification of GMAS events
FASTQ files from individuals with genotype information (from whole-genome sequencing, whole-exome sequence, or Illumina SNP Arrays) were downloaded from the GTEx database (v6p release). Library adaptors were trimmed by cutadapt (Martin 2011). We aligned the reads to the hg19 genome and transcriptome using HISAT2 (Kim et al. 2015) with parameters ‐‐mp 6,4 ‐‐no-softclip ‐‐no-mixed ‐‐no-discordant, keeping only the uniquely mapped read pairs for the following analyses. Note that the choice of genome assembly (hg19 vs. GRCh38) should not affect our conclusions as the identification of GMAS utilized genotyping data in the nuclear genome (source: GTEx; see below). Samples with fewer than 25 million uniquely aligned read pairs were considered as insufficient read coverage for detecting GMAS events and thus discarded (about 10% of all data sets). We focused on the tissues that have at least 50 samples with sufficient read coverage. In total, 7822 RNA-seq samples across 47 tissues from 515 donors were kept for the GMAS analysis.
We collected a list of high-quality SNPs from whole-genome sequencing (quality filter: GQ ≥ 20), whole-exome sequence (quality filter: GQ ≥ 20), and Illumina SNP Arrays (quality filter: IGC ≥ 0.2) provided by GTEx. In addition to the genotyped SNPs, we included all dbSNPs (version 146) that showed RNA-seq evidence of being heterozygous in at least one GTEx individual as potential candidates for the GMAS analysis. To determine which dbSNPs were heterozygous, we used the RNA-seq reads covering the candidate dbSNP position and defined the SNP to be heterozygous if it had at least three reads for each of the two alleles (with at least 20 total reads). Additionally, we further filtered out those with extreme allelic ratio (AR, defined as number of reads covering the reference allele/total number of reads), that is, AR < 0.1 or AR > 0.9. This filter removes monoallelically expressed SNPs and excludes imprinted genes and other genes with extreme allele-specific expression (ASE). In our study, this filter is necessary to focus on heterozygous SNPs observed in RNA-seq.
We applied our published method (Li et al. 2012) to predict GMAS events using the combined list of SNPs (genotyped or RNA-seq-based) and the uniquely aligned RNA-seq reads. Briefly, this method first examines ASE of all heterozygous SNPs in a gene. It then determines whether ASE is global in the specific gene, which represents gene-level ASE possibly regulated at the level of transcription or RNA decay that affects all heterozygous SNPs in the gene. Alternatively, a gene may have local ASE, that is, ASE demonstrated in only a small fraction of testable (≥20 read coverage) heterozygous SNPs. GMAS accounts for a type of such local ASE patterns, where the ASE SNP is located in an alternatively spliced exon and has significant allelic bias relative to control SNPs in the same gene (non-ASE SNPs).
Relative to the published version (Li et al. 2012), we made the following modifications in this study. First, instead of focusing solely on annotated alternatively spliced exons from GENCODE comprehensive annotation (v24lift37), we tested all internal exons for potential GMAS events. Second, we replaced the normalized expression value (NEV) by PSI calculated by the method described in Schafer et al. (2015), only keeping exons with ≥15 total reads (inclusion reads + exclusion reads) or ≥2 exclusion reads. An exon is testable if it passes the read coverage requirements for PSI calculation and has a powerful (defined as having ≥20 read coverage) non-ASE SNP in another exon of the same gene (Li et al. 2012). We further required all GMAS exons to have PSI ≤ 0.8. To avoid false positives, we only focused on GMAS events that were called in at least three samples out of the total 7822 samples we analyzed.
Estimation of tissue versus individual contributions to GMAS pattern variations
We used the lmer function from the lme4 package in R (version 3.6.0) (R Core Team 2019) to model the allelic imbalance for each GMAS exon as the following:
The allelic imbalance was calculated as the absolute difference of allelic ratio to 0.5. The fixed effects (age, ethnicity, and sex) were chosen based on the previous literature (Melé et al. 2015). The allelic imbalance variations contributed by tissues and by individuals were estimated from the above model.
Tissue specificity quantified by Jaccard index and GMAS frequency
We used the Jaccard index to quantify the extent of sharing of the GMAS pattern for an exon e between tissues i and j (seij) across individuals in whom exon e is GMAS. Specifically, , where Nei and Nej are the number of individuals with e showing GMAS pattern in tissues i and j, respectively (i ≠ j). To reliably estimate seij, we required . The final GMAS pattern shared between tissues i and j (sij) was calculated as , where E is the total number of exons with seij for tissues i and j.
Tissue and individual variability in GMAS
To assess the variability in GMAS across individuals and tissues, we used variance as a quantitative measure of dissimilarity in allelic biases. For each exon showing GMAS pattern in any given individual, we measured the variance within allelic biases of the tag SNPs in all corresponding tissues of the individual. As controls, we sampled allelic biases of the tag SNPs of the same exon in similar tissues but different individuals and calculated their variance. The distribution of variances across all individuals for the GMAS exons was then compared to that of the controls (Fig. 2B). Similarly, for each exon showing GMAS pattern in a given tissue, we calculated the variance among the allelic biases of the tag SNPs across individuals. The controls were randomly sampled allelic biases of the tag SNPs of the same exon in individuals showing GMAS pattern for the exon but different tissues. Again, we compared the distribution of variances across all tissues for the GMAS exons to the distribution of variances in controls (Fig. 2C).
Prediction of functional SNPs for GMAS
The basic rationale for our method is that a functional SNP for GMAS should show concordant relationship (cGMAS) between its genotype and the splicing pattern of the target exon across a large number of individuals. In the toy example illustrated in Supplemental Figure S5A, we first define a distance metric d = |0.5–Rtag|, where Rtag is the allelic ratio of the tag SNP defined as Nref/(Nref + Nalt). Nref and Nalt denote the number of reads harboring the reference allele and the alternative allele of the SNP, respectively. Thus, d represents the difference between the allelic ratio of the tag SNP and the expected allelic ratio of an unbiased SNP. In Supplemental Figure S5A, the candidate SNP (which is different from the tag) is assumed to be the functional SNP underlying GMAS, with the A allele causing exon inclusion and G allele causing exon skipping. Thus, for individuals with the homozygous genotype (AA or GG) at the candidate SNP, d is expected to be 0.5. On the other hand, for individuals with AG genotype at the candidate SNP, d is 0 or 1 depending on the haplotype between the tag and candidate SNPs.
Next, we define the concordance score (Si) for this example exon in individual i, similarly as used in a previous study (Lappalainen et al. 2013). Si measures the concordance level between the genotype and the splicing pattern.
For the toy example in Supplemental Figure S5A where A/G alleles of the candidate SNP cause a complete switch of exon inclusion/exclusion, the value of Si is 1. In a different scenario as illustrated in Supplemental Figure S5B where the tag SNP is considered as the candidate functional SNP, we define
Thus, in the case of a functional SNP causing a complete switch of exon inclusion/exclusion, the value of Si is also 1. In general, for true functional SNPs, Si is expected to have a distribution with a peak close to 1, whereas random neural SNPs have broadly distributed Si values (Supplemental Fig. S5C).
For more realistic cases where the two alleles of the functional SNP do not cause 100% splicing difference, the distribution of Si is multimodal. In addition to a peak close to 1, another peak in the medium Si range (>0) exists. On the other hand, a peak at 0 corresponds to nonfunctional SNPs. To unbiasedly model the distribution of Si, we fitted a Gaussian Mixture Model to identify its peaks. The number of GMM components was determined via the Bayesian information criterion (BIC). A z-test was carried out to search for peaks whose average values were significantly different from 0 (FDR ≤ 0.1). For a true functional SNP, the Si distribution should be supported by individuals with different genotypes (homozygous or heterozygous). To avoid potential false positives driven by a specific genotype in a small number of individuals, we excluded candidate SNPs where the genotype supporting the Si peak is significantly biased towards one genotype (Fisher's exact test, FDR ≤ 0.1).
To ensure that the magnitude of the peak was significant, we binned the x-axis (Si scores) into 100 bins and randomized the data points evenly across the bins to generate a background distribution. This process was repeated 500 times to estimate an average background peak level and its standard deviation. We compared the peak height to the background in the same bin and defined significant peaks by z-score > 2.58, which corresponds to P < 0.01.
For each GMAS exon, we examined all SNPs in the exon and the immediate introns as candidate functional SNPs (Supplemental Fig. S5D). SNPs that are homozygous in all individuals were not considered. The concordance score for each candidate and tag SNP pair was calculated, and the functional SNP was predicted as described above.
Power analysis for predicting functional SNPs for GMAS
To assess how many individuals our method necessitates to predict functional SNPs for GMAS, we simulated 100 functional SNPs with two alternating alleles inducing 75% difference in PSI. This allele-specific splicing difference is reflected in the allelic ratios. The total read counts of a SNP were simulated from a negative binomial distribution using parameters estimated from a real GTEx RNA-seq sample. We required all simulated SNPs to have at least 20 reads. The allelic ratios of the simulated SNPs were generated from a binomial distribution.
We simulated six groups of 200 individuals. Each group has a specific heterozygous frequency (Fig. 3B), which is defined as the fraction of individuals with heterozygous alleles at the candidate SNP position in a group. We ran the cGMAS method on the 100 SNPs by varying the number of individuals while maintaining the heterozygous frequency for prediction. Figure 3B illustrates the power of this method in the different simulations.
Analysis of ENCODE eCLIP-seq and RNA-seq data
eCLIP peaks were obtained from the ENCODE portal (https://www.encodeproject.org). The ENCODE RNA-seq data were analyzed similarly as described above for GTEx RNA-seq data. PSI values of replicated samples were averaged in Figure 4C.
Analysis of GMAS SNPs in LD with GWAS SNPs
Trait-variant associations with P-values larger than 5.0 × 10−8 were removed from the GWAS catalog (Buniello et al. 2019) (version 1.0.2, downloaded 2020-02-04). In addition, the GWAS SNPs were separated into LD blocks according to the LD information of the CEU population and further required to have R2 ≥ 0.8 and D′ ≥ 0.9. To evaluate the functional relevance of GMAS SNPs with regard to GWAS, we calculated the number of GMAS SNPs in LD with and within 200 kb of at least one GWAS SNP (referred to as GMAS-GWAS SNPs). A similar number was also calculated for the putative functional SNPs. To determine the significance of the above enrichment, we randomly sampled the same number of dbSNPs from genes that do not host GMAS events. The number of randomized dbSNPs in LD with and within 200 kb of at least one GWAS SNP was compared to that of the GMAS SNPs with a Fisher's exact test.
To investigate the enrichment of GMAS-GWAS SNPs in specific traits/diseases, we calculated the relative risk of GMAS SNPs being in LD with and within 200 kb of a GWAS SNP for the trait of interest versus control SNPs. The relative risk or risk ratio was calculated as follows:
where RRT is the relative risk for trait T, is the proportion of GMAS SNPs in LD with GWAS SNPs for trait T, and is the proportion of control SNPs in LD with GWAS traits for trait T.
As a measure of how relevant the GMAS-GWAS SNPs are to the corresponding traits, we calculated the trait-relevance ratio (TRRt) for each tissue in which the SNP showed GMAS pattern. The TRRt metric controls for the number of GMAS events identified per tissue and is calculated as
where is the trait-relevance ratio, T is the trait of interest, and t is a source tissue of a GMAS-GWAS SNP.
Cell culture
HEK293T and HeLa cells were obtained from ATCC and maintained in DMEM supplemented with 10% FBS (Thermo Fisher Scientific 10082147) and antibiotics at 37°C in 5% CO2.
Construction of minigenes
Minigenes containing SNP candidates were cloned as previously described (Yang et al. 2019). Briefly, the candidate skipped exon and ∼500 nt of each flanking intron were amplified using HeLa genomic DNA. The DNA fragment was then subcloned into the pZW1 splicing reporter using HindIII and SacII or EcoRI and SacII cloning sites. The candidates for intron retention were cloned into the pcDNA3.1 plasmid. Final constructs were confirmed by Sanger sequencing. Primers used in this study are listed in Supplemental Table S2.
Transfection, RNA extraction, reverse transcription, and PCR
Minigene constructs were transfected into >90% confluence HeLa cells using Lipofectamine 3000 (Thermo Fisher Scientific L300015). Total RNA was isolated after 24-h transfection using TRIzol (Thermo Fisher Scientific 15596018) followed by a Direct-zol RNA Miniprep plus kit (Zymo Research R2072). cDNA was produced from 2 μg of total RNA by the SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific 18091050). To amplify the candidate exons in minigene constructs, 5% of the cDNA was used as a template via 26 PCR cycles (Supplemental Table S2).
Gel electrophoresis and quantification
The PCR amplicon was loaded onto 5% polyacrylamide gel and run at 70 volts for 1.5 h. The PAGE gel was stained with SYBR Safe DNA Gel Stain (Thermo Fisher Scientific S33102) for 30 min, and the gel image was taken by the Syngene SYBR safe program (Syngene). Spliced isoforms expression level was estimated using the ImageJ software (http://imagej.nih.gov/ij/). The inclusion or intron retention rate (% inclusion) of the target exon was calculated as the intensity ratio of upper/(upper + lower) bands.
Cloning of human BUD13 and lentiviral overexpression
Human BUD13 was cloned from HeLa cDNA into the pCR 2.1-TOPO vector (Thermo Fisher Scientific 450641). After sequence confirmation, BUD13 was subcloned into the pcDNA3.1 backbone containing 3xFLAG-6HIS tag using NotI and EcoRI sites. To achieve stable overexpression, the 3xFLAG-BUD13-6HIS fragment was transferred into the pLJM1 lentiviral construct using the NdeI and EcoRI sites (Addgene plasmid #19319). We produced lentiviruses via cotransfection of pCMV-d8.91, pVSV-G, and pLJM1-3xFLAG-BUD13-6HIS into HEK293T cells using Lipofectamine 3000 (Thermo Fisher Scientific L3000015). Lentiviruses were collected from conditioned media after 48-h cotransfection and filtered through a 0.2-μm syringe filter. Lentivirus-containing medium was mixed with the same volume of DMEM containing polybrene (8 μg/mL). The lentiviruses were transduced into HEK293T cells in 10 150-mm culture plates, where they were incubated with 2 μg/mL puromycin for 48 h.
Purification of recombinant human BUD13
HEK293T cells stably expressing BUD13 were centrifuged at 1000g for 5 min at 4°C, and the pellets were resuspended with ice-cold 5 mL lysis buffer (PBS, 20 mM Imidazole, 0.5% IGEPAL CA-630, 0.5 mM DTT, 0.5× protease inhibitor cocktail, 100 U DNase I). After a 30-min incubation, the lysate was disrupted using sonication at 25% amplitude for 20 sec with 1-sec pulse. Next, the lysate was centrifuged at 13,000g for 5 min at 4°C. The supernatant was collected and filtered using a 0.45-μm syringe filter. The sample was incubated with 1 mL Ni-NTA agarose (Thermo Fisher Scientific R90110) for 6 h at 4°C followed by five times of washing with 5 mL of buffer A (PBS, 20 mM Imidazole, 0.5 mM DTT, 0.5% IGEPAL CA-630, 0.5× protease inhibitor cocktail). Proteins were eluted with 3 mL elution buffer (PBS, 250 mM Imidazole), and excess salt was removed using a desalting column according to the manufacturer's protocol (GE Healthcare 17085101). Subsequently, FLAG affinity purification was performed using 1 mL FLAG agarose beads (MilliporeSigma A2220) according to the manufacturer's protocol. Elution was performed using 100 mg/mL counter FLAG peptide. FLAG peptide and small size nonspecific proteins were removed by a 20 K Slide-A-Lyzer dialysis cassette (Thermo Fisher Scientific 66003) with 1 L binding buffer (PBS, 0.5% IGEPAL CA-630, 5% glycerol) in the cold room overnight. Recombinant BUD13 purification was confirmed by SimplyBue SafeStain (Thermo Fisher Scientific LC6060) and western blot using BUD13 antibody (Bethyl Laboratories A303-320A). Protein concentration was measured by a Pierce Coomassie (Bradford) protein assay kit (Thermo Fisher Scientific 23200) and the Turner spectrophotometer SP-830.
In vitro transcription of BUD13 target RNA
Sense and antisense oligos including T7 promoter (Supplemental Table S2) were annealed at 95°C for 5 min in a heat block, then cooled down to room temperature for 3 h. In vitro transcription was performed using a HiScribe T7 high yield RNA synthesis kit according to the manufacturer's protocol (NEB E2040S). In vitro-synthesized RNAs were treated with 10 U RNase-free DNase I (Thermo Fisher Scientific EN0525) at room temperature for 30 min, then purified by the RNA clean & concentrator-5 Kit (Zymo Research R1015). Next, RNA samples were treated with 10 U shrimp alkaline phosphatase (NEB M0371S) at 37°C for 1 h and then labeled with 0.5 μL of gamma 32P-ATP (PerkinElmer BLU502A250UC) using 20 U T4 polynucleotide kinase (NEB M0201S). Subsequently, RNA probes were purified by 5% urea PAGE extraction and an RNA clean & concentrator-5 Kit. RNA concentration was measured by Qubit 2.0 fluorometer (Thermo Fisher Scientific).
Electrophoretic mobility shift assay (EMSA)
The RNA probes and recombinant BUD13 protein (0, 0.2, 0.4, 0.8, and 1.6 μg) were incubated in 15 μL of the binding buffer (PBS, 0.5% IGEPAL CA-630, 5% glycerol, 0.1× protease inhibitor cocktail, 10 U RNase inhibitor) at 28°C for 1 h, then loaded onto a 5% TBE-PAGE gel run at 75 V for 1.5 h. The gel was processed without drying, covered with a clear folder, and exposed to X-ray film at −80°C.
Software availability
The code for predicting functional SNPs for GMAS is available as Supplemental Code and at GitHub (https://github.com/gxiaolab/cGMAS).
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank members of the Xiao laboratory for helpful discussions and comments on this work. We thank the GTEx Consortium for generating the valuable data sets used in this study. We thank the ENCODE Consortium (Graveley and Yeo groups) for generating the eCLIP and RNA-seq data. This work was supported in part by grants U01HG009417 (National Human Genome Research Institute) and R01AG056476 (National Institute on Aging) to X.X. K.A. was supported by the UC-HBCU Initiative Fellowship from the University of California Office of the President. Y.-H.E.H. was supported by the Bioengineering supplemental fellowship of UCLA.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.265637.120.
References
- Adamson SI, Zhan L, Graveley BR. 2018. Vex-seq: high-throughput identification of the impact of genetic variation on pre-mRNA splicing efficiency. Genome Biol 19: 71. 10.1186/s13059-018-1437-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre-Gamboa R, Joosten I, Urbano PCM, van der Molen RG, van Rijssen E, van Cranenbroek B, Oosting M, Smeekens S, Jaeger M, Zorro M, et al. 2016. Differential effects of environmental and genetic factors on T and B cell immune traits. Cell Rep 17: 2474–2487. 10.1016/j.celrep.2016.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. 2010. Deciphering the splicing code. Nature 465: 53–59. 10.1038/nature09000 [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Çolak R, et al. 2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 338: 1587–1593. 10.1126/science.1230612 [DOI] [PubMed] [Google Scholar]
- Bretschneider H, Gandhi S, Deshwar AG, Zuberi K, Frey BJ. 2018. COSSMO: predicting competitive alternative splice site selection using deep learning. Bioinformatics 34: i429–i437. 10.1093/bioinformatics/bty244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, et al. 2019. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47: D1005–D1012. 10.1093/nar/gky1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Nguyen TYD, Cygan KJ, Çelik MH, Fairbrother WG, Avsec ž, Gagneur J. 2019. MMSplice: modular modeling improves the predictions of genetic variant effects on splicing. Genome Biol 20: 48. 10.1186/s13059-019-1653-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung R, Insigne KD, Yao D, Burghard CP, Wang J, Hsiao Y-HE, Jones EM, Goodman DB, Xiao X, Kosuri S. 2019. A multiplexed assay for exon recognition reveals that an unappreciated fraction of rare genetic variants cause large-effect splicing disruptions. Mol Cell 73: 183–194.e8. 10.1016/j.molcel.2018.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. 2011. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res 39: D301–D308. 10.1093/nar/gkq1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deonovic B, Wang Y, Weirather J, Wang X-J, Au KF. 2017. IDP-ASE: haplotyping and quantifying allele-specific expression at the gene and gene isoform level by hybrid sequencing. Nucleic Acids Res 45: e32–e32. 10.1093/nar/gkw1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Bao S, Rahman MA, Weyn-Vanhentenryck SM, Khan A, Wong J, Shah A, Flynn ED, Krainer AR, Zhang C. 2019. Modeling RNA-binding protein specificity in vivo by precisely registering protein-RNA crosslink sites. Mol Cell 74: 1189–1204.e6. 10.1016/j.molcel.2019.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro NM, Strober BJ, Einson J, Abell NS, Aguet F, Barbeira AN, Brandt M, Bucan M, Castel SE, Davis JR, et al. 2020. Transcriptomic signatures across human tissues identify functional rare genetic variation. Science 369: eaaz5900. 10.1126/science.aaz5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2015. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao Y-HE, Bahn JH, Lin X, Chan T-M, Wang R, Xiao X. 2016a. Alternative splicing modulated by genetic variants demonstrates accelerated evolution regulated by highly conserved proteins. Genome Res 26: 440–450. 10.1101/gr.193359.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao Y-HE, Cass AA, Bahn JH, Lin X, Xiao X. 2016b. Global approaches to alternative splicing and its regulation—recent advances and open questions. In Transcriptomics and gene regulation. Translational bioinformatics (ed. Wu J), Vol. 9, pp. 37–71. Springer Netherlands, Dordrecht. [Google Scholar]
- Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, Kosmicki JA, Arbelaez J, Cui W, Schwartz GB, et al. 2019. Predicting splicing from primary sequence with deep learning. Cell 176: 535–548.e24. 10.1016/j.cell.2018.12.015 [DOI] [PubMed] [Google Scholar]
- Ke S, Shang S, Kalachikov SM, Morozova I, Yu L, Russo JJ, Ju J, Chasin LA. 2011. Quantitative evaluation of all hexamers as exonic splicing elements. Genome Res 21: 1360–1374. 10.1101/gr.119628.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. 2013. Function of alternative splicing. Gene 514: 1–30. 10.1016/j.gene.2012.07.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedländer MR, ’t Hoen PAC, Monlong J, Rivas MA, Gonzàlez-Porta M, Kurbatova N, Griebel T, Ferreira PG, et al. 2013. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501: 506–511. 10.1038/nature12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Bahn JH, Lee J-H, Peng G, Chen Z, Nelson SF, Xiao X. 2012. Identification of allele-specific alternative mRNA processing via transcriptome sequencing. Nucleic Acids Res 40: e104. 10.1093/nar/gks280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y, Pritchard JK. 2016. RNA splicing is a primary link between genetic variation and disease. Science 352: 600–604. 10.1126/science.aad9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW. 2004. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol 4: 931–940. 10.1038/nri1497 [DOI] [PubMed] [Google Scholar]
- Manning KS, Cooper TA. 2017. The roles of RNA processing in translating genotype to phenotype. Nat Rev Mol Cell Biol 18: 102–114. 10.1038/nrm.2016.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10. Next Gener Seq Data Anal 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. 2015. The human transcriptome across tissues and individuals. Science 348: 660–665. 10.1126/science.aaa0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. 2012. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338: 1593–1599. 10.1126/science.1228186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. 10.1038/nature12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Reuter JA, Spacek DV, Snyder MP. 2015. High-throughput sequencing technologies. Mol Cell 58: 586–597. 10.1016/j.molcel.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhine CL, Neil C, Glidden DT, Cygan KJ, Fredericks AM, Wang J, Walton NA, Fairbrother WG. 2019. Future directions for high-throughput splicing assays in precision medicine. Hum Mutat 40: 1225–1234. 10.1002/humu.23866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg AB, Patwardhan RP, Shendure J, Seelig G. 2015. Learning the sequence determinants of alternative splicing from millions of random sequences. Cell 163: 698–711. 10.1016/j.cell.2015.09.054 [DOI] [PubMed] [Google Scholar]
- Rowlands CF, Baralle D, Ellingford JM. 2019. Machine learning approaches for the prioritization of genomic variants impacting pre-mRNA splicing. Cells 8: 1513. 10.3390/cells8121513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer S, Miao K, Benson CC, Heinig M, Cook SA, Hubner N. 2015. Alternative splicing signatures in RNA-seq data: percent spliced in (PSI). Curr Protoc Hum Genet 87: 11.16.1–11.16.14. 10.1002/0471142905.hg1116s87 [DOI] [PubMed] [Google Scholar]
- Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, Ter Horst R, Jansen T, Jacobs L, Bonder MJ, et al. 2016. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 167: 1125–1136.e8. 10.1016/j.cell.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soemedi R, Cygan KJ, Rhine CL, Wang J, Bulacan C, Yang J, Bayrak-Toydemir P, McDonald J, Fairbrother WG. 2017. Pathogenic variants that alter protein code often disrupt splicing. Nat Genet 49: 848–855. 10.1038/ng.3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne-Weiler T, Sanford JR. 2014. Exon identity crisis: disease-causing mutations that disrupt the splicing code. Genome Biol 15: 201. 10.1186/gb4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Horst R, Jaeger M, Smeekens SP, Oosting M, Swertz MA, Li Y, Kumar V, Diavatopoulos DA, Jansen AFM, Lemmers H, et al. 2016. Host and environmental factors influencing individual human cytokine responses. Cell 167: 1111–1124.e13. 10.1016/j.cell.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ’t Hoen PAC, Friedländer MR, Almlöf J, Sammeth M, Pulyakhina I, Anvar SY, Laros JFJ, Buermans HPJ, Karlberg O, Brännvall M, et al. 2013. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat Biotechnol 31: 1015–1022. 10.1038/nbt.2702 [DOI] [PubMed] [Google Scholar]
- Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, Xiao R, Blue SM, Chen J-Y, Cody NAL, Dominguez D, et al. 2020. A large-scale binding and functional map of human RNA-binding proteins. Nature 583: 711–719. 10.1038/s41586-020-2077-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-S, Cooper TA. 2007. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 8: 749–761. 10.1038/nrg2164 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Wang Z, Jang M, Nutiu R, Wang ET, Burge CB. 2009. Splice site strength–dependent activity and genetic buffering by poly-G runs. Nat Struct Mol Biol 16: 1094–1100. 10.1038/nsmb.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RKC, Hua Y, Gueroussov S, Najafabadi HS, Hughes TR, et al. 2015. The human splicing code reveals new insights into the genetic determinants of disease. Science 347: 1254806. 10.1126/science.1254806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RY, Quan J, Sodaei R, Aguet F, Segrè AV, Allen JA, Lanz TA, Reinhart V, Crawford M, Hasson S, et al. 2018. A systematic survey of human tissue-specific gene expression and splicing reveals new opportunities for therapeutic target identification and evaluation. bioRxiv 10.1101/311563 [DOI]
- Yang E-W, Bahn JH, Hsiao EY-H, Tan BX, Sun Y, Fu T, Zhou B, Van Nostrand EL, Pratt GA, Freese P, et al. 2019. Allele-specific binding of RNA-binding proteins reveals functional genetic variants in the RNA. Nat Commun 10: 1338. 10.1038/s41467-019-09292-w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.