Abstract
Exploring the complicated relationships underlying the clinical information is essential for the diagnosis and treatment of the Coronavirus Disease 2019 (COVID-19). Currently, few approaches are mature enough to show operational impact. Based on electronic medical records (EMRs) of 570 COVID-19 inpatients, we proposed an analysis model of diagnosis and treatment for COVID-19 based on the machine learning algorithms and complex networks. Introducing the medical information fusion, we constructed the heterogeneous information network to discover the complex relationships among the syndromes, symptoms, and medicines. We generated the numerical symptom (medicine) embeddings and divided them into seven communities (syndromes) using the combination of Skip-Gram model and Spectral Clustering (SC) algorithm. After analyzing the symptoms and medicine networks, we identified the key factors using six evaluation metrics of node centrality. The experimental results indicate that the proposed analysis model is capable of discovering the critical symptoms and symptom distribution for diagnosis; the key medicines and medicine combinations for treatment. Based on the latest COVID-19 clinical guidelines, this model could result in the higher accuracy results than the other representative clustering algorithms. Furthermore, the proposed model is able to provide tremendously valuable guidance and help the physicians to combat the COVID-19.
Keywords: Medical information fusion, Coronavirus Disease 2019 (COVID-19), Diagnosis and treatment, Analysis model
1. Introduction
As the continuous growth of Coronavirus Disease 2019 (COVID-19) cases worldwide, the early diagnosis and supportive treatments are crucial to cure the patients [1]. Based on the various patients’ manifestations, the physicians always need to find the representative symptoms from the complicated information to support their diagnosis for the different individuals [2]. After giving the accurate syndrome diagnosis based on the clinical symptoms, a serial of medicines should be combined to efficiently treat the patients. Therefore, the exploration of the effective diagnosis and therapy strategies referring to the key factors, symptom distribution, and medicine combination is of the greatest significance to diagnose and treat the COVID-19 patients.
Researches in the artificial intelligence (AI) or network analysis [3], [4], [5] make great contributions to fight against COVID-19 for the aspects of diagnosis and prognosis, treatments and cures [6], [7]. Researchers have already developed numerous algorithms or models to explore the therapeutic schemes for COVID-19 [8], [9]. Although these studies are relevant to discover the relationships of medical information, higher quality researches are needed to supply effective and reliable approaches to manage the COVID-19 pandemic [10]. Moreover, there is a broad range of potential applications of AI covering diagnosis and treatment challenges posed by the COVID-19.
The most challenges of the clinical diagnosis and treatment methods for the COVID-19 are the following two aspects:
-
•
Most current researches identify the key factors (symptoms,medicines) in diagnosis and treatment by the simple statistical methods [6], [11], such as frequency count, percentage, etc., which may be trapped into the restriction and one-sidedness without considering the multi-dimensions or multi-aspects of clinical information. Besides, the inherent relationships underlying the symptoms and medicines cannot be effectively discovered, which also leads to the inaccuracy of the analysis results [12], [13].
-
•
Despite a high volume of analysis models are constructed to support the clinical diagnosis or treatment for COVID-19, most of them only focus on one of the aspects: diagnosis or treatment [8], [14]. Moreover, most of the diagnosis models just consider the X-ray or computed tomography (CT) image data only without referring to the key factors (symptoms) in the diagnosis process, which will also lead to the inaccuracy of the analysis results [15], [16].
In this paper, we build a diagnosis and treatment analysis model for the COVID-19 to address the challenges of one-sidedness and inaccuracy of clinical analysis results. Through the patients’ manifestations, we hope this model can give the accurate diagnosis of syndromes and provide the corresponding therapies. In order to improve the comprehensiveness of analysis, we use six centrality evaluation metrics to identify core nodes in symptom and medicine networks. Then, based on the skip-gram model [17], [18], we generate the symptom and medicine embeddings with conserving the hidden relationships underlying clinical information. Finally, we use spectral clustering (SC) [19], [20] algorithm to improve the division accuracy through similarity calculation between any two symptom (medicine) embeddings. Through information fusion, we summarize the regularity of the main COVID-19 syndromes and their core symptoms and symptom distribution, core medicines and medicine combination. The following lists the contributions of the proposed analysis model of diagnosis and treatment for COVID-19 pandemic:
-
•
This analysis model identifies the key symptoms and medicines based on six centrality evaluation metrics. It uses the skip-gram model to train and generate the symptom and medicine embeddings conserving the inherent information. Furthermore, combining the node embedding and spectral clustering, the performance of symptom and medicine division will be greatly improved compared to other contrast algorithms.
-
•
The analysis model can comprehensively realize the diagnosis and treatment for COVID-19 by combining with the medical information fusion of the syndromes, symptoms, and medicines together. It can effectively identify the significant syndromes, their core symptoms and symptom distribution, core medicines and medicine combinations. By the information fusion, the accuracy of diagnosis and treatment of COVID-19 will be greatly improved.
The remaining parts of the article are organized as follows. We summarize the related work in Section 2. In Section 3, we depict the architecture of diagnosis and treatment analysis model, data preparation, and model realization. We present the experiment design and process, then give the experimental results and the discussion in Section 4. Finally, we summarize the conclusion and forecast the future works in Section 5.
2. Related work
2.1. Symptom and medicine analysis
Recently, most of the analyses for manifestations and diagnosis, drug use and treatment of COVID-19 patients focus on the simple statistical analysis methods (such as frequency count, percentage) of medical information. For the early manifestations of COVID-19, Adhikari et al. [6] reported that the classical symptoms of COVID-19 include cough, fever, headache, fatigue, pneumonia, hemoptysis, diarrhea, and dyspnea following a methodological framework. Gautier et al. [12] presented the new symptoms of taste and smell loss, excepting for the major symptoms including cough, fever, trouble breathing, etc. Based on 2,450,569 UK and 168,293 US individuals, Menni et al. [11] showed the rank of significant symptoms like loss of smell and taste, shortness of breath, fever, fatigue, persistent cough, skipped meals, diarrhea, hoarse voice, abdominal pain, delirium, chest pain. Song et al. [13] summarized that the most common symptoms included dry cough, fatigue, fever, myalgia and dyspnea, the less common symptoms involved abdominal pain, headache, nausea, diarrhea, and vomiting, and few patients have the manifestations of gastrointestinal symptoms. Medicine therapy is one of the significant treatments for COVID-19 patients. Statistical methods are also applied in medicine analysis. Dong et al. [21] discovered that some medicines, such as remdesivir, chloroquine, favipiravir, and arbidol are undergoing clinical researched to test their safety and efficiency for treating the COVID-19. Romo et al. [22] presented that the antimalarial drugs chloroquine and hydroxychloroquine may have activity against COVID-19. Ren et al. [23] presented that some medicines, such as Gypsum Fibrosum, Poria, etc., play a significant role in curing COVID-19 patients. Luo et al. [24] presented that the clinical drugs used frequently most include Armeniacae Semen Amarum, Scutellariae Radix, and Glycyrrhizae Radix et Rhizome.
Most of these aforementioned studies can effectively discover the key symptoms or medicines in the manifestations and treatments of COVID-19. However, most of the key factors are identified based on the simple statistical methods, such as frequency count, percentage, etc., which cannot be considered from the multiple inherent dimensions. Moreover, the relationships underlying symptoms, medicines, or these two together are not be considered. Therefore, an effective method to explore the key factors and their relationships is still needed to be researched in-depth.
2.2. Diagnosis and treatment model
During the period of COVID-19 pandemic, AI presents a powerful ability to support COVID-19 diagnosis and treatment based on multiple clinical data extracted from electronic medical records (EMRs) [10]. For COVID-19 detection, most of the diagnostic models are designed based on the X-ray or CT image data [7], [14]. For example, based on the X-ray and CT images, Alom et al. [25] introduced the transfer learning method and presented an inception residual recurrent convolutional neural network for COVID-19 diagnosis, which can effectively segment the COVID-19 infected regions and get a highly accurate result. Gozes et al. [26] developed an AI-based image analysis approach to realize the COVID-19 automated detection and patient monitoring, which can achieve high accuracy. Through collecting a huge amount of CT slices from various hospitals, Kumar et al. [2] trained the deep learning model by a decentralized network to detect the COVID-19. AI is also applied for discovering efficient drugs to combat the COVID-19. Gao et al. [8] applied a generative network complex for drug discovery and identified novel candidate drugs and two proposed HIV drugs for curing of COVID-19. Magar et al. [9] took Extreme Gradient Boost (XGBoost) and graph embeddings to discover new therapies in which they searched for antigen-neutralizing antibodies, and proposed antibodies as potentially effective COVID-19 treatments. Bung et al. [27] used a reinforcement learning approach to generate drug compounds with desirable properties and proposed candidate inhibitors for COVID-19.
Despite the diagnosis methods have good performance for COVID-19 detection, most of the diagnosis analysis data just includes the X-ray or CT images, which cannot reflect the inherent and significant characteristics, symptoms, referring to the diagnosis. Moreover, these analysis models of COVID-19 always focused on one aspect of diagnosis or treatment. It is more meaningful if the model research refers to these two aspects. Therefore, the diagnosis and treatment analysis model combining symptoms with medicines together should be further studied.
2.3. Information fusion
Recently, with the development of AI, complex network, and data mining, the utilization of converged various data, such as syndromes, symptoms, and medicines, from clinical EMRs, is also becoming popular [28], [29]. Ang et al. [30] performed a network analysis to find guidelines that provide medical formula for medical observation based on clinical manifestation and showed that the Citri Reticulatae Pericarpium strongly paired with the Glycyrrhizae Radix et Rhizoma. Zhai et al. [1] reported that COVID-19 infection can cause a serial of severe respiratory diseases, and the treatments involving chloroquine and hydroxychloroquine, corticosteroids, antiviral agents, antibodies, convalescent plasma transfusion, and vaccines may work efficiently for COVID-19. Chan et al. [16] presented that the manifestations include dry cough, fever, upper airway congestion, fatigue, shortness of breath, sputum production, myalgia/arthralgia with lymphopenia, prolonged prothrombin time. Some conventional medicines, such as ribavirin, lopinavir/ritonavir, glucocorticoid, beta-interferon, etc., are applied in clinical trials. Jamshidi et al. [15] constructed a diagnosis and treatment platform to combat the COVID-19 through some deep learning methods, which integrates various aspects of information from the structured or unstructured data sources, such as medical imaging, medicine data, etc., and gave a good performance of service.
Although these studies can successfully realize the tentative exploration of the relationships between symptoms and medicines, the analysis model is still considering more medical information referring to the various factors with strong associations. Hence, information fusion, such as syndromes, symptoms, and medicines combined, is effective to improve the performance of the COVID-19 diagnosis and treatment model.
3. Diagnosis and treatment analysis model design
3.1. Model architecture
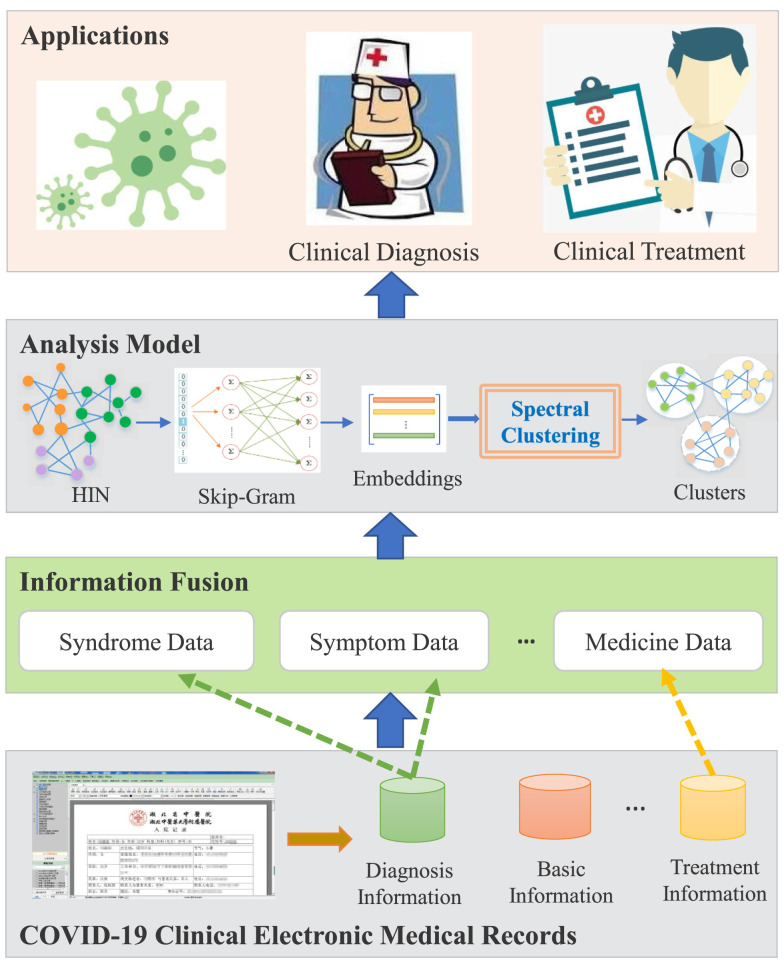
Fig. 1 shows the architecture of the COVID-19 diagnosis and treatment analysis model based on clinical EMRs, which consists of by the four components as follows:
Fig. 1.
The architecture of diagnosis and treatment analysis model.
-
•
Clinical Electronic Medical Records: COVID-19 clinical data are extracted from the EMRs in the hospital information system. Based on the research theme, we focus on the diagnosis and treatment data referring to the different databases.
-
•
Information Fusion: Each EMR corresponding to a COVID-19 patient contains the whole diagnosis and treatment information while in hospital. The data sets extracted from EMRs are desensitized, cleaned, and standardized, then, are constructed as three categories: syndrome data, symptom data, and medicine data. The relationships among homogeneous data or heterogeneous data will be conserved in each record.
-
•
Analysis Model: The three category data sets and their hidden relationships will be abstracted as the nodes and edges in a heterogeneous network. We use a serial of methodologies, including the network analysis, node centrality, node embedding [31], clustering analysis, etc., to construct the overall analytical framework. By using this framework, we can discover the core symptoms and medicines for COVID-19, and summarize the regularity of symptom distribution and medicine combination in terms of the analysis results.
-
•
Applications: In the period of COVID-19 pandemic, the clinicians can use this analysis model to realize the diagnosis and treatment: (i) Clinical diagnosis: the output results of core symptoms and common symptom distribution can offer the effective reference for the diagnosis. Based on them, clinicians can give the accurate judgment. (ii) Clinical treatment: the model will give the medicine analysis results referring to the core medicines and general medicine combination. In terms of them, the clinicians can provide the reasonable therapeutic schedule.
3.2. Data preparation
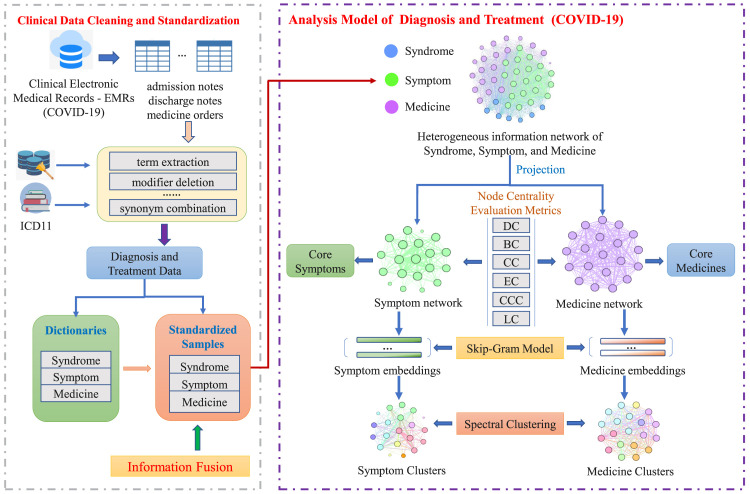
The analysis data of COVID-19 inpatients is extracted from the hospital information system in Hubei Provincial Hospital of Traditional Chinese Medicine, Wuhan, China, one of the COVID-19 designated hospitals, from January 15th to March 13th, 2020. The data preparation includes two parts (shown in Fig. 2) as follows.
Fig. 2.
Flowchart of data processing.
3.2.1. Data cleaning and standardization
The three categories of medical information is extracted from the “computerized physician order entry system”: the syndrome information is from the diagnosis conclusion in the “discharge note table”; the symptom information is from the current symptom description in the “admission note table”; the medicine information is from the “medicine order table”. Base on “International Classification of Diseases 11th Revision (ICD-11)”,1 the syndromes, symptoms, and medicines have been cleaned and standardized, of which the key processes include the term extraction, modifier deletion, terminology standardization, synonym combination, etc. Subsequently, the dictionaries of syndromes, symptoms, and medicines and their corresponding standardized data sets of COVID-19 have been constructed.
3.2.2. Information fusion
For each inpatient, the information from the EMRs has the inherent relationships among them, and the different category data should be analyzed synthetically. Hence, we fuse the syndromes, symptoms, and medicines together and try to discover the regularity of diagnosis and treatment for COVID-19.
3.3. Model realization
We used four approaches to realize the analysis model of the diagnosis and treatment for COVID-19 (Fig. 2) as follows.
3.3.1. Construction regularity of medical network
We used the heterogeneous information network (HIN) to abstract the hidden relationships underlying the medical information of COVID-19. HIN, a graph data model, can capture the relationships among entities (nodes), in which nodes and edges are annotated with class and relationship labels [32], [33], [34]. Through abstracting the syndromes, symptoms, medicines and their relationships underlying the EMRs, we constructed a heterogeneous information network model of COVID-19. In the COVID-19 HIN model, the different nodes represent the syndromes, symptoms, and medicines; the different edges denote the co-occurrence between the syndromes, symptoms, or medicines in the same EMR; the edge weights express the co-occurrence frequencies. Then, based on the theory of complex network [35], [36], we projected the nodes and edges of the COVID-19 heterogeneous information network into the symptom and medicine networks, respectively. The construction regularity of the network is set as follows: take each symptom (medicine) in the records as a node; extract the co-occurrence relation between any two symptoms (medicines) in a diagnosis (prescription) as an edge; denote the co-occurrence frequency of two symptoms (medicines) as the edge weight. Based on it, we define the undirected weighted graph of symptom or medicine, where , , and denote the set of nodes (symptoms or medicines), edges (co-occurrence relations), and weights (frequencies of co-occurrences), respectively.
3.3.2. Evaluation metrics of node centrality
In complex networks, researchers always identify the important nodes using several evaluation metrics [37]. The representative metrics includes degree centrality (BC), closeness centrality (CC), betweenness centrality (BC) [38], eigenvector centrality (EC) [39], current flow closeness centrality (CCC) [40], and load centrality (LC) [41]. In general, the DC, BC, CC, and EC are the conventional metrics to evaluate the node centrality. Considering the complexity of the medical networks, we introduced the CCC metric with the anti-noise feature and the LC metric with the characteristic of local information comparison to further evaluate. The description about these six evaluation indices are shown in Table 1.
Table 1.
Evaluation metrics of node centrality.
| Names | Abbreviations | Equations |
|---|---|---|
| Degree centrality | DC | |
| Betweenness centrality | BC | |
| Closeness centrality | CC | |
| Eigenvector centrality | EC | |
| Current flow closeness centrality | CCC | |
| Load centrality | LC | |
For DC, denotes the degree of node , represents the number of nodes; for BC, indicates the number of the shortest paths from nodes to , denotes the number of the shortest paths through node ; for CC, represents the shortest paths from to ; for EC, indicates the eigenvalues of the adjacent matrix , and is the corresponding eigenvector of ; for CCC, is a normalizing factor, is closeness index of node based on shortest paths, and corresponds to the distance between and ; for LC, is a quantity of the information that is sent from to , and denotes the overall information forwarded by .
3.3.3. Training approach of node embedding
Based on the symptom or medicine network, we used the random walk probability and the Skip-Gram model [17] to generate the symptom embeddings or medicine embeddings [42], respectively. We show the training steps as follows:
-
Step 1:
Initial the unnormalized transition probability for a symptom (medicine) node in the network. Traverse a selected node by the following rules: store the weights between this node and its neighbor nodes; summarize and normalize the weights of the current symptom (medicine) node; obtain the transition probabilities from this node to its neighbors.
-
Step 2:Based on Eqs. (1) and (2), set the unnormalized transition probabilities of each edge.
(1)
where and indicate the nodes and , separately; denotes the weight between and ; is an intermediate node from to ; expresses the search bias between and , is the breadth weight and is the depth weight; indicates the unnormalized transition probability from to ; is the shortest path from to , and .(2) -
Step 3:
Normalize the weight of each edge; obtain the transition probability from the current node to its neighbors.
-
Step 4:Acquire each node’s walk paths (). Calculate the transition probabilities from the current node to its neighbors based on Eq. (3).
where indicates the th node in a walk path; and demonstrate the nodes and ; indicates a constant for the normalization.(3) -
Step 5:
Construct a node list for all the nodes and their paths. The walk regularity is defined as: shuffle the sequence of nodes and randomly select a node; the selected node walks to the neighbor using .
-
Step 6:
Use the Skip-gram model to train the walk paths of a node, and obtain the embedding for each symptom (medicine) node.
3.3.4. Clustering approach of node embedding
The combination of graph embedding and spectral clustering (SC) algorithm works well on 20,000 node network in [20]. According to the trained symptom or medicine embeddings, we also selected the SC algorithm [19] to obtain the clustering results of symptoms or medicines, respectively. We show the clustering steps as follows:
-
Step 1:Get the weighted matrix (similarity matrix ) by Eq. (4). Present as the edge weight between and , and as the similarity of these two symptom (medicine) node embeddings. The weighted matrix is denoted as , here, is the number of nodes. Obtain the Euclidean distance between and , then get and as follows:
where is a scaling parameter to control the descending speed of as the distance descending between and . In [20], the SC algorithm can acquire the best performance when is set as , here, we also set as .(4) -
Step 2:Based on Eq. (5), obtain the degree matrix , indicates the sum of weighted edges connecting to .
(5) -
Step 3:
Denote the Laplacian matrix as , normalize as . Get the first minimum eigenvalues of and their corresponding eigenvectors. Reconstruct the normalized eigenvectors to an new eigenmatrix of size , here .
-
Step 4:
Set the cluster number as , use the K-means algorithm to divide the eigenmatrix into the symptom (medicine) clusters .
4. Experiment
4.1. Experiment design
We designed four experiments and conducted them on a single 16 GB RAM 3.6 GHz Intel (R) Core (TM) CPU. Firstly, in Section 4.2, we constructed the heterogeneous information network of syndromes, symptoms and medicines, and then projected it into the symptom and medicine networks, respectively. Then, we identified the core symptom and core medicine nodes by six evaluation metrics of node centrality in Section 4.3. Subsequently, in Section 4.4, we obtained the symptom and medicine embeddings using the skip-gram model. Then, we acquired the symptom groups and medicine combinations through the SC algorithm, respectively. Finally, we gave the experimental results and the relative analysis compared SC with other representative algorithms: K-means and hierarchical clustering algorithms in Section 4.5.
4.2. Network model construction
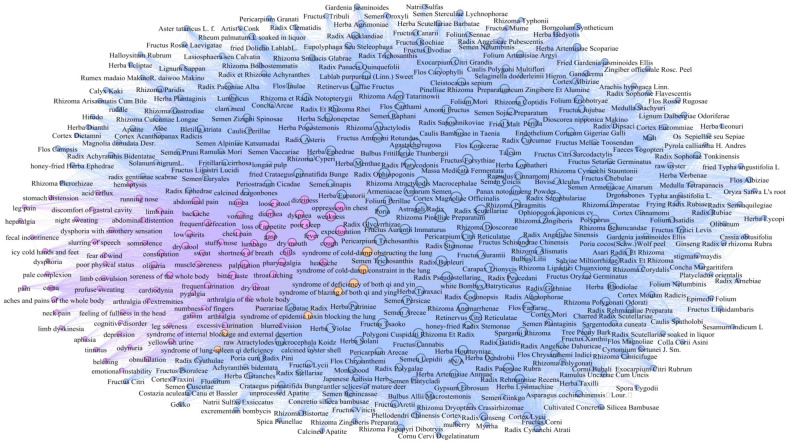
Based on the regularity of network construction in Section 3.3.1, We constructed the HIN model (Fig. 3), in which the different types of nodes represent the entities: syndromes, symptoms, and medicines with three colors; the various edges show the different relationships underlying these entities.
Fig. 3.
Heterogeneous information network of COVID-19 with syndromes (yellow nodes), symptoms (purple nodes), and medicines (blue nodes). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
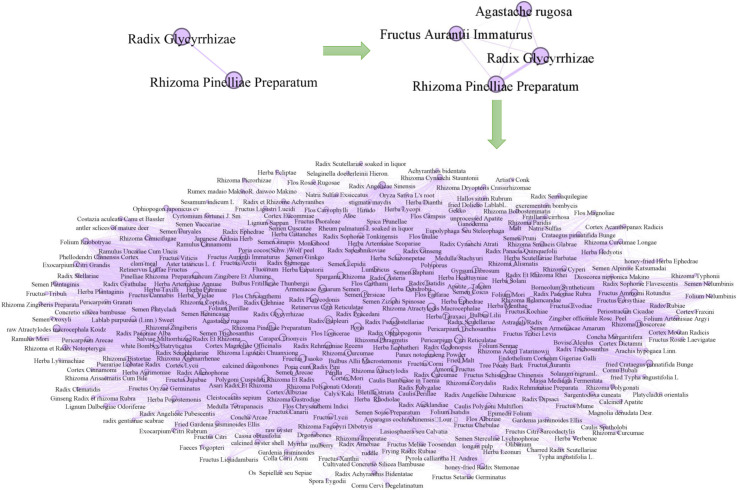
Then, we show the construction processes of COVID-19 symptom network, and medicine network in Fig. 4, Fig. 5, respectively. We initialized a network of two symptom nodes fever and cough with the relative edges. In development, the other two symptom nodes weakness and expectoration with their edges were extended into the network. Finally, we obtained a COVID-19 undirected weighted symptom network with nodes and 10,126 edges in Fig. 4. By the similar way, we also initialized a network with two medicine nodes Radix Glycyrrhizae and Rhizoma Pinelliae Preparatum, and then, extended other two medicine nodes Agastache rugosa and Fructus Aurantii Immaturus into this network. Ultimately, we acquired the medicine network with nodes and 216,327 edges in Fig. 5. We show the statistical summary of the symptom and medicine networks in Table 2, including the name, number of nodes and edges, minimum/maximum/average weights.
Fig. 4.
Construction of symptom network.
Fig. 5.
Construction of medicine network.
Table 2.
Statistical summary of the symptom and medicine networks.
| Names | Number of nodes |
Number of edges |
Minimum weights |
Maximum weights |
Average weights |
|---|---|---|---|---|---|
| Symptom network | 83 | 10,126 | 1 | 272 | 13 |
| Medicine network | 316 | 216,327 | 1 | 401 | 11 |
4.3. Core node evaluation
Based on these two networks, we used six evaluation metrics, including DC, BC, CC, EC, CCC, and LC, to get the various centrality values for the nodes, and just show the top significant symptoms and medicines in Table 3, Table 4 (the digits of the top symptoms and medicines are bolded, respectively). From Table 3 in Appendix, we show that these six metrics can identify the same top symptoms, including cough, weakness, poor sleep, expectoration, loss of appetite, oppression in chest, fever, and gasp; the DC, CC, EC, and CCC can find the same top important symptom dry mouth; the DC, CC, BC, CCC, and LC, excepting for EC, can discover the top symptom dyspnea. Based on Table 4 in Appendix, we show that these six metrics can identify the same top medicines, including Radix Glycyrrhizae, Poria, Pericarpium Citri Reticulatae, Rhizoma Atractylodis Macrocephalae, Rhizoma Pinelliae Preparatum, Astragali Radix, Radix Scutellariae, Cortex Magnoliae Officinalis, Radix Codonopsis, and Radix Ophiopogonis; excepting for the EC identified the top medicine as Radix Platycodonis, however, the centrality value 0.0912 is similar to the Radix Ophiopogonis with 0.0899.
Table 3.
Node centrality analysis of symptom network.
| No. | Symptoms | Degree | Closeness | Betweenness | Eigenvector | Current flow closeness | Load |
|---|---|---|---|---|---|---|---|
| 1 | cough | ||||||
| 2 | weakness | ||||||
| 3 | poor sleep | ||||||
| 4 | expectoration | ||||||
| 5 | loss of appetite | ||||||
| 6 | oppression in chest | ||||||
| 7 | fever | ||||||
| 8 | gasp | ||||||
| 9 | dry mouth | 0.0149 | 0.0150 | ||||
| 10 | dyspnea | 0.1718 | |||||
| 11 | diarrhea | 0.6457 | 0.0157 | 0.1740 | 0.0653 | 0.0156 | |
| 12 | headache | 0.4756 | 0.6406 | 0.0092 | 0.0650 | 0.0092 | |
| 13 | bitter taste | 0.4634 | 0.6357 | 0.0075 | 0.1717 | 0.0648 | 0.0075 |
| 14 | palpitation | 0.4512 | 0.6308 | 0.0046 | 0.1753 | 0.0645 | 0.0046 |
| 15 | loose stool | 0.4512 | 0.6308 | 0.0092 | 0.1653 | 0.0645 | 0.0093 |
| 16 | vomiting | 0.4390 | 0.6308 | 0.1510 | 0.0642 | ||
| 17 | chills | 0.4268 | 0.6212 | 0.0070 | 0.1615 | 0.0640 | 0.0070 |
| 18 | dizziness | 0.4024 | 0.6029 | 0.0059 | 0.1517 | 0.0628 | 0.0059 |
| 19 | nausea | 0.4024 | 0.6119 | 0.0030 | 0.1615 | 0.0634 | 0.0030 |
| 20 | abdominal pain | 0.4024 | 0.6119 | 0.0057 | 0.1569 | 0.0634 | 0.0056 |
Table 4.
Node centrality analysis of medicine network.
| No. | Medicines | Degree | Closeness | Betweenness | Eigenvector | Current flow closeness | Load |
|---|---|---|---|---|---|---|---|
| 1 | Radix Glycyrrhizae | ||||||
| 2 | Poria | ||||||
| 3 | Pericarpium Citri Reticulatae | ||||||
| 4 | Rhizoma Atractylodis Macrocephalae | ||||||
| 5 | Rhizoma Pinelliae Preparatum | ||||||
| 6 | Astragali Radix | ||||||
| 7 | Radix Scutellariae | ||||||
| 8 | Cortex Magnoliae Officinalis | ||||||
| 9 | Radix Codonopsis | ||||||
| 10 | Radix Ophiopogonis | 0.0899 | |||||
| 11 | Radix Platycodonis | 0.0109 | 0.1773 | 0.0109 | |||
| 12 | Pericarpium Trichosanthis | 0.0113 | 0.0908 | 0.0113 | |||
| 13 | Radix Pseudostellariae | 0.8794 | 0.8924 | 0.0108 | 0.0904 | 0.1770 | 0.0108 |
| 14 | Radix Bupleuri | 0.8635 | 0.8799 | 0.0109 | 0.0898 | 0.1763 | 0.0109 |
| 15 | Armeniacae Amarum Semen | 0.8540 | 0.8726 | 0.0100 | 0.0889 | 0.1759 | 0.0100 |
| 16 | Fructus Schisandrae Chinensis | 0.8476 | 0.8678 | 0.0103 | 0.0883 | 0.1756 | 0.0103 |
| 17 | Flos Farfarae | 0.8413 | 0.8630 | 0.0080 | 0.0892 | 0.1753 | 0.0080 |
| 18 | Agastache rugosa | 0.8381 | 0.8607 | 0.0087 | 0.0889 | 0.1752 | 0.0087 |
| 19 | Semen Trichosanthis | 0.8349 | 0.8583 | 0.0084 | 0.0885 | 0.1751 | 0.0084 |
| 20 | Bulbus Fritillariae Thunbergii | 0.8349 | 0.8583 | 0.0086 | 0.0889 | 0.1751 | 0.0086 |
We compared the identified top significant symptoms and medicines to latest clinical guidelines of COVID-19 “Diagnosis and treatment of corona virus disease-19 (7th trial edition)”.2 We show that 100% of the top symptoms (bolded in Table 3) and 80% of the top medicines (bolded in Table 4) are presented in the latest guidelines, respectively.
4.4. Symptom and medicine embedding training and clustering analysis
According to the matrices of symptom and medicine networks, we used the realization steps of node embeddings described in Section 3.3.3 to train the COVID-19 symptom and medicine embeddings (also called vectors), respectively. By transferring the one-hot vectors of symptoms and medicines into numerical vectors, we got symptom embeddings with dimensions and medicine embeddings with dimensions.
Community division [43] is introduced to discover the closely connections underlying the symptoms or medicines, which can effectively explore the special clinical syndromes of COVID-19. We used the SC algorithm to respectively divide symptom embeddings and embeddings into the their corresponding syndrome communities. The SC algorithm is well-known that the community number is confirmed as an input parameter. As the aforementioned latest clinical guidelines of COVID-19. The main syndromes of COVID-19 have been divided into seven categories, therefore, we set the input community number of SC algorithm as seven.
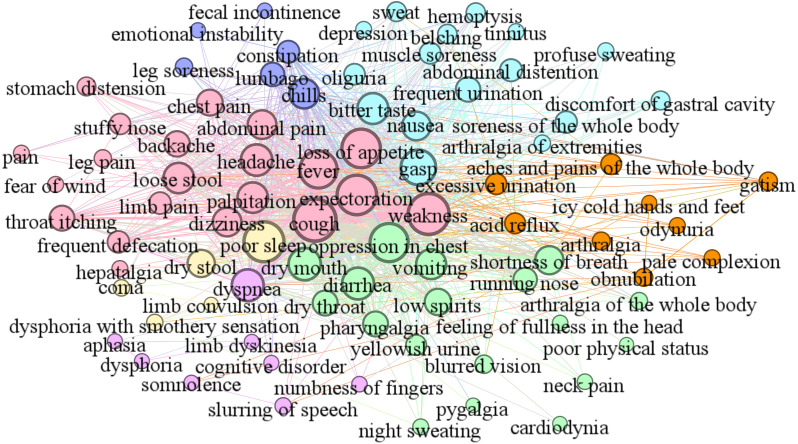
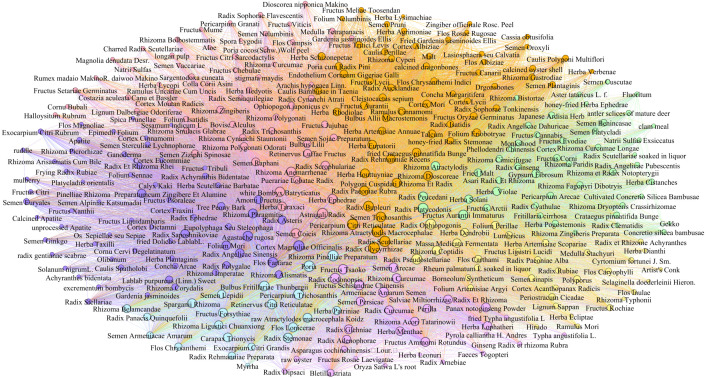
As shown in Fig. 6, Fig. 7, we divided the COVID-19 symptom network (Fig. 4) and medicine network (Fig. 5) into seven communities by the SC algorithm. The clinical syndromes (denoted as seven communities with different colors) and their corresponding symptoms and medicines are presented as follows:
Fig. 6.
Symptom communities. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
Medicine communities. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
-
•
Syndrome of cold-damp constraint in the lung (colored by rose pink) includes the core symptoms weakness, expectoration, cough, fever, and loss of appetite and other symptoms palpitation, headache, dizziness, abdominal pain, loose stool, etc. The effective treatment for this syndrome refers to the core medicine Radix Scrophulariae and other medicines Astragali Radix, Herba Taraxaci, Rhizoma Anemarrhenae, Herba Ephedrae, Rhizoma Zingiberis, etc.
-
•
Syndrome of deficiency of both qi and yin (colored by wisteria) includes the core symptom chills and other symptoms lumbago, constipation, leg soreness, emotional instability, fecal incontinence, etc. The effective treatment for this syndrome refers to the core medicine Radix Adenophorae and other medicines Fructus Schisandrae Chinensis, Salviae Miltiorrhizae Radix Et Rhizoma, Semen Arecae, Herba Lophatheri, Armeniacae Amarum Semen, etc.
-
•
Syndrome of epidemic toxin blocking the lung (colored by light blue) includes the core symptom gasp and other symptoms bitter taste, nausea, soreness of the whole body, frequent urination, abdominal distention, etc. The effective treatment for this syndrome refers to the core medicine Bulbus Fritillariae Thunbergii and other medicines Fructus Forsythiae, Pericarpium Trichosanthis, Poria, Exocarpium Citri Grandis, Semen Lepidii, etc.
-
•
Syndrome of lung–spleen qi deficiency (colored by orange) includes the core symptoms acid reflux and excessive urination and other symptoms aches and pains of the whole body, arthralgia, icy cold hands and feet, odynuria, gatism, etc. The effective treatment for this syndrome refers to the core medicine Rhizoma Dioscoreae and other medicines Pericarpium Citri Reticulatae, Polygoni Cuspidati Rhizoma Et Radix, Radix Paeoniae Rubra, Radix Glycyrrhizae, etc.
-
•
Syndrome of cold-damp obstructing the lung (colored by light green) includes the core symptom oppression in chest and other symptoms dry mouth, vomiting, diarrhea, shortness of breath, low spirits, etc. The effective treatment for this syndrome refers to the core medicine Rhizoma Atractylodis and other medicines Radix Platycodonis, Gypsum Fibrosum, Fried Malt, Radix Peucedani, Fructus Aurantii Immaturus, etc.
-
•
Syndrome of internal blockage and external desertion (colored by lilac) includes the core symptom dyspnea and other symptoms limb dyskinesia, cognitive disorder, slurring of speech, somnolence, dysphoria, etc. The effective treatment for this syndrome refers to the core medicine Tree Peony Bark and other medicines Radix Saposhnikoviae, Rhizoma Phragmitis, Flos Farfarae, Amomi Fructus, Agastache rugosa, etc.
-
•
Syndrome of blazing of both qi and ying (colored by yellow) includes the core symptom poor sleep and other symptoms dry stool, coma, limb convulsion, etc. The effective treatment for this syndrome refers to the core medicine Radix Pseudostellariae and other medicines Radix Ophiopogonis, Rhizoma Coptidis, Folium Perillae, Massa Medicata Fermentata, Periostracum Cicadae, etc.
4.5. Experimental result comparison analysis
In general, for the clinical diagnosis and treatment in real world, the numbers of symptom representations and medicines may be greater than the medical information in the latest clinical guidelines of COVID-19. Thus, we summarized the all specific syndromes and their corresponding primary symptoms and primary medicines as the evaluation standard to verify the accuracy and performance of the analysis model.
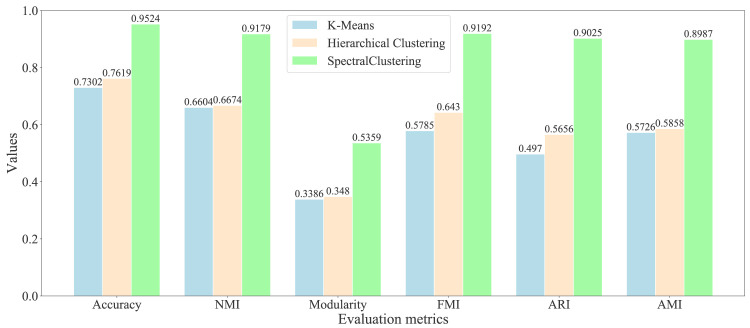
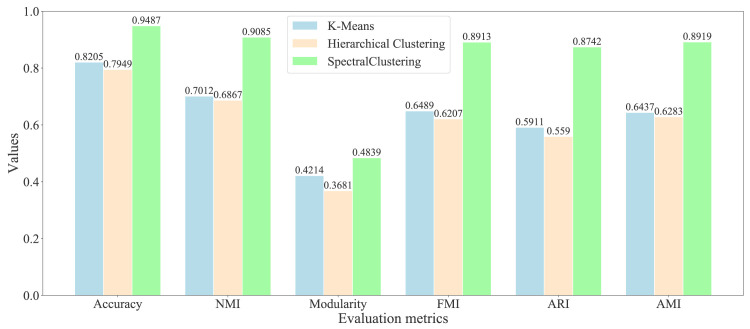
As the previous studies, we found that the non-adaptive clustering algorithms can acquire the better results than the adaptive algorithms on the graph embeddings [44]. Therefore, we selected the representative non-adaptive algorithms: K-means and hierarchical clustering algorithms as the comparison. We have used the representative evaluation metrics: accuracy, modularity [45], normalized mutual information (NMI) [46], Fowlkes–Mallows index (FMI) [47], adjusted rand index (ARI) [48], and adjusted mutual information (AMI) [49], to evaluate the quality of community division. We show the evaluation results of different symptom communities in Fig. 8, and various medicine communities in Fig. 9, respectively.
Fig. 8.
Evaluation of different clustering algorithms on the primary symptom network.
Fig. 9.
Evaluation of different clustering algorithms on the primary medicine network.
After comparative analysis, the experimental results show that the SC algorithm works better than the other two algorithms and gets more successful runs on the all metrics. From Fig. 8 of symptom division evaluation, we show that the SC algorithm can get the highest accuracy 0.9524 than other algorithms: K-means with 0.7302 and hierarchical clustering with 0.7619. For the classical metric modularity, the SC algorithm can acquire the higher value 0.5359 than K-means with 0.3386 and hierarchical clustering with 0.348. For the evaluation of community detection, it is widely believed that it is a good division when the modularity is greater than 0.3. For the other four metrics: NMI, FMI, ARI, and AMI, the SC algorithm can also obtain the highest values 0.9179, 0.9192, 0.9025, and 0.8987 than K-means with 0.6604, 0.5785, 0.497, and 0.5726, and hierarchical clustering with 0.6674, 0.643, 0.5656, and 0.5858.
Based on Fig. 9 of medicine division evaluation, the experimental results demonstrate that the SC algorithm obtain the highest accuracy 0.9487 than other algorithms: K-means with 0.8205 and hierarchical clustering with 0.7949. For the modularity, the SC algorithm can acquire the higher value 0.4839 than K-means with 0.4214 and hierarchical clustering with 0.3681. For the other four metrics: NMI, FMI, ARI, and AMI, the SC algorithm can also obtain the highest values 0.9085, 0.8913, 0.8742, and 0.8919 than K-means with 0.7012, 0.6489, 0.5911, and 0.6437, and hierarchical clustering with 0.6867, 0.6207, 0.559, and 0.6283.
5. Conclusion
In this study, we have explored an effective model to support the diagnosis and treatment of COVID-19. Through the combination of complex network and machine learning techniques, the proposed model is able to find the key factors and the clinical regularity in the process of diagnosis and treatment accurately and effectively. Firstly, we designed the symptom and medicine networks to represent the complex relationships underlying the symptoms and medicines, respectively. Secondly, we utilized six evaluation metrics to identify the core symptom and medicine nodes based on the network topology. Thirdly, we trained each symptom or medicine node using the skip-gram model to generate the numerical symptom or medicine embedding (or called a vector), which conserves the similarity relationship between any two symptoms or medicines. In order to find the symptom group or the medicine combination, we used the SC algorithm to divide the symptoms or medicines into seven communities (clusters) with the corresponding to the specific syndromes. Through checking up on the latest clinical guidelines of COVID-19 and compared to the representative clustering algorithms, we show that our model can accurately and effectively discover the symptom groups and medicine combinations, and their corresponding syndromes.
In the clinical practice of COVID-19, this model can filter the irrelevant information and acquire the key factors and important clinical regularity. Through introducing the medical information fusion, the model discovers the relative effects among the syndromes, symptoms, and medicines, which provides tremendously valuable guidance and helps physicians to give the effective diagnosis and treatment strategies for the COVID-19 patients. Furthermore, the dictionaries and embedding sets of syndromes, symptoms, and medicines will be supplied as the basic data sets for the COVID-19 researchers. The efficiency of the prediction of disease evolution for COVID-19 remains a challenging area. We will use the efficient solver such as iterative solvers to find the eigenvectors in the Spectral Clustering method in the future to improve the algorithm’s efficiency.
CRediT authorship contribution statement
Fang Hu: Methodology, Formal analysis, Writing - original draft, Writing - review & editing. Mingfang Huang: Methodology, Software, Visualization. Jing Sun: Data curation, Validation. Xiong Zhang: Conceptualization, Investigation. Jifen Liu: Resources, Conceptualization, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
General Office of National Health Commission of the People’s Republic of China, Office of National Administration of Traditional Chinese Medicine. Diagnosis and treatment of corona virus disease-19 (7th trial edition) [J]. China Medicine, 2020, 15(6): 801–805. DOI: 10. 3760/j. issn. 1673–4777. 2020. 06. 001.
Appendix.
References
- 1.Zhai P., Ding Y., Wu X., Long J., Zhong Y., Li Y. The epidemiology, diagnosis and treatment of COVID-19. Int. J. Antimicrob. Ag. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar R., Khan A.A., Zhang S., Wang W., Abuidris Y., Amin W., Kumar J. 2020. Blockchain-federated-learning and deep learning models for COVID-19 detection using CT imaging. arXiv preprint arXiv:2007.06537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu K.-H., Beam A.L., Kohane I.S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018;2(10):719–731. doi: 10.1038/s41551-018-0305-z. [DOI] [PubMed] [Google Scholar]
- 4.Davenport T., Kalakota R. The potential for artificial intelligence in healthcare. Future Healthc. J. 2019;6(2):94–98. doi: 10.7861/futurehosp.6-2-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu F., Li L., Huang X., Yan X., Huang P. Symptom distribution regularity of insomnia: Network and spectral clustering analysis. JMIR Med. Inform. 2020;8(4) doi: 10.2196/16749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adhikari S.P., Meng S., Wu Y.-J., Mao Y.-P., Ye R.-X., Wang Q.-Z., Sun C., Sylvia S., Rozelle S., Raat H., et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect. Dis. Poverty. 2020;9(1):1–12. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage covid-19 in routine clinical practice using ct images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen D., Gao K., Chen J., Wang R., Wei G. Potentially highly potent drugs for 2019-ncov. BioRxiv. 2020:1–13. [Google Scholar]
- 9.Magar R., Yadav P., Farimani A.B. 2020. Potential neutralizing antibodies discovered for novel corona virus using machine learning. arXiv preprint arXiv:2003.08447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullock J., Pham K.H., Lam C.S.N., Luengo-Oroz M., et al. 2020. Mapping the landscape of artificial intelligence applications against covid-19. arXiv preprint arXiv:2003.11336. [Google Scholar]
- 11.Menni C., Valdes A.M., Freidin M.B., Sudre C.H., Nguyen L.H., Drew D.A., Ganesh S., Varsavsky T., Cardoso M.J., Moustafa J.S.E.-S., et al. Real-time tracking of self-reported symptoms to predict potential covid-19. Nat. Med. 2020;26:1037–1040. doi: 10.1038/s41591-020-0916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautier J.-F., Ravussin Y. A new symptom of covid-19: Loss of taste and smell. Obesity. 2020;28(5):848. doi: 10.1002/oby.22809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song Y., Liu P., Shi X., Chu Y., Zhang J., Xia J., Gao X., Qu T., Wang M. Sars-cov-2 induced diarrhoea as onset symptom in patient with covid-19. Gut. 2020;69(6):1143–1144. doi: 10.1136/gutjnl-2020-320891. [DOI] [PubMed] [Google Scholar]
- 14.Kanne J.P., Little B.P., Chung J.H., Elicker B.M., Ketai L.H. Essentials for radiologists on covid-19: an update—radiology scientific expert panel. Radiology. 2020;296(2):E113–E114. doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamshidi M., Lalbakhsh A., Talla J., Peroutka Z., Hadjilooei F., Lalbakhsh P., Jamshidi M., La Spada L., Mirmozafari M., Dehghani M., et al. Artificial intelligence and covid-19: Deep learning approaches for diagnosis and treatment. IEEE Access. 2020;8:109581–109595. doi: 10.1109/ACCESS.2020.3001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan K.W., Wong V.T., Tang S.C.W. Covid-19: An update on the epidemiological, clinical, preventive and therapeutic evidence and guidelines of integrative chinese–western medicine for the management of 2019 novel coronavirus disease. Amer. J. Chin. Med. 2020;48(03):737–762. doi: 10.1142/S0192415X20500378. [DOI] [PubMed] [Google Scholar]
- 17.Mikolov T., Chen K., Corrado G., Dean J. 2013. Efficient estimation of word representations in vector space. arXiv preprint arXiv:1301.3781. [Google Scholar]
- 18.Grover A., Leskovec J. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2016. Node2vec: Scalable feature learning for networks; pp. 855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiedler M. Algebraic connectivity of graphs. Czechoslovak Math. J. 1973;23(2):298–305. [Google Scholar]
- 20.Von Luxburg U. A tutorial on spectral clustering. Stat. Comput. 2007;17(4):395–416. [Google Scholar]
- 21.Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (covid-19) Drug Discov. Ther. 2020;14(1):58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 22.Rome B.N., Avorn J. Drug evaluation during the Covid-19 pandemic. New Engl. J. Med. 2020;382:2282–2284. doi: 10.1056/NEJMp2009457. [DOI] [PubMed] [Google Scholar]
- 23.Ren J.-l., Zhang A.-H., Wang X.-J. Traditional chinese medicine for COVID-19 treatment. Pharmacol. Res. 2020;155 doi: 10.1016/j.phrs.2020.104743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo L., Jiang J., Wang C., Fitzgerald M., Hu W., Zhou Y., Zhang H., Chen S. Analysis on herbal medicines utilized for treatment of COVID-19. Acta Pharm. Sin. B. 2020;10(7):1192–1204. doi: 10.1016/j.apsb.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alom M.Z., Rahman M., Nasrin M.S., Taha T.M., Asari V.K. 2020. Covid_mtnet: covid-19 detection with multi-task deep learning approaches. arXiv reprint arXiv:2004.03747. [Google Scholar]
- 26.Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Zhang H., Ji W., Bernheim A., Siegel E. 2020. Rapid ai development cycle for the coronavirus (covid-19) pandemic: Initial results for automated detection & patient monitoring using deep learning ct image analysis. arXiv preprint arXiv:2003.05037. [Google Scholar]
- 27.Bung N., Krishnan S.R., Bulusu G., Roy A. De novo design of new chemical entities (nces) for sars-cov-2 using artificial intelligence. ChemRxiv. 2020 doi: 10.4155/fmc-2020-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muzammal M., Talat R., Sodhro A.H., Pirbhulal S. A multi-sensor data fusion enabled ensemble approach for medical data from body sensor networks. Inf. Fusion. 2020;53:155–164. [Google Scholar]
- 29.Zhang Y., Gravina R., Lu H., Villari M., Fortino G. Pea: Parallel electrocardiogram-based authentication for smart healthcare systems. J. Netw. Comput. Appl. 2018;117:10–16. [Google Scholar]
- 30.Ang L., Lee H.W., Kim A., Lee M.S. Herbal medicine for the management of covid-19 during the medical observation period: A review of guidelines. Integr. Med. Res. 2020;9(3) doi: 10.1016/j.imr.2020.100465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamilton W.L., Ying R., Leskovec J. 2017. Representation learning on graphs: Methods and applications. arXiv preprint arXiv:1709.05584. [Google Scholar]
- 32.C. Meng, R. Cheng, S. Maniu, P. Senellart, W. Zhang, Discovering meta-paths in large heterogeneous information networks, in: Proceedings of the 24th International Conference on World Wide Web, 2015, pp. 754–764.
- 33.Zhang Y., Wang R., Hossain M.S., Alhamid M.F., Guizani M. Heterogeneous information network-based content caching in the internet of vehicles. IEEE Trans. Veh. Technol. 2019;68(10):10216–10226. [Google Scholar]
- 34.Shi C., Hu B., Zhao W.X., Philip S.Y. Heterogeneous information network embedding for recommendation. IEEE Trans. Knowl. Data Eng. 2018;31(2):357–370. [Google Scholar]
- 35.Newman M.E. The structure and function of complex networks. SIAM Rev. 2003;45(2):167–256. [Google Scholar]
- 36.Hu F., Zhu Y., Liu J., Jia Y. Computing communities in complex networks using the dirichlet processing gaussian mixture model with spectral clustering. Phys. Lett. A. 2019;383(9):813–824. [Google Scholar]
- 37.Hu F., Liu Y. Multi-index algorithm of identifying important nodes in complex networks based on linear discriminant analysis. Modern Phys. Lett. B. 2015;29(03) [Google Scholar]
- 38.Freeman L.C. Centrality in social networks conceptual clarification. Social Networks. 1978;1(3):215–239. [Google Scholar]
- 39.Bonacich P. Some unique properties of eigenvector centrality. Social Networks. 2007;29(4):555–564. [Google Scholar]
- 40.Brandes U., Fleischer D. Annual Symposium on Theoretical Aspects of Computer Science. Springer; 2005. Centrality measures based on current flow; pp. 533–544. [Google Scholar]
- 41.Dolev S., Elovici Y., Puzis R. Routing betweenness centrality. J. ACM. 2010;57(4):1–27. [Google Scholar]
- 42.Hu F., Liu J., Li L., Liang J. Community detection in complex networks using node2vec with spectral clustering. Physica A. 2019;545 [Google Scholar]
- 43.Newman M.E. Finding community structure in networks using the eigenvectors of matrices. Phys. Rev. E. 2006;74(3) doi: 10.1103/PhysRevE.74.036104. [DOI] [PubMed] [Google Scholar]
- 44.Zhang N., Deng S., Sun Z., Wang G., Chen X., Zhang W., Chen H. 2019. Long-tail relation extraction via knowledge graph embeddings and graph convolution networks. arXiv preprint arXiv:1903.01306. [Google Scholar]
- 45.Newman M.E. Modularity and community structure in networks. Proc. Natl. Acad. Sci. 2006;103(23):8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danon L., Diaz-Guilera A., Duch J., Arenas A. Comparing community structure identification. J. Stat. Mech. Theory Exp. 2005;2005(09):P09008. [Google Scholar]
- 47.Fowlkes E.B., Mallows C.L. A method for comparing two hierarchical clusterings. J. Amer. Statist. Assoc. 1983;78(383):553–569. [Google Scholar]
- 48.Feizollah A., Anuar N.B., Salleh R., Amalina F. International Symposium on Biometrics and Security Technologies. IEEE; 2014. Comparative study of k-means and mini batch k-means clustering algorithms in android malware detection using network traffic analysis; pp. 193–197. [Google Scholar]
- 49.Romano S., Vinh N.X., Bailey J., Verspoor K. Adjusting for chance clustering comparison measures. J. Mach. Learn. Res. 2016;17(1):4635–4666. [Google Scholar]