Abstract
Background
Metabolism is critical for sustaining life, immunity and infection, but its role in COVID-19 is not fully understood.
Methods
Seventy-nine COVID-19 patients, 78 healthy controls (HCs) and 30 COVID-19-like patients were recruited in a prospective cohort study. Samples were collected from COVID-19 patients with mild or severe symptoms on admission, patients who progressed from mild to severe symptoms, and patients who were followed from hospital admission to discharge. The metabolome was assayed using gas chromatography–mass spectrometry.
Results
Serum butyric acid, 2-hydroxybutyric acid, l-glutamic acid, l-phenylalanine, l-serine, l-lactic acid, and cholesterol were enriched in COVID-19 and COVID-19-like patients versus HCs. Notably, d-fructose and succinic acid were enriched, and citric acid and 2-palmitoyl-glycerol were depleted in COVID-19 patients compared to COVID-19-like patients and HCs, and these four metabolites were not differentially distributed in non-COVID-19 groups. COVID-19 patients had enriched 4-deoxythreonic acid and depleted 1,5-anhydroglucitol compared to HCs and enriched oxalic acid and depleted phosphoric acid compared to COVID-19-like patients. A combination of d-fructose, citric acid and 2-palmitoyl-glycerol distinguished COVID-19 patients from HCs and COVID-19-like patients, with an area under the curve (AUC) > 0.92 after validation. The combination of 2-hydroxy-3-methylbutyric acid, 3-hydroxybutyric acid, cholesterol, succinic acid, L-ornithine, oleic acid and palmitelaidic acid predicted patients who progressed from mild to severe COVID-19, with an AUC of 0.969. After discharge, nearly one-third of metabolites were recovered in COVID-19 patients.
Conclusions
The serum metabolome of COVID-19 patients is distinctive and has important value in investigating pathogenesis, determining a diagnosis, predicting severe cases, and improving treatment.
Abbreviations: COVID-19, coronavirus disease 2019; HCs, healthy controls; AUC, area under the curve; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COPD, chronic obstructive pulmonary disease; WHO, World Health Organization; GC–MS, gas chromatography–mass spectrometry; LC-MS, liquid chromatography-mass spectrometry; OPLS-DA, orthogonal partial least squares-discriminant analysis; ALT, alanine transaminase; LDH, lactate dehydrogenase; CK-MB, creatine kinase-MB; CRP, c-reactive protein; ALB, albumin; LPV/RTV, lopinavir/ritonavir; CHOL, total cholesterol; HBDH, hydroxybutyrate dehydrogenase; GGT, gamma-glutamyl transpeptidase; CRP, C-reactive protein; PCT, procalcitonin; DB, direct bilirubin; PT, prothrombin time; INR, international normalized ratio; TCA cycle, tricarboxylic acid cycle; GMP, guanosine monophosphate; GM3s, monosialodihexosyl gangliosides; PPARα, peroxisome proliferator-activated receptor alpha
Keywords: COVID-19, SARS-CoV-2, Serum metabolome, Predictive value
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease 2019 (COVID-19) [1], has infected more than 100 million people and resulted in nearly 2.5 million deaths worldwide. The basic reproduction number (R0) of SARS-CoV-2 is nearly 2.57, which makes its prevention and control a great challenge [2]. Tests to identify infected patients are key to controlling the alarming spread of SARS-CoV-2. The diagnosis of COVID-19 primarily relies on molecular methods and serological tests [3,4]. Reliable invasive and sensitive approaches for disease diagnosis and progression prediction are urgently needed.
Metabolism is the basic characteristic and main activity of life. Metabolomics is widely used in the mechanistic study and diagnosis of various diseases, such as cancer, diabetes, cardiovascular or pulmonary disorders [5]. For chronic obstructive pulmonary disease (COPD), the levels of several serum metabolites, such as cysteinesulfonic acid, fumarate, and myoinositol, are different from healthy smokers and are used as promising signs of COPD at an early stage [6]. Certain plasma metabolites, such as ornithine, caprylic acid, azetidine, and iminodiacetic acid, are potential biomarkers for predicting and determining the severity of acute respiratory distress syndrome [7]. One small study noted that serum metabolomic and lipidomic alterations were associated with the clinical severity of COVID-19 and may be used as blood biomarkers [8]. Some metabolites, such as camostat mesylate, which is a serine protease inhibitor that blocks TMPRSS2 activity, may be considered as off-label treatments for SARS-CoV-2-infected patients [9].
The present study examined alterations in the serum metabolome of patients with COVID-19 and the associations of these alterations with COVID-19 development to identify potential biomarkers and models for distinguishing COVID-19 patients from healthy controls (HCs) and suspected patients.
2. Methods
2.1. Diagnosis
Respiratory specimens (nasopharyngeal swabs, sputum, or endotracheal aspirates) were collected daily to test for SARS-CoV-2 RNA using real-time reverse transcription polymerase chain reaction (RT-PCR) [10]. COVID-19 was diagnosed based on the WHO interim guidance [11]. The severity of COVID-19 was evaluated according to the seventh edition of the Guidelines for Diagnosis and Treatment of SARS-CoV-2 issued by the National Health Commission of the People's Republic of China [12]. Severe COVID-19 referred to cases that fulfilled any of the following three criteria: 1) dyspnea, respiratory rate ≥ 30/min; 2) blood oxygen saturation ≤ 93% and ratio of partial pressure of arterial oxygen to fraction of inspired oxygen < 300; and 3) lung infiltrates > 50% within 24 to 48 h. A mild case was defined as a confirmed case with mild symptoms that not fulfilling any of the above criteria.
Patients who had symptoms consistent with COVID-19 but were not infected with SARS-CoV-2 were defined as COVID-19-like patients according the abovementioned guidelines and who satisfied the following criteria: 1) fever or respiratory symptoms; 2) imaging manifestations of pneumonia; and 3) optional reduction in white blood cell or lymphocyte count at an early stage. Patients with exposure to COVID-19 individuals only needed to satisfy two of the three criteria for inclusion.
2.2. Participants
This study was approved by the National Health Commission of China and the Ethics Commission of the First Hospital of Zhejiang Province (IIT2020040A). Written informed consent was obtained from all participating patients. Seventy-nine patients with COVID-19 and 30 COVID-19-like patients were recruited from the First Affiliated Hospital of Zhejiang University from January 19 to March 19, 2020. Seventy-eight HCs (60 age- and gender-matched healthy, and 18 not matched with COVID-19 patients) were also recruited. The clinical data included demographic characteristics, medical comorbidities, progression, and treatment of clinical illness.
2.3. Blood sampling and testing
Blood samples were collected from COVID-19 patients, including patients with mild or severe symptoms on admission, patients who progressed from mild to severe in the hospital, and patients who were followed from hospital admission to discharge, HCs and COVID-19-like patients. Hematological biomarkers, such as full blood cell counts, inflammatory indicators, coagulation function, myocardial enzymes, renal function and liver function, were determined. IL-2, IL-4, IL-6, IL-10, and tumor necrosis factor-α (TNF-α) were detected using multiple microsphere flow immunofluorescence according to the manufacturer's instructions (BD Biosciences).
2.4. Design of metabolome studies
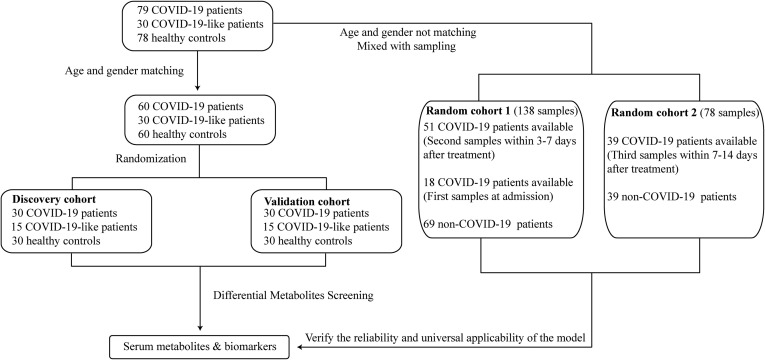
Age- and gender-matched cohorts were included to identify differences and predict biomarkers (Fig. 1 ). Among the 79 COVID-19 patients, 30 COVID-19-like patients and 78 HCs recruited in this study, 60 COVID-19 patients matched 60 HCs and 30 COVID-19-like patients in age and gender. These matched participants were randomly and evenly divided into two cohorts, the discovery and validation cohorts, of 30 COVID-19 patients, 30 HCs and 15 COVID-19-like patients. Serum samples of the COVID-19 patients used in this analysis were collected upon admission.
Fig. 1.
Study design and flow diagram. A total of 79 COVID-19 patients, 30 COVID-19-like patients and 78 HCs were included and randomly divided into the discovery or validation cohort with age- and gender-matched patients. Each cohort consisted of 30 COVID-19 patients, 30 HCs and 15 COVID-19-like patients. All serum samples were collected from patients upon admission in each cohort to test the serum metabolites for potential biomarkers. Two random cohorts containing 216 samples from COVID-19 patients and non-COVID-19 patients were created to confirm the reliability and the universal applicability of the prediction results. Random cohort 1 included the second samples from the 51 available COVID-19 patients after 3–7 days of treatment together with the first samples from 18 COVID-19 patients at admission. Random cohort 2 included the third samples of 39 available COVID-19 patients after 7–14 days of treatment. Each random cohort was matched with the same number of COVID-19-like patients and HCs who were randomly selected from all non-COVID-19 participants.
Two random cohorts were created to confirm the reliability and the universal applicability of the prediction result. These two cohorts enrolled serum samples from COVID-19 patients who did not match the 60 HCs and 30 COVID-19-like patients in age or gender and who were at different time points of the SARS-CoV-2 infection. Random cohort 1 included the second samples from 51 individuals available in the 60 age- and gender-matched COVID-19 patients mentioned above after 3–7 days of treatment together with the first samples from 18 COVID-19 patients at admission, whose samples were not used previously because they could not be age- and gender-matched to our available HCs, together with 20 COVID-19-like patients and 49 HCs who were randomly selected from all non-COVID-19 participants using the R package “MatchIt”. Random cohort 2 included the third samples of 39 available COVID-19 patients after 7–14 days of treatment together with 11 COVID-19-like patients and 28 HCs, who were randomly selected using the method mentioned above.
We obtained blood samples from 15 COVID-19 patients who provided samples at three complete time points from admission, under treatment and at discharge. Forty-five samples were used to analyze the metabolome alterations from admission to discharge compared to 78 HC samples.
2.5. Assay of the serum metabolome
The metabolome was determined as previously described with small modifications [13]. Briefly, 450 μL of methanol was added to and mixed thoroughly with 50 μL of serum for extraction. After centrifugation, the supernatant was dried, and the dried products were further methoxylated and trimethylsilylated. The concentrations of metabolites were normalized against the internal control heptadecanoic acid (Sigma-Aldrich, USA). The metabolites were analyzed using gas chromatography–mass spectrometry (GC–MS) (Agilent 7890A) coupled to an inert mass selective detector system (Agilent 5975C, Agilent Technologies, Santa Clara, CA, USA). Qualitative Analysis software (version B.07.00, Agilent, Santa Clara, CA, United States) was used for data analyses. Metabolites were identified using the NIST 17 databases. Orthogonal partial least squares-discriminant analysis (OPLS-DA) was performed to visualize differences in the metabolome profiles between groups. Differential metabolites were selected according to the statistically significant variable importance in the projection (VIP) values obtained from the OPLS-DA model and the P values from the Mann-Whitney U test on the normalized peak areas. Metabolites with VIP values > 1 and P values < 0.05 were included.
2.6. Statistical analyses
For most variables, descriptive statistics, such as the median with interquartile range (IQR; for data with skewed distribution) and the proportion (%), were calculated. The Mann-Whitney U test was used to compare any two data sets that were not normally distributed, and the Kruskal-Wallis test was used for comparisons of more than two groups. One-way ANOVA followed by the Student-Newman-Keuls method were used for all other comparisons. The patients and controls were matched by age and gender in the discovery and validation cohorts using the R package “MatchIt”. The χ2 test and Fisher's exact test were used for categorical variables. The potential predictive ability of factors for COVID-19 was tested using receiver operating characteristic (ROC) curves and random forest analysis. Spearman's rank correlation test was used to analyze the correlations between two variables. Statistical analyses were performed using SPSS software, version 20.0 and R statistical language (version R 3.0.2). For all analyses, probabilities were two-tailed, and a two-tailed P value < 0.05 was considered significant. A test power calculation of this study was 0.99 using NCSS-PASS version 11.0.7. The raw data and details of the 75 detected metabolites in all cohorts are provided in the Supplemental Material.
3. Results
3.1. Clinical characteristics of the participants
Seventy-nine COVID-19 patients, 30 COVID-19-like patients who were primarily diagnosed with influenza (Supplementary Table 1), and 78 HCs were recruited. Table 1 summarizes the demographic and clinical characteristics of the COVID-19 patients. Hypertension (n = 19, 24.1%) and diabetes mellitus (n = 11, 13.9%) were the most common comorbidities in the COVID-19 patients. Forty-eight patients in the COVID-19 group had no comorbidities, and 21 patients were in the COVID-19-like group. At admission, the levels of leukocytes, neutrophils, prothrombin time, alanine transaminase (ALT), lactate dehydrogenase (LDH), creatine kinase-MB (CK-MB), and C-reactive protein (CRP) were significantly higher in COVID-19 patients than in HCs, and lymphocyte counts and albumin levels were significantly lower. Notably, none of these parameters were significantly different between COVID-19-like patients and COVID-19 patients. Nearly 84.8% of COVID-19 patients received antiviral treatments, including 62% who received a combination of two antiviral treatments, arbidol (200 mg 3 times daily) and lopinavir/ritonavir (LPV/RTV, 400 mg twice daily and 100 mg twice daily), and 22.8% who received single antiviral treatment with arbidol or LPV/RTV (Table 1). More than half (56.9%) of COVID-19 patients received glucocorticoid treatment, and 10.1% received antibiotics.
Table 1.
Clinical characteristics of COVID-19 patients, COVID-19-like patients and healthy controls.
| Characteristic | COVID-19 patients (N = 79) |
COVID-19-like patients (N = 30) |
Healthy controls (N = 78) |
p1 (COVID-19 vs. COVID-19-like) |
p2 (COVID-19 vs. HCs) |
|---|---|---|---|---|---|
| Demographics, n (%) | |||||
| Age, median (IQR) | 51 (38, 59) | 50.5 (37.5, 68.8) | 52 (44.3, 59) | 0.24 | 0.28 |
| Male sex | 47 (59.5%) | 11 (36.7%) | 38 (55.9%) | 0.03 | 0.18 |
| Comorbidity, n (%) | |||||
| Hypertension | 19 (24.1%) | 6 (20%) | 0 | 0.65 | <0.001 |
| Diabetes | 11 (13.9%) | 2 (6.7%) | 0 | 0.29 | <0.001 |
| Fatty liver disease | 10 (12.7%) | 1 (3.3%) | 0 | 0.28 | <0.001 |
| Chronic lung diseasea | 4 (5.1%) | 2 (6.7) | 0 | 0.67 | <0.001 |
| Cardiac diseaseb | 5 (6.3%) | 1 (3.3%) | 0 | 1.0 | <0.001 |
| No comorbidity, n (%) | 48(60.7%) | 21(70%) | 78 | 0.38 | <0.001 |
| Disease severity | |||||
| Mild | 32 (40.5%) | NA | NA | NA | NA |
| Severe | 47 (59.5%) | NA | NA | NA | NA |
| Initial laboratory findings, median (IQR) | |||||
| Leukocyte count, 109/L | 6.4 (4.1, 10.1) | 7.1 (5, 10.2) | 6 (5, 6.8) | 0.14 | 0.005 |
| Neutrophil count, 109/L | 4.8 (2.8, 8.8) | 4.7 (3.1, 8.1) | 3.3 (2.6, 4.2) | 0.99 | <0.001 |
| Lymphocyte count, 109/L | 0.9 (0.5, 1.4) | 1.3 (0.9, 1.9) | 1.9 (1.5, 2.2) | 0.11 | <0.001 |
| Prothrombin time, s | 11.6 (11.3, 12.1) | 12.1 (11.6, 13.6) | 11.1(10.7,11.3) | 0.05 | 0.01 |
| Albumin, g/L | 39.1 (34.5, 43.7) | 40.2 (35.8, 43.7) | 45 (39.8, 48.2) | 0.81 | <0.001 |
| ALT, U/L | 22 (15, 40) | 17.5 (12.8, 32.3) | 17 (12, 24) | 0.27 | 0.002 |
| LDH, U/L | 229 (193, 323) | 192 (161, 228) | 185 (158, 204) | 0.67 | <0.001 |
| CK-MB, U/L | 20 (15, 23) | 16 (14, 22) | 14 (9, 17) | 0.21 | 0.007 |
| CRP, mg/L | 11.4 (3.6, 35.3) | 20.9 (1.3, 96.1) | 0.6 (0, 1.1) | 0.073 | <0.001 |
| IL-2, pg/mL | 0.95 (0.76, 1.65) | NA | NA | NA | NA |
| IL-4, pg/mL | 1.77 (1.4, 1.77) | NA | NA | NA | NA |
| IL-6, pg/mL | 15.21(5.67,34.26) | NA | NA | NA | NA |
| IL-10, pg/mL | 3.64 (2.19, 7.16) | NA | NA | NA | NA |
| TNF-α, pg/mL | 17.65(6.55,64.08) | NA | NA | NA | NA |
| IFN-γ, pg/mL | 8.95 (4.07, 24.8) | NA | NA | NA | NA |
| Treatment,- No. (%) | |||||
| Glucocorticoids | 45 (56.9%) | 0 | 0 | <0.001 | <0.001 |
| Antibiotics | 8 (10.1%) | 6 (20%) | 0 | 0.20 | <0.001 |
| Antivirals | 67 (84.8%) | 6 (20%) | 0 | <0.001 | <0.001 |
| Arbidol | 9 (11.4%) | NA | NA | NA | NA |
| LPV/RTV | 9 (11.4%) | NA | NA | NA | NA |
| Arbidol and LPV/RTV | 49 (62%) | NA | NA | NA | NA |
Abbreviations: HC, healthy control; IQR, interquartile range; ALT, alanine transaminase; LDH, lactate dehydrogenase; CK-MB, creatine kinase-MB; CRP, C-reactive protein; LPV/RTV, lopinavir and ritonavir; NA, not applicable.
Chronic lung disease includes chronic obstructive pulmonary disease and interstitial lung disease;
Cardiac disease includes congestive heart disease and coronary atherosclerotic heart disease.
3.2. The serum metabolome of COVID-19 patients is distinct from COVID-19-like patients and HCs
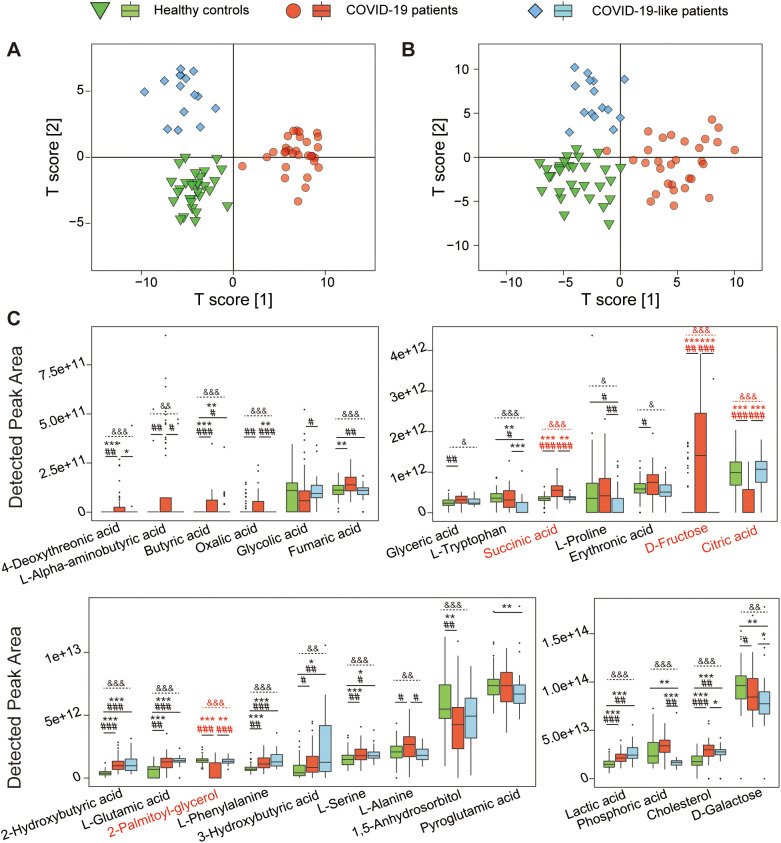
GC–MS identified 75 metabolites from all serum samples used in this study. OPLS-DA plots showed that COVID-19 patients, COVID-19-like patients and HCs were clustered separately in the discovery and validation cohorts (Fig. 2A and B), which indicated that the composition of their metabolomes was different. We created an OPLS-DA plot using the data of COVID-19 patients without comorbidities, COVID-19-like patients and HCs and found that this graphic (Supplementary Fig. 1A) was highly similar to all COVID-19 patients (Supplementary Fig. 1B), which suggests that the impact of comorbidities on the serum metabolome profile of patients with COVID-19 patients was far lower than SARS-CoV-2 infection. We also compared the serum metabolome profiles of COVID-19 patients with and without comorbidities. The results showed that none of the groups of patients with hypertension, diabetes or fatty liver disease could be distinguished from patients without comorbidities in the OPLS-DA plots (Supplementary Fig. 1C–E). This result further suggested that the effect of SARS-CoV-2 infection on the serum metabolome profile was much greater than the comorbidities.
Fig. 2.
The serum metabolome of COVID-19 patients is distinct from COVID-19-like patients and HCs. (A) OPLS-DA shows that the metabolome profiles of the COVID-19 patients (n = 30), COVID-19-like patients (n = 15) and HCs (n = 30) are clearly separated from each other in the discovery cohort. (B) OPLS-DA illustrates that the metabolome profiles of the COVID-19 patients (n = 30), COVID-19-like patients (n = 15) and HCs (n = 30) are clearly separated from each other in the validation cohort. (C) Twenty-six metabolites were significantly altered between at least two groups of COVID-19 patients (n = 60), COVID-19-like patients (n = 30) and HCs (n = 60) in the discovery and validation cohorts. Metabolites are marked in red when there is a significant difference in metabolites in the COVID-19 group compared to either of the two other groups in the discovery and validation cohorts. * and # indicate significance when comparing between two groups and “&” indicates significance in comparisons between three groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001 in the discovery cohort; #, P < 0.05; ##, P < 0.01; ###, P < 0.001 in the validation cohort. &, P < 0.05; &&, P < 0.01; &&&, P < 0.001 between the three groups.
We compared differences in the levels of each metabolite between the paired COVID-19 patients, COVID-19-like patients and HCs in the discovery cohort then validated these differences in the validation cohort. Several metabolites were changed significantly in the discovery and validation cohorts (Fig. 2C). Compared to the HCs, the serum levels of butyric acid, 2-hydroxybutyric acid, l-glutamic acid, l-phenylalanine, l-serine, l-lactic acid, and cholesterol were higher in COVID-19 and COVID-19-like patients, which suggests that respiratory tract infections widely affect these metabolites. Notably, serum d-fructose and succinic acid were enriched, and citric acid and 2-palmitoyl-glycerol were depleted in COVID-19 patients compared to COVID-19-like patients and HCs. The levels of these four serum metabolites were not significantly different between COVID-19-like patients and HCs, which suggests that these metabolites have important value in the pathogenesis and diagnosis of COVID-19. There was no significant difference in serum l-tryptophan levels between COVID-19 patients and HCs, but the level of this metabolite was higher in both of these groups than the COVID-19-like patients, which suggests that l-tryptophan metabolism is not as involved in COVID-19 patients as it is in other common respiratory infections [14]. Compared to HCs, COVID-19 patients had enriched 4-deoxythreonic acid and depleted 1,5-anhydroglucitol. Serum oxalic acid was also enriched in COVID-19 patients versus COVID-19-like patients, and phosphoric acid was depleted, which suggests different pathways in these two patient groups.
3.3. Serum metabolites distinguish COVID-19 patients from HCs and COVID-19-like patients
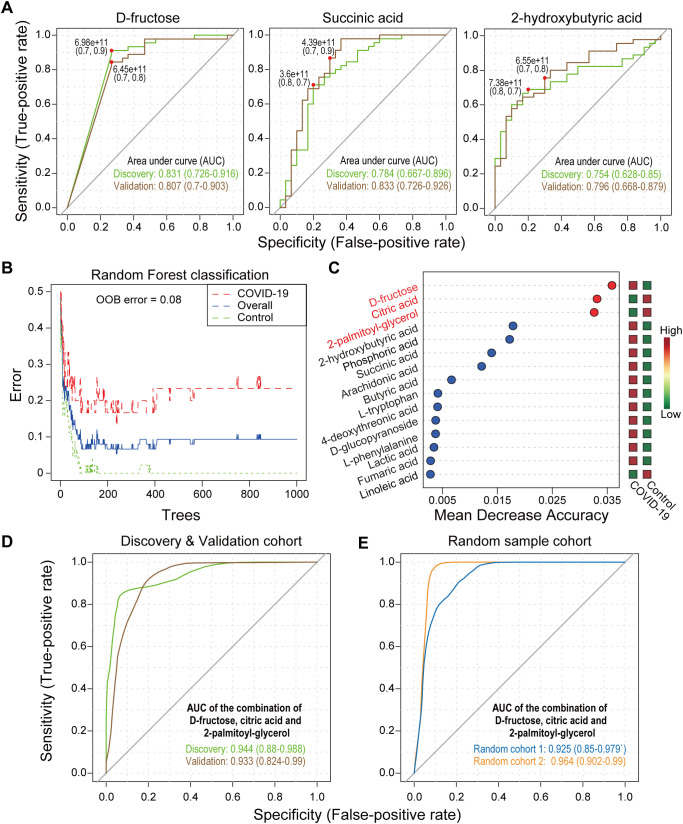
The area under the ROC curve (AUC) of d-fructose, succinic acid and 2-hydroxybutyric acid ranged from 0.7 to 0.85 in distinguishing COVID-19 patients from a group containing HCs and COVID-19-like patients in the discovery and verification cohorts (Fig. 3A), which indicates good predictive effects. In the random forest analysis of the discovery cohort, we found that three compounds, d-fructose, citric acid and 2-palmitoyl-glycerol, greatly contributed to distinguishing COVID-19 patients from the group containing HCs and COVID-19-like patients (Fig. 3B and C). Based on these three metabolites, we used binary logistic regression to create a ROC joint curve, and the AUCs were 0.944 and 0.933 in the discovery and validation cohorts, respectively, which indicates excellent distinguishing effects (Fig. 3D).
Fig. 3.
Serum metabolites distinguish COVID-19 patients from COVID-19-like patients and HCs. (A) ROC curves of d-fructose, succinic acid and 2-hydroxybutyric acid for distinguishing COVID-19 patients (n = 30) from COVID-19-like patients (n = 15) and HCs (n = 30). (B and C) Random forest analysis for biomarker screening and the top 15 metabolites ranked by the mean decrease in accuracy. (D) ROC curves of the combination of d-fructose, citric acid and 2-palmitoyl-glycerol for distinguishing COVID-19 patients (n = 30) from COVID-19-like patients (n = 15) and HCs (n = 30) in the discovery and validation cohorts. (E) ROC curves of the combination of d-fructose, citric acid and 2-palmitoyl-glycerol in the first and second random cohorts from COVID-19 patients (n = 69 in random 1; n = 39 in random 2) and non-COVID-19 patients (n = 69 in random 1; n = 39 in random 2).
We used two random cohorts to verify the predictive ability of the above biomarkers. The AUCs of d-fructose (0.775), succinic acid (0.807) and 2-hydroxybutyric acid (0.770) still indicated a good ability to distinguish COVID-19 patients from HCs and COVID-19-like patients in random cohort 1 (Supplementary Fig. 2A). Notably, the AUC of the combination of these factors was 0.925 (Fig. 3E). We performed ROC analysis in random cohort 2. Good predictions were also observed from the AUCs of d-fructose (0.894), succinic acid (0.886), and 2-hydroxybutyric acid (0.710) (Supplementary Fig. 2A) and from their joint model (0.964) (Fig. 3E). We rebuilt a cohort that included 48 COVID-19 patients without comorbidities, 30 COVID-19-like patients and 78 HCs to evaluate the predicted efficacy of these selected biomarkers. The AUCs of d-fructose (0.773), succinic acid (0.749), 2-hydroxybutyric acid (0.872) and the combination of the three compounds (0.934) also showed good prediction ability (Supplementary Fig. 2B), which indicates that comorbidities had little impact on the predictive ability of serum biomarkers.
3.4. Serum metabolites predict progression from mild to severe symptoms in COVID-19 patients
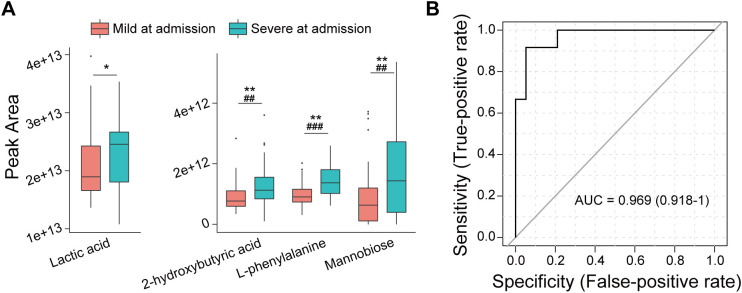
Among the 79 COVID-19 patients, 32 patients had mild disease, and 47 had severe disease at admission. Compared to the COVID-19 patients with mild disease, the serum of patients with severe symptoms was enriched with l-phenylalanine, 2-hydroxybutyric acid, mannobiose and lactic acid in the experimental and the validation groups (Fig. 4A). To investigate the potential role of serum metabolites in providing an early warning for progression from mild to severe COVID-19, we created a cohort of patients with mild disease at admission, some of whom continued to suffer from mild disease later (n = 19) and some of whom progressed to severe disease (n = 13) during hospitalization. ROC analysis revealed that only serum 3-hydroxybutyric acid (AUC = 0.708) predicted progression from mild to severe COVID-19. Notably, the combination using binary logistic regression of 7 metabolites (2-hydroxy-3-methylbutyric acid, 3-hydroxybutyric acid, cholesterol, succinic acid, l-ornithine, oleic acid and palmitelaidic acid) with AUCs greater than 0.6 in predicting mild to severe or persistent mild COVID-19 showed near-perfect prediction (AUC = 0.969) of patients who progressed from mild symptoms at admission to severe symptoms during treatment (Fig. 4B). However, this result must be verified in a larger multicenter cohort.
Fig. 4.
The combination of the levels of certain serum metabolites at admission predicts COVID-19 patients who progress from mild to severe disease in the hospital. (A) Differences in serum metabolites between COVID-19 patients with mild (n = 32) and severe (n = 47) disease at admission. (B) ROC curve based on the combination of the levels of 2-hydroxy-3-methylbutyric acid, 3-hydroxybutyric acid, cholesterol, succinic acid, L-ornithine, oleic acid and palmitelaidic acid at admission for the prediction of patients who progress from mild to severe disease in the hospital (n = 32). *, P < 0.05; **, P < 0.01; ***, P < 0.001 in the discovery cohort; #, P < 0.05; ##, P < 0.01; ###, P < 0.001 in the validation cohort.
3.5. Effects of treatment on the levels of serum metabolites in COVID-19 patients
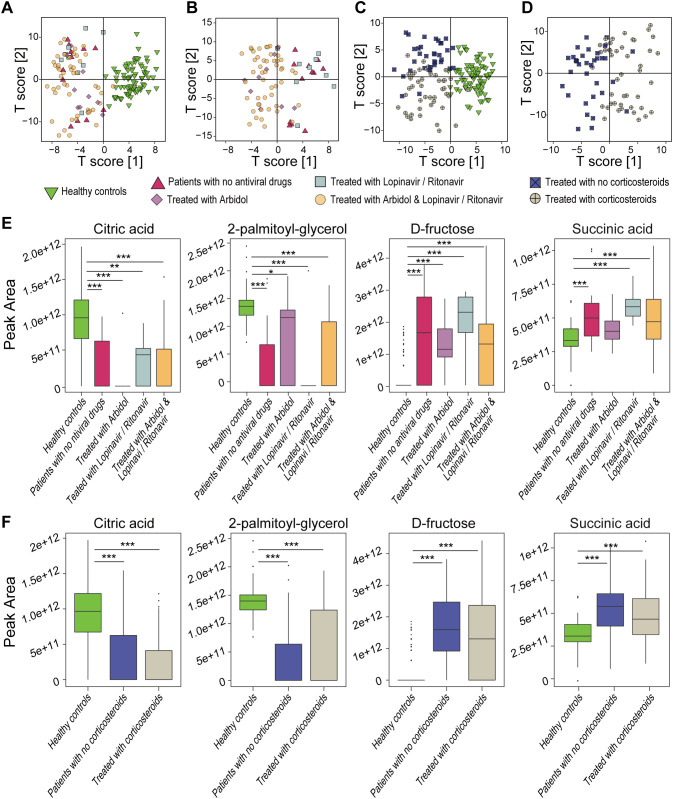
We compared alterations in serum metabolic biomarkers between 79 COVID-19 patients who received different treatments (12 without antiviral agents, 9 with oral arbidol, 9 with oral lopinavir/ritonavir, 49 with oral lopinavir/ritonavir and arbidol, 34 with glucocorticoids, and 45 without glucocorticoids) and 78 HCs. OPLS-DA analysis showed that the serum metabolic profiles were different between COVID-19 patients and HCs, regardless of the treatment used (Fig. 5A and C), but the profiles were not between patients with different antiviral treatment schemes (Fig. 5B). In contrast, the serum metabolome profile of patients treated with glucocorticoids was different (Fig. 5D). Notably, the serum levels of d-fructose, 2-palmitoyl-glycerol and citric acid, which strongly contributed to COVID-19 prediction, were significantly different between HCs and COVID-19 patients with different treatment schemes but not between patients with different treatment schemes (Fig. 5E and F).
Fig. 5.
Effects of different therapies on the serum metabolome and metabolic biomarkers. OPLS-DA plots based on the metabolome profiles of patients treated with no antiviral agents (n = 12), treated with arbidol (n = 9), treated with lopinavir and ritonavir (n = 9), or treated with arbidol together with lopinavir and ritonavir (n = 49), including (A) or not including HCs (B). OPLS-DA plots based on the metabolome profiles of patients treated with (n = 45) or without (n = 34) corticosteroids, including (C) or not including HCs (D). (E) Comparisons of the levels of d-fructose, citric acid and 2-palmitoyl-glycerol between HCs (n = 78) and COVID-19 patients (n = 79) subjected to different antiviral treatments. (F) Comparisons of the levels of d-fructose, citric acid and 2-palmitoyl-glycerol between HCs (n = 78) and COVID-19 patients (n = 79) treated with or without corticosteroids. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
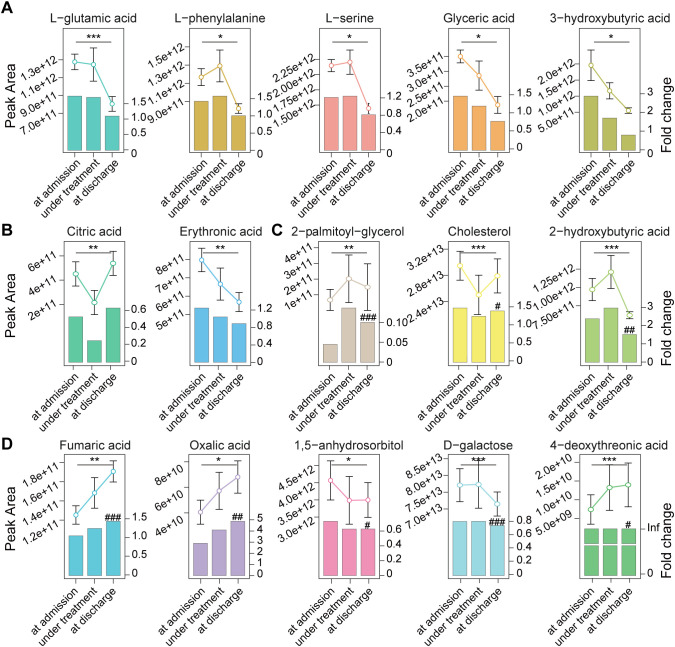
We examined alterations in serum metabolites from hospital admission to discharge. The serum levels of l-glutamic acid, l-phenylalanine, l-serine, glyceric acid, 3-hydroxybutyric acid and erythronic acid, which were enriched in COVID-19 patients compared to HCs at admission, decreased to normal at discharge (Fig. 6A and B). Citric acid, which was depleted in COVID-19 patients at admission, increased to normal levels at discharge (Fig. 6B). 2-hydroxybutyric acid and cholesterol, which were enriched in COVID-19 patients compared to HCs at admission, decreased but remained at levels higher than HCs at discharge. 2-Palmitoyl-glycerol, which was depleted in COVID-19 patients, increased but remained at levels lower than HCs at discharge (Fig. 6C). d-galactose and 1,5-anhydroglucitol, which were depleted in COVID-19 patients compared to HCs at admission, were further decreased at discharge. Fumaric acid, oxalic acid, and 4-deoxythreonic acid, which were enriched in COVID-19 patients compared to HCs at admission, were further increased at discharge (Fig. 6D). The levels of d-fructose, succinic acid, butyric acid, l-lactic acid, l-alanine, and l-alpha-aminobutyric acid were not significantly altered from admission to discharge.
Fig. 6.
Alterations in the levels of serum metabolites in COVID-19 patients from admission to discharge. Serum samples from 15 COVID-19 patients from admission, treatment to discharge and 78 samples from HCs were used. (A), (B) Serum metabolites levels that were different between COVID-19 and HCs at admission but were not significantly different between these two groups at discharge. (C) Serum metabolites levels that were different between COVID-19 and HCs at admission but tended to become not significantly different between these two groups at discharge. (D) Serum metabolites levels that were different between COVID-19 and HCs at admission and whose differences between these two groups became larger at discharge. Fold change was that of COVID-19 patients versus HCs; significant differences between patients at admission and at discharge are marked with “*”, *, P < 0.05; **, P < 0.01; ***, P < 0.001; significant differences between patients at discharge and HCs are marked with “#”, #, P < 0.05; ##, P < 0.01; ###, P < 0.001; Inf is the abbreviation of “infinite”, which indicates that the metabolite was undetectable in HCs.
3.6. Serum metabolic biomarkers are closely associated with clinical features
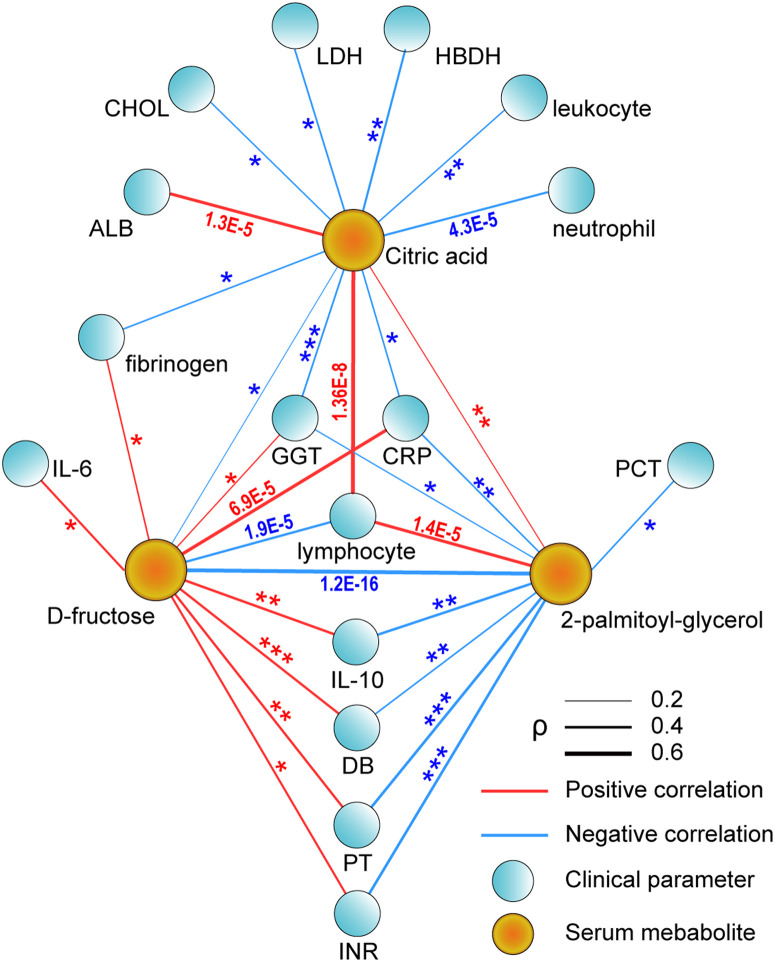
The associations of serum metabolites at admission with clinical features were investigated using Spearman's rank correlation analysis. Significant correlations were primarily observed for three biomarkers, 2-palmitoyl-glycerol, d-fructose and citric acid (Fig. 7 and Supplementary Fig. 3). The lymphocyte counts in peripheral blood positively correlated with the levels of citric acid (P = 1.36E-8) and 2-palmitoyl-glycerol (P = 1.4E-5) but negatively correlated with the levels of d-fructose (P = 1.9E-5). The levels of CRP and GGT negatively correlated with citric acid (CRP, P = 0.011; GGT, P = 7.0E-4) and 2-palmitoyl-glycerol (CRP, P = 7.5E-3; GGT, P = 0.01) but positively correlated with d-fructose (CRP, P = 6.9E-5; GGT, P = 0.024). The level of citric acid also positively correlated with ALB but negatively correlated with cholesterol, leukocytes, neutrophils, lactate dehydrogenase, hydroxybutyrate dehydrogenase, and fibrinogen. The level of d-fructose positively correlated with the international normalized ratio (INR), prothrombin time (PT), fibrinogen, direct bilirubin (DB), IL-6 and IL-10. The level of 2-palmitoyl-glycerol negatively correlated with INR, prothrombin time, direct bilirubin, IL-10 and procalcitonin.
Fig. 7.
Associations between significantly altered clinical parameters and potential metabolic biomarkers in COVID-19 patients from 78 samples at admission. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Abbreviations: ALB, albumin; CHOL, total cholesterol; LDH, lactate dehydrogenase; HBDH, hydroxybutyrate dehydrogenase; GGT, gamma-glutamyl transpeptidase; CRP, C-reactive protein; PCT, procalcitonin; DB, direct bilirubin; PT, prothrombin time; INR, international normalized ratio. The values 0.2, 0.4 and 0.6 were the correlation coefficients represented by the right segments that were used as the reference rulers of the line thickness in this figure to evaluate the correlation coefficients.
4. Discussion
Metabolism is a general term for a series of orderly chemical reactions that occur in organisms to maintain life [15]. It is the basis of life-sustaining compounds and energy. Increasing evidence shows that energy metabolism pathways, such as glycolysis and the tricarboxylic acid cycle (TCA cycle), change during the differentiation and activation of immune cells and play important roles in the functional regulation of immune cells [16]. The intermediates of metabolic pathways are also used as signaling molecules to regulate the occurrence, development and outcome of the immune response [17]. Therefore, alterations in metabolism in COVID-19 patients have attracted much attention [18,19]. Metabolome differences between COVID-19 patients and HCs were extensively examined using liquid chromatography combined with mass spectrometry [[20], [21], [22]]. For example, guanosine monophosphate (GMP) was significantly different between HCs and COVID-19 patients, and between mild and fatal COVID-19 cases [22]. Some metabolites related to kynurenine and fatty acid metabolism were altered and correlated with IL-6 levels and renal status [21]. Plasma monosialodihexosyl gangliosides (GM3s) negatively correlated with CD4+ T cell count in COVID-19 patients, and GM3-enriched exosomes positively correlated with disease severity [20]. The present study enrolled COVID-19 patients, HCs and patients with COVID-19-like symptoms. We used the GC–MS method and found that the serum metabolites of patients with COVID-19 underwent specific changes that were closely related to immune and clinical features. Taken together, these studies provide new prospective information for the pathogenesis, diagnosis and treatment of COVID-19.
Our results showed that d-fructose and succinic acid were enriched, and citric acid and 2-palmitoyl-glycerol were depleted in COVID-19 patients compared to COVID-19-like patients and HCs. The concentration of serum d-fructose is generally stable. Nearly 90% of low-dose fructose is transformed into glucose, lactic acid and glyceric acid in the intestine, and only trace fructose enters the liver. High-dose fructose leads to an increase in the proportion of fructose entering the liver, where fructose is phosphorylated to generate fructose-1-phosphate [23]. High levels of d-fructose enhance inflammation in dendritic cells (DCs) by promoting IL-6 and IL-1β production. Acute SARS-CoV-2 infection impairs DC and CD8 T-cell responses and interactions, and the immune dysfunction associated with d-fructose-induced metabolic disorders may contribute to acute COVID-19 pathogenesis [24,25]. Citric acid is a bridge between the metabolism of carbohydrates and fatty acids, and it promotes the proliferation and differentiation of immune cells, such as B cells [26]. Succinic acid is also an inflammatory signaling molecule. Succinic acid, together with citrate and fumarate, mediates the function of myelocytes during infection and inflammation, [27]. Citric acid and succinate are the key substances in the TCA cycle [28]. TCA cycle metabolites play important roles in non-metabolic signaling regulation of innate and adaptive immunity, lipid and nucleotide synthesis and DNA methylation, which may contribute to SARS-CoV-2 pathogenesis [29]. 2-palmitoyl-glycerol is a product of animal digestion of dietary fat, and it is directly absorbed by intestinal cells and participates in metabolism in vivo. It enhances the binding ability of 2-arachidonoyl-glycerol with CB1 and CB2 cannabinoid receptors to regulate cytokine release and migration in immune cells, and it inhibits adenylate cyclase [30]. 2-palmitoyl-glycerol is an endogenous activator of peroxisome proliferator-activated receptor alpha (PPARα), and the reduction in PPARα induced by the decrease in 2-palmitoyl-glycerol from COVID-19 may be an important effector of pulmonary inflammation and mechanistically involved in the pathogenesis of acute lung injury [31,32].
Our results showed that serum 1,5-anhydroglucitol and 4-deoxythreonic acid were depleted in COVID-19 patients compared to HCs, and oxalic acid and phosphoric acid were enriched in COVID-19 patients compared to COVID-19-like patients. Serum oxalic acid remained nearly 5-fold higher in COVID-19 patients at discharge compared to HCs. The non-metabolizable glucose analogue, 1,5-anhydroglucitol, is primarily derived from a plant-based diet. Its stable serum concentration reflects a steady balance between ingestion and urinary excretion. The serum concentration of 1,5-anhydroglucitol decreases when its recovery is reduced under hyperglycemic conditions. Therefore, its serum concentration reflects the average level of serum glucose in the past 1–2 weeks [33]. 4-Deoxythreonic acid is a metabolite of l-threonine, and its presence in adults negatively correlates with age. Its level increases in children with early-onset type I diabetes, but little is known about its biological functions [34]. Similar to 1,5-anhydroglucitol, oxalic acid is primarily derived from a plant-based diet. Oxalic acid is also derived from the metabolism of oxaloacetic acid, isocitrate, and ascorbic acid. It is not well metabolized in the human body. Approximately 2/3 of serum oxalic acid is excreted in urine and 1/3 in feces [35]. Therefore, the accumulation of oxalic acid in serum may be caused by an increase in endogenous production and/or the obstruction of excretion. Oxalic acid binds minerals, and it has been linked to kidney stones and other health problems [36]. Some cases of oxalate nephropathy in COVID-19 patients were reported [37].
Our results showed that serum lactic acid, butyric acid and cholesterol were enriched in COVID-19 and COVID-19-like patients compared to HCs. Lactic acid is an intermediate of anaerobic glycolysis. Its increase in serum reflects a reduced oxygen supply and hypoperfusion. Lactic acid can be oxidized to propionic acid or converted to glucose in the liver. Lactic acid plays a key role in regulating the polarization and differentiation of immune cells [38]. Butyric acid is primarily derived from the decomposition of dietary fiber by gut microbes. It is rapidly absorbed by cells and converted to acetyl-CoA via β-oxidation. Nearly 70% of the energy of intestinal epithelial cells comes from butyric acid. Butyric acid also has antibacterial and anti-inflammatory effects [39]. Nearly 80% of cholesterol comes from synthesis in the liver, which regulates cholesterol in the circulating blood. Although the cholesterol accumulated by the proinflammatory response may play a beneficial role in the response to infection, it worsens diseases related to chronic metabolic inflammation, such as atherosclerosis [40].
Our results showed that serum l-glutamic acid, l-phenylalanine, l-serine and 2-hydroxybutyric acid were enriched in COVID-19 patients and COVID-like patients compared to HCs. l-glutamic acid is converted from glutamine and may be converted into α-ketoglutaric acid. l-glutamic acid is a regulator of T cell function. Inflammatory mediators also regulate the extracellular glutamate concentration by affecting glial cells [41]. l-phenylalanine is an essential amino acid. Nearly 60% of l-phenylalanine is transformed into tyrosine by phenylalanine hydroxylase and its cofactor, 5,6,7,8-tetrahydrobiopterin, in hepatocytes. Serum l-phenylalanine was increased in patients with posttraumatic sepsis, and this increase was related to activation of the immune response. The accumulation of phenylalanine also magnified inflammation [42]. l-serine comes from nutrition absorption or the serine synthesis pathway of glycolysis. Serine is a key immune metabolite that directly regulates adaptive immunity by controlling T cell proliferation [43]. 2-hydroxybutyric acid is primarily produced in the process of L-threonine metabolism or glutathione synthesis, and it may be increased by oxidative stress or the detoxification of exogenous substances in the liver. 2-hydroxybutyric acid is often increased in patients with energy metabolism deficiency, lactic acidosis and ketoacidosis [44].
Although our investigations attempt to provide comprehensive insight into the potential contribution of metabolism to the pathogenesis, diagnosis and treatment of COVID-19, there are several limitations to be addressed in future studies. First, although our cohort included COVID-19, COVID-19-like patients and HCs, there were no asymptomatic patients with COVID-19 whose inclusion in future investigations will improve our understanding of COVID-19. Second, we used blood samples, which were easy to obtain but primarily reflected the overall metabolic alterations of patients and cannot specifically reflect the metabolic disorders of important infection sites, such as the lung. Third, our results obtained by GC–MS were limited by the intrinsic nature of this method, particularly, its sensitivity or separation and/or extraction efficiency, which also exist in LC-MS methods [45]. Finally, the importance of the data provided here and the potential for clinical application merit further investigation in purposely designed, large, confirmatory studies.
In summary, the serum metabolism of patients with COVID-19 changed specifically and reflected the influence of the lung, liver, intestine and kidney on the disease. These findings are of great significance for our understanding of the pathogenesis of COVID-19 and the diagnosis, early prognosis prediction and treatment of COVID-19.
Declaration of competing interest
None.
Acknowledgments
Acknowledgments
We thank all of the patients involved in the study and all of the front-line medical staff in the First Affiliated Hospital, School of Medicine, Zhejiang University.
Authors' contributions
DS designed the study and wrote the first draft of the manuscript. RY, HY, and YL collected the clinical data and finished the metabolic experiment. JS, JjX, WW and JfX performed the statistical analyses. KX, SG, YC, CH and JG finished the figures and tables. LxL and LjL provided data analyses, critical revision and final approval.
Research funding
This work was supported by the Science and Technology Department of Zhejiang Province (2020c03123), National Natural Science Foundation of China (81570512, 81790631, 81800457), the National Key Research and Development Program of China under Grant (2018YFC2000500), the National Science and Technology Major Project (2017ZX10204401), the Emergency Special Project of the Ministry of Science and Technology (10600100000015001206), Chinese Postdoctoral Science Foundation (2020T130101ZX) and the Zhejiang Provincial Natural Science Foundation of China (LED20H190001, LQ19H030007, LQ20H030010).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.metabol.2021.154739.
Appendix A. Supplementary data
Supplementary material 1 of raw data
Supplementary material 2 related to this article
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chinazzi M., Davis J.T., Ajelli M., Gioannini C., Litvinova M., Merler S., et al. The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak. Science (New York, NY) 2020;368:395–400. doi: 10.1126/science.aba9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beeching N.J., Fletcher T.E., Beadsworth M.B.J. Covid-19: testing times. BMJ (Clinical research ed) 2020;369 doi: 10.1136/bmj.m1403. [DOI] [PubMed] [Google Scholar]
- 4.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020;71(16):2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang A., Sun H., Wang X. Serum metabolomics as a novel diagnostic approach for disease: a systematic review. Anal Bioanal Chem. 2012;404:1239–1245. doi: 10.1007/s00216-012-6117-1. [DOI] [PubMed] [Google Scholar]
- 6.Chen Q., Deeb R.S., Ma Y., Staudt M.R., Crystal R.G., Gross S.S. Serum metabolite biomarkers discriminate healthy smokers from COPD smokers. PloS one. 2015;10 doi: 10.1371/journal.pone.0143937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin S., Yue X., Wu H., Han T.-L., Zhu J., Wang C., et al. Explore potential plasma biomarkers of acute respiratory distress syndrome (ARDS) using GC-MS metabolomics analysis. Clin Biochem. 2019;66:49–56. doi: 10.1016/j.clinbiochem.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182 doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England) 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO. Clinical management of severe acute respiratory infection when Novel coronavirus (nCoV) infection is suspected: interim guidance. https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-whennovel-coronavirus-(ncov)-infection-is-suspected (Jan 11), 2020.
- 12.National Health Commission of the People'’s Republic of China The seventh edition of the Guidelines for Diagnosis and Treatment of SARS-CoV-2. Mar 3, 2020. http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml
- 13.Li M., Wang X., Aa J., Qin W., Zha W., Ge Y., et al. GC/TOFMS analysis of metabolites in serum and urine reveals metabolic perturbation of TCA cycle in db/db mice involved in diabetic nephropathy. Am J Physiol Renal Physiol. 2013;304 doi: 10.1152/ajprenal.00536.2012. (F1317-F24) [DOI] [PubMed] [Google Scholar]
- 14.Zhou B., Lou B., Liu J., She J. Serum metabolite profiles as potential biochemical markers in young adults with community-acquired pneumonia cured by moxifloxacin therapy. Sci Rep. 2020;10:4436. doi: 10.1038/s41598-020-61290-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peradze N., Farr O.M., Mantzoros C.S. Research developments in metabolism 2018. Metabolism. 2019;91:70–79. doi: 10.1016/j.metabol.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Patel C.H., Leone R.D., Horton M.R., Powell J.D. Targeting metabolism to regulate immune responses in autoimmunity and cancer. Nat Rev Drug Discov. 2019;18:669–688. doi: 10.1038/s41573-019-0032-5. [DOI] [PubMed] [Google Scholar]
- 17.Jung J., Zeng H., Horng T. Metabolism as a guiding force for immunity. Nat Cell Biol. 2019;21:85–93. doi: 10.1038/s41556-018-0217-x. [DOI] [PubMed] [Google Scholar]
- 18.Ji D., Zhang M., Qin E., Zhang L., Xu J., Wang Y., et al. Obesity, diabetes, non-alcoholic fatty liver disease and metabolic dysfunction associated fatty liver disease are proinflammatory hypercoagulable states associated with severe disease and thrombosis in Covid-19. Metabolism. 2020;115:154437. doi: 10.1016/j.metabol.2020.154437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L., Spranger L., Soll D., Beer F., Brachs M., Spranger J., et al. Metabolic impact of weight loss induced reduction of adipose ACE-2 - potential implication in COVID-19 infections? Metabolism. 2020;113:154401. doi: 10.1016/j.metabol.2020.154401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song J.W., Lam S.M., Fan X., Cao W.J., Wang S.Y., Tian H., et al. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 2020;32:188–202. doi: 10.1016/j.cmet.2020.06.016. [e5] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas T., Stefanoni D., Reisz J.A., Nemkov T., Bertolone L., Francis R.O., et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5 doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu D., Shu T., Yang X., Song J.-X., Zhang M., Yao C., et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl Sci Rev. 2020;7(7):1157–1168. doi: 10.1093/nsr/nwaa086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang C., Hui S., Lu W., Cowan A.J., Morscher R.J., Lee G., et al. The small intestine converts dietary fructose into glucose and organic acids. Cell Metab. 2018;27:351–361. doi: 10.1016/j.cmet.2017.12.016. [e3] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaiswal N., Agrawal S., Agrawal A. High fructose-induced metabolic changes enhance inflammation in human dendritic cells. Clin Exp Immunol. 2019;197:237–249. doi: 10.1111/cei.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou R., To KK, Wong Y.C., Liu L., Zhou B., Li X., et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53:864–877. doi: 10.1016/j.immuni.2020.07.026. [e5] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams N.C., O’Neill L.A.J. A role for the Krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front Immunol. 2018;9:141. doi: 10.3389/fimmu.2018.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones R., McDonald K.E., Willson J.A., Ghesquiere B., Sammut D., Daniel E., et al. Mutations in succinate dehydrogenase B (SDHB) enhance neutrophil survival independent of HIF-1alpha expression. Blood. 2016;127:2641–2644. doi: 10.1182/blood-2016-02-696922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy M.P., O’Neill L.A.J. Krebs cycle reimagined: the emerging roles of succinate and Itaconate as signal transducers. Cell. 2018;174:780–784. doi: 10.1016/j.cell.2018.07.030. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Reyes I., Chandel N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun. 2020;11:102. doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang W., Shi R., Kang X., Zhang X., Chen P., Zhang L., et al. Monoacylglycerol lipase regulates cannabinoid receptor 2-dependent macrophage activation and cancer progression. Nat Commun. 2018;9:2574. doi: 10.1038/s41467-018-04999-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui H., Xie N., Banerjee S., Ge J., Guo S., Liu G. Impairment of fatty acid oxidation in alveolar epithelial cells mediates acute lung injury. Am J Respir Cell Mol Biol. 2019;60:167–178. doi: 10.1165/rcmb.2018-0152OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Depommier C., Vitale R.M., Iannotti F.A., Silvestri C., Flamand N., Druart C., et al. Beneficial effects of Akkermansia muciniphila are not associated with major changes in the circulating endocannabinoidome but linked to higher mono-palmitoyl-glycerol levels as new PPARalpha agonists. Cells. 2021;10 doi: 10.3390/cells10010185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sovio U., Goulding N., McBride N., Cook E., Gaccioli F., Charnock-Jones D.S., et al. A maternal serum metabolite ratio predicts fetal growth restriction at term. Nat Med. 2020;26:348–353. doi: 10.1038/s41591-020-0804-9. [DOI] [PubMed] [Google Scholar]
- 34.Lau C.E., Siskos A.P., Maitre L., Robinson O., Athersuch T.J., Want E.J., et al. Determinants of the urinary and serum metabolome in children from six European populations. BMC Med. 2018;16:202. doi: 10.1186/s12916-018-1190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Y., Zhang Y.H., Chi Z.P., Huang R., Huang H., Liu G., et al. The handling of oxalate in the body and the origin of oxalate in calcium oxalate stones. Urol Int. 2020;104:167–176. doi: 10.1159/000504417. [DOI] [PubMed] [Google Scholar]
- 36.Shihana F., Joglekar M.V., Raubenheimer J., Hardikar A.A., Buckley N.A., Seth D. Circulating human microRNA biomarkers of oxalic acid-induced acute kidney injury. Arch Toxicol. 2020;94:1725–1737. doi: 10.1007/s00204-020-02679-5. [DOI] [PubMed] [Google Scholar]
- 37.Fontana F., Cazzato S., Giovanella S., Ballestri M., Leonelli M., Mori G., et al. Oxalate nephropathy caused by excessive vitamin C administration in 2 patients with COVID-19. Kidney Int Rep. 2020;5:1815–1822. doi: 10.1016/j.ekir.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broskey N.T., Zou K., Dohm G.L., Houmard J.A. Plasma lactate as a marker for metabolic health. Exerc Sport Sci Rev. 2020;48(3):119–124. doi: 10.1249/JES.0000000000000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haak B.W., Littmann E.R., Chaubard J.L., Pickard A.J., Fontana E., Adhi F., et al. Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following Allo-HCT. Blood. 2018;131:2978–2986. doi: 10.1182/blood-2018-01-828996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon A. Cholesterol metabolism and immunity. N Engl J Med. 2014;371:1933–1935. doi: 10.1056/NEJMcibr1412016. [DOI] [PubMed] [Google Scholar]
- 41.McBrayer S.K., Mayers J.R., DiNatale G.J., Shi D.D., Khanal J., Chakraborty A.A., et al. Transaminase inhibition by 2-hydroxyglutarate impairs glutamate biosynthesis and redox homeostasis in glioma. Cell. 2018;175:101–116. doi: 10.1016/j.cell.2018.08.038. [e25] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S.S., Lin J.Y., Chen W.S., Liu M.H., Cheng C.W., Cheng M.L., et al. Phenylalanine- and leucine-defined metabolic types identify high mortality risk in patients with severe infection. Int J Infect Dis. 2019;85:143–149. doi: 10.1016/j.ijid.2019.05.030. [DOI] [PubMed] [Google Scholar]
- 43.Ma E.H., Bantug G., Griss T., Condotta S., Johnson R.M., Samborska B., et al. Serine is an essential metabolite for effector T cell expansion. Cell Metab. 2017;25:345–357. doi: 10.1016/j.cmet.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 44.Cobb J., Eckhart A., Motsinger-Reif A., Carr B., Groop L. Vol. 39. 2016. Ferrannini E alpha-Hydroxybutyric Acid is a Selective Metabolite Biomarker of Impaired Glucose Tolerance Diabetes Care; pp. 988–995. [DOI] [PubMed] [Google Scholar]
- 45.Yan R., Jiang H., Gu S., Feng N., Zhang N., Lv L., et al. Fecal metabolites were altered, identified as biomarkers and correlated with disease activity in patients with systemic lupus erythematosus in a GC-MS-based metabolomics study. Front Immunol. 2020;11:2138. doi: 10.3389/fimmu.2020.02138. [doi: 103389/fimmu] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 of raw data
Supplementary material 2 related to this article