Abstract
Background
The rs17618244 G>A β-Klotho (KLB) variant has been associated with increased risk of ballooning and inflammation in pediatric patients with metabolic associated fatty liver disease (MAFLD), by reducing KLB expression. In hepatocytes, KLB downregulation induced fat accumulation and the expression of inflammatory and lipotoxic genes. We aimed to examine firstly the impact of the KLB rs17618244 variation on liver damage in adult patients with MAFLD and secondly its effect on hepatic stellate cells (HSCs) activation.
Methods
The impact of the KLB rs17618244 variant on histological liver damage was surveyed in a retrospective cohort of 1111 adult patients with MAFLD. Subgroup analysis was performed according to the presence of obesity (BMI>35; n = 708). Immortalized HSCs (LX-2) were transfected with the KLB wild type (LX-2_KLBwt), or with the mutant one carrying the rs17618244 (LX-2_KLBmut).
Findings
At ordinal regression analysis the KLB rs17618244 variant was associated with hepatic fibrosis (OR 1.23, 95% C.I.1.004–1.51; p = 0.04), but not with steatosis, inflammation and ballooning. By stratifying patients according to the presence of obesity, the KLB A allele was further associated with lobular inflammation (OR 1.32, 95% C.I.1.02–1.72; p = 0.03) and cirrhosis (OR 2.51, 95% C.I.1.23–5.05; p = 0.01) Moreover, hepatic KLB expression correlated with that of fibrogenic genes. LX-2_KLBmut cells showed reduced KLB protein levels paralleled by an induction of pro-fibrogenic genes and enhanced proliferative rate.
Interpretation
The KLB rs17618244 variant is associated with hepatic fibrosis, inflammation and cirrhosis mainly in obese patients with MAFLD and HSCs which carry this mutation are highly proliferative and acquire a myofibroblast-like phenotype.
Funding
Ricerca Finalizzata Ministero della Salute GR-2019–12,370,172 (NP), Ricerca Corrente Fondazione IRCCS Cà Granda (PD and ALF), Ricerca Finalizzata Ministero della Salute RF-2013–02,358,319 (ALF), and Ricerca Corrente and 5 × 1000 Ministero della Salute (AA).
Keywords: MAFLD, Liver damage, HSCS activation, protein stability
Abbreviations: αSMA, Alpha-smooth muscle actin; BA, Bile acids; BMI, body mass index; CHX, cycloheximide; COL1A1, collagen type I alpha 1 chain; COL3A1, collagen type III alpha 1 chain; CYP7A1, cholesterol 7a-hydroxylase; ER, endoplasmic reticulum; FBS, fetal bovine serum; FGF19, Fibroblast Growth Factor 19; FGF21, Fibroblast Growth Factor 21; FGFR, Fibroblast Growth Factor Receptor; GAPDH, Glyceraldehyde3-Phosphate Dehydrogenase; HDL, high-density lipoprotein; HCC, hepatocellular carcinoma; HSCs, hepatic stellate cells; IGT, impaired glucose tolerance; IR, insulin resistance; KLB, β-Klotho; LDL, low-density lipoprotein; MAFLD, nonalcoholic fatty liver disease; MBOAT7, Membrane Bound O-Acyltransferase Domain Containing 7; MTTP, microsomal triglyceride transfer protein; Mut, mutant; NASH, nonalcoholic steatohepatitis; PDGF, platelet-derived growth factor; PNPLA3, Patatin-like Phospholipase domain-containing 3; PM, plasma membrane; PPARα, peroxisome proliferator-activated receptor; SHP-1, short heterodimer partner; TGFβ, Transforming Growth Factor-beta; TM6SF2, Transmembrane 6 Superfamily member gene 2; T2D, type 2 diabetes mellitus; wt, wild type
Research in Context.
Evidence before this study: Nonalcoholic fatty liver disease (NAFLD) or as recently redefined metabolic associated fatty liver disease (MAFLD) is strongly associated with obesity and type 2 diabetes, and it is the leading cause of liver disease worldwide. Inherited genetic factors play a major role in the development of MAFLD and in its progression towards end-stage conditions.
The rs17618244 G>A β-Klotho (KLB) variation has been previously associated with enhanced risk of ballooning and inflammation in pediatric MAFLD patients. The mechanism whereby KLB mutation causes liver injuries seems to be due to KLB reduced expression, that fosters the expression of inflammatory and lipotoxic genes.
Added value of this study: We examined relationship between the rs17618244 KLB variant and histologically proven liver damage in a cohort of adult MAFLD patients and in an in vitro model of hepatic stellate cells (HSCs), responsible for fibrosis. We showed that only in obese MAFLD patients, this variation is associated with lobular inflammation and fibrosis and that HSCs carrying the A allele display a higher proliferation rate and acquire a myofibroblast-like phenotype.
Implications of all the available evidence: The KLB rs17618244 genetic variant is associated with liver damage in adult patients with MAFLD and more so after stratification for the presence of obesity. Moreover, the mutated KLB modulates the expression of genes involved in collagen deposition in HSCs, partly explaining the impact of the rs17618244 variation on hepatic fibrosis. These findings may provide novel insight into the pathogenesis and progression of MAFLD.
Alt-text: Unlabelled box
1. Introduction
Nonalcoholic or more recently re-defined metabolic associated fatty liver disease (MAFLD) [1,2], defined as ectopic fat accumulation in the liver, is closely related to obesity and insulin resistance (IR) [3,4]. MAFLD is the most common chronic liver disease worldwide ranging from simple steatosis to nonalcoholic steatohepatitis (NASH), which may progress to fibrosis, cirrhosis and hepatocellular carcinoma (HCC) [5]. MAFLD is a multifactorial disease whose pathogenesis is driven by environmental and genetic factors. The common variant rs738409 I148M in the Patatin-like Phospholipase domain-containing 3 (PNPLA3) gene is the major genetic determinant of hepatic fat content, fibrosis, cirrhosis and HCC [6], [7], [8]. In the last years, strong risk variants for fatty liver, NASH and advanced fibrosis have also been identified in the Transmembrane 6 Superfamily member gene 2 (TM6SF2) and Membrane Bound O-Acyltransferase Domain Containing 7 (MBOAT7) genes [9,10].
We have recently demonstrated that the rs17618244 G>A variant in the β-Klotho (KLB) gene, which leads to reduced KLB expression, is associated with increased risk of ballooning and inflammation in pediatric patients with biopsy-proven MAFLD [11]. KLB plasma levels were lower in patients who carried the rs17618244 KLB minor A allele and were associated with lobular inflammation, ballooning and fibrosis. Finally, KLB downregulation in in vitro models induced intracellular lipid accumulation and the expression of genes involved in inflammation and lipotoxicity [11].
An association between KLB reduction and fibrosis has been recently reported by Somm et al. who demonstrated that KLB deficient mice exhibited a pro-inflammatory status and onset of fibrosis [12]. However, the role of KLB in inducing inflammation and fibrosis and the impact of the rs17618244 variant on KLB expression are not fully investigated.
KLB gene encodes for a transmembrane protein which complexes with Fibroblast Growth Factor Receptors (FGFRs) to bind the hormones FGF21 and FGF19, which are released by the liver and adipose tissue and by the intestine respectively [13]. FGF21 and FGF19 play important roles in lipid and glucose metabolism and their biosynthesis results impaired in obesity [14,15]. The former is involved in glucose and triglycerides uptake by white and brown adipose tissue through the interaction with FGFR1 [16]. FGF19, whose synthesis is induced by postprandial bile acids (BA), is transported to the liver and interacts with the FGFR4-KLB complex, leading to cholesterol 7a-hydroxylase (CYP7A1) downregulation and to the repression of BA synthesis [11]. This process is tightly regulated since BA accumulation in the liver can lead to hepatotoxicity [17,18]. Indeed, adults and children with MAFLD exhibit elevated hepatic and circulating concentrations of BA that correlate with the severity of disease, mainly with fibrosis [19,20].
Both FGF19 and FGF21 have been involved in the pathogenesis of obesity and type 2 diabetes mellitus (T2DM) representing potential therapeutic targets for the treatment of metabolic diseases [21]. FGF21 is paradoxically increased in obese patients and in those with MAFLD suggesting that obesity is a FGF21-resistance state [22,23]. Conversely, several studies reported a significant decrease of FGF19 circulating levels in obese patients with metabolic syndrome and MAFLD [24,25]. In our previous study, we found decreased plasma levels of FGF19 in children with MAFLD who carried the rs17618244 KLB minor A allele [11]. Moreover, at multivariate analysis FGF19 levels were associated not only with the KLB variant, but also with ballooning and fibrosis. However, the impact of this new genetic variant on liver damage in adults with biopsy-proven MAFLD who display a histologically more severe pattern when compared to children [26] has not been investigated yet.
Therefore, in the present study we firstly aimed to confirm the effect of the KLB rs17618244 variant on liver disease severity, previously observed in the pediatric cohort, in adult patients with biopsy-proven MAFLD, stratified according to the presence of obesity. Secondly, in order to clarify the mechanisms through which the rs17618244 polymorphism impact on liver damage, we evaluated the effect of exogenous overexpression of the wild-type or mutant form of KLB in an in vitro model of hepatic stellate cells (HSCs) whose activation represents the primary driver of hepatic fibrogenesis.
2. Materials and methods
2.1. Hepatology service cohort
We analyzed 1111 unrelated Italian patients with MAFLD (Hepatology service cohort). Patients were consecutively enrolled from the Metabolic Liver Diseases outpatient service at Fondazione IRCCS Ca’ Granda of Milan between January 1999 and December 2019 [10].
Inclusion criteria were liver biopsy for suspected NASH (Liver Clinic) or severe obesity (Bariatric Surgery) and availability of DNA samples and clinical data. Other causes of liver disease including increased alcohol intake (>30/20 g/day in males/females), viral and autoimmune hepatitis, hereditary hemochromatosis, alpha1-antitrypsin deficiency, and history infection with hepatitis B or hepatitis C were excluded.
The study conformed to the Declaration of Helsinki and was approved by the Institutional Review Board of the Fondazione Cà Granda of Milan. All patients gave written informed consent.
The demographic, anthropometric, genetic and clinical features of the Hepatology service cohort, stratified according to enrollment criteria are shown in Table S1. Analyses were also performed in the previously described cohort of pediatric patients with biopsy-proven MAFLD [9].
2.2. Biochemical and anthropometric parameters
Body mass index (BMI) and waist circumference were measured using standard procedures. Obesity was defined by BMI>35. The presence of T2DM was diagnosed when impaired fasting glucose tolerance is present and fasting glucose >110 mg/dL. Alanine aminotransferase (ALT), aspartate aminotransferase (AST), triglycerides, total cholesterol, high-density lipoprotein (HDL) and low-density lipoprotein cholesterol (LDL) were measured by standard laboratory methods.
2.3. Histological evaluation
The scoring of hepatic biopsies was performed by independent pathologists unaware of patient status and genotype. Steatosis was graded into the following four categories based on the percentage of affected hepatocytes: 0, 0%−4%; 1, 5%−32%; 2, 33%−65%; and 3, 66%−100%. Disease activity was assessed according to the MAFLD activity score (NAS), with systematic evaluation of hepatocellular ballooning and lobular inflammation; fibrosis was also staged according to the recommendations of the MAFLD Clinical Research Network. NASH was diagnosed in the presence of steatosis, lobular inflammation, and hepatocellular ballooning [27].
2.4. Genotyping
The Hepatology service cohort was genotyped for the rs738409 C>G (PNPLA3 I148M), rs58542926 C>T (TM6SF2 E167K), rs641738 C>T MBOAT7, rs17618244 A>G (KLB R728Q) variants as previously described [9,28]. Genotyping was performed in duplicate using TaqMan 5′- nuclease assays (QuantStudio 3, Thermo Fisher, Waltham, MA, USA). Results of the rs17618244 KLB genetic frequencies were compared to those obtained in not-Finnish European healthy individuals included in the 1000 Genome project Table S2.
2.5. Gene expression analysis in liver biopsies
KLB gene expression was measured in percutaneous liver biopsies of a subset of 125 severely obese patients from the Milan cohort (Bariatric surgery), whose clinical features are shown in Table S3 [29]. RNA was extracted from liver biopsies using RNeasy mini-kit (Qiagen, Hulsterweg, Netherlands). RNA quality was assessed through Agilent 2100 Bioanalyzer and samples with RNA integrity numbers (RIN) greater than or equal to 7 were used for library preparation (Ribo-reduction libraries). RNA sequencing was performed in paired-end mode with a read length of 150 nt using the Illumina HiSeq 4000 (Novogene, Hong Kong, China). The RNA sequencing detailed protocol and data analysis approach are described in the Supplementary materials and methods.
2.6. Assessment of circulating KLB and FGF19
Circulating KLB and FGF19 levels were evaluated in a subset of MAFLD adult patients (n = 175), belonging to the Hepatology service cohort, who were enrolled at the Metabolic Liver Diseases outpatient service, Fondazione IRCCS Cà Granda, Ospedale Policlinico, Milan, Italy, stratified according to the KLB rs17618244 at risk A allele (GG: n = 59; GA/AA: n = 116) and liver disease severity and matched for age and sex. KLB and FGF19 were measured on a serum aliquot collected after overnight fasting and stored at −80 °C which did not undergo any freeze-thaw cycles prior to the present study. The dosage was performed in duplicate using enzyme-linked immunosorbent assays (ELISA) colorimetric (LS-F11894 - LifeSpan BioSciences, Seattle, WA, USA; RD-191107R - BioVendor, Prague, Czech Republic, respectively), according to the manufacturer's instructions. Specifications of the kit were: detection range, 15.6–1000 pg/ml; inter-assay precision, coefficient of variability (CV) <7.9%; intra-assay precision, CV <4.4%. Our range data for intra- and inter-assay CV were 1.8–4.3% and 1.7–7.2%, respectively. The clinical characteristics of these patients are listed in Table S4.
2.7. Cell lines and treatments
LX-2 cells were purchased with certificated authentication by Merck Millipore (Darmstadt, Germany), and HepG2 cells purchased with certificated authentication by ATCC (Manassas, VA, USA). HepG2 cells were grown as previously described [11]. LX-2 cells were maintained in DMEM with FBS at different percentage (10%, 2% and 0.5%). For the experiments LX‐2 cells were grown for 24 hours in normal medium (10% FBS), and two fasting mimicking conditions (0.5% and 2% FBS).
To visualize kinetic degradation of KLB protein, LX-2 cells were treated with 20 μg/mL cycloheximide (CHX, Sigma-Aldrich, St. Louis, MO, USA) for 3 or 6 h; to induce HSCs activation, LX-2 cells were stimulated with 15 ng/mL platelet-derived growth factor (PDGF) for 24 h, while to induce the activation of FGF19/KLB signaling, LX-2 cells were exposed to 50 ng/mL recombinant FGF19 for 24 hour, both products were purchased by PeproTech (Rocky Hill, NJ, USA).
2.8. Cell transfection
For site-directed mutagenesis of KLB, was used the QuickChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). Two complimentary oligonucleotides containing the rs17618244 mutation (Forward: CTGGCGCCTCTACGACCAGCAGTTCAGGCCCTCAC; Reverse: GTGAGGGCCTGAACTGCTGGTCGTAGAGGCGCCAG) were designed to mutate specified nucleotides of a sequence within a plasmid vector containing a wild type (WT) sequence of KLB gene (RG211186, OriGene Technologies, Inc. Rockville, MD USA). PCR product was digested with DpnI enzyme and transformed into XL-10 Gold competent E. Coli cells. Transformed cells were selected on LB agar plates with 100 µg/mL ampicillin. Plasmids were extracted by HiSpeed Maxi Kit (Qiagen, Valencia, CA, USA) and the mutation were confirmed by DNA sequencing.
LX-2 cells were seeded at a density of 300,000 cells/well in a 6-wells plate and were transfected with 0.5 μg of a plasmid vector containing a WT sequence of KLB gene, of a plasmid vector containing a rs17618244 G>A KLB variant or with empty vector, using Lipofectamine Reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. All transfection experiments were performed with 80% confluent cells. Cells were collected at 48 h to perform the experiments.
2.9. Cell proliferation assay
LX-2 cells were seeded at a density of 20,000 cells/well in 96-well plates and then cultured for 48 h. The Dissociation-Enhanced Lanthanide Fluorescent Immunoassay (Delfia) Cell Proliferation Assay was performed following the manufacturer instructions (PerkinElmer, Waltham, MA, USA). The europium-labeled antibody was used to detect incorporated BrdU following Delfia Cell Proliferation Assay protocol. The dissociation of europium ions from the anti-BrdU antibody and the formation of their fluorescent chelates were obtained by DELFIA inducer reagent. The fluorescence, which is proportional to DNA synthesis, was measured by time-resolved fluorometer 2100 Envision™ Multilabel Reader (PerkinElmer).
2.10. Immunofluorescence
HepG2 and LX-2 were seeded at a density of 20,000 cells/well in 4-well chamber slides and fixed in cold methanol/acetone (3:1). The slides were incubated with the primary antibodies: KLB (dilution 1:100; Code: ab107794 Abcam, Cambridge, MA, USA), smooth muscle actin alpha (αSMA) (dilution 1:100; code:14–9760–82 Invitrogen), SMAD2/3 (dilution 1:100; code:5678 Cell Signaling, Danvers, Massachusetts, USA).
Detection of the primary antibodies was performed using goat anti-rabbit Alexa Fluor® 488 (dilution 1:500; Cat. No. A-11,070, Invitrogen) and goat anti-mouse Alexa Fluor® 555 (dilution 1:500; Cat. No. A-21,425, Invitrogen) conjugated secondary antibodies, respectively. Nuclei were counterstained with Hoechst. The images were captured and analyzed using Olympus FluoView FV1000 confocal microscope equipped with FV10-ASW version 2.0 software, using, 40x objective.
2.11. Quantitative real-time (qRT-PCR)
Total RNA was extracted using Total RNA Mini Kit Blood/Cultured Cell (Cat.No. RDB100, Geneaid, New Taipei City, Taiwan) according to the manufacturer's protocol. Reverse transcription was performed using SuperScript VILO cDNA Synthesis (Ref. 11754–050, Invitrogen). qRT-PCR amplification, detection and analysis were performed by ABI Prism 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) using SensiFast Probe HiROX Kit (2X) (Bioline, London, UK).
TaqMan gene assay (Applied Biosystems) for KLB (Hs01573147_m1), αSMA (Hs00909449_m1), collagen type I alpha 1 chain, COL1A1 (Hs00164004_m1), collagen type III alpha 1 chain, COL3A1 (Hs00943809_m1), short heterodimer partner, SHP-1 (Hs00222677_m1) and Transforming Growth Factor-beta (TGF-β) Hs.PT.49.1806981 probe by Integrated DNA Technologies (Coralville, IA, USA) were used. Glyceraldehyde3-Phosphate Dehydrogenase (GAPDH; Hs.PT.39a.22214836; by Integrated DNA Technologies) housekeeping gene was used as a reference control for normalization. Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems) was used for the analyses. Based on the ΔΔCt method, relative amounts of mRNA were expressed as fold changes versus control.
2.12. Western blotting
Total protein extraction was performed by homogenizing cells in Ripa lysis buffer (Sigma-Aldrich) and Halt Protease & Phosphate Inhibitor Cocktail (100X) (Thermo Scientific, Carlsbad, CA, USA. The homogenates were incubated on ice for 30 min and then centrifuged at 13,000 rpm for 10 min. The resulting supernatant was collected and quantified using the BCA™ Protein Assay (Thermo Scientific). The samples were then diluted in the sample buffer 4X and resolved in 8% SDS-PAGE, then transferred and immobilized onto the nitrocellulose membranes (GE Healthcare, Munich, Germany). The membranes were blocked using 5% non-fat dry milk for 30 min and incubated with the appropriate primary and secondary antibodies. Protein expression was quantified by densitometric analysis using Image J v3.91 software. The following antibodies were used: i) rabbit anti-KLB (dilution 1:1000; code: ab106794; Abcam); ii) mouse anti-αTubulin (dilution 1:5000; code: nb100–690; Novus Biologicals, Littleton, Colorado, USA).
2.13. Statistical analysis
For descriptive statistics, continuous variables were shown as mean and SD or median and interquartile range for highly skewed biological variables (i.e. ALT, AST, triglycerides, KLB and FGF19 circulating levels). Variables with skewed distributions were log-transformed before analyses in order to achieve a normal distribution. Categorical variables were presented as number and percentages. Genetic analyses were performed under additive or recessive models. Analyses were performed by fitting data into generalized linear regression models. In particular, general linear models were fit to examine continuous traits (i.e. age, BMI, total cholesterol, LDL, HDL, triglycerides, ALT, AST, insulin levels). Logistic regression models were fit to examine binary traits (NASH, severe fibrosis stage F3-F4), and ordinal regression models were fit for categorical traits (components of the MAFLD activity score: severity of steatosis, lobular inflammation and hepatocellular ballooning, stage of fibrosis). The regression models were adjusted for gender, age, BMI, PNPLA3 I148M, TM6SF2 E167K and MBOAT7 rs641738 T alleles.
In these analyses, we set the Overall Cohort (n = 1111) as the reference group to determine the primary outcome of the study. Subgroup analysis was performed according to the presence of obesity (BMI>35; n = 708 obese patients). Mean values were compared by ANOVA or by 2-tailed Student's t-test (for continuous traits) and by Pearson's chi-square test for binary (IGT/T2D) o categorical traits (genetic data), where appropriate. For the in vitro study, results were expressed as means ± SD, and differences between samples were analyzed by 2-tailed t tests or two-way ANOVA with Bonferroni correction, as post hoc test for multiple comparisons.
Statistical analyses were performed using JMP 14.0 (SAS, Cary, NC), R statistical analysis software version 3.3.2 (http://www.R-project.org/) and Prism software (version 6, GraphPad Software). P< 0.05 (two-tailed) was considered statistically significant. All data generated or analyzed during this study are included in this manuscript and in the supplementary files.
3. Results
3.1. KLB rs17618244 variant affects hepatic fibrosis in adult patients with MAFLD
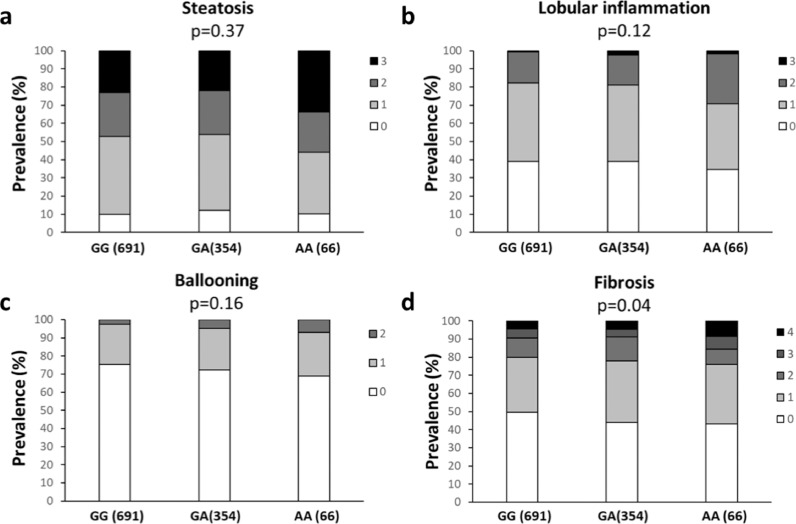
The demographic, anthropometric and clinical features of the Hepatology service cohort stratified according to the KLB rs17618244 genotype are shown in Table 1. Firstly, we analyzed the association between the rs17618244 A risk allele and the histological features of MAFLD. At ordinal regression analysis adjusted for age, gender, BMI, T2DM, PNPLA3 I148M, TM6SF2 E167K and MBOAT7 rs641738 T alleles, the KLB rs17618244 variant was associated with hepatic fibrosis (beta 1.23, 95% C.I. 1.004–1.51; p = 0.04), whereas there was no association with steatosis, lobular inflammation and ballooning (Table 2; Fig. 1a-d).
Table 1.
Demographic, anthropometric and clinical features of Hepatology service cohort (n = 1111) stratified by KLB rs17618244 G>A genotype.
| GG (n = 698) | GA (n = 351) | AA (n = 62) | P-value | |
|---|---|---|---|---|
| Sex, M | 428 (54) | 196 (50.3) | 37 (51.5) | 0.62 |
| Age, years | 47.9 ± 12.9 | 48.6 ± 13.5 | 47.8 ± 13.7 | 0.87 |
| BMI, kg/m2 | 34.6 ± 8.9 | 34.9 ± 8.6 | 33.9 ± 9.6 | 0.81 |
| IFG/T2D, yes (%) | 176 (22.2) | 96 (24.7) | 22 (29.2) | 0.73 |
| HOMA-IR | 5.10±6.70 | 4.80±3.30 | 4.20±2.80 | 0.68 |
| Insulin, IU/mL | 20.4 ± 19.4 | 19.9 ± 13.6 | 18.3 ± 13.6 | 0.87 |
| Total cholesterol, mmol/L | 5.17±1.10 | 5.17±1.06 | 5.07±1.09 | 0.66 |
| LDL cholesterol, mmol/L | 3.20±0.98 | 3.20±0.90 | 3.02±1.00 | 0.48 |
| HDL cholesterol, mmol/L | 1.27±0.35 | 1.29±0.42 | 1.24±0.35 | 0.52 |
| Triglycerides, mmol/L | 1.61±0.88 | 1.61±0.84 | 1.83±1.90 | 0.92 |
| ALT, IU/L | 31{20–53} | 31.5{19–56} | 37{23–68} | 0.15 |
| AST, IU/L | 23{18–35} | 24{18–34} | 27 {21–38} | 0.28 |
Values are reported as mean ± SD. number (%) or median {IQR}. as appropriate. BMI: body mass index. IFG: impaired fasting glucose. T2D: type 2 diabetes. Characteristics of participants were compared across KLB, rs17618244 genotypes by using ANOVA or Pearson analyses.
Table 2.
Associations between the KLB rs17618244 variant and the independent predictors of liver damage in patients from the Hepatology Service cohort (n = 1111).
| Steatosis |
Lobular inflammation |
Ballooning |
Fibrosis |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | |
| Age, years | 1.003 | 0.99–1.01 | 0.54 | 1.02 | 1.007–1.03 | 0.001 | 1.02 | 1.009–1.04 | 0.0007 | 1.03 | 1.02–1.04 | <0.0001 |
| Gender, M | 1.43 | 1.26–1.63 | <0.0001 | 1.38 | 1.19–1.57 | <0.0001 | 1.45 | 1.22–1.72 | <0.0001 | 1.58 | 1.38–1.82 | <0.0001 |
| BMI, Kg/m2 | 1.49 | 1.03–1.06 | 0.0001 | 1.34 | 1.14–1.57 | 0.0003 | 1.40 | 1.16–1.68 | 0.0002 | 1.72 | 1.46–2.01 | <0.0001 |
| IFG/T2DM, yes | 1.34 | 1.15–1.57 | 0.0002 | 1.03 | 1.02–1.05 | <0.0001 | 0.99 | 0.98–1.02 | 0.89 | 1.09 | 0.95–1.03 | 0.21 |
| PNPLA3, I148M | 1.62 | 1.36–1.90 | 0.0001 | 1.45 | 1.22–1.70 | <0.0001 | 1.23 | 1.003–1.52 | 0.045 | 1.63 | 1.38–1.95 | <0.0001 |
| TM6SF2, E167K | 2.34 | 1.68–3.22 | <0.0001 | 1.72 | 1.26–2.34 | 0.0007 | 1.28 | 0.88–1.86 | 0.18 | 1.54 | 1.13–2.10 | 0.006 |
| MBOAT7 rs641738 T | 1.09 | 0.94–1.28 | 0.22 | 1.03 | 0.88–1.21 | 0.68 | 1.01 | 0.83–1.23 | 0.89 | 1.09 | 0.93–1.28 | 0.27 |
| KLB, R728Q | 1.09 | 0.89–1.32 | 0.37 | 1.17 | 0.96–1.43 | 0.12 | 1.19 | 0.93–1.51 | 0.16 | 1.23 | 1.004–1.51 | 0.04 |
OR: odd ratio; CI: confidence interval. Values were obtained at multivariate ordinal regression analysis adjusted for sex, age, BMI (body mass index), T2DM (type 2 diabetes mellitus) and PNPLA3 I148M alleles, TM6SF2 E167K alleles and MBOAT7 rs641738 T allele. The independent variable was the KLB rs17618244 genotype (continuous trait).
Fig. 1.
The KLB rs17618244 variant is associated with fibrosis in patients with MAFLD. Association of the KLB rs17618244 variant with steatosis (a), lobular inflammation (b) ballooning grade (c) and fibrosis stage (d) in MAFLD patients from the Hepatology service cohort (n = 1111). Multivariable ordinal regression analysis adjusted for age, sex, BMI, T2D, presence of PNPLA3 I148M, TM6SF2 E167K and MBOAT7 T alleles at additive model.
3.2. The association between KLB rs17618244 variant and hepatic fibrosis seems to be obesity-related
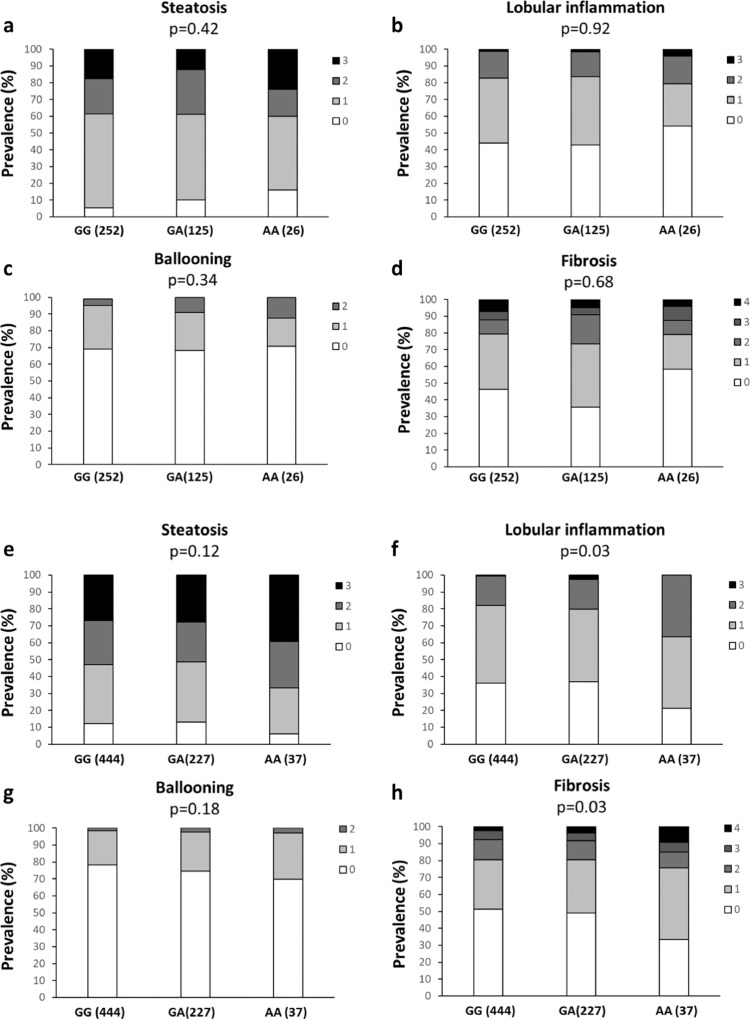
We next investigated whether the presence of obesity may modify the impact of the KLB rs17618244 variant on liver damage. By stratifying patients according to the presence of obesity (Fig. 2a-d BMI<35; Fig. 2e-h BMI>35), the association between the KLB rs17618244 variant and hepatic fibrosis was even stronger (OR 1.34, 95% C.I. 1.02–1.73; p = 0.03) (Table 3, Fig. 2h). In obese patients the KLB rs17618244 variant was further related to lobular inflammation (OR 1.32, 95% C.I. 1.02–1.72; p = 0.03) (Table 3, Fig. 2f) and higher risk of cirrhosis (OR 2.51, 95% C.I. 1.23–5.05; p = 0.01) (Table 3).
Fig. 2.
The KLB rs17618244 variant is associated with lobular inflammation and fibrosis in patients with obese MAFLD patients. Association of the KLB rs17618244 variant with steatosis, lobular inflammation, ballooning grade and fibrosis stage in MAFLD patients from the Hepatology service cohort (n = 1111) stratified according to the absence (a-d) or presence (e-h) of obesity (BMI>35). Multivariable ordinal regression analysis adjusted for age, sex, BMI, T2D, presence of PNPLA3 I148M, TM6SF2 E167K and MBOAT7 T alleles at additive model.
Table 3.
Associations between the allelic risk variants and the independent predictors of liver damage in the Hepatology service cohort stratified according to the presence of obesity (n = 403 no obese patients and n = 708 obese patients).
| Steatosis | Lobular inflammation | Ballooning | Fibrosis | Cirrhosis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Obesity no (n = 403) | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P |
| KLB, R728Q | 1.14 | 0.62–1.22 | 0.42 | 0.98 | 0.70–1.36 | 0.92 | 1.19 | 0.81–1.75 | 0.34 | 1.07 | 0.77–1.48 | 0.68 | 1.09 | 0.38–2.85 | 0.87 |
| Obesity yes (n = 708) | |||||||||||||||
| KLB R728Q | 1.21 | 0.95–1.57 | 0.12 | 1.32 | 1.02–1.72 | 0.03 | 1.25 | 0.89–1.73 | 0.18 | 1.34 | 1.02–1.73 | 0.03 | 2.51 | 1.23–5.05 | 0.01 |
| Steatosis | Lobular inflammation | Ballooning | Fibrosis | Cirrhosis | |||||||||||
| Obesity no (n = 403) | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | β | 95% CI | P |
| KLB 728Q homozygous | 0.98 | 0.64–1.49 | 0.91 | 0.99 | 0.65–1.51 | 0.98 | 1.26 | 0.76–1.97 | 0.33 | 0.89 | 0.58–1.36 | 0.59 | 2.16 | 0.72–5.42 | 0.12 |
| Obesity yes (n = 708) | |||||||||||||||
| KLB 728Q homozygous | 1.43 | 1.03–2.01 | 0.03 | 1.55 | 1.09–2.20 | 0.01 | 1.21 | 0.79–1.80 | 0.35 | 1.46 | 1.03–2.05 | 0.03 | 2.01 | 0.89–3.97 | 0.06 |
OR: odd ratio; CI: confidence interval. Values were obtained at multivariate ordinal regression analysis adjusted for sex, age, BMI (body mass index), T2DM (type 2 diabetes mellitus) and PNPLA3 I148M alleles, TM6SF2 E167K alleles and MBOAT7 rs641738 T allele. The independent variable was the KLB rs17618244 genotype (continuous trait).
Moreover, we found an association between the KLB rs17618244 variant and steatosis in obese patients by using a recessive model (OR 1.43, 95% C.I. 1.03–2.01; p = 0.03) (Table 3). The effect on lobular inflammation was even stronger by applying this model (OR 1.55, 95% C.I. 1.09–2.20; p = 0.01) whereas the impact on fibrosis remain unchanged (OR 1.46, 95% C.I. 1.03–2.05; p = 0.03) (Table 3).
To confirm whether the impact of the KLB rs17618244 variant on liver damage is related to obesity, we stratified the previously studied cohort of pediatric patients with biopsy-proven MAFLD [11], according to the presence of obesity. The KLB rs17618244 variant was even more associated with lobular inflammation (OR 2.74, 95% C.I. 1.35–5.70; p = 0.005) and remained associated with ballooning (OR 1.93, 95% C.I. 0.97–3.93; p = 0.05) in obese children whereas we didn't find any effect on fibrosis (Table S5) probably due to the less severe hepatic disease in children compared to adults.
3.3. The KLB rs17618244 gene variant impacts on hepatic and circulating expression of KLB
We previously demonstrated that the KLB 17618244 variant was associated with lower circulating KLB and with a reduced hepatic expression in pediatric patients with MAFLD. KLB gene expression was evaluated in percutaneous liver biopsies of a subset of 125 obese patients from the Hepatology service cohort who underwent bariatric surgery. Transcriptomic data were available for these patients who were stratified according to KLB rs17618244 genotype which was available in 121 subjects (GG: n = 78; GA+AA=43).
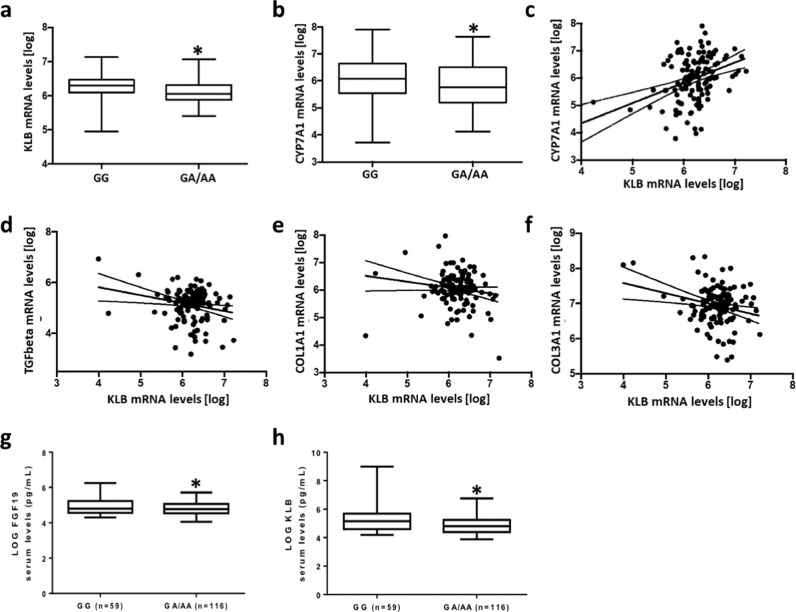
KLB hepatic mRNA levels were reduced in obese patients who carried the rs17618244 A allele compared to non-carriers (6.06±0.27 vs 6.25±0.32 p = 0.004 log transformed). Notably, also the hepatic expression of CYP7A1, the first and rate-limiting enzyme of bile acid synthesis which is a direct target of the KLB-FGFR4 complex, was reduced in subjects who carried the rs17618244 A allele (5.77±0.92 vs 6.12±0.72; p = 0.036 log transformed) (Fig. 3a-b).
Fig. 3.
KLB mRNA levels are higher in patients who carry the at risk A allele and correlate with the expression of genes involved in fibrogenesis. KLB (a) and CYP7A1 (b) mRNA levels in carriers of the rs17618244 A allele compared to non-carriers. Correlation analyses between hepatic KLB gene expression and CYP7A1 (c), TGF-β (d), COL1A1 (e) and COL3A1 (f) mRNA levels evaluated by transcriptome analysis on liver biopsies (n = 125). Circulating levels of KLB (g) and FGF19 (h) in 175 NAFLD adult patients. Bar graphs represents KLB and FGF serum levels stratified according to the presence of the KLB at risk A allele (GG=59; GA/AA=116). Data were logarithmically transformed and analyzed by 2-tailed T-tests, Adjusted *p<0.05.
Moreover, we found a strong correlation between the hepatic expression of KLB and CYP7A1 (y = 1.20+0.77x; p<0.0001 log transformed), TGF-β (y = 7.14–0.32x; p = 0.012 log transformed), COL1A1 (y = 6.44–0.0007x; p = 0.0009 log transformed) and COL3A1 (y = 7.20–0.0005x; p = 0.036 log transformed) (Fig. 3c-f). In addition, hepatic KLB correlated with MTTP (y = 5.85+0.35x; p<0.0001 log transformed) and PPARα (y = 7.47+0.0003x; p = 0.02 log transformed) (Fig. S1).
These findings confirmed the association between a low hepatic expression of KLB and pro-fibrogenic genes.
We next evaluated the concentrations of circulating KLB and FGF19 in a subgroup of MAFLD patients (n = 175), belonging to the Hepatology service cohort, stratified according to the presence of the KLB rs17618244 at risk A allele. At the bivariate analysis, we found that KLB and FGF19 serum levels were lower in patients carrying the A allele of the KLB rs17618244 variant (p = 0.019 and p = 0.048 vs non-carriers, respectively) (Fig. 3g-h). In addition, at the multivariate generalized linear model, adjusted for sex, age, BMI, T2DM and PNPLA3 genotype, KLB and FGF19 serum levels remained significantly associated with KLB rs17618244 variant (Estimate β=−0.17±0.07; 95% CI −0.32–0,02; p = 0.018 and Estimate β=−0.07±0.03; 95% CI −0.14–0,007; p = 0.029, respectively). Moreover, circulating KLB and FGF19 decreased in NASH patients compared to non-NASH (p = 0.029 and p = 0.0002, respectively; data not shown). Likewise, in a multivariate generalized linear model, adjusted for sex, age, BMI, T2DM, PNPLA3 genotype and the KLB rs17618244 variant, reduced KLB serum levels remained associated with lobular inflammation (Estimate β=−0.62±0.30; 95%CI-1.24–0.06; p = 0.039), ballooning (Estimate β=−0.75±0.30; 95%CI-1.38–0.21; p = 0.012) and fibrosis (Estimate β=−0.51±0.27; 95%CI-1.05–0.01; p = 0.048), while those of FGF19 with steatosis (Estimate β=−0.79±0.35; 95%CI-1.48–0.12; p = 0.025), lobular inflammation (Estimate β=−0.73±0.36; 95%CI-1.47–0.04; p = 0.043) and ballooning (Estimate β=−0.83±0.42; 95%CI-1.68–0.04; p = 0.047) (Table S6).
3.4. The KLB gene/protein expression is downregulated in hepatic stellate cells under fed condition
The activation of HSCs is the main driver of hepatic fibrosis [30]. Therefore, the evaluation of the KLB gene/protein expression in HSCs could explain its role in MAFLD-related fibrosis.
We investigated the expression of KLB gene/protein in LX-2 cells, a human HSC cell line which is GG homozygous for the KLB rs17618244 variant. LX-2 cells were exposed to a set of different culture conditions (10% FBS and 0.5–2% FBS to mimic a fasting condition) previously reported to affect the pro-fibrogenic activation in this cell type [31].
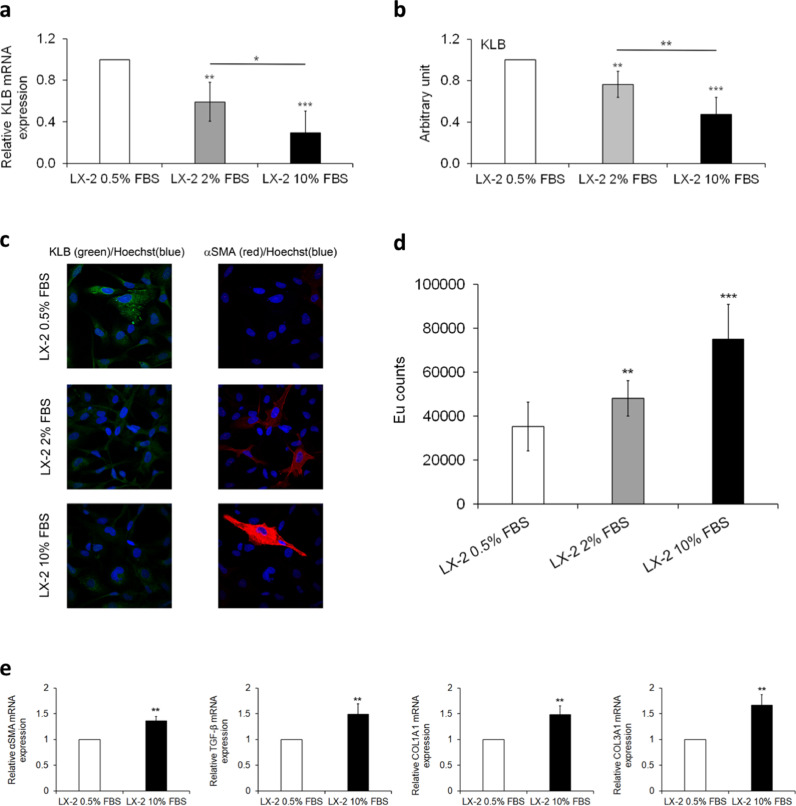
We found that KLB mRNA and protein expression levels were significantly lower in fed condition (10% FBS) than in fasting ones (Fig. 4a-b, adjusted p<0.05 and p<0.01 vs LX-2 2% FBS, and Fig. S2). The KLB expression was progressively reduced from 0.5% to 10% FBS (Fig. 4a-b, adjusted p<0.01 and p<0.001 vs LX-2 0.5% FBS; Fig. S2). The decrease in KLB protein observed under 2% and 10% FBS was confirmed also by immunofluorescence assay (Fig. 4c). Moreover, immunofluorescence images showed an increase of α-SMA expression concomitantly with KLB protein downregulation (Fig. 4c). Interestingly, the subcellular distribution of KLB in LX-2 cells was different from that observed in HepG2 cells (Fig. S3). Indeed, in LX-2 cells KLB was diffusely expressed in plasma membrane (PM) and into the cytoplasm, showing a granular pattern distribution, while in HepG2 cells KLB protein is mainly localized in PM, endoplasmic reticulum (ER) and Golgi compartments, where post-translational modifications occur.
Fig. 4.
Analysis of KLB gene/proteinr expression in LX-2 cells under normal or fasting condition. In this experimental setting LX-2 cells were cultured in fasting (0.5% and 2% FBS) or normal medium (10% FBS) for 24 h. Analysis of relative KLB gene expression evaluated by qRT-PCR (a). Relative expression of KLB protein levels against α-tubulin determined by densitometric analysis of Western blots. The intensity of the bands was analyzed by ImageJ software (b). Representative immunofluorescence images of cellular distribution of KLB (green) and αSMA (red). Nuclei are reported in blue. Magnification 60X (c). Cell proliferation evaluated by a BrdU incorporation and expressed as Europium (Eu) counts (d). Expressions of αSMA, TGF-β, COL1A1 and COL3A1 genes were measured by qRT-PCR (e). Data are the mean ± SD of 2 independent experiments repeated at least in triplicate. Data were analyzed by 2-tailed t tests, Adjusted *p<0.05; ** p<0.01; ***p<0.001.
LX-2 cells showed also a significant increase of cell proliferation rate at 2% and 10% FBS (Fig. 4d, adjusted p<0.01 and p<0.001 vs LX-2 0.5% FBS). The expression of αSMA, TGF-β, COL1A1 and COL3A1 (Fig. 4e, adjusted p<0.01 vs LX-2 0.5% FBS) was higher in LX-2 cells cultured under 10% FBS thus suggesting the activation pro-fibrogenic genes in fed status. Similarly, LX-2 cells exposed to a pro-fibrogenic stimulus, induced by PDGF [32], exhibited a myofibroblast-like phenotype, characterized by increased proliferation rate and α-SMA induction (adjusted p<0.001 and p<0.01 vs LX-2 + vehicle, respectively) paralleled by reduced expression of KLB (adjusted p<0.01 vs LX-2 + vehicle), thus suggesting that fed condition may be a good model to reproduce a physiological activation of HSCs (Fig. S4a-d).
3.5. Mutant KLB affects KLB protein expression/stability and induces a pro-fibrogenic phenotype in LX-2 cells
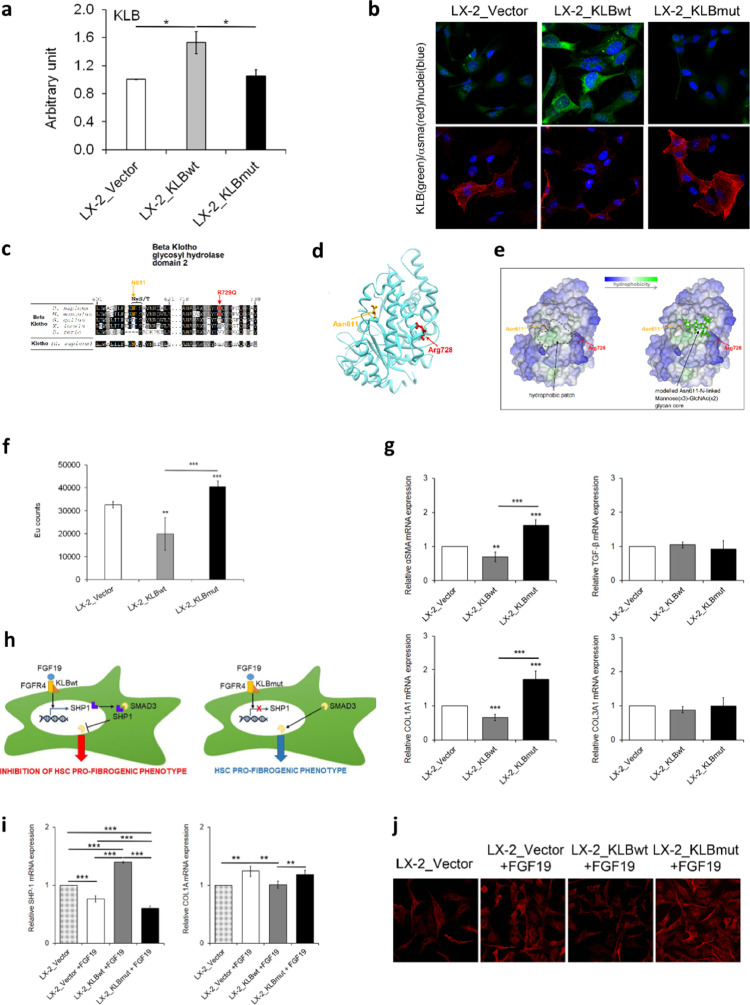
To examine the effect of the KLB rs17618244 variant on the pro-fibrogenic phenotype in HSCs, we transfected LX-2 cells maintained at 10% FBS with a control empty vector plasmid (LX-2_Vector), a plasmid containing the wild type sequence of KLB (LX-2_KLBwt), or a plasmid containing the mutant KLB (LX-2_KLBmut).
KLB mRNA levels were strongly increased after transfection of either KLBwt or KLBmut plasmids, although the higher levels were observed in cells transfected with the KLBwt one (Fig. S5a, adjusted p<0.001 vs LX-2_Vector). Protein expression was significantly upregulated in LX-2_KLBwt cells compared to empty vector, thus confirming KLB overexpression (Fig. S5b). Notably, the overexpression of KLB mutant form did not upregulate the KLB protein levels, resembling the effect of the rs17618244 variant (Fig. S5b and Fig. 5a, adjusted p<0.05 vs both LX-2_Vector and LX-2_KLBwt). To possibly explain the differences in KLB expression between wt and mutated forms, LX-2 cells were treated with a moderate dose of CHX (20 μg/mL) which led to a more pronounced protein degradation in LX-2_KLBmut (Fig. S5c). Interestingly, immunofluorescence images showed higher KLB levels, mainly distributed in the ER and Golgi compartments, and a concomitant downregulation of αSMA in LX-2_KLBwt compared to LX-2_KLBmut cells (Fig. 5b). Conversely, the overexpression of the mutated plasmid was not able to induce exogenous KLB protein levels and we didn't find differences in its localization compared to endogenous form in LX-2_Vector cells. Finally, we observed that soluble KLB is slightly released only by KLBwt cells and not by mutated ones as a possible consequence of reduced KLB protein levels in the latter (Fig. S5d).
Fig. 5.
Analysis of the effect of wild-type and mutant KLB overexpression in LX-2 cells. In this experimental setting LX-2 cells were transiently transfected with pCMV6 plasmid (LX-2_Vector), KLB wild-type plasmid (LX-2_KLBwt), or KLB mutant plasmid (LX-2_KLBmut), and all analyses were performed after 48 h. Relative expression of KLB protein levels was determined by densitometric analysis of Western blots. The intensity of the bands was analyzed by ImageJ software (a). Representative immunofluorescence images of cellular distribution of KLB (green) and αSMA (red). Nuclei are reported in blue. Magnification 60X (b). Multiple sequence alignment around the site of the Arg728Gln variant among orthologues of KLB and the human Klotho paralogue. Consensus and similar residues are respectively in black and gray background. The consensus residues NxS/T (where “x” can be any residue except proline) for the N-glycosylation of Asn611 are indicated (c). The structure of the KLB GH2 domain isolated from the Protein Data Bank entry 5VAN and the modelled to show the position of Arg728, the N-glycosylable Asn611 (d). Same KLB GH2 domain structure as in D represented as molecular surface colored by residue hydrophobicity, highlighting the surface-exposed protein hydrophobic patch (left) and showing a modelled mannose(x3)-GlcNAc(x2) glycan N-linked to Asn611 (e). Cell proliferation was evaluated by a BrdU incorporation kit and expressed as Eu counts (f). mRNA levels of αSMA, TGF-β, COL1A1 and COL3A1 were evaluated by qRT-PCR (g). Hypothesis of mechanism by which KLB exert the inhibition of pro-fibrogenic genes after LX-2 cells stimulation with FGF19 (h). mRNA levels of SHP-1 and COL1A1 were evaluated by qRT-PCR (i). Representative immunofluorescence images of cellular distribution of SMAD3 (red). Magnification 40X (j). Quantitative data are the mean ± SD of 2 independent experiments repeated at least in triplicate. Data were analyzed by two-way ANOVA with Bonferroni post hoc test for multiple comparisons. Adjusted *p<0.05; **p<0.01; ***p<0.001.
The decrease of KLB expression and the higher degradation rate in LX-2_KLBmut cells could be due to the impact of the transition from Arg728 to Gln728 on protein stability. DynaMut, a web server that assesses the impact of a mutation on protein dynamic and stability resulting from changes of vibrational entropy energy (VEE), predicted a ΔVEE between KLBwt and KLBmut that suggests a decrease of molecular flexibility [33]. However, since Arg728 is poorly conserved (Fig. 5c) and fully exposed on the surface of the glycosyl hydrolase 2 (GH2) domain (Fig. 5d), it seems to be not relevant for protein folding. Notwithstanding, Arg728 is at the border of a patch of hydrophobic residues on the surface of the GH2 domain, and also near to Asn611 in the consensus NxS/T motif (residues 611-NLS-613) targeted by N-glycosylation. We next modelled the mannose(x3)-GlcNAc(x2) core N-linked to Asn611. The resulting model supports the hypothesis that Arg728Gln substitution may alter the enzymatic insertion or trimming of the glycan at Asn611(Fig. 5e).
Finally, we investigated the mechanism though which the rs17618244 mutation may impact on liver damage. We found that the proliferation rate significantly decreased in LX-2_KLBwt cells compared to LX-2_Vector (Fig. 5f, adjusted p<0.01), whereas LX-2_KLBmut cells were highly proliferative with respect to both control and LX-2_KLBwt ones (Fig. 5f, adjusted p<0.001). The pro-fibrogenic genes αSMA and COL1A1 were specifically modulated by exogenous KLB. In particular, KLBwt induced a significant downregulation of these genes (Fig. 5g, p<0.01 and p<0.001 vs LX-2_Vector), whilst KLBmut upregulated their expression (Fig. 5g, adjusted p<0.001 vs LX-2_Vector and LX-2_KLBmut). As shown in Fig. 5h, we hypothesized that KLBwt in LX-2 cells may mediate the increased FGF19-dependent transcription of the SHP-1, which in turn may bind SMAD3 impairing its nuclear translocation and abrogating its transcriptional activity on pro-fibrogenic genes [34]. On the contrary KLBmut may block this molecular network. This hypothesis was supported by the evidence that after 24 h of treatment with FGF19, KLBwt cells exhibited upregulation of SHP-1 gene expression (Fig. 5i, adjusted p<0.001 vs LX-2_Vector with or without FGF19 and vs LX-2_KLBmut), and repressed nuclear translocation of SMAD3 (Fig. 5j), while KLBmut was unable to induce these effects (Fig. 5i-j).
4. Discussion
In our previous study we demonstrated that the KLB rs17618244 variant was associated with pediatric MAFLD, and particularly with higher risk of ballooning and lobular inflammation whereas it did not impact on fibrosis [11]. To confirm these findings, we evaluated the effect of the KLB rs17618244 variant on the entire spectrum of liver damage in adults with biopsy proven MAFLD and we found that it was associated only with hepatic fibrosis. However, by stratifying adult patients according to the presence of obesity, the rs17618244 variant was significantly associated with lobular inflammation, fibrosis and cirrhosis. Moreover, we confirmed that the impact of KLB rs17618244 variant on hepatic inflammation was even stronger in the cohort of MAFLD children stratified by obesity. It has been demonstrated that KLB expression is reduced in obese mice [35] and it may be the underlaying cause of FGF21 resistance and of MAFLD development [36]. Therefore, it could be speculated that the effect of the variant on liver damage progression could be related to obesity.
In addition, plasma levels of KLB and FGF19 were reduced in children with MAFLD and more so in those who carried the rs17618244 KLB minor A allele [11] and correlated with ballooning and fibrosis. Consistently, circulating KLB and FGF19 levels were reduced in adult patients, carrying the at-risk A allele and their concentrations were related to the histological spectrum of MAFLD. All of these findings support the association between the KLB rs17618244 variant, which leads to reduced KLB levels, and liver damage thus confirming what previously observed in pediatric patients with MAFLD [11].
Differently from children who have a less severe hepatic injury, in adults with MAFLD the KLB rs17618244 variant is associated with fibrosis. This effect is consistent with the study by Somm et al. [12], which reported an association between KLB reduction and fibrosis in mice although the role of KLB in inducing MAFLD-related fibrosis in humans remains to be clarified. In keeping with these results, transcriptomic data revealed that the KLB rs17618244 variant was associated with a reduced hepatic expression of KLB which negatively correlated with that of TGF-β, COL1A1 and COL3A1 transcripts suggesting a role of KLB downregulation in inducing pro-fibrogenic genes.
Therefore, in order to investigate whether the KLB rs17618244 variant exerts a pro-fibrogenic effect compared to the ancestor KLB protein which may have an opposite anti-fibrogenic action, we evaluated the effect of the rs17618244 G>A mutation in HSCs whose activation represents the primary driver of hepatic fibrogenesis. We previously demonstrated that the expression of the KLB rs17618244 variant in hepatocytes caused a downregulation of KLB protein and increased intracellular lipid accumulation, whereas the ancestral allele exerted a protective effect against lipids overload, lipotoxicity and inflammation [11]. In the present study, we observed that LX-2 cells which acquire a pro-fibrogenic phenotype after serum or PDGF supplementation, showed reduced KLB mRNA and protein levels thus confirming a strong correlation between low KLB expression and the trans-differentiation from resting to myofibroblast activated cells. Interestingly, in LX-2 cells KLB protein is mainly localized in plasma membrane and into the cytoplasm, with a more pronounced granular pattern distribution that resembles vesicles, compared to HepG2 cells, which showed even an ER and Golgi localization, where the post-translational modifications occur.
In order to understand the role of KLB in HSCs activation, we transfected LX-2 cells with wild type or mutated form of KLB. Similar to data obtained in HepG2 cells [11], the overexpression of exogenous KLBmut caused a downregulation of KLB protein and a concomitant increase of protein degradation, thus suggesting that rs17618244 G>A mutation may cause instability of protein, without affecting intracellular redistribution. Indeed, immunofluorescence images showed higher KLB protein in LX-2_KLBwt cells and it was mainly distributed in the ER and Golgi compartments. Conversely, the overexpression of the mutated plasmid was not able to induce exogenous KLB protein levels and we didn't find differences in its localization compared to endogenous form in LX-2_Vector cells.
Moreover, we observed that LX-2 cells don't release KLB at the extracellular level compared to hepatocytes [11] which may exert a potential paracrine effect on surrounding cells and the soluble KLB is slightly released only by KLBwt cells and not by mutated ones as a possible consequence of reduced KLB protein levels in the latter. We cannot rule out that KLB may exert an intracellular function as coreceptor of nuclear or cytoplasmic receptor, however it is also possible that the translocation of KLB to PM in LX-2 cells may be mediated by vesicles or its inactivation may be mediated by endocytosis processes. All these assumptions are merely speculative and further studies are required to better understand the role of different KLB intra- and extra-cellular distribution.
To further investigate the effect of the KLB rs17618244 mutation on protein stability we performed in silico analyses. The dramatic drop in KLB expression due to the overexpression of mutant KLB gene in LX-2 cells cannot be attributed to structural local destabilization induced by the Arg728Gln per se. Indeed, prediction modeling indicates that this residue is localized on the protein surface and fully exposed to the solvent, thus appearing to be not relevant for the domain fold. However, due to the short distance, the Arg728Gln substitution might interfere with the N-glycosylation at residue Asn611 that is located in a hydrophobic patch exposed on the protein surface. This finding suggests that the N-glycosylation at this asparagine site might be necessary to ensure hydration over the hydrophobic patch to avoid protein aggregation phenomena. Indeed, Asn611 in KLB has been effectively found N-glycosylated in liver cells [37]. Since alterations in N-glycosylation patterns are known to cause changes in proteins’ stability [38], the impaired N-glycosylation at residue Asn611 could explain the reduced KLB expression and the higher degradation rate induced by the Arg728Gln mutation although further experiments are required to confirm this hypothesis. In addition, as well as other secretory proteins, KLB that is putatively N-glycosylated in 11 residues, could be stabilized in ER and a defective N-glycosylation of protein due to the Arg728Gln substitution might alter its cellular distribution and abundance39.
Finally, in vitro data demonstrated that the proliferation rate and the expression of pro-fibrogenic genes as αSMA and COL1A1 were specifically modulated by exogenous KLB. Indeed, we found that LX-2_KLBwt cell were less proliferative and showed reduced αSMA and COL1A1 mRNA levels whilst mutated KLB upregulated both proliferation rate and the expression of the mentioned genes, thus confirming a shift towards a myofibroblast-like phenotype. Since the impairment of the SHP-1/SMAD3 network seems to be involved in fibrosis resolving in HSCs [34], we hypothesized that it could play a role also in modulating the KLB-mediated pro-fibrogenic phenotype in LX-2 cells. Preliminary data suggests that FGF19 interaction with exogenous wild type KLB induced the increased transcription of SHP-1 that binds and inhibits SMAD3 nuclear translocation, thus abrogating its transcriptional activity on pro-fibrogenic genes. Conversely, in the presence of KLBmut the SHP-1 expression is reduced and its binding with SMAD3 is abolished, thus inducing fibrogenesis. However, further studies are required to better explore this mechanism.
In conclusion, the KLB rs17618244 genetic variant is associated with liver damage in adult patients with MAFLD and more so after stratification for the presence of obesity. The Arg728Gln mutation possibly alters the N-glycosylation pattern reducing KLB stability and its levels. Moreover, the mutated KLB modulates the expression of genes involved in collagen deposition partly explaining the impact of the rs17618244 variation on hepatic fibrosis.
Disclosures
The authors declare no conflict of interest relevant to the present manuscript.
Author contributions
The authors’ responsibilities were as follows: Paola Dongiovanni, Anna Alisi: study conceptualization and design, data analysis and interpretation, manuscript drafting. Nadia Panera, Marica Meroni: execution of experiments, data analysis and interpretation, manuscript drafting. Miriam Longo, Annalisa Crudele, Erika Paolini: data analysis and execution of experiments. Emanuele Bellacchio and Valentina D'Oria: data analysis. Luca Valenti, Luca Miele, Anna Ludovica Fracanzani: data collection. Marco Maggioni: histological analysis.
Grant support
Ricerca Finalizzata Ministero della Salute GR-2019-12370172 (NP), Ricerca Corrente Fondazione IRCCS Cà Granda(PD and ALF) and Ricerca Finalizzata Ministero della Salute RF-2013-02358319 (ALF), and Ricerca Corrente and 5 × 1000 Ministero della Salute (AA).
Footnotes
All authors read and approved the final version of the manuscript.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103249.
Contributor Information
Anna Alisi, Email: anna.alisi@opbg.net.
Paola Dongiovanni, Email: paola.dongiovanni@policlinico.mi.it.
Appendix. Supplementary materials
References
- 1.Eslam M., Newsome P.N., Sarin S.K. A new definition for metabolic dysfunction-associated fatty liver disease: an international expert consensus statement. J Hepatol. 2020 doi: 10.1016/j.jhep.2020.03.039. [DOI] [PubMed] [Google Scholar]
- 2.Eslam M., Sanyal A.J., George J. MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology. 2020;158:1999–2014. doi: 10.1053/j.gastro.2019.11.312. .e1. [DOI] [PubMed] [Google Scholar]
- 3.Marchesini G., Brizi M., Bianchi G. Nonalcoholic fatty liver disease: a feature of the metabolic syndrome. Diabetes. 2001;50:1844–1850. doi: 10.2337/diabetes.50.8.1844. [DOI] [PubMed] [Google Scholar]
- 4.Meroni M., Longo M., Rustichelli A. Nutrition and genetics in NAFLD. The Perfect Binomium. 2020;21 doi: 10.3390/ijms21082986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Younossi Z.M., Koenig A.B., Abdelatif D. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 6.Romeo S., Kozlitina J., Xing C. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valenti L., Alisi A., Galmozzi E. I148M patatin-like phospholipase domain-containing 3 gene variant and severity of pediatric nonalcoholic fatty liver disease. Hepatology. 2010;52:1274–1280. doi: 10.1002/hep.23823. [DOI] [PubMed] [Google Scholar]
- 8.Dongiovanni P., Romeo S., Valenti L. Hepatocellular carcinoma in nonalcoholic fatty liver: role of environmental and genetic factors. World J Gastroenterol. 2014;20:12945–12955. doi: 10.3748/wjg.v20.i36.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dongiovanni P., Petta S., Maglio C. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015;61:506–514. doi: 10.1002/hep.27490. [DOI] [PubMed] [Google Scholar]
- 10.Mancina R.M., Dongiovanni P., Petta S. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology. 2016;150:1219–1230. doi: 10.1053/j.gastro.2016.01.032. .e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dongiovanni P., Crudele A., Panera N. β-Klotho gene variation is associated with liver damage in children with NAFLD. J Hepatol. 2020;72:411–419. doi: 10.1016/j.jhep.2019.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Somm E., Henry H., Bruce S.J. β-Klotho deficiency shifts the gut-liver bile acid axis and induces hepatic alterations in mice. Am J Physiol Endocrinol Metab. 2018;315:E833–E847. doi: 10.1152/ajpendo.00182.2018. [DOI] [PubMed] [Google Scholar]
- 13.Schumann G., Liu C., O'Reilly P., et al. KLB is associated with alcohol drinking, and its gene product β-Klotho is necessary for FGF21 regulation of alcohol preference. 2016;113:14372–7. [DOI] [PMC free article] [PubMed]
- 14.Haidari F., Asadi M. Evaluation of the effect of oral taurine supplementation on fasting levels of fibroblast growth factors, β-Klotho co-receptor, some biochemical indices and body composition in obese women on a weight-loss diet: a study protocol for a double-blind. Random Control Trial. 2019;20:315. doi: 10.1186/s13063-019-3421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang F., Yu L., Lin X. Minireview: roles of Fibroblast growth factors 19 and 21 in metabolic regulation and chronic diseases. Mol Endocrinol. 2015;29:1400–1413. doi: 10.1210/me.2015-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markan K.R., Naber M.C., Small S.M. FGF21 resistance is not mediated by downregulation of beta-klotho expression in white adipose tissue. Mol Metab. 2017;6:602–610. doi: 10.1016/j.molmet.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inagaki T., Choi M., Moschetta A. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Moschetta A., Kliewer S.A. Weaving betaKlotho into bile acid metabolism. J Clin Invest. 2005;115:2075–2077. doi: 10.1172/JCI26046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puri P., Daita K., Joyce A., et al. The presence and severity of nonalcoholic steatohepatitis is associated with specific changes in circulating bile acids. 2018;67:534–48. [DOI] [PMC free article] [PubMed]
- 20.Jahnel J., Zöhrer E., Alisi A. Serum bile acid levels in children with nonalcoholic fatty liver disease. J Pediatr Gastroenterol Nutr. 2015;61:85–90. doi: 10.1097/MPG.0000000000000774. [DOI] [PubMed] [Google Scholar]
- 21.Owen B.M., Mangelsdorf D.J., Kliewer S.A. Tissue-specific actions of the metabolic hormones FGF15/19 and FGF21. Trends Endocrinol Metab. 2015;26:22–29. doi: 10.1016/j.tem.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dushay J., Chui P.C., Gopalakrishnan G.S. Increased fibroblast growth factor 21 in obesity and nonalcoholic fatty liver disease. Gastroenterology. 2010;139:456–463. doi: 10.1053/j.gastro.2010.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gómez-Ambrosi J., Gallego-Escuredo J.M., Catalán V. FGF19 and FGF21 serum concentrations in human obesity and type 2 diabetes behave differently after diet- or surgically-induced weight loss. Clin Nutr. 2017;36:861–868. doi: 10.1016/j.clnu.2016.04.027. [DOI] [PubMed] [Google Scholar]
- 24.Gallego-Escuredo J.M., Gómez-Ambrosi J., Catalan V. Opposite alterations in FGF21 and FGF19 levels and disturbed expression of the receptor machinery for endocrine FGFs in obese patients. Int J Obes (Lond) 2015;39:121–129. doi: 10.1038/ijo.2014.76. [DOI] [PubMed] [Google Scholar]
- 25.Jiao N., Baker S.S., Chapa-Rodriguez A. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut. 2018;67:1881–1891. doi: 10.1136/gutjnl-2017-314307. [DOI] [PubMed] [Google Scholar]
- 26.Nobili V., Alisi A., Newton K.P. Comparison of the phenotype and approach to pediatric vs adult patients with nonalcoholic fatty liver disease. Gastroenterology. 2016;150:1798–1810. doi: 10.1053/j.gastro.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleiner D.E., Brunt E.M., Van Natta M. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 28.Dongiovanni P., Petta S., Mannisto V. Statin use and non-alcoholic steatohepatitis in at risk individuals. J Hepatol. 2015;63:705–712. doi: 10.1016/j.jhep.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Dongiovanni P., Meroni M., Baselli G., et al. PCSK7 gene variation bridges atherogenic dyslipidemia with hepatic inflammation in NAFLD patients. 2019;60:1144–53. [DOI] [PMC free article] [PubMed]
- 30.Lee Y.A., Wallace M.C., Friedman S.L. Pathobiology of liver fibrosis: a translational success story. Gut. 2015;64:830–841. doi: 10.1136/gutjnl-2014-306842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo Re O., Panebianco C., Porto S., et al. Fasting inhibits hepatic stellate cells activation and potentiates anti-cancer activity of Sorafenib in hepatocellular cancer cells. 2018;233:1202–12. [DOI] [PubMed]
- 32.Tsuchida T., Friedman S.L. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol. 2017;14:397–411. doi: 10.1038/nrgastro.2017.38. [DOI] [PubMed] [Google Scholar]
- 33.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46:W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carino A., Biagioli M., Marchianò S. Disruption of TFGβ-SMAD3 pathway by the nuclear receptor SHP mediates the antifibrotic activities of BAR704, a novel highly selective FXR ligand. Pharmacol Res. 2018;131:17–31. doi: 10.1016/j.phrs.2018.02.033. [DOI] [PubMed] [Google Scholar]
- 35.Fisher F.M., Chui P.C., Antonellis P.J. Obesity is a fibroblast growth factor 21 (FGF21)-resistant state. Diabetes. 2010;59:2781–2789. doi: 10.2337/db10-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji F., Liu Y., Hao J.G. KLB gene polymorphism is associated with obesity and non-alcoholic fatty liver disease in the Han Chinese. Aging (Albany NY) 2019;11:7847–7858. doi: 10.18632/aging.102293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao H., Hwang J.E., Wu R. Mass spectrometric analysis of the N-glycoproteome in statin-treated liver cells with two lectin-independent chemical enrichment methods. Int J Mass Spectrom. 2018;429:66–75. doi: 10.1016/j.ijms.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H.S., Qi Y., Im W. Effects of N-glycosylation on protein conformation and dynamics: protein data bank analysis and molecular dynamics simulation study. Sci Rep. 2015;5:8926. doi: 10.1038/srep08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.