Abstract
Objectives
To facilitate clinical and translational research, imaging and non-imaging clinical data from multiple disparate systems must be aggregated for analysis. Study participant records from various sources are linked together and to patient records when possible to address research questions while ensuring patient privacy. This paper presents a novel tool that pseudonymizes participant identifiers (PIDs) using a researcher-driven automated process that takes advantage of application-programming interface (API) and the Perl Open-Source Digital Imaging and Communications in Medicine Archive (POSDA) to further de-identify PIDs. The tool, on-demand cohort and API participant identifier pseudonymization (O-CAPP), employs a pseudonymization method based on the type of incoming research data.
Methods
For images, pseudonymization of PIDs is done using API calls that receive PIDs present in Digital Imaging and Communications in Medicine (DICOM) headers and returns the pseudonymized identifiers. For non-imaging clinical research data, PIDs provided by study principal investigators (PIs) are pseudonymized using a nightly automated process. The pseudonymized PIDs (P-PIDs) along with other protected health information is further de-identified using POSDA.
Results
A sample of 250 PIDs pseudonymized by O-CAPP were selected and successfully validated. Of those, 125 PIDs that were pseudonymized by the nightly automated process were validated by multiple clinical trial investigators (CTIs). For the other 125, CTIs validated radiologic image pseudonymization by API request based on the provided PID and P-PID mappings.
Conclusions
We developed a novel approach of an on-demand pseudonymization process that will aide researchers in obtaining a comprehensive and holistic view of study participant data without compromising patient privacy.
Keywords: Data Management, De-identification, Multimedia, PACS, Semantic Web
I. Introduction
Translational research aims to improve population health outcomes by applying and building upon the knowledge from basic scientific research to address critical medical needs. Electronic Health Record (EHR) use has become ubiquitous in clinical care, revolutionizing patient data collection and storage [1]. The data from EHRs and other related systems can be leveraged to facilitate translational research [2,3].
Frequently, the data leveraged for clinical and translational research comes from multiple disparate systems and must be aggregated for analysis, requiring that individual patient records from various sources are linked together using a unique identifier [4]. For example, the medical record number (MRN) is one of the most commonly used variables for uniquely identifying patients within the medical record, and it is often used as a link to merge clinical data from multiple sources. In addition to the MRN, other identifiers, such as patient name, date of birth, and so forth, are used to link and identify patient records using entity resolution tools, such as OYSTER [5,6].
Timely availability of medical images is also critical because their interpretation significantly improves health outcomes [7,8]. Radiologic images (e.g., computed tomography, magnetic resonance imaging) are usually stored in a system that is separate from the EHR, a picture archiving and communication system (PACS) [9]. Clinical trials may use a PACS or store images in a research image repository. Digital Imaging and Communications in Medicine (DICOM) is the international standard for medical images and related information [10]. Image metadata contains patient identifiers encoded within the DICOM file, including MRNs and participant identifiers (PIDs).
Aggregating data from heterogeneous sources regularly raises concerns because physicians and clinical researchers must comply with the Health Insurance Portability and Accountability Act (HIPAA) or the General Data Protection Regulation (GDPR) to ensure patient privacy and confidentiality [11,12]. To exchange and share data for research purposes without compromising patient privacy and confidentiality, a standardized process is required that will allow for the de-identification of the data while maintaining the linkage between records from the various datasets [11,13]. The more datasets there are that require linkage, the more cumbersome the process will be.
De-identification refers to the technical approaches used to protect privacy and facilitate the secondary use of health data by removing any association within the dataset that could tie the data back to the individual, i.e., protected health information (PHI) [14–16]. The link between the individual and the de-identified data is maintained by an authorized entity. Naive approaches to de-identification simply remove all personal data, a technique referred to as anonymization. The anonymization process fully de-identifies a dataset in a manner that does not allow the data to be re-identified because the link between individual and the de-identified data is never maintained [14,15]. This process makes it impossible to recombine or merge source data with additional heterogeneous sources later.
Another de-identification technique is pseudonymization, in which identifiers such as PIDs, service dates, and patients addresses are replaced by one or more artificial identifiers or pseudonyms [16]. In this case, the modified data cannot be identified without knowing a certain key [14,16,17]. To address the limitations brought on by anonymization, pseudonymization techniques can be used to obscure the PIDs but still allow for re-identification for the purposes of record linkage and merging datasets from disparate sources in accordance with Institutional Review Board (IRB) guidelines.
Traditionally, diagnostic images used in research are deidentified manually or by using various DICOM de-identification toolkits [18] or the Perl Open-Source Digital Imaging and Communications in Medicine Archive (POSDA) tools suite [19,20]. In addition, there are pseudonymization services that provide secure and legislation-compliant solutions to ensure that (1) participants can be identified only by the person in charge of identifying data, and (2) patients who appear in multiple studies obtain the same pseudonym and are not considered as two different persons [21]. However, existing commercial tools, such as the IBM Optim Data Privacy Solution and the Oracle Data Masking Pack assign the same pseudonym to a participant enrolled in multiple clinical studies [22], which can negatively impact privacy according to our study.
We propose a study-specific, researcher-driven pseudonymization process, housed in a clinical data warehouse (CDW), by which the researcher initiates pseudonymization using on-demand automated methods to generate pseudonyms from PIDs. We employed an application-programming interface (API)-driven method to generate pseudonyms. The CDW acts as a trusted third party, or honest broker, to generate and manage the pseudonyms needed for the de-identification of data. The representational state transfer (RESTful) API enables authorized researchers to communicate programmatically with the CDW [23].
At the University of Arkansas for Medical Sciences (UAMS), the Arkansas Image Enterprise Systems (ARIES; https://aries.uams.edu) pilot project was initiated to create a research data repository to maintain de-identified data from collaborators that can be queried for secondary use. For the research data collected from multiple studies, PIDs are first pseudonymized, and then the protected health information (PHI) is de-identified using POSDA before the data is loaded to the ARIES database.
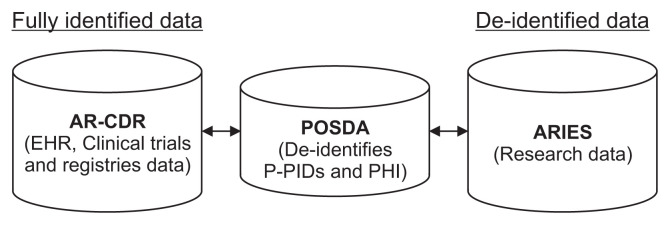
The mappings between the PIDs and pseudonyms of participant identifiers (P-PIDs) are stored in the Arkansas Clinical Data Repository (AR-CDR), UAMS’s clinical data warehouse [24]. ARIES can be linked to fully identified data using the mappings in the AR-CDR (Figure 1).
Figure 1.
ARIES, POSDA, and AR-CDR setup at the University of Arkansas for Medical Sciences. De-identified research data in ARIES can be linked back to fully identified AR-CDR data using (1) P-PID to PID mappings maintained by AR-CDR and (2) de-identified P-PID to P-PID mappings from POSDA. ARIES: Arkansas Image Enterprise Systems, POSDA: Perl Open-Source Digital Imaging and Communications in Medicine Archive, AR-CDR: Arkansas Clinical Data Repository, PID: participant identifiers, P-PID: pseudonyms of participant identifiers, EHR: electronic health record, PHI: protected health information.
ARIES integrates not only diagnostic images and image-derived features, but also motor assessments, cognitive assessments, clinical rating scales, demographics, and clinical data. ARIES manages the data and the associated metadata using shared semantic representations based on axiomatically rich ontologies [25]. Ontologies are used to instantiate a knowledge graph that combines instance data from study cohorts in a triple-store database that supports reasoning over and querying of the integrated data [25]. This approach removes obstacles to working with different source representations for the same type of information, connecting and interpreting different types of data that are about the same phenomena, and combining diverse datasets that are about the same individuals.
In this paper, we describe the on-demand cohort and API participant identifier pseudonymization (O-CAPP) tool developed at UAMS to pseudonymize PIDs automatically. In addition, we will discuss the importance of this novel approach to accomplish data integration in ARIES.
II. Methods
The IRB classified this study as exempt and approved this project. Patients’ data used were obtained under IRB approval at the University of Arkansas for Medical Sciences (No. 261743).
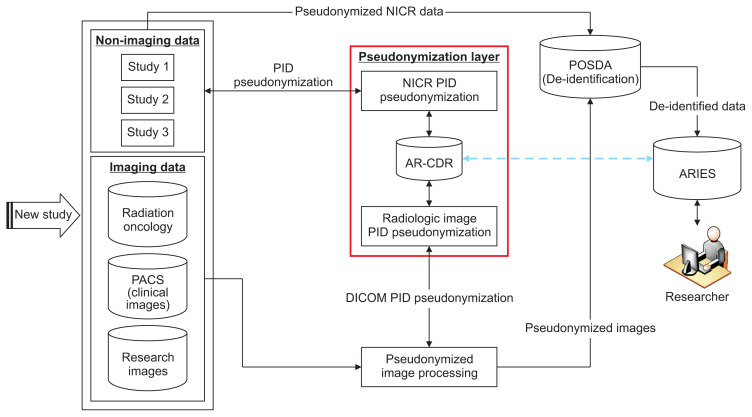
At UAMS, we developed a single-center, multi-study setup to de-identify and integrate research data from multiple disparate systems. This process of receiving longitudinal research data, O-CAPP’s pseudonymization of imaging and non-imaging clinical research (NICR) PIDs, de-identifying data using the POSDA tool suite and related procedures, and transforming it to ARIES is outlined in Figure 2. Mapping between the PIDs and P-PIDs is securely stored in AR-CDR for future associations with the ARIES database to identify participants enrolled in multiple studies.
Figure 2.
Pipeline for receiving heterogeneous-longitudinal data, pseudonymization of PIDs for NICR and diagnostic imaging data, POSDA P-PIDs and PHI de-identification, and transformation into ARIES database for secondary data use. The pseudonymization algorithm is hosted in AR-CDR. The details of pseudonymization using AR-CDR data for both NICR pseudonymization (NICR-P) and radiologic image pseudonymization (RIP) requests is shown in Figure 3. The “Pseudonymization Layer” represents O-CAPP’s framework to receive PIDs, execute the pseudonymization algorithm, and return P-PIDs. The process is presented in detail in Figure 4. The blue dotted line represents de-identified research data in ARIES that can be linked back to fully identified AR-CDR data using the mappings maintained in AR-CDR and POSDA. PID: participant identifiers, P-PID: pseudonyms of participant identifiers, AR-CDR: Arkansas Clinical Data Repository, POSDA: Perl Open-Source Digital Imaging and Communications in Medicine Archive, NICR: non-imaging clinical research, ARIES: Arkansas Image Enterprise Systems, PHI: protected health information, O-CAPP: participant identifier pseudonymization, PACS: picture archiving and communication system, DICOM: Digital Imaging and Communications in Medicine.
O-CAPP’s pseudonymization method is based on the type of incoming research data. For NICR data, pseudonymization of PIDs occurs through an automated nightly process, referred to as the NICR pseudonymization (NICR-P) process, as shown in Figure 3. Conversely, for radiologic images, PIDs are pseudonymized via API calls, referred to as the radiologic image pseudonymization (RIP) process.
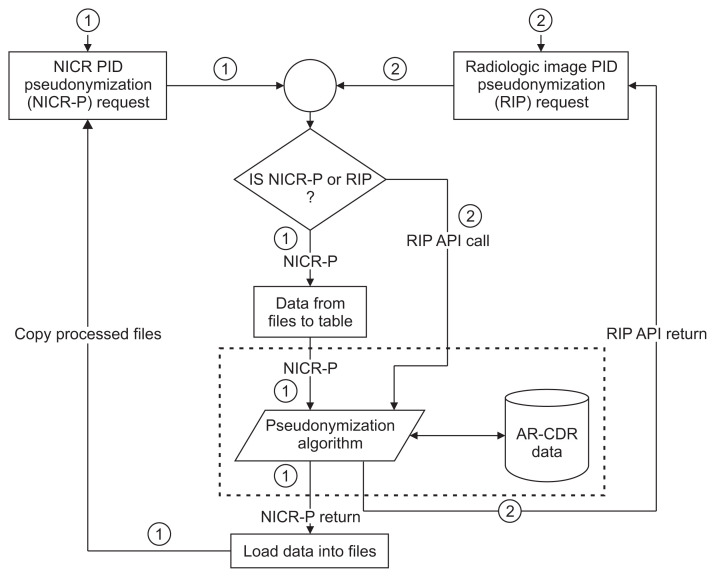
Figure 3.
Flow chart of the O-CAPP pseudonymization process based on the type of incoming research data: NICR vs. radiologic imaging data. Paths ① and ② represent the steps involved in receiving pseudonymization requests, pseudonymizing PIDs using AR-CDR data, and returning the P-PIDs for NICR and radiologic imaging data, respectively. O-CAPP: participant identifier pseudonymization, NICR: non-imaging clinical research, PID: participant identifiers, P-PID: pseudonyms of participant identifiers, AR-CDR: Arkansas Clinical Data Repository, API: application-programming interface.
(1) NICR-P process: In this process, as outlined by Path ① in Figure 3, principal investigators (PIs) provide a list of PIDs via secure data transfer, which is stored on the AR-CDR file server. O-CAPP’s nightly automated process scans the dataset for each study and generates associated P-PIDs. The NICR-P process requires the following data elements to perform pseudonymization: STUDY_ID, STUDY_NAME, MRN, and ALTERNATE_ID. O-CAPP reads data from the source files and loads the source data into a pre-process staging table. P-PIDs are then generated and stored in the post-process staging table. The PIDs and associated P-PIDs are copied from the staging table to source files in respective study directories.
(2) RIP process: In this process, radiologic images are pseudonymized by O-CAPP via a secured API call. Each API request initiates the pseudonymization algorithm by submitting the PIDs available within the image’s DICOM header. Upon completion of pseudonymization, the generated P-PIDs are returned in response to the API call, which will replace PIDs in the respective DICOM header. The process of extracting a PID from a DICOM header, initiating an API request by submitting the PID, and replacing the PID with the returned P-PID in the DICOM header is done by the “pseudonymized image processing” component. All the API requests and responses for pseudonymization are securely stored in the AR-CDR’s audit table. The table holds both PIDs received and P-PIDs returned. The full RIP process is represented by Path ② in Figure 3.
1. O-CAPP Framework Setup at UAMS
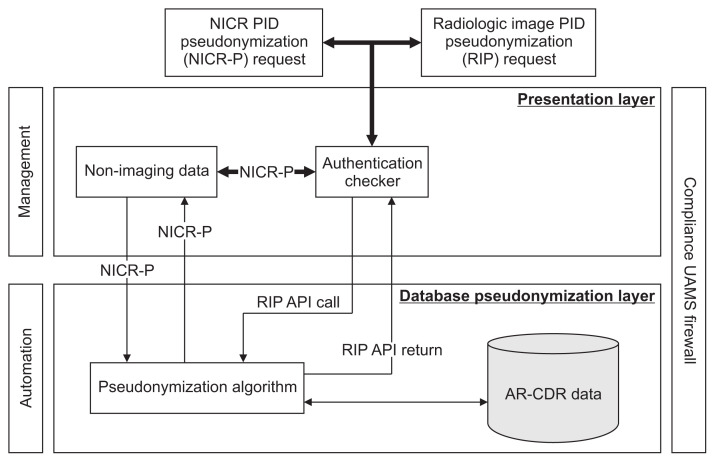
The O-CAPP framework for receiving pseudonymization requests and the process to pseudonymize PIDs (Figure 4) is divided into two layers, namely, the presentation layer (PL) and the database pseudonymization layer (DPL).
Figure 4.
O-CAPP’s framework for receiving pseudonymization requests and the process to pseudonymize PIDs. Presentation Layer authenticates requestors and submits the pseudonymization request to Database Pseudonymization Layer for P-PID generation. Presentation Layer returns P-PIDs to requestor. O-CAPP: participant identifier pseudonymization, PID: participant identifiers, P-PID: pseudonyms of participant identifiers, NICR: non-imaging clinical research, AR-CDR: Arkansas Clinical Data Repository, UAMS: University of Arkansas for Medical Sciences, API: application-programming interface.
The PL allows the requestor to communicate with the DPL based on the type of research data requiring pseudonymization. The PL facilitates two-way communication with the downstream DPL that receives PIDs and returns P-PIDs. The DPL receives the PIDs, executes the pseudonymization process, and generates P-PIDs. This layer consists of a pseudonymization unit, which contains the pseudonymization algorithm that runs on two AR-CDR tables: PATIENT and PATIENT_ID_MAP.
2. O-CAPP’s Pseudonymization Algorithm
The basic working mechanism of the pseudonymization algorithm is to check whether the requested participant record already exist in any of the studies available in the AR-CDR. This is achieved using a record-linkage toolkit built on an open-source entity resolution system, OYSTER [5,6,26]. The entity resolution component of OYSTER is configured to match participant records based on 18 custom-tailored identity rules. These rules employ participant attributes, such as first name, last name, date of birth (DOB), MRN, Social Security number (SSN), and so forth, for record linkage. For example, (1) match on participant’s first name, last name, and DOB, (2) match on SSN, DOB, and first name, and (3) match on SSN, DOB, MRN, and Soundex match on first name, etc. To link two participant records, the records should match on at-least three of the aforementioned attributes. Within the AR-CDR, the record-linkage toolkit assigns a unique identifier (UID) to each participant and stores one record per participant in the PATIENT table. The PATIENT_ID_MAP table stores all the identifiers a participant might have from multiple sources. For example, as shown in Table 1, a single participant has UID “DW01” and three P-PIDs “1009”, “1015”, and “5001”, each associated with the corresponding data sources “Clinical System”, “Clinical Trial 1”, and “Clinical Trial 2”.
Table 1.
Partial columns of PATIENT_ID_MAP table, which holds all PIDs and P-PIDs that a participant might have per clinical systems or studies along with a UID
| UID | P-PID | Study name | PID-type | PID |
|---|---|---|---|---|
| DW01 | 1009 | Clinical System | MRN | M0123 |
| DW01 | 1015 | Clinical Trial 1 | PID | CTRA901 |
| DW01 | 5001 | Clinical Trial 2 | PID | CTRB501 |
| DW02 | 2019 | Clinical System | MRN | M0977 |
| DW02 | 8020 | Clinical Trial 1 | PID | CTRX881 |
| DW02 | 7079 | Clinical Trial 2 | PID | CTRY015 |
PID: participant identifiers, P-PID: pseudonyms of participant identifiers, UID: unique identifier, MRN: medical record number.
The pseudonymization algorithm flow starts by checking whether the incoming participant record exists in the AR-CDR’s PATIENT and PATIENT_ID_MAP tables. Based on the results, one of the below three cases is executed.
(1) Case 1: In cases in which there is no record of the participant in either of the AR-CDR patient tables, the algorithm assumes it is a new participant. A record for this participant is inserted into the PATIENT and PATIENT_ID_MAP tables. A UID is generated for this participant, and a study-specific P-PID is generated and returned to the PL.
(2) Case 2: In cases in which a record of the participant is located in both AR-CDR patient tables, but there is no study-specific identifier, a P-PID for the study is generated and stored in the PATIENT_ID_MAP table and returned to the PL.
(3) Case 3: In cases in which a record already exists for the participant in both AR-CDR patient tables for the specific study, the existing P-PID for the participant is returned to the PL.
III. Results
At UAMS, three studies are actively using the O-CAPP tool to pseudonymize an enrolled participant’s PID using either of the pseudonymization methods (NICR-P and RIP). The tool has been used to pseudonymize 2,225 participants. The goal of the O-CAPP is to identify participant enrolled in multiple studies using PID and P-PID mappings, allowing researchers to gain a holistic view of participants’ data collected from multiple studies and clinical facts from EHR upon IRB approval without compromising participant privacy.
To validate the successful implementation of O-CAPP’s pseudonymization algorithm, we validated PID and P-PID mappings stored in the AR-CDR against the identifiers collected by the PIs for their studies. Of the 2,225 PIDs that were pseudonymized by O-CAPP, we randomly selected 250 PIDs (n = 2,225, confidence interval = 95%, ɛ = 5%) for validation by multiple clinical trial investigators (CTIs) based on a two-point scale: matched and not-matched. Both pseudonymization methods (NICR-P and RIP) were validated using 125 samples each from the 250-validation sample. To validate the NICR-P process, mappings of 125 participants’ PIDs and P-PIDs were extracted from the AR-CDR. For the 75 participants, P-PIDs were provided to the CTIs for verification. For these P-PIDs, CTIs provided PIDs from their study records for comparison using the aforementioned scale. For the remaining 50 participants, PIDs were provided to the CTIs, and the CTIs provided P-PIDs from their records. Both result sets received from the CTIs were then matched against the AR-CDR mappings, and we found that the accuracy was 100%. To validate the RIP process, the PIDs and P-PIDs mappings were provided to the CTIs. The CTIs made O-CAPP API requests for the given participants, the P-PIDs returned by the API calls were then matched to the provided PID and P-PID mappings. Based on the results, the accuracy was reported as 100%.
IV. Discussion
This paper presented the O-CAPP tool, which will be an integral cog in the larger scheme of creating a de-identified ARIES repository for clinical and translational research. Deidentified data from multiple disparate systems that include imaging, clinical trials, patient registries, and clinical information systems will aide researchers in gaining a comprehensive and holistic view of patient data, which is seldom available with disparate systems [24,27]. For instance, a publicly available COVID-19 clinical dataset including radiology and CT images in The Cancer Imaging Archive (TCIA) was pseudonymized and de-identified using the O-CAPP and POSDA tools, respectively [28]. The possibility of participant re-identification is also an option with this process upon IRB approval and consent. Moreover, clinical facts available in the AR-CDR for participants can be easily merged with data collected from multiple clinical trials. All P-PIDs per study can be identified using a single AR-CDR UID as shown in Table 1.
O-CAPP pseudonymizes PIDs using the API and a nightly automated process. Calling the API for single PID pseudonymization is feasible, but for batch processing with hundreds of PIDs, using a nightly automated process is more efficient. Moreover, most researchers submitting data are not technical users to call the API. PIDs are considered PHI, and the API used to initiate the pseudonymization process must be secure. Hence, a secure API was developed to protect patient privacy. The API can be accessed within the UAMS firewalls only, and every API request is logged along with the requestor’s credentials in an audit log.
Periodic validation of the P-PIDs is required in the future. If a patient is assigned an incorrect P-PID then manual correction of the error may be required in the AR-CDR. The O-CAPP tool was built to detect participant enrollment across multiple studies and trials, but currently the tool is not being used to its full extent because the datasets available in ARIES are limited to only a few clinical trial sites with a few thousand patients. Therefore, we are not able to assess the overall system performance. Moreover, we have not yet encountered a patient enrolled in multiple studies; therefore, we were unable to verify whether O-CAPP would flag such patients.
A scenario that requires further investigation is the case of a participant that is accidentally assigned more than one PID in the same study, which is common in longitudinal studies spanning several years. In this case, multiple pseudonymization requests need to be made to account for the multiple PIDs. However, O-CAPP would need to identify and match each of these requests and assign only a single P-PID to the participant for that particular study. In the future, we intend to address this issue by modifying O-CAPP to accept multiple identifiers that are separated with a comma delimiter so that patient pseudonymization can be done in a single request.
The ultimate goal of O-CAPP is to identify PIDs and replace them with P-PIDs automatically before submission to POSDA. However, we were unable to validate this scenario because the study data are currently maintained as individual files and are not in a structured database.
Currently accessing the API outside the UAMS firewall has been disabled. Upon further testing, the API will be opened to external users and will be accessible after successful authentication via the UAMS active directory server’s Lightweight Directory Access Protocol (LDAP). O-CAPP solves the single-center, multi-study disparate data linking problem; however, the problem of integrating multi-center studies is still unresolved. The problem of obtaining a comprehensive view of a patient participating in multi-center studies can be resolved by developing a unique national patient identifier, which is no longer illegal due to recent legislation passed by the United States House of Representatives [29,30].
Our study demonstrates the value of O-CAPP to pseudonymize, de-identify and integrate heterogeneous datasets into a common repository using POSDA tools. In addition, mapping between PIDs and P-PIDs helps identify diverse datasets that are about the same individual. As our work with ARIES continues to expand to include additional studies and datasets from various systems, the full applications of O-CAPP can be validated. The ARIES repository will support ontology-driven data-querying capabilities to expand the scope of secondary research.
To our knowledge, this is the first study on the generation of pseudonyms using an API and automated processes devoted to simplifying the process of merging heterogeneous sources for research in adherence to patient safety and privacy.
Acknowledgments
This work was supported in part by the National Cancer Institute, National Institute of Health (No. 3U24CA215109-02S1, 1U24CA215109) and National Center for Advancing Translational Sciences (No. UL1 TR003107). Resources for this study was provided by the Arkansas Clinical Data Repository (AR-CDR) maintained by the Department of Biomedical Informatics in the College of Medicine at the University of Arkansas for Medical Sciences.
Footnotes
Conflict of Interest
No potential conflict of interest relevant to this article was reported.
References
- 1.Evans RS. Electronic Health Records: then, now, and in the future. Yearb Med Inform. 2016;Suppl 1(Suppl 1):S48–61. doi: 10.15265/IYS-2016-s006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordo AH, Levaux HP, Becnel LB, Galvez J, Rao P, Stem K, et al. Use of EHRs data for clinical research: historical progress and current applications. Learn Health Syst. 2019;3(1):e10076. doi: 10.1002/lrh2.10076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penning ML, Blach C, Walden A, Wang P, Donovan KM, Garza MY, et al. Near real time EHR data utilization in a clinical study. Stud Health Technol Inform. 2020;270:337–41. doi: 10.3233/SHTI200178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gliklich RE, Dreyer NA, Leavy MB. Registries for evaluating patient outcomes: a user’s guide. 3rd ed. Rockville (MD): Agency for Healthcare Research and Quality; 2014. [PubMed] [Google Scholar]
- 5.Nelson E, Talburt JR. Entity resolution for longitudinal studies in education using OYSTER. Proceedings of 2011 Information and Knowledge Engineering Conference (IKE); 2011 Jul 18–20; Las Vegas, NV. pp. 286–90. [Google Scholar]
- 6.Talburt JR, Zhou Y. A practical guide to entity resolution with OYSTER. In: Sadiq S, editor. Handbook of data quality. Heidelberg, Germany: Springer; 2013. pp. 235–70. [Google Scholar]
- 7.Erickson BJ, Buckner JC. Imaging in clinical trials. Cancer Inform. 2007;4:13–8. [PMC free article] [PubMed] [Google Scholar]
- 8.Grant JB, Hayes RP, Baker DW, Cangialose CB, Kieszak SM, Ballard DJ. Informatics, imaging, and healthcare quality management: imaging quality improvement opportunities and lessons learned form HCFA’s Health Care Quality Improvement Program. Clin Perform Qual Health Care. 1997;5(3):133–9. [PubMed] [Google Scholar]
- 9.Strickland NH. PACS (picture archiving and communication systems): filmless radiology. Arch Dis Child. 2000;83(1):82–6. doi: 10.1136/adc.83.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DICOM standards [Internet] Arlington (VA): DICOM; c2020. Digital Imaging and Communications in Medicine. [cited 2020 Oct 23]. Available from: https://www.dicomstandard.org/current. [Google Scholar]
- 11.Nass SJ, Levit LA, Gostin LO. Beyond the HIPAA Privacy Rule: enhancing privacy, improving health through research. Washington (DC): National Academies Press; 2009. [PubMed] [Google Scholar]
- 12.Linden T, Khandelwal R, Harkous H, Fawaz K. The privacy policy landscape after the GDPR. Proc Priv Enhanc Technol. 2020;(1):47–64. [Google Scholar]
- 13.Nelson GS. Practical implications of sharing data: a primer on data privacy, anonymization, and de-identification. Proceedings of SAS Global Forum; 2015 Apr 26–29; Dallas, TX. pp. 1–23. [Google Scholar]
- 14.Kushida CA, Nichols DA, Jadrnicek R, Miller R, Walsh JK, Griffin K. Strategies for de-identification and anonymization of electronic health record data for use in multicenter research studies. Med Care. 2012;50(Suppl):S82–101. doi: 10.1097/MLR.0b013e3182585355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chevrier R, Foufi V, Gaudet-Blavignac C, Robert A, Lovis C. Use and understanding of anonymization and de-identification in the biomedical literature: scoping review. J Med Internet Res. 2019;21(5):e13484. doi: 10.2196/13484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kayaalp M. Modes of de-identification. AMIA Annu Symp Proc. 2018;2017:1044–50. [PMC free article] [PubMed] [Google Scholar]
- 17.Riedl B, Neubauer T, Goluch G, Boehm O, Reinauer G, Krumboeck A. A secure architecture for the pseudonymization of medical data. Proceedings of the 2nd International Conference on Availability, Reliability and Security (ARES); 2007 Apr 10–13; Vienna, Austria. pp. 318–24. [Google Scholar]
- 18.Aryanto KY, Oudkerk M, van Ooijen PM. Free DICOM de-identification tools in clinical research: functioning and safety of patient privacy. Eur Radiol. 2015;25(12):3685–95. doi: 10.1007/s00330-015-3794-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett W, Smith K, Jarosz Q, Nolan T, Bosch W. Reengineering workflow for curation of DICOM datasets. J Digit Imaging. 2018;31(6):783–91. doi: 10.1007/s10278-018-0097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perl Open Source Digital Imaging and Communications in Medicine Archive (POSDA) [Internet] [place unknown] github.com. 2019 [cited at 2020 Sep 17]. Available from: https://github.com/UAMS-DBMI/PosdaTools.
- 21.Bruland P, Doods J, Brix T, Dugas M, Storck M. Connecting healthcare and clinical research: workflow optimizations through seamless integration of EHR, pseudonymization services and EDC systems. Int J Med Inform. 2018;119:103–8. doi: 10.1016/j.ijmedinf.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 22.Meystre SM, Lovis C, Burkle T, Tognola G, Budrionis A, Lehmann CU. Clinical data reuse or secondary use: current status and potential future progress. Yearb Med Inform. 2017;26(1):38–52. doi: 10.15265/IY-2017-007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fielding RT, Taylor RN. Architectural styles and the design of network-based software architectures. Irvine (CA): University of California; 2000. [Google Scholar]
- 24.Baghal A, Zozus M, Baghal A, Al-Shukri S, Prior F. Factors associated with increased adoption of a research data warehouse. Stud Health Technol Inform. 2019;257:31–5. [PubMed] [Google Scholar]
- 25.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–5. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Syed H, Talburt J, Liu F, Pullen D, Wu N. Developing and refining matching rules for entity resolution. Proceedings of the International Conference on Information and Knowledge Engineering (IKE); 2012 Jul 16–19; Las Vegas, NV. [Google Scholar]
- 27.Foran DJ, Chen W, Chu H, Sadimin E, Loh D, Riedlinger G, et al. Roadmap to a comprehensive clinical data warehouse for precision medicine applications in oncology. Cancer Inform. 2017;16:1176935117694349. doi: 10.1177/1176935117694349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Cancer Imaging Archive. Chest imaging with clinical and genomic correlates representing a rural COVID-19 positive population (COVID-19-AR) [Internet] Fayetteville (AR): The Cancer Imaging Archive; 2020. [cited at 2020 Sep 17]. Available from: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=70226443. [Google Scholar]
- 29.Sood HS, Bates DW, Halamka JD, Sheikh A. Has the time come for a unique patient identifier for the U.S.? NEJM Catal. 2018;4(1):1–4. [Google Scholar]
- 30.Luthi S, Cohen JK. House votes to overturn ban on national patient identifier [Internet] Chicago (IL): Modern Healthcare; 2019. [cited at 2020 Oct 22]. Available from: https://www.modernhealthcare.com/politicspolicy/house-votes-overturn-ban-national-patientidentifier. [Google Scholar]