Abstract
The field of phosphoinositide signaling has expanded significantly in recent years. Phosphoinositides (also known as phosphatidylinositol phosphates or PIPs) are universal signaling molecules that directly interact with membrane proteins or with cytosolic proteins containing domains that directly bind phosphoinositides and are recruited to cell membranes. Through the activities of phosphoinositide kinases and phosphoinositide phosphatases, seven distinct phosphoinositide lipid molecules are formed from the parent molecule, phosphatidylinositol. PIP signals regulate a wide range of cellular functions, including cytoskeletal assembly, membrane binding and fusion, ciliogenesis, vesicular transport, and signal transduction. Given the many excellent reviews on phosphoinositide kinases, phosphoinositide phosphatases, and PIPs in general, in this review, we discuss recent studies and advances in PIP lipid signaling in the retina. We specifically focus on PIP lipids from vertebrate (e.g., bovine, rat, mouse, toad, and zebrafish) and invertebrate (e.g., Drosophila, horseshoe crab, and squid) retinas. We also discuss the importance of PIPs revealed from animal models and human diseases, and methods to study PIP levels both in vitro and in vivo. We propose that future studies should investigate the function and mechanism of activation of PIP-modifying enzymes/phosphatases and further unravel PIP regulation and function in the different cell types of the retina.
Supplementary key words: light activation, phosphoinositide kinases, phosphoinositide phosphatases, phosphoinositide binding proteins
Abbreviations: CNG, cyclic nucleotide-gated; CNGA1, cyclic nucleotidegated channel subunit α1; CNTF, ciliary neurotrophic factor; EPO, erythropoietin; FYCO1, FYVE and coiled-coil domain autophagy adaptor 1; GPCR, G protein-coupled receptor; Grb14, growth factor receptor-bound protein 14; GSK3β, glycogen synthase kinase 3β; IGF, insulin-like growth factor; INPP5, inositol polyphosphate 5-phosphatase; IP3, inositol triphosphate; IR, insulin receptor; IRS, insulin receptor substrate; mTOR, mammalian target of rapamycin; OCRL, oculocerebrorenal syndrome of Lowe; PDGF, platelet-derived growth factor; PH, pleckstrin homology; PI, phosphatidylinositol; PIKfyve, FYVE-type zinc finger containing phosphoinositide kinase; PI3K, phosphoinositide 3-kinase; PI4K, phosphatidylinositol 4-kinase; PIP, phosphatidylinositol phosphate; PI(3)P, phosphatidylinositol 3-phosphate; PI(4)P, phosphatidylinositol 4-phosphate; PI(5)P, phosphatidylinositol 5-phosphate; PI(3,4) P2, phosphatidylinositol 3,4-bisphosphate; PI(3,5)P2, phosphatidylinositol 3,5-bisphosphate; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; PI(3,4,5)P2, phosphatidylinositol 3,4,5-trisphosphate; PIPK, phosphatidylinositol phosphate kinase; PITP, phosphatidylinositol transfer protein; PKC, protein kinase C; PLC, phospholipase C; PTEN, phosphatase and tensin homolog; PTP1B, protein tyrosine phosphatase 1B; RA, Ras-associating; ROS, rod outer segment; RPE, retinal pigment epithelium; SH2, Src homology 2; SHIP, Src homology 2 domain-containing inositol polyphosphate 5-phosphatase; TRP, transient receptor potential; Vps, vacuolar protein sorting; VDAC, voltage-dependent anion channel
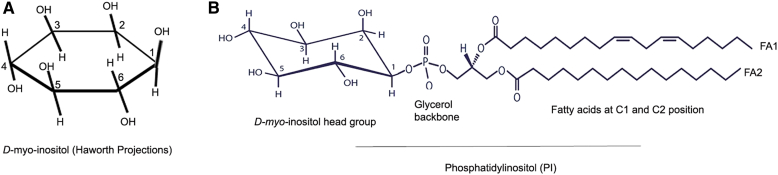
Phosphatidylinositol (PI) is a minor constituent (∼0.5–1%) of the total phospholipid pool in the cell membrane (1). Phosphatidylinositol consists of a head group of D-myo-inositol and a backbone of the trihydroxy alcohol, glycerol, in which the C1 and C2 positions of glycerol are occupied with two fatty acids (Fig. 1). The free-OH groups (3, 4, and 5) in the inositol head group undergo phosphorylation by specific phosphoinositide kinases. These phosphorylated phosphatidylinositol phosphates are collectively called phosphoinositides (PIPs).
Fig. 1.
Structure of phosphatidylinositol (PI). D-myo-inositol presented as a Haworth projection (A). Phosphatidylinositol contains D-myo-inositol attached to a glycerol backbone and two fatty acids attached to the C1 and C2 positions of the glycerol (B).
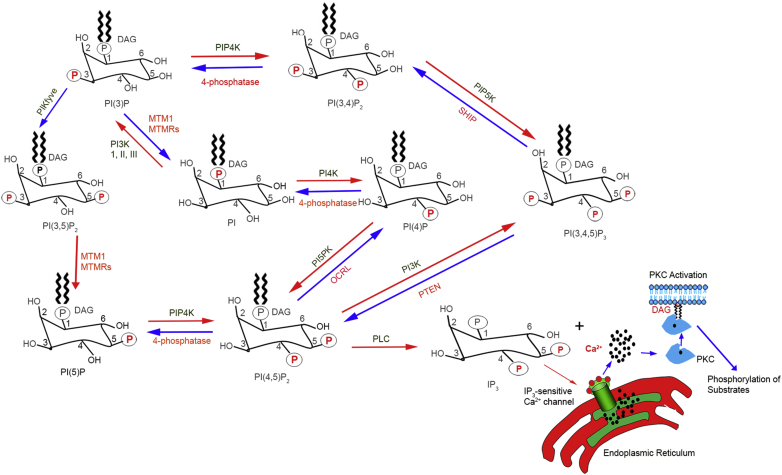
The intracellular pools of PIPs are dynamically converted from one form to the other through the action of specific phosphoinositide kinases, and phosphoinositide phosphatases can generate seven distinct PIPs (1, 2, 3) (Fig. 2). These molecules include PI(3)P, PI(4)P, PI(5)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3. Of these PIPs, the D3-phosphoinositides include PI(3)P, PI(3,4)P2, and PI(3,4,5)P3; the D4-phosphoinositides include PI(4)P and PI(4,5)P2; and the D5-phosphoinositides include PI(5)P and PI(3,5)P2. The D3-PIPs are formed by the action of the phosphoinositide 3-kinases (PI3Ks), which use PI, PI(4)P, and PI(4,5)P2 to generate PI(3)P, PI(3,4)P2, and PI(3,4,5)P3. The D4-PIPs are formed by the action of the PI4 kinases (PI4Ks), which use PI and PI(5)P to generate PI(4)P and PI(4,5)P2. The D5-PIPs are formed by the action of the enzyme PI5K, which uses PI and PI(3)P to generate PI(5)P and PI(3,5)P2 (1, 3). These seven distinct PIPs are present in all mammalian cells (4), and their formation is also present in the retina/photoreceptor cells (5, 6, 7, 8). The D3-, D4-, and D5-phosphoinositides regulate cytoskeletal organization, membrane fusion and budding, ciliogenesis, vesicular transport, and signal transduction (9, 10). PI(3)P is involved in the vesicle export from the Golgi. PI(4,5)P2 is involved in exocytosis, cytoskeletal regulation, and regulation of phospholipase D/A2. PI(3,5)P2 regulates Golgi, lysosomal/endosome trafficking, and osmoprotection. PI(3,4,5)P3 is involved in cell survival, solute transport, and regulation of ARF/Rac protein (1, 9).
Fig. 2.
Generation of seven distinct phosphoinositides. The inositol head group contains several free hydroxyls that undergo phosphorylation by phosphoinositide kinases and dephosphorylation by phosphoinositide phosphatases. Phosphorylation of free hydroxyls at the 3, 4, 5 positions facilitates the generation of seven PPIs. PI(4,5)P2 undergoes hydrolysis by phospholipase C (PLC) generates two-second messenger signaling molecules, diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3). IP3 binds to IP3-sensitive Ca2+ channels on the endoplasmic reticulum and mobilizes the release of Ca2+. Both, DAG and Ca2+ activate protein kinase C (PKC). Activated PKC phosphorylates its downstream target proteins. MTMl, myotubularin l (3’phosphatase); MTMRs, myotubularin-related phospholipid phosphatase; PI(3)P, phosphatidylinositol 3-phosphate; PI(4)P, phosphatidylinositol 4-phosphate; PI(5)P, phosphatidylinositol 5-phosphate; PI(3,4)P2, phosphatidylinositol 3,4-bisphosphate; PI(3,5)P2, phosphatidylinositol 3,5-bisphosphate; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; PI(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate. PIP4K, phosphatidylinositol 5-phosphate 4-kinase; PI3K, phosphoinositide 3-kinase; PIP5K, phosphatidylinositol 4-phosphate 5-kinase; PTEN, phosphatase and tensin homolog; SHIP, Src homology 2 (SH2) domain-containing inositol polyphosphate 5-phosphatase; PIKfyve, FYVE-type zinc finger containing phosphoinositide kinase; OCRL, Oculocerebrorenal inositol polyphosphate-5-phosphatase; PI4K, phosphatidylinositol 4-kinase.
There are many excellent reviews on phosphoinositide kinases, phosphoinositide phosphatases, and PIPs in general. This review describes the recent studies and advances in PIP lipids and their signaling in the retina. The PIP lipids from vertebrate (e.g., bovine, rat, mouse, toad, zebrafish) and invertebrate [e.g., Drosophila, limulus (horseshoe crab), squid] retina are discussed. The importance of PIPs revealed from animal models and human diseases and methods to study PIP levels in vitro and in vivo are also described.
Phosphoinositides in the retina
In 1981, Anderson and Hollyfield (11) reported that light stimulates the incorporation of inositol in the vertebrate retina. Light has also been shown to stimulate the generation of PIs in horizontal cells of the retina (12, 13). Subsequent studies showed that light adaptation of bovine retinas in situ stimulates PI synthesis in retinal rod outer segment (ROS) membranes in vitro (5). The work described in 1983 and 1984 by Anderson and colleagues revealed a functional significance in photoreceptor horizontal cell synapses (12, 13). Light stimulates the PI metabolism in these cells, and these PIPs are important for synaptic ribbon formation, glutamate release, and signaling (12, 13).
Phosphatidylinositol is a comparatively large component of the membranes of most cells in metazoans ranging from 4 to 20 mol% of total phospholipid (1, 14), and analysis of phosphatidylinositol in six different mammalian species shows 4.4–6.4% mol% of total retina phospholipid (14, 15, 16, 17). Studies have shown that retinal ROS membranes contain lower levels of PI than do retinal pigment epithelium (RPE) cells (18) and ER membranes isolated from bovine retinas (19). Recently, Wensel’s laboratory reported that ROSs and fragments of inner segments contain PI(3)P at 0.0035 mol% and PI(4)P and PI(4,5)P2 on the order of 0.04 mol%, 10-fold higher than the levels of PI(3)P of total phospholipid (14).
Phototransduction is modulated by phosphoinositides within photoreceptors (5, 20), which increase phospholipase C (PLC) activity in the outer segment membranes, increase the uptake of radiolabeled inositol and phosphoinositide turnover in photoreceptor cells (20, 21), and release inositol 1,4,5-trisphosphate (IP3) from the retina (22). Furthermore, IP3 receptors (23) and PLC (24, 25) are expressed in the outer segments. In invertebrates, phototransduction is mediated through PLC activation, while in the vertebrate retina, the phototransduction cascade is mediated through rhodopsin activation and subsequent hydrolysis of cGMP (26, 27). Interestingly, in the intrinsically photosensitive retinal ganglion cells, PLC-mediated hydrolysis is activated by the photopigment, melanopsin, which couples Gq to open the transient receptor potential (TRP) channels (28), suggesting evolutionarily conserved pathways that use PI(4,5)P2 as a substrate for the modulation of phototransduction.
One of the potential functions of PLC is involvement in the translocation of arrestin from inner to outer segments of photoreceptor cells (29). Furthermore, activators of the downstream effector of PLC, protein kinase C (PKC), and PLC facilitate arrestin translocation to outer segment membranes, independent of light (29). Physiological studies also show that phototransduction is modulated by PIPs (14, 30). The cone cyclic nucleotide-gated (CNG) (31) and olfactory (32) channels are known to be inhibited by PI3K-generated PI(3,4,5)P3.
It has been suggested that phosphoinositides play an important role in photoreceptor cell processes, such as disk morphogenesis, endocytosis, exocytosis, membrane budding, endosomal sorting, and post-Golgi vesicle trafficking. In rhodopsin trafficking, the involvement of PI(3)P and PI(4,5)P2 has been demonstrated (33, 34). The actin-nucleating proteins Arp2 and Arp3 are demonstrated to be involved in basal disc extensions (34), and local pools of PI(4,5)P2 may be important for their function (35, 36).
Metabolism of phosphoinositides in the retina
There is an active PI metabolism in the vertebrate retina and ROSs (6, 7, 8, 12, 13, 20, 21, 22, 25, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51). Light has been shown to stimulate several enzymes that are involved in PI metabolism, such as class I PI3K (7, 52), class III PI3K (53, 54), PI synthetase (5), DAG kinase (6), phosphatidylethanolamine N-methyltransferase (55), phospholipase A2 (56), phospholipase D (57), PLC (58, 59, 60, 61), PKC (62), lipid phosphate phosphatase (63), and DAG lipase (37). Light also modulates the second messengers generated in the retina from DAG, PC, and PA (37). Further, light activates proteins regulated by insulin signaling (64, 65, 66, 67).
Phosphoinositide kinases and phosphatases
Forty-seven genes encode 19 phosphoinositide kinases and phosphatidylinositol phosphate (PIP) kinases (PIPKs) and 28 PIP phosphatases in mammals (2). The action of the lipid kinases, PI3K, PI4K, and PI5K, and PI3-, PI4- and PI5-specific lipid phosphatases can generate seven distinct phosphoinositide lipids in mammalian cells (1, 2, 3). The phosphoinositide kinase isoforms are divided into three major families: the PI 3-kinases (PI3Ks), PI 4-kinases (PI4Ks), and PIPKs (68).
Phosphoinositide 3-kinases
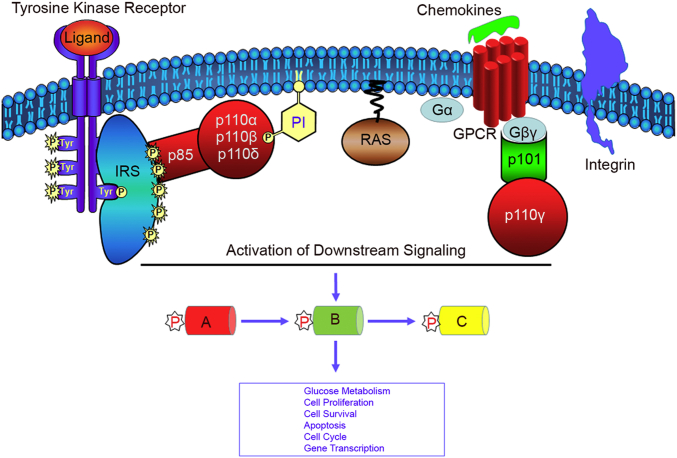
The phosphoinositide 3-kinases have been broadly divided into four classes: class I, class II, class III, and class IV. Members of class I PI3K enzymes are heterodimers and consist of a p110 kDa catalytic subunit and a p85 regulatory subunit that contains two Src-homology regions capable of binding to phosphotyrosine sequences on the growth factor receptors (10). The growth factors, such as platelet-derived growth factor (PDGF), insulin, insulin-like growth factor (IGF)-1, and nerve growth factors, bind to its cognate receptor tyrosine kinase(s) and activate class I PI3Ks (69) (Fig. 3).
Fig. 3.
Activation of phosphoinositide 3-kinases. PI3Ks are activated through tyrosine kinase receptor activation, interaction with RAS, through GPCR activation, or are integrin-mediated. The PIPs generated at the membrane attract phospholipid-binding proteins in the cytosol, activate the downstream signaling (A–C), and regulate various aspects of cellular functions. In the case of the IR, IRS adaptor proteins bind to the IR by facilitating a platform for the binding of multiple PI3K molecules and amplify the signal.
In mammals, class I PI3K-p110 catalytic subunits are encoded by four genes: Pik3ca, Pik3cb, Pik3cg, and Pik3cd, which are referred to as PI3Kα, -β, -γ, and -δ (70). The expression profile of these genes in tissues is not uniform. Ubiquitous expression of Pik3ca and Pik3cb genes has been demonstrated, whereas leukocytes specifically express Pik3cg and Pik3cd genes (70). Pik3cg is also expressed in cardiac tissues (71). All of the PI3Kα, -β, -γ, and -δ-p110 subunits have the phosphatidylinositol kinase domain, a catalytic domain, a C2 lipid-binding domain, and a GTPase Ras domain; these proteins all have a significant homology at the N-terminal end of the molecule (72). In mammalian cells, the catalytic subunits of p110α, p110β, and p110δ are associated with any of five different regulatory subunits, p85α, p85β, p55γ, p55α, and p50α, referred to as “p85 subunits,” for phosphorylation of the lipid substrates (73). The p85 regulatory subunits are encoded by three genes, Pik3r1, Pik3r2, and Pik3r3 (73). Pik3r1 encodes the p85α, p55α, and p50α subunits. Both regulatory subunits, p85α and p85β, are universally expressed in all tissues (72), including retina (74), whereas p55γ is specifically expressed in brain tissues (75). The expression of p50α and p55α has been demonstrated in fat, muscle, liver, and brain tissues (76, 77). The regulatory p85 subunits interact with a unique domain present at the N-terminal end of the p110 catalytic subunits (78). In addition to the Src homology 2 (SH2) domain, the p85 regulatory subunit also contains an SH3 domain, which is capable of binding to proline-rich sequences and also contains a region with high conservation to the breakpoint cluster region (78). Extensive studies on class I PI3K have been carried out in the retina, especially in photoreceptor cells. Functionally, class I PI3K is essential for cone photoreceptor structure and function but not for rod photoreceptor cell survival (74, 79, 80).
Research also indicates that class IA PI3Kβ activation is regulated both by the G protein-coupled receptors (GPCRs) and receptor tyrosine kinases (81). The class I PI3Kγ lacks the p85-binding motif; hence, it interacts with p101 (82) and p84/87 adaptor proteins for regulation (83, 84). The class IB PI3Kγ is activated through GPCR signaling (Fig. 3). Upon GPCR activation by the ligand(s), heterotrimeric G protein composed of α-, β-, and γ-subunits binds to the GPCR (85). The α-subunit of G protein in its bound GDP readily exchanges with the cytosolic GTP, and upon GPCR activation dissociates from the rest of the βγ-subunits and activates the downstream effector molecules (86). The free Gβγ-subunits of heterotrimeric G proteins, mostly the Gi subtype, activate PI3K signaling (71, 87) (Fig. 3). The PI3K family of lipid kinases has also been shown to be stimulated by chemokines in regulating the chemotaxis of inflammatory T lymphocytes, eosinophils, neutrophils, and macrophages, and PI3K signaling regulates the chemokine-activated cell migration (88).
The structure and function of class II PI3Ks are distinct from those of class I PI3Ks. The class II PI3Ks have a C-terminal C2 domain, which lacks a critical aspartate residue that coordinates the binding of Ca2+. The class II PI3K binds to lipid in a Ca2+-independent manner (89, 90). The class II PI3K consists of three catalytic isoforms, which include C2α, C2β, and C2γ, but is distinct from class I and III, as they do not have regulator subunits (89, 90). The class II enzyme catalyzes the conversion of PI to PI(3)P and PI(4)P to PI(3,4)P2 (89, 90). PI(3,4)P2 has been shown to play a role in the invagination step of clathrin-mediated endocytosis (91). The class II PI3K C2α and C2β isoforms are ubiquitously expressed in all tissues, but the expression of the C2γ isoform is restricted to hepatocytes (91). To date, there are no available studies of class II PI3K in the retina.
The structure and function of class III PI3K are distinct from those of class I and class II PI3K. Class III PI3K, also known as vacuolar protein sorting (Vps)34, was first identified in budding yeast, Saccharomyces cerevisiae, and screens for proteins involved in the regulation of vesicle-mediated Vps (92). Several PI(3)P binding proteins have been identified, and all are involved in protein trafficking (93). The crystal structure of Vps34 has been solved (94). Class III enzymes, which phosphorylate only PI, are heterodimers of a catalytic subunit associated with the serine/threonine-protein kinase adaptor subunit that is required for membrane recruitment (9, 10). The class III PI3K, Vps34p, is responsible for producing the majority of the cellular PI(3)P and is involved in protein trafficking through the lysosome (93). Class III PI3K Vps34 is closer to class I PI3K in terms of heterodimers of catalytic (Vps34) and regulatory (Vps15, a 150 kDa protein) subunits (95). Class III enzyme-generated PI(3)P is mainly involved in the trafficking of vesicles and proteins (96). Some studies show that PI(3)P regulates immune cell function and phagocytosis (97, 98). Class III PI3K plays an important role in rod, bipolar, and RPE cell functions in the retina (14, 53, 99, 100).
Class IV is a collection of enzymes, which include ataxia-telangiectasia mutated, ataxia telangiectasia and Rad3-related, DNA-dependent protein kinase, and mammalian target of rapamycin (mTOR). These enzymes are occasionally referred to as the class IV PI3Ks. Unlike class I, II, and III PI3Ks, which are lipid kinases, class IV PI3Ks are protein serine/threonine kinases (101). However, in vitro, class I PI3K has been shown to have protein kinase activity (102).
Insulin receptor-regulated class i PI3K in photoreceptor cells
In 1997, class I phosphoinositide 3-kinase (PI3K) was reported to be responsible for the generation of D3-PIPs in retinal ROSs (7). Furthermore, tyrosine phosphorylation in vitro was shown to stimulate the PI3K activity in isolated bovine retinal outer segment membranes (8). The class I PI3K is composed of two subunits, a p110 catalytic subunit and a p85 regulatory subunit (10). With the application of p85 regulatory subunits that contain both N-terminal and C-terminal SH2 domains, Rajala and Anderson (103) identified that the insulin receptor (IR) in the retina/ROS is the receptor that regulates P13K activity. The authors also observed that in vitro tyrosine phosphorylation enhanced the phosphorylation of the IR, which results in the recruitment of the p85-N-SH2 domain, binds to the tyrosine-phosphorylated IR, and activates PI3K (103).
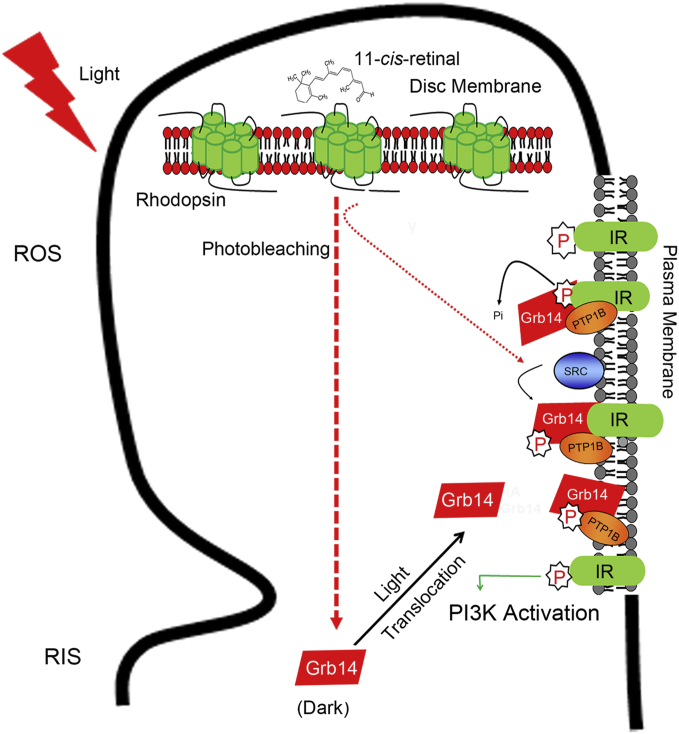
In rod photoreceptors, PI3K activation is mediated through light-dependent tyrosine phosphorylation of the IR (52, 104) (Fig. 4). The IR activation has been shown to be independent of G protein transducin activation and dependent on the photobleaching of rhodopsin (52). This IR/PI3K activation is a noncanonical rhodopsin activation (105) (Fig. 4). In the retina, the IR is constitutively phosphorylated (106, 107). However, the phosphorylation state of the IR is regulated through dark and light adaptation (108). In dark-adapted conditions, the IR is in the inactive state because of increased protein tyrosine phosphatase 1B (PTP1B) activity, which dephosphorylates the IR and results in the reduced association of PI3K with the IR (108). Upon rhodopsin activation, adaptor protein growth factor receptor-bound protein 14 (Grb14) localized to rod inner segments in the dark translocates to the outer segments in light and undergoes tyrosine phosphorylation by a nonreceptor tyrosine kinase, Src, in a rhodopsin-dependent manner (67, 109) (Fig. 4). The Src phosphorylation was abolished in animals deficient in the photobleaching of rhodopsin (109). The tyrosine-phosphorylated-Grb14 binds to PTP1B and inactivates its activity (Fig. 4). Thus, the IR becomes active, which then activates PI3K and promotes photoreceptor survival (108, 109, 110) (Fig. 4). Conditional deletion of the IR in rod photoreceptors resulted in stress-induced photoreceptor degeneration (111). The serine/threonine kinase Akt2, the downstream effector of IR, and PI3K deletion result in stress-induced photoreceptor degeneration (112). The proteins that regulate IR singing pathway in rods are also expressed in cones (113, 114). Interestingly, deletion of the IR in cones resulted in cone degeneration without added stress (113).
Fig. 4.
Noncanonical IR signaling-mediated activation of PI3K in the retina/photoreceptor cells. The IR in the retina is constitutively activated. However, the activated (phosphorylation) state of the IR is light-dependent. Under a dark-adapted state, the IR undergoes dephosphorylation by PTP1B and keeps the IR in an inactive state. Upon illumination, rhodopsin activation facilitates the translocation of Grb14 from inner segments to outer segments, where it undergoes a light- and rhodopsin-dependent phosphorylation by a nonreceptor tyrosine kinase, Src. Phosphorylated Grb14 binds to PTP1B and inhibits its activity, thus preserving the phosphorylation of the IR, which can bind to PI3K and generate PIPs. RIS, rod inner segments; Pi, inorganic phosphate; P, phosphorylation.
Interaction between phosphoinositides and phospholipid-binding proteins
Several pleckstrin homology (PH) domain-containing proteins are expressed in the retina. PIPs regulate these proteins directly or indirectly. These proteins include PH domain retinal protein 1 (PHR1) (115, 116), Evectin-1 (117), Grb14 (118), IR substrate (IRS)-1 and IRS-2 (106, 119, 120), and three isoforms of Akt (Akt1, Akt2, and Akt3) (66, 106, 112). Evectin-1 is involved in the trafficking of post-Golgi membranes in photoreceptor cells (117). The PH domain retinal protein 1 PH domain is identical to serine/threonine kinase Akt and does not bind to inositol phosphates but interacts with transducin Gβγ subunits (115, 116). In the retina, the Akt-PH domain binds to PIPs (66). The PH domain of Akt binds to the cytoskeletal protein myosin II (121). This protein is mutated in patients with Usher syndrome (122). However, the interaction between myosin and Akt has not been tested in the retina. PH domain and leucine-rich repeat protein phosphatase (PHLPP) and PH domain and leucine-rich repeat protein phosphatase-like (PHLPPL) are PH domain-containing proteins known to dephosphorylate Akt isoforms (123, 124) and are also expressed in the retina (125, 126, 127).
Downstream effects of PI3K signaling in the retina
In vivo, light stimulates the tyrosine phosphorylation of the IR, which results in the recruitment of PI3K and subsequent activation of Akt, the downstream effector of PI3K regulated through PI3K-generated PIPs (104). The addition of insulin as the ligand for the IR also stimulates the tyrosine phosphorylation of the IR, which results in the activation of PI3K ex vivo (66, 103). In the retina, light stress induces the phosphorylation of the IR, which results in the activation of the PI3K/Akt pathway through increased tyrosine phosphorylation of the IR (111). Addition of insulin growth factor-1, the ligand for IGF-1R, ex vivo to retinas stimulates the tyrosine phosphorylation of the IGF-1R, which results in the activation of the PI3K/Akt pathway (128). Interestingly, light stress, but not physiological light, stimulates the tyrosine phosphorylation of the IGF-1R that results in the activation of the PI3k/Akt downstream signaling pathway (128). In the retina, PI3K activation stimulates serine/threonine kinase Akt in vitro and in vivo (66, 106, 112).
Akt kinase exists as three isoforms: Ak1, Akt2, and Akt3. All three isoforms are expressed in retina and rod photoreceptor cells (66, 106). Membrane binding of Akt1 is mediated through its PH domain binding to PI3K-generated PIPs (66). In transgenic Xenopus laevis, the PH domain fused to GFP under the control of the Xenopus opsin promoter binds to PIPs in a light-dependent manner (66). In dark-adapted conditions, the actin exists as a stress fiber phenotype. Upon light-illumination, reorganization of the actin cytoskeleton was found to colocalize with PIPs in a X. laevis model (66). The expression of a mutant version of the PH domain in which the arginine 25 residue is substituted with cysteine failed to bind to PIPs (66).
The IR/PI3K/Akt signaling proteins are also expressed in the rod inner segments. These proteins may play an important role in cellular signaling events (105). One of the functions of PI3K-activated Akt in the inner segment is to regulate hexokinase 2 as it interacts with mitochondria in the photoreceptors (125). Growth factor-mediated activation of Akt was demonstrated to increase the association of hexokinase 2 with mitochondria in normal tissues and cells (129). Akt activation inhibits the dissociation of hexokinase 2 from mitochondria and is the primary event in the induction of apoptosis (125, 129, 130, 131). The mechanism of interaction between hexokinase 2 and mitochondria is that Akt phosphorylates glycogen synthase kinase 3β (GSK3β), which renders GSK3β inactive (132, 133). In the absence of Akt activation, GSK3β is active and phosphorylates the voltage-dependent anion channel (VDAC) on serine 51 that disrupts the binding of hexokinase 2 to the VDAC (134). The PI3K-generated PIP-activated Akt prevents the release of cytochrome c from mitochondria and inhibits apoptosis (129, 135). Retinal photoreceptors are postmitotic neurons and apoptosis is detrimental; therefore, the PIP-activated Akt promotes photoreceptor survival (112, 125).
Interaction between the rod cng channel and PI3K in rod photoreceptor cells
Class I PI3Kγ lacks a p85-binding motif. Hence, it interacts with p101 (82) and p84/87 adaptor proteins for regulation (84, 136). Class I PI3Kγ is known to be regulated through Ras proteins (137) (Fig. 3). Ras belongs to a family of related proteins called small GTPases (138). These proteins have a Ras-associating (RA) domain, and PI3K interacts with this RA domain (139). In photoreceptor cells, the C-terminal region of the rod CNG channel subunit α1 (CNGA1) has 50–70% tertiary structural homology with Ras proteins (140). This domain has been named the Ras-like domain (140). In rod photoreceptor cells, PI3Kγ activation occurs through the interaction of its RA -domain with the Ras-like domain of CNGA1 (141). The interaction of PI3Kγ with CNGA1 does not affect the channel physiology. However, PI3Kγ uses CNGA1 as an anchor to achieve a close vicinity to its substrates to generate PIPs (141).
PI3K KO phenotypes in the retina and rpe
In the retina, loss of p85α does not affect the overall morphology, but decreased PI3K activity associated with the IR is observed (142). Conditional deletion of the p85α (pik3r1) regulatory subunit in rod photoreceptor cells does not affect the structure of the retina (74). Mice with conditional deletion of p85α exhibited a slight delay in recovery kinetics and a delay in the translocation of rod arrestin from the inner segments to the outer segments (74). The absence of the disease phenotype in mouse rods lacking p85α could be explained by the expression of p85β in p85α KO mice (74). Interestingly, the deletion of the p85α-subunit of PI3K in cones results in age-related cone degeneration (79). The surviving cone-terminals of the cone-p85α KO mice exhibit progressive disorganization of synaptic structures (79). The loss of p85α in cones does not affect rod structure and function (79). Conditional deletion of the catalytic subunit of PI3K, p110α, in cones also resulted in cone degeneration (143). These studies highlight that PI3K signaling is indispensable for cone photoreceptor survival. These findings also suggest that other PIP-signaling pathways may regulate rod survival.
Conditional deletion of the class III PI3K Vps34 gene in rods results in a failure in the fusion of endosomal and autophagy-related membranes with lysosomes that prompts the buildup of anomalous membrane structures (53). These mice have normal structure and function and trafficking of rhodopsin to the outer segments; however, the mice experience progressive rod degeneration by 12 weeks of age (53). The rod degeneration accompanying Vps34 gene deletion is much faster than that of rods lacking autophagy genes, highlighting that PI(3)P is required for endosome recycling and other pathways that are necessary for rod photoreceptor survival (53). Vps34 has recently been shown to be essential for on-bipolar cell survival. Loss of this enzyme in these cells results in a significant loss of structure and function (99). This study further highlights that PI(3)P is necessary for the fusion of autophagosomes with lysosomes and maturation of late endosomes, and PI(3)P is needed for the maintenance of on-bipolar cells health (99).
In RPE, PI(3)P is essential for the fusion with autophagosomes, lysosomes, and phagosomes (100). These findings highlight that PI(3)P is essential for RPE cell health. In cone photoreceptors, the ablation of Vps34 resulted in an age-related cone degeneration (R. V. S. Rajala, unpublished observations).
Phosphatidylinositol phosphate kinases
Phosphatidylinositol 4-kinases (PI4Ks) make PI(4)P from phosphatidylinositol (3). PI(4)P is an important molecule for the generation of other phosphoinositides involved in signaling, such as PI(4,5)P2, and is the substrate for the generation of second messengers, such as IP3 and DAG, through the action of PLC (144). PI(4,5)P2 also serves as a substrate for PI3K for the generation of PI(3,4,5)P3 (10, 145). PIPKs phosphorylate the D5 position of inositol on PI(4)P and the D4 position of PI(5)P to generate PI(4,5)P2 (146). Numerous PIPKs have been identified, purified, and cloned (147, 148, 149), and are separated into type 1 and type II based on their biochemical and immunological characteristics (10, 150). Based on substrate specificity, PIPK1 (type 1) phosphorylates PI(4)P and PIPKII (type II) phosphorylates PI(5)P (151). The mechanism of activation of PIPKs is not completely understood. Type I PIPK is activated by GTPγs (152), small G proteins (153), PA (154), heparin, and spermine (155). Spermine and PA do not affect PIPKII activity, but heparin inhibits PIPKII activity (155). Tyrosine phosphorylation has been shown to influence the activity of PIPKs (156).
In most cells, the PI(4,5)P2 is generated through type I PIPKs and global KOs of the α, β, and γ isoforms of this kinase have been produced (157). This isoform is highly expressed in the retina (158) and other neurons (159, 160). Early postnatal mortality was reported when this kinase was deleted (161). The mouse genes that encode PIPKs are Pip4k2a, Pip4k2b, Pip4k2c, Pip5k1a, and Pip5k1c; all of these were detected in the retinal mouse proteome (162).
In bovine ROS membranes, type II PIPK activity is regulated by tyrosine phosphorylation (163), and PI(5)P serves as a substrate for the synthesis of PI(4,5)P2 (163). In rodents and transgenic frog retina, PIPKIIα activity is stimulated by light, and the membrane binding of PIPKIIα to ROS proteins is tyrosine phosphorylation-dependent (164). PI(4,5)P2 is a critical PIP and regulates several key biological processes, such as actin cytoskeletal organization, endocytosis, exocytosis, modulation of ion channels, gene expression, angiogenesis, vesicular transport, cell migration, and nuclear functions (165). PI(4,5)P2 has been shown to activate cGMP-phosphodiesterase in ROS membranes, which results in the inhibition of ion influx through CNG channels (30, 166). In mammalian retina, the rod CNG channels, KCN1, TPR channels, and Na+-Ca2+ exchange are known to be regulated by PI(4,5)P2 (167). In photoreceptor cells, PI(4,5)P2 regulates the biogenesis of light-sensing organelles and plays an important role in the delivery of rhodopsin-containing membranes to the ROS (33).
One of the receptor tyrosine kinases, epidermal growth factor receptor, has a cluster of basic residues in the juxtamembrane domain. PI(4,5)P2 directly binds to this region and has been shown to activate EGFR (168). These basic residues are also present in other growth factor receptors, such as IGF-1R, IR, FGFR1, PDGFR, VEGFR1, EPHB2, TRKA, and TRKB receptor (168). However, the activation of these receptors by PI(4,5)P2 has not been studied.
Phosphatidylinositol-3-phosphate 5-kinase or pikfyve
In humans, FYVE finger-containing phosphoinositide kinase is encoded by the PIKFYVE gene (169). PIKfyve phosphorylates PI(3)P to PI(3,5)P2 and PI to PI(5)P (170, 171, 172). The FYVE finger domain of PIKfyve tethers to membrane PI(3)P and is essential for membrane localization of PIKfyve to the cytosolic leaflet of endosomes (173). This binding is needed to phosphorylate PI to PI(5)P and PI(3)P to PI(3,5)P2 (170). Dysregulated enzyme activity of PIKfyve results in enlarged lysosomes due to defective synthesis of PI(3,5)P2, which results in defective lysosome fission events; thus, PIKfyve navigates all aspects of the vesicular and endocytic pathways (172, 174). Mutations in one of the alleles of PIKfyve are linked to Francois-Neetens corneal fleck dystrophy (175). The ablation of both alleles in the mouse is lethal (176). Changes in PIKfyve have been shown to inhibit insulin-mediated glucose uptake (177). Mice with selective gene ablation of PIKfyve in skeletal muscle show prediabetic symptoms, which include insulin resistance, hyperinsulinemia, glucose intolerance, and increased adiposity (178). In one study, inhibition of PIKfyve resulted in the prevention of myocardial apoptosis and cardiac hypertrophy mediated through the activation of sirtuin-3 (SIRT3), a major mitochondria NAD+-dependent deacetylase in obese mice (179). Because photoreceptor cells are highly metabolic (180, 181), the PIKfyve enzyme might regulate several important functions. However, there are no available studies of this enzyme in the retina.
It was previously reported that Vps34-generated PI(3)P regulates the canonical autophagy (182). However, Vps34-independent noncanonical autophagy has been observed in sensory neurons from the PI(3)P-generated Vps34 enzyme, T-lymphocytes, and glucose-starved cells incubated with PI3K inhibitor, Wortmannin, which inhibits the Vps34 enzyme (183). PI(5)P-dependent noncanonical autophagy has been reported in cells depleted of PI(3)P (170). PIKfyve plays an important role in generating PI(5)P upon binding to PI(3)P through its FYVE domain (170). This PI(5)P can be converted to PI(4,5)P2 through the action of the type II phosphatidylinositol 5-phosphate 4-kinase (PIP4K) enzyme, which was previously shown to express in rod photoreceptor cells (163). The generated PI(4,5)P2 can perform various functions, including serving as a substrate for class I PI3K for the generation of PI(3,4,5)P3 (10, 80, 145). In cone photoreceptor cells, the deletion of class I PI3K that makes PI(3,4,5)P3 resulted in age-related cone degeneration (79, 143).
Neuroprotective roles of PI3K in the retina
Several growth factors, such as PDGF, brain-derived neurotrophic factor, insulin, IGF-1 and -2, basic fibroblast growth factor, erythropoietin (EPO), and ciliary neurotrophic factor (CNTF), promote retinal cell survival through PI3K/Akt activation. Insulin and IGF-1 have been shown to promote photoreceptor survival (106, 184), whereas PDGF, CNTF, and EPO promote the survival of ganglion cells and RPE (185). basic fibroblast growth factor-mediated activation of PI3K has been observed in Müller cells (186). PI3K exerts its neuroprotective effect through its downstream effector, Akt (66, 106, 112). IR activity is important for PI3K/Akt activation (66, 106). For the signal to be maintained for a longer period, the protein tyrosine phosphatase, PTP1B, must be inactivated. It has been shown that activated Akt phosphorylates PTP1B on serine 50 (187), which inhibits the activity of PTP1B to facilitate a positive feedback mechanism for IR/PI3K/Akt signaling. In diabetic retinopathy, PI3K activity is downregulated (188) due to increased PTP1B activity (107). Furthermore, decreased activation of mTOR and p760S6K, and increased activation of GSK3β, the downstream effector of PI3K, are downregulated in diabetic retinopathy (189).
PI3K activation through the IR inhibits caspase-mediated cell death of retinal neurons in culture (190). 17β-Estradiol protects stressed retina through PI3K activation (191, 192). The deletion of the phosphatase and tensin homolog (PTEN) results in an increased level of PI(3,4,5)P3, which protects ganglion cells (193). Activation of PI3K through PDGF stimulation protects retinal pericytes under diabetic conditions (194). Activation of PI3K/Akt protects the RPE from oxidative stress (195, 196, 197). brain-derived neurotrophic factor-induced PI3K activation protects axotomized retinal ganglion cells (198). The neuroprotective function of CNTF is mediated through the PI3K pathway in vitro and in vivo (199). The PI3K/Akt pathway activated through cytokine EPO promotes adult CNS neuron regeneration (200). The PI3K pathway is necessary for nerve regrowth in the goldfish retina (201). PI3K also plays an important role in retinal development and survival of differentiated neurons in vivo (202). In addition, PI3K regulates the circadian output in the retina (203).
Constitutive activation of Akt has been observed in cone photoreceptor cells (204). In rods, Akt activation is transient under physiological light (66), hyperosmotic stress (205), oxidative damage (206), or light stress (112). In cone photoreceptor cells, PI metabolism is Ca2+-dependent and essential for glutamate release and synaptic ribbon formation (207). In dark-adapted cones, a rise in intracellular Ca2+ increases PI metabolism at the synaptic terminals compared with the ROS (207). These changes in cones result in the release of glutamate from the synapse, which halts PI metabolism in the adjacent horizontal cells. Under light-adapted conditions, increased PI metabolism has been observed in outer segments that support phototransduction (207). Decreased intracellular Ca2+ results in decreased PI metabolism in the synapse, which blocks the release of glutamate that releases the block on PI metabolism in the adjacent horizontal cells (207).
Inositol polyphosphates also play an important role in synaptic ribbon disassembly (207). Previous studies showed that the activation of PI metabolism and photoreceptor synapses under dark-adapted conditions is in parallel with the disappearance of synaptic ribbons (207). Further, inositol polyphosphates (IP3) have been shown to influence synaptic ribbons in cone photoreceptors but not in rods, suggesting that the cones have an active PI metabolism in cone synapses (207). Consistent with these studies, cones lacking class I PI3K exhibit disorganization of synaptic ribbons (207). In the retina/photoreceptors, a cross-talk exists between IR/PI3K and phototransduction. The PI3K pathway is activated through photobleaching of rhodopsin but is not dependent on G protein transducin activation (52). These observations suggest that other G proteins might be involved in the activation of the PI3K pathway. Such a cross-talk between tyrosine kinases and GPCR signaling has been observed outside of the retina. Mitogen-activated protein kinases, extracellular-regulated kinases, stress-activated protein kinase (208, 209), and nonreceptor protein tyrosine kinases (210) are the best examples of this cross-talk.
Phosphoinositide phosphatases
To date, over 35 mammalian phosphoinositide phosphatases that downregulate many phosphoinositide signals have been identified (211). A retina proteome shows the presence of phosphoinositide phosphatases that are encoded by Mtmr2 and Synj1, Inpp1, Inpp4a, Inpp4b, and Inpp5e genes (162). Synaptojanins regulate synaptic function (212, 213, 214), which includes endocytosis and vesicle uncoating, and profound cone defects have been observed in zebrafish when this phosphatase is lacking (215, 216, 217). In cone photoreceptor cells, the phosphoinositide phosphatase, synaptojanin 1 (SynJ1), has been shown to regulate autophagy and endosomal trafficking, and mutations in zebrafish cone photoreceptors show abnormal accumulation of late endosomes and autophagosomes (215, 217, 218, 219). Synaptojanin 1 has also been shown to regulate cognition and plays an important role in Down syndrome (220).
Defects in oculocerebrorenal syndrome of Lowe (OCRL) phosphatase are associated with glaucoma, a second leading cause of blindness worldwide (221, 222). Mutations in inositol polyphosphate 5-phosphatase (INPP5)E are associated with Joubert syndrome and Bardet-Bedi syndrome (223, 224, 225, 226), which result in retinal degeneration. Both PI(4,5)P2 and PI(3,4,5)P3 serve as substrates for INPP5E and the levels of PI(4,5)P2 are suggested to be important for the regulation of cilium (14). The 3′phosphatase PTEN regulates retinal neurogenesis and Notch signaling (227). Further inactivation of PTEN in the RPE causes retinal degeneration, highlighting the importance of this phosphatase in RPE functions (228). In an animal model of retinitis pigmentosa, the deletion of PTEN in cone photoreceptors resulted in cone photoreceptor survival (229). The retinal amacrine cell number is shown to be regulated by PTEN through the modulation of Erk, TGFβ, and serine-threonine kinase Akt (230). Retinal interneuron morphogenesis and synaptic layer formation are shown to be regulated by PTEN (231). Interestingly, the deletion of PTEN together with the retinoblastoma gene resulted in penetrant bilateral retinoblastomas (232). PI3K activation triggered by insulin or other growth factors phosphorylates PI(4,5)P2 to PI(3,4,5)P3, which can be rapidly dephosphorylated by PTEN to generate PI(4,5)P2 or by INPP5s to generate PI(3,4)P2. INPP5 also hydrolyzes PI(4,5)P2 to PI(4)P (211).
In mammalian cells, ten 5-phosphatases have been identified. These 5-phosphatases regulate several aspects of cellular function, including synaptic vesicle recycling, cell proliferation, embryonic development, control of hematopoietic development, and insulin-mediated signaling (233). The 5-phosphatases, OCRL and INPP5E, are mutated in Lowe and Joubert syndrome, respectively (234). Src homology 2 domain-containing inositol polyphosphate 5-phosphatase (SHIP)2 and skeletal muscle- and kidney-enriched inositol phosphatase (SKIP) are negative regulators of insulin signaling and glucose metabolism (235, 236). Insulin resistance is associated with polymorphisms in SHIP2 (237). INPP4A and INPP4B, the inositol 4-phosphatases, dephosphorylate PI(3,4)P2 to PI(3)P, and these phosphatases have been shown to control neuroexcitatory cell death (238, 239). The Sac phosphatases dephosphorylate PI(3)P, PI(4)P, PI(5)P, and PI(3,5)P2 to form PI (211). Degenerative neuropathy has been reported with loss of one of the Sac phosphatase genes, FIG4 gene (240). Like phosphoinositide kinases, mutations or changes in the expression of phosphatases cause many human diseases (211).
The role of phosphoinositides in the invertebrate retina
Although this review focuses on phosphoinositide metabolism in the vertebrate retina, it is important to note that PI metabolism is also important in the invertebrate retina. Visual excitation in Limulus (horseshoe crab) photoreceptors have been shown to be mediated by myo-inositol polyphosphate (241). In Drosophila photoreceptor cells, light stimulated the hydrolysis of PI(4,5)P2 by PLC into DAG and IP3 (242, 243, 244). IP3 facilitates the opening of two calcium-sensitive channels, TRP and TRP-like (218), which results in plasma membrane depolarization (245). In the vertebrate retina, cGMP modulates the channel but not the IP3 (26). In squid photoreceptor membranes, PI-PLC is activated through metarhodopsin-activated G protein, Gq (246). In invertebrates, retinal degenerations result from a mutation in various enzymes of the PI cycle (243, 244, 245). In Drosophila, phosphatidylinositol transfer protein (PITP) is essential for photoreceptor cell survival and recovery of light stimulation (250). Further, rhodopsin inactivation is mediated by the visual arrestin, Arr2. Its translocation from the rhabdomeric cell body to the microvilli is governed by myosin III-driven PI(3,4,5)P3-dependent movement (251, 252, 253, 254). Drosophila Arr2 has a PH domain that binds to PI(3,4,5)P3 (251), but mammalian visual arrestins do not. The light-induced trafficking of Arr2 to the microvilli, the termination of phototransduction, and recovery are impaired in Drosophila retinas lacking phosphoinositide biosynthesis and trafficking genes cds and rdgB or overexpression of 3′phosphoinositide phosphatase PTEN, which dephosphorylates PI(3,4,5)P3 to PI(4,5)P2 (255). In Drosophila, light-activated TRP and TRP-like channels regulate the PI signaling cascade (256). In Drosophila photoreceptor membranes, PI(4)P synthesis is catalyzed by PI4KIIIα (257). Further, PIP5K is needed for the generation of PI(4,5)P2 for GPCR signaling in Drosophila (258).
Roles of phosphosphoinositide lipids in the retina
Alterations in the metabolic flow of events in the retina can lead to retinal degeneration (see RETNET; https://sph.uth.edu/retnet/, May 2020). It has been reported that blue light-excited all-trans-retinal and 11-cis-retinal irrevocably disrupt plasma membrane-bound PI(4,5)P2 and alter its function in nonvisual cells (259). This altered PI(4,5)P2 increases cytosolic calcium and causes a change in cell morphology, leading to cell death (259). These studies suggest photoexcited chromophore-prompted PI(4,5)P2 alteration, which leads to oxidative damage to the plasma membrane (259). This altered PI(4,5)P2 function is independent of nonvisual GPCR activation (259). mTOR1 activation is mediated by lysosomal positing, while amino acids promote the recruitment of FYVE and coiled-coil domain autophagy adaptor 1 (FYCO1), a PI(3)P binding protein, to the lysosomes and facilitate the interaction between FYCO1 lysosomes and the ER that encompasses protrudin, a PI(3)P effector protein (260). These studies suggest that the Vps34-generated PI(3)P regulates mTORC1 activation through lysosomal positioning (260). In addition, the cause of congenital cataracts is due to mutations in the FYCO1 gene (261). In the retina, type 1 metabotropic glutamate receptors are coupled to the hydrolysis of polyphosphoinositide (262). Class I PI3K in the retina has a dual function in regulating cell survival and cell trafficking (263). A mathematical model of the PI signaling pathway has been constructed, and the authors argue that this model reflects the study state of the PI pathway (264). In the zebrafish retina, PITPβ is needed for the integrity of cone cell outer segments (265). PITP protein transfers the PI and PC across membranes (266). Further, mutations in the PYK2-binding domain of phosphatidylinositol transfer membrane-associated protein (PIPNM3) caused autosomal dominant cone dystrophy (CORD5) in two Swedish families (267). The cone CNG (31) and olfactory (32) channels are known to be inhibited by the PI3K-generated PI(3,4,5)P3. There are no available studies on the role of PI(3,4,5)P3 in rod CNG channels. Further, PLC and PI(4,5)P2 levels in phototransduction are regulated by ceramide kinase (268). The components of PI signaling in photoreceptor outer segments were identified in the mid-1990s (25). PIPs and proteins that are involved in phosphorylation of these lipids have been observed in the axoneme of the bovine ROSs (269). In ROSs, transducin βγ-subunits have been shown to regulate the metabolism of PI(4,5)P2 (270). Alterations in retinal structure and PI degradation by light damage and chronic lithium treatment have been reported (271).
The PI synthesis in photoreceptor cell inner segments is enhanced by light and cytidine (20, 272). In rat retina, the PI synthesis and phosphorylation are stimulated by light (41). The PI turnover is also enhanced in light (21). The lipid PI(4,5)P2 has been shown to regulate blood vessel stability; this lipid could be a viable therapeutic target for angiogenesis therapies in diseases such as AMD, cancer, and proliferative diabetic retinopathy (273). RPE-mediated photoreceptor phagocytosis is regulated by phosphoinositide lipid signaling (274). In cone photoreceptor cells, CNGA3-associated mutation potentiates the PI sensitivity of cone-CNG channels by disrupting the interaction between subunits (275). In photosensitive chicken retinal ganglion cells, light has been shown to stimulate the PI cycle (276).
Phosphoinositides in retinal cilia
PI(4)P and PI(4,5)P2 are important for the regulation of primary cilia, especially in assembly and disassembly (277, 278, 279, 280, 281). Phosphoinositide signaling has been coupled with proper protein trafficking and Hedgehog signaling in the primary cilia (280, 282). The 5′PI phosphatase, INPP5E, is paired with proper ciliogenesis (280, 282).
INPP5E dephosphorylates PI(3,4,5)P3 to PI(4,5)P2 (282). This inositol phosphatase is important in ciliary development in zebrafish (223). Mutations in INPP5E result in Joubert syndrome, a rare disorder characterized by deformation of the midbrain, retinitis pigmentosa, renal cysts, and polydactyly (283). INPP5E has been shown to regulate the phosphoinositide-dependent cilia transition zone function. IPP5E mutant embryos have dysfunctional Hedgehog signaling (284). In the retina, ciliopathy protein RPGR interacts with phosphodiesterase 6δ and regulates the ciliary localization of INPP5E (285).
In addition to INPP5E, OCRL is also important for the regulation of cilia (277, 278, 279, 280, 281, 286). Bardet-Biedl syndrome is caused by the mutations in BBSome, and this protein complex is involved in ciliary trafficking and binds to phosphoinositides (287, 288). In vitro, the PH domains of BBS5 bind to PI(3)P (287). Similar specificities in PIP binding have been observed with BBS1, -4, -5, -8, -9, and -18. A recent review of phosphoinositides mentioned the caveat that commercial PIP strips have local surface densities of PIPs, which are significantly different under physiological conditions (14). Further studies are warranted to establish that these proteins indeed bind to PIPs in vivo.
The signaling role of phosphoinositides in a cellular process
The phosphorylated phosphoinositides are formed in response to growth factor stimulation or GPCR activation or are activated by other cellular cues that attract proteins from the intracellular side of the cells through protein-lipid interactions. The proteins that interact with PIPs are called phospholipid-binding or phosphoinositide binding proteins. It is essential to establish the specific lipid involved in given cellular processes. It is also important to identify the effector protein that is regulated by the PIP signal that governs the cellular functions.
Investigators have identified several phosphoinositide-binding proteins that relay the PIP signal. Generally, two groups of interacting sites exist for the stereoselective association of PIP with effector proteins (247). The first group comprises a 10–20 co-linear sequence enriched with basic and hydrophobic residues that mediate interaction with PIP (247). The second group has proteins consisting of around 120 amino acids that share homology with a well-established tertiary structure of the PH domain (247). These PH domains are extensively characterized and are well-known PIP binding modules. The PH domain sequence was first observed in the platelet PKC substrate pleckstrin (248), which contains a collinear sequence of 120 amino acids and is present in more than 100 proteins.
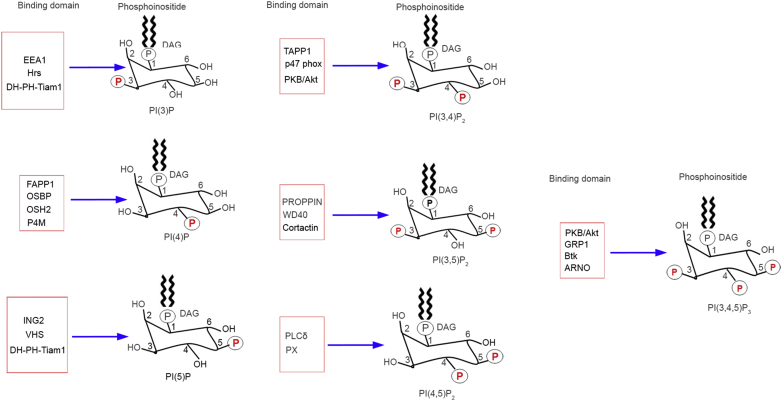
PI(3)P binds to FYVE and PX domain-containing proteins (247). PI(4,5)P2, PI(3.4)P2, and PI(3,4,5)P3 bind to proteins containing PH domains (249, 289, 290, 291). PI(5)P binds to proteins containing the plant homeodomain domain (292). Several studies showed that phosphoinositides were detected qualitatively or quantitatively using the PH, PX, and FYVE domains (291, 293). The FYVE domains of EEA1 and Hrs have been used to detect PI(3)P (173, 291, 294). The PH domains of FAPP1, OSBP, and OSH2 and the P4M domain of SidM/GOLPH3 have been used to detect PI(4)P (291). The plant homeodomain domain of ING2 (292), Tam1 DH-PHc domain (295), and the VHS domain of TOM1 have been used to detect PI(5)P (296). The PROPPIN domain of Atg18P and the WD40 domain of raptor have been used to detect PI(3,5)P2 (297). The PH domains of TAPP1, p47phox, and PKB/Akt have been used to detect PI(3,4)P2 (291). The PH domain of PLCδ1 and the PX domain of tubby have been used to detect PI(4,5)P2 (291). The PH domains of PKB/Akt, GRP1, Btk, and ARNO have been used to detect PI(3,4,5)P3 (249, 291). Some of these domains may bind to more than one PIP lipid (291).
Cortactin has been shown to bind to PI(3,5)P2 (298). It does not contain canonical PIP binding motifs, such as PH or FYVE, but contains a stretch of basic lysine residues that may bind to PIP. This region is located within the N-terminal fourth repeat domain of cortactin (298). One study showed that the lactadherin C2 domain binds to PS (299); this domain could be used as a probe to determine the levels of PS in cells/tissues. PH domains do not always bind to PIPs, but sometimes act as platforms for protein-protein interaction domains (300, 301). Recently, the PIP binding domain has been used to quantitate the levels of PI3K-generated PIs in rod photoreceptor cells (53). A list of phospholipid-binding proteins that bind to specific PIPs commonly used as probes to study protein-lipid interaction is presented in Fig. 5.
Fig. 5.
Commonly used phosphoinositide-binding protein probes (domains) to determine the levels and localization of specific PIPs in vitro and in vivo.
Methods to detect cellular levels of phosphoinositides
Mass spectrometry is commonly used to detect PIPs. The PIPs can be identified through the m/z values of their collision-induced fragments. This approach provides a “fingerprint” for each PIP (3). By including a standard such as lysophosphatidic acid, the specific PIP can be identified and quantitated (3). Mass spectrometric methods are very sensitive and overcome the need for radioactive labeling.
Effector phosphoinositide binding proteins bind to specific PIPs. The proteins fused to reporters have been used as valuable tools to analyze specific PIPs in live or fixed cells and tissues (3, 53). Fluorescently tagged phosphoinositide-binding protein (either GFP or DsRed) or antibodies against fusion proteins have been used as probes for the immunoelectron or fluorescent microscopic detection of specific localization of PIPs (302). In rod photoreceptor cells, RPE cells, zebrafish, and transgenic Xenopus retina, PIP probes have been successfully employed to determine the localization of specific PIPs (53, 66, 100, 217, 218).
The fastest way of testing the potential phosphoinositide binding proteins is through protein-lipid overlay assays using purified phosphoinositide-binding proteins in vitro (290, 293, 299). Because the PIPs are localized to membranes, sometimes they may not interact with a specific phosphoinositide-binding protein in vitro. In those cases, liposomes containing specific PIPs can be used to mimic cellular membranes (3). Incubation of these liposomes with phosphoinositide-binding protein and the associated phosphoinositide-binding proteins with liposomes can be recovered by centrifugation of heavy multilamellar liposomes or affinity isolation of small unilamellar vesicles, followed by immunoblotting or radioactive counting (3). Liposome-based assays are better in terms of measuring differential affinities of phosphoinositide-binding proteins to PIP than are protein-lipid overlay assays (3). These liposomes can be placed on the surface of the sensor chip for surface plasmon resonance spectroscopy (3). Association and dissociation constants can be calculated when the phosphoinositide-binding proteins are passed through a flow cell and bind to the immobilized PIP, which causes a change in surface refractory index (3).
Identification and measurement of phosphoinositide-metabolizing enzyme activity is another way of measuring PIPs in a given tissue or cell. These assays can be performed using radioactive ATP followed by TLC, after which the incorporated radioactivity in the PIPs can be counted (52, 104, 303). Nonradioactive enzyme assays are also available to determine the phosphoinositide-metabolizing enzymes (304). Many assays have been developed for the measurement of PI kinases and PI phosphatases in cell extracts or with purified proteins. These extracts or purified proteins can be incubated with radiolabeled PIPs, and their phosphorylated or dephosphorylated PIPs can be monitored using TLC or HPLC (305). Researchers have also developed novel mass assays to quantify phosphoinositides from cells and tissues (306, 307, 308).
Competitive ELISAs have been developed to determine the PI kinase assays (3). Several ELISAs have been developed to quantify the levels of specific PIPs using phosphoinositide-binding proteins (249, 309). The phosphoinositide-binding proteins are coupled to enzymes, peroxidase-conjugated antibodies, or antibody epitopes for the detection and quantification of various PIPs in cells and tissues (53, 309, 310). In photoreceptor cells, PI3K-generated products have been shown using phosphoinositide-binding proteins coupled with rhodopsin 1D4 antibody epitope on ELISA (53, 311). Fluorescence resonance energy transfer is another technique to determine the binding between PIP and phosphoinositide-binding proteins (3).
There are many other high-resolution techniques coupled with molecular probes to visualize the location, organization, and dynamics of PIPs (312). These techniques include single-particle tracking, fluorescence recovery after photobleaching, and fluorescence correlation spectroscopy (313). The combination of lipid probes and available microscopic techniques is very powerful, but these approaches have several potential limitations and caveats (313). There are numerous commercially available PIP-specific antibodies. A number of investigators have used these antibodies to localize PIP in cells and tissues (54, 314, 315, 316). Also, transgenic frogs (66) and zebrafish (217) models have been used to study the generation and localization of PIPs in vivo.
Conclusions and future directions
Numerous studies have explored the roles of various phosphoinositides, phosphoinositide kinases, and phosphoinositide phosphatases in the vertebrate retina. The minor lipids play an important role in various aspects of photoreceptor biochemistry and physiology. These lipids are involved in protein transport, ciliogenesis, cell survival, vesicular transport, autophagy, phagocytosis, and synaptic functions. Many researchers have demonstrated that dysfunctional PI metabolism causes retinal degenerative diseases. In other cell types, various PIPs are generated through the activation receptors. However, in the retina, light stimulates the activation of phosphoinositide-metabolizing enzymes. The mechanism of these enzyme activations is currently under investigation. Future studies should be aimed at the function and mechanism of activation of phosphoinositide-metabolizing enzymes/phosphatases and PIP function and regulation. The phosphoinositide metabolism in the cone synapse is different from that of rod synapses. Identification of phosphoinositide kinases and phosphoinositide phosphatases and identification and localization of specific PIPs in various cell types of the retina in health and disease would advance the field.
Conflict of interest
The author declares that he has no conflicts of interest with the contents of this article.
Acknowledgments
The author acknowledges Ms. Kathy J. Kyler, Staff Editor, University of Oklahoma Health Sciences Center, for editing this manuscript.
Author ORCIDs
Raju V. S. Rajala https://orcid.org/0000-0003-3783-8504
Funding and additional information
This study was supported by National Institutes of Health Grants EY00871 and EY030024 and National Eye Institute Core Grant EY12190, the BrightFocus Foundation, and an unrestricted grant from Research to Prevent Blindness, Inc. to the University of Oklahoma Health Sciences Center Department of Ophthalmology. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sasaki T., Takasuga S., Sasaki J., Kofuji S., Eguchi S., Yamazaki M., Suzuki A. Mammalian phosphoinositide kinases and phosphatases. Prog. Lipid Res. 2009;48:307–343. doi: 10.1016/j.plipres.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Rusten T.E., Stenmark H. Analyzing phosphoinositides and their interacting proteins. Nat. Methods. 2006;3:251–258. doi: 10.1038/nmeth867. [DOI] [PubMed] [Google Scholar]
- 4.Rameh L.E., Cantley L.C. The role of phosphoinositide 3-kinase lipid products in cell function. J. Biol. Chem. 1999;274:8347–8350. doi: 10.1074/jbc.274.13.8347. [DOI] [PubMed] [Google Scholar]
- 5.Ghalayini A.J., Anderson R.E. Light adaptation of bovine retinas in situ stimulates phosphatidylinositol synthesis in rod outer segments in vitro. Curr. Eye Res. 1995;14:1025–1029. doi: 10.3109/02713689508998525. [DOI] [PubMed] [Google Scholar]
- 6.Huang Z., Ghalayini A., Guo X.X., Alvarez K.M., Anderson R.E. Light-mediated activation of diacylglycerol kinase in rat and bovine rod outer segments. J. Neurochem. 2000;75:355–362. doi: 10.1046/j.1471-4159.2000.0750355.x. [DOI] [PubMed] [Google Scholar]
- 7.Guo X., Ghalayini A.J., Chen H., Anderson R.E. Phosphatidylinositol 3-kinase in bovine photoreceptor rod outer segments. Invest. Ophthalmol. Vis. Sci. 1997;38:1873–1882. [PubMed] [Google Scholar]
- 8.Guo X.X., Huang Z., Bell M.W., Chen H., Anderson R.E. Tyrosine phosphorylation is involved in phosphatidylinositol 3-kinase activation in bovine rod outer segments. Mol. Vis. 2000;6:216–221. [PubMed] [Google Scholar]
- 9.Martin T.F. Phosphoinositide lipids as signaling molecules: common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu. Rev. Cell Dev. Biol. 1998;14:231–264. doi: 10.1146/annurev.cellbio.14.1.231. [DOI] [PubMed] [Google Scholar]
- 10.Fruman D.A., Meyers R.E., Cantley L.C. Phosphoinositide kinases. Annu. Rev. Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 11.Anderson R.E., Hollyfield J.G. Light stimulates the incorporation of inositol into phosphatidylinositol in the retina. Biochim. Biophys. Acta. 1981;665:619–622. doi: 10.1016/0005-2760(81)90280-0. [DOI] [PubMed] [Google Scholar]
- 12.Anderson R.E., Maude M.B., Kelleher P.A., Rayborn M.E., Hollyfield J.G. Phosphoinositide metabolism in the retina: localization to horizontal cells and regulation by light and divalent cations. J. Neurochem. 1983;41:764–771. doi: 10.1111/j.1471-4159.1983.tb04806.x. [DOI] [PubMed] [Google Scholar]
- 13.Anderson R.E., Hollyfield J.G. Inositol incorporation into phosphoinositides in retinal horizontal cells of Xenopus laevis: enhancement by acetylcholine, inhibition by glycine. J. Cell Biol. 1984;99:686–691. doi: 10.1083/jcb.99.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wensel T.G. Phosphoinositides in retinal function and disease. Cells. 2020;9:866. doi: 10.3390/cells9040866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broekhuyse R.M. Phospholipids in tissues of the eye. I. Isolation, characterization and quantitative analysis by two-dimensional thin-layer chromatography of diacyl and vinyl-ether phospholipids. Biochim. Biophys. Acta. 1968;152:307–315. doi: 10.1016/0005-2760(68)90038-6. [DOI] [PubMed] [Google Scholar]
- 16.Anderson R.E., Feldman L.S., Feldman G.L. Lipids of ocular tissues. II. The phospholipids of mature bovine and rabbit whole retina. Biochim. Biophys. Acta. 1970;202:367–373. [PubMed] [Google Scholar]
- 17.Anderson R.E. Lipids of ocular tissues. IV. A comparison of the phospholipids from the retina of six mammalian species. Exp. Eye Res. 1970;10:339–344. doi: 10.1016/s0014-4835(70)80046-x. [DOI] [PubMed] [Google Scholar]
- 18.Anderson R.E., Lissandrello P.M., Maude M.B., Matthes M.T. Lipids of bovine retinal pigment epithelium. Exp. Eye Res. 1976;23:149–157. doi: 10.1016/0014-4835(76)90198-6. [DOI] [PubMed] [Google Scholar]
- 19.Anderson R.E., Maude M.B., Zimmerman W. Lipids of ocular tissues–X. Lipid composition of subcellular fractions of bovine retina. Vision Res. 1975;15:1087–1090. doi: 10.1016/0042-6989(75)90005-x. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt S.Y. Light- and cytidine-dependent phosphatidylinositol synthesis in photoreceptor cells of the rat. J. Cell Biol. 1983;97:832–837. doi: 10.1083/jcb.97.3.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt S.Y. Light enhances the turnover of phosphatidylinositol in rat retinas. J. Neurochem. 1983;40:1630–1638. doi: 10.1111/j.1471-4159.1983.tb08136.x. [DOI] [PubMed] [Google Scholar]
- 22.Jung H.H., Reme C.E., Pfeilschifter J. Light evoked inositol trisphosphate release in the rat retina in vitro. Curr. Eye Res. 1993;12:727–732. doi: 10.3109/02713689308995768. [DOI] [PubMed] [Google Scholar]
- 23.Day N.S., Koutz C.A., Anderson R.E. Inositol-1,4,5-trisphosphate receptors in the vertebrate retina. Curr. Eye Res. 1993;12:981–992. doi: 10.3109/02713689309029224. [DOI] [PubMed] [Google Scholar]
- 24.Ghalayini A.J., Tarver A.P., Mackin W.M., Koutz C.A., Anderson R.E. Identification and immunolocalization of phospholipase C in bovine rod outer segments. J. Neurochem. 1991;57:1405–1412. doi: 10.1111/j.1471-4159.1991.tb08307.x. [DOI] [PubMed] [Google Scholar]
- 25.Peng Y.W., Rhee S.G., Yu W.P., Ho Y.K., Schoen T., Chader G.J., Yau K.W. Identification of components of a phosphoinositide signaling pathway in retinal rod outer segments. Proc. Natl. Acad. Sci. USA. 1997;94:1995–2000. doi: 10.1073/pnas.94.5.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaupp U.B., Seifert R. Cyclic nucleotide-gated ion channels. Physiol. Rev. 2002;82:769–824. doi: 10.1152/physrev.00008.2002. [DOI] [PubMed] [Google Scholar]
- 27.Yau K.W., Hardie R.C. Phototransduction motifs and variations. Cell. 2009;139:246–264. doi: 10.1016/j.cell.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt T.M., Chen S.K., Hattar S. Intrinsically photosensitive retinal ganglion cells: many subtypes, diverse functions. Trends Neurosci. 2011;34:572–580. doi: 10.1016/j.tins.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orisme W., Li J., Goldmann T., Bolch S., Wolfrum U., Smith W.C. Light-dependent translocation of arrestin in rod photoreceptors is signaled through a phospholipase C cascade and requires ATP. Cell. Signal. 2010;22:447–456. doi: 10.1016/j.cellsig.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He F., Mao M., Wensel T.G. Enhancement of phototransduction g protein-effector interactions by phosphoinositides. J. Biol. Chem. 2004;279:8986–8990. doi: 10.1074/jbc.M311488200. [DOI] [PubMed] [Google Scholar]
- 31.Bright S.R., Rich E.D., Varnum M.D. Regulation of human cone cyclic nucleotide-gated channels by endogenous phospholipids and exogenously applied phosphatidylinositol 3,4,5-trisphosphate. Mol. Pharmacol. 2007;71:176–183. doi: 10.1124/mol.106.026401. [DOI] [PubMed] [Google Scholar]
- 32.Brady J.D., Rich E.D., Martens J.R., Karpen J.W., Varnum M.D., Brown R.L. Interplay between PIP3 and calmodulin regulation of olfactory cyclic nucleotide-gated channels. Proc. Natl. Acad. Sci. USA. 2006;103:15635–15640. doi: 10.1073/pnas.0603344103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deretic D., Traverso V., Parkins N., Jackson F., Rodriguez de Turco E.B., Ransom N. Phosphoinositides, ezrin/moesin, and rac1 regulate fusion of rhodopsin transport carriers in retinal photoreceptors. Mol. Biol. Cell. 2004;15:359–370. doi: 10.1091/mbc.E03-04-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spencer W.J., Lewis T.R., Phan S., Cady M.A., Serebrovskaya E.O., Schneider N.F., Kim K.Y., Cameron L.A., Skiba N.P., Ellisman M.H. Photoreceptor disc membranes are formed through an Arp2/3-dependent lamellipodium-like mechanism. Proc. Natl. Acad. Sci. USA. 2019;116:27043–27052. doi: 10.1073/pnas.1913518117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bucki R., Wang Y.H., Yang C., Kandy S.K., Fatunmbi O., Bradley R., Pogoda K., Svitkina T., Radhakrishnan R., Janmey P.A. Lateral distribution of phosphatidylinositol 4,5-bisphosphate in membranes regulates formin- and ARP2/3-mediated actin nucleation. J. Biol. Chem. 2019;294:4704–4722. doi: 10.1074/jbc.RA118.005552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daste F., Walrant A., Holst M.R., Gadsby J.R., Mason J., Lee J.E., Brook D., Mettlen M., Larsson E., Lee S.F. Control of actin polymerization via the coincidence of phosphoinositides and high membrane curvature. J. Cell Biol. 2017;216:3745–3765. doi: 10.1083/jcb.201704061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giusto N.M., Pasquare S.J., Salvador G.A., Ilincheta de Boschero M.G. Lipid second messengers and related enzymes in vertebrate rod outer segments. J. Lipid Res. 2010;51:685–700. doi: 10.1194/jlr.R001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson R.E., Maude M.B., Kelleher P.A., Maida T.M., Basinger S.F. Metabolism of phosphatidylcholine in the frog retina. Biochim. Biophys. Acta. 1980;620:212–226. doi: 10.1016/0005-2760(80)90203-9. [DOI] [PubMed] [Google Scholar]
- 39.Anderson R.E., Kelleher P.A., Maude M.B. Metabolism of phosphatidylethanolamine in the frog retina. Biochim. Biophys. Acta. 1980;620:227–235. doi: 10.1016/0005-2760(80)90204-0. [DOI] [PubMed] [Google Scholar]
- 40.Anderson R.E., Maude M.B., Pu G.A., Hollyfield J.G. Effect of light on the metabolism of lipids in the rat retina. J. Neurochem. 1985;44:773–778. doi: 10.1111/j.1471-4159.1985.tb12882.x. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt S.Y. Phosphatidylinositol synthesis and phosphorylation are enhanced by light in rat retinas. J. Biol. Chem. 1983;258:6863–6868. [PubMed] [Google Scholar]
- 42.Choe H.G., Ghalayini A.J., Anderson R.E. Phosphoinositide metabolism in frog rod outer segments. Exp. Eye Res. 1990;51:167–176. doi: 10.1016/0014-4835(90)90069-7. [DOI] [PubMed] [Google Scholar]
- 43.Ghalayini A.J., Guo X.X., Koutz C.A., Anderson R.E. Light stimulates tyrosine phosphorylation of rat rod outer segments In vivo. Exp. Eye Res. 1998;66:817–821. doi: 10.1006/exer.1998.0498. [DOI] [PubMed] [Google Scholar]
- 44.Brown J.E., Blazynski C., Cohen A.I. Light induces a rapid and transient increase in inositol-trisphosphate in toad rod outer segments. Biochem. Biophys. Res. Commun. 1987;146:1392–1396. doi: 10.1016/0006-291x(87)90804-7. [DOI] [PubMed] [Google Scholar]
- 45.Das N.D., Yoshioka T., Samuelson D., Shichi H. Immunocytochemical localization of phosphatidylinositol-4,5-bisphosphate in dark- and light-adapted rat retinas. Cell Struct. Funct. 1986;11:53–63. doi: 10.1247/csf.11.53. [DOI] [PubMed] [Google Scholar]
- 46.Ferreira P.A., Shortridge R.D., Pak W.L. Distinctive subtypes of bovine phospholipase C that have preferential expression in the retina and high homology to the norpA gene product of Drosophila. Proc. Natl. Acad. Sci. USA. 1993;90:6042–6046. doi: 10.1073/pnas.90.13.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gehm B.D., Mc Connell D.G. Phosphoinositide synthesis in bovine rod outer segments. Biochemistry. 1990;29:5442–5446. doi: 10.1021/bi00475a005. [DOI] [PubMed] [Google Scholar]
- 48.Gehm B.D., Pinke R.M., Laquerre S., Chafouleas J.G., Schultz D.A., Pepperl D.J., McConnell D.G. Activation of bovine rod outer segment phosphatidylinositol-4,5-bisphosphate phospholipase C by calmodulin antagonists does not depend on calmodulin. Biochemistry. 1991;30:11302–11306. doi: 10.1021/bi00111a016. [DOI] [PubMed] [Google Scholar]
- 49.Hayashi F., Amakawa T. Light-mediated breakdown of phosphatidylinositol-4,5-bisphosphate in isolated rod outer segments of frog photoreceptor. Biochem. Biophys. Res. Commun. 1985;128:954–959. doi: 10.1016/0006-291x(85)90139-1. [DOI] [PubMed] [Google Scholar]
- 50.Jelsema C.L. Regulation of phospholipase A2 and phospholipase C in rod outer segments of bovine retina involves a common GTP-binding protein but different mechanisms of action. Ann. N. Y. Acad. Sci. 1989;559:158–177. doi: 10.1111/j.1749-6632.1989.tb22607.x. [DOI] [PubMed] [Google Scholar]
- 51.Millar F.A., Fisher S.C., Muir C.A., Edwards E., Hawthorne J.N. Polyphosphoinositide hydrolysis in response to light stimulation of rat and chick retina and retinal rod outer segments. Biochim. Biophys. Acta. 1988;970:205–211. doi: 10.1016/0167-4889(88)90180-2. [DOI] [PubMed] [Google Scholar]
- 52.Rajala A., Anderson R.E., Ma J.X., Lem J., Al Ubaidi M.R., Rajala R.V. G-protein-coupled receptor rhodopsin regulates the phosphorylation of retinal insulin receptor. J. Biol. Chem. 2007;282:9865–9873. doi: 10.1074/jbc.M608845200. [DOI] [PubMed] [Google Scholar]
- 53.He F., Agosto M.A., Anastassov I.A., Tse D.Y., Wu S.M., Wensel T.G. Phosphatidylinositol-3-phosphate is light-regulated and essential for survival in retinal rods. Sci. Rep. 2016;6:26978. doi: 10.1038/srep26978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajala R.V., Rajala A., Morris A.J., Anderson R.E. Phosphoinositides: minor lipids make a major impact on photoreceptor cell functions. Sci. Rep. 2014;4:5463. doi: 10.1038/srep05463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roque M.E., Salvador G.A., Giusto N.M. Light activation of phosphatidylethanolamine N-methyltransferase in rod outer segments and its modulation by association states of transducin. Exp. Eye Res. 1999;69:555–562. doi: 10.1006/exer.1999.0738. [DOI] [PubMed] [Google Scholar]
- 56.Castagnet P.I., Giusto N.M. Properties of phospholipase A2 activity from bovine retinal rod outer segments. Exp. Eye Res. 1993;56:709–719. doi: 10.1006/exer.1993.1088. [DOI] [PubMed] [Google Scholar]
- 57.Salvador G.A., Giusto N.M. Phospholipase D from photoreceptor rod outer segments is a downstream effector of RhoA: evidence of a light-dependent mechanism. Exp. Eye Res. 2006;83:202–211. doi: 10.1016/j.exer.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 58.Giusto N.M., Pasquare S.J., Salvador G.A., Castagnet P.I., Roque M.E., Ilincheta de Boschero M.G. Lipid metabolism in vertebrate retinal rod outer segments. Prog. Lipid Res. 2000;39:315–391. doi: 10.1016/s0163-7827(00)00009-6. [DOI] [PubMed] [Google Scholar]
- 59.Ghalayini A., Anderson R.E. Phosphatidylinositol 4,5-bisphosphate: light-mediated breakdown in the vertebrate retina. Biochem. Biophys. Res. Commun. 1984;124:503–506. doi: 10.1016/0006-291x(84)91582-1. [DOI] [PubMed] [Google Scholar]
- 60.Ghalayini A.J., Anderson R.E. Activation of bovine rod outer segment phospholipase C by arrestin. J. Biol. Chem. 1992;267:17977–17982. [PubMed] [Google Scholar]
- 61.Ghalayini A.J., Weber N.R., Rundle D.R., Koutz C.A., Lambert D., Guo X.X., Anderson R.E. Phospholipase Cgamma1 in bovine rod outer segments: immunolocalization and light-dependent binding to membranes. J. Neurochem. 1998;70:171–178. doi: 10.1046/j.1471-4159.1998.70010171.x. [DOI] [PubMed] [Google Scholar]
- 62.Natalini P.M., Mateos M.V., Ilincheta de Boschero M.G., Giusto N.M. A novel light-dependent activation of DAGK and PKC in bovine photoreceptor nuclei. Exp. Eye Res. 2014;125:142–155. doi: 10.1016/j.exer.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 63.Pasquaré S.J., Salvador G.A., Roque M.E., Giusto N.M. Effect of light on phosphatidate phosphohydrolase activity of retina rod outer segments: the role of transducin. Arch. Biochem. Biophys. 2000;379:299–306. doi: 10.1006/abbi.2000.1861. [DOI] [PubMed] [Google Scholar]
- 64.Balasubramanian N., Slepak V.Z. Light-mediated activation of Rac-1 in photoreceptor outer segments. Curr. Biol. 2003;13:1306–1310. doi: 10.1016/s0960-9822(03)00511-6. [DOI] [PubMed] [Google Scholar]
- 65.Natalini P.M., Mateos M.V., Ilincheta de Boschero M.G., Giusto N.M. Insulin-related signaling pathways elicited by light in photoreceptor nuclei from bovine retina. Exp. Eye Res. 2016;145:36–47. doi: 10.1016/j.exer.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 66.Li G., Rajala A., Wiechmann A.F., Anderson R.E., Rajala R.V. Activation and membrane binding of retinal protein kinase Balpha/Akt1 is regulated through light-dependent generation of phosphoinositides. J. Neurochem. 2008;107:1382–1397. doi: 10.1111/j.1471-4159.2008.05707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajala A., Daly R.J., Tanito M., Allen D.T., Holt L.J., Lobanova E.S., Arshavsky V.Y., Rajala R.V. Growth factor receptor-bound protein 14 undergoes light-dependent intracellular translocation in rod photoreceptors: functional role in retinal insulin receptor activation. Biochemistry. 2009;48:5563–5572. doi: 10.1021/bi9000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown J.R., Auger K.R. Phylogenomics of phosphoinositide lipid kinases: perspectives on the evolution of second messenger signaling and drug discovery. BMC Evol. Biol. 2011;11:4. doi: 10.1186/1471-2148-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cantley L.C. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 70.Zhao L., Vogt P.K. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hirsch E., Lembo G., Montrucchio G., Rommel C., Costa C., Barberis L. Signaling through PI3Kgamma: a common platform for leukocyte, platelet and cardiovascular stress sensing. Thromb. Haemost. 2006;95:29–35. [PubMed] [Google Scholar]
- 72.Hirsch E., Costa C., Ciraolo E. Phosphoinositide 3-kinases as a common platform for multi-hormone signaling. J. Endocrinol. 2007;194:243–256. doi: 10.1677/JOE-07-0097. [DOI] [PubMed] [Google Scholar]
- 73.Geering B., Cutillas P.R., Nock G., Gharbi S.I., Vanhaesebroeck B. Class IA phosphoinositide 3-kinases are obligate p85-p110 heterodimers. Proc. Natl. Acad. Sci. USA. 2007;104:7809–7814. doi: 10.1073/pnas.0700373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ivanovic I., Allen D.T., Dighe R., Le Y.Z., Anderson R.E., Rajala R.V. Phosphoinositide 3-kinase signaling in retinal rod photoreceptors. Invest. Ophthalmol. Vis. Sci. 2011;52:6355–6362. doi: 10.1167/iovs.10-7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pons S., Asano T., Glasheen E., Miralpeix M., Zhang Y., Fisher T.L., Myers M.G., Jr., Sun X.J., White M.F. The structure and function of p55PIK reveal a new regulatory subunit for phosphatidylinositol 3-kinase. Mol. Cell. Biol. 1995;15:4453–4465. doi: 10.1128/mcb.15.8.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Antonetti D.A., Algenstaedt P., Kahn C.R. Insulin receptor substrate 1 binds two novel splice variants of the regulatory subunit of phosphatidylinositol 3-kinase in muscle and brain. Mol. Cell. Biol. 1996;16:2195–2203. doi: 10.1128/mcb.16.5.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Inukai K., Anai M., Van Breda E., Hosaka T., Katagiri H., Funaki M., Fukushima Y., Ogihara T., Yazaki Y., Kikuchi M. A novel 55-kDa regulatory subunit for phosphatidylinositol 3-kinase structurally similar to p55PIK Is generated by alternative splicing of the p85alpha gene. J. Biol. Chem. 1996;271:5317–5320. doi: 10.1074/jbc.271.10.5317. [DOI] [PubMed] [Google Scholar]
- 78.Hiles I.D., Otsu M., Volinia S., Fry M.J., Gout I., Dhand R., Panayotou G., Ruiz-Larrea F., Thompson A., Totty N.F. Phosphatidylinositol 3-kinase: structure and expression of the 110 kd catalytic subunit. Cell. 1992;70:419–429. doi: 10.1016/0092-8674(92)90166-a. [DOI] [PubMed] [Google Scholar]
- 79.Ivanovic I., Anderson R.E., Le Y.Z., Fliesler S.J., Sherry D.M., Rajala R.V. Deletion of the p85alpha regulatory subunit of phosphoinositide 3-kinase in cone photoreceptor cells results in cone photoreceptor degeneration. Invest. Ophthalmol. Vis. Sci. 2011;52:3775–3783. doi: 10.1167/iovs.10-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rajala R.V. Phosphoinositide 3-kinase signaling in the vertebrate retina. J. Lipid Res. 2010;51:4–22. doi: 10.1194/jlr.R000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murga C., Fukuhara S., Gutkind J.S. A novel role for phosphatidylinositol 3-kinase beta in signaling from G protein-coupled receptors to Akt. J. Biol. Chem. 2000;275:12069–12073. doi: 10.1074/jbc.275.16.12069. [DOI] [PubMed] [Google Scholar]
- 82.Stephens L.R., Eguinoa A., Erdjument-Bromage H., Lui M., Cooke F., Coadwell J., Smrcka A.S., Thelen M., Cadwallader K., Tempst P. The G beta gamma sensitivity of a PI3K is dependent upon a tightly associated adaptor, p101. Cell. 1997;89:105–114. doi: 10.1016/s0092-8674(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 83.Suire S., Hawkins P., Stephens L. Activation of phosphoinositide 3-kinase gamma by Ras. Curr. Biol. 2002;12:1068–1075. doi: 10.1016/s0960-9822(02)00933-8. [DOI] [PubMed] [Google Scholar]
- 84.Voigt P., Brock C., Nurnberg B., Schaefer. M. Assigning functional domains within the p101 regulatory subunit of phosphoinositide 3-kinase gamma. J. Biol. Chem. 2005;280:5121–5127. doi: 10.1074/jbc.M413104200. [DOI] [PubMed] [Google Scholar]
- 85.Weis W.I., Kobilka B.K. The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 2018;87:897–919. doi: 10.1146/annurev-biochem-060614-033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smrcka A.V. G protein betagamma subunits: central mediators of G protein-coupled receptor signaling. Cell. Mol. Life Sci. 2008;65:2191–2214. doi: 10.1007/s00018-008-8006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Murga C., Laguinge L., Wetzker R., Cuadrado A., Gutkind J.S. Activation of Akt/protein kinase B by G protein-coupled receptors. A role for alpha and beta gamma subunits of heterotrimeric G proteins acting through phosphatidylinositol-3-OH kinasegamma. J. Biol. Chem. 1998;273:19080–19085. doi: 10.1074/jbc.273.30.19080. [DOI] [PubMed] [Google Scholar]
- 88.Curnock A.P., Logan M.K., Ward S.G. Chemokine signalling: pivoting around multiple phosphoinositide 3-kinases. Immunology. 2002;105:125–136. doi: 10.1046/j.1365-2567.2002.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Margaria J.P., Ratto E., Gozzelino L., Li H., Hirsch E. Class II PI3Ks at the intersection between signal transduction and membrane trafficking. Biomolecules. 2019;9:104. doi: 10.3390/biom9030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gulluni F., De Santis M.C., Margaria J.P., Martini M., Hirsch E. Class II PI3K functions in cell biology and disease. Trends Cell Biol. 2019;29:339–359. doi: 10.1016/j.tcb.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 91.Posor Y., Eichhorn-Grunig M., Haucke V. Phosphoinositides in endocytosis. Biochim. Biophys. Acta. 2015;1851:794–804. doi: 10.1016/j.bbalip.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 92.Backer J.M. The regulation and function of class III PI3Ks: novel roles for Vps34. Biochem. J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 93.Marat A.L., Haucke V. Phosphatidylinositol 3-phosphates–at the interface between cell signalling and membrane traffic. EMBO J. 2016;35:561–579. doi: 10.15252/embj.201593564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miller S., Tavshanjian B., Oleksy A., Perisic O., Houseman B.T., Shokat K.M., Williams R.L. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327:1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stjepanovic G., Baskaran S., Lin M.G., Hurley J.H. Vps34 kinase domain dynamics regulate the autophagic PI 3-kinase complex. Mol. Cell. 2017;67:528–534.e3. doi: 10.1016/j.molcel.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mayinger P. Phosphoinositides and vesicular membrane traffic. Biochim. Biophys. Acta. 2012;1821:1104–1113. doi: 10.1016/j.bbalip.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gillooly D.J., Simonsen A., Stenmark H. Phosphoinositides and phagocytosis. J. Cell Biol. 2001;155:15–17. doi: 10.1083/jcb.200109001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kale S.D., Gu B., Capelluto D.G., Dou D., Feldman E., Rumore A., Arredondo F.D., Hanlon R., Fudal I., Rouxel T. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell. 2010;142:284–295. doi: 10.1016/j.cell.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 99.He F., Nichols R.M., Kailasam L., Wensel T.G., Agosto M.A. Critical role for phosphatidylinositol-3 kinase Vps34/PIK3C3 in ON-bipolar cells. Invest. Ophthalmol. Vis. Sci. 2019;60:2861–2874. doi: 10.1167/iovs.19-26586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He F., Agosto M.A., Nichols R.M., Wensel T.G. Multiple phosphatidylinositol(3)phosphate roles in retinal pigment epithelium membrane recycling. bioRxiv. 2020 doi: 10.1101/2020.01.09.899815. [DOI] [Google Scholar]
- 101.Takai H., Wang R.C., Takai K.K., Yang H., de Lange T. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131:1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 102.Thomas D., Powell J.A., Green B.D., Barry E.F., Ma Y., Woodcock J., Fitter S., Zannettino A.C., Pitson S.M., Hughes T.P. Protein kinase activity of phosphoinositide 3-kinase regulates cytokine-dependent cell survival. PLoS Biol. 2013;11:e1001515. doi: 10.1371/journal.pbio.1001515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rajala R.V., Anderson R.E. Interaction of the insulin receptor beta-subunit with phosphatidylinositol 3-kinase in bovine ROS. Invest. Ophthalmol. Vis. Sci. 2001;42:3110–3117. [PubMed] [Google Scholar]
- 104.Rajala R.V., McClellan M.E., Ash J.D., Anderson R.E. In vivo regulation of phosphoinositide 3-kinase in retina through light-induced tyrosine phosphorylation of the insulin receptor beta-subunit. J. Biol. Chem. 2002;277:43319–43326. doi: 10.1074/jbc.M206355200. [DOI] [PubMed] [Google Scholar]
- 105.Rajala A., Rajala R.V.S. A non-canonical rhodopsin-mediated insulin receptor signaling pathway in retinal photoreceptor neurons. Cell Biol. Int. 2020;44:1020–1027. doi: 10.1002/cbin.11299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Reiter C.E., Sandirasegarane L., Wolpert E.B., Klinger M., Simpson I.A., Barber A.J., Antonetti D.A., Kester M., Gardner T.W. Characterization of insulin signaling in rat retina in vivo and ex vivo. Am. J. Physiol. Endocrinol. Metab. 2003;285:E763–E774. doi: 10.1152/ajpendo.00507.2002. [DOI] [PubMed] [Google Scholar]
- 107.Rajala R.V., Wiskur B., Tanito M., Callegan M., Rajala A. Diabetes reduces autophosphorylation of retinal insulin receptor and increases protein-tyrosine phosphatase-1B activity. Invest. Ophthalmol. Vis. Sci. 2009;50:1033–1040. doi: 10.1167/iovs.08-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rajala R.V., Tanito M., Neel B.G., Rajala A. Enhanced retinal insulin receptor-activated neuroprotective survival signal in mice lacking the protein-tyrosine phosphatase-1B gene. J. Biol. Chem. 2010;285:8894–8904. doi: 10.1074/jbc.M109.070854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Basavarajappa D.K., Gupta V.K., Dighe R., Rajala A., Rajala R.V. Phosphorylated Grb14 is an endogenous inhibitor of retinal protein tyrosine phosphatase 1B, and light-dependent activation of Src phosphorylates Grb14. Mol. Cell. Biol. 2011;31:3975–3987. doi: 10.1128/MCB.05659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rajala R.V., Anderson R.E. Rhodopsin-regulated insulin receptor signaling pathway in rod photoreceptor neurons. Mol. Neurobiol. 2010;42:39–47. doi: 10.1007/s12035-010-8130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rajala A., Tanito M., Le Y.Z., Kahn C.R., Rajala R.V. Loss of neuroprotective survival signal in mice lacking insulin receptor gene in rod photoreceptor cells. J. Biol. Chem. 2008;283:19781–19792. doi: 10.1074/jbc.M802374200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li G., Anderson R.E., Tomita H., Adler R., Liu X., Zack D.J., Rajala R.V. Nonredundant role of Akt2 for neuroprotection of rod photoreceptor cells from light-induced cell death. J. Neurosci. 2007;27:203–211. doi: 10.1523/JNEUROSCI.0445-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rajala A., Wang Y., Rajala R.V. Activation of oncogenic tyrosine kinase signaling promotes insulin receptor-mediated cone photoreceptor survival. Oncotarget. 2016;7:46924–46942. doi: 10.18632/oncotarget.10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rajala A., Dighe R., Agbaga M.P., Anderson R.E., Rajala R.V. Insulin receptor signaling in cones. J. Biol. Chem. 2013;288:19503–19515. doi: 10.1074/jbc.M113.469064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu S., Ladak R., Swanson D.A., Soltyk A., Sun H., Ploder L., Vidgen D., Duncan A.M., Garami E., Valle D. PHR1 encodes an abundant, pleckstrin homology domain-containing integral membrane protein in the photoreceptor outer segments. J. Biol. Chem. 1999;274:35676–35685. doi: 10.1074/jbc.274.50.35676. [DOI] [PubMed] [Google Scholar]
- 116.Xu S., Wang Y., Zhao H., Zhang L., Xiong W., Yau K.W., Hiel H., Glowatzki E., Ryugo D.K., Valle D. PHR1, a PH domain-containing protein expressed in primary sensory neurons. Mol. Cell. Biol. 2004;24:9137–9151. doi: 10.1128/MCB.24.20.9137-9151.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Krappa R., Nguyen A., Burrola P., Deretic D., Lemke G. Evectins: vesicular proteins that carry a pleckstrin homology domain and localize to post-Golgi membranes. Proc. Natl. Acad. Sci. USA. 1999;96:4633–4638. doi: 10.1073/pnas.96.8.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rajala R.V., Chan M.D., Rajala A. Lipid-protein interactions of growth factor receptor-bound protein 14 in insulin receptor signaling. Biochemistry. 2005;44:15461–15471. doi: 10.1021/bi0513148. [DOI] [PubMed] [Google Scholar]
- 119.Rajala R.V., McClellan M.E., Chan M.D., Tsiokas L., Anderson R.E. Interaction of the retinal insulin receptor beta-subunit with the P85 subunit of phosphoinositide 3-kinase. Biochemistry. 2004;43:5637–5650. doi: 10.1021/bi035913v. [DOI] [PubMed] [Google Scholar]
- 120.Yi X., Schubert M., Peachey N.S., Suzuma K., Burks D.J., Kushner J.A., Suzuma I., Cahill C., Flint C.L., Dow M.A. Insulin receptor substrate 2 is essential for maturation and survival of photoreceptor cells. J. Neurosci. 2005;25:1240–1248. doi: 10.1523/JNEUROSCI.3664-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tanaka M., Konishi H., Touhara K., Sakane F., Hirata M., Ono Y., Kikkawa U. Identification of myosin II as a binding protein to the PH domain of protein kinase B. Biochem. Biophys. Res. Commun. 1999;255:169–174. doi: 10.1006/bbrc.1999.0162. [DOI] [PubMed] [Google Scholar]
- 122.Weil D., Blanchard S., Kaplan J., Guilford P., Gibson F., Walsh J., Mburu P., Varela A., Levilliers J., Weston M.D. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- 123.Brognard J., Sierecki E., Gao T., Newton A.C. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol. Cell. 2007;25:917–931. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 124.Gao T., Furnari F., Newton A.C. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol. Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 125.Rajala A., Gupta V.K., Anderson R.E., Rajala R.V. Light activation of the insulin receptor regulates mitochondrial hexokinase. A possible mechanism of retinal neuroprotection. Mitochondrion. 2013;13:566–576. doi: 10.1016/j.mito.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rajala R.V., Kanan Y., Anderson R.E. Photoreceptor neuroprotection: regulation of Akt activation through serine/threonine phosphatases, PHLPP and PHLPPL. Adv. Exp. Med. Biol. 2016;854:419–424. doi: 10.1007/978-3-319-17121-0_55. [DOI] [PubMed] [Google Scholar]
- 127.Kanan Y., Matsumoto H., Song H., Sokolov M., Anderson R.E., Rajala R.V. Serine/threonine kinase Akt activation regulates the activity of retinal serine/threonine phosphatases, PHLPP and PHLPPL. J. Neurochem. 2010;113:477–488. doi: 10.1111/j.1471-4159.2010.06609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dilly A.K., Rajala R.V. Insulin growth factor 1 receptor/PI3K/AKT survival pathway in outer segment membranes of rod photoreceptors. Invest. Ophthalmol. Vis. Sci. 2008;49:4765–4773. doi: 10.1167/iovs.08-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Majewski N., Nogueira V., Bhaskar P., Coy P.E., Skeen J.E., Gottlob K., Chandel N.S., Thompson C.B., Robey R.B., Hay N. Hexokinase-mitochondria interaction mediated by Akt is required to inhibit apoptosis in the presence or absence of Bax and Bak. Mol. Cell. 2004;16:819–830. doi: 10.1016/j.molcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 130.Majewski N., Nogueira V., Robey R.B., Hay N. Akt inhibits apoptosis downstream of BID cleavage via a glucose-dependent mechanism involving mitochondrial hexokinases. Mol. Cell. Biol. 2004;24:730–740. doi: 10.1128/MCB.24.2.730-740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gottlob K., Majewski N., Kennedy S., Kandel E., Robey R.B., Hay N. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 2001;15:1406–1418. doi: 10.1101/gad.889901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Feng J., Lucchinetti E., Ahuja P., Pasch T., Perriard J.C., Zaugg M. Isoflurane postconditioning prevents opening of the mitochondrial permeability transition pore through inhibition of glycogen synthase kinase 3beta. Anesthesiology. 2005;103:987–995. doi: 10.1097/00000542-200511000-00013. [DOI] [PubMed] [Google Scholar]
- 133.Pastorino J.G., Hoek J.B., Shulga N. Activation of glycogen synthase kinase 3beta disrupts the binding of hexokinase II to mitochondria by phosphorylating voltage-dependent anion channel and potentiates chemotherapy-induced cytotoxicity. Cancer Res. 2005;65:10545–10554. doi: 10.1158/0008-5472.CAN-05-1925. [DOI] [PubMed] [Google Scholar]
- 134.Cross D.A., Alessi D.R., Cohen P., Andjelkovich M., Hemmings B.A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 135.Robey R.B., Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25:4683–4696. doi: 10.1038/sj.onc.1209595. [DOI] [PubMed] [Google Scholar]
- 136.Suire S., Coadwell J., Ferguson G.J., Davidson K., Hawkins P., Stephens L. p84, a new Gbetagamma-activated regulatory subunit of the type IB phosphoinositide 3-kinase p110gamma. Curr. Biol. 2005;15:566–570. doi: 10.1016/j.cub.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 137.Rubio I., Rodriguez-Viciana P., Downward J., Wetzker R. Interaction of Ras with phosphoinositide 3-kinase gamma. Biochem. J. 1997;326:891–895. doi: 10.1042/bj3260891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wennerberg K., Rossman K.L., Der C.J. The Ras superfamily at a glance. J. Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 139.Patel M., Cote J.F. Ras GTPases’ interaction with effector domains: Breaking the families’ barrier. Commun. Integr. Biol. 2013;6:e24298. doi: 10.4161/cib.24298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rajala R.V., Rajala A., Gupta V.K. Conservation and divergence of Grb7 family of Ras-binding domains. Protein Cell. 2012;3:60–70. doi: 10.1007/s13238-012-2001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gupta V.K., Rajala A., Rajala R.V. Non-canonical regulation of phosphatidylinositol 3-kinase gamma isoform activity in retinal rod photoreceptor cells. Cell Commun. Signal. 2015;13:7. doi: 10.1186/s12964-015-0087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rajala R.V., McClellan M.E., Ash J.D., Anderson R.E. Regulation of retinal phosphoinositide 3-kinase activity in p85alpha-subunit knockout mice. Adv. Exp. Med. Biol. 2003;533:369–376. doi: 10.1007/978-1-4615-0067-4_47. [DOI] [PubMed] [Google Scholar]
- 143.Rajala R.V., Ranjo-Bishop M., Wang Y., Rajala A., Anderson R.E. The p110alpha isoform of phosphoinositide 3-kinase is essential for cone photoreceptor survival. Biochimie. 2015;112:35–40. doi: 10.1016/j.biochi.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Streb H., Irvine R.F., Berridge M.J., Schulz I. Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate. Nature. 1983;306:67–69. doi: 10.1038/306067a0. [DOI] [PubMed] [Google Scholar]
- 145.Rajala R.V., Anderson R.E. Focus on molecules: phosphatidylinositol-4,5-bisphosphate (PIP2) Exp. Eye Res. 2010;91:324–325. doi: 10.1016/j.exer.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Anderson R.A., Boronenkov I.V., Doughman S.D., Kunz J., Loijens J.C. Phosphatidylinositol phosphate kinases, a multifaceted family of signaling enzymes. J. Biol. Chem. 1999;274:9907–9910. doi: 10.1074/jbc.274.15.9907. [DOI] [PubMed] [Google Scholar]
- 147.Divecha N., Brooksbank C.E., Irvine R.F. Purification and characterization of phosphatidylinositol 4-phosphate 5-kinases. Biochem. J. 1992;288:637–642. doi: 10.1042/bj2880637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Moritz A., De Graan P.N., Ekhart P.F., Gispen W.H., Wirtz K.W. Purification of a phosphatidylinositol 4-phosphate kinase from bovine brain membranes. J. Neurochem. 1990;54:351–354. doi: 10.1111/j.1471-4159.1990.tb13322.x. [DOI] [PubMed] [Google Scholar]
- 149.Boronenkov I.V., Anderson R.A. The sequence of phosphatidylinositol-4-phosphate 5-kinase defines a novel family of lipid kinases. J. Biol. Chem. 1995;270:2881–2884. doi: 10.1074/jbc.270.7.2881. [DOI] [PubMed] [Google Scholar]
- 150.Loijens J.C., Boronenkov I.V., Parker G.J., Anderson R.A. The phosphatidylinositol 4-phosphate 5-kinase family. Adv. Enzyme Regul. 1996;36:115–140. doi: 10.1016/0065-2571(95)00005-4. [DOI] [PubMed] [Google Scholar]
- 151.Rameh L.E., Tolias K.F., Duckworth B.C., Cantley L.C. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- 152.Smith C.D., Chang K.J. Regulation of brain phosphatidylinositol-4-phosphate kinase by GTP analogues. A potential role for guanine nucleotide regulatory proteins. J. Biol. Chem. 1989;264:3206–3210. [PubMed] [Google Scholar]
- 153.Martin A., Brown F.D., Hodgkin M.N., Bradwell A.J., Cook S.J., Hart M., Wakelam M.J. Activation of phospholipase D and phosphatidylinositol 4-phosphate 5-kinase in HL60 membranes is mediated by endogenous Arf but not Rho. J. Biol. Chem. 1996;271:17397–17403. doi: 10.1074/jbc.271.29.17397. [DOI] [PubMed] [Google Scholar]
- 154.Jenkins G.H., Fisette P.L., Anderson R.A. Type I phosphatidylinositol 4-phosphate 5-kinase isoforms are specifically stimulated by phosphatidic acid. J. Biol. Chem. 1994;269:11547–11554. [PubMed] [Google Scholar]
- 155.Bazenet C.E., Ruano A.R., Brockman J.L., Anderson R.A. The human erythrocyte contains two forms of phosphatidylinositol-4-phosphate 5-kinase which are differentially active toward membranes. J. Biol. Chem. 1990;265:18012–18022. [PubMed] [Google Scholar]
- 156.Cochet C., Filhol O., Payrastre B., Hunter T., Gill G.N. Interaction between the epidermal growth factor receptor and phosphoinositide kinases. J. Biol. Chem. 1991;266:637–644. [PubMed] [Google Scholar]
- 157.Choi S., Thapa N., Tan X., Hedman A.C., Anderson R.A. PIP kinases define PI4,5P2signaling specificity by association with effectors. Biochim. Biophys. Acta. 2015;1851:711–723. doi: 10.1016/j.bbalip.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sakagami H., Katsumata O., Hara Y., Tamaki H., Fukaya M. Preferential localization of type I phosphatidylinositol 4-phosphate 5-kinase γ at the periactive zone of mouse photoreceptor ribbon synapses. Brain Res. 2014;1586:23–33. doi: 10.1016/j.brainres.2014.08.051. [DOI] [PubMed] [Google Scholar]
- 159.Wenk M.R., Pellegrini L., Klenchin V.A., Di Paolo G., Chang S., Daniell L., Arioka M., Martin T.F., De Camilli P. PIP kinase Igamma is the major PI(4,5)P(2) synthesizing enzyme at the synapse. Neuron. 2001;32:79–88. doi: 10.1016/s0896-6273(01)00456-1. [DOI] [PubMed] [Google Scholar]
- 160.Wright B.D., Loo L., Street S.E., Ma A., Taylor-Blake B., Stashko M.A., Jin J., Janzen W.P., Frye S.V., Zylka M.J. The lipid kinase PIP5K1C regulates pain signaling and sensitization. Neuron. 2014;82:836–847. doi: 10.1016/j.neuron.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Di Paolo G., Moskowitz H.S., Gipson K., Wenk M.R., Voronov S., Obayashi M., Flavell R., Fitzsimonds R.M., Ryan T.A., De Camilli P. Impaired PtdIns(4,5)P2 synthesis in nerve terminals produces defects in synaptic vesicle trafficking. Nature. 2004;431:415–422. doi: 10.1038/nature02896. [DOI] [PubMed] [Google Scholar]
- 162.Zhao L., Chen Y., Bajaj A.O., Eblimit A., Xu M., Soens Z.T., Wang F., Ge Z., Jung S.Y., He F. Integrative subcellular proteomic analysis allows accurate prediction of human disease-causing genes. Genome Res. 2016;26:660–669. doi: 10.1101/gr.198911.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Huang Z., Guo X.X., Chen S.X., Alvarez K.M., Bell M.W., Anderson R.E. Regulation of type II phosphatidylinositol phosphate kinase by tyrosine phosphorylation in bovine rod outer segments. Biochemistry. 2001;40:4550–4559. doi: 10.1021/bi002575e. [DOI] [PubMed] [Google Scholar]
- 164.Huang Z., Anderson R.E., Cao W., Wiechmann A.F., Rajala R.V. Light-induced tyrosine phosphorylation of rod outer segment membrane proteins regulate the translocation, membrane binding and activation of type ii alpha phosphatidylinositol-5-phosphate 4-kinase. Neurochem. Res. 2011;36:627–635. doi: 10.1007/s11064-010-0146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Doughman R.L., Firestone A.J., Anderson R.A. Phosphatidylinositol phosphate kinases put PI4,5P(2) in its place. J. Membr. Biol. 2003;194:77–89. doi: 10.1007/s00232-003-2027-7. [DOI] [PubMed] [Google Scholar]
- 166.Womack K.B., Gordon S.E., He F., Wensel T.G., Lu C.C., Hilgemann D.W. Do phosphatidylinositides modulate vertebrate phototransduction? J. Neurosci. 2000;20:2792–2799. doi: 10.1523/JNEUROSCI.20-08-02792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hille B., Dickson E.J., Kruse M., Vivas O., Suh B.C. Phosphoinositides regulate ion channels. Biochim. Biophys. Acta. 2015;1851:844–856. doi: 10.1016/j.bbalip.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Michailidis I.E., Rusinova R., Georgakopoulos A., Chen Y., Iyengar R., Robakis N.K., Logothetis D.E., Baki L. Phosphatidylinositol-4,5-bisphosphate regulates epidermal growth factor receptor activation. Pflugers Arch. 2011;461:387–397. doi: 10.1007/s00424-010-0904-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shisheva A., Sbrissa D., Ikonomov O. Cloning, characterization, and expression of a novel Zn2+-binding FYVE finger-containing phosphoinositide kinase in insulin-sensitive cells. Mol. Cell. Biol. 1999;19:623–634. doi: 10.1128/mcb.19.1.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Vicinanza M., Korolchuk V.I., Ashkenazi A., Puri C., Menzies F.M., Clarke J.H., Rubinsztein D.C. PI(5)P regulates autophagosome biogenesis. Mol. Cell. 2015;57:219–234. doi: 10.1016/j.molcel.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cabezas A., Pattni K., Stenmark H. Cloning and subcellular localization of a human phosphatidylinositol 3-phosphate 5-kinase. PIKfyve/Fab1. Gene. 2006;371:34–41. doi: 10.1016/j.gene.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 172.Hasegawa J., Strunk B.S., Weisman L.S. PI5P and PI(3,5)P2: minor, but essential phosphoinositides. Cell Struct. Funct. 2017;42:49–60. doi: 10.1247/csf.17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Stenmark H., Aasland R., Driscoll P.C. The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett. 2002;513:77–84. doi: 10.1016/s0014-5793(01)03308-7. [DOI] [PubMed] [Google Scholar]
- 174.Nicot A.S., Laporte J. Endosomal phosphoinositides and human diseases. Traffic. 2008;9:1240–1249. doi: 10.1111/j.1600-0854.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Boisset G., Polok B.K., Schorderet D.F. Characterization of pip5k3 fleck corneal dystrophy-linked gene in zebrafish. Gene Expr. Patterns. 2008;8:404–410. doi: 10.1016/j.gep.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 176.Ikonomov O.C., Sbrissa D., Delvecchio K., Xie Y., Jin J.P., Rappolee D., Shisheva A. The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve-/= embryos but normality of PIKfyve+/- mice. J. Biol. Chem. 2011;286:13404–13413. doi: 10.1074/jbc.M111.222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liu Y., Lai Y.C., Hill E.V., Tyteca D., Carpentier S., Ingvaldsen A., Vertommen D., Lantier L., Foretz M., Dequiedt F. Phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) is an AMPK target participating in contraction-stimulated glucose uptake in skeletal muscle. Biochem. J. 2013;455:195–206. doi: 10.1042/BJ20130644. [DOI] [PubMed] [Google Scholar]
- 178.Ikonomov O.C., Sbrissa D., Delvecchio K., Feng H.Z., Cartee G.D., Jin J.P., Shisheva A. Muscle-specific Pikfyve gene disruption causes glucose intolerance, insulin resistance, adiposity, and hyperinsulinemia but not muscle fiber-type switching. Am. J. Physiol. Endocrinol. Metab. 2013;305:E119–E131. doi: 10.1152/ajpendo.00030.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tronchere H., Cinato M., Timotin A., Guitou L., Villedieu C., Thibault H., Baetz D., Payrastre B., Valet P., Parini A. Inhibition of PIKfyve prevents myocardial apoptosis and hypertrophy through activation of SIRT3 in obese mice. EMBO Mol. Med. 2017;9:770–785. doi: 10.15252/emmm.201607096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rajala R.V.S. Aerobic glycolysis in the retina: functional roles of pyruvate kinase isoforms. Front. Cell Dev. Biol. 2020;8:266. doi: 10.3389/fcell.2020.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rajala A., Soni K., Rajala R.V.S. Metabolic and non-metabolic roles of pyruvate kinase M2 isoform in diabetic retinopathy. Sci. Rep. 2020;10:7456. doi: 10.1038/s41598-020-64487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Simonsen A., Tooze S.A. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.McAlpine F., Williamson L.E., Tooze S.A., Chan E.Y. Regulation of nutrient-sensitive autophagy by uncoordinated 51-like kinases 1 and 2. Autophagy. 2013;9:361–373. doi: 10.4161/auto.23066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Biswas S.K., Zhao Y., Nagalingam A., Gardner T.W., Sandirasegarane L. PDGF- and insulin/IGF-1-specific distinct modes of class IA PI 3-kinase activation in normal rat retinas and RGC-5 retinal ganglion cells. Invest. Ophthalmol. Vis. Sci. 2008;49:3687–3698. doi: 10.1167/iovs.07-1455. [DOI] [PubMed] [Google Scholar]
- 185.Hollborn M., Bringmann A., Faude F., Wiedemann P., Kohen L. Signaling pathways involved in PDGF-evoked cellular responses in human RPE cells. Biochem. Biophys. Res. Commun. 2006;344:912–919. doi: 10.1016/j.bbrc.2006.03.185. [DOI] [PubMed] [Google Scholar]
- 186.Hollborn M., Jahn K., Limb G.A., Kohen L., Wiedemann P., Bringmann A. Characterization of the basic fibroblast growth factor-evoked proliferation of the human Muller cell line, MIO-M1. Graefes Arch. Clin. Exp. Ophthalmol. 2004;242:414–422. doi: 10.1007/s00417-004-0879-x. [DOI] [PubMed] [Google Scholar]
- 187.Ravichandran L.V., Chen H., Li Y., Quon M.J. Phosphorylation of PTP1B at Ser(50) by Akt impairs its ability to dephosphorylate the insulin receptor. Mol. Endocrinol. 2001;15:1768–1780. doi: 10.1210/mend.15.10.0711. [DOI] [PubMed] [Google Scholar]
- 188.Reiter C.E., Wu X., Sandirasegarane L., Nakamura M., Gilbert K.A., Singh R.S., Fort P.E., Antonetti D.A., Gardner T.W. Diabetes reduces basal retinal insulin receptor signaling: reversal with systemic and local insulin. Diabetes. 2006;55:1148–1156. doi: 10.2337/diabetes.55.04.06.db05-0744. [DOI] [PubMed] [Google Scholar]
- 189.Reiter C.E., Gardner T.W. Functions of insulin and insulin receptor signaling in retina: possible implications for diabetic retinopathy. Prog. Retin. Eye Res. 2003;22:545–562. doi: 10.1016/s1350-9462(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 190.Barber A.J., Nakamura M., Wolpert E.B., Reiter C.E., Seigel G.M., Antonetti D.A., Gardner T.W. Insulin rescues retinal neurons from apoptosis by a phosphatidylinositol 3-kinase/Akt-mediated mechanism that reduces the activation of caspase-3. J. Biol. Chem. 2001;276:32814–32821. doi: 10.1074/jbc.M104738200. [DOI] [PubMed] [Google Scholar]
- 191.Yu X., Rajala R.V., McGinnis J.F., Li F., Anderson R.E., Yan X., Li S., Elias R.V., Knapp R.R., Zhou X. Involvement of insulin/phosphoinositide 3-kinase/Akt signal pathway in 17 beta-estradiol-mediated neuroprotection. J. Biol. Chem. 2004;279:13086–13094. doi: 10.1074/jbc.M313283200. [DOI] [PubMed] [Google Scholar]
- 192.Cao W., Rajala R.V., Li F., Anderson R.E., Wei N., Soliman C.E., McGinnis J.F. Neuroprotective effect of estrogen upon retinal neurons in vitro. Adv. Exp. Med. Biol. 2003;533:395–402. doi: 10.1007/978-1-4615-0067-4_50. [DOI] [PubMed] [Google Scholar]
- 193.Park K.K., Liu K., Hu Y., Smith P.D., Wang C., Cai B., Xu B., Connolly L., Kramvis I., Sahin M. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Stitt A.W., Hughes S.J., Canning P., Lynch O., Cox O., Frizzell N., Thorpe S.R., Cotter T.G., Curtis T.M., Gardiner T.A. Substrates modified by advanced glycation end-products cause dysfunction and death in retinal pericytes by reducing survival signals mediated by platelet-derived growth factor. Diabetologia. 2004;47:1735–1746. doi: 10.1007/s00125-004-1523-3. [DOI] [PubMed] [Google Scholar]
- 195.Wang K., Spector A. Alpha-crystallin prevents irreversible protein denaturation and acts cooperatively with other heat-shock proteins to renature the stabilized partially denatured protein in an ATP-dependent manner. Eur. J. Biochem. 2000;267:4705–4712. doi: 10.1046/j.1432-1327.2000.01521.x. [DOI] [PubMed] [Google Scholar]
- 196.Defoe D.M., Grindstaff R.D. Epidermal growth factor stimulation of RPE cell survival: contribution of phosphatidylinositol 3-kinase and mitogen-activated protein kinase pathways. Exp. Eye Res. 2004;79:51–59. doi: 10.1016/j.exer.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 197.Yang P., Peairs J.J., Tano R., Jaffe G.J. Oxidant-mediated Akt activation in human RPE cells. Invest. Ophthalmol. Vis. Sci. 2006;47:4598–4606. doi: 10.1167/iovs.06-0140. [DOI] [PubMed] [Google Scholar]
- 198.Nakazawa T., Tamai M., Mori N. Brain-derived neurotrophic factor prevents axotomized retinal ganglion cell death through MAPK and PI3K signaling pathways. Invest. Ophthalmol. Vis. Sci. 2002;43:3319–3326. [PubMed] [Google Scholar]
- 199.Ikeda K., Tatsuno T., Noguchi H., Nakayama C. Ciliary neurotrophic factor protects rat retina cells in vitro and in vivo via PI3 kinase. Curr. Eye Res. 2004;29:349–355. doi: 10.1080/02713680490516279. [DOI] [PubMed] [Google Scholar]
- 200.Kretz A., Happold C.J., Marticke J.K., Isenmann S. Erythropoietin promotes regeneration of adult CNS neurons via Jak2/Stat3 and PI3K/AKT pathway activation. Mol. Cell. Neurosci. 2005;29:569–579. doi: 10.1016/j.mcn.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 201.Lavie Y., Dybowski J., Agranoff B.W. Wortmannin blocks goldfish retinal phosphatidylinositol 3-kinase and neurite outgrowth. Neurochem. Res. 1997;22:373–378. doi: 10.1023/a:1027391206791. [DOI] [PubMed] [Google Scholar]
- 202.Pimentel B., Rodriguez-Borlado L., Hernandez C., Carrera A.C. A Role for phosphoinositide 3-kinase in the control of cell division and survival during retinal development. Dev. Biol. 2002;247:295–306. doi: 10.1006/dbio.2002.0703. [DOI] [PubMed] [Google Scholar]
- 203.Ko M.L., Jian K., Shi L., Ko G.Y. Phosphatidylinositol 3 kinase-Akt signaling serves as a circadian output in the retina. J. Neurochem. 2009;108:1607–1620. doi: 10.1111/j.1471-4159.2009.05931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Johnson L.E., van Veen T., Ekstrom P.A. Differential Akt activation in the photoreceptors of normal and rd1 mice. Cell Tissue Res. 2005;320:213–222. doi: 10.1007/s00441-004-1046-8. [DOI] [PubMed] [Google Scholar]
- 205.Rajala R.V., Ivanovic I., Dilly A.K. Retinal insulin receptor signaling in hyperosmotic stress. Vitam. Horm. 2009;80:583–612. doi: 10.1016/S0083-6729(08)00620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chen J., Wang L., Chen Y., Sternberg P., Cai J. Phosphatidylinositol 3 kinase pathway and 4-hydroxy-2-nonenal-induced oxidative injury in the RPE. Invest. Ophthalmol. Vis. Sci. 2009;50:936–942. doi: 10.1167/iovs.08-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Schmitz F., Drenckhahn D. Li(+)-induced structural changes of synaptic ribbons are related to the phosphoinositide metabolism in photoreceptor synapses. Brain Res. 1993;604:142–148. doi: 10.1016/0006-8993(93)90360-y. [DOI] [PubMed] [Google Scholar]
- 208.Luttrell L.M., van Biesen T., Hawes B.E., Koch W.J., Krueger K.M., Touhara K., Lefkowitz R.J. G-protein-coupled receptors and their regulation: activation of the MAP kinase signaling pathway by G-protein-coupled receptors. Adv. Second Messenger Phosphoprotein Res. 1997;31:263–277. [PubMed] [Google Scholar]
- 209.Luttrell L.M., Daaka Y., Lefkowitz R.J. Regulation of tyrosine kinase cascades by G-protein-coupled receptors. Curr. Opin. Cell Biol. 1999;11:177–183. doi: 10.1016/s0955-0674(99)80023-4. [DOI] [PubMed] [Google Scholar]
- 210.Lev S., Hernandez J., Martinez R., Chen A., Plowman G., Schlessinger J. Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein. Mol. Cell. Biol. 1999;19:2278–2288. doi: 10.1128/mcb.19.3.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Dyson J.M., Fedele C.G., Davies E.M., Becanovic J., Mitchell C.A. Phosphoinositide phosphatases: just as important as the kinases. Subcell. Biochem. 2012;58:215–279. doi: 10.1007/978-94-007-3012-0_7. [DOI] [PubMed] [Google Scholar]
- 212.Verstreken P., Koh T.W., Schulze K.L., Zhai R.G., Hiesinger P.R., Zhou Y., Mehta S.Q., Cao Y., Roos J., Bellen H.J. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- 213.Chi Y., Zhou B., Wang W.Q., Chung S.K., Kwon Y.U., Ahn Y.H., Chang Y.T., Tsujishita Y., Hurley J.H., Zhang Z.Y. Comparative mechanistic and substrate specificity study of inositol polyphosphate 5-phosphatase Schizosaccharomyces pombe Synaptojanin and SHIP2. J. Biol. Chem. 2004;279:44987–44995. doi: 10.1074/jbc.M406416200. [DOI] [PubMed] [Google Scholar]
- 214.Chang-Ileto B., Frere S.G., Chan R.B., Voronov S.V., Roux A., Di Paolo G. Synaptojanin 1-mediated PI(4,5)P2 hydrolysis is modulated by membrane curvature and facilitates membrane fission. Dev. Cell. 2011;20:206–218. doi: 10.1016/j.devcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Holzhausen L.C., Lewis A.A., Cheong K.K., Brockerhoff S.E. Differential role for synaptojanin 1 in rod and cone photoreceptors. J. Comp. Neurol. 2009;517:633–644. doi: 10.1002/cne.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Van Epps H.A., Hayashi M., Lucast L., Stearns G.W., Hurley J.B., De Camilli P., Brockerhoff S.E. The zebrafish nrc mutant reveals a role for the polyphosphoinositide phosphatase synaptojanin 1 in cone photoreceptor ribbon anchoring. J. Neurosci. 2004;24:8641–8650. doi: 10.1523/JNEUROSCI.2892-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.George A.A., Hayden S., Stanton G.R., Brockerhoff S.E. Arf6 and the 5′phosphatase of synaptojanin 1 regulate autophagy in cone photoreceptors. BioEssays. 2016;38(Suppl. 1):S119–S135. doi: 10.1002/bies.201670913. [DOI] [PubMed] [Google Scholar]
- 218.Brockerhoff S.E. Phosphoinositides and photoreceptors. Mol. Neurobiol. 2011;44:420–425. doi: 10.1007/s12035-011-8208-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.George A.A., Hayden S., Holzhausen L.C., Ma E.Y., Suzuki S.C., Brockerhoff S.E. Synaptojanin 1 is required for endolysosomal trafficking of synaptic proteins in cone photoreceptor inner segments. PLoS One. 2014;9:e84394. doi: 10.1371/journal.pone.0084394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Voronov S.V., Frere S.G., Giovedi S., Pollina E.A., Borel C., Zhang H., Schmidt C., Akeson E.C., Wenk M.R., Cimasoni L. Synaptojanin 1-linked phosphoinositide dyshomeostasis and cognitive deficits in mouse models of Down’s syndrome. Proc. Natl. Acad. Sci. USA. 2008;105:9415–9420. doi: 10.1073/pnas.0803756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Prosseda P.P., Luo N., Wang B., Alvarado J.A., Hu Y., Sun Y. Loss of OCRL increases ciliary PI(4,5)P(2) in Lowe oculocerebrorenal syndrome. J. Cell Sci. 2017;130:3447–3454. doi: 10.1242/jcs.200857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Luo N., Conwell M.D., Chen X., Kettenhofen C.I., Westlake C.J., Cantor L.B., Wells C.D., Weinreb R.N., Corson T.W., Spandau D.F. Primary cilia signaling mediates intraocular pressure sensation. Proc. Natl. Acad. Sci. USA. 2014;111:12871–12876. doi: 10.1073/pnas.1323292111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Luo N., Lu J., Sun Y. Evidence of a role of inositol polyphosphate 5-phosphatase INPP5E in cilia formation in zebrafish. Vision Res. 2012;75:98–107. doi: 10.1016/j.visres.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Travaglini L., Brancati F., Silhavy J., Iannicelli M., Nickerson E., Elkhartoufi N., Scott E., Spencer E., Gabriel S., Thomas S. Phenotypic spectrum and prevalence of INPP5E mutations in Joubert syndrome and related disorders. Eur. J. Hum. Genet. 2013;21:1074–1078. doi: 10.1038/ejhg.2012.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Plotnikova O.V., Seo S., Cottle D.L., Conduit S., Hakim S., Dyson J.M., Mitchell C.A., Smyth I.M. INPP5E interacts with AURKA, linking phosphoinositide signaling to primary cilium stability. J. Cell Sci. 2015;128:364–372. doi: 10.1242/jcs.161323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Xu W., Jin M., Hu R., Wang H., Zhang F., Yuan S., Cao Y. The Joubert syndrome protein Inpp5e controls ciliogenesis by regulating phosphoinositides at the apical membrane. J. Am. Soc. Nephrol. 2017;28:118–129. doi: 10.1681/ASN.2015080906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Jo H.S., Kang K.H., Joe C.O., Kim J.W. Pten coordinates retinal neurogenesis by regulating Notch signalling. EMBO J. 2012;31:817–828. doi: 10.1038/emboj.2011.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kim J.W., Kang K.H., Burrola P., Mak T.W., Lemke G. Retinal degeneration triggered by inactivation of PTEN in the retinal pigment epithelium. Genes Dev. 2008;22:3147–3157. doi: 10.1101/gad.1700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Venkatesh A., Ma S., Le Y.Z., Hall M.N., Ruegg M.A., Punzo C. Activated mTORC1 promotes long-term cone survival in retinitis pigmentosa mice. J. Clin. Invest. 2015;125:1446–1458. doi: 10.1172/JCI79766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Tachibana N., Cantrup R., Dixit R., Touahri Y., Kaushik G., Zinyk D., Daftarian N., Biernaskie J., McFarlane S., Schuurmans C. Pten regulates retinal amacrine cell number by modulating Akt, Tgfβ, and Erk signaling. J. Neurosci. 2016;36:9454–9471. doi: 10.1523/JNEUROSCI.0936-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sakagami K., Chen B., Nusinowitz S., Wu H., Yang X.J. PTEN regulates retinal interneuron morphogenesis and synaptic layer formation. Mol. Cell. Neurosci. 2012;49:171–183. doi: 10.1016/j.mcn.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Xie C., Lu H., Nomura A., Hanse E.A., Forster C.L., Parker J.B., Linden M.A., Karasch C., Hallstrom T.C. Co-deleting Pten with Rb in retinal progenitor cells in mice results in fully penetrant bilateral retinoblastomas. Mol. Cancer. 2015;14:93. doi: 10.1186/s12943-015-0360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hopkins B.D., Hodakoski C., Barrows D., Mense S.M., Parsons R.E. PTEN function: the long and the short of it. Trends Biochem. Sci. 2014;39:183–190. doi: 10.1016/j.tibs.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Conduit S.E., Dyson J.M., Mitchell C.A. Inositol polyphosphate 5-phosphatases; new players in the regulation of cilia and ciliopathies. FEBS Lett. 2012;586:2846–2857. doi: 10.1016/j.febslet.2012.07.037. [DOI] [PubMed] [Google Scholar]
- 235.Ijuin T., Takenawa T. SKIP negatively regulates insulin-induced GLUT4 translocation and membrane ruffle formation. Mol. Cell. Biol. 2003;23:1209–1220. doi: 10.1128/MCB.23.4.1209-1220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ijuin T., Yu Y.E., Mizutani K., Pao A., Tateya S., Tamori Y., Bradley A., Takenawa T. Increased insulin action in SKIP heterozygous knockout mice. Mol. Cell. Biol. 2008;28:5184–5195. doi: 10.1128/MCB.01990-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Hao Y.M., Liu Q.J., Wang R.Y., Cao Y.P., Zhang Y., Zuo L.F. Single nucleotide polymorphisms on SHIP2 is associated with type 2 diabetes mellitus in Chinese Han population. Eur. Rev. Med. Pharmacol. Sci. 2015;19:129–137. [PubMed] [Google Scholar]
- 238.Sasaki J., Kofuji S., Itoh R., Momiyama T., Takayama K., Murakami H., Chida S., Tsuya Y., Takasuga S., Eguchi S. The PtdIns(3,4)P(2) phosphatase INPP4A is a suppressor of excitotoxic neuronal death. Nature. 2010;465:497–501. doi: 10.1038/nature09023. [DOI] [PubMed] [Google Scholar]
- 239.Ji L., Kim N.H., Huh S.O., Rhee H.J. Depletion of inositol polyphosphate 4-phosphatase II suppresses callosal axon formation in the developing mice. Mol. Cells. 2016;39:501–507. doi: 10.14348/molcells.2016.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Lenk G.M., Berry I.R., Stutterd C.A., Blyth M., Green L., Vadlamani G., Warren D., Craven I., Fanjul-Fernandez M., Rodriguez-Casero V. Cerebral hypomyelination associated with biallelic variants of FIG4. Hum. Mutat. 2019;40:619–630. doi: 10.1002/humu.23720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Brown J.E., Rubin L.J., Ghalayini A.J., Tarver A.P., Irvine R.F., Berridge M.J., Anderson R.E. myo-Inositol polyphosphate may be a messenger for visual excitation in Limulus photoreceptors. Nature. 1984;311:160–163. doi: 10.1038/311160a0. [DOI] [PubMed] [Google Scholar]
- 242.Hardie R.C., Raghu P. Visual transduction in Drosophila. Nature. 2001;413:186–193. doi: 10.1038/35093002. [DOI] [PubMed] [Google Scholar]
- 243.Wang T., Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- 244.Borst A. Drosophila’s view on insect vision. Curr. Biol. 2009;19:R36–R47. doi: 10.1016/j.cub.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 245.Hardie R.C. Phototransduction in Drosophila melanogaster. J. Exp. Biol. 2001;204:3403–3409. doi: 10.1242/jeb.204.20.3403. [DOI] [PubMed] [Google Scholar]
- 246.Suzuki T., Narita K., Yoshihara K., Nagai K., Kito Y. Phosphatidyl inositol-phospholipase C in squid photoreceptor membrane is activated by stable metarhodopsin via GTP-binding protein. Gq. Vision Res. 1995;35:1011–1017. doi: 10.1016/0042-6989(94)00219-c. [DOI] [PubMed] [Google Scholar]
- 247.Fruman D.A., Rameh L.E., Cantley L.C. Phosphoinositide binding domains: embracing 3-phosphate. Cell. 1999;97:817–820. doi: 10.1016/s0092-8674(00)80792-8. [DOI] [PubMed] [Google Scholar]
- 248.Baig A., Bao X., Wolf M., Haslam R.J. The platelet protein kinase C substrate pleckstrin binds directly to SDPR protein. Platelets. 2009;20:446–457. doi: 10.3109/09537100903137314. [DOI] [PubMed] [Google Scholar]
- 249.Furutani M., Tsujita K., Itoh T., Ijuin T., Takenawa T. Application of phosphoinositide-binding domains for the detection and quantification of specific phosphoinositides. Anal. Biochem. 2006;355:8–18. doi: 10.1016/j.ab.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 250.Milligan S.C., Alb J.G., Jr., Elagina R.B., Bankaitis V.A., Hyde D.R. The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J. Cell Biol. 1997;139:351–363. doi: 10.1083/jcb.139.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lee S.J., Xu H., Kang L.W., Amzel L.M., Montell C. Light adaptation through phosphoinositide-regulated translocation of Drosophila visual arrestin. Neuron. 2003;39:121–132. doi: 10.1016/s0896-6273(03)00390-8. [DOI] [PubMed] [Google Scholar]
- 252.Lee S.J., Montell C. Light-dependent translocation of visual arrestin regulated by the NINAC myosin III. Neuron. 2004;43:95–103. doi: 10.1016/j.neuron.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 253.Strissel K.J., Arshavsky V.Y. Myosin III illuminates the mechanism of arrestin translocation. Neuron. 2004;43:2–4. doi: 10.1016/j.neuron.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 254.Frechter S., Minke B. Light-regulated translocation of signaling proteins in Drosophila photoreceptors. J. Physiol. Paris. 2006;99:133–139. doi: 10.1016/j.jphysparis.2005.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Stocker H., Andjelkovic M., Oldham S., Laffargue M., Wymann M.P., Hemmings B.A., Hafen E. Living with lethal PIP3 levels: viability of flies lacking PTEN restored by a PH domain mutation in Akt/PKB. Science. 2002;295:2088–2091. doi: 10.1126/science.1068094. [DOI] [PubMed] [Google Scholar]
- 256.Katz B., Minke B. The Drosophila light-activated TRP and TRPL channels - Targets of the phosphoinositide signaling cascade. Prog. Retin. Eye Res. 2018;66:200–219. doi: 10.1016/j.preteyeres.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 257.Liu C.H., Bollepalli M.K., Long S.V., Asteriti S., Tan J., Brill J.A., Hardie R.C. Genetic dissection of the phosphoinositide cycle in Drosophila photoreceptors. J. Cell Sci. 2018;131:jcs214478. doi: 10.1242/jcs.214478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Chakrabarti P., Kolay S., Yadav S., Kumari K., Nair A., Trivedi D., Raghu P. A dPIP5K dependent pool of phosphatidylinositol 4,5 bisphosphate (PIP2) is required for G-protein coupled signal transduction in Drosophila photoreceptors. PLoS Genet. 2015;11:e1004948. doi: 10.1371/journal.pgen.1004948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Ratnayake K., Payton J.L., Lakmal O.H., Karunarathne A. Blue light excited retinal intercepts cellular signaling. Sci. Rep. 2018;8:10207. doi: 10.1038/s41598-018-28254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Hong Z., Pedersen N.M., Wang L., Torgersen M.L., Stenmark H., Raiborg C. PtdIns3P controls mTORC1 signaling through lysosomal positioning. J. Cell Biol. 2017;216:4217–4233. doi: 10.1083/jcb.201611073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Chen J., Ma Z., Jiao X., Fariss R., Kantorow W.L., Kantorow M., Pras E., Frydman M., Pras E., Riazuddin S. Mutations in FYCO1 cause autosomal-recessive congenital cataracts. Am. J. Hum. Genet. 2011;88:827–838. doi: 10.1016/j.ajhg.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Romano M.R., Di Menna L., Scarselli P., Mascio G., Madonna M., Notartomaso S., Puliti A., Bruno V., Battaglia G., Nicoletti F. Type-1, but not type-5, metabotropic glutamate receptors are coupled to polyphosphoinositide hydrolysis in the retina. Neurochem. Res. 2016;41:924–932. doi: 10.1007/s11064-015-1775-y. [DOI] [PubMed] [Google Scholar]
- 263.Azadi S., Brush R.S., Anderson R.E., Rajala R.V. Class I phosphoinositide 3-kinase exerts a differential role on cell survival and cell trafficking in retina. Adv. Exp. Med. Biol. 2016;854:363–369. doi: 10.1007/978-3-319-17121-0_48. [DOI] [PubMed] [Google Scholar]
- 264.Olivença D.V., Uliyakina I., Fonseca L.L., Amaral M.D., Voit E.O., Pinto F.R. A mathematical model of the phosphoinositide pathway. Sci. Rep. 2018;8:3904. doi: 10.1038/s41598-018-22226-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Ile K.E., Kassen S., Cao C., Vihtehlic T., Shah S.D., Mousley C.J., Alb J.G., Jr., Huijbregts R.P., Stearns G.W., Brockerhoff S.E. Zebrafish class 1 phosphatidylinositol transfer proteins: PITPbeta and double cone cell outer segment integrity in retina. Traffic. 2010;11:1151–1167. doi: 10.1111/j.1600-0854.2010.01085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Hsuan J., Cockcroft S. The PITP family of phosphatidylinositol transfer proteins. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-reviews3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Köhn L., Kadzhaev K., Burstedt M.S., Haraldsson S., Hallberg B., Sandgren O., Golovleva I. Mutation in the PYK2-binding domain of PITPNM3 causes autosomal dominant cone dystrophy (CORD5) in two Swedish families. Eur. J. Hum. Genet. 2007;15:664–671. doi: 10.1038/sj.ejhg.5201817. [DOI] [PubMed] [Google Scholar]
- 268.Dasgupta U., Bamba T., Chiantia S., Karim P., Tayoun A.N., Yonamine I., Rawat S.S., Rao R.P., Nagashima K., Fukusaki E. Ceramide kinase regulates phospholipase C and phosphatidylinositol 4, 5, bisphosphate in phototransduction. Proc. Natl. Acad. Sci. USA. 2009;106:20063–20068. doi: 10.1073/pnas.0911028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Baranova L.A., Grits A.N., Volotovskii I.D. Phosphorylation of proteins and phosphoinositides in the axoneme of the bovine rod outer segments. The effect of structural factor. Biofizika. 1996;41:1258–1263. [Article in Russian] [PubMed] [Google Scholar]
- 270.Grigor’ev I.V., Grits A.I., Artamonov I.D., Volotovskii I.D. Beta-gamma-transducin controls the metabolism of phosphatidylinositide 4,5-diphosphate in rod outer segments. [Article in Russian] Dokl. Akad. Nauk. 1993;331:235–237. [PubMed] [Google Scholar]
- 271.Pfeilschifter J., Reme C., Dietrich C. Light-induced phosphoinositide degradation and light-induced structural alterations in the rat retina are enhanced after chronic lithium treatment. Biochem. Biophys. Res. Commun. 1988;156:1111–1119. doi: 10.1016/s0006-291x(88)80747-2. [DOI] [PubMed] [Google Scholar]
- 272.Schmidt S.Y. Cytidine metabolism in photoreceptor cells of the rat. J. Cell Biol. 1983;97:824–831. doi: 10.1083/jcb.97.3.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Im E., Kazlauskas A. PtdIns-4,5–P2 as a potential therapeutic target for pathologic angiogenesis. Expert Opin. Ther. Targets. 2007;11:443–451. doi: 10.1517/14728222.11.4.443. [DOI] [PubMed] [Google Scholar]
- 274.Mustafi D., Kevany B.M., Genoud C., Bai X., Palczewski K. Photoreceptor phagocytosis is mediated by phosphoinositide signaling. FASEB J. 2013;27:4585–4595. doi: 10.1096/fj.13-237537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Cioffi D.L., Rich T.C. Feedback regulation of cone cyclic nucleotide channels by phosphoinositides. Focus on “CNGA3 achromatopsia-associated mutation potentiates the phosphoinositide sensitivity of cone photoreceptor CNG channels by altering intersubunit interactions”. Am. J. Physiol. Cell Physiol. 2013;305:C131–C132. doi: 10.1152/ajpcell.00136.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Contín M.A., Verra D.M., Salvador G., Ilincheta M., Giusto N.M., Guido M.E. Light activation of the phosphoinositide cycle in intrinsically photosensitive chicken retinal ganglion cells. Invest. Ophthalmol. Vis. Sci. 2010;51:5491–5498. doi: 10.1167/iovs.10-5643. [DOI] [PubMed] [Google Scholar]
- 277.Phua S.C., Nihongaki Y., Inoue T. Autonomy declared by primary cilia through compartmentalization of membrane phosphoinositides. Curr. Opin. Cell Biol. 2018;50:72–78. doi: 10.1016/j.ceb.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Xu W., Jin M., Huang W., Wang H., Hu R., Li J., Cao Y. Apical PtdIns(4,5)P(2) is required for ciliogenesis and suppression of polycystic kidney disease. FASEB J. 2019;33:2848–2857. doi: 10.1096/fj.201800385RRR. [DOI] [PubMed] [Google Scholar]
- 279.Park J., Lee N., Kavoussi A., Seo J.T., Kim C.H., Moon S.J. Ciliary phosphoinositide regulates ciliary protein trafficking in drosophila. Cell Rep. 2015;13:2808–2816. doi: 10.1016/j.celrep.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 280.Garcia-Gonzalo F.R., Phua S.C., Roberson E.C., Garcia G., 3rd, Abedin M., Schurmans S., Inoue T., Reiter J.F. Phosphoinositides regulate ciliary protein trafficking to modulate hedgehog signaling. Dev. Cell. 2015;34:400–409. doi: 10.1016/j.devcel.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Vieira O.V., Gaus K., Verkade P., Fullekrug J., Vaz W.L., Simons K. FAPP2, cilium formation, and compartmentalization of the apical membrane in polarized Madin-Darby canine kidney (MDCK) cells. Proc. Natl. Acad. Sci. USA. 2006;103:18556–18561. doi: 10.1073/pnas.0608291103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Nakatsu F. A phosphoinositide code for primary cilia. Dev. Cell. 2015;34:379–380. doi: 10.1016/j.devcel.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 283.Lee J.E., Gleeson J.G. Cilia in the nervous system: linking cilia function and neurodevelopmental disorders. Curr. Opin. Neurol. 2011;24:98–105. doi: 10.1097/WCO.0b013e3283444d05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Dyson J.M., Conduit S.E., Feeney S.J., Hakim S., DiTommaso T., Fulcher A.J., Sriratana A., Ramm G., Horan K.A., Gurung R. INPP5E regulates phosphoinositide-dependent cilia transition zone function. J. Cell Biol. 2017;216:247–263. doi: 10.1083/jcb.201511055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Rao K.N., Zhang W., Li L., Anand M., Khanna H. Prenylated retinal ciliopathy protein RPGR interacts with PDE6delta and regulates ciliary localization of Joubert syndrome-associated protein INPP5E. Hum. Mol. Genet. 2016;25:4533–4545. doi: 10.1093/hmg/ddw281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Bielas S.L., Silhavy J.L., Brancati F., Kisseleva M.V., Al-Gazali L., Sztriha L., Bayoumi R.A., Zaki M.S., Abdel-Aleem A., Rosti R.O. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat. Genet. 2009;41:1032–1036. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peränen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 288.Jin H., White S.R., Shida T., Schulz S., Aguiar M., Gygi S.P., Bazan J.F., Nachury M.V. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Dowler S., Currie R.A., Campbell D.G., Deak M., Kular G., Downes C.P., Alessi D.R. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Lemmon M.A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 291.Idevall-Hagren O., De Camilli P. Detection and manipulation of phosphoinositides. Biochim. Biophys. Acta. 2015;1851:736–745. doi: 10.1016/j.bbalip.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Gozani O., Karuman P., Jones D.R., Ivanov D., Cha J., Lugovskoy A.A., Baird C.L., Zhu H., Field S.J., Lessnick S.L. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 293.Lemmon M.A. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 294.Stenmark H. Harald Stenmark: hands on FYVE-fingers. Interview by Caitlin Sedwick. J. Cell Biol. 2011;192:544–545. doi: 10.1083/jcb.1924pi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Viaud J., Lagarrigue F., Ramel D., Allart S., Chicanne G., Ceccato L., Courilleau D., Xuereb J.M., Pertz O., Payrastre B. Phosphatidylinositol 5-phosphate regulates invasion through binding and activation of Tiam1. Nat. Commun. 2014;5:4080. doi: 10.1038/ncomms5080. [DOI] [PubMed] [Google Scholar]
- 296.Boal F., Mansour R., Gayral M., Saland E., Chicanne G., Xuereb J.M., Marcellin M., Burlet-Schiltz O., Sansonetti P.J., Payrastre B. TOM1 is a PI5P effector involved in the regulation of endosomal maturation. J. Cell Sci. 2015;128:815–827. doi: 10.1242/jcs.166314. [DOI] [PubMed] [Google Scholar]
- 297.Tamura N., Oku M., Ito M., Noda N.N., Inagaki F., Sakai Y. Atg18 phosphoregulation controls organellar dynamics by modulating its phosphoinositide-binding activity. J. Cell Biol. 2013;202:685–698. doi: 10.1083/jcb.201302067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Hong N.H., Qi A., Weaver A.M. PI(3,5)P2 controls endosomal branched actin dynamics by regulating cortactin-actin interactions. J. Cell Biol. 2015;210:753–769. doi: 10.1083/jcb.201412127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Shirey C.M., Scott J.L., Stahelin R.V. Notes and tips for improving quality of lipid-protein overlay assays. Anal. Biochem. 2017;516:9–12. doi: 10.1016/j.ab.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Touhara K., Inglese J., Pitcher J.A., Shaw G., Lefkowitz R.J. Binding of G protein beta gamma-subunits to pleckstrin homology domains. J. Biol. Chem. 1994;269:10217–10220. [PubMed] [Google Scholar]
- 301.Rodriguez M.M., Ron D., Touhara K., Chen C.H., Mochly-Rosen D. RACK1, a protein kinase C anchoring protein, coordinates the binding of activated protein kinase C and select pleckstrin homology domains in vitro. Biochemistry. 1999;38:13787–13794. doi: 10.1021/bi991055k. [DOI] [PubMed] [Google Scholar]
- 302.Balla T., Varnai P. Visualization of cellular phosphoinositide pools with GFP-fused protein-domains. Curr. Protoc. Cell Biol. 2009 doi: 10.1002/0471143030.cb2404s42. Chapter 24: Unit 24.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Kaplan D.R., Whitman M., Schaffhausen B., Pallas D.C., White M., Cantley L., Roberts T.M. Common elements in growth factor stimulation and oncogenic transformation: 85 kd phosphoprotein and phosphatidylinositol kinase activity. Cell. 1987;50:1021–1029. doi: 10.1016/0092-8674(87)90168-1. [DOI] [PubMed] [Google Scholar]
- 304.Gray A., Olsson H., Batty I.H., Priganica L., Peter D.C. Nonradioactive methods for the assay of phosphoinositide 3-kinases and phosphoinositide phosphatases and selective detection of signaling lipids in cell and tissue extracts. Anal. Biochem. 2003;313:234–245. doi: 10.1016/s0003-2697(02)00607-3. [DOI] [PubMed] [Google Scholar]
- 305.Munnik T. Analysis of D3-,4-,5-phosphorylated phosphoinositides using HPLC. Methods Mol. Biol. 2013;1009:17–24. doi: 10.1007/978-1-62703-401-2_2. [DOI] [PubMed] [Google Scholar]
- 306.Chicanne G., Severin S., Boscheron C., Terrisse A.D., Gratacap M.P., Gaits-Iacovoni F., Tronchère H., Payrastre B. A novel mass assay to quantify the bioactive lipid PtdIns3P in various biological samples. Biochem. J. 2012;447:17–23. doi: 10.1042/BJ20120945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Ghosh A., Sharma S., Shinde D., Ramya V., Raghu P. A novel mass assay to measure phosphatidylinositol-5-phosphate from cells and tissues. Biosci. Rep. 2019;39 doi: 10.1042/BSR20192502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Letcher A.J., Schell M.J., Irvine R.F. A femtomole-sensitivity mass assay for inositol hexakisphosphate. Methods Mol. Biol. 2010;645:61–71. doi: 10.1007/978-1-60327-175-2_4. [DOI] [PubMed] [Google Scholar]
- 309.Tsujita K., Itoh T., Ijuin T., Yamamoto A., Shisheva A., Laporte J., Takenawa T. Myotubularin regulates the function of the late endosome through the gram domain-phosphatidylinositol 3,5-bisphosphate interaction. J. Biol. Chem. 2004;279:13817–13824. doi: 10.1074/jbc.M312294200. [DOI] [PubMed] [Google Scholar]
- 310.Oikawa T., Yamaguchi H., Itoh T., Kato M., Ijuin T., Yamazaki D., Suetsugu S., Takenawa T. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat. Cell Biol. 2004;6:420–426. doi: 10.1038/ncb1125. [DOI] [PubMed] [Google Scholar]
- 311.Rajala A., McCauley A., Brush R.S., Nguyen K., Rajala R.V. Phosphoinositide lipids in ocular tissues. Biology (Basel) 2020;9:125. doi: 10.3390/biology9060125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Irino Y., Tokuda E., Hasegawa J., Itoh T., Takenawa T. Quantification and visualization of phosphoinositides by quantum dot-labeled specific binding-domain probes. J. Lipid Res. 2012;53:810–819. doi: 10.1194/jlr.D019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Maekawa M., Fairn G.D. Molecular probes to visualize the location, organization and dynamics of lipids. J. Cell Sci. 2014;127:4801–4812. doi: 10.1242/jcs.150524. [DOI] [PubMed] [Google Scholar]
- 314.Chen R., Kang V.H., Chen J., Shope J.C., Torabinejad J., Dewald D.B., Prestwich G.D. A monoclonal antibody to visualize PtdIns(3,4,5)P(3) in cells. J. Histochem. Cytochem. 2002;50:697–708. doi: 10.1177/002215540205000511. [DOI] [PubMed] [Google Scholar]
- 315.Prestwich G.D. Phosphoinositide signaling; from affinity probes to pharmaceutical targets. Chem. Biol. 2004;11:619–637. doi: 10.1016/j.chembiol.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 316.Hammond G.R., Schiavo G., Irvine R.F. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2) Biochem. J. 2009;422:23–35. doi: 10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]