Abstract
Infectious diseases caused by viruses can elevate up to undesired pandemic conditions affecting the global population and normal life function. These in turn impact the established world economy, create jobless situations, physical, mental, emotional stress, and challenge the human survival. Therefore, timely detection, treatment, isolation and prevention of spreading the pandemic infectious diseases not beyond the originated town is critical to avoid global impairment of life (e.g., Corona virus disease - 2019, COVID-19). The objective of this review article is to emphasize the recent advancements in the electrochemical diagnostics of twelve life-threatening viruses namely - COVID-19, Middle east respiratory syndrome (MERS), Severe acute respiratory syndrome (SARS), Influenza, Hepatitis, Human immunodeficiency virus (HIV), Human papilloma virus (HPV), Zika virus, Herpes simplex virus, Chikungunya, Dengue, and Rotavirus. This review describes the design, principle, underlying rationale, receptor, and mechanistic aspects of sensor systems reported for such viruses. Electrochemical sensor systems which comprised either antibody or aptamers or direct/mediated electron transfer in the recognition matrix were explicitly segregated into separate sub-sections for critical comparison. This review emphasizes the current challenges involved in translating laboratory research to real-world device applications, future prospects and commercialization aspects of electrochemical diagnostic devices for virus detection. The background and overall progress provided in this review are expected to be insightful to the researchers in sensor field and facilitate the design and fabrication of electrochemical sensors for life-threatening viruses with broader applicability to any desired pathogens.
Keywords: Infectious diseases, Diagnostics, Electrochemical biosensors, Virus detection, COVID-19, Point of care (POC)
Graphical abstract
1. Introduction
Viruses are the smallest transmittable agents which cause numerous diseases such as Chikungunya, Chickenpox, Dengue, Ebola, Flu, Hepatitis, Influenza, Middle east respiratory syndrome (MERS), Severe acute respiratory syndrome (SARS), and many more (Shah and Wilkins, 2003). A transferable viral particle typically comprises nucleic acids in the core and proteins in the outer shell. Most of the reported viruses have either ribonucleic acid (RNA) or deoxyribonucleic acid (DNA) inherent material to encode proteins (Diemer and Stedman, 2012). These viruses are proficient of fast dispersal and therefore form enduring threats to the worldwide public health. Viruses employ different machineries to enter host cells that depend on their metabolism for self-replication (Tram et al., 2016). The capability of viruses to transmute speedily along with a complicated interchange amid diverse aspects like universal movement of animals/human, geographical changes, and environmental variations contribute to the development of frequent transferable diseases (Kaushik et al., 2017). Hence, fronting the encounters and menacing penalties instigated by the spread of transferable diseases, a precise, high throughput virus scrutiny and analysis to accomplish operative disease regulator have become the key apprehensions of people (Campuzano et al., 2017).
A very recent example of viral spread is the pandemic of Corona Virus Disease-19 (COVID-19) all over the world within a short duration of 3–4 months which harmed millions of lives (Singhal, 2020). Pandemic refers to the occurrence of a new disease over a wide geographic area and affecting an exceptionally high proportion of the population. A pandemic is basically a global epidemic that spreads to more than one continent and affects millions of people (Spinelli and Pellino, 2020). Other recent outbreaks that occurred in the last decade include influenza A (H1N1 subtype) in 2009 and Ebola in 2014 (Muyembe-Tamfum et al., 2012). In the past century, there were some other notable viral pandemics recorded which caused deaths of millions of people worldwide including - flu pandemic (H1N1 virus) in 1918, flu (H2N2 virus) in 1957, swine flu (H1N1 pdm09 virus) in 2009, MERS-Cov in 2012–13, Ebola during 2014–2016 and the ongoing COVID-19 from December 2019 (Ahmed et al., 2007; Glinsky, 2010; Song et al., 2012).
Classical viral diagnostic approaches comprise viral separation, immunofluorescence based on microscopy, enzyme-based antibody assay and polymerase chain reaction (PCR) based qualitative assay which is becoming superseded for repetitive clinical testing (Faria and Zucolotto, 2019). These techniques require extremely long turnaround time ranging from 2 to 14 days, which is unable to combat for virus that spreads rapidly. The existing diagnostic tests are not only taking longer time but also expensive. Therefore, fast, reliable and reproducible analytical methods are required as the need of the hour by which one can be able to identify such causative agents in various matrices (Faria and Zucolotto, 2019). Biosensors are one of the significant analytical devices emerged as an alternative to the conventional cellular and heavily biological assays using tissues, cells, and invasive approaches on organs for viral detection.
Among several types of biosensors, electrochemical biosensors have been operated for several years in diverse fields (Goud et al., 2018; Reddy et al., 2020). Such biosensors analyze any variations in dielectric properties, and charge distribution though the interaction between analyte and biorecognition element on the electrode surface. Electrochemical biosensors are categorized into amperometric (Diba et al., 2015), potentiometric (Wang et al., 2010), voltammetric (Caygill et al., 2010) and impedimetric (Simão et al., 2020) based on the method of transduction. These electrochemical biosensors have been utilized to analyze several biological agents such as proteins, nucleic acid, disease biomarkers (Premaratne et al., 2017; Reddy et al., 2020) and several others (Goud et al., 2017; GOUD et al., 2016; K. Yugender Goud et al., 2019; Kotagiri Yugender Goud et al., 2019; Satyanarayana et al., 2019; Yugender Goud et al., 2016). Electrochemical biosensors are claimed to be highly sensitive types of transducers by offering ultra-low-level sensitivity up to parts per trillion level or sub-pico/femto molar range with linear output, low power requirements and good resolution (Niroula et al., 2016; Premaratne et al., 2018; Rasouli et al., 2018; Singh and Krishnan, 2014) Additionally, they provide an excellent repeatability, accuracy and ability to be miniaturized as a very tiny device form (Kim et al., 2018). These electrochemical sensors can be used and deployed as a point of care (POC) and point of need detection devices (Jayant et al., 2015). Such POC testing empowers medical staff to make quick triage and handling verdicts when analyzing patient's treatment response (Gattani et al., 2019; Khan et al., 2020).
It is highly necessitated to have an overview of the latest innovative approaches and understand the challenges involved in the electrochemical diagnostics of infectious viral diseases. Such awareness will significantly help the researchers to come up with the best suitable biorecognition elements which successfully address the challenges involved while using electrochemical transducers.
1.1. Human viral infectious diseases
The current section provides brief information on each individual virus, core components of the molecular structure, disease symptoms of the virus, and adverse effects of infection caused to the global community.
Corona viruses are a large class of viruses that are common in people (Masters, 2006). First Corona virus disease – 2019 (COVID-19) or Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) case was reported in December 2019 (Guo et al., 2020). The World Health Organization has announced the COVID-19 outbreak as pandemic disease on March 11, 2020 (Cucinotta and Vanelli, 2020). SARS is a species of coronavirus that infects humans, bats and certain other mammals that started in 2002 (Pang, 2003). It is an enveloped positive-sense single-stranded RNA virus that enters its host cell by binding to the angiotensin-converting enzyme 2 receptors (SATIJA and LAL, 2007). MERS caused by species of corona virus, appeared in 2012 in Saudi Arabia. Humans, bats and camels were infected by MERS (Reusken et al., 2013). The molecular structure of MERS virus is an enveloped, positive-sense, single-stranded RNA which penetrates the cell by attaching to the dipeptidyl peptidase 4 (DPP4) receptor. Influenza is a virus that infects the respiratory system in nose, throat and lungs (Munster et al., 2009). Usually, people infected by influenza recover on their own (Cao et al., 2016). Although the annual influenza vaccine isn't completely percent effective, it's still the best defense strategy against flu. The human immunodeficiency virus (HIV) appeared in Congo around 1920 through crossing species from chimpanzees to humans (Sharp and Hahn, 2010). Acquired immunodeficiency syndrome (AIDS) occur after infection that cause progressive failure of the immunity leading to life-threatening opportunistic infections (Monaco et al., 2016).
Hepatitis is an inflammation of the liver. The most cases of Hepatitis are due to Viral infection. The type of hepatitis is named for the causative virus; for example, hepatitis A, hepatitis B, hepatitis C, hepatitis D, hepatitis E (HAV, HBV, HCV, HDV, and HEV) (Shin et al., 2016). Zika virus is transmitted by Aedes mosquitoes, which bite during the day. In 1952, the first human cases of Zika virus were detected and from that date outbreaks of Zika virus have been reported in tropical Africa, Southeast Asia, and the Pacific Islands. Herpes simplex virus is categorized into herpes virus 1 and human alpha herpes virus 2, that of the Herpesviridae family, cause viral infections to humans (Hogestyn et al., 2018). Chikungunya is found all over the world especially in the coastal area of Kenya in 2004 (Mayer et al., 2017). The nucleic material is a small, enveloped, virus with a positive-sense RNA genome. Mosquitoes are responsible for the infection transmission of Dengue virus (DENV) (Acosta et al., 2008). DENV is a single enveloped positive-stranded RNA virus present in five serotypes (Martial et al., 2008). Half of the world population (~3 billion people) are infected by DENV.
Rapid and selective detection of such infectious viruses is highly necessitated to improve the clinical diagnostics.
1.2. Available biomarkers to detect infectious viral diseases
Several biomarkers (RNA, DNA, glyco-proteins, peptides, antibodies, antigens, etc.) can be used as a target analyte molecule to detect the viral infectious diseases (Pashchenko et al., 2018). In particular, these biomarkers were considered as two major categories – Antigens and Antibodies. As mentioned in the introduction, most of the reported viruses have either RNA or DNA as the inherent material to encode proteins (Diemer and Stedman, 2012). Viruses contain three structural components namely - genetic material (DNA or RNA), nucleocapsid protein and capsid proteins. The genetic material is primarily covered with the envelop proteins those are called nucleocapsid proteins, followed by the covering of secondary envelop proteins i.e., capsid protein.
Virus entry mechanism begins with the attachment to cell-surface receptors and ends with the delivery of the viral genome to the cell cytoplasm (Dimitrov, 2004). This entry mechanism occurs in two types – endocytic (clathrin-mediated endocytosis and penetration) and non-endocytic (fusion at the cell surface) routes. The whole virus components (antigen components) – RNA or DNA, nucleo/capsid proteins will be readily available whenever the virus enters the host cells. White blood cells (B lymphocytes or B cells) secrete the antibodies in the human/animal body in response to the virus antigen components. Hence, the availability of both the antigens (RNA or DNA, nucleo/capsid proteins) and respective antibodies in the nearby cell atmosphere, makes it easier to detect by the electrochemical methods with the combination of appropriate bio-recognition elements (Asif et al., 2020; Kermali et al., 2020; Ponti et al., 2020).
Apart from the virus detection, monitoring the severity or stage of the viral infection is also very important to treat the diseased person. Severity of the viral infection can possibly be assessed by detecting the other biomarker components such as C-reactive protein, interleukins, glutamate, breath pH, Tumor necrosis factor (TNF-α), interferons, hematological biomarkers, and D-dimer. All these biomarker components could be detectable at the bedside by the electrochemical methods with the help of suitable bio-recognition elements (Kotru et al., 2021; Kudr et al., 2021). For example, Wei Gao et al. demonstrated the multiplexed portable, wireless electrochemical biosensor for the rapid screening of COVID-19. The proposed sensor successfully detected the antigen nucleocapsid protein, Immunoglobulin (IgM and IgG) antibodies, as well as the inflammatory biomarker C-reactive protein which in turn helped to know the status of the disease (Torrente-Rodríguez et al., 2020).
1.3. Comprehensive overview of the recent literature
Various review articles have assessed the performance of sensor systems proposed for the detection of viruses. Some of these review articles have summarized the detection of viruses such as HIV (Farzin et al., 2020; Nawaz et al., 2020; Parolo et al., 2020a; Rodrigo et al., 2014), HIV-1 (Lifson et al., 2016), Influenza (Krejcova et al., 2014), Avian Influenza (Moulick et al., 2017), Human and Animal Influenza (Dziąbowska et al., 2018), Hepatitis B (Yao, 2014), Noroviruses (Liu and Moore, 2020), bacterial, viral, and toxin bio-threats (Walper et al., 2018), pathogens (Mokhtarzadeh et al., 2017), SARS (Halfpenny and Wright, 2010; Orooji et al., 2021; Tran et al., 2020), COVID-19 (Chauhan et al., 2020; H. Chen et al., 2020; Ji et al., 2020; Jin et al., 2020; H. Li et al., 2020; Morales-Narváez and Dincer, 2020; Ravi et al., 2020; Shereen et al., 2020; Udugama et al., 2020; Wang et al., 2020; Weiss et al., 2020), Dengue, Zika (Khristunova et al., 2020), viruses in the aquatic environment (Farkas et al., 2020; Srivastava et al., 2020; Tran et al., 2020), viruses in the food, environmental samples (Yadav et al., 2010), and antibody specific to the viruses (Parolo et al., 2020a; Xu et al., 2019). Other review articles have summarized the sensor systems constructed for the detection of different viruses using simple device-based approaches (Cheng et al., 2009), nano-electronic devices (Yeom, 2011), bioanalytical microsystems (Yadav et al., 2010), integrated sensor systems (Dincau et al., 2017), microfluidic system (Sin et al., 2014), paper-based microfluidic system (Deka et al., 2020; Gong and Sinton, 2017), POC devices (Tram et al., 2016), lab-on-a-chip technologies (Zhu et al., 2020), piezoelectric, magnetostrictive (Narita et al., 2020), DNA microarray (Fesseha and Tilahun, 2020), and clustered regulatory interspaced short palindromic repeats (CRISPR) systems (Hass et al., 2020; Kostyusheva et al., 2020; Strich and Chertow, 2018). On the other hand, some review articles have focused on the recognition matrix comprising Au nanoparticles (Draz and Shafiee, 2018; Franco et al., 2015; Halfpenny and Wright, 2010), Quantum dots (QD) (Halfpenny and Wright, 2010; Yeom, 2011), carbon nanotubes/nanowires (Yeom, 2011), Aptamers (Acquah et al., 2016; Hong et al., 2012; Labib and Berezovski, 2013), label-free and labeled immuno assays (Parolo et al., 2020b; Sin et al., 2014), molecularly imprinted polymers (MIP) (Cui et al., 2020; Yang et al., 2020), and other nanomaterials (Kizek et al., 2015; Mokhtarzadeh et al., 2017; Nasrollahzadeh et al., 2020). Some of the review articles have focused on the transduction methods in virus diagnosis based on optical and/or electrochemical techniques (Cheng and Toh, 2013; Cui et al., 2020; Khristunova et al., 2020; Krejcova et al., 2014; Ozer et al., 2020; Xu et al., 2019; Yang et al., 2020). On the contrary, one review article has highlighted the applications of bacteriophages (lytic and nonlytic) of viruses in conjugation with nanomaterials (virus–nanomaterial composites) used in the analytical devices towards the detection of explosives, proteins, bacteria, viruses, spores, and toxins (Mao et al., 2009).
1.4. Scope and objectives of the present Review
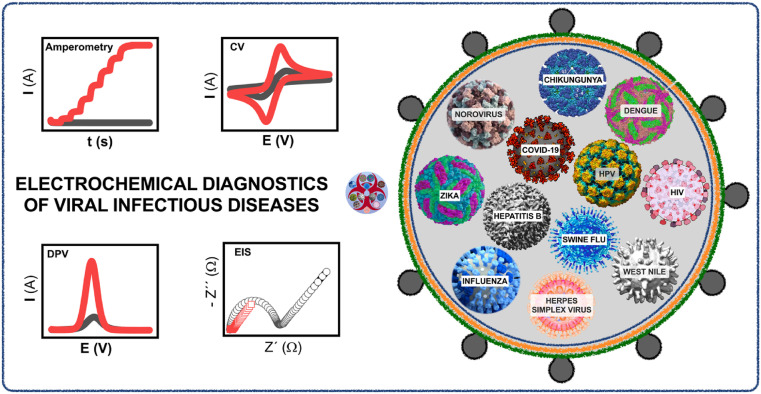
The rationale behind this review is to emphasize the latest advancements in the electrochemical diagnostics of human viral infectious diseases. Precisely, this review provides an overview of the electrochemical detection of twelve life-threatening viruses namely - COVID-19, MERS, SARS, Influenza, Hepatitis, HIV, HPV, Zika, Herpes simplex virus, Chikungunya, Dengue, and Rotavirus. All the relevant electrochemical works are explicitly segregated into relevant sub-sections for understanding the core idea/principle/working mechanism behind the sensor fabrication. The first sub-section exclusively summarizes the electrochemical/other sensor systems reported for the specific detection of COVID-19/SARS-CoV-2 with the objective of providing a critical overview of the latest research trend at a single place. The remaining three individual sub-sections meticulously discuss the electrochemical sensor systems which comprised either antibody or aptamers or direct/mediated electron transfer in the recognition matrix. The major challenges on translating laboratory research to real-world device applications are critically discussed. Future perspectives and commercialization aspects of electrochemical sensors for virus detection are also mentioned. The background and overall progress provided in this review help the researchers to come up with new innovative strategies in fabricating electrochemical sensor for not only such life-threatening viruses but also for the other pathogens/biomolecules/organisms/therapeutic drugs/environmental samples and vital biomarkers. Fig. 1 conveys the overall summary of electrochemical sensors towards the detection of the above selected infectious diseases.
Fig. 1.
Schematic representation of the role of electrochemical sensors in viral infectious diseases diagnostics.
2. Electrochemical diagnostics
Electrochemical biosensors are one of the most reliable and highly sensitive transduction systems which provide ultra-low-level detection of target analyte with low power consumption (Rasouli et al., 2018). Moreover, they deliver an exceptional reproducibility, accuracy and ability to be miniaturized as a very tiny device form (Kim et al., 2018). These electrochemical sensors can be used and deployed as a POC and point of need detection devices.
Fundamental variation among the electrochemical biosensors is the method of transduction - Amperometry, voltammetry, and impedance spectroscopy. All the electroactive molecules can be certainly recognized based on their redox characteristics whereas even nonelectroactive molecules can also be monitored by impedance/capacitance changes. Variations observed in the current/potential/impedance values indicate the pertinent changes in chemical composition of the sensor components. Electrical signal is generated when the target analyte interacts with the recognition matrix (Reddy et al., 2020).
Amperometric sensors work at a fixed redox potential and the magnitude of resultant current increases linearly with the concentration of target analyte. Most of the existing portable sensors were constructed using amperometry because highly sensitive and reproducible results can be obtained within very short time intervals.
Cyclic voltammetry (CV) provides the preliminary redox characteristics of the recognition elements or target analyte. Nature and reversibility of the electrochemical redox reaction (perfectly reversible/semi or quasi reversible/irreversible) can be easily understood from the cyclic voltammograms. Electrochemically active surface area of different materials immobilized on the electrodes can be measured to accurately interpret the reasons behind their improved electrochemical performance. CV analysis at multiple scan rates furnishes the valuable insights on redox reaction mechanism (diffusion or adsorption controlled) which in turn helps to decide the method of transduction depending on the anticipated application. Electrode surface is intact in the case of diffusion controlled redox reactions and hence the same sensor surface could be reused even after several scans. On the other hand, adsorption-controlled redox reaction leads to the deposition of materials on to the electrode surface which will be more appropriate for the applications such as removal of toxic metal ions/electrochemical deposition of materials (Elgrishi et al., 2018).
Target analytes can be detected even up to picomolar or femtomolar concentrations with differential pulse voltammetry (DPV) and square wave voltammetry (SWV). The difference among CV, DPV and SWV is the mode of applied potential.
Electrochemical impedance spectroscopy (EIS) determines the changes in capacitance/impedance of the system by imposing a sine wave (Amplitude 5–10 mV). Interfacial characteristics at the electrode-electrolyte (adsorption/desorption) can be explored with EIS. Kinetic/mechanistic aspects as well as the reaction rates of the electrochemical redox reaction can be obtained from EIS data. Several direct/label-free electrochemical immunosensors were reported in the literature using affinity based biological recognition elements and EIS as the transduction method (Reddy et al., 2020).
The current section critically reviews the performance of different electrochemical sensors which comprise either antibody or aptamer or direct/mediated electron transfer in the recognition matrix. Electrochemical/analytical biosensors of COVID-19 were separately discussed in the initial subsection to provide the readers a better overview of the latest trend in the clinical diagnostics of COVID-19. Electrochemical biosensing systems reported for other viruses are systematically discussed in the next subsections.
2.1. Specific detection of COVID-19/SARS-CoV-2
This sub-section collectively describes the analytical and electrochemical sensor systems reported for the detection of COVID-19/SARS-CoV-2 virus. Biosensors constructed to selectively detect the COVID-19/SARS-CoV-2 virus (Cui and Zhou, 2020; Fauci et al., 2020; Goodnough et al., 2020; Irfan, 2020; Qiu et al., 2020; Seo et al., 2020) based on reverse transcription polymerase chain reaction (RT PCR) (Won et al., 2020), serological testing (Sethuraman et al., 2020), whole blood antibody (Carter et al., 2020), enzyme-linked immunosorbent assay (ELISA) (M. Li et al., 2020), localized surface plasmon resonance (LSPR) (Qiu et al., 2020), CRISPR (Broughton et al., 2020), Field-effect Transistor (FET) (Seo et al., 2020) and electrochemical (Fabiani et al., 2021; González-López and Fernández Abedul, 2020; Rashed et al., 2020; Zhao et al., 2021) methods are critically discussed.
Detection of COVID-19 was reported using the ELISA and Rapid immune-chromatographic assay (RICA) approaches with the polyclonal and monoclonal antibodies (pAbs and mAbs) generated from animals. Sandwich ELISA involved biotin-conjugated rabbit anti-COVID-19 pAbs, bovine serum albumin (BSA), phosphate buffer saline (PBS), Horseradish peroxidase (HRP)-Streptavidin and 3,3′,5,5′-tetramethylbenzidine (TMB). RICA approach involved QD@SiO2 and rabbit anti-COVID-19 pAbs on nitrocellulose membrane. The proposed approaches were able to selectively detect COVID-19 as low as 100 ng mL−1 (ELISA) and 10 ng mL−1 (RICA). Practical applicability was demonstrated in the biopsy samples of patients (M. Li et al., 2020).
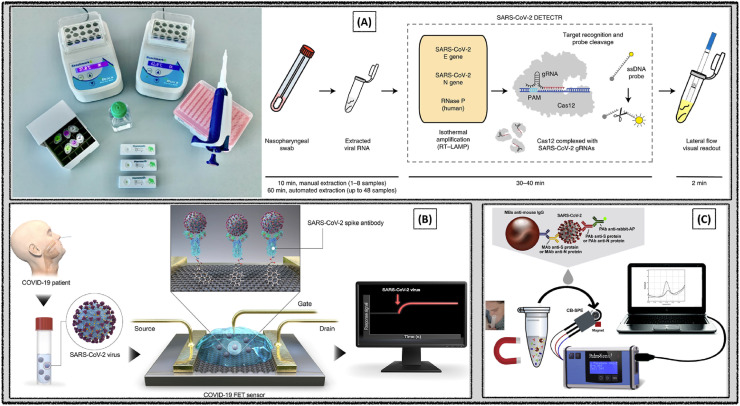
Rapid sensor system was fabricated for the highly selective detection of SARS-CoV-2 using the CRISPR–Cas12 based reverse-transcription loop-mediated, isothermal amplification (RT-LAMP) assay (Fig. 2 A). Spike (S), envelope (E), matrix (M), and nucleocapsid (N) proteins are the critical structural components of SARS-CoV-2. E and N proteins of SARS-CoV-2 were targeted to design the primer gRNAs. Required assay reaction time was 30–40 min and the method was easy-to-implement. The detection limit was found to be 10 copies μL−1. Sensor performance was validated with the clinical samples collected from the respiratory swab of COVID-19 infected patients in the United States (Broughton et al., 2020).
Fig. 2.
Biosensors constructed for the selective detection of SARS-CoV-2 using (A) CRISPR–Cas12 based RT-LAMP assay. Reproduced with permission (Broughton et al., 2020). Copyright 2020, Nature. (B) FET. Reproduced with permission (Seo et al., 2020). Copyright 2020, American Chemical Society. (C) Voltammetry. Reproduced with permission (Fabiani et al., 2021). Copyright 2021, Elsevier.
Selective detection of SARS-CoV-2 was demonstrated using LSPR method. The sensing mechanism involved the nucleic acid hybridization between the target and the recognition matrix of 2D gold nanoislands (AuNIs) functionalized with complementary DNA receptors. Thermoplasmonic heat led to the improved selectivity of the sensor by elevating the in-situ hybridization temperature which in turn helped to specifically recognize two different gene sequences. The observed detection limit was 0.22 pM of SARS-CoV-2 (Qiu et al., 2020).
Immunological FET sensor system was fabricated using the antibody coated graphene sheets to detect the SARS-CoV-2 spike protein (Fig. 2B). S protein was chosen as the diagnostic antigen considering the highly immunogenic transmembrane characteristics and the distinguishable amino acid sequence. Selective recognition of SARS-CoV-2 spike antibody was verified with ELISA prior to the FET sensor construction. Reliable I–V electrical signals were obtained with highly stable ohmic contact for the detection of SARS-CoV-2. The proposed sensor system was able to successfully detect the SARS- CoV-2 up to 1 fg mL−1 in buffer and 100 fg mL−1 in clinical transport medium. Limit of detection was 16 plaque-forming unit (PFU) mL−1 SARS-CoV-2 in culture medium whereas 242 copies mL−1 in the clinical samples. Sensor performance was demonstrated with the nasopharyngeal swab clinical specimens of COVID-19 patients. Advantages of the reported FET sensor were instantaneous measurements using small amounts of analytes as well as the ability to detect the SARS-CoV-2 spike protein without involving any sample pretreatment or labeling (Seo et al., 2020).
Fabiani et al., have proposed an electrochemical immunosensor for fast and accurate detection of coronavirus in saliva. A biosensor was developed to detect S protein or N protein by employing magnetic nanoparticles and alkaline phosphatase coupled secondary antibody as an immunological label (Fabiani et al., 2021). Upon biosensing of SARS-CoV-2 protein, the enzymatic by-product 1-naphthol was sensed on the carbon-based screen-printed electrode. The analytical performance of the developed biosensor was estimated using a standard solution of viral proteins (S and N) in buffer medium and extended to detect the same in saliva samples (Fig. 2C). The obtained detection levels in saliva were 19 and 8 ng mL−1 for S and N proteins respectively.
On the other hand, Smartphone based ultrasensitive electrochemical sensor was demonstrated for the detection of COVID-19 in patients (Zhao et al., 2021). The recognition matrix comprised of calixarene functionalized graphene oxide surface was used to detect RNA of COVID-19 by relying on a sandwich-type detection. The method is free from the amplification of nucleic acid and exhibited high specificity during actual testing of COVID patients. The detection limit of the tested samples was found to be 200 copies mL−1, the lowest till date claimed by authors by offering a simple, low-cost and useful method.
Electrochemical detection of SARS-CoV-2 antibodies was presented using an impedimetric sensing platform (Rashed et al., 2020). The method was based on a pre-incubation of receptor-binding domain (RBD) of viral spike protein in well plates and later tested with samples of anti- SARS-CoV-2 monoclonal antibody. Consequently, some blind samples were tested from a group of patients with negative and positive results. Antibodies against coronavirus are normally directed against recombinant spike protein or the minor RBD share of spike protein. The micro-wells were coated with antibodies which enabled rapid impedimetric detection of COVID traces in the collected samples of patients. This sensing method could able to differentiate spikes in impedimetric analysis from a negative control. More testings are anticipated to finalize detection level for this approach (Rashed et al., 2020).
Table 1 illustrates the summary of different analytical/electrochemical biosensing diagnostics methods for the detection SARS-CoV-2 virus. The limit of detections was 0.7 nM with Co–TiO2 nanotube based receptor by Chronoamperometry (Vadlamani et al., 2020), 0.4 pg mL−1 with ZnO NW μPADs by EIS (X. Li et al., 2020), 2.5 pM with the Au NPs/DNA/Alkaline Phosphatase by CV (Martínez-Paredes et al., 2009), 90 fM with nCovid-19Ab/AuNPs/FTO by DPV (Mahari et al., 2020). It was observed that all the electrochemical transduction methods provided promising detection limits and exhibited better sensor performance.
Table 1.
Several analytical/electrochemical biosensors reported for the diagnostics of SARS-CoV-2 viruses.
| S NO | Virus | Recognition Matrix | Method | Concentration Range | LOD | Real Samples | Ref |
|---|---|---|---|---|---|---|---|
| 1 | COVID | Cobalt-functionalized TiO2 nanotubes (Co-TNTs)- receptor binding domain (RBD) | Chronoamperometry | 14–1400 nM | 0.7 nM | Nasal secretions and saliva samples | Vadlamani et al. (2020) |
| 2 | COVID | A 16-well plate/receptor binding domain (RBD) | EIS | 0.1–10 μg mL−1 | 0.1 μg mL−1 | Serum | Rashed et al. (2021) |
| 3 | COVID | Laser-engraved graphene/Ab | Chronoamperometry | 20–250 ng mL−1 | Blood saliva | Torrente-Rodríguez et al. (2020) | |
| 4 | COVID | ZnO NW μPADs | EIS | 10 ng mL−1- 10 μg mL−1 | 0.4 pg mL−1 | Serum | X. Li et al. (2020) |
| 5 | Reactive oxygen species (ROS) | (MWCNTs) on the tip of steel needles | Voltammetry | – | – | Sputum | Miripour et al. (2020) |
| 6 | COVID | SPE/Carbon Black/magnetic beads/antibody/phosphatase as immunological label | Voltammetry | – | 19 ng mL−1 | Saliva | Fabiani et al. (2021) |
| 7 | COVID | Au NPs/Ti Electrode Single strand DNA | Electrochemical | – | – | – | Tripathy and Singh (2020) |
| 8 | COVID | Fluorine doped tin oxide electrode (FTO) (AuNPs) and/(nCovid-19Ab)/SPE | DPV CovSens-Ultrasensitive In-House Built Printed Circuit Board | 1 fM - 1 μM | 90 fM | Saliva | Mahari et al. (2020) |
| 9 | COVID | SARS COVID Ab/PBASE/Graphene/Field Effect Transistor | Chronoamperometry | 1 fg mL−1 - 10 pg mL−1 | 1.6 × 101 PFU mL−1 | Nasopharyngeal Swab | Seo et al. (2020) |
| 10 | SARS Virus | SPE/Au NPs/DNA/Alkaline Phosphatase | Cyclic Voltammetry | 5 and 100 pM | 2.5 pM | – | Martínez-Paredes et al. (2009) |
| 11 | COVID | Lanthanide-doped polystyrene NPs/nucleocapsid phosphoprotein of SARS-CoV-2 | Portable fluorescence reader excitation and emission wavelengths of 365 and 615 nm | – | – | Serum | Z. Chen et al. (2020) |
| 12 | COVID | Poly (amino ester) with carboxyl groups (PC)-coated magnetic nanoparticles (pcMNPs) | Viral RNA extraction method pcMNPs-RNA complexes extract RNA & introduce into RT-PCR reactions | 10 - 105 copies of SARS-CoV-2 | – | Serum | Zhao et al. (2020) |
| 13 | COVID | Kits | Colloidal Gold-Immunochromatographic Assay Kit & ELISA | – | – | Serum | Xiang et al. (2020) |
| 14 | COVID | Receptor-binding domain (RBD) | ELISA | – | – | Serum | Amanat et al. (2020) |
| 15 | COVID | (polymethyl methacrylate)microbeads/streptavidin/biotinylated recombinant RBD, S1 and N | Multiplexed flow cytometric bead array (C19BA) | – | – | Serum | Egia-Mendikute et al. (2020) |
| 16 | COVID | SARS-CoV-2 S1 and nucleocapsid (N)- subunits of the spike glycoprotein | ELISA | – | – | Serum | Algaissi et al. (2020) |
| 17 | COVID | Immunosensor | ELISA/Western blot | 200–1600 ng mL−1 | 100 ng mL−1 | M. Li et al. (2020) |
2.2. Electrochemical sensors based on direct/mediated electron transfer
Direct electron transfer (DET) mechanism signifies that the electron transfer between electrode and recognition element occurs directly without the help of any mediator. Sensors which work on DET mechanism are considered as third generation biosensors. Recently, DET based biosensing methods gained much attention due to the unique advantages such as eliminating the leachable artificial mediators and operation in a potential window close to the redox potential of the biorecognition element (Teymourian et al., 2020). Some of the surface imprinted or MIP based electrochemical sensors also works on the DET principle (Bosserdt et al., 2013; Peng et al., 2016; Sharma et al., 2019). This sub-section presents an overview of different electrochemical approaches based on the DET in the detection of various viruses, such as HIV-1 (Bhimji et al., 2013; Lee et al., 2013; Shin et al., 2019), HBV (Asran et al., 2020), Zika (Tancharoen et al., 2019), Influenza (Fu et al., 2014) and Dengue virus (Navakul et al., 2017).
An electrochemical sensor was reported for the detection of HIV-1 virus based on direct electron transfer signal. AuNPs were electrodeposited on Indium tin oxide (ITO)|Glass to improve the electron transfer by enhanced electrochemical surface area. Later, antibody fragments were immobilized onto the AuNPs|ITO electrode by self-assembled monolayer method with Au-thiol interaction, followed by the HIV-1 virus-like particle were applied for the direct electron determination. HIV-1 virus particles were detected using CV as transduction technique and observed the response in the concentration range of 600 fg mL−1 - 375 pg mL−1 (Lee et al., 2013).
Richard G. Compton group reported an electrochemical rapid sensor for the detection of single influenza virus tagged with the silver nanoparticles (AgNPs). Influenza virus concentration was detected while exposing the virus and AgNPs on the carbon nanofiber working electrode. The AgNPs naturally restrict to adsorb on the carbon nanofibers surface, but virus interacted AgNPs were easily adsorbed on the working electrode surface. These changes were recorded by running in-vitro stripping voltammetry technique. In the next stage, single virus detection was achieved via nano-impact technique. Here, random individual nanometer-sized virus collisions with the AgNPs were monitored by using the spikes of chronoamperogram during oxidation or reduction of AgNPs (Sepunaru et al., 2016).
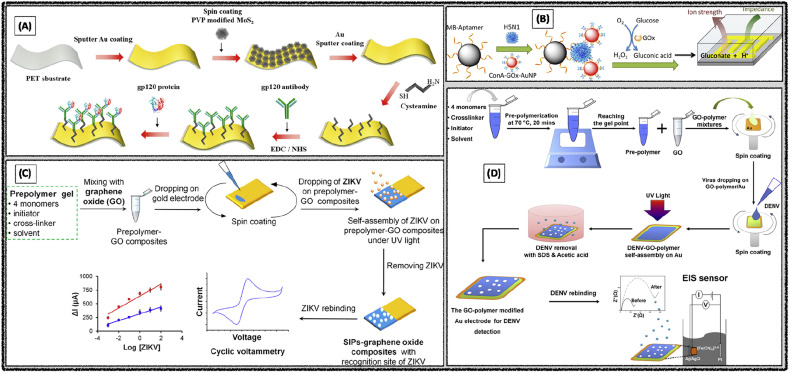
An electrochemical biosensor was developed for the detection of glycoprotein GP120, a surface protein of HIV1 by using Au/MoS2/Au sputtered monolayers on poly ethylene terephthalate (PET) substrate. Here, Au was initially sputtered on the PET substrate and followed by the MoS2 NPs and Au were spin-coated on the PET/Au surface. Then glycoprotein GP120 antibody was immobilized on Au/MoS2/Au surface using cysteamine intermediate layer (Fig. 3 A). The GP120 glycoprotein and antibody interactions were measured by SWV analysis using the Ferry-Ferro redox couple electroactive solution and observed the limit of detection 0.1 pg mL−1 (Shin et al., 2019).
Fig. 3.
Electrochemical detection of (A) HIV-1 using the Au/MoS2 NPs/Au Nanolayer|PET. Reproduced with permission (Shin et al., 2019). Copyright 2019, MDPI. (B) Avian Influenza Virus using enzyme catalysis. Reproduced with permission (Fu et al., 2014). Copyright 2014, American Chemical Society. (C) Zika Virus in serum using surface imprinted graphene oxide composite. Reproduced with permission (Tancharoen et al., 2019). Copyright 2019, American Chemical Society. (D) Dengue virus using surface imprinted GO-polymer|Au. Reproduced with permission (Navakul et al., 2017). Copyright 2017, Elsevier.
An electrochemical impedimetric biosensor was developed at an ultra-low ionic strength environment. Magnetic beads were first modified with aptamers sequence, which would capture the specific Avian influenza H5N1 virus, followed by the addition of concanavalin A, glucose oxidase (GOx), and AuNPs to create bio-nano composites (Fig. 3B). The sandwich complex facilitates glucose oxidation and results in high ionic strength which creates low impedance environment. The high susceptibility of electrochemical impedance on the ion strength endowed highly sensitive biosensor with a detection limit of 8 × 10−4 Hemagglutination units (HAU) in 200 μL sample (Fu et al., 2014).
An electrochemical sensor was developed based on surface imprinted polymers as recognition elements for the detection of Zika virus. Initially, the polymer gel was prepared by using monomers acrylamide, methacrylic acid, methyl-methacrylate, N vinylpyrrolidone and initiator azobis (isobutyronitrile) and cross-linker (N,N′-(1,2-dihydroxyethylene) bis(acrylamide)). Resultant polymer gel was mixed with graphene oxide (GO) and casted on the gold electrode surface. Later Zika virus sample was added onto the surface of the gel and exposed it to UV light for the polymerization. Then, washing steps were done with acetic acid and distilled water to remove the bonded virus particle from the polymer matrix (Fig. 3C). CV analysis of Zika virus particles in the concentration range of 10−3 to 102 PFU mL−1 produced the low detection limit of 2 × 10−4 PFU mL−1 (Tancharoen et al., 2019).
K Navakul et al. proposed a novel electrochemical method for the screening of dengue virus antibodies by using surface-assembled graphene-polymer. Surface imprinted GO-polymer was synthesized by using the suitable monomers and cross-linkers. These polymers were transferred onto the gold surface and added the dengue virus particles, then kept it under UV-light for the self-assembly process (Fig. 3D). In further step, dengue virus particles were removed from the surface imprinted polymer with the sodium dodecyl sulfate (SDS)-acetic acid washing steps. EIS analysis of the dengue virus offered the low detection limit of 0.12 PFU mL−1 (Navakul et al., 2017).
Precise and rapid EIS analysis of Zika virus was demonstrated using a 16 × 20 electrochemical complementary metal oxide semiconductor (CMOS) biosensor array as a POC device. Small sensor array (140 × 140 μm2 pixel) was constructed with on-chip sensors capable of polar-mode measurement. The sensor array was comprised of the specific capturing probe 5′‘GCTTGGCCAGGTCACTCATTGAAAATCCTC to the Zika virus. The designed architecture produced highly sensitive signal response without much noise. The sensor system was employed to measure hybridization of Zika virus oligonucleotides (Hsu et al., 2018).
ELISA based voltammetric detection of HIV1 and HIV2 viruses was reported using microelectrodes. When the antigens GP41 (HIV1) or GP36 (HIV-2) and SU-8 deposited on 3D microelectrodes react with the HIV antibodies led to the formation of redox p-aminophenol. DPV analysis in the concentration range 0.001–1 μg mL−1 of antibody offered a detection limit of 1 ng mL−1 (6.7 pM) for both HIV-1 and HIV-2. Sensor performance was successfully demonstrated in the clinical samples of HIV patients (Bhimji et al., 2013).
Table 2 illustrates the summary of different electrochemical diagnostics methods based on direct electron transfer as a recognition element for the detection of viral infectious diseases. Relevant literature reports were arranged in the table with the transduction technique, recognition element/assay, working calibration range and sensor limit of detection. Interestingly, Au/MoS2 nanoparticles/PET substrate based immunosensor exhibited the good low detection limit of 0.066 pg mL−1 for HIV detection (Shin et al., 2019).
Table 2.
Electrochemical detection of viral infectious diseases based on direct electron transfer as recognition mechanism.
| S NO | Virus | Recognition Matrix | Method | Concentration Range | LOD | Ref |
|---|---|---|---|---|---|---|
| 1 | HIV-1 | Au NPs on the Indium Tin Oxide coated glass (ITO) electrode | CV | 600 fg mL−1 - 375 pg mL−1 | 10 pg mL−1 | Lee et al. (2013) |
| 2 | HIV-1 | Au (Au/MoS2/Au nanolayer) on the polyethylene terephthalate (PET) | SWV | 0.1 pg mL−1 - 10 ng mL−1 | 0.066 pg mL−1 | Shin et al. (2019) |
| 3 | HBV | NiFe2O4-IL|CPE | SWV | 8 nM - 2.2 μM and 2.2 μM–15.5 mM | 2 nM | Asran et al. (2020) |
| 4 | Zika | Surface imprinted polymers and graphene oxide composites | CV | 10−3 - 102 PFU mL−1 | 2 × 10−4 PFU mL−1 | Tancharoen et al. (2019) |
| 5 | Avian influenza virus H5N1 | Magnetic beads aptamer, concanavalin A (ConA), glucose oxidase (GOx), (AuNPs) bionanocomposites | EIS | 0.001–1 HAU | 8 × 10−4 HAU | Fu et al. (2014) |
| 6 | Dengue | Au coated with graphene oxide reinforced polymer | EIS | 1 to 2 × 103 PFU mL−1 | 0.12 PFU mL−1 | Navakul et al. (2017) |
| 7 | HIV1 HIV2 | Electrochemical ELISA | DPV | 0.001–1 μg mL−1 | 1 ng mL−1 (6.7 pM) | Bhimji et al. (2013) |
2.3. Antibody based electrochemical biosensors
Sensors which rely on the specific non-covalent interactions between antibodies (analyte specific probes) and antigens (target analytes) are designated as immunosensors. These immunosensors are considered as gold standard biorecognition elements in several industries such as clinical diagnostics, food safety control, drug screening and development, environmental monitoring, forensic analysis, managing of biological threats, the prevention and control of epidemic diseases, etc. (Chikkaveeraiah et al., 2012; Diaconu et al., 2013; Kokkinos et al., 2016). Especially the electrochemical immunosensors have emerged as an alternative diagnostic methods for the traditional clinical assays in medical diagnostics and pandemic diseases management (Hussein et al., 2020; Ranjan et al., 2021; Torrente-Rodríguez et al., 2020). This section summarizes the sensor systems in which the recognition matrix is comprised of antibodies for the selective electrochemical detection of hepatitis A (Mandli et al., 2017), of hepatitis B (Akkapinyo et al., 2020; Wei et al., 2020), of hepatitis C (Zhao and Liu, 2016), five types of hepatitis (Tang et al., 2010), HPV (Piro et al., 2011; Valencia et al., 2016; Zari et al., 2009), HIV (Akkapinyo et al., 2020; Macchia et al., 2020), Zika (Cabral-Miranda et al., 2018; Draz et al., 2018; Faria and Mazon, 2019; Kaushik et al., 2018), Dengue (Cheng et al., 2012; Nawaz et al., 2018), Rotavirus (Attar et al., 2016; Liu et al., 2013) and Influenza virus (Zhou et al., 2013).
Electrochemical immunosensor was demonstrated for the detection of H9N2 Avian influenza virus through a combination of immunomagnetic separation and the electrochemical detection method. It was reported that the direct sensing of the virus was possible with the aid of homemade magneto Au electrode, which facilitated for the direct accumulation of the complex formed during the electrochemical process, eventually obtained the signal of hydrogen peroxide. They demonstrated the immunoreaction taking place prior to the electrochemical detection. The reported immunosensor has good selectivity, reproducibility, sensitivity and detected the virus in the range of 0.01–1000 ng mL−1 with a low detection limit of 10 pg mL−1 in the chicken dung samples (Zhou et al., 2013).
An integrated single electrochemical immunosensor was demonstrated for the combined detection of five hepatitis virus antigens. The protein A was immobilized onto six nanogold working electrodes followed by the drop-casting the HAV, HBV, HCV, HDV, and HEV monoclonal antibodies for the formation of the immune-complexes. Eventually the change in surface charge of the sensor was observed to be linearly proportional to the concentrations of the antigens. Simultaneously recorded the signals of each individual electrode for the detection of five analytes in a single step automatically. The observed low detection limit was ≤1.0 ng mL−1 for almost all types of analytes (Tang et al., 2010).
Label-free immunosensor was developed for the detection of N protein, a biomarker for the SARS. In2O3 nanowire combined with a probe of antibody mimic proteins (Fibronectin, Fn) was used for the specific binding of target analyte. (Ishikawa et al., 2009). Sub-nanomolar concentration of the analyte was detected through the fabricated immunosensor and no response was seen in the absence of Fn probe with the target N protein. Owing to the selectivity, sensitivity of the developed immunosensor demonstrates the effective practical application.
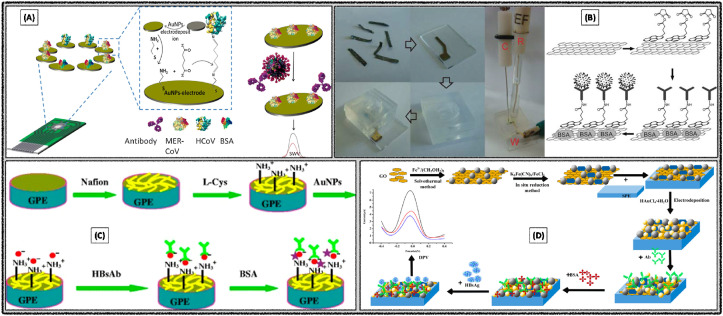
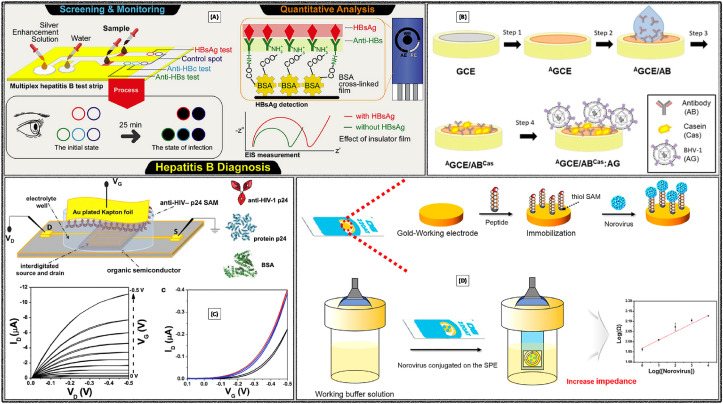
Disposable electrochemical immunosensor was developed with AuNPs modified screen-printed electrode for the detection of MERS-CoV. The immunosensor facilitated detection of different corona viruses through the biomarker recombinant spike protein S1 (Fig. 4 A) (Layqah and Eissa, 2019). It was observed that the electron transfer efficiency as well as electrode surface area increased with the aid of AuNPs, which eventually leads to the high sensitivity of the fabricated immunosensor. Biological fluids samples were examined by spiking into the artificial nasal samples and achieved a good recovery limit. Low detection limit was observed as 0.4 pg mL−1 for human corona virus (HCoV) and 1.0 pg mL−1 for MERS-CoV respectively.
Fig. 4.
Electrochemical detection of (A) MERS-CoV using modified SPCE. Reproduced with permission (Layqah and Eissa, 2019). Copyright 2019, Springer-Verlag GmbH Austria. (B) Rotavirus through the graphene film-based immuno-sensor. Reproduced with permission (F. Liu et al., 2011). Copyright 2011, Springer-Verlag GmbH Austria. (C) HBsAg using graphene paste electrode. Reproduced with permission (Huang et al., 2012). Copyright 2012, Springer-Verlag GmbH Austria. (D) human HBsAg using GO/Fe3O4/PB nanocomposite modified SPE. Reproduced with permission (Wei et al., 2020). Copyright 2020, MDPI.
A voltammetric immunosensor was developed for the detection of Rotavirus using graphene. The immunosensor was fabricated through sequential steps, primarily with the formation of graphene film by thermal annealing followed by the surface modification with pyrene derivative (Fig. 4B) (F. Liu et al., 2011). The fabricated immunosensor was used for the formation of Antibody-Antigen complex by covalent linkage, then the responses were recorded with CV. It was able to detect 105 PFU mL−1 of input cells with 30.7% sensitivity. On the other hand, electrochemical impedimetric sensor was reported for the detection of rotavirus using gold sono-nanoparticles (AuSNPs). Glassy carbon electrode (GCE) was modified initially with AuSNPs followed by the self-assembled monolayers of cysteamine, then crosslinking by cysteamine for further binding of the antibody (Attar et al., 2016). Then impedimetric response observed from the complex generated upon binding of Antigens with their specific Antibodies, with the detection limit of 2.3 PFU mL−1. Selectivity of the sensor was demonstrated in the presence of hepatitis A virus and enterovirus.
Disposable electrochemical immunosensor was developed for the detection of hepatitis B surface antigen (HBsAg) with the aid of AuNPs with graphene paste electrode and Nafion – L-Cysteine composite film (Fig. 4C) (Huang et al., 2012). Sensor performance was attributed to the synergetic effects of highly conducting nature of graphene, composite film and biocompatibility of AuNPs. DPV analysis of HBsAg in the range of 0.5–800 ng mL−1 produced the detection limit of 0.1 ng mL−1.
A label-free immunosensor was constructed for the detection of HBsAg using the nanocomposite of graphene oxide, iron oxide and Prussian blue (GO/Fe3O4/PB) (Wei et al., 2020). GO/Fe3O4/PB nanocomposites and AuNPs were prepared by the chemical route and coated onto the screen-printed carbon electrode (SPCE), which helped to increase the sensitivity and also the immobilization of HBsAg (Fig. 4D). PB acted as the redox probe. DPV as the electrochemical technique for the detection of HBsAg in the range of 0.5–200 pg mL−1 produced a low detection limit of 0.166 pg mL−1. The fabricated immunosensor was tested with several clinical serum samples and observed good reproducibility, selectivity and stability.
Ultrasensitive label-free, low-cost electrochemical immunosensor for the detection of hepatitis B virus was demonstrated using modified SPCE (Akkapinyo et al., 2020). Bovine serum albumin was immobilized followed by activation with N-ethyl-N′-(3-(dimethylamino)propyl)carbo-diimide/N-hydroxy succinimide (EDC/NHS) chemistry and hepatitis B surface antibodies (Fig. 5 A). Impedimetric response was used to detect HBsAg in a linear range of 5–3000 ng mL−1, with a low detection limit of 2.1 ng mL−1.
Fig. 5.
Electrochemical sensors constructed for the detection of (A) hepatitis B virus using modified SPCE. Reproduced with permission (Akkapinyo et al., 2020). Copyright 2020, Elsevier. (B) Bovine herpesvirus type 1 AG using AGCE/ABCas:AG. Reproduced with permission (Garcia et al., 2020). Copyright 2020, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (C) Single molecule HIV-1 p24 using the electrolyte-gated organic field-effect transistors. Reproduced with permission (Macchia et al., 2020). Copyright 2020, Elsevier. (D) Norovirus using NoroBP peptide modified SPGE. Reproduced with permission (Baek et al., 2019). Copyright 2019, Elsevier.
Development of label free, low-cost impedimetric immunosensor for the detection bovine herpesvirus type 1 antigen (BHV-1 AG) was demonstrated using a simple strategy (Garcia et al., 2020). The immunosensor was constructed with initial activation of the electrode by chronoamperometry followed by the immobilization of appropriate antibody (BHV-1 AB) which was obtained from egg yolk of immunized chickens, then blocking the bare part of electrode surface with casein (Fig. 5B). Then impedimetric method was applied for the detection of antigen in the range of 10–50 Median Tissue Culture Infectious Dose (TCID50) TCID50 mL−1, with limit of quantification of 2.00 TCID50 mL−1 and detection limit 0.66 TCID50 mL−1.
Bioelectronic sensor was demonstrated for the detection of single-molecule, HIV-1 p24 capsid protein (Macchia et al., 2020). The sensor system was a low-cost, label-free and highly sensitive platform for the detection of capsid protein HIV-1 p24. It was proposed that a single-molecule transistor (SiMoT) works based on the electrolyte-gated field-effect transistor phenomenon (Fig. 5C). Gate was bio-functionalized with antibodies and observed the low detection limit of with 30 × 10−21 M.
Electrochemical biosensor was reported for the detection of human norovirus using novel peptides (Baek et al., 2019). Eight peptides were tested as recognition elements, which initially verified individually their effectiveness in sensing the virus and identified that NoroBP peptide coated onto the gold electrode exhibited enhanced binding efficiency and observed the low detection limit 1.7 copies mL−1 (Fig. 5D). The developed sensor was successfully applied the for the detection of norovirus in oyster.
Label free biosensor was developed for the detection of Japanese encephalitis viral (JEV) antigens by the immobilization of the serum antibodies (Huy et al., 2011). The fabrication carried out with the interdigitated electrodes by sputtering Ti and Pt on thermally thick SiO2 layer. The immobilization of serum antibodies was successfully verified by Fourier transform infrared spectrometry and fluorescence microscopy. Low detection limit of the impedimetric sensor was 0.75 μg mL−1 JEV in the concentration range of 1–10 μg mL−1.
Nano bio-polymer based solid phase disposable immuno-biosensor kit was developed for the detection of E. coli bacteriophage (MS2) by employing amperometric technique. The designed micro-flow system exhibited high sensitivity (Braustein and Braustein, 2014). The principle behind the sensor performance was covalent bioconjugation of antibodies and biological support to polymeric beads. The developed immuno-biosensor was able to detect the viral concentration as low as 10 PFU mL−1 in the range of 10–1010 PFU mL−1.
Paper-based portable microfluidic platform was developed for the selective detection of antibody markers of HIV and HCV through multiplexed electrochemical technique. Sensor performance was successfully demonstrated in serum samples with the low detection limits of 300 pg mL−1 HIV and 750 pg mL−1 HCV (Zhao and Liu, 2016). Advantages of the electrochemical microfluidic paper-based immunosensor array was the usage of a handheld multi-channel potentiostat which simultaneously analyzed up to eight samples within 20 min.
A disposable screen printed ultra-sensitive immuno sensor was developed for the detection of influenza A based on the interaction with monoclonal antibodies (Dunajová et al., 2020). Calibration plot was drawn with the absolute changes of charge transfer resistance (ΔRct) of redox probe versus the logarithm concentration of the virus protein. Sensor performance was compared with or without modification of the electrode surface by human serum albumin (HSA) in buffered solution and horse blood. The fabricated immunosensor was capable of identifying influenza A virus in the range of 0.18 fM to 0.18 nM, with the low detection limit of 0.79 fM.
Ultrasensitive electrochemical detection of dengue type 2 virus (DENV-2) was reported using a nanoporous alumina-modified platinum electrode and sensitive membrane (Cheng et al., 2012). The employed biorecognition element was anti-DENV-2 monoclonal antibody (clone 3H5, isotype IgG). The fabricated nano-biosensor produced the low detection limit of 1 PFU mL−1 in the linear range from 1 to 103 PFU mL−1 DENV-2.
Label-free immunoassay platform was reported for the specific detection of human enterovirus 71 (EV 71) using poly (o-phenylenediamine) (PoPD) and AuNPs (Kong et al., 2013). PoPD film was formed on a glassy carbon electrode by using electro polymerization. CV analysis of the sensor yielded a low detection limit of 0.04 ng mL−1 in the concentration range of 0.1–80 ng mL−1 EV 71.
Paper based sensors have been emerged an alternative to the glassy carbon and PET based screen printed electrodes, due to their unique advantages of flexibility, easy to prepare, recyclability and biodegradability (Mahato et al., 2017). A microfluidic paper-based electro-analytical device integrated with micro wire Au electrode was developed for the specific detection of West Nile virus particles (Channon et al., 2018). Here the authors used antibodies as a recognition element and EIS as transduction principle. In another report a paper based electrochemical immunosensor was developed for the detection of HBV (Li et al., 2015).
Table 3 illustrates the summary of different electrochemical diagnostics methods based on antibody-based recognition elements for the detection of viral infectious diseases. All the relevant articles were arranged in the table with the transduction technique, recognition element/assay, working calibration range and sensor limit of detection. Interestingly, tremendous sensitivity was observed in the case of SPCE/GO/Fe3O4/PB/Antibody based sensor (Wei et al., 2020) for HBV detection, electrochemical microfluidic paper-based immunosensor array (E-lPIA) (Zhao and Liu, 2016) for HCV detection, interdigitated micro-electrode of gold (IDE-Au) array/ZIKV specific envelop protein antibody (Zev-Abs) (Kaushik et al., 2018) and ZnO nanostructures immobilized with ZIKV-NS1 antibody on Printed Circuit Board (PCB) (Faria and Mazon, 2019) based senor for Zika virus detection.
Table 3.
Electrochemical diagnostics methods based on antibodies as recognition element for detection of viral infectious diseases.
| S NO | Virus | Recognition Matrix | Method | Concentration Range | LOD | Ref |
|---|---|---|---|---|---|---|
| 1 | HAV | Carbon nanopowder paste electrode/antibody labeled with peroxidase | Chronoamperometry | 2 × 10−4 – 5 × 10−3 IU. mL−1 | 26 × 10−5 IU mL−1 | Mandli et al. (2017) |
| 2 | HPV | Au peptide (SPINNTKPHEAR) linked to a 6-aminohexanoic (Ahx) residue and ferrocene (Fc) | Chronoamperometry | 0.010–0.020 μg L−1 | 0.010 μg L−1 | Valencia et al. (2016) |
| 3 | HPV | GCE/anti HPV-16-L1:anti-HPV complex | SWV | - | 0.1 nmol dm−3 | Piro et al. (2011) |
| 4 | HPV | SCPGE/DNA | SWV | 0–5 ng mL−1 | 0.3 ng mL−1 | Zari et al. (2009) |
| 5 | HBV | Graphene paste electrode|Au NPs/Nafion-cysteine/Antibody | DPV | 0.5–200 ng mL−1 | 0.01 ng mL−1 | Huang et al. (2012) |
| 6 | HBV | SPCE/Graphene Oxide/Fe3O4/Prussian Blue Nanocomposites/Antibody | DPV | 0.5 pg mL−1 - 200 ng mL−1 | 0.166 pg mL−1 | Wei et al. (2020) |
| 7 | HBV | SPCE/EDC NHS/Antibody | EIS | 5–3000 ng mL−1 | 2.1 ng mL−1 | Akkapinyo et al. (2020) |
| 8 | HIV, HCV | Electrochemical microfluidic paper-based immunosensor array | Voltammetry | - | 300 pg mL−1 and 750 pg mL−1 | Zhao and Liu (2016) |
| 9 | HIV-1 | Single-molecule transistor (SiMoT) SiO2 FET/Antibody | FET | 1 to 1 × 107 zM | 30 × 10−21 M | Macchia et al. (2020) |
| 10 | HIV-1 | SPGE/Antibody | EIS | 0–105 copies mL−1 | 1.7 copies mL−1 | Baek et al. (2019) |
| 11 | Zika | Interdigitated micro-electrode of gold (IDE-Au) array/ZIKV specific envelop protein antibody | EIS | 10 pM - 1 nM | 10 pM | Kaushik et al. (2018) |
| 12 | Zika | ZnO nanostructures immobilized with ZIKV-NS1 antibody on Printed Circuit Board (PCB) | CV | 0.1–100 ng mL−1. | 1.00 pg mL−1 | Faria and Mazon (2019) |
| 13 | Zika | Paper microchips with printed electrodes/Pt NPs/Antibody | Conductivity | 101-105 particle μL−1 | 101 particle μL−1 of ZIKV in PBS | Draz et al. (2018) |
| 14 | Zika virus specific antibodies | SPCE/antigen ZIKV-derived proteins | EIS, SWV | 17 fg mL−1 of non-structural protein 1 and 53 fg mL−1 of domain III envelope protein | Cabral-Miranda et al. (2018) | |
| 15 | Dengue type 2 virus (DENV-2) | Nanoporous alumina-modified platinum electrode/Antibody | DPV | 1 - 103 PFU mL−1 | 1 PFU mL−1 | Cheng et al. (2012) |
| 16 | Dengue | SPCE/Bovine serum Albumin (BSA) Nanostructured antibody | EIS | 1–200 ng mL−1 | 0.3 ng mL−1 | Nawaz et al. (2018) |
| 17 | Rotavirus | GCE Au sononanoparticles (AuSNPs)/Antibody | EIS | 4.6 to 4.6 × 104 PFU mL−1 | 2.3 PFU mL−1 | Attar et al. (2016) |
| 18 | Rotavirus | Micropatterned reduced graphene oxide field-effect transistor (MRGO-FET)/Antibody | FET | 10 - 105 PFU mL−1 | 102 PFU mL−1 | Liu et al. (2013) |
| 19 | Avian Influenza H9N2 | MBs, Antibody horseradish peroxidase–streptavidin conjugate (HRP-SA) | DPV | 1–600 ng mL−1 | 10 pg mL−1 | Zhou et al. (2013) |
| 20 | Hepatitis A,B,C,D,E | Immunosensor array Au NPs | Potentiometry | 1.0–350 ng mL−1 | 0.5, 0.3, 0.8, 0.5, and 1.0 ng mL−1 | Tang et al. (2010) |
| 0.8–350 ng mL−1, 1.5–350 ng mL−1 | ||||||
| 1.0–350 ng mL−1 and 1.5–350 ng mL−1 | ||||||
| 21 | SARS | In2O3 nanowire Antibody mimic proteins | Voltammetry | – | 44 μM | Ishikawa et al. (2009) |
| 22 | Japanese encephalitis virus | Interdigitated electrodes by sputtering Ti and Pt on thermally thick silicon dioxide (SiO2) layer | Potentiometry | 1–10 μg mL−1 | 0.75 μg mL−1 | Huy et al. (2011) |
| 23 | MERS COVID H COV | Immunosensor Au NPs carbon electrodes | SWV | 0.001–100 ng mL−1 and 0.01–10,000 ng mL−1 | 1.04 pg mL−1 and 0.4 pg mL−1 | Layqah and Eissa (2019) |
| 24 | Rotavirus | Graphene oxide (GO) film from 1-pyrenebutyric acid N-hydroxy succinimide ester (PSE)/Antibody | CV | 103 - 105 PFU mL−1 | 103 PFU mL−1 | F. Liu et al. (2011) |
| 25 | E. coli bacteriophage (MS2) | Nanoporous oxirane-derivatized beads/Antibody | Chronoamperometry | 10 and 1010 PFU mL−1 | 10 PFU mL−1 | Braustein and Braustein (2014) |
| 26 | Influenza A | SPCE/monoclonal antibody | EIS | 0.18 fM - 0.18 nM | 0.79 fM | Dunajová et al. (2020) |
| 27 | Human EV 71 | GCE/Poly (o-phenylenediamine) (PoPD) Au NPs/Antibody | CV | 0.1–80 ng mL−1 | 0.04 ng mL−1 | Kong et al. (2013) |
| 28 | Dengue | Au/anti-NS1 | EIS, ECS | 10 − 2000 ng mL−1 5.0–1000 ng mL−1 | 0.5 ng mL−1 | Cecchetto et al. (2017) |
| 29 | Herpes virus type 1 | GCE/Antibody | EIS | 10 - 50 TCID50 mL−1 | 0.66 TCID50 mL−1 | Garcia et al. (2020) |
2.4. Aptamer based electrochemical biosensors
Aptamers are the oligonucleotide or peptide molecules that are capable of binding strongly and specifically to the targets based on affinity complexation led by their sequence-specific three-dimensional structures (Jarczewska et al., 2016). Thus, aptamers have been used in biosensors to achieve the selectivity and specificity. Due to the comparable selectivity with antibody, these have constantly been employed in various sensors to make it robust and temperature tolerant (Zhou et al., 2014). Another advantage of the aptamer-based biosensors is their relatively smaller size, which enables them to get access to the deep-seated target sites. Whereas, conventional antibody-based biosensing cannot be able to perform it due to their larger size (Banerjee and Nilsen-Hamilton, 2019), making aptamer an appealing choice to develop the biosensors for various targets including viruses. The virus contains capsid proteins, the genetic materials (either RNA or DNA), and in some viruses a lipid bilayer or envelope for their protection. The electrochemical detection of viruses was achieved by targeting these moieties, mainly using labeled and label-free techniques. In labeled formats the recognition is monitored by the target and aptamer physicochemical interactions with a specific tag, whereas in the label-free formats the detection depends on direct signal generation at the transducer surface. In the recent past, various other viruses have been detected using the aptamer-based sensitive electrochemical biosensors including SARS, hepatitis virus, HIV, bovine viral diarrhea virus, Ebola, influenza, rabies, etc. (van den Kieboom et al., 2015).
This section critically summarizes the electrochemical approaches which utilized the aptamers as bio-recognition element towards the detection of viruses, such as HAV (Manzano et al., 2018), HBV (Campos-Ferreira et al., 2013; Jampasa et al., 2018; 2014; Karimizefreh et al., 2014; Qian et al., 2020; Tran et al., 2011), HCV (Ghanbari et al., 2017; Ghanbari and Roushani, 2018; S. Liu et al., 2011), HPV(Bartosik et al., 2016; Campos-Ferreira et al., 2016; Civit et al., 2012; Huang et al., 2015; Souza et al., 2014), HIV (Chen et al., 2012; Fatin et al., 2019; Guo et al., 2013; Radhi et al., 2016; Rahim Ruslinda et al., 2013; Wang et al., 2015), Zika (Aydinlik et al., 2011; Dolai and Tabib-azar, 2020; Faria and Zucolotto, 2019; Moço et al., 2019; Steinmetz et al., 2019), Dengue (Rai et al., 2012), Chikungunya (Singhal et al., 2018b; 2018a), Avian (Dong et al., 2015; X. Liu et al., 2011), Influenza (Tam et al., 2009; Yamanaka et al., 2011) and Hepatitis (Cho et al., 2018; Li et al., 2015).
The detection of viral proteins has been reported relatively quickly and due to this reason, in the recent COVID-19 pandemic outbreak the biosensors targeting capsid-proteins have been used for preliminary screening of the patients suffering from the disease. Similarly, for the viruses, the aptamer-based detection has also been reported in the recent past, which consists of the selective, robust, specific aptamer probe that has an affinity of binding towards the proteinaceous part of viral-materials (van den Kieboom et al., 2015). For instance, Ghanbari et al. used the graphene quantum dots based electrochemical sensor probe for detecting core antigen from the HCV. Resistance changes were monitored when the affinity binding exists between the aptamer probe and HCV core antigen. In this report, the developed sensor was capable of detecting the target antigen in a wide concentration range (0–70 pg mL−1 and 70–400 pg mL−1) with the detection limit of 3.3 pg mL−1. The sensor probe was reported to be extremely selective towards the target analyte and also it worked excellently even in human serum samples (Ghanbari et al., 2017). Similarly, in another report from Kazhal Ghanbari et al. have reported another aptasensor immobilized on multi-walled carbon nanotubes (MWCNTs), Chitosan and MIP i.e., MWCNT-Chit/MIP/Apt as a probe surface to immobilize the specific aptamer to the core antigen from the HCV. This biosensor has shown a wide dynamic range of 5 fg mL−1 to 1.0 pg mL−1 with an excellent detection limit of 1.67 fg mL−1. The sensor was also capable of detecting the core antigen in the real sample following the spike-recovery method and got recovered more than 99% of the antigen (Ghanbari and Roushani, 2018). Achieving better sensitivity is one of the utmost requirements for the detection in early/onset of any viral infection, where the symptoms are rarely perceived even by the infected individual (Mahato et al., 2018). In this stage of infection, the viral loads in the sample are way less and sometimes these are found in traces. In view of this, various sensitive and effective biosensors have been reported targeting the viral moieties.
The aptamer-based viral sensing has been done in mainly labeled and label-free techniques. The labeled detections have been facilitated by the electrochemical active labels typically conjugated to the aptamers which could give the electrical/mechanical signals when reacting with the specific target molecules. Common electrochemical active label tags are methylene blue, ferrocene, etc. Generally, these labeled biorecognition elements based electrochemical sensors involve vast immobilization protocols and take little longer time for the immobilization and sensing. Label-free biosensors are an alternative to the labeled recognition elements, by avoiding the laborious labelling steps and the challenging label reaction/operations, the cost of the biosensor could be reduced as well as the analysis can be performed in short time (Rhouati et al., 2016).
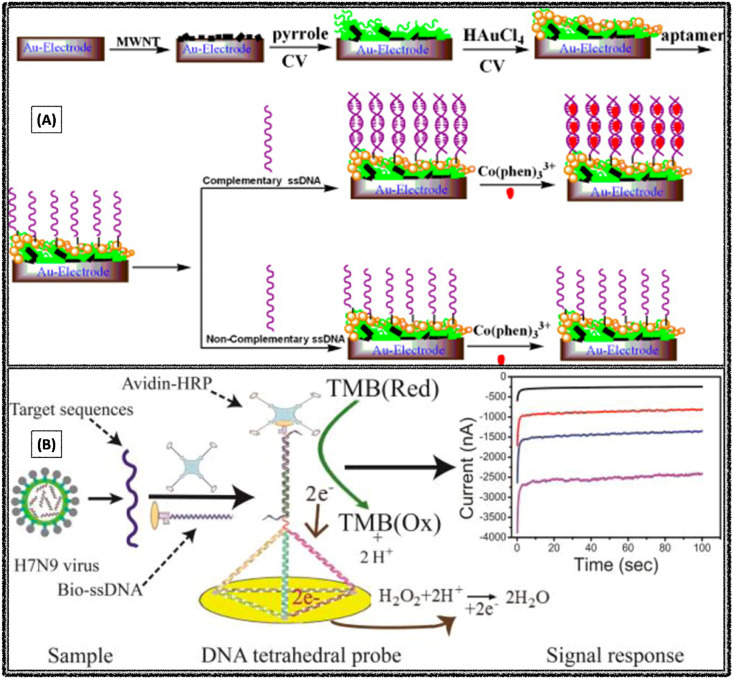
X Liu et al. reported an electrochemical aptasensor for the detection of avian influenza virus, H5N1 by adopting DPV as a transduction technique. Initially, the Au electrode surface was modified with MWCNT, poly pyrrole nanowires and gold nanoparticles interface, followed by the immobilization of the aptamers sequence (Fig. 6 A). The nanomaterial interface of electrode surface offers porous nature, large effective surface area, highly electrocatalytic activities and electronic conductivity properties, apart from this it would help in the better immobilization of aptamers. While in presence of target virus DNA, it would specifically bind with the immobilized aptamers sequence, and followed electrochemical signal indicator binds specifically to paired sequence (not to lone sequence), these changes were recorded by using DPV. Based on this principle, the limit of detection was observed as 0.43 pM H5N1 in the concentration range of 5 pM - 1 nM (X. Liu et al., 2011).
Fig. 6.
Electrochemical aptasensors constructed for the specific detection of avian influenza A viruses (A) H5N1. Reproduced with permission (X. Liu et al., 2011). Copyright 2011, Elsevier. (B) H7N9. Reproduced with permission (Dong et al., 2015). Copyright 2015, American Chemical Society.
S Dong et al. constructed a novel electrochemical biosensor with tetrahedral DNA nanostructure based aptasensor for the detection of avian influenza A (H7N9) virus through recognizing a fragment of the hemagglutinin gene sequence. Initially, the tetrahedral DNA probe was immobilized on gold electrode surface through thiolate self-assembly method, followed by the addition of longer nucleotide complementary target DNA sequences (Fig. 6B). The target DNA sequence was labeled with biotinylated detection probe, which could produce an amperometric signal in presence of avidin horseradish peroxidase. Good selectivity was observed as the electrochemical biosensor could specifically recognize the target DNA fragment of influenza A (H7N9) virus even in presence of other types of influenza viruses, such as influenza A (H1N1) and (H3N2) viruses. The achieved limit of detection was 100 fM target nucleotide sequences of H7N9 virus (Dong et al., 2015).
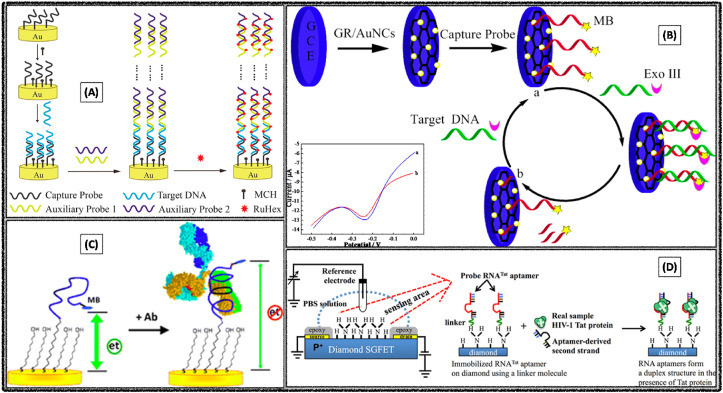
Xian Chen et al. developed an ultrasensitive electrochemical aptasensor for the detection of HIV (Chen et al., 2012). Herein, the DNA biosensor has been immobilized on the long-range self-assembly axillary probes, that would lead mico-meter range from the electrode transducer surface, which could help in the proximity changes towards the electroactive surface area while interacting with the target virus (Fig. 7 A). This could help eventually to enhancing the electrochemical signal of a great amount of redox indicator [Ru(NH3)6]3+. DPV was employed as a transduction technique and screened the HIV DNA sample in the concentration range over 2 aM to 10 pM. The aptasensor exhibited remarkable sensitivity with the detection limit of 2 aM in serum or in cell lycates.
Fig. 7.
Electrochemical detection of HIV using (A) long-range self-assembled DNA nanostructures. Reproduced with permission (Chen et al., 2012). Copyright 2012, American Chemical Society. (B) Graphene stabilized gold nanoclusters. Reproduced with permission (Wang et al., 2015). Copyright 2015, American Chemical Society. (C) Peptide-based biosensing platform (Gerasimov and Lai, 2010) Copyright 2010, Royal Society of Chemistry. (D) Diamond-FET-based RNA aptamer. Reproduced with permission (Rahim Ruslinda et al., 2013). Copyright 2013, Elsevier.
In another report Wang Yijia et al. constructed an ultrasensitive aptasensor for HIV detection based on graphene stabilized gold nanoclusters with exonuclease amplification (Wang et al., 2015). Here, the authors initially synthesized the graphene stabilized gold particle by the one-step ultrasonic method, and immobilized on the GCE surface, followed by the immobilization of exonuclease III DNA clusters on GCE/GR-AuNPs electrode surface (Fig. 7B). The developed DPV based aptasensor was able to detect as low as 30 aM. In another report, an electrochemical peptide-based senor was constructed for the specific detection of HIV (Gerasimov and Lai, 2010). Methylene blue labeled aptasensor offered the detection limit of 10 nM or 1.5 μg mL−1 (Fig. 7C).
A. Rahim Ruslinda et al. constructed a diamond-FET based RNA aptasensing platform for the specific detection of HIV-1 Tat protein (Rahim Ruslinda et al., 2013). Herein, the RNA-Tat aptamer was immobilized on the diamond electrode surface channel via linker. In the presence of HIV-1 Tat, the probe aptamer and aptamer-derived second strand form a duplex structure, whereby the hole carrier density is decreased owing to the decreased negative charge on the surface channel (Fig. 7D). Based on this principle the achieved detection limit was 100 pM HIV-Tat protein.
In another report, X Quian et al. constructed an ultrasensitive electrochemical aptasensor for hepatitis B virus detection. Here, water-soluble pillar [5]arene stabilized Pd NPs were assembled with reduced graphene oxide nanosheet (WP5–Pd/RGO) to prepare the supporting matrix for better immobilization of the probe DNA and increase the electro-catalytic performance of the transducer surface. The label was prepared with auxiliary DNA and the hydroxylatopillar [5]arene stabilized Au NPs anchored on metal-organic frameworks-derived cobalt sulfide nanobox (HP5–Au/CoS-aDNA). The principle involved in the present aptasensor was the capturing interactions of tDNA and pDNA lead to the increase in the HP5–Au/CoS-aDNA label, it would involve in more electro-catalytic activity towards H2O2, these changes were recorded by amperometry. Under the optimized conditions, the proposed sensor displayed a linear relationship between amperometric currents and the logarithm of tDNA solution from 1 fM – 1 nM, and the achieved low detection limit was 0.32 fM. The reported sensor system offered remarkable stability, reproducibility, specificity, and accuracy, which provided a potential and promising prospect for clinical diagnosis and analysis (Qian et al., 2020).
On the other hand, some viruses are used for the therapeutic agents, especially oncolytic viruses (OVs) were very promising could selectively replicate in and kill tumor cells. But repetitive administration of OVs provokes the generation of neutralizing antibodies that could diminish the anticancer properties. Excess amount of this virus particles harms the patient body, hence the detection of OVs is important. M Labib et al. reported an article for the specific detection of vesicular stomatitis virus (VSV) through electrochemical aptasensing method. Here, self-assembled thiolated primer was used as a linker to immobilize the anti-VSV aptamers. The aptamer sequences of F-CCA TCA CCC TAT TAT CTC ATT ATC TCG TTT TCC CTA TGC G-cR (ZMYK-20), F-GCG ACA ACA CGG ACG GTT GAG ACT TTA ATT CTG CTC ACG G-cR (ZMYK-22), F-GGG ACC TAT CAG GCG ATG TGA AAA CTC TTA TAC CAC TGG-cR (ZMYK-23) and F-CCA CCA TGC ACG ACC CAC GCA ATG ACA GTA ACA CAC CTC G-cR (ZMYK-28) were used for achieving the selectivity. The complementary interaction of VSV and aptamers sequences increases the impedance values, while introducing of free antibodies, which were specifically binds with the VSV led to decrease in the impedance values. Depending on this principle, the authors have developed calibration plots for four different aptamer clones ZMYK-20, ZMYK-22, ZMYK-23, and ZMYK-28 and got the highest protective properties with dissociation constants of 17, 8, 20, and 13 nM, respectively (Labib et al., 2012b).
Label-free impedimetric aptasensor was developed for the specific detection of Zika virus in real human serum samples. Here, the oxidized glassy carbon electrode was modified with silsesquioxane-functionalized gold nanoparticles to immobilize the aptamer sequences. CV, EIS and Atomic force microscopy (AFM) studies were utilized for monitoring the biosensing fabrication steps. EIS analysis of Zika virus in the concentration range of 1.0 pM to 1 μM led to the low detection limit of 0.82 pM. Moreover, the reported impedimetric aptasensor exhibited a good stability and satisfactory sensitivity with high selectivity to detect Zika virus in human serum samples, which suggests its promising clinical applications for the early diagnosis of zika virus associated pathologies (Steinmetz et al., 2019).
S Jampasa et al. developed a novel two signal on electrochemical aptasensing methodology by employing a sandwich-hybridization of pyrrolidinyl peptide nucleic acid (PNA) probes on screen printed electrodes. Here, the capture PNA probe (P1) was initially immobilized on SPCE surface followed by anthraquinone labeled signaling probe (AQ-P2), which could complementary sequence to the target DNA. Two types of interaction methodologies were followed namely at upstream (ASU) and at downstream (ASD). It was reported that higher current response values were observed at ASD compare to the ASU. Finally, ASD methodology applied for the simultaneous detection of two high-risk human papillomavirus DNA sequences. The calculated detection limits were 150 and 153 pM, for HPV type 16 and 18 sequences respectively in the concentration range of 0.5–100 nM (Jampasa et al., 2018).
In another report, an ultrasensitive DNA biosensor was developed for the detection of HPV by using EIS and DPV techniques. Initially, the capture probe was immobilized on graphene/Au nanorod/polythionine modified GCE. Two types of auxiliary probes of long size self-assembled DNA structure were immobilized after target DNA. Finally, [Ru (phen)3]2+ was used as electroactive indicative medium, which was deposited on auxiliary probes due to electrostatic interaction, these changes were recorded by DPV. Based on this principle, The calculated low detection limit from calibration plot was 0.403 pM HPV in the concentration range of 0.1 pM - 0.1 nM (Huang et al., 2015).
Paper based electrochemical aptasensor have been emerged as the alternative to PET based screen printed electrodes for the detection various viruses, such as hepatitis B (Srisomwat et al., 2021, 2020), hepatitis C (Zhao and Liu, 2016), chikungunya (Singhal et al., 2018a), zika virus (Dolai and Tabib-Azar, 2020) and papilloma virus (Teengam et al., 2017). Dolai S et al. developed a paper based microfluidic platform for the zika virus detection using potentiometric transduction method (Dolai and Tabib-Azar, 2020). Herein, the proposed aptamer-based potentiometric sensor was able to detect the whole Zika virus for the first time with a minimum sensitivity of 2.6 nV/Zika and the minimum detectable signal of 1.2 × 106 Zika. The combination of microfluidics with paper based electrochemical biosensing systems accelerates the development of POC devices for the rapid and onsite detection of viruses.
Table 4 illustrates the summary of different electrochemical diagnostic methods based on aptamers as a recognition element for the detection of viral infectious diseases. All the relevant reports were arranged in the table with their transduction technique, recognition element/assay, working calibration range and sensor limit of detection. Interestingly, the best sensitivity (fM and aM range) values were observed in the case of GCE/MWCNT-Chit/MIP/Apt (Ghanbari and Roushani, 2018) based sensor for HCV, AuE/Self-Assembled DNA Nanostructures (Chen et al., 2012) based sensor for HIV, hollow HP5–Au/CoS (Qian et al., 2020) based senor for HBV and G/Au NR/PT DNA (Huang et al., 2015) based sensor for HPV detection.
Table 4.
Electrochemical diagnostics methods based on aptamers as recognition element for detection of viral infectious diseases.
| S NO | Virus | Recognition Matrix | Method | Concentration Range | LOD | Ref |
|---|---|---|---|---|---|---|
| 1 | HCV | GCE/MWCNT-Chit/MIP/Apt | CV, DPV, EIS | 5.0–1.0 pg mL−1 | 1.67 fg mL−1 | Ghanbari and Roushani (2018) |
| 2 | HCV | GCE/AuNPs/Apt | DPV | 1 × 10−21 to 1 × 10−11 M | 5 × 10−17 M | S. Liu et al. (2011) |
| 3 | HCV | GCE/GQD/Apt | EIS | 10–400 pg mL−1 | 3.3 pg mL−1 | Ghanbari et al. (2017) |
| 4 | HIV | HRP/NF-kB/Tx-CP/GE | Amperometry | 5–500 nM | 5 nM | Guo et al. (2013) |
| 5 | HIV – 1 | Diamond FET-based RNA aptamer | FET | 1–100 nM | 1 nM | Rahim Ruslinda et al. (2013) |
| 6 | HIV 1 | Ni–Au/fMWCNT | FET | 0.2 nM - 1 μM | 600 pM | Fatin et al. (2019) |
| 7 | HIV | GCE/GR/AuNCs/Apt | DPV | 0.1 fM - 100 nM | 30 aM | Wang et al. (2015) |
| 8 | HIV | AuE/Self-Assembled DNA Nanostructures | DPV | 2 aM - 10 PM | 2 aM | Chen et al. (2012) |
| 9 | HIV 1 | Si/SiO2/Au/MWCNT | FET | 1.64–3.74 nM | 1.64 nM | Radhi et al. (2016) |
| 10 | HIV | GCE/PPI/Strep/ssDNA | SWV, EIS | 0.1–16 nM | 3.44 pM | John et al. (2014) |
| 11 | HAV | SPGEs DNA probe | CV | 10 fg μL−1 - 0.1 ng μL−1 | 0.15 fg μL−1 | Manzano et al. (2018) |
| 12 | HBV | PNA probe (P1) immobilized on a screen-printed carbon electrode (SPCE) | DPV | 0.1–1000 nM | 150 pM | Jampasa et al. (2018) |
| 13 | HBV | PANI–MWCNT platinum electrode arrays peptide aptamers | SWV | 10–50 nM | 490 pM | Tran et al. (2011) |
| 14 | HBV | Au DNA | EIS | 1 nM–1000 nM | 1 nM | Karimizefreh et al. (2014) |
| 15 | HBV | (GC) (prGO) (MoS2) RNA aptamer _ | DPV | 3.5 pM–35.3 pM | 1.75 pM | Chekin et al. (2018) |
| 16 | HBV | (acpcPNA) probe (AQ-PNA) and (G-PANI) | SWV | 10–200 nM. | 2.3 nM | Teengam et al. (2017) |
| 17 | HBV 16 | SPGE cysteine film DNA | DPV | 18.75 nM and 250 nM | 18.13 nM | Campos-Ferreira et al. (2013) |
| 18 | HBV 16 | SPCE pyrrolidinyl peptide nucleic acid probe | SWV | 0.02 and 12.0 μM | 4 nM | Jampasa et al. (2014) |
| 19 | HBV | hollow HP5–Au/CoS | Chronoamperometry | 1 fM – 1 nM | 0.32 fM | Qian et al. (2020) |
| 20 | HBV | Paper/Ag NPs-DNA modified MμBs | ASV | 100–1500 pM | 85 pM | Li et al. (2015) |
| 21 | HPV16E7p, 18E6 and 45E6 | Au arrays DNA Probe | steps and sweeps technique | 0.1–10 nM 0.1–12 nM 0.1–1 nM | 220 pM, 170 pM and 110 pM | Civit et al. (2012) |
| 22 | HPV | G/Au NR/PT DNA | EIS and DPV | 0.1 pM - 10 nM | 50 fM | Huang et al. (2015) |
| 23 | HPV16 | (PGE) DNA | DPV | 40–5000 pg L−1 | 16 pg L−1 | Campos-Ferreira et al. (2016) |
| 24 | HPV | SPEs DNA magnetic bead-modified DNA probes | chronoamperometry | 1 pM - 1 nM | 1 pM | Bartosik et al. (2016) |
| 25 | HPV 16 | pencil graphite electrode (PGE)/Methylene blue DNA | DPV | 2–10 nM | 1.49 nM | Souza et al. (2014) |
| 26 | Influenza A | Microfluidic RT-PCR chip and disposable electrical printed (DEP) | SWV | 5.36 × 102 to, 5.36 × 105 copies μL−1 | – | Yamanaka et al. (2011) |
| 27 | Influenza A | PtE/MWCNT/DNA | SWV | 1–10 nM | 0.5 nM | Tam et al. (2009) |
| 28 | Vesicular Stomatitis virus (vsv) | AuE/Apt/Ab | EIS | 800 - 2200 PFU | 600 PFU | Labib et al. (2012b) |
| 29 | Vaccinia virus (VACV) | AuE/Apt/ | EIS | 500 - 3000 PFU | 330 PFU | Labib et al. (2012a) |
| 30 | Zika | polyethylene terephthalate (PET) Au DNA | EIS | 54–340 nM | 25 nM | Faria and Zucolotto (2019) |
| 31 | Zika | silsesquioxane-functionalized gold nanoparticles (AuNPs-SiPy) | EIS | 1 pM–1 μM | 0.82 pM | Steinmetz et al. (2019) |
| 32 | Zika | graphite disk reduced graphene oxide and polytyramine-conducting polymer | DPV | 1.72 × 1010 copies mL−1 (10−16 g mL−1) to 1.72 copies mL−1 (10−16 g mL−1) | 1.72 copies mL−1 (0.1 fg mL−1) | Moço et al. (2019) |
| 33 | Zika | Paper aptamer | potentiometry | – | – | Dolai and Tabib-azar (2020) |
| 34 | Zika | poly-(3-amino- 4-hydroxybenzoic acid)-modified PGE | SWV | 84.0 pM - 1.41 nM | 25.4 pM | Aydinlik et al. (2011) |
| 35 | Chikungunya virus | (SPGEs) (MoS2 NSs) probe PDNA | EIS, CV | (0.1 nM–80 μM) (0.1 nM–100 μM) | 3.4 nM | Singhal et al. (2018b) |
| 36 | Chikungunya virus | Paper Fe3O4@Au Nanocubes based DNA | CV | 0.1 nM - 100 μM | 0.1 nM | Singhal et al. (2018a) |
| 37 | Avian influenza virus H5N1 gene | AuE/MWCNT/PPNWs/GNPs/DNA-aptamer. | DPV | 5 pM–1 nM | 0.43 pM | X. Liu et al. (2011) |
| 38 | Avian influenza A (H7N9) | AuE/tetrahedral structured DNA probe | Amperometry | 1 pM - 100 nM | 0.75 pM | Dong et al. (2015) |
| 39 | Herpes DNA | Paper Zn–Ag nanoblooms DNA | CV | 113–103 and 3 × 105 to 3 × 106 copies mL−1 | 97 copies mL−1 | Narang et al. (2018) |
| 40 | Herpes | Poly pyrrole silicon aptamer | Conductometry | 5–20 nM | 2 nM | Tam et al. (2010) |
| 41 | Hepatitis B | Graphene/CPPyNW | field-effect transistor (FET) | 10 aM to 0.1 μM | 10 aM | Cho et al. (2018) |
| 42 | Dengue Virus | Alumina over platinum wire Electrode probe DNA | DPV | 0.01 nM to 1 μM | 9.5 pm | Rai et al. (2012) |
| 43 | Ostreid herpesvirus 1 (OsHV-1) 1 | Au/DNA | Chronoamperometry | 1.50 × 102 to 3.34 × 105 OsHV-1 DNA copies/50 ng of total DNA. | 207 OsHV-1 target copies | Toldrà et al. (2020) |
AuE – gold electrode; MWCNT - multi-wall carbon nanotubes; PPNWs - polypyrrole nanowires; GNPs – gold nanoparticles; DPV – differential pulse voltammetry; ASV – anodic stripping voltammetry; SWV – square wave voltammetry; EIS – electrochemical impedance spectroscopy; SPE – screen printed electrode; MμBs - magnetic microbeads; PFU – Plaque forming units; PPI – Poly(propylene imine); CPPyNW – carboxylic polypyrrole nanowires; GCE – glassy carbon electrode; Chit – chitosan; GQD – graphene quantum dots; FET – field effect transistor; APTES – triethoxysilane on silicon substrates; Au NPs – Gold nanoparticles; PANI–MWCNT – polyaniline-multiwalled carbon nanotube; GC – Glassy carbon; rGO – reduced graphene oxide; MoS2 – molybdenum sulfide; acpcPNA – anthraquinone-labeled 28 pyrrolidinyl peptide nucleic acid; G-PANI – graphene-polyaniline; PGE – pencil graphite electrode; PET – polyethylene terephthalate; AuNPs-SiPy – silsesquioxane-functionalized gold nanoparticles.
3. Challenges and future perspectives
Electrochemical diagnostic methods are emerging as an alternative rapid sensing methodology over the conventional clinical screening methods. However, several challenges still remain unsolvable and need to be carefully addressed by the scientific research community working on sensors development. All the current challenges encountered in the electrochemical viral infectious disease diagnosis and the possible solutions to overcome such challenges are discussed in this section.
Challenge I: Screening sample separation – an appropriate screening sample selection and preparation are the utmost important steps, while transferring the biosensors systems from laboratory to onsite-clinics (Ritzi-Lehnert, 2012). Raw bio sample screening hurdles with a lot of interferences, such as proteins, vitamins, common drug intakes naturally present in the bio-fluids. Hence, several separation methods need to be performed before going to sensing. Possible Solution: Latest innovative developments in the microfluidics technology help to overcome this challenge (Whitesides, 2006). Challenge II: Fouling studies – Electrochemical biosensor system mostly relies on the binding interaction or conformational changes of biorecognition element while reacting with the target analyte, but not depends on the adsorption of the target analyte. Sometimes, it leads to false readings due to nonspecific adsorption of proteins, cells, and other biomolecules, which are present in the complex bio fluids. Possible Solution: This problem can possibly be overcome by using various antifouling layers such as polyethylene glycol (PEG), biomimetic materials, zwitterionic polymers, synthetic peptides and carbon nanofibers. Recently, Jiang et al. have critically discussed the strategies to lower the fouling and to translate these biosensor innovations to commercial opportunities (Jiang et al., 2020). Challenge III: Selectivity and sensitivity – Achieving the best selectivity and sensitivity are two prominent challenges to successfully commercialize the biosensor systems. Especially in clinical complex samples, the performance of biosensor is poor, need to improve competitiveness with the commercial clinical laboratory analysis. Possible Solution: Nanomaterials play crucial role in achieving the high sensitivity whereas the specific biorecognition elements (Antibodies, Aptamers, Imprinted polymers, CRISPR based systems, etc.) deliver the best selectivity. Hence, the synergistic combination of both these materials may yield the advanced hybrid nanocomposites thereby facilitates the construction of an ideal biosensor system.
In spite of the achieved development in biosensors for the specific detection of viruses using different bio-recognition elements, a plenty of research is yet to be done in the special areas such as utilization of different anti-fouling coating layers to reduce the fouling and adoption of advanced bio-compatible nanomaterials to enhance the sensitivity of the transducer system. Accurate detection of the other biomarker components such as C-reactive protein, interleukins, glutamate, breath pH, TNF-α, interferons, hematological biomarkers apart from the virus particle detection helps to understand the severity of the infection. This dual detection could be achieved using multiplexed electrochemical sensing models with the aid of microfluidic sensing systems, and in turn provide valuable help in the better management of viral pandemic diseases.
We believe that the manuscript gives critical insights on the latest developments and could help the researchers in designing an integrated electrochemical multi-sensing system for early, rapid, accurate detection of viral infectious diseases.
4. Summary and conclusions
Overall, this review summarized the recently reported electrochemical sensor systems for the selective detection of twelve life-threatening viruses namely - COVID-19, MERS, SARS, Influenza, Hepatitis, HIV, HPV, Zika, Herpes simplex virus, Chikungunya, Dengue, and Rotavirus. Sensor systems reported for other viruses namely - Enterovirus 71, West Nile virus, human norovirus, vesicular stomatitis virus, Japanese encephalitis virus, etc. were also critically discussed. Brief information of each individual virus (molecular structure, disease symptoms and magnitude of infection caused) was mentioned in the introduction section. Literature reports on the detection of COVID-19/SARS-CoV-2 were collectively summarized in a separate sub-section. All the electrochemical works were discussed by categorizing them into three major sub-sections depending on the recognition matrix/sensor mechanism (antibody or aptamers or direct/mediated electron transfer). The challenges involved in the real time monitoring of infectious diseases were keenly addressed by highlighting the key issues along with appropriate possible solutions. We strongly believe that this review significantly helps the researchers working in the fields of materials chemistry, nanotechnology and biotechnology to come up with innovative strategies to construct a successful portable electrochemical sensor system for rapid screening of various viral infectious diseases.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
K. Yugender Goud would like to thank the support received from the Department of NanoEngineering, University of California San Diego, USA. K. Koteshwara Reddy and Hern Kim acknowledge the support received by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A1A03038817) and by the Korea Institute of Technology Evaluation and Planning (KETEP) funded by the Ministry of Trade, Industry & Energy (MOTIE) (20194010201750), Republic of Korea. Ahmed Khorshed acknowledges the support received from the Department of Pharmaceutical Analytical Chemistry, Faculty of Pharmacy, Sohag University, Egypt. V. Sunil Kumar and Prof. K.V. Gobi acknowledge the support received from Department of Chemistry, National Institute of Technology Warangal, Telangana, India.
References
- Acosta E.G., Castilla V., Damonte E.B. Functional entry of dengue virus into Aedes albopictus mosquito cells is dependent on clathrin-mediated endocytosis. J. Gen. Virol. 2008;89:474–484. doi: 10.1099/vir.0.83357-0. [DOI] [PubMed] [Google Scholar]
- Acquah C., Danquah M.K., Agyei D., Moy C.K.S., Sidhu A., Ongkudon C.M. Deploying aptameric sensing technology for rapid pandemic monitoring. Crit. Rev. Biotechnol. 2016;36:1010–1022. doi: 10.3109/07388551.2015.1083940. [DOI] [PubMed] [Google Scholar]
- Ahmed R., Oldstone M.B.A., Palese P. Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat. Immunol. 2007;8:1188–1193. doi: 10.1038/ni1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkapinyo C., Khownarumit P., Waraho-Zhmayev D., Poo-arporn R.P. Development of a multiplex immunochromatographic strip test and ultrasensitive electrochemical immunosensor for hepatitis B virus screening. Anal. Chim. Acta. 2020;1095:162–171. doi: 10.1016/j.aca.2019.10.016. [DOI] [PubMed] [Google Scholar]
- Algaissi A., Alfaleh M.A., Hala S., Abujamel T.S., Alamri S.S., Almahboub S.A., Alluhaybi K.A., Hobani H.I., Alsulaiman R.M., AlHarbi R.H., ElAssouli M.-Z. ak, Alhabbab R.Y., AlSaieedi A.A., Abdulaal W.H., Al-Somali A.A., Alofi F.S., Khogeer A.A., Alkayyal A.A., Mahmoud A.B., Almontashiri N.A.M., Pain A., Hashem A.M. SARS-CoV-2 S1 and N-based serological assays reveal rapid seroconversion and induction of specific antibody response in COVID-19 patients. Sci. Rep. 2020;10:16561. doi: 10.1038/s41598-020-73491-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D.S., Lugo L.A., Kojic E.M., Stoever J., Liu S.T.H., Cunningham-Rundles C., Felgner P.L., Moran T., García-Sastre A., Caplivski D., Cheng A.C., Kedzierska K., Vapalahti O., Hepojoki J.M., Simon V., Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M., Ajmal M., Ashraf G., Muhammad N., Aziz A., Iftikhar T., Wang J., Liu H. The role of biosensors in coronavirus disease-2019 outbreak. Curr. Opin. Electrochem. 2020;23:174–184. doi: 10.1016/j.coelec.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asran A.M., Mohamed M.A., Ahmed N., Banks C.E., Allam N.K. An innovative electrochemical platform for the sensitive determination of the hepatitis B inhibitor Entecavir with ionic liquid as a mediator. J. Mol. Liq. 2020;302:112498. doi: 10.1016/j.molliq.2020.112498. [DOI] [Google Scholar]
- Attar A., Mandli J., Ennaji M.M., Amine A. Label-free electrochemical impedance detection of rotavirus based on immobilized antibodies on gold sononanoparticles. Electroanalysis. 2016;28:1839–1846. doi: 10.1002/elan.201600179. [DOI] [Google Scholar]
- Aydinlik S., Ozkan-Ariksoysal D., Kara P., Sayiner A.A., Ozsoz M. A nucleic acid-based electrochemical biosensor for the detection of influenza B virus from PCR samples using gold nanoparticle-adsorbed disposable graphite electrode and Meldola's blue as an intercalator. Anal. Methods. 2011;3:1607. doi: 10.1039/c1ay05146f. [DOI] [Google Scholar]
- Baek S.H., Kim M.W., Park C.Y., Choi C.-S., Kailasa S.K., Park J.P., Park T.J. Development of a rapid and sensitive electrochemical biosensor for detection of human norovirus via novel specific binding peptides. Biosens. Bioelectron. 2019;123:223–229. doi: 10.1016/j.bios.2018.08.064. [DOI] [PubMed] [Google Scholar]
- Banerjee S., Nilsen-Hamilton M. E. Coli Infection [Working Title] IntechOpen; 2019. Aptamers for infectious disease diagnosis. [DOI] [Google Scholar]
- Bartosik M., Durikova H., Vojtesek B., Anton M., Jandakova E., Hrstka R. Electrochemical chip-based genomagnetic assay for detection of high-risk human papillomavirus DNA. Biosens. Bioelectron. 2016;83:300–305. doi: 10.1016/j.bios.2016.04.035. [DOI] [PubMed] [Google Scholar]
- Bhimji A., Zaragoza A.A., Live L.S., Kelley S.O. Electrochemical enzyme-linked immunosorbent assay featuring proximal reagent generation: detection of human immunodeficiency virus antibodies in clinical samples. Anal. Chem. 2013;85:6813–6819. doi: 10.1021/ac4009429. [DOI] [PubMed] [Google Scholar]
- Bosserdt M., Gajovic-Eichelman N., Scheller F.W. Modulation of direct electron transfer of cytochrome c by use of a molecularly imprinted thin film. Anal. Bioanal. Chem. 2013 doi: 10.1007/s00216-013-7009-8. [DOI] [PubMed] [Google Scholar]
- Braustein H.E., Braustein I.E. Real time diagnostic point of care by amperometric immuno-biosensor kit by flow technology. ECS Trans. 2014;58:1–17. doi: 10.1149/05840.0001ecst. [DOI] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral-Miranda G., Cardoso A.R., Ferreira L.C.S., Sales M.G.F., Bachmann M.F. Biosensor-based selective detection of Zika virus specific antibodies in infected individuals. Biosens. Bioelectron. 2018;113:101–107. doi: 10.1016/j.bios.2018.04.058. [DOI] [PubMed] [Google Scholar]
- Campos-Ferreira D.S., Nascimento G.A., Souza E.V.M., Souto-Maior M.A., Arruda M.S., Zanforlin D.M.L., Ekert M.H.F., Bruneska D., Lima-Filho J.L. Electrochemical DNA biosensor for human papillomavirus 16 detection in real samples. Anal. Chim. Acta. 2013;804:258–263. doi: 10.1016/j.aca.2013.10.038. [DOI] [PubMed] [Google Scholar]
- Campos-Ferreira D.S., Souza E.V.M., Nascimento G.A., Zanforlin D.M.L., Arruda M.S., Beltrão M.F.S., Melo A.L., Bruneska D., Lima-Filho J.L. Electrochemical DNA biosensor for the detection of human papillomavirus E6 gene inserted in recombinant plasmid. Arab. J. Chem. 2016;9:443–450. doi: 10.1016/j.arabjc.2014.05.023. [DOI] [Google Scholar]
- Campuzano S., Yáñez-Sedeño P., Pingarrón J.M. Electrochemical biosensing for the diagnosis of viral infections and tropical diseases. ChemElectroChem. 2017;4:753–777. doi: 10.1002/celc.201600805. [DOI] [Google Scholar]
- Cao P., Wang Z., Yan A.W.C., McVernon J., Xu J., Heffernan J.M., Kedzierska K., McCaw J.M. On the role of CD8+ T cells in determining recovery time from influenza virus infection. Front. Immunol. 2016;7 doi: 10.3389/fimmu.2016.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill R.L., Blair G.E., Millner P.A. A review on viral biosensors to detect human pathogens. Anal. Chim. Acta. 2010;681:8–15. doi: 10.1016/j.aca.2010.09.038. [DOI] [PubMed] [Google Scholar]
- Cecchetto J., Fernandes F.C.B., Lopes R., Bueno P.R. The capacitive sensing of NS1 Flavivirus biomarker. Biosens. Bioelectron. 2017;87:949–956. doi: 10.1016/j.bios.2016.08.097. [DOI] [PubMed] [Google Scholar]
- Channon R.B., Yang Y., Feibelman K.M., Geiss B.J., Dandy D.S., Henry C.S. Development of an electrochemical paper-based analytical device for trace detection of virus particles. Anal. Chem. 2018 doi: 10.1021/acs.analchem.8b02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan D.S., Prasad R., Srivastava R., Jaggi M., Chauhan S.C., Yallapu M.M. Comprehensive review on current interventions, diagnostic, and nanotechnology perspectives against SARS-CoV-2. Bioconjug. Chem. acs.bioconjchem. 2020 doi: 10.1021/acs.bioconjchem.0c00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekin F., Bagga K., Subramanian P., Jijie R., Singh S.K., Kurungot S., Boukherroub R., Szunerits S. Nucleic aptamer modified porous reduced graphene oxide/MoS2 based electrodes for viral detection: application to human papillomavirus (HPV) Sensor. Actuator. B Chem. 2018;262:991–1000. doi: 10.1016/j.snb.2018.02.065. [DOI] [Google Scholar]
- Chen H., Guo J., Wang C., Luo F., Yu X., Zhang W., Li J., Zhao D., Xu D., Gong Q., Liao J., Yang H., Hou W., Zhang Y. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020 doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Hong C.-Y.Y., Lin Y.-H.H., Chen J.-H.H., Chen G.-N.N., Yang H.-H.H. Enzyme-free and label-free ultrasensitive electrochemical detection of human immunodeficiency virus DNA in biological samples based on long-range self-assembled DNA nanostructures. Anal. Chem. 2012;84:8277–8283. doi: 10.1021/ac3017828. [DOI] [PubMed] [Google Scholar]
- Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y., Lin G. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- Cheng M.S., Ho J.S., Tan C.H., Wong J.P.S., Ng L.C., Toh C.-S. Development of an electrochemical membrane-based nanobiosensor for ultrasensitive detection of dengue virus. Anal. Chim. Acta. 2012;725:74–80. doi: 10.1016/j.aca.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Cheng M.S., Toh C.-S. Novel biosensing methodologies for ultrasensitive detection of viruses. Analyst. 2013;138:6219. doi: 10.1039/c3an01394d. [DOI] [PubMed] [Google Scholar]
- Cheng X., Chen G., Rodriguez W.R. Micro- and nanotechnology for viral detection. Anal. Bioanal. Chem. 2009;393:487–501. doi: 10.1007/s00216-008-2514-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikkaveeraiah B.V., Bhirde A.A., Morgan N.Y., Eden H.S., Chen X. Electrochemical immunosensors for detection of cancer protein biomarkers. ACS Nano. 2012;6:6546–6561. doi: 10.1021/nn3023969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K.H., Shin D.H., Oh J., An J.H., Lee J.S., Jang J. Multidimensional conductive nanofilm-based flexible aptasensor for ultrasensitive and selective HBsAg detection. ACS Appl. Mater. Interfaces. 2018;10:28412–28419. doi: 10.1021/acsami.8b09918. [DOI] [PubMed] [Google Scholar]
- Civit L., Fragoso A., Hölters S., Dürst M., O'Sullivan C.K. Electrochemical genosensor array for the simultaneous detection of multiple high-risk human papillomavirus sequences in clinical samples. Anal. Chim. Acta. 2012;715:93–98. doi: 10.1016/j.aca.2011.12.009. [DOI] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020 doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F., Zhou H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020;165:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F., Zhou Z., Zhou H.S. Molecularly imprinted polymers and surface imprinted polymers based electrochemical biosensor for infectious diseases. Sensors. 2020;20:996. doi: 10.3390/s20040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deka B., Kalita R., Bhatia D., Mishra A. Applications of paper as a support material in biomedical sciences: a decadal review. Sensors Int. 2020;1:100004. doi: 10.1016/j.sintl.2020.100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaconu I., Cristea C., Hârceagă V., Marrazza G., Berindan-Neagoe I., Săndulescu R. Electrochemical immunosensors in breast and ovarian cancer. Clin. Chim. Acta. 2013;425:128–138. doi: 10.1016/j.cca.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Diba F.S., Kim S., Lee H.J. Amperometric bioaffinity sensing platform for avian influenza virus proteins with aptamer modified gold nanoparticles on carbon chips. Biosens. Bioelectron. 2015;72:355–361. doi: 10.1016/j.bios.2015.05.020. [DOI] [PubMed] [Google Scholar]
- Diemer G.S., Stedman K.M. A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol. Direct. 2012;7:13. doi: 10.1186/1745-6150-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D.S. Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dincau B.M., Lee Y., Kim J.-H., Yeo W.-H. Recent advances in nanoparticle concentration and their application in viral detection using integrated sensors. Sensors. 2017;17:2316. doi: 10.3390/s17102316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolai S., Tabib-azar M. Whole virus detection using paper potentiometry and Aptamers. Preprints. 2020;1–8 doi: 10.20944/preprints202002.0291.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolai S., Tabib‐Azar M. Whole virus detection using aptamers and paper‐based sensor potentiometry. Med. Dev. Sens. 2020 doi: 10.1002/mds3.10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Zhao R., Zhu J., Lu X., Li Y., Qiu S., Jia L., Jiao X., Song S., Fan C., Hao R., Song H. Electrochemical DNA biosensor based on a tetrahedral nanostructure probe for the detection of avian influenza A (H7N9) virus. ACS Appl. Mater. Interfaces. 2015;7:8834–8842. doi: 10.1021/acsami.5b01438. [DOI] [PubMed] [Google Scholar]
- Draz M.S., Shafiee H. Applications of gold nanoparticles in virus detection. Theranostics. 2018;8:1985–2017. doi: 10.7150/thno.23856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draz M.S., Venkataramani M., Lakshminarayanan H., Saygili E., Moazeni M., Vasan A., Li Y., Sun X., Hua S., Yu X.G., Shafiee H. Nanoparticle-enhanced electrical detection of Zika virus on paper microchips. Nanoscale. 2018 doi: 10.1039/c8nr01646a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunajová A.A., Gál M., Tomčíková K., Sokolová R., Kolivoška V., Vaněčková E., Kielar F., Kostolanský F., Varečková E., Naumowicz M. Ultrasensitive impedimetric immunosensor for influenza A detection. J. Electroanal. Chem. 2020;858:113813. doi: 10.1016/j.jelechem.2019.113813. [DOI] [Google Scholar]
- Dziąbowska K., Czaczyk E., Nidzworski D. Detection methods of human and animal influenza virus—current trends. Biosensors. 2018;8:94. doi: 10.3390/bios8040094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egia-Mendikute L., Bosch A., Prieto-Fernandez E., Lee S.Y., Jimenez-Lasheras B., Garcia del Rio A., Antonana-Vildosola A., Bruzzone C., Bizkarguenaga M., Embade N., Abrescia N.G.A., Mato J.M., Millet O., Palazon A. Sensitive detection of SARS-CoV-2 seroconversion by flow cytometry reveals the presence of nucleoprotein-reactive antibodies in Covid-19-naive individuals. medRxiv 2020. 2020;28:20162941. doi: 10.1101/2020.07.28.20162941. 07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgrishi N., Rountree K.J., McCarthy B.D., Rountree E.S., Eisenhart T.T., Dempsey J.L. A practical beginner's guide to cyclic voltammetry. J. Chem. Educ. 2018;95:197–206. doi: 10.1021/acs.jchemed.7b00361. [DOI] [Google Scholar]
- Fabiani L., Saroglia M., Galatà G., De Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria A.M., Mazon T. Early diagnosis of Zika infection using a ZnO nanostructures-based rapid electrochemical biosensor. Talanta. 2019;203:153–160. doi: 10.1016/j.talanta.2019.04.080. [DOI] [PubMed] [Google Scholar]
- Faria H.A.M., Zucolotto V. Label-free electrochemical DNA biosensor for zika virus identification. Biosens. Bioelectron. 2019;131:149–155. doi: 10.1016/j.bios.2019.02.018. [DOI] [PubMed] [Google Scholar]
- Farkas K., Mannion F., Hillary L.S., Malham S.K., Walker D.I. Emerging technologies for the rapid detection of enteric viruses in the aquatic environment. Curr. Opin. Environ. Sci. Heal. 2020;16:1–6. doi: 10.1016/j.coesh.2020.01.007. [DOI] [Google Scholar]
- Farzin L., Shamsipur M., Samandari L., Sheibani S. HIV biosensors for early diagnosis of infection: the intertwine of nanotechnology with sensing strategies. Talanta. 2020;206:120201. doi: 10.1016/j.talanta.2019.120201. [DOI] [PubMed] [Google Scholar]
- Fatin M.F., Rahim Ruslinda A., Gopinath S.C.B., Arshad M.K.M. High-performance interactive analysis of split aptamer and HIV-1 Tat on multiwall carbon nanotube-modified field-effect transistor. Int. J. Biol. Macromol. 2019;125:414–422. doi: 10.1016/j.ijbiomac.2018.12.066. [DOI] [PubMed] [Google Scholar]
- Fauci A.S., Lane H.C., Redfield R.R. Covid-19 — navigating the uncharted. N. Engl. J. Med. 2020;382:1268–1269. doi: 10.1056/nejme2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesseha H., Tilahun H. Principles and applications of deoxyribonucleic acid microarray: a review. Pathol. Lab. Med. Open J. 2020;1:54–62. doi: 10.17140/PLMOJ-I-109. [DOI] [Google Scholar]
- Franco R., Pedrosa P., Carlos F.F., Veigas B., Baptista P.V. Handbook of Nanoparticles. Springer International Publishing; Cham: 2015. Gold nanoparticles for DNA/RNA-Based diagnostics; pp. 1–25. [DOI] [Google Scholar]
- Fu Y., Callaway Z., Lum J., Wang R., Lin J., Li Y. Exploiting enzyme catalysis in ultra-low ion strength media for impedance biosensing of avian influenza virus using a bare interdigitated electrode. Anal. Chem. 2014;86:1965–1971. doi: 10.1021/ac402550f. [DOI] [PubMed] [Google Scholar]
- Garcia L.F., Silvio Batista Rodrigues E., Rocha Lino de Souza G., Jubé Wastowski I., Mota de Oliveira F., Torres Pio dos Santos W., Souza Gil E. Impedimetric biosensor for bovine herpesvirus type 1‐antigen detection. Electroanal. elan. 2020:201900606. doi: 10.1002/elan.201900606. [DOI] [Google Scholar]
- Gattani A., Singh S.V., Agrawal A., Khan M.H., Singh P. Recent progress in electrochemical biosensors as point of care diagnostics in livestock health. Anal. Biochem. 2019;579:25–34. doi: 10.1016/j.ab.2019.05.014. [DOI] [PubMed] [Google Scholar]
- Gerasimov J.Y., Lai R.Y. An electrochemical peptide-based biosensing platform for HIV detection. Chem. Commun. 2010;46:395–397. doi: 10.1039/B919070H. [DOI] [PubMed] [Google Scholar]
- Ghanbari K., Roushani M. A nanohybrid probe based on double recognition of an aptamer MIP grafted onto a MWCNTs-Chit nanocomposite for sensing hepatitis C virus core antigen. Sensor. Actuator. B Chem. 2018;258:1066–1071. doi: 10.1016/j.snb.2017.11.145. [DOI] [Google Scholar]
- Ghanbari K., Roushani M., Azadbakht A. Ultra-sensitive aptasensor based on a GQD nanocomposite for detection of hepatitis C virus core antigen. Anal. Biochem. 2017;534:64–69. doi: 10.1016/j.ab.2017.07.016. [DOI] [PubMed] [Google Scholar]
- Glinsky G.V. Genomic analysis of pandemic (H1N1) 2009 reveals association of increasing disease severity with emergence of novel hemagglutinin mutations. Cell Cycle. 2010;9:958–970. doi: 10.4161/cc.9.5.10913. [DOI] [PubMed] [Google Scholar]
- Gong M.M., Sinton D. Turning the page: advancing paper-based microfluidics for broad diagnostic application. Chem. Rev. 2017;117:8447–8480. doi: 10.1021/acs.chemrev.7b00024. [DOI] [PubMed] [Google Scholar]
- González-López A., Fernández Abedul M.T. Elsevier Inc; 2020. Genosensor on Gold Films with Enzymatic Electrochemical Detection of a SARS Virus Sequence, Laboratory Methods in Dynamic Electroanalysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnough B.A., Thomas K., Kaplan S. 2020. Testing Falls Woefully Short as Trump Seeks an End to Stay-At-Home Orders 1–4. [Google Scholar]
- Goud K.Y., Hayat A., Catanante G., Gobi K.V., Marty J.L. An electrochemical aptasensor based on functionalized graphene oxide assisted electrocatalytic signal amplification of methylene blue for aflatoxin B1 detection. Electrochim. Acta. 2017;244:96–103. doi: 10.1016/j.electacta.2017.05.089. M., S. [DOI] [Google Scholar]
- Goud K.Y., Kailasa S.K., Kumar V., Tsang Y.F., Lee S.E.E., Gobi K.V., Kim K.-H.H., Kalisa S.K., Kumar V., Tsang Y.F., Lee S.E.E., Gobi K.V., Kim K.-H.H. Progress on nanostructured electrochemical sensors and their recognition elements for detection of mycotoxins: a review. Biosens. Bioelectron. 2018;121:205–222. doi: 10.1016/j.bios.2018.08.029. [DOI] [PubMed] [Google Scholar]
- Goud K.Y., M S., Reddy K.K., Gobi K.V. Development of highly selective electrochemical impedance sensor for detection of sub-micromolar concentrations of 5-Chloro-2,4-dinitrotoluene. J. Chem. Sci. 2016;128:763–770. doi: 10.1007/s12039-016-1078-0. [DOI] [Google Scholar]
- Goud K. Yugender, Moonla C., Mishra R.K., Yu C., Narayan R., Litvan I., Wang J. Wearable electrochemical microneedle sensor for continuous monitoring of levodopa: toward Parkinson management. ACS Sens. 2019;4:2196–2204. doi: 10.1021/acssensors.9b01127. [DOI] [PubMed] [Google Scholar]
- Goud Kotagiri Yugender, Satyanarayana M., Hayat A., Gobi K.V., Marty J.L. Nanoparticles in Pharmacotherapy. Elsevier; 2019. Nanomaterial-based electrochemical sensors in pharmaceutical applications; pp. 195–216. [DOI] [Google Scholar]
- Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Mil. Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Chen J., Chen G. A label-free electrochemical biosensor for detection of HIV related gene based on interaction between DNA and protein. Sensor. Actuator. B Chem. 2013;184:113–117. doi: 10.1016/j.snb.2013.04.046. [DOI] [Google Scholar]
- Halfpenny K.C., Wright D.W. Nanoparticle detection of respiratory infection. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010;2:277–290. doi: 10.1002/wnan.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass K.N., Bao M., He Q., Park M., Qin P., Du K. bioRxiv 2020; 2020. Integrated Micropillar Polydimethylsiloxane Accurate CRISPR Detection (IMPACT) System for Rapid Viral DNA Sensing. 03.17.994137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogestyn J., Mock D., Mayer-Proschel M. Contributions of neurotropic human herpesviruses herpes simplex virus 1 and human herpesvirus 6 to neurodegenerative disease pathology. Neural Regen. Res. 2018;13:211. doi: 10.4103/1673-5374.226380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P., Li W., Li J. Applications of aptasensors in clinical diagnostics. Sensors. 2012;12:1181–1193. doi: 10.3390/s120201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.-L., Sun A., Zhao Y., Aronoff-Spencer E., Hall D.A. 2018 IEEE Custom Integrated Circuits Conference (CICC) IEEE; 2018. A 16×20 electrochemical CMOS biosensor array with in-pixel averaging using polar modulation; pp. 1–4. [DOI] [Google Scholar]
- Huang H., Bai W., Dong C., Guo R., Liu Z. An ultrasensitive electrochemical DNA biosensor based on graphene/Au nanorod/polythionine for human papillomavirus DNA detection. Biosens. Bioelectron. 2015;68:442–446. doi: 10.1016/j.bios.2015.01.039. [DOI] [PubMed] [Google Scholar]
- Huang K.-J., Li J., Liu Y.-M., Cao X., Yu S., Yu M. Disposable immunoassay for hepatitis B surface antigen based on a graphene paste electrode functionalized with gold nanoparticles and a Nafion-cysteine conjugate. Microchim. Acta. 2012;177:419–426. doi: 10.1007/s00604-012-0805-6. [DOI] [Google Scholar]
- Hussein H.A., Hassan R.Y.A., Chino M., Febbraio F. Point-of-care diagnostics of covid-19: from current work to future perspectives. Sensors. 2020 doi: 10.3390/s20154289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huy T.Q., Hanh N.T.H., Thuy N.T., Chung P. Van, Nga P.T., Tuan M.A. A novel biosensor based on serum antibody immobilization for rapid detection of viral antigens. Talanta. 2011;86:271–277. doi: 10.1016/j.talanta.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfan U. 2020. The case for ending the Covid-19 pandemic with mass testing; pp. 1–15. Vox. [Google Scholar]
- Ishikawa F.N., Chang H.-K., Curreli M., Liao H.-I., Olson C.A., Chen P.-C., Zhang R., Roberts R.W., Sun R., Cote R.J., Thompson M.E., Zhou C. Label-free, electrical detection of the SARS virus N-protein with nanowire biosensors utilizing antibody mimics as capture probes. ACS Nano. 2009;3:1219–1224. doi: 10.1021/nn900086c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jampasa S., Siangproh W., Laocharoensuk R., Yanatatsaneejit P., Vilaivan T., Chailapakul O. A new DNA sensor design for the simultaneous detection of HPV type 16 and 18 DNA. Sensor. Actuator. B Chem. 2018;265:514–521. doi: 10.1016/j.snb.2018.03.045. [DOI] [Google Scholar]
- Jampasa S., Wonsawat W., Rodthongkum N., Siangproh W., Yanatatsaneejit P., Vilaivan T., Chailapakul O. Electrochemical detection of human papillomavirus DNA type 16 using a pyrrolidinyl peptide nucleic acid probe immobilized on screen-printed carbon electrodes. Biosens. Bioelectron. 2014;54:428–434. doi: 10.1016/j.bios.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Jarczewska M., Górski Ł., Malinowska E. Electrochemical aptamer-based biosensors as potential tools for clinical diagnostics. Anal. Methods. 2016;8:3861–3877. doi: 10.1039/C6AY00499G. [DOI] [Google Scholar]
- Jayant R., Yndart A., Sagar V., Bhansali S., Nair M., Kaushik A., Atluri V. Electrochemical sensing method for point-of-care cortisol detection in human immunodeficiency virus-infected patients. Int. J. Nanomed. 2015;677 doi: 10.2147/IJN.S75514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji T., Liu Z., Wang G.Q., Guo X., Akbar khan S., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q. Detection of COVID-19: a review of the current literature and future perspectives. Biosens. Bioelectron. 2020 doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Wang G., Hein R., Liu N., Luo X., Davis J.J. Antifouling strategies for selective in vitro and in vivo sensing. Chem. Rev. 2020 doi: 10.1021/acs.chemrev.9b00739. [DOI] [PubMed] [Google Scholar]
- Jin Y., Yang H., Ji W., Wu W., Chen S., Zhang W., Duan G. Virology, epidemiology, pathogenesis, and control of covid-19. Viruses. 2020 doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S.V., Rotherham L.S., Khati M., Mamba B.B., Arotiba O.A. Towards HIV detection: novel poly(propylene imine) dendrimer-streptavidin platform for electrochemical DNA and gp120 aptamer biosensors. Int. J. Electrochem. Sci. 2014;9:5425–5437. hdl.handle.net/10204/7644. [Google Scholar]
- Karimizefreh A., Sasanpour P., Jokar E., Mohammadpour R., Vaezjalali M., Tekieh T. 2014 IEEE 34th International Scientific Conference on Electronics and Nanotechnology (ELNANO) IEEE; 2014. Human Papilloma Virus biosensor based on electrochemical impedance spectroscopy of DNA hybridization; pp. 368–370. [DOI] [Google Scholar]
- Kaushik A., Tiwari S., Jayant R.D., Vashist A., Nikkhah-Moshaie R., El-Hage N., Nair M. Electrochemical biosensors for early stage zika diagnostics. Trends Biotechnol. 2017;35:308–317. doi: 10.1016/j.tibtech.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik A., Yndart A., Kumar S., Jayant R.D., Vashist A., Brown A.N., Li C.-Z., Nair M. A sensitive electrochemical immunosensor for label-free detection of Zika-virus protein. Sci. Rep. 2018;8:9700. doi: 10.1038/s41598-018-28035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermali M., Khalsa R.K., Pillai K., Ismail Z., Harky A. The role of biomarkers in diagnosis of COVID-19 – a systematic review. Life Sci. 2020;254:117788. doi: 10.1016/j.lfs.2020.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.Z.H., Hasan M.R., Hossain S.I., Ahommed M.S., Daizy M. Ultrasensitive detection of pathogenic viruses with electrochemical biosensor: state of the art. Biosens. Bioelectron. 2020;166:112431. doi: 10.1016/j.bios.2020.112431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khristunova E., Dorozhko E., Korotkova E., Kratochvil B., Vyskocil V., Barek J. Label-free electrochemical biosensors for the determination of flaviviruses: dengue, zika, and Japanese encephalitis. Sensors. 2020;20:4600. doi: 10.3390/s20164600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Jeerapan I., Sempionatto J.R., Barfidokht A., Mishra R.K., Campbell A.S., Hubble L.J., Wang J. Wearable bioelectronics: enzyme-based body-worn electronic devices. Acc. Chem. Res. 2018;51:2820–2828. doi: 10.1021/acs.accounts.8b00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizek R., Krejcova L., Michalek P., Merlos Rodrigo M., Heger Z., Krizkova S., Vaculovicova M., Hynek D., Adam V. Nanoscale virus biosensors: state of the art. Nanobiosensors Dis. Diagnosis. 2015;47 doi: 10.2147/NDD.S56771. [DOI] [Google Scholar]
- Kokkinos C., Economou A., Prodromidis M.I. Electrochemical immunosensors: critical survey of different architectures and transduction strategies. TrAC Trends Anal. Chem. (Reference Ed.) 2016;79:88–105. doi: 10.1016/j.trac.2015.11.020. [DOI] [Google Scholar]
- Kong Y., Li J., Wu S., Cheng W., Rana R.K., Zhu J.-J. Functionalization of poly(o-phenylenediamine) with gold nanoparticles as a label-free immunoassay platform for the detection of human enterovirus 71. Sensor. Actuator. B Chem. 2013;183:187–193. doi: 10.1016/j.snb.2013.03.139. [DOI] [Google Scholar]
- Kostyusheva A., Brezgin S., Babin Y., Vasil I., Kostyushev D. Preprint.org; 2020. CRISPR-cas Systems for Diagnosing Infectious Diseases; pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotru S., Klimuntowski M., Ridha H., Uddin Z., Askhar A.A., Singh G., Howlader M.M.R. Electrochemical sensing: a prognostic tool in the fight against COVID-19. TrAC Trends Anal. Chem. (Reference Ed.) 2021;116198 doi: 10.1016/j.trac.2021.116198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejcova L., Hynek D., Michalek P., Milosavljevic V., Kopel P., Zitka O., Konecna M., Kynicky J., Adam V., Hubalek J., Kizek R. Electrochemical sensors and biosensors for influenza detection - literature survey 2012-2013. Int. J. Electrochem. Sci. 2014;9:3440–3448. [Google Scholar]
- Kudr J., Michalek P., Ilieva L., Adam V., Zitka O. COVID-19: a challenge for electrochemical biosensors. TrAC Trends Anal. Chem. (Reference Ed.) 2021:116192. doi: 10.1016/j.trac.2021.116192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib M., Berezovski M.V. Advances in Biochemical Engineering/Biotechnology. 2013. Electrochemical aptasensors for microbial and viral pathogens; pp. 155–181. [DOI] [PubMed] [Google Scholar]
- Labib M., Zamay A.S., Kolovskaya O.S., Reshetneva I.T., Zamay G.S., Kibbee R.J., Sattar S.A., Zamay T.N., Berezovski M.V. Aptamer-based viability impedimetric sensor for bacteria. Anal. Chem. 2012;84:8966–8969. doi: 10.1021/ac302902s. [DOI] [PubMed] [Google Scholar]
- Labib M., Zamay A.S., Muharemagic D., Chechik A., Bell J.C., Berezovski M.V. Electrochemical sensing of aptamer-facilitated virus immunoshielding. Anal. Chem. 2012;84:1677–1686. doi: 10.1021/ac202978r. [DOI] [PubMed] [Google Scholar]
- Layqah L.A., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta. 2019;186:224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Oh B.-K., Choi J.-W. Electrochemical sensor based on direct electron transfer of HIV-1 Virus at Au nanoparticle modified ITO electrode. Biosens. Bioelectron. 2013;49:531–535. doi: 10.1016/j.bios.2013.06.010. [DOI] [PubMed] [Google Scholar]
- Li H., Liu S.M., Yu X.H., Tang S.L., Tang C.K. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Jin R., Peng Y., Wang C., Ren W., Lv F., Gong S., Fang F., Wang Q., Li J., Shen T., Sun H., Zhou L., Cui Y., Song H., Sun L. 2020. Generation of Antibodies against COVID-19 Virus for Development of Diagnostic Tools. medRxiv 2020.02.20.20025999. [DOI] [Google Scholar]
- Li X., Qin Z., Fu H., Li T., Peng R., Li Z., Rini J.M., Liu X. Enhancing performance of paper-based electrochemical impedance spectroscopy nanobiosensors: an experimental approach. Biosens. Bioelectron. 2020;112672 doi: 10.1016/j.bios.2020.112672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Scida K., Crooks R.M. Detection of hepatitis B virus DNA with a paper electrochemical sensor. Anal. Chem. 2015;87:9009–9015. doi: 10.1021/acs.analchem.5b02210. [DOI] [PubMed] [Google Scholar]
- Lifson M.A., Ozen M.O., Inci F., Wang S., Inan H., Baday M., Henrich T.J., Demirci U. Advances in biosensing strategies for HIV-1 detection, diagnosis, and therapeutic monitoring. Adv. Drug Deliv. Rev. 2016;103:90–104. doi: 10.1016/j.addr.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Choi K.S., Park T.J., Lee S.Y., Seo T.S. Graphene-based electrochemical biosensor for pathogenic virus detection. BioChip J. 2011;5:123–128. doi: 10.1007/s13206-011-5204-2. [DOI] [Google Scholar]
- Liu F., Kim Y.H., Cheon D.S., Seo T.S. Micropatterned reduced graphene oxide based field-effect transistor for real-time virus detection. Sensor. Actuator. B Chem. 2013;186:252–257. doi: 10.1016/j.snb.2013.05.097. [DOI] [Google Scholar]
- Liu L., Moore M.D. A survey of analytical techniques for Noroviruses. Foods. 2020;9:318. doi: 10.3390/foods9030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Wu P., Li W., Zhang H., Cai C. Ultrasensitive and selective electrochemical identification of hepatitis C virus genotype 1b based on specific endonuclease combined with gold nanoparticles signal amplification. Anal. Chem. 2011;83:4752–4758. doi: 10.1021/ac200624f. [DOI] [PubMed] [Google Scholar]
- Liu X., Cheng Z., Fan H., Ai S., Han R. Electrochemical detection of avian influenza virus H5N1 gene sequence using a DNA aptamer immobilized onto a hybrid nanomaterial-modified electrode. Electrochim. Acta. 2011;56:6266–6270. doi: 10.1016/j.electacta.2011.05.055. [DOI] [Google Scholar]
- Macchia E., Sarcina L., Picca R.A., Manoli K., Di Franco C., Scamarcio G., Torsi L. Ultra-low HIV-1 p24 detection limits with a bioelectronic sensor. Anal. Bioanal. Chem. 2020;412:811–818. doi: 10.1007/s00216-019-02319-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahari S., Roberts A., Shahdeo D., Gandhi S. 2020. eCovSens-Ultrasensitive Novel In-House Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of nCovid-19 Antigen, a Spike Protein Domain 1 of SARS-CoV-2. [DOI] [Google Scholar]
- Mahato K., Kumar A., Maurya P.K., Chandra P. Shifting paradigm of cancer diagnoses in clinically relevant samples based on miniaturized electrochemical nanobiosensors and microfluidic devices. Biosens. Bioelectron. 2018;100:411–428. doi: 10.1016/j.bios.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Mahato K., Srivastava A., Chandra P. Paper based diagnostics for personalized health care: emerging technologies and commercial aspects. Biosens. Bioelectron. 2017 doi: 10.1016/j.bios.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Mandli J., Attar A., Ennaji M.M., Amine A. Indirect competitive electrochemical immunosensor for hepatitis A virus antigen detection. J. Electroanal. Chem. 2017;799:213–221. doi: 10.1016/j.jelechem.2017.05.047. [DOI] [Google Scholar]
- Manzano M., Viezzi S., Mazerat S., Marks R.S., Vidic J. Rapid and label-free electrochemical DNA biosensor for detecting hepatitis A virus. Biosens. Bioelectron. 2018;100:89–95. doi: 10.1016/j.bios.2017.08.043. [DOI] [PubMed] [Google Scholar]
- Mao C., Liu A., Cao B. Virus-based chemical and biological sensing. Angew. Chem. Int. Ed. 2009;48:6790–6810. doi: 10.1002/anie.200900231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martial J., Dussart P., Plumelle Y., Moravie V., Verlaeten O., Najioullah F., Cabié A., Fonteau C., Césaire R., Kaidomar S., Thomas L. Influence of the dengue serotype, previous dengue infection, and plasma viral load on clinical presentation and outcome during a dengue-2 and dengue-4 Co-epidemic. Am. J. Trop. Med. Hyg. 2008;78:990–998. doi: 10.4269/ajtmh.2008.78.990. [DOI] [PubMed] [Google Scholar]
- Martínez-Paredes G., González-García M.B., Costa-García A. Genosensor for SARS virus detection based on gold nanostructured screen-printed carbon electrodes. Electroanalysis. 2009;21:379–385. doi: 10.1002/elan.200804399. [DOI] [Google Scholar]
- Masters P.S. Advances in Virus Research. 2006. The molecular biology of coronaviruses; pp. 193–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer S.V., Tesh R.B., Vasilakis N. The emergence of arthropod-borne viral diseases: a global prospective on dengue, chikungunya and zika fevers. Acta Trop. 2017;166:155–163. doi: 10.1016/j.actatropica.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miripour Z.S., Sarrami-Forooshani R., Sanati H., Makarem J., Taheri M.S., Shojaeian F., Eskafi A.H., Abbasvandi F., Namdar N., Ghafari H., Aghaee P., Zandi A., Faramarzpour M., Hoseinyazdi M., Tayebi M., Abdolahad M. Real-time diagnosis of reactive oxygen species (ROS) in fresh sputum by electrochemical tracing; correlation between COVID-19 and viral-induced ROS in lung/respiratory epithelium during this pandemic. Biosens. Bioelectron. 2020;165:112435. doi: 10.1016/j.bios.2020.112435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moço A.C.R., Guedes P.H., Flauzino J.M.R., da Silva H.S., Vieira J.G., Castro A.C.H., Gomes É.V.R., Tolentino F.M., Soares M.M.C.N., Madurro J.M., Brito‐Madurro A.G. Electrochemical detection of zika virus in biological samples: a step for diagnosis point‐of‐care. Electroanalysis. 2019;31:1580–1587. doi: 10.1002/elan.201900068. [DOI] [Google Scholar]
- Mokhtarzadeh A., Eivazzadeh-Keihan R., Pashazadeh P., Hejazi M., Gharaatifar N., Hasanzadeh M., Baradaran B., de la Guardia M. Nanomaterial-based biosensors for detection of pathogenic virus. TrAC Trends Anal. Chem. (Reference Ed.) 2017;97:445–457. doi: 10.1016/j.trac.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco C.L., Gootenberg D.B., Zhao G., Handley S.A., Ghebremichael M.S., Lim E.S., Lankowski A., Baldridge M.T., Wilen C.B., Flagg M., Norman J.M., Keller B.C., Luévano J.M., Wang D., Boum Y., Martin J.N., Hunt P.W., Bangsberg D.R., Siedner M.J., Kwon D.S., Virgin H.W. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe. 2016;19:311–322. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Narváez E., Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19. Biosens. Bioelectron. 2020;163:112274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulick A., Richtera L., Milosavljevic V., Cernei N., Haddad Y., Zitka O., Kopel P., Heger Z., Adam V. Advanced nanotechnologies in avian influenza: current status and future trends – a review. Anal. Chim. Acta. 2017;983:42–53. doi: 10.1016/j.aca.2017.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., de Wit E., van den Brand J.M.A., Herfst S., Schrauwen E.J.A., Bestebroer T.M., van de Vijver D., Boucher C.A., Koopmans M., Rimmelzwaan G.F., Kuiken T., Osterhaus A.D.M.E., Fouchier R.A.M. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science. 2009;325:481–483. doi: 10.1126/science.1177127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyembe-Tamfum J.J., Mulangu S., Masumu J., Kayembe J.M., Kemp A., Paweska J.T. Ebola virus outbreaks in Africa: past and present. Onderstepoort J. Vet. Res. 2012;79 doi: 10.4102/ojvr.v79i2.451. [DOI] [PubMed] [Google Scholar]
- Narang J., Singhal C., Mathur A., Sharma S., Singla V., Pundir C.S. Portable bioactive paper based genosensor incorporated with Zn-Ag nanoblooms for herpes detection at the point-of-care. Int. J. Biol. Macromol. 2018;107:2559–2565. doi: 10.1016/j.ijbiomac.2017.10.146. [DOI] [PubMed] [Google Scholar]
- Narita F., Wang Z., Kurita H., Li Z., Shi Y., Jia Y., Soutis C. A review of piezoelectric and magnetostrictive biosensor materials for detection of COVID‐19 and other viruses. Adv. Mater. 2020;2005448:2005448. doi: 10.1002/adma.202005448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrollahzadeh M., Sajjadi M., Soufi G.J., Iravani S., Varma R.S. Nanomaterials and nanotechnology-associated innovations against viral infections with a focus on coronaviruses. Nanomaterials. 2020;10 doi: 10.3390/nano10061072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navakul K., Warakulwit C., Yenchitsomanus P. thai, Panya A., Lieberzeit P.A., Sangma C. A novel method for dengue virus detection and antibody screening using a graphene-polymer based electrochemical biosensor. Nanomed. Nanotechnol. Biol. Med. 2017;13:549–557. doi: 10.1016/j.nano.2016.08.009. [DOI] [PubMed] [Google Scholar]
- Nawaz H., Tahir M., Anwar S., Majeed M.I., Rashid N. Nanobiosensors. Wiley; 2020. Detection of HIV virus using biosensor; pp. 149–169. [DOI] [Google Scholar]
- Nawaz M.H., Hayat A., Catanante G., Latif U., Marty J.L. Development of a portable and disposable NS1 based electrochemical immunosensor for early diagnosis of dengue virus. Anal. Chim. Acta. 2018;1026:1–7. doi: 10.1016/j.aca.2018.04.032. [DOI] [PubMed] [Google Scholar]
- Niroula J., Premaratne G., Ali Shojaee S., Lucca D.A., Krishnan S. Combined covalent and noncovalent carboxylation of carbon nanotubes for sensitivity enhancement of clinical immunosensors. Chem. Commun. 2016;52 doi: 10.1039/c6cc07022a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orooji Y., Sohrabi H., Hemmat N., Oroojalian F., Baradaran B., Mokhtarzadeh A., Mohaghegh M., Karimi-Maleh H. Nano-Micro Letters. Springer Singapore; 2021. An Overview on SARS-CoV-2 (COVID-19) and Other Human Coronaviruses and Their Detection Capability via Amplification Assay, Chemical Sensing, Biosensing, Immunosensing, and Clinical Assays. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer T., Geiss B.J., Henry C.S. Review—chemical and biological sensors for viral detection. J. Electrochem. Soc. 2020;167 doi: 10.1149/2.0232003JES. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X. Evaluation of control measures implemented in the Severe acute respiratory syndrome outbreak in beijing, 2003. J. Am. Med. Assoc. 2003;290:3215. doi: 10.1001/jama.290.24.3215. [DOI] [PubMed] [Google Scholar]
- Parolo C., Greenwood A.S., Ogden N.E., Kang D., Hawes C., Ortega G., Arroyo-Currás N., Plaxco K.W. E-DNA scaffold sensors and the reagentless, single-step, measurement of HIV-diagnostic antibodies in human serum. Microsyst. Nanoeng. 2020;6:13. doi: 10.1038/s41378-019-0119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolo C., Sena-Torralba A., Bergua J.F., Calucho E., Fuentes-Chust C., Hu L., Rivas L., Álvarez-Diduk R., Nguyen E.P., Cinti S., Quesada-González D., Merkoçi A. Tutorial: design and fabrication of nanoparticle-based lateral-flow immunoassays. Nat. Protoc. 2020 doi: 10.1038/s41596-020-0357-x. [DOI] [PubMed] [Google Scholar]
- Pashchenko O., Shelby T., Banerjee T., Santra S. A comparison of optical, electrochemical, magnetic, and colorimetric point-of-care biosensors for infectious disease diagnosis. ACS Infect. Dis. 2018;4:1162–1178. doi: 10.1021/acsinfecdis.8b00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Yarman A., Jetzschmann K.J., Jeoung J.H., Schad D., Dobbek H., Wollenberger U., Scheller F.W. Molecularly imprinted electropolymer for a hexameric heme protein with direct electron transfer and peroxide electrocatalysis. 2016. Sensors (Switzerland) [DOI] [PMC free article] [PubMed]
- Piro B., Kapella A., Le V.H., Anquetin G., Zhang Q.D., Reisberg S., Noel V., Tran L.D., Duc H.T., Pham M.C. Towards the detection of human papillomavirus infection by a reagentless electrochemical peptide biosensor. Electrochim. Acta. 2011;56:10688–10693. doi: 10.1016/j.electacta.2011.04.094. [DOI] [Google Scholar]
- Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab Sci. 2020;8363:1–11. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premaratne G., Farias S., Krishnan S. Pyrenyl carbon nanostructures for ultrasensitive measurements of formaldehyde in urine. Anal. Chim. Acta. 2017;970:23–29. doi: 10.1016/j.aca.2017.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premaratne G., Niroula J., Patel M.K., Zhong W., Suib S.L., Kaan Kalkan A., Krishnan S. Electrochemical and surface-plasmon correlation of a serum-autoantibody immunoassay with binding insights: graphenyl surface versus mercapto-monolayer surface. Anal. Chem. 2018;90:12456–12463. doi: 10.1021/acs.analchem.8b01565. [DOI] [PubMed] [Google Scholar]
- Qian X., Tan S., Li Z., Qu Q., Li L., Yang L. A robust host-guest interaction controlled probe immobilization strategy for the ultrasensitive detection of HBV DNA using hollow HP5–Au/CoS nanobox as biosensing platform. Biosens. Bioelectron. 2020;153:112051. doi: 10.1016/j.bios.2020.112051. [DOI] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-Functional plasmonic photothermal biosensors for highly accurate Severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Radhi M.S., Ruslinda A.R., Fatin M.F., Hashwan S.S.B., Md Arshad M.K., Hashim U. 2016 IEEE International Conference on Semiconductor Electronics (ICSE) IEEE; 2016. HIV-1 Tat peptide detection by using RNA aptamer on MWCNT modified electrode; pp. 204–207. [DOI] [Google Scholar]
- Rahim Ruslinda A., Tanabe K., Ibori S., Wang X., Kawarada H. Effects of diamond-FET-based RNA aptamer sensing for detection of real sample of HIV-1 Tat protein. Biosens. Bioelectron. 2013;40:277–282. doi: 10.1016/j.bios.2012.07.048. [DOI] [PubMed] [Google Scholar]
- Rai V., Hapuarachchi H.C., Ng L.C., Soh S.H., Leo Y.S., Toh C.-S. Ultrasensitive cDNA detection of dengue virus RNA using electrochemical nanoporous membrane-based biosensor. PloS One. 2012;7 doi: 10.1371/journal.pone.0042346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan P., Singhal A., Yadav S., Kumar N., Murali S., Sanghi S.K., Khan R. Rapid diagnosis of SARS-CoV-2 using potential point-of-care electrochemical immunosensor: toward the future prospects. Int. Rev. Immunol. 2021 doi: 10.1080/08830185.2021.1872566. [DOI] [PubMed] [Google Scholar]
- Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N., Valdez J., Flynn J., Williams S.J. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021;171:112709. doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N., Valdez J., Flynn J., Williams S.J. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector A R T I C L E I N F O. medRxiv. 2020;5727 doi: 10.1101/2020.08.10.20171652. 2020.08.10.20171652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasouli E., Shahnavaz Z., Basirun W.J., Rezayi M., Avan A., Ghayour-Mobarhan M., Khandanlou R., Johan M.R. Advancements in electrochemical DNA sensor for detection of human papilloma virus - a review. Anal. Biochem. 2018;556:136–144. doi: 10.1016/j.ab.2018.07.002. [DOI] [PubMed] [Google Scholar]
- Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020;165:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K.K., Bandal H., Satyanarayana M., Goud K.Y., Gobi K.V., Jayaramudu T., Amalraj J., Kim H. Recent trends in electrochemical sensors for vital biomedical markers using hybrid nanostructured materials. Adv. Sci. 2020;7:1902980. doi: 10.1002/advs.201902980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B.E.M., Haagmans B.L., Müller M.A., Gutierrez C., Godeke G.-J., Meyer B., Muth D., Raj V.S., Vries L.S.-D., Corman V.M., Drexler J.-F., Smits S.L., El Tahir Y.E., De Sousa R., van Beek J., Nowotny N., van Maanen K., Hidalgo-Hermoso E., Bosch B.-J., Rottier P., Osterhaus A., Gortázar-Schmidt C., Drosten C., Koopmans M.P.G. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhouati A., Catanante G., Nunes G., Hayat A., Marty J.L. Label-free aptasensors for the detection of mycotoxins. Sensors. 2016;16:1–21. doi: 10.3390/s16122178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzi-Lehnert M. Development of chip-compatible sample preparation for diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2012;12:189–206. doi: 10.1586/erm.11.98. [DOI] [PubMed] [Google Scholar]
- Rodrigo M.A.M., Heger Z., Cernei N., Jinemez A.M.J., Zitka O., Adam V., Kizek R. HIV biosensors - the potential of the electrochemical way. Int. J. Electrochem. Sci. 2014;9:3449–3457. [Google Scholar]
- Satija N., Lal S.K. The molecular biology of SARS coronavirus. Ann. N. Y. Acad. Sci. 2007;1102:26–38. doi: 10.1196/annals.1408.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayana M., Goud K.Y., Reddy K.K., Kumar V.S., Gobi K.V. Silver nanoparticles impregnated chitosan layered carbon nanotube as sensor interface for electrochemical detection of clopidogrel in-vitro. Mater. Sci. Eng. C. 2019;101:103–110. doi: 10.1016/j.msec.2019.03.083. [DOI] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S. Il. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Sepunaru L., Plowman B.J., Sokolov S.V., Young N.P., Compton R.G. Rapid electrochemical detection of single influenza viruses tagged with silver nanoparticles. Chem. Sci. 2016;7:3892–3899. doi: 10.1039/C6SC00412A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. J. Am. Med. Assoc. 2020;323:2249. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- Shah J., Wilkins E. Electrochemical biosensors for detection of biological warfare agents. Electroanalysis. 2003;15:157–167. doi: 10.1002/elan.200390019. [DOI] [Google Scholar]
- Sharma P.S., Garcia-Cruz A., Cieplak M., Noworyta K.R., Kutner W. ‘Gate effect’ in molecularly imprinted polymers: the current state of understanding. Curr. Opin. Electrochem. 2019 doi: 10.1016/j.coelec.2019.04.020. [DOI] [Google Scholar]
- Sharp P.M., Hahn B.H. The evolution of HIV-1 and the origin of AIDS. Philos. Trans. R. Soc. B Biol. Sci. 2010;365:2487–2494. doi: 10.1098/rstb.2010.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020 doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin E.-C., Sung P.S., Park S.-H. Immune responses and immunopathology in acute and chronic viral hepatitis. Nat. Rev. Immunol. 2016;16:509–523. doi: 10.1038/nri.2016.69. [DOI] [PubMed] [Google Scholar]
- Shin M., Yoon J., Yi C., Lee T., Choi J.W. Flexible HIV-1 biosensor based on the au/MoS2 nanoparticles/au nanolayer on the PET substrate. Nanomaterials. 2019;9:1076. doi: 10.3390/nano9081076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão E.P., Silva D.B.S., Cordeiro M.T., Gil L.H.V., Andrade C.A.S., Oliveira M.D.L. Nanostructured impedimetric lectin-based biosensor for arboviruses detection. Talanta. 2020;208:120338. doi: 10.1016/j.talanta.2019.120338. [DOI] [PubMed] [Google Scholar]
- Sin M.L., Mach K.E., Wong P.K., Liao J.C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2014;14:225–244. doi: 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V., Krishnan S. An electrochemical mass sensor for diagnosing diabetes in human serum. Analyst. 2014;139:724. doi: 10.1039/c3an01542d. [DOI] [PubMed] [Google Scholar]
- Singhal C., Dubey A., Mathur A., Pundir C.S., Narang J. Paper based DNA biosensor for detection of chikungunya virus using gold shells coated magnetic nanocubes. Process Biochem. 2018;74:35–42. doi: 10.1016/j.procbio.2018.08.020. [DOI] [Google Scholar]
- Singhal C., Khanuja M., Chaudhary N., Pundir C.S., Narang J. Detection of chikungunya virus DNA using two-dimensional MoS2 nanosheets based disposable biosensor. Sci. Rep. 2018;8:7734. doi: 10.1038/s41598-018-25824-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Moon H.-J., An D.-J., Jeoung H.-Y., Kim H., Yeom M.-J., Hong M., Nam J.-H., Park S.-J., Park B.-K., Oh J.-S., Song M., Webster R.G., Kim J.-K., Kang B.-K. A novel reassortant canine H3N1 influenza virus between pandemic H1N1 and canine H3N2 influenza viruses in Korea. J. Gen. Virol. 2012;93:551–554. doi: 10.1099/vir.0.037739-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza E.V., Nascimento G.A., Santana N.A. de, Campos-Ferreira D.S., Bibiano J. de A., Arruda M.S., Bruneska D., Lima-Filho J.L. Electrochemical DNA biosensor for sequences related to the human papillomavirus type 16 using methylene blue. Biosens. J. 2014 doi: 10.4172/2090-4967.1000107. 03. [DOI] [Google Scholar]
- Spinelli A., Pellino G. COVID-19 pandemic: perspectives on an unfolding crisis. Br. J. Surg. 2020;107:785–787. doi: 10.1002/bjs.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisomwat C., Teengam P., Chuaypen N., Tangkijvanich P., Vilaivan T., Chailapakul O. Pop-up paper electrochemical device for label-free hepatitis B virus DNA detection. Sensor. Actuator. B Chem. 2020 doi: 10.1016/j.snb.2020.128077. [DOI] [Google Scholar]
- Srisomwat C., Yakoh A., Chuaypen N., Tangkijvanich P., Vilaivan T., Chailapakul O. Amplification-free DNA sensor for the one-step detection of the hepatitis B virus using an automated paper-based lateral flow electrochemical device. Anal. Chem. 2021 doi: 10.1021/acs.analchem.0c04283. [DOI] [PubMed] [Google Scholar]
- Srivastava K.R., Awasthi S., Mishra P.K., Srivastava P.K. Waterborne Pathogens. Elsevier; 2020. Biosensors/molecular tools for detection of waterborne pathogens; pp. 237–277. [DOI] [Google Scholar]
- Steinmetz M., Lima D., Viana A.G., Fujiwara S.T., Pessôa C.A., Etto R.M., Wohnrath K. A sensitive label-free impedimetric DNA biosensor based on silsesquioxane-functionalized gold nanoparticles for Zika Virus detection. Biosens. Bioelectron. 2019;141:111351. doi: 10.1016/j.bios.2019.111351. [DOI] [PubMed] [Google Scholar]
- Strich J.R., Chertow D.S. CRISPR-cas biology and its application to infectious diseases. J. Clin. Microbiol. 2018;57:1–14. doi: 10.1128/JCM.01307-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P.D., Tuan M.A., Huy T.Q., Le A.-T., Hieu N. Van. Facile preparation of a DNA sensor for rapid herpes virus detection. Mater. Sci. Eng. C. 2010;30:1145–1150. doi: 10.1016/j.msec.2010.06.010. [DOI] [Google Scholar]
- Tam P.D., Van Hieu N., Chien N.D., Le A.-T., Anh Tuan M. DNA sensor development based on multi-wall carbon nanotubes for label-free influenza virus (type A) detection. J. Immunol. Methods. 2009;350:118–124. doi: 10.1016/j.jim.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Tancharoen C., Sukjee W., Thepparit C., Jaimipuk T., Auewarakul P., Thitithanyanont A., Sangma C. Electrochemical biosensor based on surface imprinting for zika virus detection in serum. ACS Sens. 2019;4:69–75. doi: 10.1021/acssensors.8b00885. [DOI] [PubMed] [Google Scholar]
- Tang D., Tang J., Su B., Ren J., Chen G. Simultaneous determination of five-type hepatitis virus antigens in 5min using an integrated automatic electrochemical immunosensor array. Biosens. Bioelectron. 2010;25:1658–1662. doi: 10.1016/j.bios.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Teengam P., Siangproh W., Tuantranont A., Henry C.S., Vilaivan T., Chailapakul O. Electrochemical paper-based peptide nucleic acid biosensor for detecting human papillomavirus. Anal. Chim. Acta. 2017;952:32–40. doi: 10.1016/j.aca.2016.11.071. [DOI] [PubMed] [Google Scholar]
- Teymourian H., Barfidokht A., Wang J. Electrochemical glucose sensors in diabetes management: an updated review (2010-2020) Chem. Soc. Rev. 2020 doi: 10.1039/d0cs00304b. [DOI] [PubMed] [Google Scholar]
- Toldrà A., Furones M.D., O'Sullivan C.K., Campàs M. Detection of isothermally amplified ostreid herpesvirus 1 DNA in Pacific oyster (Crassostrea gigas) using a miniaturised electrochemical biosensor. Talanta. 2020;207:120308. doi: 10.1016/j.talanta.2019.120308. [DOI] [PubMed] [Google Scholar]
- Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;3:1–18. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tram D.T.N., Wang H., Sugiarto S., Li T., Ang W.H., Lee C., Pastorin G. Advances in nanomaterials and their applications in point of care (POC) devices for the diagnosis of infectious diseases. Biotechnol. Adv. 2016;34:1275–1288. doi: 10.1016/j.biotechadv.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H.N., Le G.T., Nguyen D.T., Juang R.-S., Rinklebe J., Bhatnagar A., Lima E.C., Iqbal H.M.N., Sarmah A.K., Chao H.-P. SARS-CoV-2 coronavirus in water and wastewater: a critical review about presence and concern. Environ. Res. 2020;110265 doi: 10.1016/j.envres.2020.110265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.D., Nguyen D.T., Nguyen B.H., Do Q.P., Le Nguyen H. Development of interdigitated arrays coated with functional polyaniline/MWCNT for electrochemical biodetection: application for human papilloma virus. Talanta. 2011;85:1560–1565. doi: 10.1016/j.talanta.2011.06.048. [DOI] [PubMed] [Google Scholar]
- Tripathy S., Singh S.G. Label-free electrochemical detection of DNA hybridization: a method for COVID-19 diagnosis. Trans. Indian Natl. Acad. Eng. 2020;5:205–209. doi: 10.1007/s41403-020-00103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020 doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Vadlamani B.S., Uppal T., Verma S.C., Misra M. Functionalized tio2 nanotube-based electrochemical biosensor for rapid detection of sars-cov-2. Sensors. 2020;20:1–10. doi: 10.3390/s20205871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia D.P., Dantas L.M.F., Lara A., García J., Rivera Z., Rosas J., Bertotti M. Development of a bio-electrochemical immunosensor based on the immobilization of SPINNTKPHEAR peptide derived from HPV-L1 protein on a gold electrode surface. J. Electroanal. Chem. 2016;770:50–55. doi: 10.1016/j.jelechem.2016.03.040. [DOI] [Google Scholar]
- van den Kieboom C.H., van der Beek S.L., Mészáros T., Gyurcsányi R.E., Ferwerda G., de Jonge M.I. Aptasensors for viral diagnostics. TrAC Trends Anal. Chem. (Reference Ed.) 2015;74:58–67. doi: 10.1016/j.trac.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walper S.A., Lasarte Aragonés G., Sapsford K.E., Brown C.W., Rowland C.E., Breger J.C., Medintz I.L. Detecting biothreat agents: from current diagnostics to developing sensor technologies. ACS Sens. 2018;3:1894–2024. doi: 10.1021/acssensors.8b00420. [DOI] [PubMed] [Google Scholar]
- Wang H.J., Du S.H., Yue X., Chen C.X. Review and prospect of pathological features of corona virus disease. J. Forensic Med. 2020 doi: 10.12116/j.issn.1004-5619.2020.01.004. [DOI] [PubMed] [Google Scholar]
- Wang Y., Bai X., Wen W., Zhang X., Wang S. Ultrasensitive electrochemical biosensor for HIV gene detection based on graphene stabilized gold nanoclusters with exonuclease amplification. ACS Appl. Mater. Interfaces. 2015;7:18872–18879. doi: 10.1021/acsami.5b05857. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang Z., Jain V., Yi J., Mueller S., Sokolov J., Liu Z., Levon K., Rigas B., Rafailovich M.H. Potentiometric sensors based on surface molecular imprinting: detection of cancer biomarkers and viruses. Sensor. Actuator. B Chem. 2010;146:381–387. doi: 10.1016/j.snb.2010.02.032. [DOI] [Google Scholar]
- Wei S., Xiao H., Cao L., Chen Z. A label-free immunosensor based on graphene oxide/Fe3O4/prussian blue nanocomposites for the electrochemical determination of HBsAg. Biosensors. 2020;10:24. doi: 10.3390/bios10030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss C., Carriere M., Fusco L., Fusco L., Capua I., Regla-Nava J.A., Pasquali M., Pasquali M., Pasquali M., Scott J.A., Vitale F., Vitale F., Unal M.A., Mattevi C., Bedognetti D., Merkoçi A., Merkoçi A., Tasciotti E., Tasciotti E., Yilmazer A., Yilmazer A., Gogotsi Y., Stellacci F., Stellacci F., Delogu L.G. Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS Nano. 2020;14:6383–6406. doi: 10.1021/acsnano.0c03697. [DOI] [PubMed] [Google Scholar]
- Whitesides G.M. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- Won J., Lee S., Park M., Kim T.Y., Park M.G., Choi B.Y., Kim D., Chang H., Kim V.N., Lee C.J. Development of a laboratory-safe and low-cost detection protocol for SARS-CoV-2 of the coronavirus disease 2019 (COVID-19) Exp. Neurobiol. 2020;29:107–119. doi: 10.5607/en20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Yan M., Li H., Liu T., Lin C., Huang S., Shen C. 2020. Evaluation of Enzyme-Linked Immunoassay and Colloidal Gold-Immunochromatographic Assay Kit for Detection of Novel Coronavirus (SARS-Cov-2) Causing an Outbreak of Pneumonia (COVID-19). medRxiv. [DOI] [Google Scholar]
- Xu W., Wang D., Li D., Liu C.C. Recent developments of electrochemical and optical biosensors for antibody detection. Int. J. Mol. Sci. 2019;21:134. doi: 10.3390/ijms21010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R., Dwivedi S., Kumar S., Chaudhury A. Trends and perspectives of biosensors for food and environmental virology. Food Environ. Virol. 2010;2:53–63. doi: 10.1007/s12560-010-9034-5. [DOI] [Google Scholar]
- Yamanaka K., Saito M., Kondoh K., Hossain M.M., Koketsu R., Sasaki T., Nagatani N., Ikuta K., Tamiya E. Rapid detection for primary screening of influenza A virus: microfluidic RT-PCR chip and electrochemical DNA sensor. Analyst. 2011;136:2064. doi: 10.1039/c1an15066a. [DOI] [PubMed] [Google Scholar]
- Yang Y., Yan W., Guo C., Zhang J., Yu L., Zhang G., Wang X., Fang G., Sun D. Magnetic molecularly imprinted electrochemical sensors: a review. Anal. Chim. Acta. 2020;1106:1–21. doi: 10.1016/j.aca.2020.01.044. [DOI] [PubMed] [Google Scholar]
- Yao C.-Y. Biosensors for hepatitis B virus detection. World J. Gastroenterol. 2014;20:12485. doi: 10.3748/wjg.v20.i35.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom S.-H. Nanostructures in biosensor-A review. Front. Biosci. 2011;16:997. doi: 10.2741/3731. [DOI] [PubMed] [Google Scholar]
- Yugender Goud K., Catanante G., Hayat A., Vengatajalabathy Gobi K., Marty J.L. Disposable and portable electrochemical aptasensor for label free detection of aflatoxin B1 in alcoholic beverages. Sensor. Actuator. B Chem. 2016;235:466–473. doi: 10.1016/j.snb.2016.05.112. M., S. [DOI] [Google Scholar]
- Zari N., Amine A., Ennaji M.M. Label-free DNA biosensor for electrochemical detection of short DNA sequences related to human papilloma virus. Anal. Lett. 2009;42:519–535. doi: 10.1080/00032710802421897. [DOI] [Google Scholar]
- Zhao C., Liu X. A portable paper-based microfluidic platform for multiplexed electrochemical detection of human immunodeficiency virus and hepatitis C virus antibodies in serum. Biomicrofluidics. 2016;10 doi: 10.1063/1.4945311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Liu F., Xie W., Zhou T.-C.C., OuYang J., Jin L., Li H., Zhao C.-Y.Y., Zhang L., Wei J., Zhang Y.-P.P., Li C.-P.P. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor. Actuator. B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Cui H., Song W., Ru X., Zhou W., Yu X. 2020. A Simple Magnetic Nanoparticles-Based Viral RNA Extraction Method for Efficient Detection of SARS-CoV-2 2020; pp. 6–9. [DOI] [Google Scholar]
- Zhou C.-H., Shu Y., Hong Z.-Y., Pang D.-W., Zhang Z.-L. Electrochemical magnetoimmunosensing approach for the sensitive detection of H9N2 avian influenza virus particles. Chem. Asian J. 2013;8:2220–2226. doi: 10.1002/asia.201300521. [DOI] [PubMed] [Google Scholar]
- Zhou W., Jimmy Huang P.-J., Ding J., Liu J. Aptamer-based biosensors for biomedical diagnostics. Analyst. 2014;139:2627. doi: 10.1039/c4an00132j. [DOI] [PubMed] [Google Scholar]
- Zhu H., Fohlerová Z., Pekárek J., Basova E., Neužil P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020;153:112041. doi: 10.1016/j.bios.2020.112041. [DOI] [PMC free article] [PubMed] [Google Scholar]