Significance
Meristem determinacy/indeterminacy influences flower number and seed production in crops. Two closely related cool-season cereals, barley and wheat, produce variable and defined numbers of spikelets in their inflorescences, respectively. In this study, we identify a series of allelic barley mutants named intermedium-m and double seed1 that develop wheat-like determinate inflorescences producing a terminal spikelet and a reduced number of spikelets. INT-M/DUB1 is an APETALA2-like transcription factor that promotes an active inflorescence meristem via suppression of spikelet initiation and the maintenance of meristem identity. Our work has identified key regulators that may prolong meristem activities and could be genetically engineered in barley, wheat, and other cereals to improve grain yield.
Keywords: barley and wheat, inflorescence determinacy, APETALA2/Q, MADS1, MADS3/58
Abstract
Inflorescence architecture dictates the number of flowers and, ultimately, seeds. The architectural discrepancies between two related cereals, barley and wheat, are controlled by differences in determinacy of inflorescence and spikelet meristems. Here, we characterize two allelic series of mutations named intermedium-m (int-m) and double seed1 (dub1) that convert barley indeterminate inflorescences into wheat-like determinate inflorescences bearing a multifloreted terminal spikelet and spikelets with additional florets. INT-M/DUB1 encodes an APETALA2-like transcription factor (HvAP2L-H5) that suppresses ectopic and precocious spikelet initiation signals and maintains meristem activity. HvAP2L-H5 inhibits the identity shift of an inflorescence meristem (IM) to a terminal spikelet meristem (TSM) in barley. Null mutations in AP2L-5 lead to fewer spikelets per inflorescence but extra florets per spikelet. In wheat, prolonged and elevated AP2L-A5 activity in rAP2L-A5 mutants delays but does not suppress the IM−TSM transition. We hypothesize that the regulation of AP2L-5 orthologs and downstream genes contributes to the different inflorescence determinacy in barley and wheat. We show that AP2L-5 proteins are evolutionarily conserved in grasses, promote IM activity, and restrict floret number per spikelet. This study provides insights into the regulation of spikelet and floret number, and hence grain yield in barley and wheat.
Inflorescence architecture controls flower and hence seed production and is largely defined by the fates of inflorescence shoot apical meristems (SAMs) (1–3). Inflorescence meristems (IMs) are either indeterminate if they remain undifferentiated reiteratively forming lateral primordia on their flanks or determinate if they differentiate producing a fixed number of lateral organs. Typically, the IM is determinate if it develops into a floral meristem (FM) and forms a terminal flower (3). Grass IMs produce unique structures called spikelets as basic inflorescence units and become determinate if the IM transitions to a terminal spikelet meristem (TSM) (3, 4). The spikelet meristem (SM) itself, unlike the determinate FM, may be indeterminate or determinate, producing a variable or defined number of flowers (i.e., florets), respectively (3, 5).
In eudicot snapdragon (Antirrhinum majus) centroradialis (cen) and Arabidopsis (Arabidopsis thaliana) terminal flower1 (tfl1) mutants, the inflorescences form a terminal flower (1, 6). In rice (Oryza sativa) and barley (Hordeum vulgare), cen/tfl1 mutants also show increased IM determinacy and thereby a reduced number of lateral branches/spikelets but not the formation of a terminal spikelet (TS) (7, 8). In snapdragon and Arabidopsis cen/tfl1 mutants with a terminal flower, FLORICAULA/LEAFY (FLO/LFY) is ectopically expressed in the apical domes of the inflorescences (1, 6). By contrast, loss-of-function mutations of rice LFY (RFL; aka ABERRANT PANICLE ORGANIZATION2, APO2) convert an indeterminate IM to a TSM (9, 10). APO1, the rice ortholog of Arabidopsis UNUSUAL FLORAL ORGANS, functions synergistically with RFL/APO2 to promote IM identity and spiral inflorescence phyllotaxy (10–12). Hence, the genetic control of inflorescence architecture and the formation of a terminal flower/spikelet differs between eudicots and grasses (3, 13).
Spikelet determinacy is controlled by microRNA172 (miR172)-targeted APETALA2-like (AP2L) transcription factors. In maize (Zea mays), INDETERMINATE SPIKELET1 (IDS1) and the Sister of IDS1 (SID1) promote SM determinacy. Null mutations in ids1 or ids1 sid1 mutants result in the production of additional lateral sterile bracts per spikelet (14–16). Their sister genes Q (rAP2L-A5, resistance to miR172 cleavage) in tetraploid/hexaploid wheat (Triticum spp.) and rice OsIDS1 and SUPERNUMERARY BRACT (SNB/OsSID1) are conserved in regulating SM determinacy (17–20). In addition, proteins of IDS1/SID1 orthologs also influence IM determinacy, and a reduction of their activity results in fewer lateral spikelets in wheat, and decreased primary branches but not the IM−TSM shift in maize and rice (15–21). Similarly, Arabidopsis AP2 maintains meristem activity and indeterminacy of the SAM/IM. Indeed, AP2 directly inhibits multiple flowering time and floral organ identity genes, including SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), APETALA1 (AP1), FRUITFUL (FUL), SEPALLATA3 (SEP3), and AGAMOUS (AG) and indirectly promotes TFL1 (22–25). Furthermore, AG represses WUSCHEL (WUS) and stem cell maintenance in Arabidopsis FM (26). AP2 also likely regulates the stem cell niche in Arabidopsis SAM through mechanisms independent of AG, but via modifying the WUS-CLAVATA3 (WUS-CLV3) negative feedback circuit (23). However, it remains poorly known whether and how IM and SM determinacy are regulated by meristem identity and maintenance genes in grass inflorescences.
The closely related temperate cereals barley and wheat share unbranched inflorescences (i.e., spikes) but are characterized by different determinacy of spikes and spikelets. Barley has an indeterminate IM and single-flowered spikelets, whereas wheat develops a determinate IM, but indeterminate multifloreted spikelets (5). It is not known which genes control IM indeterminacy and SM determinacy in barley and differences in IM/SM determinacy between barley and wheat. Here, we describe two allelic barley mutant groups int-m and dub1 that produce a TS and additional florets per spikelet. We determine that INT-M/DUB1 encodes an AP2L transcription factor (HvAP2L-H5) that is orthologous to maize IDS1 and wheat Q. We show that HvAP2L-H5 promotes IM indeterminacy by suppressing ectopic differentiation cues and maintaining IM activity in barley. Furthermore, we argue that regulation of AP2L-5 orthologs and floral organ identity genes has contributed to the evolution of differences in IM determinacy between barley and wheat.
Results and Discussion
int-m/dub1 Mutants Produce a Multifloreted Terminal Spikelet and Multifloreted Spikelets.
To reveal the genetic basis of meristem determinacy, we characterized two groups of induced barley mutants named intermedium-m (int-m.85 and int-m.1a) and double seed1 (dub1) (SI Appendix, Table S1). The int-m mutants were originally described as spike row-type mutants, forming an intermediate type between a two- and six-rowed spike (27). These int-m mutants were backcrossed to the wild-type Bowman to obtain near-isogenic lines (NILs) (BW429, int-m.85*BC7 and BW430, int-m.1a*BC5) (28). The dub1 mutants produce an additional grain per spikelet and a “fasciated” inflorescence tip (27).
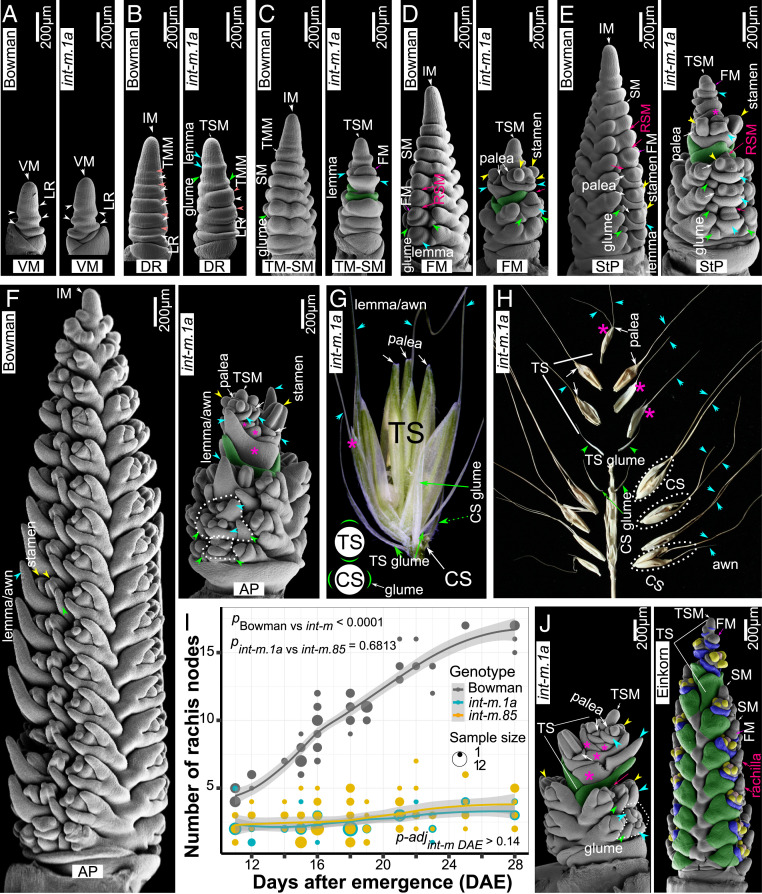
The int-m mutants produce two or more peculiar sterile bracts (i.e., without subtended axillary meristems) (Fig. 1 B–F and SI Appendix, Fig. S1) in the upper part of the inflorescence where multiple “spikelets” were thought to be “fused” (27). However, these abnormal “spikelets” are, in fact, florets, since they are neither flanked by two lateral spikelets, typical for the spikelet triplets (i.e., one central spikelet flanked by two lateral spikelets at each rachis node) in barley, nor subtended by two glumes, a diagnostic feature of grass spikelets (29) (Fig. 1 C–H and SI Appendix, Fig. S1). In addition, the two basal sterile bracts are morphologically similar to glumes of central spikelets that are less elongated and form very short awns at the distal ends (Fig. 1 C–H and SI Appendix, Fig. S1). The upper nonbasal sterile bracts expand laterally and produce awns that elongate extensively at the distal tips and are phenotypically lemma-like but lack axillary FM in the axils (Fig. 1 and SI Appendix, Fig. S1). The distinct identities of lemmas and glumes may be diagnosed by MADS1/LEAFY HULL STERILE1 (LHS1) that is only transcribed in lemmas/paleae but not in glumes in grasses (30, 31). HvMADS1 is expressed in the emerging bract-like structures in the upper part of the inflorescences except the two basal subtending bracts, indicating their lemma and glume identities, respectively (SI Appendix, Fig. S1). Therefore, these features suggest the formation of a TS that is subtended by two glumes and bears multiple florets. The paired TS glumes in int-m NIL initiate early upon the vegetative-to-reproductive phase transition (vegetative meristem [VM]−IM) as indicated by the formation of double ridges (DR) (Fig. 1 A and B). These TS glumes are oriented in the perpendicular plane to the glumes of central spikelets (Fig. 1G and SI Appendix, Fig. S1). As expected, int-m inflorescences exhibit characteristic features of apical termination, where inflorescences are shortened (SI Appendix, Fig. S1) and the final spikelet number is defined early in development (Fig. 1I).
Fig. 1.
The int-m mutants produce a multifloreted TS and spikelets with extra florets. (A–F) Developing shoot apices of the wild-type Bowman and the NIL mutant BW430 (int-m.1a*BC5). No differences are found between Bowman and int-m at the VM stage (A). Sterile leaf ridges (LRs) without subtended spikelet ridges (SPRs, aka TMMs in barley) emerge at the DR stage (B) in int-m, marking the initiation of a TSM. The paired sterile bracts (i.e., glumes; green shaded) subtending a TS become prominent at the TM-SM (C), floret meristem (FM) (D), StP (E), and awn primordium (AP) (F) stages. (G) A multifloreted TS (i.e., multiple pairs of lemmas and paleae per spikelet) in int-m shows that the plane of TS glumes is perpendicular to that of central spikelets (CS) glumes (depicted in Inset). (D–F and H) RSMs are desuppressed in int-m, which may lead to the formation of additional florets in some CSs (circled in H). (I) The int-m mutants produce fewer and a defined number of spikelets. Analysis of covariance is used to compare the predicted regression lines of rachis nodes vs. days after emergence (DAE) of Bowman, int-m.1a, and int-m.85 (n = 340). No significant difference (P = 0.6813) is found between int-m.1a and int-m.85 mutants, while the number of rachis nodes in Bowman is significantly higher than that of each int-m mutant (P < 0.0001). Tukey test finds nonsignificant differences in the number of rachis nodes (adjusted P values for multiple comparisons p-adj > 0.14) between successive time points over development in int-m mutants. (J) The int-m.1a mutants form a determinate IM and an indeterminate SM. These int-m morphologies resemble a determinate IM and multifloreted spikelets in wheat. Floral organs are overlaid with pseudocolors: green glumes, yellow stamens, pink pistils, and blue lemmas. Pink asterisks mark lemma-like sterile bracts forming elongated awns within the TS.
In the wild-type Bowman, a single floret initiates in the abaxial side of each sessile central spikelet and the SM is suppressed as a residual SM (RSM) (15) in the adaxial region (Fig. 1 D and E and SI Appendix, Fig. S1). The RSM appears to be desuppressed in both int-m NILs, leading to the formation of additional fertile or sterile florets in some central spikelets (Fig. 1 F and H and SI Appendix, Fig. S1). Therefore, INT-M promotes SM determinacy and thus restricts the floret number per spikelet.
Like int-m, dub1 mutants produce a TS and extra florets in some spikelets (SI Appendix, Fig. S2). We further demonstrate that dub1 mutants are allelic to int-m mutants. Specifically, the F1 hybrids (five plants) and two F2 populations (68 individuals) of the int-m.85 × dub1.3 crosses all formed TS (SI Appendix, Fig. S2). Therefore, the INT-M/DUB1 locus promotes IM indeterminacy and SM determinacy, leading to a variable spikelet number per inflorescence and a fixed number of florets per spikelet in barley. The int-m/dub1 inflorescence morphologies resemble those in wheat having a determinate IM, multifloreted spikelets, and opposite orientation between TS and central/lateral spikelets on the rachis (Fig. 1J and SI Appendix, Fig. S1).
INT-M/DUB1 Encodes an APETALA2-Like (HvAP2L-5H) Transcription Factor.
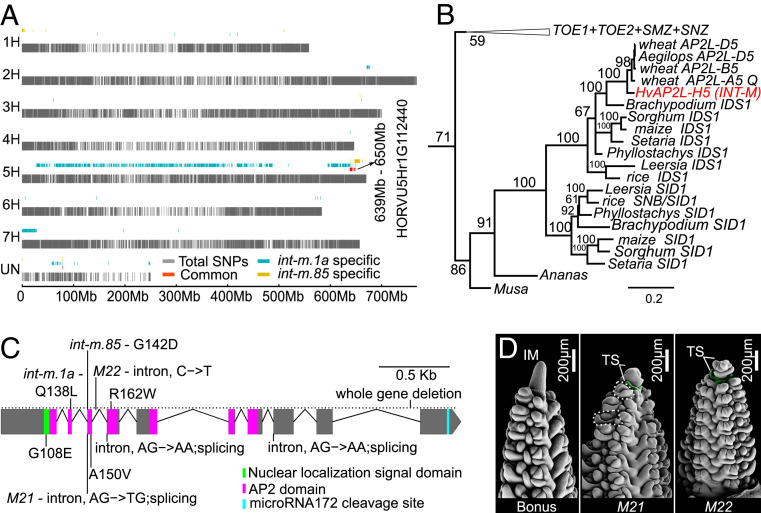
The int-m phenotype is caused by a single recessive locus (32), verified by segregation analysis in an int-m.85 × Proctor cross (mutant/wild type = 46/162, χ2(1:3), df = 1 = 0.92, P = 0.337). To identify the causal gene, we applied RNA sequencing (RNA-seq) using developing inflorescences of the NILs (int-m.1a and int-m.85) and wild-type cultivars Bowman and Bonus. We identified a single overlapping introgression region in an interval between 639 and 650 Mb on the chromosome 5H in the allelic int-m mutants (Fig. 2A and SI Appendix, Figs. S3–S5). Variant comparison between int-m and wild types found only one candidate gene (HORVU5Hr1G112440) harboring mutant-specific functional mutations (SI Appendix, Figs. S3–S5 and Dataset S1). The candidate gene (HvAP2L-H5) is orthologous to maize IDS1, rice OsIDS1, and wheat Q genes encoding an AP2L transcription factor (Fig. 2B). We confirmed that HvAP2L-H5 cosegregates with the mutant phenotype in an F3 population (158 plants) derived from an int-m.85 × Proctor cross (SI Appendix, Fig. S6). In addition, resequencing of HvAP2L-H5 in 24 independent allelic dub1 mutants found nine distinct types of mutations primarily in the first AP2 domain, including four nonsynonymous mutations, three substitutions in splicing sites, one substitution in the third intron, and a whole-gene deletion identified in one mutant (Fig. 2C and SI Appendix, Table S1). As in int-m plants, all dub1 mutants produce a determinate IM with some central spikelets producing desuppressed RSMs and extra florets in spikelets (27) (Fig. 2D and SI Appendix, Figs. S2 and S7).
Fig. 2.
INT-M encodes an APETALA2 transcription factor. (A) RNA-seq revealed a ∼10-Mb overlapping introgression region of the allelic int-m.1a and int-m.85. Functional annotation of variants within this introgression region identified a barley ortholog (HvAP2L-H5) of maize INDETERMINATE SPIKELET1 (IDS1) and wheat AP2L-A5 as the candidate gene. (B) Maximum likelihood phylogenetic tree of INT-M/IDS1 genes; bootstrap values of >50 are shown above branches. (C) INT-M gene model with position of mutations in 24 allelic mutants termed dub1 (SI Appendix, Table S1). (D) Two exemplary allelic dub1 mutants (M21 and M22 in Bonus background) produce determinate inflorescences and desuppressed RSMs (circled by dashed lines) in M21.
HvAP2L-H5 Inhibits the IM−TSM Identity Shift by Repressing Organ Differentiation and Maintaining Meristem Activity in Barley.
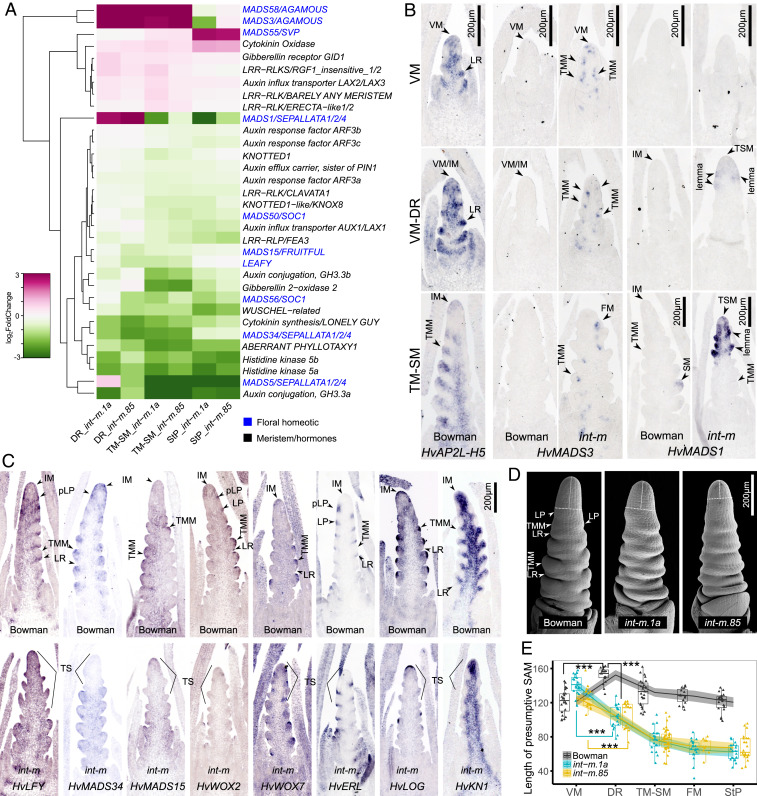
To unravel how HvAP2L-H5 regulates IM indeterminacy and SM determinacy, we profiled stage- and mutant-specific transcriptomic changes in developing inflorescences using the two int-m NIL mutants and Bowman. We concentrated our analyses on genes that were changed in the same direction in the two NIL mutants, to minimize the effects of background mutations. To identify changes primarily due to early HvAP2L-H5 activity and putatively linked to TSM formation in int-m mutants, we focused our analyses on two early stages of IM and SM formation, the DR stage (Fig. 1B) and the triple-mound SM (TM-SM) stage (Fig. 1C). A total of 312 genes were significantly changed between Bowman and the two int-m mutants during the early reproductive stages, with 136 genes at DR and 238 genes at TM-SM (Dataset S2). These misregulated genes are enriched in categories of signal transduction, reproductive structure development, and cell differentiation, including floral homeotic genes (e.g., MADS1, MADS3, and MADS58), meristem maintenance (e.g., WUS-CLV3, KNOTTED1 [KN1]), and phytohormone-related genes (e.g., cytokinins [CK], auxins) genes (Fig. 3A and SI Appendix, Fig. S8). To further identify changes that correlate with int-m morphologies, we used in situ hybridization to examine the temporal and spatial expression patterns of the misregulated genes involved in organ differentiation, IM identity, and maintenance.
Fig. 3.
INT-M suppresses organ differentiation and maintains meristem activity. (A) Genes differentially expressed between int-m mutants and Bowman before floral organ differentiation (DR and TM-SM) are enriched in activities associated with floral development, phytohormone, and meristem maintenance. These selected genes are plotted at the DR, TM-SM, and StP stages. Green and purple indicate genes down- and up-regulated in the mutant relative to the wild-type Bowman. (B) HvMADS3 and HvMADS1 are precociously and ectopically expressed in the int-m mutant prior to or concurrent with the TS initiation. (C) Expression patterns of putative IM identity and meristem genes. Specifically, HvLFY, HvMADS34, HvWOX2, and HvWOX7 are localized in incipient lateral primordia (pLP) in IM and TMM. HvLFY, HvERL, and HvWOX2 are found in bracts subtending the TMM. HvMADS15 is expressed in IM, TMM, and SM. HvLOG is confined in the L1 epidermal cells of IM and TMM, while HvKN1 is restricted to non-L1 epidermal IM and TMM cells. However, no obvious temporal or spatial changes in the expression of these genes are found to correlate with TS formation in int-m mutants. (D and E) A strong reduction of SAM length in int-m coincides with the shift of an IM to a TSM at the DR stage. Significant differences (***P < 0.001) are found between VM and DR stages within each genotype using Tukey test, and between the wild-type Bowman and int-m mutants using the Dunnett's test. Each triangle in the plot represents a measurement of a biological replicate.
INT-M suppresses precocious and ectopic spikelet specification signals in the IM and promotes IM identity. In Bowman, expression of AGAMOUS (AG)-like genes (HvMADS3 and HvMADS58, orthologous to rice OsMADS3 and OsMADS58, respectively) and HvAP2L-H5 are mutually exclusive. HvMADS3 and HvMADS58 are localized in stamens and pistils, while HvAP2L-H5 is restricted to lemmas, paleae, and RSMs (SI Appendix, Fig. S9). HvMADS3 and HvMADS58 are barely detectable before the TM-SM stage in Bowman, but significantly elevated in int-m (Fig. 3 A and B and SI Appendix, Fig. S9). Similar to precocious and ectopic expression of AG in Arabidopsis ap2 null mutants (22), HvMADS3 and HvMADS58 in int-m mutants are precociously and ectopically transcribed in meristems during VM, VM-DR, and TM-SM stages where TSM and triple-mound meristems (TMMs) subsequently emerge (Fig. 3B and SI Appendix, Fig. S9). HvMADS1, a SEP-like gene and rice OsMADS1/LHS1 ortholog, colocalizes with HvAP2L-H5 in SMs and subsequently lemmas, paleae, and RSMs, marking the SM initiation and lemma/palea identity (Fig. 3B and SI Appendix, Fig. S9). Indeed, AP2L-5/OsIDS1 and MADS1 both specify lemma/palea identity, regulating the interconversion of glume and lemma in wheat and rice (17–19, 21, 30, 33). HvMADS1 transcripts only become detectable in SMs after SM initiation but not in IMs or TMMs, whereas HvAP2L-H5 is widely transcribed in IMs, TMMs, and SMs before and during the TM-SM stage in Bowman (Fig. 3B and SI Appendix, Fig. S9). In int-m, ectopic and precocious HvMADS1 expression is found in the upper part of the inflorescence at DR and TM-SM stages, coinciding with the position and timing of TS initiation (Fig. 3B and SI Appendix, Fig. S9). Additionally, in Bowman HvMADS1 expression becomes detectable in IMs later during the stamen primordium (StP) stage (SI Appendix, Fig. S9), consistent with the IM degeneration (5) and predicted decreased activity of HvAP2L-H5 in IMs as plants get older (34). Moreover, expression levels of HvMADS1, a molecular marker for spikelet initiation, in whole inflorescence tissue of int-m mutants are significantly lower than in the wild-type Bowman after the TM-SM stage (Fig. 3A), which correlates with a reduced spikelet number in int-m mutants compared to Bowman (Fig. 1I; TM-SM is equivalent to DAE16). These observations collectively indicate that HvAP2L-H5 represses the precocious and ectopic expression of HvMADS1 in IM during early inflorescence development, while HvAP2L-H5 and HvMADS1 contribute to the SM initiation and lemma/palea identity specification during spikelet differentiation. In Arabidopsis, AP2 directly suppresses the expression of AG and SEP3 (25), and AG exhibits precocious and ectopic expression in ap2 null mutants (22–25). In addition, in Arabidopsis, AG represses WUS expression and is required for floral determinacy (26). Likewise, HvAP2L-H5 may inhibit the ectopic/precocious expression of HvMADS1 and HvMADS3/HvMADS58 genes during early spike development, thereby repressing SM initiation in the spike tip and maintaining an undifferentiated IM in barley.
In int-m mutants, genes putatively promoting VM−IM transition (i.e., IM specification) and inflorescence differentiation, including HvLFY and HvMADS15/HvFUL2, and HvMADS34, are down-regulated (Fig. 3A and SI Appendix, Fig. S10). Their sister paralogs/cofactors, including HvAPO1 and two AP1/FUL-like genes (HvMADS14/HvVRN1 and HvMADS18/HvFUL3) that are also critical in IM initiation and differentiation (10, 12, 35, 36), are not significantly altered in int-m mutants (Fig. 3A, SI Appendix, Fig. S10A, and Dataset S2). In barley, HvAPO1 transcripts are restricted to VM/IM/TSM and, subsequently, FM, but are not expressed in TMM or SM (SI Appendix, Fig. S10). In int-m mutants, AP1/FUL-like and HvMADS34 genes exhibit broad expression in VM, IM, TMM, SM, FM, and floral organs over development, supporting their pleiotropic roles in IM initiation and subsequent spikelet and floret development (SI Appendix, Fig. S10). Null mutations in the rice LFY ortholog lead to a decreased number of primary branches and the formation of a TS in the inflorescences as observed in int-m mutants (Fig. 1I) (9, 10). INT-M might therefore promote IM activity by up-regulating HvLFY expression in the IM. Furthermore, the MADS-box genes may promote IM determinacy directly via inhibiting IM activity, as Arabidopsis FUL directly represses AP2 and hence WUS expression to promote meristem arrest (34). However, LFY, MADS34, and FUL2 are broadly transcribed in SM, FM, and floral organs (SI Appendix, Fig. S10). Consequently, the reduction in transcript levels observed in transcriptome analyses of the whole inflorescences may reflect the decreased number of rachis nodes and SMs in int-m compared to Bowman (Fig. 1I). Taken together, because of their pleiotropic effects on inflorescence development, it remains unclear whether HvLFY, HvAP1/HvFUL-like, or HvMADS34 contribute to TS formation in int-m mutants.
INT-M also influences genes involved in cell proliferation and differentiation. Particularly, transcript levels of several putative stem cell-promoting factors were dampened in int-m, including three WUS-related genes (e.g., WUS, WOX2, and WOX7), KN1, KNOX8, a CK-activating enzyme (LONELY GUY [LOG]), a type-A response regulator (ABERRANT PHYLLOTAXY1), a gibberellin degradation gene GA2OX and genes impacting polarized auxin gradients (PIN1 and PINOID) (37–40) (Fig. 3A, SI Appendix, Fig. S11, and Dataset S2). Consistently, signals promoting cell differentiation were elevated in int-m (Fig. 3A), including several Leucine-Rich Repeat-like Kinases (LRR-RLK, BARELY ANY MERISTEM [BAM], ERECTA-LIKE [ERL], RGF1-insensitive 1), a gibberellin receptor (GID1) and a Cytokinin Oxidase (CKX) (40–42). HvAP2L-H5, HvWOX2, HvWOX7, HvERL, HvLOG, and HvKN1 are transcribed in different domains of the IM in Bowman (Fig. 3C). Specifically, HvERL, HvWOX2, and HvWOX7 are localized in emerging lateral primordia in IM. HvLOG is restricted to the L1 epidermal cells of IM, whereas HvKN1 is found in non-L1 epidermal cells of IM (Fig. 3C). However, we did not find obvious spatial or temporal changes in the expression of these genes in SAM prior to or at the TSM initiation in int-m mutants, suggesting that HvAP2L-H5 only influences their expression levels (Fig. 3 and SI Appendix, Fig. S11). In Arabidopsis, a suite of distinct LRR-RLK receptors function redundantly to buffer stem cell proliferation (42, 43). For instance, BAM receptors compensate for altered CLV1 activity and restrict stem cell proliferation (42), while elevated ERL receptors repress WUS expression (44). Indeed, Arabidopsis AP2 maintains stem cells in the SAM likely via regulation of the WUS-CLV3 loop (23). We, therefore, examined whether expression changes in the meristem genes correlate with differences in meristem maintenance between Bowman and int-m. To determine whether the maintenance of the SAM is altered in int-m, we quantified the length and width of the presumptive SAM (i.e., apices above the latest visible lateral primordium) (Fig. 3D) across development. The length of SAM in int-m is greatly decreased at DR stage, concurrent with the shift of SAM identity from IM to TSM (Fig. 3E and SI Appendix, Fig. S11 and Table S2). In contrast, SAM length in Bowman gradually diminishes over development (Fig. 3E). Therefore, HvAP2L-H5 inhibits the IM−TSM shift likely by suppressing spikelet initiation cues and maintaining an active IM.
AP2L-5 Proteins Have an Ancestral and Conserved Role in Prolonging IM Activity in Grasses.
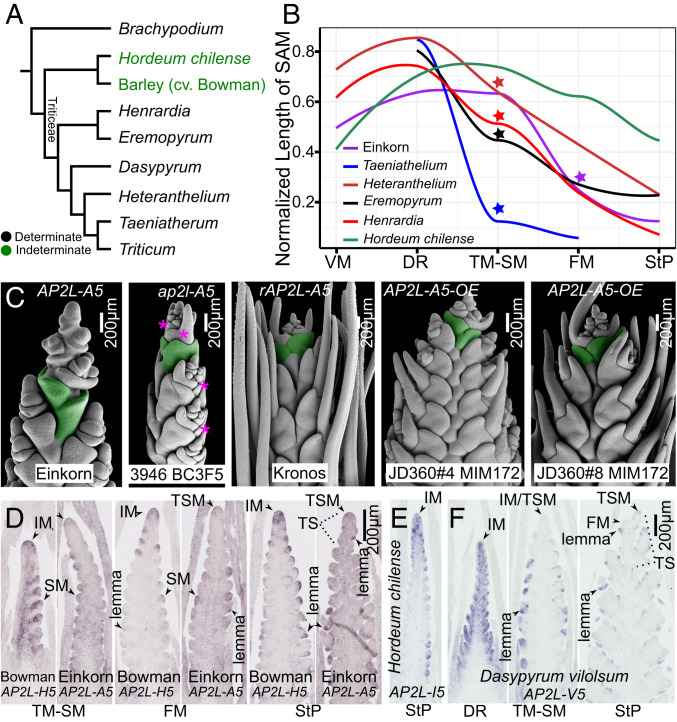
Phylogenetic analysis inferred that indeterminate inflorescences are derived in Hordeum, with determinate inflorescences being ancestral in Triticeae (Fig. 4A and SI Appendix, Fig. S12). To investigate the similarities and differences in indeterminate and determinate inflorescences, we compared SAM size changes over development within several Triticeae species. As the spikelet number is fixed after the IM−TSM transition, we estimated the stage of IM−TSM transition when the spikelet number became fixed in each species (SI Appendix, Fig. S12). As in int-m mutants and different from Hordeum species, the length of SAM sharply decreases at the IM−TSM transition in each of these Triticeae species (Fig. 4B and SI Appendix, Fig. S12). The exact timing of IM−TSM transitions differs among Triticeae species and int-m/dub1 mutants. In int-m/dub1 mutants, IM−TSM transition varies in different mutants/conditions/backgrounds, occurring extremely early at DR upon the VM−IM phase change or late after the StP stage (Fig. 1 and SI Appendix, Figs. S1, S2, and S7). By contrast, IM−TSM identity shifts occur at TM-SM stage in most examined Triticeae species and at FM stage in Einkorn (Triticum monococcum) carrying a wild-type AP2L-A5 gene. Wheat ap2l-A5 null mutations lead to early IM−TSM transition and a reduced spikelet number per spike, while prolonged and elevated levels of AP2L-A5 delay IM−TSM transition and thus increase spikelet number per spike (19–21). Furthermore, maize ids1 sid1 and rice osids1 snb double mutants produce fewer primary branches, whereas increased AP2L-5 activities in mimicry of miR172 (MIM172) or miR172-resistant mutants result in more primary branches in rice panicles and maize tassels (16, 18, 45). These observations collectively indicate an ancestral and conserved function of AP2L-5 in grasses in maintaining IM activity. Compared with wheat AP2L-A5, barley HvAP2L-H5 exhibits a stronger effect and completely inhibits the IM−TSM identity shift where int-m/dub1 null mutations revert the derived indeterminate inflorescences to the ancestral determinate state (Figs. 1 and 4).
Fig. 4.
INT-M likely contributes to divergence of inflorescence indeterminacy/determinacy in barley and wheat. (A) Unlike Hordeum, most Triticeae species have determinate IMs. Brachypodium distachyon is used as the outgroup. (B) SAM length gradually decreases over development in H. chilense, but sharply declines in several Triticeae species at TM-SM or FM stages (stars) that coincide with the transitions of an IM to a TSM. The exact timing of this transition varies between species, at TM-SM stage in four examined Triticeae species and at FM stage in Einkorn. The reduction in each species is statistically significant (P < 0.05), except in Heteranthelium using the Tukey test over development in each species; however, only one measurement of TM-SM stage is recorded for Heteranthelium (out of a total of 30 measurements for all stages). It should be noted that the meristem length is also reduced in H. chilense as the apical meristem is consumed to produce lateral organs, but the magnitude/rate is relatively lower (a more flattened curve) (SI Appendix, Table S4). Replicate number for each stage of each species is listed in SI Appendix, Table S4. (C) The cultivated tetraploid wheat Kronos harboring mutations in miR172 target site (rAP2L-A5) and two independent MIM172 transgenic lines have prolonged and elevated expression of TtAP2L-A5 (19) and form the wild-type determinate inflorescences. The null mutation ap2l-A5 leads to production of sterile lemmas as int-m mutants. (D–F) AP2L-5 orthologs in selected Triticeae species are localized in IM, SM, and lemma over development. Transcripts of AP2L-5 orthologs in Hordeum species (D and E) persist broadly in IM at FM and/or StP stages, while TmAP2L-A5 (D) and DvAP2L-V5 (F) are more localized in the SAM.
The evolution of this stronger effect of HvAP2L-H5 in IM maintenance in barley may be achieved via trans (protein)- and/or cis-regulatory changes of HvAP2L-H5 itself, and/or changes in the HvAP2L-H5−mediated genetic regulatory network. We examined protein sequences, expression levels, and domains of AP2L-5 orthologs and the expression patterns of putative downstream genes of AP2L-5 inferred from the int-m mutant analysis. We identified 21 barley-specific amino acids by comparing protein sequences of AP2L-5 orthologs from Triticum urartu A, Aegilops tauschii D, and wheatgrass (Thinopyrum elongatum) E genomes. These changes in barley HvAP2L-H5 are predicted to be functionally neutral, but might still contribute to its enhanced effects on IM activity (SI Appendix, Table S3). Furthermore, prolonged and elevated levels of the functional homeolog AP2L-A5 via a mutation in miR172 target site (Q) in cultivated wheat or MIM172 delay IM−TSM transition and increase spikelet number per spike (19–21), but do not promote the formation of a barley-like indeterminate IM (Fig. 4C). This observation thus suggests that changes in the transcript levels of AP2L-5 alone are not sufficient for the formation of an indeterminate IM. However, expression domains of AP2L-5 appear to be different between Triticeae species with determinate inflorescences and Hordeum species carrying derived indeterminate inflorescences. Like HvAP2L-H5, Einkorn TmAP2L-A5, Dasypyrum villosum DvAP2L-V5, and Hordeum chilense HcAP2L-I5 show conserved expression in IM, SM, and lemmas (Fig. 4 D–F). However, AP2L-5 transcripts in Bowman and H. chilense are more broadly localized in IM than those in Einkorn (TmAP2L-A5) or D. villosum (DvAP2L-V5) (Fig. 4 D–F and SI Appendix, Fig. S13). Furthermore, HvAP2L-H5 is inversely oriented in the genome compared to its orthologs in species with TS, such as wheat species and wheatgrass (46) (SI Appendix, Fig. S14). HvAP2L-H5 might have distinct regulatory elements compared to the functional TaAP2L-A5 in hexaploid wheat indicated by putative accessible chromatin regions (47, 48) (SI Appendix, Fig. S14). Interestingly, two AP2L-5 paralogs (IDS1/SID1), from a gene duplication early in grass family, are retained and functionally redundant in IM maintenance in maize and rice, while the SID1 paralogs were lost in Triticeae genomes (Fig. 2B and SI Appendix, Fig. S15) (16, 18). Within the Triticeae, barley HvAP2L-H5 underwent a genomic inversion and evolved an increased effect from its ancestral function in promoting IM activity to causing an indeterminate inflorescence. Therefore, our results indicate that differences in the expression domain of HvAP2L-H5 might cause its different effects on the barley spike compared to the other Triticeae. However, we cannot rule out that differences in the protein sequences, expression level, and downstream targets or interactors of HvAP2L-H5 might contribute to the formation of an indeterminate spike in barley.
In line with an enhanced effect of HvAP2L-H5 on IM activity, putative downstream genes of HvAP2L-H5 might likewise exhibit distinct expression patterns in barley compared to their orthologs in TS-carrying Triticeae species. We then tested this hypothesis and examined the expression of three most differentially up-regulated genes identified from int-m mutant analysis in selected TS-carrying Triticeae species. As expected, distinct from HvMADS1 in Bowman, Einkorn TmMADS1 and D. villosum DvMADS1 exhibit similar expression patterns to HvMADS1 in int-m mutants and are transcribed in the upper part of the inflorescence where the TSM emerges and differentiates (SI Appendix, Fig. S16). Similar to early expression of HvMADS3 in int-m but not in Bowman, MADS3 in Einkorn and bread wheat (subgenome A homeolog) is precociously transcribed at DR stage (SI Appendix, Fig. S17). TmMADS3 is localized in spikelet ridges at DR and SMs at SM stage before the TSM initiation (SI Appendix, Fig. S17). These observations suggest that the suppression of precocious spikelet initiation/differentiation cues (e.g., MADS1 and MADS3) in the IM is important for the maintenance of indeterminate inflorescences in barley, as observed in Arabidopsis ap2 or tfl1 null mutants. Likewise, genes involved in meristem activity (Fig. 3), such as LFY (9–12), LOG (41), and WOX-like genes (44), might also play critical roles for the evolution of inflorescence indeterminacy, which would be interesting to investigate in the future.
AP2L-5 Transcription Factors Repress the Lateral Organ Initiation in Spikelets of Grasses.
While int-m mutants have a reduced number of rachis nodes in spikes, they produce additional lateral sterile bracts and, occasionally, extra fertile florets in central spikelets (SI Appendix, Fig. S1). Similar to int-m, wheat ap2l-A5 null mutants have an increased floret number per spikelet (19–21), and maize ids1 sid1 and rice osids1 snb mutants produce extra sterile bracts per spikelet (15–18).
Regarding the contrasting effects of AP2L-5 on spike and spikelet determinacy, AP2L-5 orthologs, including IDS1 and the sister paralog SID1 clade, are suggested to inhibit differentiation in both spikes and spikelets (Figs. 1 and 3) (14–21). In spikes, AP2L-5 orthologs suppress IM differentiation and hence prolong IM activity and promote initiation of additional lateral branches/spikelets. Differently, in spikelets, AP2L-5 proteins influence the determinacy but not identity of SM per se and suppress the formation of axillary FMs from an SM, thereby restricting the number of lateral florets/bracts (3, 15–21). In fact, the desuppression of HvMADS1 might contribute to both IM−TSM shift in spikes and extra florets in central spikelets. Specifically, early elevated levels of MADS1 at the DR stage in int-m mutants correlate with the production of a multifloreted TS (Fig. 3). Likewise, HvMADS1 is predicted to be transcribed in the lemmas/paleae of the extra florets in central spikelets in int-m mutants. On the other hand, it could also be that AP2L-5 proteins target different meristem/floral identity genes in the IM versus SM to regulate the initiation of axillary meristems and IM−TSM identity shift in spikes, and formation of axillary florets and thus SM determinacy in spikelets. In the IM, desuppression of MADS3/MADS58 expression and regulation of meristem maintenance genes strongly correlate with IM−TSM identity shift and TS formation in int-m mutants and Einkorn (Fig. 3). Many floral homeotic MADS-box genes are significantly altered in int-m mutants during the StP stage (SI Appendix, Fig. S18); however, it remains unclear through which targets AP2L-5 proteins repress the initiation of lateral florets in spikelets.
AP2L-5 Promotes Axillary FM Development and Specifies the Identity of Lemmas.
The ap2l-5 null mutants produce additional florets in spikelets; however, extra florets may be sterile without associated axillary FM initiated in the axils of the lemmas in barley (Fig. 1 F–H and SI Appendix, Fig. S1) and wheat (21), suggesting that AP2L-5 promotes floret development. Disruption of HvAP2L-H5 causes very early defects in IM patterning and massive alterations in gene expression that might account for later developmental defects. It is therefore difficult to conclude whether differences in floret development and lemma specification are secondary effects or caused by HvAP2L-H5 itself. However, transcripts of HvAP2L-H5 and TmAP2L-A5 were detected in lemmas and paleae (SI Appendix, Figs. S9 and S12), suggesting that AP2L-5 proteins contribute to lemma specification. In addition, a number of floral homeotic MADS-box genes, including B-class, C-class, and E-class SEP-like, are misregulated in int-m mutants (SI Appendix, Fig. S18), which might influence floral development and organ specification as shown for wheat and rice (18, 21).
In this study, we identify HvAP2L-H5 as the causal gene for the int-m and dub1 mutant phenotypes and demonstrate that HvAP2L-H5 promotes IM indeterminacy by repressing floral differentiation genes and promoting the expression of meristem maintenance genes in barley. We hypothesize that changes in protein sequences, cis-regulation and/or downstream genes of AP2L-5 have contributed to the evolution of inflorescence determinacy/indeterminacy in Triticeae. We show that AP2L-5 and downstream targets regulate IM and SM determinacy that is central to increasing spikelet and fertile floret number. Our findings thus provide targets for the modification of inflorescence architecture in barley and wheat.
Materials and Methods
Plant Materials, Phenotyping, and Gene Identification.
We phenotyped and analyzed two allelic groups of barley intermedium-m (int-m) and double seed1 (dub1) mutants as described in SI Appendix, Supplementary Text.
Analysis of Gene Expression.
RNA-seq and RNA in situ hybridization were used to uncover how HvAP2L-H5 influences the indeterminacy of IM, as described in SI Appendix, Supplementary Text.
Comparative Analysis of Inflorescence Indeterminacy/Determinacy in Barley and Wheat.
We tested the hypothesis of whether and how HvAP2L-H5 may contribute to the differences in determinacy/indeterminacy of inflorescences and spikelets between barley and wheat, which is described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We are deeply indebted to Udda Lunqvist, who dedicated her work to generating and characterizing a unique collection of barley mutants that today is key to unraveling the genetics of barley development and performance. We are grateful for the support from Thea Rütjes, Caren Dawidson, Astrid Oehl, Edelgard Wendeler, Kerstin Luxa, Coral Vincent, Peter Huijser, Ivan Acosta, and George Coupland, and for grains for the study from NordGen, US Department of Agriculture-Germplasm Resources Information Network and Jorge Dubcovsky at the University of California, Davis. We thank He Gao, Toby Kellogg, the editor, and three anonymous reviewers for constructive comments on the manuscript. Computational infrastructure and support were provided by the Max Planck Institute for Plant Breeding Research Information Technology Services and the Centre for Information and Media Technology at the Heinrich Heine University Düsseldorf. This work was funded by the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy EXC-2048/1 (Project ID390686111; M.v.K.) and an Alexander von Humboldt Postdoctoral Fellowship (J.Z.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011779118/-/DCSupplemental.
Data Availability.
All the RNA-seq data presented in this paper, including the raw data, are deposited in the National Center for Biotechnology Information database with the accession number PRJNA668924.
Change History
December 21, 2021: The SI Appendix has been updated.
References
- 1.Bradley D., et al., Control of inflorescence architecture in Antirrhinum. Nature 379, 791–797 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Prusinkiewicz P., Erasmus Y., Lane B., Harder L. D., Coen E., Evolution and development of inflorescence architectures. Science 316, 1452–1456 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Bommert P., Whipple C., Grass inflorescence architecture and meristem determinacy. Semin. Cell Dev. Biol. 79, 37–47 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Weberling F., Morphology Flowers and Inflorescences (Cambridge University Press, 1992). [Google Scholar]
- 5.Bonnett O. T., “Inflorescences of maize, wheat, rye, barley, and oats: Their initiation and development” (Bulletin 721, University of Illinois, 1966). [Google Scholar]
- 6.Bradley D., Ratcliffe O., Vincent C., Carpenter R., Coen E., Inflorescence commitment and architecture in Arabidopsis. Science 275, 80–83 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Nakagawa M., Shimamoto K., Kyozuka J., Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29, 743–750 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Bi X., et al., CENTRORADIALIS interacts with FLOWERING LOCUS T-like genes to control floret development and grain number. Plant Physiol. 180, 1013–1030 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao N. N., Prasad K., Kumar P. R., Vijayraghavan U., Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc. Natl. Acad. Sci. U.S.A. 105, 3646–3651 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeda-Kawakatsu K., Maekawa M., Izawa T., Itoh J., Nagato Y., ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 69, 168–180 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Ikeda K., Ito M., Nagasawa N., Kyozuka J., Nagato Y., Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J. 51, 1030–1040 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Ikeda K., Nagasawa N., Nagato Y., ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev. Biol. 282, 349–360 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Whipple C. J., Grass inflorescence architecture and evolution: The origin of novel signaling centers. New Phytol. 216, 367–372 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Chuck G., Meeley R., Irish E., Sakai H., Hake S., The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Genet. 39, 1517–1521 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Chuck G., Meeley R. B., Hake S., The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 12, 1145–1154 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuck G., Meeley R., Hake S., Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135, 3013–3019 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Lee D.-Y., Lee J., Moon S., Park S. Y., An G., The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J. 49, 64–78 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Lee D.-Y., An G., Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 69, 445–461 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Debernardi J. M., Lin H., Chuck G., Faris J. D., Dubcovsky J., microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 144, 1966–1975 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenwood J. R., Finnegan E. J., Watanabe N., Trevaskis B., Swain S. M., New alleles of the wheat domestication gene Q reveal multiple roles in growth and reproductive development. Development 144, 1959–1965 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Debernardi J. M., Greenwood J. R., Jean Finnegan E., Jernstedt J., Dubcovsky J., APETALA 2-like genes AP2L2 and Q specify lemma identity and axillary floral meristem development in wheat. Plant J. 101, 171–187 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drews G. N., Bowman J. L., Meyerowitz E. M., Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65, 991–1002 (1991). [DOI] [PubMed] [Google Scholar]
- 23.Würschum T., Gross-Hardt R., Laux T., APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell 18, 295–307 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krogan N. T., Hogan K., Long J. A., APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139, 4180–4190 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yant L., et al., Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22, 2156–2170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenhard M., Bohnert A., Jürgens G., Laux T., Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105, 805–814 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Bregitzer P., Lundqvist U., Carollo Blake V., Eds., Barley Genet. Newsl. 37 (2007). [Google Scholar]
- 28.Druka A., et al., Genetic dissection of barley morphology and development. Plant Physiol. 155, 617–627 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clifford H., “Spikelet and floral morphology” in Grass Systematics and Evolution, Soderstrom T., Hilu K., Campbell C., Barkworth M., Eds. (Smithsonian Institution Press, 1987), pp. 21–30. [Google Scholar]
- 30.Prasad K., Parameswaran S., Vijayraghavan U., OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 43, 915–928 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Malcomber S. T., Kellogg E. A., Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16, 1692–1706 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lundqvist U., Lundqvist A., Induced intermedium mutants in barley: Origin, morphology and inheritance. Hereditas 108, 13–26 (1988). [Google Scholar]
- 33.Jeon J.-S., et al., Leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12, 871–884 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balanzà V., et al., Genetic control of meristem arrest and life span in Arabidopsis by a FRUITFULL-APETALA2 pathway. Nat. Commun. 9, 565 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikeda-Kawakatsu K., et al., Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem. Plant Physiol. 150, 736–747 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi K., et al., Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24, 1848–1859 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurakawa T., et al., Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z., Tucker E., Hermann M., Laux T., A molecular framework for the embryonic Initiation of shoot meristem stem cells. Dev. Cell 40, 264–277.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Giulini A., Wang J., Jackson D., Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430, 1031–1034 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Kitagawa M., Jackson D., Control of meristem size. Annu. Rev. Plant Biol. 70, 269–291 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Bartrina I., Otto E., Strnad M., Werner T., Schmülling T., Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23, 69–80 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nimchuk Z. L., CLAVATA1 controls distinct signaling outputs that buffer shoot stem cell proliferation through a two-step transcriptional compensation loop. PLoS Genet. 13, e1006681 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez-Leal D., et al., Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat. Genet. 51, 786–792 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimura Y., Tasaka M., Torii K. U., Uchida N., ERECTA-family genes coordinate stem cell functions between the epidermal and internal layers of the shoot apical meristem. Development 145, dev156380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L., et al., Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 15504–15509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H., et al., Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 368, eaba5435 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Lu Z., et al., The prevalence, evolution and chromatin signatures of plant regulatory elements. Nat. Plants 5, 1250–1259 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Li Z., et al., The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements. Genome Biol. 20, 139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the RNA-seq data presented in this paper, including the raw data, are deposited in the National Center for Biotechnology Information database with the accession number PRJNA668924.