Significance
Genetic studies show the founders of all living non-African populations expanded from Africa ca. 65 to 45 ka. This “late dispersal” model has been challenged by the discovery of isolated AMHs at caves in southern China suggested as early as ca. 120 ka. We assessed the age of early AMH fossils from five caves in this region using ancient DNA analysis and a multimethod geological dating strategy. We found they were much younger than previously suggested, with some remains dating to the Holocene owing to the complex depositional history at these subtropical caves. Current evidence shows AMHs settled southern China within the timeframe set by molecular data of less than ca. 50 to 45 ka and no earlier.
Keywords: anatomically modern humans, Late Pleistocene, East Asia, ancient DNA, dating
Abstract
The expansion of anatomically modern humans (AMHs) from Africa around 65,000 to 45,000 y ago (ca. 65 to 45 ka) led to the establishment of present-day non-African populations. Some paleoanthropologists have argued that fossil discoveries from Huanglong, Zhiren, Luna, and Fuyan caves in southern China indicate one or more prior dispersals, perhaps as early as ca. 120 ka. We investigated the age of the human remains from three of these localities and two additional early AMH sites (Yangjiapo and Sanyou caves, Hubei) by combining ancient DNA (aDNA) analysis with a multimethod geological dating strategy. Although U–Th dating of capping flowstones suggested they lie within the range ca. 168 to 70 ka, analyses of aDNA and direct AMS 14C dating on human teeth from Fuyan and Yangjiapo caves showed they derive from the Holocene. OSL dating of sediments and AMS 14C analysis of mammal teeth and charcoal also demonstrated major discrepancies from the flowstone ages; the difference between them being an order of magnitude or more at most of these localities. Our work highlights the surprisingly complex depositional history recorded at these subtropical caves which involved one or more episodes of erosion and redeposition or intrusion as recently as the late Holocene. In light of our findings, the first appearance datum for AMHs in southern China should probably lie within the timeframe set by molecular data of ca. 50 to 45 ka.
The fossil record suggests that Homo sapiens had evolved in Africa by 315,000 y ago (315 ka) (1), spread into West Asia before 177 ka (2), but disappeared and were seemingly replaced by Homo neanderthalensis until ca. 75 to 55 ka (3, 4). A second and final excursion from Africa by so-called anatomically modern humans (AMHs) occurred soon after and broadly coincides with the extinction of the last archaic hominins, ca. 40 to 30 ka (5, 6). This dispersal involved the ancestors of all present-day non-Africans and according to molecular data occurred ca. 65 to 45 ka (7, 8). Additional support for this “late dispersal” theory is provided by the geographical structure of contemporary DNA lineages with all non-Africans closely related to present-day and ancient eastern African populations (9, 10), as well as a clinal pattern of decreasing diversity from Africa to Eurasia, the signature of serial founder effect (10–12). Corroboration has also been provided by the estimated split time between western and eastern Eurasians of ca. 47 to 42 ka as determined by ancient DNA (aDNA) from the 46,880 to 43,210 cal y B.P. (calendar year before present, i.e., before AD1950) Ust’-Ishim femur (western Siberia, Russian Federation) and the 42,000 to 39,000 cal B.P. Tianyuan skeleton (Northeast China) (13–15). Finally, the upper age boundary for this dispersal is set by interbreeding between early AMHs and the Neanderthals estimated to have occurred ca. 65 to 47 ka and the ancestors of New Guineans with the Denisovans ca. 46 ka and again ca. 30 ka (13, 16–19).
In contrast, some paleoanthropologists have suggested that AMHs settled mainland East Asia much earlier, within the period of ca. 120 to 70 ka, in accordance with the “early dispersal” theory. This model is based largely upon the dating of isolated human teeth recovered at Huanglong, Luna, and Fuyan caves and a partial mandible from Zhirendong in southern China (20–24). Yet several researchers have raised questions about these and other sites on the basis of uncertainties surrounding the identification of some of them as AMHs, relationships between human remains and dated materials, or limited information available about their depositional context and dating (25–27).
Here, we describe the results of an investigation of the arrival time of AMHs in southern China at five apparent early AMH cave localities involving aDNA analyses of human teeth and the dating of flowstones, sediments, fossil remains, and charcoal. The five localities we studied are the following:
-
1)
Huanglong cave, located about 25 km from the town of Yunxi, northern Hubei Province (Fig. 1). Excavations by the Hubei Provincial Institute of Cultural Relics and Archaeology during three field seasons from 2004 to 2006 provided a rich mammal record, comprising 91 taxa and representing a Middle to Late Pleistocene Ailuropoda-Stegodon fauna, stone artifacts, and seven AMH teeth dated indirectly with U–Th dating on thin flowstone formations ca. 101 to 81 ka (20).
-
2)
Luna cave, situated in the karst mountains of the southeastern part of the Bubing basin, Guangxi Zhuang Autonomous Region (Fig. 1). A small sample of mammal fossils (Ailuropoda-Stegodon assemblage), stone artifacts, and two AMH teeth were recovered during excavations by the Natural History Museum of Guangxi Autonomous Region in 2004 and 2008. They have since been dated indirectly through U–Th dating of flowstone in the range ca. 127 to 70 ka (21).
-
3)
Fuyan cave, located in Daoxian County, Hunan Province (Fig. 1). Excavations from 2011 to 2013 resulted in a large sample of mammal fossils (Ailuropoda-Stegodon faunal group) and 47 AMH teeth but no associated artifacts (22). They have been dated indirectly using U–Th dating of flowstone within the range ca. 120 to 80 ka (22). Two additional (in situ) AMH teeth, stratigraphically associated with the original finds, were recovered by us during field investigations at the site during early 2019.
-
4)
Yangjiapo Cave is a large karstic chamber located in Jianshi County (Fig. 1). It was excavated during 2004 by the Hubei Provincial Institute of Cultural Relics and Archeology and yielded 11 AMH teeth found in association with the fragmentary bones of 80 species belonging to an Ailuropoda-Stegodon fauna, implying it should be of similar age to Huanglong, Luna, and Fuyan caves. No stone artifacts or other cultural remains were found.
-
5)
Sanyou Cave is a small chamber within a limestone hill at the confluence of the Yangtze River and Xiling Gorge, close to Yichang city, Hubei Province (Fig. 1). A small excavation was undertaken in 1986 by the Yichang Museum and led to the recovery of a possible Late Pleistocene age partial AMH cranial vault (Fig. 1).
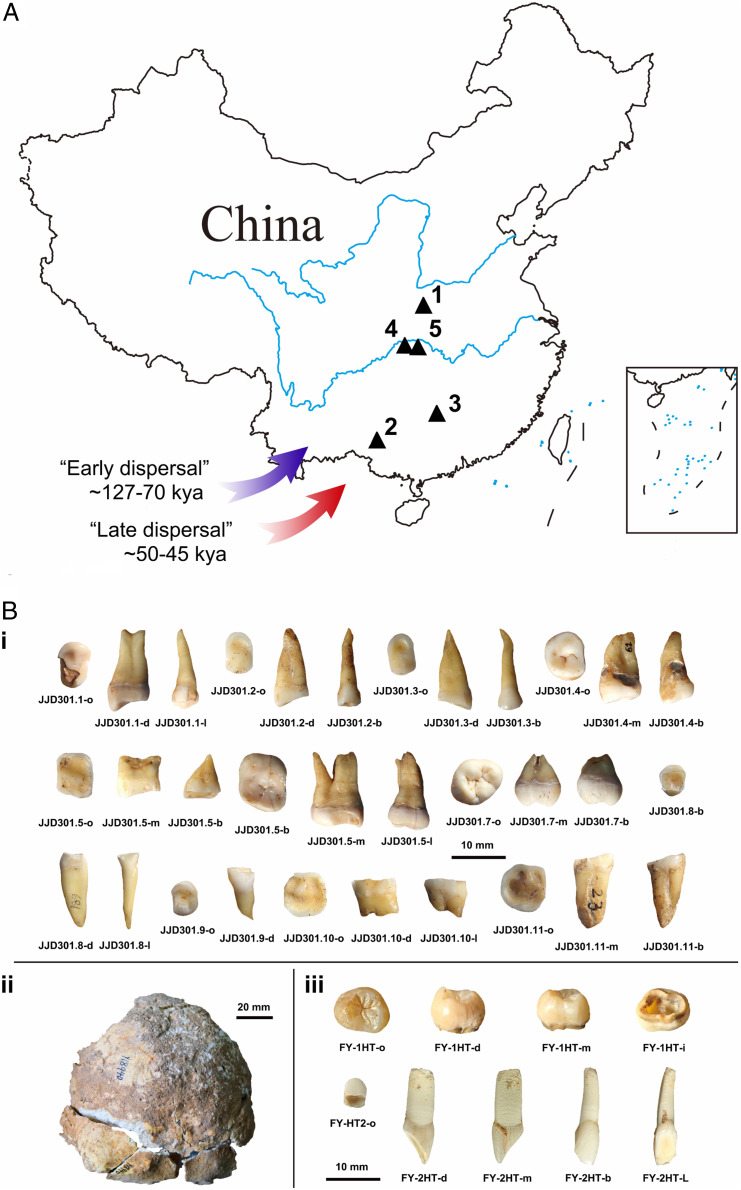
Fig. 1.
(A) Geographical location of Huanglong Cave (1), Luna Cave (2), Fuyan Cave (3), Yangjiapo Cave (4), and Sanyou Cave (5). (B) Human remains from three localities: Yangjiapo Cave (i), Sanyou Cave (ii), and Fuyan Cave (iii). b = buccal, d = distal, l = lingual, m = mesial, and o = occlusal).
Results
aDNA.
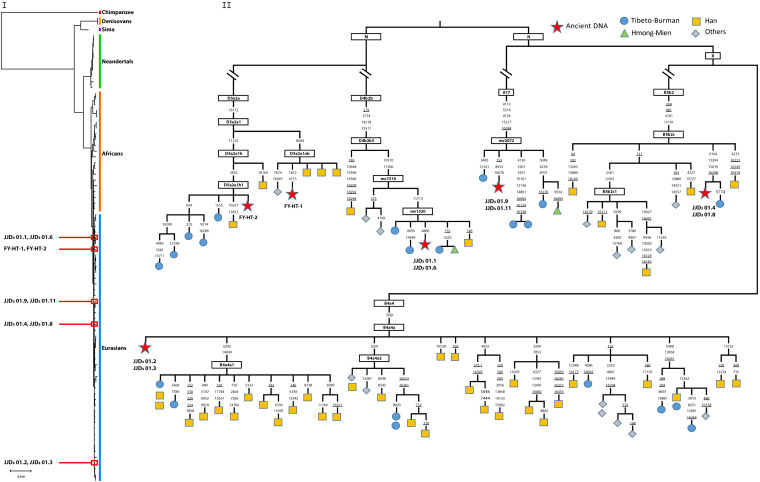
Our approach was to attempt DNA extraction on human remains from Yangjiapo, Sanyou, and Fuyan caves (samples from Huanglong and Luna caves were unavailable). We were successful in performing DNA extraction, library creation, targeted capture, and next generation sequencing on eight human teeth with nearly complete roots from Yangjiapo Cave and two collected in situ from Fuyan Cave (Fig. 2 I and II and SI Appendix, Fig. S1). Together, they yielded 410,451 to 969,814 and 3,619 to 43,679 unique mitochonridal DNA (mtDNA) fragments, respectively (Table 1). Based on aDNA sequence authenticity, we identified four unique sequences across the eight Yangjiapo teeth, which were assigned to haplogroups D4b2b5 (samples JJD301.1 and JJD301.6), B4a4a (JJD301.2 and JJD301.3), B5b2c (JJD301.4 and JJD301.8), and A17 (JJD301.9 and JJD301.11) (Fig. 2II). The Fuyan teeth represented two unique sequences, which belonged to the sub-sub-lineages of haplogroup D5a2a: D5a2a1ab (FY-HT-1) and D5a2a1h1 (FY-HT-2) (Fig. 2II). We then built a maximum parsimony tree using the four Yangjiapo and two Fuyan mtDNA sequences, sequences from 53 recent humans of diverse geographical origin, 10 ancient AMH sequences, 10 Neanderthals, 2 Denisovans, a hominin from Sima de los Huesos, and a chimpanzee. The Yangjiapo Cave and Fuyan mtDNA fell within the variation of present-day Eurasian lineages (Fig. 2I). Surprisingly, the mtDNA lineage found in the Fuyan sample FY-HT-2 has been detected in living Tibeto–Burman populations, revealing potential genetic links to them (Fig. 2II).
Fig. 2.
Phylogenetic trees incorporating complete human mtDNA sequences from Yangjiapo Cave and Fuyan Cave. (I) Phylogenetic analysis of the eight Yangjiapo Cave and two Fuyan Cave mtDNA genomes, inferred using the neighbor-joining method. The eight Yangjiapo Cave and two Fuyan Cave mtDNA genomes fall within the cluster of modern humans and outside of the cluster containing Neanderthals, Denisovans, and a hominin from Sima de los Huesos. (II) mtDNA Haplogroup of the Yangjiapo Cave and Fuyan Cave individuals (red stars) and their closest present-day relatives collected from this study and published data (SI Appendix, Table S4). The polymorphic positions are indicated on branches; recurrent mutations are underlined.
Table 1.
General characteristics of eight Yanjiapo DNA libraries and two Fuyan DNA libraries
| Sample ID | Extract ID | Library ID | Unique reads | Average depth | Contamination | 5′-end C→T ratio | 3′-end G→A ratio |
| JJD3 01.1 | A92601 | G151015R00101 | 9246 | 38.27 | 0.44% | 4.75% | 5.03% |
| JJD3 01.6 | A92602 | G151015R00201 | 21420 | 108.16 | 0.74% | 6.46% | 6.16% |
| JJD3 01.2 | AB2901 | G160122R02701 | 22649 | 129.76 | 0.97% | 9.25% | 8.26% |
| JJD3 01.3 | AB2902 | G160122R02801 | 29663 | 195.07 | 0.92% | 8.92% | 7.24% |
| JJD3 01.4 | AB2903 | G160122R02901 | 30569 | 226.25 | 1.18% | 4.48% | 4.05% |
| JJD3 01.8 | AB2904 | G160122R03001 | 24309 | 137.62 | 1.17% | 7.25% | 5.65% |
| JJD3 01.9 | AB2905 | G160122R03101 | 30346 | 222.13 | 0.55% | 7.55% | 5.89% |
| JJD3 01.11 | AB2906 | G160122R03201 | 31197 | 257.37 | 0.84% | 7.00% | 5.97% |
| FY-HT-1 | E42303 | M197149 | 43697 | 344.62 | 3.32% | 0.67% | 0.73% |
| FY-HT-2 | E42302 | M197148 | 3619 | 29.17 | 0.44% | 3.61% | 2.79% |
The coalescence time of the Yangjiapo and Fuyan lineages was estimated using maximum likelihood and ρ–statistic-based methods. Using mtDNA sequences from lineages related to those found in these samples, the clade divergence times were estimated to be 3.63/3.36 ka (D4b2b5), 11.01/10.91 ka (B4a4a), 12.40/14.38 ka (B5b2c), 15.61/12.20 ka (A17), 1.69/1.29 ka (D5a2a1ab), and 7.44/6.68 ka (D5a2a1h1) (SI Appendix, Table S1I part A). These results suggest that, together, the maximum age of the Yangjiapo and Fuyan samples is less than ca. 15.6 ka. Tip dates were also estimated under a Bayesian framework implemented in BEAST (BEAST software). The Yangjiapo sequences were dated 2.4 ka (samples JJD301.1 and JJD301.6), 2.7 ka (JJD301.2 and JJD301.3), 3.0 ka (JJD301.4 and JJD301.8), and 7.6 ka (JJD301.9 and JJD301.11), with similar ages for the Fuyan Cave teeth of 3.7 ka (FY-HT-1) and 12.0 ka (FY-HT-2) (SI Appendix, Fig. S1).
Multimethod Dating.
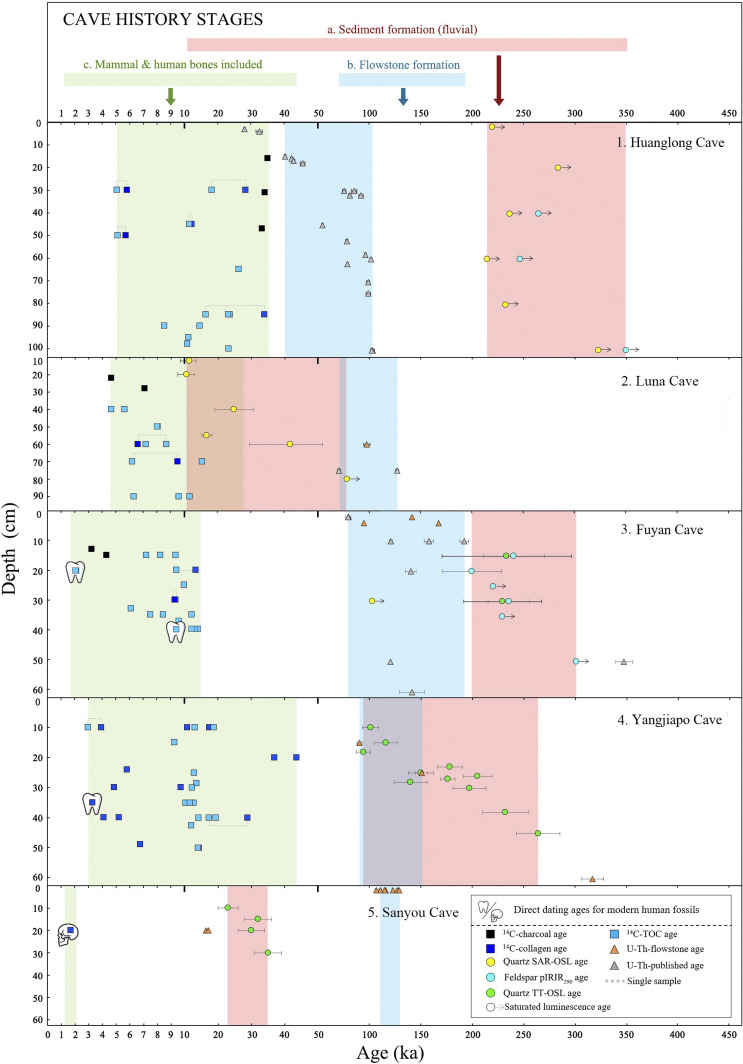
We visited the five cave sites with local archeologists who were involved in the original excavations in order to reassess the sedimentary context and stratigraphic framework of each and to collect in situ sediment, charcoal, and fossil samples for a multimethod dating analysis (see Methods). For Huanglong Cave, we performed optically stimulated luminescence (OSL) dating on six sediment samples from the human fossil-bearing unit (Layer 3 and ref. 20), and they all produced saturated ages of >215 ka (Fig. 3 and SI Appendix, Figs. S2 and S3 and Table S1). These results contrast with published Late Pleistocene U–Th ages on flowstone at the cave, which were previously used to constrain the age of the human remains to ca. 103 to 81 ka (SI Appendix, Table S2; ref. 20). Four mammal teeth (HLD-4, HLD-5, HLD-9, and JHLD-30) and one bone (HLD-45) from Layer 3 yielded collagen > weight 1% weight, judged from % N and C:N ratios, and they provided AMS 14C ages between 33,910 and 34,340 cal. y B.P. and 5,840 to 5,620 cal. y B.P. (at 68.2% probability) (Fig. 3 and SI Appendix, Fig. S2 and Table S3). Despite yielding low collagen wt.%, the remaining eight teeth from this unit provided AMS 14C ages from 25,940 to 26,700 cal. y B.P., to 8,620 to 8,450 cal. y B.P. (at 68.2% probability) (Fig. 3 and SI Appendix, Fig. S2 and Table S3). Thirteen mammal teeth dated with AMS 14C were identified to the species level and represented either an extant taxon (Bubalis bubalis) and Cervus sp. or a lineage which disappeared during the Holocene (Rhinoceros sinensis) (SI Appendix, Table S3). Three fragments of charcoal samples collected by us in situ from Layer 3 were also dated with AMS 14C within the range 34,850 to 35,540 cal. y B.P., to 33,920 to 33,290 cal. y B.P. (at 68.2% probability) (Fig. 3 and SI Appendix, Fig. S2 and Table S3), supporting ages from the mammal teeth.
Fig. 3.
Multimethod dating results for southern Chinese paleoanthropological cave localities. 1) Huanglong Cave, 2) Luna Cave, 3) Fuyan Cave, 4) Yangjiapo Cave, and 5) Sanyou Cave. Three stages (a, b, and c) in the history of each cave are also indicated (see text for discussion). Error bars are measurement errors. ka is thousands of years ago.
At Luna Cave, we collected flowstone from the same level (70 to 60 cm), as previously sampled for U–Th dating and close to the depth where the two AMH teeth were recovered (70 to 65 cm) (SI Appendix, Fig. S2; ref. 21). Our U–Th dating yielded an age of 97 ± 3 ka (Fig. 3 and SI Appendix, Fig. S2 and Table S2), which is significantly younger than the previously reported age of 127 ± 2 ka (21). We then applied OSL dating to six sediment samples collected from depths ranging 80 to 10 cm and they provided ages of >78 ka (saturated) to 11 ± 2 ka (Fig. 3 and SI Appendix, Figs. S2 and S3 and Table S1). The stratigraphically closest OSL dates to the AMH teeth were dated >78 ka (80 cm) and 42 ka (60 cm), respectively. Two of the nine mammal teeth and two bone samples yielded sufficient collagen for AMS 14C dating, and they provided ages in the range of 9,530 to 9,420 cal. y B.P. (LND-C-6-2) and 6,710 to 6,490 cal. y B.P. (LND-C-6-4) (at 68.2% probability) (Fig. 3 and SI Appendix, Fig. S2 and Table S3). Despite yielding low collagen wt.%, the remaining teeth provided ages ranging from 15,500 to 14,860 cal. y B.P. to 4,780 to 4,530 cal. y B.P. (at 68.2% probability) (Fig. 3 and SI Appendix, Fig. S2 and Table S3). The nine mammal teeth used for 14C dating were identified to the species level and represented an extant taxon (five Sus scrofa, two Bovidae, and two Cervus sp.) (SI Appendix, Table S3). Additionally, two charcoal samples collected by us, from sediments immediately above the human fossil layer (32 to 25 cm) suggested a depositional time for them between 7,160 and 7,040 cal. y B.P. and 4,780 to 4,550 cal. y B.P. (at 68.2% probability) (Fig. 3 and SI Appendix, Fig. S2 and Table S3).
We collected speleothem samples from Layer 1 of Fuyan Cave, which sits immediately above the unit which yielded 47 AMH teeth (i.e., Layer 2; SI Appendix, Fig. S4; ref. 22). Our U–Th dating on three flowstone samples provided varying ages of 142 ± 2 ka, 95 ± 1 ka, and 168 ± 2 ka (Fig. 3 and SI Appendix, Fig. S4 and Table S2). We then produced OSL ages on six sediment samples from Layer 2, and they ranged from > 302 ka (saturated age) to 200 ± 29 ka (Fig. 3and SI Appendix, Figs. S3 and S4 and Table S1). After prescreening (collagen > 1% wt.), we applied AMS 14C dating to 16 teeth, including two human teeth (FY-HT-1 and FY-HT-2) collected by us in situ and which were subjected also to aDNA analysis (see above; Fig. 1). The collagen ages of two teeth (FY3-1 and FY3-5) are 13,590 to 13,350 cal. y B.P. and 9,390 to 9,160 cal. y B.P., respectively. Reassuringly, the collagen ages are the same as the total organic carbon (TOC) ages indicating good quality for the dating results. The human teeth, which are clearly AMH and fit metrically and morphologically within the range of earlier finds from the site, provided ages of 9,479 to 9,290 cal. y B.P. to 2,670 to 2,370 cal. y B.P. (at 68.2% probability) (Fig. 3 and SI Appendix, Fig. S4 and Table S3). Human tooth FY-HT-1 was dated twice, and nearly identical results were obtained in both instances. The additional, low collagen, tooth samples provided a broadly comparable range of 15,290 to 14,660 cal. y B.P. to 6,210 to 6,050 cal. y B.P. (at 68.2% probability) but should be interpreted cautiously (Fig. 3 and SI Appendix, Fig. S4 and Table S3). A total of 14 mammal teeth besides H. sapiens used for 14C dating were identified to the species, and all of them represented an extant taxon (2 Sus scrofa, 2 Hystrix subcristata, and 10 Cervus sp.) (SI Appendix, Table S3). Two charcoal samples collected by us in situ provided AMS 14C ages of 4,410 to 4,300 cal. y B.P. and 3,330 to 3,230 cal B.P. (at 68.2% probability), supporting the AMS 14C dating results for the human and mammal samples of Holocene age (Fig. 3 and SI Appendix, Fig. S4 and Table S3).
Details of the stratigraphic section for two sampling areas within Yangjiapo Cave are provided in SI Appendix, Fig. S5. The 11 human teeth (Fig. 1) all clearly represent AMH in terms of crown and root morphology, crown metrics, and aDNA (see above). To estimate their geological age, we initially applied U–Th dating on speleothem samples from the base of the sequence (Layer 4) and from within the sediment containing the human teeth (Layer 2). They provided ages of 317 ± 11 ka (Layer 4), and 151 ± 6 ka and 90 ± 3 ka (Layer 2) (Fig. 3 and SI Appendix, Fig. S5 and Table S2). OSL dating on seven sediment samples obtained from Layer 2 provided age estimates between 205 ± 14 ka and 94 ± 7 ka (Fig. 3 and SI Appendix, Figs. S2 and S5 and Table S1). Thus, combining the results of U–Th dating of speleothem and OSL dating of sediments would place these AMH teeth within the range 205 to 90 ka. However, we assessed 16 mammal teeth for AMS 14C dating at the NTUAMS Lab but found only 2 yielded sufficient collagen, and they provided ages of 29,390 to 28,550 cal. y B.P. (YJP-1054) and 4,050 to 3,850 cal. y B.P. (YJP-2936) (at 68.2% probability) (Fig. 3 and SI Appendix, Fig. S5 and Table S3). The remaining (low wt.% collagen) samples provided a wide range between 19,800 to 19,100 cal. y B.P. and 9,410 to 9,170 cal. y B.P., (Fig. 3 and SI Appendix, Fig. S5 and Table S3). Collagen was extracted from a further 11 bone samples by Beta Analytic and yielded 14C ages from 44,370 to 43,090 cal. y B.P. to 4,900 to 4,850 cal. y B.P. We also ran AMS 14C dating on a single human tooth (HBJS-DCD-H-T-488) from Yangjiapo Cave (sample JJD301.11), which was also subject to aDNA analysis, and it was found to be of Holocene age: 3,370 to 3,280 cal. y B.P. (at 68.2% probability) (Fig. 3 and SI Appendix, Fig. S5 and Tables S3 and S4). A total of 16 mammal teeth besides H. sapiens were identified to the species level, and all of them represented either an extant taxon (Sus scrofa, Hystrix subcristata, and Cervus sp.) or lineage which disappeared during the Holocene Epoch (Rhinoceros sinensis) (SI Appendix, Table S3).
The final locality in our study is Sanyou Cave, which has provided an occipital fragment belonging to an AMH. Here, the stratigraphic sequence is straightforward and comprises just a single sedimentary unit, with the human remains (Fig. 1) recovered at a depth of 20 cm (SI Appendix, Fig. S4). We applied U–Th dating to seven flowstone samples overlying the human remains and sealing in the deposits, and they yielded ages of between 129 ± 0.9 ka and 107 ± 0.9 ka (Fig. 3 and SI Appendix, Fig. S4 and Table S2). Additionally, three small stalactites dislodged from the roof and recovered from within the sediment at a depth of 20 cm were dated with the U–Th method. The resulting ages were between 17 ± 0.1 ka to 16 ± 0.1 ka, indicating they had formed well after the capping flowstone and possibly coincident with the formation of the sediment (Fig. 3 and SI Appendix, Fig. S4 and Table S2). However, OSL dating of four sediment samples collected 150 cm away from the human recovery location produced a vertical sequence of the following ages: 35 ± 4 ka (30 cm), 30 ± 4 ka (20 cm), 32 ± 4 ka (15 cm), and 23 ± 3 ka (10 cm) (Fig. 3 and SI Appendix, Figs. S3 and S4 and Table S1). Finally, a bone sample from the human occipital was dated with AMS 14C and yielded an age of 1,730 to 1,640 cal. y B.P. (at 68.2% probability) (Fig. 3 and SI Appendix, Fig. S4 and Table S3).
Discussion
Our research highlights some of the problems which can arise from an overreliance on a single method to place a timeframe around key events in human evolution. In this instance, the use of U–Th dating on cave flowstone to infer the arrival time of AMHs in southern China has produced erroneously old ages and resulted in a false conflict between fossil and genetic data. While some paleoanthropologists believe that AMHs settled this region during the interval ca. 120 to 70 ka, our research shows otherwise. We found that in the five cave sites we investigated, U–Th dating of cave flowstones has misled researchers over the dating of human remains and the suggested early arrival time does not hold up to scrutiny. When we sequenced mtDNA from two human teeth stratigraphically associated with 47 earlier finds from Fuyan Cave and eight teeth from Yangjiapo Cave, all of them provided a maximum coalescence age <16 ka. Tip dates from them (12.0 to 2.4 ka) were also an order of magnitude younger than ages inferred from the U–Th dating of flowstones (ca. 151 to 90 ka). As would be expected, AMS 14C dates of human teeth from Fuyan Cave (2,510 ± 140 cal. y B.P., 2,540 ± 130 cal. y B.P., and 9,380 ± 90 cal. y B.P.) are somewhat younger than their molecular tip dates (3.7 ka, 3.7 ka, and 12.0 ka, respectively) (SI Appendix, Table S4). The same situation applied to a single human tooth from Yangjiapo Cave (AMS 14C age 3,310 ± 75 cal. y B.P.) (tip date, 7.6 ka) (SI Appendix, Table S4). Still, it is clear that they were all buried in cave sediments during the Holocene and not 120 to 80 ka, as suggested previously for Fuyan Cave (22).
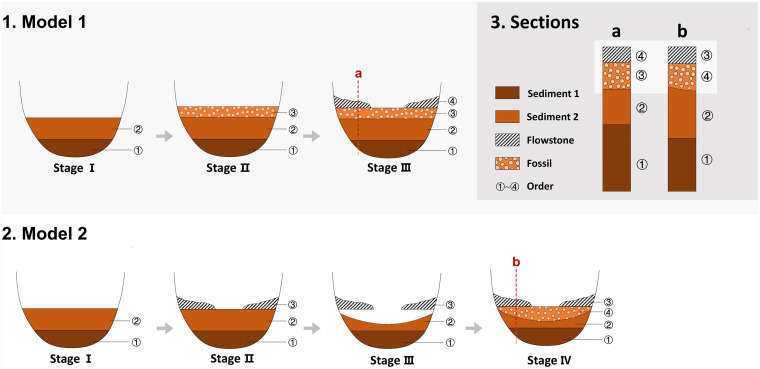
OSL dating of sediments and AMS 14C dating of fossils and charcoal from these five caves also highlighted major discrepancies with flowstone ages. At only one site did sediment ages match those from the flowstones, with major inconsistencies found at all of the other localities. Similarly, the assumed contemporaneity of the fossils, including AMH remains, and flowstones was shown to be incorrect at all five localities, with the offset between them an order of magnitude in most cases. The final assumption we tested was that the fossils and charcoal were the same age as the sediments containing them. This was found to be broadly correct at Luna Cave but was rejected for Huanglong, Fuyan, Yangjiapo, and Sanyou caves, where the sediments were substantially older than the fossils and charcoal they enclosed. Such a large offset in ages across the various samples highlights a surprisingly complex depositional history at all of these localities involving episodes of speleogenesis, sedimentation, and erosion/redeposition (Figs. 3 and 4). The formation of flowstones represents a fixed point in time and, therefore, provides a static chronological marker. They formed, for the most part, in these caves during the warm and humid Marine Isotope Stage 5 (Figs. 3 and 4). Sediment formation, on the other hand, is a much more dynamic process and represents differences in the hydrological regime operating at each locality through time (Fig. 4). The simplest case we observed was at Yangjiapo Cave, a large chamber with an underground stream, wherein the flowstones and sediments formed during the later Middle to early Late Pleistocene, but the mammal fossil remains were deposited from 43.6 ka (perhaps earlier, but beyond the range of 14C) until the late Holocene. Such a situation can only be explained by multiple episodes of relatively low energy water induced erosion and redeposition within a dark cave without resetting of the OSL clock. A complex depositional history must have prevailed at all five cave localities because the fossil teeth (and, where present, charcoal) was found to always be much younger than the flowstones, and at four of the caves, the sediments enclosing them as well. A further explanation could be the intrusion of younger fossils into older deposits through small scale erosional events, sediment collapse, bioturbation, or anthropogenic disturbance during the Holocene.
Fig. 4.
Two models of depositional history for cave sites in southern China and their effect on the exposed sedimentary profile. Sections a and b belong to the same profile, but the layers occur in alternate depositional order.
Also relevant here is the human mandible from Zhirendong in Guangzhi Zhuang Automonous Region, previously dated stratigraphically using U–Th analysis on flowstones to ≥100 ka (23) and most recently to ca. 190 to 130 ka (24). In the absence of direct dating, we believe the lessons drawn from our analysis of these five other karst caves in southern China might also apply to Zhirendong, and we await direct dating or aDNA analysis before we can be confident of its age (see also refs. 26, 27). Additionally, the classification of the Zhirendong mandible continues to be controversial despite some researchers insisting it represents an early AMH (23, 28, 29). Its two preserved molars are heavily worn, raising doubts about the identification of morphological traits and supposed affinities to AMHs. In contrast, the fossil shows many resemblances to archaic taxa, including in its body thickness and shape (23) and the absence of the distinctive anatomical components of a true AMH chin. Finally, a long-term trend toward dental gracilization, paralleling that seen in H. sapiens, has also been documented for H. erectus in Indonesia and among Middle Pleistocene hominins in China (30), and this might explain dental similarities between the Zhirendong individual and early AMHs.
Several other Late Pleistocene cave localities in southern China are of interest because they have provided evidence for either early AMH or human remains with unexpected morphologies. At Liujiang, an AMH cranium has been dated stratigraphically from U–Th analyses of flowstone within the range of ca. 139 to 68 ka despite ongoing uncertainties surrounding its provenience (31, 32). While at Longtanshan 1, two heavily worn teeth, possibly sampling AMHs, have been dated through U–Th analysis of flowstone and mammal bone to a minimum age of ca. 83 to 60 ka (33). In contrast, human remains from Maludong, Dushan Cave, and Longlin (Laomaocao) Cave exhibit either archaic or a mosaic of archaic and AMH unqiue traits and have been indirectly dated to the period ca. 15 to 11 ka (34–38). However, a recent study utilizing conventional and laser ablation U–Th dating of deer bones and teeth has determined that some fossils from Maludong are in fact Middle Pleistocene in age (39). Thus, this recent work at Maludong combined with the results of our present study strongly implies that most Pleistocene paleoanthropological caves in southern China are likely to exhibit a more complex depositional history than has been assumed (39). We would stress, therefore, that until direct dating, preferably using AMS 14C or aDNA analyses, is successfully conducted on these human remains, their age should probably be regarded as uncertain.
Further afield, human remains from Tam Pa Ling in Laos and Lida Ajer in Indonesia, as well as stone artifacts from Kota Tampan in Peninsular Malaysia, might also support an early arrival for AMHs across the broad region (40–42). The only human remains from Tam Pa Ling possibly dating >48 ka is a partial mandible (TPL3) (40). However, we would consider the OSL quartz age (48 ± 5 ka) associated with this jaw to be more reliable than the infrared stimulated luminescence (IRSL) feldspar age (70 ± 8 ka). At Lida Ajer, a sample of mammal remains, including two AMH teeth recovered by Dubois in the 1880s, have recently been dated between 73 and 63 ka using coupled U-series/ESR (electron spin resonance) dating (41). While this stratigraphic age has received guarded support (26), greater confidence would be provided by the direct AMS 14C dating or aDNA analysis of the human remains. Finally, a recent reanalysis of artifacts from Kota Tampan, dated to ca. 70 ka through OSL dating of enclosing sediments, suggests they were likely to have been made by AMHs (42). Still, we note that further work is required to understand the depositional history of the site and to assess the relationships between the artifacts and apparently reworked and dated Toba Ash deposits.
In light of our findings, we conclude that claims for an early arrival of AMHs in southern China as seemingly documented at Huanglong, Luna, and Fuyan caves cannot be substantiated at present. Instead, we find that the earliest evidence for AMHs in the region is less than ca. 35 ka, in line with molecular estimates of ca. 50 to 45 ka (7–14, 16–19, 26). It is also noteworthy that two other instances of the direct dating AMHs from China have resulted in purported Late Pleistocene remains being confirmed as Holocene in age (i.e., Sulawasu and Gaozi) (43, 44). Similar efforts in Europe have also seen skeletons once thought to be Late Pleistocene also redated to the Holocene (45). We add to this list for southern China Yangjiapo, Sanyou, and Fuyan caves. Our work provides an understanding of the challenges associated with reconstructing depositional histories in karst caves in subtropical areas like southern China, which includes episodes of erosion and redeposition, and possibly intrusion, which occurred as recently as the late Holocene and is detectible only through a comprehensive dating strategy. Moving forward, there is an urgent need for researchers working in the region to routinely adopt a multimethod strategy, including the targeting of human remains for direct AMS 14C dating and aDNA analysis.
Methods
aDNA.
aDNA work on eight Yangjiapo teeth (JJD301.1, JJD301.2, JJD301.3, JJD301.4, JJD301.6, JJD301.8, JJD301.9, and JJD301.11) and two Fuyan teeth (FY-HT-1 and FY-HT-2) was carried out in a dedicated aDNA facility at Fudan University, according to established precautions for working with ancient human DNA (46). For contamination monitoring, we included extraction negative controls (where no sample powder was used) and library negative controls (where extract was supplemented by water) in every batch of samples processed and carried them through the entire wet laboratory processing.
The Yangjiapo and Fuyan teeth were analyzed with permission from the relevant institute of Cultural Relics and Archaeology and the relevant excavators of the sites. Because human fossils are scarce and extremely valuable, any sort of destruction has to be kept to an absolute minimum. Following the removal of surface material, the root of each tooth was drilled using a Dremel tool and single-use drill bits, and about 30 mg powder was sampled for DNA analysis. No damage was done to the outer surface of the tooth. The bone powder of each sample was used to prepare 54 μL DNA extract using a silica-based DNA extraction protocol, optimized for the recovery of very short molecules from ancient biological material (47).
Next-generation sequencing libraries were prepared using a double-stranded protocol and partial uracil–DNA–glycosylase (UDG) treatment that retains some characteristic aDNA damage at the last nucleotide (48). Libraries were amplified with two indexing primers in four parallel PCRs using Herculase II Fusion DNA polymerase (Agilent). Amplification products from the same library were pooled and purified using the MinElute PCR purification kit (Qiagen). Library concentration was quantified on an Agilent 2100 Bioanalyzer DNA 1000 chip.
Target enrichment of the mitochondrial genome was performed on each amplified library using MyGenostics Human Mitochondria Capture Kit (MyGenostics Inc.), as described (49, 50), except that the hybridization and wash temperatures were lowered to 60 °C and 55 °C to facilitate enrichment of short library molecules (47). A final postenrichment amplification was performed for 15 cycles. The postenrichment amplified product was then quantified via qPCR. Sequencing was performed using an Illumina HiSEq. 2000 platform at MyGenostics Inc. The 100-bp (base pair) paired-end reads and a single index read were generated according to the manufacturer’s instructions.
Only sequences which perfectly matched the expected index combinations were retained. Sample reads were subsequently trimmed of adapter sequences and overlapping paired-end reads were merged into a single sequence using leeHom (51). We mapped the merged sequences (>35 bp) to the revised Cambridge Reference Sequence (NCBI reference sequence NC_012920.1) using BWA (Burrows-Wheeler Alignment) 0.7.12 (52) with parameters “aln –n 0.01 –o 2 –l 16500,” which disable read seeding and allow more mismatches and up to two gaps (53). Reads with mapping quality lower than 30 were discarded, and PCR duplicates were removed using SAMtools 0.1.29 (54). Only reads passing the above filtering were used for downstream analyses. A Genome Analysis Toolkit (55) was used with parameters “HaplotypeCaller BP_RESOLUTION” to obtain the coverage at each position. We used MIA (Mapping Iterative Assembler) (56) to reconstruct the complete mtDNA genome because it took into account the frequent sequence errors associated with base damage in aDNA sequences. Using these sequences and the above-mentioned parameters, we reconstructed eight Yangjiapo and two Fuyan mitochondrial genomes with average depth ranging from 29 to 344×.
The authenticity of the ancient genome sequence was mainly determined by the combination of two observations of the same specimen. First, deep sequencing of the mitochondrial genome indicates that the vast majority of the DNA fragments come from a single individual. Second, the patterns of DNA degradation, in particular nucleotide misincorporations resulting from deamination of cytosine residues at the ends of DNA fragments, indicate that the mtDNA is ancient.
The estimation of mitochondrial contamination mainly followed the method described in ref. 57. Because the mitochondrial genome is a circular molecule and BWA can only use linear references, drops in coverage are expected at the junction point (i.e., near the first and last positions of the mitochondrial genome). For eight Yangjiapo and two Fuyan samples, the average frequencies of the majority base at each position were between 99.75% and 99.88%. The low consensus support (around 50%) at some positions was due to misalignment around insertions/deletions or C-stretch. Hence, for each library, the majority of the mtDNA sequence is from the same individual. Both haplogroup defining mutations and private mutations of each sample were determined using HAPLOFIND (58). The postmortem DNA damage pattern was characterized using mapDamage 2.0 (59).
We performed phylogenetic inferences using four Yangjiapo and two Fuyan unique mitochondrial genome sequences constructed by MIA as described above. First, the D-loop (positions 16024 through 576) was removed, as it does not evolve at a constant rate across all human lineages and was usually excluded in previous studies (60, 61). Using MAFFT version 7, we aligned four constructed sequences to mtDNA sequences of 53 modern humans of diverse origins (62) together with mtDNA sequences of ten ancient modern humans (62–64), ten Neandertals (53, 56, 65–67), two Denisovans (68, 69), a hominin from Sima de los Huesos (70), and a chimpanzee (NC_001643.1) (71). We constructed a neighbor-joining tree using MEGA6 (72) and performed the test of phylogeny by 500 bootstrap replications. Mitochondrial haplogroups and defining mutations of six unique sequences were determined by using Haplofind (58).
By using previously described methods (73, 74), the coalescence time of four Yangjiapo lineages and two Fuyan lineages was estimated by using the maximum likelihood method and ρ-statistic–based method. Maximum likelihood estimates of coalescence time for major clades in each tree using PAML (Phylogenetic Analysis by Maximum Likelihood) v4.4 (75). For the ρ–statistic-based method, the time to the most recent common ancestor (TMRCA) of each lineage was estimated using the rate in ref. 76) and SDs were calculated following ref. 77). Tip dates of four Yangjiapo sequences and two Fuyan sequences were estimated under a Bayesian framework implemented in BEAST (78). We aligned ten modern humans and ten directly carbon-dated ancient modern humans mentioned above to four sequences together with all other sequences under the same lineage, respectively. Following ref. 77, we used the Hasegawa-Kishino-Yano model of nucleotide substitution with a gamma-distributed rate of heterogeneity and a fixed proportion of invariant sites (HKY+G + I). We tested models based on a strict molecular clock versus a log-normal uncorrelated relaxed clock using a constant or a coalescent Bayesian skyline population size as priors. A Markov chain Monte Carlo run with 30,000,000 iterations was carried out for each model, sampling every 1,000 steps. The first 6,000,000 iterations were discarded as burn-in. According to likelihood scores of different models, we adopted a model of strict clock and a prior of coalescent Bayesian skyline population size.
U–Th Analyses.
230Th/U dating has been conducted on flowstone and stalactite samples from the cave sites. The 230Th dating work was performed at the Isotope Laboratory, Xi’an Jaiotong University, using multicollector inductively coupled plasma mass spectrometers (Thermo Finnigan Neptune-plus). We used standard chemistry procedures to separate U and Th for dating (79). A triple spike (229Th–233U–236U) isotope dilution method was employed to correct for instrumental fractionation and determine U–Th isotopic ratios and concentrations. The instrumentation, standardization and half-lives are reported in Cheng et al. (79, 80). All U–Th isotopes were measured on a MasCom multiplier behind the retarding potential quadrupole in the peak jumping mode. We followed similar procedures of characterizing the multiplier as described in Cheng et al. (81). Uncertainties in U–Th isotopic data were calculated offline at 2 σ level, including corrections for blanks, multiplier dark noise, abundance sensitivity, and contents of the same nuclides in spike solution. Corrected 230Th ages assume the initial 230Th/232Th atomic ratio of 4.4 ± 2.2 × 10−6, the values for a material at secular equilibrium with the bulk earth 232Th/238U value of 3.8.
OSL Dating.
All measurements were carried out in the Luminescence Dating Laboratory at Nanjing University. Normal pretreatment methods were used (i.e., first, the two outer ends of the samples were removed under subdued red light and used for water content and dose rate measurements). Then, the middle unexposed section was treated with 10% HCl and 30% H2O2 to remove carbonates and organic matter, respectively. After this, the coarser grain size fraction (63 to 90 μm) was extracted by wet sieving and the pure quartz grains (no significant IRSL signals) were obtained by normally acid etch (40% hydrogen fluoride for 40 min, followed by 40 min with a 10% HCl rinse). Parts of the samples were prepared by 40% fluosilicic acid for 2 wk to obtain the pure 40 to 63 μm grains. For K-rich feldspar extraction, a portion of the initial 63 to 90 μm sieved fraction was cleaned with 10% HF for 15 min to remove coatings and the outer alpha irradiated layer and then rinsed in 10% HCl acid for 15 min to remove any precipitated fluorides. The grains were then separated in density heavy liquid. Quartz grains were mounted as large (8-mm) aliquots on stainless steel discs and K-rich feldspars as small (2-mm) aliquots in stainless steel cups.
All of the luminescence measurements were made using Risø TL/OSL readers model DA-20 (82), equipped with blue light-emitting diodes (LEDs) (470 nm, ∼80 mW cm2) and infrared (IR) LEDs (870 nm, ∼135 mW cm2). The OSL readers were also equipped with an accurately calibrated 90Sr /90Y beta source. Quartz OSL signals were collected through a 7.5-mm Schott U-340 (UV) glass filter, whereas feldspar (post-IR) IRSL was collected through a Corning 7-59 and Schott BG-39 glass filter combination.
The radionuclide concentrations were measured by the gamma spectrometry with a high purity Ge-detector (83) or neutron activation analysis. In situ water content (mass of moisture/dry mass) was determined by weighing the sample before and after drying and was assigned an absolute uncertainty of ± 7%. A small internal dose rate contribution from U and Th at 0.030 ± 0.015 and 0.06 ± 0.03 Gy/ka was also included for quartz and K-feldspar, respectively (84, 85). For the K-feldspar dose rates, K and Rb concentrations of 12.5 ± 0.5% and 400 ± 100 parts per million were also assumed for the internal dose rate calculation. Using the revised dose rate conversion factors of Guerin et al. (86) and water content attenuation factors (87), the elemental concentrations were converted into effective dose rate. Calculation of the cosmic dose rate is based on Prescott and Hutton (88).
The standard single aliquot regeneration dose optically stimulated luminescence (SAR-OSL) (89, 90) thermally transferred optically stimulated luminescence OSL (TT-OSL) (91) and post-IR IR stimulated luminescence (pIRIR290) (92–94) methods were used to dating the quartz and potassium rich feldspar grains from all these five caves. For SAR-OSL, the aliquots were primarily preheated for 10 s at 260 °C, while the response to the test dose were measured after a cut-heat to 220 °C, followed by the optical stimulation for 40 s at 125 °C. For the TT-OSL, using TT-OSL response to a test dose to correct for sensitivity changes in the TT-OSL signal, the two high temperature optical bleach (400 s at 280 °C) was also inserted in the middle and at the end of each SAR cycle to remove charge carryover. For K-feldspar pIRIR290, the aliquots were preheated at 320 °C for 60 s, followed by IR light at 290 °C after stimulation at 200 °C. All the dose–response curves were fitted using saturating exponential functions in Analyst version 4.31.7 (95).
AMS 14C.
Most of the fossil teeth were dated by Professor Hong-Chun Li in the NTUAMS Lab at National Taiwan University. An additional 14 teeth and seven charcoal samples were dated by Beta Analytic Inc. The surface of the teeth samples were initially cleaned mechanically. An ultrasound water bath and a standard acid-base acid (0.5 M HCl-0.1 M NaOH-0.5 M HCl) treatment were used to remove detritus, carbonates, and humus substances from the samples as much as possible. Then, the samples were dried and crushed into small pieces. Any detrital grains that could be observed were then removed. The crude collagen was extracted following the method of ref. 96. About 1 g treated sample was dissolved by 100 mL 1 M HCl for at least 24 h. After settlement, discarding the supernatant in the upper part of the sample, and the solution in the lower part containing “jelly like” collagen, was washed by deionized water 2 to 3 times to lower pH down to 2 to 3. Discarding the supernatant again, the solution was heated in a water bath at 80 °C, then the solution was filtered by a Whatman glass microfiber (∼1.6-μm pore size) filter (90-mm ∅ circle) to remove particles. The filtered solution was then evaporated to less than 10 mL and then transferred into a weighed vial, in which the sample turned to solid collagen by freeze drying. The whole procedure takes about one wk. Collagen percentage of the tooth sample and C% of the collagen were estimated by using the collagen weight, tooth sample weight, and carbon amount calculated from CO2 pressure. The N%, C%, δ15N, and δ13C of available collagen and tooth samples were measured by elemental analysis-isotopic ratio mass spectrometry (EA-IRMS). The TOC samples were also used for AMS 14C dating following the procedures described in ref 97). CO2 produced from the collagen and TOC was purified cryogenically on a vacuum line and sealed into combination tubes for graphitization, as described in ref. 86). The graphite sample targets were measured together with the targets made from oxalic acid standards (OXII, 4900C), backgrounds (NTUB is a pure limestone sampled from the upper Devonian stratum and BKG is an anthracite purchased from the US National Institute of Standards and Technology) and known age intercomparison samples in the NTUAMS Lab, with a 1.0 MV HVEE Tandetron Model 4110 BO- accelerator mass spectrometry. To avoid Lithium interference, we measured 14C3+ mode for 200 blocks (30 s per block). Measured 14C dates were calibrated by using the calibration curve IntCal13 (98).
Supplementary Material
Acknowledgments
We thank Guan-jun Shen, Jun-yi Ge, Qing-feng Shao, Hua Tu, Xiong-xin Yang, Wei Liao, Cheng-yun Xiao, Xu-bin Liu, Sheng-ming Qu, Qiong-xuan Zeng, Deng-ke Liu, You-feng Ning, Hong-yan Zhang, and Lu-peng Yu for their continual support, which made our work possible. X.-f.S. and H.-y.L. were supported by the Global Change Program of the Ministry of Science and Technology of China (2016YFA0600503) and the National Natural Science Foundation of China (41690111 and 41972185), S.-q.W. and H.L. were supported by the National Natural Science Foundation of China (91731303), and D.C. was supported by the Australian Research Council (FT120100168).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019158118/-/DCSupplemental.
Data Availability
The Yangjiapo and Fuyan sequences reported in this paper have been deposited in the GenBank database (accession nos. MH671321-MH671328 and MN308088-MN308089). All other study data are included in the article and/or SI Appendix.
References
- 1.Hublin J. J., et al., New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens. Nature 546, 289–292 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Hershkovitz I., et al., The earliest modern humans outside Africa. Science 359, 456–459 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Shea J. J., Transitions or turnovers? Climatically-forced extinctions of Homo sapiens and Neanderthals in the east mediterranean levant. Quat. Sci. Rev. 27, 2253–2270 (2008). [Google Scholar]
- 4.Hershkovitz I., et al., Levantine cranium from Manot Cave (Israel) foreshadows the first European modern humans. Nature 520, 216–219 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Stepanchuk V. N., 2006. The Lower and Middle Paleolithic of Ukraine (Nizhnii i srednii paleolit Ukrainy). Chernovtsy: Zelena Bukovina. (In Russian).
- 6.Hublin J. J., et al., Radiocarbon dates from the grotte du Renne and saint-césaire support a Neandertal origin for the châtelperronian. Proc. Natl. Acad. Sci. U.S.A. 109, 18743–18748 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Q., et al., A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 23, 553–559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mallick S., et al., The simons genome diversity project: 300 genomes from 142 diverse populations. Nature 538, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tishkoff S. A., et al., The genetic structure and history of Africans and African Americans. Science 324, 1035–1044 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeGiorgio M., Jakobsson M., Rosenberg N. A., Out of Africa: Modern human origins special feature: Explaining worldwide patterns of human genetic variation using a coalescent-based serial founder model of migration outward from Africa. Proc. Natl. Acad. Sci. U.S.A. 106, 16057–16062 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H., Prugnolle F., Manica A., Balloux F., A geographically explicit genetic model of worldwide human-settlement history. Am. J. Hum. Genet. 79, 230–237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran S., et al., Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. U.S.A. 102, 15942–15947 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Q., et al., Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514, 445–449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Q., et al., DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl. Acad. Sci. U.S.A. 110, 2223–2227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang H., Tong H., Zhang S., Chen F., Trinkaus E., An early modern human from Tianyuan Cave, Zhoukoudian, China. Proc. Natl. Acad. Sci. U.S.A. 104, 6573–6578 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sankararaman S., Patterson N., Li H., Pääbo S., Reich D., The date of interbreeding between Neandertals and modern humans. PLoS Genet. 8, e1002947 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moorjani P., et al., A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years. Proc. Natl. Acad. Sci. U.S.A. 113, 5652–5657 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seguin-Orlando A., et al., Paleogenomics. Genomic structure in Europeans dating back at least 36,200 years. Science 346, 1113–1118 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Jacobs G. S., et al., Multiple deeply divergent Denisovan ancestries in Papuans. Cell 177, 1010–1021.e32 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Shen G., et al., Mass spectrometric U-series dating of Huanglong cave in Hubei Province, Central China: Evidence for early presence of modern humans in eastern Asia. J. Hum. Evol. 65, 162–167 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Bae C. J., et al., Modern human teeth from late Pleistocene Luna cave (Guangxi, China). Quat. Int. 354, 169–183 (2014). [Google Scholar]
- 22.Liu W., et al., The earliest unequivocally modern humans in southern China. Nature 526, 696–699 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Liu W., et al., Human remains from Zhirendong, south China, and modern human emergence in East Asia. Proc. Natl. Acad. Sci. U.S.A. 107, 19201–19206 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge J., Deng C., Wang Y., Shao Q., Zhou X.et al., Climate-influenced cave deposition and human occupation during the Pleistocene in Zhiren Cave, southwest China. Quat. Int. 559, 14–23 (2020). [Google Scholar]
- 25.Michel V., et al., The earliest modern Homo sapiens in China? J. Hum. Evol. 101, 101–104 (2016). [DOI] [PubMed] [Google Scholar]
- 26.O’Connell J. F., et al., When did Homo sapiens first reach Southeast Asia and Sahul? Proc. Natl. Acad. Sci. U.S.A. 115, 8482–8490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun X., Lu Y., Wen S., Chronological problems in Chinese human fossil sites (In Chinese). Chin. Sci. Bull. 65, 2136–2144 (2020). [Google Scholar]
- 28.Dennell R., Palaeoanthropology: Early Homo sapiens in China. Nature 468, 512–513 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Schwartz J., What constitutes Homo sapiens? Morphology versus received wisdom. J. Anthropol. Sci. 94, 65–80 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Kaifu Y., et al., Taxonomic affinities and evolutionary history of the early Pleistocene hominids of java: Dentognathic evidence. Am. J. Phys. Anthropol. 128, 709–726 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Shen G., et al., U-Series dating of Liujiang hominid site in Guangxi, Southern China. J. Hum. Evol. 43, 817–829 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Wu X. Z., Poirier F. E., Human Evolution in China (Oxford University Press, New York, 1995). [Google Scholar]
- 33.Curnoe D., Xueping J., Hu S., Taçon P. S. C., Li Y., Dental remains from Longtanshan Cave 1 (Yunnan, China), and the initial presence of anatomically modern humans in East Asia. Quat. Int. 400, 180–186 (2016). [Google Scholar]
- 34.Curnoe D., et al., Human remains from the Pleistocene-Holocene transition of southwest China suggest a complex evolutionary history for East Asians. PLoS One 7, e31918 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xueping J., et al., Further geological and palaeoanthropolgocial investigations at the Maludong hominin locality, Yunnan Province, southwest China. (In English). Chin. Sci. Bull. 58, 4472–4485 (2013). [Google Scholar]
- 36.Curnoe D., et al., A hominin femur with archaic affinities from the Late Pleistocene of Southwest China. PLoS One 10, e0143332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Curnoe D., Ji X., Taçon P. S. C., Yaozheng G., Possible signatures of hominin hybridization from the early Holocene of Southwest China. Sci. Rep. 5, 12408 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao W., et al., Mosaic dental morphology in a terminal Pleistocene hominin from Dushan Cave in southern China. Sci. Rep. 9, 2347 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curnoe D., et al., Implications of multi-modal age distributions in Pleistocene cave deposits: Case study of Maludong palaeoanthropological locality, southern China. J. Archaeol. Sci. Rep. 25, 388–399 (2019). [Google Scholar]
- 40.Demeter F., et al., Early modern humans and morphological variation in southeast Asia: Fossil evidence from Tam Pa Ling, Laos. PLoS One 10, e0121193 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westaway K. E., et al., An early modern human presence in Sumatra 73,000-63,000 years ago. Nature 548, 322–325 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Goh H. M., et al., The palaeolithic stone assemblage of Kota Tampan, West Malaysia. Antiquity 94, e25 (2020). [Google Scholar]
- 43.Keates S. G., Hodgins G. W. L., Kuzmin Y. V., Orlova L. A., First direct dating of a presumed Pleistocene hominid from China: AMS radiocarbon age of a femur from the ordos plateau. J. Hum. Evol. 53, 1–5 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Fu R., et al., Modern Homo sapiens skeleton from Qianyang cave in Liaoning, northeastern China and its U-series dating. J. Hum. Evol. 55, 349–352 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Hoffmann A., Hublin J. J., Hüls M., Terberger T., The Homo aurignaciensis hauseri from combe-capelle: A mesolithic burial. J. Hum. Evol. 61, 211–214 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Knapp M., Clarke A. C., Horsburgh K. A., Matisoo-Smith E. A., Setting the stage–Building and working in an ancient DNA laboratory. Ann. Anat. 194, 3–6 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Dabney J., et al., Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U.S.A. 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohland N., Harney E., Mallick S., Nordenfelt S., Reich D., Partial uracil-DNA-glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20130624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.An W., et al., Mutation analysis of Chinese sporadic congenital sideroblastic anemia by targeted capture sequencing. J. Hematol. Oncol. 8, 55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ning C., et al., Ancient mitochondrial genome reveals trace of prehistoric migration in the east Pamir by pastoralists. J. Hum. Genet. 61, 103–108 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Renaud G., Stenzel U., Kelso J., leeHom: Adaptor trimming and merging for Illumina sequencing reads. Nucleic Acids Res. 42, e141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prüfer K., et al., The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and samtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKenna A., et al., The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Briggs A. W., et al., Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science 325, 318–321 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Cassidy L. M., et al., Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc. Natl. Acad. Sci. U.S.A. 113, 368–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vianello D., et al., HAPLOFIND: A new method for high-throughput mtDNA haplogroup assignment. Hum. Mutat. 34, 1189–1194 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Jónsson H., Ginolhac A., Schubert M., Johnson P. L. F., Orlando L., mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benazzi S., et al., Archaeology. The makers of the Protoaurignacian and implications for Neandertal extinction. Science 348, 793–796 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Ingman M., Kaessmann H., Pääbo S., Gyllensten U., Mitochondrial genome variation and the origin of modern humans. Nature 408, 708–713 (2000). [DOI] [PubMed] [Google Scholar]
- 62.Krause J., et al., A complete mtDNA genome of an early modern human from Kostenki, Russia. Curr. Biol. 20, 231–236 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Ermini L., et al., Complete mitochondrial genome sequence of the Tyrolean Iceman. Curr. Biol. 18, 1687–1693 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Gilbert M. T., et al., Paleo-Eskimo mtDNA genome reveals matrilineal discontinuity in Greenland. Science 320, 1787–1789 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Green R. E., et al., A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell 134, 416–426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gansauge M. T., Meyer M., Selective enrichment of damaged DNA molecules for ancient genome sequencing. Genome Res. 24, 1543–1549 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skoglund P., et al., Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc. Natl. Acad. Sci. U.S.A. 111, 2229–2234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krause J., et al., The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature 464, 894–897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reich D., et al., Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyer M., et al., A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature 505, 403–406 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Horai S., Hayasaka K., Kondo R., Tsugane K., Takahata N., Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs. Proc. Natl. Acad. Sci. U.S.A. 92, 532–536 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., Mega6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kang L., et al., mtDNA lineage expansions in Sherpa population suggest adaptive evolution in Tibetan highlands. Mol. Biol. Evol. 30, 2579–2587 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Zheng H. X., Yan S., Qin Z. D., Jin L., MtDNA analysis of global populations support that major population expansions began before Neolithic Time. Sci. Rep. 2, 745 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Soares P., et al., Correcting for purifying selection: An improved human mitochondrial molecular clock. Am. J. Hum. Genet. 84, 740–759 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saillard J., Forster P., Lynnerup N., Bandelt H. J., Nørby S., mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am. J. Hum. Genet. 67, 718–726 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Drummond A. J., Rambaut A., BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edwards R. L., Chen J. H., Wasserburg G. J., 238U, 234U, 230Th, 232Th systematics and the precise measurement of time over the past 500,000 years. Earth Planet. Sci. Lett. 81, 175–192 (1987). [Google Scholar]
- 80.Cheng H., et al., The half-lives of U-234 and Th-230. Chem. Geol. 169, 17–33 (2000). [Google Scholar]
- 81.Cheng H., et al., Improvements in 230Th dating, 230Th and 234U half-life values, and U–Th isotopic measurements by multi-collector inductively coupled plasma mass spectrometry. Earth Planet. Sci. Lett. 371, 82–91 (2013). [Google Scholar]
- 82.Murray A. S., Marten R., Johnston A., Martin P., Analysis for naturally occurring radionuclides at environmental concentrations by gamma spectrometry. J. Radioanal. Nucl. Chem. 115, 263–288 (1987). [Google Scholar]
- 83.Mejdahl V., Internal radioactivity in quartz and feldspar grains. Anc. TL 5, 10–17 (1987). [Google Scholar]
- 84.Vandenberghe D., De Corte F., Buylaert J. P., Kucera J., Van den Haute P., On the internal radioactivity in quartz. Radiat. Meas. 43, 771–775 (2008). [Google Scholar]
- 85.Zhao H., Li S. H., Internal dose rate to K-feldspar grains from radioactive elements other than potassium. Radiat. Meas. 40, 84–93 (2005). [Google Scholar]
- 86.Guérin G., Mercier N., Adamiec G., Dose-rate conversion factors: Update. Anc. TL 29, 5–8 (2011). [Google Scholar]
- 87.Aitken M. J., Thermoluminescence Dating (Academic Press, London, 1985). [Google Scholar]
- 88.Prescott J. R., Hutton J. T., Cosmic ray contributions to dose rates for luminescence and ESR dating: Large depths and long-term time variations. Radiat. Meas. 23, 497–500 (1994). [Google Scholar]
- 89.Murray A. S., Wintle A. G., Luminescence dating of quartz using an improved single-aliquot regenerative-dose protocol. Radiat. Meas. 32, 57–73 (2000). [Google Scholar]
- 90.Murray A. S., Wintle A. G., The single aliquot regenerative dose protocol: Potential for improvements in reliability. Radiat. Meas. 37, 377–381 (2003). [Google Scholar]
- 91.Stevens T., Buylaert J. P., Murray A. S., Towards development of a broadly- applicable SAR TT-OSL dating protocol for quartz. Radiat. Meas. 44, 639–645 (2009). [Google Scholar]
- 92.Li B., Li S. H., Luminescence dating of K-feldspar from sediments: A protocol without anomalous fading correction. Quat. Geochronol. 6, 468–479 (2011). [Google Scholar]
- 93.Thiel C., et al., Luminescence dating of the Stratzing loess profile (Austria) e Testing the potential of an elevated temperature post-IR IRSL protocol. Quat. Int. 234, 23–31 (2011). [Google Scholar]
- 94.Buylaert J. P., et al., A robust feldspar luminescence dating method for Middle and Late Pleistocene sediments. Boreas 41, 435–451 (2012). [Google Scholar]
- 95.Duller G. A. T., The Analyst software package for luminescence data: Overview and recent improvements. Anc. TL 33, 35–42 (2015). [Google Scholar]
- 96.Dunbar E., Cook G. T., Naysmith P., Tripney B. G., Xu S., AMS 14C dating at the scottish Universities environmental research centre (SUERC) radiocarbon dating laboratory. Radiocarbon 58, 9–23 (2016). [Google Scholar]
- 97.Xu X., et al., Modifying a sealed tube zinc reduction method for preparation of AMS graphite targets: Reducing background and attaining high precision. Nucl. Instrum. Methods Phys. Res. B 259, 320–329 (2007). [Google Scholar]
- 98.Niu M., Heaton T. J., Blackwell P. G., Buck C. E., The Bayesian approach to radiocarbon calibration curve estimation: The IntCal13, Marine13, and SHCal13 methodologies. Radiocarbon 55, 1905–1922 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Yangjiapo and Fuyan sequences reported in this paper have been deposited in the GenBank database (accession nos. MH671321-MH671328 and MN308088-MN308089). All other study data are included in the article and/or SI Appendix.