Abstract
Mitochondrial DNA mutations transmitted maternally within the oocyte cytoplasm often cause life-threatening disorders. Here we explore the use of nuclear genome transfer between unfertilized oocytes of two donors to prevent the transmission of mitochondrial mutations. Nuclear genome transfer did not reduce developmental efficiency to the blastocyst stage, and genome integrity was maintained provided that spontaneous oocyte activation was avoided through the transfer of incompletely assembled spindle–chromosome complexes. Mitochondrial DNA transferred with the nuclear genome was initially detected at levels below 1%, decreasing in blastocysts and stem-cell lines to undetectable levels, and remained undetectable after passaging for more than one year, clonal expansion, differentiation into neurons, cardiomyocytes or β-cells, and after cellular reprogramming. Stem cells and differentiated cells had mitochondrial respiratory chain enzyme activities and oxygen consumption rates indistinguishable from controls. These results demonstrate the potential of nuclear genome transfer to prevent the transmission of mitochondrial disorders in humans.
A crucial determinant of the phenotypic severity in most maternally inherited mitochondrial diseases is heteroplasmy, that is, the proportion of mutant, relative to total, mitochondrial DNA (mtDNA) in a cell. Owing to the cytoplasmic segregation of mitochondria during cell division, the level of heteroplasmy is subject to broad fluctuations, in particular during the developmental expansion of mtDNA from the premeiotic germ cell to the mature human oocyte1–4. As a result, an unaffected carrier of a mtDNA mutation may have an affected child. Although prenatal genetic diagnosis can select embryos with a reduced mutation load, variation between blastomeres in single embryos limits the effectiveness of such screening3, and considerable levels of mutant mtDNA can remain, resulting in a carrier5.
On the basis of these considerations, the Nuffield Council on Bioethics has endorsed research to prevent transmission of mtDNA mutations6, including the transfer of the nuclear genome into an enucleated oocyte containing normal mitochondria. In mice, transfer between fertilized eggs (zygotes) is effective in preventing the transmission of pathogenic mtDNA7 and in rhesus monkeys, genome exchange between unfertilized oocytes gave rise to live births8. In human cells, the transfer of pronuclei between zygotes resulted in minimal carryover of donormtDNA9. However, the exchange reduced developmental potential, possibly because the transfer introduced an abnormal centrosome number, resulting in multipolar spindles10 and aneuploidy11. As the centrosome is sperm-derived12, nuclear genome exchange before fertilization avoids this issue. Furthermore, pronuclear transfer requires the fertilization of both donor and recipient oocytes, resulting in the destruction of half of the embryos. By contrast, human oocytes would only be fertilized after successful genome exchange.
To determine the consequences of nuclear genome transfer in unfertilized oocytes, we chose parthenogenetic activation instead of fertilization, as it avoids the generation of human embryos for research (Supplementary Fig. 1). Transfer of the nuclear DNA between oocytes of two unrelated donors resulted in the exchange of the mitochondrial genotype and the elimination of mtDNA variants, including a variant found in the MT-TV gene (encoding mt-tRNAVal). However, nuclear genome transfer frequently induced premature oocyte activation and failure to extrude the second polar body normally. As manipulation-induced karyotypic abnormalities present a risk beyond those normally incurred during assisted reproductive technologies, this is probably an obstacle to the clinical translation of this technique. We found that premature activation could be prevented by partial depolymerization of the spindle–chromosome complex through cryopreservation or cooling to room temperature, allowing normal polar body extrusion, efficient development to the blastocyst stage, and the derivation of karyotypically normal stem cells. Therefore, nuclear genome transfer, rather than the transfer of intact spindle–chromosome complexes, should be effective in preventing the transmission of mtDNA mutations.
Efficient development after genome exchange
We first determined that artificial activation of unfertilized oocytes resulted in extrusion of the second polar body (Supplementary Fig. 2a) and efficient development to the blastocyst stage (Fig. 1a), allowing derivation of four stem-cell lines (parthenogenetic embryonic stem (pES) cells 2–5 (pES2–5); Supplementary Fig. 2b–o). If polar body extrusion had accurately segregated sister chromatids, a haploid zygote with 23 chromosomes should result in stem-cell lines devoid of heterozygosity. Short tandem repeat (STR) genotyping and Affymetrix single nucleotide polymorphism (SNP) arrays showed that four out of four stem-cell lines were homozygous for all chromosomes (Supplementary Fig. 2p and Supplementary Table1). All cell lines were diploid (SupplementaryFig.2q–t), suggesting that the genome had undergone endoreplication, as previously observed in mouse parthenotes13. These results suggest that parthenogenesis is appropriate for in vitro studies on the feasibility and consequences of oocyte genome exchange.
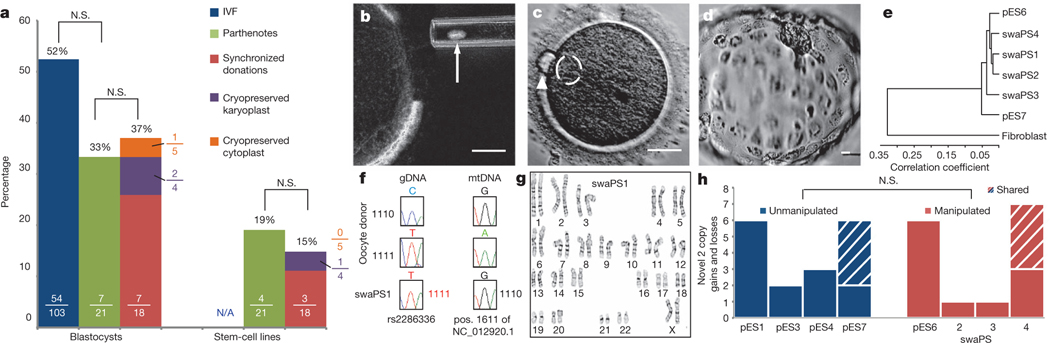
Figure 1 |. Efficient development and genomic integrity after nuclear genome exchange.
a, Developmental potential of IVF embryos, parthenogenic and genome-exchanged oocytes. Numbers in each bar displays the proportions of oocytes developing to the indicated stage. b, Removal of the karyoplast (indicated by arrow). c. Polar body extrusion (arrowhead) and pronuclear formation (dashed circle) at 4 h after activation d, Blastocyst stage at day 7 after activation. e, Cluster diagram of global gene expression profiles of swaPS cell lines. f, Sanger sequences of genomic DNA (gDNA) and mtDNA. g, Karyotype of swaPS1 cells at P3. h, Copy number variation analysis. N.S., not significant. Scale bars, 25 μm for all panels.
To exchange the nuclear genome between oocytes of two different donors, menstrual cycles were synchronized using oral contraceptives, with synchronized retrieval successful in all (four out of four) women. Four oocyte donors donated a total of 62 mature metaphase II (MII) oocytes (19, 17, 11 and 15 oocytes for the individual donors). Using microtubule birefringence, 18 of these oocytes had the nuclear genome removed (Fig. 1b) and subsequently fused to enucleated oocytes using either Sendai virus or an electrical pulse. Development after exchange was very efficient: of the 18 oocytes, seven developed to the blastocyst stage, with at least one blastocyst for each donor (Fig. 1a, d and Supplementary Fig. 3a–h). From these blastocysts, three swapped pluripotent stem (swaPS) cell lines were derived. swaPS cells expressed markers of pluripotency, had gene expression profiles comparable to those of established embryonic and parthenogenic embryonic stem-cell lines, and were able to differentiate into cell types and tissues of each germ layer (Fig. 1e and Supplementary Fig. 3i–t). Sequencing of mtDNA and nuclear DNA polymorphisms and STR genotyping confirmed that nuclear genome transfer had resulted in the exchange of the mitochondrial genotype (Fig. 1f, Supplementary Fig. 4a and Supplementary Tables 2 and 3).
To determine whether the transfer affected the integrity of the nuclear genome, karyotype analysis and high-resolution SNP arrays were used. Both swaPS1 and swaPS2 cells had normal karyotypes of 46,XX chromosomes, whereas swaPS3 cells contained an extra chromosome 12 (Fig. 1g and Supplementary Fig. 3u, v). The origin of this additional chromosome was due to a mitotic segregation error, as chromosome 12 did not contain regions of heterozygosity by SNP array analysis (Supplementary Information). Although trisomy 12 is a frequent artefact of in vitro stem-cell culture14, karyotyping of swaPS1 cells after 9 months in culture revealed no alterations (Supplementary Information). Manipulation-induced strain on chromosomes during genome exchange might result in chromosome breaks and faulty repair, as no homologous template is available after segregation of sister chromatids. Duplication during S phase would result in homozygous copy number variants (CNVs). We identified an average of 3.75 homozygous CNVs in five cell lines with spindle transfer and 4.25 in four unmanipulated parthenogenetic stem-cell lines (Fig. 1h). swaPS4 cells and the unmanipulated cell line pES7 originate from the same donor, and were found to share more than half of the CNVs, suggesting that most, if not all, CNVs originated in the female germ line, and were not generated by the manipulation. We also found no differences in the number of heterozygous CNVs (Supplementary Fig. 3x). To determine whether epigenetic alterations resulted in gene expression changes after genome transfer, gene expression in manipulated cell lines was compared to unmanipulated controls. Only one gene (THBS1, thrombospondin 1) was expressed at significantly lower levels (P < 0.01), with no genes having increased expression.
swaPS2 and swaPS3 cells were homozygous for 99.8% of the genome, consistent with accurate extrusion of a haploid genome into the polar body. The 0.2% of heterozygosity was due to CNVs that diverged in sequence (Supplementary Information). By contrast, the swaPS1cell line was homozygous for merely 43.3% of the genome (Fig. 2a and Supplementary Table 4), consistent with a cell line that had undergone the first, but not the second, meiosis15. To determine the frequency of chromosome segregation errors during the preimplantation stages, we biopsied polar bodies and blastomeres, or used morulas and blastocysts, for analysis. Using whole-genome amplification, followed by either PCR of loci on all 23 chromosomes or microarray-based analysis, we found that seven out of nine preimplantation embryos were homozygous throughout the genome, demonstrating normal polar body extrusion (Supplementary Fig. 4b–e). One was heterozygous for all chromosomes, whereas another showed a copy number gain and heterozygosity on chromosome 4, demonstrating that polar body extrusion had been inaccurate (Fig. 2b). Failed, or inaccurate extrusion of the polar body was probably due to the transfer procedure, as we did not observe any heterozygosity in unmanipulated stem-cell lines16 (Supplementary Fig. 2p).
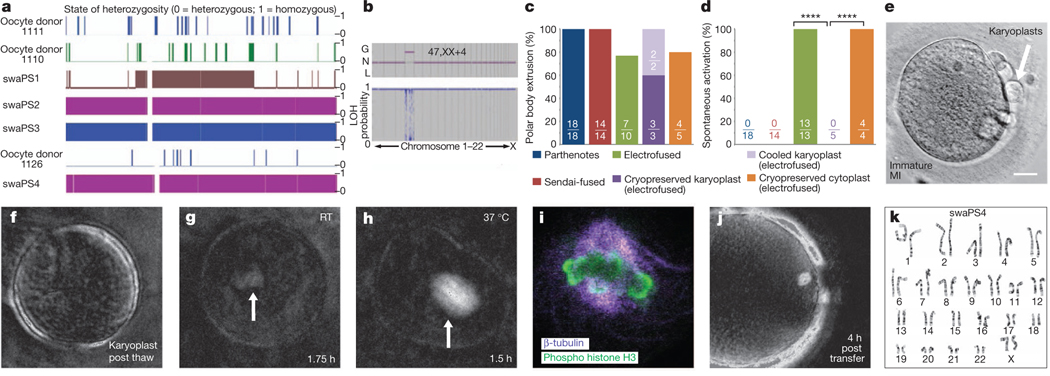
Figure 2 |. Spontaneous activation can be prevented through spindle cooling.
a, Affymetrix SNP microarray (chromosome 7). b, Array analysis at the cleavage stage, revealing trisomy on chromosome 4. G, gain; L, loss; LOH, loss of heterozygosity; N, normal. c, Frequency of polar body extrusion. d, Frequency of manipulation-induced activation. ****P < 0.0001. e, Karyoplasts after cryopreservation. Scale bar, 25 mm. f–h, Spindle birefringence at indicated temperatures and time points post thaw. Arrows indicate site of spindle. RT, room temperature. i, Confocal analysis of a thawed karyoplast after 2 h at room temperature. j, Spindle birefringence after transfer of a thawed karyoplast into an enucleated oocyte. k, Karyotype of stem-cell line derived from a cryopreserved karyoplast, swaPS4, at P4.
Immature spindles prevent spontaneous activation
Through transfer of karyoplasts into oocytes of either the same or a different donor, we determined that after transfer using electrical pulses, all oocytes (13 out of 13) formed one or two pronuclei 3–5 h after transfer, suggesting that the electrical pulse had caused premature exit from meiosis. Parthenotes with molecularly confirmed karyotypic abnormalities were all derived from oocytes that were activated as a result of the transfer. By contrast, none of the oocytes (0 out of 14) fused to karyoplasts using Sendai virus activated as a result of the manipulation. All remained at meiosis for 4–6 h after transfer, and extruded a second polar body only after artificial activation using a calcium ionophore followed by incubation in the translation inhibitor puromycin (Fig. 2c, d).
We reasoned that manipulation-induced activation was related to the spindle–chromosome complex. When oocytes without a spindle were exposed to an identical electrical fusion pulse during somatic cell nuclear transfer, they remained stable in meiosis16. Furthermore, when sperm is injected prematurely, within an hour after extrusion of the first polar body and before the assembly of a mature MII spindle, most oocytes fail to activate17. This suggests that after an activating stimulus, only mature spindles with bipolar attachment of chromosomes generate a signal promoting the exit from meiosis, thereby ensuring accurate chromosome segregation. To explore whether immature spindles prevented manipulation-induced activation, we monitored an oocyte progressing from metaphase I (MI) to MII and aspirated the nuclear genome within 1 h after extrusion of the first polar body, while the spindle was still immature (Supplementary Fig. 5a). Transfer-induced activation did not occur and only after artificial activation did the oocyte extrude the polar body and develop to the blastocyst stage, allowing derivation of a pluripotent stem-cell line (pES6) with a normal karyotype and homozygosity across the genome (Supplementary Fig. 5b–g and Supplementary Table 5).
As timing the removal of the nuclear genome relative to the extrusion of the first polar body may not be practical, we used reduced temperatures to induce partial spindle depolymerization in mature MII oocytes. As the removal of the spindle–chromosome complex relies on the presence of a birefringent spindle, we extracted the nuclear genome on the heated stage of a microscope, before exposing the karyoplast to reduced temperatures. Two karyoplasts were placed on ice for 2 h, and then re-transferred by electrofusion into enucleated oocytes kept at 37 °C. Fused oocytes remained in meiosis for more than 3 h post transfer and required an artificial activation stimulus for polar body extrusion. We next extracted karyoplasts from 30 oocytes and cryopreserved them below the zona pellucida of an immature oocyte (Fig. 2e). After thawing, 27 out of 30 karyoplasts were intact. Spindle birefringence was not detected immediately after thaw but reformed in karyoplasts kept at 37 °C for approximately 2 h (two out of two), but not when maintained at room temperature (none out of three) (Fig. 2f–h). Cryopreservation did not result in the dispersion of chromosomes: in all five karyoplasts, chromosomes remained attached to microtubules (Fig. 2i), consistent with the finding that some, but not all spindle microtubules are cold sensitive18. Within 1 h of thawing, three karyoplasts were fused to enucleated oocytes of a different donor using electrical pulses. At 4 h after transfer, we observed a birefringent spindle with perpendicular alignment to the oolemma (Fig. 2j). All (three out of three) oocytes remained stable at meiosis and extruded the second polar body only after artificial activation, forming a single pronucleus (Supplementary Fig. 6a, b). Two of the three parthenotes (66%) developed to the blastocyst stage, allowing the derivation of the karyotypically normal stem-cell line swaPS4 (Fig. 2k and Supplementary Fig. 6c–l). Homozygosity throughout the genome showed that polar body extrusion had normally segregated sister chromatids (Fig. 2a and Supplementary Tables 4 and 5). An additional thawed karyoplast was kept at 37 °C for 4 h, and only then fused to the oocyte, resulting in spontaneous activation and a failure to extrude the second polar body.
Furthermore, we vitrified MII oocytes. Immediately after thaw, birefringence of the microtubule spindle was undetectable in all oocytes (none out of six). After incubation for 1–2 h at 37°C, birefringence became visible in all oocytes, which were subsequently enucleated. Spindle–chromosome complexes from oocytes of a different donor retrieved on the day of thawing were transferred into five enucleated oocytes by electrofusion, with one karyoplast failing to fuse. One of the five oocytes developed to the blastocyst stage (Fig. 1a and Supplementary Fig. 6m–o). Unlike in oocytes after transfer of cryopreserved karyoplasts, oocytes underwent spontaneous activation (four out of four monitored during the relevant time period, Fig. 2d). These results demonstrate that the maturation of the spindle–chromosome complex has a major role in the ability of human oocytes to exit meiosis.
Stable exchange of mitochondrial genotypes
To determine mitochondrial genotypes, we identified polymorphic mtDNA variants (SNPs) in the hypervariable regions (HVR1 and HVR2) of each donor (Supplementary Table 6), and sequenced the complete mtDNA genome of two donors (Supplementary Table 7). Two polymorphisms, m.4715A>G (MT-ND2) and m.16129A>G (non-coding region), were homoplasmic by restriction fragment length polymorphism (RFLP). Moreover, one of the donors was homoplasmic for a variant at the 39 terminus of the MT-TV gene (m.1670A>T) that has been identified as a rare polymorphism19. Although no differences in the mt-tRNAVal steady-state level were observed in cultured fibroblasts, a functional significance is possible as an analogous 39 terminus polymorphism in mt-tRNAGlu has been identified as a pathogenic mutation20 (Supplementary Fig. 7a–d).
To estimate the amount of potential mtDNA carryover, we quantified mtDNA in human oocytes and karyoplasts using quantitative PCR (qPCR). Karyoplasts had an average mtDNA copy number of 1,1296±785 (mean ± s.d., n = 22), or 0.36% of the total mtDNA found within MII oocytes (311,146±6206,521, n = 5) (Fig. 3a). This corresponded with volumetric measurements of karyoplasts, which were 0.89% of that of intact oocytes (4,9616±1,964μm3 versus 559,093±245,217 μm3, respectively; n = 18 and 21) (Fig. 3a). Staining of mitochondria in oocytes and karyoplasts with MitoTracker or antibodies recognizing the complex V α-subunit indicated that the spindle–chromosome complex was devoid of mitochondria (Fig. 3b, c). We therefore expected a mtDNA carryover of less than 1%.
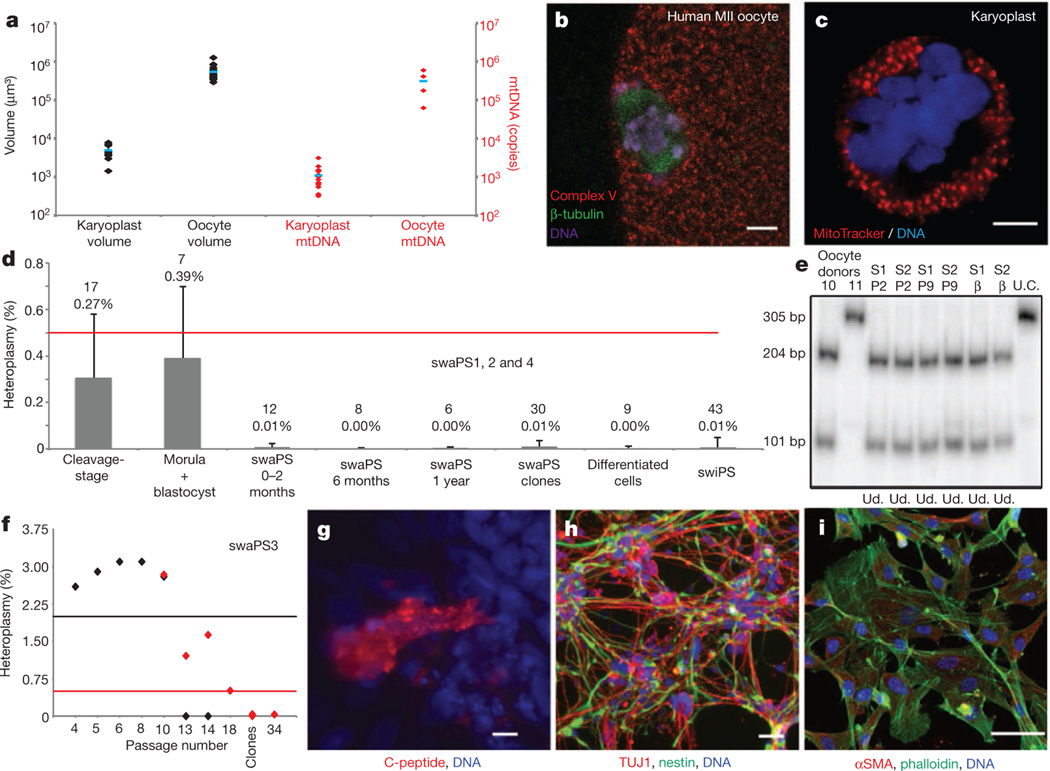
Figure 3 |. Low levels of mtDNA carryover.
a, Volume (black) and mtDNA copies (red) of karyoplasts and oocytes. Blue barsdenote themean. b, c, Distribution of mitochondria in the oocyte (b) and the karyoplast (c). d, Mean heteroplasmy quantification by ARMS-qPCR. Red line indicates limit of detection. Error bars indicate s.d., with the mean value and n number shown. e, RFLP analysis of swaPS1 and swaPS2 (S1 and S2, respectively) at P2 and P9 and as β-cells. bp, base pairs; U.C., undigested control; Ud., undetectable. f, Heteroplasmy in swaPS3 cells. ARMS-qPCR (red diamonds) and RFLP (black diamonds); black and red lines indicate detection limits. g–i, Directed differentiation into β-cells (g), neurons (h) and fibroblasts (i). αSMA, α-smooth muscle actin. Scale bars, 5 μm (b, c) and 50 μm (g–i).
Heteroplasmy was determined by last-hot cycle PCR RFLP, with a detection threshold of approximately 2%, and allele refractory mutation system (ARMS)-qPCR, with a detection threshold of approximately 0.5% (refs 21, 22; Supplementary Figs 7 and 8). As considerable fluctuations in heteroplasmy have been reported in blastomeres of mouse and monkey embryos23,24, we first quantified 17 embryos at the cleavage stage, including individual blastomeres, and found heteroplasmy to be less than 0.5%. In seven samples that reached morula or blastocyst stage, the average heteroplasmy remained less than 0.5%, and in all cases 1% or lower. Overall, preimplantation embryos had a mean heteroplasmy of 0.31% ± 0.27% (n = 24; Fig. 3d and Supplementary Fig. 8e).
With the generation of stem-cell lines, we asked whether the original mitochondrial genotype could re-emerge after extensive passaging, clonal expansion, cellular differentiation and reprogramming. Quantification at passages (P)2–P59 (1year in culture), showed that heteroplasmy in the swaPS1, swaPS2 and swaPS4 cell lines remained undetectable (Fig. 3d, e and Supplementary Figs 7e–l and 8f). mtDNA heteroplasmy was detected at low levels in swaPS3 cells at P4–P10 (2.79% ± 0.27%; range: 2.3–3.1% by RFLP), but became undetectable from P14 onward (Fig. 3f and Supplementary Fig. 7e, i–j). As heterogeneity in heteroplasmy may not be detected by population analysis, ten colonies for each swaPS cell line were grown from single cells. In all 40 colonies, heteroplasmy was undetectable (Fig. 3d and Supplementary Fig. 8f).
In mice, specific mitochondrial genotypes can selectively expand in differentiated cells, resulting in altered levels of heteroplasmy25,26. To determine whether such alterations occurred after differentiation, cells of each germ layer (pancreatic cells, neurons, fibroblasts and cardiomyocytes) were differentiated in vitro (Fig. 3g–i and Supplementary Video 1). Heteroplasmy was undetectable by either ARMS-qPCR or RFLP (Fig. 3d, e and Supplementary Fig. 8g).
A bottleneck in the mitotic inheritance of mitochondrial DNA mutations occurs during induced pluripotent stem (iPS) cell generation, resulting in iPS cell colonies with differing percentages of a mitochondrial mutation27. To test whether such a bottleneck (mimicking the one in the female germ line) could alter the ratio of the two mitochondrial genotypes, we reprogrammed fibroblasts differentiated from swaPS cells (Fig. 4a) into iPS cells (Supplementary Fig. 9a–h). In 43 iPS cell colonies and 6 cell lines, termed swiPS, heteroplasmy was undetectable (mean heteroplasmy 0.01% ± 0.04%; n = 43; Fig. 3d and Supplementary Fig. 9i). As none of these manipulations resulted in re-emergence of significant levels of heteroplasmy, we next asked whether swaPS cells retained any mitochondrial DNA transferred with the karyoplast. Using repeat-PCR amplification of a product in the HVR, with five SNPs per primer pair, we could specifically amplify the minority product. In neither swaPS1 nor swaPS2 cells could the minority product be detected beyond P2 (detection limit 0.0001%), whereas in swaPS3 cells detection was seen until P18, after which it was undetectable (detection limit 0.1%; Supplementary Fig. 9j, k). As stem cells and fibroblasts contain less than 1,500 mt DNA copies28, a considerable proportion of the cell population must be homoplasmic.
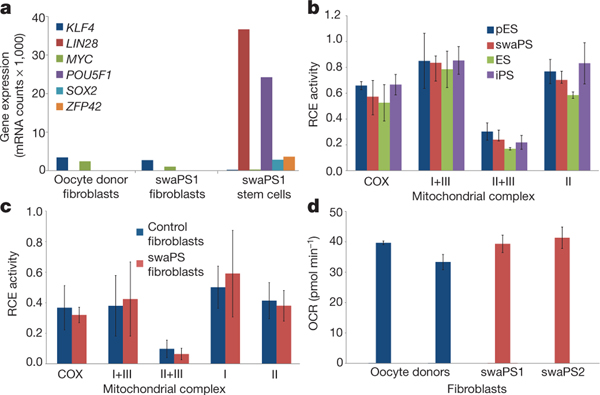
Figure 4 |. swaPS cells support a normal metabolic profile.
a, Nanostring gene expression analysis of fibroblasts derived from swaPS1 cells. mRNA counts per 100 ng RNA. b, Respiratory chain enzyme (RCE) activities in ES, pES, swaPS and iPS cell lines derived from oocyte donor skin cells. COX, cytochrome c oxidase (also known as complex IV). c, RCE activities of mitochondrial respiratory complexes in swaPS1- and swaPS2-derived fibroblasts (swaPS fibroblasts) compared withcontrol fibroblasts. d, Analysis of basal oxygen consumption rate (OCR) in swaPS1- and swaPS2-derived fibroblasts compared with control fibroblasts. Error bars denote s.d.
Normal mitochondrial activity in swaPS cells
Despite the low levels of mitochondrial heteroplasmy, we considered the possibility that the presence of two mitochondrial genotypes within the same cell might impair mitochondrial functions26. We determined that swaPS1 and swaPS2 cells had population doubling times of 33 and 31.5 h, respectively, a proliferation rate comparable to embryonic stem (ES) cell lines29, suggesting no metabolic disadvantage as a result of the exchange. We next determined the biochemical activities of mitochondrial respiratory chain enzymes. No differences were found in comparisons between undifferentiated swaPS1 and swaPS2, ES, pES and iPS cell lines generated from oocyte donor skin fibroblasts (Fig. 4b). We also assessed mitochondrial respiratory chain enzyme activities in differentiated cell types (Fig. 4c) and basal oxygen consumption in live culture (Fig. 4d), and found no differences between swaPS-derived fibroblasts and oocyte donor skin fibroblasts.
Discussion
Here we show that the transfer of nuclear genomes into human oocytes results in efficient preimplantation development, with essentially complete and mitotically stable exchange of the mitochondrial lineage, supporting normal mitochondrial activity. The low levels of heteroplasmy that are detected immediately after transfer become undetectable and do not increase after clonal expansion, cellular differentiation or when exposed to a bottleneck. Any potentially remaining heteroplasmy is far below the levels required for clinical presentation of a mitochondrial mutation, and below the levels of heteroplasmy able to cause behavioural defects in mice26. Importantly, the transfer is compatible with the integrity of the nuclear genome, as it did not result in an increase in CNVs.
We found that manipulation of an intact spindle–chromosome complex frequently induced premature activation of the oocyte, resulting in karyotype abnormalities. Preventing such abnormalities is crucial for successful clinical application of nuclear genome transfer, as it is unclear whether visual inspection or even molecular assays can reliably eliminate abnormal embryos. Exposure of the karyoplast, but not of the cytoplast, to low temperatures prevented this manipulation-induced activation. Spindle–chromosome complexes at or below room temperature showed decreased birefringence of microtubules, a measure for the regularity of microtubule alignment. Importantly, after fusion and incubation at 37°C, spindle birefringence returned, allowing polar body extrusion, normal preimplantation development, and derivation of karyotypically normal stem cells. Such reversible destabilization of spindle microtubules by low temperatures has been previously observed in human and animal oocytes30,31. During vitrification of human oocytes, partial depolymerization of the spindle occurs owing to the exposure to room temperature, rather than the vitrification itself32, but does not result in the dispersion of chromosomes and does not increase the incidence of karyotypic abnormalities33,34, allowing the adoption for clinical use35. In a concurrent study using temperatures of 37 °C to maintain a fully assembled spindle during manipulation36,37, frequent signs of premature activation were found, despite the use of Sendai virus for fusion, which is less prone to induce activation than an electrical pulse8. As a result, more than half of the oocytes were lost due to a failure in polar body extrusion and the formation of karyotypically abnormal embryos. By contrast, during our study, all oocytes were briefly exposed to room temperature (21°C), and when karyoplasts were transferred using Sendai virus, all oocytes remained at meiosis and extruded the polar body only after artificial activation. A further difference between the two studies is the timing of oocyte retrieval at 35 instead of 36 h after hormonal induction of ovulation. Both exposure to room temperature and early retrieval probably result in more oocytes with immature spindles at the time of manipulation. Bipolar attachment of chromosomes to spindle microtubules reduces the activity of aurora B kinase and of the spindle assembly checkpoint in a tension-dependent manner38. Manipulation may result in increased tension and decreased kinase target phosphorylation in fully assembled spindles, but not in spindles with incomplete bipolar attachment of chromosomes, thereby avoiding premature activation.
Although concerns have been raised that the transfer of the maternal genome between oocytes may introduce ‘epigenetic’ changes39, we did not observe meaningful differences in gene expression between genome-exchanged and unmanipulated cells. Although our methods allow maternal nuclear genome transfer without apparent adverse consequences to the embryo, the accuracy of chromosome segregation at the second meiosis will need to be determined in a large number of samples. Furthermore, animal models may be required to determine whether the transfer of incompletely assembled spindles into oocytes is compatible with normal fertilization and development to term. An analogous technique for the transfer of the paternal genome by intracytoplasmic sperm injection is performed in approximately half of all in vitro fertilization (IVF) cycles in the United States40, providing a precedent for clinical translation. Before proceeding with human clinical trials on the transfer of the maternal genome, it will be important to publicly discuss patient needs, ethical considerations, and to establish appropriate guidelines for the use of oocyte nuclear genome transfer in assisted reproduction.
METHODS SUMMARY
The nuclear genome was removed from mature MII oocytes, and was fused to an enucleated oocyte of a different donor. Oocytes were activated, allowed to develop to the blastocyst stage and stem cells were derived.
Full Methods and any associated references are available in the online version of the paper.
METHODS
Research subjects.
Human oocytes were donated by women enrolled in the reproductive egg donation program at Columbia University Medical Center (CUMC). Oocyte donors were given the option to donate for research only after they had enrolled and qualified as reproductive egg donors. Their decision to donate was therefore independent of the decision to donate for research. Compensation was equivalent to that given for reproductive donation, and did not depend on the number or quality of oocytes donated, in agreement with American Society for Reproductive Medicine (ASRM) guidelines41. The menstrual cycles of the subjects were synchronized using oral contraceptive pills. On the third day after the discontinuance of the oral contraceptive pills, ovarian hyperstimulation was initiated regardless of menstrual flow, using parenterally administered gonadotropins. Once lead follicles reached 18–22 mm diameter, 4 mg leuprolide acetate was administered to trigger final maturation and oocytes were retrieved 35 h later. With each donation, a skin biopsy and 3–4 ml of blood were taken. All human subject protocols were reviewed and approved by the CUMC institutional review board and the embryonic stem-cell research oversight committee of CUMC. All oocyte donors gave informed consent.
Manipulation of human oocytes.
Oocytes were transported in vials containing GMOPS plus (Vitro life), using a portable incubator heated to 37 °C. A total of 73 MII oocytes was used for this study. Oocytes of different donors were placed in separate drops of GMOPS plus containing 5 μg ml−1 cytochalasin B (Sigma) covered with mineral oil (Irvine Scientific). Karyoplasts were aspirated into a pipette with a diameter of 20mm (Humagen), after incubation in medium containing cytochalasin B for 3–5 min. If the karyoplast contained a larger amount of cytoplasm, the extra cytoplasm was removed by pressing the cytoplasm against the zona pellucida. Karyoplasts of donor 1 were inserted below the zona pellucida of an enucleated oocyte of donor 2, and fused using either inactivated Sendai virus HVJ-E (GenomeOne, Cosmo Bio), diluted with fusion buffer 1:40, or electrofusion, performed in cell fusion medium (0.26 M mannitol, 0.1mM MgSO4, 0.05% BSA and 0.5mM HEPES) using 2–8 fusion pulses of 20μ;s width and 1.3 Vcm−1 strength (LF201, NEPA Gene). Each exchange required approximately 10min. After aspiration of no more than two karyoplasts, transfer was undertaken, unless karyoplasts were used for cryopreservation. All manipulations were performed on a 37°C heated stage (Tokai Hit) of a Nikon TE 2000U inverted microscope, using Narishige micromanipulators. The transfer of the oocytes from the incubator to the inverted microscope required their brief exposure to room temperature (21 °C). Oocyte culture was conducted in Global medium supplemented with 10% plasmanate (Talecris), or in Global total (LifeGlobal) in a Minc incubator (Cook Medical), infused with a defined gas mixture of 6% CO2, 5% O2 and 89% N2. Parthenogenetic activation of oocytes was done using ionomycin, followed by incubation in 10 mM puromycin for 4 h. Manipulation of oocytes was completed within 3–5h after aspiration.
Vitrification and thawing of all oocytes was achieved through the use of the cryotop kit (Kitazato) and used in accordance with the manufacturer’s instructions on an unheated stage of a Nikon SMZ1500. For vitrification, oocytes were placed into basic solution with equilibrium solution added over a 15-min period. Oocytes were transferred to vitrification solution and then immediately placed onto the cryotop, with minimal solution carry over, and plunged directly into liquid nitrogen. The cryotop was placed into a protective straw for storage. After thawing, the cryotop was quickly placed into pre-warmed (37 °C) thawing solution for 1 min. Oocytes were then transferred into diluent solution (21°C) and washing solution (21 °C) over a 9-min period, after which they were transferred to regular culture medium or GMOPS plus. Ten out of eleven cryopreserved oocytes were viable after thawing. Statistical analysis of development and spontaneous activation was undertaken using the chi-square test. P < 0.05 was considered statistically significant.
Stem-cell derivation and analysis.
Parthenotes were allowed to develop to the expanded blastocyst stage, or day 6 or 7 after activation. The trophectoderm of blastocysts were ablated using the Lykos Laser (Hamilton Thorne), as previously described42. Isolated ICMs were plated on mouse embryonic fibroblast feeder (MEF) layers in stem-cell culture medium (hESm; KO-DMEM with high glucose, supplemented with 20% knockout serum replacement and bFGF; all cell culture reagents from Life Technologies). For pluripotency analysis, stem cells were fixed and stained for OCT4 (also known as POU5F1), SOX2 (both Stemgent), NANOG (Cell Signaling Technology), SSEA3, SSEA4 (both R&D Systems), TRA-1–60 and TRA-1–80 (Millipore). Images were taken using an Olympus IX71 epifluorescence microscope. Live cultures were sent to Cell Line Genetics or WiCell for karyotyping and STR genotyping. Gene expression analysis was undertaken using the Illumina HumanHT-12 Expression BeadChip and analysed using the Illumina Beadstudio software. Quantitative gene expression analysis was undertaken using the Nanostring nCounter system and analysed using nSolver (Nanostring Technologies). Teratomas were generated by subcutaneous injection into NSG mice (Jackson Laboratories) and collected after 10–15 weeks. Animal experimentation was performed under a Columbia Institutional Animal Care and Use Committee protocol.
Nuclear and mitochondrial genotyping.
Samples for both nuclear and mitochondrial genotyping were prepared using the high-pure template preparation kit as per the manufacturer’s instructions (Roche). Using a range of nuclear DNA and mtDNA primers (Supplementary Table 8), PCR products were generated using Red-Taq (Sigma) or Blue-Taq (Denville Scientific) as per the manufacturer’s instructions and purified using the high-pure PCR product purification kit (Roche). Sanger sequencing was undertaken via either Genewiz or Macrogen. Sequences were analysed using ApE (http://biologylabs.utah.edu/jorgensen/wayned/ape/). Complete mitochondrial genotyping was undertaken using the Affymetrix GeneChip human mitochondrial resequencing array 2.0 (MitoChip v.2.0), according to the manufacturer’s recommended protocols (primers and condition for the long PCR described in Supplementary Table 8). Sequences were directly compared to the revised Cambridge Reference Sequence for human mtDNA (NCBI accession NC_012920). Genotyping of nuclear DNA was done using Affymetrix GeneChip human mapping 250 K NspI arrays according to manufacturer’s instructions. Analysis was performed using Affymetrix genotyping console.
Copy number variation analysis.
Copy number analysis was performed using Affymetrix 6.0 SNP arrays as per the manufacturer’s instructions. CNVs were detected using NEXUS 6 and the SNP-FASST2 segmentation algorithm. High gains were set to the threshold of 0.7, gains at 0.1, losses at −0.15 and big losses at −1.1. A significance threshold of 5.0 × 10−7 was used, with minimum number of 10 probes required per CNV call and a minimum size of 50 kilobases (kb). Copy losses/gains were analysed to a minimum of 10 kb. After participation analysis to determine whether CNVs were new or preexisting, each call was manually inspected for visual confirmation of the call. Either a t-test or a one-way analysis of variance (ANOVA) with Bonferroni’s multiple comparison test was used for statistical analysis. P < 0.05 was considered to be statistically significant.
Quantification of mt DNA copy number.
Quantitative real-time PCR analysis of mtDNA copy number was achieved using previously designed primers and calculating unknown samples on a standard curve plotting copy number against a mean threshold value. A standard curve was generated using serial dilutions of a purified-PCR product generated using Red-Taq (Sigma) and previously published primers designed for the nucleotide positions 8259–8273 and 8475–8489 (ref. 43; Supplementary Table 8). Quantification of mtDNA copy number was achieved through the use of primers designed for the nucleotide positions 8290–8308 and 8418–8438. Samples were prepared using the high-pure template purification kit as per the manufacturer’s instructions, although samples were eluted in only 30 μl of elution buffer (Roche). Reaction mixtures were prepared (in triplicate) with 3 μl template DNA, 5 μl 2 × SYBR Green PCR master mix (Promega), 100 nM of each primer and water to a final volume of 10 μl. The reactions were performed in a Stratagene MX3000P with the following cycle: hold at 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s, 53 °C for 30 s, and 72 °C for 1 min. The copy number of an unknown sample was calculated from the standard curve and adjusted for the dilution factor.
Estimation of oocyte and karyoplast volume.
Oocytes and karyoplasts were imaged on either an Olympus IX71 or a Zeiss LSM5 PASCAL microscope following staining with Mito Tracker Red (Life Technologies) and either Hoescht (Sigma) or Draq5 (Biostatus). Single images were taken at the midpoint of each sample of interest, with complete Z stacks also imaged. Images were analysed using Zen LE (Zeiss) and based on the measured diameter, the volume was calculated (with the assumption that oocytes and karyoplasts are spherical).
Quantification of heteroplasmy levels: ARMS-qPCR and last-hot cycle RFLP.
Heteroplasmy was analysed using both last-hot cycle RFLP and ARMS-qPCR. The presence of three SNPs, m.1670A>T (MT-TV), m.4715A>G (MT-ND2) and m.16129A>G (non-coding region), was validated by Sanger sequencing and the polymorphisms were analysed by RFLP. The regions flanking the variations site were PCR-amplified (primers and PCR condition described in Supplementary Table 8). The 305-base-pair (bp) fragment containing the m.4715A variant was digested by BspEI into two fragments (204 and 101 bp), whereas the m.4715G sequence lacked the BspEI recognition site (Supplementary Fig. 7a). The 387-bp fragment containing the m.16129A variant was digested by BanI into two fragments (176 and 211 bp), whereas the m.16129G mtDNA had an additional BanI recognition site yielding three fragments (176, 130 and 81 bp) (Supplementary Fig. 7b). The 312-bp fragment containing the m.1670T variant was digested by AluI in two fragments (120 and 192 bp), whereas the m.1670A had an additional AluI recognition site yielding to three fragments (120, 60 and 132 bp) (Supplementary Fig. 7c). To assess heteroplasmy, [α−32P]deoxycytidine 5′-triphosphate (dCTP; 3,000 Ci mmol−1) (Perkin Elmer Health Science) was added to the last PCR cycle, the hot-labelled digested products were electrophoresed in a 10% non-denaturing acrylamide gel, and the bands analysed in a PhosphorImager (Typhoon TRIO variable mode imager, GE Healthcare Life Science) using ImageQuant TL v.7.01 software (GE Healthcare). The experiments were performed in duplicates using oocyte donor fibroblasts (1110 and 1111), swaPS1 and swaPS2 cells (P2, P9 and P40), cells differentiated in pancreatic β-cells, and swaPS3 cells (P4, P5, P6, P8, P10, P13, P14 and P20).
ARMS-qPCR primers were designed to amplify specifically the DNA of only one donor with two homozygous SNPS used at either end of the sequence based on the sequencing of HVR1 (Supplementary Table 6). Furthermore, the primers were designed such that the product generated would contain internal SNPs that could be verified by Sanger sequencing to confirm donor-specific product amplification (Supplementary Fig. 8a). To create a set of standards to confirm accuracy of the assay, the HVR of the various donors was amplified using Red-Taq (Sigma), and the product purified with the high-pure PCR product purification kit (Roche). The concentration of PCR product was calculated using a NanoDrop spectrophotometer (NanoDrop Technologies), and the copy number was calculated based on a standard curve (101–107 copies) generated with the assumption that DNA has a molecular mass of 650 daltons (Da) (primers and conditions in Supplementary Table 8). From this, standards were generated at various percentages of the two donors to confirm accuracy of the ARMS-qPCR assay. Expected values for the standards were matched against measured values using the equation: Mutant heteroplasmy level (%) = 1/(1 + (1/2)ΔCT) × 100%, in which ΔCT = CTwild type – CTmutant (Supplementary Fig. 8b–d). After confirmation, reactions were run under conditions d, e or f (as described in Supplementary Table 8), with samples of unknown heteroplasmy run alongside positive and negative controls and calculated using the above equation. Samples were run in triplicate in a reaction containing 0.6 ng μl−1 specific donor sample, 500 nM of each primer, 5 μl of 2× SYBR green (Promega) and water to a final volume of 10 μl and analysed using a Stratagene MX300P (Agilent). The determination of homoplasmy was undertaken using primers designed around the HVR of donors 1110 and 1111, allowing the incorporation of five SNPs per primer pair to facilitate primer specificity (Supplementary Table 6). Target DNA was amplified using Blue-Taq (Denville Scientific) under cycle conditions k (as described in Supplementary Table 8), before being analysed on a 2% agarose gel. Subsequently, 1 μl of the PCR product was diluted 1:100 and re-amplified under the same conditions for a further 30 cycles before being analysed on a 2% agarose gel.
High-resolution northern blot analysis.
Total RNA from cultured primary fibroblasts grown in a 10 cm2 dish was extracted using Trizol reagent (Life Technologies) according to the manufacturer’s instructions. Large RNA species were precipitated by the addition of 10 mol l−1 LiCl, allowing smaller RNAs to be precipitated from the resulting supernatant. Small RNAs (1.5 μg) were denaturated (70 °C for 5 min) and separated through an 8%, 8 mol l−1 urea denaturating polyacrylamide (19:1) gel using 0.5 × Tris-borate EDTA (TBE) as running buffer. Separated samples were electroblotted onto Nytran SuPerCharge TurboBlotter membrane (Whatman) using a TE77X semi-dry transfer unit (Hoefer) and immobilized by ultraviolet cross-linking. Regions of mtDNA encompassing the tRNAVal and tRNALeu(UUR) genes were amplified using specific primers (see Supplementary Table 8). Purified PCR products were radiolabelled with [α−32P] dCTP (3,000 Ci mmol−1) by random primer method, and unincorporated nucleotides were removed by gel filtration through a Sephadex G-50 DNA grade column (Amersham Pharmacia Biotech). Hybridization was performed at 42 °C overnight using a QuikHyb hybridization solution (Agilent Technologies Stratagene Products Division) containing 500,000 c.p.m. radiolabelled probes. After hybridization, two 15-min washes were performed at room temperature in 2 × SSC and 0.1% SDS, followed by one 30-min wash in 2 × SSC and 0.1% SDS at 60 °C. Blots were subjected to PhosphorImager analysis and the radioactive signal for the mt-tRNAVal probe (69 bp) normalized to the tRNALeu(UUR) signal (75 bp).
Directed differentiation of swaPS lines.
To confirm the heteroplasmy levels in terminally differentiated tissues, swaPS1 and swaPS2 were differentiated along endodermal, ectodermal and mesodermal lineages. β-cell differentiation was performed as described44, with the addition of calcium chelator, EGTA (75μM), on day1 andan activin receptor-like kinase inhibitor, SB431542 (2 μM), on days 9–12 of differentiation. Staining for SOX17, PDX1 (both R&D Systems) and C-peptide (Millipore) was undertaken at days 3, 10 and 14. DNA samples for heteroplasmy analysis were collected at day 14. The ectodermal differentiation was undertaken following the previously described dual-SMAD protocol45. After 2 weeks and two passages as neural progenitors, further differentiation was induced through the addition of BDNF (10ng ml−1, R&D Systems). After a further 3 weeks, cells were fixed for immunostaining, or DNA was collected for heteroplasmy analysis. Antibodies used for staining included TUJ1 (Sigma), MAP2 (Abcam), nestin, neurofilament (both Millipore) and SOX2 (Stemgent). Cells of the mesodermal lineage were generated using two protocols. First, the generation of contracting cardiomyocytes (from swaPS1, swaPS2 and swaPS4) was achieved using a previously published protocol46, with videos recorded using an Olympus IX71 with the DP2-BSW software. Second, the differentiation of swaPS1 and swaPS2 into fibroblasts was achieved by growing undifferentiated stem cells in human fibroblast medium (hFm; DMEM supplemented with 10% FBS, 1% penicillin/streptomycin and 1% glutamax) for 2–4 weeks with a single passage during this time. After 14–28days of growth, cells were sorted by FACS to enrich for TRA-1–60− SSEA4− CD56− CD13+ cells (Supplementary Fig. 9a). Antibodies used for staining included aSMA, phalloidin (both Sigma) and CD-13 (BD Biosciences). Gene expression analysis of the swaPS fibroblasts was undertaken using the Nanostring nCounter system as previously described. After differentiation, swaPS fibroblasts were placed into hESm and grown for 3 weeks on a feeder layer. No colonies were visible during this time period, and flow cytometry confirmed the absence of stem-cell-positive markers indicating swaPS fibroblasts could not spontaneously revert to a stem-cell state (Supplementary Fig. 9b, c).
Generation of iPS cell lines.
Biopsies were taken using a biopsy kit (AccuPunch, Accuderm) after local anaesthesia using lidocaine (1%, Hospira). Punch biopsies (3 mm) were cut into 10–15 small pieces from which fibroblasts were allowed to grow for 4 weeks. Fibroblasts were then passaged using TrypLE and plated in hFm at a density of 50,000 cells per well (6-well plate) overnight before being infected with the Cytotune iPS Sendai reprogramming kit as per the manufacturer’s instructions (all reagents from Life Technologies). Infected cells were grown in hESm containing three additional compounds47 (thiazovivin, SB431542 and PD0325901; all Stemgent) for 10 days before FACS enrichment of SSEA4+ Tra-1–60+ CD13− cells. Colonies were picked 7–14 days later and pluripotency was confirmed through the staining of pluripotency markers as described above and Nanostring gene expression analysis as previously described (Supplementary Fig. 9d–h).
Metabolic analysis.
Stem-cell lines were transferred from growing on MEFs to Matrigel (BD Biosciences)-coated plates and cultured in m-TeSR (StemCell Technologies). Cells were grown to approximately 90% confluence in 10-cm2 dishes before being collected and stored at −80°C. Biochemical activities of COX, NADH-cytochrome c reductase (complex I + III), succinate-cytochrome c reductase (complex II + III), NADH-CoQ reductase (complex I), succinate dehydrogenase (complex II) and citrate synthase were assayed spectrophotometrically as previously described48. Respiratory chain enzyme activity values were normalized to citrate synthase, an index of mitochondrial mass, and data were expressed as mean ± s.d. of at least two experiments. Biochemical activities were measured in three pES cell lines, two swaPS cell lines, two human ES cell lines and three iPS lines. Metabolic analysis was also performed in swaPS-derived fibroblasts (two cell lines) compared to control fibroblasts (six cell lines). One-way ANOVA with Bonferroni’s multiple comparison test was used to compare groups. P < 0.05 was considered to be statistically significant. Live metabolic analysis of cells was undertaken using the Seahorse stress-test kit as per the manufacturer’s instructions. In brief, 42,500 cells were seeded into the assay plate and allowed to grow overnight. The next day cells growth media was replaced with XF assay medium (Seahorse) supplemented with 25mM glucose, 0.4% BSA (both Sigma) 1% glutamax and 1% sodium pyruvate (both Invitrogen) for 1 h. After 1 h, cells were analysed using the XF24 (Seahorse). Experiments were performed in duplicates with an n of 4 per group. Results were analysed using the Seahorse XF24 software one-way ANOVA with Bonferroni’s multiple comparison test was used to compare groups. P < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgements
We thank S. Chang and K. Eggan for discussions, Z. Hall for critical reading of the manuscript, and L. Yu and O. Nahum for SNP-array preparation. We thank anonymous oocyte donors for participating in research, and M. Spencer for a Lykos laser system. This work was supported by the New York Stem Cell Foundation, the New York State Stem Cell Science award C026184, and the Bernard and Anne Spitzer Fund.
Footnotes
Supplementary Information is available in the online version of the paper.
References
- 1.Jenuth JP, Peterson AC, Fu K. & Shoubridge EA Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nature Genet. 14, 146–151 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Bolhuis PA et al. Rapid shift in genotype of human mitochondrial DNA in a family with Leber’s hereditary optic neuropathy. Biochem. Biophys. Res. Commun 170, 994–997 (1990). [DOI] [PubMed] [Google Scholar]
- 3.Cree LM et al. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nature Genet. 40, 249–254 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Wai T, Teoli D. & Shoubridge EA The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nature Genet. 40, 1484–1488 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Steffann J. et al. Analysis of mtDNA variant segregation during early human embryonic development: a tool for successful NARP preimplantation diagnosis. J. Med. Genet 43, 244–247 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nuffield Council on Bioethics. Novel Techniques for the Prevention of Mitochondrial DNA Disorders: an Ethical Review http://www.nuffieldbioethics.org/news/discussion-event-novel-techniques-prevention-mitochondrial-dna-disordersethical-review (2012).
- 7.Sato A. et al. Gene therapy for progeny of mito-mice carrying pathogenic mtDNA by nuclear transplantation. Proc. Natl Acad. Sci. USA 102, 16765–16770 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tachibana M. et al. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature 461, 367–372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craven L. et al. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature 465, 82–85 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egli D. et al. Reprogramming within hours following nuclear transfer into mouse but not human zygotes. Nature Comm. 2, 488 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganem NJ, Godinho SA & Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sathananthan AH et al. Centrioles in the beginning of human development. Proc. Natl Acad. Sci. USA 88, 4806–4810 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaufman MH, Robertson EJ, Handyside AH & Evans MJ Establishment of pluripotent cell lines from haploid mouse embryos. J. Embryol. Exp. Morphol 73, 249–261 (1983). [PubMed] [Google Scholar]
- 14.Draper JS et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nature Biotechnol. 22, 53–54 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Kim K. et al. Histocompatible embryonic stem cells by parthenogenesis. Science 315, 482–486 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Noggle S. et al. Human oocytes reprogram somatic cells to a pluripotent state. Nature 478, 70–75 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Hyun CS et al. Optimal ICSI timing after the first polar body extrusion in in vitro matured human oocytes. Hum. Reprod 22, 1991–1995 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Brinkley BR & Cartwright J Jr. Cold-labile and cold-stable microtubules in the mitotic spindle of mammalian cells. Ann. NY Acad. Sci 253, 428–439 (1975). [DOI] [PubMed] [Google Scholar]
- 19.van Oven M. & Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat 30, E386–E394 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Mimaki M. et al. Reversible infantile respiratory chain deficiency: a clinical and molecular study. Ann. Neurol 68, 845–854 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Venegas V, Li F. & Wong L-J Analysis of mitochondrial DNA point mutation heteroplasmy by ARMS quantitative PCR. Curr. Protocols Human Genet Chapter 19, Unit–19.16 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Bai R-K & Wong L-JC Detection and quantification of heteroplasmic mutant mitochondrial DNA by real-time amplification refractory mutation system quantitative PCR analysis: a single-step approach. Clinical Chem. 50, 996–1001 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Lee HS et al. Rapid mitochondrial DNA segregation in primate preimplantation embryos precedes somatic and germline bottleneck. CellRep. 1, 506–515 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meirelles FV & Smith LC Mitochondrial genotype segregation during preimplantation development in mouse heteroplasmic embryos. Genetics 148, 877–883 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenuth JP, Peterson AC & Shoubridge EA Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nature Genet. 16, 93–95 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Sharpley MS et al. Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell 151, 333–343 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujikura J. et al. Induced pluripotent stem cells generated from diabetic patients with mitochondrial DNA A3243G mutation. Diabetologia 55, 1689–1698 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Birket MJ et al. A reduction in ATP demand and mitochondrial activity with neural differentiation of human embryonic stem cells. J. Cell Sci 124, 348–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cowan CA et al. Derivation of embryonic stem-cell lines from human blastocysts. New Engl. J. Med 350, 1353–1356 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Inoue S, Fuseler J, Salmon ED & Ellis GW Functional organization of mitotic microtubules. Physical chemistry of the in vivo equilibrium system. Biophys. J 15, 725–744 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bianchi V, Coticchio G, Fava L, Flamigni C.& Borini A. Meiotic spindle imaging in human oocytes frozen with a slow freezing procedure involving high sucrose concentration. Human Reprod. 20, 1078–1083 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Larman MG, Minasi MG, Rienzi L. & Gardner DK Maintenance of the meiotic spindle during vitrification in human and mouse oocytes. Reprod. Biomed. Online 15, 692–700 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Forman EJ et al. Oocyte vitrification does not increase the risk of embryonic aneuploidy or diminish the implantation potential of blastocysts created after intracytoplasmic sperm injection: a novel, paired randomized controlled trial using DNA fingerprinting. Fertil. Steril 98, 644–649 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Gook DA, Osborn SM, Bourne H. & Johnston WI Fertilization of human oocytes following cryopreservation; normal karyotypes and absence of stray chromosomes. Hum. Reprod 9, 684–691 (1994). [DOI] [PubMed] [Google Scholar]
- 35.ASRM. Mature oocyte cryopreservation: a guideline. Fertil. Steril. 10.1016/j.fertnstert.2012.09.028 (18 October 2012). [DOI] [PubMed] [Google Scholar]
- 36.Tachibana M. et al. Towards germline gene therapy of inherited mitochondrial diseases. Nature 10.1038/nature11647 (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tachibana M, Sparman M. & Mitalipov S. Chromosome transfer in mature oocytes. Nature Protoc. 5, 1138–1147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu D, Vader G, Vromans MJ, Lampson MA & Lens SM Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 323, 1350–1353 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A mother’s gift, minus mitochondria. Nature Med. 16, 645 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Society for Assisted Reproductive Technology, American Society for Reproductive Medicine. Assisted reproductive technology in the United States: 2000 results generated from the American Society for Reproductive Medicine/Society for Assisted Reproductive Technology Registry. Fertil. Steril 81, 1207–1220 (2004). [DOI] [PubMed] [Google Scholar]
- 41.The Ethics Committee of the American Society for Reproductive Medicine. Financial compensation of oocyte donors. Fertil. Steril 88, 305–309 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Chen AE et al. Optimal timing of inner cell mass isolation increases the efficiency of human embryonic stem cell derivation and allows generation of sibling cell lines. Cell Stem Cell 4, 103–106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin DP-C et al. Comparison of mitochondrial DNA contents in human embryos with good or poor morphology at the 8-cell stage. Fertil. Steril 81, 73–79 (2004). [DOI] [PubMed] [Google Scholar]
- 44.D’Amour KA et al. Production of pancreatic hormone–expressing endocrine cells from human embryonic stem cells. Nature Biotechnol. 24, 1392–1401 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Chambers SM et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature Biotechnol. 27, 275–280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burridge PW et al. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PloS ONE 6, e18293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin T. et al. A chemical platform for improved induction of human iPSCs. Nature Methods 6, 805–808 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DiMauro S. et al. Cytochrome c oxidase deficiency in Leigh syndrome. Ann. Neurol 22, 498–506 (1987). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.