We extended the known annotation space for the NCLDV by 46%, revealing high-probability structural matches for fully 45% of the 9,671 query proteins and confirming up to 98% of existing annotations per virus. The most prevalent protein families included ankyrin repeat- and MORN repeat-containing proteins, many of which included an F-box, suggesting extensive host cell modulation among the NCLDV. Regression suggested a minimum requirement for around 36 protein structural superfamilies for a viable NCLDV, and beyond around 200 superfamilies, genome expansion by the acquisition of new functions was abruptly replaced by paralogization. We found homologs to herpesvirus surface glycoprotein gB in cytoplasmic viruses. This study provided the first prediction of an endonuclease in 10 of the 20 viruses examined; the first report in a virus of a phenolic acid decarboxylase, proteasomal subunit, or cysteine knot (defensin) protein; and the first report of a prokaryotic-type ribosomal protein in a eukaryotic virus.
KEYWORDS: NCLDV, giant virus, vaccinia, Mimivirus, HHsearch, NCLDV
ABSTRACT
We conducted an exhaustive search for three-dimensional structural homologs to the proteins of 20 key phylogenetically distinct nucleocytoplasmic DNA viruses (NCLDV). Structural matches covered 429 known protein domain superfamilies, with the most highly represented being ankyrin repeat, P-loop NTPase, F-box, protein kinase, and membrane occupation and recognition nexus (MORN) repeat. Domain superfamily diversity correlated with genome size, but a diversity of around 200 superfamilies appeared to correlate with an abrupt switch to paralogization. Extensive structural homology was found across the range of eukaryotic RNA polymerase II subunits and their associated basal transcription factors, with the coordinated gain and loss of clusters of subunits on a virus-by-virus basis. The total number of predicted endonucleases across the 20 NCLDV was nearly quadrupled from 36 to 132, covering much of the structural and functional diversity of endonucleases throughout the biosphere in DNA restriction, repair, and homing. Unexpected findings included capsid protein-transcription factor chimeras; endonuclease chimeras; enzymes for detoxification; antimicrobial peptides and toxin-antitoxin systems associated with symbiosis, immunity, and addiction; and novel proteins for membrane abscission and protein turnover.
IMPORTANCE We extended the known annotation space for the NCLDV by 46%, revealing high-probability structural matches for fully 45% of the 9,671 query proteins and confirming up to 98% of existing annotations per virus. The most prevalent protein families included ankyrin repeat- and MORN repeat-containing proteins, many of which included an F-box, suggesting extensive host cell modulation among the NCLDV. Regression suggested a minimum requirement for around 36 protein structural superfamilies for a viable NCLDV, and beyond around 200 superfamilies, genome expansion by the acquisition of new functions was abruptly replaced by paralogization. We found homologs to herpesvirus surface glycoprotein gB in cytoplasmic viruses. This study provided the first prediction of an endonuclease in 10 of the 20 viruses examined; the first report in a virus of a phenolic acid decarboxylase, proteasomal subunit, or cysteine knot (defensin) protein; and the first report of a prokaryotic-type ribosomal protein in a eukaryotic virus.
INTRODUCTION
The 2003 discovery of the first giant virus, Mimivirus (1), proved transformative to virology and added new context to the established large DNA virus families (Poxviridae, Iridoviridae, and Chlorellaviridae). A decade later, a new “giant of giants,” Pandoravirus, with its 2.7-Mb genome encoding more than 2,500 proteins (2), dwarfed the 800-kb Mimivirus genome by more than equal measure. The past decade has seen the characterization of many new large DNA virus genomes via the integration of metagenomics, next-generation nucleic acid sequencing, more proficient sequence alignment algorithms (3), and greater interconnectivity of bioinformatics resources for the fast and automated annotation of genes, proteins, protein folds, and protein domains. There are now as many as nine families of nucleocytoplasmic large DNA viruses (NCLDV), whose shared characteristics include a greater or lesser degree of cytoplasmic involvement in their replication, independence from the host replication machinery, large DNA genomes, and genes for DNA replication, DNA repair, transcription, and mRNA translation (4).
Although many NCLDV genes have been annotated for function, comprehensive genome annotation is confounded by the minimal or nonexistent conservation of amino acid sequence across a broad swath of evolutionary space. In just one example familiar to the authors, a classical Rossmann fold was revealed within the crystal structure for vaccinia protein VP39 (5), whose existence had been entirely unpredictable on the basis of sequencing despite the well-established nature of this fold and the many proteins containing it. New additions to the BLAST pipeline, including BLASTP, PSI-BLAST, and BLASTCLUST, have helped to some extent in closing the annotation gap (3). The use of tertiary structural information, however, may be a much more sensitive method for finding matches whose similarities have fully decayed at the protein sequence level. The detection of distant sequence homology has been sensitized by the use of sequence substitution “profiles” treated as hidden Markov models (HMMs) of multiple-sequence alignments (MSAs) of the growing numbers of members of various protein families. Using tools such as PfamScan (6, 7), individual sequence queries can be searched against profile MSA HMMs, driving the expansion of the Pfam database of known protein families (8). More powerful, although currently lacking in PfamScan, would be an ability to perform profile MSA versus profile MSA searches. Such searches led to the prediction of NCLDV members of the archaeoeukaryotic primase superfamily (9). In our hands, PfamScan seemed slow to update its profiles and seemed to overlook structural homologs we were otherwise able to find in the pdb70 database (unpublished data).
One powerful package, HHsuite (10), employs profile-profile alignments to identify homologous proteins, starting with the creation of MSAs for query proteins and then embellishing these with secondary structural prediction. HMM profiles of the resulting MSAs are searched against a database of HMMs derived from bona fide experimental protein structures (PDB or SCOP). The combination of sequence and secondary structural alignments and the use of real structures provides a potentially powerful tool for protein families with marginal or absent sequence similarity, and has the potential to harvest the biosphere-wide structural proteomics initiative of the earlier part of the current millennium (11). HHsuite has been applied in a number of problems, including the prediction of open reading frames (12), analysis of G protein-coupled receptors (13), identification of novel protein repeats (14), prediction of poxviral RNA polymerase homologs (15, 16), and the identification of PH domains in the S. cerevisiae proteome (17). Here, we have applied the HHsuite toolbox more comprehensively, providing the first exhaustive search of the proteomes of 20 NCLDV-type members, identifying protein superfamily members among previously uncharacterized proteins and filling gaps in the NCLDV core proteome. We have expanded our previously published work of multisubunit DNA-directed RNA polymerase (MSDDRP) subunits and predicted a number of viral protein homologs not previously identified.
RESULTS AND DISCUSSION
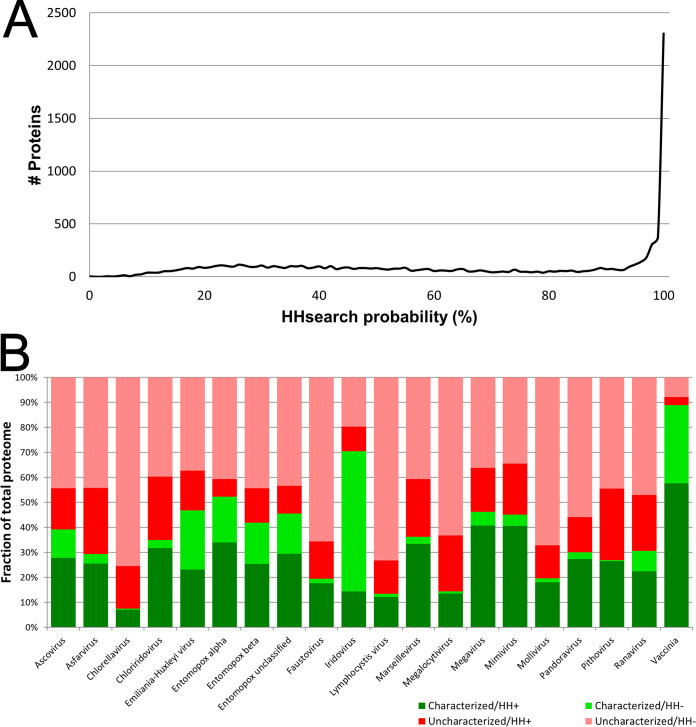
Here, we have “deep-mined” new protein annotations in a selection of 20 phylogenetically distinct NCLDV chosen to cover all known NCLDV families, key subfamilies, genera, species, and unclassified viruses therein (Table 1). Mining was based on tertiary structure homology. Proteomes of the 20 viruses comprised a total of 9,671 proteins, from each of which an HMM was derived via a combination of MSA and predicted secondary structure. Each resulting HMM was used to query an HMM database generated from actual protein tertiary structures deposited in pdb70. The search output for each query protein, showing all matching pdb70 entries/regions, was thresholded according to a probability parameter calculated by the search engine. An 80% threshold was chosen for the probability parameter based on the initial descriptions of HHsuite (10, 18, 19) and prior literature (17) in which a probability threshold of 80% yielded a false-positive rate of just 0.15%. In the current study, the best-scoring database match exceeded the 80% probability threshold for 45% of the 9,671 query proteins and fell within the topmost (99 to 100%) probability bin for fully 23.8% of proteins (Fig. 1a). This provided bootstrap confirmation of our chosen probability threshold. Apparently, our approach could successfully uncover structural homologs for nearly half of all NCLDV proteins—in the vast majority of cases covering most of the length of the query and target proteins (see Fig. S1 in the supplemental material). Where an unknown NCLDV query protein matched a pdb70 entry of known function, this annotation was transferred directly to the NCLDV query protein. Since functions are already known for a substantial proportion of proteins resident in pdb70, there were frequent opportunities for such “annotation transfer.” Raw search results have been uploaded to the following URL: https://sites.google.com/view/gershonlab-hhsearch-results/results.
TABLE 1.
The 20 phylogenetically distinct NCLDV analyzed in this study (listed alphabetically)b
| Virus (local name) | Family | Subfamily | Genus | Scientific or common name | Strain | UniProt proteome identifier | No. of proteins | UniProt reference proteomec |
|---|---|---|---|---|---|---|---|---|
| Ascovirus | Ascoviridae | Ascovirus | Heliothis virescens ascovirus 3g | UP000232493 | 194 | N | ||
| Asfarvirus | Asfarviridae | Asfivirus | African swine fever virus Georgia 2007/1 | UP000141072 | 188 | N | ||
| Chlorella virusa | Phycodnaviridae | Chlorovirus | Paramecium bursaria Chlorella virus 1 (PBCV-1) | UP000000862 | 794 | Y | ||
| Emiliania huxleyi virusa | Phycodnaviridae | Coccolithovirus | Emiliania huxleyi virus 86 (EhV-86) | Isolate United Kingdom/ English Channel/1999 | UP000000863 | 472 | Y | |
| Entomopox alpha | Poxviridae | Entomopoxvirinae | Alphaentomopoxvirus | Anomala cuprea entomopoxvirus | UP000174145 | 241 | Y | |
| Entomopox beta | Poxviridae | Entomopoxvirinae | Betaentomopoxvirus | Choristoneura biennis entomopoxvirus (CbEPV) | UP000014934 | 311 | Y | |
| Entomopox unclassified | Poxviridae | Entomopoxvirinae | Melanoplus sanguinipes entomopoxvirus (MsEPV) | Isolate Tucson | UP000172353 | 261 | Y | |
| Faustovirus | Unclassified viruses | Faustovirus sp. | E12 | UP000244833 | 492 | Y | ||
| Iridoviridae/Chloriridovirus | Iridoviridae | Betairidovirinae | Chloriridovirus | Invertebrate iridescent virus 3 (IIV-3) (mosquito iridescent virus) | UP000001358 | 126 | Y | |
| Iridoviridae/Iridovirus | Iridoviridae | Betairidovirinae | Iridovirus | Invertebrate iridescent virus 6 (IIV-6) (Chilo iridescent virus) | UP000001359 | 469 | Y | |
| Iridoviridae/Lymphocystivirus | Iridoviridae | Alphairidovirinae | Lymphocystivirus | Lymphocystis disease virus | Isolate China | UP000106699 | 239 | Y |
| Iridoviridae/Megalocytivirus | Iridoviridae | Alphairidovirinae | Megalocytivirus | Infectious spleen and kidney necrosis virus (ISKNV) | Isolate Mandarin fish/China/Nanhai/1998 | UP000008773 | 125 | Y |
| Iridioviridae/Ranavirus | Iridoviridae | Alphairidovirinae | Ranavirus | Frog virus 3 (isolate Goorha) (FV-3) | UP000008770 | 98 | Y | |
| Marseillevirus | Marseilleviridae | Marseillevirus | Marseillevirus marseillevirus (GBM) | UP000029780 | 428 | Y | ||
| Megavirus | Mimiviridae | Mimivirus | Megavirus courdo11 (unclassified Mimivirus) | UP000241137 | 1217 | Y | ||
| Mimivirus | Mimiviridae | Mimivirus | Acanthamoeba polyphaga mimivirus (APMV) | Rowbotham-Bradford | UP000201519 | 979 | N | |
| Mollivirus | Unclassified viruses | Mollivirus sibericum | UP000202709 | 514 | Y | |||
| Pandoravirus | Pandoravirus | Pandoravirus inopinatum | UP000202511 | 1839 | Y | |||
| Pithovirus | Pithoviridae | Pithovirus | Pithovirus sibericum | UP000202176 | 467 | Y | ||
| Vaccinia | Poxviridae | Chordopoxvirinae | Orthopoxvirus | Vaccinia virus | Western Reserve (WR) | UP000000344 | 217 | Y |
Two clades within the Phycodnaviridae based on RNA polymerase subunit presence/absence (16). Chlorella virus represents the following genera: Chlorovirus, Phaeovirus, Raphidovirus, Prasinovirus, Yellowstone lake phycodnaviruses 1 and 2, and Ostreococcus tauri virus 2. Emiliania huxleyi virus represents the following genera: Coccolithovirus, Prymnesiovirus, Organic lake phycodnaviruses 1 and 2, Chrysochromulina ericina virus, and Aureococcus anophagefferens virus.
A total of 9,671 proteins were analyzed among the 20 viruses. Reviewed proteomes were used where available. All taxonomic rankings (family, subfamily, genus, species, and strain) are according to UniProt’s “Lineage” fields. Where multiple alternate species or strains were available, the one with the largest proteome was chosen (with the exception of Vaccinia, where the WR strain was used in place of Tian Tan). This table represents the group of NCDV analyzed previously (16) with the following differences: two classes only of phycodnavirus are considered (see the text), the three subfamilies of Entomopoxvirinae are considered separately, and Mollivirus has been added. Although Megavirus courdo11 is an unclassified member of the Mimiviridae, it is referred to here as Megavirus. After its initial discovery, Megavirus was regarded as a new virus family (99, 100), but after phylogenetic studies of additional new Mimivirus and Megavirus genomes, Megavirus was instead grouped with the Mimiviridae (101).
N, no; Y, yes.
FIG 1.
Structure-based deep mining of functions for 20 representative NCLDV proteomes. (a) Line histogram of HHsearch “probability” score associated with best HHsearch database match to each of the 9,671 query proteins over 20 representative NCLDV proteomes. The x axis shows HHsearch probability parameter (bin width, 1%). The y axis shows number of proteins associated with each bin. (b) Extension of existing annotations. Green/red indicates the fraction of virus proteome possessing or lacking, respectively, an associated UniProt functional annotation prior to the current study. Red-group proteins were annotated in UniProt either as “uncharacterized protein” or with an annotation comprising simply the submitted gene name. Dark/light green indicates the fraction of UniProt-annotated proteomes confirmed or not confirmed, respectively, by HHsearch at or above the 80% probability threshold. Dark/light red indicates the fraction of UniProt-unannotated proteomes for which HHsearch did or did not, respectively, provide a first-time functional annotation at or above the 80% threshold. In generating the dark green region, agreement between annotated NCLDV query proteins and corresponding pdb70 hits was assessed conservatively, as either an identical stated protein function, keyword, or leaf gene ontology (GO) term. Proteomes showing low overall annotation rates prior to the current study (green region below 20% on the y axis; namely, Chlorella virus, Lymphocystis virus, Megalocytivirus, and Mollivirus) may have been handicapped by a slow synchronization between UniProt and the Pfam and InterPro databases.
Prior to the current study, the 20 query proteomes were annotated to a variable extent, the most incompletely and completely annotated being Chlorella virus (7.4%) and Vaccinia virus (89%), respectively (Fig. 1b, total green). The current study confirmed between 20% (Iridovirus) and 98% (Pithovirus) of existing annotations (Fig. 1b, dark green versus total green), validating our structure-based approach. Perhaps more interestingly, our approach provided first-time annotations for many previously uncharacterized proteins from each of the 20 selected viruses. First-time annotations covered between 15% (Entomopoxvirus alpha) and 39% (Chloriridovirus) of the previously uncharacterized segments of virus proteomes (Fig. 1b, dark red versus total red). Apparently, substantial inroads could be made into the uncharacterized proteomes of the NCLDV via structure-based homology.
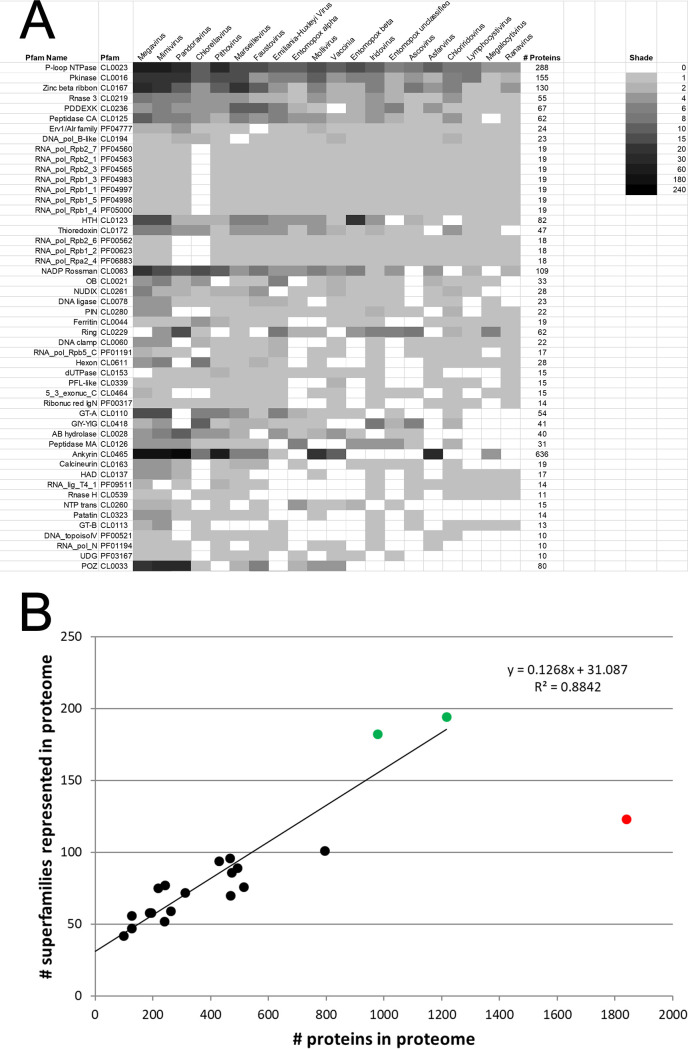
NCLDV matches to annotated pdb70 entries were formalized into functional classes by visual inspection of, in each case, the HMM homology region in the pdb70 target in order to find overlapping entries in the Pfam (8) protein domain family database. Pfam tagging in this manner accounted for 87.5% of all of the NCLDV proteins showing structural homologs, covering a total of 429 Pfams (see Fig. S2 in the supplemental material; the top 50 Pfams are shown in Fig. 2a). Pfams with the greatest overall representation among the 20 viruses comprised the ankyrin repeats (636 proteins), P-loop NTPases (288 proteins), F-box proteins (222 proteins), protein kinases (155 proteins), and membrane occupation and recognition nexus (MORN) repeat-containing proteins (149 proteins) (Fig. 2a and Fig. S2) Proteins in these five families were particularly prevalent among the giant viruses (Megavirus, Mimivirus, Marseillevirus, Pandoravirus, and Pithovirus). For the 20 viruses, good correlation (R2 = 0.88) was observed between the number of proteins in the proteome versus the number of distinct protein domain superfamilies represented therein (Fig. 2b), suggesting that NCLDV genome expansion has gone hand in hand with the acquisition of novel superfamily functions (with larger genomes being more functionally diverse). Interestingly, there was a single, and quite dramatic outlier to this correlation, namely, Pandoravirus, whose proteome is the largest by far among all currently known viruses. Despite its genome being larger than that of its closest neighbor (Megavirus) by a factor of 1.5, this was not accompanied by any net increase in the numbers of protein domain superfamilies in the Pandoravirus proteome—rather, there was actually a 20% decrease in superfamily diversity compared with that of Mimivirus (Fig. 2b). Apparently, there is a threshold above which the gain of superfamily diversity (new orthologs) has no appreciable selective advantage in relation to the diversification of existing ones (new paralogs). “Paralogization” seems to have taken over as an evolutionary driver at an apparently quite definable point in genome growth. Nonetheless, this conclusion is based on just one data point. Overall, Mimivirus and Megavirus showed the greatest proteomic diversity in terms of total protein superfamilies represented in their proteomes (Fig. 2b, green points). Conversely, extending the linear regression line back to near x-y parity (x = 39 proteins; y = 36 superfamilies), suggested a minimum requirement of around 36 core superfamilies for a viable NCLDV. This would be within range of the 47 NCLDV orthologous (core) genes uncovered by others (4, 20–23) and discussed further below.
FIG 2.
Pfams across the NCLDV. (a) From structural homology searches of all 9,671 proteins from the 20 viruses (covering both previously annotated and unannotated proteins), matches passing the 80% probability threshold were assigned to Pfam superfamilies according to Pfam tags mapping to the homology overlap region. A total of 429 protein superfamilies could be assigned, the top 50 of which are shown here. See Fig. S2 in the supplemental material for all 429 superfamily assignments and the ranking method. Grayscale indicates the number of matching proteins per superfamily per virus. Individual query proteins matching multiple superfamilies above the 80% probability threshold were included in the counts for multiple superfamilies according to the rules given in Materials and Methods. Superfamilies (here) are also referred to as “clans” by Pfam. (b) Superfamily diversity versus proteome size for the 20 NCLDV. For each virus, the total number of proteins in its proteome (x axis) is charted against the total number of superfamilies found among proteome members (summed from panel a; y axis). Green points indicate the two viruses with the greatest Pfam diversity (Megavirus and Mimivirus). Red point indicates Pandoravirus, a major outlier. The linear regression trendline (extended back to the y axis) applies to all datapoints except that for Pandoravirus.
Pan-NCLDV orthologous genes.
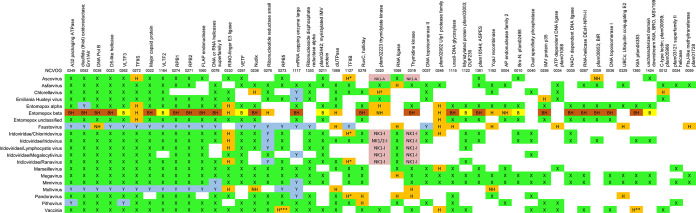
Protein sequence homology studies have identified a set of nucleocytoplasmic virus orthologous genes/groups (NCVOGs)—genes conserved across the NCLDV (4). Updated listings have accompanied the discovery of additional NCLDV (20, 22–27), and the current NCVOG count stands at 47 (24), with few changes accompanying more recent virus discoveries (21). Few of the NCVOGs are universally conserved among the NCLDV. Via our structure-based approach, viral coverage was extended in 44 of the 47 NCVOGs (21 NCVOGs if excluding entomopox beta, which was not included in prior analyses, Table 2). For two NCVOGs, namely, RING-finger E3 ligase and the “pfam02902 Ulp1 protease family” (Table 2), orthologs were found for the first time in seven distinct viruses. With the finding of structural homologs in four viruses, coverage of the transcription elongation factor TFIIS was extended to cover all 20 viruses (Table 2). The four TFIIS paralogs found in Pandoravirus alone (all of which were previously annotated as uncharacterized proteins) supported the expansion of the ultralarge Pandoravirus genome by a paralogization mechanism (Fig. 2b).
TABLE 2.
Expanded coverage by structure-based deep mining of 47 genes previously designated NCVOGsa
NCVOGs (4, 21, 22, 24), conserved among NCLDV on the basis of protein sequence homology, are ordered (left to right) by descending coverage among our 20 NCLDV. NCVOGs are named according to additional file 4 in Yutin et al. (24). “X” (green) indicates prior coverage on the basis of sequence homology (see references 21 and 25) and references therein); “Y” (lilac) indicates NCVOG additions in Koonin and Yutin (21). “H” (mustard) indicates new coverage via structure-based deep mining. H*, TFIIB structural homolog lacking zinc finger domain. H**, known KilA-N domain protein from literature (vaccinia protein p28 [67, 102, 103]). H***, known RPB5 homolog from literature (vaccinia protein RP22 [15]). “NH” indicates deep mining result supported by UniProt protein name (not present in prior NCVOG analyses). Since prior NCVOG analyses did not include entomopox beta, coverage for this virus was partially elucidated by BLASTP search (“B,” yellow). “BH” (brown) indicates combination of BLASTP and deep mining. NK1, NK2 (pink), nucleoside kinase-type (NK) proteins are dually listed between NCVOGs 0319 and 0320 to cover both the original designation and our interpretation (see Table S2 in the supplemental material). Coverage may appear low for some NCVOGs since they were designated as such on the basis of all known NCLDV (25) as opposed to the 20 representatives considered here.
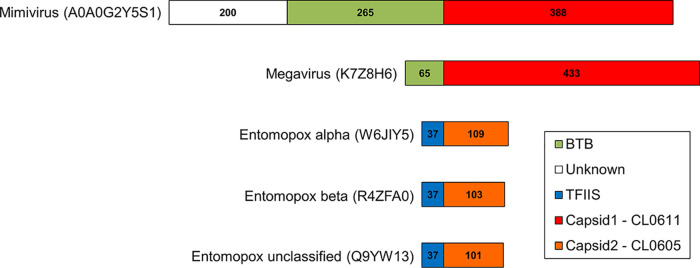
The “major capsid protein” NCVOG (Table 2) covers two distinct protein superfamilies: CL0055 (“nucleoplasmin-like viral coat and capsid proteins superfamily”), which is based on the jelly roll fold and includes the “D13-like” external scaffold of the poxviridae (28), and CL0611 (“hexon-like superfamily”), which includes beta-sandwich viral coat proteins such as the recently characterized Chlorella virus capsid (29). Here, domains belonging to CL0611 were found in proteins from Mimivirus and Megavirus, previously annotated “BTB/POZ-containing,” in which the capsid-like domain was fused to the C-terminal side of the BTB/POZ domain in a novel chimeric arrangement (Fig. 3). In addition, structural matches were found to a third capsid protein superfamily, CL0605 (“single-stranded DNA [ssDNA] viruses nucleoplasmin-like/VP coat superfamily”). Proteins in the latter superfamily comprise a beta sandwich with two sheets in a jelly roll topology, as found, for example, in coat protein VP2 of parvo-like virus AAV2. Among our 20 viruses, structural matches to CL0605 were found exclusively in the Entomopoxvirinae, and all had a TFIIS-type zinc finger fused to the protein N terminus (Fig. 3). These orthologs had no BLASTP counterparts in any organism outside the Entomopoxvirinae (not even a vaccinia ortholog, for example), and no other member of CL0605 possessed a TFIIS-type fusion. Nonetheless, examples of viral structural proteins incorporating zinc fingers have been reported, include the retrovirus nucleocapsid protein (30) and the reovirus capsid proteins delta 3, sigma 3, and lambda 1 (31–34). Notably, the entomopoxvirus proteomes were found to possess no TFIIS-like protein other than the TFIIS-capsid protein fusion (Table 3). Structure-based deep mining also revealed two novel “capsid-like” members of superfamily CL0611 from Chlorella virus, which had chitin-binding domains fused either centrally or at the C terminus. However, this was described by others in detail while the current article was in preparation (29). Consistent with sequence-based homology approaches (21), structure-based deep mining revealed no capsid-like protein of any kind in the Pandoravirus proteome despite the very large size of its proteome and the apparent central role of capsid-like proteins for viruses in general.
FIG 3.
Chimeric “capsid-like” proteins in the NCLDV. A BTB/POZ domain is fused N-terminally to the capsid-like domain (Pfam CL0611; see text) of Mimivirus and Megavirus proteins A0A0G2Y5S1 and K7Z8H6, respectively (both annotated in UniProt as “putative BTB/POZ domain-containing protein”). A TFIIS-type zinc finger is fused N-terminally to the capsid-like domain (Pfam CL0605) of entomopoxvirus alpha, beta, and unclassified proteins W6JIY5, R4ZFA0, and Q9YW13, respectively. These were annotated in UniProt as “capsid protein, polyoma VP1-like,” “uncharacterized protein,” and “uncharacterized protein MSV079,” respectively. These proteins had no BLASTP counterparts outside the Entomopoxvirinae. The Entomopoxvirinae were unique among the NCLDV examined here in possessing two capsid-like proteins each. The second one, a CL0611 superfamily member, is likely an external scaffold used during virion morphogenesis (see the text).
TABLE 3.
Orthologs of yeast MSDDRP subunits and basal transcription factors in NCLDV found by all methodsa
RPB12 homologs, which are considered separately in Fig. 4a, were omitted. Apart from Vaccinia virus, yeast nomenclature is used. Green indicates MSDDRP subunits annotated correctly prior to Mirzakhanyan and Gershon (16). Yellow indicates MSDDRP subunit annotation newly presented in Table 2 of Mirzakhanyan and Gershon (16) via sequence homology searching. Gray indicates the same as yellow, but split gene (16). Cyan indicates newly identified here by structure-based deep mining. Rows are ordered/underlined according to a phylogenetic tree inferred from a binary trait matrix of subunit/transcription factor presence/absence (see Fig. S3 in the supplemental material). Although classified appropriately according to Pfam and InterPro databases, some subunits were not annotated accordingly in UniProt (see Table S1 in the supplemental material). The yeast subunit nomenclature provides a basis for nomenclature unification across the NCLDV. *, 78% probability. **, TFIIB cyclin domain only. ***, Entomopox TFIIS is fused to the N terminus of a capsid-like protein (Fig. 3; see text).
For some NCVOG proteins, deep mining revealed additional paralogs within a virus proteome. For example, two copies each of the RPB1, RPB2, and RPB5 subunits of DNA-directed RNA polymerase (below) were found within the Megavirus proteome (data not shown).
Multisubunit DNA-dependent RNA polymerase and transcription factor orthologs.
Eukaryotes encode 12-subunit DNA-dependent RNA polymerases (DDRPs) comprising two large subunits (RPB1 and RPB2 and a number of smaller ones). As long established, the poxviruses encode an 8-subunit enzyme comparable in architecture to the eukaryotic one (35). Subunits of the vaccinia enzyme are orthologous to eukaryotic subunits (see references 15 and 35 and references therein) and other NCLDV (see reference 16 and references therein), among which the two largest, RPB1 and RPB2, are well conserved at the sequence level (16) (Table 3). So far, just one NCLDV, namely Chlorella virus, has failed to yield any known DDRP subunits at all via any search method, including the structure-based deep mining here (Table 3 and Table S1 in the supplemental material; see also below). This failure included a thorough inspection of all Chlorella virus search results in the current study for matches below our 80% probability threshold. This nondetection strongly reinforced a conclusion that, perhaps uniquely among the NCLDV, Chlorella virus does not encode a DDRP enzyme. It does, nonetheless, encode orthologs of transcription initiation factors TBP and TFIIB and transcription elongation factor TFIIS (Table 3 and Table S1).
Viral orthologs of the eukaryotic smaller subunits have proven much more elusive than those of the two large subunits, due to their much weaker protein sequence conservation. Earlier studies (15, 16) demonstrated the potential for structure-based homology searches to find small subunits among the NCLDV and highlighted some instances of their prior misannotation. Here, we have completed the structure-based mining of DDRP small-subunit genes in the NCLDV (Table 3 and Table S1). Yeast RNA polymerase II shows an RPB3-10-11-12 subassembly (36). The generally coincident presence/absence of RPB3, RPB10, and RPB11 in the NCLDV is now quite clear (Table 3), suggesting the coordinated acquisition/loss of this subassembly during viral evolution. Interestingly, the presence/absence of this subassembly seemed partially complementary to that of poxviral subunit RP35. The absence of both RPB3-10-11 and RP35 in Ascovirus, the Iridoviridae, and the giant viruses Marseillevirus, Mollivirus, Pandoravirus, and Pithovirus raises the possibility of an as-yet-undetected complementary subunit or subassembly for these viruses that is unrecognizable in the absence of functional enzyme purification. The entomopoxvirus MSDDRP seems distinct from vaccinia in containing no obvious homolog of RP22 or RP07 by structural homology or BLASTP.
An additional finding was the presence of an apparent RPB8 subunit in EhV-86, with 95% probability (Table 3). RPB8 has previously been found only in eukaryotes (37) and some archaea (hyperthermophilic Crenarchaeota and “Candidatus Korarchaeota” [38, 39]). The finding of RPB8 in a virus is therefore novel. Using the EhV-86 ortholog as a BLAST query, all additional RPB8 orthologs were from other Emiliania huxleyi viruses. The resulting protein cluster showed strong amino acid sequence conservation with no indels (see Fig. S4 in the supplemental material). In contrast, Emiliania huxleyi virus and Saccharomyces cerevisiae RPB8 protein sequences showed only weak sequence similarity, along with substantial truncation of the Emiliania huxleyi virus protein relative to yeast (Fig. S4). It is not clear why viral representation of RPB8 would be confined to just a single genus (Coccolithovirus) of a single NCLDV family (the Phycodnaviridae). A prior study (16) suggested two patterns of overall RNA pol subunit representation among the Phycodnaviridae, in which the coccolithoviruses could be grouped with the Prymnesiovirus genus, Chrysochromulina ericina virus (CeV01), Aureococcus anophagefferens virus, and unclassified Organic Lake phycodnavirus 1 and 2. However, RPB8 was not detected in any of these relatives.
In eukaryotes, basal promoter utilization by RNA polymerase II is mediated by two basal transcription factors, namely TATA binding protein (TBP), which binds the TATA element of the eukaryotic promoter, and TFIIB, whose C-terminal cyclin domains interact with TBP and whose N-terminal zinc finger interacts with RNA polymerase II. By structure-based deep mining, novel TBP and TFIIB orthologs were predicted in 8 and 10 NCLDV, respectively (Table 3), although Ascovirus and Chloriridovirus TFIIB orthologs possessed only the cyclin domains and not the zinc finger. TFIIB was previously designated an NCVOG (Table 2) via protein sequence similarity (24). Although TBP has been previously identified in specific virus clades, it has not been designated an NCVOG. Approximately 50% of Mimivirus genes contain a conserved, upstream AAAATTGA motif, which may be structurally comparable to the TATA box promoter element (40). In a cursory analysis, we found similar sequences immediately upstream of the genes of some NCLDV (e.g., Megavirus and Ascovirus) but not others (e.g., Chlorella virus; data not shown).
Novel zinc ribbon protein superfamily.
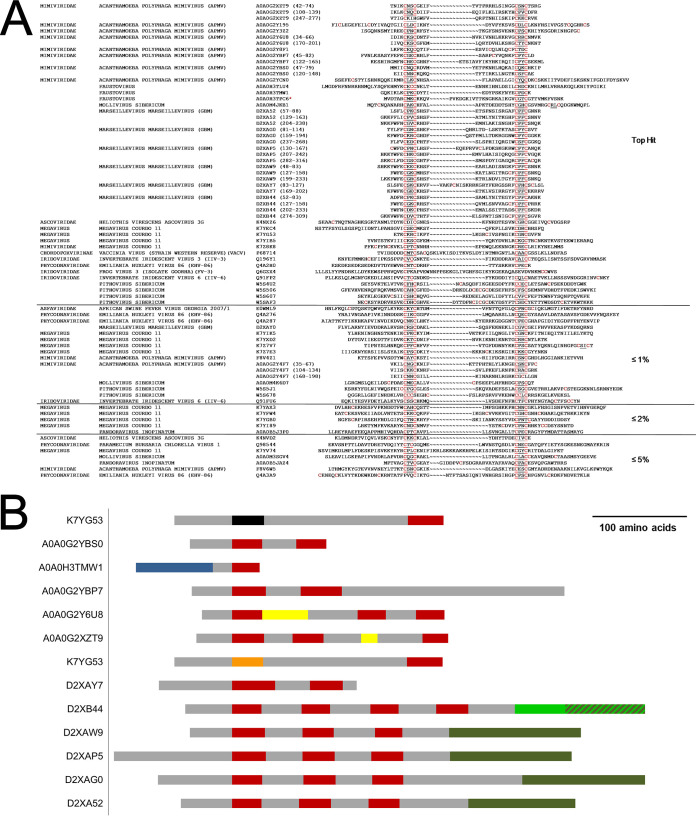
We also noted the presence among NCLDV of structural homologs to the zinc finger region of eukaryotic RNA polymerase subunit RPB12 (Fig. 4a). RPB12 is required for RNA polymerase open complex formation (41). RPB12 orthologs have not been identified previously in viruses, and the family shown in Fig. 4a may or may not represent bona fide RPB12. The N-terminal region of eukaryotic RPB12 encompassing the zinc finger region is known to form part of a larger zinc beta ribbon superfamily that includes eukaryotic transcription factors TFIIS and TFIIB and some ribosomal and other proteins (8, 42). Figure 4a may represent a broader zinc ribbon superfamily for the following reasons: (i) for 26 of the 58 NCLDV proteins shown, the top structural homolog comprised a non-RPB12 C4-type zinc finger-containing protein (Fig. 4a; marked ≤1%, ≤2%, and ≤5%), although eukaryotic RPB12 was also a structural homolog within the 80% probability threshold; (ii) all proteins of Fig. 4a lack the conserved C-terminal region characteristic of eukaryotic RPB12 (41); (iii) overall sequence conservation among the 58 NCLDV proteins was nonexistent (data not shown); (iv) unlike RPB12, vaccinia protein A19, present within the family (UniProt accession number P68714; Fig. 4a), is not a core vaccinia RNA polymerase subunit (although it associates with transcriptional components and is required for vaccinia early gene transcription [43]); (v) whereas RNA pol subunits are typically present in NCLDV proteomes in a single copy, some NCLDV proteomes were found to contain multiple zinc ribbon protein family members (Table 4); and (vi) not all NCLDV query proteins matching a C4 zinc finger protein showed eukaryotic RPB12 as a structural homolog (data not shown). Nonetheless, a bona fide RPB12 subfamily seems to exist within the zinc ribbon superfamily of Fig. 4a. Supporting this, eukaryotic RPB12 was a specific structural homolog of even NCLDV sequences that entirely lacked a consensus zinc finger: In eight of the RPB12 structural homology regions (Fig. 4a), either the first or second CxxC of the finger motif contained a nonconsensus number of “x” residues or was missing a cysteine entirely. In the most dramatic example, the Ranavirus RPB12 homolog (UniProt accession number Q6GZX4; Fig. 4a) contained no CxxC at all.
FIG 4.
Structural homology between NCLDV proteins and eukaryotic RPB12 with scores exceeding the 80% probability threshold. (a) Sequence alignment. The great majority of NCLDV structural homologs possessed a CxxC….CxxC zinc finger (cysteines are highlighted in red). In total, 77 structural homology regions are shown, within a total of 58 proteins. The majority of these proteins were annotated as transcription factors, restriction endonucleases, zinc ribbon-containing proteins, or NAD-glutamate dehydrogenase, or were unclassified. For proteins containing multiple matching regions, the individual regions are indicated by sequence position in parentheses after the accession number. In only one instance (red star) does the homology region shown comprise the entire NCLDV protein. “Top hit” indicates that eukaryotic RPB12 was the highest scoring structural match to the homology regions in this section. ≤1%, ≤2%, and ≤5% indicate that eukaryotic RPB12 was within 1, 2, or 5 percentage probability points, respectively, of the highest scoring structural homolog (and was not necessarily the number two homolog). This applied to 26 of the 58 proteins shown here. (b) RPB12 orthologs from panel a that contain additional regions of structural homology. Red, RPB12 (PF03604) homology regions; black, SH3 (PF00018) homology; blue, HypA (PF01155) homology; orange, Nab2 (PF11517) homology; dark green, VSR endonuclease (PF03852) homology; light green, type IV restriction endonuclease homology; striped light/dark green, overlapping type IV restriction endonuclease and VSR endonuclease homology; yellow, structural homology with “uncharacterized protein PF0385” (Q8U3S0_PYRFU); gray, uncharacterized region.
TABLE 4.
Number of RPB12-ZnF proteins versus total number of proteinsa
| Virus | Total no. of proteins | No. of RPB12-ZnF proteins |
|---|---|---|
| Megavirus | 1,217 | 13 |
| Mimivirus | 979 | 11 |
| Marseillevirus | 428 | 7 |
| Pithovirus | 467 | 6 |
| Emiliania huxleyi virus | 472 | 4 |
| Mollivirus | 514 | 3 |
| Faustovirus | 492 | 3 |
| Pandoravirus | 1,839 | 2 |
| Iridovirus | 469 | 2 |
| Ascovirus | 194 | 2 |
| Chlorella virus | 794 | 1 |
| Vaccinia | 217 | 1 |
| Asfarvirus | 188 | 1 |
| Chloriridovirus | 126 | 1 |
| Ranavirus | 98 | 1 |
| Entomopox beta | 311 | 0 |
| Entomopox unclassified | 261 | 0 |
| Entomopox alpha | 241 | 0 |
| Lymphocystivirus | 239 | 0 |
| Megalocytivirus | 125 | 0 |
Numbers of RPB12-type zinc finger-containing (RPB12-ZnF) proteins per NCLDV proteome by counting proteins shown in Fig. 4a (RPB12-ZnF) versus the total number of proteins per proteome (all proteins). Nearly half of the 58 proteins were from Mimivirus or Megavirus, while several virus taxa (Entomopoxvirinae, Megavirus, Lymphocystivirus, Megalocytivirus) had none at all. As a function of proteome size, representation in Chlorella virus and Pandoravirus was low.
Some of the RPB12 structural homologs of Fig. 4a contain additional domains or motifs, such as an N-terminal SH3 motif or C-terminal very short patch repair (VSR) endonuclease domain (Fig. 4b). In the majority of these proteins, multiple repeating RPB12 structural homology domains were separated by regions with no detectable structural homology. VSR endonucleases have not been previously observed with zinc finger motifs, although some group II HNH endonucleases, which share a similar catalytic core with VSR, contain a C4-type zinc finger domain upstream of the C-terminal HNH endonuclease domain (44). The RPB12-like repeats observed here may be involved in DNA binding. Overall, it seems likely that RPB12 is a subset of a larger protein superfamily.
Endonucleases.
DNA endonucleases fall into several major structural families and superfamilies (Fig. 5a) (44–47). The broadest of these is perhaps “PD-(D/E)xK”, which is defined on the basis of a conserved PD-(D/E)xK motif essential for catalysis. It encompasses, functionally, the type I to IV restriction endonucleases (REases), which cleave both DNA strands within a specific recognition sequence (48, 49), MMR (mismatch repair)-type and VSR-type nicking and other endonucleases (50, 51) (Fig. 5a), with superfamily members existing as either monomers, homodimers, or homotetramers. Of the four types of REase (Fig. 5a), type II REases are the best characterized and the most prevalent in the biosphere, with more than 3,500 known members (46, 48, 49). Although found predominantly in bacteria, they are also encoded by Chlorella virus (52), with, for example, two reported in PBCV-1 (R.CviAI [53] and R.CviAII [54]), an additional two in Chlorella virus NY-2A (55, 56) and one in Chlorella virus IL-3A (57). A large number of additional Chlorella virus type II REases can be found in the REBASE database (http://rebase.neb.com/rebase/rebase.html) (43 enzymes from 34 distinct Chlorella virus species), most of which remain unpublished. The biological roles of the Chlorella virus enzymes remain unverified (52).
FIG 5.
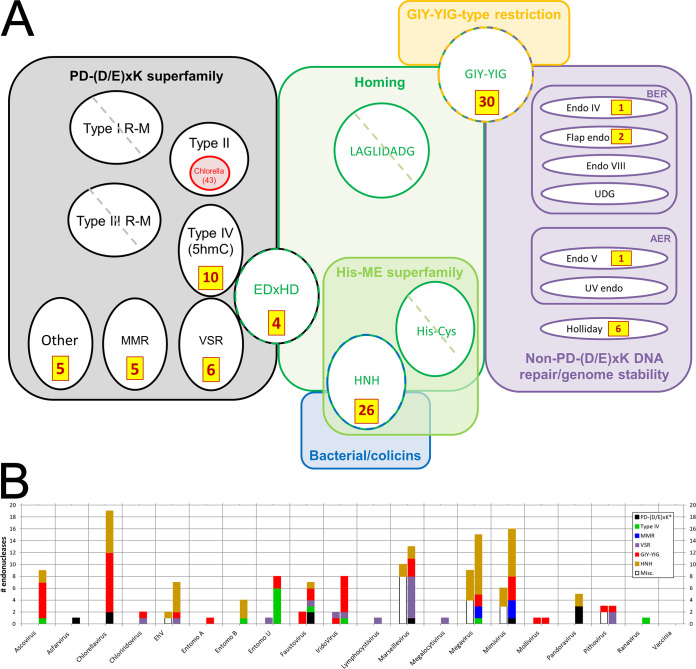
Structure-based deep mining markedly elevates numbers of endonucleases identified across the NCLDV. All accession numbers are given in Table S3 in the supplemental material. (a) Counts of newly identified NCLDV endonucleases (brown/yellow boxes) mapped onto known DNA endonuclease structural/functional classes (colored boxes/circles/ovals). Circles/ovals with no counts shown indicate major classes with no representatives reported among any NCLDV (diagonal hatch) or none newly identified here (no hatch). PD-(D/E)xK (black), structural superfamily showing the following functional classes: REases (types I to IV), nicking endonucleases for DNA mismatch repair (MMR), and very short patch repair (VSR), and one class of homing endonucleases (EDxHD). The VSR, type IV, and EDxHD groups are shown touching to illustrate their particularly close structural relationship in search results (see the text). Types I and III REase polypeptides are denoted “R-M” due to their dual function as restriction-modifying enzymes, in which we focused only on the “catalytic site for DNA cleavage” (96) and “endonuclease domain” (97), respectively. Red indicates strain depth dimensionality for Chlorella virus type II REases specifically (from REBASE rather than UniProt; see text). Other major colors indicate functional classes encompassing either structural families (green, blue, and orange) or functional classes (purple). In the latter, BER and AER represent base and alternative excision repair, respectively; UDG, uracil DNA glycosylase (an endonuclease); Endo IV, AP-endonuclease. Counts do not include our assignments/reassignments of previously identified/annotated endonucleases (see the text). (b) NCLDV endonuclease counts by virus. Entomopox A, B, and U refer to entomopox alpha, beta, and unclassified, respectively. Bars to the left and right of the central tick represent counts before and after deep mining, respectively. “Before” counts include proteins that matched an endonuclease here and were also “endonuclease” or “nuclease” according to UniProt gene_name. “After” counts represent “before” counts plus endonucleases newly identified here plus reassignments (see the text). Each bar is divided by color according to endonuclease class (see color legend). The PD-(D/E)xK* class refers to PD-(D/E)xK homing plus “Other”” (panel a). The “Misc.” class (“before”) contains, exclusively, members reassigned to other classes in the “after” sections based on primary structural homolog (see the text). Excluded from the graph are all Chlorella virus “red ring” (panel a) restriction endonucleases not from PBCV-1 (Table 1). For simplicity, the small numbers of repair endonucleases (purple section of panel a) are omitted. Counts represent “top hit only” structural matches).
Sequence is not very effective as a homology tool for the prediction of endonucleases, particularly those of the PD-(D/E)xK superfamily (58) or the homing endonuclease classes shown in Fig. 5a. However, conservation of secondary structure (59, 60) within, for example, the PD-(D/E)xK-containing catalytic domain (48, 58, 60) facilitated our structure-based deep mining approach. Here, a total of 96 new endonucleases were predicted over a number of structural and functional classes (Fig. 5a). These supplemented 36 proteins among our 20 NCLDV whose UniProt annotations already included the strings “endonuclease,” “restriction endonuclease,” or “nuclease” (these 36 included two proteins mentioned in the literature as VSR-type nucleases but missed by UniProt [61]). An updated overall total count of 132 endonuclease/nucleases was therefore yielded by these numbers (Fig. 5; see also Table S3 in the supplemental material). For 10 of our 20 NCLDV, deep mining provided the first-time prediction of any endonuclease (Fig. 5b), and for many of the remaining viruses prior endonuclease genes numbered just one or two. Totals were highly variable from virus to virus, even among comparable NCLDV (e.g., among the amoebal giant viruses Pithovirus, Pandoravirus, Mollivirus, Megavirus, and Mimivirus) or between the three entomopoxviruses (Fig. 5b), suggesting that their roles may not be central to virus replication. Endonuclease classes predicted in the NCLDV for the first time included type IV/5hmC REases, which were the primary structural homologs of 10 newly predicted endonucleases from seven NCLDV (Fig. 5). All of these were annotated by UniProt as uncharacterized, ALI motif, leucine-rich repeat, or N1R/p28-like proteins on the basis of distinct (nonnuclease) domains. Six of them were from one virus alone (entomopox unclassified; Fig. 5b)—the largest number within a single NCLDV genome. The type IV/5hmC class of REases recognize modified (typically methylated) DNA, suggesting a specific need to restrict methylated DNA among the NCLDV. In another example, UniProt showed no prior occurrence of an NCLDV VSR endonuclease (the two noted by Aravind et al. [61], above, were reassigned here). In contrast, several major classes of endonuclease remained entirely unrepresented among the NCLDV even after deep mining (Fig. 5a). These included the type I and III REases and the LAGLIDADG and His-Cys homing endonucleases (which do not appear to be underrepresented in the PDB), suggesting that modification-coupled DNA restriction and homing are profoundly redundant functions for the NCLDV. Indeed, the finding of NCLDV enzymes that restrict methylated DNA (the type IV/5hmC REases, above) would suggest a reason why methylation may be irrelevant as a self-protection mechanism.
In addition to the newly predicted endonucleases, many of the 36 previously reported nucleases (above) were assigned to a specific class or reassigned based on their primary structural homolog (Table S3). For example, nine NCLDV proteins annotated by UniProt as either “restriction endonuclease” (n = 6), “group 1 intron putative endonuclease” (n = 1), “putative nuclease” (n = 1), or “helicase nuclease” (n = 1) were assigned to the VSR subset of PD-(D/E)xK. In another example, protein 069L from IIV-6 (Table 1) and protein MSV196 from MSV (Table 1), were reassigned from VSR-type endonucleases (61) to type IV/5hmC endonuclease (47, 62, 63), since VSR appeared as only the 5th-ranked structural match for each of the two proteins, with 5hmC versus VSR probabilities of 96.9%/94.4% and 97%/94.8%, respectively. Their UniProt annotations showed them as Bro-N domain (PF02498)-containing (069L) or “ALI motif gene family” (MSV196—the “ALI” motif being a subset of Bro-N). These annotations were based on different domains within the two proteins.
No structural homologs were found above the 80% probability threshold for either of two known Chlorella virus restriction endonucleases, R.CviAI (53) and R.CviAII (54) (discussed above). Apparently, they did not align well with any structures in the PDB database, in which type II enzymes were well represented. While the HHsearch structural homology tool can find large numbers of authentic PDDExK enzymes, it can fail with PDDExK protein subfamilies with few members or more distant structural homology (58). Perhaps functional type II REases cover a wider fold space than PD-(D/E)xK alone.
Despite MMR endonucleases (EndoMS and NucS-type) being quite uncommon in the biosphere overall (64, 65), five such endonucleases were predicted here, all from Megavirus and Mimivirus, all of which were previously annotated as “uncharacterized” or “KilA-N domain-containing” proteins. EndoMS and NucS typically have an N-terminal DNA binding/dimerization domain and C-terminal catalytic domain (66). While the C-terminal regions of the Mimivirus/Megavirus homologs matched the catalytic domain, the N-terminal regions comprised an APSES or KilA-N type domain. Both APSES and KilA-N are DNA binding domains commonly found in eukaryotic viruses and in cellular LAGLIDADG endonucleases (67).
GIY-YIG family endonucleases (Fig. 5a) are typically encoded by phage and fungi (44), in which their most common function is homing/self-propagation of group 1 homing introns (68, 69). They are typically small proteins (approximately 100 amino acids) with short “GIY” and “YIG” motifs in the N-terminal region, along with extended recognition sites for their DNA targets. Ten GIY-YIGs had been previously identified in seven NCLDV. Here, we predicted an additional 30 from an additional six viruses (Fig. 5), the largest increases being in Ascovirus, Chlorella virus and Iridovirus. Additional domains were found fused to the N- and/or C termini of some of these proteins, such as a NUMOD3, Tc5 transposase DNA-binding domain, CENP-B N-terminal DNA-binding domain, KilA-N, and HIT zinc finger domains (data not shown).
Members of another endonuclease family, HNH, are found as homing enzymes within group 1 and group 2 introns, and also as bacterial restriction endonucleases (e.g., PacI), colicins (70) and/or DNA/RNA nonspecific endonucleases (44, 45). PacI, a “rare-cutting” REase, cleaves duplex DNA within the sequence 5′-TTAAT̂TAA-3′ (44, 71). Group I homing endonucleases such as I-HmuI also have a highly conserved target site, but unlike PacI, they cleave only one DNA strand. DNA/RNA nonspecific endonucleases in the HNH family are extracellular (72) and function in bacterial self-defense against neutrophil extracellular traps (73), among other functions. Here, HNH endonucleases were the primary structural homologs of 26 NCLDV proteins, almost tripling the total known among our 20 NCLDV—the largest increases being observed in Chlorella virus and Mimivirus (Fig. 5b). Some of the newly predicted HNH endonucleases (Table S3) showed internal repeats of the I-HmuI or PacI homology regions.
Two Faustovirus and two Chlorella virus proteins, annotated in UniProt as “uncharacterized,” showed the homing endonuclease I-bth0305I as a top structural hit. I-bth0305I is annotated in UniProt as a “mobile intron protein” of a lineage that has been termed the “EDxHD family” (Fig. 5a). Some endonuclease classes, such as the VSR, type IV/5hmC, and EDxHD, which cover very distinct functional roles, were found to be particularly closely related in three-dimensional structure, with members of these classes interleaved in search results for a specific NCLDV query protein. Other NCLDV queries showed only a single structural homolog within the PD-(D/E)xK superfamily. This was probably not due to a paucity of closely related structural choices within the database, since more than 73 of the 142 Pfams within the PD-(D/E)xK superfamily have yielded crystal structures. Instead, individual structural homologs seem to have been selected against quite a fine-grained structural landscape. In yet other cases, cellular REases with a highly conserved type IV/5hmC fold have been found that lack nearly all of the commonly conserved residues (62). For all of the above reasons, we hesitate to assign functional roles to specific NCLDV endonucleases on the basis of structural homology alone.
Repeat domain proteins.
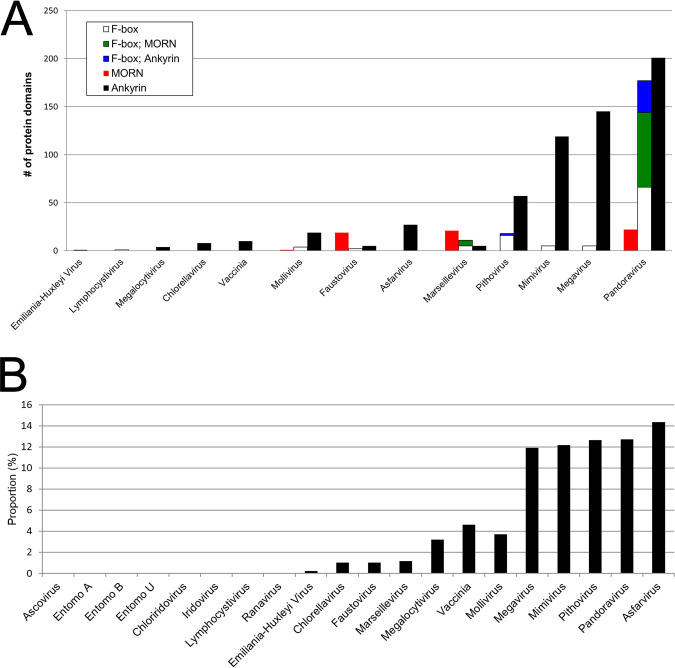
Numerous ankyrin repeat motif-containing proteins were identified in the genomes of the NCLDV, although not all NCLDV were found to encode them. Members of this protein family have recently been shown to target host defense proteins for degradation (74). Amoeba-infecting NCLDV show a correlation between genome size and the number of encoded proteins containing ankyrin, MORN, and WD40 repeat domains (75), with repeat-containing proteins in Megavirus, Mimivirus, and Pandoravirus comprising a substantial portion of their total proteomes (75, 76). Here, structure-based deep mining led to the identification of large numbers of additional repeat domain-containing proteins across the NCLDV (Fig. 6a), mostly identified with very high probability. The most substantial increases were found among the ankyrin- and MORN-repeat-containing protein families (Fig. 6a), with as many as 234 ankyrin repeat-containing proteins found in Pandoravirus. Pandoravirus, Pithovirus, Mimivirus, Megavirus, and Asfarvirus showed markedly higher proportions of their proteomes devoted to ankyrin repeat proteins than the other viruses (ranging from 11.9% to 14.4% overall; Fig. 6b). Although this group includes four amoeba-infecting viruses, three additional amoeba-infecting viruses (Faustovirus, Marseillevirus, and Mollivirus) showed substantially lower numbers (Fig. 6b), with only five ankyrin repeat-containing proteins found in Faustovirus (Fig. 6b) (75). Perhaps most surprising was the finding of 27 ankyrin repeat-containing proteins in asfarvirus (the highest proportion of all the proteomes; Fig. 6b) since UniProt contained no annotated ankyrin repeat proteins at all for this virus family (although three such proteins identified by Pfam were not autotransferred to UniProt). Structural matches comprised a diversity of ankyrin-repeat containing proteins in PDB and, as in the PDB structures, ankyrin repeats in the NCLDV queries are scattered throughout the protein.
FIG 6.
(a) Counts of ankyrin repeat (CL0465)-, MORN repeat (CL0251)-, and F-box motif (CL0271)-containing proteins per virus for 13 of the 20 representative NCLDV. The remaining seven NCLDV contained none. Blue and green, proteins containing an F-box plus repeat domains. Viruses are ordered (left to right) by overall numbers of such proteins per proteome. (b) Ankyrin repeat-containing protein counts as a proportion of total genes in the NCLDV proteome. Eight of our 20 NCLDV (left) lacked any such proteins. NCLDV are labeled as in panel a and Fig. 5b.
A total of 147 MORN (membrane occupation and recognition nexus) repeat-containing proteins were also discovered, almost entirely in the amoeba-infecting Faustovirus, Marseillevirus, and Pandoravirus (Fig. 6a). Many of the NCLDV query proteins were annotated in UniProt as “unclassified.” In contrast to the ankyrin repeat-containing queries (above), the 147 NCLDV queries matched only one MORN-repeat containing protein in PDB, namely, a histone methyltransferase. To the best of our knowledge, only a few MORN repeat-containing proteins have ever been identified in any organism. These include the junctophilins, a group of mammalian proteins found within membrane junctional complexes (77, 78), which serve to bridge membrane pairs (such as the plasma membrane and the membrane of the endoplasmic reticulum or sarcoplasmic reticulum). Within the bridge, the junctophilin’s N-terminal MORN motif, comprising eight repeats of a 14 amino-acid sequence (77), interacts with the phosphoinositides of one cell membrane (see Jiang et al. [78] and references therein), while a hydrophobic C-terminal transmembrane region anchors to the other. The NCLDV MORN repeat proteins do not appear to be acting as junctophilins. Of ∼40 NCLDV MORN repeat proteins examined at random, only one had a transmembrane region, and it was located at the protein N terminus, rather than the C terminus (data not shown). Moreover, the MORN repeat region tended to fall within the C-terminal halves of most NCLDV proteins. A second group of MORN repeat-containing proteins has been found in unicellular parasites such as Toxoplasma gondii and Toxoplasma brucei (79, 80). This group appears to interface membranes with cytoskeletal components. In the NCLDV, many of the MORN repeat-containing proteins also showed structural homology to a TCP10_C family domain (data not shown), which is centriole related. The centriole has two roles, namely, as part of the eukaryotic centrosome (a microtubule organizing center during mitosis) and as the basal body from which cilia and flagella emanate (81, 82). The NCLDV MORN repeat proteins with a TCP10_C domain may tether viral membranes to cytoskeletal structures such as those found in ciliated or flagellated amoebae.
In Pandoravirus, Pithovirus, and Marseillevirus, many of the proteins possessing C-terminal ankyrin or MORN repeat motifs also contain an N-terminal F-box (Fig. 6a). In Pandoravirus, 78 of the 100 identified MORN repeat-containing proteins and 33 of the 234 ankyrin repeat-containing proteins showed this arrangement. This orientation is novel for the NCLDV. The F-box domain of poxvirus ankyrin repeat-containing proteins, for example, is located at the protein C terminus (74, 83). Interestingly, the F-box domains of the poxvirus proteins scored well below our 80% probability threshold for structural homology (data not shown). Instances of N-terminal F-box with C-terminal ankyrin domain proteins have previously been described in Legionella pneumophila (84), but sequence alignments of NCLDV proteins with these proteins showed no obvious sequence homology (data not shown).
Structural homologs shared narrowly among NCLDV.
In addition to protein families shared broadly among the NCLDV (above), some structural homologs were shared more narrowly (see Table S4 in the supplemental material). Structural homologs were from a variety of organisms, which may simply reflect proteins amenable to structural biology or those having some specific interest rather than being a particularly relevant organism for the NCLDV. Nonetheless, the broad representation of microbes among structural homologs (Table S4) suggested the possibility of horizontal gene transfer during microbial processes such as phagocytosis. For the most part, structural homology was at the domain or fold level only, so the corresponding protein annotations tended to be structurally oriented (e.g., CHAP domain, winged helix-turn-helix) and therefore unsatisfying in deducing the overall function of the NCLDV protein. In one case, however, the probable function was clear, namely, for NCLDV homologs to herpesvirus glycoprotein B.
Herpesvirus glycoprotein B.
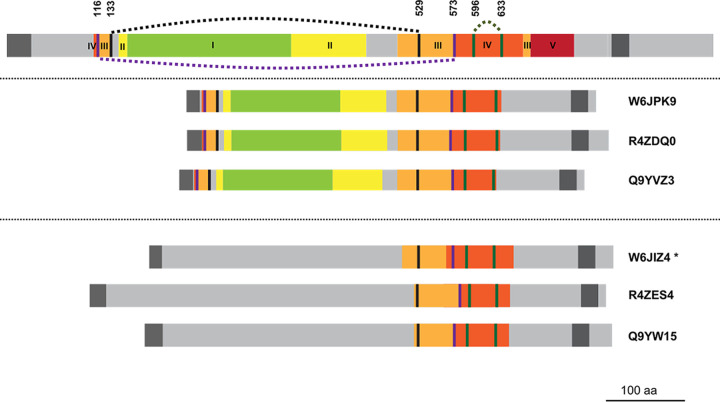
NCLDV structural homologs of herpesvirus glycoprotein B (gB) were detected, although the Herpesviridae are not considered members of the NCLDV due to their exclusively nuclear sites of replication (85). gB is an essential, trimeric herpesvirus surface glycoprotein—the most highly conserved member of the 5-protein herpesvirus host cell fusion and virus entry complex (86). It features an N-terminal signal sequence, ectodomain (comprising at least 80% of the 904-residue protein), C-terminal transmembrane anchor, and a relatively short cytoplasmic tail (Fig. 7). The crystal structure for the gB ectodomain (87) shows five distinct subdomains connected by flexible linkers, and five intramolecular disulfide bonds (87) (Fig. 7). Subdomains III and IV are each discontinuous in the linear sequence and are stabilized by a disulfide bond (87). A pair of structural homologs was found in each of three entomopoxviruses (Fig. 7). Like HSV-1 gB, the six homologs each showed a predicted N-terminal signal sequence, an apparent ectodomain, C-terminal transmembrane anchor, and a short cytoplasmic tail. One of the two protein clusters (Fig. 7, center) was highly structurally homologous to HSV-1 gB (98% probability, covering all gB subdomains except subdomain V; Fig. 7). The disulfide bonds stabilizing the discontinuous segments of subdomains III and IV were conserved (87), as was the disulfide bond within domain IV (Fig. 7).
FIG 7.
HSV-1 envelope glycoprotein B (gB) aligned with six proteins from entomopoxviruses. (Upper) HSV-1 (strain KOS) protein gB (UniProt accession P06437). (Center) Entomopox alpha, beta, and unclassified accessions W6JPK9, R4ZDQ0, and Q9YVZ3, respectively. Lower section: Entomopox alpha, beta, and unclassified accessions W6JIZ4, R4ZES4, and Q9YW15, respectively. Green, yellow, orange, red, and brown indicate subdomains of the HSV-1 gB ectodomain labeled I to V, respectively, in Heldwein et al. (87). These subdomains were localized within entomopoxvirus proteins by visual inspection of conserved residues identified by multiple sequence alignments and on the basis of secondary structural alignment (data not shown). Intervening light gray regions were not shown in the crystal structure (87). Dark gray, predicted N-terminal signal peptide and C-terminal transmembrane regions. Of the five disulfide bonds conserved among herpesviruses (87), three are shown (broken curved lines joining pairs of colored vertical lines: 133 to 529 [black]; 116 to 573 [purple]; and 596 to 633 [green]). The two remaining disulfides, in HSV-1 gB domains I and II (not shown), were missing from all six entomopox proteins. In the lower protein cluster, only the 596 to 633 cysteine pair is preserved. The starred accession was annotated “putative glycoprotein B” in UniProt, following the BLASTP homology noted in Table 1 of Mitsuhashi et al. (98), while the others remain “uncharacterized.”
The other entomopoxvirus protein cluster (Fig. 7, lower) showed lower overall structural homology to gB (96% probability), covering only the C-terminal segments of the two discontinuous subdomains—III and IV—and only one of cysteine from each pair of segment-bridging disulfides. The disulfide within domain IV was, however, preserved (Fig. 7). The subdomains that were most conserved highly between HSV-1 gB and the six entomopoxvirus proteins, namely subdomains III and IV, lie in the most exposed, membrane-distal region of the protein (87).
Unique structural homologs.
Additional structural homologs were identified in individual NCLDV only (see Supplemental Results and Table S5 in the supplemental material). As in Table S4, a homology region extending beyond just the single-domain level and accompanied by an explicit Pfam functional description were considered functionally predictive for the NCLDV query protein. Conversely, where the Pfam descriptor was generic and/or the homology region was only narrowly localized, the result was considered diagnostic of a structural fold only. A number of unique structural homologs were identified (see Supplemental Results and Table S5), some of which represented protein classes that were not, apparently, identified previously in any virus. These included a phenolic acid decarboxylase, a prokaryotic-type ribosomal protein (to our knowledge the first to be reported in a eukaryotic virus), a gasdermin-related apparent molecular decoy, a proteasomal subunit, an HIG1 domain family member (HIG1 being induced by hypoxia), and the first report to our knowledge of cysteine knot proteins in a virus, including an apparent defensin (see Supplemental Results and Table S5).
Transmembrane domains and potential signal sequences.
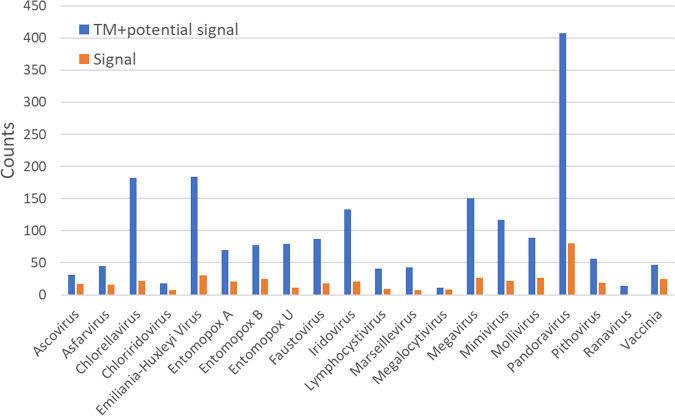
In addition to structural homology searching, we enumerated predicted transmembrane (TM)-containing (88) and potential secretory signal peptide-containing (89) proteins among the 20 viruses (Fig. 8). One of the more unexpected of the predicted TM domains/membrane anchors was located at the N terminus of the ETF1 subunit of the heterodimeric vaccinia transcription factor VETF (see Fig. S5 in the supplemental material). However, VETF is considered to be packaged in the virion core, compartmentalized away from the virion envelope by the proteinaceous virion core wall. Since the repression of either ETF1 or ETF2 synthesis during infection is known to lead to a block in virion morphogenesis (90, 91), it seems possible that this TM domain may be a membrane attachment point during virion morphogenesis—perhaps for the packaging of a vaccinia transcriptosome-based assembly (92) or “nucleoid” (93). In support of such a model, the morphogenic block upon repression of the ETF2 subunit yields immature virions lacking genomic DNA (90).
FIG 8.
Numbers of proteins per viral proteome possessing a predicted transmembrane domain (88) and potential secretory signal peptides (89) enumerated per viral proteome. Some overlap may exist between the two sets of counts.
Conclusions.
Here, structural homology was used to expand the annotation of previously unclassified proteins. This approach proved very successful. Gaps among “core” (NCVOG) proteins were filled, and additional RNAP subunits and basal transcription factor homologs were identified, along with many new endonucleases and proteins with functions not previously described in any virus.
In considering the merits of structural over sequence homology, the latter seems challenged in extending protein families with low sequence homology, such as the REases, or those with high sequence homology and therefore already essentially complete, such as the serine/threonine protein kinases. The structural approach will be as powerful as the number of annotated three-dimensional structural models present in the PDB, with the possibility of a bias in structural databases toward proteins of medical and/or economic importance.
MATERIALS AND METHODS
Version 3.0.0 of the HHsuite package was installed on the High-Performance Computing Cluster at University of California—Irvine/Research Cyber Infrastructure Center. The usage of HHsuite, including the interpretation of results, has been well-described by its developers and earlier users (https://github.com/soedinglab/hh-suite/wiki and https://toolkit.tuebingen.mpg.de/tools/hhpred) (10, 12, 17–19, 58, 94). Briefly, for each of the 20 viruses in Table 1, the UniProt complete proteome was downloaded and the resulting data set deconstructed to individual FASTA protein sequence files. For each of the resulting query protein sequences, a multiple-sequence alignment (MSA) was generated using HHblits in batch mode against “uniprot20,” a database of UniProt sequences clustered at the 20% sequence identity level provided by HHsuite. The threshold for sequence inclusion in an MSA was an E value of <10−3. After supplementing MSAs with PSIPRED-generated secondary structural information via the addss.pl tool, profile HMMs combining information from MSAs and their corresponding secondary structure predictions were generated via the tool HHmake. Via the HHsearch tool, the resulting profile HMM were used as sequential queries against a database derived from pdb70 (downloaded from the HHsuite server). Searches were made in local alignment mode with the maximum accuracy alignment algorithm (MAC) “on.” Any initial search terminating with error was rerun using the HHpred server (part of the MPI Bioinformatics Toolkit) with greater numbers of search iterations, modified MAC realignment, or MAC turned off.
One output (.hhr) file was generated per match per query protein and contained extensive header information and a text version of the structural alignment. In-house code was used to extract, from .hhr files, the PDB identifiers and chains of matching structures, statistical match scores, query homology regions, and the target homology regions, then tabulate them on a per query basis. The resulting tables were annotated with query accession number, descriptor, and protein length (from the individual protein FASTA files used as HHblits inputs), then annotated as well with the query protein’s UniProt keyword, gene ontology (GO) biological process, and GO molecular function annotations. The resulting tables were then thresholded at 80% probability in accordance with reports (10, 17) on the high specificity and accuracy of this threshold.
Filtered data were then further annotated manually with motif, domain, and/or other protein information derived from www.rcsb.org by manual lookup via the homology target’s PDB identifier. Manual annotation of homology regions was aided by visual inspection in RCSB’s “full protein feature view” of regions in the target’s primary structure covered by X-ray crystal structures and/or coincident with domains in the Pfam database, transmembrane domains, and/or other features. These annotations (notably all associated Pfams) were then transferred to the query sequence after correction for the differential sequence positions of the homology region in query and target. Query proteins with multiple distinct homology regions were annotated according to all, and query proteins with overlapping homology regions to distinct target proteins were annotated according to the highest probability score. For Pfams within higher order groupings (superfamilies or clans), the former were replaced with the latter (e.g., for heatmap figures).
Rules for the assignment of HHsearch output to multiple superfamilies (heatmap).
Five ambiguous situations were handled as follows. (i) A query structurally homologous to distinct superfamilies via distinct regions of the query (e.g., N-terminal F-box, C-terminal ankyrin) was enumerated under both superfamilies. (ii) If a query was structurally homologous to a single target protein with repeats of a superfamily match, each unique superfamily was listed only once per query and counted as a single hit for the heatmap. (iii) If a query was structurally homologous to multiple target proteins in the 80 to 100% probability range that included multiple superfamilies, only the superfamily associated with the highest-probability target was enumerated, or the target with greatest coverage if probabilities for both were similar. (iv) Query proteins with highly fragmented homology regions (e.g., collagen-like proteins and query proteins with extended coiled coil regions) were searched again via the HHpred server with a lower MAC realignment threshold or in global realignment mode to yield greater alignment length. (v) Target proteins with no Pfam identifiers across the homology region (Fig. S1, example 7) were excluded from heatmaps.
Transmembrane and secretory signal peptide search.
FASTA files of the complete UniProt proteome for each of the 20 viruses were searched for putative transmembrane helices and secretory signal peptides using TMHMM v2.0 (88) (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0) and SignalP v5.0 (89) (https://services.healthtech.dtu.dk/service.php?SignalP-5.0), respectively. TMHMM v2.0 was run with a single-line output per protein, then filtered to retain proteins with at least one predicted transmembrane domain (which may also serve as a signal peptide). For SignalP v5.0, proteomes were searched for matches in Eukarya, then filtered to retain proteins with predicted secretory peptides.
Dolpenny.
Dolpenny (95) and Consense programs were installed as part of the PHYLIP package from the University of Washington website (http://evolution.genetics.washington.edu/phylip.html). Dolpenny was run using the Dollo parsimony method with species order set to be continually reconsidered. Ancestral states for all RNA polymerase subunits of Chlorella virus and for Megalocytivirus RPB5 were represented by a question mark (“?”). For all other RNA polymerase subunits, TFIIS, TFIIB, and TBP, they were represented as 1 and 0 for presence and absence, respectively. A rooted consensus tree was built from the Dolpenny output using Consense with the consensus type “Majority rules (extended).”
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by NIH grant R01GM134144-01. Y.M. was supported by U.S. Public Health Service training grant AI007319 from the National Institutes of Health.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The High-Performance Computing Cluster was administered by Harry Mangalam. We appreciate the help of Harry Mangalam and James Anthony Walker.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D. 2003. A giant virus in amoebae. Science 299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 2.Philippe N, Legendre M, Doutre G, Coute Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie JM, Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 3.Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, Raytselis Y, Sayers EW, Tao T, Ye J, Zaretskaya I. 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41:W29–33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer LM, Aravind L, Koonin EV. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol 75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodel AE, Gershon PD, Shi X, Quiocho FA. 1996. The 1.85Å structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell 85:247–256. doi: 10.1016/S0092-8674(00)81101-0. [DOI] [PubMed] [Google Scholar]
- 6.Mistry J, Bateman A, Finn RD. 2007. Predicting active site residue annotations in the Pfam database. BMC Bioinformatics 8:298. doi: 10.1186/1471-2105-8-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, Gough J, Haft DR, Huang H, Letunic I, Lopez R, Luciani A, Madeira F, Marchler-Bauer A, Mi H, Natale DA, Necci M, Nuka G, Orengo C, Pandurangan AP, Paysan-Lafosse T, Pesseat S, Potter SC, Qureshi MA, Rawlings ND, Redaschi N, Richardson LJ, Rivoire C, Salazar GA, Sangrador-Vegas A, Sigrist CJA, Sillitoe I, Sutton GG, Thanki N, Thomas PD, Tosatto SCE, Yong SY, Finn RD. 2019. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47:D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iyer LM, Koonin EV, Leipe DD, Aravind L. 2005. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res 33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildebrand A, Remmert M, Biegert A, Soding J. 2009. Fast and accurate automatic structure prediction with HHpred. Proteins-Structure Function and Bioinformatics 77(Suppl 9):128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 11.Montelione GT. 2012. The Protein Structure Initiative: achievements and visions for the future. F1000 Biol Rep 4:7. doi: 10.3410/B4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobb B, Kurtz DA, Moreno-Hagelsieb G, Doxey AC. 2015. Remote homology and the functions of metagenomic dark matter. Front Genet 6:234. doi: 10.3389/fgene.2015.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nordstrom KJ, Sallman Almen M, Edstam MM, Fredriksson R, Schioth HB. 2011. Independent HHsearch, Needleman–Wunsch-based, and motif analyses reveal the overall hierarchy for most of the G protein-coupled receptor families. Mol Biol Evol 28:2471–2480. doi: 10.1093/molbev/msr061. [DOI] [PubMed] [Google Scholar]
- 14.O’Day DH, Suhre K, Myre MA, Chatterjee-Chakraborty M, Chavez SE. 2006. Isolation, characterization, and bioinformatic analysis of calmodulin-binding protein cmbB reveals a novel tandem IP22 repeat common to many Dictyostelium and Mimivirus proteins. Biochem Biophys Res Commun 346:879–888. doi: 10.1016/j.bbrc.2006.05.204. [DOI] [PubMed] [Google Scholar]
- 15.Knutson BA, Broyles SS. 2008. Expansion of poxvirus RNA polymerase subunits sharing homology with corresponding subunits of RNA polymerase II. Virus Genes 36:307–311. doi: 10.1007/s11262-008-0207-3. [DOI] [PubMed] [Google Scholar]
- 16.Mirzakhanyan Y, Gershon PD. 2017. Multisubunit DNA-dependent RNA polymerases from vaccinia virus and other nucleocytoplasmic large-DNA viruses: impressions from the age of structure. Microbiol Mol Biol Rev 81:e00010-17. doi: 10.1128/MMBR.00010-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fidler DR, Murphy SE, Courtis K, Antonoudiou P, El-Tohamy R, Ient J, Levine TP. 2016. Using HHsearch to tackle proteins of unknown function: a pilot study with PH domains. Traffic 17:1214–1226. doi: 10.1111/tra.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 20.Yutin N, Koonin EV. 2012. Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol J 9:161. doi: 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koonin EV, Yutin N. 2019. Evolution of the large nucleocytoplasmic DNA viruses of eukaryotes and convergent origins of viral gigantism. Adv Virus Res 103:167–202. doi: 10.1016/bs.aivir.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Koonin EV, Yutin N. 2010. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 53:284–292. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin EV, Yutin N. 2018. Multiple evolutionary origins of giant viruses. F1000Res 7:1840. doi: 10.12688/f1000research.16248.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yutin N, Wolf YI, Raoult D, Koonin EV. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J 6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yutin N, Wolf YI, Koonin EV. 2014. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology 466–467:38–52. doi: 10.1016/j.virol.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abrahao J, Silva L, Silva LS, Khalil JYB, Rodrigues R, Arantes T, Assis F, Boratto P, Andrade M, Kroon EG, Ribeiro B, Bergier I, Seligmann H, Ghigo E, Colson P, Levasseur A, Kroemer G, Raoult D, La Scola B. 2018. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat Commun 9:749. doi: 10.1038/s41467-018-03168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz F, Yutin N, Ivanova NN, Ortega DR, Lee TK, Vierheilig J, Daims H, Horn M, Wagner M, Jensen GJ, Kyrpides NC, Koonin EV, Woyke T. 2017. Giant viruses with an expanded complement of translation system components. Science 356:82–85. doi: 10.1126/science.aal4657. [DOI] [PubMed] [Google Scholar]
- 28.Bahar MW, Graham SC, Stuart DI, Grimes JM. 2011. Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure 19:1011–1020. doi: 10.1016/j.str.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang Q, Zhu D, Agarkova I, Adhikari J, Klose T, Liu Y, Chen Z, Sun Y, Gross ML, Van Etten JL, Zhang X, Rossmann MG. 2019. Near-atomic structure of a giant virus. Nat Commun 10:388. doi: 10.1038/s41467-019-08319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urbaneja MA, McGrath CF, Kane BP, Henderson LE, Casas-Finet JR. 2000. Nucleic acid binding properties of the simian immunodeficiency virus nucleocapsid protein NCp8. J Biol Chem 275:10394–10404. doi: 10.1074/jbc.275.14.10394. [DOI] [PubMed] [Google Scholar]
- 31.Shepard DA, Ehnstrom JG, Skinner PJ, Schiff LA. 1996. Mutations in the zinc-binding motif of the reovirus capsid protein delta 3 eliminate its ability to associate with capsid protein mu 1. J Virol 70:2065–2068. doi: 10.1128/JVI.70.3.2065-2068.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemay G, Danis C. 1994. Reovirus lambda 1 protein: affinity for double-stranded nucleic acids by a small amino-terminal region of the protein independent from the zinc finger motif. J Gen Virol 75:3261–3266. doi: 10.1099/0022-1317-75-11-3261. [DOI] [PubMed] [Google Scholar]
- 33.Olland AM, Jane-Valbuena J, Schiff LA, Nibert ML, Harrison SC. 2001. Structure of the reovirus outer capsid and dsRNA-binding protein sigma3 at 1.8 A resolution. EMBO J 20:979–989. doi: 10.1093/emboj/20.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartlett JA, Joklik WK. 1988. The sequence of the reovirus serotype 3 L3 genome segment which encodes the major core protein lambda 1. Virology 167:31–37. doi: 10.1016/0042-6822(88)90051-7. [DOI] [PubMed] [Google Scholar]
- 35.Moss B. 2013. Poxviridae, p 2129–2159. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 36.Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. 2000. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 37.Briand JF, Navarro F, Rematier P, Boschiero C, Labarre S, Werner M, Shpakovski GV, Thuriaux P. 2001. Partners of Rpb8p, a small subunit shared by yeast RNA polymerases I, II and III. Mol Cell Biol 21:6056–6065. doi: 10.1128/mcb.21.17.6056-6065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koonin EV, Makarova KS, Elkins JG. 2007. Orthologs of the small RPB8 subunit of the eukaryotic RNA polymerases are conserved in hyperthermophilic Crenarchaeota and “Korarchaeota.” Biol Direct 2:38. doi: 10.1186/1745-6150-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwapisz M, Beckouet F, Thuriaux P. 2008. Early evolution of eukaryotic DNA-dependent RNA polymerases. Trends Genet 24:211–215. doi: 10.1016/j.tig.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Suhre K, Audic S, Claverie JM. 2005. Mimivirus gene promoters exhibit an unprecedented conservation among all eukaryotes. Proc Natl Acad Sci U S A 102:14689–14693. doi: 10.1073/pnas.0506465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reich C, Zeller M, Milkereit P, Hausner W, Cramer P, Tschochner H, Thomm M. 2009. The archaeal RNA polymerase subunit P and the eukaryotic polymerase subunit Rpb12 are interchangeable in vivo and in vitro. Mol Microbiol 71:989–1002. doi: 10.1111/j.1365-2958.2008.06577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Treich I, Carles C, Riva M, Sentenac A. 1992. RPC10 encodes a new mini subunit shared by yeast nuclear RNA polymerases. Gene Expr 2:31–37. [PMC free article] [PubMed] [Google Scholar]
- 43.Satheshkumar PS, Olano LR, Hammer CH, Zhao M, Moss B. 2013. Interactions of the vaccinia virus A19 protein. J Virol 87:10710–10720. doi: 10.1128/JVI.01261-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoddard BL. 2005. Homing endonuclease structure and function. Q Rev Biophys 38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 45.Stoddard BL. 2011. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure 19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, Firman K, Gromova ES, Gumport RI, Halford SE, Hattman S, Heitman J, Hornby DP, Janulaitis A, Jeltsch A, Josephsen J, Kiss A, Klaenhammer TR, Kobayashi I, Kong H, Kruger DH, Lacks S, Marinus MG, Miyahara M, Morgan RD, Murray NE, Nagaraja V, Piekarowicz A, Pingoud A, Raleigh E, Rao DN, Reich N, Repin VE, Selker EU, Shaw PC, Stein DC, Stoddard BL, Szybalski W, Trautner TA, Van Etten JL, Vitor JM, Wilson GG, Xu SY. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res 31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horton JR, Borgaro JG, Griggs RM, Quimby A, Guan S, Zhang X, Wilson GG, Zheng Y, Zhu Z, Cheng X. 2014. Structure of 5-hydroxymethylcytosine-specific restriction enzyme, AbaSI, in complex with DNA. Nucleic Acids Res 42:7947–7959. doi: 10.1093/nar/gku497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pingoud A, Jeltsch A. 2001. Structure and function of type II restriction endonucleases. Nucleic Acids Res 29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pingoud A, Fuxreiter M, Pingoud V, Wende W. 2005. Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci 62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 50.Hennecke F, Kolmar H, Brundl K, Fritz HJ. 1991. The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature 353:776–778. doi: 10.1038/353776a0. [DOI] [PubMed] [Google Scholar]
- 51.Macintyre G, Doiron KM, Cupples CG. 1997. The Vsr endonuclease of Escherichia coli: an efficient DNA repair enzyme and a potent mutagen. J Bacteriol 179:6048–6052. doi: 10.1128/jb.179.19.6048-6052.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agarkova IV, Dunigan DD, Van Etten JL. 2006. Virion-associated restriction endonucleases of chloroviruses. J Virol 80:8114–8123. doi: 10.1128/JVI.00486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia YN, Burbank DE, Uher L, Rabussay D, Van Etten JL. 1986. Restriction endonuclease activity induced by PBCV-1 virus infection of a Chlorella-like green alga. Mol Cell Biol 6:1430–1439. doi: 10.1128/mcb.6.5.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Nelson M, Nietfeldt JW, Burbank DE, Van Etten JL. 1992. Characterization of Chlorella virus PBCV-1 CviAII restriction and modification system. Nucleic Acids Res 20:5351–5356. doi: 10.1093/nar/20.20.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chan SH, Zhu Z, Dunigan DD, Van Etten JL, Xu SY. 2006. Cloning of Nt.CviQII nicking endonuclease and its cognate methyltransferase: M.CviQII methylates AG sequences. Protein Expr Purif 49:138–150. doi: 10.1016/j.pep.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Nelson M, Nietfeldt J, Xia Y, Burbank D, Ropp S, Van Etten JL. 1998. Chlorella virus NY-2A encodes at least 12 DNA endonuclease/methyltransferase genes. Virology 240:366–375. doi: 10.1006/viro.1997.8936. [DOI] [PubMed] [Google Scholar]
- 57.Xia YN, Burbank DE, Uher L, Rabussay D, Van Etten JL. 1987. IL-3A virus infection of a Chlorella-like green alga induces a DNA restriction endonuclease with novel sequence specificity. Nucleic Acids Res 15:6075–6090. doi: 10.1093/nar/15.15.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laganeckas M, Margelevicius M, Venclovas C. 2011. Identification of new homologs of PD-(D/E)XK nucleases by support vector machines trained on data derived from profile-profile alignments. Nucleic Acids Res 39:1187–1196. doi: 10.1093/nar/gkq958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steczkiewicz K, Muszewska A, Knizewski L, Rychlewski L, Ginalski K. 2012. Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res 40:7016–7045. doi: 10.1093/nar/gks382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Venclovas C, Timinskas A, Siksnys V. 1994. Five-stranded beta-sheet sandwiched with two alpha-helices: a structural link between restriction endonucleases EcoRI and EcoRV. Proteins 20:279–282. doi: 10.1002/prot.340200308. [DOI] [PubMed] [Google Scholar]
- 61.Aravind L, Makarova KS, Koonin EV. 2000. Survey and summary: Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res 28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kazrani AA, Kowalska M, Czapinska H, Bochtler M. 2014. Crystal structure of the 5hmC specific endonuclease PvuRts1I. Nucleic Acids Res 42:5929–5936. doi: 10.1093/nar/gku186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H, Guan S, Quimby A, Cohen-Karni D, Pradhan S, Wilson G, Roberts RJ, Zhu Z, Zheng Y. 2011. Comparative characterization of the PvuRts1I family of restriction enzymes and their application in mapping genomic 5-hydroxymethylcytosine. Nucleic Acids Res 39:9294–9305. doi: 10.1093/nar/gkr607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishino S, Skouloubris S, Kudo H, l’Hermitte-Stead C, Es-Sadik A, Lambry JC, Ishino Y, Myllykallio H. 2018. Activation of the mismatch-specific endonuclease EndoMS/NucS by the replication clamp is required for high fidelity DNA replication. Nucleic Acids Res 46:6206–6217. doi: 10.1093/nar/gky460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakae S, Hijikata A, Tsuji T, Yonezawa K, Kouyama KI, Mayanagi K, Ishino S, Ishino Y, Shirai T. 2016. Structure of the EndoMS-DNA complex as mismatch restriction endonuclease. Structure 24:1960–1971. doi: 10.1016/j.str.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 66.Ren B, Kuhn J, Meslet-Cladiere L, Briffotaux J, Norais C, Lavigne R, Flament D, Ladenstein R, Myllykallio H. 2009. Structure and function of a novel endonuclease acting on branched DNA substrates. EMBO J 28:2479–2489. doi: 10.1038/emboj.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iyer LM, Koonin EV, Aravind L. 2002. Extensive domain shuffling in transcription regulators of DNA viruses and implications for the origin of fungal APSES transcription factors. Genome Biol 3:RESEARCH0012. doi: 10.1186/gb-2002-3-3-research0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dunin-Horkawicz S, Feder M, Bujnicki JM. 2006. Phylogenomic analysis of the GIY-YIG nuclease superfamily. BMC Genomics 7:98. doi: 10.1186/1471-2164-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kowalski JC, Belfort M, Stapleton MA, Holpert M, Dansereau JT, Pietrokovski S, Baxter SM, Derbyshire V. 1999. Configuration of the catalytic GIY-YIG domain of intron endonuclease I-TevI: coincidence of computational and molecular findings. Nucleic Acids Res 27:2115–2125. doi: 10.1093/nar/27.10.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jurica MS, Stoddard BL. 1999. Homing endonucleases: structure, function and evolution. Cell Mol Life Sci 55:1304–1326. doi: 10.1007/s000180050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen BW, Heiter DF, Chan SH, Wang H, Xu SY, Morgan RD, Wilson GG, Stoddard BL. 2010. Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI. Structure 18:734–743. doi: 10.1016/j.str.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moon AF, Midon M, Meiss G, Pingoud A, London RE, Pedersen LC. 2011. Structural insights into catalytic and substrate binding mechanisms of the strategic EndA nuclease from Streptococcus pneumoniae. Nucleic Acids Res 39:2943–2953. doi: 10.1093/nar/gkq1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. Science 303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 74.Liu R, Olano LR, Mirzakhanyan Y, Gershon PD, Moss B. 2019. Vaccinia virus ankyrin-repeat/F-box protein targets interferon-induced IFITs for proteasomal degradation. Cell Rep 29:816–828.e816. doi: 10.1016/j.celrep.2019.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shukla A, Chatterjee A, Kondabagil K. 2018. The number of genes encoding repeat domain-containing proteins positively correlates with genome size in amoebal giant viruses. Virus Evol 4:vex039. doi: 10.1093/ve/vex039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aherfi S, Colson P, La Scola B, Raoult D. 2016. Giant viruses of amoebas: an update. Front Microbiol 7:349. doi: 10.3389/fmicb.2016.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. 2000. Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell 6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 78.Jiang J, Tang M, Huang Z, Chen L. 2019. Junctophilins emerge as novel therapeutic targets. J Cell Physiol 234:16933–16943. doi: 10.1002/jcp.28405. [DOI] [PubMed] [Google Scholar]
- 79.Gubbels MJ, Vaishnava S, Boot N, Dubremetz JF, Striepen B. 2006. A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J Cell Sci 119:2236–2245. doi: 10.1242/jcs.02949. [DOI] [PubMed] [Google Scholar]
- 80.Morriswood B, Schmidt K. 2015. A MORN repeat protein facilitates protein entry into the flagellar pocket of Trypanosoma brucei. Eukaryot Cell 14:1081–1093. doi: 10.1128/EC.00094-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hatzopoulos GN, Erat MC, Cutts E, Rogala KB, Slater LM, Stansfeld PJ, Vakonakis I. 2013. Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture. Structure 21:2069–2077. doi: 10.1016/j.str.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng X, Gooi LM, Wason A, Gabriel E, Mehrjardi NZ, Yang Q, Zhang X, Debec A, Basiri ML, Avidor-Reiss T, Pozniakovsky A, Poser I, Saric T, Hyman AA, Li H, Gopalakrishnan J. 2014. Conserved TCP domain of Sas-4/CPAP is essential for pericentriolar material tethering during centrosome biogenesis. Proc Natl Acad Sci U S A 111:E354–363. doi: 10.1073/pnas.1317535111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mercer AA, Fleming SB, Ueda N. 2005. F-box-like domains are present in most poxvirus ankyrin repeat proteins. Virus Genes 31:127–133. doi: 10.1007/s11262-005-1784-z. [DOI] [PubMed] [Google Scholar]
- 84.Price CT, Al-Quadan T, Santic M, Jones SC, Abu Kwaik Y. 2010. Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J Exp Med 207:1713–1726. doi: 10.1084/jem.20100771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pellett PE, Roizman B. 2013. Herpesviridae. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th edition ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 86.Cai WH, Gu B, Person S. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol 62:2596–2604. doi: 10.1128/JVI.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 88.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 89.Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 90.Hu X, Wolffe EJ, Weisberg AS, Carroll LJ, Moss B. 1998. Repression of the A8L gene, encoding the early transcription factor 82-kilodalton subunit, inhibits morphogenesis of vaccinia virions. J Virol 72:104–112. doi: 10.1128/JVI.72.1.104-112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu X, Carroll LJ, Wolffe EJ, Moss B. 1996. De novo synthesis of the early transcription factor 70-kilodalton subunit is required for morphogenesis of vaccinia virions. J Virol 70:7669–7677. doi: 10.1128/JVI.70.11.7669-7677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Y, Ahn B-Y, Moss B. 1994. Targeting of a multicomponent transcription apparatus into assembling vaccinia virus particles requires RAP94, an RNA polymerase-associated protein. J Virol 68:1360–1370. doi: 10.1128/JVI.68.3.1360-1370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holowczak JA, Thomas VL, Flores L. 1975. Isolation and characterization of vaccinia virus “nucleoids. Virology 67:506–519. doi: 10.1016/0042-6822(75)90451-1. [DOI] [PubMed] [Google Scholar]
- 94.Steinegger M, Meier M, Mirdita M, Vohringer H, Haunsberger SJ, Soding J. 2019. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics 20:473. doi: 10.1186/s12859-019-3019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hendy MD, Penny D. 1982. Branch and bound algorithms to determine minimal evolutionary trees. Math Biosci 59:277–290. doi: 10.1016/0025-5564(82)90027-X. [DOI] [Google Scholar]
- 96.Loenen WA, Dryden DT, Raleigh EA, Wilson GG. 2014. Type I restriction enzymes and their relatives. Nucleic Acids Res 42:20–44. doi: 10.1093/nar/gkt847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wyszomirski KH, Curth U, Alves J, Mackeldanz P, Moncke-Buchner E, Schutkowski M, Kruger DH, Reuter M. 2012. Type III restriction endonuclease EcoP15I is a heterotrimeric complex containing one Res subunit with several DNA-binding regions and ATPase activity. Nucleic Acids Res 40:3610–3622. doi: 10.1093/nar/gkr1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mitsuhashi W, Miyamoto K, Wada S. 2014. The complete genome sequence of the Alphaentomopoxvirus Anomala cuprea entomopoxvirus, including its terminal hairpin loop sequences, suggests a potentially unique mode of apoptosis inhibition and mode of DNA replication. Virology 452–453:95–116. doi: 10.1016/j.virol.2013.12.036. [DOI] [PubMed] [Google Scholar]
- 99.Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. 2011. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc Natl Acad Sci U S A 108:17486–17491. doi: 10.1073/pnas.1110889108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mihara T, Koyano H, Hingamp P, Grimsley N, Goto S, Ogata H. 2018. Taxon richness of “Megaviridae” exceeds those of Bacteria and Archaea in the ocean. Microbes Environ 33:162–171. doi: 10.1264/jsme2.ME17203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Claverie JM, Abergel C. 2018. Mimiviridae: an expanding family of highly diverse large dsDNA viruses infecting a wide phylogenetic range of aquatic eukaryotes. Viruses 10:506. doi: 10.3390/v10090506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang J, Huang Q, Zhou X, Shen MM, Yen A, Yu SX, Dong G, Qu K, Huang P, Anderson EM, Daniel-Issakani S, Buller RM, Payan DG, Lu HH. 2004. The poxvirus p28 virulence factor is an E3 ubiquitin ligase. J Biol Chem 279:54110–54116. doi: 10.1074/jbc.M410583200. [DOI] [PubMed] [Google Scholar]
- 103.Nerenberg BT, Taylor J, Bartee E, Gouveia K, Barry M, Fruh K. 2005. The poxviral RING protein p28 is a ubiquitin ligase that targets ubiquitin to viral replication factories. J Virol 79:597–601. doi: 10.1128/JVI.79.1.597-601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.