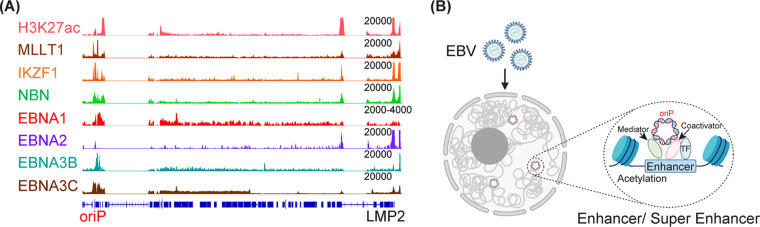
EBV is associated with ∼200,000 cancers each year. In vitro, EBV can transform primary human B lymphocytes into immortalized cell lines. EBV-encoded proteins, along with noncoding RNAs and microRNAs, hijack cellular proteins and pathways to control cell growth. EBV nuclear proteins usurp normal transcriptional programs to activate the expression of key oncogenes, including MYC, to provide a proliferation signal. EBV nuclear antigens also repress CDKN2A to suppress senescence. EBV membrane protein activates NF-κB to provide survival signals. EBV genomes are maintained by EBNA1, which tethers EBV episomes to the host chromosomes during mitosis. However, little is known about where EBV episomes are located in interphase cells. In interphase cells, EBV promoters drive the expression of latency genes, while oriP functions as an enhancer for these promoters. In this study, integrative analyses of published lymphoblastoid cell line (LCL) Hi-C data and our 4C-seq experiments position EBV episomes to host genomes with active epigenetic marks. These contact points were significantly enriched for super enhancers. The close proximity of EBV episomes and the super enhancers that are enriched for transcription cofactors or mediators in lymphoblasts may benefit EBV gene expression, suggesting a novel mechanism of transcriptional activation.
KEYWORDS: Epstein-Barr Virus, physical interaction, Hi-C, lymphoblastoid, 4C-seq
ABSTRACT
The Epstein-Barr virus (EBV) episome is known to interact with the three-dimensional structure of the human genome in infected cells. However, the exact locations of these interactions and their potential functional consequences remain unclear. Recently, high-resolution chromatin conformation capture (Hi-C) assays in lymphoblastoid cells have become available, enabling us to precisely map the contacts between the EBV episome(s) and the human host genome. Using available Hi-C data at a 10-kb resolution, we have identified 15,000 reproducible contacts between EBV episome(s) and the human genome. These contacts are highly enriched in chromatin regions denoted by typical or super enhancers and active markers, including histone H3K27ac and H3K4me1. Additionally, these contacts are highly enriched at loci bound by host transcription factors that regulate B cell growth (e.g., IKZF1 and RUNX3), factors that enhance cell proliferation (e.g., HDGF), or factors that promote viral replication (e.g., NBS1 and NFIC). EBV contacts show nearly 2-fold enrichment in host regions bound by EBV nuclear antigen 2 (EBNA2) and EBNA3 transcription factors. Circular chromosome conformation capture followed by sequencing (4C-seq) using the EBV origin of plasmid replication (oriP) as a “bait” in lymphoblastoid cells further confirmed contacts with active chromatin regions. Collectively, our analysis supports interactions between EBV episome(s) and active regions of the human genome in lymphoblastoid cells.
IMPORTANCE EBV is associated with ∼200,000 cancers each year. In vitro, EBV can transform primary human B lymphocytes into immortalized cell lines. EBV-encoded proteins, along with noncoding RNAs and microRNAs, hijack cellular proteins and pathways to control cell growth. EBV nuclear proteins usurp normal transcriptional programs to activate the expression of key oncogenes, including MYC, to provide a proliferation signal. EBV nuclear antigens also repress CDKN2A to suppress senescence. EBV membrane protein activates NF-κB to provide survival signals. EBV genomes are maintained by EBNA1, which tethers EBV episomes to the host chromosomes during mitosis. However, little is known about where EBV episomes are located in interphase cells. In interphase cells, EBV promoters drive the expression of latency genes, while oriP functions as an enhancer for these promoters. In this study, integrative analyses of published lymphoblastoid cell line (LCL) Hi-C data and our 4C-seq experiments position EBV episomes to host genomes with active epigenetic marks. These contact points were significantly enriched for super enhancers. The close proximity of EBV episomes and the super enhancers that are enriched for transcription cofactors or mediators in lymphoblasts may benefit EBV gene expression, suggesting a novel mechanism of transcriptional activation.
INTRODUCTION
Epstein-Barr virus (EBV) was the first human DNA tumor virus identified, over fifty years ago from African Burkitt’s lymphoma (1, 2). EBV causes ∼200,000 cancers each year. These cancers include Burkitt’s lymphoma, Hodgkin’s lymphoma, posttransplant lymphoproliferative disease (PTLD), AIDS central nervous system (CNS) lymphoma, nasopharyngeal carcinoma (NPC), and 10% of gastric cancers (3). In vitro, EBV transforms primary resting B lymphocytes (RBLs) into continuously proliferating lymphoblastoid cell lines (LCLs) through expression of type III EBV latency genes that include 6 EBV nuclear antigens (EBNAs), three latent membrane proteins (LMPs), noncoding RNAs, and microRNAs (miRNAs) (4). LCLs express the same EBV genes as PTLD and AIDS lymphoma. Therefore, LCLs provide a useful model system to study EBV oncogenesis. Genetic studies found that many of these genes are essential for continuous LCL growth and viral reactivation (5). EBNA2 is the major EBV transcription activator and it activates MYC and other EBV latency genes (6, 7). Epstein-Barr virus nuclear antigen leader protein (EBNALP) strongly coactivates with EBNA2 (8). EBNA3A and 3C repress senescence through inhibition of CDKN2A (9–11). LMP1 activates NF-κB to provide survival signals (12).
During primary infection, EBV membrane proteins gp350 and gp42 bind to B cell receptors CR2 and HLA class II molecules. The viral membrane is then fused to the endosomal membrane and the viral capsid is released into the cytoplasm (13). The linear viral DNA is circularized and chromatinized to form an episome in the nucleus. During mitosis, EBNA1 binds to the EBV origin of plasmid replication (oriP) to initiate replication (14). EBNA1 also tethers the episomes to the host chromosomes, allowing episomes to segregate into daughter cells with the host chromosome, maintaining a stable episome copy number (15–17). In interphase, the EBV Cp promoter drives the expression of EBNAs and the bidirectional LMP1/LMP2 promoter drives the expression of LMPs. EBNA1 strongly activates a reporter under the control of oriP, and oriP functions as an enhancer for Cp and LMPs. Many host transcription factors (TFs) can bind to the oriP and may contribute to oriP enhancer activity (18, 19).
Enhancers are marked by active histone markers H3K27ac and H3K4me1. Enhancers loop to promoters to increase the expression of their direct target genes. Clusters of dense enhancers with extraordinary strong H3K27ac signals form super enhancers (SEs). SEs are bound by clusters of TFs. These TFs recruit transcription cofactors and basal transcription factors such as BRD4 and mediator subunits. SEs are critically important for cancer cell proliferation (20) and LCL SEs control the expression of key oncogenes, including MYC. EBV TFs also converge to SEs to promote cell growth and survival (21, 22). However, SEs are more sensitive to perturbations than average enhancers and selectively targeting SEs may lead to novel therapies (23, 24).
Chromatin conformation capture (3C)-based techniques can capture the interactions between genomic DNAs in cells. The 3C technique followed by deep sequencing (Hi-C) defines the genome-wide chromatin interactions (25). Captured Hi-C (cHi-C) uses oligonucleotides to enrich a collection of specific DNA regions and their contacting genomic DNAs. Circular chromatin conformation capture followed by deep sequencing (4C-seq) captures the interactions between one specific genomic region (viewpoint) and the rest of the genome. These 3C-based assays provided powerful tools to interrogate the interactions between extrachromosomal viral episomes and the host genome. LCL GM12878 Hi-C defines high-resolution genomic interactions (26). Analysis of this data set at 1 million base-pair resolution found EBV episomes tend to interact with gene-poor chromosomes (27). Interestingly, reanalyzing the same data set at 10-kb resolution found EBV episomes tend to interact with transcriptionally active regions (28). In Burkitt’s lymphoma cell lines, 4C-seq using the EBV genome as viewpoint found that EBV episomes frequently interact with repressed genomic regions. However, in LCLs the episome 4C-seq also correlated with Hi-C, showing EBV episomes interact with active host genomic regions, even though the resolution was not as high (28). Episomal hepatitis B virus (HBV) DNA interacts with actively transcribed genomic regions (29). cHi-C found HBV episomal DNA preferentially contacts CpG islands, whereas adenovirus preferentially associates with transcriptional start sites (30). These findings indicate that the interactions between viral episomes and the host DNA vary dramatically, depending on different cell types or viral latency programs. More analyses are needed to further understand the functional significance of the interactions between viral episomes and the host genome.
We set out to further evaluate the relationship between EBV episomes in LCLs using high-resolution Hi-C data. We found that EBV episomes preferentially interact with enhancers and super enhancers. The interactions were then validated by 4C-seq. These finding add a new dimension to our understanding of EBV oncogenesis.
RESULTS
EBV contacts highly active regions of the host genome.
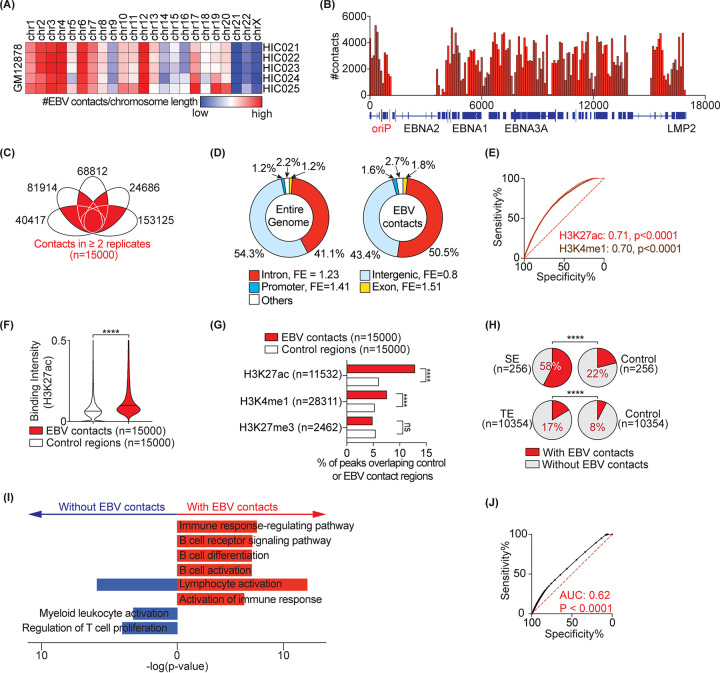
We reanalyzed currently available high-resolution Hi-C data (26) from the lymphoblastoid cell line GM12878 (GSE63525) by examining the reads supporting colocalization (i.e., contact) of EBV episome(s) and the human genome. We sourced data from 5 biological replicates with the highest sequencing coverage containing ∼76 to 247 million paired-end high-quality contacts, of which ∼24,686 to 153,125 were between the EBV episome(s) and the human genome (Table S1 in the supplemental material). In agreement with a previous study (27), at chromosomal-level resolution, larger chromosomes (e.g., chr1-4) displayed more EBV contacts (Fig. 1A, adjusted for chromosomal length). We also found higher numbers of contacts with chromosomes 12, 13, and 17. All regions of the EBV episome(s) seem to participate in contacts (Fig. 1B). In order to construct a high-resolution and reproducible EBV and human host contact map, we focused on contacts at 10-kb resolution that are shared among two or more biological replicates. We found 15,000 such contacts (Fig. 1C and Table S1), of which nearly half were located in intronic regions, showing a 1.2-fold enrichment compared to all intronic regions genome-wide (Fig. 1D). Contacts also significantly overlapped promoter and exonic regions, but failed to overlap intergenic loci compared to what would be expected given the fractions of all promoters, exons, or intergenic loci in the entire genome (Fig. 1D).
FIG 1.
EBV contacts are enriched in active regions of the host genome. (A) EBV interaction frequency with the host genome at the chromosomal level. (B) Distribution of contact frequency across the EBV genome. (C) 15,000 interactions between EBV and the host genome common to n ≥ 2 replicates. Each eclipse represents one Hi-C replicate in the GM12878 line. Data are from GSE63525. (D) Distribution of genomics elements (i.e., introns, exons, promoters, and intergenic regions) in the entire genome (left) and in the EBV contacts (right). FE, fold enrichment compared to the entire genome. (E) The receiver operating characteristics (ROC) curves distinguishing control regions (n = 15,000) from EBV contact loci (n = 15,000) based on the indicated histone marker. Numbers show the area under the curve (AUC) for each marker. Peak annotations are from ENCODE (31). (F) Violin plot comparing mean H3k27ac signal intensity in 15,000 control regions versus with EBV contacts. Median are highlighted by solid and dashed lines. (G) Bar-charts showing percentage of histone marker peaks that overlap with EBV contact or control regions. Fisher’s exact test: ****, P < 0.0001; ns, not significant. (H) EBV contact enrichment in super enhancer (SE) architectures (top) and typical enhancers (TE) (bottom). SE and TE annotations are from GM12878 (23). Percentages of regions interacting with EBV are shown. Controls are size-matched regions randomly selected across the entire genome. (I) Pathway analysis of SE-containing genes with and without EBV contacts using Metascape. The selected top 8 pathways are highlighted. (J) ROC curves distinguishing control regions from EBV-interacting regions based on all open chromatin regions annotated from ATAC-seq (GSE103301) in GM12878.
To characterize epigenetic markers at the contact loci, we first sourced 22 publicly available genome-wide histone modification data in GM12878 from the encyclopedia of DNA elements (ENCODE) (31). We used an unbiased approach examining which, if any, histone modifications can distinguish EBV contacts from control regions (i.e., size-matched noncontact loci). This was done by comparing the mean signal intensity of the histone modification in EBV contacts versus control regions using receiver operating characteristic (ROC) and the area under the ROC curves (AUC) (Table S2). Histone markers, including H3K27ac and H3K4me1, that are typically associated with active chromatin regions such as enhancers and promoters had the highest predictive power (AUC ∼0.7; Fig. 1E, Table S2) at separating EBV contacts from noncontacts. This was also evident as the mean binding intensity of H3K27ac in EBV contact regions was higher than in control loci (Fig. 1F). Conversely, histone modifications associated with polycomb repression or heterochromatin, including H3K27me3, did not show strong predictive power (AUC ∼0.5; Fig. 1G). In addition, genomic loci with active histone markers were ∼1.5- to 2.1-fold more enriched in EBV contacts compared to control regions (Fig. 1G). Importantly, for all LCL H3K27ac peaks, ∼12% of them had contacts with EBV episomes (Fig. 1G). Similarly, ∼8% of all H3K4m1 peaks had contacts with EBV episomes (Fig. 1G). In contrast, significantly fewer contacts were detected between control regions and the EBV episomes. These findings suggest that EBV episomes selectively target active enhancers in LCLs.
We next examined direct overlap between EBV contact regions and enhancer loci. Enhancers are often annotated into two broad classes based on their size and associated histone marker signals, i.e., typical enhancers (TEs) and super enhancers (SEs). We sourced 10,354 TE and 256 SE annotations in GM12878 cells from public data (23). Nearly 58% of SEs and 17% of TEs overlapped with EBV contacts compared to 22% and 8% in control regions, respectively (Fig. 1H). We specifically focused on SEs as the most active elements of the human genome and having the highest levels of H3K27ac. To understand whether SEs with or without EBV contacts belong to different functional categories, we performed pathway analysis on adjacent target genes. While genes near SEs with or without EBV contacts were enriched in lymphocyte activation, genes near SEs with EBV contacts were specifically enriched in B cell functions (e.g., receptor signaling, differentiation, and activation), suggesting that EBV contacts may affect the function of contacted SEs (Fig. 1I).
To further substantiate overrepresentation of EBV contacts in active regions of host chromatin, we obtained data from an existing assay for transposase-accessible chromatin using sequencing (ATAC-seq; GSE103301) in the GM12878 line (32). EBV contact regions had higher ATAC-seq signal than control regions and the ATAC signal had a significant predictive power at distinguishing EBV contacts and control regions (Fig. 1J; AUC = 0.62, P < 0.0001). Collectively, our data show that, in GM12878 cells, EBV episomes contact active and open chromatin regions.
EBV contacts genomic loci bound by key host and viral transcription factors.
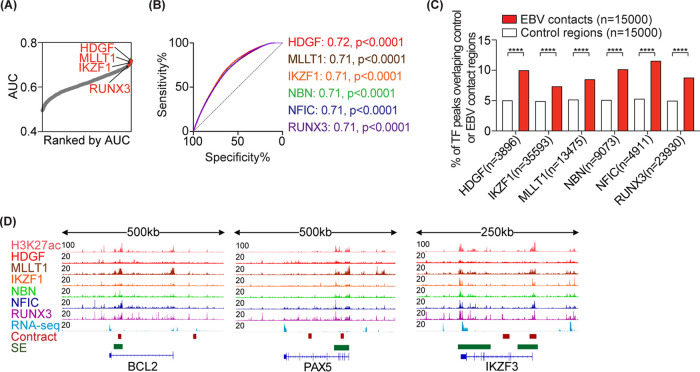
We next asked whether any of the host or viral TFs are enriched in EBV contact loci, as such information could help in identifying functional consequences and/or potential regulators of EBV contacts. To this end, we downloaded all 134 transcription factor genome-wide binding profiles (ChIP-seq) in GM12878 from ENCODE (31) (see Table S2A). We employed the above-mentioned approach to identify TFs that have higher binding intensity in EBV-contact regions than in control regions. Notably, top host transcription factors enriched in EBV contacts belonged to one of the following categories: (i) TFs that regulate B cell function (e.g., IKZF1 and RUNX3); (ii) TFs that enhance cell proliferation (e.g., HDGF); or (iii) TFs that affect viral replication (e.g., NBN and NFIC) (Fig. 2A and B, Table S2). Binding peaks for these TFs were colocalized with EBV contacts and were 1.5- to 2-fold more enriched in EBV contacts compared to control regions (Fig. 2C and D).
FIG 2.
EBV-contact regions are bound by key host transcription factors. (A) AUCs showing the power of each TF to distinguish EBV contacts (n = 15,000) from control regions (n = 15,000). The top TFs with higher binding signals in EBV contacts than in control loci are highlighted. (B) ROC curves showing the specificity and sensitivity of the indicated top TFs in discriminating EBV contacts from control loci. AUC values and their P values are indicated. (C) Bar chart showing the percentage of TF peaks that overlap with EBV contacts versus control regions. Fisher’s exact test: ****, P < 0.0001. (D) ChIP-seq tracks for three representative loci with one or more EBV contacts and SE.
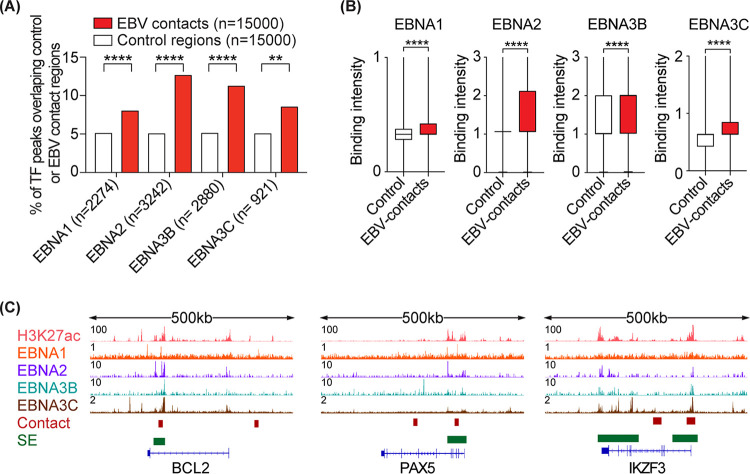
EBV-encoded TFs (e.g., EBNA1, 2, 3B, and 3C) are known to bind human host genomes (21, 22) and are speculated to be involved in tethering EBV episome(s) to the host chromatin. To test this, we first obtained ChIP-seq data available for four EBV TFs in LCLs and examined the overlap between the TF-bound loci and EBV-contact regions. EBV TF-bound regions overlapped ∼1.5 to 2.5-fold more with EBV contact loci than with control regions, with EBNA2 and EBNA3B having the largest overlap (Fig. 3A). Moreover, EBV-contact regions had higher binding intensities for these TFs compared to controls (Fig. 3B, representative loci are shown in Fig. 3C).
FIG 3.
EBV-contact host regions are bound by viral transcription factors. (A) Bar chart showing the percentage of TF peaks that overlap with EBV contacts versus control regions. Fisher’s exact test: ****, P < 0.0001. (B) Boxplots comparing the maximum binding intensity of the indicated TF in EBV contacts versus control regions. Two-tailed Mann-Whitney test: ****, P < 0.0001. (C) ChIP-seq tracks for three representative loci.
Taken together, our data show that EBV contacts were colocalized with bound regions of specific host and viral TFs.
4C-seq validates EBV contacts with host active chromatin regions.
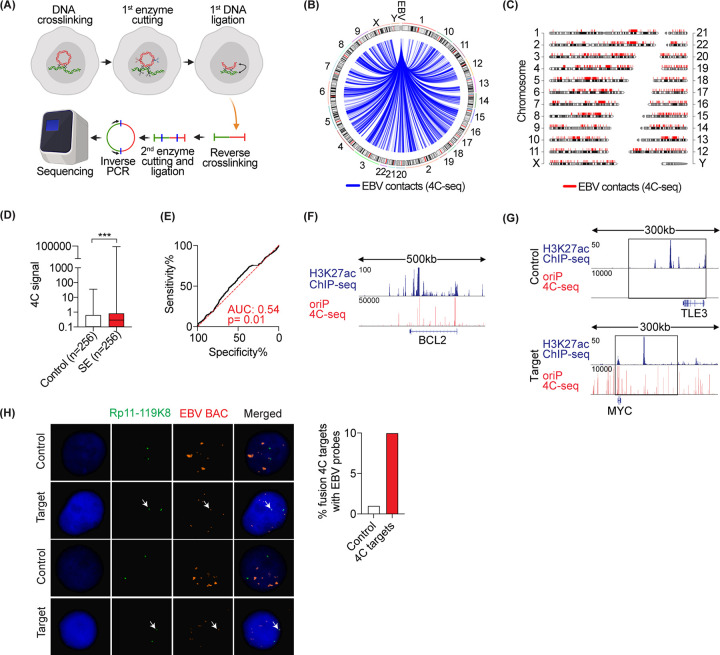
To independently verify EBV contacts with host active chromatin regions, we performed 4C-seq in GM12878 cells. We chose oriP as viewpoint to identify the host DNA in close proximity to EBV episomes. Cells were first cross-linked with formaldehyde and lysed. Cellular DNA was cut by DpnII and the DNA ends were then ligated. After reverse cross-linking, purified DNA was cut again with NlaIII and then ligated. Inverse PCR from the edges of oriP was used to amplify the host DNA ligated to oriP DNA. The amplified DNA was deep sequenced. The reads were then mapped to the host genome (Fig. 4A), similarly to what was previously reported (28). This yielded ∼8 million high-quality reads that were mapped to human or EBV genomes, respectively, providing a whole-genome tethering map of EBV episome(s) to the human genome (Fig. 4B and C). We then calculated the 4C signal at super enhancer loci. Consistent with the Hi-C data, our experiment showed higher 4C signal in SE loci compared to controls (Fig. 4D) and colocalization with H3K27ac (Fig. 4E). H3K27ac and 4C-seq tracks are shown at BCL2 representative loci (Fig. 4F).
FIG 4.
4C-seq-based map of EBV host contacts in GM12878 cells. (A) 4C-seq experimental design. (B and C) EBV-tethering sites on human host chromosomes by circus plot (B) and chromosome plot (C). (D) Boxplot comparing mean 4C signal intensities at 256 SEs versus size-matched controls. The median is highlighted as a solid line. (E) ROC curve showing the specificity and sensitivity of the 4C signal (mean signal intensities) in discriminating EBV contacts from control loci. (F) Tracks showing H3K27ac ChIP-seq and colocalization with 4C signal in GM12878 cells at the BCL2 locus. (G and H) Validation of 4C interaction sites by FISH. (G) Tracks showing H3K27ac ChIP-seq and 4C signal at MYC versus control locus. The precise position of the FISH probe is highlighted by an open box. (H) Mitotic nuclei (left) stained with dual FISH probes of EBV-BAC (red) and target/control (green). Bar chart (right) showing quantification of the percentage of colocalized FISH signals between EBV-BAC and target/control probes.
We next employed an orthogonal fluorescence in situ hybridization (FISH)-based method to visualize one of the contacts between EBV and the host genomic loci. We chose the MYC locus since it plays an important role in EBV-driven diseases (33, 34) and contains strong EBV contacts and H3K27ac signals (Fig. 4G). As a control, we selected a region with similar levels of H3K27ac signal but without any EBV contacts (Fig. 4G). EBV bacterial artificial chromosome (BAC) was labeled with red fluorescent dye and MYC- or control loci-specific BACs were labeled with green fluorescent dye. After hybridization, colocalizations of red and green FISH probes were quantified using a fluorescent microscope (Fig. 4H). As evidenced by FISH, we observed a 10-fold increase in the colocalization of MYC and EBV probes compared to control and EBV probes (Fig. 4H). Collectively, these data validate the colocalization of EBV episome(s) and active regions of the host chromatin.
DISCUSSION
The mechanisms through which EBV episomes persist during mitosis are well characterized. In interphase cells, EBV latency genes are expressed from the episomes. In LCLs, the major EBV-encoded transcription factor EBNA2 is tethered to EBV enhancers/promoters through host transcription factors, such as RBPJ, to activate the expression of other latency genes (6, 35). EBNA2 can induce the DNA binding of RBPJ to facilitate access to viral and host DNA (36, 37). Traditionally, the positions of the episomes in the nuclei can be traced using FISH. However, FISH is very low throughput. Only a couple of loci could be visualized simultaneously. Before next-generation sequencing techniques were available, little was known about the precise location of the EBV episomes in interphase cell nuclei at the molecular level. With the development of next-generation sequencing technique-based assays, it is now possible to pinpoint the exact location of viral episomes in the nucleus. By analyzing a high-resolution GM12878 LCL Hi-C map with billions of valid genomic DNA interactions, we positioned EBV episomes in close proximity to active enhancers. Furthermore, we found EBV episomes to be selectively in close proximity to super enhancers. The episomal docking sites were enriched with active enhancer markers, EBNAs, and several cellular TFs. We also used an additional 4C-seq assay to validate the findings from Hi-C.
SE complexes form phase-separated compartments to ensure efficient transcription (38). EBNAs are components of phase-separated condensates (39) and oriP functions as a strong enhancer that activates other type III latency genes. EBNAs can also bind to oriP (Fig. 5A). Therefore, the episomes may dock onto SEs though EBNAs. EBV episomes can take advantage of the existing SEs by positioning themselves in close physical proximity to use the proteins already present in condensate to ensure efficient transcription of viral genes (Fig. 5B).
FIG 5.
EBV episome TF binding and phase separation. (A) ChIP-seq reads were mapped to the EBV genome and visualized in genome browser. EBV genome annotation is shown at the bottom. The position of oriP is indicated. The peak height is indicated above each track on the right. (B) Schematic of EBV episomes and the host genome. The EBV episome is in close proximity to the TF, coactivator, and mediator in phase-separated condensate.
In Burkitt’s lymphoma cell lines, EBV episome docking sites are enriched in EBNA1 binding. Depletion of EBNA1 reduced episome docking to host sites and affected the repression of genes near the docking sites (28). In addition, cellular TFs EBF1 and RBPJ are all enriched at the docking site. These data support the notion that EBNA1 can tether episomes to host chromatin in interphase cells. In LCLs, Kim et al. determined from Hi-C data that active histone markers H3K27ac, H3K4me1, and EBNA2, but not EBNA1, are enriched at EBV-docking sites (28). However, by selecting highly reproducible Hi-C contact points, we found EBNA1 was significantly enriched in EBV episome-docking sites (Fig. 3A). These data suggest that EBNA1 may also tether EBV episomes to host chromatin in type III latency cells.
In type I EBV latency, where only EBNA1 and noncoding RNA and miRNAs are expressed, EBV episomes dock to repressed chromatin (28). Recently, Okabe et al. reported that in EBV+ gastric cancer cell lines, EBV infection induces H3k27ac at the site of interaction and EBV-interacting loci exhibit heterochromatin-to-euchromatin transitions (40). EBV+ gastric cancers express type II EBV latency, in which EBNA1, LMPs, noncoding RNAs, and miRNAs are expressed. These data suggested that EBV episomes dock to sites with different epigenetic hallmarks in cells expressing different EBV latency genes.
DNA tumor viruses transform normal human cells through distinct mechanisms. Interestingly, HBV and adenovirus both preferentially dock at active chromatin. Human papillomaviruses (HPV) integration sites frequently colocalize with SEs, suggesting that, before integration, HPV genomes are in close proximity to SEs (41). Here, we found that EBV episomes also selectively target host SEs. These findings suggest that DNA tumor viruses have evolved a common strategy to transform normal cells to cause cancer.
MATERIALS AND METHODS
Hi-C data analysis.
The analysis of Hi-C data was done by Juicer and Juicer tools (42). Chimeric reference genome hg38_ebv was generated by replacing chromosome Y of the human reference genome (GRCh38; hg38) with the EBV genome (ACC number NC_007605). The index of chimeric reference genome was built by the bwa v0.7.17 index procedure (43). Restriction sites were identified by generate_site_positions.py with digestion enzyme as MboI and genome release as hg38_ebv. Raw data were processed by juicer.sh with parameters “-z hg38_ebv.fa -D . -p hg38_ebv.chrom.sizes -y restriction_sites/hg38_ebv_MboI.txt -t 40.” EBV contacts with each host chromosome at resolution 10,000 bp were extracted from “.hic” files using Juicer tools with parameters “dump observed None chrN chrY BP 10000,” where chrN ranges from chr1 to chrX. Normalized counts of interactions between EBV episome(s) and host genome were defined as observed counts divided by chromosomal length (Fig. 1A). Shared EBV contacts were defined as contacts that were present in at least 2 replicates with exactly the same interaction sites on both human genome and EBV genome.
4C-seq assay in GM12878 cells.
The 4C libraries were prepared using previously published protocols with minor modifications (44). Briefly, 1 × 107 cells were fixed in 1% formaldehyde for 10 min at 37°C. Nuclei were resuspended in 500 μl of restriction enzyme buffer and permeabilized by incubation with 10% SDS (final concentration 0.3%) at 37°C overnight. DpnII restriction enzyme (500 units) was added and incubated overnight at 37°C with shaking. Digestion was stopped after adding 1.6% SDS and incubated at 65°C for 25 min. Samples were then diluted in 7.5 ml of 1× ligation buffer and 100 Weiss units of T4 DNA ligase was added. Samples were kept at 16°C for 24 h. The ligated chromatin was digested by proteinase K at 65°C overnight. Samples were purified by phenol-chloroform extraction and ethanol precipitation. Samples were further digested by 150 units of NlaIII and circularized using T4 DNA ligase. After purification, 16 parallel PCRs, each containing 3.2 μg of circularized DNA, were performed with primers as follows: (i) index primer for replicate 1, 5′-AAT GAT ACG GCG ACC ACC GAA CAC TCT TTC CCT ACA CGA CGC TCT TCC GAT CTGTCCGG CGC GAT TGC TGC GAT C; (ii) index primer for replicate 2, 5′-AAT GAT ACG GCG ACC ACC GAA CAC TCT TTC CCT ACA CGA CGC TCT TCC GAT CTATCAGG CGC GAT TGC TGC GAT C; and (iii) universal primer, 5′-CAA GCA GAA GAC GGC ATA CGA GAG ACA ACC AGT GGA GTC CG. The amplicons were extracted by AMPure XP beads. The bar-coded DNA libraries were sequenced as 75-bp single-end reads using the Illumina HiSeq2000 platform.
4C-seq data analysis.
The 4C-seq data analysis was performed as previously described (44). After demultiplexing, the reads that contain the reading primer were kept. The sequences before the DpnII and after the NalIII sites were trimmed to extract the captured fragment. The fragments were mapped to the hg19 assembly by Burrows-Wheeler Aligner (BWA). The ligation sites were determined. Mapped fragments were further aligned to a reduced in silico digestion “library” containing DpnII sites genome wide. A “binarization” step was performed to minimize the PCR duplication bias. Then, a window-based model was used to identify significantly interacting regions. Sites with only one read were eliminated to reduce background noise. Reproducible sites were defined as 4C sites with coverage of >1 in both biological replicates. The subsequent differential 4C-seq interactions were identified using the W4Cseq package. The regions with P values smaller than 0.05 were taken as candidates with significant differences.
ChIP-seq data analysis.
For EBV TF ChIP-seq data, all sequencing reads were aligned to a human reference genome (GRCh38; hg38) using Bowtie v1.2.2 (45) with default single-end alignment settings and additional parameters of “–chunkmbs 1000 S -m 1.” The index for reference genome was built by using “bowtie-build” with default parameters. The aligned reads were further filtered out for low-quality reads (q < 30) and then sorted and indexed by Samtools v1.9 (46). TF binding intensity profiles (i.e., bigWig files) were generated by bamCoverage v3.2.0 (47) using “--normalizeUsing BPM --minMappingQuality 30 --ignoreDuplicates --extendReads 250 --blackListFileName hg38.blacklist.bed.” Binding profiles were visualized by IGV genome browser v2.8.0 (48). Binding peaks were called by “callpeaks” procedure from MACS2 v2.1.2 (49) using default parameters but “--nomodel -t treatment.” The identified peaks were further screened against “hg38 blacklisted” genomic regions, mitochondrial DNA, and pseudochromosomes. TF binding intensities in bound regions were calculated by UCSC bigWigAverageOverBed v2 with default parameters and the mean signal intensities were visualized by PRISM v8.4.3. Selected sequencing libraries (Fig. 5A) were aligned to the EBV genome (ACC number NC_007605) using Bowtie v1.2.2 with either default single-end or paired-end alignment settings, with additional parameters “–chunkmbs 1000 -S -m 1.” The filtering of low-quality reads, sorting, indexing, and visualization of aligned reads are the same as the processes for the EBV TF ChIP-seq data.
EBV contacts analysis.
EBV contacts that are common to two or more Hi-C biological replicates were obtained (Fig. 1C, Table S1B). Peak annotations for human TFs were sourced from ENCODE database with accession numbers listed in Table S2A. Annotations of TEs and SEs were obtained from reference 23. Overlap between EBV contacts and TFs, SEs, or TEs were calculated using bedtools intersect (50).
RNA-seq data analysis.
For visualization of gene expression, transcriptome sequencing (RNA-seq) raw data were aligned to a human reference genome (GRCh38; hg38) using Bowtie v1.2.2 (45) with parameters “--chunkmbs 1000 -S -m 100 -n 1,” then sorted and indexed by Samtools v1.9 (46). The genome-wide gene expression profiles (i.e., bigWig files) were generated by bamCoverage v3.2.0 (47) using parameters “–normalizeUsing BPM --minMappingQuality 30 --ignoreDuplicates --extendReads 250 --blackListFileName hg38.blacklist.bed,” where the blacklisted regions were obtained from reference 51. The gene expression profiles were visualized using IGV genome browser v2.8.0 (48).
FISH assay protocol.
Cells were treated with 0.075 M KCl, fixed with methanol:acetic acid (at 3:1), and mounted to slides. Slides were treated with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 37°C for 30 min and dehydrated in ethanol at room temperature. Probes (10 μl each) were added to slides. Probes and cellular DNA were denatured at 73°C for 2 min and hybridization was done at 37°C for 48 h in a humidified chamber. Slides were washed in 50% formamide/2× SSC (pH 7.2) and then 2× SSC at 42°C. Cellular DNA was labeled with DAPI (4′,6-diamidino-2-phenylindole). The EBV BAC was labeled with SpectrumOrange. The other BACs were labeled with SpectrumGreen (Abbott Molecular).
Data sources.
Hi-C samples were collected from GSE63525 (HIC021, HIC022, HIC023, HIC024, and HIC025). RNA-seq data are from GSE126379. The ChIP-seq data for histone markers and TFs are from ENCODE. The ChIP-seq data for EBV TFs EBNA1, EBNA2, EBNA3B, and EBNA3C are from GSE98121, GSE29498, and GSE76166, respectively. The annotations for super enhancers and typical enhancers are from reference 23.
Data availability.
The 4C data are available in the Gene Expression Omnibus (GEO) under accession number GSE154052.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grant R35GM138283 to M.K. and grants 5R01AI123420 and 5R01CA047006 to B.Z.), the Showalter Trust (research award to M.K.), and the Intramural Research Programs of the National Institute of Diabetes and Digestive and Kidney Diseases (project number ZIA/DK075149 to B.A.) of the National Institutes of Health.
L.W. and B.Y. performed all computational work and J.L. performed all experimental work and contributed to the writing of the manuscript. B.Y. and J.L. analyzed the 4C data. All other authors contributed significantly to computational, experimental, or conceptual development of this work. B.A., B.Z., and M.K. conceptualized the study, supervised the project, and wrote the manuscript.
We have no competing interests to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lieberman PM. 2014. Virology. Epstein-Barr virus turns 50. Science 343:1323–1325. doi: 10.1126/science.1252786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young LS, Yap LF, Murray PG. 2016. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer 16:789–802. doi: 10.1038/nrc.2016.92. [DOI] [PubMed] [Google Scholar]
- 3.Cohen JI, Fauci AS, Varmus H, Nabel GJ. 2011. Epstein-Barr virus: an important vaccine target for cancer prevention. Sci Transl Med 3:107fs7. doi: 10.1126/scitranslmed.3002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saha A, Robertson ES. 2019. Mechanisms of B-cell oncogenesis induced by Epstein-Barr virus. J Virol 93:e00238-19. doi: 10.1128/JVI.00238-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakravorty S, Yan B, Wang C, Wang L, Quaid JT, Lin CF, Briggs SD, Majumder J, Canaria DA, Chauss D, Chopra G, Olson MR, Zhao B, Afzali B, Kazemian M. 2019. Integrated pan-cancer map of EBV-associated neoplasms reveals functional host-virus interactions. Cancer Res 79:6010–6023. doi: 10.1158/0008-5472.CAN-19-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao B, Zou J, Wang H, Johannsen E, Peng CW, Quackenbush J, Mar JC, Morton CC, Freedman ML, Blacklow SC, Aster JC, Bernstein BE, Kieff E. 2011. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci U S A 108:14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaiser C, Laux G, Eick D, Jochner N, Bornkamm GW, Kempkes B. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol 73:4481–4484. doi: 10.1128/JVI.73.5.4481-4484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harada S, Kieff E. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J Virol 71:6611–6618. doi: 10.1128/JVI.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maruo S, Zhao B, Johannsen E, Kieff E, Zou J, Takada K. 2011. Epstein-Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression. Proc Natl Acad Sci U S A 108:1919–1924. doi: 10.1073/pnas.1019599108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maruo S, Wu Y, Ishikawa S, Kanda T, Iwakiri D, Takada K. 2006. Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc Natl Acad Sci U S A 103:19500–19505. doi: 10.1073/pnas.0604919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skalska L, White RE, Parker GA, Turro E, Sinclair AJ, Paschos K, Allday MJ. 2013. Induction of p16(INK4a) is the major barrier to proliferation when Epstein-Barr virus (EBV) transforms primary B cells into lymphoblastoid cell lines. PLoS Pathog 9:e1003187. doi: 10.1371/journal.ppat.1003187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cahir-McFarland ED, Davidson DM, Schauer SL, Duong J, Kieff E. 2000. NF-kappa B inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc Natl Acad Sci U S A 97:6055–6060. doi: 10.1073/pnas.100119497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutt-Fletcher LM. 2007. Epstein-Barr virus entry. J Virol 81:7825–7832. doi: 10.1128/JVI.00445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gahn TA, Schildkraut CL. 1989. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell 58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 15.De Leo A, Calderon A, Lieberman PM. 2020. Control of viral latency by episome maintenance proteins. Trends Microbiol 28:150–162. doi: 10.1016/j.tim.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frappier L. 2015. EBNA1. Curr Top Microbiol Immunol 391:3–34. doi: 10.1007/978-3-319-22834-1_1. [DOI] [PubMed] [Google Scholar]
- 17.Messick TE, Smith GR, Soldan SS, McDonnell ME, Deakyne JS, Malecka KA, Tolvinski L, van den Heuvel APJ, Gu BW, Cassel JA, Tran DH, Wassermann BR, Zhang Y, Velvadapu V, Zartler ER, Busson P, Reitz AB, Lieberman PM. 2019. Structure-based design of small-molecule inhibitors of EBNA1 DNA binding blocks Epstein-Barr virus latent infection and tumor growth. Sci Transl Med 11:eaau5612. doi: 10.1126/scitranslmed.aau5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arvey A, Tempera I, Lieberman PM. 2013. Interpreting the Epstein-Barr Virus (EBV) epigenome using high-throughput data. Viruses 5:1042–1054. doi: 10.3390/v5041042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arvey A, Tempera I, Tsai K, Chen HS, Tikhmyanova N, Klichinsky M, Leslie C, Lieberman PM. 2012. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe 12:233–245. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sengupta S, George RE. 2017. Super-enhancer-driven transcriptional dependencies in cancer. Trends Cancer 3:269–281. doi: 10.1016/j.trecan.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H, Schmidt SC, Jiang S, Willox B, Bernhardt K, Liang J, Johannsen EC, Kharchenko P, Gewurz BE, Kieff E, Zhao B. 2015. Epstein-Barr virus oncoprotein super-enhancers control B cell growth. Cell Host Microbe 17:205–216. doi: 10.1016/j.chom.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunnell A, Webb HM, Wood CD, McClellan MJ, Wichaidit B, Kempkes B, Jenner RG, Osborne C, Farrell PJ, West MJ. 2016. RUNX super-enhancer control through the Notch pathway by Epstein-Barr virus transcription factors regulates B cell growth. Nucleic Acids Res 44:4636–4650. doi: 10.1093/nar/gkw085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, Reichert E, Kung AL, Rodig SJ, Young RA, Shipp MA, Bradner JE. 2013. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, Dastur A, Amzallag A, Ramaswamy S, Tesar B, Jenkins CE, Hannett NM, McMillin D, Sanda T, Sim T, Kim ND, Look T, Mitsiades CS, Weng AP, Brown JR, Benes CH, Marto JA, Young RA, Gray NS. 2014. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511:616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kempfer R, Pombo A. 2020. Methods for mapping 3D chromosome architecture. Nat Rev Genet 21:207–226. doi: 10.1038/s41576-019-0195-2. [DOI] [PubMed] [Google Scholar]
- 26.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moquin SA, Thomas S, Whalen S, Warburton A, Fernandez SG, McBride AA, Pollard KS, Miranda JL. 2017. The Epstein-Barr virus episome maneuvers between nuclear chromatin compartments during reactivation. J Virol 92:e01413-17. doi: 10.1128/JVI.01413-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim KD, Tanizawa H, De Leo A, Vladimirova O, Kossenkov A, Lu F, Showe LC, Noma KI, Lieberman PM. 2020. Epigenetic specifications of host chromosome docking sites for latent Epstein-Barr virus. Nat Commun 11:877. doi: 10.1038/s41467-019-14152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hensel KO, Cantner F, Bangert F, Wirth S, Postberg J. 2018. Episomal HBV persistence within transcribed host nuclear chromatin compartments involves HBx. Epigenetics Chromatin 11:34. doi: 10.1186/s13072-018-0204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreau P, Cournac A, Palumbo GA, Marbouty M, Mortaza S, Thierry A, Cairo S, Lavigne M, Koszul R, Neuveut C. 2018. Tridimensional infiltration of DNA viruses into the host genome shows preferential contact with active chromatin. Nat Commun 9:4268. doi: 10.1038/s41467-018-06739-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, Onate KC, Graham K, Miyasato SR, Dreszer TR, Strattan JS, Jolanki O, Tanaka FY, Cherry JM. 2018. The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 46:D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki M, Liao W, Wos F, Johnston AD, DeGrazia J, Ishii J, Bloom T, Zody MC, Germer S, Greally JM. 2018. Whole-genome bisulfite sequencing with improved accuracy and cost. Genome Res 28:1364–1371. doi: 10.1101/gr.232587.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allday MJ. 2009. How does Epstein-Barr virus (EBV) complement the activation of Myc in the pathogenesis of Burkitt's lymphoma? Semin Cancer Biol 19:366–376. doi: 10.1016/j.semcancer.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo R, Jiang C, Zhang Y, Govande A, Trudeau SJ, Chen F, Fry CJ, Puri R, Wolinsky E, Schineller M, Frost TC, Gebre M, Zhao B, Giulino-Roth L, Doench JG, Teng M, Gewurz BE. 2020. MYC controls the Epstein-Barr virus lytic switch. Mol Cell 78:653–669. doi: 10.1016/j.molcel.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McClellan MJ, Wood CD, Ojeniyi O, Cooper TJ, Kanhere A, Arvey A, Webb HM, Palermo RD, Harth-Hertle ML, Kempkes B, Jenner RG, West MJ. 2013. Modulation of enhancer looping and differential gene targeting by Epstein-Barr virus transcription factors directs cellular reprogramming. PLoS Pathog 9:e1003636. doi: 10.1371/journal.ppat.1003636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Portal D, Zhao B, Calderwood MA, Sommermann T, Johannsen E, Kieff E. 2011. EBV nuclear antigen EBNALP dismisses transcription repressors NCoR and RBPJ from enhancers and EBNA2 increases NCoR-deficient RBPJ DNA binding. Proc Natl Acad Sci U S A 108:7808–7813. doi: 10.1073/pnas.1104991108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu F, Chen HS, Kossenkov AV, DeWispeleare K, Won KJ, Lieberman PM. 2016. EBNA2 drives formation of new chromosome binding sites and target genes for B-cell master regulatory transcription factors RBP-jkappa and EBF1. PLoS Pathog 12:e1005339. doi: 10.1371/journal.ppat.1005339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. 2017. A phase separation model for transcriptional control. Cell 169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng Q, Wang L, Qin Z, Wang J, Zheng X, Wei L, Zhang X, Zhang X, Liu C, Li Z, Wu Y, Li G, Yan Q, Ma J. 2020. Phase separation of Epstein-Barr virus EBNA2 and its coactivator EBNALP controls gene expression. J Virol 94:e01771-19. doi: 10.1128/JVI.01771-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okabe A, Huang KK, Matsusaka K, Fukuyo M, Xing M, Ong X, Hoshii T, Usui G, Seki M, Mano Y, Rahmutulla B, Kanda T, Suzuki T, Rha SY, Ushiku T, Fukayama M, Tan P, Kaneda A. 2020. Cross-species chromatin interactions drive transcriptional rewiring in Epstein-Barr virus-positive gastric adenocarcinoma. Nat Genet 52:919–930. doi: 10.1038/s41588-020-0665-7. [DOI] [PubMed] [Google Scholar]
- 41.Dooley KE, Warburton A, McBride AA. 2016. Tandemly integrated HPV16 can form a Brd4-dependent super-enhancer-like element that drives transcription of viral oncogenes. mBio 7:e01446-16. doi: 10.1128/mBio.01446-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durand NC, Shamim MS, Machol I, Rao SS, Huntley MH, Lander ES, Aiden EL. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Splinter E, de Wit E, van de Werken HJ, Klous P, de Laat W. 2012. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods 58:221–230. doi: 10.1016/j.ymeth.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44:W160–5. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amemiya HM, Kundaje A, Boyle AP. 2019. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep 9:9354. doi: 10.1038/s41598-019-45839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 4C data are available in the Gene Expression Omnibus (GEO) under accession number GSE154052.