Abstract
Retinopathy of prematurity (ROP) is a vasoproliferative retinal disease affecting premature infants. In addition to prematurity itself and oxygen treatment, genetic factors have been suggested to predispose to ROP. We aimed to identify potentially pathogenic genes and biological pathways associated with ROP by analyzing variants from whole exome sequencing (WES) data of premature infants. As part of a multicenter ROP cohort study, 100 non-Hispanic Caucasian preterm infants enriched in phenotypic extremes were subjected to WES. Gene-based testing was done on coding nonsynonymous variants. Genes showing enrichment of qualifying variants in severe ROP compared to mild or no ROP from gene-based tests with adjustment for gestational age and birth weight were selected for gene set enrichment analysis (GSEA). Mean BW of included infants with pre-plus, type-1 or type 2 ROP including aggressive posterior ROP (n = 58) and mild or no ROP (n = 42) were 744 g and 995 g, respectively. No single genes reached genome-wide significance that could account for a severe phenotype. GSEA identified two significantly associated pathways (smooth endoplasmic reticulum and vitamin C metabolism) after correction for multiple tests. WES of premature infants revealed potential pathways that may be important in the pathogenesis of ROP and in further genetic studies.
Subject terms: Retinopathy of prematurity, Medical genomics
Introduction
Retinopathy of prematurity (ROP) is a retinal vascular disease affecting prematurely born infants. Although there have been many advances in neonatal care and management for ROP, ROP remains a leading cause of childhood blindness throughout the world1,2. The most significant risk factors for ROP include low birth weight (BW), early gestational age (GA) and oxygen treatment3. However, some high-risk infants with low BW and early GA do not develop ROP, whereas some low-risk infants develop severe ROP including aggressive posterior ROP (AP-ROP)4–7. In these infants at phenotypic extremes, a study from our group demonstrated that known clinical risk factors were not significantly associated with development of ROP, suggesting presence of other risk factors for ROP8.
The i-ROP consortium includes collaborators from 14 academic institutions throughout the world with the goal of developing better methods for diagnosing, understanding and treating ROP through computer-based image analysis, genetic analysis and biomedical informatics analysis. Over the past 10 years, over 1700 subjects have participated in the research where demographic data, eye exam data, images taken at the eye exams, as well as other systemic health data have been collected and stored in a large data repository. The 100 non-Hispanic Caucasian preterm infants in this study were selected from the i-ROP consortium samples with the goal of building a sample set that is enriched in phenotypic extremes.
Genetic factors have been suggested to predispose to ROP3,9. Racial differences in incidence and severity of ROP3,10,11, high concordance rate among monozygotic twins12,13, and strain differences in animal models of ROP14–16 suggest possible roles of genetic factors in ROP. However, the field of ROP genetics is still in its infancy. A number of studies investigated the frequency of specific genetic variants in premature infants with or without ROP9. However, few of them showed strong association or were replicated in other populations, although some studies reported promising gene variants17,18. Moreover, most studies examined only a few variants from a small number of candidate genes, which were mostly related to retinal angiogenesis9. Studies using updated technologies such as genome-wide association studies (GWAS) or next-generation sequencing (NGS) based approaches have not been reported in ROP.
Whole exome sequencing (WES) has revealed novel pathogenic genes or variants especially in Mendelian disorders (e.g. retinitis pigmentosa) and also in multifactorial diseases (e.g. glaucoma19–22, age-related macular degeneration23–25) in ophthalmology. Also, combining WES and pathway analysis has enabled researchers to find novel biological pathways or polygenic burdens associated with disease in small to moderate-scale sequencing studies19,26–28. This study aimed to identify potentially pathogenic genes and biological pathways associated with ROP by analyzing whole exome sequencing data from 100 preterm infants enriched in phenotypic extremes, by analyzing the variants both by rare variant methods and common variant methods.
Results
Characteristics of subjects
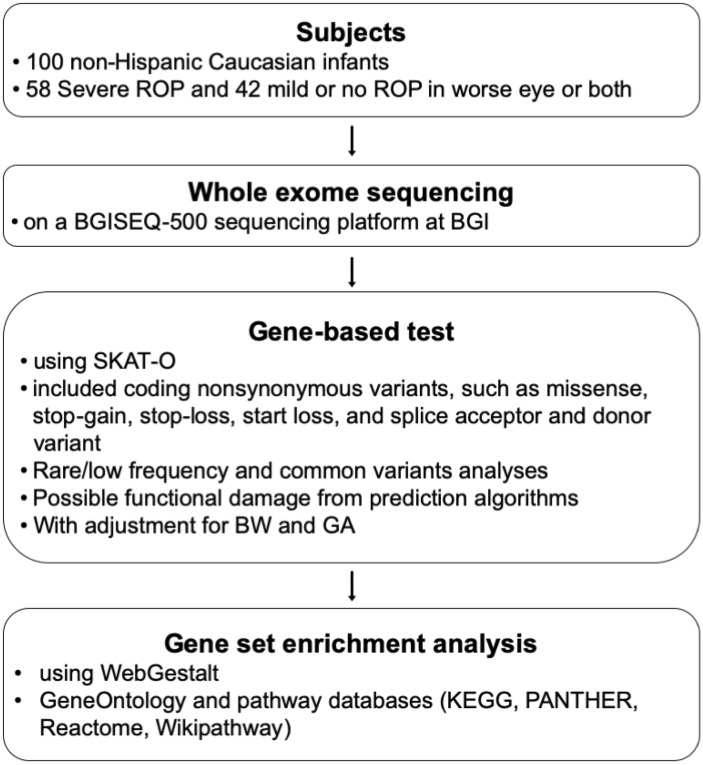
The overall scheme of this study is shown in Fig. 1. Fifty-eight premature infants with severe ROP (49 type 1 and 9 type 2 or pre-plus) and 42 with mild or no ROP (5 mild and 37 no ROP) in the worse eye or both eyes were included. Characteristics of 100 subjects including demographics, characteristics of ROP and associated morbidities of prematurity are summarized in Table 1. In the severe ROP group, 12 patients with bilateral AP-ROP were included.
Figure 1.
The overall scheme of this study. ROP retinopathy of prematurity, SKAT-O sequence kernel association optimal unified test, BW birth weight, GA gestational age, KEGG Kyoto Encyclopedia of Genes and Genomes, PANTHER Protein ANalysis THrough Evolutionary Relationships.
Table 1.
Characteristics of subjects: demographics, retinopathy of prematurity characteristics, and associated morbidities of prematurity.
| Parameters | Patient group | |
|---|---|---|
| No or mild ROP | Severe ROP | |
| No. of infants | 42 | 58 |
| Birth weight (g), mean ± SD | 995 ± 247 | 744 ± 256 |
| Gestational age (weeks), mean ± SD | 27.6 ± 2.0 | 25.2 ± 2.0 |
| Gender, male (%) | 16 (38) | 31 (53) |
| Race/ethnicity (%) | ||
| Non-Hispanic White | 100 | 100 |
| Lowest zone, no. of eyes (%) | ||
| Never Developed | 72 (86) | 0 (0) |
| Zone I | 0 (0) | 30 (26) |
| Zone II or III | 12 (14) | 86 (74) |
| Highest stage of ROP, no. of eyes (%) | ||
| Stage 0 | 72 (86) | 0 (0) |
| Stage 1 | 2 (2) | 0 (0) |
| Stage 2 | 10 (12) | 24 (21) |
| Stage 3 | 0 (0) | 92 (79) |
| Worst plus disease category, no. of eyes (%) | ||
| Plus | 0 (0) | 57 (49) |
| No plus | ||
| Pre-plus | 0 (0) | 35 (30) |
| Normal | 84 (100) | 24 (21) |
| AP-ROP, No. of eyes (%) | 0 (0) | 24 (21) |
| Associated morbidities of prematurity and management | ||
| BPD (chronic lung disease) (%) | 11 (26) | 40 (69) |
| IVH (%) | ||
| None | 36 (85) | 36 (62) |
| Grade I–II | 3 (7) | 14 (24) |
| Grade III–IV | 3 (7) | 8 (14) |
| Sepsis, overall (%) | 5 (12) | 29 (50) |
| Bacterial (%) | 3 (7) | 26 (45) |
| Fungal (%) | 0 (0) | 2 (3) |
| Unknown type (%) | 2 (5) | 1 (2) |
| NEC, surgical (%) | 1 (2) | 10 (17) |
+Two sample test of proportion.
*Student two-sample T-test.
^Fisher exact table test.
ROP retinopathy of prematurity, BPD bronchopulmonary dysplasia, IVH intraventricular hemorrhage, NEC necrotizing enterocolitis.
Sequencing results
The mean percentage of mapping on genome was 99.8% and mean sequencing depth on target 149.8 ± 36.8 (Supplemental Table S1). A minimum 1- and tenfold coverage per base was achieved on average for 95.9% and 92.0% of the target region, respectively (Supplemental Table S1). The number of each type of variant is summarized in Supplemental Table S2. Mean number of missense variants per patient was 9283 ± 271.
Gene-based analysis
Gene-based analysis using SKAT-O, which combines the effects of all rare variants across a gene, with adjustment for BW and GA revealed no genes that reached genome-wide significance (P < 2.5 × 10–6). From the rare/low-frequency variants analysis, the most strongly associated genes by SKAT-O included RNF225, PMS1, PRMT3, SPANXD, ANKRD30A and THSD4 (Table 2). Among the 263 candidate genes, CD36, NOX4, NTRK2, IGFBP7, and NTF4 were the most strongly associated with ROP (Supplemental Table S3). From the common variants analysis, which analyzes each variant separately, the most strongly associated genes by SKAT-O included ZNF341, ALG10B, OR4D6, KCNE1, and FAM198A (Table 3).
Table 2.
Results of rare/low-frequency variants analysis: the 20 most strongly associated genes in 100 non-Hispanic Caucasian preterm infants by SKAT-O.
| Rank | Official symbol | Gene name | Entrez gene ID | P value | Number of included variants | Number of variant alleles | |
|---|---|---|---|---|---|---|---|
| No or mild ROP | Severe ROP | ||||||
| 1 | RNF225 | Ring finger protein 225 | 646882 | 2.39E−04 | 1 | 10 | 2 |
| 2 | PMS1 | PMS1 homolog 1, mismatch repair system component | 5378 | 2.51E−04 | 5 | 7 | 0 |
| 3 | PRMT3 | Protein arginine methyltransferase 3 | 10196 | 2.61E−04 | 4 | 2 | 2 |
| 4 | SPANXD | SPANX family member D | 64648 | 3.29E−04 | 2 | 34 | 24 |
| 5 | ANKRD30A | Ankyrin repeat domain 30A | 91074 | 3.30E−04 | 4 | 16 | 20 |
| 6 | THSD4 | Thrombospondin type 1 domain containing 4 | 79875 | 5.04E−04 | 2 | 4 | 0 |
| 7 | ATP13A4 | ATPase 13A4 | 84239 | 6.10E−04 | 6 | 10 | 0 |
| 8 | FGD6 | FYVE, RhoGEF and PH domain containing 6 | 55785 | 6.20E−04 | 4 | 4 | 0 |
| 9 | DNALI1 | Dynein axonemal light intermediate chain 1 | 7802 | 8.26E−04 | 3 | 0 | 3 |
| 10 | PIGQ | Phosphatidylinositol glycan anchor biosynthesis class Q | 9091 | 8.28E−04 | 3 | 0 | 3 |
| 11 | OPA1 | OPA1, mitochondrial dynamin like GTPase | 4976 | 8.57E−04 | 1 | 5 | 0 |
| 12 | MYO3B | Myosin IIIB | 140469 | 9.61E−04 | 4 | 0 | 4 |
| 13 | ATAD3B | ATPase family, AAA domain containing 3B | 83858 | 9.72E−04 | 8 | 11 | 3 |
| 14 | PCSK6 | Proprotein convertase subtilisin/kexin type 6 | 5046 | 1.02E−03 | 7 | 11 | 1 |
| 15 | CHML | CHM like, Rab escort protein 2 | 1122 | 1.11E−03 | 1 | 0 | 5 |
| 16 | TRIM36 | Tripartite motif containing 36 | 55521 | 1.12E−03 | 3 | 0 | 3 |
| 17 | PPP4R1 | Protein phosphatase 4 regulatory subunit 1 | 9989 | 1.17E−03 | 4 | 0 | 5 |
| 18 | TBC1D32 | TBC1 domain family member 32 | 221322 | 1.44E−03 | 5 | 6 | 6 |
| 19 | CATSPER1 | Cation channel sperm associated 1 | 117144 | 1.45E−03 | 5 | 1 | 4 |
| 20 | RHBDF2 | Rhomboid 5 homolog 2 | 79651 | 1.45E−03 | 3 | 3 | 2 |
Table 3.
Results of common variants analysis: the 20 most strongly associated genes in 100 non-Hispanic Caucasian preterm infants by SKAT-O.
| Rank | Official Symbol | Gene name | Entrez gene ID | P value | Number of included variants | Number of variant alleles | |
|---|---|---|---|---|---|---|---|
| No or mild ROP | Severe ROP | ||||||
| 1 | ZNF341 | Zinc finger protein 341 | 84905 | 8.28E−05 | 6 | 29 | 43 |
| 2 | ALG10B | ALG10B, alpha-1,2-glucosyltransferase | 144245 | 2.82E−04 | 4 | 47 | 63 |
| 3 | OR4D6 | Olfactory receptor family 4 subfamily D member 6 | 219983 | 3.93E−04 | 5 | 77 | 90 |
| 4 | KCNE1 | Potassium voltage-gated channel subfamily E regulatory subunit 1 | 3753 | 4.14E−04 | 1 | 16 | 47 |
| 5 | FAM198A | Family with sequence similarity 198 member A | 729085 | 4.97E−04 | 7 | 145 | 216 |
| 6 | PI16 | Peptidase inhibitor 16 | 221476 | 5.23E−04 | 2 | 50 | 56 |
| 7 | CSTF2T | Cleavage stimulation factor subunit 2 tau variant | 23283 | 5.89E−04 | 3 | 11 | 32 |
| 8 | SPANXD | SPANX family member D | 64648 | 7.71E−04 | 2 | 38 | 34 |
| 9 | RNASE11 | Ribonuclease A family member 11 | 122651 | 7.88E−04 | 3 | 27 | 70 |
| 10 | COG7 | Component of oligomeric golgi complex 7 | 91949 | 7.99E−04 | 3 | 0 | 15 |
| 11 | NEDD9 | Neural precursor cell expressed, developmentally down-regulated 9 | 4739 | 8.37E−04 | 5 | 89 | 146 |
| 12 | OBSCN | Obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF | 84033 | 8.59E−04 | 80 | 495 | 634 |
| 13 | PKP3 | Plakophilin 3 | 11187 | 9.70E−04 | 11 | 177 | 223 |
| 14 | MTPN | Myotrophin | 136319 | 1.06E−03 | 1 | 2 | 12 |
| 15 | TLDC1 | TBC/LysM-associated domain containing 1 | 57707 | 1.09E − 03 | 8 | 87 | 118 |
| 16 | TNK2 | Tyrosine kinase non receptor 2 | 10188 | 1.13E−03 | 13 | 88 | 85 |
| 17 | CAPN2 | Calpain 2 | 824 | 1.25E−03 | 5 | 31 | 49 |
| 18 | FAT1 | FAT atypical cadherin 1 | 2195 | 1.29E−03 | 44 | 256 | 305 |
| 19 | KEL | Kell blood group, metallo-endopeptidase | 3792 | 1.33E−03 | 6 | 12 | 4 |
| 20 | ZUFSP | Zinc finger with UFM1 specific peptidase domain | 221302 | 1.48E−03 | 4 | 23 | 42 |
In order to check if there is confounding between genes that are contributing to prematurity and genes that are contributing to ROP specifically, we compared the twenty genes with the largest associations from the rare variant and common variant analysis sets with the largest gene list for prematurity to date29 and found no intersection.
Gene set enrichment analysis (GSEA)
GSEA identified one pathway (GO:0005790, smooth endoplasmic reticulum) from the rare/low-frequency variants analysis that was significantly enriched after correction for multiple tests (Table 4). GSEA also identified 25 pathways from databases including GO, KEGG, PANTHER, Reactome, and Wikipathways with empirical P values of less than 0.05, including the beta2 adrenergic receptor signaling pathway, dopamine receptor mediated signaling pathway, T cell activation, nicotinate/nicotinamide metabolism, and platelet-derived growth factor (PDGF) signaling pathway (Table 4). From the common variants analysis, GSEA identified one significantly enriched pathway (Reactome R-HSA-196836, Vitamin C metabolism) (Table 5). Other pathways with empirical P values of less than 0.05 are listed in Table 5.
Table 4.
Results of rare/low-frequency variants analysis: a list of the most significantly enriched pathways (P value < 0.05) from gene set enrichment analysis using WebGestalt.
| Accession | Pathway name | P value | Adjusted P value* |
|---|---|---|---|
| Gene ontology biological process | |||
| GO:1903828 | Negative regulation of cellular protein localization | 4.40E−02 | 6.42E−01 |
| GO:0002263 | Cell activation involved in immune response | 3.90E−02 | 6.43E−01 |
| GO:0044706 | Multi-multicellular organism process | 2.60E−02 | 6.50E−01 |
| Gene ontology cellular localization | |||
| GO:0005790 | Smooth endoplasmic reticulum | 5.02E−03 | 4.70E−02 |
| GO:0042579 | Microbody | 8.00E−03 | 4.46E−01 |
| GO:0044815 | DNA packaging complex | 4.60E−02 | 4.70E−01 |
| GO:0005793 | Endoplasmic reticulum-Golgi intermediate compartment | 8.00E−03 | 4.80E−01 |
| GO:0031225 | Anchored component of membrane | 2.80E−02 | 4.94E−01 |
| GO:0031201 | SNARE complex | 2.90E−02 | 5.10E−01 |
| GO:0048770 | Pigment granule | 1.50E−02 | 5.75E−01 |
| GO:0044306 | Neuron projection terminus | 4.20E−02 | 5.88E−01 |
| Gene ontology molecular function | |||
| GO:0000149 | SNARE binding | 2.00E−03 | 6.77E−01 |
| GO:0016776 | Phosphotransferase activity, phosphate group as acceptor | 1.61E−02 | 7.26E−01 |
| GO:0001085 | RNA polymerase II transcription factor binding | 1.00E−02 | 7.69E−01 |
| KEGG | |||
| hsa00760 | Nicotinate and nicotinamide metabolism: Homo sapiens | 1.01E−02 | 3.73E−01 |
| PANTHER | |||
| P04378 | Beta2 adrenergic receptor signaling pathway | 4.24E−02 | 2.69E−01 |
| P05912 | Dopamine receptor mediated signaling pathway | 1.11E−02 | 3.00E−01 |
| P00053 | T cell activation | 1.70E−02 | 3.48E−01 |
| P00047 | PDGF signaling pathway | 2.80E−02 | 5.16E−01 |
| Reactome | |||
| R-HSA-2151201 | Transcriptional activation of mitochondrial biogenesis | 3.01E−03 | 4.67E−01 |
| R-HSA-179419 | APC:Cdc20 mediated degradation of cell cycle proteins prior to satisfation of the cell cycle checkpoint | 2.11E−02 | 5.70E−01 |
| R-HSA-453276 | Regulation of mitotic cell cycle | 1.40E−02 | 5.74E−01 |
| R-HSA-2262752 | Cellular responses to stress | 2.50E−02 | 5.76E−01 |
| Wikipathways | |||
| WP481 | Insulin signaling | 1.90E−02 | 7.74E−01 |
| WP455 | GPCRs, Class A Rhodopsin-like | 8.00E−03 | 7.81E−01 |
SNARE soluble N-ethylmaleimide-sensitive factor attachment protein receptor, PDGF platelet-derived growth factor, APC/C anaphase-promoting complex, GPCR G-protein-coupled receptor.
*By Benjamini–Hochberg procedure.
Numbers in bold indicate the smallest adjusted p-values, i.e. most significant pathway enrichments among all rows in the table.
Table 5.
Results of common variants analysis: a list of the most significantly enriched pathways (P value < 0.05) from gene set enrichment analysis using WebGestalt.
| Accession | Pathway Name | P value | Adjusted P value* |
|---|---|---|---|
| Gene ontology biological process | |||
| GO:0034367 | Macromolecular complex remodeling | 8.12E−03 | 4.61E−01 |
| GO:0048857 | Neural nucleus development | 3.01E−03 | 6.05E−01 |
| GO:0060191 | Regulation of lipase activity | 3.00E−03 | 6.53E−01 |
| GO:0035902 | Response to immobilization stress | 6.15E−03 | 6.79E−01 |
| GO:0002507 | Tolerance induction | 2.97E−02 | 7.11E−01 |
| GO:0090077 | Foam cell differentiation | 1.00E−02 | 8.03E−01 |
| Gene ontology cellular localization | |||
| GO:0032994 | Protein–lipid complex | 9.02E−03 | 4.21E−01 |
| GO:0060076 | Excitatory synapse | 2.53E−02 | 4.80E−01 |
| GO:0098636 | Protein complex involved in cell adhesion | 2.80E−02 | 5.24E−01 |
| GO:1990391 | DNA repair complex | 2.71E−02 | 5.61E−01 |
| GO:0042383 | Sarcolemma | 3.00E−03 | 5.97E−01 |
| GO:0045178 | Basal part of cell | 3.10E−02 | 6.35E−01 |
| Gene ontology molecular function | |||
| GO:0005487 | Nucleocytoplasmic transporter activity | 1.21E−02 | 7.29E−01 |
| KEGG | |||
| hsa04924 | Renin secretion: Homo sapiens (human) | 2.00E−03 | 3.87E−01 |
| hsa04930 | Type II diabetes mellitus: Homo sapiens (human) | 7.00E−03 | 4.13E−01 |
| hsa03430 | Mismatch repair: Homo sapiens (human) | 1.22E−02 | 4.92E−01 |
| PANTHER | |||
| P04378 | Beta2 adrenergic receptor signaling pathway | 4.24E−02 | 2.69E−01 |
| Reactome | |||
| R-HSA-196836 | Vitamin C (ascorbate) metabolism | 1.15E−02 | 4.71E−02 |
| R-HSA-5693616 | Presynaptic phase of homologous DNA pairing and strand exchange | 0.00E+00 | 4.77E−01 |
| R-HSA-170670 | Adenylate cyclase inhibitory pathway | 3.12E−03 | 4.83E−01 |
| R-HSA-71288 | Creatine metabolism | 3.35E−03 | 5.12E−01 |
| R-HSA-997269 | Inhibition of adenylate cyclase pathway | 4.11E−03 | 5.19E−01 |
| R-HSA-3656253 | Defective EXT1 causes exostoses 1, TRPS2 and CHDS | 0.00E+00 | 5.35E−01 |
| R-HSA-448706 | Interleukin-1 processing | 1.84E−02 | 5.41E−01 |
| R-HSA-3656237 | Defective EXT2 causes exostoses 2 | 2.06E−03 | 5.55E−01 |
| R-HSA-8868766 | rRNA processing in the mitochondrion | 2.25E−03 | 5.84E−01 |
| Wikipathways | |||
| WP3407 | FTO Obesity Variant Mechanism | 1.68E−02 | 8.61E−02 |
| WP3601 | Composition of Lipid Particles | 3.39E−03 | 3.44E−01 |
| WP3943 | Robo4 and VEGF Signaling Pathways Crosstalk | 3.40E−02 | 3.65E−01 |
| WP1584 | Type II diabetes mellitus | 7.19E−03 | 4.84E−01 |
| WP3657 | Hematopoietic Stem Cell Gene Regulation by GABP alpha/beta Complex | 1.12E−02 | 5.56E−01 |
| WP3634 | Insulin signaling in human adipocytes (normal condition) | 3.45E−02 | 5.93E−01 |
| WP3635 | Insulin signaling in human adipocytes (diabetic condition) | 2.21E−02 | 6.29E−01 |
| WP2011 | SREBF and miR33 in cholesterol and lipid homeostasis | 1.35E−02 | 6.50E−01 |
| WP3301 | MFAP5-mediated ovarian cancer cell motility and invasiveness | 3.30E−02 | 6.62E−01 |
| WP3844 | PI3K-AKT-mTOR signaling pathway and therapeutic opportunities | 2.51E−02 | 6.62E−01 |
| WP727 | Monoamine Transport | 4.32E−02 | 6.66E−01 |
| WP430 | Statin Pathway | 2.01E−03 | 6.66E−01 |
EXT1 Exostosin 1, TRPS2 Trichorhinophalangeal syndrome type 2, CHDS chondrosarcoma, EXT2 Exostosin 2, Robo4 Roundabout homolog 4, VEGF vascular endothelial growth factor, SREBF Sterol regulatory element-binding transcription factor, MFAP5 Microfibril Associated Protein 5, PI3K Phosphoinositide 3-kinase, AKT Protein kinase B, mTOR mammalian target of rapamycin.
*By Benjamini–Hochberg procedure.
Numbers in bold indicate the smallest adjusted p-values, i.e. most significant pathway enrichments among all rows in the table.
Discussion
This study aimed to identify potential genes and pathways associated with ROP by analyzing rare and low-frequency variants from whole exome sequencing data of 100 preterm infants enriched in phenotypic extremes. The key findings from this study are as follows: (1) gene-based analysis with adjustment for BW and GA revealed no genes that reached genome-wide significance; and (2) GSEA identified 2 significantly enriched pathways (smooth endoplasmic reticulum and vitamin C metabolism) that may be important in the pathogenesis of ROP.
Gene-based analysis, which aggregates all the rare variants in each gene, using SKAT-O with adjustment for BW and GA identified many genes with low P values, although no genes reached formal genome-wide significance (P < 2.5 × 10–6). The top most strongly associated genes included several genes of potential interest (Table 2 and 3). THSD4 (P = 5.04 × 10–4) encodes thrombospondin type-1 domain-containing protein 4, also known as “A disintegrin and metalloproteinase with thrombospondin motifs-like protein 6” (ADAMTSL-6). ADAMTSL-6, expressed in various tissues including retina (from FANTOM5 data), is an extracellular matrix protein that promotes assembly of the fibrillin-1 matrix30. Fibrillin-1 controls activation of TGF-β31,32, which is an angiogenic activator and has been implicated in retinal vascular diseases such as diabetic retinopathy33,34. Therefore, THSD4 is one potential target for studies on ROP pathogenesis.
OPA1, the most common gene mutated in dominant optic atrophy, encodes a dynamin-related GTPase that is necessary for mitochondrial inner membrane fusion and maintenance of mitochondrial architecture35. Recently, it was suggested that diabetes resulted in reduced opa1 gene expression and mitochondrial dysfunction in an animal model of diabetic retinopathy (Verma A et al. IOVS 2016;57:ARVO E-Abstract 5446). However, the role of OPA1 in retinal angiogenesis has not been investigated.
Calpain 2, which is encoded by CAPN2, has been known to be involved in neurodegeneration. A recent study showed that inhibition of calpain 2 ameliorated retinal ischemic injury, suggesting that calpain-2 inhibitor might prevent ischemia-induced retinal degeneration36. Therefore, CAPN2 is a potential target for the treatment of ROP.
Among the 263 candidate genes, CD36 showed the lowest P value from SKAT-O test (Supplemental Table S3). CD36 encodes platelet glycoprotein 4, also known as the thrombospondin receptor. Platelet glycoprotein 4 was suggested to be involved in angiogenesis in various ways with or without mediating thrombospondin37,38. Also, as a multi-ligand scavenger receptor, it has been implicated in retina homeostasis38. Therefore, CD36 may also be a potential target for future ROP studies.
GSEA identified several pathways that may be important in the pathogenesis of ROP. Identified pathways included the β2 adrenergic receptor signaling pathway, dopamine receptor mediated signaling pathway, T cell activation, PDGF signaling pathway, Robo4 and VEGF signaling pathways crosstalk, and vitamin C (ascorbate) metabolism (Table 4 and 5). A series of studies showed that the β2 adrenergic receptors play a pivotal role in the regulation of vascular endothelial growth factor (VEGF) production and retinal neovascularization39. Also, the β1/β2 adrenergic receptor antagonist propranolol showed reduction in VEGF expression, retinal neovascularization and vascular leakage in oxygen induced retinopathy39. A recent systematic review on clinical trials which investigated the effect of beta-blockers on ROP concluded that low to moderate quality evidence suggests that prophylactic administration of oral beta-blockers might reduce progression towards stage 3 ROP and decrease the need for treatment40.
Retinal dopamine, which is synthesized in and released by subtypes of amacrine and interplexiform cells, and its receptor signaling pathway has not been associated with retinal angiogenesis41. However, studies suggested that dopamine mediates diverse functions including retina development, visual signaling, and myopic eye growth41,42. Thus, further studies to examine the relationships between dopamine receptor mediated signaling and retinal vascular development or ROP is warranted.
A growing body of evidence supports important roles of inflammation in ROP. Recently, a study showed that regulatory T cells are recruited to the retina in oxygen-induced retinopathy and reduce severe microvascular disease43. In this manner, the T cell activation pathway might be related to development of ROP. The PDGF signaling pathway was also important for pericyte viability and the subsequent prevention of VEGF/VEGFR-2 overexpression and angiogenesis in oxygen-induced retinopathy44. Although PDGF antagonists have been tried for patients with neovascular age-related macular degeneration with unclear benefits (https://clinicaltrials.gov/ct2/show/NCT01944839; Last accessed on 09/13/2019)45, the exact role of PDGF in ROP requires further investigation.
One of the identified pathways (GO:0005790, smooth endoplasmic reticulum) was significantly enriched after correction for multiple tests. In general, smooth endoplasmic reticulum is associated with production of carbohydrate and lipids such as steroid hormones, cholesterol and membrane phospholipids. It also plays an important role in protein modification and intracellular protein transport. However, whether biological processes in smooth endoplasmic reticulum have specific roles in ROP is not known.
A few studies investigated the role of vitamin C in ROP or retinal angiogenesis. An in vitro study showed that vitamin C prevented VEGF-induced increases in endothelial permeability46, and retinal level of vitamin C was reduced in the rat model of ROP47. However, a randomized controlled trial in 2005 which compared high or low dose supplementation of vitamin C on clinical outcome including ROP showed no significant effects on the development of (any stage) ROP48. Further studies to examine the relationships between vitamin C metabolism and ROP is warranted.
This study has several limitations. First, the statistical power was not high because of small sample size (58 severe ROP and 42 mild or no ROP). The small sample size may be one of the reasons why no genes reached genome-wide significance in this study. Second, although we tried to include phenotypic extremes to overcome low statistical power due to small sample size, we could not identify enough phenotypically extreme patients from the i-ROP database. In this study, the severe ROP group included 12 patients with AP-ROP and the remaining patients were selected for highest birthweight in severe ROP group and lowest birthweight in no or mild ROP group. Thus, we believe that our subjects are enriched in phenotypic extremes, which may mean possible enrichment of rare pathogenic variants in our subjects. Third, as a result of the first two limitations, identified association with pathways that are plausible biologically need to be replicated in larger studies.
This study is, to the best of our knowledge, the first NGS-based genetic study in ROP. In this study, we analyzed WES data of 100 premature infants from the large-scale multicenter i-ROP consortium. Although no genes reached genome-wide significance, the results revealed genes and pathways that may be important in development or progression of ROP. Novel genes and pathways may be the targets of future genetic studies such as GWAS.
Methods
This study was approved by the Institutional Review Board at the coordinating center (Oregon Health and Science University) and at each of 8 study centers (Columbia University, University of Illinois at Chicago, William Beaumont Hospital, Children’s Hospital Los Angeles, Cedars-Sinai Medical Center, University of Miami, Weill Cornell Medical Center, and the Genomics Institute at The Lundquist Institute / Harbor-UCLA Medical Center). This study was conducted in accordance with the Declaration of Helsinki49. Written informed consent for the study was obtained from parents of all infants enrolled.
Subjects
The subjects for this study were selected from participants of the Imaging and Informatics for ROP (i-ROP) study, a prospective multicenter cohort study which enrolled preterm infants for ROP screening and collected clinical and imaging data (retinal images obtained using a wide-angle fundus camera [RetCam; Natus Medical Incorporated, Pleasanton, CA]) and blood or saliva samples. In this study, the ROP diagnosis of each eye exam was made by combining clinical exam at each study center and image-based diagnoses by 3 trained graders, as previously described50. The i-ROP study database was reviewed to identify 100 non-Hispanic Caucasian infants enriched in phenotypic extremes (e.g. AP-ROP, non-LBW infants with severe ROP, LBW infants with no ROP) to maximize variant discovery. Among the 373 non-Hispanic Caucasian infants enrolled between July 2011 and October 2016, patients with AP-ROP in at least one eye were selected first. Remaining cases were selected for lowest birthweight in the case of no or mild ROP (defined as ROP less than type 2 ROP), and highest birthweight in severe ROP (defined as pre-plus, type-2 or type 1 ROP), with an enforced ratio of approximately 4:1:1:4 of no, mild, pre-plus/type-2, and type 1 ROP, respectively. In cases where a patient had a twin in the selected set, the most phenotypically extreme infant of the two was selected. Patients without detailed information on demographics, ROP screening, co-morbidities of prematurity, or imaging data were excluded.
Whole exome sequencing
Genomic DNA was extracted from peripheral blood samples and whole-exome sequencing was performed at Beijing Genomics Institute (BGI; Hong Kong, China). Briefly, genomic DNA was randomly fragmented and enriched for exome sequences using the SureSelect Human All Exon Kit (Agilent Technologies, Santa Clara, CA, USA) and sequencing was performed on a BGISEQ-500 sequencing platform (BGI; Hong Kong, China). Sequencing-derived raw image files were processed by BGISEQ-500 base-calling Software for base-calling and the sequence data of each sample was generated as paired-end reads51.
Bioinformatics analyses
After removing reads containing sequence adaptors and low-quality reads, reads of each sample were aligned to the reference human genome, Genome Reference Consortium Human Build 37 using Burrows-Wheeler Aligner software. Local realignment around insertions and deletions (InDels) and base quality score recalibration were performed using Genome Analysis Toolkit (GATK), with duplicate reads removed by Picard MarkDuplicates. The HaplotypeCaller of GATK was used to call SNPs. After performing hard-filtering for SNPs and Indels as previously described52, the SnpEff tool was used to annotate SNPs (reported in dbSNP v.141) and Indels. The variants were annotated with the allele frequency in 1000 Genomes Project (http://www.internationalgenome.org/)53 or ESP6500 database (http://evs.gs.washington.edu/EVS/), and with prediction algorithms including SIFT54, PolyPhen255, MutationAssessor56, FATHMM57, and MutationTaster58.
Selection of candidate genes
Previously reported candidate genes in ROP and additional genes involving pathways in retinal angiogenesis/vasculogenesis, neuronal development, neuroprotection, and retinal inflammation were selected (n = 164; Supplemental Table S4). Also, additional genes (n = 99) that are related to the 164 candidate genes were identified using Cytoscape with GeneMANIA plugin which finds related genes using large-scale functional association data including protein and genetic interactions, pathways, co-expression, co-localization and protein domain similarity (Supplemental Table S5)59.
Gene-based test
Gene and pathway analyses were run on two subsets of data. First, gene-based testing was performed to detect rare and low-frequency variants with relatively large effect. For the test, variants with a MAF of greater than 0.05 in 1000 Genomes Project and ESP6500 were excluded. In gene-based testing, in order to provide sufficient power, all rare variants within the gene are combined for the statistical analysis. Second, for the common variant analysis, variants with a MAF of ≥ 0.05 in 1000 Genomes Project or ESP6500 were selected. In common variant analysis, each variant is analyzed separately. The qualifying variants for gene-based tests included coding nonsynonymous variants, including missense, stop-gain, stop-loss, start loss, and splice acceptor and donor variants that were predicted to be functionally damaging from at least 1 of the 5 prediction algorithms (SIFT, PolyPhen2, MutationAssessor, FATHMM, and MutationTaster). These qualifying variants in individual genes from the two subsets of data (rare / low-frequency variants and common variants) were subject to sequence kernel association optimal unified test (SKAT-O)60 using the genipe (GENome-wide Imputation PipelinE) v.1.4.0 module61 with adjustment for BW and GA.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed using WebGestalt (WEB-based Gene SeT AnaLysis Toolkit, http://www.webgestalt.org) using two sets of genes from the rare/low-frequency and common variants analyses, respectively62. For GSEA, we included Gene Ontology (GO, http://geneontology.org)63, KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.kegg.jp/)64, PANTHER database (Protein ANalysis THrough Evolutionary Relationships, http://www.pantherdb.org)65, Reactome (https://reactome.org)66, and WikiPathways (https://wikipathways.org)67. The obtained P values were adjusted by Benjamini–Hochberg correction (false discovery rate, P < 0.05).
Conference presentation
Portions of this study were presented at the 2018 ARVO annual meeting in Honolulu, HI, on May 1, 2018.
Supplementary Information
Acknowledgements
This study was supported by Grant R01EY19474 and P30EY10572 from the National Institutes of Health (Bethesda, MD), by Grant SCH-1622679 from the National Science Foundation (Arlington, VA), and by unrestricted departmental funding from Research to Prevent Blindness (New York, NY). The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI Grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease diabetes Research Center (DRC) Grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
Author contributions
S.K. and K.S. wrote the main manuscript text. S.K., K.S. performed the statistical analysis. R.S., J.P.C., S.O. selected and curated the dataset. R.V.P.C., A.N., K.D., A.B., and J.H. provided clinical expertise for diagnoses. X.L., Y.D.C., and K.T. reviewed the statistical analysis and provided further insight. C.S. provided expertise in neonatology. J.R, and M.C. led the project and provided oversight. Imaging and Informatics in Retinopathy of Prematurity (i-ROP) Research Consortium provided the patient data used in the study.
Competing interests
Michael Chiang is an unpaid member of the Scientific Advisory Board for Clarity Medical Systems (Pleasanton, CA), a Consultant for Novartis (Basel, Switzerland), and an initial member of Inteleretina. R. V. Paul Chan is a Consultant for Visunex Medical Systems (Fremont, CA) and a Consultant for Alcon (Fort Worth, TX), Allergan (Irvine, CA), and Bausch and Lomb (St. Louis, MO). Rest of the authors declare to have no competing interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sang Jin Kim and Kemal Sonmez.
These authors jointly supervised this work: Jerome I. Rotter and Michael F. Chiang.
A comprehensive list of consortium members appears at the end of the paper.
Contributor Information
Jerome I. Rotter, Email: jrotter@labiomed.org
Michael F. Chiang, Email: chiangm@ohsu.edu
Imaging and Informatics in Retinopathy of Prematurity (i-ROP) Research Consortium:
Michael F. Chiang, Susan Ostmo, Sang Jin Kim, Kemal Sonmez, J. Peter Campbell, R. V. Paul Chan, Karyn Jonas, Jason Horowitz, Osode Coki, Cheryl-Ann Eccles, Leora Sarna, Anton Orlin, Audina Berrocal, Catherin Negron, Kimberly Denser, Kristi Cumming, Tammy Osentoski, Tammy Check, Mary Zajechowski, Thomas Lee, Evan Kruger, Kathryn McGovern, Charles Simmons, Raghu Murthy, Sharon Galvis, Jerome Rotter, Ida Chen, Xiaohui Li, Kent Taylor, Kaye Roll, Jayashree Kalpathy-Cramer, Deniz Erdogmus, Stratis Ioannidis, Maria Ana Martinez-Castellanos, Samantha Salinas-Longoria, Rafael Romero, Andrea Arriola, Francisco Olguin-Manriquez, Miroslava Meraz-Gutierrez, Carlos M. Dulanto-Reinoso, and Cristina Montero-Mendoza
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-83552-y.
References
- 1.Blencowe H, Lawn JE, Vazquez T, Fielder A, Gilbert C. Preterm-associated visual impairment and estimates of retinopathy of prematurity at regional and global levels for 2010. Pediatr Res. 2013;74(Suppl 1):35–49. doi: 10.1038/pr.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert C. Retinopathy of prematurity: a global perspective of the epidemics, population of babies at risk and implications for control. Early Hum Dev. 2008;84:77–82. doi: 10.1016/j.earlhumdev.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Kim SJ, et al. Retinopathy of Prematurity: A Review of Risk Factors and their Clinical Significance. Surv Ophthalmol. 2018 doi: 10.1016/j.survophthal.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn YJ, et al. Characteristic clinical features associated with aggressive posterior retinopathy of prematurity. Eye (Lond) 2017;31:924–930. doi: 10.1038/eye.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen YH, et al. Retinopathy of prematurity in neonatal patients with birth weight greater than 1500 g in Taiwan. Biomed J. 2013;36:84–89. doi: 10.4103/2319-4170.110399. [DOI] [PubMed] [Google Scholar]
- 6.Park SH, Yum HR, Kim S, Lee YC. Retinopathy of prematurity in Korean infants with birthweight greater than 1500 g. Br J Ophthalmol. 2016;100:834–838. doi: 10.1136/bjophthalmol-2015-306960. [DOI] [PubMed] [Google Scholar]
- 7.Wheeler DT, et al. Retinopathy of prematurity in infants weighing less than 500 grams at birth enrolled in the early treatment for retinopathy of prematurity study. Ophthalmology. 2011;118:1145–1151. doi: 10.1016/j.ophtha.2010.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Port AD, Chan RV, Ostmo S, Choi D, Chiang MF. Risk factors for retinopathy of prematurity: insights from outlier infants. Graefes Arch Clin Exp Ophthalmol. 2014;252:1669–1677. doi: 10.1007/s00417-014-2716-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swan R, et al. The genetics of retinopathy of prematurity: a model for neovascular retinal disease. Ophthalmol Retina. 2018;2:949–962. doi: 10.1016/j.oret.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saunders, R. A. et al. Racial variation in retinopathy of prematurity. The Cryotherapy for Retinopathy of Prematurity Cooperative Group. Arch Ophthalmol115, 604–608 (1997). [DOI] [PubMed]
- 11.Chiang MF, Arons RR, Flynn JT, Starren JB. Incidence of retinopathy of prematurity from 1996 to 2000: analysis of a comprehensive New York state patient database. Ophthalmology. 2004;111:1317–1325. doi: 10.1016/j.ophtha.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 12.Ortega-Molina JM, et al. Genetic and Environmental Influences on Retinopathy of Prematurity. Mediators Inflamm. 2015;2015:764159. doi: 10.1155/2015/764159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bizzarro MJ, et al. Genetic susceptibility to retinopathy of prematurity. Pediatrics. 2006;118:1858–1863. doi: 10.1542/peds.2006-1088. [DOI] [PubMed] [Google Scholar]
- 14.van Wijngaarden, P., Coster, D. J., Brereton, H. M., Gibbins, I. L. & Williams, K. A. Strain-dependent differences in oxygen-induced retinopathy in the inbred rat. Invest Ophthalmol Vis Sci46, 1445–1452, doi:10.1167/iovs.04-0708 (2005). [DOI] [PubMed]
- 15.van Wijngaarden, P., Brereton, H. M., Coster, D. J. & Williams, K. A. Genetic influences on susceptibility to oxygen-induced retinopathy. Invest Ophthalmol Vis Sci48, 1761–1766, doi:10.1167/iovs.06-0531 (2007). [DOI] [PubMed]
- 16.Floyd BN, et al. Differences between rat strains in models of retinopathy of prematurity. Mol Vis. 2005;11:524–530. [PubMed] [Google Scholar]
- 17.Hartnett ME, et al. Genetic variants associated with severe retinopathy of prematurity in extremely low birth weight infants. Invest Ophthalmol Vis Sci. 2014;55:6194–6203. doi: 10.1167/iovs.14-14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartnett ME, Cotten CM. Genomics in the neonatal nursery: Focus on ROP. Semin Perinatol. 2015;39:604–610. doi: 10.1053/j.semperi.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou T, et al. Whole exome sequencing implicates eye development, the unfolded protein response and plasma membrane homeostasis in primary open-angle glaucoma. PLoS ONE. 2017;12:e0172427. doi: 10.1371/journal.pone.0172427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Micheal S, et al. Identification of TP53BP2 as a Novel Candidate Gene for Primary Open Angle Glaucoma by Whole Exome Sequencing in a Large Multiplex Family. Mol Neurobiol. 2018;55:1387–1395. doi: 10.1007/s12035-017-0403-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang C, et al. Detection of mutations in MYOC, OPTN, NTF4, WDR36 and CYP1B1 in Chinese juvenile onset open-angle glaucoma using exome sequencing. Sci Rep. 2018;8:4498. doi: 10.1038/s41598-018-22337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferre-Fernandez JJ, et al. Whole-Exome Sequencing of Congenital Glaucoma Patients Reveals Hypermorphic Variants in GPATCH3, a New Gene Involved in Ocular and Craniofacial Development. Sci Rep. 2017;7:46175. doi: 10.1038/srep46175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sardell RJ, et al. Whole exome sequencing of extreme age-related macular degeneration phenotypes. Mol Vis. 2016;22:1062–1076. [PMC free article] [PubMed] [Google Scholar]
- 24.Huang LZ, et al. Whole-exome sequencing implicates UBE3D in age-related macular degeneration in East Asian populations. Nat Commun. 2015;6:6687. doi: 10.1038/ncomms7687. [DOI] [PubMed] [Google Scholar]
- 25.Duvvari MR, et al. Whole Exome Sequencing in Patients with the Cuticular Drusen Subtype of Age-Related Macular Degeneration. PLoS ONE. 2016;11:e0152047. doi: 10.1371/journal.pone.0152047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cirulli ET, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, et al. Sporadic Hirschsprung Disease: Mutational Spectrum and Novel Candidate Genes Revealed by Next-generation Sequencing. Sci Rep. 2017;7:14796. doi: 10.1038/s41598-017-14835-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knijnenburg TA, et al. Genomic and molecular characterization of preterm birth. Proc Natl Acad Sci U S A. 2019;116:5819–5827. doi: 10.1073/pnas.1716314116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsutsui K, et al. ADAMTSL-6 is a novel extracellular matrix protein that binds to fibrillin-1 and promotes fibrillin-1 fibril formation. J Biol Chem. 2010;285:4870–4882. doi: 10.1074/jbc.M109.076919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neptune ER, et al. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33:407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 32.Kaartinen V, Warburton D. Fibrillin controls TGF-beta activation. Nat Genet. 2003;33:331–332. doi: 10.1038/ng0303-331. [DOI] [PubMed] [Google Scholar]
- 33.Van Geest RJ, Klaassen I, Vogels IM, Van Noorden CJ, Schlingemann RO. Differential TGF-{beta} signaling in retinal vascular cells: a role in diabetic retinopathy? Invest Ophthalmol Vis Sci. 2010;51:1857–1865. doi: 10.1167/iovs.09-4181. [DOI] [PubMed] [Google Scholar]
- 34.Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 35.Chun BY, Rizzo JF., 3rd Dominant optic atrophy: updates on the pathophysiology and clinical manifestations of the optic atrophy 1 mutation. Curr Opin Ophthalmol. 2016;27:475–480. doi: 10.1097/icu.0000000000000314. [DOI] [PubMed] [Google Scholar]
- 36.Lam PM, Gonzalez MI. Calpain activation and neuronal death during early epileptogenesis. Neurobiol Dis. 2019;124:141–151. doi: 10.1016/j.nbd.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silverstein, R. L. & Febbraio, M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal2, re3, doi:10.1126/scisignal.272re3 (2009). [DOI] [PMC free article] [PubMed]
- 38.Febbraio M, Hajjar DP, Silverstein RL. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J Clin Invest. 2001;108:785–791. doi: 10.1172/jci14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casini G, Dal Monte M, Fornaciari I, Filippi L, Bagnoli P. The beta-adrenergic system as a possible new target for pharmacologic treatment of neovascular retinal diseases. Prog Retin Eye Res. 2014;42:103–129. doi: 10.1016/j.preteyeres.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Kaempfen, S., Neumann, R. P., Jost, K. & Schulzke, S. M. Beta-blockers for prevention and treatment of retinopathy of prematurity in preterm infants. Cochrane Database Syst Rev3, Cd011893, doi:10.1002/14651858.CD011893.pub2 (2018). [DOI] [PMC free article] [PubMed]
- 41.Zhou X, Pardue MT, Iuvone PM, Qu J. Dopamine signaling and myopia development: What are the key challenges. Prog Retin Eye Res. 2017;61:60–71. doi: 10.1016/j.preteyeres.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witkovsky P. Dopamine and retinal function. Doc Ophthalmol. 2004;108:17–40. doi: 10.1023/B:DOOP.0000019487.88486.0a. [DOI] [PubMed] [Google Scholar]
- 43.Deliyanti D, et al. Foxp3(+) Tregs are recruited to the retina to repair pathological angiogenesis. Nat Commun. 2017;8:748. doi: 10.1038/s41467-017-00751-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilkinson-Berka JL, et al. Inhibition of platelet-derived growth factor promotes pericyte loss and angiogenesis in ischemic retinopathy. Am J Pathol. 2004;164:1263–1273. doi: 10.1016/s0002-9440(10)63214-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaffe GJ, et al. Dual Antagonism of PDGF and VEGF in Neovascular Age-Related Macular Degeneration: A Phase IIb, Multicenter, Randomized Controlled Trial. Ophthalmology. 2017;124:224–234. doi: 10.1016/j.ophtha.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 46.Ulker E, Parker WH, Raj A, Qu ZC, May JM. Ascorbic acid prevents VEGF-induced increases in endothelial barrier permeability. Mol Cell Biochem. 2016;412:73–79. doi: 10.1007/s11010-015-2609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Penn, J. S., Thum, L. A. & Naash, M. I. Oxygen-induced retinopathy in the rat. Vitamins C and E as potential therapies. Invest Ophthalmol Vis Sci33, 1836–1845 (1992). [PubMed]
- 48.Darlow BA, et al. Vitamin C supplementation in very preterm infants: a randomised controlled trial. Arch Dis Child Fetal Neonatal Ed. 2005;90:F117–122. doi: 10.1136/adc.2004.056440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.World Medical Association Declaration of Helsinki ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 50.Ryan MC, et al. Development and Evaluation of Reference Standards for Image-based Telemedicine Diagnosis and Clinical Research Studies in Ophthalmology. AMIA Annu Symp Proc. 2014;2014:1902–1910. [PMC free article] [PubMed] [Google Scholar]
- 51.Huang J, et al. A reference human genome dataset of the BGISEQ-500 sequencer. Gigascience. 2017;6:1–9. doi: 10.1093/gigascience/gix024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics43, 11.10.11–33, doi:10.1002/0471250953.bi1110s43 (2013). [DOI] [PMC free article] [PubMed]
- 53.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 55.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucl. Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shihab HA, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 59.Warde-Farley D, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucl. Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee S, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lemieux Perreault, L. P., Legault, M. A., Asselin, G. & Dube, M. P. genipe: an automated genome-wide imputation pipeline with automatic reporting and statistical tools. Bioinformatics32, 3661–3663, doi:10.1093/bioinformatics/btw487 (2016). [DOI] [PMC free article] [PubMed]
- 62.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucl. Acids Res. 2017;45:W130–w137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Expansion of the Gene Ontology knowledgebase and resources Nucl. Acids Res. 2017;45:D331–d338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucl. Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mi H, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucl. Acids Res. 2017;45:D183–d189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fabregat A, et al. The reactome pathway knowledgebase. Nucl. Acids Res. 2018;46:D649–d655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Slenter DN, et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucl. Acids Res. 2018;46:D661–d667. doi: 10.1093/nar/gkx1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.