Abstract
Osteoarthritis (OA) of the knee is a major source of disability that has no known treatment or cure. Morphological and compositional MRI is commonly used for assessing the bone and soft tissues in the knee to enhance the understanding of OA pathophysiology. However, it is challenging to extend these imaging methods and their subsequent analysis techniques to study large population cohorts due to slow and inefficient imaging acquisition and postprocessing tools. This can create a bottle-neck in assessing early OA changes and evaluating the responses of novel therapeutics. The purpose of this review article is to highlight recent developments in tools for enhancing the efficiency of knee MRI methods useful to study OA. Advances in efficient MRI data acquisition and reconstruction tools for morphological and compositional imaging, efficient automated image analysis tools, and hardware improvements to further drive efficient imaging are discussed in this review. For each topic, we discuss the current challenges as well as potential future opportunities to alleviate these challenges.
MAGNETIC RESONANCE IMAGING (MRI) is pervasively utilized in musculoskeletal imaging.1 The knee is one of the most commonly imaged joints using MRI. Clinically, MRI is commonly used to assess internal derangement of the knee, especially in tissues such as cartilage, bone, menisci, and the ligaments. Beyond the clinic, knee MRI is used widely to study the degenerative disease of osteoarthritis (OA)—one of the leading causes of disability in elderly populations. Knee OA affects ~17 million adults in the US. Yet besides knee arthroplasty for endstage OA, there is no treatment or cure for the disease.2,3 Weight-bearing radiography to measure joint space loss and osteophyte incidence using the Kellgren-Lawrence grading system is the current gold-standard for diagnosing OA.4 However, this system is wholly insensitive to the soft tissues involved in this whole-joint multifactorial disease and it cannot depict early-stage changes at a point where they may be reversible.5
MRI enables assessment of OA as a whole-joint disease due to its ability to simultaneously interrogate multiple tissues in the joint using multicontrast morphological imaging. Advances in compositional imaging of tissues using quantitative MRI may also potentially provide noninvasive imaging biomarkers of OA. One of the primary objectives of imaging research studies in OA, morphological or compositional, is to develop tools to assess the early onset and progression of the disease for evaluating the efficacy of pharmacologic or lifestyle-based interventions. Given the multiplicity of OA risk factors (such as age, body mass index [BMI], gender, previous trauma, joint alignment, etc), such endeavors likely require a combination of morphological and compositional imaging in large populations that consist of healthy subjects with predispositions for OA as well as subjects with OA.2
The challenge in population-wide OA research is the lack of efficiency in current imaging and analysis techniques, which precludes systematic implementation of advanced imaging in large epidemiological studies. For example, knee MRI in the largest observational OA study, the Osteoarthritis Initiative (OAI), with ~5000 participants, required almost 1 hour of MRI scan time, where scan time constraints limited scanning both knees with the same sequences. By comparison, in the UK Biobank, an observational study of ~100,000 participants,6 the brain MRI protocol only requires 36 minutes, where every additional minute of MRI scanning was surmised to increase study costs by £1 million.7 Thus, there is a pressing need to develop time-efficient imaging tools that provide morphological and compositional imaging insights to improve understanding of OA pathophysiology and to screen the appropriate populations for inclusion in clinical trials.8 Moreover, large imaging datasets also present the challenge of developing techniques to interpret insights from images in a timely and efficient manner. Images from the OAI are still being manually analyzed, even 15 years after the inclusion of the first subjects. Going forward, there is a need to pair efficient image analysis tools with efficient acquisition methods.
Given the current challenges of imaging OA, the goals of this review article are to present advances in: 1) efficient morphological imaging of the knee to evaluate structural abnormalities; 2) compositional imaging to determine prestructural soft-tissue changes; 3) automated image analysis techniques to gain salient insights from acquired images; and 4) imaging hardware to enable more efficient MRI sequences.
Morphological Imaging
Imaging the structure of the knee is essential to enhance the understanding of longitudinal and spatial structural degradation patterns induced in OA. The heterogeneous whole-organ nature of OA necessitates imaging protocols with adequate sensitivity for whole-joint structural analysis.9 The assessment of tissue structure is fundamental in semiquantitative scoring mechanisms of OA severity and progression.10 Traditional musculoskeletal imaging entails the use of 2D fast spin echo (FSE, also known as turbo spin echo, TSE) sequences that provide excellent in-plane resolution, but at the expense of thick slices and slice gaps due to signal-to-noise ratio (SNR) efficiency limitations. This requires using multiple thick-slice 2D sequences with the same contrast in different imaging planes, which may increase susceptibility to partial volume artifacts and preclude arbitrary plane reformations. Additionally, results from the OAI have demonstrated that cartilage and meniscus morphometry is predictive of OA progression.11,12 Such changes were measured using high-resolution sequences with favorable segmentation contrasts for cartilage and menisci, such as the double-echo steady-state (DESS) or spoiled gradient recalled echo (SPGR).13 However, in the OAI the DESS sequence required 11 minutes of scan time per knee. Consequently, increasing the speed of such protocols can directly minimize the costs of clinical trials and studies, and allow for larger enrollment cohorts. The following section includes recent advances in efficient 2D imaging techniques to lower the scan time of current protocols as well as novel 3D imaging techniques for rapid near-isotropic high-resolution imaging. A summary of some of the different techniques developed recently is provided in Table 1.
TABLE 1.
Characteristics of 2D FSE Sequences and Recent Methods for More Efficient Morphologic Knee MRI
| Properties | 2D FSE | 3D FSE | 3D T2-Sh | 3D qDESS |
|---|---|---|---|---|
| In-plane resolution | High | Moderate | Moderate | High |
| Through-plane resolution | Low | High | High | Moderate |
| Slice gaps | Present | Absent | Absent | Absent |
| Blurring | Low | Moderate | Moderate | Low |
| Contrasts | Conventional | Conventional | Multiple; Conventional | Dual; Novel |
| Quantitative | No | No | No | Yes (T2) |
| Reconstruction time | Fast | Fast | Moderate | Fast |
| Normalized sampling speed* | 1.20μL*min (39), 2.25μL*min (32), 2.92μL*min (48) | 1.03μL*min (32) | 1.30μL*min (39) | 0.90μL*min (48) |
| Product sequence | Yes | Yes | No | No |
| Demonstrated acceleration | ||||
| Parallel imaging | Yes | Yes | Yes | Yes |
| Compressed sensing | Yes | Yes | Yes | No |
| Deep learning | Yes | Yes | No | Yes |
Normalized sampling speed was calculated as a product of the voxel volume and the scan time for T2-weighted sequences, where smaller values indicate higher efficiencies.
Conventional MRI Acceleration Techniques
Conventional acceleration techniques may include parallel imaging (PI), compressed sensing (CS), or a combination of the two.14–17 In PI, k-space data are systematically undersampled and recovered subsequently (either in the image domain or the frequency domain) using spatial encoding patterns achieved from multiple phased-array coil elements. However, high PI accelerations can cause aliasing artifacts or noise amplification through increased g-factors, placing a fundamental threshold on maximum accelerations. On the other hand, CS relies on the inherent compressibility (or sparsity) of the image data in a transform domain (total variation, wavelet, etc) for accelerating acquisitions.15 Such sparsity, which leads to incoherent or random noise-like aliasing artifacts, can be exploited to recover the unaliased image using sparse or low-rank models. However, CS reconstructions require hand-crafted regularization parameters, which can either lead to either overregularized (smoothed) or inadequately unaliased images. Moreover, the nonlinear iterative CS algorithms generally require long computation times, which has limited the pervasiveness of this method.
Accelerating 2D-FSE MRI
PI, routinely available on all major MRI vendor platforms, is the most common image acceleration technique. PI can typically achieve modest accelerations factors (R) of R = 2 with common knee coils, while higher R = 3–4 may lead to image artifacts.18 CS has seen a limited role in accelerating 2D-FSE since optimal sparsifying transforms for enforcing incoherence are challenging to implement with undersampling only in one phase-encoding dimension.
Advances in deep learning and convolutional neural networks (CNNs) are providing additional methods for accelerating 2D-FSE acquisitions. Hammernik et al demonstrated a data-driven variational network (VN) for undersampled complex-reconstruction that embedded CS principles into a variational model.19 Such a VN network was trained with fully-sampled and undersampled images and their accompanying sensitivity maps, where all empirical free-parameters were learned in a fully data-driven manner using an unrolled gradient-descent scheme. This technique outperformed conventional reconstruction for diagnostic knee MRI 2D FSE sequences by enabling accelerations of R = 4 for both uniform and variable density sampling while maintaining diagnostic image quality. Example images obtained with prospective VN undersampling are shown in Fig. 1.
FIGURE 1:
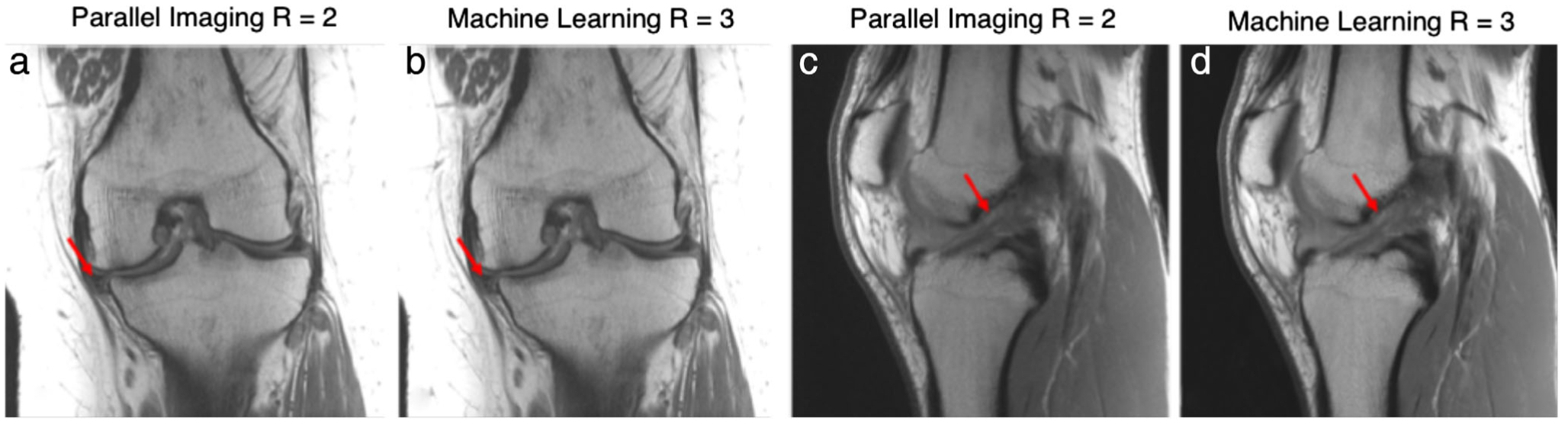
An example tear of the medial meniscus is seen with associated cartilage thinning and subchondral marrow changes imaged with a proton-density weighted coronal sequence (a,b). Similarly, an example anterior cruciate ligament (ACL) tear imaged with a sagittal proton-density-weighted sequence is also shown (c,d). Images were acquired on a 3T system (Skyra, Siemens) using a 15-channel knee coil. Two variational networks models were trained, one for coronal and one for sagittal acquisitions, each with 10 fully sampled cases. For testing, prospective accelerations were performed with parallel imaging (GRAPPA, R = 2) and the VN (R = 3). Images courtesy Dr. Florian Knoll, New York University, New York, NY.
Similarly, Liu et al demonstrated the concept of Sampling-Augmented Neural neTwork with Incoherent Structure (SANTIS) using a generative adversarial network (GAN).20,21 In this technique, a generator neural network reconstructs an image with sparse observations subject to specific undersampling operators. Additionally, a discriminator network evaluates whether the generator images resemble the ground-truth fully-sampled images and subsequently provides the generator this feedback through a separate loss function, known as adversarial loss. Such a paired generator-discriminator GAN approach has been shown to preserve perceptual image quality better than a generator network only.22 The SANTIS network demonstrated high-fidelity accelerations for R = 3 (Fig. 2); however, image sharpness was somewhat degraded for R = 5 acceleration.
FIGURE 2:
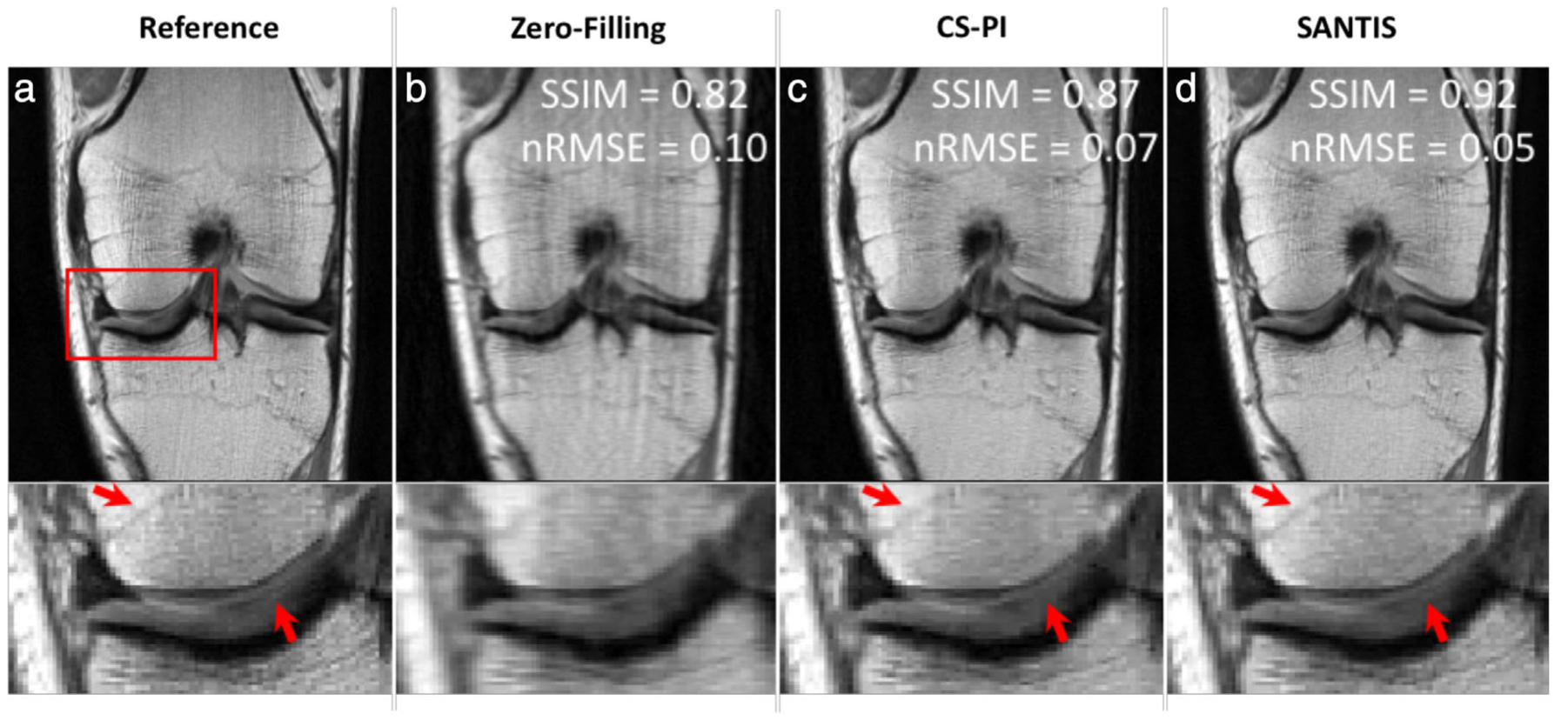
Representative examples of knee images (coronal proton-density weighted fast spin echo sequence, PD-FSE scanned on a GE 3T Premier) obtained using the different reconstruction methods at acceleration rate R = 3. Compared to the traditional method with a combination of compressed sensing and parallel imaging (CS-PI), the sampling-augmented neural network with incoherent structure (SANTIS) for MR image reconstruction provided improved removal of aliasing artifacts in the bone and cartilage and greater preservation of sharp tissue details. SANTIS reconstruction required 0.06 s/slice, compared to 2.2 s/slice for CS-PI. SSIM: structural similarity index; nRMSE: normalized root mean squared error. Images courtesy Dr. Fang Liu, University of Wisconsin, Madison, WI.
OPPORTUNITIES AND CHALLENGES.
Compared to traditional protocols with R = 2 accelerations, new promising reconstruction methods may provide additional 2-fold higher accelerations and have a high potential to capture internal derangement in the knee. These techniques could also be combined with novel acquisition strategies such as simultaneous multislice to further accelerate image acquisition.23 From a deep-learning standpoint, the large fully-sampled raw-data fastMRI dataset is an exciting resource to develop novel reconstruction methods.24 However, it is also important to simultaneously develop new techniques for evaluating the robustness of CNN reconstructions since, unlike PI or CS, it is challenging to evaluate network performance on phantoms since the algorithm is usually trained on in vivo datasets. Moreover, there is no convenient point spread function for interpreting the nonlinear network behavior outputs and generalizability. Thus, developing tools to understand CNN failure modes is essential prior to widespread implementation.
From 2D to 3D-FSE
Instead of multiple accelerated 2D-FSE sequences, there is a large interest in 3D sequences to acquire images with isotropic resolutions comparable to conventional 2D-FSE in-plane resolutions.25 3D steady-state imaging approaches have enabled rapid musculoskeletal MRI with a favorable contrast for cartilage.26,27 More recently, 3D-FSE sequences (common vendor names, GE [Milwaukee, WI]: CUBE; Siemens [Erlangen, Germany]: SPACE; Philips [Best, Netherlands]: VISTA) have also demonstrated promising diagnostic outcomes.28 However, long scan times and the resultant tradeoffs are the fundamental challenges of practical 3D-FSE acquisitions. Increasing the echo train length (ETL) of the acquisitions (~10 for 2D imaging vs. ~30+ for 3D imaging) can decrease scan time but leads to modulation of the k-space data due to T2 decay over the readout, which results in image blurring or ringing artifacts, depending on the view ordering.29 Thus, even 2D and 3D-FSE scans prescribed with identical matrix sizes may have different perceptual resolutions.
CS is a natural choice to accelerate 3D-FSE acquisitions due to the flexibility to choose varying sampling patterns to promote sparsifying requirements. Kijowski et al showed that intermediate-weighted 3D-FSE, both with and without CS, provided comparable SNR and diagnostic performance in assessing the cartilage, meniscus, and ligaments.30 Combining an R = 2 × 2 (phase and slice direction) PI approach with CS provided an additional 30% scan time reduction, resulting in 0.5 × 0.5 × 1.0 mm resolution scans in 3:16 minutes. Additionally, intermediate-weighted and T2-weighted 3D FSE sequences accelerated with CAIPIRINHA (controlled aliasing in parallel imaging results in higher acceleration) showed high image quality and high concordance with arthroscopic findings in a pediatric population.31–33 CAIPIRINHA enabled phase and slice direction undersampling (R = 2 × 2) to generate 0.5–0.6 mm isotropic resolution images in under 5 minutes, by optimizing the use of coil sensitivities, with example multiplanar images shown in Fig. 3.34
FIGURE 3:
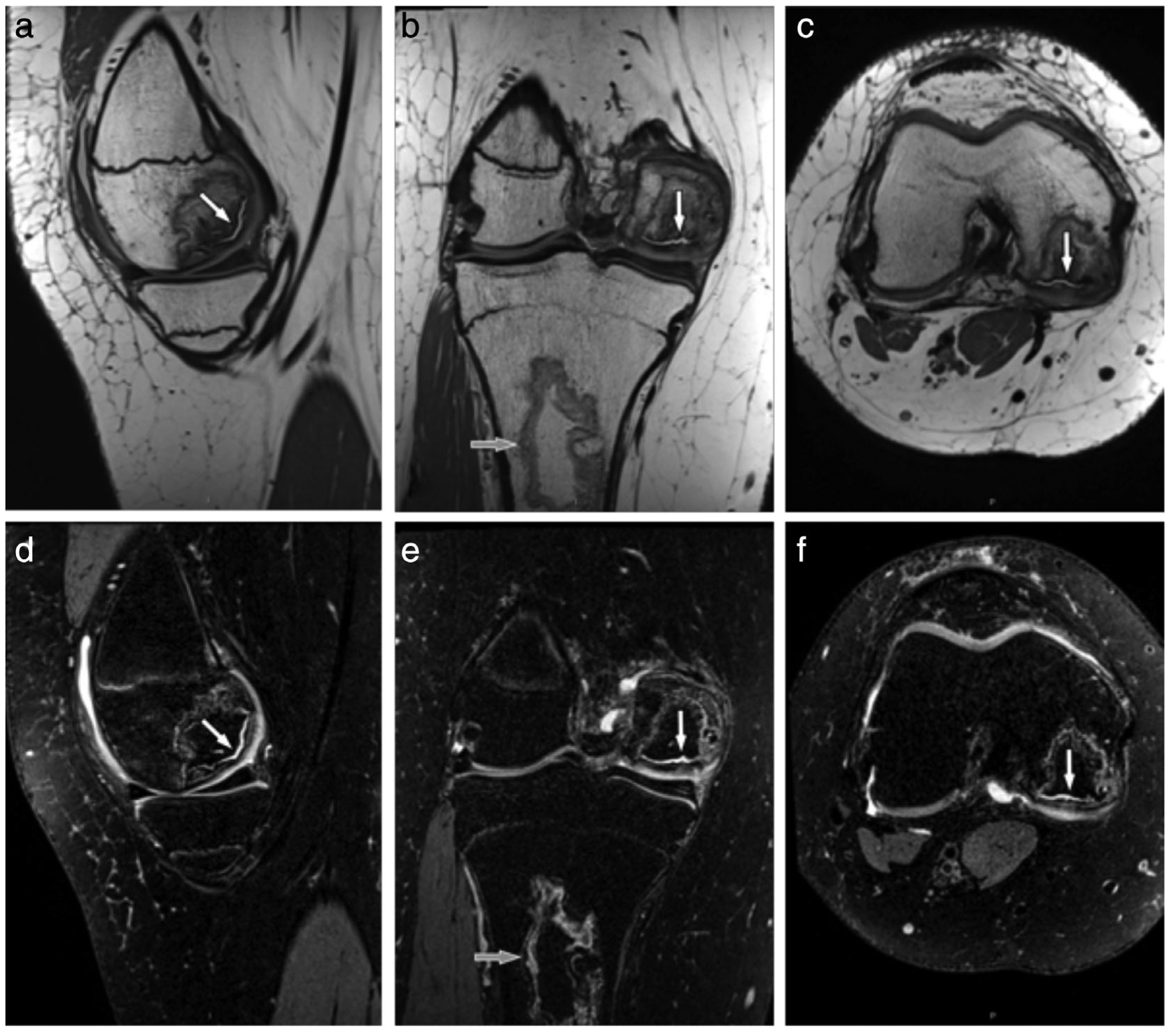
A 16-year-old female teenager with a history of leukemia and long-term glucocorticoid treatment. Isotropic sagittal intermediate-weighted (a) and fat-suppressed T2-weighted (d) CAIPIRINHA (controlled aliasing in parallel imaging results in higher acceleration) SPACE TSE MR images with coronal and axial intermediate-weighted (c,d) and fat-suppressed T2-weighted (e,f) reformation MR images show an osteonecrosis of the medial femoral condyle with separation of a large osteochondral fragment as indicated by intersecting joint fluid (white arrows). In addition, there is a medullary bone infarct (b,e; gray arrows) in the proximal tibial metaphysis. The intermediate-weighted 3D CAIPIRINHA SPACE TSE dataset was acquired with a 0.5 × 0.5 × 0.5 mm3 spatial resolution and a total acquisition time of 4 minute and 41 seconds. The fat-suppressed T2-weighted 3D CAIPIRINHA SPACE TSE dataset was acquired with a 0.6 × 0.6 × 0.6 mm3 spatial resolution and a total acquisition time of 4 minutes and 45 seconds. No interpolations techniques were used. Images courtesy Dr. Jan Fritz, Johns Hopkins University, Baltimore, MD.
Moreover, Mardani et al showed promising results for reconstructing 3D-FSE images (R = 3) using GANs to reconstruct undersampled images from a fully-sampled healthy dataset.35 Additional evaluations in a diagnostic setting and potential synergistic combinations with conventional acceleration approaches, may be beneficial to evaluate the diagnostic potential of such techniques.
OPPORTUNITIES AND CHALLENGES.
3D-FSE techniques have a high potential to mitigate several limitations of 2D-FSE imaging, but remain limited by ETL-induced blurring. Using novel variable flip-angle methods or investigating wave-encoded variable density single-shot FSE may present new methods to reduce image blurring.36,37 An additional limitation for the method is intrinsic to the acquisition, since only a single contrast is acquired per sequence. Evaluating the diagnostic utility of the image contrasts that is essential for specific abnormalities will be necessary to determine the overall diagnostic utility. Future acceleration techniques could also utilize the data acquisition advances along with deep-learning reconstructions for achieving the maximal image accelerations.
Novel 3D MRI Approaches
T2-Shuffling is a promising new extension of 3D FSE that randomly shuffles the ordering of the echo trains to mitigate blurring induced by long echo trains, especially for tissues with shorter-T2 relaxation times (<100 msec).38 It also resamples similar positions in k-space at multiple echo times (TE) that enables a 4D acquisition, which consists of a 3D images that are reconstructed with different TEs (Fig. 4). A 7-minute T2-Shuffling sequence (with 0.55 × 0.55 × 0.6 mm resolution) has been shown to have high concordance with conventional imaging for diagnostic pediatric knee MRI.39 This technique has shown promise for prospective targeted imaging protocols.40
FIGURE 4:
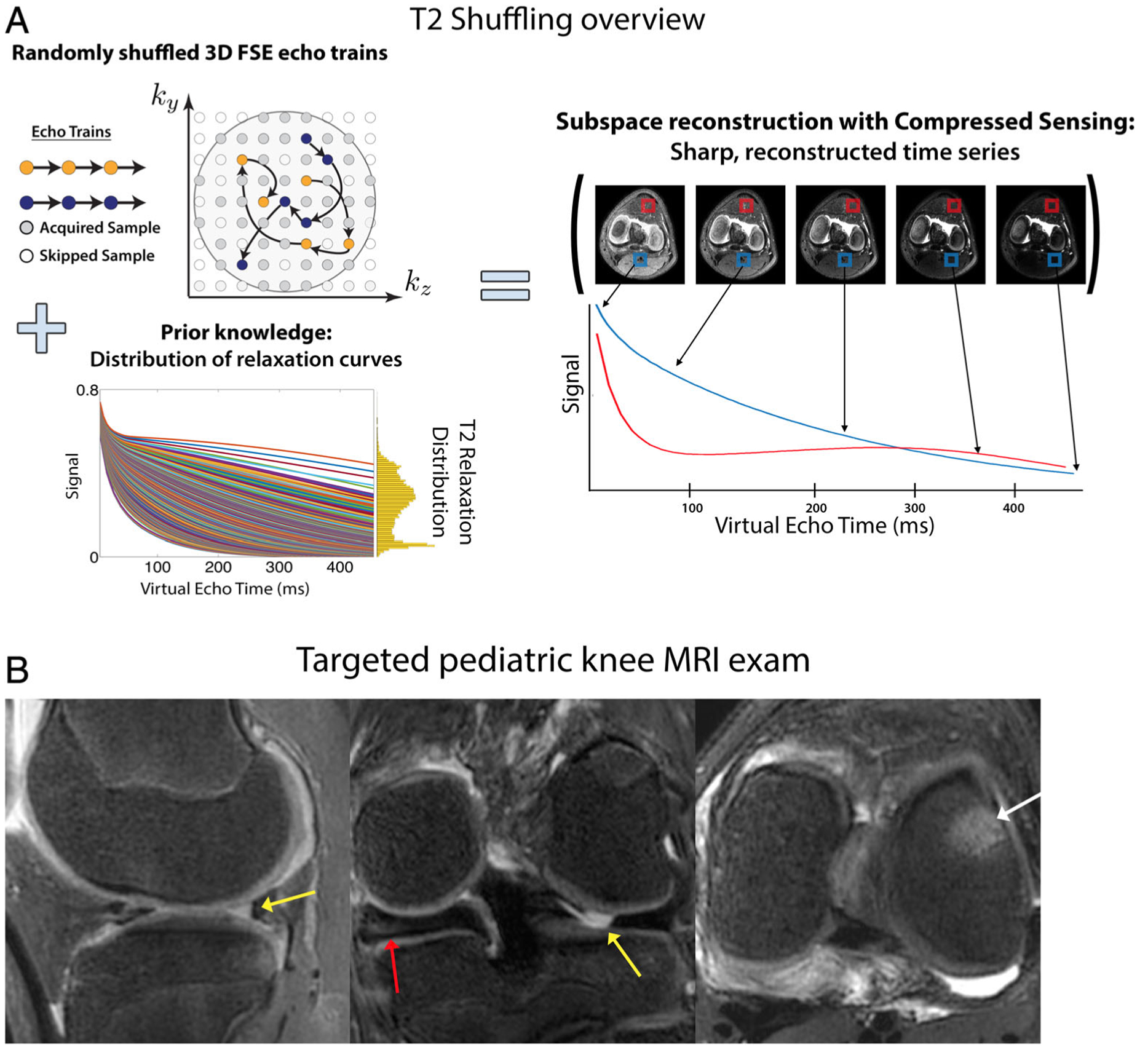
(a) An overview of the T2-Shuffling technique. A 3D FSE acquisition is modified to randomly shuffle the phase encode ordering of each echo train. Using prior knowledge of the signal relaxation curves for specific anatomy, a subspace-constraint incorporating compressed sensing was applied, leading to a 4D reconstruction along the signal relaxation curve. (b) T2-Shuffling images from a targeted pediatric knee MRI exam reformatted into (left) sagittal PD, (middle) coronal intermediate, and (right) axial T2 weighting. Clinical suspicion of lateral meniscal tear was confirmed with MRI (yellow arrows). Additional related findings were medial discoid meniscus (red arrow) and bone marrow edema (white arrow). Images courtesy Dr. Jon Tamir, University of California Berkeley, Berkeley, CA.
Similarly, the quantitative double-echo steady-state (qDESS) is an alternative approach for rapid 3D morphological imaging. An extension of balanced steady-state free precession, qDESS generates two echoes. The first qDESS echo (S+) is a gradient echo with T2/T1 weighting while the second echo (S−) is akin to a steady-state spin-echo formulated from previously spoiled echo pathways.41–43 Unlike the commercial DESS sequence used in the OAI, where the two echoes are combined using root sum-of-squares, qDESS echoes are kept separate. By analytically modeling the qDESS signal pathways using extended phase graphs (EPG),44 the separate echoes can be used for fluid nulling and to calculate quantitative T2 relaxation times (Fig. 5).45,46 A 5-minute qDESS sequence with 0.3 × 0.4 × 1.5 mm resolution has been shown to provide simultaneous T2-relaxation-time and morphometry estimates for cartilage and meniscus47 and has high concordance with conventional diagnostic 2D FSE for knee MRI.48 SNR-efficient extensions of qDESS that include 3D radial sampling with ultrashort TEs, with simultaneous fat and water separation, and with 0.5 mm isotropic resolution in 8 minutes (Fig. 6) also offer a promising approach for rapid morphological and compositional imaging.49
FIGURE 5:
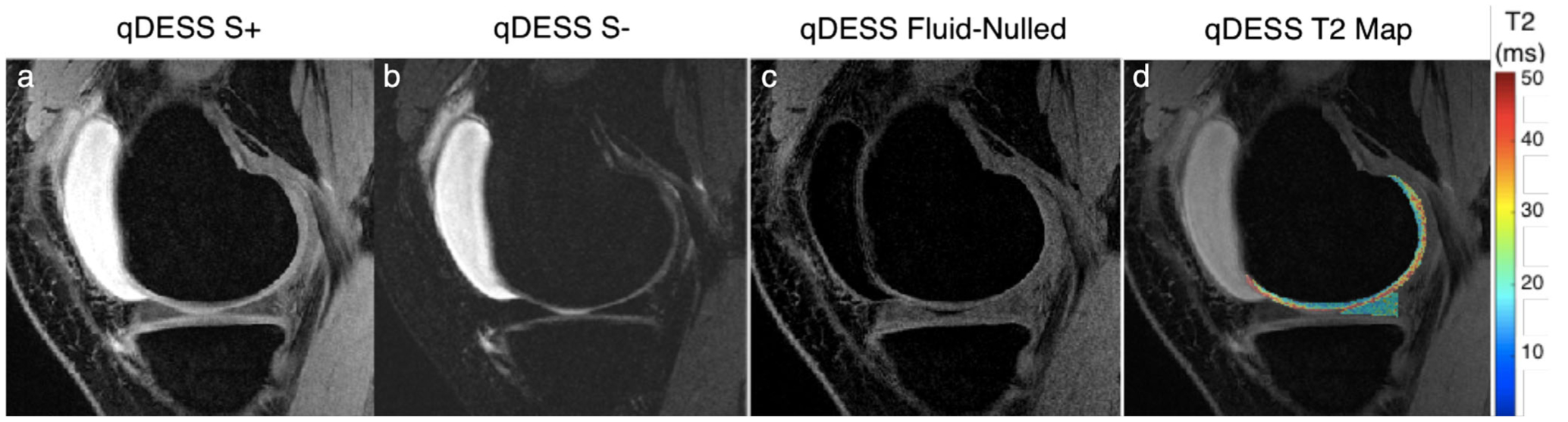
Representative images from a quantitative double-echo steady-state (qDESS) sequence that can generate two novel contrasts. The first echo (S+) generates a T2/T1-weighted contrast (a) while the second echo (S–) has a higher T2 weighting (b). By performing a weighted subtraction of the two echoes, fluid nulling can be performed to enhance visualization of soft tissues (c). Analytical modeling of the two echoes can also be used to accurately characterize the T2 relaxation times of the cartilage and meniscus (d).
FIGURE 6:
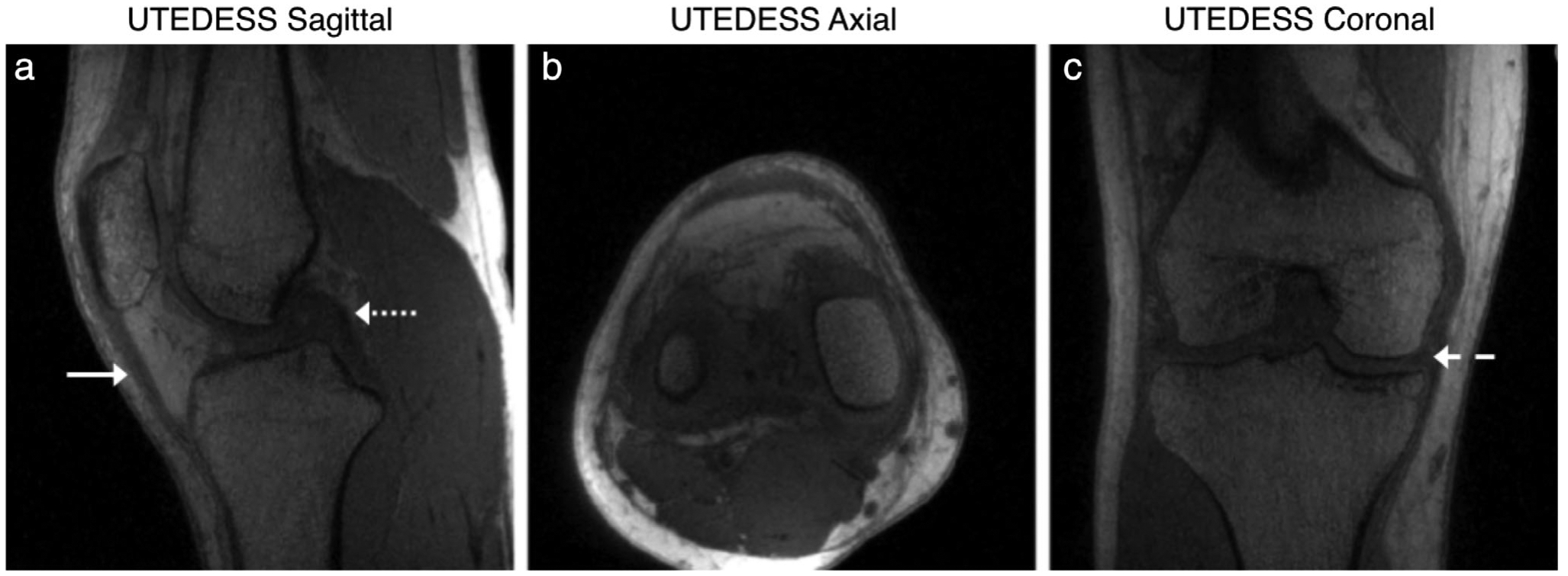
Example multiplanar images from 3D radial ultra-short echo time qDESS (UTEDESS) sequence that can generate high-isotropic resolution images with high SNR from short-T2 tissues such as the tendons (solid arrow), ligaments (dotted arrow), and the meniscus (dashed arrow).
OPPORTUNITIES AND CHALLENGES.
Both T2-Shuffling and qDESS have shown promising results in small cohorts; however, implementations across multiple sites and vendors will be necessary for pervasive adoption of these techniques. A primary challenge associated with T2-Shuffling is the computationally intensive reconstruction, even with advanced computational hardware. Reducing this duration will be essential for pervasive adoption. For qDESS, the primary challenges are unfamiliarity of contrasts and an underestimation of the size of bone marrow edema (BME).48 However, this could be addressed with abbreviated protocols where an accelerated 2D FSE sequence can be paired with qDESS to maintain familiarity with image contrasts and to garner the benefits of qDESS.48
Retrospective Image Quality Enhancement
Image super-resolution, the technique of enhancing image resolution using learned image priors, is an active area of research in computer vision. While super-resolution has had limited use for enhancing MRI previously, recent advances in deep learning (DL) have enabled considerable image quality enhancement with near real-time reconstructions.50,51 Such super-resolution CNNs have been used for improving the slice resolution of DESS MRI scans from the OAI 3-fold, without biasing quantitative image quality or qualitative radiologist image quality metrics52 (Fig. 7). That study also demonstrated that CNN super-resolution significantly outperformed Fourier interpolation (zero padding in k-space), which is commonly used directly on MRI scanners and in DICOM viewers. Additionally, these enhanced images also minimally biased subtle features such as osteophytes and cartilage.53 Such techniques may be attractive for replacing Fourier interpolations. Combined with other DL techniques such as image denoising, super-resolution may be used to enhance MRI scans independent of sequence and contrast type and may be applied to all the rapid techniques described above.
FIGURE 7:
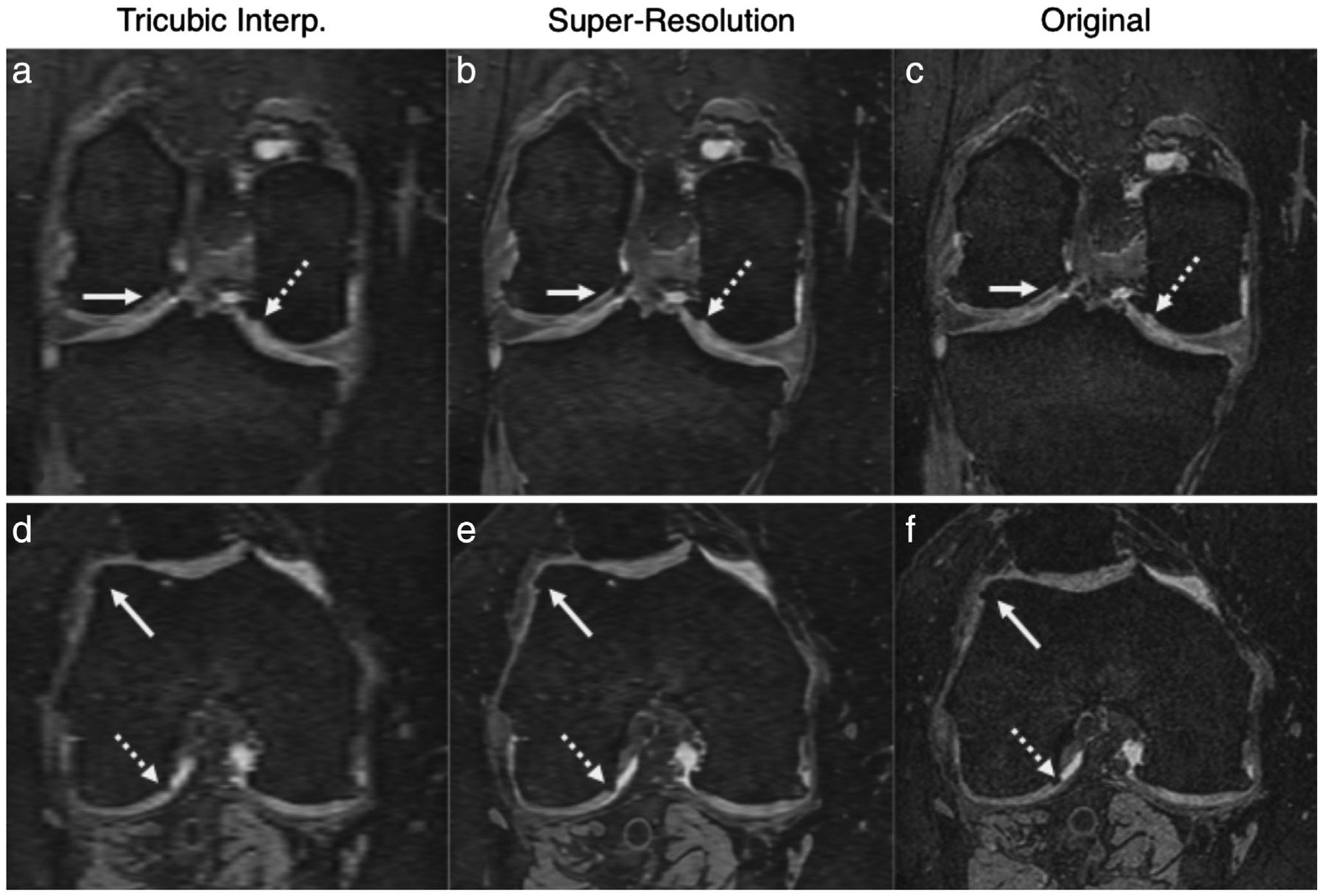
Example super-resolution images used for enhancing the slice resolution (left-right) of sagittal DESS sequences 3-fold. Comparisons between the tricubic interpolated images (a,d) show considerable blurring of the osteophytes and cartilage, as depicted by the arrows. Comparatively, the super-resolution images (b,e) show higher image sharpness and are more comparable to the original resolution images (c,f).
OPPORTUNITIES AND CHALLENGES.
DL methods such as super-resolution and reconstruction are promising for accelerating MRI acquisitions. While super-resolution provides an image-space interpretation of enhancing MRI resolution, techniques such as VN and SANTIS provide a k-space interpretation of resolution enhancement and antialiasing. Both approaches provide promising results and may be used synergistically; however, to date no head-to-head study has systematically compared the relative efficacy of the two approaches. Additionally, with many DL-based acceleration tasks, GANs tend to generate images with sharper image features, likely due to the use of nonlocal losses compared to traditional losses such as L1-norm or L2-norm. Instead, GANs try to parametrize probability distribution functions of plausible high-quality images synthesized from lower-quality inputs. While this may provide sharpness, it may also lead to image hallucinations—the existence of plausible image features incorrectly appearing in images. In the context of musculoskeletal MRI, where lesions in the cartilage and meniscus may only be a few pixels in size, such hallucinations are a large concern.
Moreover, most DL approaches have commonly been used with retrospectively undersampled data, which may not always account for image filtering following the acquisition of prospectively undersampled datasets, especially across the various MRI vendors. Evaluating the generalizability of such techniques on larger and clinically relevant datasets across vendors and field strengths will help identify optimal network architectures and loss functions. Overall, while DL approaches for accelerating image acquisition are promising, prospective use in clinical studies will ultimately define the true utility of the DL techniques.
Compositional (Quantitative) Imaging
Besides morphological imaging to assess tissue structure, MRI can generate compositional or quantitative measures that are representative of the biochemical tissue composition. Such techniques may provide indications of the early degenerative changes occurring in the tissue microstructure prior to the onset of irreversible morphological changes. Quantitative musculoskeletal MRI commonly assesses collagenous tissues, with a majority of techniques interrogating articular cartilage due to its central role in OA.54 In normal articular cartilage, water contributes ~80% of the wet weight, while collagen and proteoglycans contribute ~60% and 15% of dry weight, respectively.55 Compositional imaging techniques exploit that the tissue signal is impacted by and can be modulated using the hydration level and the collagen and proteoglycan microstructure. Despite promising results for depicting early degenerative changes, they are hindered by long acquisition durations due to accuracy constraints induced by low SNR. While the biochemical and histological properties of articular cartilage were described in detail previously,55,56 the following sections will focus on increased efficiency for acquiring a multitude of compositional markers for knee MRI.
T2 Relaxation Time
T2, or the transverse relaxation time constant, is widely used technique to assess collagen microstructure and hydration levels, with the hypothesis that tissue ultrastructure degradation leads to increased hydration in the collagen and meniscus, thereby leading to increased T2 times.57–60 2D spin-echo with multiple TEs is the most common technique for acquiring T2 relaxation times.61 The simplicity of the acquisition and the subsequent monoexponential fitting has made this sequence attractive, despite shortcomings such as stimulated echoes, imperfect refocusing radiofrequency (RF) pulses, and magnetization transfer effects.62–64 Moreover, the long acquisition durations required for sampling multiple TEs (usually 4–8 separate echoes) and the relatively coarse resolutions limit the full utility of this method.65 In addition to monoexponential fitting, biexponential T2 fitting models have also been explored to differentiate bound and free water in cartilage; however, these models require more complex fitting techniques and data with short TEs (and high SNR) to accurately quantify the rapidly decaying T2 component.66
The qDESS sequence can be used for accelerated and high-resolution T2 mapping.45,67 While the multiple TEs in traditional T2 measurement sequences are required to develop robustness to noise, diffusion, T1 relaxation, and off-resonance, these effects can explicitly be modeled in the highly SNR-efficient qDESS sequence using extended-phase-graphs (EPG) analysis.44,46 Matzat et al demonstrated that qDESS generates accurate T2 relaxation time measurements with high dynamic range and high concordance to single-echo spin-echo measurements.68 Cartesian qDESS has been shown to have accurate T2 measurements of the cartilage and meniscus, while 3D-radial ultrashort TE qDESS (UTEDESS) has been shown to produce T2 measurements of the tendons, ligaments, and meniscus.47,49 DL-based super-resolution can also enhance qDESS image quality without biasing quantitative T2 parameter maps.69 Compared to spin-echo-based T2 measurement methods, qDESS provides higher resolution, which may be used to investigate laminar differences in cartilage and menisci. However, with a dual-echo approach, qDESS may not have the sensitivity to generate multicomponent fits.
DL-based image reconstructions can also be used to accelerate parametric mapping. Liu et al demonstrated a model-augmented neural network with incoherent k-space sampling (MANTIS) method to generate T2 parameter maps from undersampled MRI data (Fig. 8).70 MANTIS utilizes a cyclic loss with two loss terms. One loss tries to ensure that the T2 parameter maps from the reconstruction produce synthetic undersampled k-space data consistent with the acquired k-space, while the other loss tries to ensure consistency between the undersampled and fully-sampled parameter maps. This method shows the potential for combining data-driven DL and MR physics signal modeling while utilizing the multiecho redundancies for ensuring additional data consistency.71
FIGURE 8:
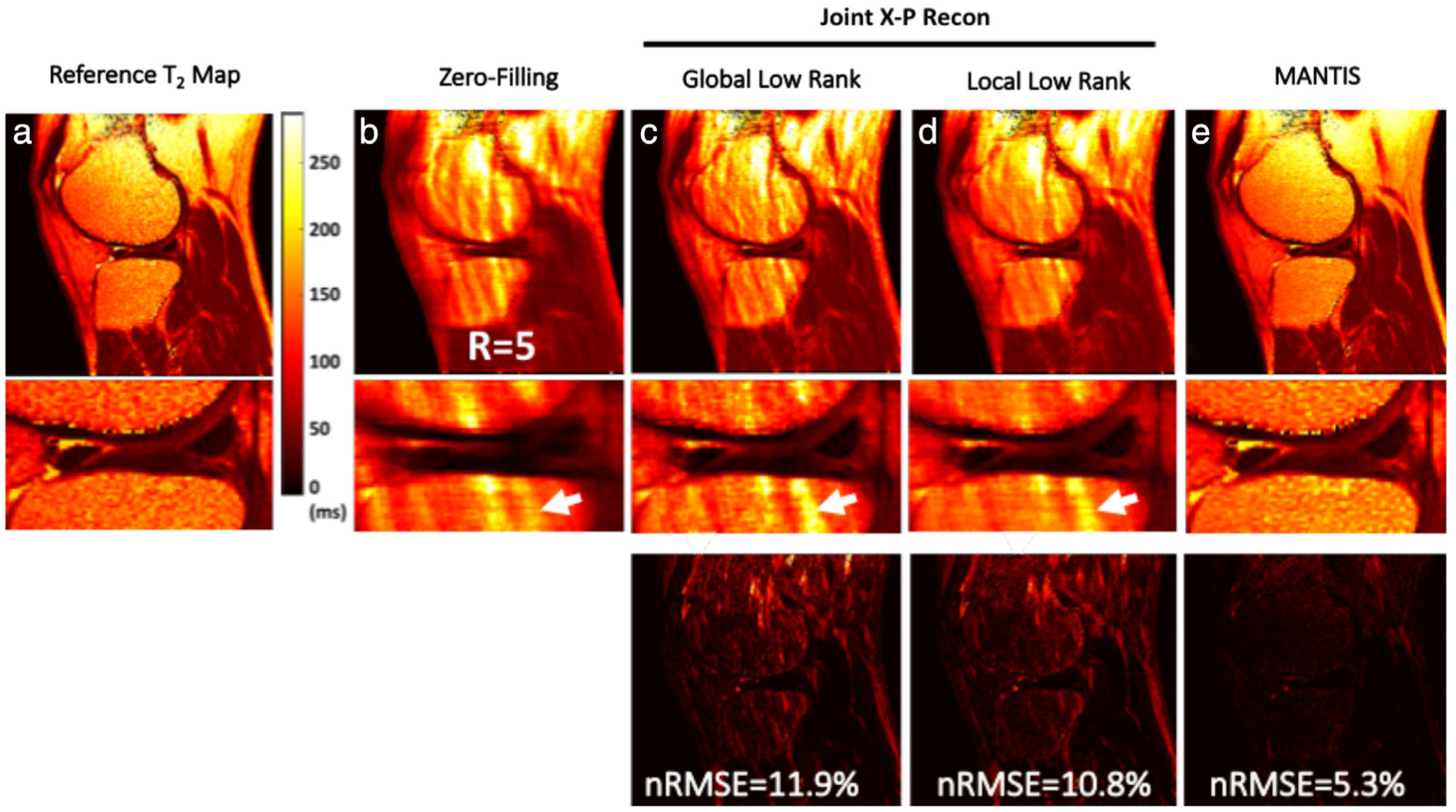
Representative T2 maps estimated from different reconstruction methods at R = 5 using an eight-echo spin-echo T2 mapping sequence (GE 3T Signa Excite Hdx), respectively, for a symptomatic patient. The global low rank (GLR) reconstruction generated images with noticeable artifacts in bone and fatty tissues (white arrows). Although the local low rank (LLR) reconstruction improved overall image quality with reduced image artifacts, it led to noticeable smoothing due to the exploitation of local sparsity at high acceleration. In contrast, model-augmented neural network with incoherent k-space sampling (MANTIS) generated nearly artifact-free T2 maps with well-preserved sharpness and texture comparable to the reference T2 maps. This qualitative observation was also confirmed using the residual error maps (displayed with the same scale) and nRMSE values. Images courtesy Dr. Fang Liu, University of Wisconsin, Madison, WI.
T1ρ Relaxation Times
T1ρ (or spin-lock) relaxation time studies spins relaxation in the influence of a constant RF field, which has shown sensitivity to cartilage proteoglycan depletion.72 In this method, spins are locked in the transverse plane with a constant RF following excitation. T1ρ relaxation times are calculated using exponential decay models of signal intensities when RF pulses with varying spin-lock durations are applied to the tissue of interest. Typically, T1ρ imaging is performed using a spin-lock preparatory pulse followed by a 3D segmented elliptic-centric spoiled gradient echo (SPGR) readout.73,74 Similar to parametric T2 relaxation time measurements, T1ρ efficiency is hindered since it requires 3D image volumes with varying spin-lock durations. Recently, T1ρ acquisitions have been accelerated using PI and CS (Fig. 9) without considerably affecting the quantitative T1ρ values.75,76 Such acquisitions have also been extended to provide biexponential models of T1ρ decay.77
FIGURE 9:
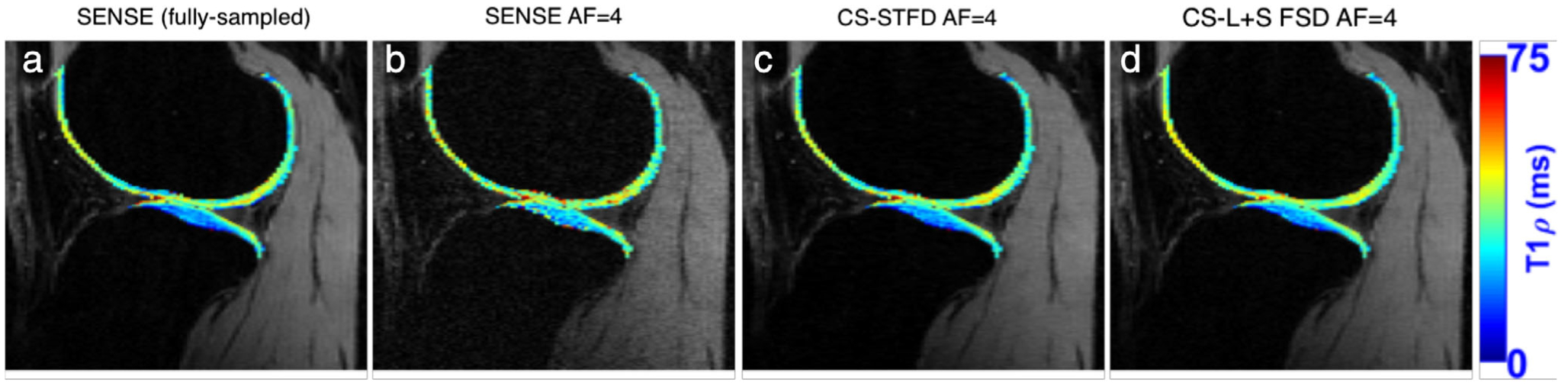
(a) A fully-sampled T1ρ map, with images reconstructed with SENSE (b) 4-fold accelerated T1ρ, with images reconstructed with SENSE (c) 4-fold accelerated T1ρ, with images reconstructed with CS and spatiotemporal finite difference (STFD) (d) 4-fold accelerated T1ρ, with images reconstructed with CS and low-rank plus sparse with spatial finite difference (L + S SFD). Images courtesy Dr. Marcelo Zibetti and Dr. Ravinder Regatte, New York University, New York NY.
Glycosaminoglycan Chemical Exchange Saturation Transfer (GagCEST)
Chemical exchange saturation transfer (CEST) is an MR contrast enhancement technique that enables the indirect detection of molecules with exchangeable protons. Glycosaminoglycans (GAGs) in cartilage exhibit a concentration-dependent CEST effect between their hydroxyl (−OH) and bulk water protons.78 CEST sensitizes MRI to GAG concentration and enables imaging GAG distributions with high spatial resolution.79 The high specificity to GAG without the need for special hardware or intravenous contrast makes GagCEST a promising method for studying cartilage matrix composition.80 However, the exchange rate and proximity of resonance frequencies between GAG hydroxyl protons and bulk water protons make efficient and specific saturation of GAG hydroxyl protons challenging at 3T.81 At 7T, studies have shown a high correlation between GagCEST and sodium (23Na) imaging, which is a direct measure of mapping the negatively charged GAG using positively charged 23Na, but suffers from poor resolution, long scan times, and the need for specialized hardware.80 New multislice and 3D GagCEST methods have demonstrated the potential of volumetric GagCEST analysis in clinically feasible scan times.82,83
Diffusion
Measurement of the diffusive properties of cartilage have been shown in limited studies, but with promising results.84,85 The random orientation of the proteoglycan molecules represents an isotropic restriction on water diffusivity, which can be probed using mean diffusivity, while highly organized collagen architecture of cartilage represents anisotropic water diffusion, which can be depicted using fractional anisotropy.84 Diffusion MRI is intrinsically limited by SNR, since increasing the diffusion sensitivity necessarily lowers tissue signal, which makes image acceleration challenging. However, including the diffusion signal model as a constraint in a model-based CS reconstruction can accelerate diffusion-weighted echo planar imaging (EPI) sequences.86 Moreover, the qDESS sequence has previously been extended to also calculate T2 and apparent diffusion coefficients simultaneously by using two sequences with varying spoiler gradients for imparting a variable T2 and diffusion weighting on the resultant images.67,87
OPPORTUNITIES AND CHALLENGES.
Novel techniques such as stimulated-echo readout-segmented EPI readouts and intrinsic SNR-based regularizers for the quantitative measurements could further accelerate compositional imaging techniques.87,88 Combining acquisition and modeling approaches from different sequences may also help parametric mapping. For example, qDESS could be extended to generate multiple TEs, as demonstrated with the 8-echo UTEDESS, in order to generate rapid multicomponent relaxation models. There also exist several DL methods to accelerate image acquisition; however, many of them have primarily been applied for morphological imaging.19,20,35 Beyond musculoskeletal imaging, DL image reconstruction methods have been utilized in undersampled neuroimaging and dynamic cardiac MRI.89–91 By combining these methods to overcome temporal redundancy and using parametric constraints for quantitative accuracy (such as those described as in MANTIS and SNR-regularization), there is potential for further accelerating parametric mapping.
Combining such DL-based reconstruction methods with traditional approaches such as parallel imaging and compressed sensing,75,92 dynamic view sharing,93 and novel 4D or multiecho acquisitions as in T2-shuffling or qDESS38,47 could enhance rapid quantitative knee MRI. These techniques could be extended to additional promising biomarkers that require multiecho curve-fitting such as T2* relaxation time mapping. Imaging with a 3D cones UTE trajectory has shown quantitative T2* sensitivity to meniscal degeneration and has been used for morphological imaging of the calcified cartilage endplate in the spine.94,95 Furthermore, to depict GAG changes, DL-based methods may allow for lower gadolinium dosage in delayed gadolinium-enhanced MRI of cartilage (dGEMRIC) or for DL-based denoising of sodium MRI.96–98 Advances in MR fingerprinting, primarily used in neuroimaging, may also provide a method to perform parametric mapping of various relaxation components in the knee.99
While accelerating parametric sequences is a necessary goal, it should not come at the expense of quantitative accuracy. Traditional acceleration approaches often reduce image SNR or introduce artifacts, which limits the utility of the quantitative parameters.14,100 Characterizing the dependency of parametric accuracy on SNR and choosing appropriate accelerations is essential to develop optimal quantitative knee MRI protocols. Furthermore, evaluating the impact of different hardware (field strengths, coils, vendors, etc) on parametric accuracy is vital to assess method robustness. Although it may be ideal to generate the exact same quantitative parameters across various hardware setups, depending on the study design, even just maintaining high method repeatability for longitudinal analyses on the same scanner would also be beneficial. Such a perspective was used in the multiyear OAI study.
Rapid Biomarker Extraction
Automated Tissue Segmentation
The development of rapid imaging techniques enables scanning larger population cohorts in shorter time periods. Such an increase in the extent of imaging data available necessitates novel techniques to parse through the images and to generate potential biomarkers for early OA onset and progression. For example, loss in medial femorotibial cartilage thickness over 24 months has been shown to be associated with radiographic and pain progression in the knee.101 Bone shape has also been associated with an increased risk of incident OA.102,103 Meniscal tears and extrusion have also been investigated as possible OA biomarkers.104 The processing pipeline of extracting these quantitative morphological measurements includes a mandatory step of image segmentation, often performed with manual or semiautomatic procedures. An accurate automatic algorithm for segmentation could establish a standardized practice for identifying morphological imaging biomarkers and efficient data analysis. Due to the lack of a fully-automated method, there is limited information on relationships between knee morphological features and OA risk factors such as age, gender, and BMI in larger populations and there is a pressing need to establish the prognostic capability of such imaging biomarkers.
DL advances have enabled automated, rapid, and accurate musculoskeletal tissue segmentation. Prasoon et al first used DL for segmentation by posing segmentation as a classification problem by utilizing a triplanar patch-based approach where three CNNs were trained to classify the central voxel of each patch as background or foreground (tibial cartilage).105 However, posing segmentation as a classification problem, as was historically done in image analysis via classical machine learning, does not intrinsically capitalize on the full potential of hierarchical convolutional feature extraction. In deep CNNs, each layer aggregates information at increasing levels of abstraction, starting with simple image elements, such as edges or contrast, to more complex and semantic aggregations, for uncovering latent patterns to accomplish pattern recognition tasks.106 The encouraging potential of CNNs is strictly related to their ability to aggregate local and global features, which is not fully optimized in approaches that leverage segmentation as a classification framework.
Shelhammer et al addressed the issue of incorporating spatial contextual cues by proposing a fully-convolutional architecture to output pixelwise predictions.107 A popular innovation in this fully-convolutional architecture that has produced accurate results is the “encoder-decoder” architecture. This architecture features a symmetric network that first learns image encoding by downsampling with convolutions with increasing feature maps and subsequently learns to decode the encoded image into a segmentation mask by upsampling with “transposed convolutions” (note that transpose convolutions are often incorrectly referred to as “deconvolutions”; however, this operation typically first upsamples the input followed by a simple convolution). Liu et al combined 2D SegNet, a popular encoder-decoder architecture, with deformable models to perform bone and cartilage segmentation to obtain performance superior to state-of-the-art image processing-based methods.108 SegNet uses an upsampling strategy using prestored upsampling indices in the downsampling max-pooling steps and offers an efficient alternative to transposed convolutions for recovering high-resolution image features. A similar approach was used by the same group to expand to additional knee tissues by using a fully connected 3D conditional random field to fine-tune the 2D segmentation results to account for 3D through-plane contextual relationships.109
Norman et al also used a encoder-decoder architecture, U-Net, to solve a multitissue knee segmentation problem.110 Unlike SegNet, the U-Net architecture features deep skip connections and transfers entire feature maps via transposed convolutions between the encoder an decoder, to obtain upsampled feature maps to combine local and global features (Fig. 10). The importance of skip connections is widely explored in biomedical image segmentation, and it has been shown how the expanding path can better recover spatial information by merging features skipped from the various resolution levels on the contracting path.111 This method demonstrated a state-of-the-art performance in cartilage and meniscus segmentation, with limited differences between the automated and manual approaches (Fig. 11). Additionally, that study evaluated segmentation performance in the context of the specific clinical and research applications. The accuracy and repeatability of cartilage morphometry features of volume and thickness and compositional markers of T1ρ and T2 were also reported, showing high machine-human concordance.
FIGURE 10:
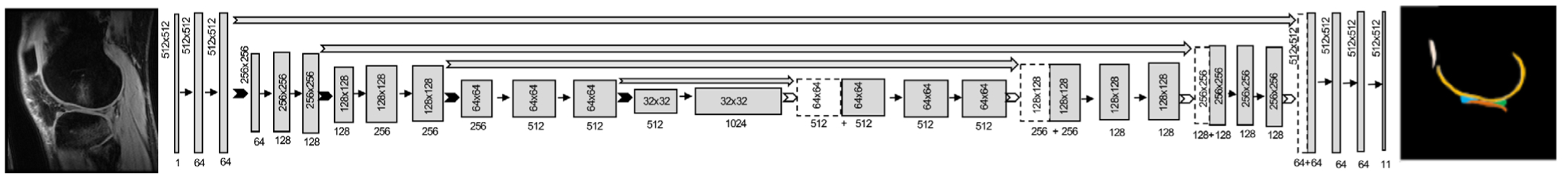
The architecture for the 2D U-Net encoder-decoder convolutional neural network. An input image with pixel dimensions of 512 × 512 is successively downsampled (dimensions in gray box) using 2 × 2 max pooling layers, but with an increasing number of feature maps (dimensions under gray box) in the decoder. In the decoder, the image is similarly upsampled using transposed convolutions to produce a segmentation mask with the same dimensions as the input image, with labels for multiple tissues in the knee.
FIGURE 11:
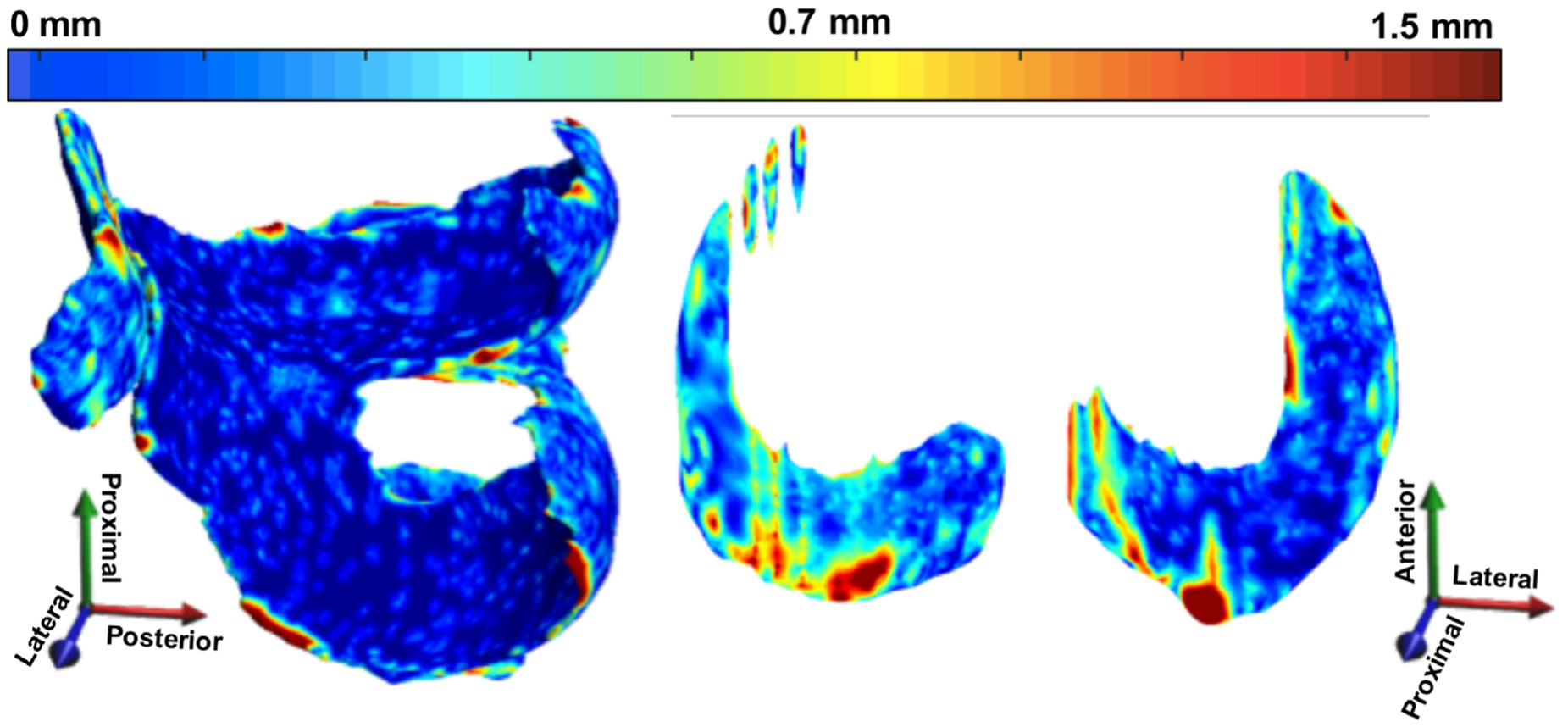
Example 3D point-by-point distance error maps (in mm) between manual and automatic segmentation for cartilage and meniscus using a DESS scan from the OAI that has a resolution of 0.34 × 0.45 × 0.7 mm3. The errors in the weight-bearing regions are minimal, with a majority of the errors lying mostly on the edge slices, which are on the order of the image resolution.
Image Interpretation
In addition to global features such as cartilage or meniscal morphometry derived from segmentation models, local morphological abnormalities have been widely studied as manifestations of joint degeneration. Semiquantitative scoring systems, such as the MRI Osteoarthritis Knee Score (MOAKS)10 and the Whole-Organ Magnetic Resonance Imaging Score (WORMS)112 have been used to grade local morphological abnormalities in subjects with OA. Such lesion severity scores have been compared with other findings such as meniscal defects, the presence of BMEs, as well as radiographic and clinical scores.113,114 These soft-tissue abnormalities have also been considered as possible predictors of total knee replacement surgery.115
The pervasive application of semiquantitative scoring is currently hampered by the time and level of expertise necessary to perform reliable grading, the lack of a true gold standard, and the intra- and interobserver variability. Automating or expediting morphological grading could enable a multitissue analysis of large cohorts in research studies such as the OAI and may potentially enhance the routine radiologist workflow and reduce costs in a clinical setting. Several promising applications of DL to automate classification of cartilage, meniscal, and ligament lesions have been proposed recently.116–119 Despite being a nascent technology, automated interpretation of tissue health using DL is summarized in considerable detail as the subject of several recent review articles.120,121
OPPORTUNITIES AND CHALLENGES.
All the above DL-based segmentation studies utilized each 2D slice as a separate input. Moving to 3D approaches may help increase network performance; however, current GPU hardware constraints either necessitate using shallower 3D models or using 3D image patches instead of the entire volumes. Decreasing network complexity and limiting large-scale contextual information may hinder the performance of such networks.122 DL approaches will also benefit from larger training corpuses, since it is hypothesized that a power law relationship exists between the size of the training data and the network performance.122,123
Gaining an ability to interpret the results from DL algorithms is essential to improve network performance and to guard against biases in algorithm outcomes or the curation of training datasets. Developing uncertainty metrics in addition to output predictions would be useful in this regard.124 Moreover, most current segmentation and classification algorithms are trained using training datasets compiled by single readers, which may also introduce reader-specific biases. Extending studies to generate probabilistic outcomes based on multiple readers would be essential to use CNN methods with confidence.
While external validation for images obtained from multiple sites and vendors will be necessary for ensuring generalizability, only a few trained models are available publicly. Desai et al proposed the open-source musculoskeletal analysis (DOSMA) tool (Fig. 12) capable of automated segmentation, registration, and quantitative fitting for multiple sequences, which may be a promising method to evaluate generalizability.125 Furthermore, borrowing lessons from traditional computer vision, a major factor in the success of DL for natural image classification is the presence of ImageNet—a large, standardized, and publicly-available dataset.126 Researchers globally can access ImageNet to train their models and to compare the performance of varying algorithms. Creating such a knee MRI dataset representative of varying biomarkers (morphological and compositional) combined with tools to share image data and corresponding labels, neural network architectures and weights, and overall results will be paramount to improve the accuracy of generalizable automated tools.
FIGURE 12:
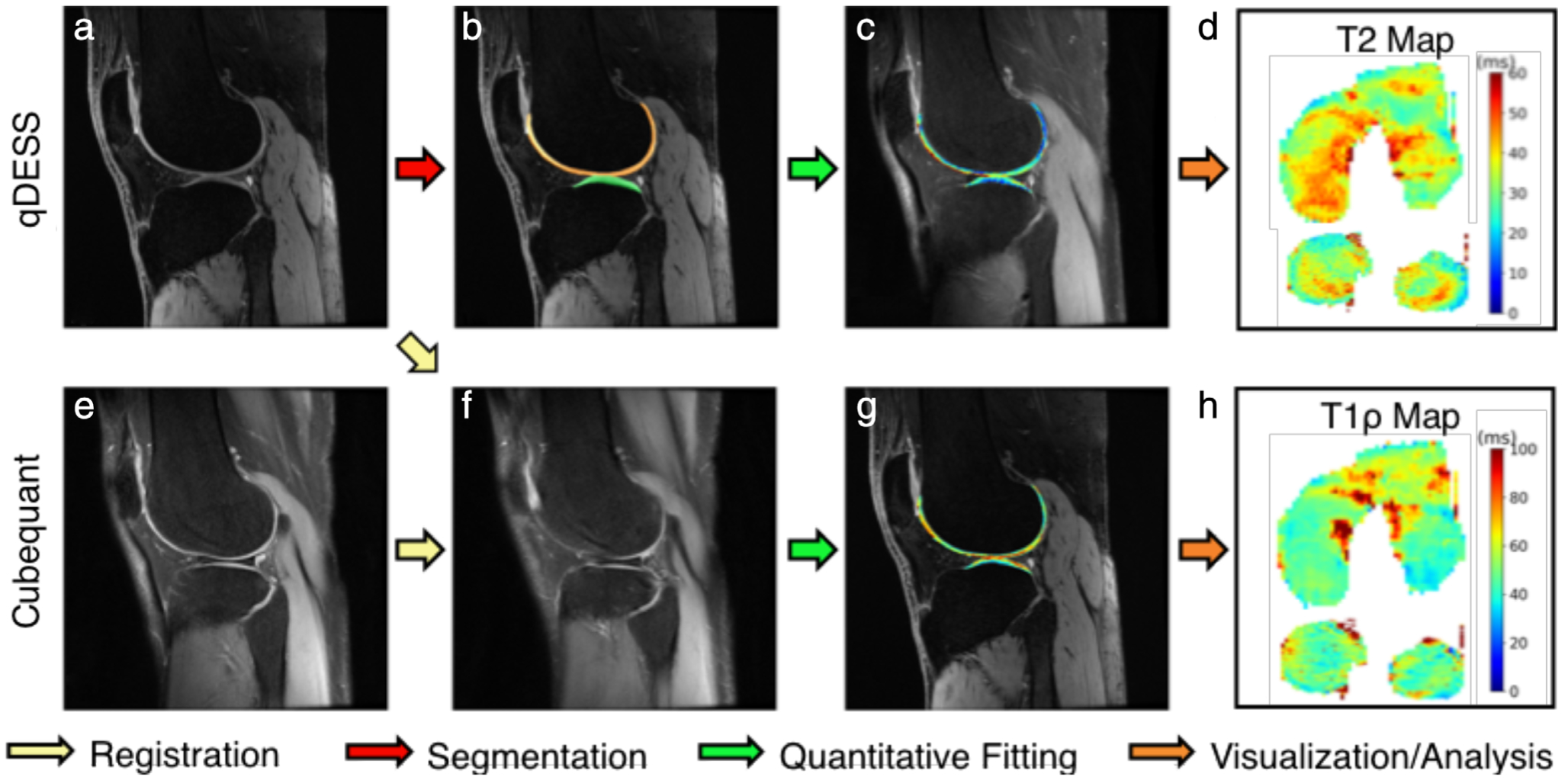
A sample analysis workflow using the fully-automated deep open source musculoskeletal analysis (DOSMA) framework. DOSMA automates tissue segmentation (red arrow), interscan registration (yellow arrow), quantitative fitting (green arrow), and visualization and region subdivision (orange arrow). In this example, qDESS scans (a) are used to automatically segment femoral and tibial cartilage (b) and analytically solve for T2 maps in each region (c,d). Corresponding CubeQuant scans for T1ρ relaxation time generation (e) are registered to the high-resolution qDESS scans (f) and used to fit T1ρ maps (g). Quantitative maps with 3D quantitative information are projected into 2D planes to visualize the femoral and tibial cartilage surfaces (d,h). Image courtesy Arjun Desai, Stanford University, Stanford, CA.
Hardware Improvements
MRI Hardware
Specialized RF coils offer the potential to greatly enhance SNR in musculoskeletal MRI, allowing for faster scanning, higher resolution, or improved diagnostic quality. Further, coils play a role in patient comfort and flexibility, which can be a determining factor in degraded image quality through patient motion. Phased-array coils that match the anatomy of interest have greatly improved MRI scanning by enabling parallel imaging while optimizing SNR.127 Current clinical hardware for extremity MRI is built around various anatomy of interest and utilizes between 4 and 18 coil-array elements. These coils mostly utilize a hard-shell design and often function to both transmit and receive RF signals, which can reduce heating. However, this design is often rigid, which necessitates a one-size-fits-all approach. Flexible coil-arrays offer the ability to more closely customize the coil fit to not only the anatomy of interest, but also specifically to each patient.128 By placing coil elements closer to the body, these coils increase their sensitivity to tissue signal. Further, the reduced size and increased flexibility in these coils can improve patient comfort and permit simultaneous use of multiple coil-arrays for high-resolution bilateral knee MRI.128
More recently, screen printed MRI coils129,130 and adaptive image receive (AIR) coils131 have been developed to further enhance the flexibility of coils to conform to multiple forms of anatomy while being inherently lightweight. In addition, new high-impedance detectors132 and stretchable RF conductors133 formed by liquid metal have been incorporated into wearable and even stretchable coils, which may not only optimize SNR for an anatomy of interest, but also optimize imaging during movement in order to assess tissue biomechanics.134 In combination with more efficient pulse sequences, improved hardware promises better access and flexibility for imaging bones and joints while improving scan times and patient comfort.
Bilateral Knee Imaging
OA is commonly a bilateral disease.135 While costs as well as long scan times have precluded separate scanning of both knees, simultaneous bilateral examinations can provide many benefits clinically, as well as in longitudinal studies.136 Clinically, the contralateral knee may serve as an internal control for evaluation of anatomy, joint injury, or sources of pain. In research exams, bilateral exams can provide an internal control of quantitative MRI values.137 Further, bilateral knee MRI can drastically reduce costs and patient motion, improve patient comfort and retention, and eliminate potential between scan sources of error in research studies that require scanning of both knees.
Flexible, phased-array coils offer the possibility to image both knees simultaneously, in similar scan times as unilateral exams, while retaining excellent image quality. These coil-arrays have minimal coil element coupling between two coil-arrays, allowing both knee volumes to be imaged without affecting image SNR compared to single knee scans, simply by exciting and encoding both sides.138 Such a bilateral coil-array setup utilizing two 16-channel flexible coil arrays can image both knees simultaneously (Fig. 13) in similar scan times to conventional unilateral knee scans, while maintaining image quality.128 Additionally, cartilage T2 relaxation times were maintained between single knee scans and simultaneous bilateral scans. This study was performed using commercially available coils and sequences demonstrating that these methods can already be utilized widely. While some challenges still remain for right–left acceleration of 2D sagittal scans and non-Cartesian k-space trajectories, new imaging methods using SMS139 and optimized trajectories140 along with the localized sensitivities of the individual coils may offer solutions,14,16,17 respectively.
FIGURE 13:
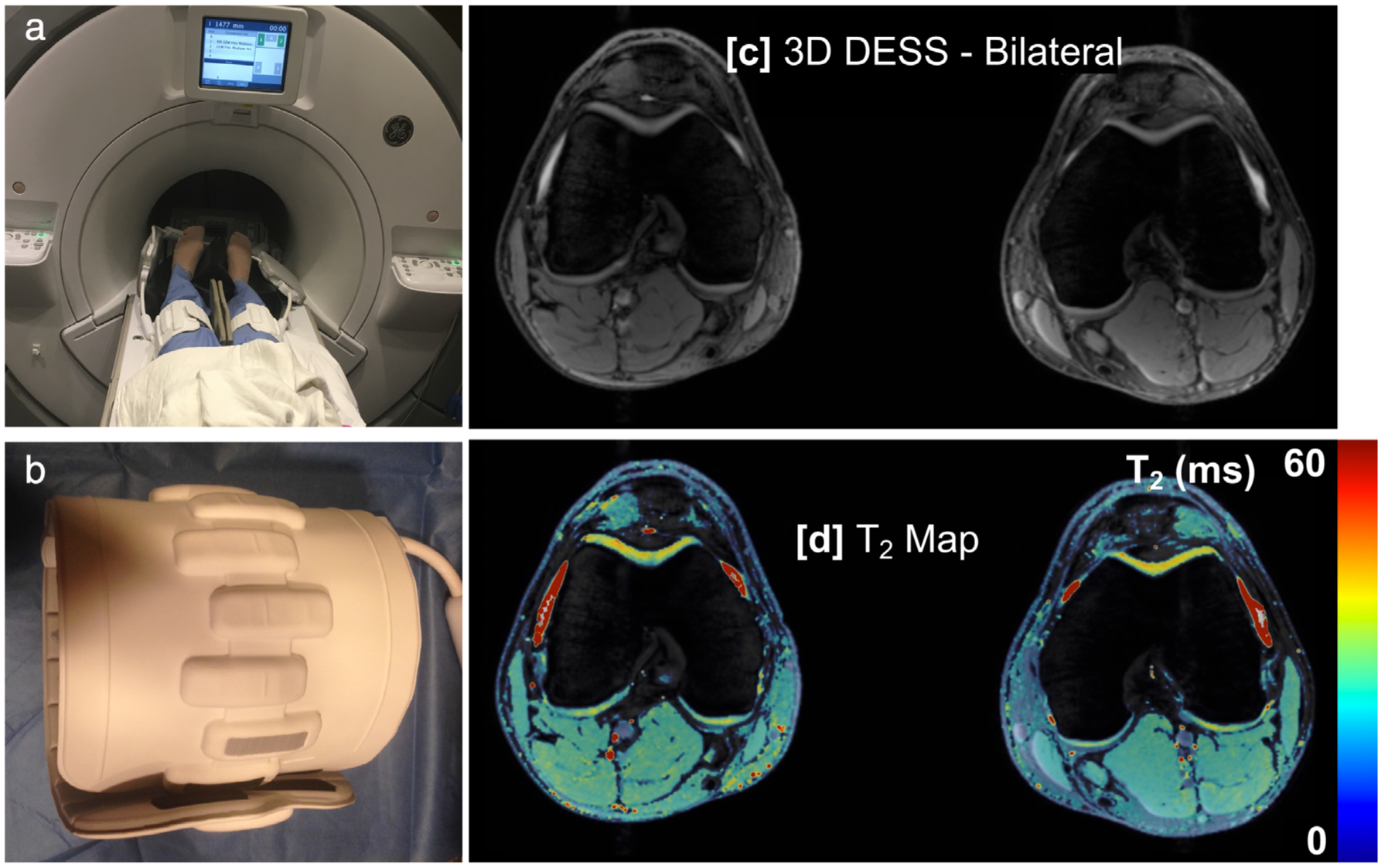
A simultaneous bilateral knee MRI setup (a) with two 16-channel flexible phased-array coils (b) that can be tightly wrapped around each knee. This allows for high-resolution imaging (c) and T2 relaxation time mapping (d) of both knees with similar scan times and SNR to single knee acquisitions, and has been shown to maintain quantitative accuracy.
Ultra-High Field MRI: 7T
Ultra-high field (UHF) imaging offers numerous opportunities for knee MRI and is one of the first FDA and CE approved applications for 7T clinical imaging. In particular, SNR increases linearly with field strength, allowing for a drastic increase in resolution while maintaining SNR at 7T compared with 3T and 1.5T.141 This high resolution may permit better visualization and characterization of anatomy and pathology of joint structures, many of which are thin, especially in advanced OA. Alternatively, the increased SNR can be utilized to decrease scan time by increasing parallel imaging factors and utilizing the increased SNR at 7T to overcome g-factor SNR losses.
OPPORTUNITIES AND CHALLENGES.
7T MRI may be utilized to increase SNR, and to reduce echo-train lengths in combination with parallel imaging, for reducing blurring in 3D-FSE sequences without added scan time. This may allow for isotropic acquisitions with clinical contrasts and subsequent multiplanar reformatting to reduce the time of research and clinical knee exams. 7T MRI also offers improved capabilities for quantitative imaging. In addition to the benefits of increased SNR for all quantitative imaging methods, including opportunities to reduce noise, increase resolution, and better visualize laminar differences, the increased chemical exchange shift at 7T offers an improvement in sensitivity and specificity to imaging proteoglycan changes that may precede structural changes. In GagCEST, this increased chemical shift offers more selective saturation of the GAG hydroxyl protons, resulting in reduced direct water saturation effects (more SNR) as well as increased saturation power for more efficient saturation of these exchanging protons.81,82 For T1ρ relaxation time mapping, the relaxation component from GAG hydroxyl protons increases quadratically with static field (B0), offering increased sensitivity and specificity to GAG at 7T.
While 7T offers many benefits, it also has many technical challenges that must be understood and overcome for widespread application. Inhomogeneities in both static magnetic field (B0) and applied magnetic field (B1) are magnified at 7T. In particular, wavelength effects in tissue at 7T cause variations in the effective transmit B1 field, resulting in large variations in SNR and image contrast (shading). Field shimming, particularly with higher-order shim fields, offer the most basic methods to improve field variations. Parallel transmit, which offers creating uniform B1 fields and can minimize RF deposition, offers considerable promise but requires careful attention to RF power deposition and currently lacks clinical approval.142 Additionally, power deposition increases quadratically with increases in magnetic field.143 As a result, tissue heating is a major concern at UHF. This is especially true of FSE diagnostic sequences. However, since deposited RF power is a function of tissue volume excited, this is a larger problem with large body areas such as the hips than smaller areas such as the knee when localized transmit coils are used.
Conclusion and Summary
In this article we reviewed recent advances for improving the efficiency of generating and analyzing morphological and compositional MRI. For image acquisition, T2-Shuffling and 3D-FSE may be promising for accelerated morphological imaging, especially if the echo-train blurring can be mitigated. Compositional imaging can be accelerated by incorporating relaxation models into novel DL-based image reconstruction methods. Quantitative measures such as T2 and T1ρ relaxometry present opportunities for use in large clinical trials due to demonstrated changes in OA and the potential for further acceleration. Sequences such as qDESS are promising for large-scale clinical trials, as they simultaneously produce morphological and compositional measures, and suitable segmentation contrasts in relatively short scan times. Advances in MRI hardware such as next-generation RF coils and UHF scanners will further accelerate image acquisition. Improvements in image analysis tools such as segmentation and classification will alleviate the burden of manual analysis and improve the repeatability of imaging studies. Together, such efficient imaging and analysis methods are essential to study large cohorts at varying stages of OA and with varying risk factors in order to improve our overall understanding of OA, particularly at an early stage of the disease where reversible changes may still be possible.
Acknowledgements
Contract grant sponsor: National Institutes of Health (NIH); Contract grant numbers: NIH R01 AR063643, R01 EB002524, K24 AR062068, R00 EB022634, P41 EB015891, K99 AR070902, R61 AR073552, P50 AR060752; Contract grant sponsor: GE Healthcare and Philips (research support).
Disclosures
A.C. has provided consulting services to Skope MR Inc., Subtle Medical, Culvert Engineering, and Image Analysis Group, has received honoraria from Chondrometrics GmBH, and is a shareholder of Subtle Medical, LVIS Corporation, and Brain Key. V.P. receives research support from GE Healthcare. S.M. receives research support from GE Healthcare and has provided consulting services for Smith Research, and receives royalties from the University of California Office of the President. G.G. receives research support from GE Healthcare and Philips. B.H. receive research support from GE Healthcare and Philips, and is a shareholder of LVIS Corporation. No commercial support has been accepted related to the development or publication of this activity.
References
- 1.Solomon DH, Katz JN, Carrino JA, et al. Trends in knee magnetic resonance imaging. Med Care 2003;41:687–692. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Jordan JM. Epidemiology of osteoarthritis. Clin Geriatr Med 2010;26:355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cross M, Smith E, Hoy D, et al. The global burden of hip and knee osteoarthritis: Estimates from the Global Burden of Disease 2010 study. Ann Rheum Dis 2014;73:1323–1330. [DOI] [PubMed] [Google Scholar]
- 4.Kellgren JH, Lawrence JS. Osteo-arthrosis and disk degeneration in an urban population. Ann Rheum Dis 1958;17:388–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guermazi A, Roemer FW, Burstein D, Hayashi D. Why radiography should no longer be considered a surrogate outcome measure for longitudinal assessment of cartilage in knee osteoarthritis. Arthritis Res Ther 2011;13:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sudlow C, Gallacher J, Allen N, et al. UK Biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller KL, Alfaro-Almagro F, Bangerter NK, et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci 2016;19:1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunter DJ, Altman RD, Cicuttini F, et al. OARSI clinical trials recommendations: Knee imaging in clinical trials in osteoarthritis. Osteoarthr Cartil 2015;23:698–715. [DOI] [PubMed] [Google Scholar]
- 9.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum 2012;64:1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter DJ, Guermazi A, Lo GH, et al. Evolution of semi-quantitative whole joint assessment of knee OA: MOAKS (MRI Osteoarthritis Knee Score). Osteoarthr Cartil 2011;19:990–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wirth W, Hunter DJ, Nevitt MC, et al. Predictive and concurrent validity of cartilage thickness change as a marker of knee osteoarthritis progression: Data from the Osteoarthritis Initiative. Osteoarthr Cartil 2017;25:2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bloecker K, Wirth W, Guermazi A, Hitzl W, Hunter DJ, Eckstein F. Longitudinal change in quantitative meniscus measurements in knee osteoarthritis—Data from the Osteoarthritis Initiative. Eur Radiol 2015; 25:2960–2968. [DOI] [PubMed] [Google Scholar]
- 13.Eckstein F, Kunz M, Schutzer M, et al. Two year longitudinal change and test-retest-precision of knee cartilage morphology in a pilot study for the osteoarthritis initiative. Osteoarthr Cartil 2007;15:1326–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: Sensitivity encoding for fast MRI. Magn Reson Med 1999;42:952–962. [PubMed] [Google Scholar]
- 15.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med 2007;58: 1182–1195. [DOI] [PubMed] [Google Scholar]
- 16.Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002;47:1202–1210. [DOI] [PubMed] [Google Scholar]
- 17.Sodickson DK, Manning WJ. Simultaneous acquisition of spatial harmonics (SMASH): Fast imaging with radiofrequency coil arrays. Magn Reson Med 1997;38:591–603. [DOI] [PubMed] [Google Scholar]
- 18.Garwood ER, Recht MP, White LM. Advanced imaging techniques in the knee: Benefits and limitations of new rapid acquisition strategies for routine knee MRI. Am J Roentgenol 2017;209:552–560. [DOI] [PubMed] [Google Scholar]
- 19.Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med 2018; 79:3055–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F, Samsonov A, Chen L, Kijowski R, Feng L. SANTIS: Sampling-augmented neural network with incoherent structure for MR image reconstruction. Magn Reson Med 2019;82:1890–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodfellow IJ, Pouget-Abadie J, Mirza M, et al. Generative adversarial networks. Adv Neural Inf Process Syst 2014;2672–2680. [Google Scholar]
- 22.Ledig C, Theis L, Huszar F, et al. Photo-realistic single image super-resolution using a generative adversarial network. In: 2017 IEEE Conf Comput Vis Pattern Recognit 2017;105–114. [Google Scholar]
- 23.Fritz J, Fritz B, Zhang J, et al. Simultaneous multislice accelerated turbo spin echo magnetic resonance imaging: Comparison and combination with in-plane parallel imaging acceleration for high-resolution magnetic resonance imaging of the knee. Invest Radiol 2017;52: 529–537. [DOI] [PubMed] [Google Scholar]
- 24.Zbontar J, Knoll F, Sriram A, et al. fastMRI: An open dataset and benchmarks for accelerated MRI. arXiv Prepr arXiv181108839 2018;1–29.
- 25.Gold GE, Busse RF, Beehler C, et al. Isotropic MRI of the knee with 3D fast spin-echo extended echo-train acquisition (XETA): Initial experience. AJR Am J Roentgenol 2007;188:1287–1293. [DOI] [PubMed] [Google Scholar]
- 26.Hargreaves BA, Gold GE, Lang PK, et al. MR imaging of articular cartilage using driven equilibrium. Magn Reson Med 1999;42:695–703. [DOI] [PubMed] [Google Scholar]
- 27.Kijowski R, Blankenbaker DG, Klaers JL, Shinki K, De Smet AA, Block WF. Vastly undersampled isotropic projection steady-state free precession imaging of the knee: Diagnostic performance compared with conventional MR. Radiology 2009;251:185–194. [DOI] [PubMed] [Google Scholar]
- 28.Kijowski R, Davis KW, Woods MA, et al. Knee joint: Comprehensive assessment with 3D isotropic resolution fast spin-echo MR imaging—diagnostic performance compared with that of conventional MR imaging at 3.0 T. Radiology 2009;252:486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Busse RF, Brau AC, Vu A, et al. Effects of refocusing flip angle modulation and view ordering in 3D fast spin echo. Magn Reson Med 2008; 60:640–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kijowski R, Rosas H, Samsonov A, King K, Peters R, Liu F. Knee imaging: Rapid three-dimensional fast spin-echo using compressed sensing. J Magn Reson Imaging 2017;45:1712–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fritz J, Ahlawat S, Fritz B, et al. 10-Min 3D turbo spin echo MRI of the knee in children: Arthroscopy-validated accuracy for the diagnosis of internal derangement. J Magn Reson Imaging 2019;49:e139–e151. [DOI] [PubMed] [Google Scholar]
- 32.Del Grande F, Delcogliano M, Guglielmi R, et al. Fully automated 10-minute 3D CAIPIRINHA SPACE TSE MRI of the knee in adults. Invest Radiol 2018;53:689–697. [DOI] [PubMed] [Google Scholar]
- 33.Fritz J, Fritz B, Thawait GG, Meyer H, Gilson WD, Raithel E. Three-dimensional CAIPIRINHA SPACE TSE for 5-minute high-resolution MRI of the knee. Invest Radiol 2016;51:609–617. [DOI] [PubMed] [Google Scholar]
- 34.Breuer FA, Blaimer M, Mueller MF, et al. Controlled aliasing in volumetric parallel imaging (2D CAIPIRINHA). Magn Reson Med 2006;55: 549–556. [DOI] [PubMed] [Google Scholar]
- 35.Mardani M, Gong E, Cheng JY, et al. Deep generative adversarial neural networks for compressive sensing MRI. IEEE Trans Med Imaging 2019;38:167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mugler JP, Epstein FH, Brookeman JR. Shaping the signal response during the approach to steady state in three-dimensional magnetization-prepared rapid gradient-echo imaging using variable flip angles. Magn Reson Med 1992;28:165–185. [DOI] [PubMed] [Google Scholar]
- 37.Bilgic B, Gagoski BA, Cauley SF, et al. Wave-CAIPI for highly accelerated 3D imaging. Magn Reson Med 2015;73:2152–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamir JI, Uecker M, Chen W, et al. T2 shuffling: Sharp, multicontrast, volumetric fast spin-echo imaging. Magn Reson Med 2017;77: 180–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bao S, Tamir JI, Young JL, et al. Fast comprehensive single-sequence four-dimensional pediatric knee MRI with T 2 shuffling. J Magn Reson Imaging 2017;45:1700–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamir JI, Taviani V, Alley MT, et al. Targeted rapid knee MRI exam using T2 shuffling. J Magn Reson Imaging 2019;49:e195–e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruder H, Fischer H, Graumann R, Deimling M. A new steady-state imaging sequence for simultaneous acquisition of two MR images with clearly different contrasts. Magn Reson Med 1988;7:35–42. [DOI] [PubMed] [Google Scholar]
- 42.Redpath TW, Jones RA. FADE—A new fast imaging sequence. Magn Reson Med 1988;6:224–234. [DOI] [PubMed] [Google Scholar]
- 43.Lee SY, Cho ZH. Fast SSFP gradient echo sequence for simultaneous acquisitions of FID and echo signals. Magn Reson Med 1988;8: 142–150. [DOI] [PubMed] [Google Scholar]
- 44.Weigel M Extended phase graphs: Dephasing, RF pulses, and echoes — pure and simple. J Magn Reson Imaging 2015;41:266–295. [DOI] [PubMed] [Google Scholar]
- 45.Welsch GH, Scheffler K, Mamisch TC, et al. Rapid estimation of cartilage T2 based on double echo at steady state (DESS) with 3 Tesla. Magn Reson Med 2009;62:544–549. [DOI] [PubMed] [Google Scholar]
- 46.Sveinsson B, Chaudhari A, Gold G, Hargreaves B. A simple analytic method for estimating T2 in the knee from DESS. Magn Reson Imaging 2017;38:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaudhari AS, Black MS, Eijgenraam S, et al. Five-minute knee MRI for simultaneous morphometry and T 2 relaxometry of cartilage and meniscus and for semiquantitative radiological assessment using double-echo in steady-state at 3T. J Magn Reson Imaging 2018;47: 1328–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaudhari AS, Stevens KJ, Sveinsson B, et al. Combined 5-minute double-echo in steady-state with separated echoes and 2-minute proton-density-weighted 2D FSE sequence for comprehensive whole-joint knee MRI assessment. J Magn Reson Imaging 2018;1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaudhari AS, Sveinsson B, Moran CJ, et al. Imaging and T2 relaxometry of short-T2 connective tissues in the knee using ultrashort echo-time double-echo steady-state (UTEDESS). Magn Reson Med 2017;78:2136–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Reeth E, Tham IIWK, Tan CH, Poh CL, van Reeth E, Tham IIWK. Super-resolution in magnetic resonance imaging: A review. Concepts Magn Reson Part A 2012;40:306–325. [Google Scholar]
- 51.Wang Y-HH, Qiao J, Li J-BB, Fu P, Chu S-CC, Roddick JF. Sparse representation-based MRI super-resolution reconstruction. Meas J Int Meas Confed 2014;47:946–953. [Google Scholar]
- 52.Chaudhari AS, Fang Z, Kogan F, et al. Super-resolution musculoskeletal MRI using deep learning. Magn Reson Med 2018;80:2139–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaudhari AS, Stevens KJ, Wood JP, et al. Utility of deep learning super-resolution in the context of osteoarthritis MRI biomarkers. J Magn Reson Imaging 2019. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baum T, Joseph GB, Karampinos DC, Jungmann PM, Link TM, Bauer JS. Cartilage and meniscal T2 relaxation time as non-invasive biomarker for knee osteoarthritis and cartilage repair procedures. Osteoarthritis Cartilage 2013;21:1474–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sophia Fox AJ, Bedi A, Rodeo SA. The basic science of articular cartilage: Structure, composition, and function. Sports Health 2009;1: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Link TM, Neumann J, Li X. Prestructural cartilage assessment using MRI. J Magn Reson Imaging 2017;45:949–965. [DOI] [PubMed] [Google Scholar]
- 57.Dardzinski BJ, Mosher TJ, Li S, Van Slyke MA, Smith MB. Spatial variation of T2 in human articular cartilage. Radiology 1997;205:546–550. [DOI] [PubMed] [Google Scholar]
- 58.Mosher TJ, Dardzinski BJ, Smith MB. Human articular cartilage: Influence of aging and early symptomatic degeneration on the spatial variation of T2—Preliminary findings at 3 T. Radiology 2000;214:259–266. [DOI] [PubMed] [Google Scholar]
- 59.David-Vaudey E, Ghosh S, Ries M, Majumdar S. T2 relaxation time measurements in osteoarthritis. Magn Reson Imaging 2004;22: 673–682. [DOI] [PubMed] [Google Scholar]
- 60.Nissi MJ, Töyräs J, Laasanen MS, et al. Proteoglycan and collagen sensitive MRI evaluation of normal and degenerated articular cartilage. J Orthop Res 2004;22:557–564. [DOI] [PubMed] [Google Scholar]
- 61.Mosher TJ, Dardzinski BJ. Cartilage MRI T2 relaxation time mapping: Overview and applications. Semin Musculoskelet Radiol 2004;8: 355–368. [DOI] [PubMed] [Google Scholar]
- 62.Maier CF, Tan SG, Hariharan H, Potter HG. T2 quantitation of articular cartilage at 1.5 T. J Magn Reson Imaging 2003;17:358–364. [DOI] [PubMed] [Google Scholar]
- 63.Smith HE, Mosher TJ, Dardzinski BJ, et al. Spatial variation in cartilage T2 of the knee. J Magn Reson Imaging 2001;14:50–55. [DOI] [PubMed] [Google Scholar]
- 64.Watanabe A, Boesch C, Obata T, Anderson SE. Effect of multislice acquisition on T1 and T2 measurements of articular cartilage at 3T. J Magn Reson Imaging 2007;26:109–117. [DOI] [PubMed] [Google Scholar]
- 65.Peterfy CG, Schneider E, Nevitt M. The osteoarthritis initiative: Report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthr Cartil 2008;16:1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reiter DA, Lin PC, Fishbein KW, Spencer RG. Multicomponent T 2 relaxation analysis in cartilage. Magn Reson Med 2009;61:803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Staroswiecki E, Granlund KL, Alley MT, Gold GE, Hargreaves BA. Simultaneous estimation of T2 and apparent diffusion coefficient in human articular cartilage in vivo with a modified three-dimensional double echo steady state (DESS) sequence at 3 T. Magn Reson Med 2012;67:1086–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matzat SJ, McWalter EJ, Kogan F, Chen W, Gold GE. T2 relaxation time quantitation differs between pulse sequences in articular cartilage. J Magn Reson Imaging 2014;37:105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaudhari A, Fang Z, Lee JH, Gold G, Hargreaves B. Deep learning super-resolution enables rapid simultaneous morphological and quantitative magnetic resonance imaging. In: Int Work Mach Learn Med Image Reconstr 2018;3–11. [Google Scholar]
- 70.Liu F, Feng L, Kijowski R. MANTIS: Model-Augmented Neural neTwork with Incoherent k-space Sampling for efficient MR parameter mapping. Magn Reson Med 2019;82:174–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cai C, Wang C, Zeng Y, et al. Single-shot T 2 mapping using overlapping-echo detachment planar imaging and a deep convolutional neural network. Magn Reson Med 2018;80:2202–2214. [DOI] [PubMed] [Google Scholar]
- 72.Duvvuri U, Reddy R, Patel SD, Kaufman JH, Kneeland JB, Leigh JS. T (1ρ)-relaxation in articular cartilage: Effects of enzymatic degradation. Magn Reson Med 1997;38:863–867. [DOI] [PubMed] [Google Scholar]
- 73.Zuo J, Li X, Banerjee S, Han E, Majumdar S. Parallel imaging of knee cartilage at 3 Tesla. J Magn Reson Imaging 2007;26:1001–1009. [DOI] [PubMed] [Google Scholar]
- 74.Witschey WRT, Borthakur A, Elliott MA, et al. T1ρ-prepared balanced gradient echo for rapid 3D T1ρ MRI. J Magn Reson Imaging 2008;28: 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zibetti MVW, Sharafi A, Otazo R, Regatte RR. Accelerating 3D-T 1ρ mapping of cartilage using compressed sensing with different sparse and low rank models. Magn Reson Med 2018;80:1475–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pandit P, Rivoire J, King K, Li X. Accelerated T1ρ acquisition for knee cartilage quantification using compressed sensing and data-driven parallel imaging: A feasibility study. Magn Reson Med 2016;75: 1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharafi A, Xia D, Chang G, Regatte RR. Biexponential T1ρ relaxation mapping of human knee cartilage in vivo at 3 T. NMR Biomed 2017; 30:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ling W, Regatte RR, Navon G, Jerschow A. Assessment of glycosaminoglycan concentration in vivo by chemical exchange-dependent saturation transfer (gagCEST). Proc Natl Acad Sci U S A 2008;105: 2266–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kogan F, Hariharan H, Reddy R. Chemical exchange saturation transfer (CEST) imaging: Description of technique and potential clinical applications. Curr Radiol Rep 2013;1:102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schmitt B, Zbýn S, Stelzeneder D, et al. Cartilage quality assessment by using glycosaminoglycan chemical exchange saturation transfer and (23)Na MR imaging at 7 T. Radiology 2011;260:257–264. [DOI] [PubMed] [Google Scholar]
- 81.Singh A, Haris M, Cai K, et al. Chemical exchange saturation transfer magnetic resonance imaging of human knee cartilage at 3 T and 7 T. Magn Reson Med 2012;68:588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kogan F, Hargreaves BA, Gold GE. Volumetric multislice gagCEST imaging of articular cartilage: Optimization and comparison with T1ρ. Magn Reson Med 2016;77:1134–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krishnamoorthy G, Nanga RPR, Bagga P, Hariharan H, Reddy R. High quality three-dimensional gagCEST imaging of in vivo human knee cartilage at 7 Tesla. Magn Reson Med 2017;77:1866–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raya JG, Melkus G, Adam-Neumair S, et al. Diffusion-tensor imaging of human articular cartilage specimens with early signs of cartilage damage. Radiology 2013;266:831–841. [DOI] [PubMed] [Google Scholar]
- 85.Raya JG, Horng A, Dietrich O, et al. Articular cartilage: in vivo diffusion-tensor imaging. Radiology 2012;262:550–559. [DOI] [PubMed] [Google Scholar]
- 86.Knoll F, Raya JG, Halloran RO, et al. A model-based reconstruction for undersampled radial spin-echo DTI with variational penalties on the diffusion tensor. NMR Biomed 2015;28:353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sveinsson B, Gold GE, Hargreaves BA, Yoon D. SNR-weighted regularization of ADC estimates from double-echo in steady-state (DESS). Magn Reson Med 2019;81:711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wengler K, Fukuda T, Komatsu DE, et al. In vivo evaluation of human patellar tendon microstructure and microcirculation with diffusion MRI. J Magn Reson Imaging 2019;49:1–11. [DOI] [PubMed] [Google Scholar]
- 89.Gibbons EK, Hodgson KK, Chaudhari AS, et al. Simultaneous NODDI and GFA parameter map generation from subsampled q-space imaging using deep learning. Magn Reson Med 2019;81:2399–2411. [DOI] [PubMed] [Google Scholar]
- 90.Schlemper J, Caballero J, Hajnal JV, Price AN, Rueckert D. A deep cascade of convolutional neural networks for dynamic MR image reconstruction. IEEE Trans Med Imaging 2018;37:491–503. [DOI] [PubMed] [Google Scholar]
- 91.Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS. Image reconstruction by domain-transform manifold learning. Nature 2018;555:487–492. [DOI] [PubMed] [Google Scholar]
- 92.Blaimer M, Breuer F, Mueller M, Heidemann RM, Griswold MA, Jakob PM. SMASH, SENSE, PILS, GRAPPA: How to choose the optimal method. Top Magn Reson Imaging 2004;15:223–236. [DOI] [PubMed] [Google Scholar]
- 93.Saranathan M, Rettmann DW, Hargreaves BA, Clarke SE, Vasanawala SS. DIfferential Subsampling with Cartesian Ordering (DISCO): A high spatio-temporal resolution Dixon imaging sequence for multiphasic contrast enhanced abdominal imaging. J Magn Reson Imaging 2012;35:1484–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Williams A, Qian Y, Golla S, Chu CR. UTE-T2* mapping detects sub-clinical meniscus injury after anterior cruciate ligament tear. Osteoarthr Cartil 2012;20:486–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim YJ, Cha JG, Shin YS, et al. 3D ultrashort TE MRI for evaluation of cartilaginous endplate of cervical disk in vivo: Feasibility and correlation with disk degeneration in T2-weighted spin-echo sequence. AJR Am J Roentgenol 2018;210:1131–1140. [DOI] [PubMed] [Google Scholar]
- 96.Bashir A, Gray ML, Hartke J, Burstein D. Nondestructive imaging of human cartilage glycosaminoglycan concentration by MRI. Magn Reson Med 1999;41:857–865. [DOI] [PubMed] [Google Scholar]
- 97.Gong E, Pauly JM, Wintermark M, Zaharchuk G. Deep learning enables reduced gadolinium dose for contrast-enhanced brain MRI. J Magn Reson Imaging 2018;48:330–340. [DOI] [PubMed] [Google Scholar]
- 98.Shapiro EM, Borthakur A, Gougoutas A, Reddy R. 23NA MRI accurately measures fixed charge density in articular cartilage. Magn Reson Med 2002;47:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature 2013;495:187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Raya JG, Dietrich O, Horng A, Weber J, Reiser MF, Glaser C. T2 measurement in articular cartilage: Impact of the fitting method on accuracy and precision at low SNR. Magn Reson Med 2010;63:181–193. [DOI] [PubMed] [Google Scholar]
- 101.Eckstein F, Collins JE, Nevitt MC, et al. Cartilage thickness change as an imaging biomarker of knee osteoarthritis progression: Data from the Foundation for the National Institutes of Health Osteoarthritis Biomarkers Consortium. Arthritis Rheumatol 2015;67:3184–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Neogi T, Bowes MA, Niu J, et al. Magnetic resonance imaging-based three-dimensional bone shape of the knee predicts onset of knee osteoarthritis: Data from the osteoarthritis initiative. Arthritis Rheum 2013;65:2048–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bredbenner TL, Eliason TD, Potter RS, Mason RL, Havill LM, Nicolella DP. Statistical shape modeling describes variation in tibia and femur surface geometry between control and incidence groups from the Osteoarthritis Initiative database. J Biomech 2010;43: 1780–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Berthiaume MJ, Raynauld JP, Martel-Pelletier J, et al. Meniscal tear and extrusion are strongly associated with progression of symptomatic knee osteoarthritis as assessed by quantitative magnetic resonance imaging. Ann Rheum Dis 2005;64:556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. Med Image Comput Comput Assist Interv 2013;16(Pt 2):246–253. [DOI] [PubMed] [Google Scholar]
- 106.Bengio Y, Lee H. Editorial introduction to the neural networks special issue on deep learning of representations. Neural Networks 2015; 64:1–3. [DOI] [PubMed] [Google Scholar]
- 107.Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell 2017;39: 640–651. [DOI] [PubMed] [Google Scholar]
- 108.Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med 2018;79:2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou Z, Zhao G, Kijowski R, Liu F. Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med 2018;80: 2759–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Norman B, Pedoia V, Majumdar S. Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology 2018;288:177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C. The importance of skip connections in biomedical image segmentation. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 2016; 10008 LNCS:179–187. [Google Scholar]
- 112.Peterfy CG, Guermazi A, Zaim S, et al. Whole-organ magnetic resonance imaging score (WORMS) of the knee in osteoarthritis. Osteoarthr Cartil 2004;12:177–190. [DOI] [PubMed] [Google Scholar]
- 113.Link TM, Steinbach LS, Ghosh S, et al. Osteoarthritis: MR imaging findings in different stages of disease and correlation with clinical findings. Radiology 2003;226:373–381. [DOI] [PubMed] [Google Scholar]
- 114.Felson DT, McLaughlin S, Goggins J, et al. Bone marrow edema and its relation to progression of knee osteoarthritis. Ann Intern Med 2003;139(5I). [DOI] [PubMed] [Google Scholar]
- 115.Eckstein F, Kwoh CK, Boudreau RM, et al. Quantitative MRI measures of cartilage predict knee replacement: A case-control study from the Osteoarthritis Initiative. Ann Rheum Dis 2013;72:707–714. [DOI] [PubMed] [Google Scholar]
- 116.Liu F, Zhou Z, Samsonov A, et al. Deep learning approach for evaluating knee MR images: Achieving high diagnostic performance for cartilage lesion detection. Radiology 2018;289:160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pedoia V, Norman B, Mehany SN, Bucknor MD, Link TM, Majumdar S. 3D convolutional neural networks for detection and severity staging of meniscus and PFJ cartilage morphological degenerative changes in osteoarthritis and anterior cruciate ligament subjects. J Magn Reson Imaging 2019;49:400–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bien N, Rajpurkar P, Ball RL, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PLoS Med 2018;15:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu F, Guan B, Zhou Z, et al. Fully automated diagnosis of anterior cruciate ligament tears on knee MR images by using deep learning. Radiol Artif Intell 2019;1:180091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hirschmann A, Cyriac J, Stieltjes B, Kober T, Richiardi J, Omoumi P. Artificial intelligence in musculoskeletal imaging: Review of current literature, challenges, and trends. Semin Musculoskelet Radiol 2019;23: 304–311. [DOI] [PubMed] [Google Scholar]
- 121.Chea P, Mandell JC. Current applications and future directions of deep learning in musculoskeletal radiology. Skeletal Radiol 2019. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 122.Desai AAD, Gold GE, Hargreaves BA, Chaudhari AS. Technical considerations for semantic segmentation in MRI using convolutional neural networks. arXiv Prepr arXiv1902 01977 2019;1–24.
- 123.Hestness J, Narang S, Ardalani N, et al. Deep learning scaling is predictable, empirically. 2017;1–19. [Google Scholar]
- 124.Gal Y, Ghahramani Z. Dropout as a Bayesian approximation: Representing model uncertainty in deep learning. In: Proc 33rd Int Conf Int Conf Mach Learn, vol. 48. JMLR.org; 2016;1050–1059 [ICML’16]. [Google Scholar]
- 125.Desai AD, Barbieri M, Mazzoli V, et al. DOSMA: A deep-learning, open-source framework for musculoskeletal MRI analysis. In: Proc 27th Annual Meeting ISMRM, Montreal; 2019. p 1135. [Google Scholar]
- 126.Deng J, Dong W, Socher R, Li L-J, Kai Li, Li Fei-Fei. ImageNet: A large-scale hierarchical image database. In: 2009 IEEE Conf Comput Vis Pattern Recognit 2009;248–255. [Google Scholar]
- 127.Asher KA, Bangerter NK, Watkins RD, Gold GE. Radiofrequency coils for musculoskeletal magnetic resonance imaging. Top Magn Reson Imaging 2010;21:315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kogan F, Levine E, Chaudhari AS, et al. Simultaneous bilateral-knee MR imaging. Magn Reson Med 2018;80:529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Corea JR, Flynn AM, Lechêne B, et al. Screen-printed flexible MRI receive coils. Nat Commun 2016;7:10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Winkler SA, Corea J, Lechêne B, et al. Evaluation of a flexible 12-channel screen-printed pediatric MRI coil. Radiology 2019;181883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McGee KP, Stormont RS, Lindsay SA, et al. Characterization and evaluation of a flexible MRI receive coil array for radiation therapy MR treatment planning using highly decoupled RF circuits. Phys Med Biol 2018;63:08NT02. [DOI] [PubMed] [Google Scholar]
- 132.Zhang B, Sodickson DK, Cloos MA. A high-impedance detector-array glove for magnetic resonance imaging of the hand. Nat Biomed Eng 2018;2:570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mehmann A, Vogt C, Varga M, et al. Automatic resonance frequency retuning of stretchable liquid metal receive coil for magnetic resonance imaging. IEEE Trans Med Imaging 2019;38:1420–1426. [DOI] [PubMed] [Google Scholar]
- 134.Mazzoli V, Schoormans J, Froeling M, et al. Accelerated 4D self-gated MRI of tibiofemoral kinematics. NMR Biomed 2017;30. [DOI] [PubMed] [Google Scholar]
- 135.Metcalfe AJ, Andersson MLE, Goodfellow R, Thorstensson CA. Is knee osteoarthritis a symmetrical disease? Analysis of a 12 year prospective cohort study. Musculoskelet Disord 2012;13:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jungmann PM, Brucker PU, Baum T, et al. Bilateral cartilage T2 mapping 9 years after Mega-OATS implantation at the knee: A quantitative 3T MRI study. Osteoarthr Cartil 2015;23:2119–2128. [DOI] [PubMed] [Google Scholar]
- 137.Bae J-H, Hosseini A, Wang Y, et al. Articular cartilage of the knee 3 years after ACL reconstruction. A quantitative T2 relaxometry analysis of 10 knees. Acta Orthop 2015;86:605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Deshmane A, Gulani V, Griswold MA, Seiberlich N. Parallel MR imaging. J Magn Reson Imaging 2012;36:55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Barth M, Breuer F, Koopmans PJ, Norris DG, Poser BA. Simultaneous multislice (SMS) imaging techniques. Magn Reson Med 2016;75: 63–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Addy NO, Ingle RR, Wu HH, Hu BS, Nishimura DG. High-resolution variable-density 3D cones coronary MRA. Magn Reson Med 2015;74: 614–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ugurbil K Imaging at ultrahigh magnetic fields: History, challenges, and solutions. Neuroimage 2018;168:7–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rietsch SHG, Orzada S, Bitz AK, Gratz M, Ladd ME, Quick HH. Parallel transmit capability of various RF transmit elements and arrays at 7T MRI. Magn Reson Med 2017;79:1116–1126. [DOI] [PubMed] [Google Scholar]
- 143.Brix G, Seebass M, Hellwig G, Griebel J. Estimation of heat transfer and temperature rise in partial-body regions during MR procedures: An analytical approach with respect to safety considerations. Magn Reson Imaging 2002;20:65–76. [DOI] [PubMed] [Google Scholar]


