Abstract
Antibiotic-producing microorganisms have developed several self-resistance mechanisms to protect them from autotoxicity. Transporters belonging to the resistance- nodulation-division (RND) superfamily commonly confer multidrug resistance in Gram-negative bacteria. Phenazines are heterocyclic, nitrogen-containing and redox-active compounds that exhibit diverse activities. We previously identified six phenazines from Lysobacter antibioticus OH13, a soil bacterium emerging as a potential biocontrol agent. Among these phenazines, myxin, a di-N-oxide phenazine, exhibited potent activity against a variety of microorganisms. In this study, we identified a novel RND efflux pump gene cluster, designated lexABC, which is located far away in the genome from the myxin biosynthesis gene cluster. We found a putative LysR-type transcriptional regulator encoding gene lexR, which was adjacent to lexABC. Deletion of lexABC or lexR gene resulted in significant increasing susceptibility of strains to myxin and loss of myxin production. The results demonstrated that LexABC pump conferred resistance against myxin. The myxin produced at lower concentrations in these mutants was derivatized by deoxidation and O-methylation. Furthermore, we found that the abolishment of myxin with deletion of LaPhzB, which is an essential gene in myxin biosynthesis, resulted in significant downregulation of the lexABC. However, exogenous supplementation with myxin to LaPhzB mutant could efficiently induce the expression of lexABC genes. Moreover, lexR mutation also led to decreased expression of lexABC, which indicates that LexR potentially positively modulated the expression of lexABC. Our findings reveal a resistance mechanism against myxin of L. antibioticus, which coordinates regulatory pathways to protect itself from autotoxicity.
Keywords: phenazine, RND pump, antibiotic resistance, LysR-type regulator, Lysobacter antibioticus
Introduction
Phenazines are a large class of heterocyclic, nitrogen-containing and redox-active natural products exhibiting a wide range of biological activities including antibiotic, antitumor, biofilm modulation and so on. Since the first blue phenazine pyocyanin (PYO) was identified, over 180 natural phenazines have been isolated and more than 6,000 phenazines have been chemically synthesized. Natural phenazines have been identified as secondary metabolites primarily from Pseudomonas and Streptomyces (Laursen and Nielsen, 2004; Cimmino et al., 2012; Guttenberger et al., 2017).
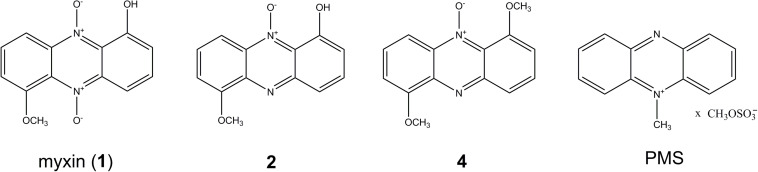
Lysobacter, a genus of Gram-negative bacteria, is emerging as a novel source of biocontrol agents against phytopathogenic microorganisms because they have the ability to produce abundant active lytic enzymes and secondary metabolites (Xie et al., 2012; Panthee et al., 2016; Puopolo et al., 2018). We previously isolated six phenazines from Lysobacter antibioticus OH13, of which myxin (Figure 1), an N-oxide phenazine, drew much attention due to its potent activity against microbes (Weigele et al., 1970; Zhao et al., 2016) and human cancer cells (Viktorsson et al., 2017).
FIGURE 1.
Structures of phenazines from L. antibioticus OH13 and PMS (phenazine methosulfate).
Our previous work revealed that myxin was derived from phenazine 1,6-dicarboxylic acid (PDC) and was biosynthesized by a LaPhz gene cluster including six core genes, LaPhzB-G, and four modification genes, LaPhzNO1, LaPhzS, LaPhzM, and LaPhzX. We first experimentally characterized LaPhzNO1, homologous to Baeyer-Villiger flavoproteins catalyzing phenazine N-oxidation (Zhao et al., 2016). The unique feature of myxin is that it not only belongs to the class of phenazine compounds but also to a class known as heterocyclic N-oxides with bioreductively activated, hypoxia-selective DNA-damaging properties (Mfuh and Larionov, 2015). Early studies showed that myxin interacted with DNA through intercalation and inhibited DNA template-controlled RNA synthesis (Hollstein and Van Gemert, 1971; Hollstein and Butler, 1972). More recent findings indicated that myxin caused bioreductively activated, radical-mediated DNA strand cleavage under both aerobic and anaerobic conditions (Chowdhury et al., 2012). The unique mode of action and strong biological activities of myxin make it an attractive target of study and development.
Antibiotic-producing microorganisms have evolved several self-resistance mechanisms to prevent autotoxicity, such as antibiotic efflux, antibiotic modification, self-sacrifice, target repair or protection (Ogawara, 2019). One of the self-resistance mechanisms is that efflux pumps transport toxic compounds outside of the cells (Yan et al., 2020). Efflux pumps have been classified into five major groups based on their protein sequences, which are RND (resistance-nodulation-cell division), MFS (major facilitator superfamily), MATE (multidrug and toxic compound extrusion), SMR (small multidrug resistance), and ABC (ATP-binding cassette) superfamilies or families (Delmar et al., 2014).
RND efflux pumps play important roles in multidrug resistance (MDR) due to their broad-spectrum substrate profile in Gram-negative bacteria. The RND efflux systems are composed of a tripartite complex: an inner membrane protein (IMP), an outer membrane protein (OMP), and a peri-plasmic membrane fusion protein (MFP) that links the IMP and OMP (Li et al., 2015; Venter et al., 2015). The typical RND efflux pump systems, such as the AcrAB-TolC pump of Escherichia coli and the Mex pumps in Pseudomonas aeruginosa, have been intensively studied (Peterson and Kaur, 2018). The AcrAB-TolC pump has an extremely wide range of substrates and can transport harmful molecules to the extracellular medium (Li et al., 2015). Recently, it was revealed that the AcrAB-TolC pump is essential in tetracycline resistance acquisition by plasmid transfer (Nolivos et al., 2019). RND pumps in P. aeruginosa including MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexXY, and MexGHI-OpmD are significant determinants of multidrug resistance against various antibiotic families such as β-lactams, aminoglycosides, fluoroquinolones and phenazines (Li et al., 2015; Sakhtaha et al., 2016). Multiple regulatory components modulate RND pumps, for example, MexAB-OprM is regulated by MexR (Evans et al., 2001), NalD (Morita et al., 2006), CpxR (Tian et al., 2016), PA3898, and PA2100 (Heacock-Kang et al., 2018). LysR-type transcriptional regulator, MexT (Fargier et al., 2012), and AdeL (Coyne et al., 2010) involve in activating the expression of MexEF-OprN and AdeFGH efflux pump, respectively.
We found that myxin showed highly effective inhibition against many bacteria, such as methicillin-resistant Staphylococcus aureus (MRSA, MIC 0.05 μg/mL) (Zhao et al., 2016), Xanthomonas oryzae pv. oryzae (MIC 2 μg/mL), and Xanthomonas oryzae pv. oryzicola (MIC 2 μg/mL). The antibiotic properties of myxin have been the subject of intense research efforts, but little is known about their effect on the producing organisms, and moreover, how L. antibioticus itself responds to and survives in the presence of myxin.
Here, we identified a gene cluster on the chromosome of L. antibioticus OH13, lexABC, encoding an RND family multidrug efflux pump that confers resistance to myxin and phenazine methosulfate (PMS) (Figure 1). We discovered that the expression of lexABC was promoted by myxin and positively regulated by a LysR-type transcriptional regulator, indicating that the LexABC pump could coordinate with myxin production to protect L. antibioticus OH13 from autotoxicity.
Results
The LexABC Efflux System and LexR Regulator Are Involved in Self-Resistance Against Myxin
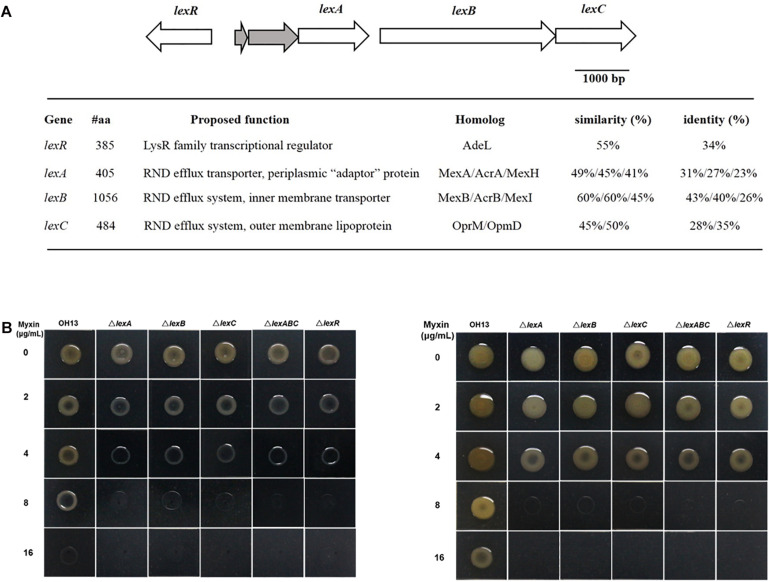
Bioinformatics analysis of L. antibioticus OH13 genomic sequence revealed three genes encoding RND efflux pump proteins, which were similar to MexAB-OprM and MexGHI-OpmD from P. aeruginosa, located distant from the phenazine biosynthetic gene cluster. We proposed naming these three genes lexA, lexB, and lexC (NCBI accession no. MW029615, MW029616, MW029617, respectively) for Lysobacter phenazine efflux system proteins. LexA had 49%/45%/41% sequence similarity to MexA/AcrA/MexH, and LexB shared 60%/60%/45% similarity with MexB/AcrB/MexI, respectively, whereas LexC was homologous to OprM and OpmD (45 and 50% similarity) but was not a putative TolC-like protein (Figure 2A and Supplementary Figure S1). Moreover, a putative gene encoding LysR family transcriptional regulator, lexR (NCBI accession no. MW029618) was found at the upstream of lexABC locus. LexR shared 55% similarity with AdeL which was a LysR family transcriptional regulator (Figure 2A and Supplementary Figure S1). To elucidate the role of the LexABC system and assumed regulator LexR in myxin resistance, mutants lacking lexA, lexB, lexC, lexABC, and lexR (ΔlexA, ΔlexB, ΔlexC, ΔlexABC, and ΔlexR) were constructed. Next, we tested the contribution of these genes in myxin resistance by growing various mutants on NA medium containing different concentrations of this compound (Figure 2B and Table 1). After culturing for 24 h, the mutants showed severe growth defects on medium with 4, 8, and 16 μg/mL myxin compared to the wild type (Figure 2B and Table 1). At lower myxin concentrations (2 and 4 μg/mL), the growth of the mutants caught up after 48 h cultivation. However, at higher myxin concentrations (8 and 16 μg/mL), the growth of these mutants was completely stopped (Figure 2B and Table 1). And the complemented strains of lexA, lexB, lexABC restored their resistance against myxin to the level of the wild type (Supplementary Figure S2A). The sensitivities of ΔlexA, ΔlexB, ΔlexC, ΔlexABC, and ΔlexR strains were also demonstrated by a filter disk assay for myxin-dependent growth inhibition. The inhibition circles of myxin against the above mentioned mutants increased with the growing concentrations of myxin (0.125, 0.25, 0.5 μg), while 0.5 μg of myxin had little effect on the growth of the wild type (Supplementary Figure S3). These results demonstrated the important roles of the RND system LexABC and the regulator LexR in self-resistance of L. antibioticus OH13 against myxin.
FIGURE 2.
Schematic representation of lex genes and susceptibility of different strains to myxin. (A) Organization of lexA, lexB, lexC, lexR in L. antibioticus OH13 and their predicted functions. (B) Growth of L. antibioticus OH13 and its derivatives on media containing different concentrations of myxin. Left, cultured for 24 h. Right, cultured for 48 h. Images are representative of three independent experiments.
TABLE 1.
Susceptibility of L. antibioticus OH13 and its derivatives to different concentrations of myxin.
|
Growth status of L. antibioticus strains | ||||||||||||
| Myxin (μg/mL) |
24 h |
48 h |
||||||||||
| OH13 | △lexA | △lexB | △lexC | △lexABC | △lexR | OH13 | △lexA | △lexB | △lexC | △lexABC | △lexR | |
| 0 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ |
| 2 | ++++ | +++ | +++ | +++ | +++ | +++ | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ |
| 4 | ++++ | ++ | ++ | ++ | ++ | ++ | +++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| 8 | +++ | + | + | + | + | + | +++++ | + | + | + | + | + |
| 16 | + | − | − | − | − | − | ++++ | − | − | − | − | − |
Corresponding to Figure 2B. “+” indicated obvious colony grown, and different numbers of “+” showed various growth level. “−” indicated no colony grown.
LexABC System and LexR Are Required for Normal Myxin Production
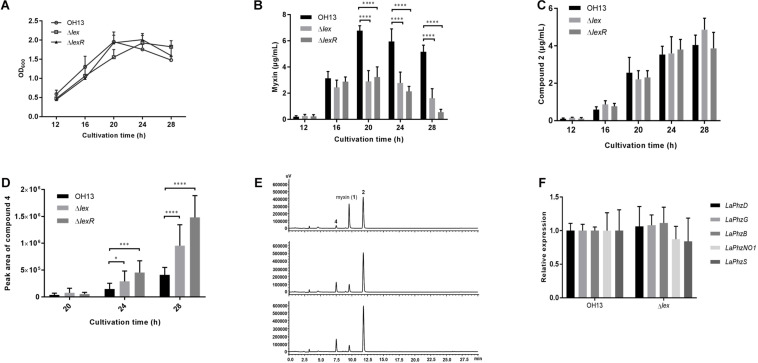
To investigate the role of the LexABC system and LexR in cell growth and myxin production, cell growth curves and myxin production of wild-type strain OH13, ΔlexABC (hereafter referring to lexABC as lex) and ΔlexR were compared in 1/10 TSB medium. At 12 h, the growth rate and myxin production were comparable between strain OH13 and the lex mutant (Figure 3A). The growth of the lex mutant was delayed at 16 and 20 h but caught up with OH13 at 24 h (Figure 3A). However, the accumulation of myxin in the lex mutant was flat since 16 h, which was over 2-fold less than that in OH13 (Figure 3B). Deletion of lexR did not affect the bacterial growth but resulted in similar decrease level of myxin production with lex mutant (Figures 3A,B). These results indicated that L. antibioticus could endure a certain concentration of myxin without the LexABC efflux pump or LexR. To explore whether myxin was converted to its derivatives in lex or lexR mutant, we detected the yield of other main phenazine compounds. The results showed that compound 2 (Figure 1) was not significantly changed in lex or lexR mutant compared with that in OH13 (Figure 3C). Meanwhile, there was an increased peak corresponding to phenazine compound 4 (Figure 1) drawing our attention (Figure 3E). Quantitative analysis revealed that compound 4 was significantly increased in lex or lexR mutant compared to OH13 at the cultivation time of 24 and 28 h (Figure 3D). Compounds 2 and 4 showed inhibitory activity against certain bacteria but not as strong as myxin in the tests we conducted (Zhao et al., 2016). The results suggested that L. antibioticus converted myxin to its derivatives to protect itself when the LexABC efflux pump or LexR was deficient. To further determine whether the missing of LexABC pump or LexR enhances the intracellular myxin accumulation, we analyzed the intracellular phenazines of OH13, Δlex and ΔlexR strains. The result showed that compound 2 but not myxin accumulated in the intracellular, but it was not as expected that lex and lexR mutants had a higher phenazine accumulation than the wild type (Supplementary Figure S4).
FIGURE 3.
LexABC pump and LexR are required for normal myxin production of L. antibioticus OH13. (A) Growth curves of OH13, Δlex and ΔlexR strains cultured in 1/10 TSB for 12, 16, 20, 24, and 28 h after inoculation. (B) Myxin production in OH13, Δlex and ΔlexR strains grown in 1/10 TSB for different time points. Supernatant of cultures was extracted and detected for myxin yield. (C) Compound 2 production in OH13, Δlex and ΔlexR strains grown in 1/10 TSB for different time points. (D) Compound 4 production in OH13 (top), Δlex (middle) and ΔlexR (bottom) strains grown in 1/10 TSB for different time points. The production of compound 4 was indicated with peak area of HPLC. (E) HPLC analysis of extract from OH13, Δlex and ΔlexR strains at cultivation time of 28 h. (F) Transcriptional expression of myxin biosynthetic genes in strain OH13 and lex mutant. Data shown in graph are the means from three independent experiments and error bars are standard deviations. Asterisks indicate statistically significant difference according to t-test in GraphPad Prism 7.0. ∗p < 0.05, ∗∗∗p < 0.001, ****p < 0.0001.
To understand whether the absence of the LexABC efflux pump influences myxin biosynthesis at the transcriptional level, the relative gene expression of myxin biosynthetic genes were compared between OH13 and the lex mutant. The detected genes included three core genes, LaPhzD, LaPhzG, and LaPhzB, for the formation of the myxin precursor, and two modification genes, LaPhzNO1, encoding Baeyer-Villiger similar flavoproteins responsible for phenazine N-oxidation, and LaPhzS, encoding FAD-dependent enzyme for catalyzing decarboxylative hydroxylation of PDC (Zhao et al., 2016). The qPCR showed that the expression of these biosynthetic genes was not significantly changed in the absence of lex (Figure 3F). These results suggested that the lack of the LexABC efflux pump affected myxin production but not by impacting its biosynthesis at the transcriptional level. The LexABC efflux pump is necessary for protecting normal cell growth and myxin production under a high level of myxin accumulation.
Expression of the LexABC Efflux Pump Is Induced by Myxin
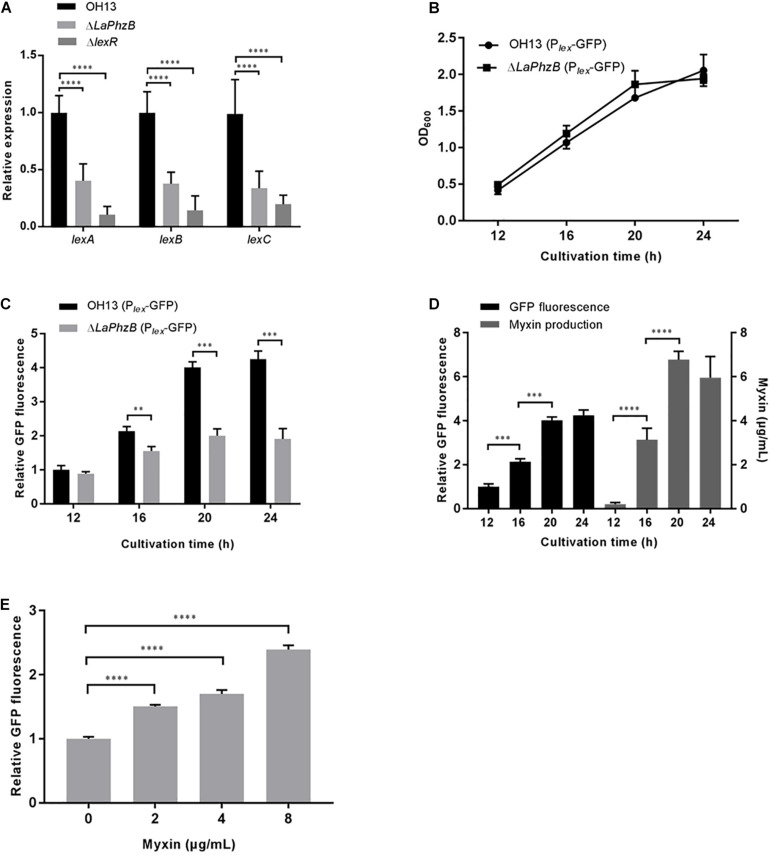
Our previous study showed that deletion of LaPhzB abolished production of all of the phenazines (Zhao et al., 2016). The transcriptional levels of the LexABC efflux system genes in strain OH13 and the LaPhzB mutant were determined by qPCR. The results indicated that the expression levels of lexA, lexB, and lexC were significantly down-regulated in the LaPhzB mutant compared to that in the wild type (Figure 4A). The decreasing expression level of lex in the myxin deficient mutant suggested that myxin possibly stimulated LexABC expression.
FIGURE 4.
Expression of LexABC was stimulated by myxin. (A) qPCR analysis of lexA, lexB, lexC genes expression in strain OH13, ΔLaPhzB, ΔlexR strains. (B) Growth curves of strains OH13 (Plex-GFP) and ΔLaPhzB (Plex-GFP) cultured in 1/10 TSB for 12, 16, 20, and 24 h after inoculation. (C) Relative GFP fluorescence in strains OH13 (Plex-GFP) and ΔLaPhzB (Plex-GFP) at different time points. The fluorescence intensity in strain OH13 (Plex-GFP) at 12 h was normalized as 1. (D) Presentation of the relationship between myxin concentration and gene expression of lexABC. Data from Figures 3B,4C of strain OH13. (E) Relative GFP fluorescence in strain ΔLaPhzB (Plex-GFP) added with 0, 2, 4, 8 μg/mL myxin. The fluorescence intensity in strain ΔLaPhzB (Plex-GFP) without myxin was normalized as 1. Data shown in graph are the means from three replicates and error bars are standard deviations. Asterisks indicate statistically significant difference according to t-test in GraphPad Prism 7.0. ∗∗p < 0.01, ∗∗∗p < 0.001, ****p < 0.0001.
To test this assumption, we constructed strains OH13 (Plex-GFP) and ΔLaPhzB (Plex-GFP) that expressed GFP under the control of the lex promoter. The growth rate of both strains was about the same (Figure 4B), but the fluorescence of strain OH13 (Plex-GFP) exhibited a substantial increase along with bacterial growth from 12 to 20 h, whereas the fluorescence in strain ΔLaPhzB (Plex-GFP) was stable since the 16th h of cultivation. The relative fluorescence of ΔLaPhzB (Plex-GFP) was approximately 50% lower than that of OH13 (Plex-GFP) at 20 and 24 h (Figure 4C). To show the relationship between myxin concentration and gene expression of lexABC, the myxin production and GFP fluorescence of strain OH13 were combined in one diagram (data from Figures 3B, 4C). Expression of lexABC genes was consistent with myxin production during cell growth (Figure 4D), suggesting that lexABC expression is influenced by myxin. To further confirm these results, the expression of lexABC was analyzed in strain ΔLaPhzB (Plex-GFP) supplemented with different concentrations of myxin. The data showed that 2, 4, and 8 μg/mL of myxin enhanced the expression of LexABC by 1. 5-, 1. 7-, and 2.4-fold, respectively, compared with the control (Figure 4E). These results suggested that the expression of LexABC was induced by myxin and was concentration dependent.
LexR Could Positively Regulate LexABC Efflux Expression
The regulation mechanism of how these transporters sense and respond to the accumulation of myxin remains unclear. As described above, deletion of lexR resulted in strains more sensitive to myxin and decreased myxin production, which was similar with the LexABC system deficient strains (Figures 2B, 3B). LexR was predicted as a LysR family transcriptional regulator, so the transcriptional level of lexABC was compared between the wild type and lexR mutant by qPCR. The gene expression level of lexA, lexB, and lexC was significantly reduced by more than 2-fold in the lexR mutant (Figure 4A). The results indicated that LexR could positively regulate the expression of LexABC system and play an essential role in myxin resistance.
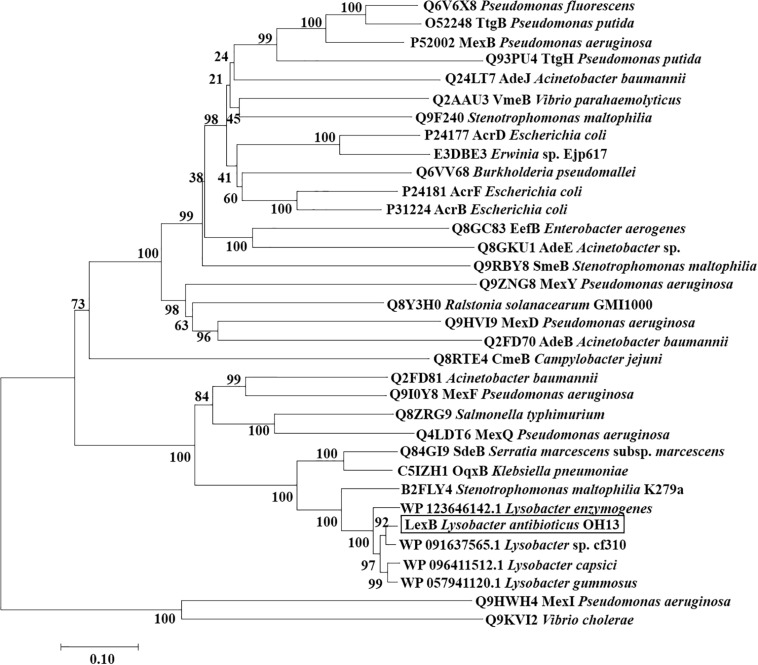
Phylogenetic Analysis of the LexABC Efflux Pump
To elucidate the evolutionary relationship of the LexABC efflux with other RND efflux complexes, a phylogenetic tree was constructed with LexB and its homolog proteins (amino acid similarity above 53%), which were inner membrane proteins and functional in the recognition and binding of substrates. The phylogenetic tree contained 34 proteins from various bacterial species. LexB clustered together with the putative RND proteins from the other four Lysobacter strains in the tree, which showed that LexB homologs were conserved in Lysobacter species. The close homologs were the RND protein of Stenotrophomonas maltophilia K279a and SdeB from Serratia marcescens subsp. marcescens, which mainly exports fluoroquinolones (Kumar and Worobec, 2005; Chen et al., 2011). MexI from P. aeruginosa, which was characterized for Pseudomonas quinolone signal (PQS) (Aendekerk et al., 2005) and phenazine efflux (Sakhtaha et al., 2016), also showed close evolutionary relationship with LexB (Figure 5). The results indicated that the closely related RND homologs presented similarity in the substrates.
FIGURE 5.
Phylogenetic tree of LexB and other RND proteins. Neighbor-joining tree based on the amino acid sequences of RND family proteins related to LexB was generated using MEGA 7.0 with 1,000 bootstrap replicates. The black box indicates the position of LexB from L. antibioticus OH13. The sequences of four Lysobacter species were from NCBI, and the other sequences were obtained from TCDB (Transporter Classification Database).
Discussion
In this study, we identified a novel RND efflux pump, LexABC, which plays an important role in myxin resistance and production. LexABC confers resistance against phenazines such as myxin and PMS. We revealed that LexABC expression was sufficiently induced by myxin and potentially regulated by the LysR-type transcriptional regulator LexR.
The RND superfamily is common in Gram-negative bacteria and is a major contributor to multidrug resistance among human pathogens such as Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter species (Li and Nikaido, 2004, 2009). Moreover, RND efflux pumps have also been reported in plant pathogens, for example, Erwinia amylovora (Pletzer and Weingart, 2014), Dickeya species (Liang et al., 2019), and Pseudomonas syringae (Stoitsova et al., 2008). Research into the RND efflux pump in biocontrol bacteria has been limited. Our study will contribute to the self-resistance of antibiotic-producing bacteria and provide new insight into pathogen multidrug resistance.
The results showed that the inactivation of any of the lexABC genes or all three genes could cause the same level of growth defect in presence of myxin. These findings demonstrated that the function of any component of LexABC is indispensable. The growth of the lex mutant lagged behind the wild type in the early stage and then caught up at a late stage in 1/10 TSB. The yield of myxin peaked at 20 h and then went down in the wild type, whereas the accumulation of compound 2 and 4 increased along with the increase of cultivation time (Figures 3B–D). The results suggested that when the accumulation of myxin reached a certain concentration which the strains can’t endure, myxin could be partly converted to its derivatives to protect bacteria from the toxicity. The production of myxin was significantly reduced in the lex and lexR mutant compared to the wild type after 20 h cultivation, and the yield of compound 2 didn’t show noteworthy change but compound 4 increased (Figures 3B–D). So the missing myxin in lex or lexR mutant was supposed to be converted into compound 4. Compound 2 can be converted into compound 4 with O-methylation by O-methyltransferase LaPhzM (Jiang et al., 2018), but how myxin is transformed into compound 2 still needs study. Myxin or its derivatives did not show an increasing accumulation in the intracellular of lex and lexR mutant compared to the wild type (Supplementary Figure S4). Moreover, lex mutant had comparative myxin biosynthetic genes expression level to OH13 (Figure 3F). These results indicated that L. antibioticus protected itself by reducing myxin production when the LexABC system was disrupt but without affecting the biosynthesis of this compound. Antibiotic-producing bacteria usually involved several resistance strategies to protect themselves from suicide, and the LexABC efflux pump may be involved in the first line of defense in L. antibioticus. With the loss of LexABC efflux pump, the conversion of myxin to its derivatives was another defense line. The gene LaPhzX encoding putative small monooxygenase in myxin biosynthetic gene cluster, which is similar to ActVA-Orf6, TcmH (Zhao et al., 2016) and PumA (Sporer et al., 2018), has not been characterized. PumA was reported to contribute to phenazine resistance and was required for the conversion of PMS to unique phenazine metabolites, but its exact function was not clear (Sporer et al., 2018). We predict LaPhzX may contribute to the defense line of myxin, but further study is needed to determine its function in myxin resistance.
The expression level of the genes lexABC were increased along with the accumulation of myxin, and deletion of the myxin biosynthetic gene LaPhzB caused a decrease of the transcriptional expression of lexABC (Figures 4A,D). Moreover, exogenous addition of 8 μg/mL myxin to the LaPhzB mutant could enhance lexABC expression by more than 2-fold compared with that of the control (Figure 4E). These results demonstrated that myxin was not only an antibiotic but also physiologically an elicitor of the LexABC RND pump. This is similar with the famous phenazine PYO and its intermediate 5-methylphenazine-1-carboxylic acid (5-Me-PCA) which induce expression of the RND efflux pump-encoding operon mexGHI-opmD. MexGHI-OpmD transporter confers resistance to 5-Me-PCA, and its structurally similar, synthetic phenazine PMS (Sakhtaha et al., 2016). Disruption of LexABC system also led to L. antibioticus strains arrest of growth in presence of 50 and 100 μg/mL of PMS (Supplementary Figure S2B). The phylogenetic tree showed that the LexB RND protein was close to MexI. LexA, LexB, and LexC showed similarity to MexH, MexI and OpmD, respectively, but there was no additional MexG homolog adjacent which is an anomalous cytoplasmic membrane component not required for the normal export of 5-Me-PCA and PMS. The induction of mexGHI-opmD by phenazine was mediated by activating the redox-active transcription factor SoxR (Dietrich et al., 2006; Sakhtaha et al., 2016). SoxR acts as a redox sensor and is activated by redox-active compounds through oxidation or nitrosylation of its [2Fe-2S] cluster (Singh et al., 2013). Phenazine/SoxR/MexGHI-OpmD system is a model for regulation of natural product efflux and self-protection in Gram-negative antibiotics producing bacteria. We hypothesize a specific pathway to sense myxin production in self-protection against the detrimental effect of this antibiotic. The gene lexR encodes LysR family transcriptional regulator, which is located upstream of the lexABC locus, was studied. Deletion of this gene resulted in growth defect of L. antibioticus in presence of myxin and decreased myxin production (Figures 2, 3B). Moreover, the gene expression of lexA, B, C was down-regulated significantly in lexR mutant (Figure 4A). Our data showed that LexR played an important role in myxin resistance by positively regulating lexABC expression. Besides SoxR, MexT also functions as a redox-responsive regulator inducing the MexEF-OprN efflux system (Fargier et al., 2012). Myxin is a redox-cycling compound with N-oxidation, however, LexR is not similar to MexT or SoxR. A different signaling mechanism might underlying the regulatory linkage between LexR and LexABC pump. Though myxin/LexABC system shares much similarity with phenazine (5-Me-PCA and PMS)/MexGHI-OpmD system, myxin is featured as N-oxidation and O-methylation, which is different from 5-Me-PCA and PMS of N-methylation. Therefore, the LexABC-based myxin resistance mechanism and its regulatory network potentially will be a novel self-resistance model in antibiotic producing bacteria.
In conclusion, we report resistance mechanisms against myxin, which is a heterocyclic N-oxides penazine with a potent broad spectrum of antimicrobial activities. The LexABC efflux pump confers myxin resistance, and myxin conversion played more important role in the resistance mechanisms after LexABC loss. The Lex RND efflux pump is potentially regulated by a LysR family regulator LexR. Significantly, this study also revealed a signaling role of myxin in the induction of lexABC expression at the transcriptional level, which further extended our perceptions about myxin.
Materials and Methods
Bacterial Strains, Growth Conditions, and General DNA Manipulations
The strains and plasmids used in this study are listed in Table 2. L. antibioticus OH13 and derivatives were routinely grown at 28°C in nutrient broth (NB, 3 g beef extract, 1 g yeast extract, 5 g tryptone, 10 g sucrose, pH 7.0–7.2, in one liter distilled water; without sucrose, NBWS), nutrient agar (NA, 1 L NB with 17 g agar; without sucrose, NAWS), 1/10 tryptic soy broth (TSB). 1/10 TSB medium is more appropriate for myxin production. E. coli strains were routinely grown at 37°C in Luria-Bertani (LB) broth. Where necessary, media were supplemented with gentamycin (25 or 50 μg/mL). Plasmid preparation and DNA extraction were carried out with Omega kits (Norcross, GA).
TABLE 2.
Strains and plasmids used in this study.
| Strains or plasmids | Description | Sources or references |
| Strains | ||
| Lysobacter antibioticus | ||
| OH13 | Wild-type strain | Zhao et al., 2016 |
| OH13(Plex-GFP) | OH13 with lex promoter driving gfp expression | This study |
| △LaPhzB | OH13 with deletion in LaPhzB | Zhao et al., 2016 |
| △LaPhzB(Plex-GFP) | △LaPhzB with lex promoter driving gfp expression | This study |
| △lexA | OH13 with deletion in lexA | This study |
| △lexB | OH13 with deletion in lexB | This study |
| △lexC | OH13 with deletion in lexC | This study |
| △lexABC | OH13 with deletion in lexABC | This study |
| △lexR | OH13 with deletion in lexR | This study |
| △lexA(pBBR) | △lexA carrying pBBR1-MCS5 vector, Gmr | This study |
| △lexA(pBBR-lexA) | △lexA carrying pBBR-lexA vector, Gmr | This study |
| △lexB(pBBR) | △lexB carrying pBBR1-MCS5 vector, Gmr | This study |
| △lexB(pBBR-lexB) | △lexB carrying pBBR-lexB vector, Gmr | This study |
| △lexABC(pBBR) | △lexABC carrying pBBR1-MCS5 vector, Gmr | This study |
| △lexABC(pBBR-lexABC) | △lexABC carrying pBBR-lexABC vector, Gmr | This study |
| Escherichia coli | ||
| DH5α | F- φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk-,mk +) phoA supE44 λ-thi-1 gyrA96 | TransGen, China |
| Plasmids | ||
| pJQ200SK | Suicide cloning vector, Gmr | Quandt and Hynes, 1993 |
| pJQ200SK-lexA | pJQ200SK with two flanking fragments of lexA | This study |
| pJQ200SK-lexB | pJQ200SK with two flanking fragments of lexB | This study |
| pJQ200SK-lexC | pJQ200SK with two flanking fragments of lexC | This study |
| pJQ200SK-lexABC | pJQ200SK with two flanking fragments of lexABC | This study |
| pJQ200SK-lexR | pJQ200SK with two flanking fragments of lexR | This study |
| pJQ200SK-Plex-gfp | pJQ200SK with two flanking fragments of lexA start site and gfp | This study |
| pBBR1-MCS5 | Broad host range cloning vector, lacZ, Gmr | Kovach et al., 1995 |
| pBBR-lexA | pBBR1-MCS5 carrying ORF of lexA and its promoter, Gmr | This study |
| pBBR-lexB | pBBR1-MCS5 carrying ORF of lexB and its promoter, Gmr | This study |
| pBBR-lexABC | pBBR1-MCS5 carrying ORF of lexABC and its promoter, Gmr | This study |
Gene Deletion and Complementation
The oligonucleotide primers used in this study are listed in Supplementary Table S1. DNA manipulation was conducted by following the methods described previously (Zhao et al., 2016). Briefly, for gene in-frame deletions, fragments containing the upstream and downstream regions of the target genes were cloned into pJQ200SK (Quandt and Hynes, 1993) and transformed into E. coli DH5α for the construction of gene in-frame deletion constructs. The final constructs were transformed into the competent cells of OH13 wild type or its derivatives by electroporation. The gentamycin-resistant colonies were screened by PCR using primers to identify the single crossover mutants. Then, the single crossover mutants were grown on NAWS medium containing 8% sucrose, which would screen for deletion mutants through a second homologous recombination of the flanking regions. The mutants were verified by PCR using primers.
For complementation, the open reading frames (ORFs) of the target genes with their own promoters were cloned into pBBR1-MCS5 (Kovach et al., 1995). The expression constructs were confirmed by PCR and DNA sequencing and introduced into wild type or corresponding mutants by electroporation. The complementation strains were screened on NAWS medium containing gentamycin and were validated by PCR.
Preparation and Analysis of Myxin
L. antibioticus OH13 was cultivated for 24 h in a 250 mL flask containing 50 mL NB medium. The seed culture was then transferred into 200 mL 1/10 TSB medium in a 1 liter flask (a total of 50 flasks). The flasks were placed in incubators for 24 h with shaking at 180 rpm at 28°C. Then, the cultures were extracted with equal volumes of ethyl acetate, and the organic phase was evaporated to dryness under vacuum to yield the crude extract. The crude extract was chromatographed over a Sephadex LH-20 column using 1:1 CH2Cl2-MeOH as the elution solvent. The fractions were further purified by reversed-phase high-performance liquid chromatography (HPLC) with CH3CN in H2O (Agilent-C18, 5 μm, 9.4 × 250 mm) under 283 nm with a 4.0 mL/min flow rate.
To analyze myxin production in OH13 and its derivatives, equal volume of ethyl acetate was used to extract 1 mL supernatant of cultures grown in 1/10 TSB. The crude extract was transferred to an Eppendorf tube and fully evaporated. Then, the residues were dissolved with 200 μL methanol and prepared for HPLC analysis. To determine the production of intracellular myxin, 1 mL culture grown for 20 h in 1/10 TSB was harvested and centrifuged at 12,000 rpm for 1 min. Supernatant was removed and the pellet was washed with 1/10 TSB for twice. The pellets were extracted with 1 mL of ethyl acetate overnight. Then the extract was dissolved in methanol and subjected to HPLC.
The detection of phenazines was through HPLC (Agilent SB-C18, 250 × 4.6 mm) with a 1.0 mL/min flow rate. The mobile phase was 20–45% CH3CN in H2O from 0 to 5 min, 45 to 50% CH3CN in H2O from 5 to 19 min, 50 to 60% CH3CN in H2O from 19 to 20 min, 60 to 100% CH3CN in H2O from 20 to 23 min, 100% CH3CN from 23 to 27 min, 100 to 20% CH3CN in H2O from 27 to 28 min, and 20% CH3CN in H2O from 28 to 30 min.
Susceptibility Test of L. antibioticus to Phenazines
The susceptibility of the OH13 wild type and mutants to phenazines was determined by agar screening tests. Myxin was dissolved in DMSO, whereas PMS was dissolved in water. Liquid precultures of each strain to be tested were grown in NB medium to 1.0 of OD600. Five microliters of cultures were spotted on NA media containing the indicated concentrations of phenazines and incubated at 28°C. After 24 and 48 h of growth, the colonies were imaged using a Canon camera. For the inhibition assays of myxin against Lysobacter strains, bacterial suspensions with OD600 1.0 were 100-fold diluted and then spread on NA plates, and subsequently different concentrations of myxin were dropped on filter papers.
RNA Isolation and qRT-PCR
Bacterial cells were cultured and harvested at an OD600 of approximately 1.0. RNA extraction was performed using the Omega bacterial RNA extraction kit (Omega biotek, Norcross, GA) by following the manufacturer’s instructions. The concentration of RNA, A260/A280 and A260/A230 ratios were determined by using an Eppendorf BioPhotometer plus. In qPCR analysis, a 250 ng RNA sample was used for genomic DNA elimination and cDNA synthesis by a Vazyme HiScript II Q RT SuperMix (with gDNA wiper) by following the manufacturer’s protocol. Specific primers for qRT-PCR (Supplementary Table S1) were designed by Primer 3 online. The housekeeping gene 16S rDNA was used as a reference. The qPCR analysis was conducted on a Quantstudio 6 Flex system (Applied Biosystems) using ChamQTM SYBR qPCR Master Mix (Vazyme) with the following cycle profile: 1 cycle at 95°C for 30 s, followed by 40 cycles at 95°C for 5 s, 60°C for 34 s, and 1 cycle at 95°C for 30 s, 95°C 60 s, and 95°C for 15 s. The experiment was repeated three times, each time with triplicates. Data were analyzed using the 2–Δ Δ CT method.
Construction of GFP Expressing Strains and Flow Cytometry Analysis
Strains expressing GFP under the control of Plex were generated by homologous recombination. The fragment of gfp was integrated into the chromosome of L. antibioticus located in front of the lexA start site. We amplified regions ∼600 bp up and downstream of the gfp insertion locus using OH13 genomic DNA as a template, whereas gfp was amplified from a synthetic plasmid. The fragments were cloned into pJQ200SK using homologous recombination with a pEASY® -Uni Seamless Cloning and Assembly Kit (TransGen, China). The construct was transferred into the OH13 wild type and LaPhzB mutant strains through electroporation. After twice homologous recombination, the gfp fragment was integrated into the chromosome of L. antibioticus, subsequently obtaining the OH13 (Plex-GFP) and ΔLaPhzB (Plex-GFP) strains. For quantification of lex expression, strains were grown in 1/10 TSB medium for different time periods. The ΔLaPhzB (Plex-GFP) strain was also cultured in 1/10 TSB medium and supplied with different concentrations of myxin when the OD600 reached 0.6. GFP fluorescence was analyzed by detecting the mean fluorescence intensity (MFI) of 10,000 cells with a CytoFLEX flow cytometer (BD FACSJazz).
Construction of the Phylogenic Tree
The amino acid sequence of LexB was applied to blast research in the Transporter Classification Database (TCDB)1 and NCBI. 29 sequences from TCDB and 4 sequences of Lysobacter species from NCBI were downloaded. The amino acid sequences of the RND family proteins related to LexB were aligned, and a neighbor-joining tree was constructed with MEGA 7.0 (Kumar et al., 2016).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YZ contributed to conceptualization, investigation, writing and editing the manuscript, and funding acquisition. JL, TJ, and RH did the experiments and analyzed data. GX and HX modified the main body of the manuscript. FL contributed to review the manuscript, supervision, and funding acquisition. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by the National Natural Science Foundation of China (31701840), China Agriculture Research System (CARS-28), and Jiangsu Agricultural Science and Technology Innovation Fund [CX (19)3102].
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.618513/full#supplementary-material
References
- Aendekerk S., Diggle S. P., Song Z. J., Høiby N., Cornelis P., Williams P., et al. (2005). The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology 151 1113–1125. 10.1099/mic.0.27631-0 [DOI] [PubMed] [Google Scholar]
- Chen C. H., Huang C. C., Chung T. C., Hu R. M., Huang Y. W., Yang T. C. (2011). Contribution of resistance-nodulation-division efflux pump operon smeU1-V-W-U2-X to multidrug resistance of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 55 5826–5833. 10.1128/AAC.00317-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury G., Sarkar U., Pullen S., William R. W., Rajapakse A., Fuchs-Knotts T., et al. (2012). DNA Strand Cleavage by the Phenazine Di-N-oxide Natural Product Myxin under Both Aerobic and Anaerobic Conditions. Chem. Res. Toxicol. 25 197–206. 10.1021/tx2004213 [DOI] [PubMed] [Google Scholar]
- Cimmino A., Evidente A., Mathieu V., Andolfi A., Lefranc F., Kornienko A., et al. (2012). Phenazines and cancer. Nat. Prod. Rep. 29 487–501. 10.1039/C2NP00079B [DOI] [PubMed] [Google Scholar]
- Coyne S., Rosenfeld N., Lambert T., Courvalin P., Périchon B. (2010). Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54 4389–4393. 10.1128/AAC.00155-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmar J. A., Su C. C., Yu E. W. (2014). Bacterial multidrug efflux transporters. Ann. Rev. Biophysics 43 93–117. 10.1146/annurev-biophys-051013-022855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich L. E., Price-Whelan A., Petersen A., Whiteley M., Newman D. K. (2006). The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61 1308–1321. 10.1111/j.1365-2958.2006.05306.x [DOI] [PubMed] [Google Scholar]
- Evans K., Adewoye L., Poole K. (2001). MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183 807–812. 10.1128/JB.183.3.807-812.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargier E., Mac Aogáin M., Mooij M. J., Woods D. F., Morrissey J. P., Dobson A. D., et al. (2012). MexT functions as a redox-responsive regulator modulating disulfide stress resistance in Pseudomonas aeruginosa. J. Bacteriol. 194 3502–3511. 10.1128/jb.06632-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenberger N., Blankenfeldt W., Breinbauer R. (2017). Recent developments in the isolation, biological function, biosynthesis, and synthesis of phenazine natural products. Bioorgan. Med. Chem. 25 6149–6166. 10.1016/j.bmc.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Heacock-Kang Y., Sun Z., Zarzycki-Siek J., Poonsuk K., McMillan I. A., Chuanchuen R., et al. (2018). Two Regulators, PA3898 and PA2100, modulate the Pseudomonas aeruginosa multidrug resistance MexAB-OprM and EmrAB efflux pumps and biofilm formation. Antimicrob. Agents Chemother. 62 e1459–e1418. 10.1128/AAC.01459-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein U., Butler P. L. (1972). Inhibition of ribonucleic acid synthesis by myxin. Biochemistry 11 1345–1350. 10.1021/bi00758a003 [DOI] [PubMed] [Google Scholar]
- Hollstein U., Van Gemert R. J. J. (1971). Interaction of phenazines with polydeoxyribonucleotides. Biochemistry 10 497–504. 10.1021/bi00779a023 [DOI] [PubMed] [Google Scholar]
- Jiang J. S., Guiza Beltran D., Schacht A., Wright S., Zhang L. M., Du L. C. (2018). Functional and Structural Analysis of Phenazine O-Methyltransferase LaPhzM from Lysobacter antibioticus OH13 and One-Pot Enzymatic Synthesis of the Antibiotic Myxin. ACS. Chem. Biol. 13 1003–1012. 10.1021/acschembio.8b00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166 175–176. 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
- Kumar A., Worobec E. A. (2005). Cloning, sequencing, and characterization of the SdeAB multidrug efflux pump of Serratia marcescens. Antimicrob. Agents Chemother. 49 1495–1501. 10.1128/AAC.49.4.1495-1501.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen J. B., Nielsen J. (2004). Phenazine Natural Products: biosynthesis, synthetic analogues, and biological activity. Chem. Rev. 104 1663–1685. 10.1021/cr020473j [DOI] [PubMed] [Google Scholar]
- Li X. Z., Nikaido H. (2004). Efflux-mediated drug resistance in bacteria. Drugs 64 159–204. 10.2165/00003495-200464020-00004 [DOI] [PubMed] [Google Scholar]
- Li X. Z., Nikaido H. (2009). Efflux-mediated drug resistance in bacteria: an update. Drugs 69 1555–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Z., Plésiat P., Nikaido H. (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28 337–418. 10.2165/11317030-000000000-00000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z. B., Huang L. H., He F., Zhou X. F., Shi Z. R., Zhou J. N., et al. (2019). A substrate-activated efflux pump, DesABC, confers zeamine resistance to Dickeya zeae. mBio 10 e713–e719. 10.1128/mBio.00713-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mfuh A. M., Larionov O. V. (2015). Heterocyclic N-oxides – An emerging class of therapeutic agents. Curr. Med. Chem. 22 2819–2857. 10.2174/0929867322666150619104007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y., Cao L., Gould V. C., Avison M. B., Poole K. (2006). nalD encodes a second repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J. Bacteriol. 188 8649–8654. 10.1128/JB.01342-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolivos S., Cayron J., Dedieu A., Page A., Delolme F., Lesterlin C. (2019). Role of AcrAB-TolC multidrug efflux pump in drug-resistance acquisition by plasmid transfer. Science 364 778–782. 10.1126/science.aav6390 [DOI] [PubMed] [Google Scholar]
- Ogawara H. (2019). Comparison of antibiotic resistance mechanisms in antibiotic-producing and pathogenic bacteria. Molecules 24:3430. 10.3390/molecules24193430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panthee S., Hamamoto H., Paudel A., Sekimizu K. (2016). Lysobacter species: a potential source of novel antibiotics. Arch. Microbiol. 198 839–845. 10.1007/s00203-016-1278-5 [DOI] [PubMed] [Google Scholar]
- Peterson E., Kaur P. (2018). Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 9:2928. 10.3389/fmicb.2018.02928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletzer D., Weingart H. (2014). Characterization and regulation of the resistance-nodulation-cell division-type multidrug efflux pumps MdtABC and MdtUVW from the fire blight pathogen Erwinia amylovora. BMC Microbiol. 14:185. 10.1186/1471-2180-14-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporer A. J., Beierschmitt C., Bendebury A., Zink K. E., Price-Whelan A., Buzzeo M. C., et al. (2018). Pseudomonas aeruginosa PumA acts on an endogenous phenazine to promote self-resistance. Microbiology 164 790–800. 10.1099/mic.0.000657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puopolo G., Tomada S., Pertot I. (2018). The impact of the omics era on the knowledge and use of Lysobacter species to control phytopathogenic micro-organisms. J. Appl. Microbiol. 124 15–27. 10.1111/jam.13607 [DOI] [PubMed] [Google Scholar]
- Quandt J., Hynes M. F. (1993). Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127 15–21. 10.1016/0378-1119(93)90611-6 [DOI] [PubMed] [Google Scholar]
- Sakhtaha H., Koyama L., Zhang Y. H., Moralesa D. K., Fieldsa B. L., Price-Whelana A., et al. (2016). The Pseudomonas aeruginosa efflux pump MexGHI-OpmD transports a natural phenazine that controls gene expression and biofilm development. Proc. Nat. Acad. Sci. U.S.A. 113 E3538–E3547. 10.1073/pnas.1600424113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. K., Shin J. H., Lee K. L., Imlay J. A., Roe J. H. (2013). Comparative study of SoxR activation by redox-active compounds. Mol. Microbiol. 90 983–996. 10.1111/mmi.12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoitsova S. O., Braun Y., Ullrich M. S., Weingart H. (2008). Characterization of the RND-type multidrug efflux pump MexAB-OprM of the plant pathogen Pseudomonas syringae. Appl. Environ. Microbiol. 74 3387–3393. 10.1128/AEM.02866-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z. X., Yi X. X., Cho A., O’Gara F., Wang Y. P. (2016). CpxR activates MexAB-OprM efflux pump expression and enhances antibiotic resistance in both laboratory and clinical nalB-type isolates of Pseudomonas aeruginosa. PLoS Pathog. 12:e1005932. 10.1371/journal.ppat.1005932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter H., Mowla R., Ohene-Agyei T., Ma S. (2015). RND-type drug efflux pumps from Gram-negative bacteria: molecular mechanism and inhibition. Front. Microbiol. 6:377. 10.3389/fmicb.2015.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viktorsson E. Ö, Melling G. B., Aesoy R., Sabir M., Snellingen S., Prandina A., et al. (2017). Total synthesis and antileukemic evaluations of the phenazine 5,10-dioxide natural products iodinin, myxin and their derivatives. Bioorgan. Med. Chem. 25 2285–2293. 10.1016/j.bmc.2017.02.058 [DOI] [PubMed] [Google Scholar]
- Weigele M., Maestrone G., Mitrovic M., Leimgruber W. (1970). Antimicrobial agents structurally related to myxin. Antimicrob. Agents Chemother. 10 46–49. [PubMed] [Google Scholar]
- Xie Y. X., Wright S., Shen Y. M., Du L. D. (2012). Bioactive natural products from Lysobacter. Nat. Prod. Rep. 29 1277–1287. 10.1039/c2np20064c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Liu N., Tang Y. (2020). Recent developments in self-resistance gene directed natural product discovery. Nat. Prod. Rep. 37 879–892. 10.1039/C9NP00050J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Y., Qian G. L., Ye Y. H., Wright S., Chen H. T., Shen Y. M., et al. (2016). Heterocyclic aromatic N-oxidation in the biosynthesis of phenazine antibiotics from Lysobacter antibioticus. Org. Lett. 18 2495–2498. 10.1021/acs.orglett.6b01089 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.