SPRED1 is a negative regulator of the MAPK pathway frequently deleted in human melanoma. Through in vivo modeling in zebrafish and mechanistic analyses in human cell lines, Ablain et al. demonstrate that SPRED1 inactivation confers resistance to targeted inhibition of V600 mutant BRAF, the most common driver of melanoma.
Abstract
Functional evaluation of genetic lesions can discover a role in cancer initiation and progression and help develop novel therapeutic strategies. We previously identified the negative MAPK regulator SPRED1 as a novel tumor suppressor in KIT-driven melanoma. Here, we show that SPRED1 is also frequently deleted in human melanoma driven by mutant BRAF. We found that SPRED1 inactivation in human melanoma cell lines and primary zebrafish melanoma conferred resistance to BRAFV600E inhibition in vitro and in vivo. Mechanistically, SPRED1 loss promoted melanoma cell proliferation under mutant BRAF inhibition by reactivating MAPK activity. Consistently, biallelic deletion of SPRED1 was observed in a patient whose melanoma acquired resistance to MAPK-targeted therapy. These studies combining work in human cells and in vivo modeling in zebrafish demonstrate a new mechanism of resistance to BRAFV600E inhibition in melanoma.
Introduction
Next-generation sequencing of human tumors performed by The Cancer Genome Atlas (TCGA) Research Network has uncovered a plethora of genomic abnormalities that may be used to identify potential novel oncogenes or tumor suppressors (McLendon et al., 2008). Most functional analyses have focused on recurrent point mutations, but the role of copy-number alterations remains understudied. Functional assessment of amplified or deleted chromosomal regions is necessary to formally implicate candidate genes in malignant processes and discover new therapeutically actionable cancer vulnerabilities.
Melanoma is one of the tumor types exhibiting the most genetic alterations (Alexandrov et al., 2013). Its formation is fueled by the hyperactivation of the MAPK pathway through activating mutations in genes like BRAF, NRAS, and KIT or via the inactivation of negative regulators of the pathway, such as NF1 (Akbani et al., 2015; Hayward et al., 2017). Close to 50% of cutaneous melanoma are driven by the BRAFV600E mutant, and specific BRAFV600E inhibitors have shown remarkable clinical efficacy (Bollag et al., 2010; Flaherty et al., 2010; Chapman et al., 2011; Hauschild et al., 2012). However, resistance to these targeted therapies invariably arises within a year of treatment start (Chapman et al., 2011; McArthur et al., 2014; Wagle et al., 2011). In most cases, resistance is due to the reactivation of the MAPK pathway through additional genomic lesions affecting BRAF itself or other players of the pathway (Wagle et al., 2011; Johannessen et al., 2010; Nazarian et al., 2010; Shi et al., 2012).
By analyzing copy-number alterations in human melanoma, we recently identified frequent biallelic deletions of SPRED1 (Sprouty-related Ena/VASP homology 1 [EVH1] domain containing 1; Ablain et al., 2018). SPRED1 is a negative regulator of the MAPK pathway (Nonami et al., 2004; Wakioka et al., 2001). In humans, inactivating germline mutations in SPRED1 cause Legius syndrome, a developmental disorder characterized by skin pigmentation abnormalities reminiscent of neurofibromatosis type 1 (Hirata et al., 2016; Brems et al., 2007). Neurofibromatosis type 1 is due to the loss of function of the neurofibromin (NF1) gene that encodes a GTPase (guanosine triphosphatase)-activating protein that catalyzes the conversion of active GTP-bound Ras into inactive guanosine diphosphate–bound Ras, resulting in the down-regulation of the MAPK pathway (Xu et al., 1990a, b; Ballester et al., 1990; Martin et al., 1990). The similarities between Legius syndrome and neurofibromatosis type 1 suggested that SPRED1 and NF1 share biological functions. Indeed, at the molecular level, SPRED1 has been shown to directly recruit NF1 to the plasma membrane, thus allowing it to inhibit Ras activity (Stowe et al., 2012). Structural insights have recently confirmed the physical interactions between SPRED1, NF1, and Ras (Yan et al., 2020). We previously demonstrated that SPRED1 acts as a tumor-suppressor gene in melanoma, especially in the context of KIT mutations (Ablain et al., 2018).
Here, we report that SPRED1 deletions can also be found in melanoma driven by BRAF mutants. Modeling SPRED1 loss in adult zebrafish melanoma and human melanoma cell lines, we show that it confers resistance to BRAF inhibition by sustaining low levels of MAPK signaling. We also found SPRED1 deletions associated with acquired resistance to MAPK inhibition in a patient with melanoma. Our data thus nominate SPRED1 loss as a new mechanism of resistance to MAPK-targeted therapy in melanoma.
Results and discussion
SPRED1 is frequently deleted in human cutaneous melanoma
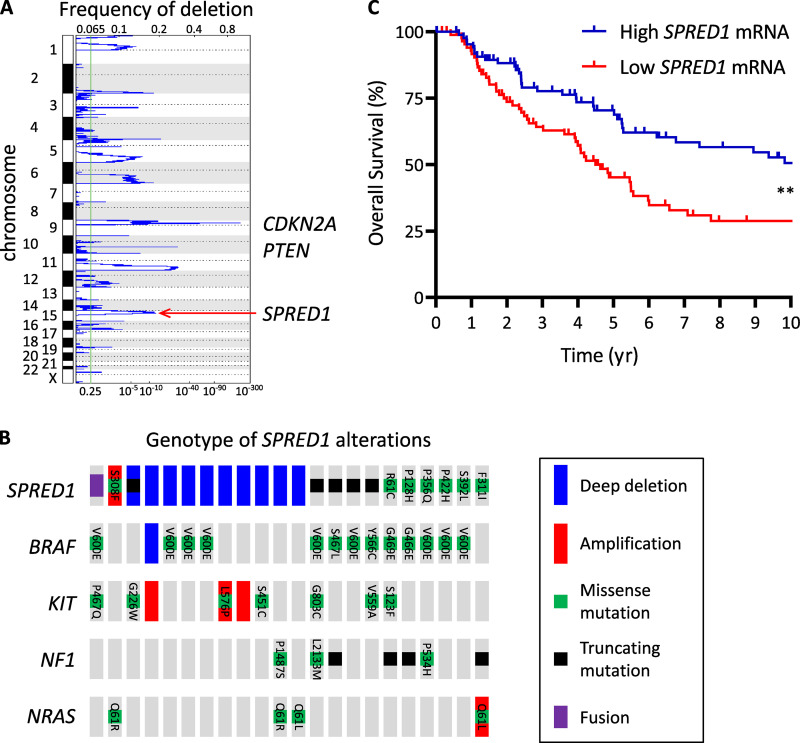
SPRED1 is the only gene found in a frequent focal deletion on chromosome 15 (chr15:38,244,770–38,745,783; overall frequency, 24%; q-value = 0.0076) identified by GISTIC analysis of a cohort of 363 human cutaneous melanoma samples with both copy-number and mutation information from the Pan-Cancer TCGA dataset (Fig. 1 A). Overall, SPRED1 was altered in 22 of 363 cases (6%). Homozygous deletions were present in 10 patients with cutaneous melanoma (2.7%), while 4 and 7 of 363 cases (1.1% and 1.9%) harbored truncating mutations and missense mutations, respectively (Fig. 1 B). A gene fusion that likely acts as a truncating event was also detected in one patient. We can thus estimate SPRED1 loss-of-function alterations at 4.1% of human cutaneous melanomas. Note that some point mutations may also result in loss of function, especially those affecting the EVH1 and Sprouty-related regions, the functional domains of SPRED1.
Figure 1.
SPRED1 is frequently deleted in human cutaneous melanoma. (A) Diagram representing copy-number losses in the TCGA cohort of human cutaneous melanomas (363 samples). Frequent deletions of CDKN2A on chromosome 9 and PTEN on chromosome 10 are highlighted. SPRED1 was identified by GISTIC analysis in a recurrent focal deletion on chromosome 15. (B) Driver lesions in oncogenes occurring with SPRED1 alterations in human cutaneous melanoma (total, 363 samples). (C) Survival curves of patients with melanoma stratified according to SPRED1 expression levels. High SPRED1, 88 samples (25%) with highest mRNA expression; low SPRED1, 88 samples (25%) with lowest mRNA expression. **, P = 0.0012, log-rank test.
SPRED1 is also homozygously deleted in 5% of mesothelioma (4 of 82 cases) and 3% of ovarian cancer (12 of 398 cases). It is inactivated in 4% of lung adenocarcinoma (19 of 507 cases), mostly through deep deletions and truncating mutations, similar to melanoma and consistent with the critical role of the MAPK pathway as an oncogenic driver of this tumor type. In addition, SPRED1 is altered in 7% of uterine corpus endometrial carcinoma (34 of 509 cases). In this tumor type, SPRED1 alterations are mostly missense or truncating mutations, thus differing from the pattern observed in melanoma. However, these alterations significantly co-occur with NF1 loss of function as well as with receptor tyrosine kinase mutations (in KIT or EGFR), suggesting that a subtype of uterine cancers is driven by MAPK activation.
Importantly, low SPRED1 expression correlates with poor prognosis in patients with melanoma (Fig. 1 C). Among the samples with SPRED1 alterations, genetic lesions in CDKN2A and TP53 accounted for the majority of tumor-suppressor losses (Fig. S1 A). While KIT mutations or amplifications were frequent, as previously noted (Ablain et al., 2018), 13 out of 22 melanoma samples with SPRED1 alterations exhibited BRAF mutations (Fig. 1 B). These genomic data indicate that SPRED1 is recurrently lost in BRAF-driven human melanoma, prompting us to test its role by genetic manipulation in human melanoma cell lines and zebrafish models.
Figure S1.
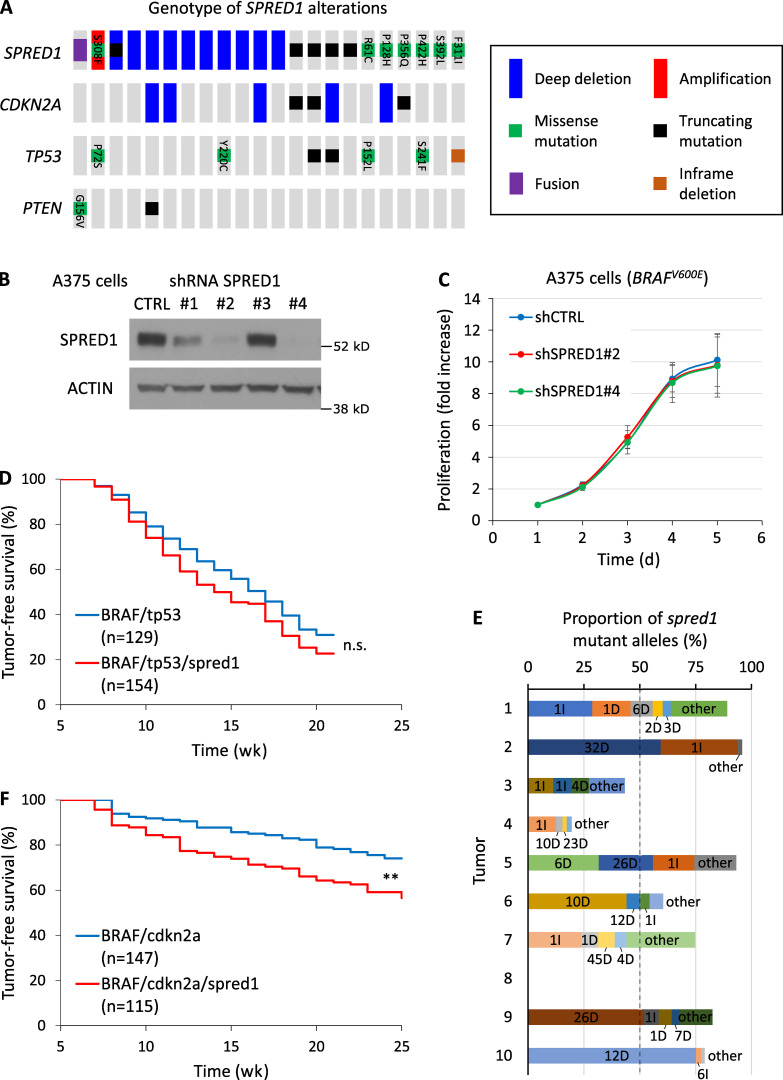
SPRED1 inactivation modestly alters melanoma growth in vitro and in vivo. (A) Driver lesions in tumor-suppressor genes occurring with SPRED1 alterations in human cutaneous melanoma (total, 363 samples). (B) Western blot analysis of SPRED1 levels in A375 melanoma cells stably transduced with shRNAs directed against SPRED1. shRNA#2 and #4 were selected for SPRED1 down-regulation experiments. Representative of two independent experiments. (C) Proliferation of BRAF-driven A375 human melanoma cells expressing either a control shRNA (shCTRL) or shRNAs against SPRED1 (shSPRED1#2 and #4). Mean ± SD of five independent experiments. P > 0.05 (not significant) compared with shCTRL, paired two-tailed t test. (D) Tumor-free survival curves of casper zebrafish injected with vectors expressing BRAFV600E and targeting either tp53 or both tp53 and spred1. Pooled data of three independent experiments. P = 0.08 (n.s., not significant), log-rank test. (E) Proportion of spred1 mutant alleles in 10 primary BRAF/tp53/spred1 zebrafish melanomas, as measured by deep sequencing of the CRISPR target loci. Indels are indicated as the number of base pairs altered followed by the type of indel: insertion (I) or deletion (D). (F) Tumor-free survival curves of casper zebrafish injected with vectors expressing BRAFV600E and targeting either cdkn2a or both cdkn2a and spred1. Pooled data of three independent experiments. **, P = 0.001, log-rank test.
SPRED1 inactivation modestly alters melanoma growth in vitro and in vivo
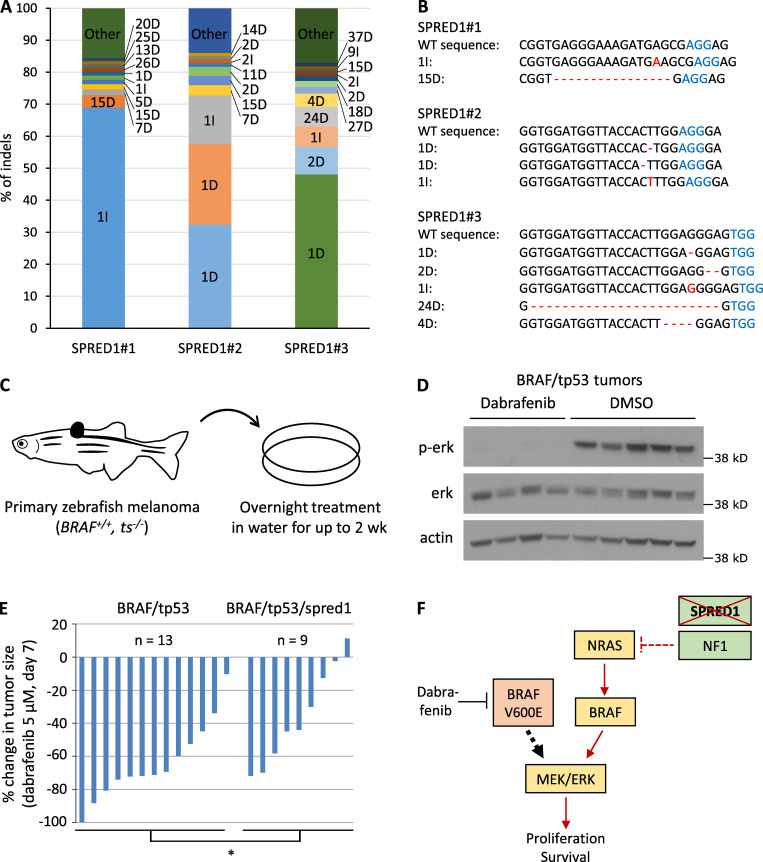
We first investigated the effects of SPRED1 loss on melanoma growth. We selected two efficient shRNAs against SPRED1 (sh#2 and sh#4; Fig. S1 B) and observed that SPRED1 down-regulation did not affect cell proliferation in BRAFV600E-driven human melanoma cell lines (Fig. S1 C). To test the role of SPRED1 in melanoma in vivo, we inactivated spred1 by CRISPR in a BRAF-driven zebrafish melanoma model using a method that we developed previously (Ablain et al., 2015, 2018; Ceol et al., 2011). This only slightly, but not significantly, affected tumor onset in conjunction with tp53 loss (Fig. S1 D), even though 70% of tumors displayed inactivating indels at spred1 CRISPR target locus, as assessed by deep sequencing (Fig. S1 E). Yet, spred1 inactivation accelerated melanoma onset in the context of cdkn2a loss that yields less aggressive tumors in zebrafish (Fig. S1 F). These in vivo data support the existence of a selective pressure for SPRED1 loss, consistent with our analyses of human melanoma genetics. Overall, these results indicate that SPRED1 loss modestly increases melanoma initiation and proliferation in the background of BRAF activating mutations.
Interestingly, we noticed that four of the BRAF mutations cooccurring with SPRED1 alterations did not affect the V600 residue that concentrates 80% of BRAF mutations in melanoma (Fig. 1 B). Three of them (G466E, S467L, and G469E) are known as class 3 BRAF mutants (Yao et al., 2017). Unlike BRAFV600E, which exhibits enhanced kinase activity and acts as a monomer, class 3 BRAF mutants display low kinase activity but increased binding to wild-type Ras and CRAF, which results in MAPK pathway activation. This genetic observation raises the intriguing possibility that the effects of SPRED1 loss of function on melanoma growth may be more evident in the context of weaker oncogenic drivers that act through the wild-type Ras/RAF pathway.
SPRED1 down-regulation reduces sensitivity to BRAF inhibition in BRAF-driven human melanoma cell lines by sustaining MAPK activity
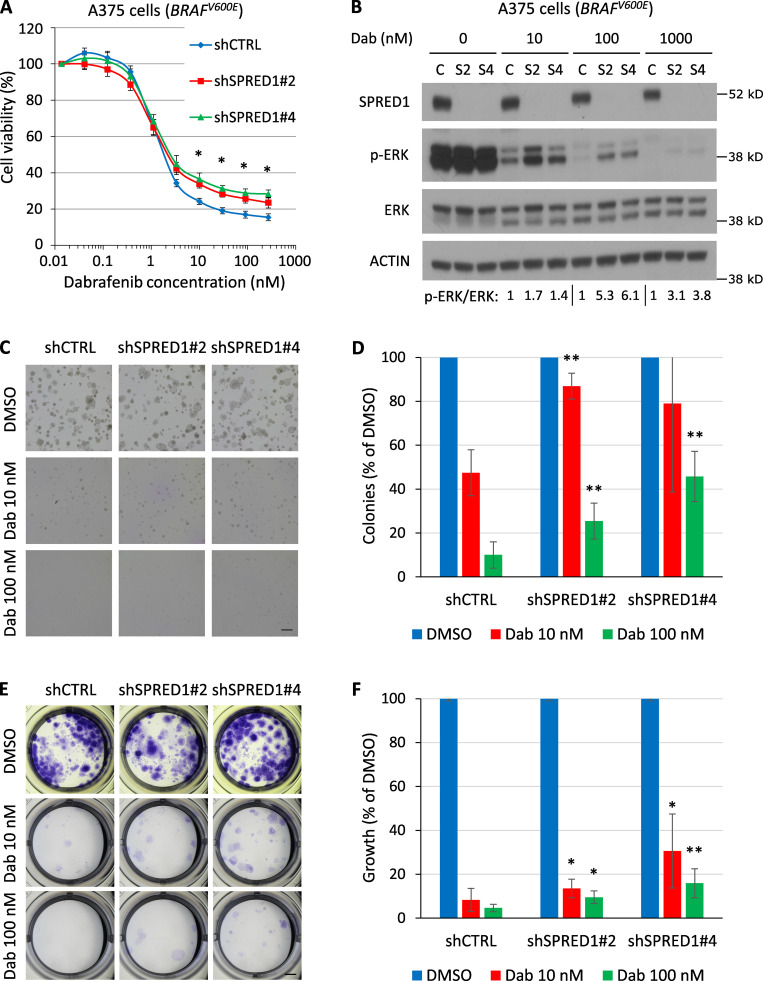
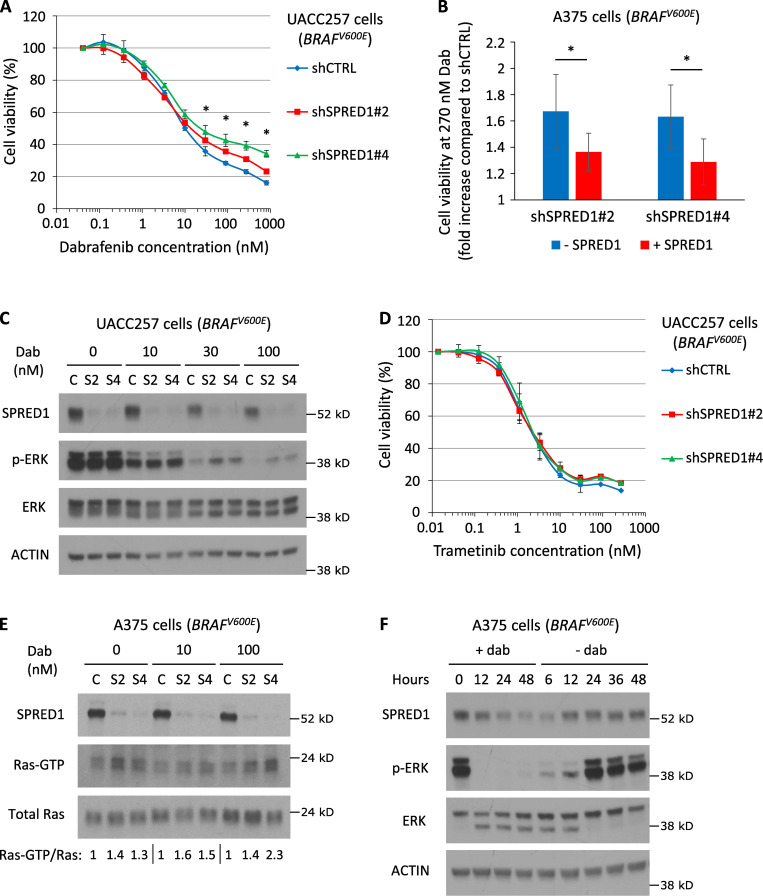
Because of the functional similarities between NF1 and SPRED1 and the fact that NF1 loss confers resistance to BRAF inhibition in BRAF mutant melanoma (Whittaker et al., 2013; Maertens et al., 2013), we examined the impact of SPRED1 loss on oncogene inhibition. SPRED1 down-regulation significantly decreased sensitivity to the BRAFV600E inhibitor dabrafenib, as assessed by cell viability over a range of drug concentrations in two different BRAF-driven melanoma cell lines, A375 and UACC257 (Fig. 2 A and Fig. S2 A). SPRED1 inactivation did not significantly alter the IC50 value for dabrafenib, suggesting that the effects of SPRED1 loss were only visible upon more complete inhibition of BRAFV600E oncogenic activity. This phenotype was specific for SPRED1 down-regulation, as SPRED1 overexpression partially restored sensitivity to the drug (Fig. S2 B). Resistance to BRAFV600E inhibition was associated with the persistence of a low but detectable level of ERK phosphorylation in dabrafenib-treated cells expressing shRNAs against SPRED1 (Fig. 2 B and Fig. S2 C). This residual MAPK activity downstream of SPRED1 down-regulation at least partially explained the observed resistance, since the latter was abrogated by treatment with the MEK inhibitor trametinib (Fig. S2 D). At the molecular level, SPRED1 down-regulation increased Ras activity both in untreated conditions and under BRAF inhibition, as evidenced by higher levels of Ras-GTP (Fig. S2 E), in line with the idea that SPRED1 modulates the MAPK pathway by inhibiting the activity of wild-type Ras. Interestingly, SPRED1 protein levels decreased following MAPK pathway inhibition by dabrafenib in BRAFV600E-driven human melanoma cells and increased in response to the restoration of MAPK activity after drug washout (Fig. S2 F), suggesting that SPRED1 itself is regulated by the output of the pathway. SPRED1 thus acts as a negative feedback regulator of the MAPK pathway. The effect of long-term treatment with dabrafenib on the colony-forming capacity (Fig. 2, C and D) and growth of melanoma cells (Fig. 2, E and F) was also significantly reduced by SPRED1 down-regulation. Together, these findings demonstrate that SPRED1 loss decreases the sensitivity of BRAF-driven melanoma cells to BRAF inhibition by sustaining partial MAPK pathway activity.
Figure 2.
SPRED1 down-regulation reduces sensitivity to BRAF inhibition in BRAF-driven human melanoma cell lines by sustaining MAPK activity in vitro. (A) Viability of BRAF-driven A375 human melanoma cells stably expressing a control shRNA (shCTRL) or shRNAs directed against SPRED1 (shSPRED1#2 and #4) and treated with increasing concentrations of the BRAFV600E inhibitor dabrafenib for 4 d. Mean ± SD of three independent experiments; *, P < 0.05 for both shRNAs against SPRED1 compared with the control shRNA, paired two-tailed t test. (B) Western blot analysis of MAPK pathway activity in the cells described in A treated with dabrafenib (Dab) for 6 h. C, shCTRL; S2, shSPRED1#2; S4, shSPRED1#4. Signal intensity ratio of p-ERK to total ERK is indicated. Note the residual p-ERK levels under strong BRAF inhibition in cells with reduced SPRED1. Representative of three independent experiments. (C) Representative images of colonies formed in Matrigel by the cells described in A after 14 d of treatment with dabrafenib or DMSO (vehicle). Scale bar, 500 µm. (D) Number of colonies quantified from the experiments described in C, relative to DMSO-treated cells. Mean ± SD of five independent experiments; **, P < 0.01 compared with shCTRL, paired two-tailed t test. (E) Representative images of clones formed by the cells described in A after 14 d of treatment with dabrafenib and stained by crystal violet. Scale bar, 2 mm. (F) Intensity of crystal violet staining from the experiments described in E relative to DMSO-treated cells. Mean ± SD of six independent experiments; *, P < 0.05; **, P < 0.01 compared with shCTRL, paired two-tailed t test.
Figure S2.
SPRED1 inactivation confers resistance to BRAF inhibition in BRAF-driven human melanoma cell lines. (A) Viability of BRAF-driven UACC257 human melanoma cells stably expressing a control shRNA (shCTRL) or shRNAs directed against SPRED1 (shSPRED1#2 and #4) and treated with increasing concentrations of the BRAFV600E inhibitor dabrafenib for 4 d. Mean ± SD of three independent experiments; *, P < 0.05 for both shRNAs against SPRED1 compared with the control shRNA, paired t test. (B) Relative viability at 270 nM dabrafenib between A375 cells expressing shRNAs against SPRED1 (shSPRED1#2 and #4) and cells expressing a control shRNA in the presence (+SPRED1) or absence (−SPRED1) of inducible SPRED1 expression. Mean ± SD of four independent experiments. shSPRED1#2: *, P = 0.03; shSPRED1#4: *, P = 0.01, paired t test. (C) Western blot analysis of MAPK pathway activity in the cells described in A treated with the indicated concentrations of dabrafenib (Dab) for 6 h. C, shCTRL; S2, shSPRED1#2; S4, shSPRED1#4. Representative of three independent experiments. (D) Viability of the cells described in A treated with increasing concentrations of the MEK inhibitor trametinib for 4 d. Mean ± SD of three independent experiments; P > 0.05 (not significant), paired t test. (E) Western blot analysis of SPRED1, active Ras (Ras-GTP) and total Ras levels in BRAF-driven A375 human melanoma cells stably expressing a control shRNA (C) or shRNAs directed against SPRED1 (S2 and S4) and treated with the indicated concentrations of dabrafenib for 6 h. Signal intensity ratio of Ras-GTP to total Ras is indicated. Representative of two independent experiments. (F) Western blot analysis of MAPK pathway activity and SPRED1 levels in BRAF-driven A375 human melanoma cells treated with 100 nM dabrafenib (+dab) for the indicated durations before drug washout and further culture in the absence of treatment (−dab) for the indicated durations. Representative of three independent experiments.
SPRED1 loss is selected for in human melanoma cells under continuous BRAF inhibition
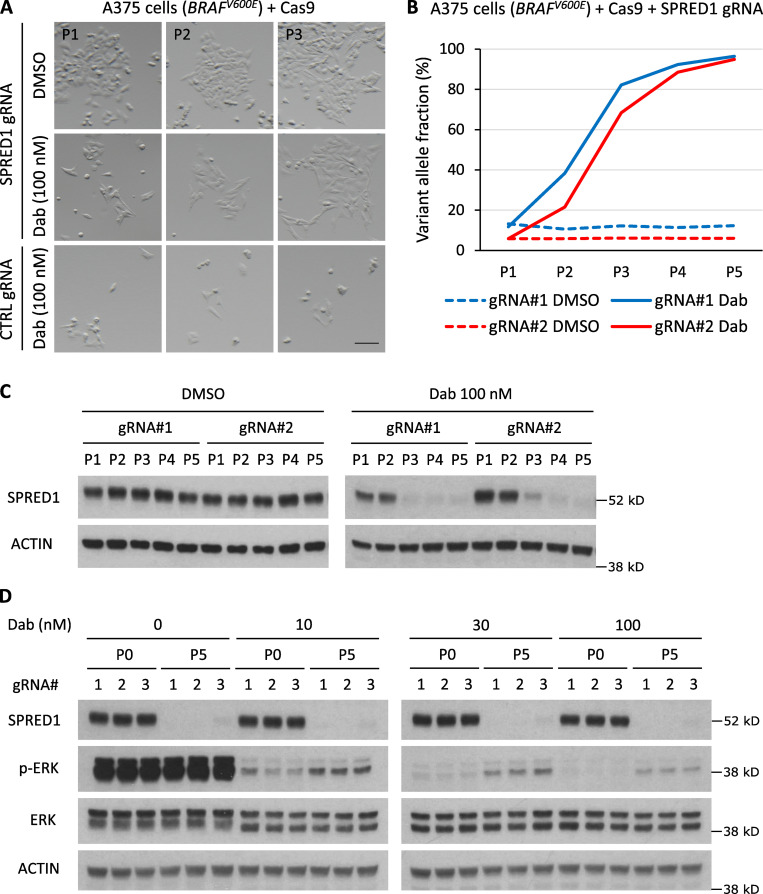
We confirmed these results with a CRISPR approach using vectors expressing Cas9 and three independent guide RNAs (gRNAs) targeting SPRED1. We suboptimally transfected BRAF-driven A375 cells to mimic the effects of mosaic acquisition of SPRED1 loss and subjected them to long-term BRAF inhibition. We then assessed the prevalence of SPRED1 inactivation overtime in these heterogeneous cultures at the genomic level by sequencing of the CRISPR target sites and at the protein level by Western blotting. Dabrafenib treatment dramatically reduced the growth of A375 cells. Yet, we observed the rapid emergence of clones resistant to dabrafenib treatment after one passage, specifically in cultures transfected with SPRED1 gRNAs (Fig. 3 A). The proportion of SPRED1 mutant alleles sharply increased in dabrafenib-treated cultures while remaining constant in vehicle-treated cultures (Fig. 3 B). Deep sequencing revealed that the vast majority of these CRISPR variants exhibited frameshift mutations (Fig. S3, A and B). Accordingly, SPRED1 protein levels remained constant over five passages in the absence of drugs but dropped within three passages of dabrafenib treatment (Fig. 3 C), indicating that SPRED1-deficient cells were rapidly selected for. Within five passages under drug pressure, the genotype of transfected cultures had thus evolved from mostly wild type to almost completely knockout for SPRED1. As expected, SPRED1 knockout cells displayed residual MAPK signaling under dabrafenib treatment compared with control cells (Fig. 3 D). These data show that SPRED1 loss confers a selective advantage to melanoma cells under continuous pharmacologic BRAF inhibition by increasing cell proliferation and/or survival in vitro.
Figure 3.
SPRED1 loss is selected for in human melanoma cells under continuous BRAF inhibition. (A) Representative pictures at indicated passages (P) of cultures of BRAF-driven A375 human melanoma cells transiently transfected with a vector expressing Cas9 and either a control gRNA or a gRNA targeting SPRED1 and treated with DMSO (vehicle) or 100 nM dabrafenib (Dab). Scale bar, 100 µm. (B) Evolution of frameshift variant allele fraction in cultured A375 human melanoma cells transiently transfected with two independent vectors targeting SPRED1 by CRISPR and treated with either DMSO or 100 nM dabrafenib over five passages. Representative of three independent experiments. (C) Western blot analysis of SPRED1 levels in the cultures described in B. Representative of three independent experiments. (D) Western blot analysis of MAPK pathway activity in A375 human melanoma cells transiently transfected with three independent vectors targeting SPRED1 by CRISPR and stimulated with 0, 10, 30, or 100 nM dabrafenib before any treatment (P0) or after five passages in 100 nM dabrafenib (P5). Representative of three independent experiments.
Figure S3.
In vivo treatment of zebrafish melanomas reveals different sensitivities to BRAF inhibition between BRAF/tp53 and BRAF/tp53/spred1 tumors. (A) Distribution of SPRED1 mutant alleles in cultures of BRAF-driven A375 human melanoma cells transiently transfected with a vector expressing Cas9 and either of three gRNAs targeting SPRED1 after five passages in 100 nM dabrafenib, as measured by deep sequencing of the CRISPR target loci. Indels are indicated as the number of base pairs altered followed by the type of indel: insertion (I) or deletion (D). (B) Most common mutant alleles found in the cells described in A. (C) Schematic representation of the procedure to treat adult zebrafish bearing primary tumors. Fish were immerged in 50 ml water containing the drug for 12 h (overnight). Treatment was repeated daily for 14 d. ts, tumor suppressor. (D) Western blot analysis of MAPK pathway activity in primary zebrafish tumors treated with DMSO (control) or 5 µM dabrafenib for 3 d following the protocol described in C. BRAF, BRAFV600E. Representative of two independent experiments. (E) Quantification of tumor size after 7 d of dabrafenib treatment relative to tumor size before treatment in zebrafish bearing BRAF/tp53 (n = 13) or BRAF/tp53/spred1 (n = 9) tumors. Pooled data of three independent experiments. *, P = 0.03, two-tailed t test. (F) Diagram summarizing the effect of SPRED1 loss in BRAF-driven melanoma. Under treatment with the BRAFV600E inhibitor dabrafenib, SPRED1 loss annihilates the inhibitory activity of NF1 on Ras, which results in the activation of the RAF/MEK/ERK cascade that fuels cell survival and proliferation.
SPRED1 loss confers resistance to BRAF inhibition in BRAF-driven zebrafish melanoma in vivo
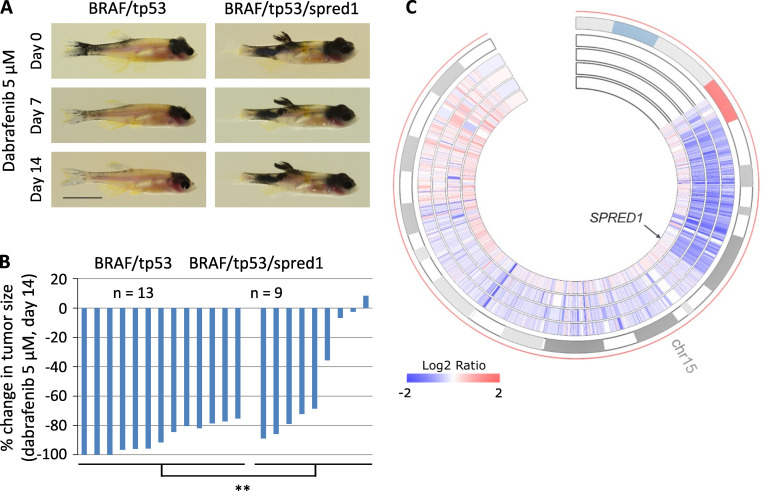
We next assessed the effect of SPRED1 inactivation on melanoma sensitivity to targeted therapy in vivo. To test the response of primary zebrafish melanoma to pharmacological challenge, we treated adult zebrafish daily for 14 d in a small volume of drugs (Fig. S3 C). Treatment of BRAF-driven melanoma with the BRAFV600E inhibitor dabrafenib abrogated MAPK signaling in tumors (Fig. S3 D). Primary spred1 CRISPR tumors in zebrafish displayed a reduced response to dabrafenib compared with control tumors after both 7 and 14 d of treatment (Fig. 4, A and B; and Fig. S3 E). These results indicate that SPRED1 loss also reduces the sensitivity of BRAF-driven melanoma cells to BRAF inhibition in vivo.
Figure 4.
SPRED1 loss confers resistance to BRAF inhibition in BRAF-driven zebrafish melanoma in vivo and is associated with acquired resistance to targeted therapy in a patient with melanoma. (A) Representative pictures of zebrafish bearing BRAF/tp53 or BRAF/tp53/spred1 tumors and treated daily with 5 µM dabrafenib for 14 d. Scale bar, 1 cm. (B) Quantification of tumor size after 14 d of dabrafenib treatment relative to tumor size before treatment in zebrafish with BRAF/tp53 (n = 13) or BRAF/tp53/spred1 (n = 9) tumors. Pooled data of three independent experiments. **, P = 0.01, two-tailed t test. (C) Circos representation of chromosome 15 copy-number changes of four tumors from a patient who was treated with and responded to dabrafenib plus trametinib. The outermost layer represents the chromosomal regions of chromosome 15, and the inner heatmaps indicate copy-number changes (log2 ratio) averaged in 1-kb genomic windows (from inner to outer: baseline, DD-DP1, DD-DP2, and DD-DP3). Arrow indicates the location of SPRED1.
SPRED1 deletion is associated with acquired resistance to MAPK inhibition in a patient with melanoma
When human melanoma cell lines and clinical melanoma are exposed to MAPK inhibitors chronically, the resulting acquired resistant cells and tumors often harbor multiple genetic and nongenetic alterations capable of driving the resistance phenotype (Hugo et al., 2015). With combinatorial targeting of the MAPK pathway using BRAFV600E and allosteric MEK inhibitors, multiple genetic alterations often converge to reactivate the MAPK pathway (Moriceau et al., 2015). In the context of the above functional data, we asked whether SPRED1 loss contributes to acquired resistance clinically by reanalyzing whole-exome sequence data for patient-derived tumors. A patient (patient 11 in Moriceau et al. [2015] and patient 22 in Hugo et al. [2015]) with metastatic BRAFV600E melanoma was treated with dabrafenib plus trametinib. Comparison of whole-exome sequencing data from a pretreatment tumor (January 2013), three distinct double-drug disease-progressive (DD-DP) tumors (October 2013), and the patient-matched normal genomic DNA detected resistance-associated copy-number loss of a region within chromosome 15 encompassing SPRED1 in all three DD-DP tumors relative to the pretreatment tumor (Fig. 4 C). This finding is consistent with a key role of MAPK reactivation, which is driven at least in part by SPRED1 deletion, underlying this patient’s disease progression.
During chronic selection with MAPK inhibitors (especially with dual MAPK pathway inhibition), multiple but convergent genetic and nongenetic alterations are likely operative in fully reactivating the MAPK pathway. Accordingly, in the patient presented here, low-level BRAFV600E copy-number gain and DUSP4 copy-number loss already coexist in all three acquired resistant tumors in addition to the SPRED1 copy-number loss (Fig. 4 C). It is not known whether these are subclonal and mutually exclusive events, but functional studies indicate that distinct MAPK alterations can cooperate quantitatively (nonredundantly) to achieve a more robust resistant phenotype (Moriceau et al., 2015). DUSP and SPROUTY family members are both transcriptional targets of phospho-ERK (p-ERK) and negative feedback regulators of the MAPK pathway, albeit at distinct steps of the signaling cascade (Pratilas et al., 2009). Mechanistically, it is conceivable that recurrent drivers of MAPK inhibitor resistance (e.g., BRAF amplification and NRAS mutations) render BRAFV600E signaling in resistant tumor cells more sensitive to negative feedback regulation. Therefore, there would be selective pressure to coacquire alterations that disable negative feedback at multiple levels of the MAPK pathway.
Collectively, our results demonstrate that deletions of the negative MAPK pathway regulator SPRED1 confer resistance to MAPK inhibition in mutant BRAF–driven melanoma in human cell lines, animal models, and patients. We found loss-of-function alterations of SPRED1 through deep deletions or truncating mutations in 4% of melanomas. In the context of BRAFV600E, SPRED1 loss modestly affected melanoma initiation in vivo or melanoma growth in the absence of treatment in vitro. While this may explain the relatively low frequency of SPRED1 deletions in human cutaneous melanoma, it also contrasts with our observations in KIT-driven melanoma, where SPRED1 inactivation significantly accelerated tumor onset and increased cell proliferation (Ablain et al., 2018). This difference may be due to the position at which SPRED1 exerts its tumor-suppressor activity in the MAPK pathway. Consistent with our data showing that SPRED1 loss enhances Ras activity in melanoma cells, previous reports have indicated that SPRED1 directly represses Ras, thus acting downstream of KIT but upstream of RAF (Stowe et al., 2012). SPRED1 loss may therefore strongly potentiate the MAPK signal induced by KIT activation that transits through wild-type Ras, while its effect on MEK and ERK phosphorylation levels is masked in the context of BRAFV600E that bypasses wild-type Ras and directly activates MEK (Fig. S3 F). Combined with the fact that SPRED1 is likely a weaker MAPK regulator than BRAFV600E, this may also explain why the effects of SPRED1 loss of function on melanoma cell proliferation are only visible upon profound inhibition of mutant BRAF (i.e., under relatively high dabrafenib concentrations). Upon BRAF inhibition, melanoma cells may rely on basal MAPK signaling for survival. Under these circumstances, we showed that SPRED1 loss by itself increases residual MAPK activity and sustains low levels of cell proliferation, thus allowing melanoma cells to resist treatment acutely (Fig. S3 F). Our findings uncover SPRED1 as one of the feedback mechanisms that are altered in melanoma and whose loss confers resistance to MAPK inhibition.
Materials and methods
DNA constructs
To down-regulate SPRED1 in human melanoma cell lines, we used pLKO shRNA lentiviral vectors (Sigma-Aldrich) with the following hairpin sequences: shSPRED1#2, 5′-CCGGGAGCATGTTGTATCATTGTATCTCGAGATACAATGATACAACATGCTCTTTTTG-3′ (TRCN0000056712); shSPRED1#4, 5′-CCGGGAATACGTACAGCGGCAAATACTCGAGTATTTGCCGCTGTACGTATTCTTTTTTG-3′ (TRCN0000417109).
To inactivate SPRED1 in human melanoma cell lines, we used the LentiCRISPR lentiviral vector (Shalem et al., 2014) with the following gRNAs: SPRED1#1, 5′-GCGGTGAGGGAAAGATGAGCG-3′; SPRED1#2, 5′-GGTGGATGGTTACCACTTGG-3′; SPRED1#3, 5′-GTGGTTACCACTTGGAGGGAG-3′.
To inactivate spred1 in zebrafish melanomas, we used the CRISPR MiniCoopR vector (Ablain et al., 2018) that targets genes specifically in the melanocytes of the zebrafish. The following gRNAs were introduced into the CRISPR MiniCoopR vector by restriction cloning using the BseRI enzyme: tp53, 5′-GGTGGGAGAGTGGATGGCTG-3′; cdkn2a, 5′-GTTCTGGCAGCGTCGTGCAG-3′; spred1, 5′-GGCGTCCGCCGGGCTCTGGA-3′.
Zebrafish handling
Zebrafish were handled according to our vertebrate animal protocol that has been approved by Boston Children’s Hospital Animal Care Committee and includes detailed experimental procedures for all in vivo experiments described in this paper. Zebrafish of casper strain were bred and embryos were collected for microinjection. The Tol2 transgenesis technology enables the stable integration of expression vectors into the fish genome (Kawakami et al., 2004). 25 pg of DNA constructs and 25 pg of Tol2 mRNA were injected into one cell–stage embryos. After microinjection, embryos were raised in E3 medium at 28.5°C.
Embryos were sorted for melanocyte rescue at 96 h after fertilization and raised to adulthood (20–25 zebrafish per 3.5-liter tank). Adult fish were scored weekly for melanoma formation starting at the first appearance of raised lesions. Tumor scoring in adult zebrafish was blinded by deidentifying each tank for the duration of the experiment and only revealing the genotype of each tank at the time of final analysis. Experiments were independently repeated at least three times. Statistical analysis on Kaplan–Meier survival curves was performed using log-rank (Mantel–Cox) test.
Adult fish treatment
Adult zebrafish were treated daily by overnight (12 h) immersion in 50 ml of water containing the drug in Petri dishes. Treatment was performed with DMSO or 5 µM dabrafenib (LC Laboratories) for 14 d. The concentration of dabrafenib was determined by a dose-escalation study starting at 100 nM dabrafenib to identify the maximum tolerated dose in adult zebrafish bearing primary melanomas.
Adult fish were photographed using a Nikon D3100 camera with an AF-S Micro NIKKOR 60-mm lens. Tumor size was measured with ImageJ.
TCGA data analysis
SPRED1 was identified in a frequent focal deletion by GISTIC analysis of the TCGA data for human melanoma (Beroukhim et al., 2007). We focused our analysis on a cohort of 363 patients with both copy-number and mutation information. The various alterations in select genes were examined using Oncoprint (Cerami et al., 2012; Gao et al., 2013). Survival data were available for 352 patients.
Cell culture
A375 and UACC257 cell lines were purchased from ATCC and cultured in DMEM (Life Technologies), 10% FBS (Atlanta Biologicals), 1X GlutaMAX (Life Technologies), and 1% penicillin-streptomycin (Life Technologies). Cells were grown at 37°C in 5% CO2. All cultures were regularly checked for the presence of mycoplasma.
Lentiviral particles were produced by cotransfection of 293T cells with pLKO or pHAGE vectors and packaging plasmids pVSV-G and psPAX2 using FuGENE HD (Promega). Viral particles were harvested 48 and 72 h after transfection, concentrated by overnight polyethylene glycol precipitation, resuspended in PBS, and stored at −80°C. Human melanoma cell lines were overlaid with viral particles diluted in medium supplemented with 5 µg/ml polybrene (Sigma-Aldrich) for 12 h at 37°C. 48 h after transduction, infected cells were selected with 1 µg/ml puromycin (Gibco) and maintained under selection by replacing the antibiotics every 48 h.
A375 cells were transfected with pLenti-CRISPR vectors using FuGENE HD (Promega).
Proliferation and drug sensitivity were measured using the CellTiter Glo (Promega) luminescent cell viability assay, according to the manufacturer’s instructions. Cells were seeded at an initial density of 1,000 cells/well (for A375 cell line) or 3,000 cells/well (for UACC257 cell line) in 394-well plates in duplicate or triplicate wells. Experiments were repeated at least three times independently, and a paired two-tailed t test was used to assess significance between groups.
To measure cell growth under long-term drug treatment, A375 cells were plated at a density of 300 cells/well in 24-well plates. Medium containing DMSO or dabrafenib was replaced every 2 d. After 14 d of treatment, cells were fixed with ice-cold methanol for 10 min, stained with a 0.5% crystal violet solution for 10 min, washed with water, and imaged using a Nikon SMZ18 microscope equipped with a DS-Ri2 camera. Staining intensity in each well was quantified with ImageJ.
To measure colony-forming capacity, cells were seeded at a density of 2,000 cells/well in 50% Matrigel containing DMSO or dabrafenib in 24-well plates. The semisolid medium (400 µl) was covered with 1 ml of DMEM and 10% FBS containing DMSO or dabrafenib that was replaced every 2 d. After 14 d, wells were imaged using a Nikon SMZ18 microscope equipped with a DS-Ri2 camera. The number of colonies in each well was quantified with ImageJ.
CRISPR sequencing
Genomic DNA from human cell lines or zebrafish tumors was extracted with QuickExtract solution (Epibio). DNA libraries were prepared by PCR amplification of the CRISPR loci using the following primers: tp53, 5′-CTGTGTTTGCCAGGAGTACTTG-3′ and 5′-TATGTGTGTGTATGCGCTTTTG-3′; cdkn2a, 5′-ATCATGACGTTACTGGCGTTTA-3′ and 5′-TCGCAGTGATCTTTTGTATTGG-3′; spred1, 5′-GAGAGTGTGTGATCTGTGGCTC-3′ and 5′-CATTCGCTAATGTTTTATGCCA-3′.
Sequencing was performed by the MGH DNA core and analyzed using the Basepair online tool (https://www.basepairtech.com).
Protein analysis
For Western blot, cells were lysed with RIPA lysis buffer containing protease inhibitors (cOmplete; Roche) and phosphatase inhibitors (PhosSTOP; Roche). Lysates were incubated for 20 min on ice and spun down for 10 min at 14,000 revolutions per minute at 4°C. Protein concentrations were normalized using the DC protein assay (Bio-Rad). Samples were denatured by adding Laemmli sample buffer (Bio-Rad) with 5% β-mercaptoethanol (Sigma-Aldrich) and boiled at 95°C for 5 min before loading. Proteins were separated on a 12% mini-PROTEAN TGX (Bio-Rad) precast gel and transferred onto a polyvinylidene fluoride membrane using the iBlot2 system (Thermo Fisher Scientific). The primary antibodies were SPRED1 (#94063; Cell Signaling Technologies), p-ERK (#9101; Cell Signaling Technologies), ERK (#9102; Cell Signaling Technologies), and β-ACTIN (A2228; Sigma-Aldrich). Secondary antibodies were HRP anti-mouse and HRP anti-rabbit (Cell Signaling Technologies). Membranes were developed using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific). Ras activity was measured by purifying GTP-bound Ras from cell lysates using Ras Activation Assay Biochem Kit (BK008; Cytoskeleton) and following the manufacturer’s instructions. Band intensity was quantified with ImageJ.
Copy-number variation analysis
Whole-exome sequence data of patient-derived tumors (n = 4) along with normal tissues of a melanoma patient were analyzed to detect the somatic copy-number variations as previously described (Moriceau et al., 2015). Circos representation of copy-number changes was generated by R package RCircos. Melanoma tissues and patient-matched normal tissue were collected with the approval of institutional review boards at University of California, Los Angeles and informed consent of the patient.
Statistical analysis
Comparison of Kaplan–Meier survival curves was performed by a log-rank (Mantel–Cox) test. Statistical differences in cell viability experiments were estimated using paired two-tailed t test. Graphs show the mean ± SD. No statistical methods were used to predetermine sample size. *, P < 0.05; **, P < 0.01; n.s., not significant (P > 0.05).
Online supplemental material
Fig. S1 shows that SPRED1 inactivation modestly alters melanoma growth in vitro and in vivo. Fig. S2 demonstrates that SPRED1 inactivation confers resistance to BRAF inhibition in BRAF-driven human melanoma cell lines by sustaining MAPK pathway activity. Fig. S3 describes the influence of SPRED1 inactivation on the sensitivity of zebrafish melanomas to long-term BRAF inhibition both in vitro and in vivo.
Acknowledgments
This work was supported by the National Institutes of Health/National Cancer Institute (grants R01CA103846 and P01CA163222 to L.I. Zon; grant K99CA201465 to J. Ablain; grants 1R01CA176111A1 and 1R21CA215910-011 to R.S. Lo; and grant P01CA244118 to G. Moriceau and R.S. Lo), the Melanoma Research Alliance (L.I. Zon, S. Liu, and R.S. Lo), and the V Foundation for Cancer Research (R.S. Lo). S. Liu is supported by a Jonsson Comprehensive Cancer Center postdoctoral fellowship and R.S. Lo by the Ressler Family Foundation. L.I. Zon is a Howard Hughes Medical Institute Investigator.
Author contributions: J. Ablain designed the study, performed cell line and zebrafish experiments, performed TCGA data analyses, analyzed results, discussed findings, and wrote the manuscript. S. Liu performed genomic analyses on patients with melanoma treated with MAPK inhibitors, discussed findings, and wrote the manuscript. G. Moriceau generated genomic data from clinical samples and discussed findings. R.S. Lo collected clinical samples, discussed findings, and wrote the manuscript. L.I. Zon discussed findings and wrote the manuscript.
References
- Ablain, J., Durand E.M., Yang S., Zhou Y., and Zon L.I.. 2015. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev. Cell. 32:756–764. 10.1016/j.devcel.2015.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablain, J., Xu M., Rothschild H., Jordan R.C., Mito J.K., Daniels B.H., Bell C.F., Joseph N.M., Wu H., Bastian B.C., et al. 2018. Human tumor genomics and zebrafish modeling identify SPRED1 loss as a driver of mucosal melanoma. Science. 362:1055–1060. 10.1126/science.aau6509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbani, R., Akdemir K.C., Aksoy B.A., Albert M., Ally A., Amin S.B., Arachchi H., Arora A., Auman J.T., Ayala B., et al. Cancer Genome Atlas Network . 2015. Genomic Classification of Cutaneous Melanoma. Cell. 161:1681–1696. 10.1016/j.cell.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov, L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A.J.R., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., et al. ICGC PedBrain . 2013. Signatures of mutational processes in human cancer. Nature. 500:415–421. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester, R., Marchuk D., Boguski M., Saulino A., Letcher R., Wigler M., and Collins F.. 1990. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 63:851–859. 10.1016/0092-8674(90)90151-4 [DOI] [PubMed] [Google Scholar]
- Beroukhim, R., Getz G., Nghiemphu L., Barretina J., Hsueh T., Linhart D., Vivanco I., Lee J.C., Huang J.H., Alexander S., et al. 2007. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. USA. 104:20007–20012. 10.1073/pnas.0710052104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag, G., Hirth P., Tsai J., Zhang J., Ibrahim P.N., Cho H., Spevak W., Zhang C., Zhang Y., Habets G., et al. 2010. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 467:596–599. 10.1038/nature09454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brems, H., Chmara M., Sahbatou M., Denayer E., Taniguchi K., Kato R., Somers R., Messiaen L., De Schepper S., Fryns J.P., et al. 2007. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat. Genet. 39:1120–1126. 10.1038/ng2113 [DOI] [PubMed] [Google Scholar]
- Ceol, C.J., Houvras Y., Jane-Valbuena J., Bilodeau S., Orlando D.A., Battisti V., Fritsch L., Lin W.M., Hollmann T.J., Ferré F., et al. 2011. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 471:513–517. 10.1038/nature09806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami, E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., et al. BRIM-3 Study Group . 2011. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364:2507–2516. 10.1056/NEJMoa1103782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty, K.T., Puzanov I., Kim K.B., Ribas A., McArthur G.A., Sosman J.A., O’Dwyer P.J., Lee R.J., Grippo J.F., Nolop K., and Chapman P.B.. 2010. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363:809–819. 10.1056/NEJMoa1002011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild, A., Grob J.J., Demidov L.V., Jouary T., Gutzmer R., Millward M., Rutkowski P., Blank C.U., Miller W.H. Jr., Kaempgen E., et al. 2012. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 380:358–365. 10.1016/S0140-6736(12)60868-X [DOI] [PubMed] [Google Scholar]
- Hayward, N.K., Wilmott J.S., Waddell N., Johansson P.A., Field M.A., Nones K., Patch A.M., Kakavand H., Alexandrov L.B., Burke H., et al. 2017. Whole-genome landscapes of major melanoma subtypes. Nature. 545:175–180. 10.1038/nature22071 [DOI] [PubMed] [Google Scholar]
- Hirata, Y., Brems H., Suzuki M., Kanamori M., Okada M., Morita R., Llano-Rivas I., Ose T., Messiaen L., Legius E., and Yoshimura A.. 2016. Interaction between a domain of the negative regulator of the ras-ERK pathway, SPRED1 protein, and the GTPase-activating protein-related domain of neurofibromin is implicated in legius syndrome and neurofibromatosis type 1. J. Biol. Chem. 291:3124–3134. 10.1074/jbc.M115.703710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo, W., Shi H., Sun L., Piva M., Song C., Kong X., Moriceau G., Hong A., Dahlman K.B., Johnson D.B., et al. 2015. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell. 162:1271–1285. 10.1016/j.cell.2015.07.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen, C.M., Boehm J.S., Kim S.Y., Thomas S.R., Wardwell L., Johnson L.A., Emery C.M., Stransky N., Cogdill A.P., Barretina J., et al. 2010. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 468:968–972. 10.1038/nature09627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, K., Takeda H., Kawakami N., Kobayashi M., Matsuda N., and Mishina M.. 2004. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell. 7:133–144. 10.1016/j.devcel.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Maertens, O., Johnson B., Hollstein P., Frederick D.T., Cooper Z.A., Messiaen L., Bronson R.T., McMahon M., Granter S., Flaherty K., et al. 2013. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 3:338–349. 10.1158/2159-8290.CD-12-0313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G.A., Viskochil D., Bollag G., McCabe P.C., Crosier W.J., Haubruck H., Conroy L., Clark R., O’Connell P., Cawthon R.M., et al. 1990. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 63:843–849. 10.1016/0092-8674(90)90150-D [DOI] [PubMed] [Google Scholar]
- McArthur, G.A., Chapman P.B., Robert C., Larkin J., Haanen J.B., Dummer R., Ribas A., Hogg D., Hamid O., Ascierto P.A., et al. 2014. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol. 15:323–332. 10.1016/S1470-2045(14)70012-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon, R., Friedman A., Bigner D., Van Meir E.G., Brat D.J., Mastrogianakis G.M., Olson J.J., Mikkelsen T., Lehman N., Aldape K., et al. Cancer Genome Atlas Research Network . 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 455:1061–1068. 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriceau, G., Hugo W., Hong A., Shi H., Kong X., Yu C.C., Koya R.C., Samatar A.A., Khanlou N., Braun J., et al. 2015. Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell. 27:240–256. 10.1016/j.ccell.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian, R., Shi H., Wang Q., Kong X., Koya R.C., Lee H., Chen Z., Lee M.K., Attar N., Sazegar H., et al. 2010. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 468:973–977. 10.1038/nature09626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonami, A., Kato R., Taniguchi K., Yoshiga D., Taketomi T., Fukuyama S., Harada M., Sasaki A., and Yoshimura A.. 2004. Spred-1 negatively regulates interleukin-3-mediated ERK/mitogen-activated protein (MAP) kinase activation in hematopoietic cells. J. Biol. Chem. 279:52543–52551. 10.1074/jbc.M405189200 [DOI] [PubMed] [Google Scholar]
- Pratilas, C.A., Taylor B.S., Ye Q., Viale A., Sander C., Solit D.B., and Rosen N.. 2009. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc. Natl. Acad. Sci. USA. 106:4519–4524. 10.1073/pnas.0900780106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem, O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., and Zhang F.. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343:84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Moriceau G., Kong X., Lee M.K., Lee H., Koya R.C., Ng C., Chodon T., Scolyer R.A., Dahlman K.B., et al. 2012. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun. 3:724. 10.1038/ncomms1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe, I.B., Mercado E.L., Stowe T.R., Bell E.L., Oses-Prieto J.A., Hernández H., Burlingame A.L., and McCormick F.. 2012. A shared molecular mechanism underlies the human rasopathies Legius syndrome and Neurofibromatosis-1. Genes Dev. 26:1421–1426. 10.1101/gad.190876.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle, N., Emery C., Berger M.F., Davis M.J., Sawyer A., Pochanard P., Kehoe S.M., Johannessen C.M., Macconaill L.E., Hahn W.C., et al. 2011. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 29:3085–3096. 10.1200/JCO.2010.33.2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakioka, T., Sasaki A., Kato R., Shouda T., Matsumoto A., Miyoshi K., Tsuneoka M., Komiya S., Baron R., and Yoshimura A.. 2001. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 412:647–651. 10.1038/35088082 [DOI] [PubMed] [Google Scholar]
- Whittaker, S.R., Theurillat J.P., Van Allen E., Wagle N., Hsiao J., Cowley G.S., Schadendorf D., Root D.E., and Garraway L.A.. 2013. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 3:350–362. 10.1158/2159-8290.CD-12-0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G.F., Lin B., Tanaka K., Dunn D., Wood D., Gesteland R., White R., Weiss R., and Tamanoi F.. 1990a. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 63:835–841. 10.1016/0092-8674(90)90149-9 [DOI] [PubMed] [Google Scholar]
- Xu, G.F., O’Connell P., Viskochil D., Cawthon R., Robertson M., Culver M., Dunn D., Stevens J., Gesteland R., White R., and Weiss R.. 1990b. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 62:599–608. 10.1016/0092-8674(90)90024-9 [DOI] [PubMed] [Google Scholar]
- Yan, W., Markegard E., Dharmaiah S., Urisman A., Drew M., Esposito D., Scheffzek K., Nissley D.V., McCormick F., and Simanshu D.K.. 2020. Structural Insights into the SPRED1-Neurofibromin-KRAS Complex and Disruption of SPRED1-Neurofibromin Interaction by Oncogenic EGFR. Cell Rep. 32:107909. 10.1016/j.celrep.2020.107909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Z., Yaeger R., Rodrik-Outmezguine V.S., Tao A., Torres N.M., Chang M.T., Drosten M., Zhao H., Cecchi F., Hembrough T., et al. 2017. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature. 548:234–238. 10.1038/nature23291 [DOI] [PMC free article] [PubMed] [Google Scholar]