Abstract
COVID-19 patients can excrete viable SARS-CoV-2 virus via urine and faeces, which has raised concerns over the possibility of COVID-19 transmission via aerosolized contaminated water or via the faecal-oral route. These concerns are especially exacerbated in many low- and middle-income countries, where untreated sewage is frequently discharged to surface waters. SARS-CoV-2 RNA has been detected in river water (RW) and raw wastewater (WW) samples. However, little is known about SARS-CoV-2 viability in these environmental matrices. Determining the persistence of SARS-CoV-2 in water under different environmental conditions is of great importance for basic assumptions in quantitative microbial risk assessment (QMRA). In this study, the persistence of SARS-CoV-2 was assessed using plaque assays following spiking of RW and WW samples with infectious SARS-CoV-2 that was previously isolated from a COVID-19 patient. These assays were carried out on autoclaved RW and WW samples, filtered (0.22 µm) and unfiltered, at 4 °C and 24 °C. Linear and nonlinear regression models were adjusted to the data. The Weibull regression model achieved the lowest root mean square error (RMSE) and was hence chosen to estimate T90 and T99 (time required for 1 log and 2 log reductions, respectively). SARS-CoV-2 remained viable longer in filtered compared with unfiltered samples. RW and WW showed T90 values of 1.9 and 1.2 day and T99 values of 6.4 and 4.0 days, respectively. When samples were filtered through 0.22 µm pore size membranes, T90 values increased to 3.3 and 1.5 days, and T99 increased to 8.5 and 4.5 days, for RW and WW samples, respectively. Remarkable increases in SARS-CoV-2 persistence were observed in assays at 4 °C, which showed T90 values of 7.7 and 5.5 days, and T99 values of 18.7 and 17.5 days for RW and WW, respectively. These results highlight the variability of SARS-CoV-2 persistence in water and wastewater matrices and can be highly relevant to efforts aimed at quantifying water-related risks, which could be valuable for understanding and controlling the pandemic.
Keywords: SARS-CoV-2, viability, persistence, water, wastewater, temperature
Graphical Abstract
1. Introduction
The ongoing COVID-19 pandemic has already led to more than 2.6 million reported deaths globally by February 2021. A novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been identified as the etiologic agent of COVID-19. Coronaviruses are enveloped RNA viruses broadly distributed among humans, other mammals, and birds, causing acute and persistent infectious (Knipe and Howley, 2013). It has been hypothesized that the disease origin could be associated with spillover transmission phenomena from a wild animal reservoir, such as pangolins (Lam et al. 2020), bats (Zhou et al. 2020), to humans.
Although COVID-19 is a respiratory disease, large amounts of SARS-CoV-2 RNA have been detected in stools (Wu et al. 2020) of patients, and subsequently in raw sewage (Ahmed et al. 2020; Chernicharo et al. 2020; Fongaro et al. 2020; La Rosa et al. 2021; Medema et al. 2020; Mota et al. 2021), sewage sludge (Peccia et al. 2020), and surface water (Guerrero-Latorre et al. 2020). Viable SARS-CoV-2 has been isolated from urine (Sun et al. 2020) and faeces (Wang et al. 2020, Zhang et al. 2020, Xiao et al. 2020) of patients, which raises concerns over the possibility of the faecal-oral or faecal-nasal transmission routes for COVID-19. Kang et al. (2020) have recently reported circumstantial evidence of SARS-CoV-2 transmission by aerosolized wastewater in a residential building in Guangzhou, China. The presence of viable SARS-CoV-2 in water and wastewater may have far-reaching consequences for public health and pandemic control strategies (Heller et al. 2020), especially in developing countries with inadequate access to sanitation and safe water. Currently, there is no evidence that COVID-19 can be transmitted via contaminated water (La Rosa et al., 2020). Nevertheless, the World Health Organization has highlighted the need for research concerning SARS-CoV-2 persistence on environmental matrices, such as surface water and wastewater (WHO, 2020).
Determining the persistence of SARS-CoV-2 in river water under different environmental conditions is of great importance for basic assumptions in quantitative microbial risk assessment (QMRA). Environmental conditions can have a strong effect on viral viability, including seasonal or temperature variations, turbidity, extent of river water contamination with wastewater, etc. In addition, virus type can affect virus survival in the environment (John and Rose 2005). Virus structure has a significant influence on virus persistence in environmental matrices, depending on whether its external layer remains intact (Mitchell and Akram, 2019). For this reason, enveloped virus such as coronaviruses, with their fragile lipid external layer, are usually much less persistent than non-enveloped viruses in water, remaining viable for a few days, as opposed to months, as is the case for the latter (Kutz and Gerba, 1988).
Due to the high risks of infection, biosafety-level 3 laboratories are required for propagation methods involving viable SARS-CoV-2. For this reason, different studies have used alternative viruses as surrogates to predict the behavior of coronavirus in environmental matrices (Aquino De Carvalho et al. 2017; Casanova et al. 2009, Gundy et al. 2009). However, the effects of different matrices and environmental parameters on the inactivation of viruses are complex and yield highly variable results, depending on the viral strain used as surrogate (Carraturo et al. 2020). Although studies based on surrogate viruses are highly relevant, surrogate viruses can respond differently to environmental stresses when compared with target pathogen viruses, emphasizing the need for tests using the actual pathogen of concern.
Bivins et al. (2020) have recently assessed how SARS-CoV-2 persistence in water and wastewater is affected by warm temperatures. They showed that at 50 °C and 70 °C, T90 values in wastewater were 15 and 2 minutes, respectively, whereas at room temperature (20 °C), T90 was 1.7 day, for the same matrix. Similarly, SARS-CoV-2 RNA showed T90 values ranging from 8.04 to 27.8 days in wastewater and from 9.4 to 58.6 days in tap water for temperatures of 37 °C and 4 °C, respectively (Ahmed et al., 2020b). Viable SARS-CoV-2 persistence in water and wastewater at low temperatures (4 °C) remains unknown. Determining the persistence of SARS-CoV-2 in water and wastewater at high and low temperatures is of great importance to the planning and implementing of pandemic control strategies.
In this study, we evaluated the persistence of SARS-CoV-2 in river water and wastewater samples under different, simulated environmental conditions by using plaque assay in Vero cells. Virus persistence assays were carried out at 4 °C and 24 °C, to simulate different seasons in the year, and with filtered and unfiltered samples, to simulate water matrices with different solids content and turbidity. Virus persistence data generated with unfiltered raw wastewater samples could be used to simulate sewer networks and rivers that are heavily impacted by raw sewage discharges. Filtered river water samples could be used to simulate river water matrices with extremely low turbity.
2. Material and methods
2.1. Sampling
The environmental matrices used in this study were river water (RW) and raw wastewater (WW). RW was collected at Rio das Velhas, in the municipality of Nova Lima, Minas Gerais State, Brazil (20°00′33.9"S 43°49′50.6"W), whereas WW was collected from Arrudas Wastewater Treatment Plant (19°53′46.9"S 43°52′41.4"W).
2.2. Environmental conditions
The persistence of SARS-CoV-2 in RW and WW was assessed at 4 °C and 24 °C for 15 days to determine the influence of seasonal temperature variations (cold and hot weather). Filtered and unfiltered RW and WW samples were used to simulate water matrices with different solids content and turbidity, going from sewer networks and rivers that are heavily impacted by raw sewage discharges (unfiltered raw WW) to river water matrices with extremely low turbidity (Filtered RW, RWF). Filtered samples were prepared by consecutively filtering through nitrate cellulose membranes with 0.45 µm and 0.22 µm pore sizes (RWF and WWF). All samples (1 L each) were autoclaved at 121 °C for 15 minutes to eliminate the effect of pathogens other than SARS-CoV-2 that could be present in the original samples. Autoclaved samples were stored at 4 °C until the cell culture infectivity assay. All assays were performed in triplicate.
2.3. Physicochemical parameters
Samples were characterized by typical water and wastewater physicochemical parameters, such as total solids (TS), volatile solids (VS), total solids suspended (TSS), Chemical Oxygen Demand (COD), nitrogen (ammonia), and orthophosphate, all measured according to APHA (2017). Additional parameters characterized in total and filtered samples are presented in Table 1 .
Table 1.
Physicochemical parameters for river water (RW) and raw wastewater (WW), filtered (RWF and WWF) and unfiltered.
| Parameters | WW | RW | WWF | RWF |
|---|---|---|---|---|
| TS (mg/L) | 455 ± 16 | 34 ± 2 | NA | NA |
| VS (mg/L) | 255 ± 11 | 10 ± 5 | NA | NA |
| TSS (mg/L) | 267 ± 17 | 8 ± 0 | NA | NA |
| VSS (mg/L) | 227 ± 24 | 2 ± 1 | NA | NA |
| VS/TS | 0.56 | 0.29 | NA | NA |
| Turbidity (NTU) | 274 ± 2 | 10 ± 1 | 6 ± 0 | 1 ± 0 |
| COD (mg/L) | 334 ± 19 | < 5 | 53 ± 3 | < 5 |
| Ammonia (mg/L) | 30.21 | 0.58 | 25.79 | 0.23 |
| Orthophosphate (mg/L) | 3.1 | <0.001 | 2.3 | <0.001 |
| pH | 7.50 | 5.50 | 7.50 | 5.50 |
NA: not applicable.
2.4. SARS-CoV-2 cell culture infectivity assays
All SARS-CoV-2 persistence tests were performed in a high-containment, biosafety level 4 facility (OIE BSL-4 – World Organization for Animal Health) at Laboratório Federal de Defesa Agropecuária, LFDA-MG, located in Pedro Leopoldo, MG, Brazil, by inoculating infectious SARS-CoV-2 in autoclaved water and wastewater samples. SARS-CoV-2 isolate SP02/BRA (SARS.CoV2/SP02.2020.HIAE.Br) was kindly provided by Dr. Edison Luiz Durigon (Department of Microbiology, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil) (Araujo et al. 2020).
SARS-CoV-2 virus stock was prepared after infection of Vero CCL-81 cells and stored at -80 °C until further use in spiking tests. SARS-CoV-2 titers were determined with plaque assay in 24 wells plates seeded with Vero CCL-81 cells at a concentration of 1 × 105 cells/well. After reaching a confluence of 80-90%, ten-fold dilutions of virus suspensions in DMEM-2% FBS were transferred (100 μL/well) to the seeded plates. After 1 h adsorption, the wells were covered with an overlay of DMEM-2% FBS containing 1% (w/v) carboxymethyl cellulose (CMC). Plates were incubated at 37 °C in 5% CO2 for 72 hours, fixed in 10% formalin solution and stained with a 0.5% Crystal Violet solution. Plaque forming units (PFU) were manually enumerated and registered as PFU/mL.
2.5. Sample inoculation with SARS-CoV-2
Virus persistence in each of the environmental matrices inoculated in triplicates with SARS-CoV-2 was also determined through plaque assay. Briefly, a 4.8 mL aliquot of each different water sample was transferred to a sterile 15 mL polypropylene conical tube. Then, 1 × 105 PFU of stock SARS-CoV-2 in 200 µL was added to each tube (Vf= 5mL; SARS-CoV-2 designed initial titer = 2 × 104 PFU/mL). Inoculated water matrices were either kept at 24 °C or stored at 4 °C, according to the experimental design. After exposure, infectious SARS-CoV-2 in suspension were tested for infectivity in Vero CCL-81 cells at time points 0, 6, 24, 48, 72, 96, 120, 240 and 360 hours post-inoculation. At each time point, including immediately following inoculation, 50 μL of each sample was collected and added to 450 μL of DMEM-2% FBS in a 1.5 mL microcentrifuge tube. Negative controls consisting of 50 μL of uninoculated wastewater added to 450 μL of DMEM-2% FBS were also prepared at each time point.
2.6. Statistical analyses
In addition to the log-linear model (Equation 1), nonlinear models were applied to describe the decay patterns and obtained by nonlinear least square (nls) method, including exponential-nls (Equation 2), exponential biphasic (exp-biphasic, Equation 3), Weibull (Equation 4), and Gompertz (Equation 5) models.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
Where k is the slope and first-order decay constant, is the concentration of the virus at time t, and is the starting virus concentration at t= 0. The exponential biphasic decay consisted of initial and final, fast and slow decay periods, in which b1 and b2 correspond to the fast and slow decay constants, whereas a1 and a2 represent the decay of the virus at the start of the fast and slow decay periods, respectively (Bivins et al. 2020). The Weibull model is described by Asym1, which represents the horizontal asymptote on the right side, Drop is a numeric parameter representing the change from Asym to the y-intercept, lrc1 is a numeric parameter representing the natural logarithm of the rate constant, pwr is a numeric parameter representing the power to which t is raised. In the Gompertz model, Asym2 represents the asymptote, b3 is related to the value of the function at t= 0 and b4 is a numeric parameter related to the scale of the x-axis.
All regressions and statistical analyses were performed using the pharmacokinetic (PK) nlstools packages in CRAN and Rstudio (Team, 2020). Linear regressions were assessed by R2 values and by checking the normality and variance homogeneity assumptions in the Q-Q and residuals vs fitted values plots. A Shapiro-Wilk test was performed to complementarily assess the assumption of normality, whereas a skew ratio (the ratio between skewness value and standard error of skewness) greater than 2 was used as a reference to regard the data as having unignorable skewness (Yan et al., 2016). The skew ratio was determined using the Skwelmm package. The occurrence of significant differences between first-decay constants was assessed using ANOVA (stats package). For nonlinear regressions, the start parameters of the exponential (exponential-nls), exponential biphasic (exp-biphasic), Weibull, and Gompertz fits were estimated using the self-start models. The fit of the models to the observed data was assessed using an extra sum-of-squares F. The Bayesian information criterion (BIC) test and the root-mean-square errors (RMSE) were used to compare the fits (stats package). Models with low BIC and RMSE were preferred over fits with high values. When similar values were obtained, the less complex model was considered as the best fit (Kauppinen and Miettinen, 2017). For the decay rate comparison, T90 and T99 (the time to achieve 90% and 99% reductions from the initial titer) were determined using the best fitting model. Spearman correlations between T90 and T99 values and physicochemical parameters were also assessed using Rstudio (stats package).
3. Results and Discussion
3.1. First-Order Decay Rate Constants
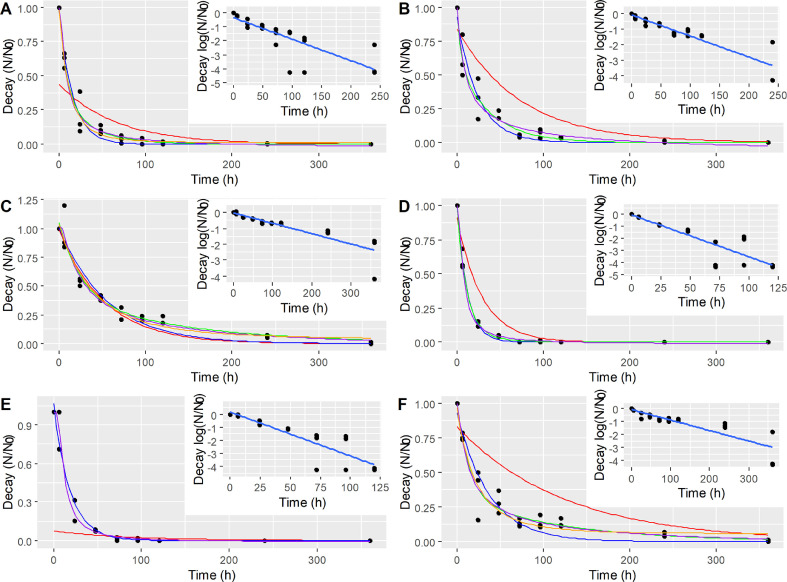
First-order log-linear models were fit to experimental data for preliminary assessment of SARS-CoV-2 decay (Table 2 ). On Fig. 1 , each inset shows the linear regressions performed for the original experimental data from triplicate assays (black dots). First-order decay rates for unfiltered samples were -0.37 d−1 and -0.83 d−1 for RW-24°C and WW-24°C, respectively. Samples filtered through 0.22 µm-pore size membranes presented slightly lower decay rates of -0.32 d−1 and -0.80 d−1 for RWF-24°C and WWF-24°C, whereas the first-order decay rates in samples at 4 °C presented even lower values of 0.16 d−1 (RW-4°C) and 0.19 d−1 (WW-4°C). ANOVA and pairwise comparisons of the decay rates showed that RW-4°C and WW-4°C exhibited significantly lower (p < 0.05) values than the other decay rates, indicating a significant increase in viral persistence at low temperature.
Table 2.
Models with the lowest RMSE and BIC values compared to log-linear regressions.
| Sample | Model | Decay rate (d−1) | RMSE | BIC |
|---|---|---|---|---|
| River water (RW-24°C) |
Log-linear (R2= 0.649) | m= -0.37 | 0.8017 | 67.0 |
| Exponential-nls | b=-1.55 | 0.0559 | -69.1 | |
| Exp-biphasic | b1 = -2.82 | 0.0478 | 71.1 | |
| b2 = -0.66 | ||||
| Gompertz | b3= 0.98 | 0.0490 | -72.8 | |
| Weibull | = -4.1 | 0.0486 | 70.19 | |
| Filtered river water (RWF-24°C) |
Log-linear (R2= 0.821) | m= -0.32 | 0.4525 | 39.6 |
| Exponential-nls | b= -0.98 | 0.0795 | -50.2 | |
| Exp-biphasic | b1 = -5.40 | 0.0633 | -55.9 | |
| b2 = -0.63 | ||||
| Gompertz | N/A | - | - | |
| Weibull | = -6.6 | 0.0665 | -53.2 | |
| River water at 4 °C (RW-4°C) |
Log-linear (R2= 0.761) | m= -0.16 | 0.4049 | 37.69 |
| Exponential-nls | b= -0.43 | 0.0827 | -48.0 | |
| Exp-biphasic | b1 = -0.96 | 0.0633 | -56.8 | |
| b2 = -0.17 | ||||
| Gompertz | b3=0.99 | 0.0728 | -51.7 | |
| Weibull | = -2.2 | 0.0636 | -55.7 | |
| Wastewater (WW-24°C) |
Log-linear (R2= 0.791) | m= -0.83 | 0.7350 | 55.8 |
| Exponential-nls | b= 1.98 | 0.0767 | -118 | |
| Exp-biphasic | b1 = -2.30 | 0.0213 | -115 | |
| b2 = -0.85 | ||||
| Gompertz | b3= 0.99 | 0.0214 | -118 | |
| Weibull | = -2.5 | 0.0224 | -112 | |
| Filtered wastewater (WWF-24°C) |
Log-linear (R2= 0.796) | m= -0.80 | 0.6959 | 53.5 |
| Exponential-nls | b= 1.29 | 0.0827 | -52.1 | |
| Exp-biphasic | N/A | |||
| Gompertz | N/A | |||
| Weibull | = -0.5 | 0.052 | -66.5 | |
| Wastewater at 4 °C (WW-4°C) |
Log-linear (R2= 0.752) | m= -0.19 | 0.5094 | 50.08 |
| Exponential-nls | b= -0.65 | 0.0867 | -49.4 | |
| Exp-biphasic | b1 = -1.71 | 0.0622 | -56.8 | |
| b2 = -0.20 | ||||
| Gompertz | b3= 0.98 | 0.0679 | -55.4 | |
| Weibull | = -4.8 | 0.0619 | -57.1 |
Fig. 1.
Log-linear and nonlinear regressions for A) river water (RW-24°C); B) filtered river water (RWF-24°C); C) river water at 4 °C (RW-4°C); D) wastewater (WW-24°C); E) filtered wastewater (WWF-24°C); F) wastewater at 4 °C (WW-4°C). Log-linear model (red), exponential-nls model (blue), exp-biphasic model (green), Weibull model (purple), Gompertz model (orange).
Arrhenius equation parameters were established in order to determine reaction coefficients for different temperatures (4 °C and 24 °C) (Supplementary material – Equation S1 and Figure S.7). The first-order decay constants followed Arrhenius relationship with activation energies of 22.8 and 50.4 KJ mol−1 for RW and WW, respectively, indicating that SARS-CoV-2 decay in WW was more sensitive to temperature than in RW. Similar energies of 39 kJ mol−1 have been reported for the inactivation of poliovirus type 1 during thermophilic (51 to 56 °C) anaerobic digestion of sludge (Popat et al. 2010) and 59.8 kJ mol−1 for poliovirus 2 inactivation by Chloramine-T in water (5 to 35 °C, Gowda et al. 1981). Furthermore, MHV (Murine coronavirus) inactivation in stainless steel showed values between 30 and 100 kJ mol−1 (Casanova et al. 2010; Roos, 2020).
3.2. Nonlinear Regressions
Log-linear regressions were a poor fit to the experimental data, as indicated by RMSE values (Table 2). Four nonlinear models were tested: exponential-nls, exponential biphasic, Weibull, and Gompertz. The regression parameters calculated for each model are presented in Table S1 (Supplementary materials). All nonlinear models showed better fits compared to linear models, as evidenced by the lower RSME values compared to those found for linear regressions. Overall, the Weibull model was a better fit for the complete dataset.
Although linear first-order kinetics is a classical model of virus decay in water and wastewater, including enveloped and non-enveloped viruses, in some cases, its fitting is not carefully assessed, hampering the ability to identify factors that can affect inactivation kinetics or determine the actual patterns of decay (Dean et al. 2020). In this study, the lack of accuracy of the log-linear model was the consequence of possible outliers and the violation of the assumptions of normality and variance homogeneity. The violation of the assumption of normality (Shapiro-Wilk test and Q-Q plots, Figure S1 to S6 – Supplementary material) occurred for all of log-normal regressions. The assessment of the skew ratio showed high skewness in all cases (except for WW-24°C and WW-4°C). Also, several matrices showed violations of the assumption of variance homogeneity (i.e., RW-4°C, WWF-24°C and WW-4°C), increasing uncertainty in linear regressions (Figure S1 to S6 – Supplementary material).
Exponential models representing a "flat tail" have been reported as providers of better representation of the decay of pathogens, including viruses. For instance, biphasic dynamics can arise as a consequence of pathogens population heterogeneity or hardening off (Brouwer et al. 2017). Similarly, biphasic inactivation kinetics has also been observed for non-enveloped viruses, which was attributed to subpopulations of viruses with varied susceptibilities to solution chemistry or temperature (Ye et al. 2016). Kauppinen and Miettinen (2017) found that Weibull and Double-Weibull models presented the best fittings for Norovirus GII genome inactivation in wastewater (3 °C) and drinking water (21 °C), respectively, which showed a high tailing effect describing the long persistence of the virus. The tailing effect represented by the Weibull model has been reported as providing a better description of inactivation of foot-and-mouth disease virus clones subjected to 50 plaque-to-plaque transfers since small differences in the virus replication, which are amplified during the course of replication, resulted in larger fluctuations and the "flat tail" in the probability distribution, which eventually develops a stretched exponential Weibull shape (Lázaro et al. 2003). The experimental data shown in the current study indicated higher fluctuations in SARS-CoV-2 survival at low viral titer and mainly at low temperature.
3.3. SARS-CoV-2 T90 and T99 in water and wastewater
T90 and T99 values were calculated for SARS-CoV-2 survival in all samples using the Weibull model (Table 3 ). RW-24°C and WW-24°C showed T90 values of 1.9 and 1.2 day and T99 values of 6.4 and 4.0 days, respectively. These T90 values were within the same range of those reported by Bivins et al. (2020), which were 2.0 and 1.6 days for tap water and wastewater samples, respectively. However, these T99 values (at 24 °C) are slightly higher than those reported by Bivins et al. (2020), at 3.9 days and 3.2 days for tap water and wastewater, respectively. The differences at longer survival times could potentially be due to the differences in the regression models used (Weibull in the current study, versus Log-linear). When samples were filtered through 0.22 µm pore size membranes, T90 values increased to 3.3 and 1.5 days, and T99 increased to 8.5 and 4.5 days, for RW-24°C and WW-24°C samples, respectively. Remarkable increases in SARS-CoV-2 survival times were observed in essays at 4 °C, which showed T90 values of 7.7 and 5.5 days, and T99 values of 18.7 and 17.5 days for river water and wastewater samples, respectively.
Table 3.
Weibull regression parameters, estimated T90 and T99 for each of the samples.
| Sample | Weibull regression Parameters |
|||||
|---|---|---|---|---|---|---|
| Asym | Drop | Lrc1 | Pwr | T90 (days) | T99 (days) | |
| River water (RW-24°C) | 0.995 | 1.031 | 1.758 | -0.981 | 1.9 | 6.4 |
| Filtered river water (RWF-24°C) | 0.998 | 1.102 | 1.295 | -0.660 | 3.3 | 8.5 |
| River water at 4 °C (RW-4°C) | 1.013 | 1.105 | 2.397 | -0.779 | 7.7 | 18.7 |
| Wastewater (WW-24°C) | 0.997 | 1.015 | 2.258 | -1.300 | 1.2 | 4.0 |
| Filtered wastewater (WWF-24°C) | 0.999 | 1.010 | 3.796 | -1.636 | 1.5 | 4.5 |
| Wastewater at 4 °C (WW-4°C) | 1.001 | 1.071 | 1.618 | -0.691 | 5.5 | 17.5 |
Temperature is recognized as having a strong effect on virus persistence. High persistence at low temperatures has been previously reported for other coronaviruses (Casanova et al. 2009; Gundy et al. 2009; Bertrand et al. 2012). SARS-CoV-2 viability has not yet been confirmed in real wastewater or natural river water. However, assessing the effect of temperature on SARS-CoV-2 persistence in water matrices could be highly relevant for quantifying risks related to exposure to SARS-CoV-2 contaminated water, since several countries are currently experiencing peaks of COVID-19 transmissions, which coincide with the boreal winter. Low seasonal temperatures may increase the water-related risk of SARS-CoV-2 transmission, raising concerns over the risk of COVID-19 transmission by aerosolized contaminated water and wastewater, or via the fecal-oral transmission.
Solids content and composition were likely a secondary factor affecting SARS-CoV-2 persistence in the water and wastewater samples tested. Longer SARS-CoV-2 survival times were observed in river water compared to wastewater, and in filtered samples compared to unfiltered samples. Although no significant correlations were observed for T90 or T99 values and physicochemical parameters (Table S.2 – supplementary material), solids composition and pH can affect SARS-CoV-2 persistence. In RW-24°C, the lower pH may have stimulated higher electrostatic interactions and viral adsorption to the solids, which presented a more mineral composition (volatile solids/total solids ratio of 0.3) compared to the solids present in wastewater samples (volatile solids/total solids ratio of 0.5). These results agree with the effect of matrix composition previously described in the literature, with a faster virus inactivation in complex rather than in simpler matrices (Bertrand et al. 2012). On the same note, viral persistence in non-sterile water and wastewater should be lower compared to viral persistence in autoclaved samples (determined in the current study), as also verified for SARS-CoV-2 RNA by Ahmed et al. (2020b). pH and organic and inorganic solids can play important roles in the formation of pH-dependent electrically charged surfaces (Michen and Graule, 2009; Scheller et al. 2020) by producing significant alterations in the virus structure proteins due to changes in its isoelectric point (IEP). A hypothetical explanation for faster inactivation in unfiltered compared to filtered river water samples is the presence of inorganic clays, due to their highly adsorptive properties, which could potentially act as SARS-CoV-2 inhibitors (Sahel N Abduljauwad et al. 2020). Table 4 shows a comparison of the SARS-CoV-2 T90 values determined in the current study with T90 values previously reported for other coronaviruses, surrogate coronaviruses and other viruses.
Table 4.
Best fitting models and T90 estimates for SARS-CoV-2, other coronaviruses, surrogate coronaviruses and other viruses in water matrices.
| Reference | Virus | Matrix | Temp | Best-fitting model | RMSE | T90 (days) |
|---|---|---|---|---|---|---|
| This work | SARS-CoV-2 | RW* | 24 °C | Weibull | 0.0486 | 1.9 |
| RW* | 4 °C | Weibull | 0.0636 | 7.7 | ||
| RWF* | 24 °C | Weibull | 0.0665 | 3.3 | ||
| WW* | 24 °C | Weibull | 0.0224 | 1.2 | ||
| WW* | 4 °C | Weibull | 0.0619 | 5.5 | ||
| WWF* | 24 °C | Weibull | 0.0520 | 1.5 | ||
| Ahmed et al. (2020b) | SARS-CoV-2 RNA | WW | 37 °C | First-order | 1.10 | 0.74 |
| WW | 25 °C | First-order | 0.67 | 12.6 | ||
| WW | 15 °C | First-order | 0.59 | 20.4 | ||
| WW | 4 °C | First-order | 0.37 | 27.8 | ||
| WW* | 37 °C | First-order | 0.59 | 5.71 | ||
| WW* | 25 °C | First-order | 0.48 | 13.5 | ||
| WW* | 15 °C | First-order | 0.32 | 29.9 | ||
| WW* | 4 °C | First-order | 0.14 | 43.2 | ||
| TW-D | 37 °C | First-order | 0.86 | 9.40 | ||
| TW-D | 25 °C | First-order | 0.68 | 15.2 | ||
| TW-D | 15 °C | First-order | 0.33 | 51.2 | ||
| TW-D | 4 °C | First-order | 0.17 | 58.6 | ||
| Bivins et al. (2020) | SARS-CoV-2 | WW | 20 °C | Log-linear | 1.8 | 1.6 |
| TW | 20 °C | Log-linear | 1.2 | 2.0 | ||
| WW | 50 °C | Log-linear | 1.4 | 15 min | ||
| WW | 70 °C | Log-linear | 1.9 | 2.2 min | ||
| Kauppinen and Miettinen (2017) | Norovirus GII_A RNA | WW | 3 °C | Double Weibull | 0.11 | 38 |
| WW | 21 °C | Log-linear | 0.03 | 58 | ||
| WW | 36 °C | Log-linear | 0.11 | 58 | ||
| DW | 21 °C | Weibull | 0.09 | 230 | ||
| DW | 36 °C | Log-linear | 0.31 | 58 | ||
| Gundy et al. (2009) | HCoV | TP | 23 °C | Log-linear | N/D | 8.1 |
| TP-F | 23 °C | Log-linear | N/D | 6.8 | ||
| TP-F | 4 °C | Log-linear | N/D | 392 | ||
| WW | 23 °C | Log-linear | N/D | 2.4 | ||
| WW-F | 23 °C | Log-linear | N/D | 1.6 | ||
| SE | 23 °C | Log-linear | N/D | 1.9 | ||
| FIPV | TW | 23 °C | Log-linear | N/D | 8.3 | |
| TW-F | 23 °C | Log-linear | N/D | 6.8 | ||
| TW-F | 4 °C | Log-linear | N/D | 87.0 | ||
| WW | 23 °C | Log-linear | N/D | 1.7 | ||
| WW-F | 23 °C | Log-linear | N/D | 1.6 | ||
| SE | 23 °C | Log-linear | N/D | 1.8 | ||
| PV-1 | TW | 23 °C | Log-linear | N/D | 47.5 | |
| TW-F | 23 °C | Log-linear | N/D | 43.3 | ||
| TW-F | 4 °C | Log-linear | N/D | 135 | ||
| WW | 23 °C | Log-linear | N/D | 7.3 | ||
| WW-F | 23 °C | Log-linear | N/D | 23.6 | ||
| SE | 23 °C | Log-linear | N/D | 3.8 | ||
| Casanova et al. (2009)* | TGEV | W | 4 °C | Log-linear | N/D | 110 |
| W | 25 °C | Log-linear | N/D | 11 | ||
| WW* | 4 °C | Log-linear | N/D | 24 | ||
| WW* | 25 °C | Log-linear | N/D | 4 | ||
| MVH | W | 4 °C | Log-linear | N/D | >365 | |
| W | 25 °C | Log-linear | N/D | 9 | ||
| WW* | 4 °C | Log-linear | N/D | 35 | ||
| WW* | 25 °C | Log-linear | N/D | 3 | ||
| Bibby et al. 2015* | Ebola | WW* | 20 °C | Log-linear | N/D | < 1 d |
Matrices abbreviations- TP-D: Dechlorinated Tap Water; AWW; Autoclaved Wastewater; DW: Drinking Water; RW: River Water, RW-F: River Water-Filtered; SE: secondary effluent (treated wastewater); TW: Tap Water; TW-F: Tap Water-Filtered WW: Wastewater; WW-F: Wastewater – Filtered; Viruses abbreviations- FIPV: Feline infectious peritonitis virus. HCoV: Human coronavirus 229E; MHV: mouse hepatitis; PV-1: Poliovirus 1 LSc-2ab. TGEV: transmissible gastroenteritis. (*) Autoclaved or pasteurized samples.
4. Conclusion
Knowing how long SARS-CoV-2 can remain viable in water and wastewater is essential for assessing the risks to public health associated with contaminated water. Our data showed that temperature had a strong effect on SARS-CoV-2 persistence, with T90 values at 4 °C of 7.7 and 5.5 days for RW and WW (respectively), which are 4 to 4.5 times T90 values determined at 24 °C for the same samples. It is important to note that faecal-oral transmission of COVID-19 has not yet been confirmed. Nevertheless, these results could be highly relevant for quantitative microbial risk assessment (QMRA) related to exposure to SARS-CoV-2 contaminated water, since several countries are currently experiencing the highest peaks to date in new COVID-19 cases. In places where adequate sanitation infrastructure is not in place, high SARS-CoV-2 loads could be reaching water bodies and remain active for relatively long periods. Low seasonal temperatures may increase the risk of water-related SARS-CoV-2 transmission, raising concerns over COVID-19 transmission via aerosolized contaminated water and wastewater. One important limitation of this study is the fact that persistence assays were carried out on sterile samples, spiked with infective SARS-CoV-2. Therefore, actual survival times in non-sterile, naturally complex environmental samples could likely be shorter.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are thankful to Marcelo Fernandes Camargos and Kelly Fagundes Nascimento for all their support. We especially thank Laboratório Federal de Defesa Agropecuária LFDA-MG, Ministry of Agriculture, Livestock and Food Supply (MAPA), for allowing and supporting our activities in the BSL-4 laboratory. This work was supported with grants from INCT Dengue (Project 465425/2014-3 financed by MCTI/CNPQ/CAPES/FAPS N° 16/2014 - PROGRAMA INCT). This work was also supported by a grant from the Royal Society (ICA\R1\191241). LCO is funded with a research scholarship from CAPES, AFTF, BCL, BSASS, EAC. The authors would like to thank COPASA (Sanitation Company for the State of Minas Gerais) for making water and wastewater samples available.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2021.117002.
Appendix. Supplementary materials
References
- Abduljauwad Sahel N., Habib T., Ahmed H.-R. Nano-clays as Potential Pseudo-antibodies for COVID-19. Nanoscale Res. Lett. 2020;15:173. doi: 10.1186/s11671-020-03403-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;138764 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environmental Research. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHA . In: Standard Methods for the Examination of Water and Wastewater. 23rd. Baird R.B., editor. Water Environment Federation, American Public Health Association, American Water Works Association; 2017. [Google Scholar]
- Aquino De Carvalho N., Stachler E.N., Cimabue N., Bibby K. Evaluation of Phi6 Persistence and Suitability as an Enveloped Virus Surrogate. Environ. Sci. Technol. 2017;51:8692–8700. doi: 10.1021/acs.est.7b01296. [DOI] [PubMed] [Google Scholar]
- Araujo D.B., Machado R.R.G., Amgarten D.E., Malta F.M., de Araujo G.G., Monteiro C.O., et al. Memórias do Instituto Oswaldo Cruz; 2020. SARS-CoV-2 Isolation From the First Reported Patients in Brazil and Establishment of a Coordinated Task Network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand I., Schijven J.F., Sánchez G., Wyn-Jones P., Ottoson J., Morin T., Muscillo M., Verani M., Nasser A., de Roda Husman A.M., Myrmel M., Sellwood J., Cook N., Gantzer C. The impact of temperature on the inactivation of enteric viruses in food and water: A review. J. Appl. Microbiol. 2012;112:1059–1074. doi: 10.1111/j.1365-2672.2012.05267.x. [DOI] [PubMed] [Google Scholar]
- Bibby K., Fischer R.J., Casson L.W., Stachler E., Haas C.N., Munster V.J. Persistence of Ebola virus in sterilized wastewater. Environmental science & technology letters. 2015;2(9):245–249. doi: 10.1021/acs.estlett.5b00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in Water and Wastewater. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00730. acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer A.F., Eisenberg M.C., Remais J.V., Collender P.A., Meza R., Eisenberg J.N. Modeling biphasic environmental decay of pathogens and implications for risk analysis. Environmental science & technology. 2017;51(4):2186–2196. doi: 10.1021/acs.est.6b04030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraturo F., Del Giudice C., Morelli M., Cerullo V., Libralato G., Galdiero E., Guida M. Persistence of SARS-CoV-2 in the environment and COVID-19 transmission risk from environmental matrices and surfaces. Environ. Pollut. 2020;265 doi: 10.1016/j.envpol.2020.115010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43:1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L.M., Jeon S., Rutala W.A., Weber D.J., Sobsey M.D. Effects of air temperature and relative humidity on coronavirus survival on surfaces. Applied and environmental microbiology. 2010;76(9):2712–2717. doi: 10.1128/AEM.02291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernicharo C.A.L., Araújo J.C., Mota Filho C.R., et al. Sewage monitoring as an epidemiological surveillance tool to control covid-19: a case study in the city of Belo Horizonte. Engenharia Sanitária e Ambiental. 2020 http://abes-dn.org.br/?page_id=35681 online. [Google Scholar]
- Dean K., Wissler A., Hernandez-Suarez J.S., Nejadhashemi A.P., Mitchell J. Modeling the persistence of viruses in untreated groundwater. Sci. Total Environ. 2020;717 doi: 10.1016/j.scitotenv.2019.134599. [DOI] [PubMed] [Google Scholar]
- Fongaro, G., Stoco, P.H., Souza, D.S.M., Grisard, E.C., Magri, M.E., Rogovski, P., Schorner, M.A., Barazzetti, F.H., Christoff, A.P., Oliveira, L.F.V. de, Bazzo, M.L., Wagner, G., Hernandez, M., Rodriguez-Lazaro, D., 2020. SARS-CoV-2 in human sewage in Santa Catalina, Brazil, November 2019. medRxiv 2020.06.26.20140731.
- Gowda N.M., Trieff N.M., Stanton G.J. Inactivation of poliovirus by chloramine-T. Applied and environmental microbiology. 1981;42(3):469–476. doi: 10.1128/aem.42.3.469-476.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Latorre L., Ballesteros I., Villacrés-Granda I., Granda M.G., Freire-Paspuel B., Ríos-Touma B. SARS-CoV-2 in river water: Implications in low sanitation countries. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food and Environmental Virology. 2009;1(1):10–14. [Google Scholar]
- Heller L., Mota C.R., Greco D.B. COVID-19 faecal-oral transmission: Are we asking the right questions? Sci. Total Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.138919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John D.E., Rose J.B. Review of Factors Affecting Microbial Survival in Groundwater. Environ. Sci. Technol. 2005;39:7345–7356. doi: 10.1021/es047995w. [DOI] [PubMed] [Google Scholar]
- Kang M., Wei J., Yuan J., Guo J., Zhang Y., Hang J., Qu Y., Qian H., Zhuang Y., Chen X., Peng X., Shi T., Wang J., Wu J., Song T., He J., Li Y., Zhong N. Probable evidence of fecal aerosol transmission of SARS-CoV-2 in a high-rise building. Annals of internal medicine. 2020;173(12):974–980. doi: 10.7326/M20-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppinen A., Miettinen I. Persistence of Norovirus GII Genome in Drinking Water and Wastewater at Different Temperatures. Pathogens. 2017;6:48. doi: 10.3390/pathogens6040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe D.M., Howley P.M., editors. Fields Virology. 6th ed. Lippincott Williams & Wilkins; Philadelphia: 2013. [Google Scholar]
- Kutz S.M., Gerba C.P. Comparison of virus survival in freshwater sources. Water Sci. Technol. 1988;20:467–471. doi: 10.2166/wst.1988.0327. [DOI] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: Occurrence, persistence and concentration methods - A scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Bonadonna L., Lucentini L., Suffredini E. SARS-CoV-2 has been circulating in northern Italy since December 2019: Evidence from environmental monitoring. Sci. Total Environ. 2021;750 doi: 10.1016/j.scitotenv.2020.141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T.-Y., Jia N., Zhang Y.-W., Shum M.H.-H., Jiang J.-F., Zhu H.-C., Tong Y.-G., Shi Y.-X., Ni X.-B., Liao Y.-S., Li W.-J., Jiang B.-G., Wei W., Yuan T.-T., Zheng K., Cui X.-M., Li J., Pei G.-Q., Qiang X., Cheung W.Y.-M., Li L.-F., Sun F.-F., Qin S., Huang J.-C., Leung G.M., Holmes E.C., Hu Y.-L., Guan Y., Cao W.-C. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lázaro E., Escarmís C., Pérez-Mercader J., Manrubia S.C., Domingo E. Resistance of virus to extinction on bottleneck passages: study of a decaying and fluctuating pattern of fitness loss. Proceedings of The National Academy of Sciences. 2003;100(19):10830–10835. doi: 10.1073/pnas.1332668100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema, G., Heijnen, L., Elsinga, G., Italiaander, R., 2020. Presence of SARS-Coronavirus-2 in sewage. Medrxiv. doi: 10.1101/2020.03.29.20045880. [DOI] [PubMed]
- Michen B., Graule T. Isoelectric points of viruses. J. Appl. Microbiol. 2009 doi: 10.1111/j.1365-2672.2010.04663.x. [DOI] [PubMed] [Google Scholar]
- Mitchell J., Akram S. In: Global Water Pathogen Project. Yates M., editor. 2017. Pathogen specific persistence modeling data. UNESCO, E. Lansing, MI. http://www.waterpathogens.org. http://www.waterpathogens.org/book/pathogen-specificpersistence-modeling-data Part 4 Management of Risk from Excreta and Wastewater, Michigan State University. [Google Scholar]
- Mota C.R., Bressani-Ribeiro T., Araújo J.C. Assessing spatial distribution of COVID-19 prevalence in Brazil using decentralized sewage monitoring. Wat Res. 2021 doi: 10.1016/j.watres.2021.117388. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popat S.C., Yates M.V., Deshusses M.A. Kinetics of inactivation of indicator pathogens during thermophilic anaerobic digestion. Water Research. 2010;44(20):5965–5972. doi: 10.1016/j.watres.2010.07.045. [DOI] [PubMed] [Google Scholar]
- Roos Y.H. Water and Pathogenic Viruses Inactivation—Food Engineering Perspectives. Food Engineering Reviews. 2020;12(3):251–267. [Google Scholar]
- Scheller C., Krebs F., Minkner R., Astner I., Gil-Moles M., Wätzig H. Physicochemical properties of SARS-CoV-2 for drug targeting, virus inactivation and attenuation, vaccine formulation and quality control. Electrophoresis. 2020;41:1137–1151. doi: 10.1002/elps.202000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhu A., Li H., Zheng K., Zhuang Z., Chen Z., Shi Y., Zhang Z., Chen S., Liu X., Dai J., Li X., Huang S., Huang X., Luo L., Wen L., Zhuo J., Li Yuming, Wang Y., Zhang L., Zhang Y., Li F., Feng L., Chen X., Zhong N., Yang Z., Huang J., Zhao J., Li Yi-min. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg. Microbes Infect. 2020;9:991–993. doi: 10.1080/22221751.2020.1760144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R.C. 2020. R: A language and environment for statistical computing. [Google Scholar]
- Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2020. Status of environmental surveillance for SARS-CoV-2 virus. Scientific brief [WWW Document]. URL https://www.who.int/publications/i/item/WHO-2019-nCoV-sci-brief-environmentalSampling-2020-1 (accessed 10.30.20).
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Chen R., Huang Y. Mixed-effects models with skewed distributions for time-varying decay rate in HIV dynamics. Commun. Stat.-Simul. Comput. 2016;45(2):737–757. doi: 10.1080/03610918.2013.873129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, Partitioning, and Recovery of Enveloped Viruses in Untreated Municipal Wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen C, Zhu S, Shu C, Wang D, Song J. Isolation of 2019-nCoV from a Stool Specimen of a Laboratory-Confirmed Case of the Coronavirus Disease 2019 (COVID-19) China CDC Weekly. 2020;2(8):123–124. [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C., Hughes A.C., Bi Y., Shi W. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol. 2020;30:2196–2203. doi: 10.1016/j.cub.2020.05.023. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.