Silver gulls carry phylogenetically diverse Escherichia coli, including globally dominant extraintestinal pathogenic E. coli (ExPEC) sequence types and pandemic ExPEC-ST131 clades; however, our large-scale study (504 samples) on silver gulls nesting off the coast of New South Wales identified E. coli ST457 as the most prevalent. A phylogenetic analysis of whole-genome sequences (WGS) of 138 ST457 samples comprising 42 from gulls, 2 from humans (Australia), and 14 from poultry farmed in Paraguay were compared with 80 WGS deposited in public databases from diverse sources and countries.
KEYWORDS: ST457, ExPEC, ESBL, AmpC, I1 plasmids
ABSTRACT
Silver gulls carry phylogenetically diverse Escherichia coli, including globally dominant extraintestinal pathogenic E. coli (ExPEC) sequence types and pandemic ExPEC-ST131 clades; however, our large-scale study (504 samples) on silver gulls nesting off the coast of New South Wales identified E. coli ST457 as the most prevalent. A phylogenetic analysis of whole-genome sequences (WGS) of 138 ST457 samples comprising 42 from gulls, 2 from humans (Australia), and 14 from poultry farmed in Paraguay were compared with 80 WGS deposited in public databases from diverse sources and countries. E. coli ST457 strains are phylogenetic group F, carry fimH145, and partition into five main clades in accordance to predominant flagella H-antigen carriage. Although we identified considerable phylogenetic diversity among the 138 ST457 strains, closely related subclades (<100 SNPs) suggested zoonotic or zooanthroponosis transmission between humans, wild birds, and food-producing animals. Australian human clinical and gull strains in two of the clades were closely related (≤80 SNPs). Regarding plasmid content, country, or country/source, specific connections were observed, including I1/ST23, I1/ST314, and I1/ST315 disseminating blaCMY-2 in Australia, I1/ST113 carrying blaCTX-M-8 and mcr-5 in Paraguayan poultry, and F2:A-:B1 plasmids of Dutch origin being detected across multiple ST457 clades. We identified a high prevalence of nearly identical I1/ST23 plasmids carrying blaCMY-2 among Australian gull and clinical human strains. In summary, ST457 is a broad host range, geographically diverse E. coli lineage that can cause human extraintestinal disease, including urinary tract infection, and displays a remarkable ability to capture mobile elements that carry and transmit genes encoding resistance to critically important antibiotics.
INTRODUCTION
The antibiotic resistance crisis is growing steadily across almost every host and environment tested worldwide (1). These trends are especially concerning in common and emerging pathogens, including extraintestinal pathogenic Escherichia coli (ExPEC) (2). ExPEC cause a wide range of opportunistic infections that have proved to be a challenge to define and diagnose with accuracy, and increasingly are resistant to multiple antimicrobial agents (3, 4). The number of E. coli sequence types (ST) identified to have caused ExPEC infections in humans and animals continues to rise (5, 6). Whole-genome sequencing programs are shedding light on the phylogeny of emerging ExPEC, improving efforts to define their virulence and antibiotic resistance gene (ARG) pool, but the emphasis to date has overwhelmingly been on pathogens of human clinical relevance (7–10).
E. coli ST457 belongs to Clermont phylogroup F (11) and was first reported in 2008 (sampled between 2004 and 2005) in the United Kingdom from a patient with a urinary tract infection (UTI) (12). ST457 has since been reported in studies from all continents and from diverse sources (13–18), including wild animals in Antarctica (19). These observations indicate that ST457 is a broad host range and globally disseminated E. coli lineage.
ST457 often carries resistance to clinically important antibiotics. Carbapenemase-producing clinical strains of E. coli ST457 from patients with bloodstream infection in Italy and Mexico (KPC-2), in Shanghai (NDM-5), and in the United States (KPC-3) have been reported (17, 20–22). ST457 is frequently associated with the production of CTX-M (mainly from the CTX-M-1 group: CTX-M-15, CTX-M-27, and CTX-M-55) and CMY-2 β-lactamases in diverse countries, including Australia, Brazil, Mexico, United States, Canada, Italy, China, and Thailand (13, 16, 17, 23–27).
E. coli ST457 is also often reported to carry plasmid-associated mcr genes that encode resistance to colistin, an antibiotic of last resort. Reports of the isolation of ST457 carrying mcr-1 from patients residing in the United States, Mexico, China, and Vietnam (16, 24, 25, 28) are concerning. Reports of mcr-1-carrying ST457 causing mastitis in dairy cattle in Japan (14) and mcr-1-positive ST457 strains in wildlife and in poultry, predominantly in Asia (26, 29), shed light on the scale by which ST457 has spread. Notably, ST457 carrying mcr-3 has been isolated from a wild bird in China (30).
In poultry production systems in Paraguay, we described a multidrug-resistant (MDR) ST457 clone carrying mcr-5, blaCTX-M-8, blaTEM-1A, aph(6)-Id and aph-Ib, and sul2 (31). mcr-5 was typically located on an I1/ST113 plasmid that coharbored blaCTX-M-8. I1/ST113 plasmids have been previously described to spread blaCTX-M-8 in Japan (32), Brazil (33), and Germany (34). I2 plasmids recovered from E. coli ST457 were found to separately carry mcr-3 in China and mcr-1 in a clinical strain in Mexico and poultry strains from China, respectively (24, 26, 30). blaCMY-2 has also been detected on I1 plasmids carried by E. coli ST457 in poultry, wildlife, and companion animals in Brazil (15, 23, 35), underscoring the important role played by I1 plasmids in capturing and spreading genes encoding resistance to clinically important antibiotics in ST457, as previously shown in other E. coli lineages (15). The I1 plasmids belonged to various STs, including ST12, which is a notable plasmid lineage spreading blaCMY-2 globally (36). blaCTX-M-55 was carried by F plasmids, specifically F18:A-:B1, in a clinical strain of ST457 from the United States and F33:A-:B- in poultry from Brazil (16, 23).
MDR E. coli are a feature of the fecal contents of silver gulls (Chroicocephalus novaehollandiae) nesting on several islands off the coast of New South Wales, Australia. In an earlier study in 2012, Dolejska et al. (37) described E. coli belonging to diverse sequence types that carried carbapenemase genes, including blaIMP-4, blaIMP-38, and blaIMP-26, and extended-spectrum-β-lactamase genes such as blaCMY-2 from the cloacal contents of 504 gull chicks inhabiting three islands off the coast of New South Wales, Australia. Subsequent analyses, described here, showed that E. coli ST457 were a feature of isolates recovered during that study. Specifically, we sought to determine if resistance to clinically important antibiotics is acquired and mobilized among the E. coli ST457 sourced from these silver gulls by conducting whole-genome sequencing. We used a combination of short- and long-read DNA sequencing and hybrid assemblies to generate complete sequences of the chromosome and plasmid cargo of an ST457 strain representative of the panel of ST457 strains from Australian gulls. We also recovered E. coli ST457 sequences from EnteroBase (https://enterobase.warwick.ac.uk/) from different hosts and countries and performed detailed phylogenetic analyses to examine clade structure and genomic features of ST457.
(Part of this work was presented in the abstract book of the 30th European Congress of Clinical Microbiology and Infectious Diseases [ECCMID] 2020 [Abstract no. 3781; event cancelled] and as a poster at the One Health EJP Annual Scientific Meeting 2020 [online event].)
RESULTS
Phylogeny.
All 138 strains of E. coli ST457 belonged to phylogenetic group F and carried fimH145. Based on our single-nucleotide polymorphism (SNP) analysis, the collection of ST457 partitioned into five main clades, largely in accordance to flagella H-antigen carriage, and included H25 South, H25 North, H6, H45, and H16 and two minor clades (identified here as MC1 and MC2) (Fig. 1; Table S1 in the supplemental material). Serogroup O11 dominated the collection (79%; 109/138), as did the H25 flagella antigen (67%; 92/138) and, unsurprisingly, serotype O11:H25 predominated (52%, 72/138). ST457 with O11:H25 was found in three out of the five main clades and it was the predominant serotype in two out of those three clades (H25 North, H25 South). Several clade-specific serotypes, including O11:H16, O11:H6, and O11:H45, were also detected. The geographical origin of the strains varied even at the clade level, including countries from North and South America, Europe, Australia, and Asia. Interestingly, even when the gull strains from Australia were excluded, four clades contained strains of Australian origin.
FIG 1.
Phylogenetic analysis of the global E. coli ST457 cohort, highlighting the source diversity. The five primary clades of ST457 are defined using H antigen from clade-typical serotype. Two clades of H25 were distinguished based on the most common origin of the strains, separating North or South hemisphere. Two minor clades (MC1 and MC2) were not evaluated separately. The white circle indicates that the specific metadata was not available.
The H25 North clade contained 28 strains and compared to the other clades the SNP analysis showed considerable diversity (>1,500 SNPs on average) (Table S2, H25 North). However, four separate subclades (up to 20 SNPs different) were detected within the clade in three bovine strains from Luxembourg (HA9247, HA9232, and HA9243), four human strains from the United States (RA8202, RA8205, RA8209, and RA8206), two clinical strains from China (XA6422 and WA4383), and two strains from the UK (TA7464 and SA4138). Notably, the UK strains, the U.S. strains, and a single strain from Cambodia (WA7349) and Australia (FB3603) created a closely related subclade with differences between 39 to 44 SNPs (87 to 92 SNPs for the Australian strain). We would like to highlight a close phylogenetic relation between human and food-producing animal strains observed in the H25 North clade. Strain GB0145 from Brazilian swine was separated by 46 SNPs from human strain FB5883 from China, while strain PA1693 from poultry in Vietnam differed by 69 SNPs from YA1066, a human clinical strain from China.
The strains of the H25 North clade differed in ARGs and virulence-associated genes (VAGs); however, the resistance gene cargo was overall higher than in the other four ST457 clades. An average strain from H25 North clade carried twice as many ARGs as an average strain from the H16 or H6 clades, and for the H25 South and H45 clades, the ratio was even higher. All strains in the H25 North clade lacked the yersiniabactin siderophore gene cluster (fyuA, irp1, irp2, and ybtAEPQSTUX) compared to the other four ST457 clades (Fig. 2).
FIG 2.
Phylogenetic analysis of the E. coli ST457 collection. The diagram depicts country of origin, year of isolation, source, serotype, carriage of yersiniabactin (pink), ESBL/AmpC and mcr genes (green), I1 plasmid STs (purple), and combination of F replicons (blue) for each strain individually. The clades are distinguished with the branch colors as follows: H16 (orange), H25 North (red), H6 (turquoise), H45 (purple), and H25 South (blue). The Australian “gulls” subset is distinguished with the text color based on the location of the isolation as follows: Montague Island (green), Five Islands (purple), and Sydney (blue). The empty fields indicate the specific metadata was not available.
The H16 clade comprised 7 strains and consisted mostly of bovine strains from the Netherlands. Strains VA5580, VA5582, and VA5574 were clonal and differed in 3 to 9 SNPs (Table S2, H16). All six Dutch strains were closely related, as the number of SNPs between the two most distinct strains was only 72. Interestingly, a single Australian human strain was part of this clade but was phylogenetically distant (approximately 3,700 SNPs) from the others in the clade. It also carried different ARGs, including blaIMP-4, and VAGs cargo and housed different plasmid replicons (Fig. S1).
The H6 clade (29 strains) was mostly of poultry origin from South and North America, in part due to the inclusion of our collection of mostly clonal Paraguayan poultry strains (n = 14; 1508e to 1527k). Surprisingly, the H6 clade contained four strains from Japan, including three human isolates. Notably, Japanese human strain QA5359 was closely related to human Peruvian strain VA5961 (69 SNPs). Moreover, Japanese human strain QA5364 showed close similarity with U.S. canine strain SA0179 (106 SNPs) and three U.S. poultry strains ZA5550 (124 SNPs), VA9753 (132 SNPs), MA1963 (146 SNPs) (Table S2, H6).
A close similarity was evident between Australian human clinical and gull strains in the H45 clade (14 strains). Gull strain CE1765 differed from the two clinical strains WA5506 and WA5541 by 21 and 33 SNPs, respectively, highlighting potential zoonotic/zooanthroponosis (reverse zoonosis) linkages with ST457 in Australia (Table S2, H45).
The H25 South clade (55 strains) of ST457 contained most of the Australian gull strains. Only five strains, gull strain CE1803A, Argentinian human strain RA2540, and three clonal U.K. human strains (GA9323, GA9192, and GA9368) displayed a large number of SNPs (>650) compared to the remaining strains in the clade (Table S2, H25 South). The other 50 strains originating from Australia, Germany, the United States, Japan, New Zealand, and the Netherlands from 2009 to 2019 differed by ≤181 SNPs. Several strains from different countries or sources were closely related within the H25 South clade (up to 80 SNPs); for example, U.S. poultry strain FB9087 possessed 76 and 77 SNPs compared to German human strain RA5006 and one of the gull strains CE1679, respectively.
Subsets of the human/clinical strains from Australia were clonal (EC0125 and EC0382, 1 SNP difference; WA5591 and WA5546, 10 SNP difference) despite being isolated from different locations (Wollongong and Sydney) or different years (2009 and 2014). Similarly, we detected many apparent clones (≤20 SNP differences) within the collection of isolates from silver gulls. Close genetic associations were also evident when comparing strains from Australian gulls with Australian human strains of ST457 (≤30 SNP differences between strains within these two categories). Human isolate WA5546 differed by up to 30 SNPs with 11 gull strains and, notably, only 10 SNPs with gull strain CE1966 (Table S2, H25 South). Of interest, ST457 strains CE1679 (gull) and IA1961 (Laughing Kookaburra Dacelo novaeguineae) isolated in Queensland showed only 20 SNP differences. The highest prevalence of ARGs and VAGs was present in 12 non-Australian ST457 strains (5.5 ARGs per non-Australian strain on average compared to 1.1 ARGs on average in Australian strains) in this clade. Phylogeny of the accessory genome showed that Australian strains (except strain CE1803A) of the H25 South clade clustered together while other strains of this clade were distant and some partitioned with different accessory clusters (Fig. S2). Interestingly, when comparing accessory genomes, all strains from H25 North clade clustered together. This was also observed for strains belonging to the H45 clade. On the other hand, all strains from the Netherlands clustered together in accessory genome phylogeny despite being part of different core genome clades and coming from different sources (Fig. S2).
Virulence-associated genes.
All ST457 strains carried diverse VAGs ranging from 43 to 75 per strain (Fig. S1). VAGs shared by all were those that play a role in heme/iron acquisition, e.g., enterobactin-related genes fepB, fes, entB, or chuY and shuA. All strains carried genes encoding fimbria and adhesins, including fimH or ecpD, the outer membrane protein gene ompA, and the invasion-associated gene aslA. However, in four ST457 strains, aslA was expected to be nonfunctional because of truncation (Fig. S1). Yersiniabactin siderophore genes fyuA, irp1, irp2, and ybtAEPQSTUX were detected in most strains (76%, 105/138). K antigens genes of group II, including kpsD (96%, 132/138) and kpsM (86%, 118/138), were found in most strains, while the capsule regions were variable. Besides kpsM and fyuA, the other commonly reported VAGs recognized as potential ExPEC or APEC markers (19, 38, 88) were present in lower number in our strains, including papC (8%; 11/138), iroN (14%; 20/138), hlyA (3%; 4/138), or sfa/focD/cnf1 (1%; 1/138).
Antibiotic resistance genes.
We detected genes encoding resistance to 13 antimicrobials groups: β-lactams (90%; 124/138), aminoglycosides (54%; 75/138), sulfonamides (54%; 75/138), tetracyclines (38%; 53/138), trimethoprim (32%; 43/138), amphenicols (22%; 31/138), colistin (18%; 26/138), macrolides (17%; 23/138), fosfomycine (5%; 7/138), quinolones (4%; 6/138), carbapenems (2%; 3/138), lincosamides (2%; 3/138), and rifampin (1%; 1/138).
The five main clades were quite diverse in resistance gene cargo both qualitatively and quantitatively. An average strain from the H25 North clade carried more ARGs (13.0 genes per strain) compared with ST457 strains from H6 (4.9) or H16 (6.1) clades. The gull-associated clades H45 (1.1) and H25 South (2.6) that were prevalent in Australia typically carried limited resistance gene cargo.
Extended spectrum β-lactamase (ESBL) and AmpC β-lactamase genes were frequently identified in the ST457 strains. The most common genes of this group were blaCMY-2 (47%, 65/138), associated mostly with the H25 South, H45, and H16 clades, and blaCTX-M-1; 2; 3; 8; 12; 14; 27; 55 variants found in 46/138 strains (33%). blaCTX-M-8 and blaCTX-M-27 were the most prevalent among these variants (15/138, 11% each). blaCTX-M-8 was found mostly in Paraguayan poultry (H6 clade) and in a Bolivian human strain (H25 North), while blaCTX-M-27 was associated only with the H25 North clade with 54% (15/28) clade prevalence. blaCTX-M-55 was found with 23% (7/28) and 3% (1/29) clade prevalence in H25 North and H6 clades, respectively.
Genes encoding resistance to sulfonamides were frequently identified as follows: sul2 47% (65/138), sul1 13% (18/138), and sul3 12% (16/138). However, sul2 was more prevalent in H25 North, H16, and H6 clades, where it was detected in 76 to 86% of ST457 strains. sul3 was found only in the H25 North clade in 57% (16/28) of strains, for which only five of them were clonal.
Carbapenemase genes were found sporadically in three strains only. Specifically, we identified blaIMP-4 in an Australian human strain, blaNDM-9 in a poultry strain from China, and blaOXA-23, which was located on the chromosome of CE1628, from the Australian gull collection (Fig. 3).
FIG 3.
Genetic environment of blaOXA-23 present in the chromosomal structure of 32,421 bp (GenBank MT468652) in the CE1628 gull strain. The region includes the fragmented ABC transporter ΔyddA from the E. coli K-12 backbone (gray), mobile genetics elements (blue), resistance genes (lime), mercury resistance operon (yellow), gene fragments (black), and other genes (pink). The labels below the sequence refer to mobile genetics elements and other full genes, while the upper labels identify fragments of genes as follows: ORF, open reading frame fragment; CP, chemotaxis protein fragment. Identified regions are marked by numbers in the colored boxes below to show the reference(s) that was highly similar and used for annotations, as follows: E. coli MN51447 plasmid (green, region 1: 3,734 bp, 99.92% identity [ID]; region 10: 899 bp, 100% ID); uncultured bacterium FJ012880 plasmid (orange, region 2: 4,034 bp, 100% ID, region 8: 3,275 bp, 100% ID); Acinetobacter spp. (red, CP020586, region 4: 2,313 bp, including blaOXA-23, 100% ID, region 3: 1,293 bp, 99.85% ID; CP048014, region 2A: 1,481 bp, 100% ID; CP044484, region 7: 189 bp, 100% ID); E. coli KJ716226 plasmid (turquoise, region 5: 2,577 bp, including blaOXA-23, 100% ID), and E. coli CP020934 plasmid (dark purple, region 9: 13,911 bp, 99.99% ID; region 6: 769 bp, 99.10% ID).
Plasmid-mediated resistance to colistin was represented by carriage of mcr-1.1 or mcr-2.1 in 46% (13/28) of strains in the H25 North clade. In the H6 clade, the occurrence of mcr-5.1 (45%, 13/39) was high but, in contrast to the diversity of strains in the H25 North clade, was due to clonal spread on Paraguayan farms.
Overall, we detected 10 strains, mostly from the H45 clade, which did not carry any ARGs. In contrast, ST457 strain HA3078 from a poultry source in China carried 29 resistance genes and 9 plasmid replicons, indicating that ST457 can carry extensive resistance gene cargo on diverse plasmids.
The Australian gull and human collections from H25 South and H45 clades were quite unified regarding resistance gene content. Most of these strains carried only the plasmid-borne gene blaCMY-2; however, CE1887 gull strain carried 13 resistance genes, including blaCMY-2 and 7 plasmid replicons.
blaOXA-23 and carbapenemase-related phenotypic characteristics.
A region of DNA from the chromosome of gull strain CE1628 containing blaOXA-23 (GenBank MT468652) carried multiple mobile elements and other resistance genes, including sul2, aph(3′)-Ib, aph(6)-Id, and aph(3′)-Ia, and a mercury resistance operon (Fig. 3). Using BLASTn, we did not find a full match of this region with sequences deposited in public databases. The genetic structure suggests it is comprised of separate regions derived from apparently unrelated sources and its path to formation remains unknown. While the region spanning blaOXA-23 is typical of those observed in Acinetobacter spp., subregions of it have been seen within E. coli plasmid KJ716226 (39) (Fig. 3).
Strain CE1628 was positive for carbapenemase activity using a matrix-assisted laser desorption ionization (MALDI) assay, however, the activity was low. MIC values showed the strain was resistant to five extended-spectrum β-lactam antibiotics or their combination with β-lactamases inhibitors as follows: ampicillin (MIC > 128 mg/liter), ampicillin-sulbactam (32 mg/liter), cefuroxime (64 mg/liter), cefotaxime (>8 mg/liter), and cefprozil (64 mg/liter). The strain was susceptible to meropenem (<0.125 mg/liter) and intermediate to ertapenem (1 mg/liter) (Table S3). In contrast to our findings, a previously described E. coli strain carrying the blaOXA-23 region on plasmid KJ716226 showed higher MIC values for carbapenems: meropenem 4 mg/liter (resistant) and ertapenem 6 mg/liter (resistant) (39). MIC values for all tested antibiotics and their interpretation using CLSI breakpoints (40) can be found in Table S3.
Plasmids carried by E. coli ST457 and their transferability.
ST457 strains carried various plasmid replicons, including different Col-type replicons, C, B/O/K/Z, FIA, FIB, FIC, FII, HI1A, HI1B, HI2, I1, I2, L/M, N, X1, X2, X4, Y, and p0111. F plasmids (61%; 84/138), particularly FIB (49%, 68/138), I1 (59%; 81/138), and I2 (18%; 25/138) predominated among large plasmids (Fig. S1). While I1 plasmids were prevalent mostly in the H25 South (82%, 45/55), H45 (71%, 10/14), and H6 (62%, 18/29) clades, F plasmids were common in the H25 North (89%, 25/28), H16 (86%, 6/7), and H6 (83%, 24/29) clades. I2 plasmids were frequently identified in H25 North (36%, 10/28) and H45 (29%, 4/14) clades.
Previously, we demonstrated transfer of I1/ST113 plasmids carrying blaCTX-M-8 and mcr-5 using Paraguayan poultry strains as a source to recipient E. coli strains via conjugation (31). Using representative ST457 from gulls, successful conjugal transfer was achieved in 9 of 13 strains that carried I1 plasmids with blaCMY-2 and in a single strain carrying an F18:A-:B1 plasmid with blaDHA-1.
I1 plasmids.
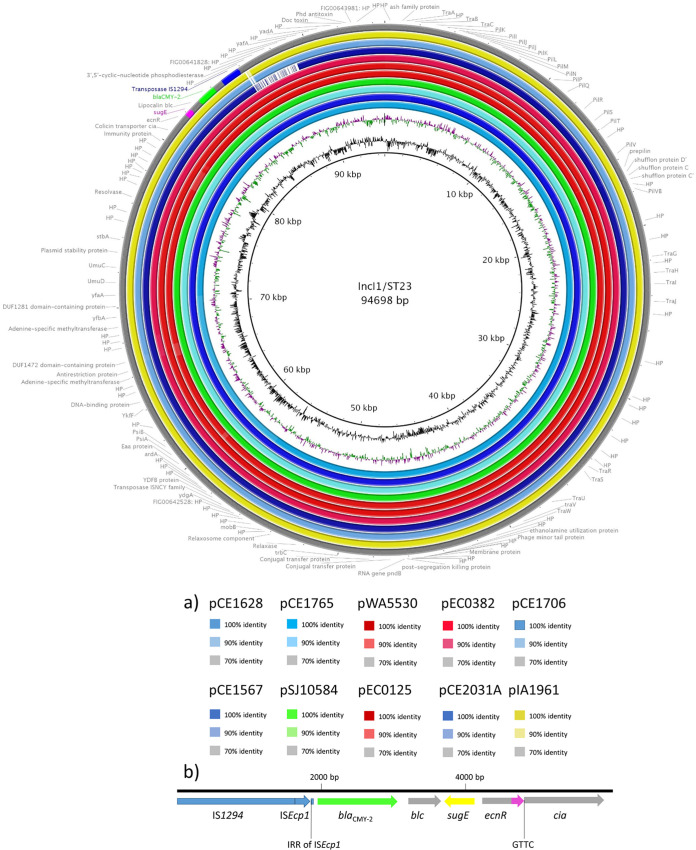
I1/ST23 plasmids disseminating blaCMY-2 showed high prevalence in Australia, as we detected their presence in most of the ST457 gull strain collection (81%; 34/42). Only one gull strain did not carry an I1 plasmid, whereas the rest carried I1-blaCMY-2 plasmids with novel ST315 (10%, 4/42) and ST314 (7%, 3/42) sequence types. From 12 Australian human and clinical strains in the H25 South and H45 clades, 67% (8/12) carried I1/ST23 plasmids. This plasmid was also detected in Australian avian strain IA1961 (Dacelo novaeguineae). The complete sequence of a 94,698-bp I1/ST23 plasmid was generated here from E. coli strain CE1628 by combining PacBio and Illumina data and performing a hybrid assembly using Unicycler (GenBank MT468651). The polished sequence was used as a reference for BRIG analyses (Figure 4a) and showed that closely related I1/ST23 plasmids are shared by a subset of human and gull strains (Fig. 4). Not all gull plasmids were identical, but all of them encoded the same resistance structure comprising blaCMY-2 and sugE (resistance to quaternary ammonium compounds) flanked by IS1294 (Figure 4b). The region spanning blaCMY-2 comprises 6,053 bp and showed 100% sequence identity with previously described plasmids with I1 and C replicons (41, 42). An I1/ST265 plasmid (pS10584; 94,697 bp) in Salmonella enterica strain SJTUF10584 isolated in Shanghai, China showed 99.9% sequence identity with our gull reference plasmid using BLASTn. Figure 4, which depicts pS10584 and selected I1/ST23 plasmids sourced from wild birds and humans, shows that identical plasmids were found in E. coli strains from human and gull sources plus S. enterica strain SJTUF10584. A BRIG comparison of all ST23, ST314, and ST315 plasmids that carried the same blaCMY-2 region brings the total to 51 related plasmids (Fig. S3).
FIG 4.
(a) BRIG comparison of selected I1/ST23 plasmids carrying blaCMY-2 (lime) coming from Australian silver gulls (blue), humans (red and pink), laughing kookaburra (yellow), and including Salmonella enterica serovar Typhimurium strain SJTUF10584 plasmid pS10584 (I1/ST265) from Shanghai (green). Gull strains carrying these plasmids came from Five Islands (CE1628, CE1567, and CE1706), Sydney (CE1765), and Montague Island (CE2031A). EC0125 (Wollongong) and EC0382 (SGH Sydney) are clinical strains. EnteroBase strains include a clinical strain from Sydney, 2013 (WA5530) and laughing kookaburra avian strain from Queensland, 2011 (IA1961). (b) Genetic context of blaCMY-2 gene (6,053 bp) in I1/ST23 was identical with the I1/ST2 plasmid pJIE512e described in Tagg et al. (42), which proposed that blaCMY-2 (lime), sugE (yellow), blc (outer membrane lipoprotein, gray), and ΔecnR (entericidin R fragment, gray) were mobilized from C plasmids (pink represents an 159-bp fragment of the C backbone) by the insertion sequence IS1294 (blue). The gene encoding colicin Ia protein Δcia came from the I1 plasmid backbone and was fragmented by the insertion. The insert was flanked by the IS1294 target site CTTG and bounded by the IS1294 terIS look-alike sequence GTTC at the other side, as was previously described (42).
We previously described the dissemination of I1/ST113 plasmids carrying blaCTX-M-8 and mcr-5 in Paraguayan farms (31) but we did not detect I1/ST113 plasmids in other strains from the ST457 collection.
I1/ST12 plasmids carrying blaCMY-2 were found only in five strains of ST457, but while other I1 plasmids were strictly country associated, this plasmid sublineage was found in different clades of ST457 internationally, including in strains from the United States (poultry), China (poultry), Brazil (swine), and Germany (human).
F plasmids.
We found F replicons (FIB, FII, FIC, FIA) in 61% (84/138) of the ST457 strains. Carriage of these plasmid replicons was dominant in the H25 North, H6, and H16 clades. We noticed that several combinations of F replicons were repetitive (Fig. 2). F29:A-:B- plasmids (n = 9) were detected in a subset of Australian gulls, all of them from the same location (Montague Island). F64:A-:B27 (n = 14) was related to Paraguayan poultry. Interestingly, F2:A-:B1 (n = 11) plasmids were more related to the country of their origin than with clonal relationships of the strains that carried them, as they were found in all strains from the Netherlands (bovine and human origin) even if they belonged to different clades (H25 South and H25 North). Moreover, F2:A:B1 plasmids were detected in two poultry strains from the United States (H6 and H25 South) and a human strain from Japan (H6).
F24:A-:B1 plasmids (n = 15) were prevalent in the H25 North clade (50%; 14/28) and found in one Japanese strain from a human in the H6 clade. A similar situation was observed for F18:A-:B1 plasmids (n = 10), which were prevalent in the H25 North clade (29%; 8/28) but present as well in H6 and H25 South clades, with one identified in a Japanese strain (water source) and one detected in a gull strain isolated in Australia.
ST457-specific chromosomal regions of difference.
We detected 141 regions of difference (RODs) in the ST457 chromosome (referred to as AA to FK, Table S4 and Table S5) which were shared by all three completed ST457 genome sequences and absent in E. coli K-12. Ninety-six of these 141 RODs were detected in 95% of our ST457 strains, suggesting they may play a role in the expression of features unique to E. coli ST457 (Table S4). RAST identified more than 300 proteins in shared RODs and many of these had functions involved in toxin/antitoxin systems, including RelE-like proteins (AC, AR, and CZ), HicAB-like proteins (AG), HigB (DS), RelB (BR), and ParDE (CN). Two RODs were associated with heme uptake (AI and ED), many encoded putative adhesins (AW, CJ, CR, DR, EP, and FI) and general secretion pathway proteins (FC). All ST457 strains also possessed a region (DY) for efflux of antibiotics and RODs with MFS-type transporters (EI, EJ, and EK). Notably, however, approximately 36% (35/96) of all the shared RODs carried genes encoding proteins which seemed to be related to metabolism and substrate uptake and transport across cell membranes. These included proteins for metabolism of sucrose (EW), cellobiose (DP), arabinose (AA and EY), galactose (AA), xylose (AA, AK, and BX), 3-phospho-d-glycerate (EY), serine (AM, BU, and EI), histidine (CW), aspartate (CW and ER), N-acetylgalactosamine (FJ), and hydroxyaromatic compounds (EU).
DISCUSSION
Tracking diverse E. coli sequence types is becoming critically important in the One Health understanding of disease epidemiology, particularly concerning the zoonotic and reverse zoonotic transfer of pathogens and antimicrobial resistance. Here, we isolated and performed whole-genome sequencing on 42 E. coli ST457 strains from Australian gulls, 2 Australian clinical strains, and 14 E. coli ST457 from chickens in Paraguay, then utilized the EnteroBase database where we recovered 80 ST457 WGS to determine their phylogenetic relationships to one another from a global context. EnteroBase currently places the oldest strain of ST457 to 2001, in an Australian wild bird. The EnteroBase ST457 strains have increased from 176 to 219 in 5 months, since we began the analysis on 30 October 2019. EnteroBase records demonstrate the global expansion of this lineage and indicate ST457 can be isolated from diverse sources, including healthy and sick humans, poultry, cattle, swine, wild animals, companion animals, water, and food. ST457 is geographically disseminated, having been isolated from countries including Australia, New Zealand, China, Japan, Vietnam, Cambodia, the United States, Argentina, Bolivia, Mexico, Ecuador, Peru, Brazil, Germany, the United Kingdom, Luxembourg, France, Italy, the Netherlands, and Spain (http://enterobase.warwick.ac.uk/).
Through the generation of a global phylogeny, we identified five primary clades for E. coli ST457. Three of these clades contained strains from our studies, clustering with other sequences in EnteroBase. We observed several cases of clonal spread defined (≤20 SNP differences in the 3,756 core genes), including strains of bovine origin from Luxembourg, human strains from the United States, clinical strains from China, human strains from the United Kingdom, bovine strains from the Netherlands, poultry strains from Paraguay, gull strains from Australia, and human and clinical strains from Australia. We would like to especially highlight the sharing of the same clones of ST457 among an Australian gull and human/clinical strains that was observed in three cases (Table S2, H25 South). Our data suggest that a clonal lineage of ST457 can occupy a different niche in New South Wales, Australia and implies gulls may be an environmental reservoir and transmission route for human pathogens. Whether our data supports evidence for zoonosis, zooanthroponosis, or both is uncertain. Gulls from Five Islands are known to visit municipal waste sites in the Wollongong region of New South Wales (43). Australian silver gulls were previously identified as carriers of virulent and antibiotic-resistant human-associated E. coli clones (44). Interestingly, a wild bird (Dacelo novaeguineae) ST457 strain coming from Queensland is closely related to one of our gull-sourced strains, which highlights that such strains may be further distributed in the environment across long distances via the flight behavior of the birds that carry them.
Moreover, we have detected closely related (≤80 SNPs) human strains of ST457 sourced from very distinct countries, e.g., Japan/Peru; United Kingdom/United States/Cambodia; New Zealand/the Netherlands; Germany/Japan; New Zealand/Germany; and the Netherlands/Australia (clinical). Similarly, closely related isolates were observed from diverse sources and countries as Brazil (swine)/China (human); China (poultry)/Vietnam (clinical); United States (poultry)/German (human); United States (poultry)/Australia (gull); United States (poultry)/Australia (human); United States (water)/Japan (human), or New Zealand (human)/United States (water) (Table S2, H25 North, H25 South, H6). These examples of highly related strains being sourced from such distinct environments highlight the international and cross-environmental transmission of microbes, and is also an indication of the capacity of E. coli ST457 to inhabit multiple and diverse niches and acquire mobile genetic elements from those niches. Similarly, E. coli strains belonging to STs which commonly cause ExPEC diseases show zoonotic potential. Some have been detected even within the most distinct environmental niches, including ST131, ST73, ST648, or ST95 in Antarctic pinnipeds (19). Moreover, a lack of specific host-adaptation markers was shown previously for ExPEC, such as ST131 or ST648, which suggests they are able to survive and proliferate in diverse hosts and are not specifically human-adapted (45). The phylogenetic analysis of the accessory genome of ST457 showed that accessory clades are in relatively good concordance with core genome clades and do not demonstrate niche-based adaptation (Fig. S2). This suggests at least some clades or subclades (H45, H25 North, Australian part of H25 South) are quite conserved and well-developed, which is interesting for this seemingly young lineage and in contrast to observations in ST648, for which accessory genome to core genome clade clustering was limited (45). Nevertheless, further sampling is needed to shed light on the distribution and evolution of ST457.
The frequently observed VAGs in ST457 encoded a range of functions suspected to play roles in cell survival during extraintestinal infections, including siderophores and heme-acquisition proteins, adhesins, capsules, and fimbria. Capsule antigens play crucial roles in many host pathogen interactions, including ExPEC, as they provide protection against complement-mediated killing (19, 46) and influence niche colonization (47). Genes encoding the siderophore enterobactin seem to be related not only to iron uptake but they may play other physiological roles, including responses to oxidative stress (48). Another siderophore, yersiniabactin, was previously described to contribute to the virulence of E. coli and possibly to copper resistance (46), and its frequent association with UPEC strains even led to attempts to use it as an antigen for vaccines against UTIs (49). Interestingly, the yersiniabactin gene cluster was not present in the H25 North clade, which otherwise seems to be a progressive clade with a propensity to carry resistance gene cargo. The reason for lacking the yersiniabactin is unclear; pathogen evolution is complex and the acquisition of virulence attributes by commensal or otherwise less-pathogenic lineages, as suggested by the acquisition of ColV plasmids and genomic islands (50), represents one path (51–53). On the other hand, loss of genomic DNA represents another path to pathogen evolution. Some E. coli may undergo a process of genome reduction to become more host-adapted and be less obvious to the host immune system (46).
One of the main factors which determine whether a bacterium will be successful in a specific niche is reflected in its metabolism capacities (46). ExPEC strains are capable of utilizing substrates not typical of commensal flora (54, 55). Some studies propose that the capacity for rapid growth linked to metabolic capacity should be considered a virulence attribute (56, 57). We detected genes in all ST457 strains that were absent in E. coli K-12 that may to be related with utilization of three (3-phospho-d-glycerate, cellobiose, and N-acetyl-d-galactosamine) out of four nutrients predicted by Monk et al. (54) to give ExPEC strains a catabolic advantage. Further studies are needed to investigate these preliminary observations.
A large-scale study focused on identifying resistance to critically important antibiotics in silver gulls in Australia revealed that 22% of the samples were positive for E. coli isolates resistant to extended-spectrum cephalosporins (44). Previous research focused on phylogenetic group F in Australia identified that some STs are more likely to be fluoroquinolone resistant (ST354, ST648, and ST3711) than others (ST59 and ST62) with unclear reasons as to why (58). Two ST457 wild bird strains described in that study were included in our collection (IA1961, referred to as B1700, and IA1962, referred to as B093) (58). Our results showed ST457 carried quinolone-resistance genes only at low frequency, while appearing better adapted to carrying ESBL/AmpC genes. Most of the Australian ST457 strains sourced from gulls and humans in our study were not MDR, carrying only blaCMY-2, but they were widespread, representing the most common chromosome type isolated from these gull samples. A recent study focusing on ESBL-producing E. coli strains from a hospital in Taiwan showed that ST457 caused 4% of UTIs (3rd after ST131 and ST38; equal to ST69 and ST405), 5% of primary bacteremia (4th after ST131, ST38, and ST405), and 8% of biliary tree infections (2nd after ST131; equal to ST38, ST12, and ST10) (38). Another study from a hospital in Nigeria with a nonbiased selection of ExPEC strains from urine, blood, and intraabdominal wounds identified ST457 as the second most prevalent type (20%; 12/60), and all of these strains carried blaCTX-M-15 (18). These findings reveal ST457 is one of the major ESBL-connected E. coli pathogens in Taiwan and in Nigeria, two countries with no known ST457 representatives in EnteroBase. These data suggest that ST457 is an emerging E. coli ST that is able to cause ExPEC disease and occupy diverse niches in humans, food animals, wild birds, and water. Moreover, ST457 seems to benefit from carriage of ESBL/AmpC genes. Whether this advantage lies specifically in the carriage of the resistance genes or from carriage of plasmids with genetic cargo that confers resistance to these antibiotics and other unknown traits requires further investigation. As these ESBL/AmpC genes are often carried on plasmids, it is conceivable that the capacity to acquire plasmids carrying these resistance genes plays a role in the success of ST457 and other pathogenic lineages.
Based on general findings and our observations, we focused on the two most common plasmid families, I1 and F plasmids, which are known to disseminate ESBL/AmpC genes (59–61). Successful conjugation experiments were performed with I1/ST23 and F18:A-:B1 plasmids from the gull collection in this study and previously with I1/ST113 plasmids from the Paraguayan collection (31). While we could not identify a single plasmid lineage which would be related to ST457 in general, we observed several country or country/source specific connections: (i) I1/ST23, I1/ST314, and I1/ST315 disseminating blaCMY-2 in Australia; (ii) I1/ST113 carrying blaCTX-M-8 and mcr-5 in Paraguayan poultry; and (iii) F2:A-:B1 plasmids of Dutch origin appearing across multiple ST457 clades.
We investigated the spread of I1/ST23 plasmids carrying blaCMY-2 among Australian strains, including gull, human, and clinical isolates. Comparisons of the sequences showed that identical plasmids are shared by isolates from gulls and humans, directly linking the microbiomes in Australia. Interestingly, these plasmids of identical structure were found predominantly in gull isolates from Sydney, with some also present in gulls from Five Islands but none from Montague Island, the most distant from large human populations of the three sites (Fig. S3, Fig. 2). This proximity of gulls to humans in urban areas is the likely contributing factor here, indicating that the gull gut microbiome can acquire β-lactam-resistant E. coli from environments where extended-spectrum β-lactams are used. These observations concur with those from a previous study in Alaska that showed that, in the case of antibiotic resistance, the proximity of the gull population to urban areas is a relevant factor influencing the level of resistance found in the gull microbial population (62).
Moreover, we identified a Salmonella strain SJTUF10584 from GenBank carrying a highly similar plasmid (100% query and 99.9% identity compared to the CE1628 I1/ST23 reference), yet with a different plasmid sequence type 265 from Shanghai. A comparison of plasmid sequences from previous studies which described a blaCMY-2 region with 100% identity with our reference I1/ST23 plasmid suggests the region may have been transferred to I1 plasmids from a C plasmid (formerly A/C) (41, 42). Moreover, this complex resistance structure was present in two novel I1 plasmid sequence types described in this study, I1/ST314 and I1/ST315. I1/ST23 plasmids are known to disseminate blaCMY-2 in American crows (36). Moreover, I1/ST23-blaCMY-2 was detected in six strains from a children’s hospital in Seattle, with one of the strains typed as E. coli ST457 (63). These observations lend weight to the hypothesis that our phylogenetic inferences of E. coli ST457 show the impact of this lineage reaching beyond Australia, despite its apparent prevalence there.
We observed four Japanese human and environmental strains in the H6 clade, which was otherwise related to isolates from poultry of North and South American origin. A previous study from Norizuki et al. (32) suggested that the importation of South American chickens may have enhanced the spread of blaCTX-M-8 on I1/ST113 or closely derived plasmids in Japan, with the detection of closely related plasmids from the imported chickens and the Japanese population. We observed a high prevalence of I1/ST113 plasmids carrying blaCTX-M-8 and mcr-5 in our chicken samples from Paraguay. Further investigation that examines phylogenetic relations between those I1/ST113 plasmids is needed to interrogate this hypothesis.
Plasmids found in E. coli with similar backbone sequences have a remarkable capacity to acquire drug resistance genes that reflect the antimicrobial selection pressures to which their host E. coli are exposed (52, 64, 65). Plasmids are also able to move about (66) within an animal species, between humans and animals (52), and between E. coli and Salmonella enterica (64). From the avian collection here, we detected some clearly successful plasmid lineages. This includes I1/ST12, which is known for the global dissemination of blaCMY-2 in various environments (36, 60, 67). Most of the detected F replicons were previously reported from diverse sources, in diverse E. coli plasmid STs, e.g., F24:A-:B1 or F18:A-:B1 (19, 61). However, F18:A-:B1 plasmids of approximately 226 kbp carrying eight resistance genes encoding resistance to six antibiotic classes, including mcr-1 and blaCTX-M-55, were previously described in ST457 (16). Plasmids with this replicon were present in one gull strain and several human strains in our study.
Notably, we detected a blaOXA-23 carbapenemase-encoding gene on the chromosome of an ST457 isolated from a silver gull. To the best of our knowledge, this is the first report of this carbapenemase gene in E. coli from a wild bird, as it is typically present in Acinetobacter spp. and generally uncommon in E. coli (39, 68). The strain showed low carbapenemase activity and its MIC values for carbapenems were much lower than in the previously described plasmid-encoded blaOXA-23 region detected in a clinical E. coli isolate (39). As the plasmid region of 2,577 bp described in GenBank accession KJ716226 (39) was identical to ours (Fig. 3), we may assume that broader gene context may influence gene expression.
In summary, this study provides insights into E. coli ST457, a lineage of E. coli that displays a propensity to acquire and retain plasmid cargo carrying resistance to clinically important antibiotics and cause extraintestinal pathogenic E. coli (ExPEC) disease. ST457 was the most frequent ESBL-carrying E. coli ST recovered from Australian silver gulls sampled from multiple islands off the coast of New South Wales in 2012. A comparison of all ST457 WGS available in public repositories with those generated in this study enabled us to readily identify shared clonal lineages in gulls and humans in Australia and worldwide, indicating that in some cases gulls may be an important vector for the transmission of extended-spectrum β-lactam resistant ST457 that is capable of causing ExPEC disease. Our analyses show that: (i) plasmids carried by E. coli ST457 play an important role in the carriage and dissemination of resistance to clinically important antibiotics, particularly ESBL/AmpC genes, and possibly mcr genes, and a conjugative I1/ST23 plasmid was completely sequenced here; (ii) ST457 may be emerging as a pathogen in humans given its recent description in several hospitals linked with ExPEC disease in countries in both the northern and southern hemispheres; (iii) ST457 has been isolated from diverse mammalian and avian hosts and from the environment, underscoring its metabolic capacity; and (iv) plasmid acquisition has played an important role in the evolution of E. coli ST457.
MATERIALS AND METHODS
E. coli ST457 included in this study.
This study compares a total of 138 WGS derived from three unrelated collections of ST457. One small collection comprises 14 strains from Paraguayan chickens from our previous study (Nesporova et al., 2019 [31]). A second collection comprises 44 strains from an ongoing study of E. coli sourced from Australian silver gulls and includes two strains from separate clinical human settings in Australia. The third and largest collection of ST457 consists of 80 WGS from EnteroBase from diverse countries and sources. Detailed metadata of the studied E. coli ST457 collection are found in the supplemental material (Table S1). Whole-genome sequencing was performed for our Australian and Paraguayan strains (57) using different platforms. However, all raw sequencing data were quality trimmed (Q ≤ 20) using Trimmomatic software (v.0.36) (69) and assembled using SPAdes software (v.3.11.1) (70).
Paraguayan poultry strains.
Cloacal swabs were collected in 2012 from 12 different chicken farms in Paraguay which purchased chickens from two breeding companies. The samples were cultivated on MacConkey agar with cefotaxime (2 mg/liter) or colistin (3.5 mg/liter) and mcr-5-positive E. coli strains were analyzed using approaches, including whole-genome sequencing (GenBank BioProject PRJNA513237), as described previously (Nesporova et al., 2019 [31]). Of 28 E. coli strains carrying mcr-5, ST457 was the predominant sequence type with 12 clonal strains carrying mcr-5 and blaCTX-M-8 on an I1/ST113 plasmid derived from 3 farms (Nesporova et al., 2019 [31]). Two more strains of ST457 from this study were included here (GenBank BioSample SAMN14840864 and SAMN14840960).
Australian gulls and clinical strains.
Cloacal swabs from a total of 504 silver gulls (Chroicocephalus novaehollandiae) were collected in 2012. Gulls were sourced from three locations (White Bay in Sydney, Five Islands, and Montague Island) (37). E. coli ST457 (43 strains) were obtained from cultivation on MacConkey agar with cefotaxime (2 mg/liter) and they were the most prevalent E. coli ST in the gull study (9%; 43/497). All strains were subjected to whole-genome sequencing. Genomic DNA for sequencing was isolated using NucleoSpin tissue kit (Macherey-Nagel GmbH & Co.). DNA libraries were prepared using Nextera XT DNA sample preparation kit with modifications (71) and sequenced on a NovaSeq (Illumina, San Diego, USA) platform. A total of 42 WGS were included in this study (GenBank BioProject accession PRJNA630096); one was excluded due to low data quality. Two human clinical strains were sequenced using the same approach as for gulls. EC0125 came from Wollongong hospital from a patient with a UTI and EC038 from the CDS Reference Lab collection at St George Hospital (SGH) in Sydney (GenBank BioProject PRJNA632868).
EnteroBase data.
Searching for E. coli ST457 in EnteroBase provided 176 results up to 30 October 2019 (http://enterobase.warwick.ac.uk/). Eighty ST457 WGS were publicly available and provided enough metadata (Table S1) to be considered for further analysis. The metadata of interest was the source of isolation, year of sampling, and country of origin, since different institutions have inconsistent strategies in obtaining and reporting metadata. The source of isolation was simplified in some strains in order to reduce complexity (Table S1). All human strains which were related to infections or came from urine or blood are referred as “clinical”; other human strains are referred as “human” and may still have been sourced from infected patients but lack the specification in the metadata. Avian strains include wild birds other than Australian gulls, which form a separate “gulls” category. “Canine,” “poultry,” “bovine,” and “swine” categories do not distinguish between healthy and infected animals or animal products, in the case of food-producing animals. “Water” includes strains isolated from environmental and wastewater bodies.
Long-read sequencing, hybrid assembly, and annotation.
The purpose of long-read sequencing was to obtain high-quality E. coli ST457 reference sequences. Long-read sequencing was performed for two of our silver gull strains CE1628 and CE1803A (GenBank Biosample SAMN14966947 and SAMN14966948) and a Paraguayan poultry strain 1517k (GenBank Biosample SAMN11130353) using single molecule real-time (SMRT) sequencing performed on a Sequel I platform (Pacific Biosciences, PacBio, USA). Whole-genome DNA of the strains was isolated with a NucleoSpin microbial DNA kit (Macherey-Nagel, Germany) and the libraries were prepared using the microbial multiplexing protocol available at https://www.pacb.com/wp-content/uploads/Procedure-Checklist-Preparing-Multiplexed-Microbial-SMRTbell-Libraries-for-the-PacBio-Sequel-System.pdf. Shearing was performed using g-tubes (Covaris, USA).
We obtained PacBio sequences using the hierarchical genome assembly process (HGAP4 in SMRT Link v.6) with minimum seed coverage of 30. To obtain complete circular plasmid sequences, hybrid assembly of short and long reads was performed using Unicycler (72). BRIG (v.0.95) (73) was used to align closely related plasmids and to generate figures. Geneious software (v.7.1.9) was used to perform the annotations and create figures.
The complete chromosomal and plasmids sequences were automatically annotated using RASTtk (74) and manually curated using BLASTn (75) and ISfinder (76) for selected regions.
Transferability, typing, and assembly of selected plasmids.
Plasmids, replicons, and their STs were analyzed using ABRicate (77) and PlasmidFinder (v.2.0) and pMLST (v.2.0), respectively (https://cge.cbs.dtu.dk/services/). We used 90% coverage of the query sequence and 90% identity as thresholds for the presence of detected replicons.
As I1/ST23 and closely related plasmids I1/ST314 and I1/ST315 carrying blaCMY-2 were highly prevalent in ST457 strains from the Australian gull collection (98%, 41/42), conjugation experiments were performed to test the transferability of I1/ST23 (n = 11) and I1/ST315 (n = 2) in selected representative strains using the filter-mating method. Plasmid-free E. coli MT102, resistant to rifampin and sodium azide, was used as a recipient strain. Transconjugants were selected on medium with sodium azide (100 mg/liter), rifampin (25 mg/liter), and cefotaxime (2 mg/liter). Crude DNA from transconjugants was isolated using a heat lysis protocol and the transfer of blaCMY-2 was confirmed using PCR. A single gull strain carried an F plasmid with blaDHA-1 and no I1 was present; we performed the conjugation experiment for this strain as described above and confirmed blaDHA-1 transfer.
Carbapenemase activity and MIC.
Strain CE1628 coming from a gull carried an unusual structure containing blaOXA-23 in its chromosome. Therefore, the carbapenemase activity was evaluated by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) assay (78). The MICs for a set of 24 antibiotics or their combinations, focusing primarily on β-lactams, were determined for this strain using the broth microdilution method and evaluated using CLSI breakpoints (40).
WGS data evaluation.
Sequencing data from all three subsets were evaluated using publicly available databases.
Sequence types were confirmed using MLST (v.2.0), serotypes were assigned using SerotypeFinder (v.2.0), and fimH type was evaluated using FimTyper (v.1.0) (available at https://cge.cbs.dtu.dk/services/). Phylogenetic groups were determined using ClermonTyping (11).
ABRicate in conjunction with the NCBI Bacterial Antimicrobial Resistance Reference Gene Database (79) and the Virulence Factor DataBase (VFDB) (80) were used to identify ARGs and VAGs, respectively. Unless specified otherwise, thresholds were set at minimum level of 95% coverage of the query sequence and 98% identity for detection of ARGs and 90% coverage of the query sequence and 95% identity for VAGs.
Phylogenetic analysis.
Single nucleotide polymorphisms in the core genome of ST457 collection were evaluated. The open reading frames were predicted in fasta files using Prokka (81) and a multi-fasta alignment was created using Roary (v.3.7.0) (82) with default settings. A total of 3,758 core genes were identified and used for building a phylogeny tree using RAxML (83) under GTR model supported by 100 bootstraps. The SNP tree was visualized using iTOL with midrooted function (84) and GrapeTree (85).
A SNP matrix was created to compare SNP difference for each pair of strains. The strains and their SNP count were divided into separate tables corresponding to the clades in the tree and outstanding values were highlighted using the heat map function in MS Excel (Table S2). The accessory genome of the ST457 collection was evaluated employing Prokka (81) and Roary (82) and its phylogeny was visualized using iTOL (84).
Comparison of genetic content of E. coli ST457 collection.
In order to determine genetic features which may be specific for E. coli ST457, our three annotated long-read sequences from gulls and poultry were compared with the reference genome E. coli K-12 strain MG1655 (GenBank accession SAMN02604091) (86) using progressiveMauve (87) alignments and BLASTn (75). Genetic regions of 200 bp and longer present in all three PacBio sequences and not present in K-12 were noted as AA to FK using RAST annotation and positions in the hybrid assembly of CE1628 chromosome. The presence of these regions was subsequently evaluated in all our strains (Table S4 and S5) to determine which regions are present in most (at least 95%) of our strains and may shed light on their role in host colonization or niche adaptation more broadly.
Data availability.
Fasta files obtained in this study were deposited in GenBank under BioProjects PRJNA513237 and PRJNA630550 (Paraguayan poultry), PRJNA630096 (Australian silver gulls), and PRJNA632868 (Australian humans). Chromosomes of three PacBio-sequenced ST457 strains (SAMN11130353, SAMN14966947, and SAMN14966948), annotated pCE1628 I1/ST23 plasmid (MT468651), and annotated chromosomal region from CE1628 carrying blaOXA-23 (MT468652) were deposited in GenBank as well. All raw sequencing data of our gull and poultry strains were uploaded into EnteroBase.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Czech Science Foundation (18-23532S), by CEITEC 2020-Central European Institute of Technology (LQ1601) from the Czech Ministry of Education, Youth and Sports within the National Program for Sustainability II, by Internal Grant Agency of University of Veterinary and Pharmaceutical Sciences Brno (project number 205/2019/FVHE), and by Charles University Research Fund PROGRES (project number Q39). This study was partly funded by the Medical Research Future Fund Frontier Health and Medical Research Program (MRFF75873) and the Australian Centre for Genomic Epidemiological Microbiology (AusGEM), a joint research initiative between the NSW Department of Primary Industries and the University of Technology Sydney.
We thank J. Lausova, D. Cervinkova, I. Jamborova, J. Palkovicova, V. Petrikova, E. Mrkvicova, and K. Anatanawad for their help in the laboratory. We thank M. Havlicek, N. Carlile, and D. Priddel for their help in the field. We acknowledge P. Newton and D. Rafferty for providing clinical isolates. We thank M. Cummins for guidance in performing several bioinformatics analyses. We acknowledge institutions which share their WGS in EnteroBase and other publicly available repositories.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Ventola CL. 2015. The antibiotic resistance crisis: part 1: causes and threats. P T 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 2.Poolman JT, Wacker M. 2016. Extraintestinal pathogenic Escherichia coli, a common human pathogen: challenges for vaccine development and progress in the field. J Infect Dis 213:6–13. doi: 10.1093/infdis/jiv429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pitout JDD. 2012. Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol 3:1–7. doi: 10.3389/fmicb.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poirel L, Madec JY, Lupo A, Schink AK, Kieffer N, Nordmann P, Schwarz S. 2018. Antimicrobial resistance in Escherichia coli. Microbiol Spectr 6. doi: 10.1128/microbiolspec.ARBA-0026-2017.5. [DOI] [PubMed] [Google Scholar]
- 5.Manges AR, Geum HM, Guo A, Edens TJ, Fibke CD, Pitout JDD. 2019. Global extraintestinal pathogenic Escherichia Coli (ExPEC) lineages. Clin Microbiol 32:e00135-18. doi: 10.1128/CMR.00135-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biran D, Ron EZ. 2018. Extraintestinal pathogenic Escherichia coli. Curr Top Microbiol Immunol 416:149–161. doi: 10.1007/82_2018_108. [DOI] [PubMed] [Google Scholar]
- 7.Smalla K, Cook K, Djordjevic SP, Klümper U, Gillings M. 2018. Environmental dimensions of antibiotic resistance: assessment of basic science gaps. FEMS Microbiol Ecol 94. doi: 10.1093/femsec/fiy195. [DOI] [PubMed] [Google Scholar]
- 8.Djordjevic SP, Stokes HW, Roy Chowdhury P. 2013. Mobile elements, zoonotic pathogens and commensal bacteria: conduits for the delivery of resistance genes into humans, production animals and soil microbiota. Front Microbiol 4:86. doi: 10.3389/fmicb.2013.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyrsch ER, Roy Chowdhury P, Chapman TA, Charles IG, Hammond JM, Djordjevic SP. 2016. Genomic microbial epidemiology is needed to comprehend the global problem of antibiotic resistance and to improve pathogen diagnosis. Front Microbiol 15:843. doi: 10.3389/fmicb.2016.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robins-Browne RM, Holt KE, Ingle DJ, Hocking DM, Yang J, Tauschek M. 2016. Are Escherichia coli pathotypes still relevant in the era of whole-genome sequencing? Front Cell Infect Microbiol 6:141. doi: 10.3389/fcimb.2016.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beghain J, Bridier-Nahmias A, Le Nagard H, Denamur E, Clermont O. 2018. ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genom 4:e000192. doi: 10.1099/mgen.0.000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau SH, Reddy S, Cheesbrough J, Bolton FJ, Willshaw G, Cheasty T, Fox AJ, Upton M. 2008. Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J Clin Microbiol 46:1076–1080. doi: 10.1128/JCM.02065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo S, Wakeham D, Brouwers HJ, Cobbold RN, Abraham S, Mollinger JL, Johnson JR, Chapman TA, Gordon DM, Barrs VR, Trott DJ. 2015. Human-associated fluoroquinolone-resistant Escherichia coli clonal lineages, including ST354, isolated from canine feces and extraintestinal infections in Australia. Microbes Infect 17:266–274. doi: 10.1016/j.micinf.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki S, Ohnishi M, Kawanishi M, Akiba M, Kuroda M. 2016. Investigation of a plasmid genome database for colistin-resistance gene mcr-1. Lancet Infect Dis 16:284–285. doi: 10.1016/S1473-3099(16)00008-6. [DOI] [PubMed] [Google Scholar]
- 15.Melo LC, Oresco C, Leigue L, Netto HM, Melville PA, Benites NR, Saras E, Haenni M, Lincopan N, Madec JY. 2018. Prevalence and molecular features of ESBL/pAmpC-producing Enterobacteriaceae in healthy and diseased companion animals in Brazil. Vet Microbiol 221:59–66. doi: 10.1016/j.vetmic.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 16.McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, Schaecher KE. 2016. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 60:4420–4421. doi: 10.1128/AAC.01103-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Accogli M, Giani T, Monaco M, Giufrè M, García-Fernández A, Conte V, D'Ancona F, Pantosti A, Rossolini GM, Cerquetti M. 2014. Emergence of Escherichia coli ST131 sub-clone H30 producing VIM-1 and KPC-3 carbapenemases, Italy. J Antimicrob Chemother 69:2293–2296. doi: 10.1093/jac/dku132. [DOI] [PubMed] [Google Scholar]
- 18.Seni J, Peirano G, Okon KO, Jibrin YB, Mohammed A, Mshana SE, DeVinney R, Pitout JDD. 2018. The population structure of clinical extra-intestinal Escherichia coli in a teaching hospital from Nigeria. Diagn Microbiol Infect Dis 92:46–49. doi: 10.1016/j.diagmicrobio.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Mora A, García-Peña FJ, Alonso MP, Pedraza-Diaz S, Ortega-Mora LM, Garcia-Parraga D, López C, Viso S, Dahbi G, Marzoa J, Sergeant MJ, García V, Blanco J. 2018. Impact of human-associated Escherichia coli clonal groups in Antarctic pinnipeds: presence of ST73, ST95, ST141 and ST131. Sci Rep 8:4678. doi: 10.1038/s41598-018-22943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aquino-Andrade A, Merida-Vieyra J, Arias de la Garza E, Arzate-Barbosa P, De Colsa Ranero A. 2018. Carbapenemase-producing Enterobacteriaceae in Mexico: report of seven non-clonal cases in a pediatric hospital. BMC Microbiol 18:1–8. doi: 10.1186/s12866-018-1166-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan F, Tian D, Wang B, Zhao W, Qin H, Zhang T, Zhang H. 2019. Fecal carriage and molecular epidemiology of carbapenem-resistant Enterobacteriaceae from outpatient children in Shanghai. BMC Infect Dis 19:678. doi: 10.1186/s12879-019-4298-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chavda KD, Chen L, Jacobs MR, Bonomo RA, Kreiswirth BN. 2016. Molecular diversity and plasmid analysis of KPC-producing Escherichia coli. Antimicrob Agents Chemother 60:4073–4081. doi: 10.1128/AAC.00452-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunha MPV, Lincopan N, Cerdeira L, Esposito F, Dropa M, Franco LS, Moreno AM, Knöbl T. 2017. Coexistence of CTX-M-2, CTX-M-55, CMY-2, FosA3, and QnrB19 in extraintestinal pathogenic Escherichia coli from poultry in Brazil. Antimicrob Agents Chemother 61:e02474-16. doi: 10.1128/AAC.02474-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vieyra J, De Colsa-Ranero A, Arzate-Barbosa P, Arias-de la Garza E, Méndez-Tenorio A, Murcia-Garzón J, Aquino-Andrade A. 2019. First clinical isolate of Escherichia coli harboring mcr-1 gene in Mexico. PLoS One 14:e0214648. doi: 10.1371/journal.pone.0214648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H, Qu F, Shan B, Huang B, Jia W, Chen C, Li A, Miao M, Zhang X, Bao C, Xu Y, Chavda KD, Tang YW, Kreiswirth BN, Du H, Chen L. 2016. Detection of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae from different hospitals in China. Antimicrob Agents Chemother 60:5033–5035. doi: 10.1128/AAC.00440-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P, Wang J, Wang X, Bai X, Ma J, Dang R, Xiong Y, Fanning S, Bai L, Yang Z. 2019. Characterization of five Escherichia coli isolates co-expressing ESBL and MCR-1 resistance mechanisms from different origins in China. Front Microbiol 10:1994. doi: 10.3389/fmicb.2019.01994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seenama C, Thamlikitkul V, Ratthawongjirakul P. 2019. Multilocus sequence typing and blaESBL characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolated from healthy humans and swine in Northern Thailand. Infect Drug Resist 12:2201–2214. doi: 10.2147/IDR.S209545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tada T, Nhung PH, Shimada K, Tsuchiya M, Phuong DM, Anh NQ, Ohmagari N, Kirikae T. 2017. Emergence of colistin-resistant Escherichia coli clinical isolates harboring mcr-1 in Vietnam. Int J Infect Dis 63:72–73. doi: 10.1016/j.ijid.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Yang QE, Tansawai U, Andrey DO, Wang S, Wang Y, Sands K, Kiddee A, Assawatheptawee K, Bunchu N, Hassan B, Rutland Walsh T, Niumsup PR. 2019. Environmental dissemination of mcr-1 positive Enterobacteriaceae by Chrysomya spp. (common blowfly): an increasing public health risk. Environ Int 122:281–290. doi: 10.1016/j.envint.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Zhai W, Li J, Liu D, Zhang Q, Shen Z, Wang S, Wang Y. 2017. Presence of an mcr-3 variant in Aeromonas caviae, Proteus mirabilis, and Escherichia coli from one domestic duck. Antimicrob Agents Chemother 62:e02106-17. doi: 10.1128/AAC.02106-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nesporova K, Jamborova I, Valcek A, Medvecky M, Literak I, Dolejska M. 2019. Various conjugative plasmids carrying the mcr-5 gene in Escherichia coli isolates from healthy chickens in Paraguay. J Antimicrob Chemother 74:3394–3397. doi: 10.1093/jac/dkz317. [DOI] [PubMed] [Google Scholar]
- 32.Norizuki C, Wachino JI, Suzuki M, Kawamura K, Nagano N, Kimura K, Arakawa Y. 2017. Specific blaCTX-M-8/IncI1 plasmid transfer among genetically diverse Escherichia coli isolates between humans and chickens. Antimicrob Agents Chemother 61:e00663-17. doi: 10.1128/AAC.00663-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferreira JC, Penha Filho RAC, Andrade LN, Berchieri A, Darini ALC. 2014. IncI1/ST113 and IncI1/ST114 conjugative plasmids carrying blaCTX-M-8 in Escherichia coli isolated from poultry in Brazil. Diagn Microbiol Infect Dis 80:304–306. doi: 10.1016/j.diagmicrobio.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 34.Eller C, Leistner R, Guerra B, Fischer J, Wendt C, Rabsch W, Werner G, Pfeifer Y. 2014. Emergence of extended-spectrum β-lactamase (ESBL) CTX-M-8 in Germany. J Antimicrob Chemother 69:562–564. doi: 10.1093/jac/dkt387. [DOI] [PubMed] [Google Scholar]
- 35.Sellera FP, Fernandes MR, Moura Q, Lopes RB, Souza TA, Cerdeira L, Lincopan N. 2018. Draft genome sequence of a blaCMY-2/IncI1-harbouring Escherichia coli D:ST457 isolated from coastal benthic organisms. J Glob Antimicrob Resist 14:83–84. doi: 10.1016/j.jgar.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Jamborova I, Dolejska M, Zurek L, Townsend AK, Clark AB, Ellis JC, Papousek I, Cizek A, Literak I. 2017. Plasmid-mediated resistance to cephalosporins and quinolones in Escherichia coli from American crows in the USA. Environ Microbiol 19:2025–2036. doi: 10.1111/1462-2920.13722. [DOI] [PubMed] [Google Scholar]
- 37.Dolejska M, Masarikova M, Dobiasova H, Jamborova I, Karpiskova R, Havlicek M, Carlile N, Priddel D, Cizek A, Literak I. 2016. High prevalence of Salmonella and IMP-4-producing Enterobacteriaceae in the silver gull on Five Islands, Australia. J Antimicrob Chemother 71:63–70. doi: 10.1093/jac/dkv306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hung WT, Cheng MF, Tseng FC, Chen YS, Lee SSJ, Chang TH, Lin HH, Hung CH, Wang JL. 2019. Bloodstream infection with extended-spectrum beta-lactamase-producing Escherichia coli: the role of virulence genes. J Microbiol Immunol Infect 52:947–955. doi: 10.1016/j.jmii.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 39.La MV, Jureen R, Lin RT, Teo JW. 2014. Unusual detection of an Acinetobacter class D carbapenemase gene, blaOXA-23, in a clinical Escherichia coli isolate. J Clin Microbiol 52:3822–3823. doi: 10.1128/JCM.01566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing; twenty-eighth informational supplement; CLSI document M100-S28. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 41.Cao G, Allard MW, Hoffmann M, Monday SR, Muruvanda T, Luo Y, Payne J, Rump L, Meng K, Zhao S, McDermott PF, Brown EW, Meng J. 2015. Complete sequences of six IncA/C plasmids of multidrug-resistant Salmonella enterica subsp. enterica serotype Newport. Genome Announc 3:e00027-15. doi: 10.1128/genomeA.00027-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tagg KA, Iredell JR, Partridge SR. 2014. Complete sequencing of IncI1 sequence type 2 plasmid pJIE512b indicates mobilization of blaCMY-2 from an IncA/C plasmid. Antimicrob Agents Chemother 58:4949–4952. doi: 10.1128/AAC.02773-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith GC, Carlile N. 1993. Food and feeding ecology of breeding silver gulls (Larus novaehollandiae) in urban Australia. Colon Waterbird 16:9–16. doi: 10.2307/1521551. [DOI] [Google Scholar]
- 44.Mukerji S, Stegger M, Truswell AV, Laird T, Jordan D, Abraham RJ, Harb A, Barton M, O'Dea M, Abraham S. 2019. Resistance to critically important antimicrobials in Australian silver gulls (Chroicocephalus novaehollandiae) and evidence of anthropogenic origins. J Antimicrob Chemother 74:2566–2574. doi: 10.1093/jac/dkz242. [DOI] [PubMed] [Google Scholar]
- 45.Schaufler K, Semmler T, Wieler LH, Trott DJ, Pitout J, Peirano G, Bonnedahl J, Dolejska M, Literak I, Fuchs S, Ahmed N, Grobbel M, Torres C, McNally A, Pickard D, Ewers C, Croucher NJ, Corander J, Guenther S. 2019. Genomic and functional analysis of emerging virulent and multidrug-resistant Escherichia coli lineage sequence type 648. Antimicrob Agents Chemother 63:e00243-19. doi: 10.1128/AAC.00243-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunne KA, Chaudhuri RR, Rossiter AE, Beriotto I, Browning DF, Squire D, Cunningham AF, Cole JA, Loman N, Henderson IR. 2017. Sequencing a piece of history: complete genome sequence of the original Escherichia coli strain. Microb Genom 3:mgen000106. doi: 10.1099/mgen.0.000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marklund BI, Tennent JM, Garcia E, Hamers A, Båga M, Lindberg F, Gaastra W, Normark S. 1992. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol Microbiol 6:2225–2242. doi: 10.1111/j.1365-2958.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 48.Peralta DR, Adler C, Corbalán NS, Paz García EC, Pomares MF, Vincent PA. 2016. Enterobactin as part of the oxidative stress response repertoire. PLoS One 11:e0157799. doi: 10.1371/journal.pone.0157799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mike LA, Smith SN, Sumner CA, Eaton KA, Mobley HL. 2016. Siderophore vaccine conjugates protect against uropathogenic Escherichia coli urinary tract infection. Proc Natl Acad Sci U S A 113:13468–13473. doi: 10.1073/pnas.1606324113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gal-Mor O, Finlay BB. 2006. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol 8:1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 51.McKinnon J, Roy Chowdhury P, Djordjevic SP. 2018. Genomic analysis of multidrug-resistant Escherichia coli ST58 causing urosepsis. Int J Antimicrob Agents 52:430–435. doi: 10.1016/j.ijantimicag.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 52.Reid CJ, McKinnon J, Djordjevic SP. 2019. Clonal ST131-H22 Escherichia coli strains from a healthy pig and a human urinary tract infection carry highly similar resistance and virulence plasmids. Microb Genom 5:e000295. doi: 10.1099/mgen.0.000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson TJ, Thorsness JL, Anderson CP, Lynne AM, Foley SL, Han J, Fricke WF, McDermott PF, White DG, Khatri M, Stell AL, Flores C, Singer RS. 2010. Horizontal gene transfer of a ColV plasmid has resulted in a dominant avian clonal type of Salmonella enterica serovar Kentucky. PLoS One 5:e15524. doi: 10.1371/journal.pone.0015524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monk JM, Charusanti P, Aziz RK, Lerman JA, Premyodhin N, Orth JD, Feist AM, Palsson BØ. 2013. Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proc Natl Acad Sci U S A 110:20338–20343. doi: 10.1073/pnas.1307797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reitzer L, Zimmern P. 2019. Rapid growth and metabolism of uropathogenic Escherichia coli in relation to urine composition. Clin Microbiol Rev 33:e00101-19. doi: 10.1128/CMR.00101-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forsyth VS, Armbruster CE, Smith SN, Pirani A, Springman AC, Walters MS, Nielubowicz GR, Himpsl SD, Snitkin ES, Mobley HLT. 2018. Rapid growth of uropathogenic Escherichia coli during human urinary tract infection. mBio 9:e00186-18. doi: 10.1128/mBio.00186-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mobley HL. 2016. Measuring Escherichia coli gene expression during human urinary tract infections. Pathogens 5:7. doi: 10.3390/pathogens5010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vangchhia B, Abraham S, Bell JM, Collignon P, Gibson JS, Ingram PR, Johnson JR, Kennedy K, Trott DJ, Turnidge JD, Gordon DM. 2016. Phylogenetic diversity, antimicrobial susceptibility and virulence characteristics of phylogroup F Escherichia coli in Australia. Microbiology (Reading) 162:1904–1912. doi: 10.1099/mic.0.000367. [DOI] [PubMed] [Google Scholar]
- 59.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roer L, Overballe-Petersen S, Hansen F, Johannesen TB, Stegger M, Bortolaia V, Leekitcharoenphon P, Korsgaard HB, Seyfarth AM, Mossong J, Wattiau P, Boland C, Hansen DS, Hasman H, Hammerum AM, Hendriksen RS. 2019. ST131 fimH22 Escherichia coli isolate with a blaCMY-2/IncI1/ST12 plasmid obtained from a patient with bloodstream infection: highly similar to E. coli isolates of broiler origin. J Antimicrob Chemother 74:557–560. doi: 10.1093/jac/dky484. [DOI] [PubMed] [Google Scholar]
- 61.Villa L, García-Fernández A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 62.Atterby C, Ramey AM, Hall GG, Järhult J, Börjesson S, Bonnedahl J. 2016. Increased prevalence of antibiotic-resistant E. coli in gulls sampled in Southcentral Alaska is associated with urban environments. Infect Ecol Epidemiol 6:32334. doi: 10.3402/iee.v6.32334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weissman SJ, Adler A, Qin X, Zerr DM. 2013. Emergence of extended-spectrum β-lactam resistance among Escherichia coli at a US academic children's hospital is clonal at the sequence type level for CTX-M-15, but not for CMY-2. Int J Antimicrob Agents 41:414–420. doi: 10.1016/j.ijantimicag.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wyrsch ER, Reid CJ, DeMaere MZ, Liu MY, Chapman TA, Roy Chowdhury P, Djordjevic SP. 2019. Complete sequences of multiple-drug resistant IncHI2 ST3 plasmids in Escherichia coli of porcine origin in Australia. Front Sustain Food Syst 3. doi: 10.3389/fsufs.2019.00018. [DOI] [Google Scholar]
- 65.Jarocki VM, Reid CJ, Chapman TA, Djordjevic SP. 2019. Escherichia coli ST302: genomic analysis of vrulence potential and antimicrobial resistance mediated by mobile genetic elements. Front Microbiol 10:3098. doi: 10.3389/fmicb.2019.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Venturini C, Hassan KA, Roy Chowdhury P, Paulsen IT, Walker MJ, Djordjevic SP. 2013. Sequences of two related multiple antibiotic resistance virulence plasmids sharing a unique IS26-related molecular signature isolated from different Escherichia coli pathotypes from different hosts. PLoS One 8:e78862. doi: 10.1371/journal.pone.0078862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hansen KH, Bortolaia V, Nielsen CA, Nielsen JB, Schønning K, Agersø Y, Guardabassi L. 2016. Host-specific patterns of genetic diversity among IncI1-Iγ and IncK plasmids encoding CMY-2 β-lactamase in Escherichia coli isolates from humans, poultry meat, poultry, and dogs in Denmark. Appl Environ Microbiol 82:4705–4714. doi: 10.1128/AEM.00495-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Y, Xu Q, Li T, Fu Y, Shi Y, Lan P, Zhao P, Chen Q, Zhou Z, Jiang Y, Peleg AY, Yu Y. 2018. OXA-23 is a prevalent mechanism contributing to sulbactam resistance in diverse Acinetobacter baumannii clinical strains. Antimicrob Agents Chemother 63:e01676-18. doi: 10.1128/AAC.01676-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reid CJ, Wyrsch ER, Roy Chowdhury P, Zingali T, Liu M, Darling AE, Chapman TA, Djordjevic SP. 2017. Porcine commensal Escherichia coli: a reservoir for class 1 integrons associated with IS26. Microb Genom 12:e000143. doi: 10.1099/mgen.0.000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Siguier P, Filée J, Chandler M. 2006. Insertion sequences in prokaryotic genomes. Curr Opin Microbiol 9:526–531. doi: 10.1016/j.mib.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 77.Seemann T. ABRicate, GitHub. https://github.com/tseemann/abricate.
- 78.Hrabak J, Walkova R, Studentova V, Chudackova E, Bergerova T. 2011. Carbapenemase activity detection by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 49:3222–3227. doi: 10.1128/JCM.00984-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, Tyson GH, Zhao S, Hsu CH, McDermott PF, Tadesse DA, Morales C, Simmons M, Tillman G, Wasilenko J, Folster JP, Klimke W. 2019. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen L, Zheng D, Liu B, Yang J, Jin Q. 2016. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res 44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 82.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, Carriço JA, Achtman M. 2018. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res 28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. 2014. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Najafi S, Rahimi M, Nikousefat Z. 2019. Extra-intestinal pathogenic Escherichia coli from human and avian origin: detection of the most common virulence-encoding genes. Vet Res Forum 10:43–49. doi: 10.30466/vrf.2019.34307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fasta files obtained in this study were deposited in GenBank under BioProjects PRJNA513237 and PRJNA630550 (Paraguayan poultry), PRJNA630096 (Australian silver gulls), and PRJNA632868 (Australian humans). Chromosomes of three PacBio-sequenced ST457 strains (SAMN11130353, SAMN14966947, and SAMN14966948), annotated pCE1628 I1/ST23 plasmid (MT468651), and annotated chromosomal region from CE1628 carrying blaOXA-23 (MT468652) were deposited in GenBank as well. All raw sequencing data of our gull and poultry strains were uploaded into EnteroBase.