Graphical Abstract
Graphical Abstract.
Recently, the World Health Organization (WHO) declared a pandemic situation due to a new viral infection (COVID-19) caused by a novel virus (Sars-CoV-2).
Abstract
Recently, the World Health Organization (WHO) declared a pandemic situation due to a new viral infection (COVID-19) caused by a novel virus (Sars-CoV-2). COVID-19 is today the leading cause of death from viral infections in the world. It is known that many elements play important roles in viral infections, both in virus survival, and in the activation of the host's immune system, which depends on the presence of micronutrients to maintain the integrity of its functions. In this sense, the metallome can be an important object of study for understanding viral infections. Therefore, this work presents an overview of the role of trace elements in the immune system and the state of the art in metallomics, highlighting the challenges found in studies focusing on viral infections.
Introduction
Infectious disease epidemics are occurring more frequently, and spreading faster in different regions of the world, resulting in significant impacts on health and economic systems.1,2 Recently, the World Health Organization (WHO) declared a state of a pandemic due to a viral infection caused by a new virus of the Coronaviridae family: Sars-CoV-2.3 Coronavirus disease (COVID-19) is currently one of the main causes of death in the world, causing about one million deaths on September 22, 2020.4 Although COVID-19 is now the leading cause of death from viral infections in a short period of time, other infectious diseases caused by viruses are also responsible for numerous deaths, such as HIV (human immunodeficiency virus),5 hepatitis (A, B, C, D, E),6 yellow fever,7 measles,8 dengue,9 zika-virus10 and others.1,4 A potentially fatal combination of newly-discovered diseases, and the re-emergence of many long-established ones, demands urgent responses. In this sense, the development of rapid diagnostics and effective control strategies for epidemic prevention are essential.1
Many trace elements play pivotal roles in viral infections, both in virus survival, since metals are important metalloproteins responsible for virus attachment to the host, and for the characterization of viral infections due to the deregulation of metal homeostasis during infection.11 For example, zinc (Zn),12 iron (Fe),13 and copper (Cu)14 are some of the most common metals that bind to proteins associated with viral infections. Metal ions participate in the maturation of the genome (RNA or DNA), activation, and catalytic mechanisms, in reverse transcription, initial integration processes, and the protection of newly synthesized DNA.15 In addition, the host's immune system depends on the presence of these micronutrients to maintain the integrity of its functions.16 In this sense, the metallome of a biological system is an important object of study for understanding the effect of metals on viral infections.
Metallomics is an important area of science that allows an understanding of the effect of biometals in the biological systems.17–19 In metallomics, metal determination requires extremely sensitive analytical tools, such as inductively coupled plasma optical emission spectroscopy (ICP OES20) and inductively coupled plasma mass spectrometry (ICP-MS21,22). Furthermore, as many metals are linked to biomolecules (proteins and metabolites), separation techniques, such as liquid chromatography (LC)23,24 and capillary electrophoresis (CE),25,26 have been required in metallomics studies. LA-ICP-MS (laser ablation inductively coupled plasma mass spectrometry),27,28 SIMS (secondary ion mass spectrometry),29,30 X-ray fluorescence spectrometry (XRF),31,32 and LIBS (laser-induced breakdown spectroscopy)33,34 are also important techniques to obtain information on the spatial distribution of metals in solid samples, such as tissues and cells.17,19,35,36
In this sense, this review presents a brief description of the state of the art in metallomics, highlighting the main challenges found from sampling to quantification. Besides, possible biochemical mechanisms involving metals are presented in this work to illustrate the relationship between trace elements and the immune system against viral infections.
Metals and viral infections: a complex interaction
Viruses are small particles of genetic material (RNA or DNA) surrounded by a lipid or protein layer that replicate quickly and in large numbers after infection of a host organism. Several types of viruses have been catalogued as pathogens that cause human diseases.37 Infectious respiratory diseases, including the influenza virus (H1N1, H2N2, H2N9, and others), are possibly the most common viral infections observed in the world.38,39 These infections are associated with inflammation of the upper airways and lungs.38 In addition, there are other lethal viruses that infect other parts of the body causing major infections, such as gastroenteritis (intestine),40 hepatitis (liver),6 meningitis (nervous system),41 zika-virus10 and rubella42 (placenta and foetus). For example, hepatitis B and C viruses can cause chronic infections in the liver, resulting in serious illnesses such as cirrhosis, liver failure and even liver cancer.6 HIV is a mutated retrovirus that acts primarily to suppress the host's immune system.5
Furthermore, novel human viruses can also be developed from mutation viruses that generally affect animals. The infection usually happens from the interaction of the host animal with a susceptible human.2 Severe acute respiratory syndrome coronaviruses (SARS), such as SARS-CoV and SARS-CoV-2, are examples of viruses of animal origin.43,44 The highly contagious respiratory virus SARS has fuelled global fears of pandemics since its first appearance in China.43 In 2002-2003, the WHO launched a global alert for a newly discovered respiratory coronavirus that was called SARS-CoV; shortly after in 2012, a new alert was given for the Middle East respiratory syndrome coronavirus (MERS-CoV). In recent years, the world has witnessed the worst Ebola outbreak of all time that began to hit West Africa in early 2014.45 Now, SARS-CoV-2 has been the cause of a new pandemic condition, with mortality rates of approximately 3%.43,46
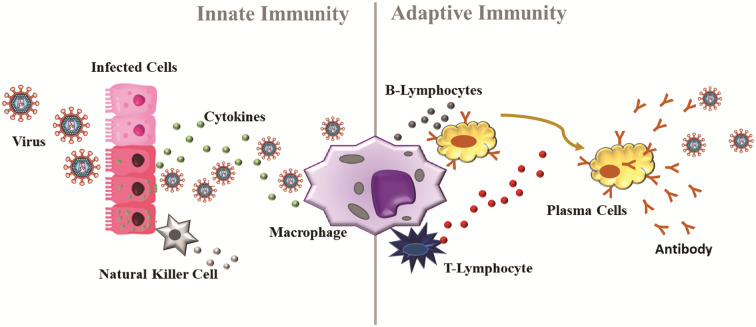
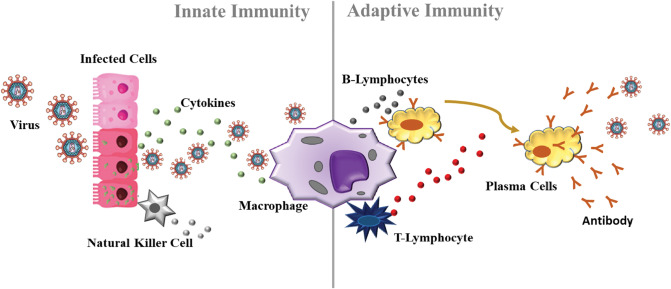
It is well known that during an infection caused by a virus, several changes occur in the biological system, such as vasodilation and vascular permeability.47,48 This change allows the arrival of immune cells. The immune system is primarily responsible for maintaining the physiological integrity of the body, eliminating foreign material and infectious viruses.47 Several antigen-presenting cells modulate the immune system (Fig. 1).48 Macrophages and neutrophils are among the cells of the first line of defence (innate response) due to their phagocytic, cytotoxic, and secretory activities. When any virus enters the body, it is phagocyted and digested by macrophages.47,48 T- and B-lymphocytes are the effector cells of the immune system.49 B-cells produce specific antibodies in response to the antigen, while T-cells assist B-cells in the production of antibodies and mediating the cellular immune response. Antibodies, such as immunoglobulin (IgA, IgM and IgG), are the second line of defence (adaptative response) of the immune system.47–49 In addition, other molecules, such as cytokines, play a pivotal role in cellular communication during the infection process.47,48 Although the effects of micronutrients on neutrophils’ functions are not clear, micronutrients such as Fe, Zn, Cu, Se, etc. can influence several components of innate immunity.50 In this sense, many studies report changes in the levels of metals in the host's biological system during viral infections.51–56 The suggested mechanism for homeostasis of the main trace elements during viral infection is shown below.
Fig. 1.
Immune system during a viral infection. When any virus enters the body, the immune system is activated by cytokines. Macrophages, natural Killer cells, and B and T cells constitute the first line of defense of the immune system (innate response). After some time of the infection (∼ 7 days), antibodies are produced to fight the infection, therefore constituting the second line of defense of the immune system (adaptive response).
Zinc homeostasis and innate immunity
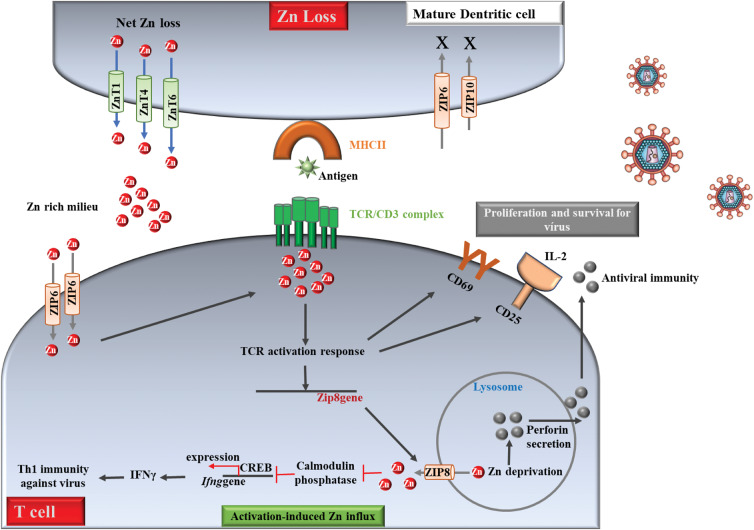
Zinc is the second most abundant element in the cell, being a cofactor in more than 3000 metalloenzymes, including copper and zinc SOD, metallothionein, and several zinc proteins (including zinc fingers).57,58 Zinc is required for the normal development and function of the organism in which it is an important component, mediating innate immunity.16 For example, it has been demonstrated that Zn deficiency is associated with reduced antibody production, impaired function of the immune system, decreased NK cell activity, reduced cytokine production and deceased chemotaxis and oxidative burst of neutrophil granulocytes.59 Zn deficiency reduces the number of peripheral and thymic T cells, their proliferation in response to phytohemagglutinin, and the functions of T helpers and cytotoxic T cells. In addition, Zn deficiency acts indirectly by reducing the levels of active serum thymulin, a zinc-dependent nonapeptide hormone that regulates the differentiation of immature T cells in the thymus and the function of mature peripheral T cells.57,59 Many studies have reported the relationship between Zn and viral infections.60–63 For example, Zn deficiency was observed in HIV infection,63 tuberculosis61 and hepatitis,56 suggesting that low levels of organism Zn predict an increased susceptibility to virus infection. On the other hand, supplemental zinc can increase the number of T-cell lymphocytes circulating in the blood, and consequently the ability of lymphocytes to fight infection improves.57,64,65 The importance of Zn modulation in the immune response to infection goes beyond the concept of nutritional immunity.66 In a previously uncharacterized role, Zn ions have emerged as signalling molecules that trigger pathways required for the activation of innate and T cell mediated immunity. Thus, innate and adaptive defences integrate Zn regulation to reinforce the pathogen clearance. Phagocytes such as neutrophils, macrophages and dendritic cells (DCs) are primary defenders at sites of pathogen encounter and must rapidly mount an immune response followed by cross-talk to induce T cell activation. Under viral infection, Zn is dynamically regulated, and the Zn modulatory approach used by immune cells is contextual depending on the cell type and the pathogen in question.57,66 In summary, Zn has antiviral properties against several viruses.59 Although mechanistic studies are lacking, Zn appears to inhibit viral protease and polymerase enzymatic processes, as well as physical processes, such as virus attachment, infection, and uncoating.12Fig. 2 shows a suggested mechanism of Zn regulation in T cell mediated antiviral defences based on evidence from several studies.57,59,66,67 From Fig. 2, it was observed that the activation of T cells is essential for proliferation, cytokine production, and virus clearance. Zinc is exported from DCs to drive the loss of Zn. This process matures them and increases surface MHCII (major histocompatibility complex II). When T cells establish physical contact with an antigen presenting MHCII, Zn is imported from the extracellular space via ZIP6. The Zn lost from DCs into the extracellular milieu can potentially be drawn into T cells.57,59,66 Imported Zn accumulates in the subsynaptic region at the contact point of the DC-T cell to drive the TCR activation response leading to an increase in proteins CD69 and CD25. These molecules support the survival and proliferation of T cells required to elicit T cell antiviral immunity. Activation enhances Zip8 expression, which may be negatively controlled by mi-RNAs. ZIP8 imports lysosomal Zn into the cytosol. Cytosolic Zn inhibits Ca2+/calmodulin phosphatase to sustain CREB mediated interferon-gamma (IFNγ) production to bolster Th1 immunity against viruses. Lysosomal Zn deprivation caused by ZIP8 induces perforin secretion from lysosomes that are essential in CD8 T cell responses to viral infection.57,59,66
Fig. 2.
Zinc regulation in T cell mediated antiviral defences. Activation of T cells is essential for cytokine production and clearance of viruses. Zn is exported from dentritic cells (DCs) to drive Zn loss. This process matures them and increases surface MHCII. When T cells establish physical contact with an antigen-presenting MHCII, Zn is imported from the extracellular space via ZIP6. The Zn lost from DCs into the extracellular milieu can potentially be drawn into T cells. Imported Zn accumulates in the sub-synaptic region at the point of DC-T cell contact to drive the TCR activation response leading to an increase in CD69 and CD25. These molecules support survival and proliferation of T cells required to elicit T cell antiviral immunity. Activation enhances Zip8 expression, which may be negatively controlled by mi-RNAs. ZIP8 imports lysosomal Zn into the cytosol. Cytosolic Zn inhibits Ca2P/calmodulin phosphatase to sustain CREB mediated IFNγ production to bolster Th1 immunity against microbes. Lysosomal Zn deprivation caused by ZIP8 induces perforin secretion from lysosomes that is essential in CD8 T cell responses to viral infection. Red dots, Zn ions; solid arrows and lines, established links; dotted arrows and lines, predicted links.
Cell culture studies have demonstrated that high Zn concentrations and the addition of pyrithione to stimulate cellular importation of Zn2+, results in inhibition of the replication of various RNA viruses, including influenza virus, respiratory syncytial virus, and various picornaviruses.67,68 It has suggested that in picornavirus and coronavirus, such an effect is realized due to the interference in the processing of viral polyprotein. Zinc effectively inhibits the activity of nidovirus RNA-synthesis (including SARS-CoV) in vitro, which is carried out by changing the activity of RdRp (RNA-dependent RNA polymerase) during the extension phase of RNA synthesis.68 Thus, it may be suggested that in coronaviruses, Zn2+ may inhibit both adequate proteolytic processing of polyproteins replicase and RdRp activity. Like other coronaviruses, SARS-CoV-2 causing COVID-19 also shares more than 75% sequence similarity at the genome level with SARS-CoV,3,44 allowing for the hypothesis that the antiviral effects of Zn can be performed on SARS-CoV-2 as well.67
Iron homeostasis and innate immunity
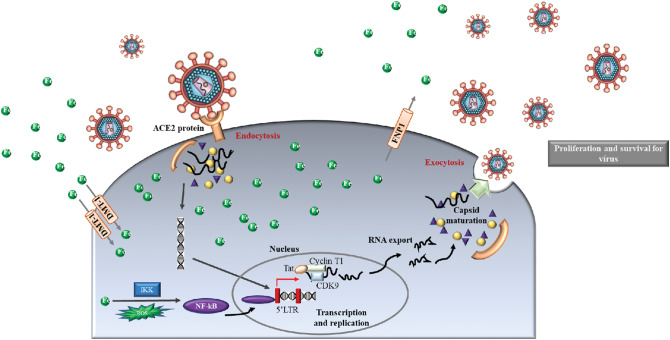
Iron is another important element with fundamental biological functions for both the host and the virus because Fe is responsible for several cellular processes including DNA replication and cell proliferation.16 It is also required for hydroxyl radical generation within phagocytes. Iron binds lactoferrin and transferrin, which interact with neutrophils to kill viruses.69 However, an imbalance (excess or deficiency) of Fe can increase susceptibility to infection. The effects of Fe deficiency on immunologic function are influenced to a significant degree by the causal factor(s) such as the reduced dietary intake or blood loss that led to Fe deficiency.70 On the other hand, excess Fe levels can potentially increase the activity of viruses, raising their mutation rates71 (Fig. 3). Elevated iron levels have been associated with increased progression of hepatitis virus (B and C) infection and poor prognosis for the patient.72 It is not clear whether the elevated Fe levels are a cause or result of high viral loads (or simply hepatitis infection). Accumulation of ions in the brain has also been reported for zika virus infection, suggesting a dysregulation of Fe metabolism during viral infection.73 Fe supplementation has been shown to increase mortality in HIV infected patients, regardless of the severity of anemia, suggesting a detrimental role for excess Fe in HIV infection.74 HIV replication depends on host cell enzymes, some of which are involved in transcription, translation of viral mRNA, and viral assembly that require iron.71 For SARS-CoV-2 infection, although it is a new infection, some clues have been suggested for this infection from another viral infection. For example, recently, some drugs used to treat HIV (e.g., lopinavir/ritonaviran), and Ebola virus, and MERS diseases (e.g., remdesivir) has been recommended for the treatment of SARS-CoV-2,75,76 suggesting similarity in the viral life cycle. Considering this fact, it is reasonable to suggest the importance of iron for SARS-Cov-2 replication. Fig. 3 shows a possible mechanism for the Fe effect on the life cycle of coronaviruses.13
Fig. 3.
Effect of iron on viral infections. The iron ion is absorbed by the cell through the divalent metallic iron transport protein 1 (DMT1). The ferroportin membrane conveyor (FPN1) is responsible for controlling iron exports. Sufficient levels of intracellular iron support virus replication. Once the virus's RNA is integrated into the host's genome, the nuclear factor (NF)-κB binds to the long terminal 5 ′repetition (LTR) mediating the transcription of viral genes. Iron can activate NF-κB generating reactive oxygen species (ROS) and activation of the IκB kinase (IKK), which leads to the location of NF-κB in the nucleus. Efficient transcription of the integrated virus genome involves the transactivating protein Tat and its interaction partners CDK9 and cyclin T1. In conditions of low cell iron, CDK9 and cyclin T1 dissociate and Tat-mediated transcription is inhibited.
In Fig. 3, it is observed that the Fe ion is absorbed by the cell through the divalent metallic iron transport protein 1 (DMT1).13 The export of iron is controlled by the ferroportin membrane transporter (FPN1). Within the cells, sufficient Fe levels support the virus replication, and participate in the heme biosynthesis in the mitochondria.70 The virus enters host cells through binding to various receptors, such as the angiotensin-converting enzyme 2 (ACE2) receptor (for SARS-CoV), then it dismounts to release the viral genome and nucleocapsid.13,70 The transcription and translation of viral genes produces viral genomic RNA and structural proteins. The whole process of viral replication requires iron-containing enzymes and consumes abundant adenosine triphosphate (ATP).13,70 Fe is a critical participant for mitochondria to produce ATP. In short, adequate Fe allows the virus to complete its replication process, and otherwise iron deficiency impairs this process.13,70,71 In this sense, the regulation of iron distribution acts as an innate immune mechanism against invading pathogens.71 Even in the absence of infection, several facets of human iron metabolism ensure that iron is difficult to access by the virus. First, most of the iron in humans is sequestered intracellularly, complexed within hemoglobin inside erythrocytes.70 Some viruses, therefore, have developed mechanisms to liberate hemoglobin through lysis of erythrocytes to ultimately extract iron from heme. However, hemolytic pathogens must subsequently compete with haptoglobin and hemopexin, the host's glycoproteins that liberated hemoglobin and heme, respectively.70,71 A second factor that limits the availability of iron to invade pathogens is the scarcity of free extracellular iron.70 Extracellular iron is bound with high affinity by transferrin, which in healthy individuals is typically less than 50% saturated with iron.78 When the transferrin binding capacity is exceeded, iron can also be chelated with less affinity for a series of molecules in the plasma, including albumin, citrate, and amino acids.15,77 Some studies have suggested iron chelators (deferoxamine, deferiprone and deferasirox) as an alternative to mitigate viral infections, such as SARS-CoV-2, through multiple mechanisms, including: (i) inhibition of viral replication; (ii) decrease of Fe availability; (iii) upregulation of B cells; (iv) prevention of pulmonary fibrosis and pulmonary decline by reducing the accumulation of pulmonary Fe; (v) improvement of the neutralizing antiviral antibody.78 Some studies indicate Fe chelators as a potential adjuvant therapy to prevent viral replication, and great care must be taken because iron deficiency can affect other systems, such as the central nervous, gastrointestinal and muscular systems, causing several other diseases, such as anemia and cancer.77
Copper homeostasis and innate immunity
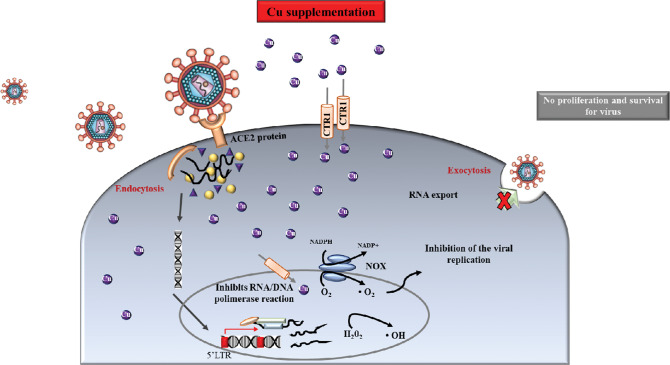
Copper homeostasis is intricately linked to Fe metabolism, which can affect the availability of Fe to host cells and virus.77,79 The dysregulation of Cu homeostasis can result in abnormal cell function or damage due to its central role in host-pathogen interaction, affecting innate and adaptive immunity.80 In this sense, humans with Cu deficiency show susceptibility to infections.80,81 However, the Cu level balance can assist to kill several infectious viruses. In fact, Cu-induced viral death can be mediated via ROS, and in this regard, Cu and hydrogen peroxide play essential roles79,80 (Fig. 4). For example, Cu is involved in inhibiting viral entry and replication, and in the degradation of mRNA and capsid proteins that are involved in the viral life cycle.80
Fig. 4.
Copper antiviral activity. Exposure to Cu generates reactive oxygen species (ROS) that can result in unreversed damage in virus morphology (i.e., envelope and surface spikes) and destruction of the viral genome, making viral replication difficult.
In addition, Cu supplementation was shown to restore the secretion and activity of IL-2 in animals with Cu deficiency. Increasing IL2 synthesis is crucial for T cell proliferation and NK cell cytotoxicity. Copper also normalized impaired immune functions by modulating neutrophil activity, the blastogenic response to T cell mitogens, and the balance between T helper cells (Th1 and Th2).80 People with weakened immune systems are always at increased risk for infectious diseases, including COVID-19.82 It is important to highlight that Cu deficiency can be the result of an imbalance of Cu and other minerals in the diet.83 In older people, Cu deficiency is more frequent, being a result of malnutrition, malabsorption, or excessive zinc intake, and can be acquired or inherited.83 Therefore, Cu supplementation can help fight viral infection,15,77,79,80,82 especially in older people where marginal or severe deficiency of Cu is a strong possibility.81,83 However, at the moment, there is not enough data or knowledge concerning the effect of therapeutic supplementation of Cu regarding the susceptibility and outcome of COVID-19.80,82
Other trace elements in viral infections
Selenium (Se) is essential for the activity of glutathione peroxidase which is involved in the degradation of hydrogen peroxide and the release of oxygen radicals.77,84 Selenium deficiency is associated with altered mitochondrial electron transport. In addition, Se deficiency, particularly in association with vitamin E deficiency, reduces the antibody response.85 Selenium supplements increase antibody titers.85 These effects are dose-dependent because higher doses of the trace element can also suppress antibody response.77,86 These interactions between Se and the immune system may be important in the pathogenesis of the single cardiomyopathy.87 Besides, Se influences the induced immunity by the vaccine against influenza.85,86 In relation to magnesium (Mg), the deficiency of this element results in low levels of serum IgG and IgM, NK, and T cells.88,89 The possibility of Mg deficiency activating a latent virus should be borne in mind.90 However, data on these aspects of cell-mediated immunity in Mg deficiency are required.89 For manganese (Mn), there is little information about the role of this element in the functioning of the immune system.77,91 Deficiencies are rare and their impact on susceptibility to infection is not known. Cobalt (Co) can influence the production of antibodies and alter susceptibility to infectious agents.92 Nickel (Ni) can have an impact on the immune system through its interactions with Fe, Zn, and Cu. Nickel depresses the production of antibody, the phagocyte function, and interferon activity.93 Arsenic (As) can alter the production of interferon and antibodies. Cell-mediated reactions are largely affected.55 Moderate exposure to heavy metals, such as lead (Pb) and cadmium (Cd) can produce impaired immune responses, including antibody deficiency, phagocyte dysfunction, and depressed cell-mediated immunity, thereby enhancing susceptibility to certain virus infections94
State of the art in metallomics: from sampling to quantification
Sample preparation: sampling, storage, and sample treatment procedure
In any chemical analysis, including metallomics studies, it is recommended that there be minimal manipulation of the sample to avoid loss of analytes and/or contamination.22,95–97 However, due to the complexity of biological samples, such as blood, urine, and tissue, this recommendation is not always followed.98–100 Therefore, in metallomics studies, the sample preparation step constitutes one of the most critical steps for obtaining accurate and precise results.22,99 It is at this stage that contamination and alterations of chemical species can occur. The sample preparation step can be divided into three important stages: (i) sampling; (ii) storage; (iii) sample treatment.
The sampling process must be highly planned and rigorously executed since it is from this stage that it will be possible to identify and quantify the chemical species present in a system.101 Thus, some precautions must be considered during the sampling, such as (i) type of container used for sample collection; (ii) nature of the sample; (iii) sample complexity; (iv) physical–chemical properties of the chemical species to be analyzed; (v) analytical technique to be used in the study.102
In many clinical studies, sterile containers are generally used to collect samples. These containers are free of DNase and protease preventing possible degradation of the biomolecules that bind to the metal. In addition, preservatives are used to prevent bacterial growth. Boric acid, sodium azide, formaldehyde, and chloral hexidine are examples of preservatives used to inhibit bacterial growth.103 However, it is important to understand that these preservatives can affect some chemical properties of the interest species, changing their oxidation states.104 Centrifugation and filtration are also common steps for removing materials such as cellular components.96 In some cases, vacuum devices are needed to increase speed. In addition, some methods of miniaturization and automation, such as 96-well plate filters and syringe filters, have also recently been used to clean biological samples. In this case, the pore size of the filters used (e.g., ultra-, micro-, and nanofiltration) plays an important role during the cleaning process.96,105 However, two main limitations affect this strategy: (i) if the viscosity of the sample is high (e.g., serum) or the level of concomitant proteins composing the sample matrix is elevated (e.g., whole blood), the membrane pores can be easily blocked, minimizing the efficiency of the method; (ii) the metalloproteins might experience unspecific absorptions on the membranes that could yield either low metalloprotein recovery or, in the worst case, disruption of the metal–protein complexes. In the first case, sample dilution before preconcentration and protein precipitation to remove interferences (e.g., hemoglobin precipitation from whole blood) are recommended strategies to improve the performance of the filters. In the second case, the use of different buffers with variable ionic strength could help to minimize the interaction of the analyte on the membrane.104
Furthermore, the chemical stability of the analytes can also be considered as critical as all aspects related to the sampling procedures.103 For example, during the storage step, some disadvantages can be pointed out, such as degradation that will depend on the species chemical nature and can be influenced by biochemical processes such as enzymatic activity.19 Besides, the temperature is an important parameter for chemical reaction rates, which can determine the rate of transformation of species.102,106 Therefore, reducing the temperature is an important decision to decrease species transformation. Lyophilization is one of the most common drying procedures used for the determination of trace elements in biological materials. Lyophilization allows water or other solvents to be removed from the sample using a sublimation process. Shock freezing is another strategy also applied for sample stabilization. Shock freezing of the desired samples in the gas phase under liquid nitrogen is the safest technique to prevent species changes and can be performed immediately at the sampling site. If this approach is not feasible within the specific project, the short-term preservation of biological material at −20 °C is recommended.103
To minimize the presence of interferent compounds that can compromise the detection sensitivity of the analytical method, the sample treatment procedure is generally required.19,22,103 Several types of sample preparation are described in the literature to remove interferents, such as solid-phase extraction and its miniaturization strategies,107–112 liquid–liquid extraction, and its miniaturization approaches,113–116 cloud point extraction,117 ionic liquid extraction,118–121 membrane-associated solvent extraction24,114 and ultrasound-assisted extraction.22,99,122 Many of these methods are examples of sample preparation approaches used in studies focusing on metallobiomolecules. For studies aiming at total determination of metals in biological matrices with the minimum presence of organic load, microwave-assisted acid digestion is generally the most recommended sample preparation procedure.23,123 It is important to highlight that the choice of the best sample preparation method must consider the nature of the analyte and the analytical technique to be used. Current sample preparation protocols employ small amounts of sample, as well as simpler methods, which are good starting points before the real analysis and lead to less experimental errors.96 In addition, to characterize an unknown metalloprotein in an environment that is rich in proteins is exceptionally challenging. The central issue that makes it difficult to characterize metalloproteins is the difficulty in preserving their native state during analysis. To accurately quantify metalloproteins, it is vital that their native state is kept intact and is not altered by denaturing conditions. The use of strong acids/bases concentrated inorganic salt, organic solvents and heat contribute to the loss of native folded states. Thus, sample preparation for metallobiomolecules should endeavor to use buffers that are of physiological pH, as this will help to prevent changes in the secondary and tertiary structures that lead to loss of metal binding. The main problem of these techniques is that when non-volatile buffers are used for sample solubilization (e.g., Tris-HCl, phosphate, etc.), they might be also preconcentrated during the concentration process, increasing sample salinity and viscosity. Ammonium acetate is recommended as a buffer for lyophilization but with dilution to about 0.2 mol L−1.104 A metalloproteomic workflow should ensure that each possible source of experimental error is mitigated to a point where its influence is negligible.36,124 New modern approaches have been developed to achieve these objectives, such as rapid analysis, low sample volume required, and easy operation. For example, nanoparticles (NPs) have demonstrated the ability to overcome the challenges and limitations associated with sample preparation of biological matrices for clinical studies in human disease, offering new tools and ideas to advance in the field of metallomics.125,126 Due to the rapid development of nanotechnology over the past decade, many new methods to detect a virus in infectious diseases have been designed. For example, a novel microfluidic device with a microbeads array using a bioconjugate oligonucleotide–Au nanoparticle labeled with multiple-enzymes to detect HPV (Human papillomavirus) was developed.127 This method uses microbeads as a detection platform that was functionalized with the capture probes and modified electron-rich proteins and uses gold nanoparticles functionalized with horseradish peroxidase (HRP) as a label with a secondary DNA probe. The functionalized microbeads were introduced independently into the arranged chambers using the chip loading plate. A single channel was used to generate barrier structures to confine the microbeads and make the beads array accessible by microfluidics. Through “sandwich” hybridization, the labels of Au nanoparticles functionalized with enzymes were brought close to the surface of the microbeads. The oxidation of biotin–tyramine by hydrogen peroxide resulted in the deposition of several portions of biotin on the surface of the beads. This deposition is markedly increased in the presence of immobilized electron-rich proteins. The streptavidin-labeled quantum dots were then allowed to bind to the deposited biotin portions and display the signal. The improved detection sensitivity was achieved where the large surface area of Au nanoparticle carriers increased the amount of HRP bound per sandwich hybridization. The on-chip genotyping method can discriminate as low as 1 fmol L−1 of synthesized HPV oligonucleotide DNA. The increase in the chip-based signal of the amplified assay resulted in a sensitivity 1000 times greater than the off-chip test. In addition, this on-chip format can discriminate and genotype 10 copies per L of genomic HPV DNA using PCR products. These results demonstrated that this on-chip approach can achieve extremely sensitive detection and target DNA genotyping and can be further developed to detect disease-related biomolecules at the lowest level in their earliest incidence.127
Techniques applied in metallomics studies focusing on viral infections
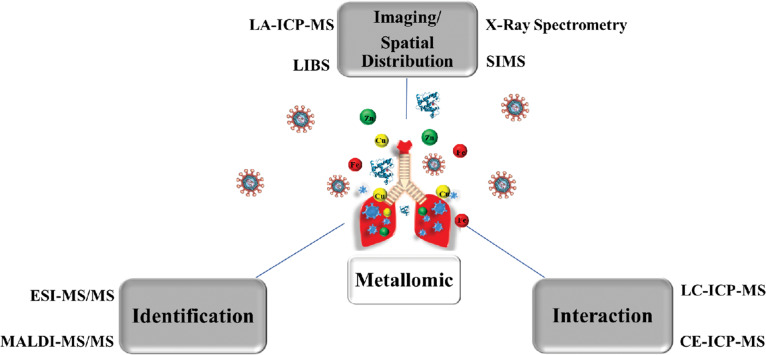
The characterization of the interaction of metals with biological systems requires analytical techniques capable of providing information about the identity and concentrations of these species in biological samples17–19 (Fig. 5). The acquisition of this type of data is possible by direct spectrometric analysis or by combining these with separation techniques. The next topics show an overview of the main analytical techniques applied to metallomics studies with a focus on studies of viral infections, highlighting the challenges associated with each one.
Fig. 5.
Main techniques applied in metallomics studies. Where, LA-ICP-MS – laser ablation inductively coupled plasma mass spectrometry; LIBS – laser induced breakdown spectroscopy; LC-ICP-MS – liquid chromatography coupled to the ICP-MS; CE-ICP-MS - Capillary electrophoresis coupled to ICP-MS; SIMS – secondary ion mass spectrometry; XRF – X-ray fluorescence spectrometry; MALDI-MS – matrix-assisted laser desorption/ionization mass spectrometry; ESI-MS- electrospray ionization mass spectrometry.
Inductively coupled plasma mass spectrometry (ICP-MS)
Certainly, plasma-based instruments are the most popular tools used in metallomics studies.17 ICP OES and ICP-MS are examples of these techniques. Although ICP OES is a multi-element technique and has a wide linear range, its detection limits are generally higher for some trace elements and it is not able to determine isotopes. On the other hand, ICP-MS is extremely sensitive, due to the efficient ionization processes by the highly energetic source and detection using a mass spectrometer.128 Besides, ICP-MS allows determining many metals, metalloids, some non-metals, and isotopes. However, ICP-MS of low-resolution suffers from spectral overlays (isobaric interferences and polyatomic interferences). To minimize these interferences, dynamic reaction cells (DRCs) are generally used, allowing a satisfactory determination. In addition, high-resolution techniques, such as multi-collectors, have been used in metallomics studies, but the high cost of this technique is still a limitation.
ICP-MS has been used to assess the concentrations of metals in fluids of patients infected by virus.129–131 For example, Cu, Zn, and Se were determined in the blood serum of patients with tuberculosis (TB). The effect of HIV co-infection was also assessed in the study. For the study, a total of 155 TB patients were evaluated, of which 74 had HIV co-infection. In addition, 31 healthy individuals were used as controls. As a result, it was observed that when compared to the control group, the concentrations of Zn and Se were significantly lower for patients with TB without HIV co-infection. While Cu levels, and the Cu/Zn ratio were significantly higher (p < 0.05) for the same group. For patients with TB and HIV, the Zn and Se concentrations were significantly lower and a Cu/Zn ratio significantly higher than those found in patients with TB and without HIV co-infection was observed. In addition, the Zn concentration increased significantly at the end of the intensive phase of anti-TB chemotherapy in patients without HIV co-infection.132 A similar finding was observed by Saod et al. who reported that Fe concentration was significantly higher in HBV infected patients as compared to those in healthy controls. However, patients with HBV had significantly lower serum Zn concentrations compared to those in healthy controls. Meanwhile, the Se concentration was significantly lower in patients infected with HBV when compared to healthy controls.133
These studies corroborate data that show the fundamental role of trace elements in the biological system during viral infection and report the efficiency of ICP-MS in this type of study. However, for studies that aim to characterize the function of metallobiomolecules, hyphenated separation techniques to ICP-MS are required.18,36,134 In addition, mass spectrometry to identify the organic fraction is also needed. The following is a brief discussion of the main hyphenated techniques applied in metallomics.
Hyphenated techniques to ICP-MS
Metallothionein (Mt) isoforms are examples of metallobiomolecules that play a key role in viral infections.135,136 However, characterizing a metalloprotein in a protein-rich environment is exceptionally challenging. Nevertheless, there are several ways in which metalloproteins can be targeted and measured. For example, the combination of separation techniques coupled with ICP-MS and organic mass spectrometry is one of the main strategies used in metallomics with a focus on metallobiomolecules.18,36,128,137
Liquid chromatography (LC) can be coupled to the ICP-MS plasma without major modifications in the sample introduction system.138 If the flow of the mobile phase is compatible with the absorption of the nebulizer solution, the column output can be connected directly to the pneumatic nebulizer.139 With recent advances in the field, new nebulizers have been built to operate at a wide variety of flow rates, allowing them to be interfaced with the standard LC configuration, or with μ-LC or nano-LC systems. The versatility of LC in its different modes combined with ICP-MS provides a sensitive response to most elements, being able to obtain isotopic information.140 However, the nature of the association between the metal and the biomolecule portion is essential for choosing the best LC configuration.36,138,139,141 For example, proteins that contain covalently bound metals (e.g., Se142 and As143) are amenable to conventional strategies, including multidimensional methods involving reverse-phase, hydrophilic liquid chromatography (HILIC), and size exclusion chromatography (SEC) and ion exchange. In general, the principle of the LC-ICP-MS technique combines the potential for high-resolution separation of metallobiomolecules by LC with the elemental detection sensitivity of ICP-MS.36,139 Although LC-ICP-MS presents diverse applications in metallomics, especially those associated to metalloproteins, the instability of the analyte signal caused by a low flow LC system, as well as the use of a mobile phase with high concentration of salt or with high composition of organic solvent, can negatively affect its stability and reduce the sensitivity of the technique, causing misunderstandings in quantification studies.36,138,141 Nguyen et al. developed a method based on LC-ICP-MS for the analysis of chemical speciation of As in serum and urine samples.144 As is known to have immunomodulatory properties and may have the potential to alter susceptibility to hepatitis E virus (HEV) infection in humans.131 In this sense, five arsenic species were studied, including arsenite (AsIII), arsenate (AsV), monomethylsonic acid (MMA), dimethylarsinic acid (DMA), and arsenobetaine (AsB). For this study, an elution with ammonium carbonate ((NH4)2CO3), and a strong anion exchange column (Hamilton PRP-X100) allowed the chromatographic separation of the five arsenic species. The extraction efficiency was greater than 91% for the urine matrix and the recoveries were in the range of 94 to 139% for the As species present in human serum. The limit of detection (LD) and the limit of quantification (LQ) of the method, calculated by the signal/noise ratio, varied between 0.3 to 1.5 and 1.0 to 5.0 ng mL−1, respectively. The main species of As found in urine and serum samples were AsB and DMA.144
Another technique used to study metallobiomolecules is capillary electrophoresis (CE) coupled with ICP-MS (CE-ICP-MS).145,146 Among their advantages, we highlight the high separation efficiency and the low sample consumption (∼100 nL), which are important factors for studies in biological samples with limited quantities.147 In addition, the use of capillary columns without coating favors the simplicity and low cost of the technique. When compared to LC, EC is less subject to the presence of artifacts due to the absence of interaction of the analytes with the stationary phase. Basically, in CE, electrically charged species are separated under the action of an electric field applied in a capillary tube containing a conductive solution, responsible for the migration of species towards the detector. The interface between the EC techniques and ICP-MS must not cause the electrophoretic band to widen and, at the same time, it must transport the sample aerosol to the plasma efficiently, favoring the processes of solvation and ionization.145,147 A resource used to implement the CE-ICP-MS interface is the introduction of the capillary into the nebulizer.145 The separation mechanisms and high resolution have made CE a very efficient technique for the separation of complex samples.146 The potential of this technique has been explored in studies of metalloprotein speciation. However, although CE-ICP-MS has gained prominence as a reliable quantitative method, this technique is limited to the analysis of proteins with high molecular weight, since it increases the risk of adsorption of a protein on the capillary wall. In addition, the interfaces designed to couple CE to the ICP-MS are necessary for the high yield analysis of metallometabolites due to the incompatibility between the limited flow rate of the CE and the higher sample capture rate of the ICP-MS.145–147 Michalke et al. proposed a method based on CE-ICP-MS for speciation of Fe (Fe2+/Fe3+) in cerebrospinal fluid (CSF).25 There is strong evidence that high levels of Fe in the biological system can potentially increase the activity of viruses in the biological system, increasing their mutation rates.74,78 In this sense, high levels of Fe have been associated with an increase in the hepatic enzyme ALT (alanine transaminase) in fluids of patients infected with hepatitis C virus.72 The method for Fe speciation used hydrochloric acid (HCl, 20 mM) as the electrolyte of conduction voltage and +25 kV separation voltage. Satisfactory LD (3 μg L−1) and LQ (150 μg L−1) were obtained with an injection volume of 13 nL. Species recoveries ranged from 98 ± 4% to 105 ± 6% (n = 6) for Fe2+ and Fe3+, respectively. The accuracy of the method was validated, using LC-ICP-MS.25
Techniques for bioimaging studies
It is known that several viral infections can affect important tissues and organs during infection, causing an imbalance in metal homeostasis and species that bind to metals, such as Mt.148,149 For example, the accumulation of the metals in fibrotic livers caused by chronic HCV infection can contribute to hepatic injury, suggesting that the liver is the main organ affected by the hepatitis virus.6,56 For SARS-CoV-2, damage to the respiratory, gastrointestinal, and cardiovascular systems has been reported as an effect of COVID-19.75,76 In this sense, for those samples in the solid-state, which require low manipulation, imaging techniques offer extensive possibilities for a better understanding of biological systems, allowing the mapping of chemical components at the tissue, cellular, and subcellular levels.35,150
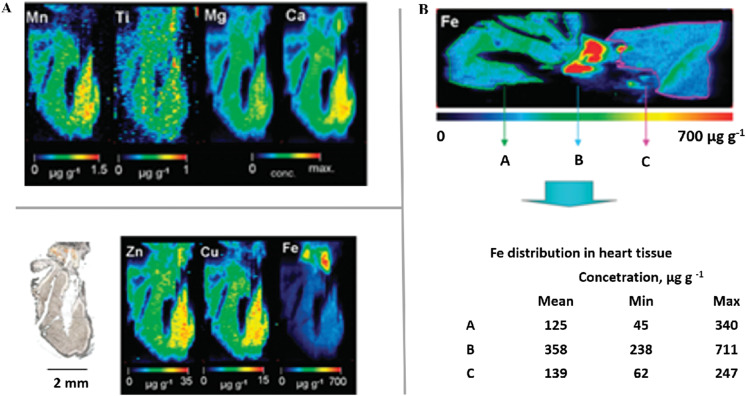
In recent years, laser ablation (LA) coupled to ICP-MS has been introduced as a promising tool for the elemental imaging of biological tissue sections.151–155 LA-ICP-MS is a feasible technique with elemental and isotopic sensitivity, allowing quantitative data to be obtained or imaging/mapping of elements to be performed directly on biological tissues.150,156 LA-ICP-MS was used to assess the spatial distribution of metals in liver tissue.154 In this study, it was observed that there is a decrease in the concentrations of Zn and Cu in the liver materials during viral infection caused by HBV.157 However, the levels of Zn and Cu increased in parallel with the improvement of inflammation in antiviral-treated HBV patients, suggesting that these metals are affected strongly during viral infection. Some studies have reported that SARS-CoV-2 can affect several organs, including the heart, resulting in complications that can lead to the patient's death. Thus, in another study, LA-ICP-MS was used to obtain the spatial distribution of trace elements (Fe, Cu, Zn, Mn, Ca, Ti,) in native mouse heart cryosections (Fig. 6).158 In Fig. 6A, it was observed that Zn and Cu were homogeneously distributed. Ti, Mn, and Ca were detected at a lower concentration than Cu and Zn. However, Fe was the element with the highest concentration in the heart (Fig. 6B).
Fig. 6.
Elemental distribution using LA-ICP-MS. (A) Bioimaging of Zn, Cu, Fe, Mn, Ti, Mg and Ca in a 40 μm section of heart tissue. (B) Iron image measured by LA-ICP-MS, including a table that summarizes the Fe concentrations determined in heart tissue. Adapted from158 with permission of Copyright (2020) American Chemical Society.
In general, analysis by LA-ICP-MS starts with a laser beam focused on the sample surface to generate fine particles.159 The ablated particles are then transported to the secondary excitation source of the ICP-MS instrument for ionization of the sampled mass. The excited ions in the plasma torch are subsequently introduced into a mass spectrometer detector for elemental and isotopic analysis.160 In general, the commonly applied laser ablation parameters differ between imaging and single spot analysis.161 For images, the laser spot size is usually a trade-off between the desired resolution, sensitivity, and concentration, as well as the required analysis time. Besides, the laser irradiance should be selected carefully, such that (i) the sample is ablated completely, (ii) no additional background from the ablation of the target substrate material is produced, and (iii) the neighbouring sections are not affected by the laser plume or shock wave.161 In the case of single spot analysis, the laser spot size is usually selected slightly larger than the target cells. The laser irradiance is usually chosen so high, that a complete cell can be ablated with a single laser pulse while keeping the background from the substrate low. Consequently, irradiance for single spot analysis is generally selected considerably higher compared to imaging. So far, a comparison of the analytical figures of merit for both laser ablation operation modes has not been investigated, yet.161 The solid sampling capability of LA-ICP-MS brings several advantages such as low sample consumption, minimal handling, low risk of contamination, reduction of polyatomic interferences due to dry plasma conditions, spatial information, and high elemental sensitivity, but the obtained quantification data can be important.162 In other words, for the quantification using LA-ICP-MS, the sensitivities achieved by the technique depend on the matrix and the applied laser ablation parameters. First, matrix-matched standards are required; second, they must be ablated using the same laser ablation parameters to ensure reliable quantification. Considering that laser spot size and irradiance differ significantly between standard imaging conditions and standard single spot analysis conditions, it is questionable whether both modes of laser ablation provide comparable quantitative results.35,155,159,161,162
The absence of certified reference material (CRM) for many matrices is also a challenge to quantify trace elements using LA-ICP-MS.163,164 However, calibration using matrix-matched standards for biological tissue analysis has been applied. In this regard, the quantification can be normalized to the 13C response or to an internal standard, e.g., Co, added to the matrices at a defined concentration. Using matrix-matched calibration based on standards spotted to the nitrocellulose membrane, Lohr et al. achieved satisfactory LD for metal content in a single cell.161 An absolute quantitative imaging strategy using isotope dilution (ID) LA-ICP-MS (ID-LA-ICP-MS) is another strategy applied to quantify metals in biological tissue.151 The advantage of this strategy is the ability to increase precision in quantitative analysis, without the need for matrix-matching. However, ID requires that the element has at least two stable isotopes.162 For the quantification of metalloproteins, this technique does not consider the homogeneous distribution of proteins. Therefore, an internal standard must be used. Sulfur present in most proteins is a natural choice provided that the target protein is known and contains either methionine or cysteine. The simultaneous detection of S and P allows, in the case of homologous proteins containing cysteine and methionine, the use of S as an internal standard for the quantification of phosphor. In addition, LA-ICP-MS can be combined with immunohistochemistry (IHC), isoelectric focusing electrophoresis (IEF), and polyacrylamide gel electrophoresis (PAGE) to measure the distributions of element and metalloprotein in tissues, increasing the versatility of the technique.128,150
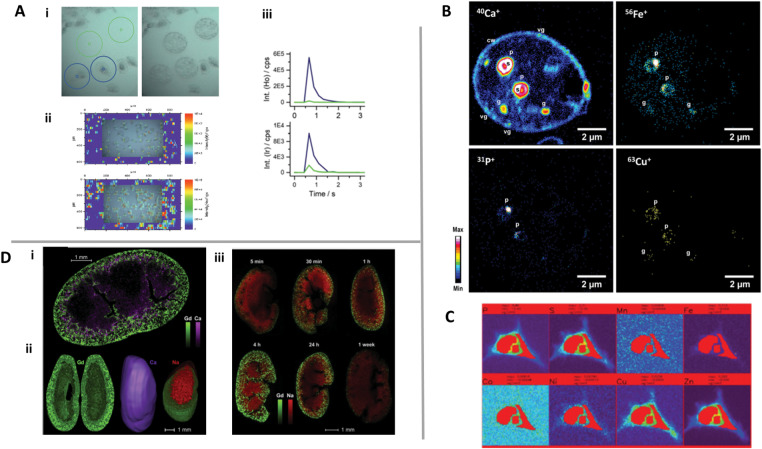
With the advancement of LA technology and the ICP-MS sensitivity, it has been possible to obtain a resolution to identify the distribution of the main elements at single cells150,161 (Fig. 7A). Other studies have employed lanthanide linked MAb staining to identify protein expression in single cells.165 These studies converge with the existing techniques such as flow cytometry and fluorescence microscopy to meet the growing need to define cell biology and immunology on a cell-by-cell basis.159
Fig. 7.
Multi-elemental imaging on single cells using: (A) LA-ICP-MS, wherein (i) and (iii) are measurements of the single cell spot (blue circle) and blanks (green circle), and (ii) is the constructed image. Adapted with permission from162 Marketplace (2020) Royal Society of Chemistry; (B) NanoSIMS with an O− RF plasma primary ion source. Adapted with permission from,170 Copyright (2020) Elsevier; (C) SR-μXRF. Adapted with permission from,170 Marketplace (2020) Royal Society of Chemistry and (D) LIBS, where (i) represents the 2-D high resolution elemental map of Gd (green) and Ca (purple); (ii) represents 3-D distribution of Gd (green) and Ca (purple); (iii) elemental images of Gd (green) and Na (red) in the kidneys at different times. Adapted with permission from173 Copyright (2020) Elsevier.
Secondary ion mass spectrometry (SIMS) is another technique that provides information about the elemental and isotopic composition of samples in situ; however, from a few micrometers to a sub-micron scale.159 SIMS is based on ion sputtering from a solid surface by focused beams of positive or negative primary ion and the subsequent analysis of the produced secondary ions by a mass spectrometer under high vacuum.159 Different combinations of primary ion sources (e.g., Ga+, Bi+, Cs+, O−) and mass spectrometers (quadrupole, time of flight, double sector) allow a wide range of applications for this technique.166 In the case of NanoSIMS, the primary ion beam (either Cs+ or O−) is scanned over the sample surface and the secondary ions are directed toward a double sector analyzer.166 This double sector is composed of an electrostatic field that homogenizes the kinetic energy and focuses the ion beam and a magnetic field that will deflect the ions according to their mass over charge ratios (m/z). Finally, the ions are counted using seven parallel detectors that allow simultaneous detection, which is a requirement for high-precision isotopic ratios.159,166
The high spatial resolution of NanoSIMS has created numerous new research possibilities.159,166 Steinhauser et al. is a good example of using stable isotopes to study stem cell division.167 The virus-host interactions and elemental transfer using the NanoSIMS technique was also reported.168 The sensitive subcellular location of trace metals was possible by the application of a recently developed RF plasma oxygen primary ion source on NanoSIMS which has shown good improvements in terms of lateral resolution (below 50 nm), sensitivity, and stability.169 Pene et al. used NanoSIMS to obtain chemical imaging of macro and trace elements with subcellular resolution169 (Fig. 7B). In this work, Ca, Mg, P, Fe, Cu, and Zn were detected in pyrenoids, contractile vacuoles, and granules at baseline levels. In this sense, this work suggests that NanoSIMS can be an important analytical strategy to assess the effect of biometals on viral infections since the distribution of metals in different cell organelles can be obtained.
Although SIMS has gained great attention in important clinical studies, this technique has some limitations, such as the intensity of the secondary ion registered for an element of interest depends on the ionization yield of each element and its local concentration in the sampling area.166 In other words, the ionization yield can vary greatly according to the chemical environment from which the ions are ejected. Thus, the “matrix effect”, for example, can avoid direct quantification deduced from the measured ion signal. Besides, NanoSIMS operates under a high vacuum, therefore, the sample must be stable and withstand the condition of ultra-high-vacuum. In this sense, biological samples must be dehydrated, and the cellular structure must be stabilized before analysis. Embedding the sample into resin increases stability and minimizes the effects of charging on the sample surface during analysis. The use of resin also increases the potential interference and matrix effects during the measurement. Therefore, quantification is difficult and remains limited due to the matrix effects.159,169
X-ray fluorescence spectrometry (XRF) is also a multi-elemental analytical technique based on X-ray energy radiation that can be used to obtain the spatial distribution of trace elements in biological samples.159 Excited atoms can emit characteristic radiation during the subsequent relaxation process and can carry information about the elemental composition of the specimen in the irradiated region. When XRF uses synchrotron radiation (SR) as a source of excitation, great sensitivity and resolution are obtained. For example, a relative LD of several ng g−1, even as low as 10 ng g−1 can be achieved with a few micrograms of sample.35,159 These characteristics of synchrotron radiation microbeam X-ray fluorescence (SR-μXRF) allow a multi-elemental analysis of larger, smaller elements and trace in a microscopic region of a biological sample. Using SR-μXRF, systematic research was done to map trace elements, such as P, S, Fe, Cu, Zn, Mn, Ni, and Co distribution at the level of cells, and subcells, mapping the location of Fe and other elements selected, in the cell170 (Fig. 7C). Good sensitivity is obtained thanks to third-generation synchrotron facilities, which provide high-intensity X-rays with a brightness of up to 11 orders of magnitude more than conventional X-ray tubes, thus offering an elemental sensitivity in the femtogram (10−15) to attogram (10−18) range.159,171 Undoubtedly, XRF nano-imaging can help understand cell pathways, but it is important to highlight that the need for micro-SXRF is still high to probe large tissues and understand the sample heterogeneity.171 In addition, due to the high penetration of X-rays into the matter, 3D maps can be obtained by translating and rotating the sample.35,159 An advantage of SRXRF compared to other techniques is the possibility of investigating samples in their frozen-hydrated state with a minimum of sample preparation due to the implementation of cryostats that limit potential metal redistribution and speciation change, particularly for soluble species and low stable complexes. However, sample degradation related to intense beam exposure and the need for a long time for analysis can also be a limitation for this technique.159
In recent years, elemental imaging using laser-induced breakdown spectroscopy (LIBS) has been a promising approach for biological and medical applications.172,173 Recently, the high-resolution 2-D elemental map in the kidney was reported to show the applicability of LIBS in clinical studies.174 LIBS was used on the organ scale to create images of Gd, Ca, and Na in a series of adjacent coronal kidney sections174 (Fig. 7D-i and iii). The consecutive images were pooled to reconstruct the organ in 3-D (Fig. 7D-ii).
LIBS is an analytical method based on optical emission spectroscopy.175 High-power laser pulses are focused on the sample surface, therefore ablating small amounts of the sample (from micrograms to a fraction of a nanogram, depending on the experimental setup) and creating plasma.174 In LIBS, the steps of ablation, atomization, and excitation occur simultaneously during a single laser pulse. Atoms and ions excited by the high plasma temperature return to lower energy levels by emitting radiation. The emitted light radiation represents a specific elemental response that can be collected by external optical systems and subsequently analyzed with one or more spectrometers equipped with detectors, generally intensified charge-coupled device (ICCD) cameras.172,173 The ability of LIBS to obtain elemental signals from a single laser pulse and a very small amount of material (fraction of a nanogram) gives this technology many advantages, such as multi-elemental capability, ease of use, and operation under atmospheric pressure.173,176 In addition, this is the only all-optical technique that is fully compatible with optical microscopy, providing elemental information resolved in space with a range of parts per million sensitivity (accessible for most metals) and a resolution on a micrometer scale.177 Another asset of the technique lies in its fast acquisition speed. These aspects make LIBS-based imaging promising for application in clinical studies, such as viral infection.173 However, this technique is not able to evaluate the isotope rate. Besides, LIBS imaging requires high levels of instrumentation controls, including at least systems to stabilize both the laser energy and the laser focus. It is also important to mention that quantitative calibration is not always possible depending on the availability of standards.172,173
Identification of organic species in metallomics and bioinformatics
MALDI-MS (matrix-assisted laser desorption/ionization mass spectrometry) and ESI-MS (electrospray ionization mass spectrometry) are important techniques that, together with ICP-MS, represent powerful tools for the identification of known or unknown metallobiomolecules.36 ESI and MALDI are smoother ionization techniques that can provide molecular information and are often used as a complementary technique to ICP-MS.18,36,134 By combining LC-ICP-MS or CE-ICP-MS with ESI-MS/MS, the analytes of interest can be quantified and identified with a high degree of mass accuracy and sensitivity.36 The successful application of this technique employs methods that introduce the analyte species into the detector in a mobile phase depleted of as many other species as possible to avoid suppression of ionization. Several studies show the efficiency of this combination for the understanding of the role of trace elements and metallobiomolecules in biological systems during viral infections. Table 1 summarizes some of these applications.
Table 1.
Summarizing metallomics studies focusing on viral infections
| Virus | Biological sample | Element/protein target | Ref. |
|---|---|---|---|
| Chikungunya virus | Cell | Mg | 178 |
| Hepatitis B virus | Liver tissue | Zn, Cu, Fe, Ag, Co, Al, Ni, Cd, Ag, Pb, Cr, Mn | 157 |
| Influenza virus (AH1N1) | Serum | Pb, Hg, Cr, Cd, As, Se, Zn, Ca, Na, Cu | 129 |
| Coxsackievirus B3 | Tissue | As | 179 |
| Coxsackievirus | Blood | AuNPs/DNA target | 180 |
| Influenza virus (AH1N1) | Lung tissue | 114Cd | 181 |
| Dengue virus | Cell | AuNPs/DNA target | 182 |
| Influenza virus (H9N2) | Serum | 111Cd | 183 |
| Influenza virus (H9N2, H1N1, H3N2) | Excretions | AuNPs/lectin Con A and glycoproteins | 184 and 185 |
| Hepatitis B virus (HBV) | Serum | AuNPs/DNA target | 186 |
| Herpes-simplex virus (HSV) | Plasma | Zn | 187 |
| Human papillomavirus (HPV) | Plasma | Zn | 187 |
| Human immunodeficiency virus (HIV) | Plasma | Zn | 187 |
| Human immunodeficiency virus (HIV) | Tissue/cell | Eu | 188 |
| Human immunodeficiency virus (HIV) | Plasma | Ga3+/hemoglobin–haptoglobin | 189 |
| Human papillomavirus (HPV) | Cell | As | 190 |
| Hepatitis B virus (HBV) | Cell | Lanthanide/DNA target | 191 |
In addition, it is important to emphasize the importance of bioinformatics for elucidation of complex data obtained in omics studies.11 Bioinformatics can establish standard formats using algorithms based on mathematical and statistical models and developing efficient methods to store, retrieve, and share high-performance data in the omics studies, providing a considerable contribution to overcome the limitation of empirical methods using predictive tools.192 For example, the prediction of macromolecules’ metal-binding domains, such as proteins, can be performed using the protein sequence data from continuously updated libraries such as Pfam193 and MASCOT23,96,98 used to identify proteins and to identify various post-translational modification sites. Besides, to create images from elemental spatial distribution in biological samples, MATLAB156 is generally used, although some groups develop in house software for this end.194 Therefore, the bioinformatics approach is an important tool to obtain complete information on omics studies, complementary to the experimental methods currently available.
Summary and outlook
Considering all the points discussed in this text, it is clear that metals play important roles in viral infections, such as helping in the activation of immune cells, production of antibodies and inhibition of virus replication. However, the relationship between trace elements and infections caused by viruses is still complex since the specific functions of several elements remain largely undefined. Therefore, there is still a great amount to explore to achieve a correct understanding of the biochemical mechanisms involved between trace elements and viral infections. In this sense, the search for complementary processes, including the exploration of strategies based on metallomics to help health professionals better understand and treat diseases, is welcome and of great importance.
Regarding possible trends, we really believe that advances in hyphenated techniques will continue to drive the area, increasing the separation resolution of metallobiomolecules and the detection sensitivity. In addition, multimodal approaches are very promising, as they can collect complementary information about the nature of the samples, allowing them to obtain qualitative, quantitative, and spatial data of different elements and isotopes in different types of biological samples. For example, LIBS imaging can be combined with LA-ICP-MS for elemental mapping in the tissue of the heart, brain and lungs, allowing an understanding of the complexity of COVID-19 infection, and its effect on the deregulation of the metal homeostasis. LA-ICP-MS allows the identification of isotopes under certain conditions and may be more sensitive. LIBS is completely optical and has no restrictions on the analysis of elements with a low atomic number. In addition, LIBS imaging or LA-ICP-MS imaging can be combined with other imaging modalities such as Raman spectroscopy or laser-induced fluorescence, increasing the amount of system information.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
The authors thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brazil, and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, São Paulo, Brazil, grant number 2018/00786-0) for financial support and the fellowship.
Abbreviation
- (NH4)2CO3
Ammonium carbonate
- ACE2
Angiotensin-converting enzyme 2
- ALT
Alanine transaminase
- As
Arsenic
- AsB
Arsenobetaine
- AsIII
Arsenite
- AsV
Arsenate
- ATP
Adenosine triphosphate
- Ca
Calcium
- Cd
Cadmium
- CE
Capillary electrophoresis
- CE-ICP-MS
Capillary electrophoresis coupled to ICP-MS
- Co
Cobalt
- COVID-19
Coronavirus disease-19
- CSF
Cerebrospinal fluid
- Cu
Copper
- DCs
Dendritic cells
- DMA
Dimethylarsinic acid
- DMT1
Divalent metallic iron transport protein 1
- DRC
Dynamic reaction cells
- ESI-MS
Electrospray ionization mass spectrometry
- Fe
Iron
- FPN1
Ferroportin membrane transporter
- HBV
Hepatitis B virus
- HCl
Hydrochloric acid
- HCV
Hepatitis C virus
- HEV
Hepatitis E virus
- HILIC
Hydrophilic liquid chromatography
- HIV
Human immunodeficiency virus
- HPV
Human papillomavirus
- HRP
Horseradish peroxidase
- ICCD
Intensified charge-coupled device
- ICP OES
Inductively Coupled Plasma Optical Emission spectroscopy
- ICP-MS
Inductively Coupled Plasma Mass Spectrometry
- ID
Isotope Dilution
- IFNγ
Interferon-gamma
- IHC
Immunohistochemistry
- LA-ICP-MS
Laser Ablation Inductively Coupled Plasma Mass Spectrometry
- LC
Liquid chromatography
- LC-ICP-MS
Liquid chromatography coupled to the ICP-MS
- LD
Limit of detection
- LIBS
Laser Induced Breakdown Spectroscopy
- LQ
Limit of quantification
- m/z
Mass over charge ratios
- MALDI-MS
Matrix-assisted laser desorption/ionization mass spectrometry
- MERS-CoV
Middle East Respiratory Syndrome coronavirus
- MHCII
Major histocompatibility complex II
- MMA
Monomethylsonic acid
- Mn
Manganese
- Mt
Metallothionein
- Ni
Nickel
- NPs
Nanoparticles
- PAGE
Polyacrylamide gel electrophoresis
- Pb
Lead
- RdRp
RNA-dependent RNA polymerase
- SARS-CoV
Severe Acute Respiratory Syndrome coronaviruses
- Se
Selenium
- SEC
Size exclusion chromatography
- SIMS
Secondary Ion Mass Spectrometry
- SR
Synchrotron radiation
- TB
Tuberculosis
- WHO
World Health Organization
- XRF
X-Ray Fluorescence spectrometry
- Zn
Zinc
References
- World Health Organization , Managing epidemics: key facts about major deadly diseases, WHO, Luxembourg, 2018 [Google Scholar]
- Grubaugh N. D., Ladner J. T., Lemey P., Pybus O. G., Rambaut A., Holmes E. C. and Andersen K. G., Nat. Microbiol., 2019, 4, 10–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W. J., Wang D., Xu W., Holmes E. C., Gao G. F., Wu G., Chen W., Shi W. and Tan W., Lancet, 2020, 395, 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Online , https://covid19.who.int/, accessed September 2020
- Li M., Liu W., Bauch T., Graviss E. A., Arduino R. C., Kimata J. T., Chen M. and Wang J., Nat. Commun., 2020, 11, 1–15 10.1038/s41467-020-17753-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A. L., El-Sayed M. H., Kao J. H., Lazarus J. V., Lemoine M., Lok A. S. and Zoulim F., Nat. Rev. Gastroenterol. Hepatol., 20201–10 10.1038/s41575-020-0332-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz J. M., Hankerson S. J., Alexandre B. R., Henry M. D., Martins A. F., Ferraz L. P., Ruiz-Miranda C. R., Sci. Rep., 2019, 9, 1–13 10.1038/s41598-019-49199-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P. A., Moss W. J., Takeda M., de Swart R. L., Thompson K. M. and Goodson J. L., Nat. Rev. Dis. Primers, 2016, 2, 1–16 10.1038/nrdp.2016.49 [DOI] [PubMed] [Google Scholar]
- Guzman M. G., Gubler D. J., Izquierdo A., Martinez E. and Halstead S. B., Nat. Rev. Dis. Primers, 2016, 2, 1–25 10.1038/nrdp.2016.55 [DOI] [PubMed] [Google Scholar]
- Giraldo M. I., Xia H., Aguilera-Aguirre L., Hage A., van Tol S., Shan C., Xie X., Sturdevant G. L., Robertson S. J., McNally K. L., Meade-White K., Azar S. R., Rossi S. L., Maury W., Woodson M., Ramage H., Johnson J. R., Krogan N. J., Morais M. C., Best S. M., Shi P. Y. and Rajsbaum R., Nature, 20201–23 10.1038/s41586-020-2457-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasapis C. T., J. Inorg. Biochem., 2018, 186, 157–161 [DOI] [PubMed] [Google Scholar]
- Kumar A., Kubota Y., Chernov M. and Kasuya H., Med. Hypotheses, 2020, 144, 1–3 10.1016/j.mehy.2020.109848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Zhang S., Nekhai S. and Liu S., Current Clinical Microbiology Reports, 2020, 7, 13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreou A., Trantza S., Filippou D., Filippou D., Sipsas N. and Tsiodras S., In Vivo, 2020, 34, 1567–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N., Chen X. and Geiger J. D., Viruses, 2020, 12, 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K. K., Verma R., Oleske J. M., Scolpino A. and Bogden J. D., Nutr. Res., 2019, 71, 21–29 [DOI] [PubMed] [Google Scholar]
- Ogra Y., Hirata T., Metallomics: Recent Analytical Techniques and Applications, Springer Nature, Switzerland, 1st edn, 2017 [Google Scholar]
- Haraguchi H., Metallomics, 2017, 9, 1001–1013 [DOI] [PubMed] [Google Scholar]
- de Jesus J. R., da Costa L. F., Lehmann E. L., Galazzi R. M., Madrid K. C. and Arruda M. A. Z., in Metallomics: The Science of Biometals, ed. Arruda M. A. Z., Springer International Publishing, Switzerland, 1st edn, 2018, ch. 8, pp. 183–211 [Google Scholar]
- Thyssen G. M., Holtkamp M., Kaulfürst-Soboll H., Wehe C. A., Sperling M., von Schaewen A. and Karst U., Metallomics, 2017, 9, 676–684 [DOI] [PubMed] [Google Scholar]
- Abduljabbar T. N., Sharp B. L., Reid H. J., Barzegar-Befroeid N., Peto T. and Lengyel I., Talanta, 2019, 204, 663–669 [DOI] [PubMed] [Google Scholar]
- Jesus J. R., Arruda M. A. Z., Rapid Commun. Mass Spectrom., 2020, 34, 1–10 10.1002/rcm.8798 [DOI] [PubMed] [Google Scholar]
- Pessôa G. D. S., de Jesus J. R., Balbuena T. S., Aurélio M. and Arruda Z., Rapid Commun. Mass Spectrom., 2020, 34, 1–10 10.1002/rcm.8698 [DOI] [PubMed] [Google Scholar]
- Møller L. H., Jensen C. S., Nguyen T. T. T. N., Stürup S. and Gammelgaard B., J. Anal. At. Spectrom., 2015, 30, 277–284 [Google Scholar]
- Michalke B., Willkommen D. and Venkataramani V., J. Visualized Exp., 2020, 159, 1–8 10.3791/61055 [DOI] [PubMed] [Google Scholar]
- Michalke B., Willkommen D. and Venkataramani V., Front. Chem., 2019, 7, 1–9 10.3389/fchem.2019.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose M. H. M., Theiner S., Kornauth C., Meier-Menches S. M., Heffeter P., Berger W., Koellensperger G. and Keppler B. K., Metallomics, 2018, 10, 388–396 [DOI] [PubMed] [Google Scholar]
- Konz I., Fernández B., Fernández M. L., Pereiro R., González-Iglesias H., Coca-Prados M., Sanz-Medel A., Anal. Bioanal. Chem., 2014, 406, 2343–2348 [DOI] [PubMed] [Google Scholar]
- Altelaar A. F. M., Klinkert I., Jalink K., de Lange R. P. J., Adan R. A. H., Heeren R. M. A. and Piersma S. R., Anal. Chem., 2006, 78, 734–742 [DOI] [PubMed] [Google Scholar]
- Gates S. D., Condit R., Moussatche N., Stewart B. J., Malkin A. J. and Weber P. K., Anal. Chem., 2018, 90, 1613–1620 [DOI] [PubMed] [Google Scholar]
- Finnegan M. E., Visanji N. P., Romero-Canelon I., House E., Rajan S., Mosselmans J. F. W., Hazrati L. N., Dobson J. and Collingwood J. F., J. Neurosci. Methods, 2019, 319, 28–39 [DOI] [PubMed] [Google Scholar]
- Arruda M. A. Z., Metallomics: The Science of Biometals, Springer International Publishing, Switzerland, 1st edn, 2018 [Google Scholar]
- Wang Z., Chu Y., Chen F., Sheng Z. and Guo L., Appl. Opt., 2019, 58, 7615–7620 [DOI] [PubMed] [Google Scholar]
- Singh V. K., Sharma J., Pathak A. K., Ghany C. T. and Gondal M. A., Biophys. Rev., 2018, 10, 1221–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler V. L., Müller E. I. and Pozebon D., in Metallomics: The Science of Biometals, ed. Arruda M. A. Z., Springer International Publishing, Switzerland, 1st edn, 2018, ch. 7, pp. 139–181 [Google Scholar]
- Montes-Bayón M., Sharar M., Corte-Rodriguez M., TrAC, Trends Anal. Chem., 2018, 104, 4–10 [Google Scholar]
- Gaudin R. and Barteneva N. S., Nat. Commun., 20141–11 10.1038/ncomms7022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Smith G. J. D., Fouchier R. A. M., Peiris M., Kedzierska K., Doherty P. C., Palese P., Shaw M. L., Treanor J., Webster R. G., García-Sastre A., Nat. Rev. Dis. Primers, 2018, 4, 1–21 10.1038/s41572-018-0002-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadi S., Gaaloul ben Hnia N., Barre R. S., Wexler A. S., Ristenpart W. D. and Bouvier N. M., Nat. Commun., 20201–9 10.1038/s41467-020-17888-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onozuka D., Sci. Rep., 2014, 4, 1–6 10.1038/srep05157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayanlade A., Nwayor I. J., Sergi C., Ayanlade O. S., di Carlo P., Jeje O. D. and Jegede M. O., Sci. Rep., 2020, 10, 1–13 10.1038/s41598-020-71094-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Zhu Z., Xu Q., Fang X., Liu X., Xiong P., Song L., Xu W. and Xu A., Sci. Rep., 2017, 7, 1–8 10.1038/s41598-017-18281-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J. and Qian Z., Nat. Commun., 20201–12 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Bert N., Tan A. T., Kunasegaran K., Tham C. Y. L., Hafezi M., Chia A., Chng M. H. Y., Lin M., Tan N., Linster M., Chia W. N., Chen M. I. C., Wang L. F., Ooi E. E., Kalimuddin S., Tambyah P. A., Low J. G. H., Tan Y. J. and Bertoletti A., Nature, 2020, 584, 457–462 [DOI] [PubMed] [Google Scholar]
- Fan H., Du X., Zhang J., Zheng H., Lu X., Wu Q., Li H., Wang H., Shi Y., Gao G., Zhou Z., Tan D. X. and Li X., Sci. Rep., 2017, 7, 1–13 10.1038/srep41226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhilan M., Suresh I. and Nesakumar N., Measurement: Journal of the International Measurement Confederation, 2021, 168, 1–15 10.1016/j.measurement.2020.108335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan S. R., Pradeu T., Masters S. L. and Mogensen T. H., Nat. Rev. Immunol., 20201–14 10.1038/s41577-020-0391-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera A., Siracusa M. C., Yap G. S. and Gause W. C., Nat. Immunol., 2016, 17, 356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mentzer A. J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., López-Camacho C., Slon-Campos J., Zhao Y., Stuart D. I., Paesen G. C., Grimes J. M., Antson A. A., Bayfield O. W., Hawkins D. E. D. P., Ker D.-S., Wang B., Turtle L., Subramaniam K., Thomson P., Zhang P., Dold C., Ratcliff J., Simmonds P., de Silva T., Sopp P., Wellington D., Rajapaksa U., Chen Y.-L., Salio M., Napolitani G., Paes W., Borrow P., Kessler B. M., Fry J. W., Schwabe N. F., Semple M. G., Baillie J. K., Moore S. C., Openshaw P. J. M., Ansari M. A., Dunachie S., Barnes E., Frater J., Kerr G., Goulder P., Lockett T., Levin R., Zhang Y., Jing R., Ho L.-P., Cornall R. J., Conlon C. P., Klenerman P., Screaton G. R., Mongkolsapaya J., McMichael A., Knight J. C., Ogg G. and Dong T., Nat. Immunol., 20201–16 10.1038/s41590-020-0782-6 [DOI] [PubMed] [Google Scholar]
- Rambhai H. K., Ashby F. J., Qing K. and Srivastava A., Mol. Ther. – Methods Clin. Dev., 2020, 18, 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feleke B. E., Feleke T. E., Mekonnen D. and Beyene M. B., Clin. Nutr. ESPEN, 2019, 31, 56–60 [DOI] [PubMed] [Google Scholar]
- Zhan J., Wallace T. C., Butts S. J., Cao S., Ansu V., Spence L. A., Weaver C. M., Gletsu-Miller N., Nutrients, 2020, 12, 1–11 10.3390/nu12051245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder-Keller A., Bolivar V. J., Zink S. and Kramer L. D., J. Neuropathol. Exp. Neurol., 2020, 79, 767–776 [DOI] [PubMed] [Google Scholar]
- Sivasubramanian S., Shenoy J., Kamath S. P., Kulkarni V., Srinivasan M., Shantharam Baliga B., HIV and AIDS Review, 2020, 9, 56–60 [Google Scholar]
- Cardenas A., Smit E., Welch B. M., Bethel J. and Kile M. L., Environ. Res., 2018, 166, 570–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read S. A., O’Connor K. S., Suppiah V., Ahlenstiel C. L. E., Obeid S., Cook K. M., Cunningham A., Douglas M. W., Hogg P. J., Booth D., George J. and Ahlenstiel G., Nat. Commun., 2017, 8, 1–15 10.1038/ncomms15245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywald M., Wessels I. and Rink L., Int. J. Mol. Sci., 2017, 18, 1–34 10.3390/ijms18102222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus J. R., Aragão A. Z. B., Arruda M. A. Z., Ramos C. H. I., Front. Chem., 2019, 7, 1–10 10.3389/fchem.2019.00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read S. A., Obeid S., Ahlenstiel C. and Ahlenstiel G., Adv. Nutr., 2019, 10, 696–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amare B., Moges B., Mulu A., Yifru S. and Kassu A., BioMed Res. Int., 20151–10 10.1155/2015/598605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feleke B. E., Feleke T. E., Mekonnen D. and Beyene M. B., Clin. Nutr. ESPEN, 2019, 31, 56–60 [DOI] [PubMed] [Google Scholar]
- Martinez S. S., Campa A., Li Y., Fleetwood C., Stewart T., Ramamoorthy V. and Baum M. K., J. Nutr., 2017, 147, 556–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatsalya V., Cave M. C., Kumar R., Srivastava S., Khanal S., Jenson A. B., Schwandt M. L., Barve S. S., Ramchandani V. A. and McClain C. J., AIDS Res. Hum. Retroviruses, 2019, 35, 92–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read S. A., Obeid S., Ahlenstiel C. and Ahlenstiel G., Adv. Nutr., 2019, 10, 696–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna-Padilla I. A., Briceño O., Aguilar-Vargas A., Rodríguez-Moguel N. C., Villazon-De la Rosa A., Pinto-Cardoso S., Flores-Murrieta F. J., Perichart-Perera O., Tolentino-Dolores M., Vargas-Infante Y., Reyes-Terán G., Nutrition, 2020, 70, 1–7 10.1016/j.nut.2019.110585 [DOI] [PubMed] [Google Scholar]
- Subramanian Vignesh K. and Deepe G. S., Arch. Biochem. Biophys., 2016, 611, 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Kubota Y., Chernov M. and Kasuya H., Med. Hypotheses, 2020, 144, 1–3 10.1016/j.mehy.2020.109848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A. J. W., van den Worml S. H. E., Sims A. C., Baric R. S., Snijder E. J. and van Hemert M. J., PLoS Pathog., 2010, 6, 1–10 10.1371/journal.ppat.1001176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell D. B., Heyden E. L. and Pretorius E., Front. Immunol., 2020, 11, 1–15 10.3389/fimmu.2020.01221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat J. E. and Skaar E. P., Cell Host Microbe, 2013, 13, 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. C., Bayeva M., Taiwo B., Palella F. J., Hope T. J. and Ardehali H., AIDS Res. Hum. Retroviruses, 2015, 31, 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çam H., Yılmaz N., Arab J. Gastroenterol., 2020, 21, 85–90 [DOI] [PubMed] [Google Scholar]
- Snyder-Keller A., Bolivar V. J., Zink S. and Kramer L. D., J. Neuropathol. Exp. Neurol., 2020, 79, 767–776 [DOI] [PubMed] [Google Scholar]
- Haider B. A., Spiegelman D., Hertzmark E., Sando D., Duggan C., Makubi A., Sudfeld C., Aris E., Chalamilla G. E. and Fawzi W. W., Am. J. Trop. Med. Hyg., 2019, 100, 1512–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J. and Cao B., Lancet, 2020, 395, 497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M. L., DeBolt C., Lindquist S., Lofy K. H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L. A., Patel A., Gerber S. I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M. A., Weldon W. C., Biggs H. M., Uyeki T. M. and Pillai S. K., N. Engl. J. Med., 2020, 382, 929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G. and Carver P. L., Clin. Microbiol. Infect., 2018, 24, 16–23 [DOI] [PubMed] [Google Scholar]
- Dalamaga M., Karampela I. and Mantzoros C. S., Metab., Clin. Exp., 2020, 108, 1–3 10.1016/j.metabol.2020.154260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., World Sci. News, 2018, 99, 148–168 [Google Scholar]
- Raha S., Mallick R., Basak S. and Duttaroy A. K., Med. Hypotheses, 2020, 142, 109814. 10.1016/j.mehy.2020.109814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaetke L. M., Chow-Johnson H. S. and Chow C. K., Arch. Toxicol., 2014, 88, 1929–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreou A., Trantza S., Filippou D., Filippou D., Sipsas N. and Tsiodras S., In Vivo, 2020, 34, 1567–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wazir S. M. and Ghobrial I., J. Community Hosp. Intern. Med. Perspect., 2017, 7, 265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler G., Kincses A., Mosolygó T., Maræ M. A., Nové M., Gajdács M., Sanmartín C., McNeil H. E., Blair J. M. A., Domínguez-Álvarez E., Molecules, 2019, 24, 1–16 10.3390/molecules24234264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivory K., Prieto E., Spinks C., Armah C. N., Goldson A. J., Dainty J. R. and Nicoletti C., Clin. Nutr., 2017, 36, 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojadoost B., Taha-Abdelaziz K., Alkie T. N., Bekele-Yitbarek A., Barjesteh N., Laursen A., Smith T. K., Shojadoost J. and Sharif S., Vet. Immunol. Immunopathol., 2020, 227, 1–5 10.1016/j.vetimm.2020.110089 [DOI] [PubMed] [Google Scholar]
- Zhang J., Taylor E. W., Bennett K., Saad R. and Rayman M. P., Am. J. Clin. Nutr., 2020, 111, 1297–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace T. C., J. Am. Coll. Nutr., 20201–9 10.1080/07315724.2020.1785971 [DOI] [PubMed] [Google Scholar]
- Chaigne-Delalande B., Li F. Y., O’Connor G. M., Lukacs M. J., Jiang P., Zheng L., Shatzer A., Biancalana M., Pittaluga S., Matthews H. F., Jancel T. J., Bleesing J. J., Marsh R. A., Kuijpers T. W., Nichols K. E., Lucas C. L., Nagpal S., Mehmet H., Su H. C., Cohen J. I., Uzel G. and Lenardo M. J., Science, 2013, 341, 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ben Ouirane K., Boulard Y. and Bressanelli S., J. Biol. Chem., 2019, 294, 7573–7587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera N. C. N., Godahewa G. I., Lee S., Kim M. J., Hwang J. Y., Kwon M. G., Hwang S. D. and Lee J., Fish Shellfish Immunol., 2017, 68, 435–442 [DOI] [PubMed] [Google Scholar]
- Silvestre M. C., dos Reis V. M. S. and Sato M. N., An. Bras. Dermatol., 2018, 93, 242–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Liu H., Jian Z., Cui H., Fang J., Zuo Z., Deng J., Li Y., Wang X., Zhao L., He R. and Tang H., Ecotoxicol. Environ. Saf., 2020, 203, 1–10 10.1016/j.ecoenv.2020.111006 [DOI] [PubMed] [Google Scholar]
- Tsatsakis A., Petrakis D., Nikolouzakis T. K., Docea A. O., Calina D., Vinceti M., Goumenou M., Kostoff R. N., Mamoulakis C., Aschner M., Hernández A. F., Food Chem. Toxicol., 2020, 141, 1–15 10.1016/j.fct.2020.111418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus J. R., da Silva Fernandes R., de Souza Pessoa G., Raimundo I. M., Arruda M. A. Z., Talanta, 2017, 170, 199–209 [DOI] [PubMed] [Google Scholar]
- Jesus J. R., Santos H. M., López-Fernández H., Lodeiro C., Arruda M. A. Z. and Capelo J. L., Talanta, 2018, 178, 864–869 [DOI] [PubMed] [Google Scholar]
- de Jesus J. R., de G., Pessôa S., Sussulini A., Martínez J. L. C., Arruda M. A. Z., J. Proteomics, 2016, 145, 187–196 [DOI] [PubMed] [Google Scholar]
- de Jesus J. R., Galazzi R. M., de Lima T. B., Banzato C. E. M., de Almeida Lima e Silva L. F., de Rosalmeida Dantas C., Gozzo F. C., Arruda M. A. Z., Clin. Biochem., 2017, 50, 1118–1125 [DOI] [PubMed] [Google Scholar]
- de Jesus J. R., in Proteomic and Ionomic Study for identification of Biomarker in Biological Fluid Samples of Patients with Psychiatric Disorders and Healthy Individuals, ed. de Jesus J. R., Springer, Switzerland, 1st edn, 2019, ch. 3, pp. 39–58 [Google Scholar]
- de Jesus J. R., de Campos B. K., Galazzi R. M., Martinez J. L. C., Arruda M. A. Z., Anal. Bioanal. Chem., 2015, 407, 661–667 [DOI] [PubMed] [Google Scholar]
- Delanghe J. and Speeckaert M., Biochem. Med., 2014, 24, 89–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesko M. F., Hartwig C. A., Bizzi C. A., Pereira J. S. F., Mello P. A., Flores E. M. M., Int. J. Mass Spectrom., 2011, 307, 123–136 [Google Scholar]
- Niu Z., Zhang W., Yu C., Zhang J. and Wen Y., TrAC, Trends Anal. Chem., 2018, 102, 123–146 [Google Scholar]
- Montes-Bayón M., Sharar M., Corte-Rodriguez M., TrAC, Trends Anal. Chem., 2018, 104, 4–10 10.1016/j.trac.2017.09.025 [DOI] [Google Scholar]
- Chutipongtanate S., Changtong C., Weeraphan C., Hongeng S., Srisomsap C. and Svasti J., Proteomics, 2015, 12, 1–8 10.1186/s12014-015-9087-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu V., Picó Y., TrAC, Trends Anal. Chem., 2019, 118, 709–721 [Google Scholar]
- Manousi N., Rosenberg E., Deliyanni E. A. and Zachariadis G. A., Molecules, 2020, 25, 1–24 10.3390/molecules25102411 [DOI] [Google Scholar]
- Fei J. J., Zhao L. Y., Wu X. H., Cui X. B., Min H., Lian H. Z. and Chen Y. J., Microchim. Acta, 2020, 187, 1–10 10.1007/s00604-020-04329-0 [DOI] [Google Scholar]
- Abou-Zeid R. E., Dacrory S., Ali K. A. and Kamel S., Int. J. Biol. Macromol., 2018, 119, 207–214 [DOI] [PubMed] [Google Scholar]
- Chen L., Wang Z., Pei J. and Huang X., Anal. Chem., 2020, 92, 2251–2257 [DOI] [PubMed] [Google Scholar]
- Ibrahim W. A. W., Nodeh H. R. and Sanagi M. M., Crit. Rev. Anal. Chem., 2016, 46, 267–283 [DOI] [PubMed] [Google Scholar]
- Zambrzycka-Szelewa E., Leśniewska B., Godlewska-Żyłkiewicz B., Compr. Anal. Chem., 2019, 86, 261–293 [Google Scholar]
- de la Calle I., Pena-Pereira F., Lavilla I. and Bendicho C., Anal. Chim. Acta, 2016, 936, 12–39 [DOI] [PubMed] [Google Scholar]
- Sajid M., Kabeer M. and Falath W., Molecules, 2020, 25, 1–13 10.3390/molecules25153395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rykowska I., Ziemblińska J. and Nowak I., J. Mol. Liq., 2018, 259, 319–339 [Google Scholar]
- Vicentino P. O., Cassella R. J., Leite D. and Resano M., Talanta, 2020, 206, 1–8 10.1016/j.talanta.2019.120230 [DOI] [PubMed] [Google Scholar]
- Khan M., Kazi T. G., Afridi H. I., Bilal M., Akhtar A., Ullah N., Khan S. and Talpur S., Ultrason. Sonochem., 2017, 39, 313–320 [DOI] [PubMed] [Google Scholar]
- Kazi T. G., Naeemullah G. K. and Tuzen M., Food Chem., 2015, 172, 161–165 [DOI] [PubMed] [Google Scholar]
- Trujillo-Rodríguez M. J., Rocío-Bautista P., Pino V. and Afonso A. M., TrAC, Trends Anal. Chem., 2013, 51, 87–106 [Google Scholar]
- Shukla S. K., Pandey S. and Pandey S., J. Chromatogr. A, 2018, 1559, 44–61 [DOI] [PubMed] [Google Scholar]
- Rykowska I., Ziemblińska J. and Nowak I., J. Mol. Liq., 2018, 259, 319–339 [Google Scholar]
- Duarte F. A., Mello P. A., Muller E. I., Diehl L. O., Flores E. M. M. and Gatiboni T. L., Ultrason. Sonochem., 2017, 40, 24–29 [DOI] [PubMed] [Google Scholar]
- de Jesus J. R., Proteomic and Ionomic Study for identification of Biomarker in Biological Fluid Samples of Patients with Psychiatric Disorders and Healthy Individuals, Springer, Switzerland, 1st edn, 2019 [Google Scholar]
- Lothian A. and Roberts B. R., J. Visualized Exp., 2020, 110, 1–18 10.3791/53737 [DOI] [Google Scholar]
- Hamdy M. E., del Carlo M., Hussein H. A., Salah T. A., El-Deeb A. H., Emara M. M., Pezzoni G. and Compagnone D., J. Nanobiotechnol., 2018, 16, 1–12 10.1186/s12951-018-0374-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu I. H., Chen W. H., Wu T. K. and Sun Y. C., J. Chromatogr. A, 2011, 1218, 1795–1801 [DOI] [PubMed] [Google Scholar]
- Zhang H., Liu L., Li C. W., Fu H., Chen Y. and Yang M., Biosens. Bioelectron., 2011, 29, 89–96 [DOI] [PubMed] [Google Scholar]
- Ogra Y. and Hirata T., Metallomics: Recent Analytical Techniques and Applications, Springer, 1st edn, 2017 [Google Scholar]
- Moya M., Bautista E. G., Velázquez-González A., Vázquez-Gutiérrez F., Tzintzun G., García-Arreola M. E., Castillejos M., Hernández A., Sci. Rep., 2013, 3, 1–7 10.1038/srep01284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J. D., Hu X., Ko E. J., Park S., Fernandes J., Lee Y. T., Orr M. L., Hao L., Smith M. R., Neujahr D. C., Uppal K., Kang S. M., Jones D. P. and Go Y. M., Environ. Int., 2019, 127, 720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney C. D., Kmush B., Navas-Acien A., Francesconi K., Gössler W., Schulze K., Fairweather D. L., Mehra S., Nelson K. E., Klein S. L., Li W., Ali H., Shaikh S., Merrill R. D., Wu L., West K. P., Christian P. and Labrique A. B., Environ. Res., 2015, 142, 273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassu A., Yabutani T., Mahmud Z. H., Mohammad A., Nguyen N., Huong B. T. M., Hailemariam G., Diro E., Ayele B., Wondmikun Y., Motonaka J. and Ota F., Eur. J. Clin. Nutr., 2006, 60, 580–586 [DOI] [PubMed] [Google Scholar]
- Saod W. M., Darwish N. T., Zaidan T. A. and Alfalujie A. W. A., AIP Conference Proceedings, American Institute of Physics Inc., 2018 [Google Scholar]
- Sussulini A. and Hauser-Davis R. A., in Metallomics: The Science of Biometals, ed. Arruda M. A. Z., Springer International Publishing, Switzerland, 1st edn, 2018, ch. 2, pp. 21–37 [Google Scholar]
- Ziller A., Fraissinet-Tachet L., Metallomics, 2018, 10, 1549–1559 [DOI] [PubMed] [Google Scholar]
- Rahman M. T. and Karim M. M., Biol. Trace Elem. Res., 2018, 182, 1–13 [DOI] [PubMed] [Google Scholar]
- Sussulini A., Kratzin H., Jahn O., Banzato C. E. M., Arruda M. A. Z. and Becker J. S., Anal. Chem., 2010, 82, 5859–5864 [DOI] [PubMed] [Google Scholar]
- Galazzi R. M., Chacón-Madrid K., Freitas D. C., da Costa L. F., Arruda M. A. Z., Rapid Commun. Mass Spectrom., 2020, 34, 1–14 10.1002/rcm.8726 [DOI] [PubMed] [Google Scholar]
- Bishop D. P., Hare D. J., Clases D. and Doble P. A., TrAC, Trends Anal. Chem., 2018, 104, 11–21 [Google Scholar]
- Quarles C. D., Macke M., Michalke B., Zischka H., Karst U., Sullivan P. and Field M. P., Metallomics, 20201–8 10.1039/d0mt00132e [DOI] [PubMed] [Google Scholar]
- Bishop D. P., Hare D. J., Clases D. and Doble P. A., TrAC, Trends Anal. Chem., 2018, 104, 11–21 [Google Scholar]
- Letsiou S., Nomikos T., Panagiotakos D. B., Pergantis S. A., Fragopoulou E., Pitsavos C., Stefanadis C. and Antonopoulou S., BioFactors, 2014, 40, 524–535 [DOI] [PubMed] [Google Scholar]
- Kazui Y., Ohta H., Watanabe D., Igawa T., Kasamatsu M., Suzuki Y. and Seto Y., Forensic Toxicol., 2019, 37, 424–431 [Google Scholar]
- Nguyen M. H., Pham T. D., Nguyen T. L., Vu H. A., Ta T. T., Tu M. B., Nguyen T. H. Y. and Chu D. B., J. Anal. Methods Chem., 20181–8 10.1155/2018/9462019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes R. M., Faresenius Journal Anal. Chem., 1998, 361, 246–251 [Google Scholar]
- Qu H., Mudalige T. K. and Linder S. W., Anal. Chem., 2014, 86, 11620–11627 [DOI] [PubMed] [Google Scholar]
- Aleksenko S. S., Matczuk M. and Timerbaev A. R., Electrophoresis, 2017, 38, 1661–1668 [DOI] [PubMed] [Google Scholar]
- Read S. A., Parnell G., Booth D., Douglas M. W., George J. and Ahlenstiel G., J. Viral Hepatitis, 2018, 25, 491–501 [DOI] [PubMed] [Google Scholar]
- O’connor K. S., Parnell G., Patrick E., Ahlenstiel G., Suppiah V., van der Poorten D., Read S. A., Leung R., Douglas M. W., Yang J. Y. H., Stewart G. J., Liddle C., George J. and Booth D. R., Genes Immun., 2014, 15, 88–94 [DOI] [PubMed] [Google Scholar]
- Stewart T. J., Metallomics, 2019, 11, 29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Wang J., Li H., Luo X. and Li J., Anal. Chim. Acta, 2017, 984, 66–75 [DOI] [PubMed] [Google Scholar]
- Sussulini A., Becker J. S. and Becker J. S., Mass Spectrom. Rev., 2017, 36, 47–57 [DOI] [PubMed] [Google Scholar]
- Adams F. and Barbante C., Comprehensive Anal. Chem., 2015, 69, 315–337 [Google Scholar]
- Susnea I. and Weiskirchen R., Mass Spectrom. Rev., 2016, 35, 666–686 [DOI] [PubMed] [Google Scholar]
- Konz I., Fernández B., Fernández M. L., Pereiro R., González-Iglesias H., Coca-Prados M., Sanz-Medel A., Anal. Bioanal. Chem., 2014, 406, 2343–2348 [DOI] [PubMed] [Google Scholar]
- Pozebon D., Scheffler G. L., Dressler V. L., Nunes M. A. G., J. Anal. At. Spectrom., 2014, 29, 2204–2228 [Google Scholar]
- Sahin M., Karayakar F., Koksal A. R., Yetim A., İyisoy M. S., Şen İ., Alkım H., Alkım C. and Colak T., Biol. Trace Elem. Res., 2019, 188, 245–250 [DOI] [PubMed] [Google Scholar]
- Becker J. S., Breuer U., Hsieh H. F., Osterholt T., Kumtabtim U., Wu B., Matusch A., Caruso J. A. and Qin Z., Anal. Chem., 2010, 82, 9528–9533 [DOI] [PubMed] [Google Scholar]
- Schaumlöffel D., Hutchinson R., Malherbe J., le Coustumer P., Gontier E. and Isaure M.-P., Metallomics: Analytical Techniques and Speciation Methods, ed. Mickalke B., John Wiley & Sons, New Jersey, 1st edn, 2016, ch. 4, pp. 83–116 [Google Scholar]
- Lee R. F. S., Theiner S., Meibom A., Koellensperger G., Keppler B. K. and Dyson P. J., Metallomics, 2017, 9, 365–381 [DOI] [PubMed] [Google Scholar]
- Löhr K., Traub H., Wanka A. J., Panne U. and Jakubowski N., J. Anal. At. Spectrom., 2018, 33, 1579–1587 [Google Scholar]
- Pozebon D., Scheffler G. L. and Dressler V. L., J. Anal. At. Spectrom., 2017, 32, 890–919 [Google Scholar]
- Voss M., Nunes M. A. G., Corazza G., Flores E. M. M., Müller E. I. and Dressler V. L., Talanta, 2017, 170, 488–495 [DOI] [PubMed] [Google Scholar]
- O’Reilly J., Douglas D., Braybrook J., So P. W., Vergucht E., Garrevoet J., Vekemans B., Vincze L., Goenaga-Infante H., J. Anal. At. Spectrom., 2014, 29, 1378–1384 [Google Scholar]
- Pichaandi J., Zhao G., Bouzekri A., Lu E., Ornatsky O., Baranov V., Nitz M. and Winnik M. A., Chem. Sci., 2019, 10, 2965–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cunha M. M. L., Trepout S., Messaoudi C., di Wu T., Ortega R., Guerquin-Kern J. L. and Marco S., Micron, 2016, 84, 23–36 [DOI] [PubMed] [Google Scholar]
- Steinhauser M. L., Bailey A. P., Senyo S. E., Guillermier C., Perlstein T. S., Gould A. P., Lee R. T. and Lechene C. P., Nature, 2012, 481, 516–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasulka A. L., Thamatrakoln K., Kopf S. H., Guan Y., Poulos B., Moradian A., Sweredoski M. J., Hess S., Sullivan M. B., Bidle K. D. and Orphan V. J., Environ. Microbiol., 2018, 20, 671–692 [DOI] [PubMed] [Google Scholar]
- Penen F., Malherbe J., Isaure M. P., Dobritzsch D., Bertalan I., Gontier E., le Coustumer P., Schaumlöffel D., J. Trace Elem. Med. Biol., 2016, 37, 62–68 [DOI] [PubMed] [Google Scholar]
- Chen Y., Lai B., Zhang Z. and Cohen S. M., Metallomics, 2017, 9, 250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceko M. J., Hummitzsch K., Hatzirodos N., Rodgers R. J. and Harris H. H., Metallomics, 2015, 7, 828–836 [DOI] [PubMed] [Google Scholar]
- Galbács G., Anal. Bioanal. Chem., 2017, 407, 1–26 10.1007/s00216-015-8855-3 [DOI] [PubMed] [Google Scholar]
- Busser B., Moncayo S., Coll J. L., Sancey L., Motto-Ros V., Coord. Chem. Rev., 2018, 358, 70–79 [Google Scholar]
- Motto-Ros V., Sancey L., Wang X. C., Ma Q. L., Lux F., Bai X. S., Panczer G., Tillement O. and Yu J., Spectrochim. Acta, Part B, 2013, 87, 168–174 [Google Scholar]
- Bardarov K., Buchvarov I., Yordanova T. and Georgiev P., SPIE-Intl. Soc. Optical Eng., 20197 [Google Scholar]
- Dell’Aglio M., Alrifai R., de Giacomo A., Spectrochim. Acta, Part B, 2018, 148, 105–112 [Google Scholar]
- Gondal M. A., Aldakheel R. K., Almessiere M. A., Nasr M. M., Almusairii J. A. and Gondal B., J. Pharm. Biomed. Anal., 2020, 183, 1–10 10.1016/j.jpba.2020.113153 [DOI] [PubMed] [Google Scholar]
- Kaur R., Mudgal R., Narwal M. and Tomar S., Virus Res., 2018, 256, 209–218 [DOI] [PubMed] [Google Scholar]
- Molin Y., Frisk P., Hjelm E., Blomberg J., Friman G., Ilbäck N. G., Microbes Infect., 2010, 12, 1027–1034 [DOI] [PubMed] [Google Scholar]
- Chen Y. S., Hung Y. C., Lin W. H. and Huang G. S., Nanotechnology, 2010, 21, 1–9 10.1088/0957-4484/21/19/195101 [DOI] [PubMed] [Google Scholar]
- Chandler J. D., Hu X., Ko E. J., Park S., Fernandes J., Lee Y. T., Orr M. L., Hao L., Smith M. R., Neujahr D. C., Uppal K., Kang S. M., Jones D. P. and Go Y. M., Environ. Int., 2019, 127, 720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu I. H., Chen W. H., Wu T. K. and Sun Y. C., J. Chromatogr. A, 2011, 1218, 1795–1801 [DOI] [PubMed] [Google Scholar]
- Xiao G., Chen B., He M. and Hu B., Anal. Chim. Acta, 2020, 1096, 18–25 [DOI] [PubMed] [Google Scholar]
- Xiao G., Chen B., He M., Shi K., Zhang X., Li X., Wu Q., Pang D. and Hu B., Spectrochim. Acta, Part B, 2017, 138, 90–96 [Google Scholar]
- Zhang X., Xiao G., Chen B., He M. and Hu B., Talanta, 2018, 188, 442–447 [DOI] [PubMed] [Google Scholar]
- Li X. M., Luo J., Zhang N. B. and Wei Q. L., Anal. Chim. Acta, 2015, 881, 117–123 [DOI] [PubMed] [Google Scholar]
- Friedland B. A., Hoesley C. J., Plagianos M., Hoskin E., Zhang S., Teleshova N., Alami M., Novak L., Kleinbeck K. R., Katzen L. L., Zydowsky T. M., Fernández-Romero J. A. and Creasy G. W., J. Acquired Immune Defic. Syndr., 2016, 73, 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevadiya B. D., Woldstad C., Ottemann B. M., Dash P., Sajja B. R., Lamberty B., Morsey B., Kocher T., Dutta R., Bade A. N., Liu Y., Callen S. E., Fox H. S., Byrareddy S. N., McMillan J. E. M., Bronich T. K., Edagwa B. J., Boska M. D. and Gendelman H. E., Theranostics, 2018, 8, 256–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. and Kaltashov I. A., J. Am. Soc. Mass Spectrom, 2016, 27, 2025–2032 [DOI] [PubMed] [Google Scholar]
- Akhtar A., Ghali L., Wang S. X., Bell C., Li D. and Wen X., Int. J. Mol. Sci., 2019, 20, 1–20 10.3390/ijms20092156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Yan X., Huang Y., Wen R., Li Z., Yang L., Yang C. J. and Wang Q., Anal. Chem., 2013, 85, 9428–9432 [DOI] [PubMed] [Google Scholar]
- Yadav D., Journal of Next Generation Sequencing & Applications, 2015, 2, 1–2 10.4172/2469-9853.1000e102 [DOI] [Google Scholar]
- Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E. L. L., Eddy S. R., Bateman A. and Finn R. D., Nucleic Acids Res., 2012, 40, 290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Fernández H., de S. Pessôa G., Arruda M. A. Z., Capelo-Martínez J. L., Fdez-Riverola F., Glez-Peña D., Reboiro-Jato M., J. Cheminf., 2016, 8, 1–10 10.1186/s13321-016-0178-7 [DOI] [PMC free article] [PubMed] [Google Scholar]