Abstract
Background
Measuring the seroprevalence of antibodies to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is central to understanding infection risk and fatality rates. We studied Coronavirus Disease 2019 (COVID-19)-antibody seroprevalence in a community sample drawn from Santa Clara County.
Methods
On 3 and 4 April 2020, we tested 3328 county residents for immunoglobulin G (IgG) and immunoglobulin M (IgM) antibodies to SARS-CoV-2 using a rapid lateral-flow assay (Premier Biotech). Participants were recruited using advertisements that were targeted to reach county residents that matched the county population by gender, race/ethnicity and zip code of residence. We estimate weights to match our sample to the county by zip, age, sex and race/ethnicity. We report the weighted and unweighted prevalence of antibodies to SARS-CoV-2. We adjust for test-performance characteristics by combining data from 18 independent test-kit assessments: 14 for specificity and 4 for sensitivity.
Results
The raw prevalence of antibodies in our sample was 1.5% [exact binomial 95% confidence interval (CI) 1.1–2.0%]. Test-performance specificity in our data was 99.5% (95% CI 99.2–99.7%) and sensitivity was 82.8% (95% CI 76.0–88.4%). The unweighted prevalence adjusted for test-performance characteristics was 1.2% (95% CI 0.7–1.8%). After weighting for population demographics, the prevalence was 2.8% (95% CI 1.3–4.2%), using bootstrap to estimate confidence bounds. These prevalence point estimates imply that 53 000 [95% CI 26 000 to 82 000 using weighted prevalence; 23 000 (95% CI 14 000–35 000) using unweighted prevalence] people were infected in Santa Clara County by late March—many more than the ∼1200 confirmed cases at the time.
Conclusion
The estimated prevalence of SARS-CoV-2 antibodies in Santa Clara County implies that COVID-19 was likely more widespread than indicated by the number of cases in late March, 2020. At the time, low-burden contexts such as Santa Clara County were far from herd-immunity thresholds.
Keywords: COVID-19, seroprevalence, infection fatality rate
Key Messages
Seroprevalence studies of Coronavirus Disease 2019 (COVID-19) provide estimates of the extent of infection that are more representative of true transmission than indicated by case numbers.
In late March 2020, the seroprevalence of antibodies to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Santa Clara County, California was estimated at 2.8% [95% confidence interval (CI) 1.3–4.2%] after weighting for county demographics and adjusting for test performance (1.5% unadjusted, 95% CI 1.1–2.0%).
These prevalence point estimates imply that 53 000 (95% CI 26 000 to 82 000 using weighted prevalence) people were infected in Santa Clara County by late March—many more than the ∼1200 confirmed cases at the time.
Using the estimated number of infections and the deaths in Santa Clara County at the time, we estimate a local infection fatality rate of 0.17%.
Introduction
The first two cases of Coronavirus Disease 2019 (COVID-19) in Santa Clara County, California were identified in returning travellers on 31 January and 1 February 2020, and the first COVID-19 death in the county was announced on 9 March.1 In the following month, nearly 1200 additional cases were identified in Santa Clara County, showing a pattern of rapid case increase that was reflective of community transmission. However, the case definition in Santa Clara and many other locations relies on polymerase chain reaction (PCR)-based tests that check for active infections.2 In addition, PCR-based tests were initially restricted to those with symptomatic disease. Thus, the true extent of infection remains unknown, as confirmed cases miss those with mild or no symptoms and those who have already recovered from infection.
Measuring the true extent of infection is key for epidemic projections and planning response to the epidemic. For example, early projections suggested that, in the absence of strict measures to reduce transmission, the COVID-19 pandemic could overwhelm existing hospital-bed and intensive-care-unit capacity throughout the USA and lead to >2 million deaths.3 In the absence of seroprevalence surveys, estimates of the fatality rate in these and other epidemic projections have relied on the number of confirmed cases multiplied by an estimated factor representing unknown or asymptomatic cases to arrive at the number of infections.4–7 However, the magnitude of that factor is highly uncertain and has been difficult to assess because of three independent processes that introduce measurement error: (i) cases have been diagnosed with PCR-based tests, which do not provide information about resolved infections; (ii) the majority of cases tested early in the course of the epidemic have been acutely ill and highly symptomatic, whereas most asymptomatic or mildly symptomatic individuals have not been tested; and (iii) PCR-based testing rates have been highly variable across contexts and over time, leading to inaccurate relationships between the numbers of cases and infections.
On 3 and 4 April 2020, we conducted a survey of residents of Santa Clara County to measure the seroprevalence of antibodies to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and better approximate the number of infections. To the best of our knowledge, this was the first SARS-CoV-2 seroprevalence survey conducted in the USA. At the time of this study, Santa Clara County had the largest number of confirmed cases of any county in Northern California. The county also had several of the earliest known cases of COVID-19 in the state—including one of the first presumed cases of community-acquired disease—making it an especially appropriate location for testing a population-level sample for the presence of active and past infections.
Methods
We conducted serologic testing for SARS-CoV-2 antibodies in 3328 adults and children in Santa Clara County using capillary blood draws and a lateral-flow immunoassay. In this section, we describe our sampling and recruitment approaches, specimen-collection methods, antibody-testing procedure, test-kit validation and statistical methods. Our protocol was informed by a World Health Organization protocol for population-level COVID-19 antibody testing.8 We conducted our study with the cooperation of the Santa Clara County Department of Public Health.
Study participants and sample recruitment
We recruited participants by placing targeted advertisements on Facebook aimed at residents of Santa Clara County. We used Facebook to quickly reach a large number of county residents and because it allows granular targeting by zip code and socio-demographic characteristics.9 We posted our advertisements targeting two populations: ads aimed at a representative population of the county by zip code and specially targeted ads to balance our sample for under-represented zip codes. In addition, we capped registrations from overrepresented areas after our registration slots filled up quickly with participants from wealthier zip codes. Individuals who clicked on the advertisement were directed to a survey hosted by the Stanford REDCap platform, which provided information about the study.10 The survey asked for six data elements: zip code of residence, age, sex, race/ethnicity, underlying co-morbidities and prior clinical symptoms. Over 24 hours, we registered 3285 adults, and each adult was allowed to bring one child from the same household with them (889 children registered). Additional details of the participant-selection process are provided below (and in Supplementary Data, available as Supplementary data at IJE online).
Specimen-collection and testing methods
We established drive-through test sites in three locations spaced across Santa Clara County: two county parks in Los Gatos and San Jose, and a church in Mountain View. Only individuals with a participant identification (participant ID) were allowed into the testing area. Verbal informed consent was obtained to minimize participant and staff exposure. With participants in their vehicles, sample collectors in personal protective equipment drew 50–200µL of capillary blood into an EDTA-coated microtainer. Tubes were barcoded and linked with the participant ID. Samples were couriered from the collection sites to a test-reading facility with steady lighting and climate conditions. Technicians drew whole blood up to a fill line on the manufacturer’s pipette and placed it in the test-kit well, followed by a buffer. Test kits were read 12–20 minutes after the buffer was placed. Technicians barcoded tests to match sample barcodes and documented all test results.
Test kit performance
The manufacturer’s performance characteristics were available prior to the study (using 85 confirmed positive and 371 confirmed negative samples). We conducted additional testing to assess the kit performance and continued collecting information from assessments of test performance to incorporate into the analysis. Broadly, test performance was assessed against gold-standard positive specimens from patients with PCR-confirmed COVID-19 (with or without additional confirmation of antibody presence) for sensitivity and gold-standard negative specimens from pre-COVID-era and early-COVID-era specimens. More details on the data provenance, procedures to assess test-performance characteristics and concordance between gold-standard and kit results are provided below (Supplementary Data, available as Supplementary data at IJE online).
Statistical analysis
Our estimation of the prevalence of COVID-19 proceeded in three steps. First, we report the raw frequencies of positive tests as a proportion of the final sample size. Second, we report the estimated sample prevalence, adjusted for test performance characteristics. Because SARS-CoV-2 lateral-flow antibody assays are relatively new, we gathered all available information on test performance characteristics (sensitivity and specificity), with a focus on test specificity, which can be of paramount importance when prevalence is not high. We use an estimate of test sensitivity and specificity based on pooling all information available to us. Details of each sample, including test-kit agreement numbers, specimen type and available information on data provenance, are provided in the Supplementary Data, available as Supplementary data at IJE online.
Third, we report the weighted prevalence after weighting for the zip code, sex, age (using four age categories: 0–19, 20–39, 40–69 and 70+) and race/ethnicity (non-Hispanic White, Asian, Hispanic and other) distributions of Santa Clara County (as measured in the 2018 American Community Survey). We use weights obtained through iterative proportional fitting, or raking.11 Raking generates weights by iteratively adjusting the marginal weights of each population control variable (e.g. sex) until convergence is achieved on all control variables. We match Santa Clara County demographics by zip, sex, age and race.
We use a bootstrap procedure to estimate confidence bounds for the unweighted and weighted prevalence, while accounting for sampling error and propagating the uncertainty in the sensitivity and specificity. We account for clustering of test results within families by drawing household clusters in the bootstrap samples. We use the basic percentile of the bootstrap distribution to construct confidence intervals.12 This procedure assumes that the community sample, negative control sample and positive control sample are drawn independently. More details on our bootstrap procedures are in the Supplementary Data, available as Supplementary data at IJE online.
Public involvement
Multiple stakeholders in Santa Clara County, including the department of public health, members of the board of supervisors, county parks and multiple community members, were engaged in the design and execution of the study.
Results
The test kit used in this study (Premier Biotech, Minneapolis, MN) was tested prior to field deployment. In all, we collected information on 3404 specimens from 14 sample sets used for assessing the specificity of this particular kit and on 187 specimens from 4 sample sets used for assessing sensitivity. These tests were performed by the test kit manufacturer, US and Chinese regulatory agencies, as well as independent labs. Additional details on the sample sets and the pooling approaches used to combine the estimates are provided in the Supplementary Data, available as Supplementary data at IJE online. The estimated specificity using data from all the samples was 99.53% [95% confidence interval (CI) 99.24–99.73%] and sensitivity was 85.56% (95% CI 79.69–90.26%).
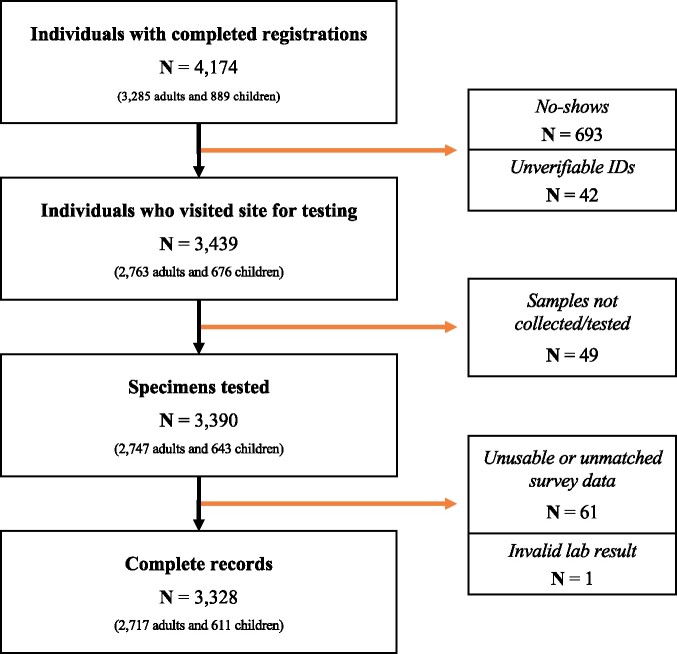
Our study included 3439 individuals who registered for the study and arrived at testing sites. We excluded observations of individuals who could not be tested (e.g. unable to obtain blood or blood clotted, N = 49), whose test results could not be used (e.g. if an incorrect participant ID was recorded onsite, N = 32), who did not reside in Santa Clara County (N = 29) and who had invalid test results (no control band, N = 1). This yielded an analytic sample of 3328 individuals with complete records including survey registration, attendance at a test site for specimen collection and lab results (Figure 1). The sample distribution meaningfully deviated from that of the Santa Clara County population along several dimensions: sex (63% in sample was female, 50% in county), race (8% of the sample was Hispanic, 26% in the county; 19% of the sample was Asian, 28% in the county) and zip distribution (Supplementary Figure 1, available as Supplementary data at IJE online). Table 1 includes demographic characteristics of our unweighted sample, the weighted sample and Santa Clara County.13Supplementary Figure 1, available as Supplementary data at IJE online, shows the geographical zip-code distribution of study participants in the county (counts and density per 1000 population).
Figure 1.
Flow diagram of participants who filled out the survey and registered, visited a site for testing and were associated with a tested specimen. We were able to associate 3328 individuals with complete survey, site and lab-result data. ‘Individuals with completed registrations’ refers to individuals who completed the initial online survey and were able to select a test site and time. ‘No-shows’ refers to participants who filled out the survey and obtained a site registration but for whom we do not have a record of attendance onsite. ‘Unverifiable IDs’ refers to records from the site data with duplicate participant identifications (IDs) for which we cannot verify which individual attended the site (this may be due to participants bringing incorrect IDs and/or technical errors in the REDCap ID assignment process). ‘Samples not collected or not tested’ includes at least 10 individuals who visited the site but did not consent to participate, as well as several children who may have decided not to have their fingers pricked after completing intake onsite. This also includes specimens that were lost before they could be tested in the lab. ‘Unusable or unmatched survey data’ includes individuals with invalid zip codes, participant IDs from the lab results that could not be matched back to the survey responses and participants who withdrew from the study. Unmatched participant IDs may be due to participants stating an incorrect participant ID at the test site or site data collectors incorrectly recording stated participant IDs. ‘Invalid lab result’ refers to one test for which the on-board control failed and the lab result could not be correctly interpreted.
Table 1.
Sample characteristics relative to Santa Clara County population estimates from the 2018 American Community Survey
| Characteristic | Sample—unweighted | Sample—weighted | County | |
|---|---|---|---|---|
| Population (N) | 3328 | 3328 | 1 943 411 | |
| Women (%) | 63.1 | 49.5 | 49.5 | |
| Men (%) | 36.9 | 50.5 | 50.5 | |
| Age (%) | 0–19 | 19.1 | 25.5 | 25.5 |
| 20–39 | 27.3 | 29.2 | 29.2 | |
| 40–69 | 51.3 | 37.1 | 37.1 | |
| ⩾70 | 2.3 | 8.3 | 8.3 | |
| Race/ethnicity (%) | Non-Hispanic White | 64.1 | 33.1 | 33.1 |
| Hispanic | 8.0 | 26.4 | 26.3 | |
| Asian | 18.7 | 27.7 | 27.8 | |
| Other | 9.2 | 12.8 | 12.8 |
Table 2 shows that the frequency of positivity in our unweighted sample was similar between men and women, was highest among Hispanic participants and ranged between 1.4% and 1.9% across ages. Positivity among those who reported recent loss of taste in the past 2 weeks (n = 59) was ∼22%.
Table 2.
Univariate frequencies of positivity along demographic and clinical features
| N in population | Portion positive, unadjusted (%; N) | |||
|---|---|---|---|---|
| Race/ethnicity | White | 2116 | 1.0 | 21 |
| Asian | 623 | 1.9 | 12 | |
| Hispanic | 266 | 4.9 | 13 | |
| Other | 306 | 1.3 | 4 | |
| Total | 3311a | 1.5 | 50 | |
| Sex | Male | 1228 | 1.5 | 19 |
| Female | 2100 | 1.5 | 31 | |
| Total | 3328 | 1.5 | 50 | |
| Age | 0–19 | 637 | 1.4 | 9 |
| 20–39 | 907 | 1.9 | 17 | |
| 40–69 | 1706 | 1.3 | 23 | |
| ⩾70 | 78 | 1.3 | 1 | |
| Symptoms in past 2 weeks | Fever | 148 | 3.4 | 5 |
| Cough | 618 | 2.6 | 16 | |
| Shortness of breath | 200 | 3.0 | 6 | |
| Runny nose | 568 | 2.1 | 12 | |
| Sore throat | 542 | 1.8 | 10 | |
| Loss of smell | 60 | 21.7 | 13 | |
| Loss of taste | 59 | 22.0 | 13 | |
| No symptoms | 2156 | 1.0 | 22 | |
| Symptoms in past 2 months | Fever | 866 | 2.0 | 17 |
| Cough | 1534 | 1.6 | 25 | |
| Shortness of breath | 542 | 2.2 | 12 | |
| Runny nose | 1329 | 1.5 | 20 | |
| Sore throat | 1397 | 1.8 | 25 | |
| Loss of smell | 188 | 11.2 | 21 | |
| Loss of taste | 187 | 10.7 | 20 | |
| No symptoms | 1029 | 0.9 | 9 | |
17 people did not indicate race.
The total number of positive cases by either IgG or IgM in our unadjusted sample was 50—a crude prevalence rate of 1.50% (exact binomial 95% CI 1.11–1.98%; Table 3). Accounting for test sensitivity and specificity and sampling error, our point estimate of the unweighted population prevalence was 1.22% (bootstrap 95% CI 0.66–1.79%). After adjusting for test performance and weighting our sample to approximate Santa Clara County demographics by zip, race, age and sex, the prevalence was 2.76% (95% CI 1.32 – 4.22%). The increase in prevalence after weighting is primarily driven by relatively higher prevalence among Hispanic participants residing in under-sampled zip codes of the county. This distribution of higher prevalence in those groups corresponds closely with the distribution of confirmed cases in Santa Clara County in late March: a disproportionate number of cases were experienced by Hispanic populations residing in the eastern portion of Santa Clara County.14
Table 3.
Prevalence estimation in Santa Clara County
| Approach | Point estimate (%) | Uncertainty (95% CI) |
|---|---|---|
| Unadjusted (%) | 50/3328 = 1.50% | 1.11–1.98% (binomial exact) |
| Adjusted for test performance (unweighted) | 1.22% | 0.66–1.79% |
| Adjusted for test performance and weighted | 2.76% | 1.32–4.22% |
We report the prevalence and uncertainty bounds of estimates from unadjusted frequency counts, estimates adjusted for test-performance characteristics and estimates adjusted for test-performance characteristics and weighted by zip code, sex, age and race/ethnicity. For adjusted prevalences, we estimate the point estimate and uncertainty using the bootstrap as described in the Methods and Supplementary Data, available as Supplementary data at IJE online.
We can use our prevalence estimates to approximate the infection fatality rate from COVID-19 in Santa Clara County. Our prevalence estimate of 2.76% applied to Santa Clara County’s population implies a little over 53 000 infections (95% CI 26 000–82 000). Since the development of antibodies takes ∼7 days from the time of infection, that estimate represents the cumulative incidence of infections in Santa Clara County up to 27 March 2020, 1 week before the first day of testing. We then examine the cumulative COVID-19-associated deaths in Santa Clara County 2, 3 (our preferred) and 4 weeks after 27 March, which allows us to estimate the range of the infection fatality rate given a lag of 2–4 weeks from infection to death.15,16 The number of people who died with COVID-19 in Santa Clara County by 11 April (2 weeks), 18 April (3 weeks) and 25 April (4 weeks) was 65, 90 and 106, respectively (Supplementary Figure 2, available as Supplementary data at IJE online, shows the time trends of cases and deaths in Santa Clara County around the time of the study).17 These estimates of deaths then correspond to an infection fatality rate of 0.12% (65/53 000), 0.17% (90/53 000; preferred) and 0.2% (106/53 000). Finally, we estimated the infection fatality rate within our four age strata (0–19, 20–39, 40–69 and 70+) using the age-specific portion of deaths in Santa Clara County and the implied number of infections from our study and the county demographics. Our data are limited to calculate with accuracy the infection fatality rate in age strata, but they suggest vast differences in fatality risk (Supplementary Table 4, available as Supplementary data at IJE online).
Discussion
After adjusting for test-performance characteristics and weighting for county demographics, we estimate that the seroprevalence of antibodies to SARS-CoV-2 in Santa Clara County in late March was 2.76%, with uncertainty bounds from 1.32% to 4.22%.
The most important implication of these findings is that, early in the pandemic, the number of infections was much greater than the reported number of cases. Using the weighted estimates, our data imply that, by 27 March (7 days prior to our survey), ∼53 000 (95% CI 26 000–82 000) people had been infected in Santa Clara County. The reported number of confirmed positive cases in the county on 27 March was 948 (~1,200 on the days of the study)—56-fold lower than the number of infections predicted by this study.14 This infection-to-case ratio, also referred to as an under-ascertainment rate, was meaningfully higher than other estimates at the time.18,19 This under-ascertainment rate is a fundamental parameter of many projection and epidemiologic models, and was used as a calibration target for understanding epidemic stage and calculating fatality rates.20,21 The under-ascertainment for COVID-19 is likely due to a combination of reliance on PCR for case identification, which misses convalescent cases; early spread in the absence of systematic testing; and asymptomatic or lightly symptomatic infections that go undetected.
The estimated infection fatality rate of 0.17% is based on the assumption that the prevalence in our study reflects the situation in Santa Clara 7 days prior to the study. If antibodies take longer to appear, or if the average duration from case identification to death is <3 weeks, then the prevalence rate at the time of the survey was higher and the infection fatality rate would be lower. On the other hand, if deaths from COVID-19 are under-reported, then the fatality rate estimates would increase. Our prevalence and fatality rate estimates can be used to update existing models, given the large upwards revision of under-ascertainment.
Whereas our weighted-prevalence estimate of 2.76% is indicative of the situation in Santa Clara County as of late March, other areas are likely to have different seroprevalence estimates based on population demographics, effective contact rates and social-distancing policies. The infection fatality rate in different locations also varies and may be substantially higher in places where the hospitals were overwhelmed (e.g. New York City or Bergamo) or where infections are concentrated among vulnerable individuals (e.g. nursing home residents).22–24 For example, in many European countries, 42–57% of deaths occurred in nursing homes and preliminary estimates for the USA are approaching the same range.25,26 infection fatality rate estimates may be substantially higher in such settings.
Our prevalence estimate also suggests that, at this time, the large majority of the population in Santa Clara County remains without IgM or IgG antibodies to SARS-CoV-2. However, repeated serologic testing in different geographies, spaced a few weeks apart, is needed to evaluate the extent of infection spread over time.
This study has several limitations. The primary limitation concerns sample selection biases. Our sample may be enriched with COVID-19 participants by selecting for individuals with a belief or curiosity concerning past infection. We discuss further and attempt to quantify the potential impact of this bias in the Supplementary Data, available as Supplementary data at IJE online (Section S4). Notably, we find that, under the most penalizing scenario of selection, the population-prevalence estimate changes from 2.76% to 2.11% (Supplementary Table 3, available as Supplementary data at IJE online). Our study may also have selected for groups of people more likely to skew our sample against COVID-19 participants. For example, our sample strategy selected for members of Santa Clara County with ready access to Facebook who viewed our advertisement early after the registration opened. Our sample ended up with an over-representation of White women between the ages of 40 and 70 years, and an under-representation of Hispanic and Asian populations, relative to our community. Those imbalances were partly addressed by weighting our sample by zip code, race, age and sex to match the county. Our survey also selected for members of the population who were able to spare the time to drive to the testing site, which may have skewed our sample against essential workers. Our study was also limited in that it could not ascertain the representativeness of SARS-CoV-2 antibodies in populations with possibly high prevalence, such as homeless populations and those in nursing homes. The overall direction and magnitude of these selection effects are hard to fully bound, and our estimates reflect the prevalence in our sample, weighted to match county demographics.
Another potential limitation is that, with relatively low prevalence estimates, the precision of seroprevalence estimates depends on the performance of the serology tests for SARS-CoV-2 antibodies. Our adjusted results depend on the current estimates of specificity and sensitivity of these tests. We estimated the specificity and sensitivity based on pooled estimates from several independent assessments that use combined IgG and IgM to identify positive-antibody presence. Lower test specificity or greater uncertainty in the test performance could, in principle, meaningfully change the study’s conclusions. The performance of the test kit used in this study has been studied extensively in comparison to other SARS-CoV-2-antibody tests, including for use with capillary blood and in comparison with tests that require venipuncture (Supplementary Data Section 2, available as Supplementary data at IJE online).
Our infection fatality rate could be biased downward if the number of COVID-19 deaths in the county has been substantially undercounted. This has some plausibility given that some early COVID-19 deaths may have gone undiagnosed due to limited testing. However, many deaths early in the spring of 2020 that were suspected of being due to COVID-19 were retrospectively evaluated, and those in which COVID-19 was implicated in the death were added to the official tally, reducing the effect of undercounting. We note that, even if deaths are 50% higher than the recorded deaths, this would still imply an infection fatality rate of 0.18–0.30%.
Over 100 teams worldwide have tested population samples for SARS-CoV-2 antibodies, with findings consistent with a large under-ascertainment of SARS-CoV-2 infections. Our study was one of the earliest to be done and the large under-ascertainment was partly driven by the limited testing in the early phases of the pandemic. Large under-ascertainment (up to several hundred-fold) was seen in other early surveys, whereas the extent of under-ascertainment decreased as more testing was being performed in most locations.27,28 An early serosurvey in Los Angeles County, California on 10–11 April estimates a seroprevalence of 4.65%.29 Our data from Santa Clara County suggest that the spread of the infection is similar to other moderately affected areas such as Los Angeles in early spring. Documented infections remained low in Santa Clara County until early summer. Santa Clara was part of a large seroprevalence survey among dialysis patients in the USA.30 In that study, conducted in the first week of July 2020, the estimated seroprevalence in Santa Clara County was 4.1%, and this corresponded to an infection fatality rate of ∼0.2%, similar to our estimate.
We conclude that, based on seroprevalence sampling of a large regional population, the best estimates for the prevalence of SARS-CoV-2 antibodies in Santa Clara County were 2.76% by late March (95% CI 1.32–4.22%), with large variability by race/ethnicity. This prevalence is far smaller than the theoretical final size of the epidemic31 and suggests that, in late March, a large majority of the population did not have IgG or IgM antibodies to the virus. Our study offers valuable data for an early period of the pandemic, when PCR testing was limited, and these data can be used as a baseline for comparison against subsequent seroprevalence studies to estimate the evolution of both seroprevalence and infection fatality rates over time. Our prevalence and infection fatality rate estimates do not advocate for or refute the usefulness of any non-pharmaceutical interventions. Instead, these new data reduce uncertainty around the size of the population that has been infected. This allows better monitoring of interventions and reducing uncertainty about the state of the epidemic also may carry intrinsic public benefits. It is important to note that the under-ascertainment of infections in Santa Clara may change over time (e.g. depending on greater availability of testing) and the under-ascertainment rate may be different in other locations. Improved test accuracy and larger sample sizes with random sampling can further reduce uncertainty in estimates. Our work demonstrates the feasibility of seroprevalence surveys of population samples to inform our understanding of this pandemic’s progression, project estimates of community vulnerability and monitor infection fatality rates in different populations over time.
Supplementary data
Supplementary data are available at IJE online.
Author contributions
E.B., N.S. and J.B. conceived of the project and were involved in every aspect of the project; B.M., S.S. and J.T. were involved in protocol development and in every aspect of the survey execution; J.P.A.I. conceptualized the data collection and analysis; R.B.D., C.L., Z.W., R.S.W. and A.B. were site directors and led critical components of the data collection and interpretation; T.K. and D.E. were instrumental in assessing, procuring and fielding the test kits; R.G. led recruitment efforts and was involved in data analysis. All authors were critically involved in interpretation of the data, drafting and revising of the manuscript.
Funding
This work was supported by gift support from the Stanford COVID-19 Seroprevalence Studies Fund. The funders had no role in the design and conduct of the study, nor in the decision to prepare and submit the manuscript for publication.
Supplementary Material
Acknowledgements
The authors acknowledge the support of the Santa Clara County Department of Public Health, the Santa Clara County community, all study participants and the many volunteers and staff without whom this study could not have been accomplished in the midst of the COVID-19 crisis. We would also like to acknowledge the following individuals for critical comments and contributions that improved the data and the paper: David Allison, Manisha Desai, Liran Einav, Julia Gross, Emilia Ling, Tom MaCurdy, Charles McCulloch, Ben Moran, Barry Nalebuff, Richard Olshen, Ken Shotts and Frank Wolak. The Institutional Review Board at Stanford University approved the study prior to recruitment (Protocol # IRB-55702). Sharing of the primary data is restricted under a human-subjects-protection agreement. Sharing of de-identified data may be available upon request and review by the authors.
Conflict of interest
None declared. Of note, test kits were purchased from Premier Biotech for this study, and none of the authors has a relationship to the test manufacturer (beyond purchasing the tests).
References
- 1. Novel Coronavirus Press Archives—Public Health Department—County of Santa Clara. https://www.sccgov.org/sites/phd/DiseaseInformation/novel-coronavirus/Pages/archives.aspx (12 April 2020, date last accessed).
- 2. Santa Clara County Public Health: COVID-19 Information for Healthcare Providers. https://www.sccgov.org/sites/phd-p/Diseases/novel-coronavirus/Pages/Responsibilities-and-Guidance.aspx (23 November 2020, date last accessed).
- 3.Report 12—The Global Impact of COVID-19 and Strategies for Mitigation and Suppression. Imperial College London. http://www.imperial.ac.uk/medicine/departments/school-public-health/infectious-disease-epidemiology/mrc-global-infectious-disease-analysis/covid-19/report-12-global-impact-covid-19/ (7 April 2020, date last accessed).
- 4. Spychalski P, Błażyńska-Spychalska A, Kobiela J.. Estimating case fatality rates of COVID-19. Lancet Infect Dis 2020:774–775. https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(20)30246-2/abstract (9 April 2020, date last accessed ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Onder G, Rezza G, Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA2020;323:1775–1776. https://jamanetwork.com/journals/jama/fullarticle/2763667 (9 April 2020, date last accessed). [DOI] [PubMed] [Google Scholar]
- 6. Li L-Q, Huang T, Wang Y-Q. et al. COVID-19 patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. J Med Virol 2020;92:1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu JT, Leung K, Bushman M. et al. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med 2020;26:506–510. https://www.nature.com/articles/s41591-020-0822-7 (8 April 2020, date last accessed). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Population-Based Age-Stratified Seroepidemiological Investigation Protocol for COVID-19 Virus Infection. https://www.who.int/publications-detail/population-based-age-stratified-seroepidemiological-investigation-protocol-for-covid-19-virus-infection (8 April 2020, date last accessed).
- 9. Shaver LG, Khawer A, Yi Y. et al. Using Facebook advertising to recruit representative samples: feasibility assessment of a cross-sectional survey. J Med Internet Res 2019;21:e14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.REDCap: Resources operated by Stanford Research IT. https://med.stanford.edu/researchit/resources.html#citations (7 April 2020, date last accessed).
- 11. Deville J-C, Särndal C-E, Sautory O.. Generalized raking procedures in survey sampling. J Am Stat Assoc 1993;88:1013–20. [Google Scholar]
- 12. Efron B. Bootstrap methods: another look at the jackknife. Ann Stat 1979;7:1–26. [Google Scholar]
- 13. Bureau UC. American Community Survey (ACS). The United States Census Bureau. https://www.census.gov/programs-surveys/acs (10 April 2020, date last accessed).
- 14.County of Santa Clara Public Health Department—Coronavirus (COVID-19) Data Dashboard. https://www.sccgov.org/sites/phd/DiseaseInformation/novel-coronavirus/Pages/dashboard.aspx#cases (11 April 2020, date last accessed).
- 15. Bhatraju PK, Ghassemieh BJ, Nichols M. et al. Covid-19 in critically ill patients in the Seattle Region—case series. N Engl J Med 2020;382:2012–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weiss P, Murdoch DR.. Clinical course and mortality risk of severe COVID-19. Lancet 2020;395:1014–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coronavirus (COVID-19) Data Dashboard—Novel Coronavirus (COVID-19)—County of Santa Clara. https://www.sccgov.org/sites/covid19/Pages/dashboard.aspx#cases (20 April 2020, date last accessed).
- 18. Li R, Pei S, Chen B. et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2). Science2020;368:489–493. https://science.sciencemag.org/content/early/2020/03/24/science.abb3221 (10 April 2020, date last accessed). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salomon JA. Defining high-value information for COVID-19 decision-making. medRxiv2020;2020.04.06.20052506. http://medrxiv.org/content/early/2020/04/08/2020.04.06.20052506.abstract (11 February 2021, date last accessed).
- 20. Verity R, Okell LC, Dorigatti I. et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. The Lancet Infectious Diseases 2020;20:669–677. https://www.medrxiv.org/content/10.1101/2020.03.09.20033357v1 (10 April 2020, date last accessed). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. IHME | COVID-19 Projections. Institute for Health Metrics and Evaluation. https://covid19.healthdata.org/ (10 April 2020, date last accessed).
- 22. Reifer J, Hayum N, Heszkel B, Klagsbald I, Streva VA.. SARS-CoV-2 IgG antibody responses in New York City. Diagnostic Microbiology and Infectious Disease 2020;98:115–128. https://www.medrxiv.org/content/10.1101/2020.05.23.20111427v1 (20 June 2020, date last accessed). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rosenberg ES, Tesoriero JM, Rosenthal EM. et al. Cumulative incidence and diagnosis of SARS-CoV-2 infection in New York. Annals of Epidemiology2020;48:23–29. https://www.medrxiv.org/content/10.1101/2020.05.25.20113050v1 https://www.medrxiv.org/content/10.1101/2020.05.23.20111427v1 (05 June 2020, date last accessed). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abrams HR, Loomer L, Gandhi A, Grabowski DC.. Characteristics of U.S. nursing homes with COVID-19 cases. J Am Geriatr Soc 2020;68:1653–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Half of coronavirus deaths happen in care homes, data from EU suggests. The Guardian. 2020. https://www.theguardian.com/world/2020/apr/13/half-of-coronavirus-deaths-happen-in-care-homes-data-from-eu-suggests (23 April 2020, date last accessed).
- 26. Girvan G. 42% of COVID-19 Deaths in Nursing Homes & Assisted Living Facilities. 2020. https://freopp.org/the-covid-19-nursing-home-crisis-by-the-numbers-3a47433c3f70 (17 June 2020, date last accessed).
- 27. Ioannidis J. The infection fatality rate of COVID-19 inferred from seroprevalence data. Bull World Health Organization. https://www.who.int/bulletin/online_first/BLT.20.265892.pdf?ua=1 (11 February 2021, date last accessed). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ioannidis JPA. Global perspective of COVID-19 epidemiology for a full-cycle pandemic. Eur J Clin Invest 2020;50: 1–9. https://onlinelibrary.wiley.com/doi/abs/10.1111/eci.13423 (11 February 2021, date last accessed). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sood N, Simon P, Ebner P. et al. Seroprevalence of SARS-CoV-2–specific antibodies among adults in Los Angeles County, California, on April 10-11, 2020. JAMA 2020;323:2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anand S, Montez-Rath M, Han J. et al. Prevalence of SARS-CoV-2 antibodies in a large nationwide sample of patients on dialysis in the USA: a cross-sectional study. Lancet 2020;396:1335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Coronavirus (COVID-19): Press Conference with Marc Lipsitch, 03/04/20. News. 2020. https://www.hsph.harvard.edu/news/features/coronavirus-covid-19-press-conference-with-marc-lipsitch-03-04-20/ (10 April 2020, date last accessed).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.