Abstract
The human genome has an almost equal distribution of unique and transposable genetic elements. Although at the transcriptome level, a relatively higher contribution from transposable elements derived RNA has been reported. This is further highlighted with evidence from pervasive transcription. Of the total RNA, noncoding RNAs (ncRNAs) are significant contributors to the transcriptome pool with sizeable fraction from repetitive elements of the human genome, inclusive of Long Interspersed Nuclear Elements (LINEs) and Short Interspersed Nuclear Elements (SINEs). ncRNAs are increasingly being implicated in diverse functional roles especially during conditions of stress. These stress responses are driven through diverse mediators, inclusive of long and short ncRNAs. ncRNAs such as MALAT1, GAS5, miR-204 and miR-199a-5p have been functionally involved during oxidative stress, endoplasmic reticulum (ER) stress and unfolded protein response (UPR). Also, within SINEs, Alu RNAs derived from primate-specific Alu repeats with ~11% human genome contribution, playing a significant role. Pathogenic diseases, including the recent COVID-19, leads to differential regulation of ncRNAs. Although, limited evidence suggests the need for an inquest into the role of ncRNAs in determining the host response towards pathogen challenge.
Keywords: ncRNAs, infections, stress response, unfolded protein response (UPR), ER stress, SARS-CoV-2
Introduction
Until recently, a great proportion of RNA not coding for any protein (~98%) was referred to as ‘junk’. This ncRNA is a new ‘treasure trove’ for their involvement in various gene regulations and disease pathogenesis. ncRNAs are classified into two broad categories with a base number cut-off of 200 nucleotides (nts) [1]. ncRNAs below 200 nts are termed as small noncoding RNAs (e.g. miRNA, piRNA, snRNA, rRNA, tRNA, snoRNA), while ncRNAs with more than 200 nts are termed as long ncRNAs (lncRNAs). lncRNAs are further classified as long intergenic ncRNA (lincRNA), natural antisense transcript, bidirectional lncRNA and intronic lncRNA [2, 3].
After RNA polymerase II mediated transcription, lncRNAs undergo specific post-transcriptional modifications similar to mRNA like 5′-capping, 3′-polyadenylation and intracellular transport [4]. Emerging experimental pieces of evidence highlight the gene regulatory activity of ncRNAs at both transcriptional and translational levels like splicing, epigenetic modulations, transcription and translation [2]. A few ncRNAs contain a short open reading frame and are associated with ribosomes, indicating their possible additional role in mRNA regulation [5, 6]. The cellular interactions of ncRNAs are mainly mediated by base pairing, secondary structures and transcriptional regulations [7]. The transcriptome wide distribution and functional classification of ncRNAs are depicted in Figure 1.
Figure 1.
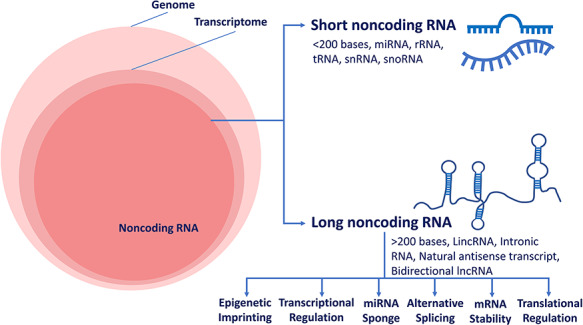
Transcriptome wide distribution and functional classification of ncRNAs. The left panel shows Venn diagram with components that constitute the human genome. The right panel shows different classes of ncRNAs.
Next-generation sequencing (NGS) enabled evidence highlighted the functional role of ncRNAs in several disease pathologies, infections, inflammation and stress [8–14]. ncRNAs are dysregulated in many disease pathologies, either as a secondary effect of disrupted mRNA expression or they may act as an active driver of the pathogenesis [7]. Augmented understanding of the ncRNA functions might facilitate molecular insight into the inflammation, stress and immune regulation, especially host–pathogen interaction. This may help identify putative therapeutic interventions towards infectious diseases. Oxidative stress, endoplasmic reticulum (ER) stress, unfolded protein response (UPR), immune action and inflammation are the major hallmarks of infection. This review aims to bring together the facets of above-mentioned signs of infection and highlights the role of ncRNAs during pathogen-induced stress response.
Oxidative stress response during infection
Normal cellular aerobic metabolism produces reactive oxygen species (ROS) as a by-product in multiple organelles such as mitochondria, ER and peroxisome. ROS are oxygen-containing reactive chemicals like hydrogenperoxide (H2O2), superoxide (O2•-) and hydroxyl radical (•OH). These are subsequently scavenged by antioxidant enzymes to maintain the redox homeostasis [15]. Generally, ROS are involved in cellular signal transduction, cytokine regulation, immunomodulation, regulation of transcription and apoptosis. An imbalance between ROS generation and scavenging disrupts the redox homeostasis, leading to the accumulation of ROS in cells, and oxidative stress. This damages tissue, protein, lipid and DNA, followed by activation of inflammatory signalling pathway and eventually, cell death [16, 17].
Role of ncRNAs during infection-induced oxidative stress
Since the first report in 1979 [18], several studies have reported viral infection induced oxidative stress, and cellular damage, including members of Flaviviridae genus such as Zika virus [19, 20], Dengue virus (DENV) [21, 22], Hepacivirus [23, 24] and West Nile virus [25, 26]. DENV induces NOX-mediated oxidative stress, activates interferon regulatory factor 3 (IRF3), interferon regulatory factor 7 (IRF7), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), signal transducer and activator of transcription 1 (STAT1) causing cell damage, particularly in dendritic cells [21]. DENV also induces CCL5 and CCAAT/enhancer-binding protein β (C/EBPβ) leading to hepatocellular oxidative stress [27]. Highly pathogenic strains of Influenza A virus (IAV) are also reported to induce oxidative stress. The H3N2 and HKx31 strains of IAV induce both cellular and mitochondrial ROS, causing cellular damage and lung injury [28–30], whereas PR8 strain induces NOX2 NADPH oxidase-mediated ROS in the endosome which modulate IAV pathogenesis [31]. Reduction of oxidative stress was found to decrease inflammation and facilitate disease recovery in both the cases [30, 31].
In human immunodeficiency virus (HIV) infected individuals, increase in oxidative stress, with an elevated level of ROS production, oxidized nucleotide (8-oxo-G), lipid peroxidation and associated formation of malondialdehyde (MDA) in plasma were detected [32–35]. Other studies also highlighted reduced total antioxidant activity [36], decreased reduced glutathione/oxidized glutathione (GSH/GSSG) ratio [37] in lung epithelial fluid and decreased GSH content in blood during HIV infection [33–35]. The majority of the Hepatitis C virus (HCV) proteins (E1, E2, NS3, NS4B, NS5A) induce oxidative stress in hepatocytes, hepatic stellate cells and peripheral blood mononuclear cells (PBMC) [38–42]. Localization of HCV core proteinsin ER and mitochondria affects Ca2+ efflux from ER to mitochondria. This causes oxidation of GSH and increased mitochondrial ROS [43]. The association of oxidative stress during infection seems to be important as several studies highlight that antioxidant treatment leads to better infection management [31, 44, 45].
Two independent research groups reported the involvement of miR-155 in oxidative stress during infection. Wang et al. [46] reported the role of miR-155 during Mycobacterium infection, while Yang et al. [47] highlighted the same in acute kidney injury during sepsis. In the latter case, the study also showed that antagonizing the miR-155 ameliorated the liver injury by decreasing the oxidative stress [47]. Another study highlighted the involvement of miR-4321 and miR-4270 in NADPH oxidase 5 (NOX5)-mediated oxidative stress in sepsis-induced acute kidney injury [48].
Role of ncRNAs during disease-induced oxidative stress
Several ncRNAs are associated with oxidative stress during different diseases. In some cases, ncRNAs were reported to regulate oxidative stress response, while their expression is also modulated by oxidative stress. Expression of miRNAs during oxidative stress has been observed in Parkinson’s disease [49], Alzheimer’s disease [50], Ageing [51], Cancer [52], Diabetes [53] and Osteoarthritis [54]. Tsai et al. [55] reviewed the role of ncRNAs in systemic lupus erythematosus (SLE)-mediated oxidative stress with functional role of miR-21, miR-29b, miR-126b and miR-146a. In other diseases, miR-500a-5p and miR-144 have been shown to regulate oxidative stress during breast cancer and anaemia, respectively [56, 57]. The role of ncRNAs during oxidative stress in different disease conditions is collated in Table 1.
Table 1.
ncRNAs involved in stress and immune response
| ncRNAs | Cellular stress response | Immune response | References |
|---|---|---|---|
| MALAT1 | MALAT1 regulates generation of ROS | MALAT1 induce IL-6, IL-1β, TNF-α, CXCL8 and acts as sponge of miR-146a | [62–64] |
| NEAT1 | Modulates superoxide in LPS-treated rat mesangial cells | Promotes activation of NLRP3, NLRC4 & AIM2 inflammasome to activate IL-1β and pyroptosis | [65, 66] |
| HULC | Upregulated by oxidative stress; regulate CCA cell migration and invasion by targeting IL-6 and CXCR4 via sponging miR-372/miR-373 | Regulates expression of proinflammatory VCAM1, ICAM1, Il-6 TNF-α & IL-8 during LPS-induced sepsis | [67–69] |
| XIST | Upregulated by oxidative stress; induces endothelial to mesenchymal transition (EMT) & promotes oxidative stress-induced Osteosarcoma cell invasion & migration | IL-6 is increased with XIST expression in CFA/LPS induced female mice | [70, 71] |
| H19 | Upregulated in oxidative stress; regulate CCA cell migration & invasion by targeting IL-6 & CXCR4 via sponging let-7a/let-7b | H19 elevate levels of TNF-α, IL-6, IL-8 & IL-1β mediated by NF-κB induced signalling cascade in H. pylori infected cells. H19 overexpression negatively regulates miR-874 and decreases proinflammatory cytokines in sepsis | [67, 72, 73] |
| GAS5 | Decreases the level of ROS & MDA; increases the level of SOD | GAS5 is associated with KLF2 regulation & NF-κB inhibition in sepsis | [74, 75] |
| UCA1 | Downregulate UCA1 reduces oxidative stress; inflammation in Parkinson’s through the inhibition of the PI3K/Akt signalling pathway | Regulates expression of proinflammatory VCAM1, ICAM1, Il-6 TNF-α & IL-8 during LPS-induced sepsis. UCA1 increase expression of MEF2C to inhibit NF-κB and proinflammatory cytokines | [68, 76, 77] |
| HOTAIR | HOTAIR overexpression prevented ischemia–reperfusion-induced oxidative stress by activating AMP-activated protein kinase alpha (AMPKα) | Regulates activation of NF-κB & its target IL-6, iNOS by degrading IκBα. HOTAIR upregulation negatively regulates miR-211 and upregulates IL-1β, IL-6R, IL16 & TNF-α in monocytes | [78–80] |
| FOXD3-AS1 | Regulates oxidative stress-induced lung epithelial cell death through negative regulation of miR-150 | FOXD3-AS1 negatively regulates IL-3 & IL-13 in CD4+ T cells in allergic rhinitis | [81, 82] |
Alu RNA, a member of SINE repeat family, is also involved in the stress response during infection. Although a small number of Alu RNAs (103–104 molecules/cell) are present in normal cells, the number increases drastically to nearly 20-fold during stress response in cancer and viral infections [58–60]. Accumulation of Alu RNA in endothelial cells promotes the production of ROS and IL-1β. This is mediated by the NF-κB signalling pathway with lower expression of endothelial nitric oxide synthase (eNOS) and superoxide dismutase 2 (SOD2) [61]. A focussed elucidation of the functional role of ncRNAs in regulating the oxidative stress response holds potential.
ER stress and UPR during infection
To establish a favourable environment for viral replication, some of the positive-strand RNA viruses (HCV, SARS-CoV and DENV) modulate viral protein driven rearrangement of host intracellular organelle membranes like ER [83, 84]. This modified organelle is called Double Membrane Vesicle (DMV), where structural and non-structural viral proteins accumulate and alter the protein and lipid content of the ER [85–87]. After processing at ER, the nascent replicated viral genome is trafficked through the Golgi complex and secretory pathway and thereafter leaves the infected cell after maturation [88]. This membrane remodelling induces ER stress by triggering a cellular signalling pathway, UPR. This pathway aims to maintain cellular homeostasis and ER protein levels. If homeostasis is not achieved, the cell progresses towards apoptosis.
Mechanism of UPR
The UPR is mediated by three transmembrane proteins which sense the viral and misfolded proteins. These sensors are PKR-like ER Protein Kinase (PERK), Inositol-Requiring Protein 1 (IRE1) and Activating Transcription Factor 6 (ATF6) [89]. Viral proteins distract the regulatory element Binding immunoglobulin Protein (BiP) from binding to UPR sensors. IRE1 activation by dimerization and phosphorylation aids in splicing of the X-box binding protein 1 (XBP1) mRNA followed by the subsequent translation of spliced XBP1 transcription factor, XBP1s [90]. This splicing enables the expression of chaperones and proteins essential for the ER-Associated Degradation (ERAD) pathway. ERAD leads to the production of proinflammatory cytokines [91], lipid synthesis [92], autophagy response [93] and regulatory protein P58IPK [94] (Figure 2). Activated IRE1 also initiates mRNA degradation to decrease the protein load on the ER by Regulated IRE1-Dependent Degradation (RIDD) pathway [95]. Moreover, IRE1 also recruits Tumor Necrosis Factor (TNF) receptor associated factor 2 (TRAF2) to the ER membrane to phosphorylate IκB and promote NF-κB activation [96]. Phosphorylated IRE1 activates c-Jun N terminal kinase (JNK) signalling pathway to maintain the balance between apoptosis and survival.
Figure 2.
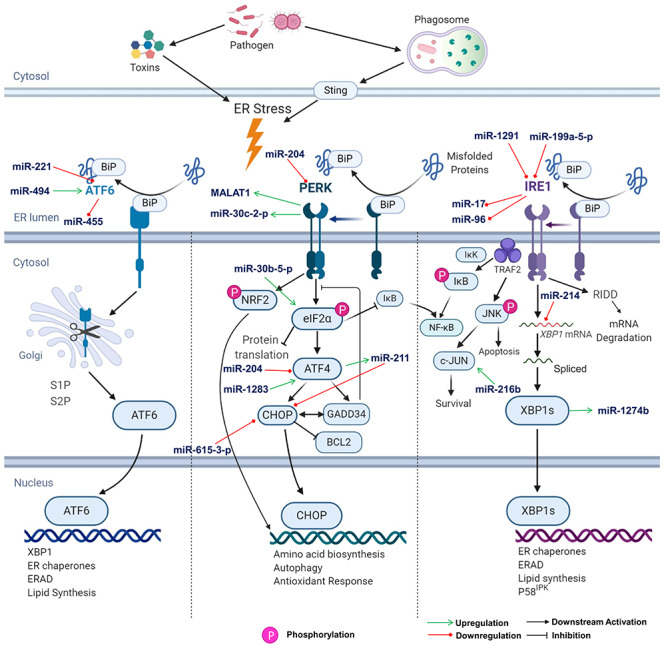
Unfolded protein response pathway. The signalling flow in a UPR pathway is shown schematically. From left to right, the panels represent ATF6, PERK and IRE1 axis of the UPR, respectively. Role of ncRNA at different stages are highlighted with arrows. (eIF2α- Eukaryotic transcription Initiation Factor 2 α, S1P- Site 1 protease, S2P- Site 2 protease, BCL2- B Cell Lymphoma 2). The acronyms for rest of the genes are included in the manuscript.
PERK is activated similarly to IRE1. Activated PERK attenuates the global protein production by phosphorylating the α-subunit of translation factor, eIF2α. Simultaneously, activated PERK induces the expression of ATF4 [97] through an alternative translation cascade. This, in turn, induces expression of transcription factor CCATT/Enhancer-Binding Protein-Homologous Protein (CHOP) and Growth Arrest and DNA Damage Inducible Protein 34 (GADD34). CHOP is a pro-apoptotic protein, which inhibits anti-apoptotic protein B cell lymphoma 2 (BCL2) [98]. GADD34 is a phosphatase that inhibits the phosphorylation of eIF2α [99]. Additionally, PERK phosphorylates NF-E2 related factor 2 (NRF2) and induces the genes involved in antioxidant response (Figure 2) [100].
Following ER stress, ATF6 translocates to the Golgi apparatus and undergoes a two-step proteolytic cleavage, with the release of transcriptionally active N-terminal domain. The truncated ATF6 then translocates to the nucleus, where it transactivates the ER stress response element (ERSE) harbouring the UPR genes such as BiP, GRP94 and P58IPK (Figure 2) [101].
UPR during infections
Several studies have reported as to how the DNA and RNA viruses modulate the UPR pathway selectively for their replication and to suppress apoptosis until viral replication is complete [102, 103]. Some pathogens directly interact with the ER functioning, causing ER stress and UPR activation [104]. Duodenal biopsy of HIV patients and liver cells of HCV patients show activated ER stress response [105, 106]. Along with overexpression of the GRP94 in severe acute respiratory syndrome coronavirus (SARS-CoV) infected cells, significant phosphorylation of PKR and PERK has also been reported [107]. Zika virus and Rhinovirus, a major cause of cystic fibrosis, were also found to induce the ER stress response and impaired UPR [108, 109]. The UPR activation by IAV [110–112] and respiratory syncytial virus (RSV) infection [113] in A549 and primary HTBE cell lines have also been reported.
UPR can also be activated by bacteria, such as facultative intracellular bacteria, Brucella melitensis and Listeria monocytogenes [114, 115], as well as bacterial toxins such as pore forming toxin, cholera toxin and shiga toxins.
Role of ncRNAs during UPR
Several examples of ncRNA interactions in the UPR pathway are known; however, they are mostly reported in diseases like diabetes, cancer and cardiovascular disease with little evidence during infectious diseases. In diabetic pancreatic β-cells, miR-204 controls the PERK activity [116]. PERK is also known to induce the expression of miR-483, which disrupts cellular ATP homeostasis during UPR [117]. In diabetic progenitor cells, miR-200 and miR-466 are degraded by RIDD activity of IRE1, resulting in abnormal angiogenesis [118]. Few miRNAs have also been reported to be involved in the IRE1-XBP1 axis, which in turn also regulate miRNAs through XBP1 and IRE1 [119]. In ATF6 axis, calreticulin, a Ca+ binding protein, is regulated by miR-455. Besides, ncRNA-RB1, which shares a bidirectional promoter region with RB1 gene, is involved in the regulation of calreticulin. CHOP and IRE1 were reported to regulate miR-216b level [120]. lncRNA-GADD7 is induced in palmitate-induced ER stress and is a general regulator of oxidative stress [121]. In hepatocellular carcinoma, lincRNA-p21 acts as a tumour suppressor. The lncRNA TUG1 was reported to reduce apoptosis through ER stress reduction in cold-induced liver injury [122]. Expression of lncRNA MALAT1 was increased in flavivirus infection through PERK-dependent transcriptional activity [123].
McMahon et al. [119] summarized the involvement of different miRNAs during UPR. Quan et al. [124] explored transcriptome-wide changes in ncRNA expression during UPR to identify potential regulators of UPR genes. Although several studies highlight the role of ncRNAs during the UPR in different diseases, there is scope for mechanistic elucidation.
Immune response to infection
In the event of pathogen invasion, the host cell quickly recognizes the pathogen-associated molecular pattern (PAMP). This happens through pattern-recognizing receptor (PRR) located on the host cell membrane (TLR-1,2, 4, 5 and 6), endosomal membrane (TLR 3, TLR7/8 and TLR 9) or cytoplasm (retinoic acid inducible gene 1-like receptor- RLR and NOD like receptor- NLR). TLR-1, 2, 4, 5 and 6 detect bacterial pathogenic components of lipopolysaccharide (LPS) and flagellin. Cytoplasmic and endosomal TLR 3, TLR 7/8 and TLR 9 detect viral nucleic acid components of dsRNA, ssRNA and viral DNA, respectively. Upon detection of the PAMP, PRR activates NF-κB, activator protein 1 (AP1) and IRF. This induces inflammatory and immune response via the expression of several proinflammatory cytokines, chemokines and interferons. These interferons bind with respective receptors and activate JAK/STAT signalling. The phosphorylated STAT translocates to the nucleus and is involved in the ISGF3 transcription complex (STAT1, STAT2, IRF9) formation. This initiates the expression of IFN-Stimulated Genes (ISG) to mediate antiviral responses [125].
Natural killer cells kill the infected cells by perforin and granzymes. Dendritic cell and macrophage phagocytize and neutralize the pathogens through lysosomal degradation by enzymes and ROS and RNS production [126]. The degraded pathogen-derived particles are presented by macrophages and dendritic cells (antigen presentation) and, in turn, initiate the immune response. Excessive production of the ROS triggers ER stress and impairs cellular function such as antigen presentation and successive T-cell activation [127]. A detailed representation of the immune responses upon pathogen infection is depicted in the Figure 3.
Figure 3.
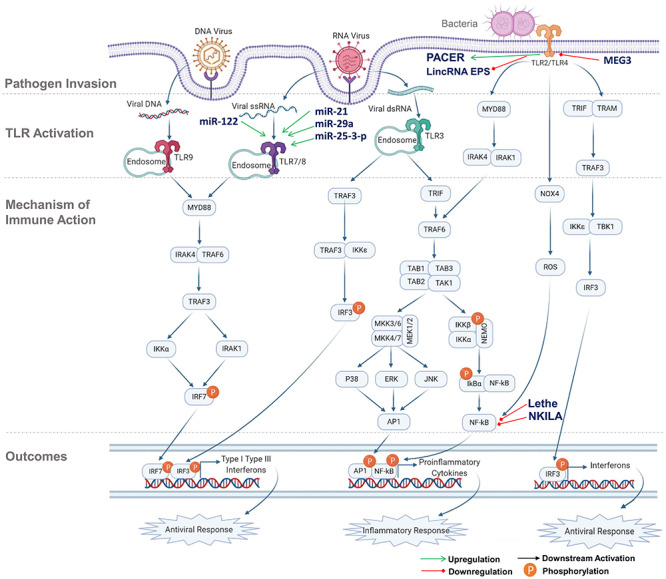
Immune response activation pathway. From top to bottom order, the panels show the pathogen invasion, TLR activation, mechanism of immune action and the outcomes, respectively. ncRNA regulations at different stages are highlighted. (MYD88- Myeloid Differentiation Primary Response 88, TRIF- Toll/Interleukin-1 Receptor) Domain-Containing Adaptor Protein, TRAM- Translocating Chain-Associated Membrane, IRAK- Interleukin-1 Receptor (IL-1R) Associated Kinase, TAB- TGF-β-Activated Kinase 1 and MAP3K7-Binding Protein, TAK (also known as MAP3K7, Mitogen-Activated Protein Kinase Kinase Kinase 7), MEK (also known as MAP2K, Mitogen-Activated Protein Kinase Kinase), NEMO- NF-κB Essential Modulator, ERK- Extracellular-signal-Regulated Kinase). The acronyms for rest of the genes are included in the manuscript.
Differential expression of ncRNAs during infection
Differential expression of ncRNAs has been reported to modulate the PRR-mediated signalling cascade, cytokine-mediated inflammatory pathway and interferon-mediated antiviral pathways [128]. For example, in HCV-infected patient’s liver, lncRNA-CMPK2 is upregulated. The knockdown of lncRNA-CMPK2 leads to the reduction of HCV replication and upregulation of IFN response in human primary hepatocytes [129]. Another study reported increased levels of lncRNA-EGOT, mediated by NF-κB, in HCV and influenza infection, although the mechanism and antiviral response is not elucidated [130]. lncRNA-GAS5 overexpression is also reported in HCV infection and GAS5 alleviates HCV replication by binding to its NS3 protein [131].
lncRNA-Negative Regulator of AntiViral response (NRAV) is reported to be associated with viral infections like IAV, RSV and Herpes Simplex virus. Downregulation of NRAV in infected cells enhances the ISG expression as a response to viral infection [132]. This might be mediated by lncRNA-NRAV associated protein ZONAB [133]. Nuclear Factor of Activated T cell (NFAT), an important modulator of T cell mediated immune response, plays a major role in HIV infection by increasing viral transcription and replication. lncRNA-Noncoding Repressor Of NFAT (NRON) acts as a protein-RNA scaffold and binds to NFAT and regulates its nuclear trafficking. Moreover, lncRNA-NRON downregulation by HIV protein, Nef increases the NFAT activity to facilitate viral protein synthesis and replication [134]. lncRNA Lethe functions as a decoy and interacts with the RelA subunit of NF-κB to negatively regulate NF-κB signalling. Additionally, lncRNA-Lethe overexpression suppresses the NF-κB mediated interleukin transcription [135]. NF-κB interacting lncRNA (lncRNA-NKILA), a post-translational regulator of NF-κB activity, negatively regulates NF-κB by blocking the IκB phosphorylation and subsequent degradation [136]. lncRNA NKILA and lncRNA-Lethe both act as negative regulators of NF-κB modulating the immune response. lncDC exclusively expressed in human dendritic cells is crucial for antigen uptake and T cell activation. lncDA binds to STAT3 and prevents its dephosphorylation and mediates transcription of genes essential for dendritic cell differentiation [137].
miRNAs can also modulate the expression of lncRNAs. miR-140 specifically binds with lncRNA NEAT1 and modulates expression of lncRNA NEAT1in adipocytes [138]. Multiple studies reported NEAT1 and miRNA-140 upregulation during both bacterial and viral infections [66, 139–141]. However, the mechanism towards miR-140 mediated regulation of NEAT1 is yet to be explored. Some of the lncRNAs act as competitive endogenous RNA (ceRNA) to sponge miRNA activity. lncRNA NRAV was reported to sponge miR-509-3p and promote the Rab5c-mediated respiratory syncytial virus replication [132]. RSV infection causes overexpression of NRAV, thereby facilitating viral replication. However, the same group reported the host antiviral response downregulated the NRAV expression to curb the viral replication. lncRNA PVT1 acts as ceRNA and adsorbs miR-203 to promote upregulation of E2F3 in RSV infection [142]. Several studies have also reported Alu RNA mediated induction of NF-κB in a TLR-independent manner activated NLRP3 inflammasome in retinal pigmented epithelial cells [143, 144]. ncRNA thus seems to be an important regulator of immune responses during infection.
Cytokines involved in inflammation
As discussed in the earlier section, upon pathogen recognition by PRRs and Toll-like receptor (TLRs), proinflammatory cytokines like IL-6, IL-1β and TNF-α are induced. This facilitates inflammatory response to clear infection by TNF-α, and ROS-mediated phagocytosis in macrophages [145]. TNF-α and IL-6 promote the acute phase response and upregulate the C reactive protein (CRP) in the liver. This, in turn, activates the complement system leading to opsonization of the pathogen. IL-1β exhibits prostaglandin-mediated immune response and activates ROS, NO and iNOS [146, 147]. In the case of Mycobacterium tuberculosis (MTB) infection, the regulation of IL-1 and type 1 interferon leads to a distinct inflammatory response [148]. Increased expression of IL-10, a signature of RSV-infected lower respiratory tract cells, attenuates acute inflammatory response and late inflammatory changes [149]. Similarly, in RSV-infected children, IFN-ɣ, IL-8, TNF-α and other eosinophil activated chemokines (GM-CSF and Eotaxin) were increased in nasal lavage fluid [150]. In IAV H1N1 infections with acute respiratory distress syndrome (ARDS), higher level of IL-1RA, IL-6, IL-8, TNF-α, Interferon gamma induced Protein 10 (IP-10) and monocyte chemoattractant protein 1 (MCP-1) were reported [151, 152].
Like other cytopathic viruses, SARS-CoV-2 induces pyroptosis, a severe inflammatory cell death cascade [153, 154]. This includes IL-1β, IFN-ɣ, IL-6, IP-10, MCP-1, CXCL3, CXCL9 and CXCL10 [155–157]. In HIV-infected patients, the levels of CRP and proinflammatory cytokines like TNF-α, IL-6, B cell Activating Factor (BAFF) are increased irrespective of antiretroviral therapy [158]. HCV core and NS3-mediated increase in TNF-α and IL-10 have been reported in dendritic cells isolated from the blood of infected patients [159]. In HCV-infected liver cells, Nod-Like Receptor P3 inflammasome (NLRP3) stimulates IL-1β production and promotesthe production of other proinflammatory cytokines [160]. The degree of inflammation and tissue damage in HIV and HCV co-infected patients was found to be greater than the two individual infections [161]. In some cases, the robust local and systemic cytokine production, inflammatory infiltration and virus-induced tissue destruction lead to a condition called ‘cytokine storm’, observed in cases of IAV, SARS-CoV-2, RSV and dengue [162–165].
ncRNAs in regulation of cytokine production
Expressions of many ncRNAs are altered during inflammatory response with a functional role. lincRNA-COX2 and PACER are upregulated in macrophages as a result of TLR4 stimulation by LPS [128, 135, 166], while, lincRNA-EPS is downregulated in LPS-stimulated TLR4 signalling [8]. lncRNA THRIL acts as a scaffold to hnRNPL and binds to TNF-α promoter inducing its transcription upon TLR2 stimulation [167]. lncRNA PACER, a decoy for binding and sequestering NF-κB p50 homodimer, regulates a key anti-inflammatory gene PTGS2 [166]. It is also reported that TLR7 signalling mediation by miR-122 activates acute pulmonary inflammation in mice [168]. Secretion of proinflammatory cytokines by macrophages is activated by miRNAs, miR-21, miR-29a and miR-25-3p through TLR8 signalling cascade [169, 170].
lncRNA ANRIL is positively associated with the expression of proinflammatory cytokines like IL-4, IL-6, IL-17, IL-13 and TNF-α in the nasal mucosa during allergic rhinitis [82]. MALAT1 has also been shown to increase the expression of IL-1β, IL-6, ICAM1, TNF-α and E-selectin in pulmonary microvascular endothelial cells [171]. lncRNA-Mirt2 acts as a checkpoint for rogue inflammatory response and prevents ubiquitinoylation of TRAF6. This inhibits the NF-κB mediated production of proinflammatory cytokines and acts as a feedback regulator of the inflammatory response [172]. lncRNA PMS2L2 was also shown to act as a negative regulator of inflammation associated miR-203 with possible involvement in LPS-induced inflammatory injury [173]. lncRNA SNHG16, by binding with miR-15a/16, modulates the TLR4-mediated signalling cascade [174]. Another lncRNA MEG3 protects the respiratory epithelial cell in RSV infection by suppressing TLR4-dependent NF-κB signalling axis [175]. Several studies reported the involvement of many other ncRNAs during inflammatory response (Table 1).
Cross-talk between ER stress, oxidative stress and immune response
All the three branches of UPR were shown to induce proinflammatory transcriptional programs, mainly governed by NF-κB and AP1 [176]. Keestra-Gounder et al. [177] reported Nucleotide-binding Oligomerization Domain (NOD) mediated (NOD1 and NOD2) increase in IL6 production, upon activation of IRE1 axis of UPR. RIDD has been associated with translational activation of RIG-1, thereby inducing NF-κB associated immune response against RNA viruses [178]. The cross-talk between ER stress, oxidative stress and immune response in the event of pathogen infection has been summarized in the Figure 4.
Figure 4.
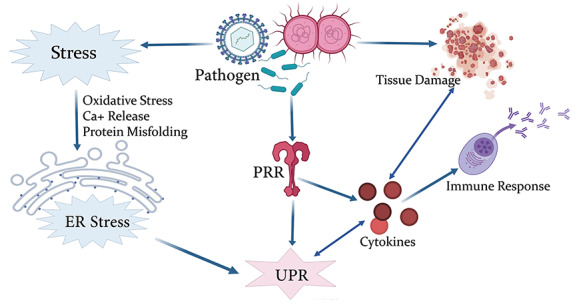
Crosstalk between ER stress, UPR and immune response. Graphical representation of the host response during pathogen challenge.
ncRNA and COVID-19
The SARS-CoV-2 virus is a positive-sense single-stranded RNA virus of the β-coronavirus family which caused the Coronavirus disease-2019 (COVID-19). SARS-CoV-2, in a similar fashion to SARS-CoV, infects the host cells by binding of angiotensin-converting enzyme 2 (ACE2) receptor and viral S protein fusion primed by transmembrane serine protease 2 (TMPRSS2) [179]. ACE2 converts angiotensin 2 to angiotensin 1 leading to ATP production and NADPH Oxidase 4 (NOX4) mediated ROS production [180, 181]. The other members of the Coronavirus family are reported to form an ER-derived DMV. SARS-CoV’s ORF-7a, ORF-8a and ORF-9b are localized in host mitochondria. Of this, ORF-7a and ORF-8a facilitate the viral replication [182, 183], while ORF-9b suppresses the innate immunity by modulating the TRAF3/TRAF6 axis [184]. These ORFs share high sequence homology with the corresponding ORFs of SARS-CoV-2. It is plausible that SARS-CoV-2 also forms an ER-derived DMV to facilitate viral replication. Besides, a machine learning model of the SARS-CoV-2 genome revealed the possibility of host-mitochondrial localization of the virus, which may lead to the formation of mitochondria-derived DMVs for the replication of the virus [185]. Although this mitochondrial localization of viral RNA/protein is proposed to be mediated by direct interaction of viral Nsp with a mitochondrial import receptor Tomm70, there is no experimental evidence. Codo et al. [186] reported elevated levels of mitochondrial ROS in SARS-CoV-2 infected cells. This was shown to stabilize the HIF-1α, which in turn promotes glycolysis and excessive production of proinflammatory cytokines. The combined effect of the two events leads to decreasedviral load in cells.
The analysis of SARS-CoV infection associated mRNA expression revealed a quintuple ceRNA network involving miR-124-3p, lncRNA-Gm16917, circRNA-Ppp1r10, circRNA-C330019G07Rik, Ddx58 mRNA and transcription factor STAT2 [157]. Viral infection triggers IFN-mediated overexpression of Ddx58 containing the helicase domain which is crucial for viral replication. The Ddx58 is also a regulator of splicing during miRNA biogenesis. Thus, the viral helicase association with Ddx58 raises the possibility of altering miRNA biogenesis with viral replication advantage. 3’UTR of the Ddx58 mRNA has a binding site for the miR-124-3p which has an impact on mRNA stability. lncRNA-G16917 acts as a sponge to miR-124-3p with possible role in viral replication. A similar ceRNA network is plausible in response to SARS-CoV-2 infection. Using RNA-seq data of mock and SARS-CoV-2 infected normal human bronchial epithelial (NBHE) cells as well as lung biopsies of healthy and SARS-CoV-2 infected patients, Vishnubalaji et al. [187] analysed differentially expressed genes and ncRNAs. They reported 155 upregulated and 195 downregulated ncRNAs in SARS-CoV-2 infected NBHE cells. Comparing the data with differentially expressed ncRNAs in COVID-19 patients’ lung biopsies, they identified five common upregulated and 57 common downregulated ncRNAs. RCC2-AS1, AC011603.3, AC145207.2, SNHG25 and AC008760.2 are the five common upregulated ncRNAs. MALAT1 and NEAT1 were slightly upregulated in SARS-CoV-2 infected NBHE cells as compared to mock. However, they were not found to be overexpressed in the lung tissues from COVID-19 patients.
HIF-1α is a key player involved in the regulation of glucose metabolism and translation of glycolysis associated genes. A possible mechanism for ncRNAs to influence the SARS-CoV-2 infection is through regulation of HIF-1α. Experimental pieces of evidence suggest the involvement of ncRNAs in the regulation of HIF-1α. The miR-130a-3p overexpression represses glycolysis by targeting the HIF-1α. HOTAIR acts as a decoy for miR-130a-3p and regulates the HIF-1αin hepatocellular carcinoma [188]. LincRNA-p21 regulates HIF-1α degradation in a hypoxic condition. Overexpression of lncRNA-Let suppresses HIF-1α expression in hepatocellular and breast cancer [189]. It would be interesting to explore the role of the differentially expressed ncRNAs in the host response to infections like SARS-CoV-2.
Discussion
There are multiple pieces of evidence that link expression of miRNAs and ncRNAs in oxidative and ER stress in a wide range of diseases. However, the involvement of ncRNAs and miRNAs in regulating the UPR during viral infection has limited evidence. Upon entering the cell, many positive-strand RNA viruses form DMV for the viral replication. The formation of DMV is dependent on various viral proteins as well as host protein. However, the involvement of ncRNAs in regulation of the host proteins involved in DMV formation during viral infection needs further investigation. Moreover, the exact mechanism of regulation of antiviral response by ncRNA is yet to be explored. ncRNAs may also be potentially targeted to alleviate stress and possibly aid in disease recovery.
In addition to the involvement in different stress responses, immune and inflammatory responses, ncRNAs are also being investigated as biomarkers and therapeutic targets in different diseases. The association of ncRNAs is well characterized in cancer. Multiple studies have also established the potential of ncRNAs as biomarkers and therapeutic targets [190, 191]. Both lncRNAs and miRNAs are detectable in body fluids, including saliva, blood, serum, urine, and thus offer a comparatively easier detection of diseases. For example urine-based detection of lncRNA-PCA3, a prostate cancer specific lncRNA, offers easier and more sensitive diagnosis than the conventional antigen-based test [192]. Similarly, MALAT1 is also detectable in urine for prostate cancer [193]. Urinary detection of UCA1 in transitional cell carcinoma, and salivary detection of HOTAIR in oral squamous cell carcinoma offers potential diagnosis methods [194, 195]. lncRNA UC001NCR and AX800134, of viral origin, are promising biomarkers in HBV-induced hepatocellular carcinoma [196]. ncRNAs are also explored as potential therapeutic targets. miRNAs can be targeted for therapeutic use by small molecules, and oligonucleotide-based approaches [191]. miR-122, which facilitates the HCV infection, is being targeted by specific inhibitor Miravirsen and is undergoing clinical trial [168]. Small molecules such as Enoxacin have been shown to regulate the miRNA expression levels in tumours [197]. miRNA expression level can also be altered by miRNA-mimics (miR-28-5p, miR-125-3p) and antagomirs (antagomir-122) [191, 198].
Although similar to other RNA-based diagnosis, the spatiotemporal expression of ncRNAs in diseases poses a challenge for identification as a biomarker. The same is true for its usage as prognostic marker. However, a deeper insight into the intricacies of ncRNA-based regulation of infection and its outcomes may provide potential diagnostic and therapeutic options in infectious diseases.
Key Points
Noncoding RNA (ncRNA) regulates the oxidative stress, induced during infections as well as diseases like Diabetes, Parkinson’s, Alzheimer’s and Cancer.
Bacterial toxins, viral proteins, infection-induced ER stress and subsequent unfolded protein response.
Differential expression of ncRNAs regulate pattern recognition receptors (PRR) mediated activation of immune response, cytokine signalling and antiviral response.
In vitro and in silico studies indicate the involvement of ncRNAs in modulating cytokine storm and viral replication during the SARS-CoV-2 infection.
Funding
Council of Scientific and Industrial Research (CSIR) Grant Number MLP-2005. Fondation Botnar Grant Number CLP-0031. Indo-US Science and Technology Forum (IUSSTF) Grant Number CLP-0033.
Conflict of Interest
Authors wish to declare no conflict of interest. Funding body did not have any role in planning of the study.
Acknowledgements
The authors acknowledge the funding from CSIR (MLP-2005), Fondation Botnar (CLP-0031) and IUSSTF (CLP-0033) towards the study. P.C. acknowledges Council of Scientific and Industrial Research (CSIR) for providing Junior Research Fellowship. The authors also acknowledge the help of Dr. Sachin Sharma, CSIR-Institute of Genomics and Integrative Biology (CSIR-IGIB) towards proofreading the manuscript.
Partha Chattopadhyay: My PhD research at CSIR-IGIB is focused on elucidating the role of ncRNA in modulating responses to pathogen challenge and developing model systems to study the underlying mechanism.
Janani Srinivasa Vasudevan: I am interested to understand the host–pathogen interaction from the Genomics perspective. This includes knowing the genome features of the pathogen as well as the host response.
Rajesh Pandey: Lab is working towards INtegrative GENomics of HOst-PathogEn (INGEN-HOPE). Elucidation of the role of ncRNA during host stress response leading to physiological homeostasis is one of the central themes.
References
- 1. Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of long noncoding rnas. Annu Rev Immunol 2017;35:177–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murphy MB, Medvedev AE. Long noncoding RNAs as regulators of toll-like receptor signaling and innate immunity. J Leukoc Biol 2016;99:839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013;154:26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011;147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bazzini AA, Johnstone TG, Christiano R, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 2014;33:981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boon RA, Jaé N, Holdt L, Dimmeler S. Long noncoding rnas: from clinical genetics to therapeutic targets? J Am Coll Cardiol 2016;67:1214–1226. [DOI] [PubMed] [Google Scholar]
- 8. Atianand MK, Hu W, Satpathy AT, et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 2016;165:1672–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou Q, Huang XR, Yu J, et al. Long noncoding RNA Arid2-IR is a novel therapeutic target for renal inflammation. Mol Ther 2015;23:1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saayman S, Ackley A, Turner A-MW, et al. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther 2014;22:1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mandlik A, Livny J, Robins WP, et al. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 2011;10:165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng X-W, Chen Z-F, Wan Y-F, et al. Long non-coding RNA H19 suppression protects the endothelium against Hyperglycemic-induced inflammation via inhibiting expression of miR-29b target gene vascular endothelial growth factor a through activation of the protein kinase B/endothelial nitric oxide synthase pathway. Front Cell Dev Biol 2019;7:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li L, Dai H, Nguyen A-P, et al. Influenza A virus utilizes non-canonical cap-snatching to diversify its mRNA/ncRNA. RNA 2020;26:1170–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Y, Xu L, Lu B, et al. LncRNA H19/microRNA-675/PPARα axis regulates liver cell injury and energy metabolism remodelling induced by hepatitis B X protein via Akt/mTOR signalling. Mol Immunol 2019;116:18–28. [DOI] [PubMed] [Google Scholar]
- 15. Roy J, Galano J-M, Durand T, et al. Physiological role of reactive oxygen species as promoters of natural defenses. FASEB J 2017;31:3729–3745. [DOI] [PubMed] [Google Scholar]
- 16. Pizzino G, Irrera N, Cucinotta M, et al. Oxidative stress: harms and benefits for human health. Oxid Med Cell Longev 2017;2017:8416763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dandekar A, Mendez R, Zhang K. Cross talk between ER stress, oxidative stress, and inflammation in health and disease. Methods Mol Biol 2015;1292:205–214. [DOI] [PubMed] [Google Scholar]
- 18. Peterhans E. Sendai virus stimulates chemiluminescence in mouse spleen cells. Biochem Biophys Res Commun 1979;91:383–392. [DOI] [PubMed] [Google Scholar]
- 19. Li G, Poulsen M, Fenyvuesvolgyi C, et al. Characterization of cytopathic factors through genome-wide analysis of the Zika viral proteins in fission yeast. Proc Natl Acad Sci U S A 2017;114:E376–E385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Amorim R, Temzi A, Griffin BD, et al. Zika virus inhibits eIF2α-dependent stress granule assembly. PLoS Negl Trop Dis 2017;11:e0005775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olagnier D, Peri S, Steel C, et al. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog 2014;10:e1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Al-Alimi AA, Ali SA, Al-Hassan FM, et al. Dengue virus type 2 (DENV2)-induced oxidative responses in monocytes from glucose-6-phosphate dehydrogenase (G6PD)-deficient and G6PD normal subjects. PLoS Negl Trop Dis 2014;8:e2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Medvedev R, Ploen D, Hildt E. HCV and oxidative stress: implications for HCV life cycle and HCV-associated pathogenesis. Oxid Med Cell Longev 2016;2016:9012580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rebbani K, Tsukiyama-Kohara K. HCV-induced oxidative stress: battlefield-winning strategy. Oxid Med Cell Longev 2016;2016:7425628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gullberg RC, Jordan Steel J, Moon SL, et al. Oxidative stress influences positive strand RNA virus genome synthesis and capping. Virology 2015;475:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Basu M, Courtney SC, Brinton MA. Arsenite-induced stress granule formation is inhibited by elevated levels of reduced glutathione in West Nile virus-infected cells. PLoS Pathog 2017;13:e1006240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin YL, Liu CC, Chuang JI, et al. Involvement of oxidative stress, NF-IL-6, and RANTES expression in dengue-2-virus-infected human liver cells. Virology 2000;276:114–126. [DOI] [PubMed] [Google Scholar]
- 28. Bernard K, Hecker L, Luckhardt TR, et al. NADPH oxidases in lung health and disease. Antioxid Redox Signal 2014;20:2838–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Imai Y, Kuba K, Neely GG, et al. Identification of oxidative stress and toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell 2008;133:235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. To EE, Erlich JR, Liong F, et al. Mitochondrial reactive oxygen species contribute to pathological inflammation during Influenza A virus infection in mice. Antioxid Redox Signal 2020;32:929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. To EE, Luong R, Diao J, et al. Novel endosomal NOX2 oxidase inhibitor ameliorates pandemic Influenza A virus-induced lung inflammation in mice. Respirology 2019;24:1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wanchu A, Rana SV, Pallikkuth S, et al. Short communication: oxidative stress in HIV-infected individuals: a cross-sectional study. AIDS Res Hum Retroviruses 2009;25:1307–1311. [DOI] [PubMed] [Google Scholar]
- 33. Watanabe LM, Barbosa Júnior F, Jordão AA, et al. Influence of HIV infection and the use of antiretroviral therapy on selenium and selenomethionine concentrations and antioxidant protection. Nutrition 2016;32:1238–1242. [DOI] [PubMed] [Google Scholar]
- 34. Aukrust P, Luna L, Ueland T, et al. Impaired base excision repair and accumulation of oxidative base lesions in CD4+ T cells of HIV-infected patients. Blood 2005;105:4730–4735. [DOI] [PubMed] [Google Scholar]
- 35. Awodele O, Olayemi SO, Nwite JA, et al. Investigation of the levels of oxidative stress parameters in HIV and HIV-TB co-infected patients. J Infect Dev Ctries 2011;6:79–85. [DOI] [PubMed] [Google Scholar]
- 36. Teto G, Kanmogne GD, Torimiro JN, et al. Lipid peroxidation and total cholesterol in HAART-naïve patients infected with circulating recombinant forms of human immunodeficiency virus type-1 in Cameroon. PLoS One 2013;8:e65126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bindoli A, Rigobello MP. Principles in redox signaling: from chemistry to functional significance. Antioxid Redox Signal 2013;18:1557–1593. [DOI] [PubMed] [Google Scholar]
- 38. Jansons J, Sominskaya I, Petrakova N, et al. The immunogenicity in mice of HCV core delivered as DNA is modulated by its capacity to induce oxidative stress and oxidative stress response. Cells 2019;8:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bureau C, Bernad J, Chaouche N, et al. Nonstructural 3 protein of hepatitis C virus triggers an oxidative burst in human monocytes via activation of NADPH oxidase. J Biol Chem 2001;276:23077–23083. [DOI] [PubMed] [Google Scholar]
- 40. Smirnova OA, Ivanova ON, Bartosch B, et al. Hepatitis C virus NS5A protein triggers oxidative stress by inducing NADPH oxidases 1 and 4 and cytochrome P450 2E1. Oxid Med Cell Longev 2016;2016:8341937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ming-Ju H, Yih-Shou H, Tzy-Yen C, et al. Hepatitis C virus E2 protein induce reactive oxygen species (ROS)-related fibrogenesis in the HSC-T6 hepatic stellate cell line. J Cell Biochem 2011;112:233–243. [DOI] [PubMed] [Google Scholar]
- 42. Li S, Ye L, Yu X, et al. Hepatitis C virus NS4B induces unfolded protein response and endoplasmic reticulum overload response-dependent NF-kappaB activation. Virology 2009;391:257–264. [DOI] [PubMed] [Google Scholar]
- 43. Korenaga M, Wang T, Li Y, et al. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J Biol Chem 2005;280:37481–37488. [DOI] [PubMed] [Google Scholar]
- 44. Liu T, Castro S, Brasier AR, et al. Reactive oxygen species mediate virus-induced STAT activation: role of tyrosine phosphatases. J Biol Chem 2004;279:2461–2469. [DOI] [PubMed] [Google Scholar]
- 45. Hosakote YM, Liu T, Castro SM, et al. Respiratory syncytial virus induces oxidative stress by modulating antioxidant enzymes. Am J Respir Cell Mol Biol 2009;41:348–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang J, Wu M, Wen J, et al. MicroRNA-155 induction by Mycobacterium bovis BCG enhances ROS production through targeting SHIP1. Mol Immunol 2014;62:29–36. [DOI] [PubMed] [Google Scholar]
- 47. Yang Z-B, Chen W-W, Chen H-P, et al. MiR-155 aggravated septic liver injury by oxidative stress-mediated ER stress and mitochondrial dysfunction via targeting Nrf-2. Exp Mol Pathol 2018;105:387–394. [DOI] [PubMed] [Google Scholar]
- 48. Ge Q-M, Huang C-M, Zhu X-Y, et al. Differentially expressed miRNAs in sepsis-induced acute kidney injury target oxidative stress and mitochondrial dysfunction pathways. PLoS One 2017;12:e0173292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prasad KN. Oxidative stress, pro-inflammatory cytokines, and antioxidants regulate expression levels of MicroRNAs in Parkinson’s disease. Curr Aging Sci 2017;10:177–184. [DOI] [PubMed] [Google Scholar]
- 50. Prasad KN. Oxidative stress and pro-inflammatory cytokines may act as one of the signals for regulating microRNAs expression in Alzheimer’s disease. Mech Ageing Dev 2017;162:63–71. [DOI] [PubMed] [Google Scholar]
- 51. Guillaumet-Adkins A, Yañez Y, Peris-Diaz MD, et al. Epigenetics and oxidative stress in aging. Oxid Med Cell Longev 2017;2017:9175806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lan J, Huang Z, Han J, et al. Redox regulation of microRNAs in cancer. Cancer Lett 2018;418:250–259. [DOI] [PubMed] [Google Scholar]
- 53. Zhang W, Xu W, Feng Y, et al. Non-coding RNA involvement in the pathogenesis of diabetic cardiomyopathy. J Cell Mol Med 2019;23:5859–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cheleschi S, De Palma A, Pascarelli NA, et al. Could oxidative stress regulate the expression of MicroRNA-146a and MicroRNA-34a in human osteoarthritic chondrocyte cultures? Int J Mol Sci 2017;18:2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tsai C-Y, Hsieh S-C, Lu C-S, et al. Cross-talk between mitochondrial dysfunction-provoked oxidative stress and aberrant noncoding RNA expression in the pathogenesis and pathophysiology of SLE. Int J Mol Sci 2019;20:5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Degli Esposti D, Aushev VN, Lee E, et al. miR-500a-5p regulates oxidative stress response genes in breast cancer and predicts cancer survival. Sci Rep 2017;7:15966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sangokoya C, Telen MJ, Chi J-T. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood 2010;116:4338–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jang KL, Collins MK, Latchman DS. The human immunodeficiency virus tat protein increases the transcription of human Alu repeated sequences by increasing the activity of the cellular transcription factor TFIIIC. J Acquir Immune Defic Syndr 1992;5:1142–1147. [PubMed] [Google Scholar]
- 59. Panning B, Smiley JR. Activation of RNA polymerase III transcription of human Alu elements by herpes simplex virus. Virology 1994;202:408–417. [DOI] [PubMed] [Google Scholar]
- 60. Zheng Y, Hlady RA, Joyce BT, et al. DNA methylation of individual repetitive elements in hepatitis C virus infection-induced hepatocellular carcinoma. Clin Epigenetics 2019;11:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang W, Wang W-H, Azadzoi KM, et al. Alu RNA accumulation in hyperglycemia augments oxidative stress and impairs eNOS and SOD2 expression in endothelial cells. Mol Cell Endocrinol 2016;426:91–100. [DOI] [PubMed] [Google Scholar]
- 62. Chen J, Ke S, Zhong L, et al. Long noncoding RNA MALAT1 regulates generation of reactive oxygen species and the insulin responses in male mice. Biochem Pharmacol 2018;152:94–103. [DOI] [PubMed] [Google Scholar]
- 63. Huang M, Wang H, Hu X, et al. lncRNA MALAT1 binds chromatin remodeling subunit BRG1 to epigenetically promote inflammation-related hepatocellular carcinoma progression. OncoImmunology 2019;8:e1518628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dai L, Zhang G, Cheng Z, et al. Knockdown of LncRNA MALAT1 contributes to the suppression of inflammatory responses by up-regulating miR-146a in LPS-induced acute lung injury. Connect Tissue Res 2018;59:581–592. [DOI] [PubMed] [Google Scholar]
- 65. Chen Y, Qiu J, Chen B, et al. Long non-coding RNA NEAT1 plays an important role in sepsis-induced acute kidney injury by targeting miR-204 and modulating the NF-κB pathway. Int Immunopharmacol 2018;59:252–260. [DOI] [PubMed] [Google Scholar]
- 66. Zhang P, Cao L, Zhou R, et al. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun 2019;10:1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang W-T, Ye H, Wei P-P, et al. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol 2016;9:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen Y, Fu Y, Song Y-F, et al. Increased expression of lncRNA UCA1 and HULC is required for pro-inflammatory response during LPS induced sepsis in endothelial cells. Front Physiol 2019;10:608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen X, Song D. LPS promotes the progression of sepsis by activation of lncRNA HULC/miR-204-5p/TRPM7 network in HUVECs. Biosci Rep 2020;40:BSR20200740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wen J-F, Jiang Y-Q, Li C, et al. LncRNA-XIST promotes the oxidative stress-induced migration, invasion, and epithelial-to-mesenchymal transition of osteosarcoma cancer cells through miR-153-SNAI1 axis. Cell Biol Int 2020;44:1991–2001. [DOI] [PubMed] [Google Scholar]
- 71. Shenoda BB, Ramanathan S, Gupta R, et al. Xist attenuates acute inflammatory response by female cells. Cell Mol Life Sci 2020. doi: 10.1007/s00018-020-03500-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang Y, Yan J, Li C, et al. LncRNA H19 induced by helicobacter pylori infection promotes gastric cancer cell growth via enhancing NF-κB-induced inflammation. J Inflamm 2019;16:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fang Y, Hu J, Wang Z, et al. LncRNA H19 functions as an aquaporin 1 competitive endogenous RNA to regulate microRNA-874 expression in LPS sepsis. Biomed Pharmacother 2018;105:1183–1191. [DOI] [PubMed] [Google Scholar]
- 74. Xie C, Wu W, Tang A, et al. lncRNA GAS5/miR-452-5p reduces oxidative stress and Pyroptosis of high-glucose-stimulated renal tubular cells. Diabetes Metab Syndr Obes 2019;12:2609–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Li F, Sun J, Huang S, et al. LncRNA GAS5 overexpression reverses LPS-induced inflammatory injury and apoptosis through up-regulating KLF2 expression in ATDC5 chondrocytes. Cell Physiol Biochem 2018;45:1241–1251. [DOI] [PubMed] [Google Scholar]
- 76. Cai L, Tu L, Li T, et al. Downregulation of lncRNA UCA1 ameliorates the damage of dopaminergic neurons, reduces oxidative stress and inflammation in Parkinson’s disease through the inhibition of the PI3K/Akt signaling pathway. Int Immunopharmacol 2019;75:105734. [DOI] [PubMed] [Google Scholar]
- 77. Yu Q, Zhao M-W, Yang P. LncRNA UCA1 suppresses the inflammation via modulating miR-203-mediated regulation of MEF2C/NF-κB Signaling pathway in epilepsy. Neurochem Res 2020;45:783–795. [DOI] [PubMed] [Google Scholar]
- 78. Meng K, Jiao J, Zhu R-R, et al. The long noncoding RNA Hotair regulates oxidative stress and cardiac Myocyte apoptosis during ischemia-reperfusion injury. Oxid Med Cell Longev 2020;2020:1645249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Obaid M, Udden SMN, Deb P, et al. LncRNA HOTAIR regulates lipopolysaccharide-induced cytokine expression and inflammatory response in macrophages. Sci Rep 2018;8:15670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chen J, Gu X, Zhou L, et al. Long non-coding RNA-HOTAIR promotes the progression of sepsis by acting as a sponge of miR-211 to induce IL-6R expression. Exp Ther Med 2019;18:3959–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen X, Hu J, Pan Y, et al. Novel noncoding RNAs biomarkers in acute respiratory distress syndrome. Expert Rev Respir Med 2020;14:299–306. [DOI] [PubMed] [Google Scholar]
- 82. Zhang H, Zhu X, Liu X, et al. Long non-coding RNA FOXD3-AS1 regulates the expression and secretion of IL-25 in nasal epithelial cells to inhibit Th2 type immunoreaction in allergic rhinitis. Mol Cell Biochem 2020;473:239–246. [DOI] [PubMed] [Google Scholar]
- 83. Paul D, Bartenschlager R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol 2013;2:32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wolff G, Melia CE, Snijder EJ, et al. Double-membrane vesicles as platforms for viral replication. Trends Microbiol 2020;28:1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Welsch S, Miller S, Romero-Brey I, et al. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 2009;5:365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Martín-Acebes MA, Blázquez A-B, Jiménez de Oya N, et al. West Nile virus replication requires fatty acid synthesis but is independent on phosphatidylinositol-4-phosphate lipids. PLoS One 2011;6:e24970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Junjhon J, Pennington JG, Edwards TJ, et al. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J Virol 2014;88:4687–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol 2005;3:13–22. [DOI] [PubMed] [Google Scholar]
- 89. Bertolotti A, Zhang Y, Hendershot LM, et al. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2000;2:326–332. [DOI] [PubMed] [Google Scholar]
- 90. Yoshida H, Matsui T, Yamamoto A, et al. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001;107:881–891. [DOI] [PubMed] [Google Scholar]
- 91. Martinon F, Chen X, Lee A-H, et al. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol 2010;11:411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lee A-H, Scapa EF, Cohen DE, et al. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 2008;320:1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Margariti A, Li H, Chen T, et al. XBP1 mRNA splicing triggers an autophagic response in endothelial cells through BECLIN-1 transcriptional activation. J Biol Chem 2013;288:859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. van Huizen R, Martindale JL, Gorospe M, et al. P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eIF2alpha signaling. J Biol Chem 2003;278:15558–15564. [DOI] [PubMed] [Google Scholar]
- 95. Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 2006;313:104–107. [DOI] [PubMed] [Google Scholar]
- 96. Hu P, Han Z, Couvillon AD, et al. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1alpha-mediated NF-kappaB activation and down-regulation of TRAF2 expression. Mol Cell Biol 2006;26:3071–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Harding HP, Novoa I, Zhang Y, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 2000;6:1099–1108. [DOI] [PubMed] [Google Scholar]
- 98. Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 2011;13:184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Novoa I, Zhang Y, Zeng H, et al. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J 2003;22:1180–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem 2004;279:20108–20117. [DOI] [PubMed] [Google Scholar]
- 101. Nadanaka S, Okada T, Yoshida H, et al. Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol Cell Biol 2007;27:1027–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Fung TS, Torres J, Liu DX. The emerging roles of Viroporins in ER stress response and autophagy induction during virus infection. Viruses 2015;7:2834–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Li S, Kong L, Yu X. The expanding roles of endoplasmic reticulum stress in virus replication and pathogenesis. Crit Rev Microbiol 2015;41:150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tattoli I, Sorbara MT, Vuckovic D, et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 2012;11:563–575. [DOI] [PubMed] [Google Scholar]
- 105. Maingat F, Halloran B, Acharjee S, et al. Inflammation and epithelial cell injury in AIDS enteropathy: involvement of endoplasmic reticulum stress. FASEB J 2011;25:2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Asselah T, Bièche I, Mansouri A, et al. In vivo hepatic endoplasmic reticulum stress in patients with chronic hepatitis C. J Pathol 2010;221:264–274. [DOI] [PubMed] [Google Scholar]
- 107. Pfefferle S, Krähling V, Ditt V, et al. Reverse genetic characterization of the natural genomic deletion in SARS-coronavirus strain Frankfurt-1 open reading frame 7b reveals an attenuating function of the 7b protein in-vitro and in-vivo. Virol J 2009;6:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Schögler A, Caliaro O, Brügger M, et al. Modulation of the unfolded protein response pathway as an antiviral approach in airway epithelial cells. Antiviral Res 2019;162:44–50. [DOI] [PubMed] [Google Scholar]
- 109. Richardson RB, Ohlson MB, Eitson JL, et al. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat Microbiol 2018;3:1214–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hassan IH, Zhang MS, Powers LS, et al. Influenza A viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway. J Biol Chem 2012;287:4679–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Roberson EC, Tully JE, Guala AS, et al. Influenza induces endoplasmic reticulum stress, caspase-12-dependent apoptosis, and c-Jun N-terminal kinase-mediated transforming growth factor-β release in lung epithelial cells. Am J Respir Cell Mol Biol 2012;46:573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Landeras-Bueno S, Fernández Y, Falcón A, et al. Chemical genomics identifies the PERK-mediated unfolded protein stress response as a cellular target for influenza virus inhibition. MBio 2016;7:e00085–e00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Hassan I, Gaines KS, Hottel WJ, et al. Inositol-requiring enzyme 1 inhibits respiratory syncytial virus replication. J Biol Chem 2014;289:7537–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Smith JA, Khan M, Magnani DD, et al. Brucella induces an unfolded protein response via TcpB that supports intracellular replication in macrophages. PLoS Pathog 2013;9:e1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Pillich H, Loose M, Zimmer K-P, et al. Activation of the unfolded protein response by listeria monocytogenes. Cell Microbiol 2012;14:949–964. [DOI] [PubMed] [Google Scholar]
- 116. Xu G, Thielen LA, Chen J, et al. Serum miR-204 is an early biomarker of type 1 diabetes-associated pancreatic beta-cell loss. Am J Physiol Endocrinol Metab 2019;317:E723–E730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hiramatsu N, Chiang K, Aivati C, et al. PERK-mediated induction of microRNA-483 disrupts cellular ATP homeostasis during the unfolded protein response. J Biol Chem 2020;295:237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wang J-M, Qiu Y, Yang Z-Q, et al. Inositol-requiring enzyme 1 facilitates diabetic wound healing through modulating MicroRNAs. Diabetes 2017;66:177–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. McMahon M, Samali A, Chevet E. Regulation of the unfolded protein response by noncoding RNA. Am J Physiol Cell Physiol 2017;313:C243–C254. [DOI] [PubMed] [Google Scholar]
- 120. Xu Z, Bu Y, Chitnis N, et al. miR-216b regulation of c-Jun mediates GADD153/CHOP-dependent apoptosis. Nat Commun 2016;7:11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Brookheart RT, Michel CI, Listenberger LL, et al. The non-coding RNA gadd7 is a regulator of lipid-induced oxidative and endoplasmic reticulum stress. J Biol Chem 2009;284:7446–7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Su S, Liu J, He K, et al. Overexpression of the long noncoding RNA TUG1 protects against cold-induced injury of mouse livers by inhibiting apoptosis and inflammation. FEBS J 2016;283:1261–1274. [DOI] [PubMed] [Google Scholar]
- 123. Bhattacharyya S, Vrati S. The Malat1 long non-coding RNA is upregulated by signalling through the PERK axis of unfolded protein response during flavivirus infection. Sci Rep 2015;5:17794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Quan H, Fan Q, Li C, et al. The transcriptional profiles and functional implications of long non-coding RNAs in the unfolded protein response. Sci Rep 2018;8:4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 2014;32:513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci U S A 2000;97:8841–8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Muralidharan S, Mandrekar P. Cellular stress response and innate immune signaling: integrating pathways in host defense and inflammation. J Leukoc Biol 2013;94:1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013;341:789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kambara H, Niazi F, Kostadinova L, et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res 2014;42:10668–10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Carnero E, Barriocanal M, Prior C, et al. Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep 2016;17:1013–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Qian X, Xu C, Zhao P, et al. Long non-coding RNA GAS5 inhibited hepatitis C virus replication by binding viral NS3 protein. Virology 2016;492:155–165. [DOI] [PubMed] [Google Scholar]
- 132. Li J, Li M, Wang X, et al. Long noncoding RNA NRAV promotes respiratory syncytial virus replication by targeting the MicroRNA miR-509-3p/Rab5c Axis To regulate vesicle transportation. J Virol 2020;94:e00113-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Ouyang J, Zhu X, Chen Y, et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 2014;16:616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Imam H, Bano AS, Patel P, et al. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci Rep 2015;5:8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Rapicavoli NA, Qu K, Zhang J, et al. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2013;2:e00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Liu B, Sun L, Liu Q, et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015;27:370–381. [DOI] [PubMed] [Google Scholar]
- 137. Wang P, Xue Y, Han Y, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014;344:310–313. [DOI] [PubMed] [Google Scholar]
- 138. Gernapudi R, Wolfson B, Zhang Y, et al. Microrna 140 promotes expression of long noncoding RNA NEAT1 in adipogenesis. Mol Cell Biol 2016;36:30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Hirose T, Virnicchi G, Tanigawa A, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell 2014;25:169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zhang Y, Shao L. Decreased microRNA-140-5p contributes to respiratory syncytial virus disease through targeting toll-like receptor 4. Exp Ther Med 2018;16:993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Zhu S, Zhang X, Guan H, et al. miR-140-5p regulates T cell differentiation and attenuates experimental autoimmune encephalomyelitis by affecting CD4+T cell metabolism and DNA methylation. Int Immunopharmacol 2019;75:105778. [DOI] [PubMed] [Google Scholar]
- 142. Yu X, Zhe Z, Tang B, et al. α-Asarone suppresses the proliferation and migration of ASMCs through targeting the lncRNA-PVT1/miR-203a/E2F3 signal pathway in RSV-infected rats. Acta Biochim Biophys Sin (Shanghai) 2017;49:598–608. [DOI] [PubMed] [Google Scholar]
- 143. Gelfand BD, Wright CB, Kim Y, et al. Iron toxicity in the retina requires alu RNA and the NLRP3 inflammasome. Cell Rep 2015;11:1686–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kerur N, Hirano Y, Tarallo V, et al. TLR-independent and P2X7-dependent signaling mediate Alu RNA-induced NLRP3 inflammasome activation in geographic atrophy. Invest Ophthalmol Vis Sci 2013;54:7395–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Pfeffer K. Biological functions of tumor necrosis factor cytokines and their receptors. Cytokine Growth Factor Rev 2003;14:185–191. [DOI] [PubMed] [Google Scholar]
- 146. Banerjee M, Saxena M. Interleukin-1 (IL-1) family of cytokines: role in type 2 diabetes. Clin Chim Acta 2012;413:1163–1170. [DOI] [PubMed] [Google Scholar]
- 147. Holohan C, Szegezdi E, Ritter T, et al. Cytokine-induced beta-cell apoptosis is NO-dependent, mitochondria-mediated and inhibited by BCL-XL. J Cell Mol Med 2008;12:591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Mayer-Barber KD, Yan B. Clash of the cytokine titans: counter-regulation of interleukin-1 and type I interferon-mediated inflammatory responses. Cell Mol Immunol 2017;14:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Sun L, Cornell TT, LeVine A, et al. Dual role of interleukin-10 in the regulation of respiratory syncitial virus (RSV)-induced lung inflammation. Clin Exp Immunol 2013;172:263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Choi J, Callaway Z, Kim H-B, et al. The role of TNF-alpha in eosinophilic inflammation associated with RSV bronchiolitis. Pediatr Allergy Immunol 2010;21:474–479. [DOI] [PubMed] [Google Scholar]
- 151. To KKW, Hung IFN, Li IWS, et al. Delayed clearance of viral load and marked cytokine activation in severe cases of pandemic H1N1 2009 influenza virus infection. Clin Infect Dis 2010;50:850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Gao R, Bhatnagar J, Blau DM, et al. Cytokine and chemokine profiles in lung tissues from fatal cases of 2009 pandemic Influenza A (H1N1): role of the host immune response in pathogenesis. Am J Pathol 2013;183:1258–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Chen I-Y, Moriyama M, Chang M-F, et al. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front Microbiol 2019;10:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Park WB, Kwon NJ, Choi SJ, et al. Virus isolation from the first patient with SARS-CoV-2 in Korea. J Korean Med Sci 2020;35:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Zhang B, Zhou X, Qiu Y, et al. Clinical characteristics of 82 death cases with COVID-19. PLoS One 2020;15:e0235458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Arora S, Singh P, Dohare R, et al. Unravelling host-pathogen interactions: ceRNA network in SARS-CoV-2 infection (COVID-19). Gene 2020;762:145057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Pérez PS, Romaniuk MA, Duette GA, et al. Extracellular vesicles and chronic inflammation during HIV infection. J Extracell Vesicles 2019;8:1687275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Dolganiuc A, Kodys K, Kopasz A, et al. Hepatitis C virus core and nonstructural protein 3 proteins induce pro- and anti-inflammatory cytokines and inhibit dendritic cell differentiation. J Immunol 2003;170:5615–5624. [DOI] [PubMed] [Google Scholar]
- 160. Negash AA, Ramos HJ, Crochet N, et al. IL-1β production through the NLRP3 inflammasome by hepatic macrophages links hepatitis C virus infection with liver inflammation and disease. PLoS Pathog 2013;9:e1003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Dong Y, Zhi X, Lei G. Changes of body immunity and inflammatory response in HIV/HCV co-infected patients. Exp Ther Med 2019;17:403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Teijaro JR, Walsh KB, Rice S, et al. Mapping the innate signaling cascade essential for cytokine storm during influenza virus infection. Proc Natl Acad Sci U S A 2014;111:3799–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Ye Q, Wang B, Mao J. The pathogenesis and treatment of the `cytokine storm’ in COVID-19. J Infect 2020;80:607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Boukhvalova MS, Prince GA, Soroush L, et al. The TLR4 agonist, monophosphoryl lipid a, attenuates the cytokine storm associated with respiratory syncytial virus vaccine-enhanced disease. Vaccine 2006;24:5027–5035. [DOI] [PubMed] [Google Scholar]
- 165. Kuczera D, Assolini JP, Tomiotto-Pellissier F, et al. Highlights for dengue Immunopathogenesis: antibody-dependent enhancement, cytokine storm, and beyond. J Interferon Cytokine Res 2018;38:69–80. [DOI] [PubMed] [Google Scholar]
- 166. Krawczyk M. Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. Elife 2014;3:e01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Li Z, Chao T-C, Chang K-Y, et al. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A 2014;111:1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Wang Y, Liang H, Jin F, et al. Injured liver-released miRNA-122 elicits acute pulmonary inflammation via activating alveolar macrophage TLR7 signaling pathway. Proc Natl Acad Sci U S A 2019;116:6162–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. He WA, Calore F, Londhe P, et al. Microvesicles containing miRNAs promote muscle cell death in cancer cachexia via TLR7. Proc Natl Acad Sci U S A 2014;111:4525–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Casadei L, Calore F, Creighton CJ, et al. Exosome-derived miR-25-3p and miR-92a-3p stimulate Liposarcoma progression. Cancer Res 2017;77:3846–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Yao M-Y, Zhang W-H, Ma W-T, et al. Long non-coding RNA MALAT1 exacerbates acute respiratory distress syndrome by upregulating ICAM-1 expression via microRNA-150-5p downregulation. Aging (Albany, NY) 2020;12:6570–6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Du M, Yuan L, Tan X, et al. The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat Commun 2017;8:2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Li X, Yu M, Chen L, et al. LncRNA PMS2L2 protects ATDC5 chondrocytes against lipopolysaccharide-induced inflammatory injury by sponging miR-203. Life Sci 2019;217:283–292. [DOI] [PubMed] [Google Scholar]
- 174. Wang W, Lou C, Gao J, et al. LncRNA SNHG16 reverses the effects of miR-15a/16 on LPS-induced inflammatory pathway. Biomed Pharmacother 2018;106:1661–1667. [DOI] [PubMed] [Google Scholar]
- 175. Tao X-W, Zeng L-K, Wang H-Z, et al. LncRNA MEG3 ameliorates respiratory syncytial virus infection by suppressing TLR4 signaling. Mol Med Rep 2018;17:4138–4144. [DOI] [PubMed] [Google Scholar]
- 176. Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature 2008;454:455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Keestra-Gounder AM, Byndloss MX, Seyffert N, et al. NOD1 and NOD2 signalling links ER stress with inflammation. Nature 2016;532:394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Lencer WI, DeLuca H, Grey MJ, et al. Innate immunity at mucosal surfaces: the IRE1-RIDD-RIG-I pathway. Trends Immunol 2015;36:401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Shi T-T, Yang F-Y, Liu C, et al. Angiotensin-converting enzyme 2 regulates mitochondrial function in pancreatic β-cells. Biochem Biophys Res Commun 2018;495:860–866. [DOI] [PubMed] [Google Scholar]
- 181. Kim S-M, Kim Y-G, Jeong K-H, et al. Angiotensin II-induced mitochondrial Nox4 is a major endogenous source of oxidative stress in kidney tubular cells. PLoS One 2012;7:e39739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Tan Y-X, Tan THP, MJ-R L, et al. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J Virol 2007;81:6346–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Chen C-Y, Ping Y-H, Lee H-C, et al. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J Infect Dis 2007;196:405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Shi C-S, Qi H-Y, Boularan C, et al. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol 2014;193:3080–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Wu KE, Fazal FM, Parker KR, et al. RNA-GPS predicts SARS-CoV-2 RNA residency to host mitochondria and nucleolus. Cell Syst 2020;11:102–108.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Codo AC, Davanzo GG, Monteiro L d B, et al. Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1α/glycolysis-dependent Axis. Cell Metab 2020;32:437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Vishnubalaji R, Shaath H, Alajez NM. Protein coding and long noncoding RNA (lncRNA) transcriptional landscape in SARS-CoV-2 infected bronchial epithelial cells highlight a role for interferon and inflammatory response. Genes (Basel) 2020;11:760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Hu M, Fu Q, Jing C, et al. LncRNA HOTAIR knockdown inhibits glycolysis by regulating miR-130a-3p/HIF1A in hepatocellular carcinoma under hypoxia. Biomed Pharmacother 2020;125:109703. [DOI] [PubMed] [Google Scholar]
- 189. Yang F, Huo X, Yuan S, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell 2013;49:1083–1096. [DOI] [PubMed] [Google Scholar]
- 190. Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer 2017;140:1955–1967. [DOI] [PubMed] [Google Scholar]
- 191. Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 2013;12:847–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Ouyang B, Bracken B, Burke B, et al. A duplex quantitative polymerase chain reaction assay based on quantification of alpha-methylacyl-CoA racemase transcripts and prostate cancer antigen 3 in urine sediments improved diagnostic accuracy for prostate cancer. J Urol 2009;181:2508–2513 discussion 2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Wang F, Ren S, Chen R, et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget 2014;5:11091–11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Srivastava AK, Singh PK, Rath SK, et al. Appraisal of diagnostic ability of UCA1 as a biomarker of carcinoma of the urinary bladder. Tumour Biol 2014;35:11435–11442. [DOI] [PubMed] [Google Scholar]
- 195. Tang H, Wu Z, Zhang J, et al. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol Med Rep 2013;7:761–766. [DOI] [PubMed] [Google Scholar]
- 196. Heo MJ, Yun J, Kim SG. Role of non-coding RNAs in liver disease progression to hepatocellular carcinoma. Arch Pharm Res 2019;42:48–62. [DOI] [PubMed] [Google Scholar]
- 197. Melo S, Villanueva A, Moutinho C, et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc Natl Acad Sci U S A 2011;108:4394–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005;438:685–689. [DOI] [PubMed] [Google Scholar]


