Abstract
Pregnancy zone protein (PZP), a member of the proteinase inhibitor I39 (‐2‐macroglobulin) family of proteins, is involved in the initiation and development of various tumors. The gene encoding PZP is hypermethylated and expressed at low levels in hepatocellular carcinoma (HCC) tissue and cells, but the function of PZP in HCC cells remains unclear. Here, we analyzed DNA methylation and mRNA expression of HCC in The Cancer Genome Atlas Liver Hepatocellular Carcinoma dataset. We identified 10 methylation‐driven genes, of which PZP was significantly hypermethylated and poorly expressed in tumor tissue. We confirmed that PZP is highly methylated and poorly expressed in HCC cell lines via quantitative real‐time PCR experiment and methylation‐specific PCR. Furthermore, PZP markedly inhibited the proliferation, invasion and migration of HCC cells. These findings may provide a basis for exploring novel therapeutic targets for HCC.
Keywords: HCC, invasion and migration, methylation, proliferation, PZP
We analyzed DNA methylation and mRNA expression of hepatocellular carcinoma (HCC) in the TCGA‐LIHC dataset. We identified 10 methylation‐driven genes, of which pregnancy zone protein (PZP) was significantly hypermethylated and poorly expressed in tumor tissue. We confirmed that PZP is highly methylated and poorly expressed in HCC cell lines via RT‐PCR and methylation‐specific PCR. Furthermore, PZP markedly inhibited the proliferation, invasion and migration of HCC cells.
Abbreviations
- 5‐aza‐dC
5‐aza‐2ʹ‐deoxycytidine
- CGI
CpG island
- DEmRNA
differentially expressed mRNA
- FC
fold change
- HCC
hepatocellular carcinoma
- MSP
methylation‐specific PCR
- MTT
3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl‐tetrazolium bromide
- NC
negative control
- PZP
pregnancy zone protein
- SD
standard deviation
- TCGA‐LIHC
The Cancer Genome Atlas Liver Hepatocellular Carcinoma
Liver cancer is the sixth most common cancer and the fourth most common cause of cancer‐related deaths in the world [1]. Hepatocellular carcinoma (HCC) is the main histological subtype of liver cancer, accounting for 90% of primary liver cancer, yet its pathogenesis remains unclear. With the deeper research on tumors, it has been reported that the two mechanisms associated with the occurrence and development of tumors are genetic variation and epigenetic modification [2]. Currently, the epigenetic mechanism that has been most frequently researched mainly focuses on DNA methylation and histone modification. DNA methylation has been the first epigenetic phenomenon [3]. People usually refer to the covalent addition of a methyl (CH3) group to the C5 position of the cytosine pyrimidine ring (usually in CpG dinucleotides) as DNA methylation [4, 5]. The CpG site exists in two forms: one being dispersed in DNA sequence, and another being highly concentrated in large clusters called CpG islands (CGIs) (CpG > 50%, length > 200 base pairs) [6]. At present, DNA methylation, especially the aberrant CGI methylation in the gene promoter region, has become the hotspot of tumor research [7]. Aberrant CGI methylation of the gene promoter region can activate proto‐oncogenes or silence tumor suppressor genes, leading to altered expression of downstream key genes and consequently promoting the aberrant proliferation of cancer cells [6]. A number of research studies have unveiled that aberrant DNA methylation is able to facilitate tumor cell proliferation, invasion and migration of colorectal cancer [8], gastric cancer [9], liver cancer [10, 11], HCC [12] and so on.
This study found that pregnancy zone protein (PZP) in HCC tissue and cells was hypermethylated and poorly expressed. PZP is a protein coding gene that is capable of suppressing all four classes of proteinases by a unique ‘trapping’ mechanism. Devriendt et al. [13] cloned the full‐length cDNA of PZP in HCC in 1991. Subsequently, research has indicated that PZP is associated with the liver fibrosis in nonalcoholic fatty liver [14], while liver fibrosis is the beginning of normal liver tissue gradually transferring to cirrhotic tissue. Nevertheless, the functional mechanism of PZP in HCC cells remains unclear, and there is an urgent need to carry out further investigation.
This study initially analyzed data of methylation and mRNA expression of HCC through The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA‐LIHC) database, finding that the down‐regulation of PZP in HCC was caused by promoter hypermethylation. After that, we explored the relationship between PZP methylation level and PZP expression in HCC cell proliferation, migration, invasion and apoptosis through in vitro cell experiments and further clarified the mechanism of PZP regulating the development of HCC.
Materials and methods
Data source and processing
HCC data, including methylation data, mRNA expression profiles and clinical information, were downloaded from the TCGA‐LIHC dataset. A total of 424 mRNA expression profiles (normal: n = 50, tumor: n = 374) and methylation data of 430 samples (normal: n = 50, tumor: n = 380) were obtained. Methylation data were standardized using the ‘limma’ package. Differential analysis was conducted to screen the differentially expressed mRNAs (DEmRNAs) using the ‘edgeR’ package (|logFC| > 2, P adj < 0.05). Candidate methylation‐driven genes were screened using the ‘MethylMix’ package (|logFC| > 0.5, P adj < 0.05, Correlation coefficient < −0.3). The ‘survival’ package was used to analyze the effect of the methylation and expression levels of the target methylation‐driven gene on a patient's prognosis.
Cell culture
This study used the human liver cell line HL‐7702 (3131C0001000200006) provided by Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, and the human HCC cell lines Hep G2 (3111C0001CCC000035), Hep3b (3111C0001CCC000376), HuH‐7 (3111C0001CCC000679), Li‐7 (3111C0001CCC000678) and Hep 3B2.1‐7 (3111C0001CCC000474) provided by Cell Source Center, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences. The cells were cultured at 37 °C in a humid environment containing 5% CO2 according to the manufacturer’s instructions.
DNA extraction and sodium bisulfite modification
QIAamp DNA Mini Kit (50) (51304; Qiagen, Hilden, Germany) was used to extract the genomic DNA of the human normal liver cell line and the five human HCC cell lines according to the instructions. Thereafter, the genomic DNA was processed by sodium bisulfite: 2 μg DNA was dissolved in 50 μL deionized water, denatured in 0.2 mol·L−1 NaOH at 50 °C for 10 min and water bathed in fresh 30 μL 10 mmol·L−1 hydroquinone and 520 μL sodium bisulfite (3.6 mol·L−1, pH 5.0) at 50 °C for 18 h. After being purified in adsorption column, it was precipitated in absolute alcohol and dissolved in 50 μL deionized water.
Methylation‐specific PCR
Methylation‐specific PCR (MSP) was used for amplification of the PZP gene sequence. Primers were designed according to the PZP gene sequence (Gene ID: 5858) and synthesized by MethPrimer. The 25 μL PCR system was as follows: 10× buffer 2.5 μL, dNTP 2.0 μL, TaqE 0.5 μL, double‐distilled H2O 17.5 μL, Pmix 1 μL, MgSO4 0.5 μL and DNA 1 μL. Thirty‐five cycles began after predenaturation at 94 °C for 3 min: 94 °C 30 s, 71 °C 1 min, 75 °C 45 s. At last, extension was processed at 72 °C for 7 min (preserved at 4 °C). PCR products were loaded on 2% agarose gel electrophoresis and then observed. Primer sequences are listed in Table S1.
Real‐time quantitative PCR
According to the manufacturer's instructions, TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA from HCC tissue and cells. Then RNA was transcribed into cDNA by using the Reverse Transcription Assay Kit (Invitrogen). Quantitative real‐time PCR was performed on an ABI 7900HT instrument (Applied Biosystems, Foster City, CA, USA) using the miScript SYBR Green PCR Kit (Qiagen). GAPDH was used as internal reference, and each measurement was normalized to GAPDH expression. The quantitative value was expressed using the method to compare the differences of target gene expression in the control group and the test group. We repeated the experiment in triplicate. Primer sequences are listed in Table S1.
Demethylation experiment
For the demethylation experiment, all the cell lines were processed using 2.5 μmol·L−1 demethylating agent 5‐aza‐2′‐deoxycytidine (5‐aza‐dC) for 6 days. Subsequently, cells were collected, and total RNA was extracted to detect PZP expression.
Construction of pCDH‐PZP recombinant plasmid
The cDNAs of HuH‐7 and Hep G2 cells were used as templates to amplify the human full‐length PZP coding sequence. pCDH‐CMV‐MCS‐EF1‐Puro (YB‐0879; Shanghai Yu Bo Biotech Co., Ltd., Shanghai, China) lentivirus vector was used to construct PZP overexpression plasmid. Packed lentiviral supernatant was added to the growth medium of HL‐7702 and Hep G2 cells, and Puromycin was used to screen stably transfected cell lines.
MTT assay
Cells in the negative control (NC) group and PZP overexpression group were digested with 0.25% trypsin and seeded in a 96‐well plate at a density of 2 × 103 cells per well. Three repeated wells were set for each treatment. Each well was added with 200 μL RPMI 1640 medium, which contained 10% FBS. At 0, 1, 2, 3, 4 and 5 days, 20 μL 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl‐tetrazolium bromide (MTT; 5 g·L−1) was added into the cells, and then the cells were incubated for an additional 4 h. Then we removed the supernatant and used 150 μL dimethyl sulfoxide to dissolve the formed formazan crystals. At last, we used a Bio‐Rad‐550 (Bio‐Rad Laboratories, Redmond, WA, USA) microplate reader to detect the absorbance of each well at 490 nm (A 490 nm). The experiment was repeated three times.
Cell migration and invasion assays
Wound healing assay
HuH‐7 and Hep G2 cells (5 × 106 cells per well) were inoculated into a six‐well plate. After cells were grown to 80% in confluence, a scratch was made on the center of cells using the tip of a 200‐μL pipette. Thereafter, cells were temporarily washed twice by medium to remove the floating cells and then cultured for an additional 24 h in a fresh medium. The cell migration distance at 0 and 24 h was measured under a microscope.
Transwell assay
A 24‐well Transwell chamber (8‐μm pores) from BD was used to identify cell invasion. We applied Matrigel to the upper chamber and placed about 2 × 104 cells in it, and then added DMEM containing 10% FBS to the lower chamber. We cultured the cells in a 37 °C incubator for 48 h, then used a cotton swab to remove noninvading cells in the upper chamber and stained the cells that invaded to the lower chamber with 0.5% crystal violet. Finally, we observed the cells under a microscope and took pictures.
Statistical analysis
We used spss 22.0 software (SPSS, Chicago, IL, USA) to process all data. All measurement data were expressed as mean ± standard deviation (SD). Student's t‐test and one‐way ANOVA were used to analyze the differences between two groups and the comparisons among more than two groups, respectively: *,# P < 0.05, **,## P < 0.01.
Results
PZP gene is hypermethylated in HCC tissue
To obtain the key gene that affects the occurrence and development of HCC in a DNA methylation manner, we downloaded methylation and mRNA expression data of HCC from the TCGA‐LIHC database. Ten candidate methylation‐driven genes were screened using the ‘MethylMix’ package after standardized treatment (Fig. 1A), among which PZP was markedly highly methylated (Fig. 1B) and its expression in HCC tissue was abnormally decreased (Fig. 1D). Further, the correlation analysis results suggested that PZP methylation level was markedly negatively correlated with its expression level (Fig. 1C). Survival analysis was performed based on the methylation and expression levels of PZP, unveiling that the survival time of patients with PZP hypermethylation and low expression was markedly lower than that of patients with low methylation and high expression (Fig. 1E). Accordingly, it can be concluded that PZP gene is hypermethylated in HCC tissue.
Fig. 1.
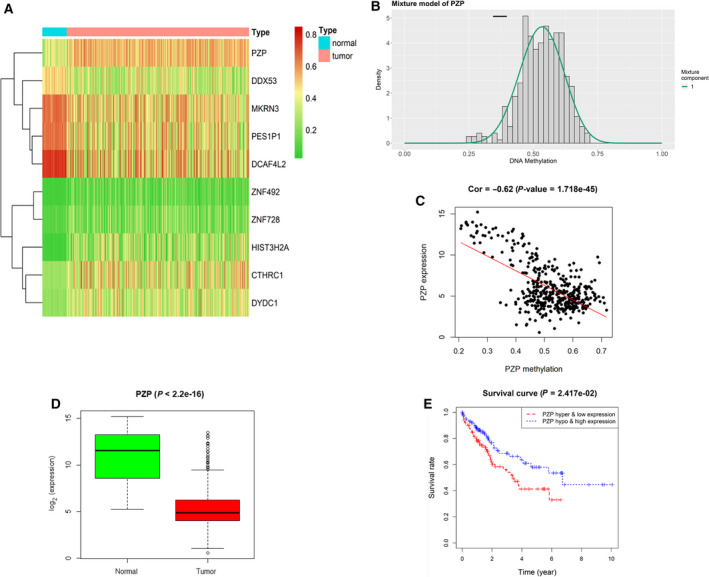
Analysis of methylation‐driven genes and prognosis of HCC. (A) Heatmap for HCC‐related methylation‐driven genes (the color from green to red stands for the tendency of methylation level from low to high). (B) Mixture model of PZP gene DNA methylation. The distribution map shows the methylation of PZP gene. The histogram and black horizontal bars, respectively, represent the distribution of methylation in tumor samples and in normal samples. (C) Correlation analysis for PZP methylation level and expression level. (D) Relative expression of PZP gene in TCGA‐LIHC database. (E) Survival analysis based on the methylation and expression levels of PZP gene.
CpG methylation of PZP gene in HCC and normal cells
Bioinformatics analysis indicated that PZP down‐regulation in HCC might be caused by its hypermethylation. Hence we further explored PZP methylation by conducting in vitro experiments. Firstly, quantitative real‐time PCR was performed to detect PZP mRNA expression in a human normal liver cell line and five HCC cell lines, and we discovered that PZP expression in HCC cells was lower than that in human normal cells, among which PZP was most markedly down‐regulated in Hep G2 and HuH‐7 cells (Fig. 2A). Subsequently, genomic DNAs of HCC cell lines were processed by sodium bisulfite, and PZP methylation was evaluated by MSP. PZP gene was amplified by methylation‐specific primers in Hep G2 and HuH‐7 cells but could not be done by nonmethylation‐specific ones, which uncovered that PZP was completely methylated in Hep G2 and HuH‐7 cells. In addition, PZP gene was amplified by both methylation‐specific primers and nonmethylation‐specific ones in Hep3b, Li‐7 and Hep 3B2.1‐7 cells, which demonstrated that PZP was partially methylated in these cells (Fig. 2B). After cell lines were treated with demethylating agents (5‐aza‐dC), quantitative real‐time PCR was performed and revealed that PZP expression was significantly increased in Hep G2, Hep3b, HuH‐7, Li‐7 and Hep 3B2.1‐7 cell lines by comparison with that in cells without 5‐aza‐dC (Fig. 2A), suggesting that PZP expression was regulated by its methylation. The experimental result was basically consistent with that of HCC methylation data in the TCGA‐LIHC dataset, which unveiled that PZP promoter hypermethylation inhibited PZP expression.
Fig. 2.

PZP expression and methylation in HCC cells. (A) Quantitative real‐time PCR was performed to detect mRNA expression of PZP in human liver cell line and five human HCC cell lines (asterisks [*] represent the comparison of PZP expression in normal liver cells and HCC cells; number signs [#] represent comparison of PZP expression between the HCC cells and that with or without 5‐Aza). (B) MSP result of PZP in HCC cell lines. Error bars indicate SD. All experiments were repeated three times. *P < 0.05 and **P < 0.01 represent a comparison with the 5‐Aza (–) treatment group of the HL‐7702 cell line; # P < 0.05 and ## P < 0.01 represent a comparison with the 5‐Aza (–) treatment group of the corresponding cell line. The significance analysis of the results in Fig. 3A used a t‐test for the comparison of the results of each cell line with the 5‐Aza (–) treatment group and used ANOVA for the comparison with the 5‐Aza (–) treatment group of the HL‐7702 cell line. M, methylated allele; NorL, lymphocyte DNA used as the reference of nonmethylation; Pos, in vitro methylated DNA used as the reference of methylation; U, unmethylated allele.
PZP inhibits HCC cell proliferation, invasion and migration upon overexpression
In the earlier experiments, we found that PZP in HCC tissue was hypermethylated and down‐regulated. Thereafter, single‐gene Gene Ontology enrichment analysis for the PZP gene found that PZP was mainly involved in the biological processes, such as protein complement cascade activation and coagulation regulation. The results are shown in Table S2. Because ‘complement and coagulation cascade’ may be associated with the metastasis of clear cell renal cell carcinoma (CCRCC) [15], and the complement system plays a key role in tumor progression and can enhance angiogenesis and promote tumor growth, as well as metastasis [16], we investigated the effect of PZP on HCC cell proliferation, invasion and migration in vitro. We performed quantitative real‐time PCR on the HepG2 and HuH‐7 cells with pCDH empty vector and pCDH‐PZP overexpression vector to detect PZP mRNA expression and found that PZP expression in HCC cells with overexpressed PZP was obviously higher than that of the control (Fig. 3A). MTT assay was conducted and indicated that compared with the control group, the increase of A 490 nm of HepG2 and HuH‐7 cells upon PZP overexpression was significantly reduced (Fig. 3B), which suggested that cell proliferation ability was significantly decreased upon PZP overexpression. Wound healing and Transwell assays were used to further explore the effect of PZP on HCC cell invasion and migration upon overexpression. Compared with the control group, the wound healing rate and the number of HepG2 and HuH‐7 cells breaking through the membrane upon PZP overexpression were remarkably reduced, uncovering that the migratory and invasive abilities of the two cells were markedly weakened upon PZP overexpression (Fig. 3C,D). Collectively, the proliferative, migratory and invasive abilities of HepG2 and HuH‐7 cells could be markedly suppressed by overexpressing PZP.
Fig. 3.
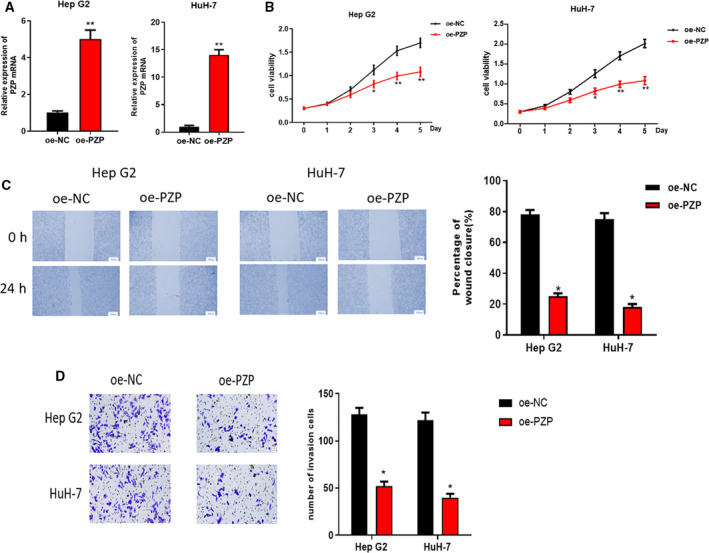
The effect of PZP on HepG2 and HuH‐7 cell growth. HepG2 and HuH‐7 cells were transfected with PZP overexpression vectors. (A) Quantitative real‐time PCR was conducted to detect PZP overexpression efficiency. (B) MTT assay was used to determine the viability of HepG2 and HuH‐7 cells transfected with pCDH‐PZP overexpression vector. (C) HepG2 and HuH‐7 cell migration upon PZP overexpression detected by wound healing assay. The statistical graph (right) shows the corresponding cell migration rate, scale: 200 µm. (D) Cell invasion graph of HepG2 and HuH‐7 cells breaking through Matrigel in Transwell assay. Scale bars: 20 µm. The statistical graph (right) shows the relative cell invasion number. Error bars indicate SD. All experiments were repeated three times. *P < 0.05 represents a comparison with the oe‐NC group, and **P < 0.01 represents a comparison with the oe‐NC group, Student's t‐test method. oe, overexpression.
Discussion
At present, aberrant CGI methylation in the gene promoter region has become the hotspot of tumor research [7]. It contributes to activation of proto‐oncogenes or silencing of tumor suppressor genes, resulting in differential expression of downstream key genes, which leads to promotion of aberrant cell proliferation [6]. Due to its reversibility, DNA methylation serving as a therapeutic target of tumors will be promising, and the research of DNA methylation in HCC also will draw extensive attention.
Currently, it has been found that many DNAs in HCC are abnormally methylated and affect the development of cancer. Research has indicated that Mybbp1a is able to combine with DNMT1 to form a complex, can induce aberrant CGI hypermethylation of Igfbp5 to inhibit the secretion of Igfbp5, and can further activate IGF1/AKT signaling pathway and facilitate the occurrence and development of HCC [17]. Besides, HOTAIR represses miR‐122 expression, activates Cyclin G1 protein expression and promotes HCC tumor formation by epigenetic inheritance of DNA methylation [18]. Besides, osteopontin changes DNA methylation by up‐regulating DNMT1, which makes CD133+/CD44+ tumor stem cells in HCC sensitive to 5‐aza‐dC [19].
In this study, 10 candidate methylation‐driven genes were screened through the TCGA‐LIHC database, among which PZP was markedly hypermethylated in HCC tissue and its methylation level was markedly negatively correlated with its expression level. Survival analysis was performed based on the methylation and expression levels of PZP, unveiling that the survival time of patients with PZP hypermethylation and low expression was markedly shorter than that of patients with low methylation and high expression. Consequently, we speculated that methylation of the PZP gene in the promoter region inhibits the binding of certain transcription factors, hence leading to transcriptional repression. Next, cells were further processed by a demethylating agent 5‐azacytidine (5‐Aza), which unveiled that demethylation inhibitor could demethylate PZP to further activate gene expression. Thereafter, we discovered that overexpressing PZP could suppress HCC cell proliferation, invasion and migration. Taken together, the results of our experiments verified that PZP acts as a tumor suppressor gene, and its hypermethylation promotes HCC cell proliferation, migration and invasion. The finding helps people better understand the role of PZP methylation in HCC and provide a basis for exploring novel therapeutic targets for HCC.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
All authors contributed to data analysis and drafting and revising the article, gave final approval of the version to be published and agreed to be accountable for all aspects of the work.
Supporting information
Table S1. Primer sequences.
Table S2. Results of GO enrichment analysis.
Minhua Wu and Hui Lan contributed equally to this article
Data accessibility
The data and materials in this study are available from the corresponding author upon reasonable request.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA and Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68, 394–424. [DOI] [PubMed] [Google Scholar]
- 2. Wu CT and Morris JR (2001) Genes, genetics, and epigenetics: a correspondence. Science 293, 1103–1105. [DOI] [PubMed] [Google Scholar]
- 3. Hotchkiss RD (1948) The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem 175, 315–332. [PubMed] [Google Scholar]
- 4. Morgan AE, Davies TJ and Mc Auley MT (2018) The role of DNA methylation in ageing and cancer. Proc Nutr Soc 77, 412–422. [DOI] [PubMed] [Google Scholar]
- 5. Holcakova J (2018) Effect of DNA methylation on the development of cancer. Klin Onkol 31, 41–45. [DOI] [PubMed] [Google Scholar]
- 6. Kulis M and Esteller M (2010) DNA methylation and cancer. Adv Genet 70, 27–56. [DOI] [PubMed] [Google Scholar]
- 7. Mersakova S, Nachajova M, Szepe P, Kasajova PS and Halasova E (2016) DNA methylation and detection of cervical cancer and precancerous lesions using molecular methods. Tumour Biol 37, 23–27. [DOI] [PubMed] [Google Scholar]
- 8. Liu X, Chen X, Zeng K, Xu M, He B, Pan Y, Sun H, Pan B, Xu X, Xu T et al. (2018) DNA‐methylation‐mediated silencing of miR‐486‐5p promotes colorectal cancer proliferation and migration through activation of PLAGL2/IGF2/beta‐catenin signal pathways. Cell Death Dis 9, 1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xin L, Liu L, Liu C, Zhou LQ, Zhou Q, Yuan YW, Li SH and Zhang HT (2020) DNA‐methylation‐mediated silencing of miR‐7‐5p promotes gastric cancer stem cell invasion via increasing Smo and Hes1. J Cell Physiol 235, 2643–2654. [DOI] [PubMed] [Google Scholar]
- 10. Cai C, Xie X, Zhou J, Fang X, Wang F and Wang M (2020) Identification of TAF1, SAT1, and ARHGEF9 as DNA methylation biomarkers for hepatocellular carcinoma. J Cell Physiol 235, 611–618. [DOI] [PubMed] [Google Scholar]
- 11. Fu Y, Feng MX, Yu J, Ma MZ, Liu XJ, Li J, Yang XM, Wang YH, Zhang YL, Ao JP et al. (2014) DNA methylation‐mediated silencing of matricellular protein dermatopontin promotes hepatocellular carcinoma metastasis by alpha3beta1 integrin‐Rho GTPase signalling. Oncotarget 30, 6701–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yao R, Jiang H, Ma Y, Wang L, Wang L, Du J, Hou P, Gao Y, Zhao L, Wang G et al. (2014) PRMT7 induces epithelial‐to‐mesenchymal transition and promotes metastasis in breast cancer. Cancer Res 74, 5656–5667. [DOI] [PubMed] [Google Scholar]
- 13. Devriendt K, Van den Berghe H, Cassiman JJ and Marynen P (1991) Primary structure of pregnancy zone protein. Molecular cloning of a full‐length PZP cDNA clone by the polymerase chain reaction. Biochim Biophys Acta 1088, 95–103. [DOI] [PubMed] [Google Scholar]
- 14. Chalasani N, Guo X, Loomba R, Goodarzi MO, Haritunians T, Kwon S, Cui J, Taylor KD, Wilson L, Cummings OW et al. (2010) Genome‐wide association study identifies variants associated with histologic features of nonalcoholic fatty liver disease. Gastroenterology 139, 1567–1576, e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wei W, Lv Y, Gan Z, Zhang Y, Han X and Xu Z (2019) Identification of key genes involved in the metastasis of clear cell renal cell carcinoma Oncol Lett 17, 4321–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao P, Wu J, Lu F, Peng X, Liu C, Zhou N and Ying M (2019) The imbalance in the complement system and its possible physiological mechanisms in patients with lung cancer. BMC Cancer 19, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weng X, Wu J, Lv Z, Peng C, Chen J, Zhang C, He B, Tong R, Hu W, Ding C et al. (2019) Targeting Mybbp1a suppresses HCC progression via inhibiting IGF1/AKT pathway by CpG islands hypo‐methylation dependent promotion of IGFBP5. EBioMedicine 44, 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng D, Deng J, Zhang B, He X, Meng Z, Li G, Ye H, Zheng S, Wei L, Deng X et al. (2018) LncRNA HOTAIR epigenetically suppresses miR‐122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine 36, 159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao X, Sheng Y, Yang J, Wang C, Zhang R, Zhu Y, Zhang Z, Zhang K, Yan S, Sun H et al. (2018) Osteopontin alters DNA methylation through up‐regulating DNMT1 and sensitizes CD133+/CD44+ cancer stem cells to 5 azacytidine in hepatocellular carcinoma. J Exp Clin Cancer Res 37, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequences.
Table S2. Results of GO enrichment analysis.
Data Availability Statement
The data and materials in this study are available from the corresponding author upon reasonable request.


