Abstract
Background & Aims
Given that gastrointestinal (GI) symptoms are a prominent extrapulmonary manifestation of COVID-19, we investigated intestinal infection with SARS-CoV-2, its effect on pathogenesis, and clinical significance.
Methods
Human intestinal biopsy tissues were obtained from patients with COVID-19 (n = 19) and uninfected control individuals (n = 10) for microscopic examination, cytometry by time of flight analyses, and RNA sequencing. Additionally, disease severity and mortality were examined in patients with and without GI symptoms in 2 large, independent cohorts of hospitalized patients in the United States (N = 634) and Europe (N = 287) using multivariate logistic regressions.
Results
COVID-19 case patients and control individuals in the biopsy cohort were comparable for age, sex, rates of hospitalization, and relevant comorbid conditions. SARS-CoV-2 was detected in small intestinal epithelial cells by immunofluorescence staining or electron microscopy in 15 of 17 patients studied. High-dimensional analyses of GI tissues showed low levels of inflammation, including down-regulation of key inflammatory genes including IFNG, CXCL8, CXCL2, and IL1B and reduced frequencies of proinflammatory dendritic cells compared with control individuals. Consistent with these findings, we found a significant reduction in disease severity and mortality in patients presenting with GI symptoms that was independent of sex, age, and comorbid illnesses and despite similar nasopharyngeal SARS-CoV-2 viral loads. Furthermore, there was reduced levels of key inflammatory proteins in circulation in patients with GI symptoms.
Conclusions
These data highlight the absence of a proinflammatory response in the GI tract despite detection of SARS-CoV-2. In parallel, reduced mortality in patients with COVID-19 presenting with GI symptoms was observed. A potential role of the GI tract in attenuating SARS-CoV-2–associated inflammation needs to be further examined.
Keywords: COVID-19, SARS-CoV-2, GI symptoms, GI infection, outcomes, host immune response
Abbreviations used in this paper: ACE, angiotensin-converting enzyme; ATE, average treatment effect; AUC, area under the curve; BMI, body mass index; BSL, biosafety level; CyTOF, mass cytometry by time of flight; DC, dendritic cell; DEG, differentially expressed gene; EC, epithelial compartment; FDR, false discovery rate; GI, gastrointestinal; ICU, intensive care unit; IEL, intraepithelial lymphocyte; IF, immunofluorescence; IL, interleukin; IRB, institutional review board; KEGG, Kyoto Encyclopedia of Genes and Genomes; LP, lamina propria; MSH, Mount Sinai Hospital; NP, nasopharyngeal; OR, odds ratio; pDC, plasmacytoid dendritic cell; RNA-seq, RNA sequencing; RT-qPCR, reverse-transcriptase quantitative polymerase chain reaction; Th, T helper; TNF, tumor necrosis factor
See editorial on page 2251.
What You Need to Know.
Background and Context
Gastrointestinal manifestations are common in COVID-19; however, to date, there is limited evidence of SARS-CoV-2 infection of human enterocytes, tissue immune responses, and relationship to clinical outcomes.
New Findings
These results demonstrate immunofluorescence and electron microscopic detection of SARS-CoV-2 in small intestinal biopsy samples obtained from patients with COVID-19. The results also reveal down-regulation of key inflammatory pathways and reduced myeloid cells in intestinal biopsy samples as well as lower severity and mortality in patients with COVID-19 with GI symptoms in a multivariable model in 2 large independent cohorts from the United States and Europe.
Limitations
Clinical documentation of gastrointestinal (GI) symptoms might vary depending on providers and on the acuity of the patients’ presentation.
Impact
These data demonstrate in vivo GI tract infection by SARS-CoV-2 and the clinical impact of GI symptoms on COVID-19 outcomes in 2 large patient cohorts.
Gastrointestinal (GI) symptoms comprising nausea, vomiting, and/or diarrhea1 are a common extrapulmonary manifestation in COVID-19. Additionally, the presence of GI involvement by SARS-CoV-2 has also been suggested by clinical,2 nonhuman primate,3 and in vitro4 , 5 data. However, to date, there is limited evidence of SARS-CoV-2 infection of human intestinal epithelial cells,6 and there are no studies on the response of the GI immune system in patients with COVID-19.
Given the immune dysregulation seen in COVID-19,7,8 we aimed to document infection of the GI tract in patients with COVID-19, to define the cellular and transcriptomic changes within the GI tract, and to determine the impact of GI symptoms on COVID-19 outcomes. Here, we present findings from well-characterized cohorts of patients with COVID-19 hospitalized in tertiary care centers from both New York City, New York, and Milan, Italy, where we conducted high-dimensional analyses of mucosal and systemic immune parameters and investigated disease outcomes associated with GI involvement in patients with COVID-19.
Materials and Methods
Clinical Cohorts
Intestinal Biopsy Cohort
Endoscopic biopsy samples were obtained from 20 patients with COVID-19 and 10 control individuals undergoing clinically indicated endoscopic procedures after informed consent with the Mount Sinai Hospital (MSH) institutional review board (IRB)–approved protocol (IRB 16-0583). The demographic characteristics of these patients and control individuals are provided in Supplementary Tables 1 and 2. COVID-19 severity is defined in Supplementary Table 3 and the Supplementary Methods (Supplementary Tables 1–5).
Discovery Cohort
A total of 634 patients with COVID-19, admitted to MSH between April 1, 2020, and April 15, 2020, who met study inclusion criteria were enrolled in a discovery cohort under an IRB approved protocol (IRB-20-03297A) (Supplementary Methods; Supplementary Table 6, Supplementary Table 7, Supplementary Table 8, Supplementary Table 9).
External Validation Cohort
We analyzed a cohort of 287 patients admitted to a tertiary care center in Milan, Italy, between February 22, 2020, and March 30, 2020, with COVID-19 (Supplementary Methods; Supplementary Table 10, Supplementary Table 11).
Internal Validation Cohort
A distinct internal validation cohort of patients who were hospitalized at MSH between April 16, 2020, and April 30, 2020, (Supplementary Methods; Supplementary Table 12, Supplementary Table 13, Supplementary Table 14, Supplementary Table 15) was analyzed using a predictive model.
Immunofluorescent Microscopy
Formalin-fixed, paraffin-embedded tissue was analyzed (Supplementary Methods). Primary and secondary antibodies are summarized in Supplementary Table 16.
Transmission Electron Microscopy (TEM), Electron Tomography (ET), and Immunoelectron Microscopy (Immuno-EM)
Biopsy specimens and infected Vero E6 cells (positive control) were examined by electron microscopy (Supplementary Methods). Immuno-EM was performed with a mouse polyclonal antiserum against SARS-CoV-2 receptor binding domain (RBD) of spike protein and 10 nm gold conjugated anti-mouse secondary antibodies.
Cell Culture Experiments, Virus Isolation, and Viral RNA Detection From Gastrointestinal Biopsy Tissues
Endoscopic biopsy tissue samples were homogenized, inoculated on Vero E6 monolayers under biosafety level (BSL) 3 conditions and monitored daily for potential cytopathic effect. Biopsy homogenate supernatants were assessed for the presence of infective particles by plaque assay (Supplementary Methods). To detect SARS-CoV-2 RNA from intestinal biopsy samples, a modified version of the Centers for Disease Control and Prevention 2019 novel coronavirus reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) was used (Supplementary Methods).
Biopsy Sample Collection and Processing for Mass Cytometry (Cytometry by time-of-flight)
Endoscopic biopsy samples were processed in BSL 3 facility within 2 hours of collection to obtain suspension of the epithelial compartment (EC) and lamina propria (LP) (Supplementary Methods).
Mass Cytometry Processing, Data Acquisition, and Data Analysis
Cells were processed as previously described,9 acquired on a Helios Mass Cytometer, and demultiplexed using the Zunder single-cell debarcoder. Debarcoded files were uploaded to Cytobank for analyses, followed by annotation using Astrolabe Cytometry Platform (Astrolabe Diagnostics, Inc) and clustering using Clustergrammer2’s interactive heatmap (Supplementary Methods).
Blood Collection and Processing for Mass Cytometry
Phlebotomy was performed on the intestinal biopsy cohort patients at the time of endoscopic evaluation. Blood samples from patients with COVID-19 were processed in enhanced BSL 2 conditions (Supplementary Methods).
Specimen Processing for Nucleic Acid Extraction and RNA Sequencing
Total RNA was extracted from the cells isolated from both the intestinal compartments, EC and LP cellular fractions, using the Direct-zol RNA Miniprep Plus (Zymo) kit according to the manufacturer’s instructions. RNA from case patients and control individuals was then used for qRT-PCR and RNA sequencing (RNA-seq) (Supplementary Methods).
RNA Sequencing
Library Preparation and Sequencing
RNA-seq was performed on RNA isolated from the EC and LP samples obtained from COVID-19 case patients and control individuals (Supplementary Methods).
Computational Analyses
Descriptive Statistics
For univariable statistical analyses, Graph Pad Prism, version 8, was used to calculate an unpaired 2-tailed t test for continuous variables and either the Fisher exact test or chi-square test for categorical variables.
Multivariate Model Based on the Discovery Cohort and External Validation Cohort
A multivariate logistic regression was used to model each outcome as a function of GI symptoms and clinical variables including age, sex, body mass index (BMI), and comorbidities. Significant associations were determined based on the 95% confidence interval based on 1000 bootstrap iterations (Supplementary Methods).
Predictive Performance Based on the Internal Validation Cohort
Only age and BMI were adjusted for, because they were the only variables significantly associated with both outcomes across different GI symptom models in the discovery cohort (Supplementary Table 9). Then, the estimated model was used to predict the outcome of patients in the internal validation cohort.
Average Treatment Effect
The average treatment effect (ATE) of GI symptoms on COVID-19 outcomes was estimated via the tmle (target maximum likelihood estimation) package available in R Cran.10
Quantification of SARS-CoV-2 Nasopharyngeal Viral Loads
SARS-CoV-2 viral loads were determined as previously reported11 (Supplementary Methods).
Inflammatory Cytokine Panel and Associations With Gastrointestinal Symptoms
The Ella (ProteinSimple) cytokine platform was used to measure tumor necrosis factor (TNF) α, interleukin (IL) 6, IL8, and IL1β.8 Unpaired 2-tailed t tests were used to compare individual cytokines quantified by the ELLA panel between GI symptomatic and asymptomatic groups. P values were adjusted via Benjamini-Hochberg.12
Multiplexed Proteomic Assay (Olink)
A multiplexed proteomic inflammation panel (Olink, 92 inflammation-related proteins) was used to quantify circulating cytokines using an antibody-mediated proximity extension-based assay. The Benjamini-Hochberg procedure was used to adjust P values for multiple testing.
Consensus Clustering of Olink Data and Defining Associations With Gastrointestinal Symptoms
Consensus clustering was performed on the abundance of the 92 cytokines across all 238 samples using the R package ConsensusClusterPlus.13 Associations between GI symptoms and Olink proteomic data were derived using unpaired t tests comparing the symptomatic and asymptomatic groups. P values were adjusted via Benjamini-Hochberg (10% false discovery rate [FDR] threshold of significance).
Data and Materials Availability
Data and materials will be made available upon request.
Results
The Gastrointestinal Tract Was Endoscopically Uninflamed in Patients With COVID-19
Twenty patients with COVID-19 and 10 uninfected control individuals underwent esophagogastroduodenoscopy, colonoscopy, or both (Supplementary Tables 1 and 2). Patient 10 was excluded after multiple negative SARS-CoV-2 nasopharyngeal (NP) PCR test and negative COVID-19 antibody test results. COVID-19 case patients and control individuals in the biopsy cohort were comparable for age, sex, rates of hospitalization, and relevant comorbidities (Supplementary Table 1). Of the patients with COVID-19, 12 were classified as asymptomatic/mild/moderate and 7 as severe (Supplementary Tables 1 and 2). GI biopsies were performed after 25.9 ± 30.3 days from last positive NP swab result. Of the 19 patients, 12 (63%) had a positive SARS-COV-2 PCR swab result most proximal to their biopsy, whereas 7 (37%) had a negative swab result (after previously being positive) (Figure 1 A and Supplementary Table 2). COVID-19 treatment regimens and presence of GI symptoms are detailed in Supplementary Table 2. Sample allocation for different assays is detailed in Supplementary Table 2 and Supplementary Figure 1.
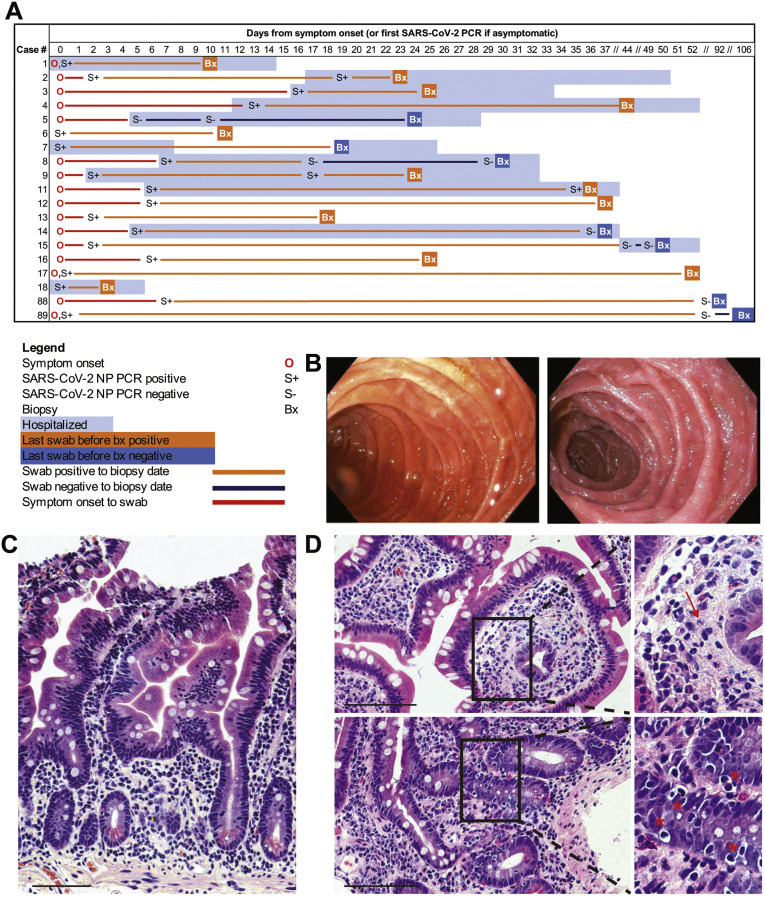
Figure 1.
Clinical timing, endoscopic findings, and histologic features in the small intestines of patients with COVID-19. (A) Timing of GI evaluation with respect to COVID-19 disease course. (B) Representative endoscopic images of the duodenum in patients with COVID-19 (left) and control individuals (right). (C) Histologically normal duodenal tissue in a patient with COVID-19. (D) Histologic signs of inflammation detected in duodenal biopsy samples of patients with COVID-19, including neutrophils (arrow) and increased intraepithelial lymphocytes (∗). Scale bar, 100 μm.
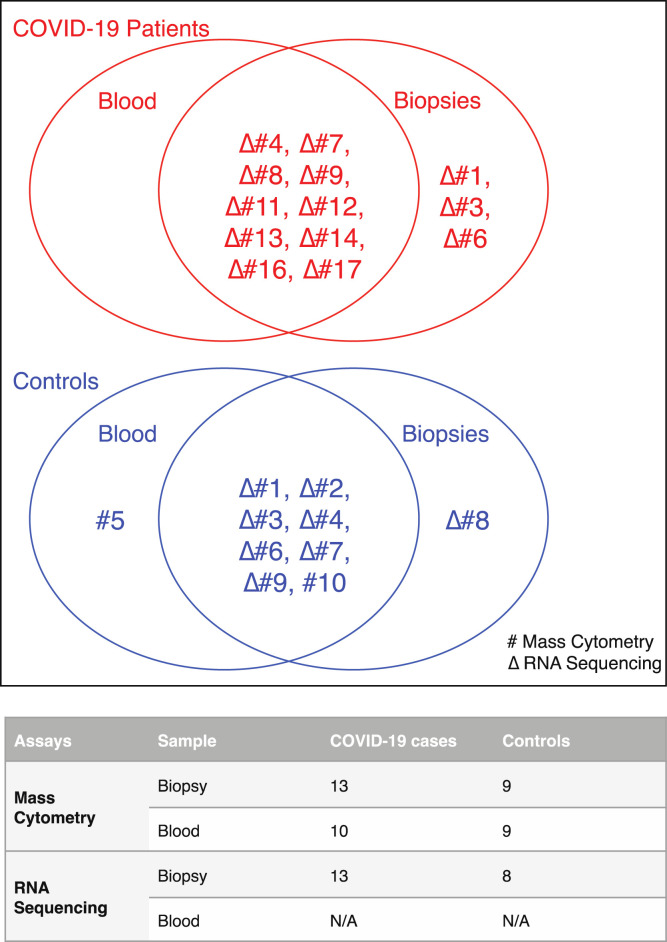
Supplementary Figure 1.
Sample allocation for different assays in patients with COVID-19 and control individuals. Venn diagrams showing blood and biopsy samples used for mass cytometry (#) and RNA sequencing (Δ) in patients with COVID-19 (red) and control individuals (blue). The numbers in the Venn diagrams refer to respective patient and control cases detailed Supplementary Table 2. The table summarizes the total number of blood and biopsy samples allocated for mass cytometry and RNA-seq. N/A, not applicable.
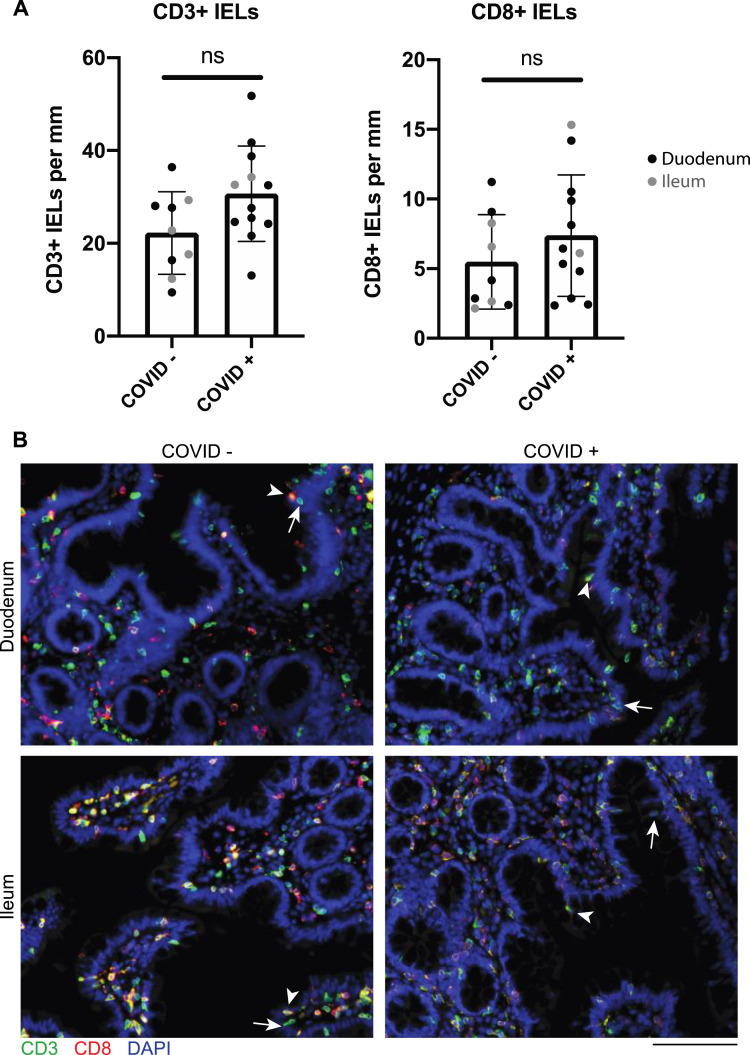
The GI mucosa was endoscopically uninflamed in all individuals (Figure 1 B), except for 1 case where inflammation was attributed to transplant rejection. Histology was normal in 7 of the 17 cases examined, and the remaining (n = 10) patients had a mild increase in intraepithelial lymphocytes (IELs) and/or a scant neutrophilic infiltration (Figure 1 C and D and Supplementary Figure 2). CD3+CD8+ IELs and CD3+CD8– IELs were not significantly different in case patients (n = 12: 10 duodenum, 2 ileum) compared to control individuals (n = 9: 5 duodenum, 4 ileum) (Supplementary Figure 3).
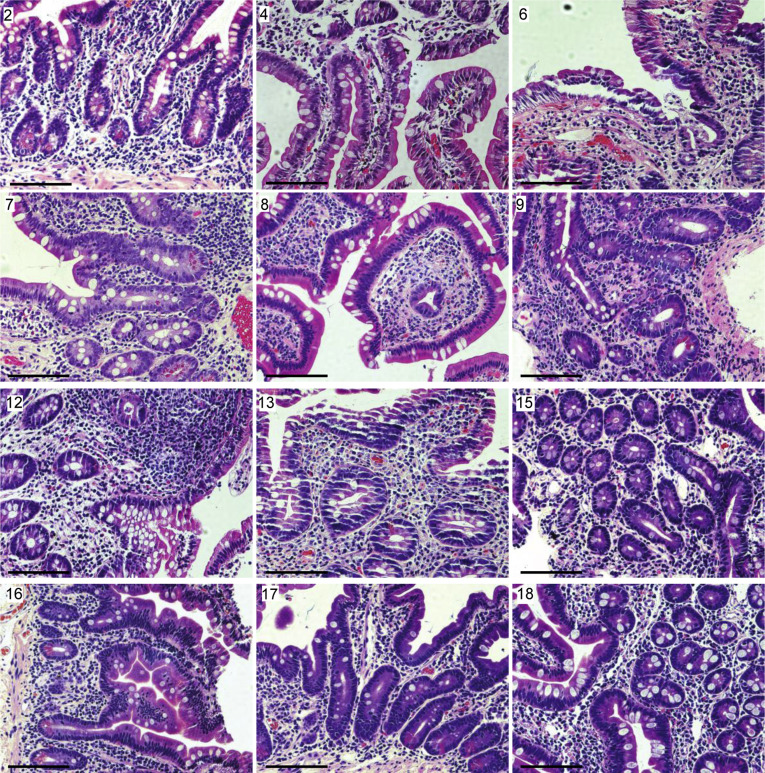
Supplementary Figure 2.
Representative H&E staining of small intestinal biopsy specimens of patients with COVID-19. Patient number in the top left corner corresponds with the patient number in Supplementary Table 2. All biopsy specimens are duodenal with the exception of patient 12, which is from the terminal ileum. Scale bar: 100 μm.
Supplementary Figure 3.
IELs are not increased in small intestinal biopsy samples from patients with COVID-19 compared to control individuals. (A) CD3+ and CD8+ IELs per millimeter of epithelium in patients with COVID-19 and uninfected control individuals in the duodenum (black) and ileum (gray). P values generated from unpaired t tests. (B) Representative IF images of small intestinal biopsy samples showing CD3 (green), CD8 (red), and DAPI (blue). Representative CD8+ IELs (arrowhead) and representative CD8– IELs (arrow) are indicated. Scale bar: 100 μm. ns, not significant.
Small Bowel Intestinal Epithelial Cells Have Robust Expression of Angiotensin-Converting Enzyme 2 and Harbor SARS-CoV-2 Antigens
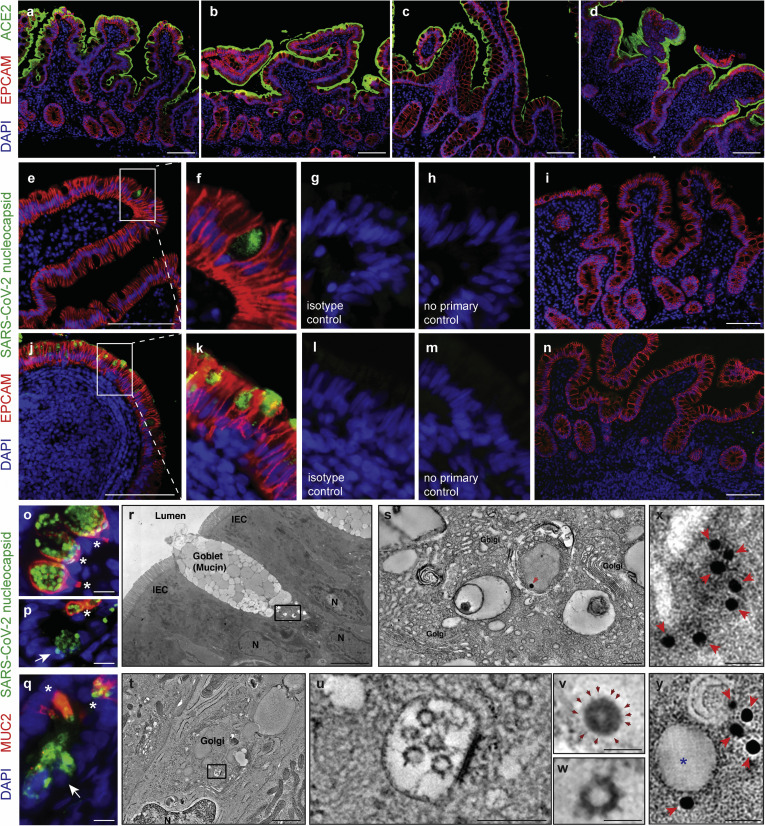
Robust expression of angiotensin-converting enzyme (ACE) 2 was noted on the small intestinal brush border in both control individuals and COVID-19 patients (Figure 2 A–D). Additionally, we detected SARS-CoV-2 nucleocapsid protein in small intestinal epithelial cells of 11 of 12 patients with COVID-19 tested (Figure 2 E–H and J–M, Supplementary Figure 4, and Supplementary Table 4), indicative of virus infection in these cells. When present, the distribution of viral antigens was exclusively seen in the epithelium and was patchy in the upper small intestines (duodenum, Figure 2 E–H) but diffuse in the lower small intestines (ileum, Figure 2 J–M). The presence of viral antigens on immunofluorescence (IF) did not correlate with the presence of histologic abnormalities. To further define viral nucleocapsid protein positive cells, costaining with MUC2 to define goblet cells14 was performed. Viral nucleocapsid primarily colocalized with MUC2, representing infected goblet cells (Figure 2 O–Q). There were a few cells positive for the viral nucleocapsid protein but negative for MUC2 that tended to be located at the base of the crypts (Figure 2 P and Q). The more diffuse viral antigen staining in the ileum as compared to the duodenum is not explained by apparent differences in ACE2 protein expression (Figure 2 A–D); however, it may be explained by increased goblet cells in the ileum,15 and these data appear to be consistent with organoid cultures.4 As negative controls, 5 duodenal and 6 ileal biopsy samples from 10 patients collected before the pandemic (Supplementary Table 5) showed no evidence of viral antigens (Figure 2 I and N and Supplementary Figure 5).
Figure 2.
SARS-CoV-2 viral particles and protein are detectable in intestinal tissues of patients with COVID-19. (A–H) IF staining of (A, B) duodenal and (C, D) ileal biopsy samples of (B, D) patients COVID-19 and (A, C) controls with ACE2 (green), EPCAM (red), and DAPI (blue). (E–N) IF staining of (E–I) duodenal and (J–N) ileal biopsy samples from (E–H, J–M) patients and (I, N) control individuals with SARS-CoV-2 nucleocapsid (green), EPCAM (red), and DAPI (blue) including (G, L) isotype and (H, M) no primary controls. (O–Q) IF staining of (O, P) duodenal and (Q) ileal biopsy samples of patients with SARS-CoV-2 nucleocapsid (green), MUC2 (red), and DAPI (blue) showing SARS-CoV-2 nucleocapsid in goblet cells (∗MUC2+) and nongoblet epithelial cells (arrows, MUC2–). (R–W) Electron tomography of a duodenal biopsy. (R) Montaged projection overview. (S) Tomographic reconstruction of the region indicated by the rectangle in R showing the goblet cell Golgi region. (V) Detail of the presumptive virion indicated by the red arrow in S. Note the dark nucleocapsid puncta and surface spikes (arrows). (T) Electron tomography of an ileal biopsy from a patient with COVID-19 , montaged tomographic reconstruction of a goblet cell Golgi region. (U) Detail of the region indicated by the rectangle in T, showing a presumptive exit compartment containing 5 presumptive SARS-CoV-2 virions. (W) Detail of a presumptive virion from U; membrane bilayer and surface spikes are evident. The virion structures in R–W are comparable with those from a SARS-CoV-2–infected cultured cell (Supplementary Figure 6 and Supplementary Movies 1 and 2). (X) Projection image of a presumptive SARS-CoV-2 virion within an intestinal epithelial cell from a biopsy obtained from a COVID-19 patient with labelled spike protein by Immuno-EM (arrows).36 Detail of the presumptive virion itself is not apparent in the projection image. (Y) A single slice (approximately 10 nm) from a tomographic reconstruction of the same area shown in X. The spherical shape and membrane bilayer of the presumptive SARS-CoV-2 virion (indicated by ∗) are discernible, with gold particles connoting anti-S labeling localized to the presumptive virion’s outer periphery. Scale bars: 100 μm (A–N), 10 μm (O–Q), 5 μm (R), 0.2 μm (S, U), 1 μm (T), 0.05 μm (V, W), and 0.025 μm (X, Y). DAPI, 4′,6-diamidino-2-phenylindole; EPCAM, epithelial cell adhesion molecule.
Supplementary Figure 4.
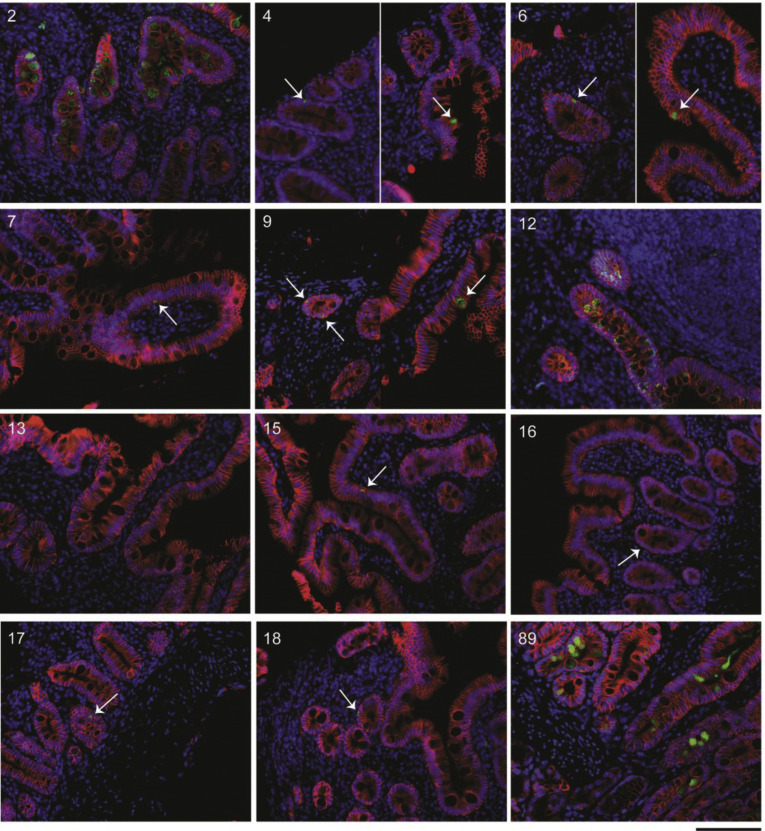
Representative IF images of small intestinal biopsy specimens of patients with COVID-19. SARS-CoV-2 nucleocapsid (green), EPCAM (red), and DAPI (blue) in all patients with COVID-19 where tissue was available for IF. Patient number in the top right corner corresponds with the patient number in Supplementary Table 2. All biopsy specimens are duodenal with the exception of patient 12, which is from the terminal ileum. Patient 8 is missing because of technical difficulties during IF staining. Scale bar: 100 μm.
Supplementary Figure 5.
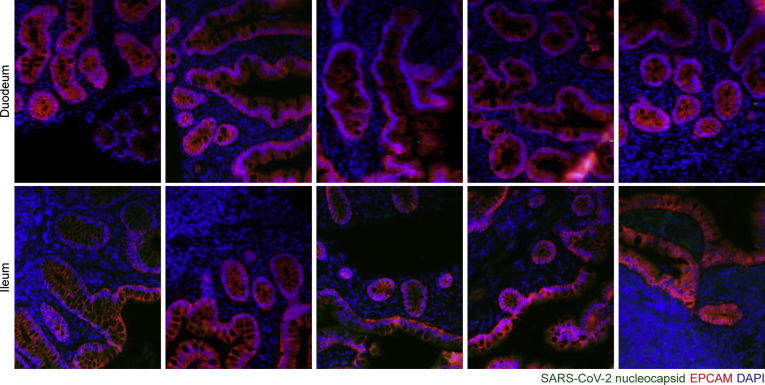
Representative immunofluorescence images of small intestinal biopsy specimens of control individuals. SARS-CoV-2 nucleocapsid (green), EPCAM (red), and DAPI (blue) in duodenal biopsy specimens (upper) and ileal biopsy specimens (lower). Scale bar: 100 μm.
Ultrastructural Analyses of Gastrointestinal Tissues Show Viral Particles in Small Intestinal Epithelial Cells
Next, we performed transmission electron microscopy in 16 patients. Eight of these patients showed presence of 70–110-nm viral particles in the intestinal epithelial cells of the duodenum and/or ileum by transmission electron microscopy (Supplementary Table 4). Representative ET images (Figure 2 R–W and Supplementary Figure 6) showed the presence of viral particles morphologically suggestive of SARS-CoV-2 in the duodenum (Figure 2 R, S, and V) and the ileum (Figure 2 T, U, and W), confirmed with Immuno-EM mouse polyclonal antiserum against SARS-CoV-2 Receptor Binding Domain (RBD) (Figure 2 X and Y). These particles in the exit vesicles of duodenal goblet cells (Figure 2 R, S, and V) are consistent with the colocalization of MUC2 staining using IF.
Supplementary Figure 6.
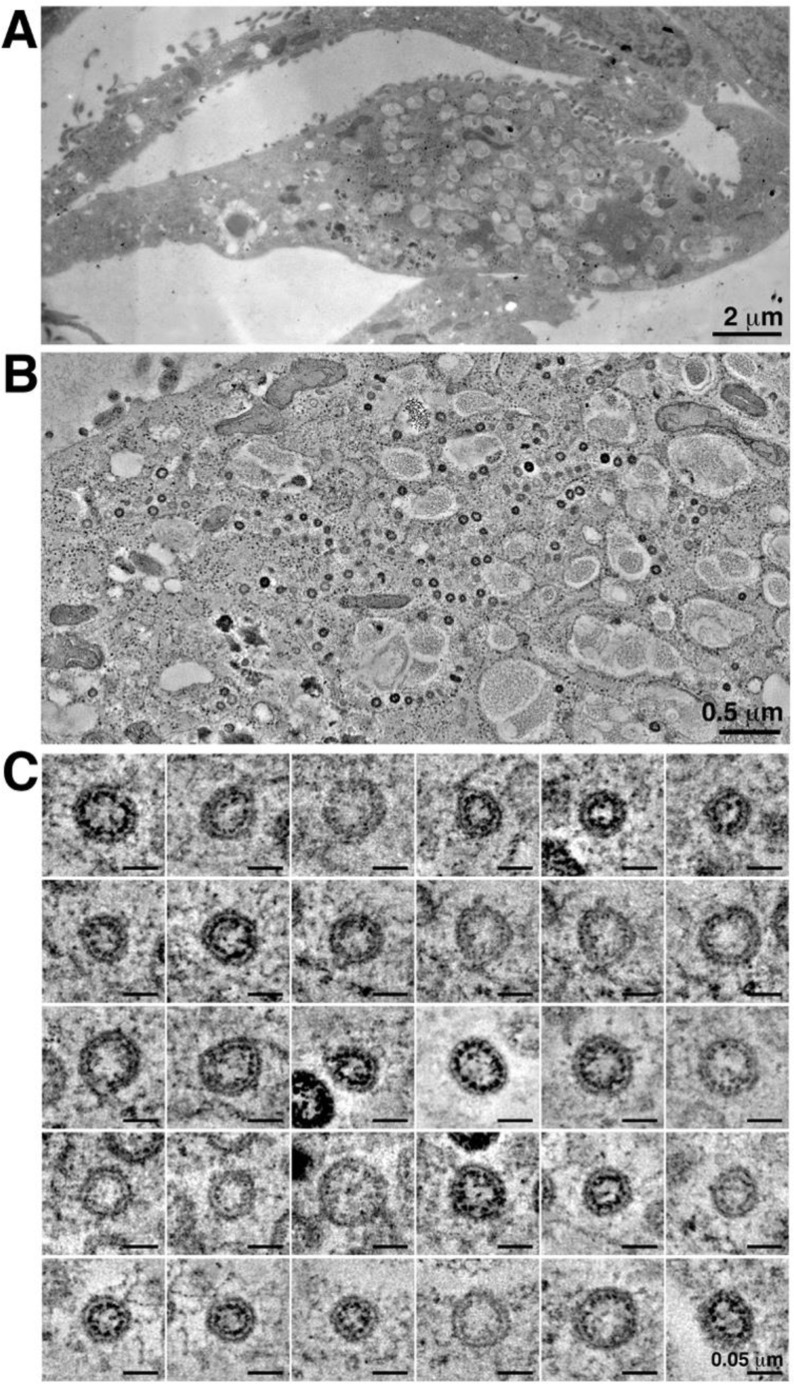
Electron microscopy by high-pressure freezing/freeze substitution fixation of presumptive SARS-CoV-2 infection in culture Vero cells. (A) Montaged overview of an infected cell (150-nm section) (presented for comparison with analogous structures found in tissue samples (Figure 2 and Supplementary Movie 1), which could not be preserved under similar optimal conditions for electron microscopy). The cell exhibits large numbers of cytoplasmic vacuoles, surface blebbing, and general cytopathogenicity. (B) Montaged tomographic reconstruction of the central portion of the cell shown in A. Large numbers of presumptive SARS-CoV-2 virions are contained within cytoplasmic compartments; most are closely adjacent to the compartment’s peripheries. (C) Gallery of 30 individual presumptive SARS-CoV-2 virions taken from the tomogram shown in B. Each example is displayed as an equatorial view with a tomographic thickness of 4.7 nm.
No Infectious Virions Identified in the Gastrointestinal Tissues of Patients With COVID-19
We inoculated Vero E6 cells with the supernatants of homogenized intestinal tissues but did not observe any apparent cytopathic effects or plaque formation after 7 days of culture. In addition, cell culture supernatants did not show the presence of viral RNA by RT-qPCR.
Gastrointestinal Lamina Propria Dendritic Cells Are Depleted in Patients With COVID-19
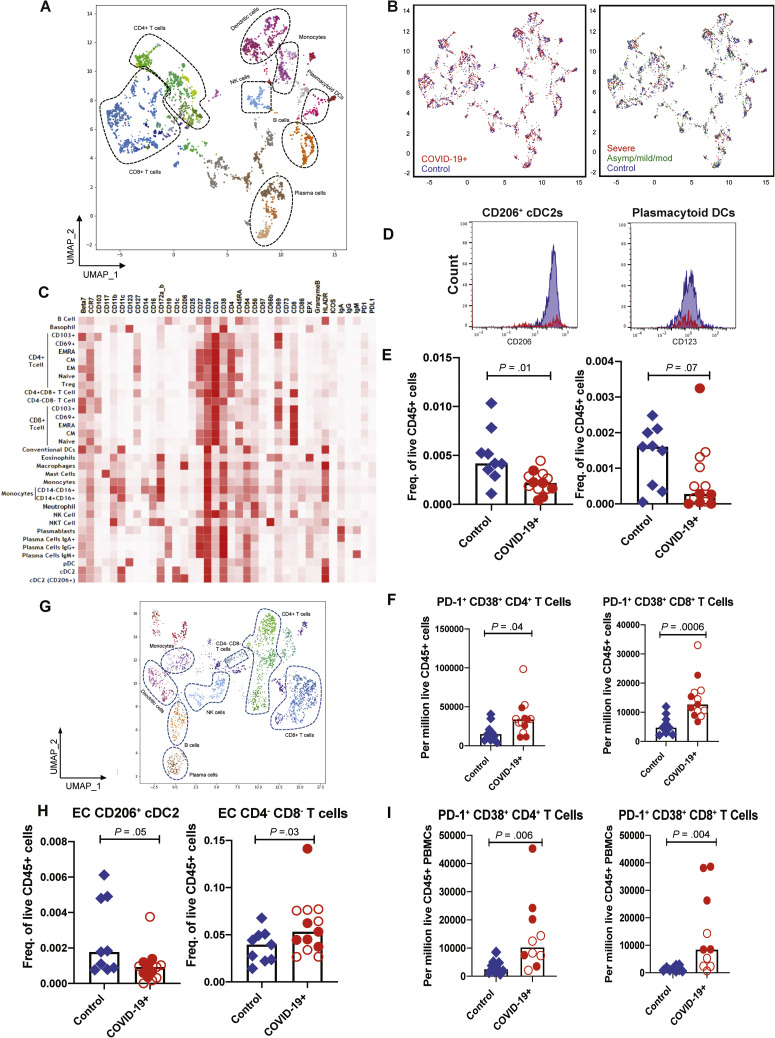
Next, we performed mass cytometry by time of flight (CyTOF) based on immunophenotypic analysis on GI tissue and peripheral blood from a subset of COVID-19 cases (GI tissue, n = 13; blood, n = 10) and control individuals (GI tissue, n = 9; blood, n = 9) (Supplementary Tables 1 and 2 and Supplementary Figure 1). The LP and EC were analyzed separately. Immune populations were clustered on the basis of cell-type specific markers for both the intestinal compartments (LP and EC) and blood (Figure 3 A, C, and G, Supplementary Figures 7 A and 8 A, and Supplementary Data File 1). Although the overall distributions of canonical immune cell subsets in the GI LP were comparable between patients and control individuals (Figure 3 A and B [left]), few immune populations showed differences, as will be detailed. No clear differences in the LP could be discerned based on severity (Figure 3 B [right] and Supplementary Data File 2).
Figure 3.
CyTOF-based analysis identified immune cell signatures in intestinal biopsy samples and blood from patients with COVID-19 and control individuals. Uniform manifold approximation and projection (UMAP) presentation of (A) the 8 clusters of LP immune populations based on 38 markers, (B, left) by infection status with COVID-19 patients (red) and controls (blue) and (B, right) by disease severity with control individuals (blue) and patients with severe (red) and asymptomatic/mild/moderate (green) COVID-19. (C) The heatmap depicting immune populations in the LP based on specific cell-type markers. (D) Representative histograms comparing CD206+ and CD123+ in DC subsets in patients (red) and control individuals (blue). (E) Relative frequencies of CD206+ cDC2 and plasmacytoid DCs in LP of patients and control individuals (unsupervised analysis). (F) Relative frequencies of PD-1+ CD38+ (effector) CD4+ and CD8+ T cells in the LP of control individuals and patients (supervised analysis). (G) UMAP presentation of the 8 clusters of immune populations based on 38 markers in the EC of intestinal biopsy samples. (H) Relative frequencies of CD206+ cDC2 and CD4–CD8– T cells in the EC of control individuals and patients (unsupervised analysis). (I) Relative frequencies of PD-1+CD38+ (effector) CD4+ and CD8+ T cells in blood of control individuals and patients (supervised analysis). Open red circles denote patients with asymptomatic/mild/moderate disease, and filled red circles denote patients with severe COVID-19. Bar plots represent median values. Freq., frequency.
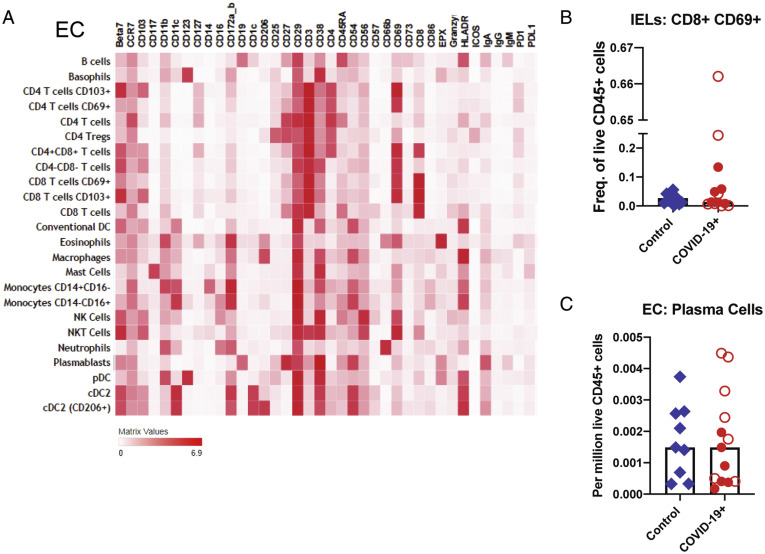
Supplementary Figure 7.
Altered immune populations in the EC of patients with COVID-19 compared to control individuals. (A) The heatmap shows the clustering and distribution of different cell types in the EC. Relative frequencies of (B) IELs and (C) plasma cells in the EC of control individuals and patients with COVID-19. Open red circles denote patients with asymptomatic/mild/moderate disease, and filled red circles denote patients with severe COVID-19. The bar plots show median frequencies. NK, natural killer; NKT, natural killer T.
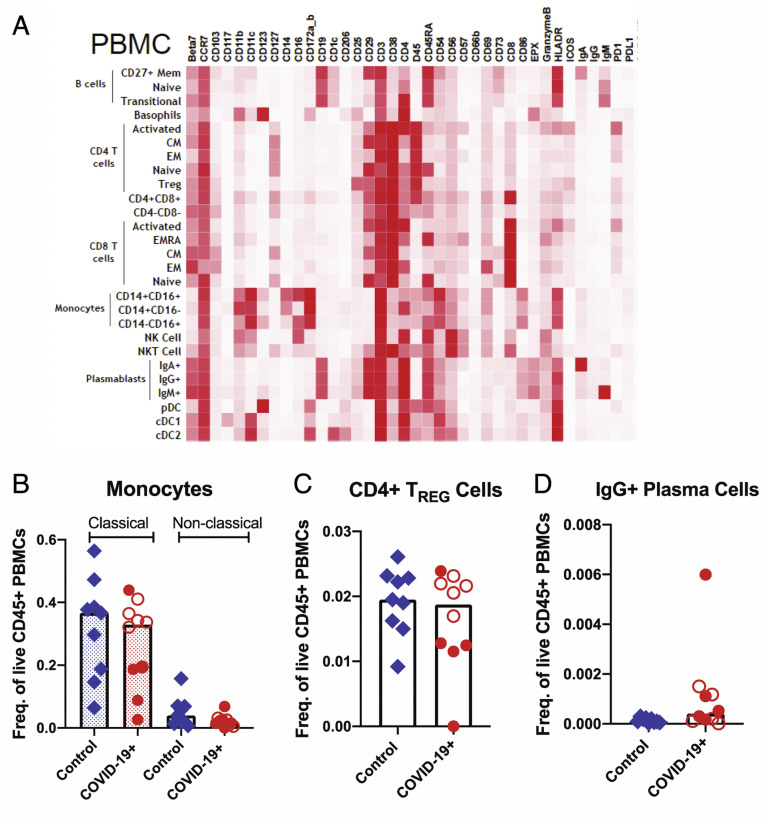
Supplementary Figure 8.
Altered immune populations in the blood of patients with COVID-19 compared to control individuals. (A) The heatmap shows the clustering and distribution of different immune cell types in the blood. Relative frequencies of (B) classical (dotted bars) and nonclassical monocytes (open bars), (C) CD4+ regulatory T cells, and (D) IgG+ plasma cells in the blood of control individuals and patients with COVID-19. Open red circles denote patients with asymptomatic/mild/moderate disease, and filled red circles denote patients with severe COVID-19. The bar plots show median frequencies. CM, central memory; EM, effector memory; PBMC, peripheral blood mononuclear cell.
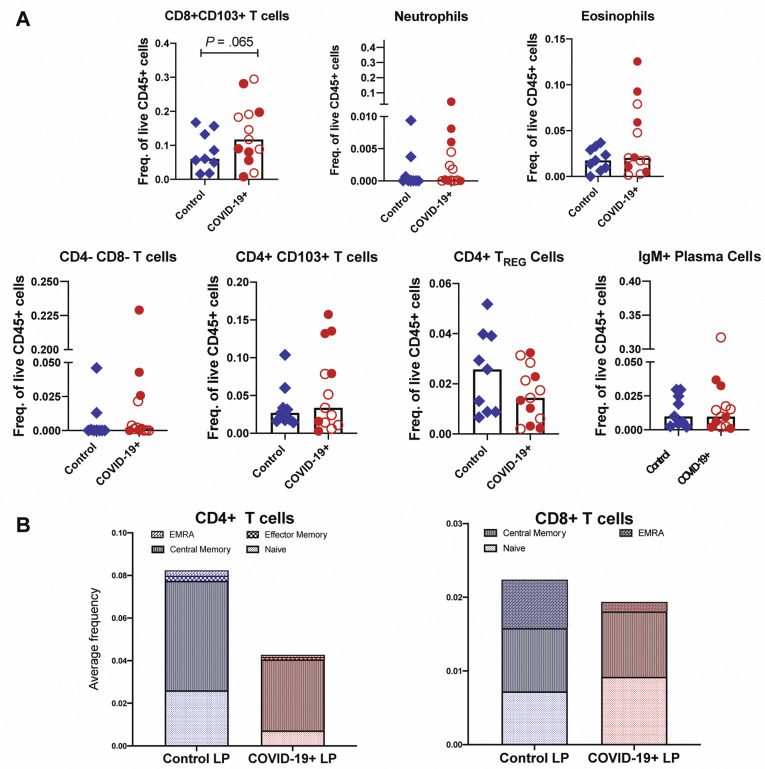
In the LP, CD206+CD1c+ cDC2 (conventional dendritic cells [DCs] 0.4-fold decrease; P = .01) and plasmacytoid DCs (pDCs) were reduced in COVID-19 cases (0.5-fold decrease; P = .07) (Figure 3 D and E), analogous to changes described in the blood.16 Effector (PD-1+CD38+) CD4+ and CD8+ T cells (Figure 3 F) and CD8+CD103+ T cells (tissue resident memory) (Supplementary Figure 9 A) were increased in patients compared to control individuals (1.7-fold increase; P = .06). In the EC, there was a decrease in CD206+cDC2 (0.4-fold decrease; P = .05) and an increase in CD4–CD8– IELs (1.6-fold increase; P = .03) in patients compared to control individuals (Figure 3 H). Alterations in other immune populations in the LP and EC are shown in Supplementary Figures 7 and 9, respectively.
Supplementary Figure 9.
Altered immune populations in the LP of patients with COVID-19 compared to control individuals. (A) Relative frequencies of LP immune cells in control individuals and patients with COVID-19. Open red circles denote patients with asymptomatic/mild/moderate disease, and filled red circles denote patients with severe COVID-19. The bar plots show median frequencies. (B) The stacked bar graphs show the distribution of average frequencies of naive and memory CD4+ and CD8+ T cells in the LP of patients with COVID-19 and control individuals. EMRA, effector memory T cells that re-express CD45RA; Freq., frequency; TREG, T regulatory.
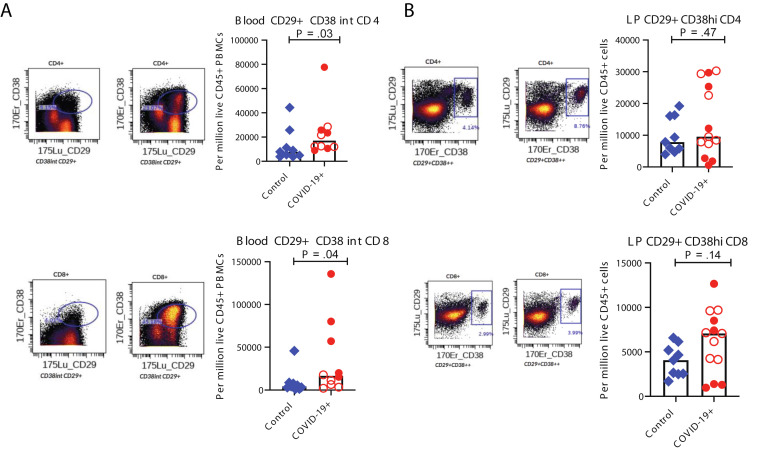
Among peripheral blood mononuclear cells, effector (PD-1+CD38+) CD4+ and CD8+ T cells were significantly increased in patients (Figure 3 I). Alterations in monocytes, regulatory T cells, and IgG+ plasma cells are shown in Supplementary Figure 8. Finally, a significant increase in activated (CD29+CD38+) CD4+ T cells was noted in PBMCs (Supplementary Figure 10 A) and a nonsignificant increase of these activated T cells in the LP of patients compared to control individuals (Supplementary Figure 10 B). Details of all immune population changes are provided in Supplementary Data File 2.
Supplementary Figure 10.
Altered T-cell populations in blood and intestinal biopsy samples of patients with COVID-19 compared to control individuals based on supervised analysis. Representative CyTOF plots and bar plots comparing the frequencies of CD29+CD38+CD4+ and CD29+CD38+CD8+ T cells in (A) the blood and (B) LP of control individuals (blue) and patients with COVID-19 (red). Open red circles denote patients with asymptomatic/mild/moderate disease, and filled red circles denote patients with severe COVID-19. The bar plots show median frequencies. CyTOF, cytometry by time of flight.
Altogether, intestinal tissues of COVID-19 patients showed altered distribution of immune cell subsets, most notable for reduced frequencies of CD206+CD1c+ cDC2 and pDCs and an increased frequency of effector T cells.
Gastrointestinal Lamina Propria Proinflammatory Pathways Are Down-regulated in Patients With COVID-19
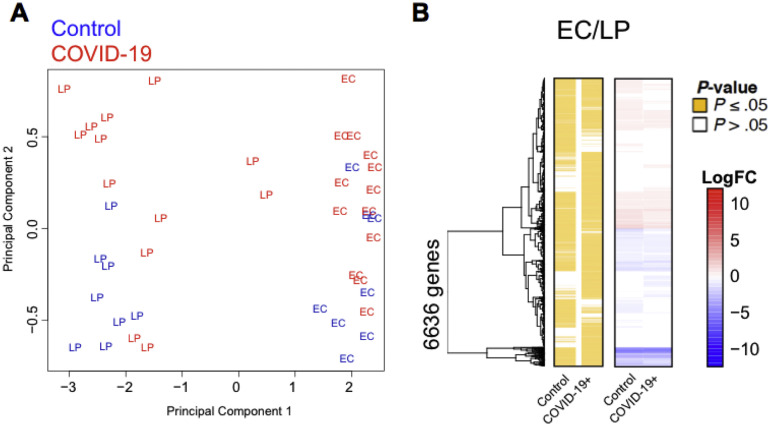
Next, we performed RNA-seq on the EC and LP in 13 patients with COVID-19 and 8 control individuals. The EC and LP clustered separately on the basis of their top transcriptional signatures, showing distinctness of the 2 compartments (Supplementary Figure 11 and Supplementary Data File 3). A total of 1063 differentially expressed genes (DEGs) were identified out of total 11,419 genes detected (Figure 4 A and Supplementary Data File 3). The majority of DEGs were detected in the LP (n = 1061; FDR,17 ≤0.05) compared to 12 DEGs in the EC that largely overlapped with the LP (Figure 4 A). Both the LP and EC showed up-regulation of the genes involved in immunomodulation, including the antimicrobial peptide LCN2 and the metallothioneins MT1E, MT1F, MT1H, MT1M, MT1X, MT2A, and TMEM107. In addition, heat shock proteins HSPA1A and HASPA1B were down-regulated in both compartments. Pathway enrichment analysis of DEGs ranked by significance showed several Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways that were depleted in patients compared to control individuals (Figure 4 B), including pathways linked to T helper (Th) type 17 cell differentiation and inflammatory bowel diseases, which are characterized by the depletion of RORA, IL4R, IFNG, IL18R1, IL1B, STAT4, and HLA-DRA. Pathways linked to antigen processing, Th1 and Th2 cell differentiation, and MAPK signaling were significantly down-regulated in the LP from patients. In contrast, genes associated with amino acid metabolism (NOS2, SMS, ALDH2, and GOT2), mineral absorption (MT1G, MT2A, and MT1E), and mucin biosynthesis (GALNT7, GALNT3, and GALNT8) were significantly up-regulated in patients compared to control individuals (Figure 4 B).
Supplementary Figure 11.
Distinct expression profiles in the intestinal EC and LP. (A) Principal component analysis of EC and LP fractions of patients with COVID-19 and control individuals. The 2 tissue fractions separate on principal component 1 (x-axis). (B) Hierarchical clustering of the average expression changes for 6636 genes (rows) characterizing the EC (red) or LP (blue) fractions (FDR, ≤0.05) in the intestinal biopsy samples of patients with COVID-19 and control individuals. The left panel indicates significant genes in yellow for each tissue compartment. The color bar (right) indicates the average log2(FC).
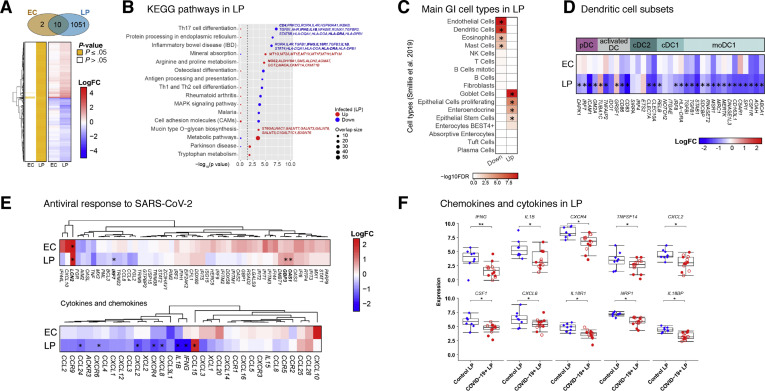
Figure 4.
Transcriptional changes in intestinal biopsy samples from patients with COVID-19 compared with control individuals. (A) Hierarchical clustering of average expression changes for 1063 genes (rows) with induced (red) or depleted (blue) expression (FDR, ≤0.05) in the EC and LP of intestinal biopsy samples from patients with COVID-19. The panel on the left indicates significant genes for each tissue fraction in yellow. The color bar indicates the average log2 fold change (FC). (B) The top enriched pathways (KEGG) that are induced (red) or depleted (blue) in the LP of patients with COVID-19 are displayed. The dashed line indicates the P ≤ .05 cutoff. Gene names are indicated for main pathways. (C) Deconvolution of main gastrointestinal cell types enriched or depleted in the LP of patients with COVID-19 compared with control individuals. Reference single cell RNA–seq cell-type signatures were taken from Smillie et al18 (P ≤ .05, Fisher exact test). (D) Average expression changes for DC markers in the EC and LP. Reference small conditional RNA–seq cell-type signatures were taken from Martin et al. The color bar indicates the average log2(FC). (E) Hierarchical clustering of average expression changes (columns) in the EC and LP for genes related to antiviral response to SARS-CoV-2 in post mortem lung tissue of patients with COVID-19, as described by Blanco-Mello et al (top) and for cytokines and chemokines (bottom). The color bar indicates the average log2 FC. (F) The gene expression levels for the top 10 significant chemokines and cytokines in the LP of patients with COVID-19 and control individuals. ∗P < .05, ∗∗P < .01. moDC, monocyte-derived dendritic cell.
We considered the possibility that the observed expression changes could imply alterations in relative cell type proportions (in addition to transcriptional alterations within cells). Therefore, we interrogated data derived from single-cell RNA-seq18 for enrichment of cell type–specific gene expression signatures. Consistent with our CyTOF data (Figure 3 and Supplementary Data Files 1 and 2), genes associated with DCs and eosinophils were reduced in patients compared to control individuals (Figure 4 C). Additionally, signatures related to the size of endothelial cell and mast cell pools were reduced, whereas genes linked to goblet cells, proliferating epithelial cells, enteroendocrine cells, and epithelial stem cells were increased, possibly reflecting the sequelae of intestinal epithelial infection by SARS-CoV-2 and subsequent recovery (Figure 4 C).
We probed myeloid gene signatures further and found significant down-regulation of genes associated with pDCs (DAPK1, IRF7, ICAM1, and GM2A), activated DCs (TNFAIP2, CD86, and CD83), cDC1 (RELB, IRF8, and HLA-DRA), and cDC2 (CLEC7A and CLEC10A). Additionally, LP genes associated with inflammatory DCs (monocyte-derived DCs) (TGFBI, TGFB1, STAB1, SDCBP, RNASET2, MSR1, MRC1, MERTK, DNASE1L3, CD163L1, C5AR1, SPI1, CSF1R, AOAH, and ABCA) were significantly reduced (Figure 4 D), consistent with our CyTOF results.
Next, we looked at the average EC and LP expression of recently reported gene signatures linked to the antiviral response against SARS-CoV-2 from post mortem lung tissue7 and human intestinal organoids.5 Although we did not observe a substantial acute SARS-CoV-2 response, there was significant up-regulation of LCN2 in both EC and LP and of OAS and GBP3 in LP only. Notably, we observed a trend toward induction of antiviral response genes in the EC, where expression levels of canonical antiviral genes such as IFI44L, IFIT1, IFITM3, IFI44, IFI6, and OAS3 was increased (Figure 4 E, top).
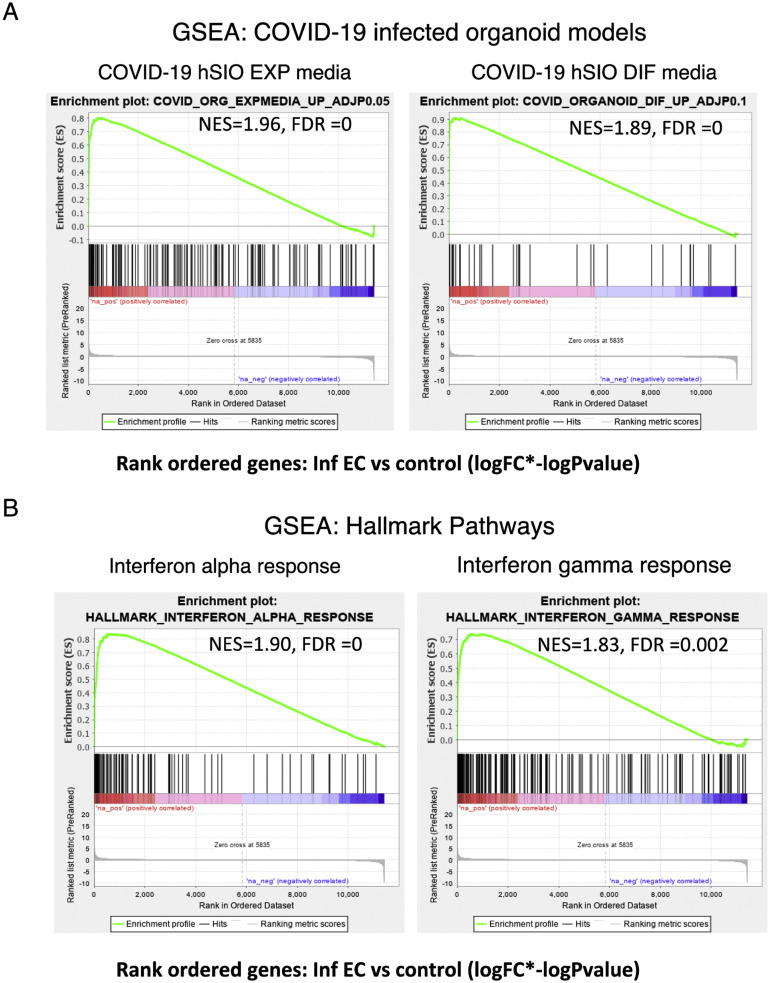
Finally, using gene set enrichment analysis, we rank-ordered the EC DEGs according to effect size (log[fold change] × –log[P value]) and tested for enrichment in the reported SARS-CoV-2–infected organoid gene signatures5 (Supplementary Figure 12 A). The genes up-regulated in the EC of patients were significantly enriched in the SARS-COV-2–infected organoid gene data sets. Hallmark pathway enrichment analyses on this ranked EC gene list showed that the top 2 processes associated with genes up-regulated in EC were the interferon alpha response (normalized enrichment score, 1.91; FDR, <0.005) and interferon gamma response (normalized enrichment score, 1.8; FDR, 0.005) (Supplementary Figure 12 B), indicative of the host antiviral response against SARS-CoV-2 in the EC.
Supplementary Figure 12.
Immune signatures in the EC of patients with COVID-19. GSEA was performed using a rank-ordered list of genes differentially expressed in the infected EC vs control EC. The metric for ranking was log(FC) × –log(P value). (A) GSEA was performed on the rank-ordered EC gene set using SARS-CoV-2–infected organoid data sets. The gene sets tested were molecular signatures curated from SARS-CoV-2–infected organoid experimental data sets using hSIOs grown in either (1) Wnt high expansion (EXP) medium (at adjP < .05) or (2) differentiation (DIF) medium (at adjP < .1). Only gene sets significantly enriched (at FDR of <0.05) are displayed. (B) GSEA was performed for the same rank-ordered EC gene set using the Hallmark pathway data sets. Two significantly enriched pathways were found to be associated with up-regulated genes in infected EC relative to control individuals (at FDR of <0.05). Normalized enrichment score (NES) and FDR values are as indicated.
Projection of our RNA-seq data set on SARS-CoV-2–infected human bronchial epithelial cells7 showed that several inflammatory cytokines and chemokines such as IL-1β, IFN-γ, CCL24, and CXCL8 were down-regulated in the intestines of patients with COVID-19 (Figure 4 E, bottom). The only chemokine significantly increased was CCL15, which is structurally similar to antimicrobial peptides and has a role in maintaining intestinal homeostasis19 (Figure 4 E, bottom). Key inflammatory genes, including IFNG, IL1B, CXCR4, TNFSF14, CXCL2, CSF-1, CXCL8, IL18R1, NRP1, and IL18BP, were down-regulated in the LP of patients compared to control individuals (Figure 4 F).
Together, these data show a dynamic remodeling of GI tissues by SARS-CoV-2, notably with a significant down-regulation of pathways associated with inflammation and antigen presentation in the LP with a concomitant activation of antiviral response signaling genes in the EC.
Clinical Impact of Gastrointestinal Involvement During COVID-19: Frequency of Gastrointestinal Symptoms in a Discovery Cohort
Given the observed down-regulation of key inflammatory genes, we hypothesized that intestinal involvement in COVID-19 is associated with a milder disease course. We tested this hypothesis in a discovery cohort consisting of 634 patients with COVID-19 hospitalized at MSH and meeting inclusion criteria (Supplementary Figure 13). Demographics (sex, age, and race/ethnicity) and clinical variables, including the presence of comorbidities and COVID-19 severity, were analyzed (Supplementary Table 6). Next, we recorded the presence of GI symptoms (diarrhea, nausea, vomiting) present at the time of hospital admission to avoid iatrogenic confounders. Overall, 299 patients (47%) reported any of the GI symptoms (nausea, vomiting, and/or diarrhea), with diarrhea being the most common (245 [39%] patients), followed by nausea (157 [25%] patients), and then vomiting (82 [13%] patients) (Supplementary Table 6).
Supplementary Figure 13.
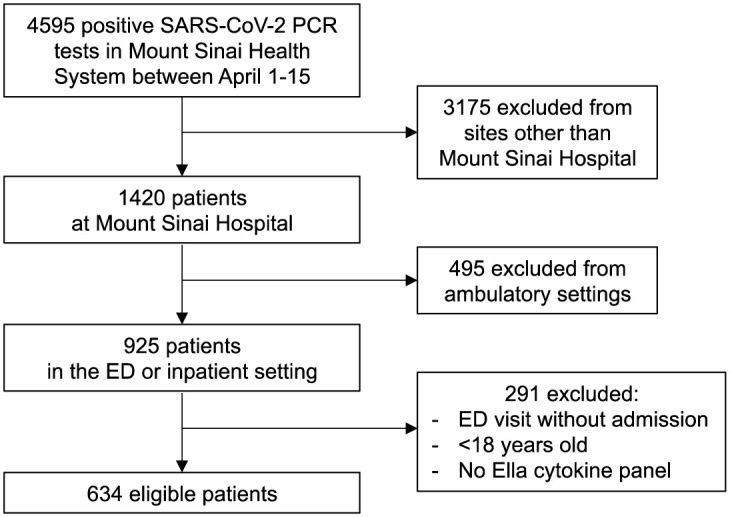
Flow diagram of the discovery cohort. The diagram shows the total number of patients admitted to the Mount Sinai Health System between April 1 and 15, 2020, and the selection process that was adopted to select patients in the discovery cohort. ED, emergency department.
COVID-19 Severity Is Significantly Reduced in Patients With Gastrointestinal Symptoms When Compared to Those Without Gastrointestinal Symptoms in Multivariate Analysis
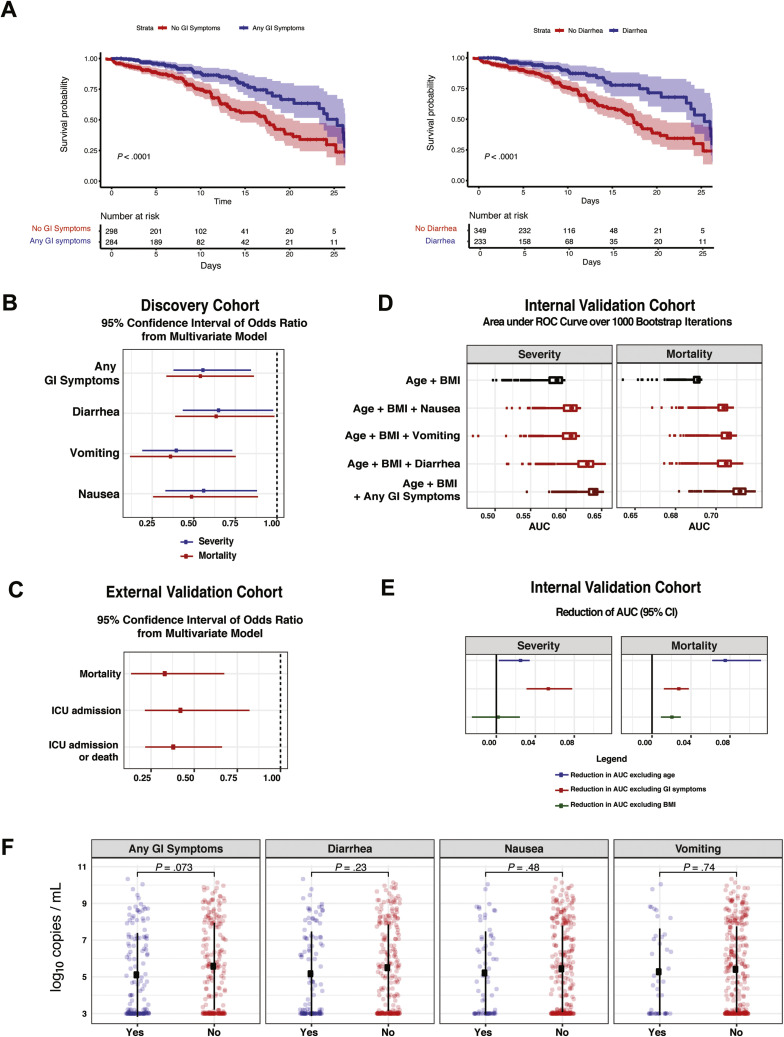
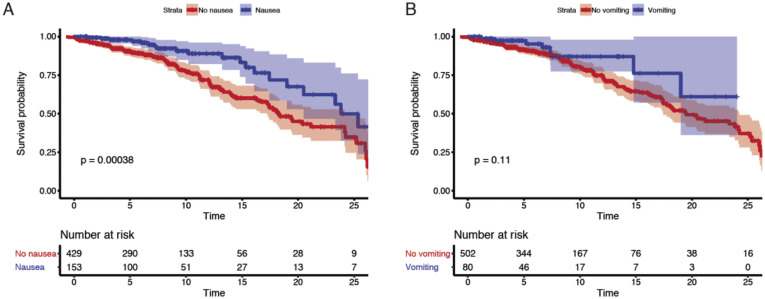
Among the discovery cohort, 54 (9%) patients had mild disease, 361 (57%) had moderate disease, 158 (25%) had severe disease, and 61 (10%) had severe COVID-19 with end organ damage (Supplementary Tables 3 and 6). A total of 110 patients were admitted to the intensive care unit (ICU) (17%), and 151 patients (24%) died by the end of data collection (Supplementary Table 6). Patients presenting with GI symptoms had less severe disease than patients without GI symptoms (P < .001 chi-square test) (Table 1 ). Notably, only 54 (9%) patients in the entire cohort (31 [10.3%] with and 23 [6.8%] without GI symptoms, respectively) had mild disease on presentation (ie, not requiring any type of supplemental oxygen [peripheral oxygen saturation of >94% on room air] and with no evidence of pneumonia); therefore, a majority of patients with GI symptoms had concomitant respiratory symptoms. Mortality was significantly lower in COVID-19 patients with GI symptoms (15.7%) than those without GI symptoms (31.0%; P < .0001, Fisher exact test) (Table 1). Furthermore, each individual GI symptom (nausea, vomiting, and diarrhea) was associated with less severe disease (P < .02, Fisher exact test) and lower mortality (P < .001, Fisher exact test) (Supplementary Table 7). These findings were further emphasized by Kaplan-Meier estimates of survival over short-term follow-up of 25 days (P < .001, log-rank test) (Figure 5 A and Supplementary Figure 14 A and B). Consistent with prior reports,8 older age and higher disease severity were associated with higher mortality (Supplementary Table 8).
Table 1.
Basic demographics, clinical characteristics and outcomes in patients with and without GI symptoms
| Variable | GI symptoms (n=299) | No GI symptoms (n=335) | P-value |
|---|---|---|---|
| Age (years) | 60.5 ± 15.0 | 67.2 ± 15.7 | <.0001 |
| Male | 168 (56.2) | 201 (60.0) | .33 |
| Race/ethnicities | |||
| Hispanic | 85 (28.4) | 92 (27.5) | .13 |
| African-American | 66 (22.1) | 95 (28.4) | |
| White | 70 (23.4) | 67 (20.0) | |
| Asian | 22 (7.4) | 13 (3.9) | |
| Other | 56 (18.7) | 68 (20.3) | |
| Comorbidities | |||
| Hypertension | 112 (37.5) | 117 (34.9) | .51 |
| Diabetes | 58 (19.4) | 83 (24.8) | .13 |
| Obesity (BMI>30)∗ | 108 (40.6) | 103 (34.1) | .12 |
| Chronic lung disease | 34 (11.4) | 25 (7.5) | .10 |
| Heart disease | 48 (16.1) | 63 (18.8) | .40 |
| Chronic kidney disease | 41 (13.7) | 54 (16.1) | .44 |
| Cancer | 27 (9.0) | 39 (11.6) | .30 |
| HIV | 5 (1.7) | 6 (1.8) | .99 |
| IBD | 4 (1.3) | 3 (0.9) | .71 |
| Disease severity | |||
| Mild | 31 (10.4) | 23 (6.9) | |
| Moderate | 188 (62.9) | 173 (51.6) | |
| Severe | 63 (21.1) | 95 (28.4) | |
| Severe with EOD | 17 (5.7) | 44 (13.1) | .0004 |
| Outcomes | |||
| ICU admission | 45 (15.1) | 65 (19.4) | .17 |
| Mortality | 47 (15.7) | 104 (31.0) | <.0001 |
NOTE. For age, an unpaired 2-tailed t test was performed. For categorical variables, the Fisher exact test or the chi-square test was used, as appropriate. Bolded values represent significant P values <.05.
EOD, end organ damage; IBD, inflammatory bowel disease; SD, standard deviation.
Figure 5.
Patients with COVID-19 with GI symptoms had reduced severity and mortality despite similar NP viral loads compared to those without GI symptoms. (A) Kaplan-Meier curves for survival stratified by any GI symptoms (left) and diarrhea (right) for patients in the discovery cohort. P values from log rank test and 95% CIs of Kaplan-Meier curves are shown. The number of patients at risk are reported for the respective timepoints. (B) 95% CIs of ORs of GI symptoms based on 1000 bootstrap iterations in a multivariate logistic regression for severity (blue) and mortality (red). (C) Validation based on the external cohort. 95% CIs of ORs of the diarrhea covariate based on 1000 bootstrap iterations to capture mortality, ICU admission, and composite outcome of ICU admission or death. Results are based on multivariate models after accounting for confounders including BMI, age, sex, lung disease, heart disease, and hypertension. (D) Validation based on the internal cohort. Boxplot of AUC over 1000 bootstrap iterations to predict mortality and disease severity in the internal validation cohort. (E) 95% CI of the reduction in AUC based on 1000 bootstrap iterations for the model “age + BMI + any GI symptoms” after removing age (blue), GI symptoms (red), and BMI (green). (F) SARS-CoV-2 viral load copies per milliliter (log10 transformed based on N2 primer with the addition of a constant) stratified by GI symptoms. The square corresponds to the average viral load, and the error bars show 1 standard deviation of uncertainty from the mean. P values from 2-tailed unpaired t tests are reported. CI, confidence interval.
Supplementary Figure 14.
Nausea and vomiting were associated with reduced mortality and severity. Kaplan-Meier curves for mortality stratified by (A) nausea and (B) vomiting for patients in the discovery cohort. P values from log rank test and 95% CIs of Kaplan-Meier curves are shown. Below each Kaplan-Meier curve, the number of patients at risk for different timepoints are reported.
Next, we created a multivariate model, adjusting for age, BMI, sex, race/ethnicity, diabetes, hypertension (HTN), chronic lung disease, and heart disease to determine the impact of GI symptoms on COVID-19 outcomes (Table 1). Consistent with the published literature,20 age and BMI were positively associated with COVID-19 severity and mortality (Supplementary Table 9). The presence of any GI symptoms, as well as diarrhea, nausea, and vomiting individually, was inversely associated with COVID-19 severity and mortality (Figure 5 B and Supplementary Table 9). Patients who presented with GI symptoms had 50% reduced odds of having severe disease (odds ratio [OR], 0.56) and death from COVID-19 (OR, 0.54) compared to the patients who presented without GI symptoms (Figure 5 B and Supplementary Table 9).
An External Validation Cohort Further Confirms Decreased Mortality in Patients With COVID-19 With Gastrointestinal Symptoms on Multivariate Testing
Next, we confirmed our findings in an external validation cohort in which GI symptoms on admission were characterized as the presence or absence of diarrhea (Supplementary Table 10). Consistent with the discovery cohort, patients with diarrhea on admission had significantly lower mortality (10.0%) compared to patients without diarrhea (23.7%; P = .008). Additionally, patients with diarrhea had a lower composite outcome of mortality or ICU admission compared to those without diarrhea (20% vs 40%; P = .001) (Supplementary Table 10). On multivariate logistic regression analyses, adjusting for age, sex, BMI, diabetes, chronic heart and lung disease, and other confounders, we observed that the presence of diarrhea on admission was significantly inversely associated with mortality with a median OR of 0.33 over 1000 bootstrap iterations (Figure 5 C). In 270 patients for whom treatment data were available, no specific treatment was associated with GI symptoms (P values, >0.05) (Supplementary Table 11). In addition, diarrhea was significantly associated with mortality after adjustment for all treatments (Supplementary Table 11). Thus, our observations from this external validation cohort were in alignment with those from the discovery cohort.
Presence of Gastrointestinal Symptoms Can Be Used to Predict Reduced Disease Severity and Mortality in Patients With COVID-19
Next, we developed a predictive model based on the discovery cohort and applied it to a distinct internal validation cohort. The inclusion of “any GI symptoms” in a model consisting of age and BMI improved the ability to predict severity and mortality with a median area under the curve (AUC) of 0.64 (age + BMI + any GI symptoms) vs 0.59 (age + BMI) for disease severity and 0.73 (age + BMI + any GI symptoms) vs 0.70 (age + BMI) for mortality (Figure 5 D and Supplementary Table 12). In addition, the effect of GI symptoms, age, and BMI on the AUC was evaluated by excluding each variable one at a time from the model and calculating the consequent reduction in AUC. The exclusion of GI symptoms resulted in a significant reduction in the AUC, with a median value of 0.054 for disease severity and 0.03 for mortality. Notably, the effect of GI symptoms on the AUC was more dramatic than that of age (AUC reduction of 0.054 vs 0.025) for disease severity (Figure 5 E and Supplementary Table 12).
Average Treatment Effect of Gastrointestinal Symptoms on COVID-19 Outcomes
Using causal inference methodology, we quantified the ATE of GI symptoms on COVID-19 outcomes while accounting for potential confounders. We performed this analysis on the MSH cohort, combining the discovery and internal validation cohorts, and on the external validation cohort. The marginal effect of GI symptoms in the MSH cohort was significant for both severity and mortality after adjustment for all confounders (Supplementary Data File 4). Additionally, based on the external validation cohort, the ATE for diarrhea was significant for mortality and the combined outcome of ICU admission or death, but not for ICU admission alone (Supplementary Data File 4). The OR for the marginal treatment effect of diarrhea was 0.9 for mortality in both the MSH and external validation cohorts.
Nasopharyngeal SARS-CoV-2 Viral Loads Are Similar in Patients With and Without Gastrointestinal Symptoms
Given recent reports suggesting that NP SARS-CoV-2 viral loads are correlated with disease outcomes,11 we compared NP viral loads in a subset of the discovery and internal validation cohorts (n = 329, where data available). Patients with and without GI symptoms had comparable SARS-CoV-2 NP viral loads (mean log10 copies/mL, 5.1 [standard deviation, 2.3]) and 5.6 [standard deviation, 2.4], respectively) (P = .07); furthermore, no significant differences were observed for each individual GI symptom (Figure 5 F).
Patients With COVID-19 With Gastrointestinal Symptoms Have Reduced Levels of Circulating Cytokines Associated With Inflammation and Tissue Damage
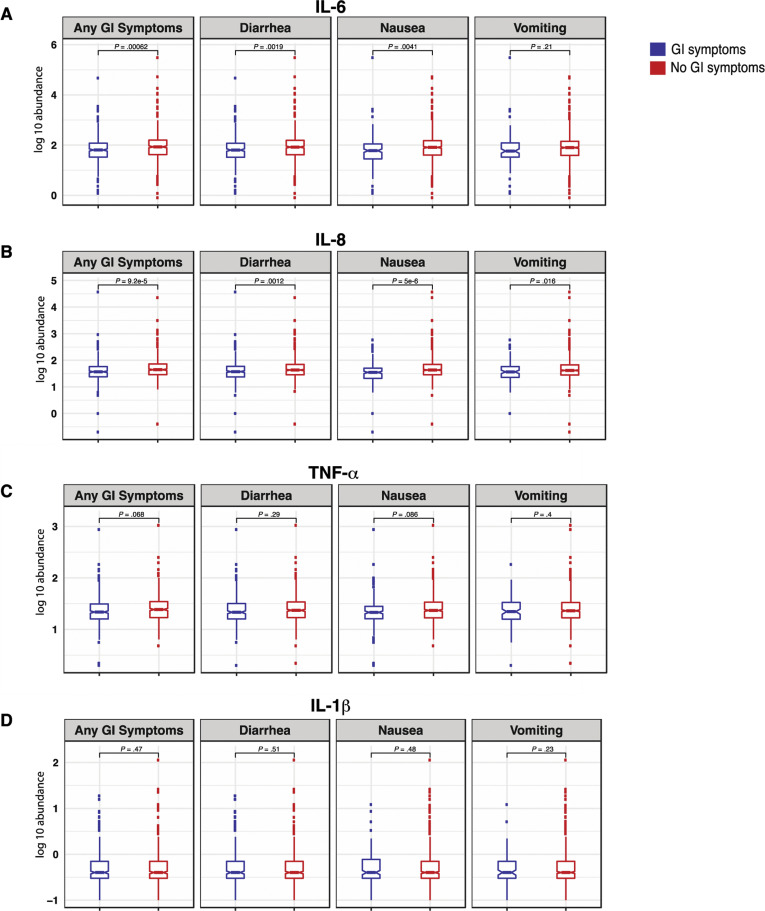
To correlate the observed mortality difference with GI symptoms with known biomarkers for severe COVID-19, we examined IL6, IL8, TNF-α, and IL1β levels measured on admission. IL6 and IL8, which are known to be directly associated with poor survival,8 were found to be significantly reduced in the circulation of patients with GI symptoms (FDR, 10%) (Supplementary Figure 15 and Supplementary Table 13).
Supplementary Figure 15.
Patients with COVID-19 with GI symptoms had reduced levels of circulating IL6 and IL8. (A) IL6, (B) IL8, (C) TNF-α, and (D) IL1β at the time of admission in patients with and without GI symptoms. Boxplots represent the median and interquartile range. P values were calculated using the unpaired 2-tailed t test.
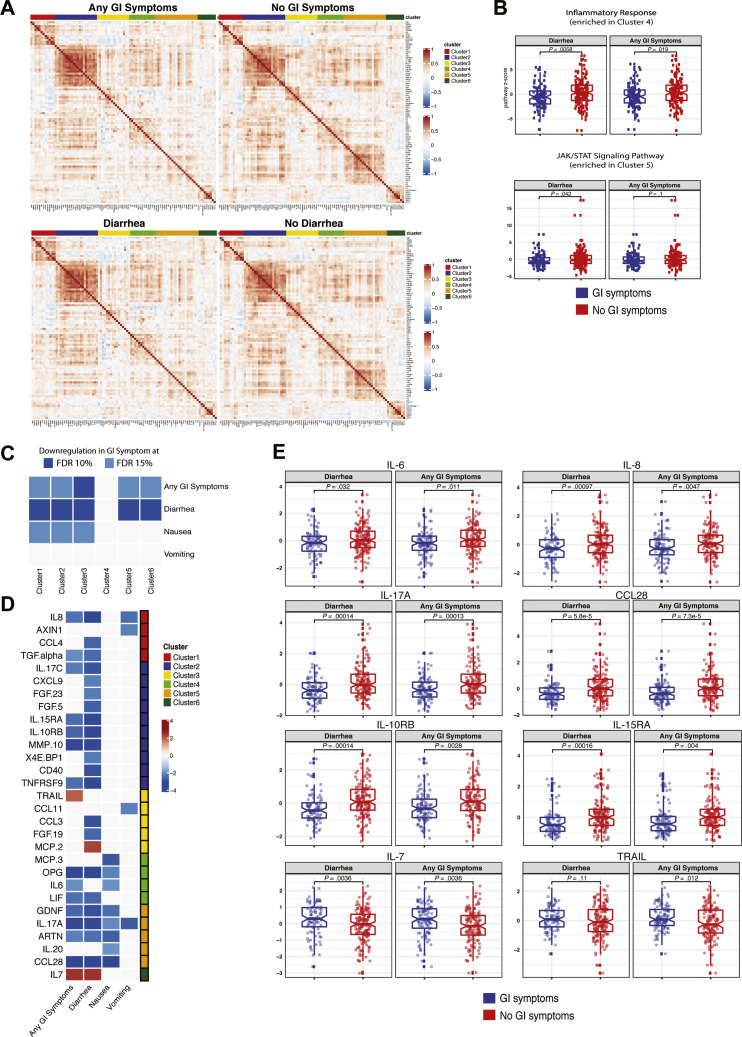
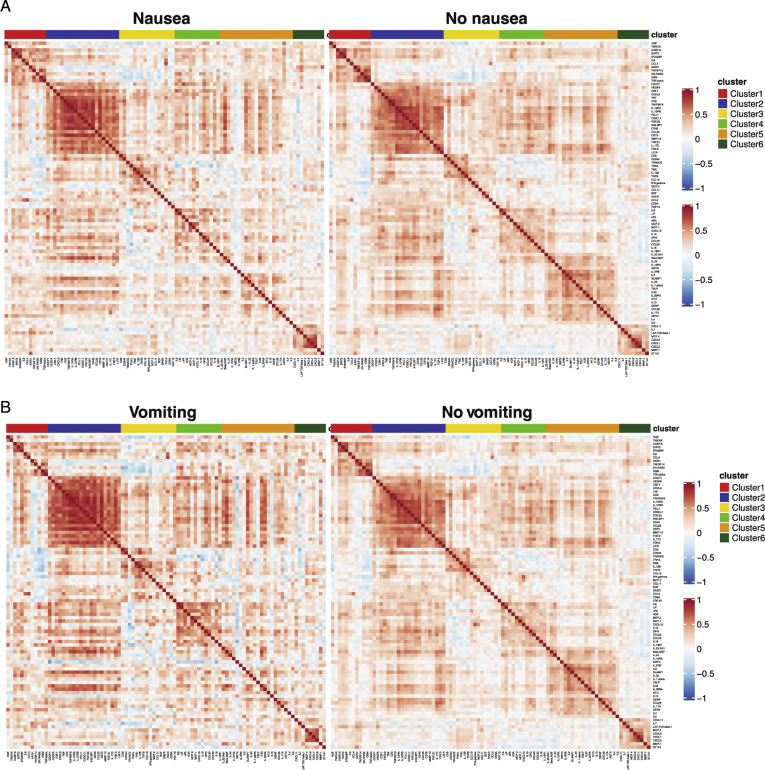
Next, we performed a validated, multiplexed proteomic assay (Olink) in 238 patients (from among the discovery and internal validation cohorts; GI symptoms, n = 104; no GI symptoms, n = 134) for whom serum samples were available for analyses. Unsupervised consensus clustering of 92 analytes showed 6 groups of analytes with similar expression patterns across all patients (Figure 6 A and Supplementary Table 14). Analytes in clusters 5 and 6 displayed less correlation in patients with GI symptoms compared to those without GI symptoms (Figure 6 A and Supplementary Figure 16). The KEGG JAK/STAT signaling pathway was significantly enriched in cluster 5, and the Hallmark inflammatory response pathway was significantly enriched in cluster 4 (Fisher exact test, 10% FDR). These pathways were down-regulated in patients with diarrhea (P < .05 from t test) (Figure 6 B), suggesting a reduced inflammatory response in patients with GI symptoms. Additionally, clusters 1, 2, 3, 5, and 6 were significantly down-regulated in patients with GI symptoms compared to those without (FDR, 15%) (Figure 6 C). This seemed to be driven mostly by diarrhea because the same clusters were significantly down-regulated in patients with diarrhea (FDR, 10%). We observed a similar, albeit reduced, signal for nausea and vomiting, likely because of the smaller sample size (n = 29 for vomiting, n = 54 for nausea).
Figure 6.
Patients with COVID-19 with GI symptoms have reduced levels of circulating inflammatory cytokines. (A) Correlation matrix (Pearson) for 92 markers in the Olink panel across patients with any GI symptoms (top left) compared with no GI symptoms (top right) and patients with diarrhea (bottom left) compared with patients without diarrhea (bottom right). Cluster assignment is reported on the top of the heatmap. (B) Boxplot of Hallmark inflammatory response and KEGG JAK/STAT signaling pathway Z-scores stratified by GI symptoms that were significantly enriched at 10% FDR in cluster 4 and cluster 5, respectively. (C) Significant associations between proteomic clusters and GI symptoms at 10% (dark blue) and 15% (light blue) FDR based on unpaired 2-tailed t test. (D) Analytes associated with GI symptoms at 10% FDR based on unpaired t test. The intensity of the color is proportional to the –log10P value. Negative associations are in blue, and positive associations are in red. On the right side of the heatmap, the cluster assignment for each marker is reported. (E) Boxplots represent the median and interquartile range of select differentially expressed markers stratified by GI symptoms. P values from the unpaired t test are reported.
Supplementary Figure 16.
Correlation matrix (Pearson) for 92 markers contained in the Olink platform. (A) Correlation matrix across patients with nausea (left) compared to patients without nausea (right) and (B) patients with vomiting (left) compared to patients without vomiting (right). Cluster assignment derived using unsupervised consensus clustering is reported on the top of the heatmap.
Key inflammatory cytokines and chemokines were significantly down-regulated (IL8, TGF-α, IL17C, IL15RA, IL10RB, MMP10, TNFRSF9, OPG, IL6, LIF, GDNF, IL-17A, ARTN, and CCL28), whereas TNF-related apoptosis inducing ligand (TRAIL), a cytokine with immune regulatory properties,21 and IL7, a cytokine associated with T-cell development,22 were significantly up-regulated in patients with GI symptoms (t test FDR, 10%) (Figure 6 D and E and Supplementary Table 15).
Overall, GI symptoms are associated with significantly reduced levels of key inflammatory cytokines including IL6, IL8, IL17, and CCL28 that are known to be associated with poor COVID-19 outcomes.
Discussion
Given the robust expression of ACE2 on the small intestinal epithelium, we hypothesized that the intestines would be susceptible to SARS-CoV-2 infection. Here, we detailed for the first time, to our knowledge, SARS-CoV-2 infection of human intestinal epithelial cells in vivo using IF and electron microscopy. Specifically, infected intestinal cells were primarily goblet cells. We also observed a mild inflammatory response in the intestinal tissues despite the presence of SARS-CoV-2 antigens. Finally, we found reduced systemic inflammation and mortality in hospitalized patients with COVID-19 presenting with GI symptoms.
Using multiple approaches, we observed evidence of reduced inflammatory response within the GI tract. This includes a lack of inflammatory monocytes and macrophages and a depletion of DC subsets in the GI tract, which is in contrast to the significant inflammatory response observed in the blood and lungs of patients with severe COVID-19.23 Additionally, the down-regulation of several proinflammatory genes that were found to be elevated in the lungs during SARS-CoV-2 infection24 was observed in GI tissues. Finally, systemic levels of IL6 and IL8, as well as IL1720 and CCL28,25 were lower in hospitalized patients presenting with GI symptoms, despite comparable NP SARS-CoV-2 viral loads. Notably, the reduced circulating IL17 and CCL28 (by Olink) is consistent with our RNA-seq data. The observed attenuation of GI inflammation is in alignment with data from the 2003 severe acute respiratory syndrome epidemic,26 autopsy studies from patients with COVID-19,27 and animal models.28 , 29
In 2 distinct and large cohorts of patients with COVID-19, we observed a significant reduction in mortality in patients presenting with GI symptoms compared to those without GI symptoms, even after adjusting for multiple confounders including age and comorbidities, which is consistent with findings in 2 smaller cohorts.30 , 31 Notably, this finding is different from early reports suggesting increased severity with GI symptoms,32 likely attributable to the inclusion of abnormal liver function test results, which are associated with poor outcomes.
We duly acknowledge some limitations of our study. GI biopsies were performed on a distinct set of patients undergoing clinically indicated procedures, and therefore, they were not all in the acute phase of illness. Furthermore, given that only 3 patients in the biopsy cohort had GI symptoms, we were unable to perform comparisons between those with and without GI symptoms. Although we could not isolate infectious virus from intestinal biopsy samples (possibly because of culture methods, low multiplicity of infection, or inactivation of virus after contact with enteric secretions), we show presence of virus in intestinal tissue using 2 parallel methods, IF and electron microscopy/electron tomography. One of the possible reasons why SARS-CoV-2 induced less severe inflammation in the gut could be through the induction of potent neutralizing IgA antibodies, which are predominantly produced in the intestines and do not fix complement, unlike IgG antibodies mainly induced in the lungs.33 , 34 Furthermore, dimeric IgA (as would be induced in the gut) is more potent in viral neutralization than IgG.35 Finally, we acknowledge that the reporting of GI symptoms can be subject to individual variation and that they have potential for being underreported.
In summary, our data detail the previously unappreciated GI tissue response to SARS-CoV-2 and provide a rationale for future mechanistic studies to understand a possible attenuation of SARS-CoV-2 pathogenicity by the intestinal environment.
Acknowledgments
The authors would like to thank the clinical staff, physicians, and patients who participated in this study. The authors also thank Dr Randy Albrecht for support with the BSL 3 facility and procedures at the Icahn School of Medicine at Mount Sinai.
CRediT Authorship Contributions
Alexandra E. Livanos, MD (Investigation: Equal); Divya Jha, PhD (Investigation: Equal); Francesca Cossarini, MD (Investigation: Equal); Ana S. Gonzalez-Reiche, PhD (Investigation: Equal); Minami Tokuyama, BS (Investigation: Equal); Teresa Aydillo, PhD (Investigation: Equal); Tommaso L. Parigi, MD (Investigation: Equal); Mark S. Ladinsky, PhD (Formal analysis: Supporting); Irene Ramos, PhD (Investigation: Supporting); Katie Dunleavy, MD (Investigation: Supporting); Brian Lee, BS (Investigation: Supporting); Rebekah Dixon, BS (Investigation: Supporting); Steven T. Chen, BS (Investigation: Supporting); Gustavo Martinez-Delgado, PhD (Investigation: Supporting); Satish Nagula, MD (Investigation: Supporting); Emily A. Bruce, PhD (Data curation: Supporting); Huaibin M. Ko, MD (Investigation: Supporting); Benjamin S. Glicksberg, PhD (Investigation: Supporting); Girish Nadkarni, MD (Investigation: Supporting); Elisabet Pujadas, MD (Investigation: Supporting); Jason Reidy, MS (Investigation: Supporting); Steven Naymagon, MD (Investigation: Supporting); Ari Grinspan, MD (Investigation: Supporting); Jawad Ahmad, MD (Investigation: Supporting); Michael Tankelevich, BS (Investigation: Supporting); Yaron Bram, PhD (Data curation: Supporting); Ronald Gordon, MD (Investigation: Supporting); Keshav Sharma, MS (Investigation: Supporting); Jane Houldsworth, BS (Investigation: Supporting); Graham J. Britton, PhD (Investigation: Supporting); Alice Chen-Liaw, BS (Investigation: Supporting); Matthew P. Spindler, BS (Investigation: Supporting); Tamar Plitt, BS (Investigation: Supporting); Pei Wang, PhD (Supporting: Supporting); Andrea Cerutti, MD, PhD (Supporting: Supporting); Jeremiah J. Faith, PhD (Supporting: Supporting); Jean-Frederic Colombel, MD (Supporting: Supporting); Ephraim Kenigsberg, PhD (Supporting: Supporting); Carmen Argmann, PhD (Supporting: Supporting); Miriam Merad, MD, PhD (Supporting: Supporting); Sacha Gnjatic, PhD (Supporting: Supporting); Noam Harpaz, MD (Supporting: Supporting); Silvio Danese, MD, PhD (Supporting: Supporting); Carlos Cordon-Cardo, MD, PhD (Supporting: Supporting); Adeeb Rahman, PhD (Supporting: Supporting); Robert E. Schwartz, MD, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting); Nikhil A. Kumta, MD (Supporting: Supporting); Alessio Aghemo, MD, PhD (Supporting: Supporting); Pamela J. Bjorkman, PhD (Formal analysis: Supporting; Investigation: Supporting); Francesca Petralia, PhD (Supporting: Supporting); Harm van Bakel, PhD (Supporting: Supporting); Adolfo Garcia-Sastre, PhD (Supervision: Supporting); Saurabh Mehandru, MD (Investigation: Lead; Writing – original draft: Lead; Writing – review & editing: Lead).
Footnotes
Conflicts of interest These authors disclose the following: Girish Nadkarni reports employment with, consultancy agreements with, and ownership interest in Pensieve Health and Renalytix AI; has received consulting fees from AstraZeneca, BioVie, GLG Consulting, and Reata; and has served as a scientific advisor or member of Pensieve Health and Renalytix AI. Jeremiah J. Faith has served on the scientific advisor board of Vedanta Biosciences and has received grants from Janssen (unrelated to this work). Jean-Frederic Colombel has served as a consultant for AbbVie, Amgen, Arena Pharmaceuticals, Boehringer Ingelheim, Celgene Corporations, Celltrion, Eli Lilly, and Enterome (all unrelated to this work). Sacha Gnjatic has received research grants from Bristol Myers Squibb, Genentech, Immune Design, Agenus, and Janssen and has served as a consultant for Merck, OncoMed, and Neon Therapeutics. Noam Harpaz has served as a consultant for Lilly USA and has received a pathology service contract with AbbVie and Celgene. Silvio Danese has served as a consultant for Ely Lilly, Enthera, Ferring Pharmaceuticals, Gilead, Hospira, Inotrem, Janssen, Johnson & Johnson, Merck Sharp & Dohme Mundipharma, Mylan, Pfizer, Roche, Sandoz, Sublimity Therapeutics, Takeda, TiGenix, UCB, and Vifor. Robert E. Schwartz has served on the scientific advisory board of Miromatrix Inc and is a consultant and speaker for Alnylam Inc. Nikhil A. Kumta has served as a consultant for Apollo Endosurgery, Boston Scientific, Gyrus AMCI, and Olympus. The remaining authors disclose no conflicts.
Funding This research was partly funded by National Institutes of Health (NIH)/National Institute of Diabetes and Digestive and Kidney Diseases 123749 0S1 (to Saurabh Mehandru). Additional support was provided by the Center for Research for Influenza Pathogenesis, a National Institute of Allergy and Infectious Diseases (NIAID)–supported Center of Excellence for Influenza Research and Surveillance (contract no. HHSN272201400008C), and NIAID R01AI113186 (to Harm van Bakel). Additionally, the work was supported by the generous support of the JPB Foundation, the Open Philanthropy Project (research grant 2020-215611 [5384]), the Defense Advanced Research Projects Agency, and anonymous donors to Adolfo Garcia-Sastre. Minami Tokuyama was funded by the Digestive Disease Research Foundation. Ana S. Gonzalez-Reiche is supported in part by a Robin Chemers Neustein Postdoctoral Fellowship Award. The research carried out by Harm van Bakel and Ana S. Gonzalez-Reiche was supported by the Office of Research Infrastructure of the NIH under awards S10OD018522 and S10OD026880. Robert E. Schwartz is funded by NCI R01CA234614, NIAID 2R01AI107301, and NIDDK R01DK121072 and is supported as an Irma Hirschl Trust Research Award Scholar. Emily A. Bruce is supported by NIH grant P20GM125498 (awarded to University of Vermont Translational Global Infectious Disease Research Center). Pamela J. Bjorkman and Mark S. Ladinsky are supported by George Mason University Fast Grants. Steven T. Chen is supported by grant F30CA243210. Graham J. Britton is supported by a Research Fellowship Award from the Crohn’s and Colitis Foundation of America. Matthew P. Spindler is supported by NIH T32 5T32AI007605. Sacha Gnjatic is supported by grants U24 CA224319, U01 DK124165, and P01 CA190174.
Author names in bold designate shared co-first authorship.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2021.02.056.
Supplementary Materials and Methods
Clinical Cohorts
Intestinal Biopsy Cohort
Participants included hospitalized patients at MSH as well as those seen in the outpatient GI practices who underwent endoscopy between April 17, 2020, and June 2, 2020. COVID-19 case patients and control individuals were defined on the basis of nasopharyngeal SARS-CoV-2 swab PCR tests. Inclusion criteria included (1) a positive NP SARS-CoV-2 PCR test result, relevant clinical symptoms, and serologic evidence of anti–SARS-CoV-2 antibodies (for case patients) and a negative NP SARS-CoV-2 test AND absence of fever, cough, shortness of breath, and relevant contact history (for control individuals); (2) clinical indication for endoscopic procedure; and (3) the patient and/or his/her health care proxy’s ability to provide informed consent. Exclusion criteria included (1) comorbid conditions including severe coagulopathy, (2) concomitant anticoagulation use, (3) critical illness and any other clinical parameter that could potentially increase the risk of additional research biopsies; and (4) failure to obtain consent. COVID-19 severity was defined based on an internal scoring system developed by the Department of Infectious Diseases at MSH. This scoring system was developed according to the World Health Organization Ordinal Clinical Progression/Improvement Scale (https://www.who.int/publications/i/item/covid-19-therapeutic-trial-synopsis) and based on oxygenation status and organ damage, with the following definitions:
-
•
mild: SpO2 of >94% on room air AND no pneumonia on imaging;
-
•
moderate: SpO2 of <94% on room air OR pneumonia on imaging;
-
•
severe: high-flow nasal cannula, non-rebreather mask, bilevel positive airway pressure (noninvasive positive airway ventilation), or mechanical ventilation AND no pressor medications AND creatinine clearance of >30 mL/min AND alanine aminotransferase of <5 times the upper limit of normal;
-
•
Severe with evidence of EOD: high-flow nasal canula, non-rebreather mask, bilevel positive airway pressure (non-invasive positive airway ventilation), or mechanical ventilation AND pressor medications OR creatinine clearance of <30 mL/min OR new renal replacement therapy OR alanine aminotransferase of >5 times the upper limit of normal (Supplementary Table 3).
Discovery Cohort
Patients admitted to MSH between April 1, 2020, and April 15, 2020, were recruited into the discovery cohort if they were SARS-CoV-2 PCR positive, if they were older than 18 years, and if the ELLA panel of cytokines (IL6, IL8, IL1β, and TNF-α) was performed as part of clinical care. Clinical details from eligible patients were extracted from the Mount Sinai Data Warehouse under an IRB-approved protocol (IRB-20-03297A North American registry of the digestive manifestations of COVID-19). Inclusion criteria included (1) a positive NP SARS-CoV-2 PCR test result within the Mount Sinai Health System between April 1 and 15, 2020, and admission to MSH; (2) age >18 years; and (3) patients who had an ELLA cytokine panel performed during hospitalization. Exclusion criteria included (1) testing at a site outside of MSH in an ambulatory setting or those who were tested in the emergency department but not admitted; (2) age <18 years; and (3) patients without an ELLA cytokine panel.
A total of 634 participants were included in the discovery cohort (Supplementary Figure 11). In addition to demographic information (including race and ethnicity, age, and sex), clinical characteristics, laboratory data, and outcomes data were extracted from the medical charts. Covariates that were studied included BMI (obesity defined as BMI of >30 kg/m2) and comorbid conditions including hypertension, diabetes, chronic lung disease (including asthma and chronic obstructive pulmonary disease [COPD]), heart disease (including coronary artery disease, atrial fibrillation, and heart failure), chronic kidney disease, cancer, HIV, and inflammatory bowel disease.
GI symptoms were defined as more than 1 episode of either diarrhea, nausea, and/or vomiting at the time of admission. If only 1 episode of either diarrhea, nausea, and/or vomiting was specifically documented, patients were not considered to have GI symptoms. Additionally, we did not consider GI symptoms that developed during the course of hospitalization, because they could reflect nosocomial or treatment-related effects, and considered only the GI symptoms that were present at the time of hospital admission so as to avoid including iatrogenic confounders (treatments or hospital-acquired illnesses that can result in diarrhea, nausea, and vomiting).
Disease severity (as described) and mortality were considered as outcomes variables. Mortality was calculated as patient status (dead or alive) at 25 days after admission. If no information was available after discharge, patients were censored at the time of hospital discharge.
External Validation Cohort
This cohort consisted of 287 patients admitted to a tertiary care center in Milan, Italy, between February 22, 2020, and March 30, 2020, with a confirmed positive SARS-CoV-2 PCR result and who did not die or were not transferred to the ICU within 24 hours from admission were studied as detailed in Aghemo et al.1 The presence of vomiting and diarrhea (defined as at least 3 loose bowel movements per day) on or before admission was recorded. Outcomes were analyzed using ICU admission, death, or the composite study endpoint of ICU admission or death within 20 days of hospitalization.
Internal Validation Cohort
The internal validation cohort is a distinct cohort of patients admitted to MSH between April 16, 2020, and April 30, 2020, used to test a predictive model for COVID-19 severity and mortality. The same inclusion and exclusion criteria as in the discovery cohort were used with the following differences: (1) a positive SARS-CoV-2 PCR test result between April 16, 2020, and April 30, 2020; (2) an additional exclusion of patients who were already included in the discovery cohort. From a total of 408 patients, 242 met inclusion criteria and were thus included in the internal validation cohort. Demographic, clinical, and outcomes-related data were extracted from patients’ medical records, as described for the discovery cohort.
SARS-CoV-2 Testing
The SARS-CoV-2 PCR was run in the Clinical Microbiology laboratory as part of routine care on the Roche cobas platform, which performs selective amplification of 2 targets, the ORF-1 gene (target 1) and the E-gene for pan-sarbecovirus (target 2) (detects SARS-CoV-2 as well as severe acute respiratory syndrome or Middle East respiratory syndrome viruses, but not routine seasonal coronavirus). A positive result indicated that either both target 1 and target 2 were detected (the majority of cases) or that target 1 alone was detected. A presumptive positive result indicates a negative target 1 result and a positive target 2 result, which, according to the manufacturer, can be a result of the following: “(1) a sample at concentrations near or below the limit of detection of the test, (2) a mutation in the target 1 target region in the oligo binding sites, or (3) infection with some other sarbecovirus (eg, SARS-CoV or some other sarbecovirus previously unknown to infect humans), or (4) other factors.” Patients with a presumptive positive SARS-CoV-2 PCR were included in the analysis if they were treated clinically as having COVID-19.
Immunofluorescent Microscopy
Sections (5 μm) of formalin-fixed, paraffin-embedded tissue were dewaxed in xylene and rehydrated in graded alcohol and then washed in phosphate-buffered saline (PBS). Heat-induced epitope retrieval was performed by incubating slides in a pressure cooker for 15 minutes on high in target retrieval solution (Dako, S1699). Once slides cooled to room temperature, they were washed twice in PBS and then permeabilized for 30 minutes in 0.1% tritonX-100 in PBS. Nonspecific binding was blocked with 10% goat serum for 1 hour at room temperature. Sections were then incubated in primary antibodies diluted in blocking solution overnight at 4°C. Primary and secondary antibodies are summarized in Supplementary Table 16. Slides were washed in 0.1% Tween 20 plus PBS thrice and then incubated in secondary antibody and 4′,6-diamidino-2-phenylindole (1 μg/mL) for 1 hour at room temperature. Sections were washed twice in 0.1% Tween 20 plus PBS and once in PBS and then mounted with Fluoromount-G (Electron Microscopy Sciences, 1798425). Controls included omitting the primary antibody (no primary control) or substituting primary antibodies with nonreactive antibodies of the same isotype (isotype control). Tissue was visualized and imaged using a Nikon Eclipse Ni microscope and digital SLR camera (Nikon, DS-Qi2).
Intraepithelial Lymphocyte Quantification
Three 10× nonoverlapping IF images were taken for each biopsy sample. Twelve biopsy samples (10 duodenum, 2 ileum) from 11 patients with COVID-19 in the biopsy cohort were analyzed along with 9 uninfected control individuals (5 duodenum, 4 ileum). CD3+ IELs and CD3+CD8+ IELs were quantified for each image. The length of epithelium in each image was measured in ImageJ (National Institutes of Heath).2 Biopsy samples from patients with COVID-19 and control individuals were compared via unpaired t test.
Routine Clinical Electron Microscopy
After pos-fixation in 1% osmium tetroxide, tissues were serially dehydrated and embedded in epoxy resin in standard fashion. One-micron toluidine-stained scout sections were prepared for light microscopic orientation; 80-nm ultrathin sections for electron microscopy were stained with uranyl acetate and lead citrate and examined in a Hitachi 7650 transmission electron microscope at 80 kV.
Infection of Cultured Cells for Electron Microscopy and Electron Tomography Analyses
Viral infections of cultured cells were conducted at the UVM BSL-3 facility under an approved institutional biosafety protocol. SARS-CoV-2 strain 2019-nCoV/USA_USA-WA1/2020 (WA1) was generously provided by Kenneth Plante and the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch and propagated in African green monkey kidney cells (Vero E6), which were kindly provided by J.L. Whitton. Vero E6 cells were maintained in complete Dulbecco’s modified Eagle medium (Thermo Fisher Scientific, catalog no. 11965–092) containing 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, catalog no. 16140–071), 1% HEPES buffer solution (15630–130), and 1% penicillin-streptomycin (Thermo Fisher Scientific, catalog no. 15140–122). Cells were grown in a humidified incubator at 37°C with 5% CO2. Vero E6 cells seeded in 6-well dishes and infected with SARS-CoV-2 at a multiplicity of infection of 0.01 for 48 hours before fixing and preparing for electron microscopy. Cells were prefixed with 3% glutaraldehyde, 1% paraformaldehyde, 5% sucrose in 0.1 mol/L sodium cacodylate trihydrate, removed from the plates, and further prepared by high-pressure freezing and freeze substitution, as will be described.
Electron Microscopy and Dual-Axis Tomography of Intestinal Biopsy Tissue
Tissue samples were fixed with 3% glutaraldehyde to meet biosafety requirements. Tissues were rinsed with cold 0.1 mol/L sodium cacodylate trihydrate plus 5% sucrose and further dissected to block sizes sufficient for high-pressure freezing. Tissues or cultured cells were rinsed with 0.1 mol/L cacodylate buffer containing 10% Ficoll (external cryoprotectant), placed into brass planchettes (Ted Pella, Inc), and ultrarapidly frozen with an HPM-010 High Pressure Freezing Machine (Bal-Tec/ABRA). Vitreously frozen samples were transferred under liquid nitrogen to Nalgene cryogenic vials (Thermo Fisher Scientific) containing a frozen mixture of 2% OsO4, 0.05% uranyl acetate in acetone. Vials were placed in an AFS-2 freeze-substitution machine (Leica Microsystems), and the samples were freeze-substituted for 72 hours at –90°C. Samples were then warmed to –20°C over 24 hours and held at that temperature for a further 12 hours before being warmed to room temperature, rinsed 3 times with acetone, and then infiltrated into Epon-Araldite resin (Electron Microscopy Sciences). Samples were flat-embedded between 2 Teflon-coated glass microscope slides, and the resin was polymerized at 60°C for 24 hours. Embedded tissue blocks were observed by light microscopy to ascertain preservation quality and select regions of interest (ie, apical epithelium). Blocks were extracted with a scalpel and glued to plastic sectioning stubs before sectioning. Semithin (150-nm) serial sections were cut with a UC6 ultramicrotome (Leica Microsystems) using a diamond knife (Diatome, Ltd). Sections were placed on formvar-coated copper-rhodium slot grids (Electron Microscopy Sciences) and stained with 3% uranyl acetate and lead citrate. Colloidal gold particles (10 nm) were placed on both surfaces of the grids to serve as fiducial markers for tomographic image alignment. Grids were placed in a dual-axis tomography holder (Model 2010, E.A. Fischione Instruments) and imaged with a Tecnai G2 T12 transmission electron microscope (120 KeV, Thermo Fisher Scientific). Images were recorded with a 2k × 2k CCD camera (XP1000, Gatan). Tomographic tilt series and large-area montages were acquired automatically using the SerialEM software package.3 For dual-axis tomography, images were collected at 1° intervals as samples were tilted ±62°. The grid was then rotated 90°, and a second tilt series was acquired about the orthogonal axis. Tomograms were calculated, analyzed, and modeled using the IMOD software package4 , 5 on MacPro and iMac Pro computers (Apple, Inc).
Presumptive SARS-CoV-2 virions were identified from tomographic reconstructions of tissue samples by observing structures resembling virions described in cryo–electron tomography studies of purified SARS-CoV-2 and SARS-CoV-2 in infected cells6, 7, 8, 9 and comparing to identified virions within SARS-CoV-2–infected cultured cells (Supplementary Figure 6). We used the following criteria for SARS-CoV-2 virion identification in tissues: (1) structures that were spherical in 3 dimensions and not continuous with other adjacent structures, with approximately 60–120-nmol/L diameters and (2) spherical structures with densities corresponding to a distinct membrane bilayer, internal puncta consistent with ribonucleoproteins,6 and densities corresponding to surface spikes on the external peripheries of the spheres. Particles resembling virions were examined in 3 dimensions by tomography before assignments (Supplementary Movies 1 and 2). We note that the inner vesicles of multivesicular bodies (MVBs) have been misidentified as SARS-CoV-2 by electron microscopy.10 We therefore compared measurements of MVB inner vesicles and presumptive coronavirus virions from what we identified as intracellular exit compartments within the same tomogram (unpublished results) with our previous tomographic reconstructions of MVBs.11 , 12 We distinguished virions inside a cytoplasmic exit compartment from the inner vesicles of an MVB based on differences in size (MVB inner virions are generally smaller in diameter than coronaviruses) and the presence of surface spikes and internal puncta (MVB inner vesicles do not present surface spikes or internal puncta).
Cell Culture Experiments and Virus Isolation
African green monkey kidney epithelial cells (Vero E6) were originally purchased from American Type Culture Collection. Cells were maintained in Dulbecco’s modified Eagle’s medium with l-glutamate, sodium pyruvate (Corning) supplemented with 10% fetal bovine serum (FBS), 100 U penicillin/mL, and 100 mg streptomycin/mL. For all experiments, the cells were always maintained in monolayers.
Several attempts were made to isolate live infectious particles from these biopsy samples. Briefly, biopsy specimens were collected and stored in PBS until homogenization. After homogenization and centrifugation (10,000g, 20 minutes, 4°C), the resulting supernatant was inoculated onto a Vero E6 monolayer maintained in optimal virus growth media for SARS-CoV-2 virus (Dulbecco’s modified Eagle medium with l-glutamate, sodium pyruvate, 2% FBS, 100 U penicillin/mL, and 100 mg streptomycin/mL; 10 mmol/L nonessential amino acids, 1 mmol/L sodium pyruvate, and 10 mmol/L HEPES). Vero E6 cells were incubated at 37°C and 5% CO2 for a week and monitored daily for potential cytopathic effect.
Cell culture supernatants were also collected and assessed for the presence of infective particles by plaque assay. Briefly, 10-fold serial dilutions were performed in infection media for SARS-CoV-2 and inoculated onto a confluent Vero E6 cell monolayer in a 6-well plate. After 1 hour of adsorption, supernatants were removed, and cell monolayers were overlaid with minimum essential media containing 2% FBS and purified agar (Thermo Scientific Oxoid) at a final concentration of 0.7%. Cells were then incubated for 3 days at 37°C. Cells were fixed overnight with 10% formaldehyde for the inactivation of potential SARS-CoV-2 virus. Overlay was removed, and cells were washed once with PBS. A 2% crystal violet solution was used for plaque visualization and count. Experiments were performed under BSL 3 conditions.
Viral SARS-CoV-2 RNA Detection in Intestinal Biopsy Tissue
To detect SARS-CoV-2 RNA from intestinal biopsy specimens, a modified version of the Centers for Disease Control and Prevention 2019-nCoV real-time RT-qPCR was used. Primers and probes were commercially available (Integrated DNA Technologies, catalog no. 10006713, RUO Kit). SARS-CoV-2 primer and probe sets consisted of 2 2019-nCoV-specific sets (N1 and N2). A third primer set was used to detect host cellular RNaseP. Reactions were run using the QuantiFast Pathogen RT-PCR +IC Kit (Qiagen, catalog no. 211454). Assays were run using USA/WA-1/2020 SARS-CoV-2 RNA as a positive control and nuclease-free water as a nontemplate control in a 384-well format. A plasmid containing the genome sequence of the N protein (Integrated DNA Technologies, catalog no. 10006625, RUO Kit) was used to calculate genome copy number from their respective cycle thresholds using the linear equation from the respective plasmid standard curves. The limit of detection was established as 1–10 copies/μL. Reactions were performed in duplicate using the following cycling conditions on the Roche LightCycler 480 Instrument II (Roche Molecular Systems, 05015243001): 50°C for 20 minutes, 95°C for 1 second, and 95°C for 5 minutes, followed by 45 cycles of 95°C for 15 seconds and 60°C for 45 seconds. The limit of detection for SARS-CoV-2 was determined by using a commercially available plasmid control (Integrated DNA Technologies, catalog no. 10006625).
Biopsy Collection and Processing for Mass Cytometry
Biopsy samples were transferred to 10 mL of dissociation buffer (1 mol/L HEPES [Lonza], 5 mmoles/L EDTA [Invitrogen], 10% FBS in HBSS buffer [Gibco]). The tubes were kept in a shaker (180 revolutions/minute, 37°C) for 20 minutes and then gently vortexed. Cell suspensions were collected after passing the biopsy specimens through 100 μm cell strainers. A second round of EDTA dissociation was performed as detailed earlier. The cell suspension was centrifuged at 1800 revolutions/minute to pellet the EC and kept on ice. The remaining tissue was transferred to fresh tubes containing a digestion buffer (2% FBS, 0.005 g Collagenase Type IV per sample [Sigma], and 100 μl DNase I [Sigma] in RPMI). Tubes were placed in the shaker (180 revolutions/minute, 37°C) for 40 minutes and thereafter gently vortexed. The digested tissues were filtered through 100 μm cell strainers, followed by another round of filtration through 40 μm cell strainers. Cell suspensions were centrifuged at 1800 revolutions/minute to obtain LP mononuclear cells. Both the EC and LP pellets were then resuspended into 500 μL of RPMI (Gibco) containing 10% FBS+ 1 μl Rh103 +1 μl 5-Iodo-2’-deoxyuridine and incubated at 37°C for 20 minutes. Then, 5 mL RPMI (+10% FBS) was added to each tube and spun at 1800 revolutions/minute to pellet cells. Next, 700 μL of Prot1 stabilizer (SmartTube Inc) was added to each tube and transferred to cryovials and incubated at room temperature for 10 minutes. Cryovials were immediately transferred to –80°C until the sample was acquired for mass cytometry, as detailed in the next section.
Blood Collection and Processing for Mass Cytometry
Briefly, 15 mL of LymphoSep lymphocyte separation medium (MP Bio) was added to each 50-mL centrifugation tube. Blood was diluted with PBS to bring the volume up to 30 mL, and diluted blood was layered gently over LymphoSep. Tubes were then centrifuged at 2000 revolutions/minute for 20 minutes with the brakes and acceleration off. After centrifugation, the buffy coat containing peripheral blood mononuclear cells was transferred to another tube and was centrifuged at 1800 revolutions/minute to pellet the cells. Pellets were resuspended in PBS, and tubes were centrifuged at 1800 revolutions/minute. Finally, the pellets were resuspended in the freezing medium (10% dimethyl sulfoxide + 44% FBS in RPMI) and cryopreserved at –80°C.
Mass Cytometry Processing and Data Acquisition
Cells were processed as previously described by Geanon et al.13 Briefly, EC and LP SmartTube proteomic stabilized samples were thawed in a 10°C water bath and washed with Cell Staining Buffer (Fluidigm). To facilitate data acquisition and doublet removal, multiple samples were also barcoded using Fluidigm Pd barcoding kits and then washed and pooled for data acquisition. Immediately before data acquisition, samples were washed with Cell Staining Buffer and Cell Acquisition Solution (Fluidigm) and resuspended at a concentration of 1 million cells/mL in Cell Acquisition Solution containing a 1:20 dilution of EQ Normalization beads (Fluidigm). The samples were then acquired on a Helios Mass Cytometer equipped with a wide-bore sample injector at an event rate of <400 events per second. After acquisition, repeat acquisitions of the were same sample concatenated and normalized using the Fluidigm software, and barcoded samples were demultiplexed using the Zunder single cell debarcoder.
Mass Cytometry Data Analysis
Debarcoded files were uploaded to Cytobank for analyses. Immune cells were identified based on Ir-193 DNA intensity and CD45 expression; Ce140+ normalization beads, CD45-low/Ir-193-low debris, and cross-sample and Gaussian ion-cloud multiplets were excluded from subsequent downstream analysis. The cytometry by time of flight antibody panel is detailed in Supplementary Data File 1. Major immune cell types were identified using the automated Astrolabe approach, the result of which largely correlated well with our manual gating approaches. The impact of each tested condition on relative staining quality was evaluated in 2 ways: (1) overall correlations were determined by calculating the Pearson correlation coefficients for the median expression of each marker across each defined immune subset and (2) a staining index (SI) was calculated using defined populations showing the highest and lowest expression levels of each marker: SI = (Medianpos – Medianneg)/2 × Std.Devneg. It has already been described that SmartTube-based fixation protocols take into account previously described mass cytometry artifacts such as cell-cell multiplets, isotopic spillover or oxidation, or mass cytometer instrument configuration.13
Statistical Analysis for Mass Cytometry
Pregated viable CD45+ cells were first clustered and annotated using the Astrolabe Cytometry Platform (Astrolabe Diagnostics, Inc), which involves using a hierarchy-based Flow Self-Organizing Maps (SOMs) algorithm for labeling cell populations in individual samples. These Astrolabe Profiling clusters from each tissue type were then meta-clustered across all samples using Clustergrammer2’s interactive heatmap as a method to interrogate antibody expression across every cluster and curate and assign cell population categories. Single-sample clusters were also visualized using UMAP. Pairwise comparisons were performed on the frequencies of each identified cell population between the patient cohorts (COVID-19 vs control, COVID-19 severe vs control, and COVID19-asymptomatic/mild/moderate vs control) to determine fold change (FC), P values and FDR-adjusted P values using the Benjamini-Hochberg14 method to account for multiple comparisons.
RNA Sequencing
Library Preparation and Sequencing
Directional RNA-seq libraries were prepared from 50 ng of total RNA from EC and LP samples with the TruSeq Stranded Total RNA prep with Ribo-Zero kit (catalog no. 20020599). Paired-end (100–base pair) sequencing was performed for DNA libraries on an Illumina NovaSeq instrument on a NovaSeq S1 Flowcell, with an average yield of 39 million paired-end reads/sample.
RNA-Sequencing Analysis
Base calling and quality scoring of sequencing data were done through Illumina’s Real-Time Analysis software. RNA-seq data processing and reference mapping were done with custom analysis scripts combining publicly available tools as previously described15 with modifications as follows, and reads were mapped to a custom reference that combined the human hg38 reference genome (release 34, GRCh38.p13) and the SARS-CoV-2 genome (RefSeq NC_045512) for simultaneous quantification of host and virus transcripts.
Differential gene expression analysis was performed with the Bioconductor edgeR package16 using, as input, a combined matrix of mapped paired-end read raw counts, with genes in rows and samples in columns. Before differential gene expression analysis, gene counts were converted to fragments per kilobase per million reads (FPKM) with the RSEM package with default settings in strand-specific mode.17
Genes with less than 1 FPKM in at least 50% of the samples were removed. The remaining gene counts were then normalized across samples using the weighted trimmed mean of M values method.18 The dispersion was estimated by fitting a generalized linear model as implemented in edgeR, and sex was fitted as a covariate on a per-patient paired design. Pairwise comparisons were performed between sample groups (ie, between tissue sections as well as between case patients and control individuals). Significant expression differences were selected based on empirical Bayes-adjusted P values corrected for multiple testing using the Benjamini-Hochberg method (q ≤ .05).
Gene Ontology and Pathway Enrichment Analysis
KEGG pathway and Gene Ontology biological process, molecular function, and/or cellular component enrichment analyses were performed using the gProfileR R version 0.6.8 package.19 The background gene set was restricted genes with detected expression (defined as genes with expression levels above 1 FPKM in at least 50% of samples). Genes with differential expression were ranked by log2 fold change and used as an ordered query. P values were corrected using the g:SCS algorithm to account for multiple comparisons.
Cell-Type Deconvolution and Gene Signature Enrichment Analysis
For cell-type deconvolution of the bulk RNA-seq data, Gene Set Enrichment Analysis (GSEA) of DEGs of case patient vs control individual comparisons was performed against cell-type gene-expression single-cell signatures from intestinal mucosa20 and gene-expression signatures from ileal DC subsets.21 Similarly, DEGs were tested for enrichment of gene signatures associated with an antiviral response, inflammation, and cytokine signaling in acutely infected post mortem tissue with SARS-CoV-222 were tested for significant (P ≤ .05) enrichment using Fisher exact tests and using Bonferroni correction for multiple comparisons.
Additionally, GSEA23 was carried out on a rank-ordered list of the infected EC vs control molecular analysis. The ranking metric used was log(FC) × –log(P value); however, the results were similar when the log(FC) metric was also used (data not shown). For the COVID-19–associated data sets, we curated 2 signatures from infected organoids,24 hSIOs-COVID-19: human small intestinal organoids (hSIOs) grown in either (1) Wnt high-expansion medium (at adjusted P < .05) or (2) differentiation medium (at adjusted P < .1). The standard GSEA settings were used, namely, meandiv for normalization mode, weighted enrichment statistic, and 1000 permutations. GSEA using the Hallmark database (version 7.125) was also performed with the same settings.
Computational Analyses
Multivariate Model Based on Discovery Cohort
For this analysis, we considered 570 patients with clinical descriptors including age, sex, race/ethnicity, BMI, comorbidities (including hypertension, diabetes, chronic lung disease (including asthma and COPD), heart disease (including coronary artery disease, atrial fibrillation and heart failure), and GI symptoms. A multivariate logistic regression was used to model severity and mortality as a function of each of the GI symptoms and clinical variables including race, age, sex, BMI, heart and lung diseases, and hypertension.
In particular, race was stratified as White, Black (African American), Hispanic, and others; lung disease was set equal to 1 if the patient was affected by either COPD or asthma and to 0 otherwise; heart disease was set equal to 1 if the patient was affected by coronary artery disease, atrial fibrillation, or heart failure and to 0 otherwise. The severity indicator was set equal to 1 for patients with severe and severe with EOD disease and to 0 for patients with mild and moderate COVID-19; mortality was set equal to 1 for deceased patients and to 0 otherwise.
Significant associations were determined based on 95% confidence intervals (CIs) based on 1000 bootstrap iterations. At each bootstrap iteration, patients were sampled with replacements, and logistic regressions were estimated considering as outcome severity and mortality. Then, 95% CIs of coefficients and ORs were estimated across bootstrap iterations.
External Validation Cohort
For this analysis, we considered 228 patients with clinical data such as age, sex, and GI symptoms, as described by Aghemo et al.1 A multivariate logistic regression was used to model mortality, ICU admission, and the composite outcome of ICU admission or mortality as a function of the presence or absence of diarrhea and clinical variables including age, sex, BMI, heart disease, COPD, diabetes, and hypertension. Heart disease was set equal to 1 if the patient was either affected by coronary artery disease or atrial fibrillation and to 0 otherwise. The 95% CI of ORs were computed based on 1000 bootstrap iterations, as described.
In 270 patients from this cohort, treatment data were available. Treatments included hydroxychloroquine; antiviral treatments, including lopinavir-ritonavir and darunavir-cobicistat; tocilizumab, steroids, antibiotics; including ceftriaxone, azithromycin, and piperacillin-tazobactam; statins; ACE inhibitors, and angiotensin II receptor blockers. Using these data, we performed the Fisher exact test to determine whether any treatments were associated with diarrhea. Additionally, we computed 95% CIs of ORs based on 1000 bootstrap iterations.
Predictive Performance Based on the Internal Validation Cohort
For this analysis, we considered 233 patients with clinical data including age, BMI, and GI symptoms. To evaluate the predictive performance of each model, bootstrapping was performed. Specifically, at each bootstrap iteration, we randomly sampled patients in the discovery cohort with replacement and estimated a logistic regression to model each outcome as function of a particular GI symptom, age, and BMI. In this analysis, only age and BMI were adjusted for because they were the only variables significantly associated with both outcomes across different GI symptoms models in the discovery cohort (Figure 5 B and Supplementary Table 9). Then, the estimated model was used to predict the outcome of patients in the internal validation cohort. This procedure was repeated for 1000 bootstrap iterations. For each iteration, the receiving operating characteristic (ROC) curve and AUC were computed. For comparison purposes, the distribution of the AUC across 1000 bootstrap iterations from the predictive model based on age and BMI only was considered. Figure 5 Dshows the boxplot of AUC values across 1000 bootstrap iterations. Then, considering the following model,
outcome = f(age + BMI + any GI symptom) Model 1
we evaluated the effect of each variable on the outcome by computing the reduction in AUC obtained after removing 1 variable at a time. For this purpose, the AUC of model 1 was compared to the following 3 models:
outcome = f(age + BMI) model 2
outcome = f(age + any GI symptom) model 3
outcome = f(BMI + any GI symptom) model 4
for 1000 bootstrap iterations. Following this strategy, at each bootstrap iteration, patients were sampled with replacement. Figure 5 E shows the 95% CIs of difference in the AUCs between model 1 and models 2, 3, and 4 (ie, AUCModel1 – AUCModel2, AUCModel1 – AUCModel3, and AUCModel1 – AUCModel4) across 1000 bootstrap iterations. The difference in AUC was computed considering both mortality and severity as the outcome.
Average Treatment Effect
The ATE for the Mount Sinai Cohort (MSH), combining the discovery and internal validation cohorts, and for the external validation cohort were calculated via the TMLE package in R.26 For the MSH cohort, the ATE was calculated for each GI symptom using as outcomes disease severity and mortality. The marginal effect was calculated after adjustment for covariates such as age, race, BMI, sex, diabetes, lung disease, heart disease, and hypertension. For the external validation cohort, the ATE was calculated for diarrhea on ICU admission, mortality, and the composite of ICU admission and mortality. The marginal effect was calculated after adjustment for covariates such as age, BMI, sex, diabetes, lung disease, heart disease, and hypertension.
Quantification of SARS-CoV-2 Nasopharyngeal Viral Loads
SARS-CoV-2 viral loads were determined as detailed in Pujadas et al.27 Briefly, viral RNA was extracted from the NP swab specimen, followed by real-time RT-PCR using N2 primers. Only specimens with N2Cpt of <38 were included. SARS-CoV-2 viral RNA was calculated with the delta CT method and a standard curve. Viral loads are presented as log base 10–transformed uncorrected N2 value + 1000 (constant added before transformation).27 For patients with multiple NP swabs available, the first swab was used for analysis.
ELLA Cytokine Panel
The ELLA platform is a method for rapid cytokine measurement using microfluidics enzyme-linked immunosorbent assays. The assay measured TNF-α, IL6, IL8, and IL1β, previously validated by the Mount Sinai Human Immune Monitoring Center using plasma from patients with multiple myeloma and recently reported for large cohort of patients with COVID-19 admitted to MSH.
Multiplexed Proteomic Assay (Olink)
For analysis of circulating cytokines, we used a multiplexed proteomic inflammation panel (Olink), which consists of 92 inflammation-related proteins quantified by an antibody-mediated proximity extension–based assay. Samples with normalized protein expression values below the limit of detection in >75% of samples were excluded from further analysis. For the remainder of analytes, any sample under the limit of detection was assigned a value of the limit of detection divided by the square root of 2. The log2(FC) over the median healthy control protein expression was then calculated, and the Benjamini-Hochberg procedure was used to adjust P values for multiple testing.
Consensus Clustering of Olink Data
For this analysis, we considered 238 samples with GI symptom annotation. Consensus clustering was performed based on the abundance of 92 cytokines across all 238 samples. Consensus clustering was performed using the R packages ConsensusClusterPlus based on z-score–normalized data. Specifically, markers were partitioned into 6 clusters using the K-means algorithm, which was repeated 1000 times. Then, markers in each cluster were considered to derive cluster z-score signatures via the package GSVA. Based on these signatures, the associations between different clusters and GI symptoms were derived via logistic regression with outcome corresponding to each GI symptom. Figure 6 C shows the signed FDR (–log10 scale). P values were adjusted via Benjamini-Hochberg. The pathway analysis for the clusters described was carried out considering the entire KEGG and Hallmark databases.
Supplementary Table 3.
Criteria for Scoring Disease Severity in Patients With COVID-19
| Severity score | Criteria |
|---|---|
| Mild | Spo2 of >94% on room air AND no pneumonia on imaging |
| Moderate | Spo2 of <94% on room air OR pneumonia on imaging |
| Severe | High-flow nasal cannula, non-rebreather mask, bilevel positive airway pressure (noninvasive positive airway ventilation), or mechanical ventilation AND no pressor medications AND creatinine clearance >30 mL/min AND alanine aminotransferase of <5 times the upper limit of normal |
| Severe with EOD | High-flow nasal cannula, non-rebreather mask, bilevel positive airway pressure (noninvasive positive airway ventilation), or mechanical ventilation AND pressor medications OR creatinine clearance of <30 OR new renal replacement therapy OR alanine aminotransferase of >5 times the upper limit of normal |
Supplementary Table 6.
Discovery Cohort Basic Demographics, Clinical Characteristics, and Outcomes
| Variable | Discovery cohort (n = 634) |
|---|---|
| Age, y, mean ± SD | 64.0 ± 15.7 |
| Male, n (%) | 369 (58.2) |
| Race/ethnicity, n (%) | |
| Hispanic | 177 (27.9) |
| African American | 161 (25.4) |
| White | 137 (21.6) |
| Asian | 35 (5.5) |
| Other | 124 (19.6) |
| Comorbidities, n (%) | |
| Hypertension | 229 (36.1) |
| Diabetes mellitus | 141 (22.2) |
| Obesity (BMI > 30)a | 211 (37.1) |
| Chronic lung disease | 59 (9.3) |
| Heart disease | 111 (17.5) |
| Chronic kidney disease | 95 (15.0) |
| Cancer | 66 (10.4) |
| HIV | 11 (1.7) |
| Inflammatory bowel disease | 7 (1.1) |
| Disease severity, n (%) | |
| Mild | 54 (8.5) |
| Moderate | 361 (56.9) |
| Severe | 158 (24.9) |
| Severe with EOD | 61 (9.6) |
| Outcomes, n (%) | |
| ICU admission | 110 (17.4) |
| Mortality | 151 (23.8) |
| GI symptoms, n (%) | |
| Nausea | 157 (24.8) |
| Vomiting | 82 (12.9) |
| Diarrhea | 245 (38.6) |
| Any GI symptoms | 299 (47.2) |
SD, standard deviation.
BMI information was available for 568 of 634 patients.
Supplementary Table 7.
COVID-19 Disease Severity and Mortality in Patients With and Without GI Symptoms in the Discovery Cohort
| GI symptom | Severity | GI symptom |
P valuea | |
|---|---|---|---|---|
| Presence, n | Absence, n | |||
| Nausea | Mild | 16 | 38 | .0112 |
| Moderate | 102 | 259 | ||
| Severe | 32 | 126 | ||
| Severe EOD | 7 | 54 | ||
| Vomiting | Mild | 10 | 44 | .0156 |
| Moderate | 56 | 305 | ||
| Severe | 11 | 147 | ||
| Severe EOD | 5 | 56 | ||
| Diarrhea | Mild | 26 | 28 | .0102 |
| Moderate | 152 | 209 | ||
| Severe | 52 | 106 | ||
| Severe EOD | 15 | 46 | ||
| Any GI symptoms | Mild | 31 | 23 | .0003 |
| Moderate | 188 | 173 | ||
| Severe | 63 | 95 | ||
| Severe EOD | 17 | 44 | ||
| Mortality | ||||
| Nausea | Nonsurvivor | 21 | 130 | .0003 |
| Survivor | 136 | 347 | ||
| Vomiting | Nonsurvivor | 8 | 143 | .0008 |
| Survivor | 74 | 409 | ||
| Diarrhea | Nonsurvivor | 39 | 112 | .0002 |
| Survivor | 206 | 277 | ||
| Any GI symptoms | Nonsurvivor | 47 | 104 | <.0001 |
| Survivor | 252 | 231 | ||
Fisher exact test was used to calculate P values.
Supplementary Table 8.
Basic Demographics in Survivors and Nonsurvivors in the Discovery Cohort
| Variable | Survivors (n = 483) | Nonsurvivors (n = 151) | P value |
|---|---|---|---|
| Age, y, mean ± SD | 61.3 ± 15.2 | 72.6 ± 14.1 | <.0001 |
| Male, n (%) | 287 (59.4) | 82 (54.3) | .30 |
| Disease severity, n (%) | |||
| Mild | 48 (9.9) | 6 (4.0) | |
| Moderate | 318 (65.8) | 43 (28.5) | |
| Severe | 95 (19.7) | 63 (41.7) | |
| Severe with EOD | 22 (4.6) | 39 (25.8) | <.0001 |
NOTE. For age, an unpaired 2-tailed t test was performed. For categorical variables, the Fisher exact test or chi-square test was used as appropriate. Bold values indicate P values that are significant (<.05).
SD, standard deviation.
Supplementary Table 9.
CI of ORs Based on 1000 Bootstrap Iterations for Severity, Mortality, and ICU Admission in the Discovery Cohort
| Variable | Quantile | ||
|---|---|---|---|
| 2.5% | 50% | 97.5% | |
| Severity | |||
| (Intercept) | 0.015 | 0.071 | 0.320 |
| Any GI symptom | 0.378 | 0.559 | 0.844 |
| baseline.GenderMale | 0.939 | 1.380 | 2.090 |
| baseline.Age | 1.004 | 1.016 | 1.031 |
| baseline.DIABETES | 0.600 | 0.995 | 1.729 |
| baseline.BMI | 1.009 | 1.039 | 1.069 |
| baseline.RACE_ETHNICITY_BLACK OR AFRICAN-AMERICAN | 0.293 | 0.503 | 0.876 |
| baseline.RACE_ETHNICITY_HISPANIC | 0.688 | 1.188 | 2.035 |
| baseline.RACE_ETHNICITY_OTHER | 0.771 | 1.286 | 2.245 |
| baseline.HTN | 0.636 | 0.990 | 1.543 |
| baseline.Lung.Disease | 0.272 | 0.562 | 1.063 |
| baseline.Heart.Disease | 0.673 | 1.095 | 1.842 |
| (Intercept) | 0.013 | 0.060 | 0.259 |
| Diarrhea | 0.433 | 0.653 | 0.978 |
| baseline.GenderMale | 0.963 | 1.413 | 2.124 |
| baseline.Age | 1.005 | 1.017 | 1.033 |
| baseline.DIABETES | 0.604 | 1.012 | 1.747 |
| baseline.BMI | 1.008 | 1.037 | 1.067 |
| baseline.RACE_ETHNICITY_BLACK OR AFRICAN-AMERICAN | 0.300 | 0.521 | 0.920 |
| baseline.RACE_ETHNICITY_HISPANIC | 0.707 | 1.215 | 2.098 |
| baseline.RACE_ETHNICITY_OTHER | 0.777 | 1.311 | 2.280 |
| baseline.HTN | 0.634 | 0.977 | 1.529 |
| baseline.Lung.Disease | 0.267 | 0.546 | 1.042 |
| baseline.Heart.Disease | 0.686 | 1.113 | 1.826 |
| (Intercept) | 0.013 | 0.058 | 0.271 |
| Nausea | 0.329 | 0.563 | 0.880 |
| baseline.GenderMale | 0.932 | 1.364 | 2.089 |
| baseline.Age | 1.005 | 1.018 | 1.032 |
| baseline.DIABETES | 0.609 | 1.015 | 1.738 |
| baseline.BMI | 1.007 | 1.036 | 1.065 |
| baseline.RACE_ETHNICITY_BLACK OR AFRICAN-AMERICAN | 0.322 | 0.543 | 0.946 |
| baseline.RACE_ETHNICITY_HISPANIC | 0.749 | 1.264 | 2.169 |
| baseline.RACE_ETHNICITY_OTHER | 0.816 | 1.340 | 2.338 |
| baseline.HTN | 0.615 | 0.950 | 1.477 |
| baseline.Lung.Disease | 0.248 | 0.516 | 0.980 |
| baseline.Heart.Disease | 0.676 | 1.091 | 1.830 |
| (Intercept) | 0.012 | 0.054 | 0.236 |
| Vomiting | 0.190 | 0.399 | 0.732 |
| baseline.GenderMale | 0.938 | 1.395 | 2.107 |
| baseline.Age | 1.006 | 1.018 | 1.032 |
| baseline.DIABETES | 0.615 | 1.032 | 1.744 |
| baseline.BMI | 1.006 | 1.036 | 1.065 |
| baseline.RACE_ETHNICITY_BLACK OR AFRICAN-AMERICAN | 0.330 | 0.558 | 0.962 |
| baseline.RACE_ETHNICITY_HISPANIC | 0.766 | 1.286 | 2.191 |
| baseline.RACE_ETHNICITY_OTHER | 0.802 | 1.337 | 2.307 |
| baseline.HTN | 0.610 | 0.950 | 1.511 |
| baseline.Lung.Disease | 0.261 | 0.526 | 0.981 |
| baseline.Heart.Disease | 0.703 | 1.123 | 1.886 |
| Mortality | |||
| (Intercept) | 0.000 | 0.003 | 0.022 |
| Any GI symptom | 0.335 | 0.544 | 0.861 |
| baseline.GenderMale | 0.679 | 1.049 | 1.703 |
| baseline.Age | 1.036 | 1.053 | 1.074 |
| baseline.DIABETES | 0.527 | 0.930 | 1.605 |
| baseline.BMI | 1.010 | 1.043 | 1.081 |
| baseline.RACE_ETHNICITY_BLACK OR AFRICAN-AMERICAN | 0.529 | 1.035 | 1.959 |
| baseline.RACE_ETHNICITY_HISPANIC | 0.867 | 1.597 | 2.899 |
| baseline.RACE_ETHNICITY_OTHER | 0.450 | 0.878 | 1.630 |
| baseline.HTN | 0.600 | 1.007 | 1.665 |
| baseline.Lung.Disease | 0.342 | 0.778 | 1.577 |
| baseline.Heart.Disease | 0.652 | 1.154 | 2.028 |
| (Intercept) | 0.000 | 0.003 | 0.017 |
| Diarrhea | 0.388 | 0.638 | 0.985 |
| baseline.GenderMale | 0.702 | 1.076 | 1.737 |
| baseline.Age | 1.038 | 1.055 | 1.076 |
| baseline.DIABETES | 0.531 | 0.945 | 1.608 |
| baseline.BMI | 1.007 | 1.041 | 1.079 |
| baseline.RACE_ETHNICITY_BLACK OR AFRICAN-AMERICAN | 0.562 | 1.071 | 2.020 |
| baseline.RACE_ETHNICITY_HISPANIC | 0.901 | 1.640 | 2.898 |
| baseline.RACE_ETHNICITY_OTHER | 0.468 | 0.900 | 1.709 |
| baseline.HTN | 0.595 | 1.001 | 1.665 |
| baseline.Lung.Disease | 0.333 | 0.752 | 1.561 |
| baseline.Heart.Disease | 0.660 | 1.166 | 2.020 |
| (Intercept) | 0.000 | 0.003 | 0.019 |
| Nausea | 0.255 | 0.490 | 0.886 |
| baseline.GenderMale | 0.669 | 1.041 | 1.719 |
| baseline.Age | 1.038 | 1.055 | 1.075 |
| baseline.DIABETES | 0.528 | 0.938 | 1.623 |
| baseline.BMI | 1.006 | 1.040 | 1.077 |
| baseline.RACE_ETHNICITY_BLACK OR AFRICAN-AMERICAN | 0.591 | 1.112 | 2.116 |
| baseline.RACE_ETHNICITY_HISPANIC | 0.956 | 1.718 | 3.085 |
| baseline.RACE_ETHNICITY_OTHER | 0.474 | 0.916 | 1.703 |
| baseline.HTN | 0.579 | 0.960 | 1.571 |
| baseline.Lung.Disease | 0.313 | 0.699 | 1.457 |
| baseline.Heart.Disease | 0.655 | 1.155 | 2.007 |
| (Intercept) | 0.000 | 0.002 | 0.016 |
| Vomiting | 0.116 | 0.364 | 0.753 |
| baseline.GenderMale | 0.690 | 1.070 | 1.759 |
| baseline.Age | 1.038 | 1.055 | 1.076 |
| baseline.DIABETES | 0.558 | 0.970 | 1.652 |
| baseline.BMI | 1.006 | 1.040 | 1.077 |
| baseline.RACE_ETHNICITY_BLACK OR AFRICAN-AMERICAN | 0.609 | 1.145 | 2.144 |
| baseline.RACE_ETHNICITY_HISPANIC | 1.004 | 1.746 | 3.086 |
| baseline.RACE_ETHNICITY_OTHER | 0.463 | 0.907 | 1.697 |
| baseline.HTN | 0.586 | 0.974 | 1.620 |
| baseline.Lung.Disease | 0.318 | 0.717 | 1.489 |
| baseline.Heart.Disease | 0.670 | 1.186 | 2.025 |
| ICU admission | |||
| (Intercept) | 0.03 | 0.17 | 1.11 |
| Any GI symptom | 0.46 | 0.75 | 1.19 |
| baseline.GenderMale | 1.23 | 2.00 | 3.26 |
| baseline.Age | 0.98 | 0.99 | 1.01 |
| baseline.DIABETES | 0.91 | 1.70 | 3.23 |
| baseline.BMI | 0.98 | 1.02 | 1.05 |
| baseline.RACE_ETHNICITY_BLACK OR AFRICAN-AMERICAN | 0.26 | 0.57 | 1.20 |
| baseline.RACE_ETHNICITY_HISPANIC | 0.79 | 1.50 | 2.95 |
| baseline.RACE_ETHNICITY_OTHER | 0.48 | 0.98 | 1.99 |
| baseline.HTN | 0.34 | 0.61 | 1.08 |
| baseline.Lung.Disease | 0.15 | 0.63 | 1.45 |
| baseline.Heart.Disease | 0.20 | 0.51 | 0.97 |
| (Intercept) | 0.02 | 0.16 | 1.10 |
| Diarrhea | 0.45 | 0.76 | 1.25 |
| baseline.GenderMale | 1.23 | 2.00 | 3.27 |
| baseline.Age | 0.98 | 0.99 | 1.01 |
| baseline.DIABETES | 0.89 | 1.70 | 3.25 |
| baseline.BMI | 0.98 | 1.02 | 1.05 |
| baseline.RACE_ETHNICITY_BLACK OR AFRICAN-AMERICAN | 0.26 | 0.56 | 1.19 |
| baseline.RACE_ETHNICITY_HISPANIC | 0.78 | 1.50 | 2.96 |
| baseline.RACE_ETHNICITY_OTHER | 0.48 | 0.98 | 1.98 |
| baseline.HTN | 0.34 | 0.61 | 1.09 |
| baseline.Lung.Disease | 0.15 | 0.62 | 1.44 |
| baseline.Heart.Disease | 0.20 | 0.52 | 0.99 |
| (Intercept) | 0.02 | 0.14 | 0.95 |
| Nausea | 0.45 | 0.85 | 1.42 |
| baseline.GenderMale | 1.24 | 2.00 | 3.33 |
| baseline.Age | 0.98 | 0.99 | 1.01 |
| baseline.DIABETES | 0.91 | 1.74 | 3.31 |
| baseline.BMI | 0.98 | 1.02 | 1.05 |
| baseline.RACE_ETHNICITY_BLACK OR AFRICAN-AMERICAN | 0.27 | 0.60 | 1.25 |
| baseline.RACE_ETHNICITY_HISPANIC | 0.81 | 1.54 | 3.00 |
| baseline.RACE_ETHNICITY_OTHER | 0.49 | 0.99 | 1.97 |
| baseline.HTN | 0.33 | 0.59 | 1.06 |
| baseline.Lung.Disease | 0.14 | 0.60 | 1.40 |
| baseline.Heart.Disease | 0.20 | 0.51 | 0.98 |
| (Intercept) | 0.02 | 0.15 | 0.98 |
| Vomiting | 0.16 | 0.53 | 1.08 |
| baseline.GenderMale | 1.21 | 1.98 | 3.26 |
| baseline.Age | 0.98 | 0.99 | 1.01 |
| baseline.DIABETES | 0.92 | 1.74 | 3.31 |
| baseline.BMI | 0.98 | 1.02 | 1.05 |
| baseline.RACE_ETHNICITY_BLACK OR AFRICAN-AMERICAN | 0.27 | 0.60 | 1.26 |
| baseline.RACE_ETHNICITY_HISPANIC | 0.82 | 1.55 | 3.01 |
| baseline.RACE_ETHNICITY_OTHER | 0.48 | 0.99 | 1.96 |
| baseline.HTN | 0.34 | 0.61 | 1.07 |
| baseline.Lung.Disease | 0.15 | 0.62 | 1.43 |
| baseline.Heart.Disease | 0.20 | 0.52 | 0.99 |
HTN, hypertension.
Supplementary Table 10.
Age, Sex, and Mortality in an External Validation (Italian) Cohort Stratified by the Presence or Absence of Diarrhea on Admission
| Variable | Diarrhea on admission (n = 80) | No diarrhea on admission (n = 207) | P value |
|---|---|---|---|
| Age, y, mean ± SD | 60.6 ± 13.9 | 65.5 ± 13.3 | .006 |
| Male, n (%) | 46 (57.5) | 149 (72.0) | .024 |
| ICU admission, n (%) | 9 (11.3) | 43 (20.8) | .06 |
| Mortality, n (%) | 8 (10.0) | 49 (23.7) | .008 |
| Death or ICU admission, n (%) | 16 (20.0) | 83 (40.1) | .001 |
NOTE. For age, an unpaired 2-tailed t test was performed. For categorical variables, the Fisher exact test or chi-square test was used as appropriate. Bold values indicate P values that are significant (<.05).
SD, standard deviation.
Supplementary Table 11.
Association Between Diarrhea and Treatment in External Validation Cohort Using the Fisher Exact Test
| Treatment | Fisher exact P value | ||
|---|---|---|---|
| Hydroxychloroquine | .409 | ||
| Lopinavir.ritonavir | .466 | ||
| Darunavir.cobicistat | .473 | ||
| Tocilizumab | > .999 | ||
| Steroids | .698 | ||
| Ceftriaxone | .871 | ||
| Azithromycin | .209 | ||
| Piperacillin.Tazobactam | .837 | ||
| Statins | .609 | ||
| ACE.inhibitors | .052 | ||
| ARBs | .844 | ||
| Outcome | Quantiles | ||
| 2.50% | 50% | 97.50% | |
| Death | |||
| (Intercept) | 0.025 | 0.138 | 0.434 |
| diarrhea | 0.111 | 0.313 | 0.699 |
| treatment.Hydroxychloroquine | 0.170 | 0.706 | 2.965 |
| treatment.Lopinavir.ritonavir | 0.740 | 2.397 | 11.006 |
| treatment.Darunavir.cobicistat | 0.477 | 1.723 | 8.455 |
| treatment.Tocilizumab | 0.000 | 0.472 | 2.692 |
| treatment.Steroids | 0.000 | 1.626 | 7.849 |
| treatment.Ceftriaxone | 0.415 | 1.079 | 3.740 |
| treatment.Azithromycin | 0.252 | 0.651 | 1.462 |
| treatment.Piperacillin.Tazobactam | 0.961 | 3.014 | 11.740 |
| treatment.Statins | 0.981 | 2.261 | 5.127 |
| treatment.ACE.inhibitors | 0.588 | 1.503 | 3.413 |
| treatment.ARBs | 0.502 | 1.389 | 3.645 |
| ICU | |||
| (Intercept) | 8.89E–10 | 1.15E–08 | 3.40E–08 |
| diarrhea | 0.122 | 0.407 | 1.083 |
| treatment.Hydroxychloroquine | 1.52E–09 | 0.093814 | 1.265738 |
| treatment.Lopinavir.ritonavir | 33,992,289 | 318,000,000 | 4.11E+16 |
| treatment.Darunavir.cobicistat | 9669004 | 96256078 | 1.13E+16 |
| treatment.Tocilizumab | 3.17E–08 | 1.93167 | 11.84107 |
| treatment.Steroids | 0.557 | 4.866 | 46.290 |
| treatment.Ceftriaxone | 0.474 | 1.606 | 8.810 |
| treatment.Azithromycin | 0.264 | 0.719 | 1.756 |
| treatment.Piperacillin.Tazobactam | 0.097 | 0.807 | 6.591 |
| treatment.Statins | 0.140 | 0.580 | 1.655 |
| treatment.ACE.inhibitors | 0.192 | 0.683 | 2.022 |
| treatment.ARBs | 0.381 | 1.362 | 3.893 |
| ICU or death | |||
| (Intercept) | 0.054 | 0.219 | 0.674 |
| diarrhea | 0.126 | 0.297 | 0.585 |
| treatment.Hydroxychloroquine | 0.000 | 0.144 | 0.729 |
| treatment.Lopinavir.ritonavir | 4.142 | 22.183 | 215,000,000 |
| treatment.Darunavir.cobicistat | 2.143 | 11.491 | 102,000,000 |
| treatment.Tocilizumab | 0.000 | 1.198 | 7.012 |
| treatment.Steroids | 1.424 | 8.464 | 75,358,718 |
| treatment.Ceftriaxone | 0.559 | 1.304 | 4.167 |
| treatment.Azithromycin | 0.224 | 0.510 | 1.003 |
| treatment.Piperacillin.Tazobactam | 0.656 | 2.090 | 7.553 |
| treatment.Statins | 0.696 | 1.595 | 3.529 |
| treatment.ACE.inhibitors | 0.615 | 1.409 | 2.987 |
| treatment.ARBs | 0.668 | 1.642 | 4.449 |
NOTE. Association between diarrhea and outcome after adjustment for treatment based on the external validation cohort. Quantile of OR based on 1000 bootstrap iterations is reported.
Supplementary Table 12.
95% CI of AUC Based on 1000 Bootstrap Iterations for Severity, Mortality, and ICU Admission in the Internal Validation Cohort
| Variable | Quantile | ||
|---|---|---|---|
| 2.50% | 50% | 97.50% | |
| Severity | |||
| Age + BMI | 0.539 | 0.587 | 0.598 |
| Age + BMI + nausea | 0.567 | 0.608 | 0.619 |
| Age + BMI + vomiting | 0.558 | 0.607 | 0.618 |
| Age + BMI + diarrhea | 0.574 | 0.630 | 0.651 |
| Age + BMI + any GI symptoms | 0.605 | 0.640 | 0.651 |
| Mortality | |||
| Age + BMI | 0.685 | 0.700 | 0.702 |
| Age + BMI + nausea | 0.698 | 0.717 | 0.722 |
| Age + BMI + vomiting | 0.702 | 0.719 | 0.724 |
| Age + BMI + diarrhea | 0.697 | 0.718 | 0.726 |
| Age + BMI + any GI symptoms | 0.708 | 0.727 | 0.736 |
| ICU admission | |||
| Age + BMI | 0.534 | 0.560 | 0.599 |
| Age + BMI + nausea | 0.496 | 0.523 | 0.650 |
| Age + BMI + vomiting | 0.488 | 0.515 | 0.626 |
| Age + BMI + diarrhea | 0.562 | 0.649 | 0.667 |
| Age + BMI + any GI symptoms | 0.570 | 0.647 | 0.670 |
Supplementary Table 13.
IL6, IL8, TNF-α, and IL1β Concentrations on Admission in Patients With and Without GI Symptoms
| Cytokine | Nausea | Vomiting | Diarrhea | Any GI symptoms |
|---|---|---|---|---|
| IL6 | –1.958 | –0.473 | –2.226 | –2.484 |
| IL8 | –4.098 | –1.440 | –2.302 | –3.133 |
| TNF-α | –0.815 | –0.311 | –0.406 | –0.864 |
| IL1β | –0.295 | –0.473 | –0.295 | –0.295 |
NOTE. Benjamini-adjusted P values (signed –log10 scale) from t test are reported. Associations passing a 10% FDR are highlighted in yellow.
Supplementary Table 14.
Cluster Assignment for Each of the 92 Olink Analytes
| Marker | Cluster | Marker | Cluster |
|---|---|---|---|
| IL8 | 1 | MCP.3 | 4 |
| AXIN1 | 1 | OPG | 4 |
| OSM | 1 | uPA | 4 |
| CCL4 | 1 | IL6 | 4 |
| TGF.alpha | 1 | MCP.1 | 4 |
| TNFSF14 | 1 | IL18 | 4 |
| HGF | 1 | IL.18R1 | 4 |
| SIRT2 | 1 | IL10 | 4 |
| EN.RAGE | 1 | CCL23 | 4 |
| CASP.8 | 1 | CXCL10 | 4 |
| TWEAK | 1 | LIF | 4 |
| STAMBP | 1 | CCL20 | 4 |
| VEGFA | 2 | ADA | 4 |
| CDCP1 | 2 | GDNF | 5 |
| IL.17C | 2 | IL.17A | 5 |
| CXCL9 | 2 | IL.20RA | 5 |
| CST5 | 2 | IL.2RB | 5 |
| FGF.23 | 2 | IL.1.alpha | 5 |
| FGF.5 | 2 | IL2 | 5 |
| LIF.R | 2 | TSLP | 5 |
| FGF.21 | 2 | SLAMF1 | 5 |
| IL.15RA | 2 | IL.10RA | 5 |
| IL.10RB | 2 | IL.22.RA1 | 5 |
| PD.L1 | 2 | Beta.NGF | 5 |
| MMP.10 | 2 | IL.24 | 5 |
| TNF | 2 | IL13 | 5 |
| CD5 | 2 | ARTN | 5 |
| X4E.BP1 | 2 | IL.20 | 5 |
| CD40 | 2 | CCL28 | 5 |
| CCL25 | 2 | IL33 | 5 |
| CX3CL1 | 2 | IL4 | 5 |
| TNFRSF9 | 2 | NRTN | 5 |
| CSF.1 | 2 | NT.3 | 5 |
| CD8A | 3 | IL5 | 5 |
| CD244 | 3 | IL7 | 6 |
| TRAIL | 3 | LAP.TGF.beta.1 | 6 |
| CD6 | 3 | CXCL11 | 6 |
| SCF | 3 | CXCL1 | 6 |
| CCL11 | 3 | MCP.4 | 6 |
| CCL19 | 3 | MMP.1 | 6 |
| TRANCE | 3 | CXCL5 | 6 |
| IL.12B | 3 | CXCL6 | 6 |
| CCL3 | 3 | ST1A1 | 6 |
| Flt3L | 3 | ||
| DNER | 3 | ||
| IFN.gamma | 3 | ||
| FGF.19 | 3 | ||
| MCP.2 | 3 | ||
| TNFB | 3 |
Supplementary Table 15.
Olink Analytes in Patients With and Without GI Symptoms
| Analyte | Any GI symptoms | Nausea | Vomiting | Diarrhea |
|---|---|---|---|---|
| IL8 | –1.315 | –0.654 | –1.345 | –1.562 |
| VEGFA | 0.006 | 0.014 | –0.356 | –0.027 |
| CD8A | –0.823 | –0.175 | 0.193 | –0.823 |
| MCP.3 | –0.524 | –1.437 | –0.422 | –0.254 |
| GDNF | –1.355 | –1.345 | –0.014 | –1.490 |
| CDCP1 | –0.449 | –0.309 | –0.175 | –0.524 |
| CD244 | –0.023 | 0.110 | 0.126 | –0.017 |
| IL7 | 1.345 | 0.175 | –0.123 | 1.345 |
| OPG | –2.183 | –1.209 | –0.014 | –2.183 |
| LAP.TGF.beta.1 | 0.356 | –0.006 | –0.626 | 0.407 |
| uPA | –0.156 | –0.009 | 0.126 | –0.385 |
| IL6 | –1.063 | –1.022 | –0.254 | –0.747 |
| IL.17C | –1.209 | –0.287 | –0.023 | –1.455 |
| MCP.1 | –0.175 | –0.654 | –0.458 | –0.187 |
| IL.17A | –2.183 | –1.097 | –1.419 | –2.183 |
| CXCL11 | 0.058 | –0.195 | –0.004 | 0.027 |
| AXIN1 | –0.009 | –0.031 | –1.209 | –0.026 |
| TRAIL | 1.063 | 0.626 | 0.314 | 0.458 |
| IL.20RA | –0.548 | –0.573 | –0.563 | –0.618 |
| CXCL9 | –0.573 | –0.367 | 0.023 | –1.209 |
| CST5 | –0.626 | –0.187 | –0.004 | –0.969 |
| IL.2RB | –0.156 | –0.028 | 0.009 | –0.044 |
| IL.1.alpha | 0.046 | –0.023 | 0.001 | 0.162 |
| OSM | –0.178 | –0.117 | –0.175 | –0.242 |
| IL2 | –0.341 | –0.424 | –0.287 | –0.264 |
| CXCL1 | –0.058 | –0.533 | –0.733 | –0.164 |
| TSLP | 0.009 | 0.001 | –0.031 | –0.009 |
| CCL4 | –0.287 | –0.022 | 0.022 | –1.355 |
| CD6 | –0.001 | 0.027 | 0.363 | 0.001 |
| SCF | –0.245 | –0.022 | –0.114 | –0.082 |
| IL18 | –0.254 | –0.068 | 0.332 | –0.434 |
| SLAMF1 | –0.618 | –0.707 | –0.190 | –0.708 |
| TGF.alpha | –1.087 | –0.626 | –0.675 | –1.345 |
| MCP.4 | 0.191 | –0.175 | –0.461 | 0.058 |
| CCL11 | –0.327 | –0.440 | –1.209 | –0.260 |
| TNFSF14 | 0.014 | –0.014 | –0.434 | –0.012 |
| FGF.23 | –0.556 | –0.058 | 0.036 | –1.209 |
| IL.10RA | 0.027 | 0.218 | –0.044 | –0.026 |
| FGF.5 | –0.823 | –0.164 | –0.027 | –1.365 |
| MMP.1 | 0.110 | 0.156 | –0.218 | 0.027 |
| LIF.R | –0.347 | –0.009 | 0.310 | –0.358 |
| FGF.21 | –0.495 | –0.003 | 0.056 | –0.880 |
| CCL19 | –0.044 | –0.227 | –0.156 | –0.175 |
| IL.15RA | –1.345 | –0.175 | 0.022 | –2.177 |
| IL.10RB | –1.355 | –0.389 | –0.079 | –2.183 |
| IL.22.RA1 | –0.073 | –0.022 | –0.009 | –0.254 |
| IL.18R1 | –0.208 | –0.054 | 0.175 | –0.156 |
| PD.L1 | –0.441 | –0.195 | 0.031 | –0.702 |
| Beta.NGF | –0.573 | –0.458 | –0.022 | –0.495 |
| CXCL5 | –0.014 | –0.144 | –0.424 | –0.042 |
| TRANCE | 0.933 | 0.458 | 0.236 | 0.654 |
| HGF | –0.626 | –0.333 | –0.009 | –0.529 |
| IL.12B | 0.060 | 0.058 | –0.009 | 0.038 |
| IL.24 | –0.536 | –0.218 | 0.023 | –0.618 |
| IL13 | –0.007 | 0.075 | 0.068 | –0.126 |
| ARTN | –1.209 | –1.365 | –0.618 | –1.345 |
| MMP.10 | –1.562 | –0.967 | –0.156 | –2.073 |
| IL10 | –0.379 | –0.377 | 0.058 | –0.270 |
| TNF | –0.643 | –0.175 | –0.027 | –0.932 |
| CCL23 | –0.023 | –0.218 | 0.001 | 0.001 |
| CD5 | –0.823 | –0.357 | –0.036 | –0.933 |
| CCL3 | –0.933 | –0.553 | –0.576 | –1.490 |
| Flt3L | 0.031 | –0.009 | –0.377 | –0.009 |
| CXCL6 | –0.156 | –0.385 | –0.347 | –0.385 |
| CXCL10 | –0.001 | –0.441 | –0.012 | 0.031 |
| X4E.BP1 | –0.733 | –0.211 | 0.064 | –1.223 |
| IL.20 | –0.247 | –1.022 | –0.270 | –0.195 |
| SIRT2 | –0.123 | –0.164 | –0.332 | –0.162 |
| CCL28 | –2.183 | –2.183 | –0.737 | –2.183 |
| DNER | 0.175 | 0.317 | 0.332 | 0.202 |
| EN.RAGE | –0.357 | –0.193 | –0.166 | –0.526 |
| CD40 | –0.823 | –0.079 | 0.001 | –1.490 |
| IL33 | –0.236 | –0.164 | 0.023 | –0.287 |
| IFN.gamma | 0.218 | 0.044 | 0.175 | 0.270 |
| FGF.19 | –0.458 | –0.001 | –0.036 | –1.355 |
| IL4 | –0.020 | –0.175 | –0.027 | –0.009 |
| LIF | –1.345 | –0.526 | –0.573 | –1.355 |
| NRTN | 0.001 | –0.168 | –0.036 | –0.012 |
| MCP.2 | 0.319 | –0.327 | –0.036 | 1.231 |
| CASP.8 | –0.438 | –0.377 | –0.264 | –0.731 |
| CCL25 | –0.264 | 0.025 | –0.075 | –0.626 |
| CX3CL1 | –0.556 | –0.164 | –0.009 | –0.810 |
| TNFRSF9 | –1.345 | –0.175 | –0.012 | –2.029 |
| NT.3 | –0.012 | –0.363 | 0.009 | –0.122 |
| TWEAK | –0.014 | 0.014 | –0.113 | –0.012 |
| CCL20 | –0.737 | –0.576 | 0.187 | –0.823 |
| ST1A1 | 0.156 | –0.025 | –0.175 | 0.068 |
| STAMBP | –0.201 | –0.171 | –0.175 | –0.377 |
| IL5 | 0.156 | –0.009 | 0.156 | –0.009 |
| ADA | –0.259 | –0.270 | 0.175 | –0.434 |
| TNFB | 0.009 | –0.009 | –0.036 | 0.014 |
| CSF.1 | –0.458 | –0.028 | 0.012 | –0.450 |
NOTE. Values are P values from t test comparing patients with and without GI symptoms. Signed Benjamini-Hochberg–adjusted P value (–log10 scale) are reported.
Supplementary Table 16.
List of Antibodies Used for Microscopy Studies
| Antigen | Clone | Vendor | Catalog number | Host | Conjugation | Dilution |
|---|---|---|---|---|---|---|
| ACE2 | Polyclonal | Abcam | ab15348 | Rabbit | Unconjugated | 1:1000 |
| EPCAM | SPM491 | GeneTex | GTX34693 | Mouse | Unconjugated | 1:100 |
| SARS-CoV-2 nucleocapsid | Polyclonal | NA | NA | Rabbit | Unconjugated | 1:2000 |
| CD3 | Polyclonal | Abcam | ab5690 | Rabbit | Unconjugated | 1:500 |
| CD8a | C8/468-C8/144B | Abcam | ab199016 | Mouse | Unconjugated | 1:200 |
| MUC2 | SPM512 | Abcam | ab231427 | Mouse | Unconjugated | 1:200 |
| No known specificity (isotype control) | Polyclonal | Abcam | ab37415 | Rabbit | Unconjugated | variable |
| Yeast GAL4 (isotype control) | 15-6E10A7 | Abcam | ab170190 | Mouse | Unconjugated | variable |
| Mouse IgG Heavy and light chains | Polyclonal | Abcam | ab150116 | Goat | Alexa Fluor 594 | 1:1000 |
| Rabbit IgG Heavy and light chains | Polyclonal | Abcam | ab150077 | Goat | Alexa Fluor 488 | 1:1000 |
NA, not applicable.
Supplementary Material
Individual Patient Characteristics From Biopsy Cohort
Montaged tomographic reconstruction of Golgi region of a goblet cell in the apical epithelium of the terminal ileum of a patient with COVID-19. The movie begins with an overview of the goblet cell Golgi complex within a reconstructed volume of a 150-nm section. Intestinal epithelial cells border the goblet cell on the left and right side, and the apical portion of the epithelium is in the upper right, outside of the field of view. The movie progressively zooms in to detail a membrane-bound compartment located at the trans-side of the Golgi stack. This compartment contains 5 presumptive SARS-CoV-2 virions that closely resemble the structures illustrated in Supplementary Figure 6 and in similar data sets of presumptive SARS-CoV-2 virions in SARS-CoV-2–infected cultured cells in Supplementary Movie 2.
Tomographic reconstruction of SARS-CoV-2–infected cultured cells, prepared by high-pressure freezing and freeze-substitution fixation. The movie, which is presented for comparison with analogous structures found in tissue samples (Figure 2 and Supplementary Movie 1) that could not be preserved under similar optimal conditions, shows an overview of a central portion of an infected cell, featuring large numbers of presumptive SARS-CoV-2 virions within membrane-bound compartments, and then increases in magnification to show details of groups of presumptive virions and then individual presumptive virions themselves. Note the consistent presence of dense nucleocapsid puncta arranged within the core of each presumptive virion, membrane bilayers, and occasional surface spike densities visible through the volume.
References
- 1.Gupta A., Madhavan M.V., Sehgal K., et al. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- 2.Sultan S., Altayar O., Siddique S.M., et al. AGA Institute rapid review of the GI and liver manifestations of COVID-19, meta-analysis of international data, and recommendations for the consultative management of patients with COVID-19. Gastroenterology. 2020;159:320–334. doi: 10.1053/j.gastro.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munster V.J., Feldmann F., Williamson B.N., et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585(7824):268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zang R., Gomez Castro M.F., McCune B.T., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5(47) doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamers M.M., Beumer J., van der Vaart J., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao F., Tang M., Zheng X., et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Valle DM, Kim-Schulze S, Hsin-Hui H, et al. An inflammatory cytokine signature helps predict COVID-19 severity and death. medRχiv. Preprint posted online May 30, 2020. 10.1101/2020.05.28.20115758. [DOI]
- 9.Geanon D, Lee B, Kelly G, et al. A streamlined CyTOF workflow to facilitate standardized multi-site immune profiling of COVID-19 patients. medRχiv. Preprint posted online June 29, 2020. 10.1101/2020.06.26.20141341. [DOI]
- 10.Gruber S., van der Laan M. tmle: an R package for targeted maximum likelihood estimation. J Stat Softw. 2012;51:1–35. [Google Scholar]
- 11.Pujadas E., Chaudhry F., McBride R., et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med. 2020;8(9):e70. doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 13.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang S.K., Dohrman A.F., Basbaum C.B., et al. Localization of mucin (MUC2 and MUC3) messenger RNA and peptide expression in human normal intestine and colon cancer. Gastroenterology. 1994;107:28–36. doi: 10.1016/0016-5085(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 15.Barker N., van de Wetering M., Clevers H. The intestinal stem cell. Genes Dev. 2008;22:1856–1864. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laing A.G., Lorenc A., del Molino del Barrio I., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smillie C.S., Biton M., Ordovas-Montanes J., et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 2019;178:714–730. doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotarsky K., Sitnik K.M., Stenstad H., et al. A novel role for constitutively expressed epithelial-derived chemokines as antibacterial peptides in the intestinal mucosa. Mucosal Immunol. 2010;3:40–48. doi: 10.1038/mi.2009.115. [DOI] [PubMed] [Google Scholar]
- 20.Rao X., Wu C., Wang S., et al. The importance of overweight in COVID-19: a retrospective analysis in a single center of Wuhan, China. Medicine (Baltimore) 2020;99(43) doi: 10.1097/MD.0000000000022766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falschlehner C., Schaefer U., Walczak H. Following TRAIL’s path in the immune system. Immunology. 2009;127:145–154. doi: 10.1111/j.1365-2567.2009.03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackall C.L., Fry T.J., Gress R.E. Harnessing the biology of IL-7 for therapeutic application. Nat Rev Immunol. 2011;11:330–342. doi: 10.1038/nri2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H., Zhou P., Wei Y., et al. Histopathologic changes and SARS-CoV-2 immunostaining in the lung of a patient with COVID-19. Ann Intern Med. 2020;172:629–632. doi: 10.7326/M20-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao M., Liu Y., Yuan J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 25.Lucas C., Wong P., Klein J., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leung W.K., To K.F., Chan P.K., et al. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skok K., Vander K., Setaffy L., et al. COVID-19 autopsies: procedure, technical aspects and cause of fatal course. Experiences from a single-center. Pathol Res Pract. 2021;217:153305. doi: 10.1016/j.prp.2020.153305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sia S.F., Yan L.M., Chin A.W.H., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583(7818):834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi J., Wen Z., Zhong G., et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368(6494):1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nobel Y.R., Phipps M., Zucker J., et al. Gastrointestinal symptoms and coronavirus disease 2019: a case-control study from the United States. Gastroenterology. 2020;159:373–375. doi: 10.1053/j.gastro.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aghemo A., Piovani D., Parigi T.L., et al. COVID-19 digestive system involvement and clinical outcomes in a large academic hospital in Milan, Italy. Clin Gastroenterol Hepatol. 2020;18:2366–2368. doi: 10.1016/j.cgh.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao R., Qiu Y., He J.S., et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2020;5:667–678. doi: 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerutti A., Chen K., Chorny A. Immunoglobulin responses at the mucosal interface. Annu Rev Immunol. 2011;29:273–293. doi: 10.1146/annurev-immunol-031210-101317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerutti A. The regulation of IgA class switching. Nat Rev Immunol. 2008;8:421–434. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Lorenzi JCC, Muecksch F, et al. Enhanced SARS-CoV-2 neutralization by secretory IgA in vitro. bioRχiv. Preprint posted online June 9, 2020. 10.1101/2020.09.09.288555. [DOI]
- 36.Cohen A.A., Gnanapragasam P.N.P., Lee Y.E., et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371(6530):735–741. doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 1.Aghemo A., Piovani D., Parigi T.L., et al. COVID-19 digestive system involvement and clinical outcomes in a large academic hospital in Milan, Italy. Clin Gastroenterol Hepatol. 2020;18:2366–2368. doi: 10.1016/j.cgh.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Mastronarde D.N., Held S.R. Automated tilt series alignment and tomographic reconstruction in IMOD. J Struct Biol. 2017;197:102–113. doi: 10.1016/j.jsb.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mastronarde D.N. Correction for non-perpendicularity of beam and tilt axis in tomographic reconstructions with the IMOD package. J Microsc. 2008;230:212–217. doi: 10.1111/j.1365-2818.2008.01977.x. [DOI] [PubMed] [Google Scholar]
- 6.Yao H., Song Y., Chen Y., et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ke Z., Oton J., Qu K., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588(7838):498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein S, Cortese M, Winter SL, et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. bioRχiv. Preprint posted online June 23, 2020. 10.1101/2020.06.23.167064. [DOI] [PMC free article] [PubMed]
- 9.Turoňová B., Sikora M., Schürmann C., et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370(6513):203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calomeni E., Satoskar A., Ayoub I., et al. Multivesicular bodies mimicking SARS-CoV-2 in patients without COVID-19. Kidney Int. 2020;98:233–234. doi: 10.1016/j.kint.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He W., Ladinsky M.S., Huey-Tubman K.E., et al. FcRn-mediated antibody transport across epithelial cells revealed by electron tomography. Nature. 2008;455(7212):542–546. doi: 10.1038/nature07255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ladinsky M.S., Huey-Tubman K.E., Bjorkman P.J. Electron tomography of late stages of FcRn-mediated antibody transcytosis in neonatal rat small intestine. Mol Biol Cell. 2012;23:2537–2545. doi: 10.1091/mbc.E12-02-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geanon D, Lee B, Kelly G, et al. A streamlined CyTOF workflow to facilitate standardized multi-site immune profiling of COVID-19 patients. medRχiv. Preprint posted online June 29, 2020. 10.1101/2020.06.26.20141341. [DOI]
- 14.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 15.Holmes G., Gonzalez-Reiche A.S., Lu N., et al. Integrated transcriptome and network analysis reveals spatiotemporal dynamics of calvarial suturogenesis. Cell Rep. 2020;32(1):107871. doi: 10.1016/j.celrep.2020.107871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reimand J., Kull M., Peterson H., et al. Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35(Suppl 2):W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smillie C.S., Biton M., Ordovas-Montanes J., et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 2019;178:714–730. doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin J.C., Chang C., Boschetti G., et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 2019;178:1493–1508. doi: 10.1016/j.cell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamers M.M., Beumer J., van der Vaart J., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liberzon A., Birger C., Thorvaldsdottir H., et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruber S., van der Laan M. tmle: an R package for targeted maximum likelihood estimation. J Stat Softw. 2012;51:1–35. [Google Scholar]
- 27.Pujadas E., Chaudhry F., McBride R., et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med. 2020;8(9):e70. doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Individual Patient Characteristics From Biopsy Cohort
Montaged tomographic reconstruction of Golgi region of a goblet cell in the apical epithelium of the terminal ileum of a patient with COVID-19. The movie begins with an overview of the goblet cell Golgi complex within a reconstructed volume of a 150-nm section. Intestinal epithelial cells border the goblet cell on the left and right side, and the apical portion of the epithelium is in the upper right, outside of the field of view. The movie progressively zooms in to detail a membrane-bound compartment located at the trans-side of the Golgi stack. This compartment contains 5 presumptive SARS-CoV-2 virions that closely resemble the structures illustrated in Supplementary Figure 6 and in similar data sets of presumptive SARS-CoV-2 virions in SARS-CoV-2–infected cultured cells in Supplementary Movie 2.
Tomographic reconstruction of SARS-CoV-2–infected cultured cells, prepared by high-pressure freezing and freeze-substitution fixation. The movie, which is presented for comparison with analogous structures found in tissue samples (Figure 2 and Supplementary Movie 1) that could not be preserved under similar optimal conditions, shows an overview of a central portion of an infected cell, featuring large numbers of presumptive SARS-CoV-2 virions within membrane-bound compartments, and then increases in magnification to show details of groups of presumptive virions and then individual presumptive virions themselves. Note the consistent presence of dense nucleocapsid puncta arranged within the core of each presumptive virion, membrane bilayers, and occasional surface spike densities visible through the volume.