Abstract
Understanding the persistence of antibody in convalescent COVID-19 patients may help to answer the current major concerns such as the risk of reinfection, the protection period of vaccination and the possibility of building an active herd immunity. This retrospective cohort study included 172 COVID-19 patients who were hospitalized in Wuhan. A total of 404 serum samples were obtained over six months from hospitalization to convalescence. Antibodies in the specimens were quantitatively analyzed by the capture chemiluminescence immunoassays (CLIA). All patients were positive for the anti-SARS-CoV-2 IgM/IgG at the onset of COVID-19 symptoms, and the IgG antibody persisted in all the patients during the convalescence. However, only approximately 25% of patients can detect the IgM antibodies, IgM against N protein (N-IgM) and receptor binding domain of S protein (RBD-IgM) at the 27th week. The titers of IgM, N-IgM and RBD-IgM reduced to 16.7%, 17.6% and 15.2% of their peak values respectively. In contrast, the titers of IgG, N-IgG and RBD-IgG peaked at 4–5th week and reduced to 85.9%, 62.6% and 87.2% of their peak values respectively at the end of observation. Dynamic behavior of antibodies and their correlation in age, gender and severity groups were investigated. In general, the COVID-19 antibody was sustained at high levels for over six months in most of the convalescent patients. Only a few patients with antibody reducing to an undetectable level which needs further attention. The humoral immune response against SARS-CoV-2 infection in COVID-19 patients exhibits a typical dynamic of acquired immunity.
Electronic supplementary material
The online version of this article (10.1007/s12250-021-00360-4) contains supplementary material, which is available to authorized users.
Keywords: Coronavirus disease 2019 (COVID-19), Humoral Responses, Sustainability, Convalescent
Introduction
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has already become a global threat to human health and dramatically paralyzed socio-economic life by causing hundreds and thousands of mortalities and millions of morbidities worldwide. With no specified established antiviral treatment against SARS-CoV-2, adaptive immunity and supportive remedies are currently considered the main source of COVID-19 therapy. Adaptive immunity is triggered by an antigen and further imputed by cell-mediated immunity and antibody production. Nearly all patients will produce antibodies on the exposure to SARS-CoV-2 infection (Long et al. 2020a). However, according to the short-term analysis (Ibarrondo et al. 2020; Long et al. 2020a), the sustainability of specific antibodies against SARS-CoV-2 was shown to last for few weeks or a bit more. We understand these papers are timely reports. Nevertheless, these early records raise an immediate question: how long can the antibodies produced by acquired immunity in COVID-19 patients last? The question is undeviatingly linked to several major concerns: the chance of reinfection by the same serum type of SARS-CoV-2 (Okhuese et al. 2020), the duration of efficacy of the vaccines (Krause et al. 2020) and the pragmatism of herd immunity (Eleni et al. 2020).
Aiming to answer this question and respond to the concerns, we conducted a retrospective study upon a cohort of the COVID-19 patients in Wuhan to accurately measure the longevity and intensity of specific humoral immune responses to SARS-CoV-2 infection from the onset of symptoms to convalesce phase, by using the commercially available chemiluminescence immunoassays kits and detector (CLIA). Based on the investigation, we depict the time-dependent pattern of the anti-SARS-CoV-2 antibody response dynamics and reveal that, with a few exceptions, the antibody level of most patients during convalescence could be maintained at a high level for at least 6 months.
Materials and Methods
Patients
This retrospective cohort study involved 404 serum specimens obtained from 172 COVID-19 patients who were hospitalized in Wuhan during the epidemic peak period and discharged. These serum specimens were collected at onset, hospitalization and rehabilitation period. All the participants were recorded with clinical information such as epidemiology history, exposure history, signs and symptoms, laboratory findings, typical CT image, positive with RT-PCR for SARS-CoV-2. The proportions of age, sex and disease severity of the cohort patients are shown in Table 1.
Table 1.
Constitution of the COVID-19 patient cohort in this study.
| Number | Proportion of the total (%) | ||
|---|---|---|---|
| Age (years old) | < 65 | 117 | 68.0 |
| ≥ 65 | 55 | 32.0 | |
| Sex | Male | 91 | 52.9 |
| Female | 81 | 47.1 | |
| Severity | Mild | 107 | 62.2 |
| Severe | 58 | 33.7 | |
| Critical severe | 7 | 4.1 |
Serologic Testing
In order to analyze the dynamic and distribution of positive rate and antibody titer, the samples were divided into two categories, diseased and convalescent period. The samples of the diseased period were collected in the first 6 weeks with the onset of symptoms to be the initial time point. While the samples collected in the period when the patients discharged from the hospital with two consecutive negative RT-PCR results, were categorized as convalescent period, the duration of this period was mainly from the 12th to the 26th week.
All serum samples were inactivated at 56 °C for 30 min and stored at − 20 °C before testing. The antibodies against SARS-CoV-2 were measured using SARS-CoV-2 chemiluminescent immunoassay (CLIA) kits supplied by Shenzhen Mindray Bio-Medical Electronics Co., Ltd. with a chemiluminescence immunoassays analyzer (CL-900i, DE/CA05/IvD-238321–0297-00). The SARS-CoV-2 chemiluminescent immunoassay kits used in this study were SARS-CoV-2 IgM kit, SARS-CoV-2 IgG kit, SARS-CoV-2 N-IgM kit, SARS-CoV-2 RBD- IgM kit, SARS-CoV-2 N-IgG kit, SARS-CoV-2 RBD-IgG kit, and SARS-CoV-2 Total Antibody kit. All kits were validated with the standard protocol. To simplify the description, the antibodies against SARS-CoV-2 detected in this study are referred to as IgM, N-IgM, RBD-IgM, IgG, N-IgG, RBD-IgG, and S1-total antibody.
According to the manufacturer’s instructions, IgG and IgM detections were developed based on an indirect method. The recombinant antigens containing the nucleoprotein (N protein) or/and receptor-binding domain (RBD) of the spike protein (S protein) of SARS-CoV-2 were coated on paramagnetic microparticles used to capture corresponding antibodies. The anti-human IgG/IgM antibody-conjugated alkaline phosphatase was used respectively as the detection antibody. IgG and IgM detections consist of two steps. In the first step, antigen-coated paramagnetic microparticles captured specific antibodies in the sample. After a wash step to remove unbound substances, anti-human IgG/IgM antibody-conjugated alkaline phosphatase was added to bind to antibodies captured by paramagnetic microparticles. After the second wash step to remove unbound substances, 3-(2′-spiroadamantyl)-4-methoxy-4-(3′’-phosphoryloxy)-phenyl-1,2-dioxetane (AMPPD) was added and catalyzed by alkaline phosphatase to emit light at 540 nm. The resulting chemiluminescent reaction was measured as Relative Light Units (RLUs) by a photomultiplier in the instrument.
The total antibody kit was developed based on a double-antigen sandwich immunoassay. The recombinant antigens containing S1 protein were coated on paramagnetic microparticles and conjugated alkaline phosphatase. The detection is a one-step method. Antigen-coated paramagnetic microparticles and antigen-conjugated alkaline phosphatase bound the specific antibodies in the sample. After a wash step to remove unbound substances, AMPPD was added and catalyzed by alkaline phosphatase to emit light at 540 nm. The resulting chemiluminescent reaction was measured as RLUs.
IgG antibodies and S1-total antibodies were calculated as AU/mL and the Cutoff value is 10 AU/mL. IgM antibodies were presented as the measured Relative Light Units (RLUs) divided by the cutoff value (cutoff index, COI): COI ≥ 1 was defined as positive and COI < 1 as negative. The cutoff value of the positive antibody titer was determined according to the manufacturer's instructions.
Statistical Analysis
Based on the antibody level changed over time from the first to the 26th week (with exclusive of 6th to 12th week), dynamics of antibodies were described by locally weighted regression and smoothing scatterplots (LOESS) model. Pearson correlation coefficient analysis (Pearson correlation) was applied to analyze the correlation of antibody dynamics. The similarity of antibody dynamic in different groups was quantitatively analyzed by the Kolmogorov–Smirnov test, including antibody level (distance) and distribution pattern (P value). Unless otherwise stated, statistical analysis was conducted by the software R (The R Foundation, http://www.r-project.org, version 4.0.0) and GraphPad Prism 8.0.
Results
Antibody Positive Rate
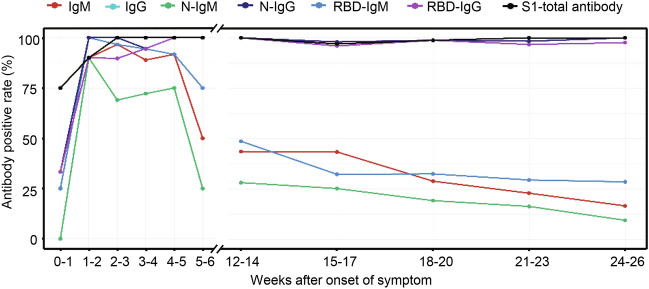
The antibody-positive rate reflects the proportion of the people who develop humoral immune response after being infected by SARS-CoV-2. As shown in Fig. 1, at the onset of the symptoms, the anti–SARS-COV-2 antibodies IgM/IgG positive rates peaked at the 2nd week. Likewise, the peak value of positive rate in N-IgG was observed at the 2nd week, but the RBD-IgG positive rate peaked at the 5th week. The positive rates of RBD-IgM and the N-IgM peaked at the 2nd week and then began to decline rapidly. During the first 6 weeks, the peak positive rates of IgM, N-IgM and RBD-IgM were 96.6%, 90% and 100%, respectively. Subsequently, at the convalescence period, no obvious decline in the positive rate of the total antibody was witnessed. Similarly, the positive rates of IgG, N-IgG, and RBD-IgG sustained a high level. However, the positive rates of the IgM, N-IgM and RBD-IgM continued to decline and dropped to approximately 25% at the 27th week.
Fig. 1.
The positive rate of antibodies from the onset of symptoms to convalesces. Demonstrating the positive rate of different antibody appearances, peak point, intensities and durability from the onset of the symptoms (on hospitalization) to the convalescent phase till 26th week. Note: IgG line is overlapped by N-IgG.
Antibody Titer
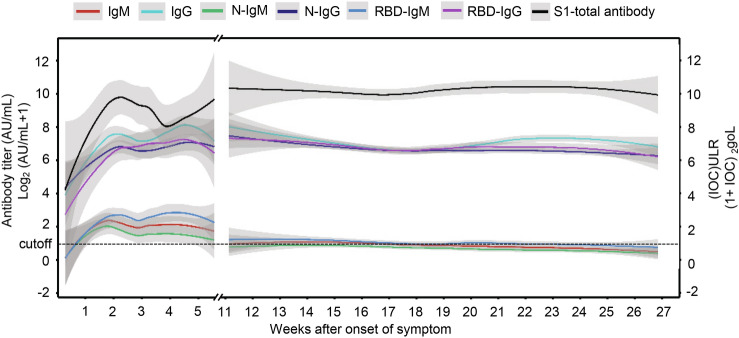
Figure 2 presents the fitting dynamics of the magnitude of the antibodies induced by SARS-CoV-2 through the locally weighted scatterplot smoothing (LOESS) analysis method. The titer of IgG, N-IgG and RBD-IgG all peaked at the 4th to 5th week. Similarly, the peaks of IgM, N-IgM and RBD- IgM titers were observed at the 4th week. Moreover, a significant difference was noticed in the waning of IgG and IgM in the convalescent period. No obvious decline was found in the titer of IgG, N-IgG and RBD-IgG throughout this period, while the IgM level, N and RBD-IgM waned off quickly. Disparately, the average value of IgM and the N-IgM titers decreased below the cutoff value before the 11th week, and RBD -IgM at the 16th–17th week. During the observation period, the titers of IgM, N-IgM and RBD-IgM only left 16.7%, 17.6% and 15.2%, respectively. On the contrary, the intensity of IgG, N-IgG and RBD-IgG titers retained 85.9%, 62.6% and 87.2%, respectively. Interestingly, the total antibody level was found sustained at a high level for over 6 months, with the average value of 1961 AU/mL from 4th–6th month. All measured invidious data with trend line LOESS fitting can be seen in Supplementary Figure S1.
Fig. 2.
The titer of different antibodies from the onset of symptoms to the convalescent phase. The titer of different antibodies starts rising from the onset of symptoms to their peak value during the infection with notable disparities in their intensities, and the most sustainable antibodies can maintain up to 6 months. The dotted line is the cutoff value, and the gray shading is the 95% confident interval.
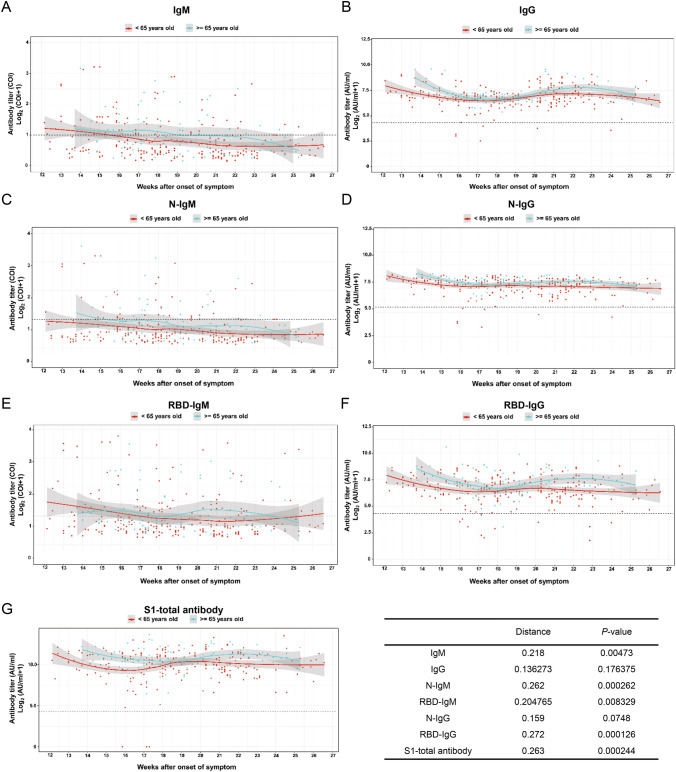
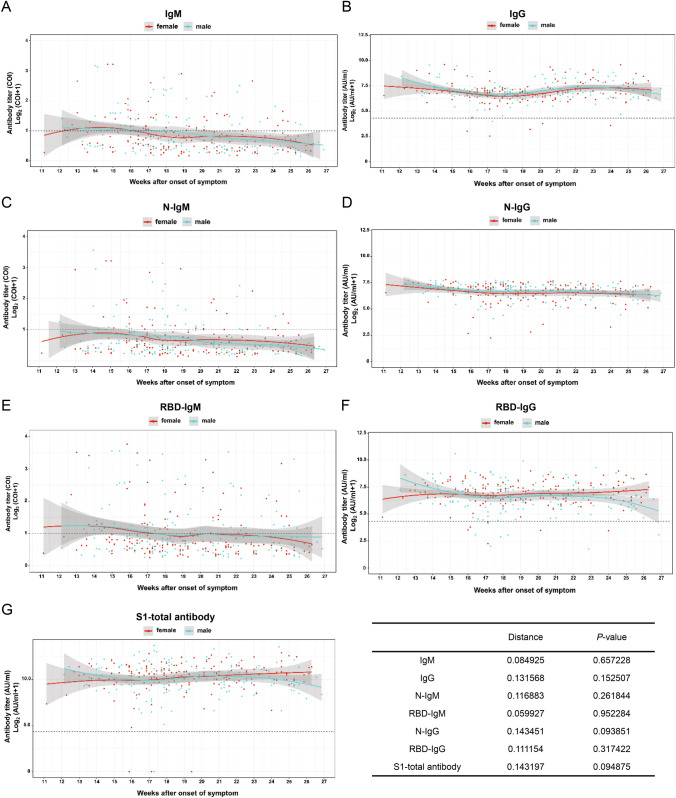
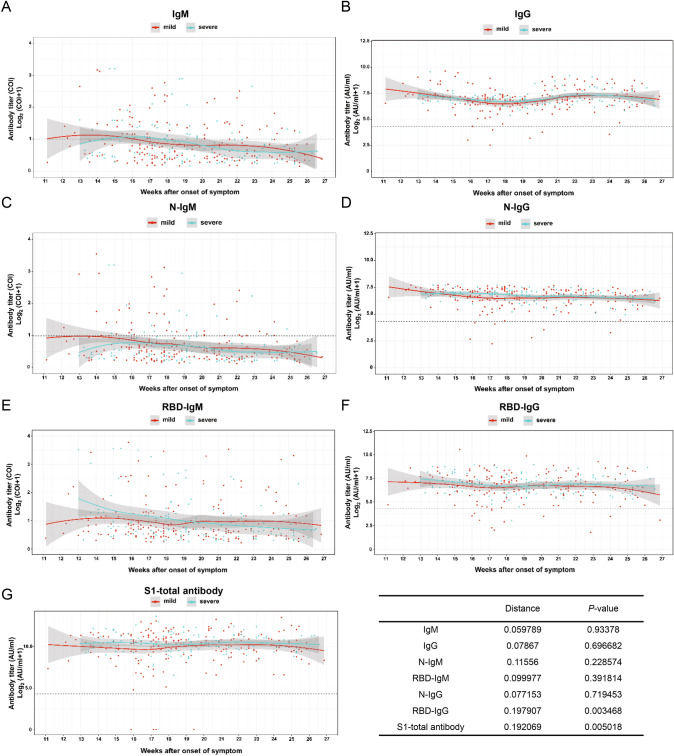
To investigate the correlation of age, gender and disease severity with antibody dynamic pattern, the test results were classified and the data were fitted by the software R. Further, the Kolmogorov–Smirnov test (Manoel et al. 1996), which is a nonparametric test of the equality of continuous, one-dimensional probability distribution, is employed to quantify the level similarity of the antibody dynamics among the comparison groups, including both antibody level (distance) and distribution pattern (P-value) during convalescence. The smaller K-S distance derived from Cumulative Distribution Functions among subgroups is equal to the measurement of their similarity of titer (Baselice et al. 2019). The test results indicate the distribution pattern is positively correlated to the distance value. Figure 3 presents that the dynamic patterns of IgM, N-IgM, RBD-IgM, RBD-IgG and S1-total antibody have significant differences (P < 0.05) in age groups. All antibodies reveal less variation (P > 0.05) between male and female patients (Fig. 4). The dynamic patterns of S1-total antibody and RBD-IgG have obvious variation between mild and severe groups (P < 0.05), while the dynamic patterns of the other five antibodies reveal less variation (Fig. 5). Further investigations are necessary to understand the clinical significance of these differences and similarities in antibody magnitude in COVID-19 patients.
Fig. 3.
Comparison of humoral responses of the patients with different ages in convalescence. A total of 326 samples was tested, they are divided into two age groups (< 65 yr and ≥ 65 yr). The titer levels of seven kinds of antibodies show a slight difference between the groups. Except for IgG and N-IgG, the titer trends of IgM, N-IgM, RBD-IgM, RBD-IgG and S1-total antibody are significantly different between the two age groups (P < 0.05). The trend line is fitted by LOESS and 95% confidence interval was shown in gray. The dashed line is the cutoff value. The inserted table summarizes the statistical data of the age group.
Fig. 4.
Comparison of humoral responses of male and female patients in convalescence. A total of 326 samples was tested, they are divided into two gender groups (female and male). The titer trends of all seven kinds of antibodies show no significant difference between the two gender groups (P > 0.05). The trend line is fitted by LOESS and 95% confidence interval was shown in gray. The dashed line is the cutoff value. The inserted table summarizes the statistical data of the gender group.
Fig. 5.
Comparison of humoral response in patients with mild and severe symptoms in convalescence A total of 326 samples was divided into mild and severe (including critical severe) groups. Except for IgM, IgG, N-IgM, N-IgG, RBD-IgM, the titer trends of RBD-IgG and S1-total antibody are significantly different between the two severity groups (P < 0.05). The trend line is fitted by LOESS and 95% confidence interval was shown in gray. The dashed line is the cutoff value. The inserted table summarizes the statistic data of the severity group.
Special Cases
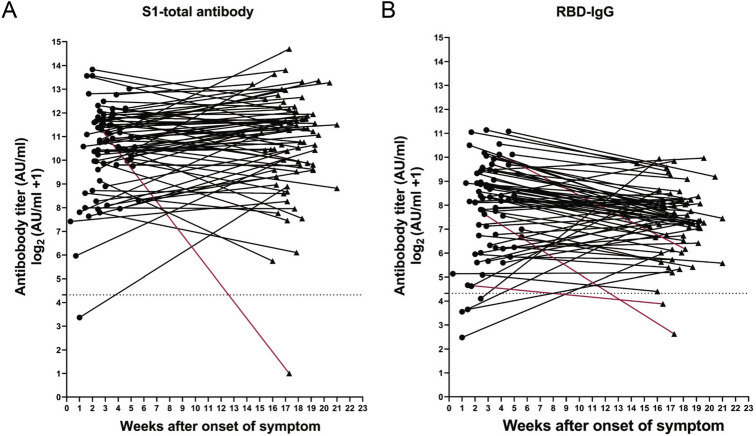
In overall evaluation, our results unveiled a few special cases that exhibited unique humoral responses after confronted by COVID-19. Of these cases, one developed the total antibody at the onset of the disease but it rapidly declined to the almost unmeasurable range at 17th–18th week (Fig. 6A), and the other one did not develop the total antibody response at all (data not shown). The IgG level remained at a low level in one convalescent patient (only less than 10% of the average level) while another individual did not generate IgG at all. Curiously, two patients’ RBD-IgG was at undetectable levels and one patient remained at a low level (less than 10% of the average level) within 3–4 months, and two did not develop RBD-IgG while they generated IgG (Fig. 6B).
Fig. 6.
Patients with unusual immune responses. A total of 78 patients were twice sampled during the convalescent period. The titer of S1-total and RBD-IgG antibodies were evaluated and some individuals exhibited unusual immune responses. A one case with a rapid decline in S1-total antibody; B RBD-IgG levels in three cases dropped rapidly. Abnormal antibody trends marked by red.
Correlation of Antibodies
We used the change in antibody titer as a reference to analyze the correlations of those antibodies. As shown in Fig. 7, N-IgM and IgM present the highest degree of positive correlation, and there is also a high positive correlation between N-IgG and IgG. There are weak correlations between RBD-IgM and IgM, RBD-IgG and IgG. This means that N antigen can cause the body to produce IgG and IgM easier than RBD antigen. However, it is interesting that the change of RBD-IgG titer is positively correlated with the change of S1-total antibody titer. Combine with the above phenomena, this would be interpreted as RBD may more induce the production of other forms of antibodies directly, such as IgA. The phenomenon and speculation need to be further confirmed.
Fig. 7.
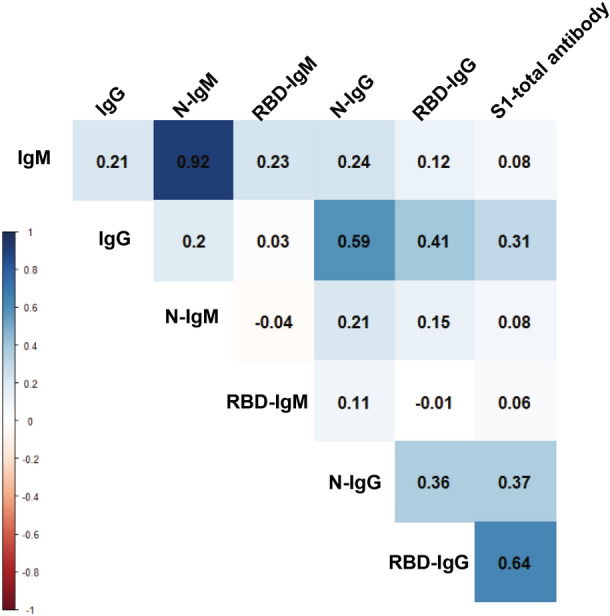
The correlation amongst different humoral responses in convalescence. There are great correlations between N-IgM and IgM, N-IgG and IgG, RBD-IgG and S1-total antibody, analyzed in 326 samples.
Discussion
The main finding of this study is that the levels of S1-total antibodies against SARS-CoV-2 and RBD-IgG antibodies in COVID-19 patients can be stably maintained for at least 6 months. The antibody waning pattern during the onset of the disease and rapid decline of IgM is similar to many previous observations (Long et al. 2020a; Kellam et al. 2020). Therefore, we will not go into the details of the onset phase but focus on the convalescent period. Previous reports described the short-term pattern of antibody level in a small number of population samples (Liu et al. 2020) and the sustainability of antibodies in asymptomatic (Long et al. 2020b) and mild patients (Ibarrondo et al. 2020). In this study, we found that the antibody titer, especially the S1-total antibody and RBD-IgG levels can continue to exist at high levels, which provides a positive signal for the anti-secondary infection. Studies on infections induced by the SARS virus show that the positive rate of humoral immunity produced in the human body remained above 80% within 3 years (Cao et al. 2007). Although the levels of IgG and neutralizing antibodies continue to decline, their residuals are still abundant, which has reference value for the study of SARS-CoV-2 immunity. However, SARS-CoV-2 is new to us, and the monitoring of the anti-SARS-CoV-2 antibodies will continue.
A report about SARS-CoV-2 reinfection enflames people’s trepidations regarding the recurrence COVID-19 (To et al. 2020). Our current findings further disclose that most of the convalescent COVID-19 patients have had a stabilized antibodies level after being discharged from the hospital and should be able to resist the risk of re-infection by the same serum type SARS-CoV-2 virus at least, and during a follow-up visit, nobody was found recurrence or re-infected. According to WHO, there are hundreds of COVID-19 vaccine candidates under development, with a few already in the market. With no established specified anti-SARS-CoV-2 therapy to this date, vaccines against SARS-CoV-2 are highly anticipated, or even thought to be the ultimate solution. However, the rapid decline of antibody levels in mild patients has escalated the public’s anxiety about the long-term effectiveness of the COVID-19 vaccines (Ibarrondo et al. 2020). Since vaccines and natural viral infections have similar immunological mechanisms, the findings of this study provide some reference for the evaluation of the long-term efficacy of vaccines. These uncertainties can only be elucidated on the completion of the human trials and large-scale vaccination. Another fact is that the majority of the SARS-CoV-2 infected people (> 98% in this study) can retain their S1-total antibody and RBD-antibody at a high level (> 1900 AU/mL) for over 6 months. This could be also a reference for the vaccination areas to build active herd immunity.
Remarkably, we found some special cases with unusual immune responses, which are predominantly focused currently. These special groups exhibited unique humoral responses after confronted by SARS-CoV-2. The individuals were either unable to provoke specific immune response timely or their antibody level dropped abruptly in an unusual manner. These special cases are of high importance with an immunological perspective and might be significantly vulnerable to reinfection. Until now, there is only one definitive report of reinfection within 4 months, proven by genetic sequencing of the SARS-CoV-2 (Cao et al. 2007). In that case, the re-infection possibly occurred due to the early decay of antibodies with no protective immunity, accentuating that the special patients without proper humoral immunity are at the risk of re-infection. Although currently a lot of attention has been placed on antibody-based immunity, there are increasing supportive evidences that T cells can play an important role against COVID-19 (To et al. 2020). Based on our findings, some patients did not produce antibodies or the antibody titer was under the undetectable level. Surprisingly, those patients successfully recovered from COVID-19, emphasizing the potential role of T-cell immunity. However, it is not clear whether SARS-CoV-2 can induce long-term T memory cell responses, and the importance of these responses in persistent immunity is to be investigated.
To our knowledge, this study is one of the longest longitudinal evaluations, based on a large number of specimens of the convalescent COVID-19 patients. The humoral immune response induced by SARS-CoV-2 infection exhibits typical dynamic behavior of acquired immunity. We classified these responses based on their magnitude, titer intensities, waning abilities and sustainability over time. It is clear that the S1-total antibody and RBD-IgG can continue to maintain a high level for more than 6 months. The findings of this study are helpful, to some extent, to sort out the issues of re-infection, herd immunity, and the efficacy of antiviral vaccines, which are major concerns in the current stage of the COVID-19 pandemic.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgement
This work was supported by the National Key Research and Development Program of China (Grant No. 2020YFC0861100 and 2020YFC0000), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB29050100), the Key Technology Development Program of Shenzhen (Grant No. JSGG20200225153042494), the Youth Innovation Promotion Association of CAS (Grant No. 2014308), and the Interdisciplinary Innovation Team of CAS. We thank Dr. Ding GAO in the core facility and the Technical support of the Wuhan Institute of Virology. We are also thankful to Prof. Fei Deng in the National Virus Resource Center for her technical support.
Author Contributions
QZ, XLZ, ZQY, XMC, LHD, XYS, QZ, YL, YKL, BY, and HW cared for the patient and extracted data in the hospital. YGC, XLT, DM, X.E.Z conceived and designed the experiments. YZ, NS, SSC, JZ, JC, AA, ML, MMC performed laboratory experiments. KL, QFY, DM developed the detection method. NS, DBW and BDH are involved in data analysis. YZ, AA, DM, X.E.Z wrote the manuscript. X.E.Z. supervised the whole project.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
The study was approved by the Ethics Commission of Hubei Provincial Hospital of Traditional Chinese Medicine and Wuhan Institute of Virology, Chinese Academy of Sciences.
Footnotes
Yang Zheng and Qing Zhang contributed equally to this work.
Contributor Information
Yegang Cheng, Email: chengyegang@hbhtcm.com.
Xiaolin Tong, Email: tongxiaolin@vip.163.com.
Dong Men, Email: d.men@wh.iov.cn.
Xian-En Zhang, Email: zhangxe@ibp.ac.cn.
References
- Baselice F, Ferraioli G, Pascazio SA. Denoising of MR images using Kolmogorov-Smirnov distance in a non-local framework. Magn Reson Imaging. 2019;57:176–193. doi: 10.1016/j.mri.2018.11.022. [DOI] [PubMed] [Google Scholar]
- Cao WC, Liu W, Zhang PH, Zhang F, Richardus JH. Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med. 2007;357:1162–1163. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- Eleni P, Loukas K, Konstantinos P, George P. Long-term and herd immunity against sars-cov-2: implications from current and past knowledge. Pathog Dis. 2020;78:ftaa025. doi: 10.1093/femspd/ftaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarrondo FJ, Fulcher JA, Goodman-Meza D, Elliott J, Yang OTTO. Rapid decay of anti-sars-cov-2 antibodies in persons with mild covid-19. N Engl J Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellam P, Barclay W. The dynamics of humoral immune responses following sars-cov-2 infection and the potential for reinfection. J Gen Virol. 2020;101:791–797. doi: 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause P, Fleming TR, Longini I, Henao-Restrepo AM, Henao-Restrepo AM. Covid-19 vaccine trials should seek worthwhile efficacy. Lancet. 2020;396:741–742. doi: 10.1016/S0140-6736(20)31821-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang J, Xu X, Liao G, Hu CH. Patterns of IgG and IgM antibody response in covid-19 patients. Emerg Microbes Infect. 2020;9:1269–1274. doi: 10.1080/22221751.2020.1773324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, Liao P, Qiu JF, Lin Y, Cai XF, Wang DQ, Hu Y, Ren JH, Tang N, Xu YY, Yu LH, Mo Z, Gong F, Zhang XL, Tian WG, Hu L, Zhang XX, Xiang JL, Du HX, Liu HW, Lang CH, Luo XH, Wu SB, Cui XP, Zhou Z, Zhu MM, Wang J, Xue CJ, Li XF, Wang L, Li ZJ, Wang K, Niu CC, Yang QJ, Tang XJ, Zhang Y, Liu XM, Li JJ, Zhang DC, Zhang F, Liu P, Yuan J, Li Q, Hu JL, Chen J, Huang AL (2020a) Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 26:845–848 [DOI] [PubMed]
- Long QX, Tang XJ, Shi QL, Li Q, Huang AL (2020b) Clinical and immunological assessment of asymptomatic sars-cov 2 infections. Nat Med 26:1200–1204 [DOI] [PubMed]
- Okhuese AV. Estimation of the probability of reinfection with covid-19 by the susceptible-exposed-infectious-removed-undetectable-susceptible model. JMIR Pub Health Surveill. 2020;6:e19097. doi: 10.2196/19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoel AW, PachecoAndrew H. Testing association between species abundance and a continuous variable with kolmogorov-smirnov statistics. Plant Ecol. 1996;124:95–99. doi: 10.1007/BF00045147. [DOI] [Google Scholar]
- To KW, Hung FN, Ip JD, Chu WH, Yuen KY (2020) Covid-19 re-infection by a phylogenetically distinct sars-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 10.1093/cid/ciaa1275 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.