Abstract
How plant cells expand and gain shape remains enigmatic. Current models consider that these processes are driven by turgor pressure acting upon the cell wall. Using nanoimaging, we show that the cell wall contains pectin nanofilaments that possess an intrinsic expansion capacity. We show, by growth models containing such structures, that a complex plant cell shape can derive from a chemically induced local and polarised expansion of the pectin nanofilaments, without turgor-driven growth. Thus the plant cell wall, outside of the cell itself, is an active participant in shaping plant cells. Extracellular matrix function in multiple kingdoms, including animals, may similarly guide cell shape.
One Sentence Summary:
Methylation of ordered pectin drives changes in plant cell wall shape.
The plant cell wall, a component of the plant extracellular matrix, responds to extra- and intracellular cues to affect cell shape, cell size, and cell division (1–3).
It is an intricate composite of polysaccharides, such as cellulose, hemicellulose, and pectins. The cell wall is considered biphasic: crystalline cellulose microfibrils tethered by hemicellulose are submerged in a gel-like matrix of pectins and proteins. How its components associate into a coherent, strong and dynamic material remains unknown. Cellulose microfibrils, the main load-bearing components that drive growth anisotropy, are deposited directly in the cell wall by plasma membrane-localized cellulose synthase complexes that are guided by cortical microtubules (4–7). Pectins constitute a structurally diverse family of polysaccharides with the defining feature of 1,4-linked α-D-galactosyluronic acid (GalpA). We focus on homogalacturonan (HG) polysaccharides containing exclusively linear chains of GalpA. Homogalacturonan also exists as HG glycan domains in heteroglycans containing more complex and branched pectins (rhamnogalacturonan type I and II), and in glycoconjugates such as the proteoglycan APAP1 (8, 9). Homogalacturonans are synthesized in methylated form and undergo a demethylesterification only after insertion into the wall (2). The demethylated HG and the resulting negatively charged acidic carboxyl groups it contains, is present in rapidly expanding cells and demethylation correlates with wall elasticity change (2, 3). However, the relationship between HG methylesterifiction and growth remains elusive. In the primary cell wall, matrix polysaccharides may form a continuous cross-linked network with covalent bonds to structural glycoproteins. Nuclear Magnetic Resonance indicates numerous intermolecular links between wall polysaccharides, notably direct association of cellulose and pectins, though their supramolecular complexes are yet to be determined (10).
In the currently accepted model, plant cell growth is driven by turgor pressure (11) causing strain in the wall and leading to irreversible deformation or creep (12). On the molecular level this is thought to be mediated by reorganizing the cellulose microfibrils (wall remodeling) (11). Anisotropic expansion is caused by ordered arrays of cellulose microfibrils that restrict growth parallel to their orientation (13), however, some evidence suggests that microfibril alignment is not sufficient for anisotropic expansion (14).
Here we study growth and morphogenesis of epidermal cells, the pavement cells, in the Arabidopsis cotyledon (Fig. 1A). These cells possess an undulatory pattern in their lateral (anticlinal) walls, called “lobes”, with mathematical order that can be perceptualized by data sonification (fig. S1, Audio files S1 and S2). Lobes are initiated by ROP GTPases concentrating microtubules at the future convex position (15, 16) associated with increased cell wall thickness, radial microfibril distribution, and HG demethylesterification (16, 17). Turgor-induced buckling of anticlinal walls due to tension in periclinal walls and their local reinforcement linked to pectin demethylation has been proposed to explain pavement cell shape (18).
Fig. 1. The 3D-dSTORM nanoscopy and cryoSEM reveals homogalacturonan nanofilaments.
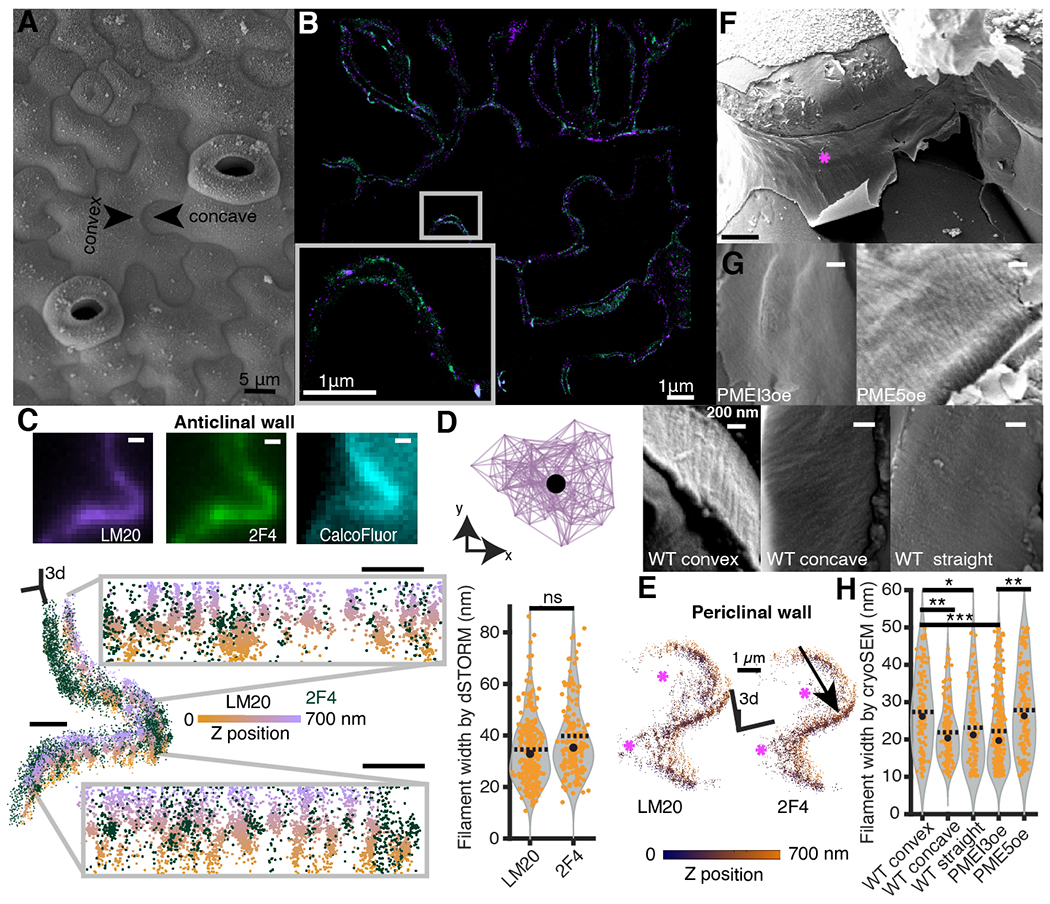
(A) A surface view of cotyledon epidermal pavement cells using cryoSEM. (B) A high magnification epidermal region observed by dSTORM; methylated (violet) and demethylated (green) HG. Pixel size, 10 nm. (C) Diffraction-limited image of the lobed region of the anticlinal wall (top), and the same region imaged with 3D-dSTORM shown as a 3D scatterplot of the coordinates of localized emitters (bottom); demethylated (green) and methylated (orange-violet colormap encoding the Z-position) HG nanofilaments. Grey insets represent two wall segments in the orthogonal view. Scale bars, 500 nm. (D) Lateral view of segmented nanofilament represented as a bidirectional graph (top); the filament width is estimated as the median pairwise distance between centroid (black dot) and each point in the graph. The nanofilament width (bottom) representing respectively 159 and 96 methylated and demethylated filaments. (E) The 3D-dSTORM imaging of lobed wall segment showing junction between the anticlinal wall (black arrow) and periclinal walls (magenta stars). The orange-blue colormap encodes Z-position. (F) Representative gross-scale cryoSEM picture with the fracture exposing convex anticlinal wall (magenta star). Pixel size, 20 nm. (G) Fine-scale pictures of the convex anticlinal walls in PMEI3oe and PME5oe (top row) and WT convex, concave and straight anticlinal walls (bottom row). Pixel size, 4 nm. (H) The filament width distribution observed by CryoSEM, representing 109, 137 and 104 filaments measured in respectively WT convex, concave and straight walls. For PME5oe and PMEI3oe, the convex and concave wall data were pooled together; 139 and 322 filaments were measured in PME5oe and PMEI3oe. P-values: * - p < 0.05, ** - p < 0.001, *** - p < 0.0001, obtained with multiple group comparison Kruskal-Wallis test and Bonferroni correction. In (D) and (H), black dot and dashed line represent median and mean respectively.
The prevailing view of pectin tertiary structure in the wall is as an amorphous collection of polymers. However, there is a rich literature proposing a crystalline structure for HG (19, 20 and references within). Prior in vitro X-ray diffraction studies identified HG tertiary structures as helices with three galacturonic acid residues per helical turn. These helices arrange uniaxially in fibrous quaternary structures packed in hexagonal and rectangular lattices for methylated and demethylated HG, respectively (19, 20). Yet, little is known about HG higher-order structure in intact tissues.
Here we present the in muro nanostructure of the homoglycan polymeric form of HG using super-resolution three-dimensional direct STochastic Optical Reconstruction Microscopy (3D-dSTORM) (21–25) and cryo-Scanning Electron Microscopy (cryoSEM). We show that in the cotyledon anticlinal walls HG assembles into discrete nanofilaments rather than a continuously interlinked network. Although the fine structure of these nanofilaments is not discerned with our techniques, we propose that they may be a quaternary structure similar to that observed by X-ray diffraction. This led us to formulate an intrinsic cell wall expansion ‘expanding beam’ model of pavement cell morphogenesis. In this model, local homogalacturonan demethylesterification leads to nanofilament radial swelling, caused by conversion between quaternary structures with different packaging. We further test this hypothesis by showing that demethylesterification of HG alone is sufficient to induce tissue expansion. Finally, we formalize the model as a three-dimensional nonlinear finite element method (FEM) model predicting tissue topology, local cell wall thickness, tension, and growth.
Results
Quaternary structure of polymeric homogalacturonans as nanofilaments
The 3D-dSTORM nanoscopy provides insight into biological structures at the nanometer scale (21–25). Combining 3D-dSTORM and immuno-labeling using antibodies against highly methylesterified (LM20) and low- or non-esterified (2F4) HG on 4 µm thick tissue sections we obtained ~40–50 nm lateral and ~80 nm axial resolution and depth reconstruction of ~800 nm (26, 27), (figs. S2 and S3). Both antibodies bind in the cell wall near the plasma membrane, but rarely in the middle lamella, suggesting limited epitope accessibility to an antibody or lack of such epitopes in this location, (Fig. 1B). 3D-dSTORM revealed that in the anticlinal walls HG forms aligned filaments perpendicular to the cotyledon surface, which we hereafter call HG nanofilaments (Fig. 1C, figs. S4 and S5A, and Movie S1). Their estimated width is ~40 nm (Fig. 1D and fig. S5B), which is the limit of our reported lateral resolution. In the periclinal walls, in contrast, we didn’t detect a filamentous pattern, suggesting distinctive HG organization in different walls of the same cells (Fig. 1E and fig. S5C and Movies S2 to S4). Different photo-switching properties of fluorescent probes used in this study, Alexa647 and CF568, makes the nanofilament organization more apparent when using Alexa647 (28), (fig. S3C). Swapping the fluorophore confirmed that HG in anticlinal walls organizes into parallel and often interspaced nanofilaments, either predominantly methylated or demethylated (fig. S5D, and Movies S5 and S6). These observations let us hypothesize that HG nanofilaments are the quaternary structures predicted by X-ray diffraction studies. However, 3D-dSTORM resolution was insufficient to discern any difference in LM20 and 2F4 tagged nanofilaments.
HG methylation affects cell wall filament diameter
To gain better insight into the HG nanofilaments architecture, we used cryo-fracture SEM, which, by using hydrated and ultra-frozen samples, permits examination of the native tissue structure. The cryogenically prepared tissue is fractured, allowing the observation of otherwise hidden anticlinal walls (Fig. 1F). Similarly to 3D-dSTORM, cryoSEM revealed organized filaments in anticlinal, but not periclinal walls (Fig. 1G, fig. S6A), with median (± median absolute deviation) full width at half-maximum (FWHM in nm) of 26 ± 8, 20 ± 4 and 21 ± 7 and a median filament interspacing of 58 ± 18, 42 ±10 and 46 ± 12 in convex and concave sides of lobed walls, and in straight walls, respectively (Fig. 1H and fig. S6, B and C). To test the potential impact of HG differential methylation on the cryoSEM structures, we used plants with inducible overexpression of PECTIN METHYLESTERASE 5 (PME5oe) to promote HG demethylation and inducible overexpression of PECTIN METHYLESTERASE INHIBITOR 3 (PMEI3oe) to inhibit demethylation (2, 29, 30). We observed filaments with a median width of 26 ± 9 nm and 19 ± 7 nm and median interspacing of 49 ± 17 nm and 39 ± 15 nm respectively for PME5oe and PMEI3oe (Fig. 1, G and H). Filaments in HG-demethylated walls (PME5oe) were approximately 1.4 times the cross-section of filaments in HG-methylated (PMEI3oe), consistent with in vitro differential packaging of methylated and demethylated HG in distinct quaternary structures (19, 20). While the cryoSEM lacks chemical information, the filament position, size, and reaction to PME activity suggest they contain HG. These data and the close association of cellulose with pectin supported by Nuclear Magnetic Resonance let us speculate that cellulose and HG may coexist in the same higher-order structures (10).
Demethylation-mediated nanofilament inflation underlie anisotropic growth
We propose that lobes in the anticlinal wall could emerge by spatially varying HG demethylation along and across the wall causing local radial swelling of the HG nanofilaments, thus local wall expansion. To formalize this hypothesis we refer to the in vitro quaternary structure of homogalacturonans (19) where uniaxially oriented helices of methylated HG pack in a hexagonal net with a = 0.837 nm side length whereas demethylated and calcium-bound HG in a rectangular lattice with unit cell dimensions b = 1.23 nm and c = 0.99 nm. We hypothesize that, in vivo, HG demethylation converts between these quaternary structures, leading to the expansion ratio 1.42 in average lateral dimension and shortening in the axial dimension of -0.013 nm per helical repeating unit (Materials and Methods). The ~1.42-fold expansion is compatible with our cryoSEM measurements of HG nanofilaments width going predominantly from methylated (PMEI3oe) to demethylated (PME5oe) form (Fig. 1H).
Finite Element Method implementation of the model
To test the above hypothesis, we developed a 3D nonlinear finite element method model that simulates lobe growth and morphogenesis by local swelling of oriented HG nanofilaments (31, 32). The two-cell junction is comprised of the bottom and top periclinal walls connected by a perpendicular anticlinal wall (fig. S7). At each growth iteration, exocytosis deposits an amount of methylated HG proportional to the area of the element’s face directly in contact with the cytoplasm. The change of the element’s edge relaxation length is proportional to the HG demethylation rate obtained from 3D-dSTORM data and follows the local HG anisotropic organization (Fig. 2, and figs. S8 and S9). HG exocytosis rate was the only non-experimental parameter and was set initially to 0.05. The primary outputs of the model are the shape prediction - the lobes in anticlinal walls, the topography of the periclinal walls - and the local cell wall thickness. Starting from a straight wall after around 200 iterations, we observe maturing lobes in the anticlinal wall, corresponding in shape to 2–3 days after germination in plant tissue (Fig. 3, A to C).
Fig. 2. HG methylation asymmetry affects lobe formation.
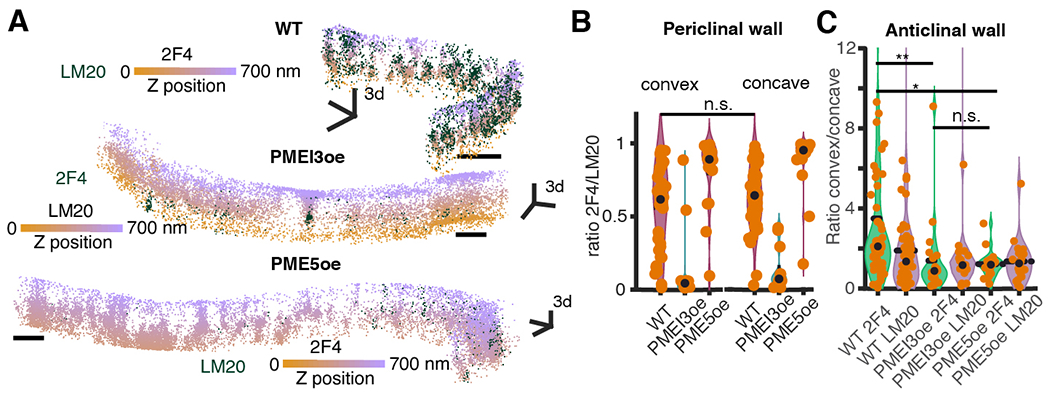
(A) Representative lobed wall segments imaged with 3D-dSTORM in WT, PME5oe and PMEI3oe cotyledons. The orange-violet colormap encodes Z-position. Scale bars, 500 nm. (B) HG methylation asymmetry between convex and concave regions of a lobe in the periclinal walls and (C) in the lobed anticlinal walls for WT, PME5oe and PMEI3oe and straight walls for WT plants. P-values: * - p < 0.05, ** - p < 0.001, *** - p < 0.0001, obtained with multiple group comparison Kruskal-Wallis test and Bonferroni correction. The number of analyzed regions: 100, 39, and 23 (B), 53, 22 and 25 (C) for WT, PME5oe, and PMEI3oe cotyledons respectively.
Fig. 3. The expanding beam model of pavement cell shape.
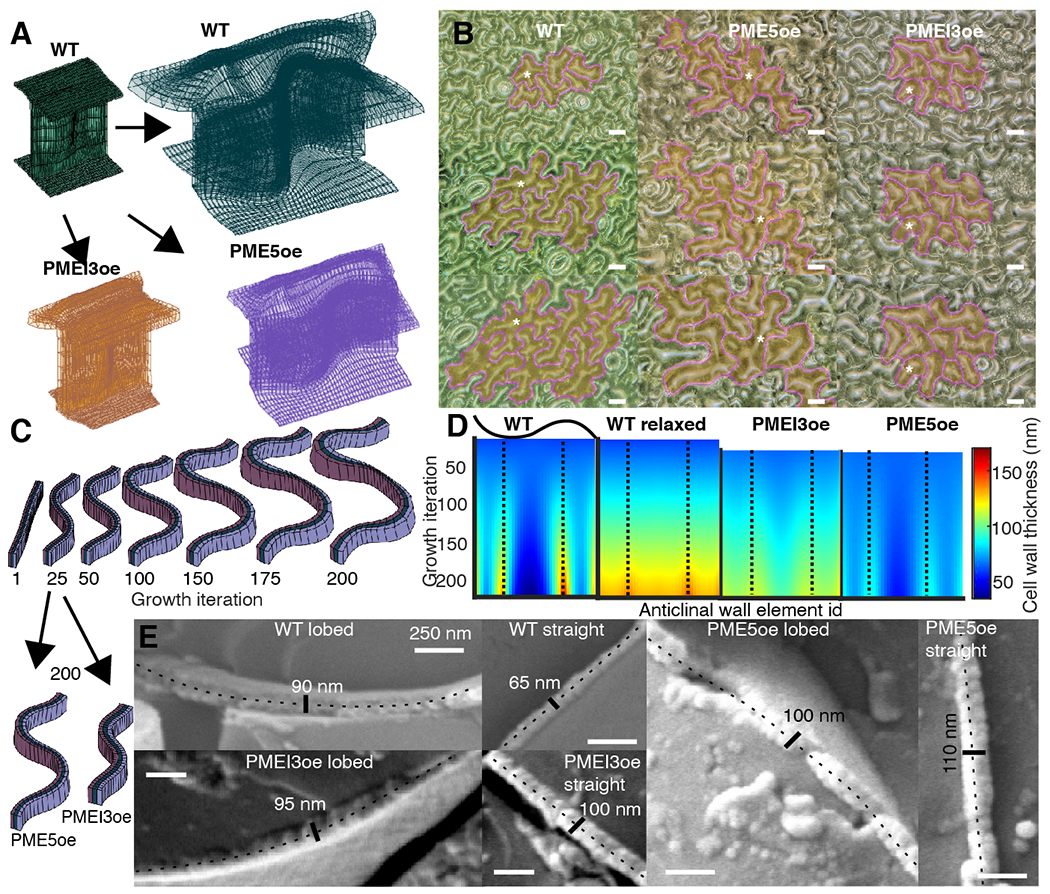
(A) Finite element method implementation of the model in WT (green), PME5oe (violet) and PMEI3oe (orange) cotyledons. (B) Digital microscope images of a time-course of WT, PME5oe and PMEI3oe cotyledons at 1, 2, and 3-days post-germination. Tracked cells are highlighted in yellow with magenta contours; white stars, selected tracked cells. (C) Single element-high cross-section of the anticlinal wall at selected iteration steps showing the development of lobes and the cell wall thickness. Black arrows point from the 25th iteration (the start of PME5oe/PMEI3oe induction) to the 200th iteration. (D) A spatiotemporal heat map of cell wall thickness output from the computational model, obtained using a single element-height middle segment of the anticlinal wall. Each color represents the wall thickness at a particular local portion or segment of the wall used in the model, termed an anticlinal wall element (X-axis), for any given growth iteration of the model (Y-axis). The wall begins with uniform thickness (Iteration 0 at the top of each image) that increases in variability between elements with increasing iterations. Dashed lines represent the center of the lobes. The WT relaxed corresponds to stress-free incompatible growth state, before elastic transformation is applied (fig. S7B). (E) CryoSEM images of cryo-fractured lobed and straight walls in WT, PME5oe and PMEI3oe cotyledons. In each image, the fractured anticlinal wall is labelled with a dotted line together with the measured thickness. Scale bars, 250 nm.
One outcome of the model is variability in the thickness along the length of the anticlinal wall, (Fig. 3, C and D), that does not exceed 250 nm. This is less than that measured in dSTORM images (~250–500 nm) and significantly less than the >1 micron figures forming the basis of other models and based on TEM data (17). These (dSTORM and TEM) experimental methods rely on sections and pretreatments that may relax and distort tissues. CryoSEM, with cryo-fracture, is expected to preserve the native constrained state of the walls and imaging of the wall in WT and overexpressor lines demonstrate wall thicknesses (< 250 nm) that are within the range of those in the model (Fig. 3E).
Next, to model the impact of HG methylation on lobe formation, we induced in silico PME and PMEI overexpression starting at iteration 25, corresponding to one-day post-germination induction in the cotyledon. Simulation of PMEI induction agreed with observations leading to growth impairment (Fig. 3, A, to C and fig. S10A). To achieve size concordance for PME induction between the model and experiment, we had to decrease the exocytosis rate by 60 %, suggesting the existence of a regulatory loop between wall composition and exocytosis. As exocytosis of new wall material is expected to impact wall thickness, we compared outputs of the model (Fig. 3D) with cryo-fractured walls (Fig. 3E). Thickness variability along the anticlinal wall was more pronounced for WT (compare thicknesses for model, Fig. 3D, with cryo-fractures lobed vs straight, Fig. 3E) than for the PME5oe and PMEI3oe lines where anticlinal wall thickness was found to be more homogenous between lobed and straight portions (Fig. 3, D and E).
Testing model predictions by reversible dehydration
In contrast to models that correlate tissue tension to growth, our model predicts a limited amount of local tension and compression in the anticlinal walls. If this is similar to the situation in vivo, then lobes should not change their shape in cotyledons when turgor pressure is reduced. On the other hand, the model does predict that the periclinal walls are under tension in the lobed regions, and under compression in indentations. This agrees with the observed shape of periclinal walls over lobes and indentations and with the microtubules aligned along the direction of maximal tensional stress in the periclinal walls (33), Fig. 4A. To test for the presence of wall tension, we followed cell shape changes after partial and reversible cotyledon dehydration (Fig. 4, B, and C, and Movie S7). Drying is characterized by wrinkling and inversion of the periclinal wall, indicating that the turgor was reduced enough to return wall regions to their resting length. We observed no change in the outline of the anticlinal walls, despite the drying being inhomogeneous throughout the tissue. Therefore, alleviating turgor in anticlinal walls does not lead to observable strain which is in contrast to the lobed periclinal walls which flatten/invert upon dehydration, consistent with the computational model Fig. 4D.
Fig. 4. Model tests by dehydration.
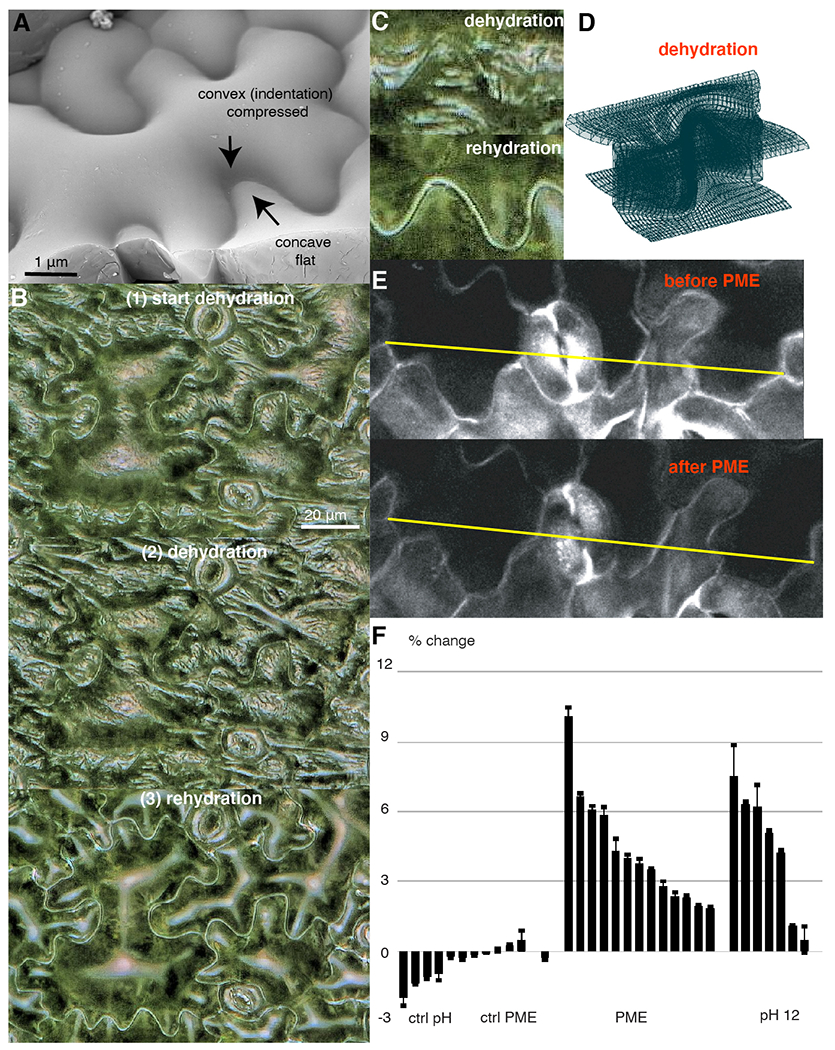
(A) The topography of pavement cell by cryoSEM. (B) Digital microscope images of a WT cotyledon at the inset (top), during (middle) and after (bottom) dehydration. (C) Enlargement of undulating walls from (B). (D) FEM model of a cell wall upon dehydration. (E) Confocal images of a WT cotyledon before (top) and after (bottom) PME enzyme treatment. (F) Tissue expansion quantification upon HG demethylation with PME enzyme or with a pH 12 buffer measured as the %-change in the distance between two reference points (yellow line in (C)); bars (mean ± standard deviation) showing different measurements from different cotyledon regions ordered according to the amount of expansion.
Homogalacturonan demethylation alone leads to tissue expansion
Finally, we tested whether demethylation of HG could lead to cell wall expansion in the absence of turgor pressure. Plasmolyzed cotyledons were maintained at a high calcium concentration, and HG demethylation was achieved either by adding PME enzyme or by incubating the tissue at high pH, causing HG demethylation (fig. S11). Assuming that polymeric HG constitutes 10 % of the wall and upon demethylation it expands by a factor of 1.42 in lateral dimension (Fig. 5A), for tissue length Δ one gets 10 %·Δ·1.42 – 10 %·Δ = ~4.2 % expected expansion if all HG were demethylated. As presented in Fig. 4, E and F and fig. S10B, both treatments led to tissue expansion within this range; however, this may be further influenced by contaminating cell wall remodeling enzymes.
Fig. 5. Cell wall model including homogalacturonan nanofilaments.
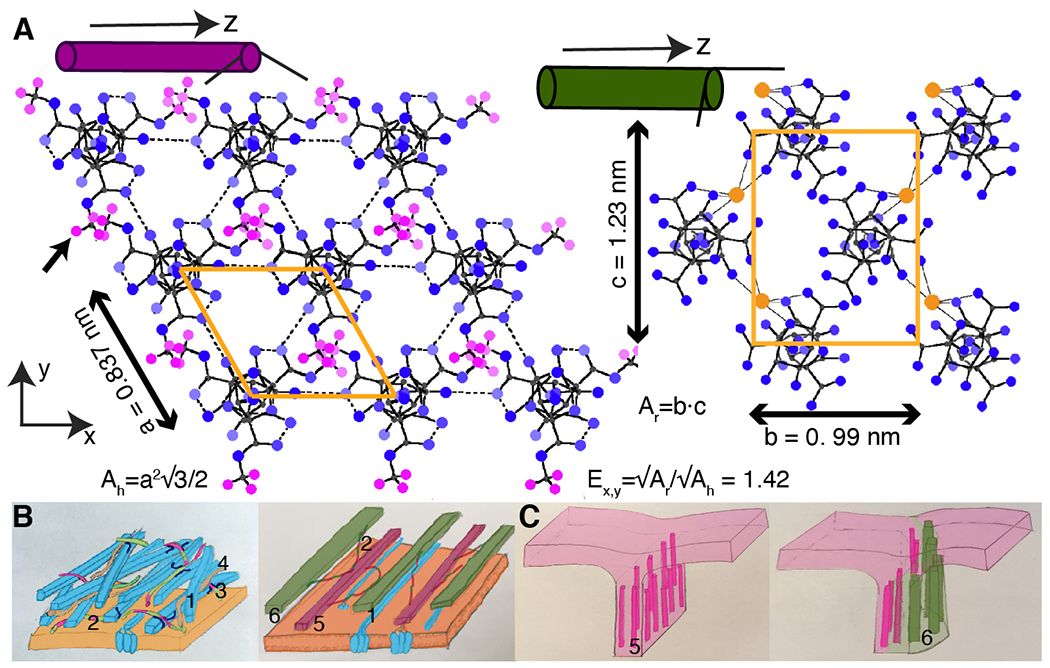
(A) HG nanofilaments as quaternary structures composed of helical chains uniaxially arranged on a hexagonal or a rectangular net for (left) methylated and (right) demethylated HG with unit cell areas (orange polygons) Ah and Ar, respectively; Ex,y, expansion in a lateral dimension. Color circles: blue – oxygen, small grey – carbon, magenta – methyl hydrogen (black arrow), large orange - calcium ion. Color scheme is based on the monochrome diagram of Walkinshaw and Arnott (1981a,b) (19, 20). (B) The cell wall models (left) without and (right) with homogalacturonan nanofilaments. (1) Cellulose microfibrils (blue) embedded in pectin matrix containing: (2) homogalacturonans methylated and demethylated (violet and green segments), (3) and (4) rhamnogalacturonans type II and I (red). Updated cell wall model contains (5) methylated (violet), and (6) demethylated (green) HG nanofilaments. (C) The expanding beam model: (left) young wall comprised of (5) methylated HG; (right) local HG nanofilament demethylation causes asymmetric cell wall expansion and lobe formation (green).
Discussion
Here we observe that, in the anticlinal walls of pavement cells, HG organizes into nanofilaments, either methylated or demethylated, that are oriented perpendicular to the cotyledon surface. We propose that these nanofilaments are the quaternary structures of homogalacturonan previously observed in vitro by Walkinshaw and Arnott (19, 20), composed of individual 3-screw symmetry helical chains arranged on a hexagonal (methylated HG) or a rectangular (demethylated HG) lattice (Fig. 5A). Based on our observations and the prior X-ray characterization, we propose an 'expanding beam' model for lobe formation. In this framework, the undulating form of anticlinal walls is initiated by demethylation-induced and spatially-varying radial swelling and axial contraction of HG nanofilaments, causing local and oriented wall expansion in the direction perpendicular to the dominant nanofilament orientation (Fig. 5, B, and C). The HG methylation influences the width of filaments detected with cryoSEM, supporting the hypothesis that nanofilament swelling is due to the conversion between two quaternary forms of HG. Our model indicates that local methylation asymmetry alone may not be sufficient to explain lobe formation, suggesting that HG organization in nanofilaments is necessary for lobe morphogenesis (30).
It remains to be determined if cellulose and other more complex associations of heteroglycans and glycoconjugates reside in the filaments. The HG nanofilaments appear to be absent in the periclinal walls, suggesting that HG forms an amorphous mesh, as depicted in current cell wall models (Fig. 5B), (6). The nanofilament orientation is compatible with the spring-like behavior of epidermis, since filaments would stiffen the tissue along their axis and permit reversible lateral extension (34).
In our model, wall thickening and cell expansion are inevitably linked, even though thickening is not always observed in rapidly expanding cells (4). In addition, the model focuses on the most recently deposited material, assuming that previous layers adapt to expansion, thus clearly stating the importance of cell wall remodeling to permit cell expansion.
While the basis for the proposal of the expanding beam model for Arabidopsis pavement cells is an observation of HG nanofilaments, similar self-expansion may well apply to other types of extracellular matrix polymers in which organized structures could be changed post-synthesis (35). For example, the carrageenan of red algae (36) and the polarized calcium-dependent alginates in brown algae (37) have the requisite attributes. Thus, biochemical processes similar to those that act upon pectins could underlie growth in organisms that have cell walls but do not have HG. Our observations could inspire the development of smart materials with the regulated capacity to expand. It may even be the case that localized swelling of the extracellular matrix in non-walled organisms such as animals could alter morphogenetic or metastatic processes.
Supplementary Material
Acknowledgments:
The dSTORM was performed at the MRC Laboratory of Molecular Biology, Cambridge, and we thank Nick Barry and Jonathan Howe for their support. We thank Aram Gurzadyan for the critical evaluation of our FEM model and Herman Höfte for help with the fundraising and discussion of the results.
Funding: AP has received the support of the French National Research Agency (ANR) GoodVibration ANR-17-CE13–0007 and of the EU in the framework of the Marie-Curie FP7 COFUND People Program, through the award of an AgreenSkills+ fellowship (under grant agreement n° 201310). The IJPB benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007). The Microscopy Facility at the Sainsbury Laboratory is supported by the Gatsby Charitable Foundation, and work at the Sainsbury Laboratory was supported by the Gatsby Charitable Foundation (through fellowship GAT3395/DAA). The Meyerowitz Laboratory has support from the Howard Hughes Medical Institute. This work has benefited from the support of IJPB's Plant Observatory technological platforms.
Footnotes
Competing interests: Authors declare no competing interests. EMM is a member of the Management Board of the Sainsbury Laboratory at Cambridge University.
Data and materials availability: All data, codes, and materials used in this study are available. Please contact Alexis Peaucelle for the material request. All data are available in the manuscript or the supplement or at DOI: 10.17632/xb3f5jrbwv.1. All codes used in this study are available: DOI: 10.5281/zenodo.3581144, DOI:10.5281/zenodo.3581097, DOI: 10.5281/zenodo.3581129, DOI:10.5281/zenodo.3581151.
References
- 1.Hamant O, Heisler MG, Jönsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM, Couder Y, Traas J, Developmental patterning by mechanical signals in Arabidopsis. Science 322, 1650–1655 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Peaucelle A, Wightman R, Höfte H, The Control of Growth Symmetry Breaking in the Arabidopsis Hypocotyl. Curr. Biol 25, 1746–1752 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Bosch M, Cheung AY, Hepler PK, Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 138, 1334–1346 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H, A plasma membrane-bound putative endo-1,4-β-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17, 5563–5576 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bichet A, Desnos T, Turner S, Grandjean O, Höfte H, BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 25, 137–148 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Zhang T, Di. Vavylonis, D. M. Durachko, D. J. Cosgrove, Nanoscale movements of cellulose microfibrils in primary cell walls. Nat. Plants 3, 17056 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paredez AR, Somerville CR, Ehrhardt DW, Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312, 1491–1495 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, Yuan C, Hao Z, Zhu X, Avci U, Miller JS, Baldwin D, Pham C, Orlando R, Darvill A, Hahn MG, Kieliszewski MJ, Mohnen D, An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell. 25, 270–287 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atmodjo MA, Hao Z, Mohnen D, Evolving Views of Pectin Biosynthesis. Annu. Rev. Plant Biol 64, 747–779 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Hong M, Solid-state NMR investigations of cellulose structure and interactions with matrix polysaccharides in plant primary cell walls. J. Exp. Bot 67, 503–514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peaucelle A, Cell wall expansion: Case study of a biomechanical process (Springer, Cham, 2018; http://link.springer.com/10.1007/978-3-319-69944-8_7), vol. 23. [Google Scholar]
- 12.Cosgrove DJ, Plant cell wall extensibility: Connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot 67, 463–476 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Baskin TI, ANISOTROPIC EXPANSION OF THE PLANT CELL WALL. Annu. Rev. Cell Dev. Biol 21, 203–222 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Wiedemeier AMD, Judy-March JE, Hocart CH, Wasteneys GO, Williamson RE, Baskin TI, Mutant alleles of Arabidopsis RADIALLY SWOLLEN 4 and 7 reduce growth anisotropy without altering the transverse orientation of cortical microtubules or cellulose microfibrils. Development. 129, 4821–4830 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Fu Y, Xu T, Zhu L, Wen M, Yang Z, A ROP GTPase Signaling Pathway Controls Cortical Microtubule Ordering and Cell Expansion in Arabidopsis. Curr. Biol 19, 1827–1832 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sapala A, Runions A, Smith RS, Mechanics, geometry and genetics of epidermal cell shape regulation: different pieces of the same puzzle. Curr. Opin. Plant Biol 47, 1–8 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Majda M, Grones P, Sintorn IM, Vain T, Milani P, Krupinski P, Zagórska-Marek B, Viotti C, Jönsson H, Mellerowicz EJ, Hamant O, Robert S, Mechanochemical Polarization of Contiguous Cell Walls Shapes Plant Pavement Cells. Dev. Cell 43, 290–304.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Altartouri B, Bidhendi AJ, Tani T, Suzuki J, Conrad C, Chebli Y, Liu N, Karunakaran C, Scarcelli G, Geitmann A, Pectin Chemistry and Cellulose Crystallinity Govern Pavement Cell Morphogenesis in a Multi-Step Mechanism. Plant Physiol. 181, 127–141 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walkinshaw MD, Arnott S, Conformations and interactions of pectins. II. Models for junction zones in pectinic acid and calcium pectate gels. J. Mol. Biol 153, 1075–1085 (1981). [DOI] [PubMed] [Google Scholar]
- 20.Walkinshaw MD, Arnott S, Conformations and interactions of pectins. I. X-ray diffraction analyses of sodium pectate in neutral and acidified forms. J. Mol. Biol 153, 1055–1073 (1981). [DOI] [PubMed] [Google Scholar]
- 21.Huang B, Wang W, Bates M, Zhuang X, Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF, Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Rust MJ, Bates M, Zhuang X, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods. 3, 793–796 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bates M, Huang B, Dempsey GT, Zhuang X, Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317, 1749–1753 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heilemann M, Linde S van de, Schüttpelz M, Kasper R, Seefeldt B, Mukherjee A, Tinnefeld P, Sauer M, Subdiffraction-Resolution Fluorescence Imaging with Conventional Fluorescent Probes. Angew. Chemie Int. Ed 47, 6172–6176 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Verhertbruggen Y, Marcus SE, Haeger A, Ordaz-Ortiz JJ, Knox JP, An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr. Res 344, 1858–1862 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Liners F, Letesson J-J, Didembourg C, Van Cutsem P, Monoclonal Antibodies against Pectin. Plant Physiol. 91, 1419–1424 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X, Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods 8, 1027–1040 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Höfte H, Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol 21, 1720–1726 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Rui Y, Chen Y, Yi H, Purzycki T, Puri VM, Anderson CT, Synergistic pectin degradation and guard cell pressurization underlie stomatal pore formation. Plant Physiol. 180, 66–77 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim N, NAFEMS Introduction to Nonlinear Finite Element Analysis (2014). [Google Scholar]
- 32.Rodriguez EK, Hoger A, McCulloch AD, Stress-dependent finite growth in soft elastic tissues. J. Biomech 27, 455–467 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Sampathkumar A, Krupinski P, Wightman R, Milani P, Berquand A, Boudaoud A, Hamant O, Jönsson H, Meyerowitz EM, Subcellular and supracellular mechanical stress prescribes cytoskeleton behavior in Arabidopsis cotyledon pavement cells. Elife. 3 (2014), doi: 10.7554/eLife.01967.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sotiriou P, Giannoutsou E, Panteris E, Galatis B, Apostolakos P, Local differentiation of cell wall matrix polysaccharides in sinuous pavement cells: its possible involvement in the flexibility of cell shape. Plant Biol. 20, 223–237 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Popper ZA, Michel G, Hervé C, Domozych DS, Willats WGT, Tuohy MG, Kloareg B, Stengel DB, Evolution and Diversity of Plant Cell Walls: From Algae to Flowering Plants. Annu. Rev. Plant Biol 62, 567–590 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Ficko-Blean E, Hervé C, Michel G, Sweet and sour sugars from the sea: the biosynthesis and remodeling of sulfated cell wall polysaccharides from marine macroalgae. Perspect. Phycol. 2, 51–64 (2015). [Google Scholar]
- 37.Terauchi M, Nagasato C, Inoue A, Ito T, Motomura T, Distribution of alginate and cellulose and regulatory role of calcium in the cell wall of the brown alga Ectocarpus siliculosus (Ectocarpales, Phaeophyceae). Planta. 244 (2016), doi: 10.1007/s00425-016-2516-4.. [DOI] [PubMed] [Google Scholar]
- 38.Wightman R, Wallis S, Aston P, Hydathode pit development in the alpine plant Saxifraga cochlearis. Flora. 233, 99–108 (2017). [Google Scholar]
- 39.Haas KT, Lee M, Esposito A, Venkitaraman AR, Single-molecule localization microscopy reveals molecular transactions during RAD51 filament assembly at cellular DNA damage sites. Nucleic Acids Res. 46, 2398–2416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


