Abstract
Ilex is a monogeneric plant group (containing approximately 600 species) in the Aquifoliaceae family and one of the most commonly used medicinal herbs. However, its taxonomy and phylogenetic relationships at the species level are debatable. Herein, we obtained the complete chloroplast genomes of all 19 Ilex types that are native to Hong Kong. The genomes are conserved in structure, gene content and arrangement. The chloroplast genomes range in size from 157,119 bp in Ilex graciliflora to 158,020 bp in Ilex kwangtungensis. All these genomes contain 125 genes, of which 88 are protein-coding and 37 are tRNA genes. Four highly varied sequences (rps16-trnQ, rpl32-trnL, ndhD-psaC and ycf1) were found. The number of repeats in the Ilex genomes is mostly conserved, but the number of repeating motifs varies. The phylogenetic relationship among the 19 Ilex genomes, together with eight other available genomes in other studies, was investigated. Most of the species could be correctly assigned to the section or even series level, consistent with previous taxonomy, except Ilex rotunda var. microcarpa, Ilex asprella var. tapuensis and Ilex chapaensis. These species were reclassified; I. rotunda was placed in the section Micrococca, while the other two were grouped with the section Pseudoaquifolium. These studies provide a better understanding of Ilex phylogeny and refine its classification.
Subject terms: Genome, Plant sciences
Introduction
Ilex, a monogeneric plant group in the family Aquifoliaceae, is a widespread genus. It can either be evergreen or deciduous and can be found throughout subtropical regions. There are approximately 600 species in total, and some of them have medical uses1. Some kinds of Ilex are commonly used herbs in traditional Chinese medicine (TCM), and they are effective in treating influenza, relieving pain and anti-inflammation2,3. The mechanism has been recently proposed. Asprellcosides from I. asprella can significantly inhibit the replication of influenza A virus, while rotundarpene from I. rotunda var. microcarpa can inhibit the TNF-α-mediated pathway4,5.
Ilex species have a variety of pharmacological properties, and accurate classification of them is needed. Cuénoud and colleagues used the rbcL and atp-rbcL spacer to study the phylogenetic relationship of Ilex. A total of 116 Ilex species were classified into the American clade, Asian/North American clade, deciduous clade and Eurasian clade6. Manen and colleagues used both chloroplast markers and the 5S RNA spacer to assign 105 Ilex into the 4 clades. After that, Ilex was further assigned to different subgenera, sections and alliances7. Nuclear DNA ITS and nepGS were also included in recent research1. In addition to the commonly used DNA regions, Lin and colleagues have also used the nuclear segment gapC to study the speciation of Ilex8. According to Flora of China, Ilex can be divided into three subgenera, Byronia, Ilex and Prinos9. These three subgenera can be further classified into taxonomic sections and series. The classification was generally based on morphological analysis, and only short universal DNA markers of some species were included in the studies.
Chloroplasts are responsible for multiple functions, such as photosynthesis10, amino acid synthesis and nitrogen metabolism in plants11,12. As most DNA markers, such as rbcL, matK and psbA, are located in the chloroplast genome, the development of chloroplast whole-genome sequencing has contributed to solving the phylogenetic relationship of plants13–15. In addition to classical DNA barcoding regions, novel DNA markers can also be found16.
Here, we obtained the complete chloroplast genomes of 19 Ilex species native to Hong Kong. Divergence hotspot investigation, repeat analysis and polymorphism studies were performed. Through including other published Ilex chloroplast genomes, topologies of the Ilex phylogenies were constructed.
Results
Chloroplast genome assembly
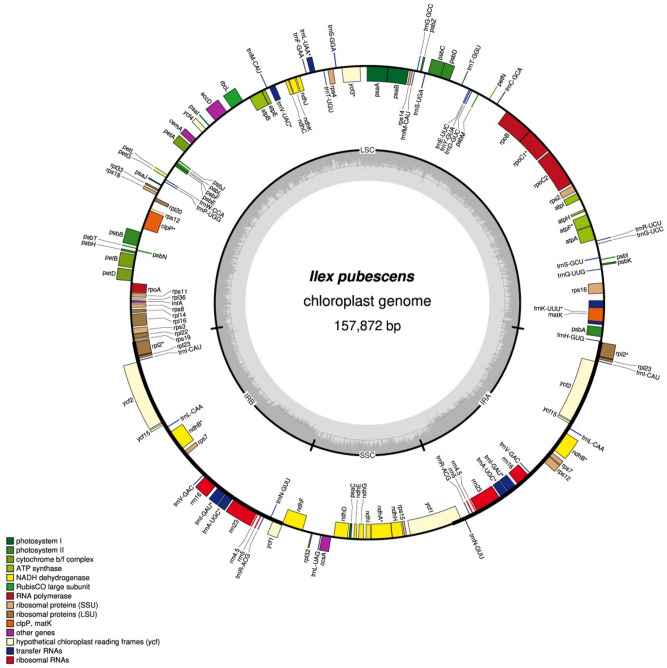
By using the Illumina NovaSeq 6000 system, the complete chloroplast genomes of 19 Ilex species were sequenced. Raw data were generated with an average read length of 150 bp. Complete chloroplast sequences were assembled by de novo assembly and validated by mapping the raw reads into the assembled contigs. Sequences of the reads and contigs were compared, and the final plastomes were submitted to GenBank with accession numbers according to the supplementary material (Supplementary Table S1). After that, 19 Ilex genomes were used for indel, SSR and REPuter analyses. The corresponding genome map of I. pubescens is shown as a reference (Fig. 1).
Figure 1.
Chloroplast genome map of I. pubescens. The genome structure and gene arrangement in the 19 Ilex species were conserved. Introns containing coding regions are indicated with asterisks. The inner grey circle represents the GC content of each region. Genes shown in the inner circle are transcribed clockwise and vice versa. Genes belonging to different functional groups are indicated in different colours.
Genome structure and gene content
These genomes followed the typical quadripartite structure of angiosperms, which consisted of one LSC Sect. (86,506–87,400 bp), one SSC Sect. (18,380–18,442 bp) and a pair of IR regions (26,065–26,125 bp). The GC content ranged from 37.6% to 37.7%, which was consistent with a previous report of Ilex chloroplast genomes17–19. All of the 19 Ilex species have the same gene content and gene order. They contain 125 genes, of which 88 are protein-coding genes, 37 are tRNA genes and 8 are encode rRNA (Table 1). Seventeen genes contain one or more introns, of which 12 (ycf3, atpF, petB, petD, ndhA, ndhB, rpoC1, rps12, rps16, rpl2, rpl16, and clpP) are protein-coding, and five are responsible for tRNA (trnA-UGC, trnI-GAU, trnL-UAA, trnQ-UUG, and trnV-UAC). Several genes are duplicated in the IR regions, among which 8 (ndhB, rps7, rps12, rpl2, rpl23, ycf1, ycf2, and ycf15) are protein coding, 7 (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) encoded tRNAs, and 4 (rrn4.5, rrn5, rrn16, and rrn23) encode rRNA (Table 2). Only one pseudogene, ycf1, was found in all Ilex genomes. This observation was consistent with the seven Ilex genomes in Yao’s study17. The similarities in the genome structure and genetic content indicate that the chloroplast genomes of all 19 Ilex species are highly conserved.
Table 1.
Summary of the assembly data for Ilex chloroplast genomes.
| Species | Genome size (bp) | LSC (bp) | IR (bp) | SSC (bp) | Number of genes | rRNA | tRNA | Protein-coding genes | A% | C% | G% | T% | GC% |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I. latifolia | 157,558 | 86,945 | 26,093 | 18,427 | 125 | 8 | 37 | 88 | 30.9 | 19.1 | 18.5 | 31.5 | 37.6 |
| I. lohfauensis | 157,469 | 86,873 | 26,078 | 18,440 | 125 | 8 | 37 | 88 | 30.8 | 19.2 | 18.5 | 31.5 | 37.6 |
| I. kwangtungensis | 158,020 | 87,400 | 26,104 | 18,412 | 125 | 8 | 37 | 88 | 30.9 | 19.1 | 18.5 | 31.5 | 37.6 |
| I. triflora | 157,706 | 87,183 | 26,065 | 18,393 | 125 | 8 | 37 | 88 | 30.8 | 19.2 | 18.5 | 31.5 | 37.7 |
| I. ficoidea | 157,536 | 86,922 | 26,094 | 18,426 | 125 | 8 | 37 | 88 | 30.8 | 19.2 | 18.5 | 31.5 | 37.6 |
| I. rotunda var. microcarpa | 157,780 | 87,094 | 26,125 | 18,436 | 125 | 8 | 37 | 88 | 30.8 | 19.2 | 18.5 | 31.5 | 37.6 |
| I. asprella | 157,856 | 87,265 | 26,075 | 18,441 | 125 | 8 | 37 | 88 | 30.8 | 19.1 | 18.5 | 31.6 | 37.6 |
| I. pubescens | 157,872 | 87,285 | 26,073 | 18,441 | 125 | 8 | 37 | 88 | 30.8 | 19.1 | 18.5 | 31.6 | 37.6 |
| I. asprella var. tapuensis | 157,671 | 87,161 | 26,065 | 18,380 | 125 | 8 | 37 | 88 | 30.8 | 19.2 | 18.5 | 31.5 | 37.7 |
| I. hanceana | 157,478 | 86,889 | 26,074 | 18,441 | 125 | 8 | 37 | 88 | 30.8 | 19.2 | 18.5 | 31.5 | 37.6 |
| I. cinerea | 157,215 | 86,601 | 26,094 | 18,426 | 125 | 8 | 37 | 88 | 30.8 | 19.2 | 18.5 | 31.5 | 37.7 |
| I. championii | 157,468 | 86,878 | 26,074 | 18,442 | 125 | 8 | 37 | 88 | 30.8 | 19.2 | 18.5 | 31.5 | 37.6 |
| I. graciliflora | 157,119 | 86,506 | 26,093 | 18,427 | 125 | 8 | 37 | 88 | 30.8 | 19.2 | 18.5 | 31.5 | 37.7 |
| I. memecylifolia | 157,842 | 87,249 | 26,076 | 18,441 | 125 | 8 | 37 | 88 | 30.8 | 19.1 | 18.5 | 31.6 | 37.6 |
| I. chapaensis | 157,665 | 87,155 | 26,065 | 18,380 | 125 | 8 | 37 | 88 | 30.8 | 19.2 | 18.5 | 31.5 | 37.7 |
| I. cornuta | 157,216 | 86,607 | 26,091 | 18,427 | 125 | 8 | 37 | 88 | 30.8 | 19.2 | 18.5 | 31.5 | 37.7 |
| I. lancilimba | 157,998 | 87,382 | 26,105 | 18,406 | 125 | 8 | 37 | 88 | 30.9 | 19.2 | 18.5 | 31.5 | 37.6 |
| I. dasyphylla | 158,009 | 87,388 | 26,105 | 18,411 | 125 | 8 | 37 | 88 | 30.9 | 19.1 | 18.5 | 31.5 | 37.6 |
| I. viridis | 157,661 | 87,147 | 26,065 | 18,384 | 125 | 8 | 37 | 88 | 30.8 | 19.2 | 18.5 | 31.5 | 37.7 |
Table 2.
Gene content and functional classification of the 19 Ilex chloroplast genomes.
| Gene category | Gene function | Gene name |
|---|---|---|
| Photosynthesis-related genes | Rubisco | rbcL |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Assembly/stability of photosystem I | **ycf3, ycf4 | |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| ATP synthase | atpA, atpB, atpE, *atpF, atpH, atpI | |
| Cytochrome b/f complex | petA, *petB, *petD, petG, petL, petN | |
| Cytochrome c synthesis | ccsA | |
| NADPH dehydrogenase | *ndhA, *ndhB(× 2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Transcription- and translation-related genes | Transcription | rpoA, rpoB, *rpoC1, rpoC2, |
| Ribosomal protein | rps2, rps3, rps4, rps7(× 2), rps8, rps11, *rps12(× 2), rps14, rps15, *rps16, rps18, rps19, *rpl2(× 2), rpl14, *rpl16, rpl20, rpl22, rpl23(× 2), rpl32, rpl33, rpl36 | |
| Translation initiation factor | infA | |
| RNA genes | Ribosomal RNA | rrn4.5(× 2), rrn5(× 2), rrn16(× 2), rrn23(× 2) |
| Transfer RNA | *trnA-UGC(× 2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC, trnH-GUG, trnI-CAU(× 2), *trnI-GAU(× 2), trnK-UUU, trnL-CAA(× 2), *trnL-UAA, trnL-UAG, trnM-CAU, trnfM-CAU, trnN-GUU(× 2), trnP-UGG, *trnQ-UUG, trnR-ACG(× 2), trnR-UCU, trnS-GGA, trnS-GCU, trnS-UGA, trnT-UGU, trnT-GGU, trnV-GAC(× 2), *trnV-UAC, trnW-CCA, trnY-GUA | |
| Miscellaneous group | Maturase | matK |
| Inner membrane protein | cemA | |
| ATP-dependent protease | **clpP | |
| Acetyl-CoA carboxylase | accD | |
| Unknown functions | ycf1(× 2), ycf2(× 2), ycf15(× 2) |
Intron-containing genes are labelled with asterisks. The number of introns corresponds to the number of asterisks.
Simple and complex repeats analysis
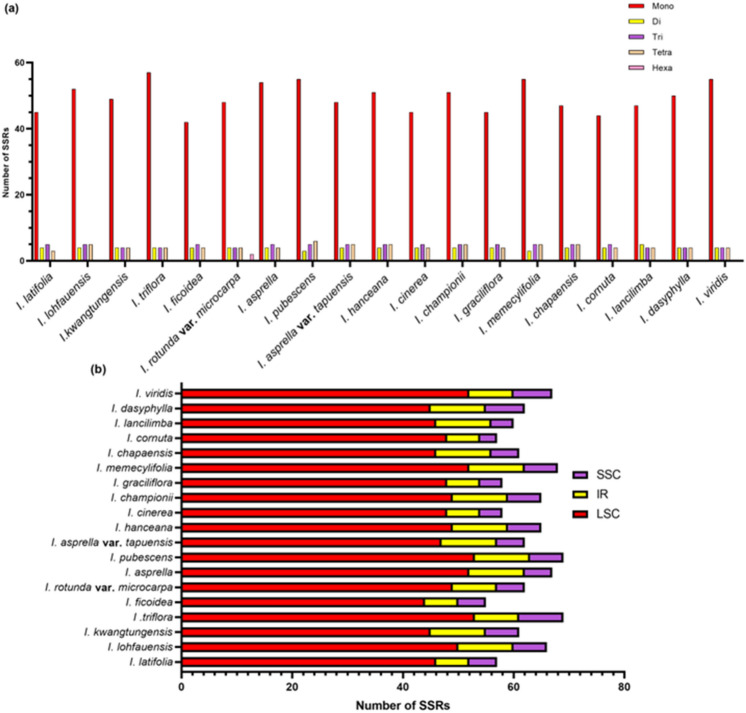
Simple sequence repeats (SSRs) are repeating DNA motifs that range from one to six nucleotides20. The number and position of repeated motifs are thought to be useful in population genetic studies and have been widely used in finding polymorphisms in chloroplast genomes21,22. By using MISA analysis tools, the number and position of different SSR motifs in 19 Ilex were investigated. All of the Ilex species do not have penta-nucleotide motifs, while Ilex rotunda var. microcarpa is the only species containing hexa-nucleotide repeat motifs. Mono-nucleotide repeat motifs account for the largest proportion, ranging from 44 to 56 (Fig. 2a). Most of the mono-nucleotide repeat motifs are A/T repeats, while only 5 species, I. triflora, I. viridis, I. asprella var. tapuensis, I. cinerea and I. graciliflora, have C/G repeat motifs. A/T repeats occupy at least 75.9% of the total SSR, and the phenomenon of AT richness in the SSR of terrestrial plants has been reported in previous studies23,24. The numbers and types of di-, tri- and tetra-nucleotide motifs are mostly conserved among Ilex, except for the ACAT/ATGT tetra-nucleotide motif, which is present only in I. ficoidea and I. pubescens (Supplementary Table S2). Regarding the SSR distribution, most of the SSRs are found in the LSC and SSC regions but not the IR regions, which is consistent with studies in angiosperms (Fig. 2b)25–27. Owing to the variation in the LSC, IR and SSC lengths, densities of SSRs in different regions were also calculated (Supplementary Table S3). Only a few SSRs were found in the two IR regions; therefore, only slight variation was observed in these regions. On the other hand, relatively large variation in the SSR density was noticed in the SSC regions, which varied from 1.63E−04 in I. cornuta to 4.35E-04 in I. triflora.
Figure 2.
Classification of simple sequence repeats (SSRs) in the chloroplast genomes of 19 Ilex species based on (a) repeat motifs of SSRs and (b) the distribution of SSRs.
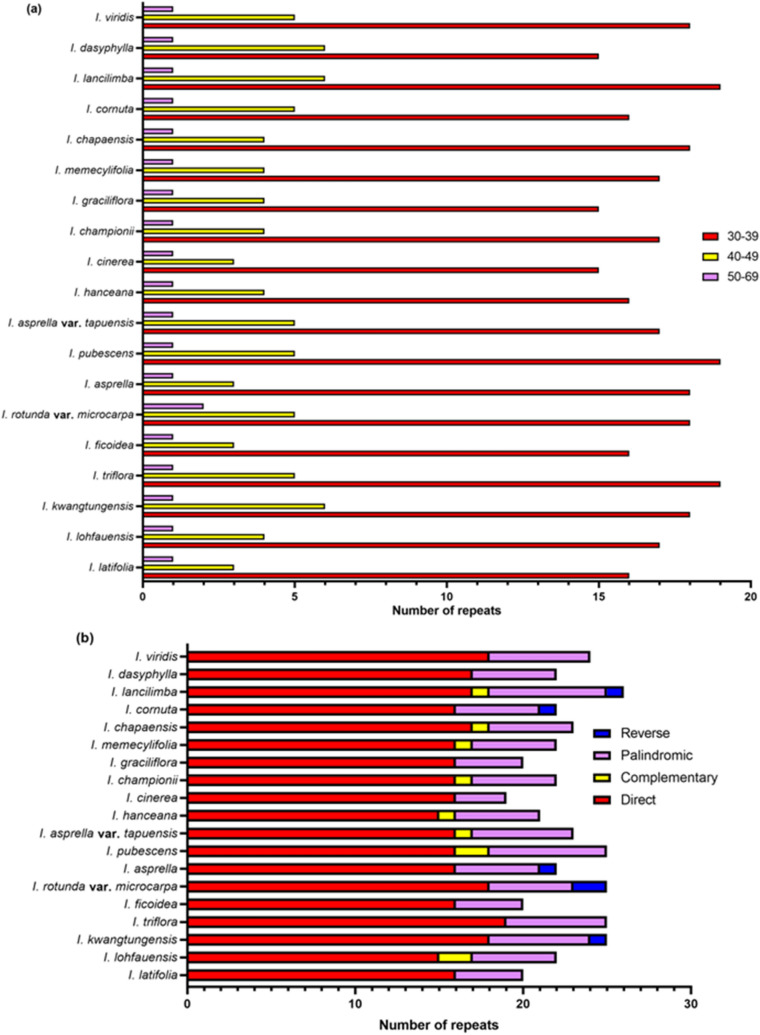
Complex repeat regions are important in recombination and variation in chloroplast genomes28. By using the REPuter algorithm, complex repeats of Ilex were analysed. The length of repeats is 30–66 bp, which falls into the typical range of other angiosperms29,30. There are only slight variations in the total number of repeats, which ranged from 19 to 26, but the number of each repeating motif is different. The most abundant repeats are direct repeats, while palindromic repeats are secondary. Complementary repeats can be observed in only eight Ilex species, and reverse repeats can be seen only in five Ilex species (Fig. 3). The variation in the repeating motifs of SSRs and complex repeats can be used as DNA markers to identify these species.
Figure 3.
(a) Length of repeats and (b) frequency of repeating motifs of the 19 Ilex chloroplast genomes determined by REPuter.
Interspecies comparison of chloroplast genomes
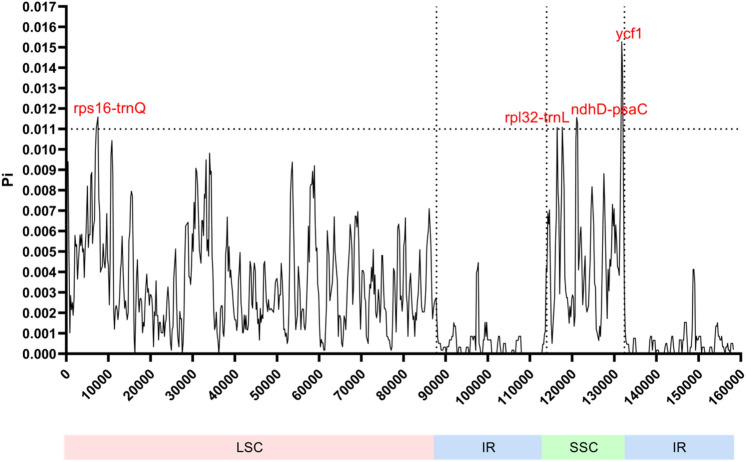
Single-nucleotide polymorphisms (SNPs) and DNA insertion-deletion mutations (indels) are important for discovering DNA markers and barcodes in medicinal herbs31,32. Therefore, the number of SNPs and indels were calculated by pair-wise alignment of the 19 Ilex chloroplast genomes. In brief, the 19 Ilex genomes were first aligned with each other. The number of polymorphisms was identified by an in-house developed Perl algorithm. By comparing the numbers, several groupings could be seen among the Ilex species. Most of the Ilex species have 200 to 600 SNPs and 60 to 150 indels. The average numbers of SNPs and indels are 436 and 100, respectively. On the other hand, fewer than 80 SNPs and 20 indels were discovered among I. lohfauensis, I. championii and I. hanceana. Relatively small numbers of SNPs and indels were also observed in the group of I. kwangtungensis, I. dasyphylla, and I. lancilimba and the group of I. latifolia, I. cornuta, I. cinerea, and I. graciliflora (Supplementary Table S4). In fact, the three groups mentioned were classified as sections of Pseudoaquifolium, Lioprinus and Ilex in Flora of China. The same classification was given in our phylogenetic analysis. To locate the divergent hotspots, sliding window analysis was performed. The average Pi (π) value among the 19 Ilex species was 0.00268, which indicated that there were only slight variations in the chloroplast genomes. However, we still found four highly varied hotspots (Pi > 0.01), rps16-trnQ, rpl32-trnL, ndhD-psaC and ycf1 (Fig. 4). rps16-trnQ belongs to the LSC region, while the other three are located in the SSC region. The two IR regions showed the smallest divergence, which is consistent with the characteristics of other chloroplast genomes33–35.
Figure 4.
Nucleotide diversity values between 19 Ilex species determined by using whole chloroplast genomes. Variation hotspots (Pi > 0.011) are labelled above the corresponding gene position.
To investigate the correlation between the SNPs/indels and the Pi distance, the suggested hotspot sequences of the 19 Ilex chloroplast genomes were first aligned. The numbers of SNPs and indels were then compared by using I. latifolia as a reference. Special features were seen in all four hotspots. First, there are 68 bp gap regions in rps16-trnQ of I. triflora and I. viridis. We also located 5 bp deletions in rps16-trnQ of I. kwangtungensis, I. triflora, I. lanchilimba I. dasyphylla and I. viridis. Second, 5 bp truncations were also discovered in rpl32-trnL of I. latifolia, I. ficoidea, I. cinerea and I. cornuta. All of these species were members in the Ilex section. However, as the deletions are small in size, the divergence distance of this region is relatively lower than that of the other suggested hotspots. Third, although the ndhD-psaC intergenic spacer is relatively short (approximately 130 bp) compared with that of the other suggested hotspots, 12 SNPs were located in I. kwangtungensis, I. triflora, I. lanchilimba I. dasyphylla and I. viridis. The grouping was consistent with the observation in rps16-trnQ. Fourth, 29 bp deletions were observed in ycf1 of I. asprella var. tapuensis and I. chapaensis, which may explain the unexpected position of these two species in the phylogenetic tree. All of the observations suggested that the SNPs/indels play an important role in the divergence distance, which is potentially useful for section classification. After sliding window analysis, a sequence identity plot was also constructed by using the online program mVISTA. Similar to in Yao’s study, the complete chloroplast genome of Helwingia himalaica was used as the reference genome for comparison17. We found that the gene arrangement and contents of the Ilex genus genomes are similar to those of the Helwingia genome (Supplementary Fig. S1). Several divergence hotspots, trnK-rps16-trnQ, ndhF-rpl32-trnL and ycf1, were identified, and the IR regions are mostly conserved. On the other hand, groupings in the gene arrangement could also be observed in the sequence identity plot. Compared with the other Ilex genomes, truncation of rpoB-trnC was observed in I. lohfauensis, I. hanceana and I. championii. Truncation of ycf4-cemA was also discovered in the group of I. latifolia, I. ficoidea, I. cinerea, I. graciliflora and I. cornuta. As expected, the groupings were similar to our findings in the SNP investigation and phylogenetic analysis, which suggested that the deletions in rpoB-trnC and ycf4-cemA may act as DNA markers in the Hanceanae series and the Ilex section, respectively.
Genetic interspecies divergence and intraspecific variation
To validate the reliability of the suggested variation hotspots, interspecific divergence and intraspecific variation of these regions were compared. In brief, all 19 Hong Kong Ilex plastomes were aligned. Interspecies divergence (π) of the hotspots was then compared by DnaSP (Supplementary Table S5). In the aspect of intraspecific comparison, the published reference genomes of Ilex were first retrieved from public resources. They were then aligned with our corresponding Ilex plastomes (MT704834, MT764239, and MT764252), and their numbers of SNPs and indels were compared (Table 3). Among the four suggested hotspots, the ndhD-psaC intergenic spacer showed the highest interspecific distance. The interspecific distances of rps16-trnQ and rpl32-trnL were comparable. The divergence of ycf1 was lower than expected, as the upstream sequences of ycf1 were mostly conserved. Variations were commonly seen only at the 3′ end since the 3′ end overlapped with the SSC/IRb junction. All of the suggested hotspots had a higher divergence than the interspecific distance of the whole cp genome. Moreover, compared with that of the available published genomes, the intraspecific variation of the complete plastomes was also extremely low. There are 10 SNPs in the plastomes of I. asprella and I. cornuta. There are also 2 and 14 indels in these two species, respectively. Only one SNP and no indel were observed in the plastomes of I. latifolia. We showed that the chloroplast genomes are highly conserved within the same species, and four suggested hotspots, rps16-trnQ, rpl32-trnL, ndhD-psaC and ycf1, can potentially be used for species classification.
Table 3.
Intraspecific comparison of the single-nucleotide polymorphisms (SNPs) and indels of Hong Kong Ilex and reference genomes.
IR expansion and contraction investigation
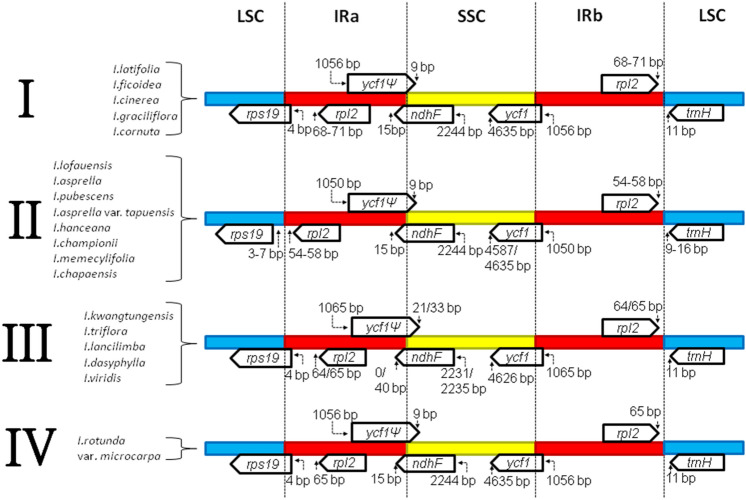
In addition to the investigation of nucleotide divergence, expansion and contraction of the border regions were also analysed for the 19 Hong Kong Ilex species (Fig. 5). The gene arrangement of all Ilex species is highly conserved, in which rps19, rpl2, ycf1, ndhF and trnH are present at the junction of LSC/IRa, IRa/SSC, SSC/IRb, and IRb/LSC. Most of the junctions are stable; however, we could still classify the features into four distinct groups (groups I–IV) (Fig. 5; Supplementary Table S6). In brief, the lengths of the ycf1 pseudogene (ycf1Ψ) in group I and group III Ilex are 1056 bp and 1065 bp, respectively. The rps19 gene of group II Ilex is found within the LSC region, while it is located across the LSC/IR boundary in other groups of Ilex. The sizes of rpl2 and ndhF also varied in these three groups of Ilex. Group IV Ilex possess features observed in both group I, group II and group III Ilex. On the other hand, subgrouping was observed in group II and group III Ilex. For instance, 4587 bp of the ycf1 gene is located in the SSC region of I. asprella var. tapuensis and I. chapaensis; in contrast, 4635 bp of the ycf1 gene of the other members in group II Ilex is located in the SSC region. The IR regions of I. kwangtungensis and I. dasyphylla show no expansion into the ndhF gene; in contrast, a 40 bp expansion was observed in the other members of group III Ilex. Moreover, the ndhF genes of these two species are 2231 bp but not 2235 bp across the SSC region.
Figure 5.
Comparison of the junction regions among 19 Hong Kong Ilex species. Ψ indicates pseudogenes. Slashes indicate the location on the genomes. Numbers above the genes denote the distance between the end of the gene and the border sites. The figure is not to scale.
Phylogenetic analysis
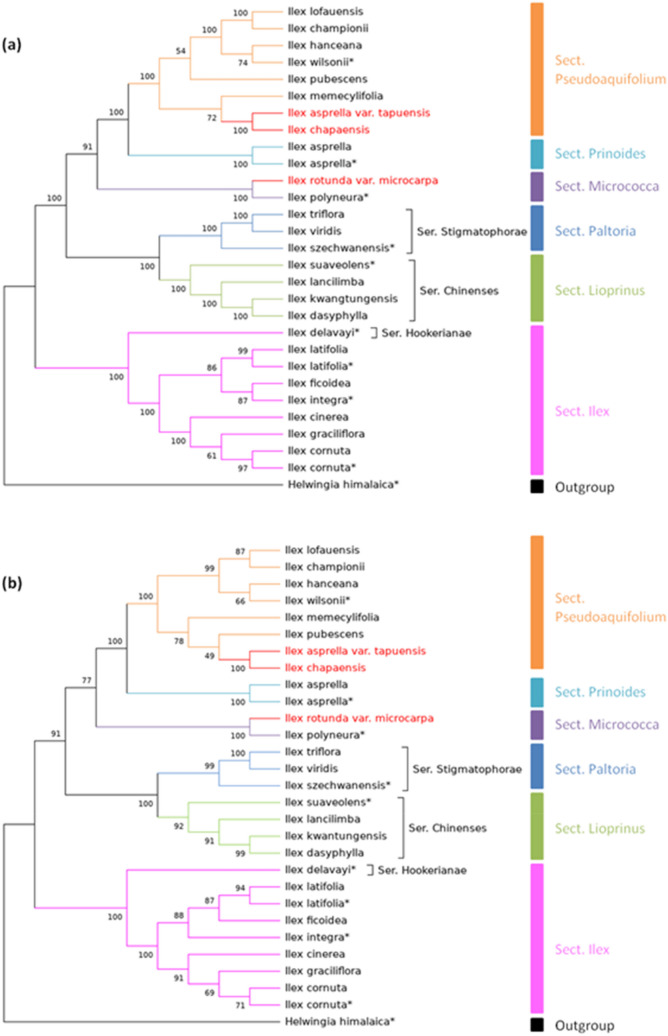
To obtain a more accurate analysis of the Ilex phylogeny, available Ilex genomes in NCBI were also included in our study. By using MEGA software, model testing was performed, and the maximum likelihood phylogenetic tree is illustrated (Fig. 6). The phylogeny matches the studies of Cuénoud and colleagues6. In brief, I. triflora and I. viridis were grouped together and put into the North American clade, while I. pubescens, I. wilsonii, I. asprella and I. rotunda var. microcarpa were assigned to the deciduous clade. I. latifolia, I. ficoidea, I. cornuta and I. integra were classified as the Eurasian clade. Similar results have been shown in research using psbA-trnH spacers and ITS regions7. Our studies were also consistent with the classification in Flora of China and Flora Reipublicae Popularis Sinicae. As expected, I. lohfauensis, I. championii and I. hanceana were grouped together, as well as I. kwangtungensis, I. dasyphylla and I. lancilimba, in which they were classified as the Hanceanae and Chinensis S. Y. Hu series, respectively. The Hookerianae and Stigmatophore series were also correctly assigned. As three out of four variation hotspots are located in the SSC region of the Ilex chloroplast genomes, phylogenetic studies were also carried out by using only SSC sequences. As expected, the classification was similar to the mentioned results, although some of the nodes had lower bootstrap values. All of the sections and series were assigned similarly to those with the use of the whole chloroplast genomes. The results illustrate that the SSC region, with three located DNA hotspots, can provide sufficient information for Ilex identification and phylogenetic investigation.
Figure 6.
Phylogenetic tree of the Ilex species constructed via maximum likelihood (ML) with 1000 bootstraps by using (a) whole chloroplast genomes and (b) SSC regions. The numbers above indicate the corresponding bootstrap values, and the corresponding sections and series are labelled according to Flora of China and Flora Reipublicae Popularis Sinicae, respectively. Genomes obtained from previous research are marked with asterisks, while unexpected species classifications are labelled in red.
Discussion
Here, we present the complete cp genomes of 19 native Ilex species in Hong Kong.
Although the organization of the chloroplast genome is highly similar among these 19 Ilex species and other angiosperms, significant variations in the total length, boundary of the regions and repeat distribution can be observed in the 19 plastomes (Table 1; Figs. 2, 3, 5). The genome sizes vary from 157,119 in I. graciliflora to 158,020 bp in I. kwangtungensis, which show a nearly 1 kb difference. Previous studies of seven Ilex chloroplast genomes found at most 300 bp differences17. Therefore, it is unexpected to observe more variations in our study. In many genera, including Nicotiana and Quercus, the phenomenon is due to the contraction and expansion of the IR regions34,36. Nonetheless, only slight variation of approximately 60 bp was observed in the Ilex IR regions. On the other hand, large differences were observed in the LSC regions. I. graciliflora and I. kwangtungensis possess the shortest and the longest LSC regions, respectively, with a difference of 900 bp. We then found two massive deletions in the trnT-trnL and ycf4-cemA spacers of I. graciliflora. The two deletions are approximately 700 bp in total. This finding may provide an explanation for the variation in genome size. In addition to the deletions in I. graciliflora, the trnT-trnL deletion was also noticed in I. cinerea and I. cornuta, while the ycf4-cemA deletion was observed in I. latifolia, I. ficoidea, I. cinerea and I. cornuta. These results were illustrated by mVISTA analysis (Supplementary Fig. S1). By referring to the sequence identity plot of mVISTA, a deletion in the rpoB-trnC spacer was found in I. lohfauensis, I. hanceana and I. championii. In fact, deletions in these regions have been widely used for species identification. The trnT-trnL deletion, which is 350 bp, is unique in the subfamily Cactoideae but not the other closely related cactus species37. Deletion in the ycf4-cemA intergenic region also provides a novel marker, LYCE, to discriminate between Angelica polymorpha and its adulterant Ligusticum officinale38. I. lohfauensis, I. hanceana and I. championii are members the Hanceanae series. Deletion in the rpoB-trnC intergenic region can serve as a molecular marker for species identification in these species.
All of the Ilex chloroplast genomes show a similar arrangement in the boundary regions (Fig. 5). In brief, SSC/IR boundaries were found within ycf1 in all of the Ilex species. For this reason, the pseudogene ycf1Ψ was created and overlapped with ndhF. This phenomenon has also been found in other angiosperms, such as Salvia and Prunus39,40. Based on the variation of the IR expansion, we were able to divide the Ilex studied into four different groups. Most of the Ilex species in group II and group III were classified as members of the deciduous clade and the North American clade, respectively7. I. latifolia, I. ficoidea, and I. cornuta of the group I Ilex were also assigned to the Eurasian clade in Cuénoud’s study and the Ilex section in Flora of China6,9. Group IV Ilex possess features observed in group I, group II and group III Ilex, which may explain the unexpected classification of I. rotunda var. microcarpa in our phylogenetic studies. In addition to the general grouping of Ilex, subgrouping was also observed in group II and group III Ilex (Supplementary Table S6). Compared with that in the other members of group II Ilex, the length of ycf1 in I. asprella var. tapuensis and I. chapaensis are especially different. This result may be the reason for the unexpected position of these two species in our phylogenetic analysis. On the other hand, the boundary arrangements of I. kwangtungensis and I. dasyphylla are more similar to one another, suggesting that these two species may have a closer taxonomic relationship than the other members of group III Ilex. The results indicate that the phenomenon of IR expansion and the features of genome boundaries can provide useful information for Ilex and other angiosperm classifications.
According to our sliding window analysis, rps16-trnQ, rpl32-trnL, ndhD-psaC and ycf1 show the greatest variations (Fig. 4). However, traditional barcoding regions, such as rbcL, matK and the trnH-psbA spacer, show only a low Pi value (Pi < 0.005). This finding illustrated that traditional DNA barcoding regions are not able to provide sufficient differences in classifying the species at a low taxonomic level. In fact, rps16-trnQ, rpl32-trnL, ndhD-psaC and ycf1 are divergent hotspots in other chloroplast genomes41. The rpl32-trnL spacer is a fast-evolving sequence and is used to discriminate between Lactuca and Helianthus42. rps16-trnQ has also been suggested for DNA barcoding for 12 different genera of angiosperms43. ycf1 has been proposed as the most promising cp DNA barcode, as it can distinguish the species much better than the combination of matK, rbcL and the trnH-psbA spacer44. As mentioned above, significant deletions were observed in rpoB-trnC, trnT-trnL and ycf4-cemA (Supplementary Fig. S1). These regions can also be developed for authentication purposes.
By using the complete cp genomes and SSC regions, phylogenetic analysis was performed (Fig. 6). Nearly all of the species were correctly classified in their corresponding sections, except I. rotunda var. microcarpa, I. chapaensis and I. asprella var. tapuensis. There are two discrepancies. First, according to Flora Reipublicae Popularis Sinicae, I. rotunda var. microcarpa should be in the Lioprinus section. However, I. rotunda var. microcarpa has been grouped to the Micrococca section in our genomic study. As the Lioprinus and Micrococca sections were classified as two different subgenera, distinct morphological variations could be observed. Some common features in the Micrococca section, such as the number of lateral veins and pyrenes, can be found in I. rotunda var. microcarpa. On the other hand, according to Flora Reipublicae Popularis Sinicae, the leaf surface of the Micrococca section is in membranous form. However, in our I. rotunda var. microcarpa specimen, the leaf surface appeared thin and leathery. In addition to Flora Reipublicae Popularis Sinicae, other references were considered for the phylogeny of I. rotunda var. microcarpa. Cuénoud and colleagues assigned I. rotunda and I. pubescens into the same deciduous clade, and the most closely related species with I. rotunda was I. micrococca6. The same conclusion was drawn by Manen and colleagues7. Interestingly, I. micrococca was also a member of the Micrococca section, which is consistent with our phylogenetic analysis. Combining these observations, we conclude that I. rotunda should be more closely related to the Micrococca section instead of the Lioprinus section. However, additional samples are needed to further confirm this observation. Another interesting finding was also noticed in I. chapaensis and I. asprella var. tapuensis. They were assigned as members of the Pseudoaquifolium section in our phylogenetic analysis. However, Flora Reipublicae Popularis Sinicae assigned them as members of the Prinoides section and grouped them together with I. asprella. This discrepancy needs further address by analysing nuclear and mitochondrial markers.
Materials and methods
Plant materials
Fresh young leaves of Ilex plants were collected in Hong Kong, China. Species were identified by Dr. David T.W. LAU (Curator of Shiu-Ying Hu Herbarium, School of Life Sciences, CUHK), with Flora of Hong Kong and Flora of China. Specimens with vouchers listed in Supplementary Table S1 were deposited in Shiu-Ying Hu Herbarium, School of Life Sciences, the Chinese University of Hong Kong. All Ilex species are not controlled under Protection of Endangered Species of Animals and Plants Ordinance (Cap. 586), Forests and Countryside Ordinance (Cap. 96), and not listed as rare and precious plants of Hong Kong. Their status are shown in Agriculture, Fisheries and Conservation Department (AFCD) Herbarium webpage (https://www.herbarium.gov.hk/result_list.aspx). Moreover, all of the individuals are collected with AFCD staff. All collections are permitted and legal.
DNA extraction and chloroplast genome sequencing
Fresh Ilex leaves (100 mg) were used to extract total genomic DNA by using a DNeasy Plant Pro Kit (Qiagen Co., Hilden, Germany) following the manufacturer’s protocol. Extracted DNA was quantified in a NanoDrop Lite (Thermo Fisher Scientific, Massachusetts, USA; quality cut-off, OD 260/280 ratio between 1.7 – 2.0) and Qubit 2.0 (Invitrogen, Carlsbad, USA). DNA was visualized by 1% agarose gel electrophoresis for quality checks. Illumina 150 bp paired-end (PE) libraries for each Ilex species were constructed and sequenced on the NovaSeq 6000 platform (Illumina Inc., San Diego, CA, USA) by Novogene Bioinformatics Technology Co., Ltd. (https://en.novogene.com/, Beijing, China). Poor-quality reads (Phred score < 33) were removed by quality trimming in the CLC Assembly Cell package v5.1.1 (CLC Inc., Denmark).
Genome assembly and annotation
Clean filtered reads were assembled into contigs using the CLC de novo assembler in CLC Assembly Cell package and SOAPdenovo v3.23 with default parameters. Gaps were filled by the Gapcloser module in SOAP package. Contigs were then aligned to the reference genome, Ilex cornuta (NC044416), and assembled into a complete chloroplast genome. Genome annotation was performed on the GeSeq platform by using complete cp genomes of Ilex cornuta (NC044416) and Ilex integra (NC044417) as references. A few adjustments for protein-coding genes and start and stop codons were performed manually. Chloroplast circular maps were then drawn in OGDRAW v1.3.1 (http://ogdraw.mpimp-golm.mpg.de/) according to the adjusted genome annotation45. All of the annotated genomes were deposited in GenBank with the accession numbers listed in Table 1.
Repeat sequence identification
Repeat motifs in Ilex species were identified by two different programs. Regarding microsatellites, the positions and motifs of simple sequence repeats (SSRs) were analysed by MISA software (https://webblast.ipk-gatersleben.de/misa/). To screen for the SSRs, they were identified with thresholds of 10, 5, 4, 3, 3, and 3 repeat units for mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides, respectively. To identify the long repeat motifs, REPuter v1.0 was used to locate forward, reverse, complementary and palindromic sequences, with a minimum repeat size of 30 bp and 90% identity. Statistical analysis was performed by GraphPad Prism v9.0.0 (GraphPad Software, La Jolla, CA, USA).
Chloroplast genome comparison
All 19 Ilex complete chloroplast genomes were aligned using MAFFT v7.0 and manually adjusted in BioEdit v7.2. The number and position of indels and SNPs were then investigated by an in-house python code developed by Seoul National University. Sliding window analysis was also performed to compare the nucleotide divergence (π) among the 19 Ilex species using DnaSP v6.0. The window length was set as 600 bp with a 200 bp step size. To illustrate the interspecific variations, full alignments of the complete chloroplast genomes of all 19 Ilex species and Helwingia himalaica (NC031370) were visualized in the mVISTA program under Shuffle-LAGAN mode.
Phylogenetic analysis
Complete chloroplast genomes and SSC sequences of the 19 Ilex species together with those of Ilex species available in NCBI, which were Ilex asprella (NC045274), Ilex cornuta (NC044416), Ilex integra (NC044417), Ilex latifolia (NC047291), Ilex szechwanensis (KX426466), Ilex suaveolens (MN830249), Ilex polyneura (KX426468), Ilex delavayi (KX426470) and Ilex wilsonii (KX426471), were used for phylogenetic analysis. The chloroplast sequence of Helwingia himalaica (NC031370) was also included as an outgroup. All of the chloroplast sequences were then aligned with MAFFT v7.0. The best nucleotide substitution model (GTR + R + I) was tested. Maximum likelihood (ML) with 1000 bootstrap replicates was constructed with MEGA-X software. Most of the taxonomic sections were labelled according to Flora of China. The Micrococca Prinoides sections and taxonomic series were labelled according to Flora Reipublicae Popularis Sinicae.
Conclusion
The chloroplast genomes of 19 Ilex species were sequenced, among which 14 genomes were reported for the first time in this study. Variations in the repeating motifs were illustrated, and four divergence hotspots were located. These can be potentially useful to develop authentication markers for medicinal Ilex. Together with other published Ilex chloroplast genomes, the phylogenetic relationship of 25 Ilex plastomes was studied. This work gives us a better understanding of Ilex phylogeny.
Supplementary Information
Acknowledgements
The authors thank the Agriculture, Fisheries and Conservation Department of HKSAR for assisting in sample collection. The work was supported in part by a Health and Medical Research Fund (project No. 19180302) of Hong Kong SAR.
Author contributions
B.L.H.K. performed the experiments, specimen collection, DNA extraction, genome assembly and analysis and wrote the paper. T.W.D.L. was responsible for field collection, species identification and morphological analysis of specimens. T.J.Y. and H.S.P. provided the genome assembly technique, performed data analysis and provided technical support in designing algorithms. Z.L. and P.C.S. designed the experiments, contributed materials/reagents/analysis tools, and reviewed the draft of the paper.
Data availability
The complete chloroplast sequences generated and analysed during the current study are available in GenBank (MT704834, MT764239-MT764254 and MT767004-MT767005).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-84705-9.
References
- 1.Manen J, Barriera G, Naciri Y. The history of extant Ilex species (Aquifoliaceae): Evidence of hybridization within a Miocene radiation. Mol. Phylogenet. Evol. 2010;57:961. doi: 10.1016/j.ympev.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Dai W, et al. The roots of Ilex asprella extract lessens acute respiratory distress syndrome in mice induced by influenza virus. J. Ethnopharmacol. 2014;155:1575. doi: 10.1016/j.jep.2014.07.051. [DOI] [PubMed] [Google Scholar]
- 3.Wu P, Gao H, Liu J, Liu L, Zhou H, Liu Z. Triterpenoid saponins with anti-inflammatory activities from Ilex pubescens roots. Phytochemistry. 2017;134:122. doi: 10.1016/j.phytochem.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Zhang W, et al. Asprellcosides B of Ilex asprella inhibits influenza A virus infection by blocking the hemagglutinin-mediated membrane fusion. Front. Microbiol. 2019;9:3325. doi: 10.3389/fmicb.2018.03325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim YJ, et al. Rotundarpene inhibits toll-like receptor 2 activation-induced production of inflammatory mediators in keratinocytes by suppressing the Akt and NF-κB pathways. Int. Immunopharmacol. 2014;18:325–332. doi: 10.1016/j.intimp.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 6.Cuénoud P, Martienz M, Loizeau P, Spichiger R, Andrews S, Manen J. Molecular phylogeny and biogeography of the genus Ilex L (Aquifoliaceae) Ann. Bot. 2000;85:111–122. doi: 10.1006/anbo.1999.1003. [DOI] [Google Scholar]
- 7.Manen J, Boulter MC, Naciri-Graven Y. The complex history of the genus Ilex L. (Aquifoliaceae): evidence from the comparison of plastid and nuclear DNA sequences and from fossil data. Plant Syst. Evol. 2002;235:79–98. doi: 10.1007/s00606-002-0225-x. [DOI] [Google Scholar]
- 8.Shi L, et al. Molecular evidence for the hybrid origin of Ilex dabieshanensis (Aquifoliaceae) PLoS ONE. 2016 doi: 10.1371/journal.pone.0147825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S, Ma H, Feng Y, Barriera G, Loizeau P. Flora of China:Aquifoliaceae. New York: Science Press; 2008. [Google Scholar]
- 10.Jensen RG, Bassham JA. Photosynthesis by isolated chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 1966;56:1095. doi: 10.1073/pnas.56.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlin-Neumann GA, Tobin EM. Transit peptides of nuclear-encoded chloroplast proteins share a common amino acid framework. EMBO J. 1986;5:9–13. doi: 10.1002/j.1460-2075.1986.tb04170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makino A, Osmond B. Effects of nitrogen nutrition on nitrogen partitioning between chloroplasts and mitochondria in pea and wheat. Plant Physiol. 1991;96:355–362. doi: 10.1104/pp.96.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuruta S, Ebina M, Kobayashi M, Takahashi W. Complete chloroplast genomes of Erianthus arundinaceus and Miscanthus sinensis: Comparative genomics and evolution of the Saccharum complex. PLoS ONE. 2017 doi: 10.1371/journal.pone.0169992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao M, Song Y, Ni J, Yao X, Tan Y, Xu Z. Comparative chloroplast genomics and phylogenetics of nine Lindera species (Lauraceae) Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, Yue M, Niu C, Ma X, Li Z. Comparative analysis of the complete chloroplast genome of four endangered herbals of Notopterygium. Genes. 2017;8:124. doi: 10.3390/genes8040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang C, Wu W, Liu Y, Jheng C, Chen T, Lin B, Lin J, Chen T, Lee Y. In: Orchid Biotechnology III. Chen WH, editor. Singapore: World Scientific; 2017. pp. 61–90. [Google Scholar]
- 17.Yao X, Tan Y, Liu Y, Song Y, Yang J, Corlett RT. Chloroplast genome structure in Ilex (Aquifoliaceae) Sci. Rep. 2016;6:1–10. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park J, Kim Y, Nam S, Kwon W, Xi H. The complete chloroplast genome of horned holly, Ilex cornuta Lindl. & Paxton (Aquifoliaceae) Mitochondrial DNA Part B. 2019;4(1):1275–1276. doi: 10.1080/23802359.2019.1591212. [DOI] [Google Scholar]
- 19.Park J, Kim Y, Kwon W, Nam S, Xi H. The complete chloroplast genome of Nepal Holly, Ilex integra Thunb.(Aquifoliaceae) Mitochondrial DNA Part B. 2019;4(1):1257–1258. doi: 10.1080/23802359.2019.1591235. [DOI] [Google Scholar]
- 20.Richard G, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol. Mol. Biol. Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Provan J, Corbett G, Powell W, McNicol JW. Chloroplast DNA variability in wild and cultivated rice (Oryza spp.) revealed by polymorphic chloroplast simple sequence repeats. Genome. 1997;40:104–110. doi: 10.1139/g97-014. [DOI] [PubMed] [Google Scholar]
- 22.Xue J, Wang S, Zhou S. Polymorphic chloroplast microsatellite loci in Nelumbo (Nelumbonaceae) Am. J. Bot. 2012;99:240–244. doi: 10.3732/ajb.1100547. [DOI] [PubMed] [Google Scholar]
- 23.Kaila T, et al. Chloroplast genome sequence of clusterbean (Cyamopsis tetragonoloba L.): Genome structure and comparative analysis. Genes. 2017;8:212. doi: 10.3390/genes8090212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, et al. Comparison of four complete chloroplast genomes of medicinal and ornamental meconopsis species: genome organization and species discrimination. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim K, Lee H. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11:247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- 26.Huotari T, Korpelainen H. Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene. 2012;508:96–105. doi: 10.1016/j.gene.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 27.Yi D, Lee H, Sun B, Chung MY, Kim K. The complete chloroplast DNA sequence of Eleutherococcus senticosus (Araliaceae); comparative evolutionary analyses with other three asterids. Mol. Cells. 2012;33:497–508. doi: 10.1007/s10059-012-2281-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogihara Y, Terachi T, Sasakuma T. Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc. Natl. Acad. Sci. U.S.A. 1988;85:8573–8577. doi: 10.1073/pnas.85.22.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Lin F, Huang P, Guo W, Zheng Y. Complete chloroplast genome sequence of Decaisnea insignis: Genome organization, genomic resources and comparative analysis. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greiner S, et al. The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. Sequence evaluation and plastome evolution. Nucleic Acids Res. 2008;36:2366–2378. doi: 10.1093/nar/gkn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Liao B, Song J, Pang X, Han J, Chen S. A fast SNP identification and analysis of intraspecific variation in the medicinal Panax species based on DNA barcoding. Gene. 2013;530:39–43. doi: 10.1016/j.gene.2013.07.097. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Wang X, Wang L, Chen X, Pang X, Han J. A nucleotide signature for the identification of American ginseng and its products. Front. Plant Sci. 2016;7:319. doi: 10.3389/fpls.2016.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong Z, Wu Z, Zhao K, Yang Z, Zhang N, Guo J, Tembrock LR, Xu D. Comparative analyses of five complete chloroplast genomes from the genus Pterocarpus (Fabacaeae) Int. J. Mol. Sci. 2020;21:11. doi: 10.3390/ijms21113758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, Zhou T, Duan D, Yang J, Feng L, Zhao G. Comparative analysis of the complete chloroplast genomes of five quercus species. Front. Plant Sci. 2016;7:959. doi: 10.3389/fpls.2016.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song Y, Dong W, Liu B, Xu C, Yao X, Gao J, Corlett RT. Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae. Front. Plant Sci. 2015;6:662. doi: 10.3389/fpls.2015.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asaf S, Khan AL, Khan AR, Waqas M, Kang SM, Khan MA, Lee SM, Lee IJ. Complete chloroplast genome of Nicotiana otophora and its comparison with related species. Front. Plant Sci. 2016;7:843. doi: 10.3389/fpls.2016.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Applequist WL, Wallace RS. Deletions in the plastid trnT–trnL intergenic spacer define clades within Cactaceae subfamily Cactoideae. Plant Syst. Evol. 2002;231:153–162. doi: 10.1007/s006060200017. [DOI] [Google Scholar]
- 38.Park I, Yang S, Kim WJ, Song JH, Lee HS, Lee HO, Lee JH, Ahn SN, Moon BC. Sequencing and comparative analysis of the chloroplast genome of Angelica polymorpha and the development of a novel Indel marker for species identification. Molecules. 2019;24:1038. doi: 10.3390/molecules24061038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian J, et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE. 2013 doi: 10.1371/journal.pone.0057607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao, X., Yan, M., Ding, Y., Huo, Y. & Yuan, Z. Characterization and comparative analysis of the complete chloroplast genome sequence from Prunus avium. PeerJ. https://peerj.com/articles/8210/ (2019). [DOI] [PMC free article] [PubMed]
- 41.Li Y, Xu W, Zou W, Jiang D, Liu X. Complete chloroplast genome sequences of two endangered Phoebe (Lauraceae) species. Bot. Stud. 2017;58:37. doi: 10.1186/s40529-017-0192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timme RE, Kuehl JV, Boore JL, Jansen RK. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of divergent regions and categorization of shared repeats. Am. J. Bot. 2007;94:302–312. doi: 10.3732/ajb.94.3.302. [DOI] [PubMed] [Google Scholar]
- 43.Dong W, Liu J, Yu J, Wang L, Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE. 2012 doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong W, et al. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015;5:8348. doi: 10.1038/srep08348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47(W1):W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Y, Liu B. Complete chloroplast genome sequence of Ilex latifolia (Aquifoliaceae), a traditional Chinese tea. Mitochondrial DNA Part B. 2020;5(1):190–191. doi: 10.1080/23802359.2019.1698985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete chloroplast sequences generated and analysed during the current study are available in GenBank (MT704834, MT764239-MT764254 and MT767004-MT767005).