SUMMARY
Dicistrovirus intergenic region internal ribosomal entry sites (IGR IRESs) do not require initiator tRNA, an AUG codon, or initiation factors and jump-start translation from the middle of the elongation cycle via formation of IRES/80S complexes resembling the pre-translocation state. eEF2 then translocates the [codon-anticodon]-mimicking pseudoknot I (PKI) from ribosomal A sites to P sites, bringing the first sense codon into the decoding center. Halastavi árva virus (HalV) contains an IGR that is related to previously described IGR IRESs but lacks domain 2, which enables these IRESs to bind to individual 40S ribosomal subunits. By using In vitro reconstitution and cryoelectron microscopy (cryo-EM), we now report that the HalV IGR IRES functions by the simplest initiation mechanism that involves binding to 80S ribosomes such that PKI is placed in the P site, so that the A site contains the first codon that is directly accessible for decoding without prior eEF2-mediated translocation of PKI.
In Brief
Initiation on previously described IGR IRESs requires eEF2-mediated pseudo-translocation to bring the first codon into the decoding center. Abaeva et al. find that binding of the HalV IGR IRES to 80S ribosomes places PKI in the P site, making the A site codon directly accessible for decoding without prior translocation.
INTRODUCTION
The canonical process of translation initiation in eukaryotes begins with separated ribosomal subunits and involves the coordinated activities of >10 eukaryotic initiation factors (eIFs) (Jackson et al., 2010). In outline, the 43S preinitiation complex, comprising the 40S ribosomal subunit, initiator tRNA, and eIFs 2, 3, 1, and 1A, is recruited to the capped 5′ end of mRNA by eIF4F, eIF4A, and eIF4B and scans downstream to the initiation codon, where it stops and forms a 48S initiation complex with established codon-anticodon base pairing in the ribosomal P site. After recognition of the initiation codon, eIF5 and eIF5B promote dissociation of factors from the 40S subunit and its joining with a 60S subunit, forming an elongation-competent 80S ribosome. This process is regulated in response to physiological changes. For example, cells respond to viral infection by phosphorylation of eIF2, leading to global downregulation of translation (Mohr and Sonenberg, 2012).
Although the majority of cellular mRNAs initiate translation by the 5′ end-dependent scanning mechanism, initiation on a substantial proportion of viral mRNAs is mediated by so-called internal ribosomal entry sites (IRESs), which contain cis-elements that mediate 5′ end-independent ribosomal recruitment to internal locations within mRNAs. Viral IRESs are grouped into several major classes, each with its own common structural core, conserved sequence motifs, and characteristic factor requirements (Mailliot and Martin, 2018). Although structurally unrelated IRESs use distinct initiation mechanisms, all of them are based on non-canonical interactions of IRESs with canonical components of translational apparatus (e.g., eIF4G, eIF3, or 40S and 60S ribosomal subunits) (e.g., Pestova et al., 1996, 1998b; Wilson et al., 2000; de Breyne et al., 2009; Imai et al., 2016). As a rule, initiation on viral IRESs requires only a subset of canonical eIFs, which allows them to circumvent various cellular regulatory mechanisms (Jackson et al., 2010; Mailliot and Martin, 2018).
The most streamlined translation mechanism identified to date is used by dicistrovirus intergenic region (IGR) IRESs, which exploit direct interaction with the ribosome to jumpstart translation from the elongation stage, skipping requirements for initiator tRNA, an AUG initiation codon, and initiation factors (Sasaki and Nakashima, 2000; Wilson et al., 2000; Jan et al., 2003; Pestova and Hellen, 2003). IGR IRESs are ~190 nucleotides long and form two closely related subclasses, epitomized by Cricket paralysis virus (CrPV) (herein designated type VIa) and Taura syndrome virus (TSV) (type VIb). They fold into three pseudoknots (PKs): two nested PKs (PKII and PKIII) and a third PK (PKI), which forms an independent domain (Kanamori and Nakashima, 2001; Nakashima and Uchiumi, 2009). PKII contains an internal loop (L1.1) that binds to the L1 stalk of the 60S subunit, whereas PKIII has two exposed stem-loops, SLIV and SLV, containing conserved apical motifs that bind to the ribosomal proteins eS25, uS7, and uS11 on the head of the 40S subunit (Muhs et al., 2011; Fernández et al., 2014; Koh et al., 2014). PKI mimics the anticodon stem-loop of tRNA base paired to a cognate codon (Costantino et al., 2008). The first sense codon is located at the 3′ edge of PKI, making PKI responsible for the correct placement of this codon into the ribosomal decoding center. Type VIa and type VIb IGR IRESs have similar compact structures, but in the latter, PKI contains an additional hairpin (stemloop III [SLIII]).
IGR IRESs bind either to individual 40S subunits followed by recruitment of 60S subunits, or directly to 80S ribosomes. In IGR IRES-80S complexes, which alternate between canonical and rotated states, the IRES is located in the intersubunit space, with PKI mimicking the tRNA-mRNA interaction in the decoding center of the ribosomal A site (Fernández et al., 2014; Koh et al., 2014). Thus, before the first sense codon can be decoded, PKI has to be removed from the A site. For this, eEF2-GTP binds to the IRES/80S complex in the rotated state and induces translocation of PKI to the P site, thereby bringing the first sense codon into the A site where it is decoded by a cognate eEF1A·GTP/aa-tRNA ternary complex (Yamamoto et al., 2007; Fernández et al., 2014; Muhs et al., 2015; Abeyrathne et al., 2016; Murray et al., 2016). This aa-tRNA is released from eEF1A, accommodates in the A site, and eEF2 then promotes another translocation event, moving aa-tRNA to the P site and the IRES to the E site, thereby yielding a ribosomal complex that is competent to begin elongation. Initiation mediated by IGR IRESs thus proceeds via formation of an IRES/80S complex that mimics the pre-translocation state and starts in the middle of an elongation cycle.
Advances in metagenomics have revealed a plethora of novel dicistrovirus-like viruses (e.g., Culley et al., 2007; Boros et al., 2011; Shi et al., 2016), some of which contain IGR sequences that are shorter than canonical IGR IRESs and lack SLIV and SLV-like motifs. These differences suggest that if these divergent IGRs function as IRESs, they may initiate translation by novel mechanisms. Here, we identified a distinct class of such IGRs epitomized by the IGR from Halastavi árva virus (HalV), which was isolated from the intestinal contents of freshwater carp (Cyprinus carpio) (Boros et al., 2011). HalV has a dicistronic RNA genome with an IRES in the 5′ UTR that promotes translation of the open reading frame (ORF) 1 nonstructural protein precursor (Abaeva et al., 2016) and an IGR preceding ORF2, which encodes the capsid protein precursor. By using biochemical analysis and cryoelectron microscopy (cryo-EM) to characterize the HalV IGR, we determined that it contains an IRES that promotes translation by a novel mechanism that is even simpler than that of previously described IGR IRESs.
RESULTS
Factor-Independent Internal Ribosomal Entry on the HalV IGR
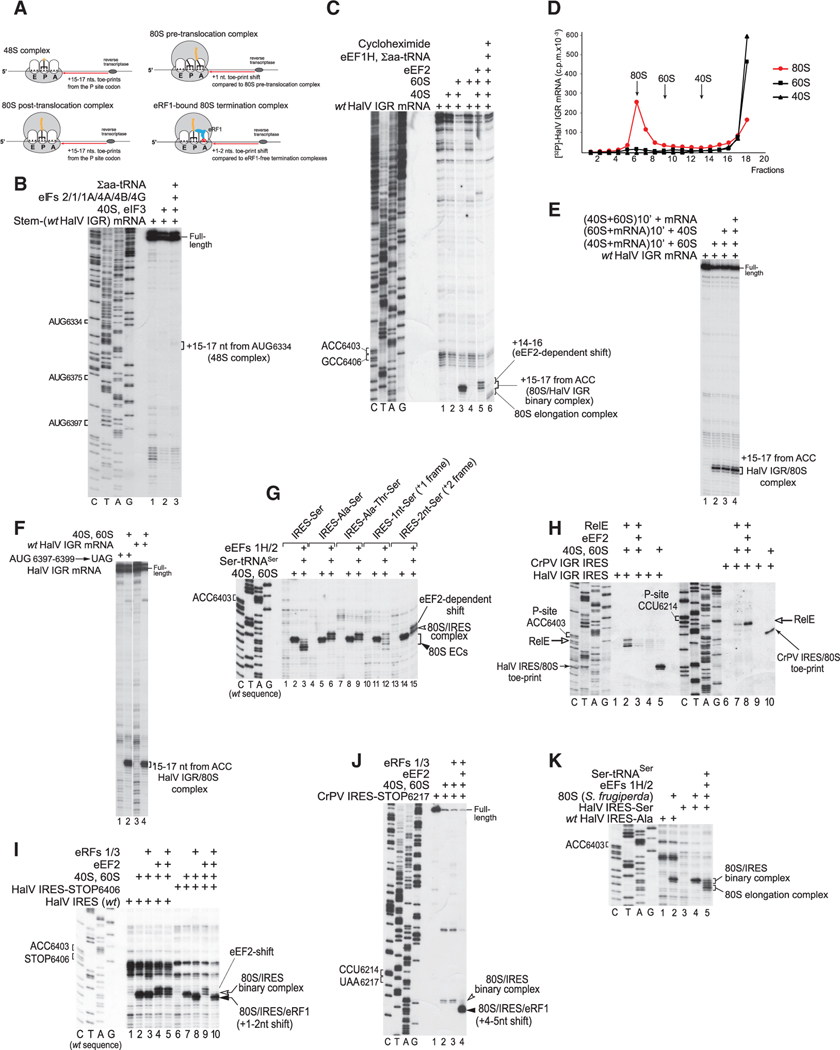
The homology between HalV and dicistrovirus genomes (Boros et al., 2011) suggests that the HalV IGR also promotes initiation on ORF2 by a non-canonical IRES-dependent mechanism. The stop codon of HalV ORF1 is UAA6276–6278. The HalV ORF2 is in frame with the UAA6382–6384 stop codon downstream of ORF1, and initiation on ORF2 must therefore start after this triplet. The first AUG triplet in ORF2 is AUG6397–6399. To determine the start site and the mechanism of initiation on the HalV ORF2, we used an In vitro reconstitution approach. Ribosomal complexes were assembled from individual mammalian translational components (ribosomal subunits, translation factors, and aa-tRNAs) on mRNA comprising a 5′-terminal stable hairpin (ΔG = −25.80 kcal/mol) to block the 5′ end-dependent initiation, followed by nucleotides 6211–7460 of the HalV genome. The ribosomal position was then determined by toe-printing, which involves extension by reverse transcriptase of a primer annealed to the ribosome-bound mRNA. cDNA synthesis is arrested by the leading edge of the 40S subunit, yielding characteristic toe-prints +15–17 nt from the P site codon (Figure 1A).
Figure 1. The Mechanism of Initiation on the HalV IGR IRES.
(A) Schematic representation of various ribosomal complexes showing the relative positions of their toe-prints.
(B) 48S complex formation on HalV IGR mRNA comprising a 5′-terminal stem (ΔG = −25.80 kcal/mol) followed by nucleotides 6211–7460 of the HalV genome in the presence of 40S subunits, Σaa-tRNA, and canonical eIFs, as indicated, assayed by toe-printing. The positions of AUG codons in the HalV IGR and assembled ribosomal complexes are indicated.
(C) Direct binding of the HalV IGR mRNA to 80S ribosomes followed by elongation upon addition of eEF1H, eEF2, Σaa-tRNA, and cycloheximide, assayed by toeprinting. The positions of the A site and P site codons of the HalV IGR IRES are shown on the left. The positions of ribosomal complexes and the eEF2-mediated toe-print shift are indicated on the right. Separation of lanes by white lines indicates that they were juxtaposed from the same gel.
(D) Association of 32P-labeled HalV IGR-containing mRNA with individual 40S and 60S ribosomal subunits and 80S ribosomes, assayed by sucrose density gradient centrifugation.
(E) Toe-printing analysis of ribosomal association of the HalV IGR IRES depending on the order of incubation of mRNA, 40S ribosomal subunits, and 60S ribosomal subunits. The position of IRES/80S complexes is indicated.
(F) Ribosome-binding activity of the AUG6397-6399→UAG stop codon HalV IGR mRNA mutant, assayed by toe-printing. The position of IRES/80S complexes is indicated.
(G) The fidelity of reading frame selection on the HalV IGR IRES investigated by the ability of 80S/IRES complexes formed on GCC6406-6408(Ala)→UCU(Ser), ACU6409-6411(Thr)→UCU(Ser), and AUU6412-6414(Ile)→UCU(Ser) HalV IGR variants and IGR mutants with UCU(Ser) placed in the +1 or +2 reading frame by insertion of one (G) or two (GC) nucleotides. between ACC6403–6405 and UCU6406–6408(Ser) codons to undergo one-cycle elongation in the presence of eEF1H, eEF2, and Ser-tRNASer, assayed by toe-printing. The positions of IRES/80S binary complexes and 80S elongation complexes (80S ECs) are shown on the right.
(H) Comparison of the eEF2 dependency of the A site accessibility in 80S ribosomal complexes assembled on HalV and CrPV IGR IRESs, assayed by RelE cleavage. Sites of RelE cleavages were determined by primer extension. The positions of P site codons, RelE cleavages, and IRES/80S control toe-prints are indicated.
(I and J) Comparison of the eEF2 dependency of the A site accessibility in 80S ribosomal complexes formed on HalV and CrPV IGR IRESs by binding of eRF1 and eRF3 to 80S ribosomes assembled on (I) HalV IRES-STOP(UAA) and (J) CrPV IRES-STOP(UAA) mutant mRNAs, assayed by toe-printing. The positions of ribosomal complexes and P site and stop codons are indicated.
(K) The ability of HalV IGR IRES to form elongation-competent complexes with insect (Spodoptera frugiperda) 80S ribosomes, assayed by toe-printing. The positions of the A site codon and ribosomal complexes are indicated. (B, C, and G–K) Lanes C, T, A, and G depict CrPV or HalV sequences, as indicated.
In the presence of 40S subunits, Met-tRNAiMet, and all canonical eIFs, we observed faint toe-prints that could correspond to 48S complexes formed at the out-of-frame AUG6334 triplet, but no toe-prints that could be consistent with 48S complex formation at any of ORF2’s in-frame codons (Figure 1B). Strikingly, although in contrast to the CrPV IGR IRES, HalV mRNA did not bind to individual 40S subunits (Figure 1C, lane 2; Figure 1D, black triangles), it bound efficiently to 80S ribosomes, yielding prominent toe-prints +15-+17 nt downstream of the ACC6403–6405 codon in ORF2 (Figure 1C, lane 3; Figure 1D, red circles). To confirm that HalV mRNA binds directly to pre-assembled 80S ribosomes, we performed preincubation experiments. Thus, mRNA was preincubated with 40S or 60S subunits and then 60S or 40S subunits were added, respectively, or 40S and 60S subunits were first preincubated to form 80S ribosomes and then mRNA was added to the mixture. The highest yield of toe-prints +15-+17 nt downstream of the ACC6403–6405 codon was observed in the last case (Figure 1E), indicating that HalV mRNA binds directly to 80S ribosomes. The position of toe-prints suggested that ACC6403–6405 and the following ORF2’s GCC6406–6408 (Ala) triplet occupied the ribosomal P and ribosomal A sites, respectively, and that ribosomes did not recognize AUG6397–6399, the first AUG in ORF2. Moreover, the AUG6397–6399→UAG stop codon mutant HalV mRNA bound 80S ribosomes as efficiently as the WT HalV mRNA (Figure 1F). Addition of eEF1H, eEF2, and total aa-tRNA to 80S/HalV mRNA complexes enabled ribosomes to undergo elongation, which was arrested by cycloheximide (Figure 1C, lane 6). Notably, in the absence of eEF1H and aa-tRNA, eEF2 induced a −2 nt upward shift of the toe-print (Figure 1C, lane 5) that may reflect its trapping of the ribosome in the rotated state or with the head in a swiveled position (e.g., Flis et al., 2018). In conclusion, the HalV IGR contains an IRES that binds productively to 80S ribosomes in a factor-independent manner.
To facilitate investigation of the mechanisms of delivery of the first aa-tRNA and initial translocation events on the HalV IGR IRES, the presumed A site GCC6406–6408(Ala) codon was replaced by the UCU6406–6408(Ser) codon (a GCC6406-6408(Ala)→UCU(Ser) HalV IGR IRES variant) because in-vitro-transcribed Ser-tRNASer-AGA is readily available and active in mammalian elongation (Zinoviev et al., 2018). Addition of Ser-tRNASer, eEF1H, and eEF2 to 80S ribosomes bound to a GCC6406(Ala)→UCU(Ser) IGR variant resulted in a forward toe-print shift by precisely 3 nt (Figure 1G, lane 3), which would be consistent with one, but not two, cycles of translocation as in the case of the CrPV IRES. Such translocation did not occur when the UCU(Ser) codon replaced the second or third triplet downstream of the IRES, and in this case we observed only the eEF2-induced upward toe-print shift (Figure 1G, lanes 6 and 9). However, a low level of templated translocation was apparent in the +1, but not in the +2, frame (Figure 1G, lanes 12 and 15), indicating that the fidelity of initiation is high, but not absolute. Notably, in the case with the UCU(Ser) codon in the +1 position, we observed not only the +4 nt toe-print shift that corresponds to one cycle of translocation but also a more prominent toe-print at the +2 position indicative of binding of Ser-tRNASer (Zinoviev et al., 2015) in the pre-translocation state, reflecting inefficient translocation in this reading frame after delivery of aa-tRNA to the A site.
To confirm that binding of the HalV IGR IRES to 80S ribosomes places the GCC6406–6408 triplet directly in the ribosomal A site without a prior eEF2-dependent pseudo-translocation event, we used the bacterial toxin RelE, which cleaves mRNA in the A site (Pedersen et al., 2003; Neubauer et al., 2009; Pisareva et al., 2011). RelE efficiently cleaved the 80S-bound HalV IGR IRES within the GCC6406–6408 triplet (Figure 1H, lane 2). Importantly, in contrast to cleavage of the CrPV IRES that occurred efficiently only on inclusion of eEF2 (Figure 1H, compare lanes 7 and 8), consistent with the requirement for eEF2 to translocate PKI from the A site (Fernández et al., 2014), cleavage of the HalV IRES not only occurred independently of eEF2 but also was even inhibited by it (Figure 1H, lane 3), likely reflecting competition between RelE and eEF2 for binding to the A site. The A site in HalV IRES-bound 80S ribosomes is therefore vacant and can accept aa-tRNA directly. To further prove that the A site is vacant, we replaced it by a stop codon. The 80S ribosomes associated with the GCC6406→stop codon HalV IRES variant efficiently bound eRF1·eRF3 independently of eEF2, evident by the characteristic +1–2 nt toe-print shift caused by the A site mRNA compaction (Figure 1I, lanes 7 and 8) (Alkalaeva et al., 2006; Brown et al., 2015). By contrast, 80S ribosomes associated with the analogous CrPV IRES mutant did require eEF2 to bind eRF1·eRF3, and binding induced a +4–5 nt toe-print shift that included eEF2-dependent initial IRES translocation and mRNA compaction caused by binding of eRF1·eRF3 (Figure 1J; Muhs et al., 2015).
The HalV IGR IRES also efficiently bound to insect (Spodoptera frugiperda) 80S ribosomes, yielding prominent toe-prints +15– +17 nt downstream of ACC6403–6405 and underwent one cycle of translocation on the GCC6406–6408(Ala)→UCU(Ser) HalV IRES variant in the presence of eEFs and Ser-tRNASer (Figure 1K).
In conclusion, In vitro reconstitution revealed that the HalV IGR contains an IRES that binds directly to 80S ribosomes and jumpstarts translation from the elongation stage by accepting aa-tRNA to the ribosomal A site, containing the Ala codon GCC6406–6408. In contrast to dicistrovirus CrPV-like IGR IRESs, this step does not require prior eEF2-mediated translocation of a portion of the IRES from A sites to P sites. However, similarly to CrPV-like IRESs (e.g., Petrov et al., 2016), the HalV IRES is not wholly specific in determining the reading frame for translation, with a minor fraction of initiation events occurring in the +1 frame.
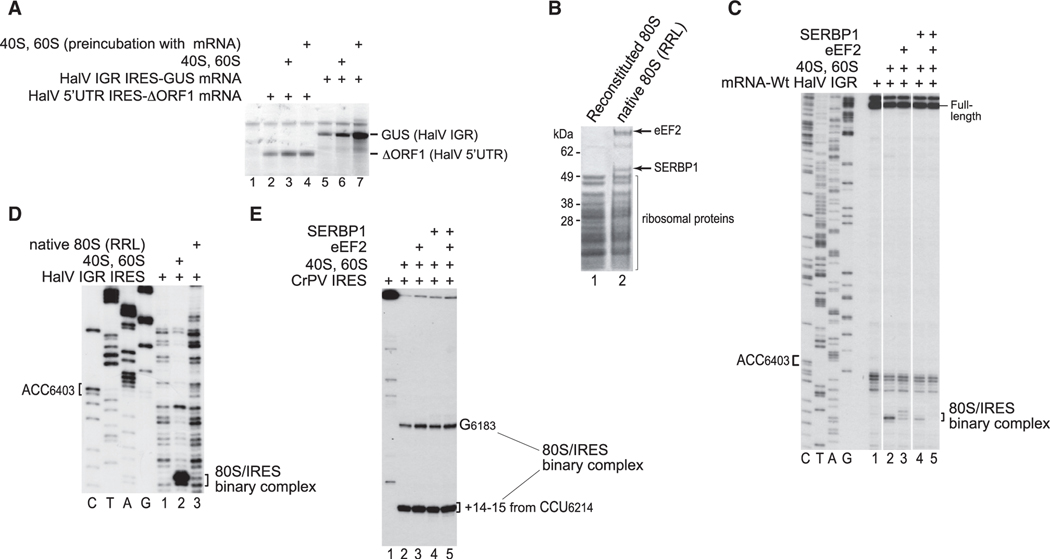
Inhibition of Ribosomal Binding of the HalV IGR IRES by SERBP1 and eEF2
Although the HalV IGR IRES bound productively to 80S ribosomes assembled In vitro from individual salt-washed ribosomal subunits, it did not promote efficient translation in rabbit reticulocyte lysate (RRL) (Figure 2A, lane 5). However, the translational efficiency was increased by addition of assembled In vitro vacant 80S ribosomes, and particularly, by preincubation of mRNA with such ribosomes before the addition of both to the translation mixtures (Figure 2A, lanes 6 and 7). By contrast, translation promoted by the 5′-terminal HalV IRES (Abaeva et al., 2016) was not significantly affected by addition of 80S ribosomes, irrespective of preincubation (Figure 2A, lanes 2–4). Non-programmed 80S ribosomes in RRL, human peripheral blood mononuclear cells, and Drosophila melanogaster embryonic extracts are associated with SERPINE1 mRNA binding protein 1 (SERBP1) and eEF2 (Anger et al., 2013; Zinoviev et al., 2015; Brown et al., 2018; Figure 2B). SERBP1 binds to the head of the 40S subunit and then enters the mRNA-binding channel and follows the mRNA-binding path until the A site where it interacts with domain IV of eEF2 (Anger et al., 2013; Brown et al., 2018). It prompted us to investigate whether ribosomal association with SERBP1 and eEF2 is responsible for the low translational activity of the HalV IGR IRES in RRL. Preincubation of 80S ribosomes with SERBP1 strongly reduced their ability to bind the HalV IGR IRES, whereas preincubation with SERBP1 and eEF2 nearly abrogated formation of IRES/80S complexes (Figure 2C). Consistently, SERBP1/eEF2-associated native purified RRL 80S ribosomes (Figure 2B) did not bind the HalV IGR IRES (Figure 2D). By contrast, SERBP1 and eEF2 did not affect ribosomal association of the CrPV IRES (Figure 2E). Thus, the presence of SERBP1 and eEF2 on 80S ribosomes specifically prevents their binding to the HalV IGR IRES and is responsible for the low activity of the IRES in RRL.
Figure 2. The Influence of SERBP1 on Initiation on the HalV IRES.
(A) Translation in RRL driven by HalV 5′ UTR and IGR IRESs, depending on addition of 40S and 60S subunits with or without their preincubation with mRNA.
(B) Protein composition of 80S ribosomes reconstituted from individual purified 40S and 60S subunits, and native 80S ribosomes purified from RRL, assayed by SDS-PAGE followed by SYPRO staining.
(C) The influence of SERBP1 with/without eEF2 on ribosomal binding of the HalV IGR IRES, assayed by toe-printing. The positions of the A site codon and ribosomal complexes are indicated. Separation of lanes by white lines indicates that they were juxtaposed from the same gel.
(D) Comparison of the binding of the HalV IGR IRES to reconstituted and native 80S ribosomes, assayed by toe-printing. The positions of the A site codon and ribosomal complexes are indicated. (C and D) Lanes C, T, A, and G depict HalV sequence.
(E) The influence of SERBP1 with/without eEF2 on ribosomal binding of CrPV IGR IRES, assayed by toe-printing. The positions of toe-prints corresponding to IRES/80S complexes are indicated.
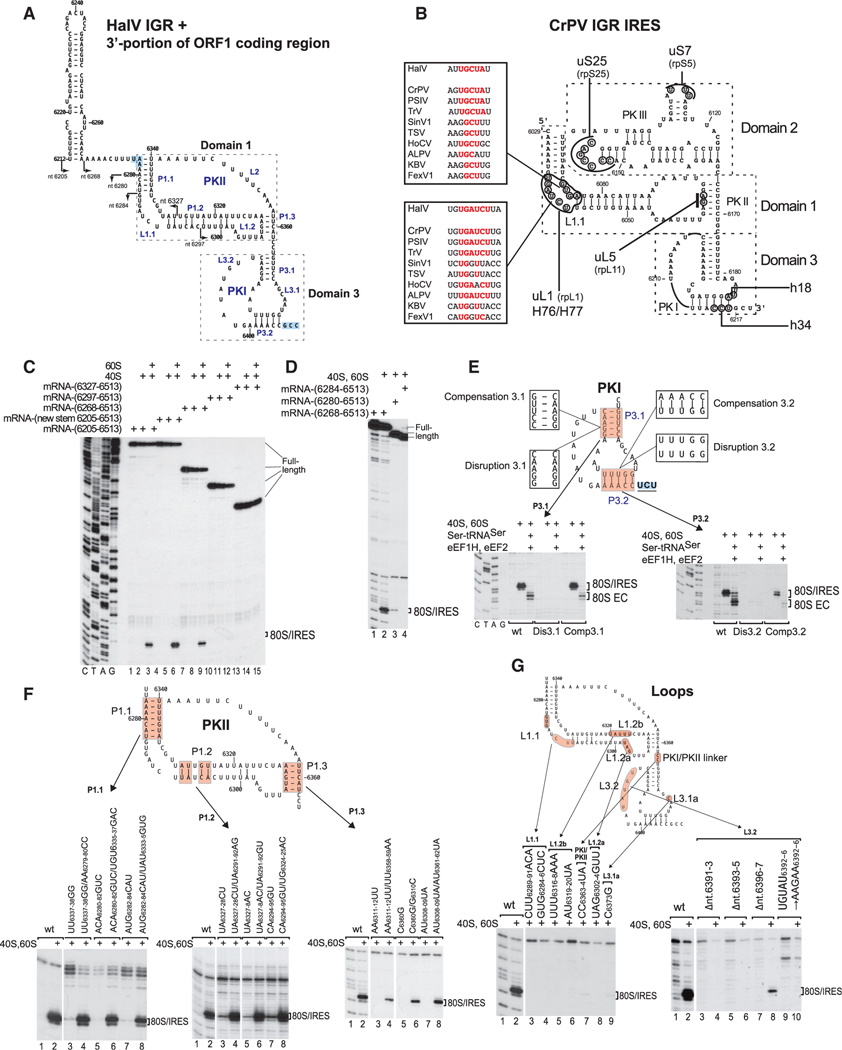
Structural Model of the HalV IGR IRES
To aid understanding of the mechanism of the HalV IGR IRES function, a structural model of the HalV IGR and the adjacent 3′-terminal region of ORF1 (Figure 3A) was derived on the basis of computational analysis (see STAR Methods) and chemical/enzymatic foot-printing (Figures S1A and S1B). The 3′-terminal region of ORF1 (nucleotides 6211–6267) forms a large hairpin (Figures 3A and S1C). The HalV IGR consists of two domains that are comparable to domains 1 and 3 of the CrPV-like IGR IRESs (compare Figures 3A and 3B). Domain 1 is represented by a PK (PKII), comprising helical elements P1.1 (which contains the ORF1 stop codon), P1.2, and P1.3, and internal loops, including loops (e.g., L1.1) with sequences like those in corresponding loops in CrPV-like IGR IRESs (Figures 3A and 3B and S2). Domain 3 (named in accordance with the nomenclature for CrPV-like IGR IRESs [Costantino and Kieft, 2005]) comprises a second PK, PKI, at the 3′ border of the IGR and ends with the A-site GCC6406–6408(Ala) codon. There is no equivalent of domain 2 comprising PKIII of CrPV-like IGR IRESs or of the conserved stem-loops IV and V in them that bind to the 40S subunit.
Figure 3. Structure and Mutational Analysis of the HalV IGR IRES.
(A) Model of the HalV IGR IRES and the adjacent 3'-terminal region of ORF1, derived on the basis of computational analysis (see STAR Methods) and chemical/ enzymatic foot-printing (Figures S1A and S1B). It is annotated to show nucleotides at 20 nt intervals, IRES domains and secondary structural elements (based on the nomenclature proposed by Costantino and Kieft (2005)), the ORF1 stop codon (UAA6276) and the ORF2 A site codon (GCC6406) (both boxed blue). Arrows indicate the 5′ borders of truncated HalV IGR mRNAs used in experiments to assay IRES activity in ribosome binding (C and D).
(B) Model of the CrPV IGR IRES, showing IRES domains and secondary structure elements, nucleotides that interact with ribosomal proteins and elements of 18Sand 28S rRNAs, and conserved motifs in the L1.1 loop in dicistrovirus IGR IRESs (Nishiyama et al., 2003; Pfingsten et al., 2006).
(C and D) The 5′-terminal border of the HalV IGR IRES assayed by toe-printing of IRES/80S complex formation. The position of IRES/80S binary complexes is indicated. Lanes C, T, A, and G depict HalV sequence.
(E and F) Analysis of the influence of disruptive and compensatory substitutions in helical elements of domain 3/PKI (E) and domain 1/PKII (F) on the HalV IGR IRES’s ribosome-binding (E and F) and elongation (E) activity, analyzed by toe-printing. The positions of IRES/80S binary complexes and 80S ECs are indicated.
(G) Analysis of the influence of substitutions in single-stranded elements of the HalV IGR IRES on its ribosome-binding activity, analyzed by toe-printing. The position of IRES/80S binary complexes is indicated. (F and G) Separation of lanes by white lines indicates that they were juxtaposed from the same gel.
The HalV IGR IRES Epitomizes a Novel Class of IGR IRESs
Database searches identified HalV-like IGRs in several uncategorized viral genomes from arthropods, including Changjiang picorna-like virus 14 (CPLV14), Shahe arthropod virus 1 (SAV1) (Shi et al., 2016), Kuiper virus, and the identical Drellivirus. A partial HalV-like IGR sequence (Proasellus solanasi transcriptome shotgun assembly [TSA]-1) was identified in a TSA sequence from the crustacean P. solanasi. The nucleotide sequence of this IRES fragment and the amino acid sequence of the 82-aa-long ORF2 fragment are 86% and 57% identical to CPLV14, respectively.
In these genomes, the IGRs are flanked by ORF1 that encodes dicistrovirus-like nonstructural proteins and ORF2 that encodes capsid proteins in the order VP2-VP4-VP3-VP1 that is characteristic of dicistroviruses. Amino acid sequence identity in the 3C protease/3D polymerase segment of ORF1 of these viruses ranges from 32.7% to 71.2% and in ORF2 from 43.2% to 60.2% (Tables S1 and S2). Identity between ORF1 and ORF2 of these viruses and of representative members of the Cripavirus genus (CrPV), Triatovirus genus (Triatoma virus) and the two clades of insect- and crustacean-infecting viruses (Acute bee paralysis virus [ABPV] and TSV, respectively) in the Aparavirus genus of Dicistroviridae (www.ictv.global/report/dicistroviridae) did not exceed 27.4% for the 3CD region and 25.1% for the ORF2 region, respectively (Tables S1 and S2). Phylogenetic analysis of ORF2 amino acid sequences confirmed that HalV, CPLV14, SAV1, and Kuiper virus form a clade, here designated “Halárvirus,” that is distinct from the Aparavirus, Cripavirus, and Triatovirus genera (Figure S1D).
The HalV IGR IRES is 129 nt long from the 5′ border of P1.1 to the 3′ border of PKI, and these putative Halávirus IGR IRESs are similar (SAV1: 123 nt; CPLV14: 126 nt; Kuiper virus: 129 nt; Figure S2). By extension of the nomenclature for CrPV-like IGR IRESs (type VIa) and TSV-like IGR IRESs (type VIb), we suggest that HalV-like IGR IRESs be designated type VIc (Figure S2). Pairwise sequence identity between them is high (45%–66%), and they all have a HalV IGR-like structure comprising two domains with a large loop in domain 1 instead of a PKIII-like element. Conserved nucleotides are concentrated in the L1 loop, in P3.2 and in the adjacent L3.1 and L3.2 elements of PK1 (Figure S2).
Mutational Analysis of the HalV IRES
Progressive 5′-terminal deletions were made to determine the 5′ border of the HalV IGR IRES (indicated in Figure 3A). Deletion of the ORF1 hairpin and its replacement by a mirror-image version were tolerated, whereas deletion to nucleotide 6280 and nucleotide 6284 strongly reduced and abrogated ribosomal binding of the IRES, respectively (Figures 3C and 3D). The ORF1 hairpin is therefore dispensable, nucleotides 6268–6405 are sufficient, and the integrity of P1.1 is important for IRES function in ribosome binding. Thus, the HalV IGR IRES is 129 nt and is consequently one-third smaller than previously characterized IGR IRESs (~190 nt).
To verify the HalV IGR IRES model, we performed mutagenesis to confirm the ability of certain regions to form the predicted functionally important secondary structure elements. For this, destabilizing and compensatory mutations were introduced into PKI and PKII. Ribosomal binding and elongation activities of the GCC6406–6408(Ala)→UCU(Ser) IRES variant were abrogated by substitutions in P3.1 and P3.2 of PKI but were restored fully or partially, respectively, by compensatory second-site substitutions (Figure 3E). Similarly, in the case of PKII, IRES function was abrogated by destabilizing substitutions in P1.1 and P1.3 and strongly impaired by destabilizing substitutions in the proximal region of P1.2 but was fully restored by compensatory substitutions (Figure 3F).
Whereas helical elements function to support the structure of the IRES, single-stranded regions might engage in specific interactions with the ribosome to ensure efficient, stable binding or correct orientation of functionally important motifs. Mutational analysis revealed that whereas the long L2 loop (nucleotides 6341–6357) in PKII that replaces PKIII/domain 2 of CrPV-like IGR IRESs was quite tolerant of substitutions (Figure S3), other unpaired elements, such as the L1.1, L1.2a, and L1.2b loops in PKII, L3.1a and L3.2 loops in PKII, and the PKI/PKII linker, had critical functions that were strongly sequence dependent (Figure 3G). These elements contribute to or are essential for the function of previously described IGR IRESs, and many of them interact with components of the ribosome (Figure 3B). Thus, the L1.1 loop binds to uL1 and H76/H77 of the L1 stalk (e.g., Schüler et al., 2006 ; Fernández et al., 2014) and is important for ribosomal attachment to and translocation of the IRES (Jang et al., 2009; Pfingsten et al., 2006, 2010); the L3.1a loop interacts with h18 (Schüler et al., 2006 ) and substitutions in it reduce IRES-mediated translation (Costantino et al., 2008); and the L3.2 loop, which interacts with uS7 (Abeyrathne et al., 2016; Acosta-Reyes et al., 2019), plays a role in translocation (Ruehle et al., 2015).
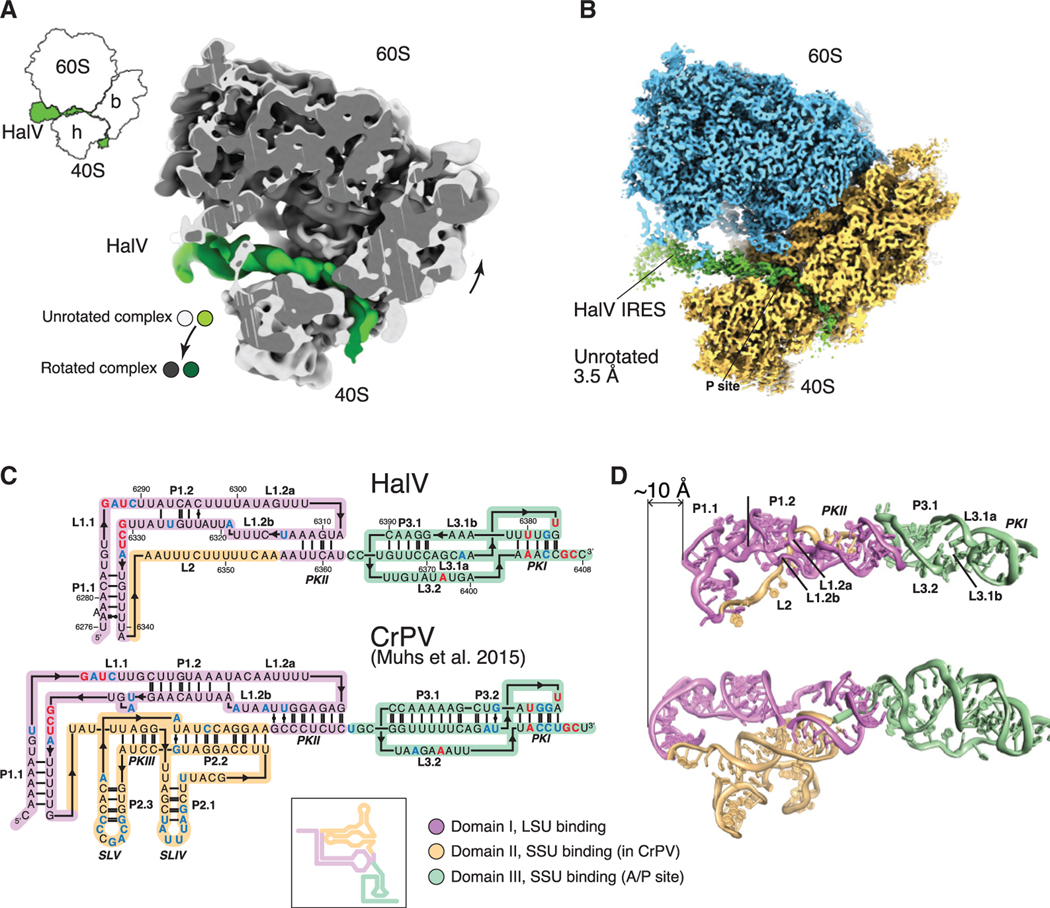
Structure of the HalV IGR IRES Bound to the Rabbit 80S Ribosome
Cryo-EM of the HalV IRES/rabbit 80S ribosome complex yielded two classes (Figure S4), one with rotated ribosomes and the other with ribosomes in the unrotated (classical) conformation (Figure 4A). Refinement of the rotated class yielded an average resolution of 3.6 Å . However, the 40S subunit is likely to be oscillating between several close rotated conformations, as its densities appear scanter than those of the 60S subunit. The unrotated class appears sturdier and presents similarly solid cryoEM densities for both ribosomal subunits, yielding a 3.49 Å resolution reconstruction (Figure 4B). Ribosomes in the unrotated state resembled those in CrPV IGR IRES/80S complexes after the first or second translocation steps, when the IRES is either in the P site or E site, or the hybrid P/E state (Figure S5) (Muhs et al., 2015; Pisareva et al., 2018). Hence, the head of the 40S subunit is not swiveled as in the complex with the CrPV IRES bound to the A site of the 80S ribosome (Fernández et al., 2014) and in ribosomes in the post-translocation (POST) state (Budkevich et al., 2014). The rotated state is similar to that observed in the presence of A-site-bound CrPV IRES (Fernández et al., 2014) and is closely related to the pre-translocation 1 (PRE-1) state (Figure S5) (Budkevich et al., 2014). In both classes, PKI mimics the anticodon stem-loop of tRNA base paired to a mRNA codon in the P site, leaving the A site vacant, consistent with the biochemical data described above.
Figure 4. Overview of the HalV IGR IRES Bound to the O. cuniculus 80S Ribosome.
(A) Superimposition of the 60S subunit from the unrotated and rotated complexes, to emphasize 40S and IRES movement.
(B) Cryo-EM map of the unrotated complex. The 40S is semi-transparent to reveal the path of the IRES.
(C) The 3D-based secondary structure diagrams of the IGR IRES from HalV (top) and CrPV (bottom). Color coding by domain according to functional role. Residueconservation: >80%, red; 60%–80%, blue (within a set of 19 IGR IRES sequences [see Figure S2]). Inset, classical representation of a secondary structure diagram for the CrPV IRES.
(D) Comparison of the overall structures of HalV and CrPV. Color coding as in (C).
Modeling of the HalV IGR IRES was based on the predicted secondary structure and tertiary structure elements described above (Figure 3A). The structure of the HalV IGR IRES was mainly built by homology modeling using various previously determined structures of CrPV and Plautia stali intestine virus (PSIV) IGR IRESs (see STAR Methods; Pfingsten et al., 2006; Schüler et al., 2006). Because of the lower resolution of the map at the IRES except at PKI (Figures S4 and S6; Table S3), low-pass-filtered maps were used to aid with modeling (see STAR Methods). The 3D structure of the HalV IGR IRES is similar in the unrotated and rotated states of the IRES/80S complex (Figures S6A–S6D) and is shorter than the CrPV IGR IRES by ~10 Å (~3 base pairs) (Figure 4D). In sum, the conserved L1.1 loop and PKI (Figures S2 and 4C), which bind to the L1 stalk and to the P site, respectively (Figure 4B), are closer to one another in the HalV than in the CrPV IGR IRES (Figure 4D).
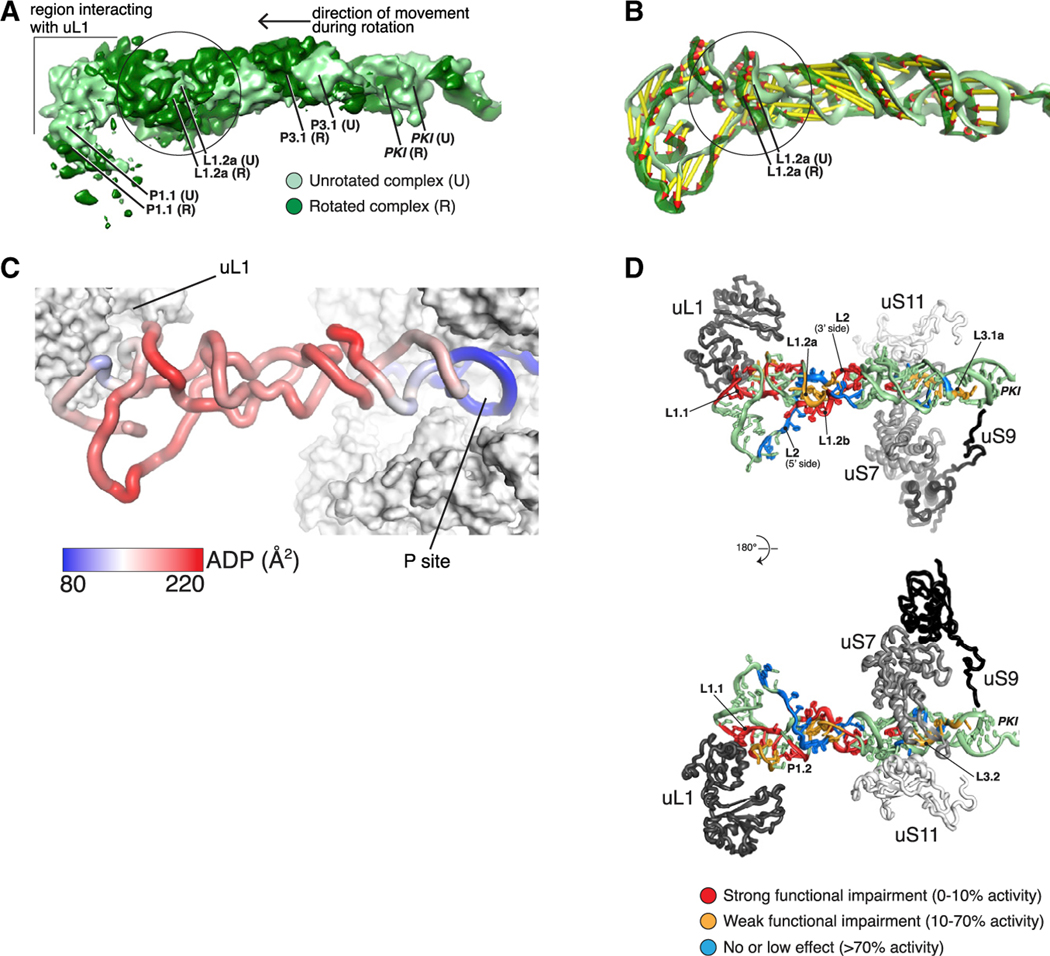
The HalV IRES also differs from the CrPV IRES in containing fewer Watson-Crick base pairs in conserved helical segments, except for the peripheral P1.1 and P3.2 (Figure 4C). Although domain 3 is 60 Å long in both IRESs, the longest continuous stack of base pairs is 11 in CrPV, but only 5 in HalV (Figures 4C and 4D). The atomic model of the unrotated 80S bound to HalV IRES was fitted (using molecular dynamics flexible fitting, see STAR Methods) into its rotated counterpart. Although the rotated HalV RES/80S map is relatively poorly resolved in the 40S and IRES regions, it was nevertheless possible to analyze the conformational changes of the IRES upon rotation of the 80S ribosome. The implication that the HalV IRES is more flexible was confirmed by the density maps, which revealed a bulge in the central region of the rotated state (Figure 5A). This area of the structure comprises three single-stranded regions: L1.2a, L1.2b, and L2. Movement of IRES domain 3 upon subunit rotation (vectors with larger amplitude, Figure 5B) results in a compression of the RNA in that area (vectors with smaller amplitude, Figure 5B). Analysis of the atomic displacement parameters (ADPs) indicated that the central region of the IRES, which does not interact with the 80S, is the most dynamic (Figure 5C). Notably, mutations in the HalV-specific L2 loop had a relatively low effect on the activity (Figure S3), whereas substitutions in this central region and in the region that binds uL1 were among the most deleterious (Figures 3G and 5D).
Figure 5. Flexibility of the HalV IRES Is Key to Its Function.
(A) Compression of the IRES in its central region during rotation. Superimposition of filtered density maps (Gaussian filter 1.5) of the unrotated and rotated states of the 80S-bound IRES. The circle indicates the region where additional bulging density is observed in the rotated complex.
(B) Superimposition as in (A) of the IRES 3D models (color coding as in A). Vectors help visualize the direction of the movement as well as its amplitude. Vectors were calculated by measuring the distance between phosphate atoms.
(C) The central region of the IRES is the most dynamic. Color coding of the unrotated IRES by atomic displacement parameter (ADP).
(D) Mutations that impair function cluster to regions interacting with ribosomal proteins and to the central region. Color coding of the IRES according to the effect of mutations on its function. Percentage of activity in comparison with wild-type RNA.
The HalV IRES Interacts with a Subset of the Ribosomal Proteins Bound by Canonical IGR IRESs
Interactions with ribosomal proteins occur at the extremities of the HalV IGR IRES, i.e., the conserved L1.1 elbow, and the PKI/loop L3.2 region (Figure 6). The downstream ORF2 also interacts with ribosomal protein uS9 at the mRNA entry site. Protein-RNA interactions mostly involve backbone atoms of the viral RNA and basic amino acids from ribosomal proteins. HalV L1.1 interacts with uL1 similarly to previously characterized IGR IRESs (e.g., Schüler et al., 2006 ; Fernández et al., 2014; Abeyrathne et al., 2016), i.e., with the a helix of uL1 formed by residues 121–127, as well as loop residues Arg48 (unrotated)/Gln44 (rotated state) and Lys161. The significance of the IRES residues that are predicted to be involved into interaction with uL1 was confirmed by extensive mutagenesis of the L1.1 loop (Figure 3G, left). However, as with prior structures of IGR IRESs, precise details concerning the interactions could not be established due to the lower resolution of the map in this area. PKI and L3.2 interact with the ribosomal proteins uS9 (Arg146 interacting with G6377), uS7 (Arg135 and Arg136 within interacting distance of nucleotides 6396–6400), and uS11 (Lys63 and Asp67 near nucleotides 6395–6397). The latter interaction disappears in the rotated state (compare corresponding panels in Figures 6A and 6C). The predicted interactions are consistent with the inactivating mutations in L3.2 loop and PKI (G6377) (Figures 3G, right, and 7A). Nucleotides 6411–6420 in ORF2 bend around uS3, interacting mostly with Arg residues. Many of these interactions are also seen with type VIa and type VIb IGR IRESs (e.g., Abeyrathne et al., 2016; Acosta-Reyes et al., 2019; Muhs et al., 2015), but several of their other characteristic interactions were not observed for the HalV IRES. Thus, although interaction of uL5 with the PKII region is a hallmark of CrPV IRES-80S ribosome complexes (Schüler et al., 2006), the HalV IRES does not come closer than ~18 Å to uL5. Because of the absence of SLIV and SLV-like elements (Figure 4C), the HalV IGR IRES does not interact with either eS28 or with eS25, which remain ~20 Å away in the unrotated state and ~13 Å away in the rotated state.
Figure 6. Contacts between the HalV IRES and Ribosomal Proteins.

Overview of a cross section of the ribosome showing the 80S-bound HalV IGR IRES in the unrotated state. The exposed section is indicated on the small 80S/HalV IRES schema on the right by a red rectangle.
(A) Blow-up figures on the HalV IRES interaction with the 80S ribosome in the unrotated state.
(B) Interactions between PKI and 18S rRNA in the unrotated state.
(C) View as in (A) for the rotated state. Note the missing interaction between the HalV IRES and uS11 in this state.
(D) View as in (B) for the rotated state. The exposed area is indicated on the small 80S/HalV IRES schema below (B) and (D) by a black circle. IRES nucleotides,which interact with ribosomal proteins and 18S rRNA and which were shown by mutational analysis to be important for IRES function (Figures 3G and 7A), are marked with asterisks. Note that the PK1 region in the rotated state is more flexible than in the unrotated state, as demonstrated by the scanter local densities and resolution compared with the unrotated state. Color coding for all panels: 40S, dark/light yellow; 60S, dark/light blue; HalV, dark/light green; uL1, magenta; uS11, yellow; uS7, red; uL5, teal; uS3, light blue; uS9, purple.
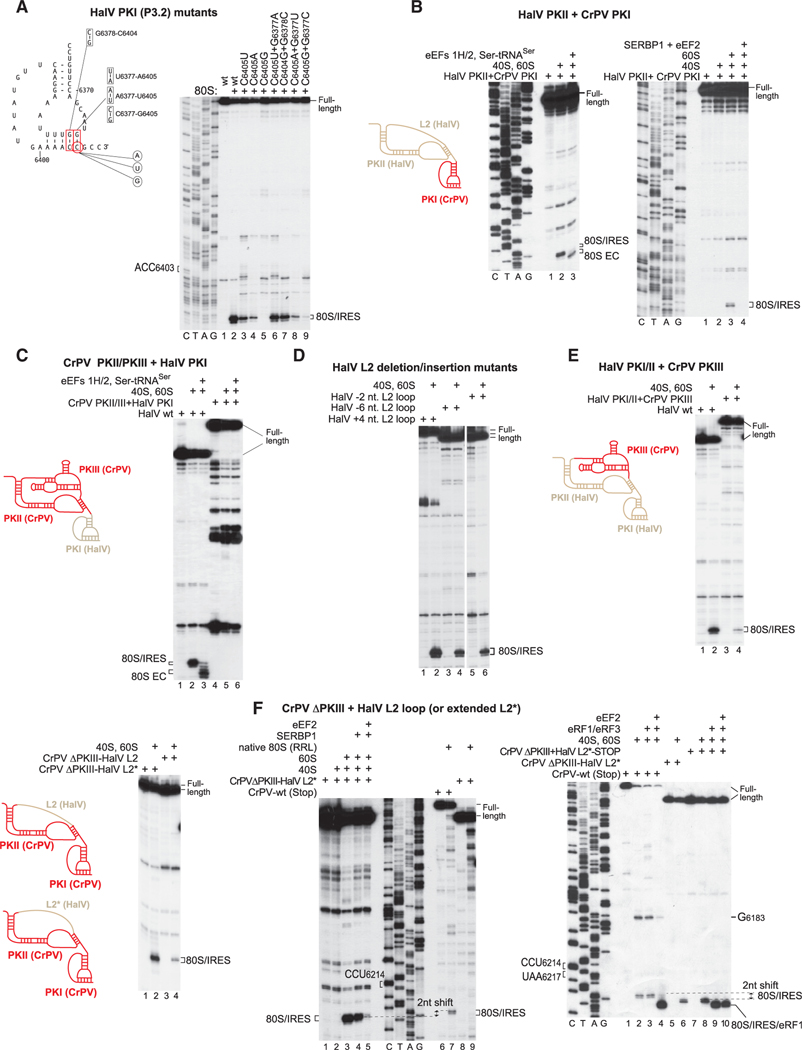
Figure 7. The P-Site-Stabilizing Interaction of PKI and Activities of Hybrid HalV/CrPV IGR IRESs.
(A) Left: model of the HalV IGR PKI structure, showing nucleotides targeted for substitution to assay a stabilizing interaction of PKI in the P site. Right: binding of 80S ribosomes to the indicated HalV IGR IRES mutants, assayed by toe-printing.
(B, C, E, and F) Ribosomal binding, one-cycle elongation and release factor binding on WT and hybrid HalV/CrPV IGR IRESs (shown schematically in each panel) in the presence of the indicated translational components, assayed by toe-printing. The positions of ribosomal complexes are shown.
(D) Ribosome-binding activity of the L2 deletion/insertion HalV IGR IRES mutants, assayed by toe-printing. Separation of lanes by white lines indicates that they were juxtaposed from the same gel. (A–F) The positions of ribosomal complexes are indicated. (A, B, and F) Lanes C, T, A, and G depict CrPV or HalV sequences.
PKI of the HalV IGR IRES Mimics the tRNA-mRNA Interaction in the P Site
In the unrotated HalV IGR IRES/80S structure, PKI’s conformation and its interactions with the ribosomal RNA (rRNA) and ribosomal proteins in the P site are like those of previously characterized IGR IRESs in post-translocated complexes (Figures S6E and S7; Costantino et al., 2008; Muhs et al., 2015; Fernández et al., 2014; Zhu et al., 2011; Koh et al., 2014). Thus, G6377 in the PKI base pair adjacent to ORF2 stacks against C1701 and interacts with U1248 of 18S rRNA (Figure S6E). The importance of these interactions is discussed below. It is interesting to note that in the rotated state, the PKI of the HalV IRES presents scanter densities than the more stable unrotated conformation (Figures 6B and 6D).
In the HalV IGR IRES/80S complex in its rotated state, A1058 of 18S rRNA interacts in the minor groove of the HalV U6380-A6402 pair, and U1830 contacts C6404 (Figure 6D). In this state, the U6381-A6401 base pairing has been disrupted, so that PKI contains four instead of five base pairs (Figure S7, solid box). The structural changes in PKI are accompanied by conformational changes in L3.2, although the density for the loop residues is too weak to pinpoint actual nucleotide conformations, especially in the rotated state. As suggested for CrPV and TSV IRESs (Ruehle et al., 2015; Abeyrathne et al., 2016), these changes could influence the conformation of domain 3, affecting translocation or stabilizing PKI in the P site. Like other canonical IGR IRESs, the HalV IGR IRES should be able to interact with different ribosomal binding sites during the initiation to elongation transition. Our structures thus illustrate the dynamic nature of PKI in the HalV IGR IRES, which can adjust the number of base pairs it contains that eventually could lead to complete disruption of helix P3.2 on exiting the ribosome as in the CrPV IGR IRES (Pisareva et al., 2018; Figure S7, dashed box).
Stabilizing Interactions of PKI in the P Site
The interactions of G6377 in PKI with U1248 and C1701 of 18S rRNA (Figures 6B and 6D and S6E) parallel stabilizing interactions established by CrPV and PSIV PKI (Zhu et al., 2011; Muhs et al., 2015) and stacking of 16S rRNA C1400 on the wobble base of the tRNA anticodon and packing of G966 against its ribose moiety (Selmer et al., 2006; Korostelev et al., 2006). The observation that compensatory substitutions in P3.2 of PKI failed to fully restore IRES function (Figure 3E) suggests that the interaction of PKI with the P site may impose specific sequence requirements on one or more of the base pairs in PKI that mimic the initiating codon-anticodon base pair. This hypothesis was tested by mutational analysis (Figure 7A). Disruption of the G6377-C6405 base pair by mutating C6405 reduced or even abrogated IRES function (Figure 7A, lanes 3–5). Activity was fully restored by A6377-U6405 substitutions, but not by U6377-A6405 or C6377-G6405 substitutions (Figure 7A, lanes 6, 8, and 9). By contrast, an IGR mutant with a G6378-C6403/C6378-G6403 switch in the adjacent base pair retained nearly full activity (Figure 7A, lane 7). The requirement for a purine at the position of G6377 in PKI is consistent with the conservation of base-paired purines at this position in HalV-like and canonical IGR IRESs (Figure S2; Nakashima and Uchiumi, 2009).
The Activities of Chimeric HalV/CrPV IGR IRESs
IGR IRESs have a modular organization (Jang and Jan, 2010; Pfingsten et al., 2007) and it is thus feasible that their evolution involved recombinational exchange of domains. We therefore investigated the activities of various HalV/CrPV IRES hybrids. Replacement of PKI in the GCC6406(Ala)→UCU(Ser) HalV IRES by CrPV PKI yielded an IRES that could bind to 80S ribosomes, accept cognate aa-tRNA into the A site, and undergo translocation (Figure 7B, left). Its binding to 80S ribosomes was abrogated by eEF2/SERBP1 (Figure 7B, right), which is characteristic of HalV, but not CrPV, IRESs (Figures 2C and 2E). Interestingly, the reverse exchange of PKI in the CrPV IRES by HalV PKI did not yield an active hybrid (Figure 7C). Thus, the specific mode of ribosomal binding of CrPV IRES that does not involve initial direct placement of PKI in the P site might not be compatible with the shorter P3.1 stem of HalV PKI.
Next, we investigated the effect of replacement of HalV L2 loop by CrPV PKIII and vice versa. In addition to substitutions (Figure S3), the HalV L2 loop tolerated small insertions and deletions (Figure 7D). However, replacement of the L2 loop by CrPV PKIII yielded a chimeric IRES with a more strongly reduced ribosomal binding activity (Figure 7E). The reverse exchange of CrPV PKIII by nucleotides 6342–6354 of the HalV L2 loop yielded the CrPVΔPKIII/HalV L2 hybrid that had low ribosome binding activity, but extending the loop to 18 nt (the CrPVΔPKIII/HalV L2* variant) substantially increased the activity of the hybrid IRES (Figure 7F, left). This chimeric CrPVΔPKIII/HalV L2* IRES became sensitive to inhibition by SERBP1 and eEF2 like the HalV IRES and also lost the ability to bind to 40S subunits and native 80S ribosomes (Figure 7F, middle). Strikingly, it bound directly to the P site, leaving the A site accessible without prior eEF2-mediated pseudo-translocation. Thus, 80S ribosomal complexes with the CrPVΔPKIII/HalV L2* IRES yielded toe-prints that were shifted forward by 2 nt compared with the WT CrPV IRES (Figure 7F, middle, compare lanes 3 and 7, and right, compare lanes 2 and 6). Moreover, 80S complexes formed on a variant of this IRES with the A site GCU6217–6219→UAA stop codon substitution were able to bind eRF1·eRF3 independently of eEF2, yielding a 2 nt toe-print shift like the HalV IRES instead of the WT CrPV IRES (Figure 7F, right panel, compare lanes 3 and 4 and 9 and 10). A change in the mode of ribosomal binding of the CrPVΔPKIII/HalV L2* IRES was also evidenced by the disappearance of the G6183 toe-print (Figure 7, right, compare lanes 1–4 with lanes 5–10), which is a hallmark of ribosomal binding of the WT CrPV IGR IRES to 80S ribosomes (e.g., Wilson et al., 2000; Pestova and Hellen, 2003; Muhs et al., 2015). Thus, the difference in the ribosomal binding of CrPV and HalV IRESs is determined by the presence of the PKIII domain in the former.
DISCUSSION
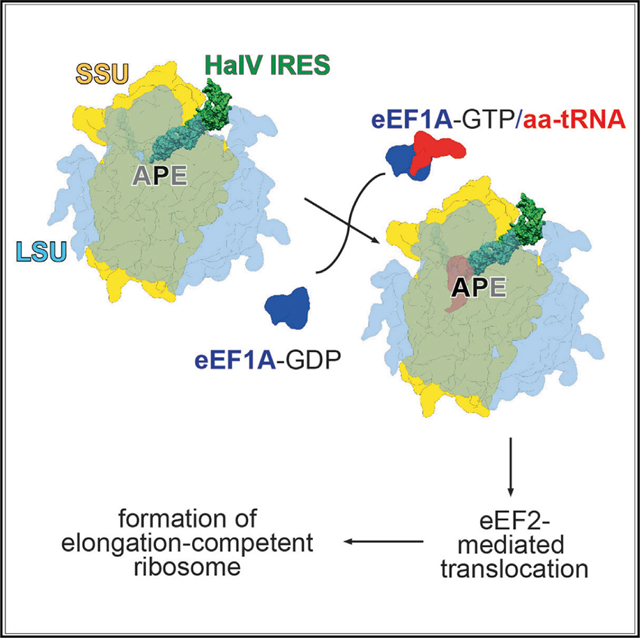
We report that the Halastavi árva virus IGR contains an IRES that is related to CrPV-like dicistrovirus IGR IRESs but differs from them structurally and functionally in key respects. Thus, the HalV IGR IRES comprises equivalents of domains 1 and 3 of CrPV-like IRESs, but it lacks the equivalent of domain 2, which enables CrPV-like IRESs to bind stably to the 40S subunit (e.g., Costantino and Kieft, 2005). As a result, the HalV IGR IRES binds directly only to 80S ribosomes. More importantly, PKI of this IRES, which mimics the codon-anticodon interaction, is placed in the ribosomal P site, so that the A site containing the first codon is directly accessible for decoding by eEF1A/aa-tRNA without the prior eEF2-mediated movement of the PKI from the A-to-P site that is required for initiation on CrPV-like IGR IRESs (Fernández et al., 2014). Thus, the HalV IRES functions by the simplest initiation mechanism known to date: binding of the IRES to 80S ribosomes places the [codon-anti-codon]-mimicking PKI directly into the ribosomal P site, which is followed by eEF1A-mediated decoding of the A site codon.
The elements of the HalV IGR IRES that interact with the 80S ribosome are restricted to the IRES’s extremities. Thus, the L1.1 loop contains conserved motifs like those in CrPV-like IGR IRESs and, analogously, interacts with ribosomal protein uL1 in the L1 stalk of the 60S subunit, whereas at the opposite end, elements in PKI interact with protein and rRNA constituents of the ribosomal P site. Binding of PKI in the P site is stabilized by G6377, which interacts directly with universally conserved elements of the P site, and by the L3.2 loop, as has been suggested for the post-translocation canonical IGR IRES (Abeyrathne et al., 2016). Consistent with direct binding to the P site, the HalV IGR IRES is 10 Å shorter than CrPV-like IGR IRESs, reflecting that the P site is closer than the A site to the L1 stalk. The HalV IGR IRES is also intrinsically less ordered, with fewer base pairs in most conserved helical segments, and more flexible than canonical IGR IRESs. The higher flexibility was already apparent from the density maps, which revealed a bulge in the central region of the rotated state of the IRES, an area that comprises three single-stranded regions: L1.2a, L1.2b, and L2. The flexibility appears to be important for IRES function, because substitutions in L1.2a and L1.2b strongly reduced IRES function, potentially by introducing stabilizing base pairs that could increase rigidity. However, the biggest difference between the HalV IRES and previously characterized IGR IRESs is the lack in the former of the equivalent of domain 2, which contains the conserved stemloops IV and V that interact with uS7 and eS25 in the 40S subunit (e.g., Schüler et al., 2006 ; Abeyrathne et al., 2016). Although it is well established that domain 2 enables stable binding of type VIa and type VIb IGR IRESs to 40S subunits (Jan and Sarnow, 2002; Nishiyama et al., 2003; Costantino and Kieft, 2005), we found that it is also responsible for positioning the PKI outside the P site. Thus, deletion of domain 2 from the CrPV IGR IRES resulted in direct binding of PKI to the P site, which eliminated the necessity for the initial step of eEF2-dependent translocation of PKI of the IRES. In the absence of domain 2, even the greater length of the CrPV IGR IRES did not induce slippage of PKI into the A site. Notably, lengthening the linker that replaced domain 2 in the CrPV IGR IRESΔPKIII mutant increased its efficiency of action, likely reflecting the importance of the IRES flexibility for the new mode of ribosomal binding.
Importantly, we observed that ribosomal binding of the HalV IGR IRES is inhibited by SERBP1/eEF2, which associate with non-programmed eukaryotic 80S ribosomes (e.g., Anger et al., 2013; Zinoviev et al., 2015; Brown et al., 2018). As a result, the translational efficiency of the HalV IRES in RRL was very low. By contrast, the WT CrPV IGR IRES was resistant to SERBP1/eEF2, but deletion of domain 2 rendered the CrPV IGR IRES susceptible to such inhibition. Thus, the presence of domain 2 on an IGR IRES not only determines the ribosomal position of PKI but also confers resistance to SERBP1/eEF2 inhibition. This raises the question of how the HalV IGR IRES overcomes such inhibition in vivo. HalV was derived from the intestinal content of carp, and one possibility is that its natural host, which has not yet been established, may lack SERBP1 or encodes a homolog that does not impair translation in this way. Another possibility is that the viral infection could influence the cytoplasmic conditions, leading to re-lief of this inhibition. In this respect it is worth mentioning that although initial studies had suggested that domain 2 contains elements that are critical for the function of canonical IGR IRESs (e.g., Jan and Sarnow, 2002), dicistrovirus infection is now known to alter the intracellular environment in a manner that suppresses the defect in IRES function caused by mutations in this domain (Kerr et al., 2016). The nature of this change has not been established, but it is possible that stress-induced changes in the localization of SERBP1 (e.g., Lee et al., 2014), its dissociation from ribosomes, or even its degradation during infection could lead to activation of IRES function. In this context, it would be of interest to determine the potential influence of other 80S-binding proteins such as interferon-related developmental regulator 2 (IFRD2) and coiled-coil domain containing 124 (CCDC124) (Brown et al., 2018; Wells et al., 2020) on the activity of HalV and canonical IGR IRESs.
Our metagenomic analyses determined that HalV is only one of a family of related IGR elements that lack an equivalent of domain 2. Although we cannot exclude the possibility that they derive from canonical IGR IRES progenitors as a result of loss of a functional element (i.e., domain 3), a more likely scenario is that HalV-like IRESs form an ancestral class (VIc). According to that model, the presence of domain 3 in type VIa and type VIb IRESs would represent an evolutionary acquisition, favored by a resulting gain of function. Perhaps this gain was the switch to initiation from P sites to A sites, which has been suggested to facilitate post-binding events by partitioning the energetic penalty for the required dissociation of the IRES after initial recruitment to the ribosome (Muhs et al., 2015). It could also have been resistance to inhibition by SERBP1/eEF2 or other ribosome-binding proteins. In this model, IRES evolution involves accretion of domains, as suggested for the evolution of rRNA (Petrov et al., 2014) and hepatitis C virus-like IRESs (Asnani et al., 2015) and their subsequent dissemination to other viral genomes by recombination. The number of novel, often divergent, dicistroviruses is increasing rapidly (e.g., Shi et al., 2016), and it will be interesting to determine whether there are IGR IRESs that lack domain 2 but nevertheless still bind stably to 40S subunits or that contain some equivalent of domain 2 but nevertheless bind in the P site. An exciting possibility is that IGR-like elements with divergent domain 1 or domain 2 elements may have evolved the ability to exploit other potential sites of interaction, leading to utilization of divergent mechanisms of initiation.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tatyana Pestova (tatyana.pestova@downstate.edu).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Data and Code Availability
The atomic coordinates of the rotated and unrotated IRES/80S complexes have been deposited in the Protein Data Bank (PDB). The accession numbers for the atomic models of the [80S-HalV IGR IRES] in the rotated and unrotated conformation reported in this paper are: 7A01 and 6ZVK, respectively.
The cryo-EM maps of the rotated and unrotated IRES/80S complexes have been deposited in the Electron Microscopy Data Bank (EMDB) with the accession codes: EMD-11590 and EMD-11459, respectively.
Gel images are available at: https://data.mendeley.com/datasets/wx2ypvtxh8/draft?a=8017d354-6b04-4735-a5f8-464205fbbad9
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Plasmids were propagated in Escherichia coli DH5a cells (F– φ80lacZΔ M15 Δ (lacZYA-argF) U169 recA1 endA1 hsdR17 (rK– mK+) phoA supE44 λ- thi–1 gyrA96 relA1) and recombinant proteins were expressed in E. coli BL21(DE3) cells (F– ompT hsdSB (rB−, mB−) gal dcm (DE3)), respectively, both grown in LB medium at 37°C. Overnight cultures of transformed E. coli BL21(DE3) cells were inoculated into flasks containing 1000 – 2000 mL LB medium with antibiotic, incubated with shaking at 37°C until OD260 = 0.5, followed by addition of 1mM IPTG and further incubation for 3 hours at 37°C.
METHOD DETAILS
Construction of Plasmids
Vectors for expression of His6-tagged eIF1 and eIF1A (Pestova et al., 1998a), eIF4A and eIF4B (Pestova et al., 1996), eIF4G736–1115 (Lomakin et al., 2000), Escherichia coli methionyl tRNA synthetase (Lomakin et al., 2006), Serbp1 (Heaton et al., 2001) and eRF1 (Seit-Nebi et al., 2001) have been described. A vector for expression of His6-tagged eRF3a (aa 139–499) lacking the N-terminal 138 aa (referred to as eRF3 in the text) was made by inserting DNA between BglII and Nde1 sites of pET23(b) (GenScript, Piscataway, NJ).
The transcription vectors for tRNASer (UCU codon) (Zinoviev et al., 2015) has been described. New transcription vectors were made by inserting DNA into pUC57 (GeneWiz, South Plainfield, NJ) or by subsequent site-directed mutagenesis (NorClone Biotech, London, ON, Canada; 96 Proteins, San Francisco, CA). The vector monocistronic Stem-HalV nt.6211–7460 contained DNA corresponding to a T7 promoter, a stable hairpin (GGCCGACCCGGTGACGGGTCGGCC) (ΔG = −25.80 kcal/mol), HalV nt. 6211–7460 and a linker comprising AvrII, SmaI, ClaI and XbaI sites. The vectors for truncated HalV IGR IRES mutants contained a T7 promoter followed by the corresponding HalV sequences (nt.6205–6513, 6268–6513, 6280–6513, 6284–6513, 6297–6513 and 6327–6513) and restriction sites for EcoRV and EcoRI. The vectors for HalV IGR IRES substitution/deletion/insertion mutants were made using a base construct containing nt. 6268–6483.
The transcription vector Stem-HalV IGR-GUS (employed for synthesis of mRNA used for translation in rabbit reticulocyte lysate (RRL)) included a T7 promoter, the hairpin described above, HalV nt.6268–6513 (with a GCC6406–8UCU substitution, as well as a AUG6397–9UAG substitution that eliminates a possibility of detecting a product originated from non-specific AUG-dependent initiation within the IRES), a modified fragment of the E. coli beta-glucuronidase (GUS) ORF (GenBank: AAA68923.1), HalV nt. 7404–7460 and an [AvrII-Sma1-Cla1-Xba1] linker. The transcribed mRNA encodes a 286 aa-long (31.7 kDa) fusion protein comprising a Serine residue and aa 2–36 of the HalV ORF2 linked to a modified form of aa 351–602 of the GUS ORF in which AUG codons 407 and 446 had been substituted and codons 528, 548, 560, 569, 583 and 596 had been replaced by AUG triplets. The MC Stem-HalV transcription vector (employed for synthesis of mRNA used for translation in RRL) containing the HalV 5′UTR and an adjacent 256 aa-long DORF1 (nt. 1–1682) has been described (Abaeva et al., 2016).
Vectors for transcription of CrPV IGR IRES mRNAs contained CrPV nt. 5997–6320 or a variant (CrPV5997–6320-Stop) with GCT6217–9TAA substitutions inserted between BamH1 and EcoR1 sites of pUC57 (GeneWiz). The vectors HalV PKII+CrPV PKI/Ser-Stop and CrPV PKII/PKIII+HalV PKI(SerStop) for transcription of hybrid HalV-CrPV IGR IRESs contained HalV nt.6268–6365 + CrPV nt.6174–6320 (GCTACA6217–6222→TCTTAA) and CrPV nt. 5997–6173 + HalV nt. 6366–6483(GCCACT6406–6411→TCTTAA) down-stream of T7 promoters, respectively. The vectors for transcription of HalV L2 −2 nt. L2 loop, HalV −6 nt L2 loop and HalV +4 nt. L2 loop mutant mRNAs were made by replacing the L2 loop sequence AAUUUCUUUUUCAA by the sequences AAUUCUUUUCAA, AAUCUCAA and AAUUUUUCUUUUUUUCAA, respectively. The vector for transcription of HalV PKI/II + CrPV PKIII mRNA was made by replacing HalV nt. 6342–6353 inclusive by CrPV nt. 6110–6162. The transcription vectors CrPV ΔPKIII + HalV L2 loop and CrPV ΔPKIII + HalV extended L2* loop were made by replacing CrPV nt. 6110–6161 by HalV nt. 6343–6353 and by the HalV-related sequence AAUUUUUCUUUUUUUUC, respectively.
All RNAs were transcribed using T7 RNA polymerase (Thermo Scientific).
Purification of factors, ribosomal subunits and aminoacylation of tRNA
Native mammalian 40S and 60S subunits, eIF2, eIF3, eEF1H, eEF2 and total aminoacyl-tRNA synthetases were purified from rabbit reticulocyte lysate (RRL), and insect 40S and 60S subunits were purified from S. frugiperda cell-free extract (Promega) as described (Pisarev et al., 2007). Native 80S ribosomes were purified from 200 μL RRL (Promega), which was layered onto 10%–30% sucrose density gradients in buffer A (20 mM Tris [pH 7.5], 100 mM KAc, 2.5 mM MgCl2, 2 mM DTT and 0.25 mM spermidine) and centrifuged in a Beckman SW55 rotor at 53,000 rpm for 90 min at 4C. Fractions were collected across the gradient, and the position of 80S ribosomes was determined by monitoring the absorbance at 260 nm. Recombinant eIF1, eIF1A, eIF4A, eIF4B, eIF4G736–1115, eRF1, eRF3, Escherichia coli methionyl tRNA synthetase and SERBP1 were expressed and purified from E. coli as described (Pisarev et al., 2007; Alkalaeva et al., 2006; Zinoviev et al., 2015). RelE was a gift from Venki Ramakrishnan. Native total calf liver tRNA (Promega) and In vitro transcribed tRNASer were aminoacylated using Escherichia coli methionyl tRNA synthetase (for obtaining Met-tRNAiMet) or total aminoacyl-tRNA synthetases (for aminoacylation of elongator tRNAs) as described (Pisarev et al., 2007; Zinoviev et al., 2015).
Assembly and analysis of ribosomal complexes by toe-printing
For assembly of 48S initiation complexes, 2 pmol Stem-(wt HalV IGR) mRNA, 3 pmol 40S subunits, 6 pmol eIF2, 4.5 pmol eIF3, 8 pmol eIF1, 8 pmol eIF1A, 10 pmol eIF4A, 5 pmol eIF4B, 5 pmol eIF4G736–1115 and total tRNA containing 7 pmol Met-tRNAiMet were incubated in 40 μL buffer A supplemented with 1 mM ATP and 0.1 mM GTP for 10 min at 37°C. For studying ribosomal asociation of HalV, CrPV and hybrid HalV/CrPV IGR IRESs, 2 pmol mRNA was incubated with 5 pmol 40S subunits, 8 pmol 60S subunits or both in the presence or absence of indicated combinations of 10 pmol eRF1, 10 pmol eRF3, 10 pmol eEF2 and 10 pmol SERBP1 in 40 μL buffer A supplemented with 1 mM ATP and 0.1 mM GTP for 10 min at 37°C. To examine the elongation competence of assembled IRES/80S complexes, reaction mixtures were supplemented with combinations of 10 pmol eEF2, 10 pmol eEF1H, 500 μg/mL of cycloheximide, 15 μg appropriately aminoacylated native total tRNA or 6 pmol Ser-tRNASer, and incubation was continued for 10 min at 37°C. Resulting ribosomal complexes were analyzed by toe-printing as described (Pisarev et al., 2007) using avian myeloblastosis virus reverse transcriptase (AMV RT) (Promega) and [32P]-labeled oligonucleotide primers complementary to HalV nt. 6458–72 or CrPV nt. 6304–19, as appropriate. cDNA products were resolved in 6% polyacrylamide sequencing gels followed by autoradiography or phosphoimager analysis.
Assembly and analysis of ribosomal complexes by sucrose density gradient centrifugation
Ribosomal complexes were formed by incubating 10 pmol [32P]-labeled HalV IGR IRES mRNA with 30 pmol 40S subunits, 40 pmol 60S subunits or both, as indicated, in buffer A supplemented wth 1 mM ATP and 0.1 mM GTP for 10 min at 37°C, and then resolved by centrifugation through 10%–30% sucrose density gradients in buffer A in a Beckman SW55 rotor at 53,000 rpm for 90 min at 4°C. The optical density of fractionated gradients was measured at 260 nm, and the presence of [32P]-labeled mRNA was monitored by Cherenkov counting.
Assembly and analysis of ribosomal complexes by RelE cleavage
Analysis of cleavage of ribosome-bound mRNA by RelE was done as described (Skabkin et al., 2013). 80S ribosomal complexes were assembled in the presence or absence of eEF2 as described above for preparation of complexes for toe-printing analysis, and then incubated with 20 pmol RelE for 10 min at 37°C. After that, mRNA was phenol-extracted and analyzed by primer extension using AMV RT and the same [32P]-labeled primers, as described above for toe-printing analysis. cDNA products were resolved in 6% polyacrylamide sequencing gels followed by autoradiography or phosphoimager analysis.
Viral nucleotide sequences
Sequences were retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/nuccore) using the following accession numbers: Acute bee paralysis virus (ABPV), NC_002548.1; Aphid lethal paralysis virus 1 (ALPV), NC_004365.1; Beihai mantis shrimp virus 5 (NC_032434.1); Black queen-cell virus (BQCV) (NC_003784.1); Changjiang picorna-like virus 14 (NC_032773.1); Cricket paralysis virus (CrPV), NC_003924.1; Drellivirus strain 93C3 (KX924637); Drosophila C virus (DCV) strain EB (NC_001834.1); Empeyrat virus (KU754505.1); Formica exsecta virus 1 (NC_023021.1); Goose dicistrovirus (NC_029052.1); Halastavi árva RNA virus (NC_016418.1); Himetobi P virus (HiPV) (NC_003782.1); Homalodisca coagulata virus 1 (NC_008029.1); Israel acute paralysis virus (IAPV), NC_009025.1; Kashmir bee virus (KBV) (NC_004807.1); Kuiper virus (KX657785.1); Macrobrachium rosenbergii Taihu virus strain cn-taihu100401(NC_018570.2; Mud crab virus (NC_014793.1); Nilaparvata lugens C virus (NlCV), KM270560.1; Plautia stali intestine virus (PSIV) (NC_003779.1); Rhopalosiphum padi virus (RhPV) (NC_001874.1); Shahe arthropod virus 1 strains SHWC01c3692 (KX883988), SHWCII5326 (KX883671), SHWC0209c12762 (NC_032422) and SHWC0209c12762 (KX883641); Solenopsis invicta virus 1 (SinV), NC_006559.1; Taura syndrome virus (TSV), NC_003005.1; Triatoma virus (TriV) (NC_003783.1); Wenzhou shrimp virus 7 (NC_032420.1) and the TSA Sequence Proasellus solanasi HAFJ01006557.1.
Phylogenetic analysis (Figure S1D)
Dicistrovirus ORF2 capsid protein precursor amino acid sequences were obtained by conceptual translation of viral sequences downstream of the IGR IRES, submitted to the web server http://phylogeny.lirmm.fr/phylo_cgi/index.cgi (Dereeper et al., 2008) for alignment using CLUSTAL-W (default parameters) and elimination of positions containing gaps, and then used for inference of the maximum likelihood (ML) phylogenetic tree using IQ-TREE (Nguyen et al., 2015), applying the WAG+FO model for amino acid substitution and using 10,000 ultrafast bootstraps (Hoang et al., 2018). Statistical support for individual nodes was estimated using the bootstrap value.
Identification of candidate IGR IRESs
Candidate IGR sequences were identified using BLASTN (https://www.ncbi.nlm.nih.gov/BLAST/) searches of nucleotide collection (nr/nt) and TSA sequences, and BLASTX searches of non-redundant protein (nr) and Transcriptome Shotgun Assembly (TSA) protein (tsa_nr) sequences in the NCBI database. Nucleotide searches used the parameters: E, 1000; word size, 11; match/mismatch scores, 1/1; gap costs, 2/1, and polypeptide searches used the parameters: E, 1000; word size, 6; Matrix: BLOSUM62; gap costs, 9/1. TSA searches were limited to Arthropoda and Mollusca, and ‘hits’ were characterized by 6-frame translation. Polypeptide sequences were used in BLAST searches to verify that the C-terminal region of ORF1 encoded the 3D polymerase and that ORF2 encoded capsid proteins. IGR sequences were aligned using EMBOSS Matcher (https://www.ebi.ac.uk/Tools/psa/emboss_matcher/nucleotide.html) and Clustal Omega. Viral 3CD and P1 capsid protein sequences were aligned using Clustal Omega with default parameters to determine pairwise sequence identities.
In vitro translation (Figure 2A)
MC Stem-HalV and GUS-(HalV IGR) mRNAs (3 pmol) were translated in 20 μL reaction volume of Flexi rabbit reticulocyte lysate (Promega) in the presence of [35S]methionine (> 37.0 TBq/mmol; Perkin Elmer) for 30 minutes at 30°C. When indicated, reaction mix-tures were supplemented with 6 pmol 40S and 8 pmol 60S subunits with or without prior preincubation with mRNAs for 5 minutes at 37°C in buffer A supplemented with 1 mM ATP and 0.1 mM GTP. Translation products were resolved by electrophoresis using Nu-PAGE 4%–12% Bis-Tris precast gels (Invitrogen), followed by autoradiography.
Chemical and enzymatic probing (Figures S1A–S1C)
HalV IGR-containing mRNA was enzymatically digested with RNase V1 or chemically modified with 1-cyclohexyl-(2-morpholinoethyl) carbodiimide metho-p-toluene sulphonate (CMCT), dimethylsulfate (DMS) or N-methylisatoic anhydride (NMIA) exactly as described (Abaeva et al., 2016). Cleaved or modified sites were identified by primer extension, using AMV RT and primers complementary to HalV nt. 6353–70 or nt. 6458–72. cDNA products were resolved in 6% polyacrylamide sequencing gels followed by autoradiography.
Nucleotide sequence alignment (Figure S2)
IGR sequences were aligned automatically using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) (https://www.ebi.ac.uk/Tools/msa/clustalo/) and then manually using Ugene v. 1.22 (Okonechnikov et al., 2012), relying on published sequence alignments as a guide (Jan, 2006; Kanamori and Nakashima, 2001; Pfingsten et al., 2006).
Initial HalV IGR structure modeling
Structural elements were initially modeled using CentroidFold (https://www.ncrna.org/centroidfold) (Sato et al., 2009) and Mfold (http://www.unafold.org/) (Zuker, 2003). Tertiary structures were modeled using pKiss (https://bibiserv.cebitec.uni-bielefeld.de/pkiss) (Janssen and Giegerich, 2015), in all instances using default parameters.
Grid preparation
15 pmol HalV mRNA (nt. 6268–6483) was incubated with 7.5 pmol 40S subunits and 7.5 pmol 60S subunits in 40 μL buffer B (20 mM Tris [pH 7.5], 100 mM KAc, 5 mM MgCl2, 1 mM DTT, 0.25 mM spermidine, 0.5 mM ATP and 0.5 mM GTP) for 5 minutes at 37°C to allow IRES/ 80S complex formation. After incubation, the reaction mixture was diluted using buffer B to achieve the concentration of 80S ribosomes of 70 nM, and 4 μL of the sample was applied onto the Quantifoil R2/2 400-mesh holey carbon grid, which had been coated with thin carbon film and glow-discharged. The sample was incubated on the grid for 30 s and then blotted with filter paper for 1.5 s in a temperature and humidity controlled Vitrobot Mark IV (T = 4°C, humidity 100%, blot force 5) followed by vitrification in liquid ethane.
Image acquisition
Data collection was performed on a spherical aberration corrected Titan Krios S-FEG instrument (FEI Company) at 300 kV using the EPU software (FEI Company) for automated data acquisition. Data were collected at a nominal underfocus of −0.6 to −4.5 μm at a magnification of 127,272 X yielding a pixel size of 1.1 Å . Micrographs were recorded as a movie stack on a Falcon II direct electron detector (FEI Company), each movie stack were fractionated into 17 frames for a total exposure of 1 s corresponding to an electron dose of 60 ē/Å2.
Image processing
Drift and gain correction and dose weighting were performed using MotionCor2 (Zheng et al., 2017). A dose weighted average image of the whole stack was used to determine the contrast transfer function with the software Gctf (Zhang, 2016).The following process was done using RELION 3.0 (Zivanov et al., 2018). Particles were picked using a Laplacian of Gaussian function (min diameter 300 Å , max diameter 320 Å ). 337,268 particles were extracted with a box size of 360 pixels and binned three-fold for 3D classification into 5 classes. Two sets of subclasses depicting high-resolution features were selected for refinement, one “unrotated” (55589 particles) and one “rotated” (42135 particles). Refinement of unrotated and rotated classes yielded an average resolution of 3.6 Å and 3.5 Å , respectively. The unrotated class has been focused refined with a mask on the 60S, the body and the head of the 40S, yielding respectively 3.49, 3.52 and 4.13 Å resolution. Determination of the local resolution of the final density map was performed using ResMap (Kucukelbir et al., 2014).
Map fitting and model refinement
A cryo-EM structure of the 80S ribosome from Oryctolagus cuniculus (PDB ID 4UJE; (Budkevich et al., 2014)) was fitted in the density map using Chimera v. 1.10.2 (Pettersen et al., 2004). RNA regions that were not seen in this reference structure were built within Assemble v. 2 (Jossinet et al., 2010). Molecular dynamics flexible fitting (MDFF; (Trabuco et al., 2008)) with explicit solvent was performed for the complete 80S ribosome in VMD v. 1.9.2 (Humphrey et al., 1996), using NAMD v. 2 (gscale 0.3, numsteps 500,000, minsteps 2,000) (Phillips et al., 2005). RNA geometry and fit in density were improved by running Erraser within Phenix v. 1.10.1–2155 (Adams et al., 2010; Afonine et al., 2013; Chou et al., 2013; Jain et al., 2015), for rRNA fragments of ~990 nt. This model was further minimized using NAMD.
The IGR IRES from HalV within the unrotated complex was principally built by homology with various CrPV and PSIV IRES structures (earlier 3D models based on only partially correct 2D models had been built ab initio). More specifically, the IGR IRES was assembled from the following modules: ab initio modeled PKI and L3.2; P3.1 and L3.1 from residues 113–120 and 201–207 in PDB ID 1HR2 (double helical region of the P4-P6 domain from the T. thermophila group I intron that contains a tandem of purine-purine base pairs (Juneau et al., 2001)); PKII and P1.2 from PDB ID 2IL9 (X-ray crystal structure of the PSIV IRES lacking PKI at 3.1 Å resolution (Pfingsten et al., 2006)); P1.1 and L1.1 from PDB ID 2NOQ (first cryo-EM reconstruction of a complete 80S-bound CrPV IRES (Schüler et al., 2006 )). P1.2 required the most attention during manual rebuilding as it is shorter and less rich in Watson-Crick pairs than its counterpart in PSIV.
These modules were connected to one another and placed in filtered density maps (using Gaussian filters with 1.5 and 2.0 widths), using the ‘fit in map’ option in Chimera v. 1.10.2 (Pettersen et al., 2004), and the ‘real space refine’ and ‘regularize zone’ options in Coot v. 0.8.2 (Emsley and Cowtan, 2004). For real-space refinement and subsequent manual rebuilding, the IRES was stripped off of the 80S ribosome, except for RNA and protein residues within ~15 Å of any IRES residue. Upon assembling this partial model, the path for the L2 single strand that replaces the larger SSU-binding domain in CrPV and PSIV became straightforward. This 13-nucleotide long U and C-rich RNA segment wraps around L1.2a and L1.2b and could be modeled using the first residues of the corresponding domain in the CrPV IRES, that were extended using the ‘add terminal residue’ functionality in Coot with HalV-specific residues, in order to connect to PKII. The sequence of the IRES was manually edited using the ‘swapna’ command in Chimera and the ‘simple mutate’ functionality in Coot.
Manual fitting with geometry correction was carried out throughout the entire IRES, with a particular attention to the L1.1/L1 stalk interface, the central region of the IRES that comprises L1.2a, L1.2b, and L2, the L3.2 joining region that interacts with protein eS7, as well as the two pseudoknots. Assigning the correct sequence register for the IRES was made possible by first building the PKI region, which is the most conserved across IGR IRESs (Figures S2 and S6F), and where the density allows purines to be distinguished from pyrimidines (Figure S6). The resulting model was used as input for real-space refinement in Phenix v. dev-2474 (Adams et al., 2010; Afonine et al., 2013), using the unfiltered density map. The refinement procedure included simulated annealing (starting temperature = 600 K) and global minimization for 5–10 macro-cycles and took into account RNA and protein secondary structure restraints (search_method = from_ca).
A close-to-final model of the HalV IRES in the unrotated state was used as the starting model for the IRES in the rotated state. The rotated IRES structure was refined using the same procedure as the unrotated state. Both structures were placed back into the refined 80S ribosomes using rRNA and ribosomal proteins as a guide, further real-space refined using Phenix v. dev-3885, and validated using Phenix validation tools which include the Molprobity suite (Williams et al., 2018). The deposited structures were obtained after real space refinement and model validation in Phenix v. dev-3885 (Afonine et al., 2018), as well as removing clashes > 1.0 in Coot v. 0.9-pre EL, part of the CCP-EM suite (Burnley et al., 2017). Figures were generated using Chimera v. 1.10.2 (Pettersen et al., 2004), Chimera X 1.0, and Pymol v. 1.8.0.5 and v. 2.0.6. (Schrödinger). Validation statistics are in Table S3.
QUANTIFICATION AND STATISTICAL ANALYSIS
All In vitro experiments were repeated at least three times, and representative gel images and sucrose density gradient graphs were shown. Gel quantifications were obtained by overnight phosphoimaging with a BAS-IP SR 2040 E Super Resolution Storage phosphor screen (GE Healthcare) followed by imaging using an Amersham Typhoon IP biomolecular imager (GE Healthcare). Toeprints were quantified as a percentage of total radiolabeled cDNA using ImageQuant TL v8.2. Data (Figure S3) is presented as the mean of three independent experiments.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E.coli BL21 Star (DE3) | Invitrogen | C601003 |
| E.coli DH5α | Invitrogen | 18265017 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| T7 RNA polymerase | Thermo Scientific | EP0111 |
| Avian Myeloblastosis Virus Reverse Transcriptase |
Promega | M5108 |
| T4 Polynucleotide Kinase | New England Biolabs | M0201S |
| tRNA calf liver | Promega | Y209X |
| Rabbit reticulocyte lysate | Green Hectares | n/a |
| Flexi Rabbit Reticulocyte Lysate System | Promega | L4540 |
| TNT® T7 Insect Cell Extract Protein Expression System | Promega | L1101 |
| Isopropyl-beta-D-thiogalactoside | Gold Biotechnology | I2481C100 |
| Ni-NTA Agarose | QIAGEN | 30230 |
| MonoS 5/50 GL column | Cytiva Life Sciences | 17516801 |
| MonoQ 5/50 GL column | Cytiva Life Sciences | 17516601 |
| Superdex 200 Increase 5/150 GL | Cytiva Life Sciences | 28990945 |
| RiboLock RNase inhibitor | Thermo Scientific | EO0381 |
| [35S]-methionine | Perkin Elmer | NEG009T |
| [γ−32P]ATP | Perkin Elmer | BLU002Z |
| [α−32P]-UTP | Perkin Elmer | BLU007H |
| BamHI | New England Biolabs | R0136S |
| BstN1 | New England Biolabs | R0607S |
| EcoRV | New England Biolabs | R0195S |
| EcoRI | New England Biolabs | R0101S |
| HindIII | New England Biolabs | R0104S |
| XbaI | New England Biolabs | R0145S |
| Deposited Data | ||
| Raw data | Mendeley Data | https://data.mendeley.com/datasets/wx2ypvtxh8/draft?a=8017d354-6b04-4735-a5f8-464205fbbad9 |
| Structure of [80S-HalV IGR IRES] complex(rotated conformation) | This paper | PDB: 7A01 |
| Structure of [80S-HalV IGR IRES] complex (unrotated conformation) | This paper | PDB: 6ZVK |
| Cryo-electron microscopy map of [80S-HalV IGR IRES] complex (rotated conformation) | This paper | EMDB: EMD-11590 |
| Cryo-electron microscopy map of [80S-HalV IGR IRES] complex (unrotated conformation) | This paper | EMDB: EMD-11459 |
| Oligonucleotides | ||
| ssDNA (GATTGCTTGTTTAG) HalV | Thermo Fisher | N/A |
| ssDNA (GTAATTCCTTCGCTAAC) CrPV | Thermo Fisher | N/A |
| Recombinant DNA | ||
| pQE31-His6-eIF1 | Pisarev et al., 2007 | N/A |
| pET28-His6-eIF1A | Pisarev et al., 2007 | N/A |
| pET15b-His6-eIF4A | Pisarev et al., 2007 | N/A |
| pET15b-His6-eIF4B | Pisarev et al., 2007 | N/A |
| pET28-eIF4G736–1115 | Pisarev et al., 2007 | N/A |
| pET23b-eRF1-His6 | Pisarev et al., 2007 | N/A |
| pET23b-His6-eRF3a(139–499) | This work | N/A |
| pET-15b PAI-RBP | Heaton et al., 2001 | N/A |
| pMetRS | Lomakin et al., 2006 | N/A |
| pUC57-tRNASer (UCU) | Zinoviev et al., 2015 | N/A |
| pUC57-T7-Stem-HalV nt.6211–7460 | This work | N/A |
| pUC57-T7-Stem-HalV nt.6205–6513 | This work | N/A |
| pUC57-T7-Stem-HalV nt.6268–6513 | This work | N/A |
| pUC57-T7-Stem-HalV nt.6280–6513 | This work | N/A |
| pUC57-T7-Stem-HalV nt.6284–6513 | This work | N/A |
| pUC57-T7-Stem-HalV nt.6297–6513 | This work | N/A |
| pUC57-T7-Stem-HalV nt.6327–6513 | This work | N/A |
| pUC57-T7-Stem-HalV nt.6268–6483 | This work | N/A |
| pUC57-T7-Stem-HalV IGR-GUS | This work | N/A |
| pUC57-T7-HalV(2nt. L2 loop) | This work | N/A |
| pUC57-T7-HalV(6nt. L2 loop) | This work | N/A |
| pUC57-T7-HalV(+4nt. L2 loop) | This work | N/A |
| MC Stem-HalV | Abaeva et al., 2016 | N/A |
| pUC57-CrPV nt. 5997–6320 | This work | N/A |
| pUC57-CrPV5997–6320-Stop | This work | N/A |
| HalV PKII+CrPV PKI/SerStop | This work | N/A |
| pUC57-T7-CrPV PKII/PKIII+HalV PKI(SerStop) | This work | N/A |
| pUC57-T7-HalV PKI/II + CrPV PKIII | This work | N/A |
| pUC57-T7-CrPV ΔPKIII + HalV L2 loop | This work | N/A |
| pUC57-T7-CrPV ΔPKIII+HalV extended L2* loop | This work | N/A |
| Software and Algorithms | ||
| BLASTN | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome | RRID:SCR_001598 |
| BLASTX | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastx&PAGE_TYPE=BlastSearch&BLAST_SPEC=&LINK_LOC=blasttab&LAST_PAGE=blastn | RRID:SCR_001653 |
| EMBOSS Matcher | https://www.ebi.ac.uk/Tools/psa/emboss_matcher/ | RRID:SCR_017252 |
| Clustal Omega | https://www.ebi.ac.uk/Tools/msa/clustalo/ | RRID:SCR_001591 |
| CentroidFold | https://www.ncrna.org/centroidfold | RRID:SCR_017253 |
| Mfold | http://www.unafold.org/ | RRID:SCR_008543 |
| pKISS | https://bibiserv.cebitec.uni-bielefeld.de/pkiss | RRID:SCR_017256 |
| ImageQuant TL version 8.1 | Cytiva | RRID:SCR_014246 |
| Ugene v. 1.22 | http://ugene.net/ | RRID:SCR_005579 |
| MotionCor | https://emcore.ucsf.edu/ucsf-software | N/A |
| Gctf | https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/ | RRID:SCR_016500 |
| RELION 3.0 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page | RRID:SCR_016274 |
| ResMap | http://resmap.sourceforge.net/ | N/A |
| UCSF Chimera | https://www.cgl.ucsf.edu/chimera/ | RRID:SCR_004097 |
| UCSF Chimera X | https://www.cgl.ucsf.edu/chimerax/ | RRID:SCR_015872 |
| Pymol v. 1.8.0.5, v. 2.0.6 | https://pymol.org/2/ | RRID:SCR_000305 |
| Assemble v. 2 | http://bioinformatics.org/S2S/ | https://pubmed.ncbi.nlm.nih.gov/20562414/ |
| Molecular Dynamic Flexible Fitting | https://www.ks.uiuc.edu/Research/mdff/ | https://www.sciencedirect.com/science/article/pii/S0969212608001330 |
| Visual Molecular Dynamics | https://www.ks.uiuc.edu/Research/vmd/ | https://nlm.nih.gov/8744570/ |
| NAMD v. 2 | http://www.ks.uiuc.edu/Research/namd/ | RRID:SCR_014894 |
| Phenix v. 1.10.1–2155, v. dev-2474 | http://www.phenix-online.org/ | RRID:SCR_014224 |
| Phenix v. dev.3885 | http://www.phenix-online.org/ | https://journals.iucr.org/d/issues/2018/09/00/kw5139/index.html |
| Coot v. 0.8.2 and v. 0.9-pre EL | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ | RRID:SCR_014222 https://pubmed.ncbi.nlm.nih.gov/29872001/ |
| EPU software | FEI Company | n/a |
| Molprobity | http://molprobity.biochem.duke.edu | RRID:SCR_014226 |
| Phylogeny.fr | http://phylogeny.lirmm.fr/phylo_cgi/index.cgi | RRID:SCR_010266 |
| IQ-TREE | http://www.iqtree.org/ | RRID:SCR_017254 |
Highlights.
Halastavi árva virus (HalV) intergenic region (IGR) contains an IRES
In contrast to previously described IGR IRESs, HalV IGR IRES lacks domain 2
HalV IGR IRES functions by binding to assembled 80S ribosomes
It places PKI in the P site, making the A site codon directly accessible for decodin
ACKNOWLEDGMENTS
We thank Cajetan Neubauer and Venki Ramakrishnan for their gift of RelE and Alexander Myasnikov for assistance in data acquisition. This work was supported by NIH grants GM122602 to T.V.P. and AI123406 to C.U.T.H. and ANR grants ANR-14-ACHN-0024 @RAction program “ANR CryoEM80S,” ANR-11-LABX-0057_NETARN, and ERC-2017-STG #759120 “TransTryp” to Y.H. This project was supported by Institut National de la Santé et de la Recherche Médicale (INSERM), Centre National de la Recherche Scientifique (CNRS), and Universitéde Bordeaux.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108476.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abaeva IS, Pestova TV, and Hellen CU (2016). Attachment of ribosomal complexes and retrograde scanning during initiation on the Halastavi árva virus IRES. Nucleic Acids Res. 44, 2362–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeyrathne PD, Koh CS, Grant T, Grigorieff N, and Korostelev AA (2016). Ensemble cryo-EM uncovers inchworm-like translocation of áviral IRES through the ribosome. eLife 5, e14874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-Reyes F, Neupane R, Frank J, and Fernández IS (2019). The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs. EMBO J. 38, e102226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P, Headd J, Terwilliger T, and Adams P. (2013). New tool: phenix. real_space_refine. Comp. Crystallog. Newsletter 4, 43–44. [Google Scholar]
- Afonine PV, Klaholz BP, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, Adams PD, and Urzhumtsev A. (2018). New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, and Pestova TV (2006). In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125, 1125–1136. [DOI] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, and Beckmann R. (2013). Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85. [DOI] [PubMed] [Google Scholar]
- Asnani M, Kumar P, and Hellen CU (2015). Widespread distribution and structural diversity of Type IV IRESs in members of Picornaviridae. Virology 478, 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros Á, Pankovics P, Simmonds P, and Reuter G. (2011). Novel positivesense, single-stranded RNA (+ssRNA) virus with di-cistronic genome from intestinal content of freshwater carp (Cyprinus carpio). PLoS ONE 6, e29145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Shao S, Murray J, Hegde RS, and Ramakrishnan V. (2015). Structural basis for stop codon recognition in eukaryotes. Nature 524, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Baird MR, Yip MC, Murray J, and Shao S. (2018). Structures of translationally inactive mammalian ribosomes. eLife 7, e40486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budkevich TV, Giesebrecht J, Behrmann E, Loerke J, Ramrath DJ, Mielke T, Ismer J, Hildebrand PW, Tung CS, Nierhaus KH, et al. (2014). Regulation of the mammalian elongation cycle by subunit rolling: a eukaryotic-specific ribosome rearrangement. Cell 158, 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnley T, Palmer CM, and Winn M. (2017). Recent developments in the CCP-EM software suite. Acta Crystallogr. D Struct. Biol. 73, 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou FC, Sripakdeevong P, Dibrov SM, Hermann T, and Das R. (2013). Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nat. Methods 10, 74–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino D, and Kieft JS (2005). A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA 11, 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino DA, Pfingsten JS, Rambo RP, and Kieft JS (2008). tRNAmRNA mimicry drives translation initiation from a viral IRES. Nat. Struct. Mol. Biol. 15, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley AI, Lang AS, and Suttle CA (2007). The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities. Virol. J. 4, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breyne S, Yu Y, Unbehaun A, Pestova TV, and Hellen CU (2009). Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. USA 106, 9197–9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fernández IS, Bai XC, Murshudov G, Scheres SH, and Ramakrishnan V. (2014). Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 157, 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flis J, Holm M, Rundlet EJ, Loerke J, Hilal T, Dabrowski M, Bürger J, Mielke T, Blanchard SC, Spahn CMT, and Budkevich TV (2018). tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis. Cell Rep. 25, 2676–2688.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton JH, Dlakic WM, Dlakic M, and Gelehrter TD (2001). Identification and cDNA cloning of a novel RNA-binding protein that interacts with the cyclic nucleotide-responsive sequence in the Type-1 plasminogen activator inhibitor mRNA. J. Biol. Chem. 276, 3341–3347. [DOI] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, and Vinh LS (2018). UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, and Schulten K. (1996). VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Imai S, Kumar P, Hellen CU, D’Souza VM, and Wagner G. (2016). An accurately preorganized IRES RNA structure enables eIF4G capture for initiation of viral translation. Nat. Struct. Mol. Biol. 23, 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, and Pestova TV (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Richardson DC, and Richardson JS (2015). Computational methods for RNA structure validation and improvement. Methods Enzymol. 558, 181–212. [DOI] [PubMed] [Google Scholar]
- Jan E. (2006). Divergent IRES elements in invertebrates. Virus Res. 119, 16–28. [DOI] [PubMed] [Google Scholar]
- Jan E, and Sarnow P. (2002). Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 324, 889–902. [DOI] [PubMed] [Google Scholar]
- Jan E, Kinzy TG, and Sarnow P. (2003). Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc. Natl. Acad. Sci. USA 100, 15410–15415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang CJ, and Jan E. (2010). Modular domains of the Dicistroviridae intergenic internal ribosome entry site. RNA 16, 1182–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang CJ, Lo MC, and Jan E. (2009). Conserved element of the dicistrovirus IGR IRES that mimics an E-site tRNA/ribosome interaction mediates multiple functions. J. Mol. Biol. 387, 42–58. [DOI] [PubMed] [Google Scholar]
- Janssen S, and Giegerich R. (2015). The RNA shapes studio. Bioinformatics 31, 423–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossinet F, Ludwig TE, and Westhof E. (2010). Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics 26, 2057–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau K, Podell E, Harrington DJ, and Cech TR (2001). Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure 9, 221–231. [DOI] [PubMed] [Google Scholar]
- Kanamori Y, and Nakashima N. (2001). A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7, 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr CH, Ma ZW, Jang CJ, Thompson SR, and Jan E. (2016). Molecular analysis of the factorless internal ribosome entry site in Cricket Paralysis virus infection. Sci. Rep. 6, 37319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CS, Brilot AF, Grigorieff N, and Korostelev AA (2014). Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center. Proc. Natl. Acad. Sci. USA 111, 9139–9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Laurberg M, and Noller HF (2006). Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126, 1065–1077. [DOI] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, and Tagare HD (2014). Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Wei HM, Chen LY, and Li C. (2014). Localization of SERBP1 in stress granules and nucleoli. FEBS J. 281, 352–364. [DOI] [PubMed] [Google Scholar]
- Lomakin IB, Hellen CU, and Pestova TV (2000). Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20, 6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, and Pestova TV (2006). The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 25, 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailliot J, and Martin F. (2018). Viral internal ribosomal entry sites: four classes for one goal. Wiley Interdiscip. Rev. RNA 9, e1458. [DOI] [PubMed] [Google Scholar]
- Mohr I, and Sonenberg N. (2012). Host translation at the nexus of infection and immunity. Cell Host Microbe 12, 470–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhs M, Yamamoto H, Ismer J, Takaku H, Nashimoto M, Uchiumi T, Nakashima N, Mielke T, Hildebrand PW, Nierhaus KH, and Spahn CM (2011). Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res. 39, 5264–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhs M, Hilal T, Mielke T, Skabkin MA, Sanbonmatsu KY, Pestova TV, and Spahn CM (2015). Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES. Mol. Cell 57, 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J, Savva CG, Shin BS, Dever TE, Ramakrishnan V, and Fernández IS (2016). Structural characterization of ribosome recruitment and translocation by type IV IRES. eLife 5, e13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N, and Uchiumi T. (2009). Functional analysis of structural motifs in dicistroviruses. Virus Res. 139, 137–147. [DOI] [PubMed] [Google Scholar]
- Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, and Brodersen DE (2009). The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139, 1084–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, and Minh BQ (2015). IQTREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Yamamoto H, Shibuya N, Hatakeyama Y, Hachimori A, Uchiumi T, and Nakashima N. (2003). Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 31, 2434–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, and Fursov M; UGENE team (2012). Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28, 1166–1167. [DOI] [PubMed] [Google Scholar]
- Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, and Ehrenberg M. (2003). The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140. [DOI] [PubMed] [Google Scholar]
- Pestova TV, and Hellen CU (2003). Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 17, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, and Shatsky IN (1996). Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16, 6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, and Hellen CU (1998a). Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394, 854–859. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, and Hellen CU (1998b). A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Bernier CR, Hsiao C, Norris AM, Kovacs NA, Waterbury CC, Stepanov VG, Harvey SC, Fox GE, Wartell RM, et al. (2014). Evolution of the ribosome at atomic resolution. Proc. Natl. Acad. Sci. USA 111, 10251–10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A, Grosely R, Chen J, O’Leary SE, and Puglisi JD (2016). Multiple parallel pathways of translation initiation on the CrPV IRES. Mol. Cell 62, 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pfingsten JS, Costantino DA, and Kieft JS (2006). Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science 314, 1450–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten JS, Costantino DA, and Kieft JS (2007). Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J. Mol. Biol. 370, 856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten JS, Castile AE, and Kieft JS (2010). Mechanistic role of structurally dynamic regions in Dicistroviridae IGR IRESs. J. Mol. Biol. 395, 205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, and Schulten K. (2005). Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CU, and Pestova TV (2007). Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 430, 147–177. [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, and Pisarev AV (2011). Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 30, 1804–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, and Fernández IS (2018). Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES. eLife 7, e34062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruehle MD, Zhang H, Sheridan RM, Mitra S, Chen Y, Gonzalez RL, Cooperman BS, and Kieft JS (2015). A dynamic RNA loop in an IRES affects multiple steps of elongation factor-mediated translation initiation. eLife 4, e08146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J, and Nakashima N. (2000). Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. USA 97, 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Hamada M, Asai K, and Mituyama T. (2009). CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 37, W277–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, and Spahn CM (2006). Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 13, 1092–1096. [DOI] [PubMed] [Google Scholar]
- Seit-Nebi A, Frolova L, Justesen J, and Kisselev L. (2001). Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 29, 3982–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV 4th, Weixlbaumer A, Petry S, Kelley AC, Weir JR, and Ramakrishnan V. (2006). Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942. [DOI] [PubMed] [Google Scholar]
- Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, Qin XC, Li J, Cao JP, Eden JS, et al. (2016). Redefining the invertebrate RNA virosphere. Nature 540, 539–543. [DOI] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Hellen CU, and Pestova TV (2013). Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol. Cell 51, 249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, and Schulten K. (2008). Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JN, Buschauer R, Mackens-Kiani T, Best K, Kratzat H, Berninghausen O, Becker T, Gilbert W, Cheng J, and Beckmann R. (2020). Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes. PLoS Biol. 18, e3000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CU, and Sarnow P. (2000). Initiation of protein synthesis from the A site of the ribosome. Cell 102, 511–520. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Nakashima N, Ikeda Y, and Uchiumi T. (2007). Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus internal ribosome entry site. J. Biol. Chem. 282, 7770–7776. [DOI] [PubMed] [Google Scholar]
- Zhang K. (2016). Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Korostelev A, Costantino DA, Donohue JP, Noller HF, and Kieft JS (2011). Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome. Proc. Natl. Acad. Sci. USA 108, 1839–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A, Hellen CUT, and Pestova TV (2015). Multiple mechanisms of reinitiation on bicistronic calicivirus mRNAs. Mol. Cell 57, 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A, Goyal A, Jindal S, LaCava J, Komar AA, Rodnina MV, Hellen CUT, and Pestova TV (2018). Functions of unconventional mammalian translational GTPases GTPBP1 and GTPBP2. Genes Dev. 32, 1226–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates of the rotated and unrotated IRES/80S complexes have been deposited in the Protein Data Bank (PDB). The accession numbers for the atomic models of the [80S-HalV IGR IRES] in the rotated and unrotated conformation reported in this paper are: 7A01 and 6ZVK, respectively.
The cryo-EM maps of the rotated and unrotated IRES/80S complexes have been deposited in the Electron Microscopy Data Bank (EMDB) with the accession codes: EMD-11590 and EMD-11459, respectively.
Gel images are available at: https://data.mendeley.com/datasets/wx2ypvtxh8/draft?a=8017d354-6b04-4735-a5f8-464205fbbad9