Abstract
Patients with hereditary tyrosinemia type I (HT1) present acute and irreversible liver and kidney damage during infancy. CRISPR-Cas9-mediated gene correction during infancy may provide a promising approach to treat patients with HT1. However, all previous studies were performed on adult HT1 rodent models, which cannot authentically recapitulate some symptoms of human patients. The efficacy and safety should be verified in large animals to translate precise gene therapy to clinical practice. Here, we delivered CRISPR-Cas9 and donor templates via adeno-associated virus to newborn HT1 rabbits. The lethal phenotypes could be rescued, and notably, these HT1 rabbits reached adulthood normally without 2-(2-nitro-4-trifluoromethylbenzyol)-1,3 cyclohexanedione administration and even gave birth to offspring. Adeno-associated virus (AAV)-treated HT1 rabbits displayed normal liver and kidney structures and functions. Homology-directed repair-mediated precise gene corrections and non-homologous end joining-mediated out-of-frame to in-frame corrections in the livers were observed with efficiencies of 0.90%–3.71% and 2.39%–6.35%, respectively, which appeared to be sufficient to recover liver function and decrease liver and kidney damage. This study provides useful large-animal preclinical data for rescuing hepatocyte-related monogenetic metabolic disorders with precise gene therapy.
Keywords: hereditary tyrosinemia type I, rabbit models, CRISPR-Cas9-mediated precise gene therapy, adeno-associated virus
Graphical Abstract
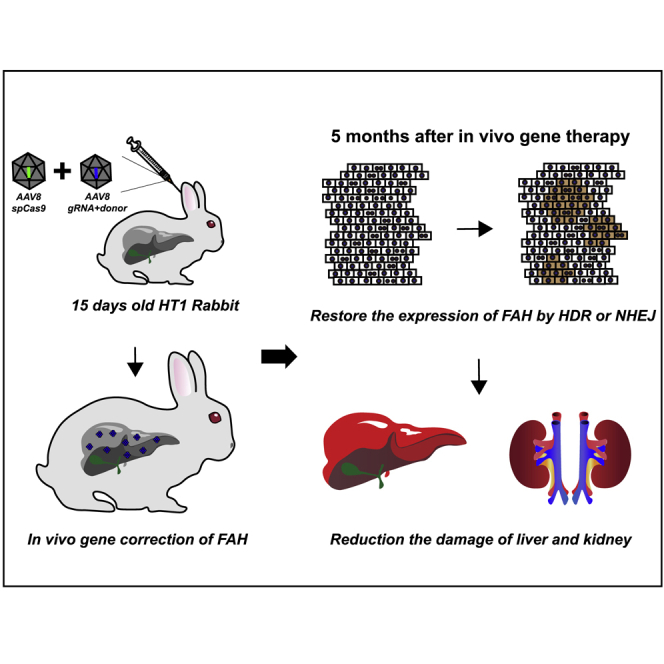
CRISPR-Cas9-mediated precise FAH gene correction is a promising strategy to treat hereditary tyrosinemia type I (HT1). In this study, Wang and colleagues rescued the lethal phenotypes and recovered the liver and kidney function of HT1 rabbits by CRISPR-Cas9-mediated precise FAH gene correction. This study provides useful large-animal preclinical data for rescuing hepatocyte-related monogenetic metabolic disorders with precise gene therapy.
Introduction
Hereditary tyrosinemia type I (HT1) is a rare autosomal-recessive inborn error of metabolism that occurs in approximately 1 case in every 100,000 births in Sweden and 1 per 1,846 in a restricted area in Canada.1 In humans, this disease is caused by the deficiency of the metabolic enzyme fumarylacetoacetate hydrolase (FAH), which is required for the last step in the metabolism of the amino acid tyrosine (Tyr).2 Abnormal Tyr metabolism results in the accumulation of toxic metabolites, such as fumarylacetoacetate and succinylacetone (SA), in hepatocytes and renal proximal tubules. Patients with HT1 develop symptoms within the first few weeks of life if left untreated. The acute onset of HT1 is characterized by severe liver failure, renal tubular dysfunction, and most frequently, death.3 The chronic form of HT1 is characterized by a progressive liver disease, typically severe fibrosis and hepatocellular carcinoma. The most common treatment for HT1 is the combination of pharmacological blockades and low-Tyr diet. 2-(2-Nitro-4-trifluoromethylbenzyol)-1,3 cyclohexanedione (NTBC) is an effective chemical drug for patients with HT1 that blocks the upstream Tyr metabolic enzymes (inhibitor of 4-hydroxyphenylpyruvate dioxygenase).1 However, NTBC therapy has numerous drawbacks, such as having no therapeutic effect on some patients with HT1, incomplete inhibition of the target enzyme, off-target effects, and drug interactions.4 Orthotopic liver transplantation is another effective clinical method for curing patients with HTI but is limited by donor organ shortage and immune rejection responses.5 These concerns and limitations emphasize the clinical need to find alternative therapies to cure this monogenic liver disease, and one example is gene therapy.
The CRISPR-Cas9 technology provides a powerful tool for the gene correction of monogenic diseases.6, 7, 8, 9, 10, 11 HT1 is a monogenic liver disease that is particularly suitable for gene-repair-mediated therapy, because gene-corrected hepatocytes can expand and repopulate the liver.12 The CRISPR-Cas9-mediated precise correction of Fah gene mutation has been previously performed only in mouse and rat HT1 models.13, 14, 15, 16 However, mouse and rat HT1 models fail to fully reproduce the disease phenotype observed in humans; for instance, no evident kidney damage was observed in mouse HT1 models.17 In addition, the livers of mice and rats are considerably smaller than those of humans at only approximately 1 g in an adult mouse and 10 g in an adult rat compared with 1,500 g in an adult human. Whether CRISPR-Cas9-mediated precise FAH gene correction in vivo is effective in large animals with a liver size similar to that of human remains unverified.
As large animals, rabbits are widely used as models for human metabolic diseases because rabbits are closer to humans than rodents in terms of physiological characteristics.18, 19, 20 Rabbits have relatively large livers at approximately 60 g, which is 60 and 6 times more than those of mice and rats, respectively. In addition, kidney damage similar to that in human patients has been observed in the FAH-knockout rabbit model.21 Therefore, the treatment effectiveness of correcting the gene mutation of hepatocytes in rabbits can be easily translated for the clinical treatment of HT1 in humans.
HT1 large-animal models, including rabbit and pig, have been previously reported by our team21 and Hickey et al.22,23 An HT1 pig model was produced by disrupting FAH exon 5 with a neomycin resistance expression cassette,22 which is unsuitable for testing the CRISPR-Cas9-mediated precise correction of FAH gene with point mutations or insertion and deletion (indel) mutations in vivo. Thus, ex vivo gene therapy in autologous hepatocytes or CRISPR-Cas9-mediated knockin of FAH cDNA in vivo was applied to prevent liver failure and restore metabolic function in this pig model.24,25 Our HT1 rabbit model harbored 10-bp deletions in exon 2, which is suitable for CRISPR-Cas9-mediated precise FAH gene correction in vivo.
Fah gene correction was previously conducted in the adult stage of mice and rats.13, 14, 15, 16 Here, we performed CRISPR-Cas9-mediated precise gene correction in vivo in 15-day-old rabbits with HT1. Adeno-associated virus (AAV) 8-packaged Cas9, FAH-single guide RNA (sgRNA), and donor templates were delivered to the livers of HT1 rabbits via ear vein injection. Homology-directed repair (HDR)- and non-homologous end joining (NHEJ)-mediated (out-of-frame to in-frame mutations) gene corrections occurred in the livers of the treated HT1 rabbits with efficiencies of 0.90%–3.71% and 2.39%–6.35%, respectively. The rabbits treated with gene correction exhibited normal structure and function of both livers and kidneys. Notably, these rabbits were able to grow up to adult normally without NTBC treatment and give birth to offspring. This study provides preclinical data to bridge the gap between rodents and humans for rescuing hepatocyte-related genetic metabolic disorders with point mutations or indel mutations.
Results
Screening of Optimal sgRNA that Targets Exon 2 of FAH Gene in Rabbit Fatal Fibroblasts (RFFs)
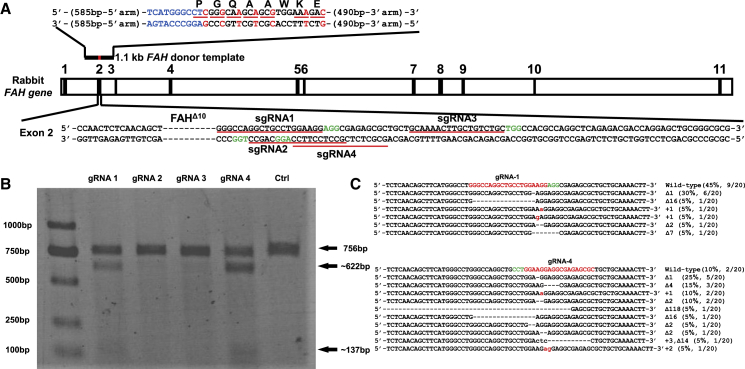
Four sgRNAs, namely, sgRNA1–4, which target exon 2 of rabbit FAH gene, were designed and constructed into U6-sgRNA-expressing vectors to screen the optimal sgRNA for the gene correction of FAHΔ10/Δ10 rabbits (Figure 1A). The constructed sgRNA and Cas9-expressing vectors were co-transfected into RFFs derived from 15-day-old New Zealand rabbit fetus. Genomic DNAs were extracted 5 days post-transfection and initially evaluated via polymerase chain reaction (PCR) and T7 endonuclease I (T7EN1) cleavage assay to identify the formation of indels at the desired sites. The results of the T7EN1 cleavage assay showed that different sgRNAs exhibited varying cleavage efficiencies, and the cleavage bands of sgRNA1 and sgRNA4 were darker than those of sgRNA2 and sgRNA3 (Figure 1B). The Sanger amplicon sequencing results were consistent with those of the T7EN1 cleavage assay (Figure S1). The amplicons of sgRNA1 and sgRNA4 were sub-cloned into the T vector and further subjected to Sanger sequencing. Out of 20 sub-clones each for sgRNA1 and sgRNA4, 11 (11/20, 55.0%) and 18 (18/20, 90.0%) exhibited the intended mutant alleles, respectively (Figure 1C). For FAHΔ10/Δ10 rabbit, 3N+2 bp deletions or 3N+1 bp insertions could transform out-of-frame mutations into in-frame mutations and also result in truncated or lengthened FAH protein expression. The preceding Sanger sequencing results showed that 15.0% (3/20) of the sub-clones of sgRNA1 and 35.0% (7/20) of those of sgRNA4 harbored 3N+1 bp insertions or 3N+2 bp deletions (Figure 1C). Thus, we selected sgRNA4 to target exon 2 of rabbit FAH gene for the gene therapy of HT1 rabbit models.
Figure 1.
Selection of Optimal sgRNA that Targets Exon 2 of Rabbit FAH Gene
(A) Schematic of the strategy for correcting the FAH gene of HT1 rabbits. The 5′ arm (585 bp) and 3′ arm (490 bp) of the donor template are labeled with a black line, and the corrected loci are labeled with a red line, above FAH exon 2. Base pairs labeled in blue indicate the correction of 10-bp deletions. Synonymous mutations are labeled in red. The dashed line in exon 2 represents the 10-bp deletions in the FAHΔ10/Δ10 rabbit. The four sgRNA-targeting sites and PAM sequences are marked with red and green underlines, respectively. (B) Detection of sgRNA1–4 and Cas9-mediated cleavage of exon 2 of the rabbit FAH gene by PCR and T7EN1 cleavage assay. (C) Summary of the Sanger sequencing results of the sgRNA1 and sgRNA4 TA clones (20 clones for each PCR product). For wild-type sequences, the PAM and targeting sequences are highlighted in green and red, respectively.
In Vivo Delivery of the CRISPR-Cas9 System and Donor Templates Can Rescue the Lethal Phenotypes of FAH-Deficient Rabbits
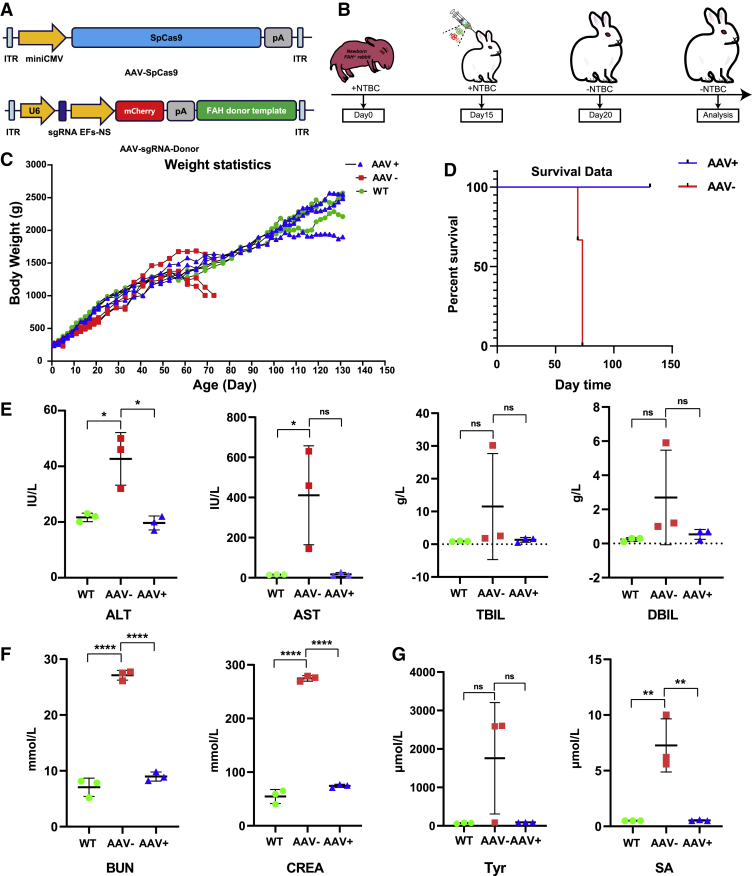
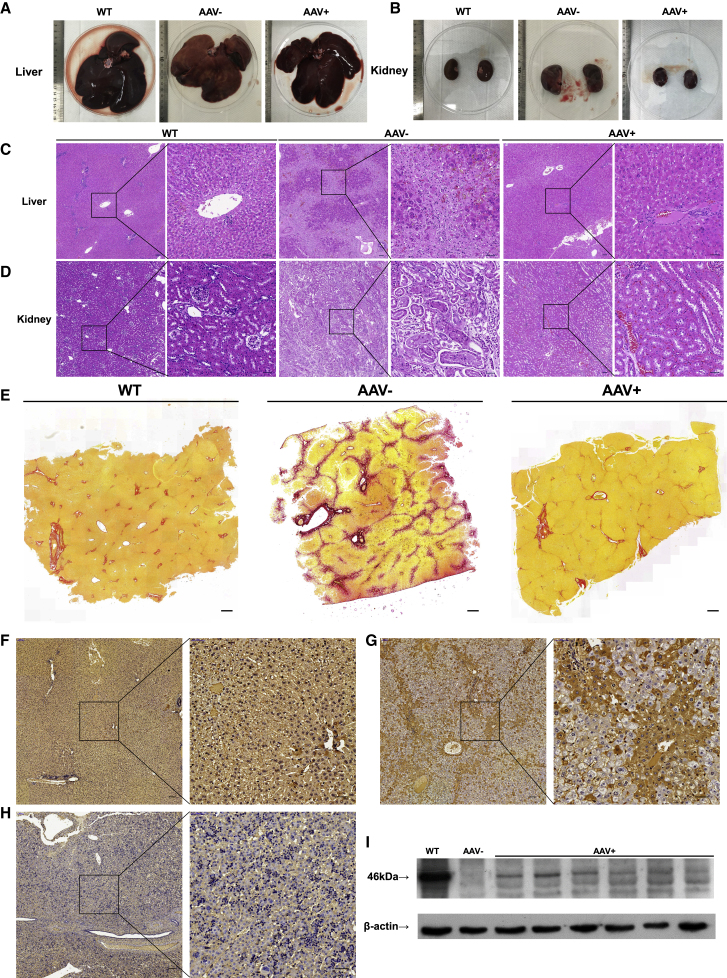
At present, the most promising and safe delivery method to the liver is based on AAV, which has already been clinically approved.26 FAH-sgRNA4 and corrected donor template with 585-bp left homology arm and 490-bp right homology arm were cloned into AAV A3 vectors (referred to as A3-sgRNA4-Donor) (Figures 1A and 2A). The AAV A3 vector contains a U6 promoter-directed sgRNA expression cassette and an EF-1α short promoter (EFS-NS)-driven mCherry expression cassette (Figure 2A). To quickly determine the HDR-mediated precise gene corrections by PCR and prevent the cleavage of the donor itself or the repetitive digestion of the repaired genome, we introduced seven synonymous point mutations (one at the protospacer adjacent motif (PAM) site of sgRNA4, five at the sgRNA4 targeting site, and one upstream of the sgRNA4 targeting site), namely, CTGGGCCAGGCTGCCTGGAAGGAG > CTCGGGCAAGCAGCGTGGAAAGAC, into the donor templates (Figure 1A). Among the investigated AAV serotypes, AAV serotype 8 (AAV8) has higher affinity for hepatocytes and targets the liver specifically.27,28 The constructed A3-sgRNA4-Donor and commercial SpCas9-expressing AAV plasmid (A2 vector) were used for packaging AAV8 (referred to as AAV8-sgRNA4-Donor and AAV8-SpCas9, respectively). HT1 rabbit models were produced by mating one heterozygous FAHΔ10/+ male rabbit with four FAHΔ10/+ female rabbits. At 15 days post-mating, the four pregnant female rabbits were provided with NTBC-containing water (0.2 L/day, 7.5 mg/L). After approximately 30 days of gestation, 28 bunnies were born at term from the four pregnant mothers. Among the 28 rabbits, 6 (FAH1-6, FAH2-3, FAH3-2, FAH3-6, FAH3-8, and FAH4-2) harbored homozygous 10-bp deletions in exon 2 of the FAH gene, 13 were heterozygotes, and 9 were wild type (Figure S2). NTBC was continuously administered to the six homozygotes until the rabbits were 15 days old. To investigate whether CRISPR-Cas9 could correct HT1 rabbit models in vivo, we administered 3 × 1012 genome copies of packaged AAV8-SpCas9 and 6 × 1012 genome copies of AAV8-sgRNA4-Donor into three 15-day-old HT1 rabbits (referred to as FA1, FA2, and FA3) through ear vein injection (Figure 2B). The three control HT1 rabbits (referred to as FK-1, FK-2, and FK-3) were injected with only phosphate buffered saline (PBS). At 5 days post-injection, the HT1 rabbits injected with AAV8 and PBS were withdrawn from NTBC (Figure 2B). The AAV-treated HT1 rabbits and wild-type rabbits steadily reached sexual maturation (Figures 2C and S3). By contrast, the PBS-treated HT1 rabbits survived for only less than 11 weeks after birth and exhibited weight loss at the time of death (Figure 2D). To verify the presence of liver and kidney lesions in the HT1 rabbits, we extracted serum from blood samples and analyzed the levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (TBIL), and direct bilirubin (DBIL), which were elevated as a consequence of liver damage. In addition, blood urea nitrogen (BUN) and creatinine (CREA) indicated kidney damage. All six parameters of the HT1 rabbits that underwent gene therapy were at normal levels as age-matched wild-type rabbits, whereas AAV-untreated HT1 rabbits had high levels of kidney and/or liver damage indices (Figures 2E, 2F, and S4A). The concentrations of Tyr and its metabolic product SA also remained at normal levels (Figure 2G). The serum of one rabbit without AAV treatment contained particularly high levels of lipids, such as triglyceride (TG) and cholesterol (CHOL), whereas the TG and CHOL of AAV-treated HT1 rabbits were rescued to normal levels (Figures S4A–S4D). Two HT1 rabbits (FA1 and FA2) treated with AAV for 5 months, one treated with AAV for 9 months, and age-matched wild-type rabbits (n = 3) were euthanized. No substantial structural differences were found in the liver and kidney between the wild-type rabbits and AAV-treated HT1 rabbits (Figures 3A, 3B, and S4E). Liver damage, such as prominent liver swelling, hemorrhage, and yellow/green discoloration, was not observed in the wild-type rabbits and AAV-treated HT1 rabbits but was found in the untreated HT1 rabbits (Figure 3A). Notably, renal dysplasia, renal calculus, and renal medullary cystic lesion were found in some untreated HT1 rabbits but were rescued in AAV-treated HT1 rabbits (Figures 3B and S4E). The collected liver and kidney tissues were further analyzed via hematoxylin and eosin (H&E) staining. The results showed that the treated livers and kidneys were normalized compared with the livers and kidneys of the untreated animals and had fewer dysplastic hepatocytes, tubule interstitial nephritis, glomerular and renal tubular structure anomalies, and regions of inflammatory cellular infiltration (Figures 3C and 3D). The chronic features of human disorders, especially liver fibrosis, have been clinically observed in patients with HT1.29 In HT1 rabbits with gene therapy, the results of Sirius red staining showed that the signs of fibrosis were very mild and similar to that of age-matched wild-type rabbits, whereas remarkable liver fibrosis was observed in untreated HT1 rabbits (Figure 3E). These results suggested that CRISPR-Cas9-mediated gene correction in the liver could recover liver function, decrease liver and kidney damages, and rescue the lethal phenotype of HT1 rabbits.
Figure 2.
Delivery of AAV8-SpCas9 and AAV8-sgRNA4-Donor to Cure HT1 Rabbits
(A) Schematic views of the AAV vectors of SpCas9 and sgRNA4-Donor. (B) Key time points of AAV delivery and NTBC withdrawal. (C and D) Kaplan-Meier body weights (C) and survival (D) curves of the wild-type, AAV-treated, and AAV-untreated HT1 rabbits. (E–G) Biochemical analysis of the serum from the AAV-treated and AAV-untreated HT1 rabbits for the comparison of liver damage markers, namely, aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin (TBIL), and direct bilirubin (DBIL) (E); renal damage markers, namely, blood urea nitrogen (BUN) and creatinine (CREA) (F); and the main biomarker, namely, succinylacetone (SA) and tyrosine (Tyr) (G). Error bars: mean ± SD. Asterisk indicates statistical significance as follows: ns, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001.
Figure 3.
CRISPR-Cas9-Mediated FAH Gene Correction Decreased the Liver and Kidney Damage in HT1 Rabbits
(A and B) Pictures of the livers (A) and kidneys (B) of wild-type, AAV-treated, and AAV-untreated rabbits. (C and D) H&E staining of the liver (C) and kidney (D) sections from the wild-type rabbits, AAV-treated rabbits, and AAV-untreated rabbits. Black squares are amplified in the right images. Scale bars, 100 μm (left panels); 50 μm (right panels). (E) Liver fibrosis was detected by Sirius red staining in the liver tissues of wild-type, AAV-treated, and AAV-untreated HT1 rabbits. Scale bars, 500 μm. (F–H) FAH IHC of the liver sections from the wild-type rabbits (F) and HT1 rabbits injected with AAV8 (G) or PBS (H). The right panel shows a high-magnification view (box with black dashed line). Scale bars: 100 μm (left panels); 50 μm (right panels), respectively. (I) Western blot analysis of the FAH expression in the livers of the wild-type, AAV-treated, and AAV-untreated HT1 rabbits.
CRISPR-Cas9-Mediated Gene Editing Corrects FAH Mutation in the Rabbit Liver
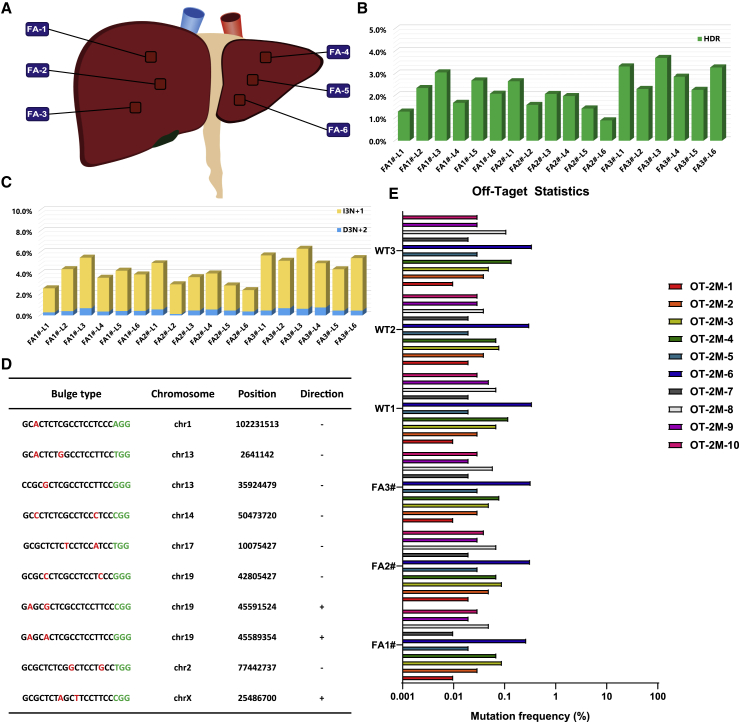
We first verified the status and copy number of AAV vector in treated liver tissues. Droplet digital PCR (ddPCR) results showed that 496–17,300 copies of AAV8-SpCas9 and 2,212–14,820 copies of AAV8-sgRNA4-Donor exist in 100 ng of whole DNAs from AAV-treated liver tissues (Figure S5). In addition, immunofluorescence staining by a mCherry-specific antibody confirmed that only the AAV-treated liver tissues expressed exogenous mCherry fluorescent protein (Figure S6A) and not the heart, kidney, lung, and spleen (Figure S6B). We next examined the liver tissues of treated rabbits via western blot analysis, and immunohistochemical (IHC) staining with FAH-specific antibody was performed to determine whether Cas9-mediated genome editing generates FAH+ hepatocytes in vivo. Western blot analysis confirmed that full-length or truncated/lengthened FAH proteins were expressed in the livers of the treated HT1 rabbits (Figure 3I). FAH+ hepatocytes (4.21% ± 3.49%) were detected in AAV-treated HT1 rabbits at the time of euthanasia, but not in the livers of the untreated HT1 rabbits, by IHC staining (Figures 3F–3H). These corrected hepatocytes formed widespread patches in the livers because of the expansion of the initially repaired hepatocytes. This finding was similar to CRISPR-Cas9-mediated Fah correction in mice and rats.13,14,16 To measure FAH gene repair frequency, the liver and kidney tissues of each euthanized treated rabbit from six positions in each liver or kidney (the six liver tissues of FA1, FA2, and FA3 rabbits, three from the right lobe and three from the left lobe, referred to as FA1#-L1–6, FA2#-L1–6, and FA3#-L1–6, respectively; the six kidney tissues of FA1, FA2, and FA3 rabbits, referred to as FA1#-K1–6, FA2#-K1–6, and FA3#-K1–6, respectively) were collected to extract genomic DNAs for PCR analysis with specific HDR-F and HDR-R primers (Figures 4A and S7A). The PCR results suggested that HDR-mediated gene therapy specifically occurred in the livers of the AAV-treated rabbits, but not in the kidneys (Figure S7B). The efficiency of gene repair and the patterns of gene modifications in the livers of AAV-treated rabbits were further analyzed by amplicon deep sequencing. The quantification of the high-throughput sequencing reads showed that HDR-mediated precise FAH gene correction was observed with efficiencies ranging from 0.90% to 3.71% (1.29% for FA1#-L1, 2.34% for FA1#-L2, 3.07% for FA1#-L3, 1.68% for FA1#-L4, 2.71% for FA1#-L5, 2.09% for FA1#-L6, 2.67% for FA2#-L1, 1.59% for FA2#-L2, 2.08% for FA2#-L3, 1.98% for FA2#-L4, 1.43% for FA2#-L5, 0.90% for FA2#-L6, 3.33% for FA3#-L1, 2.30% for FA3#-L2, 3.71% for FA3#-L3, 2.87% for FA3#-L4, 2.26% for FA3#-L5, and 3.28% for FA3#-L6) (Figure 4B). In addition, indel mutations ranging from 38-bp deletion to 68-bp insertions at the predicted cutting site were also found with efficiencies of 5.31%, 6.61%, 8.61%, 6.14%, 6.32%, and 5.98% for FA1#-L1–6; 8.05%, 5.69%, 5.85%, 6.25%, 4.87%, and 4.75% for FA2#-L1–6; and 9.15%, 9.06%, 9.92%, 8.34%, 7.33%, and 8.28% for FA3#-L1–6, respectively (Figures S8 and S9). In these indel mutations, the out-of-frame to in-frame mutations (3N+1 bp insertions [I3N+1] or 3N+2 bp deletions [D3N+2]) can remove the premature stop codon, resulting in truncated/lengthened FAH protein expression with deletions or insertions for several amino acids, which have no evident functional effects on FAH protein. We suggested that NHEJ-mediated in-frame editing may also contribute to the therapeutic effects of CRISPR-Cas9. The efficiencies of these out-of-frame to in-frame mutations in each collected tissue ranged from 2.39% to 6.35% (2.57%, 4.39%, 5.51%, 3.57%, 4.23%, and 3.89% for FA1#-L1–6; 4.97%, 2.94%, 3.63%, 3.97%, 2.84%, and 2.39% for FA2#-L1–6; and 5.73%, 5.22%, 6.35%, 4.95%, 4.38%, and 5.47% for FA3#-L1–6, respectively) (Figures 4C, S8, and S9). In total, the efficiencies of the CRISPR-Cas9-mediated corrections, including precise and in-frame corrections, were 3.86%, 6.73%, 8.58%, 5.25%, 6.94%, and 5.98% for FA1#-L1–6; 7.64%, 4.53%, 5.71%, 5.95%, 4.27%, and 3.29% for FA2#-L1–6; and 9.06%, 7.52%, 10.06%, 7.82%, 6.64%, and 8.75% for FA3#-L1–6, respectively. These results correspond to the results of the western blot analysis and IHC staining (Figure S8C). The efficiencies of the HDR-mediated precise FAH gene corrections and NHEJ-mediated out-of-frame to in-frame corrections in FA3 (2.26%–3.71% and 4.38%–6.35%) were higher than those in FA1 (1.29%–3.07% and 2.57%–5.51%) and FA2 (0.90%–2.67% and 2.39%–4.97%); thus, gene-corrected hepatocytes could expand and repopulate in the liver.
Figure 4.
CRISPR-Cas9-Mediated In Vivo Genome Editing Partially Corrects FAH Mutation in the Livers of HT1 Rabbits without Off-Target Effects
(A) Schematic views of the six positions of the HT1 rabbit livers injected with AAV (three for the right lobe and three for the left lobe) were collected for deep sequencing. (B and C) Histogram of the gene-corrected patterns, including precise HDR (B) and out-of-frame to in-frame mutations (3N+1 bp insertion and 3N+2 bp deletion) (C). (D) List of the top 10 potential off-target sites for sgRNA4. Mismatched nucleotides are indicated in red, and PAM sequences are labeled in green. (E) Targeted deep sequencing results for the top 10 potential off-target sites in the livers of the wild-type rabbits and AAV-treated HT1 rabbits.
Although the corrected FAH protein could rescue the lethal phenotypes of HT1, the efficiencies of gene correction were relatively low. A previous report showed that HDR is preferentially utilized in neonatal mammalian livers.30 To further improve the initial efficiency of HDR-mediated precise gene correction in the liver, three 1-week-old HT1 rabbits (FA01, FA02, and FA03) were injected with AAV8-sgRNA4-Donor and AAV8-SpCas9. These three injected HT1 rabbits were euthanized 7 days post-injection, and their liver tissues were collected for amplicon deep sequencing. The deep sequencing results showed that the efficiencies of HDR-mediated precise gene corrections ranged from 1.71% to 4.13% (1.71%–4.13% for FA01, 1.97%–3.01% for FA02, and 2.64%–5.83% for FA03), which were higher than the correction efficiencies observed 5 months after the AAV treatment of 15-day-old rabbits (FA1 and FA2) (Figure S10).
To analyze the off-target effects of the CRISPR-Cas9 system in vivo, we predicted 10 potential off-target sites (OTSs) with 2-bp mismatches (Figure 4D) in the rabbit genome for sgRNA4 using a published prediction tool, Cas-OFFinder.31 Genomic DNAs from six positions in each liver of FA1, FA2, FA3, and three age-matched wild-type rabbits were mixed together and used for amplifying the potential OTSs via PCR. PCR amplicons were further analyzed by high-throughput sequencing. More than 99.9% of the amplicons in OTS-1–5 and OTS-7–10 were wild-type sequences, and the indel frequency of these sites was negligible (Figure 4E). In OTS-6, 0.27%–0.35% of the amplicons carried different sequences with a C deletion at the outside of the sgRNA4 potential targeting site, and these sequences were also detected in wild-type rabbits; therefore, these different sequences may be caused by PCR or sequence errors. These results indicated that none of these potential OTSs exhibited more sequence alterations than the background of wild-type animals.
Global Gene Expression Pattern of Gene-Corrected HT1 Rabbits
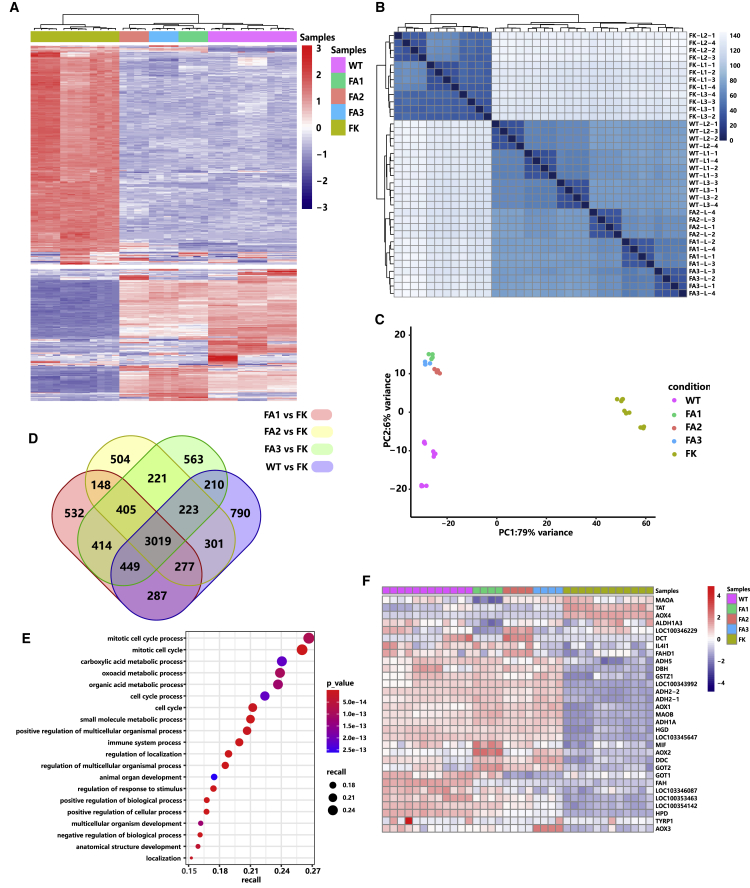
RNA sequencing (RNA-seq) analysis was performed to identify global gene expression patterns and define whether CRISPR-Cas9-mediated FAH gene correction could rescue global transcriptional profiles in the liver. Total RNAs were extracted from the liver tissues of the sacrificed three wild-type rabbits, three AAV-treated HT1 rabbits, and three untreated HT-1 rabbits for RNA-seq analysis. To support repeatability and reliability, we measured four replicates from each liver (two for the right lobe and two for the left lobe, referred to as WT-L1-1–4, WT-L2-1–4, and WT-L3-1–4 in wild-type rabbits; FA1-L-1–4, FA2-L-1–4, and FA3-L-1–4 in AAV-treated HT1 rabbits; and FK-L1-1–4, FK-L2-1–4, and FK-L3-1–4 in untreated HT1 rabbits). Data clustering indicated that the livers of the wild-type rabbits and AAV-treated HT1 rabbits were closer to each other in terms of global gene expression, whereas the livers of the untreated HT1 rabbits showed a more distinct expression (Figures 5A–5C). Compared with the livers of the AAV-untreated HT1 rabbits, the wild-type, FA1, FA2, and FA3 rabbits had 5,556, 5,531, 5,098, and 5,504 differentially expressed genes, respectively (Figure 5D). We specifically examined 3,019 commonly differentially expressed genes in the livers of the wild-type rabbits and AAV-treated HT1 rabbits. Gene Ontology (GO) enrichment analysis showed that the 3,019 commonly differentially expressed genes were primarily enriched in the metabolic signaling pathways, cell-cycle process, development process, immune system process, and biological and cellular processes (Figure 5E). The enriched metabolic signaling pathways include carboxylic acid metabolic process, oxoacid metabolic process, organic acid metabolic process, and small-molecule metabolic process. We further analyzed the commonly 1,238 upregulated (Figure S11A) and 1,756 downregulated genes (Figure S11B). GO enrichment analysis showed that the commonly upregulated genes were enriched in the metabolic signaling pathways, including terpenoid, steroid, alcohol, carboxylic acid, monocarboxylic acid, oxoacid, organic acid, lipid, small molecule, and cellular lipid metabolic process (Figure S11C), whereas the commonly downregulated genes were enriched in DNA replication, cell division, and immune system process (Figure S11D). This finding suggested that AAV-mediated gene therapy improves liver metabolism function. Steroid lipid and cellular lipid processes became normal, which may explain the high lipid and CHOL levels in the serum of the untreated HT1 rabbits and the low lipid and CHOL levels in the wild-type rabbits and AAV-treated HT1 rabbits (Figure S11C). Subsequently, we focused on examining the expression patterns of Tyr metabolic genes in the livers of the wild-type, AAV-treated, and AAV-untreated HT1 rabbits. Compared with the untreated HT1 rabbits, the AAV-treated HT1 rabbits exhibited upregulated to normal or close to normal levels of p-hydroxyphenylpyruvate dioxygenase (HPD, second step), homogentisate oxidase (HGD, third step), and maleylacetoacetate isomerase (GSTZ1, fourth step) in the livers. By contrast, Tyr aminotransferase (TAT, first step) was downregulated to normal or close to normal levels (Figure 5F). In addition, the bypass genes of Tyr metabolism, such as alcohol dehydrogenase 1A and 2 (ADH1A, ADH2), L-3,4-dihydroxyphenylalanine (DOPA) decarboxylase (DDC), glutamate oxalate transaminase 2 (GOT2), alternative oxidase 1, 2, 3, and 4 (AOX1, AOX2, AOX3, and AOX4), and monoamine oxidase A and B (MAOA, MAOB), were upregulated or downregulated to normal or close to normal expression levels. These results suggested that the repaired hepatocytes rescued the global gene expression pattern, particularly the genes of metabolic signaling pathways.
Figure 5.
Global Transcriptome Patterns of the Gene-Corrected HT1 Rabbits
(A) Hierarchical cluster analysis of the top 500 highest variance genes shows the transcriptome differences in the livers of the AAV-treated HT1 rabbits (FA1, FA2, and FA3), the PBS-treated HT1 rabbits (n = 3), and the wild-type rabbits (n = 3). Red and blue color intensities represent gene upregulation and downregulation, respectively. (B) Sample-to-sample distance analysis of the livers of the wild-type (n = 3), AAV-treated (n = 3), and PBS-treated (n = 3) HT1 rabbits. (C) Principal-component analysis of the wild-type rabbits (n = 3), AAV-treated HT1 rabbits (n = 3), and PBS-treated HT1 rabbits (n = 3). Points with different colors represent various sample groups. The percentage of variability explained by each component is shown on the axis. (D) Number of differentially expressed genes in the livers of the AAV-treated HT1 rabbits (n = 3) and the wild-type rabbits (n = 3) compared with the AAV-untreated HT1 rabbits (n = 3) (Venn diagram). (E) Gene Ontology (GO) enrichment analysis of the 3,019 commonly differentially expressed genes in (D). (F) Heatmap of the relative expression levels of genes related to the tyrosine metabolism pathway between wild-type rabbits (n = 3), AAV-treated HT1 rabbits (n = 3), and PBS-treated HT1 rabbits (n = 3).
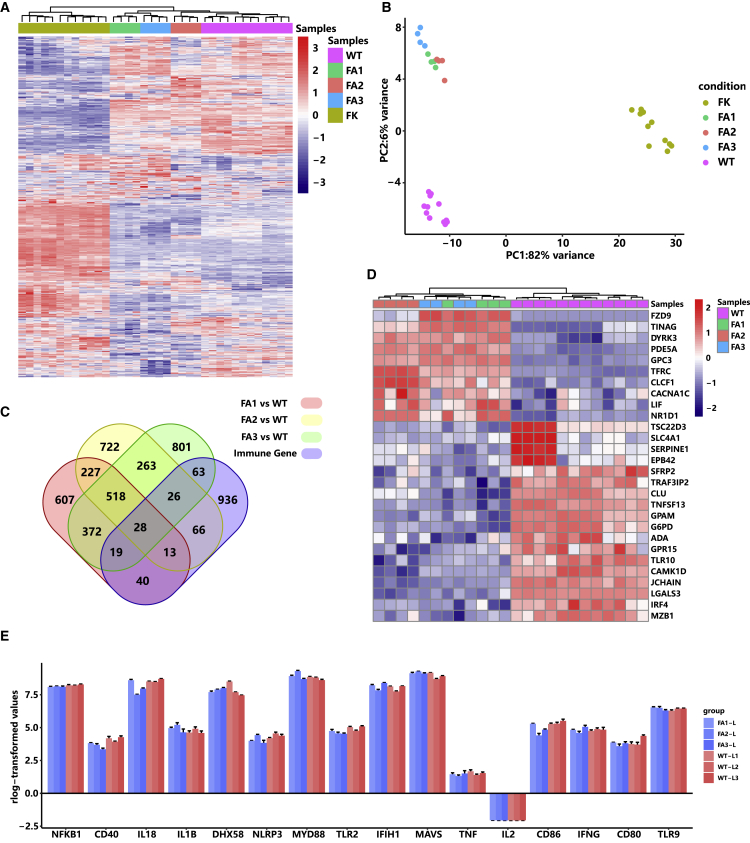
One main concern in AAV-mediated gene therapy is the immunogenicity of AAV protein capsid, its DNA genome, and the protein products of the transgenes. We further analyzed the liver transcriptomes of wild-type rabbits and AAV-treated and untreated HT1 rabbits to verify whether these antigens interact with host immune responses. Data clustering of the genes of the immune system process (GO: 0002376, including the process of the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats) showed that AAV-treated HT1 rabbits were closer to wild-type rabbits than AAV-untreated HT1 rabbits (Figures 6A and 6B). Compared with wild-type rabbits, only 28 commonly differentially expressed genes of the immune system process were identified in AAV-treated HT1 rabbits (Figure 6C). Among these 28 genes, only one key immune-related gene, ILR10, was involved, which suggested that mild immune responses occurred in AAV-treated rabbits (Figure 6D). The AAV-untreated rabbits had severe immune responses, which may be caused by liver and kidney damage and were consistent with the IHC results. We specifically analyzed the expression levels of inflammation-related genes, including NLRP3, IL1β, IL2, IL18, IFNγ, TNF, NFKB1, TLR2, TLR9, CD40, CD80, CD86, MAVS, IFIH1, and MYD88 (Figure 6E). Compared with those in wild-type rabbits, the expression levels of these genes did not change in AAV-treated rabbits, suggesting AAV treatment did not induce inflammatory responses.
Figure 6.
Gene Transcriptome Patterns of the Immune System Process in the Gene-Corrected HT1 Rabbits
(A and B) Hierarchical cluster analysis (A) and principal-component analysis (B) of the wild-type rabbits (n = 3), AAV-treated HT1 rabbits (n = 3), and PBS-treated HT1 rabbits (n = 3) using immune system process gene (n = 1,300). (C) Number of differentially expressed genes between AAV-treated HT1 rabbits and wild-type rabbits. There are 28 common differentially expressed genes of the immune system process. (D) Heatmap of the 28 common differentially expressed genes in the immune system process in (C). (E) Expression levels of the genes that have been reported to be involved in host immune responses to AAV.
Offspring of Gene-Corrected HT1 Rabbits
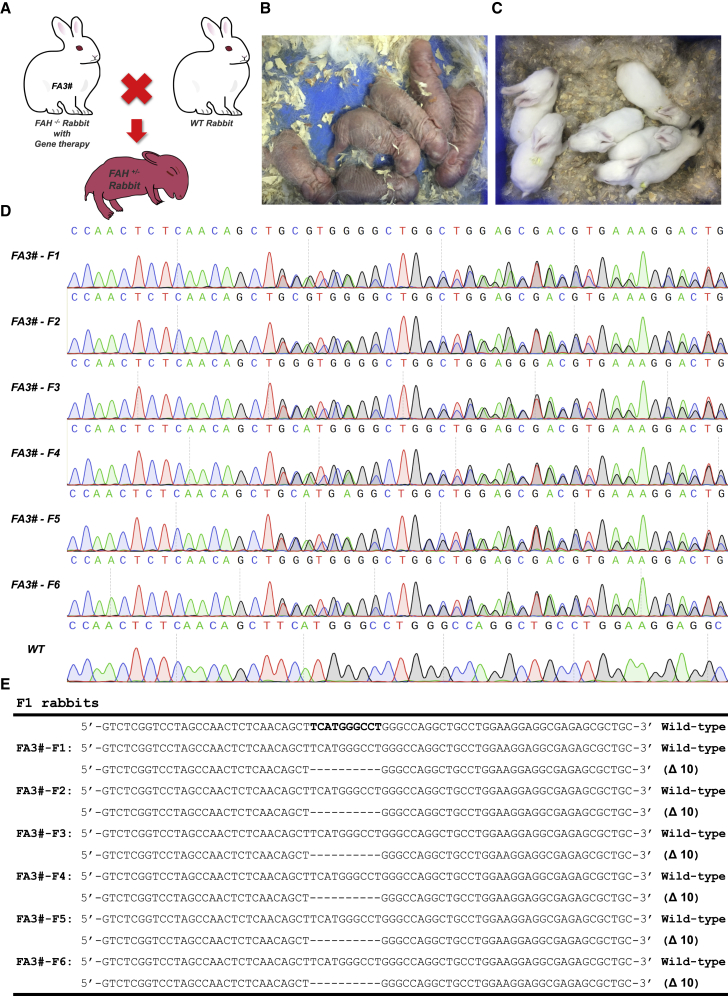
Before euthanasia, the AAV-treated FA3 HT1 rabbit was healthy and reached maturation age without overt abnormalities. The adult FA3 rabbit was mated with wild-type male rabbits (Figure 7A). The female FA3 rabbit became pregnant and delivered six bunnies without NTBC administration (Figures 7B and 7C). The ear tissues of the six rabbits were collected for the extraction of genomic DNAs. Gene modification was confirmed via the Sanger sequencing of PCR products that covered the FAH mutation sites. All six rabbits had one allele with 10-bp deletions at the FAH locus and the other remained intact (Figures 7D and 7E). All the offspring grew well, and no obvious differences were detected compared with the wild-type control. These data suggested that the AAV-treated HT1 rabbits can produce healthy heterozygous offspring.
Figure 7.
Gene-Corrected HT1 Rabbits Can Give Birth to Normal Offspring
(A) Overview of mating of a wild-type male rabbit with a gene-corrected female HT1 rabbit (FA3). (B and C) Photographs of the 1-day-old (B) and 20-day-old (C) F1 rabbits. (D) Sanger sequencing results of the PCR products of the FAH gene amplified from six F1 bunnies. (E) Summary of the genotypes of all six F1 bunnies.
Discussion
Patients with HT1 manifest with irreversible liver and kidney damage at a young age.3 In mice, the CRISPR-Cas9-mediated precise correction of Fah mutation was conducted in the adult stage.13, 14, 15, 16 However, the treatment of adult HT1 animal models has several disadvantages: (1) the underlying genetic defect may have already caused irreversible pathological changes; (2) the sufficient protein expression level for ameliorating or preventing the disease requires prohibitively large amounts of gene delivery vectors; (3) adult tissues may be poorly infected by vectors; and (4) the well-established immune responses in adult animals may produce additional antibodies for vectors, including Cas9 protein and AAV.32 These disadvantages will reduce treatment effectiveness. Early gene transfer in the neonatal or even fetal period may overcome these obstacles. Therefore, we started treatment in the newborn stage in our study on rabbits, during which gene corrections could be achieved in mutant cells before the development of the disease. For the gene therapy of the newborn rabbits, a dose of AAV with approximately 3 × 1012 genome copies of AAV8-SpCas9 and 6 × 1012 genome copies of AAV8-sgRNA4-Donor was applied. This dose was considerably lower than the required dose for adult rabbits (approximately 6 × 1013 genome copies of AAV8-SpCas9 and 1.2 × 1014 genome copies of AAV8-sgRNA4-Donor).
No HT1 symptoms were observed in all the AAV-treated rabbits, which reached sexual maturation age and produced offspring when mated with wild-type rabbits. Serum biochemical results showed that the common indices of hepatic function, namely, AST, ALT, TBIL, and DBIL, were within normal range at 11 weeks after treatment. No prominent liver swelling, hemorrhage, yellow/green discoloration, dysplastic hepatocytes, and inflammatory cellular infiltration (which occurred in the untreated HT1 rabbits) were found in the euthanized HT1 rabbits treated with AAV for 5 or 9 months. Whole mRNA deep sequencing showed that the metabolic pathways of the liver were rescued to a normal functional level. Interestingly, the metabolic processes of steroid, lipid, and cellular lipid were also rescued, which was consistent with the downregulation of CHOL and lipids to normal level in the serum of AAV-treated rabbits. Such a finding has not yet been described in similar treatment trials using mouse and rat models. The preceding results suggested that liver function was recovered at the molecular level.
In similar previous studies on mice, the information on whether kidney damage occurred in HT1 models or whether treatment was effective for kidney damage was not available, because the HT1 mouse model cannot recapitulate the renal symptoms of patients with HT1. The HT1 rabbit models exhibit typical kidney damages, including renal dysplasia, renal calculus, and renal medullary cystic lesion, which are similar to those in human patients with HT1. After AAV treatment, we found that the renal symptoms and the indices of renal function, namely, BUN and CREA, were normal compared with age-matched wild-type rabbits. The H&E staining results also confirmed that the kidneys of AAV-treated rabbits presented normal histology. Kidney damage is supposed to be caused by the accumulation of toxic metabolites, such as fumarylacetoacetate and SA, as a result of FAH gene mutation in patients. CRISPR-Cas9-mediated partial liver gene correction could recover the metabolic function of the liver. Because the gene correction was carried out in newborn rabbits, the reduction of toxic metabolites in the early stage of development for an individual patient should reduce kidney damage, although the FAH gene correction did not occur in the kidneys of AAV-treated HT1 rabbits. To our knowledge, this study is the first to simultaneously evaluate the effects of gene therapy treatment on the liver and kidney.
About 30% of human pathogenic mutations are caused by insertions or deletions.33 HT1 can be caused by different mutation patterns in the different exons of the FAH gene, including indels and point mutations. About 20% of patients harbor an insertion or deletion of a sequence. The mutation pattern in our HT1 rabbits was a 10-bp deletion in exon 2 of the FAH gene, which occurs among 20% of patients. The delivery of the CRISPR-Cas9 system and donor templates via AAV8 to HT1 rabbit hepatocytes resulted in HDR- and NHEJ-mediated (out-of-frame to in-frame) FAH gene correction. Theoretically, both mutation patterns could rescue full or truncated/lengthened FAH protein expression, although a previous paper reported that HDR is preferentially utilized in neonatal mammalian livers.30 The correction efficiency achieved by HDR and out-of-frame to in-frame mutations in HT1 rabbits treated with AAV for 5 months varied in different parts of the liver and ranged from 0.90% to 3.07% for HDR and from 2.39% to 5.51% for out-of-frame to in-frame mutations. The efficiencies of HDR-mediated precise gene correction and NHEJ-mediated out-of-frame to in-frame corrections ranged from 2.26% to 3.71% and from 4.38% to 6.35%, respectively, in the HT1 rabbits treated with AAV for 9 months. The corrected hepatocyte population would expand during the growth of an individual treated rabbit, and the rate of normal hepatocytes would become higher with the increase of time after treatment. Given that the younger newborn animal would achieve higher editing efficiency when tested in mice, we used 1-week-old HT1 rabbit instead of 15-day-old rabbit for gene therapy. Amplicon deep sequencing results showed that the initial proportion of cells in the liver through HDR-mediated gene correction could reach 2.93%, 2.68%, and 4.22%, which are even higher than we had observed in 15-day-old HT1 rabbits that were treated for 5 months. Therefore, we speculated that choosing an early stage of development, particularly right after birth, to perform gene therapy may be a better alternative approach to increase the effectiveness of HDR-mediated precise gene therapy and improve liver function.
We used deep sequencing to verify off-target events, which are a concern in CRISPR-Cas9 system-mediated gene therapy, and we did not discern indel mutations at the predicted OTSs in the AAV-treated HT1 rabbits. Our treated rabbits and their offspring with heterozygous mutation were able to grow normally. All of the evidence indicated that CRISPR-Cas9-mediated gene correction is a safe approach for treating HT1.
In summary, this study was the first attempt to demonstrate the efficacy and safety of the CRISPR-Cas9-mediated gene correction of monogenetic liver metabolic disease in large mammals in vivo. Our results demonstrated that the delivery of Cas9, sgRNA, and donor templates via AAV induced sufficient FAH+ hepatocytes and rescued liver and kidney damage in FAH knockout rabbits. This study provides preclinical efficacy and safety data in large animals for translating gene therapy into clinical practice.
Materials and Methods
Animals
New Zealand white rabbits were used in this study. All the experiments that involved rabbits were conducted under the approval of the Institutional Animal Care and Use Committee of Guangzhou Institutes of Biomedicine and Health (GIBH), Chinese Academy of Sciences (Animal Welfare Assurance #A5748-01).
The FAHΔ10/+ male and female rabbits obtained in the previous study were used as parents.21 To obtain FAHΔ10/Δ10 rabbits, we bred one FAHΔ10/+ male rabbit with four female rabbits. After 15 days of pregnancy, the pregnant FAHΔ10/+ female rabbits received 200 mL of drinking water per day with 7.5 mg/L NTBC. All the homozygotes were administered with NTBC treatment with the same dose as that in the pregnant FAHΔ10/+ female rabbits for 15 days, at which time point they were injected with AAV8-SpCas9 and AAV8-sgRNA4-Donor.
Plasmid Construction
sgRNA1, sgRNA2, sgRNA3, and sgRNA4 were designed in accordance with the locations of 10-bp deletions in exon 2 of rabbit FAH gene. The complementary oligonucleotides of sgRNAs were synthesized and then annealed to double-stranded DNAs. The annealed products were then cloned into the BbsI-digested U6-sgRNA cloning vector to form sgRNA-expressing plasmid. A repaired donor template with seven synonymous mutations was constructed by using the ClonExpress MultiS One Step Cloning Kit (C113-02; Vazyme Biotech). Synonymous mutations were introduced into the PCR primers (FAH-HDR-R1: 5′-CGCGTCTTTCCACGCTGCTTGCCCGAGGCCCATGAAGCTGTTGAGAG-3′ and FAH-HDR-F2: 5′-CCTCGGGCAAGCAGCGTGGAAAGACGCGAGAGCGCTGCTGCAAAACT-3′). The optimal sgRNA-expressing cassette and the repaired donor template were then cloned into the A2 vector. The primers used in this study are listed in Table S1.
RFFs Culture and Transfection
RFFs were isolated from the 15-day-old fetuses of wild-type New Zealand white rabbits. After removing the heads, tails, limbs, and viscera, the remaining fetuses were digested with 0.5 mg/mL collagenase IV for 2 h in a cell incubator at 38°C. The isolated RFFs were cultured in the RFF culture medium (Dulbecco’s modified Eagle’s medium; HyClone), supplemented with 15% fetal bovine serum (FBS; GIBCO), 1% nonessential amino acids (GIBCO), 2 mM GlutaMAX (GIBCO), 1 mM sodium pyruvate (GIBCO), and 2% penicillin-streptomycin (HyClone) for 12 h and then frozen in a cell freezing medium (90% FBS and 10% dimethyl sulfoxide). The RFFs were thawed and cultured in 10-cm plates to sub-confluence a day before transfection. Then 1 × 106 RFFs were electroporated with pcDNA3.1-SpCas9 (10 μg) and sgRNA-expressing vectors (3 μg of each sgRNA) at 1,350 V, 30 ms, and 1 pulse using the Neon transfection system (Life Technology). The transfected RFFs were collected after 4 days of G418 selection and used for genomic DNAs extractions using TIANamp Genomic DNA Kit (TIANGEN). Genomic DNAs were used as templates for PCR. The PCR products were subjected to restriction enzyme digestion using T7EN1 (Vazyme Biotech), followed by Sanger sequencing. The PCR products of sgRNA1 and sgRNA4 were further sub-cloned into the pMD18-T vector (Takara) and sequenced to determine the mutation patterns and efficiencies.
T7EN1 Cleavage Assay
PCR was performed to amplify the genomic sequences around the target sites by using the KOD One PCR Master Mix (KMM-201; TOYOBO). The PCR products were purified by using the HiPure Gel Pure DNA Mini Kit (Magen, D2111-03). Approximately 200 ng of each PCR product was annealed under the following conditions: 95°C for 5 min, 95–85°C at −2°C/s, 85°C–25°C at −0.1°C/s, and held at 4°C. The annealed products were incubated with 10 U T7EN1 (EN303-01/02; Vazyme Biotech) at 37°C for 15 min. The reaction was quenched by adding 1.5 μL of 0.25 M EDTA, and the digested PCR products were separated with 2% agarose gel.
AAV8 Production and Purification
AAV8 was produced and purified by PackGene Biotech. Donor template and optimal sgRNA with U6 promoter were packaged into an AAV8 with mCherry fluorescent reporter, referred to as AAV8-sgRNA4-Donor. SpCas9 with the miniCMV promoter was packaged into the other AAV8, referred to as AAV8- SpCas9. The purified AAV8 was stored in a freezer at −80°C.
Virus Delivery
AAV8-SpCas9 (0.1 mL per rabbit, 3 × 1013 genome copies/mL) and AAV8-sgRNA4-Donor (0.15 mL per rabbit, 4 × 1013 genome copies/mL) were thawed and mixed. The virus mixture was injected into three 15-day-old FAHΔ10/Δ10 rabbits via intravenous injection. The control group (the three remaining HT1 rabbits) was injected with 0.25 mL of PBS.
Genomic DNA Extraction and Genotyping
Genomic DNAs were extracted from the ear of newborn rabbits and the liver tissues of wild-type rabbits, AAV-treated rabbits, and untreated HT1 rabbits by using TIANamp Genomic DNA Kit (TIANGEN). The newborn rabbits were genotyped by PCR and Sanger sequencing. The PCR primers used were as follows: FAH-F: 5′-CAGGTCTCAGGTTACAGAGC-3′ and FAH-R: 5′-AGGTGCATCGTGGCAACAGC-3′. The PCR products were subjected to target deep sequencing by Guangzhou IGE Biotechnology to genotype liver tissues.
Hematological Analysis
Blood was collected by puncturing the ear artery. Serum was separated by centrifugation at 900 × g for 20 min and stored at −80°C prior to biochemistry and amino acid analyses. Blood biochemical indicators were measured by the Guangdong Provincial Animal Quality Monitoring Institute.
H&E Staining, Sirius Red Staining, and IHC
The rabbits were euthanized by injecting air intravenously. The liver and kidney tissues obtained from wild-type, AAV-treated, and AAV-untreated rabbits were fixed in situ with 4% buffered paraformaldehyde for 2 days. The fixed tissues were dissected, embedded into paraffin wax, and then cross-sectioned at 3 μm to a slide by using a vibratome. The liver and kidney slides were deparaffinized with xylene and subsequently rehydrated in a graded series of alcohol (100%, 90%, 80%, 70%, and 50%), followed by distilled water (dH2O) for H&E staining. Lastly, the rehydrated slides were stained with H&E for routine histology and with Sirius red to analyze for liver fibrosis, and then coverslipped. Liver sections were dewaxed, rehydrated, and stained with anti-FAH antibody (ab140167, 1:400 dilution; Abcam) and anti-mCherry antibody (ab125096, 1:500 dilution; Abcam) for IHC staining.
Western Blot Analysis
The total proteins of the liver tissues obtained from the wild-type, AAV-treated, and AAV-untreated rabbits were extracted using a Minute total protein extraction kit (for animal cultured cells and tissues) (SD-001/SN-002; Invent Biotechnologies) in accordance with the protocol. The extracted total proteins were quantified by BCA assay and boiled with sodium dodecyl sulfate (SDS) loading buffer (62.6 mM Tris-HCl, 10% glycerol, 0.01% bromophenol blue, and 2% SDS [pH 6.8]) for 15 min. Equal amounts of proteins were resolved by 10% SDS-polyacrylamide gel electrophoresis and then transferred to polyvinylidene fluoride membranes (Millipore). The membranes were blocked with 5% skim milk in Tris-buffered saline with Tween (TBST) for 2 h, then incubated with primary antibodies against FAH protein for 2 h at room temperature (ab140167, 1:1,000 dilution; Abcam) and rinsed three times with TBST. The membranes were added with a horseradish-conjugated secondary antibody (sc-2004, 1:5,000 dilution; Santa Cruz Biotechnology) applied to incubate the membranes for 1 h at room temperature after rewashing three times with TBST. Then the protein signal was visualized using ECL Plus (Amersham) in accordance with the manufacturer’s instructions. In addition, the membranes were re-probed with an actin antibody (sc-47778, 1:5,000 dilution; Santa Cruz Biotechnology) as the loading control.
RNA-Seq and Data Analysis
The total mRNA of the livers of the wild-type, AAV-treated, and AAV-untreated rabbits was isolated with RNAiso Plus (TaKaRa) and sent to Annoroad Gene Technology Corporation (Beijing) for database construction and sequencing. A total of 2 μg of RNA per sample was used as input material for the RNA sample preparation. Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (E7530L; NEB) following the manufacturer’s recommendations, and index codes were added to attribute the sequences to each sample. After cluster generation, the libraries were sequenced on an Illumina platform, and 150-bp paired-end reads were generated.
Off-Target Analysis
The potential OTSs for sgRNA4 were predicted in the rabbit genome in accordance with an online design tool (http://www.rgenome.net/cas-offinder/),31 which allows the un-gapped alignment with up to two mismatches in the sgRNA target sequence. All potential OTSs were amplified by PCR and then subjected to deep sequencing to confirm the off-target effects. The primers used for amplifying the OTSs are listed in Table S1.
Statistical Analysis
The GraphPad Prism software (version 8) was used for data analysis. The data of serum biochemical analysis are presented as the mean values ± SD. The two-tailed Student’s t test was used to assess a significant difference. p <0.05 was considered statistically significant.
Data Availability
The high-throughput sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) and the accession number is GEO: GSE142722 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142722).
Acknowledgments
We thank lab animal center of GIBH for assistance with animal feeding and care. We also thank all the members in the laboratory of Prof. Liangxue Lai. This work was financially supported by National Key Research and Development Program of China (2017YFA0105103); National Natural Science Foundation of China (81941004, 81702115, 81672317, and 81800555); the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16030503); Key Research & Development Program of Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory) (2018GZR110104004); the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2019347); China Postdoctoral Science Foundation (2020M682945); Science and Technology Planning Project of Guangdong Province, China (2020B1212060052, 2017B020231001, and 2014B020225003); Science and Technology Program of Guangzhou, China (201704030034, 201907010039, 202002030382, and 202007030003); and Research Unit of Generation of Large Animal Disease Models, Chinese Academy of Medical Sciences (2019-I2M-5-025).
Author Contributions
N.L. and J.W. performed most of the experiments and analyzed the data. S.G. performed the informatics analyses. Q.Z. and Z.O. participated in AAV injection and provided animal care. J.X., L.L., Q.J., F.C., W.G., H.S., Y.L., H.X., Z.Z., X.Z., M.L., Y.Y., and L.Q. provided technical assistance. K.W., L.L., and H.W. conceived, designed, and supervised the project. K.W. and L.L. prepared the manuscript.
Declaration of Interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.11.023.
Contributor Information
Han Wu, Email: wu_han@gibh.ac.cn.
Liangxue Lai, Email: lai_liangxue@gibh.ac.cn.
Kepin Wang, Email: wang_kepin@gibh.ac.cn.
Supplemental Information
References
- 1.Lindstedt S., Holme E., Lock E.A., Hjalmarson O., Strandvik B. Treatment of hereditary tyrosinaemia type I by inhibition of 4-hydroxyphenylpyruvate dioxygenase. Lancet. 1992;340:813–817. doi: 10.1016/0140-6736(92)92685-9. [DOI] [PubMed] [Google Scholar]
- 2.St-Louis M., Tanguay R.M. Mutations in the fumarylacetoacetate hydrolase gene causing hereditary tyrosinemia type I: overview. Hum. Mutat. 1997;9:291–299. doi: 10.1002/(SICI)1098-1004(1997)9:4<291::AID-HUMU1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 3.Russo P., O’Regan S. Visceral pathology of hereditary tyrosinemia type I. Am. J. Hum. Genet. 1990;47:317–324. [PMC free article] [PubMed] [Google Scholar]
- 4.Mayorandan S., Meyer U., Gokcay G., Segarra N.G., de Baulny H.O., van Spronsen F., Zeman J., de Laet C., Spiekerkoetter U., Thimm E. Cross-sectional study of 168 patients with hepatorenal tyrosinaemia and implications for clinical practice. Orphanet J. Rare Dis. 2014;9:107. doi: 10.1186/s13023-014-0107-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adam R., Karam V., Delvart V., O’Grady J., Mirza D., Klempnauer J., Castaing D., Neuhaus P., Jamieson N., Salizzoni M., All contributing centers (www.eltr.org); European Liver and Intestine Transplant Association (ELITA Evolution of indications and results of liver transplantation in Europe. A report from the European Liver Transplant Registry (ELTR) J. Hepatol. 2012;57:675–688. doi: 10.1016/j.jhep.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 6.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amoasii L., Hildyard J.C.W., Li H., Sanchez-Ortiz E., Mireault A., Caballero D., Harron R., Stathopoulou T.R., Massey C., Shelton J.M. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362:86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y., Wang L., Bell P., McMenamin D., He Z., White J., Yu H., Xu C., Morizono H., Musunuru K. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beyret E., Liao H.K., Yamamoto M., Hernandez-Benitez R., Fu Y., Erikson G., Reddy P., Izpisua Belmonte J.C. Single-dose CRISPR-Cas9 therapy extends lifespan of mice with Hutchinson-Gilford progeria syndrome. Nat. Med. 2019;25:419–422. doi: 10.1038/s41591-019-0343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santiago-Fernández O., Osorio F.G., Quesada V., Rodríguez F., Basso S., Maeso D., Rolas L., Barkaway A., Nourshargh S., Folgueras A.R. Development of a CRISPR/Cas9-based therapy for Hutchinson-Gilford progeria syndrome. Nat. Med. 2019;25:423–426. doi: 10.1038/s41591-018-0338-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paulk N.K., Wursthorn K., Wang Z., Finegold M.J., Kay M.A., Grompe M. Adeno-associated virus gene repair corrects a mouse model of hereditary tyrosinemia in vivo. Hepatology. 2010;51:1200–1208. doi: 10.1002/hep.23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song C.-Q., Jiang T., Richter M., Rhym L.H., Koblan L.W., Zafra M.P., Schatoff E.M., Doman J.L., Cao Y., Dow L.E. Adenine base editing in an adult mouse model of tyrosinaemia. Nat. Biomed. Eng. 2020;4:125–130. doi: 10.1038/s41551-019-0357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao Y., Wang L., Guo N., Wang S., Yang L., Li Y., Wang M., Yin S., Han H., Zeng L. Cas9-nickase-mediated genome editing corrects hereditary tyrosinemia in rats. J. Biol. Chem. 2018;293:6883–6892. doi: 10.1074/jbc.RA117.000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs S.M.M., van Beurden D.H.A., Klomp L.W.J., Berger R., van den Berg I.E.T. Kidneys of mice with hereditary tyrosinemia type I are extremely sensitive to cytotoxicity. Pediatr. Res. 2006;59:365–370. doi: 10.1203/01.pdr.0000198810.57642.b4. [DOI] [PubMed] [Google Scholar]
- 18.Bosze Z., Houdebine L.M. Application of rabbits in biomedical research: A review. World Rabbit Sci. 2006;14:1–14. [Google Scholar]
- 19.Fan J., Watanabe T. Transgenic rabbits as therapeutic protein bioreactors and human disease models. Pharmacol. Ther. 2003;99:261–282. doi: 10.1016/s0163-7258(03)00069-x. [DOI] [PubMed] [Google Scholar]
- 20.Fan J., Kitajima S., Watanabe T., Xu J., Zhang J., Liu E., Chen Y.E. Rabbit models for the study of human atherosclerosis: from pathophysiological mechanisms to translational medicine. Pharmacol. Ther. 2015;146:104–119. doi: 10.1016/j.pharmthera.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L., Zhang Q., Yang H., Zou Q., Lai C., Jiang F., Zhao P., Luo Z., Yang J., Chen Q. Fumarylacetoacetate Hydrolase Knock-out Rabbit Model for Hereditary Tyrosinemia Type 1. J. Biol. Chem. 2017;292:4755–4763. doi: 10.1074/jbc.M116.764787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hickey R.D., Lillegard J.B., Fisher J.E., McKenzie T.J., Hofherr S.E., Finegold M.J., Nyberg S.L., Grompe M. Efficient production of Fah-null heterozygote pigs by chimeric adeno-associated virus-mediated gene knockout and somatic cell nuclear transfer. Hepatology. 2011;54:1351–1359. doi: 10.1002/hep.24490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hickey R.D., Mao S.A., Glorioso J., Lillegard J.B., Fisher J.E., Amiot B., Rinaldo P., Harding C.O., Marler R., Finegold M.J. Fumarylacetoacetate hydrolase deficient pigs are a novel large animal model of metabolic liver disease. Stem Cell Res. (Amst.) 2014;13:144–153. doi: 10.1016/j.scr.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hickey R.D., Mao S.A., Glorioso J., Elgilani F., Amiot B., Chen H., Rinaldo P., Marler R., Jiang H., DeGrado T.R. Curative ex vivo liver-directed gene therapy in a pig model of hereditary tyrosinemia type 1. Sci. Transl. Med. 2016;8:349ra99. doi: 10.1126/scitranslmed.aaf3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicolas C.T., Hickey R.D., Allen K.L., Du Z., VanLith C.J., Guthman R.M., Amiot B., Suksanpaisan L., Han B., Francipane M.G. Ectopic hepatocyte transplantation cures the pig model of tyrosinemia. bioRxiv. 2019 doi: 10.1101/648493. [DOI] [Google Scholar]
- 26.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sands M.S. AAV-mediated liver-directed gene therapy. Methods Mol. Biol. 2011;807:141–157. doi: 10.1007/978-1-61779-370-7_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakai H., Fuess S., Storm T.A., Muramatsu S., Nara Y., Kay M.A. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J. Virol. 2005;79:214–224. doi: 10.1128/JVI.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L., Shao Y., Li L., Tian F., Cen J., Chen X., Hu D., Zhou Y., Xie W., Zheng Y. Efficient liver repopulation of transplanted hepatocyte prevents cirrhosis in a rat model of hereditary tyrosinemia type I. Sci. Rep. 2016;6:31460. doi: 10.1038/srep31460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anguela X.M., Sharma R., Doyon Y., Wechsler T., Paschon D., Davidson R.J., Zhou S., Gregory P.D., Holmes M.C., High K.A. In Vivo Genome Editing in Neonatal Mouse Liver Preferentially Utilizes Homology Directed Repair. Blood. 2015;126:4422. [Google Scholar]
- 31.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waddington S.N., Kennea N.L., Buckley S.M.K., Gregory L.G., Themis M., Coutelle C. Fetal and neonatal gene therapy: benefits and pitfalls. Gene Ther. 2004;11(Suppl 1):S92–S97. doi: 10.1038/sj.gt.3302375. [DOI] [PubMed] [Google Scholar]
- 33.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The high-throughput sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) and the accession number is GEO: GSE142722 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142722).