Abstract
Antimicrobial resistance (AMR) continues to exert a substantial toll on the global health and world economy and is now expected to be hidden by COVID-19 for a while. The wrong consumption of antibiotics during the COVID-19 pandemic will raise disastrous effects on AMR management and antibiotic stewardship programs. This is related to the concerns extrapolated due to an increase in mortality rates in patients with bacterial coinfections. Importantly, the immune system of COVID-19 patients in regions with high AMR may be fighting on two fronts altogether, the virus and MDR bacteria. Current control policies to manage AMR and prioritization of antibiotic stewardship plans are mandatory during this pandemic. This review aims to discuss the rising concerns of the excess use of antibiotics in COVID-19 patients highlighting the role of bacterial coinfections in these patients. Types of prescribed antibiotics and the development of antibiotic resistance is addressed as well.
Keywords: Antimicrobial resistance, COVID-19, Secondary bacterial infection, Stewardship
Introduction
The escalating increase in numbers of multidrug-resistant bacterial infections that are undetected, undiagnosed, and increasingly uncurable infections menace the health of people worldwide [1]. In the last two decades, antimicrobial resistance (AMR) is considered a global health and development threat. WHO has declared that AMR is one of the top 10 global public health threats facing humanity. Since the antibiotic era, with the discovery of the first antibiotics that were considered miracle drugs that saved lives, the misuse and abuse of antimicrobials in veterinary and human medicine have augmented the rising worldwide spectacle of AMR [2]. Moreover, morbidity and mortality from infectious diseases are significant consequences of AMR affecting patients [3,4]. A review paper published in 2014 showed that the estimated number of deaths due to infections with multiple drug resistant pathogens by the year 2050 is expected to reach 10 million/year in case of not applying a strict action plan to combat AMR [5]. Additionally, the estimated mortality rates according to the geographical regions were also reported by this review as follows; Asia, Africa, Europe, Latin America, North America, and Oceania 4,730,000, 4,150,000, 390,000, 392,000, 317,000, and 22,000, respectively [5]. At this time, the SARS-CoV-2 pandemic is ruling every aspect of health care across the world, putting other longer-term public health issues—taking in the increase of antimicrobial resistance [6]. Many health problems will be exacerbated by this pandemic and will persist for a time after COVID-19. It is noteworthy to mention that the deadly nature of influenza during the 1918 pandemic were explained by secondary bacterial pneumonias [7,8]. In case of respiratory viral illness as influenza or COVID-19; both innate and adaptive antibacterial host defences are impaired, colonising bacteria take advantage of this temporary compromise of a physical and immunological barrier to cause secondary bacterial pneumonias, causing worse outcomes and deadly illness particularly in patients with comorbidities and previously healthy people [9]. The rationale for prescribing antibiotics in patients with COVID-19 might be due to the previous experience with increased mortality in patients due to bacterial superinfection in influenza, where many studies demonstrated initial co-infection or secondary bacterial pneumonia in hospitalized patients [10] as well as difficulty in ruling out bacterial co-infection on patient's presentation. Moreover, physicians will and should on the side of treating critically ill hospitalized COVID-19 patients whom the secondary bacterial infections are not certain with broad spectrum antibiotics. Therefore, decline in adhering to stewardship programs is noticed as the health care providers struggle to save the COVID-19 patients [9]. Though, the difficulties in differentiating pneumoniae due to bacterial infection or COVID-19 infection and the absence of antiviral therapy with confirmed efficacy and consequently the urgent need of using antibiotics as part of empirical strategy in treating COVID-19 patients, particularly the critically ill. It is extremely important to reasonably use the antibiotics even during the pandemic to avoid much complicated consequences. However, this hypothesis raises concerns of the outcomes due to the antibiotic overuse and increased rates of bacterial resistance. Here, we review the impact of current COVID-19 pandemic on escalating the threat of AMR highlighting the association of bacterial coinfections in COVID-19 patients.
Antimicrobial resistance and its role in COVID-19
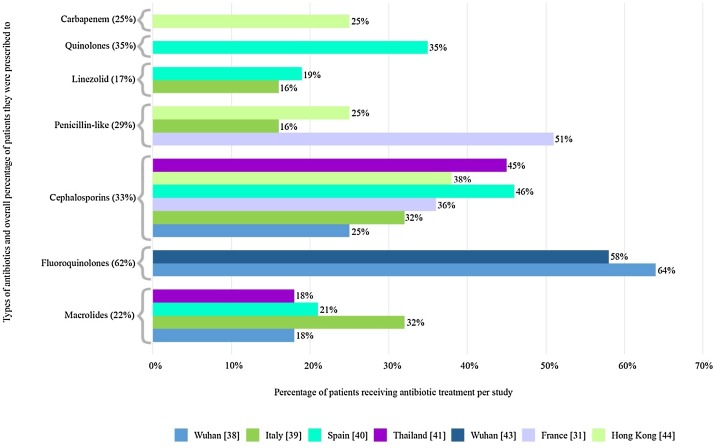
Monitoring and understanding the prevalence, mechanisms and spread of AMR are priorities for both individual patient care and global infection control strategies [11]. With regards to COVID-19 patients, it has been reported that up to 72% of patients admitted to hospitals received antibiotic treatment [12]. The information on the use of antibiotics to treat COVID-19 infection is easily accessible to the public on the internet beside the increased pressure exerted on clinical settings and hospitals [13]. The lack of sufficient knowledge of the value of antibiotics together with the fear of COVID-19 infection has directly impacted the increased access to the over-the-counter antibiotics, particularly in low- and middle-income countries with poor antibiotic control actions and restricted access to health settings [13]. In this context, it has been reported that 68.9% of COVID-19 patients had received antibiotics (mostly azithromycin and ceftriaxone) with a self-medication rate of 33.0% before hospital admission [14]. The gigantic use of biocides and disinfectants during the pandemic will cause unusual liberation of such antimicrobials into environmental resources and further select antibiotic resistant organisms [15]. Collectively, the selection and the development of highly resistant bacteria due to the aggravated use of antibiotics as well as disinfectants due to the above-mentioned reasons will affect the clinical prognosis of severe COVID-19 patients receiving care at hospitals and lead to poor patients' outcomes. In this context, extremely and pan drug-resistant organisms have been reported to cause serious coinfections in COVID-19 patients [16] and unfortunately, death of cases who encountered bacterial coinfections together with COVID-19 is recently reported [17]. A retrospective cohort analysis of 191 patients from two hospitals in Wuhan, Zhou et al. reported that 95% of patients were treated with antibiotics along with 21% treated with antivirals [18]. Death rate of 11% among 99 patients was also reported in a study in Wuhan, and 71% were treated with antibiotics; however, only 1% experienced co-infection with bacteria and 4% had fungal co-infections [36]. Moreover, antibiotic therapy was administered to all patients in a single-centre study conducted in Wuhan which involved 36 non-survivors with COVID-19, and 61.1% of the cases used combined antibiotic therapy whereas 38.9% received single antibiotic [19]. With regards to the recommendations of the use of antibiotics in patients with suspected or confirmed COVID-19 differ by country, with some recommendations encouraging antibiotics administration in a large proportion of patients, particularly because it is not well understood how ‘pneumonia’ may cause radiological alterations even in asymptomatic patients in the context of a viral infection, consequently, many documents recommend antibiotic therapy for patients with COVID-19 and ‘pneumonia’ [20]. It is noteworthy to mention that macrolides (azithromycin) was the second most prescribed treatment for COVID-19, and 41% of respondents reported that they had prescribed azithromycin for COVID-19 [21]. Azithromycin is an existing medication that is repurposed for the current pandemic since novel Coronavirus treatments are being developed. However, other study advised against using azithromycin except in treating community acquired pneumonia (CAP) as they declared that ” there is no clear evidence that azithromycin may exert beneficial effects in COVID-19 beyond antibacterial activity in bacterial superinfection” [22]. It has also been recommended that certain antibacterials, for example teicoplanin (a glycopeptide antibiotic) could be promising in treating COVID-19 after previously exhibiting antiviral activity against coronaviruses (amongst others) [23]. Data from 7 countries confirmed that COVID-19 patients had received antibiotics, these data are shown in Table 1 . Fig. 1 also groups the different studies per antibiotic from Table 1, displaying their respective percentages as well as the total percentage as represented in Table 1. Despite this, excessive caution should be performed given that misuse or overuse of antibiotics is well-known to be a significant driver of the prevalence of AMR. A group of European clinicians state that it can be difficult to distinguish COVID-19 from bacterial pneumonia, which means that some patients with no bacterial infections are getting unnecessary antibiotics [6]. AMR affects all countries, but the burden is disproportionately higher in low middle income countries (LMICs) [24]. The incidence of COVID-19 in LMICs is increasing now, raising serious concerns given the weak health systems, low awareness, and limited preparedness for outbreaks in many LMICs. Rise in AMR in LMICS could be amplified by a large COVID-19 pandemic [25]. The authors of a recent summary suggested that the empirical antibacterial therapy for hospitalized COVID-19 patients with suspected bacterial coinfections should not exceed 5 days upon improvement of signs and symptoms [26].The authors added that shortening of the duration of antibiotics in patients with sepsis can be achieved by the support of procalcitonin levels [26]. As mentioned above, it is difficult to differentiate between bacterial and viral pneumonia, the Infectious Disease Society of America (IDSA) guideline on community acquired pneumonia (CAP) concluded that procalcitonin cannot be used in the decision to start or withhold antibiotics in patients with CAP [31]. As for hospital acquired pneumonia (HAP) and ventilator associated pneumonia (VAP), the IDSA guideline conducted broad evidence summaries to assess the added value of using procalcitonin, CRP, or the Modified Clinical Pulmonary Infection Score plus clinical criteria for the diagnosis of HAP or VAP [12]. They declared that none of these modalities were of extra value when compared to clinical data alone.
Table 1.
Different antibiotics prescribed to COVID-19 patients and their known AMR threats.
| Prescribed antibiotics in cases of Covid-19 | Antimicrobial resistance (known) | No. of patients and geographical location | Total | References |
|---|---|---|---|---|
| Azithromycin, clarithromycin (macrolides) | Helicobacter sp., Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes | 18% Wuhan (25/138) | 54/250 (22%) | [[36], [37], [38], [39], [40], [41]] |
| 32% Italy (17/53) | ||||
| 21% Spain (10/48) | ||||
| 18% Thailand (2/11) | ||||
| Moxifloxacin (fluoroquinolones) | Clostridium difficile, Mycobacterium tuberculosis,Neisseria gonorrhoeae, Pseudomonas Aeruginosa | 64% Wuhan (88/138) | 127/205 (62%) | [38,42,43] |
| 58% Wuhan (39/67) | ||||
| Ceftriaxone, Cefoxatime (cephalosporins) | ESBL-Producing Enterobacteriaceae, N. gonorrhoeae, P. aeruginosa | 25% Wuhan (35/138) | 112/338 (33%) | [31,[38], [39], [40], [41], [42],44] |
| 32% Italy (17/53) | ||||
| 36% France (14/39) | ||||
| 46% Spain (22/48) | ||||
| 38% Hong Kong (19/49) | ||||
| 45% Thailand (5/11) | ||||
| Piperacillin-tazobactam, Oxacillin, Amoxicillin (penicillin-like) | ESBL-Producing Enterobacteriaceae, S. aureus, S. pneumoniae | 51% France (20/39) | 41/141 (29%) | [11,39,42,31,44] |
| 16% Italy (9/53) | ||||
| 25% Hong Kong (12/49) | ||||
| Linezolid (oxazolidinone) | Enterococcus faecium, Enterococcus faecalis, Staphylococcus epidermidis | 16% Italy (9/53) | 18/101 (17%) | [39,40,45,46] |
| 19% Spain (9/48) | ||||
| levofloxacin (quinolone) | Escherichia coli | 35% Spain (17/48) | 17/48 (35%) | [40,47] |
| Meropenem (carbapenem) | Enterobacteriaceae sp., Klebsiella pneumoniae | 25% Hong Kong (12/49) | 12/49 (25%) | [44,48,49] |
Fig. 1.
Percentage (per study) of patients receiving specific antibiotics and total percentage of patients they were prescribed to.
Bacterial Co-infections
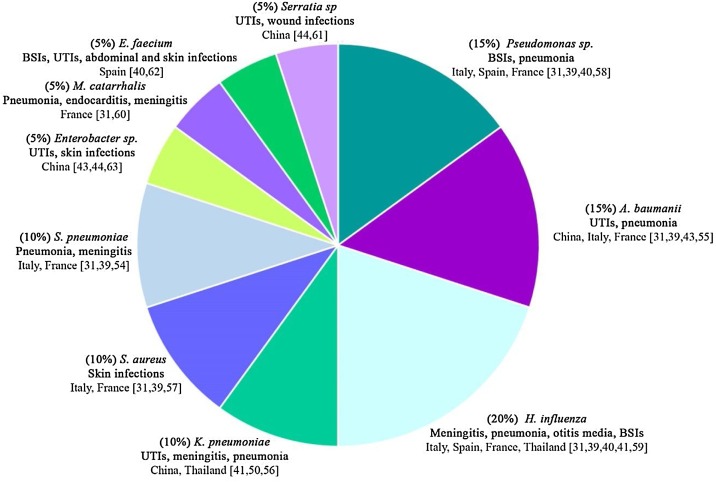
Patients presented with respiratory viral infections are predisposed to secondary bacterial infections and these result in increased disease severity and mortality [27].COVID-19 presents with a spectrum of disease severity, ranging from symptomless and non-specific flu-like symptoms, to pneumonia, and life-threatening complications such as acute respiratory distress syndrome (ARDS) and multiple organ failure [28]. Moreover, secondary bacterial infections are proven to represent a major risk factor for adverse COVID-19 outcomes. In spite of the proven importance of co-infections in the severity of respiratory illness presentations, they are unexplored during large pandemics of respiratory infections [27]. Data of patients with confirmed bacterial coinfection with COVID-19 are demonstrated by several studies and shown in Table 2 and Fig. 2 . The results of a meta-analysis and systemic review identified 24 studies focusing on bacterial coinfections in hospitalized COVID-19 patients revealed that 3.5% of patients experienced coinfections and secondary infection was detected in 14.3% of COVID-19 patients with overall bacterial infection 6.9%. Similar findings were also reported by other meta-analysis and systemic review that included 30 studies evaluating coinfections among COVID-19 patients; they reported that 7% of the patients had bacterial coinfections [29]. The findings of narrative review showed that percentage of coinfections in COVID-19 patients extensively varying from no co-infections to 100% coinfections in patients who died, and broad inconsistency of antibiotic use by severity of illness, ranging from 20% to 100% for antibiotics [30]. The findings of a retrospective monocenter study that was conducted in France revealed that a rate of 28% bacterial coinfections were detected in patients with severe SARS-CoV-2 and the authors highly encouraged the use of systemic empiric antibiotic treatment with a third generation cephalosporins. The bacterial coinfections detected in their study were mostly due to due to methicillin-sensitive Staphylococcus aureus, Haemophilus influenzae, Streptococcus pneumoniae and Enterobacteriaceae [31].
Table 2.
Percentage of patients receiving antibiotics and percentage of confirmed secondary infections.
| % Covid-19 patients treated with antibiotics | Confirmed secondary bacterial infection | Infecting agent (where reported) | Geographical location | References |
|---|---|---|---|---|
| 41/41 (100%) | 4/41 (10%) | – | Wuhan, China | [19] |
| 49/52 (94%) | 1/94 (1%) | K. pneumoniae | Wuhan, China | [50] |
| 26/53 (49%) | 16/53 (30%) | A. baumannii, Haemophilus influenza, P. aeruginosa, S. aureus, S. pneumoniae | Milan, Italy | [39] |
| 39/92 (42%) | 26/92 (28%) | A. baumannii, H. influenza, Moraxella catarrhalis, P. aeruginosa, S. aureus, S. pneumoniae | France | [31] |
| 181/191 (95%) | 28/191 (15%) | – | Wuhan, China | [18] |
| 143/150 (95%) | 12/150 (8%) | – | Wuhan China | [51] |
| 42/48 (88%) | 6/48 (13%) | E. faecium, H. influenza, P. aeruginosa | Vitoria, Spain | [40] |
| 37/298 (12%) | 30/298 (10%) | – | Shenzhen City, China | [52] |
| 319/476 (67%) | 35/410 (9%) | – | Wuhan, Shanghai, and Anhui (China) | [53] |
| 49/49 (100%) | 2/49 (4%) | Serratia sp., Enterobacter sp. | Hong Kong, China | [44] |
| 6/11(55%) | 6/11 (55%) | H. influenza, K. pneumoniae | Bangkok, Thailand | [41] |
| 66/67 (99%) | 3/28 (11%) | A. baumanii, Enterobacter cloacae | Wuhan, China | [43] |
| 1824/2536 (72%) | 190/1923 (10%) |
Fig. 2.
Different infecting agents' prevalence and location they appeared.
It represents the different bacteria that were found when testing patients in the respective studies as outlined in Table 2, giving the study locations, types of infections caused by and prevalence of each bacteria [[54], [55], [56], [57], [58], [59], [60], [61], [62], [63]].
A recent retrospective multicenter study in china stated that 96% of the total patients with secondary bacterial infections died due to sepsis. Half of non-survivors experienced a secondary infection (27 out of 54); ventilator-associated pneumonia occurred in 10 of 32 patients (31%) requiring invasive mechanical ventilation [18]. The authors stated an overall occurrence of 15% secondary bacterial infections most of them in patients who died. Elske et al., stated that in the overall cohort, 5% developed VAP during hospitalization. However, VAP was diagnosed in up to 31% of those who had mechanical ventilation [26]. A retrospective study was conducted to assess the prevalence of VAP in COVID-19 patients admitted to the ICU, they stated that VAP was mainly diagnosed by clinical presentation and BAL fluid was tested by direct microscopic examination with Gram staining, microbiologic culture, and susceptibility testing. Consequently, empirical antibiotic therapy is started as soon as VAP is suspected [32]. Almost 71% of COVID-19 patients receive empirical antibiotic therapy upon admission which is known for its potential to inhibit pathogen growth in culture-based testing required for conventional in-vitro diagnostics [27]. In this context, molecular diagnostic approaches which detect the pathogen’s DNA must be performed to ensure accurate early diagnosis and appropriate antimicrobial therapy for secondary bacterial infections in order to avoid fatal consequences. In addition, by knowing this data, modifying empirical antibiotic management guidelines for COVID-19 patients could be achieved. Furthermore, identifying mixed infections using the advanced molecular methods will provide significant data on infectious pathogens causing coinfections and AMR in healthcare settings.
Antimicrobial stewardship
Antimicrobial stewardship refers to collaborative, coordinated programs and interventions designed to improve antimicrobial prescribing by promoting the selection of the optimal antimicrobial drug regimen including dosing, duration of therapy and route of administration and minimize unintended consequences [33]. Despite the fact that reducing emergence of AMR and drug-resistant infections is the major aim of antimicrobial stewardship. In normal situations, in advanced healthcare facilities, antimicrobial stewardship is mainly a behavior change challenge of healthcare professionals and healthcare organizations. In healthcare facilities in LMICs, antimicrobial stewardship is further irritated by limited access to medicine, concerns regarding counterfeit antibiotics and inadequate hospital infection prevention programs infrastructure. On March 1, 2020, an informal Twitter poll asked the antimicrobial stewardship communities whether antimicrobial stewardship programs at their healthcare facilities had been involved in SARSCoV-2/COVID-19 pandemic response or preparation. This generated 254 responses: 30% stated direct involvement, 28% noted indirect involvement, and 39% reported no involvement in emergency response efforts or planning [34]. Respiratory virus pandemics associated with secondary bacterial complications such as pneumonia and ARDS provide an opportunity for antimicrobial stewardship programs to monitor and enhance compliance with treatment guidelines; broad spectrum antibiotics have been administered to severe COVID-19 cases [35]. Given the challenges of detecting COVID-19 from bacterial pneumonia, the doubt about bacterial superinfections, the absence of specific antiviral treatment for the current pandemic, and the high rates of mortality; consideration of antibiotics as part of the empirical therapy for the most severe suspected or confirmed COVID-19 cases should not be neglected (e.g patients requiring mechanical ventilation). Yet, even during a pandemic antibiotics should be used wisely and cautiously [6].
Conclusion and recommendations
The global threat of AMR will persist beyond the COVID-19 crisis. Data on the prevalence of AMR infections before and after the outbreak should be gathered to determine the dissemination of AMR pathogens particularly from the developing countries. Comparative genomic analyses of clinical infectious pathogens before, through and after the pandemic is one potential procedure that could elucidate various mechanisms underlying the acquisition of AMR genes circulating in healthcare settings. Reserving antibiotic therapy for patients presented with the most severe COVID-19 illness and limiting duration of antibiotic treatment to five days as generally recommended in most guidelines.
Even though it is challenging to employ a research in the mid of a pandemic, deep-monitoring research is now required to improve management of COVID-19 patients.
Funding
No funding sources.
Competing interests
None declared.
Ethical approval
Not required.
References
- 1.Strathdee S.A., Davies S.C., Marcelin J.R. Confronting antimicrobial resistance beyond the COVID-19 pandemic and the 2020 US election. Lancet. 2020;396:1050–1053. doi: 10.1016/S0140-6736(20)32063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferri M., Ranucci E., Romagnoli P., Giaccone V. Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr. 2017;57:2857–2876. doi: 10.1080/10408398.2015.1077192. [DOI] [PubMed] [Google Scholar]
- 3.Chokshi A., Sifri Z., Cennimo D., Horng H. Global contributors to antibiotic resistance. J Global Infect Dis. 2019;11:36. doi: 10.4103/jgid.jgid_110_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shrestha P., Cooper B.S., Coast J., Oppong R., Do Thi Thuy N., Phodha T., et al. Enumerating the economic cost of antimicrobial resistance per antibiotic consumed to inform the evaluation of interventions affecting their use. Antimicrob Resist Infect Control. 2018;7:98. doi: 10.1186/s13756-018-0384-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.2014. Antimicrobial Resistance: tackling a crisis for the health and wealth of nations.https://amr-review.org/sites/default/files/AMRReviewPaper-Tacklingacrisisforthehealthandwealthofnations_1 [Google Scholar]
- 6.Huttner B.D., Catho G., Pano-Pardo J.R., Pulcini C., Schouten J. COVID-19: don’t neglect antimicrobial stewardship principles! Clin Microbiol Infect. 2020;26:808–810. doi: 10.1016/j.cmi.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chien Y.-W., Klugman K.P., Morens D.M. Bacterial pathogens and death during the 1918 influenza pandemic. N Engl J Med. 2009;361:2582–2583. doi: 10.1056/NEJMc0908216. [DOI] [PubMed] [Google Scholar]
- 8.Sheng Z.-M., Chertow D.S., Ambroggio X., McCall S., Przygodzki R.M., Cunningham R.E., et al. Autopsy series of 68 cases dying before and during the 1918 influenza pandemic peak. Proc Natl Acad Sci U S A. 2011;108:16416–16421. doi: 10.1073/pnas.1111179108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginsburg A.S., Klugman K.P. COVID-19 pneumonia and the appropriate use of antibiotics. Lancet Glob Health. 2020;8:e1453–e1454. doi: 10.1016/S2214-109X(20)30444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein E.Y., Monteforte B., Gupta A., Jiang W., May L., Hsieh Y., et al. The frequency of influenza and bacterial coinfection: a systematic review and meta-analysis. Influenza Other Respir Viruses. 2016;10:394–403. doi: 10.1111/irv.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boolchandani M., D’Souza A.W., Dantas G. Sequencing-based methods and resources to study antimicrobial resistance. Nat Rev Genet. 2019;20:356–370. doi: 10.1038/s41576-019-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X.-W., Wu X.-X., Jiang X.-G., Xu K.-J., Ying L.-J., Ma C.-L., et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruiz J. Enhanced antibiotic resistance as a collateral COVID-19 pandemic effect? J Hosp Infect. 2021;107:114–115. doi: 10.1016/j.jhin.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zavala-Flores E., Salcedo-Matienzo J. Medicación prehospitalaria en pacientes hospitalizados por COVID-19 en un hospital público de Lima-Perú. Acta Med Peru. 2020;37 doi: 10.35663/amp.2020.373.1277. [DOI] [Google Scholar]
- 15.Pal C., Bengtsson-Palme J., Kristiansson E., Larsson D.G.J. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics. 2015;16:964. doi: 10.1186/s12864-015-2153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharifipour E., Shams S., Esmkhani M., Khodadadi J., Fotouhi-Ardakani R., Koohpaei A., et al. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect Dis. 2020;20:646. doi: 10.1186/s12879-020-05374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J., Zhang K., Zhao L., Ma W., Liu T. Adsorption and polymerization of aniline on a carboxylic group-modified fibrous substrate. Synth Met. 2014;188:6–12. doi: 10.1016/j.synthmet.2013.10.028. [DOI] [Google Scholar]
- 18.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:10. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sermo, Breaking results: Sermo’s COVID-19 real time barometer study WAVE I: March 25–27, (n.d.). https://publiccdn.sermo.com/covid19/c8/be4e/4edbd4/dbd4ba4ac5a3b3d9a479f99cc5/wave-i-sermo-covid-19-global-analysis-final.pdf.
- 22.Sultana J., Cutroneo P.M., Crisafulli S., Puglisi G., Caramori G., Trifirò G. Azithromycin in COVID-19 patients: pharmacological mechanism, clinical evidence and prescribing guidelines. Drug Saf. 2020;43:691–698. doi: 10.1007/s40264-020-00976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baron S.A., Devaux C., Colson P., Raoult D., Rolain J.-M. Teicoplanin: an alternative drug for the treatment of COVID-19? Int J Antimicrob Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Neill J, Tackling Drug resistant infections globally: final report and recommendations, (n.d.). https://amrreview.org/sites/default/files/160518_Final%20paper_with%20cover.pdf.
- 25.Antimicrobial resistance in the age of COVID-19. Nat Microbiol. 2020;5:779. doi: 10.1038/s41564-020-0739-4. [DOI] [PubMed] [Google Scholar]
- 26.Sieswerda E., de Boer M.G.J., Bonten M.M.J., Boersma W.G., Jonkers R.E., Aleva R.M., et al. Recommendations for antibacterial therapy in adults with COVID-19 — an evidence based guideline. Clin Microbiol Infect. 2021;27:61–66. doi: 10.1016/j.cmi.2020.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox M.J., Loman N., Bogaert D., O’Grady J. Co-infections: potentially lethal and unexplored in COVID-19. Lancet Microbe. 2020;1:e11. doi: 10.1016/S2666-5247(20)30009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zafer M.M., El-Mahallawy H.A., Ashour H.M. Severe COVID-19 and sepsis: immune pathogenesis and laboratory markers. Microorganisms. 2021;9:159. doi: 10.3390/microorganisms9010159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lansbury L., Lim B., Baskaran V., Lim W.S. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81:266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clancy C.J., Nguyen M.H. COVID-19, superinfections and antimicrobial development: What can we expect? Clin Infect Dis. 2020:ciaa524. doi: 10.1093/cid/ciaa524. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Contou D., Claudinon A., Pajot O., Micaëlo M., Longuet Flandre P., Dubert M., et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann Intensive Care. 2020;10:119. doi: 10.1186/s13613-020-00736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luyt C.-E., Sahnoun T., Gautier M., Vidal P., Burrel S., Pineton de Chambrun M., et al. Ventilator-associated pneumonia in patients with SARS-CoV-2-associated acute respiratory distress syndrome requiring ECMO: a retrospective cohort study. Ann Intensive Care. 2020;10:158. doi: 10.1186/s13613-020-00775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fishman N. Society for Healthcare Epidemiology of America, Infectious Diseases Society of America, Pediatric Infectious Diseases Society, Policy Statement on Antimicrobial Stewardship by the Society for Healthcare Epidemiology of America (SHEA), the Infectious Diseases Society of America (IDSA), and the Pediatric Infectious Diseases Society (PIDS) Infect Control Hosp Epidemiol. 2012;33:322–327. doi: 10.1086/665010. [DOI] [PubMed] [Google Scholar]
- 34.Stevens M.P., Patel P.K., Nori P. Involving antimicrobial stewardship programs in COVID-19 response efforts: all hands on deck. Infect Control Hosp Epidemiol. 2020;41:744–745. doi: 10.1017/ice.2020.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323:1488. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Firth A., Prathapan P. Azithromycin: the first broad-spectrum therapeutic. Eur J Med Chem. 2020;207 doi: 10.1016/j.ejmech.2020.112739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stasi C., Fallani S., Voller F., Silvestri C. Treatment for COVID-19: an overview. Eur J Pharmacol. 2020;889 doi: 10.1016/j.ejphar.2020.173644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coppola S., Ciabattoni A., Pozzi T., Castagna V., Bassi G.L., Chiumello D. Hazardous mismatch between pulmonary pathogens and antibiotic treatments in COVID-19 patients. Br J Anaesth. 2020;125:e380–e382. doi: 10.1016/j.bja.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrasa H., Rello J., Tejada S., Martín A., Balziskueta G., Vinuesa C., et al. SARS-CoV-2 in Spanish intensive care units: early experience with 15-day survival in Vitoria. Anaesth Crit Care Pain Med. 2020;39:553–561. doi: 10.1016/j.accpm.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pongpirul W.A., Mott J.A., Woodring J.V., Uyeki T.M., MacArthur J.R., Vachiraphan A., et al. Clinical characteristics of patients hospitalized with coronavirus disease, Thailand. Emerg Infect Dis. 2020;26:1580–1585. doi: 10.3201/eid2607.200598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ventola C.L. The antibiotic resistance crisis. P T. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z., Yang B., Li Q., Wen L., Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020;71:769–777. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling L, So C, Shum HP, Chan PK, Lai CK, Kandamby DH, et al., Critically ill patients with COVID-19 in Hong Kong: a multicentre retrospective observational cohort study, Crit Care Resusc. (n.d.) 7. [DOI] [PMC free article] [PubMed]
- 45.Kosecka-Strojek M., Sadowy E., Gawryszewska I., Klepacka J., Tomasik T., Michalik M., et al. Emergence of linezolid-resistant Staphylococcus epidermidis in the tertiary children’s hospital in Cracow, Poland. Eur J Clin Microbiol Infect Dis. 2020;39:1717–1725. doi: 10.1007/s10096-020-03893-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osman K., Zolnikov T.R., Badr J., Naim H., Hanafy M., Saad A., et al. Vancomycin and florfenicol resistant Enterococcus faecalis and Enterococcus faecium isolated from human urine in an Egyptian urban–rural community. Acta Trop. 2020;201 doi: 10.1016/j.actatropica.2019.105209. [DOI] [PubMed] [Google Scholar]
- 47.Girijan S.K., Paul R., R.K. V.j, Pillai D. Investigating the impact of hospital antibiotic usage on aquatic environment and aquaculture systems: a molecular study of quinolone resistance in Escherichia coli. Sci Total Environ. 2020;748 doi: 10.1016/j.scitotenv.2020.141538. [DOI] [PubMed] [Google Scholar]
- 48.Candevir Ulu A., Güven Gökmen T., Kibar F., Kurtaran B., Önlen C., Kuşçu F., et al. Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae at a Turkish centre: is the increase of resistance a threat for Europe? J Glob Antimicrob Resist. 2017;11:10–16. doi: 10.1016/j.jgar.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 49.Malchione M.D., Torres L.M., Hartley D.M., Koch M., Goodman J.L. Carbapenem and colistin resistance in Enterobacteriaceae in Southeast Asia: review and mapping of emerging and overlapping challenges. Int J Antimicrob Agents. 2019;54:381–399. doi: 10.1016/j.ijantimicag.2019.07.019. [DOI] [PubMed] [Google Scholar]
- 50.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020:1–3. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai Q., Huang D., Ou P., Yu H., Zhu Z., Xia Z., et al. COVID-19 in a designated infectious diseases hospital outside Hubei Province, China. Allergy. 2020;75:1742–1752. doi: 10.1111/all.14309. [DOI] [PubMed] [Google Scholar]
- 53.Feng Y., Ling Y., Bai T., Xie Y., Huang J., Li J., et al. COVID-19 with different severities: a multicenter study of clinical features. Am J Respir Crit Care Med. 2020;201:1380–1388. doi: 10.1164/rccm.202002-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Centers for Disease Control and Prevention, Pneumococcal disease (2017). https://www.cdc.gov/pneumococcal/clinicians/clinical-features.html.
- 55.Centers for Disease Control and Prevention, Healthcare-associated infections — Acinetobacter (2019). https://www.cdc.gov/hai/organisms/acinetobacter.html.
- 56.Centers for Disease Control and Prevention, Healthcare-associated infections — Klebsiella (2010). https://www.cdc.gov/hai/organisms/klebsiella/klebsiella.html.
- 57.Centers for Disease Control and Prevention, Healthcare-associated infections — Staphylococcus aureus (2011). https://www.cdc.gov/hai/organisms/staph.html.
- 58.Centers for Disease Control and Prevention, Healthcare-associated infections — Pseudomonas aeruginosa (2019). https://www.cdc.gov/hai/organisms/pseudomonas.html.
- 59.Centers for Disease Control and Prevention, Haemophilus influenzae disease (including Hib) (2020). https://www.cdc.gov/hi-disease/.
- 60.Centers for Disease Control and Prevention, Gonorrhea — Moraxella catarrhalis (2017). https://www.cdc.gov/std/gonorrhea/lab/mcat.htm.
- 61.Nazzaro G. Etymologia: Serratia marcescens. Emerg Infect Dis. 2019;25:2012. doi: 10.3201/eid2511.ET2511. [DOI] [Google Scholar]
- 62.Halpern M., Waissler A., Dror A., Lev-Yadun S. Advances in applied microbiology. Elsevier; 2011. Biological warfare of the spiny plant; pp. 97–116. [DOI] [PubMed] [Google Scholar]
- 63.Cunningham D.J., Leber A. Principles and practice of pediatric infectious diseases. Elsevier; 2018. Enterobacter, Cronobacter, and Pantoea Species. pp. 824–7.e1. [DOI] [Google Scholar]