Abstract
Transmitter signalling is the universal chemical language of any nervous system, but little is known about its early evolution. Here, we summarize data about the distribution and functions of neurotransmitter systems in basal metazoans as well as outline hypotheses of their origins. We explore the scenario that neurons arose from genetically different populations of secretory cells capable of volume chemical transmission and integration of behaviours without canonical synapses. The closest representation of this primordial organization is currently found in Placozoa, disk-like animals with the simplest known cell composition but complex behaviours. We propose that injury-related signalling was the evolutionary predecessor for integrative functions of early transmitters such as nitric oxide, ATP, protons, glutamate and small peptides. By contrast, acetylcholine, dopamine, noradrenaline, octopamine, serotonin and histamine were recruited as canonical neurotransmitters relatively later in animal evolution, only in bilaterians. Ligand-gated ion channels often preceded the establishment of novel neurotransmitter systems. Moreover, lineage-specific diversification of neurotransmitter receptors occurred in parallel within Cnidaria and several bilaterian lineages, including acoels. In summary, ancestral diversification of secretory signal molecules provides unique chemical microenvironments for behaviour-driven innovations that pave the way to complex brain functions and elementary cognition.
This article is part of the theme issue ‘Basal cognition: multicellularity, neurons and the cognitive lens'.
Keywords: evolution, synapse, nervous system, neurotransmitters, Metazoa, volume transmission
1. Introduction: behaviour is a pacemaker of evolution
This paper summarizes some older and novel insights about evolutionary aspects of neurotransmitter functions. The narrative is not designed to provide a systematic comparative review dealing with the distribution of chemical signalling in the brain. Rather we would like to ask some simple but difficult questions, which might help to decipher fundamental principles of neural organization in historical context following Dobzhansky's famous motto: ‘Nothing in biology makes sense except in the light of evolution’ [1]. Rephrasing this, we can also add: ‘Nothing in neuroscience makes sense except in the light of transmitters’. Both statements illuminate the topic of this essay and should be united to understand our brain genealogy.
Origins of neural systems, brains, eusociality and cognition are the major transitions in planetary evolution [2]. There is no shortage of hypotheses of neuronal origins [3–24]. But we still do not know why, how and when it happened? The vast majority of organisms on the Tree of Life survive without neurons and brains. Indeed, 85% of biological evolution on the Earth (more than 3 billion years!) occurred without nervous systems as we know it. So, the origin(s) of neurons is a rare event. As with any biological adaptation, neural systems (and neurotransmitters) are passive products of evolution. The term ‘passive’ is used here to emphasize the lack of goal-oriented evolution of nervous systems. There is no a priori goal or trends, which ‘requires’ the origin of neurons in multicellular organisms. However, certain traits (preadaptations) might set the stage and ‘facilitate’ development of innovations in pre-existing integrative systems, which would eventually lead to neural systems as we know them today in the majority of Metazoa. This approach also implies that there were alternative neural/integrative systems in the past; some of them are extinct and some are still present in extant animals, including some nerveless basal metazoans. What kind of ‘preadaptations and factors triggered the formation of early neural systems?
‘Behaviour is the pacemaker of evolution’ [25] and, as indicated by Ernst Mayr, ‘behavioural shifts have been involved in most evolutionary innovations’. ‘Any behaviour that turns out to be of evolutionary significance is likely to be reinforced by the selection of genetic determinant of such behaviour known as the “Baldwin effect”’ [25, p. 137].
Our working hypothesis is that ancestral neural systems evolved from genetically heterogeneous populations of polarized secretory cells without canonical synapses and capable of volume (non-synaptic [26–35]) transmission (figure 1). We further propose that integrations of behaviours by transmitters [36,37], originally related to novel feeding ecology, injury and defense, created the molecular playground for the parallel development of neuroid-type signalling in early metazoans (figure 1).
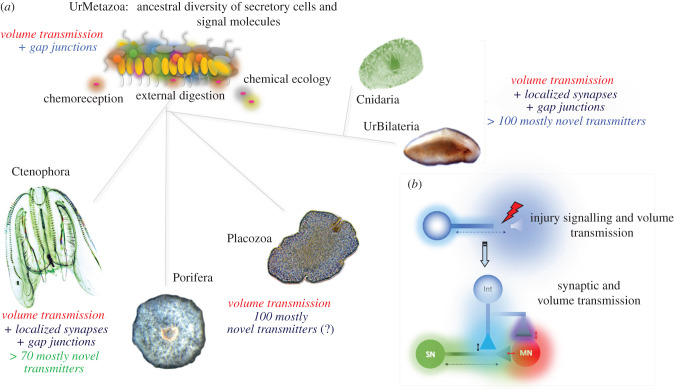
Figure 1.
(a) Hypothetical ancestor of all metazoans (Urmetazoa) with a diversity of secretory cells capable of volume transmission and early radiation of basal metazoan lineages preserving both non-synaptic transmission and the parallel development of systems for intercellular communications including synapses. (b) Non-specific injury response and repair signalling as an evolutionary predecessor of neurotransmitters [19]. Shadows show gradients of neurotransmitters from their release sites.
Volume transmission can be long-distance or more localized [26–35], tuned by different chemical natures (reactivity, stability, size) of messenger molecules and the presence of sufficient intercellular space.
The emergence of extracellular digestion and relevant elaborated secretion in metazoan ancestors was an important preadaptation to form intercellular regions with unique chemical microenvironments of signal molecules. Indeed, in a multicellular organism, such a biologically significant goal as feeding requires substantial integration of multiple effectors: ciliated, contractile and secretory cells. Feeding also includes innate immune protection against potential pathogens (e.g. using nitric oxide and toxins) and injury-induced regenerative responses.
Under this scenario, the genetic determinants providing immediate behavioural output could be mutations affecting putative signal molecules and their reception and inactivation systems. The molecular targets for natural selection would be secreted metabolites, proteins (including proteolytic digestive enzymes), small peptides and toxins with changeable three-dimensional structures, their interactions with orphan receptors, modulators, transporters, inactivation and uptake mechanisms, etc. The resulting sea of potential chemical messengers would be, in a direct sense, a hotspot for the selection of molecular modules capable of generating system innovations.
In contrast with highly stereotyped electrical signalling and wiring, these chemically and purely combinatorial transmitter innovations are more adaptable and simpler. The behavioural coordination of effectors can be easily tuned by dynamic changes of concentrations in a mixture of freely diffusible intercellular messengers. Such functional reconstruction provides a versatile platform for selecting and recruiting molecular modules at all levels of the biological organization with traceable mechanisms and measurable benefits for survival.
In summary, we suggest not that the nervous system evolved transmitters, but rather that transmitters made the nervous system by integrating ancestral populations of secretory cells for behavioural coordinations without synapses. The diversity of ancestral transmitters (signal molecules) may have been the most profound of pre-neuronal metazoan adaptations. This idea was originally discussed by D. A. Sakharov in 1970 [36–40] when comparative studies of neurotransmitters were in their infancy. Regrettably, we still know very little about the diversity of transmitters and their functional roles among early branching metazoans. Only five quite derived phyla (out of 35) have been sufficiently investigated in terms of the transmitter organization of their neural circuits and behaviours. These are chordates, arthropods, nematodes, annelids and molluscs. In the next sections, we will take advantage of the burgeoning genomic data to identify shared signal molecules between metazoan lineages with and without neural systems. Animal phylogeny is significantly revised compared to the situation a decade ago. Thus, it would be important to incorporate the distribution of transmitters within the novel phylogenomic framework.
2. Convergent evolution and diversity of neural systems in Metazoa
It is well accepted that dramatic environmental changes at the Precambrian–Cambrian boundary [41,42] caused profound diversification of body plans both in Ediacaran and early Cambrian biota. Multicellularity and bigger body sizes, supported by oceanic oxygenation more than 550 Mya, facilitated the transition in modes of feeding on pino- and phagocytosis toward extracellular digestion characteristics of metazoans. Predation or escape from predation [23] may have been subsequential behavioural innovations that contributed to multiple origins of neuroid and other integrative systems in early metazoans.
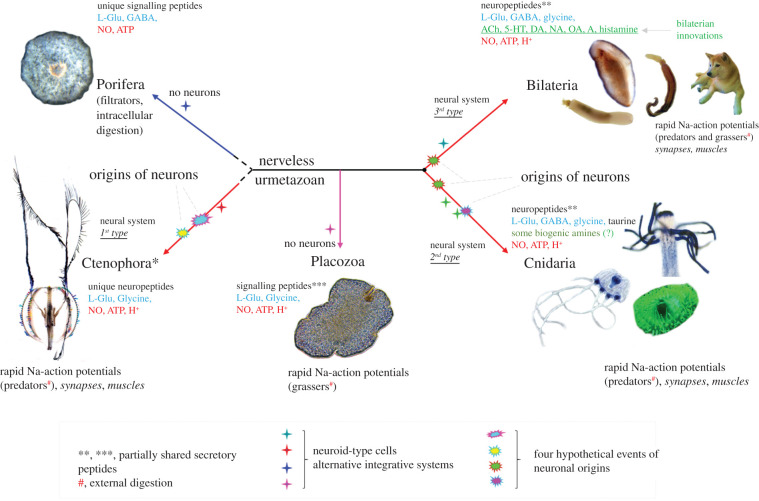
As a result, three out of five basal Metazoa lineages (Ctenophora, Cnidaria and Bilateria) evolved neural systems with remarkably different spatio-temporal organizations (figure 2). Two lineages derived from the common ancestor of all Metazoa (Urmetazoan), Porifera and Placozoa, have remained nerveless. This might be attributable to their success with distinct ecological niches, alternative life strategies and alternative integrative systems.
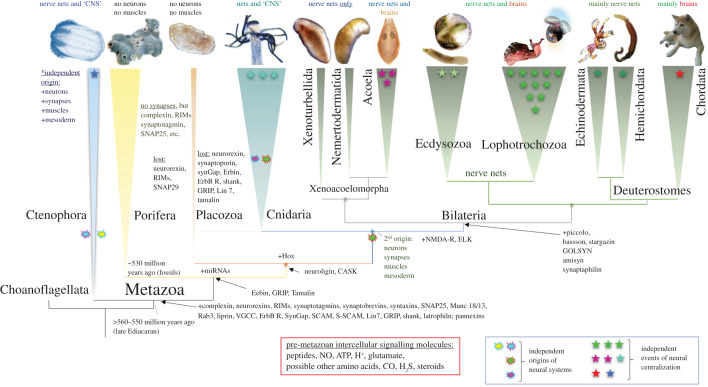
Figure 2.
Parallel evolution of neural systems across Metazoa. Reconstruction of phylogenetic relationships among major clades of basal Metazoa (the consensus tree according to [43–49]). The schematic diagram illustrates the hypothetical events of neuronal origins (cloud marks) and neuronal centralizations (stars). The presence/absence of the central nervous system (CNS), nerve nets or brains is indicated above each phyletic lineage. Ctenophores have been recovered as the lineage sister to all other metazoans. Ctenophores have two distinct neural systems (the epidermal and mesogleal nerve nets [50–54]). Porifera and Placozoa have no recognized neurons and muscles. Cnidaria and Bilateria might have a shared ancestry of their neuronal systems (the 2nd independent origin event). However, it is also possible that some neuronal populations in cnidaria evolved independently compared to bilaterians. Events of neuronal centralization (stars) and the formation of complex brains occurred more than twenty times across animal lineages [19,55,56] with the greatest diversity of neural system types in Mollusca [19]. Arrows show gains (+) of selected synaptic proteins, microRNAs machinery and Hox genes. The red box outlines the proposed ancestral set of pre-metazoan transmitters.
Notably, representatives of two groups of obligate and morphologically highly simplified parasites secondarily lost their neural systems [55]. The first phylum is Dicyemida [57–59], which belongs to the clade Lophotrochozoa. These parasites inhabit the renal system of cephalopods, and their body is reduced to just a few dozen cells with ultra-compact genomes [57]. The second group is a specialized lineage of parasitic cnidarians—Myxozoa [60]. Myxozoans also have highly simplified morphology, a very small number of cells (but some still preserved muscles [61]) and compact genomes [62,63]. Also notably, the rest of the known parasitic groups (more than 30% of extant animal taxa, including microscopic or sessile animals) did not ‘lose’ their neural systems. Once the nervous system was gained, it was never lost in all the representatives of the 32 phyla over a half billion years of evolution. The nervous system is an expensive but highly ‘valuable’ trait supporting adaptive behaviours and developmental programs.
Porifera and Placozoa (figures 1 and 2) are macroscopic and complex animals with hundreds of thousands to many millions of cells. Their elaborate behaviours (e.g. predation in some sponges and rapid locomotion in placozoans) are performed in the absence of neuro-muscular organization. Both phyla have a very rich repertoire of genes, many with some synteny to bilaterians [64–69]. There is no creditable evidence that sponges and Placozoan ancestors, as free-living and free behaving organisms, secondarily lose neural systems from their respective common ancestors. These descendants of early nerveless metazoans have survived well for 540 million years.
Ctenophores or comb jellies have not one but two distinct neural systems [50]. Ctenophores are very distantly related to Cnidaria [43–45,70]. These two phyla of gelatinous predators are remarkably different in virtually all aspects of their anatomical, developmental and genomic composition (figure 2). Based on the uniqueness of the neuro-muscular organization across these phyla and bilaterians, we suggested that neural systems, synapses, muscles and mesoderm evolved independently in ctenophores [46,56,71,72]. These hypotheses have initially been met with some caution [73–75]. The primary scepticism was based on selected evolutionary models, which recovered sponges' traditionally most basal in the animal phylogeny [76,77]. However, both the Sponge-first scenario [47] and the Ctenophore-first scenario [45] are equally compatible with the independent origins of neural systems, as discussed elsewhere [24,72,74,76].
As soon as neurons arose, there was immediately parallel diversification of neuronal cell types and countless examples of the convergent evolution for similar behaviours. Figure 2 also shows that events of neuronal centralization and the formation of complex brains may have occurred more than twenty times independently across animal lineages [19,55,56], with the greatest diversity of neural system types in Mollusca [19].
Intriguingly, among about 100 classes of bilaterians, representatives of only three classes (insects, crustaceans and mammals) developed eusociality—the most evolutionarily successful form of social organization [78]. According to Edward Wilson [78], eusociality arose 18 times: once in the lineages leading to termites, ants, ambrosia beetles, aphids, thrips and humans, respectively. Eusociality arose twice in naked mole rats, three times independently in both wasps and shrimps, and at least four times in bees. Naked descendants of one group of apes, 1–3 million years ago, evolved consciousness of our kind and conquered the planet [78] after 3.5 billion years of experimenting by Nature.
Remarkably, both the formation of social relationships and complex behavioural integrations were performed by balancing similarly diverse subsets of transmitters [37,79–86] as in early branching animal lineages. Thus, the broadest spectrum of nearly all studied behaviours is inherently traced to multiple transmitter pathways, and the transmitter organization as well as synaptic and non-synaptic integration of behaviors are intrinsically embedded in every neural circuit from a comb jelly to Octopus. What is the actual diversity of transmitters across metazoans?
3. The diversity of synapses and (neuro)transmitters
The Neuron Doctrine, established by Santiago Ramón y Cajal, more than 100 years [87,88], was a serious conceptual challenge to the ‘pure’ electrical paradigm of neuronal information transmission and functions. The Neuron doctrine postulates no syncytium in the brain; rather, neurons are individual entities that are physically separated by synapses and extracellular space. The challenge of finding mechanisms of intraneuronal communications has been partially resolved by the discovery of chemical transmitters in the 1920–1930s [89]. The principle was simple and powerful: a chemical messenger must be released from one presynaptic neuron and interacts with one or more postsynaptic cells (e.g. muscle, glands, neurons, etc.). The battle of ‘spikes and soups’ [90] was settled in 1953–1956 when synapses were visualized by electron microscopy. It happened at the same time as the double helix structure of DNA was revealed.
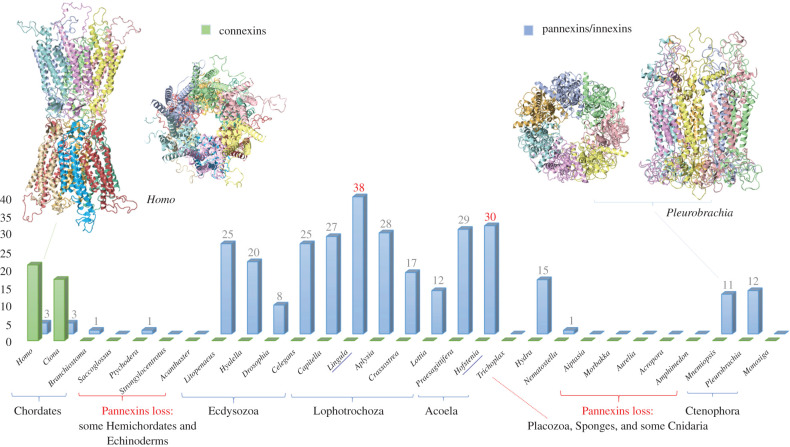
However, following the triumph of the theory of chemical transmission, the electrical synapses (=gap junctions) were discovered, which are mediated by two independently evolved groups of proteins with similar transmembrane topologies: pannexins (=innexins) and connexins. Connexins have been found only in tunicates and vertebrates, whereas pannexins [91,92] are broadly distributed across Metazoa (figure 3). This is a perfect example of the convergent evolution of synapses! Pannexins were independently lost in placozoans, sponges and selected lineages of Cnidaria, Hemichordata and Echinodermata. The loss of pannexins does not always correlate with simpler behaviour repertoires. For example, there are no recognized pannexins in the genome of cubomedusa, Morbakka—an active predator with complex visual systems and elaborated behaviours. The largest number of genes encoded pannexins (38) was found in the sessile brachiopod, Lingula, which shows a highly reduced spectrum of movements (figure 3). The functional explanation of such molecular diversity of pannexins is unclear, and different pannexins might be responsible for distinct electrophysiological properties of various electrical synapses. Nevertheless, pannexins might have other ‘secretory’ functions. It was shown that in mammals, pannexins do not form classical gap junctions. By contrast, they might work as channels for the non-synaptic release of certain signal molecules [94–109] and therefore participate in volume transmission.
Figure 3.
The distribution of gap junction proteins across Metazoa. Pannexins (innexins) and connexins are two unrelated families of proteins [91] with similar transmembrane topology, which illustrate unique examples of convergent evolution of electrical synapses. Note multiple examples of gene gains and losses across Metazoa. Y-axis indicates the presence and the number of genes encoding connexins or pannexins in selected reference species. The two representative three-dimensional structures are shown at the top of the figure using examples from humans (model of connexin 7jkc) and the ctenophore Pleurobrachia bachei (model of pannexin 6wbi), respectively. The three-dimensional reconstruction was performed using Phyre2 modelling [93].
The chemical synapses and transmitters are incredibly diverse. Initially, only two neurotransmitters were identified: acetylcholine and noradrenaline for the parasympathetic and sympathetic parts of the nervous system, respectively [89,90]. Over the subsequent decades, many novel messengers have been discovered both in vertebrates and invertebrates. By the end of the twentieth century, the 200 year-old electrical brain paradigm [110] had been gradually transformed into the chemical brain paradigm with about two dozen low molecular weight neurotransmitters and thousands of neuropeptides—all acting within synaptic clefts and beyond.
Extrasynaptic diffusion of neuronal and glial messengers was also named volume transmission [31,32,35,111–114], where a complex and dynamic mixture of signal molecules creates unique microchemical regions within the extracellular space of the brain. In such cases, the boundary between the transmission within a confined space of a synapse and long-distance hormonal signalling is now removed. These cases represent two opposite poles of the same very ancient and fundamental intercellular chemical signalling process in multicellular organisms.
Many neurons, both in vertebrates and invertebrates, might have one neuronal terminal forming a classical synapse with a classical neurotransmitter, and the second axon of the very same neuron might release the identical transmitter in a circulatory system, where it acts as a hormone. The classic examples are serotonin-containing interneurons controlling feeding and associated behavioural arousal in the leech (Retzius cells [115,116]) and gastropod molluscs (MCC neurons [16,117–121]. Similar functions of serotonin in annelids and molluscs as the evolutionarily conserved regulator of behavioural arousal illustrate a given transmitter's role as an integrator of multiple effectors leading to a biologically relevant behaviour or behavioural state(s) [17,117,122].
The experimentally confirmed low molecular weight transmitters include polar amino acids, their derivatives and gases. They are (1) L-glutamate, (2) D- and (3) L-aspartate, (4) glycine, (5) gamma-aminobutyric acid (GABA), (6) D-serine, (7) serotonin, (8) dopamine, (9) noradrenaline, (10) adrenaline, (11) octopamine, (12) tyramine, (13) histamine, (14) acetylcholine, (15) taurine, (16) protons, (17) ATP, (18) nitric oxide (NO), (19) carbon monoxide (CO) and (20) H2S. The list might incorporate short lipids (derivatives of arachidonic acid, acting on cannabinoid receptors), trace amines, purines such as adenosine, and nicotinamide adenine dinucleotide, etc.
Considering that these intercellular chemical messengers can also be produced and operate as signal molecules in non-neuronal cells, the correct term would be transmitter. The term neurotransmitter refers to signal molecules with a confirmed neuronal release. In fact, 95% of the serotonin in our body is not located in neurons; it is produced in the intestine where it has been increasingly recognized for its hormonal, autocrine and paracrine actions [123] (e.g. enterochromaffin cells, enterocytes, platelets). Furthermore, non-neuronal serotonin can significantly contribute to such neuronal functions as learning and memory [124]. This note is very important for comparative studies. There is widespread usage of the words ‘neurotransmitters’ or ‘neuropeptides’ by describing signal molecules in organisms without neural systems and synapses (e.g. for primarily nerveless placozoans or sponges). Similarly, the presence of a given molecule (or a relevant synthetic enzyme) such as serotonin or catecholamines or many others is often misinterpreted as evidence of neural systems and neuronal signalling. Such an assumption is incorrect and misleading.
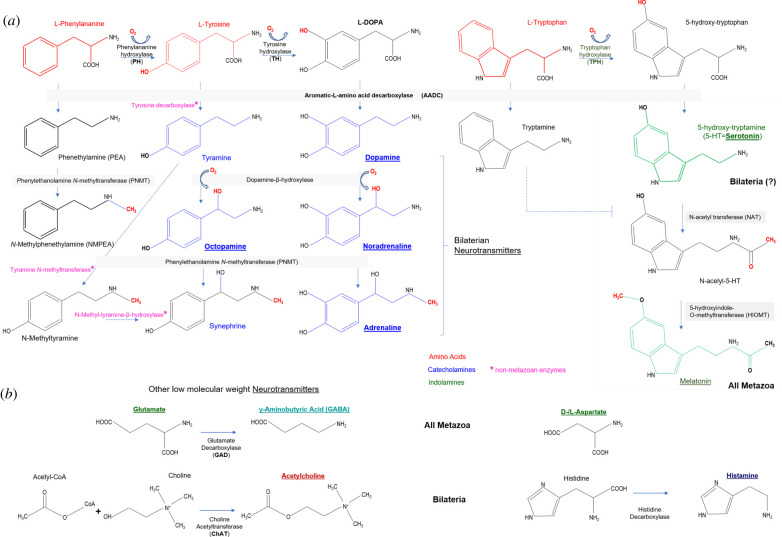
Figure 4 provides an overview of synthetic pathways for biogenic amines, GABA and acetylcholine, with several comments. This diagram shows the enzymes identified in animals. However, there are alternative pathways of synthesis and catabolism of monoamines and acetylcholine in other eukaryotes and prokaryotes. The detection of monoamines or acetylcholine in some tissues might be owing to bacterial contamination. In fact, some bacteria names are dedicated to the high synthesis of acetylcholine—Bacillus acetylcholini; these and related bacteria acetylate choline and present in fermented sauerkraut and silos [89]. The presence and usage of transmitters in particular animals might reflect enzymatic activities of symbionts and associated microbiota. Finally, the presence of a given chemical does not provide direct evidence for its transmitter function, which should be experimentally proved by careful functional studies.
Figure 4.
Enzymes and synthetic pathways for low molecular weight transmitters. (a) Synthesis of biogenic amines; (b) synthesis of GABA and acetylcholine. The respective enzymes and their abbreviations are the same as used in the text. Asterisks (*) indicate synthetic enzymes from non-metazoan species.
The entire scope of the comparative diversity of neurotransmitters, especially in minor phyla, is unknown. A prevailing view is that most, if not all, (neuro)transmitters should be the same in different animal lineages. This is not the case. Although there are very limited data dealing with direct microchemical analyses of the transmitter candidates in representatives of early branched metazoan lineages, the distribution of low molecular weight transmitters is not uniform across basal Metazoa (figure 5). Both neuropeptides and low molecular weight transmitters show a noticeable degree of lineage-specific adaptations.
Figure 5.
The presence of major classes of transmitters in five basal metazoan lineages. The diagram shows the unrooted metazoan tree with hypothetical events of neuronal origins and the occurrence of other integrative systems together with representative types of feeding. See the text for details. Abbreviations: 5-HT, serotonin; DA, dopamine; NO, noradrenaline; A, adrenaline; OA, octopamine.
4. Why are there so many transmitters? A case for parallel evolution?
Regardless of the current limitations of comparative data, there is a strong consensus that every neural system consists of many types of transmitter systems. In other words, in any neural system, a significant fraction of neurons have different types of transmitter(s)/secretory specificity. Why are there so many transmitters? This fundamental question was addressed about 60 years ago [40]. The logic was simple. If a chemical messenger acts only as a pure transmitter(=messenger) at the synaptic cleft within a specific wiring diagram, only two neurotransmitters are needed (e.g. for excitation and inhibition, respectively). If there are two different receptors for the same transmitter (to induce excitation and inhibition)—then even one transmitter might be sufficient. Again, this is not the case, and two situations might explain the observed transmitter diversity in extant neural systems.
The first, functional, scenario: different transmitters have different molecular functions both in pre- and postsynaptic cells as well as different systemic functions beyond the synaptic cleft (volume and hormonal transmissions). In this case, a transmitter might also act as an integrator of behaviours such as fight versus flight [125], feeding [116,122], respiration [126–128], aggression [79,80,82] and reproduction [129–137]. Different transmitters might help to recruit different circuits and behaviours and be responsible for the hierarchy of behavioural outputs or behavioural choices [122]. Thus, different transmitters evolved within neural systems or recruited from non-neuronal cells to support novel functions and innovations. More likely, some transmitter systems performed distinct systemic functions and acted as integrators of behaviours in early nerveless animals (e.g. before they were recruited as neurotransmitters' by neurons). This corollary is essential to reconstruct the dawn of neuronal evolution.
The second, evolutionary, scenario proposes that extant neuronal cell types preserved a primordial diversity of ancestral types of secretory specificity from early animals [16,36]. The deep evolutionary conservations of certain transmitter phenotypes in individually identified homologous neurons have been reported. One of the most famous examples is the MCCs (metacerebral cells), a pair of giant serotonin-containing interneurons involved in feeding arousal [119,138,139]. The homologues of MCC can be recognized across all Euthyneura: from Lymnaea, Helix, Clione and Aplysia to Pleurobranchaea and Tritonia [16,119–121,140]—it is the level of molluscan subclasses separated by more than 380 million years of evolution in each direction! It is probably the most distant homological lineage of any single neuron identified to date.
The two scenarios explaining the observed diversity of transmitters are not mutually exclusive. Probably both explanations are valid to a different degree within specific constraints of various body plans and environments.
Moreover, we propose that the scope of chemical transmitter diversity in the early metazoans was not only comparable or, perhaps, even greater than can be found in extant bilaterians. Considering that in Precambrian metazoans, we might expect fewer morphological and microanatomical constraints (e.g. no neuronal centralization, no synapses, no muscles), the overall ancestral secretory diversity of early animal cell types might be greater than in today's animals. Some transmitters might be lost rather than gained with the formation of more compartmentalized and localized brain circuits (e.g. for fast locomotion, life on land, etc.). Comparative studies will be able to clarify these questions.
Figure 5 summarizes the large-scale distribution of known transmitter candidates across all five basal Metazoa clades, in respect of shared and lineage-specific innovations. This diagram represents about a dozen confirmed classical transmitters and hundreds of neuropeptides. Why did natural selection recruit these particular transmitters in the first place? And why were they preserved throughout half a billion years of biological evolution? To what degree would the distribution of transmitters in neural systems of early branching metazoans help to resolve these questions? We will begin with the parallel and divergent evolution of peptidergic systems. Then we will discuss a stabilizing selection of classical transmitters in greater detail.
Neuropeptides or short signalling peptides occur in every studied nervous system. However, few neuropeptides or prohormones have been shared across phyla. There are no pan-neuronal neuropeptides for the entire animal kingdom (figure 5). Some prohormones (e.g. neuropeptide Y) are conserved across bilaterians and a few can also be found in cnidarians and placozoans (e.g. insulins), but not in ctenophores and sponges. This type of cross-phyla distribution suggests that the majority of neuropeptide prohormones might either evolve independently or be highly derived beyond the recognition of their ancestors. Clearly, final peptide products are even more diverged across species reflecting their rapid evolutionary rate and high adaptability of peptide-mediated signalling.
As a result, the diversity of secretory peptides is overwhelming. The number of signal molecules of the peptide nature might exceed one hundred in any animal species (considering multiple products of the same prohormone and posttranslational modifications) regardless of the presence or absence of neural functions. It looks as though ctenophores [46,72] and sponges [66] independently evolved separate and not overlapping sets of small secretory signalling peptides. There are comparable numbers of secretory peptides in Cnidaria and Bilateria, with few shared homologous prohormones such as insulins. The major conclusion is simple.
(a). First neural systems were peptidergic
This situation is preserved in the chemical design of all extant neural systems. In other words, peptides are the earliest and most widespread neurotransmitters as well as hormones that predominantly act beyond the synaptic cleft. This statement does not mean that neurons and neural systems are homologous. Neurons might evolve many times from different populations of secretory (including peptidergic) cells as it was originally proposed more than 60 years ago [6,7,9,12,16,19,24,38].
Several factors might contribute to selecting peptides as intercellular messengers in the first place [19]. Short secreted peptides are easy to synthesize in any cell, both using ribosomal and non-ribosomal synthesis. Oligopeptides have a wide range of conformations and can be employed to activate many receptor types (e.g. G-protein-coupled and acid-sensing ionotropic (amiloride) receptors) and potential cellular targets. Because of this, similar peptide motifs can be evolved independently and convergently many times (as with RFamide type peptides across the tree of life). Various protoneuronal cells might develop similar types of peptidergic signalling in parallel.
Signalling peptides are widely used in virtually all eukaryotic organisms for a myriad of functions: pheromones and repellents, control of life cycle and differentiation, injury—regeneration responses, immunity, toxins, chaperones, etc. These first ‘true’ peptide transmitters could also act as growth factors controlling trans-differentiation and cell divisions. We proposed that these peptide-based signals/growth factors were originally ‘preadapted’ to repairing injuries, regeneration and re-establishment of integrative systems for intercellular directional communications [19] (figure 1b).
An enormous diversity of G-protein-coupled receptors (GPCR) and acid-sensing ionotropic (amiloride) receptors can also be viewed as important preadaptation, which facilitated recruitments of peptide messengers into early neural systems. These receptors can be directly gated by small peptides and protons, providing the pathway for fast transmission. Because the precise ligand specificity is not absolutely critical for activation of GPCRs and acid-sensitive amiloride type and many other receptors, it might not be surprising that the evolutionary occurrence of the majority of receptor families including ligand-gated ion channels often preceded the establishment of novel neurotransmitter systems.
By contrast with neuropeptides, the group of low molecular weight transmitters is relatively small. Even smaller is the subset of shared transmitters across basal metazoans. There are only 5–6 of them (figure 5): glutamate, GABA, glycine, ATP, NO and protons. Why were these molecules preserved for signalling, and what was the relevant preadaptation to recruit them in the first place as transmitters? We think it was the injury/regeneration response.
5. Adaptive injury/regeneration signalling as generalizing neurogenic factors in evolution
This hypothesis was proposed in 2009 in the search for natural causes that would lead to the development of neuroid elements and the first transmitters [19]. We postulated that induction of massive, well-coordinated gene expression events could be by itself an integrating factor triggering the formation of such multifunctional and information processing cells as neurons. Adaptive genome-wide responses, especially in secretory cells, naturally evolved as a result of stress or injury. Thus, a non-specific injury induced by natural causes or predation or as part of immune responses to pathogens from the food might be the major neurogenic factor in evolution.
Indeed, damaging and high threshold nociceptive stimuli that are not sufficient to kill a cell can induce an integrated repair process, distant chemical signalling and re-growth of asymmetric proneuronal processes, thereby acting as inductors of a novel neuronal-like phenotype initially. Neurons might have evolved in ancestral metazoans as a result of development in the adaptive cellular regenerative response (wound healing) to localized injury and stress, leading to a coordinated (potentially defensive) reaction and behaviour of the entire organism.
Furthermore, an injury by its nature might lead to the release of an array of evolutionarily conserved and highly abundant intermediates that can act as primordial signal molecules. For example, such widespread cellular metabolites as ATP, NO, protons and glutamate (Glu) have already been selected in unicellular eukaryotes or colonies for the same reasons. The classical example is the role of glutamate in wound signalling in plants [141]. Besides, protons and ATP are components of the majority of secretory vesicles during exocytosis. The widespread distribution of respective receptors supports this idea: P2X for ATP, pH-gated channels, iGluRs for polar amino acids (glutamate, aspartate and glycine), soluble guanylyl cyclases for NO-in all domains of life.
We think this is a functional reason why these molecules (and their receptors) acted as the first, initially non-specific, transmitters in early animals for the fast signalling associated with the injury. These are metabolically cheap and abundant chemical intermediates co-opted to induce rapid localized repair and defensive responses as preserved in most of the metazoans today, including humans. Extensive pharmacology has been developed on these four transmitter systems to control nociceptive pathways and pain in medicine.
In parallel, different cells in early metazoans also used peptides as signal molecules for slower, more specific, and coordinated regenerative, morphogenic and behavioural responses—which therefore act similarly to extant neuropeptides and morphogenic factors in cnidarians or bilaterians [142].
The adaptive injury regeneration signal cascade set the stage for the evolution of the first ‘true’ transmitters and growth factors controlling neuroplasticity as in the form of memory of injury [143]. The signal transduction pathways of this form of long-term memory of injury are apparently preserved and operate in both molluscan and vertebrate neuronal circuits underlying learning and nonassociative memory [143–146].
We also can anticipate possibly autocatalytic processes of the modular recruitment of other signalling components and transmitters in injury circuits. One such evolutionarily conserved molecular module is related to a massively polarized relocalization of gene products [RNAs or proteins] to different cell compartments [147]. This is also the universal process widely used today by different neural circuits as part of synaptic and plasticity mechanisms. The process of polarized RNA transport to neuronal processes or cilia is at the core of directional signalling, regeneration and interactions with multiple targets, including synaptogenesis.
6. Parallel recruitments of non-neuronal transmitters to neuronal functions
There are multiple examples of independent recruitment of ancestral non-neuronal signals (i.e. ATP, glutamate, NO) for neurotransmitter functions making purinergic, glutaminergic, nitrergic nerves and pathways.
The free radical, gaseous nitric oxide (NO) is one of the earliest and the most versatile transmitters: from archaea to plants, fungi and animals. NO is also the simplest signal molecule and produced by one of the most evolutionarily conservative and complex synthetic enzymes (figure 6a). Across all domains of life, NO is associated with a response to injury, innate immunity, repairing and differentiation, and acts as an intermediate of the nitrogen cycle [154]. NO is a perfect illustrative example of recruiting metabolites and paracrine signal molecules to be a neurotransmitter, but only in some lineages and some cell types.
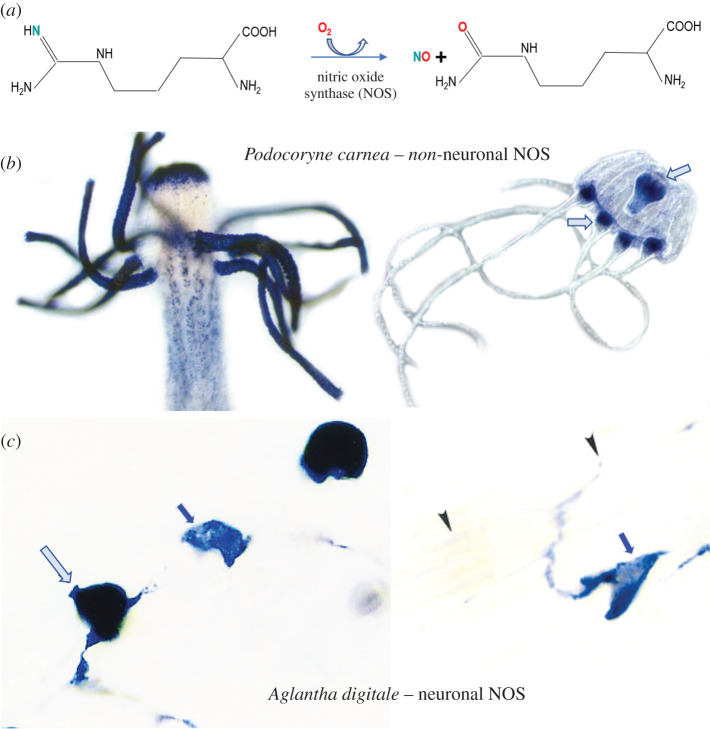
Figure 6.
Nitric oxide signalling in Cnidaria. (a) Schematic representation of the enzymatic synthesis of NO from arginine and molecular oxygen with NO and citrulline as co-products of this reaction, which requires multiple cofactors [148,149]. Non-neuronal (Podocoryne -B) and neuronal (Aglantha – C) localization of NOS in different hydrozoans. (b) The distribution of putative nitrergic cells is revealed by fixative-resistant NADPH-diaphorase histochemistry [150–152] in both polyp and medusa stages of the hydrozoan, Podocoryne carnea. Despite its widespread distribution and abundance, NOS in Podocoryne is expressed in non-neuronal cells, predominantly around the mouth and tentacles. (c) By contrast, nitric oxide synthase (NOS) is specifically expressed in neurons (arrows) of tentacles of the trachymedusa, Aglantha digitale [153].
Nitrergic (NO-releasing) neurons constitute about 2% of cell populations in our brain, where NO operates as a classical highly localized neurotransmitter sensu stricto. It is an apparent paradox since NO is free radical gas, can diffuse beyond synaptic cleft and can act as a volume transmitter [155]. The paradox is resolved by modelling and chemical quantification of presynaptic NO release. J. Garthwaite showed [114] that when small numbers of nitric oxide synthase (NOS) molecules localized at spines, its diffusion and action within the synapse was similar to other canonical neurotransmitters. For example, at the synapse, if there are 49 postsynaptic nNOS molecules, 1 per NMDA receptor, then each NOS enzyme produces 20 NO molecules per second. The calculated gradient of NO concentration was very steep from a synaptic release source, reaching 1 nM at a distance of 60 nm away within the presynaptic terminal [114]). This is sufficient to transmit a signal to its target cell because each NO, by binding to soluble guanylate cyclase, can result in the synthesis of 5000 molecules of cGMP per second at steady state [114]. Under a picomolar concentration range, NO operates within the disc of about 400 nm, which is comparable to a single mammalian synapse. However, a slight increase in a number of NOS molecules or greater NO production mediated by high-frequency stimulation of Ca2+ influx, lipopolysharachides or different microenvironments can easily convert NO action to a hormonal-type volume transmitter affecting thousands of targets. Under this mode of operation, NO can now act as a principal secretory molecule in mechanisms of innate immunity. NO can be released from macrophages or other phagocytotic cells in micromolar concentrations sufficient to kill invasive bacteria or other cells. Similarly, NO controls systemic blood pressure, reproductive functions and coordinates numerous molecular, cellular and tissue targets by integrating organism-wide processes of development, synaptic growth, differentiation, learning and memory (reviewed in [154]). In other words, the action of the very same messenger in the same organism can differently coordinate behaviours of many cells, irrespective of their identity, electrical and anatomical connectivity.
Comparative biology and evolution of NO-mediated signalling flawlessly reflect these versatile functions of NO in all animal lineages studied so far [154]. NOS/NO is present in many neural systems, but it is not everywhere. Some lineages, such as nematodes, completely lost NOS. There are many species where nitrergic neurons are absent, but NOS is widely distributed in non-neuronal tissues. There are multiple examples when within the same phyletic lineage, we can see a gradual increase in the representation of neuronally located NOS, often viewed as a transition functions from peripheral non-neuronal sources to the CNS, as in some gastropod molluscs [140,150,156–158].
In most Cnidaria, NOS is not located in neurons, but NOS is widely expressed in non-neuronal cells across species (figure 6b). In Hydra, non-neuronal NO signalling controls regeneration, feeding behaviour and chemosensation [159–161]. To the best of our knowledge, there is only one documented case in Cnidaria, when non-neuronal functions of NO, as a transmitter and an integrator of behaviours, have been ‘delegated’ to neurons. It was discovered in the trachymedusa Aglantha digitale—a highly advanced hydrozoan with giant axons and annulus type of the central nervous system [162,163]. At least 14 functional conductive systems have been physiologically identified in Aglantha including endodermal and ectodermal epithelial pathways [163–167]—an amazing example of neuroid complexity within a ‘simple’ jellyfish! We should view Aglantha as an example of what cnidarian nervous systems are ‘capable of achieving’ with distinct adaptations to a mid-water pelagic lifestyle. In Aglantha, NOS is uniquely expressed in two subpopulations of tentacle neurons (figure 6c) and control both cilia beating and feeding types of locomotion [153].
Similar ‘delegation’ of functions from non-neuronal cells to neurons can easily be reconstructed for other low molecular weight neurotransmitters, including amino acids, GABA, acetylcholine and monoamines. Non-neuronal and neuronal functions of the same transmitter often coexist in the same organism (including humans), as we illustrate for serotonin and NO above. We can postulate such a situation for all twenty known low molecular weight transmitters.
7. Lineage-specific recruitments of transmitters
A snapshot of comparative transmitter diversity in five superclades of basal metazoans is a tip of the iceberg of lineage-specific signalling innovations (figure 5). The majority of knowledge about (neuro)transmitters is derived from bilaterians, which represent textbook cases of transmitter distribution and functions. Cnidarians are surprisingly different in terms of their overall transmitter context and only partially share the overall transmitter complement with bilaterians (figures 5 and 7). Regrettably, endogenous signalling molecules are mostly unknown for three other superclades of basal metazoans (ctenophores, sponges and placozoans—see also §8).
Figure 7.
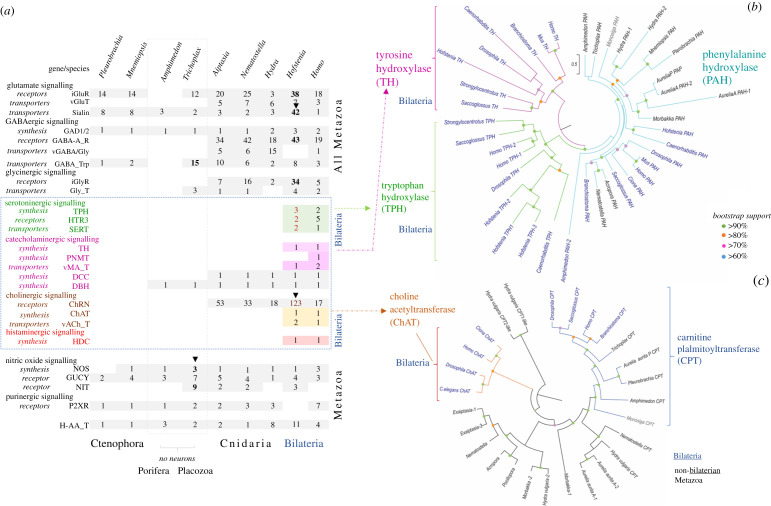
The phylogenetic distribution of canonical transmitter systems in Metazoa. (a) The table summarizes synthetic pathways for low molecular weight transmitters (see figure 4 for details and abbreviations), their transporters and receptors. Numbers indicate genes of respective molecular components in the reference genome of basal metazoans. Classical monoaminergic and cholinergic systems (including respective vesicular transporters and synthetic enzymes) have been identified only in bilaterian animals. Bold numbers and arrowheads indicate species with the greatest lineage-specific diversification of particular gene families. (b) Phylogenetic relationships within the superfamily of tryptophan- (TPH), tyrosine- (TH) and phenylalanine (PAH) hydroxylases, key monoamine synthetic enzymes (see figure 4 for details). Note that canonical TH and TPH have been only identified in bilaterians. (c) Phylogenetic relationships within the superfamily of transferases. Of note, the canonical choline-acetyltransferase (ChAT), the key enzyme for the synthesis of acetylcholine (figure 4), is present in bilaterians. On the other hand, cnidarians have related enzymes; their catalytic activity, and their ability to synthesize acetylcholine is unknown. See sections 7e and 7f for details.
(a). Nitric oxide and ATP
Genomes of ctenophores, placozoans and sponges encode purinergic P2X receptors and may use ATP as a signal molecule, but their systemic functional roles are unknown. There is one described truncated NOS gene in two ctenophore species, Mnemiopsis and Cestum, with unknown function [168]. Other studied ctenophores apparently lost NOS. NO signalling is confirmed in the control of rhythmic movements, development and metamorphosis of nerveless sponges [169,170] as well as in placozoans.
In contrast with other invertebrates studied, placozoans of genera Trichoplax and Hoilungia have three distinct NOS genes, including a PDZ domain-containing NOS [168]. Distinct NOSs are expressed in different subpopulations of cells, with a noticeable distribution close to the edge regions of Trichoplax. These data suggest both the compartmentalized release of NO and a greater diversity of cell types in placozoans than anticipated. The most surprising discovery was in receptor machinery for NO. It includes both canonical and novel NIT-domain-containing soluble guanylate cyclases as putative NO/nitrite/nitrate sensors [168]. Thus, although Trichoplax and Hoilungia exemplify the morphologically simplest free-living animals, the complexity of NO-cGMP-mediated signalling in Placozoa is greater than in vertebrates. This situation further illuminates multiple lineage-specific diversifications of NOSs and NO/nitrite/nitrate sensors from Metazoa's common ancestor and the preservation of conservative NOS architecture from prokaryotic ancestors.
(b). D-/L- glutamate and aspartate
D-amino acids are less studied but essential signalling molecules in neural, hormonal and immune systems. The comparative survey of L-/D-aspartate and L-/D-glutamate has been performed in representatives of four phyla of early branching Metazoa [171]: cnidarians (Aglantha); placozoans (Trichoplax), sponges (Sycon) and ctenophores (Pleurobrachia, Mnemiopsis, Bolinopsis and Beroe). It was shown that the placozoans, cnidarians and sponges had high micromolar concentrations of D-aspartate, whereas D-glutamate was not detectable. By contrast, in ctenophores, D-glutamate was the dominant enantiomer with no or trace amounts of D-aspartate.
D-glutamate also depolarized muscle cells, elevated intercellular calcium and induced muscle contractions, acting synergistically to L-glutamate [46].
These data suggest lineage-specific diversifications in the recruitment of D-amino acids and imply distinct signalling functions of these molecules early in the animal evolution. In ctenophores, L-glutamate operates as an excitatory neuro-muscular transmitter [46,71,72], but glutamate packing in synaptic vesicles might occur using different sialin-type transporters (similarly to placozoans and sponges) rather than vesicular glutamate transporters as in cnidarians and bilaterians (figure 7). Synthetic enzymes for glutamate are also different, suggesting that L-/D-glutamate might be recruited to neuronal functions in ctenophores differently compared to the cnidarian/bilaterian ancestor [72]. Ionotropic glutamate receptors (iGluRs) are very abundant and differentially expressed in Pleurobrachia [46]. Glycine might also activate iGluRs [172,173] and therefore act as (co)transmitter/modulator in ctenophores, but cellular and systemic functions of glycine are unknown. There is no vesicular glycine transporter either.
(c). Gamma-aminobutyric acid
GABA was not detected in ctenophore neurons and did not induce noticeable effects on ctenophore behaviours [46]. GABA was found in ctenophore muscles, which led to the hypothesis that GABA evolved as a by-product of glutamate's metabolic inactivation at the neuro-muscular synapses, with the potential use of GABA for muscle energetics [71]. Ctenophores, sponges and placozoans have neither GABA vesicular transporters nor ionotropic GABA receptors (figure 7). We might expect that transmitter molecular functions of GABA in these animals are different from cnidarians and bilaterians. In sponges, GABA induces coordinated contractions and changes in water flow. These pharmacological effects are different from actions of glutamate and NO at the same cell populations and behaviours [169,174,175].
(d). Bilaterian innovations
Serotonin, dopamine, noradrenaline, adrenaline, octopamine, tyramine, histamine and acetylcholine neurotransmitter pathways were not convincingly detected in ctenophores, placozoans, sponges and most of the cnidarians [176]. Respective synthetic enzymes and respective vesicular transporters have not been identified in the sequenced genomes of ctenophores and sponges, placozoans and cnidarians (figure 7). The pharmacological data about the presence of these transmitters are controversial and often generated using protocols that are difficult to compare.
(e). Cholinergic systems
In discussing the importance of transmitters for functional evolution, we will consider the lineage-specific diversification of cnidarian neural systems, starting with the quest for acetylcholine within these diverse groups of animals. Acetylcholine was the first transmitter discovered more than 100 years ago [89]. It is in humans the most studied neuro-muscular transmitter, with many cognitive and visceral functions. This simple molecule appears to be everywhere. However, its presence in neurons, endogenous synthesis and functions of acetylcholine in Cnidaria has not been confirmed [177–179].
Completion of more cnidarian genomes [180] has allowed the re-examination of cholinergic signalling. Phylogenetic analysis of the only biosynthetic enzyme for acetylcholine, choline-acetyltransferase (ChAT), shows that none of the studied cnidarian species contains a canonical ChAT (figure 7a,c). All the predicted bilaterian ChAT sequences cluster together and the related carnitine palmitoyltransferases (CPT) also cluster together. Interestingly, all cnidarian sequences, except the Aurelia aurita from the Pacific, form two separate clades. One appears to be more closely related to the CPT enzyme, and the other is cnidarian-specific. Further examination of the predicted cnidarian ChAT/CPT-like sequences shows that many of the known critical substrate interacting residues as well as residues shown experimentally by crystallography to be important for catalytic activity are not present in studied Cnidaria [181,182].
This analysis does not exclude a possibility for the non-canonical synthesis of acetylcholine (its recruitments from food, pathogens, or symbionts) or the presence of other related enzymes in cnidarians. However, it appears that only bilaterians have an endogenous functional canonical ChAT, classical neuronal cholinergic signalling and presynaptic acetylcholine release. Neuronal vesicular acetylcholine transporters have not been found in the sequenced cnidarian genomes and not reported in any cnidarian species (figure 7a). Although hypothetical nicotine-like and muscarine-like receptors are present in Cnidaria genomes and specifically expressed, the behavioural threshold concentration of acetylcholine is 10 mM [183], which suggests that this action is less specific and other endogenous ligands for the nicotinic receptors exist in Cnidaria. Indeed, recent work on Octopus clearly confirmed the possibility that at least some nicotinic receptors in the suckers are not sensitive to acetylcholine, but might be activated by chemosensory stimuli [184]. These findings also elaborate on the scenario that the origin of ligand-gated nicotine-like receptors channels predate the recruitment of acetylcholone as (neuro)transmitter in Metazoa.
(f). Biogenic amines
The diversity of biogenic amines in animals has been produced by several evolutionarily conserved enzymes (figure 4). Ctenophores [185], sponges [66,186–191], placozoans and cnidarians [192–210] might synthesize additional catecholamines and indolamines, but their locations and potential transmitter functions should be subject to careful investigations. There are several, mostly early, reports for the presence of serotonin, dopamine and noradrenaline in cnidarians [197,200,211–220], but it was not consistent with genomic data about predicted synthetic enzymes [176,177,179]. Although some gene orthologues were found, the complete canonical pathways for synthesis of dopamine, noradrenaline, octopamine, adrenaline, serotonin and histamine have not been detected (figures 4 and 7a,b). A few specific details that led to this conclusion are summarized below.
The superfamily of aromatic amino acid hydroxylase enzymes contains three major categories based on substrate specificity (see figures 4 and 7 for details): tryptophan hydroxylase (TPH: EC 1.14.16.4), tyrosine hydroxylase (TH: EC 1.14.16.2) and phenylalanine hydroxylase (PAH: EC 1.14. 16.1) All contain the biopterin-dependent aromatic amino acid hydroxylase (AAAH) catalytic domain (PF00351), which is critical to function [221,222]. In addition to the catalytic domain, the ACT domain, which is linked to metabolic enzymes regulated by amino acid concentration, was also sporadically detected but not necessary for function [223]. TPH and TH enzymes catalyse the rate-limiting steps involved in the synthesis of serotonin and catecholamines (figure 4).
The AAAH family of predicted proteins is highly conserved and found across eukaryotes and prokaryotes [223]. Phylogenetic analysis shows bacterial AAAH homologues form a separate clade to eukaryotic AAAH [223]. Some bacterial AAAH homologues have been expressed in Chromobacterium violaceum, which was shown to be a PAH-like enzyme and shares 22% identity to the human PAH catalytic domain [224]. The bacterial AAAH homologues are enzymatically different from the eukaryotic AAAHs because they do not form homotetramers, and some use different metals for catalysis [225].
At the base of the eukaryotic radiation, putative PAH-like enzymes are detected in unicellular eukaryotic genomes as well as choanoflagellate genomes [223]. The protozoan parasite Leishmania major was shown to have a functional PAH [226]. Phylogenetically and functionally, all predicted AAAH enzymes from protists, choanoflagellates, amoebozoa as well as algae and even mosses cluster with the PAH family of AAAHs [223].
There are three distinct eukaryotic clusters for the AAAH family of enzymes detected that represent PAH, TPH and TH sequences, with the PAH being most basal [223]. It was shown that the cnidarian Nematostella vectensis and the placozoan Trichoplax adhaerens H1 genomes both contain only one gene with the AAAH domain and clusters in the PAH family [223]. Besides, it was also shown that the predicted AAAH protein of Nematostella clustered, shared the highest identity with the PAH family of genes [46,72,176]. Our phylogenetic analysis also shows that all of the predicted cnidarian AAAH enzymes cluster with PAH. Few contain more than one predicted protein with biopterin-dependent AAAH catalytic domain (PF00351).
The cnidarian genomes analysed to date do not contain the specific rate-limiting enzymes TH and TPH involved in the synthesis of catecholamines (dopamine/noradrenaline/octopamine) and serotonin. This situation suggests that if conventional biogenic amines are indeed produced (as suggested by some immunohistochemical data), some other non-canonical, possible cnidaria-specific enzymes might exist. This situation is also consistent with the notion that TPH and TH are bilaterian innovations (figures 4 and 7).
To further support this assertion, we investigated the site-directed mutagenesis performed on both TH and TPH to see if any non-bilaterian metazoans contain critical amino acid residues to confer any TH or TPH enzymatic activity. Surprisingly, in TH, a single mutation of Asp425 (D425 V) nearly abolishes the enzymatic activity for the enzyme to produce L-DOPA [227]. And the relative specificity of TH for phenylalanine versus tyrosine, as measured by the (V/Kphe)/(V/Ktyr) value, increased by 80 000-fold in the D425 V enzyme [228]. Only predicted TH enzymes contain this critical amino acid residue, and none of the non-bilaterian putative PAH contains this residue but contain nonpolar residues. Similarly, with TPH, Tyr235 is conserved in all known TPH enzymes, whereas both TH and PAH contain a smaller hydrophobic amino acid residue [229,230]. The TPH (Y235A) or (Y235 L) mutants reduce the specific activity of TPH [229,230]. Only predicted TPH enzymes contain this critical amino acid residue and none of the non-bilaterian putative PAH contains this residue (figure 7).
The most sensitive analytical methods of capillary electrophoresis did not confirm the presence of serotonin or dopamine in sponges, placozoans and ctenophores [46,168,171,231]. Of note, the predicted PAH enzymes in two ctenophores, Pleurobrachia and Mnemiopsis, both cluster with the cnidarian PAH enzymes. By contrast, glutamate, GABA, glycine, taurine and other amino acids were found with attomolar to nanomolar limits of detection.
In our opinion, the data reported above indicate that the majority of canonical biogenic amines (serotonin, dopamine, noradrenaline, adrenaline, octopamine, tyramine and histamine) and acetylcholine as neurotransmitters are bilaterian (but not cnidarian) innovations.
Remarkably, acoels (known as basal bilaterian lineage) have an unexpected diversity of synthetic, vesicle packing and receptive pathways for all known low molecular weight transmitter pathways [232,233]. Amazingly, Hofstenia has a more complex serotonergic system than humans. Hofstenia also exceeds humans in the diversity of nicotinic-like/cholinergic, glycine, GABA and glutamate ionotropic receptors. It might not be surprising considering the dynamic behaviours of these acoels, but their nervous system is numerically very small and simpler than in the majority of bilaterians. Here, we might have a fundamentally important situation related to cellular bases of behaviour: the apparent morphological simplicity reversely correlates to the cryptic molecular complexity of transmitter and signalling pathways. In the next section, we will explore this scenario using Placozoa as one of the most promising models for deciphering the origin of a neuronal organization.
8. Volume transmission in placozoans as the most ancestral mode of behavioural integration
In previous sections, we summarized data about the distribution and functions of neurotransmitter systems in basal metazoans as well as outlined hypotheses of their origins. We explored the scenario that neurons arose from genetically different secretory cells capable of volume transmission and integration of behaviours without canonical synapses. We think that the closest representation of this primordial organization is currently found in Placozoa (figures 1 and 3), disk-like animals (figure 8) with the simplest known cell composition but complex behaviours [236–246].
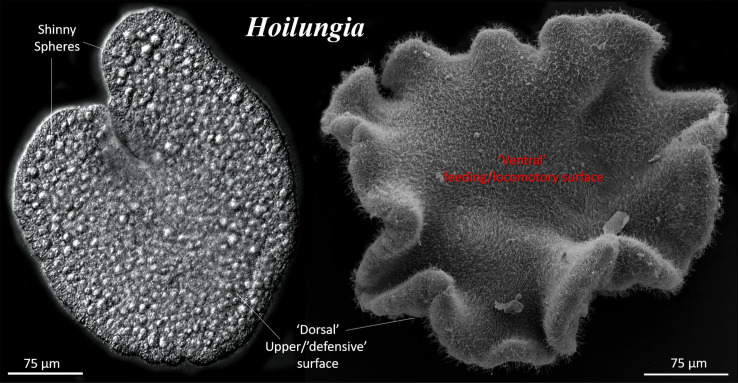
Figure 8.
General morphology of the placozoan, Hoilungia [234]. (a) Differential interference contrast (DIC) image of a living animal. Shiny spheres are large specialized cells on the ‘dorsal’ surface, releasing antipredatory toxins [235]. (b) Scanning electron microscopy of the entire animal (see details in [236]). The lower (‘ventral’) side faces the substrate. This site is primarily responsible for active ciliated locomotion and feeding. The upper (‘dorsal’) side is significantly different and contains a few subpopulations of upper epithelium and shiny sphere cells, which might be necessary for the animals' defense.
There are three formally described genera of Placozoa (Trichoplax, Hoilungia and Polyplacotoma [234,247,248]), but likely more than one hundred species live in warm tropical and subtropical shallow waters of the world's oceans [249]. These are not the most basally branching metazoan lineage (figure 2). The current consensus stands that Placozoa is the sister group to the clade Cnidaria+Bilarteria [43,45,46], although some authors consider Placozoa as highly derived and secondarily simplified cnidarians [48]. Regardless of the proposed phylogenies, Placozoa represents a very attractive model, not only to better understand the origin and evolution of animals and the nervous system in particular [250] but to decipher the logic of volume transmission and chemical integration of multiple effectors without neurons, synapses, muscles—all traits, and characteristics for the rest of basal Metazoa.
Placozoa is an idealized window to the animal past, present and future. Placozoans are not living fossils but animals that might look like Ediacaran organisms, such as Dickinsonia [251]. The type of external digestion on algal and bacterial mats looks the same as the conceptual modes for metazoan feeding in late Proterozoic [23,76,241,251,252]. Placozoans are the simplest known free-living animals with six morphologically recognized cell types [253] organized in three layers [253]. There are no tissues, no organs. But there are exceptionally complex and fast behaviours [231,239,240,242–244], including social behaviour [238] and ultrafast contractions [237,242,243,245]. The cellular bases of behaviour in placozoans are unknown, but we expect rapid progress and a conceptual breakthrough in this direction.
It was recently shown that placozoans exhibited all-or-none sodium-dependent action potentials, perhaps to support the rapid propagation and integration of electrical and chemical signals across the animal and cell layers [254]. A specialized meshwork of fibre cells (figure 9d), located in the middle layer of cells, was considered to be an analogue of the neural and muscular systems [255]. Still, no synapses and no gap junctions have been described morphologically [253,255]. The pannexins and connexins—the canonical gap junction proteins—are not encoded in the sequenced genome of T. adhaerens and its kin [64,234,248]. Adherent junctions do facilitate diffusion of potential nutrients into the animals [256], but it is unknown if they participate in the propagation of any electrical signals. Both emphaptic electrical coupling [257–260] and non-synaptic/volume-type chemical transmission might contribute to the observed behavioural integration in Placozoa. Testing and separation of these, apparently complementary, mechanisms would be important questions to be addressed experimentally.
Figure 9.
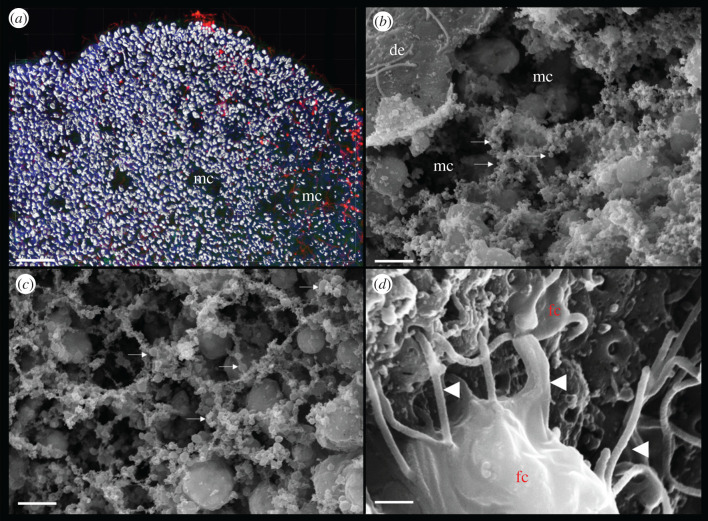
Internal morphology of Trichoplax adhaerens. (a) Three-dimensional reconstruction of cell boundaries using laser scanning microscopy (approx. 20 µm stock of images) shows regions with fewer cell densities and even distinct microcavities (mc). Nuclei are labelled by DAPI (red). (b) The same types of microcavities can also be observed under scanning electron microscopy (SEM). Special preparation by drying through the critical point can preserve fragile cell organization. (c) Arrows indicate various varicosity type structures, which might be secretory sites or exosomes. It appears that the meshwork of these varicosities surrounds cells and microcavities. See the section 8 for details. (d) Fibre cells (fc) in the middle cell layer of Trichoplax. Fibre cells might be heterogeneous with a few subpopulations. Fibre cells have many elongated long processes spread in all directions. They might be functional analogues of neurons and muscles [255] with large mitochondrial clusters [253,255] and endosymbiotic bacteria [261]. Abbreviations: de, ‘dorsal’/upper epithelium; mc, microcavities; fc, fibre cells. Scale bars: (a) 20 µm, (b,c) 5 µm, (d) 1 µm.
In summary, Placozoa's cellular architecture ideally corresponds to the proposed body plan of early animals at a ‘pre-neuronal’ stage of evolution (figure 1a). Most of the cells in Placozoa are secretory. There is highly developed external secretion both for feeding (Trichoplax looks like ‘gliding pancreatic gland’) and chemical defense (chemical ecology). There are profound chemoreceptive, targeting and exploratory behaviours [239]. Trichoplax and its kin apparently have an interior milieu in complex and highly dynamic mixtures of internal metabolites, signal molecules, its own and bacterial toxins, complex symbiotic interactions with bacteria. The known and many more features of placozoans provide a powerful paradigm for the future exploration of electrochemical/emphaptic or chemical mechanisms of behavioural integration without synapses. This is a pure chemical computational integrative machinery without neurons and any anatomically defined integrative centre or core.
The most interesting feature of Placozoa's microanatomy is the presence of an extensive system of internal microcavities, which we can observe both in fixed and living preparations (figure 9a,b). It appears that these regions of extracellular space are surrounded by multiple secretory-like vesicles (figure 9b,c), which can be both parts of cellular processes and exosomes. Exosomes are extracellular vesicles of different sizes and might present complex molecular packages to deposit and release all kinds of biomolecules from metabolites and transmitters to DNA and RNA [262–264].
Therefore, in Placozoa, we can imagine the playground for the natural selection of transmitters and the development of alternative integrative systems using complex and highly dynamic chemical gradients and changeable molecular maps (figure 10). It is a chemical hub of innovations. Such an idealized system might explain the origin of complexity and novel properties from chaos and unrelated components, as we hypothesized at the beginning of this paper. This conceptual theoretical model can be tested experimentally.
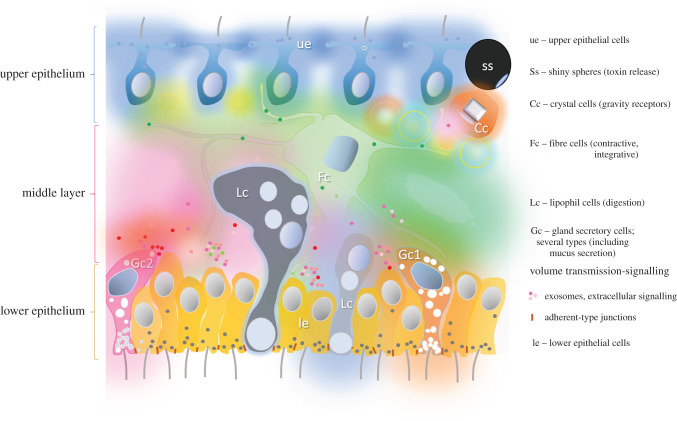
Figure 10.
Volume transmission in Placozoa as the hypothetical ancestral model of chemical behavioural integration without synapses. The cross-section of a generalized Placozoan with known cell types (their abbreviations shown on the right side of the figure). The reconstruction is based on several electron microscopic and functional studies as well as our unpublished data [168,234,240,241,245,253,255,256,265–267]; see text for details. Different colours reflect the secretory activity of many cell types with multiple overlapping chemical gradients of signal molecules, coupled with electrical and/or metabolic activity of certain endogenous pacemakers and receptive components. There is also a diversity of secretory vesicles in various cell types and, perhaps, of extracellular vesicles (exosomes)—all participating in the Claude Bernard's milieu intérieur. Under this scenario, a complex mixture (a soup) of 20–100 transmitters and modulators coordinate multiple targets and integrate behavioural outputs. This chemical combinatorial system represents a foundation for generating behavioural and, sub-sequentially, cellular innovations and setting up conditions for basal cognition.
The emerging experimental data apparently support the feasibility of a versatile ‘Solaris’-type chemical brain analogue without a classical hard-wired neuron-rich brain. We already know that Trichoplax uses a diversity of secretory peptides [268] to coordinate and achieve peptidergic integration of behaviour [240,245]. Importantly, the distribution of secretory/peptidergic cells is relatively random; it occurs mostly at the ridge of the animal. Such topographical arrangement allows efficient coordination of sensor and effector cellular populations. Behavioural integration might not be limited by peptides. There is evidence that glycine [231], NO [168], glutamate, GABA [171] and pH might also be involved in intercellular signalling and behavioural coordination. It would be important to determine whether these transmitters integrate natural feeding or mimic the behavioural reactions to tissue damage (as recapitulations of possible ancestral functions early transmitters). In Placozoa, we might have a representative set of first transmitters for non-neuronal integration of behaviours.
Under this scenario of chemical multi-transmitter integration, we may predict and experimentally test emerging novel systemic properties. For example, quantitative observations of free-moving Trichoplax identified very slow behavioural oscillations of over 600 s [244]. The observed dynamic patterns in this animal without synapses might reflect the action of multiple chemical gradients of signal molecules, potentially coupled with some electrical/emphaptic and/or metabolic activity of certain endogenous pacemakers and receptive components. The concept of the electrical syntax recently developed by G. Buzsaki for mammalian neuronal assemblies [269] can be expanded to incorporate the chemical grammar of the transmitter sea. We anticipate that the experimental sociobiology of secretory cells may open unprecedented opportunities to understand the origin of basal cognition in the complex animal with the simplest cellular and tissue organization.
9. Conclusion and perspectives
This paper explored the hypothesis that neurons primarily evolved from secretory cells [6,7,9,16,19,24,36,38]. Under this scenario, we define six complementary systemic ‘proneuronal’ adaptations:
-
(1)
The substantial diversity of secretory cell types in the nerveless ancestor of all Metazoa as seen in the present-day placozoans. It could be at least a couple of dozen such cell types, including cells involved in digestion, immunity, cell–cell recognition, differentiation, chemical ecology, defense, etc.
-
(2)
This situation provided conditions to form an enormous mixture (greater than 100–300) of potential signal molecules as playgrounds for future evolutionary selection in the late Precambrian.
-
(3)
Both low molecular weight and peptide transmitters were present, targeted multiple receptors, effectors and, consequently, led to modulation and improved coordination of behaviours in early animals without synapses.
-
(4)
Synapses were not needed for such behavioural integration. Secretory and receptive components could function separately at large distances using volume-type transmission [30,32,37]. Synapses and additional synaptic cleft components evolved later in evolution convergently after early neurons [72].
-
(5)
Hundreds of pre-existing G-protein-coupled and ligand-gated receptors (a key preadaptation) provided a broad diversity of targets for combinatorial selection of chemosensory complexes and their sub-sequential cell-specific expression.
-
(6)
Ion channels and gap junction proteins were recruited in parallel (or sometimes secondarily) as triggers to speed up intra- and intercellular communications, to localize and further synchronize cell ensembles involved in the generation of behaviours. The original functions of these hemichannels might be associated with non-synaptic secretion of metabolites and signal molecules.
Thus, any given transmitter not only acts as a pure messenger, but it integrates signalling pathways and behaviours [37]. Transmitters made the nervous system. The sociobiology of heterogeneous secretory cells was transformed into chemical sociobiology of heterogeneous neurons. Morphologically ‘simpler’ nerve nets can be examples of secondarily simplifications evolved to control stereotyped cnidarian behaviours. The electrical wiring diagrams and some reflex circuits might also represent examples of secondary simplification.
Placozoans represent a powerful model for alternative integrative systems, where waves of chemical gradients and oscillations form an internal dynamic microenvironment with multiple cryptic chemical pacemakers and temporal integrators of novel functions and behaviours.
Acknowledgements
We thank two anonymous reviewers for very important comments and suggestions. We also would like to thank Drs. M. Eitel and F. Varoqueaux for Hoilungia/H13 haplotype access.
Data accessibility
This article has no additional data.
Authors' contribution
L.L.M. was involved in concept, data analysis, imaging, writing the manuscript and funding; A.B.K. and D.Y.R. were involved in protein modelling and molecular analysis of selected protein families; D.Y.R. was involved in Placozoa imaging; A.B.K. was involved in phylogenetic trees.
Competing interests
We declare we have no competing interests
Funding
This work was supported by the Human Frontiers Science Program (RGP0060/2017), National Science Foundation (1146575, 1557923, 1548121 and 1645219) and NIH R01NS114491 grants to L.L.M.
References
- 1.Dobzhansky T. 1973. Nothing in biology makes sense except in the light of evolution. Am. Biol. Teach. 35, 125-129. ( 10.2307/4444260) [DOI] [Google Scholar]
- 2.Maynard Smith J, Szathmary EM. 1995. The major transitions in evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 3.Jennings HS. 1906. Behavior of the lower organisms. New York, NY: Macmillan. [Google Scholar]
- 4.Parker GH. 1919. The elementary nervous system. Philadelphia, PA: Lippincott. [Google Scholar]
- 5.Haldane JBC. 1954. La signalization animal. Anne. Biol. 58, 89-98. [Google Scholar]
- 6.Clark RB. 1956. On the transformation of neurosecretory cells into ordinary nerve cells. Fysiogr. Sallsk. Lund. Forth. 26, 82-89. [Google Scholar]
- 7.Clark RB. 1956. On the origin of neurosecretory cells. Ann. Sci. Nat. Zool. 18, 199-207. [Google Scholar]
- 8.Pantin CFA. 1956. The origin of the nervous system. Pubbl. Staz. Zool. Napoli 28, 171-181. [Google Scholar]
- 9.Grundfest H. 1959. Evolution of conduction in the nervous system. In Evolution of nervous control from primitive organisms to Man (ed. Bass AD), pp. 43-86. Washington, DC: American Association for Advancement of Science. [Google Scholar]
- 10.Passano LM. 1963. Primitive nervous systems. Proc. Natl Acad. Sci. USA 50, 306-313. ( 10.1073/pnas.50.2.306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bullock TH, Horridge GA. 1965. Structure and function in the nervous systems of invertebrates. San Francisco, CA: Freeman. [Google Scholar]
- 12.Grundfest H. 1965. Evolution of electrophysiological properties among sensory receptor systems. In Essays on physiological evolution (ed. Pringle JWS), pp. 107-138. Oxford, UK: Pergamon Press. [Google Scholar]
- 13.Horridge GA. 1968. The origin of the nervous system. In Structure and function of nervous tissue (ed. Bourne GH), pp. 1-33. New York, NY: Academic Press. [Google Scholar]
- 14.Lentz TL. 1968. Primitive nervous systems. New Haven, CC: Yale University Press. [Google Scholar]
- 15.Mackie GO. 1970. Neuroid conduction and the evolution of conducting tissues. Q. Rev. Biol. 45, 319-332. ( 10.1086/406645) [DOI] [PubMed] [Google Scholar]
- 16.Sakharov DA. 1974. Genealogy of neurons. Moscow, Russia: Nauka. [Google Scholar]
- 17.Mackie GO. 1990. The elementary nervous systems revisited. Am. Zool. 30, 907-920. ( 10.1093/icb/30.4.907) [DOI] [Google Scholar]
- 18.Holland ND. 2003. Early central nervous system evolution: an era of skin brains? Nat. Rev. Neurosci. 4, 617-627. ( 10.1038/nrn1175) [DOI] [PubMed] [Google Scholar]
- 19.Moroz LL. 2009. On the independent origins of complex brains and neurons. Brain Behav. Evol. 74, 177-190. ( 10.1159/000258665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan TJ, Grant SG. 2009. The origin and evolution of synapses. Nat. Rev. Neurosci. 10, 701-712. ( 10.1038/nrn2717) [DOI] [PubMed] [Google Scholar]
- 21.Keijzer F, van Duijn M, Lyon P. 2013. What nervous systems do: early evolution, input–output, and the skin brain thesis. Adapt. Behav. 21, 67-85. ( 10.1177/1059712312465330) [DOI] [Google Scholar]
- 22.Jorgensen EM. 2014. Animal evolution: looking for the first nervous system. Curr. Biol. 24, R655-R658. ( 10.1016/j.cub.2014.06.036) [DOI] [PubMed] [Google Scholar]
- 23.Monk T, Paulin MG. 2014. Predation and the origin of neurones. Brain Behav. Evol. 84, 246-261. ( 10.1159/000368177) [DOI] [PubMed] [Google Scholar]
- 24.Moroz LL. 2014. The genealogy of genealogy of neurons. Commun. Integr. Biol. 7, e993269. ( 10.4161/19420889.2014.993269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayr E. 2001. What evolution is. New York, NY: Basic Books. [Google Scholar]
- 26.Agnati LF, Bjelke B, Fuxe K. 1995. Volume versus wiring transmission in the brain: a new theoretical frame for neuropsychopharmacology. Med. Res. Rev. 15, 33-45. ( 10.1002/med.2610150104) [DOI] [PubMed] [Google Scholar]
- 27.Agnati LF, Fuxe K. 2014. Extracellular-vesicle type of volume transmission and tunnelling-nanotube type of wiring transmission add a new dimension to brain neuro-glial networks. Phil. Trans. R. Soc. B 369, 20130505. ( 10.1098/rstb.2013.0505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agnati LF, Guidolin D, Guescini M, Genedani S, Fuxe K. 2010. Understanding wiring and volume transmission. Brain Res. Rev. 64, 137-159. ( 10.1016/j.brainresrev.2010.03.003) [DOI] [PubMed] [Google Scholar]
- 29.Agnati LF, Leo G, Zanardi A, Genedani S, Rivera A, Fuxe K, Guidolin D. 2006. Volume transmission and wiring transmission from cellular to molecular networks: history and perspectives. Acta Physiol. 187, 329-344. ( 10.1111/j.1748-1716.2006.01579.x) [DOI] [PubMed] [Google Scholar]
- 30.Borroto-Escuela DO, Agnati LF, Bechter K, Jansson A, Tarakanov AO, Fuxe K. 2015. The role of transmitter diffusion and flow versus extracellular vesicles in volume transmission in the brain neural glial networks. Phil. Trans. R. Soc. B 370, 20140183. ( 10.1098/rstb.2014.0183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuxe K, et al. 2007. From the Golgi–Cajal mapping to the transmitter-based characterization of the neuronal networks leading to two modes of brain communication: wiring and volume transmission. Brain Res. Rev. 55, 17-54. ( 10.1016/j.brainresrev.2007.02.009) [DOI] [PubMed] [Google Scholar]
- 32.Sykova E. 2004. Extrasynaptic volume transmission and diffusion parameters of the extracellular space. Neuroscience 129, 861-876. ( 10.1016/j.neuroscience.2004.06.077) [DOI] [PubMed] [Google Scholar]
- 33.Trueta C, De-Miguel FF. 2012. Extrasynaptic exocytosis and its mechanisms: a source of molecules mediating volume transmission in the nervous system. Front. Physiol. 3, 319. ( 10.3389/fphys.2012.00319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoli M, Agnati LF. 1996. Wiring and volume transmission in the central nervous system: the concept of closed and open synapses. Prog. Neurobiol. 49, 363-380. ( 10.1016/0301-0082(96)00020-2) [DOI] [PubMed] [Google Scholar]
- 35.Zoli M, Torri C, Ferrari R, Jansson A, Zini I, Fuxe K, Agnati LF. 1998. The emergence of the volume transmission concept. Brain Res. Rev. 26, 136-147. ( 10.1016/s0165-0173(97)00048-9) [DOI] [PubMed] [Google Scholar]
- 36.Sakharov DA. 1974. Evolutionary aspects of transmitter heterogeneity. J. Neural Transm. Suppl. 11, 43-59. [PubMed] [Google Scholar]
- 37.Dyakonova VA, Sakharov DA. 2019. Post-reflex neurobiology of behavior. Moscow, Russia: Yask Press. [Google Scholar]
- 38.Sakharov DA. 1970. Cellular aspects of invertebrate neuropharmacology. Annu. Rev. Pharmacol. 10, 335-352. ( 10.1146/annurev.pa.10.040170.002003) [DOI] [PubMed] [Google Scholar]
- 39.Sakharov DA. 1970. Principal approaches to the systematization of nerve cells (in Russian). Zhurnal Obschey Biologii (Journal General Biology - Russian 31, 449-457. [Google Scholar]
- 40.Sakharov DA. 1972. Why are neurons different? (in Russian). Priroda 52–62.
- 41.Erwin DH, Laflamme M, Tweedt SM, Sperling EA, Pisani D, Peterson KJ. 2011. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science 334, 1091-1097. ( 10.1126/science.1206375) [DOI] [PubMed] [Google Scholar]
- 42.Erwin DH, Valentine JW. 2013. The Cambrian explosion. The construction of animal biodiversity, p. 406. Greenwood Village, CO: Roberts and Company. [Google Scholar]
- 43.Laumer CE, et al. 2019. Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc. R. Soc. B 286, 20190831. ( 10.1098/rspb.2019.0831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whelan NV, Kocot KM, Moroz LL, Halanych KM. 2015. Error, signal, and the placement of Ctenophora sister to all other animals. Proc. Natl Acad. Sci. USA 112, 5773-5778. ( 10.1073/pnas.1503453112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whelan NV, Kocot KM, Moroz TP, Mukherjee K, Williams P, Paulay G, Moroz LL, Halanych KM. 2017. Ctenophore relationships and their placement as the sister group to all other animals. Nat. Ecol. Evol. 1, 1737-1746. ( 10.1038/s41559-017-0331-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moroz LL, et al. 2014. The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109-114. ( 10.1038/nature13400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simion P, et al. 2017. A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr. Biol. 27, 958-967. ( 10.1016/j.cub.2017.02.031) [DOI] [PubMed] [Google Scholar]
- 48.Laumer CE, Gruber-Vodicka H, Hadfield MG, Pearse VB, Riesgo A, Marioni JC, Giribet G. 2018. Support for a clade of Placozoa and Cnidaria in genes with minimal compositional bias. Elife 7, e36278. ( 10.7554/eLife.36278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kocot KM, et al. 2017. Phylogenomics of Lophotrochozoa with consideration of systematic error. Syst. Biol. 66, 256-282. ( 10.1093/sysbio/syw079) [DOI] [PubMed] [Google Scholar]
- 50.Hernandez-Nicaise M-L. 1991. Ctenophora. In Microscopic anatomy of invertebrates: Placozoa, Porifera, Cnidaria, and Ctenophora (eds Harrison FWFW, Westfall JA), pp. 359-418. New York, NY: Wiley. [Google Scholar]
- 51.Jager M, Manuel M. 2016. Ctenophores: an evolutionary-developmental perspective. Curr. Opin. Genet. Dev. 39, 85-92. ( 10.1016/j.gde.2016.05.020) [DOI] [PubMed] [Google Scholar]
- 52.Norekian TP, Moroz LL. 2019. Neural system and receptor diversity in the ctenophore Beroe abyssicola. J. Comp. Neurol. 527, 1986-2008. ( 10.1002/cne.24633) [DOI] [PubMed] [Google Scholar]
- 53.Norekian TP, Moroz LL. 2019. Neuromuscular organization of the Ctenophore Pleurobrachia bachei. J. Comp. Neurol. 527, 406-436. ( 10.1002/cne.24546) [DOI] [PubMed] [Google Scholar]
- 54.Norekian TP, Moroz LL. 2020. Comparative neuroanatomy of ctenophores: neural and muscular systems in Euplokamis dunlapae and related species. J. Comp. Neurol. 528, 481-501. ( 10.1002/cne.24770) [DOI] [PubMed] [Google Scholar]
- 55.Moroz LL. 2018. NeuroSystematics and periodic system of neurons: model vs reference species at single-cell resolution. ACS Chem. Neurosci. 9, 1884-1903. ( 10.1021/acschemneuro.8b00100) [DOI] [PubMed] [Google Scholar]
- 56.Moroz LL. 2012. Phylogenomics meets neuroscience: how many times might complex brains have evolved? Acta Biol. Hung. 63(Suppl 2), 3-19. ( 10.1556/ABiol.63.2012.Suppl.2.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zverkov OA, et al. 2019. Dicyemida and Orthonectida: two stories of body plan simplification. Front. Genet. 10, 443. ( 10.3389/fgene.2019.00443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brusca RC, Brusca GJ.. 2003. Invertebrates, 2nd edn. Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- 59.Nielsen C. 2012. Animal evolution: interrelationships of the living phyla. Oxford, UK: Oxford University Press. [Google Scholar]
- 60.Chang ES, Neuhof M, Rubinstein ND, Diamant A, Philippe H, Huchon D, Cartwright P. 2015. Genomic insights into the evolutionary origin of Myxozoa within Cnidaria. Proc. Natl Acad. Sci. USA 112, 14 912-14 917. ( 10.1073/pnas.1511468112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gruhl A, Okamura B. 2012. Development and myogenesis of the vermiform Buddenbrockia (Myxozoa) and implications for cnidarian body plan evolution. EvoDevo 3, 10. ( 10.1186/2041-9139-3-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kyger R, et al. 2020. Myxosporea (Myxozoa, Cnidaria) lack DNA cytosine methylation. Mol. Biol. Evol., msaa214. ( 10.1093/molbev/msaa214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yahalomi D, Haddas-Sasson M, Rubinstein ND, Feldstein T, Diamant A, Huchon D. 2017. The multipartite mitochondrial genome of Enteromyxum leei (Myxozoa): eight fast-evolving megacircles. Mol. Biol. Evol. 34, 1551-1556. ( 10.1093/molbev/msx072) [DOI] [PubMed] [Google Scholar]
- 64.Srivastava M, et al. 2008. The Trichoplax genome and the nature of Placozoans. Nature 454, 955-960. ( 10.1038/nature07191) [DOI] [PubMed] [Google Scholar]
- 65.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455, 1193-1197. ( 10.1038/nature07415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Srivastava M, et al. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720-726. ( 10.1038/nature09201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fortunato SA, Adamski M, Ramos OM, Leininger S, Liu J, Ferrier DE, Adamska M. 2014. Calcisponges have a ParaHox gene and dynamic expression of dispersed NK homeobox genes. Nature 514, 620-623. ( 10.1038/nature13881) [DOI] [PubMed] [Google Scholar]
- 68.Fernandez-Valverde SL, Degnan BM. 2016. Bilaterian-like promoters in the highly compact Amphimedon queenslandica genome. Sci. Rep. 6, 22496. ( 10.1038/srep22496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riesgo A, Farrar N, Windsor PJ, Giribet G, Leys SP. 2014. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol. Biol. Evol. 31, 1102-1120. ( 10.1093/molbev/msu057) [DOI] [PubMed] [Google Scholar]
- 70.Whelan NV, Kocot KM, Halanych KM. 2015. Employing phylogenomics to resolve the relationships among Cnidarians, Ctenophores, Sponges, Placozoans, and Bilaterians. Integr. Comp. Biol. 55, 1084-1095. ( 10.1093/icb/icv037) [DOI] [PubMed] [Google Scholar]
- 71.Moroz LL. 2015. Convergent evolution of neural systems in ctenophores. J. Exp. Biol. 218, 598-611. ( 10.1242/jeb.110692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moroz LL, Kohn AB. 2016. Independent origins of neurons and synapses: insights from ctenophores. Phil. Trans. R. Soc. B 371, 20150041. ( 10.1098/rstb.2015.0041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marlow H, Arendt D. 2014. Evolution: ctenophore genomes and the origin of neurons. Curr. Biol. 24, R757-R761. ( 10.1016/j.cub.2014.06.057) [DOI] [PubMed] [Google Scholar]
- 74.Telford MJ, Moroz LL, Halanych KM. 2016. Evolution: a sisterly dispute. Nature 529, 286-287. ( 10.1038/529286a) [DOI] [PubMed] [Google Scholar]
- 75.Jekely G, Paps J, Nielsen C. 2015. The phylogenetic position of ctenophores and the origin(s) of nervous systems. Evodevo 6, 1. ( 10.1186/2041-9139-6-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nielsen C. 2019. Early animal evolution: a morphologist's view. R. Soc. Open Sci. 6, 190638. ( 10.1098/rsos.190638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Daley AC, Antcliffe JB. 2019. Evolution: the battle of the first animals. Curr. Biol. 29, R257-R259. ( 10.1016/j.cub.2019.02.031) [DOI] [PubMed] [Google Scholar]
- 78.Wilson EO. 2012. The social conquest of earth. New York, NY: Liveright. [Google Scholar]
- 79.Weiger WA. 1997. Serotonergic modulation of behaviour: a phylogenetic overview. Biol. Rev. Camb. Philos. Soc. 72, 61-95. ( 10.1017/s0006323196004975) [DOI] [PubMed] [Google Scholar]
- 80.Bacque-Cazenave J, Cattaert D, Delbecque JP, Fossat P. 2017. Social harassment induces anxiety-like behaviour in crayfish. Sci. Rep. 7, 39935. ( 10.1038/srep39935) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou C, Rao Y, Rao Y. 2008. A subset of octopaminergic neurons are important for Drosophila aggression. Nat. Neurosci. 11, 1059-1067. ( 10.1038/nn.2164) [DOI] [PubMed] [Google Scholar]
- 82.Rittschof CC, Vekaria HJ, Palmer JH, Sullivan PG. 2019. Biogenic amines and activity levels alter the neural energetic response to aggressive social cues in the honey bee Apis mellifera. J. Neurosci. Res. 97, 991-1003. ( 10.1002/jnr.24443) [DOI] [PubMed] [Google Scholar]
- 83.Momohara Y, Aonuma H, Nagayama T. 2018. Tyraminergic modulation of agonistic outcomes in crayfish. J. Comp. Physiol. A 204, 465-473. ( 10.1007/s00359-018-1255-3) [DOI] [PubMed] [Google Scholar]
- 84.Kravitz EA. 1988. Hormonal control of behavior: amines and the biasing of behavioral output in lobsters. Science 241, 1775-1781. ( 10.1126/science.2902685) [DOI] [PubMed] [Google Scholar]
- 85.Gellner AK, Voelter J, Schmidt U, Beins EC, Stein V, Philipsen A, Hurlemann R. 2020. Molecular and neurocircuitry mechanisms of social avoidance. Cell. Mol. Life Sci. ( 10.1007/s00018-020-03649-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kendrick KM, Guastella AJ, Becker B. 2018. Overview of human oxytocin research. Curr. Top Behav. Neurosci. 35, 321-348. ( 10.1007/7854_2017_19) [DOI] [PubMed] [Google Scholar]
- 87.Shepherd GM. 2015. Foundations of the neuron doctrine. Oxford, UK: Oxford University Press. [Google Scholar]
- 88.Bullock TH, Bennett MV, Johnston D, Josephson R, Marder E, Fields RD. 2005. Neuroscience. The neuron doctrine, redux. Science 310, 791-793. ( 10.1126/science.1114394) [DOI] [PubMed] [Google Scholar]
- 89.Valenstein ES. 2002. The discovery of chemical neurotransmitters. Brain Cogn. 49, 73-95. ( 10.1006/brcg.2001.1487) [DOI] [PubMed] [Google Scholar]
- 90.Valenstein ES. 2005. The war of the soups and the sparks: The discovery of neurotransmitters and the dispute over how nerves communicate. New York, NY: Columbia University Press. [Google Scholar]
- 91.Panchin YV. 2005. Evolution of gap junction proteins—the pannexin alternative. J. Exp. Biol. 208, 1415-1419. ( 10.1242/jeb.01547) [DOI] [PubMed] [Google Scholar]
- 92.Slivko-Koltchik GA, Kuznetsov VP, Panchin YV. 2019. Are there gap junctions without connexins or pannexins? BMC Evol. Biol. 19, 46. ( 10.1186/s12862-019-1369-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845-858. ( 10.1038/nprot.2015.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang N, et al. 2013. Paracrine signaling through plasma membrane hemichannels. Biochim. Biophys. Acta 1828, 35-50. ( 10.1016/j.bbamem.2012.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taruno A. 2018. ATP release channels. Int. J. Mol. Sci. 19, 308. ( 10.3390/ijms19030808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ruan Z, Orozco IJ, Du J, Lu W. 2020. Structures of human pannexin 1 reveal ion pathways and mechanism of gating. Nature 584, 646-651. ( 10.1038/s41586-020-2357-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Orellana JA, Martinez AD, Retamal MA. 2013. Gap junction channels and hemichannels in the CNS: regulation by signaling molecules. Neuropharmacology 75, 567-582. ( 10.1016/j.neuropharm.2013.02.020) [DOI] [PubMed] [Google Scholar]
- 98.Nurse CA. 2014. Synaptic and paracrine mechanisms at carotid body arterial chemoreceptors. J. Physiol. 592, 3419-3426. ( 10.1113/jphysiol.2013.269829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Montero TD, Orellana JA. 2015. Hemichannels: new pathways for gliotransmitter release. Neuroscience 286, 45-59. ( 10.1016/j.neuroscience.2014.11.048) [DOI] [PubMed] [Google Scholar]
- 100.Momboisse F, Olivares MJ, Baez-Matus X, Guerra MJ, Flores-Munoz C, Saez JC, Martinez AD, Cardenas AM. 2014. Pannexin 1 channels: new actors in the regulation of catecholamine release from adrenal chromaffin cells. Front. Cell Neurosci. 8, 270. ( 10.3389/fncel.2014.00270) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Locovei S, Bao L, Dahl G. 2006. Pannexin 1 in erythrocytes: function without a gap. Proc. Natl Acad. Sci. USA 103, 7655-7659. ( 10.1073/pnas.0601037103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang YJ, Maruyama Y, Dvoryanchikov G, Pereira E, Chaudhari N, Roper SD. 2007. The role of pannexin 1 hemichannels in ATP release and cell–cell communication in mouse taste buds. Proc. Natl Acad. Sci. USA 104, 6436-6441. ( 10.1073/pnas.0611280104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gaete PS, Contreras JE. 2020. Taking a close look at a large-pore channel. Elife 9, e56114. ( 10.7554/eLife.56114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deng Z, He Z, Maksaev G, Bitter RM, Rau M, Fitzpatrick JAJ, Yuan P. 2020. Cryo-EM structures of the ATP release channel pannexin 1. Nat. Struct. Mol. Biol. 27, 373-381. ( 10.1038/s41594-020-0401-0) [DOI] [PubMed] [Google Scholar]
- 105.Dahl G, Qiu F, Wang J. 2013. The bizarre pharmacology of the ATP release channel pannexin1. Neuropharmacology 75, 583-593. ( 10.1016/j.neuropharm.2013.02.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dahl G, Locovei S. 2006. Pannexin: to gap or not to gap, is that a question? IUBMB Life 58, 409-419. ( 10.1080/15216540600794526) [DOI] [PubMed] [Google Scholar]
- 107.Dahl G. 2015. ATP release through pannexon channels. Phil. Trans. R. Soc. B 370, 20140191. ( 10.1098/rstb.2014.0191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bao L, Locovei S, Dahl G. 2004. Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Lett. 572, 65-68. ( 10.1016/j.febslet.2004.07.009) [DOI] [PubMed] [Google Scholar]
- 109.Abudara V, Retamal MA, Del Rio R, Orellana JA. 2018. Synaptic functions of hemichannels and pannexons: a double-edged sword. Front. Mol. Neurosci. 11, 435. ( 10.3389/fnmol.2018.00435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Piccolino M. 2006. Luigi Galvani's path to animal electricity. C. R. Biol. 329, 303-318. ( 10.1016/j.crvi.2006.03.002) [DOI] [PubMed] [Google Scholar]
- 111.Nieuwenhuys R. 2000. Comparative aspects of volume transmission, with sidelight on other forms of intercellular communication. Prog. Brain Res. 125, 49-126. ( 10.1016/S0079-6123(00)25006-1) [DOI] [PubMed] [Google Scholar]
- 112.Ridet I, Privat A. 2000. Volume transmission. Trends Neurosci. 23, 58-59. ( 10.1016/s0166-2236(99)01523-4) [DOI] [PubMed] [Google Scholar]
- 113.Taber KH, Hurley RA. 2014. Volume transmission in the brain: beyond the synapse. J. Neuropsychiatry Clin. Neurosci. 26, iv, 1–4. ( 10.1176/appi.neuropsych.13110351) [DOI] [PubMed] [Google Scholar]
- 114.Garthwaite J. 2016. From synaptically localized to volume transmission by nitric oxide. J. Physiol. 594, 9-18. ( 10.1113/JP270297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carrettam M 1988. The Retzius cells in the leech: a review of their properties and synaptic connections. Comp. Biochem. Physiol. A: Physiol. 91, 405-413. ( 10.1016/0300-9629(88)90611-1) [DOI] [PubMed] [Google Scholar]
- 116.Lent CM. 1974. Neuronal control of mucus secretion by leeches: toward a general theory for serotonin. Am. Zool. 14, 931-942. ( 10.1093/icb/14.3.931) [DOI] [Google Scholar]
- 117.Hatcher NG, Zhang X, Stuart JN, Moroz LL, Sweedler JV, Gillette R. 2008. 5-HT and 5-HT-SO4, but not tryptophan or 5-HIAA levels in single feeding neurons track animal hunger state. J. Neurochem. 104, 1358-1363. ( 10.1111/j.1471-4159.2007.05084.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gillette R, Davis W. 1977. The role of the metacerebral giant neuron in the feeding behavior of Pleurobranchaea. J. Comp. Physiol. 116, 129-159. ( 10.1007/BF00605400) [DOI] [Google Scholar]
- 119.Weiss KR, Kupfermann I. 1976. Homology of the giant serotonergic neurons (metacerebral cells) in Aplysia and pulmonate molluscs. Brain Res. 117, 33-49. ( 10.1016/0006-8993(76)90554-0) [DOI] [PubMed] [Google Scholar]
- 120.Moroz LL, Sudlow LC, Jing J, Gillette R. 1997. Serotonin-immunoreactivity in peripheral tissues of the opisthobranch molluscs Pleurobranchaea californica and Tritonia diomedea. J. Comp. Neurol. 382, 176-188. () [DOI] [PubMed] [Google Scholar]
- 121.Sudlow LC, Jing J, Moroz LL, Gillette R. 1998. Serotonin immunoreactivity in the central nervous system of the marine molluscs Pleurobranchaea californica and Tritonia diomedea. J. Comp. Neurol. 395, 466-480. () [DOI] [PubMed] [Google Scholar]
- 122.Gillette R, Huang RC, Hatcher N, Moroz LL. 2000. Cost–benefit analysis potential in feeding behavior of a predatory snail by integration of hunger, taste, and pain. Proc. Natl Acad. Sci. USA 97, 3585-3590. ( 10.1073/pnas.97.7.3585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Terry N, Margolis KG. 2017. Serotonergic mechanisms regulating the GI tract: experimental evidence and therapeutic relevance. Handb. Exp. Pharmacol. 239, 319-342. ( 10.1007/164_2016_103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nautiyal KM, Dailey CA, Jahn JL, Rodriquez E, Son NH, Sweedler JV, Silver R. 2012. Serotonin of mast cell origin contributes to hippocampal function. Eur. J. Neurosci. 36, 2347-2359. ( 10.1111/j.1460-9568.2012.08138.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Watanabe K, Chiu H, Pfeiffer BD, Wong AM, Hoopfer ED, Rubin GM, Anderson DJ. 2017. A circuit node that integrates convergent input from neuromodulatory and social behavior-promoting neurons to control aggression in Drosophila. Neuron 95, 1112-1128 e1117. ( 10.1016/j.neuron.2017.08.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moroz LL, Winlow W. 1992. Respiratory behaviour in Lymnaea stagnalis: pharmacological and cellular analyses. Acta Biol. Hung. 43, 421-429. [PubMed] [Google Scholar]
- 127.Moroz LL. 1991. Monoaminergic control of the respiratory behaviour in freshwater pulmonate snail, Lymnaea stagnalis (L.). In Signal molecules and behaviour (eds Winlow W, Vinogradova OV, Sakharov DA), pp. 101-123. Manchester, UK: Manchester University Press. [Google Scholar]
- 128.Winlow W, Moroz LL, Syed NI. 1992. Mechanisms of behavioral selection in Lymnaea stagnalis. In Neurobiology of motor programme selection (eds Kien J, McCrohan CR, Winlow W), pp. 52-72. Oxford, UK: Pergamon Press. [Google Scholar]
- 129.Hatcher NG, Sweedler JV. 2008. Aplysia bag cells function as a distributed neurosecretory network. J. Neurophysiol. 99, 333-343. ( 10.1152/jn.00968.2007) [DOI] [PubMed] [Google Scholar]
- 130.Bernheim SM, Mayeri E. 1995. Complex behavior induced by egg-laying hormone in Aplysia. J. Comp. Physiol. A 176, 131-136. ( 10.1007/BF00197759) [DOI] [PubMed] [Google Scholar]
- 131.Painter SD. 1992. Coordination of reproductive activity in Aplysia: peptide neurohormones, neurotransmitters, and pheromones encoded by the egg-laying hormone family of genes. Biol. Bull. 183, 165-172. ( 10.2307/1542419) [DOI] [PubMed] [Google Scholar]
- 132.Sossin WS, Sweet-Cordero A, Scheller RH. 1990. Dale's hypothesis revisited: different neuropeptides derived from a common prohormone are targeted to different processes. Proc. Natl Acad. Sci. USA 87, 4845-4848. ( 10.1073/pnas.87.12.4845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ferguson GP, Ter Maat A, Pinsker HM. 1989. Egg laying in Aplysia. II. Organization of central and peripheral pathways for initiating neurosecretory activity and behavioral patterns. J. Comp. Physiol. A 164, 849-857. ( 10.1007/BF00616756) [DOI] [PubMed] [Google Scholar]
- 134.Ferguson GP, Ter Maat A, Parsons DW, Pinsker HM. 1989. Egg laying in Aplysia. I. Behavioral patterns and muscle activity of freely behaving animals after selectively elicited bag cell discharges. J. Comp. Physiol. A 164, 835-847. ( 10.1007/BF00616755) [DOI] [PubMed] [Google Scholar]
- 135.McAllister LB, Scheller RH, Kandel ER, Axel R. 1983. In situ hybridization to study the origin and fate of identified neurons. Science 222, 800-808. ( 10.1126/science.6356362) [DOI] [PubMed] [Google Scholar]
- 136.Kupfermann I. 1970. Stimulation of egg laying by extracts of neuroendocrine cells (bag cells) of abdominal ganglion of Aplysia. J. Neurophysiol. 33, 877-881. ( 10.1152/jn.1970.33.6.877) [DOI] [PubMed] [Google Scholar]
- 137.Kupfermann I. 1967. Stimulation of egg laying: possible neuroendocrine function of bag cells of abdominal ganglion of Aplysia californica. Nature 216, 814-815. ( 10.1038/216814a0) [DOI] [PubMed] [Google Scholar]
- 138.Rosen SC, Kupfermann I, Goldstein RS, Weiss KR. 1983. Lesion of a serotonergic modulatory neuron in Aplysia produces a specific defect in feeding behavior. Brain Res. 260, 151-155. ( 10.1016/0006-8993(83)90778-3) [DOI] [PubMed] [Google Scholar]
- 139.Kupfermann I, Weiss KR. 1982. Activity of an identified serotonergic neuron in free moving Aplysia correlates with behavioral arousal. Brain Res. 241, 334-337. ( 10.1016/0006-8993(82)91072-1) [DOI] [PubMed] [Google Scholar]
- 140.Moroz LL, Norekian TP, Pirtle TJ, Robertson KJ, Satterlie RA. 2000. Distribution of NADPH-diaphorase reactivity and effects of nitric oxide on feeding and locomotory circuitry in the pteropod mollusc, Clione limacina. J. Comp. Neurol. 427, 274-284. () [DOI] [PubMed] [Google Scholar]
- 141.Toyota M, Spencer D, Sawai-Toyota S, Jiaqi W, Zhang T, Koo AJ, Howe GA, Gilroy S. 2018. Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361, 1112-1115. ( 10.1126/science.aat7744) [DOI] [PubMed] [Google Scholar]
- 142.Takahashi T. 2020. Comparative aspects of structure and function of Cnidarian neuropeptides. Front. Endocrinol. 11, 339. ( 10.3389/fendo.2020.00339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Walters ET, Moroz LL. 2009. Molluscan memory of injury: evolutionary insights into chronic pain and neurological disorders. Brain Behav. Evol. 74, 206-218. ( 10.1159/000258667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kandel ER. 2001. The molecular biology of memory storage: a dialogue between genes and synapses. Science 294, 1030-1038. ( 10.1126/science.1067020) [DOI] [PubMed] [Google Scholar]
- 145.Walters ET, Williams ACC. 2019. Evolution of mechanisms and behaviour important for pain. Phil. Trans. R. Soc. B 374, 20190275. ( 10.1098/rstb.2019.0275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Walters ET. 2018. Nociceptive biology of molluscs and arthropods: evolutionary clues about functions and mechanisms potentially related to pain. Front. Physiol. 9, 1049. ( 10.3389/fphys.2018.01049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Puthanveettil SV, et al. 2013. A strategy to capture and characterize the synaptic transcriptome. Proc. Natl Acad. Sci. USA 110, 7464-7469. ( 10.1073/pnas.1304422110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ignarro LJ. 2000. Nitric oxide: biology and pathobiology, p. 1003. San Diego, CA: Academic Press. [Google Scholar]
- 149.Moncada S, Palmer RM, Higgs EA. 1991. Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacol. Rev. 43, 109-142. [PubMed] [Google Scholar]
- 150.Moroz LL. 2000. Giant identified NO-releasing neurons and comparative histochemistry of putative nitrergic systems in gastropod molluscs. Microsc. Res. Tech. 49, 557-569. () [DOI] [PubMed] [Google Scholar]
- 151.Dawson TM, Bredt DS, Fotuhi M, Hwang PM, Snyder SH. 1991. Nitric oxide synthase and neuronal NADPH diaphorase are identical in brain and peripheral tissues. Proc. Natl Acad. Sci. USA 88, 7797-7801. ( 10.1073/pnas.88.17.7797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hope BT, Michael GJ, Knigge KM, Vincent SR. 1991. Neuronal NADPH diaphorase is a nitric oxide synthase. Proc. Natl Acad. Sci. USA 88, 2811-2814. ( 10.1073/pnas.88.7.2811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Moroz LL, Meech RW, Sweedler JV, Mackie GO. 2004. Nitric oxide regulates swimming in the jellyfish Aglantha digitale. J. Comp. Neurol. 471, 26-36. ( 10.1002/cne.20023) [DOI] [PubMed] [Google Scholar]
- 154.Moroz LL, Kohn AB. 2011. Parallel evolution of nitric oxide signaling: diversity of synthesis and memory pathways. Front. Biosci. (Landmark Ed) 16, 2008-2051. ( 10.2741/3837) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Steinert JR, et al. 2008. Nitric oxide is a volume transmitter regulating postsynaptic excitability at a glutamatergic synapse. Neuron 60, 642-656. ( 10.1016/j.neuron.2008.08.025) [DOI] [PubMed] [Google Scholar]
- 156.Moroz LL, Park JH, Winlow W. 1993. Nitric oxide activates buccal motor patterns in Lymnaea stagnalis. Neuroreport 4, 643-646. ( 10.1097/00001756-199306000-00010) [DOI] [PubMed] [Google Scholar]
- 157.Moroz LL, Gillette R. 1996. NADPH-diaphorase localization in the CNS and peripheral tissues of the predatory sea-slug Pleurobranchaea californica. J. Comp. Neurol. 367, 607-622. () [DOI] [PubMed] [Google Scholar]
- 158.Moroz LL, Gillette R. 1995. From Polyplacophora to Cephalopoda: comparative analysis of nitric oxide signalling in mollusca. Acta Biol. Hung. 46, 169-182. [PubMed] [Google Scholar]
- 159.Colasanti M, Venturini G, Merante A, Musci G, Lauro GM. 1997. Nitric oxide involvement in Hydra vulgaris very primitive olfactory-like system. J. Neurosci. 17, 493-499. ( 10.1523/JNEUROSCI.17-01-00493.1997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Colasanti M, Mazzone V, Mancinelli L, Leone S, Venturini G. 2009. Involvement of nitric oxide in the head regeneration of Hydra vulgaris. Nitric Oxide 21, 164-170. ( 10.1016/j.niox.2009.07.003) [DOI] [PubMed] [Google Scholar]
- 161.Cristino L, Guglielmotti V, Cotugno A, Musio C, Santillo S. 2008. Nitric oxide signaling pathways at neural level in invertebrates: functional implications in cnidarians. Brain Res. 1225, 17-25. ( 10.1016/j.brainres.2008.04.056) [DOI] [PubMed] [Google Scholar]
- 162.Norekian TP, Moroz LL. 2020. Atlas of the neuromuscular system in the Trachymedusa Aglantha digitale: insights from the advanced hydrozoan. J. Comp. Neurol. 528, 1231-1254. ( 10.1002/cne.24821) [DOI] [PubMed] [Google Scholar]
- 163.Mackie GO. 2004. Central neural circuitry in the jellyfish Aglantha: a model 'simple nervous system'. Neurosignals 13, 5-19. ( 10.1159/000076155) [DOI] [PubMed] [Google Scholar]
- 164.Mackie G, Meech R. 1995. Central circuitry in the jellyfish Aglantha. II: the ring giant and carrier systems. J. Exp. Biol. 198, 2271-2278. [DOI] [PubMed] [Google Scholar]
- 165.Mackie G, Meech R. 1995. Central circuitry in the jellyfish Aglantha. I: the relay system. J. Exp. Biol. 198, 2261-2270. [DOI] [PubMed] [Google Scholar]
- 166.Mackie GO, Marx RM, Meech RW. 2003. Central circuitry in the jellyfish Aglantha digitale IV. Pathways coordinating feeding behaviour. J. Exp. Biol. 206, 2487-2505. ( 10.1242/jeb.00450) [DOI] [PubMed] [Google Scholar]
- 167.Mackie GO, Meech RW. 2000. Central circuitry in the jellyfish Aglantha digitale. III. The rootlet and pacemaker systems. J. Exp. Biol. 203, 1797-1807. [DOI] [PubMed] [Google Scholar]
- 168.Moroz LL, Romanova DY, Nikitin MA, Sohn D, Kohn AB, Neveu E, Varoqueaux F, Fasshauer D. 2020. The diversification and lineage-specific expansion of nitric oxide signaling in Placozoa: insights in the evolution of gaseous transmission. Sci. Rep. 10, 13020. ( 10.1038/s41598-020-69851-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ellwanger K, Nickel M. 2006. Neuroactive substances specifically modulate rhythmic body contractions in the nerveless metazoon Tethya wilhelma (Demospongiae. Porifera). Front. Zool. 3, 7. ( 10.1186/1742-9994-3-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Say TE, Degnan SM. 2019. Molecular and behavioural evidence that interdependent photo- and chemosensory systems regulate larval settlement in a marine sponge. Mol. Ecol. 29, 247-261. ( 10.1111/mec.15318) [DOI] [PubMed] [Google Scholar]
- 171.Moroz LL, Sohn D, Romanova DY, Kohn AB. 2020. Microchemical identification of enantiomers in early-branching animals: lineage-specific diversification in the usage of D-glutamate and D-aspartate. Biochem. Biophys. Res. Commun. 527, 947-952. ( 10.1016/j.bbrc.2020.04.135) [DOI] [PubMed] [Google Scholar]
- 172.Alberstein R, Grey R, Zimmet A, Simmons DK, Mayer ML. 2015. Glycine activated ion channel subunits encoded by ctenophore glutamate receptor genes. Proc. Natl Acad. Sci. USA 112, E6048-E6057. ( 10.1073/pnas.1513771112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yu A, Alberstein R, Thomas A, Zimmet A, Grey R, Mayer ML, Lau AY. 2016. Molecular lock regulates binding of glycine to a primitive NMDA receptor. Proc. Natl Acad. Sci. USA 113, E6786-E6795. ( 10.1073/pnas.1607010113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Elliott GR, Leys SP. 2010. Evidence for glutamate, GABA and NO in coordinating behaviour in the sponge, Ephydatia muelleri (Demospongiae, Spongillidae). J. Exp. Biol. 213, 2310-2321. ( 10.1242/jeb.039859) [DOI] [PubMed] [Google Scholar]
- 175.Leys SP, Mah JL, McGill PR, Hamonic L, De Leo FC, Kahn AS. 2019. Sponge behavior and the chemical basis of responses: a post-genomic view. Integr. Comp. Biol. 59, 751-764. ( 10.1093/icb/icz122) [DOI] [PubMed] [Google Scholar]
- 176.Moroz LL, Kohn AB. 2015. Unbiased view of synaptic and neuronal gene complement in Ctenophores: are there pan-neuronal and pan-synaptic genes across Metazoa? Integr. Comp. Biol. 55, 1028-1049. ( 10.1093/icb/icv104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kass-Simon G, Pierobon P. 2007. Cnidarian chemical neurotransmission, an updated overview. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 146, 9-25. ( 10.1016/j.cbpa.2006.09.008) [DOI] [PubMed] [Google Scholar]
- 178.Oren M, Brickner I, Appelbaum L, Levy O. 2014. Fast neurotransmission related genes are expressed in non nervous endoderm in the sea anemone Nematostella vectensis. PLoS ONE 9, e93832. ( 10.1371/journal.pone.0093832) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Anctil M. 2009. Chemical transmission in the sea anemone Nematostella vectensis: a genomic perspective. Comp. Biochem. Physiol. Part D: Genomics Proteomics 4, 268-289. ( 10.1016/j.cbd.2009.07.001) [DOI] [PubMed] [Google Scholar]
- 180.Khalturin K, et al. 2019. Medusozoan genomes inform the evolution of the jellyfish body plan. Nat. Ecol. Evol. 3, 811-822. ( 10.1038/s41559-019-0853-y) [DOI] [PubMed] [Google Scholar]
- 181.Govindasamy L, Pedersen B, Lian W, Kukar T, Gu Y, Jin S, Agbandje-McKenna M, Wu D, McKenna R. 2004. Structural insights and functional implications of choline acetyltransferase. J. Struct. Biol. 148, 226-235. ( 10.1016/j.jsb.2004.06.005) [DOI] [PubMed] [Google Scholar]
- 182.Lian W, Gu Y, Pedersen B, Kukar T, Govindasamy L, Agbandje-McKenna M, Jin S, McKenna R, Wu D. 2004. Crystallization and preliminary X-ray crystallographic studies on recombinant rat choline acetyltransferase. Acta Crystallogr. D Biol. Crystallogr. 60, 374-375. ( 10.1107/S090744490302818X) [DOI] [PubMed] [Google Scholar]
- 183.Faltine-Gonzalez DZ, Layden MJ. 2019. Characterization of nAChRs in Nematostella vectensis supports neuronal and non-neuronal roles in the cnidarian–bilaterian common ancestor. Evodevo 10, 27. ( 10.1186/s13227-019-0136-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bronson PG, Chaivorapol C, Ortmann W, Behrens TW, Graham RR. 2012. The genetics of type I interferon in systemic lupus erythematosus. Curr. Opin Immunol. 24, 530-537. ( 10.1016/j.coi.2012.07.008) [DOI] [PubMed] [Google Scholar]
- 185.Townsend JP, Sweeney AM. 2019. Catecholic compounds in Ctenophore colloblast and nerve net proteins suggest a structural role for DOPA-like molecules in an early-diverging animal lineage. Biol. Bull. 236, 55-65. ( 10.1086/700695) [DOI] [PubMed] [Google Scholar]
- 186.Kenny NJ, et al. 2020. Tracing animal genomic evolution with the chromosomal-level assembly of the freshwater sponge Ephydatia muelleri. Nat. Commun. 11, 3676. ( 10.1038/s41467-020-17397-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Einarsdottir E, Liu HB, Freysdottir J, Gotfredsen CH, Omarsdottir S. 2016. Immunomodulatory N-acyl dopamine glycosides from the icelandic marine sponge Myxilla incrustans collected at a hydrothermal vent site. Planta Med. 82, 903-909. ( 10.1055/s-0042-105877) [DOI] [PubMed] [Google Scholar]
- 188.Alghazwi M, Kan YQ, Zhang W, Gai WP, Yan XX. 2016. Neuroprotective activities of marine natural products from marine sponges. Curr. Med. Chem. 23, 360-382. ( 10.2174/0929867323666151127201249) [DOI] [PubMed] [Google Scholar]
- 189.Bifulco G, Bruno I, Riccio R, Lavayre J, Bourdy G. 1995. Further brominated bis- and tris-indole alkaloids from the deep-water New Caledonian marine sponge Orina Sp. J. Nat. Prod. 58, 1254-1260. ( 10.1021/np50122a017) [DOI] [PubMed] [Google Scholar]
- 190.Lee HS, Yoon KM, Han YR, Lee KJ, Chung SC, Kim TI, Lee SH, Shin J, Oh KB. 2009. 5-Hydroxyindole-type alkaloids, as Candida albicans isocitrate lyase inhibitors, from the tropical sponge Hyrtios sp. Bioorg. Med. Chem. Lett. 19, 1051-1053. ( 10.1016/j.bmcl.2009.01.017) [DOI] [PubMed] [Google Scholar]
- 191.Salmoun M, Devijver C, Daloze D, Braekman JC, van Soest RW. 2002. 5-hydroxytryptamine-derived alkaloids from two marine sponges of the genus Hyrtios. J. Nat. Prod. 65, 1173-1176. ( 10.1021/np020009+) [DOI] [PubMed] [Google Scholar]
- 192.Gregson RP, Lohr RR, Marwood JF, Quinn RJ. 1981. 3-Hydroxy-4-methoxyphenethylamine, the cardioactive constituent of a soft coral. Experientia 37, 493-494. ( 10.1007/BF01986157) [DOI] [PubMed] [Google Scholar]
- 193.Carlberg M. 1983. Evidence of dopa in the nerves of sea anemones. J. Neural Transm. 57, 75-84. ( 10.1007/BF01250049) [DOI] [PubMed] [Google Scholar]
- 194.Van Marle J, Van Weeren-Kramer J, Lind A. 1983. Properties of a catecholaminergic system in some coelenterates. A histochemical and autoradiographic study. Comp. Biochem. Physiol. C: Comp. Pharmacol. 76, 193-197. ( 10.1016/0742-8413(83)90062-2) [DOI] [PubMed] [Google Scholar]
- 195.Carlberg M, Jergil B, Lindbladh C, Rosengren E. 1984. Enzymatic 5-hydroxylation of L-dopa by a tyrosinase isolated from the sea anemone Metridium senile. Gen. Pharmacol. 15, 301-307. ( 10.1016/0306-3623(84)90005-3) [DOI] [PubMed] [Google Scholar]
- 196.Venturini G, Silei O, Palladini G, Carolei A, Margotta V. 1984. Aminergic neurotransmitters and adenylate cyclase in hydra. Comp. Biochem. Physiol. C Comp. Pharmacol. Toxicol. 78, 345-348. ( 10.1016/0742-8413(84)90095-1) [DOI] [PubMed] [Google Scholar]
- 197.Kolberg KJ, Martin VJ. 1988. Morphological, cytochemical and neuropharmacological evidence for the presence of catecholamines in hydrozoan planulae. Development 103, 249-258. [DOI] [PubMed] [Google Scholar]
- 198.Carlberg M. 1990. 3,4-Dihydroxyphenylethylamine, L-3,4-dihydroxyphenylalanine and 3,4,5-trihydroxyphenylalanine: oxidation and binding to membranes. A comparative study of a neurotransmitter, a precursor and a neurotransmitter candidate in primitive nervous systems. J. Neural Transm. Gen. Sect. 81, 111-119. ( 10.1007/BF01245831) [DOI] [PubMed] [Google Scholar]
- 199.Pani AK, Anctil M. 1994. Evidence for biosynthesis and catabolism of monoamines in the sea pansy Renilla koellikeri (Cnidaria). Neurochem. Int. 25, 465-474. ( 10.1016/0197-0186(94)90023-x) [DOI] [PubMed] [Google Scholar]
- 200.Walker RJ, Brooks HL, Holden-Dye L. 1996. Evolution and overview of classical transmitter molecules and their receptors. Parasitology 113, S3-S33. ( 10.1017/s0031182000077878) [DOI] [PubMed] [Google Scholar]
- 201.McCauley DW. 1997. Serotonin plays an early role in the metamorphosis of the hydrozoan Phialidium gregarium. Dev. Biol. 190, 229-240. ( 10.1006/dbio.1997.8698) [DOI] [PubMed] [Google Scholar]
- 202.Anctil M, Hurtubise P, Gillis MA. 2002. Tyrosine hydroxylase and dopamine-β-hydroxylase immunoreactivities in the cnidarian Renilla koellikeri. Cell Tissue Res. 310, 109-117. ( 10.1007/s00441-002-0601-4) [DOI] [PubMed] [Google Scholar]
- 203.Ostroumova TV, Markova LN. 2002. The effects of dopamine synthesis inhibitors and dopamine antagonists on regeneration in the hydra Hydra attenuata. Neurosci. Behav. Physiol. 32, 293-298. ( 10.1023/a:1015066424928) [DOI] [PubMed] [Google Scholar]
- 204.Zega G, Pennati R, Fanzago A, De Bernardi F. 2007. Serotonin involvement in the metamorphosis of the hydroid Eudendrium racemosum. Int. J. Dev. Biol. 51, 307-313. ( 10.1387/ijdb.062195gz) [DOI] [PubMed] [Google Scholar]
- 205.Markova LN, Ostroumova TV, Akimov MG, Bezuglov VV. 2008. N-arachidonoyl dopamine is a possible factor of the rate of tentacle formation in freshwater hydra. Ontogenez 39, 66-71. [PubMed] [Google Scholar]
- 206.Ostroumova TV, Markova LN, Akimov MG, Gretskaia NM, Bezuglov VV. 2010. Docosahexaenoyl dopamine in freshwater hydra: effects on regeneration and metabolic changes. Ontogenez 41, 199-203. [PubMed] [Google Scholar]
- 207.Hwang DS, Masic A, Prajatelistia E, Iordachescu M, Waite JH. 2013. Marine hydroid perisarc: a chitin- and melanin-reinforced composite with DOPA-iron(III) complexes. Acta Biomater. 9, 8110-8117. ( 10.1016/j.actbio.2013.06.015) [DOI] [PubMed] [Google Scholar]
- 208.Isomura N, Yamauchi C, Takeuchi Y, Takemura A. 2013. Does dopamine block the spawning of the acroporid coral Acropora tenuis? Sci. Rep. 3, 2649. ( 10.1038/srep02649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Taira J, Higa I, Tsuchida E, Isomura N, Iguchi A. 2018. Neurotransmitters in hermatypic coral, Acropora spp., and its contribution to synchronous spawning during reproductive event. Biochem. Biophys. Res. Commun. 501, 80-84. ( 10.1016/j.bbrc.2018.04.170) [DOI] [PubMed] [Google Scholar]
- 210.Moeller M, Nietzer S, Schupp PJ. 2019. Neuroactive compounds induce larval settlement in the scleractinian coral Leptastrea purpurea. Sci. Rep. 9, 2291. ( 10.1038/s41598-019-38794-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mathias AP, Ross DM, Schachter M. 1960. The distribution 5-hydroxytryptamine, tetramethyammonium, homarine, and other substances in sea anemones. J. Physiol. 151, 296-311. ( 10.1113/jphysiol.1960.sp006439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Welsh JH. 1960. 5-Hydroxytryptamine in coelenterates. Nature 186, 811-812. ( 10.1038/186811a0) [DOI] [PubMed] [Google Scholar]
- 213.Wood JG, Lentz TL. 1964. Histochemical localization of amines in hydra and the sea anemone. Nature 201, 88-90. ( 10.1038/201088b0) [DOI] [PubMed] [Google Scholar]
- 214.Kline ES, Weissbach H. 1965. Hydroxyindoleamines in the nematocysts of Hydra littoralis. Life Sci. (1962) 4, 63-67. ( 10.1016/0024-3205(65)90034-2) [DOI] [PubMed] [Google Scholar]
- 215.Welsh JH. 1968. Distribution of serotonin in the nervous system of various animal species. Adv. Pharmacol. 6, 171-188. ( 10.1016/s1054-3589(08)61171-0) [DOI] [PubMed] [Google Scholar]
- 216.Umbriaco D, Anctil M, Descarries L. 1990. Serotonin-immunoreactive neurons in the cnidarian Renilla koellikeri. J. Comp. Neurol. 291, 167-178. ( 10.1002/cne.902910202) [DOI] [PubMed] [Google Scholar]
- 217.Hay-Schmidt A. 2000. The evolution of the serotonergic nervous system. Proc. R. Soc. B 267, 1071-1079. ( 10.1098/rspb.2000.1111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Mayorova TD, Kosevich IA. 2013. Serotonin-immunoreactive neural system and contractile system in the hydroid Cladonema (Cnidaria, Hydrozoa). Invert. Neurosci. 13, 99-106. ( 10.1007/s10158-013-0152-2) [DOI] [PubMed] [Google Scholar]
- 219.Carlyle RF. 1969. The occurrence of catecholamines in the sea anemone. Br. J. Pharmacol. 36, 182P. [PMC free article] [PubMed] [Google Scholar]
- 220.Lenicque PM, Toneby MI, Doumenc D. 1977. Demonstration of biogenic amines and localization of monoamine oxidases in the sea anemone Metridium senile (Linne). Comp. Biochem. Physiol. C: Comp. Pharmacol. 56, 31-34. ( 10.1016/0306-4492(77)90045-4) [DOI] [PubMed] [Google Scholar]
- 221.Fitzpatrick PF. 1999. Tetrahydropterin-dependent amino acid hydroxylases. Annu. Rev. Biochem. 68, 355-381. ( 10.1146/annurev.biochem.68.1.355) [DOI] [PubMed] [Google Scholar]
- 222.Grenett HE, Ledley FD, Reed LL, Woo SL. 1987. Full-length cDNA for rabbit tryptophan hydroxylase: functional domains and evolution of aromatic amino acid hydroxylases. Proc. Natl Acad. Sci. USA 84, 5530-5534. ( 10.1073/pnas.84.16.5530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Cao J, Shi F, Liu X, Huang G, Zhou M. 2010. Phylogenetic analysis and evolution of aromatic amino acid hydroxylase. FEBS Lett. 584, 4775-4782. ( 10.1016/j.febslet.2010.11.005) [DOI] [PubMed] [Google Scholar]
- 224.Chen D, Frey PA. 1998. Phenylalanine hydroxylase from Chromobacterium violaceum. Uncoupled oxidation of tetrahydropterin and the role of iron in hyroxylation. J. Biol. Chem. 273, 25 594-25 601. ( 10.1074/jbc.273.40.25594) [DOI] [PubMed] [Google Scholar]
- 225.Fitzpatrick PF. 2003. Mechanism of aromatic amino acid hydroxylation. Biochemistry 42, 14 083-14 091. ( 10.1021/bi035656u) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Lye LF, Kang SO, Nosanchuk JD, Casadevall A, Beverley SM. 2011. Phenylalanine hydroxylase (PAH) from the lower eukaryote Leishmania major. Mol. Biochem. Parasitol. 175, 58-67. ( 10.1016/j.molbiopara.2010.09.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Daubner SC, Melendez J, Fitzpatrick PF. 2000. Reversing the substrate specificities of phenylalanine and tyrosine hydroxylase: aspartate 425 of tyrosine hydroxylase is essential for L-DOPA formation. Biochemistry 39, 9652-9661. ( 10.1021/bi000493k) [DOI] [PubMed] [Google Scholar]
- 228.Daubner SC, et al. 2013. Mutagenesis of a specificity-determining residue in tyrosine hydroxylase establishes that the enzyme is a robust phenylalanine hydroxylase but a fragile tyrosine hydroxylase. Biochemistry 52, 1446-1455. ( 10.1021/bi400031n) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jiang GC, Yohrling GJT, Schmitt JD, Vrana KE. 2000. Identification of substrate orienting and phosphorylation sites within tryptophan hydroxylase using homology-based molecular modeling. J. Mol. Biol. 302, 1005-1017. ( 10.1006/jmbi.2000.4097) [DOI] [PubMed] [Google Scholar]
- 230.Wang L, Erlandsen H, Haavik J, Knappskog PM, Stevens RC. 2002. Three-dimensional structure of human tryptophan hydroxylase and its implications for the biosynthesis of the neurotransmitters serotonin and melatonin. Biochemistry 41, 12 569-12 574. ( 10.1021/bi026561f) [DOI] [PubMed] [Google Scholar]
- 231.Romanova DY, Heyland A, Sohn D, Kohn AB, Fasshauer D, Varoqueaux F, Moroz LL. 2020. Glycine as a signaling molecule and chemoattractant in Trichoplax (Placozoa): insights into the early evolution of neurotransmitters. Neuroreport 31, 490-497. ( 10.1097/WNR.0000000000001436) [DOI] [PubMed] [Google Scholar]
- 232.Hulett RE, Potter D, Srivastava M. 2020. Neural architecture and regeneration in the acoel Hofstenia miamia. Proc. R. Soc. B 287, 20201198. ( 10.1098/rspb.2020.1198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Gehrke AR, et al. 2019. Acoel genome reveals the regulatory landscape of whole-body regeneration. Science 363, aau6173. ( 10.1126/science.aau6173) [DOI] [PubMed] [Google Scholar]
- 234.Eitel M, et al. 2018. Comparative genomics and the nature of Placozoan species. PLoS Biol. 16, e2005359. ( 10.1371/journal.pbio.2005359) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Jackson AM, Buss LW. 2009. Shiny spheres of Placozoans (Trichoplax) function in anti-predator defense. Invert. Biol. 128, 205-212. ( 10.1111/j.1744-7410.2009.00177.x) [DOI] [Google Scholar]
- 236.Romanova DY, Varoqueaux F, Daraspe J, Nikitin MA, Eitel M, Fasshauer D, Moroz LL. 2020. Hidden cell diversity in Placozoa: ultrastructural insights from Hoilungia hongkongensis. Biorxiv 2020.11.13.382382. ( 10.1101/2020.11.13.382382) [DOI] [PMC free article] [PubMed]
- 237.Armon S, Bull MS, Aranda-Diaz A, Prakash M. 2018. Ultrafast epithelial contractions provide insights into contraction speed limits and tissue integrity. Proc. Natl Acad. Sci. USA 115, E10333-E10341. ( 10.1073/pnas.1802934115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Fortunato A, Aktipis A. 2019. Social feeding behavior of Trichoplax adhaerens. Front. Ecol. Evol. 7, 19. ( 10.3389/fevo.2019.00019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Heyland A, Croll R, Goodall S, Kranyak J, Wyeth R. 2014. Trichoplax adhaerens, an enigmatic basal metazoan with potential. Methods Mol. Biol. 1128, 45-61. ( 10.1007/978-1-62703-974-1_4) [DOI] [PubMed] [Google Scholar]
- 240.Senatore A, Reese TS, Smith CL. 2017. Neuropeptidergic integration of behavior in Trichoplax adhaerens, an animal without synapses. J. Exp. Biol. 220, 3381-3390. ( 10.1242/jeb.162396) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Smith CL, Mayorova TD. 2019. Insights into the evolution of digestive systems from studies of Trichoplax adhaerens. Cell Tissue Res. 377, 353-367. ( 10.1007/s00441-019-03057-z) [DOI] [PubMed] [Google Scholar]
- 242.Smith CL, Pivovarova N, Reese TS. 2015. Coordinated feeding behavior in Trichoplax, an animal without synapses. PLoS ONE 10, e0136098. ( 10.1371/journal.pone.0136098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Smith CL, Reese TS, Govezensky T, Barrio RA. 2019. Coherent directed movement toward food modeled in Trichoplax, a ciliated animal lacking a nervous system. Proc. Natl Acad. Sci. USA 116, 8901-8908. ( 10.1073/pnas.1815655116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ueda T, Koya S, Maruyama YK. 1999. Dynamic patterns in the locomotion and feeding behaviors by the Placozoan Trichoplax adhaerence. Biosystems 54, 65-70. ( 10.1016/s0303-2647(99)00066-0) [DOI] [PubMed] [Google Scholar]
- 245.Varoqueaux F, Williams EA, Grandemange S, Truscello L, Kamm K, Schierwater B, Jekely G, Fasshauer D. 2018. High cell diversity and complex peptidergic signaling underlie Placozoan behavior. Curr. Biol. 28, 3495-3501 e3492. ( 10.1016/j.cub.2018.08.067) [DOI] [PubMed] [Google Scholar]
- 246.Zuccolotto-Arellano J, Cuervo-Gonzalez R. 2020. Binary fission in Trichoplax is orthogonal to the subsequent division plane. Mech. Dev. 162, 103608. ( 10.1016/j.mod.2020.103608) [DOI] [PubMed] [Google Scholar]
- 247.Eitel M, Osigus HJ, DeSalle R, Schierwater B. 2013. Global diversity of the Placozoa. PLoS ONE 8, e57131. ( 10.1371/journal.pone.0057131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Osigus HJ, Rolfes S, Herzog R, Kamm K, Schierwater B. 2019. Polyplacotoma mediterranea is a new ramified Placozoan species. Curr. Biol. 29, R148-R149. ( 10.1016/j.cub.2019.01.068) [DOI] [PubMed] [Google Scholar]
- 249.Schierwater B, DeSalle R. 2018. Placozoa. Curr. Biol. 28, R97-R98. ( 10.1016/j.cub.2017.11.042) [DOI] [PubMed] [Google Scholar]
- 250.Varoqueaux F, Fasshauer D. 2017. Getting nervous: an evolutionary overhaul for communication. Annu. Rev. Genet. 51, 455-476. ( 10.1146/annurev-genet-120116-024648) [DOI] [PubMed] [Google Scholar]
- 251.Sperling EA, Vinther J. 2010. A Placozoan affinity for Dickinsonia and the evolution of late Proterozoic metazoan feeding modes. Evol. Dev. 12, 201-209. ( 10.1111/j.1525-142X.2010.00404.x) [DOI] [PubMed] [Google Scholar]
- 252.Steinmetz PRH. 2019. A non-bilaterian perspective on the development and evolution of animal digestive systems. Cell Tissue Res. 377, 321-339. ( 10.1007/s00441-019-03075-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Smith CL, Varoqueaux F, Kittelmann M, Azzam RN, Cooper B, Winters CA, Eitel M, Fasshauer D, Reese TS. 2014. Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens. Curr. Biol. 24, 1565-1572. ( 10.1016/j.cub.2014.05.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Romanova DY, Smirnov IV, Nikitin MA, Kohn AB, Borman AI, Malyshev AY, Balaban PM, Moroz LL. 2020. Sodium action potentials in placozoa: insights into behavioral integration and evolution of nerveless animals. Biochem. Biophys. Res. Commun. 532, 120-126. ( 10.1016/j.bbrc.2020.08.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Grell KG, Ruthmann A. 1991. Placozoa. In Microscopic anatomy of invertebrates (ed Harrison F. W.), pp. 13-27. New York, NY: Wiley-Liss. [Google Scholar]
- 256.Smith CL, Reese TS. 2016. Adherens junctions modulate diffusion between epithelial cells in Trichoplax adhaerens. Biol. Bull. 231, 216-224. ( 10.1086/691069) [DOI] [PubMed] [Google Scholar]
- 257.Anastassiou CA, Perin R, Markram H, Koch C. 2011. Ephaptic coupling of cortical neurons. Nat. Neurosci. 14, 217-223. ( 10.1038/nn.2727) [DOI] [PubMed] [Google Scholar]
- 258.Faber DS, Pereda AE. 2018. Two forms of electrical transmission between neurons. Front. Mol. Neurosci. 11, 427. ( 10.3389/fnmol.2018.00427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Grundfest H. 1967. Synaptic and ephaptic transmission. In The neurosciences. A study program (eds Quarton GC, Melnechuk T, Schmitt FO), pp. 353-372. New York, NY: The Rockefeller University Press. [Google Scholar]
- 260.Grundfest H. 1971. The general electrophysiology of input membrane in electrogenic excitable cells. In Principles of receptor physiology (ed. Loewenstein WR), pp. 135-165. Berlin, Germany: Springer-Verlag. [Google Scholar]
- 261.Gruber-Vodicka HR, Leisch N, Kleiner M, Hinzke T, Liebeke M, McFall-Ngai M, Hadfield MG, Dubilier N. 2019. Two intracellular and cell type-specific bacterial symbionts in the Placozoan Trichoplax H2. Nat. Microbiol. 4, 1465-1474. ( 10.1038/s41564-019-0475-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Hoshino D, Kirkbride KC, Costello K, Clark ES, Sinha S, Grega-Larson N, Tyska MJ, Weaver AM. 2013. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 5, 1159-1168. ( 10.1016/j.celrep.2013.10.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Bobrie A, Colombo M, Raposo G, Thery C. 2011. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12, 1659-1668. ( 10.1111/j.1600-0854.2011.01225.x) [DOI] [PubMed] [Google Scholar]
- 264.Morton DJ, Kuiper EG, Jones SK, Leung SW, Corbett AH, Fasken MB. 2018. The RNA exosome and RNA exosome-linked disease. RNA 24, 127-142. ( 10.1261/rna.064626.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Guidi L, Eitel M, Cesarini E, Schierwater B, Balsamo M. 2011. Ultrastructural analyses support different morphological lineages in the phylum placozoa Grell, 1971. J. Morphol. 272, 371-378. ( 10.1002/jmor.10922) [DOI] [PubMed] [Google Scholar]
- 266.Mayorova TD, Hammar K, Winters CA, Reese TS, Smith CL. 2019. The ventral epithelium of Trichoplax adhaerens deploys in distinct patterns cells that secrete digestive enzymes, mucus or diverse neuropeptides. Biol. Open 8, bio045674. ( 10.1242/bio.045674) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Mayorova TD, Smith CL, Hammar K, Winters CA, Pivovarova NB, Aronova MA, Leapman RD, Reese TS. 2018. Cells containing aragonite crystals mediate responses to gravity in Trichoplax adhaerens (Placozoa), an animal lacking neurons and synapses. PLoS ONE 13, e0190905. ( 10.1371/journal.pone.0190905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Nikitin M. 2015. Bioinformatic prediction of Trichoplax adhaerens regulatory peptides. Gen. Comp. Endocrinol. 212, 145-155. ( 10.1016/j.ygcen.2014.03.049) [DOI] [PubMed] [Google Scholar]
- 269.Buzsaki G. 2010. Neural syntax: cell assemblies, synapsembles, and readers. Neuron 68, 362-385. ( 10.1016/j.neuron.2010.09.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.