Cancer is second leading cause of death in the United States, and part of improving survival outcomes is early detection and treatment. Assessment of molecular pathology is critical for cancer treatment, as tumor grade and metastatic potential determines therapeutic modality and likelihood of efficacy. Tissue-based analyses using tumor biopsies are limited by their availability and narrow scope of sampling (i.e., only one small section of a large, heterogeneous or metastatic tumor). Recent technology uses circulating tumor DNA, termed cell-free tumor DNA (ctDNA), that is present in body fluids (blood plasma, urine, spinal fluid) from dead cancer cells to assay for cancer type and grade. The advantage of liquid biopsies like ctDNA is that the DNA is reflective of both local tumors and distant metastatic sites, and that multiple samples can be taken non-invasively. However, these assays are only effective for tumor types with a high mutational burden, require complex analysis and are associated with high false positive/negative rates. Recent work in the microbiome space has suggested that fecal microbial composition and metabolites have predictive features in both colorectal cancer (CRC) and response to chemotherapy or immunotherapy (1). Previous efforts have been made to use the fecal microbiota signature as a non-invasive diagnostic and prognostic tool for CRC, however, the application to broader cancer types has not been attempted. Thus, the diagnostic power of microbiota still remains unclear, but represents an area of particular interest for both identifying biomarkers of disease and therapeutic response.
In a recent manuscript published in Nature, Poore et al. address the possibility of using microbial DNA (mbDNA) to discriminate between cancer and healthy patients (2). They mined The Cancer Genome Atlas (TCGA), a compendium of whole-genome and whole-transcriptome sequencing studies that were performed on over 20,000 primary cancer and normal-matched samples of 33 cancer types, for microbial sequences. Microbial reads were examined from 18,116 samples from 10,481 patients, of which 6.4 x 1012 reads were classified as non-human (7.2% of total). Of this subset, 35.2% were identified as bacteria (3.5% of total), and 12.6% of these bacterial sequences were resolved to the genus level using a publicly available reference database. Despite rigorous standards for sample quality by the TCGA, there were several limitations to the use of these sequences. The tumor-associated microbiome DNA is of low biomass compared to host tissue DNA, which makes identifying and eliminating low-abundance and batch-specific contaminants challenging. The samples were sequenced at several centers using different platforms, a documented confounding variable in microbial studies, and healthy individuals were excluded from sampling. Additionally, the TCGA did not require negative blank extraction controls to be sequenced, therefore limiting ability to identify contaminants from the RNA/DNA isolation process. To address these challenges, Poore et al. implemented stringent, state-of-the-art bioinformatics pipelines. Their statistical framework was designed to identify potential contaminants, removing taxa only found in single batches or sequencing centers. This process identified 283 potential contaminating microbes, which were eliminated along with up to 92% of total reads using a strict filtering schema.
To assess the predictive ability of the resulting tumor-associated microbial signatures, the group trained a machine learning (ML) model using the normalized data to perform 1) cancer vs normal discrimination, 2) identification of cancer type, and 3) distinction between stage I and IV cancers. This model performed well for outcomes 1 and 2, with an area under the precision-recall curve P-value of 0.0089 for distinguishing one cancer type versus all others. The model also was able to discriminate between stage I and stage IV tumors in three cancer types, including CRC, but not for other types or for intermediate stages, a noted limitation. Cancers with published associations to the presence of specific microorganisms (i.e., Fusobacterium spp. in gastrointestinal tumors (1), human papillomavirus infection in cervical squamous cell carcinoma and head and neck squamous cell carcinoma samples (3)) were also examined to ensure that the analysis could detect these specific microbial signals in their associated cancer samples. The risk of using such stringent parameters is that real signals reflecting the normal commensal microbiota of the body sites may be discarded with cancer-associated signal; Poore et al. addressed this problem by re-evaluating the ML models pre and post-decontamination, and found that while two of the 33 may be unreliable, for all other cancer types, data that underwent this rigorous decontamination performed as well or better in tissue-vs-normal distinction. Cancer type identification was less accurate, and thus a universal decontamination pipeline may not be appropriate in all situations. These findings indicate that there are unique microbial signatures that belong to discrete tumor type tissues, and that these signatures may be useful for diagnosis.
Spurred by these findings, the group then sought to compare the ability of blood-based mbDNA in discriminating between normal and healthy tissue and between cancer types. They focused on using plasma-based signatures as opposed to whole blood, and developed a ML model based on the sequences from the matched blood samples in the TCGA cohort. The resulting model performed well in distinguishing between types of cancer, but again struggled to distinguish between cancer grades, a significant limitation for predictive biomarkers. Interestingly, the algorithm was able to discriminate healthy vs tumor in two cancer types that have a low mutational burden, glioblastoma multiforme at 95% accuracy and pancreatic adenocarcinoma at 93%, which opens up new interest in examining a microbial component in development or progression of these diseases. Poore et al. sought to validate their findings through analyzing an additional cohort of 69 subjects (cancer and HIV-free) and 100 patients with one of three cancer types: prostate, lung and melanoma. Cell free DNA was extracted from these samples using gold-standard microbiology practices and whole metagenomic sequencing was performed. The same decontamination and normalization pipeline was applied to the analysis of these reads, followed by the ML protocol used earlier (Figure 1). The resulting algorithm was able to discriminate between healthy and cancer patients in all cancer types tested except for melanoma (also the smallest cohort), and this performance was repeated using a second analysis pipeline and taxonomic classification database. Their results suggest that mbDNA is a potential biomarker for cancer detection and diagnosis in liquid biopsies like plasma, and excitingly that their model may outperform current technology for finding low mutational burden cancers like glioblastoma multiforme and pancreatic adenocarcinoma.
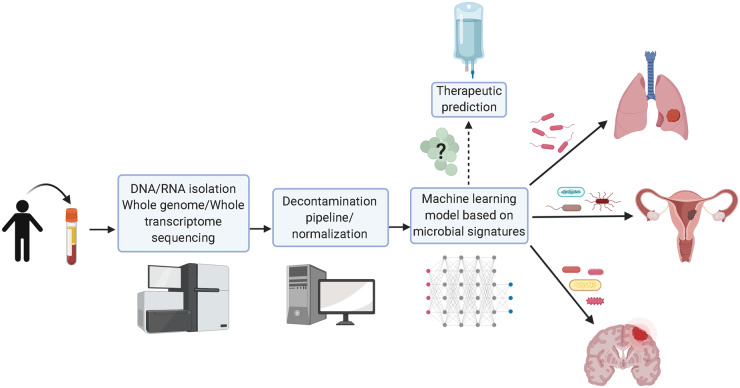
Figure 1.
A machine learning model based on circulating microbial blood-based DNA is a potential diagnostic tool for discriminating between healthy vs normal patients and cancer type. Microbial blood-based DNA in plasma from patients is sequenced and then analyzed via a rigorous bioinformatics pipeline. The microbial signature is used to diagnose cancer, with potential applications for predicting appropriate treatment and therapeutic response. Created with BioRender.com
The study by Poore et al. suggests that there are existing associations between specific cancers and microbiota, and that these relationships could be valuable for non-invasive cancer detection and diagnosis. However, only patient samples with known cancer diagnoses were enrolled in the TCGA study, and therefore the predictive power of mbDNA will require further testing using a longitudinal, randomized study. Since microbiota represent a complex ecosystem with members from bacteria, fungi and virus, it would be of great interest to investigate the predictive diagnostic power of the mycobiome/virome in cancer since 64% of the genomic reads identified in the study were from non-bacterial origin. Another tremendous opportunity for using mbDNA is the possibility to predict treatment response and to design a microbiota-based precision medicine approach. It would be important to address this possibility by linking microbiota signals with therapeutic response and survival data currently available for this cohort.
How applicable is the mbDNA approach to the clinical setting? The Poore et al. study relies on next generation sequencing (NGS) and sophisticated bioinformatic analysis. The application of NGS in oncology is quite standard (whole genome, exome, transcriptome) but clinical laboratories mostly utilize targeted gene sequencing for cancer diagnosis. A larger NGS approach such as the one described by Poore et al. is more compatible with academic hospitals or commercial laboratories than mainstream clinics. Another critical aspect of the technology is whether mbDNA could diagnose cancer in an individual at pre-cancer stage or early stage 1, especially in hard-to-screen cancers such as pancreatic, stomach and brain cancer. The Poore et al. model had limited success in discriminating between stages, and thus this remains an unknown relationship. The rate of false positive/negatives is a concern for any screening modalities. In a study using serum bacterial DNA to identify patients with hepatocellular carcinoma, ML was able to distinguish patients with hepatocellular carcinoma only 79.8% of the time, meaning two out of every 10 patients would have received either a false or a missed diagnosis (4). Even current liquid biopsy technique has been shown to have high variability between samples and a lack of reproducibility, serious concerns which have prevented inclusion of this approach as mainstream clinical diagnostic tool (5). Large multi-center studies are needed to validate a mbDNA-based model for diagnostic use in the clinic, and the microbiota signature may be more appropriate as a part of a comprehensive biomarker panel than a singular diagnostic. In summary, Poore et al. opens up a new path to cancer diagnostics by bringing the microbiome as a potential cancer biomarker, and highlights the benefits of using big data to find clinically relevant associations.
Author Contributions
All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
Authors’ Disclosures or Potential Conflicts of Interest
Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership
None declared.
Consultant or Advisory Role
None declared.
Stock Ownership
None declared.
Honoraria
None declared.
Research Funding
C. Jobin, NIH grant R01DK073338, the University of Florida Health Cancer Center Funds, and University of Florida Department of Medicine Gatorade Fund.
Expert Testimony
None declared.
Patents
None declared.
References
- 1. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA.. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 2018;33:570–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020;579:567–74. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Cancer Genome Atlas Research Network, Albert Einstein College of Medicine, Analytical Biological Services, Barretos Cancer Hospital, Baylor College of Medicine, Beckman Research Institute of City of Hope, et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cho EJ, Leem S, Kim SA, Yang J, Lee YB, Kim SS, et al. Circulating Microbiota-Based Metagenomic Signature for Detection of Hepatocellular Carcinoma. Sci Rep 2019;9:7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Rubis G, Rajeev Krishnan S, Bebawy M.. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci 2019;40:172–86. [DOI] [PubMed] [Google Scholar]