Abstract
Noncoding RNAs (ncRNAs) are involved in various biological processes, including gene expression, development, and disease. Here, we identify a novel consensus sequence of a cis-element involved in long ncRNA (lncRNA) transcription and demonstrate that lncRNA transcription from this cis-element activates meiotic recombination via chromatin remodeling. In the fission yeast fbp1 gene, glucose starvation induces a series of promoter-associated lncRNAs, referred to as metabolic-stress-induced lncRNAs (mlonRNAs), which contribute to chromatin remodeling and fbp1 activation. Translocation of the cis-element required for mlonRNA into a well-characterized meiotic recombination hotspot, ade6-M26, further stimulates transcription and meiotic recombination via local chromatin remodeling. The consensus sequence of this cis-element (mlon-box) overlaps with meiotic recombination sites in the fission yeast genome. At one such site, the SPBC24C6.09c upstream region, meiotic double-strand break (DSB) formation is induced in an mlon-box-dependent manner. Therefore, mlonRNA transcription plays a universal role in chromatin remodeling and the regulation of transcription and recombination.
Subject terms: Long non-coding RNAs, Chromatin remodelling
Senmatsu et al. show that transcription of long-noncoding RNAs (lncRNA) previously identified in the fission yeast fbp1 gene and found to induce chromatin opening, also activates meiotic recombination by inducing local chromatin remodeling at a meiotic recombination hotspot. This study furthers our understanding of the connection between lncRNA transcription and DNA break activity in yeast meiosis.
Introduction
Transcriptome analyses have revealed that numerous noncoding RNAs (ncRNAs) are transcribed in eukaryotic cells1–3. Long ncRNAs (lncRNAs) that are more than 200 nucleotides are transcribed from regions with little protein-coding potential such as intergenic and promoter regions, and the relationship between such pervasive transcriptions and various biological processes, such as gene expression, development, disease, and the control of meiotic recombination is of great biological importance4–13.
In meiosis, homologous recombination between homologous chromosomes is pivotal for meiotic progression and contributes to proper segregation of homologous chromosomes during gametogenesis14–17. This process is initiated by the formation of double-strand breaks (DSBs) catalyzed by the Spo11 protein18. Distribution of meiotically induced DSBs is not uniform but is clustered at “hotspots” where meiotic recombination is frequently induced19,20. The global distribution of DSB has been extensively studied in the highly diverged yeasts Saccharomyces cerevisiae (budding yeast) and Schizosaccharomyces pombe (fission yeast)21–24. High resolution mapping of DSB sites in budding yeast has provided insights into DSB sites and hierarchical context, such as chromosome structures, chromatin, transcription factors, and local sequence composition25. Most DSB hotspots are located in promoter regions for mRNA26 or ncRNA13, which are characterized by transcription factor binding, histone modifications and low nucleosome density, suggesting a close relationship between meiotic recombination and transcription. In fission yeast, the ade6-M26 hotspot, which contains a G-to-T substitution in the ade6 open reading frame (ORF) has been intensively investigated as a model locus of meiotic recombination hotspot27,28. The M26-mutation is a nonsense mutation creating a cyclic adenosine monophosphate (cAMP)-responsive element (CRE)-like heptanucleotide sequence 5′-ATGACGT-3′, to which the transcription factors Atf1-Pcr1 bind and activate meiotic recombination29–32. Atf1-Pcr1 binding induces M26 transcription from the M26-mutation site in the ade6 ORF, followed by histone acetylation and chromatin remodeling around the M26 site, thereby inducing meiotic recombination31–33. Taken together, these observations suggest that local chromatin configuration, histone modifications and transcriptional activity play key roles in the location of meiotic DSB sites in budding yeast and fission yeast13,32–36. However, mechanisms underlying the determinants of this DSB distribution have not been fully elucidated.
Previously, we found a stepwise transcription of lncRNAs in the upstream region of fission yeast fbp1 gene37. The fbp1 gene is activated upon glucose starvation38, and this activation is mediated by two transcription factors, Atf1 and Rst239,40. These transcription factors bind to critical cis-acting binding sequences located upstream of the fbp1 promoter, (upstream activating sequences 1 [UAS1] and 2 [UAS2])39 (Fig. 1a). During glucose starvation stress, several species of lncRNAs, referred to as metabolic stress-induced lncRNAs (mlonRNAs), are transcribed from the upstream region of the fbp1 promoter37,41. Stepwise mlonRNA transcription (mlonRNA-a, mlonRNA-b, and mlonRNA-c initiating progressively closer to the fbp1 transcription start site [TSS]) induces chromatin remodeling at the fbp1 promoter (Fig. 1a), and subsequently facilitates transcription factor binding to activate fbp1 expression37. These observations indicate that mlonRNA transcription mediates chromatin remodeling and thereby contribute to the robust induction of fbp1 transcription upon glucose starvation. However, whether or not the role played by this lncRNA transcription is limited to the fbp1 locus has not been addressed. In this study, we investigated whether mlonRNA transcription plays a universal role in chromatin remodeling in the fission yeast genome, leading to the discovery that this lncRNA transcription induces meiotic recombination through inducing chromatin remodeling.
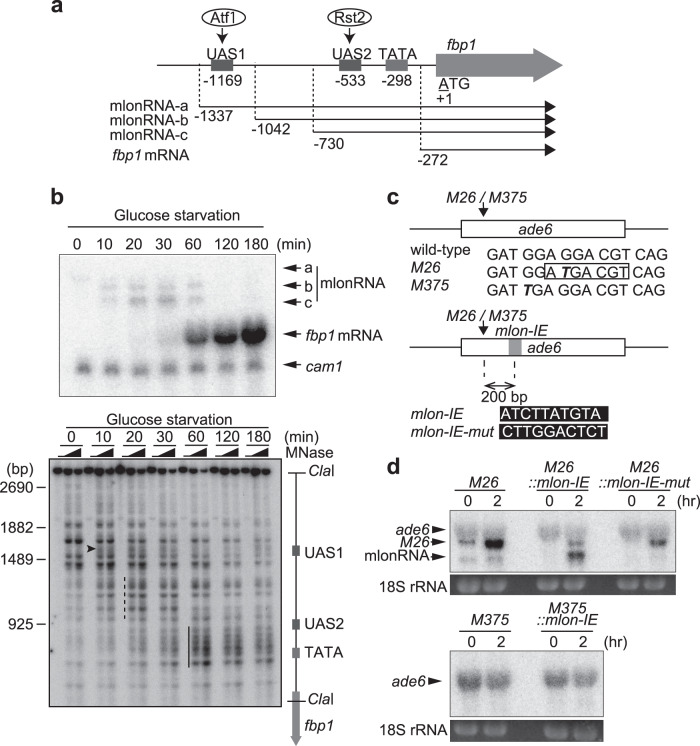
Fig. 1. mlon-IE works at other loci and conditions in addition to its role in the fbp1 upstream region.
a Schematic representation of the fbp1 upstream region containing upstream activating sequence 1 and 2 (UAS1 and UAS2), the binding sites for Atf1 and Rst2, respectively. The numbers indicate the transcription start site of the fbp1 transcripts and the distances of UAS1, UAS2 and the TATA box from the first ATG of fbp1 open reading frame (ORF). b Representative images of northern blot showing stepwise expression of mlonRNAs and fbp1 mRNA and blot of the chromatin analysis showing stepwise chromatin remodeling during glucose starvation. Wild-type haploid cells were grown to 2.0 × 107 cells/ml in YER medium, then transferred to YED medium. Cells were harvested at the indicated times. cam1 transcript was used as an internal control. c M26 and M375 mutations in ade6 (bold italic) and inserted mlon-IE sequences are shown. d ade6 transcription in indicated cells. Diploid cells were cultured to induce meiosis, as described in the Methods section. 18S rRNA stained by ethidium bromide is shown as the loading control.
Results
mlonRNA initiation element (mlon-IE) works at other loci and conditions in addition to its role in the fbp1 upstream region
We previously identified lncRNAs expressed in the fission yeast fbp1 upstream region in the process of massive transcriptional activation, naming these RNAs mlonRNA-a, mlonRNA-b, and mlonRNA-c (Fig. 1b). These transcripts initiate from far upstream of the fbp1 promoter and the initiation site progressively shifts closer to the fbp1 TSS (Fig. 1a, b). The chromatin configuration was analyzed by the distribution of micrococcal nuclease (MNase) sensitive site in fbp1 upstream region (Fig. 1b). MNase-sensitive sites appear 10 min after glucose starvation (Fig. 1b, arrowheads), coincident with the appearance of mlonRNA-b transcription. Another MNase-sensitive region appears around UAS1-UAS2 at 20 to 30 min after glucose starvation (Fig. 1b, dashed lines), when mlonRNA-c transcription is first observed. Finally, intense MNase-sensitive sites appear around the TATA box at 60 to 180 min after glucose starvation (Fig. 1b, thick lines), when massive fbp1 mRNA transcription occurs. Our previous study including these observations indicated that mlonRNA transcription induces chromatin remodeling in the transcribed tract in fbp1 upstream during the transcriptional activation processes37. We further identified the cis-element (mlonRNA initiation element [mlon-IE]) (5′-ATCTTATGTA-3′) required for initiation of mlonRNA-c transcription42. To investigate the generality of the role of mlonRNA transcription in chromatin modulation and to further assess the role of this lncRNA transcription in other aspects of chromosomal regulation, we inserted this cis-element into the ade6-M26 meiotic recombination hotspot (Fig. 1c)27,28. The M26-mutation is a nonsense mutation creating a CRE-like Atf1-Pcr1 bind site29–32. Since mlon-IE is located 200 bp downstream from UAS1 comprising an Atf1-Pcr1 binding sequence in fbp1, and Atf1 is required for mlonRNA-c expression37, the 65 bp fbp1 upstream sequence containing mlon-IE and a flanking sequence was placed 200 bp downstream from the M26 mutation site by replacing the same length of the ade6 ORF sequence (M26::mlon-IE cells). During meiosis, an additional shorter transcript is induced in M26::mlon-IE cells, but it does not appear in the M26::mlon-IE-mut control carrying mutated mlon-IE (Fig. 1c, d). The TSS of this shorter transcript is +426 from the ade6 ORF ATG, located 94 bp downstream of the mlon-IE core 10 bp (Supplementary Fig. 1a). These findings show that mlon-IE works in other loci and conditions, in addition to its role in the fbp1 locus under glucose starvation stress.
We also translocated the cis-element into a control mutant allele of ade6, ade6-M375, which has a similar G-to-T substitution without creating a CRE-like sequence (Fig. 1c). M375::mlon-IE cells do not show transcription from mlon-IE (Fig. 1d). Similar to what is observed during meiosis, osmotic stress induces a shorter transcript downstream of the mlon-IE insertion site in M26::mlon-IE cells but not in M26::mlon-IE-mut or M375::mlon-IE cells (Supplementary Fig. 1b, c). These findings suggest that mlon-IE can also work during osmotic stress, and depends on the binding of the Atf1-Pcr1 transcription factors upstream of mlon-IE. This result is consistent with what is observed in the mlonRNA-c initiation during fbp1 activation, which is dependent on UAS1 comprising Atf1-Pcr1 binding sequence37.
mlon-IE-induced transcription activates meiotic recombination via inducing local chromatin remodeling
We next examined whether transcription from mlon-IE activates meiotic recombination. Briefly, we measured the recombination rate in the ade6 locus using tester strain ade6-469, which has a nonsense mutation in the 3’ region of the ade6 gene (Supplementary Fig. 2a)32. M26::mlon-IE cells and parental ade6-M26 cells exhibit a similar number of recombinant Ade+ colonies in which a functional ade6+ gene is reconstituted by homologous recombination (Fig. 2a). Unfortunately, we cannot directly compare the number of Ade+ recombinants between ade6-M26 and M26::mlon-IE cells for the following reasons. Insertion of mlon-IE drastically changes the amino acid sequence of the Ade6 protein and thereby disrupts the protein function (Supplementary Fig. 2b). This insertion makes the distances between M26::mlon-IE/469 shorter than that between M26/469 (Supplementary Fig. 2a). Moreover, the M26::mlon-IE allele has multiple mutations that would require longer recombination tracts for the reconstitution of a functional ade6+ gene than needed for the ade6-M26 allele. However, M26::mlon-IE cells show a significantly higher meiotic recombination rate compared to M26::mlon-IE-mut cells (Fig. 2a). In contrast, M375::mlon-IE and M375::mlon-IE-mut cells show a similar recombination rate (Fig. 2a). These findings indicate that mlon-IE induces mlonRNA transcription even at 200 bp downstream of the M26 mutation point in the ade6-M26 gene and that this expression may augment meiotic recombination in comparison to mutated mlon-IE inserted cells.
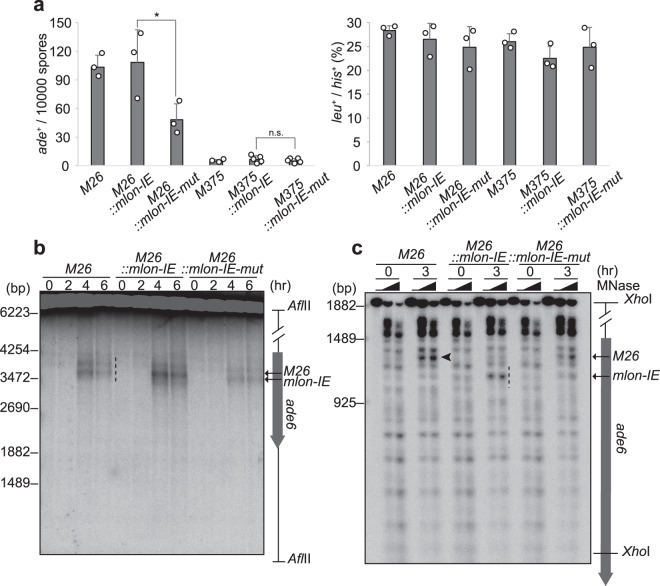
Fig. 2. Transcription from mlon-IE activates meiotic DSB formation and recombination in ade6-M26, the meiotic recombination hotspot.
a Recombination rates at the M26 or M375 control allele (indicated as ade+/104 spores) were examined, as described in the Methods section (left). Recombination between leu1-32 and his3-D1 (indicated as leu+/his+ percentage) was measured as the control (right). Error bars represent standard deviations. n = 3 or 5 biologically independent experiments. P values were calculated using the unpaired one-sided Student’s t-test: *P < 0.05, n.s. (not significant). b Indicated haploid cells possessing the pat1-114 rad50s genotype were cultured to induce meiosis, and DNA was prepared as described in the Methods section. Meiotic DSBs were detected by Southern blotting. The dotted line indicates DSBs around M26 and inserted mlon-IE. c Meiotic chromatin remodeling in indicated cells. Diploid cells were cultured as in Fig. 1c. The black arrowhead and the dotted line indicate MNase-sensitive sites at the M26 mutation and the inserted mlon-IE, respectively.
Considering the role mlon-IE-induced transcription plays in meiotic recombination, we examined meiotic DSBs, which are introduced by Rec12 (Spo11 ortholog in fission yeast) and initiate meiotic recombination43. In M26 and M26::mlon-IE-mut cells, we detected two DSB sites of comparable intensity (Fig. 2b dotted lines and Supplementary Fig. 3a, b). Remarkably, M26::mlon-IE cells show increased intensity of DSBs at the mlon-IE site, and the DSB distribution changes with marked bias toward the mlon-IE site (Fig. 2b and Supplementary Fig. 3a, b). As genome-wide DSBs assessed by pulse-field gel electrophoresis (PFGE) in these strains are indistinguishable, global DSB formation activity is not affected by the insertion of mlon-IE (Supplementary Fig. 4). To determine whether the effect of mlon-IE-induced transcription on DSB formation is due to transcription-coupled chromatin remodeling, we analyzed the chromatin configuration by MNase digestion as in Fig. 1b. In all cells carrying the M26 mutation, the chromatin at M26 is protected from MNase before meiosis (Fig. 2c arrowhead and Supplementary Fig. 3c), although some sensitive sites are observed nearby. At 3 h after onset of meiosis, MNase-sensitive sites appear at the M26 site (Fig. 2c arrowhead and Supplementary Fig. 3c)31,33. Meanwhile, M26::mlon-IE cells show an additional intense MNase-sensitive band at the mlon-IE-inserted site (Fig. 2c dotted line and Supplementary Fig. 3d), with a slight reduction in the MNase-sensitive band at the M26 site. These findings show that the M26 mutation induces alteration of the chromatin configuration into the open state in meiosis. More importantly, transcription induced from mlon-IE further induces chromatin opening around the transcriptional initiation site. This local alteration of the chromatin geometry around the mlon-IE site might influence adjacent chromatin configuration at M26. Taken together, mlon-IE-mediated transcription induces chromatin remodeling, thereby facilitating meiotic DSB formation and augmenting the meiotic recombination rate.
mlon-IE works downstream of transcription factor–binding sequences
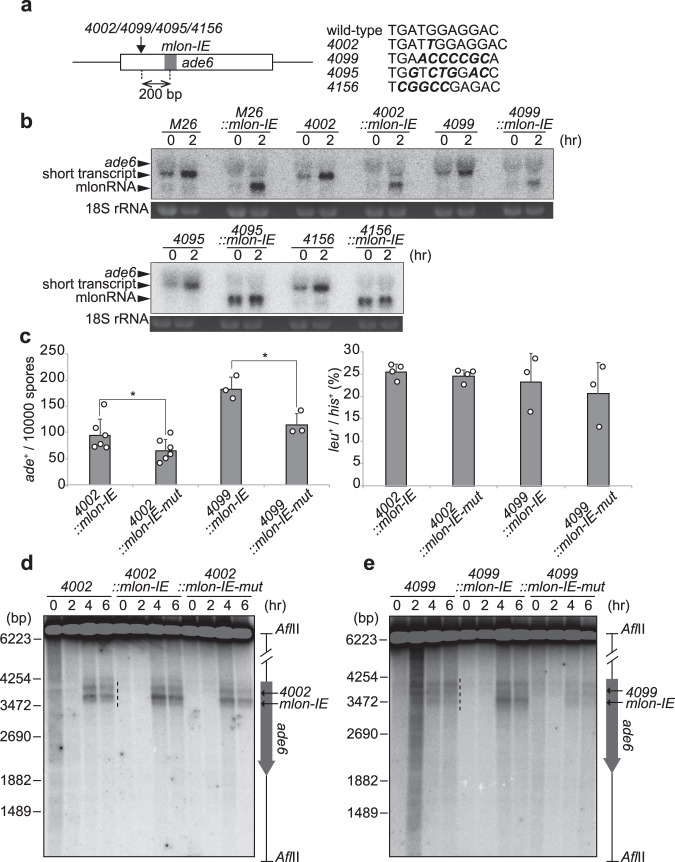
Similar to M26 sequence, other mutations at ade6 that create transcription factor–binding sequences, including CCAAT, oligo-C, 4095, and 4156 motifs, activate meiotic recombination in the ade6 locus44. Next, we tested whether mlon-IE works together with these transcription factor–binding sequences instead of the M26 sequence. In ade6-4002 (CCAAT-motif) and ade6-4099 (oligo-C-motif), hotspot activity depends on transcription factors, CCAAT-binding factor CBF (heterotrimer composed of Php2, Php3, and Php5), and C2H2 zinc finger protein (Rst2), respectively45. Regulators of ade6-4095 and ade6-4156 hotspot activity are unclear, although the ade6-4156 sequence has the potential to bind the Rds1 protein19. We inserted mlon-IE 200 bp downstream of ade6-4002, 4099, 4095, and 4156 sequences (Fig. 3a). These sequences induce a shorter transcript from inside the ade6 ORF, similarly to that seen in ade6-M26 cells (Fig. 3b). More importantly, mlon-IE inserted cells (4002::mlon-IE, 4099::mlon-IE, 4095::mlon-IE, and 4156::mlon-IE cells) show an additional shorter transcript (Fig. 3b), indicating that mlon-IE also works in the presence of other transcription factor–binding site, in addition to Atf1-Pcr1 binding site. We observed the additional shorter transcript in 4002::mlon-IE or 4099::mlon-IE cells only in meiosis (2 h), while that in 4095::mlon-IE or 4156::mlon-IE cells was strongly transcribed before the onset of meiosis (0 h; Fig. 3b). This difference might be due to the different binding pattern of transcription factors to the mutated ade6 loci (4002, 4099, 4095, and 4156). i.e., 4002 and 4099 are bound by transcription factors in a meiosis-specific manner, while 4095 and 4156 are constitutively bound by some transcription factors. Similar to M26::mlon-IE cells, additional transcriptional initiation from inserted mlon-IE activates recombination in ade6-4002 and ade6-4099 (Fig. 3c). In addition, the DSB distribution changes with a bias toward the mlon-IE-inserted site in 4002::mlon-IE and 4099::mlon-IE cells (Fig. 3d, e and Supplementary Fig. 5a–d). Again, DSB activity on a genome-wide scale is not affected by the insertion of mlon-IE in these strains (Supplementary Fig. 4).
Fig. 3. mlon-IE works downstream of binding sequences for transcription factors, CCAAT-binding factor and Rst2.
a Schematic representation of ade6-4002, ade6-4099, ade6-4095 and ade6-4156. 4002 insertion, 4099 mutation, 4095 mutation and 4156 mutation are shown (bold italic). mlon-IE was translocated 200 bp downstream from the 4002/4099/4095/4156 mutation. b ade6 transcripts of indicated cells in meiosis. Cells were cultured, as described in Fig. 1c. c Recombination rates of indicated cells were assessed, as described in Fig. 2a. Error bars represent standard deviations. n = 3 or 6 biologically independent experiments. P values were calculated using the unpaired one-sided Student’s t-test: *P < 0.05. d, e Meiotic DSBs of indicated cells were detected by Southern blotting, as described in Fig. 2b.
Natural mlonRNA transcription from an intergenic region activates meiotic DSB formation via inducing local chromatin remodeling
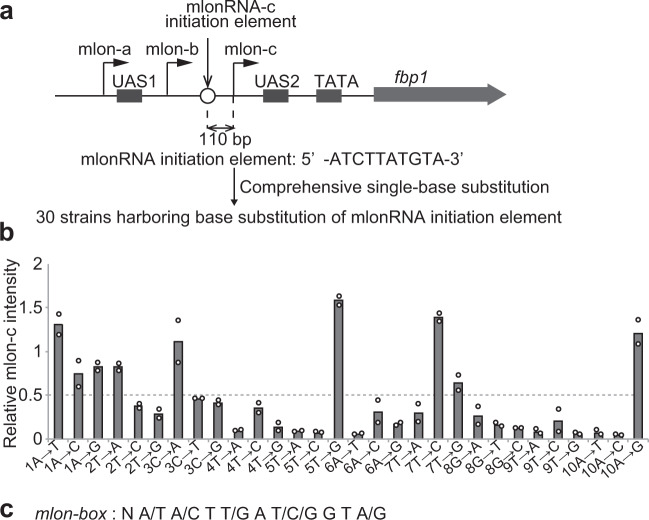
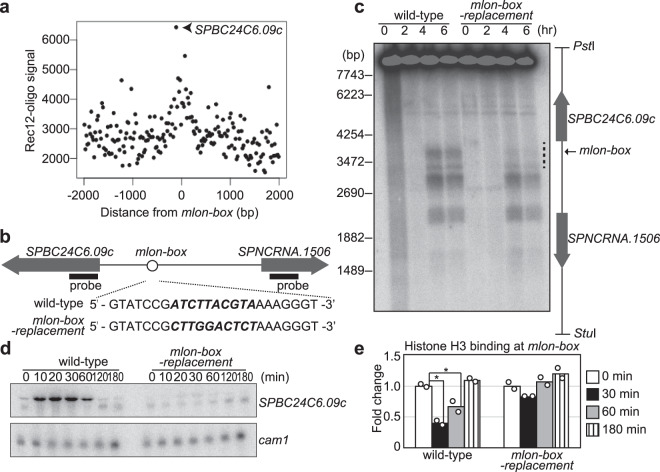
Having established the role of mlonRNA transcription from mlon-IE in the activation of meiotic recombination in the ade6 locus, we next asked if mlonRNA transcription plays a role in the regulation of meiotic recombination through chromatin modulation in the S. pombe genome. To this end, we determined the consensus sequence of mlon-IE by comprehensively mutating each of the 10 nucleotides existing in mlon-IE (Fig. 4a). By excluding mutations that decrease mlonRNA transcription <50% (Fig. 4b and Supplementary Fig. 6), we defined a consensus sequence for mlon-IE as 5′-N A/T A/C T T/G A T/C/G G T A/G-3′, and refer to this consensus element as the mlon-box (Fig. 4c). Next, we searched for mlon-box in the S. pombe genome and analyzed the positional relationship between mlon-box sites and meiotic recombination sites by using a previously reported Rec12 covalently associated oligonucleotide sequence for identifying meiotic DSB sites24 (Fig. 5a). This analysis demonstrates that meiotic recombination tends to occur near mlon-box sequences. The strongest recombination site that overlapped with the mlon-box sequence is located in the intergenic region between SPBC24C6.09c and SPNCRNA.1506 (Fig. 5b). In this region, several meiotic DSBs are observed near the mlon-box (Fig. 5c). We inactivated the mlon-box by replacing this sequence with a 10 nucleotide sequence from act1 ORF and to generate mlon-box-replacement cells (Fig. 5b)42. The act1 ORF sequence is here used as neutral sequence carrying no DSB activity. In these mutant cells, DSBs closest to the mlon-box site are selectively lost (Fig. 5c dotted line, Supplementary Fig. 7a), indicating that the natural mlon-box induces meiotic DSB formation. In addition, SPBC24C6.09c transcription, but not SPNCRNA.1506 transcription, is activated early after the onset of meiosis in an mlon-box-dependent manner (Fig. 5d and Supplementary Fig. 7b). Also, an MNase-sensitive site appears at the mlon-box, and the pattern of MNase-sensitive bands surrounding the mlon-box changes in 0.5 and 1 h after the onset of meiosis (Supplementary Fig. 7c). These results indicate that the chromatin configuration around the mlon-box changes during meiosis (Supplementary Fig. S7c arrowhead and arrows) and that these changes are dependent on the presence of the mlon-box (Supplementary Fig. 7c dotted line). Consistent with these observations, the amount of histone H3 binding at the mlon-box decreases in meiosis, and this histone eviction depends on the presence of the mlon-box (Fig. 5e). These results indicate that transcriptional initiation from the natural mlon-box in the intergenic region activates meiotic DSB formation via chromatin opening.
Fig. 4. Determination of a consensus sequence of mlon-IE.
a Schematic representation of the fbp1 upstream region, including three distinct lncRNAs (mlonRNA-a, mlonRNA-b, and mlonRNA-c) and UAS1 and UAS2. The open circle indicates mlon-IE at 110 bp upstream from the mlonRNA-c TSS. The ten nucleotides of the mlon-IE (5′-ATCTTATGTA-3′) sequence were comprehensively replaced with each of the three other nucleotides. b Relative mlonRNA-c transcript levels of indicated cells at 30 min after glucose starvation compared to wild-type cells. The band intensity of mlonRNA-c transcripts in Supplementary Fig. 6 was quantified. n = 2 biologically independent experiments. c mlon-box, a consensus sequence of mlon-IE.
Fig. 5. DSB formation at the natural meiotic recombination hotspot in the SPBC24C6.09c upstream region is mediated by mlon-box-dependent intergenic transcription.
a Relationship between mlon-box sites and meiotic DSB distribution (Rec12-oligos). The X axis indicates the distance from the nearest mlon-box, and the Y axis indicates copy numbers of the Rec12-oligos-signal. The strongest plot (arrowhead) represents the meiotic recombination hotspot in the SPBC24C6.09c upstream region. b Schematic representation of the mlon-box (open circle) in the intergenic region between SPBC24C6.09c and SPNCRNA.1506. The entire mlon-box sequence comprising 10 nucleotides was replaced with a 10 nucleotide sequence from act1 ORF in mlon-box-replacement cells (bold italic). Location of probes for northern blot was indicated by bold bar. c Detection of meiotic DSBs in the intergenic region between SPBC24C6.09c and SPNCRNA.1506. Indicated cells were cultured and analyzed, as described in Fig. 2b. The dotted line indicates DSB sites around the mlon-box. d Northern analysis to detect the SPBC24C6.09c transcript in wild-type and mlon-box-replacement cells. The left probe indicated in Fig. 4b was used. Indicated cells were cultured to induce meiosis, as described in Fig. 2b. The cam1 transcript is shown as a loading control. e, ChIP analysis to examine histone H3 binding at the mlon-box in indicated cells. Cells were cultured to induce meiosis, as described in Fig. 2b. The relative increase in the ratio at the indicated time after the onset of meiosis is indicated. n = 2 biologically independent experiments. P-values were calculated using Student’s t-test: *P < 0.05.
Previous genome-wide transcriptome analysis has shown that Atf1 regulates SPBC24C6.09c transcription46. However, we did not find a canonical Atf1-binding motif (TGACGT) in ±1 kbp from SPBC24C6.09c-mlon-box. We noticed a similar palindromic Atf1-binding-like motif (TTACGT: a single G-to-T substitution) in SPBC24C6.09c-mlon-box (Supplementary Fig. 8a). In fact, Atf1 binding at SPBC24C6.09c-mlon-box increases approximately threefold during meiosis, while the replacement of the entire mlon-box (10 bp) results in a pronounced decrease in Atf1 binding (Supplementary Fig. 8b). To selectively disrupt the mlon-box while preserving this Atf1-binding sequence, the first three nucleotides (ATC) in the mlon-box were replaced with CCT, generating mlon-box-3bp-replacement cells (Supplementary Fig. 8a). This strain harboring a mutated mlon-box shows little Atf1 binding (Supplementary Fig. 8b). Consistent with this, the transcription level in mlon-box-3bp-replacement cells is significantly decreased compared to wild-type cells (Supplementary Fig. 8c). These results indicate the importance of the mlon-box in Atf1 binding and later transcriptional activation of SPBC24C6.09c. The SPBC24C6.09c-mlon-box might enhance Atf1 affinity for the Atf1-binding motif-like sequence. The possible mechanism underlying mlon-box-mediated strengthening of Atf1 binding is that mlonRNA molecules initiated from the mlon-box bind to the Groucho-Tup1-type transcriptional corepressors Tup11-Tup12 and thereby locally antagonize their repressive functions on Atf1 binding as described previously47. An mlonRNA transcription amplification circuit such as this might play a critical role in the massive induction of SPBC24C6.09c from an incomplete Atf1-binding site.
Discussion
In this study, we identified a novel cis-element, the mlon-box, which is required for mlonRNA transcriptional initiation. mlonRNA transcription plays a critical role in the massive induction of fbp1 via chromatin opening. In addition, mlon-box-mediated transcription activates meiotic DSB formation and recombination in ade6-M26, ade6-4002, and ade6-4099 via chromatin opening. Such mlon-box transcription-associated meiotic DSB formation is observed in the natural meiotic recombination hotspot located in the SPBC24C6.09c upstream region.
It remains to be determined how mlonRNA transcription induces chromatin remodeling. Three mlonRNAs (mlonRNA-a to mlonRNA-c) are initiated far upstream from the fbp1 promoter and stepwise chromatin remodeling is induced in their transcribed tract37 (Fig. 1). Each transcriptional initiation efficiently induces local chromatin remodeling only within 290 bp of the initiation site42, and thus three consecutive mlonRNA transcription events induce stepwise chromatin remodeling over the entire regulatory upstream region of fbp1, thereby allowing the binding of transcription factors as well as RNA polymerase II (RNAPII). An intriguing possibility is that the RNAPII that initiates mlonRNA transcription binds unique accessory subunit(s) that possess histone acetyltransferase activity to induce local histone acetylation around these transcription-start sites and dissociate from the initiation complex after promoter clearance48. Such histone modifications might recruit an ATP-dependent chromatin remodeler49–51. This hypothesis is supported by the observation that histone acetylation is gradually induced from upstream of fbp1 during glucose starvation and a histone acetyltransferase, Gcn5, and an ATP-dependent chromatin remodeler, Snf22, are required for chromatin remodeling in the upstream from fbp137,52. If such novel RNAPII complexes do exist, it will be important to learn how they are selectively targeted to sites of mlon-RNA transcription initiation.
Many lncRNAs are located in the promoter region of genes and referred to as promoter-associated RNAs53. mlon-box sequences are enriched upstream of TSSs in the S. pombe genome (Supplementary Fig. 9), suggesting that mlon-box-induced transcription might regulate neighboring gene expression. Consistent with this, several studies have also reported the contribution of ncRNA transcription in gene expression control7,54,55. In addition, meiotic recombination sites in fission yeast are directed preferentially to loci expressing ncRNA13. It is thus possible that these pervasive lncRNA expressions are involved in the DSB formation as well as with later stages of meiotic recombination via facilitating chromatin remodeling. Taken together, this current study suggests a model that numerous human lncRNAs identified in association with developmental processes11,12 and diseases including cancer10,56, are traces of mlonRNA-mediated regulation of genome function. This study provides important insights into the function of widespread lncRNA transcription in the regulation of DNA-associated reactions on chromatin geometry. A further understanding of the roles of lncRNA transcription might provide important clues to treating lncRNA-associated diseases.
Methods
Fission yeast strains, genetic methods, and cell culture
Supplementary Tables 1 and 2 list the fission yeast strains and primers used in this study, respectively. YEL medium (0.5% yeast extract and 2% glucose), synthetic dextrose (SD) medium, and minimal medium (MM) were used for cell culture57. YER medium (yeast extract containing 6% glucose) and YED medium (yeast extract containing 0.1% glucose and 3% glycerol) were used for glucose repression and starvation, respectively. To induce meiosis, diploid cells were grown to a density of 0.5 × 107 cells/mL in MM medium supplemented with 5 mg/mL of NH4Cl and 1% YEL medium, then diluted with three times the amount of YEL medium, and further cultured for 4 h. Next, the cells were collected and washed with distilled water twice and transferred to MM medium lacking a nitrogen source. To detect meiotic DSBs, synchronous meiosis using a pat1-114 mutation was employed together with a rad50S mutation to efficiently detect DSBs58. The haploid pat1-114 rad50S cells were precultured in MM medium containing 5 mg/mL of NH4Cl at 25 °C, washed with distilled water twice, transferred to MM medium lacking a nitrogen source for arresting the cell cycle at the G1 phase, and cultured further at 25 °C for 16 h. G1-arrested cells were collected by centrifugation, suspended in prewarmed MM medium containing 0.5 mg/mL of NH4Cl and 1% YEL medium, and cultured at 34 °C to induce meiosis. The transformation was performed using the lithium acetate method as follows59. 1 × 108 cells from exponentially growing culture were collected by centrifugation 3000 rpm. Cells were washed by distilled water and 0.1 M Li-acetate in TE (pH 7.5). Then, cells were resuspended in 40 µl of 0.1 M Li-acetate in TE (pH 7.5), added constructed DNA and 300 µl of 40% PEG 4000 in 0.1 M Li-acetate in TE (pH 7.5). The sample was agitated at 30 °C for 30 min, followed by addition of 40 µl of DMSO and incubation at 42 °C for 15 min. After centrifugation, the cell pellet was resuspended in distilled water and plated on an appropriate selective medium. Mating and sporulation were performed on sporulation agar (SPA) medium57, followed by random spore analysis.
Construction of mutant strains carrying mlon-IE 200 bp downstream of ade6-M26, ade6-M375, ade6-4002, ade6-4009, ade6-4095, and ade6-4156
The ade6 gene region carrying M26, M375, 4002, 4099, 4095, or 4156 mutations was amplified using primer sets (p1/p2 and p3/p4). p2 and p4 were flanked by the fbp1 upstream sequence comprising 65 nucleotides containing mlon-IE (−870 to −806 from the first ATG of fbp1) and its complementary sequence, respectively. The resultant products were purified using the QIAquick gel extraction kit (Qiagen, Germany). These fragment pairs were combined at the complementary sequence flanking in p2 and p4 by polymerase chain reaction (PCR) using the p1/p3 primer set and then cloned at ApaI and SpeI sites in the pBluescript plasmid. Fission yeast cells carrying the ura4 marker gene in the HindIII site in the ade6 ORF were transformed, and the transformants were selected for uracil auxotrophy using SD plates containing 5-fluoroorotic acid (5-FOA) and uracil.
Construction of strains with single-base substitution in mlon-IE upstream at fbp1
Comprehensive mlon-IE replacement strains were constructed by PCR amplification according to the method of Senmatsu et al.42. Briefly, fbp1 upstream region was amplified using primers p5/p6 to p35 and p36/p37 to p66. The resultant products were purified using the QIAquick gel extraction kit (Qiagen, Germany). Pairs of fragments were used as templates for PCR amplification using the primer set p5 and p36. Fission yeast cells carrying the ura4 maker gene in the HpaI site in the fbp1 promoter were transformed, and the transformants were selected for uracil auxotrophy using SD plates containing 5-fluoroorotic acid (5-FOA) and uracil.
Construction of strains with mlon-box replacement in SPBC24C6.09c
For mlon-box-10bp replacement in SPBC24C6.09c, the mlon-box sequence (ATCTTACGTA) was replaced with the act1 sequence (CTTGGACTCT) by PCR amplification using primers p67/p68 and p69/p70. For mlon-box-3bp replacement in SPBC24C6.09c, mlon-box sequence was replaced (ATCTTACGTA to CCTTTACGTA) by PCR amplification using primers (p67/p71 and p69/p72).
Northern blotting
Northern blotting was performed as follows. Total RNA was prepared from S. pombe cells. Briefly, 5 × 107 cells were suspended in 0.3 ml of RNA extraction buffer (0.5 M NaCl, 0.2 M Tris-HCl [pH 7.5], 0.01 M ethylenediaminetetraacetic acid (EDTA), 1% SDS) and disrupted with 0.5 g of glass beads and 0.3 ml of phenol-CHCl3 using multi beads shocker (Yasui kikai, Osaka Japan) at conditions 2300 rpm for 30 s three times. After centrifugation of disrupted materials, S. pombe total RNA was isolated from 0.2 ml of supernatant by ethanol precipitation and dissolved in TE buffer (0.01 M Tris-HCl [pH 8.0], 0.001 M EDTA). 10 μg of total RNA was denatured in a buffer (0.02 M MOPS [pH 7.0], 0.005 M sodium acetate, 0.001 M EDTA, 5.7% formaldehyde, 50% formamide) at 60 °C for 5 min. The denatured sample was separated on 1.5% agarose gels containing formaldehyde by electrophoresis in a buffer (0.02 M MOPS [pH 7.0], 0.005 M sodium acetate, 1 mM EDTA) at 100 V for 2 h and transferred to a nylon membrane (Biodyne B, PALL, NY). To detect ade6 transcripts, an EcoRI-XhoI restriction fragment of the ade6 gene was used as a template for random-primer labeling (GE healthcare) with 32P α-dCTP (PerkinElmer, MA)31. To detect fbp1, cam1, SPBC24C6.09c, and SPNCRNA.1506 transcripts, we amplified the template fragments for random-primer labeling by PCR using primer sets p73/74, p75/76, p77/78, and p79/p80, respectively. Hybridization was performed in a buffer (1% BSA, 7% SDS, 0.5 M Na2HPO4 [pH 7.4], 1 mM EDTA) at 62 °C for 12 h, and extensively washed with wash buffer (1% SDS, 1 mM EDTA, 0.04 M Na2HPO4 [pH 7.4]). Signal was detected by a phosphor imager (FLA7000, Fuji film, Tokyo).
ChIP analysis
Chromatin immunoprecipitation (ChIP) analyses were performed according to the method of Senmatsu et al.42 with slight modifications as follows. Fifty ml of culture was incubated with 1.4 ml of 37% formaldehyde solution for 20 min at room temperature, and then 2.5 ml of 2.5 M glycine was added. After centrifugation, collected cells were washed twice with cold TBS buffer (150 mM NaCl, 20 mM Tris HCl [pH 7.5]). The cells were mixed with 400 µl of lysis 140 buffer (0.1% Na-deoxycholate, 1 mM EDTA, 50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1% Triton X100) supplemented with protease inhibitor, cOmplete (Roche), and 0.6 ml of zirconia beads were added. After disruption of the cells using a multi beads shocker (Yasuikikai, Osaka), the suspension was sonicated six times for 30 s each on ice to shear chromosomal DNA into around 500 bp fragment, and centrifuged at 4 °C. The supernatant was collected as a whole-cell extract. Two microliters of Anti-Histone H3 antibody (abcam) or 1 μl of Anti-Atf1 antibody (abcam), and 20 µl or 10 µl of Dynabeads Protein A (Invitrogen) were mixed at 4 °C to conjugate antibody and beads, and then washed twice with PBS (138 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) containing 0.1% BSA. Finally, 300 μl of the whole-cell extract was mixed with pretreated beads and allowed to immunoprecipitate at 4 °C for 16 h. The precipitates were washed twice with lysis 140 buffer, once with lysis 500 buffer (0.1% Na-deoxycholate, 1 mM EDTA, 50 mM HEPES-KOH [pH 7.5], 500 mM NaCl, 1% Triton X100), and further washed once with wash buffer (0.5% Na-deoxycholate, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 10 mM Tris-HCl [pH 8.0]) followed by once with TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The well-washed precipitates were mixed with 40 μl of elution buffer (10 mM EDTA, 1% SDS, and 50 mM Tris-HCl [pH 8.0]) and allowed to elute the immunoprecipitated protein-DNA complexes at 65 °C for 15 min (immunoprecipitation [IP] sample). To elute IP sample completely, 100 µl of elution buffer and 150 µl of TE containing 0.67% SDS in remaining beads, then incubated at 65 °C for 15 min again. Three microliters of the whole-cell extract was mixed with 97 µl of lysis 140 buffer and 400 μl of TE buffer containing 1% SDS (Input sample). IP and Input sample were added 84 µg of proteinase K (Merck, Darmstadt, Germany), and incubated at 37 °C for 16 hr. After incubation, the temperature was shifted to 65 °C and the sample was further incubated for 6 h. After incubation, DNA was phenol/chloroform extracted from each of the samples and quantified by quantitative PCR using Thermal Cycler Dice Real Time (Takara Bio, Shiga, Japan) and the THUNDERBIRD® SYBR qPCR Mix (TOYOBO, Osaka, Japan). Primer sets p81/p82 at SPBC24C6.09c-mlon-box and p83/p84 at the prp3 locus were used for quantitative PCR analysis.
Micrococcal nuclease digestion assay
Analysis of chromatin structure by indirect end-labeling was performed according to the method of Asada et al.60 with modifications as follows. 5 × 108 cells from 100 ml of the culture was harvested. Cells were incubated in 0.25 ml of preincubation solution (20 mM Tris-HCl at pH 8.0, 0.7 M 2-mercaptoethanol, 3 mM EDTA) at 30 °C for 10 min and washed once in 1 ml of ice-cold 1 M sorbitol with 10 mM EDTA. Cells were then centrifuged and resuspended in 1 ml of freshly prepared Zymolyase solution (37.5 mM Tris-HCl at pH 7.5, 0.75 M sorbitol, 1.25% glucose, 0.1% (wt/vol) Zymolyase-100T, 6.25 mM EDTA). Cells were incubated for 5 min at 30 °C and resulting spheroplasts were pelleted. All subsequent steps were done at 4 °C. The spheroplasts were then washed once in 1 ml of ice-cold 1 M Sorbitol and resuspended well by pipetting in 1 ml of freshly prepared lysis buffer (18% Ficoll-400, 10 mM KH2PO4, 10 mM K2HPO4 [pH 6.8], 1 mM MgCl2, 0.25 mM EGTA, 0.25 mM EDTA, 1 mM PMSF). After centrifugation at 13,000 rpm for 40 min, the crude nuclear pellet was resuspended well in 1.5 ml of buffer A (10 mM Tris-HCl at pH 8.0, 150 mM NaCl, 5 mM KCl, 1 mM EDTA, 1 mM PMSF). 0.5 ml aliquots of the crude chromatin suspension were digested with different amounts of MNase (0, 20, and 50 U/ml) for 5 min at 37 °C in the presence of 5 mM CaCl2. The reaction was terminated by adding 40 mM EDTA, 1% SDS, and 200 μg of proteinase K and incubated at 50 °C for 16 h. Insoluble material was removed by centrifugation at 4 °C. The supernatants were extracted with phenol/chloroform, digested with RNase A, and then extracted once with phenol/chloroform. The extracted DNA was precipitated by 2-propanol, rinsedin 70% ethanol, and resuspended in 50 μl of TE buffer. DNA samples were digested by ClaI, XhoI or ApaI for analysis of fbp1, ade6 or SPBC24C6.09c region respectively, and separated by electrophoresis on 1.5% for fbp1or 1.2% for ade6 or SPBC24C6.09c agarose gel in Tris base, acetic acid and EDTA (TAE) buffer (0.48% Tris base, 0.11% acetic acid and 0.037% EDTA) at 60 V for 20 h, followed by Southern blotting using the same probe as in the detection of fbp1, ade6 or SPBC24C6.09c transcripts. The methods for hybridization and signal detection were the same as in Northern blot analysis.
Detection of meiotic DSBs
DNA samples were prepared in agarose plugs from cells from a synchronous meiotic culture according to the method by Hirota et al.61 with modifications as follows. Briefly, 50 ml of culture was harvested and washed twice using 500 µl of 20 mM citrate-phosphate (pH 5.6), 1.2 M sorbitol, and 40 mM EDTA (CSE) buffer. After resuspension with 500 µl of CSE buffer containing 1.5 mg/mL Zymolyase 20T, cells were incubated at 37 °C for 60 min, centrifuged, and resuspended in 500 µl of 10 mM Tris-Cl (pH 7.5), 0.9 M sorbitol, and 45 mM EDTA (TSE) buffer. An equal volume of low-melting-point agarose (1%) in TSE buffer was added, and the mixture was poured into agarose plug molds (Bio-Rad Laboratories, Hercules, CA, USA). After 5 min on ice, the agarose plugs were collected into a buffer (0.25 M EDTA, 50 mM Tris-Cl [pH 7.5], and 1% sodium dodecyl sulfate [SDS]) and incubated at 55 °C for 90 min. Then, the buffer was removed, and a second buffer (1% lauryl sarcosine, 0.5 M EDTA [pH 9.5], and 1 mg/mL of proteinase K) was added. The mixture was incubated at 55 °C for 2 days. Next, agarose plugs were soaked in 1 ml of Tris-EDTA (TE) containing 0.5 mM phenylmethylsulfonyl fluoride (PMSF) at room temperature for 60 min and washed twice with TE at room temperature for 60 min. The agarose plugs were equilibrated using CutSmart buffer (New England Biolabs, Ipswich, MA, USA) overnight. After changing CutSmart buffer, the agarose plugs were incubated at room temperature for 60 min. DNA in the agarose plugs was digested by AflII or StuI and PstI-HF (New England Biolabs) to detect meiotic DSBs in ade6 or SPBC24C6.09c, respectively. The digested DNA fragments were separated by electrophoresis on 0.8% agarose gel in TAE buffer at 60 V for 20 h, followed by Southern blotting. DNA probes for ade6 and SPBC24C6.09c were amplified by PCR using primer sets p85/p86 and p87/p88, respectively. To detect whole meiotic DSBs, the agarose plugs were washed with TE for 60 min, and the chromosomal DNA and DSBs in agarose plugs were separated by pulse-field gel electrophoresis (PFGE) and stained by ethidium bromide. PFGE was carried out in 0.8% chromosomal grade agarose (Biorad) on a Biorad CHEF-DRIII system and re-circulated at 14 °C. Electrophoresis was performed for 48 h at 2 V/cm in 1 × TAE (Tris-acetate-EDTA) buffer, with a switch time of 30 min at an included angle of 100°.
Determination of the recombination rate
The recombination rate was determined according to the method by Hirota et al.32 with modifications as follows. Briefly, each strain was crossed with a tester strain (ade6-469) on SPA medium at 30 °C for 2 days. Sporulating asci were suspended in 1 ml of distilled water containing 115 units of β-glucuronidase and agitated at 30 °C for 30 min to release spores from asci and kill vegetative cells. After incubation, 500 µl of ethanol was added, and incubation was again performed at room temperature for 15 min. Spores were collected by centrifugation and resuspended in 1 mL of distilled water. Then, 104, 103, and 200 spores were spread onto SD-lacking adenine, SD-lacking leucine and histidine, and YE-containing adenine, respectively. The recombination rate in ade6 was represented as the colony number on SD-lacking adenine normalized by the colony number on YE-containing adenine. The recombination frequency between leu1 and his3 was calculated as the percentage of the colony number on SD-lacking leucine and histidine normalized by the colony number on YE-containing adenine.
5′ rapid amplification of cDNA ends experiment
The rapid amplification of complementary DNA (cDNA) ends (RACE) experiment was performed using the SMARTer® RACE 5′/3′ Kit (Takara Bio) according to the manufacturer’s instructions. The 5′ ends of the transcripts were amplified by PCR using the universal primer mix included in the kit and the gene-specific primer p89. PCR products were purified using the QIAquick gel extraction kit (Qiagen) and cloned into pCR-BluntII-TOPO (Invitrogen, Carlsbad, CA, USA). Finally, the sequences were determined using the M13 primer p90.
Statistics and reproducibility
Throughout the manuscript, all individual data points are plotted. Sample mean and standard deviation are also shown on each bar graph. All statistical analyses were conducted using a one-sided Student’s t-test.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Dr. Walter W. Steiner (Department of Biology, Niagara University) for the gift of ade6-4002, −4099, −4095, and −4156 background cells. We also thank all the members of the Hirota laboratory for their help. We acknowledge the Radioisotope Research Center in Tokyo Metropolitan University for support in the use of isotopes. This work was supported in part by JSPS KAKENHI (16H01314 to K.H. and 19J20773 to S.S.) and the Takeda Science Foundation and Yamada Science Foundation (to K.H.). Correspondence and requests for materials should be addressed to Kouji Hirota (khirota@tmu.ac.jp).
Author contributions
Conceived and designed the experiments: S.S., R.A. and K.H. performed most of the experiments: S.S. Performed the bioinformatics analysis: O.A. Wrote the paper: S.S., R.A., C.S.H., K.O. and K.H.
Data availability
S. pombe genome sequence and Transcription start sites (TSSs) can be found in PomBase as “Schizosaccharomyces_pombe.ASM294v2.30.dna.genome.fa” and “schizosaccharomyces_pombe.chr.gff3” (2017/3/22, ftp://ftp.pombase.org/pombe).
Rec12 covalently associated oligonucleotide sequence can be found as Rec12-oligo (WT_SOLID_313) under accession No. G7E49977. All uncropped image data used for all figures in the paper are present in Supplementary Fig. 10. Raw data used for all histograms in this paper are present in Supplementary data 1.
Code availability
Emboss fuzznuc (http://embossgui.sourceforge.net/demo/manual/fuzznuc.html) was used to search mlon-box sequences.
Bedtools (v.2.17.0; https://github.com/arq5x/bedtools2) was used for enrichment analysis around TSS.
R code (https://www.r-project.org/) used for visualization in this study is available at https://www.datamentor.io/r-programming/histogram/.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-01798-8.
References
- 1.David L, et al. A high-resolution map of transcription in the yeast genome. Proc. Natl Acad. Sci. USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dutrow N, et al. Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nat. Genet. 2008;40:977–986. doi: 10.1038/ng.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moretto F, Wood NE, Kelly G, Doncic A, Van Werven FJ. A regulatory circuit of two lncRNAs and a master regulator directs cell fate in yeast. Nat. Commun. 2018;9:1–12. doi: 10.1038/s41467-018-03213-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci. Adv. 2017;3:1–13. doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson KM, et al. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bester AC, et al. An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell. 2018;173:649–664. doi: 10.1016/j.cell.2018.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibb EA, et al. Human cancer long non-coding RNA transcriptomes. PLoS ONE. 2011;6:1–10. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sigova AA, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl Acad. Sci. USA. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahls WP, Siegel ER, Davidson MK. Meiotic recombination hotspots of fission yeast are directed to loci that express non-coding RNA. PLoS ONE. 2008;3:1–8. doi: 10.1371/journal.pone.0002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lichten M, Goldman AS. Meiotic recombination hotspots. Annu. Rev. Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 15.Wahls WP. Meiotic recombination hotspots: shaping the genome and insights into hypervariable minisatellite DNA change. Curr. Top. Dev. Biol. 1997;37:37–75. doi: 10.1016/S0070-2153(08)60171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petes TD. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2001;2:360–369. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 17.Vader G, Lens SMA. Chromosome segregation: taking the passenger seat. Curr. Biol. 2010;20:R879–R881. doi: 10.1016/j.cub.2010.09.037. [DOI] [PubMed] [Google Scholar]
- 18.Lam I, Keeney S. Mechanism and control of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 2014;16:1–24. doi: 10.1101/cshperspect.a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wahls WP, Davidson MK, Discrete DNA. sites regulate global distribution of meiotic recombination. Trends Genet. 2010;26:202–208. doi: 10.1016/j.tig.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tock AJ, Henderson IR. Hotspots for initiation of meiotic recombination. Front. Genet. 2018;9:1–11. doi: 10.3389/fgene.2018.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerton JL, et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A. Mapping of meiotic single-stranded DNA Reveals double-strand-break hotspots near centromeres and telomeres. Curr. Biol. 2007;17:2003–2012. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 23.Buhler C, Borde V, Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 2007;5:2797–2808. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 2014;24:1650–1664. doi: 10.1101/gr.172122.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan J, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamada S, et al. Correlation of meiotic DSB formation and transcription initiation around fission yeast recombination hotspots. Genetics. 2017;206:801–809. doi: 10.1534/genetics.116.197954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponticelli AS, Sena EP, Smith GR. Genetic and physical analysis of the M26 recombination hotspot of Schizosaccharomyces pombe. Genetics. 1988;119:491–497. doi: 10.1093/genetics/119.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szankasi P, Heyer W-D, Schuchert P, Kohli J. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe wild-type and mutant alleles including the recombination hot spot allele ade6-M26. J. Mol. Biol. 1988;204:917–925. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]
- 29.Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA. 1997;94:13765–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steiner WW, Schreckhise RW, Smith GR. Meiotic DNA Breaks at the S. pombe Recombination Hot Spot M26. Mol. Cell. 2002;9:847–855. doi: 10.1016/S1097-2765(02)00489-6. [DOI] [PubMed] [Google Scholar]
- 31.Mizuno KI, et al. The meiotic recombination hot spot created by the single-base substitution ade6-M26 results in remodeling of chromatin structure in fission yeast. Genes Dev. 1997;11:876–886. doi: 10.1101/gad.11.7.876. [DOI] [PubMed] [Google Scholar]
- 32.Hirota K, Mizuno K, Shibata T, Ohta K. Distinct chromatin modulators regulate the formation of accessible and repressive chromatin at the fission yeast recombination hotspot ade6-M26. Mol. Biol. Cell. 2008;19:1162–1173. doi: 10.1091/mbc.e07-04-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamada T, et al. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 2004;23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohta K, Shibata T, Nicolas A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adam C, et al. The PHD finger protein Spp1 has distinct functions in the Set1 and the meiotic DSB formation complexes. PLoS Genet. 2018;14:1–22. doi: 10.1371/journal.pgen.1007223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu T, Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 37.Hirota K, et al. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- 38.Vassarottis A, Friesens JD. Isolation of the Fructose-l,6-bisphosphatase gene of the yeast Schizosaccharomyces pombe. J. Biol. Chem. 1985;260:6348–6353. doi: 10.1016/S0021-9258(18)88978-5. [DOI] [PubMed] [Google Scholar]
- 39.Neely LA, Hoffman CS. Protein kinase A and mitogen-activated protein kinase pathways antagonistically regulate fission yeast fbp1 transcription by employing different modes of action at two upstream activation sites. Mol. Cell. Biol. 2000;20:6426–6434. doi: 10.1128/MCB.20.17.6426-6434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higuchi T, Watanabe Y, Yamamoto M. Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Mol. Cell. Biol. 2002;22:1–11. doi: 10.1128/MCB.22.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galipon J, Miki A, Oda A, Inada T, Ohta K. Stress-induced lncRNAs evade nuclear degradation and enter the translational machinery. Genes Cells. 2013;18:353–368. doi: 10.1111/gtc.12042. [DOI] [PubMed] [Google Scholar]
- 42.Senmatsu S, et al. lncRNA transcriptional initiation induces chromatin remodeling within a limited range in the fission yeast fbp1 promoter. Sci. Rep. 2019;9:1–8. doi: 10.1038/s41598-018-36049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keeney. Scott. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn. Stab. 2007;2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steiner WW, Steiner EM, Girvin AR, Plewik LE. Novel nucleotide sequence motifs that produce hotspots of meiotic recombination in Schizosaccharomyces pombe. Genetics. 2009;182:459–469. doi: 10.1534/genetics.109.101253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steiner WW, Davidow PA, Bagshaw ATM. Important characteristics of sequence-specific recombination hotspots in Schizosaccharomyces pombe. Genetics. 2011;187:385–396. doi: 10.1534/genetics.110.124636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen D, et al. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell. 2003;14:214–229. doi: 10.1091/mbc.e02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takemata N, et al. Local potentiation of stress-responsive genes by upstream noncoding transcription. Nucleic Acids Res. 2016;44:5174–5189. doi: 10.1093/nar/gkw142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chandy M, Gutiérrez JL, Prochasson P, Workman JL. SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryot. Cell. 2006;5:1738–1747. doi: 10.1128/EC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grant PA, Sterner DE, Duggan LJ, Workman JL, Berger SL. The SAGA unfolds: convergence of transcription regulators in chromatin modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/S0962-8924(98)01263-X. [DOI] [PubMed] [Google Scholar]
- 51.Hassan AH, Awad S, Prochasson P. The Swi2/Snf2 bromodomain is required for the displacement of SAGA and the octamer transfer of SAGA-acetylated nucleosomes. J. Biol. Chem. 2006;281:18126–18134. doi: 10.1074/jbc.M602851200. [DOI] [PubMed] [Google Scholar]
- 52.Adachi A, et al. Interplay between chromatin modulators and histone acetylation regulates the formation of accessible chromatin in the upstream regulatory region of fission yeast fbp1. Genes Genet. Syst. 2017;92:267–276. doi: 10.1266/ggs.17-00018. [DOI] [PubMed] [Google Scholar]
- 53.Lepoivre C, et al. Divergent transcription is associated with promoters of transcriptional regulators. BMC Genomics. 2013;14:1–20. doi: 10.1186/1471-2164-14-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 55.Latos PA, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 56.Iyer MK, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirota K, Tanaka K, Ohta K, Yamamoto M. Gef1p and Scd1p, the two GDP-GTP exchange factors for Cdc42p, form a ring structure that shrinks during cytokinesis in Schizosaccharomyces pombe. Mol. Biol. Cell. 2003;14:3617–3627. doi: 10.1091/mbc.e02-10-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young JA, Schreckhise RW, Steiner WW, Smith GR. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell. 2002;9:253–263. doi: 10.1016/S1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
- 59.Hirota K, Tanaka K, Watanabe Y, Yamamoto M. Functional analysis of the C-terminal cytoplasmic region of the M-factor receptor in fission yeast. Genes Cells. 2001;6:201–214. doi: 10.1046/j.1365-2443.2001.00415.x. [DOI] [PubMed] [Google Scholar]
- 60.Asada R, Takemata N, Hoffman CS, Ohta K, Hirota K. Antagonistic controls of chromatin and mRNA start site selection by Tup family corepressors and the CCAAT-binding factor. Mol. Cell. Biol. 2015;35:847–855. doi: 10.1128/MCB.00924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirota K, Steiner WW, Shibata T, Ohta K. Multiple modes of chromatin configuration at natural meiotic recombination hot spots in fission yeast. Eukaryot. Cell. 2007;6:2072–2080. doi: 10.1128/EC.00246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
S. pombe genome sequence and Transcription start sites (TSSs) can be found in PomBase as “Schizosaccharomyces_pombe.ASM294v2.30.dna.genome.fa” and “schizosaccharomyces_pombe.chr.gff3” (2017/3/22, ftp://ftp.pombase.org/pombe).
Rec12 covalently associated oligonucleotide sequence can be found as Rec12-oligo (WT_SOLID_313) under accession No. G7E49977. All uncropped image data used for all figures in the paper are present in Supplementary Fig. 10. Raw data used for all histograms in this paper are present in Supplementary data 1.
Emboss fuzznuc (http://embossgui.sourceforge.net/demo/manual/fuzznuc.html) was used to search mlon-box sequences.
Bedtools (v.2.17.0; https://github.com/arq5x/bedtools2) was used for enrichment analysis around TSS.
R code (https://www.r-project.org/) used for visualization in this study is available at https://www.datamentor.io/r-programming/histogram/.