Significance
Several techniques have identified ELF3 as particularly important in regulating trophoblast-specific HLA-C expression. This results from a single-nucleotide difference in the promotor of the HLA-C gene at the RFX5 binding site that creates an ELF3 binding site not found in the HLA-A or HLA-B promoters. We discovered a superenhancer and a positive autoregulatory feedback loop that promotes expression of the ELF3 gene in trophoblasts. Disruption of either the superenhancer by (+)-JQ1 or interference with the positive-feedback loop by wrenchnolol decreased ELF3 levels, and thus HLA-C expression. Aberrations of this complex regulatory system could be involved in control of infection, miscarriage, preterm birth, preeclampsia, as well as parturition in normal pregnancy and in development of choriocarcinoma.
Keywords: HLA-C, ELF3, superenhancer, (+)-JQ1, autoregulatory feedback loop
Abstract
HLA-C arose during evolution of pregnancy in the great apes 10 to 15 million years ago. It has a dual function on placental extravillous trophoblasts (EVTs) as it contributes to both tolerance and immunity at the maternal–fetal interface. The mode of its regulation is of considerable interest in connection with the biology of pregnancy and pregnancy abnormalities. First-trimester primary EVTs in which HLA-C is highly expressed, as well as JEG3, an EVT model cell line, were employed. Single-cell RNA-seq data and quantitative PCR identified high expression of the transcription factor ELF3 in those cells. Chromatin immunoprecipitation (ChIP)-PCR confirmed that both ELF3 and MED1 bound to the proximal HLA-C promoter region. However, binding of RFX5 to this region was absent or severely reduced, and the adjacent HLA-B locus remained closed. Expression of HLA-C was inhibited by ELF3 small interfering RNAs (siRNAs) and by wrenchnolol treatment. Wrenchnolol is a cell-permeable synthetic organic molecule that mimics ELF3 and is relatively specific for binding to ELF3’s coactivator, MED23, as our data also showed in JEG3. Moreover, the ELF3 gene is regulated by a superenhancer that spans more than 5 Mb, identified by assay for transposase-accessible chromatin using sequencing (ATAC-seq), as well as by its sensitivity to (+)-JQ1 (inhibitor of BRD4). ELF3 bound to its own promoter, thus creating an autoregulatory feedback loop that establishes expression of ELF3 and HLA-C in trophoblasts. Wrenchnolol blocked binding of MED23 to ELF3, thus disrupting the positive-feedback loop that drives ELF3 expression, with down-regulation of HLA-C expression as a consequence.
HLA-C evolved from a duplication of the HLA-B gene, as evidenced by its genomic proximity and sequence conservation. It first appeared 10 to 15 million years ago in the radiation between the two Asian apes, gibbons and orangutans. Gibbons do not have an HLA-C gene, while orangutans have only a single HLA-C1 allotype that appears to serve as an ordinary antigen-presenting molecule. The two human group allotypes HLA-C1 and -C2, distinguished by a dimorphism at residues 77 and 80, first appeared in the great apes: gorillas, bonobos, chimpanzees, and humans. At approximately the same period in evolution, HLA-G became restricted to expression on trophoblasts (1–4). Thus, the two major HLA molecules expressed on extravillous trophoblasts (EVTs), HLA-C and HLA-G, arose in evolution at about the same time, presumably in response to the evolving needs of pregnancy. HLA-C and HLA-G play important roles at the maternal–fetal interface in immunity to infections and tolerance to allogeneic tissues (5, 6).
The regulation of HLA-C expression and identification of factors that regulate it are of immense interest because of its multiple physiological and pathological roles in human disease: 1) as the major molecule that can provide for immunity at the maternal–fetal interface (6, 7); 2) as the antithesis, the major fetal alloantigen to which tolerance must be established at the maternal–fetal interface (5, 8); 3) as a principal cause of inflammatory pathology in bone marrow or hematopoietic stem cell transplantation (9); 4) its linkage in various studies to an array of inflammatory conditions, that may occur in pregnancy, including preeclampsia, spontaneous preterm birth, and infections at the maternal–fetal interface; these conditions emphasize the importance of studying the mode of regulation of expression of HLA-C; 5) its important role in NK cells in limiting progression of HIV and possibly other viral diseases (7, 10, 11); 6) its influence on the anticancer immune response to check point blockade immunotherapy (12); and 7) its unique down-regulation in childhood B cell acute lymphoblastic leukemia (13).
EVTs form the fetal side of the maternal–fetal interface. Uniquely, they express a triad of HLA molecules, HLA-C, HLA-G, and HLA-E, but they do not express the classic polymorphic HLA-A and HLA-B proteins (14). Little is known about what regulates expression of the HLA-C gene. A detailed analysis of the promoter region of HLA-A, -B, and -C genes showed a single-base pair difference—G/A—in the HLA-C proximal promoter (15). Analysis of this HLA-C–specific nucleotide sequence suggested that it could no longer bind RFX5 at this site as do HLA-A and -B promoters. ELF3, a member of the E26 transformation-specific (ETS) transcription factor family (16), was included among many such factors whose binding motifs were described in the annotated sequence. Analysis of JAR, a choriocarcinoma cell line, confirmed that the substitution of G/A at this site resulted in loss of function of the HLA-A gene promoter and gain of the HLA-C gene promoter activity in trophoblasts (15). Moreover, an electrophoretic mobility shift assay (EMSA) showed a supershift induced by either an ELF1 or ELF3 antibody (15). Our own microarray analysis of EVTs had shown expression of ELF2 and ELF3, but not ELF1. ETS transcription factors are downstream targets of the Ras/Erk pathway and thus regulate many processes, including differentiation, proliferation, apoptosis, cell cycle, tissue remodeling, and angiogenesis (17–19). The ETS family of transcription factors encompasses 27 members in humans (16). They fall into four groups, ETS class I, II (IIa, IIb), III, and IV, based on the similarity between their DNA binding motifs. ELF3, together with ELF1, ELF2, ELF4, ELF5, and EHF, was grouped into ETS class IIa. ELF6 and ELF7 fall into ETS class IIb (16). The action of Elf3 may also be important for invasion of trophoblasts (20) and plays a role in early embryonic development, both preimplantation and postimplantation, in mice (21). These clues focused our attention on ELF3 as a candidate factor that controls trophoblast-specific expression of HLA-C.
Results
ELF3 Is Highly Expressed in a Human Choriocarcinoma Cell Line and in Primary EVTs at the Maternal–Fetal Interface.
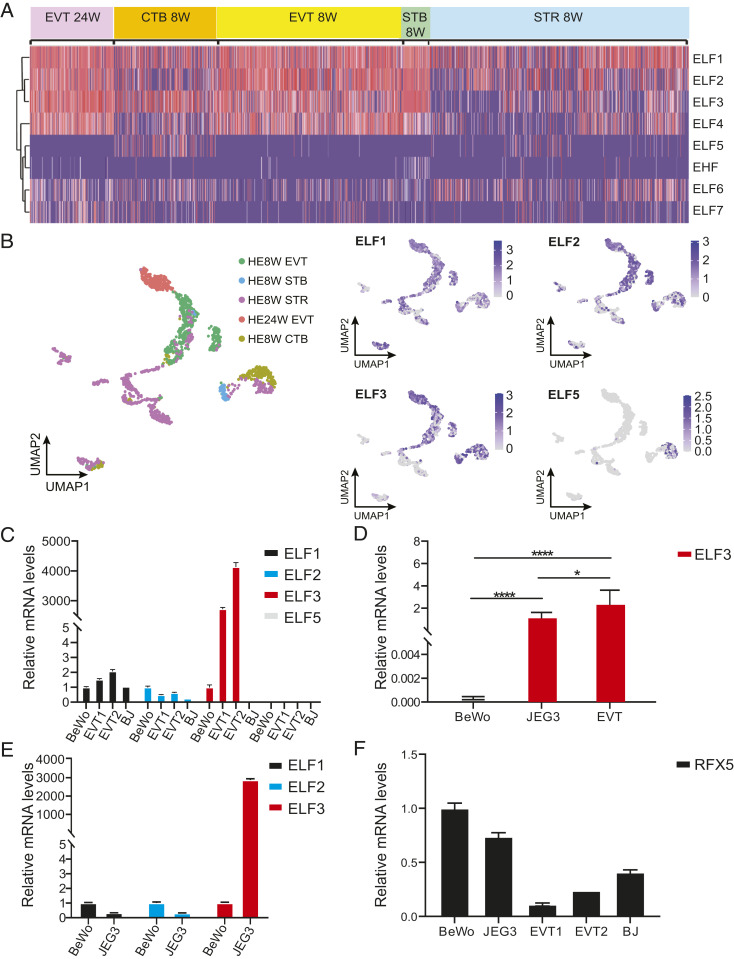
To examine the gene expression pattern of ELF3 and the closely related ETS class II family members at the maternal–fetal interface, we analyzed single-cell RNA-seq (scRNA-seq) data from human placenta across different cell types of the placenta from a publicly available database (22). Cluster analysis revealed that ELF1 and ELF2 have gene expression patterns most similar to that of ELF3. All the three ETS family members are highly expressed in both first- and second-trimester primary EVTs (EVT_8W and EVT_24W, respectively), as well as in first-trimester syncytiotrophoblasts (STB_8W). However, their expression is relatively low in first-trimester cytotrophoblasts (CTB_8W) and villous stromal cells (STR_8W) (Fig. 1A). Neither first- nor second-trimester EVTs expressed ELF5 (Fig. 1A). Umap analysis is consistent with the cluster analysis: ELF3 is highly expressed in trophoblasts, very low in stromal cells (HE8W_STR), and ELF1, ELF2 show a similar expression pattern, but higher expression levels in stromal cells than ELF3; ELF5 expression is very low at the maternal–fetal interface (Fig. 1B). Next, we confirmed ELF1, ELF2, ELF3, and ELF5 gene expression levels in the trophoblast model cell lines BeWo and JEG3 as well as in primary EVTs. Notably, ELF3 expression was much higher in primary EVTs or JEG3 than in BeWo. In the three primary EVT isolates tested, ELF3 expression was even higher than in JEG3 and 3,000 to 4,000 times higher than in BeWo (Fig. 1 C–E). The somatic cell type BJ fibroblasts express little or no ELF3. In contrast, expression levels of ELF1 were slightly higher in two different primary EVT isolates than in BeWo and JEG3. ELF2 expression was lower in primary EVTs than in BeWo (Fig. 1 C and E). ELF5 expression was absent in BeWo, EVTs, or BJ fibroblasts (Fig. 1C).
Fig. 1.
Trophoblast expression of ETS class II genes and RFX5. (A) Clustering of ETS class II genes in placental cells from single-cell sequencing data. Placental cells are clustered into five subpopulations: 24-wk extravillous trophoblasts (EVT_24W), 8-wk cytotrophoblasts (CTB_8W), 8-wk EVTs (EVT_8W), 8-wk syncytiotrophoblasts (STB_8W), and 8-wk stromal cells (STR_8W). (B) Umap analysis of the scRNA-seq data. Expression levels of ELF1, ELF2, ELF3, and ELF5 genes in human placenta cells are shown. (C) ELF1, ELF2, ELF3, and ELF5 mRNA levels in BeWo, BJ fibroblasts, and two primary EVTs by qPCR. (D) ELF3 mRNA levels in BeWo, JEG3, and six primary EVT samples. (E) ELF1, ELF2, and ELF3 mRNA levels in BeWo and JEG3 by qPCR. (F) RFX5 mRNA levels in BeWo, JEG3, primary EVTs, and BJ fibroblasts by qPCR. qPCR was done in triplicate. Data are reported as the mean ± SD. Unpaired t test. *P < 0.05; ****P < 0.0001.
In the classical model of MHC-I gene regulation, RFX5 binds to the X1 box in the proximal promoter region. We thus also examined the levels of RFX5 in primary EVTs. In contrast to ELF3, RFX5 levels in EVTs were lower than in BeWo or JEG3 (Fig. 1F). A single-nucleotide variation in the HLA-C core promoter will reduce or eliminate the binding of RFX5 at this site (15). The ELF3 binding site is adjacent to and partially overlaps with the RFX5 site. Thus, competition between ELF3 and RFX5 for binding to this site may occur.
ELF3 Small Interfering RNA Inhibits Expression of HLA-C in JEG3.
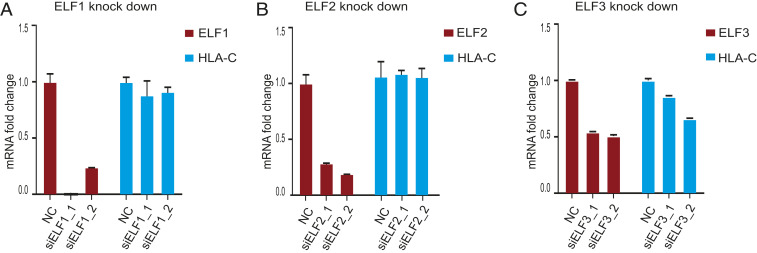
To examine whether the three most closely related ETS IIa family members, ELF1, ELF2, and ELF3, all of which are expressed in EVTs, regulate expression of HLA-C in JEG3, we performed small interfering RNA (siRNA) knockdown experiments in JEG3 cells (Fig. 2). Both ELF1 and ELF2 siRNAs reduced ELF1 (by 77% and 99%) and ELF2 (by 72% and 82%) mRNA levels in JEG3 cells (Fig. 2 A and B). Neither knockdown of ELF1 nor that of ELF2 affected expression of HLA-C (Fig. 2 A and B). The two ELF3 siRNAs down-regulated ELF3 levels only by 46% and 50%. Still, we saw a 15% reduction in HLA-C expression for siRNA1, and a 35% reduction for siRNA2 (Fig. 2C). In summary, among the closely related ETS IIa family members, only a reduction of ELF3 affected HLA-C levels in human EVT model cell lines.
Fig. 2.
HLA-C expression after ETS gene knockdown. (A) ELF1 and HLA-C mRNA levels in JEG3 after ELF1 knockdown by indicated siRNAs. The result shown here is a representative of two independent experiments. (B) ELF2 and HLA-C mRNA levels in JEG3 after ELF2 knockdown. The result shown here is a representative of two independent experiments. (C) ELF3 and HLA-C mRNA levels in JEG3 after ELF3 knockdown. The result shown here is a representative from three independent experiments. qPCR was done in triplicate. Data are reported as the mean ± SD.
ELF3 Binds to the HLA-C Proximal Promoter in JEG3.
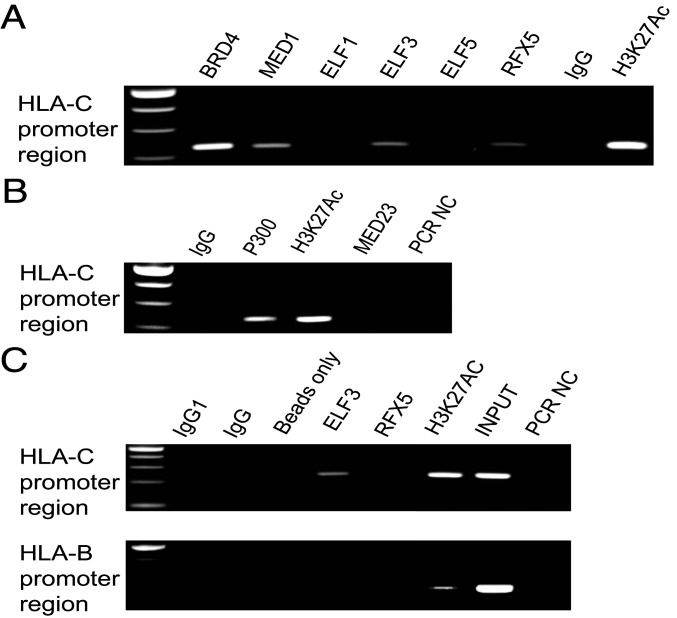
To determine whether ELF3 binds to the HLA-C promoter to regulate HLA-C expression in EVTs, we performed chromatin immunoprecipitation (ChIP)-PCR in JEG3 cells (Fig. 3A). Indeed, an ELF3 antibody immunoprecipitated material that included DNA from the HLA-C proximal promoter region (Fig. 3A). Material from the RFX5 immunoprecipitation was entirely negative in two of the four experiments performed (Fig. 3C). However, we detected binding of RFX5 to the HLA-C promoter region in the other two experiments (Fig. 3A). Since MED23 binds to ELF3 and coactivates transcription of its target gene (23), MED23 occupancy at the HLA-C promoter region was examined by ChIP-PCR. However, no MED23 occupancy at this site was detected (Fig. 3B). Another component of the Mediator complex, MED1, was also tested. It did show occupancy at the HLA-C promoter region (Fig. 3A). Since physical association between the Mediator complex and BRD4 has been demonstrated in several studies (24), we also examined BRD4 occupancy at HLA-C promoter region. Similar to MED1, BRD4 also showed occupancy at the HLA-C promoter region (Fig. 3A). H3K27Ac marks active gene regulatory elements and thus anti-H3K27Ac ChIP-PCR was used as a positive control (Fig. 3A). In summary, H3K27Ac, BRD4, and MED1 were all found to occupy the HLA-C proximal promoter region, but we did not detect binding of MED23.
Fig. 3.
Transcription factor binding at the HLA-C promoter in JEG3. Chromatin obtained from JEG3 was immunoprecipitated using antibodies against the indicated transcription factors and IgG as control. (A and B) ChIP-PCR at HLA-C promoter region. (C) ChIP-PCR at HLA-C and HLA-B promoter in JEG3.
In addition, ELF1, the closest relative to ELF3 with the highest amino acid identity (25), and ELF5 that was not expressed in EVTs, were used as specificity and negative controls (Fig. 3A). As further evidence of specificity, the proximal promoter of the HLA-B gene, adjacent to the HLA-C locus, remained silent. Neither ELF3 nor RFX5 antibodies immunoprecipitated the HLA-B promoter region (Fig. 3C). Thus, ELF3, together with H3K27Ac, BRD4, and MED1, are components of a transcription factor complex that regulates expression of HLA-C in EVTs. Importantly, the ELF3 coactivator MED23 is missing from this complex.
Wrenchnolol Inhibits HLA-C Expression in JEG3.
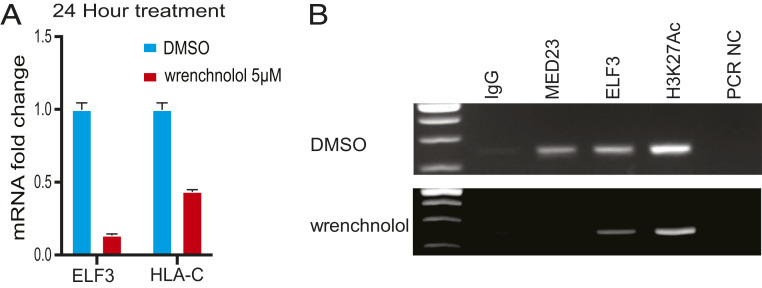
Wrenchnolol is a cell-permeable synthetic molecule that binds to the MED23 (SUR-2) subunit of the human mediator complex by mimicking the potent activation domain of ELF3 (ESE-1/ESX/ERT/Jen) (26). To examine whether blocking the interaction between ELF3 and MED23, its coactivator, would affect HLA-C expression in EVTs, we treated JEG3 cells with wrenchnolol. Indeed, treatment of JEG3 cells with 5 μM wrenchnolol for 24 h caused a 57% decrease in HLA-C transcript levels (Fig. 4A). Surprisingly, wrenchnolol treatment also strongly reduced the expression of ELF3 in JEG3 cells by 87% (Fig. 4A).
Fig. 4.
Wrenchnolol regulates both ELF3 and HLA-C expression in JEG3 cells. (A) ELF3 and HLA-C mRNA fold change after wrenchnolol treatment of JEG3 cells at 5 μM for 24 h. (B) Transcription factor binding at ELF3 promoter region in JEG3 treated with DMSO or wrenchnolol at 5 μM for 24 h. qPCR was performed in triplicate. Data are reported as the mean ± SD.
Thus, the decrease in ELF3 levels by wrenchnolol could be responsible for down-regulation of HLA-C. To understand how wrenchnolol is able to down-regulate ELF3 expression in trophoblasts, we performed MED23 ChIP-PCR. As mentioned above, we detected no MED23 binding at the HLA-C promoter region in JEG3 (Fig. 3B). However, both MED23, and ELF3 bound to the promoter of the ELF3 gene itself (Fig. 4B). Following wrenchnolol treatment, MED23 binding at the ELF3 promoter region disappeared, whereas binding of ELF3 persisted (Fig. 4B). The decrease in HLA-C levels might therefore be an indirect consequence of the disruption of the ELF3–MED23 interaction at the ELF3 promoter region. Under normal conditions, ELF3 binding to its own promoter establishes an autoregulatory feedback loop that reinforces its expression and thereby promotes the expression of HLA-C in JEG3.
ELF3 Expression Is Regulated by a Trophoblast-Specific Superenhancer.
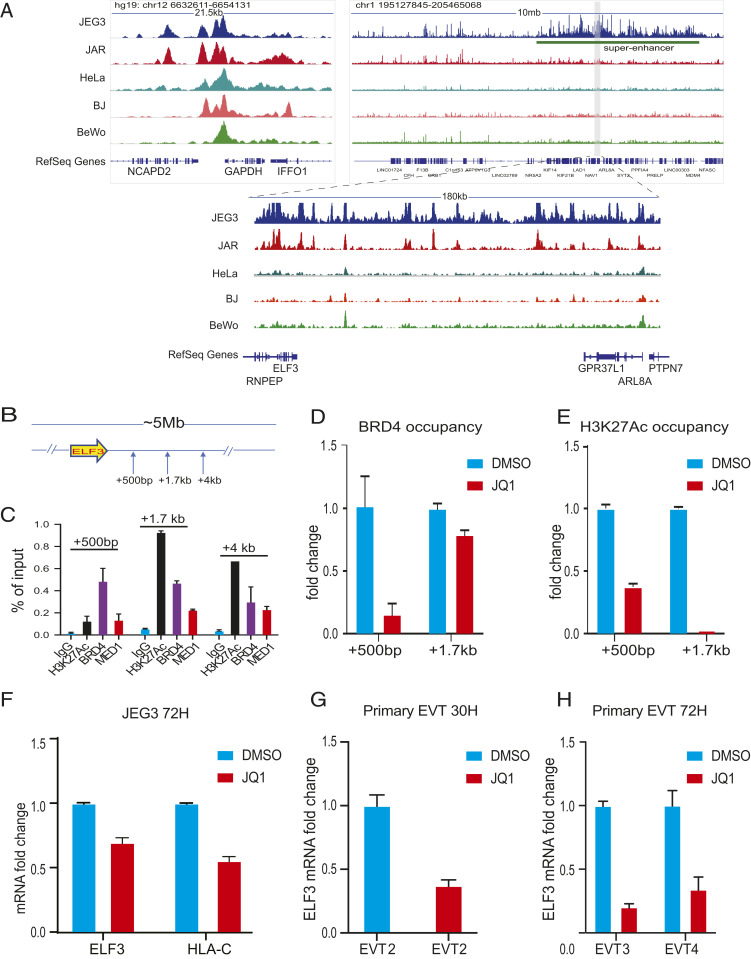
Superenhancers are clusters of enhancers that cooperatively assemble a high density of transcriptional apparatus to drive robust expression of genes with prominent roles in cell identity (27). They are typically associated with a strong occupancy signal of the Mediator complex, e.g., MED1, a member of the human bromodomain and extraterminal (BET) subfamily of proteins, binding of BRD4, and histone modification marks indicative of open chromatin, such as H3K27Ac (28). The size of superenhancers is an order of magnitude larger than that of typical enhancers and varies from several kilobases to several megabases. To further interrogate the chromatin status of the ELF3 locus in trophoblasts and control cells, we performed assay for transposase-accessible chromatin using sequencing (ATAC-seq) in JEG3, JAR, BJ fibroblasts, and HeLa cells. Human ELF3 maps to 1q32.2 (25). A >5-Mb region around ELF3 displayed a specific ATAC-seq signature in JEG3, characteristic of a superenhancer including peaks upstream and downstream of the ELF3 gene. Similar peaks yet of lower magnitude were found in JAR. No such broad peaks around the ELF3 locus were observed in the somatic cell types, BJ fibroblasts and HeLa cells (Fig. 5A). Of note, no such peaks indicative of a superenhancer were found in the promoter regions of other ETS IIa members, ELF1 and ELF2.
Fig. 5.
ELF3 is regulated by an EVT-specific superenhancer. (A) ATAC-seq of JEG3, JAR, HeLa, BJ fibroblasts, and BeWo upstream and downstream of ELF3 gene. (B) Schematic diagram showing the location of the primers used for ELF3 ChIP-qPCR analysis. (C) Chromatin obtained from JEG3 was immunoprecipitated using antibodies against H3K27Ac, BRD4, MED1, and IgG, and analyzed by qPCR with specific primer sets as indicated. (D) BRD4 occupancy fold change at +500-bp and +1.7-kb region downstream of ELF3 gene with and without JQ1 treatment at 500 nM, 72 h by ChIP-qPCR. (E) H3K27Ac occupancy fold change at +500-bp and +1.7-kb region downstream of ELF3 gene with and without JQ1 treatment at 500 nM, 72 h by ChIP-qPCR. ELF3 and HLA-C mRNA fold change after 500 nM JQ1 treatment in (F) JEG3 for 72 h; ELF3 mRNA fold change after 500 nM JQ1 treatment in (G) primary EVTs for 30 h and (H) primary EVTs for 72 h. qPCR was performed in triplicate. Data are reported as the mean ± SD.
A key characteristic of superenhancers is their sensitivity to the bromodomain and extraterminal (BET) inhibitor (+)-JQ1 (JQ1) (28). To confirm the existence of a trophoblast-specific superenhancer covering the ELF3 locus, we therefore treated JEG3 with JQ1, a BRD4 inhibitor (28, 29). In DMSO-treated JEG3 cells, ChIP-qPCR data showed the presence of the typical superenhancer markers, BRD4, MED1, and H3K27Ac at +500 bp, +1.7 kb, and +4.0 kb in the ELF3 superenhancer region (Fig. 5 B and C). After JQ1 treatment at 500 nM for 72 h, BRD4 occupancy at the two randomly selected regions within the ELF3 superenhancer, +500 bp and +1.7 kb, was examined; only the first was noticeably decreased (Fig. 5D). H3K27Ac marks at the 500-bp region was also reduced and the one at 1.7 kb disappeared completely. These data suggest the disruption of the superenhancer upon treatment with JQ1 (Fig. 5E). Next, we assessed the effect of JQ1 treatment on ELF3 and HLA-C gene expression in JEG3. As expected, JQ1 treatment reduced ELF3 expression in JEG3, by about 30%, and the HLA-C mRNA level was also decreased, by 45% (Fig. 5F). The effect of JQ1 on primary EVTs was also tested. Primary EVTs were more sensitive to the impact of JQ1 on ELF3 expression than JEG3 (Fig. 5 G and H). Three first-trimester primary EVT samples were treated with JQ1: EVT2 was treated with JQ1 for 30 h, and EVT3 and EVT4 were each treated for 72 h at a concentration of 500 nM. All three samples displayed a strong reduction of ELF3 transcripts, by 63% in EVT2, 80% in EVT3, and 66% in EVT4 (Fig. 5 G and H). In summary, both the ATAC-seq data and the BRD4 inhibitor experiment support the existence of a trophoblast-specific superenhancer, the disruption of which strongly reduces the expression of ELF3 in human EVTs.
Discussion
Considerable effort has been expended in studying the expression of human MHC class I genes, but this applies mainly to HLA-A and HLA-B, as well as to the class II gene HLA-DR. A striking feature of HLA-C is the fact that it is expressed at very high levels on EVTs, the most invasive cell of the fetal placenta. By flow cytometry analysis, its expression on peripheral somatic cells is 10 to 20% of that on EVTs. The expression of class I and class II MHC proteins is distinguished by the requirement for a “master regulator,” NLRC5 in the case of class I MHC, and CIITA in the case of class II MHC. Strikingly, however, EVTs that express very high levels of HLA-C express neither NLRC5 nor CIITA (4). Thus, an unidentified factor must replace it.
In the classical model of MHC-I gene regulation, the RFX complex binds to the X1 box of the SXY module at the proximal promoter region. Subsequently, the class I transcriptional activator, NLRC5, is then recruited to the RFX complex to promote MHC-I gene expression. The RFX complex consists of at least four subunits, RFX5 present as a dimer, RFXAP and RFXANK. Among these three factors, RFX5 is the only subunit with a known DNA binding domain. Whether RFXANK binds to RFX5 requires further exploration (30, 31). The analysis of RFXAP did not reveal any DNA-binding domain, although it is likely that charged regions of RFXAP will interact with DNA (30). Moreover, how the RFX complex assembles remains controversial. In one model, the formation of the RFX heterotetramer is initiated by the RFX5/RFXAP interaction, which results in a conformational change that enables RFXANK to bind to the subcomplex (32). In another earlier model, RFX5 binds to a preassembled RFXAP/RFXANK complex (33). RFX5 is central to multiprotein complex formation on the MHC-I and MHC-II promoters (34). Interestingly, JEG3 trophoblast cells displayed relatively low expression of RFX5 (34), and RFX5 mRNA levels in primary EVTs were even two to five times lower than in JEG3 cells (Fig. 1F). An alternative enhanceosome in which ELF3 replaces RFX5 may thus drive expression of HLA-C in EVTs. RFX5 independent gene expression has previously been reported for CIITA target genes. In fact, genome-wide studies showed that 60% of all CIITA binding sites outside of the MHC locus were not associated with RFX5 binding (35), one prominent example being CD74—the class II MHC-associated invariant chain—which has recently been identified as a host restriction factor for Ebola virus and SARS-CoV-2 (36).
As shown by detailed promoter analysis, ELF3 and RFX5 had adjacent overlapping binding sites, both occurring within the X1 box of the HLA-C core promoter (15). Intriguingly, EVTs display high levels of the ETS transcription factor ELF3, and siRNA-mediated knockdown of ELF3 decreased HLA-C expression. Thus, ELF3 competes with RFX5 for binding to the X1 box in the HLA-C proximal promoter region in JEG3. ChIP-PCR showed that both RFX5 positive and negative enhancesomes are present at the HLA-C promoter region. The identity of the immunoprecipitated PCR fragment was confirmed as derived from HLA-C by DNA sequencing. Its binding may depend on the relative concentrations of ELF3 and RFX5. RFX5 may bind to the HLA-C promoter when its level is stochastically at a relatively high level. At the other end of the spectrum, in peripheral somatic cells, which express low levels of HLA-C and do not express ELF3, RFX5 may bind with low affinity to the variant ELF3 binding site. This low-affinity binding may be sufficient to allow low-level expression of HLA-C in somatic cells driven by the same mechanism that leads to HLA-A and B expression. The finding that ELF3 can take up the role of RFX5 in regulating the trophoblast-specific expression of HLA-C is unusual. However, whether such function of ELF3 is unique to EVTs has not yet been investigated. Further quantification of RFX5 and ELF3 binding to the HLA-C promoter in somatic cells versus trophoblast cells, by EMSA or by quantitative ChIP-PCR, could be performed to support this model. It is possible that in cells that express a high level of ELF3, e.g., in mammary epithelial cells, ELF3 may play a similar role in regulating HLA-C expression as in EVTs, which still needs further exploration.
In this study, mining of scRNA-seq data was applied to identify other ETS family members with similar gene expression pattern across the different cell types at the maternal–fetal interface. scRNA-seq data are known to be limited by their low sensitivity. To identify robust candidate genes, we first aggregated cells of the same cluster as pseudobulk samples. We only focused on the top differential genes between pseudobulk samples to avoid selecting false-positive genes caused by the sparsity of the data. Other candidate genes, such as ETS family members not detected in our scRNA-seq analysis, may be further investigated separately in future studies.
ELF3 has at least three independent domains that contribute to its binding to AT-rich target sequences: a highly conserved ETS domain and two AT-hooks (37). AT-hooks are short DNA-binding motifs characterized by two arginine residues, which bind to the minor groove of AT-rich DNA (38). An AT-hook has also been described for RFX5 (39) and might assist in its binding to AT-rich ETS binding sites in cells that express little or no ELF3. Moreover, one of the ELF3 AT-hooks is required for selective gene expression in a promoter-specific manner (37), which could be one of the discriminating factors that favors preferential binding of ELF3 to the promoter of HLA-C over that of other HLA genes. Other factors that affect the selective binding may include the higher-order chromatin structure and chromatin accessibility. In JEG3, RFX5 does not bind to the HLA-B promoter, which may be due to methylation at the HLA-B promoter region. In JEG3 and trophoblast subpopulations, CpG sites are partially methylated, including those in the X box in HLA-B, but they are unmethylated in the HLA-C promoter region (40). Our ATAC-seq data showed that chromatin accessibility at the HLA-B promoter region was very low, which could explain the absence of ELF3/RFX5 binding to this region.
The present study identified ELF3 as a factor essential for trophoblast-specific expression of HLA-C. Moreover, we uncovered that the ELF3 gene is controlled by a superenhancer of >5 Mb spanning the entire ELF3 gene (Fig. 5A). Our ATAC-seq data show that there is a large region, >5 Mb of accessible chromatin, spanning the entire ELF3 locus and beyond, where MED1, BRD4, and H3K27Ac could all be detected by ChIP-PCR. Treatment with the bromodomain inhibitor JQ1 disrupted the occupancy by these transcription factors and also decreased ELF3 expression, all supporting the existence of a superenhancer. Our findings are consistent with a recent report that expression of Elf3 is associated with superenhancers in mouse trophoblast (41). The association of ELF3 with superenhancers is significantly up-regulated in preeclampsia and plays a role in preeclampsia development during pregnancy as indicated by human preeclampsia tissue sequencing data (20, 41). Moreover, ELF3 is also involved in trophoblast invasion during pregnancy, as shown in a mouse model (20). Abnormal expression of ELF3 has been implicated in invasion and metastasis in several types of cancer, including prostate cancer, and hepatocellular carcinoma (42, 43). Here, we demonstrated that JQ1 could down-regulate the expression of ELF3 by disrupting the ELF3 superenhancer. However, since there is also BRD4 occupancy at the HLA-C promoter region itself, it is possible that HLA-C expression may also be affected by JQ1 treatment directly. Whether JQ1 also regulates HLA-C expression directly by inhibiting BRD4 occupancy at the HLA-C promoter region still needs to be further explored.
Using the small-molecule inhibitor wrenchnolol, we discovered an autoregulatory feedback loop in which ELF3 binds to its own promoter and recruits MED23, thereby reinforcing its own expression (Figs. 4 and 6). Wrenchnolol is a second-generation derivative of adamanolol, a unique synthetic molecule whose wrench-like structure allows the control of gene expression by modulating a transcription factor–coactivator interaction. Wrenchnolol inhibited the ELF3-MED23 (ESX-SUR-2) interaction in vitro more strongly than did adamanolol and was no less active than adamanolol in killing SK-BR3 cells (IC50, 6.9 µM) (26). In our study, wrenchnolol started to show strong cytotoxicity at concentrations higher than 5 µM, so in our ChIP-PCR experiment, 5 μM wrenchnolol was applied, although 50% inhibition of ELF3-MED23 binding had previously been achieved at 10 µM and 100% inhibition at 30 µM wrenchnolol treatment in in vitro experiments (26). As shown by our ChIP-PCR data, wrenchnolol disrupts the ELF3–MED23 interaction at the ELF3 promoter region in JEG3. Wrenchnolol thus both regulates the expression of ELF3 as well as reduces the expression of downstream genes, such as HLA-C, which is affected by ELF3. At lower concentrations, e.g., at 1 μM wrenchnolol treatment of JEG3, no cell death or cell morphology changes were observed. However, we still detected a substantial, although lesser, decrease in ELF3 and HLA-C expression (ELF3 approximately decreased by 30% and HLA-C by 40%). So, although we cannot exclude the possibility of cell stress caused by wrenchnolol treatment, the effect of cell stress on target gene expression may be minor, yet the specificity of wrenchnolol needs more exploration. Although our ChIP-PCR data failed to detect MED23 occupancy at the HLA-C promoter region, we did, however, observe binding of MED1 to the HLA-C promoter, which implicates the Mediator complex directly in the regulation of HLA-C expression. The sensitivity of the ELF3 promoter to wrenchnolol treatment and the lack of sensitivity of the HLA-C promoter is due to the presence of MED23 in the former, and its absence from the latter.
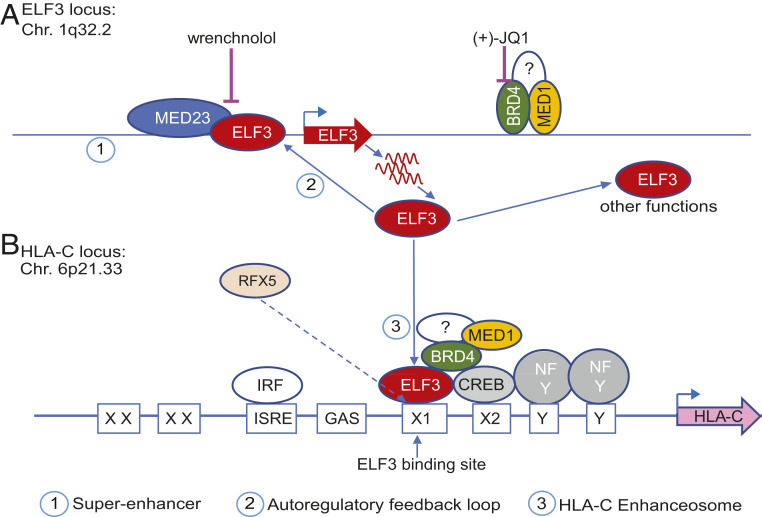
Fig. 6.
ELF3::HLA-C axis in EVTs. (A) The ELF3 gene is regulated by a superenhancer and an autoregulatory feedback loop. ELF3 reinforces its own gene expression by binding to its own promoter and recruiting the Mediator component MED23/SUR-2. Wrenchnolol disrupts the interaction of ELF3 and MED23/SUR-2, and (+)-JQ1 disrupts the superenhancer. (B) ELF3 regulates HLA-C expression. RFX5 competes with ELF3 for the binding to the ETS motif in the HLA-C promoter. The Mediator component MED1, BRD4, and other factors, together with ELF3, form the HLA-C enhanceosome and establish HLA-C expression at the maternal–fetal interface.
ELF3 was first described as a critical regulator of epithelial cell differentiation (44). More recently, ELF3 has been implicated in maintaining cell identity. Knockdown of ELF3 in keratinocytes resulted in an epithelial-to-mesenchymal transition (EMT) (45). It is thus conceivable that the autoregulatory feedback loop that reinforces high ELF3 expression might therefore be involved in maintaining trophectoderm identity, a process described in several model organisms and cell lines (46). Of note, JEG3, the trophoblast model cell line primarily used in this study, is a choriocarcinoma cell line. Given the important role of ELF3 in many cancers, including cancers of the reproductive system, including breast cancer, ovarian cancer, and prostate cancer (17), it may not be surprising that ELF3 forms a positive autoregulatory loop in JEG3. Disruption of the interaction between ELF3 and MED23 by short cell-permeable synthetic peptides in breast cancer cell lines significantly reduced expression of the oncogene HER2, typically overexpressed in patients with aggressive disease, enhanced metastasis, and increased resistance to chemotherapy (23). Whether a similar autoregulatory feedback loop exists in primary EVTs where it might establish high levels of ELF3 expression and thus promote cell proliferation and cell invasion remains to be explored.
In summary, these studies have uncovered the complex mechanism that leads to high-level expression of HLA-C in EVTs and may explain the low-level expression of HLA-C on somatic cells. ELF3 plays a central role in the process (Fig. 6). Central to its role is the single-nucleotide variation in the X1 box that distinguishes the HLA-C promoter region from that of its paralogs, particularly from its genomic neighbor, HLA-B, from which it arose by gene duplication. This variation leads to a dramatic change in the principal transcription factor that binds to the X1 box in the proximal promoter of the HLA-C gene. This means by which paralogs acquire distinct patterns of expression is unusual. Activation of ELF3 requires both its coactivator MED23, a component of the Mediator complex, and an autoregulatory feedback mechanism. The details of such “activation,” although essential for HLA-C production, remain obscure. HLA-C plays a central role in both tolerance and immunity at the maternal–fetal interface. High-level HLA-C may be critical for production of cytokines required for development of the embryo. It may also be important for protection against infection, since it is the only highly polymorphic protein expressed at the maternal–fetal interface. On the other hand, high-level expression of HLA-C may lead to allogeneic rejection of the developing fetus. Pregnancy can involve many complications. Ordered from early to later occurrence, they include infertility, miscarriage, preterm birth, preeclampsia, and delayed labor. The expression level of HLA-C may be similarly timed. To exploit this knowledge, much further work remains for cellular and molecular biologists, as well as clinicians.
Materials and Methods
Cell Culture and Treatment.
JEG3, BeWo, JAR, HeLa, and BJ fibroblasts were cultured in Dulbecco’s modified Eagle medium (DMEM) (Life Technologies) containing 10% fetal bovine serum and 1% penicillin/streptomycin solution at 37 °C with 5% CO2. For JQ1 treatment experiments, (+)-JQ1 (catalog no. 27402; BPS Bioscience) at 500 nM or an equivalent volume of DMSO were resuspended in fresh medium, added to cells, and incubated for the indicated duration. DMSO was used as control. For wrenchnolol treatment experiments, wrenchnolol (a gift from Prof. Motonari Uesugi, Kyoto University, Kyoto, Japan) was used at indicated concentrations or DMSO (as a control) was resuspended in fresh medium, added to cells, and incubated for the indicated duration.
siRNA Transfection.
Before transfection, 2 × 105 cells per well were plated into six-well plates and grown for 1 d. When the cells reached a confluency of 30 to 40%, cells were transfected with ELF1-specific siRNAs (siELF1_1: catalog no. 4392420, assay ID no. s4617; siELF1_2: catalog no. 4392420, assay ID no. s4618); ELF2-specific siRNAs (siELF2_1: catalog no. 4392420, assay ID no. s4620; siELF2_2: catalog no. 4392420, assay ID no. s4621), ELF3-specific siRNAs (catalog no. 4392420, assay ID no. s4623; catalog no. 4392420, assay ID no. s4624), and negative control siRNA (catalog no. 4390843; Thermo Fisher Scientific) using Lipofectamine RNAiMAX reagent (catalog no. 13778-030; Life Technologies), respectively. Transfected cells were grown at 37 °C. The cells were harvested for RNA extraction 72 h posttransfection.
First-Trimester Primary EVT Isolation and Treatment.
EVTs were isolated as previously described (47, 48). Discarded human placental and decidual tissue (gestational age, 6 to 12 wk) was obtained from women undergoing elective pregnancy termination at a local reproductive health clinic. All of the human tissues used for this research was deidentified, discarded clinical material. The Committee on the Use of Human Subjects [the Harvard institutional review board (IRB)] determined that this use of all the human material is exempt from the requirements of IRB review. In total, ∼20K HLA-G+ EVTs were plated in 96-well cell culture plates (Costar) precoated with 50 μL of 20 ng/mL fibronectin for 45 min (BD), in Trophoblast Medium, which consisted of DMEM/F12 medium (Gibco) supplemented with 10% (vol/vol) NCS, Glutamax, insulin, transferrin, selenium (100×; Gibco), 5 ng/mL EGF (Peprotech), and 400 units of human gonadotropic hormone (Sigma). Approximately 12 h post-plating, EVTs were treated with JQ1 at indicated concentration and indicated duration.
qRT-PCR Analysis.
Total RNA was isolated using RNeasy Mini Kit (catalog no./ID no. 74104; Qiagen), according to manufacturer’s instructions. A total of 1,000 ng of RNA was used for cDNA synthesis with the High-Capacity cDNA Reverse Transcription kit (catalog no. 4368814; Applied Biosystems) with RNase Inhibitor (catalog no. 4374966; Life Technologies) for reverse transcription. We performed qRT-PCR detection using a TaqMan Gene Expression Assays and TaqMan Fast Advanced Master Mix (catalog no. 4444557; Life Technologies). Target gene Ct values were normalized to GAPDH Ct value, and the average normalized target gene Ct values are presented. Moreover, the relative expression level was determined as 2−ΔΔCt, and data are given as the expression level relative to the calibrator (control sample). TaqMan assays were run on a Quantstudio 12k Flex real-time PCR system (Thermo Fisher Scientific) or Quantstudio 6 Flex real-time PCR system (Thermo Fisher Scientific). TaqMan Gene expression assays applied were as follows (Thermo Fisher Scientific): GAPDH, assay ID no. Hs02786624_g1, catalog no. 4331182; HLA-C, assay ID no. Hs03044135_m1, catalog no. 4331182; ELF1, assay ID no. Hs01111177_m1, catalog no. 4331182; ELF2, assay ID no. Hs00959420_g1, catalog no. 4331182; ELF3, assay ID no. Hs00963882_g1, catalog no. 4351372; and ELF5, assay ID no. Hs01063023_g1, catalog no. 4351372.
Due to the limited availability of primary EVTs, experiments including EVTs were performed in 96-well plates and analyzed with Cells-to-CT 1-Step TaqMan Kit (catalog no. A25603; Thermo Fisher) and TaqMan gene expression assays listed above.
ChIP-PCR/qPCR.
ChIP-PCR/qPCR was performed using the ChIP kit (ab500) from Abcam according to the manufacturer’s instructions. JEG3 cells were harvested, resuspended in PBS, and cross-linked using 1% formaldehyde. Glycine was added to stop the cross-linking reaction, and cells were washed twice with ice-cold PBS. Nuclei were isolated and then lysed using ice-cold Cell Lysis Buffer containing protease inhibitors and PMSF. Cross-linked chromatin was sheared using a Covaris S220 to 200- to 500-bp fragments and assessed by TapeStation assay D1000 (Agilent Technologies). Samples were then diluted with ChIP Dilution Buffer and incubated with antibodies anti-: Mock IgG1, IgG, ELF1, ELF5, RFX5 antibodies (Santa Cruz Biotechnology), ELF3 (A13489; Abclonal), BRD4 (A301-985A50) and MED1 (A300-793A) from Bethyl Laboratories, and MED23 (clone D27-1805; BD Biosciences), P300 (ab14984; Abcam), and H3K27Ac (ab4729, Abcam) overnight at 4 °C. The antibody/chromatin samples were pelleted and incubated with protein A beads. After incubation, the antibody/chromatin/beads were washed, and DNA was purified with DNA purifying slurry included in the ChIP kit. A total of 2 μL of DNA was used for qRT-PCR analysis using SYBR Green (Thermo Fisher Scientific) or 2 μl DNA/reaction was applied to PCR using platinum PCR SuperMix High Fidelity (Thermo Fisher Scientific) and analyzed by gel electrophoresis.
Primers.
Primers were as follows: HLA-C promoter region: HLA-C‐Fwd, GGCTCCAAGGGCCGTGTCTGCAC, and HLA-C‐Rev, GTCTGGGGAGAATCTGAGTCC; HLA-B promoter region: HLA_B Fwd, GGCTCCGAGGGCCGCGTCTGCAATG, and HLA_B Rev, GTCTGAGGAGACTCTGAGTCC.
ELF3 superenhancer region: +500 bp Fwd, CCCTCGGGGTCCTTCTATGA; +500 bp Rev, GGTGGGTAGTTCAGTGGAGC; +1.7 kb Fwd, GCTCTCCCTGGACCACAATC; +1.7 kb Rev, CCACACAGGTACTGCATGGT; +4 kb Fwd, GGGAGCATTGCCCTCTCATT; and +4 kb Rev, CAGGACACTGGGTGTGGTTT.
scRNA-Seq Data Analysis.
The scRNA-seq data were retrieved from publicly available datasets cataloged in GEO (Gene Expression Omnibus) with accession no. GSE89497 (22). The single-cell transcriptome profiles of stromal cells and trophoblast cells from 8- and 24-wk gestation of placenta were generated by next-generation sequencing using Illumina Hiseq4000. Hierarchical clustering was performed using the module of Hierarchical Clustering in GenePattern from the Broad Institute and heatmap was generated using the module of HierarchicalClusteringViewer in GenePattern from the Broad Institute.
The scRNA-seq data were also processed using Seurat (49). Top 2,000 variable genes were used for PCA dimensionality reduction (n = 10). The data were visualized using Umap and colored by normalized gene expression level.
ATAC-Seq.
Cell lines were grown to 70 to 80% confluency, trypsinized, and counted, and 50,000 cells were lysed and subjected to “tagmentation” reaction and library construction as previously described (50). Libraries were run on an Illumina Hi-Seq2500. The reads were trimmed and aligned to the human reference genome (hg19) with Bowtie2aligner (51) using the option -X2000. Then, we discarded reads with alignment quality < Q30, improperly paired, mapped to the unmapped contigs, chrY, and mitochondria. Duplicates were removed using MarkDuplicates in Picard tools (broadinstitute.github.io/picard/). To visualize the tracks, the reads were extended by 250 bp using igv tools (https://software.broadinstitute.org/software/igv/igvtools).
Acknowledgments
We thank Eva M. Fast and Yi Zhou for help with the ATAC-seq experiment, Hidde Ploegh and Max Friesen for insightful comments on the manuscript, and Joyce Lavecchio and Nema Kheradmand for assistance with cell sorting. This study was funded by National Institute of Allergy and Infectious Diseases Grants R21AI138019 and R01AI145862.
Footnotes
The authors declare no competing interest.
Data Availability
All study data are included in the article. Previously published data were used for this work (GEO accession no. GSE89497). ATAC-seq data have been deposited in the GEO database (accession no. GSE165511).
References
- 1.Carter A. M., Comparative studies of placentation and immunology in non-human primates suggest a scenario for the evolution of deep trophoblast invasion and an explanation for human pregnancy disorders. Reproduction 141, 391–396 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Guethlein L. A., Norman P. J., Hilton H. G., Parham P., Co-evolution of MHC class I and variable NK cell receptors in placental mammals. Immunol. Rev. 267, 259–282 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crosley E. J., Elliot M. G., Christians J. K., Crespi B. J., Placental invasion, preeclampsia risk and adaptive molecular evolution at the origin of the great apes: Evidence from genome-wide analyses. Placenta 34, 127–132 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Papúchová H., Meissner T. B., Li Q., Strominger J. L., Tilburgs T., The dual role of HLA-C in tolerance and immunity at the maternal-fetal interface. Front. Immunol. 10, 2730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tilburgs T., et al., Fetal-maternal HLA-C mismatch is associated with decidual T cell activation and induction of functional T regulatory cells. J. Reprod. Immunol. 82, 148–157 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Tilburgs T., Strominger J. L., CD8+ effector T cells at the fetal-maternal interface, balancing fetal tolerance and antiviral immunity. Am. J. Reprod. Immunol. 69, 395–407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crespo A. C., Strominger J. L., Tilburgs T., Expression of KIR2DS1 by decidual natural killer cells increases their ability to control placental HCMV infection. Proc. Natl. Acad. Sci. U.S.A. 113, 15072–15077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meuleman T., et al., HLA-C antibodies in women with recurrent miscarriage suggests that antibody mediated rejection is one of the mechanisms leading to recurrent miscarriage. J. Reprod. Immunol. 116, 28–34 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Israeli M., et al., Association between CTL precursor frequency to HLA-C mismatches and HLA-C antigen cell surface expression. Front. Immunol. 5, 547 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apps R., et al., Influence of HLA-C expression level on HIV control. Science 340, 87–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apps R., et al., Relative expression levels of the HLA class-I proteins in normal and HIV-infected cells. J. Immunol. 194, 3594–3600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowell D., et al., Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359, 582–587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reusing S. B., et al., Selective downregulation of HLA-C and HLA-E in childhood acute lymphoblastic leukaemia. Br. J. Haematol. 174, 477–480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moffett A., Loke C., Immunology of placentation in eutherian mammals. Nat. Rev. Immunol. 6, 584–594 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Johnson J. K., Wright P. W., Li H., Anderson S. K., Identification of trophoblast-specific elements in the HLA-C core promoter. HLA 92, 288–297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei G. H., et al., Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 29, 2147–2160 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sizemore G. M., Pitarresi J. R., Balakrishnan S., Ostrowski M. C., The ETS family of oncogenic transcription factors in solid tumours. Nat. Rev. Cancer 17, 337–351 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Findlay V. J., LaRue A. C., Turner D. P., Watson P. M., Watson D. K., Understanding the role of ETS-mediated gene regulation in complex biological processes. Adv. Cancer Res. 119, 1–61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavoie H., Gagnon J., Therrien M., ERK signalling: A master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 21, 607–632 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Tuteja G., Chung T., Bejerano G., Changes in the enhancer landscape during early placental development uncover a trophoblast invasion gene-enhancer network. Placenta 37, 45–55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng A. Y., et al., Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology 122, 1455–1466 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Liu Y., et al., Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 28, 819–832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asada S., et al., External control of Her2 expression and cancer cell growth by targeting a Ras-linked coactivator. Proc. Natl. Acad. Sci. U.S.A. 99, 12747–12752 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhagwat A. S., et al., BET bromodomain inhibition releases the mediator complex from select cis-regulatory elements. Cell Rep. 15, 519–530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tymms M. J., et al., A novel epithelial-expressed ETS gene, ELF3: Human and murine cDNA sequences, murine genomic organization, human mapping to 1q32.2 and expression in tissues and cancer. Oncogene 15, 2449–2462 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Shimogawa H., et al., A wrench-shaped synthetic molecule that modulates a transcription factor-coactivator interaction. J. Am. Chem. Soc. 126, 3461–3471 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Sabari B. R., et al., Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lovén J., et al., Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filippakopoulos P., et al., Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeSandro A. M., Nagarajan U. M., Boss J. M., Associations and interactions between bare lymphocyte syndrome factors. Mol. Cell. Biol. 20, 6587–6599 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshihama S., Vijayan S., Sidiq T., Kobayashi K. S., NLRC5/CITA: A key player in cancer immune surveillance. Trends Cancer 3, 28–38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burd A. L., et al., Assembly of major histocompatibility complex (MHC) class II transcription factors: Association and promoter recognition of RFX proteins. Biochemistry 43, 12750–12760 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Nekrep N., Jabrane-Ferrat N., Peterlin B. M., Mutations in the bare lymphocyte syndrome define critical steps in the assembly of the regulatory factor X complex. Mol. Cell. Biol. 20, 4455–4461 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Elsen P. J., Gobin S. J., van der Stoep N., Datema G., Viëtor H. E., Transcriptional control of MHC genes in fetal trophoblast cells. J. Reprod. Immunol. 52, 129–145 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Wong D., et al., Genomic mapping of the MHC transactivator CIITA using an integrated ChIP-seq and genetical genomics approach. Genome Biol. 15, 494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruchez A., et al., MHC class II transactivator CIITA induces cell resistance to Ebola virus and SARS-like coronaviruses. Science 370, 241–247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kopp J. L., Wilder P. J., Desler M., Kinarsky L., Rizzino A., Different domains of the transcription factor ELF3 are required in a promoter-specific manner and multiple domains control its binding to DNA. J. Biol. Chem. 282, 3027–3041 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Huth J. R., et al., The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 4, 657–665 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Stavride P., Arampatzi P., Papamatheakis J., Differential regulation of MHCII genes by PRMT6, via an AT-hook motif of RFX5. Mol. Immunol. 56, 390–398 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Guillaudeux T., et al., Methylation status and transcriptional expression of the MHC class I loci in human trophoblast cells from term placenta. J. Immunol. 154, 3283–3299 (1995). [PubMed] [Google Scholar]
- 41.Lee B. K., et al., Super-enhancer-guided mapping of regulatory networks controlling mouse trophoblast stem cells. Nat. Commun. 10, 4749 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longoni N., et al., ETS transcription factor ESE1/ELF3 orchestrates a positive feedback loop that constitutively activates NF-κB and drives prostate cancer progression. Cancer Res. 73, 4533–4547 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Zheng L., et al., ELF3 promotes epithelial-mesenchymal transition by protecting ZEB1 from miR-141-3p-mediated silencing in hepatocellular carcinoma. Cell Death Dis. 9, 387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oettgen P., et al., Isolation and characterization of a novel epithelium-specific transcription factor, ESE-1, a member of the ets family. Mol. Cell. Biol. 17, 4419–4433 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sengez B., et al., The transcription factor Elf3 is essential for a successful mesenchymal to epithelial transition. Cells 8, E858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leyva-Díaz E., Hobert O., Transcription factor autoregulation is required for acquisition and maintenance of neuronal identity. Development 146, dev177378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tilburgs T., et al., Human HLA-G+ extravillous trophoblasts: Immune-activating cells that interact with decidual leukocytes. Proc. Natl. Acad. Sci. U.S.A. 112, 7219–7224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreira L. M., et al., A distant trophoblast-specific enhancer controls HLA-G expression at the maternal–fetal interface. Proc. Natl. Acad. Sci. U.S.A. 113, 5364–5369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuart T., et al., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article. Previously published data were used for this work (GEO accession no. GSE89497). ATAC-seq data have been deposited in the GEO database (accession no. GSE165511).