Abstract
Ribonucleotides are the most abundant non-canonical nucleotides in the genome. Their vast presence and influence over genome biology is becoming increasingly appreciated. Here we review the recent progress made in understanding their genomic presence, incorporation characteristics and usefulness as biomarkers for polymerase enzymology. We also discuss ribonucleotide processing, the genetic consequences of unrepaired ribonucleotides in DNA and evidence supporting the significance of their transient presence in the nuclear genome.
Keywords: ribonucleotide incorporation, DNA replication, DNA repair, genome stability, genome-wide sequencing
Introduction
The eukaryotic DNA replication machinery is responsible for faithful duplication of genetic information stored in genomic DNA during S phase of each cell cycle. DNA Polymerases (Pols) α, δ and ε conduct the bulk of DNA synthesis by effectively selecting and incorporating correct nucleotides. However, nucleotide selectivity by these replicative polymerases (replicases) is imperfect. They insert roughly one nucleotide with an incorrect base for each hundred thousand correct nucleotides incorporated, resulting in a base-base mispair (Kunkel 2004; McCulloch and Kunkel 2008). These mispairs may be corrected by either the exonucleolytic proofreading activities of Pols δ and ε or by the DNA mismatch repair (MMR) pathway. In addition, imperfect nucleotide selectivity means that replicases also occasionally make errors by incorporating a nucleotide with an incorrect sugar moiety, with the most common type being a ribonucleotide (Nick McElhinny, Watts, et al. 2010; Clausen et al. 2013).
Ribonucleotides embedded in DNA were first identified in mammalian mitochondrial genomes (Grossman et al. 1973), but their biological significance was not appreciated until fairly recently, when their prevalence, mechanism of removal and immediate consequences were reported in yeast and mammals (Rydberg and Game 2002; Nick McElhinny, Kumar, et al. 2010; Nick McElhinny, Watts, et al. 2010; Reijns MA et al. 2012). Ribonucleotides are incorporated during genome replication at a remarkably high rate (Rydberg and Game 2002; Nick McElhinny, Watts, et al. 2010). Furthermore, mutations in genes encoding subunits of RNase H2, the main enzyme that initiates ribonucleotide removal, were found in Aicardi-Goutières Syndrome (AGS) patient families (Crow et al. 2006). Replicases incorporate incorrect sugars orders of magnitude more frequently than they do incorrect bases (Nick McElhinny, Watts, et al. 2010; Clausen et al. 2013). Many DNA polymerase active sites select against ribonucleotide incorporation via a steric clash between the 2’-OH on the incoming sugar moiety and a bulky steric gate residue (reviewed in (Joyce 1997; Brown and Suo 2011)). Several studies using archaeal, bacterial and eukaryotic model organisms have demonstrated the importance of such a steric gate residue for ribonucleotide discrimination during DNA replication and repair (DeLucia et al. 2003; McDonald et al. 2012; Vaisman et al. 2012; Donigan et al. 2014; Donigan et al. 2015; Nevin et al. 2015; Diaz-Talavera et al. 2019; Walsh et al. 2019; Zatopek et al. 2020). The majority of incorporated ribonucleotide monophosphates (rNMPs) are removed by the RNase H2-dependent ribonucleotide excision repair (RER) pathway (Rydberg and Game 2002; Nick McElhinny, Kumar, et al. 2010; Hiller et al. 2012; Reijns MA et al. 2012). High cellular concentrations of ribonucleotide triphosphates (rNTPs), relative to deoxyribonucleotide triphosphates (Nick McElhinny, Watts, et al. 2010; Wanrooij et al. 2017; Balachander et al. 2020), guarantee that all transactions involving genomic synthesis incorporate ribose into DNA with some frequency. Individual ribonucleotides dynamically disturb the local duplex, according to simulations (Fu et al. 2019), and can disrupt nucleosome binding (Dunn and Griffith 1980; Hovatter and Martinson 1987). Some critically important questions include where and when ribonucleotides are incorporated and what biological consequences ensue. In this review, we discuss recent developments in pursuit of answers to these questions, with an emphasis on ribonucleotide incorporation in the nuclear genome during DNA replication.
Genome-wide mapping of ribonucleotides
Here we discuss the progress in using these technologies to answer the “where” and “when” questions regarding ribonucleotide incorporation in the nuclear and mitochondrial genome. We also discuss the progress in using the NMP mapping to study DNA polymerase enzymology during DNA replication.
The realization that ribonucleotides are frequently incorporated into the genome during replication sparked interest in developing genome-mapping technologies to define their genomic locations. Major progress was made in 2015 when four groups independently reported sequencing-based ribonucleotide maps of various eukaryotic nuclear and mitochondrial genomes (Clausen et al. 2015; Daigaku et al. 2015; Koh et al. 2015; Reijns MAM et al. 2015).
Using replicase variants that are promiscuous for ribonucleotide incorporation, some of these studies were able to use rNMPs as footprints of replicase usage during normal DNA replication (Clausen et al. 2013; Clausen et al. 2015; Daigaku et al. 2015; Reijns MAM et al. 2015). These studies presented powerful in vivo evidence for the division of labor between Pols ε and δ. They were assigned the majority of leading and lagging strand synthesis, respectively, in confirmation of more limited mutational studies in yeast (Pursell et al. 2007; Nick McElhinny et al. 2008; Larrea et al. 2010; Miyabe et al. 2011). This demonstrated the power of rNMP mapping to study polymerase enzymology during DNA synthesis.
Ribonucleotide incorporation into the nuclear genome
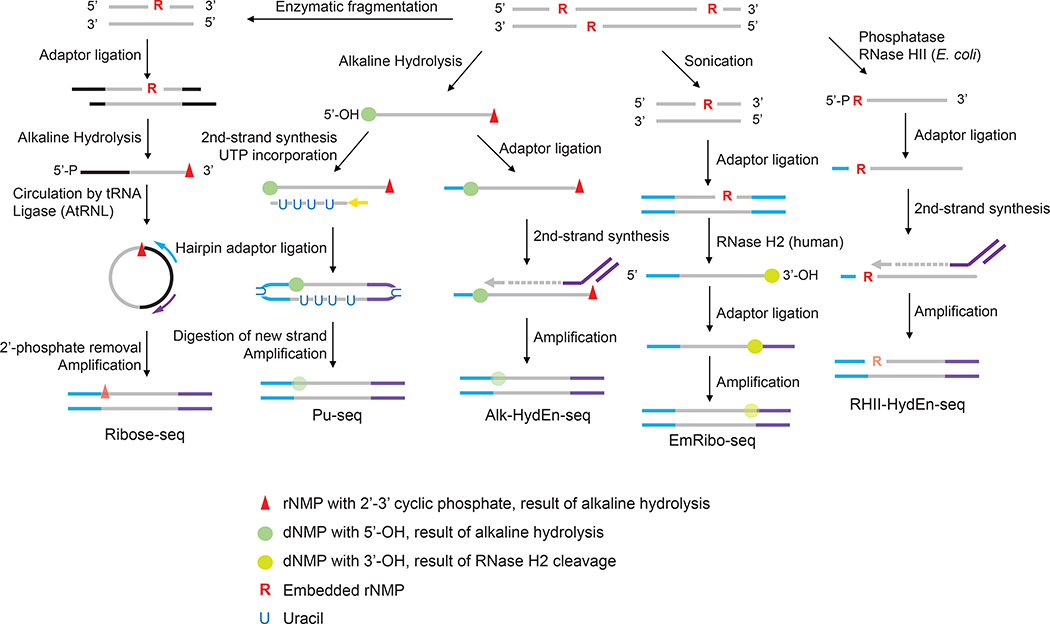
These studies simultaneously describing independent methods for genome-wide rNMP mapping were all based on high-throughput sequencing platforms. Three groups using Illumina sequencing (Clausen et al. 2015; Daigaku et al. 2015; Koh et al. 2015) and one group used Ion Torrent sequencing (Reijns MAM et al. 2015). The unifying principle was incision at genomic rNMPs, either by rNMP-recognizing RNase H2 or by alkaline hydrolysis. Nicked DNA ends were then tagged with strand-specific DNA adaptors (Figure 1).
Figure 1.
The current iterations of ribonucleotide mapping technologies. These strategies can be classified by the methods used to incise at embedded ribonucleotides. Ribose-seq, Pu-seq and Alk-HydEn-seq use alkaline hydrolysis which catalyzes hydrolysis on the 3’ side of the rNMP, resulting in 2’,3’-cyclic phosphate and a 5’-hydroxyl DNA ends. EmRibo-seq uses recombinant human RNase H2 and RHII-HydEn-seq uses E. coli type II RNase H (RNase HII), both of which cleave on the 5’ side of the rNMP, resulting in 5’-phosphate and 3’-OH ends. These technologies also differ in the location of ribonucleotide with respect to the sequencing read. Ribose-seq maps the rNMP site to the first position of the mapped read but on the opposite strand. Pu-seq and Alk-HydEn-seq identify the rNMP as located one nucleotide upstream of the mapped the read. In EmRibo-seq, the rNMP is similarly positioned one base upstream of the mapped read but on the opposite strand. For RHI-HydEn-seq, the rNMP is the first nucleotide of the mapped read. The symbols denoting the DNA/RNA ends resulting from alkaline hydrolysis or RNase cleavage are kept throughout the steps to help track their locations in the sequencing reads. It is worth noting that after the DNA amplification step during the library preparation, these symbols no longer represent the original form of the nucleotides but rather their base identities. Their opacity to emphasize this difference.
All four studies confirmed widespread rNMP incorporation with rNMPs detected universally from early to late replicating regions. Two studies in the budding yeast Saccharomyces cerevisiae (Clausen et al. 2015; Reijns MAM et al. 2015) and one in the fission yeast Schizosaccharomyces pombe (Daigaku et al. 2015) also used their respective rNMP mapping technologies to show the division of labor among replicases.
Pol α-primase synthesizes an RNA-DNA primer required for initiation of DNA synthesis. The short DNA portion synthesized by Pol α was thought to be largely removed by Pol δ-dependent strand displacement during Okazaki fragment maturation (Maga et al. 2001). However, the Pol α variants incorporates significant rNMPs, suggesting that at least some Pol α-synthesized DNA is retained after Okazaki fragment maturation (Clausen et al. 2013; Clausen et al. 2015; Reijns MAM et al. 2015). Rejins et al. estimated that Pol α synthesizes at ~1.5% of the yeast genome. Because Pol α lacks exonucleolytic proofreading activity and is the least accurate eukaryotic replicase (McCulloch and Kunkel 2008), these results indicate that Pol α contributes significantly to the spontaneous mutagenesis across the genome, especially during the lagging strand synthesis (Lujan et al. 2014). Rejins, et al. proposed that retention of DNA synthesized by Pol α may be related to inhibition of Pol δ strand displacement by nucleosome occupancy or other DNA binding proteins such as transcription factors (Reijns MAM et al. 2015). Nucleosomes appear to load before Okazaki fragment maturation (Smith and Whitehouse 2012) and to inhibit other transactions, such as MMR (Lujan et al. 2014). Thus, these protein-binding sites tend to have higher spontaneous mutation rates in both yeast and human cells (Reijns MAM et al. 2015).
These studies also revealed that rNMP incorporation by the replicases is not uniform. Ribose-seq showed that rC and rG are preferred substrates for incorporation over rA and rU in RNase H2-deficient S. cerevisiae cells (Koh et al. 2015). HydEn-seq revealed distinctive rNMP preferences for the Pol α, δ and ε variants (Clausen et al. 2013; Clausen et al. 2015). The Pol ε and δ variants favor rC and rG, the Pol α variant prefers rA, and all three disfavor rU. The Storici group recently expanded these initial observations by conducting an expansive analysis of rNMP identity and sequence context in both budding and fission yeast strains using an improved version of Ribose-seq (Balachander et al. 2020). Confirming the imbalanced rNMP incorporation observed previously, they show that rC incorporation is highly preferred followed by rG in RNase H2-deficient S. cerevisiae and S. paradoxus. rU is the least abundant ribonucleotide in the genome. This preference only partially reflects the imbalanced cellular rNTP and dNTP pools, with the most notable discrepancy being rATP relative to dATP. In S. cerevisiae, rATP is the most abundant ribonucleotide and the rATP/dATP ratio is the highest (Nick McElhinny, Watts, et al. 2010; Balachander et al. 2020). The low rate of rA and rU incorporation is likely due to the high discrimination against rATP by the replicases (Nick McElhinny, Watts, et al. 2010). In contrast, rU is the least frequently incorporated ribonucleotide in S. pombe, with rA being the most abundant, perhaps due to the extreme rATP/dATP ratio (Balachander et al. 2020). The Storici group further showed that rNMP incorporation is influenced by the immediate upstream DNA sequence, suggesting certain sequences help to stabilize the incoming ribonucleotide (Balachander et al. 2020). Because a ribonucleotide at the 3’-end of a DNA primer is an impediment to DNA polymerase extension (Watt et al. 2011; Goksenin et al. 2012), it is possible some base-pairing combinations are favored for extension. It is worth noting that different yeast strain backgrounds also impact the ribonucleotide identity and distribution in the genomes (Balachander et al. 2020). The Storici group has developed a computational toolkit named Ribose-Map that streamlined upstream read alignment and certain downstream analyses including ribonucleotide identity, genomic distribution and sequence contexts (Gombolay et al. 2019). This toolkit can be adapted for different rNMP mapping technologies.
rNMP incorporation in the mitochondrial genome
The first genome-wide ribonucleotide mapping studies also confirmed that rNMPs are stably incorporated into mitochondrial genomes (Clausen et al. 2015; Koh et al. 2015). Similar to the nuclear genome, rNMP incorporation in the mitochondrial genome is nonuniform (Clausen et al. 2015; Koh et al. 2015; Berglund et al. 2017; Wanrooij et al. 2017; Balachander et al. 2020). These studies in budding yeast disagree upon which rNMP is the most frequently incorporated (rC or rG) but agree that rU is strongly selected against. This is similar to human mitochondria and seems to be driven by imbalances in nucleotide pools (Berglund et al. 2017). The ribonucleotide spectrum in mitochondrial DNA from solid human tissues is mostly explainable by the nucleotide pools and ribonucleotide discrimination by Pol γ (Moss et al. 2017). The mitochondrial genome rNMP distribution is also nonuniform with regard to location and DNA sequence context (Clausen et al. 2015; Koh et al. 2015; Berglund et al. 2017; Balachander et al. 2020). DNA replication by Pol γ and remnants of unremoved RNA primers could both contribute to rNMP incorporation (Cerritelli Susana M. et al. 2003; Holmes et al. 2015). rNMP peaks corresponding to sites of replication initiation were observed in both yeast and human mitochondrial genomes (Clausen et al. 2015; Berglund et al. 2017). Since RNase H1-dependent removal of an RNA primer leaves two consecutive rNMPs behind (Cerritelli Susana M. et al. 2003), it is possible some of the observed peaks are from residual RNA primers. So far, mitochondria seem to lack the ability to remove single embedded ribonucleotides (Wanrooij et al. 2017). It remains unclear what consequences embedded ribonucleotides may have on mitochondrial biology.
Using rNMPs as a biomarker to map polymerase enzymology
During DNA replication, rNMPs are incorporated at a remarkably high rate. Over a million rNMPs are incorporated into the mouse genome each cell cycle (Reijns MA et al. 2012; Reijns MAM et al. 2015)). The Pol α, δ and ε active site mutants (e.g. pol1-Y869A, pol3-L612G and pol2-M644G in budding yeast) incorporate rNMPs even more frequently than their wild-type counterparts, but can still support relatively normal DNA replication (Nick McElhinny, Kumar, et al. 2010; Clausen et al. 2015). In RNase H2-deficient yeast cells, most of the embedded rNMPs are retained, leaving footprints by which the DNA synthesized by each replicase may be tracked. Replication profiles deduced from rNMP mapping in both budding and fission yeasts provided powerful support for the currently accepted model that Pol ε is the main leading strand replicase, and that Pol δ conducts the majority of lagging strand synthesis (Clausen et al. 2015; Daigaku et al. 2015; Reijns MAM et al. 2015). The profiles also yield tremendous information regarding sites of replication initiation, termination and replication timing. However, localized excursions from the canonical division of labor are occasionally observed (Clausen 2015; Daigaku et al. 2015).
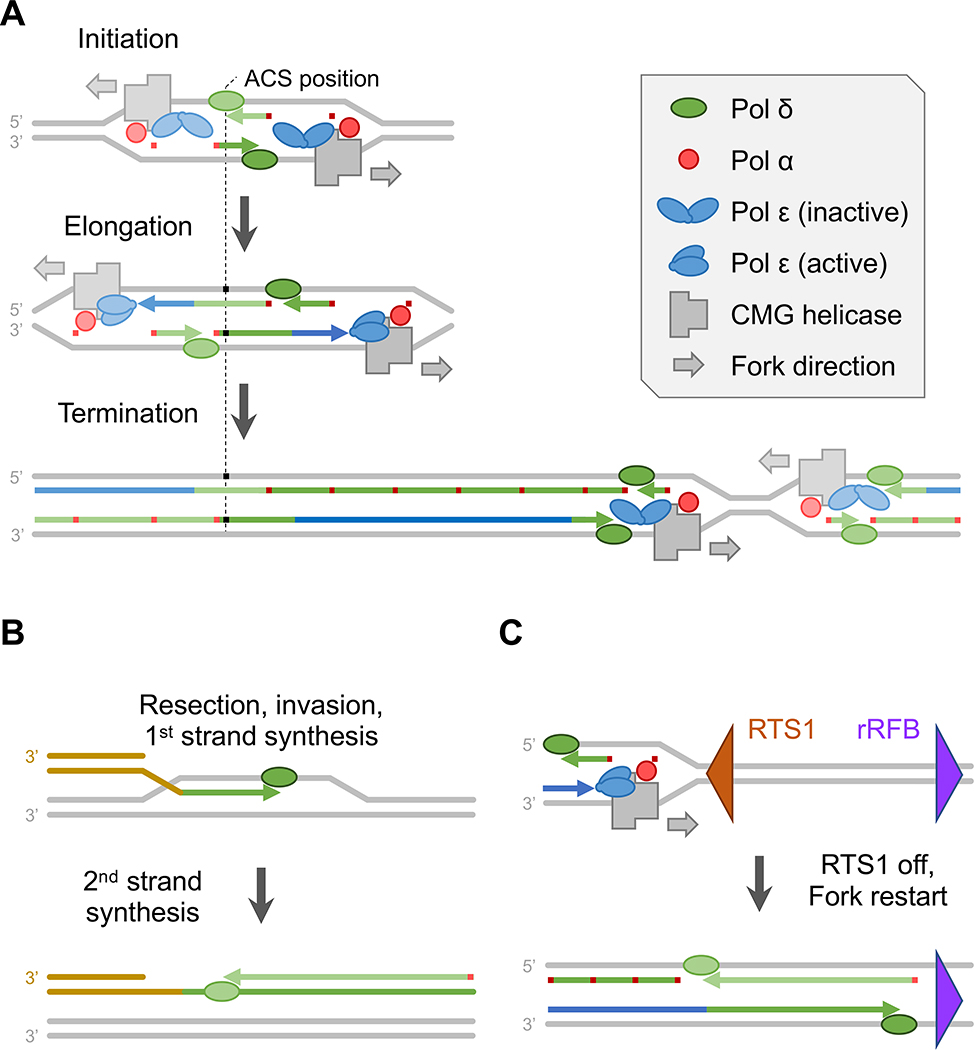
Reductions in background noise in subsequent allowed opened windows on local variations in the division of labor among the replicases (Garbacz et al. 2018; Zhou et al. 2019). Restriction digestion of genomic DNA prior to HydEn-seq sample library preparation provided consistent DNA ends to act as internal standards (Garbacz et al. 2018). This improved normalization allowed better background subtraction and quantitation. Meta-analysis of S. cerevisiae replication origins, which further reduces the effect of background noise, showed that Pol δ synthesizes both DNA strands at replication origins, suggesting that both leading and lagging strand synthesis are initiated by the first Okazaki fragment (Figure 2A) (Garbacz et al. 2018). Evidence for this model was later supported by an in vitro reconstituted system of replication (Aria and Yeeles 2018). Continued modification of the HydEn-seq protocol and the analysis pipeline, including a switch from alkaline hydrolysis to RNase H2 cleavage and measures that reduces background noise, further increased specificity and decreased noise (Zhou et al. 2019). The RHII-HydEn-seq method allowed discernment of polymerase usage down in individual regions as small as 50 bp. Thus, it was shown that nearly all active S. cerevisiae origins show an increase in the Pol δ footprint proportional to their efficiency. This provides strong support for nearly all replication initiation events in S. cerevisiae involving the mechanism where the first Okazaki fragment primes both leading and lagging strand synthesis. Similar patterns were found in S. pombe, which has more diffusive origin activities across the genome, similar to metazoans, including in human cells (Vashee et al. 2003; Dai et al. 2005; Xu et al. 2012; Tubbs et al. 2018). The RHII-HydEn-seq study also provided credible evidence suggesting that Pol δ synthesizes both strand during replication termination (Figure 2A) (Zhou et al. 2019).
Figure 2.
Polymerase usage during normal and stressed conditions. (A) Polymerase usage differs during replication initiation, elongation and termination. The first Okazaki fragments on both strands initiate leading strand synthesis. Their overlap means that Pol δ synthesize the short stretches of DNA on both strands at replication initiation sites. Pol ε and Pol δ follows the canonical division of labor for the majority of DNA replication. During termination, leading strand replication switches from Pol ε to Pol δ for the last a few Kbps. (B) Polymerase usage after homologous recombination-dependent replication restart. In S. pombe, when the replication fork is forced to stall at the RTS1 site and restart following RTS1 removal, the restarted replication predominantly uses Pol δ for synthesis of both strands. (C) Polymerase using during break-induced replication (BIR). The 3’ end of a single-ended DSB can invade the homologous sequence on the donor chromosome. DNA synthesis is forced to copy the rest of the donor chromosome. In contrast to normal DNA replication, the nascent first strand serves as the template for the second strand synthesis. Both strands are predominantly synthesized by Pol δ.
Some specialized DNA polymerases such the translesion synthesis (TLS) polymerases assist canonical DNA replication when faced with a damaged template or difficult-to-replicate region (Yang and Gao 2018). The potential role and bias of TLS polymerases in leading versus lagging strand synthesis can also be examined by rNMP mapping. HydEn-seq with an rNMP-permissive Pol η variant, showed that Pol η is primarily engaged in PCNA-dependent lagging strand synthesis (Kreisel et al. 2019). Given that Pol η can help bypass lesions on both strands in vitro (Guilliam and Yeeles 2020), the HydEn-seq in vivo data suggests that our understanding of Pol η is incomplete. Lagging strand lesions may be more accessible to Pol η, or Pol η has additional roles in lagging strand synthesis.
Most DNA repair processes require DNA synthesis as one of the last steps to fully “heal” the DNA. The roles of the major polymerases in many repair pathways are still unclear. Theoretically, rNMP mapping may help answer these questions, at least for DNA repair processes that require substantial DNA synthesis. One such process is the recombination-dependent break-induced replication (BIR), a specialized DNA double-strand break (DSB) repair pathway. BIR occurs when, following 5’-to-3’ end resection, the 3’ protruding end of a DSB invades a homologous template and primes extensive DNA synthesis (Sakofsky and Malkova 2017). Such events are studied in yeast using experimental systems wherein the centromere-proximal end of an induced DSB is homologous to one locus on another, donor chromosome. This compels DNA synthesis to proceed to the end of the donor chromosome (Donnianni and Symington 2013). HydEn-seq of purified repair products showed that Pol δ is unequivocally the main polymerase conducting DNA synthesis during BIR, with only minimal Pol α contribution (Figure 2C) (Donnianni et al. 2019). Likewise, in a recent BioRxiv manuscript, the Pu-seq approach was used to show that Pol δ is predominantly used to synthesizing both strands following homologous recombination-dependent replication restart (Figure 2C) (Naiman et al. 2020, under review, available on bioRxiv). These studies demonstrate that rNMP mapping can be used to track polymerase usage during DNA repair. It remains to be tested whether rNMP mapping has the resolution to discern polymerase usage during the small patch repair synthesis that occurs during processes such as nucleotide excision repair or base excision repair.
The biological impacts of ribonucleotides incorporated into genomic DNA during eukaryotic replication
The initial demonstration that RNase H2 can incise a single ribonucleotide in DNA (Rydberg and Game 2002) launched numerous studies of Ribonucleotide Excision Repair (RER) and the biological impacts of unrepaired genomic ribonucleotides. These include both negative and positive outcomes, and additional pathways of genomic ribonucleotide removal. The latter include exonucleolytic proofreading by the 3’−5’ exonuclease activities of Pols δ and ε, albeit inefficiently, and, in the absence of RER, incision by topoisomerase 1 (Top1) (Sekiguchi and Shuman 1997; Kim et al. 2011; Williams J. S. et al. 2013).
Negative biological consequences
Several genome instability phenotypes can result from the failure to properly remove genomic ribonucleotides. These include direct impacts on the stability of DNA, as well as the release of problematic intermediates that can be formed during ribonucleotide-processing and removal. These effects have been the subject of several recent reviews (Cerritelli S. M. and Crouch 2016; Williams J. S. et al. 2016; Klein 2017; Kellner and Luke 2020).
Structural effects on DNA.
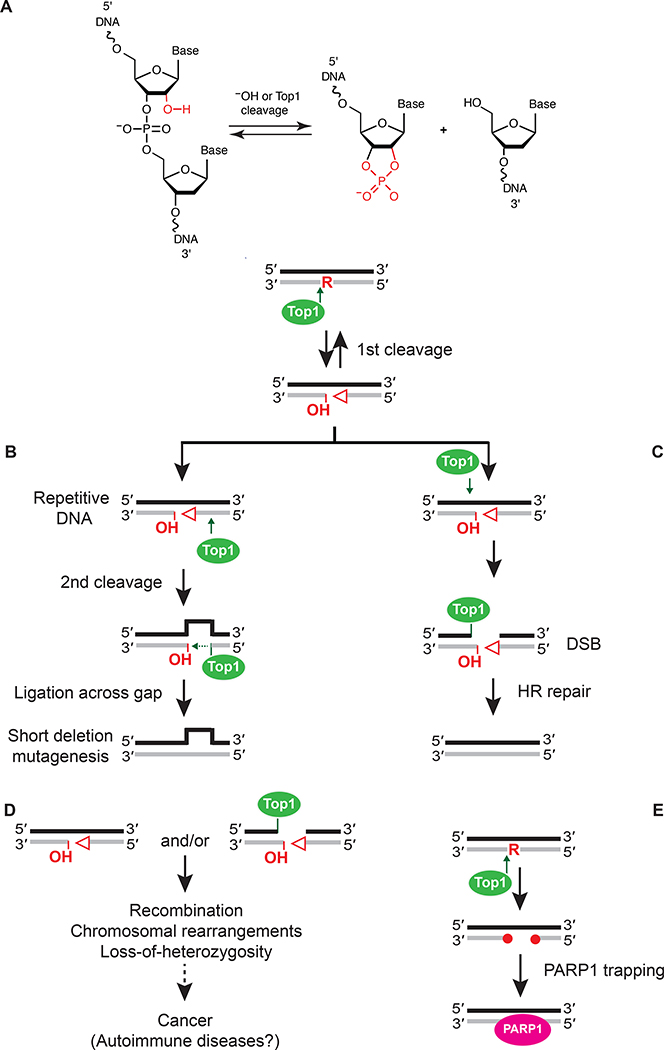
The presence of a single ribonucleotide in DNA has multiple effects on its structure. These effects have been studied in detail using a variety of approaches, including molecular dynamics simulation (Fu et al. 2019), atomic force microscopy (AFM) (Chiu et al. 2014), nuclear magnetic resonance (NMR) (DeRose et al. 2012) and X-ray crystallography (Egli et al. 1993). These investigations revealed that DNA electrostatic potential, elasticity, deoxyribose pucker, minor groove width, Watson-Crick pairing, and duplex unwinding are all impacted by the presence of a single ribonucleotide. These structural perturbations change the character of DNA and have the potential to impact DNA transactions that include replication, transcription and DNA repair. One example of this is during bypass of template ribonucleotides by DNA polymerases, a process that becomes increasingly difficult as the number of consecutive ribonucleotides increases from one to four (Watt et al. 2011; Goksenin et al. 2012; Clausen et al. 2013). Finally, at physiological pH, RNA is 100,000 times more susceptible to spontaneous hydrolysis when compared to DNA (Li Y and Breaker 1999) and therefore the presence of ribonucleotides in DNA increases the rate of formation of single-strand DNA breaks (Figure 3A).
Figure 3.
The negative physiological consequences associated with processing of ribonucleotides in DNA. (A) A single ribonucleotide in DNA can lead to formation of a single strand break (SSB) containing unligatable 5’-OH and 2’,3’-cyclic phosphate DNA ends (open triangle). This can occur following spontaneous hydrolysis or Top1 cleavage. (B) Short deletions in repeat DNA sequences are likely generated by two consecutive Top1 cleavage events, the first at the site of a ribonucleotide and the second 5’ to this initial cleavage site. This allows for strand realignment and Top1-mediated ligation across the short gap, resulting in loss of a repeat DNA sequence. (C) A DNA DSB can be directly generated following two Top1 cleavage events. If the initial Top1 cleavage event at an unrepaired ribonucleotide occurs in close proximity to a Top1 cleavage event on the opposite DNA strand, this generates a DSB that is repaired by Rad51- and Rad52-mediated HR. (D) Processing of genomic ribonucleotides results in SSBs and DSBs with unligatable DNA ends that promote various types of genome instability and may contribute to diseases such as cancer and autoimmune disorders. (E) Aberrant DNA ends produced following Top1-cleavage at unrepaired genomic ribonucleotides are recognized and bound by PARP1 to initiate DNA repair.
Ribonucleotide-dependent mutagenesis.
Much of what is currently known regarding mutations that arise during processing of ribonucleotides in DNA comes from studies performed in budding yeast. Qiu et al. first demonstrated a moderate increase in spontaneous mutation rate in a strain deleted for the gene encoding the catalytic subunit of RNase H2 (RNH201) (Qiu et al. 1999). Later frameshift reversion assays found that these mutations were primarily 4 bp deletions (Chen et al. 2000). Reversion and forward mutation reporter assays show that the primary mutations associated with loss of RNase H2 activity in yeast are short deletions (2–5 bp) in perfect or imperfect repeat sequences (Nick McElhinny, Kumar, et al. 2010; Clark et al. 2011; Kim et al. 2011), and the rate of these mutations increases in a yeast strain expressing a ribonucleotide permissive variant of Pol ε (pol2-M644G) (Nick McElhinny, Kumar, et al. 2010). These mutations are generated when Top1 cleaves at an unrepaired ribonucleotide (Sekiguchi and Shuman 1997; Kim et al. 2011). Extensive genetic and biochemical analyses demonstrate that these deletions likely result from two cleavage events by Top1, followed by strand realignment and Top1-dependent ligation across the gap (Figure 3B) (Cho et al. 2015; Huang SY et al. 2015; Sparks and Burgers 2015). In the absence of RNase H2 activity, the 2–5 bp deletion rate also depends on polymerase status. It is moderately elevated in strains with wild type polymerases or lagging strand mutator variants (pol3-L612M and pol1-L868M), but greatly elevated with a leading strand mutator polymerase (pol2-M644G) (Williams J. S. et al. 2013; Cho et al. 2015; Williams J. S. et al. 2015). This suggests that unrepaired leading strand ribonucleotides are frequent targets for Top1, possibly due to a requirement for relief of topological stress via Top1 cleavage. This Top1-induced 2–5 bp deletion mutational specificity occurs across the yeast genome (Williams Jessica S. et al. 2019).
In the absence of RNase H2 activity, Top1 cleavage at an unrepaired ribonucleotide generates aberrant 5’-OH and 2’,3’-cyclic phosphate DNA ends (Figure 3A) (Sekiguchi and Shuman 1997; Kim et al. 2011) that can be nucleolytically processed. The Srs2 helicase can unwind the 5’ end to create a flap for processing by the Exo1 nuclease (Potenski and Klein 2014). This generates a gap that can then be filled in by a DNA polymerase. The 2’,3’-cyclic phosphate and its hydrolyzed products can be removed by the Apn2 nuclease (Li F et al. 2019).
RNase H2 is also responsible for removing R-loops that arise when a nascent RNA transcript anneals with its complementary DNA template. This creates a bulky DNA structure that impedes both replication and transcription and can cause DSBs if not properly removed. RNase H2 can cleave at stretches of ribonucleotides and thereby provides a critical means of R-loop removal. In order to distinguish between mutagenic pathways, a separation-of-function variant of the gene encoding the RNase H2 catalytic subunit (RNH201 in S. cerevisiae) was engineered. This variant, Rnh201-RED (ribonucleotide excision defective), retains activity on stretches of ribonucleotides but is unable to incise at a single genomic ribonucleotide (Chon et al. 2009). The Rnh201-RED mutant has now been used in multiple model systems to demonstrate that both single unrepaired ribonucleotides and R-loops promote genome instability (reviewed in (Cerritelli S. M. and Crouch 2019)). However, it is Top1-cleavage at single ribonucleotides that initiates short deletion mutagenesis in yeast (Chon et al. 2013; Williams J. S. et al. 2017).
Replicative stress and checkpoint activation.
As discussed above, replicative stress arises during DNA polymerase bypass of ribonucleotides in template DNA (Watt et al. 2011; Goksenin et al. 2012; Clausen et al. 2013). Yeast lacking one of the RNase H2 subunits (Rnh201, Rnh202, and Rnh203 in S. cerevisiae) cannot repair genomic ribonucleotides, resulting in replication stress in a ribonucleotide permissive background (i.e. pol2-M644G). The pol2-M644G rnh201Δ strain has elevated dNTP pools (Nick McElhinny, Kumar, et al. 2010; Williams J. S. et al. 2013) and immunoblotting shows Rad53 phosphorylation (Lazzaro et al. 2012) and Rnr3 protein level increases (Williams J. S. et al. 2013), all indicators of S-phase checkpoint activation. Furthermore, the pol2-M644G rnh201Δ cells accumulate in S and G2 phases of the cell cycle (Nick McElhinny, Kumar, et al. 2010), highlighting the fact that ribonucleotides left unrepaired can impede replication and cause cellular stress. In human cells, RNase H2 silencing also causes replication stress and the accumulation of cells in S and G2, resulting in chronic activation of the post-replication repair pathway (Pizzi et al. 2015).
Chromosomal instability due to processing of single ribonucleotides in DNA.
The presence of unrepaired ribonucleotides in DNA can generate multiple types of instability, including recombination, chromosome rearrangements and chromosome loss. The first demonstration that loss of RNase H2 was associated with increased mitotic recombination in yeast came from a study of hyper-recombination (hyper-rec) mutants (Aguilera and Klein 1988). Since this initial demonstration, loss of RNase H2 activity has been shown to cause elevated gross chromosomal rearrangements (Allen-Soltero et al. 2014) and non-allelic homologous recombination (NAHR) (Conover et al. 2015). RNase H2 deficiency causes increased loss-of-heterozygosity in diploid yeast strains (Conover et al. 2015; O’Connell et al. 2015), suggesting that DSBs are formed when genomic ribonucleotides are not properly removed. DSB formation can be initiated by failure to repair R-loops or single unrepaired ribonucleotides incorporated during replication, both of which contribute to increased chromosomal instability in RNase H2-deficient strains (Cornelio et al. 2017). Unrepaired single ribonucleotides may trigger DSB formation in multiple ways. Spontaneous hydrolysis or Top1 cleavage would generate a DNA single strand break (Figure 3A) that could then be converted into a DSB. In addition, a DSB can be generated directly by a second Top1 cleavage event proximal to the initial Top1-incised ribonucleotide but on the opposite DNA strand (Figure 3C) (Huang SN et al. 2017). Consistent with Top1-dependent DSB formation in RNase H2-deficient strains, the homologous recombination (HR) repair proteins Rad51 and Rad52 are critical for cell viability in RER-deficient yeast cells (Huang SN et al. 2017). Furthermore, recombination rates are reduced upon deletion of TOP1 in RNase H2-deficient yeast strains (Potenski and Klein 2014; Conover et al. 2015).
Chromosomal instability associated with RER failure has also been observed in mouse and human cells. RNase H2-null mouse embryonic fibroblasts have increased γ-H2AX foci, substantial levels of micronuclei, and chromosomal rearrangements (Hiller et al. 2012; Reijns MA et al. 2012). Micronuclei and the DNA damage response are also observed in human cells depleted of RNase H2 (Pizzi et al. 2015).
Ribonucleotides in vertebrates: connections to human disease.
RNase H2 is essential for viability in mice (Hiller et al. 2012; Reijns MA et al. 2012). The RNase H2-RED variant also confers lethality, suggesting that removal of single ribonucleotides from genomic DNA is essential (Uehara et al. 2018). Early embryonic arrest in both variants was traced to activation of the p53-dependent DNA damage response (Reijns MA et al. 2012; Uehara et al. 2018).
There are now several lines of evidence demonstrating a connection between RNase H2 deficiency and human disease. The first was the discovery that mutations in any of the three subunits of RNase H2 can cause Aicardi-Goutieres Syndrome (AGS), a severe auto-inflammatory disorder (Crow et al. 2006). Potenski et al. (2019) modelled AGS-associated mutations in the yeast RNase H2 enzyme and found that the yeast AGS mutants have a variety of phenotypes, including a strongly reduced genome stability. Those mutant strains with the most instability had elevated levels of unrepaired ribonucleotides in their DNA (Potenski et al. 2019). Depletion of RNase H2 in human cells causes checkpoint activation and genome instability (Pizzi et al. 2015). Genomes in cells derived from AGS patients accumulate ribonucleotides and damage, suggesting that ribonucleotide removal by RNase H2 contributes to AGS etiology and is critically important for genome maintenance (Figure 3D) (Pizzi et al. 2015).
RNase H2 mutations have also been associated with Systemic Lupus Erythrematosis, another autoimmune disorder (Ramantani et al. 2010; Gunther et al. 2015). The precise link between loss of RNase H2 activity and autoimmune disorders has not been identified. However, mutations in other DNA-processing enzymes such as TREX1 and ADAR1 have also been linked to AGS (reviewed in (Crow 2015)), suggesting that it may involve improper nucleic acid processing. In addition, CRISPR screens of chemotherapy-treated human cells have identified and characterized and a molecular connection between unrepaired genomic ribonucleotides and cancer. All three RNase H2 genes were identified in a targeted screen for mutations that cause sensitivity to inhibitors of poly (ADP-ribose) polymerase (PARP) (Zimmermann et al. 2018). Top1 processing of unrepaired single ribonucleotides causes DNA lesions that trap PARP (Figure 3E). Thus, RNase H2 inactivation could harbor therapeutic potential for some cancers. In addition, a genome-wide CRISPR screen in cells treated with an inhibitor of the ATR checkpoint kinase showed that RNase H2-deficiency was synthetic lethal with ATR inhibition (Wang et al. 2019). Wang et al. also found reduced levels of RNASE H2 in prostate adenocarcinoma patient-derived samples. Together, these observations suggest that the use of ATR inhibition as a chemotherapeutic may be beneficial in cancer patients with RNase H2-deficiency. Recently, ribonucleotide removal by RNase H2 was shown to be critical for prevention of intestinal tumorigenesis in mice and as a colorectal tumor suppressor in human tumor specimens (Aden et al. 2019). RER inactivation in the mouse epidermis promotes squamous cell carcinoma (Hiller et al. 2018), thereby further highlighting the connection between ribonucleotide removal, genome instability and cancer (Figure 3D).
Positive biological consequences
In addition to the negative effects that ribonucleotides incorporated into DNA have on genome stability, there are several lines of evidence in support of positive signaling roles for ribonucleotides in DNA that include promoting DNA repair and providing important cellular signals. For example, in the active site of Pol ε, a strictly conserved methionine buttresses the steric gate tyrosine that acts to exclude ribonucleotides from polymerization. This buttressing hydrophobic amino acid is always a leucine in Pols α and δ (Lujan et al. 2013). Substitution of a leucine for the methionine in Pol ε confers higher discrimination against ribonucleotide incorporation without increasing mismatch incorporation (Nick McElhinny, Watts, et al. 2010). This suggests that ribonucleotide incorporation into leading strand DNA by Pol ε serves one or more critical cellular functions.
Signaling for strand discrimination during DNA Mismatch Repair.
One potential positive signaling function provided by leading strand ribonucleotides is during mismatch repair (MMR) of errors introduced by Pol ε during replication (Ghodgaonkar et al. 2013; Lujan et al. 2013). Nicks generated when RNase H2 cleaves at a ribonucleotide may allow an entry point for the MMR machinery to repair base-base mismatches or insertion/deletion mutations (Figure 4A). In support of this, a single ribonucleotide can initiate MMR of an adjacent mismatch in human cell extracts in a RNase H2-dependent manner (Ghodgaonkar et al. 2013). RNase H2-deficient (rnh201Δ) yeast strain shows moderate increase of mutation rates (Nick McElhinny, Kumar, et al. 2010; Ghodgaonkar et al. 2013; Lujan et al. 2013). Moreover, there is an elevated rate of MMR-dependent single base pair deletions in the pol2-M644G rnh201Δ strain containing a high density of unrepaired genomic ribonucleotides (Lujan et al. 2013).
Figure 4.
The positive consequences of ribonucleotide incorporation into DNA. (A) RNase H2 nicking at unrepaired nascent strand ribonucleotides acts as a strand-discrimination signal for MMR. (B) During NHEJ of DNA DSBs, ribonucleotides are frequently incorporated by Pol μ or TdT to promote efficient ligation by DNA Ligase 4. These incorporated ribonucleotides can later be removed during RER. (C) In S. pombe, a di-nucleotide imprint in the lagging strand is required for initiation of the mating-type switch. During replication of the mating-type locus, the imprint is suggested to consist of two consecutive ribonucleotides that are preserved after Okazaki fragment maturation. Imprint formation and preservation requires the concerted effort of several factors. The incoming fork is paused by the unknown factor X. The unknown DNA-binding factor Y is thought to protect the imprint from processing. RTS1 is a replication terminator that blocks the incoming fork from opposite direction. The imprint stalls Pol ε during leading strand synthesis in the next round of replication to promote recombination and allow a mating type switch. (D) RNase H2 nicking at ribonucleotides incorporated by Pol ε on the leading strands results in rotational freedom of the newly synthesized DNA strand and may provide relief of torsional stress.
Although this review is focused on ribonucleotides incorporated by the replicases, they are also frequently incorporated by the DNA repair polymerases (reviewed in (Vaisman and Woodgate 2018)). For example, during bypass of DNA lesions, the specialized TLS polymerases can replicate damaged DNA with reduced base and sugar selectivity. Also, during DNA DSB repair by non-homologous end joining (NHEJ), both Pol μ and TdT efficiently incorporate ribonucleotides into DNA to promote efficient ligation by DNA ligase IV (Nick McElhinny and Ramsden 2003; Ruiz et al. 2003; Brown et al. 2010; Martin et al. 2013; Moon et al. 2017; Pryor et al. 2018). This suggests that ribonucleotides incorporated by Pol μ and TdT may be preferred over deoxyribonucleotides for NHEJ, especially when dNTP concentrations are low, e.g., in non-replicating cells.
Additional cellular signaling roles for ribonucleotides.
One example of a putative positive signaling role for unrepaired genomic ribonucleotides involves the epigenetic imprint formed during mating type switching in S. pombe. During this process, an imprint is formed by two ribonucleotides left by incomplete Okazaki fragment processing at a defined location in the lagging strand (Figure 4C) (Vengrova and Dalgaard 2006). These adjacent ribonucleotides cause Pol ε to stall during the next round of replication, thereby triggering recombination between two genomic loci to facilitate a mating type switch. The possibility also exists that ribonucleotides in DNA may play a signaling role during embryonic development in higher eukaryotes.
Ribonucleotides in DNA may also signal for the relief of torsional stress in leading strand DNA (Cerritelli S. M. and Crouch 2016). The discontinuously synthesized lagging strand has free DNA ends that allow for rotation, relieving superhelical stress. In contrast, superhelical stress builds during continuous leading strand synthesis. Ribonucleotides are more abundant in the leading strand than they are in the lagging strand thanks to wild type Pol ε (Clausen et al. 2015; Daigaku et al. 2015). One reason for this may be to promote nicking by RNase H2 to relieve supercoiling (Figure 4E).
Ribonucleotide incorporation beyond animals and fungi.
Ribonucleotides are incorporated into DNA across all three kingdoms of cellular life. Like opisthokonts (animals, fungi and their close relatives), other eukaryotic lineages possess RNase H2 and synthesize their nuclear genomes with B-family replicases Pols α, δ and ε. For example, RNase H2 deficiency causes nuclear genome ribonucleotide accumulation and instability in the plant Arabidopsis thaliana (Kalhorzadeh et al. 2014). Unlike opisthokonts, most other eukaryotes do not use A-family Pol γ for mitochondrial or plastid genome synthesis. They instead they use A-family plant organellar polymerase (POP), which is more similar to bacterial DNA Polymerase I (Moriyama et al. 2011). Apicomplexans are an exception, in that their plastid genomes are replicated by a primase/helicase-fused A-family polymerase, the plastidic DNA replication/repair enzyme complex (PREX) (Seow et al. 2005). The first reported ribonucleotide mapping in a plastid-bearing eukaryote, the unicellular green alga Chlamydomonas reinhardtii, found disproportionately abundant ribonucleotides in the mitochondrial and chloroplast genomes (El-Sayed et al. 2020, under review, available on bioRxiv). Ribonucleotide ratios in these organelles implicate replication by POPs in the presence of highly imbalanced NTP pools.
Genomic ribonucleotide biology is less well defined in prokaryotes. In bacteria, different combinations of C-family polymerases are responsible for the bulk of genome synthesis (Timinskas et al. 2014), assisted by A-family Pol I and a host of repair and TLS polymerases. The C-family replicative polymerases (Yao et al. 2013) and Y-family TLS polymerases (McDonald et al. 2012; Ordonez et al. 2014) are known to incorporate ribonucleotides. In Escherichia coli, ribonucleotides are primarily removed via RER with help from Pol I (Vaisman et al. 2014), with NER as an efficient and apparently accurate backup mechanism (Vaisman et al. 2013). Pol I can incorporate ribonucleotides in vitro (Ide et al. 1993; Astatke et al. 1998). In Bacillus subtilis, loss of RER is mutagenic (Yao et al. 2013), with a spectrum that implicates error-prone re-synthesis by the C-family DnaE replicative polymerase after NER (Schroeder et al. 2017). In Archaea, DNA replication systems vary widely, using different combinations of D and B-family polymerases (Makarova et al. 2014). Examples from both groups can incorporate ribonucleotides at rates similar to the eukaryotic replicases (Gardner et al. 2004; Schermerhorn and Gardner 2015), which can be removed by RER (Heider et al. 2017). Though the details differ between kingdoms, ribonucleotide incorporation and RER appear to be conserved across cellular life.
Conclusions and perspectives
Tremendous progress has been made regarding the when, where and how of genomic ribonucleotide incorporation. It is now safe to state that rNMPs can be incorporated by any DNA polymerase during any DNA synthesis. The rate at which rNMPs are incorporated is partly determined by the nucleotide pool imbalance, the extent of which is regulated by cell cycle and DNA damage and replication checkpoint status via ribonucleotide reductase. DNA polymerases are also vastly different in their ability to discriminate against ribonucleotide incorporation. Pols ε and δ incorporate slightly less than one ribonucleotide per kilobase. Some specialized repair polymerases, such as Pols η and μ, incorporate ribonucleotides into DNA at extremely high rates. In fact, Pol μ behaves almost like an RNA polymerase. Thus, the question of why ribonucleotides are so abundantly incorporated into DNA remains. In this review, we discussed evidence of positive roles for ribonucleotides in mating type switching in S. pombe, mismatch repair, and non-homologous end joining. Widespread genomic ribonucleotides may influence nucleic acid transactions such as gene expression, nucleosome and histone marks, DNA repair, chromosome architecture and DNA mutagenesis. In contrast to the transient majority of rNMPs in the nuclear genome, embedded rNMPs are not likely removed from mitochondrial DNA (Wanrooij et al. 2017). This is particularly interesting, as the 2’-hydroxyl group makes ribonucleotides naturally less stable. It is tempting to speculate that the lack of selection against ribonucleotides in the mitochondrial genome suggests a positive role for their presence. In both nuclear and mitochondrial genomes, rNMP incorporation is not uniform. There are biases in the identity of the embedded rNMP, locus of incorporation, and in the surrounding sequence context. The biological significance of these biases remains to be determined. The use of embedded rNMPs as biomarkers for DNA polymerase action has been fruitful. Their abundance enables high resolution mapping and provides a means of examining polymerase roles in any processes that involves DNA synthesis. It remains to be seen if current techniques have sufficient resolution to detect rare events like short DNA synthesis tracts from repair events in individual cells. Genetic tools for studying the vast number of specialized polymerases will also be needed.
Acknowledgements
We thank all Kunkel lab members for thoughtful discussions.
Declaration of Interest
This work was supported by Project Z01 ES065070 to T.A.K. from the Division of Intramural Research of the NIH, NIEH.
References
- Aden K, Bartsch K, Dahl J, Reijns MAM, Esser D, Sheibani-Tezerji R, Sinha A, Wottawa F, Ito G, Mishra N et al. 2019. Epithelial RNase H2 Maintains Genome Integrity and Prevents Intestinal Tumorigenesis in Mice. Gastroenterology. 156(1):145–159 e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Klein HL. 1988. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 119(4):779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen-Soltero S, Martinez SL, Putnam CD, Kolodner RD. 2014. A saccharomyces cerevisiae RNase H2 interaction network functions to suppress genome instability. Mol Cell Biol. 34(8):1521–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aria V, Yeeles JTP. 2018. Mechanism of bidirectional leading-strand synthesis establishment at eukaryotic DNA replication origins. Mol Cell. 73(2):199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astatke M, Ng K, Grindley ND, Joyce CM. 1998. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc Natl Acad Sci U S A. 95(7):3402–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachander S, Gombolay AL, Yang T, Xu P, Newnam G, Keskin H, El-Sayed WMM, Bryksin AV, Tao S, Bowen NE et al. 2020. Ribonucleotide incorporation in yeast genomic DNA shows preference for cytosine and guanosine preceded by deoxyadenosine. Nat Commun. 11(1):2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund AK, Navarrete C, Engqvist MK, Hoberg E, Szilagyi Z, Taylor RW, Gustafsson CM, Falkenberg M, Clausen AR. 2017. Nucleotide pools dictate the identity and frequency of ribonucleotide incorporation in mitochondrial DNA. PLoS genetics. 13(2):e1006628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Fiala KA, Fowler JD, Sherrer SM, Newmister SA, Duym WW, Suo Z. 2010. A novel mechanism of sugar selection utilized by a human X-family DNA polymerase. J Mol Biol. 395(2):282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Suo Z. 2011. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 50(7):1135–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, Crouch RJ. 2016. The balancing act of ribonucleotides in DNA. Trends Biochem Sci. 41(5):434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, Crouch RJ. 2019. RNase H2-RED carpets the path to eukaryotic RNase H2 functions. DNA Repair (Amst). 84:102736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ. 2003. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell. 11(3):807–815. [DOI] [PubMed] [Google Scholar]
- Chen JZ, Qiu J, Shen B, Holmquist GP. 2000. Mutational spectrum analysis of RNase H(35) deficient Saccharomyces cerevisiae using fluorescence-based directed termination PCR. Nucleic Acids Res. 28(18):3649–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu HC, Koh KD, Evich M, Lesiak AL, Germann MW, Bongiorno A, Riedo E, Storici F. 2014. RNA intrusions change DNA elastic properties and structure. Nanoscale. 6(17):10009–10017. [DOI] [PubMed] [Google Scholar]
- Cho JE, Kim N, Jinks-Robertson S. 2015. Topoisomerase 1-dependent deletions initiated by incision at ribonucleotides are biased to the non-transcribed strand of a highly activated reporter. Nucleic Acids Res. 43(19):9306–9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H, Sparks JL, Rychlik M, Nowotny M, Burgers PM, Crouch RJ, Cerritelli SM. 2013. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 41(5):3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H, Vassilev A, DePamphilis ML, Zhao Y, Zhang J, Burgers PM, Crouch RJ, Cerritelli SM. 2009. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res. 37(1):96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AB, Lujan SA, Kissling GE, Kunkel TA. 2011. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase epsilon. DNA Repair (Amst). 10(5):476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen AR, Lujan SA, Burkholder AB, Orebaugh CD, Williams JS, Clausen MF, Malc EP, Mieczkowski PA, Fargo DC, Smith DJ et al. 2015. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struct Mol Biol. 22(3):185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen AR, Zhang S, Burgers PM, Lee MY, Kunkel TA. 2013. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase delta. DNA Repair (Amst). 12(2):121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover HN, Lujan SA, Chapman MJ, Cornelio DA, Sharif R, Williams JS, Clark AB, Camilo F, Kunkel TA, Argueso JL. 2015. Stimulation of Chromosomal Rearrangements by Ribonucleotides. Genetics. 201(3):951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelio DA, Sedam HN, Ferrarezi JA, Sampaio NM, Argueso JL. 2017. Both R-loop removal and ribonucleotide excision repair activities of RNase H2 contribute substantially to chromosome stability. DNA Repair (Amst). 52:110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ. 2015. Type I interferonopathies: mendelian type I interferon up-regulation. Curr Opin Immunol. 32:7–12. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R et al. 2006. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 38(8):910–916. [DOI] [PubMed] [Google Scholar]
- Dai J, Chuang RY, Kelly TJ. 2005. DNA replication origins in the Schizosaccharomyces pombe genome. Proc Natl Acad Sci U S A. 102(2):337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigaku Y, Keszthelyi A, Muller CA, Miyabe I, Brooks T, Retkute R, Hubank M, Nieduszynski CA, Carr AM. 2015. A global profile of replicative polymerase usage. Nat Struct Mol Biol. 22(3):192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLucia AM, Grindley ND, Joyce CM. 2003. An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a ‘steric gate’ residue for discrimination against ribonucleotides. Nucleic Acids Res. 31(14):4129–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose EF, Perera L, Murray MS, Kunkel TA, London RE. 2012. Solution structure of the Dickerson DNA dodecamer containing a single ribonucleotide. Biochemistry. 51(12):2407–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Talavera A, Calvo PA, Gonzalez-Acosta D, Diaz M, Sastre-Moreno G, Blanco-Franco L, Guerra S, Martinez-Jimenez MI, Mendez J, Blanco L. 2019. A cancer-associated point mutation disables the steric gate of human PrimPol. Sci Rep. 9(1):1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donigan KA, Cerritelli SM, McDonald JP, Vaisman A, Crouch RJ, Woodgate R. 2015. Unlocking the steric gate of DNA polymerase eta leads to increased genomic instability in Saccharomyces cerevisiae. DNA Repair (Amst). 35:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donigan KA, McLenigan MP, Yang W, Goodman MF, Woodgate R. 2014. The steric gate of DNA polymerase iota regulates ribonucleotide incorporation and deoxyribonucleotide fidelity. J Biol Chem. 289(13):9136–9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnianni RA, Symington LS. 2013. Break-induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci U S A. 110(33):13475–13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnianni RA, Zhou ZX, Lujan SA, Al-Zain A, Garcia V, Glancy E, Burkholder AB, Kunkel TA, Symington LS. 2019. DNA polymerase delta synthesizes both strands during break-induced replication. Mol Cell. 76(3):371–381 e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn K, Griffith JD. 1980. The presence of RNA in a double helix inhibits its interaction with histone protein. Nucleic Acids Res. 8(3):555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli M, Usman N, Rich A. 1993. Conformational influence of the ribose 2’-hydroxyl group: crystal structures of DNA-RNA chimeric duplexes. Biochemistry. 32(13):3221–3237. [PubMed] [Google Scholar]
- Fu I, Smith DJ, Broyde S. 2019. Rotational and translational positions determine the structural and dynamic impact of a single ribonucleotide incorporated in the nucleosome. DNA Repair (Amst). 73:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbacz MA, Lujan SA, Burkholder AB, Cox PB, Wu Q, Zhou ZX, Haber JE, Kunkel TA. 2018. Evidence that DNA polymerase delta contributes to initiating leading strand DNA replication in Saccharomyces cerevisiae. Nat Commun. 9(1):858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner AF, Joyce CM, Jack WE. 2004. Comparative kinetics of nucleotide analog incorporation by vent DNA polymerase. J Biol Chem. 279(12):11834–11842. [DOI] [PubMed] [Google Scholar]
- Ghodgaonkar MM, Lazzaro F, Olivera-Pimentel M, Artola-Boran M, Cejka P, Reijns MA, Jackson AP, Plevani P, Muzi-Falconi M, Jiricny J. 2013. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol Cell. 50(3):323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goksenin AY, Zahurancik W, LeCompte KG, Taggart DJ, Suo Z, Pursell ZF. 2012. Human DNA polymerase epsilon is able to efficiently extend from multiple consecutive ribonucleotides. J Biol Chem. 287(51):42675–42684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombolay AL, Vannberg FO, Storici F. 2019. Ribose-Map: a bioinformatics toolkit to map ribonucleotides embedded in genomic DNA. Nucleic Acids Res. 47(1):e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman LI, Watson R, Vinograd J. 1973. The presence of ribonucleotides in mature closed-circular mitochondrial DNA. Proc Natl Acad Sci U S A. 70(12):3339–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliam TA, Yeeles JTP. 2020. Reconstitution of translesion synthesis reveals a mechanism of eukaryotic DNA replication restart. Nat Struct Mol Biol. 27(5):450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther C, Kind B, Reijns MA, Berndt N, Martinez-Bueno M, Wolf C, Tungler V, Chara O, Lee YA, Hubner N et al. 2015. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J Clin Invest. 125(1):413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider MR, Burkhart BW, Santangelo TJ, Gardner AF. 2017. Defining the RNaseH2 enzyme-initiated ribonucleotide excision repair pathway in Archaea. J Biol Chem. 292(21):8835–8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller B, Achleitner M, Glage S, Naumann R, Behrendt R, Roers A. 2012. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J Exp Med. 209(8):1419–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller B, Hoppe A, Haase C, Hiller C, Schubert N, Muller W, Reijns MAM, Jackson AP, Kunkel TA, Wenzel J et al. 2018. Ribonucleotide excision repair is essential to prevent squamous cell carcinoma of the skin. Cancer Res. 78(20):5917–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes JB, Akman G, Wood SR, Sakhuja K, Cerritelli SM, Moss C, Bowmaker MR, Jacobs HT, Crouch RJ, Holt IJ. 2015. Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication. Proc Natl Acad Sci U S A. 112(30):9334–9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovatter KR, Martinson HG. 1987. Ribonucleotide-induced helical alteration in DNA prevents nucleosome formation. Proc Natl Acad Sci U S A. 84(5):1162–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SN, Williams JS, Arana ME, Kunkel TA, Pommier Y. 2017. Topoisomerase I-mediated cleavage at unrepaired ribonucleotides generates DNA double-strand breaks. EMBO J. 36(3):361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SY, Ghosh S, Pommier Y. 2015. Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites. J Biol Chem. 290(22):14068–14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide H, Yagi R, Yamaoka T, Kimura Y. 1993. Misincorporation of ribonucleotides by DNA polymerase during in vitro DNA replication. Nucleic Acids Symp Ser.(29):133–134. [PubMed] [Google Scholar]
- Joyce CM. 1997. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc Natl Acad Sci U S A. 94(5):1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhorzadeh P, Hu Z, Cools T, Amiard S, Willing EM, De Winne N, Gevaert K, De Jaeger G, Schneeberger K, White CI et al. 2014. Arabidopsis thaliana RNase H2 deficiency counteracts the needs for the WEE1 checkpoint kinase but triggers genome instability. Plant Cell. 26(9):3680–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner V, Luke B. 2020. Molecular and physiological consequences of faulty eukaryotic ribonucleotide excision repair. The EMBO journal. 39(3):e102309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. 2011. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 332(6037):1561–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL. 2017. Genome instabilities arising from ribonucleotides in DNA. DNA Repair (Amst). 56:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KD, Balachander S, Hesselberth JR, Storici F. 2015. Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. Nat Methods. 12(3):251–257, 253 p following 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisel K, Engqvist MKM, Kalm J, Thompson LJ, Bostrom M, Navarrete C, McDonald JP, Larsson E, Woodgate R, Clausen AR. 2019. DNA polymerase eta contributes to genome-wide lagging strand synthesis. Nucleic acids research. 47(5):2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. 2004. DNA replication fidelity. The Journal of biological chemistry. 279(17):16895–16898. [DOI] [PubMed] [Google Scholar]
- Larrea AA, Lujan SA, Nick McElhinny SA, Mieczkowski PA, Resnick MA, Gordenin DA, Kunkel TA. 2010. Genome-wide model for the normal eukaryotic DNA replication fork. Proc Natl Acad Sci U S A. 107(41):17674–17679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro F, Novarina D, Amara F, Watt DL, Stone JE, Costanzo V, Burgers PM, Kunkel TA, Plevani P, Muzi-Falconi M. 2012. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol Cell. 45(1):99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang Q, Seol JH, Che J, Lu X, Shim EY, Lee SE, Niu H. 2019. Apn2 resolves blocked 3’ ends and suppresses Top1-induced mutagenesis at genomic rNMP sites. Nat Struct Mol Biol. 26(3):155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Breaker RR. 1999. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2’-hydroxyl group. J Am Chem Soc. 121(23):5364–5372. [Google Scholar]
- Lujan SA, Clausen AR, Clark AB, MacAlpine HK, MacAlpine DM, Malc EP, Mieczkowski PA, Burkholder AB, Fargo DC, Gordenin DA et al. 2014. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 24(11):1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. 2013. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol Cell. 50(3):437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G, Villani G, Tillement V, Stucki M, Locatelli GA, Frouin I, Spadari S, Hubscher U. 2001. Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase delta by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1. Proc Natl Acad Sci U S A. 98(25):14298–14303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Krupovic M, Koonin EV. 2014. Evolution of replicative DNA polymerases in archaea and their contributions to the eukaryotic replication machinery. Front Microbiol. 5:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MJ, Garcia-Ortiz MV, Gomez-Bedoya A, Esteban V, Guerra S, Blanco L. 2013. A specific N-terminal extension of the 8 kDa domain is required for DNA end-bridging by human Polmu and Pollambda. Nucleic Acids Res. 41(19):9105–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch SD, Kunkel TA. 2008. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 18(1):148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Vaisman A, Kuban W, Goodman MF, Woodgate R. 2012. Mechanisms employed by Escherichia coli to prevent ribonucleotide incorporation into genomic DNA by Pol V. PLoS Genet. 8(11):e1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabe I, Kunkel TA, Carr AM. 2011. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 7(12):e1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AF, Pryor JM, Ramsden DA, Kunkel TA, Bebenek K, Pedersen LC. 2017. Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu. Nucleic Acids Res. 45(15):9138–9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama T, Terasawa K, Sato N. 2011. Conservation of POPs, the plant organellar DNA polymerases, in eukaryotes. Protist. 162(1):177–187. [DOI] [PubMed] [Google Scholar]
- Moss CF, Dalla Rosa I, Hunt LE, Yasukawa T, Young R, Jones AWE, Reddy K, Desai R, Virtue S, Elgar G et al. 2017. Aberrant ribonucleotide incorporation and multiple deletions in mitochondrial DNA of the murine MPV17 disease model. Nucleic Acids Res. 45(22):12808–12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevin P, Engen JR, Beuning PJ. 2015. Steric gate residues of Y-family DNA polymerases DinB and pol kappa are crucial for dNTP-induced conformational change. DNA Repair (Amst). 29:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. 2008. Division of labor at the eukaryotic replication fork. Mol Cell. 30(2):137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. 2010. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 6(10):774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Ramsden DA. 2003. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol Cell Biol. 23(7):2309–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. 2010. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 107(11):4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell K, Jinks-Robertson S, Petes TD. 2015. Elevated genome-wide instability in yeast mutants lacking RNase H activity. Genetics. 201(3):963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez H, Uson ML, Shuman S. 2014. Characterization of three mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation. Nucleic Acids Res. 42(17):11056–11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzi S, Sertic S, Orcesi S, Cereda C, Bianchi M, Jackson AP, Lazzaro F, Plevani P, Muzi-Falconi M. 2015. Reduction of hRNase H2 activity in Aicardi-Goutieres syndrome cells leads to replication stress and genome instability. Hum Mol Genet. 24(3):649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potenski CJ, Epshtein A, Bianco C, Klein HL. 2019. Genome instability consequences of RNase H2 Aicardi-Goutieres syndrome alleles. DNA Repair (Amst). 84:102614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potenski CJ, Klein HL. 2014. How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res. 42(16):10226–10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor JM, Conlin MP, Carvajal-Garcia J, Luedeman ME, Luthman AJ, Small GW, Ramsden DA. 2018. Ribonucleotide incorporation enables repair of chromosome breaks by nonhomologous end joining. Science. 361(6407):1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. 2007. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 317(5834):127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Qian Y, Frank P, Wintersberger U, Shen B. 1999. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol Cell Biol. 19(12):8361–8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramantani G, Kohlhase J, Hertzberg C, Innes AM, Engel K, Hunger S, Borozdin W, Mah JK, Ungerath K, Walkenhorst H et al. 2010. Expanding the phenotypic spectrum of lupus erythematosus in Aicardi-Goutieres syndrome. Arthritis Rheum. 62(5):1469–1477. [DOI] [PubMed] [Google Scholar]
- Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F et al. 2012. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 149(5):1008–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MAM, Kemp H, Ding J, de Proce SM, Jackson AP, Taylor MS. 2015. Lagging-strand replication shapes the mutational landscape of the genome. Nature. 518(7540):502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JF, Juarez R, Garcia-Diaz M, Terrados G, Picher AJ, Gonzalez-Barrera S, Fernandez de Henestrosa AR, Blanco L. 2003. Lack of sugar discrimination by human Pol mu requires a single glycine residue. Nucleic Acids Res. 31(15):4441–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B, Game J. 2002. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc Natl Acad Sci U S A. 99(26):16654–16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakofsky CJ, Malkova A. 2017. Break induced replication in eukaryotes: mechanisms, functions, and consequences. Crit Rev Biochem Mol Biol. 52(4):395–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermerhorn KM, Gardner AF. 2015. Pre-steady-state kinetic analysis of a family D DNA polymerase from Thermococcus sp. 9 degrees N reveals mechanisms for Archaeal genomic replication and maintenance. J Biol Chem. 290(36):21800–21810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JW, Randall JR, Hirst WG, O’Donnell ME, Simmons LA. 2017. Mutagenic cost of ribonucleotides in bacterial DNA. Proc Natl Acad Sci U S A. 114(44):11733–11738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi J, Shuman S. 1997. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol Cell. 1(1):89–97. [DOI] [PubMed] [Google Scholar]
- Seow F, Sato S, Janssen CS, Riehle MO, Mukhopadhyay A, Phillips RS, Wilson RJ, Barrett MP. 2005. The plastidic DNA replication enzyme complex of Plasmodium falciparum. Mol Biochem Parasitol. 141(2):145–153. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Whitehouse I. 2012. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 483(7390):434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks JL, Burgers PM. 2015. Error-free and mutagenic processing of topoisomerase 1-provoked damage at genomic ribonucleotides. EMBO J. 34(9):1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinskas K, Balvociute M, Timinskas A, Venclovas C. 2014. Comprehensive analysis of DNA polymerase III alpha subunits and their homologs in bacterial genomes. Nucleic Acids Res. 42(3):1393–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs A, Sridharan S, van Wietmarschen N, Maman Y, Callen E, Stanlie A, Wu W, Wu X, Day A, Wong N et al. 2018. Dual roles of poly(dA:dT) tracts in replication initiation and fork collapse. Cell. 174(5):1127–1142 e1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Cerritelli SM, Hasin N, Sakhuja K, London M, Iranzo J, Chon H, Grinberg A, Crouch RJ. 2018. Two RNase H2 mutants with differential rNMP processing activity reveal a threshold of ribonucleotide tolerance for embryonic development. Cell Rep. 25(5):1135–1145 e1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, Kuban W, McDonald JP, Karata K, Yang W, Goodman MF, Woodgate R. 2012. Critical amino acids in Escherichia coli UmuC responsible for sugar discrimination and base-substitution fidelity. Nucleic Acids Res. 40(13):6144–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, McDonald JP, Huston D, Kuban W, Liu L, Van Houten B, Woodgate R. 2013. Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair. PLoS Genet. 9(11):e1003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, McDonald JP, Noll S, Huston D, Loeb G, Goodman MF, Woodgate R. 2014. Investigating the mechanisms of ribonucleotide excision repair in Escherichia coli. Mutat Res. 761:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, Woodgate R. 2018. Ribonucleotide discrimination by translesion synthesis DNA polymerases. Crit Rev Biochem Mol Biol. 53(4):382–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC. 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 17(15):1894–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengrova S, Dalgaard JZ. 2006. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 7(1):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E, Henrikus SS, Vaisman A, Makiela-Dzbenska K, Armstrong TJ, Lazowski K, McDonald JP, Goodman MF, van Oijen AM, Jonczyk P et al. 2019. Role of RNase H enzymes in maintaining genome stability in Escherichia coli expressing a steric-gate mutant of pol VICE391. DNA Repair (Amst). 84:102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wang G, Feng X, Shepherd P, Zhang J, Tang M, Chen Z, Srivastava M, McLaughlin ME, Navone NM et al. 2019. Genome-wide CRISPR screens reveal synthetic lethality of RNASEH2 deficiency and ATR inhibition. Oncogene. 38(14):2451–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanrooij PH, Engqvist MKM, Forslund JME, Navarrete C, Nilsson AK, Sedman J, Wanrooij S, Clausen AR, Chabes A. 2017. Ribonucleotides incorporated by the yeast mitochondrial DNA polymerase are not repaired. Proc Natl Acad Sci U S A. 114(47):12466–12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt DL, Johansson E, Burgers PM, Kunkel TA. 2011. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair (Amst). 10(8):897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Clausen AR, Lujan SA, Marjavaara L, Clark AB, Burgers PM, Chabes A, Kunkel TA. 2015. Evidence that processing of ribonucleotides in DNA by topoisomerase 1 is leading-strand specific. Nat Struct Mol Biol. 22(4):291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Gehle DB, Kunkel TA. 2017. The role of RNase H2 in processing ribonucleotides incorporated during DNA replication. DNA Repair (Amst). 53:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Lujan SA, Kunkel TA. 2016. Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat Rev Mol Cell Biol. 17(6):350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Lujan SA, Zhou Z-X, Burkholder AB, Clark AB, Fargo DC, Kunkel TA. 2019. Genome-wide mutagenesis resulting from topoisomerase 1-processing of unrepaired ribonucleotides in DNA. DNA Repair. 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Smith DJ, Marjavaara L, Lujan SA, Chabes A, Kunkel TA. 2013. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol Cell. 49(5):1010–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yanagisawa Y, Tsankov AM, Hart C, Aoki K, Kommajosyula N, Steinmann KE, Bochicchio J, Russ C, Regev A et al. 2012. Genome-wide identification and characterization of replication origins by deep sequencing. Genome Biol. 13(4):R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Gao Y. 2018. Translesion and repair DNA polymerases: diverse structure and mechanism. Annu Rev Biochem. 87:239–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao NY, Schroeder JW, Yurieva O, Simmons LA, O’Donnell ME. 2013. Cost of rNTP/dNTP pool imbalance at the replication fork. Proc Natl Acad Sci U S A. 110(32):12942–12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatopek KM, Alpaslan E, Evans TC, Sauguet L, Gardner AF. 2020. Novel ribonucleotide discrimination in the RNA polymerase-like two-barrel catalytic core of Family D DNA polymerases. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZX, Lujan SA, Burkholder AB, Garbacz MA, Kunkel TA. 2019. Roles for DNA polymerase delta in initiating and terminating leading strand DNA replication. Nat Commun. 10(1):3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Murina O, Reijns MAM, Agathanggelou A, Challis R, Tarnauskaite Z, Muir M, Fluteau A, Aregger M, McEwan A et al. 2018. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature. 559(7713):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]