Abstract
Exogenous mechanical cues are transmitted from the extracellular matrix to the nuclear envelope (NE), where mechanical stress on the NE mediates shuttling of transcription factors and other signaling cascades that dictate downstream cellular behavior and fate decisions. To systematically study how nuclear morphology can change across various physiologic microenvironmental contexts, we cultured mesenchymal progenitor cells (MSCs) in engineered 2D and 3D hyaluronic acid hydrogel systems. Across multiple contexts we observed highly ‘wrinkled’ nuclear envelopes, and subsequently developed a quantitative single-cell imaging metric to better evaluate how wrinkles in the nuclear envelope relate to progenitor cell mechanotransduction. We determined that in soft 2D environments the NE is predominately wrinkled, and that increases in cellular mechanosensing (indicated by cellular spreading, adhesion complex growth, and nuclear localization of YAP/TAZ) occurred only in absence of nuclear envelope wrinkling. Conversely, in 3D hydrogel and tissue contexts, we found NE wrinkling occurred along with increased YAP/TAZ nuclear localization. We further determined that these NE wrinkles in 3D were largely generated by actin impingement, and compared to other nuclear morphometrics, the degree of nuclear wrinkling showed the greatest correlation with nuclear YAP/TAZ localization. These findings suggest that the degree of nuclear envelope wrinkling can predict mechanotransduction state in mesenchymal progenitor cells and highlights the differential mechanisms of NE stress generation operative in 2D and 3D microenvironmental contexts.
Keywords: Nuclear envelope, hydrogels, lamina, YAP/TAZ, mechanotransduction, super resolution imaging
INTRODUCTION
The micromechanical niche of the cell modulates phenotype and function, from proliferation and extracellular matrix deposition to downstream lineage specification 1–4. During these processes, exogenous forces are transmitted through the cytoskeleton, where they converge at the nucleus and are converted to changes in gene expression through a variety of mechanisms2, 5–7. One well-described mechanotransductive mechanism involves the force-induced translocation of proteins into the nucleus, where they initiate downstream transcriptional programs. This paradigm has been best described for the YAP/TAZ protein complex, which functions as a transcriptional co-activator that initiates gene expression patterns that define both development and disease2, 8. Outside of YAP/TAZ, similar force-sensitive nuclear translocation mechanisms have been reported for other factors, including RARγ6, MRTF-A (also known as Mkl1)5, 9, HDAC310, β-catenin11, TWIST12, and NKX2.513. For some of these factors, the mechanisms driving force-sensitive nuclear translocation are well described, such as the role of G-actin binding to MRTF-A causing its sequestration in the cytosol5, while for others, the mechanism governing force-induced transcriptional co-activator translocation has not yet been fully elucidated.
The nuclear envelope is itself a mechano-responsive element of the cell, and may regulate force-induced nuclear translocation of transcription factors 6, 14, 15. Lamin-A/C, an intermediate filament that forms an extensive network at the inner nuclear envelope, functions to maintain nuclear morphology, organization, and stiffness 16–18. Lamin-A/C is regulated at the protein level, with the ratio of lamin-A/C to lamin-B1/B2 scaling with the stiffness of the cellular niche. This expression-based regulation is enabled by a feedback circuit, wherein mechanical force causes RARγ nuclear translocation and binding to the promoter of lamin-A/C, increasing its expression6. Upregulation of lamin A/C promotes its incorporation into the nuclear envelope in a force-dependent fashion. Persistent mechanical stimulation of progenitor cells, in the form of increased substrate stiffness or dynamic mechanical loading, directs accumulation of lamin-A/C at the nuclear periphery, thereby stiffening the nucleus and sensitizing the cell to additional exogenous mechanical inputs19. This force-induced assembly of lamin-A/C results from structural polarization of the lamina, where cryptic lamin-A/C domains become exposed or buried as a consequence of force generation within the cell 20, 21. These cryptic residues within lamin-A/C contain binding sites for other inner nuclear proteins, including emerin, which is phosphorylated in a mechano-responsive fashion and is key for mechano-adaptation and nuclear stiffening of isolated nuclei with applied force 14. Besides emerin, lamin-A/C can also act as an anchoring scaffold to other proteins within the nuclear envelope, allowing for the transmission of mechanical force to complexed proteins. In addition to regulating nuclear mechanics, mechanical stress in the NE can control mechano-signaling directly. Recent work shows that mechanical engagement of the nuclear envelope (i.e., direct compression via atomic force microscopy, “AFM”) can elicit a mechano-signaling response. Mechanical stretch of the NE increased the permissivity of nuclear pore complexes, resulting in increased nuclear import of YAP/TAZ, independent of the polymerization state of the actin cytoskeleton22. These findings suggest that the nuclear envelope (NE) is a key mechano-responsive signaling hub, providing structural support for the nucleus while also playing key roles in mechano-adaptation that ultimately dictate how extracellular forces are sensed and transmitted.
Despite this important role in cellular mechanotransduction, how the morphology and function of the nuclear envelope varies in response to inputs from the microenvironment is not well defined. Commonly, the nucleus is represented as an ellipsoidal structure, with a radius of curvature that varies slowly along its perimeter. However, this ellipsoidal morphology is not present in every cell type and microenvironmental context. Rather, recent work across a variety of biomaterial platforms has illustrated several scenarios in which the nuclear envelope can show a markedly “wrinkled” morphology, including on soft substrates 6, 23,21, 24, after treatment with pharmacologic agents that alter nuclear structure or cytoskeletal contractility25, during cellular attachment and detachment 20, 21, 25–27, in breast tissue and engineered acini28, during confined migration29, 30, and following lamin-A/C knockdown 27, 31. Super resolution and electron microscopy of cells cultured on 2D glass substrates has further shown that various elements of the cytoskeleton can physically occupy these invaginations, though it is unclear if these nuclear invaginations are caused by cytoskeletal interactions or if these cytoskeletal elements are simply anchored to the nuclear envelope and deform along with the nucleus 28, 32. To date, most observations of a wrinkled nuclear morphology have been qualitative in nature, and there is a clear gap of knowledge with respect to understanding how wrinkled nuclear envelope conformations arise in response to exogenous cues, and critically, how the wrinkled nuclear morphology may regulate progenitor cell behavior and mechano-adaptation. Previous studies have proposed that the wrinkled nuclear lamina is unfurled during cellular spreading, and bulk nuclear flattening reaches steady state only when the wrinkled lamina becomes maximally-unfurled26, 27, 33. Here, we systematically examined the functionality of this nuclear envelope morphology and correlated this feature with progenitor cell mechano-responses across a variety of microenvironmental contexts using engineered hyaluronic acid hydrogel systems.
To accomplish this, we first developed quantitative metrics to define the degree of nuclear envelope wrinkling and used these tools to assess the morphological state of the nuclear envelope in an unbiased fashion. Short-term perturbations of cellular contractility in MSCs cultured on soft 2D hydrogels rapidly altered nuclear envelope morphology. When coupling single cell wrinkling metrics with the visualization of mechano-active transcription factor localization, strong correlations were observed between wrinkling state and accumulation of these factors in the nucleus. These data suggest that wrinkling engenders laxity in the nuclear envelope and results in a ‘toe-region’ in cellular mechano-sensing, wherein the nuclear envelope must be pulled taut prior to mechanically active nuclear shuttling of transcriptional activators. We further probed this relationship in 3D microenvironments and showed that the wrinkled morphology of the nuclear envelope can regulate these same responses, albeit in a different manner. In these 3D environments, increased cytoskeletal tension and mechano-sensitive signaling in MMP-degradable hydrogels correlated with increased nuclear envelope wrinkling. These wrinkles arose from impingements of the contractile actin cytoskeleton on the nucleus, in both engineered and native 3D environments. Accordingly, nuclear envelope wrinkling was the only significant predictor of mechano-sensitive transcription factor shuttling in 3D as compared to traditional 3D nuclear shape or volume morphometrics (which did not predict mechano-sensitive YAP shuttling). Together, these results indicate that the nuclear envelope morphology of a cell is a strong predictor of cellular mechanotransduction, and that may play a central role in how exogenous signals are experienced by the nucleus to regulate mechanotransductive signaling.
MATERIALS AND METHODS
Methacrylated Hyaluronic Acid (MeHA) Hydrogel Synthesis and Casting.
MeHA was synthesized as previously reported34, where methacrylic anhydride was reacted with 1% w/v sodium hyaluronate (70 kDa, Lifecore Biosciences) in dH2O with pH maintained at 8 ± 0.5. After reacting for 6h, the macromer solution was purified via dialysis (MW cutoff of 6–8 kDa) and then lyophilized for storage. Methacrylation level was confirmed to be ~108% by 1H NMR (Supplemental Figure #12) unless noted otherwise. 1mM of small peptide sequences (Genscript) from adhesive domains for fibronectin (GCGYGRGDSPG) were covalently conjugated to the HA backbone via Michael-type addition reactions of the cysteine residues on these peptides with the methacrylate on the HA backbone. Peptide coupling occurred at room temperature for 45 minutes in TEA buffer (pH 10.5, Sigma). Thin hydrogel films (thickness = 100 μm) of 3% w/v MeHA were cast and polymerized on methacrylated glass coverslips (as in 35) that allowed for covalent attachment of the MeHA hydrogel to the coverslip hydrogel during UV polymerization. Irgacure-2959 was added to this MeHA precursor solution at a final concertation of 0.05% v/v and then polymerized using a UV polymerization box with an output of 4.5 mW/cm2 at a wavelength of 365nm. Variation in polymerization times was used to alter hydrogel mechanics, as previously described36–38.
Atomic Force Microscopy.
Atomic force microscopy was performed as previously described to verify hydrogel mechanics36–38. To summarize this procedure in brief, an MFP-3D AFM (Asylum Research, Santa Barbara, CA) was utilized to perform nanoindentation using microspherical tips (R ≈ 2.25 μm, nominal k ≈ 0.03 N/m, HQ:CSC38/tipless/Cr-Au, Cantilever B NanoAndMore). For each set of indentation experiments, a 20 μm × 20 μm measurement grid (40 × 40 regions) was taken across each hydrogel. Average elastic moduli at each indentation point were determined through fitting these force-extension curves with an elastic Hertzian indentation model39.
Cell Isolation, Culture, and Pharmacologic Inhibition.
Juvenile bovine MSCs were harvested from tibio-femoral bone marrow as previously described by Huang et al 40. All MSCs were cultured in standard growth media (HG-DMEM, 10% fetal bovine serum (FBS), and 1% penicillin, streptomycin, fungizone (PSF)) for all experiments, and were cultured on tissue culture plastic (TCP) for one passage prior to re-plating on the MeHA hydrogels. For all 2D studies, cells were seeded at a density of 3,000 cells/cm2 to prevent spurious cell-cell interactions. Once seeded, MSCs were cultured on MeHA hydrogels for 18 hrs before subsequent fixation. For pharmacologic inhibition studies, inhibitors were added into the culture media prior to fixation and subsequent analyses as follows, unless noted otherwise: 25 μM ML7 for 60min, 50 μM LPA for 30min, 6 μM Cytochalasin D for 60min.
Immunostaining and Quantification of Confocal Imaging.
For normal immunostaining, MSCs were fixed in room temperature 4% PFA for 18 min, and then washed three times with PBS, followed by 10 min permeabilization at 4°C with 0.05% Triton X-100 in PBS supplemented with 320 mM sucrose and 6 mM magnesium chloride. Primary antibodies were diluted in 1% BSA in PBS and added overnight at 4°C. Antibodies and dilutions used in this study included anti-YAP/TAZ (1:200; Santa Cruz #sc-101199), anti-YAP (1:200; Santa Cruz #sc-15407), anti-LMNA (1:400; Abcam#133256), anti-LMNA (1:100; Thermo#MA3–1000), anti-RARγ1 (1:200; Cell Signaling #8965), anti-MKL1 [MRTF-A] (1:200; Abcam#49311), anti-pan-Nucleoporin [Mab414] (1:1000, Abcam#ab24609), anti-LMNB1 (1:500, Abcam#ab16048), anti-LMNA-pSer22(1:500 CST#2026). After three PBS rinses, AlexaFluor-488/546 [H+L] secondary antibodies (Molecular Probes) were added for 1hr at room temperature. F-Actin staining was performed using AlexaFluor-conjugated phalloidin (1:1000; Molecular Probes #A22283) added in with the secondary antibodies and incubated for 1hr at room temperature. Following this incubation, three additional PBS rinses were followed by DAPI staining and mounting using ProLong Gold AntiFade (Life Technologies #P36935). Images were taken on a Nikon A1R Confocal Microscope at 100× 1.4 NA (0.082 μm/px) and processed using the FIJI distribution of ImageJ41. Nuclear-to-cytoplasmic localization ratios of YAP/TAZ, MRTF-A, and RARγ were quantified through immunostaining with a pinhole diameter of 100 μm (3 AU). Nuclear masks from LMNA or DAPI staining were utilized to delineate the nuclear area from the cytoplasmic region of the cell, and the average fluorescent intensity over each region was calculated using ImageJ as in 42. Analysis of focal adhesion morphometrics was performed on paxillin staining images using the FAAS analysis platform43 with input parameters of 5 threshold and a minimum adhesion size of 0.17 μm.
Wrinkling Index Calculation.
In order to quantify the degree of nuclear envelope wrinkling, we took advantage of the fact that the brightest areas of lamin-A/C staining in a wrinkled cell are nuclear wrinkles, as these represent 2 planes of the lamin-A/C network next to each other in a sub-diffractional radius (STED super resolution microscopy revealed these could be as close as ~30nm). High resolution z-stacks of lamin-A/C immunostaining were taken as described above, with a pixel size ~100nm/px and a z-stack height of around ~100nm/slice. A high image resolution is important to provide ample contour boundaries for the eventual wrinkle index calculation. A custom macro was written for ImageJ/FIJI (Supplemental File #1/2) to take inputs from the user for filename and other details, open the z-stack, make a maximum projection image of the z-stack, auto-contrast that image, and then convert to a mask. Next the maximum projection image was binarized and details for min/max/mean/median intensity, intensity standard deviation, and nuclear aspect ratio were recorded. The sensitivity parameter for FeatureJ edge contour tracking (http://imagescience.org/meijering/software/featurej/) for Sobel-based edge detection was calculated as:
Where μ is the mean intensity, σ is the standard deviation of the intensity profile across the nucleus, and Bsf is the brightness regularization factor which allows for scaling of the sensitivity of edge-tracking based on the brightness for a given imaging session (and should remain the same through comparisons across the imaging session). Analysis confirmed that total wrinkling contour pixel count did not correlate with nuclear area (ie. large nuclei could have many wrinkles, small nuclei could have few wrinkles). The binarized output of the FeatureJ edge tracking algorithm was turned into a wrinkle index by calculating the mean bright pixel intensity, dividing by 255 (the maximum white pixel intensity), and multiplying by 100 to convert this to a percent of nuclear spread area covered by wrinkle contours. For all MSCs tested, this parameter varied from ~0–25%. The ImageJ macro used to generate this wrinkling index is included in a supplemental text file.
Lentiviral Delivery of miRNA.
Knockdown of nesprin-1G was accomplished as described previously42. In short, inhibitory miRNA were delivered to MSCs using lentiviral particles packaged with the Block-it Lentiviral Pol II miR RNAi Expression System with EmGFP (Invitrogen) targeted towards the N-terminal actin binding calponin homology domain. We tested three different target sequences, confirmed knockdown levels of Nesprin-1G by dot blot following a 1MDa size filtration, and have chosen the sequence that resulted in the highest levels of Nesprin-1G knockdown (TGCCGAGGACCTTCATCTTCT) and also cloned a separate non-targeting negative control sequence that has been previously validated, miRneg control, into a separate lentiviral vector (5’-GAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT-3’). Following production of lentiviral particles 42, virus in growth media were added to MSCs at a 1:10 dilution with 8 μg/mL polybrene, and left on the cells for 24 hours before the virus was removed. Cells were left on TCP for two additional days before being transferred to 10 kPa hydrogels and subjected to normal experimental procedures. Prior to nuclear wrinkling analysis, cells were binned out for infection by the N1G knockdown construct by screening for only GFP+ cells. For experiments on polyacrylamide gels, cells were infected overnight followed by four days on TCP prior to reseeding on polyacrylamide gels.
Polyacrylamide Gel Preparation and Traction Force Microscopy.
Traction force microscopy and related experiments were performed as described previously 42. In brief, first polyacrylamide (PA) hydrogels (Young’s modulus, E = 10kPa) were prepared as in Aratyn-Schaus44 and used for both TFM experiments and the subsequent analysis of YAP/TAZ nuclear localization after knockdown of nesprin-1G. Prior to polymerization, the polyacrylamide precursor solution was doped with 0.2 μm diameter fluorescent microspheres at 1% v/v (#F8810; Invitrogen, Carlsbad, CA). PA hydrogels were polymerized with APS/TEMED. Fibronectin (20 mg/mL) coating of gel surfaces was accomplished with 2 mg/mL sulfo-SANPAH (No. 22589; Pierce Protein Biology/Life Technologies, Rockford, IL). Small drops of a UV-curable fixative (NOA68; Norland Products, Cranbury, NJ), secured the MeHA-covered glass coverslips in a live cell imaging bath. MeHA hydrogels were subsequently washed three times with PBS and sterilized under germicidal UV light for 1 hr. MSCs were seeded on MeHA hydrogels at 3,000 cells/cm2 and allowed to culture for 18 hour before TFM analysis was performed. Phase contrast and fluorescent images of multiple cells and embedded beads were captured at 40× magnification on a DeltaVision Deconvolution Microscope (GE Healthcare Life Sciences, Marlborough, MA). Image sequences for each cell were taken before and after cell lysis with SDS (sodium dodecyl sulfate). Traction force microscopy data analysis (stack alignment, particle image velocimetry, and Fourier transform traction cytometry) was performed using a freely available plugin suite for ImageJ, created by Tseng and colleagues45, which was adapted from Dembo and colleagues46. For FTTC variables, the Poisson’s ratio of the PA hydrogel was assumed to be 0.45 and a regularization parameter of 1e-9 was used. Using a custom MATLAB script, traction force vector maps were analyzed to determine the average stress generation by each cell on the underlying substrate.
Mouse Tissue Isolation, Cryosectioning, and Immunostaining.
All animal procedures were approved by IACUC at the University of Pennsylvania. Col1a1(2.3kb)-GFPemd mice of CD1 background were generated as previously described47. Animals were scarified at 3 or 14 days postnatal (“P3”, “P14”), tissue specimens from two mice were embedded in Cryomatrix (Thermo Scientific, Waltham, MA) and 7–8 μm tissue sections were created using a cryofilm technique 48. Tissue sections were rehydrated with PBS prior to immunostaining. Sections were blocked for 1hr at RT in a blocking buffer (2%BSA/0.25% TritonX-100) prior to labelling with primary antibodies in blocking buffer overnight at 4C in a humidified box. Slides were rinsed 3×15min with PBS before secondary labelling for 2hr at RT in blocking buffer, followed by another 3×15min series of PBS rinses before mounting the coverslips in DAPI ProLong AntiFade. Slides were imaged on the AxioScan.Z1 microscope (Carl Zeiss Microscopy, Thornwood, NY) at 20× for the low-resolution stitched images, and sections were subsequently imaged on a Nikon A1R confocal microscope with a 60× 1.4NA objective for the cellular images. Representative wrinkling images were from sum of slice projections from Lamin-A/C immunostaining, and YAP/TAZ nuclear-to-cytoplasmic ratios were calculated from single confocal slices of the midplane of cells as previously described36.
Synthesis of Norbornene-modified HA (NorHA) and MMP-degradable Peptide Crosslinkers.
Modification of HA with norbornene groups as performed as previously described 49. First, sodium HA (Lifecore, 75 kDa) was converted to a tetrabutylammonium (TBA) salt by ion exchange using Dowex 50W resin. NorHA was synthesized by reacting the TBA salt of HA with 5-norbornene-2-carboxylic acid using 4-(dimethylamino)pyridine (DMAP) and ditertbutyl dicarbonate (BoC2O). The reaction was performed under nitrogen at 45 °C for 20 h. The product was purified by dialysis (SpectraPor, 6–8 kDa molecular weight cutoff) and lyophilized for storage. The degree of modification was ~20% as measured by 1H NMR (Supplemental Figure #11). Protease-degradable (GCNSVPMS↓MRGGSNCG) peptide linkers were synthesized using standard solid peptide synthesis methods as previously described50. Peptides were synthesized off a glycinol 2-cholorotrityl resin with FMOC-protected amino acids and subsequently cleaved from the resin with trifluoroacetic acid and water. Purified peptides were confirmed by Matrix Assisted Laser Desorption/Ionization (MALDI) (Supplemental Figure #12).
3D NorHA Hydrogel Preparation and Cell Culture.
NorHA hydrogels were fabricated via ultraviolet (UV)-light mediated thiol-ene addition reactions 49. NorHA hydrogels (4 wt%) were photo-crosslinked (2 mW cm−2) for 3 min in the presence of 0.05 wt% 2-hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone (I2959) photo-initiator and the degradable di-thiol peptides at a thiol-norbornene ratio of 0.3, 0.35, or 0.45 to adjust crosslinking density. For all experiments, a thiol-norbornene ratio of 0.3 was used unless specified otherwise. For all conditions, MSCs were encapsulated at a density of 4 million cells per mL, and in the presence of 2 mM thiolated RGD peptide to promote cell adhesion (GCGYGRGDSPG, GenScript). To obtain thin constructs (ca. 300 μm) hydrogels were photo-crosslinked underneath coverslips, cut into 5×5 mm gels and cultured individually in 48 well plates. Culture media (DMEM 4.5 g/L glucose with 10% FBS, 1% penicillin/streptomycin) was changed every second day for 5 days prior to staining and analysis. For cytochalasin D experiments, CytoD was added at 6 μM for 1 hour prior to fixation.
3D Hydrogel Immunostaining, Imaging, and Quantification.
Hydrogel constructs were fixed in 10% buffered formalin for 25 min at RT, permeabilized for 2 h at 4 °C with 1% triton X-100, 0.6 mM magnesium chloride and 0.1 g/mL sucrose. Prior to staining, gels were blocked with bovine serum albumin (BSA 2wt%) for 1 h at room temperature. Gels were stained overnight at 4°C in the presence of 2wt% BSA with the following antibodies: LMNA (Abcam, ab133256, 1:400), YAP/TAZ (Santa Cruz, #sc101199 1:200), and with secondary anti-mouse (Abcam, #ab150113 1:200), secondary anti-rabbit (Abcam, ab150080 1:200) and Phalloidin (Thermo, #A22287, 1:100). For analysis, z-stacks (step-size = 0.3 μm) of single cells were taken with a 60× 1.4NA PlanApo oil immersion objective using a Nikon A1R Confocal Microscope; all imaged cells were >50–100 μm from the surface of the gel to avoid an edge bias. Wrinkle indices were calculated as listed for 2D, with a regularization factor of 6 shared between all groups. YAP/TAZ nuclear-to-cytoplasmic values were calculated using Volocity, wherein the entire cell was automatically thresholded and a 2 pixel erode was applied, LMNA staining signal was used to automatically threshold the nucleus with a 1 pixel erode, the nucleus was subtracted from the cytosolic mask, and the ratio was reported as the mean pixel intensity for the nuclear region divided by the mean pixel intensity for the cytosolic region. Actin anisotropy ratios around the nucleus were calculated using the FibrilTool plugin51. For this quantification maximum projection nuclear masks were dilated by 125% around the centroid, and this area mask was applied to maximum projection F-Actin staining images and anisotropy ratios were calculated using FibrilTool in ImageJ.
Statistical Analysis.
All experiments were performed using 2–3 MSC or mouse donors and at least 3 replicate hydrogels per condition unless otherwise noted, with all data from each separate experiment binned together. Statistical comparisons were performed using a Student’s t-test or Mann-Whitney U-test when only two groups were being compared, and Kruskal-Wallis one-way ANOVA testing with Dunn’s post-hoc used to make pairwise comparisons between multiple groups unless otherwise noted (depending on sample normality). Statistical significance was set to p < 0.05.
RESULTS
The nuclear envelope (NE) is wrinkled on soft 2D hydrogels.
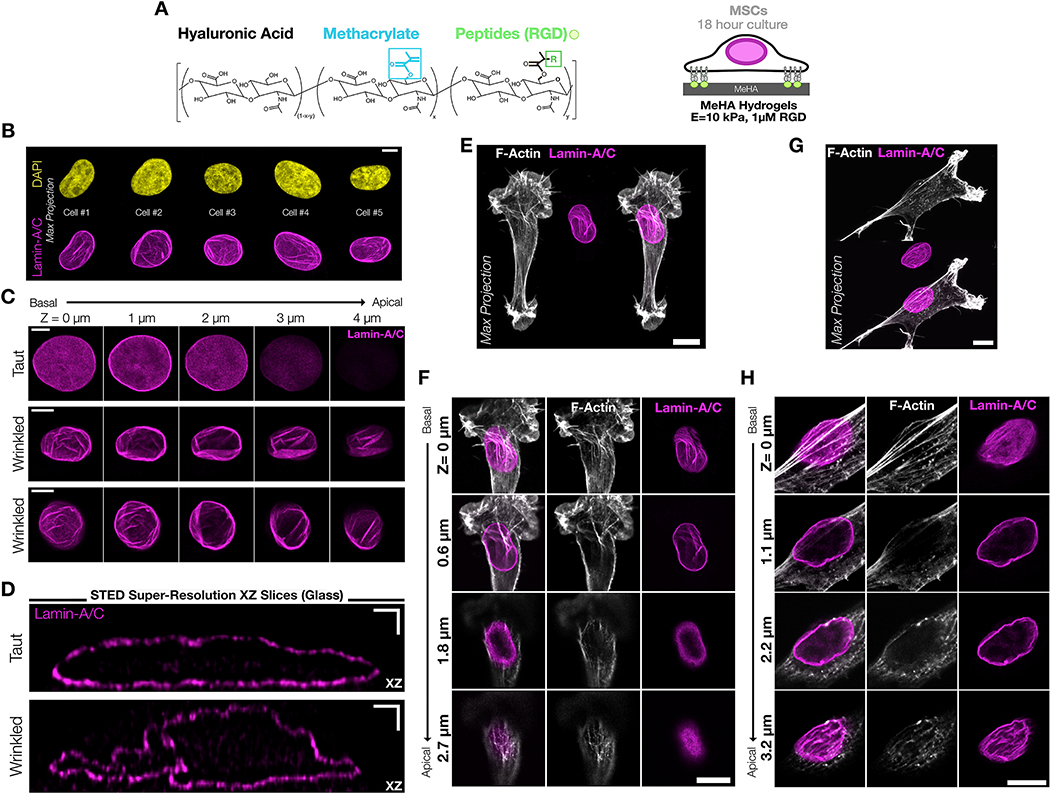
To characterize nuclear envelope morphology in response to various microenvironmental perturbations, mesenchymal stem cells (MSCs) were seeded onto thin (h=100 μm) 2D methacrylated hyaluronic acid hydrogels (MeHA), functionalized with RGD to enable cell-ECM adhesion, and UV polymerized to a Young’s Modulus of 10 kPa (verified by AFM) (Figure #1A). We chose this modulus as it represents a mechanical threshold at which mechanosensitive transcription factor shuttling to the nucleus begins in this system36, 52. Immunostaining for lamin-A/C and subsequent confocal imaging showed prominent nuclear wrinkles in this context, which were not easily detectable via DAPI staining (Figure #1B). To confirm that wrinkles included the entire inner and outer nuclear envelope, and not just the filamentous nuclear lamina, we also stained with antibodies for lamin-B1 and a pan-nucleoporin antibody (“NUP”) that binds to FXFG repeats found in many nuclear pore sub-components (Supplemental Figure #1). These images showed co-localization of both lamin-B1 and nuclear pores within wrinkles, which suggests that these wrinkles are present throughout the entire nuclear envelope and are not limited to only the inner or outer nuclear envelope. NE wrinkling showed no consistent polarity and could be found on both the apical and basal planes of the nucleus (Figure #1C).
Figure #1 – The nuclear envelope (NE) of mesenchymal progenitors on soft 2D hydrogels can have a wrinkled morphology.
(A) Schematic of methacrylated hyaluronic acid (MeHA) hydrogels modified with RGD peptides. All hydrogels were UV crosslinked to 10 kPa unless otherwise noted. Mesenchymal stem cells (MSCs) were seeded at low density and cultured for 18–24 hours prior to analysis. (B) Comparison of nuclear wrinkling in DAPI and Lamin-A/C stained nuclei from representative maximum projection confocal images of MSC nuclei cultured on 10kPa MeHA hydrogels for 24hrs. SB = 5 μm. (C) Representative lamin-A/C stained images from confocal z-stacks of one taut and two wrinkled MSC nuclei cultured on 10kPa MeHA hydrogels for 24hrs. These wrinkles varied in scale and orientation and persisted from the basal to the apical planes of the cell. SB = 5 μm. (D) Single XZ slices of lamin-A/C following STED super resolution microscopy of MSCs cultured on fibronectin-coated glass revealed that wrinkles could be as deep as ~1 μm, and in some cases fold onto themselves. SB = 1 μm. (E) Maximum projection and (F) corresponding individual slice z-stack images of MSCs on a 10 kPa hydrogel that shows wrinkling and strong actin impingements on only the basal plane of the cell. SB = 10 μm. (G) Maximum projection and (H) corresponding individual slice z-stack images of an MSC on a 10 kPa hydrogel that exhibits wrinkling across all planes of the nuclear envelope with a small degree of actin impingement on the basal surface. SB = 10 μm.
On most high-resolution confocal images, nuclear wrinkles appear as bright spots and are diffraction-limited in the XZ axis. To better visualize these wrinkled NE structures, we employed STED (stimulated emission depletion) super-resolution microscopy to visualize lamin-A/C through the nuclear depth (Figure #1D, Supplemental Figure #2). XZ slices showed that while taut nuclei had an ellipsoidal morphology, wrinkled nuclei had clear folds that penetrated as much as 0.75 – 1 μm into the nucleus. Previously, others have shown that on stiff/rigid substrates, a polarized perinuclear F-actin cap can impinge on the apical surface of the nucleus on highly-spread cells23, 25, 53. We found perinuclear actin caps in a small fraction of the MSCs on 2D substrates, and when present these actin caps did produce a small amount of a wrinkled morphology via actin impingements into the nucleus on the apical or basal planes (as seen in Figure #1E/F, and Figure #1F/G on the basal surface). However, these interactions were not common, and the majority of nuclear wrinkles in cells on 2D hydrogels were not the result of direct cytoskeletal impingement and instead were the result of nuclear crumpling/infolding. These wrinkled nuclei had a tortuous overall shape, and the majority of wrinkled nuclei lacked obvious cytoskeletal interactions (Figure #1C middle & bottom rows, Figure #1G/H on the basal plane). We additionally noted that these nuclear lamina wrinkles looked similar to the wrinkled nuclear morphology found in cells during early attachment or detachment 6, 20, 21, 24, 27, which suggested that these wrinkles might reflect a low degree of contractility in these cells. Additionally, we found microtubules occupying wrinkles in a small fraction of nuclei (Supplemental Figure #3). While others have noted that microtubules can lead to tortuous nuclear envelopes in myeloid cells that had mostly cortical actin54, it was unclear whether this microtubule entrapment within nuclear wrinkles in MSCs was the result of an active deformation or if microtubules were passively entrapped in the nuclear envelope as the nucleus deformed through other mechanisms. As a whole, it is likely that nuclear envelope wrinkling results from the summation of a variety of cellular boundary conditions that combine to shape the nuclear envelope.
Nuclear envelope wrinkling is altered following short-term perturbations in cell contractility.
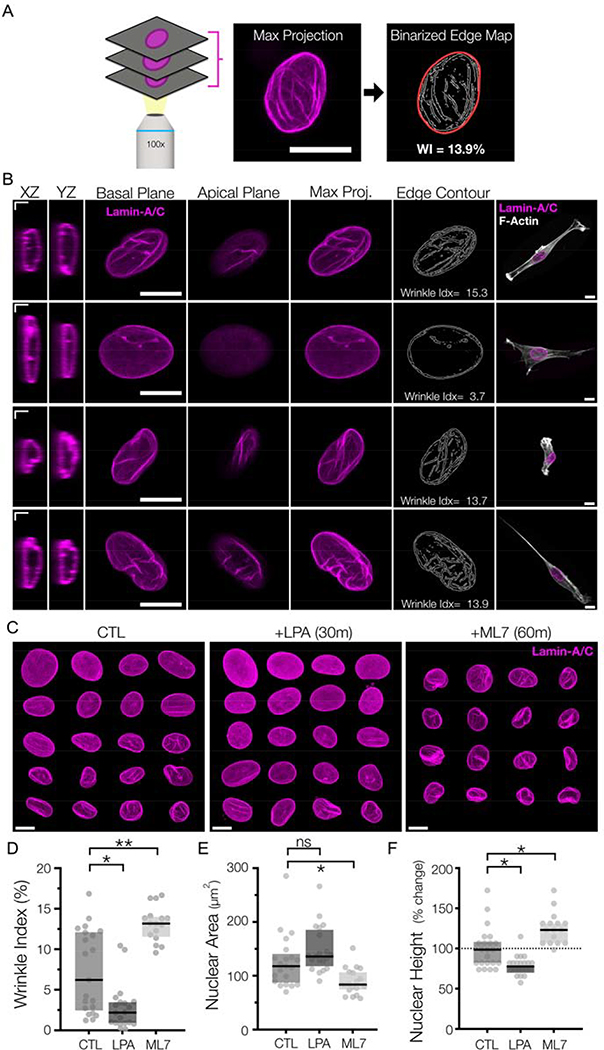
To quantify these changes in NE morphology, we developed an edge-based detection algorithm to quantify the number of contrast boundaries in maximum projections of lamin-A/C stained nuclei, as wrinkles appear as bright contours in these images. The strong changes in labeling intensity provided an objective measurement method to detect and quantify the extent of nuclear wrinkling (as a fraction the overall nuclear area) (Figure #2A, right). This metric is similar in concept to other edge detection-based measurements of chromatin condensation55–57. Across all datasets, this ‘Wrinkle Index’ ranged from 0 to ~25% of the nuclear area, with 25% representing a very wrinkled nucleus and 0% representing a completely smooth nuclear envelope (Figure #2A/B). Using this measure, we found that the majority of cells plated onto rigid glass or tissue culture plastic (TCP) substrates had smooth nuclei with little to no wrinkling. Conversely, over half of the nuclei on 2D MeHA hydrogels (modulus = 10 kPa) had some degree of wrinkling, with an average wrinkling index of ~7.5%. We also noted considerable heterogeneity between cells, likely reflecting the temporal and cell-to-cell variations in contractility commonly seen in MSC populations.
Figure #2-. Rapid alterations in NE morphology following pharmacologic changes in actomyosin contractility.
(A) Overview of the nuclear wrinkling metric based on edge detection. Maximum projection images of lamin-A/C z-stacks were taken and subjected to contrast-based Sobel edge detection. These resulting edge maps were binarized, and the fraction of wrinkled edges of these maps was normalized to the nuclear boundary area to generate the nuclear wrinkling index. SB = 5 μm. (B) Representative immunostaining of lamin-A/C from MSCs on 10 kPa hydrogels. Single confocal slices are shown of XZ/YZ slices (SB = 2 μm), as well as individual slices of the apical and basal sides of the nuclei. Maximum projection images are utilized to generate an edge-detection based contour map, which is used to generate a wrinkling index. SB = 10 μm. (C) Mosaic representations of Lamin-A/C max projection images for MSCs cultured on 10 kPa MeHA gels for 18 hours (“CTL”) or following treatment with LPA for 30m or ML7 for 60m at the end of this culture window. SB = 10 μm. Corresponding quantifications of (D) wrinkle index, (E) spread nuclear area, and (F) nuclear height of the nuclei from (C) (n>19 nuclei/group, Box plot represents median and 25th/75th quartiles, *= p<0.05, **= p<0.01, by Kruskal-Wallis one-way ANOVA testing with Dunn’s post-hoc).
Having established this baseline, we next perturbed the contractile state of cells over a short time window and evaluated its effect on nuclear wrinkling. For this, pharmacologic agonists (LPA, lysophosphatiditc acid) and inhibitors (ML7) of contractility were used to modulate the cell mechanical state following 30 or 60 minutes of treatment. Importantly, we have previously found that these two agents do not alter nuclear-actin connectivity at these time scales42. Recent work has shown how LMNA-pSer22 is a potent marker for force-sensitive lamin-A/C degradation21.
We found that immunostaining for LMNA-pSer22 showed no significant change in LMNA-pSer22 to lamin-A/C ratios after reducing contractility for 60 min, and we found no correlation between wrinkling index and LMNA-pSer22 to lamin-A/C levels (Supplemental Figure #4), supporting that during these short time windows, lamin assembly or degradation did not significantly impact nuclear envelope structure19, 21. We also previously reported that traction forces decreased by ~90% in MSCs following ML7 treatment for 60 min, whereas treatment with LPA for 30 min increased traction by ~60%42. With respect to the NE, we found that LPA treatment markedly reduced wrinkling within 15 minutes of exposure and decreased nuclear height and increased cell spread area by 30 minutes of treatment (Figure #2C). Conversely, treatment with the contractility antagonist (ML7) increased nuclear wrinkling and nuclear height, while decreasing cell spread area (Figure #2D–F). Taken together, these data show that altering cytoskeletal tension acutely alters NE wrinkling morphology, with increased contractility removing the wrinkles normally present in the nuclear envelope of mesenchymal progenitor cells cultured on soft 2D hydrogels. This data also further supports our previous observations that nuclear envelope wrinkling in 2D was not the result of cytoskeletal impingement, but rather represented a “slack” state of the nuclear envelope.
Nuclear envelope wrinkling provides a strain energy sink for the contracting cytoskeleton.
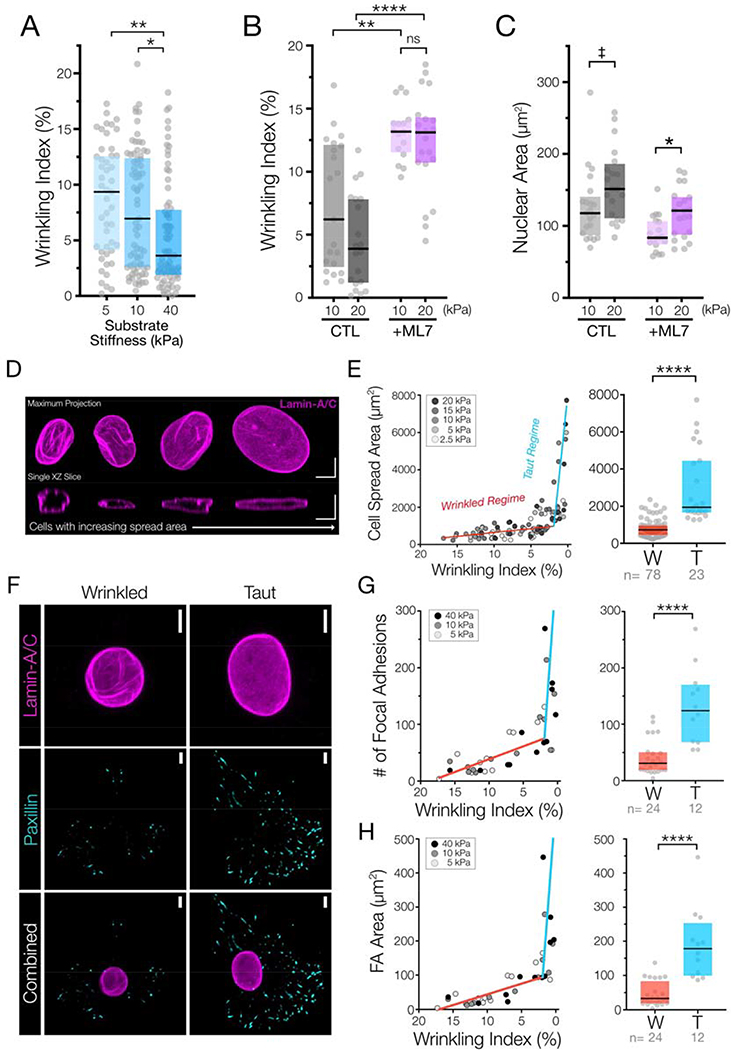
In addition to changes in nuclear envelope wrinkling with alteration in contractility, we were also interested in how this translated into a downstream mechanobiologic response. To accomplish this, we first examined how nuclear envelope morphology changed with variations in 2D substrate stiffness36. We chose to evaluate the nuclear envelope wrinkling on a wide physiologic range of substrate stiffnesses. With culture on different stiffness 2D MeHA hydrogels, nuclear wrinkling decreased significantly as hydrogel stiffness increased (Figure #3A, Supplemental Figure #5). However, the change in wrinkling over this 18h culture period was less marked than that observed with short treatment windows of contractility-modulating agents. This difference is likely due to cell heterogeneity as well as homeostatic processes and other nuclear remodeling events that occur over these longer culture windows. Interestingly, we found that the decreased nuclear wrinkling caused by increased hydrogel stiffness could be completely countermanded by inhibiting cellular contractility for a short time-period; treatment with ML7 for one hour following 23 hours of culture on 10 and 20 kPa hydrogels induced similar degrees of nuclear wrinkling in all cells, regardless of the underlying hydrogel stiffness (Figure #3B/C). This observation suggests that nuclear envelope morphology remains highly responsive to short-term changes in cellular contractility despite remodeling that might occur over these longer timeframes. Other microenvironmental cues can also alter cellular contractility, including the degree of cell-cell contact present. We recently developed a material system that incorporated RGD ligands to regulate cell-ECM signaling as well as HAVDI ligands to trigger cell-cell signaling via N-cadherin. Presentation of 1 mM of the HAVDI N-cadherin adhesive moiety in addition to 1 mM RGD ligand was previously found to decreased contractility by ~50% 36, 58. Similar amounts of HAVDI presentation (1mM) markedly increased nuclear wrinkling following 18 hours of culture (Supplemental Figure #6). These data suggest that mechanical and chemical cues from the microenvironment that serve to regulate the mechanical state of the cell also dictate the degree of nuclear envelope wrinkling.
Figure #3 – Nuclear envelope wrinkling provides a strain energy sink for the contracting cytoskeleton.
(A) Wrinkling indices of MSCs cultured on three different stiffness MeHA hydrogels for 18 hours (n=47–73 nuclei, Box plot represents median and 25th/75th quartiles, * = p<0.05, ** = p<0.01, by Kruskal-Wallis one-way ANOVA testing with Dunn’s post-hoc). (B) Wrinkling indices and (C) nuclear projected areas of MSCs cultured on 10 or 20kPa hydrogels for 18 hours, followed by treatment with DMSO (“CTL”) or with ML7 for 60m prior to fixation and analysis (n=16–21 nuclei/group, Box plot represents median and 25th/75th quartiles, * = p<0.05, ** = p<0.01, **** = p<0.0001, ‡ indicates p=0.0693, by Kruskal-Wallis one-way ANOVA testing with Dunn’s post-hoc). Wrinkling data for 10 kPa hydrogel perturbations is also represented in the previous figure. (D) Representative XY and corresponding XZ sections of the nuclear lamina from multiple MSCs with increasingly higher cell spread areas suggests that increased contractile energy removes wrinkles from the nuclear envelope along with nuclear flattening and wrinkle unfurling as the nuclei become taut. SB = 5 μm. (E) (left) Plots of wrinkling index (shown from high to low) versus cell spread areas of MSCs cultured on five stiffness conditions with moduli ranging from 2.5 – 20 kPa for 18 hours (n= 101 nuclei over 5 stiffness conditions). Bilinear fit lines represent the “wrinkled regime” (W, red) or a “taut regime” (T, blue) and (right) re-plots of this wrinkling index vs. cell spread area data that are categorically separated using this bilinear fit. Box plot represents median and 25th/75th quartiles. (n=23 or 78 cells/group, **** = p<0.0001 by Mann-Whitney U-test). (F) Representative max projection images of lamin-A/C and paxillin of wrinkled and taut nuclei in MSCs cultured on 10 kPa hydrogels for 18 hours. SB = 5 μm. (G) (left) Plots of wrinkling index (from high to low) versus number of focal adhesions and (H) (left) total focal adhesion area per cell from MSCs cultured on three different HA hydrogel stiffnesses for 18 hours (n= 36 cells). Bilinear fit lines represent the “wrinkled regime” (W, red) or a “taut regime ” (T, blue) and re-plots of the wrinkling index vs. number of focal adhesions (G)(right) or wrinkling index vs. focal adhesion area data (H)(right) that are categorically separated using this bilinear fit. Box plot represents median and 25th/75th quartiles. (n=12 or 24 cells/group, **** = p<0.0001 by Mann-Whitney U-test).
On intermediate stiffness hydrogels (10 kPa), we noted that cells with low spread area had tall nuclei with a large amount of wrinkling, whereas spread cells had nuclei that were “pulled taut”, with a higher nuclear spread area, a decreased nuclear height, and very little NE wrinkling (Figure #3D). Based on these observations, we next compared cellular spread area to the wrinkling index across a range of five different matrix elasticities, spanning 2.5 kPa to 20 kPa. Despite the inherent heterogeneity of this cell population in each condition (Supplemental Figure #5), a strong correlation between nuclear spread area and nuclear wrinkling was observed. Specifically, across all stiffness hydrogels, cell spreading was high only in cells whose nuclei were taut (with a low wrinkling index, Figure #3E). Applying bi-linear curve-fits to this data identified two distinct response regions in nuclei: 1) a wrinkling regime where there were large changes in NE wrinkling with only small changes in cell spread area, and 2) a taut regime, where nuclear wrinkling was low with a sharp increase in cell spreading. This response is analogous to the non-linear tensile stress-strain behavior of collagenous tissues, where the initial energy provided to the system is dissipated via changes in organization (toe region) prior to the rapid development of stress following further deformation after all elements are have been pulled into tension (linear region) 58.
This biphasic relationship suggests that on 2D hydrogels of physiologic stiffness, the non-ellipsoidal wrinkled morphology of the nuclear envelope imbues the system with a “toe region”, wherein input energy from the contractile cytoskeleton functions to first remove nuclear wrinkles and pull the envelope taut. Once this is accomplished, the nucleus reaches a transition point (at a wrinkling index of ~2%) after which increases in cell spreading can occur. Because cell area can be influenced by a number of factors, we extended this analysis to query the relationship between nuclear wrinkling and focal adhesion characteristics that help govern the contractile state of the cell (Figure #3F/G). Using the same bilinear thresholds defined above, both the number of focal adhesions and total focal adhesion area per cell clearly exhibited this biphasic relationship with the state of nuclear envelope wrinkling (Figure #3F/G). These data support the idea that nuclear envelope wrinkles can indeed act as a ‘strain energy sink’ 59, where robust focal adhesion maturation and cell spreading only occur once the free surface area in the nuclear envelope within these wrinkles is removed and a critical threshold of nuclear envelope tautness is reached.
Nuclear envelope tensioning through the LINC complex defines thresholds for nuclear shutting of transcriptional activators.
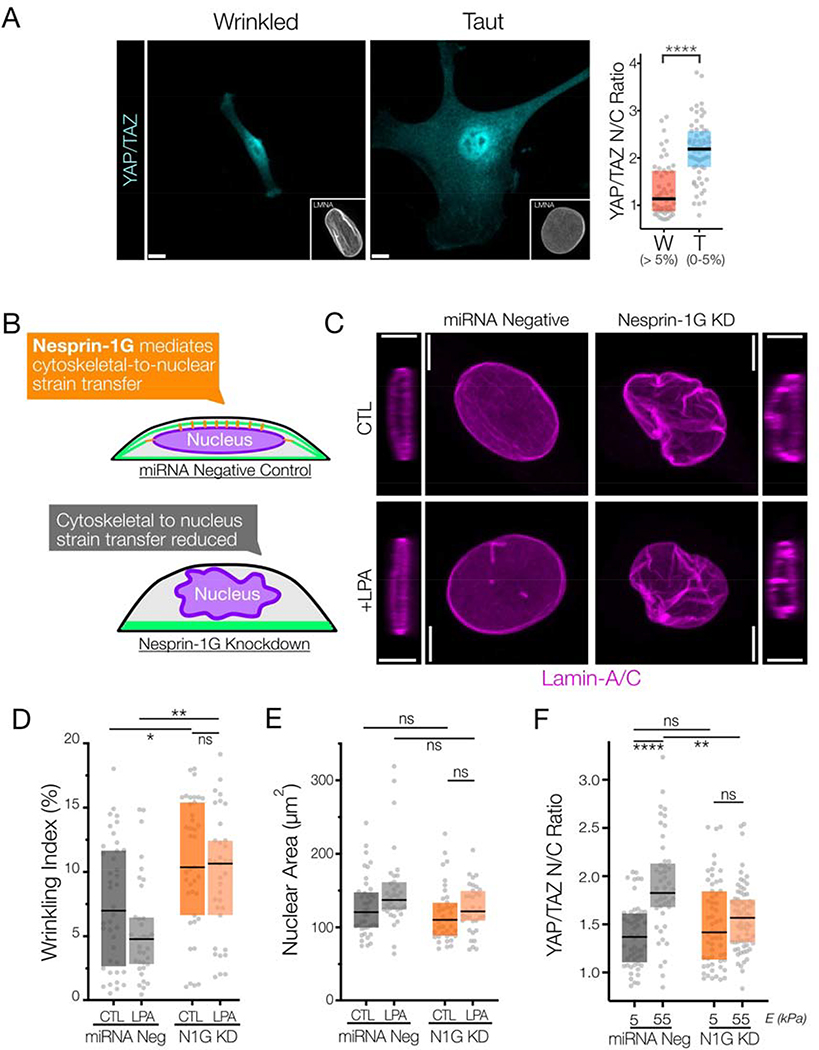
One important downstream effect of cytoskeletal stress is the force-induced nuclear translocation of transcription factors and transcriptional co-activators. In particular, YAP/TAZ, MRTF-A, and RARγ are all potent transcriptional (co-)activators associated with a contractile phenotype, and once in the nucleus they activate transcriptional programs that further feedback to regulate the mechanical state of the cell (e.g. increased stress fiber formation from MRTF-A/SRF, increased lamin-A/C expression from RARγ, and increased matrix production and focal adhesion maturation from YAP/TAZ) 2, 5, 6, 9. To determine how nuclear wrinkling changed along with the nuclear-to-cytoplasmic ratios (“N/C ratio”) of these proteins, we simultaneously quantified wrinkling indices and the N/C ratios in individual cells across a range of hydrogel stiffness (2.5 – 20 kPa, Figure #4A). For this comparison, we conservatively set the threshold for binning wrinkled nuclear (‘W’) as those having a wrinkling index of > 5% based on bilinear curve fits from the cell spreading and focal adhesion maturation data in Figure #3. We found a strong dependence of the nuclear localization of YAP/TAZ (Figure #4A) on nuclear envelope wrinkling state, where wrinkled nuclei had significantly lower nuclear-to-cytoplasmic ratios of all three factors, indicating that shuttling preferentially occurs when nuclear envelope wrinkling is low and the nuclei are taut. Similar trends were observed with MRTF-A, and RARγ, though the mechanisms that define this trend for these factors are unclear (Supplemental Figure #7). Recent studies have suggested that YAP only translocates into the nucleus when the NE is under stress, where the mechanical stress functions to increased import through nuclear pores22, 42. Our data suggests that this may occur with multiple transcriptional co-activators in addition to YAP, and further that these proteins begin to translocate into the nucleus after nuclear wrinkles are removed.
Figure #4 – Nuclear envelope wrinkling depends on cytoskeletal-to-nuclear connectivity and regulates nuclear shuttling of mechano-sensitive transcriptional co-activators.
(A) (left) Images of YAP/TAZ immunostaining for representative MSCs with wrinkled or taut nuclei (as visualized by Lamin-A/C immunostaining). (A)(right) Nuclear-to-cytoplasmic ratios of YAP/TAZ from Figure #3E, where MSCs were cultured on five stiffness conditions with moduli ranging from 2.5 – 20 kPa for 18 hours (n = 101 nuclei over 5 stiffness conditions) using categorical wrinkling data separated by a wrinkling index cutoff of 5% (W = wrinkled regime, T = taut regime). Box plot represents median and 25th/75th quartiles (n=36–45 nuclei/group YAP/TAZ, Box plot represents median and 25th/75th quartiles, **** = p<0.0001 by Mann-Whitney U-test). SB = 5 μm. (B) Overview of experimental approach to reduce nuclear envelope to cytoskeleton connectivity via Nesprin 1 Giant (N1G) knockdown. (C) Representative XY maximum projection images and XZ slices of lamin-A/C immunostaining of MSCs three days post knockdown, with or without treatment with LPA for 30m prior to fixation and staining (SB = 5 μm). Corresponding quantification of the (D) wrinkling indices and (E) nuclear spread areas of the cells from part C (n> 34 cells/group, Box plot represents median and 25th/75th quartiles, * indicates p<005 and ** indicates p<0.01 by Kruskal-Wallis one-way ANOVA testing with Dunn’s post-hoc). (F) YAP/TAZ nuclear ratios of MSCs three days post knockdown with negative control or N1G-targeting miRNA that where subsequently cultured on 5 or 55kPa polyacrylamide hydrogels for 18 hours prior to fixing and staining (n> 45 cells/group, Box plot represents median and 25th/75th quartiles shown, ** = p<0.01, **** = p<0.0001 by Kruskal-Wallis one-way ANOVA testing with Dunn’s post-hoc).
To further investigate the impact of nuclear wrinkling state on nuclear YAP translocation, we decoupled the nucleus from the cytoskeleton via miRNA-mediated knockdown of Nesprin-1 giant (N1G), a large actin-binding molecular component of the linker of nucleus and cytoskeleton (LINC) complex42 (Figure #4B). Compared to negative controls, knockdown of N1G increased NE wrinkling and prevented the changes in nuclear wrinkling and spread area that occurred in control cells with addition of LPA (Figure #4C–E). We validated that N1G knockdown did not decrease cell contractility by performing traction force microscopy of miRNA negative control cells and N1G knockdown cells on 5 kPa polyacrylamide gels. Rather, knockdown of Nesprin-1G increased traction force compared to controls (Supplemental Figure #8), consistent with previous reports 60. Immunostaining for YAP/TAZ on these polyacrylamide substrates showed that, despite the 2.5-fold increase in traction force in N1G knockdown cells in basal conditions, there was no significant change in YAP/TAZ localization between N1G knockdown cells cultured on 5 kPa and 55 kPa substrate groups (Figure #4F). Conversely, we noted robust increases in YAP/TAZ nuclear localization in control miRNA negative cells on stiff substrates (Figure #4F). These data support that YAP nuclear translocation does not depend on the contractile state of the cell per se, but rather depends on the transfer of contractile strain energy to the nucleus and generation of stress in the nuclear envelope by removal of nuclear wrinkles and tensioning of the nuclear envelope.
Nuclear envelope wrinkling is caused by actin impingement in MMP-degradable 3D HA hydrogels.
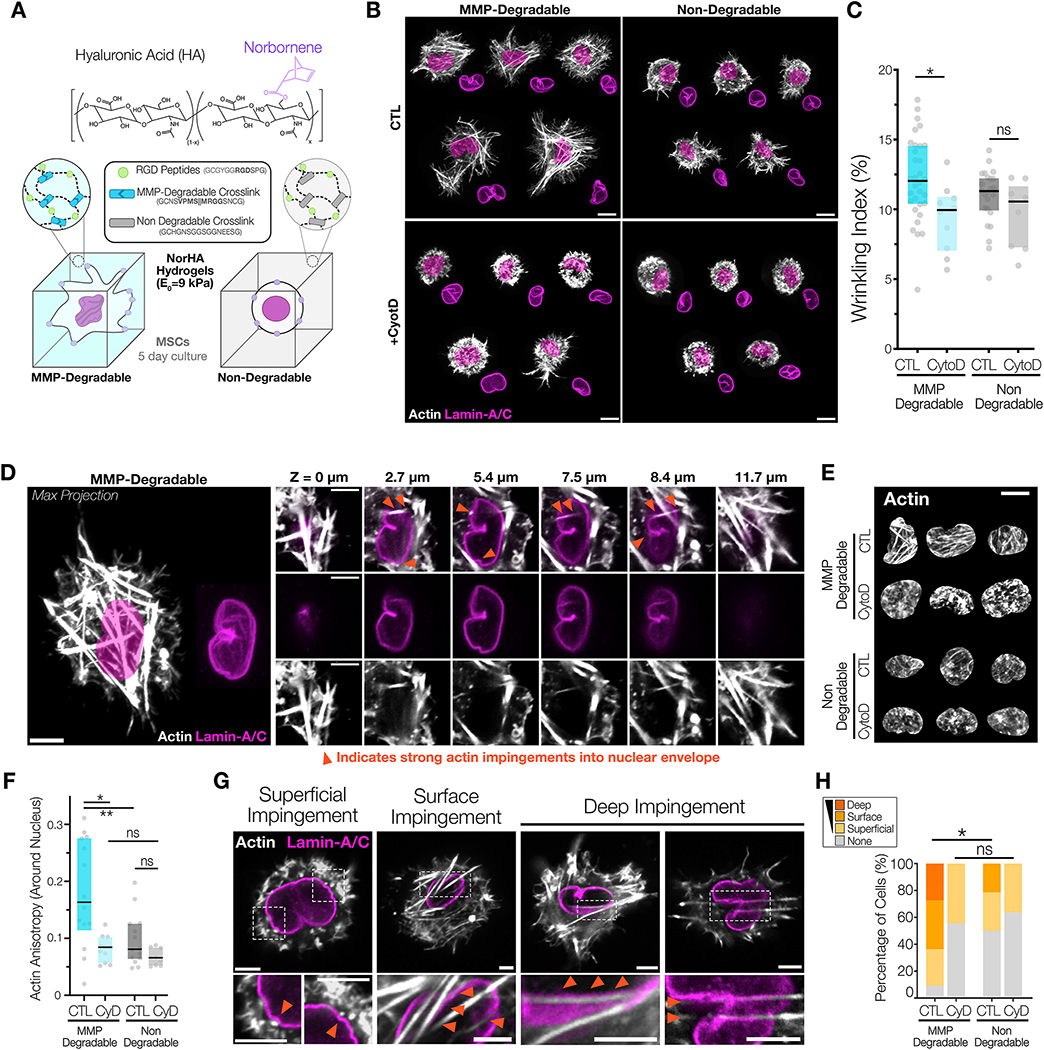
Examining nuclei on 2D hydrogel platforms helped to elucidate the principles governing nuclear envelope wrinkling in response to exogenous mechanical cues. However, in 2D culture, there exists a defined apical-to-basal cell polarity, and as such the nucleus is subjected to both tensile and compressive loads with increases in contractility 23, 25, 61. In 3D microenvironments, however, cell-ECM adhesive interactions are not constrained to a single plane, but can instead occur in any direction across all three axes62. Many recent engineered 3D environments have enabled cellular remodeling of the microenvironment, including the use of engineered fibrillar networks62, stress-relaxing hydrogels63, 64, and MMP-mediated degradability65–68. The use of these material platforms has uncovered new mechanisms of mechanosensing that arise only in these more native-like environments where cell-based remodeling of the material is possible. Using one of these advanced systems, we queried how dimensionality, remodeling, and traction generation that occur in 3D alters NE morphology and impacts mechanosensing. To do so, we used a norbornene-modified HA (NorHA) backbone that was crosslinked with MMP-degradable peptides to allow embedded cells to spread and generate force against the matrix. We have previously shown that cell spreading and traction generation in this 3D system depends on MMP-degradability of the network66, and that the inclusion of MMP-degradable crosslinks enables cell spreading and YAP nuclear translocation65. Here, we used both MMP-degradable peptide crosslinks and non-degradable peptide crosslinks as tools to modulate and control cellular traction generation and spreading in 3D hydrogels, while simultaneously evaluating nuclear morphology. To examine nuclear morphology in this context, MSCs were distributed in an RGD-modified NorHA solution that was gelled using either MMP-degradable or non-degradable peptide crosslinks to form the 3D hydrogels (Figure #5A). The elastic modulus of these hydrogels was verified to be 9 ± 0.7 kPa following crosslinking. After the five days of culture required for robust cell spreading in these 3D hydrogel systems, immunostaining for YAP/TAZ, F-actin, and lamin-A/C was performed. Cells in MMP-degradable hydrogels were well spread with significant increases in cellular volume compared to cells in hydrogels with non-degradable crosslinks (Figure #5B) (Supplemental Figure #9). Additionally, MSCs in MMP-degradable hydrogels had prominent stress fibers with a significantly more polarized shape compared to cells in non-degradable hydrogels, which had primarily cortical actin and a spherical shape (Figure #5B, Supplemental Movies #1&2).
Figure #5 – Nuclear envelope wrinkling is a strong predictor of nuclear YAP/TAZ localization in 3D MMP-degradable hydrogels.
(A) Schematic of the 3D culture of MSCs in norbornene hyaluronic acid (NorHA) hydrogels modified with RGD peptides and either MMP-degradable or non-degradable peptide crosslinkers that are used to set the initial modulus of the hydrogel to 9 kPa. For all studies MSCs were encapsulated in these NorHA hydrogels prior to polymerization, and after polymerization, MSCs were cultured for 5d prior to fixation and immunostaining. (B) Representative maximum projection images of F-actin and Lamin-A/C immunostaining in MSCs cultured in NorHA hydrogels crosslinked with either MMP-degradable or non-degradable peptides, with and without treatment of cytochalasin D (‘Cyto D’) for 1 hour prior to fixation. SB = 10 μm. (C) Quantification of wrinkling indices for cells in these conditions from part (B) (n=9–32 cells/group, Box plot represents median and 25th/75th quartiles,* = p<0.05 by Kruskal-Wallis one-way ANOVA testing with Dunn’s post-hoc). (D) Representative XY sections of a cellular nuclei in 3D MMP-Degradable NorHA hydrogel shows how the contractile actin cytoskeleton impinges on the nucleus across multiple planes. SB = 5 μm. Representative mosaic images of the maximum projection of actin around the nuclear mask (E) and quantification (F) of actin anisotropy (with higher values representing more anisotropic actin) around the nuclei of cells cultured in 3D NorHA hydrogels from part B/C(n=9–13 cells/group, Box plot represents median and 25th/75th quartiles, * = p<0.05 by Brown-Forsythe and Welch ANOVA testing with Dunnett’s T3 post-hoc). SB = 10 μm (G) In the context of actin impingement, three main categories of impingement were classified for the nuclei shown in B/C: nuclei that exhibited superficial deformations (small deformations), nuclei with surface wrinkles and grooves (that were clearly shaped by an actin fiber) and nuclei with deep impingements where stress fibers induced invaginations reaching >1 μm into the nucleus. Each panel is made from the summation of 10 z-slices from a stack, representing a 3 μm projection slice. Zoomed images shown in the middle panel, with orange arrows indicating regions of strong actin impingement into the nuclear envelope. SB = 5 μm. (H) Quantification of categorical changes with perturbations highlighted in (G) (n=9–12 cells/group, * = p<0.05 by via Fisher’s exact test between no deformation and all deformed groups).
Having established different cellular behaviors, we next evaluated nuclear features in these 3D systems. We noted higher baseline NE wrinkling for all groups compared to the same cells on soft 2D MeHA hydrogels (Figure #5B/C). Treatment with cytochalasin-D (‘CytoD’, ‘CyD’) to depolymerize the actin cytoskeleton abrogated most F-actin based structures in both groups, and revealed the significant contribution of the actin cytoskeleton to the wrinkled NE morphology in the degradable hydrogels (Figure #5B/C). Conversely, no change in NE morphology occurred following CytoD treatment of cells in non-degradable hydrogels, suggesting minimal influence of the actin cytoskeleton on nuclear envelope wrinkling morphology in this context (Figure #5B/C).
Given that reducing actin polymerization reduced nuclear wrinkling in 3D MMP-degradable hydrogels, we wondered whether these wrinkles were due to stress-generating nuclear impingements by the contractile actin cytoskeleton. Closer examination of XZ/YZ transverse slices, and XY slices highlighted multiple locations where filamentous actin impinged on the nuclear envelope in these MMP-degradable NorHA hydrogels (Figure #5D)(Supplemental Figure #10)(Supplemental Movies #1ߝ2). High degrees of actin anisotropy (indicating fibrillar and polarized actin) around the nucleus were measured for cells in MMP-degradable 3D hydrogels, with lower values witnessed for cells embedded in the non-degradable 3D hydrogels (Figure #5E/F). As expected, following cytochalasin D treatment, the anisotropy of this peri-nuclear fibrillar actin decreased, with significant changes observed only in MMP-degradable hydrogels. This suggests that, in 3D, formation of contractile actin-based stress fibers is critical for the production of the wrinkled NE morphology. To quantify these actin-based interactions with the nucleus, we categorized these impingements into distinct classes: 1) no impingement, 2) superficial impingement (small deformations), 3) surface wrinkles and grooves (clearly shaped by an actin fiber), and 4) deep impingements where stress fibers induced invaginations reaching >1 μm into the nucleus (Figure #5G). Quantification of these interactions showed that MSCs in MMP-degradable hydrogels had a higher fraction of surface and deep impingements, compared to cells in non-degradable gels, and that these nuclear envelope impingements were sensitive to actin disruption by cytochalasin D (Figure #5H).
Nuclear envelope wrinkling is a strong predictor of nuclear YAP/TAZ localization in 3D MMP-degradable hydrogels.
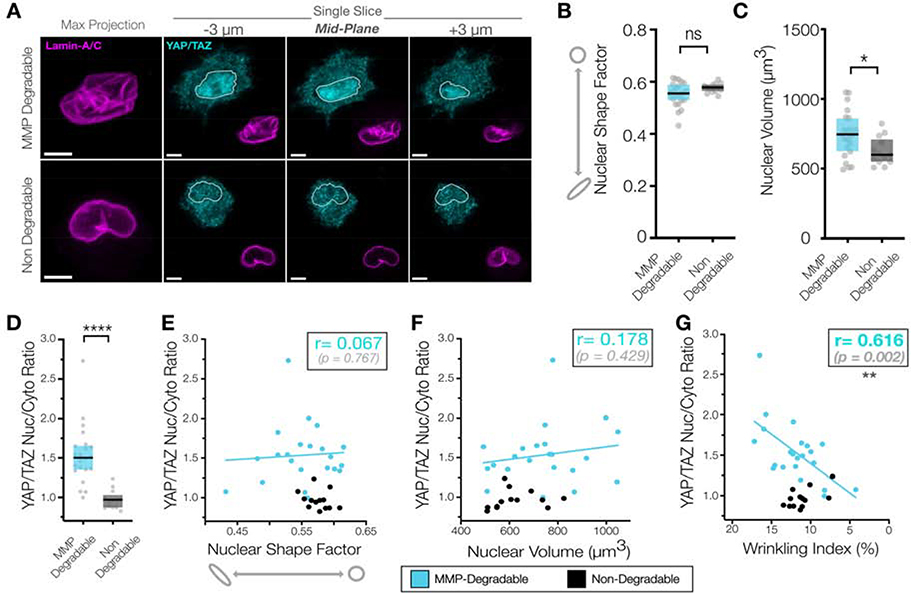
Having established that a wrinkled nuclear morphology was generated by nuclear actin impingements in 3D, we hypothesized that a wrinkled nuclear envelope morphology would correlate with increased nuclear envelope stress and consequently increase YAP nuclear translocation. Following immunostaining and imaging, enhanced nuclear YAP/TAZ signal with a wrinkled NE morphology was evident in cells in MMP-degradable hydrogels (Figure #6A). Nuclear volume was slightly increased in the MMP-degradable context (Figure #6B), with no significant differences in nuclear shape factor between conditions (Figure #6C). We next evaluated the predictive power of various other 3D nuclear morphology metrics (nuclear volume/nuclear shape factor) on YAP/TAZ nuclear localization in 3D compared to the nuclear wrinkling index. Nuclear volume and nuclear shape index were not significantly correlated with nuclear YAP/TAZ localization in MSCs cultured in MMP-degradable hydrogels (r=0.066 & p = 0.767 and r=0.178 & p = 0.429, respectively) (Figure #6D/E). However, we found that an increasing nuclear wrinkling index significantly correlated with increased nuclear YAP/TAZ localization (r=0.616 & p = 0.002), highlighting the predictive capacity of the nuclear envelope wrinkling index metric (Figure #6F). Together, our findings indicate that actin impingement into the nuclear envelope in 3D generates a highly wrinkled nuclear envelope morphology, which results in increased YAP/TAZ nuclear localization. This impingement of actin into the nuclear envelope in multiple planes in 3D hydrogels is similar to a balloon being squeezed against a grate, and this impingement likely drives local stress concentrations enabling the translocation of YAP that ultimately regulates downstream transcriptional responses that define phenotypic switches in differentiation and proliferation that arise from extracellular mechanical inputs.
Figure #6 – Nuclear envelope wrinkling in MMP-degradable 3D NorHA hydrogels is correlated with increased nuclear YAP.
(A) Representative images of YAP/TAZ and Lamin-A/C immunostaining of MSCs cultured in 9 kPa MMP-degradable and non-degradable NorHA hydrogels for 5d prior to fixation and staining, along with the corresponding quantifications of (B) nuclear shape factor and (C) nuclear volume (n=13–23 nuclei/group, * = p <0.05 by Student’s t-test). SB = 5 μm. (D) YAP/TAZ nuclear-to-cytoplasmic ratios of MSCs from A-C (n=13–22 cells, Box plot represents median and 25th/75th quartiles, **** indicates p<0.0001 by Mann-Whitney U-test). (E-G) Quantifications of 3D nuclear metrics including nuclear shape factor (E), nuclear volume (F), and nuclear wrinkling index (G) as compared to YAP/TAZ nuclear-to-cytoplasmic ratios of MSCs from A-C (n=13–22 cells, ** indicates p<0.01 from Pearson’s correlation test, r and p-values shown for MMP-degradable hydrogel fit).
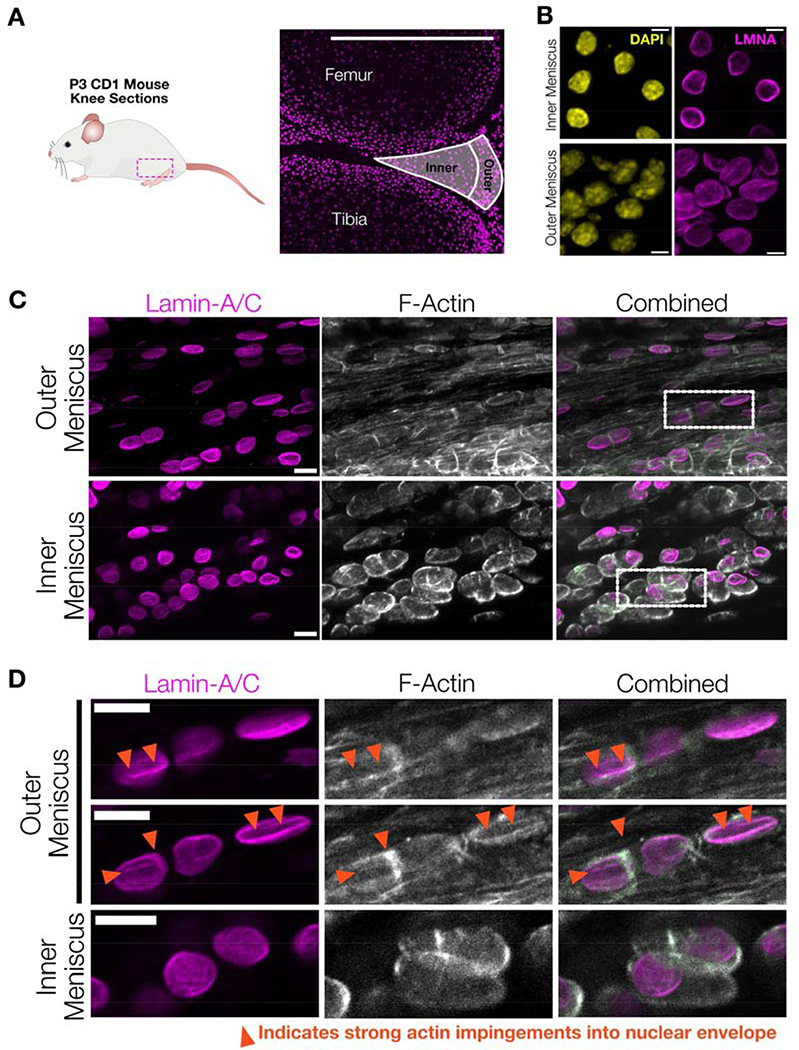
Nuclear wrinkling occurs in developing dense mesenchymal tissues.
Having observed differences in correlation between nuclear wrinkling state and YAP translocation in 2D and 3D, we next sought out to determine which phenomenon was active in native mesenchymal tissues. To accomplish this, we examined the knee meniscus, a tissue that exhibits strong regionality in its composition; with a fibrous, aligned, and collagen-rich ‘outer zone’ that gradually transitions to a more isotropic, proteoglycan-rich cartilaginous ‘inner zone’ 69–76. Immunostaining of nuclei within neonatal mouse knee joint sections (postnatal day 3) (Figure #7A) showed that nuclei in cells in the inner zones exhibited an ellipsoidal morphology, while those in the outer region had a highly wrinkled morphology (Figure #7B). This wrinkled morphology was not easily visible in DAPI stained sections but was readily apparent via lamin-A/C immunostaining. YAP/TAZ immunostaining and quantification confirmed that cells in the outer meniscus had elevated active mechano-signaling, as indicated by increased nuclear to cytoplasmic YAP/TAZ ratios (Supplemental Figure #11).
Figure #7 – Mesenchymal cells in juvenile mouse menisci have wrinkled nuclear envelopes with local actin impingement.
(A) To examine in vivo nuclear morphology of mesenchymal progenitor cells, 7.5 μm-thick sections were taken from the knee of P3 CD1 mice and immunostained. Cells from the inner and outer menisci were analyzed to characterize cell and nuclear morphology in the varying mechanical microenvironments present in the developing meniscus. SB = 500 μm. (B) Representative maximum projection z-stack images of chromatin (DAPI) and lamin-A/C across the inner and outer meniscus regions highlight increased nuclear envelope wrinkling in the outer meniscus region. SB = 5 μm. (C) Representative images taken from outer and inner meniscal regions highlighting fibrillar actin interactions with outer meniscus nuclei in P14 juvenile CD1 mice. SB = 10 μm. (D) Zoomed images from the dotted highlights in C, with two XY-sections from the same z-stack in the outer meniscus and one XY-section from the inner meniscus. These images highlight how fibrillar actin in mesenchymal progenitor cells residing in the outer meniscus can impinge on the nuclear envelope to create a wrinkled morphology (arrows), while mesenchymal progenitors residing in the inner meniscus exhibited more cortical actin structures and less wrinkled nuclei. SB = 10 μm.
To determine if the origin of nuclear wrinkling in the developing meniscus was similar to that seen in engineered 3D microenvironments, we examined actin and lamin-A/C immunostaining in juvenile mouse menisci (postnatal day 14). Filamentous actin staining revealed clear differences in the cytoskeletal architecture between the outer and inner regions, with cells in the outer meniscus region having highly polarized F-actin staining and increased filamentous actin (Figure #7C), while inner meniscus cells exhibited a predominately cortical actin organization. Individual z-stack slices revealed clear instances in the outer meniscus where wrinkles in the nuclear envelope arose along with local actin impingements (Figure #7D). Together with our 3D hydrogel data, these results suggest differences in the manner and mode of nuclear envelope deformation in 3D microenvironments compared to on 2D hydrogels. In engineered 3D hydrogels and in outer meniscal tissue, impingement of the contractile stress fiber network into the nucleus results in increased nuclear deformation and generates increasingly wrinkled nuclear envelopes. While we noted a strong similarity between 3D MMP-degradable hydrogels and mesenchymal progenitor cells within the outer meniscus in juvenile mice, it is likely that the exact relationship between nuclear envelope wrinkling and mechanotransduction will differ greatly across different tissue contexts and cell-types. The exact alignment and arrangement of the cells and ECM in a 3D system likely plays a critical role in regulating polarized actin structures and thereby impingement into the nuclear envelope in 3D77.
DISCUSSION
Our work in 2D hydrogel platforms revealed that the degree of nuclear envelope (NE) wrinkling in mesenchymal progenitor cells can predict their focal adhesion maturation state and YAP/TAZ nuclear localization. Nuclear envelope wrinkling was evident when mesenchymal stem cells were cultured on soft planar 2D substrates (on which cells exhibit apical-basal polarity, Figure #1). The degree of NE wrinkling changed rapidly in response to modulation of cell contractility (with changes seen as quickly as 30 min following perturbation, Figure #2C–F). We found that nuclear envelope wrinkling could change in the absence of lamin-A/C degradation (Supplemental Figure #4) and other groups have shown changes in nuclear envelope wrinkling without transcription or translation30, suggesting that short-term changes in wrinkling occur primarily through deformation rather than through other signaling pathways. Increases in substrate stiffness generally reduced the wrinkling index, though there was considerable heterogeneity across the population (Figure #3A). However, when the wrinkling index for individual cells was compared to cell area or focal adhesion number or size, a clear biphasic pattern was observed that fit cells across multiple substrate moduli. Specifically, cellular spreading and focal adhesion maturation remained low until a critical threshold of nuclear wrinkling was reached (Figure #3D–F), suggesting a link between the nuclear envelope wrinkling state and cellular adhesion and spreading in 2D. This may suggest that removal of nuclear wrinkling through increased actomyosin contractility proceeds until a transition point, wherein the nucleus becomes taut and adhesions mature, leading to increased cell spreading. This biphasic response is similar to that seen in recent work exploring how force-dependent talin unfolding regulates the mechanotransductive response to material stiffness78.
We further perturbed the coupling of the ECM to the nucleus by knocking down nesprin-1G, a key component of the LINC complex. Knockdown of nesprin-1G resulted in a dramatic increase in nuclear envelope wrinkling and this wrinkling was not rescued by increases in actomyosin contractility resulting from LPA treatment (Figure #4B–F). Recent work has shown that exogenous mechanical inputs from the microenvironment are transduced through the actin cytoskeleton to the nuclear envelope, and that stress in the nuclear envelope physically stretches nuclear pore complexes to bias nuclear import of key transcriptional co-activators like YAP22. We found that nuclear envelope wrinkling in cells on 2D planar hydrogels was a good predictor of increased YAP/TAZ nuclear localization, wherein cells with taut nuclear envelopes had higher YAP/TAZ nuclear (Figure #4A). In cells depleted of nesprin-1G (which have high wrinkling levels that are not altered by increased actomyosin contractility), increased substrate stiffness did not result in YAP/TAZ nuclear localization (Figure #4F). This is consistent with the idea that nuclear envelope wrinkling represents a state of low tension of the nuclear envelope. Importantly, the lack of nuclear envelope tension present in wrinkled nuclei may also regulate the function and signaling of other NE-anchored proteins, such as emerin14, 79, nuclear pores, and calcium channels. Recent work showed that deformation of the nuclear envelope can trigger Ca2+ release from the ER, which then regulates repressive histone mark deposition of H3K9me3 and H3K27me380, and other studies have found microtubule-induced nuclear wrinkling in non-adhesive cells was correlated with a local loss of H3K9me354. A pair of recent studies also examined how nuclear envelope wrinkling could modulate calcium-sensitive cPLA2 signaling 30, 81. Together, this work highlights how alterations in nuclear envelope morphology may feedback to dictate other signaling events critical to regulating gene expression and downstream cell behavior.
Our data in a 2D context supports and extends a previously described model, wherein basal levels of nuclear envelope wrinkling are actively removed during events involving increased actomyosin contractility. Increased contractile energy in the actin cytoskeleton works to deform and flatten the nucleus, resulting in two regimes of nuclear signaling (Figure #3E/G/H) 26, 33. First, in wrinkled nuclei, actomyosin contractility functions to initially remove the free surface area of the nuclear envelope without tensioning the nuclear envelope itself. Given the lack of tension in this regime, YAP nuclear translocation remains low. However, once a threshold of nuclear envelope deformation and unfurling of wrinkles is reached (such that there is no more free surface area and the nuclear envelope becomes taut), increases in actomyosin contractility results in additional deformation of the nucleus and increased stress within the nuclear envelope, which have previously been shown to promote YAP translocation into the nucleus by biasing import through nuclear pores22. Increased mechanical feedback in this state likely comes from both the transcriptional feedback of YAP82 as well as through mechanical engagement of the nucleus83. As such, nuclear envelope wrinkles likely function as a geometric reserve that must be depleted prior to robust cellular mechano-responses.
This observation of a biphasic nuclear mechano-response in cells cultured on 2D hydrogels is also in accordance with modeling-based predictions regarding the diverse roles of chromatin and lamin in nuclear mechanics26, 84. Polymeric modeling of lamin-A/C in cells suggests that the nucleus behaves like a buckled polymer shell at low strains, with the lamin-A/C network insufficiently engaged and providing little mechanical strength84. This model aligns with our data, which showed that in 2D, the entrance into a new nuclear mechanical regime was mediated by the removal of free surface area of the nuclear envelope stored in wrinkles (Figure 2). Micropipette aspiration of isolated nuclei has also shown that chromatin and lamin can coordinate to produce different mechanical responses, where chromatin appears to be the main determinant of the nuclear mechanical response at low strains85. Our data suggests that the transition from a chromatin-dominated to a lamin-dominated regime may be mediated by availability of nuclear envelope slack (in the form of wrinkles). This would imply that, until nuclear envelope wrinkles are removed, and tension is able to develop, chromatin would resist the majority of applied load. The ‘toe region’ of nuclear envelope wrinkling may therefore represent a condition in which exogenous mechanical forces are able to readily alter chromatin topology. This is consistent with both micropipette aspiration data86 and recent studies employing new genomic techniques7. Both indicate that specific regions of the chromatin can be physically deformed, and in some cases this mechanical deformation is sufficient to alter the transcriptional state of the deformed region.
While nuclear wrinkling appears to be a good predictor of mechanical signaling in cells in our system, it is important to note that nuclear wrinkling is regulated by a variety of interconnected factors across cell types and niches that could influence the transfer of mechanical forces. This may include: lamin-A/C and lamin-B levels 31, the inherent contractile state of the cell, the patency of cytoskeletal-to-nuclear coupling, actin organization/alternative mechanisms of traction generation26, and cellular polarization32. There are additional cases where various perturbations to these factors may change the setpoints of this response. For example, depletion of Lamin-A/C qualitatively increases baseline nuclear wrinkling 24, 31, 87, which may suggest that the amount of lamin-A/C in a given cell could tune the amount of wrinkling (and as such, reserve capacity of the mechanical buffer that wrinkling provides) in a given microenvironment to regulate its mechano-response87. Additionally, cells can still spread and flatten their nuclei with inhibition of contractility via Y-2763226, and so it is possible that nuclear envelope wrinkles may be removed if contractile inhibition still allows for robust spreading.
In contrast to our findings in 2D, nuclear envelope morphology in 3D environments showed a different response, where increased actomyosin activity resulted in increased wrinkling and YAP nuclear localization. In a non-degradable 3D environment (where cell spreading is not possible and stress fibers do not develop), nuclei were generally smooth and had low levels of nuclear YAP/TAZ (Figure #5B/C, Figure #6A/D). However, when cell spreading was enabled (via crosslinking with MMP-degradable peptides65, 88), an isotropic filamentous actin cytoskeleton developed and elements of this network impinged on the nuclear envelope, resulting in highly wrinkled nuclear envelopes in 3D (Figure #5B–H). Surprisingly, in 3D, the nuclear wrinkling index was the only good morphometric predictor of nuclear YAP localization (with increased nuclear wrinkling correlating with increased nuclear/cytoplasmic YAP/TAZ, Figure #6). In fibrous native tissues (such as early post-natal mouse menisci), we also noted filamentous actin fibers deforming the nuclear envelope in the outer meniscus regions, where cells also showed increased nuclear YAP/TAZ localization (Figure #6D–G, Supplemental Figure #11).
Consistent with our findings in 3D, other groups have shown increased nuclear curvature in cells cultured in stiff 3D alginate IPNs compared to soft IPNs (assessed via DAPI staining and TEM), which is comparable to our findings in MMP-degradable hydrogels.89 We hypothesize that, in 3D, contractile actomyosin impingements may act to ‘pinch off’ portions of the nucleus, tensioning the free surface area of the nuclear envelope locally. This local impingement would thereby allow for sufficient nuclear envelope tension to develop resulting in increased YAP nuclear translocation. It remains to be seen if the actin impingements in 3D environments cause local or global increases in tension in the nuclear envelope; direct evaluation of cytoskeletal-to-nuclear force transfer90 and tension in the nuclear envelope will be necessary to confirm this hypothesis. However, recent advances in membrane tension sensor design may enable some of these measurements to be made.91 That said, the filamentous sub-structure of lamin92 combined with the evidence of stress-generated nuclear blebs93 followed by envelope rupture, and the force-dependent opening of cryptic lamin-A/C sites in nuclear sub-domains,20 all support the idea that local stress concentrations may be present in the nuclear lamina.
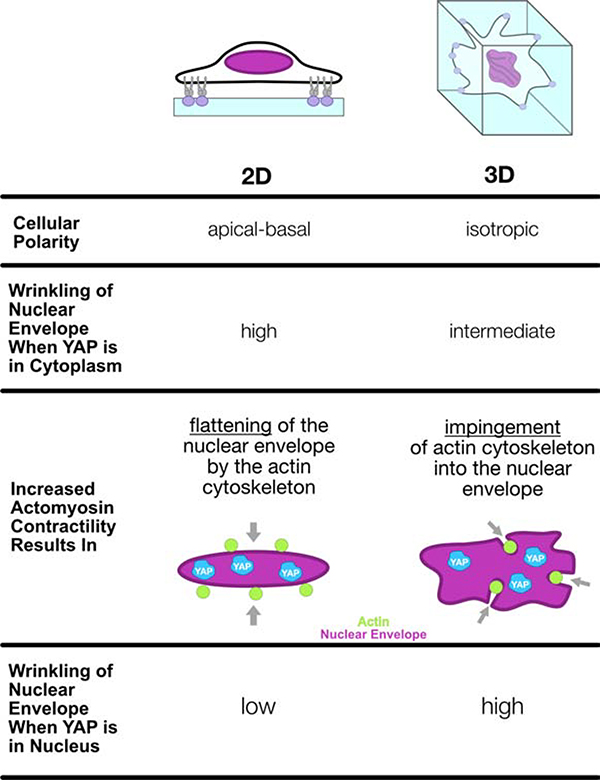
This marked difference in nuclear envelope mechano-response based on material dimensionality and degradability can be explained in part by the boundary conditions of each system (Figure #8). In 2D, there exists an anisotropic set of forces on the nucleus with a clear directionality (exhibiting apical-basal polarity) where the nucleus is sandwiched between apical and basal layers of actin filaments and then additionally tensioned through lateral actin connections. The result of this set of mechanical inputs is a nucleus that resists actin stress fiber impingement as the cell spreads and a subsequent flattening/tautening of the nuclear envelope. However, in 3D cellular spreading we find actin filament formation that intersect with the nucleus in different directions and planes. Thus, when actin stress fibers do form in 3D between two points of matrix connection, they may readily deform and impinge into the nucleus and instigate YAP/TAZ translocation. As such, this mechanical reserve provided by the nuclear wrinkles is engaged in different mechanisms in 2D and 3D, where the nuclear envelope is pulled taut and flattened in 2D as compared to being locally unfurled and tensioned by local nuclear impingements in 3D. Our observations in 3D (in both native tissue and engineered systems) also implicate the importance of the ECM organization and alignment in dictating nuclear envelope deformation and subsequent nuclear shuttling of transcription factors. These differences in the microenvironmental context of the cell are likely strong contributors to observed differences in histone mark deposition and other critical mechano-signaling events resulting from nuclear deformation across various cell types and environments. Further study of nuclear function and signaling in biomaterial platforms that reproduce physiologic ECM dimensionality, degradability, and stiffness may recapitulate the complicated mechanotransductive pathways operative in native tissue and provide new insight in mechanobiology that can be leveraged to improve tissue formation or alter disease progression.
Figure #8 – A summary of how microenvironment dimensionality influences morphologic changes of the nuclear envelope in response to increased actomyosin contractility.
MSCs cultured in a variety of physiologically-soft engineered material platforms exhibit a nuclear envelope with free surface area that manifests as a wrinkled morphology. Increasing actomyosin contractility first functions to deform and remove this free surface area prior to robust mechano-responses. (2D, left) On 2D MeHA hydrogels, MSCs exhibit clear apical-basal polarity, and increases in actomyosin contractility remove free surface area from the wrinkled nuclear envelope through flattening and tensioning by the actin cytoskeleton. This eventually results in a taut nuclear envelope and conditions where YAP/TAZ is mostly in the nucleus. (3D, right) Conversely, 3D NorHA hydrogels crosslinked with MMP-degradable peptides allow for spreading of embedded MSCs without a specific plane of polarization of filamentous actin. Increases in actomyosin contractility result in deep actin impingements into the nuclear envelope, thereby generating highly tortuous and wrinkled nuclei in cells with nuclear YAP/TAZ localization. As such, the mechanical reserve provided by nuclear wrinkles is engaged in two different mechanisms in 2D and 3D, where the nuclear envelope is pulled taut and flattened in 2D as compared to being locally unfurled and tensioned by local actin impingements in 3D.
Supplementary Material
Supplemental Movie #1- Representative z-stack of F-Actin (white) and lamin-A/C immunostaining of an MSC cultured in a MMP-degradable NorHA hydrogel.
Supplemental Movie #2- Representative z-stack of F-Actin (white) and lamin-A/C immunostaining of an MSC cultured in a non-degradable NorHA hydrogel.
Supplemental File #1- ImageJ Macro for Determining Wrinkle Index from high-resolution z-stacks of the nuclear lamina.
Supplemental File #2- Test z-stack of LMNA immunostaining in MSCs to be used for testing and configuring the ImageJ Macro.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (R01 EB008722, R01 AR056624, R01 AR071399) and the National Science Foundation-sponsored Center for Engineering Mechanobiology (CMMI-1548571). The authors would like to thank the CDB microscopy core at Penn for the use of core equipment and assistance with STED microscopy, Dr. Xi Jiang for help with tissue sectioning, Ryan Daniels for help with preliminary studies, and Dr. Steven Caliari, Dr. Benjamin Freedman, and Dr. Brianne Connizzo for helpful discussions.
Footnotes
COMPETING FINANCIAL INTERESTS STATEMENT
The authors have no competing financial interests.
DATA AVAILABILITY STATEMENT
All data are available upon request.
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Engler AJ, Sen S, Sweeney LH & Discher DE Matrix elasticity directs stem cell lineage specification. Cell 126 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Dupont S et al. Role of YAP/TAZ in mechanotransduction. Nature Publishing Group 474 (2011). [Google Scholar]
- 3.Gilbert PM et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. American Association for the Advancement of Science 329 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu F et al. Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. The Journal of cell biology 190, 693–706 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho C, Jaalouk DE, Vartiainen MK & Lammerding J Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature 497, 507–511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swift J et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tajik A et al. Transcription upregulation via force-induced direct stretching of chromatin. Nature Materials 15, 1287–1296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panciera T, Azzolin L, Cordenonsi M & Piccolo S Mechanobiology of YAP and TAZ in physiology and disease. Nat Rev Mol Cell Biol 18, 758–770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li CX et al. MicroRNA-21 preserves the fibrotic mechanical memory of mesenchymal stem cells. Nature materials (2016). [DOI] [PubMed] [Google Scholar]
- 10.Jain N, Iyer KV, Kumar A & Shivashankar GV Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proceedings of the National Academy of Sciences of the United States of America 110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avvisato CL et al. Mechanical force modulates global gene expression and beta-catenin signaling in colon cancer cells. J Cell Sci 120, 2672–2682 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Wei SC et al. Matrix stiffness drives epithelial–mesenchymal transition and tumour metastasis through a TWIST1–G3BP2 mechanotransduction pathway. Nat Cell Biol 17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dingal DPCP et al. Fractal heterogeneity in minimal matrix models of scars modulates stiff-niche stem-cell responses via nuclear exit of a mechanorepressor. Nature Materials 14, 951–960 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guilluy C et al. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat Cell Biol 16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heo S-J, Cosgrove BD, Dai EN & Mauck RL Mechano-Adaptation of the Stem Cell Nucleus. Nucleus (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pajerowski DJ, Dahl K, Zhong FL, Sammak PJ & Discher DE Physical plasticity of the nucleus in stem cell differentiation. Proceedings of the National Academy of Sciences 104, 15619–15624 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lammerding J et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. Journal of Clinical Investigation 113, 370 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK & Wilson KL The nuclear lamina comes of age. Nat Rev Mol Cell Biol 6, 21–31 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Heo S-JJ et al. Differentiation alters stem cell nuclear architecture, mechanics, and mechano-sensitivity. eLife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ihalainen TO et al. Differential basal-to-apical accessibility of lamin A/C epitopes in the nuclear lamina regulated by changes in cytoskeletal tension. Nature Materials 14, 1252–1261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buxboim A et al. Matrix Elasticity Regulates Lamin-A,C Phosphorylation and Turnover with Feedback to Actomyosin. Current Biology 24, 1909–1917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elosegui-Artola A et al. Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 171, 1397–1410 e1314 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Kim D-HH & Wirtz D Cytoskeletal tension induces the polarized architecture of the nucleus. Biomaterials 48, 161–172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buxboim A et al. Coordinated increase of nuclear tension and lamin-A with matrix stiffness outcompetes lamin-B receptor that favors soft tissue phenotypes. Mol Biol Cell 28, 3333–3348 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D-HH et al. Volume regulation and shape bifurcation in the cell nucleus. Journal of cell science 129, 457 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y et al. Moving Cell Boundaries Drive Nuclear Shaping during Cell Spreading. Biophys J 109, 670–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neelam S, Hayes PR, Zhang Q, Dickinson RB & Lele TP Vertical uniformity of cells and nuclei in epithelial monolayers. Sci Rep 6, 19689 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorgens DM et al. Deep nuclear invaginations are linked to cytoskeletal filaments – integrated bioimaging of epithelial cells in 3D culture. Journal of Cell Science 130, 177–189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katiyar A et al. Nuclear size changes caused by local motion of cell boundaries unfold the nuclear lamina and dilate chromatin and intranuclear bodies. Soft Matter 15, 9310–9317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lomakin AJ et al. The nucleus acts as a ruler tailoring cell responses to spatial constraints. bioRxiv (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JK et al. Nuclear lamin A/C harnesses the perinuclear apical actin cables to protect nuclear morphology. Nat Commun 8, 2123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Versaevel M et al. Super-resolution microscopy reveals LINC complex recruitment at nuclear indentation sites. Scientific Reports 4, 7362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lele TP, Dickinson RB & Gundersen GG Mechanical principles of nuclear shaping and positioning. J Cell Biol 217, 3330–3342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burdick JA, Chung C, Jia X, Randolph MA & Langer R Controlled degradation and mechanical behavior of photopolymerized hyaluronic acid networks. Biomacromolecules 6 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marklein RA & Burdick JA Controlling stem cell fate with material design. WILEY-VCH Verlag 22 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Cosgrove BD et al. N-cadherin adhesive interactions modulate matrix mechanosensing and fate commitment of mesenchymal stem cells. Nature materials 15, 1297–1306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loebel C et al. Metabolic Labeling to Probe the Spatiotemporal Accumulation of Matrix at the Chondrocyte–Hydrogel Interface. Advanced Functional Materials n/a, 1909802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vega SL et al. Combinatorial hydrogels with biochemical gradients for screening 3D cellular microenvironments. Nat Commun 9, 614 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dimitriadis EK, Horkay F, Maresca J, Kachar B & Chadwick RS Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophys J 82, 2798–2810 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang AH, Yeger-McKeever M, Stein A & Mauck RL Tensile properties of engineered cartilage formed from chondrocyte- and MSC-laden hydrogels. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society 16 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Driscoll TP, Cosgrove BD, Heo S-J, Shurden ZE & Mauck RL Cytoskeletal to Nuclear Strain Transfer Regulates YAP Signaling in Mesenchymal Stem Cells. Biophysical journal 108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berginski ME & Gomez SM The Focal Adhesion Analysis Server: a web tool for analyzing focal adhesion dynamics. F1000Res 2, 68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aratyn-Schaus Y, Oakes PW, Stricker J, Winter SP & Gardel ML Preparation of complaint matrices for quantifying cellular contraction. Journal of visualized experiments : JoVE (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tseng Q et al. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. National Acad Sciences 109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dembo M, Oliver T, Ishihara A & Jacobson K Imaging the traction stresses exerted by locomoting cells with the elastic substratum method. Biophysical journal (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalajzic I et al. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res 17, 15–25 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Dyment NA et al. High-Throughput, Multi-Image Cryohistology of Mineralized Tissues. J Vis Exp (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gramlich WM, Kim IL & Burdick JA Synthesis and orthogonal photopatterning of hyaluronic acid hydrogels with thiol-norbornene chemistry. Biomaterials 34, 9803–9811 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wade RJ, Bassin EJ, Rodell CB & Burdick JA Protease-degradable electrospun fibrous hydrogels. Nat Commun 6, 6639 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boudaoud A et al. FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat Protoc 9, 457–463 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Elosegui-Artola A et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol (2016). [DOI] [PubMed] [Google Scholar]
- 53.Khatau SB et al. A perinuclear actin cap regulates nuclear shape. Proceedings of the National Academy of Sciences of the United States of America 106, 19017–19022 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biedzinski S et al. Microtubules deform the nucleus and force chromatin reorganization during early differentiation of human hematopoietic stem cells. bioRxiv (2019). [Google Scholar]
- 55.Heo S-J et al. Biophysical Regulation of Chromatin Architecture Instills a Mechanical Memory in Mesenchymal Stem Cells. Scientific Reports, 16895 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Irianto J, Lee DA & Knight MM Quantification of chromatin condensation level by image processing. Medical engineering & physics 36, 412–417 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Irianto J et al. Osmotic challenge drives rapid and reversible chromatin condensation in chondrocytes. Biophysical journal 104, 759–769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Connizzo BK, Yannascoli SM & Soslowsky LJ Structure–function relationships of postnatal tendon development: A parallel to healing. Matrix Biology, 106–116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dahl KN, Kahn SM, Wilson KL & Discher DE The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci 117, 4779–4786 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Chancellor TJ, Lee J, Thodeti CK & Lele T Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced …. Biophysical journal (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Belaadi N, Aureille J & Guilluy C Under Pressure: Mechanical Stress Management in the Nucleus. Cells 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baker BM & Chen CS Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. Journal of cell science, 3015–3024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chaudhuri O et al. Substrate stress relaxation regulates cell spreading. Nat Commun 6, 6364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chaudhuri O et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater 15, 326–334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caliari SR, Vega SL, Kwon M, Soulas EM & Burdick JA Dimensionality and spreading influence MSC YAP/TAZ signaling in hydrogel environments. Biomaterials 103, 314–323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khetan S et al. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nature Materials 12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Madl CM et al. Maintenance of neural progenitor cell stemness in 3D hydrogels requires matrix remodelling. Nat Mater 16, 1233–1242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trappmann B et al. Matrix degradability controls multicellularity of 3D cell migration. Nat Commun 8, 371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Q et al. Biomechanical properties of murine meniscus surface via AFM-based nanoindentation. Journal of Biomechanics 48, 1364–1370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Q et al. Micromechanical anisotropy and heterogeneity of the meniscus extracellular matrix. Acta Biomaterialia, 356–366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McNulty AL & Guilak F Mechanobiology of the meniscus. Journal of Biomechanics 48, 1469–1478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qu F, Holloway JL, Esterhai JL, Burdick JA & Mauck RL Programmed biomolecule delivery to enable and direct cell migration for connective tissue repair. Nat Commun 8, 1780 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanchez-Adams J, Willard VP & Athanasiou KA Regional variation in the mechanical role of knee meniscus glycosaminoglycans. Journal of Applied Physiology, 1590–1596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skaggs DL, Warden WH & Mow VC Radial tie fibers influence the tensile properties of the bovine medial meniscus. Journal of orthopaedic research : official publication of the Orthopaedic Research Society, 176–185 (1994). [DOI] [PubMed] [Google Scholar]
- 75.Upton ML, Chen J & Setton LA Region-specific constitutive gene expression in the adult porcine meniscus. Journal of Orthopaedic Research, 1562–1570 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Vanderploeg EJ, Wilson CG, Imler SM, Ling C & Levenston ME Regional variations in the distribution and colocalization of extracellular matrix proteins in the juvenile bovine meniscus. Journal of anatomy, 174–186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bao M, Xie J, Piruska A & Huck WTS 3D microniches reveal the importance of cell size and shape. Nat Commun 8, 1962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Elosegui-Artola A et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol 18, 540–548 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Le H et al. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nature Cell Biology 18, 864–875 (2016). [DOI] [PubMed] [Google Scholar]
- 80.Nava MM et al. Heterochromatin-Driven Nuclear Softening Protects the Genome against Mechanical Stress-Induced Damage. Cell (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Venturini V et al. The nucleus measures shape deformation for cellular proprioception and regulates adaptive morphodynamics. bioRxiv (2019). [Google Scholar]
- 82.Nardone G et al. YAP regulates cell mechanics by controlling focal adhesion assembly. Nat Commun 8, 15321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cao X et al. A Chemomechanical Model of Matrix and Nuclear Rigidity Regulation of Focal Adhesion Size. Biophys J 109, 1807–1817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Banigan EJ, Stephens AD & Marko JF Mechanics and Buckling of Biopolymeric Shells and Cell Nuclei. Biophysical Journal 113, 1654–1663 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stephens AD, Banigan EJ, Adam SA, Goldman RD & Marko JF Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Molecular biology of the cell 28, 1984–1996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maniotis AJ, Chen CS & Ingber DE Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proceedings of the National Academy of Sciences 94, 849–854 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koushki N et al. Lamin A redistribution mediated by nuclear deformation determines dynamic localization of YAP. bioRxiv (2020). [Google Scholar]
- 88.Khetan S et al. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat Mater 12, 458–465 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stowers RS et al. Matrix stiffness induces a tumorigenic phenotype in mammary epithelium through changes in chromatin accessibility. Nat Biomed Eng 3, 1009–1019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arsenovic PT et al. Nesprin-2G, a Component of the Nuclear LINC Complex, Is Subject to Myosin-Dependent Tension. Biophysical Journal 110, 34–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Colom A et al. A fluorescent membrane tension probe. Nat Chem 10, 1118–1125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turgay Y et al. The molecular architecture of lamins in somatic cells. Nature 543, 261–264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Denais CM et al. Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Movie #1- Representative z-stack of F-Actin (white) and lamin-A/C immunostaining of an MSC cultured in a MMP-degradable NorHA hydrogel.
Supplemental Movie #2- Representative z-stack of F-Actin (white) and lamin-A/C immunostaining of an MSC cultured in a non-degradable NorHA hydrogel.
Supplemental File #1- ImageJ Macro for Determining Wrinkle Index from high-resolution z-stacks of the nuclear lamina.
Supplemental File #2- Test z-stack of LMNA immunostaining in MSCs to be used for testing and configuring the ImageJ Macro.