Abstract
BACKGROUND.
The TRansgenic Adenocarcinoma of the Mouse Prostate (TRAMP) model remains one of the most widely used transgenic mouse models of prostate cancer. This is due to its ability to recapitulate with ~100% penetrance multiple aspects of the human disease such as prostatic intraepithelial neoplasia lesions, invasive carcinoma, progression to castration-resistant prostate cancer including aggressive neuroendocrine prostate cancer and metastasis. Despite its popularity, the use of TRAMP mice is limited/slowed by the inability to distinguish the zygosity of the TRAMP transgene. This is especially true for breeding strategies implementing multiple crosses and alleles and when the rapid generation of large animal cohorts with the desired genotype is needed.
METHODS.
We developed a quantitative PCR (qPCR) approach to determine the relative TRAMP transgene copy number of mice.
RESULTS.
This method was validated by three independent laboratories across two institutions, which successfully identified the genotype of the mice 98.2% of the time (165/168) in the first attempt. The genotypes of the uncertain mice were correctly identified in the repeated experiments.
CONCLUSIONS.
We develop the first straightforward, quantitative PCR (qPCR) approach to reliably determine the TRAMP transgene zygosity. The development of this qPCR-based genotyping method enables researchers to streamline breeding strategies when creating complex genetic mouse models involving TRAMP mice; thus, ultimately reducing the required animal numbers, cost, and investigator time.
Prostate cancer remains the second leading cause of death among men with 31,620 estimated deaths in 2019 (1). Conventional approaches and large-scale genomic studies in both primary prostate tumors and metastatic castration-resistant prostate cancers (mCRPC) have identified recurrent DNA copy-number changes, mutations, rearrangements, and gene fusions (2). Many genetically engineered mouse models (GEMMs) in which the prostate epithelium has been engineered to express oncogenic elements (e.g. Large T antigen, Myc, ERG) and/or delete various tumor suppressors have been developed to model prostate cancer progression in mice, elucidate molecular and cellular mechanisms underlying tumor progression and therapy resistance, and serve as preclinical models for testing cancer prevention and therapeutic approaches (2, 3). TRAMP (TRansgenic Adenocarcinoma of the Mouse Prostate) is one of the most well-known prostate cancer mouse models. The TRAMP model uses the minimal rat probasin promoter to drive the tissue-specific expression of the large and small SV40 tumor antigens in the prostatic epithelium (4–6), resulting in the inactivation of p53 and Rb1 as well as PP2A tumor suppressors in the prostate (7, 8). This model recapitulates many features of human prostate cancer, such as different grades of lesions, transition from androgen-dependence to androgen-independence upon hormone therapy, and distant metastasis (8–10). Importantly, TRAMP mice, in a strain-influenced manner, can also develop poorly differentiated neuroendocrine prostate cancer (NEPC) in aged or castrated mice at a higher frequency than other models (8–10).
In recent years, due to the heightened use of highly potent, second-generation androgen receptor pathway inhibitors (e.g., enzalutamide, apalutamide and abiraterone) in patients with CRPC, treatment-induced, neuroendocrine prostate cancer (t-NEPC) is increasingly observed in the clinic (11–13). Recent clinical and preclinical studies suggest that t-NEPCs are enriched for RB and TP53 mutations (14–16) and involve neuroendocrine transdifferentiation (17–19). While many genes have been found to be amplified, mutated, or deleted in NEPC, it remains to be determined which are the driver genes that may cooperate with Rb1/p53 loss/mutations or are required for the Rb1/p53-deficient tumors. Further, even though the complex multi-allele Trp53/Rb1 prostate-specific knockout mouse model develops NEPC (18), it is easier to model NEPC in GEMMs by crossing new alleles into TRAMP mice, as shown in a recent report (20). Importantly, the availability of TRAMP mice in pure C57BL/6 or FVB/NJ backgrounds facilitates the study of the tumor microenvironment in NEPC progression and resistance to immunotherapy. To facilitate the use of TRAMP mice further, we sought to develop a method that could determine the zygosity of the TRAMP transgene and therefore streamline breeding strategies.
Current genotyping methods for TRAMP transgene (Tg) use melting curve analysis or standard PCR followed by gel electrophoresis with primers from the Jackson Laboratory protocol for Tg (TRAMP)8247Ng stock mice or prior reports (8). However, these methods cannot differentiate heterozygous Tg from homozygous Tg. The incorrect determination of TRAMP transgene copy number will confound interpretation of the results from any complex GEMM model involving the TRAMP mice and limit the ability to use the TRAMP model for preclinical studies. Because quantitative PCR (qPCR) is able to accurately determine DNA copy number, we decided to perform qPCR to determine the copy number of Tg by normalizing Tg Ct value to an internal control gene, Gabra1, which we have found to be a faithful internal control across diverse mouse genetic backgrounds (21). PerfeCTa SYBR Green FastMix (Quantabio Inc.) was used for qPCR. To ensure the robust performance of this assay, we performed genotyping in a single-blind fashion in two laboratories. Except for the control wild type (WT), heterozygous (Het), and homozygous (Homo) mouse samples, the persons who performed the genotyping did not know the genotypes of the breeding pairs who gave rise to these litters. The genotypes for all homozygous control mice used in the study were confirmed by breeding the putative homozygous mice to wild type mice, which generated litters that were all positive for the Tg confirmed by standard PCR genotyping method. The control heterozygous mice were litters from the confirmed homozygous mice bred to wildtype animals. The detailed protocol was described in Supplementary Materials and Methods.
To test whether our qPCR approach could accurately determine the TRAMP Tg copy number, we first extracted gDNA prepared from tail tip or ear punch using the DNeasy Blood & Tissue Kits (Qiagen Inc.) and performed qPCR analysis of control mice using 2–10 ng gDNA as templates in 10 μl reaction volumes. Our qPCR assay correctly identified the genotypes of the control mice: no PCR product was detected from the PCR reactions using WT mice and the copy number of TRAMP transgene in the Homo mice was 2-fold of the Het mice when Gabra1 was used as a control for normalization (Fig. 1A). We then performed qPCR using purified gDNA to determine the copy number of 14 TRAMP Tg positive mice and 1 TRAMP negative mouse that were from various litters and kept for experimental or breeding cohorts and were previously genotyped by standard PCR method from the Wang laboratory. Genotypes for these mice were correctly identified based on the normalized results, with copy numbers close to 0, 1, and 2 for WT, Het, and Homo respectively (Fig. 1B). We then determined whether crude gDNA prepared by NaOH extraction (22) worked in our qPCR assay. Consistent with the results using purified gDNA, qPCR using crude gDNA correctly identified the genotypes of the control mice (Fig. 1C). In addition, qPCR using crude gDNA from 31 mice (n = 3 replicates) from the Wang laboratory correctly identified (WT, Het, Homo) the first time in all but two of the unknown samples (Fig. 1D). The two uncertain mice were re-genotyped and conformed to be Het (Fig. 1E). Although the mendelian ratio was slightly skewed towards more Het (Het: Homo=1.54: 1), it could be due to the small litter size or due to the breeding of TRAMP mice to the Probasin4-Cre and a conditional allele that the Wang lab was working on. Moreover, this qPCR assay (n = 2 replicates) was able to correctly identify the genotypes of a total of 93 mice from the Frigo laboratory (45 WT, 44 Het, and 4 Homo; 4 gDNA samples from Homo mice confirmed through breeding were seeded in a single-blind manner amongst the 45 WT and 44 Het gDNA samples) and 12 mice which were previously confirmed by breeding in the Foster laboratory (4 WT, 4 Het, and 4 Homo). Furthermore, to test the robustness of this assay, qPCR analysis of 5 WT, 8 Het, and 4 Homo mice, whose genotypes have been previously confirmed by experimental breeding in the Foster lab, was performed in the Wang lab in a single-blind fashion. The results correctly identified the genotypes the first time for all except two mice (data not shown). Of these two mice, one Homo mouse and one Het mouse were identified as Het and WT, respectively. Of note, no WT or Het mice were misidentified as Homo indicating mice identified as Homo were correct every time. A repeated PCR correctly identified the genotypes for the two uncertain mice. Thus, this protocol was tested in three different laboratories across two institutions to screen 168 mice and was able to successfully identify the genotype of the mice 98.2% of the time (165/168).
Figure 1.
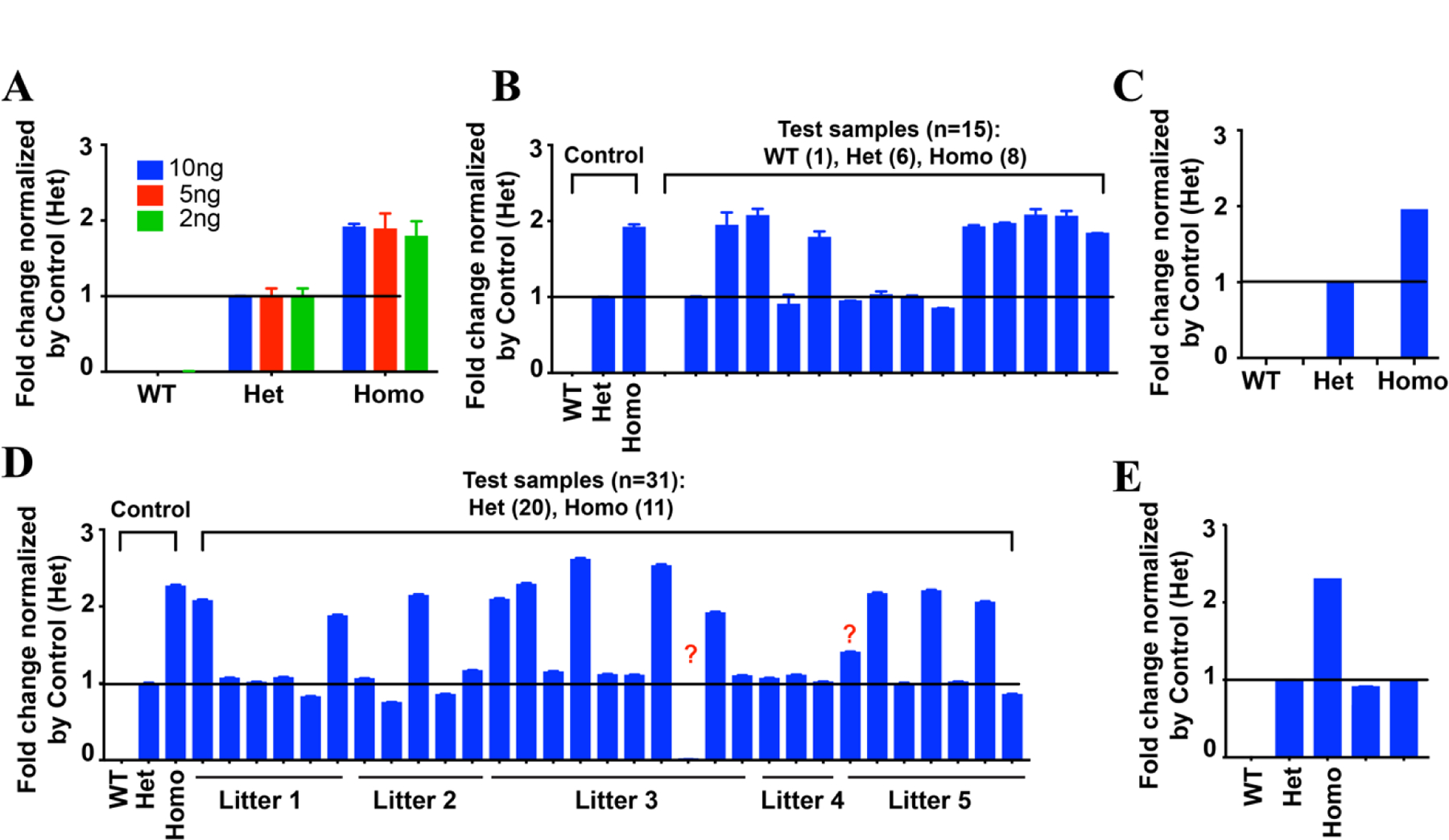
(A) TRAMP Tg copy number was confirmed by qPCR of 3 mice with known genotypes (confirmed by breeding) (WT, Het, Homo) using various amounts of purified gDNA. (B) qPCR genotyping of purified gDNA from 14 TRAMP Tg+ mice from multiple litters and 1 WT mouse. Mice were genotyped using a conventional PCR method and used in experimental cohorts or breeding cohorts in the Wang lab. (C) qPCR of 3 mice with known genotypes (WT, Het, Homo) using crude gDNA. (D) qPCR genotyping of 31 mice with unknown genotypes from 5 litters: 4 of the breeding pairs were TRAMP Homo crossed with TRAMP Het except litter 4, which was TRAMP Het crossed with WT. The copy number for the mouse denoted with “?” is 0.0000608 and 1.4, respectively. (E) The genotypes of the mice marked with “?” in D were re-tested and found to be heterozygous with copy number of 0.914 and 1.007, respectively.
Taken together, this qPCR genotyping assay for the TRAMP transgene is robust and useful for genotyping compound mutant mice involving the crossing of TRAMP mice with other alleles. Further, we anticipate that this approach can be easily adapted to determine the copy number of other Tg mice by substituting the primers for the TRAMP transgene with primers specific for other transgenes. Moreover, a small reaction volume of 10 μl combined with the use of crude genomic DNA will significantly reduce the cost and time for genotyping.
Supplementary Material
ACKNOWLEDGEMENTS
G.W. and D.E.F. are supported by the Prostate Cancer Moon Shot and Institutional Research Grant (IRG) Programs at the University of Texas MD Anderson Cancer Center. G.W. and D.E.F. are also supported by the National Institutes of Health (NIH) Prostate Cancer SPORE (P50 CA140388). G.W. is additionally supported by a University of Texas Star Award and NIH grant R00CA194289. D.E.F. is further supported by NIH grant R01CA184208 and a grant from the American Cancer Society (RSG-16-084-01 — TBE). B.A.F is supported by National Cancer Institute (NCI) grant P30CA016056 involving the use of Roswell Park Comprehensive Cancer Center’s Genomic Shared Resource.
Footnotes
CONFLICT OF INTEREST
D.E.F. has received research funding from GTx, Inc and has a familial relationship with Maia Biotechnology, Alms Therapeutics, Hinova Pharmaceuticals and Barricade Therapeutics. The other authors report no potential conflicts of interest.
REFERENCE:
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 2.Wang G, Zhao D, Spring DJ, DePinho RA. Genetics and biology of prostate cancer. Genes Dev 2018;32(17–18):1105–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R, et al. Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer research. 2013;73(9):2718–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gingrich JR, Barrios RJ, Kattan MW, Nahm HS, Finegold MJ, Greenberg NM. Androgen-independent prostate cancer progression in the TRAMP model. Cancer research. 1997;57(21):4687–91. [PubMed] [Google Scholar]
- 5.Gingrich JR, Barrios RJ, Morton RA, Boyce BF, DeMayo FJ, Finegold MJ, et al. Metastatic prostate cancer in a transgenic mouse. Cancer research. 1996;56(18):4096–102. [PubMed] [Google Scholar]
- 6.Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, et al. Prostate cancer in a transgenic mouse. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(8):3439–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiaverotti T, Couto SS, Donjacour A, Mao JH, Nagase H, Cardiff RD, et al. Dissociation of epithelial and neuroendocrine carcinoma lineages in the transgenic adenocarcinoma of mouse prostate model of prostate cancer. Am J Pathol 2008;172(1):236–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurwitz AA, Foster BA, Allison JP, Greenberg NM, Kwon ED. The TRAMP mouse as a model for prostate cancer. Curr Protoc Immunol 2001;Chapter 20:Unit 20 5. [DOI] [PubMed] [Google Scholar]
- 9.Berman-Booty LD, Knudsen KE. Models of neuroendocrine prostate cancer. Endocr Relat Cancer 2015;22(1):R33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gelman IH. How the TRAMP Model Revolutionized the Study of Prostate Cancer Progression. Cancer research. 2016;76(21):6137–9. [DOI] [PubMed] [Google Scholar]
- 11.Grabowska MM, Matusik RJ. Therapy-induced small-cell disease: from mouse to man and back. Nat Rev Urol 2018;15(11):662–3. [DOI] [PubMed] [Google Scholar]
- 12.Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas GV, et al. Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J Clin Oncol 2018;36(24):2492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies AH, Beltran H, Zoubeidi A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nature reviews Urology. 2018;15(5):271–86. [DOI] [PubMed] [Google Scholar]
- 14.Aparicio AM, Shen L, Tapia EL, Lu JF, Chen HC, Zhang J, et al. Combined Tumor Suppressor Defects Characterize Clinically Defined Aggressive Variant Prostate Cancers. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22(6):1520–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nature medicine. 2016;22(3):298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov 2011;1(6):487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355(6320):84–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355(6320):78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou M, Toivanen R, Mitrofanova A, Floch N, Hayati S, Sun Y, et al. Transdifferentiation as a Mechanism of Treatment Resistance in a Mouse Model of Castration-Resistant Prostate Cancer. Cancer Discov 2017;7(7):736–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee DK, Liu Y, Liao L, Li W, Danielpour D, Xu J. Neuroendocrine prostate carcinoma cells originate from the p63-expressing basal cells but not the pre-existing adenocarcinoma cells in mice. Cell Res 2019;29(5):420–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470(7333):269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques. 2000;29(1):52, 4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


