Abstract
Background
The coronavirus disease-19 (COVID-19) pandemic has cost lives and economic hardships globally. Various studies have found a number of different factors, such as hyperinflammation and exhausted/suppressed T cell responses to the etiological SARS coronavirus-2 (SARS-CoV-2), being associated with severe COVID-19. However, sieving the causative from associative factors of respiratory dysfunction has remained rudimentary.
Methods
We postulated that the host responses causative of respiratory dysfunction would track most closely with disease progression and resolution and thus be differentiated from other factors that are statistically associated with but not causative of severe COVID-19. To track the temporal dynamics of the host responses involved, we examined the changes in gene expression in whole blood of 6 severe and 4 non-severe COVID-19 patients across 15 different timepoints spanning the nadir of respiratory function.
Findings
We found that neutrophil activation but not type I interferon signaling transcripts tracked most closely with disease progression and resolution. Moreover, transcripts encoding for protein phosphorylation, particularly the serine-threonine kinases, many of which have known T cell proliferation and activation functions, were increased after and may thus contribute to the upswing of respiratory function. Notably, these associative genes were targeted by dexamethasone, but not methylprednisolone, which is consistent with efficacy outcomes in clinical trials.
Interpretation
Our findings suggest neutrophil activation as a critical factor of respiratory dysfunction in COVID-19. Drugs that target this pathway could be potentially repurposed for the treatment of severe COVID-19.
Funding
This study was sponsored in part by a generous gift from The Hour Glass. EEO and JGL are funded by the National Medical Research Council of Singapore, through the Clinician Scientist Awards awarded by the National Research Foundation of Singapore.
Keywords: SARS-CoV-2, COVID-19 recovery, COVID-19 pathogenesis, Transcriptome profiling
Research in context.
Evidence before this study
The COVID-19 pandemic has caused significant mortality and morbidity worldwide, but the principal drivers leading to severe disease progression and resolution remain poorly understood. Studies on severe COVID-19 patients have indicated that increased dysregulation and hyperactivation of the innate and acquired immune systems are associated with severity of the disease. However, it is unclear which of these responses are causative or merely associative.
Added value of this study
Through daily transcriptomic profiling of severe and non-severe COVID-19 patients spanning the nadir of respiratory function, this study reveals that neutrophil activation but not type I interferon signaling transcripts, tracked most closely with severe disease progression and resolution. This study thus highlights neutrophil activation as a critical factor of respiratory dysfunction in COVID-19.
Implications of all the available evidence
Our findings can guide the future development of therapeutics against COVID-19. As neutrophil activation is functionally involved in COVID-19 resolution, drugs that target this pathway could be repurposed for the treatment of severe COVID-19. Moreover, our data may provide a valuable resource towards our understanding of the host-pathogen dynamics involved in severe COVID-19 disease progression and resolution.
Alt-text: Unlabelled box
Introduction
The novel coronavirus, SARS-CoV-2, has emerged to threaten global health and economic security. Following viral infection, individuals can be asymptomatic or present with mild symptoms such as fever, dry cough and lethargy. In some cases, patients develop severe pulmonary dysfunction, including acute respiratory distress syndrome (ARDS), which is associated with poor prognosis. Clinical management of patients that progress to respiratory failure involves intensive care management and reliance on ventilators, which can impose a significant burden on healthcare systems that are already overstretched [1]. Pathogenic mechanism guided approach to expand the extremely limited therapeutic options for COVID-19 could mitigate the present negative impact of this pandemic on global health and economies.
Rapid progress in research has identified several factors that are associated with severe COVID-19. Increased inflammatory monocytes and neutrophils, accompanied by a sharp decline in lymphocytes, including T cell count and activation, have been found to be associated with severe COVID-19 [2], [3], [4], [5]. Severe COVID-19 patients have also shown heightened production of proinflammatory cytokines (TNF, IL-6 and IL-1β) and interferon (IFN) [5]. However, these responses are highly dynamic and changes daily following illness onset [6]. Their statistical association with severe COVID-19 may be heavily influenced by sampling points in the course of acute illness. Consequently, efficacy outcome of trials that tested inhibitors of hyperinflammation have been, at best, mixed. For instance, despite the consistent findings of elevated IL-6 expression in COVID-19 patients, IL-6 inhibitors (tocilizumab and sarilumab) failed to show efficacy in preventing severe COVID-19 [7,8]. IFN therapy reduced mortality if administered early, but late administration potentially worsens disease outcome [9]. IL-1 receptor antagonist (Anakinra) and TNF antagonists are promising candidates [10,11] but their effectiveness have yet to be evaluated in randomised, placebo-controlled trials. Among the licensed corticosteroids, only dexamethasone has shown efficacy in reducing COVID-19 mortality [12]. Moreover, T cell related therapies, such as the use of checkpoint inhibitors could be useful in preventing respiratory dysfunction given the findings of exhausted T cell phenotype in COVID-19 patients [13]. However, overcoming T cell exhaustion risks exacerbating the hyperinflammatory state. The mixed clinical trial outcomes of anti-inflammatory therapy and the uncertain clinical risk-benefit of T cell related therapies collectively underscore a lack of resolution on the causative factors of severe COVID-19. Sieving the causative from the associative factors of pulmonary dysfunction thus remains an unmet need.
To define the principal drivers of pulmonary dysfunction in COVID-19 patients, we focused our attention on the host response to SARS-CoV-2 infection around the period of the nadir of respiratory function. We hypothesised that the principal drivers of COVID-19 severity must track tightly with respiratory function and must resolve in sync with respiratory improvement. Thus, instead of time interval analysis [14], [15], [16], we performed daily transcriptomic profiling to obtain a detailed continuum of the host immune response in COVID-19 patients before, during and after the nadir of respiratory function. We found that the neutrophil activation pathway, but not type-I IFN signaling pathways, showed a direct inverted relationship with respiratory function – genes in this pathway increased and peaked during the nadir of respiratory function and then declined during the recovery phase. On the other hand, transcripts associated with protein phosphorylation pathways, particularly the serine threonine kinases involved in cell cycle progression and T cell development increased at convalescence. The understanding of the temporal dynamics involved in severe COVID-19 progression and recovery provided insights into the potential drug candidates that can be considered to reduce the disease burden caused by SARS-CoV-2 infection.
Methods
Ethical approval
COVID-19 patients included 8 male patients and 2 female patients. The average age of patients was 57 years (s.d.: 12.8, range: 37–76 years). Detailed subject demographics are detailed in Supplemental Table 1. Following written informed consent, all patients were recruited under the Novel Pathogen Study Protocol. This study was approved by the SingHealth Combined Institutional Review Board (2013/397/F) and the patients were enrolled for the study at Singapore General Hospital. Healthy controls consisted of baseline samples obtained from participants of a measles, mumps and rubella re-vaccination study [17]. Ten healthy control individuals (7 females, 3 males) were included, with average age of 44.1 years (s.d.: 11.8, range: 30–61 years). All participants gave written informed consent, and study approval was obtained from the SingHealth Combined Institutional Review Board (CIRB 2017/2374).
Patients and clinical data collection
Patients hospitalised at Singapore General Hospital (SGH) testing PCR-positive for SARS-CoV-2 were approached to seek informed consent to participate in the study. Samples were collected from recruited patients between January 2020 to April 2020. Clinical data was obtained from the patients’ medical records and clinical charts.
Microarray analysis
RNA was isolated from whole blood collected in Tempus Blood RNA tubes (Applied Biosystems Cat# 4342792) using the Tempus Spin RNA Isolation Kit (Life Technologies Cat# 4380204) according to manufacturer's instructions. Bioanalyzer assessment was performed on RNA samples before microarray was performed using the Affymetrix GeneChip Human Gene 2.0 ST Array (Applied Biosystems Cat# 902113) at the Duke-NUS Genome Biology Core Facility. Briefly, 50 ng of each RNA sample was processed using Affymetrix GeneChip Whole Transcript (WT) PLUS Reagent Kit (Applied Biosystems Cat# 902280), according to manufacturer's instructions. 15 μg of cRNA was used for the second cycle cDNA reaction and 5.5 μg of ss-cDNA was used for fragmentation. DNA fragment was end-labeled with biotin using terminal deoxynucleotidyl transferase before hybridisation on the arrays. Hybridisation cocktails containing fragmented, end-labeled cDNA were prepared and applied to GeneChip Human Gene 2.0 ST arrays. Hybridisation was performed at 60 rpm for 16h at 45°C using the FS450_0001 fluidics protocol. Arrays were scanned using Affymetrix GeneChip Command Console Software (AGCC) to produce .CEL intensity files. The raw .CEL intensity files and the processed files are deposited in ArrayExpress.
NanoString profiling
NanoString profiling was performed using the nCounter Human Immunology v2 Panel (NanoString Technologies, Inc. Cat# XT-CSO-HIM2-12). 50 ng of RNA was mixed with 8 ul of reporter probeset and 2ul of capture probeset, and incubated at 65 °C for 24 h. Hybridised samples were loaded onto an nCounter Sprint cartridge and imaged on a nCounter Sprint system. Raw and normalised counts were derived with the NanoString nSolver 4.0 software as per manufacturer's protocols. nCounter probe sets included negative and positive controls that were used for background thresholding, and normalizing samples for differences in sample input or hybridisation.
Plaque reduction neutralisation test
Plaque reduction neutralisation test (PRNT) was carried out on Vero E6 cells (RRID:CVCL_0574). Briefly, serial two-fold dilution of serum samples in DMEM media (Gibco Cat# 11995-040) containing 2% FCS (Hyclone Cat# SH30396.83) was incubated with 40 pfu of SARS-CoV-2 clinical isolate in equal volumes for 1 h. Thereafter, 100 μL of this serum/virus mix was added to Vero E6 cells in 24-well plates and incubated for 1 hour at 37 °C, rocking every 15 min. After 1 h, serum/virus mix was removed and cells overlaid with 500ul of 0.8% methylcellulose (Calbiochem Cat# 17851) in DMEM media (DMEM, 2% FCS, 25 mM HEPES, 1x penicillin/streptomycin). After incubating at 37 °C for 4 days, the cells were fixed with 4% formaldehyde for 30 min to inactivate the virus. Plates were rinsed with water and stained with 0.5% crystal violet for 5 min to visualise plaques. Excess crystal violet was removed and plates were rinsed with water, and allowed to dry before counting plaques.
CIBERSORT and cross-referencing with COVID-19 databases
The CIBERSORT tool was used to digitally quantify the relative frequencies of different immune cell types across time (https://cibersort.stanford.edu) [18]. To assess if the upregulated proteins in the protein phosphorylation pathway had altered phosphorylation levels, genes from the protein phosphorylation pathway were cross-referenced with the phosphoproteome study described by Bouhaddou et al. [19]. Differential levels of phosphorylation modulated by SARS-CoV-2 infection in Vero E6 were extracted from the data analysis by Bouhaddou et al. For the analysis of the potential drugs that can target module A genes, we used the Drug Gene Interaction Database (DGIdb) to query the genes represented in module A. As clusters 3 and 4 are most associated with severe disease progression and resolution, the drugs that exclusively target only clusters 1 or 2 were filtered out. Subsequently, the top drug candidates were ranked based on the total number of genes targeted in module A [20]. The clinical usage information was obtained from Health Sciences Authority (HSA) at the website:
(https://eservice.hsa.gov.sg/prism/common/enquirepublic/Search DRBProduct.do?action=load&_ga=2.183810082.563179921. 1554083187-551332391.1551944793). Other information pertaining to the drugs such as route of administration and inflammatory properties were obtained via UptoDate and clue.io.
Statistical analysis
The Transcriptome Analysis Console (Thermo Fisher) was used to analyse transcriptomic changes between the different days relative to respiratory nadir function (Day 0). Differentially expressed genes (DEGs) were defined based on fold-change > 1.5 with respect to day 0, with false discovery rate adjusted p-value < 0.05 using the Benjamini-Hochberg Step-Up FDR-controlling Procedure (α=0.05). For pathway analysis, the identified DEGs were used as input data and analysed against the Gene Ontology (GO) Biological Processes database using the Enrichr tool [21]. Adjusted p-values for each enriched pathway were obtained from the Enrichr tool analysis, and REVIGO was used to summarise the non-redundant GO terms [22]. Volcano plots and heatmaps were constructed using the Prism 8.4.3 software.
Temporal gene expression was analysed using EDGE on log2 intensity counts [23]. A total of 4,831 genes were temporally altered in severe COVID-19 patients, p-value < 0.05 and q-value < 0.05. Co-clustering of significant genes was analysed using Self-Organizing Maps (SOM) from Partek® Genomics Suite® based on the tabulated Least Square Means (LSMeans). 8 main clusters were identified which can be classified into module A and module B, representing 99.7% of the total significant genes. We excluded cluster 9 from further analysis as it contained only 13 genes. The genes in the different SOM clusters were then queried using the Enrichr tool to identify significantly enriched pathways (based on adjusted p-value < 0.05). To visualise the expression changes of pathway genes across time in severe and mild COVID-19 patients, normalised expression was tabulated by taking the average LSMeans values of all significant genes in the indicated pathways, such as cellular response to type I IFN pathway, regulation of immune effector process, neutrophil mediated immunity and protein phosphorylation pathways. Graphs and heatmaps were constructed using the Prism 8.4.3 software. To visualise the functional network and interacting partners involved in the protein phosphorylation pathway, the RegPhos database [24] and Ingenuity Pathway Analysis were used. For Principal Component Analysis (PCA), normalised gene transcript counts profiled using the NanoString Human Immunology Panel v2 were used for analysis, where the R packages used are factoextra, ggplot2 and FactoMineR. Pearson correlation analysis was performed using Prism 8.4.3 software.
Data and code availability
The raw data for microarray profiling is available at Array Express (E-MTAB-9721). The raw data and normalised counts for NanoString profiling of immune responses is available at Array Express (E-MTAB-9719).
Role of funding source
The funders have no role in study design, data collection, data analyses, interpretation or writing of report.
Results
Clinical profile of recruited patients
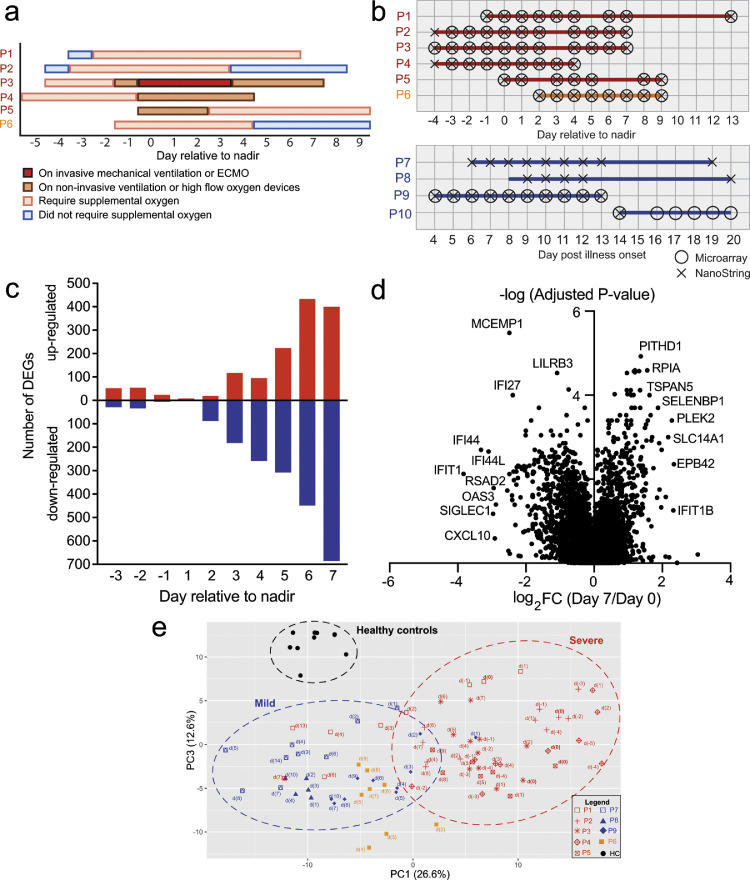
We prospectively enrolled 10 SARS-CoV-2 confirmed patients admitted to the Singapore General Hospital after written informed consent. Full blood count, C-reactive protein (CRP) and lactate dehydrogenase (LDH) were measured as part of routine COVID-19 clinical management. These data are shown in Supplemental Figure 1a-d, with normal reference range for healthy subjects shaded in grey. The severe and moderately severe COVID-19 patients displayed lymphopenia despite normal white blood cell (WBC) counts, and exhibited higher CRP and LDH levels compared to the mild COVID-19 patients. These laboratory parameters were tracked longitudinally, and were consistent with previously reported associations with severe COVID-19 [25,26]. Supplemental oxygen was administered if arterial oxygen saturation dropped below 95%. We used oxygen requirement to stratify patients into mild, moderately severe or severe COVID-19. Of the 10 patients, 4 were classified as mild as they did not require supplemental oxygen over the course of hospitalisation. Of the remaining 6 patients, all patients required supplemental oxygen; 5 patients needed more than 2 liters/minute of supplemental oxygen, requiring either non-invasive or invasive mechanical ventilation (MV) while the remaining patient received not more than 2 liters/minute of supplemental oxygen throughout hospitalisation (Supplemental Figure 1e). All 6 patients were considered to have moderately severe or severe COVID-19, and were considered as a group in our analysis. In addition, patients who required supplemental oxygen were tracked using a 4-point ordinal scale, providing an overview of the disease progression and recovery over time (Fig. 1a). For the 5 patients that needed non-invasive ventilation (NIV) or MV, the day of respiratory function nadir was defined as the day the patient was initiated on NIV or MV. For P6, the patient with moderately severe COVID-19, day of respiratory function nadir was defined as the day when arterial oxygen saturation was lowest at 93% (Supplemental Figure 1e).
Fig. 1.
Suppression of IFN and Toll-like receptor signaling pathways precede the increase in T cell proliferation and activation pathways during the recovery phase. (a) Overview of clinical status for severe (P1, P2, P3, P4 and P5) and moderately severe (P6) patients. (b) Summary of sampling points for whole blood collection from severe (red), moderately severe (orange) and mild (blue) patients. Circles indicate the time-points analysed by microarray and crosses indicate the time-points used for NanoString analysis. (c) Waterfall plot representing the number of differentially upregulated and downregulated genes (Fold change > 1.5, adjusted p-value < 0.05; Benjamini-Hochberg step-up procedure) relative to nadir (day 0) in the severe patients (n=6). (d) Volcano plot displaying genes that were differentially expressed at day 7 post-nadir relative to day 0 in the severe patients (n = 6). Genes that were most differentially regulated were annotated on the volcano plot. (e) PCA of the NanoString data for healthy controls (black, n = 9), mild (blue, n = 3), moderately severe (orange, n = 1) and severe (red, n = 5) COVID-19 patients at the various days relative to respiratory function nadir. The top contributing genes in the respective PC is shown in Supplemental Figure 2b.
Suppression of type-I IFN and Toll-like receptor signaling pathways preceded the increase in T cell proliferation and activation pathways during convalescence
To interrogate the transcriptomic changes spanning the nadir of respiratory function nadir, we sampled daily, where possible, whole blood from COVID-19 patients ranging from -4 days to 13 days, with the nadir being day 0 (Fig. 1b, Supplemental Figure 2a). Healthy controls consisted of baseline samples obtained from participants of a measles, mumps and rubella re-vaccination study (CIRB 2017/2374), as previously reported [17].
We first sought to attain an overview of whole genome expression changes before, during and after the nadir of respiratory function. Measurement of 29,405 annotated gene transcripts by microarray showed that the number of differentially expressed genes (DEGs) increased with time post-nadir (false discovery rate [FDR]-adjusted p < 0.05 and fold change [FC] > 1.5; Benjamini-Hochberg step-up procedure) (Fig. 1c). Functional annotations of the DEGs revealed that cytokine and IFN-dependent immune responses, inflammatory signaling pathways, neutrophil mediated immunity and necrotic cell death pathways were most significantly reduced with time (Supplemental Figure 2a). Notably, the most downregulated genes belonging to these pathways were IFIT1, IFI27, IFI44, IFI44L, CXCL10, RSAD2 and OAS3 (Fig. 1d). In contrast, no consistent pathway was found among the upregulated genes, with the most upregulated genes being involved in metabolism and transport (RPIA, SLC14A1, SELENBP1), cytoskeleton (PLEK2, EPB42) and Notch signaling (TSPAN5) (Fig. 1d).
Changes in expression of the innate and adaptive immune response genes were validated using the nCounter assay (NanoString Technologies), which allows simultaneous digital quantification of 579 immune-related mRNA transcripts. Mild COVID-19 patients and healthy controls were included in this analysis. Principal component analysis (PCA) confirmed that COVID-19 patients clearly segregated from healthy subjects in the PCA space (Fig. 1e). Moreover, distinct temporal immune signatures between mild and severe patients could also be largely explained by the variance in principal component (PC) 1 (Fig. 1e). The top contributors in PC1 were transcripts related to TLR signaling (TLR2, TLR4, TLR5, TLR8) and inflammatory responses (PRKCD, MAPK14) (Supplemental Figure 2b). Critically, these transcripts were over-expressed in severe COVID-19 patients before or on the day of respiratory function nadir; these genes were universally downregulated after the nadir of respiratory function (Supplemental Fig. 2b). On the other hand, PC3 segregated the healthy subjects from the infected patients. The top contributors are genes related to cytokine and IFN responses such as CXCL10, MX1, BST2, LAMP3 and IFI35, where higher levels of these transcripts were observed in both mild and severe COVID-19 patients, but not in healthy subjects (Supplemental Figure 2b). Of note, PC2 did not segregate the healthy from the infected patients (Supplemental Fig. 2c-d). Consistent with the microarray observations, the most significant immune pathways that showed reduced expression with time were the type-I IFN and TLR signaling pathways (Supplemental Figure 2e-f). On the other hand, immune pathways that were increased during the recovery phase included pathways related to T cell proliferation and activation (Supplemental Figure 2e, g). Collectively, these findings highlight that the suppression of the type-I IFN and TLR signaling pathways precedes restoration of the T cell proliferation and activation pathways.
Identification of eight distinct gene expression clusters that are temporally modulated in severe COVID-19 patients
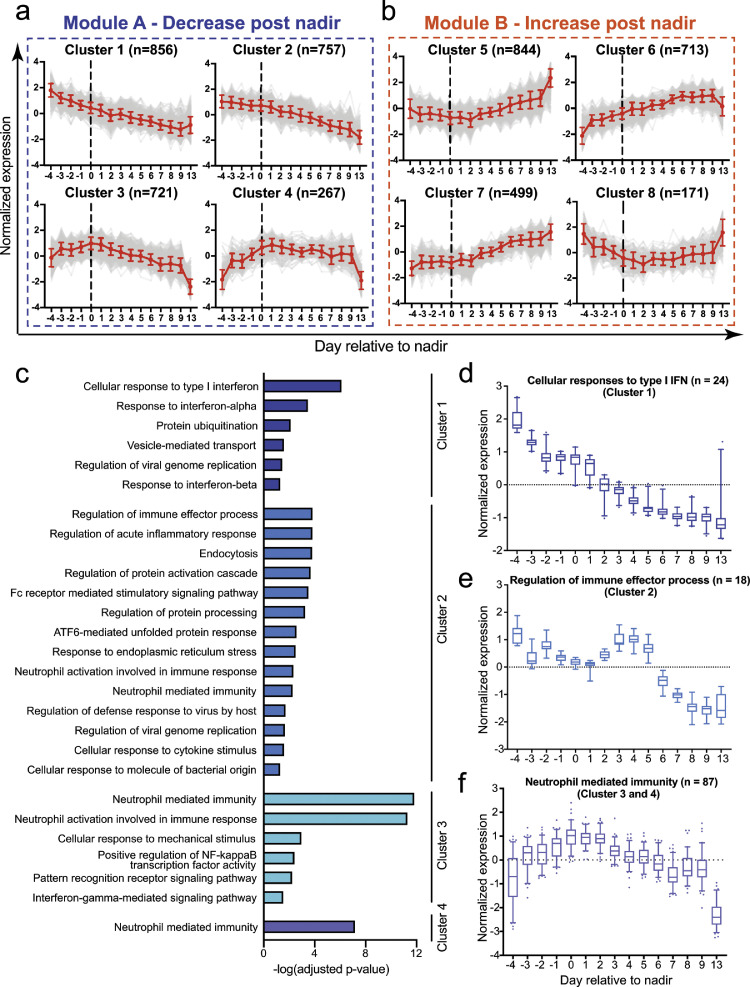
To determine the genes that tracked with disease progression and resolution, we used extraction and analysis of differential gene expression (EDGE) to obtain greater insight on temporally regulated transcripts over the course of illness in severe patients. We identified 4,831 genes (FDR-adjusted p-value < 0.05; q-value < 0.05; Benjamini-Hochberg step-up procedure), that were significantly altered over time in the 6 severe COVID-19 patients (Supplemental Fig. 3a-b). Self-organizing map (SOM) analysis of the 4,831 genes further revealed that these genes could be grouped into 8 distinct expression clusters– linear plots of expression of genes within each of these clusters against time are shown in Fig. 2a-b. These clusters can be broadly segregated into 2 main modules: Module A highlights the genes with reduced expression after respiratory nadir; module B highlights the genes with increased expression after respiratory nadir (Fig. 2a-b). In module A, clusters 1 and 2 showed a decreasing trend in gene expression from day -4 to day 13. Expression of genes in clusters 3 and 4 trended with respiratory dysfunction - expression increased from day -4 to day 0 and declined after day 0 during the recovery phase (Fig. 2a). In module B, increased expression of the majority of the genes were observed after day 0 (Clusters 5-7), albeit at different rates in the different clusters (Fig. 2b). The identity of the genes and heatmaps of the top 150 genes in each cluster are displayed in Supplemental Figure 3c-d.
Fig. 2.
Self-organizing map (SOM) clusters reveal distinct transcriptional dynamics in severe COVID-19 patients. (a-b) Average expression of significant transcripts (shown in red line) modulated by severe and moderately severe COVID-19 patients determined by EDGE (p-value < 0.05; q-value < 0.05; likelihood ratio test), in the respective SOM clusters. Module A (Clusters 1-4) highlights clusters with reduced gene expression post-nadir, and module B (Clusters 5-8) highlights clusters with increased gene expression post-nadir. n indicates the total number of transcripts in each cluster and error bars indicate standard deviation. The top 150 genes in each cluster is shown in Supplemental Figure 3c-d. (c) Pathway analysis of genes in clusters 1-4 in severe and moderately severe COVID-19 patients (n = 6). Pathways were analysed against the Gene Ontology Biological Processes database using Enrichr. All pathways with adjusted p-values < 0.05 (hypergeometric test) are displayed. (d-f) Normalised expression levels for all significant genes in the GOBP pathway for cellular response to type I IFN, regulation of immune effector process, and neutrophil mediated immunity over time in severe and moderately severe COVID-19 patients (n = 6). n refers to the number of significant genes involved in the respective pathways. Data are represented as mean ± SD. The individual genes and least squared mean (LSmean) expression levels are displayed in Supplemental Figure 3e, g and i.
We next examined the molecular pathways that were enriched in module A. Consistent with the observations from the nCounter assay, the top enriched pathway in cluster 1 is the cellular response to type I IFN (Fig. 2c), where a monotonic decrease in gene expression was observed from day -4 to day 13 (Fig. 2d). Cluster 1 contains 24 genes involved in the IFN response pathway, the majority of which are IFN-stimulated genes (ISGs) (Supplemental Figure 3e). Similar trends were also observed in mild COVID-19 patients although changes in the expression of these genes were smaller compared to severe cases (Supplemental Fig. 3f). In cluster 2, the most enriched pathway is the regulation of immune effector process (Fig. 2c), where the majority of these transcripts encodes for the variable and constant regions of the immunoglobulin heavy and light chains (Supplemental Figure 3g). These genes were less abundant in whole blood from day -4 to day 0, increased transiently from day 0-4, and then declined thereafter in the severe patients (Fig. 2e). In contrast, expression of these genes increased over time in mild patients (Supplemental Figure 3h). Along with the differences in immunoglobulin transcript temporal profiles, severe cases developed higher titers of neutralizing antibodies to SARS-CoV-2 compared to mild cases (Supplemental Table 1). Taken collectively, our data suggests that the reduction in immunoglobulin transcripts may be explained by greater proportion of B cell migration to lymphoid organs in severe compared to mild cases.
The expression of genes that tracked most closely in temporal terms relative to disease progression and resolution were in clusters 3 and 4 (Fig. 2a). Neutrophil mediated immunity and neutrophil activation were the top 2 enriched pathways in cluster 3 (Fig. 2c). Other enriched pathways in cluster 3 were cellular response to mechanical stimulus, regulation of NFkB transcription factor activity, pattern recognition receptor signaling and IFN-gamma-mediated signaling pathways (Fig. 2c). Cluster 4 only had neutrophil mediated immunity pathway being significantly modulated. Combining both clusters 3 and 4, we identified 87 genes involved in neutrophil mediated immunity that showed peak expression during the nadir of respiratory function; expression waned with upswing in respiratory function (Fig. 2f, Supplemental Figure 3i). Critically, the expression of these genes remained unchanged throughout the period of monitoring in mild patients (Supplemental Figure 3j). Collectively, these data support the notion that neutrophils may play a key role in shaping disease severity.
The abundance of genes identified in the SOM analysis could be influenced by changes in cell counts during acute and convalescent phase of infection. We thus analysed our data using CIBERSORT, which deconvolutes bulk gene expression data to enumerate immune cell type frequencies [27]. The plasma cell frequencies significantly correlated with the transcript levels of immunoglobulins (IGHV4-39, IGKV2D-40) from cluster 2, indicating that the reduced abundance of the immunoglobulin transcripts involved in immune effector processes reflected lower plasma cell frequencies (Supplemental Figure 4a). In contrast, monocyte, neutrophil and naïve CD4+ T cell frequency levels did not correlate with temporally regulated genes in the IFN response, neutrophil activation and T cell activation pathways (Supplemental Figure 4b-d), suggesting that transcriptional changes observed in these pathways in severe COVID-19 patients reflected the activation status of immune cells.
Increased protein phosphorylation signatures after respiratory function nadir in severe COVID-19 patients
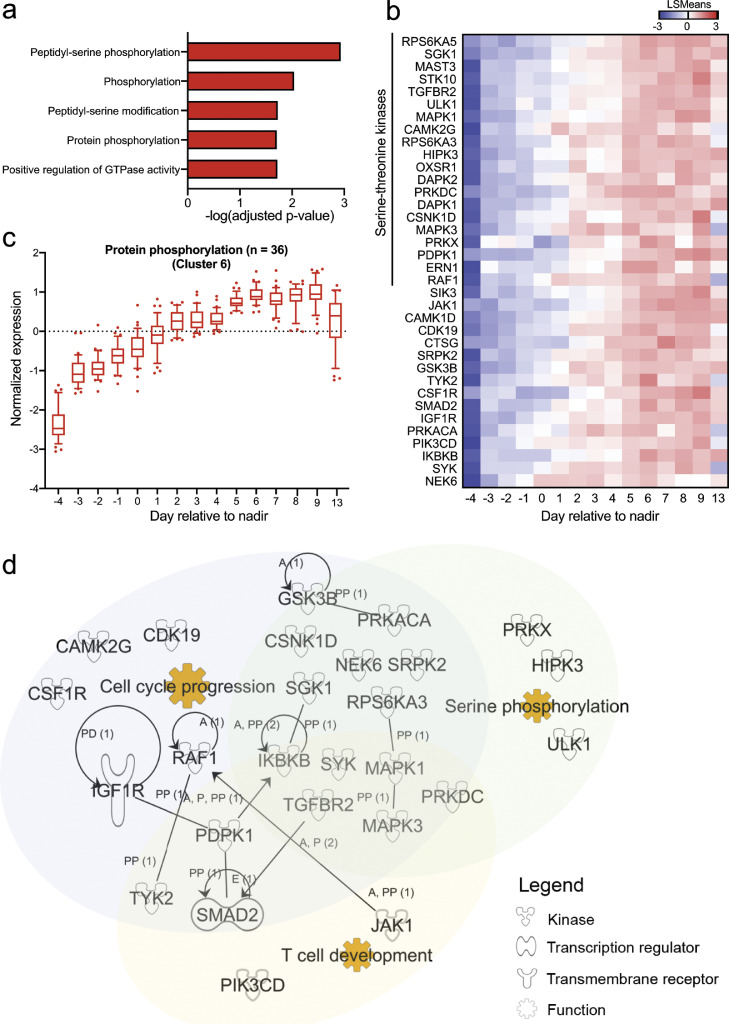
Pathway enrichment on module B genes only led to significant enrichment of protein phosphorylation, particularly peptidyl-serine phosphorylation, in cluster 6 (Fig. 3a). No significant pathways were found in the other clusters (adjusted p-value > 0.05; hypergeometric test). We identified 36 genes that were significantly upregulated in the protein phosphorylation pathway, of which 20 were serine-threonine kinases (Fig. 3b-c). When these genes were cross-referenced against RegPhos, which allows for exploration of the human protein phosphorylation regulatory network [24], many of these kinases were identified as interacting partners (Supplemental Figure 4e) in the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) pathway. Critically, these kinases are involved in cell growth, cell survival and T cell development (Fig. 3d), which could be crucial for renewal and regeneration of immune cells after viral infection. We also found that 12 of the 36 genes (33.3%) were differentially phosphorylated during SARS-CoV-2 infection (Supplemental Table 2). Of note, genes associated with the protein phosphorylation pathway were similarly increased in mild patients over time, albeit to a lesser extent as compared to severe patients (Supplemental Figure 4f). Collectively, increased expression of serine-threonine kinases over time suggests a protective role for these kinases in COVID-19 disease resolution.
Fig. 3.
Increased protein phosphorylation signatures in COVID-19 patients recovering from severe disease. (a) Pathway analysis of genes in cluster 6 in severe and moderately severe COVID-19 patients (n = 6). Pathways were analysed against the Gene Ontology Biological Processes (GOBP) database with Enrichr. All pathways with adjusted p-values < 0.05 (hypergeometric test) are displayed. (b) Heatmap showing the LSmean expression values of significant genes related to protein phosphorylation in severe COVID-19 patients (n = 6). Data are represented as mean ± SD. (c) Normalised expression levels for the significant genes shown in (b) over time in severe and moderately severe COVID-19 patients (n = 6). (d) Network map showing interactions between significant genes, and their overlapping functional annotations. Functional annotation for genes was determined using Ingenuity Pathway Analysis.
Potential molecular targets for drug repurposing
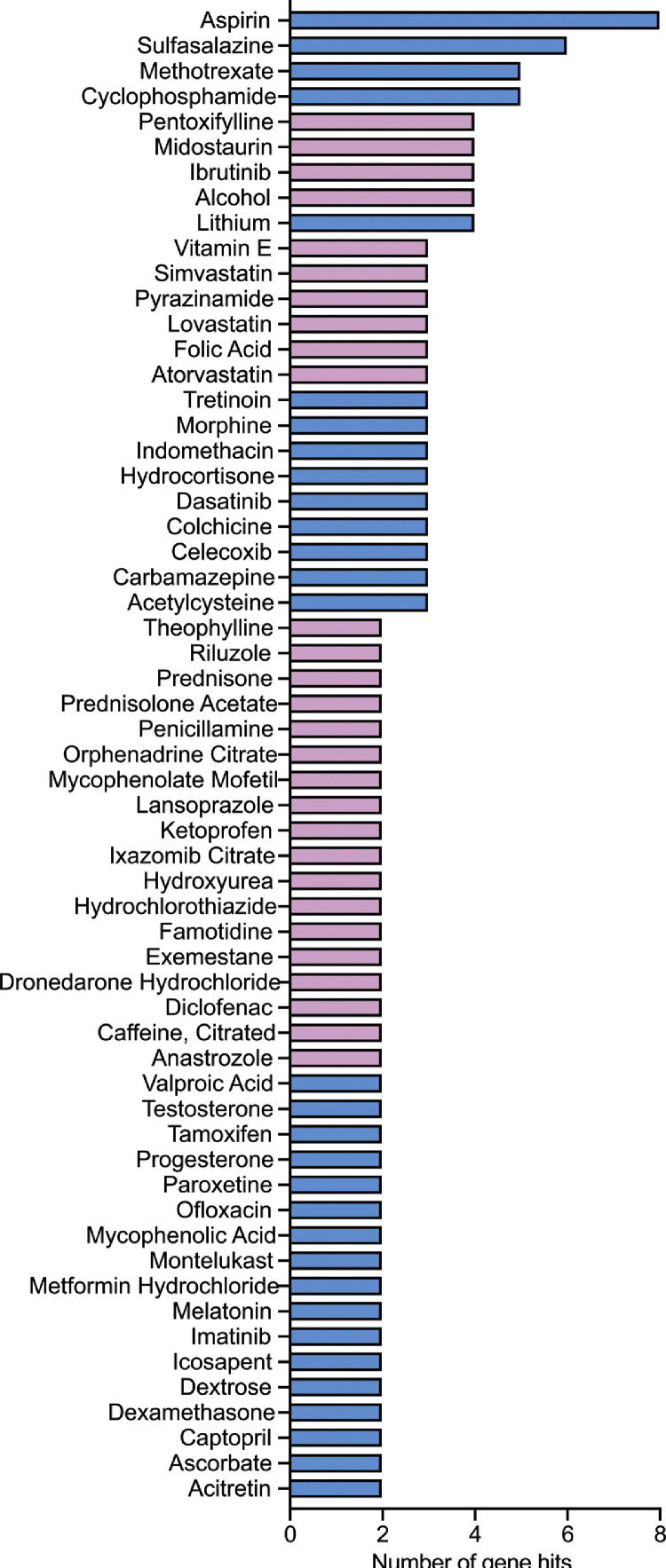
Our analyses thus far suggest that genes in clusters 3 and 4 that are involved in neutrophil activation, pattern recognition receptor signaling and NFκB-mediated responses are functionally involved in COVID-19 resolution, as these genes are most strongly associated with severe COVID-19 disease progression and resolution (Fig. 2a, c). To validate this notion, we cross-referenced the transcripts in clusters 3 and 4 with the Drug Gene Interaction Database (DGIdb), which is a curated resource that can be used to search for druggable targets of the human genome [20]. If indeed our analysis was clinically pertinent, we should identify therapies with demonstrated efficacy in reducing COVID-19 severity. One such drug is dexamethasone. This corticosteroid has been found to reduce mortality rates in severe COVID-19 patients [28]. We found that amongst the module A genes, 6 were known targets of dexamethasone. Of the 6 genes targeted by dexamethasone, 2 were in clusters 3 and 4 (Fig. 4). Specifically, dexamethasone was found to act on JUNB and IRS2 (insulin receptor substrate 2) of clusters 3 and 4, respectively (Supplemental Table 3). Both these genes are involved in the cytokine-mediated signaling pathway and are most highly expressed in neutrophils compared to other blood cells [29]. Notably, aspirin was identified as the top drug that inhibits the most genes in clusters 3 and 4, which corroborates a recent cohort study showing that aspirin use is associated with less mechanical ventilation, ICU admissions and mortality in hospitalised COVID-19 patients [30]. In contrast, host targets of a different corticosteroid, methylprednisolone [12,31], and other drugs such as azithromycin, lopinavir-ritonavir and hydroxychloroquine [31], [32], [33], [34], all of which have shown no efficacy in improving clinical outcomes in COVID-19 patients, were absent from the hits in clusters 3 and 4. Likewise, host-directed anti-IL-6 therapies such as tocilizumab and sarilumab were also not highlighted in this cross referencing. Our findings are thus consistent with previously published efficacy trials for severe COVID-19.
Fig. 4.
Candidate drugs that target clusters 3 and 4. Clinically approved oral drugs that target genes in clusters 3 and 4. Data obtained by cross-referencing with DGIdb. Drugs that also target cluster 6 genes are indicated in blue, whereas drugs do not interfere with cluster 6 genes are indicated in purple. The genes that the drugs target are shown in Supplemental Table 3.
Given how we were able to identify targets of dexamethasone but not other host-directed therapies in our analysis, we next sought to identify other drugs that may be suitable for repurposing as anti-COVID-19 therapies (Fig. 4; Supplemental Table 3). Moreover, as the cluster 6 genes from module B consist of serine-threonine kinases that could be important for convalescence, we also highlighted the drug candidates that inhibited module A genes without known activity on cluster 6 genes (Fig. 4). Many of these drugs are anti-inflammatory, such as aspirin, sulfasalazine, celecoxib and indomethacin. Interestingly, several statins such as lovastatin, atorvastatin and simvastatin can specifically target module A genes without affecting the expression of genes from cluster 6. The full list of the potential drug candidates and their corresponding gene targets are detailed in Supplemental Table 3.
Discussion
Longitudinal sampling of blood and other samples from COVID-19 patients has enabled the identification of multiple associative factors of respiratory dysfunction. However, associative factors that could also drive pulmonary dysfunction in COVID-19 patients have not been systematically defined. A major limitation to identifying drivers of SARS-CoV-2 infection induced pulmonary dysfunction is the lack of an animal model that accurately recapitulates COVID-19 pathogenesis in humans. To overcome this hurdle, we postulated that the host response to SARS-CoV-2 infection around the period of the nadir of respiratory function could provide us with a window into distinguishing the pathogenic from the associative factors of respiratory dysfunction. The principal drivers of COVID-19 severity should resolve before or track tightly with respiratory function.
To test our postulate, we coupled daily sampling with full genome gene expression measurement in combination with EDGE and SOM analyses. We identified 8 distinct gene expression clusters that were altered over the course of disease in severe patients. Cluster 1 and 2 contained transcripts, many of which are IFN-stimulated genes (ISGs) [35] and other immune effectors, all of which showed a sustained decrease in gene expression starting from day -4 relative to the nadir of respiratory function. That these pathways did not track tightly with respiratory function suggest that the pro-inflammatory and other immune responses may initiate the cascade of processes necessary for disease pathogenesis but are not the principal drivers of respiratory dysfunction.
Transcripts that tracked most closely with progression and resolution of respiratory dysfunction were in clusters 3 and 4. These transcripts belong to neutrophil-mediated activation, pattern recognition receptor signaling and NF-kB activation pathways. They were significantly enriched in severe COVID-19 patients but not in patients that did not develop progressive pulmonary deterioration. As the bone marrow is the major site of neutrophil production, the increase in cytokine and chemokine levels before respiratory function nadir (cluster 1 and 2) could have led to an increased neutrophil-to-lymphocyte ratio in severe COVID-19 patients [36], leading to the accumulation of activated neutrophils in the lung. Our finding is congruent with previous studies showing elevated neutrophil infiltration, activation and neutrophil extracellular trap (NET) markers in the lung of COVID-19 patients at post mortem examination [37,38]. Similarly, RNA-seq analysis of bronchoalveolar lavage fluid (BALF) from COVID-19 patients identified an upregulation of neutrophil enriched and chemotaxis genes (e.g. CD177, FPR2, CXCR1, CXCR2, CTSZ, S100A8, S100A9) in COVID-19 BALF cells [39,40]. Of note, S100A8 and S100A9, which were identified in cluster 3 (Supplemental Figure 3i), are also known to form a stable heterodimer during inflammation known as calprotectin [41] – plasma calprotectin levels have been found to discriminate between mild and severe COVID-19 patients [42]. Besides COVID-19, neutrophils are also enriched in the lungs of patients with ARDS caused by other viral infections [43]. For example, pathogenic influenza viral strains can cause increased neutrophil mobilisation and recruitment to the lungs in mice and humans, that can result in excessive inflammation and fatality [44,45]. Taken together, neutrophil activation may thus be an important underpinning of respiratory dysfunction in COVID-19 patients.
An additional new finding was the increased transcript levels of serine-threonine kinases (cluster 6) during the recovery from severe COVID-19, raising the possibility that these kinases play an important role in convalescence. Interestingly, many of these are serine-threonine kinases involved in cell cycle progression, which is congruent with the phosphoproteomics analysis conducted by Bouhaddou et al that demonstrated that SARS-CoV-2 infection in Vero E6 cells influences the phosphorylation levels of cell cycle proteins such as PDPK1, PRKACA, RAF1 and RPS6KA3 (Supplemental Table 2) [19]. Increased expression and activation of these genes during recovery could facilitate cell cycle progression and aid in restoring the frequencies of immune cell populations, particularly the T cells, that are decreased over the course of the disease [4,6,37,38]. This explanation is further supported by the observed increase in T cell transcripts starting from day ~6 after respiratory function nadir. Thus, although the transcripts in cluster 6 did not track with progression of respiratory dysfunction, the coincidence of their expression in respiratory function recovery suggests that T cell proliferation and development may be important for convalescence.
The tracking of genes in clusters 3 and 4 with respiratory dysfunction are consistent with multiple transcriptomics and proteomics studies in COVID-19 patients [46], [47], [48], [49] (Supplemental Table 4). That we observed these genes to be significantly altered in a high-resolution time series analysis further highlights the potential for these genes to be targeted for treating severe COVID-19. Indeed, the finding of dexamethasone [12] and aspirin [30] when we cross-referenced our list with DGIdb, suggests the clinical validity of this approach. We also identified sulfasalazine, an anti-inflammatory drug, as another potential drug candidate that targets the most number of genes from clusters 3 and 4. Interestingly, the third candidate, methotrexate is an FDA-approved inhibitor of purine synthesis that can inhibit SARS-CoV-2 replication [50]. Clinical trials to test the translatability of these drug candidates could thus be particularly fruitful.
Our present study has some limitations. While this study provides high resolution on the temporal dynamics of the host response involved in disease progression and recovery, gene expression was profiled from peripheral blood rather than infected organs. Direct assessment of gene expression in infected organs would require invasive procedures like bronchoalveolar lavage, which is ethically challenging for mild COVID-19 cases, and risks aerosolization of infectious respiratory fluids that could compromise the safety of healthcare workers. Measuring gene expression in peripheral blood hence provides an opportunity to understand how the host response to SARS-CoV-2 could impact clinical outcome. Furthermore, our findings were based on a small sample size involving 10 COVID-19 patients, of which 8 were male (Supplemental Table 1). As males and females may respond to COVID-19 differently [51], [52], [53], these temporal host responses may not be fully generalisable to females. As such, increasing the sample size, as well as a deeper temporal characterisation of neutrophil gene expression changes, phenotype and function at the target sites of infection could provide further mechanistic insights into the molecular processes that drive disease pathogenesis and recovery from severe COVID-19.
In conclusion, our results reveal the host responses involved in the progression and resolution of respiratory dysfunction in COVID-19 patients, which could be useful in guiding the choice of drugs for repurposing as COVID-19 therapeutics.
Contributors
E.Z.O., E.E.O., J.G.L. and K.R.C. conceptualised the study and wrote the manuscript. S.K. and J.G.L. conducted the clinical investigations. E.Z.O. conducted the RNA extraction and Nanostring profiling. K.R.C., E.Z.O., E.E.O., J.G.L., W.C.C., S.H.O. and C.W.T. analysed the microarray and Nanostring profiling data. H.C.T. and S.L.Z. performed the plaque reduction neutralisation tests for the sera. K.R.C., E.Z.O., W.C.C, S.H.O. and C.W.T. verified the data sets. All authors have read and approved the final version of the manuscript.
Data sharing statement
Data in this study is available upon request from the corresponding author at kuanrong.chan@duke-nus.edu.sg. Individual participant data will be made available, study protocol, statistical analysis plan, the raw and processed transcriptomics data for all participants will be made available for anyone who wishes to access the data. The data can be used for any purpose and the data will be made available indefinitely in the ArrayExpress and Expression website:
http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9721
http://wwwdev.ebi.ac.uk/gxa/experiments/E-MTAB-9721?access Key=edad7f29-fb8e-4a51-87d3-661176fa6e5e
Declaration of Competing Interests
Dr Eng Eong Ooi reports grants from National Medical Research Council, Singapore, during the conduct of the study; personal fees from Tychan Private Limited, outside the submitted work; the other authors declare no competing interests.
Acknowledgements
We thank all doctors and nurses in the isolation facility in the Singapore General Hospital. ViREMiCS was established through generous support from the Tanoto Foundation. This study was sponsored in part by a generous gift from The Hour Glass. EEO and JGL are funded by the National Medical Research Council of Singapore, through the Clinician Scientist Awards.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103262.
Contributor Information
Jenny G Low, Email: jenny.low@singhealth.com.sg.
Eng Eong Ooi, Email: engeong.ooi@duke-nus.edu.sg.
Kuan Rong Chan, Email: kuanrong.chan@duke-nus.edu.sg.
Appendix. Supplementary materials
References
- 1.Phua J, Weng L, Ling L, Egi M, Lim CM, Divatia JV. Intensive care management of coronavirus disease 2019 (COVID-19): challenges and recommendations. Lancet Respir Med. 2020;8(5):506–517. doi: 10.1016/S2213-2600(20)30161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508) doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe. 2020;27(6):883–890. doi: 10.1016/j.chom.2020.04.017. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong EZ, Chan YFZ, Leong WY, Lee NMY, Kalimuddin S, Haja Mohideen SM. A dynamic immune response shapes COVID-19 progression. Cell Host Microbe. 2020;27(6):879–882. doi: 10.1016/j.chom.2020.03.021. .e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Della-Torre E, Campochiaro C, Cavalli G, De Luca G, Napolitano A, La Marca S. Interleukin-6 blockade with sarilumab in severe COVID-19 pneumonia with systemic hyperinflammation: an open-label cohort study. Ann Rheum Dis. 2020;79(10):1277–1285. doi: 10.1136/annrheumdis-2020-218122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guaraldi G, Meschiari M, Cozzi-Lepri A, Milic J, Tonelli R, Menozzi M. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020;2(8):e474–ee84. doi: 10.1016/S2665-9913(20)30173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang N, Zhan Y, Zhu L, Hou Z, Liu F, Song P. Retrospective multicenter cohort study shows early interferon therapy is associated with favorable clinical responses in COVID-19 patients. Cell Host Microbe. 2020;28(3):455–464. doi: 10.1016/j.chom.2020.07.005. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cauchois R, Koubi M, Delarbre D, Manet C, Carvelli J, Blasco VB. Early IL-1 receptor blockade in severe inflammatory respiratory failure complicating COVID-19. Proc Natl Acad Sci U S A. 2020;117(32):18951–18953. doi: 10.1073/pnas.2009017117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldmann M, Maini RN, Woody JN, Holgate ST, Winter G, Rowland M. Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. Lancet. 2020;395(10234):1407–1409. doi: 10.1016/S0140-6736(20)30858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sterne JAC, Murthy S, Diaz JV, Slutsky AS, Villar J, Angus DC. Association between administration of systemic corticosteroids and mortality among critically Ill patients with COVID-19: a meta-analysis. JAMA. 2020;324(13):1330–1341. doi: 10.1001/jama.2020.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Biasi S, Meschiari M, Gibellini L, Bellinazzi C, Borella R, Fidanza L. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020;11(1):3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arunachalam PS, Wimmers F, Mok CKP, Perera RAPM, Scott M, Hagan T. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369(6508):1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laing AG, Lorenc A, Del Molino Del Barrio I, Das A, Fish M, Monin L. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020 doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 16.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong EZ, Gan ES, de Alwis R, Wijaya L, Ong XM, Zhang M. Genomic signature of early T-cell response is associated with lower antibody titer threshold for sterilizing immunity. Antiviral Res. 2019;166:35–41. doi: 10.1016/j.antiviral.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Correa Marrero M. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182(3):685–712.e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cotto KC, Wagner AH, Feng YY, Kiwala S, Coffman AC, Spies G. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46(D1) doi: 10.1093/nar/gkx1143. D1068-D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci USA. 2005;102(36):12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee TY, Bo-Kai Hsu J, Chang WC, Huang HD. RegPhos: a system to explore the protein kinase-substrate phosphorylation network in humans. Nucleic Acids Res. 2011;39(Database issue):D777–D787. doi: 10.1093/nar/gkq970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poggiali E, Zaino D, Immovilli P, Rovero L, Losi G, Dacrema A. Lactate dehydrogenase and C-reactive protein as predictors of respiratory failure in CoVID-19 patients. Clin Chim Acta. 2020;509:135–138. doi: 10.1016/j.cca.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan L, Wang Q, Zhang D, Ding J, Huang Q, Tang YQ. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5(1):33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L. Dexamethasone in hospitalized patients with COVID-19 - preliminary report. N Engl J Med. 2020 doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uhlen M, Karlsson MJ, Zhong W, Tebani A, Pou C, Mikes J. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science. 2019;366(6472) doi: 10.1126/science.aax9198. [DOI] [PubMed] [Google Scholar]
- 30.Chow JH, Khanna AK, Kethireddy S, Yamane D, Levine A, Jackson AM. Aspirin use is associated with decreased mechanical ventilation, icu admission, and in-hospital mortality in hospitalized patients with COVID-19. Anesth Analg. 2020 doi: 10.1213/ANE.0000000000005292. [DOI] [PubMed] [Google Scholar]
- 31.Jeronimo CMP, Farias MEL, Val FFA, Sampaio VS, Alexandre MAA, Melo GC. Methylprednisolone as adjunctive therapy for patients hospitalized with COVID-19 (Metcovid): a randomised, double-blind, phase IIb, placebo-controlled trial. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furtado RHM, Berwanger O, Fonseca HA, Corrêa TD, Ferraz LR, Lapa MG. Azithromycin in addition to standard of care versus standard of care alone in the treatment of patients admitted to the hospital with severe COVID-19 in Brazil (COALITION II): a randomised clinical trial. Lancet. 2020;396(10256):959–967. doi: 10.1016/S0140-6736(20)31862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Group RC. Lopinavir-ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)32013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann M, Mösbauer K, Hofmann-Winkler H, Kaul A, Kleine-Weber H, Krüger N. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585(7826):588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- 35.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1(6):519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Li S, Liang B, Wang X, Wang H, Li W. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55 doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barnes BJ, Adrover JM, Baxter-Stoltzfus A, Borczuk A, Cools-Lartigue J, Crawford JM. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J Exp Med. 2020;217(6) doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuo Y, Yalavarthi S, Shi H, Gockman K, Zuo M, Madison JA. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5(11) doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Li Q, Yin Y, Zhang Y, Cao Y, Lin X. Excessive neutrophils and neutrophil extracellular traps in COVID-19. Front Immunol. 2020;11:2063. doi: 10.3389/fimmu.2020.02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S, Song R, Wang Z, Jing Z, Ma J. S100A8/A9 in inflammation. Front Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silvin A, Chapuis N, Dunsmore G, Goubet AG, Dubuisson A, Derosa L. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182(6):1401–1418. doi: 10.1016/j.cell.2020.08.002. .e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camp JV, Jonsson CB. A role for neutrophils in viral respiratory disease. Front Immunol. 2017;8:550. doi: 10.3389/fimmu.2017.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang BM, Shojaei M, Teoh S, Meyers A, Ho J, Ball TB. Neutrophils-related host factors associated with severe disease and fatality in patients with influenza infection. Nat Commun. 2019;10(1):3422. doi: 10.1038/s41467-019-11249-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vidy A, Maisonnasse P, Da Costa B, Delmas B, Chevalier C, Le Goffic R. The influenza virus protein PB1-F2 increases viral pathogenesis through neutrophil recruitment and NK cells inhibition. PLoS One. 2016;11(10) doi: 10.1371/journal.pone.0165361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gardinassi LG, Souza COS, Sales-Campos H, Fonseca SG. Immune and metabolic signatures of COVID-19 revealed by transcriptomics data reuse. Front Immunol. 2020;11:1636. doi: 10.3389/fimmu.2020.01636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu M, Chen Y, Xia H, Wang C, Tan CY, Cai X. Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc Natl Acad Sci U S A. 2020;117(45):28336–28343. doi: 10.1073/pnas.2018030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia Z, Song X, Shi J, Wang W, He K. Transcriptome-based drug repositioning for coronavirus disease 2019 (COVID-19) Pathog Dis. 2020;78(4) doi: 10.1093/femspd/ftaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alsamman AM, Zayed H. The transcriptomic profiling of SARS-CoV-2 compared to SARS, MERS, EBOV, and H1N1. PLoS One. 2020;15(12) doi: 10.1371/journal.pone.0243270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caruso A, Caccuri F, Bugatti A, Zani A, Vanoni M, Bonfanti P. Methotrexate inhibits SARS-CoV-2 virus replication "in vitro". J Med Virol. 2020 doi: 10.1002/jmv.26512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi T, Ellingson MK, Wong P, Israelow B, Lucas C, Klein J. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588(7837):315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peckham H, de Gruijter NM, Raine C, Radziszewska A, Ciurtin C, Wedderburn LR. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat Commun. 2020;11(1):6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin JM, Bai P, He W, Wu F, Liu XF, Han DM. Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data for microarray profiling is available at Array Express (E-MTAB-9721). The raw data and normalised counts for NanoString profiling of immune responses is available at Array Express (E-MTAB-9719).