Abstract
The constitutive photomorphogenesis 9 (COP9) signalosome (CSN) is a versatile regulator of plant growth, development, and response to diverse pathogens. However, little research has been done to understand the function of those CSN genes in broad‐spectrum resistance to pathogens. In this study, we found that the transcript levels of wheat TaCSN5 were induced in response to inoculation with Puccinia striiformis f. sp. tritici (Pst) and treatment with salicylic acid (SA). Overexpression of TaCSN5 in Arabidopsis resulted in increased susceptibility to Pseudomonas syringae pv. tomato DC3000 infection accompanied by down‐regulation of AtPR1 expression. Overexpression of TaCSN5 in wheat lines significantly increased susceptibility to Pst accompanied by decreased SA accumulation, whereas TaCSN5‐RNAi wheat lines exhibited opposite trends. Moreover, we found that TaCSN5 negatively regulated TaG3NPR1 genes involved in the SA signalling pathway. In addition, TaCSN5‐RNAi lines showed increased resistance to multiple races of Pst. Taken together, we demonstrate that TaCSN5 contributes to negative regulation of wheat resistance to Pst in an SA‐dependent manner.
Keywords: broad spectrum, CSN subunit, transgenic plants, Triticum aestivum
Stable T4 generation transgenic TaCSN5‐silenced plants exhibited broad‐spectrum resistance to multiple races of stripe rust fungus.
1. INTRODUCTION
During pathogen–host interactions, host assistance is necessary for successful establishment of a compatible interaction (Boller & Felix, 2009; Monaghan & Zipfel, 2012; Torres, 2010). Pathogens use some components of the host to regulate their own growth, and negatively regulate plant immune responses to absorb nutrients from the host, thereby resulting in susceptibility (van Schie & Takken, 2014). All host genes that promote pathogen infection and support compatible interactions are considered susceptibility genes (Lapin & Van den Ackerveken, 2013). Knockdown or knockout of susceptibility genes can limit the virulence of pathogens, resulting in enhanced host resistance (Lapin & Van den Ackerveken, 2013; van Schie & Takken, 2014). In recent years, much progress has been made in research on mechanisms of plant susceptibility. Mildew resistance locus O (MLO), BAX inhibitor‐1 (BI‐1), lifeguard (LFG), RAC/ROP (small G proteins), and GTPase‐activating proteins (GAP) participate in membrane‐related cytoskeletal rearrangement and vesicle transport, mediating susceptibility to many kinds of pathogens (van Schie & Takken, 2014). Mitogen‐activated protein kinase (MAPK) phosphatases, MKP1, MKP2, and tyrosine phosphatase (PTP1), inhibit the MAPK cascade pathway of pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) by targeting inactive MPK3 and MPK6 in Arabidopsis (Lumbreras et al., 2010). Knockout of sugar transporters (OsSWEET11 and OsSWEET14) in rice produces broad‐spectrum resistance to bacterial blight (Xu et al., 2019).
Puccinia striiformis f. sp. tritici (Pst) is one of the most important, highly prevalent, widely distributed, and harmful wheat diseases in the world (Chen et al., 2014; Kang et al., 2017). The most economical, effective, and environment‐friendly strategy to control this disease is breeding wheat‐resistant varieties (Fisher et al., 2012). Therefore, a deep understanding of the molecular mechanism of Pst–wheat interaction will allow us to develop new strategies for durably controlling stripe rust (Staples, 2000). Many Yr (yellow rust resistance) genes have been identified in wheat, particularly those conferring broad‐spectrum disease resistance (Wu et al., 2018). Numerous functional genes in wheat have been intensively reported to be involved in the interaction between wheat and Pst (Kang et al., 2017), but there have been no reports yet of stable silencing of susceptibility genes to confer broad‐spectrum resistance to Pst.
Constitutive photomorphogenesis 9 (COP9) signalosome (CSN), an evolutionarily conserved protein complex in all eukaryotes, is a versatile regulator of plant growth and development (Deng et al., 2000). A typical CSN consists of eight subunits, named CSN1 to CSN8 (Deng et al., 2000; Qin et al., 2020). The role of CSN is related to the regulation of the ubiquitin‐26S proteasome system (UPS) pathway, which plays an important regulatory role in plant growth and development (Hua & Vierstra, 2011). CSN directly interacts with Cullin‐RING‐based E3‐ligases (CRLs), a class of E3 ubiquitin ligases (Wei et al., 2008). The regulation of protein synthesis and degradation by CSN are involved in all processes of plant growth and development (Emberley et al., 2012; Hua & Vierstra, 2011). CSN plays a key role in photomorphogenesis of plants and was discovered originally as members of a class of Constitutive Photomorphogenic/DE‐Etiolated/Fusca (COP/DET/FUS) proteins acting as repressors of photomorphogenesis in Arabidopsis. In response to the biotic stress of pathogenic insects and fungi, CSN responds by interacting with SCFCOI (COR‐insensitive 1) ligase to mediate jasmonic acid (JA) responses (Chamovitz et al., 1996). The key component of salicylic acid (SA)‐mediated disease resistance signalling pathway, NPR1 (nonexpressor of pathogenesis‐related genes), is degraded by Cullin‐3 (CUL3) ubiquitin ligase and this degradation requires a suitable CSN (Chamovitz et al., 1996). In plants, CSN5 is best known as the fifth subunit of CSNs. In Arabidopsis, CSN5 mediates the deconjugation of Neural‐precursor‐cell‐Expressed Developmentally Down‐regulated 8 (NEDD8) from the cullin subunit of E3 ubiquitin ligases (Dohmann et al., 2005). A wheat COP9 subunit 5‐like gene negatively regulates response to Puccinia recondita f. sp. tritici (Zhang et al., 2017). SA is an important signalling hormone for plant disease resistance, and it plays a key role in plant systemic acquired resistance (SAR) signal transduction and disease resistance (Verberne et al., 2000). CSN5 negatively regulates disease resistance through an SA pathway that has not been reported yet.
CSNs have been shown to play an important role in previous studies, but little research has been done to understand the functions of those CSN genes in the interaction between wheat and Pst. In the present study, we discovered an important potential susceptibility factor, TaCSN5, from TaCSNs. A previous study used transient silencing and an ethyl methane sulfonate (EMS)‐mutagenized single genome mutant to verify the negative regulation of resistance to the leaf rust pathogen by a TaCSN5‐like gene (Zhang et al., 2017). Here, we obtained stable T4 generation transgenic wheat TaCSN5 overexpression lines and TaCSN5‐RNAi lines and T3 generation TaCSN5 overexpression lines of Arabidopsis. Our results indicated that TaCSN5 participates in susceptibility of wheat to Pst in an SA‐dependent manner and that TaCSN5‐RNAi wheat lines confer broad‐spectrum resistance to Pst.
2. RESULTS
2.1. TaCSN5 is significantly induced by Pst infection and SA
The CSN is an evolutionarily conserved regulator of proteolysis in eukaryotic development (Deng et al., 2000). We used AtCSNs as seed sequences and found eight TaCSNs at the EnsemblPlants database (http://plants.ensembl.org/common/Tools/Blast?db=core), all of which exist on the wheat A, B, and D chromosomes (Table S1). To determine the roles of TaCSNs in disease resistance, we first analysed the gene transcript levels in time series dual RNA‐Seq data using wheat plants inoculated with Pst. Fragments per kilobase of exon per million mapped reads (FPKM) data of TaCSNs indicated that TaCSN4 and TaCSN5 were down‐regulated in the early stages of the incompatible interaction, whereas only TaCSN5 was up‐regulated in early stages of the compatible interaction (Figure S1). Therefore, we chose TaCSN4 and TaCSN5 for further study. TaCSN6‐7D, TaCSN8‐2B, and TaCSN8‐2D were removed due to very low transcript levels.
To test whether TaCSN4 and TaCSN5 participate in wheat responses to Pst, the transcript levels were determined by quantitative reverse transcription PCR (RT‐qPCR). The identities of the open reading frame (ORF) region of TaCSN4 and TaCSN5 on the A, B, and D chromosomes were 98.9% and 99.7%, respectively (Figure S2). We designed primers to detect transcript profiles of TaCSN4 and TaCSN5 on the three chromosomes simultaneously. The results indicated that the TaCSN4 transcript levels were hardly changed (Figure S3). In the incompatible interaction, the TaCSN5 transcripts did not show significant change. However, in the compatible interaction, TaCSN5 transcripts were significantly induced at 24 and 48 hr postinoculation (hpi) (Figure 1a). Next, we evaluated the transcript levels of TaCSN5 in response to treatment with SA. The transcript level of TaCSN5 was significantly induced at 2–8 hr posttreatment (hpt) (Figure 1b). Thus, we speculated that TaCSN5 participates in the wheat–Pst interaction in an SA‐dependent manner.
FIGURE 1.
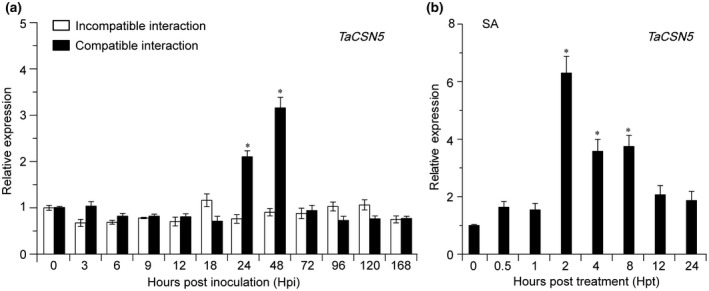
Transcript profiles of TaCSN5 in wheat leaves in response to Puccinia striiformis f. sp. tritici (Pst) infection and salicylic acid (SA) treatment. (a) The transcript levels of TaCSN5 in wheat leaves were induced during Pst infection. (b) The transcript levels of TaCSN5 in response to SA treatment. Wheat leaves treated with 0.1% ethanol were included as a control. Error bars represent the variation among three independent replicates. Quantitative reverse transcription PCR values were normalized to those for TaEF‐1α. Differences between time‐course points were assessed using Student's t test (*p < .05)
2.2. TaCSN5 negatively regulates SA‐induced disease resistance in Arabidopsis
To investigate whether the TaCSN5 promoter (TaCSN5p) responds to SA treatment, we fused the TaCSN5p with the β‐glucuronidase (GUS) gene to transform into Arabidopsis and produced six T1 generation lines (TaCSN5p‐GUS‐1, ‐2, ‐3, ‐4, ‐5, and ‐6). Three independent T1 transgenic lines (TaCSN5p‐GUS‐2, TaCSN5p‐GUS‐3, and TaCSN5p‐GUS‐6) were initially validated using PCR (Figure S4a and Table S2) and selected for further study. These three T3 transgenic lines were further confirmed by PCR assay (Figure S4b and Table S2). The histological GUS activity was determined using staining. As shown in Figure S5a, the leaves of the transgenic Arabidopsis expressed more blue signals after SA treatment. GUS expression levels were confirmed by RT‐qPCR (Figure S5b). Our results indicated that TaCSN5p can be activated by SA.
To further explore TaCSN5 function, TaCSN5 was transformed into Arabidopsis under the control of the CaMV 35S promoter and produced seven T1 generation lines (35S::TaCSN5‐OE1, ‐OE2, ‐OE3, ‐OE4, ‐OE5, ‐OE6, and ‐OE7). Three independent T1 transgenic lines (35S::TaCSN5‐OE1, 35S::TaCSN5‐OE3, and 35S::TaCSN5‐OE7) were initially validated using PCR (Figure S6a and Table S2) and RT‐qPCR assays (Figure S6b and Table S2), and selected for further study. The three T3 transgenic lines were further confirmed by PCR and RT‐qPCR assays (Figures S6c and 6d, and Table S2). We inoculated them with Pseudomonas syringae pv. tomato (Pto) DC3000; the overexpression lines had a more severe disease phenotype (Figure 2a), and the transcript level of AtPR1 was significantly down‐regulated in overexpression plants at 1 hpi (Figure 2b). The SA‐dependent pathway was found to increase the expression of the PR1 gene (Niki et al., 1998). Therefore, we treated the wild type (WT) and transgenic lines with SA, which significantly induces the AtPR1 transcript (Figure 2c). However, compared with WT plants, the transcript levels of AtPR1 in 35S::TaCSN5 plants were significantly down‐regulated at 1, 2, 4, and 8 hpt, suggesting that TaCSN5 negatively regulates SA‐induced AtPR1 expression (Figure 2c).
FIGURE 2.
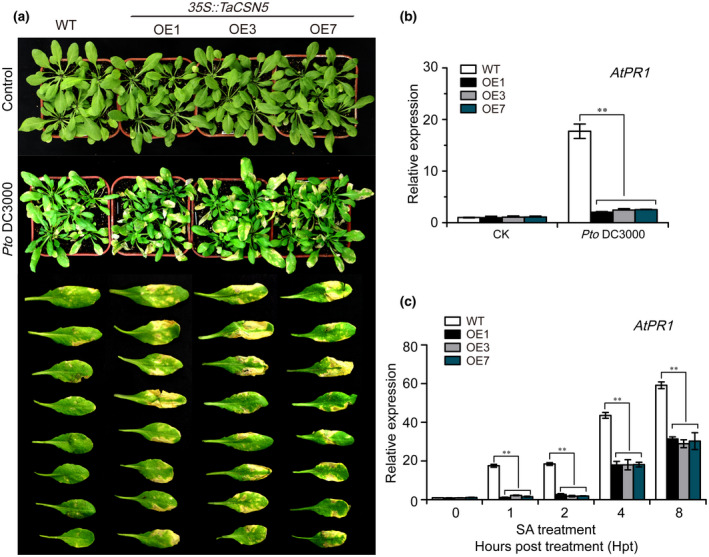
TaCSN5 negatively regulates salicylic acid (SA)‐induced disease resistance in Arabidopsis. (a) Pseudomonas syringae pv. tomato (Pto) DC3000 defence phenotypes on 4‐week‐old T3 35S::TaCSN5 Arabidopsis (OE1, OE3, and OE7) and wild type (WT) (Col‐0) leaves. (b) The transcript levels of AtPR1 in transgenic Arabidopsis and WT response to Pto DC3000 at 1 hr postinoculation (hpi). (c) The transcript levels of AtPR1 in 35S::TaCSN5 Arabidopsis plants in response to SA treatment were significantly reduced compare to WT at 1, 2, 4, and 8 hpi. Quantitative reverse transcription PCR values were normalized for AtActin2. Error bars represent the variations among three independent replicates. Asterisks indicate significant differences between transgenic and control plants at the same time points using Student's t test (**p < .01)
2.3. Overexpression of TaCSN5 significantly increases wheat susceptibility to Pst infection
To further verify the function of TaCSN5, we developed transgenic wheat plants in the Fielder background in which TaCSN5, driven by the maize ubiquitin promoter, was overexpressed and produced five T1 generation lines (TaCSN5‐OE1, ‐OE2, ‐OE3, ‐OE4, and ‐OE5). Two independent T1 transgenic lines (TaCSN5‐OE2 and TaCSN5‐OE5) were initially validated using PCR (Figure S7a and Table S2) and RT‐qPCR assays (Figure S7b and Table S2), and selected for further study. The two T4 transgenic lines were further confirmed by PCR and RT‐qPCR assays (Figures S7c and 3a, and Table S2). No significant differences were observed between the TaCSN5 overexpression plants and Fielder plants under normal growth conditions (Figure 3b). At 14 days postinoculation, hypersensitive response (HR) symptoms appeared on all leaves after inoculation with CYR23. A few fungal uredia surrounding the necrotic areas were observed on leaves of TaCSN5 overexpression lines (Figure 3c). In contrast, all leaves infected with the virulent strain CYR33 produced extensive uredia (Figure 3c). Compared with Fielder plants, fungal biomass was significantly increased by 9% to 21% in overexpression plants (Figure 3d). These results suggest that elevated expression of TaCSN5 enhanced wheat susceptibility to Pst infection.
FIGURE 3.
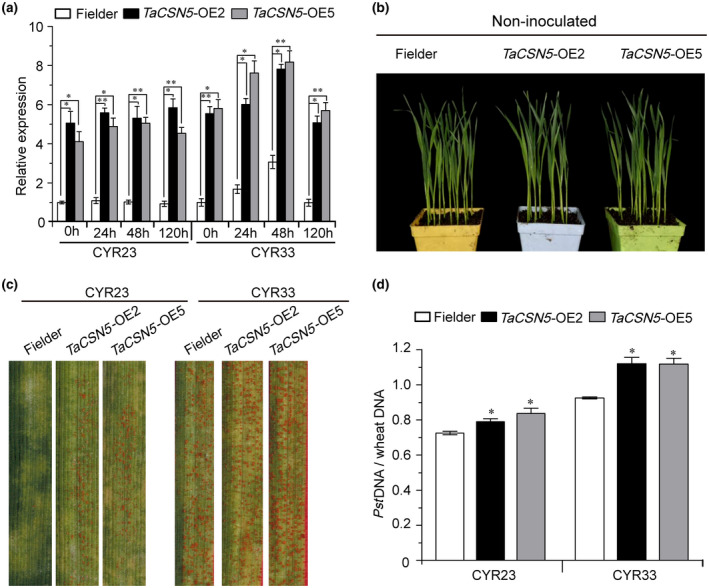
Overexpression of TaCSN5 enhances wheat susceptibility to Puccinia striiformis f. sp. tritici (Pst) infection. (a) The transcript levels of TaCSN5 in the leaves of T4 generation TaCSN5‐OE2 and TaCSN5‐OE5 lines during Pst infection. The data were normalized to those for TaEF‐1α and presented as fold changes relative to Fielder plants. (b) Phenotypes of noninoculated TaCSN5 overexpression (OE2 and OE5) and Fielder plants. (c) Foliar parts of TaCSN5‐OE2 and TaCSN5‐OE5 lines inoculated with Pst isolate CYR23 or CYR33. (d) Ratio of fungal to wheat nuclear DNA content determined using the contents of fungal Pst EF1 and wheat TaEF‐1α genes, respectively. Genomic DNA was extracted from three different plants at 7 days postinoculation. Error bars represent the variation among three independent replicates. Asterisks indicate significant differences between that in transgenic plants and Fielder plants at the same time points using Student's t test (*p < .05; **p < .01)
2.4. Suppression of TaCSN5 improves wheat resistance to Pst infection
To further investigate the function of TaCSN5 in response to Pst, we generated wheat transgenic lines silencing TaCSN5 using RNAi technology and produced 10 T1 generation lines (TaCSN5‐Ri1, ‐Ri2, ‐Ri3, ‐Ri4, ‐Ri5, ‐Ri6, ‐Ri7, ‐Ri8, ‐Ri9, and ‐Ri10). Two independent T1 transgenic lines (TaCSN5‐Ri8 and TaCSN5‐Ri10) were initially validated using PCR (Figure S8a and Table S2) and RT‐qPCR assays (Figure S8b and Table S2), and selected for further study. The two T4 transgenic lines were further confirmed by PCR and RT‐qPCR assays (Figures S8c,d and 4a, and Table S2). It is worth noting that only TaCSN5 and TaCSN6 in wheat contain an MPN domain, and the 225 bp‐RNAi fragment was designed on the MPN domain (Figure S9). The identity of the ORF region of TaCSN6 in the A, B, and D chromosome is 98.82% (Figure S10), so we designed primers to detect TaCSN6 on three chromosomes simultaneously (Table S2). No significant difference was found on the transcript level of TaCSN6 in TaCSN5‐RNAi lines compared with that in the Fielder plants (Figure S8d). No obvious differences were observed in the growth between TaCSN5‐RNAi and Fielder plants under normal growth conditions (Figure 4b). The second leaf of all wheat plants was inoculated with fresh urediospores of Pst races CYR23 and CYR33. At 14 days after inoculation, conspicuous HR symptoms appeared on all leaves after inoculation with CYR23 (Figure 4c). The TaCSN5‐RNAi lines expressed enhanced resistance with a significant reduction in sporulation compared to Fielder plants inoculated with CYR33 (Figure 4c). Moreover, compared with Fielder plants, the fungal biomass was significantly reduced by 8% to 20% (Figure 4d). These results suggest that reduction in TaCSN5 expression improves wheat resistance to Pst infection.
FIGURE 4.
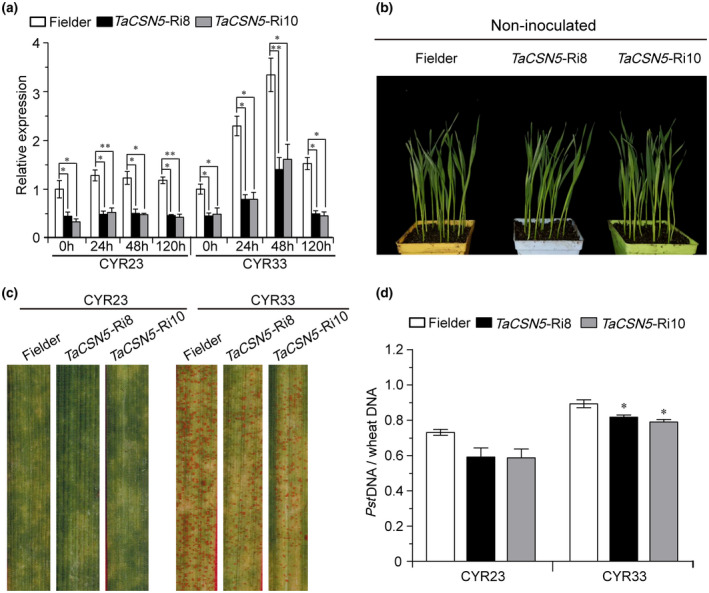
Silencing of TaCSN5 enhances wheat resistance against Puccinia striiformis f. sp. tritici (Pst) infection. (a) The transcript levels of TaCSN5 in the leaves of T4 generation TaCSN5‐Ri8 and TaCSN5‐Ri10 lines during Pst infection. The data were normalized to those for TaEF‐1α and presented as fold changes relative to Fielder. (b) Phenotypes of noninoculated TaCSN5‐RNAi (Ri8 and Ri10) and Fielder plants. (c) Foliar parts of TaCSN5‐Ri8 and TaCSN5‐Ri10 lines inoculated with Pst isolate CYR23 or CYR33. (d) Ratio of fungal to wheat nuclear DNA content determined using the contents of fungal Pst EF1 and wheat TaEF‐1α genes, respectively. Genomic DNA was extracted from three different plants at 7 days postinoculation. Error bars represent the variation among three independent replicates. Asterisks indicate significant differences between that in transgenic plants and Fielder plants at the same time points using Student's t test (*p < .05; **p < .01)
2.5. Histological changes in Pst growth in TaCSN5‐RNAi and TaCSN5 overexpression plants
To determine whether the phenotypic changes in wheat leaves are associated with fungal growth and development when TaCSN5 was knocked down, leaf segments inoculated with CYR33 were examined microscopically. We found that the length of hyphae, and the number of haustorial mother cells and infection areas in TaCSN5‐RNAi plants showed a slight but nonsignificant decrease compared to Fielder plants at 48 hpi. In addition, the infection areas per infection site were obviously smaller in TaCSN5‐RNAi plants compared with the Fielder plants at 120 hpi (Figure 5). We tested TaCSN5 overexpression wheat plants in the same way after inoculation with avirulent race CYR23. We found that the length of hyphae, and the number of haustorial mother cells and infection areas of TaCSN5 overexpression plants compared with the Fielder plants at 48 hpi showed a slight but nonsignificant increase. In addition, the infection areas per infection site were obviously larger in TaCSN5 overexpression plants compared with the Fielder plants at 120 hpi (Figure S11). These histological results indicated that TaCSN5 promotes Pst growth.
FIGURE 5.
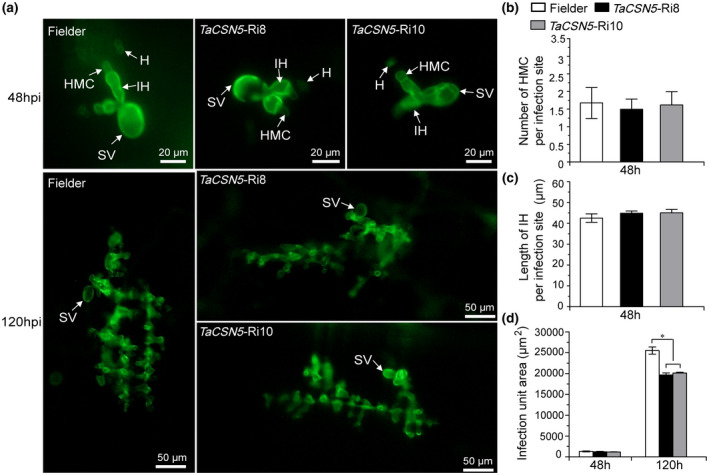
Growth of Puccinia striiformis f. sp. tritici (Pst) virulent race CYR33 was reduced in TaCSN5‐RNAi plants. (a) The fungal structures were stained with wheat germ agglutinin (WGA) in wheat leaves inoculated with Pst and observed under a fluorescence microscope. SV, substomatal vesicle; HMC, haustorial mother cell; IH, infection hypha; H, haustoria. (b) The average number of haustorial mother cells in each infection site was determined. (c) Hyphal length of CYR33, which is the average distance from the junction of the substomatal vesicle and the hypha to the tip of the hypha, was measured with DP‐BSW software. (d) Infection area of CYR33, the average of expanding hyphae, was calculated by DP‐BSW software. All results were obtained from 30–50 infection sites and three biological replications were performed. Asterisks indicate significant differences between that in transgenic plants and Fielder plants at the same time points using Student's t test (*p < .05)
2.6. TaCSN5 negatively regulates multiple pathogenesis‐related genes during Pst infection
PR1 is considered to be a marker for infection resistance in plants to improve disease resistance (Van Loon & Van Strien, 1999). PR2 has an antibacterial effect due to its hydrolytic enzyme function (Mauch et al., 1988). Several studies have reported that PR5 can improve the resistance of cereal crops to fungal diseases (Pritsch et al., 2000; Schaffrath et al., 1997). To confirm the increased resistance in TaCSN5‐RNAi plants, the expression of three TaPR genes were examined by RT‐qPCR. As shown in Figure 6, TaPR1, TaPR2, and TaPR5 were up‐regulated during Pst infection (Figure 6a–c). At the same time, a corresponding decrease was shown in the TaCSN5 overexpression plants (Figure S12a–c). Therefore, the results support the conclusion that TaCSN5 enhances the susceptibility of wheat to the stripe rust fungus.
FIGURE 6.
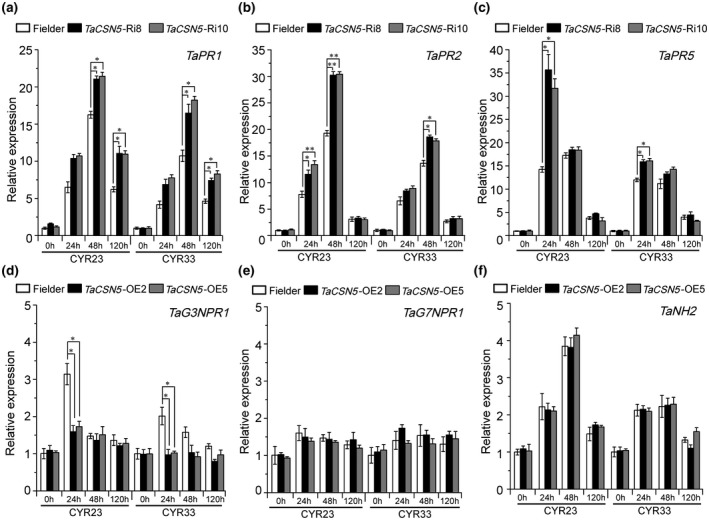
TaCSN5 down‐regulates transcription of PR genes in wheat. The transcript levels of TaPR1 (a), TaPR2 (b), and TaPR5 (c) in TaCSN5‐RNAi lines during Puccinia striiformis f. sp. tritici (Pst) infection. The transcript levels of TaG3NPR1 (d), TaG7NPR1 (e), and TaNH2 (f) in TaCSN5 overexpression lines during Pst infection. Quantitative reverse transcription PCR values were normalized to those for TaEF‐1α. The transcript levels of genes in Fielder plants at time 0 was standardized as 1. Error bars represent the variation among three independent replicates. Asterisks indicate significant differences between that in transgenic plants and Fielder plants at the same time points using Student's t test (*p < .05; **p < .01)
In Arabidopsis, CSN assists CUL3 ubiquitin ligase to degrade NPR1, the key component of the SA‐mediated disease resistance signalling pathway (Chamovitz et al., 1996). Therefore, we further explored the possibility that TaCSN5 affects the NPR1‐like genes of wheat. Wheat contains wNPR1, TaNH2, TaG3NPR1, and TaG7NPR1 genes including the NPR1‐like domain. wNPR1 is a member of the TaG3NPR1 gene family, which are required for the SA‐mediated expression of PR genes (Wang et al., 2020). We determined the transcript levels of TaG3NPR1 genes, TaG7NPR1 genes, and TaNH2 in TaCSN5 overexpression plants during Pst infection. We found that the transcript levels of TaG3NPR1 in TaCSN5 overexpression lines decreased significantly at 24 hpi (Figure 6d), but the transcript levels of TaG7NPR1 and TaNH2 showed little change relative to the Fielder plants (Figure 6e,f). Therefore, TaCSN5 negatively regulates PR genes through TaG3NPR1 genes.
2.7. TaCSN5 negatively regulates SA accumulation during Pst infection
To determine whether TaCSN5 expression can affect SA balance in wheat and thereby lead to increased susceptibility to Pst, we further quantified the SA content in TaCSN5 overexpression, TaCSN5‐RNAi, and Fielder plants during Pst infection. The SA concentration in TaCSN5 overexpression plants was significantly decreased, but in TaCSN5‐RNAi plants it showed a significant increase at 48 hpi during incompatible and compatible interactions (Figure 7). Our results indicated that TaCSN5 negatively regulates wheat resistance against Pst by affecting SA accumulation.
FIGURE 7.
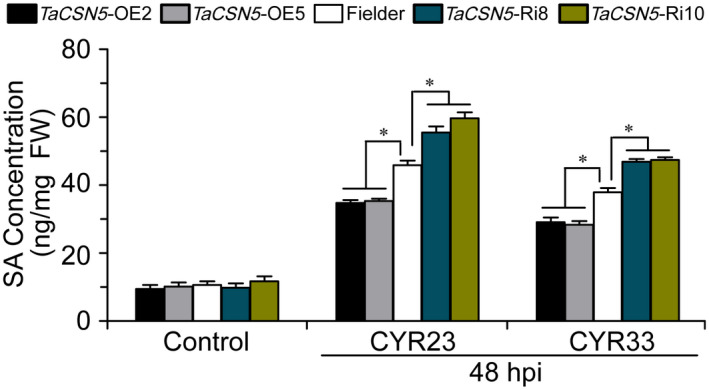
Quantification of salicylic acid (SA) in TaCSN5 overexpression, TaCSN5‐RNAi, and Fielder plants. Samples were isolated from the second leaves of the wheat seedlings infected with CYR23 (incompatible interaction) and CYR33 (incompatible interaction) at 48 hr postinoculation (hpi). Noninoculated wheat leaves were included as a control. Error bars represent the variation among three independent replicates. Asterisks indicate significant differences between transgenic and Fielder plants using Student's t test (*p < .05)
2.8. TaCSN5‐RNAi lines express broad‐spectrum resistance to Pst
The previous steps confirmed that TaCSN5‐RNAi lines have a clearly resistant phenotype to CYR33. To verify whether TaCSN5 function is specific to an individual Pst race, we used the CYR20, CYR22, CYR25, CYR29, CYR31, CYR32, and CYR34 races to examine the resistance of TaCSN5‐RNAi lines. It is worth noting that the current prevalent Pst races in China are CYR32, CYR33, and CYR34 (Liu et al., 2017). We found that both Fielder plants and transgenic lines were susceptible to CYR31 (Figure 8a), CYR32 (Figure 8a), and CYR34 (Figure 8c), and only a few fungal uredia surrounding the necrotic areas were observed on leaves inoculated with CYR29 (Figure 8d). Compared with Fielder plants, fungal biomass was significantly reduced in TaCSN5‐RNAi lines (Figure 8a–d). The Fielder plants and transgenic lines expressed disease‐resistance phenotypes to CYR20, CYR22, and CYR25. Their phenotypes were not obviously different, and there was no significant difference in fungal biomass (Figure S13a–c). In addition, we determined the expression of PR genes at 48 hpi and found that the transcript levels of TaPR1 and TaPR2 were significantly up‐regulated in TaCSN5‐RNAi lines (Figure S13a–c). Thus, our results indicate that TaCSN5‐RNAi lines confer broad‐spectrum resistance to Pst.
FIGURE 8.
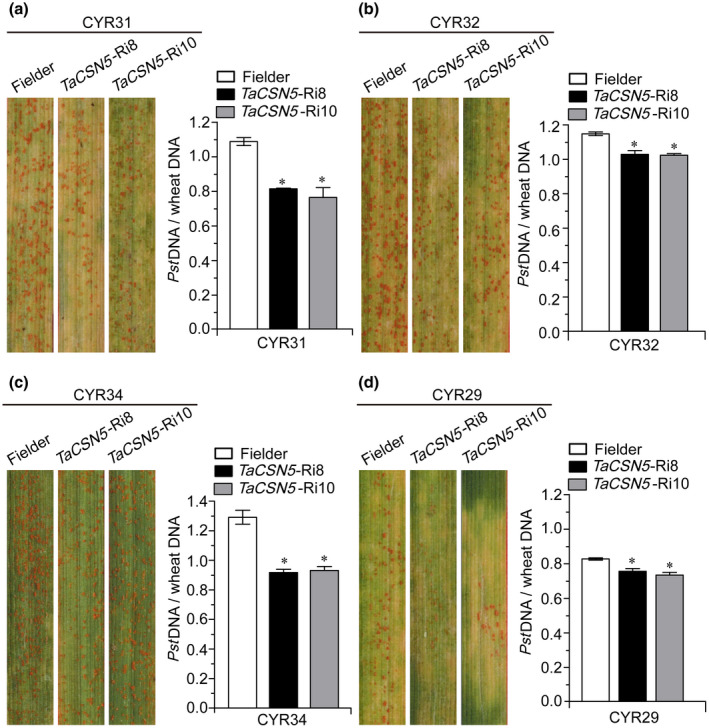
TaCSN5‐RNAi lines expressed broad‐spectrum resistance to Puccinia striiformis f. sp. tritici (Pst). Foliar parts of T4 generation TaCSN5‐Ri8 and TaCSN5‐Ri10 lines inoculated with Pst isolate CYR31 (a), CYR32 (b), CYR34 (c), and CYR29 (d). Ratio of fungal to wheat nuclear DNA content determined using the contents of fungal Pst EF1 and wheat TaEF‐1α genes, respectively. Genomic DNA was extracted from three different plants at 7 days postinoculation. Error bars represent the variation among three independent replicates. Asterisks indicate significant differences between that in transgenic and Fielder plants using Student's t test (*p < .05)
3. DISCUSSION
Pathogenesis‐related (PR) proteins are of great interest for engineering plant disease resistance (Sels et al., 2008; Sinha et al., 2014). During host–pathogen interactions, PR proteins not only accumulate locally in the infected leaf, but are also induced systemically (Hamamouch et al., 2011). In this study, we determined that TaCSN5 performs a susceptibility function; the expression of PR genes and TaG3NPR1 genes was significantly regulated (Figure 6d). Overexpression of AtNPR1 in other plant species (e.g., rice and wheat) enhances their resistance against multiple pathogens (Gao et al., 2013; Quilis et al., 2008; Xu et al., 2017). Wheat wNPR1 and barley HvNPR1 positively regulate resistance against powdery mildew and stripe rust (Gao et al., 2018). The TaG3NPR1 genes are required for SA‐mediated PR expression, and TaG7NPR1‐7A negatively regulates resistance to stem rust (Wang et al., 2020), indicating that the number of NPR1 genes in wheat is higher than in Arabidopsis, and the mechanism is more complicated. In this study, we showed that TaCSN5 negatively regulated SA‐induced AtPR1 expression (Figure 2d). Therefore, we speculate that TaG3NPR1 genes function similarly to the AtNPR1 gene. CSN assists CUL3 ubiquitin ligase to degrade NPR1 in Arabidopsis (Chamovitz et al., 1996). We therefore speculate that TaCSN5 is involved in the degradation of TaG3NPR1. Overall, TaCSN5 contributes to negative regulation of wheat resistance in an SA‐dependent manner by affecting TaG3NPR1 genes expression.
CSN is an evolutionarily conserved protein complex of 450–500 kDa in all eukaryotes and typically CSN consists of eight subunits (Deng et al., 2000; Qin et al., 2020). In this study, AtCSN1 to AtCSN8 were used as seed sequences to identify wheat TaCSNs. The preliminary analysis of transcriptome data showed that TaCSN5 was down‐regulated in the early stage of an incompatible interaction during Pst infection and was up‐regulated in the early stage of a compatible interaction, and thus may play a role in susceptibility (Table S2). The functional verification also confirmed our speculation. In Arabidopsis, both AtCSN5 and AtCSN6 have two members (Pick et al., 2009), but the BLAST search results of AtCSN5a and AtCSN5b were both TaCSN5, the same as TaCSN6 (Table S1). It is worth noting that TaCSN5‐5B was not retrieved from the EnsemblPlants database. We obtained the sequence of TaCSN5‐5B through genomic sequence alignment and the local transcriptome database, which indicates that TaCSN5‐5B is present in wheat. Based on the phenotype and histological data of TaCSN5‐RNAi lines, we found that TaCSN5 and other CSNs have no functional redundancy, indicating that TaCSN5 may be specific for the susceptibility function in wheat (Figure 3).
Previous reports have shown that a CSN5‐like gene can negatively regulate disease resistance to Puccinia triticina, and that TaPR1 is significantly inhibited (Zhang et al., 2017). We found that it is the same as the gene in this study. TaCSN5‐RNAi lines produced disease‐resistant phenotypes to five races, suggesting that TaCSN5 regulates disease resistance to other rust fungi. Overexpression of TaCSN5 in Arabidopsis resulted in susceptibility to the bacterial pathogen Pto DC3000. We therefore speculate that TaCSN5 confers susceptibility to diverse pathogens. In Arabidopsis, accumulation of the plant hormone SA and transcriptional activation of PR genes are associated with SAR (Zheng & Dong, 2013). In this study, TaCSN5 contributed to negative regulation of wheat resistance in an SA‐dependent manner, and PR genes were also significantly regulated, so we speculate that TaCSN5 may negatively regulate SAR. Therefore, TaCSN5 is a candidate susceptibility gene and will not participate in wheat resistance. CRISPR has become a revolutionary tool for plant genome editing (Wright et al., 2016). In recent years, it has been reported that gene‐editing technology has been successfully applied in many crops and has improved important traits such as heat resistance, cold tolerance, disease resistance, and yield (Zaidi et al., 2018). Further study will be performed on editing TaCSN5 with CRISPR.
4. EXPERIMENTAL PROCEDURES
4.1. Gene expression analysis
The transcript level of all TaCSN genes was determined using dual RNA‐Seq data in our laboratory. We sequenced two groups of wheat–rust interaction combinations named NIL_R versus CYR32 and NIL_S versus CYR32, and selected a series of time points at 0, 18, 24, 48, and 96 hpi. The specific method is the same as our previous study (Guo et al., 2018).
The wheat cultivar Suwon 11 was used to amplify cDNA sequences of TaCSN5. Suwon 11, having the YrSu gene, is resistant to race CYR23 but highly susceptible to race CYR31. For RNA extraction, the second leaves inoculated with CYR23 or CYR31 or treated with sterile distilled water (control) were harvested at 0, 3, 6, 9, 12, 18, 24, 48, 72, 96, 120, and 168 hpi. Total RNA was isolated using a Quick RNA isolation Kit (Huayueyang) according to the manufacturer's protocol and was quantified using a spectrometer (NanoDrop). First‐strand cDNA was synthesized from 3 mg of total RNA, using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific), followed by RT‐qPCR using ChamQ SYBR qPCR Master Mix (Vazyme). RT‐qPCR was performed with a CFX Connect Real‐Time System (Bio‐Rad). A 107‐bp fragment of wheat housekeeping gene TaEF‐1α (GenBank accession number M90077.1) was amplified as an internal reference for the RT‐qPCR analysis, and the data were calculated by the comparative 2−ΔΔ C t method (Pfaffl, 2001).
4.2. SA treatment
For SA treatments, 10‐day‐old wheat seedlings were sprayed with 2 mM SA dissolved in 0.1% (vol/vol) ethanol. Control plants were sprayed with 0.1% ethanol (Liu et al., 2019). Samples were collected at 0, 0.5, 1, 2, 4, 8, 12, and 24 hpt for RNA extraction and RT‐qPCR.
The T3 generation transgenic Arabidopsis thaliana seeds were germinated and grown on Murashige & Skoog (MS) medium for 5 days, and then exposed to MS medium containing 50 μM SA (Lu et al., 2018) under a day/night 16/8 hr cycle at 22 °C. A. thaliana Col‐0 plants were used as the control. They were sampled at 0, 1, 2, 4, and 8 hpt, and then used to extract RNA.
4.3. Production of transgenic plants
The wheat cultivar Fielder was used as the receptor material to generate transgenic plants. To produce TaCSN5 transgenic wheat plants, the coding sequence of TaCSN5 was cloned into the plant transformation vector pWMB110 driven by the maize ubiquitin promoter. The 225‐bp TaCSN5‐specific forward fragment, the 146 bp intermediate intron sequence, and the 225 bp reverse sequence of the specific fragment were synthesized by Beijing AuGCT. To generate the pWMB110‐TaCSN5‐RNAi construct, the recombinant DNA was inserted into the pWMB110 vector. For genetic transformations, an Agrobacterium‐mediated transformation system was used (Hayta et al., 2019). For Arabidopsis, the coding sequence of TaCSN5 was introduced into the plant transformation vector pCAMBIA1302 under the control of the CaMV 35S promoter. The resultant constructs were confirmed by sequencing and then transformed into Col‐0 plants via the vacuum infiltration method (Bechtold & Pelletier, 1998). Transgenic lines were initially validated by amplifying the insertion of transgene in the genomic DNA using a PCR assay (Table S2). The transcript levels of TaCSN5 in corresponding wheat transgenic lines were determined by RT‐qPCR assay (Table S2).
4.4. GUS activity assay and Pto DC3000 inoculation
For the GUS activity assay, the promoter of TaCSN5 was amplified from wheat cultivar Suwon 11, cloned into pCAMBIA1305, and transformed into Col‐0 plants as described in the previous study (Lu et al., 2018).
The Pto DC3000 was grown on King's B (KB) medium (King et al., 1954) supplemented with 25 mg/L rifampicin for 48 hr at 28 °C and resuspended in 10 mM MgCl2 to OD600 = 0.002. Leaves of 4‐week‐old plants were infected with the bacterial suspension by pressing a 1‐ml syringe (without a needle) against the abaxial side of the leaves and forcing the suspension through the stomata into the intercellular spaces as described in the previous study (Lu et al., 2018). Plants were sampled at 0 and 1 hr and then used to extract RNA.
4.5. Fungal isolates and Pst inoculation
Pst races CYR20, CYR22, CYR23, CYR25, CYR29, CYR31, CYR32, CYR33, and CYR34 were used in this study. Fielder contains Yr6 and Yr20 genes (Chen, 2007) and shows an incompatible interaction with Pst races CYR20, CYR22, CYR23, CYR25, and CYR29, and compatible interaction with Pst races CYR31, CYR32, CYR33, and CYR34. Wheat plants were grown in a greenhouse at 25/23 °C day/night temperatures and long‐day conditions (16 hr light/8 hr dark photoperiod). The second leaf of T4 generation TaCSN5 overexpression and TaCSN5‐RNAi wheat plants were inoculated with urediospores of CYR23 or CYR33. Leaves were collected at 0, 24, 48, and 120 hpi for RNA extraction and histological study. At 14 days after inoculation with Pst, the phenotypes were photographed and the inoculated leaves were collected at 7 days to measure fungal biomass by quantitative PCR (Qi et al., 2018). Transgenic wheat and Fielder plants were inoculated with CYR20, CYR22, CYR25, CYR29, CYR31, CYR32, and CYR34, and fungal biomass was measured by the same method (Qi et al., 2018).
4.6. Histochemical observation
For histochemical analysis, the leaves inoculated with CYR23 or CYR33 were sampled at 48 and 120 hpi. Wheat germ agglutinin conjugated to Alexa‐488 (Invitrogen) was used to obtain high‐quality images of Pst infection structures in wheat leaves. Stained tissues were observed under blue light excitation (excitation wavelength 450–480 nm, emission wavelength 515 nm). Infection sites (30–50) were observed to record the hyphal length, number of haustorial mother cells, and infection areas of hyphae in infected wheat per unit. The presence of a substomatal vesicle was defined as an established infection unit. The hyphal length of Pst was measured from the base of the substomatal vesicle to the apex of the longest infection hypha. A BX‐51 microscope (Olympus) was used for all microscopic observations. Experiments were repeated three times.
4.7. SA quantification
For the quantification of SA, approximately 0.1 g of fresh leaf tissue of transgenic wheat and Fielder plants was used to extract SA at 48 hpi during Pst infection, and analysed with HPLC‐MS (Qtrap 5500; AB SCIEX) with the protocol previously described (Segarra et al., 2006).
Supporting information
ACKNOWLEDGEMENTS
This study was financially supported by the National Transgenic Key Project of the Ministry of Agriculture of China (2020ZX08009‐15B), the National Natural Science Foundation of China (31972224), the National Key R&D Program of China (2018YFD0200402), the Natural Science Basic Research Program of Shaanxi (2020JZ‐13), and the 111 Project from the Ministry of Education of China (no. B07049).
Bai X, Huang X, Tian S, et al. RNAi‐mediated stable silencing of TaCSN5 confers broad‐spectrum resistance to Puccinia striiformis f. sp. tritici . Mol Plant Pathol. 2021;22:410–421. 10.1111/mpp.13034
Xingxuan Bai and Xueling Huang contributed equally to this work.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Bechtold, N. & Pelletier, G. (1998) In planta Agrobacterium‐mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods in Molecular Biology, 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Boller, T. & Felix, G. (2009) A renaissance of elicitors: Perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annual Review of Plant Biology, 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Chamovitz, D.A. , Wei, N. , Osterlund, M.T. , von Arnim, A.G. , Staub, J.M. , Matsui, M. et al. (1996) The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell, 86, 115–121. [DOI] [PubMed] [Google Scholar]
- Chen X.M. (2007) Challenges and solutions for stripe rust control in the United States. Australian Journal of Agricultural Research, 58, 648–655. [Google Scholar]
- Chen, W.Q. , Wellings, C. , Chen, X.M. , Kang, Z.S. & Liu, T.G. (2014) Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici . Molecular Plant Pathology, 15, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X.W. , Dubiel, W. , Wei, N. , Hofmann, K. & Mundt, K. (2000) Unified nomenclature for the COP9 signalosome and its subunits: An essential regulator of development. Trends in Genetics, 16, 289. [DOI] [PubMed] [Google Scholar]
- Dohmann, M.N. , Kuhnle, C. & Schwechheimer, C. (2005) Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis . The Plant Cell, 17, 1967–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberley, E.D. , Mosadeghi, R. & Deshaies, R.J. (2012) Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1‐F‐box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism. Journal of Biological Chemistry, 287, 29679–29689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, M.C. , Henk, D.A. , Briggs, C.J. , Brownstein, J.S. , Madoff, L.C. , McCraw, S.L. et al. (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature, 484, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, C.S. , Kou, X.J. , Li, H.P. , Zhang, J.B. , Saad, A.S.I. & Liao, Y.C. (2013) Inverse effects of Arabidopsis NPR1 gene on fusarium seedling blight and fusarium head blight in transgenic wheat. Plant Pathology, 62, 383–392. [Google Scholar]
- Gao, J. , Bi, W. , Li, H. , Wu, J. , Yu, X. , Liu, D. et al. (2018) WRKY transcription factors associated with NPR1‐mediated acquired resistance in barley are potential resources to improve wheat resistance to Puccinia triticina . Frontiers in Plant Science, 9, 1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Islam M.A., Lin H., Ji C., Duan Y., Liu P., Zeng Q., Day B., Kang Z., Guo J. (2018) Genome‐wide identification of cyclic nucleotide‐gated Ion channel gene family in wheat and functional analyses of TaCNGC14 and TaCNGC16. Frontiers in Plant Science, 9, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamouch, N. , Li, C.Y. , Seo, P.J. , Park, C.M. & Davis, E.L. (2011) Expression of Arabidopsis pathogenesis‐related genes during nematode infection. Molecular Plant Pathology, 12, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayta, S. , Smedley, M.A. , Demir, S.U. , Blundell, R. , Hinchliffe, A. , Atkinson, N. et al. (2019) An efficient and reproducible Agrobacterium‐mediated transformation method for hexaploid wheat (Triticum aestivum L.). Plant Methods, 15, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, Z. & Vierstra, R.D. (2011) The cullin‐RING ubiquitin‐protein ligases. Annual Review of Plant Biology, 62, 299–334. [DOI] [PubMed] [Google Scholar]
- Kang, Z.S. , Tang, C.L. , Zhao, J. , Cheng, Y.L. , Liu, J. , Guo, J. et al. (2017) Wheat–Puccinia striiformis interactions. In: Chen, X.M. & Kang, Z.S. (Eds.) Stripe rust. Springer, pp. 155–282. [Google Scholar]
- King, E.O. , Ward, M.K. & Raney, D.E. (1954) Two simple media for the demonstration of pyocyanin and fluorescin. Journal of Laboratory and Clinical Medicine, 44, 301–307. [PubMed] [Google Scholar]
- Lapin, D. & Van den Ackerveken, G. (2013) Susceptibility to plant disease: More than a failure of host immunity. Trends in Plant Science, 18, 546–554. [DOI] [PubMed] [Google Scholar]
- Liu P., Guo J., Zhang R., Zhao J., Liu C., Qi T., Duan Y., Kang Z., Guo J. (2019) TaCIPK10 interacts with and phosphorylates TaNH2 to activate wheat defense responses to stripe rust. Plant Biotechnology Journal, 17, 956–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Liu, T.G. , Zhang, Z.Y. , Jia, Q.Z. , Wang, B.T. , Gao, L. , Peng, Y.L. , Jin, S.L. & Chen, W.Q. (2017) Discovery and pathogenicity of CYR34, a new race of Puccinia striiformis f. sp. tritici in China. Acta Phytopathologica Sinica, 47, 681–687. [Google Scholar]
- Lu, P.‐P. , Yu, T.‐F. , Zheng, W.‐J. , Chen, M. , Zhou, Y.‐B. , Chen, J. et al. (2018) The wheat bax inhibitor‐1 protein interacts with an aquaporin TaPIP1 and enhances disease resistance in Arabidopsis . Frontiers in Plant Science, 9, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras, V. , Vilela, B. , Irar, S. , Solé, M. , Capellades, M. , Valls, M. et al. (2010) MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. The Plant Journal, 63, 1017–1030. [DOI] [PubMed] [Google Scholar]
- Mauch, F. , Mauch‐Mani, B. & Boller, T. (1988) Antifungal hydrolases in pea tissue. II. Inhibition of fungal growth by combinations of chitinase and β‐1,3‐glucanase. Plant Physiology, 88, 936–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan, J. & Zipfel, C. (2012) Plant pattern recognition receptor complexes at the plasma membrane. Current Opinion in Plant Biology, 15, 349–357. [DOI] [PubMed] [Google Scholar]
- Niki, T. , Mitsuhara, I. , Seo, S. , Ohtsubo, N. & Ohashi, Y. (1998) Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis‐related (PR) protein genes in wounded mature tobacco leaves. Plant and Cell Physiology, 39, 500–507. [Google Scholar]
- Pfaffl M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Research, 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick, E. , Hofmann, K. & Glickman, M.H. (2009) PCI complexes: Beyond the proteasome, CSN, and e IF3 Troika. Molecular Cell, 35, 260–264. [DOI] [PubMed] [Google Scholar]
- Pritsch, C. , Muehlbauer, G.J. , Bushnell, W.R. , Somers, D.A. & Vance, C.P. (2000) Fungal development and induction of defense response genes during early infection of wheat spikes by Fusarium graminearum . Molecular Plant‐Microbe Interactions, 13, 159–169. [DOI] [PubMed] [Google Scholar]
- Qi, T. , Zhu, X.G. , Tan, C.L. , Liu, P. , Guo, J. , Kang, Z.S. et al. (2018) Host‐induced gene silencing of an important pathogenicity factor PsCPK1 in Puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust. Plant Biotechnology Journal, 16, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, N.X. , Xu, D.Q. , Li, J.G. & Deng, X.W. (2020) COP9 signalosome: Discovery, conservation, activity, and function. Journal of Integrative Plant Biology, 62, 90–103. [DOI] [PubMed] [Google Scholar]
- Quilis, J. , Peñas, G. , Messeguer, J. , Brugidou, C. & Segundo, B.S. (2008) The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Molecular Plant‐Microbe Interactions, 21, 1215–1231. [DOI] [PubMed] [Google Scholar]
- Schaffrath, U. , Freydl, E. & Dudler, R. (1997) Evidence for different signaling pathways activated by inducers of acquired resistance in wheat. Molecular Plant‐Microbe Interactions, 10, 779–783. [Google Scholar]
- van Schie, C.N. & Takken, F.W. (2014) Susceptibility genes 101: How to be a good host. Annual Review of Phytopathology, 52, 551–581. [DOI] [PubMed] [Google Scholar]
- Segarra G., Jáuregui O., Casanova E., Trillas I. (2006) Simultaneous quantitative LC–ESI‐MS/MS analyses of salicylic acid and jasmonic acid in crude extracts of Cucumis sativus under biotic stress. Phytochemistry, 67, 395–401. [DOI] [PubMed] [Google Scholar]
- Sels, J. , Mathys, J. , De Coninck, B.M. , Cammue, B.P. & De Bolle, M.F. (2008) Plant pathogenesis‐related (PR) proteins: A focus on PR peptides. Plant Physiology and Biochemistry, 46, 941–950. [DOI] [PubMed] [Google Scholar]
- Sinha, M. , Singh, R.P. , Kushwaha, G.S. , Iqbal, N. , Singh, A. , Kaushik, S. et al. (2014) Current overview of allergens of plant pathogenesis related protein families. The Scientific World Journal, 19, 543195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples, R.C. (2000) Research on the rust fungi during the twentieth century. Annual Review of Phytopathology, 38, 49–69. [DOI] [PubMed] [Google Scholar]
- Torres, M.A. (2010) ROS in biotic interactions. Physiologia Plantarum, 138, 414–429. [DOI] [PubMed] [Google Scholar]
- Van Loon, L.C. & Van Strien, E.A. (1999) The families of pathogenesis‐related proteins, their activities, and comparative analysis of PR‐1 type proteins. Physiological and Molecular Plant Pathology, 55, 85–97. [Google Scholar]
- Verberne, M.C. , Verpoorte, R. , Bol, J.F. , Mercado‐Blanco, J. & Linthorst, H.J. (2000) Overproduction of salicylic acid in plants by bacterial transgenes enhances pathogen resistance. Nature Biotechnology, 18, 779–783. [DOI] [PubMed] [Google Scholar]
- Wang, X.J. , Zhang, H.T. , Nyamesorto, B. , Luo, Y. , Mu, X.Q. , Wang, F.Y. et al. (2020) A new mode of NPR1 action via an NB‐ARC‐NPR1 fusion protein negatively regulates defense response to stem rust pathogen in wheat. New Phytologist, 228, 959–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N. , Serino, G. & Deng, X.W. (2008) The COP9 signalosome: More than a protease. Trends in Biochemical Sciences, 33, 592–600. [DOI] [PubMed] [Google Scholar]
- Wright, A.V. , Nuñez, J.K. & Doudna, J.A. (2016) Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell, 164, 29–44. [DOI] [PubMed] [Google Scholar]
- Wu, J.H. , Huang, S. , Zeng, Q.D. , Liu, S.J. , Wang, Q.L. , Mu, J.M. et al. (2018) Comparative genome‐wide mapping versus extreme pool‐genotyping and development of diagnostic SNP markers linked to QTL for adult plant resistance to stripe rust in common wheat. Theoretical and Applied Genetics, 131, 1777–1792. [DOI] [PubMed] [Google Scholar]
- Xu, G.Y. , Yuan, M. , Ai, C.R. , Liu, L.J. , Zhuang, E. , Karapetyan, S. et al. (2017) ORF mediated translation allows engineered plant disease resistance without fitness costs. Nature, 545, 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , Xu, X. , Gong, Q. , Li, Z. , Li, Y. , Wang, S. et al. (2019) Engineering broad‐spectrum bacterial blight resistance by simultaneously disrupting variable TALE‐Binding elements of multiple susceptibility genes in rice. Molecular Plant, 12, 1434–1446. [DOI] [PubMed] [Google Scholar]
- Zaidi, S.S. , Mukhtar, M.S. & Mansoor, S. (2018) Genome editing: Targeting susceptibility genes for plant disease resistance. Trends in Biotechnology, 36, 898–906. [DOI] [PubMed] [Google Scholar]
- Zhang, H.T. , Wang, X.J. , Giroux, M.J. & Huang, L. (2017) A wheat COP9 subunit 5‐like gene is negatively involved in host response to leaf rust. Molecular Plant Pathology, 18, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Q.F. & Dong, X. (2013) Systemic acquired resistance: Turning local infection into global defense. Annual Review of Plant Biology, 64, 839–863. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


