Cadherin-17, a 7D-cadherin, has N-terminal extracellular cadherin (EC) repeats similar to those of classical cadherins but with several key differences that are relevant to its biological function. The structure presented here demonstrates that cadherin-17 is incapable of forming the EC1 tryptophan-mediated strand-swap conformation that classical cadherins use to adhere cells together.
Keywords: cadherin-17, cell adhesion, calcium binding, intestinal epithelia, LI-cadherin
Abstract
The cadherin superfamily of calcium-dependent cell-adhesion proteins has over 100 members in the human genome. All members of the superfamily feature at least a pair of extracellular cadherin (EC) repeats with calcium-binding sites in the EC linker region. The EC repeats across family members form distinct complexes that mediate cellular adhesion. For instance, classical cadherins (five EC repeats) strand-swap their N-termini and exchange tryptophan residues in EC1, while the clustered protocadherins (six EC repeats) use an extended antiparallel ‘forearm handshake’ involving repeats EC1–EC4. The 7D-cadherins, cadherin-16 (CDH16) and cadherin-17 (CDH17), are the most similar to classical cadherins and have seven EC repeats, two of which are likely to have arisen from gene duplication of EC1–2 from a classical ancestor. However, CDH16 and CDH17 lack the EC1 tryptophan residue used by classical cadherins to mediate adhesion. The structure of human CDH17 EC1–2 presented here reveals features that are not seen in classical cadherins and that are incompatible with the EC1 strand-swap mechanism for adhesion. Analyses of crystal contacts, predicted glycosylation and disease-related mutations are presented along with sequence alignments suggesting that the novel features in the CDH17 EC1–2 structure are well conserved. These results hint at distinct adhesive properties for 7D-cadherins.
1. Introduction
Cadherins form a large superfamily of calcium-dependent cell-adhesion proteins, with around 100 structurally diverse members encoded in the human genome alone (Brasch et al., 2012 ▸; Sotomayor et al., 2014 ▸). These proteins have an extracellular domain of variable length, a transmembrane domain and a cytoplasmic domain that often facilitates interactions with the cytoskeleton and other signalling proteins (Pokutta & Weis, 2007 ▸). The sequence and structural diversity of cadherins allow them to perform a wide variety of functions in development and morphogenesis (Takeichi, 1990 ▸; Brasch et al., 2012 ▸; Araç et al., 2016 ▸), mechanosensation (Gillespie & Müller, 2009 ▸; Leckband et al., 2011 ▸; Pruitt et al., 2014 ▸; Leckband & de Rooij, 2014 ▸; Katta et al., 2015 ▸; Jaiganesh et al., 2018 ▸), and neuronal recognition (Hirano & Takeichi, 2012 ▸; Biswas et al., 2012 ▸; Weiner & Jontes, 2013 ▸; Light & Jontes, 2017 ▸; Canzio & Maniatis, 2019 ▸). The most studied and nearly ubiquitous member of the family, E-cadherin (CDH1), forms the adherens junction in epithelia, is essential in embryogenesis and is considered to be a tumour suppressor (van Roy & Berx, 2008 ▸). Other cadherins have more specific functions. For instance, cadherin-23 and protocadherin-15 form a bond that is essential for sound mechanotransduction (Kazmierczak et al., 2007 ▸; Sotomayor et al., 2012 ▸), while clustered protocadherins use their diverse extracellular domains to mediate neuronal recognition (Schreiner & Weiner, 2010 ▸; Nicoludis et al., 2015 ▸; Brasch et al., 2019 ▸). Less is known, however, about nonclassical and nonclustered members of the superfamily.
Cadherin-16 (CDH16; Ksp-cadherin) and cadherin-17 (CDH17; LI-cadherin or BILL-cadherin) are the only members of the small subfamily of 7D-cadherins in vertebrates (Dantzig et al., 1994 ▸; Thomson et al., 1995 ▸; Wendeler et al., 2006 ▸). CDH16 is found in kidney epithelia (Thomson et al., 1995 ▸, 1998 ▸), while CDH17 is expressed in human and mouse intestinal cells (Dantzig et al., 1994 ▸; Angres et al., 2001 ▸), in rat liver (Berndorff et al., 1994 ▸), in mature B cells that have localized to the spleen (Ohnishi et al., 2000 ▸, 2005 ▸; Funakoshi et al., 2015 ▸) and in various cancers (Grötzinger et al., 2001 ▸; Hinoi et al., 2002 ▸; Takamura et al., 2003 ▸; Wong et al., 2003 ▸; Su et al., 2008 ▸; Ding et al., 2009 ▸; Liu et al., 2009 ▸; Park et al., 2011 ▸; Kuhlmann et al., 2017 ▸). CDH16 and CDH17 are produced in cells that are typically responsible for water absorption, but their function has yet to be clearly elucidated. In intestinal epithelia, CDH17 is mainly localized on the lateral and basolateral membrane of the cells, but is excluded from the adherens junctions and the desmosomes containing CDH1 and the desmosomal cadherins, respectively (Berndorff et al., 1994 ▸; Kreft et al., 1997 ▸). The interactions of CDH17 across cells are thought to help to maintain the width of the interstitial cleft between epithelial cells to regulate water transport (Ahl et al., 2011 ▸; Weth et al., 2017 ▸).
Cadherins are able to mediate adhesion using their extracellular domain, which has tandem extracellular cadherin (EC) repeats (Brasch et al., 2012 ▸; Sotomayor et al., 2014 ▸). The EC repeats have ∼110 residues arranged in a seven-β-strand Greek-key fold, with several conserved sequence motifs of residues involved in binding three calcium ions at the linker regions between the EC repeats (Nagar et al., 1996 ▸). Calcium binding gives rigidity to the extracellular domain and facilitates adhesion (Pokutta et al., 1994 ▸; Cailliez & Lavery, 2005 ▸; Sotomayor & Schulten, 2008 ▸). Various subfamily members utilize their EC repeats differently to mediate trans adhesion between two cellular membranes. For instance, CDH1 uses a tryptophan on the N-terminal strand of EC1 that is exchanged and inserted into the hydrophobic binding pocket of another EC1 (Shapiro et al., 1995 ▸; Boggon et al., 2002 ▸). This is called the strand-swap mechanism and all classical cadherins use it for their adhesive bond (Brasch et al., 2012 ▸). In contrast, members of the α, β and γ clustered protocadherins form antiparallel trans interactions mediated by EC1–4, and the δ1 and δ2 nonclustered protocadherins use the same set of EC repeats, forming a similar antiparallel interaction (Cooper et al., 2016 ▸; Modak & Sotomayor, 2019 ▸; Rubinstein et al., 2015 ▸; Nicoludis et al., 2015 ▸; Harrison et al., 2020 ▸). Cadherins are also capable of forming complexes on the surface of the same cell (cis interactions). Classical cadherins use this cis interaction to form adhesive patches on the cellular surface (Harrison et al., 2011 ▸), while clustered protocadherins use a heterophilic mechanism of cis interactions to direct neuronal self-avoidance (Schreiner & Weiner, 2010 ▸; Goodman et al., 2017 ▸; Brasch et al., 2019 ▸).
CDH16 and CDH17 are unique among cadherins as they have seven EC repeats and their EC1–2 and EC3–4 repeats share similarity with one another as well as with EC1–2 in classical cadherins (Berndorff et al., 1994 ▸; Thomson et al., 1995 ▸; Kreft et al., 1997 ▸; Wendeler et al., 2004 ▸, 2006 ▸; Jung et al., 2004 ▸). Gene-structure analyses suggest that CDH16 and CDH17 are likely to have arisen from a classical ancestor when the two N-terminal repeats, EC1 and EC2, were duplicated. This is further supported by analyses showing that the EC2–3 linker lacks sequence motifs with the residues necessary to bind calcium ions and might be calcium-free altogether (Berndorff et al., 1994 ▸; Angres et al., 2001 ▸; Jung et al., 2004 ▸). Additionally, the remainder of the repeats, EC5–7, share sequence similarity with EC3–5 from classical cadherins (Berndorff et al., 1994 ▸; Jung et al., 2004 ▸).
In vitro experiments have revealed that CDH17 forms homophilic trans and cis complexes in a calcium-dependent manner (Kreft et al., 1997 ▸; Wendeler et al., 2007 ▸; Baumgartner et al., 2008 ▸; Bartolmäs et al., 2012 ▸; Weth et al., 2017 ▸). Experiments have also revealed that CDH17 is capable of forming heterophilic trans interactions with the classical cadherin CDH1, which is expressed in the same cell types as CDH17 (Baumgartner et al., 2008 ▸). However, the structural details of cis and trans interactions mediating homophilic and heterophilic adhesion by CDH16 and CDH17 are not known.
To better understand the mechanism of adhesion in the 7D-cadherin family, we determined the X-ray crystal structure of the N-terminal EC1–2 repeats of Homo sapiens CDH17 (hs CDH17), which revealed unique features at its N-terminus that are relevant for its adhesion mechanism. The structure suggests that CDH17 is unable to undergo the strand-swap in EC1 and uses a mechanism of adhesion that differs from those used by classical cadherins.
2. Materials and methods
2.1. Macromolecule production
2.1.1. Cloning of CDH17 constructs
DNA encoding hs CDH17 (Harvard PlasmidID HsCD00419124, UniProt ID Q12864) with the natural variation Lys93Glu was used as a template in polymerase chain reaction amplification experiments. The sequence encoding EC1–2 was amplified and subcloned into the NdeI and XhoI restriction sites of the pET-21a vector. Additional information can be found in Supplementary Table S1.
2.1.2. Bacterial expression and purification of CDH17 fragments
Escherichia coli Rosetta2 (DE3) cells (Novagen) were transformed with the sequence-verified hs CDH17 EC1–2 construct, cultured in Terrific Broth (TB) and induced at an OD600 of ∼0.45 with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 30°C overnight. The cells were then lysed by sonication using denaturing buffer [20 mM Tris–HCl pH 7.5, 6 M guanidine hydrochloride (GuHCl), 10 mM CaCl2, 20 mM imidazole] and centrifuged. The resulting cleared lysate was incubated with Ni-Sepharose beads (GE Healthcare), washed twice with denaturing buffer and eluted with denaturing buffer supplemented with 500 mM imidazole. The purified hs CDH17 EC1–2 protein was refolded overnight at 4°C using membranes with molecular-weight cutoff (MWCO) of 2000 Da in a dialysis buffer containing 20 mM Tris–HCl pH 8.0, 5 mM CaCl2, 150 mM KCl, 50 mM NaCl, 400 mM arginine. The refolded protein was further purified on a Superdex 200 16/600 column (GE Healthcare) in 20 mM Tris–HCl pH 8.0, 2 mM CaCl2, 150 mM KCl, 50 mM NaCl. The protein was concentrated to ∼8 mg ml−1 (Vivaspin, 10 kDa MWCO), as determined by the Bradford assay, and used in crystallization trials.
2.2. Crystallization
Crystals were grown via vapour diffusion using the sitting-drop method at 4°C with 0.1 M HEPES pH 7, 0.1 M KCl, 15% PEG 5000 MME. Crystals were harvested and cryocooled in crystallization buffer supplemented with 25% glycerol as a cryoprotectant. Information regarding crystallization is provided in Supplementary Table S2.
2.3. Data collection and processing
A data set was obtained on the NECAT 24-ID-E beamline at the Advanced Photon Source (APS). The diffraction data were indexed and scaled in HKL-2000 (Otwinowski & Minor, 1997 ▸) in space group P212121 to 2.15 Å resolution. Details of data collection and processing are provided in Table 1 ▸.
Table 1. Data collection and processing.
Values in parentheses are for the highest resolution shell.
| Diffraction source | Beamline 24-ID-E, APS |
| Wavelength (Å) | 0.97918 |
| Temperature (K) | 100 |
| Detector | ADSC Quantum 315 CCD |
| Crystal-to-detector distance (mm) | 300 |
| Rotation range per image (°) | 0.75 |
| Total rotation range (°) | 90 |
| Exposure time per image (s) | 1 |
| Space group | P212121 |
| a, b, c (Å) | 58.39, 81.61, 106.81 |
| Mosaicity (°) | 0.317 |
| Resolution range (Å) | 50.00–2.15 (2.19–2.15) |
| Total no. of reflections | 260436 |
| No. of unique reflections | 28521 (1406) |
| Completeness (%) | 99.5 (99.9) |
| Multiplicity | 3.60 (3.50) |
| 〈I/σ(I)〉 | 10.46 (2.05) |
| R r.i.m. † | 0.155 (0.864) |
| Overall B factor from Wilson plot (Å2) | 30.46 |
Estimated R r.i.m. = R merge[N/(N − 1)]1/2, where N is the data multiplicity.
2.4. Structure solution and refinement
The structure was solved with Phaser (McCoy et al., 2007 ▸) and refined with REFMAC 5.8.0257 (Murshudov et al., 2011 ▸) from the CCP4 suite (Winn et al., 2011 ▸) using an initial model of hs CDH17 EC1–2 generated from a lower resolution data set. The initial model from the lower resolution data set was solved using MrBUMP with the structure of E-cadherin EC1–5 deposited in the Protein Data Bank (PDB entry 3q2v, chain A; Harrison et al., 2011 ▸; Berman et al., 2000 ▸). The sequence identity for the EC1–2 segment was 34.1%. Refinement data are provided in Table 2 ▸.
Table 2. Structure solution and refinement.
Values in parentheses are for the highest resolution shell.
| Resolution range (Å) | 47.53–2.15 (2.21–2.15) |
| Completeness (%) | 99.5 |
| σ Cutoff | F > 0.000σ(F) |
| No. of reflections, working set | 26913 (1939) |
| No. of reflections, test set | 1412 (105) |
| No. of molecules in asymmetric unit | 2 |
| Matthews coefficient (Å3 Da−1) | 2.54 |
| Final R cryst | 0.206 (0.265) |
| Final R free | 0.257 (0.282) |
| Cruickshank DPI | 0.196 |
| No. of non-H atoms | |
| Protein | 3192 |
| Ion | 6 |
| Water | 203 |
| Total | 3401 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Angles (°) | 1.269 |
| Average B factors (Å2) | |
| Protein | 32.94 |
| Ion | 24.33 |
| Water | 30.89 |
| Ramachandran plot | |
| Most favoured (%) | 87.5 |
| Allowed (%) | 12.5 |
| PDB code | 6ulm |
3. Results
3.1. The N-terminal repeats of CDH17 are structurally unique
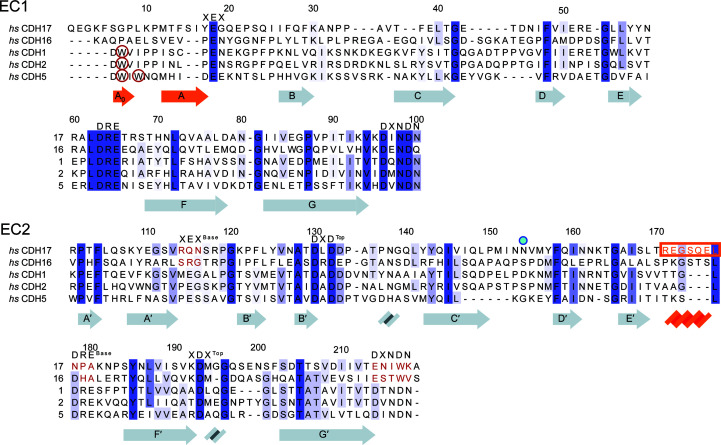
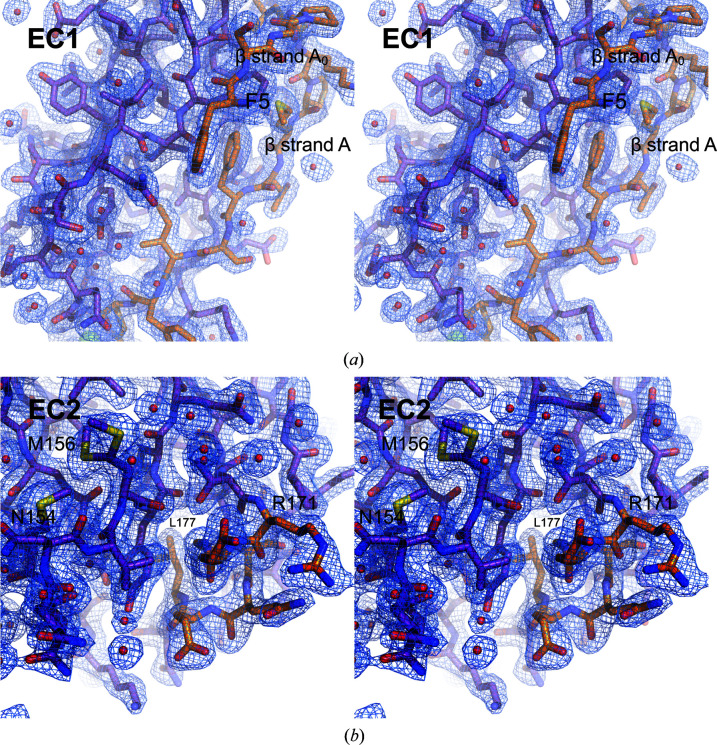
Sequence analyses suggest that CDH17 has classical-like cadherin repeats but uses a different mechanism of adhesion to the strand-swap utilized by classical cadherins. Human CDH17 and CDH16 lack the tryptophan residues exchanged by classical EC1 repeats and instead have longer N-termini (∼5–8 residues in CDH17 across vertebrates; Fig. 1 ▸, Supplementary Fig. S1, Supplementary Table S3). In addition, EC2 of CDH17 lacks the calcium-binding residues that are needed to form a canonical EC2–3 linker (Fig. 1 ▸, Supplementary Fig. S1, Supplementary Table S3) and sequences of CDH16 similarly lack these residues. To understand how 7D-cadherins structurally differ from classical cadherins in their N-terminal repeats, we produced and purified hs CDH17 EC1–2 (Gln1–Ala219), crystallized it (Supplementary Tables S1 and S2) and solved its structure refined to 2.15 Å resolution (Tables 1 ▸ and 2 ▸). This structure has two monomers in the asymmetric unit, with chain A encompassing residues Pro11–Asn215 and chain B encompassing residues Phe5–Thr213, where the standard residue numbering for cadherins refers to the protein without the signal peptide. These residues are equivalent to Pro33–Asn237 (chain A) and Phe27–Thr235 (chain B) using UniProt ID Q12864. The text below and all figure labels use the numbering without the signal peptide. The associated electron-density map (Fig. 2 ▸) allowed modelling of all of the listed residues except for residues Asn32–Val36 and Asp78–Ile83 in chain A. Since the root-mean-square deviation between the two monomers is low (0.92 Å for Cα atoms), we describe the structural features as seen in the most complete monomer (chain B) unless otherwise stated.
Figure 1.
Sequence similarity among CDH17, CDH16 and classical cadherins. Protein sequence alignments of the EC1 and EC2 repeats of the 7D-cadherins (CDH17 and CDH16) and three classical cadherins (CDH1, CDH2 and CDH5) show how these differ at their N-termini and at their EC2 C-terminal tails. Calcium-binding residues are highlighted with their respective sequences at the top. Missing calcium-binding motifs are highlighted in red for CDH17 and CDH16 in EC2. Tryptophan residues that classical cadherins utilize for the strand-swap mechanism are circled in red. The Asn154 residue implicated in disease is denoted by a blue dot (Smith et al., 2017 ▸). Secondary-structure elements of the hs CDH17 EC1–2 structure are highlighted below the sequence, with unique structural elements in orange. The sequence of the α-helix in CDH17 EC2 is highlighted by an orange box and text.
Figure 2.
Electron density (2F o − F c) for unique elements of hs CDH17 EC1–2. (a) Stereoview of EC1 detail showing β-strands A0 and A in orange, folded back against EC1. (b) Stereoview of the C-terminal side of EC2 with a two-turn α-helix, shown in orange. Residue Asn154 implicated in disease and a double conformer for residue Met156 are highlighted. Electron density is shown at 1σ with a carve radius of 1.6 Å. Red spheres indicate water molecules fitted in the electron density.
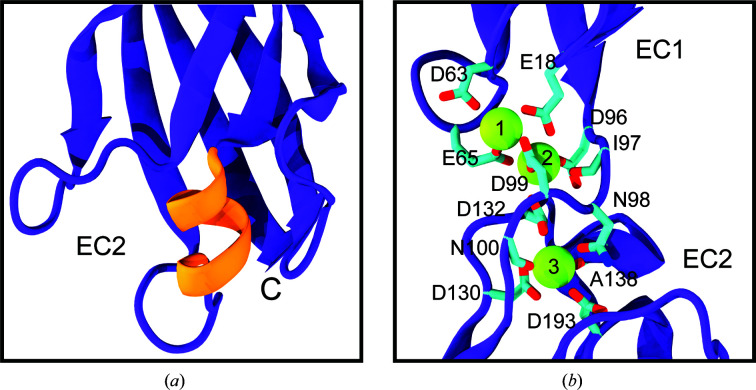
The hs CDH17 EC1–2 structure revealed both canonical and unique features relevant for function. Repeats EC1 and EC2 fold in typical seven-β-strand Greek-key motifs (β-strands labelled A–G for EC1 and A′–G′ for EC2; Figs. 3 ▸ a and 3 ▸ b), but EC1 has an extra N-terminal β-strand (Fig. 2 ▸ a) and EC2 has an additional α-helix (Figs. 2 ▸ b and 4 ▸ a; see below). As expected, the EC repeats are arranged in a linear configuration and there are three calcium ions bound (sites 1, 2 and 3) in a canonical EC1–2 linker region (Fig. 4 ▸ b). This is consistent with the canonical sequence motifs XEX and DRE present in the first repeat (17YEG19 and 63DRE65 in EC1), the DXNDN sequence motif at the linker (96DINDN100) and the sequence motifs DXD (130DLD132) and XDX (192KDM194) present in the second EC repeat (Fig. 1 ▸ and Supplementary Fig. S1). Other loops and secondary-structure elements are canonical, highlighting overall similarities with classical cadherin N-terminal EC repeats.
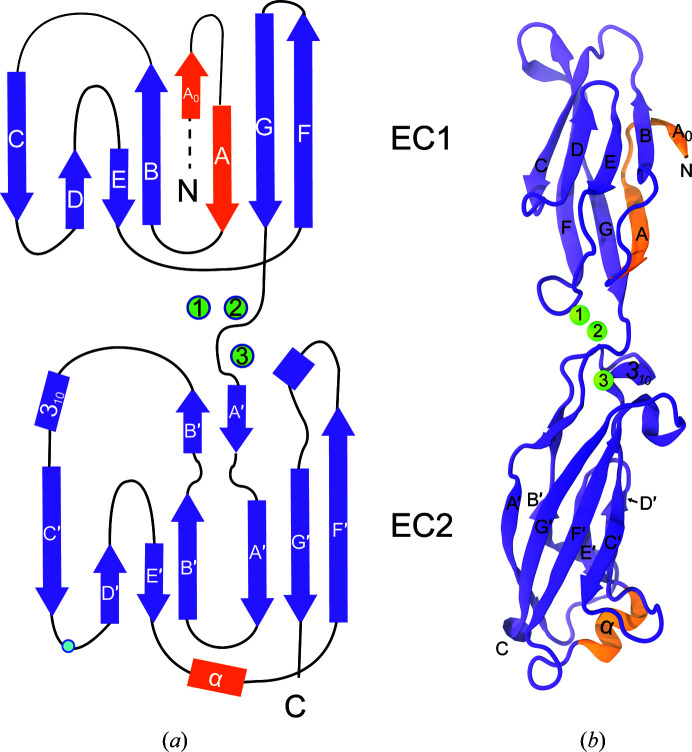
Figure 3.
Structure of hs CDH17 EC1–2. (a) Topology of hs CDH17 EC1–2. Repeats EC1 and EC2 have canonical Greek-key motifs with atypical features in orange. The β-strands are labelled A–G for EC1 and A′–G′ for EC2, and α-helices are labelled by type. N and C denote the N- and C-terminus, respectively. Residue Asn154 is denoted by a blue dot. (b) Ribbon representation of the hs CDH17 EC1–2 structure with calcium ions in green and atypical features in orange. Labels are as in (a).
Figure 4.
Structural highlights of hs CDH17 EC1–2. (a) Detail of the two-turn α-helix in the EC2 E′–F′ loop, highlighted by the orange box in Fig. 1 ▸. (b) Detail of the hs CDH17 EC1–2 canonical calcium-binding linker. Side chains of calcium-coordinating residues are shown as sticks. Some backbone atoms are omitted for visualization purposes.
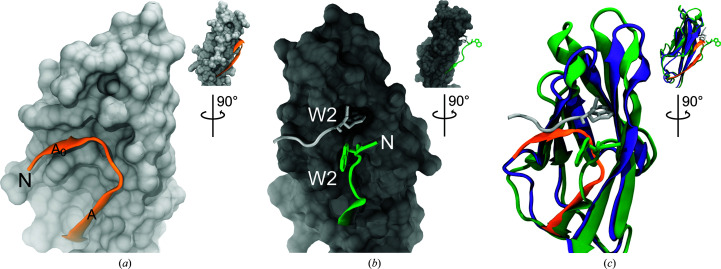
Among the unique, noncanonical features observed in the hs CDH17 EC1–2 structure is an additional N-terminal β-strand in EC1 (Phe5–Pro8), which we labelled A0 as it precedes the canonical β-strand A (Figs. 3 ▸ a and 3 ▸ b). The electron-density map at the N-terminus of the EC1 monomers is not good enough to model residues Gln1–Lys4 in the structure, suggesting some conformational variability at this end. Nevertheless, residue Phe5 and the remainder of the A0 β-strand backbone are clearly visible, with side chains that were unambiguously modelled (Fig. 2 ▸ a). The A0 β-strand runs parallel to and interacts with β-strand B, ending in a turn facilitated by two proline residues: Pro8 (highly conserved) and Pro11 (highly conserved among mammals). This double proline A0 turn leads to β-strand A, which runs antiparallel to, but does not interact with, β-strand B (Figs. 3 ▸ and 5 ▸ a). In contrast to the classical hs CDH1 EC1 (Figs. 5 ▸ b and 5 ▸ c), which uses a short N-terminus that protrudes from the monomer to engage in a strand-swap mechanism involving β-strand A, the extended N-terminus of CDH17 lays against and interacts with its own EC1 (Figs. 5 ▸ a and 5 ▸ c). These results suggest that CDH17 EC1 does not engage in classical-like strand-swapping interactions.
Figure 5.
Comparison of CDH17 and CDH1 EC1 repeats. (a) Detail of the hs CDH17 EC1 N-terminus showing residues Phe5–Glu18 in orange ribbon representation over the EC1 surface. (b) Detail of the hs CDH1 EC1 N-terminus (PDB entry 2o72; Parisini et al., 2007 ▸). The green strand shows EC1 residues Asp1–Glu11 extending out from the surface of the rest of EC1, while the silver strand shows the N-terminus of another monomer engaged in the strand-swap mechanism. A tryptophan residue (Trp2) is inserted in the hydrophobic pocket. (c) Rendering of hs CDH17 EC1 (purple and orange) structurally aligned with hs CDH1 EC1 (green) showing details of their contrasting N-termini. Insets in all panels show rotated views of EC1 highlighting the N-terminal strand location.
A second unique feature observed in the hs CDH17 EC1–2 structure is a two-turn α-helix located in EC2 between β-strands E′ and F′ (Arg171–Leu177). This EC2 E′F′ α-helix faces the EC2–3 linker region and its sequence is well conserved among mammals, with a ∼50-residue insertion in various fish species (inset in Supplementary Fig. S1 and Supplementary Fig. S2). Interestingly, the EC2 repeat lacks the XEX, DRE and DXNDN motifs of the canonical calcium-binding EC2–3 linker region (Fig. 1 ▸ and Supplementary Fig. S1), and the EC2 E′F′ α-helix is in the loop that would normally contain the DRE motif (Figs. 1 ▸ and 4 ▸ a), suggesting that the EC2–3 linker region lacks calcium-binding sites 1 and 2. The EC3 sequence has modified DXD (244DPG246) and XDX (291KDE293) motifs, indicating that it may retain calcium-binding site 3 at the top of EC3. The EC2 E′F′ α-helix located at this noncanonical linker region might help to rigidify an otherwise flexible, partial calcium-free joint.
3.2. Crystallographic contacts reveal interfaces that might be impaired by glycosylation
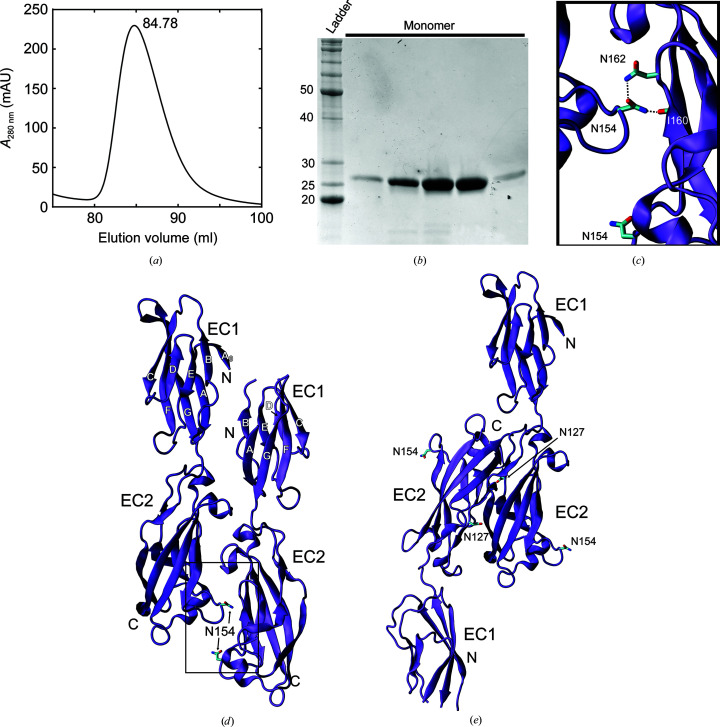
Previous crystallographic structures of cadherin extracellular domains have revealed crystal contacts as possible interfaces for cis and trans interactions, which in some cases have been validated using in vitro biophysical assays as well as ex vivo and in vivo experiments (Shapiro et al., 1995 ▸; Boggon et al., 2002 ▸; Ciatto et al., 2010 ▸; Harrison et al., 2010 ▸, 2011 ▸, 2016 ▸; Patel et al., 2006 ▸; Sotomayor et al., 2012 ▸; Geng et al., 2013 ▸; Rubinstein et al., 2015 ▸; Nicoludis et al., 2015 ▸; Cooper et al., 2016 ▸; Modak & Sotomayor, 2019 ▸; Brasch et al., 2019 ▸). Some of these interactions are dependent on glycosylation (Pinho et al., 2011 ▸; Langer et al., 2012 ▸; Brasch et al., 2011 ▸). Size-exclusion chromatography (SEC) experiments with hs CDH17 EC1–2 suggest that this fragment is monomeric in solution (Figs. 6 ▸ a and 6 ▸ b), yet similar tests have failed to detect weak, physiologically relevant interactions such as those mediating cis interfaces in classical cadherins (Harrison et al., 2011 ▸). We used the Proteins, Interfaces, Structures and Assemblies (PISA) server (Krissinel & Henrick, 2007 ▸) to identify all possible crystal contacts and evaluate their possible physiological relevance. Seven crystal contacts were identified (Figs. 6 ▸ d and 6 ▸ e, and Supplementary Fig. S3), with interface areas that range from ∼100 to ∼583 Å2, too small compared with an empirical threshold (856 Å2) that distinguishes biologically relevant interactions (Ponstingl et al., 2000 ▸). However, physiologically relevant protocadherin interfaces often involve multiple small contacts between various EC repeats (200–400 Å2 per EC repeat; Nicoludis et al., 2019 ▸; Cooper et al., 2016 ▸; Modak & Sotomayor, 2019 ▸), and the two largest CDH17 EC1–2 interfaces observed in our crystal structure would implicate EC repeats beyond EC2. Thus, these two interfaces, which are discussed below, could become large enough to be physiologically relevant when considering contact contributions from all EC repeats.
Figure 6.
Purification and crystal contacts for hs CDH17 EC1–2. (a) Elution profile of an SEC experiment for hs CDH17 EC1–2 showing a likely monomeric peak at an elution volume of 84.78 ml using a Superdex S200 column. (b) SDS–PAGE analysis of the hs CDH17 EC1–2 SEC peak. Lane 1 shows molecular-weight standards. Lanes 2–6 show fractions from the SEC peak. The band intensity corresponds to the peak intensity as equal volumes for each fraction were run on the gel. (c) Detail of the Asn154 residue implicated in disease (Smith et al., 2017 ▸) forming hydrogen bonds to residues Asn162 and Ile160. (d, e) Crystal contacts between two monomers of hs CDH17 EC1–2 as identified by PISA (Krissinel & Henrick, 2007 ▸). Residue Asn154, implicated in disease, is highlighted. A predicted N-glycosylation site, Asn127, is highlighted in (e). Interface areas are 582.9 Å2 (d) and 565.6 Å2 (e).
The first and largest interface from crystal contacts is formed by two monomers in the asymmetric unit of the crystal arranged in a parallel configuration with an interface area of 582.9 Å2 (Fig. 6 ▸ d). The orientation of the monomers is such that β-strands A0, A and B in the EC1 repeats are facing towards each other, and β-strands F and G face the outside. The monomers are, however, slightly shifted with respect to each other, and the C-terminal part of EC1 of one monomer interacts with the N-terminal part of EC1 of the next monomer, with a similar arrangement observed for the EC2 repeats, and potentially for further EC repeats that would contribute to a large cis interface if these were present in the structure. To maintain this arrangement between two parallel membranes, the complex would need to be tilted, as observed for classical cadherins that engage in cis interactions in which EC1 interacts in parallel with EC2 (594.8 Å2; Harrison et al., 2011 ▸). The possible physiological relevance of the CDH17 cis interface is supported by a Asn154Ser mutation involved in disease (Smith et al., 2017 ▸) located at the interface in EC2 (Figs. 6 ▸ c and 6 ▸ d). However, analysis of predicted glycosylation sites (Supplementary Fig. S1) suggests that sugars stemming from one of the Asn162 residues would interfere with this arrangement (Fig. 6 ▸ c), as has been observed for other cadherins (Brasch et al., 2011 ▸).
The second largest interface from crystal contacts observed in the hs CDH17 EC1–2 structure is formed by an antiparallel overlap of EC2 repeats with an interface area of 565.6 Å2 (Fig. 6 ▸ e). In this crystal contact, the EC1 repeat would interact with repeat EC3, if it was present, to form an EC1–EC3 antiparallel interface. The interface area calculated for this crystal contact is for EC2 alone, and it is similar to or larger than the interface area values per repeat observed in other structures of protocadherin interfaces (Cooper et al., 2016 ▸). Thus, an EC1–EC3 interface would have a larger buried surface area that is more likely to indicate physiological relevance. However, analysis of predicted glycosylation sites indicates that sugars stemming from Asn127 could interfere with the EC2–EC2 contact (Fig. 6 ▸ e and Supplementary Fig. S1), suggesting that such an interface is not physiologically relevant. Other crystal contacts detected by PISA had small interface areas and were incompatible with simple cis and trans interactions given the arrangement of the EC repeats (Supplementary Figs. S3a–S3e).
4. Discussion
Previous work on CDH17 focused on understanding its adhesive functions at the cellular level, with some insights into the underlying molecular mechanisms stemming from comparisons with classical cadherins and from single-molecule experiments testing its homophilic interactions with itself and its heterophilic interactions with CDH1 (Baumgartner et al., 2008 ▸; Bartolmäs et al., 2012 ▸; Baumgartner, 2013 ▸; Weth et al., 2017 ▸). The structure of EC1–2 demonstrates that CDH17 is incapable of forming the EC1 tryptophan-mediated strand-swap, the mechanism used by all classical cadherins, as its EC1 N-terminus is extended and forms the A0 strand. Additionally, the two largest crystallographic interfaces that we observed have incompatible predicted glycosylation sites for potential cis and trans interactions.
Recent work presented a structure of CDH17 EC1–4 (PDB entry 7cym) forming an antiparallel complex in which the EC2 repeat of one monomer interacts with EC4 from the other, and EC3 is not directly involved in the binding interface (Yui et al., 2020 ▸). Features observed in the EC1–2 structure presented here are also found in the EC1–4 structure. Most importantly, the N-terminal strand A0 is also folded back and interacts with strand B in EC1. The E′–F′ α-helix in EC2 is present and appears to stabilize EC2, but not the calcium-free EC2–3 linker. The EC1–4 structure, which was obtained using protein produced in a mammalian expression system, has glycans attached to residues Asn127 and Asn162 in EC2. These glycans are likely to interfere with the interfaces observed in our structure of CDH17 EC1–2. Glycosylation can modify the biophysical properties of various cadherins (Pinho et al., 2011 ▸; Langer et al., 2012 ▸), and several glycosylation sites have been experimentally identified for CDH17 (Bernhard et al., 2013 ▸; Yui et al., 2020 ▸), yet the role of glycosylation in altering CDH17 function remains to be determined.
Intriguingly, the EC1–4 protein fragment produced by Yui and coworkers was mostly monomeric in SEC multi-angle light-scattering experiments at low protein concentration and dimeric in analytical ultracentrifugation experiments at higher concentrations (Yui et al., 2020 ▸). A mutation at Phe224, at the proposed interface, abolished dimerization and cell aggregation, but also compromised protein stability. Additionally, the EC1–5 and EC3–7 protein fragments did not facilitate cell aggregation (Yui et al., 2020 ▸). This is consistent with our own bead-aggregation assays suggesting that EC7 is required for adhesion (Gray, 2020 ▸). It is likely that CDH17 mediates adhesion through a mechanism unique to the 7D-cadherins, despite their sequence similarity to classical cadherins.
5. Related literature
The following references are cited in the supporting information for this article: Almagro Armenteros et al. (2019 ▸), Edgar (2004 ▸) and Waterhouse et al. (2009 ▸).
Supplementary Material
PDB reference: human CDH17 EC1–2, 6ulm
Supplementary Methods, Figures and Tables. DOI: 10.1107/S2053230X21002247/pg5088sup1.pdf
Acknowledgments
We thank members of the Sotomayor laboratory for assistance and discussions. Use of the APS NECAT beamlines was supported by the National Institutes of Health (P41 GM103403 and S10 RR029205) and the Department of Energy (DE-AC02-06CH11357) through grants GUP 49774 and 59251.
Funding Statement
This work was funded by Ohio State University grant . National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grant R01 DK095811 to Matthew J. Tyska. Alfred P. Sloan Foundation grant FR-2015-65794 to Marcos Sotomayor.
References
- Ahl, M., Weth, A., Walcher, S. & Baumgartner, W. (2011). Theor. Biol. Med. Model. 8, 18. [DOI] [PMC free article] [PubMed]
- Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., von Heijne, G. & Nielsen, H. (2019). Nat. Biotechnol. 37, 420–423. [DOI] [PubMed]
- Angres, B., Kim, L., Jung, R., Gessner, R. & Tauber, R. (2001). Dev. Dyn. 221, 182–193. [DOI] [PubMed]
- Araç, D., Sträter, N. & Seiradake, E. (2016). Handb. Exp. Pharmacol. 234, 67–82. [DOI] [PubMed]
- Bartolmäs, T., Hirschfeld-Ihlow, C., Jonas, S., Schaefer, M. & Gessner, R. (2012). Cell. Mol. Life Sci. 69, 3851–3862. [DOI] [PMC free article] [PubMed]
- Baumgartner, W. (2013). Tissue Barriers, 1, e23815. [DOI] [PMC free article] [PubMed]
- Baumgartner, W., Wendeler, M. W., Weth, A., Koob, R., Drenckhahn, D. & Gessner, R. (2008). J. Mol. Biol. 378, 44–54. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Berndorff, D., Gessner, R., Kreft, B., Schnoy, N., Lajous-Petter, A. M., Loch, N., Reutter, W., Hortsch, M. & Tauber, R. (1994). J. Cell Biol. 125, 1353–1369. [DOI] [PMC free article] [PubMed]
- Bernhard, O. K., Greening, D. W., Barnes, T. W., Ji, H. & Simpson, R. J. (2013). Biochim. Biophys. Acta, 1834, 2372–2379. [DOI] [PubMed]
- Biswas, S., Emond, M. R. & Jontes, J. D. (2012). Neuroscience, 219, 280–289. [DOI] [PMC free article] [PubMed]
- Boggon, T. J., Murray, J., Chappuis-Flament, S., Wong, E., Gumbiner, B. M. & Shapiro, L. (2002). Science, 296, 1308–1313. [DOI] [PubMed]
- Brasch, J., Goodman, K. M., Noble, A. J., Rapp, M., Mannepalli, S., Bahna, F., Dandey, V. P., Bepler, T., Berger, B., Maniatis, T., Potter, C. S., Carragher, B., Honig, B. & Shapiro, L. (2019). Nature, 569, 280–283. [DOI] [PMC free article] [PubMed]
- Brasch, J., Harrison, O. J., Ahlsen, G., Carnally, S. M., Henderson, R. M., Honig, B. & Shapiro, L. (2011). J. Mol. Biol. 408, 57–73. [DOI] [PMC free article] [PubMed]
- Brasch, J., Harrison, O. J., Honig, B. & Shapiro, L. (2012). Trends Cell Biol. 22, 299–310. [DOI] [PMC free article] [PubMed]
- Cailliez, F. & Lavery, R. (2005). Biophys. J. 89, 3895–3903. [DOI] [PMC free article] [PubMed]
- Canzio, D. & Maniatis, T. (2019). Curr. Opin. Neurobiol. 59, 213–220. [DOI] [PMC free article] [PubMed]
- Ciatto, C., Bahna, F., Zampieri, N., VanSteenhouse, H. C., Katsamba, P. S., Ahlsen, G., Harrison, O. J., Brasch, J., Jin, X., Posy, S., Vendome, J., Ranscht, B., Jessell, T. M., Honig, B. & Shapiro, L. (2010). Nat. Struct. Mol. Biol. 17, 339–347. [DOI] [PMC free article] [PubMed]
- Cooper, S. R., Jontes, J. D. & Sotomayor, M. (2016). eLife, 5, e18529. [DOI] [PMC free article] [PubMed]
- Dantzig, A. H., Hoskins, J. A., Tabas, L. B., Bright, S., Shepard, R. L., Jenkins, I. L., Duckworth, D. C., Sportsman, J. R., Mackensen, D., Rosteck, P. R. & Skatrud, P. L. (1994). Science, 264, 430–433. [DOI] [PubMed]
- Ding, Z.-B., Shi, Y.-H., Zhou, J., Shi, G.-M., Ke, A.-W., Qiu, S.-J., Wang, X.-Y., Dai, Z., Xu, Y. & Fan, J. (2009). Cancer, 115, 4753–4765. [DOI] [PubMed]
- Edgar, R. C. (2004). Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed]
- Funakoshi, S., Shimizu, T., Numata, O., Ato, M., Melchers, F. & Ohnishi, K. (2015). PLoS One, 10, e0117566. [DOI] [PMC free article] [PubMed]
- Geng, R., Sotomayor, M., Kinder, K. J., Gopal, S. R., Gerka-Stuyt, J., Chen, D. H.-C., Hardisty-Hughes, R. E., Ball, G., Parker, A., Gaudet, R., Furness, D., Brown, S. D., Corey, D. P. & Alagramam, K. N. (2013). J. Neurosci. 33, 4395–4404. [DOI] [PMC free article] [PubMed]
- Gillespie, P. G. & Müller, U. (2009). Cell, 139, 33–44. [DOI] [PMC free article] [PubMed]
- Goodman, K. M., Rubinstein, R., Dan, H., Bahna, F., Mannepalli, S., Ahlsén, G., Aye Thu, C., Sampogna, R. V., Maniatis, T., Honig, B. & Shapiro, L. (2017). Proc. Natl Acad. Sci. USA, 114, E9829–E9837. [DOI] [PMC free article] [PubMed]
- Gray, M. E. (2020). Thesis. The Ohio State University. Columbus, Ohio, USA.
- Grötzinger, C., Kneifel, J., Patschan, D., Schnoy, N., Anagnostopoulos, I., Faiss, S., Tauber, R., Wiedenmann, B. & Gessner, R. (2001). Gut, 49, 73–81. [DOI] [PMC free article] [PubMed]
- Harrison, O. J., Bahna, F., Katsamba, P. S., Jin, X., Brasch, J., Vendome, J., Ahlsen, G., Carroll, K. J., Price, S. R., Honig, B. & Shapiro, L. (2010). Nat. Struct. Mol. Biol. 17, 348–357. [DOI] [PMC free article] [PubMed]
- Harrison, O. J., Brasch, J., Katsamba, P. S., Ahlsen, G., Noble, A. J., Dan, H., Sampogna, R. V., Potter, C. S., Carragher, B., Honig, B. & Shapiro, L. (2020). Cell. Rep. 30, 2655–2671. [DOI] [PMC free article] [PubMed]
- Harrison, O. J., Brasch, J., Lasso, G., Katsamba, P. S., Ahlsen, G., Honig, B. & Shapiro, L. (2016). Proc. Natl Acad. Sci. USA, 113, 7160–7165. [DOI] [PMC free article] [PubMed]
- Harrison, O. J., Jin, X., Hong, S., Bahna, F., Ahlsen, G., Brasch, J., Wu, Y., Vendome, J., Felsovalyi, K., Hampton, C. M., Troyanovsky, R. B., Ben-Shaul, A., Frank, J., Troyanovsky, S. M., Shapiro, L. & Honig, B. (2011). Structure, 19, 244–256. [DOI] [PMC free article] [PubMed]
- Hinoi, T., Lucas, P. C., Kuick, R., Hanash, S., Cho, K. R. & Fearon, E. R. (2002). Gastroenterology, 123, 1565–1577. [DOI] [PubMed]
- Hirano, S. & Takeichi, M. (2012). Physiol. Rev. 92, 597–634. [DOI] [PubMed]
- Jaiganesh, A., Narui, Y., Araya-Secchi, R. & Sotomayor, M. (2018). Cold Spring Harb. Perspect. Biol. 10, a029280. [DOI] [PMC free article] [PubMed]
- Jung, R., Wendeler, M. W., Danevad, M., Himmelbauer, H. & Gessner, R. (2004). Cell. Mol. Life Sci. 61, 1157–1166. [DOI] [PMC free article] [PubMed]
- Katta, S., Krieg, M. & Goodman, M. B. (2015). Annu. Rev. Cell Dev. Biol. 31, 347–371. [DOI] [PubMed]
- Kazmierczak, P., Sakaguchi, H., Tokita, J., Wilson-Kubalek, E. M., Milligan, R. A., Müller, U. & Kachar, B. (2007). Nature, 449, 87–91. [DOI] [PubMed]
- Kreft, B., Berndorff, D., Böttinger, A., Finnemann, S., Wedlich, D., Hortsch, M., Tauber, R. & Gessner, R. (1997). J. Cell Biol. 136, 1109–1121. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Kuhlmann, L., Nadler, W. M., Kerner, A., Hanke, S. A., Noll, E. M., Eisen, C., Espinet, E., Vogel, V., Trumpp, A., Sprick, M. R. & Roesli, C. P. (2017). Pancreas, 46, 311–322. [DOI] [PubMed]
- Langer, M. D., Guo, H., Shashikanth, N., Pierce, J. M. & Leckband, D. E. (2012). J. Cell Sci. 125, 2478–2485. [DOI] [PubMed]
- Leckband, D. E. & de Rooij, J. (2014). Annu. Rev. Cell Dev. Biol. 30, 291–315. [DOI] [PubMed]
- Leckband, D. E., le Duc, Q., Wang, N. & de Rooij, J. (2011). Curr. Opin. Cell Biol. 23, 523–530. [DOI] [PubMed]
- Light, S. E. W. & Jontes, J. D. (2017). Semin. Cell Dev. Biol. 69, 83–90. [DOI] [PMC free article] [PubMed]
- Liu, L. X., Lee, N. P., Chan, V. W., Xue, W., Zender, L., Zhang, C., Mao, M., Dai, H., Wang, X. L., Xu, M. Z., Lee, T. K., Ng, I. O., Chen, Y., Kung, H. F., Lowe, S. W., Poon, R. T. P., Wang, J. H. & Luk, J. M. (2009). Hepatology, 50, 1453–1463. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Modak, D. & Sotomayor, M. (2019). Commun. Biol. 2, 354. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nagar, B., Overduin, M., Ikura, M. & Rini, J. M. (1996). Nature, 380, 360–364. [DOI] [PubMed]
- Nicoludis, J. M., Green, A. G., Walujkar, S., May, E. J., Sotomayor, M., Marks, D. S. & Gaudet, R. (2019). Proc. Natl Acad. Sci. USA, 116, 17825–17830. [DOI] [PMC free article] [PubMed]
- Nicoludis, J. M., Vogt, B. E., Green, A. G., Schärfe, C. P., Marks, D. S. & Gaudet, R. (2015). eLife, 5, e18449. [DOI] [PMC free article] [PubMed]
- Ohnishi, K., Melchers, F. & Shimizu, T. (2005). Eur. J. Immunol. 35, 957–963. [DOI] [PubMed]
- Ohnishi, K., Shimizu, T., Karasuyama, H. & Melchers, F. (2000). J. Biol. Chem. 275, 31134–31144. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Parisini, E., Higgins, J. M. G., Liu, J. H., Brenner, M. B. & Wang, J. H. (2007). J. Mol. Biol. 373, 401–411. [DOI] [PMC free article] [PubMed]
- Park, J. H., Seol, J., Choi, H. J., Roh, Y. H., Choi, P. J., Lee, K. E. & Roh, M. S. (2011). Histopathology, 58, 315–318. [DOI] [PubMed]
- Patel, S. D., Ciatto, C., Chen, C. P., Bahna, F., Rajebhosale, M., Arkus, N., Schieren, I., Jessell, T. M., Honig, B., Price, S. R. & Shapiro, L. (2006). Cell, 124, 1255–1268. [DOI] [PubMed]
- Pinho, S. S., Seruca, R., Gärtner, F., Yamaguchi, Y., Gu, J., Taniguchi, N. & Reis, C. A. (2011). Cell. Mol. Life Sci. 68, 1011–1020. [DOI] [PMC free article] [PubMed]
- Pokutta, S., Herrenknecht, K., Kemler, R. & Engel, J. (1994). Eur. J. Biochem. 223, 1019–1026. [DOI] [PubMed]
- Pokutta, S. & Weis, W. I. (2007). Annu. Rev. Cell Dev. Biol. 23, 237–261. [DOI] [PubMed]
- Ponstingl, H., Henrick, K. & Thornton, J. M. (2000). Proteins, 41, 47–57. [DOI] [PubMed]
- Pruitt, B. L., Dunn, A. R., Weis, W. I. & Nelson, W. J. (2014). PLoS Biol. 12, e1001996. [DOI] [PMC free article] [PubMed]
- Roy, F. van & Berx, G. (2008). Cell. Mol. Life Sci. 65, 3756–3788. [DOI] [PMC free article] [PubMed]
- Rubinstein, R., Thu, C. A., Goodman, K. M., Wolcott, H. N., Bahna, F., Mannepalli, S., Ahlsen, G., Chevee, M., Halim, A., Clausen, H., Maniatis, T., Shapiro, L. & Honig, B. (2015). Cell, 163, 629–642. [DOI] [PMC free article] [PubMed]
- Schreiner, D. & Weiner, J. A. (2010). Proc. Natl Acad. Sci. USA, 107, 14893–14898. [DOI] [PMC free article] [PubMed]
- Shapiro, L., Fannon, A. M., Kwong, P. D., Thompson, A., Lehmann, M. S., Grübel, G., Legrand, J. F., Als-Nielsen, J., Colman, D. R. & Hendrickson, W. A. (1995). Nature, 374, 327–337. [DOI] [PubMed]
- Smith, A. R., Rota, I. A., Maio, S., Massaad, M. J., Lund, T. C., Notarangelo, L. D., Holländer, G. A. & Blazar, B. R. (2017). Blood Adv. 1, 2083–2087. [DOI] [PMC free article] [PubMed]
- Sotomayor, M., Gaudet, R. & Corey, D. P. (2014). Trends Cell Biol. 24, 524–536. [DOI] [PMC free article] [PubMed]
- Sotomayor, M. & Schulten, K. (2008). Biophys. J. 94, 4621–4633. [DOI] [PMC free article] [PubMed]
- Sotomayor, M., Weihofen, W. A., Gaudet, R. & Corey, D. P. (2012). Nature, 492, 128–132. [DOI] [PMC free article] [PubMed]
- Su, M.-C., Yuan, R.-H., Lin, C.-Y. & Jeng, Y.-M. (2008). Mod. Pathol. 21, 1379–1386. [DOI] [PubMed]
- Takamura, M., Sakamoto, M., Ino, Y., Shimamura, T., Ichida, T., Asakura, H. & Hirohashi, S. (2003). Cancer Sci. 94, 425–430. [DOI] [PMC free article] [PubMed]
- Takeichi, M. (1990). Annu. Rev. Biochem. 59, 237–252. [DOI] [PubMed]
- Thomson, R. B., Igarashi, P., Biemesderfer, D., Kim, R., Abu-Alfa, A., Soleimani, M. & Aronson, P. S. (1995). J. Biol. Chem. 270, 17594–17601. [DOI] [PubMed]
- Thomson, R. B., Ward, D. C., Quaggin, S. E., Igarashi, P., Muckler, Z. E. & Aronson, P. S. (1998). Genomics, 51, 445–451. [DOI] [PubMed]
- Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. (2009). Bioinformatics, 25, 1189–1191. [DOI] [PMC free article] [PubMed]
- Weiner, J. A. & Jontes, J. D. (2013). Front. Mol. Neurosci. 6, 4. [DOI] [PMC free article] [PubMed]
- Wendeler, M. W., Drenckhahn, D., Gessner, R. & Baumgartner, W. (2007). J. Mol. Biol. 370, 220–230. [DOI] [PubMed]
- Wendeler, M. W., Jung, R., Himmelbauer, H. & Gessner, R. (2006). Cell. Mol. Life Sci. 63, 1564–1673. [DOI] [PMC free article] [PubMed]
- Wendeler, M. W., Praus, M., Jung, R., Hecking, M., Metzig, C. & Gessner, R. (2004). Exp. Cell Res. 294, 345–355. [DOI] [PubMed]
- Weth, A., Dippl, C., Striedner, Y., Tiemann-Boege, I., Vereshchaga, Y., Golenhofen, N., Bartelt-Kirbach, B. & Baumgartner, W. (2017). Tissue Barriers, 5, e1285390. [DOI] [PMC free article] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Wong, B. W., Luk, J. M., Ng, I. O., Hu, M. Y., Liu, K. D. & Fan, S. T. (2003). Biochem. Biophys. Res. Commun. 311, 618–624. [DOI] [PubMed]
- Yui, A., Caaveiro, J. M. M., Kuroda, D., Nakakido, M., Nagatoishi, S., Goda, S., Maruno, T., Uchiyama, S. & Tsumoto, K. (2020). bioRxiv, 2020.09.18.291195.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human CDH17 EC1–2, 6ulm
Supplementary Methods, Figures and Tables. DOI: 10.1107/S2053230X21002247/pg5088sup1.pdf