Abstract
Antibiotic-resistant bacteria are a major global health threat that continues to rise due to a lack of effective vaccines. Of concern are Klebsiella pneumoniae that fail to induce in vivo germinal center B cell responses, which facilitate antibody production to fight infection. Immunotherapies using antibodies targeting antibiotic-resistant bacteria are emerging as promising alternatives, however, they cannot be efficiently derived ex vivo, necessitating the need for immune technologies to develop therapeutics. Here, PEG-based immune organoids were developed to elucidate the effects of polymer end-point chemistry, integrin ligands, and mode of K. pneumoniae antigen presentation on germinal center-like B cell phenotype and epigenetics, to better define the lymph node microenvironment factors regulating ex vivo germinal center dynamics. Notably, PEG vinyl sulfone or acrylate failed to sustain primary immune cells, but functionalization with maleimide (PEG-4MAL) led to B cell expansion and germinal center-like induction. RNA sequencing analysis of lymph node stromal and germinal center B cells showed niche associated heterogeneity of integrin-related genes. Incorporation of niche-mimicking peptides revealed that collagen-1 promoted germinal center-like dynamics and epigenetics. PEG-4MAL organoids elucidated the impact of K. pneumoniae outer membrane-embedded protein antigen versus soluble antigen presentation on germinal centers and preserved the response across young and aged mice.
Keywords: hydrogel, B cell, bacteria, histone, EZH2, aging
1. Introduction
Bacterial infections remain a major global health problem, causing more than 5 million deaths annually1. Antibiotics are currently the standard medical intervention for several bacterial pathogens, including Mycobacterium tuberculosis, Clostridioides difficile, Escherichia coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae, among others, for which we do not yet have vaccines. Even more concerning are the rising antibiotic resistance threats in the United States and worldwide. According to the 2019 Antibiotic Resistance Threats Report by the U.S. Center for Disease Control2, more than 2.8 million antibiotic-resistant infections occur in the U.S. each year, and more than 35,000 people die as a result. In addition, 223,900 cases of Clostridioides difficile occurred in 2017 and at least 12,800 people died. According to WHO’s 2019 Global tuberculosis report, in 2018, >500,000 people worldwide developed multidrug-resistant tuberculosis (MDR-TB), and an additional >180,000 cases with rifampicin-resistant TB. In recent years, the emergence of broad MDR strains of K. pneumoniae, which not only causes pneumonia and infections of the bloodstream and urinary tract, but also are not cleared by β-lactam antibiotics, including carbapenem and cephalosporins, or by non-β-lactam antibiotics, such as aminoglycosides and tetracycline, has made K. pneumoniae one of the most concerning threats3. Vaccines are still unavailable for several bacterial infections, including most antibiotic-resistant bacteria.
Vaccines to prevent such infections can only be best developed on time based on our increasing insights into the immune response. Effective vaccination requires identification of bacterial antigens, their immunogenicity, and the mode of presentation of these antigens – as embedded bacterial antigens in outer membranes or in soluble purified forms, making successful vaccination challenging. New immunotherapies using monoclonal antibodies targeting bacterial antigens are emerging as promising alternatives to passive immunization to tackle antibiotic-resistant bacteria, including K. pneumoniae4, 5, but often fail to mediate broad protection against different serotypes. Developing antibodies with high cross-reactivity requires an enhanced understanding of the underlying immune response, which typically involves a germinal center B cell response in lymph nodes that makes high-affinity antibodies through iterative somatic hypermutation and affinity maturation processes6, 7. Unfortunately, the polysaccharide antigens on many gram-negative bacteria fail to induce robust germinal center responses in vivo unless conjugated with carrier proteins that stimulate T and B cells, therefore necessitating the development of an ex vivo immune technology, where the germinal center reaction can be induced irrespective of antigen type and form. However, the development of such immune technologies is non-trivial as factors that spatially and temporally regulate the ex vivo germinal center response, ranging from the microenvironment to antigen format, are poorly understood. The goal of the current study is to identify these factors and engineer a materials-based immune organoid technology.
During an immune response to bacterial and viral infections, naïve B cells in the lymph node and spleen, encounter antigen and form sub-anatomical structures within B cell follicles in secondary lymphoid organs, referred to as germinal centers8, 9. Antigen-activated germinal center B cells undergo rapid proliferation and somatic hypermutation of their immunoglobulin variable genes. The proliferating B cells mutate their B cell receptors to produce mutant germinal center B cell clones with a range of affinities against the activating antigen10–12, which regulates cross-reactivity against serotypes. The germinal center response is a complex process involving multiple cell types and environmental cues13–15. B cell follicles are composed of B cells, CD40 ligand (CD40L) presenting follicular T helper (TFH) cells, B cell activating follicular dendritic cells (FDCs)16–18, extracellular matrix (ECM) proteins such as Arg-Gly-Asp (RGD)-presenting vitronectin15, 19, and several other factors. For example, integrin-mediated interactions between B cells and FDCs influence germinal center B cells15. B cell activation is marked by epigenetic regulation and immune receptor signaling, which come into play at critical transitional stages of the germinal center reaction20, 21. In the germinal center reaction, there is transient suppression of enhancers and promoters of genes that regulate immune signaling pathways, antigen presentation, and checkpoints, which revert to the active state when germinal center B cells are signaled to exit the germinal center reaction. Exit from the germinal center reaction occurs by differentiating into antibody-producing plasma cells or memory B cells. Currently, the tissue and microenvironment factors governing B cell dynamics, temporal expression of phenotypic and epigenetic markers, and exit from the germinal center reaction are not well understood.
Early efforts to elucidate B cell differentiation involved a controlled environment of naïve B cells co-cultured in 2D with stromal cells, called 40LB22. These cells were engineered to provide critical signals for B cell survival and differentiation, including the T cell signal CD40L and B cell activating factor (BAFF), secreted in vivo by FDCs. However, the in vitro 2D presentation of these signals is not sufficient for the induction of a germinal center response that resembles an in vivo-like response21, suggesting a critical role for the 3D microenvironment. Indeed, we have previously shown that incorporation of naïve murine B cells and 40LB in 3D organoids engineered from gelatin and silicate nanoparticles23, 24 can significantly drive germinal center B cell phenotype, transcriptome, and somatic hypermutation just after 4 days ex vivo21, similar to in vivo immunized mice. To better decouple the signals driving this response, we further engineered modular organoids with tunable microenvironments using a four-arm polyethylene glycol presenting maleimide (PEG-4MAL) functionalized with thiolated ECM-mimicking peptides and enzymatically degradable crosslinkers25. Using this system, we have shown that interactions between integrins and VCAM-1-mimicking peptide regulated phosphorylation of key proteins in murine B cell receptor (BCR) signaling, and demonstrated the ability of immune organoids to generate antigen-specific IgG1 antibodies through the addition of soluble antigen and incorporation of T cell signals, such as Fas ligand25. However, to date, the role of PEG end-group chemistry in regulating the germinal center response and contributions of microenvironment-epigenome interactions have not yet been explored, despite that such interactions would be expected to regulate cell behavior.
In addition, tissue-specific and age-related differences have been reported for B cell selection26, 27. B cells from aged mice exhibit decreased expansion in response to antigen28, 29 and elderly individuals have an inability towards brisk and sustained responses to new antigens. Aged individuals have limited ability to induce germinal center reactions and B-cell competition, partly attributable to suboptimal CD40–CD40L interactions between B and dysfunctional T cells30. Consequently, even the immunogenic vaccine candidates need to rely on T cells and CD40L functioning in vivo. Therefore, an ex vivo immune organoid that is capable of recapitulating germinal center responses to antigen in aged B cells represents a powerful tool for understanding patient-specific B cell dynamics and generating antigen-specific antibodies for aged populations, which can then be infused into patients.
In the current work, our goal was to identify factors, including polymer end-point chemistry, integrin ligands, and mode of Klebsiella Pneumoniae antigen presentation, in a synthetic immune organoid that regulates the phenotype and epigenetics of ex vivo germinal centers from young and aged murine B cells. The organoids provide a future opportunity to derive age-specific germinal center B cells, and subsequently antibody-secreting cells and antibodies against a large set of bacterial antigens that can be injected into patients as therapeutics. We provide the first comparison of advanced PEG-4MAL hydrogels with those functionalized with vinyl sulfone and acrylate in regulating the fate of primary germinal center B cells. We provide a rational methodology to develop immune organoids, driven by single-cell RNA sequencing of uniquely identified lymph node stromal cells and bulk RNA sequencing of germinal center B cells from immunized mice to define microenvironment factors to be tested for germinal center induction. We further elucidate the mechanisms by which PEG-4MAL facilitates the germinal center reaction by comparing the time-dependent effects of integrin-binding peptides on regulating the kinetics of B cell EZH2, H3k27Me3 epigenetics, and BCL6 expression. Subsequently, we demonstrate the application of PEG-4MAL as a platform to probe the potential for inducing germinal center responses and regulating epigenetics with bacterial membrane antigens from a clinical K. Pneumoniae isolate in their soluble form or membrane-embedded form. Finally, the current study sets out to demonstrate the effect of K. Pneumoniae antigens in regulating the germinal center phenotype and histone modification in B cells from aged mice >2 years old. These findings describe, for the first time, a direct comparison of PEG end-point chemistry in the engineering of immune organoids and highlight the potential of synthetic germinal center-like organoids as a platform for the future development of antigen- and age-specific antibodies for therapeutic intervention, as well developing therapeutics for aged individuals.
2. Results and Discussion
2.1. RNA sequencing of mouse lymph node stromal cells and germinal center B cells reveals expression of cell-cell and cell-ECM binding genes
Stromal cells establish the compartmentalization of lymphoid tissues, which are critical to the immune response (Figure 1A). Recently, we reported single-cell RNA sequencing of lymph node stromal cells using barcoded droplets (Figure 1A) that revealed nine peripheral lymph node non-endothelial stromal cell clusters18. Included were the established subsets, C-C Motif Chemokine Ligand 19 (Ccl19)hi T-zone reticular cells (TRCs), marginal reticular cells (MRCs), and follicular dendritic cells (FDCs). The study further identified Ccl19lo TRCs, C-X-C Motif Chemokine Ligand 9 (Cxcl9+) TRCs in the T-zone and interfollicular region, and clusters corresponding to medullary, perivascular and capsular stromal cells. In these studies, a droplet-based single-cell RNA sequencing was performed on isolated peripheral lymph node CD45-CD31- cells from mice immunized intravenously with 2.5 × 105 pfu of Lymphocytic choriomeningitis virus (LCMV)-Armstrong infected mice (day 15 post-infection)18, thereafter performing sequencing and analysis on > 14,000 single cells.
Figure 1: RNA sequencing of lymph node stromal and germinal center B cells shows niche associated heterogeneity of integrin-related genes.
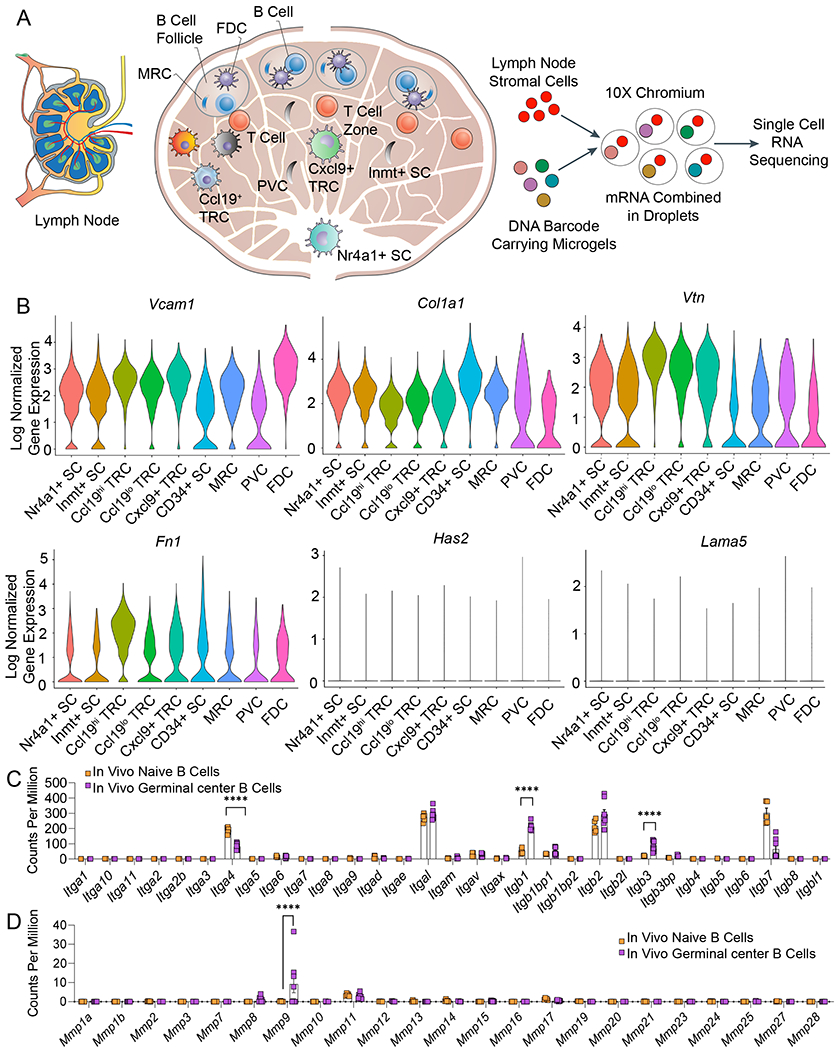
A) Schematic of lymph node structure comprised of distinct stromal cell populations inside and outside of B cell follicle. Non-endothelial stromal cells isolated from the peripheral lymph node are subjected to droplet-based single-cell RNA sequencing workflow. B) Single-cell RNA sequencing of nine uniquely identified clusters of peripheral lymph node non-endothelial stromal cells reveals the expression of mRNA encoding for proteins that bind to integrins. Violin plots represent the log-normalized gene expression (unique molecular identifier counts) for each cell grouped by the cluster. C) RNA sequencing comparison of in vivo integrin expression between naïve and germinal center B cells. D) RNA sequencing comparison of in vivo matrix metalloproteinase (MMP) expression between naïve and germinal center B cells. In C-D, data represent mean ± S.E.M. Significant differences determined using a 2-way ANOVA with Sidak’s multiple comparison test. ****p<0.0001; N=5-8.
We analyzed this single-cell RNA sequencing dataset of murine lymph node stromal cells, including cells located in the B cell follicle, to determine various genes that bind to integrins. We observed that a large fraction of FDCs and MRCs expressed high levels of collagen 1 (Col1a1), vascular cell adhesion molecule 1 (Vcam1), vitronectin (Vtn), and fibronectin (Fn1) genes (Figure 1B, Supplementary Fig. S1). VCAM1 protein binds to integrin α4β1 on B cells and is involved in B cell activation, however, the role of collagen-1 that binds to α1β1, α2β1, α10β1 and α11β1 integrins, and α5β1 and αvβ3-binding fibronectin and vitronectin are less known. In contrast, laminin (Lama5) and hyaluronan (Has2) were not abundant in these stromal cells (Figure 1B). We further performed RNA sequencing analysis on naïve versus germinal center B cells isolated from immunized mice, as reported in our recent study21, and observed that β1 and β3 were significantly upregulated in in vivo germinal center B cells compared to primary naïve B cells (Figure 1C). Integrins containing the β1 subunit can engage with collagen, fibronectin, VCAM-1, and several other matrices. Integrins α4, which forms a heterodimer with β1 subunit, can bind to fibronectin and VCAM-1, and were expressed in both naïve and germinal center B cells. In addition, we also observed high expression of αL integrin subunit, which forms a heterodimer with β2 subunit and binds to ICAM-1 (Intercellular Adhesion Molecule 1). These findings motivate the need to investigate hydrogel networks that can present bio-adhesive ligands of interest, such as collagen 1 and VCAM-1 mimics, to understand the role played by various cell-cell and cell-matrix binding proteins on germinal center phenotype and epigenetics. Another important observation from the germinal center B cell RNA sequencing was the upregulation of matrix metalloproteinase (MMP)-9 compared to naïve B cells (Figure 1D). This observation supports the need for developing immune organoids that can be remodeled by MMP-9 secreted by activated B cells, as demonstrated in the next section.
2.2. The end group functionality in PEG hydrogels impacts B cell survival and induction of germinal center response
To study the effect of various extracellular matrices and integrin-binding proteins found in the B cell follicle, we sought to use hydrogel platforms where integrin-binding ligands of interest can be functionalized for presentation to B cells and supporting stromal cells. Although matrices from naturally derived materials have been attempted to build immune tissues, as reviewed by us31, including our work using a gelatin-based organoid system23, 24, which presented RGD motifs, natural materials do not afford easy control over the microenvironment and presentation of various distinct integrin ligands, may induce confounding effects on immune cell activation due to non-integrin receptor-ligand interactions, such as CD44 binding in hyaluronic acid-based hydrogels32, and may suffer from viscosity issues and batch-to-batch variability. Furthermore, we have previously shown that the photopolymerization of gelatin methacrylate does not generate germinal center response23, 24. Therefore, we chose to use a milder polymerization reaction that does not require toxic radical photoinitiators for the UV curing. Several alternative chemistries exist for cell encapsulation and cross-linking under physiological conditions, including Schiff-base formation33, 34, amine-NHS ester coupling35, Diels–Alder reaction36, 37, radical-mediated thiol-ene38 and thiol–yne39, and nucleophilic additions of thiols to alkenes or alkyne esters or epoxy groups 40–46. However, among these, as discussed elsewhere47, Schiff-base reactions are reversible under physiological conditions and the reaction kinetics of amine-NHS ester, Diels–Alder, chemistries are not highly suitable for homogeneous distribution of cells throughout the hydrogel. In contrast, thiol-based moieties, such as peptides and polymeric macromers, can react with activated alkenes or alkynes using photogenerated radicals (e.g. acrylate and norbornene groups) or through Michael addition chemistry with maleimides (MAL), vinylsulfones (VS), and acrylates (ACR). Michael-addition hydrogel networks form through a step-growth polymerization of the thiol and an electron-deficient group (e.g. VS), stemming from a two-part propagation process where a thiolate ion reacts with the VS group to form a carbon-based anion. The resulting anion reacts with another thiol group to form the next thiolate ion, leading to a repetitive process that generates a covalently crosslinked network48.
In our studies, we chose to test Poly(ethylene glycol) (PEG) hydrogels based on 4-arm PEG macromers with either terminal vinyl sulfone (PEG-4VS), acrylate (PEG-4ACR), or maleimide groups (PEG-4MAL) (Figure 2A). These polymers were chosen because they have the same PEG macromer backbone, similar Molecular weight ~20,000 Da, can be functionalized to present integrin-specific peptides that are cysteine-terminated via a similar Michael-type addition reaction, and are crosslinked using di-thiolated crosslinkers at physiological pH and temperature under mild conditions41, as opposed to UV crosslinking (Figure 2B). While all three chemical functionalities in PEG hydrogels have been studied for a wide range of mammalian cells (PEG-MAL40, 49, 50, PEG-VS51–53, PEG-Acr54), they have not been compared directly to each other for their impact on primary B cells for the induction of germinal centers. This is an important biomaterial consideration given the growing interest in building immune tissues31, 55, 56, and the limited ability of natural matrices to control the microenvironment and integrin ligand presentation.
Figure 2: Hydrogel macromer end-modification chemistry regulates the survival and differentiation of primary immune cells.
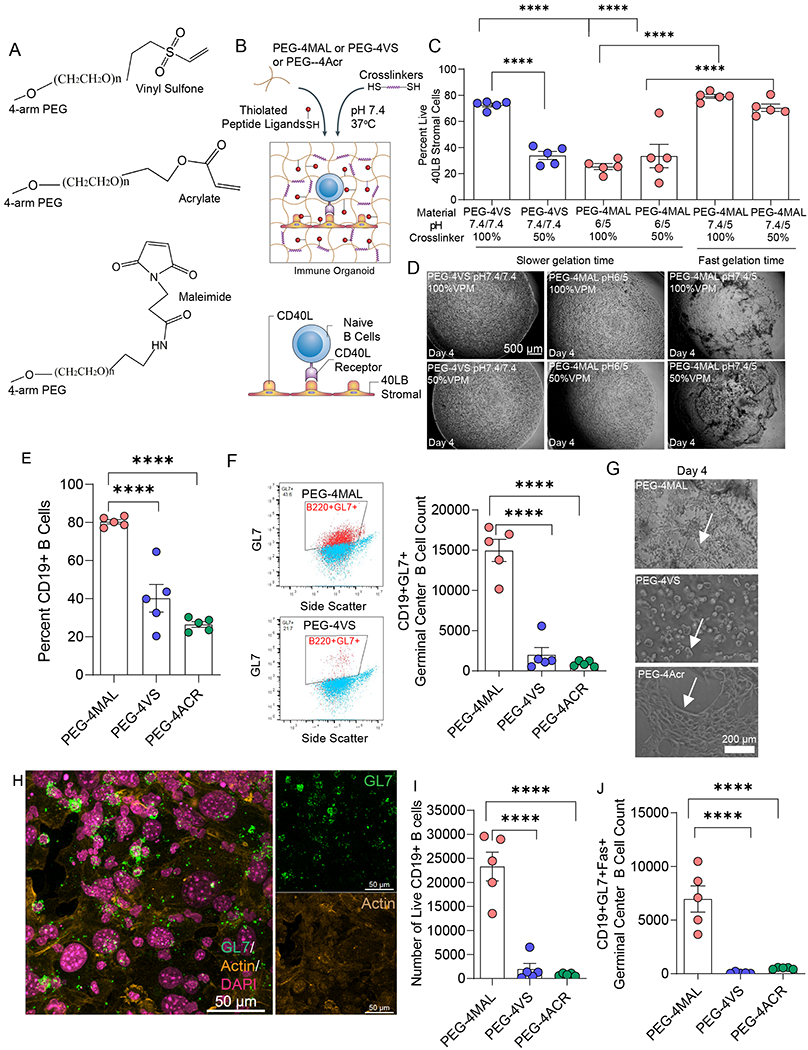
A) Chemical structure of the vinyl sulfone, acrylate, and maleimide functional groups at the end of four-arm PEG. B) Schematic of immune organoid formation with co-cultured CD40L- and BAFF-presenting stromal cells and primary, naive B cells. C) Effects of pH and degradability on 40LB cell viability in PEG-4VS and PEG-4MAL hydrogels. D) Representative brightfield images illustrating the effects of pH and degradability on stromal cell network formation in PEG-4VS and PEG-4MAL organoids. E) Quantitative flow cytometric analysis of CD19+ B cell in four-arm PEG hydrogels modified with maleimide, vinyl sulfone, or acrylate, indicating survival in hydrogels. F) Flow cytometric gating strategy and germinal center-like B cell (CD19+GL7) formation in four-arm PEG hydrogels modified with maleimide, vinyl sulfone, and acrylate. G) Effects of maleimide, vinyl sulfone, and acrylate functionalization on stromal cell network formation and survival of B cells in PEG-based organoids. H) Confocal imaging of germinal center-like B cell (GL7+) and its relative localization with 40LB stromal in PEG-4MAL organoids. Green: GL7, Yellow: Actin (false color from red), Magenta: DAPI (false color from blue). I) Effect of four-arm PEG end-modification chemistry on cell viability of CD19+ B cells cultured in organoids for 4 days. J) Quantitative flow cytometric analysis of CD19+GL7+Fas+ germinal center-like B cell formation after 4 days of culture in four-arm PEG hydrogels modified with maleimide, vinyl sulfone, or acrylate. All data represent mean ± S.E.M. and were analyzed via one-way ANOVA with Tukey’s posthoc multiple comparisons test. ****p<0.0001, ns=not significant; N=5.
We first characterized the survival of our engineered stromal cell 40LB23, 57 (developed and provided by Daisuke Kitamura22), which are BALB/c 3T3 fibroblasts stably transduced with both T cell-derived CD40 ligand (CD40L) and FDC-secreted B cell activation factor. We tested conditions where the hydrogel macromers formed fast gels (< 1min) versus slow gels (>1 min) by tuning the pH of the PEG macromer and crosslinker type. Specifically, we tested the impact of the ratio of MMP-9 degradable crosslinker peptide (GCRDVPM↓SMRGGDRCG, referred to as VPM) with non-degradable crosslinker dithiothreitol (DTT). The rationale for using VPM was that our RNA sequencing has revealed a significantly higher expression of MMP-9 in germinal center B cells as compared to naïve B cells, but other MMPs were indifferent (Figure 1D). We observed high viability among fast gelling PEG-4MAL hydrogels irrespective of the crosslinker pH and VPM:DTT ratio (50 or 100% VPM) (Figure 2C). In contrast, PEG-4VS at 50% VPM or PEG-4MAL at either 50% or 100% VPM, both of which gelled slowly at lower PEG macromer (pH 6), showed cytotoxic effects on 40LB stromal cells (Figure 2C). There were no major differences between the stability of hydrogels (Figure 2D), except that slow gelling hydrogels formed a more uniform hydrogel than fast gelling ones, which can be attributed to wrinkling due to rapid pipetting of fast gelling hydrogels. We, therefore, continued with formulations that showed high viability so that the effects can be delineated between B cells and 40LBs.
We next examined the effect of polymer chemistry on B cell viability and induction of germinal center by co-encapsulating freshly isolated naïve murine B cells and 40LBs in the hydrogel organoid and culturing in the presence of 10 ng/mL IL-4, regular RPMI with 10% FBS and 1% antibiotics. In these initial studies, we chose a VCAM-1 mimicking peptide REDV, recently reported by us25. Interestingly, unlike 40LB cells, primary B cells failed to survive and proliferate in both vinyl sulfone and acrylate functionalized organoids (Figure 2E, F, Supplementary Fig. S2A). Using CD19 as the surface biomarker of B cells and GL7 as surface epitope protein indicative of activated germinal center B cells, we found that B cells cocultured in PEG-4MAL organoids yielded a significantly higher number of CD19+ B cells and germinal center-like (CD19+GL7+) B cells (Figure 2E, F, G). The GL7 expressing B cells (green) co-localized with 40LB stromal cells (yellow, spread), as revealed by confocal imaging studies (Figure 2H). Collectively, this is the first study, to the best of our knowledge, that establishes the superiority of advanced PEG-4MAL hydrogels with those functionalized with vinyl sulfone and acrylate, at least within the conditions tested, in regulating the fate of primary B cells towards induction of germinal centers.
To understand the molecular impact of the three hydrogels with similar underlying crosslinking mechanisms but different end-point functionalities, we examined the viability of B cells and generation of the apoptotic phenotype of germinal center-like B cells, i.e. CD19+GL7+Fas+ cells. The number of viable B cells in PEG-4MAL organoids were significantly higher as determined using a live/dead flow cytometry assay (Figure 2I), in contrast to PEG-4VS and PEG-4ACR, alluding to the toxic effects of vinyl sulfone and acrylates. In addition, proliferative germinal center B cells exhibit a phenotype that is characteristic of apoptosis-sensitive cells through increased levels of death-inducing molecule Fas58. Apoptosis through the expression of Fas is critical to the negative selection of germinal center B cells in vivo and for maintaining the efficacy of these specialized cells. We observed that the PEG-4MAL organoids generated a significantly higher number of CD19+GL7+Fas+ B cells (Figure 2J), in contrast to PEG-4VS and PEG-4ACR organoids. The expression of Fas was, however, similar across all three polymer matrices (Supplementary Fig. S2B), suggesting that the molecular impact of PEG-4MAL is through enhanced survival of primary B cells, in contrast to the toxicity induced by PEG-4VS and PEG-4ACR. Further temporal analysis of proliferative and apoptotic signals would be needed to understand the molecular impact of PEG-4-VS and PEG-4ACR on B cell signaling. It should be noted that the differences in the degree of macromer functionalization, the hydrolysis rate of the end group, and reactivity towards thiols could vary among the 3 macromers that are evaluated in the current study, and can impact final crosslink density and stiffness of the hydrogels 41, which may, in turn, affect B cell viability and germinal center induction. Further analysis of the impact of these factors on the proliferative and apoptotic signals is warranted.
2.3. Collagen 1 mimicking peptide regulates germinal center phenotype and BCL6 expression towards germinal center exit-like phenotype
As indicated in Figure 1, various follicle-associated stromal cells, including FDCs, express the collagen 1 gene. This is further supported by the presence of collagen conduits in B cell follicles and whole lymph node59. How collagen-mimicking matrices compare to fibronectin, vitronectin, or VCAM-1 ligands in inducing germinal centers remains unknown. Here, we examined three synthetic adhesive peptides with different integrin-binding specificities (Figure 3A). GFOGER is a triple-helical synthetic peptide derived from type I collagen with high binding affinity for α1β1, α2β1, α10β1 and α11β160, 61. REDV is a tetrapeptide Arg-Glu-Asp-Val that mimics VCAM-1 in its ability to bind α4β1 integrins62 on B cells, and RGD is a short linear peptide present in vitronectin, fibronectin and other ECM proteins that bind several integrins, including αvβ3, αvβ1, and α5β149, 63, 64. We have previously shown that scrambled inactive peptides, such as GRDGSPC, do not engage integrins on B cells and fail to impact B cell receptors that are critical to downstream signaling and immune response 25, and therefore RDG only groups were not included in the current study. We did, however, incorporate RDG scramble peptides in the organoids as a filler to overcome solubility and viscosity issues when 100% GFOGER is used. Therefore, we used the 0.3 molar ratio of GFOGER, REDV, or RGD and 0.7 molar ratio of RDG.
Figure 3: Collagen-mimicking GFOGER peptide ligand differentially regulates germinal center-like B cell activation and signaling.
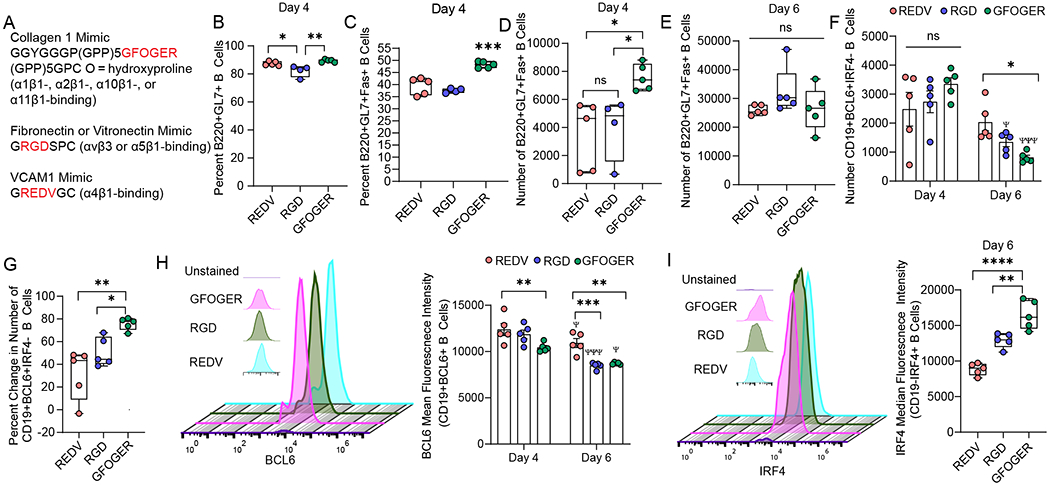
A) Sequences of integrin-binding peptides incorporated into PEG-4MAL organoids. B-C) Effects of GFOGER peptide ligand on the percent of B) B220+GL7+ B cells and C) proliferative B220+GL7+Fas+ B cells, in contrast to VCAM-1-like and RGD peptide ligands in organoid grown cultures of naïve B cells with 40LB and IL-4. D-E) Time-dependent effects of varying integrin ligand on the number of B220+GL7+Fas+ cells on D) Day 4 and E) Day 6. F-G) Temporal effect of the integrin-binding peptide on F) the number of and G) percent change in CD19+BCL6+IRF4- B cells over time. H) BCL6 mean fluorescence intensity over time in response to varying integrin ligand. The left side flow cytometry histogram indicates the expression of BCL6 on Day 4. I) IRF4 mean fluorescence intensity over time in response to varying integrin ligand. The left side flow cytometry histogram indicates the expression of IRF4 on Day 6. Data represent mean ± S.E.M. Significance determined via one-way ANOVA with Tukey’s posthoc multiple comparisons test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; N=5.
While all three peptides supported induction of nearly similar percentage of CD19+GL7+ germinal center phenotype cells by day 4 in presence of 40LB cells (Figure 3B), GFOGER peptide-functionalized PEG-4MAL organoids significantly increased the hallmark proliferative CD19+GL7+Fas+ phenotype by ~ 10% as compared to REDV and RGD peptides (Figure 3C). There were no significant differences between REDV and RGD peptides in the induction of CD19+GL7+Fas+ population. Similarly, we observed a significant increase in the number of CD19+GL7+Fas+ cells in GFOGER-organoids than in other matrices (Figure 3D). By day 6, the differences between matrices were lost, suggesting that matrices can play a role in the initial time course of induction of the germinal center phenotype (Figure 3E). Such studies are difficult to perform in vivo due to the migratory behavior of B cells that manifest unique cell cycle stages and emphasize the need for ex vivo immune organoids where nearly all cells are synchronized to similar states. By day 6, the expression of GL7 significantly reduced compared to day 4 (Supplementary Fig 3A). There were no differences across matrices on day 6, possibly suggesting further differentiation of germinal center-like B cells towards a germinal center-exit phenotype.
We next studied the temporal expression of the hallmark transcription factor, BCL6, in germinal center-like B cells differentiated in organoids. At the transcriptional level, maintenance of the germinal center B cell fate is regulated by germinal center promoting transcription factors, in particular Bcl6, and B cell exit to plasma cell fate is controlled by transcription factors such as Irf465, 66. Towards the exit of germinal centers, plasma-cell-precursor-like cells express lower Bcl667. There were no differences in the number of CD19+BCL6+IRF4- B cells on day 4 of differentiation across the three matrices (Figure 3F). In contrast, the number of CD19+BCL6+IRF4- B cells reduced by day 6 (Figure 3F), with a decrease by 31 ± 22 % in REDV, 50 ± 12 % in RGD, and maximum reduction in GFOGER-organoids with a drop by 76 ± 5 % (Figure 3G). The decrease in CD19+BCL6+IRF4- cells does not indicate a decrease in differentiated B cells because there are > 90% B220+GL7+ B cells on day 6 (Supplementary Fig 3B). Similar to CD19, B220 is a pan B cell marker in mice and both are used interchangeably in this work to identify activated murine B cells21,57 depending on conjugated-antibody availability for flow cytometry. The decrease in CD19+BCL6+IRF4- cells could reflect that the activated B cells that acquired a GL7 phenotype are differentiating into an advanced stage by day 6, possibly progressing towards a germinal center-exit phenotype ex vivo that has a marked decrease in BCL6 expression. To test this, we evaluated the expression of BCL6 in CD19+BCL6+ B cells and observed a significant decrease in BCL6 expression on day 6 and a matrix-dependent manner with cells grown in GFOGER and RGD organoids acquiring more reduction than REDV organoids (Figure 3H). These results suggest that GFOGER-presenting immune organoids induce high differentiation of germinal center B cells within 4 days of differentiation with a rapid transition to germinal center exit-like phenotype by day 6. Our hypothesis is further supported by higher expression of IRF4 in B220-IRF4+ B cells in the GFOGER-functionalized organoids as compared to REDV- or RGD-functionalized organoids (Figure 3I). IRF4 is a transcription factor that is required for the terminal differentiation of B cells to plasma cells66.
GFOGER-like sequences in collagens can bind to α1β1, α2β1, α10β1, and α11β1 integrins with high affinity. Our RNA sequencing results suggest that both naïve and in vivo germinal center B cells lack α1, α2, α10 and α11 integrins, however, express high levels of β1 integrins. Yet, other studies have shown that integrins α1β1 and α2β1 are present on activated T-cells and fibroblasts (α1β1, α2β1), among other cell types 61. In addition, integrin α11β1 is also present on a subset of fibroblasts 61. Because 40LBs are fibroblasts, it is unclear whether GFOGER binds directly to B cells in organoids or functions via 40LB cells, or whether the effect is mediated by high expression of β1 and its interaction with the triple-helical GFOGER peptide as compared to the linear REDV and RGD peptides. Therefore, further investigation is needed to determine the mode of functioning of GFOGER in organoids.
2.4. GFOGER and REDV regulate germinal center B cell epigenetic hallmarks EZH2 and H3k27Me3 in a CD40L- and BCL6-dependent manner
Germinal center B cells feature upregulation of Polycomb protein Enhancer of zeste homolog 2 (EZH2) histone methyltransferase21, 68, a core component of Polycomb repressive complex (PRC) 2 that methylates lysine 27 of histone 3 to generate H3K27me3, a histone mark associated with gene repression. EZH2 converts H3K4me3 active promoters to H3K4me3/H3K27me3 bivalent promoters for transient repression of target genes, which is reversed upon the germinal center exit. We and others have previously shown that EZH2 is required for the germinal center formation and conditional deletion of EZH2 results in failure to form germinal centers in mice21, 69. We have also shown that germinal center B cells undergo massive proliferation due to the silencing of cell cycle checkpoint genes, such as CDKN1A 21, at the epigenetic level, mediated by EZH2, which catalyzes H3k27Me3. EZH2 is absent from naïve B cells but highly induced in germinal centers. While it is known that α4β1 integrin-mediated T cell adhesion to VCAM-1 drives histone 3 methylation through a methyltransferase, the role of various integrin ligands, including collagen 1, in modulating the EZH2 and H3k27Me3 kinetics of the germinal center differentiation remains unknown. Unfortunately, the crucial EZH2 kinetics experiments cannot be easily performed in vivo because A) in mice, germinal centers are heterogeneous with many B cells entering, leaving and recycling at any given time, and B) knockdown of integrins and their ligands can be lethal. Developing biomaterials to recapitulate the process of germinal centers could improve the mechanistic investigation into signaling and epigenetic mechanisms that regulate germinal center B cells.
Along these lines, we had previously reported a gelatin-silicate nanoparticle-based immune organoid that presented RGD motifs to B cells21, 24. In these studies, the immune organoids showed remarkable similarity to in vivo germinal center B cells transcriptome, somatic hypermutation, EZH2, and H3k27Me3 protein expression, and proliferative phenotype. Analysis of the EZH2 gene from RNA sequencing results show that RGD organoids with primary B cells, 40LBs, and IL-4 manifest similar EZH2 upregulation on day 4 as immunized mice on day 10 (typical time for the formation of in vivo germinal centers) (Figure 4A). In contrast, 2D cocultures of primary naïve B cells and 40LBs in the presence of IL-4 failed to induce expression of EZH2 and were similar to the levels in naïve B cells, which do not have high expression of EZH2 (Figure 4A). To determine whether REDV and GFOGER induced differential expression of EZH2 than in RGD, we evaluated the impact of these peptides on germinal center-like B cells differentiated in PEG-4MAL organoids.
Figure 4: Expression of germinal center-like B cell epigenetics is regulated by Maleimide-presented integrin ligands.
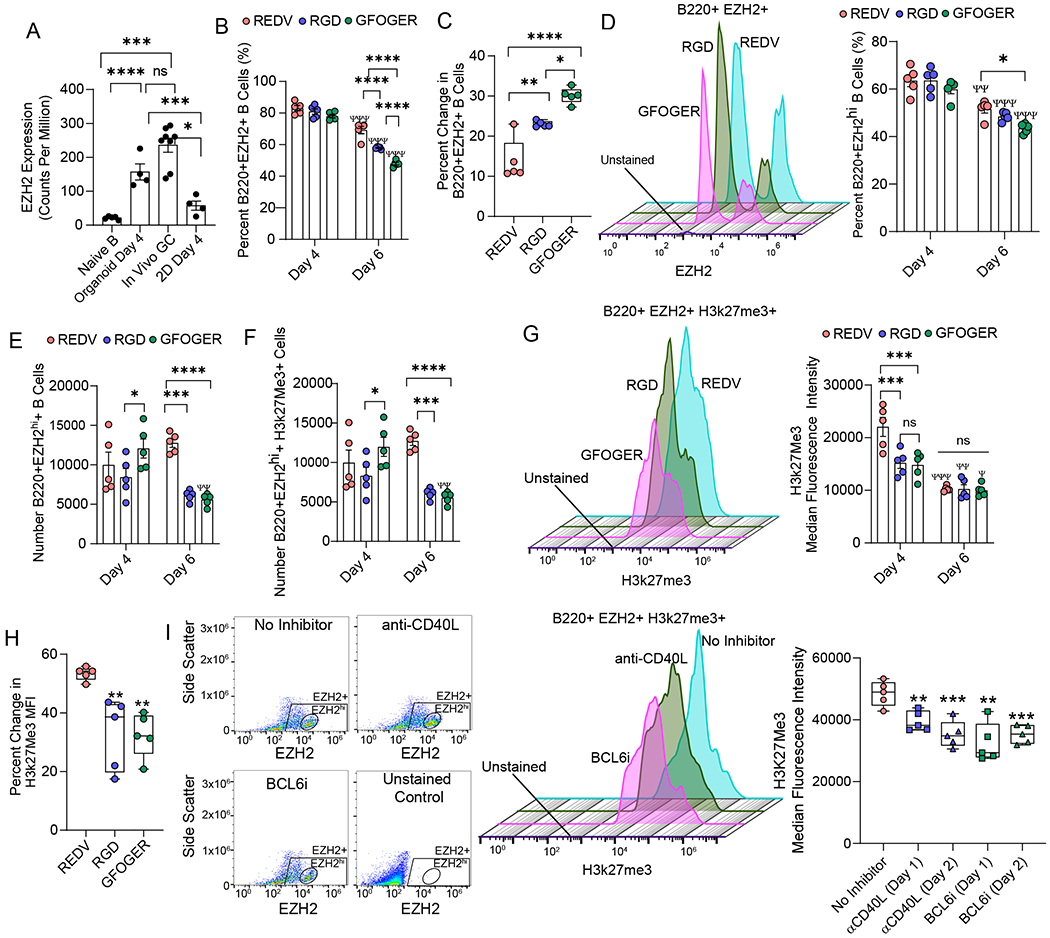
A) EZH2 expression in naïve murine B cells, generation 1, ex vivo 3D organoids made of RGD-rich matrix, in vivo germinal centers in immunized mice, and ex vivo 2D co-cultures. B) Effect of integrin-binding peptides on the percentage of B220+EZH2+ B cells over time in organoid grown cultures of naïve B cells with 40LB and IL-4. C) Change in B220+EZH2+ B cells between days 4 and 6. D-E) Effect of integrin-binding peptides on D) percentage and E) the number of B220+ B cells with high EZH2 expression. Left side flow cytometry histogram in D) indicates the expression of EZH2 on Day 6. F-G) Effect of integrin-binding peptides on histone methylation in terms of F) the number of positive cells and G) median fluorescence intensity. Left side flow cytometry histogram in G) indicates the expression of H3k27Me3 on Day 6. H) Percent change in H3k27 methylation over time in response to varying integrin ligand. I) Effect of blocking CD40L and BCL6 on days 1 or 2 of ex vivo organoid culture on H3k27 methylation. Left side flow cytometry scatter plot and histogram indicates EZH2 populations and expression of H3k27Me3. Data represent mean ± S.E.M. Significance determined via one- or two-way ANOVA with Tukey’s posthoc multiple comparisons test; N=5. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; Ψp<0.05, ΨΨp<0.01, ΨΨΨp<0.001, ΨΨΨΨp<0.0001 relative to day 4 for corresponding ligand.
We observed no significant difference in B220+EZH2+ B cells on day 4 across the matrices, however, the EZH2+ cells decreased in a matrix dependent manner by day 6 (Figure 4B). Although the decrease in all three matrices was significant compared to day 4, REDV only reduced the cells by 14% and RGD by 24%, as compared to a 30% reduction in EZH2 in GFOGER-organoids (Figure 4C). Notably, ~60–65% of the B220+ B cells were positive for EZH2hi on day 4. Percent and number of B220+EZH2hi cells followed a similar trend with matrices and temporal response, and the number of cells did not change for REDV and RGD over 6 days but only for GFOGER-functionalized organoids, as determined by 2-way ANOVA (Figure 4D, E). Similarly, the number of B220+EZH2+H3k27Me3+ cells did not change for REDV and RGD but only for GFOGER-functionalized organoids. Nearly all EZH2hi+ cells were also positive for H3k27Me3 (Figure 4F, Supplementary Fig. S4), which is expected as these cells express the highest levels of EZH2 within the cell population and EZH2 catalyzes H3k27me321. Next, we evaluated the expression of H3k27Me3, as it represents the methyltransferase activity of EZH2. On day 4, VCAM-1 mimicking REDV-organoids expressed the highest levels of H3k27Me3 (Figure 4G). In contrast, differences between ECMs were insignificant on day 6. The drop in H3k27Me3 expression by day 6 was significant across all matrices with a ~53% drop in REDV compared to a significantly lower 32% drop in GFOGER and RGD-organoids (Figure 4H).
Finally, we sought to determine whether the change in H3k27Me3 expression was regulated by the presence of T cell signal CD40L and BCL6. To understand this, we inhibited the CD40-CD40L interaction by initiating the germinal center response and then supplementing the organoid media with 50 μg/mL mouse anti-CD40L antibody on day 1 or day 2. Likewise, we inhibited BCL6 expression on day 1 or day 2 of culture by the addition of 50 μM BCL6 inhibitor to the media. After inhibition, on day 4, we observed a significant decrease in the expression of H3k27Me3 (Figure 4I), clearly indicating that the H3k27Me3 was regulated by the induction of the germinal center response in organoids.
2.5. Effect of bacterial antigen recognition on germinal center B cell differentiation in young mice
The rise of antibiotic resistance among clinically relevant bacteria is one of the most prominent public health and economic challenges70, with ~2.4 million people in Europe, North America, and Australia predicted to die from infections with resistant microorganisms in the next 30 years70, 71. Klebsiella pneumoniae, in particular, is one of the most frequent causes of nosocomial infections that causes many severe diseases including pneumonia, bacteremia, and urinary tract infection, and some recent isolates have developed pan-drug resistance72, 73. Klebsiella species have been identified as the third leading cause of hospital-acquired infections in the United States (9.9%) behind Clostridium difficile ((12.1%) and Staphylococcus aureus (10.7%)73. Among those at risk are infants (who have immature immune systems) and the elderly (who have waning immune defenses); both are subject to heightened incidence and severity of infections72, 74. Carbapenem-resistant K. pneumoniae infections have an approximate 42–48% mortality rate75, 76, and require intensive antibiotic cocktails of last-resort antibiotics with toxic side effects. With options for post-exposure antibiotic treatment dwindling, there is an urgent need for alternative therapies against antibiotic-resistant bacteria, such as K. pneumoniae, to prevent infection and treat infected patients. The external leaflet of the outer membrane of K. pneumoniae is formed by lipopolysaccharide (LPS), a glycolipid composed of lipid A, core oligosaccharide, and O-antigen that serves as a defense barrier in bacteria77. Recently, O-antigen-specific monoclonal antibodies against K. pneumoniae surface polysaccharides have been characterized as pre-emptive therapy4 but are still limited to a narrow spectrum of serotypes, necessitating the need to develop advanced therapeutics.
As a classic T cell-independent antigen, K. pneumoniae’s glycan antigens fail to induce strong germinal center reactions to generate and select high-affinity antibody variants by affinity maturation. In contrast, T cell-dependent responses are typically directed against protein antigens and although K. pneumoniae harbors several surface proteins that could potentially serve as germinal center inducing agents, these are not well studied in the context of humoral immunity. A potential solution is to identify antigens and induce ex vivo germinal center responses to generate antibody-secreting cells and subsequently, antibodies against these antigens. However, it remains unclear whether such antigenic proteins from Klebsiella would need to be presented as a soluble antigen or embedded within the bacterial membrane.
Here, as a proof of concept, we attempted to understand the latter question by testing the ability of K. pneumoniae membrane-embedded OmpA protein (KlebOM) versus soluble, purified OmpA protein (sOmpA) in inducing ex vivo germinal center responses in PEG-4MAL organoids (Figure 5A). The use of immune organoids allows us to bypass the need for these antigens to induce CD40L expression and differentiation of TFH cells in vivo, and yet test the hypothesis that membrane-embedded protein antigens may induce stronger or weaker germinal center responses than soluble antigens (both antigen forms are expected to bind to the B cell receptor). Importantly, immune organoids provide a useful platform to potentially generate ex vivo antibodies against strains of bacteria. This proof of concept study focuses on the former question to optimize the mode of presentation in immune organoids.
Figure 5: Mode of bacterial antigen presentation from a clinical isolate of Klebsiella pneumoniae determines the fate of germinal center B cell differentiation in young mice.
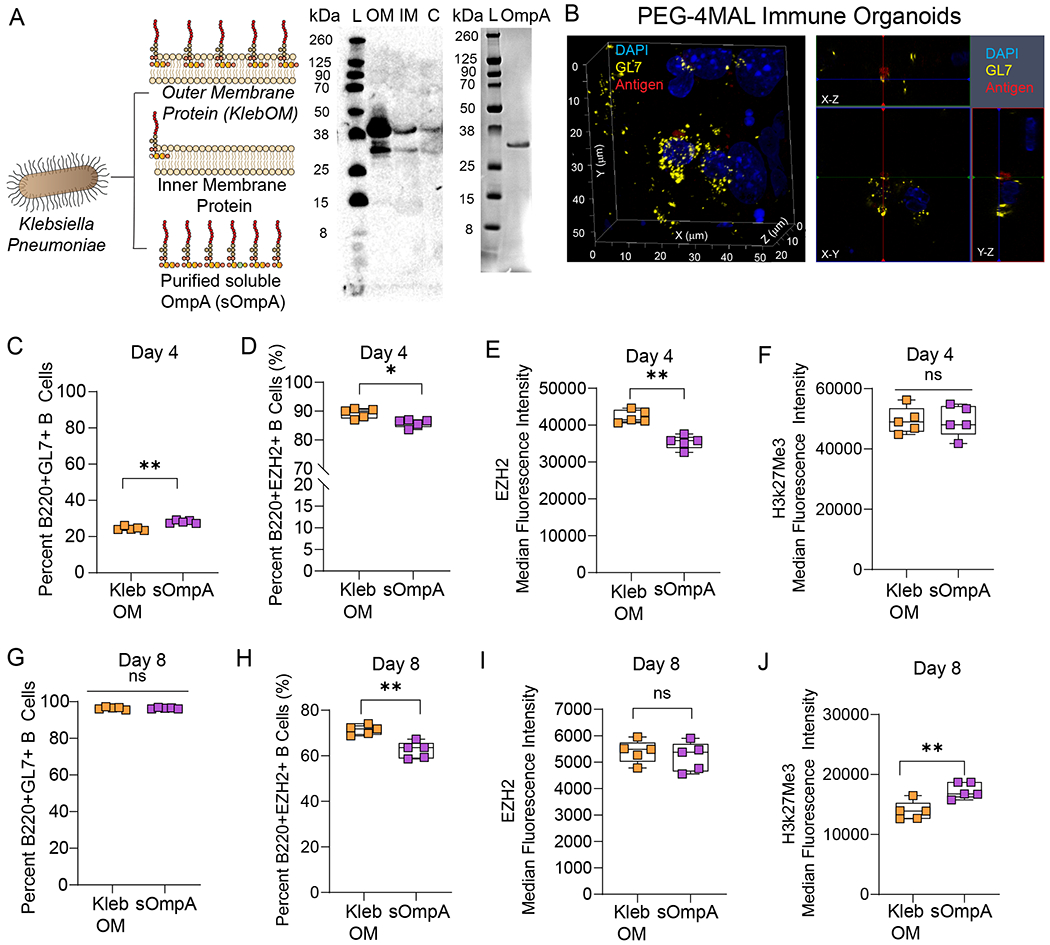
A) Schematic of bacterial outer membrane-bound antigen (KlebOM) derived from Klebsiella pneumoniae and soluble OmpA (sOmpA) protein extracted from the outer membrane. Western blot showing the band of an outer membrane protein in the total outer membrane fraction, identified using an anti-OmpA antibody. Adjacent western blot image shows the band of purified OmpA protein. L: loading control, OM: outer membrane, IM: inner membrane, C: cytosolic fraction of K. pneumoniae prep. B) Confocal microscopy image of localization of antigen NP-OVA (red) relative to germinal center-like B cell (GL7) in PEG-4MAL organoids (Left). Organoids acquire a hemispherical shape with ~4500 µm diameter and the image represent B cell cluster at the bottom of the hemisphere ~50 µm above the imaging coverslip. Confocal acquisitions with orthogonal projections indicating that the antigen colocalizes with the GL7 on the surface of B cells differentiated for 4 days in the presence of 40LB and IL-4. Antigen was added immediately after hydrogel fabrication and organoids were treated for 4 days with the antigen. Yellow: GL7 (false color from green), Blue: DAPI, Red: NP-OVA. Image representative of 3 organoids, scanned at 3-5 depths per organoid. C-F) B cell response to membrane-embedded and soluble OmpA antigen in terms of C) percent B220+GL7+ cells, D) percent B220+EZH2+ cells, E) intensity of EZH2 expression, and F) intensity of histone methylation on day 4 of ex vivo PEG-4MAL organoid culture with 40LB and IL-4. G-J) B cell response to membrane-bound and soluble antigen in terms of G) percent B220+GL7+ cells, H) percent B220+EZH2+ cells, I) intensity of EZH2 expression, and J) intensity of histone methylation on day 8 of ex vivo organoid culture. Data represent mean ± S.E.M. Significance determined via unpaired, non-parametric Mann Whitney t-test; N=5. *p<0.05, **p<0.01, ns denotes no significance.
To determine the depth of penetration of antigen in the organoids, we performed confocal imaging on PEG-4MAL organoids treated with fluorescently tagged 4-Hydroxy-3-nitrophenylacetyl hapten antigen conjugated to ovalbumin protein (NP-OVA). The molecular weight of NP-OVA is ~45 kDa, as compared to 35–50 kDa KelbOM and soluble OmpA based on the western blot (Figure 5A). The antigen transported to the core of organoids and colocalized with GL7 expressing germinal center-like B cell surface (Figure 5B). These studies are further supported by our previous report that demonstrated successful transport and interactions of antigen and BCR crosslinking anti-IgM antibodies with B cells in the organoids, leading to enhanced phosphorylation signaling25.
To determine the effect of Klebsiella antigen, we supplemented the media for each organoid with either sOmpA or KlebOM (3.2 µg); the dose of KlebOM was chosen to match, as closely as possible, the dose of highly pure sOmpA that could be isolated in a single batch. The soluble protein antigen, sOmpA, of K. pneumoniae induced modestly higher B220+GL7+ B cells than membrane-embedded OmpA, KlebOM, which also contains other membrane-bound components such as LPS (Figure 5C). Notably, the overall percentage of B220+GL7+ B cells were markedly low (~20–30%), which is possibly attributable to the antigen-mediated BCR crosslinking. It is known that soluble antigen can cause enhanced apoptosis of germinal center B cells78. Notably, both antigen formats are considered soluble in the organoids because they are not presented by follicular dendritic cells and simply added to the cell culture media. Nearly 90% of these B220+GL7+ B cells expressed EZH2 with significantly higher EZH2 expressed by cells exposed to KlebOM than sOmpA (Figure 5D, E). Interestingly, the differences did not lead to a change in the expression of H3k27Me3 by day 4 (Figure 5F). In contrast to day 4, by day 8, >90% of cells became B220+GL7+, suggesting a delayed induction of germinal centers (Figure 5G). We chose day 8 to allow a sufficient number of germinal center-like B cells in organoids. The percentage of B220+GL7+EZH2+ cells, however, declined to 60–75% (Figure 5H) and the differences in EZH2 expression became indistinguishable (Figure 5I). More importantly, the expression of H3k27Me3 by day 8 increased in organoids exposed to sOmpA (Figure 5J). These findings suggest that at least ex vivo, histone methylation is more robust with sOmpA than membrane-embedded antigen, highlighting the potential for future purified protein antigens without added LPS components. It is expected that more robust histone methylation would lead to increased output of antibody-producing cells and, subsequently, increased antibody production. This will be tested in future work using our previously established anti-Fas methodology25 and sOmpA to determine our ability to generate antigen-specific antibody formation.
2.6. Organoids predict the efficacy of bacterial antigen recognition on germinal center B cell differentiation in aged mice
We next sought to investigate the efficacy of KlebOM and sOmpA on germinal center B cell differentiation in aged mice and determine whether antigen mediated immune response can be generated in B cells from aged individuals, who are at increased risk of K. pneumoniae infections72. Aging mice display many similar features of human immune aging - including reduced humoral immune responses to vaccination, which has been attributed to a defective TFH cell function in germinal centers79 and immunosenescence of B cells including reduced expression of genes crucial to differentiation and somatic hypermutation80. However, the underlying immunological mechanisms remain unclear, and therefore a more reliable, higher throughput approach to generate antibodies is using organoids. We tested the ability of organoids to induce germinal center B cell responses in primary B cells derived from aged mice. Remarkably, we saw a reduction in the number of B220+GL7+ B cells, expression of EZH2, and the number of B220+GL7+H3kme273+ B cells in organoids without any antigen simply under the effect of 40LBs and IL-4 (Figure 6A–C). An important observation in this study is that when aged B cells are provided similar levels of CD40L (i.e. TFH signal), they still do not get activated and proliferate as much as B cells from young mice. Nevertheless, the organoids generated a sufficient number of germinal center B cells to perform a comparison between KlebOM and sOmpA in an aged setting. While both antigen forms led to a similar percentage of GL7+ B cells (Figure 6D), the proportion of B220+EZH2+ and H3k27Me3+ B cells was lower in sOmpA conditions (Figure 6E, F). Nevertheless, the expression of H3k27Me3 in B220+EZH2+H3k27Me3+ B cells was higher in conditions with sOmpA (Figure 6G) and correlated with a concomitant increase in BCL6 (Figure 6H). Therefore, it is evident that by using organoids, we can generate germinal center-like B cells in aged mice, that are initially low in numbers but follow a trajectory similar to young B cells in predicting the response of sOmpA versus KlebOM.
Figure 6: PEG-4MAL organoids induce a germinal center reaction in aged B cells and predict the immunogenic performance of Klebsiella pneumoniae protein antigen.
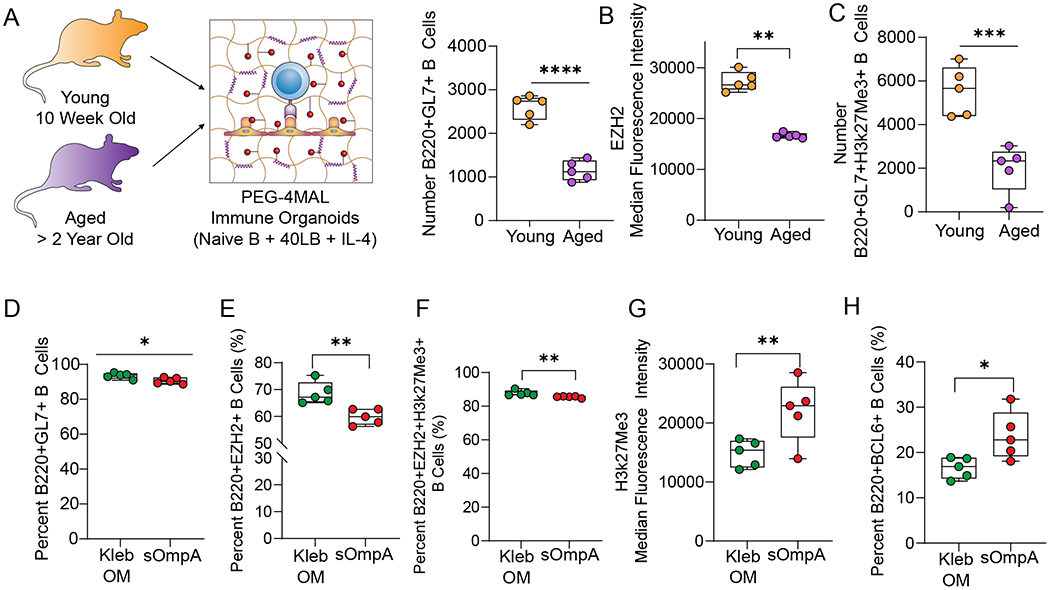
A-C) B cell response in the absence of an antigen in terms of A) number B220+GL7+ cells, B) intensity of EZH2 expression and C) the number of B220+EZH2+H3k27Me3+ B cells on day 4 of ex vivo PEG-4MAL organoid culture with 40LB and IL-4. D-H) Response to membrane-bound and soluble antigen in terms of D) percent B220+GL7+ cells, E) percent B220+EZH2+ cells, F) percent B220+EZH2+H3k27me3+ cells, G) intensity of histone methylation, and H) percent B220+BCL6+ cells on day 8 of ex vivo organoid culture using B cells derived from aged mice. Data represent mean ± S.E.M. Significance determined via unpaired, non-parametric Mann Whitney t-test; N=5. *p<0.05, **p<0.01, ns denotes no significance.
The current study used murine cells to reproducibly establish B cell dynamics in young and aged B cells ex vivo, with control over the age and minimized variability that may occur in human donor cells during the proof-of-concept stage of these studies. The murine findings can further be validated in vivo through OmpA vaccination before translating to human studies. Nevertheless, these findings can be translatable to human B cell dynamics and we are in the process of developing a human-based system81. The human studies could be guided by integrin expression as reported recently82. Finally, antibodies that are obtained through an ex vivo germinal center system can be amplified for large scale production using previously reported yeast or phage display technologies. However, there are known differences between the human and mouse immune systems83, such as the frequency of B cell subsets in different tissue compartments, that may need to be considered while developing human germinal center organoids to produce human antibodies, a work currently in progress84. When evaluating other antigens, factors such as the charge of the protein, size, and hydrophobicity of the membrane could limit diffusion of the protein into the organoid and will need to be evaluated.
3. Conclusion
This work brings together several innovative concepts, ranging from the effect of polymer end-chemistry on primary immune cell survival and differentiation, single-cell, and bulk RNA sequencing to define lymph node environment factors and the effect of these factors on the temporal kinetics of B cell differentiation. The work is the first investigation of the mode of antigen formulation from a clinical isolate of antibiotic-resistant Klebsiella pneumoniae on the temporal kinetics of ex vivo B cell differentiation in young and aged mice, and, to the best of our knowledge, the first demonstration of ex vivo B cell differentiation in polymeric organoids from primary immune cells derived from > 2-year-old aged mice. Ex vivo organoid studies offer a unique opportunity to identify protein antigens and conditions that may induce a more uniform response across both young and aged populations. This platform also offers an opportunity to derive age-specific germinal center-like B cells, and subsequently antibody-secreting cells and antibodies, which can be injected into patients as therapeutics. This aspect may avoid a vaccine development that still requires induction of germinal centers in vivo and could, in the long term, provide alternative solutions to overcoming antibiotic-resistant and regular bacterial infections. Future work will focus on determining whether the two antigen forms can induce distinct or comparable somatic hypermutation and affinity maturation, generate protective antibodies against K. pneumoniae and be useful in aged mice with a defective TFH cell function. Although polysaccharide-protein conjugate vaccines have been designed to facilitate the necessary T cell support for germinal center formation, the ensuing immune response is against both the carrier and antigen. An ex vivo immune technology where T cell support can be provided without prior activation of B cell opens the possibility for studying how B cells respond to only the antigen without confounding effects from the carrier protein. If in some instances polysaccharide antigens still do not activate B cells, an ex vivo technology can also elucidate the effect of various carriers, location of polysaccharide on the protein carrier, and valency of the polysaccharide to better design new conjugate vaccines.
4. Materials and Methods
Biomaterials and peptides
PEG-4MAL with 20 kDa molecular weight and ≥90% purity and PEG-4ACR with 21 kDa and >97% were purchased from Laysan Bio. PEG-4VS with 21 kDa molecular weight and >90% purity was purchased from Sigma. Integrin αvβ3-binding RGD peptide (GRGDSPC, >90% purity), scrambled peptide (GRDGSPC, >90% purity), integrin α4β1-binding REDV peptide (GREDVGC, >90% purity), GFOGER peptide (GYGGGPGPPGPPGPPGPPGPPGFOGERGPPGPPGPP GPPGPPGPC, ≥90% purity), and matrix metalloproteinase (MMP)-9 degradable VPM peptide (GCRDVPMSMRGGDRCG, >90% purity) were purchased from Aapptec. Non-degradable dithiothreitol (DTT) crosslinker was purchased from Sigma Aldrich. All components were reconstituted in 0.01 M HEPES buffer with pH 7.4.
Stromal cell culture and mitomycin-c treatment
40LB stromal cells, genetically modified from NIH/3T3 fibroblasts to express CD40 ligand and produce B cell activating factor, as described previously21–23, 25, 57, 85, were obtained from Dr. Daisuke Kitamura22. 40LB cells were cultured in Dulbecco’s Modified Eagle Medium containing 10% fetal bovine serum and 1% penicillin-streptomycin (P/S) and routinely passaged at 90% confluency. Before encapsulation in organoids, 40LB were mitotically inhibited via incubation with 0.01 mg/mL mitomycin-C at 37°C for 45 min.
Naïve B cell isolation from mice
For experiments involving wildtype (WT) B cells, spleens were harvested from female C57BL/6 mice, aged 8-15 weeks, from the Jackson Laboratory. Where indicated, mice aged >2 years were used to compare the effects of aging. Spleens were dissociated using a sterile plunger, as previously described 57, and the cell suspension was incubated in RBC lysis buffer for 5 min at room temperature to remove red blood cells. Naïve B cells were purified from the splenocyte suspension by negative isolation using an EasySep Mouse B Cell Isolation Kit from Stem Cell Technologies, according to the manufacturer’s instructions. All animals were handled in compliance with the procedures approved by the Institutional Animal Care and Use Committee (IACUC) at Cornell University and the University of California, San Francisco.
Organoid fabrication
Synthetic immune organoids containing 7.5% w/v of PEG-4MAL or PEG-4VS or PEG-4ACR were fabricated using the corresponding PEG macromer, adhesives peptides, and crosslinkers. PEG-4MAL was functionalized at pH 7.4 with thiolated adhesive peptides RGD, REDV, or GFOGER at a 4:0.3 MAL-to-peptide molar ratio, and RDG scramble at a 4:0.7 MAL-to-peptide molar ratio for 30 min at 37°C. MMP9-degradable VPM peptide and non-degradable DTT crosslinkers were combined at a 1:1 VPM-to-DTT molar ratio and a 4:1.5 MAL-to-crosslinker molar ratio and adjusted to pH 5-6. 40,000 Naïve B cells and 40,000 40LB stromal cells were suspended in the crosslinker solution, and 5 µL of crosslinker-cell suspension was injected into 5 µL functionalized PEG-MAL in each well of a non-treated 96 well plate. The droplet was mixed rapidly via pipet and cured at 37°C for 15 min. Post-incubation, Roswell Park Memorial Institute (RPMI) 1640 media containing 10% FBS, 1% P/S, and 10 ng/mL interleukin-4 (IL-4, Peprotech) was added to each organoid. The media was replenished every 3 days.
Inhibition of CD40L and BCL6
Where indicated, organoid media was replenished on days 1 or 2 with 50 μg/mL mouse anti-CD40L (Clone MR-1, Bio X Cell #BE0017-1), 50 μM BCL6 inhibitor 79-6 (Sigma #197345), or 10 ng/mL IL-4 alone. Changes in germinal center-like B cell dynamics were assessed on day 4 of organoid culture using flow cytometry.
Enrichment of K. pneumoniae outer membranes (KlebOM)
Outer membranes were enriched from a clinical Klebsiella pneumoniae stool isolate, designated at B308-2, provided and profiled for antibiotic resistance by Michael Satlin at Weill Cornell Medicine. This isolate is here referred to as KpB308-2. KpB308-2 was inoculated in 500mL Nutrient Broth with 100ug/mL ampicillin and incubated overnight at 37°C, shaking 220 rpm. Following incubation, cells were pelleted at 5,000xg for 40 minutes at 4°C and resuspended in 36 mL lysis buffer (20mM Tris Buffer pH 8.0, 750mM Sucrose,) with 10 mg/mL egg white lysozyme and incubated on ice shaking 180 rpm for 1 hour. Ice cold 1.5mM EDTA was slowly poured into the lysozyme-treated cells and a 1x protease inhibitor was added (Roche cat no. 04693159001). The suspensions were sonicated in an ice/water slurry for eight minutes total with the following parameters: 20 seconds on/20 seconds off, 70% amplitude. 1mM PMSF was added to the lysates, which were then spun at 1,500xg for 15 minutes at 4°C to remove large debris and unlysed cells. The supernatant was collected and spun at 100,000xg for 1 hour, 4°C to pellet membranes. The supernatant containing the soluble cytosolic fraction was removed and saved for analysis, and the pelleted membranes were resuspended in 2mL membrane wash buffer (20mM Tris Buffer pH 8.0, 300mM NaCl, 1x protease inhibitor, 1mM fresh PMSF) passaged through an 18g needle, washed in an additional 10mL supplemented wash buffer, and spun again at 100,00xg, 1 hour, 4°C. Total membrane pellets were resuspended via an 18g needle in 1% N-Lauroylsarcosine (Sigma #L5125) in supplemented wash buffer and incubated rolling at room temperature for 1 hour to selectively solubilize inner membranes 86. The remaining insoluble outer membrane fraction was pelleted at 100,000xg for 1 hour, 4°C. The supernatant containing solubilized inner membranes was removed and saved for analysis, and the enriched outer membrane pellet was resuspended once more in 10 mL supplemented membrane wash buffer, and pelleted a final time at 100,000xg, 4°C, for 1 hour. Finally, the enriched outer membrane pellet was resuspended in 3 mL of 20mM Tris-HCl, pH 8.0, 150mM NaCl, passed through a 0.22uM filter, and flash-frozen until further use. Total protein concentration was measured using a Qubit 2. Approximately 3.2 μg KlebOM were supplemented to the media for each organoid, this dose was chosen to match, as closely as possible, the amount of sOmpA tested.
Western Blotting
To identify enrichment of KpB308-2 outer membranes, western blot was against the outer membrane marker Outer Membrane Protein A (OmpA). 15ug of total protein was mixed with 1X NuPage LDS sample buffer (NP0007) and 2.5% beta-mercaptoethanol, heated at 95°C for 5 minutes, then loaded into a 4-12% NuPage Bis-Tris gel. Protein was transferred to a nitrocellulose membrane using an iBlot 2 and blocked in 5% Skim milk powder in TBST for 1 hour at room temperature. The membrane was washed 3 times with TBST between each incubation. Membranes were incubated with a 1:5000 dilution of rabbit-anti-Klebsiella OmpA antibody (Antibody Research Corporation #111226) for 1 hour at room temperature, followed by incubation in a 1:10,000 dilution of HRP-conjugated goat-anti-rabbit secondary antibody (Thermo Fisher #65-6120). Blots were treated with Pierce ECL Western Blotting Substrate (Thermo Fisher #32106) and chemiluminescence was detected using the Azure Biosystems c300 Imaging system.
Purification of K. pneumoniae Outer membrane protein A (sOmpA)
K. pneumoniae Outer membrane protein A (OmpA) was PCR amplified from genomic DNA from the clinical isolate B308-2 using the following primers: 5’ GATATACATATGAAAAAGACAGCTATCGCG 3’ and 5’ GTGGTGCTCGAGAGCCGCTGGCTGAGTTAC 3’ for insertion into the vector pET21b containing a C-terminal hexahistidine tag using the NdeI and XhoI restriction sites. Insertion was confirmed via Sanger sequencing using the primers: 5’ TAATACGACTCACTATAGGG 3’, 5’ GCTAGTTATTGCTCAGCGG 3’. The KpOmpA_pET21b construct was transformed into BL21 Star DE3 and plated on LB-Amp for overnight incubation at 37°C. Colonies were then scraped into 10 mL Terrific Broth with 100μg/mL ampicillin and grown for 2 hours before inoculating 1L Terrific Broth with 100μg/mL ampicillin. 1L cultures were grown shaking at 37°C to OD600 = 0.7 before adding 0.5mM IPTG to induce protein expression. Following IPTG addition, cultures were moved to shaking at 24°C overnight. After overnight incubation, cultures were pelleted 5,000xg, 40 minutes, 4°C and resuspended in membrane wash buffer (20mM Tris Buffer pH 8.0, 300mM NaCl) and pelleted again. To lyse cells, the pellet was resuspended in 50 mL membrane wash buffer supplemented with 1X protease inhibitor (Roche cat no. 04693159001) and 10mg/mL lysozyme and let shake on ice for 30 minutes before sonication at 70% amplitude, 20 seconds on/ 20 seconds off for a total of 8 minutes. 1mM PMSF and 5mM Beta- mercaptoethanol were added to the lysates following sonication. Unlysed cells were removed via centrifugation at 1,500xg for 10 minutes, 4°C. Cleared lysates were spun at 100,000xg, 4°C for 1 hour, washed once with membrane wash buffer supplemented with 1x protease inhibitor and 1mM fresh PMSF and spun again with the same settings. The pelleted total membrane fraction was resuspended once more in 3mL supplemented membrane wash buffer. To solubilize membranes and isolate protein, the resuspended membranes were mixed with 5% of 3:1 Styrene-maleic acid copolymer (3:1 SMA) in wash buffer and solubilized rolling at room temperature for two hours to allow the formation of membrane nanodiscs 87. Following incubation, insoluble material was pelleted at 10,000xg, 4°C, for 10 minutes and the nanodisc-solubilized membranes were mixed with 1mL of Qiagen Ni-NTA Superflow resin equilibrated with protein wash buffer (20mM Tris Buffer, pH 8.0, 500mM NaCl, 20mM Imidazole, 1x protease inhibitor, 10mM PMSF) was added to the cleared lysate and incubated rolling at room temperature for 1 hour to allow for protein binding. Lysates were loaded onto the column and washed with 40 resin volumes protein wash buffer before elution with 20mM Tris Buffer, 500mM EDTA, and 400mM Imidazole. Following elution, proteins were buffer exchanged and concentrated into 20mM Tris Buffer pH 8.0, 150mM NaCl using an Amicon 10 MWCO spin filter, passed through a 0.22uM filter to sterilize, and flash-frozen until further use. 10 μL of purified protein was run on a 4-12% NuPage Bis-Tris gel with WesternSure Chemiluminescent protein ladder (Li-Cor 926-98000) and stained for total protein with SimplyBlue Safe Stain to confirm the purity of the sample. Approximately 3.2 μg sOmpA were supplemented to the media for each organoid; this dose was experimentally limited by the amount of highly pure sOmpA that could be isolated and the number of organoids tested in a single study.
Flow cytometry and imaging analysis
On days 4 or 6, organoids were rinsed in 1X PBS and enzymatically digested in 125 U/mL collagenase type 1 (Worthington Biochemical) for 1 h at 37°C. Enzyme activity was terminated by the addition of buffer containing serum, and the cells were filtered to remove organoid debris using 96 well MultiScreen Mesh Filter Plates (EMD Millipore). Cells were resuspended in FACS Buffer (PBS++ with 2% FBS, 1% P/S, and 0.005 mM EDTA) containing antibodies against cell surface antigens and incubated in the dark on ice for 1 h. Post-incubation, the cells were washed and resuspended in FACS Buffer. For intracellular antigens, the cells were fixed for 30 min in Fixation/Permeabilization Buffer (eBioscience Foxp3/Transcription Factor Staining Buffer) and incubated in permeabilization buffer containing antibodies for 1 h on ice, protected from light. Anti-mouse monoclonal antibodies targeting cell surface antigens included anti-GL7 (FITC, PE, eFluor660; clone GL 7; Thermo Fisher), anti-B220 (PE-Cy7; clone RA3-6B2; Thermo Fisher), anti-Fas (APC; clone 15A7; Thermo Fisher), anti-CD19 (FITC; clone 1D3; Thermo Fisher), anti-CD138 (FITC; clone 300506; Thermo Fisher), anti-IgD (PE; clone 11-26C; Thermo Fisher), and anti-CD38 (APC; clone 90; Thermo Fisher). Anti-mouse monoclonal antibodies targeting intracellular antigens included anti-H3k27me3 (AF488, AF647; clone C36B11; Cell Signaling), anti-EZH2 (eFluor 660; clone AC22; Thermo Fisher), anti-CBP (FITC, AF488; clone C-1; Santa Cruz), anti-H3k27ac (AF647; clone D5E4; Cell Signaling), anti-IRF4 (FITC, PE, PE-Cy7; clone 3E4; Thermo Fisher), and anti-BCL6 (PE, APC; clone BCL-DWN; Thermo Fisher).
For imaging, organoids were cultured in glass-bottom 35 mm dishes. After 4 days in culture, organoids were fixed in 4% paraformaldehyde in PBS for 15 minutes, permeabilized with 0.5% Triton-X-100 in PBS for 30 min, and blocked with normal 20% goat serum for 30 min. Organoids were then incubated with fluorescence-conjugated primary antibodies (anti-GL7-AF488, Thermo Fisher, 1:200 dilution; anti-NP-AF647, Novus Biologicals, 1:200 dilution) for 1 hour. After washing with PBS, organoids were incubated with actin stain (phalloidin-AF568, Thermo Fisher) and nuclear stain (DAPI) for 1 hour. Organoids were washed and resuspended in PBS. The organoids were imaged on a Zeiss LSM710 confocal microscope.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software. Data analysis used an unpaired two-tailed t-test or one-way analysis of variance (ANOVA) with Tukey’s post hoc test or two-way ANOVA with Sidak’s multiple comparison test. Quantitative analyses as scatter or bar graphs are presented as means ± SEM. In all studies, *P < 0.05, **P < 0.01, and ***P < 0.001 unless otherwise stated. Non-significance is denoted by “ns.”
Supplementary Material
Acknowledgments
The authors acknowledge financial support from the National Institute of Allergy and Infectious Diseases of the US National Institutes of Health (5R01AI132738-03 awarded to A.S.), a US National Science Foundation CAREER award (DMR-1554275 awarded to A.S.), and the Innovative Molecular Analysis Technology program of the US National Cancer Institute (NIH R33-CA212968-01 awarded to A.S.). The authors acknowledge financial support from the Immunoengineering T32 training grant to S.P. (NIH and NIBIB, 1T32EB023860-01A1) and NIH 1S10RR025502 funding for the shared Zeiss LSM 710 Confocal Microscope, awarded to the Cornell University Biotechnology Resource Center. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the funding agency.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or the author.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Single-cell RNA sequence data have been deposited by Jason Cyster18 in GEO under ID code GSE112903. Bulk RNA sequencing on germinal center B cells and naïve B cells have been deposited in GEO with the primary accession codes GSE95491, as reported by us21.
References
- 1.Guilbert JJ The World Health Report 2006: working together for health. Educ Health (Abingdon) 19, 385–387 (2006). [DOI] [PubMed] [Google Scholar]
- 2.CDC. Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2019. https://www.cdc.gov/drugresistance/Biggest-Threats.html. (2019). [Google Scholar]
- 3.McKenna M Antibiotic resistance: the last resort. Nature 499, 394–396 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Pennini ME et al. Immune stealth-driven O2 serotype prevalence and potential for therapeutic antibodies against multidrug resistant Klebsiella pneumoniae. Nat Commun 8, 1991 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rollenske T et al. Cross-specificity of protective human antibodies against Klebsiella pneumoniae LPS O-antigen. Nature immunology 19, 617–624 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Cyster JG Shining a light on germinal center B cells. Cell 143, 503–505 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Allen CD et al. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nature immunology 5, 943–952 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Mesin L, Ersching J & Victora GD Germinal Center B Cell Dynamics. Immunity 45, 471–482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witzel II et al. Deconstructing Immune Microenvironments of Lymphoid Tissues for Reverse Engineering. Adv Healthc Mater 8, e1801126 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Allen CD, Okada T & Cyster JG Germinal-center organization and cellular dynamics. Immunity 27, 190–202 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen CD, Okada T, Tang HL & Cyster JG Imaging of germinal center selection events during affinity maturation. Science 315, 528–531 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Tas JM et al. Visualizing antibody affinity maturation in germinal centers. Science 351, 1048–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mintz MA et al. The HVEM-BTLA Axis Restrains T Cell Help to Germinal Center B Cells and Functions as a Cell-Extrinsic Suppressor in Lymphomagenesis. Immunity 51, 310–323 e317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phan TG, Grigorova I, Okada T & Cyster JG Subcapsular encounter and complement-dependent transport of immune complexes by lymph node B cells. Nature immunology 8, 992–1000 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Rodda LB, Bannard O & Cyster JG Integrin-mediated interactions between B cells and follicular dendritic cells influence germinal center B cell fitness. J Immunol 192, 4601–4609 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiemann B et al. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science 293, 2111–2114 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Heesters BA, Myers RC & Carroll MC Follicular dendritic cells: dynamic antigen libraries. Nature Reviews Immunology 14, 495–504 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Rodda LB et al. Single-Cell RNA Sequencing of Lymph Node Stromal Cells Reveals Niche-Associated Heterogeneity. Immunity 48, 1014–1028 e1016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen CD & Cyster JG Follicular dendritic cell networks of primary follicles and germinal centers: phenotype and function. Semin Immunol 20, 14–25 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo M et al. EZH2 Represses the B Cell Transcriptional Program and Regulates Antibody-Secreting Cell Metabolism and Antibody Production. J Immunol 200, 1039–1052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beguelin W et al. EZH2 enables germinal centre formation through epigenetic silencing of CDKN1A and an Rb-E2F1 feedback loop. Nat Commun 8, 877 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nojima T et al. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun 2, 465 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Purwada A et al. Ex vivo engineered immune organoids for controlled germinal center reactions. Biomaterials 63, 24–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purwada A & Singh A Immuno-engineered organoids for regulating the kinetics of B-cell development and antibody production. Nat Protoc 12, 168–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purwada A et al. Ex vivo synthetic immune tissues with T cell signals for differentiating antigen-specific, high affinity germinal center B cells. Biomaterials 198, 27–36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee M, Mehr R, Belelovsky A, Spencer J & Dunn-Walters DK Age- and tissue-specific differences in human germinal center B cell selection revealed by analysis of IgVH gene hypermutation and lineage trees. Eur J Immunol 32, 1947–1957 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Stedra J & Cerny J Relative contribution of T and B cells to hypermutation and selection of the antibody repertoire in germinal centers of aged mice. J Exp Med 183, 959–970 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blaeser A, McGlauchlen K & Vogel LA Aged B lymphocytes retain their ability to express surface markers but are dysfunctional in their proliferative capability during early activation events. Immun Ageing 5, 15 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dailey RW, Eun SY, Russell CE & Vogel LA B cells of aged mice show decreased expansion in response to antigen, but are normal in effector function. Cell Immunol 214, 99–109 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Siegrist CA & Aspinall R B-cell responses to vaccination at the extremes of age. Nature reviews. Immunology 9, 185–194 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Kim S, Shah SB, Graney P & Singh A Multiscale engineering of immune cells and lymphoid organs. Nature Reviews Materials 4, 355–378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bian L, Guvendiren M, Mauck RL & Burdick JA Hydrogels that mimic developmentally relevant matrix and N-cadherin interactions enhance MSC chondrogenesis. Proceedings of the National Academy of Sciences of the United States of America 110, 10117–10122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKinnon DD, Domaille DW, Cha JN & Anseth KS Biophysically defined and cytocompatible covalently adaptable networks as viscoelastic 3D cell culture systems. Adv Mater 26, 865–872 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grover GN, Lam J, Nguyen TH, Segura T & Maynard HD Biocompatible hydrogels by oxime Click chemistry. Biomacromolecules 13, 3013–3017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boere KWM et al. Biofabrication of reinforced 3D-scaffolds using two-component hydrogels. J Mater Chem B 3, 9067–9078 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith LJ et al. Diels-Alder Click-Cross-Linked Hydrogels with Increased Reactivity Enable 3D Cell Encapsulation. Biomacromolecules 19, 926–935 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Alge DL, Azagarsamy MA, Donohue DF & Anseth KS Synthetically tractable click hydrogels for three-dimensional cell culture formed using tetrazine-norbornene chemistry. Biomacromolecules 14, 949–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin XH, Wang X, Rottmar M, Nelson BJ & Maniura-Weber K Near-Infrared Light-Sensitive Polyvinyl Alcohol Hydrogel Photoresist for Spatiotemporal Control of Cell-Instructive 3D Microenvironments. Adv Mater 30 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Skardal A et al. A hydrogel bioink toolkit for mimicking native tissue biochemical and mechanical properties in bioprinted tissue constructs. Acta biomaterialia 25, 24–34 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Lee TT et al. Light-triggered in vivo activation of adhesive peptides regulates cell adhesion, inflammation and vascularization of biomaterials. Nat Mater 14, 352–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phelps EA et al. Maleimide cross-linked bioactive PEG hydrogel exhibits improved reaction kinetics and cross-linking for cell encapsulation and in situ delivery. Adv Mater 24, 64–70, 62 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phelps EA, Headen DM, Taylor WR, Thule PM & Garcia AJ Vasculogenic bio-synthetic hydrogel for enhancement of pancreatic islet engraftment and function in type 1 diabetes. Biomaterials 34, 4602–4611 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macdougall LJ, Perez-Madrigal MM, Arno MC & Dove AP Nonswelling Thiol-Yne Cross-Linked Hydrogel Materials as Cytocompatible Soft Tissue Scaffolds. Biomacromolecules 19, 1378–1388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Truong VX, Ablett MP, Richardson SM, Hoyland JA & Dove AP Simultaneous orthogonal dual-click approach to tough, in-situ-forming hydrogels for cell encapsulation. J Am Chem Soc 137, 1618–1622 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Truong VX, Tsang KM & Forsythe JS Nonswelling Click-Cross-Linked Gelatin and PEG Hydrogels with Tunable Properties Using Pluronic Linkers. Biomacromolecules 18, 757–766 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Huynh CT, Liu F, Cheng Y, Coughlin KA & Alsberg E Thiol-Epoxy “Click” Chemistry to Engineer Cytocompatible PEG-Based Hydrogel for siRNA-Mediated Osteogenesis of hMSCs. ACS Appl Mater Interfaces 10, 25936–25942 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paez JI, Farrukh A, Valbuena-Mendoza R, Wlodarczyk-Biegun MK & Del Campo A Thiol-Methylsulfone-Based Hydrogels for 3D Cell Encapsulation. ACS Appl Mater Interfaces 12, 8062–8072 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Rydholm AE, Bowman CN & Anseth KS Degradable thiol-acrylate photopolymers: polymerization and degradation behavior of an in situ forming biomaterial. Biomaterials 26, 4495–4506 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Clark AY et al. Integrin-specific hydrogels modulate transplanted human bone marrow-derived mesenchymal stem cell survival, engraftment, and reparative activities. Nat Commun 11, 114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cruz-Acuna R et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat Cell Biol 19, 1326–1335 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gjorevski N et al. Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Raeber GP, Lutolf MP & Hubbell JA Molecularly engineered PEG hydrogels: a novel model system for proteolytically mediated cell migration. Biophys J 89, 1374–1388 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kraehenbuehl TP et al. Three-dimensional extracellular matrix-directed cardioprogenitor differentiation: systematic modulation of a synthetic cell-responsive PEG-hydrogel. Biomaterials 29, 2757–2766 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Park Y, Lutolf MP, Hubbell JA, Hunziker EB & Wong M Bovine primary chondrocyte culture in synthetic matrix metalloproteinase-sensitive poly(ethylene glycol)-based hydrogels as a scaffold for cartilage repair. Tissue Eng 10, 515–522 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Goldberg MS Improving cancer immunotherapy through nanotechnology. Nat Rev Cancer 19, 587–602 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Polini A et al. Towards the development of human immune-system-on-a-chip platforms. Drug Discov Today 24, 517–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Purwada A & Singh A Immuno-engineered Organoids for Regulating the Kinetics of B cell Development and Antibody Production. Nature Protocols 12, 168–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hao Z et al. Fas receptor expression in germinal-center B cells is essential for T and B lymphocyte homeostasis. Immunity 29, 615–627 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cyster JG B cell follicles and antigen encounters of the third kind. Nature immunology 11, 989–996 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Knight CG et al. Identification in collagen type I of an integrin alpha2 beta1-binding site containing an essential GER sequence. The Journal of biological chemistry 273, 33287–33294 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Zeltz C & Gullberg D The integrin-collagen connection--a glue for tissue repair? Journal of Cell Science 129, 653–664 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Massia SP & Hubbell JA Vascular endothelial cell adhesion and spreading promoted by the peptide REDV of the IIICS region of plasma fibronectin is mediated by integrin alpha 4 beta 1. The Journal of biological chemistry 267, 14019–14026 (1992). [PubMed] [Google Scholar]
- 63.Arnaout MA, Mahalingam B & Xiong JP Integrin structure, allostery, and bidirectional signaling. Annu Rev Cell Dev Biol 21, 381–410 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Bellis SL Advantages of RGD peptides for directing cell association with biomaterials. Biomaterials 32, 4205–4210 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Basso K & Dalla-Favera R Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev 247, 172–183 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Klein U et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nature immunology 7, 773–782 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Li X et al. Cbl Ubiquitin Ligases Control B Cell Exit from the Germinal-Center Reaction. Immunity 48, 530–541 e536 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Raaphorst FM et al. Cutting edge: polycomb gene expression patterns reflect distinct B cell differentiation stages in human germinal centers. J Immunol 164, 1–4 (2000). [DOI] [PubMed] [Google Scholar]
- 69.Béguelin W et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer cell 23, 677–692 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hofer U The cost of antimicrobial resistance. Nat Rev Microbiol 17, 3 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Cassini A et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis 19, 56–66 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Podschun R & Ullmann U Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11, 589–603 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Magill SS et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370, 1198–1208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gorrie CL et al. Antimicrobial-Resistant Klebsiella pneumoniae Carriage and Infection in Specialized Geriatric Care Wards Linked to Acquisition in the Referring Hospital. Clin Infect Dis 67, 161–170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ben-David D et al. Outcome of carbapenem resistant Klebsiella pneumoniae bloodstream infections. Clin Microbiol Infect 18, 54–60 (2012). [DOI] [PubMed] [Google Scholar]
- 76.Xu L, Sun X & Ma X Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob 16, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.March C et al. Klebsiella pneumoniae outer membrane protein A is required to prevent the activation of airway epithelial cells. The Journal of biological chemistry 286, 9956–9967 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pulendran B, Kannourakis G, Nouri S, Smith KG & Nossal GJ Soluble antigen can cause enhanced apoptosis of germinal-centre B cells. Nature 375, 331–334 (1995). [DOI] [PubMed] [Google Scholar]
- 79.Sage PT, Tan CL, Freeman GJ, Haigis M & Sharpe AH Defective TFH Cell Function and Increased TFR Cells Contribute to Defective Antibody Production in Aging. Cell reports 12, 163–171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cancro MP et al. B cells and aging: molecules and mechanisms. Trends Immunol 30, 313–318 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Quizon N, Kwak K, Shah SB, Singh A & Pierce SK Bioengineered organoid models of human germinal centers. J Immunol 200 (1 Supplement) 120.14 (2018). [Google Scholar]
- 82.Kwak K et al. Intrinsic properties of human germinal center B cells set antigen affinity thresholds. Sci Immunol 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benitez A et al. Differences in mouse and human nonmemory B cell pools. J Immunol 192, 4610–4619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Quizon N, Kwak K, Shah S, Singh A & Pierce S Bioengineered organoid models of human germinal centers. The Journal of Immunology 200, 120.114 (2018). [Google Scholar]
- 85.Purwada A, Shah SB, Beguelin W, Melnick A & Singh A Modular Immune Organoids with Integrin Ligand Specificity Differentially Regulate Ex vivo B Cell Activation. ACS Biomater. Sci. Eng 3, 214–225 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Hobb RI, Fields JA, Burns CM & Thompson SA Evaluation of procedures for outer membrane isolation from Campylobacter jejuni. Microbiology 155, 979–988 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee SC et al. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nature Protocols 11, 1149–1162 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


