Abstract
This article aims to introduce the inception and operation of the COVID-19 International Collaborative Research Project, the world’s first coronavirus disease 2019 (COVID-19) open data project for research, along with its dataset and research method, and to discuss relevant considerations for collaborative research using nationwide real-world data (RWD). COVID-19 has spread across the world since early 2020, becoming a serious global health threat to life, safety, and social and economic activities. However, insufficient RWD from patients was available to help clinicians efficiently diagnose and treat patients with COVID-19, or to provide necessary information to the government for policy-making. Countries that saw a rapid surge of infections had to focus on leveraging medical professionals to treat patients, and the circumstances made it even more difficult to promptly use COVID-19 RWD. Against this backdrop, the Health Insurance Review and Assessment Service (HIRA) of Korea decided to open its COVID-19 RWD collected through Korea’s universal health insurance program, under the title of the COVID-19 International Collaborative Research Project. The dataset, consisting of 476 508 claim statements from 234 427 patients (7590 confirmed cases) and 18 691 318 claim statements of the same patients for the previous 3 years, was established and hosted on HIRA’s in-house server. Researchers who applied to participate in the project uploaded analysis code on the platform prepared by HIRA, and HIRA conducted the analysis and provided outcome values. As of November 2020, analyses have been completed for 129 research projects, which have been published or are in the process of being published in prestigious journals.
Keywords: COVID-19, Universal health insurance, Interdisciplinary research, Insurance claim review, Health Insurance Review and Assessment Service, Korea
INTRODUCTION
The advancement of globalization has promoted vigorous exchanges of people and material resources across nations and continents, and coronavirus disease 2019 (COVID-19) was no exception to the trend. Since the first outbreak in Wuhan, China in December 2019, there have been 57 639 671 confirmed cases of COVID-19, including 1 374 284 deaths reported to the World Health Organization as of November 22, 2020. In Korea, as of the same date, there have been 30 733 confirmed cases, including 505 deaths [1]. The spread of this global pandemic is expected to continue for a protracted period as there is no definite cure or vaccine for COVID-19 and many countries are heading towards flu season [2,3].
Providing proper care to patients with COVID-19 is resource-intensive because of the requirement for isolation, while the threat of community infection and potential upsurge of the number of patients are ever present. As such, it is essential to produce real-world data (RWD) and evidence through research in order to treat patients effectively and to improve the efficiency of resource allocation. Unfortunately, and as paradoxical as it may seem, countries severely affected by COVID-19 have to prioritize patient care above everything else, and it is often difficult to collect and process treatment data for clinical evidence production and policy-making.
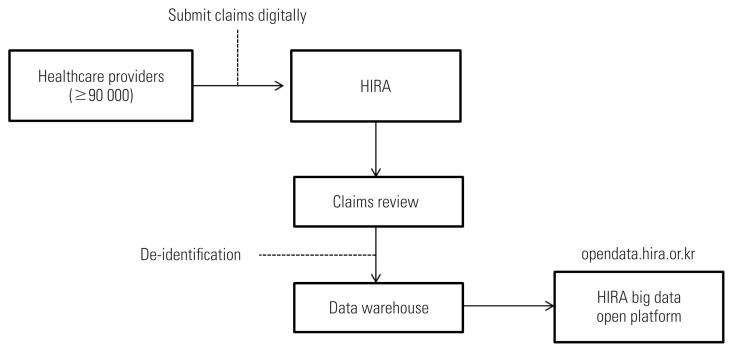
Korea has successfully established and operated universal health coverage covering the entire population and laid the ground of information technology (IT) infrastructure to collect and utilize RWD. The National Health Insurance (NHI) system of Korea is a type of social insurance program, where all Koreans and all healthcare service providers must subscribe to NHI upon their birth or inception under the National Health Insurance Act. NHI covers medically necessary services, and the Health Insurance Review and Assessment Service (HIRA) is in charge of claims review for reimbursement. To receive NHI reimbursement, healthcare service providers must submit e-claims to the HIRA web portal (Figure 1). Access to care is readily available for confirmed and suspected patients, as the expenses used for isolation and treatment of infectious diseases like COVID-19 are fully covered by the public sector (e.g., the insurer and local governments) under the Infectious Disease Control and Prevention Act.
Figure 1.
Health Insurance Review and Assessment Service (HIRA)’s data flow: from collecting data from healthcare providers to opening data to the public.
Thanks to the aforementioned healthcare system and IT infrastructure in Korea, RWD on COVID-19 were swiftly collected, processed, and de-identified in the form of standardized structured data, requiring an efficient research methodology for production of clinical evidence and policies. As the international community is suffering from COVID-19 and it takes more than just one country’s research and efforts to produce evidence, Korea decided to carry out the COVID-19 International Collaborative Research Project using new research methods to share the benefits of the data collected by HIRA with researchers around the world.
This article aims to introduce the database, research methodology, and deliverables of the COVID-19 International Collaborative Research Project, which has been run by HIRA since March 27, 2020, and thereby to contribute to the production of national-level clinical evidence based on RWD and to promote the development of international cooperative mechanisms in the face of a global health threat like COVID-19.
DATA ATTRIBUTES AND STRUCTURE
The COVID-19 International Collaborative Research Project leveraged NHI benefit claims data consisting of time-series healthcare use records of the entire population. Claims data do not have much clinical information such as test results, but offer a high value for national cohort research as they contain details of healthcare service usage of individuals based on a fee-for-service payment system. The primary purpose of claims data collection is to reimburse benefit to healthcare providers. An understanding of the NHI benefit policy and claim submission method is necessary in order to use the collected and processed data for research purposes, not to mention proficiency in statistics and statistical packages to analyze massive big data (over 1.5 billion claim statements a year).
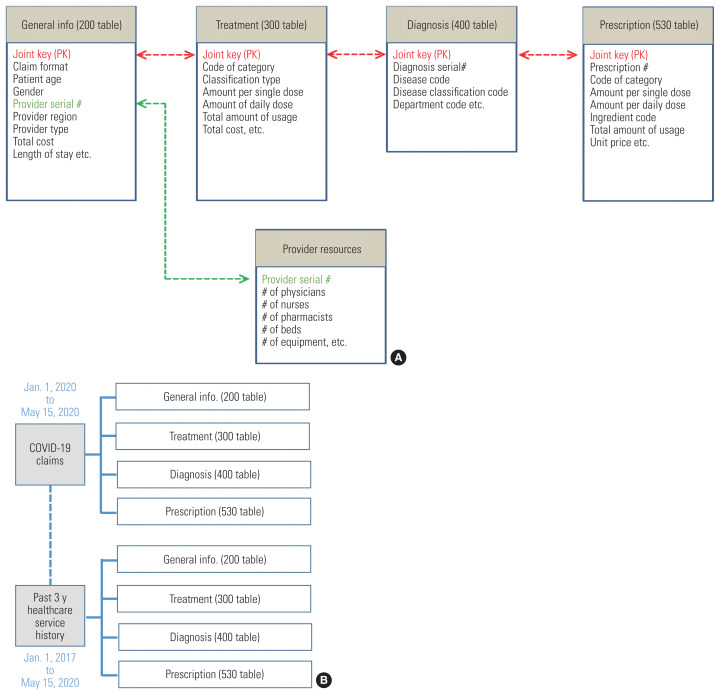
When the claims review process is completed, the variables of the data are saved in each table of the Data Warehouse (DW). Among the tables for different tasks, research-related tables include general summary information (the 200 table), treatments (the 300 table), diagnoses (the 400 table), and prescriptions (the 530 table) (Figure 2).
Figure 2.
(A) Composition of Health Insurance Review and Assessment Service’s main data tables and (B) the structure of the coronavirus disease 2019 (COVID-19) research dataset.
DATA EXTRACTION AND USED VARIABLES
The dataset used for the research was extracted as of May 15, 2020 and consists of COVID-19 treatment data and patients’ healthcare service use history for the past 3 years. The COVID-19 treatment data are composed of real-time reverse transcription polymerase chain reaction (RT-PCR) tests and claim statements, which have relevant diagnosis and fee codes. To examine patients’ underlying conditions, their healthcare service use history was collected for the period from January 1, 2017 to May 15, 2020. The composition of HIRA’s main data tables for the research project is depicted in Figure 2, with 8 tables consisting of 4 COVID-19 related medical service use tables and 4 tables presenting the 3-year history of healthcare service use of suspected and confirmed patients.
In the final dataset, there were 476 508 benefit claims for 234 427 patients, of whom 7590 were confirmed cases of COVID-19 (Table 1). In Korea, there were more female patients than male patients (3095 vs. 4495), and the 20–29 age group had the most patients (2844 persons, 37.5%). Furthermore, the diagnosis and medical service items (treatment, medical materials, and drugs) showed discrepancies between all patients and confirmed patients (Table 1). It should be noted that, under the fee-for-service payment system, a single claim statement can contain multiple diagnoses and medical services, and the figures of Table 1 refer only to simple numeric records and are not statistically meaningful values. As such, it is essential to have a prior knowledge of the NHI benefit system and claim method to properly utilize claims data for research purposes.
Table 1.
General characteristics of the COVID-19 International Collaborative Research Database
| Classification | Values | ||
|---|---|---|---|
|
| |||
| Total | Confirmed cases only | ||
| No. of claim (case) | 476 508 | 13 510 | |
|
| |||
| No. of patient (person) | 234 427 | 7590 | |
|
| |||
| Sex | Male | 111 947 | 3095 |
| Female | 122 480 | 4495 | |
|
| |||
| Age (y) | 0–9 | 8897 | 149 |
| 10–19 | 7428 | 346 | |
| 20–29 | 40 109 | 2844 | |
| 30–39 | 39 552 | 1432 | |
| 40–49 | 33 418 | 1795 | |
| 50–59 | 31 809 | 2673 | |
| 60–69 | 27 930 | 2178 | |
| 70–79 | 23 134 | 1229 | |
| 80–89 | 18 445 | 742 | |
| 90–99 | 3590 | 122 | |
| ≥100 | 116 | NA | |
|
| |||
| Top 5 diagnostic codes (%) | Special screening examination for other viral disease (44.3) | COVID-19 virus identified (72.5) | |
| Observation for other suspected diseases and conditions (6.0) | Special screening examination for other viral disease (14.5) | ||
| Fever, unspecified (4.0) | Contact with and exposure to other communicable diseases (1.7) | ||
| Contact with and exposure to other communicable diseases (4.0) | Isolation (1.5) | ||
| COVID-19 virus identified (3.7) | Coronavirus infection, unspecified site (1.2) | ||
|
| |||
| Top 5 healthcare services used (%) | Treatment | RT-PCR (2.7) | Inpatient medication preparation (1.8) |
| Safe hospital fee for infection prevention and control (management fee) (1.6) | RT-PCR (1.6) | ||
| Outpatient care – new patient (1.3) | General diet_dietitians (1.6) | ||
| Chest [X-ray] (1.2) | General diet_cook (1.5) | ||
| Full Picture Archiving and Communications System (1.0) | Chest [X-ray] (1.5) | ||
| Medical materials | Electrode & EKG (12.4) | Blood & solution for injection (20.9) | |
| Blood and solution for injection (10.4) | Electrode & EKG (20.3) | ||
| Fixed price items (9.2) | Continuous drainage (8.5) | ||
| Continuous drainage (8.8) | Genitourinary items (7.9) | ||
| Genitourinary items (7.3) | Endotracheal intubation (7.8) | ||
| Drugs | Sodium chloride (0.9) 0.9 g (9 mg/mL) (2.8) | Acetaminophen (encapsulated) (6.1) | |
| Acetaminophen (encapsulated) (2.5) | Lopinavir & ritonavir (4.2) | ||
| Sodium chloride (0.9) 9 g (9 mg/mL) (2.2) | Acetylcysteine (3.2) | ||
| Oxygen gas (1.9) | Ammonium chloride & chlorpheniramine maleate & dihydrocodeine tartrate & DL-methylephedrine hydrochloride (3.0) | ||
| Sodium chloride (0.9) 4.5 g (9 mg/mL) (1.7) | Hydrochloride sulfate (2.9) | ||
COVID-19, coronavirus disease 2019; NA, not applicable; RT-PCR, real time reverse transcription polymerase chain reaction; EKG, electrocardiogram.
Table 2 shows the variables used for analysis. The 200 table contains summarized information variables of patients’ medical service use, such as patient information, provider information, primary/secondary diagnosis, number of treatment days, inpatient/outpatient status, the patient’s route to the provider, and payment amount. The 300 table contains detailed treatment information (procedure codes, drug codes, etc.) and costs. The 400 table has diagnostic information for primary and secondary diagnoses. The 530 table contains information on outpatient prescriptions. A joint key was given to integrate the tables under a single system to enable an efficient search across tables.
Table 2.
Data extraction criteria and variables included
| Classification | Contents | |
|---|---|---|
| Data extraction criteria | COVID-19-related diseases1 | B343, B927, Z208, Z290, U18, U181, Z038, Z115, U071, U072 |
| COVID-19-related treatment received2 | RT-PCR, other COVID-19 related fee codes | |
|
| ||
| Variables included | 200 Table (summary information) | claim alternative key/patient alternative key/insurance type/gender/patient b-day/patient age_1/patient age_2/provide ID serial no./provider type/region code/healthcare service type/primary diagnosis code/secondary diagnosis code/medical department/service start date/service end date/first diagnosis date/no. of treatment days/no. of treatment & drug administration days/no. of days outpatient prescription/total drug expenditure of prescription/total no. of outpatient prescription cases/total treatment amount after review/co-payment amount/insure payment amount/100% co-payment amount/diagnosis with surgery/code of disease or injury related to official duties/code of disaster-related/specific code of injury by exogenous factors/patient status of discharge/patient’s route to the provider/type of medical aid/claim method type/claim type/month and year of review/medical department code indicated by provider/code of COVID-19 confirmed case/code of death by COVID-19 (39 variables) |
| 300 Table (treatment information) | claim alternative key/patient alternative key/code of category/code of classification type/code of classification/amount per single dose/amount per daily dose/total usage of day/total days of medication/number of total usage/unit price/medical fee/total amount after application of additional charge/code of generic drug name/exception classification code/additional charge code (16 variables) | |
| 400 Table (diagnosis information) | claim alternative key/patient alternative key/serial no. of disease/code of disease/disease classification code/medical department/code of internal medicine subspecialty department (7 variables) | |
| 530 Table (prescription information) | claim alternative key/patient alternative key/prescription issuance no./code of classification type/code of classification/single dose/number of administration per day/total days of medication/number of total usage/unit price/amount of prescribed drug/code of generic drug name (12 variables) | |
COVID-19, coronavirus disease 2019; RT-PCR, real time reverse transcription polymerase chain reaction.
Korean Standard Classification of Disease codes.
Electronic Data Interchange codes.
DATA SHARING METHOD
Two types of datasets were offered to researchers across the globe. The first type was formed based on Electronic Data Interchange (EDI) codes for treatments, drugs, and medical materials and Korean Standard Classification of Diseases (KCD) codes for diagnosis, which can be analyzed with SAS and R packages. The other type is the Common Data Model (CDM), converted from EDI and KCD code to map with international standards, which can be analyzed with ATLAS based on an R package. The CDM is used to standardize data terminology and structures between institutions and share analysis codes without data disclosure to generate result values (evidence). This research project used the OMOP (Observational Medical Outcomes Project) CDM. Researchers could choose the dataset type when they apply for the project, and produce analysis code using the disclosed data schema and sample dataset.
RESEARCH METHOD
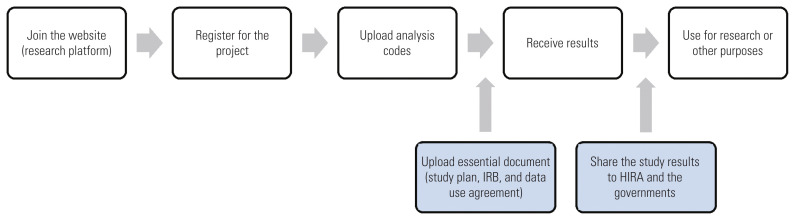
The COVID-19 International Collaborative Research Project was designed as an online-based platform (https://covid19data.hira.or.kr). There were no specific qualifications required to apply for the project. As Figure 3 illustrates, applicants only needed to submit their name, company/organization, and an email address of the company/organization, along with an online data use agreement. The required documents included a 1-page research plan and institutional review board document and written consent for data use. Applying researchers were instructed to produce and submit analysis code using the data schema and sample dataset on the online platform. Upon reception of the code, HIRA researchers ran the analysis code on the dataset established on the inhouse server, and returned the result values to the researcher. Because medical treatment data for infectious diseases are highly sensitive health information, only the analysis code and statistical result values were exchanged. This approach leveraged a distributed research method based on CDM to exchange only the produced evidence, not the actual data. This is considered an innovative attempt for collaborative research to open a dataset to more researchers without disclosing sensitive personal information.
Figure 3.
Participation process of the coronavirus disease 2019 (COVID-19) International Collaborative Research Project. IRB, institutional review board; HIRA, Health Insurance Review and Assessment Service.
PARTICIPATION STATUS AND DELIVERABLES
Participation status
As of late July 2020, after the research project stopped receiving new applications, there were 1587 researchers from 58 countries who joined the online platform. Sixty-six percent of researchers were from Korea and the United States, followed by the United Kingdom, Italy, Israel, and Canada (Table 3). Among the total members, 412 applications from 32 countries were submitted, with the majority coming from Korea and the Unite States. Around 10 applications came from the United Kingdom, Canada, Israel, and Italy, respectively.
Table 3.
Participation status of the coronavirus disease 2019 (COVID-19) International Collaborative Research Project
| Classification | Contents |
|---|---|
| Participants (person) | Total: 1587 from 58 countries |
| Korea: 527 (33.2) | |
| USA: 525 (32.2) | |
| UK: 82 (5.2) | |
| Italy: 63 (4.0) | |
|
| |
| Projects applied (case) | Total: 412 from 32 countries |
| Korea: 181 (44.0) | |
| USA: 125 (30.3) | |
| UK: 19 (4.6) | |
|
| |
| Dataset type chosen (case) | Total: 129 (analysis code provided) |
| SAS Package: 56 (43.4) | |
| R Studio: 41 (31.8) | |
| Atlas (OMOP CDM): 32 (24.8) | |
|
| |
| Research topics (case) | Total: 85 (submitted to academic journals) |
| Disease characteristics: 44 (51.8) | |
| Drugs: 34 (40.0) | |
| Others: 7 (8.2) | |
Values are presented as number (%)
OMOP, Observational Medical Outcomes Project; CDM, Common Data Model.
Among the applications, 129 studies submitted analysis code. Table 3 classified the applications by analysis tool, and showed that 56 projects (43%) requested to use SAS, 41 projects (32%) to use R Studio, and 32 projects (25%) to use Atlas using CDM. Among the 85 projects that are published or are in the process of publishing, 69 projects opted for SAS, followed by R Studio (11) and Atlas (5).
The majority of applicants chose to use SAS because it is the most commonly used statistical package to analyze large-scale HIRA data in Korea. Projects that applied to use R Studio showed a lower rate of result production. It seems that the closed analysis environment of HIRA, a public organization, was the reason behind the lower rate, despite the popularity of open-source R Studio among researchers. In Korea, public organizations that hold big data must disclose the data to the public under the law, but sensitive information like NHI claims data should remain in a closed environment and are not available for Internet connections or remote use under the guideline of the National Intelligence Service. As such, open source-based statistical analysis packages like R Studio and Python can be used in a much restricted manner as they require near real-time package updates in an Internet-supported environment.
Deliverables From the Research Project
The immediate deliverable of the COVID-19 International Collaborative Research Project is to produce scientific papers to be published in prestigious journals, so that clinical experts and governments can leverage the evidence in patient treatment and disease control policy preparation. Among the 129 projects that uploaded analysis code, 85 papers have been either published or are expected to be published, as of late November 2020. Regarding the research topic of the 85 projects, 44 projects (52%) investigated the characteristics of COVID-19, and 34 projects (40%) studied relationships between COVID-19 and medications taken for underlying conditions. Other topics included AI (artificial intelligence) analyses of COVID-19, and relationships between COVID-19 and aspects of the healthcare system (8 projects, 8%) (Table 3). A more comprehensive and final evaluation of the deliverables is expected to be available in the first half of 2021, because some research projects are still underway and some studies are dealing with complex topics within the framework of a single project.
As of late November, 2020, 21 papers have been published in academic journals (23, when including MedRxiv). Most of these are original articles, showing that nationwide RWD can be utilized in a meaningful way when the world is faced with a healthcare crisis. In particular, it is noteworthy to point out that most papers have been published in SCI (Science Citation Index) journals with a high H-index [4–9].
IMPLICATIONS AND FURTHER CONSIDERATIONS OF THIS PROJECT
In response to the global pandemic of the new coronavirus, the COVID-19 International Collaborative Research Project attempted to use a new method of research. The method achieved and produced some meaningful results, including the publication of academic research papers. Nonetheless, some considerations should be factored in to further develop and utilize the method for the production of national-level evidence.
First of all, HIRA’s data are a collection of time-series medical service use data covering all individuals in Korea, but the data were not collected for research purposes and it is difficult to preprocess and understand the data for utilization. Over 1500 researchers joined the platform and some 400 projects applied, but only 129 projects uploaded analysis code and completed the analysis, showing that the accessibility was limited due to the level of difficulty of the data. As such, data standardization is a key issue to be tackled to facilitate easier and smoother international collaborative research. HIRA began mapping its EDI claim codes to international standards. There is an increasing number of papers published in renowned academic journals that use this research method, proving the quality of such data [10–12].
Secondly, this research project was possible because it adopted a method to share only the data schema and sample data for the production of analysis code and evidence, without disclosing the highly-sensitive infectious disease related raw data stored in HIRA. The merit of this method is in data protection and security, but researchers experienced difficulties as they did not have firsthand access to the data to explore and produce more concrete analysis code. Therefore, research projects that would like to use a similar approach in the future need to consider providing basic statistics of the cohort data and a larger de-identified sample dataset to researchers.
Lastly, an effective analysis environment needs to be built, where open source-based R Studio and Python are supported for big data analysis. HIRA’s benefit claims data are representative RWD. To utilize the data through various big data analyses, research methods that strike an appropriate balance between security and utilization should be explored and examined, such as building a cloud analysis environment.
CONCLUSION
With its new approach, the short-range goal of the COVID-19 International Collaborative Research Project was to publish as many papers as possible in celebrated academic journals. However, the long-range aim of this project was to establish a shared mechanism for the international community where national-scale evidence based on a cohort study is effectively applied to patient treatment and policy-making. It is valuable and important to create research platforms and provide data, but it is no less important to build a distributed research network and promote the environment to share evidence and policy implications.
Ethis Statement
This study was approved by Institutional Review Board of the Korean National Institute for Bioethics Policy (IRB No. P01-202003-23-016).
ACKNOWLEDGEMENTS
None.
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest associated with the material presented in this paper.
FUNDING
None.
AUTHOR CONTRIBUTIONS
Conceptualization: YR, SCY, RWP, JYL. Data curation: DYC, YJL, JWK, HJL. Funding acquisition: None. Methodology: YR, SCY, RWP, YS. Writing – original draft: YR, DYC, YS, YJL, JWK, HJL. Writing – review & editing: JYL, SCY, RWP.
REFERENCES
- 1.World Health Organization WHO coronavirus disease (COVID-19) dashboard. [cited 2020 Nov 22]. Available from: https://covid19.who.int/?gclid=Cj0KCQiA8dHBRD_ARIsAC24uma-QyZn1rXWY2ll21RgY8AHUAea-uHdkcS5LJncKcOYXFy0KWfWP1YUaAh6EEALw_wcB.
- 2.Engelbrecht FA, Scholes RJ. Test for Covid-19 seasonality and the risk of second waves. One Health. 2021;12:100202. doi: 10.1016/j.onehlt.2020.100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Wang X, Nair H. Global seasonality of human seasonal coronaviruses: a clue for postpandemic circulating season of severe acute respiratory syndrome coronavirus 2? J Infect Dis. 2020;222(7):1090–1097. doi: 10.1093/infdis/jiaa436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burn E, You SC, Sena AG, Kostka K, Abedtash H, Abrahão MT, et al. Deep phenotyping of 34,128 adult patients hospitalised with COVID-19 in an international network study. Nat Commun. 2020;11(1):5009. doi: 10.1038/s41467-020-18849-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho SI, Kim YE, Jo SJ. Association of COVID-19 with skin diseases and relevant biologics: a cross-sectional study using nationwide claim data in South Korea. Br J Dermatol. 2020 doi: 10.1111/bjd.19507. [DOI] [PMC free article] [PubMed]
- 6.Jung SY, Choi JC, You SH, Kim WY. Association of renin-angiotensin-aldosterone system inhibitors with coronavirus disease 2019 (COVID-19)-related outcomes in Korea: a nationwide population-based cohort study. Clin Infect Dis. 2020;71(16):2121–2128. doi: 10.1093/cid/ciaa624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SW, Ha EK, Yeniova AÖ, Moon SY, Kim SY, Koh HY, et al. Severe clinical outcomes of COVID-19 associated with proton pump inhibitors: a nationwide cohort study with propensity score matching. Gut. 2021;70(1):76–84. doi: 10.1136/gutjnl-2020-322248. [DOI] [PubMed] [Google Scholar]
- 8.Lee SW, Yang JM, Moon SY, Yoo IK, Ha EK, Kim SY, et al. Association between mental illness and COVID-19 susceptibility and clinical outcomes in South Korea: a nationwide cohort study. Lancet Psychiatry. 2020;7(12):1025–1031. doi: 10.1016/S2215-0366(20)30421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeong HE, Lee H, Shin HJ, Choe YJ, Filion KB, Shin JY. Association between nonsteroidal antiinflammatory drug use and adverse clinical outcomes among adults hospitalized with coronavirus 2019 in South Korea: a nationwide study, clinical infectious diseases. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1056. [DOI] [PMC free article] [PubMed]
- 10.You SC, Rho Y, Bikdeli B, Kim J, Siapos A, Weaver J, et al. Association of ticagrelor vs clopidogrel with net adverse clinical events in patients with acute coronary syndrome undergoing percutaneous coronary intervention. JAMA. 2020;324(16):1640–1650. doi: 10.1001/jama.2020.16167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suchard MA, Schuemie MJ, Krumholz HM, You SC, Chen R, Pratt N, et al. Comprehensive comparative effectiveness and safety of first-line antihypertensive drug classes: a systematic, multinational, large-scale analysis. Lancet. 2019;394(10211):1816–1826. doi: 10.1016/S0140-6736(19)32317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee H, Lee JR, Jung H, Lee JY. Power of universal health coverage in the era of COVID-19: a nationwide observational study. Lancet Reg Health West Pac. 2021;7:100088. doi: 10.1016/j.lanwpc.2020.100088. [DOI] [PMC free article] [PubMed] [Google Scholar]