Abstract
Artemisinin resistance (ART) has been confirmed in Greater Mekong Sub-region countries. Currently, C580Y mutation on Pfkelch13 gene is known as the molecular marker for the detection of ART. Rapid and accurate detection of ART in field study is essential to guide malaria containment and elimination interventions. A simple method for collection of malaria-infected blood is to spot the blood on filter paper and is fast and easy for transportation and storage in the field study. This study aims to evaluate LAMP-SNP assay for C580Y mutation detection by introducing an extra mismatched nucleotide at the 3’ end of the FIP primer. The LAMP-SNP assay was performed in a water bath held at a temperature of 56°C for 45 min. LAMP-SNP products were interpreted by both gel-electrophoresis and HNB-visualized changes in color. The method was then tested with 120 P. falciparum DNA from dried blood spot samples. In comparing the LAMP-SNP assay results with those from DNA sequencing of the clinical samples, the 2 results fully agreed to detect C580Y. The sensitivity and specificity of the LAMP-SNP assay showed 100%. There were no cross-reactions with other Plasmodium species and other Pfkelch13 mutations. The LAMP-SNP assay performed in this study was rapid, reliable, and useful in detecting artemisinin resistance in the field study.
Keywords: Plasmodium falciparum, Pfkelch13 gene, artemisinin resistance, C580Y mutation, LAMP-SNP assay
INTRODUCTION
There are 87 countries in tropical and sub-tropical regions of the world suffering the tremendous malaria burden where an estimated 219 million cases and 435,000 related deaths occur every year. The most prevalent malaria parasite is P. falciparum, found in Africa, Western Pacific, Eastern Mediterranean, and Southeast Asia [1]. Since 2001, the World Health Organization had recommended artemisinin combination therapy (ACT) for uncomplicated falciparum malaria treatment. P. falciparum strains resistant to artemisinin were reported in western Cambodia [2] since 2009, where they have spread to other parts of Southeast Asia, especially Greater Mekong Sub-region countries (GMS) [3,4]. In Thailand, ACT resistant P. falciparum is established markedly in joining border areas, for which it has subsequently responded by changing the drug policy from artesunate-mefloquine to dihydroartemisinin-piperaquine in 2015 [1]. The molecular marker associated with delayed parasite clearance was the single nucleotide polymorphisms (SNPs) in the Kelch13 propeller domain of P. falciparum gene (Pfkelch13) [5]. More than 20 SNPs of the Pfkelch13 gene associated with artemisinin resistance have been identified in Southeast Asia [6–8], and at least 12 of the Pfkelch13 SNPs emerged locally in the GMS. C580Y is one of the SNPs strongly associated with artemisinin resistance, and it has reached a state of fixation in many parts of the GMS, especially Cambodia, Thailand, and Myanmar [9]. Polymerase chain reaction (PCR) amplification followed by Sanger sequencing is the commonly used technique to confirm C580Y mutation. However, these techniques are not suitable in malaria-endemic areas, particularly in developing countries, due to their high cost requiring sophisticated laboratories. They are also not suitable for field-based study. The LAMP assay is a rapid and relatively simple technique to amplify the nucleic acids and detect SNP while not requiring sophisticated equipment. Loop-mediated isothermal amplification (LAMP) and single nucleotide polymorphism (SNP) or LAMP-SNP assay has been successful in detecting SNP in Pfcrt gene for chloroquine resistance (K76T) [10] and Pfdhfr gene for pyrimethamine resistance (N51I) [11] and other organisms [12–16]. Recently, 2 studies had successfully detected C580Y using LAMP assay, but special devices were required in those studies. MinION nanopore sequencer and other expensive tools [17] were not suitable for a field study. In addition, the detection of fluorescence color change [18] requires adding a dye after completing the reaction. Thus, there is a potential concern for carry-over contamination of amplification products. In the current study, we present the LAMP-SNP assay to detect C580Y mutation among the samples from a field study, comparing the result with those from DNA sequencing. The method was simple, efficient, and only required an instrument with a constant incubation temperature under isothermal conditions. The metal-ion indicator, hydroxynaphthol blue (HNB), was incorporated into the LAMP mixture before incubation. The amplification is to be completed without opening the tube and the color change from violet to sky blue can be visualized immediately. This method could be used for a diagnosis of artemisinin resistance during the field-work.
MATERIALS AND METHODS
Ethical statement
The study obtained ethical approval from the Research Ethics Committee of Faculty of Medicine, Prince of Songkla University (REC60-096-19-2).
Sample collection and Deoxyribonucleic Acid Extraction
A total of 120 P. falciparum dried blood spots were collected between 2012 and 2017 from the malaria clinics at the Office of Disease Prevention and Control 11 and 12, Thailand. The samples covered the southern provinces of Thailand, such as Chumphon, Ranong, Phang-nga, Suratthani, and Yala provinces. The blood samples (approximately 50–80 μl) were collected by finger-prick on filter paper (Whatman 3MM, GE Healthcare, UK). They were positive for P. falciparum, and the geometric mean of the parasite density using the assumed WBCs (8,000/μl of blood) was 12,756 parasites/μl (95% CI, 67–200,000 parasites/μl). Individual samples were stored in their plastic bags at room temperature and sent to the Faculty of Medical Technology, Prince of Songkla University, Thailand. According to the manufacturer's instructions, DNA isolation was performed by a QIAamp DNA Blood Mini Kit (QIAGEN GmbH, Hilden, Germany). Genomic DNA was eluted into 100 μl of PCR-grade water and kept at −20°C until processing.
Amplification of Pfkelch13 gene and Sequencing
This study used 120 samples; Khammanee et al. [19] had performed the DNA sequencing of the 104 of these samples while the rest 16 samples were received from the Office of Disease Prevention and Control, Thailand, between 2013–2014. Detection of Pfkelch13 propeller gene and sequencing of clinical dried samples used in this study have been previously described in Khammanee et al. [19]. Briefly, the first round of PCR amplification used a primer pair K13-1F/K13-2283R. The size of the PCR product was 2,283 bp. For the second round of PCR amplification, 3 amplicons were performed on the first PCR product, consisting of 3 pairs of oligonucleotide primers that bind to the different target sequences. The product sizes were 781, 917, and 786 bp from amplification with primer pairs K13-1F/K13c694-R, K13c621F/K13c1538R, and K13c1344F/K13c2129R, respectively. The total reaction mixture in the first-round PCR was 25 μl, and the second-round was 100 μl. Each PCR mixture contained 10× buffer, 2 mM MgCl2, and 0.25 μM each of the primers, 0.25 mM each of dNTPs, 0.5 units of Platinum Taq DNA polymerase (Invitrogen Life Technologies, Carlsbad, California, USA), and 3 μl of genomic DNA template or 5 μl of PCR product of the first-round PCR in the second-round PCR. A total of 30 PCR cycles for the first round PCR and 35 cycles for the second round PCR were undertaken using Eppendorf Master Cycler (Eppendorf AG, Hamburg, Germany). The temperature cycles consisted of pre-denaturation at 94°C for 5 min followed by 94°C for 1 min, 58°C for 1 min in the first round PCR and 2 min in the second-round PCR, 72°C for 2 min, and a final extension at 72°C for 10 min. DNA template from the 3D7 strain was used as a positive control and sterile distilled water as a template for a negative control in each PCR reaction. DNA sequencing was performed by Macrogen (South Korea). Multiple nucleotide sequence alignments were analyzed with BioEdit version 7.1.3.0, using 3D7 strain of P. falciparum (GenBank accession number: PF3D7_ 1343700) as a reference to detect C580Y mutations in the gene.
Preparation of plasmid DNA as LAMP-SNP C580Y positive control
In this study, DNA fragments carrying C580Y were cloned into the pGEM®-T Easy vector (Promega Corporation, Madison, Wisconsin, USA) to generate recombinant plasmids pGEMPfkelch13C580Y. The Escherichia coli JM109 carrying recombinant plasmid was inoculated in 5 ml of LB broth containing ampicillin resistance and incubated at 37°C overnight with shaking. The recombinant plasmids were purified using the GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA) and checked by sequencing (Macrogen, Korea). Optimization of LAMP reactions used the recombinant plasmids of pGEMPfkelch13C580Y as a positive control.
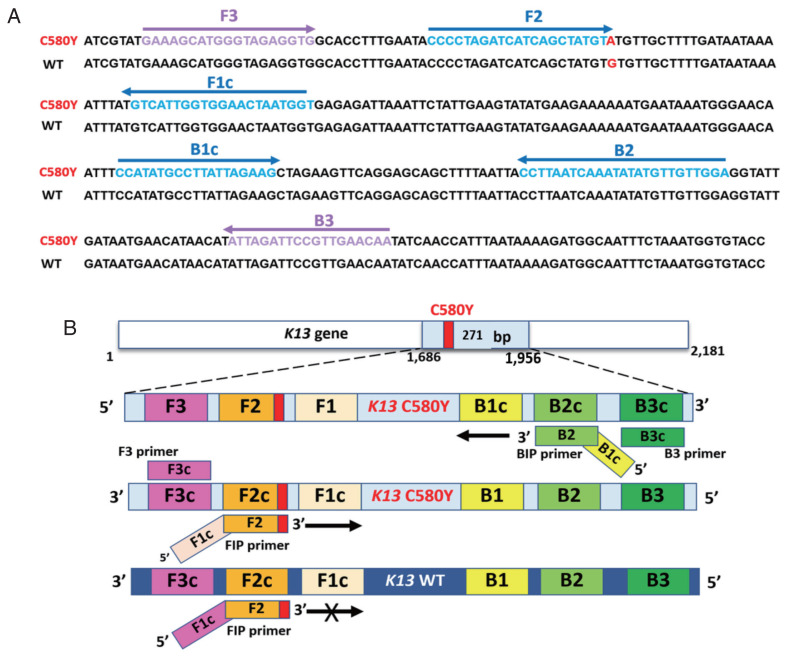
Optimization of LAMP-SNP C580Y detection
LAMP primers in the study were designed using the Primer Explorer v4 (https://primerexplorer.jp/lamp4.0.0/index.html) and were shown in Supplementary Table S1. The structure and their complementary region of LAMP-SNP primers with C580Y mutation were targeted on the DNA sequence, as shown in Fig. 1. The primer set consisted of 4 primers and recognized 6 distinct regions on the Pfkelch13 gene. The outer primers were F3 and B3 primers recognized at nucleotide positions 1686–1704 and 1938–1956. The product size was 271 bp. F1c and F2 represented a forward inner primer (FIP) and B1c and B2 for a backward inner primer (BIP). They are unique in that they each recognize 2 different regions on the target DNA. To detect a single nucleotide C580Y (TGT>TAT), the primer FIP was constructed as a specific primer with the mutation at the 3’ end of the primer with one mismatched nucleotide was added at the penultimate position as shown in Supplementary Table S1. The conditions for optimization of LAMP-SNP detection were various concentrations of parameters, including the concentrations of MgSO4 (New England Biolabs, Inc. Ipswich, Massachusetts, USA) (4 to 8 mM), inner primer (1.4 to 1.8 μM), outer primer (0.2–0.4 μM) and Bst DNA polymerase (New England Biolabs, Inc.) (4–10 U). Before amplification, hydroxynaphthol blue (HNB) (Sigma-Aldrich, St. Louis, Missouri, USA) was added to the reaction mixture at a final concentration of 120 μM. The reaction was incubated was at different temperatures and times between 55–60°C for 45–60 min, followed by heating at 80°C for 5 min to inactivate the reaction. Sterile water was used as a negative control, and a plasmid carrying pGEMPfkelch13C580Y mutation was a positive control. The 2 methods, HNB-visualization and gel electrophoresis, were used to assess the experiments. The results indicated a positive reaction if the amplification product changes in color from violet to sky blue. They showed the ladder-like pattern bands on electrophoresis, whereas for a negative result, the color remained violet, and there were no LAMP product bands on the gel.
Fig. 1.
Sequence alignment of part of the Pfkelch13 gene and schematic representation of LAMP-SNP primers used in the study. (A) Partial Pfkelch13 gene alignment between Pfkelch13 C580Y mutation and Pfkelch13 wild type sequence and location of primers, FIP (F1c-F2), BIP (B1c-B2), F3, and B3. The PCR amplification obtained from F3 and B3 was inserted into the plasmid. The mutated nucleotide of artemisinin resistance is shown in bold type, G for the wild type, and A for the Pfkelch13 C580Y mutation. (B) Schematic representation of the Pfkelch13 C580Y mutation LAMP-SNP primers. For C580Y mutation detection, a single nucleotide changed from guanine to adenine (TGT>TAT). This study was constructed of FIP primer as a specific primer with the mutation at the 3’ end of the primer and one mismatched nucleotide was added at the penultimate position. LAMP-SNP assay was negative for the Pfkelch13 wild type strain.
Assessment of the sensitivity and specificity of LAMP-SNP C580Y detection
The sensitivity of LAMP-SNP primers was conducted using the 10-fold dilutions of pGEMPfkelch13C580Y, from 5×107 to 10−1copies, and compared with the conventional PCR.
PCR reaction was conducted in a 25 μl total reaction volume containing 0.25 μM of F3 and B3 primers, 2 mM MgCl2, 0.25 mM of each dNTPs 1U of Taq DNA polymerase (Invitrogen Life Technologies) and one μl of pGEMPfkelch13C580Y were added. PCR was performed at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 56°C for 1 min, 72°C for 2 min and a final extension step at 72°C for 5 min. The PCR products were separated by electrophoresis on a 2% agarose gel and observed under UV transillumination. LAMP product was determined by visualizing the HNB post-reaction and running the products on a 2% agarose gel to identify characteristic ladder patterns. The specificity was determined using non-target DNA templates from Pfkelch13 wild-type, Pfkelch13 P574L mutation, and other Plasmodium species such as P. vivax, P. malariae, and human DNA.
Detection of C580Y mutation in Pfkelch13 gene clinical specimens
Five μl of DNA isolates from P. falciparum dried blood spots were performed with LAMP-SNP C580Y detection. The LAMP product was assessed by observing the color change of HNB and showing the ladder-like pattern bands on electrophoresis. Each sample was tested in triplicate.
Data analysis
The LAMP-SNP C580Y detection results were compared with the results from DNA sequencing, and the detectability of LAMP-SNP for Pfkelch13 C580Y mutation was measured against the parasite densities.
RESULTS
DNA sequencing results indicated that 89/120 samples were wild type and 3 of 89 samples carried point mutations at P574L, H366L+C447Y+V510G and N554S+I590T, respectively. There were 31/120 samples containing C580Y mutation. All the DNA samples were subjected to LAMP-SNP assay test.
Optimization, Sensitivity and Specificity assay
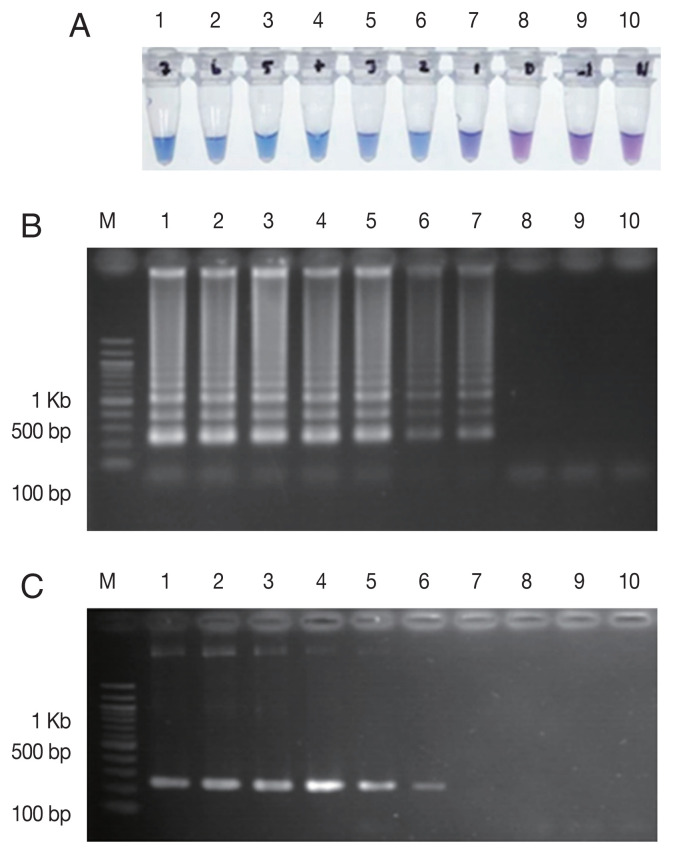
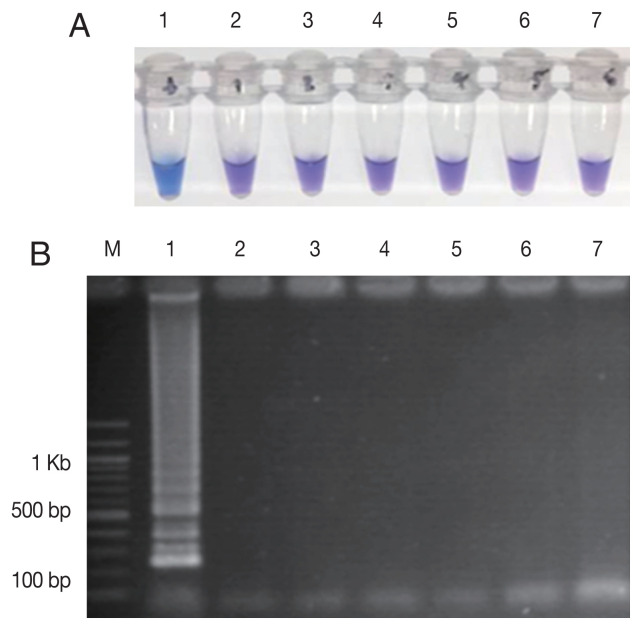
The LAMP-SNP assay was carried out at a temperature of 56°C for 45 min. The reaction used 8 U/tube of Bst DNA polymerase (New England Biolabs), 5.0 mM Mg2+, 1 M betaine (Sigma-Aldrich) and 0.8 mM dNTPs mix (Thermo Scientific, Vilnius, Lithuania), respectively. The concentrations of inner and outer primers were 1.6 μM of each FIP and BIP primers and 0.2 μM of each F3 and B3 primers, respectively. The analytical sensitivity of the LAMP-SNP assay was conducted using the 10-fold serial dilutions of plasmid DNA as a template for the LAMP-SNP and PCR experiments. The lower detection limit of LAMP-SNP assay was 50 copies of plasmid, whereas those of the conventional PCR was 500. This result showed that the sensitivity of LAMP-SNP assay was 10 times higher than that of the conventional PCR detection under the experimental conditions (Fig. 2). The LAMP-SNP assay specificity was examined by testing Pfkelch13 wild-type, P. vivax, P. malariae, and other Pfkelch13 P574L mutation as well as with the human DNA. The only plasmid carrying C580Y showed the positive color change with HNB dye (Fig. 3A) and showed the ladder-like pattern on the agarose gel (Fig. 3B). The LAMP assay did not amplify the DNA of the other organisms and human DNA.
Fig. 2.
Sensitivity comparison between LAMP-SNP assay and conventional PCR using the plasmid contained Pfkelch13C580Y. (A) LAMP-SNP products were detected base on HNB color change, (B) Detection of LAMP-SNP products by agarose gel electrophoresis, (C) Amplified PCR products were visualized by agarose gel electrophoresis using F3 and B3 primer. In panels A, B and C: M, 100 bp DNA Ladder (Thermo Scientific); Lanes 1–9: 5×107 to 5×10−1 copies of 10-fold serial dilutions of purified plasmid DNA contained Pfkelch13C580Y. Lane 10: No template control.
Fig. 3.
The specificity of the LAMP-SNP C580Y detection. (A) The specificity of LAMP-SNP assay using HNB (note the sky-blue color for a positive sample). (B) Confirmation of results of the LAMP-SNP assay products using agarose gel (1.5%) electrophoresis. In panels A and B: M, 100 bp DNA Ladder (Thermo Scientific); Label 1: plasmid pGEMPfkelch13C580Y; Label 2: plasmid pGEMPfkelch13 wild type strain; Label 3: gDNA of P. vivax; Label 4: gDNA of P. malariae; Label 5; gDNA of Pfkelch13 P574L mutation; Label 6: human DNA; Label 7: no template control.
Performance of LAMP-SNP assay for Pfkelch13 C580Y mutation detection in clinical dried blood spot samples
To validate the applicability of the LAMP-SNP assay for Pfkelch13 C580Y mutation, the assay was compared with the results from DNA sequencing. The LAMP-SNP assay detected Pfkelch13 C580Y mutation in 31 samples which were showed as positive by DNA sequencing (sensitivity=100%), while the rest 89 samples carrying C580 showed negative LAMP assay (specificity=100%). Positive and negative predictive values of detection were 100%. For samples, agarose gel indicated a positive result (the ladder-like pattern bands) though the HNB dye did not change color. This would be due to the amplification of the target sequence. Speed vacuum centrifuge was done to increase the concentration of DNA. Twenty microliters of PCR-grade water were added into an individual DNA sample, providing approximately a 5-fold increase in DNA concentration (from mean DNA concentration of 5.9 ng/μl to 28.9 ng/μl). The results of the LAMP-SNP assay were in concordance with those from the un-concentrated genomic DNA. All positive samples had products that exhibited a color of HNB dry changed from violet to sky blue, as shown in Supplementary Fig. S1. Table 1 shows the parasite density of 120 clinical samples. It ranged from 67–200,000 parasites/μl. From 31/120 (25.8%) samples positive for C580Y LAMP-SNP detection, 21 of them (67.7%) had no parasitemia data available and only 10 samples (32.3%) had parasitemia ranging from 67–200,000 parasites/μl (mean 26,286.1 parasites/μl). Therefore, in this study, LAMP-SNP assay detected the parasitemia level as low as 67 parasites/μl (>2 ng/μl of DNA). Among the remaining 89 samples which were negative for C580Y, 40 samples (44.9%) had no parasitemia data available, whereas 49 samples (55.1%) had parasitemia levels of 78–200,000 parasites/μl (mean 7,878.3 parasites/μl). This highlighted that LAMP-SNP detection sensitivity was not different regardless of lower or higher parasitemia among Pfkelch13 C580Y and wild-type C580 samples. The low parasite densities of 67 (mutant) vs. 78 (wild-type) parasites/μl and high parasite densities of 200,000 parasites/μl (equally in both groups) could be detected by this method. There were no false-positive and false-negative due to high and low parasitemia, respectively.
Table 1.
Detectability of LAMP-SNP for Pfkelch13 C580Y mutation in clinical samples
| LAMP-SNP C580Y result | Mutationa | ||
|---|---|---|---|
| C580Y | C580 | Other mutations | |
| mutation | Wild type | ||
| Positive | 31 | 0 | 0 |
| Negative | 0 | 86 | 3b |
| Parasite densities range (rang, parasite/μl) | 67–200,000 | 78–200,000 | 159–11,840 |
Results from DNA sequencing of the Pfkelch13 gene.
Other mutations: P574L, H366L+C447Y+V510G, and N554S+I590T.
DISCUSSION
C580Y mutation in the Pfkelch13 gene is one of the molecular markers of artemisinin resistance spreading throughout the greater Mekong region [6]. It is paramount important to identify the key mutation with simplicity and rapidity for treatment and epidemiological surveillance. LAMP has been previously studied to detect C580Y mutation, with the LAMP combined with the MinION sequencing method [17] and SNP-LAMP method [18]. Both studies are useful for the detection of C580Y mutation but require the equipment of high cost. In this study, the FIP primer was constructed by creating one mismatched nucleotide at the 3’ end of the primer at the penultimate position supposed to bind with C580Y mutation. The LAMP-SNP primer sets for Pfkelch13 C580Y mutation detection successfully amplified target plasmid DNA. The reaction conditions were optimized to prevent nonspecific amplification, including titration of MgSO4 and reaction temperature parameters. The lower detection limit of this procedure was 50 copies of the reference pGEMPfkelch13C580Y. This result was concordant with a previous study [17] but our method produced the result faster and applied in a common laboratory with simple instruments. The reaction was completed in less than 1 hr to amplify 271 bp LAMP product at 56°C by incubation in a water bath or heat block.
The applicability of the method was evaluated using the clinical dried blood spot samples. Our method detected the mutation in 31 out of 120 samples, which was in agreement with DNA sequencing as the gold-standard method for detecting mutation. The sensitivity of detection was 67 parasites/μl and the assay achieved 100% sensitivity and specificity for the detection of C580Y. The LAMP-SNP assay procedure in this study would be invaluable for public health due to its rapid detection of artemisinin resistance in the malaria-endemic region.
In the study, genomic DNA was extracted from dried blood spot and the purified DNA was eluted in 100 μl of elution buffer. The LAMP-SNP assay showed the sensitivity to detect approximately 2 ng/μl of DNA from dried blood spots (67 parasites/μl) detected on the agarose gel. Simultaneously, this was not visualized in detection with the naked eye for HNB color changes. We tried to optimize the concentration of magnesium and dNTPs in various concentrations but did not observe the color changes of HNB. This problem may have occurred from the source of the sample. Dried blood spots were collected between 2012–2017 with a small blood volume, approximately 50–80 μl. After drying, these samples were kept on filter paper (Whatman 3MM) and stored at room temperature with silica beads of an individual plastic bag. The humidity in a plastic bag would be higher after keeping a filter paper for a longer period of time because it was not replaced with a new silica bead, rendering the DNA degradation. To overcome this potential problem, we used a speed vacuum centrifuge to reduce the DNA solution volume. The reduction of the volume of the DNA solution yielded higher DNA concentration and it is sufficient to synthesize the larger fragment of DNA by Bst DNA polymerase., In the meantime, the concentration of the Mg2+ ion decreased, and the color of HNB changes to sky blue, indicating a positive reaction. Nevertheless, the method from this study would apply to the detection of more than one SNP. The other important mutations associated with artemisinin resistance, including N458Y, Y493H, R539T, and I543T, may be subjected to detection by multiplex-LAMP.
In conclusion, the LAMP-SNP assay to detect Pfkelch13 C580Y was highly sensitive, specific, and rapid for detecting artemisinin resistance. The method will be useful for field study even in less well-equipped laboratories and can be used to detect C580Y in mobile populations who are the likely source of transnational spread and carry parasites asymptomatically.
ACKNOWLEDGMENT
Funding for this study was provided by a grant from Prince of Songkla University, contract no. MET600576S.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Supplementary Information
REFERENCES
- 1.World Malaria Report. 2018 [Internet]; [cited 2019 May 16]. Available: https://www.who.int/malaria/publications/world-malaria-report-2018/en/
- 2.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B, Anderson JM, Duong S, Chuor CM, Taylor WR, Suon S, Mercereau-Puijalon O, Fairhurst RM, Menard D. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis. 2013;13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Straimer J, Gnädig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, Gregory PD, Urnov FD, Mercereau-Puijalon O, Benoit-Vical F, Fairhurst RM, Ménard D, Fidock DA. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski DP, Bozdech Z, Jeeyapant A, Cheah PY, Sakulthaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Das D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin PJ, Dondorp AM, Day NP, White NJ. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fairhurst RM. Understanding artemisinin-resistant malaria: what a difference a year makes. Curr Opin Infect Dis. 2015;28:417–425. doi: 10.1097/QCO.0000000000000199. https://doi.org/0.1097/QCO.0000000000000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menard D, Ariey F. Towards real-time monitoring of artemisinin resistance. Lancet Infect Dis. 2015;15:367–368. doi: 10.1016/S1473-3099(15)70046-0. [DOI] [PubMed] [Google Scholar]
- 9.Imwong M, Suwannasin K, Kunasol C, Sutawong K, Mayxay M, Rekol H, Smithuis FM, Hlaing TM, Tun KM, van der Pluijm RW, Tripura R, Miotto O, Menard D, Dhorda M, Day NPJ, White NJ, Dondorp AM. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: a molecular epidemiology observational study. Lancet Infect Dis. 2017;17:491–497. doi: 10.1016/S1473-3099(17)30048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chahar M, Mishra N, Anvikar A, Dixit R, Valecha N. Establishment and application of a novel isothermal amplification assay for rapid detection of chloroquine resistance (K76T) in Plasmodium falciparum . Sci Rep. 2017;7:41119. doi: 10.1038/srep41119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yongkiettrakul S, Kampeera J, Chareanchim W, Rattanajak R, Pornthanakasem W, Kiatpathomchai W, Kongkasuriyachai D. Simple detection of single nucleotide polymorphism in Plasmodium falciparum by SNP-LAMP assay combined with lateral flow dipstick. Parasitol Int. 2017;66:964–971. doi: 10.1016/j.parint.2016.10.024. [DOI] [PubMed] [Google Scholar]
- 12.Duan Y, Yang Y, Wang Y, Pan X, Wu J, Cai Y, Li T, Zhao D, Wang J, Zhou M. Loop-Mediated Isothermal Amplification for the Rapid Detection of the F200Y Mutant Genotype of Carbendazim-Resistant Isolates of Sclerotinia sclerotiorum. Plant Dis. 2016;100:976–983. doi: 10.1094/PDIS-10-15-1185-RE. [DOI] [PubMed] [Google Scholar]
- 13.Duan Y, Zhang X, Ge C, Wang Y, Cao J, Jia X, Wang J, Zhou M. Development and application of loop-mediated isothermal amplification for detection of the F167Y mutation of carbendazim-resistant isolates in Fusarium graminearum. Sci Rep. 2014;4:7094. doi: 10.1038/srep07094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan YB, Yang Y, Wang JX, Liu CC, He LL, Zhou MG. Development and application of loop-mediated isothermal amplification for detecting the highly benzimidazole-resistant isolates in Sclerotinia sclerotiorum. Sci Rep. 2015;5:17278. doi: 10.1038/srep17278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badolo A, Bando H, Traoré A, Ko-Ketsu M, Guelbeogo WM, Kanuka H, Ranson H, Sagnon N, Fukumoto S. Detection of G119S ace-1 (R) mutation in field-collected Anopheles gambiae mosquitoes using allele-specific loop-mediated isothermal amplification (AS-LAMP) method. Malar J. 2015;14:477. doi: 10.1186/s12936-015-0968-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badolo A, Okado K, Guelbeogo WM, Aonuma H, Bando H, Fukumoto S, Sagnon N, Kanuka H. Development of an allele-specific, loop-mediated, isothermal amplification method (AS-LAMP) to detect the L1014F kdr-w mutation in Anopheles gambiae s. l. Malar J. 2012;11:227. doi: 10.1186/1475-2875-11-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai K, Tarumoto N, Runtuwene LR, Sakai J, Hayashida K, Eshita Y, Maeda R, Tuda J, Ohno H, Murakami T, Maesaki S, Suzuki Y, Yamagishi J. An innovative diagnostic technology for the codon mutation C580Y in kelch13 of Plasmodium falciparum with MinION nanopore sequencer. Malar J. 2018;17:217. doi: 10.1186/s12936-018-2362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohon AN, Menard D, Alam MS, Perera K, Pillai DR. A novel single-nucleotide polymorphism loop-mediated isothermal amplification assay for detection of artemisinin-resistantPlasmodium falciparum malaria. Open Forum Infect Dis. 2018;5:ofy011. doi: 10.1093/ofid/ofy011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khammanee T, Sawangjaroen N, Buncherd H, Tun AW, Thanapongpichat S. Molecular Surveillance of Pfkelch13 and Pfmdr1 Mutations in Plasmodium falciparum Isolates from Southern Thailand. Korean J Parasitol. 2019;57:369–377. doi: 10.3347/kjp.2019.57.4.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.