Abstract
While the COVID-19 pandemic caused by SARS-CoV-2 is continuing, it may become worse in the coming winter months with a high potential for the emergence and spread of escape variants of SARS-CoV-2. SARS-related CoVs (SARSr-CoVs) from bats may also cause outbreaks of emerging coronavirus diseases in the future. These predictions call for the development of broad-spectrum anti-coronavirus vaccines and therapeutics to combat the current COVID-19 pandemic and future emerging coronavirus disease epidemics. In this review, we describe advances and challenges in the development of broad-spectrum vaccines and neutralizing antibodies against lineage B betacoronaviruses (β-CoV-Bs), including SARS-CoV-2, SARS-CoV, and SARSr-CoVs, as well as peptide-based pan-CoV fusion inhibitors and their potential in the prevention and treatment of COVID-19 and other human coronavirus infections.
Key words: broad-spectrum, vaccine, therapeutic, coronavirus, COVID-19, spike protein
In this article, Jiang and colleagues describe advances and challenges in the development of broad-spectrum vaccines and neutralizing antibodies against lineage B betacoronaviruses (β-CoV-Bs), including SARS-CoV-2, SARS-CoV, and SARSr-CoV, and peptide-based pan-CoV fusion inhibitors, and discuss their potential in the prevention and treatment of COVID-19 and other human coronavirus infectious diseases.
Main Text
Introduction
As of December 15 of 2020, about 75 million confirmed cases of COVID-19, caused by SARS-CoV-2 infection, and 1.66 million related deaths in 235 countries, areas, or territories were reported to WHO (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). The COVID-19 outbreak may become worse in the coming winter months with a high potential for the emergence and spread of escape variants of SARS-CoV-2, which may be resistant to vaccines and therapeutics now under development (Li et al., 2020). Furthermore, SARS-related CoVs (SARSr-CoVs) from bats, such as SARSr-CoV-WIV1, can use human ACE2 as a receptor to infect human cells, and this may contribute to a new outbreak of coronavirus disease in the near future (Zhou et al., 2020b). These predictions call for the development of broad-spectrum anti-coronavirus vaccines and therapeutics to combat the current COVID-19 pandemic caused by the wild-type and mutant SARS-CoV-2, as well as any future emerging and re-emerging coronavirus disease epidemics (Jiang et al., 2020a; Wang et al., 2020d; Yu et al., 2020).
Coronavirus (CoV) is a family of enveloped single-stranded positive-sense RNA viruses comprised of genus α (α-CoV), β (β-CoV), γ (γ-CoV), and δ (δ-CoV). β-CoV can be further divided into lineage A (β-CoV-A), B (β-CoV-B), C (β-CoV-C), and D (β-CoV-D) (Woo et al., 2012). Human SARS-CoV-2 and SARS-CoV, which infect human target cells by utilizing ACE2 as its receptor, belong to β-CoV-B. Bat SARSr-CoVs also belong to β-CoV-B, and most of them can also use human ACE2 as their receptor to infect human target cells, thus having the potential to cause emerging CoV infectious diseases in the future (Zhou et al., 2020b). Besides SARS-CoV and SARS-CoV-2, another highly pathogenic emerging coronavirus caused the first outbreak of viral respiratory disease in Saudi Arabia in 2012, termed "Middle East respiratory syndrome coronavirus” (MERS-CoV), which belongs to β-CoV-C and utilizes human DPP4 as a receptor for infecting human target cells (Lu et al., 2013).
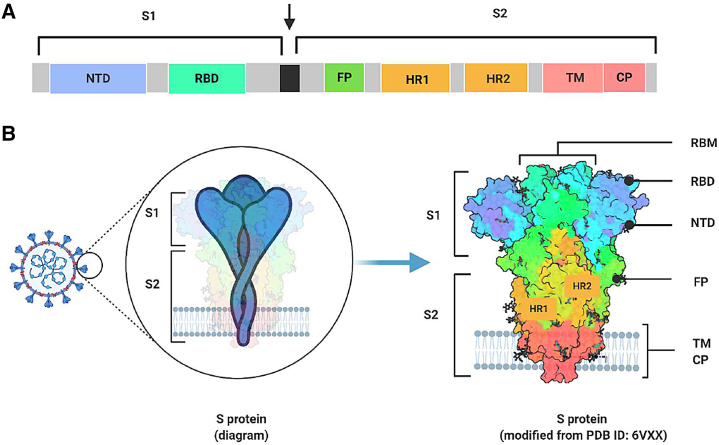
On the coronavirus envelope surface, three important structural proteins can be identified: spike protein (S), envelope protein (E), and membrane protein (M). S protein is a 180–200 kDa transmembrane (TM) glycoprotein that can be cleaved into S1 and S2 subunits by some proteases, such as transmembrane protease serine 2 (TMPRSS2), on the target cell membrane (Hoffmann et al., 2020; Xia et al., 2020a). The S1 subunit contains the N-terminal domain (NTD) and the C-terminal receptor-binding domain (RBD). Both SARS-CoV and SARS-CoV-2 utilize the RBD to bind the ACE2 receptor (Chi et al., 2020; Li et al., 2003). The ring structure in the RBD that directly contacts ACE2 is named the receptor-binding motif (RBM) (Li et al., 2005). The S2 subunit, which is composed of a fusion peptide (FP), heptad repeats 1 and 2 (HR1 and HR2), a TM region, and a cytoplasmic region, mediates the membrane fusion process. If no suitable protease exists on the cell membrane, the virus may enter the cell through endocytosis and fuse with the endosomal membrane with the assistance of acidic pH and proteases, such as cathepsin L (Hoffmann et al., 2020; Xia et al., 2020a). During the membrane fusion process, FP at the N terminus of S2 inserts into the cytoplasmal or endosomal membrane to form the pre-hairpin fusion intermediate conformation. Then, the three HR1 regions are assembled into coiled-coil trimers, and the three HR2 regions bind to hydrophobic grooves of the HR1 trimer in an anti-parallel manner, forming a six-helix bundle (6-HB), which brings the viral envelope and the cell membrane into close proximity for fusion (Xia et al., 2020d) (Figure 1).
Figure 1.
Sequence Diagram and Structure of β-CoV-B Spike Protein
(A) Sequence diagram of β-CoV-B spike (S) protein.
(B) Structure diagram of S protein position on coronavirus surface and its crystal structure (modified from PDB: 6VXX).
RBD, receptor-binding domain; RBM, receptor binding motif; NTD, N-terminal domain; HR1, heptad repeat 1; HR2, heptad repeat 2; FP, fusion peptide; TM, transmembrane domain; CP, cytoplasmic region. Illustration created by the authors using http://www.biorender.com.
Studies have shown that SARS-CoV-2 has a genome similarity of 79% and 88% with SARS-CoV and bat SARSr-CoVs (bat-SL-CoVZC45 and bat-SL-CoVZXC21), respectively. SARS-CoV, SARS-CoV-2, and SARSr-CoVs are all β-CoV-Bs, are on the same branch in the phylogenetic tree, and use the same receptor (ACE2) for infection (Cui et al., 2019; Ho, 2011; Hoffmann et al., 2020; Hu et al., 2020). This makes the development of pan-β-CoV-B vaccines and neutralizing antibodies (nAbs) feasible. We have recently designed and developed two peptide-based pan-CoV fusion inhibitors, EK1 (Xia et al., 2019) and EK1C4 (Xia et al., 2020d). Both are highly effective in inhibiting infection by all human CoVs tested, including SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-OC43, HCoV-NL63, and HCoV-229E, as well as some bat SARSr-CoVs. In this review, we discuss advances in the research and development of pan-β-CoV-B vaccines and nAbs, as well as pan-CoV fusion inhibitors, including the challenges in translating them into clinical applications.
Research and Development of Pan-β-CoV-B Vaccines
To combat the SARS-CoV-2 pandemic, a variety of vaccine candidates against SARS-CoV-2 have been developed. According to WHO, as of December 8, 2020, 162 candidate vaccines were in preclinical studies and 52 candidate vaccines were in clinical evaluation, including 13 in phase III, 16 in phase II, and 23 in phase I clinical trials (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines). The data from phase III clinical trials showed that a two-dose regimen of BNT162b2, an mRNA vaccine from Pfizer and BioNTech, conferred 95% protection against COVID-19 in persons 16 years or older (Polack et al., 2020). Several countries, including the UK and the US, have granted emergency use authorization to this vaccine. Moderna's mRNA vaccine was also reported to have roughly 95% efficacy, and will likely pass regulatory muster over the next few weeks (Cohen, 2020). In the phase III clinical trials conducted in the UK, Brazil, and South Africa, and in the UK of the ChAdOx1 nCoV-19 vaccine (AZD1222) developed by the University of Oxford and AstraZeneca (van Doremalen et al., 2020), the vaccine efficacy in participants who received two standard doses was 62%, while that of those who received a low dose followed by a standard dose was 90%, and the overall vaccine efficacy across both groups was ~70% (Voysey et al., 2020). Two vaccines have achieved regulatory authorization or approval by the Ministry of Health of the Russian Federation, including Sputnik V (formerly known as Gam-COVID-Vac), an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine developed by the Gamaleya Research Institute in Moscow (Logunov et al., 2020), and a peptide vaccine named EpiVacCorona developed by the Federal Budgetary Research Institution State Research Center of Virology and Biotechnology in Russia. Three inactivated vaccines from Beijing Institute of Biological Products/Sinopharm (named BBIBP-CorV) (Wang et al., 2020b; Xia et al., 2020c) and Wuhan Institute of Biological products/Sinopharm (no name announced), as well as Sinovac (named CoronaVac) (Zhang et al., 2020b) were approved in China as part of an emergency use program in the country for "high-risk" individuals (https://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-vaccine-tracker).
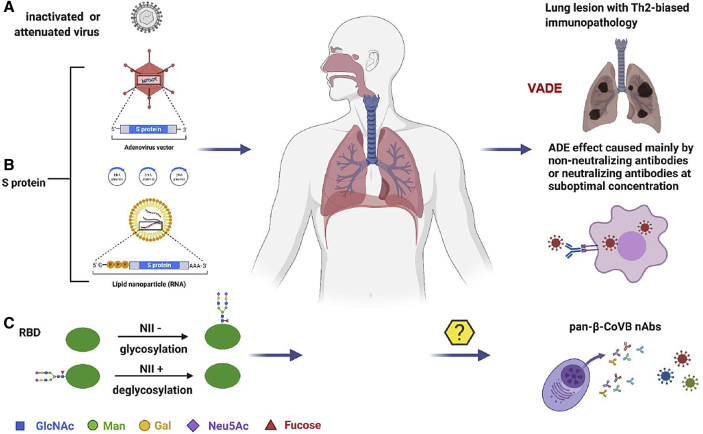
Most COVID-19 vaccine candidates, as listed above in clinical trials, are safe for use in humans because of the absence of any reported serious adverse reaction. This does not mean that vaccine safety is not a continuing critical concern in phase III clinical trials and in the future application of vaccines (Jiang, 2020). In particular, vaccine-associated disease enhancement (VADE) may occur when vaccinated people become naturally infected (Su et al., 2020) (Figure 2). VADE includes Th2-biased immunopathology and antibody-dependent enhancement (ADE) mediated by antibody-Fc receptor (FcR)-associated entry of virus into cells with or without viral replication, thus leading to the release of inflammatory cytokines or chemokines (Su et al., 2020). In general, antibodies with ADE are non-nAbs, but some nAbs can also mediate ADE at suboptimal concentration (Figure 2).
Figure 2.
Current COVID-19 Vaccine Candidates and Strategies for Developing Pan-β-CoV-B Vaccines
The antigens in the current COVID-19 vaccine candidates under clinical development consist of (A) whole viral particles either inactivated or attenuated, (B) spike (S) protein, or (C) receptor-binding domain (RBD). The vaccine-associated disease enhancement (VADE), including Th2-biased immunopathology and antibody-dependent enhancement (ADE), may occur when vaccinated people become naturally infected (Su et al., 2020). Pan-βCoVB vaccines can be designed using the strategies of glycosylation of NII-negative (NII−) sites and/or deglycosylation of the NII-positive (NII+) sites on S protein RBD. These optimized RBDs can also be used for the development of pan-β-CoV-B nAbs. Illustration created by the authors using http://www.biorender.com.
Presently, vaccine antigens against coronaviruses can be divided into several types: (1) inactivated or attenuated whole virus particles, (2) full-length S protein ectodomains, and (3) RBDs (Figure 2). Most viral vector-, mRNA-, or DNA-based vaccine candidates encode full-length S protein ectodomains of SARS-CoV-2 because the S protein contains both T and B cell epitopes that can induce cellular and humoral immune responses against viral infection. Most recently, Ahmed et al. have identified 115 T cell epitopes and 298 B cell epitopes from SARS-CoV S and N proteins and found that 27 T cell epitopes and 49 B cell epitopes are identical to those in SARS-CoV-2 S and N proteins, suggesting that the antigen containing these shared T and B cell epitopes in SARS-CoV and SARS-CoV-2 S and N proteins could be applied to the design of pan-β-CoV-B vaccines(Ahmed et al., 2020).
Indeed, S protein is highly immunogenic (Du et al., 2009), and SARS-CoV S protein-based vaccines can induce nAbs and protect mice from SARS-CoV challenge (Deming et al., 2006; Yang et al., 2004). Similarly, recent studies have shown that SARS-CoV-2 S protein-based vaccines, including adenovirus-vector- and mRNA-based vaccines, can protect mice or rhesus macaques from SARS-CoV-2 challenge (Erasmus et al., 2020; Feng et al., 2020; Holland et al., 2020).
However, based on our previous studies on SARS-CoV subunit vaccines, we found that the RBD contains the major nAb epitopes in S protein, but fewer immunodominant epitopes capable of inducing non-neutralizing antibodies or harmful immune responses than S protein. Therefore, the RBD is a better antigen than S protein in the development of SARS-CoV vaccines (Chen et al., 2005; Du et al., 2009; He et al., 2004, 2005a, 2005b). Yang et al. (2020a) have shown that a SARS-CoV-2 vaccine based on the RBD can elicit a much higher titer of neutralizing antibodies (NT50 ~1:2,400) than the full-length S ectodomain (NT50 ~1:300), S1 (NT50 ~1:1,100), or S2 (NT50 ~1:10), respectively.
Furthermore, sera from mice and rabbits immunized with SARS-CoV RBD exhibited cross-neutralizing activities against SARS-CoV-2 pseudovirus infection (Tai et al., 2020a; Zhu et al., 2020) (Table 1). Du and colleagues have designed and developed a COVID-19 vaccine candidate based on lipid nanoparticle (LNP)-encapsulated mRNA encoding the RBD (RBD mRNA-LNP). After prime-boost immunization of mice with RBD mRNA-LNP, they found that mouse antisera potently neutralized SARS-CoV-2 pseudovirus infection with an NT50 (50% neutralizing antibody titer) of ~1:10,000 and cross-neutralized infection by human SARS-CoV strains, including Tor2 and GD03, as well as palm civet SARS-CoV strain SZ3 (Tai et al., 2020b). Our group has demonstrated that immunization of mice with an RBD-Fc subunit vaccine candidate containing SARS-CoV-2 RBD and a human immunoglobulin G (IgG) Fc fragment of human IgG results in the production of high-titer nAb responses against infection by both live and pseudotyped SARS-CoV-2 with NT50 values of 1:7,166 and 1:10,523, respectively. Notably, IgG purified from the antisera could effectively cross-neutralize infection by SARS-CoV and SARSr-CoV WIV1 strains (Liu et al., 2020b). Zang et al. (2020) also used a SARS-CoV-2 RBD-Fc-based subunit vaccine to immunize mice and obtained mouse antisera with strong neutralizing nAb responses against pseudotyped SARS-CoV-2 and SARS-CoV infection with NT50 values of 1:12,764 and 1:835, respectively. These results suggest that the RBD from either SARS-CoV-2 S or SARS-CoV S protein has the necessary potential to be further developed as a pan-β-CoV-B vaccine.
Table 1.
Representative β-CoV-B Vaccines that Induce Antibodies with Cross-Neutralizing Activity against SARS-CoV-2, SARS-CoV, and SARSr-CoVs
| Name | Vaccine Format | Animal Immunized | Neutralizing and Cross-Neutralizing Activity | Ref. |
|---|---|---|---|---|
| RBD mRNA-LNP | lipid nanoparticle (LNP)-encapsulated mRNA encoding SARS-CoV-2 RBD | mice | PsV SARS-CoV-2: NT50 = ~1:10,000; live SARS-CoV-2: NT50 = 1:540; PsV SARS-CoV-Tor2: NT50 = ~1:175; PsV SARS-CoV-GD03: NT50 = ~1:125; PsV SARS-CoV-SZ-2: NT50 = ~1:75; |
Tai et al. (2020b) |
| RBD-Fc | SARS-CoV-2 RBD-Fc-based subunit vaccine | mice | PsV SARS-CoV-2: NT50 = 1:7,166; live SARS-CoV-2: NT50 = 1:10,523; PsV SARS-CoV: IC50 = 50 μg/mL; PsV SARSr-CoV-WIV1: IC50 = 25 μg/mL; |
Liu et al. (2020b) |
| INO-4800 | DNA vaccine expressing SARS-CoV-2 S protein | mice and guinea pigs | mice: limited cross-reactivity to SARS-CoV; PsV SARS-CoV-2: NT50 = 1:92 live SARS-CoV-2: NT50 = 1:340 guinea pigs: PsV SARS-CoV-2: NT50 = 1:574 live SARS-CoV-2: NT50 = >1:320 |
Smith et al. (2020) |
| RBD-Fc | SARS-CoV-2 RBD-Fc-based subunit vaccine | mice | PsV SARS-CoV: NT50 = 1:835 PsV SARS-CoV-2: NT50 = 1:12,764 live SARS-CoV-2: inhibit viral infection by 83% at 1:5,120 |
Zang et al. (2020) |
| S protein | SARS-CoV S-based subunit vaccine | mice | PsV SARS-CoV: NT50 = 1:640 PsV SARS-CoV-2: NT50 = 1:40 to 1:160 |
Zhu et al. (2020) |
| RBD | SARS-CoV GD03 RBD-based subunit vaccine | mice | PsV SARS-CoV: NT50 = 1:640 PsV SARS-CoV-2: NT50 = 1:40 to 1:160 |
Zhu et al. (2020) |
| RBD | SARS-CoV SZ16 RBD-based subunit vaccine | mice | PsV SARS-CoV: NT50 = >1:640 PsV SARS-CoV-2: NT50 = 1:40 to 1:160 |
Zhu et al. (2020) |
| RBD | SARS-CoV GD03 RBD-Fc-based subunit vaccine | rabbit | PsV SARS-CoV: NT50 = 1:160 to 1:640 PsV SARS-CoV-2: NT50 = <1:40 |
Zhu et al. (2020) |
| RBD | SARS-CoV SZ16 RBD-Fc-based subunit vaccine | rabbit | PsV SARS-CoV: NT50 = >1:640 PsV SARS-CoV-2: NT50 = 1:40 to 1:160 |
Zhu et al. (2020) |
PsV, pseudovirus.
As well as the main neutralizing epitopes of S protein, the RBD also contains some immunodominant epitopes (Kulp and Schief, 2013; Kwong et al., 2011), which may elicit non-nAbs or harmful immune responses. In general, the epitopes that are highly conserved and well exposed on the surface proteins of the enveloped viruses are able to induce high levels of broad nAbs (Sok et al., 2013). However, many of the conserved epitopes are not well exposed or are covered by glycans. As a remedy, we can either remove the glycan(s) at the conserved region to expose the neutralizing epitopes or mask the non-neutralizing epitope(s) in the immunodominant region with glycan(s). For example, Chen et al. (2014) constructed an RBD variant (RBD219-N1) by deleting the first Asn (N-1) in the RBD in the SARS-CoV S protein. Using RBD-N1 as a SARS-CoV vaccine candidate to immunize mice, they found that it could induce significantly stronger RBD-specific neutralizing antibody responses in immunized mice than RBD219-WT RBD, suggesting that deglycosylation is an important strategy for improving the immunogenicity of a neutralizing antibody epitope covered by glycan(s) in a vaccine candidate. Alternatively, Du et al. (2016) have used a strategy to reduce the immunogenicity of the predominant non-neutralizing epitopes capable of diverting host immune responses, thereby enhancing the immunogenicity of the neutralizing epitopes on the RBD in S protein of MERS-CoV. At the same time, they introduced a new concept termed the “neutralizing immunogenicity index” (NII). They first masked a selected epitope in the RBD with a glycan probe and then used the modified RBD vaccine to immunize mice, followed by testing the neutralizing titer of mouse antisera, thereby forming the basis for calculating the NII. A negative NII value suggests the negative contribution of the original unmodified epitope to the overall neutralizing immunogenicity of the RBD vaccine. Using this method, they found that the epitope containing T579 made a negative contribution. Interestingly, the introduction of a glycan through T579N mutation resulted in a significant enhancement of immunogenicity to elicit higher titers of nAbs.
Similarly, our group has proposed a concept termed the “broad neutralizing immunogenicity index” (BNII). This involves modifying the RBD vaccine by either removing the glycan(s) in the conserved region to expose the neutralizing epitopes or masking the non-neutralizing epitope(s) in the immunodominant region with glycan(s). In brief, mice are immunized with the optimized RBDs with removed or added glycan(s), and the mouse sera are then collected for testing the neutralizing activity against divergent SARS-CoV-2 mutants or different β-CoV-B strains. Finally, BNII is calculated based on the number of viral variants to be neutralized. The vaccines with higher BNII values, indicating greater potential to induce broad-spectrum nAbs, will be designed and developed.
To develop an effective and broad-spectrum vaccine, appropriate immunization routes and adjuvants are also indispensable (Zhang et al., 2014). Adjuvants can effectively enhance the immune response and reduce the amount of antigen used. Commonly used vaccine adjuvants, such as alum and MF59, are usually administered through intramuscular or intradermal injection. However, through intranasal administration, studies have shown that RBD-based vaccines can induce a stronger systemic cellular immune response and a higher local mucosal immune response in mice lungs compared with subcutaneous injection (Ma et al., 2014). Previous studies demonstrated that the stimulator of interferon genes (STING) in cells can effectively enhance the production of type I interferon and mimic the immune response process caused by viral infection (Ablasser et al., 2013). However, the delivery of STNG agonists to the cytoplasm of lung epithelial cells (AECs) without destroying the integrity of the pulmonary surfactant (PS) layer is still challenging. In recent studies, PS biomimetic liposomes (PS-GAMP) were utilized to encapsulate the natural and effective STING agonist 2′,3′-cyclic guanosine monophosphate-adenosine monophosphate (cGAMP) (Li et al., 2013; Wu et al., 2013). Owing to the similarity between PS-GAMP and PS, PS-GAMP could escape immune surveillance after intranasal immunization, enter alveolar macrophages (AMs), and flow into AECs through the gap junctions between AECs and AMs, thus activating the STING pathway in AMs and AECs without destroying the PS and the alveolar epithelial barrier. Further studies have confirmed that the combination of adjuvant PS-GAMP and inactivated H1N1 vaccine could generate broad-spectrum cross-protection. Within 2 days after a single immunization, it not only effectively combated H1N1, but also different influenza virus subtypes, including H3N2, H5N1, and H7N9 (Wang et al., 2020c). Therefore, PS-GAMP is a promising “universal” mucosal adjuvant, which can be adopted to develop pan-β-CoV-B vaccines.
Research and Development of Pan-β-CoV-BnAbs
Because of the presence of high-titer nAbs in convalescent patients, convalescent plasma therapy has been applied in clinics to treat several contagious diseases, including SARS, MERS, and COVID-19. However, it is limited by the availability of patient plasma, thus calling for research and development of nAbs against SARS-CoV-2 and SARS-CoV (Krishna et al., 2020).
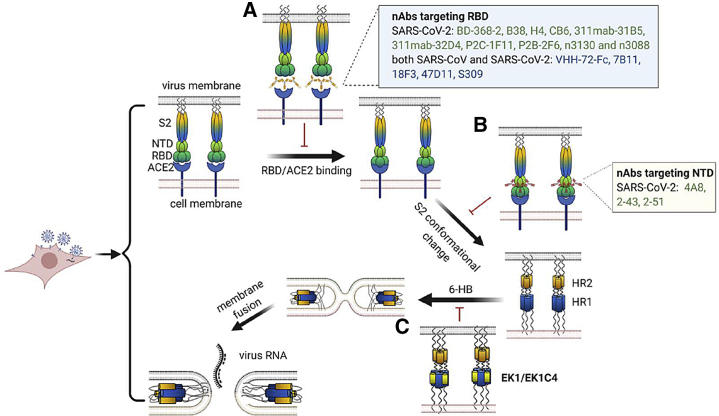
We have previously shown that the RBD contains the main neutralizing epitopes in the S protein of SARS-CoV and that these epitopes can elicit a series of potent nAbs in RBD-immunized animals (He et al., 2005b, 2006a, 2006b, 2006c). A number of these SARS-CoV RBD-specific nAbs, such as 7B11 and 18F3, could cross-neutralize SARS-CoV-2 infection (Tai et al., 2020c) (Figure 3; Table 2). Mechanistic studies have demonstrated that 7B11 neutralizes SARS-CoV and SARS-CoV-2 by recognizing epitopes close to the ACE2-binding site, thus preventing RBD-ACE2 binding. Interestingly, 18F3 could recognize the conserved region of RBD, even though its epitope does not overlap with the ACE2-binding site in RBD (Tai et al., 2020c).
Figure 3.
Mechanisms of Pan-β-CoV-B nAbs and Peptide-Based Pan-CoV Fusion Inhibitors
(A) An RBD-specific pan-β-CoV-B nAb can bind to RBD and block RBD/ACE2 binding with or without mimicking ACE2.
(B) An NTD-specific pan-β-CoV-B nAb may interfere with the conformational change of the S2 subunit and exposes the HR1 and HR2.
(C) HR1 and HR2 interact to form 6-HB, bringing the cell and virus membranes together for fusion. EK1/EK1C4 can bind with HR1 and block viral HR1 and HR2 interaction to form 6-HB. Illustration created by the authors using http://www.biorender.com.
Table 2.
Representative β-CoV-B Antibodies with Cross-Neutralizing Activity against SARS-CoV-2, SARS-CoV, and SARSr-CoVs
| Name | Identification Method | Target | Neutralizing Activities | Ref. |
|---|---|---|---|---|
| VHH-72 and bivalent VHH72-Fc | immunizing llamas with prefusion-stabilized β-CoV S proteins | RBD | VHH-72: PsV SARS-CoV: IC50 = ~3 μg/mL PsV SARS-CoV-2: IC50 = ~10 μg/mL VHH72-Fc: PsV SARS-CoV-2: IC50 = ~0.2 μg/mL |
Wrapp et al. (2020) |
| 7B11 | immunizing mice with SARS-CoV RBD-Fc fusion protein | RBD | PsV SARS-CoV: ~60% neutralization at 10 μg/mL; PsV SARS-CoV-2: 80% neutralization at 10 μg/mL |
Tai et al. (2020c) |
| 18F3 | immunizing mice with SARS-CoV RBD-Fc fusion protein | RBD | PsV SARS-CoV: ~60% neutralization at 10 μg/mL; PsV SARS-CoV-2: 80% neutralization at 10 μg/mL |
Tai et al. (2020c) |
| 47D11 | immunizing transgenic H2L2 mice with SARS-CoV S protein | RBD | PsV SARS-CoV: IC50 = 0.061 μg/mL PsV SARS-CoV-2: IC50 = 0.061 μg/mL live SARS-CoV: IC50 = 0.19 μg/mL live SARS-CoV-2: IC50 = 0.57 μg/mL |
Wang et al. (2020a) |
| S309 | identified from memory B cells of an individual infected with SARS-CoV | RBD | PsV SARS-CoV: IC50 values between 0.12 and 0.18 μg/mL PsV SARS-CoV-2: IC50 = ~0.14 μg/mL live SARS-CoV-2: IC50 = 0.08 μg/mL PsV SARSr-CoV: 80% neutralization at 10 μg/mL |
Pinto et al. (2020b) |
| ADI-55689 ADI-56046 |
identified from memory B cell repertoire from a convalescent SARS donor | RBD | PsV SARS-CoV-2: IC50 = 0.05–1.4 μg/mL PsV SARS-CoV: IC50 = 0.004–0.06 μg/mL |
Wec et al. (2020) |
Using the B cell-based Epstein-Barr virus transformation system and the SARS-CoV S-RBD as antigen, Zhu et al. (2007) have identified a human nAb, m396, which is highly potent in neutralizing SARS-CoV strains GD03, Urbani, and Tor2 isolated from infected patients, as well as strains SZ3 and SZ16 isolated from palm civets. However, m396 could not bind to the RBD of the SARS-CoV-2 S protein and had no cross-neutralizing activity against SARS-CoV-2 infection (Tian et al., 2020).
By screening the semisynthetic antibody phage display libraries, van den Brink et al. (2005) identified an RBD-specific human nAb, CR3014, which could effectively block RBD-ACE2 interaction and neutralize SARS-CoV infection at nM concentration. This nAb could efficiently protect ferrets against challenge of SARS-CoV at high dose (ter Meulen et al., 2004). The combination of two noncompeting human nAbs, CR3014 and CR3022, exhibited synergistic antiviral effect, resulting in the control of immune escape and extended breadth of protection. This synergism may allow for a lower total antibody dose for use in passive immune prophylaxis of SARS (ter Meulen et al., 2006). Interestingly, CR3022 bound potently with the SARS-CoV-2 RBD, as determined by ELISA and bio-layer interferometry (BLI), while CR3014 was unable to bind the SARS-CoV-2 RBD (Tian et al., 2020). However, CR3022 could not cross-neutralize SARS-CoV-2 at a concentration as high as 400 μg/mL. Although CR3022 can bind to a highly conserved, but cryptic, epitope in the RBD distal from the receptor-binding site between SARS-CoV and SARS-CoV-2, it binds the RBD of SARS-CoV more tightly than that of SARS-CoV-2 because the epitope in the RBD of SARS-CoV contains a glycan not present in the RBD of SARS-CoV-2. This may partly explain why CR3022 could cross-react with, but not cross-neutralize, SARS-CoV-2 infection in vitro (Yuan et al., 2020).
Screening from two non-immune phage libraries of human antibodies, Sui et al. (2004) identified eight recombinant human single-chain variable fragments (scFvs). One of them, 80R scFv, could compete with soluble ACE2 for association with the S1 domain and bound S1 with high affinity. The 80R IgG1 could effectively neutralize infection of SARS-CoV strains Tor2 and SZ2 and protected mice from SARS-CoV infection (Sui et al., 2005). Meanwhile, the 80R scFv could neither cross-react with, nor cross-neutralize, SARS-CoV-2 (Tian et al., 2020).
Cao et al. (2020) identified 14 potent neutralizing monoclonal antibodies (mAbs) from 60 convalescent COVID-19 patients by high-throughput single-cell RNA and VDJ (V, D and J genes in IgG heavy chain) sequencing of antigen-enriched B cells. Among them, an antibody designated BD-368-2 proved to be the most potent nAb against pseudotyped and live SARS-CoV-2 infection with half maximal inhibitory concentration (IC50) values of 1.2 and 1.5 ng/mL, respectively. In addition, BD-368-2 has good therapeutic and preventive effects in human ACE2 (hACE2) transgenic mice through binding to the RBD epitope which overlaps the ACE2 binding site. Similarly, two mAbs, B38 and H4, isolated from convalescent COVID-19 patients, could compete with ACE2 to bind RBD with IC50 values of 0.18 and 0.9 μg/mL against live SARS-CoV-2 infection (BetaCoV/Shenzhen/SZTH-003/2020), respectively (Wu et al., 2020b). Both B38 and H4 were able to reduce lung viral load significantly in hACE2 transgenic mice after a single administration. Notably, they recognize different epitopes on RBD, suggesting that they could be combined for clinical use.
Another nAb, named CB6, isolated from a convalescent COVID-19 patient (Shi et al., 2020), could also block RBD-ACE2 binding and showed potent and specific neutralizing activities against pseudotyped and live SARS-CoV-2 in vitro. In vivo results showed that CB6 could prevent rhesus macaques from contracting SARS-CoV-2 infection. Moreover, 311mab-31B5 and 311mab-32D4 (Chen et al., 2020), with heavy-chain variable region (VH) and light-chain variable region (VL) of single memory B cell IgG derived from three COVID-19 recovered patients, could block SARS-CoV-2 RBD interaction with ACE2 and efficiently neutralize pseudotyped SARS-CoV-2. Another study reported 206 RBD-specific mAbs derived from single B cells of 8 patients infected with SARS-CoV-2 (Ju et al., 2020). Two antibodies, P2C-1F11 and P2B-2F6, from patient 2, were able to neutralize pseudotyped SARS-CoV-2, as well as live SARS-CoV-2, with only P2B-2F6 competing with ACE2 to bind S protein directly. Antibodies HTS0422, HTS0433, HTS0446, and HTS0483, reported by Lou et al. (2020), inhibited interaction between RBD and ACE2 and neutralized five SARS-CoV-2 variants with RBD mutations, including R408I, W463R, N354D, V367F, and N354D/D364Y.
The single-domain antibody, also known as VHH, or nanobodies from camelid immunoglobulins, are promising antiviral therapeutic proteins because of their small size and numerous epitopes (Muyldermans, 2013; Wu et al., 2017). Wu et al. (2020a) developed a phage-displayed single-domain antibody library by grafting naive complementarity-determining regions into framework regions of a human germline immunoglobulin heavy-chain variable region allele and found that two antibodies, n3130 and n3088, could bind the RBD, overlapping with the CR3022 epitope, and inhibit SARS-CoV-2 pseudovirus infection. Furthermore, n3130 and n3088 could neutralize two live SARS-CoV-2 strains with IC50 values of 4.0 and 2.6 μg/mL, respectively.
While SARS-CoV-2 RBD is responsible for recognizing and binding ACE2, the function of the NTD in the S1 subunit is still elusive. It was reported that the NTD in a given CoV S protein may recognize specific glycans on the cell membrane surface to initiate viral attachment (Lu et al., 2015). Similar to the RBD, the NTD is quite immunogenic and may contain neutralizing epitope(s) (Lu et al., 2015). Chi et al. (2020) successfully isolated nAb 4A8 from peripheral blood mononuclear cell (PBMC) of COVID-19 convalescent patients using fluorescence-activated cell sorting. This nAb could bind SARS-CoV-2-NTD and neutralize both pseudotyped and live SARS-CoV-2 infection, possibly by restraining the conformational changes of S protein. Liu et al. (2020a) have isolated 19 nAbs against SARS-CoV-2 from 5 COVID-19 patients. Nine of them can neutralize live SARS-CoV-2 (strain USA-WA1/2020) infection with IC50 values ranging from 0.7 to 9 ng/mL, with four of them (2-15, 2-7, 1-57, and 1-20) targeting the RBD, three of them (2-17, 5-24, and 4-8) targeting the NTD, and two of them (2-43 and 2-51) targeting the undetermined regions on the S protein trimer. However, the mechanism underlying the neutralization of SARS-CoV-2 infection by NTD-targeting antibodies remains elusive.
Interestingly, VHH-72, a single-domain camelid antibody, which was isolated from SARS-CoV and MERS-CoV S protein-immunized llamas (Wrapp et al., 2020), showed reactivity with both SARS-CoV and SARS-CoV-2 RBD, the epitope of which overlaps with that of CR3022. After fusion with Fc, the bivalent VHH-72-Fc fusion protein could neutralize SARS-CoV-2 pseudovirus with an IC50 value of approximately 0.2 μg/mL (Table 2). Therefore, this engineered bivalent nAb targeting the conserved epitopes in RBDs of SARS-CoV and SARS-CoV-2 with cross-neutralizing activity against both SARS-CoV and SARS-CoV-2 has good potential to be developed as a pan-β-CoV-B nAb for clinical use.
The nAb 47D11 derived from transgenic H2L2 mice immunized with SARS-CoV S protein could bind to RBDs of SARS-CoV and SARS-CoV-2 with high affinity (Wang et al., 2020a). Moreover, it could cross-neutralize both SARS-CoV and SARS-CoV-2 with IC50 values of 0.2 and 0.6 μg/mL, respectively. However, the neutralization mechanism is unclear and requires further studies. Pinto et al. (2020b) have reported that S309, which binds a region distinct from that of the receptor-binding motif on RBD, can neutralize SARS-CoV-2 with an IC50 value of 79 ng/mL. Structural analysis revealed that S309 recognizes a 24-residue, glycan-containing epitope on SARS-CoV and that 19 residues are strictly conserved in SARS-CoV-2 (Pinto et al., 2020a). S309 can also recognize residues conserved within sarbecovirus isolated from human and animals, indicating its promise as a candidate for development as a broad-spectrum nAb against coronaviruses.
Peptide-Based Pan-CoV Fusion Inhibitors
It has been reported that peptides derived from the HR2 region in gp41 of HIV-1 or S2 protein of a CoV (e.g., SARS-CoV, MERS-CoV, or SARS-CoV-2) can interact with the HR1 region of the corresponding virus to form a 6-HB fusion core, thus effectively inhibiting viral S protein-mediated membrane fusion (Jiang et al., 1993; Liu et al., 2004; Lu et al., 2014; Wild et al., 1994; Xia et al., 2020d). However, none of these peptides can inhibit the infection of heterologous coronaviruses. To identify a peptide-based pan-CoV fusion inhibitor, our group designed and tested a series of peptides derived from the HR1 and HR2 regions of two α-CoVs (HCoV-NL63 and HCoV-229E) and three β-CoVs (SARS-CoV, MERS-CoV, and HCoV-OC43), including NL63-HR1P, NL63-HR2P, 229E-HR1P, 229E-HR2P, SARS-HR1P, SARS-HR2P, MERS-HR1P, MERS-HR2P, OC43-HR1P, and OC43-HR2P. Surprisingly, we found that only peptide OC43-HR2P was highly effective in inhibiting the cell-cell fusion mediated by S protein of all the above HCoVs. We then modified the sequence of OC43-HR2P by introducing double EK mutations to allow them to form stable salt bridges. One of the mutant peptides, EK1, exhibited improved potency over OC43-HR2P against infection by pseudotyped human coronaviruses, including SARS-CoV, MERS-CoV, HCoV-NL63, HCoV-229E, and HCoV-OC43. In animal models, EK1 exhibited in vivo preventive and therapeutic effect on HCoV-OC43 and MERS-CoV infection with no side effects at concentrations as high as 100 mg/kg. Also, EK1 could be widely distributed in the upper and lower respiratory tracts through nasal administration, thus allowing the prevention and treatment of coronavirus infections through the lower respiratory tract (Xia et al., 2019).
It has been reported that lipid modification can effectively improve the antiviral activity of peptides (Chong et al., 2017; Mathieu et al., 2018). Xia et al. (2020c) modified the EK1 peptide at its C terminus with palmitic acid (EK1P) or cholesterol (EK1C) and proved that EK1C had higher inhibitory activity against SASRS-CoV-2 than EK1P. Based on the sequence of EK1C, seven derivative peptides (EK1C1-EK1C7) were designed and synthesized by adding different lengths of GSG or PEG linkers between EK1 and cholesterol. Among them, EK1C4 showed the highest anti-SARS-CoV-2 activity. Its IC50 values for inhibition of spike protein-mediated cell fusion and pseudotyped and live SARS-CoV-2 infection were 1.3, 15.8, and 36.5 nM, respectively, which are 241-, 150-, and 67-fold more potent than that of EK1. EK1C4 is also much more potent than EK1 against infection of other human coronaviruses, including SARS-CoV, MERS-CoV, HCoV-OC43, HCoV-NL63, and HCoV-229E, as well as infection of two bat coronavirus, SARSr-WIV1 and SARSr-Rs3367 (Figure 3). These results suggest that EK1C4 is a highly potent pan-CoV fusion inhibitor with potential to be further developed for prevention and treatment of highly pathogenic emerging and re-emerging coronavirus infections (Wang et al., 2021).
Conclusion and Perspective
Although a variety of vaccine candidates, nAbs, and inhibitors against SARS-CoV-2 infection have been reported up to now, very few have been tested clinically. Even if they are effective against the SARS-CoV-2 now circulating worldwide, they may be unable to contain pandemics caused by SARS-CoV-2 with significant mutations in the key functional domains of SARS-CoV-2 proteins. They may also prove ineffective against newly emerging or re-emerging coronaviruses in the future. Therefore, development of broad-spectrum anti-CoV vaccines and therapeutics is urgently needed.
We first discussed the research and development of vaccines against pan-β-CoV-B viruses, including SARS-CoV-2, SARS-CoV, and bat-SARSr-CoVs. We and others have shown that RBD-based vaccines are able to induce significantly stronger nAb responses than S protein- or viral particle-based vaccines since RBD contains the main nAb epitopes in S protein (Chen et al., 2005; He et al., 2004, 2005a, 2005b; Yang et al., 2020a). Therefore, RBD-based vaccines are more effective in eliciting pan-β-CoV-B nAbs (Tai et al., 2020a) with correspondingly greater potential for development as pan-β-CoV-B vaccines (Liu et al., 2020b; Tai et al., 2020b; Zang et al., 2020; Zhu et al., 2020) (Table 1). In addition, two new concepts, NII and BNII, can be used to optimize the structure of the RBD by either removing glycan(s) at the conserved region to expose the neutralizing epitopes (Chen et al., 2014) or mask the non-neutralizing epitope(s) in the immunodominant region with glycan(s) (Du et al., 2016) to optimize the structure of the RBD for induction of stronger cross-nAb responses against pan-β-CoV-B viruses.
We then described the research and development of pan-β-CoV-B nAbs. So far, most nAbs against SARS-CoV and SARS-CoV-2 have targeted different conformational epitopes in their respective RBDs (Chen et al., 2020; He et al., 2006a; He et al., 2006b; He et al., 2006c; He et al., 2005b; Ju et al., 2020; Pinto et al., 2020b; Sui et al., 2004; Tai et al., 2020c; ter Meulen et al., 2006; Wang et al., 2020a; Zhu et al., 2007). Interestingly, many SARS-CoV RBD-specific nAbs, such as 47D11 (Wang et al., 2020a), S309 (Pinto et al., 2020b), ADI-55689 (Wec et al., 2020), and 7B11 (Tai et al., 2020c), could potently cross-neutralize SARS-CoV-2 infection (Table 2) (Yu et al., 2020). However, only a few SARS-CoV-2 RBD-specific nAbs, such as CC6.33 (Rogers et al., 2020), COVA2-02 (Brouwer et al., 2020), and COV2-2514 (Zost et al., 2020), could weakly cross-neutralize SARS-CoV infection (Jiang et al., 2020b). These results suggest that these nAbs may target a communal epitope on the RBD in S proteins of SARS-CoV and SARS-CoV-2, which could then be used for the development of more effective pan-β-CoV-B nAbs. Engineering bivalent nAbs targeting RBDs of SARS-CoV and SARS-CoV-2, such as VHH72-Fc (Wrapp et al., 2020), with cross-neutralizing activity against both SARS-CoV and SARS-CoV-2, is another approach toward the development of pan-β-CoV-B nAbs. Alternatively, a pan-β-CoV-B nAb cocktail can be designed by combining nAbs targeting different neutralizing epitopes in the RBD of SARS-CoV-2 or targeting the neutralizing epitopes in the RBDs of SARS-CoV and SARS-CoV-2. For example, a SARS-CoV-2 nAb cocktail (REGN-COV2), which consists of REGN10933 and REGN10987, that target different neutralizing epitopes in RBD, is highly effective against SARS-CoV-2 infection and in preventing viral escape mutations (Baum et al., 2020; Hansen et al., 2020).
Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs), have been widely applied in development of human disease modeling. Since hPSCs are able to differentiate into functional human cells, tissues, and organoids (Yang et al., 2020b), they are more reliable than the commonly used immortalized cell lines and more convenient and economic than animal models for drug discovery and evaluation. Several studies have shown that the organoids of blood vessel, intestine, kidney, brain, eye, lung, and liver can be infected by SARS-CoV-2 (Dickson, 2020; Makovoz et al., 2020; Ramani et al., 2020; Tindle et al., 2020; Yang et al., 2020b; Zhang et al., 2020a), and therefore they can be utilized for evaluation of nAbs and antiviral agents against SARS-CoV-2 infection. Monteil et al. (2020) have demonstrated that clinical-grade soluble human ACE2, which has a similar action mechanism as some nAbs, can inhibit SARS-CoV-2 infection in hiPSC-derived capillary organoids and hESC-derived kidney organoids. Similarly, a humanized decoy antibody (ACE2-Fc fusion protein) was reported to effectively block SARS-CoV-2 infection both in ACE2-expressing lung cells and lung organoids (Huang et al., 2020). Interestingly, Zhou et al. (2020a) have reported that both bat and human intestinal organoids can be infected by SARS-CoV-2, suggesting that the human intestinal tract is possibly a transmission route of SARS-CoV-2.
Finally, we summarized the research and development of pan-CoV fusion inhibitors. Since HR1 and HR2 sequences in the S2 subunit are highly conserved among coronaviruses, they can serve as important targets for the development of pan-CoV fusion inhibitors (Xia et al., 2019, 2020b, 2020d). Most recently, our group has successfully designed and identified a series of peptide-based pan-CoV fusion inhibitors, such as EK1 and EK1C4, which are highly effective in inhibiting infection against pseudotyped human coronaviruses tested, including SARS-CoV, MERS-CoV, SARS-CoV-2, HCoV-NL63, HCoV-229E, and HCoV-OC43, as well as some SARSr-CoVs, and against some live human coronaviruses, such as MERS-CoV, SARS-CoV-2, HCoV-NL63, HCoV-229E, and HCoV-OC43 (Xia et al., 2020a, 2020b, 2020d). Compared with antibody-based drugs with the high cost of production and transportation, peptide-based drugs are relatively economical and convenient to produce, as well as easy to store and transport at regular temperature. On October 2, 2020, New York-based Regeneron Pharmaceuticals confirmed that the company provided a single 8-g dose of nAb cocktail (REGN-COV2) to US President Donald J. Trump, who tested positive for SARS-CoV-2 (https://www.precisionvaccinations.com/8-gram-dose-covid-19-antibody-cocktail-provided-president-trump). Considering the estimated cost (US$2,593 for 200 mg/1.33 mL) of Trogarz intravenous solution (https://www.drugs.com/price-guide/trogarzo), the only antiviral antibody approved for use in clinics to treat HIV infection, the cost of a single 8-g dose of REGN-COV2, in comparison, could meet, or exceed, US$100,000. Therefore, most people in developed countries, such as the US, cannot afford it, not to mention nationals of developing countries. However, a 90-mg dose of Fuzeon (T20 peptide-based HIV fusion inhibitor) subcutaneous powder for injection costs only US$62.55 (https://www.drugs.com/price-guide/fuzeon#). Based on the half maximal effective nanomolar concentration, EK1C4 against SARS-CoV-2 infection is more effective than Fuzeon against HIV infection. Therefore, daily usage of EK1C4 is expected to be less than that of Fuzeon with the attendant cost implications. Also different from Fuzeon, EK1C4 can only be used for 2–3 weeks at the early stage of SARS-CoV-2 infection. We believe that governments of some countries, such as China, may be able to provide peptide drugs (e.g., EK1C4) to patients at low cost, or even for free, during the pandemic stage.
Therefore, pan-β-CoV-B vaccines and nAbs, as well as pan-CoV fusion inhibitors, have the necessary potential to be developed as broad-spectrum vaccines and therapeutics to combat the current COVID-19 pandemic and future outbreaks of emerging and re-emerging coronavirus diseases.
Author Contributions
M.C. and X.S. performed literature search, prepared tables and figures, and wrote the manuscript. S.J. conceived the project and revised the manuscript. All authors reviewed the manuscript and all approved of the final version.
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (82041025 to S.J.).
References
- Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Rohl I., Hopfner K.P., Ludwig J., Hornung V. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses. 2020;12 doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink E.N., Ter Meulen J., Cox F., Jongeneelen M.A., Thijsse A., Throsby M., Marissen W.E., Rood P.M., Bakker A.B., Gelderblom H.R. Molecular and biological characterization of human monoclonal antibodies binding to the spike and nucleocapsid proteins of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:1635–1644. doi: 10.1128/JVI.79.3.1635-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhang L., Qin C., Ba L., Yi C.E., Zhang F., Wei Q., He T., Yu W., Yu J. Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J. Virol. 2005;79:2678–2688. doi: 10.1128/JVI.79.5.2678-2688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.H., Du L.Y., Chag S.M., Ma C.Q., Tricoche N., Tao X.R., Seid C.A., Hudspeth E.M., Lustigman S., Tseng C.T.K. Yeast-expressed recombinant protein of the receptor-binding domain in SARS-CoV spike protein with deglycosylated forms as a SARS vaccine candidate. Hum. Vaccine Immunother. 2014;10:648–658. doi: 10.4161/hv.27464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Li R., Pan Z., Qian C., Yang Y., You R., Zhao J., Liu P., Gao L., Li Z. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol. Immunol. 2020;17:647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong H., Xue J., Xiong S., Cong Z., Ding X., Zhu Y., Liu Z., Chen T., Feng Y., He L. A lipopeptide HIV-1/2 fusion inhibitor with highly potent in vitro, ex vivo, and in vivo antiviral activity. J. Virol. 2017;91 doi: 10.1128/JVI.00288-17. e00288–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Shots of hope. Science. 2020;370:1392–1394. doi: 10.1126/science.370.6523.1392. [DOI] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming D., Sheahan T., Heise M., Yount B., Davis N., Sims A., Suthar M., Harkema J., Whitmore A., Pickles R. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 2006;3:e525. doi: 10.1371/journal.pmed.0030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson I. Organoids demonstrate gut infection by SARS-CoV-2. Nat. Rev. Gastroenterol. Hepatol. 2020;17:383. doi: 10.1038/s41575-020-0317-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020 doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Tai W., Yang Y., Zhao G., Zhu Q., Sun S., Liu C., Tao X., Tseng C.K., Perlman S. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines. Nat. Commun. 2016;7:13473. doi: 10.1038/ncomms13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erasmus J.H., Khandhar A.P., O'Connor M.A., Walls A.C., Hemann E.A., Murapa P., Archer J., Leventhal S., Fuller J.T., Lewis T.B. An alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci. Transl. Med. 2020;12:eabc9396. doi: 10.1126/scitranslmed.abc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Wang Q., Shan C., Yang C., Feng Y., Wu J., Liu X., Zhou Y., Jiang R., Hu P. An adenovirus-vectored COVID-19 vaccine confers protection from SARS-COV-2 challenge in rhesus macaques. Nat. Commun. 2020;11:4207. doi: 10.1038/s41467-020-18077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324:773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhu Q., Liu S., Zhou Y., Yang B., Li J., Jiang S. Identification of a critical neutralization determinant of severe acute respiratory syndrome (SARS)-associated coronavirus: importance for designing SARS vaccines. Virology. 2005;334:74–82. doi: 10.1016/j.virol.2005.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.X., Lu H., Siddiqui P., Zhou Y.S., Jiang S.B. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J. Immunol. 2005;174:4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- He Y., Li J., Du L., Yan X., Hu G., Zhou Y., Jiang S. Identification and characterization of novel neutralizing epitopes in the receptor-binding domain of SARS-CoV spike protein: revealing the critical antigenic determinants in inactivated SARS-CoV vaccine. Vaccine. 2006;24:5498–5508. doi: 10.1016/j.vaccine.2006.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Li J., Heck S., Lustigman S., Jiang S. Antigenic and immunogenic characterization of recombinant baculovirus-expressed severe acute respiratory syndrome coronavirus spike protein: implication for vaccine design. J. Virol. 2006;80:5757–5767. doi: 10.1128/JVI.00083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Li J., Li W., Lustigman S., Farzan M., Jiang S. Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein. J. Immunol. 2006;176:6085–6092. doi: 10.4049/jimmunol.176.10.6085. [DOI] [PubMed] [Google Scholar]
- Ho M.-S. Severe acute respiratory syndrome (SARS) In: Guerrant R., Walker D.H., Weller P.F., editors. Tropical Infectious Diseases: Principles, Pathogens and Practice. 3rd ed. Saunders Elsevier; Philadelphia: 2011. pp. 392–397. [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L.A., Kaelin E.A., Maqsood R., Estifanos B., Wu L.I., Varsani A., Halden R.U., Hogue B.G., Scotch M., Lim E.S. An 81 nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (Jan–Mar 2020) J. Virol. 2020;94:e00711-20. doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.Y., Lin M.S., Kuo T.C., Chen C.L., Lin C.C., Chou Y.C., Chao T.L., Pang Y.H., Kao H.C., Huang R.S. Humanized COVID-19 decoy antibody effectively blocks viral entry and prevents SARS-CoV-2 infection. EMBO Mol. Med. 2020:e12828. doi: 10.15252/emmm.202012828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S. Don't rush to deploy COVID-19 vaccines and drugs without sufficient safety guarantees. Nature. 2020;579:321. doi: 10.1038/d41586-020-00751-9. [DOI] [PubMed] [Google Scholar]
- Jiang S., Lin K., Strick N., Neurath A.R. HIV-1 inhibition by a peptide. Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- Jiang S., Du L., Shi Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China: calling for developing therapeutic and prophylactic strategies. Emerg. Microbes Infect. 2020;9:275–277. doi: 10.1080/22221751.2020.1723441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Zhang X., Du L. Therapeutic antibodies and fusion inhibitors targeting the spike protein of SARS-CoV-2. Expert Opin. Ther. Targets. 2020 doi: 10.1080/14728222.2020.1820482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Krishna G., Pillai V.S., Veettil M.V. Approaches and advances in the development of potential therapeutic targets and antiviral agents for the management of SARS-CoV-2 infection. Eur. J. Pharmacol. 2020;885:173450. doi: 10.1016/j.ejphar.2020.173450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp D.W., Schief W.R. Advances in structure-based vaccine design. Curr. Opin. Virol. 2013;3:322–331. doi: 10.1016/j.coviro.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Mascola J.R., Nabel G.J. Rational design of vaccines to elicit broadly neutralizing antibodies to HIV-1. Cold Spring Harb. Perspect. Med. 2011;1:a007278. doi: 10.1101/cshperspect.a007278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li X.D., Wu J., Gao D., Wang H., Sun L., Chen Z.J. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.W., Xiao G.F., Chen Y.B., He Y.X., Niu J.K., Escalante C.R., Xiong H.B., Farmar J., Debnath A.K., Tien P. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu Z., Xu W., Xia S., Gu C., Wang X., Wang Q., Zhou J., Wu Y., Cai X., Qu D. The recombinant subunit vaccine RBD-Fc, consisting of SARS-CoV-2 RBD and human IgG Fc as an immunopotentiator, elicits robust neutralizing antibody responses against SARS-CoV-2 infection. Res. Square. 2020 doi: 10.21203/rs.3.rs-61074/v1. [DOI] [Google Scholar]
- Logunov D.Y., Dolzhikova I.V., Zubkova O.V., Tukhvatullin A.I., Shcheblyakov D.V., Dzharullaeva A.S., Grousova D.M., Erokhova A.S., Kovyrshina A.V., Botikov A.G. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020;396:887–897. doi: 10.1016/S0140-6736(20)31866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y., Zhao W., Wei H., Chu M., Chao R., Yao H., Su J., Li Y., Li X., Cao Y. Cross-neutralization antibodies against SARS-CoV-2 and RBD mutations from convalescent patient antibody libraries. bioRxiv. 2020 doi: 10.1101/2020.06.06.137513. [DOI] [Google Scholar]
- Lu L., Liu Q., Du L., Jiang S. Middle East respiratory syndrome coronavirus (MERS-CoV): challenges in identifying its source and controlling its spread. Microbes Infect. 2013;15:625–629. doi: 10.1016/j.micinf.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Liu Q., Zhu Y., Chan K.H., Qin L., Li Y., Wang Q., Chan J.F., Du L., Yu F. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G.W., Wang Q.H., Gao G.F. Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.Q., Li Y., Wang L.L., Zhao G.Y., Tao X.R., Tseng C.T.K., Zhou Y.S., Du L.Y., Jiang S.B. Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: implication for designing novel mucosal MERS vaccines. Vaccine. 2014;32:2100–2108. doi: 10.1016/j.vaccine.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovoz B., Møller R., Eriksen A.Z., tenOever B.R., Blenkinsop T.A. SARS-CoV-2 infection of ocular cells from human adult donor eyes and hESC-derived eye organoids. SSRN. 2020 doi: 10.2139/ssrn.3650574. [DOI] [Google Scholar]
- Mathieu C., Porotto M., Figueira T.N., Horvat B., Moscona A. Fusion inhibitory lipopeptides engineered for prophylaxis of Nipah virus in primates. J. Infect. Dis. 2018;218:218–227. doi: 10.1093/infdis/jiy152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., Bakker A.B.H., van den Brink E.N., Weverling G.J., Martina B.E.E., Haagmans B.L., Kuiken T., de Kruif J., Preiser W., Spaan W. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363:2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., del Pozo C.H., Prosper F. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A. Structural and functional analysis of a potent sarbecovirus neutralizing antibody. bioRxiv. 2020 doi: 10.1101/2020.04.07.023903. [DOI] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A., Muller L., Ostermann P.N., Gabriel E., Abida-Islam P., Muller-Schiffmann A., Mariappan A., Goureau O., Gruell H., Walker A. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020;39:e106230. doi: 10.15252/embj.2020106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F.Z., Huang D.L., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X.Z., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D., Moldt B., Burton D.R. SnapShot: broadly neutralizing antibodies. Cell. 2013;155:728–728.e1. doi: 10.1016/j.cell.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Du L., Jiang S. Learning from the past: development of safe and effective COVID-19 vaccines. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J.H., Li W.H., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S.C., Olurinde M., Choe H. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U S A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Roberts A., Matthews L.J., Murakami A., Vogel L., Wong S.K., Subbarao K., Farzan M., Marasco W.A. Evaluation of human monoclonal antibody 80R for immunoprophylaxis of severe acute respiratory syndrome by an animal study, epitope mapping, and analysis of spike variants. J. Virol. 2005;79:5900–5906. doi: 10.1128/JVI.79.10.5900-5906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., Zhang X., Drelich A., Shi J., Hsu J.C., Luchsinger L., Hillyer C.D., Tseng C.K., Jiang S., Du L. A novel receptor-binding domain (RBD)-based mRNA vaccine against SARS-CoV-2. Cell Res. 2020;30:932–935. doi: 10.1038/s41422-020-0387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., Zhang X., He Y., Jiang S., Du L. Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2. Antiviral Res. 2020;179:104820. doi: 10.1016/j.antiviral.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindle C., Fuller M., Fonseca A., Taheri S., Ibeawuchi S.-R., Beutler N., Claire A., Castillo V., Hernandez M., Russo H. Adult stem cell-derived complete lung organoid models emulate lung disease in COVID-19. bioRxiv. 2020 doi: 10.1101/2020.10.17.344002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2020 doi: 10.1016/S0140-6736(20)32661-132661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang Y.T., Huang B.Y., Deng W., Quan Y.R., Wang W.L., Xu W.B., Zhao Y.X., Li N., Zhang J. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182:713–721 e9. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li P.Y., Yu Y., Fu Y.H., Jiang H.Y., Liu M., Sun Z.P., Jiang S.B., Lu L., Wu Y.M. Pulmonary surfactant-biomimetic nanoparticles potentiate heterosubtypic influenza immunity. Science. 2020;367 doi: 10.1126/science.aau0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shang J., Jiang S., Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front Microbiol. 2020;11:298. doi: 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xia S., Zhu Y., Lu L., Jiang S. Pan-coronavirus fusion inhibitors as the hope for today and tomorrow. Protein Cell. 2021 doi: 10.1007/s13238-020-00806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild C.T., Shugars D.C., Greenwell T.K., McDanal C.B., Matthews T.J. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U S A. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Team V.-C.C.-R., Hoffmann M. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015.e5. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Sun L., Chen X., Du F., Shi H., Chen C., Chen Z.J. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.L., Jiang S.B., Ying T.L. Single-domain antibodies as therapeutics against human viral diseases. Front Immunol. 2017;13:1802. doi: 10.3389/fimmu.2017.01802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li C., Xia S., Tian X., Kong Y., Wang Z., Gu C., Zhang R., Tu C., Xie Y. Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27:891–898.e5. doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.K., Wang Q., Du L., Tan W., Wilson I.A. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Lan Q., Su S., Wang X., Xu W., Liu Z., Zhu Y., Wang Q., Lu L., Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct. Target. Ther. 2020;5:92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu M.Q., Wang C., Xu W., Lan Q.S., Feng S.L., Qi F.F., Bao L.L., Du L.Y., Liu S.W. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhang Y., Wang Y., Wang H., Yang Y., Gao G.F., Tan W., Wu G., Xu M., Lou Z. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2020;21:39–51. doi: 10.1016/S1473-3099(20)30831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol. Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffre F. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Xiang R., Deng X., Wang L., Yu Z., Tian S., Liang R., Li Y., Ying T., Jiang S. Receptor-binding domain-specific human neutralizing monoclonal antibodies against SARS-CoV and SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5:212. doi: 10.1038/s41392-020-00318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X.Y., Lee C.C.D., So R.T.Y., Lv H.B., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang J., Gu C., Zhou B., Zhang C., Yang Y., Xu S., Zhang X., Zhou Y., Bai L., Wu Y. Immunization with the receptor-binding domain of SARS-CoV-2 elicits antibodies cross-neutralizing SARS-CoV-2 and SARS-CoV without antibody-dependent enhancement. Cell Discov. 2020;6:61. doi: 10.1038/s41421-020-00199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Jiang S., Du L. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev. Vaccin. 2014;13:761–774. doi: 10.1586/14760584.2014.912134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.Z., Chu H., Han S., Shuai H., Deng J., Hu Y.F., Gong H.R., Lee A.C., Zou Z., Yau T. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020;30:928–931. doi: 10.1038/s41422-020-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zeng G., Pan H., Li C., Hu Y., Chu K., Han W., Chen Z., Tang R., Yin W. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Liu X.J., Chiu M.C., Zhao X.Y., Wang D., Wei Y.X., Lee A.R., Zhang A.J., Chu H. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chakraborti S., He Y., Roberts A., Sheahan T., Xiao X., Hensley L.E., Prabakaran P., Rockx B., Sidorov I.A. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. U S A. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Yu D., Han Y., Yan H., Chong H., Ren L., Wang J., Li T., He Y. Cross-reactive neutralization of SARS-CoV-2 by serum antibodies from recovered SARS patients and immunized animals. Sci. Adv. 2020;6:eabc9999. doi: 10.1126/sciadv.abc9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]