Abstract
Long non-coding RNAs (lncRNAs) are involved in shaping chromosome conformation and regulation of preimplantation development. However, the role of lncRNA during somatic cell nuclear transfer (SCNT) reprogramming remains largely unknown. In the present study, we identified 114 upregulated lncRNAs in the 8-cell SCNT embryos as candidate key molecules involved in nuclear reprogramming in goat. We found that H3K4me3 was an epigenetic barrier in goat nuclear reprogramming that and injection of Kdm5b mRNA greatly improved SCNT embryos development through removal of H3K4me3. We further reported that knockdown of lnc_3712 increased the expression of Kdm5b, which led to H3K4me3 demethylation. Of note, the development of goat SCNT embryos was improved when lnc_3712 was knocked down, whereas the blastocyst rate showed no difference in lnc_3712 and Kdm5b double knockdown SCNT embryos compared with the negative control SCNT embryos. Specifically, in lnc_3712 knockdown SCNT embryos, partial of the transcriptional activity and the expression of critical embryonic genes (Wee1, Ctsb, and Ybx1) were similar with that of in vitro fertilization embryos. Therefore, our results elucidate the critical role of lnc_3712 in regulating the development of goat SCNT embryos via repressing Kdm5b, which advances our current understanding of the role of lncRNAs during nuclear reprogramming.
Keywords: somatic cell nuclear transfer, reprogramming, long non-coding RNA, Kdm5b, H3K4me3, goat
Graphical Abstract
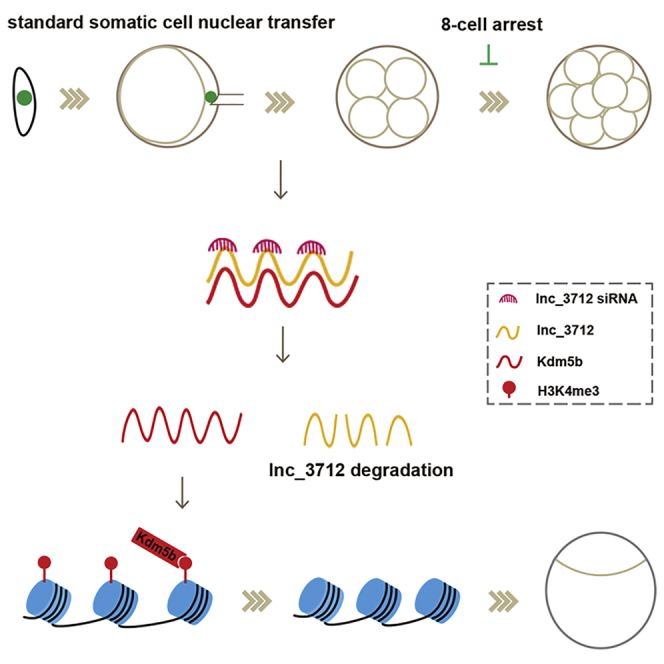
Deng and colleagues provide the lncRNA profile in goat SCNT embryos and show that knockdown of lncRNA lnc_3712 increases the expression of Kdm5b and demethylates H3K4me3, resulting in improvement of goat SCNT embryo development, which broadens the current understanding of the role of lncRNAs during the stochastic reprogramming events.
Introduction
Somatic cell nuclear transfer (SCNT) allows terminally differentiated somatic cells to be reprogrammed to a totipotent state, enabling them to generate a whole organism.1 Since the first mammal, Dolly the sheep, was cloned by SCNT, successful cloning of more than 20 mammalian species has been reported.2,3 In addition, nuclear transfer embryonic stem cells (ESCs) from aged adult and patient cells have been successfully generated.4 Therefore, the SCNT technology holds great potential in agriculture, endangered species conservation, and human therapeutics.5 Recently, nuclear-transfer ESCs from adult-aged and patient cells have been successfully generated,6,7 which expands the application of SCNT for reproductive and therapeutic cloning. Unfortunately, the low efficiency of nuclear reprogramming hampers the application of SCNT in the related fields.
Previous studies revealed that undefined epigenetic barriers preexisting in the genome of donor cells caused difficulties in zygotic/embryonic genome activation (ZGA/EGA) in SCNT-generated embryos. Developmental defects of SCNT-generated embryos first appear at the time of ZGA/EGA,8 which occurs at the 2-cell stage in mice and the 4- to 8-cell stage in human, pig, bovine, and goat.9,10 In in vitro fertilized (IVF) embryos, tri-methylation of H3 lysine 4 (H3K4me3) and H3 lysine 9 (H3K9me3) undergo dynamic change during the ZGA process,11, 12, 13 whereas H3K4me3 and H3K9me3 are retained during ZGA in SCNT-generated embryos, which have been identified as major epigenetic roadblocks of transcriptional reprogramming.8,14, 15, 16 Removal of these histone modifications greatly improved the nuclear reprogramming efficiency.8,16, 17, 18
While efforts to improve transcriptional reprogramming following SCNT have focused on preexisting histone modification in somatic cells,19 roles for non-coding RNA in the regulation of nuclear reprogramming have been fairly investigated.20,21 Long non-coding RNAs (lncRNAs) were reported to play critical roles in organizing chromatin architecture and epigenetic memory, including the recruitment of chromatin modifiers in cis or trans and modulation of histone lysine methylation and DNA methylation.22,23 lncRNA expression exhibits dynamic expression profiles during the early embryonic development of mouse, pig, goat, and human.13,24, 25, 26 Specifically, the lncRNAs interleukin 17d (il17d) and lincGET are indispensable for mouse embryos developing to late blastocyst stage.25,27,28 Our previous study reported that knockdown of lnc_137 caused development arrest at the 8-cell stage in goat.13 These studies demonstrate that lncRNA is crucial for early embryo development. Nevertheless, to date, an established contribution to nuclear reprogramming has been determined for only one unusual lncRNA, X-inactive specific transcript (Xist),29, 30, 31 in contrast with the vital roles of lncRNA during embryogenesis and enormous number in the genome.
Using RNA sequencing (RNA-seq), we identified 114 highly expressed lncRNAs in the 8-cell SCNT embryos as candidate key molecules involved in nuclear reprogramming in goat. We further reported that knockdown of lnc_3712 increased the expression of Kdm5b and demethylated H3K4me3, resulting in improvement of SCNT embryo development. Our data will broaden the current understanding of the role of lncRNAs during the stochastic reprogramming events.
Results
Dynamic changes of lncRNAs during EGA in goat SCNT embryos
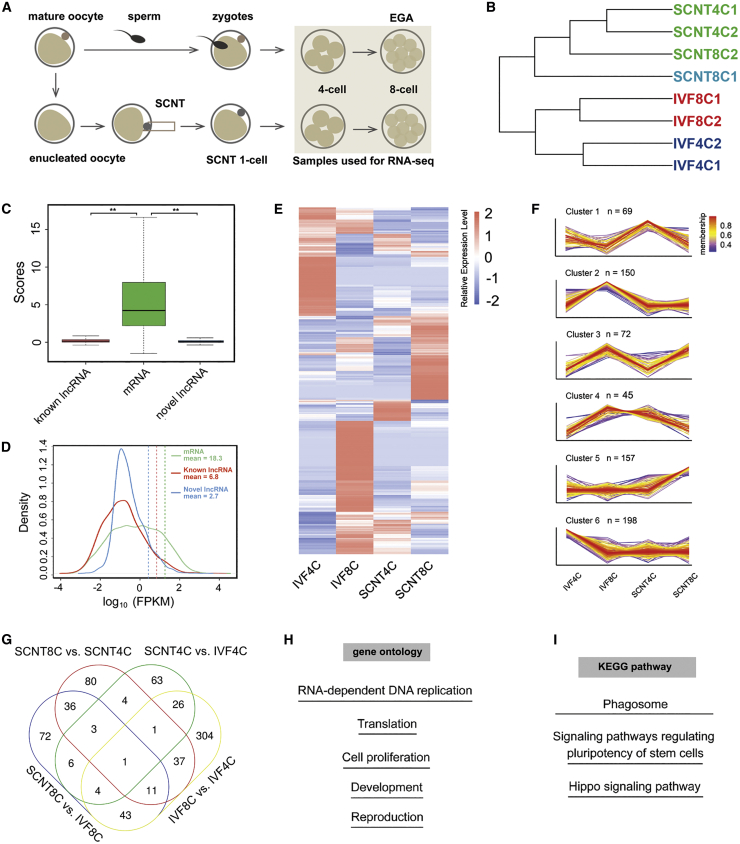
To identify the transcriptional differences of lncRNA between goat embryos derived from IVF and SCNT, we performed RNA-seq experiments using pooled embryos (10 embryos/sample) at the 4- and 8-cell stages (Figure 1A). Unsupervised hierarchical clustering showed that global lncRNA expression profiles of individual cells at the same stage are very similar, whereas a diversity of the expression profiles for different types was observed (Figure 1B; Table S1). Scores of known and novel lncRNA coding potentials were close to 0, while those of mRNA were significantly higher (Figure 1C). Fragments per kilobase of exon model per million mapped fragments (FPKM) was used to access the expression level. As shown in Figure 1D, the mean FPKM of mRNA, known lncRNA, and novel lncRNA was 18.3, 6.8, and 2.7, respectively, suggesting that the expression of lncRNA was relatively lower compared to that of mRNA.
Figure 1.
The expression profiles of lncRNAs during EGA in goat SCNT embryo
(A) Schematic illustration of the experimental procedure. (B) Unsupervised clustering of lncRNAs from the 4- and 8-cell SCNT and IVF embryos. (C) Coding potential of mRNA, known lncRNAs, and novel lncRNAs. ∗∗p < 0.01. (D) Density plot of the expression of mRNA, known lncRNAs, and novel lncRNAs. (E) Heatmap illustration showing differentially expressed lncRNAs identified by a pairwise comparison between the 4- and 8-cell IVF and SCNT embryos. (F) Six clusters of the differentially expressed lncRNAs. (G) Venn diagram of the differentially expressed lncRNAs. (H) Representative GO terms of the differentially expressed lncRNAs. (I) Representative KEGG pathways of the differentially expressed lncRNAs.
Next, we used DEseq2 to identify differentially expressed lncRNAs (|FoldChange| > 2 and p value < 0.05) during EGA of goat SCNT embryos. Compared to the 4-cell IVF embryos, 51 lncRNAs were upregulated, while 57 were downregulated in the 4-cell SCNT embryos. In the 8-cell SCNT embryos, the expression of 114 lncRNAs was increased, whereas that of 62 lncRNA was decreased compared to the IVF embryos (Figures 1E–1G; Table S2). The differentially expressed lncRNAs were further subdivided into six clusters (Figure 1F; Table S3). Specifically, 51 of the lncRNAs were both differentially expressed between the 8-cell SCNT and IVF embryos and between the 4- and 8-cell SCNT embryos (Figure 1G; Tables S1 and S2). Interestingly, Gene Ontology (GO) revealed that the differentially expressed lncRNAs were enriched in RNA-dependent DNA replication, translation, cell proliferation, development, and reproduction (Figure 1H). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed that the differentially expressed lncRNAs were enriched in phagosome, signaling pathways regulating pluripotency of stem cells, and hippo signaling pathway (Figure 1I).
Improved development of lnc_3712 knockdown SCNT embryos
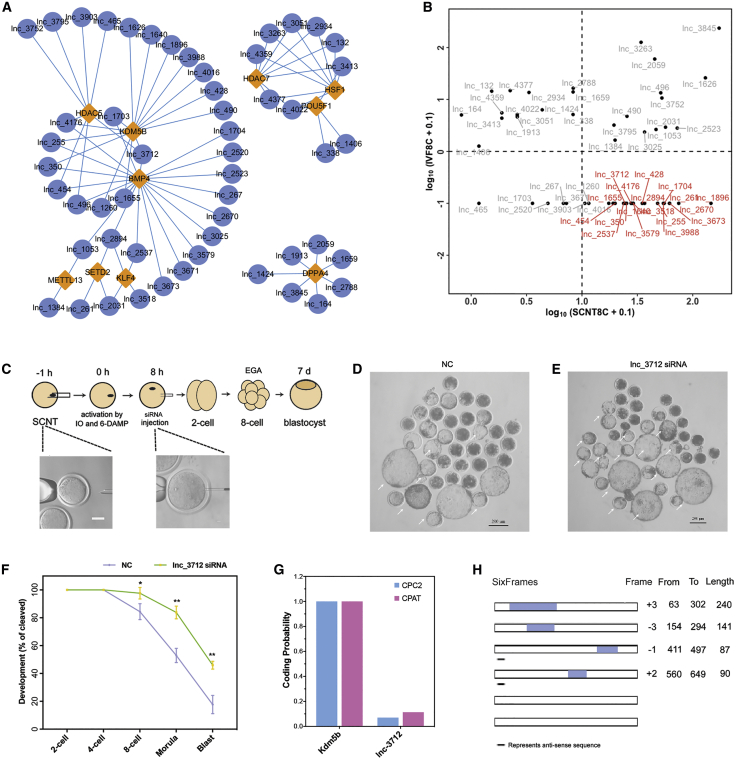
Target gene prediction for the differentially expressed lncRNAs was conducted using lncTar. We selected BMP4, KDM5B, HSF1, HDAC5, and DPPA4 as target genes, and 53 lncRNAs were predicted to target these genes (Figure 2A; Table S4). Of note, the expression of lnc_1655, lnc_255, lnc_350, lnc_3712, and lnc_4176 was significantly increased in the 8-cell SCNT embryos compared to the IVF embryos (Figure 2B). To investigate the functions of these five lncRNA transcripts, we then knocked down the expression of the five lncRNAs in SCNT embryos via injection of small interfering RNAs (siRNAs) against them (Figure 2C). No difference was observed in the percentage of blastocyst in lnc_1655, lnc_255, lnc_350, and lnc_4176 knockdown SCNT embryos compared to that of the negative control (NC) embryos (Figure S1; Tables S5 and S10). However, the percentage of blastocyst in lnc_3712 knockdown SCNT embryos was significantly increased compared to the NC embryos (Figures 2D–2F; 45.94% ± 1.3% versus 17.67% ± 3.09%; p < 0.01), suggesting that lnc_3712 impaired the development of goat SCNT embryo.
Figure 2.
Improved development of lnc_3712 knockdown SCNT embryos
(A) Representative highly correlated lncRNA-gene network. (B) Dot plot of the lncRNAs in (A); differentially expressed lncRNAs are labeled in red. (C) Schematic illustration of the microinjection procedure. (D and E) Representative images of NC and lnc_3712 knockdown embryos. Blastocysts are indicated by the white arrows. Scale bar, 200 μm. (F) Development of NC and lnc_3712 knockdown embryos. Shown are percentages of embryos that reach the indicated stages. Error bars represent SEM. ∗p < 0.05, ∗∗p < 0.01. (G) Coding potential scores of lnc_3712 and Kdm5b. (H) Prediction of putative proteins encoded by lnc_3712 using ORF Finder.
To confirm lnc_3712 is a true lncRNA transcript, we utilized the CPC2 and Coding-Potential Assessment Tool (CPAT) software to analyze the coding potential. As shown in Figure 2G, the coding probability of lnc_3712 was close to 0, while the coding probability of Kdm5b was 1. Moreover, ORF Finder revealed that the longest open reading frame of lnc_3712 was 240 bp (Figure 2H), which is less than the cutoff value (300 bp) for the frame of mRNA. These data suggest that lnc_3712 does not encode proteins.
Increased transcriptional activity in lnc_3712 knockdown SCNT embryos
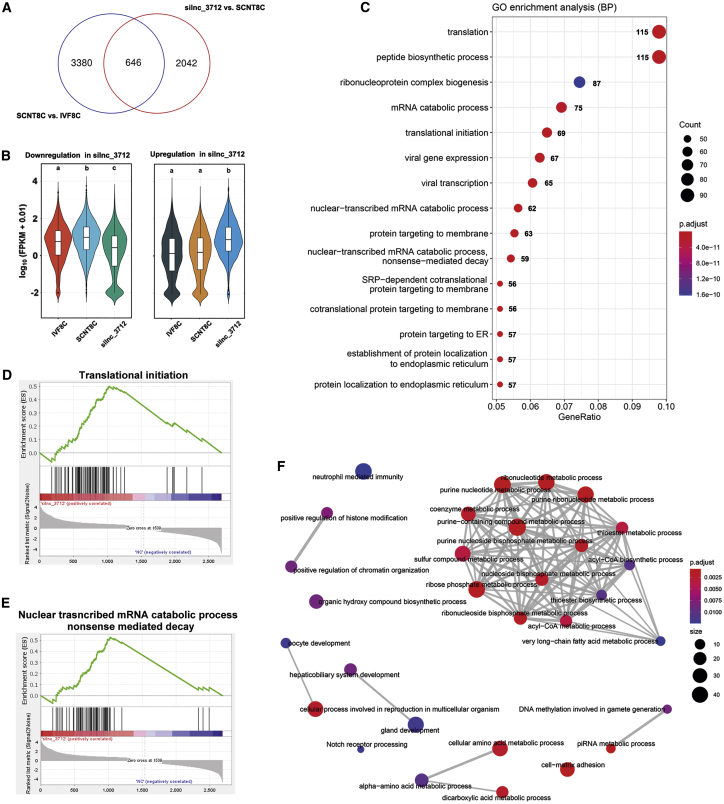
Transcriptome analysis revealed that 4,026 mRNAs were differentially expressed in the 8-cell SCNT embryos compared to the IVF embryos (Figure 3A; Table S6). In the 8-cell SCNT embryos in which lnc_3712 was knocked down, 2,688 mRNAs were differentially expressed when compared to the 8-cell NC embryos. Specifically, 646 of the differentially expressed genes (DEGs) between the 8-cell IVF and SCNT embryos were also differentially expressed in lnc_3712 knockdown SCNT embryos compared with the NC (Figure 3A; Table S7). Of the 2,688 DEGs, 1,224 DEGs were downregulated, while 1,464 DEGs were upregulated in lnc_3712 knockdown embryos compared with the SCNT controls (Figure 3B).
Figure 3.
Increased transcriptional activity in lnc_3712 knockdown SCNT embryos
(A) Venn diagram of DEGs between NC and lnc_3712 knockdown embryos and between the 8-cell SCNT and IVF embryos. (B) Violin plot of downregulated (left) and upregulated (right) DEGs in lnc_3712 knockdown embryos compared to the NC embryos. (C) GO enrichment analysis of the upregulated DEGs in lnc_3712 knockdown embryos. (D and E) GO items of the DEGs, as revealed by gene set enrichment analysis. (F) GO enrichment analysis of the downregulated DEGs in lnc_3712 knockdown embryos.
It is critical to know which pathways were repressed and de-repressed in knockdown analysis. As shown in Figure 3C, the upregulated DEGs were enriched in translation, translational initiation, mRNA catabolic membrane, nuclear-transcribed mRNA catabolic process, protein targeting to membrane and endoplasmic reticulum (ER), and protein localization to ER under biological process of GO items. Gene set enrichment analysis (GSEA) confirmed that the DEGs were enriched in translational initiation and nuclear-transcribed mRNA catabolic process (Figures 3D and 3E; Table S8). The downregulated DEGs were enriched in oocyte development, cellular processes involved in reproduction in multicellular organisms, DNA methylation involved in gamete generation, positive regulation of chromatin organization, and positive regulation of histone modification (Figure 3F). These data suggest that knockdown of lnc_3712 improved transcription of SCNT embryos during EGA, which might subsequently contribute to goat SCNT embryo development.
Restored transcriptional reprogramming in lnc_3712 knockdown SCNT embryos
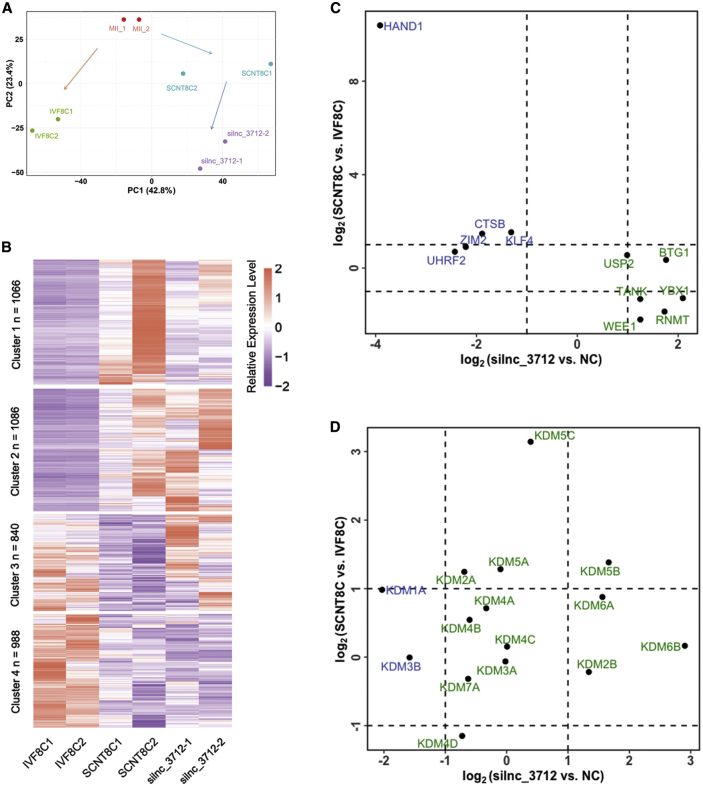
To confirm the effects of lnc_3712 knockdown on the transcription of goat SCNT embryo, we performed principal-component analysis (PCA) and constructed a heatmap of the DEGs. PCA indicated that the gene expression profile in mature oocytes, 8-cell IVF and SCNT embryos, and lnc_3712 knockdown embryos were distinct from each other in general. Moreover, the transcriptome profile of lnc_3712 knockdown embryos showed a closer relationship to that of the IVF embryo at principal component 2 (Figure 4A; Tables S7 and S8). A heatmap and series test of clusters revealed that the expression of 840 genes (cluster 3) and partial expression of genes in cluster 1 were restored in lnc_3712 knockdown embryos (Figure 4B).
Figure 4.
Restored transcriptional reprogramming in lnc_3712 knockdown SCNT embryos
(A) PCA showing the genome-wide expression road map in mature oocytes, the 8-cell IVF and SCNT embryos, and the lnc_3712 knockdown embryos. (B) Heatmap illustration revealed increased expression of genes in clusters 1 and 3 in lnc_3712 knockdown embryos. (C) The expression of critical zygotic genes in lnc_3712 knockdown embryos. (D) Boxplot of the expression of histone methylation modifiers in lnc_3712 knockdown embryos.
We next sought to investigate the expression of several genes responsible for ZGA. As shown in Figure 4C, the expression of Btg1 and Usp2 was increased in lnc_3712 knockdown embryos compared to the NC embryos but showed no statistical difference in the 8-cell SCNT and IVF embryos. The expression of Hand1, Ctsb, Klf4, and Zim2 was increased in the 8-cell SCNT embryos compared to the IVF embryos but decreased in lnc_3712 knockdown embryos compared to the NC embryos. Wee1, Rnmt, Ybx1, and Tank were downregulated in the 8-cell SCNT embryos compared to the IVF embryos but upregulated in lnc_3712 knockdown embryos compared to the NC embryos. Taken together, these data revealed that transcriptional reprogramming and the expression of three critical ZGA genes were restored in SCNT embryos in which lnc_3712 was knocked down.
Increased expression of Kdm5b in lnc_3712 knockdown SCNT embryos
Kdm1a, Kdm3a/b, and Kdm4a-d were responsible for the removal of H3K9 methylation. Expression of Kdm1a was increased (p < 0.05), while KDM4D was downregulated (p < 0.05) in the 8-cell SCNT embryos compared to the IVF embryos. However, the expression of Kdm1a (p < 0.05) and Kdm3b (p < 0.05) was decreased in lnc_3712 knockdown SCNT embryos compared to the NC embryos. Kdm2a and Kdm2b catalyzed the demethylation of H3K36. The expression of Kdm2a was increased (p < 0.05) in the 8-cell SCNT embryos compared to the IVF controls, while that of Kdm2b was upregulated (p < 0.05) in lnc_3712 knockdown SCNT embryos compared to the NC embryos. Kdm5a-c and Kdm7a are associated with the demethylation of H3K4 methylation. The expression of Kdm5a-c was increased in the 8-cell SCNT embryos compared to the IVF controls (p < 0.05). Moreover, the expression of Kdm5b was increased (p < 0.05) in lnc_3712 knockdown SCNT embryos compared to the NC embryos. The expression of Kdm6a and Kdm6b, which were able to remove H3K27 methylation, was increased (p < 0.05) in lnc_3712 knockdown embryos compared to the NC embryos (Figure 4D).
Kdm5b was a critical epigenetic factor for nuclear reprogramming
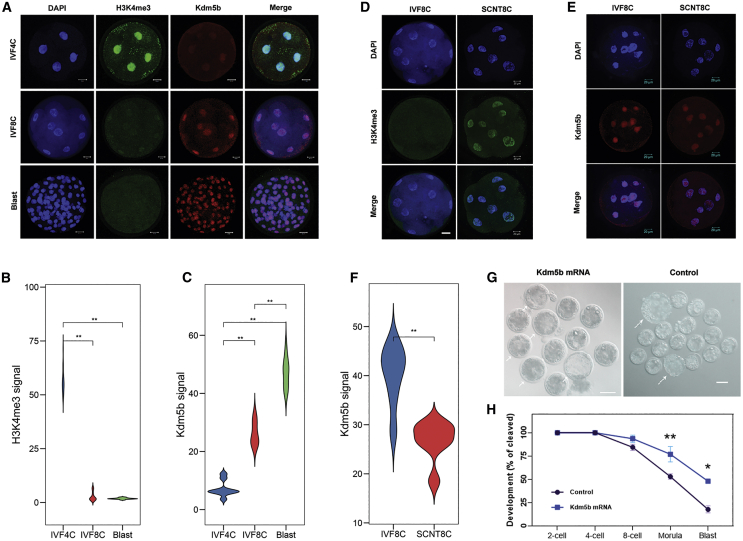
To confirm H3K4me3 is an epigenetic barrier during goat nuclear reprogramming, we next characterized the H3K4me3 modification during preimplantation development. The H3K4me3 modification was gradually removed from the 4-cell IVF embryos to blastocyst stage in goat. Meanwhile, the signal intensity of Kdm5b was increased during EGA and reached its highest level in the blastocysts (Figures 5A–5C). However, in the 8-cell SCNT embryos, the signal intensity of H3K4me3 was higher, while the level of Kdm5b was decreased compared to that of the IVF embryos (Figures 5D–5F; p < 0.01), suggesting that H3K4me3 might be an epigenetic roadblock of nuclear reprogramming. To further explore the exact effect of Kdm5b on goat nuclear reprogramming, we synthesized Kdm5b mRNA and injected it into 1-cell SCNT embryos. As shown in Figure 5G, SCNT embryos injected with Kdm5b mRNA rarely arrested during the 8-cell to morula and morula-to-blastocyst-stage transitions and developed to the blastocyst stage with higher efficiency (47.91% ± 0.86%, Figures 5G and 5H; p < 0.05). These data suggest that Kdm5b is a critical epigenetic factor that improves the development of SCNT embryos through the removal of H3K4me3.
Figure 5.
Kdm5b was a critical epigenetic factor for nuclear reprogramming
(A) Representative images of the 4- and 8-cell-stage embryos and blastocysts stained with anti-H3K4me3, anti-Kdm5b, and DAPI. Scale bar, 20 μm. (B and C) Statistical analysis of Kdm5b and H3K4me3 signal intensity in the 4- and 8-cell- and blastocyst-stage IVF embryos. ∗∗p < 0.01. (D) Representative images of the 8-cell IVF and SCNT embryos stained with anti-H3K4me3. Scale bar, 20 μm. (E) Representative images of the 8-cell IVF and SCNT embryos stained with anti-Kdm5b. Scale bar, 20 μm. (F) Statistical analysis of Kdm5b signal intensity at the 8-cell-stage IVF and SCNT embryos. Student’s t test, ∗∗p < 0.01. (G) Representative images of control and Kdm5b-injected SCNT embryos. Scale bar, 100 μm. (H) Development of control and Kdm5b-injected SCNT embryos. Shown are percentages of embryos that reached the indicated stages. Error bars represent SEM. ∗p < 0.05, ∗∗p < 0.01, Student’s t test.
Knockdown of lnc_3712 demethylated H3K4me3 via upregulating Kdm5b
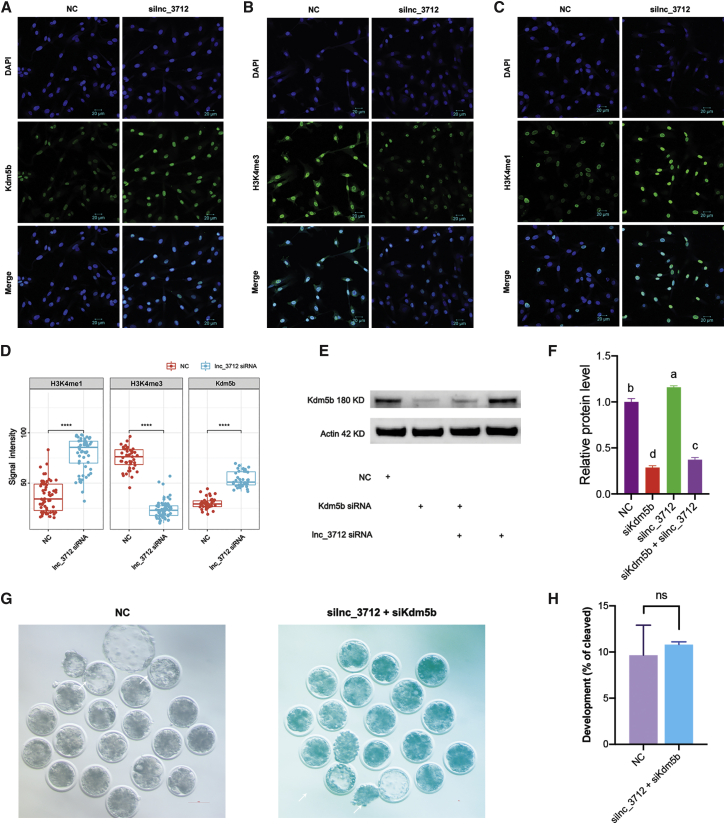
Since lnc_3712 was predicted to target Kdm5b and repressed the transcriptional level of Kdm5b (Figures 2A and 4D), we next sought to investigate whether lnc_3712 could affect the level of H3K4me3. Our immunofluorescence staining results indicated that the signal intensity of Kdm5b and H3K4me1 was significantly increased (p < 0.01), while the level of H3K4me3 was decreased (p < 0.01) in goat fetal fibroblast cells (GFFs) in which lnc_3712 was knocked down (Figures 6A–6D). Western blot results also revealed upregulation of Kdm5b in lnc_3712 knocked-down GFFs (Figures 6E and 6F; p < 0.01). In addition, we performed double knockdown of lnc_3712 and Kdm5b in 1-cell SCNT embryos. As expected, no difference was observed in the percentage of blastocyst between the double knockdown and NC groups (Figures 6G and 6H). These data suggest that lnc_3712 positively regulates the H3K4me3 modification through Kdm5b.
Figure 6.
Knockdown of lnc_3712 demethylated H3K4me3 via upregulating Kdm5b
(A) Kdm5b staining in lnc_3712 knockdown GFF cells. Scale bar, 20 μm. (B) H3K4me3 staining in lnc_3712 knockdown GFF cells. Scale bar, 20 μm. (C) H3K4me1 staining in lnc_3712 knockdown GFF cells. Scale bar, 20 μm. (D) Statistical analysis of Kdm5b, H3K4me3, and H3K4me1 signal intensity between lnc_3712 knockdown and NC GFF cells. Student’s t test, ∗∗∗∗p < 0.0001. (E) Western blot of Kdm5b in GFF cells that lnc_3712 was knocked down. (F) Statistical analysis of Kdm5b in lnc_3712 knockdown GFF cells. LSD test, statistical differences among groups were labeled with different letters. (G) Representative images of NC and lnc_3712 and Kdm5b double knockdown SCNT embryos. Blastocysts are indicated by the white arrows. Scale bar, 200 μm. (H) Statistical analysis of blastocyst rate in NC and double knockdown SCNT embryos.
Discussion
lncRNAs play vital roles during cellular reprogramming.32 However, only one unusual lncRNA, Xist, is reported to be associated with the nuclear reprogramming.29, 30, 31 In the present study, we revealed dynamic expression of lncRNAs during EGA and identified 114 upregulated lncRNAs in the 8-cell SCNT embryos as candidate key molecules involved in nuclear reprogramming in goat. Notably, we reported that knockdown of lnc_3712 increased the expression of Kdm5b and demethylated H3K4me3 and subsequently improved the development of goat SCNT embryos. Our data demonstrates that lnc_3712 serves as an epigenetic barrier during nuclear reprogramming via repressing Kdm5b.
Defects of SCNT embryos first appeared at the time of ZGA in mice8 and EGA in farm animals,15 during which a proportion of genes failed to establish normal expression patterns, hindering the proper development of SCNT embryos.33 Previous studies reported 811, 707, and 4,103 genes that were aberrantly expressed during ZGA/EGA in mouse, human, and bovine SCNT embryos,6,8,15 respectively. In line with these results, we detected 4,026 differentially expressed mRNAs during EGA of goat SCNT embryos. Our RNA-seq data suggest that the gene expression was also aberrant at the time of EGA in goat SCNT embryos. Of note, Zhang et al.34 reported that 882 genes were differentially expressed in the 2-cell IVF and SCNT embryos in bovine. Consistently, we also found differences between the 4-cell IVF and SCNT embryos. Given that our recent study revealed downregulation of maternal mRNAs in the 4-cell IVF embryos,35 the 4-cell SCNT embryos would also downregulate mRNAs from donor cells during the process of reprogramming. In addition, RNA polymerase II (RNAPII) and RNAPII-ser5 were weakly stained in the 4-cell IVF embryos,13 suggesting that the 4-cell stage is when the minor EGA occurs in goat. Therefore, the differences might result from the degradation of mRNAs and failure initiation during EGA in SCNT embryos.
Histone modifications, such as H3K4me3,14,36 H3K9me3,6,8,15 and H3K27me3,18,37 have been identified as epigenetic barriers during nuclear reprogramming in mice. High levels of H3K4me3 modification are retained at the ZGA stage in mouse and the EGA in porcine SCNT embryos.14,36,38 Consistent with these studies, H3K4me3 was also strongly detected in the 8-cell SCNT embryos in the present study. Although H3K4me3 leads to gene transcription, H3K4me3 preferred enriched in the genes highly expressed in donor cells, preventing silencing of somatic cell signature genes.16 Kdm5b was reported to catalyze the demethylation of H3K4me3 modification and activates the expression of self-renewal-associated genes in ESCs.39 Recently, Liu et al.36 reported that injection of Kdm5b mRNA improved the transition of 4-cell-to-blastocyst development in mice. Consistent with the result, we found overexpression of Kdm5b cleared H3K4me3 modification and improved the development of goat SCNT embryos. Therefore, the H3K4me3 reprogramming barrier is conserved in mammalian species, and removal of H3K4me3 by injection of Kdm5b may be a strategy for improving SCNT embryo development in goat.
In addition to the protein-coding genes, lncRNA also changed dynamically during the development of SCNT embryos. Recently, Wu et al.40 reported that most (>90%) of the differentially expressed lncRNAs were upregulated in the 2-cell SCNT embryos in mice. Consistently, we identified 177 differentially expressed lncRNAs between the 8-cell SCNT and IVF embryos, and 114 were highly expressed in the 8-cell SCNT embryos, suggesting a failure in the reprogramming of lncRNA in SCNT embryos. Of note, KEGG enrichment analysis revealed that these differentially expressed lncRNAs were enriched in many processes, including the hippo signaling pathway, which was reported to be crucial for the early development.41,42 Since lncRNAs were reported to control the hippo signaling pathway in cancer cells,43 it is highly tempting to hypothesize that lncRNA regulates nuclear reprogramming via the hippo signaling pathway. Moreover, lncRNAs were reported to play critical roles during the early embryo development, as development block occurred in lncRNAs il17d and lincGET knockdown embryos in mice.25,27,28 All together, these studies and our data support the idea that lncRNAs contribute to the nuclear reprogramming efficiency of SCNT embryos.
To this end, we identified lnc_3712 as a potential functional lncRNA that hampers the nuclear reprogramming. Knockdown of lnc_3712 greatly improved the development of goat SCNT embryos and increased the expression of 1,464 DEGs, which were enriched in transcription regulator activity, nuclear transcribed mRNA catabolic process, and translational initiation, suggesting transcription repression by lnc_3712. Considering that goat EGA initiates at the 8-cell stage and the gene expression was aberrant during ZGA/EGA in SCNT embryos, knockdown of lnc_3712 might restore the transcription repression during the activation of embryonic genes in goat. Specifically, the expression of several genes responsible for ZGA, such as Wee1, Ybx1, and Klf4, were restored in lnc_3712 knockdown embryos. Previous studies have demonstrated that Wee1, Ybx1, and Klf4 are crucial for early development in mammalian embryos, and knockout of these transcription factors resulted in developmental defects.44, 45, 46 Therefore, it is likely that knocking down expression of lnc_3712 allows the expression of zygotic genes and transcription factors, subsequently improving the development of SCNT embryos. In addition, the expression of Kdm6a and Kdm6b was significantly increased when lnc_3712 was knocked down in the 8-cell SCNT embryos. Previous studies reported that overexpression of Kdm6a removed the H3K27me3 reprogramming barrier and improved nuclear reprogramming efficiency in mouse and bovine.18,47 Given that knockdown of Kdm5b impaired the preimplantation development through disturbance of bivalent H3K4me3 and H3K27me3 histone modification,48 it is reasonable that Kdm6a and Kdm6b were also upregulated in lnc_3712 knockdown embryos as lnc_3712 targeted Kdm5b.
lncRNAs have been demonstrated to act as cis and trans elements with neighboring or distal protein-coding genes.49 In the present study, lnc_3712 was predicted to target Kdm5b. Our RNA-seq, immunofluorescence staining, and western blot data further revealed that the mRNA and protein expression of Kdm5b was upregulated, while the H3K4me3 histone modification was reduced upon lnc_3712 knockdowns in SCNT embryos and donor cells. Interestingly, no difference in the percentage of blastocyst was observed in lnc_3712 and Kdm5b double knockdown embryos and the NC embryos. Our data demonstrated that lnc_3712 is an epigenetic barrier by repressing the expression of Kdm5b. Since lnc_3712 and Kdm5b share sequence complementarity and the expression of Kdm5b was increased in lnc_3712 knockdown GFFs but decreased in the double knockdown GFFs compared to the NC GFFs, it is highly likely that lnc_3712 serves as an antisense RNA of Kdm5b.
In summary, we characterized lncRNA expression profiles of in SCNT embryos during EGA and identified a novel lncRNA, lnc_3712, as epigenetic barriers for goat SCNT embryo development. We further revealed that knockdown of lnc_3712 led to the upregulation of Kdm5b and removal of the H3K4me3 reprogramming barrier. Our efforts to elucidate the crosstalk between lncRNAs and histone methylation during EGA will facilitate the understanding of the stochastic reprogramming events and help to optimize the development of SCNT embryos.
Materials and methods
All materials were obtained from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise stated. All experimental procedures involving animals were conducted in accordance with the National Research Council’s publication “Guide for the Care and Use of Laboratory Animals” and approved by the Institutional Animal Care and Use Committee at Nanjing Agricultural University, China.
Preparation of donor cells
GFFs were isolated from the ears of a two-month-old female fetus and cultured as previously described.50 In brief, GFFs at passage 7 with 80% confluence were cultured under serum-starved conditions in DMEM medium (Gibco) containing 0.5% (v/v) fetal bovine serum (FBS; Gibco) for 3 days at 37.5°C with 5% CO2 and saturated humidity to induce entry into the G0/G1 phase of the cell cycle. GFFs were harvested with trypsin and resuspended in DMEM at 30 min before donor cell injection.
SCNT
SCNT was carried out as described previously.50 In brief, recipient MII oocytes were collected from superovulated adult female goats by brief treatment with 0.3% bovine testicular hyaluronidase. Isolated MII oocytes were enucleated in M2 medium after treatment with 2.0 μg/mL demecolcine solution for 30 min to expose the nuclei. Synchronized GFFs with a smooth shape were selected for injection into the perivitelline space. The GFFs were fused with enucleated oocytes by exposure to two 1.2-kv/cm direct current (DC) pulses, 20 μs in fusion medium. Finally, reconstructed embryos were washed three times with M2 and activated with 5 mM ionomycin for 5 min and 2 mM 6-DMAP for 4 h.
IVF
IVF was performed as described previously.51 In brief, 20 MII-stage oocytes were transferred to a 75-μL microdrop of BO-IVF medium (IVF Bioscience, Falmouth, UK) with mineral oil covering the surface, and freshly collected sperm was diluted and suspended to 2 to 9 × 106 spermatozoa/mL. Then, 50 μL of the sperm suspension was added to the microdrops and further cultured for 16 h at 38.5°C, 5% CO2, and saturated humidity. The remaining spermatozoa and cumulus cells were dispersed by gentle blowing with a pipette.
In vitro culture
The IVF-generated zygotes and SCNT-generated 1-cell embryos were transferred into BO-IVC medium (IVF Bioscience) and cultured at 38.5°C, 5% CO2, and saturated humidity. The 4- and 8-cell embryos were picked randomly and transferred into lysate buffer using a mouth pipette for RNA-seq.
RNA-seq
RNA-seq was conducted as described in a previously published study.13 In brief, directly lysed embryos were used for cDNA synthesis following the protocol for the SMARTer ultra-low input RNA cDNA preparation kit. The cDNA samples were fragmented using a Bioruptor sonication system (Diagenode, Denville, NJ, USA). Sequencing libraries were constructed from the fragmented cDNA using a NEBNext ultra DNA library prep kit according to the manufacturer’s instructions (E7370, New England Biolabs, Ipswich, MA, USA). Paired-end 150-bp sequencing was performed on HiSeq X ten by Annoroad Gene Technology Company. The accession number of the RNA-seq data reported in this paper is GEO: GSE165054.
Read quality control, alignment, and gene expression analysis
All sequenced reads were trimmed to remove adaptors and low-quality bases using fastp (v0.19.6). All the reads that passed quality control were mapped to the goat reference genome (ARS1, NCBI) using HISAT2 (v2.2.1) software with default settings. Uniquely mapped reads were subsequently assembled into transcripts guided by the reference annotation using HTSeq. Gene expression levels were quantified by calculating normalized FPKM values.
Identification of novel lncRNA
The novel lncRNAs were obtained as described in a previously published study.52 In brief, Cuffcompare was used to generate a unique set of assembled isoforms. Next, we ran the unique transcript set through the following filters: (1) size selection >200 bp, (2) read coverage threshold >3, (3) filter of mRNA transcript annotations, (4) calculating coding potential score by integrating the results from CPAT, Coding-Non-Coding Index (CNCI), and Coding Potential Calculator (CPC), (5) known protein domain filter, and (6) intergenic classification.
GO, KEGG pathway enrichment analyses, and GSEA
GO annotation, KEGG pathway enrichment analyses, and GSEA of differentially expressed transcripts were performed using clusterProfiler R package (v3.12.0) and GSEA software (v4.1.0). Data were analyzed and visualized using R project.
In vitro transcription of Kdm5b mRNA
Goat Kdm5b coding sequence was amplified from goat testis cDNA library using the phanta max super-fidelity DNA polymerase (#P505-d1, Vazyme, Nanjing, China). PCR products were cloned into the pcDNA3-mRFP plasmid (Addgene, plasmid #13032) using NEBuilder HiFi DNA assembly master mix (E2621S, NEB). mRNA was synthesized from linearized template plasmids by in vitro transcription using a mMESSAGE mMACHINE T7 ultra kit (#AM1345, Thermo Fisher Scientific), following the manufacturer’s instructions. The synthesized mRNA was precipitated using a MEGAclear kit (#AM1908, Thermo Fisher Scientific) and dissolved in nuclease-free water. Primers used in this experiment are listed in Table S9.
Microinjection
Two siRNAs were formulated using the BLOCK-iT RNAi designer tool (Table S10) and synthesized at GeneParma (Shanghai, China). In general, 5–10 pL of 20 μM lnc_3712 siRNA mixture was microinjected into the cytoplasm of SCNT-generated embryos at 8 h after onset of treatment with 6-DAMP using a piezo-driven micromanipulator (Primetech). MISSION siRNA universal NC was used to serve a NC for siRNA experiments. To overexpress Kdm5b in SCNT embryos, Kdm5b mRNA (400 ng/μL) was microinjected into the cytoplasm of SCNT embryos at 8–10 h after onset of treatment with 6-DAMP, and SCNT embryos were injected with 10 pL of nuclease-free water as the control.
Knockdown experiment in donor cells
GFFs at passage 7 with 60% confluence were transfected with two siRNA mixtures against lnc_3712 and/or one Kdm5b siRNA using lipofectamine 3000 (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. GFFs were collected at 72 h for protein expression analysis using immunofluorescence staining and western blot. The sequence of siRNA against Kdm5b is listed in Table S10.
Immunofluorescence staining
Immunofluorescence staining was performed as described previously.13 Fluorescent signals were observed using a LSM710 laser scanning confocal microscope (Carl Zeiss, Oberkochen, Germany) with the same conditions. Details of the primary and secondary antibodies are listed in Table S11.
Western blot
Western blot was performed as described previously.53 Details of the primary and secondary antibodies are listed in Table S11.
Statistical analysis
Data are presented as mean ± standard error of the mean. Differences between two groups were analyzed using the independent-samples t test, and multiple-comparison tests were analyzed by one-way ANOVA with post hoc Turkey honest significant difference test using R software (v3.5.0). Differences with p values <0.05 were considered statistically significant.
Data visualization
R programming language, including the Biocoductor software packages, was mainly used in statistical analysis and data visualization. Heatmap and boxplot of coding potential score, volcano, PCA, gene expression, GO, and signal intensity were generated using R package pheatmap (v1.0.12) and ggplot2 (v3.3.2), respectively. Rectangle tree diagrams were generated using Hiplot (https://hiplot.com.cn/basic/dendrogram) with “canberra.”
Acknowledgments
We are grateful to Dr. Rihong Guo from the Jiangsu Academy of Agricultural Sciences for technical assistance with the in vitro transcription experiment, Dr. Li Meng from South China Agricultural University for critical reading of the manuscript, and the High-Performance Computing Platform of the Bioinformatics Center, Nanjing Agricultural University, for their data analysis support. This work was financially supported by the National Natural Science Foundation of China (nos. 31672422 and 31972569 to F.W. and Y.W.), and also by the Natural Science Foundation of Jiangsu Province, China (BK20200563) and the China Postdoctoral Science Foundation (2019M661869 to M.D.).
Author contributions
M.D. and F.W. conceived and designed the study. M.D. and Y.W. performed the experiments and analysis of IVF, in vitro culture, immunofluorescence staining, western blotting, and RNA-seq. B.C. and Z.L. conducted embryo manipulation experiments. M.D. and X.D. wrote and revised the manuscript. Y.C. and Y.Y. prepared the donor cells and performed in vitro transcription of Kdm5b mRNA. F.W. and Y.Z. supervised the study and administrated the project.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2021.02.016.
Contributor Information
Yanli Zhang, Email: zhangyanli@njau.edu.cn.
Feng Wang, Email: caeet@njau.edu.cn.
Supplemental information
References
- 1.Matoba S., Zhang Y. Somatic Cell Nuclear Transfer Reprogramming: Mechanisms and Applications. Cell Stem Cell. 2018;23:471–485. doi: 10.1016/j.stem.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez-Osorio N., Urrego R., Cibelli J.B., Eilertsen K., Memili E. Reprogramming mammalian somatic cells. Theriogenology. 2012;78:1869–1886. doi: 10.1016/j.theriogenology.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 3.Wilmut I., Beaujean N., de Sousa P.A., Dinnyes A., King T.J., Paterson L.A., Wells D.N., Young L.E. Somatic cell nuclear transfer. Nature. 2002;419:583–586. doi: 10.1038/nature01079. [DOI] [PubMed] [Google Scholar]
- 4.Tachibana M., Amato P., Sparman M., Gutierrez N.M., Tippner-Hedges R., Ma H., Kang E., Fulati A., Lee H.S., Sritanaudomchai H. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153:1228–1238. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X., Smith S.L., Tian X.C., Lewin H.A., Renard J.P., Wakayama T. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat. Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- 6.Chung Y.G., Matoba S., Liu Y., Eum J.H., Lu F., Jiang W., Lee J.E., Sepilian V., Cha K.Y., Lee D.R., Zhang Y. Histone Demethylase Expression Enhances Human Somatic Cell Nuclear Transfer Efficiency and Promotes Derivation of Pluripotent Stem Cells. Cell Stem Cell. 2015;17:758–766. doi: 10.1016/j.stem.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Chung Y.G., Eum J.H., Lee J.E., Shim S.H., Sepilian V., Hong S.W., Lee Y., Treff N.R., Choi Y.H., Kimbrel E.A. Human somatic cell nuclear transfer using adult cells. Cell Stem Cell. 2014;14:777–780. doi: 10.1016/j.stem.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Matoba S., Liu Y., Lu F., Iwabuchi K.A., Shen L., Inoue A., Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159:884–895. doi: 10.1016/j.cell.2014.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee M.T., Bonneau A.R., Giraldez A.J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 2014;30:581–613. doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pálfy M., Joseph S.R., Vastenhouw N.L. The timing of zygotic genome activation. Curr. Opin. Genet. Dev. 2017;43:53–60. doi: 10.1016/j.gde.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Ding B., Cao Z., Hong R., Li H., Zuo X., Luo L., Li Y., Huang W., Li W., Zhang K., Zhang Y. WDR5 in porcine preimplantation embryos: expression, regulation of epigenetic modifications and requirement for early development. Biol. Reprod. 2017;96:758–771. doi: 10.1093/biolre/iox020. [DOI] [PubMed] [Google Scholar]
- 12.Cao Z., Li Y., Chen Z., Wang H., Zhang M., Zhou N., Wu R., Ling Y., Fang F., Li N., Zhang Y. Genome-Wide Dynamic Profiling of Histone Methylation during Nuclear Transfer-Mediated Porcine Somatic Cell Reprogramming. PLoS ONE. 2015;10:e0144897. doi: 10.1371/journal.pone.0144897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng M., Liu Z., Ren C., Zhang G., Pang J., Zhang Y., Wang F., Wan Y. Long noncoding RNAs exchange during zygotic genome activation in goat. Biol. Reprod. 2018;99:707–717. doi: 10.1093/biolre/ioy118. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z., Zhai Y., Ma X., Zhang S., An X., Yu H., Li Z. Down-Regulation of H3K4me3 by MM-102 Facilitates Epigenetic Reprogramming of Porcine Somatic Cell Nuclear Transfer Embryos. Cell. Physiol. Biochem. 2018;45:1529–1540. doi: 10.1159/000487579. [DOI] [PubMed] [Google Scholar]
- 15.Liu X., Wang Y., Gao Y., Su J., Zhang J., Xing X., Zhou C., Yao K., An Q., Zhang Y. H3K9 demethylase KDM4E is an epigenetic regulator for bovine embryonic development and a defective factor for nuclear reprogramming. Development. 2018;145:dev158261. doi: 10.1242/dev.158261. [DOI] [PubMed] [Google Scholar]
- 16.Hörmanseder E., Simeone A., Allen G.E., Bradshaw C.R., Figlmüller M., Gurdon J., Jullien J. H3K4 Methylation-Dependent Memory of Somatic Cell Identity Inhibits Reprogramming and Development of Nuclear Transfer Embryos. Cell Stem Cell. 2017;21:135–143.e6. doi: 10.1016/j.stem.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jullien J., Vodnala M., Pasque V., Oikawa M., Miyamoto K., Allen G., David S.A., Brochard V., Wang S., Bradshaw C. Gene Resistance to Transcriptional Reprogramming following Nuclear Transfer Is Directly Mediated by Multiple Chromatin-Repressive Pathways. Mol. Cell. 2017;65:873–884.e8. doi: 10.1016/j.molcel.2017.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou C., Wang Y., Zhang J., Su J., An Q., Liu X., Zhang M., Wang Y., Liu J., Zhang Y. H3K27me3 is an epigenetic barrier while KDM6A overexpression improves nuclear reprogramming efficiency. FASEB J. 2019;33:4638–4652. doi: 10.1096/fj.201801887R. [DOI] [PubMed] [Google Scholar]
- 19.Niemann H. Epigenetic reprogramming in mammalian species after SCNT-based cloning. Theriogenology. 2016;86:80–90. doi: 10.1016/j.theriogenology.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 20.Wang M., Gao Y., Qu P., Qing S., Qiao F., Zhang Y., Mager J., Wang Y. Sperm-borne miR-449b influences cleavage, epigenetic reprogramming and apoptosis of SCNT embryos in bovine. Sci. Rep. 2017;7:13403. doi: 10.1038/s41598-017-13899-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J., Qu P., Zhou C., Liu X., Ma X., Wang M., Wang Y., Su J., Liu J., Zhang Y. MicroRNA-125b is a key epigenetic regulatory factor that promotes nuclear transfer reprogramming. J. Biol. Chem. 2017;292:15916–15926. doi: 10.1074/jbc.M117.796771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flynn R.A., Chang H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouckenheimer J., Assou S., Riquier S., Hou C., Philippe N., Sansac C., Lavabre-Bertrand T., Commes T., Lemaître J.M., Boureux A., De Vos J. Long non-coding RNAs in human early embryonic development and their potential in ART. Hum. Reprod. Update. 2016;23:19–40. doi: 10.1093/humupd/dmw035. [DOI] [PubMed] [Google Scholar]
- 24.Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 25.Hamazaki N., Uesaka M., Nakashima K., Agata K., Imamura T. Gene activation-associated long noncoding RNAs function in mouse preimplantation development. Development. 2015;142:910–920. doi: 10.1242/dev.116996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., Gao Z., Wang X., Liu H., Zhang Y., Liu Z. Identification and functional analysis of long intergenic noncoding RNA genes in porcine pre-implantation embryonic development. Sci. Rep. 2016;6:38333. doi: 10.1038/srep38333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., Li X., Wang L., Li J., Zhao Y., Bou G., Li Y., Jiao G., Shen X., Wei R. A novel long intergenic noncoding RNA indispensable for the cleavage of mouse two-cell embryos. EMBO Rep. 2016;17:1452–1470. doi: 10.15252/embr.201642051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J., Wang L., Feng G., Wang Y., Li Y., Li X., Liu C., Jiao G., Huang C., Shi J. Asymmetric Expression of LincGET Biases Cell Fate in Two-Cell Mouse Embryos. Cell. 2018;175:1887–1901.e18. doi: 10.1016/j.cell.2018.11.039. [DOI] [PubMed] [Google Scholar]
- 29.Matoba S., Inoue K., Kohda T., Sugimoto M., Mizutani E., Ogonuki N., Nakamura T., Abe K., Nakano T., Ishino F., Ogura A. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc. Natl. Acad. Sci. USA. 2011;108:20621–20626. doi: 10.1073/pnas.1112664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue K., Kohda T., Sugimoto M., Sado T., Ogonuki N., Matoba S., Shiura H., Ikeda R., Mochida K., Fujii T. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science. 2010;330:496–499. doi: 10.1126/science.1194174. [DOI] [PubMed] [Google Scholar]
- 31.Ruan D., Peng J., Wang X., Ouyang Z., Zou Q., Yang Y., Chen F., Ge W., Wu H., Liu Z. XIST Derepression in Active X Chromosome Hinders Pig Somatic Cell Nuclear Transfer. Stem Cell Reports. 2018;10:494–508. doi: 10.1016/j.stemcr.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D.H., Marinov G.K., Pepke S., Singer Z.S., He P., Williams B., Schroth G.P., Elowitz M.B., Wold B.J. Single-cell transcriptome analysis reveals dynamic changes in lncRNA expression during reprogramming. Cell Stem Cell. 2015;16:88–101. doi: 10.1016/j.stem.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S., Li Y., Du W., Zhang L., Yu S., Dai Y., Zhao C., Li N. Aberrant gene expression in organs of bovine clones that die within two days after birth. Biol. Reprod. 2005;72:258–265. doi: 10.1095/biolreprod.104.029462. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L., Yu M., Xu H., Wei X., Liu Y., Huang C., Chen H., Guo Z. RNA sequencing revealed the abnormal transcriptional profile in cloned bovine embryos. Int. J. Biol. Macromol. 2020;150:492–500. doi: 10.1016/j.ijbiomac.2020.02.026. [DOI] [PubMed] [Google Scholar]
- 35.Deng M., Chen B., Liu Z., Cai Y., Wan Y., Zhang G., Fan Y., Zhang Y., Wang F. YTHDF2 Regulates Maternal Transcriptome Degradation and Embryo Development in Goat. Front. Cell. Dev. Biol. 2020;8:580367. doi: 10.3389/fcell.2020.580367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W., Liu X., Wang C., Gao Y., Gao R., Kou X., Zhao Y., Li J., Wu Y., Xiu W. Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell Discov. 2016;2:16010. doi: 10.1038/celldisc.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie B., Zhang H., Wei R., Li Q., Weng X., Kong Q., Liu Z. Histone H3 lysine 27 trimethylation acts as an epigenetic barrier in porcine nuclear reprogramming. Reproduction. 2016;151:9–16. doi: 10.1530/REP-15-0338. [DOI] [PubMed] [Google Scholar]
- 38.Zhou C., Zhang J., Zhang M., Wang D., Ma Y., Wang Y., Wang Y., Huang Y., Zhang Y. Transcriptional memory inherited from donor cells is a developmental defect of bovine cloned embryos. FASEB J. 2020;34:1637–1651. doi: 10.1096/fj.201900578RR. [DOI] [PubMed] [Google Scholar]
- 39.Xie L., Pelz C., Wang W., Bashar A., Varlamova O., Shadle S., Impey S. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011;30:1473–1484. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu F., Liu Y., Wu Q., Li D., Zhang L., Wu X., Wang R., Zhang D., Gao S., Li W. Long non-coding RNAs potentially function synergistically in the cellular reprogramming of SCNT embryos. BMC Genomics. 2018;19 doi: 10.1186/s12864-018-5021-2. 631–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anani S., Bhat S., Honma-Yamanaka N., Krawchuk D., Yamanaka Y. Initiation of Hippo signaling is linked to polarity rather than to cell position in the pre-implantation mouse embryo. Development. 2014;141:2813–2824. doi: 10.1242/dev.107276. [DOI] [PubMed] [Google Scholar]
- 42.Cockburn K., Biechele S., Garner J., Rossant J. The Hippo pathway member Nf2 is required for inner cell mass specification. Curr. Biol. 2013;23:1195–1201. doi: 10.1016/j.cub.2013.05.044. [DOI] [PubMed] [Google Scholar]
- 43.Hao Q., Zong X., Sun Q., Lin Y.C., Song Y.J., Hashemikhabir S., Hsu R.Y., Kamran M., Chaudhary R., Tripathi V. The S-phase-induced lncRNA SUNO1 promotes cell proliferation by controlling YAP1/Hippo signaling pathway. eLife. 2020;9:e55102. doi: 10.7554/eLife.55102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun J., Yan L., Shen W., Meng A. Maternal Ybx1 safeguards zebrafish oocyte maturation and maternal-to-zygotic transition by repressing global translation. Development. 2018;145:dev166587. doi: 10.1242/dev.166587. [DOI] [PubMed] [Google Scholar]
- 45.Tominaga Y., Li C., Wang R.H., Deng C.X. Murine Wee1 plays a critical role in cell cycle regulation and pre-implantation stages of embryonic development. Int. J. Biol. Sci. 2006;2:161–170. doi: 10.7150/ijbs.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao Q., Zhang X., Lu L., Yang L., Gao J., Gao Y., Ma H., Cao Y. Klf4 is required for germ-layer differentiation and body axis patterning during Xenopus embryogenesis. Development. 2012;139:3950–3961. doi: 10.1242/dev.082024. [DOI] [PubMed] [Google Scholar]
- 47.Yang L., Song L., Liu X., Bai L., Li G. KDM6A and KDM6B play contrasting roles in nuclear transfer embryos revealed by MERVL reporter system. EMBO Rep. 2018;19:e46240. doi: 10.15252/embr.201846240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J., Zhang H., Wang X., Dobbs K.B., Yao J., Qin G., Whitworth K., Walters E.M., Prather R.S., Zhao J. Impairment of preimplantation porcine embryo development by histone demethylase KDM5B knockdown through disturbance of bivalent H3K4me3-H3K27me3 modifications. Biol. Reprod. 2015;92:72. doi: 10.1095/biolreprod.114.122762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao R.W., Wang Y., Chen L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019;21:542–551. doi: 10.1038/s41556-019-0311-8. [DOI] [PubMed] [Google Scholar]
- 50.Deng M., Liu Z., Chen B., Wan Y., Yang H., Zhang Y., Cai Y., Zhou J., Wang F. Aberrant DNA and histone methylation during zygotic genome activation in goat cloned embryos. Theriogenology. 2020;148:27–36. doi: 10.1016/j.theriogenology.2020.02.036. [DOI] [PubMed] [Google Scholar]
- 51.Deng M., Zhang G., Cai Y., Liu Z., Zhang Y., Meng F., Wang F., Wan Y. DNA methylation dynamics during zygotic genome activation in goat. Theriogenology. 2020;156:144–154. doi: 10.1016/j.theriogenology.2020.07.008. [DOI] [PubMed] [Google Scholar]
- 52.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cai Y., Deng M., Liu Z., Zhang G., Pang J., An S., Wang Z., Zhang Y., Wang F. EZH2 expression and its role in spermatogonial stem cell self-renewal in goats. Theriogenology. 2020;155:222–231. doi: 10.1016/j.theriogenology.2020.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.