Abstract
Background
RhoB is a Rho family GTPase that is highly homologous to RhoA and RhoC. RhoA and RhoC have been shown to promote tumor progression in many cancer types; however, a distinct role for RhoB in cancer has not been delineated. Additionally, several well‐characterized studies have shown that small GTPases such as RhoA, Rac1, and Cdc42 are induced in vitro under hypoxia, but whether and how hypoxia regulates RhoB in breast cancer remains elusive.
Aims
To determine whether and how hypoxia regulates RhoB expression and to understand the role of RhoB in breast cancer metastasis.
Methods
We investigated the effects of hypoxia on the expression and activation of RhoB using real‐time quantitative polymerase chain reaction and western blotting. We also examined the significance of both decreased and increased RhoB expression in breast cancer using CRISPR depletion of RhoB or a vector overexpressing RhoB in 3D in vitro migration models and in an in vivo mouse model.
Results
We found that hypoxia significantly upregulated RhoB mRNA and protein expression resulting in increased levels of activated RhoB. Both loss of RhoB and gain of RhoB expression led to reduced migration in a 3D collagen matrix and invasion within a multicellular 3D spheroid. We showed that neither the reduction nor overexpression of RhoB affected tumor growth in vivo. While the loss of RhoB had no effect on metastasis, RhoB overexpression led to decreased metastasis to the lungs, liver, and lymph nodes of mice.
Conclusion
Our results suggest that RhoB may have an important role in suppressing breast cancer metastasis.
Keywords: breast cancer, cancer metastasis, hypoxia, Rho GTPase, RhoB
1. INTRODUCTION
Rho proteins are small molecules (~21 kDa) that belong to the Ras superfamily and function as binary switches in a wide variety of signaling pathways.1 They consist of a group of 20 intracellular signaling molecules that are most well known for their role in regulating the actin cytoskeleton. Ras homolog gene family, member B (RhoB), is a key regulator of several cellular processes, such as cytoskeletal organization and vesicle and membrane receptor trafficking.2, 3, 4 RhoB has several additional features that are distinct among the Rho proteins. All Rho proteins are prenylated; however, RhoB contains a unique C‐terminal region that can undergo both farnesylation or geranylgeranylation, whereas other members of this family are typically only geranylgeranylated. These distinct posttranslational modifications affect RhoB localization and function.5 In contrast to RhoA and RhoC, RhoB is localized not only at the plasma membrane but also in endosomes, multivesicular bodies, and has even been reported in the nucleus.6, 7, 8 Unlike most small GTPases, which are relatively stable, RhoB has a high rate of turnover at the protein level. Its synthesis is rapidly upregulated by many stimuli, including cellular stress, growth factors, and cytokines, which ultimately helps RhoB regulate many cellular responses like proliferation, survival, and apoptosis.3, 9, 10, 11, 12, 13
Rho GTPases play an important role in regulating cell migration and proliferation, and thus have been widely studied for their role in cancer progression and metastasis.14, 15, 16 Although Rho GTPases RhoA, RhoB, and RhoC are highly conserved and share more than 85% amino acid sequence identity, they seem to play distinct roles in tumor progression. RhoA and RhoC have been implicated in promoting tumor progression, while the role of RhoB in cancer is not as clearly defined. RhoB was first described to contribute to Ras‐induced fibroblast transformation.17 Additional studies also revealed an oncogenic role for RhoB. For example, RhoB depletion led to cell cycle arrest, apoptosis, and reduced tumorigenic potential of glioblastoma cells in vivo.18 Likewise, increased levels of RhoB were found in breast tumors when compared with the corresponding normal tissue.19 Low levels of RhoB expression in the primary tumor of patients was correlated with a favorable response to epidermal growth factor receptor tyrosine kinase inhibitor treatment, while high levels of RhoB corresponded to a poor response in lung cancer patients.20 On the other hand, emerging studies have also indicated a tumor suppressive role for RhoB. For example, the loss of RhoB expression occurs frequently in lung cancer21, 22 and has been correlated with poor patient outcomes.21 Furthermore, RhoB expression is reduced in melanoma cells when compared with primary human melanocytes.23 RhoB overexpression in vitro resulted in both a decrease in ovarian cancer cell proliferation as well as an increase in apoptosis.24 Glioblastoma cells constitutively expressing high levels of RhoB had significantly reduced cell motility, invasion through Matrigel, and growth rate compared with the empty vector containing control cells.25 The difference in these findings suggests that RhoB may function in a context‐dependent and spatially dependent manner, responding to specific signals in the tumor microenvironment.
Using hypoxia‐inducible factor 1‐alpha (HIF1‐α) as a marker, approximately 25% to 40% of invasive breast cancers contain regions of hypoxia.26 Rapid cancer cell proliferation, combined with structural and functional abnormalities in tumor blood vessels, results in regions within solid tumors that have reduced oxygen availability.27 During this process, hypoxic cells obtain invasive and metastatic properties as well as resistance to chemotherapy and radiation therapy, which together lead to a more lethal cancer phenotype.28, 29 There has been an increasing amount of evidence that the O2 content of tumor tissue is an important determinant of metastasis.30 Extensive studies have shown that small GTPases such as RhoA, RhoC, Ras‐related C3 botulinum toxin substrate 1 (Rac1), and cell division control protein 42 homolog (Cdc42) are induced in vitro under hypoxia to promote a metastatic phenotype.31, 32, 33 Hypoxic induction and activation of RhoA and Rho‐associated protein kinase 1 (ROCK1) by HIFs in breast cancer cells were shown to stimulate cell contraction, cell‐induced matrix contraction, formation of focal adhesions, FAK activation, and thus an increased cell motility.31 Moreover, activation of this signaling pathway in breast cancer patients was associated with decreased survival. The effect of hypoxia on RhoB has not been reported yet in breast cancer cells, but it has been studied in other cancer cell lines with paradoxical results. It has been reported that hypoxia induces the expression of Cdc42, Rac1, and RhoA but not RhoB in renal carcinoma cells.34 In glioblastoma cells, hypoxia did not affect RhoB expression but rather enhanced RhoB activation.35 Therefore, it remains in question whether and how hypoxia regulates RhoB in breast cancer cells and if it contributes to the overall role of RhoB in breast cancer metastasis.
We found that hypoxia upregulates the expression of RhoB at both the mRNA and protein levels leading to increased levels of activated RhoB. We further showed that both knockdown and overexpression of RhoB result in a decreased migration of breast cancer cells in a 3D collagen matrix and decreased invasion from a 3D multicellular spheroid. Finally, we assessed the role of RhoB in tumor progression. The knockout or overexpression of RhoB had no effect on tumor growth. However, overexpression of RhoB led to a significant decrease in metastasis to the lung, liver, and lymph nodes. Likewise, in tumor samples from breast cancer patients, RhoB expression positively correlates with survival.
2. RESULTS
2.1. Hypoxia induces RhoB mRNA, protein, and activation
We recently performed an RNA sequencing analysis of 31 cell lines exposed to 20% or 1% O2 conditions.36 The RhoB transcript was reliably detected in 9 out of 31 cell lines analyzed by RNA sequencing. Eight of the nine cell lines showed an induction of RhoB (FC > 1.5), with the exception of the MCF‐7 cells. To confirm these findings, we measured RhoB gene expression and verified that it was increased in MDA‐MB‐175, MDA‐MB‐231, SUM159, SUM225, and HCC1954 breast cancer cell lines by exposure to hypoxic (1% O2) or normal tissue culture (20% O2) conditions for 24 hours (Figure 1A). Subsequently, the cell lines with the highest RhoB induction at the RNA level, MDA‐MB‐175, SUM159, and MDA‐MB‐231 were then tested and verified for corresponding induction at the protein level (Figure 1B). Since Rho proteins operate by switching between an active GTP‐bound state and an inactive GDP‐bound state, we investigated whether hypoxia‐induced increases in RhoB protein levels would lead to an increase in active RhoB levels by performing a RhoB‐GTP pulldown assay. These results confirmed that hypoxia upregulates both RhoB expression and activation (Figure 1C). An immunoblot of SUM159 (Figure 1D) and MDA‐MB‐231 (Figure 1E) cells was performed to assess the levels of HIF‐1α, HIF‐2α, and RhoB protein over a 48‐hour time period. HIF‐1α and HIF‐2α expression peaked after 4 hours under 1% O2 conditions and then decreased to a steady state level following 24 hours of continuous exposure. RhoB increased steadily under hypoxic stimulation for the duration of the 48‐hour time course.
Figure 1.
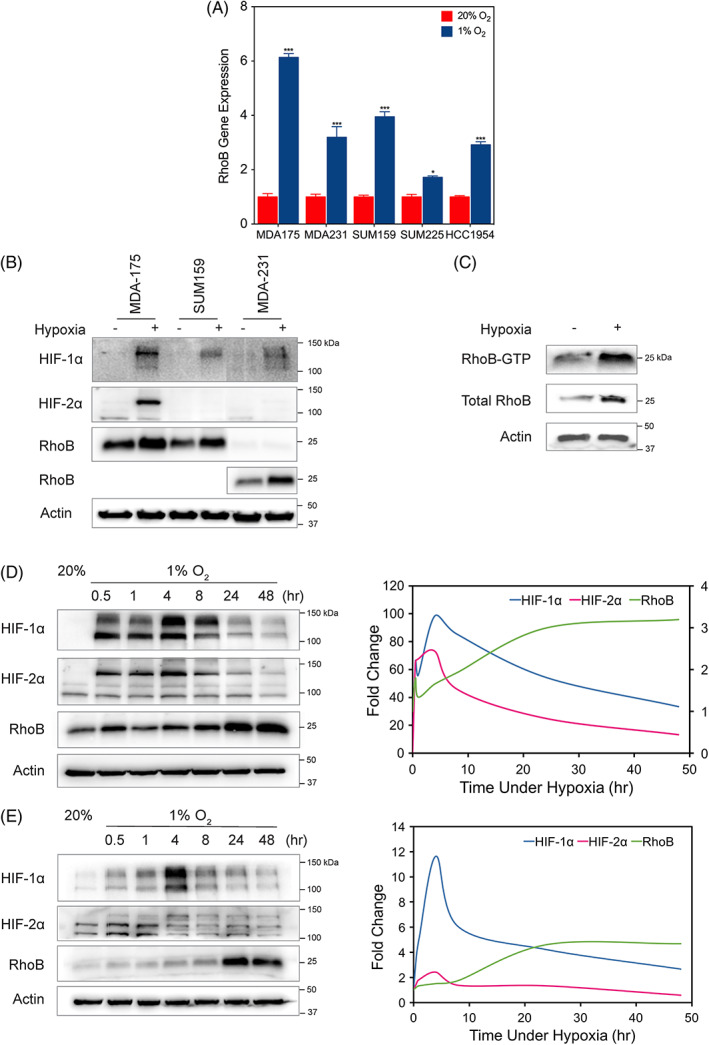
Hypoxia induces RhoB mRNA, protein, and activated RhoB protein levels. A, RhoB mRNA levels were analyzed by quantitative polymerase chain reaction in MDA‐MB‐175, MDA‐MB‐231, SUM159, SUM225, and HCC1954 breast cancer cell lines and exposed to 20% or 1% O2 for 24 h, normalized to the mean value for each of the cell lines at 20% O2. Data are shown as mean ± standard error of mean, n = 3; *, P < 0.05; ***, P < 0.001 vs. 20% O2 conditions (Student's t test). B, Immunoblot assay was performed using lysates prepared from MDA‐MB‐175, SUM159, and MDA‐MB‐231 cells exposed to 20% or 1% O2 for 48 h to assess the levels of HIF‐1α, HIF‐2α, and RhoB. Note that a 10x higher exposure time was used for RhoB protein expression in MDA‐MB‐231 cells. C, RhoB‐GTP pulldown assay was performed to detect levels of activated RhoB in MDA‐MB‐231 cells exposed to either 20% O2 or 1% O2 conditions. The amount of activated or total RhoB was detected by immunoblotting with a RhoB antibody. Immunoblot assay of (D) SUM159 or (E) MDA‐MB‐231 lysates exposed to 20% or 1% O2 for various time points over a 48‐h time course (left panel). HIF‐1α, HIF‐2α, and RhoB densitometry measurements normalized by actin levels were plotted (right panel). Molecular weights of adjacent ladder bands are indicated
2.2. Loss and gain of RhoB result in a decrease in 3D cell motility under 20% O2 conditions
RhoB has been reported to contribute to changes in cell morphology and motility, two crucial steps in the process of tumor progression and metastasis. Phase contrast images of MDA‐MB‐231 subclones were taken for qualitative observation of morphologies between the cell lines. Slight morphological changes were observed between the subclones (Figures S2A and S2C); however, these observations did not result in quantifiable differences (Figures S2B and S2D).
To determine the role of RhoB on cell motility, we generated MDA‐MB‐231 subclones that were stably transfected with a vector encoding either of two different CRISPR gRNAs targeted against RhoB (RhoB‐1 and RhoB‐2) or a vector overexpressing RhoB (RhoB+), as well as a nontarget control (NTC) cell line (Figures 4A and 5A). RhoA and RhoC levels were not altered by modifying the levels of RhoB (Figures S1). To determine whether RhoB levels could alter cell motility in 3D, each subclonal cell line was then embedded into a 3D collagen matrix and individual cell movements were tracked over a 16‐hour time period under 20% O2 conditions (Figure 2A and Movies S1–S3). Velocity over time represents the rate at which each cell moves, and its persistence time is a measure of the average time period between significant changes in the direction of movement. Both velocity and persistence time are used to calculate diffusivity (D = S2P/4), which indicates overall cell movement within a certain time frame. A small decrease in cell velocity was noted both when RhoB was abrogated as well as overexpressed (Figure 2B). Larger reductions were seen for persistence time (Figure 2C), total cell diffusivity (Figure 2D), and mean squared displacement (MSD; Figure 2E). Given that diffusivity incorporates both velocity and persistence, it follows that there is only a slight difference in the velocity of the subclones, and it is the decrease in persistence that ultimately contributes to the reduction in total cell diffusivity. These results suggest that the level of RhoB expression has a modest but statistically significant effect on modulating diffusivity.
Figure 4.
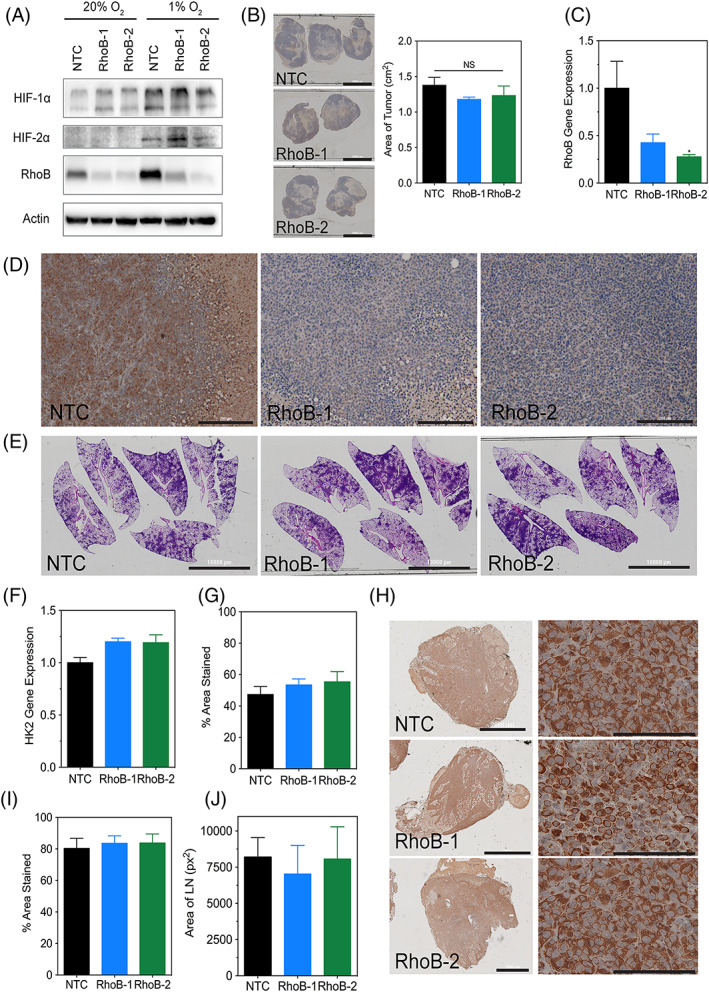
RhoB knockdown does not affect tumor growth or metastasis in vivo. A, Immunoblot assays were performed using lysates prepared from MDA‐MB‐231 nontarget control, RhoB‐1, and RhoB‐2 subclones exposed to 20% or 1% O2 for 48 h and to assess the levels of HIF‐1α, HIF‐2α, and RhoB. Molecular weights of adjacent ladder bands are indicated. B, Tumor sections for the indicated subclones of MDA‐MB‐231 cells that were injected into the mammary fat pad of NSG mice were stained for hematoxylin (scale bar = 10 000 μm) and their respective areas were measured using ImageJ software. Data are shown as mean ± standard error of mean; n = 5 (one‐way analysis of variance with Bonferroni posttest). C, Human RhoB content in the tumors was quantified by quantitative polymerase chain reaction. Data are shown as mean ± standard error of mean; n = 5. *, P < 0.05 vs. nontarget control (one‐way analysis of variance with Bonferroni posttest). D, Tumor sections (scale bar = 200 μm) were subjected to IHC using an antibody against RhoB. E. Whole‐mount inflated lungs (scale bar = 10 000 μm) were stained with hematoxylin and eosin to identify metastatic foci. F, Human genomic DNA content in the lungs was quantified by quantitative polymerase chain reaction for human HK2 gene sequences. G, The percent area stained for hematoxylin for each lung was measured using ImageJ software (see SF5A). Data are shown as mean ± standard error of mean; n = 5 (one‐way analysis of variance with Bonferroni posttest). H, Ipsilateral lymph node sections (scale bar = 2000 μm) were subjected to IHC using an antibody specific for human vimentin. Magnified images (scale bar = 100 μm) are shown on the right. (I) The percent area stained for vimentin and (J) the total area for each lymph node were measured using ImageJ software (see SF5B). Data are shown as mean ± standard error of mean; n = 4–7 (one‐way analysis of variance with Bonferroni posttest)
Figure 5.
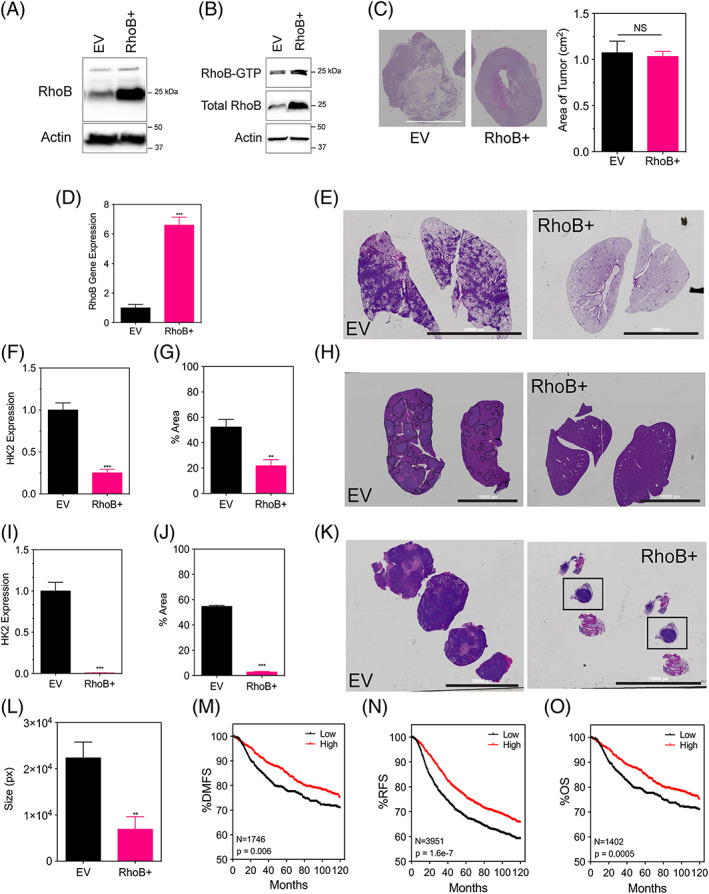
RhoB overexpression does not affect tumor growth in vivo but does contribute to a decreased metastasis. A. Immunoblot assays were performed using lysates prepared from MDA‐MB‐231 EV (empty vector) and a RhoB+ overexpressing subclone under 20% O2. B, RhoB‐GTP pulldown assay was performed to detect levels of activated RhoB in MDA‐MB‐231 EV and RhoB+ subclones under 20% O2. The amount of activated or total RhoB was detected by immunoblotting the pulldown samples and the whole cell lysates with a RhoB antibody. C, Tumor sections for the indicated subclones of MDA‐MB‐231 cells that were injected into the mammary fat pad of NSG mice were stained with hematoxylin in order to measure their area using ImageJ software (scale bar = 10 000 μm). D, Human RhoB content in the tumors was quantified by quantitative polymerase chain reaction. E, H, K. E, Whole‐mount inflated lungs (scale bar = 10 000 μm), (H) livers, and (K) lymph nodes were stained with hematoxylin and eosin to identify metastatic foci. Human genomic DNA content in the (F) lungs and (I) liver was quantified by quantitative polymerase chain reaction for human HK2 gene sequences. The percent area stained for hematoxylin for each (G) lung section and (J) liver section was measured using ImageJ software. L, The size or perimeter of each lymph node section was measured using ImageJ software. All data are shown as mean ± standard error of mean; n = 8. **, P < 0.01; ***, P < 0.001 vs. EV (student t test). Kaplan‐Meier analysis of (M) distant‐free metastasis (n = 1746), (N) relapse‐free survival (n = 3951), and (O) overall survival (n = 1402) of breast cancer patients stratified by RhoB mRNA expression above (high) or below (low) the median level is presented
Figure 2.
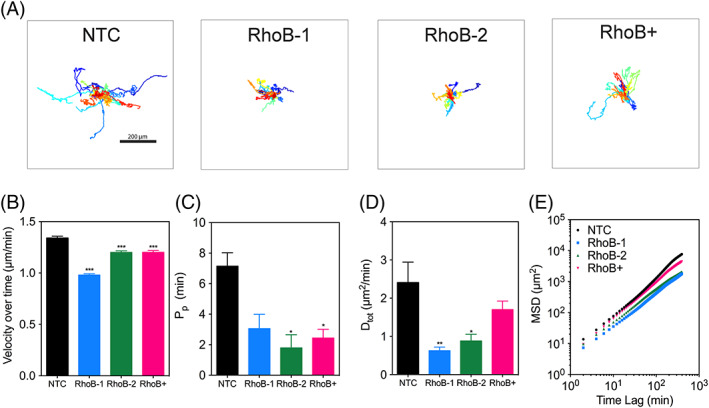
Both loss and gain of RhoB contribute to a decrease in 3D cell motility. A, MDA‐MB‐231 subclones expressing a CRISPR gRNA targeting either a non‐targeting control (NTC), RhoB (RhoB‐1 and RhoB‐2), or containing a vector overexpressing RhoB (RhoB+) were tracked and their trajectories (n = 30–50, N = 2) were plotted using x, y coordinates obtained at 2‐min intervals over a 16‐h time course. Scale bar = 200 μm. B‐E, The (B) average cell velocities, (C) persistence time, (D) total cell diffusivities, and (E) mean squared displacements of the migrating cells were calculated using x, y coordinates obtained from the Metamorph tracking software. Data are shown as mean ± standard error of mean; n = 30–50, N = 2. *, P < 0.05; **, P < 0.01; ***, P < 0.001 vs. NTC (one‐way analysis of variance with Bonferroni posttest for all comparisons)
2.3. Loss and gain of RhoB contribute to a decreased invasion in a 3D multicellular spheroid under 20% O2 conditions
Next we measured, the invasion distance of MDA‐MB‐231 spheroids cultured under 20% versus 1% O2 for 5 days using live cell microscopy (Figure S3A). Spheroids cultured under hypoxia invaded significantly more than those cultured under 1% O2 (Figure S3B). To determine whether overexpressing RhoB is sufficient to phenocopy hypoxia, we generated spheroids composed of NTC, RhoB depleted, and RhoB overexpressing cells and embedded them into 2‐mg/mL collagen gels. The spheroids were imaged on days 0, 2, 4, and 6 under 20% O2 conditions to determine invasion distance over time (Figures 3A and S4A). The spheroids were then traced, and the invaded distance was determined by calculating the spheroid radius and then normalized to the initial radius (Figures 3B and S4B). The results show that both increased and reduced levels of RhoB decrease spheroid invasion when compared with the NTC (Figure 3C). Taken together, the results show that RhoB overexpression is not sufficient to phenocopy hypoxia and suggests that the hypoxic induction of alternate genes such as RhoA and ROCK131 are important for the regulation of the migratory phenotype.
Figure 3.
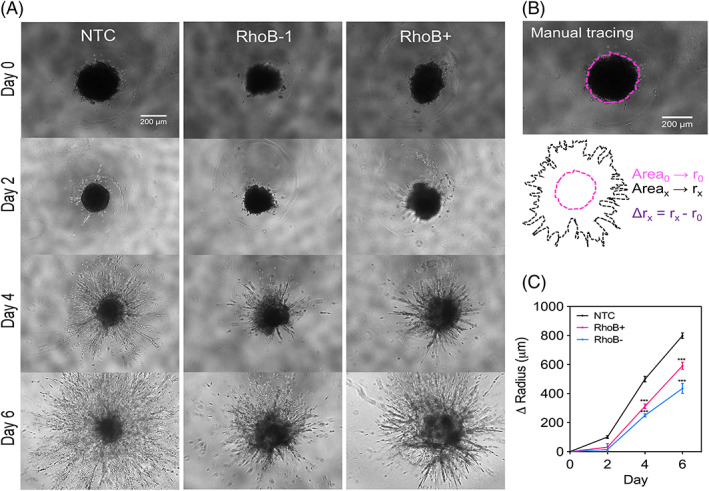
Both loss and gain of RhoB expression contribute to a decreased invasion in a multicellular spheroid model. A, The invasion of tumor spheroids composed of 10 000 nontarget control, RhoB‐1, or RhoB+ cells fully embedded into 2‐mg/mL collagen gels were imaged by phase contrast (n = 10–12, N = 2). B, The area of the spheroids was obtained by manual tracing using the NIS‐Elements software and converted into radius assuming a perfect circle. The invaded distance was determined by subtracting the initial radius to each time point. C, The change in spheroid radius as a result of invasion was measured and plotted at days 0, 2, 4, and 6. Data are shown as mean ± standard error of mean; n = 10–12, N = 2. ***, P < 0.001 vs. nontarget control (two‐way analysis of variance with Bonferroni posttest)
2.4. RhoB knockdown does not affect tumor growth or metastasis in vivo
To further investigate the impact of reducing RhoB expression in tumor progression in vivo, we injected RhoB knockdown or control MDA‐MB‐231 breast cancer cells (Figure 4A) into the MFP (mammary fat pad) of NSG mice. Tumor size, as determined by image analysis, was similar between the NTC and the RhoB knockdown subclones (Figure 4B), suggesting that RhoB does not play an essential role in tumor growth. Tumor tissue was assessed by quantitative polymerase chain reaction (qPCR) (Figure 4C) and immunohistochemistry (IHC) staining (Figure 4D) to confirm that tumors maintained a decreased level of RhoB. Lung metastases were evaluated histologically by hematoxylin and eosin (H&E) staining (Figures 4E and 4G) (method of quantification shown in Figure S5A), as well as by isolating genomic DNA from the mouse lung and quantifying human DNA content using qPCR to detect the human hexokinase 2 (HK2) gene (Figure 4F). Both methods showed no difference in terms of lung metastatic burden in the control versus the RhoB knockdown group. We also assessed breast cancer cell infiltration of the ipsilateral axillary lymph nodes to evaluate the metastatic burden at this site by performing IHC using an antibody that specifically recognizes human vimentin (method of quantification shown in Figure S5B). The lymph nodes of mice bearing both control tumors and RhoB knockdown tumors were entirely infiltrated with breast cancer cells (Figures 4H and 4I). Additionally, both groups of mice had similar sized enlarged lymph nodes (Figure 4J). These data suggest that RhoB is not required for breast cancer progression and metastasis.
2.5. RhoB overexpression does not affect tumor growth in vivo but does decrease metastasis, which is consistent with survival data in patients
Next, we aimed to determine the impact of increased levels of RhoB in tumor progression. We demonstrated that hypoxia increases the amount of activated RhoB (Figure 1C). MDA‐MB‐231 RhoB overexpressing subclones (Figures 5A and 5B) were injected into the MFP of NSG mice to assess tumor growth and metastasis. Tumor growth did not differ between mice bearing control tumors or tumors overexpressing RhoB (Figure 5C). qPCR analysis of tumor tissue, to quantify RhoB gene expression, confirmed that tumors maintained an increased level of RhoB (Figure 5D). Lung and liver metastases were evaluated histologically by H&E staining (Figures 5E, 5G, 5H, and 5J) as well as by quantifying the amount of HK2 human DNA content using qPCR (Figures 5F and 5I). Both methods show that elevated levels of RhoB in the primary tumor leads to significantly decreased lung and liver metastasis in vivo. Lymph node metastasis was analyzed by H&E staining (Figure 5K) and demonstrated that the mice bearing control tumors had enlarged and completely infiltrated lymph nodes, whereas normal follicular lymph node structure and size were maintained in the mice with overexpressed RhoB tumors (Figure 5L). These results suggest that RhoB may act as a suppressor of metastasis in breast cancer. We further analyzed primary human breast tumors,37 stratifying patients by high (above the median level) or low (below the median level) expression of RhoB using Kaplan Meier methods. High levels of RhoB expression were associated with increased distant‐free metastasis (Figure 5M), increased relapse‐free survival (Figure 5N), and increased overall survival (Figure 5O). Both our animal study and RhoB expression data from breast cancer patient tumors suggest a possible association of RhoB with a better prognosis.
3. DISCUSSION
RhoB possesses a significantly different and more complex role in tumorigenesis than RhoA or RhoC despite their close homology.38 We found that hypoxia induces RhoB expression at the mRNA and protein levels as well as increases the levels of activated RhoB. Additionally, we observed that both loss and gain of RhoB resulted in decreased 3D migration and invasion from a multicellular spheroid in vitro but did not phenocopy hypoxia. Finally, overexpression of RhoB in vivo contributed to decreased metastasis to the lungs, liver, and lymph nodes.
3.1. Hypoxia and RhoB
The presence of intratumoral hypoxia, or reduced O2 availability, is associated with an increased risk of invasion and metastasis and therefore an overall worse patient prognosis.39, 40 In renal carcinoma cells (Caki‐1), RhoB protein expression was unaffected by exposure to less than 6 hours of hypoxia.34 In a glioblastoma cell line (U87 cells), hypoxia did not alter the levels of RhoB but did enhance RhoB activation (GTP‐bound RhoB).35 However, in nontransformed cells such as human pulmonary artery smooth muscle cells, endothelial cells, and macrophages, hypoxia has been reported to increase both RhoB gene expression and protein levels.13, 41 This suggests that hypoxia may regulate RhoB differently in distinct cell types. We clearly demonstrate that in several breast cancer cell lines, hypoxia significantly upregulates both RhoB expression and protein levels within 24 hours (Figure 1). However, it should be noted that not all breast cancer cell lines show an induction of RhoB under hypoxic conditions (ie, MCF‐7 cells). It would be interesting to determine why RhoB is induced by hypoxia in only a subset of breast cancer cell lines and to determine whether HIF‐1α or HIF‐2α are required. Further downstream pathway analysis should be considered to elucidate the mechanism of hypoxia‐driven RhoB activation and its consequences on metastasis in breast cancer.
3.2. Threshold for RhoB expression
Our results show that both loss and gain of RhoB lead to a decrease in 3D cell migration and decreased invasion in a 3D multicellular spheroid (Figures 2 and 3). Our previous publications as well as publications by independent groups have shown that hypoxia increases cell motility.31, 40, 42, 43 We show that under hypoxia, spheroids invade significantly more in a 3D collagen matrix (Figure S3). However, our results also demonstrate that increased levels of RhoB expression are not sufficient to recapitulate hypoxia‐induced invasion and migration in 3D. Given these results, we propose that hypoxia induces proteins, such as RhoA, that are responsible for hypoxia‐induced migration and invasion. We also show that overexpression of RhoB inhibits metastasis, whereas abrogating RhoB expression does not promote tumor growth or metastasis (Figures 4 and 5). Based on these observations, migration in vitro and metastasis in vivo are uncoupled and 3D migration in vitro does not appear to play a deterministic role in metastasis. Further research is warranted to investigate the level of RhoB required to inhibit metastasis. Moreover, parameters such as localization of RhoB in the cell as well as the prenylation status of RhoB may also be an important future consideration.
3.3. Opposing roles of RhoB
Several studies have focused on trying to identify a definitive role for RhoB in the tumor microenvironment and its function in the metastatic cascade (reviewed thoroughly elsewhere44). The review44 highlights studies that report RhoB having tumor suppressive functions as well as oncogenic functions. Previous studies show that the tumor microenvironment plays a critical role in determining whether RhoB is able to function as an oncogene or a tumor suppressor depending on the cellular context. For example, Kazerounian et al demonstrated that RhoB differentially regulates the protein kinase B (AKT) pathway in tumor versus stromal endothelial cells, resulting in an overall positive influence of RhoB on angiogenesis and tumor progression that superseded its role as a negative modifier in cancer cells themselves.45 The group demonstrated that orthotopic implantation of shRhoB MDA‐MB‐231 cells exhibited a delay in overall tumor progression. We found no significant difference in tumor growth or metastasis in the control versus the CRISPR‐depleted RhoB in MDA‐MB‐231 cells or RhoB overexpressing cells (Figure 4). Importantly, the overexpression of RhoB blocked breast cancer metastasis in our study (Figure 5). Taken together, further studies focusing on the potential mechanism of RhoB as a metastasis suppressor are warranted.
3.4. Complex role of GTPases in metastasis
The Rho family of GTPases is very diverse and contributes to various steps of cancer progression, including proliferation, survival, invasion, and metastasis. RhoA, RhoC, and Rac1 expression and/or activity are frequently increased in human tumors, whereas RhoB is many times downregulated.22, 46, 47 The well‐established Rho GTPases, RhoA, Rac1, and Cdc42, have been shown to be crucial for the progression and metastasis of various cancers such as breast cancer, ovarian cancer, testicular cancer, and melanoma.48, 49, 50, 51 More recently, RhoA activation was shown to promote cancer stem‐like cell phenotypes that were able to resist chemotherapy in diffuse‐type gastric adenocarcinoma.52 RhoC has also been found to promote tumor progression in several cancer types. For example, in pancreatic cancer, HIF‐3α was shown to transcriptionally regulate RhoC‐Rho‐associated protein kinase 1 signaling to promote invasion and metastasis in a Balb/c mouse model.33 Further, induction of RhoC and TNF‐α expression by downregulation of miR‐509 significantly enhanced brain metastasis of breast cancer cells in mice.53 Rac1 was shown to be required for two different mTORC2 signaling pathways in HER2+ breast cancer to drive metastasis.54 Transcriptional inhibition of miR‐124 resulted in the activation of Rac1, which further activated the JNK pathway to drive pancreatic cancer cell proliferation, invasion, and metastasis.55 In contrast, it has been proposed that RhoB can either act as a tumor suppressor or tumor promoter based on the circumstance.44 Several studies show that RhoB is activated in response to various stress stimuli, such as DNA damage or hypoxia, to affect tumor growth, cell migration, and invasion.11, 14, 35, 56 We demonstrate that RhoB is activated by hypoxia, RhoB overexpression also correlates to increased levels of activated RhoB, and by overexpressing RhoB in vivo, we see a significantly decreased metastatic burden. It would be useful to test whether the in vivo results we see directly correlate to the hypoxic activation of RhoB and further understand the mechanism by which hypoxia regulates RhoB.
4. MATERIALS AND METHODS
4.1. Cell culture
MDA‐MB‐175, MDA‐MB‐231, and HCC1954 cells were obtained from ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM, Sigma Aldrich, St. Louis, MO) supplemented with 10% (v/v) fetal bovine serum (Corning) and 1% penicillin‐streptomycin (Invitrogen). SUM159 and SUM225 cells were kindly provided by the Sukumar lab and were cultured in Ham's F12 medium supplemented with 5% (v/v) fetal bovine serum, 1% penicillin‐streptomycin, and 5% insulin/hydrocortisone. Cells were maintained in a humidified environment at 37°C and 5% CO2 during culture and live cell imaging. Hypoxic cells were maintained at 37°C in a modular incubator chamber (Billups‐Rothenberg) flushed with a gas mixture containing 1% O2, 5% CO2, and 94% N2.
4.2. Knockdown by CRISPR/Cas9
LentiCRISPR v2 plasmid used for generating a CRISPR‐Cas9 endonuclease was a gift from Feng Zhang (Broad Institute, Massachusetts Institute of Technology, Cambridge, MA, obtained via Addgene; Addgene plasmid #52961). RhoB knockout by CRISPR/Cas9 was performed as previously described with slight modifications.57 Insert oligonucleotides that include a guide RNA sequence were designed as shown in Table S1. The NTC is an NTC sgRNA sequence cloned into the CRISPR vector used to clone the sgRNAs targeting RhoB. The NTC was used as the control cell line for the CRISPR‐depleted RhoB cell lines. After annealing, these oligos were inserted into the BsmBI cloning site. After bacterial transformation and DNA purification, all plasmid constructs were confirmed by Sanger sequencing. The LentiCRISPR v2 plasmids were cotransfected with 4 μg PsPAX2 and 1 μg pMD2.G into a 10‐cm dish of 293T cells using PolyJet transfection reagent (SignaGen Laboratories, Rockville, MD) according to the manufacturer's instructions. Media was refreshed 16 to 24 hours following initial transfection. Filtered viral supernatant was collected 48‐hour postmedia change and added to MDA‐MB‐231 cells. Puromycin (0.5 μg/mL) was added to the medium of cells transduced for selection. After selection, cells were expanded and used for experiments.
4.3. Overexpression by gateway cloning
R77‐E279 Hs.RHOB (provided by Dominic Esposito, Addgene plasmid #70563) was subcloned into Gateway destination vector pLenti CMV/TO Puro Dest (670‐1) (Addgene plasmid #17293)58 via LR recombination reaction. The EV vector is an empty vector used to clone RhoB and served as a control for the RhoB expression. After bacterial transformation and DNA purification, all plasmid constructs were confirmed by Sanger sequencing. This expression plasmid was transfected into 293T cells using PolyJet transfection reagent (SignaGen) according to the manufacturer's instructions. Media was refreshed 16 to 24 hours following initial transfection. Filtered viral supernatant was collected 48‐hour postmedia change and added to MDA‐MB‐231 cells. Puromycin (0.5 μg/mL) was added to the medium of cells transduced for selection. After selection, cells were expanded and used for experiments.
4.4. Reverse transcription and qPCR
Total RNA was extracted using TRI Reagent (Zymo Research, Irvine, CA) and the Direct‐zol RNA Mini Prep Plus kit (Zymo Research) according to the manufacturer's instructions. One microgram of total RNA was used for first‐strand DNA synthesis with the iScript cDNA synthesis kit (Bio‐Rad Laboratories, Hercules, CA). qPCR was performed using human‐specific primers and iTaq SYBR Green Universal Master Mix (Bio‐Rad Laboratories). The expression of each target mRNA relative to 18S rRNA was calculated based on the threshold cycle (Ct) as 2‐Δ(ΔCt), where ΔCt = Cttarget − Ct18S and Δ (ΔCt) = ΔCttest − ΔCtcontrol. Primer sequences are shown in Table S2.
4.5. Immunoblot assays
Cells were lysed in IGEPAL CA‐630 buffer (150 mM NaCl, 1% IGEPAL CA‐630, 50 mM Tris‐HCl, pH 8.0, and protease inhibitors) for 10 minutes on ice, centrifuged for 10 minutes at 13 000 rpm at 4°C, and the insoluble debris were discarded. Whole cell lysates were fractionated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane (Bio‐Rad). The membrane was incubated for 1 hour with 5% milk in TBS‐T (Tris‐buffered saline and 0.1% Tween‐20) and then incubated overnight with primary antibodies diluted in blocking buffer. Antibodies against the following proteins were used: HIF‐1α (BD Biosciences, San Jose, CA), HIF‐2α (Novus Biologicals, Littleton, CO), RhoB (Santa Cruz Biotechnology, Dallas, TX), and Actin‐HRP (ProteinTech, Rosemont, IL). The membrane was then washed and incubated with the corresponding HRP‐conjugated secondary antibody (Azure Biosystems, Dublin, CA) for 2 hours. After washing, the chemiluminescence signal was detected on an AZURE C300 using ECL (PerkinElmer, Waltham, MA).
4.6. RhoB activation assay
The levels of activated RhoB were measured by using a RhoA Pull‐down Activation Assay Biochem Kit (Cytoskeleton, Denver, CO, USA), but instead with a RhoB antibody, according to the manufacturer's instructions. MDA‐MB‐231 cells and overexpressed subclones were cultured under appropriate conditions before collecting and treating lysates. Briefly, 400 μg of protein from each sample was incubated with 50 μg of rhotekin‐RBD to capture GTP‐bound Rho proteins. Beads were then isolated, washed, and boiled before loading for sodium dodecyl sulfate polyacrylamide gel electrophoresis. A 60‐μg untreated lysate of each sample was also loaded to probe for total RhoB and beta‐actin. To immunoblot for GTP‐bound RhoB, a RhoB antibody (Santa Cruz Biotechnology, Dallas, TX) was applied instead of the RhoA antibody supplied with the kit.
4.7. Cell migration
Collagen matrices were prepared with soluble rat tail type I collagen in acetic acid (Corning) to achieve a final concentration of 1‐mg/mL collagen. 1M NaOH was then added to normalize the pH to about 7.0. Remaining volume filled with a 1:1 ratio of reconstitution buffer (0.2 HEPES [Sigma‐Aldrich], 0.26 M NaHCO3 [Sigma‐Aldrich], and water as solvent) and culture medium. All matrices were plated in 24‐well culture plates, and collagen gels were solidified for 1 hour in an incubator at 5% CO2 and 37°C. Immediately following solidification, 500 μL of cell culture medium was added on top of the gel.
Cells were incubated for 1 hour before time‐lapse movies were acquired. Cell movements over time were imaged using Biotek's Lionheart automated microscope at 10X. Images were taken every 2 minutes for 16 hours. Cells in the time‐lapse movies were tracked using MetaMorph software to calculate x and y coordinates at each time interval and construct cell trajectory maps. The cell trajectories were fit using an anisotropic persistent random walk (APRW) model of cell motility to calculate average cell velocity (S), persistence time (P p), and total cell diffusivities (D tot).59 MSDs were also calculated and fit to the same APRW model.
4.8. APRW modeling for motility analysis
APRW model analysis was performed as described in detail using MATLAB (code available).59 Three‐dimensional cell trajectory data were used to statistically profile cell migration using the MSD, which can be obtained from (x[t], y[t]) coordinates of cells with time (t). MSD (𝜏) = (x[t + 𝜏] − x[t])2 + (y[t + 𝜏] – y[t])2 where 𝜏 = 2 min * frame number. Values of persistence and speed are obtained from APRW model fitting and expressed as speed (S) and persistence (P) of cells, which can be used to calculated total cell diffusivity (D tot). D tot = (S p 2 P p + S np 2 P np)/4 where both speed (S) and persistence (P) are calculated along both the primary and nonprimary axes.
4.9. Spheroid invasion
Cell spheroids were formed in round‐bottom 96‐well tissue culture plates as described previously.60 Cells (1 × 104 cells/mL) were resuspended in spheroid formation media, (DMEM and Methocult H4100 [3:1]) and centrifuged at 1800 rpm for 20 minutes. Following 72‐hour incubation, spheroids were embedded into 2 mg/mL collagen containing DMEM and soluble rat tail type I collagen (Corning). Spheroids were placed under the appropriate conditions (20% O2 or 1% O2) for each time point and imaged using the Cytation 5 (BioTek) at 10X.
4.10. Animal studies
Female 5‐ to 7‐week‐old NSG (Charles Rivers Laboratories, Wilmington, MA) mice were used according to protocol approved by the Johns Hopkins University Animal Care and Use Committee. Mice were anesthetized, and 2 × 106 MDA‐MB‐231 cells resuspended in a 50:50 PBS:Matrigel solution were injected into the mammary fat pad. Tumors, ipsilateral axillary lymph nodes, livers, and lungs were harvested, formalin fixed, paraffin embedded, and used for IHC staining. Lung and liver tissues were also used to isolate genomic DNA for qPCR to quantify human HK2 and mouse 18S rRNA gene sequences. Tumor tissue was also used to isolate RNA for qPCR to quantify human RhoB and 18S rRNA expression.
4.11. IHC
Tumors were fixed in 10% formalin and were embedded in paraffin. Tumor, lung, liver, and lymph node sections were dewaxed with xylene and rehydrated with graded ethanol, followed by antigen retrieval using citrate‐EDTA buffer (10 mM citric acid [pH 6.1], 2 mM EDTA, and 0.05% Tween‐20) by heating at 85°C for 40 minutes and subsequently cooling at room temperature for 30 minutes. IHC staining of vimentin and RhoB was performed using the LSAB+ System HRP kit (Dako, Agilent Technologies, Santa Clara, CA) according to the manufacturer's instructions. H&E slides of the lungs, livers, lymph nodes, and tumors were scanned using the Lionheart (BioTek) automated microscope at 4X.
4.12. Kaplan‐Meier analysis
Breast cancer data were obtained from the online data portal (https://kmplot.com/) using Affymetrix id 212099_at and JetSet best probe set for RhoB.37 Pearson correlation and Kaplan‐Meier analyses were conducted using GraphPad Prism 6 software (San Diego, CA).
4.13. Statistical analysis
The mean values ± standard error of mean were calculated and plotted using GraphPad Prism 6 software. When appropriate, statistical analysis were performed to compare means, namely, two‐tailed unpaired t tests, one‐way and two‐way analysis of variance followed by Bonferroni posttests to determine statistical significance, which is indicated in the graphs as ***, P < 0.001, **, P < 0.01, and *, P < 0.05.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHORS' CONTRIBUTIONS
All authors had full access to the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Conceptualization: J. A. J. and D. M. G.; Methodology: J.A.J, I.G., and J.W.D; Investigation: J.A.J, I.G, and J.W.D.; Formal analysis : J.A.J., I.G. and J.W.D.; Resources : D.M.G; Writing‐Original Draft, J.A.J.; Writing–Review & Editing, I.G., J.W.D. and D.M.G.; Supervision; D.M.G.; Funding acquisition : D.M.G.
Supporting information
Movie S1: Migration of MDA‐MB‐231 NTC subclones through a 3D collagen matrix, related to Figure 2 . Migration of MDA‐MB‐231 subclones expressing a CRISPR expressing a non‐targeting gRNA (NTC) over a 16 h time course. Images were acquired by phase‐contrast microscopy at 10X magnification with 2‐minute time intervals. Scale bar = 100 μm. Time shown in hours.
Movie S2: Migrating MDA‐MB‐231 RhoB‐1 subclones through a 3D collagen matrix, related to Figure 2 . Migration of MDA‐MB‐231 subclones expressing a CRISPR gRNA targeting RhoB over a 16 h time course. Images were acquired by phase‐contrast microscopy at 10X magnification with 2‐minute time intervals. Scale bar = 100 μm. Time shown in hours.
Movie S3: Migrating MDA‐MB‐231 RhoB+ subclones through a 3D collagen matrix, related to Figure 2 . Migration of MDA‐MB‐231 subclones containing a vector overexpressing RhoB over a 16 h time course. Images were acquired by phase‐contrast microscopy at 10X magnification with 2‐minute time intervals. Scale bar = 100 μm. Time shown in hours.
Movie S4: Tracking example of a migrating MDA‐MB‐231 NTC cell through a 3D collagen matrix, related to Figure 2 . Tracking example of the migration of MDA‐MB‐231 NTC cells using MetaMorph software over a 16 h time course. Images were acquired by phase‐contrast microscopy at 10X magnification with 2‐minute time intervals.
Table S1. Insert Oligonucleotide Sequences used for CRISPR/Cas‐9 knockdown
Table S2. Primer Sequences used for Real‐Time qPCR
Figure S1: RhoA and RhoC RNA levels are not affected by altering RhoB expression. A‐B. A) RhoA and B) RhoC mRNA levels were analyzed by qPCR in the MDA‐MB‐231 knockdown subclones RhoB‐1 and RhoB‐2 normalized to the mean value for NTC. Data are shown as mean ± SEM; n = 2, 6. NS, p > 0.05 vs. NTC (one‐way ANOVA with Bonferroni posttest). C‐D. C) RhoA and D) RhoC mRNA levels were analyzed by qPCR in the MDA‐MB‐231 overexpressed subclone, RhoB+, normalized to the mean value for EV. Data are shown as mean ± SEM; n = 1, 3. NS, P > 0.05 vs. EV (Student t test).
Figure S2: Altering RhoB expression does not affect cell morphology. A. Phase contrast images taken at 10X magnification using the Cytation 5 automated microscope (BioTek) of MDA‐MB‐231 NTC (left), RhoB‐1 knockdown (middle) or RhoB‐2 knockdown (right) (scale bar = 200 μm). B. Area (left), circularity (middle), and axis ratio (right) measurements were taken for the NTC and RhoB knockdown cell lines in the ImageJ software. Data are shown as mean ± SEM; n = 31–34, N = 1. NS, P > 0.05 vs. EV (Student t test). C. Phase contrast images taken at 10X magnification using the Cytation 5 automated microscope (BioTek) of MDA‐MB‐231 EV (left) or RhoB+ overexpression (right) (scale bar = 200 μm). D. Area (left), circularity (middle), and axis ratio (right) measurements were taken for the EV and RhoB overexpressed cell line in ImageJ software. Data are shown as mean ± SEM; n = 33–43, N = 1. NS, P > 0.05 vs. EV (Student t test).
Figure S3: Hypoxia increases migration and invasion in spheroids. A. The invasion of tumor spheroids composed of 10,000 MDA‐MB‐231 cells fully embedded into 2 mg/mL collagen gels and placed under 20% O2 or 1% O2 for 5 days were imaged by phase contrast (scale bar = 500 μm). B. The change in spheroid radius as a result of invasion was measured on day 5, normalized to the spheroid radius at day 0, and plotted. Data are shown as mean ± SEM; n = 17–19, N = 3.2***, P < 0.001 vs. 20% (Student t test).
Figure S4: RhoB knockdown does not affect migration or invasion in spheroids A. The invasion of tumor spheroids composed of 10,000 RhoB‐1 or RhoB‐2 cells fully embedded into 2 mg/mL collagen gels were imaged by phase contrast on days 0, 2, 4, and 6 (scale bar = 200 μm). B. The change in spheroid radius as a result of invasion was measured on day 6, normalized to the spheroid radius at day 0, and plotted. Data are shown as mean ± SEM; n = 17–34, N = 1. NS, P > 0.05 vs. RhoB‐1 (Student t test).
Figure S5: Thresholding method to quantify stained areas in ImageJ. The color deconvolution plug‐in was downloaded for ImageJ and the original file was opened. The menu path was as follows: Image > Color > Colour Deconvolution using either the A. H&E or B. H DAB options. The images indicated were thresholded (Image > Adjust > Threshold) and a region of interest (ROI) was drawn over each individual A. lung or B. lymph node. These ROIs were analyzed and the percent area stained was measured.
Supplemental Figure 1: RhoA and RhoC RNA levels are not affected by altering RhoB expression.
Supplemental Figure 2: Altering RhoB expression does not affect cell morphologies.
Supplemental Figure 3: Hypoxia increases spheroid invasion and migration.
Supplemental Figure 4: RhoB knockdown does not affect migration or invasion in spheroids.
Supplemental Figure 5: Thresholding method to quantify stained areas.
ACKNOWLEDGEMENTS
The authors thank Joon Eoh for reviewing and editing the manuscript. Work in the Gilkes lab is supported by National Institute of Health (National Cancer Institute, U54‐CA210173 and R00‐CA181352), V Scholar Foundation, Susan G. Komen Foundation (CCR17483484), the Jayne Koskinas Ted Giovanis Foundation for Health and Policy, and the Breast Cancer Research Foundation.
Ju JA, Godet I, DiGiacomo JW, Gilkes DM. RhoB is regulated by hypoxia and modulates metastasis in breast cancer. Cancer Reports. 2020;3:e1164. 10.1002/cnr2.1164
REFERENCES
- 1. Parri M, Chiarugi P. Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal. 2010;8(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jaffe AB, Hall A. RHO GTPASES: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21(1):247‐269. 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- 3. Kroon J, Tol S, van Amstel S, Elias JA, Fernandez‐Borja M. The small GTPase RhoB regulates TNFα signaling in endothelial cells. PLoS One. 2013;8(9):e75031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang M, Duhadaway JB, Prendergast GC, Laury‐Kleintop LD. RhoB regulates PDGFR‐beta trafficking and signaling in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2007;27(12):2597‐2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang L, Yang L, Luo Y, Zheng Y. A novel strategy for specifically down‐regulating individual Rho GTPase activity in tumor cells. J Biol Chem. 2003;278(45):44617‐44625. [DOI] [PubMed] [Google Scholar]
- 6. Adamson P, Paterson HF, Hall A. Intracellular localization of the P21(rho) proteins. J Cell Biol. 1992;119(3):617‐627. 10.1083/jcb.119.3.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wherlock M, Gampel A, Futter C, Mellor H. Farnesyltransferase inhibitors disrupt EGF receptor traffic through modulation of the RhoB GTPase. J Cell Sci. 2004;117(15):3221‐3231. [DOI] [PubMed] [Google Scholar]
- 8. Gerald D, Adini I, Shechter S, et al. RhoB controls coordination of adult angiogenesis and lymphangiogenesis following injury by regulating VEZF1‐mediated transcription. Nat Commun. 2013;4(1):2824. 10.1038/ncomms3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prendergast GC. Actin' up: RhoB in cancer and apoptosis. Nat Rev Cancer. 2001;1(2):162‐168. [DOI] [PubMed] [Google Scholar]
- 10. Fritz G, Kaina B. Transcriptional activation of the small GTPase gene rhoB by genotoxic stress is regulated via a CCAAT element. Nucleic Acids Res. 2001;29(3):792‐798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fritz G, Kaina B, Aktories K. The Ras‐related small GTP‐binding protein RhoB is immediate‐early inducible by DNA damaging treatments. J Biol Chem. 1995;270(42):25172‐25177. [DOI] [PubMed] [Google Scholar]
- 12. Jähner D, Hunter T. The ras‐related gene rhoB is an immediate‐early gene inducible by v‐Fps, epidermal growth factor, and platelet‐derived growth factor in rat fibroblasts. Mol Cell Biol. 1991;11(7):3682‐3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wojciak‐Stothard B, Zhao L, Oliver E, et al. Role of RhoB in the regulation of pulmonary endothelial and smooth muscle cell responses to hypoxia. Circ Res. 2012;110(11):1423‐1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang M, Prendergast GC. RhoB in cancer suppression. Histol Histopathol. 2006;21(2):213‐218. [DOI] [PubMed] [Google Scholar]
- 15. Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582(14):2093‐2101. [DOI] [PubMed] [Google Scholar]
- 16. Ridley AJ. RhoA, RhoB and RhoC have different roles in cancer cell migration. J Microsc. 2013;251(3):242‐249. [DOI] [PubMed] [Google Scholar]
- 17. Prendergast GC, Khosravi‐Far R, Solski PA, Kurzawa H, Lebowitz PF, der C. Critical role of Rho in cell transformation by oncogenic Ras. Oncogene. 1995;10(12):2289‐2296. [PubMed] [Google Scholar]
- 18. Ma Y, Gong Y, Cheng Z, et al. Critical functions of RhoB in support of glioblastoma tumorigenesis. Neuro Oncol. 2015;17(4):516‐525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br J Cancer. 2002;87(6):635‐644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calvayrac O, Mazières J, Figarol S, et al. The RAS‐related GTPase RHOB confers resistance to EGFR‐tyrosine kinase inhibitors in non‐small‐cell lung cancer via an AKT‐dependent mechanism. EMBO Mol Med. 2017;9(2):238‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato N, Fukui T, Taniguchi T, et al. RhoB is frequently downregulated in non‐small‐cell lung cancer and resides in the 2p24 homozygous deletion region of a lung cancer cell line. Int J Cancer. 2007;120(3):543‐551. [DOI] [PubMed] [Google Scholar]
- 22. Mazieres J, Antonia T, Daste G, et al. Loss of RhoB expression in human lung Cancer progression. Clin Cancer Res. 2004;10(8):2742‐2750. [DOI] [PubMed] [Google Scholar]
- 23. Wen S‐J, Zhang W, Ni NN, et al. Expression of rho GTPases family in melanoma cells and its influence on cytoskeleton and migration. Oncotarget. 2017;8(18):30112‐30122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Couderc B, Pradines A, Rafii A, et al. In vivo restoration of RhoB expression leads to ovarian tumor regression. Cancer Gene Ther. 2008;15(7):456‐464. [DOI] [PubMed] [Google Scholar]
- 25. Baldwin RM, Parolin D a E, Lorimer I a J. Regulation of glioblastoma cell invasion by PKC iota and RhoB. Oncogene. 2008;27(25):3587‐3595. [DOI] [PubMed] [Google Scholar]
- 26. Lundgren K, Holm C, Landberg G. Hypoxia and breast cancer: prognostic and therapeutic implications. Cell Mol Life Sci. 2007;64(24):3233‐3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. 2014;14(6):430‐439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Z‐J, Semenza GL, Zhang H‐F. Hypoxia‐inducible factor 1 and breast cancer metastasis. J Zhejiang Univ Sci B. 2015. 10.1631/jzus. B1400221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Semenza GL. Hypoxia‐inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33(4):207‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gilkes DM, Semenza GL. Role of hypoxia‐inducible factors in breast cancer metastasis. Future Oncol. 2013;9(11):1623‐1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gilkes DM, Xiang L, Lee SJ, et al. Hypoxia‐inducible factors mediate coordinated RhoA‐ROCK1 expression and signaling in breast cancer cells. Proc Natl Acad Sci U S A. 2014;111(3):E384‐E393. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32. Xue Y, Bi F, Zhang X, et al. Role of Rac1 and Cdc42 in hypoxia induced p53 and von Hippel‐Lindau suppression and HIF1α activation. Int J Cancer. 2006;118(12):2965‐2972. 10.1002/ijc.21763 [DOI] [PubMed] [Google Scholar]
- 33. Zhou X, Guo X, Chen M, Xie C, Jiang J. HIF‐3α promotes metastatic phenotypes in pancreatic cancer by transcriptional regulation of the RhoC–ROCK1 signaling pathway. Mol Cancer Res. 2018;16(1):124‐134. 10.1158/1541-7786.MCR-17-0256 [DOI] [PubMed] [Google Scholar]
- 34. Turcotte S, Desrosiers RR, Béliveau R. HIF‐1alpha mRNA and protein upregulation involves Rho GTPase expression during hypoxia in renal cell carcinoma. J Cell Sci. 2003;116(11):2247‐2260. 10.1242/jcs.00427 [DOI] [PubMed] [Google Scholar]
- 35. Skuli N, Monferran S, Delmas C, et al. Activation of RhoB by hypoxia controls hypoxia‐inducible factor‐1α stabilization through glycogen synthase kinase‐3 in U87 glioblastoma cells. Cancer Res. 2006;66(1):482‐489. [DOI] [PubMed] [Google Scholar]
- 36. Ye IC, Fertig EJ, DiGiacomo JW, Considine M, Godet I, Gilkes DM. Molecular portrait of hypoxia in breast Cancer: a prognostic signature and novel HIF‐regulated genes. Mol Cancer Res. 2018;16(12):1889‐1901. 10.1158/1541-7786.MCR-18-0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lánczky A, Nagy Á, Bottai G, et al. miRpower: a web‐tool to validate survival‐associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160(3):439‐446. 10.1007/s10549-016-4013-7 [DOI] [PubMed] [Google Scholar]
- 38. Karlsson R, Pedersen ED, Wang Z, Brakebusch C. Rho GTPase function in tumorigenesis. Biochimica et Biophysica Acta ‐ Reviews on Cancer. 2009;1796(2):91‐98. 10.1016/j.bbcan.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 39. Gilkes DM. Implications of hypoxia in breast Cancer metastasis to bone. Int J Mol Sci. 2016;17(10):1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ju JA, Godet I, Ye IC, et al. Hypoxia selectively enhances integrin α 5 β 1 receptor expression in breast cancer to promote metastasis. Mol Cancer Res. 2017;15(6):723‐734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang G, Su J, Zhang M, et al. RhoB regulates the function of macrophages in the hypoxia‐induced inflammatory response. Cell Mol Immunol. 2017;14(3):265‐275. 10.1038/cmi.2015.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krishnamachary B, Berg‐Dixon S, Kelly B, et al. Regulation of colon carcinoma cell invasion by hypoxia‐inducible factor 1. Cancer Res. 2003;65(7):2761‐2769. 10.1158/0008-5472.can-04-4122 [DOI] [PubMed] [Google Scholar]
- 43. Chen J, Imanaka N, Chen J, Griffin JD. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E‐cadherin expression and increased cell migration and invasion. Br J Cancer. 2010;102(2):351‐360. 10.1038/sj.bjc.6605486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ju JA, Gilkes DM. Rhob: team oncogene or team tumor suppressor? Genes. 2018;9(2). 10.3390/genes9020067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kazerounian S, Gerald D, Huang M, et al. RhoB differentially controls akt function in tumor cells and stromal endothelial cells during breast tumorigenesis. Cancer Res. 2013;73(1):50‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fritz G, Just I, Kaina B. Rho GTPases are over‐expressed in human tumors. Int J Cancer. 1999;81(5):682‐687. [DOI] [PubMed] [Google Scholar]
- 47. Adnane J, Muro‐Cacho C, Mathews L, Sebti SM, Munoz‐Antonia T. Suppression of rho B expression in invasive carcinoma from head and neck cancer patients. Clin Cancer Res. 2002;8(7):2225‐2232. [PubMed] [Google Scholar]
- 48. Gómez Del Pulgar T, Benitah SA, Valerón PF, Espina C, Lacal JC. Rho GTPase expression in tumourigenesis: evidence for a significant link. Bioessays. 2005;27(6):602‐613. 10.1002/bies.20238 [DOI] [PubMed] [Google Scholar]
- 49. Kamai T, Yamanishi T, Shirataki H, et al. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin Cancer Res. 2004;10(14):4799‐4805. 10.1158/1078-0432.CCR-0436-03 [DOI] [PubMed] [Google Scholar]
- 50. Li XR, Ji F, Ouyang J, Wu W, Qian LY, Yang KY. Overexpression of RhoA is associated with poor prognosis in hepatocellular carcinoma. Eur J Surg Oncol. 2006;32(10):1130‐1134. 10.1016/j.ejso.2006.05.012 [DOI] [PubMed] [Google Scholar]
- 51. Bid HK, Roberts RD, Manchanda PK, Houghton PJ. RAC1: an emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol Cancer Ther. 2013;12(10):1925‐1934. 10.1158/1535-7163.MCT-13-0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yoon C, Cho SJ, Aksoy BA, et al. Chemotherapy resistance in diffuse‐type gastric adenocarcinoma is mediated by RhoA activation in cancer stem‐like cells. Clin Cancer Res. 2016;22(4):971‐983. 10.1158/1078-0432.CCR-15-1356 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53. Xing F, Sharma S, Liu Y, et al. MiR‐509 suppresses brain metastasis of breast cancer cells by modulating RhoC and TNF‐α. Oncogene. 2015;34(37):4890‐4900. 10.1038/onc.2014.412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Morrison Joly M, Williams MM, Hicks DJ, et al. Two distinct mTORC2‐dependent pathways converge on Rac1 to drive breast cancer metastasis. Breast Cancer Res. 2017;19(1):74. 10.1186/s13058-017-0868-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang P, Chen L, Zhang J, et al. Methylation‐mediated silencing of the miR‐124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene. 2014;33(4):514‐524. 10.1038/onc.2012.598 [DOI] [PubMed] [Google Scholar]
- 56. Vega FM, Ridley AJ. The RhoB small GTPase in physiology and disease. Small GTPases. 2016;9(5):1‐10. 10.1080/21541248.2016.1253528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sanjana NE, Shalem O, Zhang F. Improved vectors and genome‐wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783‐784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Campeau E, Ruhl VE, Rodier F, et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4(8):e6529. 10.1371/journal.pone.0006529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu P‐H, Giri A, Wirtz D. Statistical analysis of cell migration in 3D using the anisotropic persistent random walk model. Nat Protoc. 2015;10(3):517‐527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Valencia AMJ, Wu PH, Yogurtcu ON, et al. Collective cancer cell invasion induced by coordinated contractile stresses. Oncotarget. 2015;6(41):43438‐43451. 10.18632/oncotarget.5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1: Migration of MDA‐MB‐231 NTC subclones through a 3D collagen matrix, related to Figure 2 . Migration of MDA‐MB‐231 subclones expressing a CRISPR expressing a non‐targeting gRNA (NTC) over a 16 h time course. Images were acquired by phase‐contrast microscopy at 10X magnification with 2‐minute time intervals. Scale bar = 100 μm. Time shown in hours.
Movie S2: Migrating MDA‐MB‐231 RhoB‐1 subclones through a 3D collagen matrix, related to Figure 2 . Migration of MDA‐MB‐231 subclones expressing a CRISPR gRNA targeting RhoB over a 16 h time course. Images were acquired by phase‐contrast microscopy at 10X magnification with 2‐minute time intervals. Scale bar = 100 μm. Time shown in hours.
Movie S3: Migrating MDA‐MB‐231 RhoB+ subclones through a 3D collagen matrix, related to Figure 2 . Migration of MDA‐MB‐231 subclones containing a vector overexpressing RhoB over a 16 h time course. Images were acquired by phase‐contrast microscopy at 10X magnification with 2‐minute time intervals. Scale bar = 100 μm. Time shown in hours.
Movie S4: Tracking example of a migrating MDA‐MB‐231 NTC cell through a 3D collagen matrix, related to Figure 2 . Tracking example of the migration of MDA‐MB‐231 NTC cells using MetaMorph software over a 16 h time course. Images were acquired by phase‐contrast microscopy at 10X magnification with 2‐minute time intervals.
Table S1. Insert Oligonucleotide Sequences used for CRISPR/Cas‐9 knockdown
Table S2. Primer Sequences used for Real‐Time qPCR
Figure S1: RhoA and RhoC RNA levels are not affected by altering RhoB expression. A‐B. A) RhoA and B) RhoC mRNA levels were analyzed by qPCR in the MDA‐MB‐231 knockdown subclones RhoB‐1 and RhoB‐2 normalized to the mean value for NTC. Data are shown as mean ± SEM; n = 2, 6. NS, p > 0.05 vs. NTC (one‐way ANOVA with Bonferroni posttest). C‐D. C) RhoA and D) RhoC mRNA levels were analyzed by qPCR in the MDA‐MB‐231 overexpressed subclone, RhoB+, normalized to the mean value for EV. Data are shown as mean ± SEM; n = 1, 3. NS, P > 0.05 vs. EV (Student t test).
Figure S2: Altering RhoB expression does not affect cell morphology. A. Phase contrast images taken at 10X magnification using the Cytation 5 automated microscope (BioTek) of MDA‐MB‐231 NTC (left), RhoB‐1 knockdown (middle) or RhoB‐2 knockdown (right) (scale bar = 200 μm). B. Area (left), circularity (middle), and axis ratio (right) measurements were taken for the NTC and RhoB knockdown cell lines in the ImageJ software. Data are shown as mean ± SEM; n = 31–34, N = 1. NS, P > 0.05 vs. EV (Student t test). C. Phase contrast images taken at 10X magnification using the Cytation 5 automated microscope (BioTek) of MDA‐MB‐231 EV (left) or RhoB+ overexpression (right) (scale bar = 200 μm). D. Area (left), circularity (middle), and axis ratio (right) measurements were taken for the EV and RhoB overexpressed cell line in ImageJ software. Data are shown as mean ± SEM; n = 33–43, N = 1. NS, P > 0.05 vs. EV (Student t test).
Figure S3: Hypoxia increases migration and invasion in spheroids. A. The invasion of tumor spheroids composed of 10,000 MDA‐MB‐231 cells fully embedded into 2 mg/mL collagen gels and placed under 20% O2 or 1% O2 for 5 days were imaged by phase contrast (scale bar = 500 μm). B. The change in spheroid radius as a result of invasion was measured on day 5, normalized to the spheroid radius at day 0, and plotted. Data are shown as mean ± SEM; n = 17–19, N = 3.2***, P < 0.001 vs. 20% (Student t test).
Figure S4: RhoB knockdown does not affect migration or invasion in spheroids A. The invasion of tumor spheroids composed of 10,000 RhoB‐1 or RhoB‐2 cells fully embedded into 2 mg/mL collagen gels were imaged by phase contrast on days 0, 2, 4, and 6 (scale bar = 200 μm). B. The change in spheroid radius as a result of invasion was measured on day 6, normalized to the spheroid radius at day 0, and plotted. Data are shown as mean ± SEM; n = 17–34, N = 1. NS, P > 0.05 vs. RhoB‐1 (Student t test).
Figure S5: Thresholding method to quantify stained areas in ImageJ. The color deconvolution plug‐in was downloaded for ImageJ and the original file was opened. The menu path was as follows: Image > Color > Colour Deconvolution using either the A. H&E or B. H DAB options. The images indicated were thresholded (Image > Adjust > Threshold) and a region of interest (ROI) was drawn over each individual A. lung or B. lymph node. These ROIs were analyzed and the percent area stained was measured.
Supplemental Figure 1: RhoA and RhoC RNA levels are not affected by altering RhoB expression.
Supplemental Figure 2: Altering RhoB expression does not affect cell morphologies.
Supplemental Figure 3: Hypoxia increases spheroid invasion and migration.
Supplemental Figure 4: RhoB knockdown does not affect migration or invasion in spheroids.
Supplemental Figure 5: Thresholding method to quantify stained areas.


