ABSTRACT
Legionnaires’ disease (LD) is an established cause of pneumonia, and the disease remains largely underdiagnosed. Even though LD has been reported from many parts of the world, only sporadic cases have been reported in India. During February 2015–January 2020, we enrolled 597 patients with radiographically confirmed pneumonia and tested respiratory secretions for Legionella spp. by using real-time PCR, and culture. A commercial urinary antigen test (UAT) was also used to detect the Legionella pneumophila (Lp) serogroup 1 antigen in urine. An LD case was defined as a patient with pneumonia and positive results for Legionella spp. infections determined by real-time PCR (from any respiratory specimen) or culture or UAT. Demographic data, risk factors, clinical, radiological, and outcome data of Lp-positive and Lp-negative patients were compared using logistic regression. Over the study period, 14 (2.3%) patients were positive for Legionella spp. infections by real-time PCR and UAT; eight (57%) were admitted to the intensive care unit, and four (28.6%) in-hospital deaths occurred. Bivariate analysis showed that renal disease, neurological conditions, confusion, leukocytosis, and requirement of oxygen support were more common in the Lp-positive group than in the Lp-negative group. However, multivariate analysis failed to confirm most of these differences; renal disease was the only independent variable remaining significant. All test methods have intrinsic limitations in identifying Legionella; therefore, more than one testing method should be used. Application of molecular assays including real-time PCR has great value because of its high sensitivity, specificity, and rapid diagnostic potency. Increased awareness and improved diagnostic testing could facilitate early detection of cases, pathogen-directed therapy, and improved outcomes for patients.
INTRODUCTION
Legionnaires’ disease (LD) is a form of atypical pneumonia caused by bacteria belonging to the genus Legionella.1,2 Most of the clinical cases are attributable to Legionella pneumophila serogroup 1 (Lp1), and therefore, most of the diagnostic tests are specific for the detection and diagnosis of this serogroup.3–5 Risk factors for LD include exposure to whirlpool spa, recent overnight travel, immunosuppression, alcohol misuse, diabetes mellitus (DM), malignancy, hepatic or renal failure, chronic obstructive pulmonary disease (COPD), cigarette smoking, and older age.6–8 On clinical grounds, it is difficult to distinguish Legionella pneumonia from other bacterial causes of pneumonia because of overlapping clinical manifestations and radiographic findings; therefore, microbiological diagnosis is warranted.9 However, the disease is underdiagnosed and underreported in many countries because of lack of common definitions, diagnostic assays, surveillance, and reporting systems.10,11 Several methods exist for the diagnosis of legionellosis, including culture, serological tests, and urinary antigen test (UAT), albeit; none of these assays provide the desired quality with respect to sensitivity and specificity and diagnosis of all Legionella spp. in a clinically useful time frame.4,12–14 Currently, Legionella-specific PCR and their real-time counterparts have emerged as effective diagnostic strategies enabling rapid and reliable identification of all Legionella species.10,15–17
In India, only limited data are available on the prevalence of Legionella spp. that causes pneumonia, despite the high proportion of the vulnerable and at-risk populations. Legionellosis is not a notifiable disease in this country and is rarely reported.18,19 In this study, we investigated the prevalence of Legionella spp. in patients with pneumonia who were given a diagnosis at a referral hospital in India and describe the epidemiological and clinical features of LD case-patients.
MATERIALS AND METHODS
Study design and study population.
A prospective, hospital-based, observational study was conducted during February 2015–January 2020, at the All India Institute of Medical Sciences (AIIMS), a referral hospital that represents the highest level of health care in New Delhi, India. The Ethical Committee of the AIIMS has provided approval for this study (IECPG-169/15). A case of pneumonia was defined according to the American Thoracic Society and Infectious Diseases Society of America (IDSA) criteria.20 The participants were assessed for their eligibility based on the following inclusion and exclusion criteria.
Inclusion criteria.
The study included patients with radiographic evidence of pneumonia and three or more of the following signs or symptoms: fever (> 37.8°C), new or increased cough, sputum production, breathlessness, pleuritic chest pain or signs consistent with pneumonia on auscultation, abnormal white blood cell (WBC) count (either leukocytosis [elevated WBC count > 11,000 cells/μL] or leukopenia [reduction in the WBC count to < 4,000 cells/μL]), or C-reactive protein values greater than the local upper limit (> 30 mg/L). The presence of consolidation, other infiltrates, or pleural effusion, etc. was defined as radiographic evidence of pneumonia.
Patients with physician-diagnosed community-acquired pneumonia (CAP) and hospital-acquired pneumonia (HAP) were enrolled in the study. Community-acquired pneumonia is defined as pneumonia in a person in a community setting, who has not been hospitalized in the preceding 14 days or is not a resident in a nursing home or long-term care facility. Hospital-acquired pneumonia is defined as pneumonia not incubating at the time of hospital admission and occurring 48 hours or more after admission.
Demographic and clinical details were collected from each enrolled patient using a standardized questionnaire.
Exclusion criteria.
Patients without radiographic evidence of pneumonia were excluded from this study. Patients who did not give consent were also excluded.
Specimen collection and laboratory testing.
Respiratory specimens and urine were obtained from each patient for Legionella spp. testing using multiple methods as previously described (Supplementary Material). In brief, respiratory secretions were tested for Legionella spp. by using a real-time PCR assay targeting the ssrA gene, as previously described.15 Specimens determined as positive for Legionella spp. were further tested by two different real-time PCR assays to identify Lp and Lp1.15 In addition, respiratory samples were streaked onto buffered charcoal yeast extract (BCYE, Becton Dickinson, Sparks, MD) supplemented with BMPA-α (cefamandole, polymyxin B, and anisomycin) selective supplements (Oxoid, Basingstoke, United Kingdom) for Legionella culture. Urine samples were tested for Lp1 using the BinaxNOW Legionella urinary antigen card test (Alere, Scarborough, ME). An LD case was defined as a clinically compatible patient with positive results for Legionella spp. infections, determined by real-time PCR or culture or UAT.
Statistical analysis.
The descriptive analysis of the patient’s demographic and clinical features including the illness severity and laboratory findings were performed using STATA software (15.1) (StataCorp LLC, College Station, TX). Qualitative variables were expressed in numbers and percentages and quantitative variables as mean ± SD or median (interquartile range). We performed a bivariate analysis comparing patients with and without Lp, the differences of qualitative variables between the two groups; that is, Lp-positive and Lp-negative were assessed by using Pearson chi-square or Fisher’s exact tests. Differences in quantitative variables between the two groups were assessed by either t-test or rank-sum tests depending on the normality assumption, as appropriate. In addition, binary logistic regression was also used to present the results in terms of the unadjusted odds ratio under the bivariate analysis.
Variables shown to be marginally associated (P < 0.20) with the outcome (status of L. pneumophila) under bivariate analysis were entered into a multivariable logistic regression model to get the adjusted odds ratio (aOR). Results were expressed in the form of odds ratio (OR) with corresponding 95% CI. P-value < 0.05 was considered statistically significant.
RESULTS
Study population.
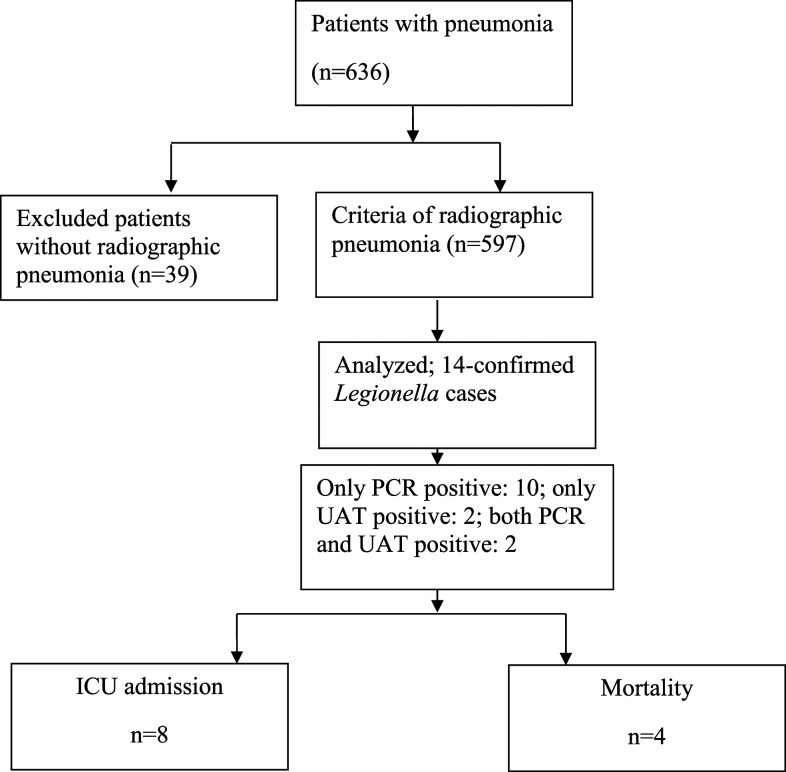
During the study period, a total of 636 patients with pneumonia were eligible, of which, 597 (93.8%) met the criteria of radiographic pneumonia, and were enrolled (Figure 1). Of these, 413 (69.2%) were CAP patients, and the remaining (184 [30.8%]) were HAP patients. Among the enrolled patients (n = 597), 349 (58.4%) were males, and 443 (74.2%) were adults > 15 years of age. Of these, 244 patients (40.8%) had a bronchoalveolar lavage (BAL) fluid tested only, 232 (38.8%) had sputum tested only, 95 (15.9%) had endotracheal (ET) aspirations tested only, and eight (1.3%) had pleural fluid tested for Legionella spp. For 18 patients from whom sputum could not be obtained, nasopharyngeal aspirates (n = 14) and throat swabs (n = 4) were tested. Underlying medical conditions including cancer, DM, hypertension, chronic lung diseases, renal diseases, cardiovascular diseases, and neurological conditions were known for 78% (466/597) of patients. Risk factors such as cigarette smoking and/or alcohol intake were known for 20.4% (122/597) patients. A history of exposure to air conditioners and potential water sources was known for 42.2% (252/597) of persons.
Figure 1.
Patient disposition and selection for Legionella testing in a referral hospital, India, February 2015–January 2020 (n = 597). ICU = intensive care unit; UAT = urinary antigen test.
Detection rate of Legionella spp.
Of the 597 patients, 12 (2%) tested positive for Legionella spp. by real-time PCR on sputum (n = 6), BAL fluid (n = 5), and ET aspirate (n = 1). Further testing by real-time PCR for these specimens (n = 12) was positive for L. pneumophila and Lp1. Among the 12 patients tested positive by real-time PCR, the UAT was performed for eight, and two were positive. Overall, the UAT was performed for 363 (61%) specimens, and four (1.1%) were positive. For two patients who tested negative by real-time PCR, the UAT indicated positive results; so in total, 14 (2.3%) patients with Legionella infections were identified. There was no isolation in culture on BCYE-α.
Demographic characteristics.
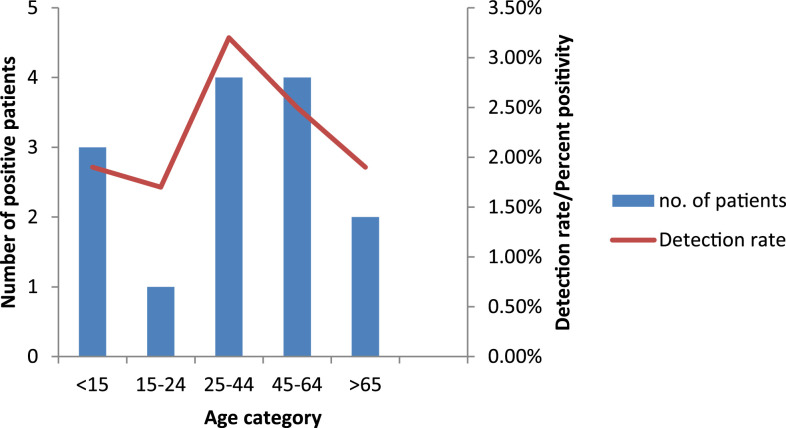
Among the 14 patients with laboratory-confirmed Legionella infections (13 cases of community and one of nosocomial origin), eight (57.1%) were male and 11 (78.6%) were adults (> 15 years); the male–female ratio was 1.3:1. The median age of the patients was 30 years (range 12–85 years; Figure 2). Of the 14 patients, eight (57.1%) were admitted to intensive care units (ICUs). More than half of the patients had underlying diseases (11 [78.6%]), including lung disease (six [42.9%]), renal disease (five [35.7%]), neurological conditions (two [14.3%]), DM (one [7.1%]), and malignancy (one [7.1%]). Four patients (28.6%) had additional risk factors including cigarette smoking or alcohol use. Exposure to potential sources of infection including air conditioners and water systems was known for six (42.9%) patients. None of their family members developed fever or any respiratory symptoms, and there was no epidemiological link between the LD-case patients.
Figure 2.
Number of case-patients and detection rates (percent positivity) for Legionella spp. infections by age-group, in a referral hospital, India, February 2015–January 2020 (n = 597). This figure appears in color at www.ajtmh.org.
Clinical features and laboratory results.
The initial clinical presentation of all patients was pneumonia; confusion was present in seven (50%) patients, pleuritic chest pain in five (35.7%), hemoptysis in two (14.3%), and diarrhea in five (35.7%) patients. Overall, extrapulmonary manifestations were found in 10 (71.4%) patients. Four (28.5%) of 14 patients had symptoms ≥ 7 days before hospital admission. The median time from the onset of symptoms to hospital admission was 5 days (range: 1–29 days). The most common chest radiographic finding was infiltrations seen in nine (64.29%) patients. Lung air space consolidations were seen in three (21.43%) patients, and ground-glass opacities and reticular opacities were present in one (7.1%) each patient. The laboratory findings included leukocytosis (n = 11 [78.6%]), elevated liver enzymes (aspartate aminotransferase (AST)/alanine aminotransferase (ALT); n = 9 [64.3%]), hyponatremia (n = 4 [28.6%]), and hypophosphatemia (n = 1 [7.1%]).
Treatment, complications, and outcome.
Common complications seen in Legionella-positive patients included acute respiratory distress syndrome (ARDS, two [14.3%] of 14 patients), sepsis (four [28.6%] patients), and secondary infections (two [14.3%] patients). Oxygen support was required for 10 (71.4%) patients.
Eleven of 14 (78.5%) patients were already on anti-Legionella therapy before the test results were available. For two patients, a change in therapy was made after testing; antibiotics active against Legionella spp. were added. Overall, six patients (42.9%) were treated with levofloxacin, six (42.9%) with azithromycin, and one (7.1%) with doxycycline. One patient did not receive anti-Legionella therapy because LD was retrospectively identified in this case. Therefore, the test results were not available to treating clinicians. The duration of hospital stay was 4–24 days (median 12 days) for Legionella-positive patients. Ten (71.4%) patients survived the illness, and four (28.6%) died despite antibiotic treatment and supportive therapies. The demographic, epidemiologic, and clinical characteristics of LD-case patients are shown in Table 1.
Table 1.
Demographic, epidemiologic, and clinical characteristics of patients positive for Legionella pneumophila, using detection by real-time PCR, urinary antigen testing, and culture on BCYE
| Case no | Hospital admission date | Gender, age (years) | Symptom duration (days) | Potential exposure to contaminated water source | Risk factors and/or underlying disease | Real-time PCR/UAT/BCYE | Intensive care unit admission | Duration of hospital stay (days) (treatment) | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| 1 | August 25, 2015 | Male, 30 | 20 | Yes, visited hospital | Smoking, AKI | +/NP/− | No | 14 (levofloxacin) | Survived |
| 2 | September 24, 2015 | Male, 56 | 30 | Yes, visited hospital | Smoking, alcoholism, COPD | +/+/− | Yes | 23 (levofloxacin) | Expired |
| 3 | September 28, 2015 | Male, 54 | 14 | Yes, visited hospital | Smoking, malignancy (acute myeloid leukemia) | +/−/− | No | 10 (meropenem, cefoperazone–sulbactam) | Expired |
| 4 | October 25, 2015 | Male, 72 | 7 | None | COPD, AKI | +/NP/− | Yes | 16 (levofloxacin) | Expired |
| 5 | May 26, 2016 | Female, 17 | 12 | Not known | None | +/−/− | No | 12 (levofloxacin) | Survived |
| 6 | June 11, 2016 | Female, 13 | 10 | Unknown | Neurological condition | +/−/− | No | 17 (azithromycin, rifampicin) | Survived |
| 7 | June 13, 2016 | Female, 85 | 4 | Not known | Diabetes mellitus, HTN | +/−/− | No | 4 (azithromycin) | Survived |
| 8 | July 14, 2016 | Male, 12 | 14 | Yes, visited hospital | Bronchial asthma | +/NP/− | Yes | 24 (azithromycin) | Survived |
| 9 | August 15, 2016 | Female, 13 | 6 | Not known | ARDS | +/NP/− | Yes | 10 (azithromycin) | Survived |
| 10 | July 18, 2017 | Male, 51 | 30 | Not known | Alcoholism | +/−/− | No | 15 (azithromycin) | Survived |
| 11 | October 1, 2017 | Female, 25 | 7 | Yes, visited hospital | AKI | −/+/− | Yes | 9 (levofloxacin) | Survived |
| 12 | October 14, 2017 | Male, 30 | 9 | None | Neurological condition | −/+/− | Yes | 4 (doxycycline) | Survived |
| 13 | October 28, 2017 | Female, 28 | 5 | Not known | ARDS, AKI | +/−/− | Yes | 9 (azithromycin) | Survived |
| 14* | July 5, 2019 | Male, 57 | 3 | Yes, healthcare exposure | HTN, interstitial lung disease | +/+/− | Yes | 11 (levofloxacin) | Expired |
AKI = acute kidney injury; ARDS = acute respiratory distress syndrome; BCYE = buffered charcoal yeast extract agar; COPD = chronic obstructive pulmonary disease; HTN = hypertension; NP = not performed; UAT = urinary antigen test.
Sequence-based typing on direct BAL fluid showed infection due to ST1.
The association of patient’s demographic and clinical features including disease severity and laboratory parameters with Lp positivity is shown in terms of odds ratio using bivariate and multivariate analysis in Tables 2 and 3, respectively. On bivariate analysis, neurological conditions, renal disease, confusion, leukocytosis, and the requirement of oxygen support were significantly more common (P < 0.05) in the Lp-positive group than in the Lp-negative group (Table 2). However, the multivariate analysis failed to confirm most of these differences. The only independent factor associated with Lp-positive patients using this model was renal disease (aOR: 3.40; 95% CI: 0.99–11.5; P = 0.05; Table 3).
Table 2.
Demographic characteristics, comorbidities, and clinical and laboratory findings of patients having pneumonia with and without Lp
| Characteristic | Lp positive (n = 14), n (%) | Lp negative (n = 583), n (%) | Unadjusted odds ratio (95% CI) | P-value |
|---|---|---|---|---|
| Population, adults | 11 (78.5) | 432 (74.10) | 0.78 (0.21–2.8) | 0.706 |
| Gender, male | 8 (57.14) | 341 (58.49) | 1.05 (0.36–3.08) | 0.919 |
| Exposure history | 6 (42.86) | 246 (42.20) | 1.02 (0.35–2.9) | 0.960 |
| Cigarette smoking | 3 (21.43) | 105 (18.01) | 1.24 (0.34–4.5) | 0.743 |
| Alcohol intake | 2 (14.29) | 74 (12.69) | 1.14 (0.25–5.22) | 0.860 |
| Comorbid conditions | ||||
| Any comorbid conditions | 11 (78.57) | 455 (78.04) | 1.03 (0.28–3.75) | 0.962 |
| Diabetes | 1 (7.14) | 90 (15.44) | 0.42 (0.05–3.26) | 0.408 |
| Malignancy | 1 (7.14) | 83 (14.24) | 0.46 (0.05–3.58) | 0.461 |
| Hypertension | 2 (14.29) | 102 (17.50) | 0.78 (0.17–3.5) | 0.755 |
| Chronic obstructive pulmonary disease | 3 (21.43) | 59 (10.12) | 2.4 (0.65–8.9) | 0.184 |
| Bronchial asthma | 1 (7.14) | 22 (3.77) | 1.96 (0.24–15.6) | 0.525 |
| Neurological disorder | 2 (14.29) | 21 (3.60) | 4.46 (0.93–21.2) | 0.040 |
| Renal disease | 5 (35.71) | 60 (10.29) | 4.84 (1.57–14.9) | 0.006 |
| Clinical presentation | ||||
| Fever | 14 (100) | 525 (90.05) | 1 | 0.214 |
| Cough | 12 (85.71) | 505 (86.62) | 0.92 (0.2–4.2) | 0.922 |
| Hemoptysis | 2 (14.29) | 50 (8.58) | 1.77 (0.38–8.1) | 0.460 |
| Dyspnoea | 13 (92.86) | 477 (81.82) | 2.88 (0.37–22.3) | 0.309 |
| Pleuritic chest pain | 5 (35.71) | 118 (20.24) | 2.18 (0.72–6.65) | 0.167 |
| Confusion | 7 (50) | 141 (24.19) | 3.13 (1.08–9.09) | 0.035 |
| Myalgia | 4 (28.57) | 121 (20.75) | 1.52 (0.47–4.9) | 0.481 |
| Abdominal pain | 4 (28.57) | 77 (13.21) | 2.62 (0.8–8.58) | 0.110 |
| Diarrhoea | 5 (35.71) | 95 (16.30) | 2.85 (0.93–8.7) | 0.065 |
| Chest radiographic findings | ||||
| Pulmonary infiltrations | 9 (64.29) | 303 (51.97) | 1.66 (0.55–5.02) | 0.367 |
| Lung consolidations | 3 (21.43) | 162 (27.79) | 0.71 (0.19–2.57) | 0.601 |
| Ground glass opacities | 1 (7.14) | 25 (4.29) | 1.71 (0.21–13.64) | 0.609 |
| Reticular opacities | 1 (7.14) | 22 (3.77) | 1.96 (0.24–15.67) | 0.525 |
| Laboratory findings | ||||
| Leukocytosis | 11 (78.57) | 285 (48.89) | 3.83 (1–13.8) | 0.041 |
| Leukopenia | 1 (7.14) | 77 (13.21) | 0.50 (0.06–3.9) | 0.514 |
| Elevated liver enzymes | 9 (64.29) | 251 (43.05) | 2.38 (0.78–7.19) | 0.124 |
| Hyponatremia | 4 (28.57) | 130 (22.30) | 1.39 (0.43–4.5) | 0.580 |
| Hypophosphatemia | 1 (7.14) | 76 (13.04) | 0.51 (0.06–3.9) | 0.523 |
| Complications | ||||
| Acute respiratory distress syndrome | 2 (14.29) | 71 (12.18) | 1.2 (0.26–5.4) | 0.812 |
| Admission to intensive care unit | 8 (57.14) | 344 (59.01) | 0.92 (0.31–2.7) | 0.889 |
| Ventilatory support | 10 (71.43) | 241 (41.34) | 3.5 (1–11.4) | 0.034 |
| Duration of hospital stay | 12.57 ± 5.95 | 18.70 ± 18.96 | 0.965 (0.914–1.02) | 0.212 |
| Survived | 10 (71.43) | 393 (67.87) | – | 0.778 |
| Expired | 4 (28.57) | 187 (32.13) | ||
| Antibiotic pretreatment | 14 (100) | 478 (81.99) | 1 | 0.080 |
Lp = Legionella pneumophila. Bold text indicates statistical significance.
Table 3.
Multivariate analysis of factors influencing Legionella pneumophila positivity
| Variable | Adjusted odds ratio | 95% CI | P-value |
|---|---|---|---|
| Neurological conditions | 4.17 | 0.79–21.9 | 0.093 |
| Renal disease | 3.40 | 0.99–11.5 | 0.050 |
| Confusion | 1.80 | 0.57–5.63 | 0.310 |
| Abdominal pain | 1.28 | 0.31–5.23 | 0.735 |
| Diarrhoea | 2.38 | 0.64–8.76 | 0.191 |
| Mechanical ventilation | 2.23 | 0.64–7.6 | 0.203 |
| Leukocytosis | 2.91 | 0.76–11.10 | 0.118 |
| Elevated liver enzymes | 1.69 | 0.53–5.38 | 0.368 |
Bold text indicates statistical significance.
The majority (92.8% [13/14]) of the LD cases were community-acquired sporadic cases, and a common exposure source was not identified. Hence, epidemiological investigations were not performed. Besides, the sites were geographically distant from one another, so the respective cases were unlikely to be related. For one patient, who was previously admitted in another hospital, Legionella infection could have possibly acquired from that facility (HAP); therefore, water sampling and testing were not conducted.
Legionella spp. and coinfections.
Among all positive cases, Legionella spp. was found to be the only pathogen detected in 13 (92.3%) cases. Coinfecting bacterial pathogen (Klebsiella pneumoniae) was seen in only one (7.1%) case. Data regarding the viral pathogens in our study population were not available.
By reviewing the medical records and laboratory reports, it was found that among the Lp-negative group (n = 583), 173 (29.7%) patients had positive cultures for Acinetobacter spp. (68/583 [11.7%]), K. pneumoniae (46/583 [7.9%]), Pseudomonas spp. (37/583 [6.3%]), Escherichia coli (9/583 [1.5%]), Staphylococcus aurues (4/583 [0.7%]), and other Gram-negative bacilli (9/583 [1.5%]) from respiratory specimens. PCR for Mycoplasma pneumoniae was positive for 8/583 (1.4%) patients on respiratory secretions.
DISCUSSION
Many legionellosis cases have been reported worldwide, including thousands of cases in the United States and Europe each year.21–23 This increase in the number of reported LD cases is mainly related to the widespread use of Legionella UAT as a predominant testing method. But these tests target only the most prevalent serogroup (Lp1) thus precluding the diagnosis of cases due to non-Lp1 stains. Legionnaires’ disease is usually diagnosed as unexplained pneumonia, and the difficulty of various assays to diagnose this low prevalence disease has been reported in former studies.24 In this study, we identified Legionella spp. infections in 2.3% of patients with pneumonia by using real-time PCR and UAT.
Legionella is not a part of normal human flora; therefore, the presence of Legionella DNA in patient samples is considered to be clinically significant. Nucleic acid amplification tests including PCR and real-time PCR has good sensitivity and specificity for the diagnosis of Legionella spp. in respiratory secretions, but the main criticism against these assays is the lack of standardization in performance.4,25 In this study, the percent positivity of real-time PCR alone was around 2%. Among the two patients who tested positive by UAT, the real-time PCR indicated negative results. This discordance could be attributable to an inadequate respiratory sample, or improper specimen collection and handling, and/or due to the presence of PCR inhibitors in the specimen.13,24
The current IDSA practice guidelines for CAP recommends the UAT as the primary diagnostic test for patients having risk factors for LD. In our study, among six patients who tested positive by real-time PCR, the UAT indicated negative results. This discrepancy could be due to the degradation of the urine antigen in the stored samples, and therefore warrants the critical need for immediate testing of urine specimens.26 In addition, this could be also related to the lower sensitivity of the UAT in milder infections which demands a higher concentration of urine sample while performing the assay.27 False-negative results by the UAT have been reported in previous studies.24,28
We could not isolate Legionella in culture from PCR-positive respiratory specimens, and this may be due to the administration of broad-spectrum antibiotics in patients before Legionella testing. Nine of 12 PCR-positive patients had empirically received a macrolide (azithromycin, n = 4) or a fluoroquinolone (levofloxacin, n = 5) antibiotic. Antibiotic exposure might have affected pathogen detection by culture. In addition to this, a delay in specimen collection and processing in a few patients might have hampered the ability to grow these organisms. Legionella culture is rarely used, and the low sensitivity of this method compared with the PCR and UAT has been previously reported.6,24 Legionella bacteria may survive poorly in the respiratory secretions. Therefore, to improve the success rate of culturing Legionella spp., specimens should be collected before antibiotic treatment, transported immediately to the laboratory, and processed promptly. Bronchoscopic samples are reported to produce a greater diagnostic yield than expectorated sputum samples.16
Legionella pneumophila detection rates in this study were in line with those reported from other countries.11,29 Lower detection rates for Legionella spp. have also been reported from South Africa (1.2%), China (3.9%), and Canada (3%).24,29,30 In the United States, detection rates of Legionella spp. ranged from 2% to 9% among patients with pneumonia.4 Legionella pneumophila serogroup 1 was the most common serotype identified in our patient population. However, in environmental surveys, non-Lp1 serogroups were also recovered from hospital water samples.31,32 Continuous clinical surveillance is required to identify the significance of these serotypes in LD patients.
In this study, regarding the age-wise distribution of LD case-patients, both younger adults and the elderly population were equally infected. This is in agreement with a recent study from South Africa29; however, in most of the international studies, elderly populations are the most affected.4,11 Males and females were almost evenly distributed among Legionella-positive cases in this study with a slight male predominance. However, a substantial male predominance is generally seen in association with legionellosis.11,33,34 These variations may be due to differences in study population, temporal or geographical differences in Legionella activity, or the time duration of the study.
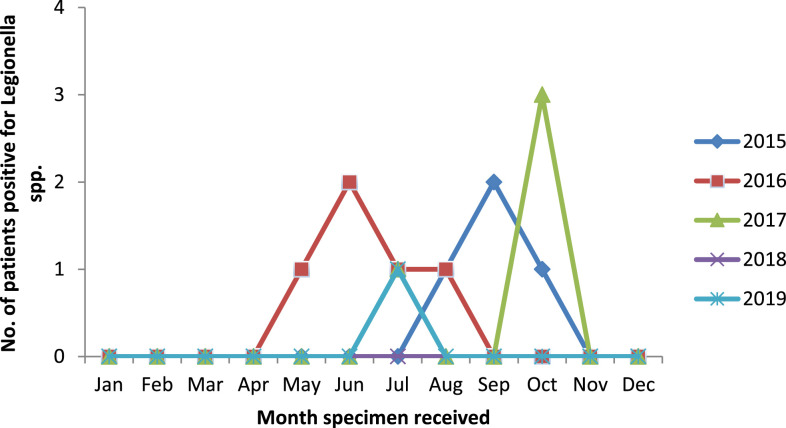
Clinical presentations of legionellosis are nonspecific and varied in different studies. It is suggested that patients having signs and symptoms of pneumonia along with extrapulmonary manifestations may have LD. In our cohort, more than half of the detected cases had extrapulmonary manifestations. This is in agreement with the study by Kao et al.34 Among the extrapulmonary manifestations, confusion and gastrointestinal symptoms were predominant. In the present study, the case fatality ratio was found to be 28.6%, which is in agreement with the overall mortality rate (5–30%) reported in other studies.11,29,34,35 All deaths were among males aged ≥ 50 years, having risk factors and coexisting conditions including cigarette smoking, malignancy, renal disease, COPD, and interstitial lung disease. In previous studies, the peak of reported LD cases usually occurred in late summer to fall, which is possibly connected to warmer, higher relative humidity and wetter weather conditions.11,33,36 In this study, all cases were observed from May to October (Figure 3); however, to establish the seasonality of this disease in India, a prolonged period of surveillance is required.
Figure 3.
Number of case-patients tested positive for Legionella infections by month, in a referral hospital, India, February 2015–January 2020 (n = 597). This figure appears in color at www.ajtmh.org.
Legionellosis is underdiagnosed and easily overlooked in many hospital settings; however, environmental surveillance can increase the index of suspicion for hospital-acquired LD.37 In previous studies, we reported Legionella colonization in the hospital water supply.31,32 The results of our environmental study increased the index of suspicion for nosocomial LD among clinicians and recommended Legionella diagnostic testing for suspected cases of nosocomial pneumonia. According to the U. S. Centers for Disease Control and Prevention (CDC), Legionella testing in patients at risk for LD with nosocomial pneumonia should always be pursued; however, environmental cultures should be reserved for clearly documented cases of hospital-acquired Legionnaires’ disease.38 Therefore, hospitalized patients with CAP and those with presumed nosocomial pneumonia should undergo diagnostic testing for Legionella.
The diagnosis of LD has major implications in patient care. Pathogen-specific treatment can be expedited in patients as many of these cases may be treated empirically with beta-lactam drug monotherapy which does not cover Legionella spp. In this study, 79% of patients were already on anti-Legionella therapy, and two patients had antibiotic treatment altered to azithromycin after Legionella detection. Among these two, one patient was given a combination therapy consisted of rifampicin and azithromycin. The survival chances increase when a combination therapy directed at Legionella spp. is implemented after LD is diagnosed.39 A recent study from the United States reported a limited benefit from a cost/benefit standpoint in an area with a low incidence of Legionella pneumonia where all patients have empirically received antibiotics with coverage for Legionella spp.40 Therefore, Legionella testing in a low prevalent area would not be cost effective.
The limitations of this study include the LD cases reported here may not reflect the true prevalence of this pathogen in our region. Some of the enrolled patients were too sick for specimens to be collected, and few patients were discharged before a sample could be obtained. Besides, because of financial constraints, we could not perform different assays consistently during the study period, which prevents from conducting the comparison of different laboratory testing methods. For a few patients who tested positive by real-time PCR, urine samples were not collected or the UAT was not performed. Many of the patients received antibiotics with Legionella activity before collecting a respiratory secretion, which might have affected Legionella detection. Last, because of the lack of a culture isolate, we were not able to perform L. pneumophila genotyping on clinical strains.
However, we applied a nested PCR-derived sequence-based typing directly on the DNA extracted from BAL fluid of a LD case-patient (case no. 14 of Table 1), amplified fragments of seven genes including two housekeeping genes (asd and neuA) and five virulence genes (flaA, pilE, mip, mompS, and proA).41,42 The purified PCR products were then sequenced (Dr. KPC Life Sciences Pvt. Ltd., Kolkata, India) following a protocol established by the European Study Group of Legionella Infections and assigned allele and finally a sequence type (ST) via the online Legionella Sequence Quality Tool (www.hpa-bioinformatics.org.uk/cgi-bin/legionella/sbt/seq_assemble_legionella 1. cgi). Legionella spp. detected in the BAL fluid of the patient was then identified as ST1. As this patient had acquired infection from another facility, we could not conduct environmental sampling to investigate the source of legionellosis.
CONCLUSION
Overall, the diagnostic tests have inherent shortcomings for the detection of Legionella spp.; hence it is recommended that whenever LD is suspected, more than one specimen type should be submitted, and more than one testing method should be used. In India, Legionella infections remain undiagnosed as most of these patients are treated empirically with antibiotics without prospective or retrospective analysis into the disease etiology. The low index of suspicion by the clinicians for Legionella pneumonia is likely the major predisposing factor because highly effective antibiotic treatment exists for LD. Increased awareness and improved laboratory testing could result in early diagnosis and more pathogen-targeted antibacterial therapy. Physicians should consider Legionella testing for patients with pneumonia especially when there is a suboptimal response to beta-lactam agents. This study provides baseline data on Legionella spp. infections in our geographical region that can be used for future surveillance programs.
Supplemental material
ACKNOWLEDGMENTS
We thank Arti Kapil, Bijay Ranjan Mirdha, Benu Dhawan, Urvashi B Singh, and Jyotsna Punj for their valuable inputs and suggestions. We also thank the resident doctors in Department of Medicine, Pediatrics, Pulmonary medicine, and Microbiology for their helpful coordination. K. S. acknowledges AIIMS and the Indian Council for Medical Research (ICMR) for providing Senior Research Fellowship (SRF) during the study period. The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
Note: Supplemental material appears at www.ajtmh.org.
REFERENCES
- 1.Diederen BM, 2008. Legionella spp. and Legionnaires’ disease. J Infect 56: 1–12. [DOI] [PubMed] [Google Scholar]
- 2.Nazarian EJ, Bopp DJ, Saylors A, Limberger RJ, Musser KA, 2008. Design and implementation of a protocol for the detection of Legionella in clinical and environmental samples. Diagn Microbiol Infect Dis 62: 125–132. [DOI] [PubMed] [Google Scholar]
- 3.Stout JE, Yu VL, 1997. Legionellosis. N Engl J Med 337: 682–687. [DOI] [PubMed] [Google Scholar]
- 4.Mercante JW, Winchell JM, 2015. Current and emerging Legionella diagnostics for laboratory and outbreak investigations. Clin Microbiol Rev 28: 95–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunha BA, Burillo A, Bouza E, 2016. Legionnaires’ disease. Lancet 387: 376–385. [DOI] [PubMed] [Google Scholar]
- 6.Chen DJ, Procop GW, Vogel S, Yen-Lieberman B, Richter SS, 2015. Utility of PCR, culture, and antigen detection methods for diagnosis of legionellosis. J Clin Microbiol 53: 3474–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention , 2013. Legionella (Legionnaires Disease and Pontiac Fever). Atlanta, GA: Centers for Disease Control and Prevention. Available at: http://www.cdc.gov/legionella/clinicians.html. Accessed April 22, 2020. [Google Scholar]
- 8.Benin AL, Benson RF, Besser RE, 2002. Trends in Legionnaires’ disease, 1980–1998: declining mortality and new patterns of diagnosis. Clin Infect Dis 35: 1039–1046. [DOI] [PubMed] [Google Scholar]
- 9.Cunha BA, 2010. Legionnaires’ disease: clinical differentiation from typical and other atypical pneumonias. Infect Dis Clin N Am 24: 73–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cristovam E, Almeida D, Caldeira D, Ferreira JJ, Marques T, 2017. Accuracy of diagnostic tests for Legionnaires’ disease: a systematic review. J Med Microbiol 66: 485–489. [DOI] [PubMed] [Google Scholar]
- 11.Phin N, Parry-Ford F, Harrison T, Stagg HR, Zhang N, Kumar K, Lortholary O, Zumla A, Abubakar I, 2014. Epidemiology and clinical management of Legionnaires’ disease. Lancet Infect Dis 14: 1011–1021. [DOI] [PubMed] [Google Scholar]
- 12.Luck C, 2010. Legionella-a case for culture. Indian J Med Res 131: 736–739. [PubMed] [Google Scholar]
- 13.Fields BS, Benson RF, Besser RE, 2002. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev 15: 506–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pierre DM, Baron J, Victor LY, Stout JE, 2017. Diagnostic testing for Legionnaires’ disease. Ann Clin Microbiol Antimicrob 16: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benitez AJ, Winchell JM, 2013. Clinical application of a multiplex real-time PCR assay for simultaneous detection of Legionella species, Legionella pneumophila, and Legionella pneumophila serogroup 1. J Clin Microbiol 51: 348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murdoch DR, 2003. Diagnosis of Legionella infection. Clin Infect Dis 36: 64–69. [DOI] [PubMed] [Google Scholar]
- 17.Mentasti M, Fry NK, Afshar B, Palepou-Foxley C, Naik FC, Harrison TG, 2012. Application of Legionella pneumophila-specific quantitative real-time PCR combined with direct amplification and sequence-based typing in the diagnosis and epidemiological investigation of Legionnaires’ disease. Eur J Clin Microbiol Infect Dis 31: 2017–2028. [DOI] [PubMed] [Google Scholar]
- 18.Chaudhry R, et al. 2017. Detection of Mycoplasma pneumoniae and Legionella pneumophila in patients having community-acquired pneumonia: a multicentric study from New Delhi, India. Am J Trop Med Hyg 97: 1710–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhry R, Sreenath K, Agrawal SK, Valavane A, 2018. Legionella and Legionnaires’ disease: time to explore in India. Indian J Med Microbiol 36: 324–333. [DOI] [PubMed] [Google Scholar]
- 20.American Thoracic Society (ATS) , Infectious Diseases Society of America (IDSA) , 2013. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med 171: 388–416. [DOI] [PubMed] [Google Scholar]
- 21.Shah PP, et al. 2018. Legionnaires’ Disease Surveillance Summary report, United States: 2014–2015. [Google Scholar]
- 22.European Centre for Disease Prevention and Control , 2018. Legionnaires’ disease. ECDC. Annual Epidemiological Report for 2016. Stockholm, Sweden: ECDC. [Google Scholar]
- 23.Vicente D, Marimón JM, Lanzeta I, Martin T, Cilla G, 2019. Fatal case of nosocomial Legionella pneumophila pneumonia, Spain, 2018. Emerg Infect Dis 25: 2097–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peci A, Winter AL, Gubbay JB, 2016. Evaluation and comparison of multiple test methods, including real-time PCR, for Legionella detection in clinical specimens. Front Public Health 4: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang G, Benson R, Pelish T, Brown E, Winchell JM, Fields B, 2010. Dual detection of Legionella pneumophila and Legionella species by real-time PCR targeting the 23S-5S rRNA gene spacer region. Clin Microbiol Infect 16: 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rigby EW, Plouffe JF, Hackman BA, Hill DS, Benson RF, Breiman RF, 1997. Stability of Legionella urinary antigens over time. Diagn Microbiol Infect Dis 28: 1–3. [DOI] [PubMed] [Google Scholar]
- 27.Garbino J, Bornand JE, Uckay I, Fonseca S, Sax H, 2014. Impact of positive legionella urinary antigen test on patient management and improvement of antibiotic use. J Clin Pathol 57: 1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Javed S, Chaudhry R, Passi K, Sharma S, Padmaja K, Dhawan B, 2010. Sero diagnosis of Legionella infection in community acquired pneumonia. Indian J Med Res 131: 92–96. [PubMed] [Google Scholar]
- 29.Wolter N, et al. 2016. Legionnaires’ disease in South Africa, 2012–2014. Emerg Infect Dis 22: 131–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin T, Ren H, Chen D, Zhou H, Jiang L, Wu D, Shen J, Pei F, 2019. National surveillance of Legionnaires’ disease, China, 2014–2016. Emerg Infect Dis 25: 1218–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sreenath K, Chaudhry R, Vinayaraj EV, Thakur B, 2019. Antibiotic susceptibility of environmental Legionella pneumophila isolated in India. Future Microbiol 14: 661–669. [DOI] [PubMed] [Google Scholar]
- 32.Chaudhry R, Sreenath K, Arvind V, Vinayaraj EV, Tanu S, 2017. Legionella pneumophila serogroup 1 in the water facilities of a tertiary healthcare center, India. Emerg Infect Dis 23: 1924–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marston BJ, Lipman HB, Breiman RF, 1994. Surveillance for Legionnaires’ disease: risk factors for morbidity and mortality. Arch Intern Med 154: 2417–2422. [PubMed] [Google Scholar]
- 34.Kao WF, Wang JT, Sheng WH, Chen YC, 2019. Community-acquired Legionnaires’ disease at a medical center in northern Taiwan. J Microbiol Immunol Infect 52: 465–470. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization , 2018. Legionellosis. Available at: https://www.who.int/news-room/fact-sheets/detail/legionellosis. Accessed April 22, 2020. [Google Scholar]
- 36.Ng V, Tang P, Jamieson F, Guyard C, Low DE, Fisman DN, 2009. Laboratory-based evaluation of legionellosis epidemiology in Ontario, Canada, 1978 to 2006. BMC Infect Dis 9: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabria M, Victor LY, 2002. Hospital-acquired legionellosis: solutions for a preventable infection. Lancet Infect Dis 2: 368–373. [DOI] [PubMed] [Google Scholar]
- 38.Eison R, 2014. Legionella pneumonia: when to suspect, diagnostic considerations, and treatment strategies for hospital-based clinicians. Curr Emerg Hosp Med Rep 2: 205–213. [Google Scholar]
- 39.Cecchini J, et al. 2017. Antimicrobial strategy for severe community-acquired legionnaires’ disease: a multicentre retrospective observational study. J Antimicrob Chemother 72: 1502–1509. [DOI] [PubMed] [Google Scholar]
- 40.Henry C, Boethel C, Copeland LA, Ghamande S, Arroliga AC, White HD, 2017. Clinical utility of testing for Legionella pneumonia in central Texas. Ann Am Thorac Soc 14: 65–69. [DOI] [PubMed] [Google Scholar]
- 41.Mentasti M, Fry NK, 2012. Sequence-Based Typing Protocol for Epidemiological Typing of Legionella pneumophila, Version 5.0. Basel, Switzerland: ESCMID Study Group for Legionella Infections, European Society of Clinical Microbiology and Infectious Diseases. [Google Scholar]
- 42.Quero S, Párraga-Niño N, Sabria M, Barrabeig I, Sala MR, Jané M, Mateu L, Sopena N, Pedro-Botet ML, Garcia-Nuñez M, 2019. Legionella SBT applied directly to respiratory samples as a rapid molecular epidemiological tool. Sci Rep 9: 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.