Abstract
For many traits, human variation is less a matter of categorical differences than quantitative variation, such as height, where individuals fall along a continuum from short to tall. Most recent studies utilize large population-based samples with whole-genome sequences to study the evolution of these traits and have made significant progress implementing a broad spectrum of techniques. However, relatively few studies of quantitative trait evolution include ethnically diverse populations, which often harbor the highest levels of genetic and phenotypic diversity. Thus, our ability to draw inferences about quantitative trait adaptation has been limited. Here we review recent studies examining human quantitative trait adaptation, and argue that including ethnically diverse populations, particularly from Africa, will be especially informative for our understanding of how humans adapt to the world around them.
Keywords: quantitative traits, adaptation, polygenic traits, natural selection, human diversity, population sampling
Genomic signatures of adaptation for quantitative traits
In the early 20th century, R.A. Fisher described how the inheritance of continuous, or quantitative, traits could be explained by the coordinated contributions of many independent loci of small effect [1]. Modern human genetic studies have since confirmed that quantitative traits are governed by many loci of varying effect sizes [2,3,4]. The process of local adaptation for quantitative traits requires the interplay of multiple components: genetic variation creates variation in heritable traits, natural selection on trait variation leads to differential fitness effects in a local environment, and differential fitness effects alter the composition of the genetic variation present in subsequent generations. Establishing local adaptation in human populations requires one to be able to identify the relationships between these attributes. There are multiple approaches to discovering these relationships, each with its own strengths and weaknesses.
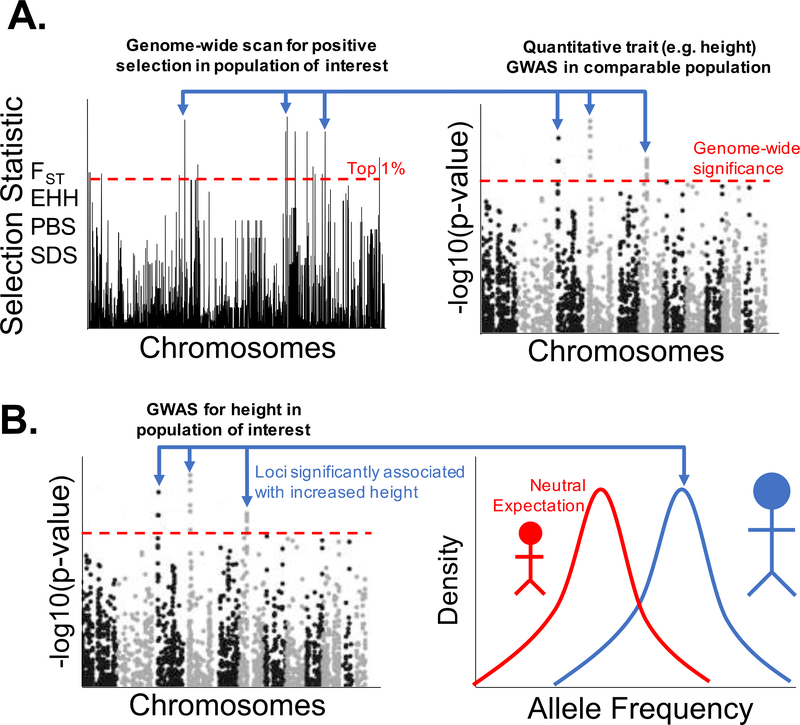
One approach is to establish an association between natural selection and genetic variation and subsequently relate the variants under selection with a quantitative trait. This can be accomplished by applying genomic scans of natural selection to identify the targets of natural selection, and then performing statistical or functional follow up analyses to connect these signals with quantitative traits (Fig. 1A). An alternative approach is to identify an association between a quantitative trait and genetic variation, and then follow up with tests for signals of natural selection in aggregate at these trait-associated loci (Fig. 1B). Here, we review each of these approaches and discuss their strengths and weaknesses. One weakness common to many studies employing either of these approaches is a disproportionate focus on populations that are not representative of global human genetic diversity. Therefore, for both of these scenarios, we describe ways in which our understanding of quantitative trait evolution could be improved by including ethnically diverse populations currently under-represented in the human genetics literature.
Fig. 1.
Two complementary approaches for studying the evolution of quantitative traits. (A) Left panel: Genome-wide scan for positive selection reveals candidate loci showing extreme values for some selection statistic (‘FST’ - Fixation Index; ‘EHH’ - Extended Haplotype Homozygosity; ‘PBS’ - Population Branch Statistic; ‘SDS’ - Singleton Density Score). Right panel: Loci showing strong signals of positive selection are also significantly associated with a quantitative trait (height) in a GWAS in a comparable population. (B) Loci significantly associated with a quantitative trait in a GWAS (left) show a correlated frequency change in the same direction (right), indicating polygenic selection acting on increased height.
Associating targets of selection with quantitative traits
One method to study quantitative trait evolution is to start by identifying targets of selection, followed by association with phenotypes. These studies begin by pinpointing genetic variants exhibiting statistical signatures of being shaped by the forces of natural selection. This can be established either by showing evidence of a rapid rise in allele frequency that is not consistent with neutral genetic drift, or by evidence of correlation with environmental or life-style conditions that is not consistent with neutral demographic spread. Follow-up analyses, be they functional genomic assays, genome-wide association studies (GWAS) in comparable populations, and/or functional annotation enrichment studies, are used to determine the quantitative traits that selection is acting on (Fig. 1A).
These kinds of studies work best when the evidence of natural selection acting on individual loci is clear. This is typically true when strong selection pressure is applied to traits for which there are segregating genetic variants with relatively large phenotypic effects. Where selection acts on traits with hundreds or thousands of segregating variants, each contributing a minor effect, no variants by themselves will show discernible evidence of selection and a different approach is warranted.
The earliest studies of genetic signatures of natural selection focused on traits influenced by one or a few loci of strong effect. One classic example is the evolution of lactose tolerance. LCT encodes the lactase enzyme which metabolizes the milk sugar, lactose, and levels of this enzyme typically decline after weaning. However, populations with a history of traditional dairy farming show high lactase levels into adulthood and, therefore display the ‘lactase persistence’ (LP) phenotype. Studies of genetic association with the LP phenotype and functional genomic studies of gene expression identified mutations that regulate expression of LCT [5,6]. These regulatory variants, located approximately 14,000 base pairs upstream of the LCT gene, show some of the strongest signals of natural selection in the human genome in both European [7] and African [6,8] populations. These signals are evident using multiple statistical tests, including global comparisons of allele frequency (FST-based analyses) as well as tests of extended haplotype homozygosity (EHH), a statistic that detects reductions in haplotype diversity and extended linkage disequilibrium surrounding a positively selected locus [9,10]. Furthermore, these regulatory mutations arose independently in Europeans and Africans, an example of convergent evolution in populations that adopted dairying practices. While LP is thought to be largely influenced by few variants of large effect, it should be noted that no more than 45% of the phenotypic variation is explained by the currently identified variants in Africa [8], suggesting other loci and/or environmental factors contribute to this trait.
Another example of local adaptation is the evolution of lipid metabolism, a quantitative trait, in the indigenous Greenlandic Inuit. Fumagalli et al. [11] used the population branch statistic (PBS), which identifies alleles exhibiting large frequency changes in a focal population compared to two reference populations, to scan the genome for positive selection. Contrasting a sample of 191 Greenlandic Inuit with European and Chinese reference populations, the strongest signal of adaptation occurred in a cluster of genes that code for fatty acid desaturases (FADS), which are important for determining levels of polyunsaturated fatty acids (PUFAs). This signal may reflect local adaptation to an arctic environment where a large portion of the Inuit diet is composed of marine fats high in PUFAs. The authors then tested for associations between the loci showing the strongest signals of selection with anthropometic and metabolic traits in a larger Greenlandic cohort. Positively selected alleles in the FADS region were significantly associated with shorter stature and lower body weight. Notably, these associations would not have been found in other Europeans cohorts, as the frequencies of the selected alleles are very low in non-Greenlandic Europeans. Both of these examples reinforce the benefits of including ethnically diverse populations in studies of adaptive trait evolution.
More recently, novel methods have been developed to scan the genome and detect signatures of recent allele frequency changes, suggesting positive selection within the last 2000 to 3000 years. The singleton density score (SDS) is one such methodology that calculates the distance to the nearest singleton mutation on either side of a test SNP [12]. Haplotypes carrying a favored allele are expected to have fewer singleton mutations in the near vicinity of the selected site. Field et al. [12] calculated this statistic for ~4.5 million SNPs in 3195 European genomes and found strong selection signals at many loci, including the LCT locus and the major histocompatibility complex (MHC) region. Subsequent analysis demonstrated significant associations between SDS scores and loci derived from European GWAS previously identified as contributing to height, skin pigmentation, infant head circumference, birth weight, and some metabolic traits.
Associating polygenic trait architecture with natural selection
Another way to identify genetic variants that play a role in quantitative trait evolution is to first identify a set of variants contributing to a quantitative trait through GWAS, which is typically performed on large genomic datasets provided by national biobanks or consortiums. Once a set of variants underlying a trait are identified, it is possible to look for statistical signatures of natural selection on aggregate across all contributing loci, even if there is not sufficient power to detect signatures of selection at individual loci. A correlated frequency change across populations at variants contributing to a quantitative trait in the same direction is a unique signature of polygenic adaptation (Fig. 1B). These approaches, however, can only be as good as the inferred genetic model of the quantitative trait. Studies of polygenic adaptation in humans have faced substantial issues, particularly regarding an inability to completely account for population structure in large-scale GWAS.
Several recent studies of this form have focused on adaptive changes in human height, a classic quantitative trait. The patterns identified in many of these studies are consistent with selection acting to increase the height of Northern Europeans and decrease the height of Southern Europeans. Signatures of selection have typically been identified by showing that population allele frequencies at height-increasing GWAS loci derived from the Genetic Investigation of Anthropometric Traits (GIANT) Consortium, which genotyped over 180,000 European individuals [3], are significantly higher in northern than southern Europe. These differences in frequency do not fit a model of neutral genetic drift, suggesting the action of polygenic selection (e.g. [13,14,15,16,17,18]). However, there is reason to believe that these results may be an artifact of uncorrected population structure in the original GWAS [19,20], and researchers will need to revisit these analyses.
Other methodologies to study polygenic adaptation depend less on measuring frequency differences at trait-associated loci between populations, but instead examine inferred genealogies at these loci. For example, Edge and Coop [21] developed a method where genealogical trees at trait-associated GWAS SNPs are estimated and used to reconstruct the historical time-course of allele-frequency change at these loci. This method generates a mean polygenic score for a trait of interest within a population, which is a sum of allele frequencies at trait-associated SNPs weighted by their marginal effect sizes. The resulting time-course of this score is then tested for evidence of natural selection by comparing it against an empirically generated null distribution. When used to investigate polygenic selection in height in Europeans in the 1000 genomes dataset, this method detected evidence of selection for increased height using GWAS SNPs from GIANT, but not from the UK Biobank, a larger and more genetically homogenous dataset [22]. A different method for estimating genealogical trees genome-wide in very large samples [23] found similar evidence of selection acting on height, BMI, and blood pressure in samples from the 1000 genomes dataset. These and other computationally efficient methods for detecting polygenic selection will become more important as genome-wide datasets become larger.
While these methods are and will continue to be useful, current studies of polygenic adaptation must be interpreted with caution. One difficulty lies in correctly annotating the effect an allele has on the trait in question. Just because a SNP contributes to a phenotype in one population does not mean that the SNP has the same, or even any, effect in another population. This could be because the SNP’s phenotypic effect depends on the local environment (Gene × Environment [G×E] interaction) or on the genetic background due to epistasis (Gene × Gene [G×G] interaction). Differences in fine-scale patterns of linkage disequilibrium (LD) among populations may also affect how well SNP associations and effect sizes transfer across groups. Further, the effects of population structure have a high probability of affecting the original GWAS. Loci identified as significantly associated with a trait of interest in a GWAS might simply be proxies for environmental variables or other background genetic effects that are not adequately controlled for in the statistical model [24]. For example, as referenced above, a pair of recent studies show that the European selection signal on height polygenic scores does not replicate when using a trait model built from GWAS summary statistics from the UK Biobank [19,20]. These studies show that uncorrected population structure within the height GWAS data drives the majority of these previously identified selection signals. The degree to which uncorrected population structure affects polygenic selection signals for other traits remains to be determined.
Importance of including ethnically diverse African populations
While these and similar studies have been invaluable for our understanding of quantitative trait adaptation, even more can be gained from the inclusion of ethnically diverse populations. African populations are especially important, as they have the highest levels of global human genetic diversity but are among the least represented in human genetic studies [25].
Modern humans originated in Africa, and only relatively recently did a small number migrate out of Africa and populate the rest of the globe [26]. This deep population history and the lack of a population bottleneck associated with the out-of-Africa expansion means that African populations have the largest effective population sizes and the highest levels of genetic and phenotypic variation of all humans [27]. Additionally, African populations inhabit a diversity of environmental conditions, including deserts, savannas, tropical rain forests, and high altitudes. This has resulted in substantial variation at traits thought to have an adaptive basis, including height [28,29], skin pigmentation [30], and response to infectious disease [31]. Selection scans in populations inhabiting extreme environments, or those at either end of a phenotypic extreme, could be used to identify signals of local adaptation (e.g. [32,33]). Loci showing particularly strong selection signals could be explored in follow-up functional assays to determine which selected loci contribute to traits of interest. Together, the high genetic and phenotypic diversity within Africa coupled with extensive environmental heterogeneity is particularly informative for detecting signatures of quantitative trait adaptation.
Furthermore, the few polygenic adaptation studies that include African populations only include those representing a small amount of the total African genetic and phenotypic variation. In addition to the issues described above, the application of polygenic adaptation models across diverse populations is complicated by the reliance of these studies on GWAS summary statistics aggregated by large consortia or biobanks that are overwhelmingly composed of European participants. A recent study estimates that 78% of all the participants in human GWAS are of European ancestry [25]. This raises issues pertaining to the generalization of trait associations and effect sizes across populations, and variants associated with complex traits and diseases in European GWAS often do not replicate in African or other Non-European populations [34,35,36]. With models not representative of global genetic diversity, the utility of these data for studying polygenic adaptation at a global scale suffers.
Including ethnically diverse African populations will also benefit our understanding of quantitative trait evolution in other ways. First, our ability to fine-map genetic associations is higher in African cohorts than in Non-Africans. Stretches of linkage disequilibrium are shorter in Africans than non-Africans [37], which increases the likelihood that an identified trait association represents a true causal SNP, or is in closer proximity to the true causal SNP, because of better tagging. Therefore, the inclusion of diverse African populations in trans-ethnic mapping studies will facilitate the discovery of true genotype-phenotype associations (e.g. [38]).
Next, Africa contains populations at the phenotypic extremes of many quantitative traits, which is particularly informative for performing GWAS. For example, many central and west-African hunter-gatherer populations display the ‘pygmy’ phenotype, with an average adult male height <150cm [28,29]. In contrast, Nilo-Saharan speaking populations from Sudan are among the tallest in the world [39]. Skin pigmentation shows similar extremes, with San hunter-gatherers from Botswana possessing relatively light skin and Nilo-Saharan pastoralists from East Africa having some of the darkest skin globally [30]. A recent GWAS examining skin pigmentation within Africa identified several genomic regions previously associated with pigmentation, including a newly described gene, MFSD12, never previously known to impact skin color [30]. This study used sample sizes that are small by today’s standards, reinforcing the benefits of including diverse African populations in modern genetic studies. More GWAS performed in African populations in this manner will pave the way for tests of polygenic adaptation.
Finally, because of the out-of-Africa bottleneck, non-African genomes contain a subset of the genetic variation found within Africa. While there is certainly genetic variation specific to non-Africans due to subsequent mutation, local adaptation, and genetic drift, alleles identified as functional in African populations are more likely to be relevant globally. However, few studies of polygenic adaptation utilize GWAS summary statistics generated in African populations. Thus, the inclusion of ethnically diverse African populations will not only fundamentally improve our ability to detect polygenic adaptation within the continent, but may also allow for better detection of these signals in all global populations through improved generalization of GWAS summary statistics and polygenic scores.
Conclusions and future directions
There are several limitations of the studies reviewed thus far that must be taken into account in future analyses. The first is that these methods rely on statistical correlations between genetic variants, selection signals, and trait values. Functional validation of loci thought to underlie a quantitative trait will be key in order to truly understand human adaptation. This will require functional genomics approaches where candidate genetic variants are introduced into cell lines in vitro, or into model organisms in vivo. Furthermore, many candidate variants under selection will likely not fall in gene coding regions, but will be regulatory in nature [40]. Thus, integrating candidate loci with loci known to play a role in gene expression, for example from the Genotype-Tissue expression project (GTEx) [41], ENCODE [42], and/or NIH Roadmap [43] databases, will be crucial.
Importantly, more widespread inclusion of indigenous populations in human adaptation studies will require substantial ethical considerations. Studies of indigenous populations require collaboration and trust between researchers, participants, and institutions, as well as the sharing of resources and benefits. Every research proposal must be thoroughly reviewed by local ethics committees, and all stakeholders should be included in every step of the research. Whenever possible researchers should consult and include bioethicists in the planning stages of any genetics study incorporating indigenous populations. This will be particularly important for ensuring the research is conducted in an ethical manner that takes the concerns of the population(s) being studied into account. Additionally, efforts should be made to return the results of these studies to participants. This could be accomplished by conducting general community outreach or by developing educational programs within the studied communities. Further, while human geneticists have typically supported the open sharing of data, this practice may be more complicated with indigenous populations. During the process of seeking ethical approval, researchers should explicitly define their data sharing policy, and directly ensure that the populations being studied consent to this policy. This will be particularly important if the data being shared could lead to commercial gain or be used in for-profit applications by other parties. Finally, national funding agencies must make the inclusion of ethnically diverse populations a priority, and make appropriate investments in the training and infrastructure needed for this kind of work.
Acknowledgements
We thank our funding sources: MM is supported by National Institutes of Health Training grant 5T32ES019851-08 and the Center of Excellence in Environmental Toxicology (CEET) at the University of pennsylvania. CZ, ST, and AP are funded by NIH grant R35 GM134957-01 and American Diabetes Association Pathway to Stop Diabetes grant #1-19-VSN-02.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* Special Interest
** Outstanding Interest
- [1].Fisher RA The correlation between relatives on the supposition of mendelian inheritance. Trans. R. Soc. Edinb 1918, 53: 399–433. [Google Scholar]
- [2].Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P, Purcell (Leader) SM, Stone JL, Sullivan PF, et al. : Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009, 460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Allen HL, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Wile CJ, Jackson AU, Vedantam S, Raychaudhuri S, et al. : Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 2010, 467:832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Weiner DJ, Wigdor EM, Ripke S, Walters RK, Kosmicki JA, Grove J, Samocha KE, Goldstein JI, Okbay A, Bybjerg-Grauholm J, et al. : Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet 2017, 49:978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Järvelä I: Identification of a variant associated with adult-type hypolactasia. Nat Genet 2002, 30:233–237. [DOI] [PubMed] [Google Scholar]
- [6].Tishkoff SA, Reed FA, Ranciaro A, Voight B, Babbit CC, Silverman JS, Powell K, Mortensen HM, Hirbo JB, Osman M, et al. : Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet, 2007, 39:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. : Genome-wide detection and characterization of positive selection in human populations. Nature 2007, 449:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ranciaro A, Campbell MC, Hirbo JB, Ko WY, Froment A, Anagnostou P, Kotze MJ, Ibrahim M, Nyambo T Omar SA, et al. : Genetic origins of lactase persistence and the spread of pastoralism in africa. Am J Hum Genet 2014, 94:496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R: Localizing Recent Adaptive Evolution in the Human Genome. PLOS Genet 2007, 3:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Voight BF Kudaravalli S, Wen X, Pritchard JK: A Map of Recent Positive Selection in the Human Genome. PLOS Biol 2006, 4:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fumagalli M, Moltke I, Grarup N, Racimo F, Bjerregaard P, Jørgensen ME, Korneliussen TS, Gerbault P, Skotte L, Linneberg A, et al. : Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science (80- ) 2015, 349:1343 LP–1347. [DOI] [PubMed] [Google Scholar]
- [12].Field Y, Boyle EA, Telis N, Gao Z, Gaulton KJ, Golan D, Yengo L, Rocheleau G, Froguel P, McCarthy MI, et al. : Detection of human adaptation during the past 2000 years. Science (80-) 2016, 354:760 LP–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Turchin MC, Chiang CWK, Palmer CD, Sankararaman S, Reich D, Hirschhorn JN: Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat Genet 2012, 44:1015–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Berg JJ, Coop G: A Population Genetic Signal of Polygenic Adaptation. PLoS Genet 2014, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zoledziewska M, Sidore C, Chiang CWK, Sanna S, Mulas A, Steri M, Busonero F, Marcus JH, Marongiu M, Maschio A, et al. : Height-reducing variants and selection for short stature in Sardinia. Nat Genet 2015, 47:1352–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, et al. : Genome-wide patterns of selection in 230 ancient Eurasians. Nature 2015, 528:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Robinson MR, Hemani G, Medina-Gomez C, Mezzavilla M, Esko T, Shakhbazov K, Powell JE, Vinkhuyzen A, Berndt SI, Gustafsson S, et al. : Population genetic differentiation of height and body mass index across Europe. Nat Genet 2015, 47:1357–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Racimo F, Berg JJ, Pickrell JK: Detecting Polygenic Adaptation in Admixture Graphs. Genetics 2018, 208:1565–1584.*This study presents a novel method to use admixture graphs to infer polygenic selection among populations. This method is applied to human populations, and evidence for polygenic selection is found acting on multiple complex traits.
- [19].Berg JJ, Harpak A, Sinnott-Armstrong N, Joergensen AM, Mostafavi H, Field Y, Boyle EA, Zhang X, Racimo F Pritchard JK, et al. : Reduced signal for polygenic adaptation of height in UK bioban. Elite 2019, 8:1–47.**One of two studies showing that previous estimates of polygenic selection on European height do not replicate when using GWAS summary statistics from the UKbiobank. This study also shows that previous height polygenic selection signals are confounded by uncorrected population structure in the discovery GWAS.
- [20].Sohail M, Maier RM, Ganna A, Bloemendal A, Martin AR, Turchin MC, Chiang CWK, Hirshhorn J, Daly MJ, Patterson N, et al. : Polygenic adaptation on height is overestimated due to uncorrected stratification in genome-wide association studies. Elife 2019, 8:1–17**One of two studies showing that previous estimates of polygenic selection on European height do not replicate when using GWAS summary statistics from the UKbiobank. This study also shows that previous height polygenic selection signals are confounded by uncorrected population structure in the discovery GWAS.
- [21].Edge MD, Coop G: Reconstructing the history of polygenic scores using coalescent trees. Genetics 2019, 211:235–262.*This study develops a set of methods to estimate the time-course of a population’s mean polygenic score through history, and whether this time course is consistent with neutral evolution or selection. This method is also applied to estimate time courses for human height polygenic scores in Europe.
- [22].Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. : The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Speidel L, Forest M, Shi S, Myers SR: A method for genome-wide genealogy estimation for thousands of samples. Nat Genet 2019, 51:1321–1329.*This study develops a novel method for estimating genealogies at each site in the genome, capable of scaling to multiple populations and tens of thousands of sequences. A novel method for using these genealogies to detect signatures of polygenic adaptation is also presented.
- [24].Novembre J, Barton NH: Tread lightly interpreting polygenic tests of selection. Genetics 2018, 208:1351–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sirugo G, Williams SM, Tishkoff SA: The Missing Diversity in Human Genetic Studies. Cell 2019, 177:26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nielsen R, Akey JM, Jakobsson M, Pritchard JK, Tishkoff S, Willerslev E: Tracing the peopling of the world through genomics Nature 2017, 541:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tishkoff SA, Reed FA, Friedk ender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo J-M, Doumbo O, et al. : The Genetic Structure and History of Africans and African Americans. Science (80- ) 2009, 324:1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jarvis JP, Scheinfeld LB Soi S, Lambert C, Omberg L, Ferwerda B, Froment A, Bodo JM, Beggs W, Hoffman G, et al. : Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African pygmies. PLoS Genet 2012, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mendizabal I, Marigorta UM, Lao O, Comas D: Adaptive evolution of loci covarying with the human African Pygmy phenotype. Hum Genet 2012, 131:1305–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Crawford NG, Kelly DE, Hansen MEB, Beltrame MH, Fan S, Bowman SL, Jewett E, Ranciaro A Thompson S, Lo Y, et al. : Loci associated with skin pigmentation identified in African populations. Science (80- ) 2017, 358:1–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Karlsson EK, Kwiatkowski DP, Sabeti PC: Natural selection and infectious disease in human populations. Nat Rev Genet 2014, 15:379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Scheinfeldt LB, Soi S, Thompson S, Ranciaro A, Woldemeskel D, Beggs W, Lambert C, Jarvis JP, Abate D, Belay G, et al. : Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol 2012, 13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fan S, Hansen MEB, Lo Y, Tishkoff SA: Going global by adapting local: A review of recent human adaptation. Science (80-) 2016, 354:54 LP–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Curtis D: Polygenic risk score for schizophrenia is more strongly associated with ancestry than with schizophrenia. Psychiatr Genet 2018, 28:85–89. [DOI] [PubMed] [Google Scholar]
- [35].Reisberg S, Iljasenko T, Lall K, Fischer K, Vilo J: Comparing distributions of polygenic risk scores of type 2 diabetes and coronary heart disease within different populations. PLoS One 2017, 12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Duncan L, Shen H, Gelaye B, Meijsen J, Ressler K, Feldman M Peterson R Domingue B, Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun 2019, 10.**This study analyzes polygenic score usage over the last decade, and finds that European participants are overwhelmingly represented. This study also shows that European ancestry-derived polygenic scores have significantly lower predictive performance when applied to nonEuropean populations.
- [37].Tishkoff SA, Dietzsch E, Speed W, Pakstis AJ Kidd JR, Cheung K, Bonne-Tamir B, Santachiara-Benerecetti AS, Moral P, Krings M et al. : Global Patterns of Linkage Disequilibrium at the CD4 Locus and Myden Human Origins. Science (80-) 1996, 271:1380 LP–1387. [DOI] [PubMed] [Google Scholar]
- [38].Kilpelainen TO, Bentley AR, Noordam R, Sung YJ, Schwander K, Winkler TW, Jakupovic H, Chasman DI, Manning A Ntalla I, et al. : Multi-ancestry study of blood lipid levels identifies four loci interacting with physical activity. Nat Commun 2019, 10:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chali D: Anthropome ric measurements of the Nilotic tribes in a refugee camp. Ethiop Med J 1995, 33, 211–217. [PubMed] [Google Scholar]
- [40].Necsulea A, Kaessmann H: Evolutionary dynamics of coding and non-coding transcriptomes. Nat Rev Genet 2014, 15:734–748. [DOI] [PubMed] [Google Scholar]
- [41].Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, Mohammadi P, Park Y, Parsana p, Segrj AV, et al. : Genetic effects on gene expression across human tissues. Nature 2017, 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, et al. : The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 2017, 46:D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. : The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 2010, 28:1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]