Abstract
The mammalian immune system evolved to tightly regulate the elimination of pathogenic microbes and neoplastic transformed cells while tolerating our own healthy cells. Here, we summarize experimental evidence for the role of Siglecs—in particular CD33-related Siglecs—as self-receptors and their sialoglycan ligands in regulating this balance between recognition of self and non-self. Sialoglycans are found in the glycocalyx and extracellular fluids and matrices of all mammalian cells and can be considered as self-associated molecular patterns (SAMPs). We also provide an overview of the known interactions of Siglec receptors and sialoglycan-SAMPs. Manipulation of the Siglec-SAMP axis offers new therapeutic opportunities for the treatment of inflammatory conditions, autoimmune diseases and also cancer immunotherapy.
Keywords: Pattern recognition, Self-receptor, Autoimmunity, Tolerance
Introduction
Carbohydrates (glycans) are one of the key building blocks of life [1, 2]. Post-translational modification of proteins with glycans significantly influences function e.g., surface glycosylation of cells modifies signaling transduction, changes the physical properties of cells, and mediates cell–cell interactions. In fact, the glycome—the aggregate of all cell surface glycans influences many interactions of immune cells with pathogens, host cells and also neoplastic cells [3, 4]. Moreover, secreted glycoproteins and glycosaminoglycans (GAGs) within the extracellular matrix (ECM) influence interactions of immune cells and migratory patterns.
Although glycans are basic molecules of living organisms, their function in physiological processes and also their role in disease are grossly understudied. This is in part due to methodological difficulties in studying them, and technologies to elucidate their complexity have only been developed in the last decades. While nucleic acids and proteins are relatively easy to analyze and synthetize, glycan molecules can be linked through various ways and the natural diversity is enormous (see Table 1 for common methods used to study Siglec-sialoglycan interactions). Vertebrate proteins are mainly post-translationally decorated with glycans through the linkage of a preformed oligosaccharides to an asparagine (N-glycoslyation) or the sequential addition of monosaccharides to serine or threonine (O-glycosylation). Lipids are also modified by the addition of glycans (glycolipids). Large chains of glycans (GAGs) are produced with smaller protein cores as a key component of the ECM along with freestanding GAG polymers like hyaluronan. Glycans of cell surface and secreted glycoconjugates of mammalian cells are often terminated with sialic acids (Sias) to form sialoglycans [5, 6]. The Sia family of alpha-keto acids consist of a characteristic 9-carbon chain backbone with a carboxylic acid at C1, and the anomeric center at C2 [7]. While over 50 different Sias exist in nature, there are two dominant Sias in most mammalian systems, i.e. N-gycolyl-neuraminic acid (Neu5Gc) and N-acetyl-neuraminic acid (Neu5Ac) [8]. The two Sias differ only by an oxygen atom and humans have lost the ability to synthesize the Neu5Gc due to a mutation in the CMAH gene, but can metabolically incorporate Neu5Gc from external sources including red meat, apparently facilitating cancer progression and atherosclerosis via a humoral inflammatory response [9–11].
Table 1.
Methods used to study Siglec-sialoglycan interactions
| Methodology | Description | Key literature |
|---|---|---|
| Analysis of glycan composition | Enzymatic release of glycans and analysis by multiple methods including chromatography or mass spectrometry | [44, 137, 138] |
| Glycan microarrays | Printing of different glycan structures on glass slides, can be for analysis of binding properties of different Siglecs | [31, 33] |
| Cell-based glycan array on CHO cells | Display of different glycans on the surface of CHO cell that are genetically engineered | [139] Narimatsu 2019 Molecular Cell |
| Structural analysis of sialoglycan-Siglec interaction | Analysis by NMR spectrometry, crystallography or electron microscopy | [24, 27] |
| Use of high-affinity ligands | Chemical modifications to produce high affinity ligands for Siglecs, which can be used to target Siglecs or probe signaling function | [77, 117, 140, 141] |
| In vitro genetically manipulated cells | Use of cells deficient for Siglecs or for Sia synthesis, can be used for in vitro or also in vivo studies | [42, 134] |
| Mouse models | Genetic models of overexpression and deficiency of Siglec receptors, deficiency of Sia processing enzymes/sialyltransferases | [42, 133, 142] |
| Naturally occurring variants, association studies | Association of genetic polymorphisms with outcome/frequency of disease | [31, 61, 143–145] |
Changes in sialoglycan presentation and the density of Sias in different tissues and diseased states are regulated by many factors. The complement of sialoglycans present depends in changes in transcript expression of glycoproteins decorated with Sias, as well as expression of Sia generating and processing enzymes including biosynthesis genes, lysosomal and Golgi transporters, sialyltransferases and sialidases [12]. The biosynthetic pathway of Sia includes enzymes of the hexosamine synthesis pathway and the internal production of Sia includes the 2-GlcNAc-epimerase (GNE), which is the rate limiting enzyme of the intracellular Sia biosynthetic pathway [5]. Sias are transferred to underlying glycan structures by sialyltransferases. Mammalian sialyltransferases are 20 rather conserved enzymes that can be subdivided into four families based on the resulting Sia linkage in the product, and general underlying structure of the substrate [13].
Sialic acid-binding Immunoglobin-like receptors (Siglecs)
Lectins are proteins that bind to glycan ligands through a carbohydrate recognition domain (CRD). Relatively few mammalian Sia-binding lectins have been discovered. Selectins that are vascular cell adhesion molecules mediating trafficking and tethering of leukocytes during vascular extravasation processes bind to a selective set of ligands (selective lectins) [14, 15]. Sia-binding immunoglobulin-like lectins (Siglecs) are a large family of I-type lectins, immune-modulatory receptors within the mammalian immune system with a major subset that underwent rapid evolutionary changes.
During the past 30 years 17 members of Siglecs were described in hominoid primates with 14 Sia-binding members in humans (Fig. 1) [16–18]. Depending on their evolutionary history, Siglecs can be divided into conserved Siglecs with orthologues in different species; and, a rapidly evolving CD33-related Siglecs (CD33rSiglecs) that do not always have clear orthologues in all mammalian species [16–18]. This is also why most CD33rSiglecs have no numbers in mice but are assigned letters (Fig. 1).
Fig. 1.
Schematic drawing of human and murine Siglec receptors. Conserved Siglecs can be found in different mammals and have orthologues between mice and humans. CD33-related Siglecs underwent rapid evolutionary changes and no orthologues can be found between mice and humans, but rather functional paralogues (e.g. Siglec-E and Siglec-9). Reprinted with permission from [136]
Siglecs are single-pass type I transmembrane proteins belonging to the immunoglobulin superfamily of proteins. Their extracellular domains consist of the V-set domain that recognizes siaologlycans and has high similarity to the variable domain of immunoglobulins. The V-set domain contains the CRD of Siglecs. It is followed by a different number of C2-set Ig-like domains. While most CD33rSiglecs have intracellular domains with inhibitory ITIM or ITIM-like motifs, the transmembrane domain bears a positively charged amino acid in the less common activating Siglecs [16–18]. Siglec-1 is a special case with many C2 domains and no intracellular signaling domain [19]. While conserved Siglecs are distributed across different chromosomes in humans, the rapidly evolving CD33rSiglecs are located largely in a cluster on human chromosome 19. Siglec diversification goes back to early mammals probably due to a ‘Red Queen’ effect resulting from interactions between hosts and pathogens [19, 20]. CD33rSiglecs developed within a chromosomal area where other polymorphic receptors are also located [21, 22]. Activating CD33rSiglecs are likely to have evolved via duplications of inhibitory receptors with a similar expression pattern on immune cells and they are considered paired receptors (see below).
The CRD of Siglecs (V-set domain) has a pocket that engages the glycan and within the pocket there is a conserved arginine residue required for Sia-binding [23–27]. Mutation of this “essential arginine” leads to complete loss of binding to sialoglycans [28] e.g., Siglec-XII in humans [28–30]. SNPs in other positively charged amino acids including lysine close to the binding pocket have also shown to influence the binding to sialoglycan ligands [31, 32]. For example, lysine at the position 131 in Siglec-9 enhances binding to sialoglycans. [31, 32]. The binding specificity varies between the different Siglecs and in different species. Specificity has been tested with glycan microarrays [31, 33, 34]. Also, binding analysis have been employed [35, 36]. Some Siglecs such as Siglec-9 bind to a broad range of sialoglycans [31]. Siglec-9 has even been found to bind hyaluronan [37]. On the other extreme, there are CD33rSiglecs with a much more limited binding spectrum such as Siglec-8 [27, 34] or also Siglec-7 [38, 39]. In addition, protein ligands have been identified for Siglecs including endogenous [40, 41] and exogenous proteins from pathogens (see below). As we will discuss below, broadly binding Siglecs such as Siglec-9 can act as receptors to recognize sialoglyans as self-associated molecular patterns (SAMPs) [42, 43]. Siglecs such as Siglec-8 have probably a more circumscribed role including the fine regulation of eosinophils in the airways [35, 44].
Most CD33rSiglecs have intracellular immune receptor tyrosine-based inhibitory motifs (ITIMs) or ITIM-like domains that lead upon activation and phosphorylation of the receptors to recruitment of SHP phosphatases (SHP1 and SHP2), which then inhibit immune cell activation [16, 45–48]. Activating Siglecs in contrast have a positively charged amino acid in the transmembrane domain that leads to association with DAP12 that contains an activating immune receptor tyrosine-based activating motif (ITAM) [41, 49–53].
Inhibitory CD33rSiglecs recognize endogenous sialoglycans as self-associated molecular patterns
The functional study of sialoglycan-Siglec interactions is strongly linked to the use of various analytical approaches (summarized in Table 1). CD33rSiglecs are mostly widely expressed on leukocytes [16, 18, 54]. Receptor expression is a dynamic process and some immune cells have been found to upregulate Siglec receptors upon activation [42, 55]. For example, T cells rapidly upregulate Siglec-5 upon T cell engagement, but over the time of longer stimulation this receptor is again downregulated [42]. Sialoglycans are found on all mammalian cells to build the glycocalyx of the cell [5, 56]. Although these sialoglycans are heterogeneous with different underlying structures and also different linkages, they could be regarded as molecular patterns in the setting of the intact cell. Since most microbes and pathogens have no sialoglycans on their surface, these patterns can be considered to be self-associated molecular patterns (SAMPs) with analogy to molecular patterns that signal foreign or pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs) [57–59]. In this context, broadly sialoglycan-binding Siglecs could be considered as pattern recognition receptors that recognize sialoglycan-SAMPs and inhibit immune cells in the vicinity of a sialoglycan-rich environment. For example, the CD33rSiglec receptor Siglec-9 on neutrophils keeps these immune cells quiet within the blood that contains high-density sialoglycan ligands on erythrocytes [60]. This finding also explains, why isolation of neutrophils and elimination of erythrocytes leads to an activation of neutrophils in many in vitro assays. As mentioned earlier, some Siglecs with a higher specificity for certain ligands such as Siglec-8 might have a more circumscribed role compared to a more broadly binding Siglec-9 [31]. Similar to other self-receptors including KIRs, CD33rSiglecs are also highly polymorphic indicating also a selection pressure and interactions with pathogens that exploit this receptor system to avoid immune control [32, 49, 61]. Siglecs act as self-receptors to recognize sialoglycans as SAMPs. However, interactions with pathogenic microbes have led a rapid diversification of this receptor system in a race between pathogens exploiting inhibitory Siglecs and protection of self from immune-mediated damage providing tolerance (this race is an example of a ‘Red Queen’ effect according to novel ‘Alice in Wonderland’ by Carroll Lewis, in which the Red Queen has to run all the time to stay in the same place). In this context, activating Siglec receptors such as human Siglec-14 or Siglec-16 have likely evolved as paired receptors with inhibitory Siglec-5 or Siglec-11 respectively to counteract bacteria exploiting inhibitory receptors to evade immune control [41, 49, 52, 53, 61, 62].
Siglecs on immune cells
As previously described, CD33rSiglec expression is found on hematopoietic cells and immune cells with a some exceptions (see below). The recognition of SAMPs by Siglec receptors importantly influence myeloid cells. Several Siglecs are expressed on myeloid precursor cells and also myeloid leukemias [63]. In humans, neutrophils express various CD33rSiglecs including CD33, Siglec-5, its paired receptor Siglec-14, Siglec-7 and Siglec-9 [30, 64]. Human monocytes and macrophages also express various Siglecs, although the expression seems to be also somewhat tissue-specific. For example, the conserved Siglec-1 (sialadhesin, CD169) is expressed only on subtypes of macrophages and upregulated by type I interferons [65, 66]. Also, conserved Siglec-15 is expressed on some subspecies of macrophages including very specialized macrophages in the bone, i.e. osteoclasts [67, 68] but has recently been reported as broadly upregulated on human cancer cells and tumor-infiltrating myeloid cells [69]. Monocytes and macrophages also express various CD33rSiglecs including Siglec-3, Siglec-5 and its paired receptor Siglec-14, Siglec-9. Siglec-10 and its functional paralogue Siglec-G is not only expressed on B cells but also on some subsets for macrophages [70–72]. Siglec-8 is a major inhibitory receptor on eosinophils [73, 74]. Siglec-8 is also expressed on human mast cells and basophils [73]. While there is no direct ortholog of this inhibitory CD33rSiglec in mice, Siglec-F is a functional paralog with similar expression pattern and function in mice [75, 76]. A recent study also showed an important immunomodulatory role of Siglec-3 as an inhibitory receptor in IgE signaling of mast cells [77]. Various Siglecs are expressed on different subtypes of DCs [78–81]. Classical, human myeloid-derived classical DCs (cDCs) are expressing various Siglecs including the conserved Siglec-15 and CD33rSiglecs Siglec-3, Siglec-5/-14, Siglec-7, Siglec-9, and Siglec-10 [80, 81]. The expression of plasmacytoid DCs (pDCs) in humans have more restricted expression with mainly Siglec-5 being present [81]. Mice also show an expression of different Siglecs on subgroups of DCs. For example, Siglec-E is expressed on some cDCs for example in the spleen [82, 83]. Also, Siglec-G is expressed on cross-presenting CD8+ cDC1 [84]. In addition, murine pDCs express Siglec-H, which is even used to identify pDCs in mice [85, 86]. NK cells express a multitude of immune modulatory receptors including the CD33rSiglec, Siglec-7 and some subsets also Siglec-9 [87–91]. While the conserved Siglec-2 (CD22) is an important B cell marker [16, 92], subtypes of B cells, the B1 cells also express the inhibitory CD33rSiglec-10 in humans and Siglec-G in mice [92, 93]. Compared to other closely-related primates, humans have a low Siglec expression on resting T cells [94]. In peripheral blood, only a small population of Siglec-7 or Siglec-9 positive T cells can be found [48]. However, CD33-related inhibitory Siglecs are upregulated in pathological conditions including chronic infection and cancer [42, 55, 95].
Siglecs on epithelial cells
While most work focuses on expression of Siglecs on immune cells, recent evidence has demonstrated that Siglecs can also be expressed on epithelial cells. Human Siglec-6 is found to be expressed on the trophoblast in the placenta [96]. In addition, human placenta also expresses ligands for Siglec-6 [96]. Ligands were also found on the uterine endometrium and Siglec-6-mediated interactions could influence the labor process [96]. A recent work has significantly expanded the knowledge about expression of Siglec within the female genital tract [97]. The inhibitory Siglec-10 was found to be expressed by the human endometrium [97]. On human endometrial cell lines, also Siglec-11/-16 was detected. Binding of human sperm to Siglec-10 could be demonstrated suggesting that interaction between sialoglycans on sperm and Siglec-10 on endometrium could influence sperm survival [97]. On the other side, Siglec receptors have also been described on sperm of different species including humans [98]. While Siglec-5/-14 has an important regulatory role on myeloid cells, Siglec-5/-14 has surprisingly been found on human amnion [61]. Interactions of group B streptococci (GBS) that bind to Siglec-5/-14 can thereby influence virulence of GBS and induction of premature birth [61]. Siglec-7 was found to be expressed on β-cells of pancreatic islets [99]. Overexpression of Siglec-7 on β-cells led to a reduction of β-cells dysfunction and Siglec-7 was downregulated in type 1 and 2 diabetes [99]. Expression of Siglec-11/-16 paired receptors was described on cervical epithelium of the female genital tract [62]. Neisseria gonnorhoeae can interact within the female genital tract by interaction with Siglec-11/-16 [62]. Siglec-XII has lost the ability to bind to Sia-containing ligands in humans due to a loss of the essential arginine [29]. Siglec-XII has been described to be expressed on multiple epithelia in different organs including the prostate and kidney [29]. Accordingly, Siglec-XII has been found on different epithelial cancers including prostate cancer [29].
Pathogen-host interactions are driving Siglec evolution
As previously discussed, CD33rSiglecs likely evolved rapidly because of multiple interactions with pathogens that abuse these inhibitory self-receptors to evade immune control. Pathogens including bacteria and viruses can mimic sialoglycan-SAMPs by producing them themselves or also scavenge it from their host. Several human-pathogenic bacteria can display sialoglycan-SAMPs on their surface including Neisseria species, Haemophilus influenzae, C. jejuni, certain strains of pathogenic Eschericha coli, and group B streptococci (GBS) [100]. Some GBS strains produce also a protein that is able to bind Siglec-5/Siglec-14 in a Sia-independent way [61, 101]. The cell wall-bound β-protein can engage inhibitory Siglec-5 on neutrophils, which is counterbalanced the paired receptor Siglec-14 [61]. Some E. coli strains including K1 have polysialic acid capsules that can engage Siglec-11 on microglia during CNS infection and evade immune control [53]. Engagement of Siglec-11 is supposedly counter-regulated in individuals carrying a functional paired Siglec-16 receptor [53]. Similar to GBS, Neisseria gonorrhoeae can express porins to serve as Sia-independent ligands for Siglec-11 and Siglec-16, which are also expressed on innate immune cells in the female genito-urinary tract [62].
Siglec-SAMP interactions in hypersensitivity and autoimmunity
Sialoglycan-SAMPs that bind to Siglec self-receptors are important modules to regulate immunity. Several lines of evidence have been provided that interruption of the sialoglycan-Siglec axis can lead overshooting reactions to antigens as in allergies and also to breakage of peripheral tolerance and autoimmunity.
Siglecs play also a major role in preventing hyperinflammation in sepsis [102]. Defects in this pathway of sialoglycan-Siglec interactions for example mediated by sialidase-producing bacteria can, therefore, lead to overshooting immune responses [103, 104]. Siglecs such as Siglec-10 might be important to protect from tissue damage in inflammatory conditions and might be a therapeutic target in this setting [105].
Siglec-8 in humans and Siglec-F in mice have been demonstrated to regulate major immune subsets involved in allergic reactions [18, 106]. Expression of these inhibitory CD33Siglecs on eosinophils and mast cells make them to an interesting therapeutic target for allergic diseases including asthma. Siglec-F has been involved in eosinophilic lung inflammation, models of food intolerance and also inflammation of the esophagus [107–109]. Blockade of Siglec-F with an antibody has shown improvement in asthma models and models of eosinophil esophagitis [75, 108]. To develop inhibitors of human Siglec-8 for allergic eosinophil-mediated diseases, an eosinophil-specific human Siglec-8 transgenic mouse has been generated [110]. Recently, Siglec-3 signaling was also implicated in regulation of activation and IgE signaling in mast cells [77]. Mice expressing transgenic human Siglec-3 in mast cells were less prone for anaphylaxis [77].
Interruption of immune inhibition mediated by sialglycan-SAMPs can lead to autoimmunity. Mainly the role of B cell Siglecs have been studied, but as mentioned earlier, Siglecs can also significantly influence antigen presentation and processing at the level of APCs including macrophages and DCs [16, 18, 78, 92]. Recent evidence also arises that Siglecs on T cells might directly influence peripheral tolerance [111]. Pathogenic autoantibodies can be seen in mice with Siglec-2 and/or Siglec-G deficiency on B cells [112–114]. Since Siglec-2/Siglec-G mediate peripheral tolerance, targeting antigens to B cells together with trans ligands for Siglec-2 and Siglec-G/-10 could be used to induce tolerance [115, 116]. This approach could also be used in patients with rheumatoid arthritis [117]. Humans with mutations in the gene coding for Sia 9-O-acetyl esterase have been found to be more prone for autoimmune diseases including rheumatoid arthritis and diabetes mellitus type 1 [118]. The esterase mediates the cleavage of an acetyl group that prevents sialgoglycan-SAMPs to be bound by B cell Siglecs [118]. Siglecs on T cells have also been involved in autoimmunity [111]. CD52 could engage Siglec-G in a model of autoimmune diabetes mellitus and reduce the severity of hyperglycemia [111]. In humans, the sialylated glycoform of CD52 binds to HMBG1 to engage Siglec-10 and suppress T cell activation [119].
Siglec-SAMP interactions in cancer immune escape
The fact that neoplastic cells can upregulate sialoglycans was noted many years ago [120–122]. Even trials have been performed, mainly in acute myeloid leukemia (AML) patients with sialidase treatment [121, 123]. Recent experimental evidence has provided molecular explanations for the effect seen upon sialidase treatment in early experiments [3, 124, 125].
Two independent groups have shown that NK cell mediated killing of cancer cells was dependent on the interaction of Siglec-7 and Siglec-9 with sialoglycan-SAMP ligands on tumor cells [88, 126]. Blockade of these interactions led to an increased killing of tumor cells [88, 126]. Hudak and colleagues have used glycopolymers containing Sia to increase the sialoglycan density of target cells [126]. NK cell mediated killing of these hypersialylated target cells was inhibited by Siglec-7 demonstrated by blocking these interactions [126]. Siglec-9 expression was found on a subset of NK cells in patients with cancer including melanoma [88]. Blocking of Siglec-9 on this subpopulation also increased killing of tumor cells, defining both Siglec-7 and Siglec-9 interactions as potential therapeutic target for improving NK cell-based cancer immunotherapy [88]. Siglec-9 is also involved in the polarization of macrophages by MUC1 decorated with sialylated Tn antigen (sTn) [127]. Non-sialylated MUC1 had no effect and blocking the interaction of Siglec-9 with sTn-MUC1 abrogated alternative macrophage polarization [127]. Additional experiments have suggested a role for Siglec-E in macrophage polarization also in mice [31]. Recent work has identified engagement of Siglec-10 on macrophages by CD24 can inhibit phagocytosis [128]. Evidence also shows that tumor-infiltrating lymphocytes (TILs) upregulate certain Siglecs including Siglec-9 [42, 55]. Siglec-9 was expressed on PD-1 high positive, tumor-specific TILs with an increased proliferation potential in patients with non-small cell lung cancer, epithelial ovarian cancer and colorectal cancer [42]. Reduction of the sialoglycan density on tumor cells enzymatically or genetically has increased T cell-mediated tumor cell killing [42]. Similar findings have been made in patients with melanoma [55]. Siglec-9 was shown to be present at the binding site of the T cell receptor influencing TCR-mediated signaling [55]. Siglec-15 has also been implicated as inhibitor of T cell activation in cancer and therapeutic targeting of Siglec-15 has led to a reduced tumor growth in mouse models [69]. Generally, both innate and adaptive antitumor immunity can be stimulated at the same time by targeting sialolglycan-Siglec interactions, although further investigations are needed to understand the exact contributions of different subtypes of cells including myeloid derived suppressor cells, cDCs, and regulatory T cells.
Outlook and therapeutic opportunity of targeting Siglec-SAMP interactions
As described in this review, Siglec-SAMP interactions are essentially involved in balancing the immune system preventing damage of healthy self-tissue and overshooting inflammatory reactions. Manipulation and enhancement of Siglec signaling could be used to treat inflammatory diseases including autoimmunity and allergies. As presented in a recent publication, engagement of Siglec-3 on mast cells could be used to desensitize from allergens and for the treatment of allergic diseases [77]. Interesting approaches including using of nanoparticles decorated with sialoglycan-SAMP have demonstrated encouraging activity in mouse models of overshooting immune reaction in sepsis [129].
On the other sides, pathogens can exploit the Siglec-SAMP axis by mimicking sialoglycans or evolving proteins that can engage inhibitory Siglecs. Interference with Siglec-SAMP interactions could also improve immunity against these pathogens. Blockade of Siglec-5 or Siglec-9 engagement by pathogenic bacteria induces improved anti-bacterial activity of myeloid cells [101, 130]. Inhibition of the Siglec-SAMP interaction strongly increased the immune control of GBS in murine infection models [131].
Finally, targeting Siglec-SAMP interactions is a potential new way to improve anti-tumor immunotherapy and current investigations are focusing on moving the intriguing preclinical findings into clinical applications. To target Siglec-SAMP interactions for anti-tumor immune stimulation, two approaches can be made. First, blocking antibodies against inhibitory Siglecs could improve immune cell function. Antibodies can also lead to endocytosis of the Siglec receptor. But also, reversing the immune suppression by reduction of the sialoglycan density within a tumor could be a valid approach. Normalization of sialoglycan density or even hyposialylation has been shown to induce anti-tumor immunity [132]. The use of a fluoro-Sia mimetic let to a hyposialylated tumor microenvironment in subcutanous murine tumors and a T cell dependent inhibition of tumor growth [132]. Genetic reduction of sialoglycan density also led to an inhibition of tumor growth [42, 133]. In contrast, complete ablation of sialoglycans on the surface of tumor cells led to an enhanced tumor growth [134]. Alterations of the glycocalyx could introduce complex changes and the complete lack of sialoglycans could potentially induce tumor cell intrinsic advantage for tumor growth. A therapeutic approach that will not induce complete absence of sialoglycans but rather intratumoral hyposialylation is the use of sialidases linked to tumor-targeted antibodies [135]. In vitro evidence shows that the linkage of a bacterial sialidase to the anti-HER2 antibody trastuzumab increases the killing by NK cells [135]. However, in vivo data is needed to validate this approach further.
Acknowledgements
Figure 2 was made with graphs provided by SMART (Servier Medical Art).
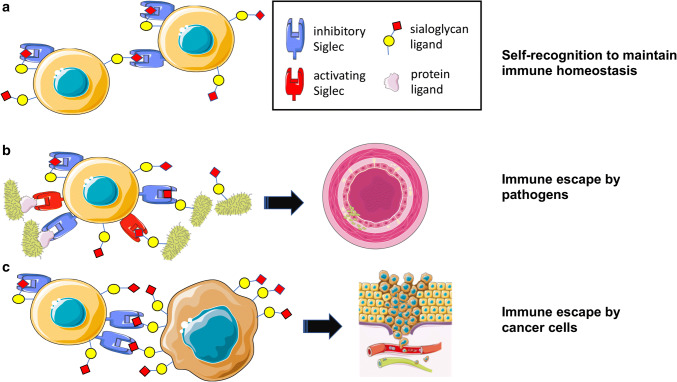
Fig. 2.
Interactions of Siglec receptors with sialoglycans mediate immune escape of pathogens and tumor cells. a Under physiological conditions, sialoglycan-Siglec interactions are inhibiting immune activation and mediate peripheral tolerance. b Pathogens can exploit the sialoglycan-Siglec pathway and bind via sialoglycan-mimicking or protein ligands to inhibitory Siglec receptors and evade immune control. Activating Siglec receptors evolved to counteract this exploitation. Pathogens engaging inhibitory Siglec receptors can escape immune control resulting in more severe infections. c Tumor cells can exploit the sialoglycan-Siglec axis in a similar way as pathogens. The hypersialylated glycocalyx of tumor cells can engage Siglec receptors on different immune cells and mediate immune escape, cancer progression and metastasis
Author contributions
HL and AV wrote the manuscript.
Funding
This work was supported by funding to HL from the Goldschmidt-Jacobson Foundation, the Swiss National Science Foundation (Grant 310030_184720 to HL), the Schoenemakers Foundation (to HL), and to NIH Grant (GM32373 AV).
Compliance with ethical standards
Conflict of interest
HL received travel grants and consultant fees from Bristol-Myers Squibb (BMS), Merck, Sharp and Dohme (MSD), and Roche. HL received research support from BMS and Palleon Pharmaceuticals.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Heinz Läubli, Email: heinz.laeubli@unibas.ch.
Ajit Varki, Email: a1varki@ucsd.edu.
References
- 1.Varki A, Kornfeld S, et al. Historical background and overview. In: Varki A, Cummings RD, et al., editors. Essentials of glycobiology. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2015. pp. 1–18. [Google Scholar]
- 2.Varki A. Biological roles of glycans. Glycobiology. 2017;27(1):3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez E, Schetters STT, van Kooyk Y. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat Rev Immunol. 2018;18(3):204–211. doi: 10.1038/nri.2018.3. [DOI] [PubMed] [Google Scholar]
- 4.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 5.Schauer R. Achievements and challenges of sialic acid research. Glycoconj J. 2000;17(7–9):485–499. doi: 10.1023/A:1011062223612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varki NM, Varki A. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab Invest. 2007;87(9):851–857. doi: 10.1038/labinvest.3700656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng L, Chen X, Varki A. Exploration of sialic acid diversity and biology using sialoglycan microarrays. Biopolymers. 2013;99(10):650–665. doi: 10.1002/bip.22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varki A. Loss of N-glycolylneuraminic acid in humans: mechanisms, consequences, and implications for hominid evolution. Am J Phys Anthropol Suppl. 2001;33:54–69. doi: 10.1002/ajpa.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samraj AN, Pearce OM, Laubli H, Crittenden AN, Bergfeld AK, Banda K, Gregg CJ, Bingman AE, Secrest P, Diaz SL, Varki NM, Varki A. A red meat-derived glycan promotes inflammation and cancer progression. Proc Natl Acad Sci USA. 2015;112(2):542–547. doi: 10.1073/pnas.1417508112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varki A. Colloquium paper: uniquely human evolution of sialic acid genetics and biology. Proc Natl Acad Sci USA. 2010;107(Suppl 2):8939–8946. doi: 10.1073/pnas.0914634107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawanishi K, Dhar C, Do R, Varki N, Gordts P, Varki A. Human species-specific loss of CMP-N-acetylneuraminic acid hydroxylase enhances atherosclerosis via intrinsic and extrinsic mechanisms. Proc Natl Acad Sci USA. 2019;116(32):16036–16045. doi: 10.1073/pnas.1902902116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boligan KF, Mesa C, Fernandez LE, von Gunten S. Cancer intelligence acquired (CIA): tumor glycosylation and sialylation codes dismantling antitumor defense. Cell Mol Life Sci. 2015;72(7):1231–1248. doi: 10.1007/s00018-014-1799-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dall’Olio F, Malagolini N, Trinchera M. Chiricolo M (2014) Sialosignaling: sialyltransferases as engines of self-fueling loops in cancer progression. Biochim Biophys Acta. 1840;9:2752–2764. doi: 10.1016/j.bbagen.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Laubli H, Borsig L. Selectins as mediators of lung metastasis. Cancer Microenviron. 2010;3(1):97–105. doi: 10.1007/s12307-010-0043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley K. The role of selectins in inflammation and disease. Trends Mol Med. 2003;9(6):263–268. doi: 10.1016/S1471-4914(03)00071-6. [DOI] [PubMed] [Google Scholar]
- 16.Macauley MS, Crocker PR, Paulson JC. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol. 2014;14(10):653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7(4):255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 18.Pillai S, Netravali IA, Cariappa A, Mattoo H. Siglecs and immune regulation. Annu Rev Immunol. 2012;30:357–392. doi: 10.1146/annurev-immunol-020711-075018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angata T. Molecular diversity and evolution of the Siglec family of cell-surface lectins. Mol Divers. 2006;10(4):555–566. doi: 10.1007/s11030-006-9029-1. [DOI] [PubMed] [Google Scholar]
- 20.Angata T. Possible influences of endogenous and exogenous ligands on the evolution of human siglecs. Front Immunol. 2018;9:2885. doi: 10.3389/fimmu.2018.02885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz F, Fong JJ, Varki A. Human-specific evolutionary changes in the biology of siglecs. Adv Exp Med Biol. 2015;842:1–16. doi: 10.1007/978-3-319-11280-0_1. [DOI] [PubMed] [Google Scholar]
- 22.Angata T, Margulies EH, Green ED, Varki A. Large-scale sequencing of the CD33-related Siglec gene cluster in five mammalian species reveals rapid evolution by multiple mechanisms. Proc Natl Acad Sci USA. 2004;101(36):13251–13256. doi: 10.1073/pnas.0404833101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Attrill H, Takazawa H, Witt S, Kelm S, Isecke R, Brossmer R, Ando T, Ishida H, Kiso M, Crocker PR, van Aalten DM. The structure of siglec-7 in complex with sialosides: leads for rational structure-based inhibitor design. Biochem J. 2006;397(2):271–278. doi: 10.1042/BJ20060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Attrill H, Imamura A, Sharma RS, Kiso M, Crocker PR, van Aalten DM. Siglec-7 undergoes a major conformational change when complexed with the alpha(2,8)-disialylganglioside GT1b. J Biol Chem. 2006;281(43):32774–32783. doi: 10.1074/jbc.M601714200. [DOI] [PubMed] [Google Scholar]
- 25.Alphey MS, Attrill H, Crocker PR, van Aalten DM. High resolution crystal structures of Siglec-7. Insights into ligand specificity in the Siglec family. J Biol Chem. 2003;278(5):3372–3377. doi: 10.1074/jbc.m210602200. [DOI] [PubMed] [Google Scholar]
- 26.Ereno-Orbea J, Sicard T, Cui H, Mazhab-Jafari MT, Benlekbir S, Guarne A, Rubinstein JL, Julien JP. Molecular basis of human CD22 function and therapeutic targeting. Nat Commun. 2017;8(1):764. doi: 10.1038/s41467-017-00836-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Propster JM, Yang F, Rabbani S, Ernst B, Allain FH, Schubert M. Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8. Proc Natl Acad Sci USA. 2016;113(29):E4170–E4179. doi: 10.1073/pnas.1602214113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angata T, Varki NM, Varki A. A second uniquely human mutation affecting sialic acid biology. J Biol Chem. 2001;276(43):40282–40287. doi: 10.1074/jbc.M105926200. [DOI] [PubMed] [Google Scholar]
- 29.Mitra N, Banda K, Altheide TK, Schaffer L, Johnson-Pais TL, Beuten J, Leach RJ, Angata T, Varki N, Varki A. SIGLEC12, a human-specific segregating (pseudo)gene, encodes a signaling molecule expressed in prostate carcinomas. J Biol Chem. 2011;286(26):23003–23011. doi: 10.1074/jbc.M111.244152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angata T, Varki A. Cloning, characterization, and phylogenetic analysis of siglec-9, a new member of the CD33-related group of siglecs. Evidence for co-evolution with sialic acid synthesis pathways. J Biol Chem. 2000;275(29):22127–22135. doi: 10.1074/jbc.m002775200. [DOI] [PubMed] [Google Scholar]
- 31.Laubli H, Pearce OM, Schwarz F, Siddiqui SS, Deng L, Stanczak MA, Deng L, Verhagen A, Secrest P, Lusk C, Schwartz AG, Varki NM, Bui JD, Varki A. Engagement of myelomonocytic Siglecs by tumor-associated ligands modulates the innate immune response to cancer. Proc Natl Acad Sci USA. 2014;111(39):14211–14216. doi: 10.1073/pnas.1409580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheong KA, Chang YS, Roh JY, Kim BJ, Kim MN, Park YM, Park HJ, Kim ND, Lee CH, Lee AY. A novel function of Siglec-9 A391C polymorphism on T cell receptor signaling. Int Arch Allergy Immunol. 2011;154(2):111–118. doi: 10.1159/000320225. [DOI] [PubMed] [Google Scholar]
- 33.Padler-Karavani V, Hurtado-Ziola N, Chang YC, Sonnenburg JL, Ronaghy A, Yu H, Verhagen A, Nizet V, Chen X, Varki N, Varki A, Angata T. Rapid evolution of binding specificities and expression patterns of inhibitory CD33-related Siglecs in primates. FASEB J. 2014;28(3):1280–1293. doi: 10.1096/fj.13-241497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bochner BS, Alvarez RA, Mehta P, Bovin NV, Blixt O, White JR, Schnaar RL. Glycan array screening reveals a candidate ligand for Siglec-8. J Biol Chem. 2005;280(6):4307–4312. doi: 10.1074/jbc.M412378200. [DOI] [PubMed] [Google Scholar]
- 35.Schleimer RP, Schnaar RL, Bochner BS. Regulation of airway inflammation by Siglec-8 and Siglec-9 sialoglycan ligand expression. Curr Opin Allergy Clin Immunol. 2016;16(1):24–30. doi: 10.1097/ACI.0000000000000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu H, Gonzalez-Gil A, Wei Y, Fernandes SM, Porell RN, Vajn K, Paulson JC, Nycholat CM, Schnaar RL. Siglec-8 and Siglec-9 binding specificities and endogenous airway ligand distributions and properties. Glycobiology. 2017;27(7):657–668. doi: 10.1093/glycob/cwx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Secundino I, Lizcano A, Roupe KM, Wang X, Cole JN, Olson J, Ali SR, Dahesh S, Amayreh LK, Henningham A, Varki A, Nizet V. Host and pathogen hyaluronan signal through human siglec-9 to suppress neutrophil activation. J Mol Med (Berl) 2016;94(2):219–233. doi: 10.1007/s00109-015-1341-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawasaki Y, Ito A, Withers DA, Taima T, Kakoi N, Saito S, Arai Y. Ganglioside DSGb5, preferred ligand for Siglec-7, inhibits NK cell cytotoxicity against renal cell carcinoma cells. Glycobiology. 2010;20(11):1373–1379. doi: 10.1093/glycob/cwq116. [DOI] [PubMed] [Google Scholar]
- 39.Avril T, North SJ, Haslam SM, Willison HJ, Crocker PR. Probing the cis interactions of the inhibitory receptor Siglec-7 with alpha2,8-disialylated ligands on natural killer cells and other leukocytes using glycan-specific antibodies and by analysis of alpha2,8-sialyltransferase gene expression. J Leukoc Biol. 2006;80(4):787–796. doi: 10.1189/jlb.1005559. [DOI] [PubMed] [Google Scholar]
- 40.Aalto K, Autio A, Kiss EA, Elima K, Nymalm Y, Veres TZ, Marttila-Ichihara F, Elovaara H, Saanijoki T, Crocker PR, Maksimow M, Bligt E, Salminen TA, Salmi M, Roivainen A, Jalkanen S. Siglec-9 is a novel leukocyte ligand for vascular adhesion protein-1 and can be used in PET imaging of inflammation and cancer. Blood. 2011;118(13):3725–3733. doi: 10.1182/blood-2010-09-311076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fong JJ, Sreedhara K, Deng L, Varki NM, Angata T, Liu Q, Nizet V, Varki A. Immunomodulatory activity of extracellular Hsp70 mediated via paired receptors Siglec-5 and Siglec-14. EMBO J. 2015;34(22):2775–2788. doi: 10.15252/embj.201591407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stanczak MA, Siddiqui SS, Trefny MP, Thommen DS, Boligan KF, von Gunten S, Tzankov A, Tietze L, Lardinois D, Heinzelmann-Schwarz V, von Bergwelt-Baidon M, Zhang W, Lenz HJ, Han Y, Amos CI, Syedbasha M, Egli A, Stenner F, Speiser DE, Varki A, Zippelius A, Laubli H. Self-associated molecular patterns mediate cancer immune evasion by engaging Siglecs on T cells. J Clin Invest. 2018 doi: 10.1172/JCI120612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varki A. Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology. 2011;21(9):1121–1124. doi: 10.1093/glycob/cwr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gonzalez-Gil A, Porell RN, Fernandes SM, Wei Y, Yu H, Carroll DJ, McBride R, Paulson JC, Tiemeyer M, Aoki K, Bochner BS, Schnaar RL. Sialylated keratan sulfate proteoglycans are Siglec-8 ligands in human airways. Glycobiology. 2018;28(10):786–801. doi: 10.1093/glycob/cwy057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doody GM, Justement LB, Delibrias CC, Matthews RJ, Lin J, Thomas ML, Fearon DT. A role in B cell activation for CD22 and the protein tyrosine phosphatase SHP. Science. 1995;269(5221):242–244. doi: 10.1126/science.7618087. [DOI] [PubMed] [Google Scholar]
- 46.Taylor VC, Buckley CD, Douglas M, Cody AJ, Simmons DL, Freeman SD. The myeloid-specific sialic acid-binding receptor, CD33, associates with the protein-tyrosine phosphatases, SHP-1 and SHP-2. J Biol Chem. 1999;274(17):11505–11512. doi: 10.1074/jbc.274.17.11505. [DOI] [PubMed] [Google Scholar]
- 47.Angata T, Kerr SC, Greaves DR, Varki NM, Crocker PR, Varki A. Cloning and characterization of human Siglec-11. A recently evolved signaling molecule that can interact with SHP-1 and SHP-2 and is expressed by tissue macrophages, including brain microglia. J Biol Chem. 2002;277(27):24466–24474. doi: 10.1074/jbc.m202833200. [DOI] [PubMed] [Google Scholar]
- 48.Ikehara Y, Ikehara SK, Paulson JC. Negative regulation of T cell receptor signaling by Siglec-7 (p70/AIRM) and Siglec-9. J Biol Chem. 2004;279(41):43117–43125. doi: 10.1074/jbc.M403538200. [DOI] [PubMed] [Google Scholar]
- 49.Cao H, Lakner U, de Bono B, Traherne JA, Trowsdale J, Barrow AD. SIGLEC16 encodes a DAP12-associated receptor expressed in macrophages that evolved from its inhibitory counterpart SIGLEC11 and has functional and non-functional alleles in humans. Eur J Immunol. 2008;38(8):2303–2315. doi: 10.1002/eji.200738078. [DOI] [PubMed] [Google Scholar]
- 50.Angata T, Hayakawa T, Yamanaka M, Varki A, Nakamura M. Discovery of Siglec-14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates. FASEB J. 2006;20(12):1964–1973. doi: 10.1096/fj.06-5800com. [DOI] [PubMed] [Google Scholar]
- 51.Blasius AL, Cella M, Maldonado J, Takai T, Colonna M. Siglec-H is an IPC-specific receptor that modulates type I IFN secretion through DAP12. Blood. 2006;107(6):2474–2476. doi: 10.1182/blood-2005-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayakawa T, Khedri Z, Schwarz F, Landig C, Liang SY, Yu H, Chen X, Fujito NT, Satta Y, Varki A, Angata T. Coevolution of Siglec-11 and Siglec-16 via gene conversion in primates. BMC Evol Biol. 2017;17(1):228. doi: 10.1186/s12862-017-1075-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwarz F, Landig CS, Siddiqui S, Secundino I, Olson J, Varki N, Nizet V, Varki A. Paired Siglec receptors generate opposite inflammatory responses to a human-specific pathogen. EMBO J. 2017 doi: 10.15252/embj.201695581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Angata T, Nycholat CM, Macauley MS. Therapeutic targeting of siglecs using antibody- and glycan-based approaches. Trends Pharmacol Sci. 2015;36(10):645–660. doi: 10.1016/j.tips.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haas Q, Frias Boligan K, Jandus C, Schneider C, Simillion C, Stanczak MA, Haubitz M, Seyed Jafari SM, Zippelius A, Baerlocher GM, Laubli H, Hunger RE, Romero P, Simon HU, von Gunten S. Siglec-9 regulates an effector memory CD8 + T-cell subset that congregates in the melanoma tumor microenvironment. Cancer Immunol Res. 2019 doi: 10.1158/2326-6066.CIR-18-0505. [DOI] [PubMed] [Google Scholar]
- 56.Varki A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. 2007;446(7139):1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- 57.Schaefer L. Complexity of danger: the diverse nature of damage-associated molecular patterns. J Biol Chem. 2014;289(51):35237–35245. doi: 10.1074/jbc.R114.619304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trinchieri G, Sher A. Cooperation of Toll-like receptor signals in innate immune defence. Nat Rev Immunol. 2007;7(3):179–190. doi: 10.1038/nri2038. [DOI] [PubMed] [Google Scholar]
- 59.Liston A, Masters SL. Homeostasis-altering molecular processes as mechanisms of inflammasome activation. Nat Rev Immunol. 2017;17(3):208–214. doi: 10.1038/nri.2016.151. [DOI] [PubMed] [Google Scholar]
- 60.Lizcano A, Secundino I, Dohrmann S, Corriden R, Rohena C, Diaz S, Ghosh P, Deng L, Nizet V, Varki A. Erythrocyte sialoglycoproteins engage Siglec-9 on neutrophils to suppress activation. Blood. 2017;129(23):3100–3110. doi: 10.1182/blood-2016-11-751636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ali SR, Fong JJ, Carlin AF, Busch TD, Linden R, Angata T, Areschoug T, Parast M, Varki N, Murray J, Nizet V, Varki A. Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J Exp Med. 2014;211(6):1231–1242. doi: 10.1084/jem.20131853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Landig CS, Hazel A, Kellman BP, Fong JJ, Schwarz F, Agarwal S, Varki N, Massari P, Lewis NE, Ram S, Varki A. Evolution of the exclusively human pathogen Neisseria gonorrhoeae: human-specific engagement of immunoregulatory Siglecs. Evol Appl. 2019;12(2):337–349. doi: 10.1111/eva.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen DH, Ball ED, Varki A. Myeloid precursors and acute myeloid leukemia cells express multiple CD33-related Siglecs. Exp Hematol. 2006;34(6):728–735. doi: 10.1016/j.exphem.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 64.von Gunten S, Yousefi S, Seitz M, Jakob SM, Schaffner T, Seger R, Takala J, Villiger PM, Simon HU. Siglec-9 transduces apoptotic and nonapoptotic death signals into neutrophils depending on the proinflammatory cytokine environment. Blood. 2005;106(4):1423–1431. doi: 10.1182/blood-2004-10-4112. [DOI] [PubMed] [Google Scholar]
- 65.Klaas M, Crocker PR. Sialoadhesin in recognition of self and non-self. Semin Immunopathol. 2012;34(3):353–364. doi: 10.1007/s00281-012-0310-3. [DOI] [PubMed] [Google Scholar]
- 66.Hartnell A, Steel J, Turley H, Jones M, Jackson DG, Crocker PR. Characterization of human sialoadhesin, a sialic acid binding receptor expressed by resident and inflammatory macrophage populations. Blood. 2001;97(1):288–296. doi: 10.1182/blood.V97.1.288. [DOI] [PubMed] [Google Scholar]
- 67.Angata T, Tabuchi Y, Nakamura K, Nakamura M. Siglec-15: an immune system Siglec conserved throughout vertebrate evolution. Glycobiology. 2007;17(8):838–846. doi: 10.1093/glycob/cwm049. [DOI] [PubMed] [Google Scholar]
- 68.Kameda Y, Takahata M, Komatsu M, Mikuni S, Hatakeyama S, Shimizu T, Angata T, Kinjo M, Minami A, Iwasaki N. Siglec-15 regulates osteoclast differentiation by modulating RANKL-induced phosphatidylinositol 3-kinase/Akt and Erk pathways in association with signaling Adaptor DAP12. J Bone Miner Res. 2013;28(12):2463–2475. doi: 10.1002/jbmr.1989. [DOI] [PubMed] [Google Scholar]
- 69.Wang J, Sun J, Liu LN, Flies DB, Nie X, Toki M, Zhang J, Song C, Zarr M, Zhou X, Han X, Archer KA, O’Neill T, Herbst RS, Boto AN, Sanmamed MF, Langermann S, Rimm DL, Chen L. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat Med. 2019;25(4):656–666. doi: 10.1038/s41591-019-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nitschke L. CD22 and Siglec-G regulate inhibition of B-cell signaling by sialic acid ligand binding and control B-cell tolerance. Glycobiology. 2014;24(9):807–817. doi: 10.1093/glycob/cwu066. [DOI] [PubMed] [Google Scholar]
- 71.Pfrengle F, Macauley MS, Kawasaki N, Paulson JC. Copresentation of antigen and ligands of Siglec-G induces B cell tolerance independent of CD22. J Immunol. 2013;191(4):1724–1731. doi: 10.4049/jimmunol.1300921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Munday J, Kerr S, Ni J, Cornish AL, Zhang JQ, Nicoll G, Floyd H, Mattei MG, Moore P, Liu D, Crocker PR. Identification, characterization and leucocyte expression of Siglec-10, a novel human sialic acid-binding receptor. Biochem J. 2001;355(Pt 2):489–497. doi: 10.1042/bj3550489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kikly KK, Bochner BS, Freeman SD, Tan KB, Gallagher KT, D’Alessio KJ, Holmes SD, Abrahamson JA, Erickson-Miller CL, Murdock PR, Tachimoto H, Schleimer RP, White JR. Identification of SAF-2, a novel siglec expressed on eosinophils, mast cells, and basophils. J Allergy Clin Immunol. 2000;105(6 Pt 1):1093–1100. doi: 10.1067/mai.2000.107127. [DOI] [PubMed] [Google Scholar]
- 74.Floyd H, Ni J, Cornish AL, Zeng Z, Liu D, Carter KC, Steel J, Crocker PR. Siglec-8. A novel eosinophil-specific member of the immunoglobulin superfamily. J Biol Chem. 2000;275(2):861–866. doi: 10.1074/jbc.275.2.861. [DOI] [PubMed] [Google Scholar]
- 75.Song DJ, Cho JY, Lee SY, Miller M, Rosenthal P, Soroosh P, Croft M, Zhang M, Varki A, Broide DH. Anti-Siglec-F antibody reduces allergen-induced eosinophilic inflammation and airway remodeling. J Immunol. 2009;183(8):5333–5341. doi: 10.4049/jimmunol.0801421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang M, Angata T, Cho JY, Miller M, Broide DH, Varki A. Defining the in vivo function of Siglec-F, a CD33-related Siglec expressed on mouse eosinophils. Blood. 2007;109(10):4280–4287. doi: 10.1182/blood-2006-08-039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duan S, Koziol-White CJ, Jester WF, Jr, Nycholat CM, Macauley MS, Panettieri RA, Jr, Paulson JC. CD33 recruitment inhibits IgE-mediated anaphylaxis and desensitizes mast cells to allergen. J Clin Invest. 2019;129(3):1387–1401. doi: 10.1172/JCI125456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lubbers J, Rodriguez E, van Kooyk Y. Modulation of immune tolerance via siglec-sialic acid interactions. Front Immunol. 2018;9:2807. doi: 10.3389/fimmu.2018.02807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Crespo HJ, Lau JT, Videira PA. Dendritic cells: a spot on sialic Acid. Front Immunol. 2013;4:491. doi: 10.3389/fimmu.2013.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bax M, Garcia-Vallejo JJ, Jang-Lee J, North SJ, Gilmartin TJ, Hernandez G, Crocker PR, Leffler H, Head SR, Haslam SM, Dell A, van Kooyk Y. Dendritic cell maturation results in pronounced changes in glycan expression affecting recognition by siglecs and galectins. J Immunol. 2007;179(12):8216–8224. doi: 10.4049/jimmunol.179.12.8216. [DOI] [PubMed] [Google Scholar]
- 81.Lock K, Zhang J, Lu J, Lee SH, Crocker PR. Expression of CD33-related siglecs on human mononuclear phagocytes, monocyte-derived dendritic cells and plasmacytoid dendritic cells. Immunobiology. 2004;209(1–2):199–207. doi: 10.1016/j.imbio.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 82.Zhang JQ, Biedermann B, Nitschke L, Crocker PR. The murine inhibitory receptor mSiglec-E is expressed broadly on cells of the innate immune system whereas mSiglec-F is restricted to eosinophils. Eur J Immunol. 2004;34(4):1175–1184. doi: 10.1002/eji.200324723. [DOI] [PubMed] [Google Scholar]
- 83.Perdicchio M, Ilarregui JM, Verstege MI, Cornelissen LA, Schetters ST, Engels S, Ambrosini M, Kalay H, Veninga H, den Haan JM, van Berkel LA, Samsom JN, Crocker PR, Sparwasser T, Berod L, Garcia-Vallejo JJ, van Kooyk Y, Unger WW. Sialic acid-modified antigens impose tolerance via inhibition of T-cell proliferation and de novo induction of regulatory T cells. Proc Natl Acad Sci USA. 2016;113(12):3329–3334. doi: 10.1073/pnas.1507706113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ding Y, Guo Z, Liu Y, Li X, Zhang Q, Xu X, Gu Y, Zhang Y, Zhao D, Cao X. The lectin Siglec-G inhibits dendritic cell cross-presentation by impairing MHC class I-peptide complex formation. Nat Immunol. 2016;17(10):1167–1175. doi: 10.1038/ni.3535. [DOI] [PubMed] [Google Scholar]
- 85.Durai V, Murphy KM. Functions of murine dendritic cells. Immunity. 2016;45(4):719–736. doi: 10.1016/j.immuni.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Loschko J, Heink S, Hackl D, Dudziak D, Reindl W, Korn T, Krug AB. Antigen targeting to plasmacytoid dendritic cells via Siglec-H inhibits Th cell-dependent autoimmunity. J Immunol. 2011;187(12):6346–6356. doi: 10.4049/jimmunol.1102307. [DOI] [PubMed] [Google Scholar]
- 87.Nicoll G, Ni J, Liu D, Klenerman P, Munday J, Dubock S, Mattei MG, Crocker PR. Identification and characterization of a novel siglec, siglec-7, expressed by human natural killer cells and monocytes. J Biol Chem. 1999;274(48):34089–34095. doi: 10.1074/jbc.274.48.34089. [DOI] [PubMed] [Google Scholar]
- 88.Jandus C, Boligan KF, Chijioke O, Liu H, Dahlhaus M, Demoulins T, Schneider C, Wehrli M, Hunger RE, Baerlocher GM, Simon HU, Romero P, Munz C, von Gunten S. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J Clin Invest. 2014;124(4):1810–1820. doi: 10.1172/JCI65899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang JQ, Nicoll G, Jones C, Crocker PR. Siglec-9, a novel sialic acid binding member of the immunoglobulin superfamily expressed broadly on human blood leukocytes. J Biol Chem. 2000;275(29):22121–22126. doi: 10.1074/jbc.M002788200. [DOI] [PubMed] [Google Scholar]
- 90.Falco M, Biassoni R, Bottino C, Vitale M, Sivori S, Augugliaro R, Moretta L, Moretta A. Identification and molecular cloning of p75/AIRM1, a novel member of the sialoadhesin family that functions as an inhibitory receptor in human natural killer cells. J Exp Med. 1999;190(6):793–802. doi: 10.1084/jem.190.6.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fong JJ, Tsai CM, Saha S, Nizet V, Varki A, Bui JD. Siglec-7 engagement by GBS beta-protein suppresses pyroptotic cell death of natural killer cells. Proc Natl Acad Sci USA. 2018;115(41):10410–10415. doi: 10.1073/pnas.1804108115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meyer SJ, Linder AT, Brandl C, Nitschke L. B cell Siglecs-news on signaling and its interplay with ligand binding. Front Immunol. 2018;9:2820. doi: 10.3389/fimmu.2018.02820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hoffmann A, Kerr S, Jellusova J, Zhang J, Weisel F, Wellmann U, Winkler TH, Kneitz B, Crocker PR, Nitschke L. Siglec-G is a B1 cell-inhibitory receptor that controls expansion and calcium signaling of the B1 cell population. Nat Immunol. 2007;8(7):695–704. doi: 10.1038/ni1480. [DOI] [PubMed] [Google Scholar]
- 94.Nguyen DH, Hurtado-Ziola N, Gagneux P, Varki A. Loss of Siglec expression on T lymphocytes during human evolution. Proc Natl Acad Sci USA. 2006;103(20):7765–7770. doi: 10.1073/pnas.0510484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Soto PC, Karris MY, Spina CA, Richman DD, Varki A. Cell-intrinsic mechanism involving Siglec-5 associated with divergent outcomes of HIV-1 infection in human and chimpanzee CD4 T cells. J Mol Med (Berl) 2013;91(2):261–270. doi: 10.1007/s00109-012-0951-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brinkman-Van der Linden EC, Hurtado-Ziola N, Hayakawa T, Wiggleton L, Benirschke K, Varki A, Varki N. Human-specific expression of Siglec-6 in the placenta. Glycobiology. 2007;17(9):922–931. doi: 10.1093/glycob/cwm065. [DOI] [PubMed] [Google Scholar]
- 97.Tecle E, Reynoso HS, Wang R, Gagneux P. The female reproductive tract contains multiple innate sialic acid-binding immunoglobulin-like lectins (Siglecs) that facilitate sperm survival. J Biol Chem. 2019 doi: 10.1074/jbc.RA119.008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alkhodair K, Almhanna H, McGetrick J, Gedair S, Gallagher ME, Fernandez-Fuertes B, Tharmalingam T, Larsen PB, Fitzpatrick E, Lonergan P, Evans ACO, Carrington SD, Reid CJ. Siglec expression on the surface of human, bull and ram sperm. Reproduction. 2018;155(4):361–371. doi: 10.1530/REP-17-0475. [DOI] [PubMed] [Google Scholar]
- 99.Dharmadhikari G, Stolz K, Hauke M, Morgan NG, Varki A, de Koning E, Kelm S, Maedler K. Siglec-7 restores beta-cell function and survival and reduces inflammation in pancreatic islets from patients with diabetes. Sci Rep. 2017;7:45319. doi: 10.1038/srep45319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chang YC, Nizet V. The interplay between Siglecs and sialylated pathogens. Glycobiology. 2014;24(9):818–825. doi: 10.1093/glycob/cwu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carlin AF, Chang YC, Areschoug T, Lindahl G, Hurtado-Ziola N, King CC, Varki A, Nizet V. Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5. J Exp Med. 2009;206(8):1691–1699. doi: 10.1084/jem.20090691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu YC, Yu MM, Chai YF, Shou ST. Sialic Acids in the Immune Response during Sepsis. Front Immunol. 2017;8:1601. doi: 10.3389/fimmu.2017.01601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chang YC, Uchiyama S, Varki A, Nizet V. Leukocyte inflammatory responses provoked by pneumococcal sialidase. MBio. 2012 doi: 10.1128/mbio.00220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen GY, Chen X, King S, Cavassani KA, Cheng J, Zheng X, Cao H, Yu H, Qu J, Fang D, Wu W, Bai XF, Liu JQ, Woodiga SA, Chen C, Sun L, Hogaboam CM, Kunkel SL, Zheng P, Liu Y. Amelioration of sepsis by inhibiting sialidase-mediated disruption of the CD24-SiglecG interaction. Nat Biotechnol. 2011;29(5):428–435. doi: 10.1038/nbt.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen GY, Tang J, Zheng P, Liu Y. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science. 2009;323(5922):1722–1725. doi: 10.1126/science.1168988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bochner BS, Zimmermann N. Role of siglecs and related glycan-binding proteins in immune responses and immunoregulation. J Allergy Clin Immunol. 2015;135(3):598–608. doi: 10.1016/j.jaci.2014.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Suzukawa M, Miller M, Rosenthal P, Cho JY, Doherty TA, Varki A, Broide D. Sialyltransferase ST3Gal-III regulates Siglec-F ligand formation and eosinophilic lung inflammation in mice. J Immunol. 2013;190(12):5939–5948. doi: 10.4049/jimmunol.1203455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rubinstein E, Cho JY, Rosenthal P, Chao J, Miller M, Pham A, Aceves SS, Varki A, Broide DH. Siglec-F inhibition reduces esophageal eosinophilia and angiogenesis in a mouse model of eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2011;53(4):409–416. doi: 10.1097/MPG.0b013e3182182ff8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Song DJ, Cho JY, Miller M, Strangman W, Zhang M, Varki A, Broide DH. Anti-Siglec-F antibody inhibits oral egg allergen induced intestinal eosinophilic inflammation in a mouse model. Clin Immunol. 2009;131(1):157–169. doi: 10.1016/j.clim.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.O’Sullivan JA, Wei Y, Carroll DJ, Moreno-Vinasco L, Cao Y, Zhang F, Lee JJ, Zhu Z, Bochner BS. Frontline science: characterization of a novel mouse strain expressing human Siglec-8 only on eosinophils. J Leukoc Biol. 2018;104(1):11–19. doi: 10.1002/JLB.2HI0917-391R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bandala-Sanchez E, Zhang Y, Reinwald S, Dromey JA, Lee BH, Qian J, Bohmer RM, Harrison LC. T cell regulation mediated by interaction of soluble CD52 with the inhibitory receptor Siglec-10. Nat Immunol. 2013;14(7):741–748. doi: 10.1038/ni.2610. [DOI] [PubMed] [Google Scholar]
- 112.O’Keefe TL, Williams GT, Davies SL, Neuberger MS. Hyperresponsive B cells in CD22-deficient mice. Science. 1996;274(5288):798–801. doi: 10.1126/science.274.5288.798. [DOI] [PubMed] [Google Scholar]
- 113.Muller J, Lunz B, Schwab I, Acs A, Nimmerjahn F, Daniel C, Nitschke L. Siglec-G deficiency leads to autoimmunity in aging C57BL/6 mice. J Immunol. 2015;195(1):51–60. doi: 10.4049/jimmunol.1403139. [DOI] [PubMed] [Google Scholar]
- 114.Jellusova J, Wellmann U, Amann K, Winkler TH, Nitschke L. CD22 x Siglec-G double-deficient mice have massively increased B1 cell numbers and develop systemic autoimmunity. J Immunol. 2010;184(7):3618–3627. doi: 10.4049/jimmunol.0902711. [DOI] [PubMed] [Google Scholar]
- 115.Macauley MS, Paulson JC. Siglecs induce tolerance to cell surface antigens by BIM-dependent deletion of the antigen-reactive B cells. J Immunol. 2014;193(9):4312–4321. doi: 10.4049/jimmunol.1401723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Duong BH, Tian H, Ota T, Completo G, Han S, Vela JL, Ota M, Kubitz M, Bovin N, Paulson JC, Nemazee D. Decoration of T-independent antigen with ligands for CD22 and Siglec-G can suppress immunity and induce B cell tolerance in vivo. J Exp Med. 2010;207(1):173–187. doi: 10.1084/jem.20091873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bednar KJ, Nycholat CM, Rao TS, Paulson JC, Fung-Leung WP, Macauley MS. Exploiting CD22 To selectively tolerize autoantibody producing B-cells in rheumatoid arthritis. ACS Chem Biol. 2019;14(4):644–654. doi: 10.1021/acschembio.8b01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Surolia I, Pirnie SP, Chellappa V, Taylor KN, Cariappa A, Moya J, Liu H, Bell DW, Driscoll DR, Diederichs S, Haider K, Netravali I, Le S, Elia R, Dow E, Lee A, Freudenberg J, De Jager PL, Chretien Y, Varki A, MacDonald ME, Gillis T, Behrens TW, Bloch D, Collier D, Korzenik J, Podolsky DK, Hafler D, Murali M, Sands B, Stone JH, Gregersen PK, Pillai S. Functionally defective germline variants of sialic acid acetylesterase in autoimmunity. Nature. 2010;466(7303):243–247. doi: 10.1038/nature09115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bandala-Sanchez E, Bediaga NG, Goddard-Borger ED, Ngui K, Naselli G, Stone NL, Neale AM, Pearce LA, Wardak A, Czabotar P, Haselhorst T, Maggioni A, Hartley-Tassell LA, Adams TE, Harrison LC. CD52 glycan binds the proinflammatory B box of HMGB1 to engage the Siglec-10 receptor and suppress human T cell function. Proc Natl Acad Sci USA. 2018;115(30):7783–7788. doi: 10.1073/pnas.1722056115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sedlacek HH, Meesmann H, Seiler FR. Regression of spontaneous mammary tumors in dogs after injection of neuraminidase-treated tumor cells. Int J Cancer. 1975;15(3):409–416. doi: 10.1002/ijc.2910150307. [DOI] [PubMed] [Google Scholar]
- 121.Currie GA, Bagshawe KD. The role of sialic acid in antigenic expression: further studies of the Landschutz ascites tumour. Br J Cancer. 1968;22(4):843–853. doi: 10.1038/bjc.1968.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bagshawe KD, Currie GA. Immunogenicity of L 1210 murine leukaemia cells after treatment with neuraminidase. Nature. 1968;218(5148):1254–1255. doi: 10.1038/2181254a0. [DOI] [PubMed] [Google Scholar]
- 123.Sanford BH. An alteration in tumor histocompatibility induced by neuraminidase. Transplantation. 1967;5(5):1273–1279. doi: 10.1097/00007890-196709000-00005. [DOI] [PubMed] [Google Scholar]
- 124.Adams OJ, Stanczak MA, von Gunten S, Laubli H. Targeting sialic acid-Siglec interactions to reverse immune suppression in cancer. Glycobiology. 2018;28(9):640–647. doi: 10.1093/glycob/cwx108. [DOI] [PubMed] [Google Scholar]
- 125.Bull C, Stoel MA, den Brok MH, Adema GJ. Sialic acids sweeten a tumor’s life. Cancer Res. 2014;74(12):3199–3204. doi: 10.1158/0008-5472.CAN-14-0728. [DOI] [PubMed] [Google Scholar]
- 126.Hudak JE, Canham SM, Bertozzi CR. Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat Chem Biol. 2014;10(1):69–75. doi: 10.1038/nchembio.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Beatson R, Tajadura-Ortega V, Achkova D, Picco G, Tsourouktsoglou TD, Klausing S, Hillier M, Maher J, Noll T, Crocker PR, Taylor-Papadimitriou J, Burchell JM. The mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nat Immunol. 2016 doi: 10.1038/ni.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barkal AA, Brewer RE, Markovic M, Kowarsky M, Barkal SA, Zaro BW, Krishnan V, Hatakeyama J, Dorigo O, Barkal LJ, Weissman IL. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature. 2019 doi: 10.1038/s41586-019-1456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Spence S, Greene MK, Fay F, Hams E, Saunders SP, Hamid U, Fitzgerald M, Beck J, Bains BK, Smyth P, Themistou E, Small DM, Schmid D, O’Kane CM, Fitzgerald DC, Abdelghany SM, Johnston JA, Fallon PG, Burrows JF, McAuley DF, Kissenpfennig A, Scott CJ. Targeting Siglecs with a sialic acid-decorated nanoparticle abrogates inflammation. Sci Transl Med. 2015;7(303):303ra140. doi: 10.1126/scitranslmed.aab3459. [DOI] [PubMed] [Google Scholar]
- 130.Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, Varki A. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. 2009;113(14):3333–3336. doi: 10.1182/blood-2008-11-187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chang YC, Olson J, Beasley FC, Tung C, Zhang J, Crocker PR, Varki A, Nizet V. Group B Streptococcus engages an inhibitory Siglec through sialic acid mimicry to blunt innate immune and inflammatory responses in vivo. PLoS Pathog. 2014;10(1):e1003846. doi: 10.1371/journal.ppat.1003846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bull C, Boltje TJ, Balneger N, Weischer SM, Wassink M, van Gemst JJ, Bloemendal VR, Boon L, van der Vlag J, Heise T, den Brok MH, Adema GJ. Sialic acid blockade suppresses tumor growth by enhancing T-cell-mediated tumor immunity. Cancer Res. 2018;78(13):3574–3588. doi: 10.1158/0008-5472.CAN-17-3376. [DOI] [PubMed] [Google Scholar]
- 133.Perdicchio M, Cornelissen LA, Streng-Ouwehand I, Engels S, Verstege MI, Boon L, Geerts D, van Kooyk Y, Unger WW. Tumor sialylation impedes T cell mediated anti-tumor responses while promoting tumor associated-regulatory T cells. Oncotarget. 2016;7(8):8771–8782. doi: 10.18632/oncotarget.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cornelissen LAM, Blanas A, van der Horst JC, Kruijssen L, Zaal A, O’Toole T, Wiercx L, van Kooyk Y, van Vliet SJ. Disruption of sialic acid metabolism drives tumor growth by augmenting CD8(+) T cell apoptosis. Int J Cancer. 2019;144(9):2290–2302. doi: 10.1002/ijc.32084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xiao H, Woods EC, Vukojicic P, Bertozzi CR. Precision glycocalyx editing as a strategy for cancer immunotherapy. Proc Natl Acad Sci USA. 2016;113(37):10304–10309. doi: 10.1073/pnas.1608069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Varki A, Schnaar RL, Crocker PR, et al. I-Type Lectins. In: Varki A, Cummings RD, et al., editors. Essentials of glycobiology. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2015. pp. 453–467. [Google Scholar]
- 137.Ramya TN, Weerapana E, Liao L, Zeng Y, Tateno H, Liao L, Yates JR, 3rd, Cravatt BF, Paulson JC. In situ trans ligands of CD22 identified by glycan-protein photocross-linking-enabled proteomics. Mol Cell Proteom. 2010;9(6):1339–1351. doi: 10.1074/mcp.M900461-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Malaker SA, Pedram K, Ferracane MJ, Bensing BA, Krishnan V, Pett C, Yu J, Woods EC, Kramer JR, Westerlind U, Dorigo O, Bertozzi CR. The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins. Proc Natl Acad Sci USA. 2019;116(15):7278–7287. doi: 10.1073/pnas.1813020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Briard JG, Jiang H, Moremen KW, Macauley MS, Wu P. Cell-based glycan arrays for probing glycan-glycan binding protein interactions. Nat Commun. 2018;9(1):880. doi: 10.1038/s41467-018-03245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rillahan CD, Schwartz E, McBride R, Fokin VV, Paulson JC. Click and pick: identification of sialoside analogues for siglec-based cell targeting. Angew Chem Int Ed Engl. 2012;51(44):11014–11018. doi: 10.1002/anie.201205831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Edgar LJ, Kawasaki N, Nycholat CM, Paulson JC. Targeted delivery of antigen to activated CD169(+) macrophages induces bias for expansion of CD8(+) T cells. Cell Chem Biol. 2019;26(1):131–136. doi: 10.1016/j.chembiol.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bednar KJ, Shanina E, Ballet R, Connors EP, Duan S, Juan J, Arlian BM, Kulis MD, Butcher EC, Fung-Leung WP, Rao TS, Paulson JC, Macauley MS. Human CD22 inhibits murine B Cell receptor activation in a human CD22 transgenic mouse Model. J Immunol. 2017;199(9):3116–3128. doi: 10.4049/jimmunol.1700898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ishii T, Angata T, Wan ES, Cho MH, Motegi T, Gao C, Ohtsubo K, Kitazume S, Gemma A, Par EP, Lomas DA, Silverman EK, Taniguchi N, Kida K. Influence of SIGLEC9 polymorphisms on COPD phenotypes including exacerbation frequency. Respirology. 2016 doi: 10.1111/resp.12952. [DOI] [PubMed] [Google Scholar]
- 144.Yamanaka M, Kato Y, Angata T, Narimatsu H. Deletion polymorphism of SIGLEC14 and its functional implications. Glycobiology. 2009;19(8):841–846. doi: 10.1093/glycob/cwp052. [DOI] [PubMed] [Google Scholar]
- 145.Hollingworth Paul, Harold Denise, Sims Rebecca, Gerrish Amy, Lambert Jean-Charles, Carrasquillo Minerva M, Abraham Richard, Hamshere Marian L, Pahwa Jaspreet Singh, Moskvina Valentina, Dowzell Kimberley, Jones Nicola, Stretton Alexandra, Thomas Charlene, Richards Alex, Ivanov Dobril, Widdowson Caroline, Chapman Jade, Lovestone Simon, Powell John, Proitsi Petroula, Lupton Michelle K, Brayne Carol, Rubinsztein David C, Gill Michael, Lawlor Brian, Lynch Aoibhinn, Brown Kristelle S, Passmore Peter A, Craig David, McGuinness Bernadette, Todd Stephen, Holmes Clive, Mann David, Smith A David, Beaumont Helen, Warden Donald, Wilcock Gordon, Love Seth, Kehoe Patrick G, Hooper Nigel M, Vardy Emma R L C, Hardy John, Mead Simon, Fox Nick C, Rossor Martin, Collinge John, Maier Wolfgang, Jessen Frank, Rüther Eckart, Schürmann Britta, Heun Reiner, Kölsch Heike, van den Bussche Hendrik, Heuser Isabella, Kornhuber Johannes, Wiltfang Jens, Dichgans Martin, Frölich Lutz, Hampel Harald, Gallacher John, Hüll Michael, Rujescu Dan, Giegling Ina, Goate Alison M, Kauwe John S K, Cruchaga Carlos, Nowotny Petra, Morris John C, Mayo Kevin, Sleegers Kristel, Bettens Karolien, Engelborghs Sebastiaan, De Deyn Peter P, Van Broeckhoven Christine, Livingston Gill, Bass Nicholas J, Gurling Hugh, McQuillin Andrew, Gwilliam Rhian, Deloukas Panagiotis, Al-Chalabi Ammar, Shaw Christopher E, Tsolaki Magda, Singleton Andrew B, Guerreiro Rita, Mühleisen Thomas W, Nöthen Markus M, Moebus Susanne, Jöckel Karl-Heinz, Klopp Norman, Wichmann H-Erich, Pankratz V Shane, Sando Sigrid B, Aasly Jan O, Barcikowska Maria, Wszolek Zbigniew K, Dickson Dennis W, Graff-Radford Neill R, Petersen Ronald C, van Duijn Cornelia M, Breteler Monique M B, Ikram M Arfan, DeStefano Anita L, Fitzpatrick Annette L, Lopez Oscar, Launer Lenore J, Seshadri Sudha, Berr Claudine, Campion Dominique, Epelbaum Jacques, Dartigues Jean-François, Tzourio Christophe, Alpérovitch Annick, Lathrop Mark, Feulner Thomas M, Friedrich Patricia, Riehle Caterina, Krawczak Michael, Schreiber Stefan, Mayhaus Manuel, Nicolhaus S, Wagenpfeil Stefan, Steinberg Stacy, Stefansson Hreinn, Stefansson Kari, Snædal Jon, Björnsson Sigurbjörn, Jonsson Palmi V, Chouraki Vincent, Genier-Boley Benjamin, Hiltunen Mikko, Soininen Hilkka, Combarros Onofre, Zelenika Diana, Delepine Marc, Bullido Maria J, Pasquier Florence, Mateo Ignacio, Frank-Garcia Ana, Porcellini Elisa, Hanon Olivier, Coto Eliecer, Alvarez Victoria, Bosco Paolo, Siciliano Gabriele, Mancuso Michelangelo, Panza Francesco, Solfrizzi Vincenzo, Nacmias Benedetta, Sorbi Sandro, Bossù Paola, Piccardi Paola, Arosio Beatrice, Annoni Giorgio, Seripa Davide, Pilotto Alberto, Scarpini Elio, Galimberti Daniela, Brice Alexis, Hannequin Didier, Licastro Federico, Jones Lesley, Holmans Peter A, Jonsson Thorlakur, Riemenschneider Matthias, Morgan Kevin, Younkin Steven G, Owen Michael J, O'Donovan Michael, Amouyel Philippe, Williams Julie. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nature Genetics. 2011;43(5):429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]