Abstract
A family of eleven glycosylphosphatidylinositol-anchored aspartyl proteases, commonly referred to as CgYapsins, regulate a myriad of cellular processes in the pathogenic yeast Candida glabrata, but their protein targets are largely unknown. Here, using the immunoprecipitation-mass spectrometry approach, we identify the flavodoxin-like protein (Fld-LP), CgPst2, to be an interactor of one of the aspartyl protease CgYps1. We also report the presence of four Fld-LPs in C. glabrata, which are required for survival in kidneys in the murine model of systemic candidiasis. We further demonstrated that of four Fld-LPs, CgPst2 was solely required for menadione detoxification. CgPst2 was found to form homo-oligomers, and contribute to cellular NADH:quinone oxidoreductase activity. CgYps1 cleaved CgPst2 at the C-terminus, and this cleavage was pivotal to oligomerization, activity and function of CgPst2. The arginine-174 residue in CgPst2 was essential for CgYps1-mediated cleavage, with alanine substitution of the arginine-174 residue also leading to elevated activity and oligomerization of CgPst2. Finally, we demonstrate that menadione treatment led to increased CgPst2 and CgYps1 protein levels, diminished CgYps1-CgPst2 interaction, and enhanced CgPst2 cleavage and activity, thereby implicating CgYps1 in activating CgPst2. Altogether, our findings of proteolytic cleavage as a key regulatory determinant of CgPst2, which belongs to the family of highly conserved, electron-carrier flavodoxin-fold-containing proteins, constituting cellular oxidative stress defense system in diverse organisms, unveil a hidden regulatory layer of environmental stress response mechanisms.
Author summary
Fungal bloodstream infections in immunodeficient patients are a serious clinical problem. Infections caused by the opportunistic fungal pathogen Candida glabrata are difficult to treat owing to its low susceptibility to widely used azole antifungals and emerging co-resistance to azole and echinocandin drugs. Despite encountering oxidative stress in host macrophages, C. glabrata is able to replicate intracellularly. Here, we show for the first time that CgYps1 aspartyl protease-mediated cleavage regulates the structural state, activity and functions of a flavodoxin-like protein CgPst2, which belongs to the family of highly conserved electron-carrier proteins that are pivotal to oxidative stress response in organisms ranging from bacteria to humans. We also show that CgPst2 is required for oxidative stress survival in vitro, and is sufficient to rescue, in vivo, the attenuated survival of a C. glabrata mutant lacking four flavodoxin-like proteins. Further, our data suggest that CgYps1 protease-mediated cleavage of CgPst2 is a dynamic event which may be coupled closely with intracellular quinone accumulation and detoxification. This strategy may aid C. glabrata establish successful infections in the human host. While advancing our understanding of fungal pathogenesis, our findings also provide insights into hitherto unknown regulatory mechanisms for flavodoxin-like fold-containing proteins.
Introduction
Opportunistic invasive mycoses pose a serious threat to global economy and human health [1]. Candida species are the most prevalent cause of life-threatening fungal bloodstream infections, that are associated with a mortality rate of up to 72% [2–5]. Although C. albicans is still the most frequently isolated Candida spp., the incidence of bloodstream infections due to non-albicans Candida spp. has increased substantially in last two decades [1,2,4,6]. C. glabrata is the second to fourth most common Candida bloodstream pathogen depending upon the geographical region, and accounts for up to 30% of Candida bloodstream infections [3,5–8]. C. glabrata differs from other prevalent pathogenic Candida species in several respects including its haploid genome, lack of secreted proteolytic activity and hyphae formation, and close phylogenetic relationship with the non-pathogenic yeast Saccharomyces cerevisiae [9–11].
A family of eleven putative glycosylphosphatidylinositol (GPI)-linked aspartyl proteases, commonly referred to as yapsins, plays an essential role in virulence of C. glabrata [12]. Other pathogenic traits of C. glabrata include the ability to adhere to biotic and abiotic surfaces probably owing to the presence of a large number of cell surface adhesins, survive high level of diverse stresses, proliferate in host macrophages and remodel chromatin and metabolic pathways in response to environmental cues [10,11]. Of eleven CgYPS1-11 genes encoding CgYapsins, CgYPS1 is essential for survival of acid stress, maintenance of pH and vacuole homeostasis, biofilm formation and survival in macrophage and mouse models [12–15]. CgYapsins are also required for suppression of the host innate immune response [15]. Intracellular proliferation of C. glabrata in macrophages is dependent upon its ability to limit activation of the NLRP3 inflammasome and Syk (spleen tyrosine kinase) signaling pathways [15]. The lack of CgYapsins resulted in higher activation of, and consequent increased production of the pro-inflammatory cytokine IL-1β in human macrophages, which led to intracellular killing of the Cgyps1-11Δ mutant [15]. Importantly, CgYps1 expression restored the intracellular survival and replication defect of the Cgyps1-11Δ mutant, pointing towards a pivotal role for CgYps1 in interaction with host immune cells [15]. However, despite the centrality of CgYapsins to C. glabrata physiology and virulence, their target proteins remain largely unidentified.
A characteristic trait of C. glabrata is the capacity to survive well under oxidative environmental conditions [11]. C. glabrata is known to exhibit about 3- and 10- fold higher resistance to hydrogen peroxide compared to C. albicans and S. cerevisiae, respectively [10,16]. Flavodoxins are highly conserved electron-carrier proteins which are implicated in oxidative stress response in bacteria and algae [17,18]. These proteins convert quinone to hydroquinone through two-electron reduction, thereby bypassing formation of the unstable and reactive semi-quinone species [17]. Flavodoxins possess a characteristic flavodoxin-like α-β-α fold with a twisted β-sheet, made of five parallel β-strands, surrounded by five α-helices, and require FMN (flavin mononucleotide) or FAD (flavin adenine dinucleotide) as cofactors for their redox activity [17,19,20]. Eukaryotes have a large number of proteins with flavodoxin-type fold domain [17,19,21]. S. cerevisiae and C. albicans contain three and four flavodoxin-like proteins (Fld-LPs), respectively, which are required for survival of the oxidative environment [22–25]. C. albicans Fld-LPs are also important for virulence in mice [25].
In the current study, we characterize, through phenotypic analysis, four Fld-LPs in C. glabrata, and show that CgPst2 is uniquely required for survival of menadione (MD) and benzoquinone (BQ) stress. Further, we identify, through immunoprecipitation and mass spectrometry analysis, 19 interactors, including CgPst2, of the CgYps1 protease, and demonstrate that CgPst2 undergoes CgYps1-dependent cleavage at the C-terminus. Arginine-174 in CgPst2 was found to be essential for cleavage, with C-terminally cleaved CgPst2 forming homo-oligomers and possessing higher activity. Altogether, our data unravel the hitherto unknown mechanism by which C. glabrata responds to, and, survive an oxidative environment by regulating the activity and structural state of a Fld-LP via proteolytic cleavage.
Results
CgPst2 is required for survival of MD and BQ stress
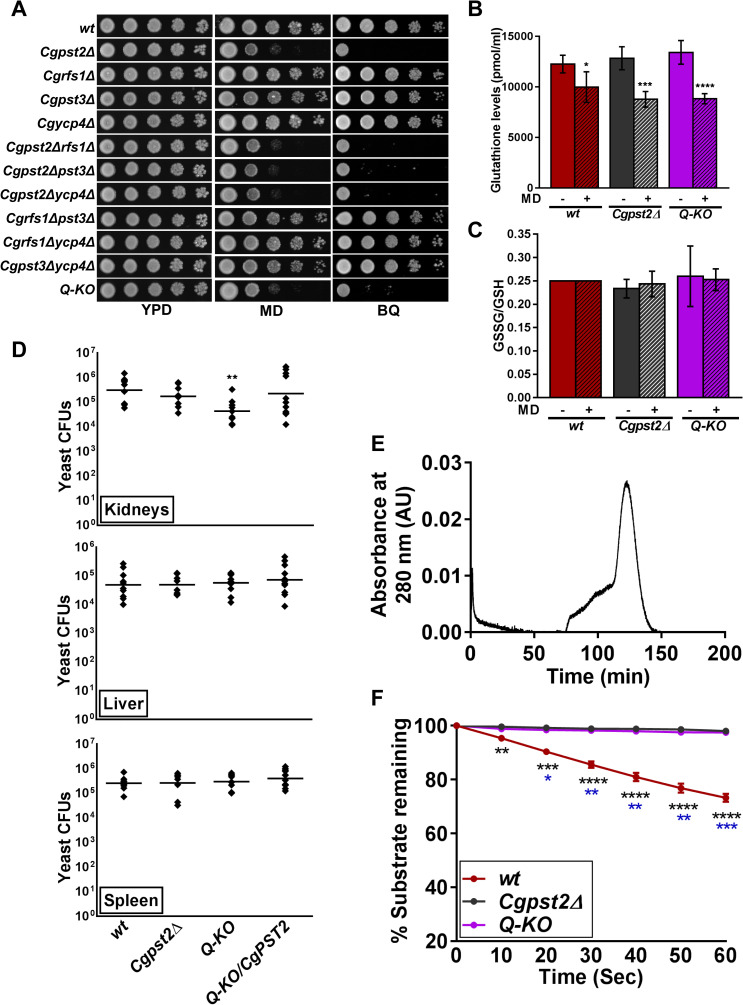
Towards deciphering the functions of Fld-LPs in C. glabrata, we first identified, using BLASTP analysis, orthologs of S. cerevisiae [22] and C. albicans [25] Fld-LPs, and found four C. glabrata proteins, CgPst2, CgRfs1, CgPst3 and CgYcp4, to exhibit varied levels of sequence similarity with one another, and with their S. cerevisiae and C. albicans counterparts (S1A Fig). CgPst2, CgRfs1, CgPst3 and CgYcp4 all possessed FMN-binding sites (S1B Fig). Notably, CgPST2 and CgRFS1 ORFs were most similar, sharing 75% identity at the nucleotide level. Next, we generated strains deleted for single, double or multiple Fld-LP genes and profiled the created mutants phenotypically. We found growth of the Cgpst2Δ mutant to be highly attenuated in the presence of MD and BQ (Fig 1A). Of note, sensitivity to other oxidative stressors, duroquinone, and cumene hydroperoxide, and thermal stress was not observed for any mutant (S2A Fig). Notably, all mutants, Cgpst2Δ single, Cgpst2Δrfs1Δ, Cgpst2Δpst3Δ and Cgpst2Δycp4Δ double, and the quadruple knockout lacking all four Fld-LPs (CgPst2, CgRfs1, CgPst3 and CgYcp4; referred as Q-KO from hereon), showed similar growth attenuation in MD and BQ-containing medium, indicating that of four Fld-LPs, CgPst2 is the sole regulator of MD and BQ stress survival (Fig 1A). Consistently, ectopic expression of CgPST2 complemented the quinone sensitivity of Cgpst2Δ and Q-KO strains (S2B Fig).
Fig 1. The flavodoxin-like protein CgPst2 plays an essential role in menadione and benzoquinone resistance.
A. Serial dilution spotting assay illustrating MD (95 μM) and BQ (4 mM) sensitivity of indicated C. glabrata mutants. The Q-KO strain lacks four flavodoxin-like proteins, CgPst2, CgRfs1, CgPst3 and CgYcp4. B. Total intracellular glutathione content of log-phase cells (10.0 OD600). Data (mean ± SEM; n = 4–5) represent total glutathione concentration normalized to one ml of cell lysate. *, p ≤ 0.05, ***, p ≤ 0.001, ****, p ≤ 0.0001; unpaired two-tailed Student’s t test. C. Ratio of oxidized (GSSG) to reduced (GSH) glutathione. 4-vinylpyridine was used to derivatize/block all free thiols in cell lysates, followed by GSSG estimation using DTNB [5,5-dithio-bis-(2-nitrobenzoic acid); Ellman’s Reagent]. Reduced GSH levels were calculated by subtracting GSSG levels from total glutathione levels. Data represent (mean ± SEM; n = 3–5). D. Survival of indicated C. glabrata strains in the murine model of systemic candidiasis. Six to eight-week-old female BALB/c mice were infected with cells (4X107 cells; 100 μl cell suspension) of overnight YPD/CAA-grown C. glabrata strains, wt, Cgpst2Δ, Q-KO and Q-KO expressing CgPST2, through tail vein injections. At 7th day post infection, mice were sacrificed, target organs (kidneys, liver and spleen) collected, homogenized in sterile PBS and appropriate homogenate dilutions were plated on YPD medium containing penicillin and streptomycin antibiotics. Total fungal burden in each mouse organ was determined, and plotted. Diamonds and bars indicate CFUs recovered from an individual mouse, and CFU geometric mean (n = 8–12), respectively, for each organ. **, p < 0.01; Mann-Whitney U-test. E. Size exclusion chromatogram of purified CgPst2 protein. 300 μg 6XHIS-FLAG-CgPst2 was applied to a Sephacryl S-200 column, and protein elution profiles were determined using the absorbance values at 280 nm. The peak corresponds to 29 kDa CgPst2 protein. AU, Arbitrary Units. F. NADH:quinone oxidoreductase activity in cell lysates (500 μg) of log-phase wt, Cgpst2Δ and Q-KO cells, as measured using menadione (500 μM) and NADH (500 μM) substrates. Absorbance of the substrate NADH was considered as 100 at 0 h time point, and NADH oxidation was deduced from the formula: [(absorbance at each time point/0 h absorbance) X 100]. Data represent mean ± SEM. Black and blue asterisks indicate statistically significant differences in activity between wt and Cgpst2Δ, and wt and Q-KO strains, respectively. *, p < 0.0332; **, p < 0.0021; ***, p < 0.0002; ****, p < 0.0001; Grouped multiple t-test (n = 3 to 4).
Menadione (2-methyl-l,4-naphthoquinone; vitamin K3), induces oxidative stress in cells, mainly due to superoxide radical and hydrogen peroxide production through the process of oxygen-dependent redox cycling [26]. Therefore, we next checked the effect of CgPst2 loss and MD treatment on intracellular levels of the antioxidant glutathione. We found similar levels of total glutathione, with unchanged GSSG/GSH ratio, in all three strains, wt, Cgpst2Δ and Q-KO (Fig 1B and 1C). Further, MD treatment also led to similar 20–35% reduction in total intracellular glutathione content in all strains, without any alteration in GSSG/GSH ratio (Fig 1B and 1C). These data suggest that neither CgFld-LP loss nor MD treatment had any appreciable impact on glutathione redox status.
To examine the role of Fld-LPs in pathogenesis of C. glabrata, we infected wt, Cgpst2Δ and four-Fld-LP-deficient (Q-KO) strains to BALB/c mice. We found Q-KO to exhibit attenuated survival, compared to wt cells, in kidneys of infected mice, which was rescued upon ectopic expression of CgPST2 (Fig 1D), indicating that CgPst2 expression is sufficient to rescue the mutant survival defect. Further, despite strong MD and BQ sensitivity in vitro, Cgpst2Δ mutant exhibited wt-like virulence in mice (Fig 1D), which could reflect functional redundancy among CgFld-LPs in vivo. Collectively, these data suggest that CgPst2 is required for survival of MD and BQ stress in vitro, and CgFld-LPs modulate organ-dependent survival in the murine systemic candidiasis model.
CgPst2 contributes to cellular NADH:quinone oxidoreductase activity
Similar to other Fld-LPs, CgPst2 is predicted to contain five-stranded parallel beta sheet surrounded by five alpha helices. WrbA, the founding member of Fld-LP family in Escherichia coli, and S. cerevisiae Pst2 are known to exist in a tetrameric form, and implicated in quinone detoxification [27–30]. Therefore, to determine if CgPst2 also forms a tetramer, we tagged CgPst2 with 6X-Histidine-FLAG epitope at the N-terminus, and purified the recombinant protein from E. coli (S3A Fig). Size exclusion chromatography (SEC) revealed CgPst2 to be a monomeric protein of 29 kDa (Fig 1E). The recombinant CgPst2 was non-functional, as it showed no NADH:quinone oxidoreductase activity in vitro (S3B Fig). Therefore, to examine if CgPst2 is a bonafide quinone-oxidoreductase, we checked the NADH:quinone oxidoreductase activity in cell extracts of wt and CgPST2-deleted strains. We monitored oxidation of NADH spectrophotometrically, and found 30% of total NADH to be oxidized within 1 min of incubation with wt cell extracts, while this oxidation was absent in Cgpst2Δ and Q-KO extract-incubated reaction mixtures (Fig 1F). Of note, the exogenous addition of FAD or FMN to the enzyme assay mixture had no effect on NADH:quinone oxidoreductase activity in wt and mutant cell extracts (S3C Fig), indicating that cell lysates have adequate amount of these cofactors. Ectopic expression of CgPST2 in Cgpst2Δ and Q-KO strains rescued the NADH oxidation defect (S3D Fig), suggesting that no NADH:quinone oxidoreductase activity in mutant cell extracts is due to the lack of CgPst2. Collectively, these data show that CgPst2 possesses NADH:quinone oxidoreductase activity, which may play a part in MD and BQ stress survival.
Immunoprecipitation (IP) and mass spectrometry (MS) analysis identified CgPst2 as a CgYps1 interactor
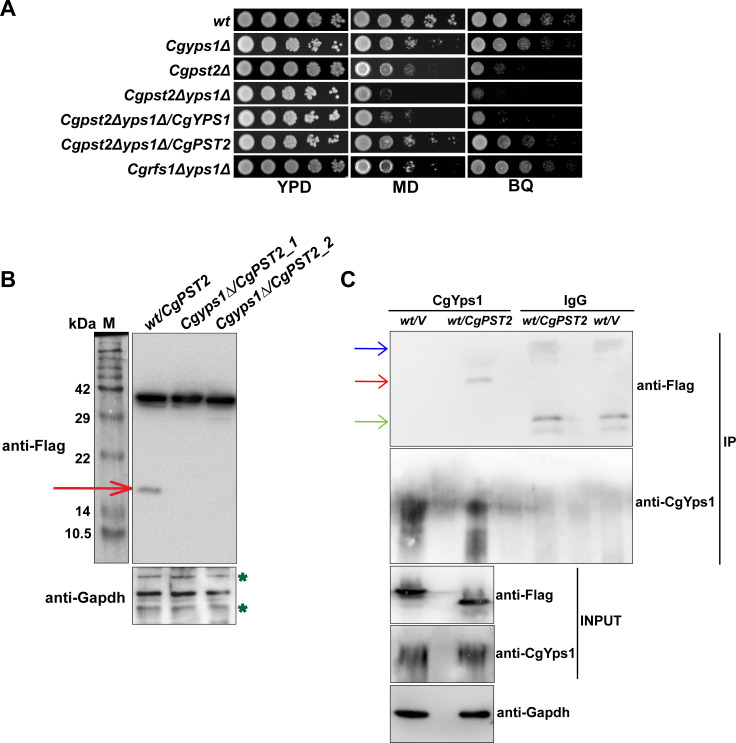
During the course of this study, we found that, compared to C. albicans and S. cerevisiae, CgPst2 was uniquely found in the secretome of C. glabrata [31]. Additionally, its secretion was enhanced in the Cgyps1-11Δ mutant, that lacks all eleven CgYapsins [31], indicating a role of CgYapsins in proper sorting/localization of CgPst2. Since CgYps1 has previously been implicated in MD tolerance [15], and the Cgyps1Δ mutant contained high reactive oxygen species (ROS) levels [14], we reasoned that CgPst2 and CgYps1 may act in conjunction during MD stress response. Thus, to decipher the relationship, if any, between CgPst2 and CgYps1, we performed three experiments. First, we generated a double mutant that lacked both CgPst2 and CgYps1. The Cgpst2Δyps1Δ mutant displayed enhanced sensitivity to MD, compared to single Cgpst2Δ and Cgyps1Δ mutants (Fig 2A). As a control, we also generated Cgrfs1Δyps1Δ mutant, and found its MD sensitivity to be similar to that of Cgyps1Δ mutant (Fig 2A). Further, ectopic expression of CgYPS1 or CgPST2 restored MD sensitivity slightly and substantially, respectively, of the Cgpst2Δyps1Δ mutant (Fig 2A). Expectedly, the Cgpst2Δyps1Δ mutant showed no NADH:quinone oxidoreductase activity (S4 Fig). Altogether, these results highlight a major and minor role for CgPst2 and CgYps1, respectively, in MD stress survival.
Fig 2. CgPst2 interacts with CgYps1.
A. Serial dilution spotting assay illustrating MD (95 μM) and BQ (4 mM) sensitivity of indicated CgYPS1-deleted strains. B. Immunoblot analysis of CgPst2-SFB in cell extracts (60 μg) of wt and Cgyps1Δ transformants expressing CgPST2-SFB. The red arrow marks the small (~ 16 kDa) cleaved fragment of CgPst2. Cgyps1Δ/CgPST2_1 and Cgyps1Δ/CgPST2_2 represent two independent Cgyps1Δ transformants. M, Protein Marker. C. Immunoblot analysis of CgPst2-CgYps1 interaction. Lysates (3.0 mg) of wt cells expressing either SFB-GFP or CgPst2-SFB were immunoprecipitated with anti-CgYps1 or anti-mouse immunoglobulin G (IgG) antibody, resolved on 4–20% polyacrylamide gradient, 12% polyacrylamide and 12% polyacrylamide gels for CgYps1, CgPst2-SFB and CgGapdh, respectively, and were probed with anti-CgYps1, anti-Flag and anti-Gapdh antibodies. CgGapdh was used as loading control. Please note that for Input samples, 200, 75 and 75 μg protein was loaded for detection of CgYps1, CgPst2-SFB and CgGapdh proteins, respectively. Red, blue and green arrows mark the CgPst2-SFB, Ig heavy chain and Ig light chain, respectively. ‘V’ refers to the vector expressing SFB-GFP protein.
Second, we generated a fusion protein by tagging CgPst2 with GFP at the C-terminus. Confocal imaging revealed the fusion protein to localize at the plasma membrane in both wt and Cgyps1Δ, and MD treatment did not alter CgPst2 localization in either strain (S5 Fig). These results preclude any role of CgYps1 in cellular localization of CgPst2.
Third, we identified interactors/probable substrates of CgYps1 protease through IP-MS analysis. For this, we immunoprecipitated CgYps1, using anti-CgYps1 polyclonal antibody [31], from log-phase wt and Cgyps1-11Δ cells, in duplicates, and sent to Taplin Mass Spectrometry Facility for protein identification. By employing three criteria, (a minimum of two unique peptides, presence in both wt replicate samples, and absence in both Cgyps1-11Δ samples), we identified 19 proteins, including CgPst2, that interacted with CgYps1 (Table 1). The functional classification and predicted localization of CgYps1 interactors is listed in Table 1. Gene Ontology (GO) analysis revealed CgYps1 interactors to primarily belong to carbohydrate metabolism, response to chemical, translation, transport and anaerobic respiration processes (S1 Table), underscoring the role of CgYps1 in regulation of diverse cellular processes. Of note, this is the first report of a fungal yapsin interacting with metabolic proteins. However, since IP-MS-based analysis can produce false positives, further validation is required to establish these highly abundant cellular proteins as bonafide CgYps1 interactors.
Table 1. List of CgYps1 interacting proteins, as identified by immunoprecipitation-mass spectrometry analysis.
| Sl. No | Interactor-encoding ORF (Systematic ID) | S. cerevisiae ortholog | Number of unique peptides (in two replicate samples) | Predicted functions¶ | Protein size (kDa) | Predicted localization# | |
|---|---|---|---|---|---|---|---|
| Oxidoreduction process and energy homeostasis | |||||||
| 1 | CAGL0A00495g | PMA1 | 16 | 2 | Plasma membrane P2-type H+-ATPase; a major regulator of cytoplasmic pH and plasma membrane potential. | 98.3 | Endoplasmic reticulum, Membrane type |
| 2 | CAGL0J00451g | TDH3 | 6 | 5 | Putative glyceraldehyde-3-phosphate dehydrogenase. | 35.9 | Cytoplasm |
| 3 | CAGL0D01298g | TKL1 | 4 | 7 | Transketolase; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway. | 73.7 | Peroxisome |
| 4 | CAGL0M09581g | ATP1 | 5 | 5 | Alpha subunit of the F1 sector of mitochondrial F1F0 ATP synthase. | 58.5 | Mitochondrion |
| 5 | CAGL0F01947g | LPD1 | 3 | 5 | Dihydrolipoamide dehydrogenase; the lipoamide dehydrogenase component (E3) of the pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase multi-enzyme complexes. | 53.1 | Mitochondrion |
| 6 | CAGL0F04213g | PET9 | 2 | 5 | Major ADP/ATP carrier of the mitochondrial inner membrane; exchanges cytosolic ADP for mitochondrially synthesized ATP. | 32.8 | Mitochondrion, Membrane type |
| 7 | CAGL0H08327g | TPI1 | 4 | 2 | Triose phosphate isomerase, abundant glycolytic enzyme. | 26.9 | Cytoplasm |
| 8 | CAGL0C02343g | ARB1 | 3 | 3 | ATPase of the ATP-binding cassette (ABC) family. | 81.1 | Plastid |
| 9 | CAGL0M13343g | GND1 | 3 | 3 | 6-phosphogluconate dehydrogenase; catalyzes an NADPH regenerating reaction in the pentose phosphate pathway. | 53.5 | Cytoplasm |
| 10 | CAGL0K11858g | PST2 | 2 | 2 | Protein with similarity to a family of flavodoxin-like proteins; induced by oxidative stress. | 21.0 | Cytoplasm |
| 11 | CAGL0I03916g | ARF2 | 2 | 2 | Putative ADP-ribosylation factor; involved in invasive growth. | 20.6 | Golgi apparatus, Membrane type |
| Ribosome biogenesis | |||||||
| 12 | CAGL0J07238g | RPS12 | 3 | 2 | Protein component of the small (40S) ribosomal subunit. | 15.5 | Cytoplasm |
| 13 | CAGL0K11748g | RPS11A | 2 | 2 | Protein component of the small (40S) ribosomal subunit. | 17.9 | Cytoplasm |
| 14 | CAGL0L04840g | RPS23 | 2 | 2 | Putative ribosomal protein. | 16.0 | Cytoplasm |
| 15 | CAGL0A00979g | RPL38 | 2 | 2 | 60S ribosomal ribosomal protein subunit. | 8.9 | Cytoplasm |
| Other functions | |||||||
| 16 | CAGL0I04356g | TIF1 | 3 | 2 | Translation initiation factor eIF4A. | 44.8 | Nucleus |
| 17 | CAGL0C04389g | HTB2 | 3 | 2 | Histone H2B; core histone protein required for chromatin assembly | 14.0 | Nucleus |
| 18 | CAGL0H02057g | GAR1 | 2 | 3 | Protein component of the H/ACA snoRNP pseudouridylase complex; modify 18S pre-rRNA. | 19.5 | Nucleus |
| 19 | CAGL0D01034g | PSA1 | 2 | 2 | GDP-mannose pyrophosphorylase involved in the synthesis of GDP-mannose for protein glycosylation | 39.4 | Cytoplasm |
¶These functions are predicted based on functions of the corresponding S. cerevisiae orthologs (www.yeastgenome.org).
#Subcellular localization of proteins was determined using DeepLoc server (http://www.cbs.dtu.dk/services/DeepLoc/).
CgYps1 cleaves CgPst2 at the C-terminus
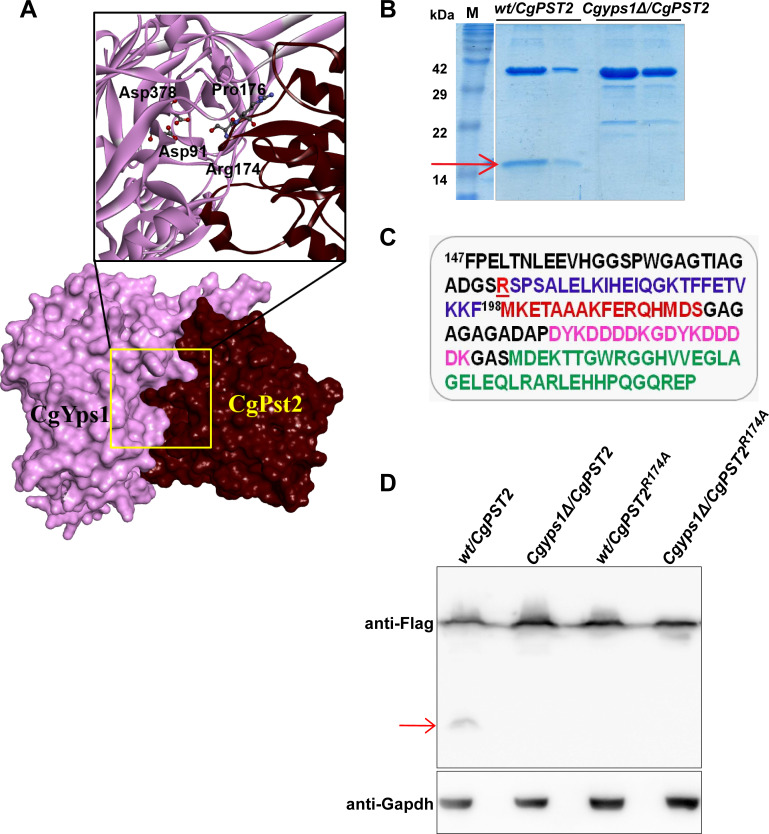
Of 19 identified interactors, one protein was CgPst2. To characterize CgPst2-CgYps1 interaction, we tagged CgPst2 with SFB epitope at the C-terminus. Immunoblot analysis revealed full length CgPst2-SFB protein (~ 40 kDa) in both wt and Cgyps1Δ strains, however, a shorter fragment of ~ 16 kDa was observed uniquely in wt cells (Fig 2B). Its absence in Cgyps1Δ mutant indicates that CgYps1 may be required for CgPst2 cleavage at the C-terminus. We also validated CgPst2-CgYps1 interaction through immunoblot (Fig 2C) and affinity purification analysis (S6 Fig). Of note, we could not check the processing of CgPst2-GFP, as multiple bands were detected by anti-GFP antibody in the Western analysis.
Next, we performed molecular docking analysis to predict the potential CgYps1-cleavage sites on CgPst2. For this, CgPst2 and CgYps1 sequences were retrieved from the Candida genome database, and submitted to the online tool I-TASSER [32]. The models with best Confidence (C)-score were selected and analyzed by Z-DOCK [33]. This structural modelling analysis predicted Aspartate-91 and Aspartate-378 of CgYps1 to interact with Proline-176 and Arginine-174 residues, present at the C-terminus of CgPst2, respectively (Fig 3A). The predicted arginine-174 residue lies in a highly conserved DGSR sequence in CgPst2, that CgPst2 shares with several Fld-LPs (S7 Fig). Importantly, D91 and D378, predicted catalytic residues of CgYps1, are required for its functions in cell wall stress [15]. Since Yapsins are known to cleave C-terminal to monobasic or dibasic residues, with S. cerevisiae Yps1 also reported to cleave at the monobasic residue [34,35], we hypothesized that CgYps1 may cleave CgPst2 at/after R174 residue. To test this, we extracted the ~16 kDa cleaved fragment of CgPst2-SFB (Fig 3B) and analyzed by peptide mass fingerprinting. We identified a peptide corresponding to the terminal sequence of CgPst2, 175SPSALELKIHEIQGKTFFETVK196 (Fig 3C), along with SFB epitope peptides (Fig 3C). Further, mutation of R174 in CgPst2 to alanine-174 (CgPst2R174A) led to disappearance of the shorter CgPst2-SFB fragment in CgPst2R174A-SFB-expressing wt (Fig 3D) and Cgpst2Δ (S8 Fig) samples, suggesting that R174 in CgPst2 is required for its cleavage at C-terminus. Of note, since we were unable to obtain good expression of N-terminally-SFB-tagged CgPst2 protein in C. glabrata, we could not use SFB-CgPst2 for cleavage analysis. Altogether, these data point towards CgPst2 being a bonafide substrate of CgYps1.
Fig 3. CgPst2 is cleaved at the C-terminus.
A. Molecular surface and Ribbon models depicting CgYps1 (Length: 601 aa; Predicted MW: 64 kDa) with CgPst2 (Length: 198 aa; Predicted MW: 21 kDa). D91 and D378 in CgYps1 represent catalytically active aspartic acid residues. The 171DGSRSPSA178 region in CgPst2 contains predicted CgYps1-binding residues, Arginine-174 and Proline-176. Hydrogen bond-forming amino acid residues are highlighted in bold letters in the predicted interaction region. The molecular surfaces of receptor (CgYps1) and substrate (CgPst2) are coloured purple and maroon, respectively. B. Coomassie-blue-stained 18% SDS-PAGE gel images indicating expression of the full-length (~ 40 kDa) and the C-terminal cleaved fragment (~ 16 kDa; marked with a red arrow) of CgPst2-SFB, after two-step affinity purification of cell extracts of wt and Cgyps1Δ strains. C. The C-terminus amino acid sequence of CgPst2-SFB protein starting with the phenylalanine (F) residue at 147th position of CgPst2 protein. CgPst2 is 198 amino acid long. The predicted cleavage residue R174 is underlined and indicated in red. The SFB tag (85 aa) consists of S-protein (MKETAAAKFERQHMDS; brown), two copies of the FLAG tag (DYKDDDDK; pink) and streptavidin-binding-peptide (MDEKTTGWRGGHVVEGLAGELEQLRARLEHHPQGQREP; green) sequences. MS analysis of the shorter CgPst2-SFB fragment identified peptides corresponding to SFB and CgPst2 sequences. D. Immunoblot analysis of wt cell extracts expressing either wild-type CgPst2-SFB or alanine-substituted-CgPst2-SFB (CgPST2R174A), using anti-Flag antibody. The red arrow marks the small cleaved fragment of wild-type CgPst2. CgGapdh was used as a loading control.
MD treatment increases NADH:quinone oxidoreductase activity
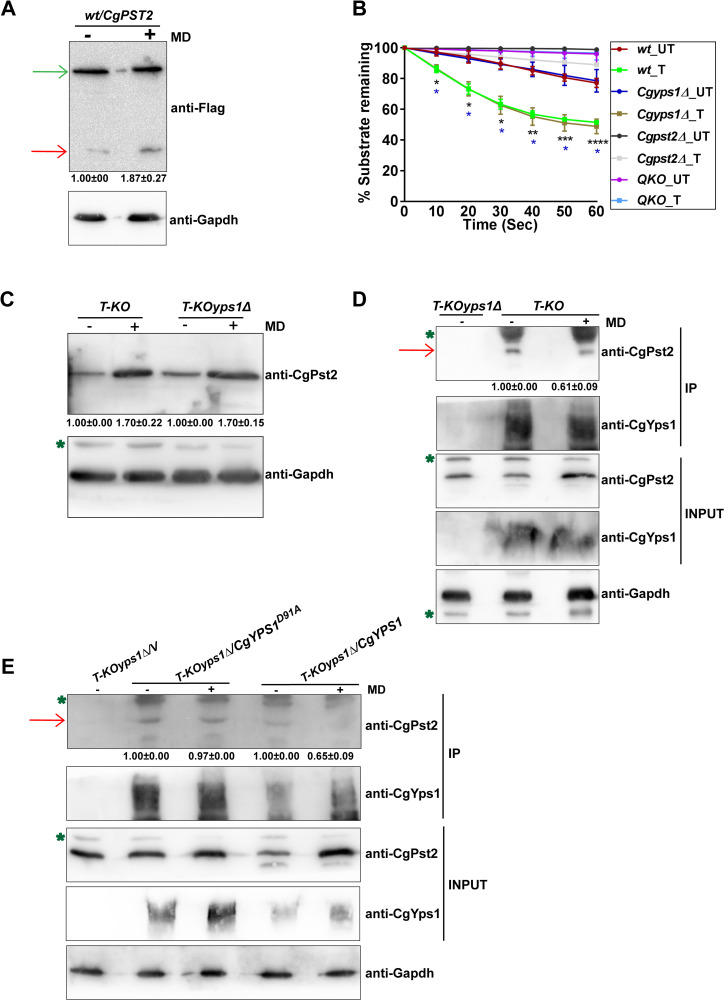
Since CgPst2 is pivotal to MD detoxification (Fig 1A), we next checked the effect of MD treatment on CgYps1-mediated processing of CgPst2. For this, we performed four experiments. First, we measured CgPst2 cleavage, and found about 1.9-fold increase in the levels of the C-terminal cleaved fragment of CgPst2 in MD-treated wt cells, compared to untreated cells (Fig 4A). Second, we determined the effect of MD-induced increase in cleavage of CgPst2 on CgPst2 activity. We found that the NADH:quinone oxidoreductase activity of CgPst2 was also higher upon MD treatment (Fig 4B). These data indicate that MD treatment enhances the C-terminal processing of CgPst2 and stimulates its activity. Intriguingly, Cgyps1Δ cells also responded to MD treatment by elevating NADH:quinone oxidoreductase activity (Fig 4B), indicating that CgPst2 is functional in this mutant, consistent with the mild MD sensitivity of the Cgyps1Δ mutant (Fig 2A). This result also suggests that CgYps1 is unlikely to be the sole mediator of MD-induced increase in CgPst2 activity, and CgPst2 functions.
Fig 4. The cleavage and NADH:quinone oxidoreductase activity of CgPst2 is increased upon menadione treatment.
A. Immunoblot analysis of CgPst2 expression, using anti-Flag antibody, in cell extracts of untreated or menadione-(MD; 90 μM for 90 min)-treated log-phase cells of the wt strain expressing CgPst2-SFB, using anti-Flag antibody. CgGapdh was used as a loading control. The intensity of the shorter 16 kDa band in 4 independent Western blots was quantified using the ImageJ densitometry software, and this signal was normalized to the corresponding CgGapdh-normalized total CgPst2 signal, as total CgPst2 levels were also elevated, upon MD treatment, compared to CgGapdh signal. Data (mean ± SEM) represent the fold-change in levels of the C-terminal cleaved fragment of CgPst2-SFB in treated cells, compared to untreated cells (considered as 1.0), and are shown underneath the blot. p ≤ 0.05; paired two-tailed Student’s t test. The red and green arrows indicate the C-terminal cleaved fragment of CgPst2 and full length CgPst2-SFB, respectively. B. NADH:quinone oxidoreductase activity in log-phase cultures of indicated C. glabrata strains that were either treated with 90 μM menadione (T) for 90 min, or left untreated (UT), as measured using menadione (500 μM) and NADH (500 μM) substrates. Absorbance of the substrate NADH was considered as 100 at 0 h time point, and NADH oxidation was deduced from the formula: [(absorbance at each time point/0 h absorbance) X 100]. Black and blue asterisks indicate statistically significant differences in activity of treated wt and Cgyps1Δ samples, respectively, compared to respective untreated lysates. The Q-KO strain lacks four flavodoxin-like proteins, CgPst2, CgRfs1, CgPst3 and CgYcp4. *, p < 0.0332; **, p < 0.0021; ***, p < 0.0002; ****, p < 0.0001; Grouped multiple t-test (n = 3 to 5). C. Immunoblot analysis of CgPst2 levels in T-KO (Cgrfs1Δpst3Δycp4Δ) and T-KOyps1Δ (Cgrfs1Δpst3Δycp4Δyps1Δ) cells, using anti-CgPst2 antibody. The intensity of individual bands in 4 independent Western blots was quantified using ImageJ densitometry software, and CgPst2 signal was normalized to the corresponding CgGapdh signal. Fold-change (mean ± SEM) in CgPst2 levels in treated cells, compared to untreated cells (considered as 1.0), is shown underneath the blot. p ≤ 0.05; paired two-tailed Student’s t test. The green asterisk indicates non-specific band. D-E. Immunoblot analysis showing CgYps1-CgPst2 (D) and CgYps1-CgYps1D91A-CgPst2 (E) interaction upon MD treatment (90 μM for 90 min) in T-KO (Cgrfs1Δpst3Δycp4Δ) and T-KOyps1Δ (Cgrfs1Δpst3Δycp4Δyps1Δ) cells, using anti-CgPst2 antibody. 6 mg precleared cell lysates were incubated with anti-CgYps1 antibody-conjugated beads for 12 h at 4°C. After bead washing, bead-bound proteins were boiled in 2X-SDS loading buffer and resolved on 4–20% polyacrylamide gradient and 12% polyacrylamide gels for CgYps1 and CgPst2 samples, respectively. For input samples, 60, 60 and 200 μg protein were loaded to detect CgPst2, CgGapdh and CgYps1 proteins, respectively. Two to three independent Western blots were quantified using the ImageJ densitometry software, and CgPst2 signal was normalized to the corresponding CgGapdh signal. Fold-change in CgPst2 levels in treated cells, compared to untreated cells (considered as 1.0), is shown underneath the blot. p ≤ 0.05; paired two-tailed Student’s t test. The red arrow marks CgPst2 band, while the green asterisk denotes non-specific band.
Third, to check if MD treatment has any effect on CgPST2 transcript and protein levels, we first generated antibody against E. coli-purified CgPst2 protein, and checked its specificity. This antibody recognized both CgPst2 and CgRfs1, however, no signal was seen in Q-KO cells lacking all four Fld-LPs (S9A Fig). Next, we generated Cgrfs1Δpst3Δycp4Δ (T-KO from hereon) and Cgrfs1Δpst3Δycp4Δyps1Δ (T-KOyps1Δ from hereon) strains to specifically amplify the CgPST2 gene, as CgPST2-amplification-specific primers showed cross-reactivity, probably, owing to high sequence similarity among CgFld-LP genes. Measurement of endogenous CgPST2 transcript and protein amounts revealed no change in transcript (S9B Fig) and about 1.7-fold higher protein levels (Fig 4C) of CgPst2 in MD-treated T-KO and T-KOyps1Δ cells, respectively, compared to corresponding untreated cells. Of note, we could not verify CgYps1-mediated CgPst2 cleavage with anti-CgPst2 antibody due to two reasons. First, anti-CgPst2 antibody did not recognize the 3 kDa C-terminal cleaved fragment of CgPst2. Second, the size difference between the cleaved N-terminal CgPst2 fragment and the full-length CgPst2, probably owing to other posttranslational modifications, was not conspicuous on SDS-PAGE analysis of cell extracts. However, whether CgPst2 undergoes any posttranslational modification is yet to be determined. In this context, it is noteworthy that Ycp4 in S. cerevisiae [36] and Pst2 in C. albicans [37] are known to undergo palmitoylation and ubiquitination, respectively. Alternatively, it is also possible that CgPst2 cleavage occurs at a low rate, and the cleaved N-terminal CgPst2 fragment represents a minor species of CgPst2 under in vitro conditions, thereby rendering detection very difficult. Altogether, these data suggest that MD treatment elevates CgPst2 protein levels without affecting its transcriptional regulation.
Fourth, we checked the effect of MD treatment on CgYps1-CgPst2 interaction. We observed that despite increased levels of CgPst2 (Fig 4C) and CgYps1 (S9C Fig) proteins upon MD treatment, the interaction between CgPst2-CgYps1 was less, as anti-CgYps1 antibody pulled down 40% lower amount of CgPst2 in MD-treated cells, compared to untreated cells (Fig 4D). Consistently, Q-KO cells expressing CgPST2 ectopically showed weaker CgYps1-CgPst2 interaction upon MD treatment (S9D Fig). This diminished CgYps1-CgPst2 interaction in MD-treated cells could be due to enhanced cleavage of CgPst2 by CgYps1, and the consequent release of CgPst2 from CgYps1. We reasoned, if this is true, the interaction of catalytically-dead CgYps1 with CgPst2, should not change upon MD treatment, as the catalytically inactive CgYps1 will not be able to process CgPst2, thereby continuing to remain associated with CgPst2. To test this notion, we checked the interaction of CgYps1D91A [one catalytic aspartate (D91) substituted with alanine] [15] with CgPst2 through co-immunoprecipitation analysis. We found that MD treatment had no significant effect on CgYps1D91A-CgPst2 interaction, as it was similar between untreated and MD-treated C. glabrata cells (Fig 4E), indicating that the release of CgPst2 from CgYps1, upon MD treatment, requires CgYps1’s proteolytic activity. Of note, to demonstrate that CgYps1D91A is catalytically inactive, we purified CgYps1 and CgYps1D91A from Pichia pastoris (S10A Fig), and checked their proteolytic activity through gelatin hydrolysis (S10B Fig) and hemoglobin digestion (S10C Fig) assays. We found CgYps1D91A to be enzymatically inactive (S10B and S10C Fig).
Altogether, these results suggest that the loss of CgYps1-CgPst2 interaction upon MD treatment is likely to be due to enhanced CgYps1-mediated cleavage of CgPst2 and subsequent release of CgPst2. These data also reinforce that MD treatment enhances CgYps1-dependent cleavage of CgPst2, which may stimulate CgPst2 activity by removing the C-terminal region. Thus, we hypothesized that the C-terminally truncated CgPst2 will be more active than CgPst2.
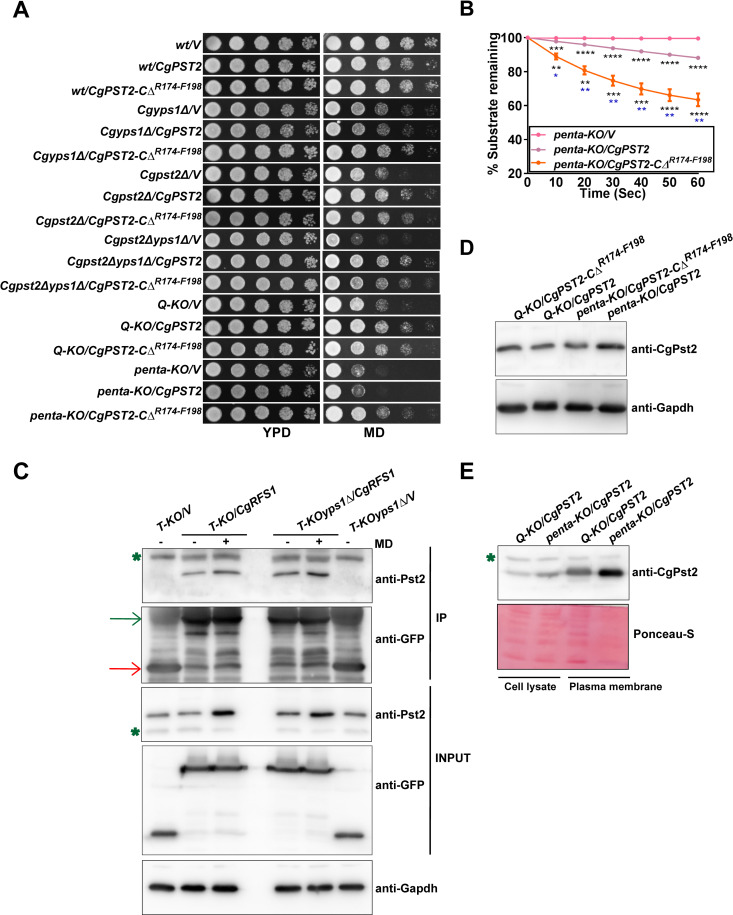
CgPst2-CΔR174-F198 complements MD sensitivity of CgPST2-deleted strains
CgPst2 is a 198 aa long protein. To test our hypothesis, we generated C-terminally truncated CgPst2 (CgPst2-CΔR174-F198), that lacked last 25 amino acids from R174 onwards, and performed two experiments: Activity determination of CgPst2 and CgPst2-CΔR174-F198 in cell lysates of mutants lacking or containing CgYps1, and Complementation analysis of MD sensitivity of different mutants. We found CgPst2-CΔR174-F198 to rescue MD sensitivity of all CgPST2-deleted strains, Cgpst2Δ, Cgpst2Δyps1Δ, Q-KO and Q-KOyps1Δ (referred as penta-KO from hereon) (Fig 5A), suggesting that CgPst2-CΔR174-F198 is functionally active. Surprisingly, we did not observe good rescue of MD sensitivity of penta-KO cells that were expressing CgPST2 (Fig 5A). Further, consistent with our hypothesis and MD growth phenotypes, we found a much faster rate of NADH oxidation in cell extracts of penta-KO expressing CgPST2-CΔR174-F198, compared to CgPST2-expressing penta-KO strain (Fig 5B), thereby suggesting that CgPst2-CΔR174-F198 is more active than CgPst2.
Fig 5. CgPst2-CΔR174-F198 possesses higher activity than CgPst2 in penta-KO.
A. Serial dilution spotting assay showing CgPST2-CΔR174-F198-mediated rescue of MD (80 μM) sensitivity. V, pRK74 vector. The Q-KO strain lacks four flavodoxin-like proteins (Fld-LPs), CgPst2, CgRfs1, CgPst3 and CgYcp4, while the penta-KO strain lacks CgYps1 along with four Fld-LPs, CgPst2, CgRfs1, CgPst3 and CgYcp4. B. NADH:quinone oxidoreductase activity in cell extracts of penta-KO expressing CgPST2 or CgPST2-CΔR174-F198. Data represent mean ± SEM (n = 3). Black and blue asterisks represent statistically significant activity differences in indicated strains compared to penta-KO/V and penta-KO/CgPST2, respectively. *, p < 0.0332; **, p < 0.0021; ***, p < 0.0002; ****, p < 0.0001; Grouped multiple t-test. V, pRK74 vector. C. Immunoblot analysis showing interaction between CgPst2 and CgRfs1-GFP. 3 mg precleared lysates of untreated and MD (90 μM for 90 min)-treated T-KO (Cgrfs1Δpst3Δycp4Δ) and T-KOyps1Δ (Cgrfs1Δpst3Δycp4Δyps1Δ) cells expressing CgRfs1-GFP were incubated with anti-GFP antibody-conjugated beads, followed by Western analysis. The red and green arrows mark GFP and CgRfs1-GFP protein bands, respectively, in IP samples, while the green asterisk denotes non-specific band. D. Immunoblot analysis of CgPst2 levels in Q-KO and penta-KO strains expressing CgPST2 or CgPST2-CΔR174-F198. E. Immunoblot analysis showing enrichment of CgPst2 in plasma membrane fractions of the Q-KO and penta-KO strains expressing CgPST2. Whole-cell lysates and plasma membrane fractions (60 μg), prepared by glass bead lysis and sucrose gradient ultracentrifugation, respectively, were resolved on 12% polyacrylamide gel, and probed with anti-CgPst2 antibody. The ponceau S-stained membrane is shown as loading control. The green asterisk denotes non-specific band.
Notably, CgPst2-mediated reversal of MD sensitivity of Cgpst2Δyps1Δ mutant but not of penta KO strain (Fig 5A) implicate other Fld-LPs (CgRfs1, CgPst3 and CgYcp4) in the regulation of CgPst2 activity. Therefore, to check if CgPst2 associates with other Fld-LPs, we tagged CgRfs1, CgPst3 and CgYcp4 proteins with the GFP epitope at their C-termini, and expressed in the T-KO strain which contains only CgPst2. Immunoprecipitation analysis revealed CgPst2 to interact with CgRfs1 under normal and MD treatment conditions in both T-KO and T-KOyps1Δ cells (Fig 5C). Notably, we could not detect CgPst2 and CgYcp4 interaction (S11 Fig), which could either be due to low CgYcp4 protein expression (S11 Fig), or CgPst2-CgYcp4 interaction may require the presence of CgRfs1. Despite multiple efforts, we could not check CgPst2-CgPst3 interaction, as we were unable to obtain good expression of CgPst3-GFP.
Further, the reduced functioning of CgPst2 in penta-KO cells raised three possibilities. First, CgPst2, compared to CgPst2-CΔR174-F198, is not expressed well in penta-KO cells. Second, CgPst2 is mislocalized in penta-KO. Third, CgYps1-mediated processing of CgPst2/CgYps1-CgPst2 interaction is important for CgPst2 functions in MD stress survival. To determine which of these possibilities accounts for the reduced functions of CgPst2 in the penta-KO strain, we first checked CgPst2 expression in Q-KO and penta-KO cells expressing CgPST2 or CgPST2-CΔR174-F198. We observed good expression of CgPst2 and CgPst2-CΔR174-F198 in both Q-KO and penta-KO (Fig 5D). In fact, the amount of CgPst2 was higher in penta-KO cells, compared to Q-KO cells (Fig 5D), indicating that MD sensitivity of the penta-KO/CgPST2 strain is not due to diminished CgPst2 protein levels.
Next, we checked the localization of CgPst2 by subcellular fractionation analysis in CgPST2-expressing Q-KO and penta-KO strains. For this, we collected plasma membrane (PM) fractions via sucrose density gradient ultracentrifugation of cell extracts of Q-KO and penta-KO strains expressing CgPST2. The purity of the obtained PM fraction was evaluated by immunoblot analysis using anti-CgPma1 antibody (S12 Fig), which detects the plasma membrane proton pump CgPma1 [13]. Notably, the signal for cytosolic CgGapdh was also largely absent in the PM fraction (S12 Fig). Anti-CgPst2 Western analysis revealed CgPst2 to be enriched in the PM fraction of both strains, with the penta-KO/CgPST2 strain also showing higher amounts of CgPst2 in cell lysates and PM samples, compared to that of the Q-KO/CgPST2 strain (Fig 5E). These data indicate that the localization of CgPst2 to the plasma membrane is not dependent upon CgYps1, and the reduced functions of CgPst2 in penta-KO/CgPST2 strain is unlikely to be owing to CgPst2 mislocalization.
Further, the enrichment of CgPst2 in the PM fraction prompted us to examine the presence of CgYps1 in this fraction, with the rationale that CgPst2 and CgYps1 interaction may occur at the plasma membrane. Notably, we detected a good signal with anti-CgYps1 antibody in the PM sample of the Q-KO/CgPST2 strain, which was absent in the penta-KO strain (S12 Fig), thereby localizing CgYps1 to the plasma membrane. Given that both CgYps1 and CgPst2 were enriched in the PM fraction (S12 Fig), the CgYps1-mediated processing of CgPst2 is likely to take place at the plasma membrane. Altogether, these data also implicate CgYps1 in regulation of CgPst2 protein levels. Moreover, the higher activity of CgPst2-CΔR174-F198 than CgPst2 in the penta-KO strain (Fig 5B) suggests that CgYps1-dependent cleavage may relieve the inhibitory effect of the C-terminal region on CgPst2 activity.
Overall, we draw six major conclusions from these data. First, CgPst2 interacts with at least one other Fld-LP, CgRfs1. Second, CgRfs1, CgPst3 and CgYcp4 are important for CgPst2 functions in MD resistance, as MD treatment could stimulate CgPst2 activity in the Cgyps1Δ mutant, despite the lack of CgPst2 activation owing to CgYps1-mediated processing. This increase in CgPst2 activity in Cgyps1Δ mutant may arise from association of CgPst2 with other flavodoxin-like proteins. Third, CgYps1-mediated processing appears to be essential for CgPst2 functions in MD detoxification only in the absence of other Fld-LPs, underscoring that CgYps1-mediated cleavage of CgPst2 may reflect one of many mechanisms controlling CgPst2 activity. Fourth, both CgYps1 and CgPst2 localize to the plasma membrane, and the plasma membrane localization of CgPst2 is not dependent on CgYps1. Fifth, despite the presence of a functional CgPst2, MD sensitivity of the Cgyps1Δ mutant points to a dual role for CgYps1 in MD resistance: CgPst2-dependent, and a yet to be identified CgPst2-independent function. Finally, since the C-terminal truncation resulted in an active CgPst2 enzyme, CgYps1-mediated cleavage of CgPst2 is likely to be a regulatory event that modulates CgPst2 activity in response to internal and external cues.
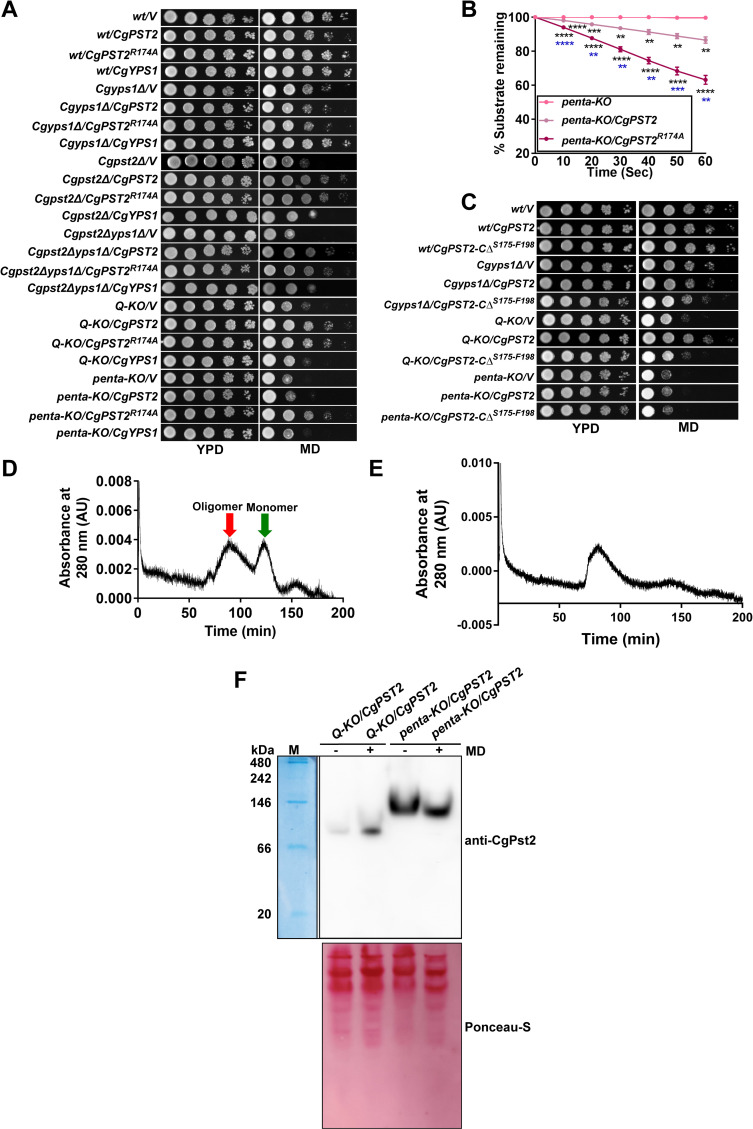
CgPst2R174A is proficient in MD detoxification
Arg-174 is essential for CgPst2 cleavage (Fig 3D). Therefore, to determine the functional role of R174 residue in regulating CgPst2 activity, we expressed CgPst2 carrying alanine substitution of the R174 residue (CgPst2R174A), and found it to complement MD sensitivity of Cgpst2Δ, Cgpst2Δyps1Δ, Q-KO and penta-KO strains (Fig 6A). Similar to CgPst2-CΔR174-F198, CgPst2R174A also possessed higher NADH:quinone oxidoreductase activity than CgPst2 in penta KO (Fig 6B). These results were unexpected, as CgPst2R174A is not cleaved by CgYps1, and thus, should retain the C-terminal region. Therefore, to check whether CgPst2R174A is hyperactive or expressed at a higher level, we performed Western analysis and assessed the protein amounts in extracts of the Q-KO and penta-KO cells expressing CgPST2 or CgPST2R174A. We found similar amounts of CgPst2R174A in both Q-KO and penta-KO strains (S13A Fig). Notably, CgPst2, which was unable to rescue MD sensitivity of the penta-KO strain (Fig 5A), showed 6-fold higher abundance than CgPst2R174A (S13A Fig), indicating that the better functioning of CgPst2R174A in penta-KO cells is not due to high protein levels.
Fig 6. CgPst2R174A rescues MD sensitivity of CgPST2-deleted strains.
A. Serial dilution spotting assay showing CgPST2R174A-mediated rescue of MD (80 μM) sensitivity. V, pRK74 vector. The Q-KO strain lacks four flavodoxin-like proteins (Fld-LPs), CgPst2, CgRfs1, CgPst3 and CgYcp4, while the penta-KO strain lacks CgYps1 along with four Fld-LPs, CgPst2, CgRfs1, CgPst3 and CgYcp4. B. NADH:quinone oxidoreductase activity in cell extracts of penta-KO expressing CgPST2 or CgPST2R174A. Data represent mean ± SEM (n = 3). Black and blue asterisks represent statistically significant activity differences in indicated strains compared to penta-KO and penta-KO/CgPST2, respectively. **, p < 0.0021; ***, p < 0.0002; ****, p < 0.0001; Grouped multiple t-test. C. Serial dilution spotting assay showing MD (80 μM) sensitivity of Q-KO and penta-KO strains expressing CgPST2-CΔS175-F198. D-E. Size exclusion chromatograms of purified CgPst2R174A (D) and CgPst2-CΔR174-F198 (E). After loading on the Sephacryl S-200 column, protein elution profiles were determined using the absorbance values at 280 nm. The CgPst2R174A showed two peaks corresponding to 296 kDa (red arrow; oligomeric form) and 29 kDa (green arrow; monomeric form) sizes, and CgPst2-CΔR174-F198 showed one peak of 296 kDa. F. Native PAGE analysis showing increased oligomer formation upon MD treatment. 200 μg whole cell lysates of untreated and MD (90 μM for 90 min)-treated Q-KO and penta-KO expressing CgPST2 were resolved in a discontinuous Tris-glycine buffer system under non-denaturing conditions, and probed with anti-CgPst2 antibody. The native protein molecular weight marker (M) was stained with coomassie brilliant blue, and is shown on the right side of the blot. The ponceau S-stained membrane is shown as loading control.
Further, to probe deeper into the active nature of CgPst2R174A in the penta-KO strain, we generated and expressed CgPst2-CΔS175-F198 protein that lacked last 24 amino acids after R174. CgPst2-CΔS175-F198 and CgPst2-CΔR174-F198 differ from each other, only in one residue, with CgPst2-CΔS175-F198 containing R174. As expected, compared to the full-length CgPst2, both these C-terminally truncated proteins ran faster on SDS-PAGE, corresponding to a molecular weight of about 19 kDa (S13B Fig). However, unlike CgPst2-CΔR174-F198, CgPst2-CΔS175-F198 expression neither reversed MD sensitivity of Q-KO and penta KO cells (Fig 6C) nor exhibited NADH:quinone oxidoreductase activity (S14A Fig), despite good protein expression (S14B Fig). Altogether, these data suggest that CgPst2-CΔS175-F198 is non-functional, and underscore the importance of R174 removal in activation of CgPst2, with CgPst2-CΔS175-F198, CgPst2-CΔR174-F198 and CgPst2R174A possessing no, higher and higher activity, respectively, compared to the full-length CgPst2.
CgPst2 processing regulates structural state
Fld-LPs are known to exist in tetramer forms, with oligomeric forms exhibiting elevated stability and/or activity [27,30,38]. The recombinant full-length CgPst2 existed in a monomeric form (Fig 1E), and was inactive (S3B Fig). Owing to differential activities of the C-terminally truncated CgPst2 proteins, we decided to examine the assembly state of these proteins. For this, we expressed and purified 6XHis-FLAG-CgPst2R174A, CgPst2-CΔR174-F198 and CgPst2-CΔS175-F198 from E. coli (S15A–S15C Fig). Despite being functional in C. glabrata, CgPst2R174A and CgPst2-CΔR174-F198 proteins showed no NADH:quinone oxidoreductase activity (S15D and S15E Fig), suggesting that these recombinant proteins probably lost enzymatic activity during purification, are misfolded, or require post-translational modification/s for activity. SEC analysis revealed two forms of CgPst2R174A, a monomer of 29 kDa and an oligomer of 296 kDa (Fig 6D), indicating that R174 residue may impede CgPst2 oligomerization. Notably, CgPst2-CΔR174-F198 was found only in the oligomeric form of 296 kDa (Fig 6E), while CgPst2-CΔS175-F198 formed aggregates and could not enter the column (S15F Fig). These data suggest that the role of C-terminal region in proper folding and function of CgPst2 is governed by R174 residue. Overall, the SEC analysis of different CgPst2 variants raises the prospect that CgPst2 exists as a monomer, and R174 removal, and by extension, the proteolytic cleavage of CgPst2 by CgYps1, may stimulate CgPst2 oligomerization. However, since all CgPst2 (wild-type, C-terminally truncated and R174-mutated) proteins, purified from E. coli, were enzymatically inactive, there is a good possibility that the misfolded CgPst2 contributes to their varied structural states.
We, therefore, next asked the question whether CgPst2 exists in a homo-oligomeric form in C. glabrata and if so, whether this oligomeric state is altered by the presence of menadione and/or CgYps1. For this, we subjected whole cell extracts of Q-KO and penta-KO strains expressing CgPST2 to native PAGE analysis, followed by Western blotting with anti-CgPst2 antibody. We observed about a 90 kDa band, probably representing CgPst2 tetramer, in CgPST2-expressing Q-KO cells, whose intensity was significantly increased upon MD treatment (Fig 6F). Intriguingly, this CgPst2 oligomer species was neither present in untreated nor MD-treated CgPST2-expressing penta-KO cells (Fig 6F). Instead, a higher-order CgPst2 band of about 130 kDa was observed in untreated and MD-treated CgPST2-expressing penta-KO cells (Fig 6F), which may represent the non-functional aggregated form of CgPst2. Further, the penta-KO cells expressing functionally active, C-terminally truncated CgPst2-CΔR174-F198 and arginine-mutated CgPst2R174A protein displayed the 90 kDa CgPst2 band, suggesting that these proteins are able to form a tetramer in C. glabrata cells, which may contribute to CgPst2 activity (S16 Fig). Altogether, three key findings emerge from these data. First, CgPst2 exists in a homo-oligomeric form. Second, MD treatment enhances CgPst2 homo-oligomerization. Third, CgYps1 is required for homo-oligomerization of CgPst2. Of note, although, we could neither detect monomeric nor higher-order oligomeric (296 kDa) form of CgPst2 in native PAGE immunoblot analysis, as observed in SEC analysis, it is possible that these CgPst2 species either represent a minor form of CgPst2 in C. glabrata or misfolding of CgPst2 in E. coli accounts for its inability to form tetramers. Further studies are warranted to address these possibilities as well as to examine higher-order homo-oligomers, formed by recombinant CgPst2-CΔR174-F198 and CgPst2-CΔR174A proteins, and CgPst2 in penta-KO/CgPST2 strain. Additionally, the possibility of post-translational modifications and/or binding of CgPst2 with other proteins contributing to the higher molecular weight CgPst2 bands in C. glabrata is yet to be precluded.
Discussion
Yapsins represent a family of GPI-anchored aspartyl proteases that is unique to fungi [35]. These proteases consist of two subunits α and β, with each subunit providing a catalytic aspartate residue to the active site cleft, predominantly cleave C-terminally to monobasic or dibasic residues, and are pivotal to fungal physiology and virulence [12,35,39]. Despite invasive fungal pathogens killing about one and a half million people every year [1], our understanding of the stress survival mechanisms of human pathogenic fungi remains woefully inadequate. CgYps1, an aspartyl protease, occupies a central stage in the regulation of many physiological processes, that are modulated by the CgYapsin family in the pathogenic yeast C. glabrata. Here, we have identified the potential interactors of a fungal Yapsin, through a pull-down assay, and characterized, in detail, the flavodoxin-like protein CgPst2, of 19 CgYps1-interacting proteins identified. We show that CgPst2 possesses NADH:quinone oxidoreductase activity, and is required for MD and BQ resistance. We report CgPst2 as the first physiological target of the CgYps1 protease, as it undergoes processing in a CgYps1-dependent manner in C. glabrata. This inference was further strengthened by an in vitro cleavage assay wherein CgYps1, purified from P. pastoris culture supernatants, could cleave the recombinant CgPst2 protein (S17 Fig), thereby underscoring the significance of CgYps1-CgPst2 interaction identified in the current study.
Flavodoxin-like fold-containing proteins, which adopt the conserved three-dimensional α/β twisted open-sheet fold, possess two-electron NAD(P)H:quinone oxidoreductase activity, and play a major role in stress response in diverse organisms including bacteria, yeasts, plants and mammals, by inhibiting redox cycling [17,18,22,25,27,28,30]. These have also been implicated in metabolic gene regulation, stress tolerance and virulence [23,25,30]. Several distinct mechanisms are used to control the activity of Fld-LPs which differ, in their oligomeric state, amino acid sequence, flavin cofactor and redox partners, from one another [17–20]. Fld-LPs use ping-pong mechanism, with FMN or FAD as cofactor, and NADH and quinone as electron donor and acceptor, respectively, to detoxify quinones via two-electron reduction pathway [17–19]. We report for the first time that CgYps1-mediated cleavage is a key determinant of activity and oligomeric state of cellular CgPst2, that adds another wholly unexpected regulatory mechanism to the Fld-LP stress defense system.
Structural studies on E. coli wrbA and S. cerevisiae Pst2 show that they form tetramer, irrespective of the FMN cofactor binding [27,30,40]. Multimerization brings many regions together and creates a complex active site [29]. An extended C-terminus has been postulated to inhibit the oligomerization of Fld-LPs, with S. cerevisiae Pst2 forming a tetramer, while Ycp4, containing additional 49 amino acids at the C-terminus, probably existing in a dimeric form, with both proteins implicated in quinone tolerance [22–24,30]. Similarly, rat and human NQO1 [NAD(P)H:quinone oxidoreductase 1)] are dimeric enzymes [41], while E. coli wrbA, which lacks the C-terminal sub-region of NQO1, forms a tetramer [27]. Furthermore, serine substitution of the proline-187 residue, a common cancer-associated polymorphism, in the C-terminal region of NQO1 resulted in NQO1 dimer destabilization and loss of catalytic activity [42,43]. The flavodoxin-like C-terminal domain of E. coli catalase HPII has also been implicated in its stability [44]. Consistent with all this, we found R174 residue in the C-terminus of CgPst2 to be essential for CgYps1-dependent cleavage, and consequent modulation of CgPst2 oligomerization and activity. The reduced CgPst2 activity and the lack of CgPst2 tetrameric form in penta KO, and contrasting effects of two C-terminal-truncations, lacking or containing R174, on CgPst2 activity, highlight the functional relevance of CgYps1-mediated CgPst2 processing in MD stress survival. However, given the conserved nature of R174 across several Fld-LPs (S7 Fig), and with this single residue mutation being sufficient to alter the activity of CgPst2, it will be intriguing to investigate if this residue is a protease target in other organisms as well.
Despite increased expression upon MD exposure (Fig 4E), localization of C. glabrata Pst2 at the plasma membrane remains unchanged under regular and MD stress conditions (S5 Fig). Notably, while the high throughput studies revealed S. cerevisiae Pst2, which is induced upon oxidative stress, to be localized to the mitochondria [45,46], the GFP fusion proteins of Pst2, Rfs1 and Ycp4 in S. cerevisiae have been reported to be localized to the eisosome domain in the plasma membrane [47]. Through confocal imaging and biochemical analysis, we showed that CgPst2 is present at the plasma membrane in C. glabrata. Similarly, C. albicans Pst2 is also located at the plasma membrane [25]. Altogether, these findings raise the possibility of cell membrane-localized Fld-LPs conferring growth advantage to fungal cells under oxidative stress-generating environmental conditions.
Regulated proteolysis is a common mechanism to modulate protein structure, function, localization and turnover [48]. Here, we demonstrate the essentiality of R174 residue in CgPst2 for CgYps1-dependent cleavage, which releases the C-terminal region. Conceivably, like other proteolysis events, multiple cellular factors including growth-phase, ROS abundance, expression of other Fld-LPs, environmental pH and CgYps1 availability, may impact the processing of CgPst2. In our analysis, the short cleaved C-terminal fragment was detected in small and variable amounts which could be due to its unstable nature. Alternatively, a small amount and non-detection of the cleaved C-terminal and N-terminal fragment of CgPst2, respectively, may indicate that only a minor fraction of CgPst2 undergoes cleavage under laboratory growth conditions.
Of note, of four Fld-LPs, the sole requirement for CgPst2 for MD and BQ resistance places CgPst2 at the forefront of hydrogen peroxide and superoxide anion radical detoxification mechanisms. Therefore, the existence of an exquisite control for CgPst2 activity is not unexpected in the pathogenic yeast C. glabrata, which encounters, and successfully counteracts the host-macrophage-elicited oxidative stress, and proliferates in macrophages [12,49,50]. In this context, it is noteworthy that the phagolysosomes of activated neutrophils have been shown to contain elevated amounts of ubiquinone [51]. So besides detoxifying endogenous ubiquinone of the electron transport chain, CgFld-LPs may aid C. glabrata survive attack of the host immune system.
Intriguingly, our genetic data suggest that MD sensitivity of the Cgyps1Δ mutant is independent of CgPst2, as the active CgPst2 constructs, CgPst2-CΔR174-F198 and CgPst2R174A, could not restore growth of the Cgyps1Δ mutant on MD medium, and the double Cgpst2Δyps1Δ mutant exhibited MD sensitivity higher than single mutants. Moreover, the MD-induced increase in CgPst2 activity in Cgyps1Δ mutant indicates that CgYps1 is unlikely to be the sole regulator of CgPst2 activity. This notion was further corroborated with CgYps1-mediated processing being important for CgPst2 functions in MD detoxification only in the absence of other Fld-LPs.
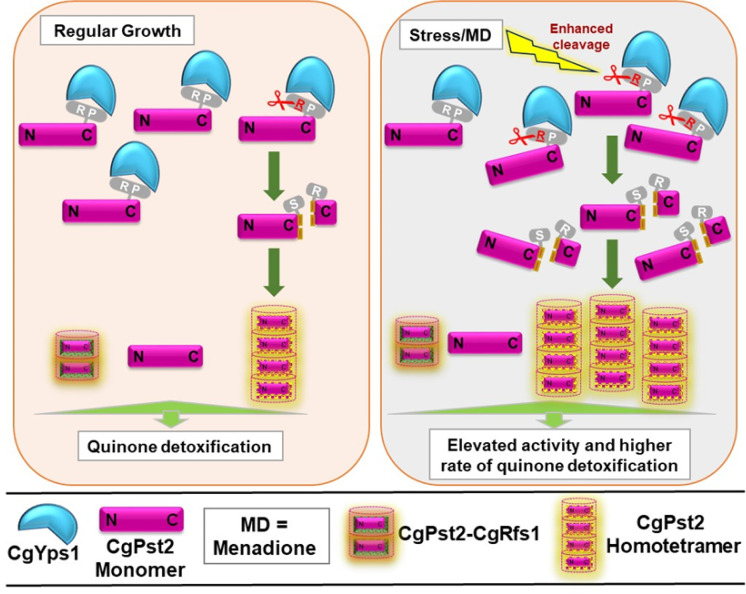
Cleavage and oligomerization, resulting in CgPst2 activation, are likely to be linked events that may occur at the plasma membrane and constitute cellular response to the quinone stress. Of note, the tetrameric form of E. coli wrbA is shown to be more thermostable than monomeric or dimeric forms [38]. Three particularly intriguing findings of our study, the homo-tetrameric form of CgPst2 in Q-KO/CgPST2 cells, CgPst2-mediated rescue of MD sensitivity of Q-KO and Cgpst2Δyps1Δ mutant, but not of penta-KO (summarized in S2 Table), that lacks CgPst2 tetramer, and the interaction of CgPst2 with CgRfs1, suggest that CgPst2 is capable of forming both homo-oligomer, and heteromers, with CgYps1 and CgFld-LP availability and/or cellular context probably dictating the type of oligomers formed. Therefore, we propose a complex multilayered regulation for NADH:quinone oxidoreductase activity of CgPst2, that could be governed by CgPst2 assembly. Given the absence of CgPst2 tetramer, and reduced CgPst2 activity in penta-KO/CgPST2 cells, we posit that CgYps1-mediated processing releases the C-terminal domain and induces CgPst2 homo-tetramerization, with the homo-tetrameric state being more active (Fig 7). In light of this, it is tempting to speculate that CgYps1-CgPst2 interaction aids in keeping a readily available cellular pool of CgPst2 for activation, when the need arises. Thus, the R174 residue, and, by extension, CgYps1-mediated cleavage, is likely to be pivotal to maintain a balance between differentially active forms of CgPst2, based on the cellular requirement. However, it must be noted that an experimental demonstration of menadione-induced, CgYps1-dependent increase in the ratio of oligomeric to monomeric form of CgPst2 is required to prove the proposed model unequivocally.
Fig 7. A schematic summarizing key findings of the study.
Arginine-174 (R) and Proline-176 (P) residues of CgPst2 are predicted to interact with CgYps1 at the plasma membrane, and CgYps1 processes R174 residue in the C-terminus of CgPst2. This cleavage, which is elevated upon menadione treatment, leads to removal of the C-terminal domain, resulting in CgPst2 homo-tetramerization, higher activity and efficient quinone detoxification. Of note, the signal, that stimulates CgYps1-mediated cleavage of CgPst2, is not known. CgPst2 also interacts with CgRfs1, however, CgPst2-CgRfs1 association is not dependent on CgYps1, and occurs under both regular and MD treatment conditions. Altogether, CgPst2 functions are regulated at multiple levels, and CgYps1-mediated cleavage of CgPst2 may reflect one of many mechanisms controlling CgPst2 activity.
Further, whether CgPst2 heteromers contain CgPst3 or CgYcp4, is yet to be determined. Additionally, the lack of in vitro phenotype for mutants deleted for CgRFS1, CgPST3 and CgYCP4 genes does not exclude their possible participation in oxidative stress-counteracting mechanisms, particularly in light of Q-KO showing attenuated survival in the mice infection model. How other Fld-LPs function, and if CgYps1 or other ten CgYapsins regulate their structural states and activity, remain to be investigated. In this context, it is worth noting that this is the first report of an aspartyl protease interacting with, and, regulating the activity of a Fld-LP through processing.
CgYps1 has a GPI-anchor region at the C-terminus, and is predicted to be present at the plasma membrane or the cell wall in C. glabrata [52] (www.candidagenome.org). Given the cell membrane localization of CgPst2 and CgYps1 (S12 Fig), CgYps1 and CgPst2 are likely to interact at the plasma membrane. However, since these are probably present on opposite sides of the plasma membrane, further studies are warranted to characterize, in-depth, the CgYps1-CgPst2 interaction at the plasma membrane. Similarly, the interaction of CgYps1 with the plasma membrane proton pump CgPma1, which has been identified as CgYps1 interactor in our IP-MS analysis (Table 1), is likely to take place at the plasma membrane, with CgYps1 being previously implicated in activity regulation of CgPma1 [13]. Moreover, Yps1 in S. cerevisiae is located at the plasma membrane [53], and CgYps1 ectopic expression rescued cell wall-related phenotypes of the S. cerevisiae yps1Δ mutant [54], indicating some molecular targets to be common to Yps1 protease of these two yeasts.
Substrate identification is necessary to elucidate the real functions of CgYapsins. Our IP-MS analysis identified, through a pull-down assay, 19 CgYps1-interacting proteins which are involved in both oxidoreduction process and energy homeostasis, and translation processes (Table 1). DeepLoc-based subcellular localization analysis revealed potential CgYps1 interactors to have varied unanticipated location ranging from the nucleus to the peroxisome (Table 1), indicating that the CgYps1-target protein repertoire is large, and not confined to the cell wall or the cell membrane proteins. However, the possibility that CgYps1 interacts non-enzymatically with some identified proteins and/or few highly abundant proteins are non-specific interactors, is yet to be ruled out. In this context, it is noteworthy that the cell wall targets of CgYps1 are likely to be underrepresented in this analysis of whole cell lysate samples. Moreover, since we could detect CgYps1 in the plasma membrane fraction (S12 Fig), it is possible that some of these identified interactions, including CgPst2-CgYps1, may occur at the plasma membrane. Additionally, the possibility of identified cytosolic proteins displaying cue-dependent dual subcellular localization is currently being investigated.
In conclusion, in addition to identifying intracellular interactors of a fungal yapsin for the first time, we report a novel regulatory mechanism for NADH:quinone oxidoreductase activity of a flavodoxin-like protein in C. glabrata, whose homologs are present in bacteria, plants and humans.
Materials and methods
Ethics statement
Mice infection experiments were performed at the Animal House Facility of Centre for DNA Fingerprinting and Diagnostics (CDFD), Hyderabad, India in accordance with guidelines of the Committee for the Purpose of Control and Supervision of Experiments on Animals, Government of India. Procedures were designed to minimize animal suffering, and approved by the Institutional Animal Ethics Committee [EAF/RK/CDFD/22]. Mice infection studies were performed, as described previously [50].
Strains and media
All C. glabrata strains are derivatives of the vaginal isolate BG2, and routinely grown at 30°C on standard YPD medium. Logarithmic (log)-phase cells were obtained by growing overnight cultures for 4–5 h at 30°C in the fresh medium.
Gene cloning and disruption
C. glabrata mutants were generated by replacing the ORF with the nat1 gene, which confers nourseothricin resistance, using the homologous recombination-based approach, and gene replacements were confirmed by PCR, as described previously [55]. Multiple gene deletions were carried out in the same strain using the recyclable nat1 selection marker, as described previously [56]. For mutant complementation and overexpression studies, C. glabrata genes were expressed from PGK1 (pRK74 plasmid) and PDC1 (pRK999 plasmid) promoter, respectively. For specific amplification of CgPST2, either the plasmid (pRK) containing CgPST2 gene along with 500 bp of 5’and 3’ UTR or genomic DNA of the Cgrfs1Δ mutant, was used as template. The CgPST2R174A allele was generated through two-fragment PCR approach, consisting of two separate PCR amplifications using mutagenesis primers, followed by fusion PCR and restriction digestion. The CgPST2, CgPST2-CΔR174-F198, CgPST2R174A and CgPST2-CΔS175-F198 genes were cloned in XmaI-Xba1, Xba1-Spe1, XmaI-Xba1, and Xba1-Xho1 sites, respectively, in the pGRB2.1 plasmid for analysis in C. glabrata, while genes were cloned in EcoR1-Xho1 sites in pET28a(+) plasmid for studies in E.coli. For CgPst2 tagging with the triple epiotpe SFB (S protein-Flag-Streptavidin-binding peptide) and GFP at the C-terminus, CgPST2 ORF (CAGL0K11858g; 0.597 kb) was cloned in XbaI-SpeI sites in pRK1349 and pRK1000 plasmid, respectively, carrying the CgPDC1 promoter. CgYPS1D91A was generated using mutagenic primers, as described previously [15]. All plasmid clones were confirmed by sequencing, functional complementation or protein expression. Strains, plasmids and primers used in this study are listed in S3–S5 Tables, respectively.
NADH:quinone oxidoreductase activity measurement
The NADH:quinone oxidoreductase activity in C. glabrata cell extracts was measured, as described previously for E. coli. purified proteins [57]. Briefly, log-phase cultures of C. glabrata strains were either treated with 90 μM MD for 90 min, or left untreated, and cell lysates were prepared by glass bead lysis method. Cell lysates (500 μg) were added to the reaction buffer [10 mM Tris-HCl (pH 7.4), 150 mM NaCl] containing NADH (500 μM) and menadione (500 μM). The blank control contained no NADH. The reaction was started with NADH addition, and the absorbance was recorded at 340 nm in a 1-cm-path-length quartz cuvette over a period of 60 seconds at 10 seconds interval on the Spectramax M5 plate reader. For CgPst2 and its variants purified from E. coli, the purified protein (50 μg) was incubated for 1 h with FMN (100 μM) or FAD (100 μM) on ice, followed by addition of other reaction components, as described above. The blank control contained no protein. The commercially available NAD(P)H:FMN oxidoreductase (1 Unit, Roche, # 10476480001) was used as positive control. Absorbance of the substrate NADH was considered as 100 at 0 h time point, and NADH oxidation was calculated by dividing the absorbance at each time point by 0 h absorbance, and multiplying the number by 100.
Immunoprecipitation, mass spectrometry and Western blot analysis
For IP, log-phase C. glabrata cell extracts were prepared by glass bead lysis. For IP-MS analysis, using anti-CgYps1 antibody, rProtein A-Sepharose Fast Flow beads (GE Healthcare) were first incubated with cell extracts (2 mg) of wt and Cgyps1-11Δ strains in lysis buffer (15 mM Na2HPO4, 150 mM NaCl, 2% Triton X- 100, 0.1% SDS, 0.5% DOC, 10 mM EDTA, 0.02% NaN3) containing protease inhibitors for 4 h at 4°C, to remove proteins binding non-specifically to beads. These precleared lysate supernatants were incubated with antibody (20 μl)-coated rProtein A-Sepharose beads for 2–3 h at 4°C with gentle rocking on an end-to-end rotator. Beads were washed once with lysis buffer and thrice with wash buffer (50 mM NaCl, 10 mM Tris, 0.02% NaN3), and centrifuged at 1500 rpm for 2 min. After boiling in 2X SDS dye for 5 min, samples containing beads were run on 10% SDS-PAGE gel, until the dye front entered the resolving gel. After coomassie brilliant blue staining, gel sections containing protein bands were cut and sent for protein identification to the Taplin Biological Mass Spectrometry Facility, Harvard Medical School, Boston. The Sequest software was used to identify peptides, and peptides, filtered to 1% false discovery rate, were mapped to the C. glabrata reference proteome database (www.candidagenome.org/). Proteins identified with ≥ 2 unique peptides in both biological replicate samples of wt and Cgyps1-11Δ strains were selected for further analysis. For identification of CgYps1-specific interactors, proteins identified in the immunoprecipitated samples of Cgyps1-11Δ mutant were removed from the CgYps1 interactor list. The raw mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [58] partner repository with the dataset identifier PXD022766.
For affinity purification, CgPst2-SFB expressing log-phase cells were grown in CAA medium for 4–5 h and collected. Cell lysates (3 mg), were incubated with streptavidin beads for 2 h at 4°C. After washing, beads were boiled in 2X SDS dye, proteins resolved on a 12% and 8% SDS-PAGE gel for CgPst2 and CgYps1 detection, respectively, and immunoblotted with anti-Flag and anti-CgYps1 antibodies. For CgPst2 protein detection, log-phase cells were either left untreated or treated with MD (90 μM) for 90 min, and cell lysates were prepared in protein extraction buffer [50 mM Tris-HCl (pH 7.5), 2 mM EDTA, 2% glucose, 10 mM sodium fluoride, 1 mM sodium orthovanadate and 1 X protease inhibitor] using glass beads. Total protein (200 μg for CgYps1 and 60 μg for CgPst2) were resolved on SDS-PAGE gel, followed by transfer to the polyvinylidene difluoride (PVDF) membrane and probing with anti-CgPst2 or anti-CgYps1 antibody.
For native PAGE analysis, cell lysates (200 μg) were separated on a discontinuous Tris-glycine polyacrylamide gel system consisting of a 4.0% stacking and a 10% resolving gel, prepared in 0.375 M Tris-HCl, (pH 8.8), under non-denaturing conditions. The running buffer contained 25 mM Tris (pH 8.0) and 192 mM glycine. The Gel was pre-run for 30 min, prior to sample loading in 2X Native PAGE dye [Tris-HCl (62.5 mM; pH 6.8), glycerol (25%) and bromophenol blue (1%)]. Gels were run for 3–4 h at 100 V at 4οC to avoid excessive heating, and proteins were transferred to PVDF membrane in the transfer buffer (25 mM Tris, 192 mM glycine, pH 8.0) containing 10% methanol for 14 h at 4οC. The membrane was processed for immunoblot analysis using anti-CgPst2 antibody. The gel lane containing the protein molecular weight marker was cut, and stained with coomassie brilliant blue.
In silico protein analysis
For molecular docking studies, CgPst2 and CgYps1 sequences were retrieved from CGD (Candida genome database; http://www.candidagenome.org), and submitted to the online tool I-TASSER (Iterative Threading ASSEmbly Refinement; https://zhanglab.ccmb.med.umich.edu/I-TASSER/). The top structure models with best Confidence (C)-score were selected and analyzed by another online tool Z-DOCK (http://zdock.umassmed.edu/) to acquire CgYps1-CgPst2 docking image.
C-terminal-cleaved CgPst2 fragment analysis
Affinity purification using streptavidin beads was used to retrieve the small C-terminal cleaved fragment of CgPst2. Cell extracts (2 mg) of wt and Cgyps1Δ strains expressing CgPst2-SFB were incubated overnight with prewashed streptavidin beads at 4οC, and centrifuged at 1500 rpm for 5 min. After 3 washes, beads were incubated with biotin solution (2 mg/ml) for 4 h at 4οC, followed by incubation of the biotin solution with S-protein beads for 2 h at 4οC. S-protein beads were washed, boiled in 2X SDS sample buffer and resolved on 18% SDS-PAGE. The gel was stained with Coomassie Brilliant Blue and destained. The band corresponding to the smaller cleaved fragment of ~16 kDa was cut and sent to the Sandor Life Sciences, Hyderabad, India (https://sandorlifesciences.co.in) for peptide mass fingerprinting analysis using MALDI Autoflex II TOF/TOF (Bruker Daltonics Inc. MA). The CgPst2 fragment was identified using parameters of peptide mass tolerance of ± 300 and fragment mass tolerance of ± 2 Da with maximum 2 missed cleavages.
CgPst2 purification and size-exclusion chromatography analysis
The CgPST2 (CAGL0K11858) ORF was cloned in EcoR1 and Xho1 sites in the pET28a(+) plasmid and transformed into E. coli BL21 (DE3) strain, followed by selection for kanamycin resistance (50 μg/ml). A purified transformant carrying pET28a-6XHis-FLAG-CgPST2 was grown in LB medium containing kanamycin and induced with IPTG (0.5 mM) at 18οC for 16 h. After cell collection via centrifugation at 8000 rpm for 10 min, cells were suspended in lysis buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl and 10 mM Imidazole] and sonicated for 30 cycles of 30 sec ON/OFF (Biorupter, Diagenode). Cell lysates were centrifuged at 18000 rpm for 20 min, and the soluble recombinant CgPst2 protein was purified from the supernatant using the TALON metal affinity resin via affinity purification. Briefly, after overnight incubation of the supernatant with TALON beads at 4°C, this mix was added to the Polypropylene Column (Thermo Fisher Scientific #R64050), washed with the wash buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl and 50 mM Imidazole), and the column-bound 6XHis-CgPst2 protein was eluted in the 250 mM imidazole-containing buffer. After removing imidazole through Amicon Ultra centrifugal filter units (3 kDa cut off), the protein purity was determined by SDS-PAGE and Western blot analysis. For size-exclusion chromatography, the column (1 cm radius and 48 cm height) was packed manually with Sephacryl S-200 matrix beads and run on the FPLC system (Bio-Rad). After pre-equilibrating the column with two column-volumes of buffer containing 50 mM Tris-HCl (pH 8.0) and 150 mM NaCl, about 300–500 μg of 6XHIS-FLAG-CgPst2 was applied to the injection ring with the volume of 300 μl. Samples were run with a flow rate of 0.22 ml/min for a total time of 5 h with at room temperature, and fractions of 0.5 ml were collected and analyzed with anti-His antibody. Blue dextran (1 mg/ml) was used to calculate the void volume. Marker proteins, thyroglobulin (A; 670 kDa), gammaglobulin (B; 158 kDa), ovalbumin (C; 44 kDa), myoglobin (D; 17kDa) and vitamin B12 (E; 1.35 kDa), of different sizes were used to calibrate the column. The calibration graph was plotted using the gel-phase distribution coefficient (Kav) against logarithm of the molecular weight (Log MW) of marker proteins. For each sample, at least two independent experiments were performed, and the molecular mass of eluted CgPst2 was calculated from the calibration plot using linear regression (R2 ≥ 0.96). CgPst2R174A, CgPst2-CΔR174-F198 and CgPst2-CΔS175-F198 were purified and analyzed in the similar fashion.
Plasma membrane isolation
Log-phase C. glabrata cells (100 ml; 2.0 OD600) were harvested, washed with MQ water, suspended in homogenization buffer [50 mM Tris (pH 7.5), EDTA (2.5 mM)] and were lysed with glass beads. An aliquot of the supernatant was saved as the whole cell lysate fraction, and the remainder supernatant was subjected to ultracentrifugation (SW 41 Ti rotor) at 25000 rpm for 30 min at 4°C. The pellet was suspended in the suspension buffer [10 mM Tris (pH 7.5), 0.5 mM EDTA and 10% glycerol] containing protease inhibitors, followed by discontinuous sucrose gradient [43.5 and 53.5% (w/v)] ultracentrifugation at 35000 rpm for 5 h. The middle layer containing the plasma membrane was collected very carefully and again subjected to ultracentrifugation at 38000 rpm for 30 min. After discarding the supernatant, the plasma membrane pellet was suspended in suspension buffer containing protease inhibitors. Protein concentration in cell lysate and plasma membrane fractions was estimated by BCA method, and 60–200 μg samples were run on SDS-PAGE and probed with anti-CgYps1, anti-CgPst2, anti-Gapdh and anti-Pma1(Santa Cruz Biotechnology; # sc-33735) antibodies. Anti-Pma1 antibody, that recognizes the C. glabrata plasma membrane ATPase CgPma1 [13], was used to check the quality of plasma membrane preparation.
Functional and statistical analysis
The Candida Genome Database (CGD)-GO Slim Mapper tool for BP (biological process) (http://www.candidagenome.org/cgi-bin/GO/goTermMapper) was used to functionally annotate C. glabrata genes. The FungiFun tool (https://elbe.hki-jena.de/fungifun/), with C. glabrata CBS138 as the reference strain, was used for GO functional enrichment analysis. The experimental data were statistically analysed using Student’s t-test. Mice data were analysed using Mann-Whitney U test. The p-values *p < 0.05; **p < 0.01; ***p < 0.001 and ****p < 0.0001 were used to determine statistical significance. Raw numerical data are presented in S6 Table.
Other protocols are provided in S1 Text.
Supporting information
A. Amino acid sequence identity (%) among Fld-LPs of C. glabrata, S. cerevisiae and C. albicans. Systematic ORF names of Fld-LPs, taken from Candida (http://www.candidagenome.org/) and Saccharomyces (https://www.yeastgenome.org/) Genome Databases, are indicated, with common gene names in brackets. Amino acid identity among protein sequences of these ORFs was determined using BLASTP analysis, with C. glabrata protein as a query sequence, against C. glabrata CBS138, C. albicans SC5314 (Assembly 19) and S. cerevisiae (S288C) reference strains. B. Schematic illustration of CgPst2, CgRfs1, CgPst3 and CgYcp4 protein domain structures, as predicted by the SMART tool 12 (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1). Amino acids spanning the predicted domains are indicated. SMART tool predicted the FMN RED and Flavodoxin_2 domains. Proteins with Flavodoxin-2 domain include bacterial and eukaryotic NAD(P)H dehydrogenase (quinone) enzymes. These enzymes catalyze the NAD(P)H-dependent two-electron reduction of quinones and protect cells against damage by free radicals and reactive oxygen species. FMN RED domain is found in several flavoproteins such as FMN-dependent NADPH-azoreductases, which catalyze the reductive cleavage of azo bond in aromatic azo compounds to the corresponding amines, and NAD(P)H:quinone oxidoreductases, which reduce quinone to the hydroquinone state to prevent interaction of the semiquinone with O2 and production of superoxide. The figure is not drawn to scale.
(TIF)
A. Serial dilution spotting analysis showing growth of C. glabrata mutants deleted for single or multiple CgFld-LP genes on indicated stress conditions. Duroquinone (DQ) and cumene hydroperoxide (CHP) were used at a concentration of 450 μM and 40 μM, respectively. Plates were incubated at 30οC unless indicated otherwise, and images were captured after 24 h for thermal stress (42οC), and 48 h for DQ and CHP. The Q-KO strain lacks four flavodoxin-like proteins, CgPst2, CgRfs1, CgPst3 and CgYcp4. B. Serial dilution spotting analysis showing CgPST2-mediated complementation of menadione (MD; 95 μM) and benzoquinone (BQ; 4 mM) sensitivity of Cgpst2Δ and Q-KO strains. ‘V’ refers to empty vector. Plates were incubated at 30οC, and images were captured after 24 h and 48 h for MD and BQ stresses, respectively.
(TIF)
A. 6X-Histidine-FLAG-CgPst2 purification from Escherichia coli. The E. coli BL21 (DE3) strain transformant carrying pET28a(+)-6XHIS-FLAG-CgPST2 plasmid was grown in LB medium, induced with IPTG (0.5 mM) at 18°C for 16 h, and cells were collected. After cell lysis, the recombinant CgPst2 protein was purified using TALON metal affinity resin via affinity purification. 30 μl eluates along with flow through were resolved on 12% SDS-PAGE, and stained with Coomassie Brilliant Blue. The black arrow marks CgPst2 band. Immunoblot analysis of these fractions with anti-His antibody (1:5000 dilution) is shown at the bottom. M, Protein Marker. B. NADH:quinone oxidoreductase activity measurement of recombinant CgPst2. 50 μg of purified CgPst2 was incubated for 1 h with FMN (100 μM) or FAD (100 μM) on ice, followed by addition of menadione (500 μM) in buffer containing Tris-HCl (10 mM; pH 7.4) and NaCl (150 mM). The reaction was started with NADH (500 μM) addition, and absorbance was recorded at 340 nm in a 1-cm-path-length quartz cuvette over a period of 60 seconds at 10 seconds interval on Spectramax M5 plate reader. The commercially available NAD(P)H:FMN oxidoreductase (1 Unit, Roche, # 10476480001) was taken as positive control. Blank contained no protein. Absorbance of the substrate NADH was considered as 100 at 0 h time point, and NADH oxidation was calculated by dividing the absorbance at each time point by 0 h absorbance, and multiplying the number by 100. Data represent mean ± SEM. Grouped multiple t-test was performed, with n = 3 to 5. **, p < 0.0021; ****, p < 0.0001. C. NADH:quinone oxidoreductase activity measurement in cell extracts of wt, Cgpst2Δ and Q-KO strains in enzymatic reaction mixtures that contained exogenously added FMN (50 μM) or FAD (50 μM). Data represent mean ± SD (n = 2–3). The Q-KO strain lacks four flavodoxin-like proteins, CgPst2, CgRfs1, CgPst3 and CgYcp4. D. NADH:quinone oxidoreductase activity measurement in extracts of Cgpst2Δ and Q-KO cells expressing CgPST2. CgPST2 expression restored the activity deficit in Cgpst2Δ and Q-KO strains. Data represent mean ± SEM. Black and blue asterisks indicate statistically significant differences in activity of QKO and Cgpst2Δ samples, respectively, expressing CgPST2, compared to corresponding strains carrying vector. Grouped multiple t-test was performed, with n = 3. *, p < 0.0332; **, p < 0.0021.
(TIF)
NADH:quinone oxidoreductase activity measurement in extracts of wt, Cgyps1Δ, Cgpst2Δ and Cgpst2Δyps1Δ strains. Data represent mean ± SEM. Grouped multiple t-test was performed, with n = 3. Black and blue asterisks indicate statistically significant activity differences in wt strain, compared to Cgpst2Δ and Cgpst2Δyps1Δ strains, respectively. Brown and orange asterisks indicate statistically significant activity differences in Cgyps1Δ mutant compared to Cgpst2Δ and Cgpst2Δyps1Δ mutants, respectively. *, p < 0.0332; **, p < 0.0021; ***, p < 0.0002.
(TIF)
Confocal images illustrating cell membrane localization of CgPst2-GFP in log-phase cells of indicated strains grown in CAA or CAA medium containing menadione (90 μM; MD) for 90 min. DIC, Differential interference contrast. Bar = 2 μm.
(TIF)
Cell extracts of wt and Cgyps1Δ strains expressing CgPst2-SFB (3 mg protein) were incubated with streptavidin beads for 2 h at 4οC and centrifuged at 2000 rpm for 5 min. Washed Beads were boiled in 2X SDS sample buffer, loaded on 8% (for CgYps1) and 12% (For SFB-tagged CgPst2) SDS-PAGE, and probed with anti-CgYps1 and anti-Flag antibodies, respectively. Please note that CgYps1 was not detectable in Input samples.
(TIF)
Protein sequences of Fld-LPs of C. glabrata, S. cerevisiae, C. albicans, Aspergillus fumigatus, Paracoccidioides brasiliensis, Bacillus cereus, E. coli, Homo sapiens and Arabidopsis thaliana were retrieved from the UniProt database (https://www.uniprot.org/uniprot/), and aligned and coloured using the Clustal Omega server (https://www.ebi.ac.uk/Tools/msa/clustalo/). Sequence alignment, corresponding to 127–198 amino acids, in the C-terminal region of CgPst2 is shown. The red arrow points to the conserved R174 residue in CgPst2. UniProtKB accession numbers are as follows: Q6FM13_CANGA (CgPst2), Q6FRB1_CANGA (CgRfs1), Q6FXR3_CANGA (CgPst3), Q6FS27_CANGA (CgYcp4), PST2_YEAST (S. cerevisiae Pst2), RFS1_YEAST (S. cerevisiae Rfs1), YCP4_YEAST (S. cerevisiae Ycp4), LOT6_YEAST (S. cerevisiae Lot6), A0A1D8PHR5_CANAL (C. albicans Pst1), Q59Y37_CANAL (C. albicans Pst2), A0A1D8PT02_CANAL (C. albicans Pst3), A0A1D8PT03_CANAL (C. albicans Ycp4), Q4WKD8_ASPFU (Aspergillus fumigatus Pst2), Q8X194_PARBR (Paracoccidioides brasiliensis y20), Q81G57_BACCR (Bacillus cereus Nrdl), NQOR_ECOLI (E. coli wrbA), NQO1_HUMAN (Homo sapiens NQO1), FQR1_ARATH (Arabidopsis thaliana FQR1).
(TIF)
CgGapdh was used as a loading control. M, Protein Marker.
(TIF)
A. Immunoblot analysis of CgPst2 expression in cell extracts of indicated C. glabrata strains to check the specificity of anti-CgPst2 polyclonal antibody. YPD-grown log-phase cells were lysed using glass beads, and 60 μg protein was resolved on 12% SDS-PAGE. After transferring proteins to the PVDF membrane, the blot was probed with antibody (1:1000 dilution) raised against purified CgPst2 protein in BALB/c mice. This mouse anti-CgPst2 sera yielded no and good signal in cell lysates of Q-KO (lacks all four flavodoxin-like proteins, CgPst2, CgRfs1, CgPst3 and CgYcp4), and Cgpst2Δ and Cgrfs1Δ mutants, respectively, indicating that the generated antibody binds to both CgPst2 and CgRfs1 proteins. CgGapdh was used as a loading control. B. qPCR-based determination of CgPST2 transcript levels. Log-phase T-KO (Cgrfs1Δpst3Δycp4Δ) and T-KOyps1Δ (Cgrfs1Δpst3Δycp4Δyps1Δ) cells were either left untreated (UT) or treated with 90 μM menadione for 90 min (T). Data (mean ± SEM, n = 3) were normalized against the CgACT1 mRNA control, and represent fold change in CgPST2 expression in treated samples, compared to corresponding untreated samples (taken as 1.0). C. Immunoblot analysis of CgYps1 expression in cell extracts of untreated or menadione-(MD; 90 μM for 90 min)-treated log-phase cells of the wt strain, using anti-CgYps1 antibody. CgGapdh was used as a loading control. Of note, CgYps1 is predicted to be highly glycosylated which may account for diffuse nature of the CgYps1 band. Consistent with predicted posttranslational modifications, the CgYps1 band corresponds to about 135 kDa, as compared to the expected size of 64 kDa. D. Immunoblot analysis showing CgYps1-CgPst2 interaction in log-phase untreated and MD (90 μM for 90 min)-treated Q-KO cells expressing CgPST2. Immunoprecipitation was carried out with anti-CgYps1 antibody, and blots were probed with anti-CgYps1 and anti-CgPst2 antibodies. Untreated penta-KO strain expressing vector (V) was used as negative control. The green asterisk denotes non-specific band. The Q-KO strain lacks four flavodoxin-like proteins (Fld-LPs), CgPst2, CgRfs1, CgPst3 and CgYcp4, while the penta-KO strain lacks CgYps1 along with four Fld-LPs, CgPst2, CgRfs1, CgPst3 and CgYcp4.
(TIF)
A. The Pichia pastoris GS115 strain expressing secretory forms of either catalytically active (rCgYps1) and inactive CgYps1 (rCgYps1D91A) proteins was grown in the BMGY medium at 30°C for 24 h and incubated in the BMMY medium containing 2% methanol for 48 h. Methanol was used to induce the AOX1 promoter, which drives the expression of CgYps1 proteins. 2% Methanol was added every 24 h to maintain the induction and expression of CgYps1 proteins. After 48 h, 1 ml supernatant was precipitated with 80% ammonium sulphate, pellet was suspended in PBS, and CgYps1 protein induction was checked by SDS PAGE. Induced proteins were purified by anion exchange chromatography using DEAE cellulose resin. The eluate fractions were resolved on 12% SDS-PAGE. The orange asterisk marks CgYps1 band. The protein samples were also probed with anti-His antibody (1:5000 dilution) and shown underneath the gel images. M, protein marker. B. CgYps1-mediated proteolysis of gelatin. The YNB-agar medium containing 1% gelatin was incubated either with 100 μg of purified proteins (rCgYps1 and rCgYps1D91A) or citrate buffer (pH 4.0; used for enzyme elution) for overnight at 37°C. The plate was stained with Coomassie Brilliant Blue G250 and imaged. The zone of hydrolysis was observed only with the CgYps1 enzyme. C. Proteolytic activity of DEAE cellulose-purified rCgYps1 and rCgYps1D91A proteins (20 μg) was measured using 2.5% hemoglobin as a substrate in the citrate buffer (100 mM; pH 4.0) for 30 min at 37°C. TCA (10%) was added to stop the enzymatic reaction and precipitate the undigested hemoglobin and CgYps1 proteins. Cleaved peptides were collected from the supernatant and incubated with sodium bicarbonate (5 mM) and Folin-Ciocalteu reagent at 37°C for 30 min. The absorbance was read at 660 nm and converted into the μ moles of tyrosine released per mg of the protein. Control indicates the enzymatic reaction mixture containing buffer and substrate, but no CgYps1 protein. Data represent mean ± SEM of 4 independent samples. Of note, rCgYps1D91A showed no proteolytic activity, indicating that Aspartate-91 in CgYps1 is required for enzyme activity. **, p < 0.01; unpaired two-tailed Student’s t test.
(TIF)
Immunoblot analysis showing no interaction between CgPst2 and CgYcp4-GFP. 6 mg precleared lysates of untreated and MD (90 uM for 90 min)-treated T-KO (Cgrfs1Δpst3Δycp4Δ) and T-KOyps1Δ (Cgrfs1Δpst3Δycp4Δyps1Δ) cells expressing CgYCP4-GFP were incubated with anti-GFP antibody-conjugated beads, followed by Western analysis with anti-GFP and anti-CgPst2 antibodies. The red and green arrows mark GFP and CgYcp4-GFP protein bands, respectively, in IP samples, while the green asterisk denotes non-specific band. Please note that Ycp4-GFP expression was not observed in Input samples, despite loading 200 μg protein.
(TIF)
Immunoblot analysis showing enrichment of CgYps1 and CgPst2 in the plasma membrane fraction of the Q-KO strain expressing CgPST2. Whole-cell lysates and plasma membrane fractions (60 μg and 200 μg for CgPst2 and CgYps1, respectively), prepared by glass bead lysis and sucrose gradient ultracentrifugation, respectively, were resolved on 4–20% polyacrylamide gradient, 12% polyacrylamide, 12% polyacrylamide and 12% polyacrylamide gels for CgYps1, CgPst2, CgPma1 and CgGapdh, respectively, and probed with anti-CgYps1, anti-CgPst2, anti-Pma1 and anti-Gapdh antibodies. The penta-KO strain was used as a negative control.
(TIF)
A. Immunoblot analysis of CgPst2 levels in Q-KO and penta-KO strains expressing CgPST2 or CgPST2R174A. Whole-cell extracts (60 μg), prepared by glass bead lysis, were resolved on 12% SDS-PAGE and probed with anti-CgPst2 and anti-Gapdh antibodies. The intensity of individual bands in 4 independent Western blots was quantified using the ImageJ densitometry software, and CgPst2 signal was normalized to the corresponding CgGapdh signal. Fold-change (mean ± SEM) in CgPst2 levels in CgPST2R174A-expressing cells, compared to CgPST2-expressing cells (considered as 1.0), is shown underneath the blot. p ≤ 0.01; paired two-tailed Student’s t test. The green asterisk indicates non-specific band. B. Immunoblot analysis of CgPst2 expression in penta-KO strains expressing CgPST2ΔR174-F198 or CgPST2ΔS175-F198. Whole-cell extracts (60 μg) of indicated strains were prepared by glass bead lysis and resolved on 18% SDS-PAGE for 4–5 h. The blots were probed with anti-CgPst2 and anti-Gapdh antibodies.
(TIF)
A. NADH:quinone oxidoreductase activity measurement in extracts of Q-KO expressing either CgPST2 or CgPST2-CΔS175-F198. Data represent mean ± SEM. Grouped multiple t-test was performed, with n = 3 to 4. *, p < 0.0332; **, p < 0.0021; ***, p < 0.0002; ****, p < 0.0001. The Q-KO strain lacks four flavodoxin-like proteins, CgPst2, CgRfs1, CgPst3 and CgYcp4. B. Immunoblot analysis of CgPst2 levels in Q-KO strains expressing CgPST2 or CgPST2-CΔS175-F198. The green asterisk denotes non-specific band.
(TIF)
A-C. 6X-Histidine-FLAG-CgPst2R174A A., 6X-Histidine-FLAG-CgPst2-CΔR174-F198 B. and 6X-Histidine-FLAG-CgPst2-CΔS175-F198 C. protein purification from E. coli. The E. coli BL21 (DE3) strain transformants carrying pET28a(+)-6XHIS-FLAG-CgPST2R174A, pET28a(+)-6XHIS-FLAG-CgPST2-CΔR174-F198 and pET28a(+)-6XHIS-FLAG-CgPST2-CΔS175-F198 plasmids were grown in LB medium, induced with IPTG (0.5 mM) at 18οC for 16 h, and cells were collected. After cell lysis, the recombinant proteins were purified using TALON metal affinity resin via affinity purification. 30 μl eluates, representing purified rCgPst2R174A A. rCgPst2-CΔR174-F198 B. and rCgPst2-CΔS175-F198 C. proteins, along with the flow through, were resolved on 12% SDS-PAGE, and stained with Coomassie Brilliant Blue. The green arrow marks CgPst2 band. M, Protein Marker. D-E. NADH:quinone oxidoreductase activity measurement of the recombinant CgPst2R174A D. and CgPst2-CΔR174-F198 E. protein (250 μg) was measured, as described in the legend of S3B Fig. Blank contained no protein. Data represent mean ± SEM. Grouped multiple t-test was performed, with n = 3. Black and blue asterisks indicate statistically significant activity differences between the positive control [NAD(P)H:FMN oxidoreductase (1 Unit, Roche, # 10476480001)] and recombinant proteins incubated with FMN (100 μM) and FAD (100 μM), respectively. *, p < 0.0332. Please note that the enzymatic activity of the purified recombinant CgPst2-CΔS175-F198 protein could not be determined, as it formed precipitates during the assay reaction. F. Size exclusion chromatogram of CgPst2-CΔS175-F198. After loading 300 μg of purified 6XHIS-FLAG-CgPst2-CΔS175-F198 protein on the Sephacryl S-200 column, protein elution profiles were determined using the absorbance values at 280 nm. The CgPst2-CΔS175-F198 protein could not bind to the column, and appeared as aggregates in the column’s void volume, as determined by column calibration with blue dextran. AU, Arbitrary Units.
(TIF)
300 μg whole cell lysates of Q-KO- expressing vector pRK74 (V) or CgPST2, and penta-KO expressing CgPST2-CΔR174-F198, CgPST2R174A and CgPST2 were resolved in a discontinuous Tris-glycine buffer system under non-denaturing conditions, and probed with anti-CgPst2 antibody. The native protein molecular weight marker (M) was stained with coomassie brilliant blue, and is shown on the right side of the blot. The ponceau S-stained membrane is shown as loading control.
(TIF)
In vitro cleavage assay. The Pichia pastoris GS115 strain expressing either CgYps1 or CgYps1D91A was induced with 2% methanol for 48 h, and pelleted down at 14000 rpm for 10 min at 4°C. The supernatants containing CgYps1 and CgYps1D91A were filtered through a 0.22 μM filter (Millipore, USA), and concentrated using Amicon Ultra centrifugal filter unit (3 kDa cutoff). The retentate was precipitated with ammonium sulphate (100% saturation) for 10 min at 4°C, followed by centrifugation and pellet suspension in citrate buffer (pH 4.0). For the cleavage assay, 30 μg of r6XHis-Flag-CgPst2 was incubated with 60 μg of partially purified rCgYps1 or rCgYps1D91A enzymes at 37°C for 4 h in citrate buffer (pH 4.0). Digested samples were run on 18% SDS-PAGE and probed with anti-His antibody. The assay samples containing both CgYps1 and CgPst2 displayed an additional faster migrating CgPst2 protein band (~ 24 kDa; marked with orange asterisk), probably representing the N-terminal fragment of the cleaved form of CgPst2, which was absent in reaction mixtures containing either CgPst2 alone or both CgPst2 and CgYps1D91A. These data indicate that the catalytically active CgYps1 can cleave CgPst2. Of note, we could not detect the small C-terminal-cleaved CgPst2 fragment, as hexa-His epitope is present at the N-terminus. To estimate the size difference between the cleaved and un-cleaved form of CgPst2, E-coli-purified full-length CgPst2 and CgPst2-CΔR174-F198 (50 μg) were run on 18% SDS-PAGE as a control, and probed with anti-his antibody. M, Protein Marker.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
We are indebted to Dr. Reshma Chowdary Alokam and Mr. Shaffiqu T S for their help in CgYps1 purification and structural modeling, and size-exclusion chromatography analyses, respectively. We thank Mubashshir Rasheed, Arvindh Kumar J, Abhijeet Singh T (CDFD-SEF), and Navitha Bedarakota and P Pranjali Milind [CDFD-Animal House Facility], for their help in CgPST2 gene cloning, SEC, confocal microscopy, and mice infections and antibody generation, respectively.
Data Availability
The raw mass spectrometry proteomics data are deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD022766. All other relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was fully supported by the DBT/Wellcome Trust India Alliance Senior Fellowship to RK [IA/S/15/1/501831; www.indiaalliance.org/]. Research in Kaur laboratory is also supported by grants from the Department of Biotechnology [BT/HRD/NBA/37/01/2014 to RK; www.dbtindia.gov.in/], and Science and Engineering Research Board, Department of Science and Technology [EMR/2016/005375 to RK; www.serb.gov.in/], Government of India. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4: 165rv13. 10.1126/scitranslmed.3004404 [DOI] [PubMed] [Google Scholar]
- 2.Bassetti M, Merelli M, Righi E, Diaz-Martin A, Rosello EM, Luzzati R, et al. Epidemiology, Species Distribution, Antifungal Susceptibility, and outcome of Candidemia across five sites in Italy and Spain. J Clin Microbiol. 2013;51: 4167–4172. 10.1128/JCM.01998-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamoth F, Lockhart SR, Berkow EL, Calandra T. Changes in the epidemiological landscape of invasive candidiasis. J Antimicrob Chemother. 2018;73: i4–i13. 10.1093/jac/dkx444 [DOI] [PubMed] [Google Scholar]
- 4.Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ. Invasive candidiasis. Nat Rev Dis Prim. 2018;4: 1–20. 10.1038/s41572-018-0001-z [DOI] [PubMed] [Google Scholar]
- 5.Pfaller MA, Diekema DJ, Turnidge JD, Castanheira M, Jones RN. Twenty years of the SENTRY antifungal surveillance program: Results for Candida species from 1997–2016. Open forum Infect Dis. 2019;6: S79–S94. 10.1093/ofid/ofy358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakrabarti A, Sood P, Rudramurthy SM, Chen S, Kaur H, Capoor M, et al. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med. 2015;41: 285–95. 10.1007/s00134-014-3603-2 [DOI] [PubMed] [Google Scholar]
- 7.Astvad KMT, Johansen HK, Røder BL, Rosenvinge FS, Knudsen JD, Lemming L, et al. Update from a 12-year nationwide fungemia surveillance: Increasing intrinsic and acquired resistance causes concern. J Clin Microbiol. 2018;56: 1–15. 10.1128/JCM.01564-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng ZR, Tian G, Ding YH, Yang K, Liu JB, Deng J. Surveillance study of the prevalence, species distribution, antifungal susceptibility, risk factors and mortality of invasive candidiasis in a tertiary teaching hospital in Southwest China. BMC Infect Dis. 2019;19: 1–12. 10.1186/s12879-018-3567-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, et al. Genome evolution in yeasts. Nature. 2004;430: 35–44. 10.1038/nature02579 [DOI] [PubMed] [Google Scholar]
- 10.Galocha M, Pais P, Cavalheiro M, Pereira D, Viana R, Teixeira MC. Divergent Approaches to virulence in C. albicans and C. glabrata: Two sides of the same coin. Int J Mol Sci. 2019;20: 2345. 10.3390/ijms20092345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar K, Askari F, Sahu MS, Kaur R. Candida glabrata: A lot more than meets the eye. Microorganisms. 2019;7: 39. 10.3390/microorganisms7020039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaur R, Ma B, Cormack BP. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc Natl Acad Sci 2007;104: 7628–33. 10.1073/pnas.0611195104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bairwa G, Kaur R. A novel role for a glycosylphosphatidylinositol-anchored aspartyl protease, CgYps1, in the regulation of pH homeostasis in Candida glabrata. Mol Microbiol. 2011;79: 900–913. 10.1111/j.1365-2958.2010.07496.x [DOI] [PubMed] [Google Scholar]
- 14.Bairwa, Rasheed M, Taigwal R, Sahoo R Kaur R. GPI (glycosylphosphatidylinositol)-linked aspartyl proteases regulate vacuole homoeostasis in Candida glabrata. Biochem J. 2014;458: 323–334. 10.1042/BJ20130757 [DOI] [PubMed] [Google Scholar]
- 15.Rasheed M, Battu A, Kaur R. Aspartyl proteases in Candida glabrata are required for suppression of the host innate immune response. J Biol Chem. 2018;293: 6410–6433. 10.1074/jbc.M117.813741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuéllar-Cruz M, Briones-Martin-del-Campo M, Cañas-Villamar I, Montalvo-Arredondo J, Riego-Ruiz L, Castaño I, et al. High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryot Cell. 2008;7: 814–25. 10.1128/EC.00011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sancho J. Flavodoxins: Sequence, folding, binding, function and beyond. Cell Mol Life Sci. 2006;63: 855–864. 10.1007/s00018-005-5514-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lodeyro AF, Ceccoli RD, Pierella Karlusich JJ, Carrillo N. The importance of flavodoxin for environmental stress tolerance in photosynthetic microorganisms and transgenic plants. Mechanism, evolution and biotechnological potential. FEBS Lett. 2012;586: 2917–2924. 10.1016/j.febslet.2012.07.026 [DOI] [PubMed] [Google Scholar]
- 19.Carey J, Brynda J, Wolfová J, Grandori R, Gustavsson T, Ettrich R, et al. WrbA bridges bacterial flavodoxins and eukaryotic NAD(P)H:quinone oxidoreductases. Protein Sci. 2007;16: 2301–2305. 10.1110/ps.073018907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farías-Rico JA, Schmidt S, Höcker B. Evolutionary relationship of two ancient protein superfolds. Nat Chem Biol. 2014;10: 710–715. 10.1038/nchembio.1579 [DOI] [PubMed] [Google Scholar]
- 21.Wang M, Roberts DL, Paschke R, Shea TM, Masters BSS, Kim J-JP. Three-dimensional structure of NADPH-cytochrome P450 reductase: Prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci. 1997;94: 8411–8416. 10.1073/pnas.94.16.8411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grandori R, Carey J. Six new candidate members of the α/β twisted open-sheet family detected by sequence similarity to flavodoxin. Protein Sci. 1994;3: 2185–2193. 10.1002/pro.5560031204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardona F, Orozco H, Friant S, Aranda A, Del Olmo M. The Saccharomyces cerevisiae flavodoxin-like proteins Ycp4 and Rfs1 play a role in stress response and in the regulation of genes related to metabolism. Arch Microbiol. 2011;193: 515–525. 10.1007/s00203-011-0696-7 [DOI] [PubMed] [Google Scholar]
- 24.North M, Tandon VJ, Thomas R, Loguinov A, Gerlovina I, Hubbard AE, et al. Genome-wide functional profiling reveals genes required for tolerance to Benzene metabolites in Yeast. Polymenis M, editor. PLoS One. 2011;6: e24205. 10.1371/journal.pone.0024205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Naseem S, Sharma S, Konopka JB. Flavodoxin-Like proteins protect Candida albicans from oxidative stress and promote virulence. PLOS Pathog. 2015;11: e1005147. 10.1371/journal.ppat.1005147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassan HM, Fridovich I. Intracellular production of superoxide radical and of hydrogen peroxide by redox active compounds. Arch Biochem Biophys. 1979;196: 385–395. 10.1016/0003-9861(79)90289-3 [DOI] [PubMed] [Google Scholar]
- 27.Grandori R, Khalifah P, Boice JA, Fairman R, Giovanielli K, Carey J. Biochemical characterization of WrbA, founding member of a new family of multimeric flavodoxin-like proteins. J Biol Chem. 1998;273: 20960–20966. 10.1074/jbc.273.33.20960 [DOI] [PubMed] [Google Scholar]
- 28.Patridge E V, Ferry JG. WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:Quinone oxidoreductase. J Bacteriol. 2006;188: 3498–3506. 10.1128/JB.188.10.3498-3506.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrade SLA, Patridge E V., Ferry JG, Einsle O. Crystal structure of the NADH:quinone oxidoreductase WrbA from Escherichia coli. J Bacteriol. 2007;189: 9101–9107. 10.1128/JB.01336-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch K, Hromic A, Sorokina M, Strandback E, Reisinger M, Gruber K, et al. Structure, biochemical and kinetic properties of recombinant Pst2p from Saccharomyces cerevisiae, a FMN-dependent NAD(P)H:quinone oxidoreductase. Biochim Biophys Acta—Proteins Proteomics. 2017;1865: 1046–1056. 10.1016/j.bbapap.2017.05.005 [DOI] [PubMed] [Google Scholar]
- 31.Rasheed M, Kumar N, Kaur R. Global secretome characterization of the pathogenic yeast Candida glabrata. J Proteome Res. 2020;19: 49–63. 10.1021/acs.jproteome.9b00299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9: 1–8. 10.1186/1471-2105-9-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z. ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30: 1771–1773. 10.1093/bioinformatics/btu097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bourbonnais Y, Ash J, Daigle M, Thomas DY. Isolation and characterization of S. cerevisiae mutants defective in somatostatin expression: cloning and functional role of a yeast gene encoding an aspartyl protease in precursor processing at monobasic cleavage sites. EMBO J. 1993;12: 285–294. 10.1002/j.1460-2075.1993.tb05655.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gagnon-Arsenault I, Tremblay J, Bourbonnais Y. Fungal yapsins and cell wall: A unique family of aspartic peptidases for a distinctive cellular function. FEMS Yeast Res. 2006;6: 966–978. 10.1111/j.1567-1364.2006.00129.x [DOI] [PubMed] [Google Scholar]
- 36.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, et al. Global analysis of protein palmitoylation in yeast. Cell. 2006;125: 1003–1013. 10.1016/j.cell.2006.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leach MD, Stead DA, Argo E, Maccallum DM, Brown AJP. Molecular and proteomic analyses highlight the importance of ubiquitination for the stress resistance, metabolic adaptation, morphogenetic regulation and virulence of Candida albicans. Mol Microbiol. 2011;79: 1574–1593. 10.1111/j.1365-2958.2011.07542.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Natalello A, Doglia SM, Carey J, Grandori R. Role of flavin mononucleotide in the thermostability and oligomerization of Escherichia coli stress-defense protein WrbA. Biochemistry. 2007;46: 543–553. 10.1021/bi061769c [DOI] [PubMed] [Google Scholar]
- 39.Albrecht A, Felk A, Pichova I, Naglik JR, Schaller M, De Groot P, et al. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J Biol Chem. 2006;281: 688–694. 10.1074/jbc.M509297200 [DOI] [PubMed] [Google Scholar]
- 40.Wolfová J, Mesters JR, Brynda J, Grandori R, Natalello A, Carey J, et al. Crystallization and preliminary diffraction analysis of Escherichia coli WrbA in complex with its cofactor flavin mononucleotide. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63: 571–575. 10.1107/S1744309107026103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li R, Bianchet MA, Talalay P, Amzel LM. The three-dimensional structure of NAD(P)H:quinone reductase a flavoprotein involved in cancer chemoprotection and chemotherapy: Mechanism of the two-electron reduction. Proc Natl Acad Sci. 1995;92: 8846–8850. 10.1073/pnas.92.19.8846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lienhart WD, Gudipati V, Uhl MK, Binter A, Pulido SA, Saf R, et al. Collapse of the native structure caused by a single amino acid exchange in human NAD(P)H:Quinone oxidoreductase. FEBS J. 2014;281: 4691–4704. 10.1111/febs.12975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Encarnación MC, Palomino-Morales RJ, Fuchs JE, Esperanza PG, Noel MT, Salido E, et al. Conformational dynamics is key to understanding loss-of-function of NQO1 cancer-associated polymorphisms and its correction by pharmacological ligands. Sci Rep. 2016;6: 1–13. 10.1038/s41598-016-0001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chelikani P, Donald LJ, Duckworth HW, Loewen PC. Hydroperoxidase II of Escherichia coli exhibits enhanced resistance to proteolytic cleavage compared to other catalases. Biochemistry. 2003;42: 5729–5735. 10.1021/bi034208j [DOI] [PubMed] [Google Scholar]
- 45.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci 2003;100: 13207–13212. 10.1073/pnas.2135385100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. Toward the complete yeast mitochondrial proteome: Multidimensional separation techniques for mitochondrial proteomics. J Proteome Res. 2006;5: 1543–1554. 10.1021/pr050477f [DOI] [PubMed] [Google Scholar]
- 47.Grossmann G, Malinsky J, Stahlschmidt W, Loibl M, Weig-Meckl I, Frommer WB, et al. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J Cell Biol. 2008;183: 1075–1088. 10.1083/jcb.200806035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.López-Otín C, Bond JS. Proteases: Multifunctional enzymes in life and disease. J Biol Chem. 2008;283: 30433–30437. 10.1074/jbc.R800035200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seider K, Brunke S, Schild L, Jablonowski N, Wilson D, Majer O, et al. The Facultative intracellular pathogen Candida glabrata subverts macrophage cytokine production and phagolysosome maturation. J Immunol. 2011;187: 3072–3086. 10.4049/jimmunol.1003730 [DOI] [PubMed] [Google Scholar]
- 50.Rai MN, Balusu S, Gorityala N, Dandu L, Kaur R. Functional genomic analysis of Candida glabrata-macrophage interaction: role of chromatin remodeling in virulence. PLoS Pathog. 2012;8: e1002863. 10.1371/journal.ppat.1002863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crawford DR, Schneider DL. Ubiquinone content and respiratory burst activity of latex-filled phagolysosomes isolated from human neutrophils and evidence for the probable involvement of a third granule. J Biol Chem. 1983;258: 5363–5367. 10.1016/S0021-9258(20)81897-3 [DOI] [PubMed] [Google Scholar]
- 52.Weig M, Jänsch L, Groß U, De Koster CG, Klis FM, De Groot PWJ. Systematic identification in silico of covalently bound cell wall proteins and analysis of protein-polysaccharide linkages of the human pathogen Candida glabrata. Microbiology. 2004;150: 3129–3144. 10.1099/mic.0.27256-0 [DOI] [PubMed] [Google Scholar]
- 53.Ash J, Dominguez M, Bergeron JJM, Thomas DY, Bourbonnais Y. The yeast proprotein convertase encoded by YAP3 is a glycophosphatidylinositol-anchored protein that localizes to the plasma membrane. Journal of Biological Chemistry. 1995. pp. 20847–20854. 10.1074/jbc.270.35.20847 [DOI] [PubMed] [Google Scholar]
- 54.Krysan DJ, Ting EL, Abeijon C, Kroos L, Fuller RS. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4: 1364–1374. 10.1128/EC.4.8.1364-1374.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borah S, Shivarathri R, Kaur R. The Rho1 GTPase-activating Protein CgBem2 Is Required for Survival of Azole Stress in Candida glabrata. J Biol Chem. 2011;286: 34311–34324. 10.1074/jbc.M111.264671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar K, Moirangthem R, Kaur R. Histone H4 dosage modulates DNA damage response in the pathogenic yeast Candida glabrata via homologous recombination pathway. PLoS Genet. 2020;16: e1008620. 10.1371/journal.pgen.1008620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herrou J, Czyz DM, Willett JW, Kim HS, Chhor G, Babnigg G, et al. WrpA is an atypical flavodoxin family protein under regulatory control of the Brucella abortus general stress response system. J Bacteriol. 2016;198: 1281–1293. 10.1128/JB.00982-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47: D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Amino acid sequence identity (%) among Fld-LPs of C. glabrata, S. cerevisiae and C. albicans. Systematic ORF names of Fld-LPs, taken from Candida (http://www.candidagenome.org/) and Saccharomyces (https://www.yeastgenome.org/) Genome Databases, are indicated, with common gene names in brackets. Amino acid identity among protein sequences of these ORFs was determined using BLASTP analysis, with C. glabrata protein as a query sequence, against C. glabrata CBS138, C. albicans SC5314 (Assembly 19) and S. cerevisiae (S288C) reference strains. B. Schematic illustration of CgPst2, CgRfs1, CgPst3 and CgYcp4 protein domain structures, as predicted by the SMART tool 12 (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1). Amino acids spanning the predicted domains are indicated. SMART tool predicted the FMN RED and Flavodoxin_2 domains. Proteins with Flavodoxin-2 domain include bacterial and eukaryotic NAD(P)H dehydrogenase (quinone) enzymes. These enzymes catalyze the NAD(P)H-dependent two-electron reduction of quinones and protect cells against damage by free radicals and reactive oxygen species. FMN RED domain is found in several flavoproteins such as FMN-dependent NADPH-azoreductases, which catalyze the reductive cleavage of azo bond in aromatic azo compounds to the corresponding amines, and NAD(P)H:quinone oxidoreductases, which reduce quinone to the hydroquinone state to prevent interaction of the semiquinone with O2 and production of superoxide. The figure is not drawn to scale.
(TIF)
A. Serial dilution spotting analysis showing growth of C. glabrata mutants deleted for single or multiple CgFld-LP genes on indicated stress conditions. Duroquinone (DQ) and cumene hydroperoxide (CHP) were used at a concentration of 450 μM and 40 μM, respectively. Plates were incubated at 30οC unless indicated otherwise, and images were captured after 24 h for thermal stress (42οC), and 48 h for DQ and CHP. The Q-KO strain lacks four flavodoxin-like proteins, CgPst2, CgRfs1, CgPst3 and CgYcp4. B. Serial dilution spotting analysis showing CgPST2-mediated complementation of menadione (MD; 95 μM) and benzoquinone (BQ; 4 mM) sensitivity of Cgpst2Δ and Q-KO strains. ‘V’ refers to empty vector. Plates were incubated at 30οC, and images were captured after 24 h and 48 h for MD and BQ stresses, respectively.
(TIF)
A. 6X-Histidine-FLAG-CgPst2 purification from Escherichia coli. The E. coli BL21 (DE3) strain transformant carrying pET28a(+)-6XHIS-FLAG-CgPST2 plasmid was grown in LB medium, induced with IPTG (0.5 mM) at 18°C for 16 h, and cells were collected. After cell lysis, the recombinant CgPst2 protein was purified using TALON metal affinity resin via affinity purification. 30 μl eluates along with flow through were resolved on 12% SDS-PAGE, and stained with Coomassie Brilliant Blue. The black arrow marks CgPst2 band. Immunoblot analysis of these fractions with anti-His antibody (1:5000 dilution) is shown at the bottom. M, Protein Marker. B. NADH:quinone oxidoreductase activity measurement of recombinant CgPst2. 50 μg of purified CgPst2 was incubated for 1 h with FMN (100 μM) or FAD (100 μM) on ice, followed by addition of menadione (500 μM) in buffer containing Tris-HCl (10 mM; pH 7.4) and NaCl (150 mM). The reaction was started with NADH (500 μM) addition, and absorbance was recorded at 340 nm in a 1-cm-path-length quartz cuvette over a period of 60 seconds at 10 seconds interval on Spectramax M5 plate reader. The commercially available NAD(P)H:FMN oxidoreductase (1 Unit, Roche, # 10476480001) was taken as positive control. Blank contained no protein. Absorbance of the substrate NADH was considered as 100 at 0 h time point, and NADH oxidation was calculated by dividing the absorbance at each time point by 0 h absorbance, and multiplying the number by 100. Data represent mean ± SEM. Grouped multiple t-test was performed, with n = 3 to 5. **, p < 0.0021; ****, p < 0.0001. C. NADH:quinone oxidoreductase activity measurement in cell extracts of wt, Cgpst2Δ and Q-KO strains in enzymatic reaction mixtures that contained exogenously added FMN (50 μM) or FAD (50 μM). Data represent mean ± SD (n = 2–3). The Q-KO strain lacks four flavodoxin-like proteins, CgPst2, CgRfs1, CgPst3 and CgYcp4. D. NADH:quinone oxidoreductase activity measurement in extracts of Cgpst2Δ and Q-KO cells expressing CgPST2. CgPST2 expression restored the activity deficit in Cgpst2Δ and Q-KO strains. Data represent mean ± SEM. Black and blue asterisks indicate statistically significant differences in activity of QKO and Cgpst2Δ samples, respectively, expressing CgPST2, compared to corresponding strains carrying vector. Grouped multiple t-test was performed, with n = 3. *, p < 0.0332; **, p < 0.0021.
(TIF)
NADH:quinone oxidoreductase activity measurement in extracts of wt, Cgyps1Δ, Cgpst2Δ and Cgpst2Δyps1Δ strains. Data represent mean ± SEM. Grouped multiple t-test was performed, with n = 3. Black and blue asterisks indicate statistically significant activity differences in wt strain, compared to Cgpst2Δ and Cgpst2Δyps1Δ strains, respectively. Brown and orange asterisks indicate statistically significant activity differences in Cgyps1Δ mutant compared to Cgpst2Δ and Cgpst2Δyps1Δ mutants, respectively. *, p < 0.0332; **, p < 0.0021; ***, p < 0.0002.
(TIF)
Confocal images illustrating cell membrane localization of CgPst2-GFP in log-phase cells of indicated strains grown in CAA or CAA medium containing menadione (90 μM; MD) for 90 min. DIC, Differential interference contrast. Bar = 2 μm.
(TIF)
Cell extracts of wt and Cgyps1Δ strains expressing CgPst2-SFB (3 mg protein) were incubated with streptavidin beads for 2 h at 4οC and centrifuged at 2000 rpm for 5 min. Washed Beads were boiled in 2X SDS sample buffer, loaded on 8% (for CgYps1) and 12% (For SFB-tagged CgPst2) SDS-PAGE, and probed with anti-CgYps1 and anti-Flag antibodies, respectively. Please note that CgYps1 was not detectable in Input samples.
(TIF)
Protein sequences of Fld-LPs of C. glabrata, S. cerevisiae, C. albicans, Aspergillus fumigatus, Paracoccidioides brasiliensis, Bacillus cereus, E. coli, Homo sapiens and Arabidopsis thaliana were retrieved from the UniProt database (https://www.uniprot.org/uniprot/), and aligned and coloured using the Clustal Omega server (https://www.ebi.ac.uk/Tools/msa/clustalo/). Sequence alignment, corresponding to 127–198 amino acids, in the C-terminal region of CgPst2 is shown. The red arrow points to the conserved R174 residue in CgPst2. UniProtKB accession numbers are as follows: Q6FM13_CANGA (CgPst2), Q6FRB1_CANGA (CgRfs1), Q6FXR3_CANGA (CgPst3), Q6FS27_CANGA (CgYcp4), PST2_YEAST (S. cerevisiae Pst2), RFS1_YEAST (S. cerevisiae Rfs1), YCP4_YEAST (S. cerevisiae Ycp4), LOT6_YEAST (S. cerevisiae Lot6), A0A1D8PHR5_CANAL (C. albicans Pst1), Q59Y37_CANAL (C. albicans Pst2), A0A1D8PT02_CANAL (C. albicans Pst3), A0A1D8PT03_CANAL (C. albicans Ycp4), Q4WKD8_ASPFU (Aspergillus fumigatus Pst2), Q8X194_PARBR (Paracoccidioides brasiliensis y20), Q81G57_BACCR (Bacillus cereus Nrdl), NQOR_ECOLI (E. coli wrbA), NQO1_HUMAN (Homo sapiens NQO1), FQR1_ARATH (Arabidopsis thaliana FQR1).
(TIF)
CgGapdh was used as a loading control. M, Protein Marker.
(TIF)
A. Immunoblot analysis of CgPst2 expression in cell extracts of indicated C. glabrata strains to check the specificity of anti-CgPst2 polyclonal antibody. YPD-grown log-phase cells were lysed using glass beads, and 60 μg protein was resolved on 12% SDS-PAGE. After transferring proteins to the PVDF membrane, the blot was probed with antibody (1:1000 dilution) raised against purified CgPst2 protein in BALB/c mice. This mouse anti-CgPst2 sera yielded no and good signal in cell lysates of Q-KO (lacks all four flavodoxin-like proteins, CgPst2, CgRfs1, CgPst3 and CgYcp4), and Cgpst2Δ and Cgrfs1Δ mutants, respectively, indicating that the generated antibody binds to both CgPst2 and CgRfs1 proteins. CgGapdh was used as a loading control. B. qPCR-based determination of CgPST2 transcript levels. Log-phase T-KO (Cgrfs1Δpst3Δycp4Δ) and T-KOyps1Δ (Cgrfs1Δpst3Δycp4Δyps1Δ) cells were either left untreated (UT) or treated with 90 μM menadione for 90 min (T). Data (mean ± SEM, n = 3) were normalized against the CgACT1 mRNA control, and represent fold change in CgPST2 expression in treated samples, compared to corresponding untreated samples (taken as 1.0). C. Immunoblot analysis of CgYps1 expression in cell extracts of untreated or menadione-(MD; 90 μM for 90 min)-treated log-phase cells of the wt strain, using anti-CgYps1 antibody. CgGapdh was used as a loading control. Of note, CgYps1 is predicted to be highly glycosylated which may account for diffuse nature of the CgYps1 band. Consistent with predicted posttranslational modifications, the CgYps1 band corresponds to about 135 kDa, as compared to the expected size of 64 kDa. D. Immunoblot analysis showing CgYps1-CgPst2 interaction in log-phase untreated and MD (90 μM for 90 min)-treated Q-KO cells expressing CgPST2. Immunoprecipitation was carried out with anti-CgYps1 antibody, and blots were probed with anti-CgYps1 and anti-CgPst2 antibodies. Untreated penta-KO strain expressing vector (V) was used as negative control. The green asterisk denotes non-specific band. The Q-KO strain lacks four flavodoxin-like proteins (Fld-LPs), CgPst2, CgRfs1, CgPst3 and CgYcp4, while the penta-KO strain lacks CgYps1 along with four Fld-LPs, CgPst2, CgRfs1, CgPst3 and CgYcp4.
(TIF)
A. The Pichia pastoris GS115 strain expressing secretory forms of either catalytically active (rCgYps1) and inactive CgYps1 (rCgYps1D91A) proteins was grown in the BMGY medium at 30°C for 24 h and incubated in the BMMY medium containing 2% methanol for 48 h. Methanol was used to induce the AOX1 promoter, which drives the expression of CgYps1 proteins. 2% Methanol was added every 24 h to maintain the induction and expression of CgYps1 proteins. After 48 h, 1 ml supernatant was precipitated with 80% ammonium sulphate, pellet was suspended in PBS, and CgYps1 protein induction was checked by SDS PAGE. Induced proteins were purified by anion exchange chromatography using DEAE cellulose resin. The eluate fractions were resolved on 12% SDS-PAGE. The orange asterisk marks CgYps1 band. The protein samples were also probed with anti-His antibody (1:5000 dilution) and shown underneath the gel images. M, protein marker. B. CgYps1-mediated proteolysis of gelatin. The YNB-agar medium containing 1% gelatin was incubated either with 100 μg of purified proteins (rCgYps1 and rCgYps1D91A) or citrate buffer (pH 4.0; used for enzyme elution) for overnight at 37°C. The plate was stained with Coomassie Brilliant Blue G250 and imaged. The zone of hydrolysis was observed only with the CgYps1 enzyme. C. Proteolytic activity of DEAE cellulose-purified rCgYps1 and rCgYps1D91A proteins (20 μg) was measured using 2.5% hemoglobin as a substrate in the citrate buffer (100 mM; pH 4.0) for 30 min at 37°C. TCA (10%) was added to stop the enzymatic reaction and precipitate the undigested hemoglobin and CgYps1 proteins. Cleaved peptides were collected from the supernatant and incubated with sodium bicarbonate (5 mM) and Folin-Ciocalteu reagent at 37°C for 30 min. The absorbance was read at 660 nm and converted into the μ moles of tyrosine released per mg of the protein. Control indicates the enzymatic reaction mixture containing buffer and substrate, but no CgYps1 protein. Data represent mean ± SEM of 4 independent samples. Of note, rCgYps1D91A showed no proteolytic activity, indicating that Aspartate-91 in CgYps1 is required for enzyme activity. **, p < 0.01; unpaired two-tailed Student’s t test.
(TIF)
Immunoblot analysis showing no interaction between CgPst2 and CgYcp4-GFP. 6 mg precleared lysates of untreated and MD (90 uM for 90 min)-treated T-KO (Cgrfs1Δpst3Δycp4Δ) and T-KOyps1Δ (Cgrfs1Δpst3Δycp4Δyps1Δ) cells expressing CgYCP4-GFP were incubated with anti-GFP antibody-conjugated beads, followed by Western analysis with anti-GFP and anti-CgPst2 antibodies. The red and green arrows mark GFP and CgYcp4-GFP protein bands, respectively, in IP samples, while the green asterisk denotes non-specific band. Please note that Ycp4-GFP expression was not observed in Input samples, despite loading 200 μg protein.
(TIF)
Immunoblot analysis showing enrichment of CgYps1 and CgPst2 in the plasma membrane fraction of the Q-KO strain expressing CgPST2. Whole-cell lysates and plasma membrane fractions (60 μg and 200 μg for CgPst2 and CgYps1, respectively), prepared by glass bead lysis and sucrose gradient ultracentrifugation, respectively, were resolved on 4–20% polyacrylamide gradient, 12% polyacrylamide, 12% polyacrylamide and 12% polyacrylamide gels for CgYps1, CgPst2, CgPma1 and CgGapdh, respectively, and probed with anti-CgYps1, anti-CgPst2, anti-Pma1 and anti-Gapdh antibodies. The penta-KO strain was used as a negative control.
(TIF)
A. Immunoblot analysis of CgPst2 levels in Q-KO and penta-KO strains expressing CgPST2 or CgPST2R174A. Whole-cell extracts (60 μg), prepared by glass bead lysis, were resolved on 12% SDS-PAGE and probed with anti-CgPst2 and anti-Gapdh antibodies. The intensity of individual bands in 4 independent Western blots was quantified using the ImageJ densitometry software, and CgPst2 signal was normalized to the corresponding CgGapdh signal. Fold-change (mean ± SEM) in CgPst2 levels in CgPST2R174A-expressing cells, compared to CgPST2-expressing cells (considered as 1.0), is shown underneath the blot. p ≤ 0.01; paired two-tailed Student’s t test. The green asterisk indicates non-specific band. B. Immunoblot analysis of CgPst2 expression in penta-KO strains expressing CgPST2ΔR174-F198 or CgPST2ΔS175-F198. Whole-cell extracts (60 μg) of indicated strains were prepared by glass bead lysis and resolved on 18% SDS-PAGE for 4–5 h. The blots were probed with anti-CgPst2 and anti-Gapdh antibodies.
(TIF)
A. NADH:quinone oxidoreductase activity measurement in extracts of Q-KO expressing either CgPST2 or CgPST2-CΔS175-F198. Data represent mean ± SEM. Grouped multiple t-test was performed, with n = 3 to 4. *, p < 0.0332; **, p < 0.0021; ***, p < 0.0002; ****, p < 0.0001. The Q-KO strain lacks four flavodoxin-like proteins, CgPst2, CgRfs1, CgPst3 and CgYcp4. B. Immunoblot analysis of CgPst2 levels in Q-KO strains expressing CgPST2 or CgPST2-CΔS175-F198. The green asterisk denotes non-specific band.
(TIF)
A-C. 6X-Histidine-FLAG-CgPst2R174A A., 6X-Histidine-FLAG-CgPst2-CΔR174-F198 B. and 6X-Histidine-FLAG-CgPst2-CΔS175-F198 C. protein purification from E. coli. The E. coli BL21 (DE3) strain transformants carrying pET28a(+)-6XHIS-FLAG-CgPST2R174A, pET28a(+)-6XHIS-FLAG-CgPST2-CΔR174-F198 and pET28a(+)-6XHIS-FLAG-CgPST2-CΔS175-F198 plasmids were grown in LB medium, induced with IPTG (0.5 mM) at 18οC for 16 h, and cells were collected. After cell lysis, the recombinant proteins were purified using TALON metal affinity resin via affinity purification. 30 μl eluates, representing purified rCgPst2R174A A. rCgPst2-CΔR174-F198 B. and rCgPst2-CΔS175-F198 C. proteins, along with the flow through, were resolved on 12% SDS-PAGE, and stained with Coomassie Brilliant Blue. The green arrow marks CgPst2 band. M, Protein Marker. D-E. NADH:quinone oxidoreductase activity measurement of the recombinant CgPst2R174A D. and CgPst2-CΔR174-F198 E. protein (250 μg) was measured, as described in the legend of S3B Fig. Blank contained no protein. Data represent mean ± SEM. Grouped multiple t-test was performed, with n = 3. Black and blue asterisks indicate statistically significant activity differences between the positive control [NAD(P)H:FMN oxidoreductase (1 Unit, Roche, # 10476480001)] and recombinant proteins incubated with FMN (100 μM) and FAD (100 μM), respectively. *, p < 0.0332. Please note that the enzymatic activity of the purified recombinant CgPst2-CΔS175-F198 protein could not be determined, as it formed precipitates during the assay reaction. F. Size exclusion chromatogram of CgPst2-CΔS175-F198. After loading 300 μg of purified 6XHIS-FLAG-CgPst2-CΔS175-F198 protein on the Sephacryl S-200 column, protein elution profiles were determined using the absorbance values at 280 nm. The CgPst2-CΔS175-F198 protein could not bind to the column, and appeared as aggregates in the column’s void volume, as determined by column calibration with blue dextran. AU, Arbitrary Units.
(TIF)
300 μg whole cell lysates of Q-KO- expressing vector pRK74 (V) or CgPST2, and penta-KO expressing CgPST2-CΔR174-F198, CgPST2R174A and CgPST2 were resolved in a discontinuous Tris-glycine buffer system under non-denaturing conditions, and probed with anti-CgPst2 antibody. The native protein molecular weight marker (M) was stained with coomassie brilliant blue, and is shown on the right side of the blot. The ponceau S-stained membrane is shown as loading control.
(TIF)
In vitro cleavage assay. The Pichia pastoris GS115 strain expressing either CgYps1 or CgYps1D91A was induced with 2% methanol for 48 h, and pelleted down at 14000 rpm for 10 min at 4°C. The supernatants containing CgYps1 and CgYps1D91A were filtered through a 0.22 μM filter (Millipore, USA), and concentrated using Amicon Ultra centrifugal filter unit (3 kDa cutoff). The retentate was precipitated with ammonium sulphate (100% saturation) for 10 min at 4°C, followed by centrifugation and pellet suspension in citrate buffer (pH 4.0). For the cleavage assay, 30 μg of r6XHis-Flag-CgPst2 was incubated with 60 μg of partially purified rCgYps1 or rCgYps1D91A enzymes at 37°C for 4 h in citrate buffer (pH 4.0). Digested samples were run on 18% SDS-PAGE and probed with anti-His antibody. The assay samples containing both CgYps1 and CgPst2 displayed an additional faster migrating CgPst2 protein band (~ 24 kDa; marked with orange asterisk), probably representing the N-terminal fragment of the cleaved form of CgPst2, which was absent in reaction mixtures containing either CgPst2 alone or both CgPst2 and CgYps1D91A. These data indicate that the catalytically active CgYps1 can cleave CgPst2. Of note, we could not detect the small C-terminal-cleaved CgPst2 fragment, as hexa-His epitope is present at the N-terminus. To estimate the size difference between the cleaved and un-cleaved form of CgPst2, E-coli-purified full-length CgPst2 and CgPst2-CΔR174-F198 (50 μg) were run on 18% SDS-PAGE as a control, and probed with anti-his antibody. M, Protein Marker.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
The raw mass spectrometry proteomics data are deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD022766. All other relevant data are within the manuscript and its Supporting Information files.