ABSTRACT
Metabolic network reconstructions have become an important tool for probing cellular metabolism in the field of systems biology. They are used as tools for quantitative prediction but also as scaffolds for further knowledge contextualization. The yeast Saccharomyces cerevisiae was one of the first organisms for which a genome-scale metabolic model (GEM) was reconstructed, in 2003, and since then 45 metabolic models have been developed for a wide variety of relevant yeasts species. A systematic evaluation of these models revealed that—despite this long modeling history—the sequential process of tracing model files, setting them up for basic simulation purposes and comparing them across species and even different versions, is still not a generalizable task. These findings call the yeast modeling community to comply to standard practices on model development and sharing in order to make GEMs accessible and useful for a wider public.
Keywords: genome-scale metabolic models, yeast species, systems biology, accessibility, usability, interoperability
Review on computational models of metabolism for diverse yeast species, their development history, applications and critical assessment of their accessibility, usability and interoperability.
INTRODUCTION
Genome-scale metabolic model reconstruction has been established as one of the major modeling approaches for systems-level metabolic studies (Gu et al. 2019). These models are mainly built in a bottom-up approach, in which genome information is combined with the accumulated knowledge about the metabolic capabilities of a living organism to reconstruct a complete metabolic map (Nielsen 2017). Another widely used approach for model reconstruction consists of the use of one or multiple well-curated networks as scaffolds, due to the high degree of conservation of metabolism for phylogenetically close species. Metabolic models have been proven to be useful as knowledge databases (Herrgård et al. 2008), tools for contextualization of omics data (Kerkhoven et al. 2016) and for guiding metabolic engineering projects (Meadows et al. 2016), enabling systematic explorations of the relationship between genotypes and phenotypes.
The metabolic model iFF708 (Förster et al. 2003) of Saccharomyces cerevisiae, the genome of which was the first eukaryotic one to be sequenced (Goffeau et al. 1996), was the first published GEM for its entire domain in 2003. This model has been used as a scaffold for further network refinements (Duarte, Herrgård and Palsson 2004; Kuepfer, Sauer and Blank 2005; Herrgård et al. 2006; Nookaew et al. 2008), which has facilitated the development of metabolic models for several other budding yeast species over the years, due to their well evolutionarily-conserved metabolic capabilities (Shen et al. 2018).
Multiple model reconstructions exist, not just for S. cerevisiae, but for several other yeast species. These reconstructions have usually been carried out by different research groups, resulting in specific network improvements according to their scientific interests, but at the same time yielding incompatible identifiers for reactions and metabolites hampering any systematic comparison and evaluation across models (Herrgård et al. 2008).
As GEMs are valuable tools for a wide variety of applications, their end users vary from academic researchers with different backgrounds and levels of computational skills, to professionals in the biotechnology and pharmaceutical industries. Therefore, there is a strong need for computational metabolic models to be accessible and published in a ready-to-use format, which facilitates their utilization by non-expert users. Additionally, the use of consistent and standardized identifiers for their components enables comparisons across models, thus simplifying the process of finding the best model for a given application.
Latest developments on yeasts GEMs
The development, interconnections and applications of metabolic models for different yeast species have been reviewed extensively (Sánchez and Nielsen 2015; Lopes and Rocha 2017; Castillo, Patil and Jouhten 2019; Chen, Li and Nielsen 2019) however, the list of yeast GEM models is continuously increasing both in number of GEMs and encompassed species. Here we briefly summarize the development history of all models for diverse yeast species that are currently available in the scientific literature. The validation strategies and main applications of these models, as described in their original publications, are provided in Table S3 (Supporting Information), indicating the type of biological data and computational methods used for each case.
S. cerevisiae is one of the most studied organisms in the Eukarya domain, which has resulted in a long modeling history with 18 networks currently available. The models iND750 (Duarte, Herrgård and Palsson 2004), iLL672 (Kuepfer, Sauer and Blank 2005) and iIN800 (Nookaew et al. 2008) were directly derived from iFF708 (Förster et al. 2003) and subsequently used as templates for iMH805/775 (Herrgård et al. 2006), iMM904 (Mo, Palsson and Herrgård 2009), iAZ900 (Zomorrodi and Maranas 2010) and iTO977 (Österlund et al. 2013) reconstructions.
As these multiple reconstructions added new knowledge and gap-fills to the network, a first attempt of unification was carried out by the knowledge base Yeast1, published in 2008 (Herrgård et al. 2008). The concept of standardized identifiers for reactions and metabolites was first implemented in this reconstruction, but simulation capabilities were not achieved. Sequential curation iterations were performed (Yeast2 and Yeast3) until the publication of Yeast4, which notably increased the network connectivity and the number of included metabolites, making it a suitable model for simulation purposes (Dobson et al. 2010). Further updates to the consensus metabolic network have shown to improve predictions on gene essentiality, induced auxotroph phenotypes and cellular growth on diverse environments (Yeast5 (Heavner et al. 2012), Yeast6 (Heavner et al. 2013) and Yeast7 (Aung, Henry and Walker 2013)). In 2019, a new version of the consensus metabolic network, Yeast8, was published (Lu et al. 2019), its reconstruction process combined information from previous GEMs, different curated databases such as KEGG (Kanehisa et al. 2016), SGD (Hellerstedt et al. 2017), BioCyc (Karp et al. 2019), Reactome (Fabregat et al. 2018) and UniProt (The UniProt Consortium 2017) and experimental data on substrate usage. Furthermore, Yeast8 provides an ecosystem of multilayer models suited for different kinds of phenotype predictions, ranging from 1011 strain-specific models to incorporation of enzyme constraints (ecYeast8) and protein 3D structures (proYeast; Lu et al. 2019).
In parallel with the development of the consensus network, iSce926 (Chowdhury, Chowdhury and Maranas 2015) was derived from Yeast7 (Aung, Henry and Walker 2013) in 2015, incorporating gene essentiality and synthetic lethality information to curate gene-reaction rules. The model iSc-AMRS-1 (Wichmann et al. 2016) was developed from iLL672 (Kuepfer, Sauer and Blank 2005) in 2016, mainly by curation of proton balancing for mitochondrial ATP production and reaction reversibility, aiming to improve flux distribution predictions in order to investigate production of isopropenoids.
The model SpoMBEL1693 for Schizosaccharomyces pombe, a model organism for eukaryotic cell cycle studies, was developed in 2012 using annotated genes and reactions from the KEGG database as a draft network (Sohn et al. 2012). iNX804, a metabolic model for Candida glabrata, known as a platform organism for pyruvate production, was reconstructed in 2013 and used for identification of gene targets for enhanced production of pyruvate-derived fine chemicals (Xu et al. 2013). The metabolism of Candida tropicalis, known as a promising host for α, ω-dicarboxylic acids production, has been studied with the model iCT646, reconstructed through the collection of multiple database information in 2016 (Mishra et al. 2016). The model iOD907, a metabolic network for Kluyveromyces lactis, a yeast commonly used in the dairy industry, was published in 2014 (Dias et al. 2014). Its reconstruction process used iMM904, for S. cerevisiae, as a scaffold and merged it with annotation for metabolic genes and transporters from KEGG (Kanehisa et al. 2017) and TCDB (Saier et al. 2016), respectively. This model was validated with data for growth on diverse carbon sources and used to investigate phenotypic differences for single gene knockout strains between K. lactis and S. cerevisiae (Dias et al. 2014).
Pichia pastoris is an established workhorse in biotechnology for heterologous protein production, as it shows superior protein secretion efficiency compared with other yeasts (Schmidt 2004). Additionally, humanized N-glycosylation patterns for recombinant protein production can be obtained by engineering its metabolism. The first two GEMs for P. pastoris, PpaMBEL1254 (Sohn et al. 2010) and iPP668 (Tomàs-Gamisans, Ferrer and Albiol 2016), were both developed in 2010 using genome annotation information from databases and literature. In 2015, ihGlycopastoris (Irani et al. 2016) was specially developed for simulation of recombinant protein production as a target, by combining the previously established iLC915 (Caspeta et al. 2012) model with humanized N-glycosylation pathways. This allowed the investigation of the influence of N-glycosylation processes on protein production and the model was used for the prediction of gene overexpression targets for improving protein yields. The model Kp.1.0 was published in 2017, in which 12 different biomass compositions were tested under different growth conditions, showing minor effects on growth and gene essentiality predictions, but drastic changes in flux distributions (Cankorur-Cetinkaya, Dikicioglu and Oliver 2017). A total of three previous P. pastoris reconstructions (Chung et al. 2010; Sohn et al. 2010; Caspeta et al. 2012) were merged into iMT1026 (Tomàs-Gamisans, Ferrer and Albiol 2016), expanding the representation of fatty acid and sphingolipid metabolism, intact N-glycosylation, O-glycosylation and glycosylphosphatidylinositol(GPI)-anchor pathways. iMT1026 was then curated to iMT1026.v3 in 2018, leading to a refinement of predictions for cellular growth on glycerol and methanol as carbon sources (Tomàs-Gamisans, Ferrer and Albiol 2018). Additionally, the model iRY1243 was created in 2017 by merging iPP668, PpaMBEL1254, iLC915 and iMT1026, also incorporating curation of biosynthesis of vitamins and cofactors, which added more than 200 metabolic genes to the network. This model was validated with the use of RNAseq data for different conditions, utilization of carbon and nitrogen sources and 13C-labeled derived fluxomics, yielding an overall high consistency of predictions for essential genes, flux distributions and different mutant phenotypes (Ye et al. 2017).
The yeast Scheffersomyces stipitis (formerly known as Pichia stipitis) has raised interest due to its great native potential for xylose utilization. In 2012, three models were published for this species: iTL885 (Liu et al. 2012) and iSS884 (Caspeta et al. 2012) were derived from previous S. cerevisiae’s models, whilst iBB814 (Balagurunathan et al. 2012) was reconstructed from genome annotation extracted from various databases. A modified version of iBB814, the model iDH814, was published in 2016 and used to elucidate the redox balance shift response to reduced oxygen supply conditions (Hilliard et al. 2018). As these four reconstructions just account for the cytoplasm, mitochondria and peroxisome as cellular compartments, a fully compartmentalized model for this relevant organism is still missing.
The oleaginous yeast Yarrowia lipolytica, is another organism for which multiple GEMs already exist. Its first model, iNL895 developed in 2012 (Loira et al. 2012) and other two following models iMK735 (Kavšcek et al. 2015) and iYali4 (Kerkhoven et al. 2016), were derived from previous networks of the phylogenetically distant yeast S. cerevisiae, in contrast to iYL619_PCP (Pan and Hua 2012), reconstructed directly from Y. lipolytica specific information available in public databases and literature. In 2018, iYLI647 (Mishra et al. 2018) was developed using a previous reconstruction for the same species, iMK735 (Kavšcek et al. 2015), as a scaffold and expanded to include the ω-oxidation pathway that converts fatty acids to long-chain dicarboxylic acids (DCAs), the subsequent fatty-acid degrading β-oxidation pathway and branched-chain amino acid degradation pathways, in order to guide simulation of metabolic engineering strategies for enhanced DCA production.
During these years, other non-conventional yeasts have gained more attention due to their fascinating and diverse phenotypes. Several GEMs have been constructed as an attempt to understand their particular traits. Rhodotorula toruloides is an oleaginous yeast, which can accumulate lipids up to 70% of its dry mass (Ratledge and Wynn 2002). Previous modeling approaches have explored the use of constraint-based methods together with a reduced metabolic network for this organism to assess lipid accumulation on different substrates (Bommareddy et al. 2015; Castañeda et al. 2018), but its first genome-scale model, rthoGEM (Tiukova et al. 2019), was published in 2019. Cell growth data using glucose, xylose and glycerol as substrates were used to validate the model, while gene targets for triacylglycerol and carotenoid production were predicted with the use of the FSEOF algorithm (Choi et al. 2010). That same year, iRhto1108 (Dinh et al. 2019), was developed using Yeast7 and the Kbase fungal metabolic network (Arkin et al. 2018) as model templates. This model increased the metabolic gene coverage in comparison to rthoGEM (from 926 to 1108) and enabled growth simulations using arabinose and cellobiose as carbon sources.
Zygosaccharomyces bailii has been described to have high tolerance towards acetic acid (Palma et al. 2017; Palma, Guerreiro and Sá-Correia 2018). It has been suggested that the Zygosaccharomyces clade diverged from Saccharomyces ancestors just before the whole genome duplication event (WGD; Kurtzman 2003), which took place approximately 100 million years ago, making the Zygosaccharomyces genus the closest pre-WGD ancestral group of relatives to study the genome evolution of S. cerevisiae (Hagman et al. 2013; Solieri et al. 2013). The model ZyPa1 (Filippo et al. 2018) was reconstructed using homology information from 20 different yeasts belonging to the Saccharomycetaceae family, and was then connected to the KEGG database to obtain a draft network. Stoichiometry and localization information for the reactions were extracted from the models Yeast7 (Aung, Henry and Walker 2013) and iOD907 (Dias et al. 2014). ZyPa1 contains 2413 genes, more than twice the number of genes in Yeast8 (Lu et al. 2019), being the metabolic model for a yeast species with the highest number of genes. This GEM has been applied to the study of cellular growth under co-consumption of lactate and glucose.
Kluyveromyces marxianus is a thermotolerant yeast that can even tolerate temperatures as extreme as 52°C (Nonklang et al. 2008), making it a specially interesting organism host for industrial bioproduction. The first GEM for K. marxianus, iSM996, was built in 2019 (Marcišauskas, Ji and Nielsen 2019) by using a draft model generated with the RAVEN Toolbox (Wang et al. 2018), aided by the KEGG database and the models iOD907 (Dias et al. 2014) and Yeast7 (Aung, Henry and Walker 2013) as sources for the network gap-filling process. iSM996 was validated using data on carbon and nitrogen source usage, and transcriptome datasets were integrated in order to simulate growth under different temperatures (Marcišauskas, Ji and Nielsen 2019).
Lachancea kluyveri is a weak Crabtree positive yeast of industrial relevance due to its capabilities for ethyl-acetate secretion, when cultivated in aerobic batch conditions, and usage of urea and uracil as sole nitrogen sources for growth. In 2020, the model iPN730 (Ghosh et al. 2020) was built on a Kbase workspace (Arkin et al. 2018) using iMM904 (Mo, Palsson and Herrgård 2009) for S. cerevisiae as a template network and other 13 fungi models as references for homologous reactions searches. The model was validated by simulating cellular growth on diverse environments (Ghosh et al. 2020).
A repository for yeast species metabolic models
All aforementioned yeasts GEMs, together with the previously published models, were used to query the literature using the keyword ‘yeast’ together with ‘metabolic model’, ‘GSM’, ‘GEM’ or ‘GENRE’ (genome-scale network reconstruction). In total, 43 model files for 12 different organisms were found either as part of publications in peer-reviewed journals, supplementary files for preprint articles in bioRxiv, or in the yeastnet model database (https://sourceforge.net/projects/yeast) when no specific publication about their reconstruction was found (as in the case of Yeast2, Yeast3 and Yeast4). Most of these yeast species belong to the Saccharomycetales order in the Ascomycota phylum, but some of them have been classified as part of other classes, as Schizosaccharomyces pombe (Schizosaccharomycetes) or even phyla, such as the Basidiomycota fungus Rhodotorula toruloides (Table S1, Supporting Information).
As expected, S. cerevisiae is the yeast species for which the most GEMs have been reconstructed, however multiple models are also available for P. pastoris, Y. lipolytica and S. stipitis (Fig. 1A). This collection of model files has been stored in a publicly available GitHub repository at https://github.com/SysBioChalmers/YeastsModels, together with the necessary scripts for their further analysis. The search and exploration processes for these models pointed out several aspects that can be classified into three main categories: accessibility, usability and interoperability.
Figure 1.
Accessibility of metabolic models for diverse yeast species. (A) Number of published models per species. (B) Number of published models per file format. Models available in several formats are counted multiple times. *NA indicates model files that were not available in either their original publications or external model repositories (C) Proportion of models provided as an SBML file in their original source or publication. (D) Proportion of yeast models stored in different public databases. Models stored in several databases are just accounted as part of the one that uploaded them first. (E) Proportion of models with continuous development tracked on public repositories.
Model accessibility
The analyzed models in this review span more than 17 years of active research, in which standards for file formats and sharing practices in the field of systems biology have changed, making the retrieval of their original files a time-consuming and not automatable task. Even though the Systems Biology Markup Language (SBML) was released in 2002 (Hucka et al. 2003), and since then has evolved to become the standard file format for metabolic modeling, 27% of the analyzed models were shared in a different format in their original publications, such as .txt, .XLS and .pdf (Fig. 1B and C), which limits scientific exchange and reproducibility of results on different setups due to their dependence on specific software applications (Ravikrishnan and Raman 2015).
As not all models could be successfully obtained from their original sources, models were also sought in other public repositories such as Biomodels (Chelliah et al. 2015), Biomet (Garcia-Albornoz et al. 2014) and openCOBRA models (Ebrahim et al. 2015; Fig. 1D), which contain curated metabolic reconstructions not just for yeast species but for all key phylogenetic groups (Monk, Nogales and Palsson 2014). The models from the last decade present in this catalogue reflect the trend of referring to unambiguous entries in such databases instead of uploading model files as supplementary material to their respective journal websites.
Notably, a novel methodology for model sharing and development has been proposed by the Yeast8 project (Lu et al. 2019) and the Memote model test suite (Lieven et al. 2020), which with the aid of version control tools, such as Git and GitHub, provides not just the final snapshot of a GEM but its whole development history, offering also a web platform for open and continuous development. These version control tools have also been implemented for Y. lipolytica, K. marxianus and R. toruloides GEMs (iYali4 (Kerkhoven et al. 2016), iSM996 (Marcišauskas, Ji and Nielsen 2019), rthoGEM (Tiukova et al. 2019) and iRhto1108 (Dinh et al. 2019)), which represent 11% of the collected models (Fig. 1E). More community-driven modeling efforts are expected to emerge in the next years as a way to circumvent the drawback of having multiple independent reconstructions available for some of these yeast species.
Model usability
In order to evaluate the complexity of the process of getting started when utilizing a GEM, a testing pipeline was developed using the RAVEN (Wang et al. 2018), COBRA (Heirendt et al. 2019) and COBRApy (Ebrahim et al. 2013) toolboxes, which in a series of sequential steps aims to obtain feasible flux balance analysis simulations (Orth, Thiele and Palsson 2010), with cellular growth maximization as an objective function, assuming that no prior knowledge about the model´s specific structure and identifiers was available. In total, SBML files for 37 models were found available in this study, and therefore analyzed by the mentioned pipeline.
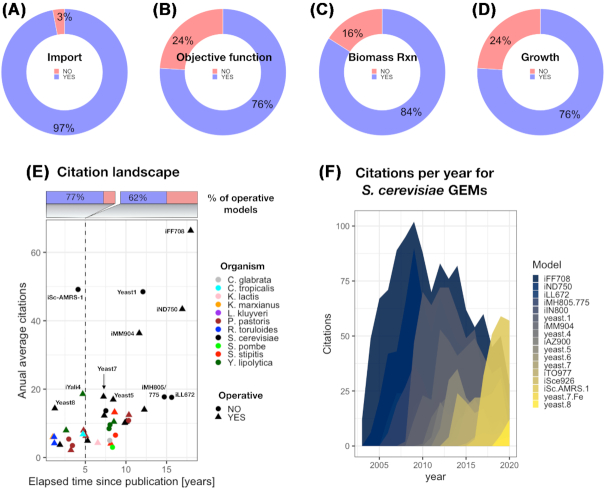
The first tested functionality was the importability of each SBML model into a non-empty MATLAB structure (Table S2, Supporting Information). This was satisfactorily achieved for the majority of these models, 97%. The only non-loadable SBML file was also tested with the COBRApy toolbox, but its import could not be accomplished due to parsing errors. Secondly, a default objective function was sought in the model structure by retrieving any non-zero coefficient in the objective function field or so called ‘c vector’. Of the analyzed models, 76% showed a predefined objective function. Further exploration found that all of these objectives are maximization of the growth rate, ‘biomass exchange’ or ‘biomass formation’. Taking this into account, traceability of a biomass pseudoreaction was also evaluated. For doing so, the presence of the substrings ‘growth’, ‘biomass’ and ‘vgro’ was explored in the model.rxns and model.rxnNames fields. In total 84% of the tested models contain a biomass pseudoreaction identifiable with the used patterns. This does not imply that a biomass reaction is absent for the 16% remaining models, but that the search for it would require a customized manual procedure for each of them.
For all of these models, maximization of the found biomass reaction was set as an objective function and all of their exchange reactions were opened in both directions (lower and upper bounds of −1000 and 1000 mmol/gDw h, respectively) to check in silico cellular growth capabilities. In total, 76% of the tested subset (28 models) showed a non-zero growth rate when subject to these constraints. We consider these models as available in a ready-to-use setup, as no further steps or manual inspection was needed to simulate growth. Detailed information for the evaluated metrics and features can be found in Table S2 (Supporting Information).
In order to assess the utilization of these models by the scientific community, the total and average annual citations were used as proxy metrics. Figure 2E shows that a larger proportion of the cited models that were recently published (<5 years ago) have been made available in a ready-to-use format (77%) in comparison to those that were published a longer time ago (62%). For the S. cerevisiae network reconstructions, it is clear that older models are on average more used or referred to in the scientific literature. However, as time has passed more models have become available and decays on citations for older models usually coincide with publication and rise of newer ones (Fig. 2F). This might suggest that scientific interest shifts towards more recent models as they accumulate the knowledge gathered by previous reconstruction iterations.
Figure 2.
Model usability. (A) Proportion of tested SBML models successfully imported with the RAVEN, COBRA or COBRApy toolboxes (total = 37 models). (B) Proportion of tested models with a default objective function. (C) Proportion of tested models with a biomass pseudoreaction identifiable with the substrings ‘biomass’, ‘growth’ or ‘vgro’. (D) Proportion of models yielding a non-zero growth rate according to the developed testing pipeline. (E) Citation landscape of models of yeasts metabolism. Annual average citations vs elapsed time since publication per species, the proportion of ‘operative models’ (available in a ready-to-use format, according to the developed testing pipeline) is indicated in the upper part for models that have been published more or less than 5 years ago. (F) Evolution of the annual citations for models of S. cerevisiae metabolism. Citations were queried from Google scholar, accessed on September 4th, 2020.
Interoperability
As described above and repeatedly concluded (Dräger and Palsson 2014; Ebrahim et al. 2015; Heavner and Price 2015; Sánchez and Nielsen 2015; Mendoza et al. 2019), the lack of identifier consistency and connection to external databases for all of the relevant components of GEMs (metabolites, reactions, genes and cellular compartments) together with the use of non-standardized file formats, are the main obstacles for direct model comparison and assessment, even across reconstructions for a single species.
In order to aid systematic model development, according to community-agreed practices, a standardized set of metabolic model tests (Memote) has recently been developed as an open-source software suite (Lieven et al. 2020). Memote tests are divided into organism- and model-specific ones, not applicable to all reconstructions, and a section of independent tests, which check for model consistency (in terms of mass and charge balance, metabolite connectivity and stoichiometric consistency), and annotation, or connection to external databases, for metabolites, reactions, genes and SBO terms (systems biology ontology terms; Courtot et al. 2011). This pipeline assigns a numerical score, based on the specific model characteristics, to each of the independent tests, relevant for comparing evolution of particular model features across versions.
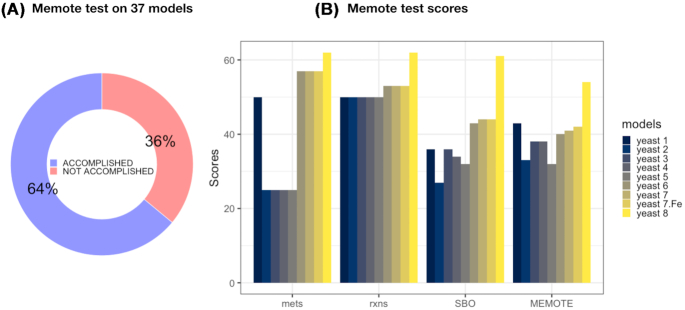
The 37 SBML model files analyzed above were furthermore tested by the Memote suite. As this software relies on the latest version of the SBML Level 3 Flux Balance Constraints package (Olivier and Bergmann 2018), not all of the models could be tested due to parsing errors for those available in previous or conflicting SBML versions (36%), as shown in Fig. 3A. Noteworthy, this is not an indicator of model quality or predictive performance, but rather one of compliance with model format standards. Further details for all of the individual tests and computed scores are available as HTML reports and also as part of Table S2 (Supporting Information), both stored in the aforementioned GitHub repository.
Figure 3.
Memote tests results. (A) Proportion of models for which the automated Memote test was accomplished. (B) Memote test scores for the consensus reconstructions of the S. cerevisiae metabolic network. Scores for metabolites, reactions and SBO terms evaluate the degree of annotation for such components with external databases identifiers that can facilitate the traceability of a component across different model versions. The Memote global score takes into account the structure, consistency, annotation and functionality of metabolic models.
The community-driven series of consensus metabolic network reconstructions for S. cerevisiae has tried to overcome some of the obstacles mentioned above by keeping consistency of identifiers across the subsequent model refinement iterations. However, this approach has not yet been applied to any of the other yeast species models analyzed in this review. Such consistency allows to interpret Memote standardized test results as an evolution of the network in different regards, offering a systematic guidance for further development. Annotation of metabolites, reactions and SBO terms has been improved throughout the different versions of the S. cerevisiae model (Fig. 3B). Resultingly, Yeast8 shows the most complete degree of annotation for all of these features, even though standardized gene identifiers that are traceable to an external database are still missing.
CONCLUSIONS
Here we reviewed, collected and evaluated the usability of the available GEMs for different yeasts species, offering a valuable concentrated resource for the community. The model recollection process evidenced that not all of them are easily accessible and multiple sources were needed to be queried. Even though specialized databases for curated GEMs exist, connections between them are still missing, which might hamper large-scale multi-species studies. We also found that GEM files have been shared in a wide variety of file formats, making the utilization of some of them dependent on specific software tools. Storing and sharing models using the latest version of the standard SBML format will facilitate scientific exchange and enable reproducibility of results, avoiding platform dependent parsing issues.
As part of this review, a simplified model test pipeline was developed and run for all of the yeast GEMs with an available SBML file. With the aim of obtaining feasible FBA simulations with the minimal number of steps, we simulated the initial familiarization process of a non-expert user with a new model. It was found that 28 of the tested models (representing 62% of the models in this catalogue) were available in a ready-to-use format, as in-silico growth was obtained without any further knowledge or utilization experience on them. This result must not be interpreted as a measurement of model quality, as biological meaningfulness or consistency of predictions were not evaluated. More robust tests were performed with the aid of the Memote suite. Nonetheless, this was not possible for all of the analyzed models due to outdated file formats. For such cases, update of their respective SBML files is recommended in order to ensure compatibility with the latest modeling and analysis tools and to facilitate further development. The results of the Memote standardized tests illustrated a progressive evolution concerning the annotation of model components for the different versions of the S. cerevisiae metabolic network, highlighting the advantages of community-driven model development.
The total or partial lack of cross-references of model components to widely used external databases is still a common trait of the models in this catalogue. GEMs are usually described as valuable scientific resources not just for quantitative predictions but as genome-scale knowledgebases of living organisms. However, as their usability and exploration are still hindered by the lack of format consistency, cross-references and continuous community development, the full exploitation of their potential remains restricted to expert users.
ACKNOWLEDGEMENTS
We are grateful to Benjamin D. Heavner for sharing his repository of S. cerevisiae models.
Contributor Information
Iván Domenzain, Department of Biology and Biological Engineering, Chalmers University of Technology, Kemivägen 10, SE-412 96, Gothenburg, Sweden; Novo Nordisk Foundation Center for Biosustainability, Chalmers University of Technology, Kemivägen 10, SE-412 96, Gothenburg, Sweden.
Feiran Li, Department of Biology and Biological Engineering, Chalmers University of Technology, Kemivägen 10, SE-412 96, Gothenburg, Sweden; Novo Nordisk Foundation Center for Biosustainability, Chalmers University of Technology, Kemivägen 10, SE-412 96, Gothenburg, Sweden.
Eduard J Kerkhoven, Department of Biology and Biological Engineering, Chalmers University of Technology, Kemivägen 10, SE-412 96, Gothenburg, Sweden; Novo Nordisk Foundation Center for Biosustainability, Chalmers University of Technology, Kemivägen 10, SE-412 96, Gothenburg, Sweden.
Verena Siewers, Department of Biology and Biological Engineering, Chalmers University of Technology, Kemivägen 10, SE-412 96, Gothenburg, Sweden; Novo Nordisk Foundation Center for Biosustainability, Chalmers University of Technology, Kemivägen 10, SE-412 96, Gothenburg, Sweden.
FUNDING
This work has received funding from the European Union's Horizon 2020 Framework Programme for Research and Innovation—Grant Agreement Number 720 824 [to I.D.] and Grant Agreement Number 686 070 [to F.L.].
Conflicts of Interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
- Arkin AP, Cottingham RW, Henry CSet al. KBase: the United States department of energy systems biology knowledgebase. Nat Biotechnol. 2018, DOI: 10.1038/nbt.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung HW, Henry SA, Walker LP. Revising the representation of fatty acid, glycerolipid, and glycerophospholipid metabolism in the consensus model of yeast metabolism. Ind Biotechnol. 2013, DOI: 10.1089/ind.2013.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagurunathan B, Jonnalagadda S, Tan Let al. Reconstruction and analysis of a genome-scale metabolic model for Scheffersomyces stipitis. Microb Cell Fact. 2012;11, DOI: 10.1186/1475-2859-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommareddy RR, Sabra W, Maheshwari Get al. Metabolic network analysis and experimental study of lipid production in Rhodosporidium toruloides grown on single and mixed substrates. Microb Cell Fact. 2015, DOI: 10.1186/s12934-015-0217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cankorur-Cetinkaya A, Dikicioglu D, Oliver SG. Metabolic modeling to identify engineering targets for Komagataella phaffii: the effect of biomass composition on gene target identification. Biotechnol Bioeng. 2017;114:2605–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspeta L, Shoaie S, Agren Ret al. Genome-scale metabolic reconstructions of Pichia stipitis and Pichia pastoris and in silico evaluation of their potentials. BMC Syst Biol. 2012, DOI: 10.1186/1752-0509-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda MT, Nuñez S, Garelli Fet al. Comprehensive analysis of a metabolic model for lipid production in Rhodosporidium toruloides. J Biotechnol. 2018, DOI: 10.1016/j.jbiotec.2018.05.010. [DOI] [PubMed] [Google Scholar]
- Castillo S, Patil KR, Jouhten P. Yeast genome-scale metabolic models for simulating genotype-phenotype relations. Prog Mol Subcell Biol. 2019, DOI: 10.1007/978-3-030-13035-0_5. [DOI] [PubMed] [Google Scholar]
- Chelliah V, Juty N, Ajmera Iet al. BioModels: ten-year anniversary. Nucleic Acids Res. 2015, DOI: 10.1093/nar/gku1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li G, Nielsen J. Genome-scale metabolic modeling from yeast to human cell models of complex diseases: latest advances and challenges. Methods Mol Biol. 2019;2049:329–45. [DOI] [PubMed] [Google Scholar]
- Choi HS, Lee SY, Kim TYet al. In silico identification of gene amplification targets for improvement of lycopene production. Appl Environ Microbiol. 2010;76:3097–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Chowdhury A, Maranas CD. Using gene essentiality and synthetic lethality information to correct yeast and CHO cell genome-scale models. Metabolites. 2015;5:536–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BKS, Selvarasu S, Andrea Cet al. Genome-scale metabolic reconstruction and in silico analysis of methylotrophic yeast Pichia pastoris for strain improvement. Microb Cell Fact. 2010;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtot M, Juty N, Knüpfer Cet al. Controlled vocabularies and semantics in systems biology. Mol Syst Biol. 2011;7:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias O, Pereira R, Gombert AKet al. iOD907, the first genome-scale metabolic model for the milk yeast Kluyveromyces lactis. Biotechnol J. 2014, DOI: 10.1002/biot.201300242. [DOI] [PubMed] [Google Scholar]
- Dinh H V., Suthers PF, Chan SHJet al. A comprehensive genome-scale model for Rhodosporidium toruloides IFO0880 accounting for functional genomics and phenotypic data. Metab Eng Commun. 2019, DOI: 10.1016/j.mec.2019.e00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson PD, Smallbone K, Jameson Det al. Further developments towards a genome-scale metabolic model of yeast. BMC Syst Biol. 2010;4:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dräger A, Palsson B. Improving collaboration by standardization efforts in systems biology. Front Bioeng Biotechnol. 2014;2, DOI: 10.3389/fbioe.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Herrgård MJ, Palsson B. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res. 2004, DOI: 10.1101/gr.2250904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim A, Almaas E, Bauer Eet al. Do genome-scale models need exact solvers or clearer standards?. Mol Syst Biol. 2015, DOI: 10.15252/msb.20156157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim A, Lerman JA, Palsson BOet al. COBRApy: constraints-based reconstruction and analysis for python. BMC Syst Biol. 2013;7:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Jupe S, Matthews Let al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippo M Di, Ortiz-Merino RA, Damiani Cet al. Genome-scale metabolic reconstruction of the stress-tolerant hybrid yeast Zygosaccharomyces parabailii. bioRxiv. 2018:373621. DOI: 10.1101/373621. [Google Scholar]
- Förster J, Famili I, Fu Pet al. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003, DOI: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Albornoz M, Thankaswamy-Kosalai S, Nilsson Aet al. BioMet Toolbox 2.0: genome-wide analysis of metabolism and omics data. Nucleic Acids Res. 2014, DOI: 10.1093/nar/gku371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Nanda P, Patra Pet al. Reconstruction and Analysis of Genome-Scale Metabolic Model of Weak Crabtree Positive Yeast Lachancea kluyveri. 2020. DOI: 10.21203/rs.2.16651/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell G, Bussey Het al. Life with 6000 genes. Science (80-). 1996, DOI: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Gu C, Kim GB, Kim WJet al. Current status and applications of genome-scale metabolic models. Genome Biol. 2019;20:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman A, Säll T, Compagno Cet al. Yeast “Make-Accumulate-Consume” life strategy evolved as a multi-step process that predates the whole genome duplication. PLoS One. 2013, DOI: 10.1371/journal.pone.0068734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavner BD, Price ND. Comparative analysis of yeast metabolic network models highlights progress, opportunities for metabolic reconstruction. PLoS Comput Biol. 2015, DOI: 10.1371/journal.pcbi.1004530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavner BD, Smallbone K, Barker Bet al. Yeast 5 - an expanded reconstruction of the Saccharomyces cerevisiae metabolic network. BMC Syst Biol. 2012, DOI: 10.1186/1752-0509-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavner BD, Smallbone K, Price NDet al. Version 6 of the consensus yeast metabolic network refines biochemical coverage and improves model performance. Database. 2013, DOI: 10.1093/database/bat059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heirendt L, Arreckx S, Pfau Tet al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat Protoc. 2019;14:639–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellerstedt ST, Nash RS, Weng Set al. Curated protein information in the Saccharomyces genome database. Database (Oxford). 2017;2017:bax011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrgård MJ, Lee BS, Portnoy Vet al. Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae. Genome Res. 2006, DOI: 10.1101/gr.4083206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrgård MJ, Swainston N, Dobson Pet al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol. 2008;26:1155–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard M, Damiani A, He QPet al. Elucidating redox balance shift in Scheffersomyces stipitis’ fermentative metabolism using a modified genome-scale metabolic model. Microb Cell Fact. 2018;17:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HMet al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–31. [DOI] [PubMed] [Google Scholar]
- Irani ZA, Kerkhoven EJ, Shojaosadati SAet al. Genome-scale metabolic model of Pichia pastoris with native and humanized glycosylation of recombinant proteins. Biotechnol Bioeng. 2016;113:961–9. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe Met al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2016;45:D353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe Met al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Billington R, Caspi Ret al. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform. 2019;20:1085–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavšcek M, Bhutada G, Madl Tet al. Optimization of lipid production with a genome-scale model of Yarrowia lipolytica. BMC Syst Biol. 2015, DOI: 10.1186/s12918-015-0217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoven EJ, Pomraning KR, Baker SEet al. Regulation of amino-acid metabolism controls flux to lipid accumulation in Yarrowia lipolytica. npj Syst Biol Appl. 2016;2:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuepfer L, Sauer U, Blank LM. Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res. 2005, DOI: 10.1101/gr.3992505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 2003;4:233–45. [DOI] [PubMed] [Google Scholar]
- Lieven C, Beber ME, Olivier BGet al. MEMOTE for standardized genome-scale metabolic model testing. Nat Biotechnol. 2020, DOI: 10.1038/s41587-020-0446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zou W, Liu Let al. A constraint-based model of Scheffersomyces stipitis for improved ethanol production. Biotechnol Biofuels. 2012;5:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loira N, Dulermo T, Nicaud JMet al. A genome-scale metabolic model of the lipid-accumulating yeast Yarrowia lipolytica. BMC Syst Biol. 2012;6, DOI: 10.1186/1752-0509-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes H, Rocha I. Genome-scale modeling of yeast: chronology, applications and critical perspectives. FEMS Yeast Res. 2017, DOI: 10.1093/femsyr/fox050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Li F, Sánchez BJet al. A consensus S. cerevisiae metabolic model Yeast8 and its ecosystem for comprehensively probing cellular metabolism. Nat Commun. 2019;10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcišauskas S, Ji B, Nielsen J. Reconstruction and analysis of a Kluyveromyces marxianus genome-scale metabolic model. BMC Bioinformatics. 2019;20:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows AL, Hawkins KM, Tsegaye Yet al. Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature. 2016, DOI: 10.1038/nature19769. [DOI] [PubMed] [Google Scholar]
- Mendoza SN, Olivier BG, Molenaar Det al. A systematic assessment of current genome-scale metabolic reconstruction tools. Genome Biol. 2019, DOI: 10.1186/s13059-019-1769-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Lee NR, Lakshmanan Met al. Genome-scale model-driven strain design for dicarboxylic acid production in Yarrowia lipolytica. BMC Syst Biol. 2018, DOI: 10.1186/s12918-018-0542-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Park GY, Lakshmanan Met al. Genome-scale metabolic modeling and in silico analysis of lipid accumulating yeast Candida tropicalis for dicarboxylic acid production. Biotechnol Bioeng. 2016, DOI: 10.1002/bit.25955. [DOI] [PubMed] [Google Scholar]
- Mo ML, Palsson BO, Herrgård MJ. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst Biol. 2009;3:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk J, Nogales J, Palsson BO. Optimizing genome-scale network reconstructions. Nat Biotechnol. 2014, DOI: 10.1038/nbt.2870. [DOI] [PubMed] [Google Scholar]
- Nielsen J. Systems biology of metabolism. Annu Rev Biochem. 2017;86:245–75. [DOI] [PubMed] [Google Scholar]
- Nonklang S, Abdel-Banat BMA, Cha-aim Ket al. High-temperature ethanol fermentation and transformation with linear DNA in the thermotolerant yeast Kluyveromyces marxianus DMKU3-1042. Appl Environ Microbiol. 2008, DOI: 10.1128/AEM.01854-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nookaew I, Jewett MC, Meechai Aet al. The genome-scale metabolic model iIN800 of Saccharomyces cerevisiae and its validation: a scaffold to query lipid metabolism. BMC Syst Biol. 2008, DOI: 10.1186/1752-0509-2-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier BG, Bergmann FT. SBML Level 3 package: flux balance constraints version 2 SBML Level 3 package: flux balance constraints ('fbc’). J Integr Bioinform. 2018, DOI: 10.1515/jib-2017-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Thiele I, Palsson BØ. What is flux balance analysis?. Nat Biotechnol. 2010;28:245–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Österlund T, Nookaew I, Bordel Set al. Mapping condition-dependent regulation of metabolism in yeast through genome-scale modeling. BMC Syst Biol. 2013;7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma M, Dias PJ, Roque F de Cet al. The Zygosaccharomyces bailii transcription factor Haa1 is required for acetic acid and copper stress responses suggesting subfunctionalization of the ancestral bifunctional protein Haa1/Cup2. BMC Genomics. 2017, DOI: 10.1186/s12864-016-3443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma M, Guerreiro JF, Sá-Correia I. Adaptive response and tolerance to acetic acid in Saccharomyces cerevisiae and Zygosaccharomyces bailii: a physiological genomics perspective. Front Microbiol. 2018, DOI: 10.3389/fmicb.2018.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan P, Hua Q. Reconstruction and in silico analysis of metabolic network for an oleaginous yeast, Yarrowia lipolytica. PLoS One. 2012, DOI: 10.1371/journal.pone.0051535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratledge C, Wynn JP. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv Appl Microbiol. 2002. DOI: 10.1016/s0065-2164(02)51000-5. [DOI] [PubMed] [Google Scholar]
- Ravikrishnan A, Raman K. Critical assessment of genome-scale metabolic networks: the need for a unified standard. Brief Bioinform. 2015, DOI: 10.1093/bib/bbv003. [DOI] [PubMed] [Google Scholar]
- Saier MHJ, Reddy VS, Tsu BVet al. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res. 2016;44:D372–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt FR. Recombinant expression systems in the pharmaceutical industry. Appl Microbiol Biotechnol. 2004;65:363–72. [DOI] [PubMed] [Google Scholar]
- Shen XX, Opulente DA, Kominek Jet al. Tempo and mode of genome evolution in the budding yeast subphylum. Cell. 2018, DOI: 10.1016/j.cell.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn SB, Graf AB, Kim TYet al. Genome-scale metabolic model of methylotrophic yeast Pichia pastoris and its use for in silico analysis of heterologous protein production. Biotechnol J. 2010;5:705–15. [DOI] [PubMed] [Google Scholar]
- Sohn SB, Kim TY, Lee JHet al. Genome-scale metabolic model of the fission yeast Schizosaccharomyces pombe and the reconciliation of in silico/in vivo mutant growth. BMC Syst Biol. 2012;6:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solieri L, Chand Dakal T, Croce MAet al. Unravelling genomic diversity of Zygosaccharomyces rouxii complex with a link to its life cycle. FEMS Yeast Res. 2013, DOI: 10.1111/1567-1364.12027. [DOI] [PubMed] [Google Scholar]
- Sánchez BJ, Nielsen J. Genome scale models of yeast: towards standardized evaluation and consistent omic integration. Integr Biol (United Kingdom). 2015, DOI: 10.1039/c5ib00083a. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium . UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiukova IA, Prigent S, Nielsen Jet al. Genome-scale model of Rhodotorula toruloides metabolism. Biotechnol Bioeng. 2019, DOI: 10.1002/bit.27162. [DOI] [PubMed] [Google Scholar]
- Tomàs-Gamisans M, Ferrer P, Albiol J. Fine-tuning the P. pastoris iMT1026 genome-scale metabolic model for improved prediction of growth on methanol or glycerol as sole carbon sources. Microb Biotechnol. 2018;11:224–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomàs-Gamisans M, Ferrer P, Albiol J. Integration and validation of the genome-scale metabolic models of Pichia pastoris: a comprehensive update of protein glycosylation pathways, lipid and energy metabolism. PLoS One. 2016, DOI: 10.1371/journal.pone.0148031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Marcišauskas S, Sánchez BJet al. RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLoS Comput Biol. 2018;14:e1006541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann G, Hawkins KM, Jackson Pet al. Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature. 2016;537:694–7. [DOI] [PubMed] [Google Scholar]
- Xu N, Liu L, Zou Wet al. Reconstruction and analysis of the genome-scale metabolic network of Candida glabrata. Mol Biosyst. 2013, DOI: 10.1039/c2mb25311a. [DOI] [PubMed] [Google Scholar]
- Ye R, Huang M, Lu Het al. Comprehensive reconstruction and evaluation of Pichia pastoris genome-scale metabolic model that accounts for 1243 ORFs. Bioresour Bioprocess. 2017, DOI: 10.1186/s40643-017-0152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomorrodi AR, Maranas CD. Improving the iMM904 S. cerevisiae metabolic model using essentiality and synthetic lethality data. BMC Syst Biol. 2010;4:178. [DOI] [PMC free article] [PubMed] [Google Scholar]